
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 13 June 2017
Sec. Plant Abiotic Stress
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01010
 Zhen Li1,2†
Zhen Li1,2† Xiaojiao Han1,2†
Xiaojiao Han1,2† Xixi Song1,2†
Xixi Song1,2† Yunxing Zhang1,2
Yunxing Zhang1,2 Jing Jiang1,2
Jing Jiang1,2 Qiang Han1,2
Qiang Han1,2 Mingying Liu1,2
Mingying Liu1,2 Guirong Qiao1,2
Guirong Qiao1,2 Renying Zhuo1,2*
Renying Zhuo1,2*Superoxide dismutase (SOD) is a very important reactive oxygen species (ROS)-scavenging enzyme. In this study, the functions of a Cu/Zn SOD gene (SaCu/Zn SOD), from Sedum alfredii, a cadmium (Cd)/zinc/lead co-hyperaccumulator of the Crassulaceae, was characterized. The expression of SaCu/Zn SOD was induced by Cd stress. Compared with wild-type (WT) plants, overexpression of SaCu/Zn SOD gene in transgenic Arabidopsis plants enhanced the antioxidative defense capacity, including SOD and peroxidase activities. Additionally, it reduced the damage associated with the overproduction of hydrogen peroxide (H2O2) and superoxide radicals (O2•-). The influence of Cd stress on ion flux across the root surface showed that overexpressing SaCu/Zn SOD in transgenic Arabidopsis plants has greater Cd uptake capacity existed in roots. A co-expression network based on microarray data showed possible oxidative regulation in Arabidopsis after Cd-induced oxidative stress, suggesting that SaCu/Zn SOD may participate in this network and enhance ROS-scavenging capability under Cd stress. Taken together, these results suggest that overexpressing SaCu/Zn SOD increased oxidative stress resistance in transgenic Arabidopsis and provide useful information for understanding the role of SaCu/Zn SOD in response to abiotic stress.
Cadmium (Cd), a non-essential trace element, is highly phytotoxic to all living cells through interference with DNA repair machinery, generation of reactive oxygen species (ROS) and disruption of regular gene expression (Valko et al., 2005; Jomova and Valko, 2011). Cd can also result in oxidative stress through interacting with the antioxidative defense system and favoring lipid peroxidation and production of ROS (Rodríguez-Serrano et al., 2006; Woertink et al., 2009).
Plants maintain intracellular ROS levels using a sophisticated antioxidative defense system that includes the antioxidative enzymes superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), peroxiredoxins (Prxs), and glutathione peroxidase (GPX) during their growth and development (Dietz, 2003; Kishikawa and Kuroda, 2014). When exposed to different abiotic stress conditions, plants will increase the accumulation of ROS, leading to enhanced tolerance to oxidative stress by disturbing protein synthesis and stability and damaging cellular macromolecules and membrane lipids in plant cells (Wang et al., 2005). However, plants have the ability to scavenge ROS, allowing control over ROS toxicity and use of ROS as signaling molecules through a complicated gene regulation network (Mittler et al., 2004; Baxter et al., 2014). SOD is one of most important ROS-scavenging enzymes, constituting the first cellular defense against oxidative stress and playing crucial roles in ROS homeostasis due to its conversion of the superoxide anion (O2•-) to H2O2 (Gupta et al., 1993; Apel and Hirt, 2004). SODs are classified into three isoforms based on their metal cofactors: manganese (Mn)-SOD, iron (Fe)-SOD and copper (Cu)/zinc (Zn)-SOD. Mn-SOD is primarily localized in mitochondria and peroxisomes; Fe-SOD primarily in chloroplast; Cu/Zn SOD in the cytosol, chloroplasts and peroxisomes (Bueno et al., 1995). Cu/Zn SOD, the most important of the three SOD enzymes, is composed of two subunits combined with Cu and Zn atoms, respectively, that increase the activity and stability of the enzyme (Lin et al., 1995). SODs can alter the O2•- to H2O2 ratio (Chabory et al., 2010), and H2O2 can modulate the expression of multiple genes, such as transcription factors, antioxidative genes and some stress-related genes. H2O2 is catalyzed to H2O by CAT, Prxs and GPX (Gupta et al., 1993). SODs, along with other enzymes, maintain intracellular ROS homeostasis. SOD overexpression can endow a certain level of tolerance to several stress conditions. For instance, transgenic cassava (Manihot esculenta) overexpressing both MeCu/Zn SOD and MeCAT1 showed improved tolerance against oxidative, cold and drought stresses through enhancing ROS-scavenging capability (Xu et al., 2013, 2014). The expression of PutCu/Zn SOD was observed in leaves and roots exposed to salt, and oxidative stresses. Transgenic yeast and Arabidopsis plants overexpressing PutCu/Zn SOD showed increased resistance to multiple abiotic stresses (Wu et al., 2016). In addition, a novel Cu/Zn SOD gene from Saussurea involucrata Kar. et Kir., SiCSD, was induced by cold, drought and oxidative stresses. Overexpression of SiCSD in transgenic tobacco improved tolerance to drought, freezing and oxidative stresses (Zhang et al., 2017).
Phytoremediation is considered a gentle, eco-friendly clean-up method. Hyperaccumulator plants can extract metals from the soil and concentrate these metals in their aerial parts. To date, approximately 450 hyperaccumulator species have been identified, but most of them are limited by their biomass and growth rate. Sedum alfredii Hance is a new Cd hyperaccumulator that was found inhabiting an abandoned lead (Pb)/Zn mine in southeast China and is an ideal material for investigating the mechanism of heavy metal stress tolerance. Due to its fast growth, the potential application value of S. alfredii is tremendous. However, the exact mechanisms that trigger ROS in S. alfredii treated with heavy metals are still unclear (Gratão et al., 2005). In our previous study, the transcriptomes from roots, stems and leaves of S. alfredii treated with 400 μM CdCl2 were identified by high-throughput sequencing (Han et al., 2016), and we found that SaCu/Zn SOD expression was highly induced by CdCl2 stress. Here, a SaCu/Zn SOD gene from S. alfredii plants was isolated and functionally characterized. Furthermore, the internal relationships between SaCu/Zn SOD and some oxidative genes under Cd stress were explored using a co-expression network. Our results will provide valuable information on the underlying mechanism and possibilities for cultivating forest tree varieties with high resistance level to oxidative stress in the future.
The Cd-hyperaccumulator plant S. alfredii was originally obtained from a Pb/Zn mine in Zhejiang Province, China, cleaned with double-distilled water and grown in a plant growth chamber at 25°C with a 16 h light/8 h dark photoperiod. Thirty-day-old uniform and robust seedlings were treated with 400 μM CdCl2 for 0, 0.5, 6, 12, 24, 48, 72 and 96 h. Leaves, roots and stems of seedlings from each sample were harvested at the indicated times, quickly frozen in liquid nitrogen and stored at -80°C for further analyses.
Seeds of Arabidopsis were surface-sterilized in 75% (v/v) ethanol for 10 min and then plated on 1/2 Murashige and Skoog (MS) solid medium. One-week-old seedlings were transferred from plates into pots containing a mixture of perlite/soil in a greenhouse at 22°C with a 16 h light/8 h dark photoperiod and 70–75% relative humidity. Four-week-old seedlings were treated with 5 mM CdCl2 for 1, 2 and 3 weeks, after which the whole plants were immediately harvested and used for cDNA microarray and physiological analysis.
Based on S. alfredii transcriptome data, SaCu/Zn-SOD was identified and isolated from a cDNA library of S. alfredii using the primers SOD-F and SOD-R (Supplementary Table S1). The deduced SaCu/Zn-SOD protein sequence and other plant SODs were aligned using CLUSTALX 2.0. A phylogenetic analysis was performed using MEGA 5.2 software with the neighbor-joining method, and the internal branch support was estimated with 1,000 bootstrap replicates.
Total RNA was extracted from S. alfredii using a RNA purification kit (NORGEN, Thorold, ON, Canada), and removed genomic DNA contamination. One microgram of total RNA was used for first-strand cDNA synthesis by the Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, Switzerland). qRT-PCR was performed on a 7300 Real-Time PCR System (Applied Biosystems, United States) using a SYBR PrimeScriptTM RT-PCR Kit (TaKaRa, Dalian, China). The PCR program was as follows: 95°C for 10 s, then 40 cycles of 95°C for 5 s and 60°C for 31 s, followed by melting curve analysis. The relative expression levels were calculated according to the method described by Livak and Schmittgen (2001). All the primers are listed in the Supplementary Table S1. For each sample, three biological replicates, each with three technical replicates were performed.
The full-length cDNA of SaCu/Zn SOD was cloned into the pBI121G vector driven by the CaMV 35S promoter and introduced into Arabidopsis (ecotype Columbia) by the floral dip method (Clough and Bent, 1998). Positive transformants were selected on 1/2 MS solid medium containing 50 mg⋅L-1 kanamycin and confirmed by genomic DNA PCR. The expression levels of the transformants were also determined by qRT-PCR. T3 homozygous lines were identified by screening for non-segregation from each independent transformant and selected for further biological functional analyses.
Seeds of wild-type (WT) and T3 homozygous overexpression (OE) plants were placed on 1/2 MS agar medium with or without 1.5 mM CdCl2 and incubated at 22°C for 3 and 7 days. The seed germination rates were determined. Four-week-old seedlings were irrigated with 5 mM CdCl2 for 0, 1, 2 and 3 weeks. After the different treatment times, the fresh weight of each plant was calculated.
For the determination of enzyme activities, 1 g leaf samples were harvested from the above Cd-treated plants, homogenized in 8 ml of 50 mM sodium phosphate buffer (PBS, pH 7.8) using a prechilled mortar and pestle, and then centrifuged at 10,000 g for 15 min at 4°C. The supernatant was used to measure SOD and POD activities, and O2•- levels (Rao et al., 1996). In addition, 0.05 g of leaf tissue was cut into pieces and submerged in a mixture of acetone and ethanol (1:1). The absorbance was measured in a spectrophotometer at 645 and 663 nm to calculate the total chlorophyll contents (Arnon, 1949). Electrolyte leakage was calculated through conductivity measurements (Fan et al., 1997). The malondialdehyde (MDA) and H2O2 contents were determined according to Velikova et al. (2000).
Rosette leaves harvested from WT and OE Arabidopsis plants were immediately soaked in 3,3′ diaminobenzidine (DAB) solution (1 mg⋅mL-1, DAB-HCl, pH 3.8) or NBT (1 mg⋅mL-1 NBT in 10 mM sodium azide and 10 mM phosphate buffer, pH 7.8), respectively, for 12 h in the dark to detect H2O2 and O2•- contents in situ, and then the chlorophyll was eliminated by boiling in 96% ethanol. Images of leaves were acquired using a stereoscopic microscope (LEICA M125, Germany) after chlorophyll elimination. At least three biological replicates were performed for each experiment and approximately 20 leaves harvested from multiple seedlings were inspected.
Fifteen-day-old seedlings were treated with 50 μM CdCl2 for 24 h and soaked in test liquid [0.1 mM KCl, 0.1 mM CaCl2, 0.05 mM CdCl2, 0.3 mM 2-(N-morpholino) ethanesulfonic acid, pH 5.8] for 15 min. The roots were immobilized on the bottom of a measuring dish in fresh test liquid. The measuring site was 200 μm from the root apex, and the net flux of Cd2+ was detected using a non-invasive micro-test technique (NMT; BIO-001A, Younger United States Science and Technology Corp., Beijing, China). The ion flux of Cd2+ was calculated according to Fick’s law of diffusion, J0 = -D × (dC/dX), where J0 is the net ion flux (in μmol⋅cm-2 per second), D is the self-diffusion coefficient for the ion (in cm2⋅s-1), dC is the difference in the ion concentrations between the two positions, and dX is the 10 μm excursion over which the electrode moved in these experiments.
Total RNA was isolated from transgenic Arabidopsis and WT plants using a RNeasy Plant Mini kit (Qiagen, Hilden, Germany) and purified for microarray data analysis by QuanMai (Shanghai, China). Three OE line constituted a mixed pool as the transgenic group, and three independent biological replicates were carried out. Briefly, double-stranded cDNA was synthesized from 0.2 μg total RNA using a Quick Amp Labeling Kit, One-Color (Agilent). The products were labeled with Cyanine-3-CTP (Cy3) and subsequently hybridized onto the microarray using an Agilent Gene Expression Hybridization Kit. The array was incubated at 65°C for 17 h and then automatically washed and stained using the Gene Expression Wash Pack (Agilent). The probe array was scanned by an Agilent Microarray Scanner (G2505C).
Array images were analyzed to obtain raw data using Feature Extraction software (version10.7.1.1, Agilent Technologies). The data were normalized using the quantile algorithm. Differentially expressed genes were then identified through their fold changes. Gene ontology (GO) was applied to determine the roles of these differentially expressed genes. Based on the microarray data, a co-expression network was constructed using the R package WGCNA1 (Langfelder and Horvath, 2008). All the parameters were set at the default values except the default power, which was set to six (R Core Team, 2014). The outputted, co-expressed genes having strong interconnections were defined as hub genes. We then chose specific genes from the hub genes for further analyses with FunNet2 and AmiGO 23. The co-expression network was constructed using Cytoscape software (Saito et al., 2012).
At least 10 plants were analyzed in each treatment. All the experiments were repeated at least three times. The results are shown as the means ± SD. Statistical determinations were carried out using a one-way ANOVA test, and differences between two groups of data for comparisons in all the experiments were evaluated as statistically significant (∗p < 0.05) or extremely significant (∗∗p < 0.01).
The full-length cDNA of SaCu/Zn SOD (KF806556) is 1,001 bp in length, encodes a 210 amino acid protein with a predicted molecular weight of 39.1 kDa and an isoelectric point of 6.41, which is flanked by stretches of 48 and 53 bp at the 5′- and 3′-untranslated regions, respectively. A multiple sequence alignment with Cu/Zn SODs from other plants was carried out and showed that the SaCu/Zn SOD protein exhibited the same familial signature, including a highly conserved active site (Figure 1A). The evolutionary relationships among SaCu/Zn SOD and other plant Cu/Zn SODs showed that SaCu/Zn SOD was most similar to the Cu/Zn SOD from Prunus persica and had a long evolutionary distance from the Cu/Zn SODs of P. trichocarpa (Figure 1B). We further investigated the genomic structure of SaCu/Zn SOD and found that SaCu/Zn SOD was approximately 1,985 bp, containing seven introns and eight exons, which is similar to the characteristics of the gene structures in P. trichocarpa and Arabidopsis (Supplementary Figure S1).
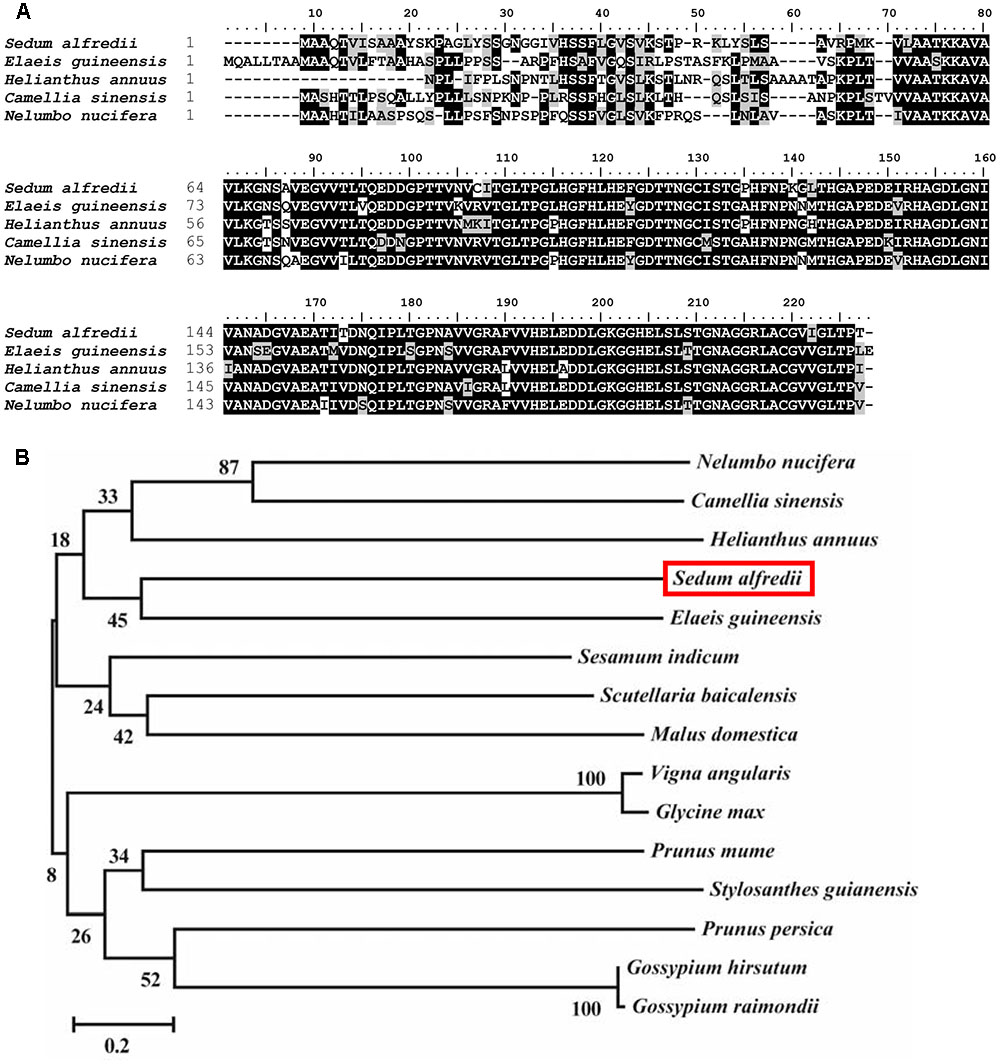
FIGURE 1. Comparison of amino acid sequences of SaCu/Zn SOD and closely related Cu/Zn SODs from other plants. (A) Alignment of the amino acid sequence of SaCu/Zn SOD with Cu/Zn SOD proteins from different plants species. (B) The phylogenetic relationships between SaCu/Zn SOD and other plant Cu/Zn SOD proteins. The neighbor-joining phylogenetic tree was created with ClustalW using MEGA 5.2. All amino acid sequences were retrieved from GenBank: S. alfredii Cu/Zn SOD (AII25435.1); Elaeis guineensis Cu/Zn SOD (XP_010940533.1); Helianthus annuus Cu/Zn SOD (CAH06449.1); Camellia sinensis Cu/Zn SOD (AKN10571.1); Nelumbo nucifera Cu/Zn SOD (XP_010267263.2); Scutellaria baicalensis Cu/Zn SOD (AEO27875.1); Prunus persica Cu/Zn SOD (AFG19614.1); Vigna angularis Cu/Zn SOD (XP_017433773.1); Sesamum indicum Cu/Zn SOD (XP_011088797.1); Glycine max Cu/Zn SOD (XP_003538169.1); Gossypium hirsutum Cu/Zn SOD (XP_016671933.1); G. raimondii Cu/Zn SOD (XP_012485000.1); P. mume Cu/Zn SOD (XP_008226630.1); Stylosanthes guianensis Cu/Zn SOD (AKO62676.1); Malus domestica Cu/Zn SOD (XP_008365087.1).
To determine the potential functions of SaCu/Zn SOD, the relative expression levels of SaCu/Zn SOD in roots, stems and leaves of S. alfredii seedlings were tested using qRT-PCR. The expression levels of SaCu/Zn SOD in the leaves, stems and roots were different. The SaCu/Zn SOD was expressed at a higher level in the leaves and at a lower level in the roots (Figure 2A). To further determine the effects of Cd stress on the expression of SaCu/Zn SOD, 30-day-old seedlings were exposed to 400 μM CdCl2 for different time periods. The expression of SaCu/Zn SOD was initially reduced, then gradually increased and peaked at 12 h of stress in the stems and 24 h in the leaves, respectively. The SaCu/Zn SOD expression did not change significantly in the roots during CdCl2 stress (Figure 2B).
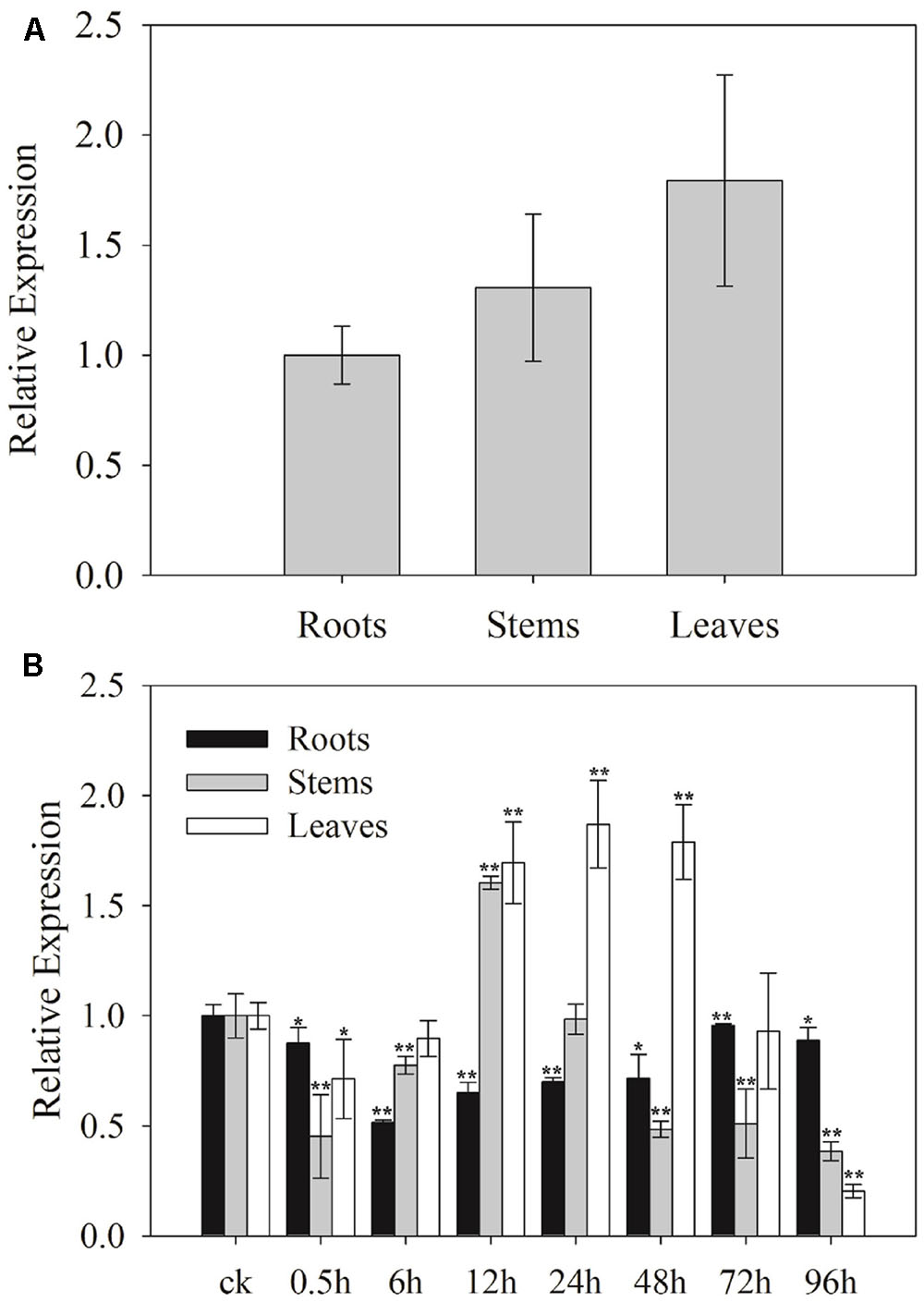
FIGURE 2. Expression profiles of SaCu/Zn SOD in different tissues under CdCl2 stress. (A) Tissue-specific expression of SaCu/Zn SOD by qRT-PCR. Total RNA was extracted from the roots, stems and leaves of 25-day-old seedlings. (B) Expression levels of SaCu/Zn SOD in the roots, stems and leaves under CdCl2 stress. The 25-day-old seedlings were obtained by hydroponic culturing with 400 μM CdCl2 for the indicated times. For each sample, three technical replicates were performed. The error bars were calculated from the multiple replicates of qRT-PCR. Bars indicate means ± SD (n = 10). Asterisks indicate significant differences at ∗p < 0.05 and ∗∗p < 0.01 based on a t-test.
To further determine the effects of SaCu/Zn SOD under Cd stress, seven T3 homozygous transgenic Arabidopsis plants constitutively expressing SaCu/Zn SOD were generated and confirmed by genomic DNA PCR and qRT-PCR (Supplementary Figure S2). Three representative transgenic lines (OE2, OE3, and OE4) were selected for further research. There was no difference in the seed germination rate between transgenic and WT Arabidopsis plants without CdCl2 treatment, but transgenic lines showed higher germination rates than WT plants under CdCl2 stress (Figure 3A and Supplementary Figure S3). After 1 week of Cd treatment, WT plants showed visual Cd toxicity symptoms, such as wilting and etiolation. The toxicity symptoms became more severe with increasing exposure time, but the toxicity symptoms in transgenic Arabidopsis were less severe than those of WT during the same exposure time (Figure 3C). Meanwhile, the chlorophyll contents decreased in transgenic and WT plants under Cd stress, but all OE lines had higher contents than WT plants (Figure 3B). Additionally, the fresh weights of transgenic Arabidopsis decreased by 10.2, and 14.0% under 2 and 3 weeks of treatment, respectively; however, greater decreases of 13.7 and 19.6%, respectively, were observed in WT (Supplementary Figure S4).
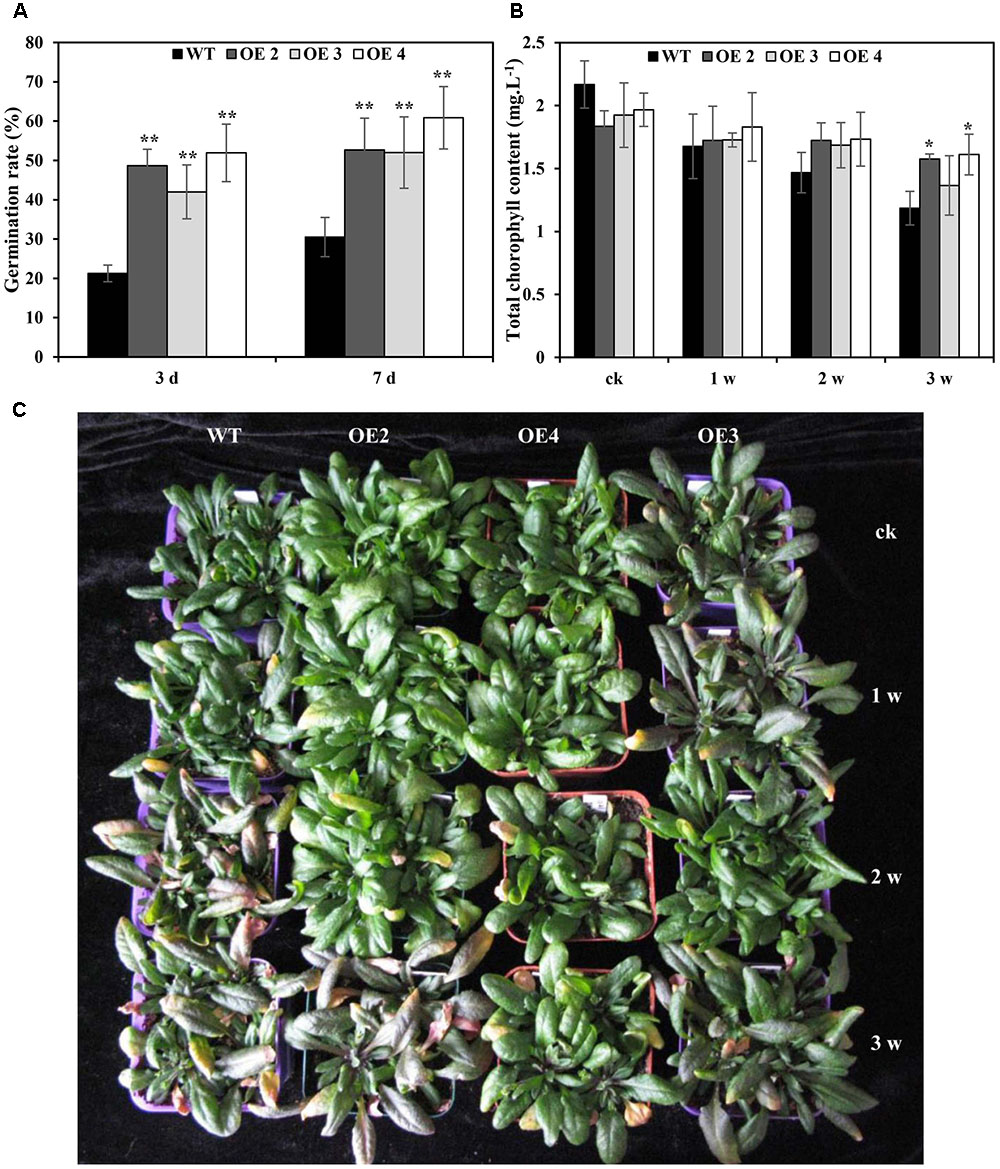
FIGURE 3. Overexpression of SaCu/Zn SOD in transgenic plants enhanced Cd tolerance. (A) Germination rates of SaCu/Zn SOD transgenic (OE2, OE3, and OE4) and WT plants on 1/2 MS agar medium supplemented with 1.5 mM CdCl2. Germination rates were determined after incubation at 22°C for 3 and 7 days. (B,C) Comparison of the chlorophyll contents and phenotypes between SaCu/Zn SOD transgenic and WT Arabidopsis plants. Four-week old seedlings were irrigated with 5 mM CdCl2 for 0, 1, 2 and 3 weeks, and the photographs of seedlings were taken. The error bars represent the standard deviations of the average values. Asterisks indicate significant differences at ∗p < 0.05 and ∗∗p < 0.01.
A non-invasive micro-test (NMT) technique was used to investigate the Cd2+ uptake in root tips of OE and WT plants. Under normal conditions, the net Cd2+ fluxes of OE and WT were within a relatively small range, but the Cd2+ influxes of OE lines were slightly higher than those of WT plants. With 50 μM CdCl2 supplementation, the net Cd2+ flux was affected (Figure 4), while OE lines exhibited higher influx than did WT plants. These results indicated that a greater Cd uptake capacity existed in these roots than in WT plants.
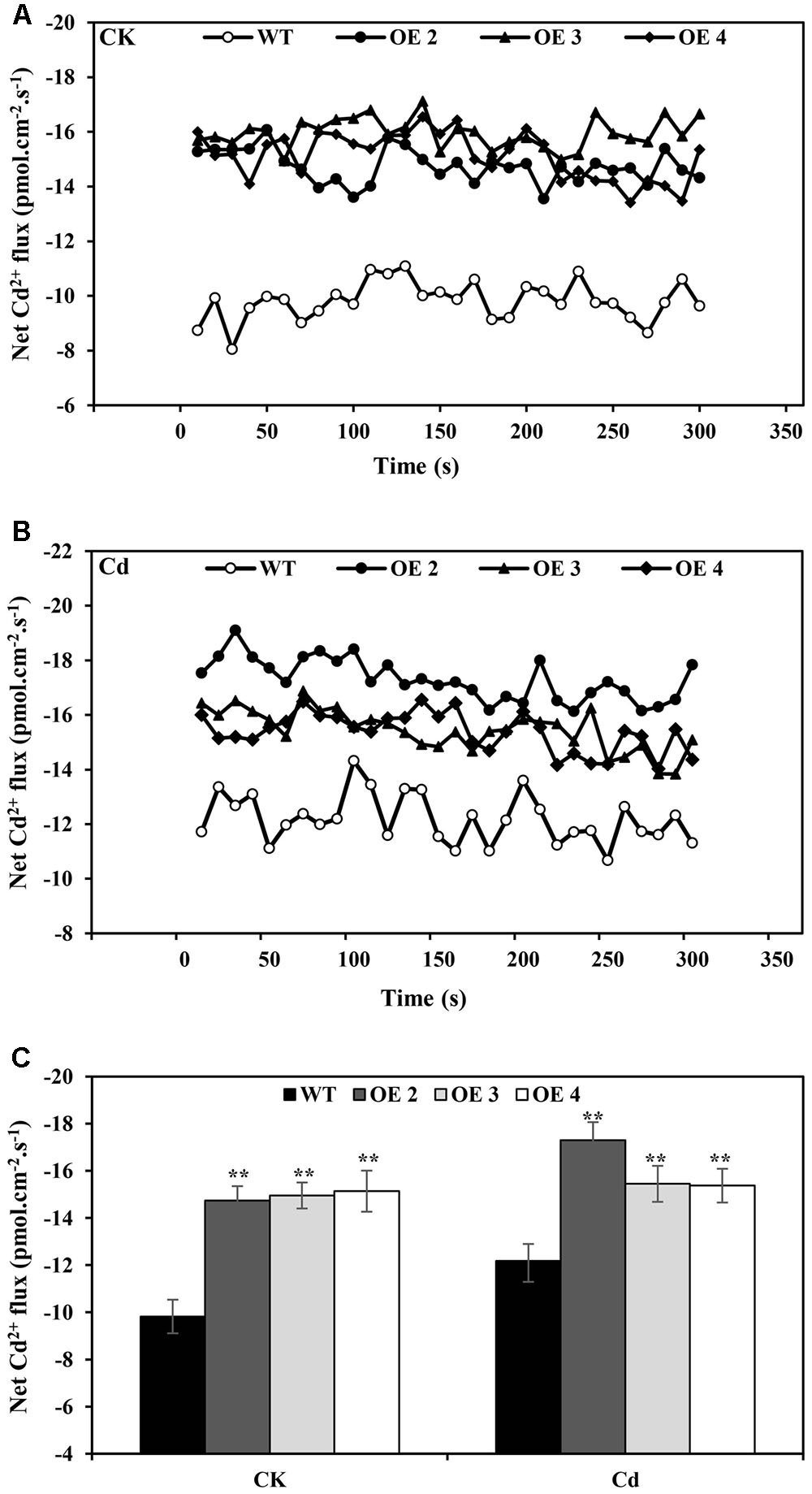
FIGURE 4. Net Cd2+ fluxes. Net Cd2+ fluxes in the roots of transgenic (OE2, OE3, and OE4) and WT plants treated without (A) or with CdCl2 stress (B), respectively. The average 300 s net Cd2+ fluxes (C) are illustrated to highlight the trend differences. Bars indicate means ± SD. Asterisks indicate significant differences at ∗p < 0.05 and ∗∗p < 0.01.
The effects of Cd toxicity on the production of ROS were analyzed by NBT and DAB staining, which produced dark blue and deep brown products, respectively. Both NBT and DAB staining showed that the O2•- and H2O2 levels in transgenic and WT plants exhibited no differences under normal growth conditions. However, under Cd stress conditions, the accumulation of O2•- and H2O2 in transgenic lines was obviously lower than that in WT plants (Figures 5A,B). Additionally, compared with WT plants, the O2•- and H2O2 contents in transgenic Arabidopsis plants overexpressing SaCu/Zn SOD were lower under Cd stress condition (Figures 5C,D). We next tested the activities of SOD and POD. After Cd treatment, these enzyme activities in the OE lines were significantly greater than those in WT plants (Figures 6A,B). This indicated that overexpression of SaCu/Zn SOD in Arabidopsis might enhance the ability to scavenge ROS. The degrees of oxidative damage in the leaves were determined by testing MDA levels and electrolyte leakage. Transgenic and WT plants showed similar growth levels without Cd stress. However, MDA levels and electrolyte leakage in the transgenic plants were significantly lower than those in WT plants under Cd stress (Figures 6C,D). These results suggested that SaCu/Zn SOD confers Cd stress tolerance through increasing the ability of ROS-scavenging and reducing membrane injury.
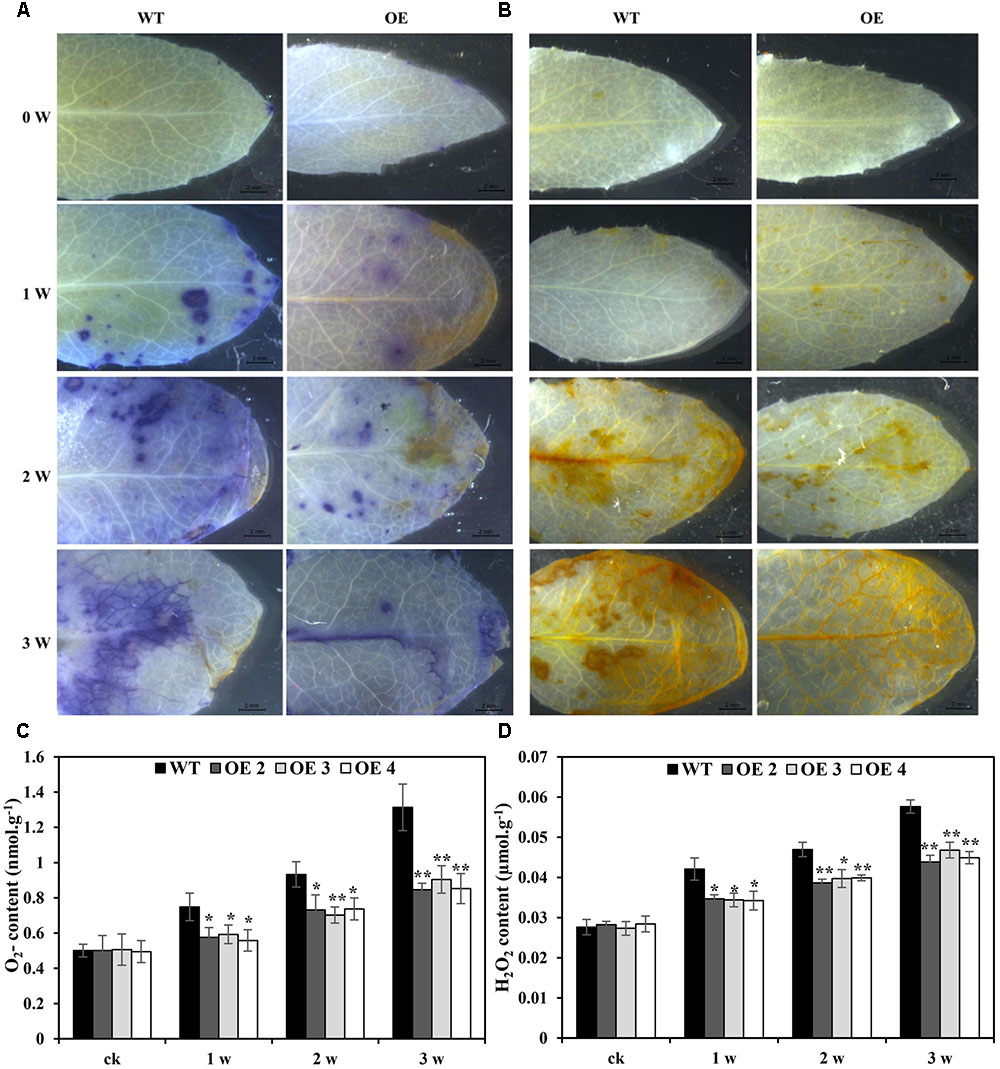
FIGURE 5. Comparison of Cd-induced O2- and H2O2 levels between WT and transgenic Arabidopsis plants. Four-week old seedlings were irrigated with 5 mM CdCl2 for 0, 1, 2, and 3 weeks. Leaves were detached from transgenic (OE2, OE3, and OE4) and WT Arabidopsis plants and the O2- and H2O2 levels were visualized by NBT (A) and DAB staining (B). (C,D) Comparison of the O2- and H2O2 contents between WT and transgenic Arabidopsis plants. Bars indicate means ± SE. Asterisks indicate significant differences at ∗p < 0.05 and ∗∗p < 0.01.
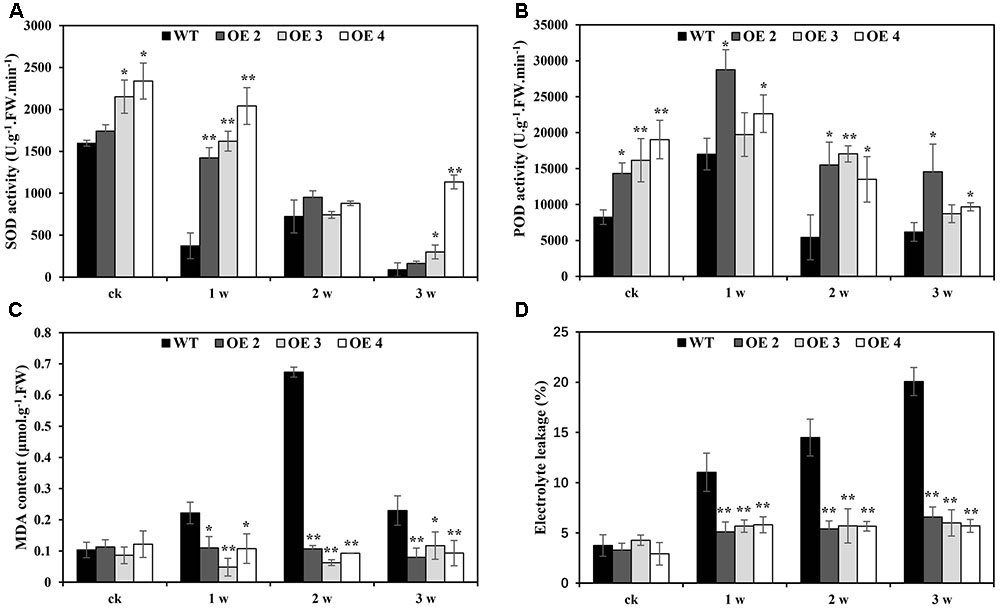
FIGURE 6. Physiological analysis of WT and SaCu/Zn SOD transgenic Arabidopsis plants. Four-week old seedlings were irrigated with 5 mM CdCl2 for 0, 1, 2, and 3 weeks and the leaves were collected to measure SOD (A) and POD (B) activities, MDA contents (C) and electrolyte leakage (D). Bars indicate means ± SE (n = 20–25 plants for each genotype). Asterisks indicate significant differences at ∗p < 0.05 and ∗∗p < 0.01.
Cadmium stress rapidly induced a marked increase in ROS and altered the antioxidant contents in WT and OE plants. To further investigate the transcriptional regulatory profiles of SaCn/Zn SOD, an Agilent Arabidopsis Oligo Microarray assay was employed. The microarray data showed that 2,966–6,030 genes were involved in this process during different treatment durations (Supplementary Table S2). Based on the microarray data, GO classification was conducted for the pathways significantly (p < 0.05) enriched in the genes that were differentially regulated under Cd stress. Numerous genes with known function were classified into the GO terms: “Biological process,” “Cellular component,” and “Molecular function” (Supplementary Figure S5). Among these, 61 cadmium stress-responsive genes involved in metal transport, binding to the cell wall, metal chelation in the cytosol, repair of stress-damaged proteins and antioxidative enzyme activity were selected to illustrate the molecular/cellular mechanisms in plants (Yang et al., 2005). As shown in Figure 7, these 61 genes were differentially up- and down-regulated under different treatment time points (Figure 7). Four members of the glutaredoxin family were up-regulated to different levels at the different time points. Furthermore, 16 peroxidase genes were found in the microarray data. Five of them [peroxidase 38 (AT4G08780), peroxidase 39 (AT4G11290), peroxidase 66 (AT5G51890), peroxidase 67 (AT5G58390), and peroxidase 70 (AT5G64110)] were significantly up-regulated in the 3rd week. By contrast, another five peroxidase genes [peroxidase 11 (AT1G68850), peroxidase 50 (AT4G37520), peroxidase 52 (AT5G05340), peroxidase 54 (AT5G06730), and peroxidase 55 (AT5G14130)] were down-regulated in the 3rd week. In addition to these two gene families, two ferritin genes [ferritin 1 (AT5G01600) and ferritin 4 (AT3G11050)], one catalase gene [CAT1 (AT1G20630)] and one thioredoxin-dependent peroxidase gene [Type 2 PrxR C (AT1G65970)] showed higher expression levels. The genes involved in these pathways were highly activated, indicating that these pathways are important for cadmium stress response.
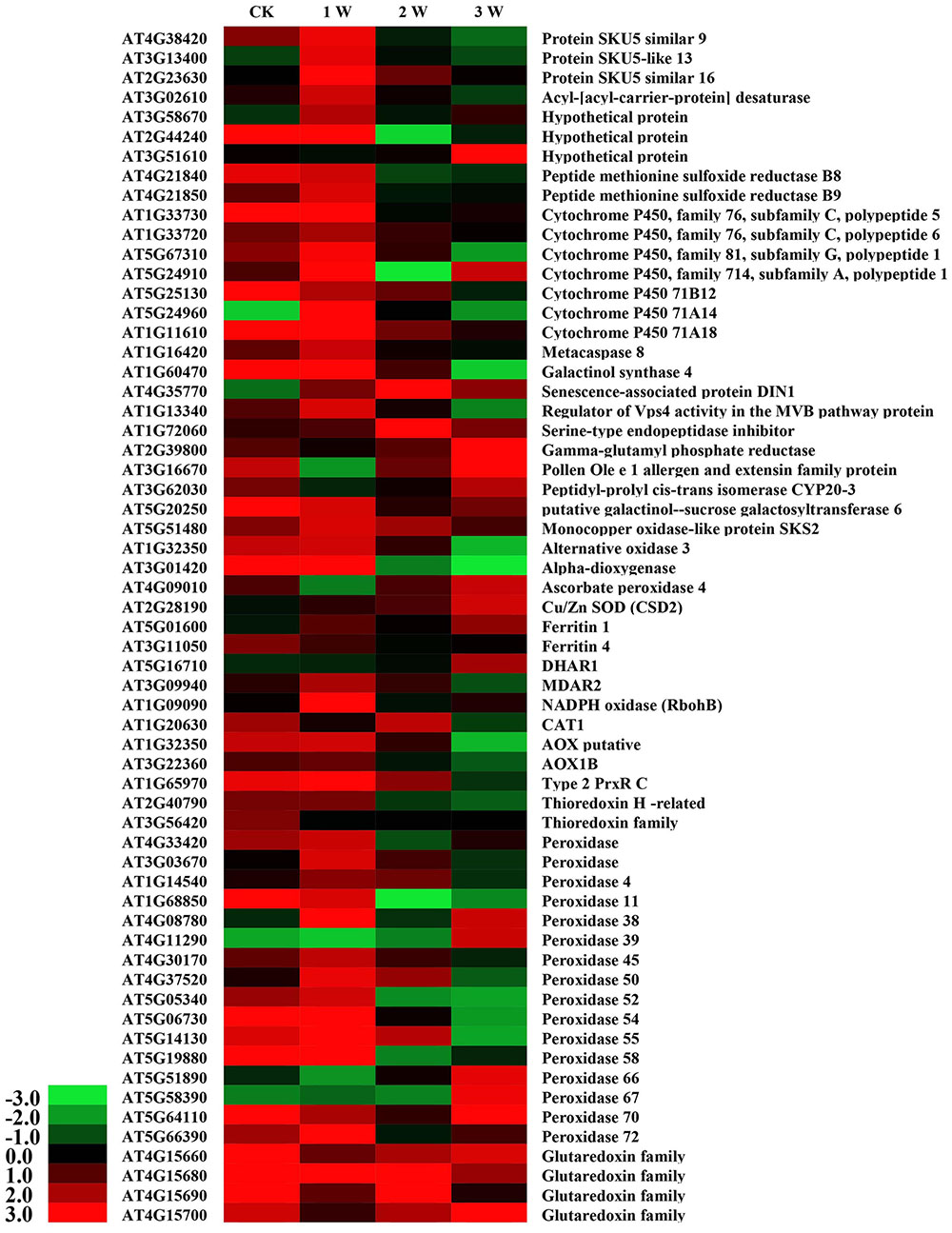
FIGURE 7. DEG expression profiles in Arabidopsis under CdCl2 stress at different time points. All the ratios are log2 transformed so that inductions and repressions of identical magnitude are numerically equal but opposite in sign. Log ratios of 0 (ratios of 1) are colored black, and increasingly positive (induction) or negative (repression) log ratios are colored red or green, respectively, with increasing intensity.
To determine the genes affected by SaCu/Zn SOD in Arabidopsis, we constructed a co-expression network based on the normalized microarray data. After excluding non- and lowly expressed genes, we identified 45 gene co-expression modules. Ultimately, the resulting network was consisted of 21 distinct gene modules, and then the top 50 hub genes were selected based on coefficients of variance (Supplementary Table S3). From the top 50 hub genes, two major genes, oxygen-evolving enhancer protein 2–1 (PSBP-1, A_84_P20343, AT1G06680) and phospholipid hydroperoxide GPX1 (A_84_P815195, AT2G25080), that might be directly relate to oxidative stress (Supplementary Table S4) were selected to constructed a stress-response co-expression subnetwork. As shown in Figure 8, PSBP-1 was regarded as the key co-expression gene, being related to 95% of edges and another hub gene, GPX1 (At1G55760 and At3G50650 interacted with GPX1 only). The functions of 11 edges (unknown genes) were not clear, and the other edges were classified into 6 categories. Of the 40 edges, 11 (∼27.5%) had catalytic activities, while 16 (∼40.0%) had binding functions. The binding targets were nucleotides, chromatins, fatty acids, metal ions, adenosine triphosphate and nicotinamide adenine dinucleotide phosphate. Red arrows indicated the metal binding genes, and PSBP-1, PSBO1, and VSR4 interacted with calcium ions. AT5G36001 interacted with zinc ions, while AT3G17250 bound to Mn2+/Mg2+ for further processing. PSBO1, LGALDH and these two hub genes were oxidative stress-related genes (blue arrows). GDI2 and AT5G51770 (purple arrows) participated in signal transduction. In addition, we classified six stress-response genes (yellow arrows), which were involved in catalysis, metal binding, and structural integrity.
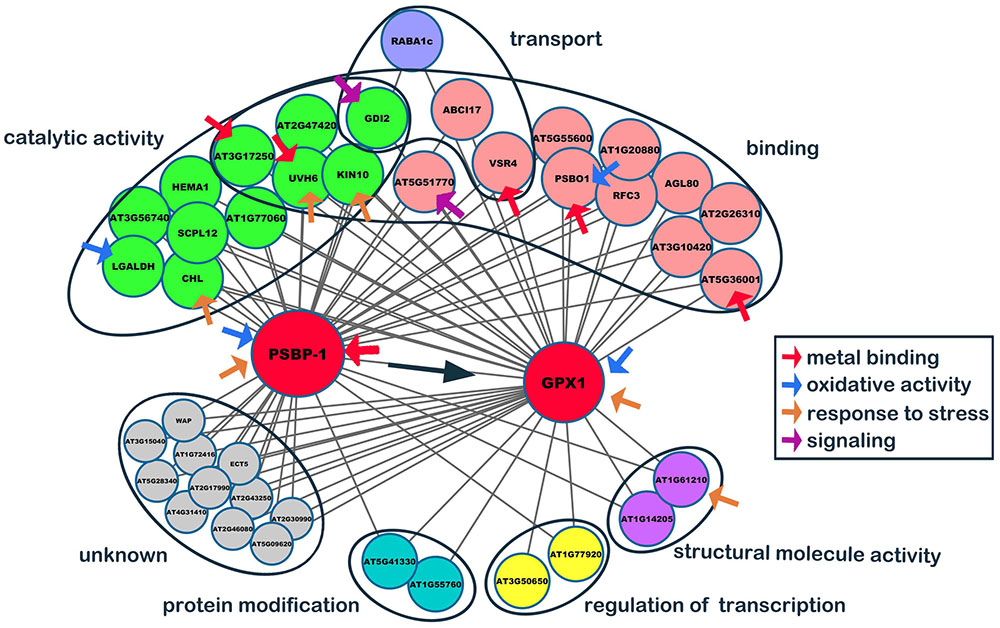
FIGURE 8. Co-expression network of PSBP2-1 and GPX1. From the top 50 hub genes, two major genes, oxygen-evolving enhancer protein 2-1 (PSBP2-1) and phospholipid hydroperoxide glutathione peroxidase 1 (GPX1), which might directly relate to oxidative stress, were selected, and a stress-response co-expression subnetwork was constructed by the Cytoscape software.
Cadmium is known to generate and lead to accumulation of ROS in plants, resulting in accelerated cell death (Foyer and Noctor, 2005). At moderate levels, ROS play an important role as signal molecules in the regulation of numerous biological processes (Baxter et al., 2014). Cd concentrations in the leaves and stems of S. alfredii reached maximums of 9,000 and 6,500 mg/kg, respectively, at a treatment level of 400 μM Cd (Yang et al., 2004). These values indicated that aerial plant parts could accumulate Cd and suffer from severely Cd stress. Initially, Cd2+ is readily taken up by the roots and translocated into stems and leaves. Then, ROS, as a molecular signal, cause toxicity in plants. In this study, the SaCu/Zn SOD expression initially decreased after 0.5 and 6 h of Cd treatment. However, to protect stem and leaf tissues from ROS damage, its expression level gradually increased and finally reached a peak (12 and 24 h).
Cadmium can enter plant cells, producing oxidative stress, inhibiting chlorophyll synthesis and disturbing photosynthetic metabolism, destroying membrane integrity, decreasing nutrient uptake and inhibiting plant growth (Schützendübel et al., 2001; Pietrini et al., 2003). In this study, the etiolation of WT was severe due to Cd exposure, and the chlorophyll content confirmed this phenomenon. However, transgenic Arabidopsis plants overexpressing SaCu/Zn SOD showed improved seed germination and increased plant growth under Cd stress conditions (Figure 3 and Supplementary Figure S4), indicating that Cd inhibited plant growth through inhibiting of chlorophyll synthesis and impairing photosynthesis. Consistently, overexpression of pea chloroplast Cu/Zn-SOD in tobacco improved photosynthetic performance under moderate stress and maintained the photosynthetic capacity after severe oxidative stress (Gupta et al., 1993).
Plants possess very efficient enzymatic antioxidant defense systems, which work in concert to control cascades of excessive oxidation and protect plant cells from oxidative damage by scavenging ROS (Gill and Tuteja, 2010). In the present study, the enhanced SOD and POD enzyme activities, decreased damage to membrane integrity, and lowered O2•- and H2O2 contents in transgenic Arabidopsis under Cd stress (Figures 5, 6) indicating that these plants were more resistant to oxidation than WT plants. We hypothesize that transgenic tissues have lower O2•- and H2O2 levels, leading to higher net rates of photosynthesis and reduced oxidative damage under stressful conditions. The protective effects of overexpression of SaCu/Zn SOD in Arabidopsis plants may involve two strategies: enhancing ROS scavenge efficiency and net rates of photosynthesis and/or decreasing damage caused by lipid peroxidation and damage affecting plasma membrane integrity.
Cadmium uptake and plasma membrane integrity have internal correlations in transgenic and WT plants (Figures 4–6). Histochemical staining and the quantification of the loss of plasma membrane integrity suggested that Cd exposure increased the levels of membrane damage in both WT and transgenic plants. It would be interesting to analyze results showing that the Cd2+ influx of OE lines increased under normal growth conditions or Cd stress, but the Cd accumulation of transgenic and WT plants was not significantly different (data not shown). Mühling and Läuchli (2003) reported that disturbed membrane functions may cause elevated Cd concentrations. Cd stress may damage plasma membrane integrity, resulting in more Cd2+ being transported into the cell and accumulating in WT plants. The Cd2+ flux increased, implying the possibility of more Cd accumulation in OE lines; then at last the Cd contents of WT and OE were similar. The Cd accumulation due to cell damage influenced the state of WT plants, but the effects of SaCu/Zn SOD overexpression in transgenic Arabidopsis decreased the damage.
Several reports have indicated that a high antioxidative capacity is required for heavy metal hyperaccumulation in plants (Cho and Seo, 2005; Wang et al., 2008). The antioxidative system is composed of non-enzymatic (ascorbate, glutathione, α-tocopherol, and carotenoid) and ROS-scavenging enzymes (SOD, POD, CAT, ascorbate peroxidase) (Mittler et al., 2004). In this study, transgenic Arabidopsis accumulated less H2O2 and O2•- than WT plants (Figure 5), which suggested that the former have a high ROS-scavenging capacity. According to microarray analysis, the most highly expressed antioxidant-related genes were the member of glutaredoxin and peroxidase family (Figure 7). In addition to prominent antioxidant enzymes, we also found many other enzymes in the microarray data, such as alternative oxidase (AOX), dioxygenases, and peptidyl proyl cis–trans isomerases (PPIase). AOX is a terminal oxidases catalyzing the reduction of oxygen to water in the plant mitochondrial electron transport chain (Millar et al., 2011). The ability of AOX to maintain critical mitochondrial and chloroplastic functions during extreme drought is likely due to its ability to reduce oxidative damage (Dahal and Vanlerberghe, 2017). Dioxygenases are non-heme iron-containing enzymes important in the biosynthesis of plant signaling compounds such as abscisic acid, gibberellins, and ethylene and of secondary metabolites, notably flavonoids and alkaloids (Prescott and John, 1996). Overexpression of a dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance (Qin and Zeevaart, 2002). PPIases are ubiquitous proteins that are found in the cytosols of both prokaryotic and eukaryotic cells, and have proposed functions in protein folding, protein degradation, stress response and signal transduction (Laxa et al., 2007). In this study, a gene similar to Arabidopsis AOX3 was significantly down-regulated at the 3rd week, but the expression levels of dioxygenase and PPIase were not sharply changed. We speculate that overexpression of SaCu/Zn SOD gene may mainly affect the high expression of antioxidative-related genes such as glutaredoxin and peroxidase, which interact with Cu/Zn SOD in Arabidopsis and work together to scavenge ROS, rather than AOX, dioxygenases and PPIase.
The co-expression network may reveal a possible model of SaCu/Zn SOD action in transgenic Arabidopsis after Cd-induced oxidative stress. Cd can affect many vital processes, including a series of genes, especially stress-related genes, interact with oxidative genes under Cd toxicity. Subsequently, catalysis and binding play important roles in Cd-exposed Arabidopsis, and many metal ions participate in these activities. Iron-sulfur (Fe-S) cluster proteins, such as UVH6, with Fe as a cofactor are essential to photosynthesis and oxidation-reduction reactions (Zhang, 2015). Zn2+ is also a cofactor for many enzymes, such as AT5G36001, which plays an essential role in transcription of many antioxidant genes (Hansen et al., 2007). The binding of Ca2+, Fe2+, Mn2+ and Mg2+ is also closely related to Cd response (Groten et al., 1991). The genetic and physiological processes of plants are strongly associated due to genes having two or more simultaneous functions at the same time, and we hypothesize that the overexpression of SaCu/Zn SOD may participate in this co-expression network against Cd-induced oxidative stress in some unclear way.
Based on the microarray data and co-expression network, we developed a schematic model for transgenic Arabidopsis with enhanced oxidation under Cd stress (Figure 9). Cd stress induces the production of ROS, which are highly reactive molecules, leading to oxidative stress. Under normal conditions, the lower levels of ROS in WT and OE plants play important roles as signaling molecules in numerous biological processes. However, Cd stress induces ROS overproduction and oxidative stress. Transgenic Arabidopsis plants have a greater ability to scavenge ROS compared with WT plants, so they have better growth than WT plants. Additionally, the expression and regulation of photosystem II and reduction peroxides were mediated by oxidative stress induced by Cd stress. Thus, SaCu/Zn SOD in Arabidopsis might act on the two hub genes, and together these genes regulate central cell differentiation, signal transduction, protein modification and secondary metabolites.
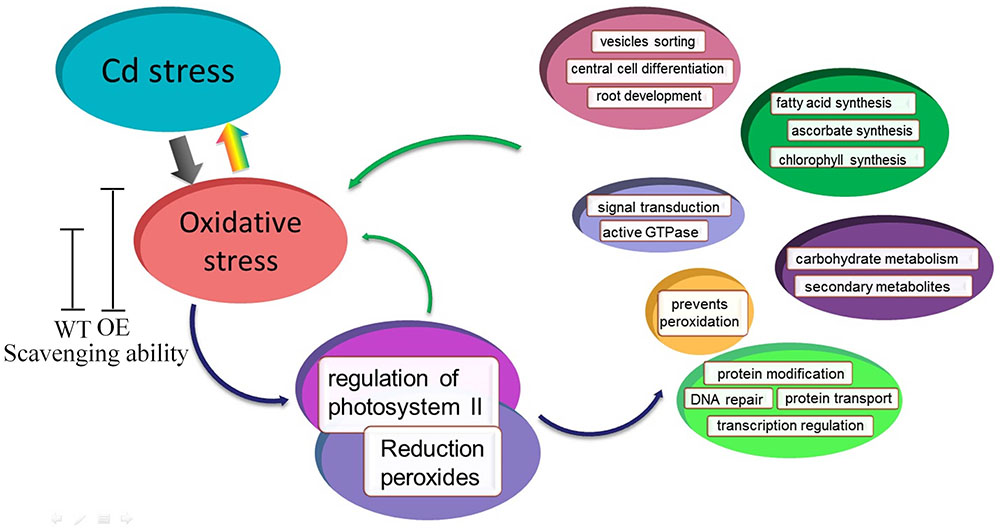
FIGURE 9. Model of SaCu/Zn SOD mediating Cd stress response in transgenic Arabidopsis. Transgenic Arabidopsis overexpressing SaCu/Zn SOD exhibit increased the expression of SaCu/Zn SOD, and as a result, these plants have greater ROS scavenging ability than WT plants. Under Cd stress, plants suffer from oxidative stress and then regulate the photosystems and peroxides. The stress response involves six categories of biological process such as signal transport, material synthesis, metabolism, protein modification, DNA repair, etc.
In summary, SaCu/Zn SOD may be responsible for conferring Cd tolerance. Additional modifications of other active-oxygen-scavenging system components in plants will help to elucidate the interactions among these protective mechanisms. We remain hopeful that investigations of this type will eventually provide significant improvements in the tolerance of cultivated plants to Cd stress.
RZ and ZL conceived and designed the experiments. ZL, XS, and XH performed the experiments. JJ, QH, YZ, ML, and GQ analyzed the data. ZL, XH, and XS wrote the manuscript. All of the authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the National High Technology Research and Development Program of China (No. 2013AA102701-3), the National Non-profit Institute Research Grant of CAF (No. TGB2013008 and RISF2014010), and the National Natural Science Foundation of China (No. 31200465).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01010/full#supplementary-material
FIGURE S1 | Comparison of the genomic DNA sequences of SaCu/Zn SOD and other plant Cu/Zn SOD genes.
FIGURE S2 | PCR detection of transgenic Arabidopsis. (A) SaCu/Zn SOD expression levels in seven independent lines of SaCu/Zn SOD transgenic and WT Arabidopsis plants. (B,C) PCR analysis of SaCu/Zn SOD -transformed Arabidopsis using vector primers (B) and gene primers (C). M: DNA Marker DL2000; 1–2: The WT line and water were used as negative control; 3–10: SaCu/Zn SOD-transformed Arabidopsis plant lines; 11: The recombinant plasmid pBI121G-SaCu/Zn SOD.
FIGURE S3 | Germination status of transgenic Arabidopsis seeds after 7 days on a culture dish. The seeds of SaCu/Zn SOD transgenic (OE2, OE3, and OE4) and WT plants were surface-sterilized and plated on 1/2 MS agar medium plates supplemented with 1.5 mM CdCl2 and grown 7 days.
FIGURE S4 | Fresh weights of transgenic and WT Arabidopsis exposed to Cd for different time periods. Bars indicate means ± SD. Asterisks indicate significant differences at ∗p < 0.05 and ∗∗p < 0.01.
FIGURE S5 | Gene ontology analyses of the differentially regulated genes identified by microarray analysis under normal growth and Cd stress conditions. (A–D) indicate 0 week (ck), 1 week, 2 weeks, and 3 weeks, respectively. GO analysis of each period includes “Biological process,” “Cellular component,” and “Molecular function”.
TABLE S1 | Primers used in this study.
TABLE S2 | Differentially expressed genes that are up- and down-regulated at least 2-fold in SaCn/Zn SOD transgenic Arabidopsis compared with WT plants under CdCl2 stress.
TABLE S3 | A complete list of top 50 hub gene module assignments.
TABLE S4 | Edge genes annotated in the co-expression subnetwork of PSBP-1 and GPX1.
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Baxter, A., Mittler, R., and Suzuki, N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. doi: 10.1093/jxb/ert375
Bueno, P., Varela, J., Gimenez-Gallego, G., and Del Rio, L. A. (1995). Peroxisomal copper, zinc superoxide dismutase. Characterization of the isoenzyme from watermelon cotyledons. Plant Physiol. 108, 1151–1160. doi: 10.1104/pp.108.3.1151
Chabory, E., Damon, C., Lenoir, A., Henry-Berger, J., Vernet, P., Cadet, R., et al. (2010). Mammalian glutathione peroxidases control acquisition and maintenance of spermatozoa integrity. J. Anim. Sci. 88, 1321–1331. doi: 10.2527/jas.2009-2583
Cho, U. H., and Seo, N. H. (2005). Oxidative stress in Arabidopsis thaliana exposed to cadmium is due to hydrogen peroxide accumulation. Plant Sci. 168, 113–120. doi: 10.1016/j.plantsci.2004.07.021
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dahal, K., and Vanlerberghe, G. C. (2017). Alternative oxidase respiration maintains both mitochondrial and chloroplast function during drought. New Phytol. 213, 560–571. doi: 10.1111/nph.14169
Dietz, K. J. (2003). Plant peroxiredoxins. Annu. Rev. Plant Biol. 54, 93–107. doi: 10.1146/annurev.arplant.54.031902.134934
Fan, L., Zheng, S., and Wang, X. (1997). Antisense suppression of phospholipase D alpha retards abscisic acid-and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell 9, 2183–2196. doi: 10.1105/tpc.9.12.2183
Foyer, C. H., and Noctor, G. (2005). Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17, 1866–1875. doi: 10.1105/tpc.105.033589
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Gratão, P. L., Polle, A., Lea, P. J., and Azevedo, R. A. (2005). Making the life of heavy metal-stressed plants a little easier. Funct. Plant Biol. 32, 481–494. doi: 10.1071/FP05016
Groten, J., Sinkeldam, E., Muys, T., Luten, J., and Van Bladeren, P. (1991). Interaction of dietary Ca, P, Mg, Mn, Cu, Fe, Zn and Se with the accumulation and oral toxicity of cadmium in rats. Food Chem. Toxicol. 29, 249–258. doi: 10.1016/0278-6915(91)90022-Y
Gupta, A. S., Heinen, J. L., Holaday, A. S., Burke, J. J., and Allen, R. D. (1993). Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 90, 1629–1633. doi: 10.1073/pnas.90.4.1629
Han, X., Yin, H., Song, X., Zhang, Y., Liu, M., Sang, J., et al. (2016). Integration of small RNAs, degradome and transcriptome sequencing in hyperaccumulator Sedum alfredii uncovers a complex regulatory network and provides insights into cadmium phytoremediation. Plant Biotechnol. J. 14, 1470–1483. doi: 10.1111/pbi.12512
Hansen, B. H., Rømma, S., Garmo, A., Pedersen, S. A., Olsvik, P. A., and Andersen, R. A. (2007). Induction and activity of oxidative stress-related proteins during waterborne Cd/Zn-exposure in brown trout (Salmo trutta). Chemosphere 67, 2241–2249. doi: 10.1016/j.chemosphere.2006.12.048
Jomova, K., and Valko, M. (2011). Advances in metal-induced oxidative stress and human disease. Toxicology 283, 65–87. doi: 10.1016/j.tox.2011.03.001
Kishikawa, N., and Kuroda, N. (2014). Chemiluminescence assay for the investigation of reactive oxygen species generator. Yakugaku Zasshi 135, 191–196. doi: 10.1248/yakushi.14-00213-1
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Laxa, M., König, J., Dietz, K. J., and Kandlbinder, A. (2007). Role of the cysteine residues in Arabidopsis thaliana cyclophilin CYP20-3 in peptidyl-prolyl cis–trans isomerase and redox-related functions. Biochem. J. 401, 287–297. doi: 10.1042/BJ20061092
Lin, C. T., Lin, M. T., Chen, Y. T., and Shaw, J. F. (1995). Subunit interaction enhances enzyme activity and stability of sweet potato cytosolic Cu/Zn-superoxide dismutase purified by a His-tagged recombinant protein method. Plant Mol. Biol. 28, 303–311. doi: 10.1007/BF00020249
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Millar, A. H., Whelan, J., Soole, K. L., and Day, D. A. (2011). Organization and regulation of mitochondrial respiration in plants. Annu. Rev. Plant Biol. 62, 79–104. doi: 10.1146/annurev-arplant-042110-103857
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Mühling, K. H., and Läuchli, A. (2003). Interaction of NaCl and Cd stress on compartmentation pattern of cations, antioxidant enzymes and proteins in leaves of two wheat genotypes differing in salt tolerance. Plant Soil 253, 219–231. doi: 10.1023/A:1024517919764
Pietrini, F., Iannelli, M. A., Pasqualini, S., and Massacci, A. (2003). Interaction of cadmium with glutathione and photosynthesis in developing leaves and chloroplasts of Phragmites australis (Cav.) Trin. ex Steudel. Plant Physiol. 133, 829–837. doi: 10.1104/pp.103.026518
Prescott, A. G., and John, P. (1996). Dioxygenases: molecular structure and role in plant metabolism. Annu. Rev. Plant Biol. 47, 245–271. doi: 10.1146/annurev.arplant.47.1.245
Qin, X., and Zeevaart, J. A. D. (2002). Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 128, 544–551. doi: 10.1104/pp.900019
R Core Team (2014). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rao, M. V., Paliyath, G., and Ormrod, D. P. (1996). Ultraviolet-B-and ozone-induced biochemical changes in antioxidant enzymes of Arabidopsis thaliana. Plant Physiol. 110, 125–136. doi: 10.1104/pp.110.1.125
Rodríguez-Serrano, M., Romero-Puertas, M. C., Zabalza, A., Corpas, F. J., Gómez, M., Del Ro, L. A., et al. (2006). Cadmium effect on oxidative metabolism of pea (Pisum sativum L.) roots. Imaging of reactive oxygen species and nitric oxide accumulation in vivo. Plant Cell Environ.. 29, 1532–1544. doi: 10.1111/j.1365-3040.2006.01531.x
Saito, R., Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P. L., Lotia, S., et al. (2012). A travel guide to cytoscape plugins. Nat. Methods 9, 1069–1076. doi: 10.1038/NMETH.2212
Schützendübel, A., Schwanz, P., Teichmann, T., Gross, K., Langenfeld-Heyser, R., Godbold, D. L., et al. (2001). Cadmium-induced changes in antioxidative systems, hydrogen peroxide content, and differentiation in Scots pine roots. Plant Physiol. 127, 887–898. doi: 10.1104/pp.010318
Valko, M., Morris, H., and Cronin, M. (2005). Metals, toxicity and oxidative stress. Curr. Med. Chem. 12, 1161–1208. doi: 10.2174/0929867053764635
Velikova, V., Yordanov, I., and Edreva, A. (2000). Oxidative stress and some antioxidant systems in acid rain-treated bean plants: protective role of exogenous polyamines. Plant Sci. 151, 59–66. doi: 10.1016/S0168-9452(99)00197-1
Wang, F. Z., Wang, Q. B., Kwon, S. Y., Kwak, S. S., and Su, W. A. (2005). Enhanced drought tolerance of transgenic rice plants expressing a pea manganese superoxide dismutase. J. Plant Physiol. 162, 465–472. doi: 10.1016/j.jplph.2004.09.009
Wang, Z., Zhang, Y., Huang, Z., and Huang, L. (2008). Antioxidative response of metal-accumulator and non-accumulator plants under cadmium stress. Plant Soil 310, 137–149. doi: 10.1007/s11104-008-9641-1
Woertink, J. S., Smeets, P. J., Groothaert, M. H., Vance, M. A., Sels, B. F., Schoonheydt, R. A., et al. (2009). A [Cu2O]2+ core in Cu-ZSM-5, the active site in the oxidation of methane to methanol. Proc. Natl. Acad. Sci. U.S.A. 106, 18908–18913. doi: 10.1073/pnas.0910461106
Wu, J., Zhang, J., Li, X., Xu, J. J., and Wang, L. (2016). Identification and characterization of a PutCu/Zn-SOD gene from Puccinellia tenuiflora (Turcz.) Scribn. et Merr. Plant Growth Regul. 79, 55–64. doi: 10.1007/s10725-015-0110-6
Xu, J., Duan, X. G., Yang, J., Beeching, J. R., and Zhang, P. (2013). Coupled expression of Cu/Zn-superoxide dismutase and catalase in cassava improves tolerance against cold and drought stresses. Plant Signal. Behav. 8:e24525. doi: 10.4161/psb.24525
Xu, J., Yang, J., Duan, X. G., Jiang, Y. M., and Zhang, P. (2014). Increased expression of native cytosolic Cu/Zn superoxide dismutase and ascorbate peroxidase improves tolerance to oxidative and chilling stresses in cassava (Manihot esculenta Crantz). BMC Plant Biol. 14:208. doi: 10.1186/s12870-014-0208-4
Yang, X. E., Jin, X. F., Feng, Y., and Islam, E. (2005). Molecular mechanisms and genetic basis of heavy metal tolerance/hyperaccumulation in plants. J. Integr. Plant Biol. 47, 1025–1035. doi: 10.1111/j.1744-7909.2005.00144.x
Yang, X. E., Long, X. X., Ye, H. B., He, Z. L., Calvert, D. V., and Stoffella, P. J. (2004). Cadmium tolerance and hyperaccumulation in a new Zn-hyperaccumulating plant species (Sedum alfredii Hance). Plant Soil 259, 181–189. doi: 10.1023/B:PLSO.0000020956.24027.f2
Zhang, C. (2015). Involvement of iron-containing proteins in genome integrity in Arabidopsis thaliana. Genome Integr. 6:2. doi: 10.4103/2041-9414.155953
Keywords: Cd stress, co-expression network, Cu/Zn SOD, oxidative stress, Sedum alfredii
Citation: Li Z, Han X, Song X, Zhang Y, Jiang J, Han Q, Liu M, Qiao G and Zhuo R (2017) Overexpressing the Sedum alfredii Cu/Zn Superoxide Dismutase Increased Resistance to Oxidative Stress in Transgenic Arabidopsis. Front. Plant Sci. 8:1010. doi: 10.3389/fpls.2017.01010
Received: 05 February 2017; Accepted: 26 May 2017;
Published: 13 June 2017.
Edited by:
Erik Harry Murchie, University of Nottingham, United KingdomReviewed by:
Manoj Kumar, University of Technology, Sydney, AustraliaCopyright © 2017 Li, Han, Song, Zhang, Jiang, Han, Liu, Qiao and Zhuo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renying Zhuo, emh1b3J5QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.