- 1Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China
- 2State Key Laboratory of Forest Genetics and Tree Breeding, Northeast Forestry University, Harbin, China
Histone modifications play important roles in regulating the expression of C4 photosynthetic genes. Given that all enzymes required for the C4 photosynthesis pathway are present in C3 plants, it has been hypothesized that this expression regulatory mechanism has been conserved. However, the relationship between histone modification and the expression of homologs of C4 photosynthetic enzyme genes has not been well determined in C3 plants. In the present study, we cloned nine hybrid poplar (Populus simonii × Populus nigra) homologs of maize (Zea mays) C4 photosynthetic enzyme genes, carbonic anhydrase (CA), pyruvate orthophosphate dikinase (PPDK), phosphoenolpyruvate carboxykinase (PCK), and phosphoenolpyruvate carboxylase (PEPC), and investigated the correlation between the expression levels of these genes and the levels of promoter histone acetylation modifications in four vegetative tissues. We found that poplar homologs of C4 homologous genes had tissue-dependent expression patterns that were mostly well-correlated with the level of histone acetylation modification (H3K9ac and H4K5ac) determined by chromatin immunoprecipitation assays. Treatment with the histone deacetylase inhibitor trichostatin A further confirmed the role of histone acetylation in the regulation of the nine target genes. Collectively, these results suggest that both H3K9ac and H4K5ac positively regulate the tissue-dependent expression pattern of the PsnCAs, PsnPPDKs, PsnPCKs, and PsnPEPCs genes and that this regulatory mechanism seems to be conserved among the C3 and C4 species. Our findings provide new insight that will aid efforts to modify the expression pattern of these homologs of C4 genes to engineer C4 plants from C3 plants.
Introduction
The C3 photosynthetic pathway is the predominant method of carbon fixation and is used by over 90% of terrestrial plant species. C3 photosynthesis is relatively inefficient, due to the inefficiency of the carbon-fixing enzyme, Rubisco, which catalyzes both carboxylation (carbon fixation) and oxygenation (photorespiration). The evolution of C4 photosynthesis about 35–40 million years ago provided a natural solution to remedy the inefficiency of Rubisco, and in angiosperms C4 photosynthesis has emerged independently at least 66 times from C3 plants (Sage et al., 2012).
Unlike C3 plants, C4 plants have a Kranz-type anatomy of altered vascular bundles, and two separate cell types, bundle sheath and mesophyll, are required for photosynthesis (Slack and Hatch, 1967; Hattersley, 1984). The light dependent reactions and the first carbon fixation step occur in the mesophyll cells, whereas the second carbon fixation step by Rubisco occurs in the lower oxygen environment of the bundle sheath cells. In addition to altered anatomical structures, the evolution of C4 photosynthesis required changes in the expression of essential enzymes such as PEPC and CA that catalyze the CO2 concentration reactions, and PPDK, NADP- or NAD-dependent malic enzymes (NADP-ME or NAD-ME, respectively) and PCK that shuttle four-carbon acids between the mesophyll and bundle sheath cells (Hatch et al., 1975).
All of the enzymes required for the C4 photosynthesis pathway are present in C3 plants, albeit with much lower activities (Ku et al., 1996). In plants, these C4 photosynthetic enzymes or their isoforms are important for central metabolism. β-CA activity is found in the cytosol of mesophyll cell of C4 plants, where it catalyzes the first reaction in the C4 photosynthesis CO2-concentrating mechanisms (CCM), the conversion of atmospheric CO2 to HCO3- (Gutierrez et al., 1974; Hatch and Burnell, 1990). In C3 plants, β-CA is localized in leaf mesophyll chloroplasts of higher C3 plants, where CA can make up 1 to 2% of total leaf protein (Okabe et al., 1984; Fett and Coleman, 1994; Peltier et al., 2006). PEPC is the primary carboxylase in the mesophyll cells, whereas a CO2 pump is established in C4 plants (Westhoff and Gowik, 2004). In C3 plants, PEPC plays a critical role in modulating the balance of carbon and nitrogen metabolism, and amino acid synthesis (Shi et al., 2015). PPDK controls amino acid metabolism and starch biosynthesis in seeds, provides PEP to the shikimate pathway for lignin biosynthesis in the mid-vein of leaves, and plays an important role in the transport of amino acids during natural leaf senescence in Arabidopsis thaliana (Taylor et al., 2010). Besides an important role in gluconeogenesis (Leegood and Ap Rees, 1978; Eastmond et al., 2000; Rylott et al., 2003; Malone et al., 2007), PCK also involves in pH stability and nitrogen and amino acid metabolism in many C3 plants (Walker et al., 1999, 2001; Lea et al., 2001; Taylor et al., 2010). As decarboxylating enzyme, PCK is found in bundle sheath cells of C4 plants, where it plays a role in decarboxylating C4 acids (Walker and Leegood, 1996). Interestingly, there is support for evidence that PCK plays a role in decarboxylating C4 acids via a partial C4 cycle in the vascular system of some C3 plants (Hibberd and Quick, 2002).
Additionally, there are a few function-specific differences between C4 photosynthetic genes and their homologous genes in C3 and C4 plants, which are mostly governed by regulatory properties of gene specific expression (Sheen, 1999). Recent studies have confirmed that C4-CA, C4-PEPC, and C4-PPDK are localized in mesophyll cells, whereas C4-NAD(P)-ME, C4-PCK, and Rubisco (RbcS) are expressed at high levels in bundle sheath cells of leaves of the C4 plant maize (Langdale, 2011). Several studies in other species have identified the regulatory mechanisms controlling the expression patterns of these C4 photosynthetic enzymes (Offermann et al., 2006, 2008; Heimann et al., 2013). In plants, the acetylation of histone H3 lysine residue 9 (H3K9ac) and H4 lysine residue 5 (H4K5ac) is associated with gene transcriptional activation and is considered a marker of euchromatin (Lee et al., 2010; Heimann et al., 2013; Hou et al., 2015; Wang et al., 2015). Current evidence suggests that increases in the level of H3K9ac and H4K5ac in the promoters of C4 photosynthetic enzyme genes upon illumination are linked to transcriptional activation (Heimann et al., 2013). A recent study showed that C4-PPDK, C4-PCK, and their non-C4 homologous genes have organ-specific expression patterns, and in maize the level of H3K9ac and H4K5ac modifications correlates well with the mRNA level of most PPDK and PCK genes (Dong et al., 2016). Given that genes encoding C4 photosynthetic enzyme evolved from their C3 counterparts, it is hypothesized that similar regulatory mechanisms may underlie the regulation of these homologous gene expressions in C3 plants.
To assess the relationship between homologous C4 photosynthetic enzyme gene expression and histone acetylation modification in C3 woody plants, we cloned nine C4 photosynthetic enzyme gene homologs from the hybrid poplar Populus simonii × Populus nigra. We investigated their expression patterns as well as the enrichment of H3K9ac and H4K5ac modifications in the promoters of these genes in leaves, stem chlorenchyma (Sc), stem vascular tissue (Sv) and roots. Overall, our results show that PsnCAs, PsnPPDKs, PsnPCKs, and PsnPEPCs have tissue-dependent expression patterns, and most of these genes’ transcript abundances are well-correlated with the levels of H3K9ac and H4K5ac. Our results and previous research on the C3 herbaceous plant Arabidopsis thaliana (Zhou et al., 2010) and the C4 gramineous plants sorghum and maize (Heimann et al., 2013) suggest that the regulation of the expression of C4 photosynthetic genes and their C3 homologs by histone acetylation is conserved. Based on this similar regulatory mechanism it may be possible to modify C3 photosynthetic gene expression to be more C4-like as a part of future efforts to engineer C3 plants that are capable of C4 photosynthesis.
Materials and Methods
Plant Materials and Treatment
The poplar hybrid P. simonii × P. nigra (hereafter referred to as poplar) was used for all studies. Poplar seedlings were grown for 1 month under a 16 h day/8 h night cycle at 25°C in a tissue culture vessel containing MH medium, pH 5.8, plus 2% sucrose and 0.7% agar. Subsequently, healthy poplar seedlings were transferred to a mixture of soil and vermiculite (1:1) and cultivated in a greenhouse under a 16 h day (25°C)/8 h night (22°C) cycle for 5 months. Leaves from the fourth internode to the sixteenth internode, stem chlorenchyma (Sc) and stem vascular tissue (Sv) from the sixth internode to the eighteenth internode, and root tissues were harvested from 20 independent 5-month-old poplar plants 4 h after the onset of illumination (Figure 1).
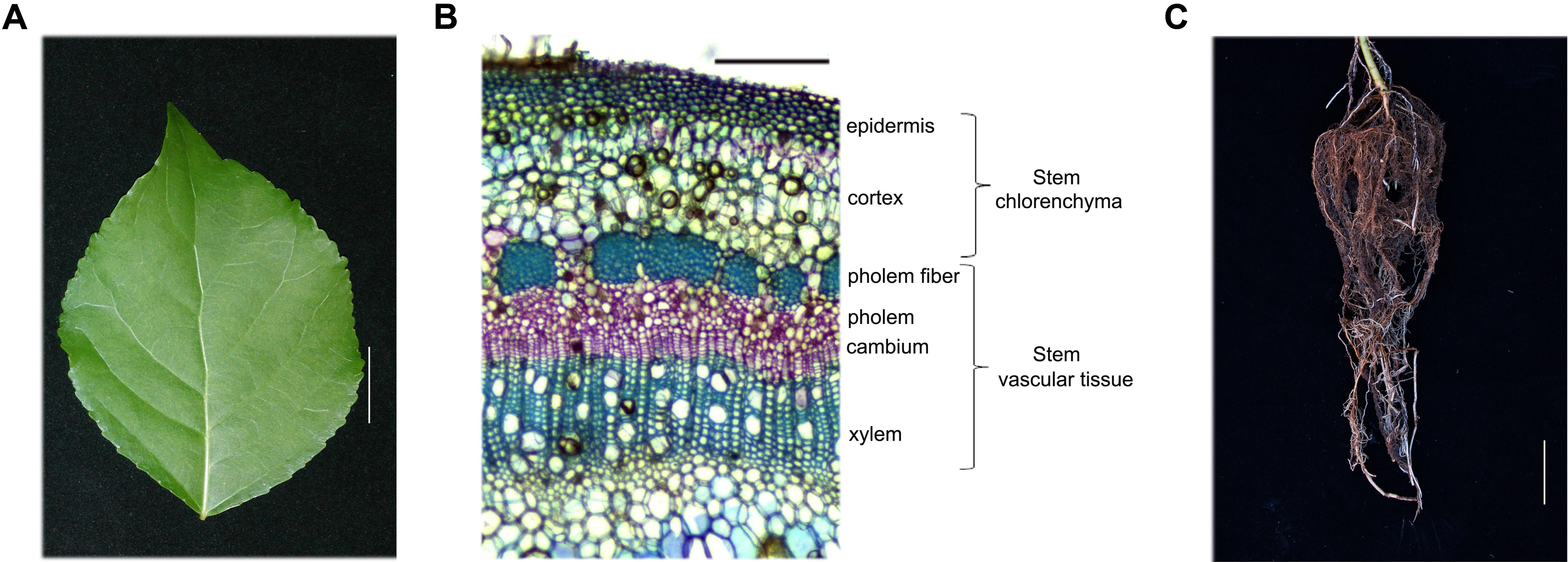
FIGURE 1. Hybrid poplar tissue and cell types used in this study. (A–C) Tissues collected from 5-month-old P. simonii × P. nigra grown in a greenhouse. (A) Leaf morphology. Scale bar, 2 cm. (B) Cross-section of a stem stained with toluidine blue. The cell types are labeled on the right. Many chloroplasts can be seen in the epidermis and cortex cells, which are called the stem chlorenchyma in our study. Phloem fiber, phloem, cambium and xylem cells are called stem vascular tissue. Scale bar, 200 μm. (C) Root morphology. Scale bar, 5 cm.
Twelve additional 5-month-old poplar plants grown in a greenhouse were sprayed with a low concentration (2.5 and 5 μM) of TSA (Sigma, St. Louis, MO, United States) for 2 days to study the correlation between histone acetylation and gene expression. All tissues were immediately frozen in liquid N2 and completely ground using the 6870 Freezer Mill (SPEX SamplePrep, Metuchen, NJ, United States).
Microscopic Observation
Free-hand cross-sections, 25–30 μm thick, of the 12th internode of fresh stems were stained with 0.1% toluidine blue (Sigma, St. Louis, MO, United States). Sections were observed under an OLYMPUS microscope (OLYMPUS, Japan) equipped with a computer-assisted digital camera MODEL ARTCAM-1400MI-WOM (ARTRAY, Japan).
Chlorophyll Content Measurement
Chlorophyll content was measured using a previously published method (Ni et al., 2009). 300 mg of leaf, Sc and Sv tissues from 5-month-old poplar were ground in a mortar and pestle under liquid nitrogen. The ground tissue was transferred to 15 ml Falcon tubes, 5 ml 80% acetone was added, and samples were then mixed in the dark for 30 min. Subsequently, the tubes were centrifuged at 3,000 rpm at 4°C for 15 min, and the supernatant was transferred to a new tube. These procedures were repeated twice. A spectrophotometer (MAPADA, Shanghai, China) was used to measure to the absorbance due to chlorophyll at 663 and 645 nm. Chlorophyll content was determined for at least six independent samples per tissue.
RNA Extraction, DNase I Digestion, and cDNA Synthesis
Total RNA was extracted from 100 mg of powdered leaves, Sc, Sv, and roots of poplar using the pBIOZOL Plant Total RNA Extraction Reagent according to the manufacturer’s instructions (BioFlux, Tokyo, Japan). The concentration of RNA was determined using the Nanodrop 2000 (Thermo Fisher, Waltham, MA, United States), and the sample was resolved on a 1.2% agarose gel to check the integrity of the RNA. To remove genomic DNA contamination, 1 μg of RNA was digested using the DNase I digestion Reagent Kit (Invitrogen, Carlsbad, CA, United States). Total cDNA was synthesized using the M-MLV Reverse Transcriptase Reagent Kit (Invitrogen, Carlsbad, CA, United States) and an oligo(dT) primer. A working sample of cDNA was prepared by diluting five-fold with sterile water and storing at -20°C.
Gene Cloning and Vector Construction
The genomic sequences of poplar genes homologous to maize C4 photosynthesis enzyme genes CA, PPDK, PCK, and PEPC were obtained from the poplar sequence database1 (Supplemental Table S1). Full-length sequences of these genes were amplified from cDNA synthesized from poplar leaf or root RNA with KOD polymerase (NEB) according to the manufacture’s protocol using the primers listed in Supplemental Table S2. PCR products were purified with the TIANgel mini purification kit (TIAN GEN, Beijing, China), cloned into the pEasy-blunt cloning vector (Trans Gene, Beijing, China), and sequenced. The upstream sequences (about 2 kb in length) corresponding to the promoters of these genes were amplified from poplar genomic DNA using the primers listed in Supplemental Table S3.
Selection of Reference Genes
Quantitative real-time polymerase chain reaction is a reliable technique for quantifying gene expression, and requires stable reference genes for data normalization (Xiao et al., 2015). However, no single reference gene has stable expression under all experimental conditions (Podevin et al., 2012). Therefore, it is necessary to choose a suitable reference gene which is expressed at a relatively stable level across the conditions being tested.
In our study, five housekeeping genes, ACTIN2, TUBLIN, UBIQUITIN (UBQ), Elongation Factor 1a (EF1a), and 18S ribosomal RNA (18S rRNA) were tested for suitability as reference genes for gene expression studies in four vegetative tissues, including leaves, Sc, Sv, and roots, as well as in TSA-treated P. simonii × P. nigra. These gene sequences were obtained from the Populus trichocarpa genome annotation v3.02. QRT-PCR was performed using the primers listed in Supplemental Table S4 to evaluate the expression variation of these candidates across tissues. The average cycle threshold (Cq) values of candidate genes ranged from 10.68 to 22.76 in the four tissues (Supplemental Figure S2A). Except for 18S rRNA, all genes showed low variability in Cq value between leaves, Sc, Sv, and roots.
The average expression stability value (M-value) is a parameter used by the geNorm software program to identify the best reference genes. The lower the M-value is, the more stable the gene expression (Bustin et al., 2009). We also used geNorm software to select the most stable reference genes (Dong et al., 2016). Based on geNorm analysis, ACTIN2 and EF1a were the most stable genes among the five candidate reference genes (Supplemental Figure S2B). In a previous study, PtACTIN2 was used as reference gene for diverse tissues of 1-year-old P. trichocarpa, including differentiated or mature xylem and phloem (Yu et al., 2013). Therefore, we chose to use ACTIN2 as a reference gene in our study.
Chromatin Immunoprecipitation (ChIP)
We carried out ChIP as previously described (Bowler et al., 2004; Gendrel et al., 2005; Li et al., 2014) with the following modifications: 1.5 g of leaves, Sc, Sv, and roots from 5-month-old poplar plants grown in a greenhouse were harvested separately and cross-linked with 1% (wt/wol) formaldehyde for 15 min at 4°C. The purified chromatin was sheared to 0.3–0.7 kb fragments by sonicating under cooling for 4 min (4 s-on, 10 s-off) at 15% amplitude using a Vibra-cell VCX-505 sonicator (Sonics, Newtown, CT, United States). The sheared chromatin was diluted with ChIP dilution buffer and pre-cleared with 100 μl of protein A agarose (Millipore, Billerica, MA, United States) (Bowler et al., 2004; Gendrel et al., 2005).
For each sample, 1 ml of diluted pre-cleared chromatin was used for immunoprecipitation, and a 10 μl aliquot was used to quantify the amount of input. To immunoprecipitate chromatin containing histone modifications, 100 μl of protein A agarose and 5 μl anti-acetyl H3K9 (07-352, Millipore, Billerica, MA, United States) or 5 μl anti-acetyl H4K5 (07-327, Millipore, Billerica, MA, United States) were added to the diluted pre-cleared chromatin. Inmunocomplexes were washed with buffer in the following order: salt buffer, LiCl buffer, and TE buffer (Li et al., 2014). After discarding TE buffer, the immunoprecipitated chromatin was eluted with elution buffer (Li et al., 2014). After formaldehyde cross-linking, ChIP-DNA was purified by phenol/chloroform/isoamyl alcohol extraction followed by ethanol precipitation (Li et al., 2014).
Quantitative Real-Time Polymerase Chain Reaction (QRT-PCR)
Quantitative real-time polymerase chain reaction was performed to profile the expression patterns of reference genes, poplar homologs of C4 photosynthetic enzyme genes and the changes in expression of these genes in TSA-treated poplar. Specific primers were designed using the online Integrated DNA Technologies software3 and are listed in Supplemental Tables S4, S5. QRT-PCR reactions were performed using SYBR Premix Ex Taq (Takara, Shiga, Japan) on a LightCycler 480 system (Roche, Basel, Sweden). Two microliters of diluted cDNA sample was used as template in an amplification reaction volume of 10 μl. The amplification program consisted of 30 s of initial denaturation at 95°C, followed by 40 cycles of 10 s at 95°C, 20 s at 60°C and 20 s at 72°C, and ended with a final extension step at 72°C for 20 s. Three replicated reactions per sample were done. For the analysis of the expression of poplar homologs of C4 photosynthetic enzyme genes, all samples were normalized to the reference gene PsnACTIN2. The final relative expression was calculated using the formula F = 2-ΔΔCt (Chen et al., 2014).
For ChIP assays, 2 μl ChIP-DNA sample was used as template in an amplification reaction volume of 10 μl. Primer positions and sequences are listed in Supplemental Figures S3, S4 and Table S6. The amplification program consisted of 30 s of initial denaturation at 95°C, followed by 45 cycles of 10 s at 95°C, 20 s at 60°C and 20 s at 72°C. Relative enrichment of H3K9ac and H4K5ac in the promoters of the PsnCA, PsnPPDK, PsnPCK, and PsnPEPC genes was calculated by normalizing to the value of these marks in the promoter of the PsnACTIN2 housekeeping gene. Relative enrichment values are based on the average of three PCR reactions for each sample.
Western Blotting
Total proteins from leaves, Sc, Sv, and roots of control and TSA-treated poplar were extracted using a phenol (Sigma, St. Louis, MO, United States) extraction procedure as described previously (Hurkman and Tanaka, 1986; Wang et al., 2011). Protein concentrations were measured using the Bio-Rad Protein Assay (Bio-Rad, United States). Thirty micrograms of protein was separated by 15% SDS-PAGE gel and transferred by electroblotting (180 mA for 3 h) to a PVDF membrane (Millipore, Billerica, MA, United States). Membranes were blocked with 5% skim milk and probed with anti-H3K9ac (07-352, Millipore, Billerica, MA, United States) and anti-H4K5ac (07-327, Millipore, Billerica, MA, United States) antibodies to detect histone marks. Polyclonal anti-actin (EASYBIO, Beijing, China) antibody was used as a control for protein loading. Fluorescence goat anti-rabbit (GAR) antibody (Odyssey, United States) was used as the secondary antibody. Membranes were digitized using a two-photo far infrared scanner (Odyssey, United States) and analyzed with Image Studio software4. Western blotting experiments were repeated three times for each protein sample.
Results
Isolation of Poplar Genes Homologous to C4 Photosynthetic Enzyme Genes
The P. simonii × P. nigra genome has not been sequenced. Therefore, we used the known protein sequences of maize C4-CA (GRMZM2G121878), C4-PPDK (GRMZM2G306345), C4-PCK (GRMZM2G001696), and C4-PEPC (GRMZM2G083841) as queries to blast against the P. trichocarpa genome database1. Using primers specific to the P. trichocarpa sequences and cDNA from P. simonii × P. nigra leaves or roots, we cloned nine P. simonii × P. nigra genes homologous to the maize C4-CA, C4-PPDK, C4-PCK, and C4-PEPC genes (Supplemental Table S1). Sequence alignments showed that the PsnCA1 and PsnCA2 proteins shares 91.6% identity, and both are different from PsnCA3. Compared with PsnCA3, the N-terminal ends of both the PsnCA1 and PsnCA2 proteins are shorter by 76 amino acids and both C-terminal ends are 8 amino acids shorter (Supplemental Figure S1A). We cloned two transcripts (named PsnPPDK1-1 and PsnPPDK1-2) for a single PsnPPDK gene, and the protein sequences share 88.08% identity (Supplemental Figure S1B). Note that these transcripts are transcribed from different promoters; therefore, for the purposes of our analysis, we treat them as independently transcribed genes. There are two PsnPCK genes, the protein sequences of which share 82.76% identity (Supplemental Figure S1C). In addition, we also identified two PsnPEPC genes, the proteins sequences of which share 94.32% identity (Supplemental Figure S1D).
PsnCA, PsnPPDK, PsnPCK, and PsnPEPC Genes Have Tissue-Dependent Expression Patterns
To better understand the potential functions of poplar genes homologous to C4 photosynthetic enzyme genes, we profiled their expression patterns in leaves, Sc (includes epidermis and cortex), Sv (includes phloem fiber, phloem, cambium and xylem), and roots (Figures 1, 2). Based on the average cycle threshold (Cq) value and the average expression stability value (M-value), we chose to use ACTIN2 as a reference gene in this study (Supplemental Figure S2). Among the CA genes, both PsnCA1 and PsnCA2 were highly expressed in roots and Sc (Figures 2A,B). However, PsnCA3 was most highly expressed in leaves (Figure 2C). Surprisingly, the level of PsnCA3 transcript was nearly 15,000-fold higher than PsnCA1 and PsnCA2 in leaves (Figure 2D). The PPDK and PCK gene pairs showed similar differences in expression pattern. For example, both PsnPPDK1-1 and PsnPCK1 were highly expressed in leaves, whereas PsnPPDK1-2 and PsnPCK2 were highly expressed in Sc and Sv (Figures 2E–J). Unlike the CA, PPDK, and PCK genes, neither PEPC gene was most highly expressed in leaves. The abundance of both transcripts was high in Sc, but the expression level of PsnPEPC1 in roots was much higher than PsnPEPC2(Figures 2K–M).
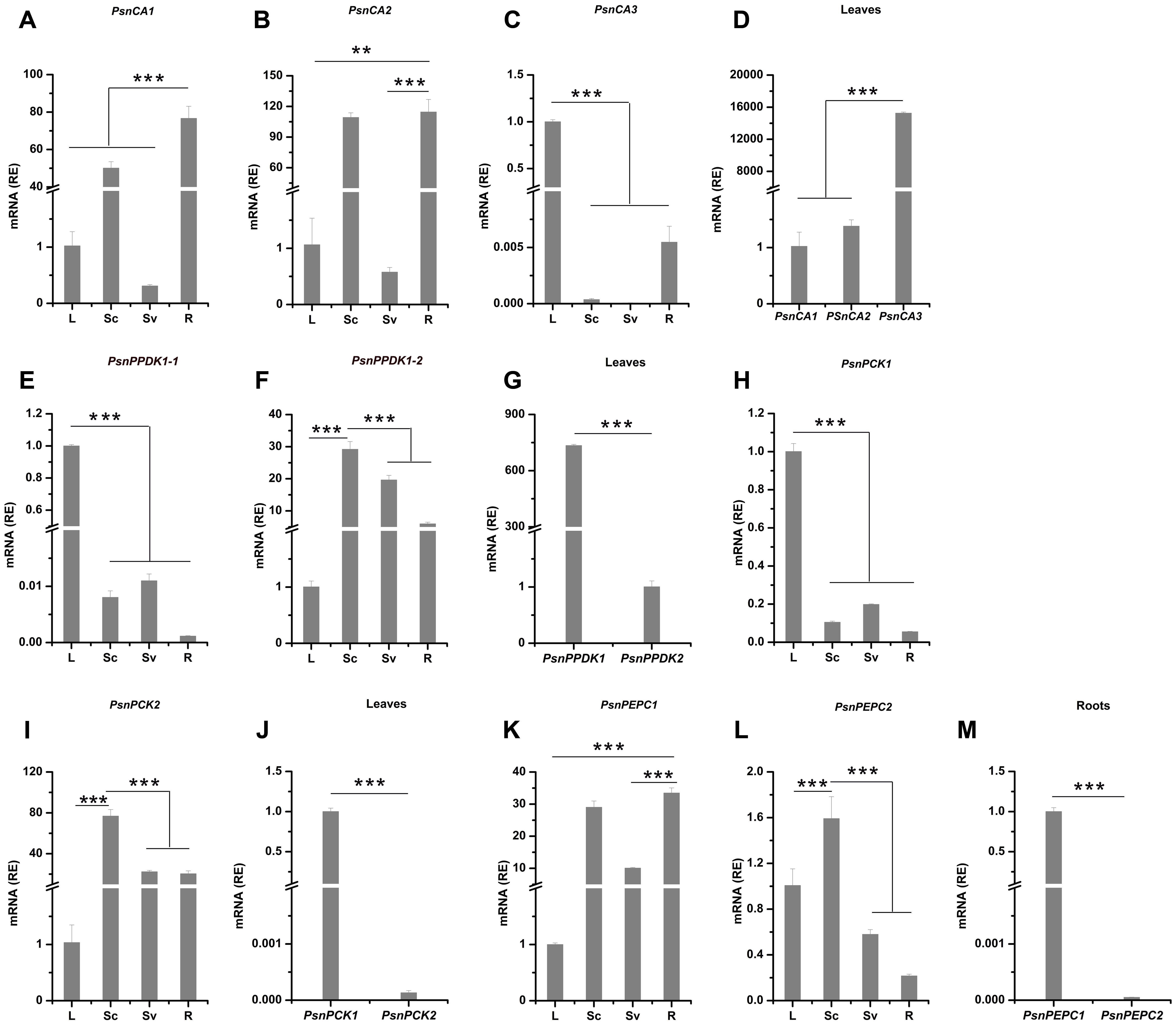
FIGURE 2. The relative expression levels of poplar genes homologous to C4 photosynthetic enzyme genes in different tissues of poplar. (A–M) All tissues were harvested from 5-month-old poplar plants grown in a greenhouse 4 h after the onset of illumination. The expression levels of PsnCA genes (A–D), PsnPPDK genes (E–G), PsnPCK genes (H–J) and PsnPEPC genes (K–M) genes. Expression levels were normalized to PsnACTIN2 expression. All relative enrichment (RE) values are means of at least three independent experiments. Bars indicate SE. Leaves (L), stem chlorenchyma (Sc), stem vascular tissue (Sv), and roots (R). Asterisks indicated significantly different means (∗∗∗p < 0.001) as determined with a t-test.
Based on the presence or absence of tissue chlorophyll (Figure 1 and Supplemental Figure S3) and gene-specific expression patterns (Figure 2), we generally classified these nine genes into two clusters. Genes in one cluster, including PsnCA3, PsnPPDK1-1, PsnPPDK1-2, PsnPCK1, PsnPCK2, and PsnPEPC2, are highly expressed in photosynthetic tissues (chlorophyll content in the leaves and Sc is 1.42 and 0.32 mg/g, respectively). Genes in the second cluster, including PsnCA1, PsnCA2, and PsnPEPC1, are highly expressed in non-photosynthetic tissues (chlorophyll content in Sv and roots is 0.002 and 0 mg/g).
The Levels of H3K9ac and H4K5ac in the Promoters of the PsnCA, PsnPPDK, PsnPCK, and PsnPEPC Genes Correlate Well with Tissue-Dependent Expression Patterns
Previous studies have found that for many plant loci, histone modification within the promoter and gene body is involved in the tissue-specific regulation of gene expression (Cai et al., 2003; Heintzman et al., 2009; Zhou et al., 2010; Ong and Corces, 2011; Heimann et al., 2013), and histone acetylation has been extensively correlated with transcriptional activation (Wolffe and Hayes, 1999; Turner, 2000). Therefore, we asked whether histone acetylation modification is correlated with the tissue-dependent expression of the PsnCA, PsnPPDK, PsnPCK, and PsnPEPC genes. We analyzed the levels of H3K9ac and H4K5ac marks in the promoters of these genes using a ChIP assay. Promoter sequences upstream of the transcription initiation site (TIS) of these genes (∼2 kb) were divided into three regions, including the distal region (P1), middle region (P2) and proximal region (P3) (Supplemental Figure S4). Immunoprecipitation efficiency of the PsnACTIN2 gene promoter was used to correct for variation in the amount of chromatin prepared from leaves, Sc, Sv, and roots (Supplemental Figure S5).
As shown in Figure 3, for almost genes, the level of H3K9ac in the P3 region (close to the TIS) was higher than in the P1 and P2 regions. In photosynthetic tissues, the highest enrichment of H3K9ac in the P3 region of PsnCA3, PsnPPDK1-1, and PsnPCK1 was detected in leaves (Figures 3A–C). This correlates well with the greater transcript abundance of these genes in leaves (Figures 2C,E,H). In Sc, relatively high enrichment of H3K9ac was observed in the promoters of the PsnPPDK1-2, PsnPCK2, and PsnPEPC2 genes, which are highly expressed in this tissue (Figures 3D–F). In the non-photosynthetic root tissue, we found that H3K9ac was strongly enriched in the promoters of PsnCA1, PsnCA2, and PsnPEPC1 which correlates with the high level of expression of these genes in roots (Figures 3G–I). We also found enrichment of H3K9ac in the P1 and P2 regions of these genes, but there was no obvious regular changes in both two regions between tissues (Figures 3A–I).
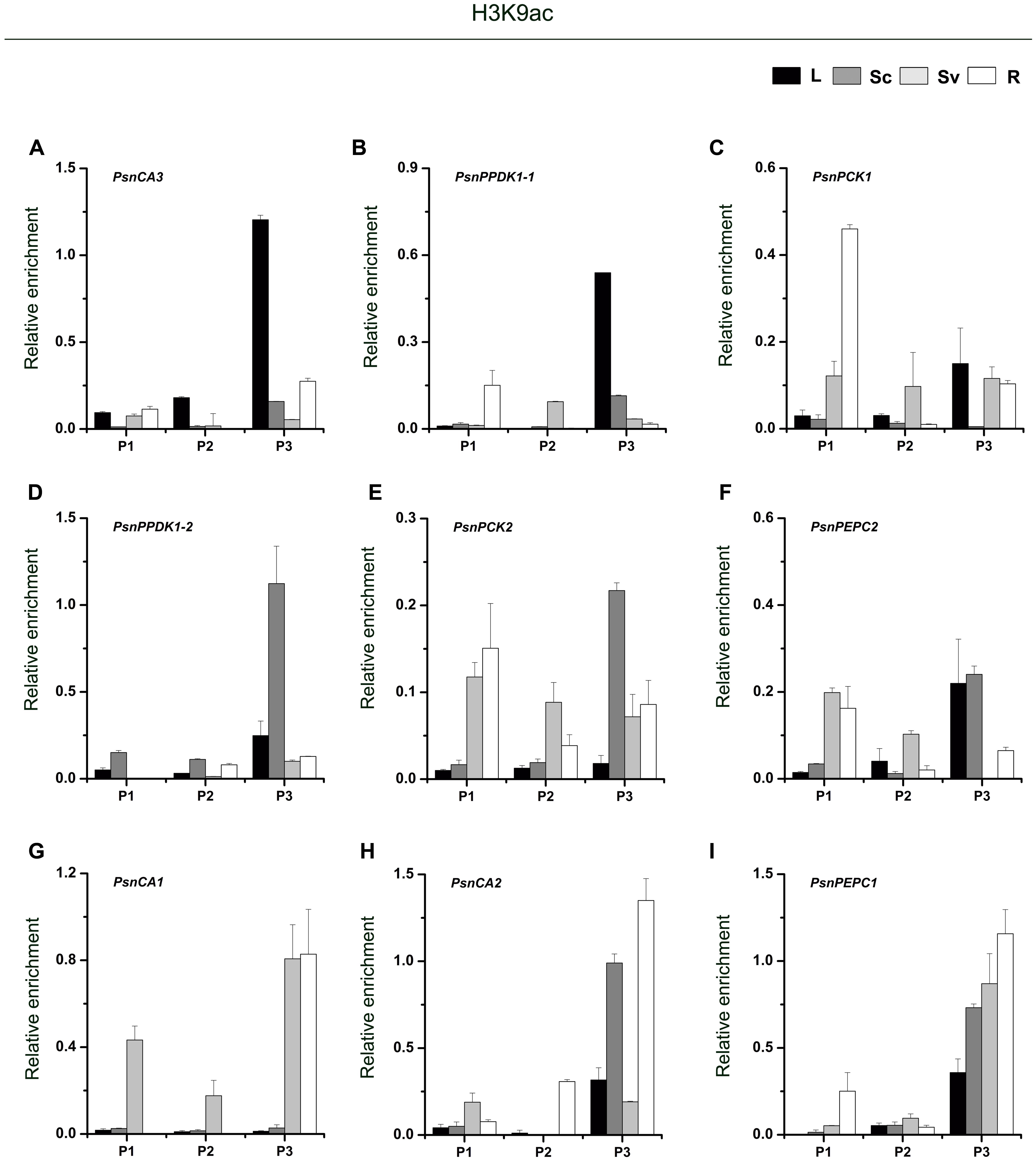
FIGURE 3. ChIP analysis of the PsnCA, PsnPPDK, PsnPCK, and PsnPEPC gene promoters using the H3K9ac antibody. (A–I) All tissues were harvested from 5-month-old poplar plants grown in a greenhouse 4 h after the onset of illumination. ChIP was used to detect H3K9ac levels in the upstream promoters of the PsnCA, PsnPPDK, PsnPCK, and PsnPEPC genes in leaves (L), stem chlorenchyma (Sc), stem vascular tissue (Sv), and roots (R). P1, P2, and P3 represent the distal, middle, and proximal promoter regions (Supplemental Figure S4). Levels of H3K9ac were standardized by the accumulation of H3K9ac in the promoter of the PsnACTIN2 gene (Supplemental Figure S5). Values are the means from three independent experiments. Bars indicate SE.
As shown in Figure 4, we found high levels of H4K5ac in the P3 region of almost target genes, which is similar to the levels of H3K9ac in that region. In terms of H4K5ac modification, we also found that strong enrichment in the P3 region of most genes was correlated with high transcript accumulation. For example, H4K5ac was highly enriched in the P3 region of the PsnCA3, PsnPPDK1-1, and PsnPCK1 genes in leaves (Figures 4A–C), and in the PsnCA1, PsnCA2, and PsnPEPC1 genes in roots (Figures 4G–I). For the PsnPCK2 and PsnPEPC2 genes, which are highly expressed in the photosynthetic tissue Sc, strong enrichment of H4K5ac in the P3 region was observed in both Sc and Sv (Figures 4E,F). However, we found the highest enrichment of H4K5ac in the P1 region of the PsnPPDK1-2 gene in Sc, not in the P3 region (Figure 4D).
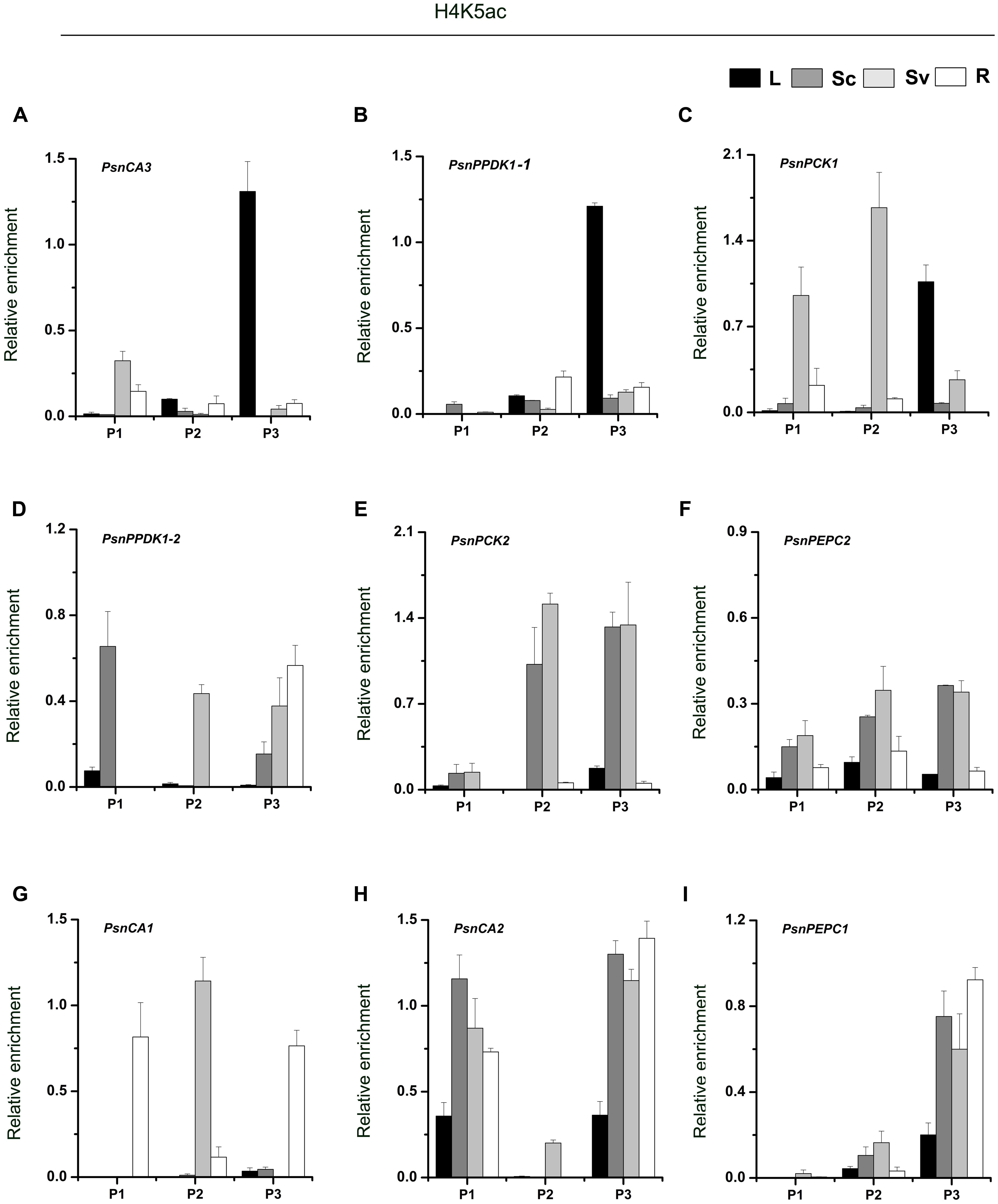
FIGURE 4. ChIP analysis of the PsnCA, PsnPPDK, PsnPCK, and PsnPEPC gene promoters using the H4K5ac antibody. (A–I) All tissues were harvested from 5-month-old poplar plants grown in a greenhouse 4 h after the onset of illumination. ChIP was used to detect H3K9ac levels in the upstream promoters of the PsnCA, PsnPPDK, PsnPCK, and PsnPEPC genes in leaves (L), stem chlorenchyma (Sc), stem vascular tissue (Sv), and roots (R). P1, P2, and P3 represent the distal, middle, and proximal promoter regions (Supplemental Figure S4). Levels of H3K9ac were standardized by the accumulation of H3K9ac in the promoter of the PsnACTIN2 gene (Supplemental Figure S5). Values are the means from three independent experiments. Bars indicate SE.
The correlation between H3K9ac and H4K5ac levels and expression levels of the PsnCA, PsnPPDK, PsnPCK, and PsnPEPC genes indicates that histone acetylation modification may regulate the tissue-dependent expression of genes homologous to C4 photosynthetic enzyme genes in C3 woody plants.
Application of Exogenous HDAC Inhibitors Alters Histone Acetylation and the Expression of PsnCA, PsnPPDK, PsnPCK, and PsnPEPC Genes
Treatment with the HDAC inhibitor, TSA, results in the accumulation of acetylated histones in the genome (Bernstein et al., 2000). In order to further study the relationship between histone acetylation and tissue-dependent expression of the PsnCA, PsnPPDK, PsnPCK, and PsnPEPC genes, we applied low concentrations (2.5 and 5 μM) of TSA to 5-month-old poplar plants and measured levels of histone acetylation and gene expression in leaves, Sc, Sv and roots (Supplemental Figures S6, S7). As we expected, western blot analysis showed that application of TSA induced a slight increase in both H3K9ac and H4K5ac in leaves, Sc, Sv, and roots (Figure 5A and Supplemental Figure S7A). Moreover, qRT-PCR results showed that TSA altered the level of gene expression to different extents in different tissues (Supplemental Figures S7B–J). In general, TSA significantly increased the abundance of PsnCA3 and PsnPPDK1-1 transcripts in leaves (Figures 5B,C), PsnPPDK1-2, PsnPCK2, and PsnPEPC2 transcripts in Sc (Figures 5E–G), and PsnCA1, PsnCA2, and PsnPEPC1 transcripts in roots (Figures 5H–J) compared to untreated tissues. However, application of TSA significantly reduced the transcript level of PsnPCK1 in leaves (Figure 5D), suggesting that TSA probably has pleiotropic effects on other regulatory networks linked to this gene.
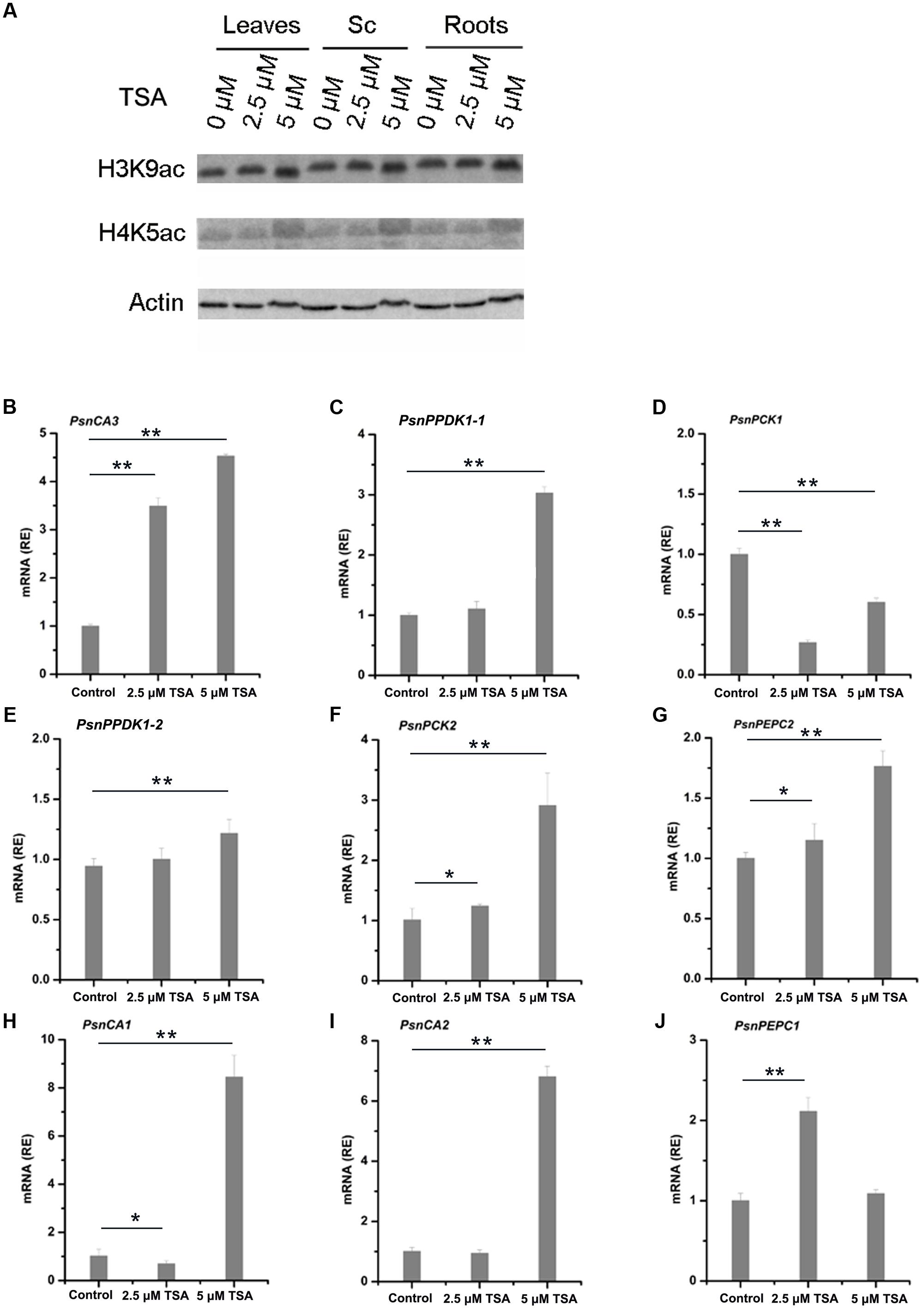
FIGURE 5. TSA affects H3K9ac and H4K5ac protein levels and mRNA levels of poplar homologs of C4 photosynthetic enzyme genes. (A) Western blot analysis of H3K9ac and H4K5ac protein levels in leaves (L), stem chlorenchyma (Sc), and roots (R) of poplar treated with TSA for 2 days. ACTIN was used as a control for equal loading. Expression of the PsnCA3, PsnPPDK1-1, and PsnPCK1 genes in leaves (B–D), the PsnPPDK1-2, PsnPCK2, and PsnPEPC2 genes in Sc (E–G), and the PsnCA1, PsnCA2, and PsnPEPC1 genes in roots (H–J) of poplar treated with TSA for 2 days. All expression levels were normalized to PsnACTIN2 expression. Values are means from three independent experiments. Bars indicate SE. Asterisks indicated significantly different means (∗p < 0.05; ∗∗p < 0.005) as determined with a t-test.
Enrichment of H3K9ac and H4K5ac Is Strongly Correlated with the Level of Tissue-Dependent Expression of the Poplar PsnCA, PsnPPDK, PsnPCK2, and PsnPEPCs Genes
In order to further confirm that H3K9ac and H4K5ac regulate the tissue-dependent expression patterns of the PsnCA, PsnPPDK, PsnPCK, and PsnPEPC genes, we performed ChIP assays to detect the enrichment of H3K9ac and H4K5ac in the promoters of these genes in TSA-treated poplar. The accumulation of H3K9ac and H4K5ac was altered by TSA to different extents in the P3 regions of almost all genes detected in different tissues (Supplemental Figures S8, S9). In general, the enrichment of both H3K9ac and H4K5ac in the P3 regions of PsnCA3 in leaves, PsnPPDK1-2, PsnPCK2 and PsnPEPC2 in Sc, as well as PsnCA2 and PsnPEPC1 in roots was enhanced to different degrees (Figures 6A,D,E,F,H,I). Additionally, the enrichment of H3K9ac in the P3 regions of PsnPPDK1-1 in leaves and PsnCA1 in roots was significantly induced by TSA (Figures 6B,G). Moreover, the levels of H3K9ac and H4K5ac were in agreement with the transcript levels observed for these genes under TSA treatment (Figure 5). In contrast, no obvious change in H3K9ac or H4K5ac level was detected for PsnPCK1 (Figure 6C), indicating that in leaves H3K9ac and H4K5ac do not play important roles in the regulation of PsnPCK1 gene expression. Taken together, these results reveal that histone acetylation modification is correlated with the tissue-dependent expression of most poplars homologs of C4 photosynthesis genes.
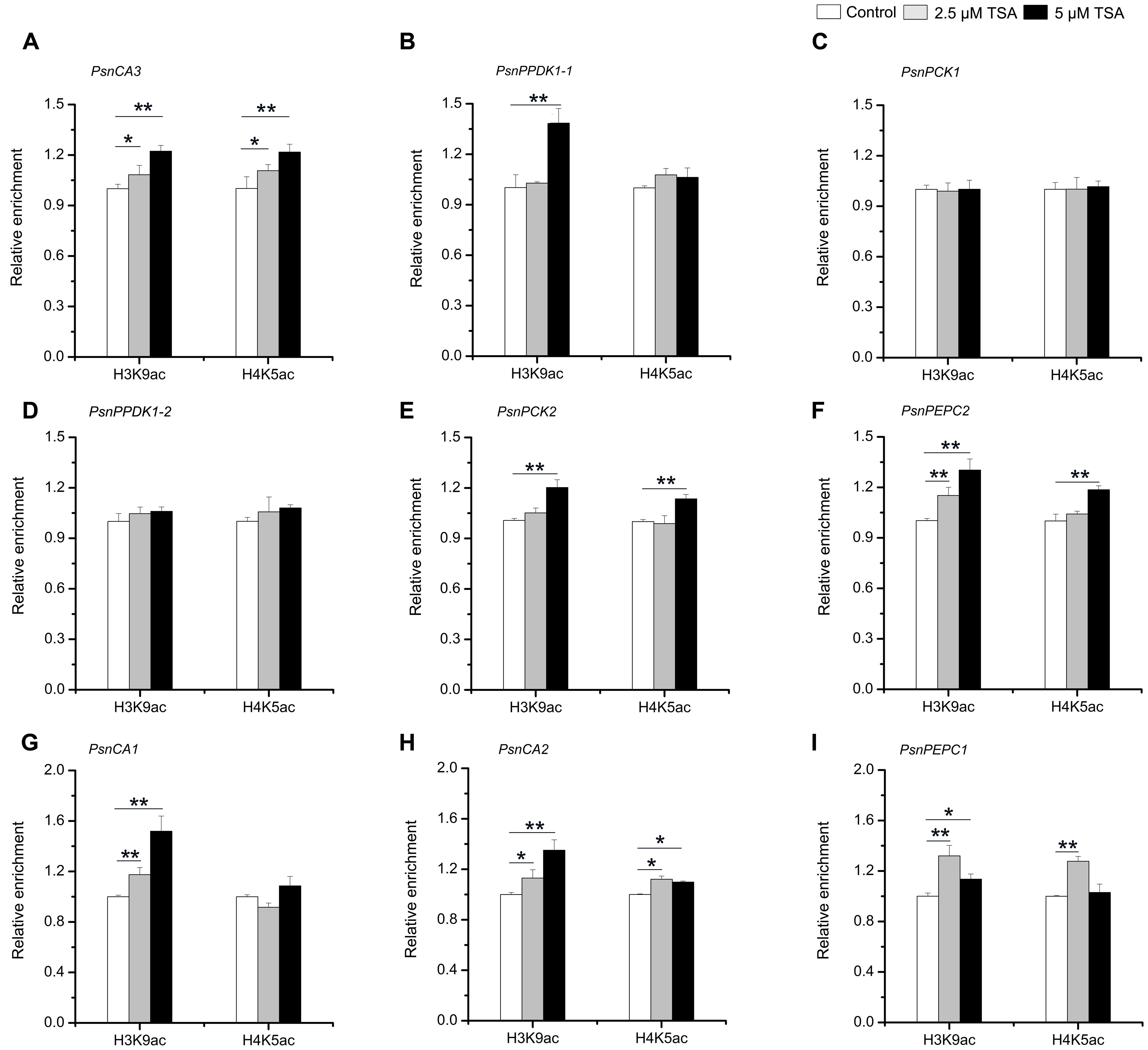
FIGURE 6. TSA affects the level of H3K9ac and H4K5ac in the promoters of the poplar PsnCA, PsnPPDK, PsnPCK, and PsnPEPC genes. ChIP was used to detect H3K9ac and H4K5ac levels in the P3 (close to TIS) promoter region of the PsnCA3, PsnPPDK1-1, and PsnPCK1 genes in leaves (L) (A–C), the PsnPPDK1-2, PsnPCK2, and PsnPEPC2 genes in stem chlorenchyma (Sc) (D–F), and the PsnCA1, PsnCA2, and PsnPEPC1 genes in roots (R) (G–I) of poplar treated with TSA for 2 days. Levels of H3K9ac and H4K5ac were standardized by the accumulation of H3K9ac and H4K5ac in the promoter of the PsnACTIN2 gene (Supplemental Figure S4). Values are the means from three independent experiments. Bars indicate SE. Asterisks indicated significantly different means (∗p < 0.05; ∗∗p < 0.005) as determined with a t-test.
Discussion
Analysis of genome sequences for a growing number of species has indicated that all of the enzymes required for C4 photosynthesis (such as CA, PPDK, PCK, and PEPC) are present in C3 plants, and even in algae and microorganisms. Moreover, C3 homologs of C4 photosynthetic enzyme genes share high sequence identity with their C4 counterparts. During the evolution from C3 to C4 plants, photosynthetic genes acquired new regulatory features, such as cell type-specific expression in mesophyll or bundle sheath cells. It is likely that duplication of genes encoding C4 photosynthesis proteins allowed ancestral functions to be maintained in one duplicate copy, while also permitting neofunctionalization of the other copy, leading to C4-specific roles and expression patterns (Ludwig, 2013).
Much of the regulation of C4 photosynthetic enzyme expression takes place at the promoter (Sheen, 1999; Taniguchi et al., 2000; Hibberd and Covshoff, 2010), and increasing evidence indicates that histone modification plays an important role in the regulation of tissue-specific expression of CA, PPDK, PCK, PEPC, and RbcS2 in C4 plants (Offermann et al., 2008; Heimann et al., 2013; Dong et al., 2016). Except for studies on the role of histone acetylation modifications in the regulation of the RbcA and RbcS genes during Pinus radiate needle development (Charron et al., 2009; Valledor et al., 2015), the role of histone modifications in the regulation of photosynthesis genes in C3 plants has not been well studied. Given that there is evidence that the C4 forms of photosynthetic enzymes have evolved from C3 counterparts, for example CA3 (β-CA in C4 plant Flaveria bidentis) and the C4 form of PPDK, evolved from chloroplastic C3 ancestors (Langdale, 2011; Ludwig, 2012), we hypothesized that histone modification plays a significant role in the regulation of photosynthetic genes expression in C3 plants. To test this hypothesis, we analyzed the relationship between histone acetylation modification and expression for poplar homologs of C4 photosynthetic genes.
We first isolated nine poplar homologs of the maize C4-CA, C4-PPDK, C4-PCK, and C4-PEPC genes and investigated the expression of these genes in photosynthetic and non-photosynthetic tissues. We found these nine poplar homologs of C4 genes had tissue-dependent expression pattern. For example, PsnCA3, PsnPPDK1-1, and PsnPCK1 are highly expressed in photosynthetic tissue leaves (Figure 2), which is consistent with the reported expression patterns of their homologous genes in C3 herbaceous species and C4 gramineous species (Fett and Coleman, 1994; Malone et al., 2007; Taylor et al., 2010; Langdale, 2011; Dong et al., 2016). However, not all poplar photosynthetic gene expression patterns are conserved with their homologs in other species. In contrast to AtPEPC1 and AtPEPC2, which account for nearly 93% of the total PEPC activity in leaves (Shi et al., 2015), PsnPEPC1 and PsnPEPC2 was highly expressed in roots, and Sc, respectively (Figure 2).
Reviewed in DiMario’s paper, various studies have shown that plants have many genes encoding CA, PPDK, PCK, and PEPC, which are found in most tissues and many intracellular compartments. In addition to uncertain roles in photosynthesis, the functions of these homologous genes are also required for many metabolic, signaling, and developmental pathways in C3 species. There is evidence that C3 β-CAs function in carbon-concentration, nitrogen-fixation, stomatal movement and development, biotic and abiotic stress responses, and amino acid as well as lipid biosynthesis (DiMario et al., 2017). A single PPDK gene that possesses a dual promoter giving rise to two transcripts is found in many C3 herbaceous species in addition to poplar, including Arabidopsis, wheat, rice, Flaveria, and even in the C4 species maize. In all cases the longer PPDK transcript (homologous to PsnPPDK1-1) encodes a protein that is targeted to the chloroplast of leaves, and the smaller protein (homologous to PsnPPDK1-2) is cytosolic (Aoyagi and Bassham, 1984; Glackin and Grula, 1990; Rosche and Westhoff, 1995; Imaizumi et al., 1997; Parsley and Hibberd, 2006). PCK is localized in phloem and trichome tissues, oil and resin ducts, developing seeds, and ripening tomato fruit (Leegood and Walker, 1999; Walker et al., 1999; Bahrami et al., 2001), suggesting it has various roles in plant development, such as gluconeogenesis, nitrogen, and amino acid metabolism. Our finding that the PsnPEPC2 gene is highly expressed in Sc is consistent with the finding that in woody plants PEPC enzyme activity is higher in current-year stems and also with PEPC’s function in sustaining the carbon flux (Berveiller et al., 2007). High expression of PEPC in the stem was possibly selected for because in addition to leaves, a strong stem is another major source of assimilated carbon in woody plants. Similar to what has been observed in some reported C4 plants, the poplar C4 genes homologs have similar expression patterns in that closely related genes, which likely have divergent roles in plant development, such as carbon and nitrogen metabolism.
We next asked whether the tissue-dependent expression patterns of the poplar CA, PPDK, PCK, and PEPC genes were correlated with histone acetylation modification. Our finding that for almost all genes, the peak of H3K9ac and H4K5ac accumulation was highest in the P3 region close to the TIS (Figures 3, 4) suggests that the nucleosomes adjacent to the TIS of active genes are hypoacetylated compared to the surrounding regions. Similar observations were also made in Arabidopsis and Maize, where peaks of H3K9ac and H4K5ac accumulation were found around the ATG position, especially in photosynthesis genes (Zhou et al., 2010; Perduns et al., 2015; Dong et al., 2016). In the C4 plant maize, PPDK and PCK gene expression is regulated by histone modifications (Dong et al., 2016). Our findings that H3K9ac and H4K5ac modifications correlate well with tissue-dependent expression of the PsnCA, PsnPPDK, PsnPCK, and PsnPEPC genes and that, with the exception of PsnPCK1, treatment with HDAC inhibitor, TSA led to tissue-specific increases in H3K9ac or H4K5ac that were correlated with mRNA levels provide evidence that these C4 homologous genes are also regulated by histone modification. The similarities between histone acetylation regulation of photosynthetic enzyme genes in C3 and C4 plants suggests that this regulatory system seems conserved in diverse C3 and C4 species. It also provides evidences that a preexisting epigenetic mechanism for promoter control was probably recruited during the evolution of C4 plants.
The continued deep sequencing of more and more closely related C3 and C4 species will allow the functions of C4 photosynthetic enzyme homologs to be analyzed in lineages of C3 species that are closely related to C4 species and give insight into how the regulation and functions of these genes changed during the transition from C3 to C4 photosynthesis. Understanding the mechanism behind the recruitment of photosynthetic genes into new biochemical pathways and identification of key factors controlling C4 gene expression will enable us to engineer “real-C4” plants from C3 plants in the future.
Author Contributions
YL and B-CW conceived and designed this work. YL performed the gene cloning, qRT-PCR, Western blotting, and TSA treatment. YL and X-MD performed the ChIP experiment. FJ performed morphological and anatomic experiments. ZS and QC provided technical help and suggestions. YL and B-CW wrote the article.
Funding
This work was supported by the National High-Tech Research and Development Program of China (grant no. 2013AA102701) and the State Key Program of National Natural Science of China (grant no. 31030017).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00950/full#supplementary-material
Abbreviations
CA, carbonic anhydrase; ChIP, chromatin immunoprecipitation; H3K9ac, acetylation of histone H3 lysine 9; H4K5ac, acetylation of histone H4 lysine 5; HDAC, histone deacetylase; NAD-ME, NAD malic enzyme; NADP-ME, NADP malic enzyme; PCK, phosphoenolpyruvate carboxykinase; PEPC, phosphoenolpyruvate carboxylase; PPDK, pyruvate orthophosphate dikinase; qRT-PCR, quantitative real-time polymerase chain reaction; Rubisco, ribulose 1,5-bisphosphate carboxylase/oxygenase; TSA, trichostatin A.
Footnotes
- ^http://www.phytozome.net/poplar
- ^https://phytozome.jgi.doe.gov/pz/portal.html
- ^http://www.idtdna.com/scitools/Applications/RealTimePCR/
- ^https://www.licor.com/bio/products/software/image_studio/
References
Aoyagi, K., and Bassham, J. A. (1984). Pyruvate orthophosphate dikinase of C3 seeds and leaves as compared to the enzyme from maize. Plant Physiol. 75, 387–392. doi: 10.1104/pp.75.2.387
Bahrami, A. R., Chen, Z.-H., Walker, R. P., Leegood, R. C., and Gray, J. E. (2001). Ripening-related occurrence of phosphoenolpyruvate carboxykinase in tomato fruit. Plant Mol. Biol. 47, 499–506. doi: 10.1023/A:1011842828723
Bernstein, B. E., Tong, J. K., and Schreiber, S. L. (2000). Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. U.S.A. 97, 13708–13713. doi: 10.1073/pnas.250477697
Berveiller, D., Vidal, J., Degrouard, J., Ambard-Bretteville, F., Pierre, J. N., Jaillard, D., et al. (2007). Tree stem phosphoenolpyruvate carboxylase (PEPC): lack of biochemical and localization evidence for a C4-like photosynthesis system. New Phytol. 176, 775–781. doi: 10.1111/j.1469-8137.2007.02283.x
Bowler, C., Benvenuto, G., Laflamme, P., Molino, D., Probst, A. V., Tariq, M., et al. (2004). Chromatin techniques for plant cells. Plant J. 39, 776–789. doi: 10.1111/j.1365-313X.2004.02169.x
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Cai, S., Han, H.-J., and Kohwi-Shigematsu, T. (2003). Tissue-specific nuclear architecture and gene expession regulated by SATB1. Nat. Genet. 34, 42–51. doi: 10.1038/ng1146
Charron, J.-B. F., He, H., Elling, A. A., and Deng, X. W. (2009). Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21, 3732–3748. doi: 10.1105/tpc.109.066845
Chen, H., Xu, X. L., Li, Y. P., and Wu, J. X. (2014). Characterization of heat shock protein 90, 70 and their transcriptional expression patterns on high temperature in adult of Grapholita molesta (Busck). Insect Sci. 21, 439–448. doi: 10.1111/1744-7917.12057
DiMario, R. J., Clayton, H., Mukherjee, A., Ludwig, M., and Moroney, J. V. (2017). Plant carbonic anhydrases: structures, locations, evolution, and physiological roles. Mol. Plant 10, 30–46. doi: 10.1016/j.molp.2016.09.001
Dong, X.-M., Li, Y., Chao, Q., Shen, J., Gong, X.-J., Zhao, B.-G., et al. (2016). Analysis of gene expression and histone modification between C4 and non-C4 homologous genes of PPDK and PCK in maize. Photosynth. Res. 129, 71–83. doi: 10.1007/s11120-016-0271-9
Eastmond, P. J., Germain, V., Lange, P. R., Bryce, J. H., Smith, S. M., and Graham, I. A. (2000). Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc. Natl. Acad. Sci. U.S.A. 97, 5669–5674. doi: 10.1073/pnas.97.10.5669
Fett, J. P., and Coleman, J. R. (1994). Characterization and expression of two cDNAs encoding carbonic anhydrase in Arabidopsis thaliana. Plant Physiol. 105, 707–713. doi: 10.1104/pp.105.2.707
Gendrel, A.-V., Lippman, Z., Martienssen, R., and Colot, V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2, 213–218. doi: 10.1038/nmeth0305-213
Glackin, C. A., and Grula, J. W. (1990). Organ-specific transcripts of different size and abundance derive from the same pyruvate, orthophosphate dikinase gene in maize. Proc. Natl. Acad. Sci. U.S.A. 87, 3004–3008. doi: 10.1073/pnas.87.8.3004
Gutierrez, M., Huber, S., Ku, S., Kanai, R., and Edwards, G. (1974). “Intracellular localization of carbon metabolism in mesophyll cells of C4 plants,” in Proceedings of the Third International Congress on Photosynthesis, (Amsterdam: Elsevier Science Publishers), 1219–1230.
Hatch, M., Kagawa, T., and Craig, S. (1975). Subdivision of C4-pathway species based on differing C4 acid decarboxylating systems and ultrastructural features. Funct. Plant Biol. 2, 111–128. doi: 10.1071/PP9750111
Hatch, M. D., and Burnell, J. N. (1990). Carbonic anhydrase activity in leaves and its role in the first step of C4 photosynthesis. Plant Physiol. 93, 825–828. doi: 10.1104/pp.93.2.825
Hattersley, P. (1984). Characterization of C4 type leaf anatomy in grasses (Poaceae). Mesophyll: bundle sheath area ratios. Ann. Bot. 53, 163–180. doi: 10.1093/oxfordjournals.aob.a086678
Heimann, L., Horst, I., Perduns, R., Dreesen, B., Offermann, S., and Peterhansel, C. (2013). A common histone modification code on C4 genes in maize and its conservation in sorghum and Setaria italica. Plant Physiol. 162, 456–469. doi: 10.1104/pp.113.216721
Heintzman, N. D., Hon, G. C., Hawkins, R. D., Kheradpour, P., Stark, A., Harp, L. F., et al. (2009). Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112. doi: 10.1038/nature07829
Hibberd, J. M., and Covshoff, S. (2010). The regulation of gene expression required for C4 photosynthesis. Annu. Rev. Plant Biol. 61, 181–207. doi: 10.1146/annurev-arplant-042809-112238
Hibberd, J. M., and Quick, W. P. (2002). Characteristics of C4 photosynthesis in stems and petioles of C3 flowering plants. Nature 415, 451–454. doi: 10.1038/415451a
Hou, H., Wang, P., Zhang, H., Wen, H., Gao, F., Ma, N., et al. (2015). Histone acetylation is involved in GA-regulated sodCp gene expression in maize aleurone layers. Plant Cell Physiol. 56, 2139–2149. doi: 10.1093/pcp/pcv126
Hurkman, W. J., and Tanaka, C. K. (1986). Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 81, 802–806. doi: 10.1104/pp.81.3.802
Imaizumi, N., Ku, M. S., Ishihara, K., Samejima, M., Kaneko, S., and Matsuoka, M. (1997). Characterization of the gene for pyruvate, orthophosphate dikinase from rice, a C3 plant, and a comparison of structure and expression between C3 and C4 genes for this protein. Plant Mol. Biol. 34, 701–716. doi: 10.1023/A:1005884515840
Ku, M., Kano-Murakami, Y., and Matsuoka, M. (1996). Evolution and expression of C4 photosynthesis genes. Plant Physiol. 111, 949. doi: 10.1104/pp.111.4.949
Langdale, J. A. (2011). C4 cycles: past, present, and future research on C4 photosynthesis. Plant Cell 23, 3879–3892. doi: 10.1105/tpc.111.092098
Lea, P., Chen, Z.-H., Leegood, R., and Walker, R. (2001). Does phosphoenolpyruvate carboxykinase have a role in both amino acid and carbohydrate metabolism? Amino Acids 20, 225–241. doi: 10.1007/s007260170041
Lee, J.-S., Smith, E., and Shilatifard, A. (2010). The language of histone crosstalk. Cell 142, 682–685. doi: 10.1016/j.cell.2010.08.011
Leegood, R., and Ap Rees, T. (1978). Phosphoenolpyruvate carboxykinase and gluconeogenesis in cotyledons of Cucurbita pepo. Biochim. Biophys. Acta 524, 207–218. doi: 10.1016/0005-2744(78)90119-5
Leegood, R. C., and Walker, R. (1999). “Phosphoenolpyruvate carboxykinase in plants: its role and regulation,” in Plant Carbohydrate Biochemistry, eds J. A. Bryant, M. M. Burrell, and N. J. Kruger (Oxford: BIOS Scientific Publishers), 201–213. doi: 10.1016/S0003-9861(03)00093-6
Li, W., Lin, Y.-C., Li, Q., Shi, R., Lin, C.-Y., Chen, H., et al. (2014). A robust chromatin immunoprecipitation protocol for studying transcription factor–DNA interactions and histone modifications in wood-forming tissue. Nat. Protocols 9, 2180–2193. doi: 10.1038/nprot.2014.146
Ludwig, M. (2012). Carbonic anhydrase and the molecular evolution of C4 photosynthesis. Plant Cell Environ. 35, 22–37. doi: 10.1111/j.1365-3040.2011.02364.x
Ludwig, M. (2013). Evolution of the C4 photosynthetic pathway: events at the cellular and molecular levels. Photosynth. Res. 117, 147–161. doi: 10.1007/s11120-013-9853-y
Malone, S., Chen, Z.-H., Bahrami, A. R., Walker, R. P., Gray, J. E., and Leegood, R. C. (2007). Phosphoenolpyruvate carboxykinase in Arabidopsis: changes in gene expression, protein and activity during vegetative and reproductive development. Plant Cell Physiol. 48, 441–450. doi: 10.1093/pcp/pcm014
Ni, Z., Kim, E.-D., Ha, M., Lackey, E., Liu, J., Zhang, Y., et al. (2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457, 327–331. doi: 10.1038/nature07523
Offermann, S., Danker, T., Dreymüller, D., Kalamajka, R., Töpsch, S., Weyand, K., et al. (2006). Illumination is necessary and sufficient to induce histone acetylation independent of transcriptional activity at the C4-specific phosphoenolpyruvate carboxylase promoter in maize. Plant Physiol. 141, 1078–1088. doi: 10.1104/pp.106.080457
Offermann, S., Dreesen, B., Horst, I., Danker, T., Jaskiewicz, M., and Peterhansel, C. (2008). Developmental and environmental signals induce distinct histone acetylation profiles on distal and proximal promoter elements of the C4-Pepc gene in maize. Genetics 179, 1891–1901. doi: 10.1534/genetics.108.087411
Okabe, K., Yang, S.-Y., Tsuzuki, M., and Miyachi, S. (1984). Carbonic anhydrase: its content in spinach leaves and its taxonomic diversity studied with anti-spinach leaf carbonic anhydrase antibody. Plant Sci. Lett. 33, 145–153. doi: 10.1016/0304-4211(84)90004-X
Ong, C.-T., and Corces, V. G. (2011). Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 12, 283–293. doi: 10.1038/nrg2957
Parsley, K., and Hibberd, J. M. (2006). The Arabidopsis PPDK gene is transcribed from two promoters to produce differentially expressed transcripts responsible for cytosolic and plastidic proteins. Plant Mol. Biol. 62, 339–349. doi: 10.1007/s11103-006-9023-0
Peltier, J.-B., Cai, Y., Sun, Q., Zabrouskov, V., Giacomelli, L., Rudella, A., et al. (2006). The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell. Proteomics 5, 114–133. doi: 10.1074/mcp.M500180-MCP200
Perduns, R., Horst-Niessen, I., and Peterhänsel, C. (2015). Photosynthetic genes and genes associated with the C4 trait in maize are characterized by a new class of highly regulated histone acetylation peaks on upstream promoters. Plant Physiol. 168, 1378–1388. doi: 10.1104/pp.15.00934
Podevin, N., Krauss, A., Henry, I., Swennen, R., and Remy, S. (2012). Selection and validation of reference genes for quantitative RT-PCR expression studies of the non-model crop Musa. Mol. Breed. 30, 1237–1252. doi: 10.1007/s11032-012-9711-1
Rosche, E., and Westhoff, P. (1995). Genomic structure and expression of the pyruvate, orthophosphate dikinase gene of the dicotyledonous C4 plant Flaveria trinervia (Asteraceae). Plant Mol. Biol. 29, 663–678. doi: 10.1007/BF00041157
Rylott, E. L., Gilday, A. D., and Graham, I. A. (2003). The gluconeogenic enzyme phosphoenolpyruvate carboxykinase in Arabidopsis is essential for seedling establishment. Plant Physiol. 131, 1834–1842. doi: 10.1104/pp.102.019174
Sage, R. F., Sage, T. L., and Kocacinar, F. (2012). Photorespiration and the evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 63, 19–47. doi: 10.1146/annurev-arplant-042811-105511
Sheen, J. (1999). C4 gene expression. Annu. Rev. Plant Biol. 50, 187–217. doi: 10.1146/annurev.arplant.50.1.187
Shi, J., Yi, K., Liu, Y., Xie, L., Zhou, Z., Chen, Y., et al. (2015). Phosphoenolpyruvate carboxylase in Arabidopsis leaves plays a crucial role in carbon and nitrogen metabolism. Plant Physiol. 167, 671–681. doi: 10.1104/pp.114.254474
Slack, C. R., and Hatch, M. D. (1967). Comparative studies on the activity of carboxylases and other enzymes in relation to the new pathway of photosynthetic carbon dioxide fixation in tropical grasses. Biochem. J. 103, 660–665. doi: 10.1042/bj1030660
Taniguchi, M., Izawa, K., Ku, M. S., Lin, J.-H., Saito, H., Ishida, Y., et al. (2000). The promoter for the maize C4 pyruvate, orthophosphate dikinase gene directs cell-and tissue-specific transcription in transgenic maize plants. Plant Cell Physiol. 41, 42–48. doi: 10.1093/pcp/41.1.42
Taylor, L., Nunes-Nesi, A., Parsley, K., Leiss, A., Leach, G., Coates, S., et al. (2010). Cytosolic pyruvate, orthophosphate dikinase functions in nitrogen remobilization during leaf senescence and limits individual seed growth and nitrogen content. Plant J. 62, 641–652. doi: 10.1111/j.1365-313X.2010.04179.x
Turner, B. M. (2000). Histone acetylation and an epigenetic code. Bioessays 22, 836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9<3.0.CO;2-X
Valledor, L., Pascual, J., Meijón, M., Escandón, M., and Cañal, M. J. (2015). Conserved epigenetic mechanisms could play a key role in regulation of photosynthesis and development-related genes during needle development of Pinus radiata. PLoS ONE 10:e0126405. doi: 10.1371/journal.pone.0126405
Walker, R. P., Chen, Z. H., Johnson, K. E., Famiani, F., Tecsi, L., and Leegood, R. C. (2001). Using immunohistochemistry to study plant metabolism: the examples of its use in the localization of amino acids in plant tissues, and of phosphoenolpyruvate carboxykinase and its possible role in pH regulation. J. Exp. Bot. 52, 565–576. doi: 10.1093/jexbot/52.356.565
Walker, R. P., Chen, Z.-H., Técsi, L. I., Famiani, F., Lea, P. J., and Leegood, R. C. (1999). Phosphoenolpyruvate carboxykinase plays a role in interactions of carbon and nitrogen metabolism during grape seed development. Planta 210, 9–18. doi: 10.1007/s004250050648
Walker, R. P., and Leegood, R. C. (1996). Phosphorylation of phosphoenolpyruvate carboxykinase in plants. Studies in plants with C4 photosynthesis and Crassulacean acid metabolism and in germinating seeds. Biochem. J. 317, 653–658. doi: 10.1042/bj3170653
Wang, H., Alvarez, S., and Hicks, L. M. (2011). Comprehensive comparison of iTRAQ and label-free LC-based quantitative proteomics approaches using two Chlamydomonas reinhardtii strains of interest for biofuels engineering. J. Proteome Res. 11, 487–501. doi: 10.1021/pr2008225
Wang, P., Zhao, L., Hou, H., Zhang, H., Huang, Y., Wang, Y., et al. (2015). Epigenetic changes are associated with programmed cell death induced by heat stress in seedling leaves of Zea mays. Plant Cell Physiol. 56, 965–976. doi: 10.1093/pcp/pcv023
Westhoff, P., and Gowik, U. (2004). Evolution of C4 phosphoenolpyruvate carboxylase. Genes and proteins: a case study with the genus Flaveria. Ann. Bot. 93, 13–23. doi: 10.1093/aob/mch003
Wolffe, A. P., and Hayes, J. J. (1999). Chromatin disruption and modification. Nucleic Acids Res. 27, 711–720. doi: 10.1093/nar/27.3.711
Xiao, X., Ma, J., Wang, J., Wu, X., Li, P., and Yao, Y. (2015). Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Front. Plant Sci. 5:788. doi: 10.3389/fpls.2014.00788
Yu, Q., Liu, J., Wang, Z., Nai, J., Lü, M., Zhou, X., et al. (2013). Characterization of the NADP-malic enzymes in the woody plant Populus trichocarpa. Mol. Biol. Rep. 40, 1385–1396. doi: 10.1007/s11033-012-2182-y
Zhou, J., Wang, X., He, K., Charron, J.-B. F., Elling, A. A., and Deng, X. W. (2010). Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol. Biol. 72, 585–595. doi: 10.1007/s11103-009-9594-7
Keywords: poplar, C4 genes, homologs, transcriptional regulation, histone acetylation modification
Citation: Li Y, Dong X-M, Jin F, Shen Z, Chao Q and Wang B-C (2017) Histone Acetylation Modifications Affect Tissue-Dependent Expression of Poplar Homologs of C4 Photosynthetic Enzyme Genes. Front. Plant Sci. 8:950. doi: 10.3389/fpls.2017.00950
Received: 18 January 2017; Accepted: 22 May 2017;
Published: 08 June 2017.
Edited by:
Saman Seneweera, University of Southern Queensland, AustraliaReviewed by:
Gaurav Zinta, Shanghai Center for Plant Stress Biology (PSC), CAS, ChinaAutar Krishen Mattoo, United States Department of Agriculture, United States
Copyright © 2017 Li, Dong, Jin, Shen, Chao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bai-Chen Wang, d2FuZ2JjQGliY2FzLmFjLmNu
 Yuan Li1,2
Yuan Li1,2 Bai-Chen Wang
Bai-Chen Wang