- 1Ministry of Agriculture Key Laboratory of Plant Pathology, College of Plant Protection, China Agricultural University, Beijing, China
- 2State Key Laboratory of Agrobiotechnology, China Agricultural University, Beijing, China
Wide quantitative variation in plant disease resistance across Arabidopsis wild populations has been documented and the underlying mechanisms remain largely unknown. To investigate the genetic and molecular basis of this variation, Arabidopsis recombinant inbred lines (RILs) derived from Aa-0 × Col-0 and Gie-0 × Col-0 crosses were constructed and used for inoculation with Pseudomonas syringae pathovars maculicola ES4326 (ES4326) and tomato DC3000 (DC3000). Bacterial growth assays revealed continuous distribution across the large differences between the most and the least susceptible lines in the RILs. Quantitative trait locus (QTL) mapping analyses identified a number of QTLs underpinning the variance in disease resistance, among which Qpm3.1, a major QTL on chromosome III from both Aa-0 and Gie-0 accessions, preferentially restricted the growth of ES4326. A genetic screen for the ES4326 gene selectively leading to bacterial growth inhibition on accession Aa-0 uncovered the effector gene hopW1-1. Further QTL analysis of disease in RILs inoculated with DC3000 carrying hopW1-1 showed that the genetic interaction between Qpm3.1 and hopW1-1 determined Arabidopsis resistance to bacterial infection. These findings illustrate the complexity of Arabidopsis-Pseudomonas interaction and highlight the importance of pathogen effectors in delineating genetic architectures of quantitative variation in plant disease resistance.
Introduction
Plants rely on a complex innate immune system to ward off pathogen attacks. Perception of pathogen-associated molecular patterns (PAMPs) by plant cell surface distributed receptors elicits the first line of defense-PAMP triggered immunity (PTI). To succeed in infection pathogens secrete effectors to the apoplast or inside plant cells to suppress PTI and reprogram host physiology in favor of disease, a process often referred to as effector-triggered susceptibility (ETS) (Jones and Dangl, 2006; Toruno et al., 2016). Plants have evolved a second line of defense, also known as effector-triggered immunity (ETI), by encoding diverse resistance (R) proteins to detect such effectors and activating high levels of immune responses such as hypersensitive cell death (Jones and Dangl, 2006). R gene-mediated resistance is typically qualitative and effective to restrict pathogen strains carrying specific effectors (avirulence proteins, Avr); in absence of a successful R-Avr interaction, however, quantitative disease resistance reduces the growth and spread of virulent pathogens, thereby protecting plants from further infection (Dangl and Jones, 2001; Poland et al., 2009).
The phenomena of quantitative disease resistance, also referred to as partial disease resistance or basal resistance, has been widely observed in many plant pathosystems, in which the distribution of resistance values in a genetically segregating population is typically continuous and does not follow a Mendelian pattern of inheritance (Poland et al., 2009). Genetic dissection of quantitative disease resistance in many crop plants with quantitative trait locus (QTL) mapping has uncovered genetic loci conferring resistance to a wide range of diseases (St. Clair, 2010). Recently, a number of genes mediating quantitative disease resistance have been successfully cloned from major crop plants including wheat (Fu et al., 2009; Krattinger et al., 2009), rice (Fukuoka et al., 2009; Hayashi et al., 2010), maize (Hurni et al., 2015; Zuo et al., 2015) and soybean (Cook et al., 2012). Notably, many of these genes provide durable resistance in the field and encode proteins distinct to typical R proteins, indicating that quantitative disease resistance and R-gene mediated resistance may recruit different components to deploy.
The model plant Arabidopsis has been used intensively to investigate the genetic and molecular basis of disease resistance to virulent pathogens with diverse lifestyles. Major progress has been achieved on how the signal molecules including salicylic acid (SA), jasmonic acid, and ethylene orchestrate plant defenses (Pieterse et al., 2012). In addition, multiple pattern recognition receptors have been uncovered in the plant to elicit PTI in response to various PAMPs (Gómez-Gómez and Boller, 2000; Zipfel et al., 2006; Miya et al., 2007; Albert et al., 2015). Nevertheless, most of these findings are based on experiments with frequently used Arabidopsis accessions such as Col-0. Several studies have demonstrated wide quantitative variation in disease resistance across Arabidopsis accessions collected from wild populations at different geographical locations, and many accessions are much less susceptible than Col-0 to pathogen infection, indicating the presence of important unknown mechanisms mediating disease resistance in these accessions (Kover and Schaal, 2002; Fan et al., 2008). Therefore, further investigation on the basis of this variation will not only broaden our knowledge on the complex genetic architectures of plant defense networks but also may help us design crops with durable resistance to a broad range of phytopathogens.
The bacterial effector HopW1-1, also known as HopPmaA, has been found in several strains of Pseudomonas syringae pathovars (pv) maculicola and phaseolicola, but is absent from the strain P. syringae pv tomato DC3000 (DC3000) (Guttman et al., 2002; Lindeberg et al., 2006). When delivered into plant cells by P. syringae, HopW1-1 targets the actin cytoskeleton and inhibits endocytosis, thereby promoting bacterial virulence in susceptible Arabidopsis plants (Kang et al., 2014). However, in some accessions such as Ws, hopW1-1 restricts bacterial growth and elicits resistance responses including the accumulation of SA (Lee et al., 2008). Three HopW1-1-interacting (WIN) proteins have been identified from Arabidopsis. Overexpression or downregulation of WIN proteins has diverse effects on plant disease resistance induced by HopW1-1 or other bacterial effectors, indicating that WIN proteins may have different roles in modulating bacteria-plant interactions (Lee et al., 2008). Variation in resistance among Arabidopsis accessions to bacterial strains carrying hopW1-1 has been revealed in several studies (Lee et al., 2008; Rant et al., 2013), but the genetic region underlying plant resistance specific to HopW1-1 remains to be revealed.
In this study, Arabidopsis accessions Aa-0 and Gie-0 were found to support different levels of bacterial growth compared with Col-0 plants. Two populations of recombinant inbred lines (RILs) derived from Aa-0 × Col-0 and Gie-0 × Col-0 crosses were constructed and used for genetic dissection of the variance in disease resistance to bacterial strain P. syringae pv maculicola ES4326 (ES4326) and DC3000, respectively. QTL analyses of the levels of bacterial growth continuously distributed across these two RILs showed that genetic variation in Aa-0 and Gie-0 at a locus on chromosome III, designated as Qpm3.1, preferentially restricted the growth of bacterial strain ES4326. Further screen for the genetic factor in ES4326 that limited bacterial growth on these Arabidopsis accessions successfully identified the effector gene hopW1-1. These findings highlight the role of pathogen effectors in quantitative natural variation in plant resistance to bacterial infection. The possible mechanism is also discussed on how Qpm3.1-hopW1-1 interaction led to enhanced resistance.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis accessions Aa-0, Gie-0, and Col-0 were used in this study. To construct RILs, F2 plants derived from Aa-0 × Col-0 and Gie-0 × Col-0 crosses with Col-0 as the female parent were self-fertilized and taken to F9 generation by single-seed descent method. Seeds of F9 plants were collected for subsequent disease assays. Heterogeneous inbred families (HIFs) were obtained by self-crossing the F6-RIL lines heterogeneous at markers cosegregating with Qpm3.1 locus, 1.3Mb and NGA172, in Col-Aa and Col-Gie populations, respectively. Arabidopsis seeds were incubated in 0.1% of agarose at 4°C for 2 days to synchronize germination. Seedlings were grown in a growth room under a 9-h photoperiod at 23°C.
Bacterial Strains and Disease Assay
The wild type and luxCDABE–tagged P. syringae strains ES4326-lux and DC3000-lux were grown on King’s B plates containing kanamycin (25 μg ml-1) and rifampicin (25 μg ml-1) as described (Fan et al., 2008). To inoculate Arabidopsis, bacterial suspensions prepared from overnight grown plates were diluted to levels as indicated and pressure-infiltrated with a needleless syringe into freshly expanded leaves of five-week-old Arabidopsis plants. To get a larger difference in bacterial growth between Col-0 and Aa-0, plants were covered after the inoculation, and for the same reason, plants were not covered when Col-0 and Gie-0 accessions were analyzed for variation in bacterial growth. To perform phenotypic analyses efficiently, bioluminescence assay was used to determine the bacterial growth on RILs (Fan et al., 2008). Briefly, leaf discs of 5 mm in diameter were excised from the inoculated leaves and put into 96-well plates to measure luminescence with the MP180 detector (Beijing Yikang Baifang Co., Ltd.). For each line, at least three individual plants were inoculated and four disks of each plant were measured. Colony counting method was also used to analyze bacterial number in leaf disks (Whalen et al., 1991).
Construction of Linkage Maps and QTL Mapping
Genomic DNA was extracted from F9 seedlings of each RIL, and the resulting DNA samples were amplified with SSLP and dCAPS (Supplementary Tables S2, S3) primer pairs in 10 μl PCR reactions. PCR products (for SSLP markers) or enzyme-digested PCR products (for dCAPS markers) were resolved by agarose gel electrophoresis (3%, w/v). Genotype data of RILs were analyzed by QTL IciMapping software and the algorithm of nnTwoOpt was chosen to locate markers on the chromosomes (Meng et al., 2015). Levels of bioluminescence of inoculated leaf disks of a RIL plant were subjected to logarithmic transformation and then an average of the resulting data was taken as the value for the disease phenotype of the line. Phenotypes of all the RILs were analyzed with the same software and the method of ICIM-Add was used to calculate the additive effect, while the method of ICIM-EPI was used to check the possible interactions among the QTLs. The threshold for statistical significance (95% confidence interval) of LOD was calculated by permutation test (n = 1000). The amount of heritability (Hˆ2) was calculated by ANOVA in QTL IciMapping software.
Construction of Cosmid Genomic Library of P. syringae pv. maculicola ES4326
The cosmid vector pLAFR5 (Keen et al., 1988) was digested with BamHI followed by dephosphorylation and ScaI digestion. Meanwhile, size-fractionated (18-27 Kb) genomic DNA fragments of ES4326 were generated via partial digestion with Sau3AI and retrieved from 0.7% agarose gel as previously described (Zhen and Swank, 1993). The DNA fragments were then ligated into the BamHI site of pLAFR5 and packaged into lambda phage using the MaxPlaxTM Lambda Packaging Extracts (Epicentre, Wisconsin, USA) according to manufacturer’s protocol. After infection of E. coli DH5α strain with the packaged products, bacterial cells were plated on King’s B agar medium supplemented with tetracycline (10 μg ml-1) and incubated overnight at 37°C. Colonies were picked and stored at –70°C to build up the library. The cosmid clones in DH5α were subsequently mobilized into the DC3000-lux strain by triparental mating with the help of plasmid pRK2013 using 96-well plates. The exconjugants were recovered on King’s B agar plates supplemented with tetracycline (5 μg ml-1), kanamycin (15 μg ml-1), and rifampicin (25 μg ml-1) after incubation for 3–5 days at 28°C.
Library Screening for Cosmids that Reduce Virulence of DC3000-Lux Specifically on Aa-0 and Cloning of the hopW1-1
The DC3000-lux clones bearing the ES4326 genomic cosmid library were used to inoculate plants of Arabidopsis accessions Aa-0 and Col-0. By using luminescence assay, clones growing poorly on Aa-0 but normally on Col-0 plants, compared with the growth of DC3000-lux, were selected and saved for further analysis. Cosmids of the selected clones were extracted and transformed into E. coli for propagation. The resulting cosmids were isolated and sequenced with M13 primers to obtain the sequences of insert ends.
The hopW1-1 gene with native promoter was cloned through Gateway recombinational technology. A pair of primers 5′-AAAAAGCAGGCTCAAAGGTTGGAGTCTCCGAGAAAC-3′ and 5′- AGAAAGCTGGGTCGATGCAGACCTGGCGTATCATAT-3′ were used to amplify the genomic DNA of ES4326. The PCR product was subsequently amplified with primers that reconstitute the complete attB sites and cloned into a modified pME6031 vector compatible with Gateway cloning, which can be stably maintained in DC3000 (Heeb et al., 2000).
Results
Natural Variation in Disease Resistance of Arabidopsis Accessions Against Diverse P. syringae Pathovars
Our and others’ studies have documented substantial variation in disease resistance to P. syringae infection among Arabidopsis natural populations (Kover and Schaal, 2002; Perchepied et al., 2006; Fan et al., 2008; Hossain and Sultana, 2015). Based on our previous work (Fan et al., 2008), we used two Arabidopsis accessions, Aa-0 and Gie-0, in combination with the reference accession Col-0, to further investigate the genetic and molecular basis of this variation. We first characterized the disease outcome of the interactions between these accessions and two bacterial strains. Results showed that 48 hours after inoculation of the plants with P. syringae strains DC3000-lux and ES4326-lux, leaf tissues of Col-0 plants developed clear disease symptoms for both strains, whereas Aa-0 plants displayed only weak chlorosis closely around injection sites for ES4326-lux and hardly any visible symptom for DC3000-lux (Figure 1A). Consistent with the symptom observed, bacterial counts of ES4326-lux and DC3000-lux in Aa-0 leaf tissue distant to the injection sites were about three and two orders of magnitude lower than that in Col-0 leaves, respectively (Figures 1B,C), indicating that Aa-0 plants were significantly more resistant to both strains than Col-0 plants.
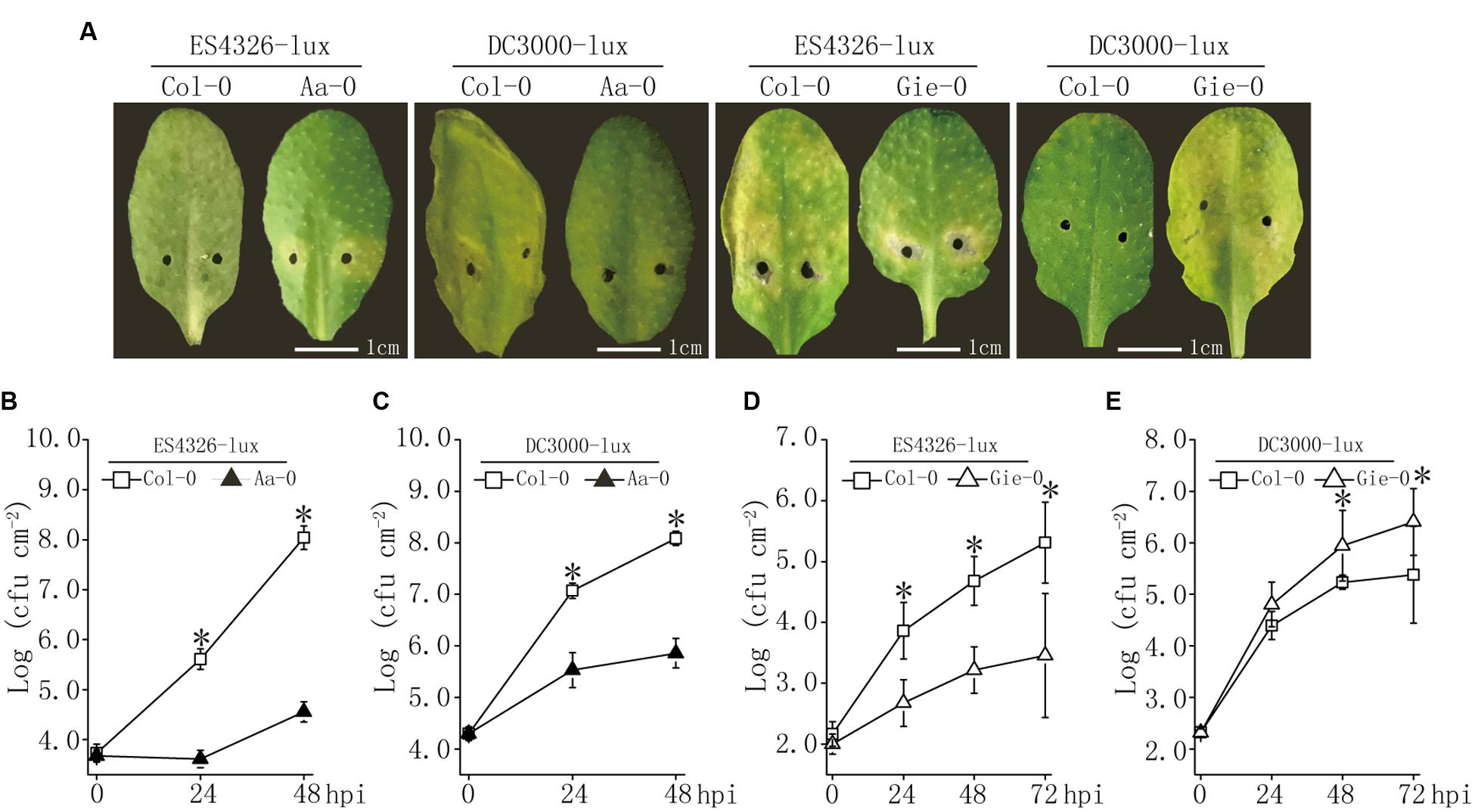
FIGURE 1. Variation in disease resistance to Pseudomonas syringae among Arabidopsis accessions Aa-0, Gie-0, and Col-0. (A) Disease symptoms on leaves infected with lux-tagged P. syringae pathovars maculicola ES4326 (ES4326-lux) or tomato DC3000 (DC3000-lux). (B–E) Bacterial growth in planta after inoculation with ES4326-lux or DC3000-lux. Bacterial suspensions were pressure infiltrated into fully expanded leaves of 4∼5 week old Arabidopsis plants using a 1 ml needleless syringe. Bacterial inocula of optical density (OD600) at 0.003 and 0.0002 were used to compare the difference in disease levels between Col-0 and Aa-0, and between Col-0 and Gie-0 plants, respectively. After inoculation, Col-0/Aa-0 plants were covered to keep high relative humidity, whereas Col-0/Gie-0 plants were left uncovered. Pictures of inoculated leaves were taken 48 and 72 hours post inoculation (hpi) for Col-0/Aa-0 and Col-0/Gie-0 plants, respectively. Asterisks indicate significant differences between the genotypes (Student’s t-test, P < 0.05). Data shown are means ± SD (n = 8). Experiments were repeated three times with similar results.
Similar to accession Aa-0, Gie-0 plants displayed attenuated symptom to ES4326-lux infection and supported two orders of magnitude lower bacterial growth than Col-0; however, for DC3000-lux inoculation, the disease symptom was enhanced and bacterial counts were 10-fold higher in Gie-0 than that in Col-0 plants (Figures 1A,D,E). These observations demonstrated that the natural variation in disease resistance was not only associated with Arabidopsis accessions but also influenced by the invading bacterial strains.
Natural Variation in Disease Resistance of Arabidopsis to Bacterial Infection was Quantitatively Determined
To further investigate genetics underlying the variation in disease resistance between these Arabidopsis accessions, we constructed two populations of RILs derived from Col-0 × Aa-0 and Col-0 × Gie-0 crosses. Ninety-six individual lines of each F9 population were inoculated with ES4326-lux or DC3000-lux, and bioluminescence assay of bacterial growth revealed continuous distribution between the most and the least susceptible lines in the two RIL populations (Figure 2, Supplementary Figure S1 and Table S1), which strongly indicated that variation in disease resistance was quantitative and likely the result of multigene-effect. Transgressive segregation could also be observed among the two tested RIL populations (Figure 2), suggesting that parents of each RIL population may carry genes associated with quantitative disease resistance at different genetic loci.
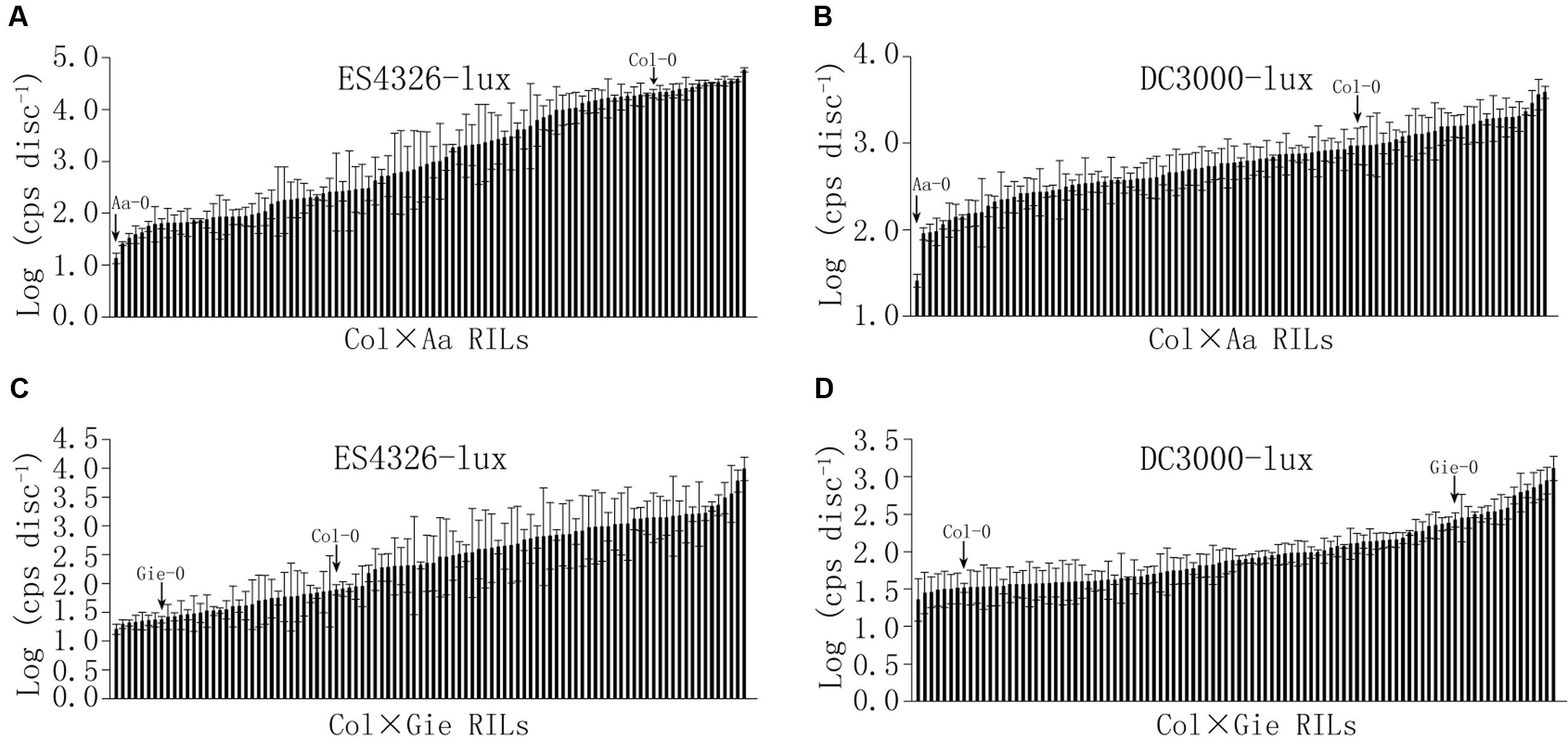
FIGURE 2. Continuous distribution of sizes of bacterial population among Col × Aa and Col × Gie RILs. Bacterial inocula of optical density (OD600) at 0.003 and 0.0002 were used for Col × Aa RILs and Col × Gie RILs, respectively. Leaf discs from four leaves per line were taken to measure luminescence (photon counts per second) at 48 hpi for Col × Aa RILs (A,B) and 72 hpi for Col × Gie RILs (C,D). Bacterial titers of parents are indicated by arrows. The experiment was repeated three times and the average levels of luminescence of each line from three experiments are shown. Error bars denote standard deviations (n = 3).
QTL Mapping Revealed Genetic Loci with Large Effect on Arabidopsis Resistance to Bacterial Infection
Simple sequence length polymorphism (SSLP) markers and derived cleaved amplified polymorphic sequence (dCAPS) markers (Supplementary Tables S2, S3) were used to analyze the genotype of each line of the two RIL populations to construct linkage maps used for QTL analysis (Supplementary Figure S2). The QTL analysis uncovered several Arabidopsis genomic regions dominating the variation in disease resistance to ES4326-lux (QTL to P. syringae pv. maculicola, Qpm) and DC3000-lux (QTL to P. syringae pv. tomato, Qpt). The amount of heritability (Hˆ2) for DC3000-lux growth in Col × Aa and Col × Gie RILs was 0.76 and 0.88, respectively, whereas that for ES4326-lux growth in Col × Aa and Col × Gie RILs was 0.88 and 0.69, respectively. For DC3000-lux, a QTL on chromosome V that conferred nearly a quarter of the variance in resistance was identified in Col × Aa RILs, and another QTL at the top arm of chromosome I was revealed in Col × Gie RILs, accounting for 60% of the variance in resistance (Figures 3A,B and Table 1). Analysis of ES4326-lux infected RILs, however, revealed a region on chromosome III highly overlapped between both RILs, accounting for over 60% of the variance in resistance (Figures 3C,D and Table 1). In addition, a number of QTLs with smaller effect were also detected in the tested RIL and bacterial interactions (Table 1).
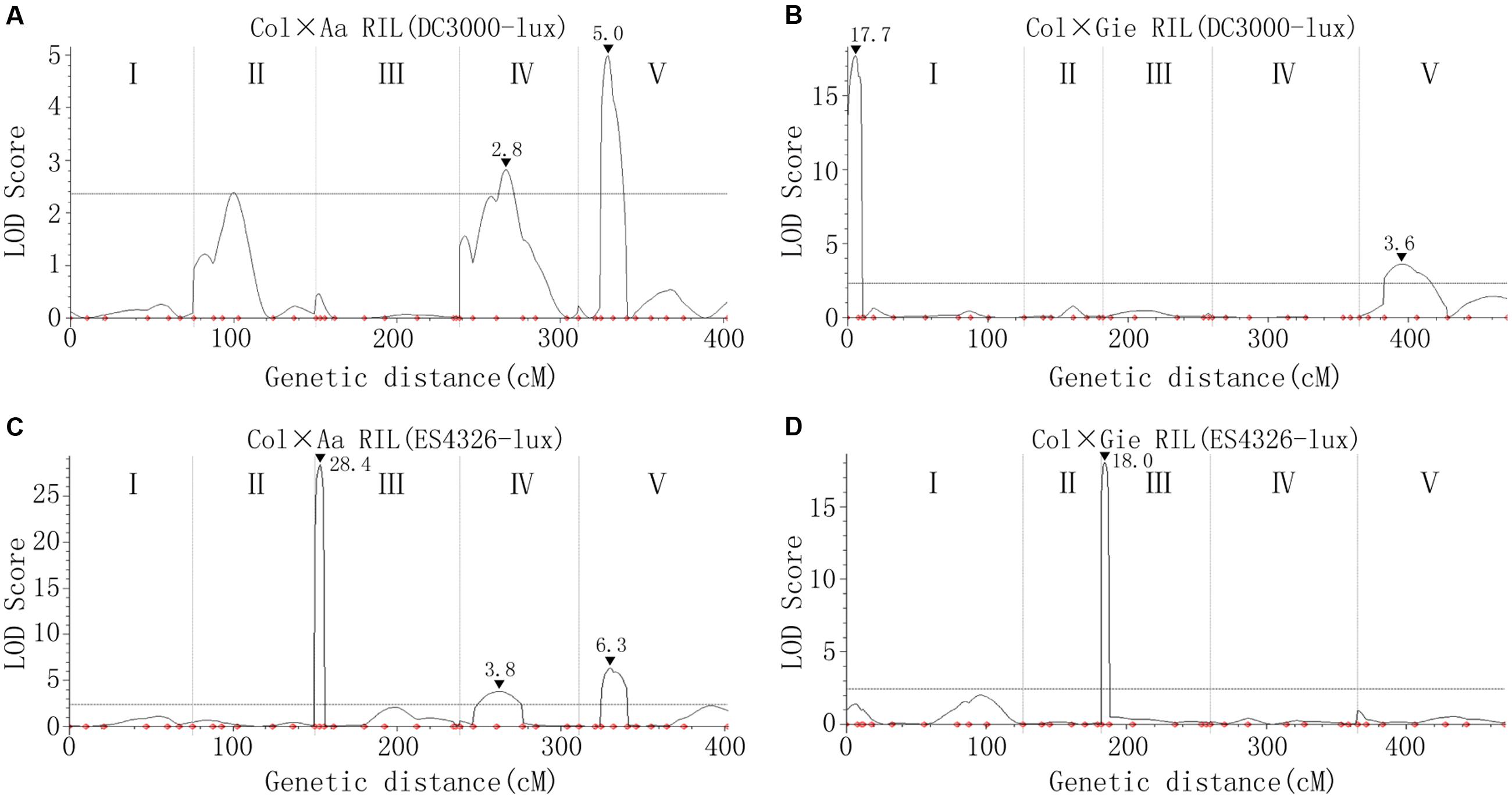
FIGURE 3. Arabidopsis quantitative trait loci (QTLs) controlling variation in disease resistance to P. syringae infection. (A,B) QTLs underlying the resistance variance to DC3000-lux in Col × Aa (A) and Col × Gie (B) RILs. (C,D) QTLs underlying the resistance variance to ES4326-lux in Col × Aa (C) and Col × Gie (D) RILs. QTL analyses are based on genotypic data and levels of bacterial luminescence of the RILs. The curved lines indicate likelihood statistics (scores of the likelihood of odds, LOD) for the positions of QTLs and the significance threshold at 95% confidence interval calculated by permutation tests (n = 1000) is plotted as horizontal lines. I–V denote five individual linkage groups. Positions of the markers used for analysis are indicated as small dots. Peaks of LOD scores are indicated by filled triangles along with the values.
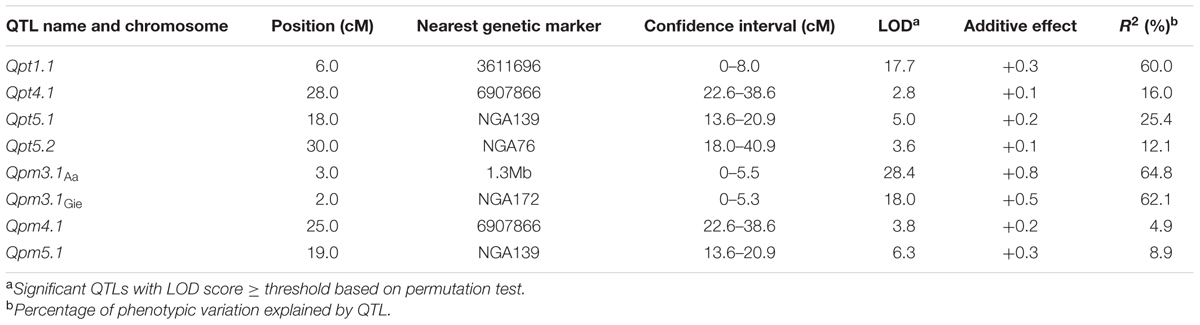
TABLE 1. Summary of Arabidopsis quantitative trait locus (QTL) conferring resistance to lux tagged Pseudomonas syringae pv. tomato DC3000 (Qpt) and pv. maculicola ES4326 (Qpm).
To test if the detected major QTLs on chromosome III (Qpm3.1Aa and Qpm3.1Gie) were truly conferring the variation in resistance to ES4326-lux, heterogeneous inbred families (HIFs) were obtained by selfing F6-RIL plants heterozygous at the QTL, and the resultant HIFs, ACHIF8 (from Col × Aa RILs) and GCHIF10 (from Col × Gie RILs), were subsequently analyzed for resistance to ES4326-lux. Results showed that plants with homozygous Aa-0 genotype at marker 1.3Mb or with Gie-0 genotype at marker NGA172 were significantly more resistant than those with homozygous Col-0 genotype (Figure 4 and Supplementary Figure S3), indicating that the variation in disease resistance was tightly linked to the genetic variation at this locus. We thus designate this highly overlapped region as Qpm3.1.
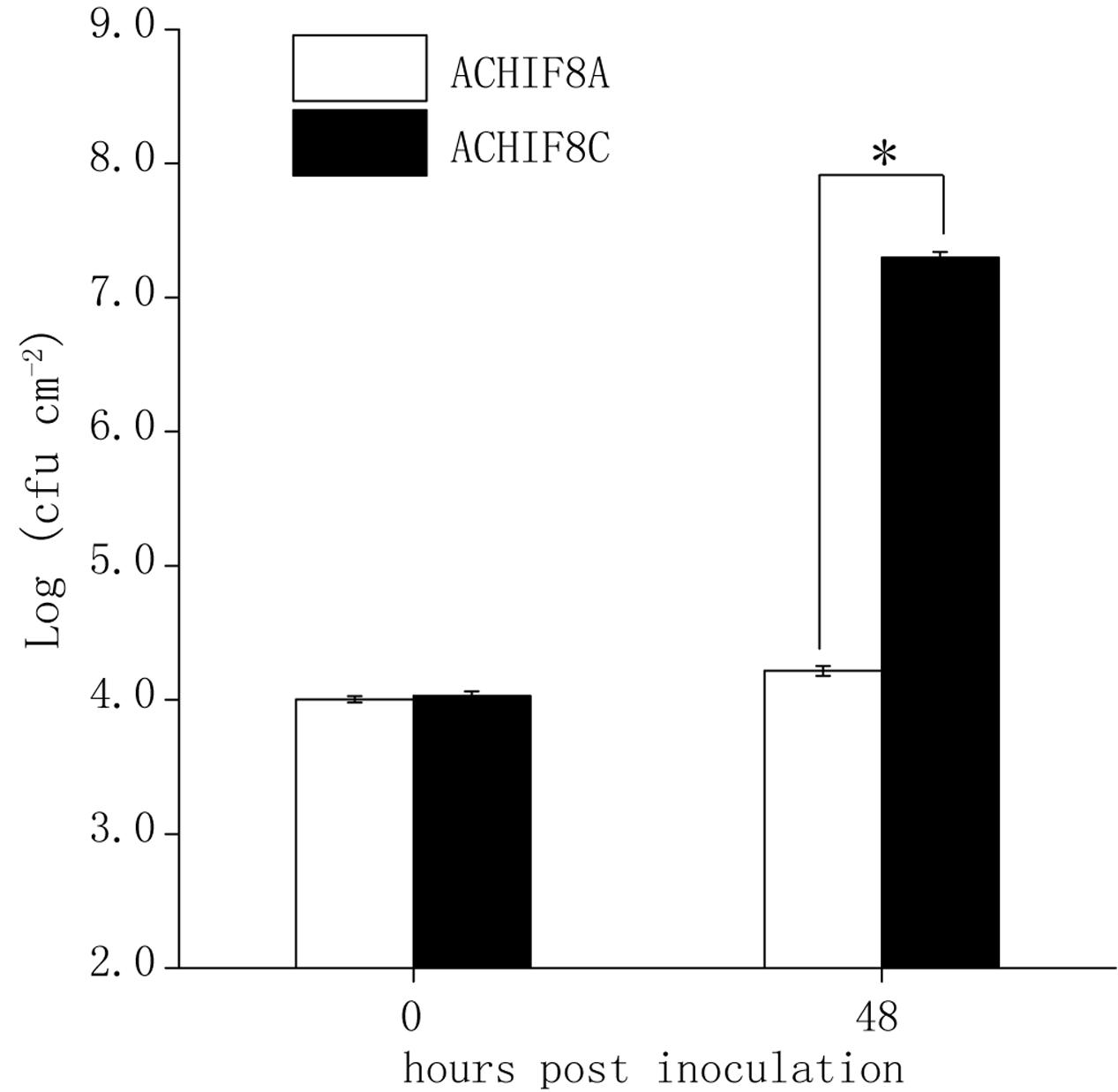
FIGURE 4. Validation of the major QTL Qpm3.1 in Aa-0 with the heterogeneous inbred family (HIF). Forty-eight individuals of a Col × Aa HIF (ACHIF8) were inoculated with ES4326-lux at OD600 = 0.003. The inoculated plants were covered to keep high relative humidity. Genotypes of individual plants were determined with the tightly associated marker 1.3 Mb and sizes of the bacterial population were compared between plants with homozygous Aa-0 (ACHIF8A) and plants with homozygous Col-0 (ACHIF8C) genotypes. Data shown are means ± SD (n = 8). The asterisk indicates a significant difference between treatments (Student’s t-test, P < 0.05). The experiment was repeated three times with similar results.
Identification of the Effector Gene hopW1-1 from Pathovar Maculicola that Reduced Bacterial Growth in Arabidopsis Accession Aa-0 and Gie-0
The above results showed that levels of ES4326-lux growth on Aa-0 and Gie-0 accessions were significantly lower compared to DC3000-lux (Figure 1) and that the prominent QTL Qpm3.1 appeared to confer resistance specifically to strain ES4326-lux and was ineffective when DC3000-lux was used for inoculation (Figure 3). Therefore, we hypothesized that certain genetic component in bacteria may also be involved in the quantitative disease resistance of Arabidopsis plants. To search for this putative genetic factor we constructed a cosmid library of ES4326 genomic DNA and mobilized the resultant library into DC3000-lux by conjugation to screen for clones impaired in virulence on Aa-0 but not on Col-0 plants. Out of nearly 500 clones screened, five cosmids were found to be capable of reducing the growth of DC3000-lux preferentially on Aa-0 plants. The resulting cosmids share a 7 kb region of the plasmid B of ES4326 (Figure 5A), in which an effector gene hopW1-1 has been described previously (Figure 5B) (Guttman et al., 2002; Lee et al., 2008). Subsequently, we cloned the hopW1-1 and found that plasmids harboring this gene significantly attenuated growth of DC3000-lux on Aa-0 and Gie-0, but not on Col-0 plants (Figure 5C and Supplementary Figure S4), suggesting that hopW1-1 may be a key player sufficient in conditioning disease resistance in Arabidopsis to ES4326.
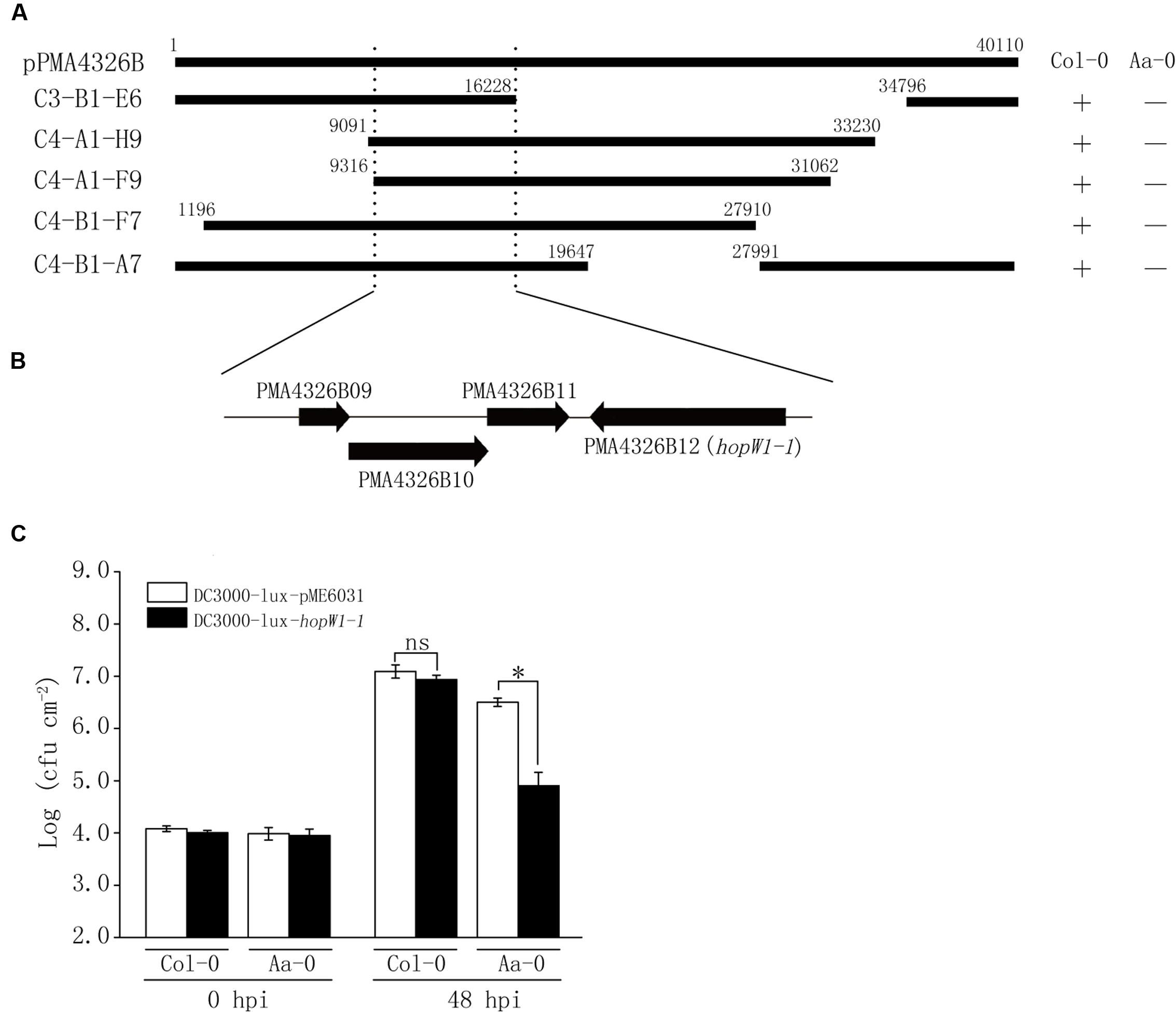
FIGURE 5. Identification from ES4326 of the effector gene hopW1-1 that selectively reduced the bacterial growth on Aa-0. (A) Cosmid clones capable of reducing bacterial growth on Aa-0 share a common region of pPMA4326B from ES4326. Top bar denotes the linearized pPMA4326B in the full length of 40,110 base pairs. Each cosmid is marked with numbers showing the positions of insert ends. “+” indicates that the growth of bacterial cosmid clone in planta was similar to that of empty vector control. “–” indicates that the growth of bacterial cosmid clone in planta was significantly reduced compared with vector control. (B) Annotated genes in the minimal region of pPMA4326B shared by all five cosmid clones. (C) The effector gene hopW1-1 selectively reduced the growth of DC3000-lux in Aa-0 but not Col-0 plants. Plants were pressure inoculated with bacteria at OD600 = 0.01. Bacterial growth was determined by counting colony forming unit (cfu) of three leaf disks from each plant and at least four plants were used in each treatment. Data shown are means ± SD (n = 12). The asterisk indicates a significant difference between treatments (Student’s t-test, P < 0.05). “ns” denotes no significant difference. This experiment was repeated three times with similar results.
Genetic Interaction between Qpm3.1 and hopW1-1 Underpinned Arabidopsis Quantitative Disease Resistance to Bacterial Infection
To further elucidate the genetic component in Arabidopsis involved in the interaction with hopW1-1, DC3000-lux transformed with hopW1-1 (DC3000-lux-hopW1-1) or empty vector (DC3000-lux-pME6031) were used to inoculate Col-Aa and Col-Gie RILs. Luminescence assay revealed continuous distribution of bacterial growth within each RIL population (Figures 6A,B, Supplementary Figure S5A, and Table S1), and QTL analysis uncovered prominent extra peaks at Qpm3.1 locus specifically associated with infections by strain DC3000-lux-hopW1-1 (compare Figure 6C with Figure 6D, and Figure 3B with Supplementary Figure S5B), indicating that the genetic interaction between Qpm3.1 and hopW1-1 may determine the resistance in Arabidopsis against bacterial infection. This speculation was further corroborated by observations that plants carrying Qpm3.1 in ACHIF8 family were significantly more resistant to strain DC3000-lux-hopW1-1, but not to DC3000-lux strain with the empty vector (Figure 6E). However, the Qpm3.1 mediated resistance was substantially compromised when higher bacterial inocula were used for infection (compare Figure 6E with Supplementary Figure S6).
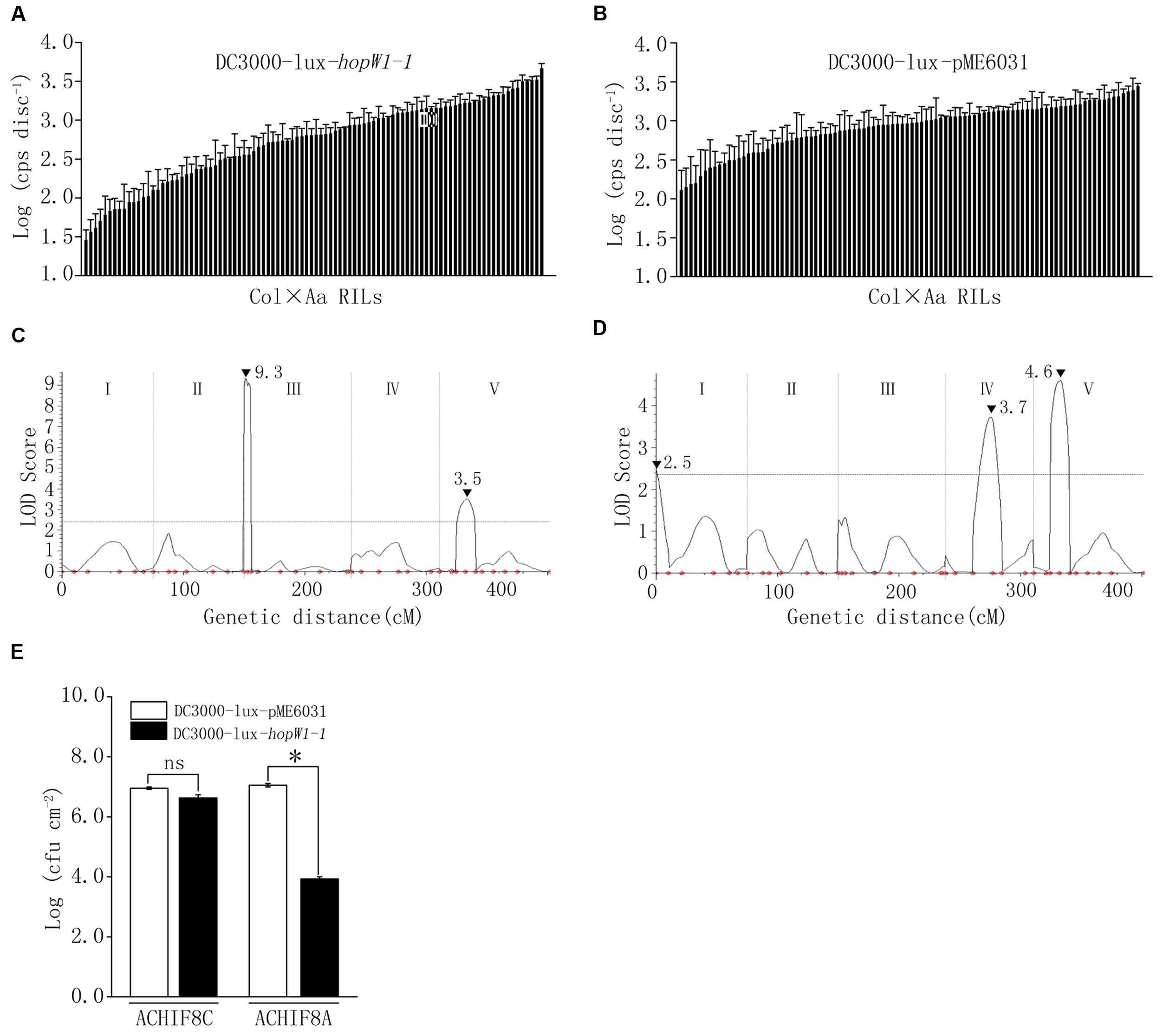
FIGURE 6. The interaction between Qpm3.1 and hopW1-1 contributed to the quantitative disease resistance. (A,B) Continuous distribution of luminescence levels of DC3000-lux-hopW1-1 (A) and DC3000-lux-pME6031 (B) in Col × Aa RILs. Leaves were pressure inoculated with the bacteria at OD600 = 0.003. Inoculated plants were covered and four leaf discs from each plant were sampled 48hpi for luminescence assay. Data shown are means of results of three repeated experiments. Error bars denote standard deviations (n = 3). (C,D) QTLs mediating quantitative resistance to DC3000-lux-hopW1-1 (C) and DC3000-lux-pME6031 (D) in Col × Aa RILs. Peak LOD scores are indicated by filled triangles along with their values. (E) Genetic interaction between Qpm3.1 and hopW1-1 restricted bacterial growth in planta. Individuals of HIF family ACHIF8 were genotyped with marker 1.3 Mb tightly associated with locus Qpm3.1. Plants with Aa-0 genotype (ACHIF8A) and plants with Col-0 genotype (ACHIF8C) were pressure infiltrated with DC3000-lux-hopW1-1 and DC3000-lux-pME6031 at OD600 = 0.0005. The inoculated plants were covered to keep high relative humidity. Levels of the bacterial population were determined at 72 hpi. Data shown are means ± SD (n = 8). The asterisk indicates a significant difference between treatments (Student’s t-test, P < 0.05). “ns” denotes no significant difference. This experiment was repeated three times with similar results.
The Epistatic Effect of Loci Qpm3.1 and Qpt1.1 on Variation in Quantitative Disease Resistance was hopW1-1 Dependent
In Col × Gie RILs major QTLs Qpm3.1 and Qpt1.1 were detected to condition variation in resistance to ES4326 and DC3000, respectively (Figures 3B,D). Interestingly, however, the peak of Qpt1.1 was absent and only Qpm3.1 was detected when DC3000-lux-hopW1-1 was used to inoculate the RILs (Supplementary Figure S5B), which strongly indicated the existence of epistasis between the two QTLs. We compared levels of bacterial growth in planta and found that on RILs with the Qpm3.1 genotype the variation in population size was apparently reduced when hopW1-1 was present in DC3000-lux (Figure 7A). Subsequently, Col × Gie RILs were subgrouped into lines with or without Qpm3.1 and used for QTL analysis of its impact on Qpt1.1. As shown in Figure 7, on lines without Qpm3.1 (Col-0 genotype at the locus) the peak of Qpt1.1 was detectable regardless of the presence of hopW1-1 in DC3000-lux (Figures 7B,C); however, on lines with Qpm3.1 the peak of Qpt1.1 was only detectable when hopW1-1 was absent in bacteria (Figure 7D), and the effector gene completely abolished the Qpt1.1-mediated variation in bacterial levels of DC3000-lux (Figure 7E). Thus, the effect of Qpm3.1 is epistatic to Qpt1.1 and the epistasis requires bacterial hopW1-1.
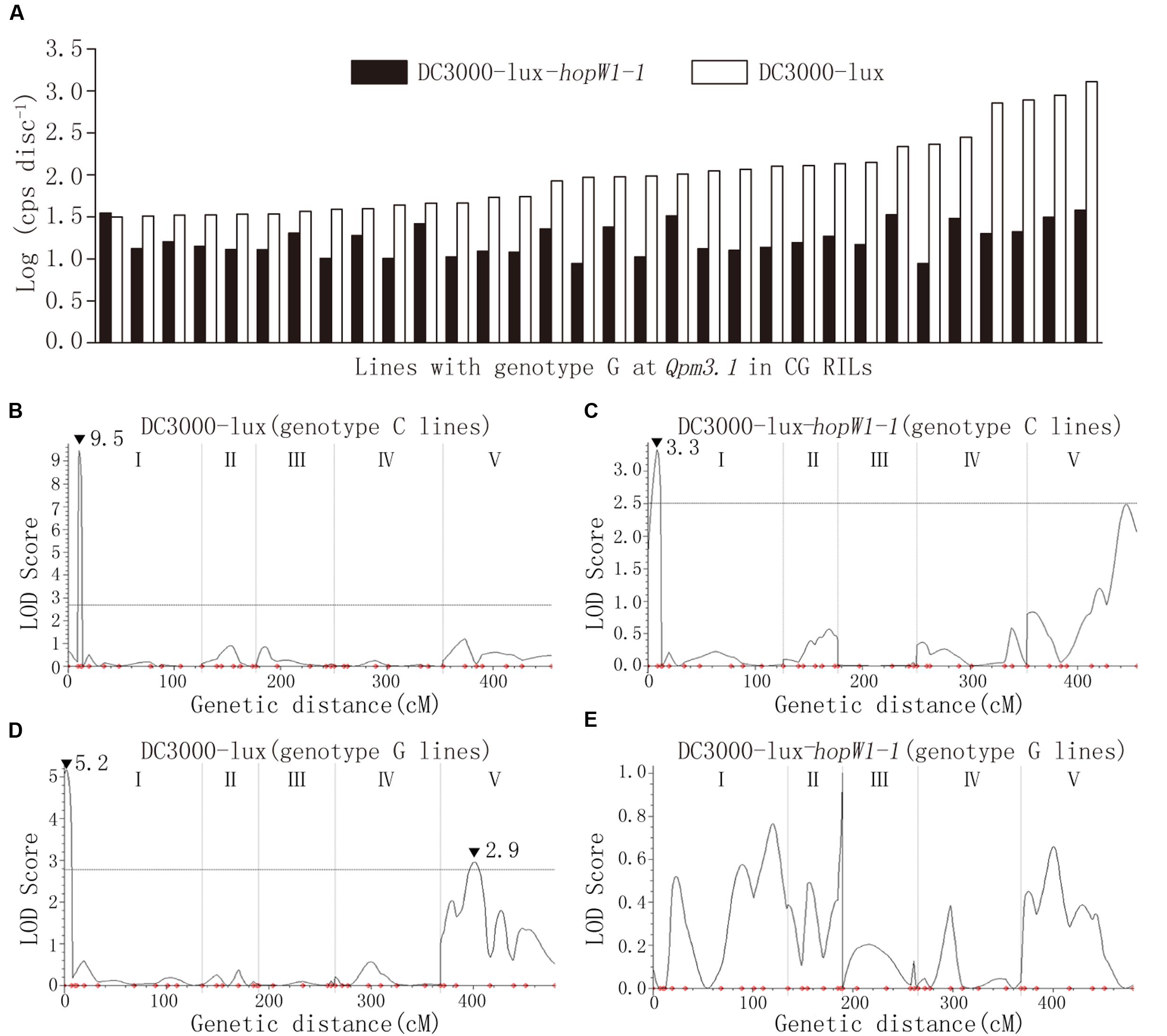
FIGURE 7. The epistatic effect between Qpm3.1 and Qpt1.1. (A) Comparison of the variation in levels of the bacterial population between DC3000-lux strains with or without the effector gene hopW1-1 on Col × Gie RILs carrying Qpm3.1. (B–E) QTL analysis of resistance variation in subgroups of Col × Gie RILs. Col × Gie RILs were inoculated with DC3000-lux strains with or without hopW1-1. Plant lines with genotypes homozygous for Col-0 (B,C) or Gie-0 (D,E) at markers NGA172 and NGA76 juxtaposing Qpm3.1 were subgrouped and used for QTL analysis. Peak LOD scores are indicated by filled triangles along with their values.
Discussion
In this study, we identified several QTLs mediating quantitative disease resistance to the lux-tagged DC3000 and ES4326 strains, respectively (Figure 3 and Table 1). The patterns of QTL distribution apparently differed in association with the strains used for inoculation. Among Qpm loci, the prominent effect of Qpm3.1 was detected from both Col × Aa and Col × Gie RILs challenged with ES4326 but was absent when inoculated with DC3000. This locus also coincides with the major QTL QRps.JIC-3.1 that has been identified during infection of Col × Ler RILs with the strain ES4326 (Rant et al., 2013), indicating that these two QTLs may be the same and potentially strain-specific and broadly exist in Arabidopsis accessions.
In contrast, the distribution of Qpt loci seemed largely distinct between the two RILs although marginal overlap of genetic region exists between Qpt5.1 and Qpt5.2 (Figure 3 and Table 1), suggesting that the genetic basis dominating variance in resistance of Aa-0 and Gie-0 against virulent DC3000 differed substantially. The strain DC3000 has been used in other studies for dissection of natural variation in disease resistance in Arabidopsis, and multiple major QTLs have been uncovered to restrict the size of the bacterial population in planta. The genetic location of Qpt5.1, one of the large effect QTLs detected in this study, is close to that of PRP-Ps2, a major QTL identified with the population of Bayreuth × Shahdara RILs (Perchepied et al., 2006), but is distant from another major QTL previously identified at the bottom of chromosome V that explains most of the variation in resistance between accessions Columbia and San Feliu-2 (Kover and Cheverud, 2007). The location of Qpt1.1, another major QTL identified in this study determining the variation in susceptibility between Col-0 and Gie-0, is clearly distinct from the QTL previously identified on the same chromosome (Fan et al., 2008). These studies demonstrate the complexity of natural variation in Arabidopsis resistance to strain DC3000. We expect that more major QTLs could be uncovered when other accessions or alternative inoculation methods are used for analysis.
Notably, we also found that genetic regions closely overlap between Qpt4.1 and Qpm4.1, and between Qpt5.1 and Qpm5.1 as well (Table 1), implying the existence of QTLs effective against both bacterial strains. However, it remains to be elucidated whether the same genes in the overlapped QTLs mediate disease resistance to both strains. The diversity of QTLs detected in disease resistance, the sensitivity of Qpm3.1 to higher bacterial inocula (Supplementary Figure S6), and the epistasis between Qpm3.1 and Qpt1.1 (Figure 7) revealed in this study collectively indicate that the genetic architecture of the quantitative disease resistance is highly complex and further molecular dissection is necessary to unravel the underlying mechanisms.
In Arabidopsis, the molecular basis of the quantitative natural variation in disease resistance to bacterial infection is still poorly understood since few of the identified QTLs has been cloned. It has been proposed that PTI, ETI, and ETS collectively determine the plant resistance to a virulent pathogen (Jones and Dangl, 2006). Hence, the genetic variation modulating the outcome of any of these processes may lead to the quantitative variation in disease resistance. It has been shown that natural allelic variation at the locus ACCELERATED CELL DEATH 6 causes marked differences in accumulation of SA and in resistance to infections by a panel of phylogenetically distinct pathogens including P. syringae pv tomato DC3000 (Todesco et al., 2009). Other studies have shown that disruption of genes required for ETI compromises resistance in Arabidopsis against both virulent and non-adapted of P. syringae strains, indicating that ETI protects the plant during these seemingly distinct interactions (Zhang et al., 2010). In addition, the genetic variation at locus FLS2, which encodes the flagellin receptor that mediates PTI responses, has also been shown to account for the variation in Arabidopsis resistance to non-adapted P. syringae pv phaseolicola (Forsyth et al., 2010). A more recent study discovered that natural variation in Arabidopsis responses to the type III effector HopAM1 secreted by disarmed DC3000 is quantitatively determined although the HopAM1-induced cell death and chlorosis do not affect bacterial growth in planta (Iakovidis et al., 2016). Our study clearly demonstrated that the locus Qpm3.1 was effective to restrict the growth of bacterial strains carrying the effector gene hopW1-1, a reminiscent of specificity commonly exhibited in R-gene mediated resistance. Several genes around the Qpm3.1 locus are indeed annotated to encode R-like proteins, including one TIR-NBS-LRR gene (At3g04220), one TIR-NB gene (At3g04210); and four receptor-like kinase/protein genes (At3g05360, At3g05370, At3g05650, and At3g05660); several major R-genes have been shown to be responsible for race-specific QTLs conditioning quantitative resistance to Xanthomonas bacteriain Arabidopsis and rice plants (Li et al., 1999; Debieu et al., 2015). However, the narrow spectrum of Qpm3.1 does not necessarily mean that a “defeated” or “weak” R-gene is responsible for the observed quantitative resistance (see discussion below) as diverse molecular mechanisms other than R-avr interaction have been found to underpin the quantitative resistance in multiple pathosystems (Poland et al., 2009; French et al., 2016). In fact, apart from the R-like genes mentioned above, 12 more genes are annotated to confer defense responses among the predicted 384 genes within the Qpm3.1 locus. It is interesting to observe that higher inocula (OD600 0.03) compromised Qpm3.1s mediated resistance (Supplementary Figure S6), and the exact underlying mechanism is still unknown. A possible explanation is that an increased amount of effectors suppressing plant defense were delivered into host cells by the type three secretion system at a higher inoculum (Alfano and Collmer, 2004), which reduced directly or indirectly the effect of Qpm3.1 and hopW1-1 interaction on the restriction of bacterial growth. This finding also implies that the interaction may be effective at the early stage of infection when the bacterial load is still low. Further cloning of the allele responsible for Qpm3.1 will help elucidate the phenomenon.
The discovery of effector-associated QTLs highlights the role of bacterial effectors in delineating natural variation in plant quantitative disease resistance. Since many agriculturally important bacterial pathogens are sequenced and effector coding genes can be efficiently predicted (da Silva et al., 2002; Guttman et al., 2002; Salanoubat et al., 2002), effector-based approaches can be thus designed to scan Arabidopsis accessions for common or rare genetic variations with large effect on pathogenesis of bacterial disease, thereby providing new insights on the fundamental bacterial effector biology. The study on Qpm3.1 also indicated that RILs and HIFs are particularly useful for QTL identification and mendelization, which will markedly facilitate subsequent positional cloning of the responsible gene.
The effector gene hopW1-1, first identified from strain ES4326, is narrowly distributed in a few P. syringae strains (Guttman et al., 2002). This effector disrupts actin cytoskeleton and inhibits endocytosis to promote bacterial virulence when delivered into plant cells (Kang et al., 2014), but elicits resistance responses and limits bacterial growth on some Arabidopsis accessions including Ws (Lee et al., 2008). Infection of Ws plants with the strain DC3000 carrying hopW1-1 specifically triggers an enhanced accumulation of SA at the early stage of interaction; a number of genes involved in SA pathways or encoding the HopW1-interacting proteins are required for the effector-triggered resistance in Ws; two loci, on chromosome III and V, have been identified using Col × Ws F2 population (Lee et al., 2008). Nevertheless, none of these described genes, loci locate at the identified Qpm3.1 locus, and no gene product was annotated in the region as actin-associated linking with the proposed function of HopW1-1; therefore, additional gene(s) conditions natural variation in Arabidopsis response to HopW1-1. Interestingly, by measuring the growth of DC3000 in planta, Dobon et al. have identified four QTLs underpinning natural variation in exogenous SA-induced resistance across Arabidopsis accessions, among which SAQ3, detected in Col-0xLer RILs, locates closely to Qpm3.1 (Dobón et al., 2011). Further investigation on the possible linkage between these two loci may shed new light on the role of SA-dependent pathways in effector-triggered immunity against plant bacterial pathogens.
Author Contributions
JF and Y-LP designed the research; QL, W-WL, and K-DP performed the research, collected and analyzed the data; QL, JF, and Y-LP wrote the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Liqun Zhang for providing the cosmid vector pLAFR5. This research was supported partly by Chinese University Scientific Fund (2012RC007) to JF, the Program for Changjiang Scholars and Innovative Research Team Project IRT1042, the 111 Project from the Ministry of Education, China, to Y-LP, and by a special fund for collaboration from the State Key Laboratory of Agrobiotechnology to Y-LP and JF.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00695/full#supplementary-material
References
Albert, I., Böhm, H., Albert, M., Feiler, C. E., Imkampe, J., Wallmeroth, N., et al. (2015). An RLP23–SOBIR1–BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1:15140. doi: 10.1038/nplants.2015.140
Alfano, J. R., and Collmer, A. (2004). TYPE III SECRETION SYSTEM EFFECTOR PROTEINS: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. doi: 10.1146/annurev.phyto.42.040103.110731
Cook, D. E., Lee, T. G., Guo, X., Melito, S., Wang, K., Bayless, A. M., et al. (2012). Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338, 1206–1209. doi: 10.1126/science.1228746
da Silva, A. C. R., Ferro, J. A., Reinach, F. C., Farah, C. S., Furlan, L. R., Quaggio, R. B., et al. (2002). Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417, 459–463. doi: 10.1038/417459a
Dangl, J. L., and Jones, J. D. G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Debieu, M., Huard-Chauveau, C., Genissel, A., Roux, F., and Roby, D. (2015). Quantitative disease resistance to the bacterial pathogen Xanthomonas campestris involves an Arabidopsis immune receptor pair and a gene of unknown function. Mol. Plant Pathol. 17, 510–520. doi: 10.1111/mpp.12298
Dobón, A., Canet, J. V., Perales, L., and Tornero, P. (2011). Quantitative genetic analysis of salicylic acid perception in Arabidopsis. Planta 234, 671–684. doi: 10.1007/s00425-011-1436-6
Fan, J., Crooks, C., and Lamb, C. (2008). High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 53, 393–399. doi: 10.1111/j.1365-313X.2007.03303.x
Forsyth, A., Mansfield, J. W., Grabov, N., de Torres, M., Sinapidou, E., and Grant, M. R. (2010). Genetic dissection of basal resistance to Pseudomonas syringae pv. phaseolicola in accessions of Arabidopsis. Mol. Plant Microbe Interact. 23, 1545–1552. doi: 10.1094/mpmi-02-10-0047
French, E., Kim, B.-S., and Iyer-Pascuzzi, A. S. (2016). Mechanisms of quantitative disease resistance in plants. Semin. Cell Dev. Biol. 56, 201–208. doi: 10.1016/j.semcdb.2016.05.015
Fu, D., Uauy, C., Distelfeld, A., Blechl, A., Epstein, L., Chen, X., et al. (2009). A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323, 1357–1360. doi: 10.1126/science.1166289
Fukuoka, S., Saka, N., Koga, H., Ono, K., Shimizu, T., Ebana, K., et al. (2009). Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325, 998–1001. doi: 10.1126/science.1175550
Gómez-Gómez, L., and Boller, T. (2000). FLS2: an LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell. 5, 1003–1011. doi: 10.1016/S1097-2765(00)80265-8
Guttman, D. S., Vinatzer, B. A., Sarkar, S. F., Ranall, M. V., Kettler, G., and Greenberg, J. T. (2002). A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295, 1722–1726. doi: 10.1126/science.295.5560.1722
Hayashi, N., Inoue, H., Kato, T., Funao, T., Shirota, M., Shimizu, T., et al. (2010). Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 64, 498–510. doi: 10.1111/j.1365-313X.2010.04348.x
Heeb, S., Itoh, Y., Nishijyo, T., Schnider, U., Keel, C., Wade, J., et al. (2000). Small, stable shuttle vectors based on the minimal pVS1 replicon for use in Gram-negative, plant-associated bacteria. Mol. Plant Microbe Interact. 13, 232–237. doi: 10.1094/mpmi.2000.13.2.232
Hossain, M. M., and Sultana, F. (2015). Genetic variation for induced and basal resistance against leaf pathogen Pseudomonas syringae pv. tomato DC3000 among Arabidopsis thaliana accessions. SpringerPlus 4:296. doi: 10.1186/s40064-015-1070-z
Hurni, S., Scheuermann, D., Krattinger, S. G., Kessel, B., Wicker, T., Herren, G., et al. (2015). The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad. Sci. U.S.A. 112, 8780–8785. doi: 10.1073/pnas.1502522112
Iakovidis, M., Teixeira, P. J. P. L., Exposito- Alonso, M., Cowper, M. G., Law, T. F., Liu, Q., et al. (2016). Effector triggered immune response in Arabidopsis thaliana is a quantitative trait. Genetics 204, 337–353. doi: 10.1534/genetics.116.190678
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kang, Y., Jelenska, J., Cecchini, N. M., Li, Y., Lee, M. W., Kovar, D. R., et al. (2014). HopW1 from Pseudomonas syringae disrupts the actin cytoskeleton to promote virulence in Arabidopsis. PLoS Pathog. 10:e1004232. doi: 10.1371/journal.ppat.1004232
Keen, N. T., Tamaki, S., Kobayashi, D., and Trollinger, D. (1988). Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70, 191–197. doi: 10.1016/0378-1119(88)90117-5
Kover, P. X., and Cheverud, J. (2007). The genetic basis of quantitative variation in susceptibility of Arabidopsis thaliana to Pseudomonas syringae (Pst DC3000): evidence for a new genetic factor of large effect. New Phytol. 174, 172–181. doi: 10.1111/j.1469-8137.2007.01985.x
Kover, P. X., and Schaal, B. A. (2002). Genetic variation for disease resistance and tolerance among Arabidopsis thaliana accessions. Proc. Natl. Acad. Sci. U.S.A. 99, 11270–11274. doi: 10.1073/pnas.102288999
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Lee, M. W., Jelenska, J., and Greenberg, J. T. (2008). Arabidopsis proteins important for modulating defense responses to Pseudomonas syringae that secrete HopW1-1. Plant J. 54, 452–465. doi: 10.1111/j.1365-313X.2008.03439.x
Li, Z.-K., Luo, L., Mei, H., Paterson, A., Zhao, X., Zhong, D., et al. (1999). A “defeated” rice resistance gene acts as a QTL against a virulent strain of Xanthomonas oryzae pv. oryzae. Mol. Gen. Genet. 261, 58–63. doi: 10.1007/s004380050941
Lindeberg, M., Cartinhour, S., Myers, C. R., Schechter, L. M., Schneider, D. J., and Collmer, A. (2006). Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol. Plant Microbe Interact. 19, 1151–1158. doi: 10.1094/mpmi-19-1151
Meng, L., Li, H., Zhang, L., and Wang, J. (2015). QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. doi: 10.1016/j.cj.2015.01.001
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Perchepied, L., Kroj, T., Tronchet, M., Loudet, O., and Roby, D. (2006). Natural variation in partial resistance to Pseudomonas syringae is controlled by two major QTLs in Arabidopsis thaliana. PLoS ONE 1:e123. doi: 10.1371/journal.pone.0000123
Pieterse, C. M. J., Does, D. V. D., Zamioudis, C., Leon-Reyes, A., and Wees, S. C. M. V. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Poland, J. A., Balint-Kurti, P. J., Wisser, R. J., Pratt, R. C., and Nelson, R. J. (2009). Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 14, 21–29. doi: 10.1016/j.tplants.2008.10.006
Rant, J. C., Arraiano, L. S., Chabannes, M., and Brown, J. K. (2013). Quantitative trait loci for partial resistance to Pseudomonas syringae pv. maculicola in Arabidopsis thaliana. Mol. Plant Pathol. 14, 828–837. doi: 10.1111/mpp.12043
Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415, 497–502. doi: 10.1038/415497a
St. Clair, D. A. (2010). Quantitative disease resistance and quantitative resistance loci in breeding. Annu. Rev. Phytopathol. 48, 247–268. doi: 10.1146/annurev-phyto-080508-081904
Todesco, M., Balasubramanian, S., Hu, T. T., Traw, M. B., Horton, M., Epple, P., et al. (2009). Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465, 632–636. doi: 10.1038/nature09083
Toruno, T. Y., Stergiopoulos, I., and Coaker, G. (2016). Plant pathogen effectors: cellular probes interfering with plant defenses in a spatial and temporal manner. Annu. Rev. Phytopathol. 54, 419–441. doi: 10.1146/annurev-phyto-080615-100204
Whalen, M. C., Innes, R. W., Bent, A. F., and Staskawicz, B. J. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell Online 3, 49–59. doi: 10.1105/tpc.3.1.49
Zhang, J., Lu, H. B., Li, X. Y., Li, Y., Cui, H. T., Wen, C. K., et al. (2010). Effector-triggered and pathogen-associated molecular pattern-triggered immunity differentially contribute to basal resistance to Pseudomonas syringae. Mol. Plant Microbe Interact. 23, 940–948. doi: 10.1094/mpmi-23-7-0940
Zhen, L., and Swank, R. T. (1993). A simple and high yield method for recovering DNA from agarose gels. Biotechniques 14, 894–898.
Zipfel, C., Kunze, G., Chinchilla, D., Caniard, A., Jones, J. D. G., Boller, T., et al. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760. doi: 10.1016/j.cell.2006.03.037
Keywords: Arabidopsis, hopW1-1, natural variation, Pseudomonas syringae, quantitative disease resistance, quantitative trait locus
Citation: Luo Q, Liu W-W, Pan K-D, Peng Y-L and Fan J (2017) Genetic Interaction between Arabidopsis Qpm3.1 Locus and Bacterial Effector Gene hopW1-1 Underlies Natural Variation in Quantitative Disease Resistance to Pseudomonas Infection. Front. Plant Sci. 8:695. doi: 10.3389/fpls.2017.00695
Received: 26 January 2017; Accepted: 18 April 2017;
Published: 04 May 2017.
Edited by:
Vincenzo Lionetti, Sapienza University of Rome, ItalyReviewed by:
Brian H. Kvitko, University of Georgia, GeorgiaWilliam Underwood, United States Department of Agriculture – Agricultural Research Service, USA
Copyright © 2017 Luo, Liu, Pan, Peng and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Fan, amZhbkBjYXUuZWR1LmNu
†These authors are joint first authors.
 Qi Luo1,2†
Qi Luo1,2† Wei-Wei Liu
Wei-Wei Liu Jun Fan
Jun Fan