- 1National Key Laboratory of Rice Biology, Institute of Crop Sciences, Zhejiang University, Hangzhou, China
- 2Hubei Collaborative Innovation Center for Grain Industry, Jingzhou, China
- 3Department of Chemistry, Zhejiang University, Hangzhou, China
- 4Institute of Nuclear Agricultural Sciences, Zhejiang University, Hangzhou, China
The xantha marker trait, which is controlled by a down-regulating epi-mutation of OsGUN4, has been applied to the production of hybrid rice. However, the molecular basis for the ability of xantha mutants to attain high photosynthetic capacity even with decreased chlorophyll contents has not been characterized. In the present study, we observed that the total chlorophyll content of the xantha mutant was only 27.2% of that of the wild-type (WT) plants. However, the xantha mutant still accumulated 59.9% of the WT δ-aminolevulinic acid content, 72.8% of the WT Mg-protoporphyrin IX content, and 63.0% of the WT protochlorophyllide a content. Additionally, the protoporphyrin IX and heme contents in the mutant increased to 155.0 and 160.0%, respectively, of the WT levels. A search for homologs resulted in the identification of 124 rice genes involved in tetrapyrrole biosynthesis and photosynthesis. With the exception of OsGUN4, OsHO-1, and OsHO-2, the expression levels of the genes involved in tetrapyrrole biosynthesis were significantly higher in the xantha mutant than in the WT plants, as were all 72 photosynthesis-associated nuclear genes. In contrast, there were no differences between the xantha mutant and WT plants regarding the expression of all 22 photosynthesis-associated chloroplast genes. Furthermore, the abundance of 1O2 and the expression levels of 1O2-related genes were lower in the xantha mutant than in the WT plants, indicating 1O2-mediated retrograde signaling was repressed in the mutant plants. These results suggested that the abundance of protoporphyrin IX used for chlorophyll synthesis decreased in the mutant, which ultimately decreased the amount of chlorophyll in the xantha mutant. Additionally, the up-regulated expression of photosynthesis-associated nuclear genes enabled the mutant to attain a high photosynthetic capacity. Our findings confirm that OsGUN4 plays an important role in tetrapyrrole biosynthesis and photosynthesis in rice. GUN4, chlorophyll synthesis pathways, and photosynthetic activities are highly conserved in plants and hence, novel traits (e.g., xantha marker trait) may be generated in other cereal crops by modifying the GUN4 gene.
Introduction
Hybrid rice is important for rice production in China and other countries (reviewed by Zhang, 2016). The leaf color marker trait has been proposed as a genetic marker for male-sterile lines, which can simplify the differentiation between bred-true and off-type (unwanted) plants, thus achieving high varietal purity during rice production (Shu et al., 1996). To date, male-sterile lines with marker traits, such as chlorina (Dong et al., 1995), xantha (Zhou et al., 2006a), green-revertible albino seedlings (Wu et al., 2003, 2011), and purple plants (Mou et al., 1995), have been developed and used to produce hybrid rice.
We previously generated a xantha marker trait through γ-irradiation of Longtefu B (LTB), which is a rice cytoplasmic male-sterile maintainer line (Zhou et al., 2006a). Plants of the xantha mutant line, named Huangyu B (HYB), have yellow leaves, and can be visually distinguished from normal green rice plants. Although HYB contains lower levels of chlorophyll a (Chl a), chlorophyll b (Chl b), and carotenoids (Car) than LTB, it retains a high photosynthetic rate comparable to that of wild-type (WT) plants at high-light intensities (Zhou et al., 2006b; Wu et al., 2007). Thus, HYB differs from most chlorophyll-deficient mutants, which typically exhibit relatively low photosynthetic activities (Liu et al., 2007, 2016; Yoo et al., 2009; Tian et al., 2013; Wang et al., 2014; Zhang et al., 2015; Yang et al., 2016). Therefore, the xantha marker trait is very unique and valuable for breeding hybrid rice varieties. It has been introduced into several cytoplasmic male-sterile lines and thermosensitive genic male-sterile lines, including Huangyu A (Zhou et al., 2006a), Jiazhe91A (Li et al., 2010), and LFS (Fu Haowei, personal communication).
Chlorophyll biosynthesis involves many enzymes (Beale, 2005; Tanaka and Tanaka, 2006) that catalyze the conversion of the first committed precursor, δ-aminolevulinic acid (ALA), to the final products in higher plants, namely Chl a and Chl b (Figure 1). Mutations to any of these genes may reduce or even eliminate chlorophyll biosynthesis, leading to leaf color changes (Liu et al., 2007, 2016; Yoo et al., 2009; Tian et al., 2013; Wang et al., 2014; Zhang et al., 2015; Yang et al., 2016). Through gene mapping and additional studies, the mutant gene responsible for the xantha marker trait of HYB was identified as a mutated OsGUN4 (genomes uncoupled 4) (Chi et al., 2010; Li et al., 2014).
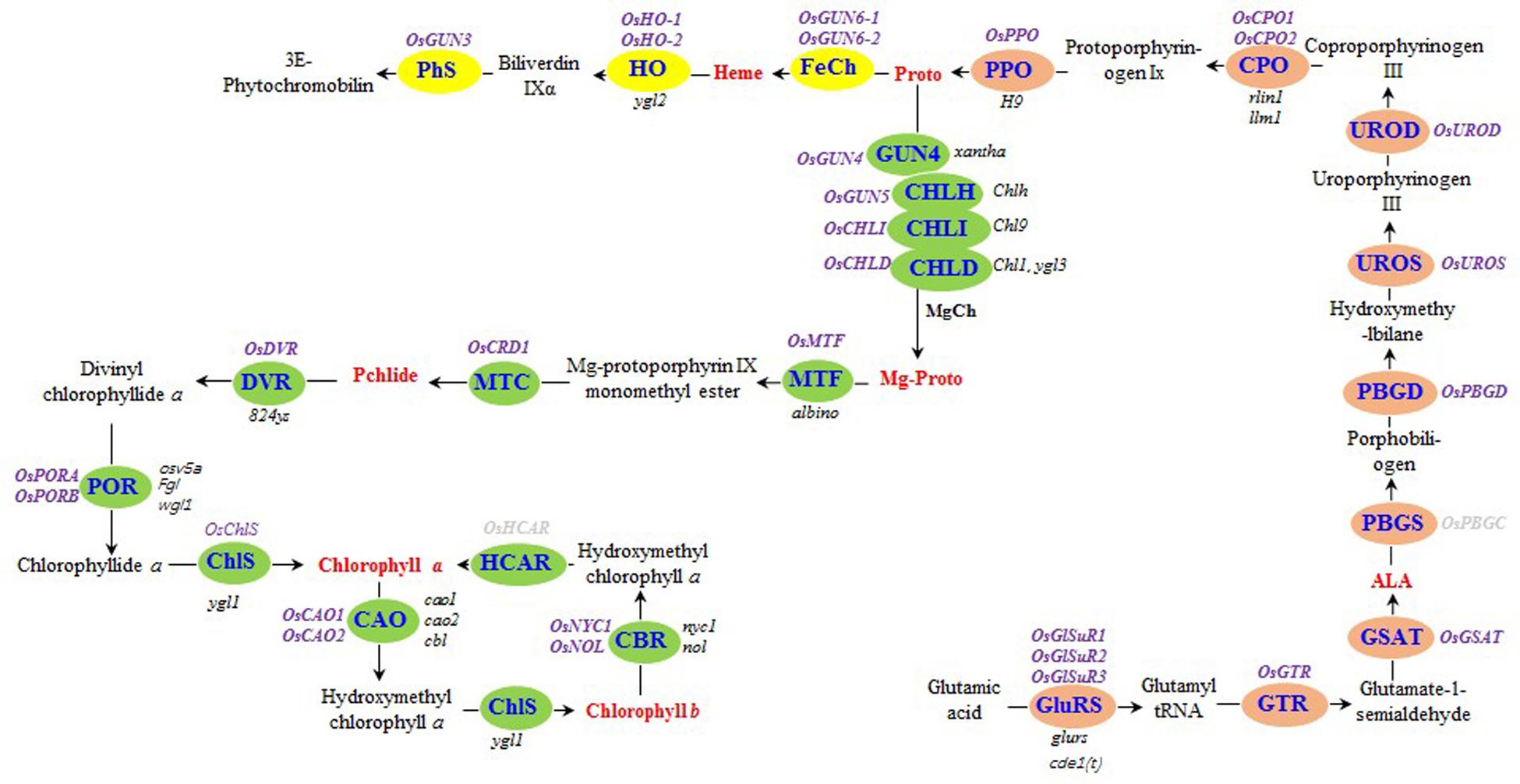
FIGURE 1. Tetrapyrrole biosynthesis pathway in rice. The enzymes of the tetrapyrrole biosynthesis pathway are indicated by blue text. The steps before the tetrapyrrole branch are presented in orange. The Mg and Fe branches are indicated in green and yellow, respectively. The rice genes encoding tetrapyrrole biosynthesis-associated enzymes are listed in purple (bold), while the rice mutants are listed in black (italics). The seven tetrapyrrole intermediates detected in this study are presented in red (bold). Rice genes encoding hydroxymethyl chlorophyll a reductase and porphobilinogen synthase were not detected, and are indicated in gray (bold). GluRS, glutamyl-tRNA synthetase; GTR, glutamyl-tRNA reductase; GSAT, glutamate-1-semialdehyde aminotransferase; ALA, δ-aminolevulinic acid; PBGS, porphobilinogen synthase; PBGD, porphobilinogen deaminase; UROS, uroporphyrinogen III synthase; UROD, uroporphyrinogen III decarboxylase; CPO, coproporphyrinogen III oxidase; PPO, protoporphyrinogen IX oxidase; Proto, protoporphyrin IX; MgCh, protoporphyrin IX Mg-chelatase; ChlH, Mg-chelatase H subunit; ChlI, Mg-chelatase I subunit; ChlD, Mg-chelatase D subunit; Mg-Proto, Mg-protoporphyrin IX; MTF, Mg-protoporphyrin IX methyltransferase; MTC, monomethyl ester oxidative cyclase; DVR, divinyl chlorophyllide a 8-vinyl reductase; POR: NADPH, protochlorophyllide oxidoreductase; ChlS, chlorophyll synthase; CAO, chlorophyllide a oxygenase; CBR, chlorophyll b reductase; HCAR, hydroxymethyl chlorophyll a reductase; FeCh, protoporphyrin IX Fe-chelatase; HO, heme oxygenase; PhS, phytochromobilin synthase; Pchlide, protochlorophyllide a.
In higher plants, more than 95% of chloroplast proteins are encoded by nuclear genes, and chlorophyll biosynthesis is closely associated with the expression of photosynthesis-associated nuclear genes (PhANGs) (Sawchuk et al., 2008; Pogson et al., 2008). The coordinated expression between chloroplast genes and PhANGs is achieved through plastid-to-nucleus retrograde signaling pathways (reviewed by Chan et al., 2016; Larkin, 2016). The GUN4 protein is involved in chlorophyll biosynthesis and retrograde signaling in Arabidopsis thaliana (Larkin et al., 2003; Strand et al., 2003; Peter and Grimm, 2009; Adhikari et al., 2011; Richter et al., 2016), Chlamydomonas reinhardtii (Formighieri et al., 2012; Brzezowski et al., 2014; Tabrizi et al., 2015), Synechocystis sp. (Sobotka et al., 2008; Chen et al., 2015) and rice (Zhou et al., 2012). It has been documented that GUN4 stimulates Mg chelatase activity in rice (Zhou et al., 2012) and other angiosperms (Richter et al., 2016). In HYB, mutations to OsGUN4 result in a significant decrease in chlorophyll content (Li et al., 2014), which also demonstrated the role of OsGUN4 in chlorophyll biosynthesis.
The xantha marker also is associated with a very unique trait, in which decreased chlorophyll content is coupled with a high photosynthetic capacity (Zhou et al., 2006b; Wu et al., 2007). To elucidate the biological mechanisms underlying the xantha marker trait, we examined the differences in tetrapyrrole synthesis, photosynthesis, and retrograde signaling molecules between the xantha mutant and its WT parent. We revealed that altered tetrapyrrole biosynthesis and deregulated transcription of PhANGs might be responsible for the xantha marker trait.
Materials and Methods
Plant Growth Conditions
The xantha mutant line HYB and its WT parental line LTB were previously developed by our group (Zhou et al., 2006a). Seedlings of the two lines were planted in plastic barrels filled with soil from a rice paddy at the Zhejiang University experimental farm. The seedlings were grown in a growth chamber under a 16-h light (1,000 μmol photons m-2 s-1; 30°C):8-h dark (26°C) photoperiod.
Analysis of Chlorophyll and Tetrapyrrole Intermediates
We maintained 32-day-old seedlings in 1× Murashige and Skoog liquid medium for 3 days. Leaf tissue was collected at midday to analyze chlorophyll and tetrapyrrole intermediate contents. The Chl and Car contents were determined using 0.5 g fresh leaf tissue according to a published method (Peter and Grimm, 2009). The Chl a, Chl b, and Car concentrations were calculated according to the method described by Lichtenthaler (1987).
The ALA content was measured using a slightly modified version of the procedure described by Czarnecki et al. (2012). Briefly, 0.5 g fresh leaf tissue was homogenized in liquid nitrogen and suspended in 500 μL acetate buffer (57 μL acetic acid, 100 μM sodium acetate, 200 μL acetoacetic ester, pH 4.6, and double-distilled H2O up to 500 μL). The sample was boiled in a water bath for 15 min and then cooled to room temperature. We added 1 mL acetyl ethyl ester to the cooled solution, which was then incubated at room temperature for 5 min. After a 5-min centrifugation at 16,000 ×g, the supernatant was transferred to a new tube, and mixed with 200 μL reagent (1 g 4-dimethylaminobenzaldehyde, 5 mL perchloric acid, 5 mL H2O, and acetic acid up to 50 mL). The optical density of the solution was measured at 553 nm, and the ALA concentration was determined using a standard curve generated with commercial ALA (Sigma–Aldrich Inc., Shanghai, China).
The protoporphyrin IX (Proto), Mg-protoporphyrin IX (Mg-Proto), and protochlorophyllide a (Pchlide) contents were determined using a commercial enzyme-linked immunosorbent assay kit (Jingmei Tech., China) (Engvall and Perlmann, 1971). Briefly, 0.5 g fresh leaf tissue ground in liquid nitrogen was suspended in 1 mL cold alkaline acetone containing 0.1 N NH4OH (9:1; v/v) and then centrifuged (16,000 ×g for 5 min). We extracted Proto (Papenbrock et al., 1999), Mg-Proto (Papenbrock et al., 1999), and Pchlide (Koski and Smith, 1948) from the supernatant. The pellets were suspended in 1 mL acetone/HCl/DMSO (10:0.5:2, v/v/v) and centrifuged (16,000 ×g for 5 min) three times. The heme content of the supernatant was determined as previously described by Peter and Grimm (2009).
Identification of Genes Involved in Tetrapyrrole Biosynthesis and Photosynthesis
The rice genes encoding enzymes involved in tetrapyrrole biosynthesis were identified either through BLAST searches of A. thaliana protein sequences in the National Center for Biotechnology Information database or using gene names (Supplementary Table S1) in the Rice Annotation Project database1. We identified 30 genes (Figure 1 and Supplementary Table S1).
Plant photosynthesis takes place in Photosystems I and II (PSI and PSII) and in the cytochrome (cyt) b6f complex (Supplementary Figure S1A). PSI consists of the PsaA–L, PsaN, and PsaO subunits, and the light-harvesting chlorophyll-binding proteins Lhca1–4 (Supplementary Figure S1A). PSII contains major intrinsic proteins (Supplementary Figure S1A), such as PsbA (D1), PsbB (CP47), PsbC (CP43), and PsbD (D2), as well as the PsaE and PsbF polypeptides, cyt b559, and the O2 evolution activity unit (PsaO, PsaP, and PsbQ). The PsbS subunit and other polypeptides (PsbH–N, PsbR, PsbT, and PsbW–Z) are also included. The electron transport chain of rice chloroplasts includes (Mn)4, P680, Phe, QA, and QB from PS II; PQH2 and PC from cyt b6f; and P700, A0, A1, Fx, FA, FB, Fd, and FNR from PS I. The Rubisco (Rbc) enzyme consists of five small subunits (RbcS) and one large subunit (RbcL) (Supplementary Figure S1B). We identified 90 photosynthesis-associated genes based on BLAST searches (Supplementary Table S1).
There are two types of RNA polymerase in plastids, i.e., nuclear-encoded plastid RNA polymerase (NEP) and plastid-encoded plastid polymerase (PEP). The sole NEP encoding gene (OsRopTp), two plastid genes that encode the subunits of PEP (OsrpoA and OsrpoB), two nuclear genes that encode PEP associated proteins (OsSIG5 and OsSIG6) were also identified (Supplementary Table S1).
Quantitative Real-Time Polymerase Chain Reaction Analysis
Total RNA was extracted from fresh leaf tissue using the Spin Plant RNA Mini Kit (Qiagen, Germany). The leaf tissue was collected at midday from seedlings that had been grown in liquid medium for 3 days after being transferred from soil. The cDNA was reverse transcribed using 1 μg total RNA, an oligo-dT18 primer, and the GoScriptTM Reverse Transcription System (Promega). The quantitative real-time polymerase chain reaction (RT-q PCR) was conducted using a SYBR Green GoTaq® qPCR Master Mix containing the ROX reference dye (Promega, Beijing, China). The rice ACTIN gene (Os03g0718100) was used as an internal control, and the relative expression levels were calculated according to the 2-ΔΔCt method (Livak and Schmittgen, 2001). Gene details and qRT-PCR primer sequences are provided in Supplementary Table S1.
Determination of Singlet Oxygen Content
The singlet oxygen (1O2) content was determined using the singlet oxygen sensor green fluorescent probe (Hideg et al., 2002). Leaves sampled at midday from seedlings grown in liquid medium were stored at -70°C until used. Frozen rice leaves were ground in liquid nitrogen and suspended with 1 mL pre-chilled lysis buffer (GENMED Scientifics Inc., United States). The mixture was briefly vortexed and immediately centrifuged at 4°C (5,000 ×g for 10 min). The supernatant was transferred to a new pre-chilled 1.5-mL tube and mixed with a solution consisting of 10 μmol singlet oxygen sensor green probe (GENMED Scientifics Inc.) and 0.02% methanol. The fluorescence spectra were analyzed at excitation and emission wavelengths of 485 and 520 nm, respectively, using a 359S fluorescence spectrophotometer (Lengguang Tech., China).
Statistical Analysis
All measurements were completed using six biological replicates, each consisting of pooled tissue from three seedlings. Statistical analyses were conducted using the F-tests and Student’s t-test.
Results
The xantha Mutation Altered the Biosynthesis of Tetrapyrrole
Similar to previous observations, field-grown xantha mutant plants exhibited yellow leaf color phenotypes (Supplementary Figures S2) and had significantly lower Chl a, Chl b, and Car contents than the WT controls (Figures 2A–C). Consequently, the Chl a/b and Car/Chl ratios were 1.8- and 11.3-fold higher for the xantha mutant than for the WT plants, respectively.
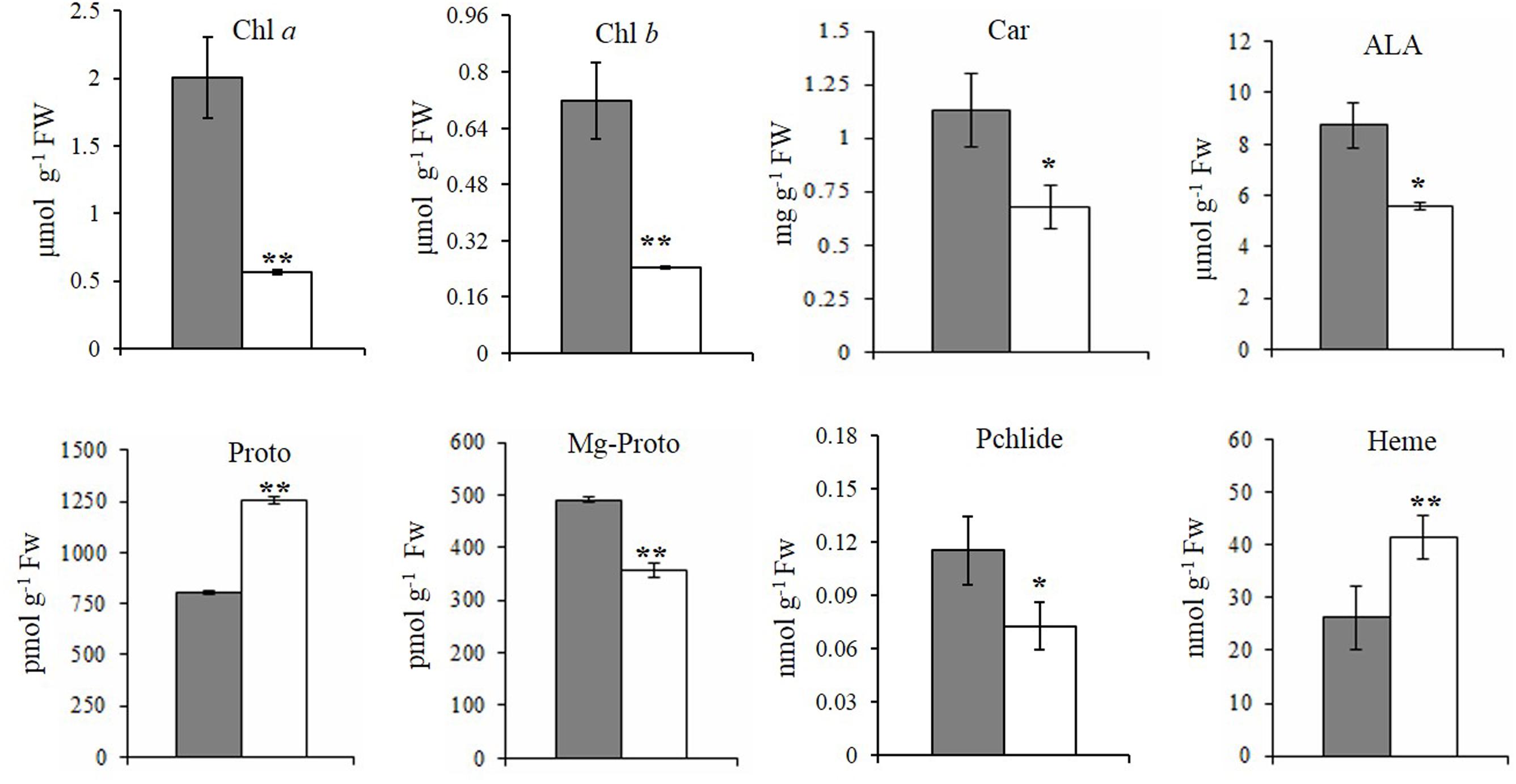
FIGURE 2. Steady-state levels of tetrapyrrole intermediates in rice. The steady-state levels of intermediates in wild-type plants (gray bar) and xantha mutants (open bar) were analyzed using six biological replicates. Error bars represent the standard error. ∗, ∗∗, and ∗∗∗ represent significant differences between wild-type and xantha mutant values at P < 0.05, P < 0.01, and P < 0.001, respectively.
To clarify how chlorophyll contents decreased, the contents of five tetrapyrrole intermediates were also analyzed. First, the ALA and Proto contents exhibited the opposite trends in the xantha mutant, with ALA levels decreasing by 40.1% (Figure 2D) and Proto levels increasing by 55.0% (Figure 2E) relative to the WT levels. In the Mg-Proto branch, the contents of Mg-Proto (Figure 2F) and Pchlide (Figure 2G) decreased by 27.2 and 37.0%, respectively. In the Fe-Proto branch, the xantha mutant accumulated 1.6-fold more heme than the WT controls (Figure 2H). These results indicated that the xantha mutation altered tetrapyrrole biosynthesis, with more intermediates diverted to the Fe-Proto branch.
To determine how the xantha mutation altered tetrapyrrole biosynthesis, we investigated the effects of the xantha mutation on the transcription of 30 rice genes involved in different tetrapyrrole biosynthesis steps (Figure 1). Consistent with the results of our previous studies, the abundance of OsGUN4 transcripts in the xantha mutant was only 0.02% of that of the WT plants (Figure 3). Surprisingly, the abundance of all transcripts except for those of OsHO-1 and OsHO-2 was significantly greater in the xantha mutant than in the WT controls (Figure 3). Additionally, the transcripts of genes involved in shared steps of the tetrapyrrole biosynthesis pathway were 1.3–8.6-fold more abundant in the xantha mutant than in the WT plants (Figure 3A), while the transcripts of genes in the Mg-Proto branch were 1.2–3.2-fold more abundant in the xantha mutant (Figure 3B). In the Fe-Proto branch, the OsGUN3, OsGUN6-1, and OsGUN6-2 expression levels were up-regulated in the xantha mutant, with 1.5–2.9-fold higher transcript levels than in the WT plants. In contrast, OsHO-1 and OsHO-2 expression was down-regulated (Figure 3C).
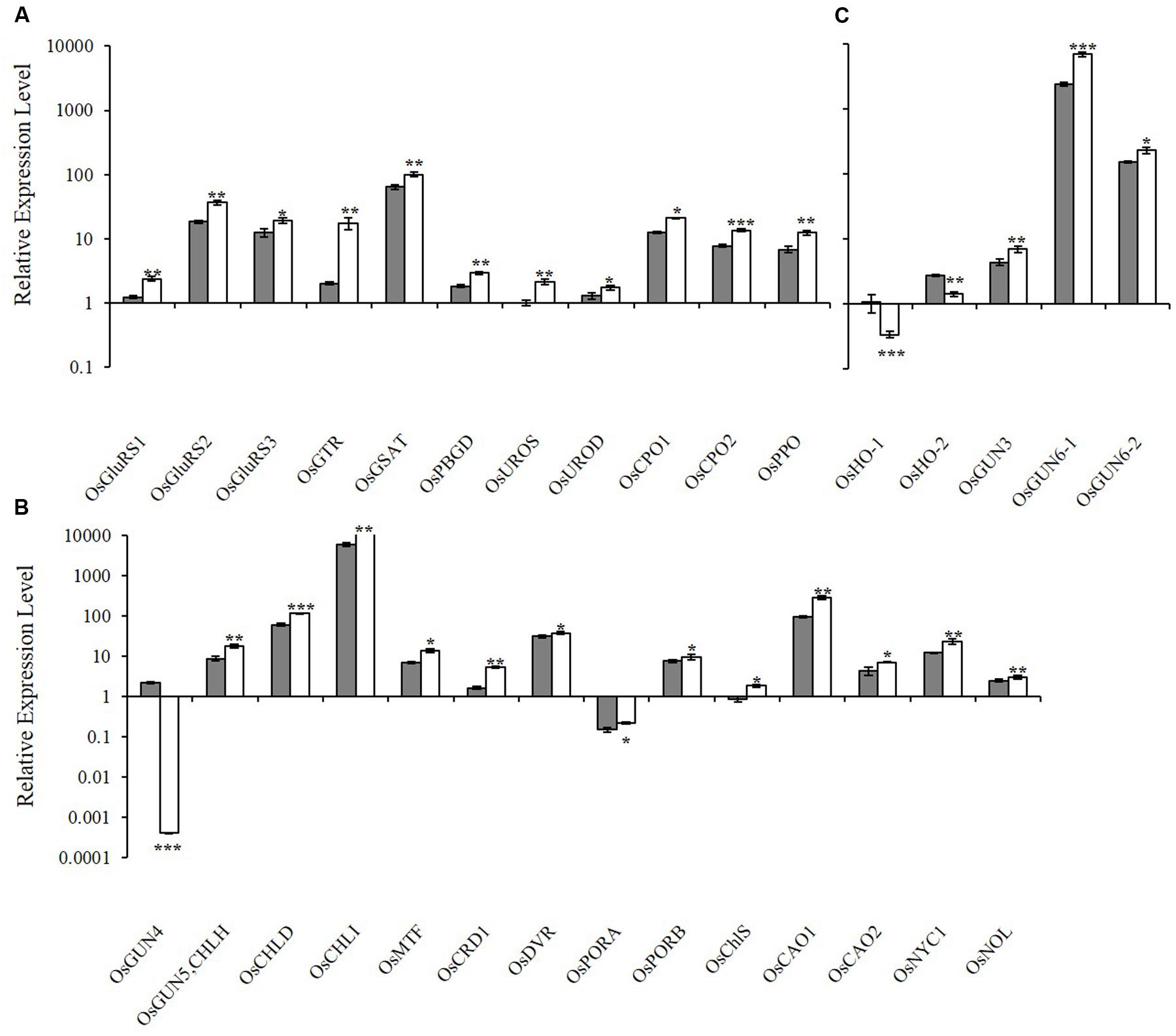
FIGURE 3. Relative expression levels of tetrapyrrole biosynthesis-associated genes in rice. (A) Expression levels of genes encoding enzymes for the shared steps of the tetrapyrrole biosynthesis pathway. (B) Expression levels of genes from the Mg branch. (C) Expression levels of the genes from the Fe branch. The transcript levels in wild-type plants (gray bar) and xantha mutants (open bar) were first normalized to the internal control gene OsACTIN and reported relative to the OsUROS level in wild-type plants (assigned a value of 1). Error bars represent the standard error. ∗, ∗∗, and ∗∗∗ represent significant differences between wild-type and xantha mutant values at P < 0.05, P < 0.01, and P < 0.001, respectively.
The xantha Mutation Up-regulated the Transcription of PhANGs, But Had No Effect on Chloroplast Genes
To characterize the biological basis underlying the high photosynthetic efficiency of the xantha mutant, the transcription of photosynthesis-associated genes (i.e., PhANGs and chloroplast genes) was investigated. Overall, the expression levels of all PhANGs were significantly higher in the xantha mutant than in the WT, while there were no differences in the chloroplast gene expression levels (Figure 4).
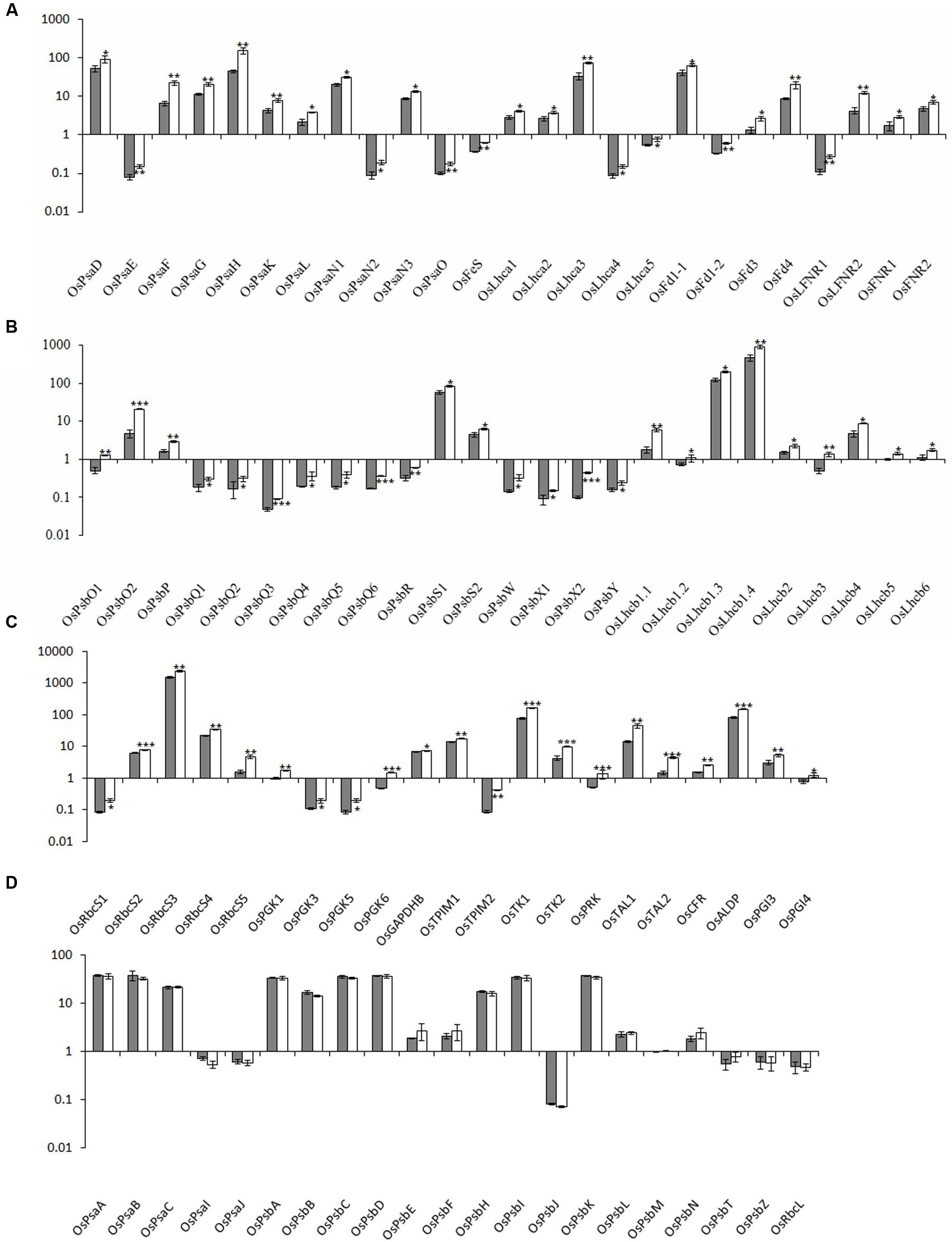
FIGURE 4. Relative expression levels of photosynthesis-associated nuclear genes in rice. The transcript levels of PhANGs for PSI (A), PSII (B), and the CO2 fixation system (C) in wild-type plants (gray bar) and xantha mutants (open bar) were analyzed using six biological replicates. (D) The transcript levels of photosynthesis-associated chloroplast genes (PhACGs) for PSI, PSII, and the CO2 fixation system in wild-type plants (gray bar) and xantha mutants (open bar). The transcript levels were first normalized to the internal control gene OsACTIN and reported relative to the OsLhcb5 level for nuclear genes and to OsPsbM for plastid genes in wild-type plants (assigned a value of 1), respectively. Error bars represent the standard error. ∗, ∗∗, and ∗∗∗ represent significant differences between wild-type and xantha mutant values at P < 0.05, P < 0.01, and P < 0.001, respectively.
We analyzed five chloroplast genes (OsPsaA–C, OsPsaI, and OsPsaJ) and 25 PhANGs in PSI (Supplementary Table S1 and Figure S1A). The PhANG expression levels were 1.5–3.5-fold higher in the xantha mutant than in the WT plants, whereas there were no significant differences in the chloroplast gene expression levels between the mutant and WT plants (Figures 4A,D). Additionally, the expression levels of five Lhca genes (OsLhca1–5) and the genes encoding electron transport chain components were up-regulated by 1.4–2.2-fold and 1.4–2.9-fold, respectively (Figure 4A).
We identified 41 PSII genes (16 chloroplast genes and 25 PhANGs) (Supplementary Table S1 and Figure S1A). The transcription level changes of PSII genes were similar to those of PSI genes. The transcripts for the genes encoding subunit and Lhcb proteins were 1.4–4.4-fold and 1.4–3.4-fold higher in the xantha mutant than in the WT plants, respectively (Figure 4B). However, there was no difference between the xantha mutant and WT plants regarding the expression of chloroplast genes encoding PSII subunits (Figure 4D).
One chloroplast gene (OsRbcL) and 22 nuclear genes (five OsRbcSs and 17 enzyme-encoding genes) were identified for the CO2 fixation system. Of these genes, only three (OsGAPD, OsTK1, and OsCFR) were previously identified in rice (Supplementary Table S1 and Figure S1B). The transcript levels for the RbcS genes and the 17 enzyme-encoding genes were 1.3–3.0-fold and 1.1–4.8-fold higher in the xantha mutant than in the WT plants, respectively (Figure 4C). However, similar to the other chloroplast genes, there was no difference in the transcription of RbcL between the xantha mutant and WT plants (Figure 4D). Thus, compared with the WT levels, the expression levels of all of the PhANGs were up-regulated in the xantha mutant, while there were no differences in the expression of the chloroplast genes.
The transcription of NEP/PEP and PEP associated genes were also investigated. No significant differences of transcript level were observed for the two plastid PEP genes, while the three nuclear genes had higher levels of transcript in the xantha mutant than in the WT plants (Supplementary Figure S3).
The xantha Mutation Decreased 1O2 Production and Down-regulated 1O2-Dependent Retrograde Signaling
Because 1O2 is believed to be important for retrograde signaling, we measured the 1O2 concentration. We observed that the 1O2 concentration in the xantha mutant was 38.0% lower than that of the WT seedlings (Figure 5A). Decreases in the 1O2 level should affect the expression of the genes in the 1O2-dependent signaling pathway. Thus, 11 such genes were identified and their transcription levels were examined. Consistent with the lower 1O2 content, the transcript abundance for these genes in the xantha mutant was only 11.0–50.0% of that of the WT plants (Figure 5B).
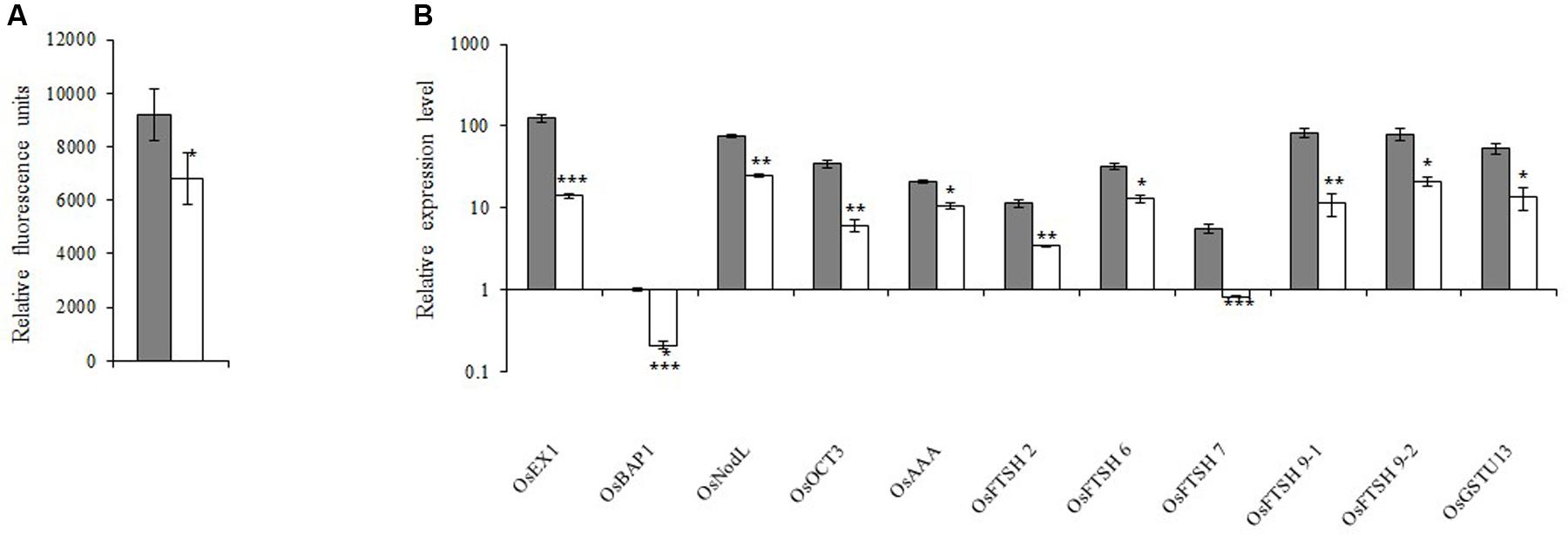
FIGURE 5. 1O2-inducible retrograde signaling pathway in rice. (A) 1O2 concentrations in xantha mutant and wild-type plants. The accumulation of 1O2 in six biological replicates was quantified based on singlet oxygen sensor green fluorescence. Error bars represent the standard error. ∗Represents a significant difference between wild-type and mutant values at P < 0.05. (B) Expression levels of genes involved in the 1O2-inducible retrograde signaling pathway in wild-type plants (gray bar) and xantha mutants (blank bar) were first normalized to the OsActin internal control and were reported relative to the wild-type OsBAP expression level (assigned a value of 1). Error bars represent the standard error. ∗, ∗∗, and ∗∗∗ represent significant differences between wild-type and xantha mutant values at P < 0.05, P < 0.01, and P < 0.001, respectively.
Discussion
The function of GUN4 during the regulation of chlorophyll synthesis and in plastid-to-nucleus retrograde signaling has been studied in A. thaliana, C. reinhardtii and Synechocystis sp., but little is known regarding the GUN4 homologs in rice and other higher plants. Based on our previous observation that an epi-mutation of OsGUN4 results in the xantha marker trait, we investigated the effects of OsGUN4 on tetrapyrrole biosynthesis and the regulation of genes involved in photosynthetic activities by comparing the xantha mutant and its WT parent. Our results may be useful for elucidating the molecular mechanism responsible for the xantha phenotype in rice plants. They may also provide novel insights into the function of GUN4 in plants beyond the model plant species A. thaliana.
Effect of the xantha Mutation on Tetrapyrrole Synthesis
The GUN4 protein was first identified as a regulator of chlorophyll biosynthesis (Larkin et al., 2003) in A. thaliana, mainly at the posttranslational level (Peter and Grimm, 2009). Many GUN4 mutants have been identified in A. thaliana (Larkin et al., 2003; Adhikari et al., 2011) and C. reinhardtii (Brzezowski et al., 2014). Knockout mutants [e.g., gun4-2 (Larkin et al., 2003) and line 026911 (Peter and Grimm, 2009)] exhibit an albino phenotype and do not accumulate chlorophyll under dark–light growth conditions. However, other A. thaliana mutants, such as gun4-1 (L88F amino acid change; Larkin et al., 2003) and line 011461 (truncated GUN4), contain chlorophyll, but only at approximately 40% (Mochizuki et al., 2001) and 50% (Peter and Grimm, 2009) of the WT level, respectively. The differences in chlorophyll content between gun4 mutants and WT A. thaliana plants diminish with decreasing light intensity (Peter and Grimm, 2009). There are also fewer differences in C. reinhardtii under dark vs. continuous light conditions (Brzezowski et al., 2014). Although the epi-mutation almost eliminates OsGUN4 expression at the transcript and protein levels (Li et al., 2014), rice xantha mutant plants still accumulate chlorophyll, albeit at 37.2% of the WT level (Figures 2A,B). This suggests that OsGUN4 and GUN4 have similar effects on chlorophyll biosynthesis.
In vitro activity studies have already demonstrated that OsGUN4 promotes magnesium chelatase activity in rice (Zhou et al., 2012). However, when its C-terminal domain was absent or phosphorylated (Zhou et al., 2012; Richter et al., 2016), it no more activates magnesium chelatase. This not only demonstrated the essential role of its C-terminal domain, but also explained different gun4 mutations could have different effects on chlorophyll biosynthesis.
The observation that the xantha mutant accumulated significantly less ALA and more Proto than the WT plants (Figure 1) is consistent with the results for the gun4 mutants of A. thaliana (Peter and Grimm, 2009; Czarnecki et al., 2012) and C. reinhardtii (Brzezowski et al., 2014). However, the xantha mutant accumulated significantly more heme than the WT control plants (Figure 2H), which is in contrast to what was observed for the mutant A. thaliana line 011461 (Peter and Grimm, 2009). In C. reinhardtii gun4 mutants, the reported changes in the heme level were not consistent. For example, Formighieri et al. (2012) concluded the per-cell heme level did not increase, while Brzezowski et al. (2014) reported that significantly more heme accumulated in the gun4 mutant than in the WT plants under continuous light conditions, but no differences were observed in plants grown in darkness.
In the Mg-Proto branch, the Mg-Proto and Pchlide contents were significantly lower in the xantha mutant than in the WT plants (Figures 2F,G). The abundance of all tetrapyrrole intermediates increased in transgenic tobacco plants overexpressing the A. thaliana GUN4 gene (Peter and Grimm, 2009). Meanwhile, the changes to Mg-Proto and Pchlide levels were only analyzed for one A. thaliana gun4 knockout mutant grown on medium supplemented with 250 μM ALA. The mutant plants accumulated more Mg-Proto and less Pchlide than the WT controls after a 24-h incubation in darkness. However, after a 24-h incubation under dim light, the trend was reversed (Peter and Grimm, 2009).
The chlorophyll deficiency in the xantha mutant may be due to the decreased abundance of Mg-Proto and Pchlide. The OsGUN4 epi-mutation resulted in an extremely low level of OsGUN4 in the xantha mutant (Li et al., 2014). Hence the mutation would greatly reduce its stimulatory effect on Mg-chelatase activity, and consequently lead to a decrease in the amount of Proto being diverted to the Mg-Proto branch and an increase of Proto accumulation (Figures 1, 2). The decreased accumulation of ALA in the xantha mutant was also consistent with the increased heme level, because heme reportedly inhibits ALA synthesis (Apitz et al., 2016). Other metabolites, e.g., heme catabolites may also play a role in the regulation of ALA synthesis.
Mutated OsGUN4 Deregulates the Transcription of Nuclear Genes Involved in Tetrapyrrole Synthesis and Photosynthesis
In the tetrapyrrole biosynthesis pathway, there are 11 genes encoding proteins/enzymes for shared steps, 14 for the Mg-Proto branch and five for the Fe-Proto branch (Figure 1). The transcription of all tetrapyrrole biosynthesis genes except for OsHO-1 and OsHO-2 was significantly up-regulated in the xantha mutant (Figure 3). A similar observation was reported for a C. reinhardtii gun4 mutant, although the HO genes were not examined (Brzezowski et al., 2014). These results suggest that the effects of GUN4 on tetrapyrrole biosynthesis are highly conserved. This phenomenon has yet to be examined in gun4 mutants of A. thaliana and other higher plants.
The expression of PhANGs is closely associated with chlorophyll biosynthesis through plastid-to-nucleus retrograde signaling mediated by genomes uncoupled genes (Chi et al., 2010; Chan et al., 2016). Similar to the tetrapyrrole biosynthesis genes, the transcription of all 122 PhANGs was significantly higher in the xantha mutant than in the WT plants, implying that the expression of PhANGs was deregulated (Figure 4). A similar observation was reported for a C. reinhardtii gun4 mutant (Brzezowski et al., 2014). In sharp contrast, there were no significant differences between the xantha mutant and WT plants regarding the transcript abundance for the photosynthesis-associated chloroplast genes (PhACGs) (Figure 4), suggesting that PhACGs and PhANGs are independently regulated.
There are two types of RNA polymerase in chloroplast but only PEP transcribes PSI and PSII genes (Kremnev and Strand, 2014; Borner et al., 2015). The two PEP encoding plastid genes (i.e., OsrpoA and OsrpoB) had similar transcript levels in the xantha mutant and in the WT plants (Supplementary Figure S3). These results are not only consistent with the present observation that the xantha mutant and WT plants had similar transcriptional levels of PhACGs (Figure 4), but also support previous conclusions (Borner et al., 2015). The core PEP enzyme requires a sigma factor for promoter recognition and initial of transcription; OsSig5 and OsSig6 are the two genes potentially encoding potential plastid sigma factors of RNA polymerase in rice (Kubota et al., 2007), their transcript levels were significantly greater in the xantha mutant than in the WT plants (Supplementary Figure S3), which suggests that it is the core units of PEP, not the sigma factor, that determine the transcription of PhACGs.
Role of OsGUN4 in Plastid-to-Nucleus Retrograde Signaling
When the GUN4 gene was first identified in A. thaliana, its function in retrograde signaling was confirmed (Larkin et al., 2003). However, there is still a lack of consensus about its exact role in this process. Two contrasting hypotheses have been proposed based on in vitro and in vivo studies involving A. thaliana and C. reinhardtii. Most studies suggested that GUN4 ‘shields’ Proto and Mg-Proto from collisions with O2, thereby decreasing the production of reactive oxygen species (ROS) (e.g., 1O2) (Larkin et al., 2003; Sobotka et al., 2008), which are important retrograde signaling molecules. Brzezowski et al. (2014) proposed that C. reinhardtii GUN4 functions as a potential sensor of excess Proto resulting from perturbations in the tetrapyrrole biosynthesis pathway, and as a mediator of the Proto-triggered 1O2-dependent retrograde signaling pathway. In contrast, Tabrizi et al. (2016) suggested that the light-dependent generation of 1O2 by GUN4–Proto may represent the first step of retrograde signaling from the chloroplast to the nucleus. In this proposal, GUN4 is involved in increasing rather than decreasing 1O2 contents.
The xantha mutant accumulated less Mg-Proto and more Proto than the WT plants (Figures 2B,C). Because OsGUN4 is almost undetectable in the xantha mutant (Li et al., 2014), if OsGUN4 is mainly responsible for shielding Proto and MgProto as suggested by Brzezowski et al. (2014), then an increase in ROS and 1O2 production would be expected. However, if Tabrizi et al. (2016) are correct, a decrease in the 1O2 content of the xantha mutant would be expected because of a lack of or a very limited amount of GUN4-MgProto. Therefore, the significantly lower 1O2 abundance in the xantha mutant than in the WT plants (Figure 5A) supports the GUN4 function proposed by Tabrizi et al. (2016). The effects of decreased 1O2 abundance in the xantha mutant are also reflected by the down-regulated transcription of genes associated with the 1O2-dependant signaling pathway (Figure 5B). Thus, these results help to explain the deregulation of PhANGs due to chlorophyll deficiencies in the xantha mutant.
A new model of tetrapyrrole-dependent plastid-to-nucleus signaling was recently proposed by Larkin (2016). It suggests that heme accumulates in active chloroplasts, and that the accumulation or export of heme to the cytosol or nucleus activates a mechanism that induces the expression of PhANGs. The observed elevated heme content (Figure 2H) and increased transcription of PhANGs (Figures 4A,B) in the xantha mutant are consistent with this model.
High Photosynthetic Capacity in gun4 Mutants
Although its chlorophyll content is only 37.2% of that of WT plants, the xantha mutant retains a high photosynthetic capacity (Zhou et al., 2006a; Wu et al., 2007). The effects of gun4 mutations on photosynthetic activities were briefly discussed for C. reinhardtii (Formighieri et al., 2012) and in more detail for A. thaliana (Daloso et al., 2014).
An earlier study (Zhou et al., 2006a) involving field-grown plants revealed that the xantha mutant had a higher net photosynthetic rate (PN) than WT plants, possibly because of its lower non-photochemical quenching (qN) value. Additionally, the maximal photochemical efficiency (FV/FM), excitation energy capture efficiency of PSII reaction centers (FV′/FM′), photochemical quenching (qP), effective PSII quantum yield (ΦPS2), and non-cyclic electron transport rate (ETR) were all higher in the xantha mutant than in the WT line. Wu et al. (2007) conducted a diurnal study on the photosynthetic activities of the xantha mutant under field conditions at high temperatures and irradiance. Photo-inhibition was observed for the WT line above 1,043 μmol photons m-2 s-1, but not for the mutant (the highest light intensity during the day was 1,235 μmol photons m-2 s-1). During the photo-inhibitory period for the WT line, the mutant exhibited higher PN, qN, FV/FM, ΦPS2, and ETR values. These observations can be partially explained by chlorophyll changes in the mutant. For example, the lower Chl b content in the mutant is likely associated with a lower capacity for photon capture. Thus, PSII would be more stable under high-light intensities, resulting in an enhanced ΦPS2 (Wu et al., 2007). The present study revealed that the transcription of PhANGs was significantly higher in the xantha mutant than in the WT plants (Figures 4A,B), which may have contributed to the high photosynthetic capacity of the mutant. For example, the up-regulated expression of LHCB genes may optimize the transfer of light energy from light-harvesting complex II to the reaction center of PSII, thereby contributing to the ETR, as suggested by Daloso et al. (2014) for A. thaliana.
Previous studies involving A. thaliana and C. reinhardtii gun4 mutants produced in consistent results regarding the effect of GUN4 on photosynthesis. In A. thaliana, Daloso et al. (2014) reported higher ΦPS2 and qP values for a gun4 mutant. This suggests the mutant transfers the excess absorbed light energy more efficiently than the WT plants despite a decrease in chlorophyll content. The mutant also likely has a greater capacity to dissipate excess absorbed energy. Although the FV/FM values were similar, the ΦPS2 value was higher for the WT C. reinhardtii (0.46) than for the gun4 knockout mutant (0.32) (Formighieri et al., 2012). The differences between the gun4 mutant and WT plants are often dependent on environmental conditions. For example, they become significant under high-light intensities (in rice) or after exposure to continuous light stress (in A. thaliana), which is probably because of the photo-protective function of GUN4 (Formighieri et al., 2012; Brzezowski et al., 2014). Therefore, the inconsistencies in the reported results are not unexpected.
Future Studies on GUN4 in Crop Plants
Since the initial discovery of A. thaliana GUN4 in 2003 (Larkin et al., 2003), follow-up studies have been conducted only for C. reinhardtii, A. thaliana, and Synechocystis sp. There were no reports describing GUN4 from other plant species until we revealed that an epi-allele of OsGUN4 is responsible for the xantha marker trait (Li et al., 2014). This previous study remains the only one involving a GUN4 from a crop plant. Because GUN4 is important for regulating chlorophyll and tetrapyrrole syntheses, as well as photosynthetic activities (Larkin, 2016), additional studies on GUN4 in crop plants, including rice, may help to more comprehensively characterize GUN4 functions. They may also provide new insights into crop productivity and environmental adaptability. Furthermore, the genetic manipulation of the GUN4 gene may generate new GUN4 alleles useful for improving the ability of crops to adapt to particular environments.
Although the GUN4 proteins of crop plants likely function similarly to the GUN4 of A. thaliana and C. reinhardtii, they may also have additional or different roles. For example, we observed that the xantha mutant had a significantly increased heme level (Figure 2H), which was not observed in A. thaliana and C. reinhardtii gun4 mutants (Peter and Grimm, 2009; Formighieri et al., 2012; Brzezowski et al., 2014). Most of the studies of A. thaliana GUN4 were performed under artificial conditions, with relatively low light intensities. Therefore, the photo-protective role of GUN4 needs to be examined in crop plants grown under field conditions with natural sunlight.
In summary, our present study extended the research on GUN4 to a crop plant, and elucidated the molecular basis of the first agronomic trait regulated by an epi-allele of the GUN4 gene. We observed that a mutated OsGUN4 altered tetrapyrrole biosynthesis and deregulated the transcription of PhANGs. Our data confirm that OsGUN4 is important for tetrapyrrole biosynthesis and photosynthetic activities in rice plants.
Author Contributions
Q-YS and J-MW conceived the study. R-QL and MJ conducted the RT-q PCR experiments. R-QL and Y-CZ completed the bioinformatic analyses. R-QL, Y-HL, and MJ analyzed the tetrapyrrole intermediates and 1O2. R-QL, Q-YS, and J-ZH analyzed the data. R-QL and J-ZH drafted the manuscript, and Q-YS prepared the final version. All authors reviewed and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by a grant from the National Key Research and Development Program of China (2016YFD0102103).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00901/full#supplementary-material
Footnotes
References
Adhikari, N. D., Froehlich, J. E., Strand, D. D., Buck, S. M., Kramer, D. M., and Larkin, R. M. (2011). GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of Mg-Chelatase and promote chlorophyll biosynthesis in Arabidopsis. Plant Cell 23, 1449–1467. doi: 10.1105/tpc.110.082503
Apitz, J., Nishimura, K., Schmied, J., Wolf, A., Hedtke, B., van, Wijk KJ, et al. (2016). Posttranslational control of ALA synthesis includes GluTR degradation by Clp protease and stabilization by GluTR-binding protein. Plant Physiol. 170, 1289–1298. doi: 10.1104/pp.15.01945
Beale, S. I. (2005). Green genes gleaned. Trends Plant Sci. 10, 301–312. doi: 10.1016/j.tplants.2005.05.005
Borner, T., Aleynikova, A. Y., Zubo, Y. O., and Kusnetsov, V. V. (2015). Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim. Biophys. Acta 1847, 761–769. doi: 10.1016/j.bbabio.2015.02.004
Brzezowski, P., Schlicke, H., Richter, A., Dent, R. M., Niyogi, K. K., and Grimm, B. (2014). The GUN4 protein plays a regulatory role in tetrapyrrole biosynthesis and chloroplast-to-nucleus signalling in Chlamydomonas reinhardtii. Plant J. 79, 285–298. doi: 10.1111/tpj.12560
Chan, K. X., Su, Y. P., Crisp, P., Mcquinn, R., and Pogson, B. J. (2016). Learning the languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 67, 25–53. doi: 10.1146/annurev-arplant-043015-111854
Chen, X., Hua, P., Xiao, W., Long, W., Lin, R., and Lin, L. (2015). Crystal structures of GUN4 in complex with porphyrins. Mol. Plant 8, 1125–1127. doi: 10.1016/j.molp.2015.04.013
Chi, X. F., Zhou, X. S., and Shu, Q. Y. (2010). Fine mapping of a xantha mutation in rice (Oryza sativa L.). Euphytica 172, 215–220. doi: 10.1007/s10681-009-0048-8
Czarnecki, O., Gläßer, C., Chen, J. G., Mayer, K. F. X., and Grimm, B. (2012). Evidence for a contribution of ALA synthesis to plastid-to-nucleus signaling. Front. Plant Sci. 3:236. doi: 10.3389/fpls.2012.00236
Daloso, D. D. M., Antunes, W. C., Santana, T. A., Pinheiro, D. P., Ribas, R. F., and Sachetto-Martins, G. (2014). Arabidopsis gun4 mutant have greater light energy transfer efficiency in photosystem II despite low chlorophyll content. Theor. Exp. Plant Physiol. 26, 177–187. doi: 10.1007/s40626-014-0025-z
Dong, F. G., Zhu, X. D., Xiong, Z. M., Cheng, S. H., Sun, Z. X., and Min, S. K. (1995). Breeding of a chlorina leaf marked indica photoperiod/temperature sensitive genic male sterile line M2S. Chin. J. Rice Sci. 9, 65–67.
Engvall, E., and Perlmann, P. (1971). Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry 8, 871–874. doi: 10.1016/0019-2791(71)90454-X
Formighieri, C., Ceol, M., Bonente, G., Rochaix, J. D., and Bassi, R. (2012). Retrograde signaling and photoprotection in a gun4 mutant of Chlamydomonas reinhardtii. Mol. Plant 5, 1242–1262. doi: 10.1093/mp/sss051
Hideg, E., Barta, C., Kálai, T., Vass, I., Hideg, K., and Asada, K. (2002). Detection of singlet oxygen and superoxide with fluorescent sensors in leaves under stress by photoinhibition or UV radiation. Plant Cell Physiol. 43, 1154–1164. doi: 10.1093/pcp/pcf145
Koski, V. M., and Smith, J. H. (1948). The isolation and spectral absorption properties of protochlorophyll from etiolated barley seedlings. J. Am. Chem. Soc. 70, 3558–3562. doi: 10.1021/ja01191a006
Kremnev, D., and Strand, A. (2014). Plastid encoded RNA polymerase activity and expression of photosynthesis genes required for embryo and seed development in Arabidopsis. Front. Plant Sci. 5:385. doi: 10.3389/fpls.2014.00385
Kubota, Y., Miyao, A., Hirochika, H., Tozawa, Y., Yasuda, H., Tsunoyama, Y., et al. (2007). Two novel nuclear genes, OsSig5 and OsSig6, encoding potential plastid sigma factors of RNA polymerase in rice: tissue-specific and light-responsive gene expression. Plant Cell Physiol. 48, 186–192. doi: 10.1093/pcp/pcl050
Larkin, R. M. (2016). Tetrapyrrole signaling in plants. Front. Plant Sci. 7:e60305. doi: 10.3389/fpls.2016.01586
Larkin, R. M., Alonso, J. M., Ecker, J. R., and Chory, J. (2003). GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906. doi: 10.1126/science.1079978
Li, R. Q., Huang, J. Z., Zhao, H. J., Fu, H. W., Li, Y. F., Liu, G. Z., et al. (2014). A down-regulated epi-allele of the genomes uncoupled 4 gene generates a xantha marker trait in rice. Theor. Appl. Genet. 127, 491–501. doi: 10.1007/s00122-014-2393-9
Li, Y., Ma, X., and Fu, H. (2010). High yield technology of Indica three-line sterile lines Jiazhe 91a. Bull. Agric. Sci. Technol. 5, 162–163.
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods. Enzymol. 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Liu, H., Li, Q., Feng, Y., Zhu, F., Yi, S., and Tao, Y. (2016). Differential regulation of protochlorophyllide oxidoreductase abundances by virescent 5A (OsV5A) and virescent 5B (OsV5B) in rice seedlings. Plant Cell Physiol. 51, 2392–2402. doi: 10.1093/pcp/pcw151
Liu, W., Fu, Y., Hu, G., Si, H., Zhu, L., and Wu, C. (2007). Identification and fine mapping of a thermo-sensitive chlorophyll deficient mutant in rice (Oryza sativa L.). Planta 226, 785–795. doi: 10.1007/s00425-007-0525-z
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mochizuki, N., Brusslan, J. A., Larkin, R., Nagatani, A., and Chory, J. (2001). Arabidopsis genomes uncoupled 5 (gun5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. U.S.A. 98, 2053–2058. doi: 10.1073/pnas.98.4.2053
Mou, T., Li, C., Yang, G., and Lu, X. (1995). Genetical studies on seedling leaf color in purple rice. Chin. J. Rice 9, 45–48.
Najafpour, M. M., and Allakhverdiev, S. I. (2011). Recent progress in the studies of structure and function of photosystems I and II. J. Photochem. Photobiol. B 152(Pt B), 173–175.
Papenbrock, J., Mock, H.-P., Kruse, E., and Grimm, B. (1999). Expression studies in tetrapyrrole biosynthesis: inverse maxima of magnesium chelatase and ferrochelatase activity during cyclic photoperiods. Planta 208, 264–273. doi: 10.1007/s004250050558
Peter, E., and Grimm, B. (2009). GUN4 is required for posttranslational control of plant tetrapyrrole biosynthesis. Mol. Plant 2, 1198–1210. doi: 10.1093/mp/ssp072
Pogson, B. J., Woo, N. S., Förster, B., and Small, I. D. (2008). Plastid signalling to the nucleus and beyond. Trends Plant Sci. 13, 602–609. doi: 10.1016/j.tplants.2008.08.008
Richter, A. S., Hochheuser, C., Fufezan, C., Heinze, L., Kuhnert, F., and Grimm, B. (2016). Phosphorylation of genomes uncoupled 4 alters stimulation of Mg chelatase activity in angiosperms. Plant Physiol. 172, 1578–1595. doi: 10.1104/pp.16.01036
Sawchuk, M. G., Donner, T. J., Head, P., and Scarpella, E. (2008). Unique and overlapping expression patterns among members of photosynthesis-associated nuclear gene families in Arabidopsis. Plant Physiol. 148, 1908–1924. doi: 10.1104/pp.108.126946
Shu, Q. Y., Xia, Y. W., Zuo, X. X., and Liu, G. F. (1996). Marker-assisted elimination of contamination in two-line hybrid rice seed production and multiplication. J. Zhejiang Agric. Univ. 22, 56–60.
Sobotka, R., Dühring, U., Komenda, J., Peter, E., Gardian, Z., Tichy, M., et al. (2008). Importance of the cyanobacterial GUN4 protein for chlorophyll metabolism and assembly of photosynthetic complexes. J. Biol. Chem. 283, 25794–25802. doi: 10.1074/jbc.M803787200
Strand,Å., Asami, T., Alonso, J., Ecker, J. R., and Chory, J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421, 79–83. doi: 10.1038/nature01204
Tabrizi, S. T., Langley, D. B., Harrop, S. J., Duff, A. P., and Willows, R. D. (2015). Structure of GUN4 from Chlamydomonas reinhardtii. Acta Crys. 71, 1094–1099. doi: 10.1107/S2053230X15012248
Tabrizi, S. T., Sawicki, A., Zhou, S., Luo, M., and Willows, R. D. (2016). GUN4-protoporphyrin ix is a singlet oxygen generator with consequences for plastid retrograde signaling. J. Biol. Chem. 291, 8978–8984. doi: 10.1074/jbc.C116.719989
Tanaka, A., and Tanaka, R. (2006). Chlorophyll metabolism. Curr. Opin. Plant Biol. 9, 248–255. doi: 10.1016/j.pbi.2006.03.011
Tian, X., Ling, Y., Fang, L., Du, P., Sang, X., and Zhao, F. (2013). Gene cloning and functional analysis of yellow green leaf 3 (ygl3) gene during the whole-plant growth stage in rice. Genes Genomics 35, 87–93. doi: 10.1007/s13258-013-0069-5
Wang, P., Li, C., Wang, Y., Huang, R., Sun, C., and Xu, Z. (2014). Identification of a geranylgeranyl reductase gene for chlorophyll synthesis in rice. Springerplus 3:201. doi: 10.1186/2193-1801-3-201
Wu, D., Shen, S., Cui, H., Xia, Y., and Shu, Q. (2003). A novel thermo/photoperiod-sensitive genic male-sterile (T/PGMS) rice mutant with green-revertible albino leaf color marker induced by gamma irradiation. Field Crops Res. 81, 141–147. doi: 10.1016/S0378-4290(02)00218-6
Wu, L., Li, R., Shu, Q., Zhao, H., Wu, D., Li, J., et al. (2011). Characterization of a new green-revertible albino mutant in rice. Crop Sci. 51, 2706–2715. doi: 10.2135/cropsci2011.01.0008
Wu, L., Shen, S., Wang, R., and Shu, Q. (2007). The diurnal variation of photosynthesis of a xantha mutant in rice. J. Nucl. Agric. Sci. 21, 425–429.
Yang, Y., Xu, J., Huang, L., Leng, Y., Dai, L., and Rao, Y. (2016). PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J. Exp. Bot. 67, 1297–1310. doi: 10.1093/jxb/erv529
Yoo, S.-C., Cho, S.-H., Sugimoto, H., Li, J., Kusumi, K., Koh, H.-J., et al. (2009). Rice Virescent3 and Stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol. 150, 388–401. doi: 10.1104/pp.109.136648
Zhang, H., Liu, L., Cai, M., Zhu, S., Zhao, J., and Zheng, T. (2015). A point mutation of magnesium chelatase OsChlI gene dampens the interaction between ChlI and ChlD subunits in rice. Plant Mol. Biol. Rep. 33, 1975–1987. doi: 10.1007/s11105-015-0889-3
Zhou, S., Sawicki, A., Willows, R. D., and Luo, M. (2012). C-terminal residues of Oryza sativa GUN4 are required for the activation of the ChlH subunit of magnesium chelatase in chlorophyll synthesis. FEBS Lett. 586, 205–210. doi: 10.1016/j.febslet.2011.12.026
Zhou, X. S., Shen, S. Q., Wu, D. X., Sun, J. W., and Shu, Q. Y. (2006a). Introduction of a xantha mutation for testing and increasing varietal purity in hybrid rice. Field Crops Res. 96, 71–79. doi: 10.1016/j.fcr.2005.05.008
Keywords: chlorophyll, tetrapyrrole, photosynthesis, retrograde signaling, PhANGs, hybrid rice, seed purity
Citation: Li R-Q, Jiang M, Liu Y-H, Zheng Y-C, Huang J-Z, Wu J-M and Shu Q-Y (2017) The xantha Marker Trait Is Associated with Altered Tetrapyrrole Biosynthesis and Deregulated Transcription of PhANGs in Rice. Front. Plant Sci. 8:901. doi: 10.3389/fpls.2017.00901
Received: 27 March 2017; Accepted: 15 May 2017;
Published: 31 May 2017.
Edited by:
Soren K. Rasmussen, University of Copenhagen, DenmarkReviewed by:
Yanjun Dong, Shanghai Normal University, ChinaRobert Drant Willows, Macquarie University, Australia
Copyright © 2017 Li, Jiang, Liu, Zheng, Huang, Wu and Shu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing-Yao Shu, cXlzaHVAemp1LmVkdS5jbg==
†These authors have contributed equally to this work.
 Rui-Qing Li1,2,3†
Rui-Qing Li1,2,3† Qing-Yao Shu
Qing-Yao Shu