
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 February 2017
Sec. Plant Cell Biology
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.00158
De novo shoot regeneration is widely used in fundamental studies and agricultural applications. Actin microfilaments are involved in many aspects of plant cell division, cell morphogenesis and cell signal transduction. However, the function of actin microfilaments during de novo shoot regeneration is poorly understood. Here, we investigated the organization of actin microfilaments during this process and found that stem cell formation was associated with microfilament depolymerization. Furthermore, inhibition of microfilament depolymerization by phalloidin treatment or downregulation of actin depolymerizing factors (ADFs) restrained stem cell initiation and shoot regeneration. Inhibition of ADF expression affected the architecture of microfilaments during stem cell formation, and the polar transport and distribution of auxin were also disrupted. Together, our results demonstrate that organization of the microfilament cytoskeleton play important roles in stem cell formation and shoot meristem induction during shoot regeneration.
Plant somatic cells can be reprogrammed to generate various organs under defined physical and chemical conditions, a process called de novo organogenesis. This phenomenon is not only critical for in vitro plant propagation and biotechnology, but also useful for understanding plant developmental regulatory mechanisms (Sugiyama, 2000). The patterns of plant de novo organogenesis depend on the specific balance of applied exogenous hormones. High ratios of auxin/cytokinin lead to root regeneration, while high cytokinin/auxin ratios induce shoot regeneration. With high concentrations of both auxin and cytokinin, callus can be generated (Skoog and Miller, 1957; Bhojwani and Razdan, 1996; Che et al., 2002).
A number of studies have been performed to understand the molecular mechanisms underlying de novo shoot regeneration in Arabidopsis (Gordon et al., 2007; Li et al., 2011; Cheng et al., 2013). Auxin is the best-known hormone exhibiting local accumulation and responses during shoot regeneration. Dynamic distribution patterns of the auxin response were clearly shown in the in vitro shoot initiation (Gordon et al., 2007). During callus formation on auxin-rich callus induction medium (CIM), auxin responsive signals represented by the DR5rev signals are uniformly present in clusters of proliferating callus cells at the edges of the callus. After transfer to cytokinin-rich shoot induction medium (SIM), low auxin-responsive signals are required for initiation of the shoot apical meristem (SAM) in the callus. WUSCHEL (WUS), a transcription factor, plays a key role in de novo shoot regeneration (Gallois et al., 2004; Gordon et al., 2007). WUS is required for shoot stem-cell formation and maintenance in SAM, on which many signaling pathways converge (Dodsworth, 2009). During de novo shoot regeneration in Arabidopsis, WUS is upregulated in the center of the regenerated SAM, which is required for stem cell induction and subsequent shoot formation (Gordon et al., 2007; Cheng et al., 2013). WUS promotes the initiation and maintenance of the overlying stem cells by stimulating the activity of CLAVATA3 (CLV3) therein, which is a small secreted peptide as a stem cell marker (Mayer et al., 1998; Fletcher et al., 1999; Brand et al., 2000). Induction of the WUS expression during shoot regeneration was regulated by the master phytohormone auxin (Cheng et al., 2013). Auxin polar transport mediated by the PINFORMED efflux carrier proteins (PINs) contributes to the initiation and maintenance of auxin-responsive gradients in specific callus tissues during shoot regeneration (Cheng et al., 2013). Polarized membrane localization of PIN1 at initiation sites of de novo SAMs can be induced by SIM incubation, which contributes to the spatially distributed auxin response. Shoot regeneration is severely reduced in the plants expressing antisense PIN1. Application of the auxin transport inhibitor naphthylphthalamic acid (NPA) disrupts the spatiotemporal auxin response and shoots regeneration, demonstrating that auxin polar transport and the asymmetric distribution of the auxin response are required for initiation of SAMs during shoot regeneration (Cheng et al., 2013).
The microfilament cytoskeleton, a major component of the plant cell cytoskeleton, is involved in many aspects of cell division, cell morphogenesis, and the establishment and maintenance of cell polarity by filament polymerization and depolymerization (Vantard and Blanchoin, 2002). Interestingly, the polar transport and distribution of auxin play critical roles in regulating cytoskeletal organization, which is important for cell polarization and morphogenesis (Pan et al., 2015; Wu et al., 2015). Proteins involved in the actin organization and dynamics have been characterized in Arabidopsis roots (Takáč et al., 2017). As an actin binding protein, actin depolymerizing factor (ADF) controls the actin organization and regulates various plant cell development and morphogenesis processes (Jiang et al., 1997; Dong et al., 2001; Allwood et al., 2002; Henty-Ridilla et al., 2014). Previous studies have demonstrated that ADF9 is expressed in the SAM and controls multicellular development including callus formation generated by root explants via both cytoplasmic and nuclear processes in Arabidopsis (Burgos-Rivera et al., 2008). Furthermore, ADF4 plays a role in pathogen perception, defense activation and transcription through the regulation of actin cytoskeletal dynamics and R-gene transcription (Porter et al., 2012). In addition, the actin cytoskeleton and its dynamics have roles in responses to abiotic and biotic stimuli.
During in vitro shoot regeneration in Arabidopsis, a group of cells in callus can develop into organizing center cells through transdifferentiation (Gordon et al., 2007; Cheng et al., 2013). It has been reported that the coexpression of ACT7 with other actin proteins is required for normal callus formation (Kandasamy et al., 2001). However, little is known about the organization of the microfilament cytoskeleton of callus cells during this process. In this study, we showed that stem cell formation in callus is associated with the process of microfilament depolymerization. Inhibition of microfilament depolymerization by phalloidin treatment or downregulation of ADFs inhibited stem cell formation and shoot induction. Furthermore, repression of ADF expression disrupted the polar transport and distribution of auxin in callus. Depolymerization of the microfilament cytoskeleton is thus required for stem cell formation during shoot regeneration, through mediating the polar transport and distribution of auxin.
Arabidopsis thaliana plants used in this study were of the Columbia (Col) and Landsberg erecta (Ler) ecotypes. The origins and ecotypes of the transgenic lines and mutants were as follows: pWUS::DsRed-N7 (Ler; Gordon et al., 2007), pCLV3::GFP-ER reporter lines (Ler; Lenhard and Laux, 2003), pWUS::GUS and pCLV3::GUS (Col; Su et al., 2009), DR5rev::GFP and pPIN1::PIN1-GFP (Col; Xu et al., 2006), 35S::GFP-ABD2-GFP (Col; Wang et al., 2008), CYCB1;1::GUS (Col; Colón-Carmona et al., 1999). The pWUS::DsRED-N7 plants were crossed with 35S::GFP-ABD2-GFP plants. The estradiol-inducible XVE binary vector (Zuo et al., 2000) was kindly provided by Dr. Nam-Hai Chua (Rockefeller University, New York, NY, USA).
Arabidopsis seeds were sterilized and plated on germination medium (Murashige and Skoog, 1962). The plates were kept at 4°C for 2 days to overcome dormancy, and then transferred to a culture room at 22°C with a 16-h-light/8-h-dark cycle for 7 days. Young seedlings were transplanted into vermiculite and grown under the conditions as described above until harvesting of siliques.
De novo shoot regeneration was performed as described by Li et al. (2011). After cold treatment, the seeds were vertically cultivated in the plate under sterile conditions (light intensity of 40 μmol photons m-2 s-1, 22°C, under a 16-h-light/8-h-dark cycle) for approximately 7 days, and root explants of 5 mm length were excised from the seedlings. Then, the explants were transferred onto CIM consisting of Gamborg’s B5 medium (Gamborg et al., 1968) containing 2% glucose, 0.5 g/L MES, 0.2 μmol/L kinetin, and 2.2 μmol/L 2,4-dichlorophenoxyacetic acid (2,4-D) with 0.8% agar, and incubated for 6 days to induce callus production. Finally, the calli were transferred onto SIM with 2% glucose, 0.5 g/L MES, 0.9 μmol/L 3-indoleacetic acid, and 0.5 μmol/L 2-isopentenyladenine to induce shoot regeneration.
Actin filaments in callus were stained with Alexa-488 phalloidin as previously described (Zheng et al., 2013). To observe the organization of the actin cytoskeleton during stem cell formation in callus of shoot regeneration, callus from pWUS::DsRed-N7 plants were subsequently subjected to fixation and staining with Alexa-488 phalloidin. Actin filaments in the callus were observed with a confocal laser scanning microscope (Leica TCS SP5, Germany) equipped with a 40× oil objective. The fluorescent phalloidin was excited with the 488-nm line of an argon laser.
The artificial microRNA (amiRNA) silencing procedure was performed as described by Schwab et al. (2006). The WMD31 (Web MicroRNA Designer3) software was used to design specific primers for ADF1 (AT3G46010), ADF2 (AT3G46000), ADF3 (AT5G59880), and ADF4 (AT5G59890). The plasmid pRS300 containing the endogenous Arabidopsis MIR319a precursor (Schwab et al., 2006) was used as a template, and ath-MIR319a was replaced by overlapping PCR with the primers A, B and adf1 adf2 adf3 adf4 I–IV (Supplementary Table S1). Then, the amiRNA cloned into the plasmid was enzymatically digested and ligated into the pER8 expression vector, which was then transformed into plants through Agrobacterium-mediated floral dip method. Hygromycin was used to screen positive transgenic lines.
Total RNAs were isolated from various calli using Trizol reagent (Invitrogen). The qRT-PCR reactions were performed for each cDNA dilution using SYBR Green Master mix with Chromo4 according to the manufacturer’s protocol (Bio-Rad). All primers used for qRT-PCR are shown in Supplementary Table S1. The relative expression level of each gene was standardized to that of the housekeeping gene TUBULIN2, and all measurements were carried out in three biological replicates. The results were analyzed using the comparative CT method, and means and standard deviations were calculated.
To investigate the expression patterns of pWUS::GUS during shoot initiation, a GUS histochemical staining assay was performed following Cheng et al. (2014). Then, the materials were destained with 70% ethanol for imaging.
To determine the effect of phalloidin on shoot induction, calli were cultured on SIM containing various concentrations of phalloidin (prepared in DMSO as a 20 mmol/L stock; Sigma) for 20 days. The regeneration frequencies of shoots under treatments of various concentrations of phalloidin were statistically analyzed, and 90 samples for each treatment were collected. Means and standard deviations were calculated (shown as ‘mean ± SD’ in Table 1). Callus cultured on SIM containing 5 μmol/L phalloidin for 0, 4, 8, 12, and 16 days were individually harvested to isolate total RNA for qRT-PCR analysis.
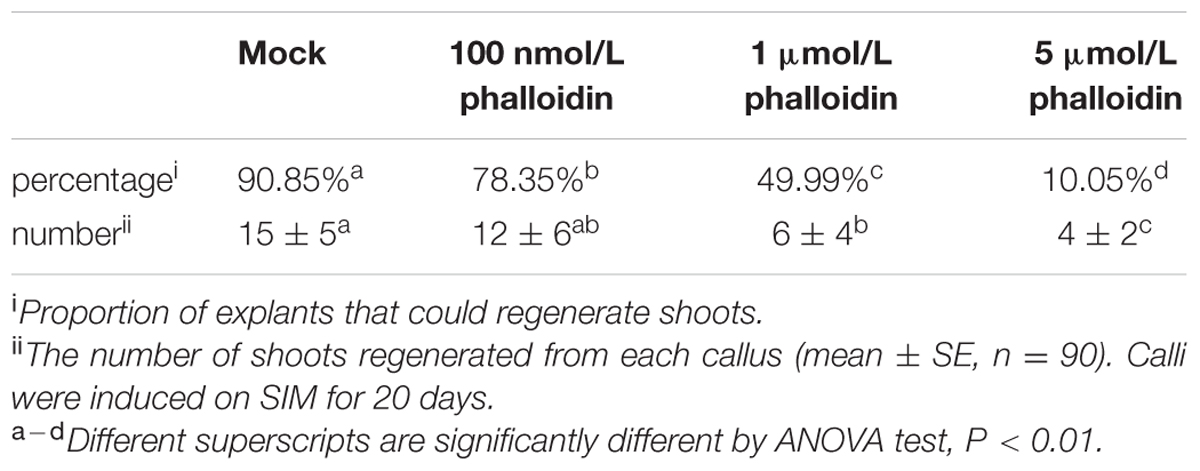
TABLE 1. Regeneration frequencies of shoots with treatment of different concentrations of phalloidin.
To induce transcription of artificial microRNAs, the calli were cultured on SIM with 10 μmol/L estradiol (prepared in DMSO as a 10 mmol/L stock; Sigma) for 20 days and estradiol was added every 2 days. Total RNA was isolated from the callus on SIM with estradiol at 12 days and used for qRT-PCR to determine the expression of ADF genes.
An Olympus JM dissecting microscope was used to photograph the callus morphologies. To investigate the green fluorescence images for actin staining and live cell imaging, samples were observed using a Leica TCS SP5 confocal microscope with a 40× oil lens. For each stage, at least 40 samples were imaged to confirm the structure of actin. To investigate red and green fluorescence images for co-localization between WUS and ABD2 in callus cells and expression patterns of WUS, CLV3, PIN1, and DR5 reporter lines, samples were observed using a 40× oil lens. For each gene marker line at various stages, at least 30 samples were imaged to confirm the expression pattern of a particular marker at each stage. The specific sets of filters used for each marker were similar to those described by Heisler et al. (2005). Leica LAS AF Lite software was used for confocal images processing.
The density and skewness of actin filaments were quantified by ImageJ according to Higaki et al. (2010). To estimate actin filaments density, we defined the occupancy of the fluorescence signal as calculated from the skeletonized image of the callus cells. The occupancy becomes lower as the actin filaments are depolymerized or fragmented. To estimate actin filaments polymerization and bundling, we used the skewness of the intensity distribution of the microfilament pixels as an indicator of its polymerization and bundling. The skewness decreases as the fluorescence intensity in the pixels decreases as a result of actin filaments depolymerized or fragmented. Individual cells were segmented manually and actin filaments at the cell border were eliminated. For ABD2 reporter lines and actin staining samples at each developmental stage, more than 50 callus cells were used for the analysis.
A de novo shoot regeneration system was established using root explants in Arabidopsis. In a two-step regeneration process, the explants were cultured on an auxin-rich CIM for 6 days and then incubated on a cytokinin-rich SIM to induce shoot regeneration (Gordon et al., 2007). Stem cell initiation and SAM formation is developmentally induced during the shoot regeneration process, and the expression of WUS is the earliest event to mark stem cell initiation (Gordon et al., 2007; Cheng et al., 2013). Fimbrin actin binding domain 2 (ABD2)-based filamentous actin (F-actin) reporters have emerged as powerful tools to study microfilament organization in living plants. To observe the organization of the microfilament cytoskeleton in stem cell initiation during shoot regeneration, pWUS::DsRED-N7/35S::GFP-ABD2-GFP double reporter lines were used to obtain explants. After incubation on CIM for 6 days (SIM for 0 day), neither WUS expression nor stem cell initiation was induced in the callus (Figure 1A). Strong GFP signals of polymerized and bundled filaments were detected both in the epidermal callus cells (Figures 1A,B) and in the inner layers of callus cells close to the vascular tissue (Figures 1A,C), suggesting that the microfilaments were mainly polymerized and bundled in these callus cells. However, regional expression of the WUS gene was detected in callus cultured on SIM for 6 days, implying stem cells were initiated (Figure 1D). At this time, microfilaments became fragmented in the WUS-expressing cells (Figures 1D,E), and were less bundled compared with the callus cells surrounding these WUS-expressing cells (Figures 1D,F). After the callus was cultured on SIM for 9 days, shoot primordia were formed (Figure 1G). The microfilaments mainly showed fragmented distributions in the WUS-expressing organizing center cells of the de novo shoot meristem (Figures 1G,H). However, the microfilaments in the callus cells close to the shoot primordia were still polymerized and bundled filaments (Figures 1G,I). To quantitatively analyze the actin architecture, we measured the skewness and density (Higaki et al., 2010) parameters to determine the extent of actin filament bundling and the percentage of occupancy of actin filaments in the WUS-expressing cells and callus cells. Consequently, the WUS-expressing cells had a lower density value than the callus cells (Figure 1J). Moreover, the actin filaments were bundled with higher skewness in the callus cells than the WUS-expressing cells (Figure 1K). However, the density and skewness values showed no significant difference in callus cells cultured on SIM for 6 days and 9 days compared with those cultured on SIM for 0 day (Figures 1J,K). We further confirmed the actin architecture observations in the stem cell initiation process using fixation and staining of the microfilament cytoskeleton. Callus derived from pWUS::DsRED-N7 plants were subjected to F-actin staining with Alexa-488 phalloidin. Similar to the ABD2 marker used to visualize actin architecture, the fluorescence signals associated with F-actin staining by Alexa-488 phalloidin were weaker in WUS-expressing cells compared with the callus cells (Supplementary Figure S1). The density and skewness values were much lower in WUS-expressing cells. These observations indicate that stem cell initiation in the induced callus is associated with the depolymerization and fragmentation of the microfilament cytoskeleton during in vitro shoot regeneration.
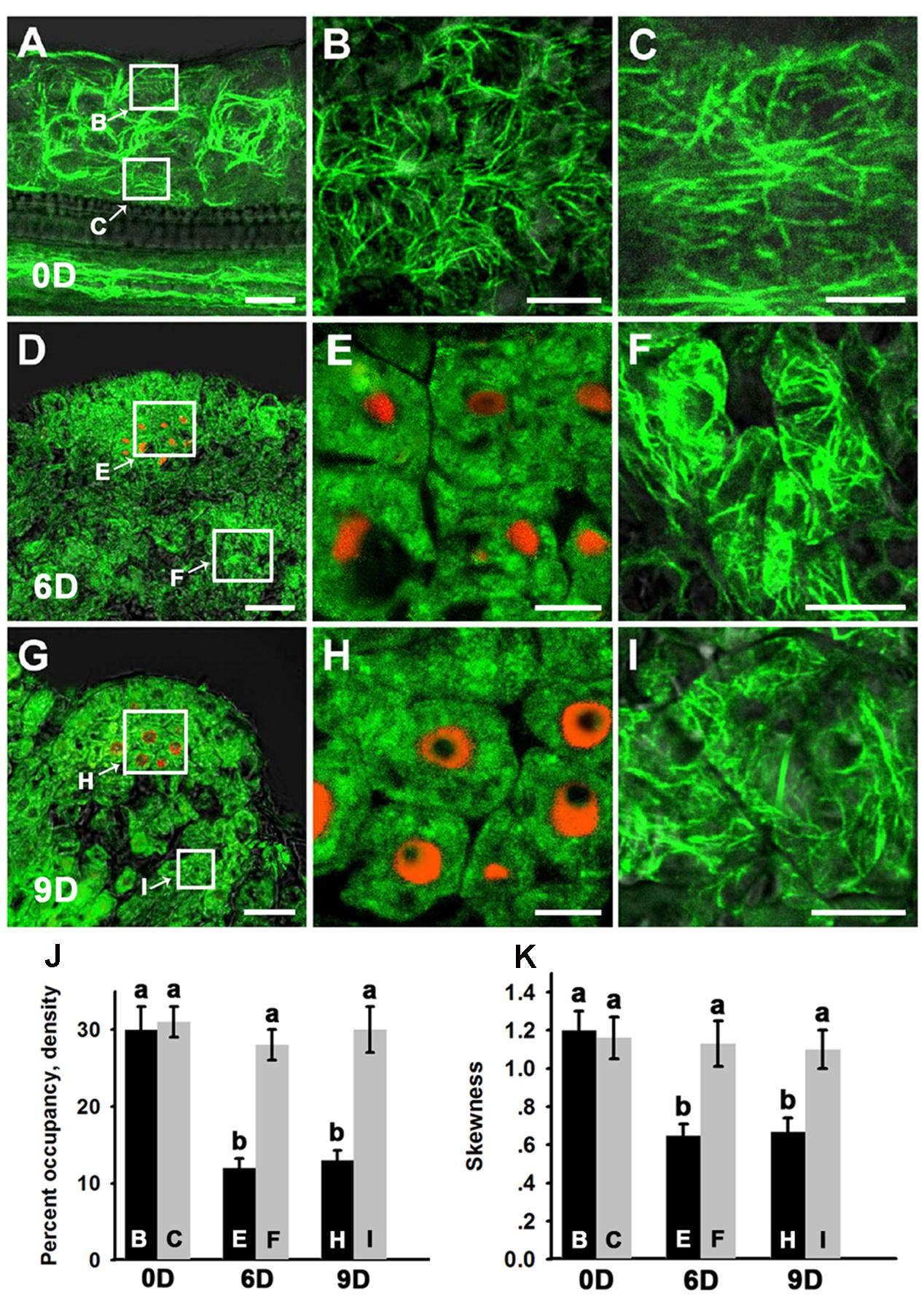
FIGURE 1. Organization of actin filaments during shoot regeneration. (A) Callus cultured on SIM for 0 days. (B,C) Magnification of the areas indicated by the arrows in (A). Strong green signals of polymerized and bundled filaments were detected both in the epidermal callus cells (B) and in the inner layers of callus cells close to the vascular tissue (C). (D) Callus cultured on SIM for 6 days. (E,F) Magnification of the areas indicated by the arrows in (D). Microfilaments became more fragmented and less bundled in the WUS-expressing cells (E) compared with the callus cells surrounding these WUS-expressing cells (F). (G) Callus cultured on SIM for 9 days. (H,I) Magnification of the areas indicated by the arrows in (G). The green signals of microfilaments mainly showed fragmented distributions in the WUS-expressing organizing center cells of the de novo shoot meristem (H) compared with the callus cells close to the shoot primordial (I). Green signal represents the fluorescence of 35S::GFP-ABD2-GFP, red signal represents the fluorescence of pWUS::DsRED-N7. Bars = 10 μm. (J) The average filament density measured in the callus cells shown in (B,C,E,F,H,I). The WUS-expressing cells had a lower density value than the callus cells. (K) The extent of filament bundling (skewness) measured in the callus cells shown in (B,C,E,F,H,I). The WUS-expressing cells had a lower skewness value than the callus cells. Different lowercases in (J,K) are significantly different by ANOVA test, P < 0.01. Error bars represent standard deviations from triplicate measurements.
To further examine the function of microfilament cytoskeleton organization during shoot regeneration, phalloidin, an inhibitor of microfilament depolymerization, was added to the SIM at different concentrations. As shown in Figures 2A–H and Table 1, 100 nmol/L phalloidin had minor effects on the induction of shoots. A high concentration of 1 μmol/L phalloidin caused remarkable decreases in the production of shoots with severe abnormal morphologies (Figures 2I–L; Table 1). Treatment with 5 μmol/L phalloidin almost completely inhibited shoot regeneration (Figures 2M–P; Table 1). In order to assess the status of cell growth and division during the phalloidin experiments, the fresh weight of callus was measured (Supplementary Figure S2A). Even treated with 5 μmol/L phalloidin, the callus continually increased their fresh weight during shoot induction on SIM. The Arabidopsis mitotic cyclin CYCB1;1 is excellent marker for cells undergoing mitosis, which is expressed around the G2/M transition (Colón-Carmona et al., 1999). The expression of CYCB1;1::GUS was retained in the callus cells under treatment of 5 μmol/L phalloidin (Supplementary Figures S2C,D). These results eliminated the general impairment of cell growth and division during phalloidin treatment. We further detected the organization of the actin filaments in the callus cells on SIM treated with 5 μmol/L phalloidin. The actin filaments were still polymerized and bundled with a filamentous distribution in these callus cells (Figures 2Q–T), and no overt differences were observed in the density and skewness values (Figures 2U,V). These results indicated that phalloidin treatment leading to inhibition of microfilament depolymerization in the callus cells inhibited shoot induction.
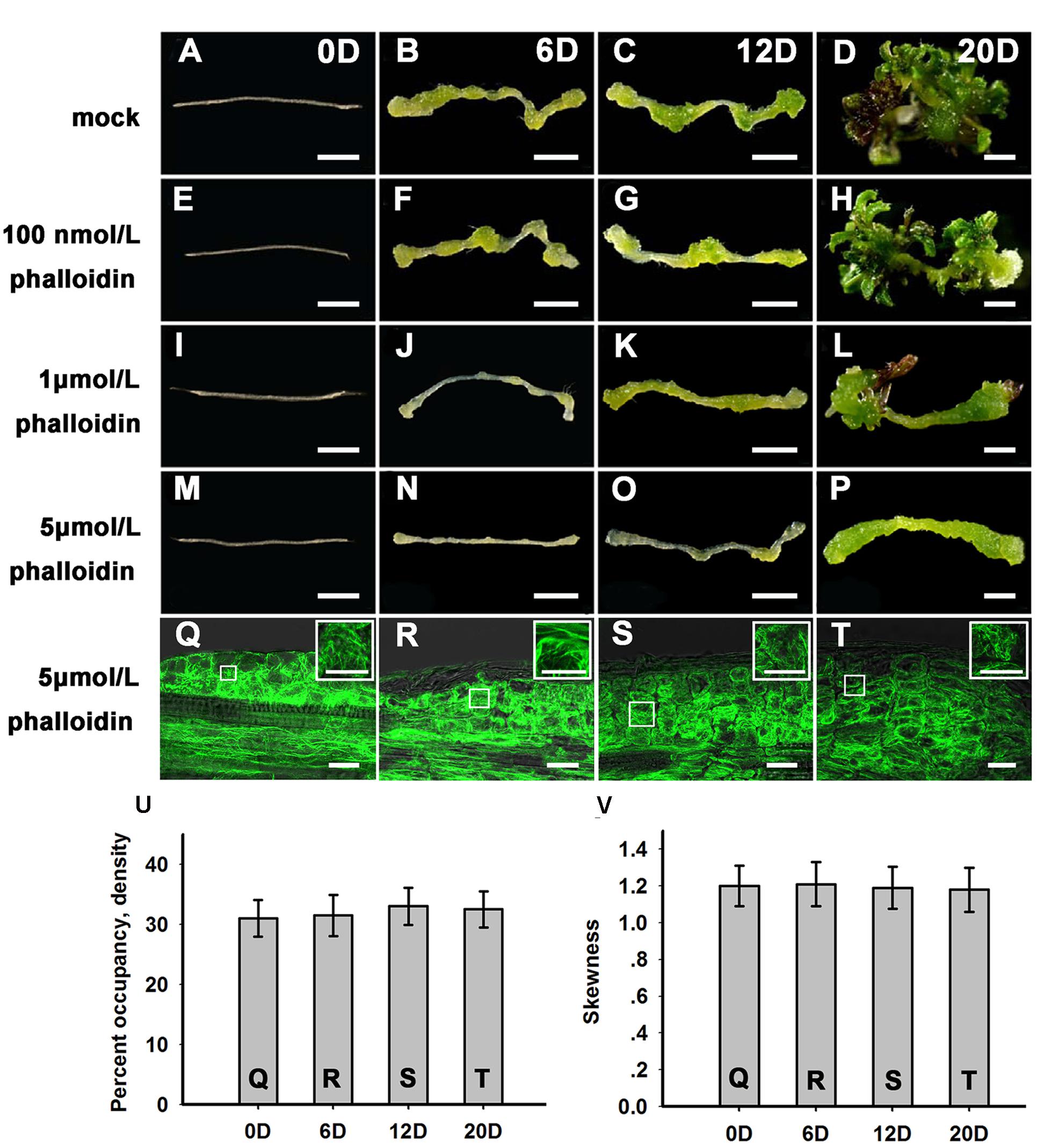
FIGURE 2. Shoot regeneration under treatment with different concentrations of phalloidin. (A–D) Shoot regeneration of wild type plants when cultured on SIM with DMSO for 0, 6, 12, and 20 days. (E–H) Shoot regeneration of wild type plants when cultured on SIM containing 100 nmol/L phalloidin (prepared in DMSO) for 0, 6, 12, and 20 days, which had minor effects on the induction of shoots. (I–L) Shoot regeneration of wild type plants when cultured on SIM containing 1 μmol/L phalloidin for 0, 6, 12, and 20 days, which caused remarkable decreases in the production of shoots. (M–P) Shoot regeneration of wild type plants when cultured on SIM containing 5 μmol/L phalloidin for 0, 6, 12, and 20 days, which almost completely inhibited shoot regeneration. Bars = 1 mm. (Q–T) 35S::GFP-ABD2-GFP signals in callus when cultured on SIM containing 5 μmol/L phalloidin for 0, 6, 12, and 20 days. The actin filaments were still polymerized and bundled with a filamentous distribution in these callus cells. Bars = 20 μm. (U) The average filament density measured in the callus cells shown in (Q,R–T). (V) The extent of filament bundling (skewness) measured in the callus cells shown in (Q,R–T). No overt differences were observed in the density and skewness values of these cells. Error bars represent standard deviations from triplicate measurements.
Previous studies have reported that WUS and CLV3 expression marks stem cell formation in shoot meristem (Laux et al., 1996; Mayer et al., 1998; Schoof et al., 2000; Weigel and Jürgens, 2002). WUS-expressing cells in the organizing center establish and maintain stem cell populations markerd by CLV3 expression within the central zone of the shoot meristem. Therefore, we studied the expression patterns of WUS and CLV3 during shoot meristem induction under phalloidin treatment using pWUS::GUS and pCLV3::GUS reporter lines. As shown in Figures 3A,B, WUS transcription was induced in several restricted regions of the callus grown on SIM without phalloidin for 6 days. Subsequently, shoot primordia appeared in these WUS-expressing regions and the WUS signals were maintained in the organizing center cells of the shoot primordia (Figure 3C). By contrast, WUS expression was not detected in callus after 9 days of culture on SIM with 5 μmol/L phalloidin (Figures 3D–F). Consequently, the suppression of WUS expression was accompanied by inhibition of shoot formation in the phalloidin-treated callus. In shoot regeneration, the induction of WUS expression specifies stem cells that are marked by CLV3 expression in callus (Cheng et al., 2010). Similar to WUS. CLV3 expression was either not detected in callus on SIM treated with 5 μmol/L phalloidin (Figures 3G–L). The expression patterns of WUS and CLV3 under 5 μmol/L phalloidin treatment were further determined by qRT-PCR analysis. As expected, the transcript levels of both WUS and CLV3 were reduced in callus treated with phalloidin compared with those in untreated tissues within 16 days culture on SIM (Figure 3M), suggesting that inhibition of microfilament depolymerization in callus cells might disturb shoot regeneration by suppressing the expression of WUS and CLV3 genes.
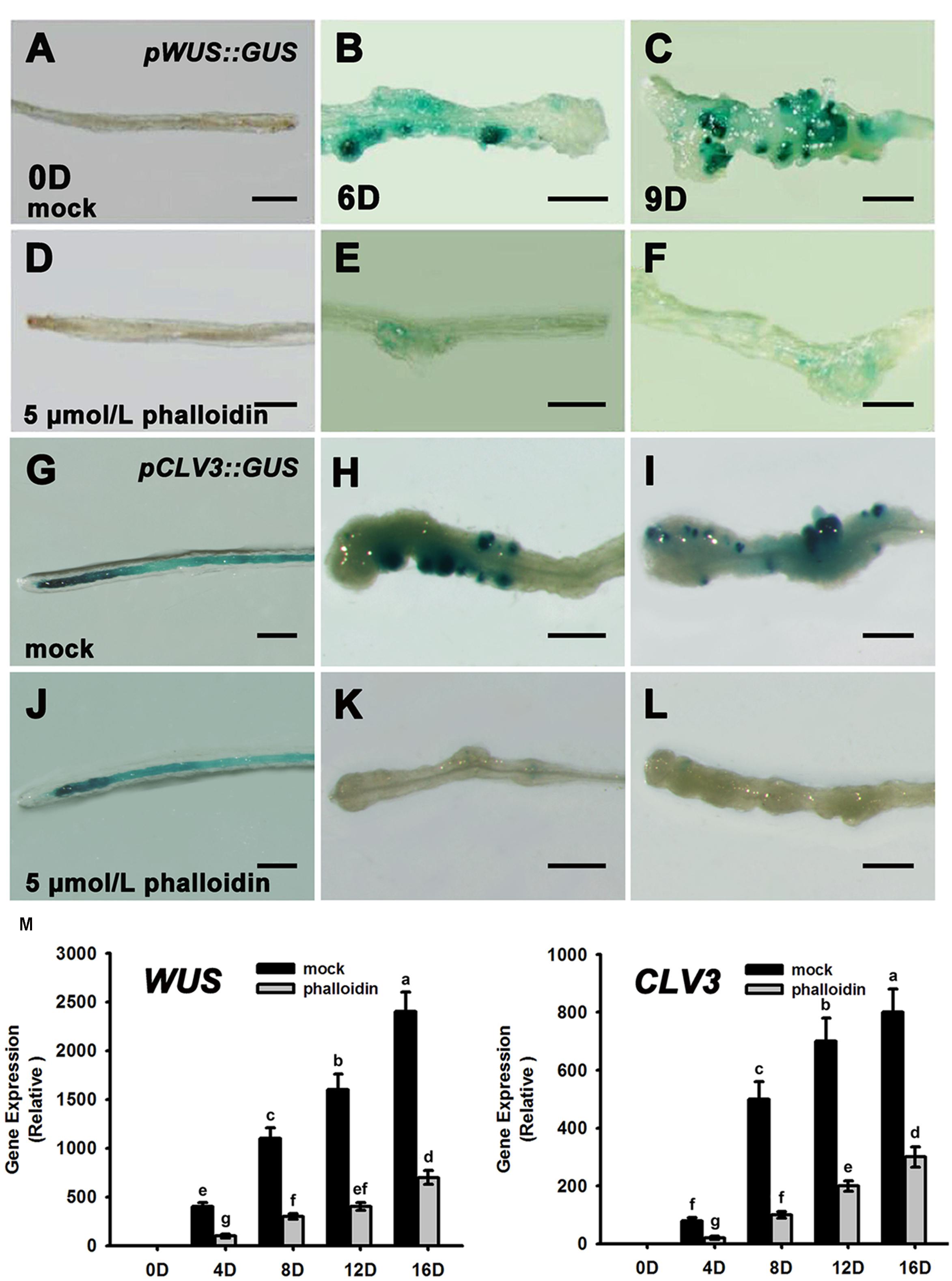
FIGURE 3. Expression patterns of WUS and CLV3 in calli treated with phalloidin. (A–C) Expression patterns of pWUS::GUS in callus cultured on SIM with DMSO for 0, 6, and 9 days. (D–F) Expression patterns of pWUS::GUS in callus cultured on SIM containing 5 μmol/L phalloidin for 0, 6, and 9 days. WUS expression was not detected in callus. (G–I) Expression patterns of pCLV3::GUS in callus cultured on SIM with DMSO for 0, 6, and 9 days. (J–L) Expression patterns of pCLV3::GUS in callus cultured on SIM containing 5 μmol/L phalloidin for 0, 6, and 9 days. CLV3 expression was not detected in callus. Bars = 1 mm. (M) Relative expression levels of WUS and CLV3 in calli as determined by qRT-PCR. The transcript levels of both WUS and CLV3 were reduced in callus treated with phalloidin. Mock, callus cultured on SIM with DMSO; phalloidin, callus cultured on SIM containing 5 μmol/L phalloidin. Different lowercases are significantly different by ANOVA test, P < 0.01. Error bars represent standard deviations from triplicate measurements.
Actin depolymerizing factors (ADFs) are a kind of actin-binding proteins of low molecular weight that exist widely in eukaryotes and play important roles in microfilament depolymerization and polymerization (Andrianantoandro and Pollard, 2006; Pei et al., 2012). We analyzed the expression patterns of ADFs during de novo shoot regeneration after cultured on SIM for 12 days using RT-PCR. Most of the ADF genes were upregulated at the early stages of shoot induction, especially ADF1. ADF2. ADF3, and ADF4, while the actin polymerizing factor ADF9 was significantly downregulated (Figure 4). These results were consistent with the depolymerization of microfilaments during shoot meristem formation.
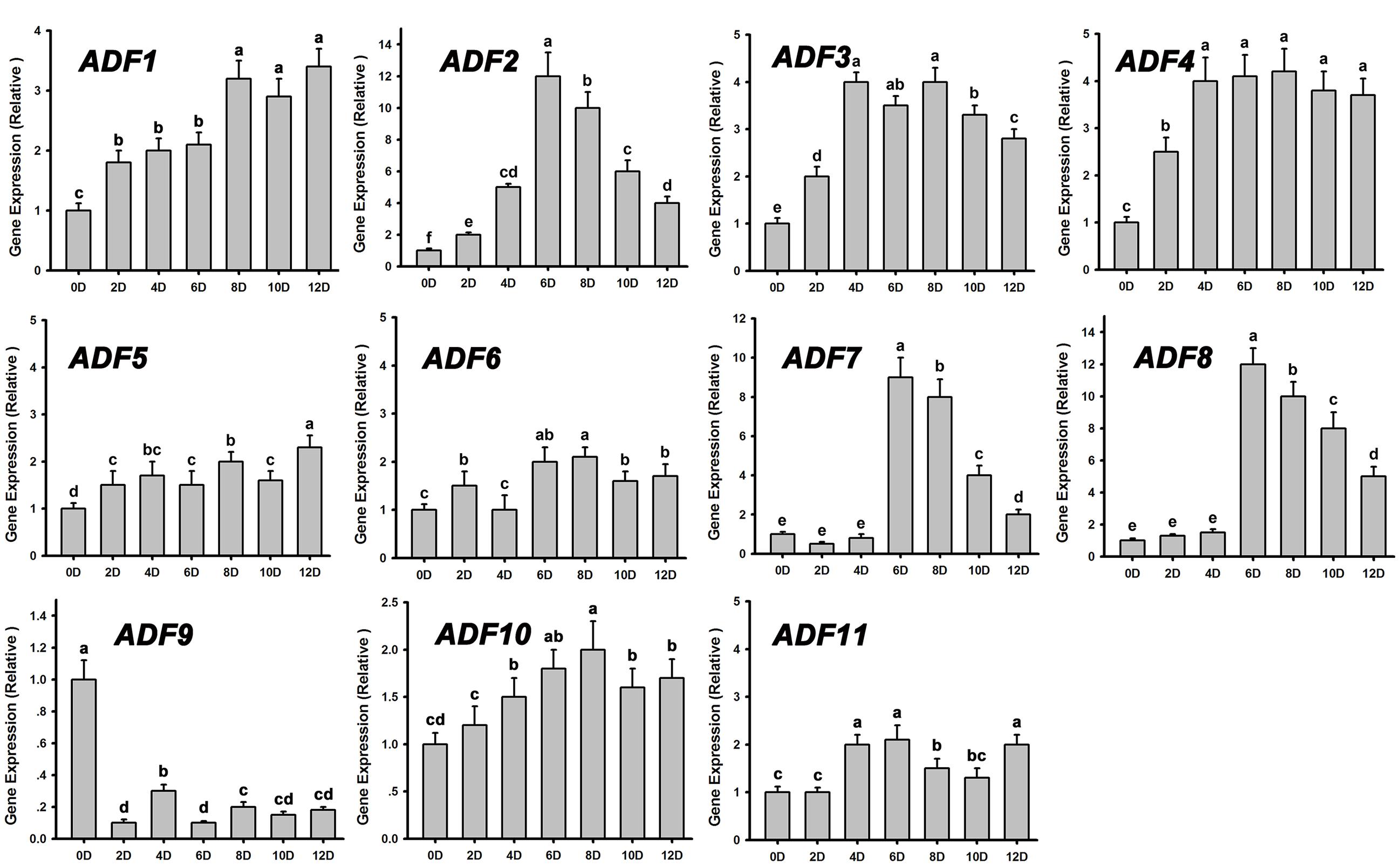
FIGURE 4. Relative expression levels of ADFs during shoot regeneration. Different lowercases are significantly different by ANOVA test, P < 0.01. Error bars represent standard deviations from triplicate measurements.
The adf single mutant did not show obvious phenotypes during shoot regeneration (data not shown), implying that the ADF family genes were functionally redundant. ADF subclass I, including ADF1. ADF2. ADF3 and ADF4, is constitutively and highly expressed in various reproductive and vegetative organs except pollen tubes (Ruzicka et al., 2007). Here, the artificial microRNA technology was used to knock down these four ADF genes simultaneously. Three lines of T2 generation plants (amiR-ADF1 2 3 4) were selected to analyze the expression levels of the ADFs. As shown in Figure 5A, ADF1. ADF2. ADF3, and ADF4 were all significantly downregulated in the three estradiol-induced transgenic lines, while the expression of the other ADF genes was almost the same as in the non-induced transgenic plants. As the transcript levels of ADF1. ADF2. ADF3, and ADF4 were downregulated most obviously in transgenic line 3, we selected this line for further analysis.
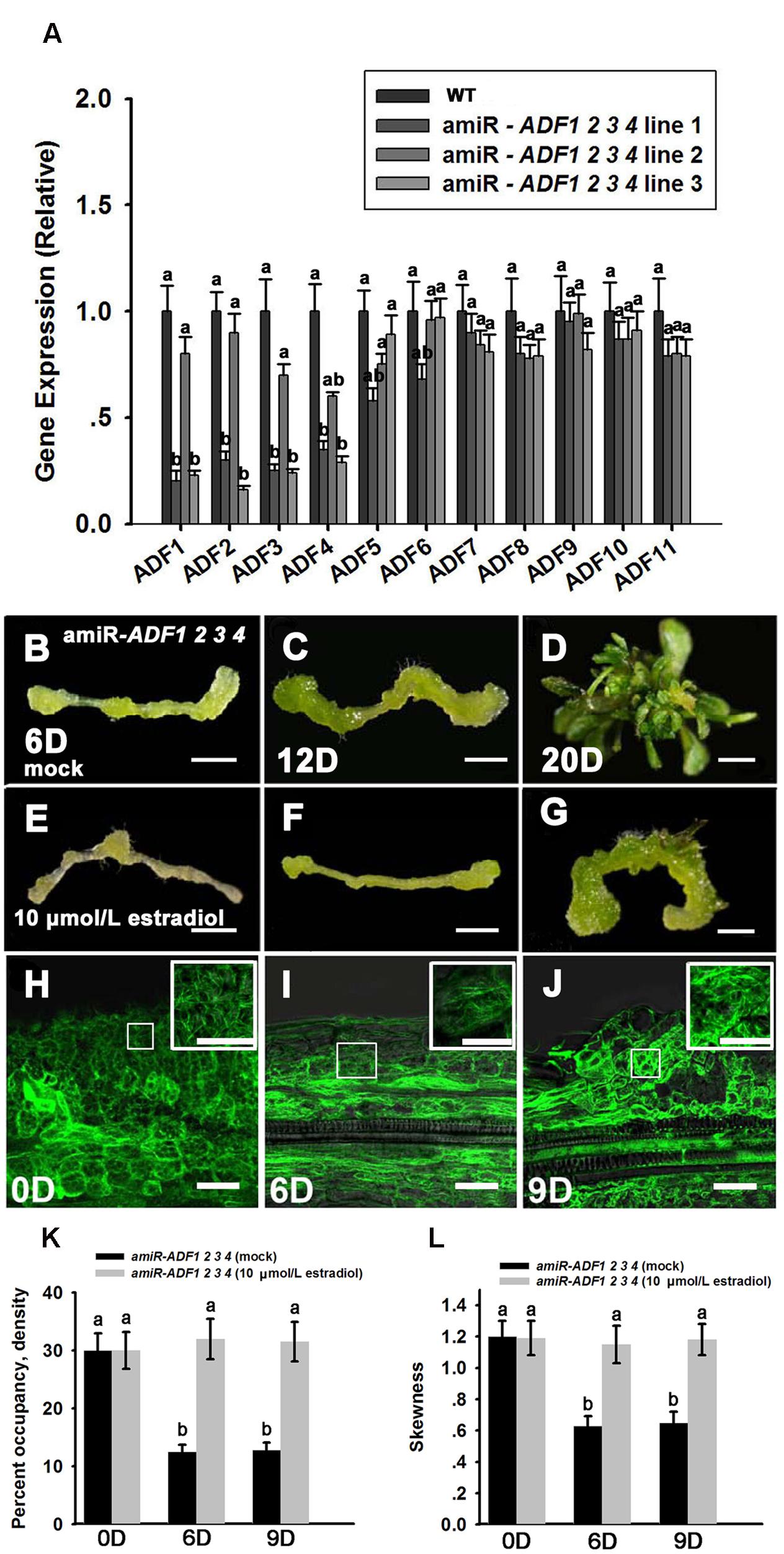
FIGURE 5. Shoot regeneration from amiR-ADF1 2 3 4 transgenic lines induced by estradiol. (A) Relative expression levels of ADFs in wild type (WT) and estradiol-induced different transgenic lines by qRT-PCR analysis. The transcript levels of ADF1. ADF2. ADF3, and ADF4 were downregulated most obviously in transgenic line 3. Different lowercases are significantly different by ANOVA test, P < 0.01. Error bars represent standard deviations from triplicate measurements. (B–D) Shoot regeneration of non-induced transgenic plants when cultured on SIM without estradiol for 6, 12, and 20 days. (E–G) Shoot regeneration of induced transgenic plants when cultured on SIM containing 10 μmol/L estradiol for 6, 12, and 20 days. Few shoot primordia emerged from the estradiol-induced transgenic plants. Bars = 1 mm. (H–J) 35S::GFP-ABD2-GFP signals in callus from induced transgenic plants when cultured on SIM containing 10 μmol/L estradiol for 0, 6, and 9 days. The actin filaments were still polymerized and bundled with a filamentous distribution in the callus cells. Bars = 20 μm. (K) The average filament density measured in the stem cells under mock control and callus cells shown in (H–J). (L) The extent of filament bundling (skewness) measured in the stem cells under mock control and callus cells shown in (H–J). No overt differences were observed in the density and skewness values in the callus cells cultured with estradiol. Different lowercases are significantly different by ANOVA test, P < 0.01. Error bars represent standard deviations from triplicate measurements.
To analyze shoot regeneration in the transgenic plants, estradiol was added to the SIM to induce the expression of the artificial microRNA that inhibited ADF1. ADF2. ADF3, and ADF4 expressions. Several green shoot primordia regenerated from non-induced transgenic plants were observed after growth on SIM for 12 days, and numerous shoots were regenerated after 20 days in the SIM (Figures 5B–D; Table 2). By contrast, few shoot primordia emerged from the estradiol-induced transgenic plants grown on SIM for 12 days (Figures 5E,F). Even if the induced callus cultured for 20 days, the number of shoots was much less than that in non-induced callus (Figure 5G; Table 2). Thus, we confirmed that simultaneously suppressing the expression of ADF1. ADF2. ADF3, and ADF4 could reduce the rates and frequencies of shoot regeneration. Like the wild type plants, the callus of the amiR-ADF1 2 3 4 transgenic plants still increased their fresh weight during shoot induction on SIM, eliminating the general impairment of cell growth and division (Supplementary Figure S2B). To further analyze the organization of the microfilament cytoskeleton in the amiR-ADF1 2 3 4 transgenic plants during shoot regeneration, 35S::GFP-ABD2-GFP was transformed into the amiR-ADF1 2 3 4 transgenic lines. As shown in Figures 5H–J, the actin filaments were still polymerized and bundled with a filamentous distribution in the callus cells of amiR-ADF1 2 3 4 plants cultured on SIM with estradiol for 9 days. Furthermore, no overt differences were observed in the density and skewness values (Figures 5K,L). These results suggest that inhibition of ADF1. ADF2. ADF3, and ADF4 functions can inhibit the normal depolymerization of microfilaments, resulting in the repression of shoot regeneration.
Next, pWUS::DsRED-N7 reporter lines were used to investigate the expression pattern of WUS in the amiR-ADF1 2 3 4 transgenic plants,. Without the induction of artificial microRNA expression by estradiol, the WUS signal was regionally distributed after the callus was grown on SIM for 9 days (Figures 6A,B). At 12 days, an intense WUS signal was detected in the shoot primordia (Figure 6C). Nevertheless, the WUS signal could not be detected in the callus of estradiol-induced amiR transgenic lines (Figures 6D–F), indicating there was no formation of the organizing center cells. Furthermore, stem cell formation was also detected in the shoot regeneration of amiR-ADF1 2 3 4 plants using pCLV3::GFP-ER reporter (Figures 6G–L). In contrast to the control without the induction of artificial microRNA expression, expression of CLV3 was completely repressed in the callus of estradiol-induced amiR lines on SIM, indicating that no stem cell was initiated.
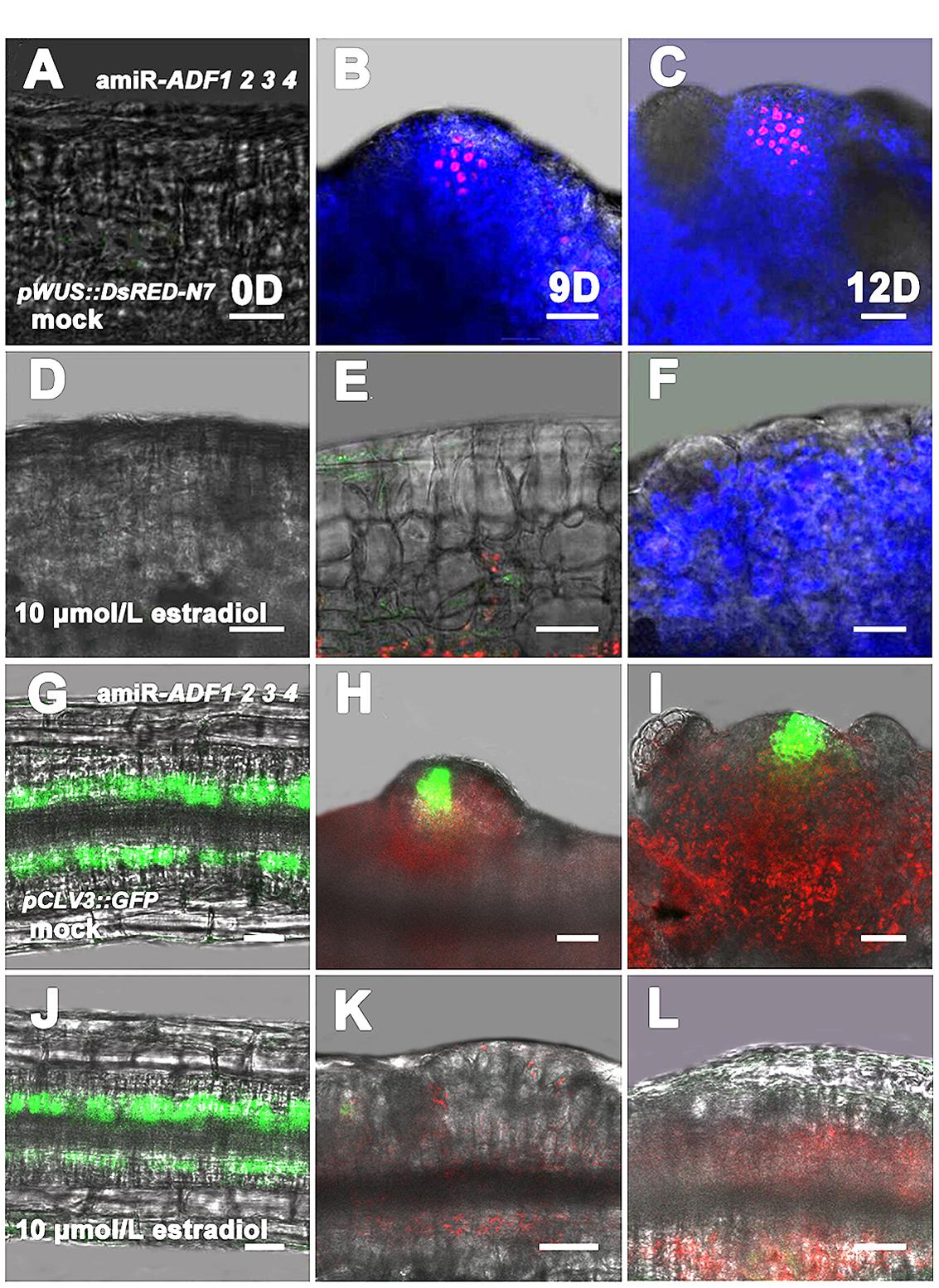
FIGURE 6. Expression patterns of pWUS::DsRED-N7 and pCLV3::GFP-ER in callus. (A–C) Expression patterns of pWUS::DsRED-N7 in callus from non-induced transgenic plants when cultured on SIM without estradiol for 0, 9, 12 days. (D–F) Expression patterns of pWUS::DsRED-N7 in callus from induced transgenic plants when cultured on SIM with 10 μmol/L estradiol for 0, 9, 12 days. The WUS signal could not be detected in the callus of estradiol-induced amiR transgenic lines. (G–I) Expression patterns of pCLV3::GFP-ER in callus from non-induced transgenic plants when cultured on SIM without estradiol for 0, 9, 12 days. (J–L) Expression patterns of pCLV3::GFP-ER in callus from induced transgenic plants when cultured on SIM with 10 μmol/L estradiol for 0, 9, 12 days. The CLV3 signal could not be detected in the callus of estradiol-induced amiR transgenic lines. Bars = 20 μm.
Previously, our study revealed that auxin polar distribution in callus is essential for Arabidopsis shoot regeneration (Cheng et al., 2013). To examine whether the establishment of auxin gradients in shoot regeneration was affected by the organization of microfilament cytoskeleton in callus cells, DR5rev::GFP signals were examined in the amiR-ADF1 2 3 4 callus. Spatially restricted distribution of DR5rev::GFP signals was firstly identified in some edge regions of non-induced callus on SIM for 6 days (Figures 7A,B,Q; Table 3). Subsequently, the DR5 signal gradually became more intense in the promeristem region and extended to the region of the shoot primordia after 12 days on SIM (Figures 7C,D,Q; Table 3). With the induction of artificial microRNA expression by estradiol, the DR5 signal was detected after 9 days on SIM but had a dispersed distribution pattern (Figure 7G,Q; Table 3). Even after 12 days on SIM, little intensive distribution of auxin was observed (Figure 7H,Q; Table 3).
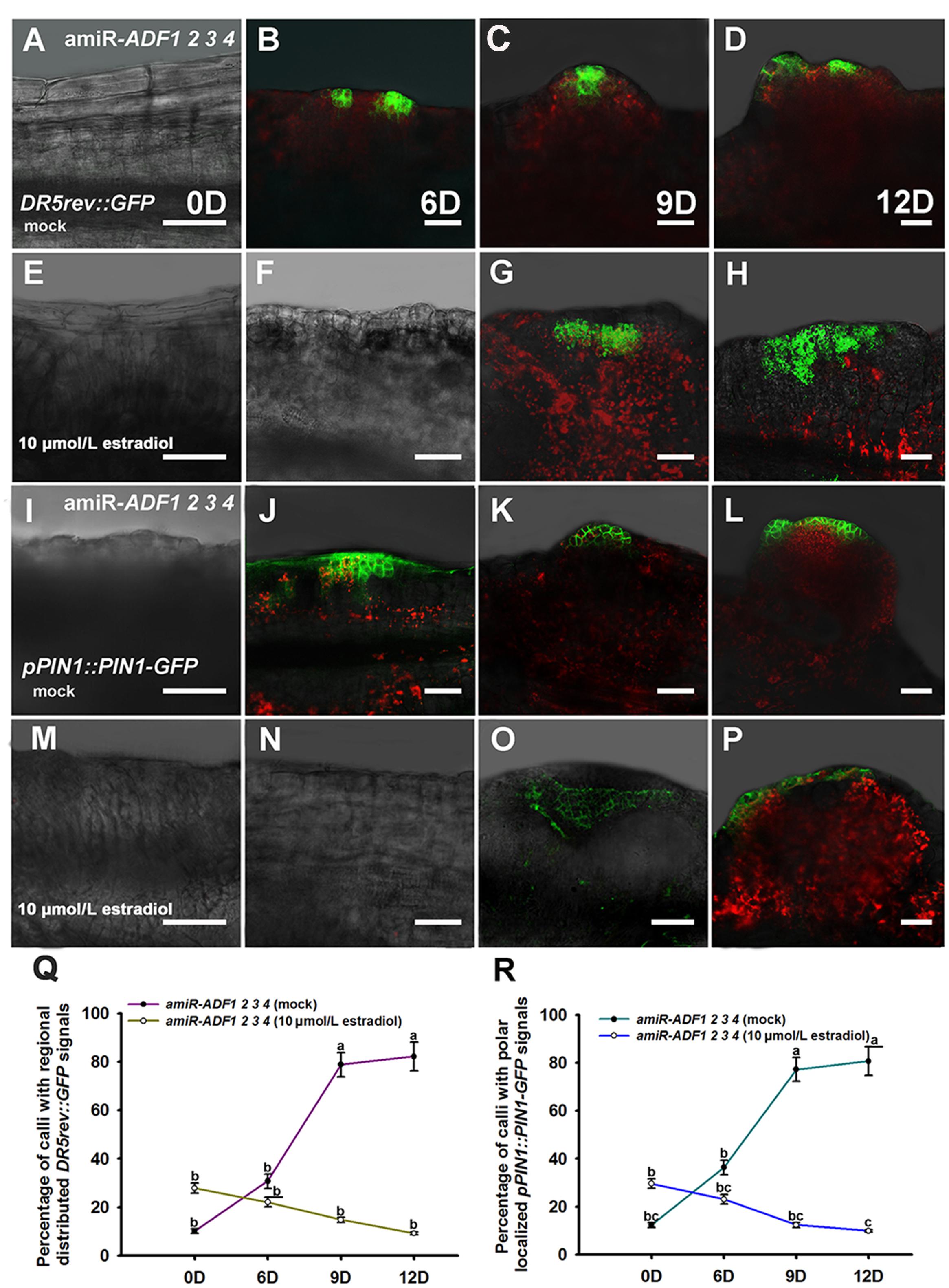
FIGURE 7. Auxin distribution and polar transport in callus from amiR-ADF1 2 3 4 transgenic lines indicated by DR5rev::GFP and pPIN1::PIN1-GFP expression. (A–D) Expression patterns of DR5rev::GFP in callus from non-induced transgenic plants when cultured on SIM without estradiol for 0, 6, 9, and 12 days. (E–H) Expression patterns of DR5rev::GFP in callus from induced transgenic plants when cultured on SIM with 10 μmol/L estradiol for 0, 6, 9, and 12 days. The DR5 signal had a dispersed distribution pattern. (I–L) Expression patterns of pPIN1::PIN1-GFP in callus from non-induced transgenic plants when cultured on SIM without estradiol for 0, 6, 9, and 12 days. (M–P) Expression patterns of pPIN1::PIN1-GFP in callus from induced transgenic plants when cultured on SIM with 10 μmol/L estradiol for 0, 6, 9, and 12 days. Weak and dispersed signals of pPIN1::PIN1-GFP were detected within estradiol–induced callus of amiR-ADF1 2 3 4 lines. Bars = 50 μm. (Q) Percentage of calli with regional distributed DR5rev::GFP signals. (R) Percentage of calli with polar localized pPIN1::PIN1-GFP signals. Different lowercases in (Q,R) are significantly different by ANOVA test, P < 0.01.
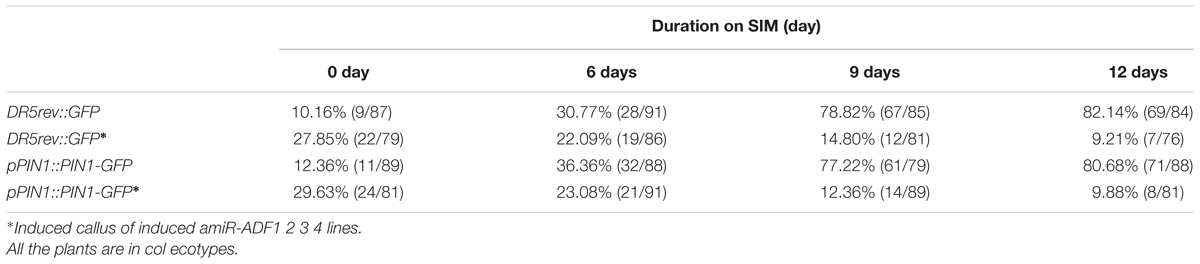
TABLE 3. Percentage of calli with regional distributed DR5rev::GFP signals or polar localized pPIN1::PIN1-GFP signals.
To determine whether auxin polar transport was disturbed when the ADFs were inhibited, the pPIN1::PIN1-GFP lines were crossed with the artificial microRNA transgenic plants. Polar localization of PIN1 was clearly observed at edge regions of some non-induced calli for 6 days after transfer of the calli to SIM (Figures 7I,J,R; Table 3). PIN1 became more polarized in calli for 9 days and 12 days during incubation on SIM without estradiol (Figures 7K,L,R; Table 3). However, weak and dispersed signals of pPIN1::PIN1-GFP were detected within estradiol–induced callus of amiR-ADF1 2 3 4 lines (Figures 7M–P,R; Table 3). These results suggested that inhibition of ADFs disrupted the auxin distribution and polar transport during in vitro shoot regeneration.
Actin filament dynamics are essential for multiple developmental processes in plant cells, especially polarized growth and pattern establishment (Vantard and Blanchoin, 2002). In the process of maize somatic embryogenesis, rearrangement of the cytoskeleton is important for the most critical switch from non-polar to polar units in the embryogenic cells (Samaj et al., 2003), indicating that cytoskeleton organization are essential for the induction of somatic embryogenesis. In animal cells, the cytoskeleton is associated with stem cell specification and the cell division direction (Mammoto and Ingber, 2009; Yima et al., 2009). The cytoskeleton-associated proteins function in both mitosis and cytokinesis in plant cells (Li et al., 2015). For example, Myosin VIII, which is an actin-based molecular motor, plays a role in guiding phragmoplast expansion to the cortical division site by association with both microtubule ends and actin (Wu and Bezanilla, 2014). Moreover, the polarity and axes de novo induction during cell wall formation are controlled by dynamic actin in protoplasts (Zaban et al., 2013). Further, in Arabidopsis nuclei, actin subclass I variants ACT2 and ACT8 were sub-localized throughout the nucleoplasm while subclass II variant ACT7 was found more concentrated in nuclear speckles, which confirm the existence of actin in plant nucleus (Kandasamy et al., 2010). Auxin polar transport has been identified as a central element of pattern formation. It is interesting that auxin controls its own transport by changing the construction of actin filaments (Maisch and Nick, 2007). In this study, we showed that stem cell formation was associated with the organization of the microfilament cytoskeleton during in vitro shoot regeneration. With phalloidin treatment or suppression of ADFs which are involved in actin polymerization at the shoot induction stage, normal actin filament depolymerization was inhibited and stem cell formation was disrupted. As the important roles of actin dynamics in mitosis and cytokinesis, polarity induction during cell wall formation and nuclear transport, we inferred that a general impairment of cell growth or division due to actin stabilization might occur with phalloidin treatment or suppression of ADFs during in vitro formation of the shoot meristem.
Auxin is a multi-functional phytohormone that regulates nearly all aspects of plant growth and development (Teale et al., 2006). Recent studies have shown that auxin modulates cell polarization by activating the ROP GTPase signaling pathway, which directly regulates cytoskeletal dynamics and organization (Lin et al., 2015). Furthermore, actin filaments participate in auxin transport by enabling actin-dependent trafficking of auxin transport components (Muday, 2000; Dhonukshe et al., 2008; Nick et al., 2009). A polarized auxin distribution is also critical for initiation of the shoot primordium during the de novo formation of a shoot meristem (Gordon et al., 2007). We showed here that both the auxin polar transport and auxin gradient distribution were disrupted by inhibiting actin depolymerization through phalloidin treatment or simultaneous suppression of ADF1. ADF2. ADF3, and ADF4 expressions, resulting in low frequencies and rates of shoot regeneration. These results suggest that inhibition of microfilament depolymerization disrupts the normal polar transport and distribution of auxin, which are required for the in vitro formation of the shoot meristem.
A total of 11 ADF genes have been characterized in Arabidopsis and are divided into four subclasses (Ruzicka et al., 2007). ADF1 has been reported to control the actin organization and affect multiple cellular and tissue morphogenesis processes, such as root hair growth, flowering timing and hypocotyl growth (Dong et al., 2001; Gilliland et al., 2002; Bamburg and Bernstein, 2008). ADF7 is specifically expressed in pollen and regulates actin cable turnover to promote normal pollen tube growth by severing actin filaments (Zheng et al., 2013). Moreover, knockdown of ADF genes in the moss Physcomitrella patens leads to a star-like radiating distribution of the cytoskeleton and results in the inhibition of tip growth. However, complementation of the ADF genes recovers the cytoskeleton distribution to a normal “fringe” structure and enables normal tip growth (Augustine et al., 2008). In this study, we showed that a single adf mutation had nearly no effect on shoot regeneration, while simultaneously suppressing ADF1. ADF2. ADF3, and ADF4 using an artificial microRNA method resulted in irregular frequencies and rates of shoot regeneration. These results suggest that ADF genes are functionally redundant in regulating microfilament organization and important for in vitro formation of the shoot meristem.
We showed the organization of the microfilament cytoskeleton during in vitro shoot regeneration and demonstrated that ADFs are critical for actin filament depolymerization, which are required for stem cell formation. Furthermore, the polar transport and distribution of auxin which play important roles during normal shoot organogenesis are involved in actin filament depolymerization. These results provide new molecular insight into the regulation of the organization of the microfilament cytoskeleton in stem cell initiation and de novo shoot regeneration.
LT and YS designed the research. LT and XL performed the research. YD analyzed the data. YS and XZ wrote the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was supported by grants from the National Natural Science Foundation of China (91217308 and 31170272).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00158/full#supplementary-material
Allwood, E. G., Anthony, R. G., Smertenko, A. P., Reichelt, S., Drobak, B. K., Doonan, J. H., et al. (2002). Regulation of the pollen-specific actin-depolymerizing factor LIADF1. Plant Cell 14, 2915–2927. doi: 10.1105/tpc.005363
Andrianantoandro, E., and Pollard, T. D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24, 13–23. doi: 10.1016/j.molcel.2006.08.006
Augustine, R. C., Vidali, L., Kleinman, K. P., and Bezanilla, M. (2008). Actin depolymerizing factor is essential for viability in plants, and its phosphoregulation is important for tip growth. Plant J. 54, 863–875. doi: 10.1111/j.1365-313X.2008.03451.x
Bamburg, J. R., and Bernstein, B. W. (2008). ADF/cofilin. Curr. Biol. 18, R273–R275. doi: 10.1016/j.cub.2008.02.002
Bhojwani, S. S., and Razdan, M. K. (1996). Plant Tissue Culture: Theory and Practice, A Revised Edition. New York, NY: Elsevier Press.
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. doi: 10.1126/science.289.5479.617
Burgos-Rivera, B., Ruzicka, D. R., Deal, R. B., McKinney, E. C., King-Reid, L., and Meagher, R. B. (2008). ACTIN DEPOLYMERIZING FACTOR9 controls development and gene expression in Arabidopsis. Plant Mol. Biol. 68, 619–632. doi: 10.1007/s11103-008-9398-1
Che, P., Gingerich, D. J., Lall, S., and Howell, S. H. (2002). Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 14, 2771–2785. doi: 10.1105/tpc.006668
Cheng, Z. J., Wang, L., Sun, W., Zhang, Y., Zhou, C., Su, Y. H., et al. (2013). Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytoskenin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol. 161, 240–251. doi: 10.1104/pp.112.203166
Cheng, Z. J., Zhao, X. Y., Shao, X. X., Wang, F., Zhou, C., Liu, Y. G., et al. (2014). Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 26, 1053–1068. doi: 10.1105/tpc.113.121566
Cheng, Z. J., Zhu, S. S., Gao, X. Q., and Zhang, X. S. (2010). Cytokinin and auxin regulates WUS induction and inflorescence regeneration in vitro in Arabidopsis. Plant Cell Rep. 29, 927–933. doi: 10.1007/s00299-010-0879-8
Colón-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Spatio-temporal analysis of mitotic activity with a labile cyclin–GUS fusion protein. Plant J. 20, 503–508. doi: 10.1046/j.1365-313x.1999.00620.x
Dhonukshe, P., Grigoriev, I., Fischer, R., Tominaga, M., Robinson, D. G., Hašek, J., et al. (2008). Auxin transport inhibitors impair vesicle motility and actin cytoskeleton dynamics in diverse eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 105, 4489–4494. doi: 10.1073/pnas.0711414105
Dodsworth, S. (2009). A diverse and intricate signalling network regulates stem cell fate in the shoot apical meristem. Dev. Biol. 336, 1–9. doi: 10.1016/j.ydbio.2009.09.031
Dong, C. H., Xia, G. X., Hong, Y., Ramachandran, S., Kost, B., and Chua, N. H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13, 1333–1346. doi: 10.1105/tpc.13.6.1333
Fletcher, J. C., Brand, U., Running, M. P., Simon, R., and Meyerowitz, E. M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. doi: 10.1126/science.283.5409.1911
Gallois, J. L., Nora, F. R., Mizukami, Y., and Sablowski, R. (2004). WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Gene. Dev. 18, 375–380. doi: 10.1101/gad.291204
Gamborg, O. L., Miller, R. A., and Ojima, K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50, 151–158. doi: 10.1016/0014-4827(68)90403-5
Gilliland, L. U., Kandasamy, M. K., Pawloski, L. C., and Meagher, R. B. (2002). Both vegetative and reproductive actin isovariants complement the stunted root hair phenotype of the Arabidopsis act2-1 mutation. Plant Physiol. 130, 2199–2209. doi: 10.1104/pp.014068
Gordon, S. P., Heisler, M. G., Reddy, G. V., Ohno, C., Das, P., and Meyerowitz, E. M. (2007). Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134, 3539–3548. doi: 10.1242/dev.010298
Heisler, M. G., Ohno, C., Das, P., Sieber, P., Reddy, G. V., Long, J. A., et al. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899–1911. doi: 10.1016/j.cub.2005.09.052
Henty-Ridilla, J. L., Li, J., Day, B., and Staiger, C. J. (2014). ACTIN DEPOLYMERIZING FACTOR4 regulates actin dynamics during innate immune signaling in Arabidopsis. Plant Cell 26, 340–352. doi: 10.1105/tpc.113.122499
Higaki, T., Kutsuna, N., Sano, T., Kondo, N., and Hasezawa, S. (2010). Quantification and cluster analysis of actin cytoskeletal structures in plant cells: role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J. 61, 156–165. doi: 10.1111/j.1365-313X.2009.04032.x
Jiang, C. J., Weeds, A. G., and Hussey, P. J. (1997). The maize actin-depolymerizing factor, ZmADF3, redistributes to the growing tip of elongating root hairs and can be induced to translocate into the nucleus with actin. Plant J. 12, 1035–1043. doi: 10.1046/j.1365-313X.1997.12051035.x
Kandasamy, M. K., Gilliland, L. U., McKinney, E. C., and Meagher, R. B. (2001). One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell 13, 1541–1554. doi: 10.1105/TPC.010026
Kandasamy, M. K., McKinney, E. C., and Meagher, R. B. (2010). Differential sublocalization of actin variants within the nucleus. Cytoskeleton 67, 729–743. doi: 10.1002/cm.20484
Laux, T., Mayer, K. F., Berger, J., and Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96.
Lenhard, M., and Laux, T. (2003). Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 130, 3163–3173. doi: 10.1242/dev.00525
Li, L., Rao, K. N., Zheng-Le, Y., Hurd, T. W., Lillo, C., and Khanna, H. (2015). Loss of retinitis pigmentosa 2 (RP2) protein affects cone photoreceptor sensory cilium elongation in mice. Cytoskeleton (Hoboken) 72, 447–454. doi: 10.1002/cm.21255
Li, W., Liu, H., Cheng, Z. J., Su, Y. H., Han, H. N., Zhang, Y., et al. (2011). DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet. 7:e1002243. doi: 10.1371/journal.pgen.1002243
Lin, D., Ren, H., and Fu, Y. (2015). ROP GTPase-mediated auxin signaling regulates pavement cell interdigitation in Arabidopsis thaliana. J. Integr. Plant Biol. 57, 31–39. doi: 10.1111/jipb.12281
Maisch, J., and Nick, P. (2007). Actin is involved in auxin-dependent patterning. Plant Physiol. 143, 1695–1704. doi: 10.1104/pp.106.094052
Mammoto, A., and Ingber, D. E. (2009). Cytoskeletal control of growth and cell fate switching. Curr. Opin. Cell Biol. 21, 864–870. doi: 10.1016/j.ceb.2009.08.001
Mayer, K. F., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. doi: 10.1016/S0092-8674(00)81703-1
Muday, G. K. (2000). Maintenance of asymmetric cellular localization of an auxin transport protein through interaction with the actin cytoskeleton. J. Plant Growth Regul. 19, 385–396. doi: 10.1007/s003440000041
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plantarum 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nick, P., Han, M. J., and An, G. (2009). Auxin stimulates its own transport by shaping actin filaments. Plant Physiol. 151, 155–167. doi: 10.1104/pp.109.140111
Pan, X., Chen, J., and Yang, Z. (2015). Auxin regulation of cell polarity in plants. Curr. Opin. Plant Biol. 28, 144–153. doi: 10.1016/j.pbi.2015.10.009
Pei, W., Du, F., Zhang, Y., He, T., and Ren, H. (2012). Control of the actin cytoskeleton in root hair development. Plant Sci. 187, 10–18. doi: 10.1016/j.plantsci.2012.01.008
Porter, K., Shimono, M., Tian, M., and Day, B. (2012). Arabidopsis actin-depolymerizing factor-4 links pathogen perception, defense activation and transcription to cytoskeletal dynamics. PLoS Pathog. 8:e1003006. doi: 10.1371/journal.ppat.1003006
Ruzicka, D. R., Kandasamy, M. K., McKinney, E. C., Burgos-Rivera, B., and Meagher, R. B. (2007). The ancient subclasses of Arabidopsis actin depolymerizing factor genes exhibit novel and differential expression. Plant J. 52, 460–472. doi: 10.1111/j.1365-313X.2007.03257.x
Samaj, J., Baluska, F., Pretová, A., and Volkmann, D. (2003). Auxin deprivation induces a developmental switch in maize somatic embryogenesis involving redistribution of microtubules and actin filaments from endoplasmic to cortical cytoskeletal arrays. Plant Cell Rep. 21, 940–945. doi: 10.1007/s00299-003-0611-z
Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F., Jürgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. doi: 10.1016/S0092-8674(00)80700.x
Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18, 1121–1133. doi: 10.1105/tpc.105.039834
Skoog, F., and Miller, C. O. (1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 11, 118–130.
Su, Y. H., Zhao, X. Y., Liu, Y. B., Zhang, C. L., O’Neil, S. D., and Zhang, X. S. (2009). Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J. 59, 448–460. doi: 10.1111/j.1365-313X.2009.03880.x
Sugiyama, M. (2000). Genetic analysis of plant morphogenesis in vitro. Int. Rev. cytol. 196, 67–84. doi: 10.1016/S0074-7696(00)96002-9
Takáč, T., Bekešová, S., and Šamaj, J. (2017). Actin depolymerization-induced changes in proteome of Arabidopsis roots. J. Proteomics 153, 89–99. doi: 10.1016/j.jprot.2016.06.010
Teale, W. D., Paponov, I. A., and Palme, K. (2006). Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7, 847–859. doi: 10.1038/nrm2020
Vantard, M., and Blanchoin, L. (2002). Actin polymerization processes in plant cells. Curr. Opin. Plant Biol. 5, 502–506. doi: 10.1016/S1369-5266(02)00300-X
Wang, Y. S., Yoo, C. M., and Blancaflor, E. B. (2008). Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol. 177, 525–536. doi: 10.1111/j.1469-8137.2007.02261.x
Weigel, D., and Jürgens, G. (2002). Stem cells that make stems. Nature 415, 751–754. doi: 10.1038/415751a
Wu, S., Xie, Y., Zhang, J., Ren, Y., Zhang, X., Wang, J., et al. (2015). VLN2 regulates plant architecture by affecting microfilament dynamics and polar auxin transport in rice. Plant Cell 27, 2829–2845. doi: 10.1105/tpc.15.00581
Wu, S. Z., and Bezanilla, M. (2014). Myosin VIII associates with microtubule ends and together with actin plays a role in guiding plant cell division. Elife 3:e03498. doi: 10.7554/eLife.03498
Xu, J., Hofhuis, H., Heidstra, R., Sauer, M., Friml, J., and Scheres, B. (2006). A molecular framework for plant regeneration. Science 311, 385–388. doi: 10.1126/science.1121790
Yima, E. K., Darling, E. M., Kulangara, K., Guilak, F., and Leong, K. W. (2009). Nanotopography-induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomaterials 31, 1299–1306. doi: 10.1016/j.biomaterials.2009.10.037
Zaban, B., Maisch, J., and Nick, P. (2013). Dynamic actin controls polarity induction de novo in protoplasts. J. Integr. Plant Biol. 55, 142–159. doi: 10.1111/jipb.12001
Zheng, Y., Xie, Y., Jiang, Y., Qu, X., and Huang, S. (2013). Arabidopsis actin-depolymerizing factor 7 severs actin filaments and regulates actin cable turnover to promote normal pollen tube growth. Plant Cell 25, 3405–3423. doi: 10.1105/tpc.113.117820
Keywords: microfilament depolymerization, stem cell formation, auxin distribution, auxin polar transport, shoot regeneration, Arabidopsis
Citation: Tang LP, Li XM, Dong YX, Zhang XS and Su YH (2017) Microfilament Depolymerization Is a Pre-requisite for Stem Cell Formation During In vitro Shoot Regeneration in Arabidopsis. Front. Plant Sci. 8:158. doi: 10.3389/fpls.2017.00158
Received: 23 October 2016; Accepted: 25 January 2017;
Published: 14 February 2017.
Edited by:
Elison B. Blancaflor, Samuel Roberts Noble Foundation, USAReviewed by:
Abidur Rahman, Iwate University, JapanCopyright © 2017 Tang, Li, Dong, Zhang and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Hua Su, c3V5aEBzZGF1LmVkdS5jbg==
†These authors have contributed equally to this work and shared first authorship.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.