- National Engineering Research Center of Plant Space Breeding, South China Agricultural University, Guangzhou, China
The regulation of innate immunity and plant growth, along with the trade-off between them, affects the defense and recovery mechanisms of the plant after it is attacked by pathogens. Although it is known that hormonal crosstalk plays a major role in regulating interaction of plant growth and PAMP-triggered immunity, the relationship between plant growth and effector-triggered immunity (ETI) remains unclear. In a large-scale yeast two-hybrid screening for Pik-H4-interacting proteins, a homeodomain transcription factor OsBIHD1 was identified, which is previously known to function in biotic and abiotic stress responses. The knockout of OsBIHD1 in rice lines carrying Pik-H4 largely compromised the resistance of the rice lines to Magnaporthe oryzae, the fungus that causes rice blast. While overexpression of OsBIHD1 resulted in enhanced expression of the pathogenesis-related (PR) and ethylene (ET) synthesis genes. Moreover, OsBIHD1 was also found to directly bind to the promoter region of ethylene-synthesis enzyme OsACO3. In addition, OsBIHD1 overexpression or deficiency provoked dwarfism and reduced brassinosteroid (BR) insensitivity through repressing the expression of several critical genes involved in BR biosynthesis and BR signaling. During M. oryzae infection, transcript levels of the crucial BR catabolic genes (CYP734A2, CYP734A4, and CYP734A6) were significantly up-regulated in OsBIHD1-OX plants. Furthermore, OsBIHD1 was found to be capable of binding to the sequence-specific cis-elements on the promoters of CYP734A2 to suppress the plant growth under fungal invasion. Our results collectively suggest a model that OsBIHD1 is required for Pik-H4-mediated blast resistance through modulating the trade-off between resistance and growth by coordinating brassinosteroid-ethylene pathway.
Introduction
Plant growth and disease resistance have been regarded as two distinct and divergent systems. However, to fend off pathogens, plants must effectively integrate multiple signals including biotic and abiotic stressors to protect themselves from pathogen invasion (Wang and Wang, 2014). Plant defense responses such as pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) depend upon critical switch, selectively repress growth and focus the energy on resisting pathogen invasion(Liu et al., 2014). The molecular trade-off between the growth and immunity is crucial to the health and survival of plants, which requires integration of the growth and immunity pathways with developmental process (Chandran et al., 2014).
Pik-H4 is an allele of the major resistance (R) gene Pi-k which consists of two adjacent nucleotide-binding domain and leucine-rich repeat (NLR) genes, Pik1-H4 and Pik2-H4 (Xiao et al., 2011). Previous studies have suggested that Pikh-1 directly interacts with the M. oryzae effector Avr-Pik and acts as an adaptor to relay signals between Avr-Pik and Pikh-2 (Zhai et al., 2014). The Avr-PikD and Pikp-1 interaction has been recently dissected from the crystal structure (Lu et al., 2010). These studies illustrate the detailed molecular mechanism of an initial recognition event mediated by NLR proteins that integrate an immunity response to rice blast resistance. On the other hand, there is very little evidence for downstream resistance mechanisms induced by the R proteins under Magnaporthe oryzae attack. The panicle blast resistance protein Pb1 specifically interacts with WRKY45 to regulate the SA immunity pathway. Pb1 overexpression enhances WRKY45 accumulation and shields it from ubiquitin-mediated proteasomal degradation. Accordingly, WRKY45 is an essential downstream regulator involved in Pb1-dependent blast resistance (Inoue et al., 2013).
In a previous yeast two-hybrid screening for Pik-H4 interacting proteins, we identified a homeodomain-containing protein, OsBIHD1, which has been previously found in suppression subtractive hybridization (SSH) assay for different BTH-responsive cDNA clones (Luo et al., 2005a). Here, we further illustrate the function of the OsBIHD1 in Pik-H4-mediated blast resistance. The results show that OsBIHD1 physically interacts with Pik-H4 and is required for Pik-H4-mediated resistance. During M. oryzae invasion, OsBIHD1 regulates blast resistance through direct activation of ET signaling pathway. At the same time, OsBIHD1 suppresses plant growth through directly activating the BR catabiotic genes. This study demonstrates that OsBIHD1, served as a critical molecular switch, coordinates the tradeoff between growth and ETI-triggered immunity in rice.
Materials and Methods
Plant Materials and Treatments
Oryzae sativa japonica cultivar Pik-H4 NIL was used as the wild-type rice strain in this study (Xiao et al., 2011). Pik-H4 NIL contains the Pik-H4 resistance gene (an allele of Pik locus) in the susceptible cultivar LTH background. The M. oryzae race GDYJ7, one of the primary M. oryzae races found in Guangdong Province, China, is incompatible with Pik-H4.
Eight-week-old rice seedlings grown under natural light in a greenhouse at 26°C were used for inoculation of rice blast fungus. For fungal inoculation, freshly prepared M. oryzae spores (1 × 105 conidia/mL 0.02% v/v gelatin) were sprayed onto the rice leaves using an air sprayer. Inoculated plants were kept in a humidity chamber in the dark at 28°C for 24 h, and the plants were then transferred to the normal growth condition. The local lesions were observed 5 days later. The total local lesions area of whole single plant was calculated, and the total area of all the investigated leaves in the whole plant was counted. Differences in blast resistance were determined by the proportion of the lesion area divided by the total leaf area on the same leave. All the experiments were performed in triplicate.
Yeast Two-Hybrid Assay
Coding sequences of Pik1-H4 and were Pik2-H4 cloned into the BD (binding domain) plasmid pGBKT7 by homologous recombination in yeast strain Y2H gold. Yeast cells containing the resulting constructs BD-Pik1-H4 were used as bait to screen for interacting-proteins from a rice yeast two-hybrid cDNA library, according to the manufacturer’s instructions of Clontech yeast two-hybrid handbook. The transformed yeast cells were cultured on SD/-Trp/-Leu and SD/-Trp/-Leu/-His/-Ade+3AT+X-α- GAL plates and results were scored after 3 days incubation at 30°C.
Bimolecular Fluorescence Complementation (BiFC) Assay
For BiFC assays, the coding regions of Pik1-H4 and OsBIHD1 were separately cloned into the AgeI/NheI sites of BiFC vectors pUC-NE1L2L-nsI and pUC-CE1RL2R-nsI, to generate Pik1-H4-nYFP and OsBIHD1-cYFP constructs labeled at their amino and carboxyl termini, respectively (Luo et al., 2013). Rice protoplasts were isolated based on the methods reported by Yang et al. (2014) with slight modifications. Briefly, 50 rice seedlings were cut into approximately 0.5 mm strips, and then incubated in an enzyme solution (1.5% Cellulase RS, 0.75% Macerozyme R-10, 0.6 M mannitol, 10 mM MES at pH 5.7, 10 mM CaCl2 and 0.1% BSA) for 4–5 h in the dark with gentle shaking (60–80 rpm). After washing twice with W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES at pH 5.7), the residues were resuspended in MMG solution (0.4 M mannitol, 15 mM MgCl2, and 4 mM MES at pH 5.7). The recombinant constructs in pairs were co-transfected into rice protoplasts and the fluorescent signals were examined by confocal microscopy Carl Zeiss LSM780.
GST Pull Down Assay
The full length Pik1-H4 CC domain cDNA sequence including stop codon was cloned into the BamHI/EcoRI sites of pGEX4p-1 and the OsBIHD1207-527 aa cDNA insert was cloned into the BamHI sites of pET28a. Expression of the Pik1-H4 CC-GST and OsBIHD1207-527 aa-His fusion proteins were induced with 0.5 mM IPTG (isopropyl β-D-thioglucosidase) for 12 h at 37°C in Escherichia coli strain BL21.The His-tagged proteins were incubated with purified GST-Pik1-H4 CC or GST alone bound to glutathione beads. After 4 h of incubation at 4°C, the beads were extensively washed four times with GST binding buffer (PBS, pH 7.2). Components bound to the beads were eluted by boiling in SDS sample buffer, and then separated on a SDS-PAGE gel and immune-blotted with anti-His and anti-GST antibodies.
Total RNA Extraction, Real-Time PCR Analysis of Gene Expression
Total RNA was extracted from 100 mg of fourth-leaf-stage rice seedling with Trizol Reagent (Invitrogen, Beijing, China) and reverse-transcribed using PrimeScript RT reagent Kit (Takara, Dalian, China) according to the manufacturer’s instructions. The cDNA was quantified by real-time PCR using a 20 μl reaction volume and SYBR Premix ExTaqTM (TaKaRa, Dalian, China) on an ABI StepOne Plus system. Primers used for PCR analysis are shown in Supplementary Table S2. Differences in gene expression were expressed as fold change relative to control and were calculated using the 2-ΔΔCT method. Each measurement was carried out in triplicate, and the error bars represent SE of the mean of fold changes for three biological replicates.
Generation of the OsBIHD1-OX and osbihd1-ko Transgenic Plants
The full-length of OsBIHD1 cDNA was isolated by RT-PCR from the leaves of fourth-leaf-stage rice plants using the cDNA F/R primers (Supplementary Table S1) encompassing the translation start and stop codons. This cDNA insert was digested with BamHI and cloned between the maize ubiquitin promoter and the Nos terminator in the plant expression vector pOX containing the hygromycin resistance gene as selection maker. CRISPR/Cas9 technology was used to generate osbihd1-ko plants. As reported by Ma et al. (2015), a 20 bp DNA fragment including a protospacer-adjacent motif (PAM) of the first exon of OsBIHD1 nucleotide sequence was fused with a U6a-gRNA box, and the resulting DNA insert digested with BsaI was inserted into the pYLCRISPR/Cas9PUbi-Hi vector. pOX-OsBIHD1 and pYLCRISPR/Cas9-OsBIHD1 were then introduced into agrobacterium strain EHA105 and then transformed to wild-type (Pik-H4 NIL) calli, as described previously. Transgenic rice plants were regenerated from the transformed calli on selection media containing 50 mg/L hygromycin and 250 mg/L cefotaxime. OsBIHD1 levels in the transgenic rice plants were further confirmed with target site sequencing and real-time PCR.
Yeast One-Hybrid Assay
The 2 Kb promoter sequences of OsACO3 and CYP734A2 were cloned into the EcoRI/MluI sites of pHIS2 (Clontech) and full length cDNA of OsBIHD1 was inserted into the NdeI/EcoRI sites of AD2 (Clontech). The constructs were co-transformed into the yeast strain AH109 (Clontech). The positive transformants were grown on SD/-Trp/-Leu/-His plates containing 100 mM 3-AT and 10 mM X-α-gal for 3 days at 30°C.
EMSA Assay
DNA binding activity of the recombinant OsBIHD1207-527aa protein was analyzed by an electrophoretic mobility shift assay (EMSA) assay. Labeled probe contained the OsBIHD1 binding site (TGTCA) and the competitor oligonucleotide contained a mutated OsBIHD1 binding motif (TCTCA). Synthesized probe and competitor fragments from the 3′ terminus were annealed and labeled with biotin (Invitrogen). DNA binding reactions were performed at 25°C for 30 min in binding buffer (Chemiluminescent EMSA Kit GS009, Beyotime biotechnology) and subjected to EMSA assay using 10% polyacrylamide gels in 0.5x Tris-borate-EDTA buffer.
Lamina Joint Test
The lamina joint tests were performed as described by Chen et al. (2015) with slight modifications. Sterilized seeds were germinated in water for 3 days and well-germinated plants were transferred onto 1/2 × Murashige and Skoog medium containing 1% agar and grown for an additional 5 days. Following this, a 500 ng/mL solution of brassinolide (Sigma–Aldrich) in 100% ethanol was applied to the tip of the second leaf blade. Three days later the lamina joint angles of the second leaves were measured.
Transcriptional Activity Assay in Tobacco Leaves and ChIP-qPCR
The promoter sequences of OsACO3 and CYP734A2 were cloned into the BamHI/NcoI sites of pCAMBIA1305, the last constructs were co-transformed into tobacco leaves with 35S:OsBIHD1 induced by Agrobacterium (strain: EHA105). The tobacco leaves were incubated in a solution containing 50 mM NaPO4 buffer (pH 7.0), 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 0.1% Triton X-100, and 1 mM X-Gluc at 37°C. CHIP-qPCR was performed as described previously (Yang et al., 2014) Rice protoplast isolated from 200 rice seedlings that were cut into approximately 0.5 mm strips, and then incubated in an enzyme solution (1.5% Cellulase RS, 0.75% Macerozyme R-10, 0.6 M mannitol, 10 mM MES at pH 5.7, 10 mM CaCl2 and 0.1% BSA) for 4–5 h in the dark with gentle shaking (60–80 rpm). OsBIHD1-GFP and empty GFP were transiently co-expressed in rice protoplasts by 40% PEG induction. Then, harvested and extracted the total protoplast chromatin. The chromatin preparations were sonicated into 0.2–0.5 kb fragments. Specific antibodies against GFP (Cat.No.11814460001, Roche) were added to the chromatin solution, which was precleared with salmon sperm DNA/Protein A-agarose beads. The precipitates were eluted from the beads. Cross-links were reversed, and residual proteins were removed by incubation with proteinase K. DNA was recovered using the QIAquick spin column (Qiagen, Valencia, CA, USA). Quantitative PCR was used to determine the amounts of genomic DNA enriched in the chromatin samples. The primers were designed to amplify DNA fragments of 150–250 bp (Yun et al., 2012).
Results
Pik-H4 Interacts with OsBIHD1 through Its CC Domain
To investigate rice proteins interacting with Pik1-H4 (one protein of the resistance protein pair Pik1-H4/Pik2-H4), we previously performed a yeast two-hybrid screen using a rice cDNA library and identified a homeodomain transcription factor OsBIHD1. The HD domain (207–527 aa) of OsBIHD1 was sufficient for binding to Pik1-H4 (Figure 1A), while this domain didn’t show any interacting with Pik2-H4 (Supplementary Figure S1). To confirm the specificity of this interaction, we sought to identify the OsBIHD1-interacting sites in the Pik1-H4 molecule which consists of four domains: CC (coiled-coil: 1–266 aa), NBS (nucleotide-binding: 267–634 aa), LRR (Leucine-rich repeats: 635–1040 aa), non-LRR domain (carboxyl-terminus: 1041–1114 aa). The strongest binding to OsBIHD1 or HD domain was achieved with the full-length Pik1-H4 and CC domain, whereas the NBS, LRR and non-LRR domains did not show any interaction (Figure 1A).
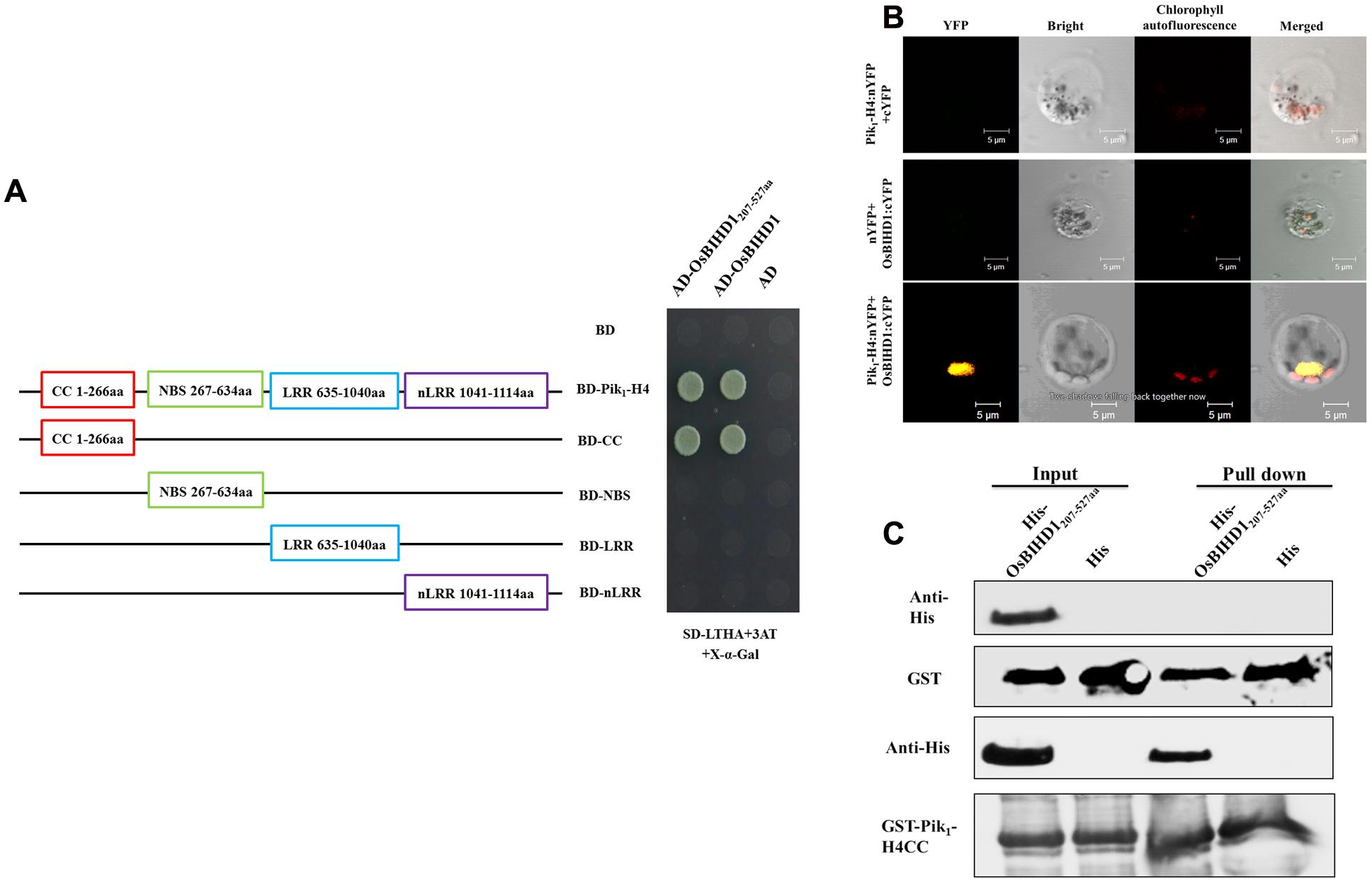
FIGURE 1. Interaction between Pik1-H4 and OsBIHD1. (A) Yeast two-hybrid analysis to confirm the specificity of the interaction between Pik1-H4 and OsBIHD1. CC domain (1–266 aa), NBS domain (367–634 aa), LRR domain (635–1040 aa) and nLRR domain (1041–1114 aa) of Pik1-H4 were fused to GAL4 DNA-binding domain, respectively, and expressed in combination with truncation construct of OsBHD1 (207–527 aa) fused to the GAL4 activation domain (AD) in yeast strain Y2H Gold. (B) Bimolecular fluorescence complementation (BiFC) analysis of Pik1-H4-OsBIHD1 interaction in rice protoplasts. cDNA coding for the full length Pik1-H4 protein was fused to the N-terminal half of YFP and co-transformed in rice protoplasts together with the cDNA coding for OsBIHD1 fused to the C-terminal moiety of YFP. The yellow fluorescence (YFP), a bright field image and chlorophyll autofluorescence (Chl) were recorded and the resulting images were merged. Scale bar is 5 μm. (C) In vitro GST pull-down assay between the Pik1-H4 CC and OsBIHD1207-527 aa. Hexahistidine-tagged OsBIHD1207-527aa (His-OsBIHD1207-527aa) and GST-fused Pik1-H41-266aa (GST-Pik1-H4 CC) were expressed in Escherichia coli and used for the analysis.
To confirm our yeast two-hybrid results, we used bimolecular fluorescence complementation (BiFC) assay to test the interaction between Pik1-H4 and OsBIHD1 in rice protoplast cells. Rice protoplast cells that were co-transfected with the vectors expressing Pik1-H4:nYFP and OsBIHD1:cYFP displayed YFP fluorescence under laser confocal scanning microscopy (Figure 1B). Further subcellular localization assay showed that Pik1-H4 and OsBIHD1 co-localized in the nucleus (Supplementary Figure S2A), indicating that Pik1-H4 interacts with OsBIHD1 in the nucleus. Direct binding was also observed between recombinant Pik1-H4 CC and OsBIHD1 HD in vitro in a GST pull-down assay (Figure 1C). Taken together, these results confirm a direct interaction between Pik1-H4 and OsBIHD1, especially between Pik1-H4 CC domain and OsBIHD1 HD domain, and the interaction may happen in the nucleus of rice cells.
To determine whether the OsBIHD1 HD region was sufficient for transcriptional activation, we tested three truncated mutants of OsBIHD1. The transcriptional activity assay indicated that the HD domain was not active in this assay whereas the full-length protein was (Supplementary Figure S2B). This result indicates that the HD region is responsible for interacting with Pik1-H4 and the activity domain is contained within OsBIHD11-180 aa.
Pik-H4 Mediated Blast Resistance Depends on OsBIHD1
We firstly examined the expression pattern of OsBIHD1 over a time course of 72 h after inoculation with M. oryzae by quantitative RT-PCR (qRT-PCR). The OsBIHD1 expression at the mRNA level was significantly increased at 6 h and reached its lowest level at 36 h, and then it was decreased at a relatively low level from 60 to 72 h after inoculation in wild-type plants. In addition, the transcript abundances of Pik1-H4 and Pik2-H4 also were up-regulated over a time course of 48 h after inoculation of blast fungus (Supplementary Figure S3).
To understand the function of OsBIHD1, we introduced OsBIHD1 overexpression (OsBIHD1-OX) and OsBIHD1 knock-out (osbihd1-ko) constructs, respectively, into wild type carrying Pik-H4, generating the rice lines Pik-H4+/OsBIHD1-OX and Pik-H4+/osbihd1-ko, respectively (Supplementary Figure S4 and Figure 2A). The expression levels of Pik1-H4 and Pik2-H4 (resistance gene cluster Pik1-H4/Pik2-H4) in Pik-H4+/OsBIHD1-OX and Pik-H4+/osbihd1-ko rice lines were similar to those in the wild-type lines (Supplementary Figure S5), indicating that the OsBIHD1 overexpression or knock-out did not affect Pik1-H4 expression in these transformants. A blast resistance test with M. oryzae race GDYJ7 (carrying Avr-PikH4) showed that the resistance was compromised in osbihd1-ko plants (Figures 2B,C), in which OsBIHD1 transcript levels in leaves were very low (Supplementary Figure S4), while the levels of resistance in Pik-H4+/OsBIHD1-OX plants were significantly increased compared with the wild type plants (Figure 2C).
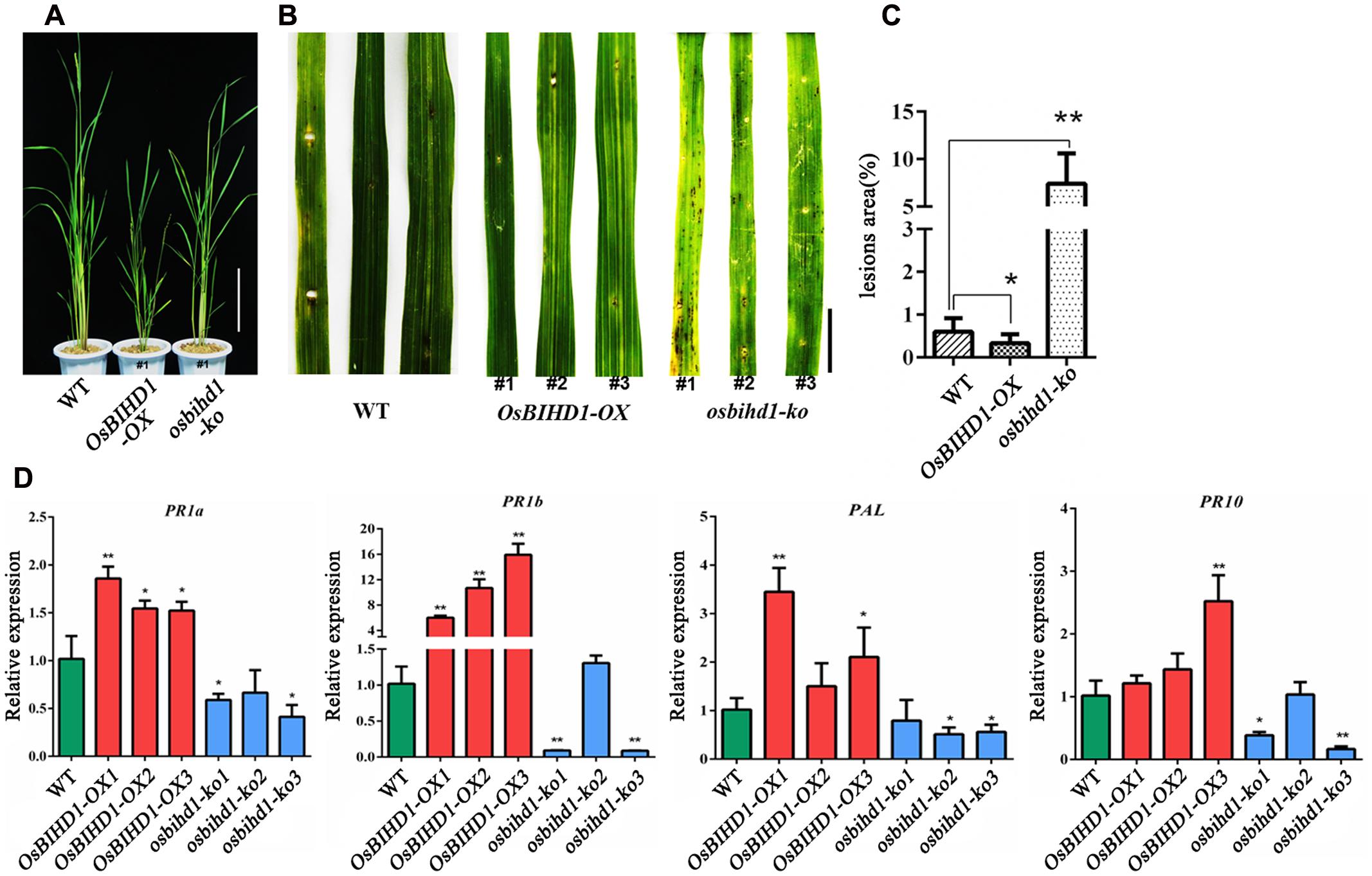
FIGURE 2. Pik1-H4-mediated blast resistance requires OsBIHD1. (A) Phenotype of Wild-type and OsBIHD1 transgenic plants. Scale bar is 20 cm. (B) Photographs of blast fungus-inoculated fourth leaves of OsBIHD1 transgenic plants and wild-type at the six-leaf stage. A conidial suspension of blast fungus race GDYJ7 was sprayed on the leaf surfaces, and local lesions were observed 5 days later. Bar is 2 cm. (C) Lesion and total leaf areas measured in 10 independent plants. The indicated values show the ratio of lesion area to leaf area mean ± SD. Asterisks indicate a significant difference according to the t-test (∗P < 0.05, ∗∗P < 0.01) compared with wild-type. (D) Relative expression of pathogen-related genes (PR1a, PR1b, PR10, and PAL) in wild-type, OsBIHD1-OX, and osbihd1-ko plants. Values are mean ± SD of three biological replicates, and asterisks indicate a significant difference according to the t-test (P < 0.05) compared with WT.
Previous studies have shown that expression of OsBIHD1 was activated on treatment with benzothiadiazole (BTH) and OsBIHD1 overexpression resulted in an elevated level of defense-related PR-1gene expression in tobacco leaves (Luo et al., 2005b). To further test whether OsBIHD1 is involved in induction or accumulation of PR-protein mRNAs in rice, the transcript levels of four PR genes: PR1a, PR1b, PR10, and PAL, were investigated in OsBIHD1-OX and osbihd1-ko plants using qRT-PCR. The results showed that the expression of all these genes was down-regulated in osbihd1-ko lines in comparison with wild-type plants under normal growth condition, while that of all these genes were significantly up-regulated in OsBIHD1-OX lines (Figure 2D). These results indicate that OsBIHD1 is required Pik-H4-mediated blast resistance and acts as a positive regulator downstream of defense signaling transduction through affecting the expression of PR genes.
OsBIHD1 Activates the ET-Dependent Defense Pathway
Accumulating evidences have illustrated that exogenous hormones such as SA, JA, and ET precursor 1-amino-cyclopropane-1-carboxylic acid (ACC) are involved in inducing the expression PR genes (Takeuchi et al., 2011). It is possible that OsBIHD1 may regulate the expression of PR genes through affecting the hormone-regulated pathway. The expression of a subset of key genes involved in ET biosynthesis including ACO1 (Iwamoto et al., 2010), ACO2 (Chae et al., 2000), ACO3 (Iwai et al., 2006), and ACS1 (Iwai et al., 2006) were analyzed with qRT-PCR in Pik-H4+/OsBIHD1-OX and Pik-H4+/osbihd-ko transgenic plants. The results showed that the transcript levels of ACO family genes were all up-regulated in OsBIHD1-OX plants while that of the ACS family gene were only slightly alerted (Figures 3A–D; Supplementary Figure S6). In addition, OsBIHD1 expression could be induced by exogenous application of ACC (Figure 3E). These results suggest that OsBIHD1 is most likely involved in ET-mediated immunity.
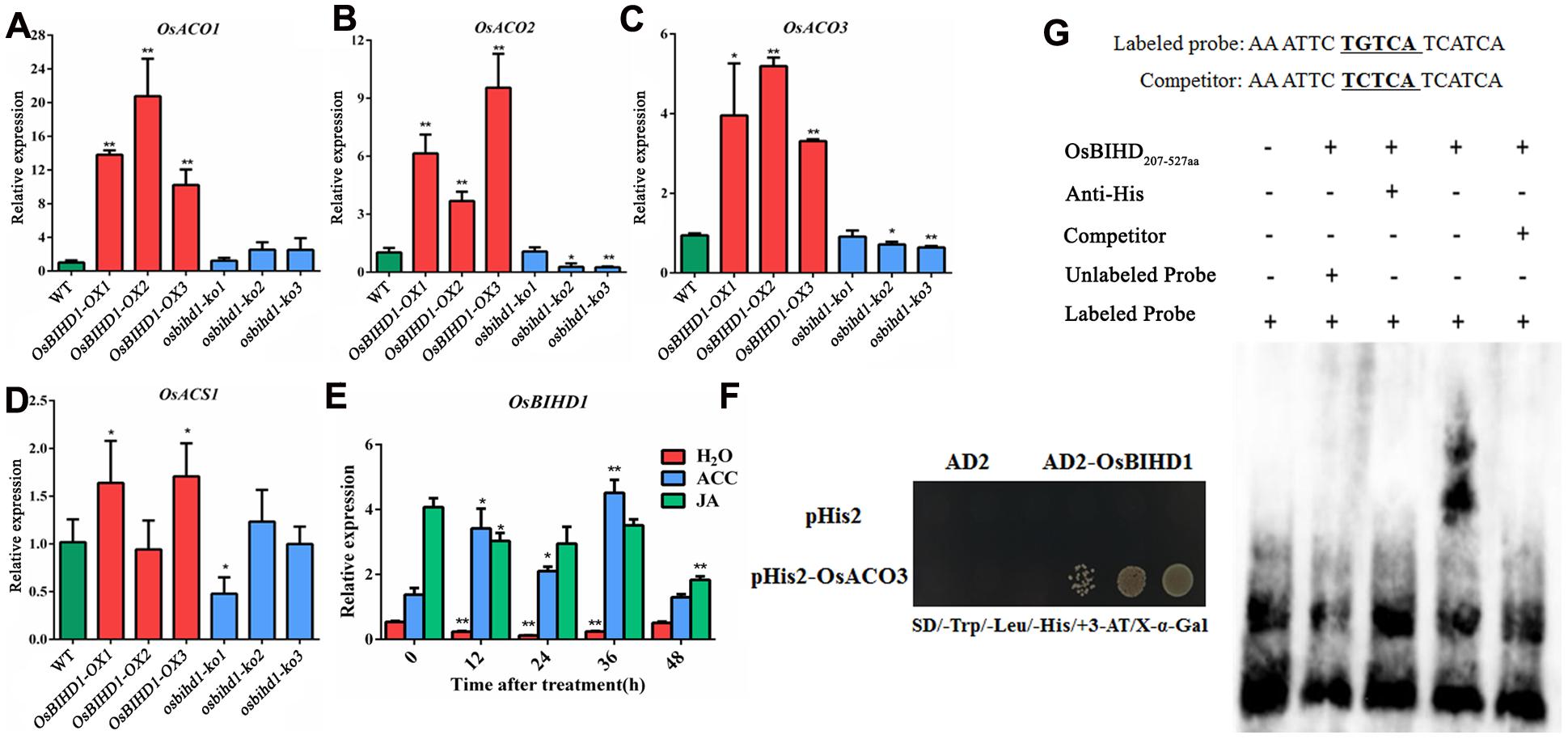
FIGURE 3. OsBIHD1 up-regulates ET synthetic gene expression and directly binds the OsACO3 promoter sequence. (A–D) Quantitative real-time PCR analyses of OsACS1, OsACO1, OsACO2, and OsACO3 transcripts in WT and OsBIHD1 transgenic plants, and asterisks indicate a significant difference according to the t-test (P < 0.05) compared with WT. (E) OsBIHD1 expression induced by exogenous ACC (1 mM) after 48 h.Sterile H2O and exogenous JA (100 μM) were used as controls. Values shown are means ± SD from three independent experiments, and asterisks indicate a significant difference according to the t-test (P < 0.05) compared with 0 h. (F) OsBIHD1 binds the OsACO3 promoter sequence in vivo. Yeast one-hybrid assay results using an X-Gal assay. (G) EMSA assay illustrating that the OsBIHD1207-527 aa protein binds to the promoter cis-element TGTCA of OsACO3 in vitro.
OsBIHD1 encodes a homeodomain protein with DNA binding activity and directly binds to TGTCA motif in the cis-element sequence. To identify whether the motif is present in the promoters of the ET biosynthesis genes, we used the plant cis-acting regulatory DNA elements (PLACE) database (Higo et al., 1999) and found there were many potential OsBIHD1 binding sites in the promoter region of OsACO3 (Supplementary Figure S7). To further elucidate whether OsBIHD1 directly activated the expression of OsACO3, yeast one-hybrid assay and an EMSA were carried out. Our results showed that OsBIHD1 physically bound to the cis-acting elements of OsACO3 in vivo (Figure 3F) and OsBIHD1 protein caused a mobility shift in the labeled probes from the cis-acting elements of OsACO3, which migrated more slowly than the free probes (Figure 3G). These results demonstrate that OsBIHD1 is involved in activating the ET-dependent defense pathway through regulating OsAOC3’s expression by directly binding to the OsAOC3 promoter region.
Overexpression of OsBIHD1 Leads to BR Insensitivity
Homeodomain-containing proteins are involved in BR phytohormone signaling transduction through activation of BR biosynthesis or catabolism in rice (Ito et al., 2002; Tsuda and Hake, 2015). In this study, both OsBIHD1 overexpression and knock-out has obvious effects on plant seedlings such as dwarfing, increasing lamina joint angles and erect leaves (Figures 4A,B; Supplementary Figure S4). In order to investigate the possible roles of OsBIHD1 in BR biosynthesis, the expression of BR biosynthesis genes D2 (Hong et al., 2003), D11 (Tanabe et al., 2005), DWARF (Hong et al., 2002), and DWARF4 (Sakamoto et al., 2006) were analyzed with qRT-PCR. The results showed overexpression of OsBIHD1 did not result in any obvious up-regulation of BR biosynthetic genes, indicating that OsBIHD1 is not associated with BR biosynthesis (Figure 4C).
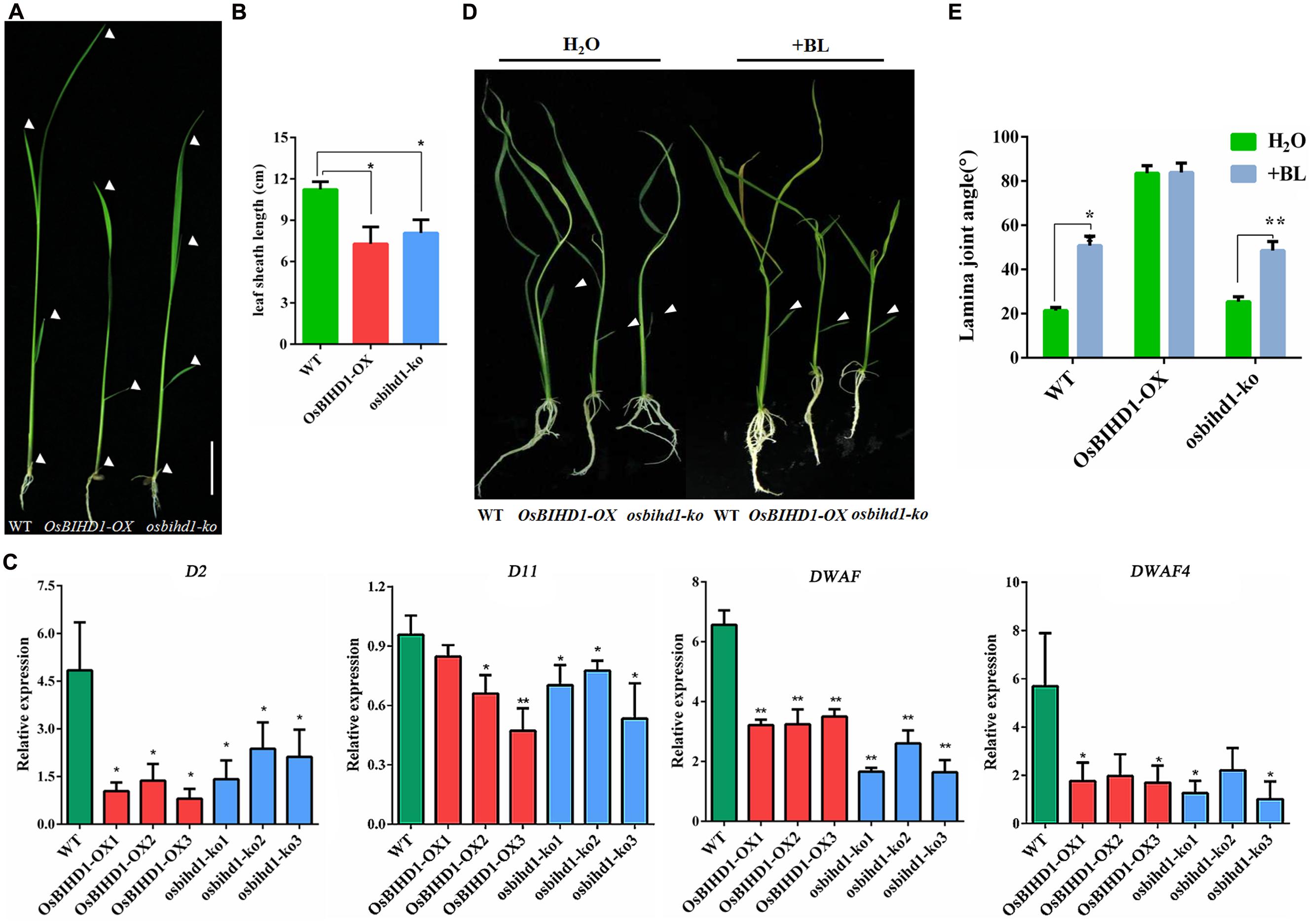
FIGURE 4. OsBIHD1 alters the expression of BR biosynthesis genes. (A) Phenotype of OsBIHD1 transgenic plants and wild-type seedlings. Scale bar is 4 cm. (B) Quantification of fourth leaf sheath length (n = 10). Values shown are means ± SD, and asterisks indicate a significant difference according to the t-test (∗P < 0.05, ∗∗P < 0.01) compared with wild-type. (C) Relative expression of BR biosynthesis genes D2, D11, DWARF4, and DWARF in wild-type, OsBIHD1-OX, osbihd1-ko. Values shown are means ± SD with three independent replicates, and asterisks indicate a significant difference according to the t-test (P < 0.05) compared with WT. (D) Phenotypes of wild type, OsBIHD1-OX, and osbihd1-ko plants after BR treatment. Twelve days old plants were treated with 2 μl BL (500 ng/L) by the micro-drop method. Controls received H2O. BL is brassinolide. (E) Quantification of lamina joint angle of second leaves (n = 10). Values shown are means ± SD, and asterisks indicate a significant difference according to the t-test (∗P < 0.05, ∗∗P < 0.01) compared with control group.
Brassinosteroid plays important roles in plant growth and development and particularly in leaf morphology (Saini et al., 2015). Considering the increase in leaf joint angle and the erect phenotype in the OsBIHD1 transgenic lines, we suspected that OsBIHD1 overexpression might lead to BR insensitivity. We then performed a lamina joint test for BR sensitivity as described previously (Chen et al., 2015). When wild type and osbihd1-ko seedlings were treated with brassinolide (500 ng/L), their lamina joint angles greatly increased (Figures 4D,E). By contrast, the angle of OsBIHD1-OX plants barely increased and the leaf blades were kept erect. Therefore, OsBIHD1 overexpression resulted in BR insensitivity, suggesting that OsBIHD1 suppresses the BR pathway by repressing BR signaling or catabolism, but not by suppressing BR biosynthesis. In addition, we measured transcript levels of BR signaling genes in OsBIHD1 transgenic plants and wild-type (Supplementary Figure S8), but we could not find clear evidence to explain whether or not OsBIHD1 modulates the expression of BR signaling-related genes to regulate leaf morphology.
OsBIHD1 Positively Regulates the Expression of BR Catabolic Genes to Coordinate Growth-Resistance Crosstalk
The brassinosteroid-deficient phenotypes of OsBIHD1 transgenicplants might be caused by the activation of BR catabolic genes, which would lead to an increase in the endogenous level of bioactive BR and a more rapid transformation into an inactivate form of BR (Yang et al., 2014). Therefore, the expression of the BR catabolic genes CYP734A2, CYP734A4, and CYP734A6 (Sakamoto et al., 2011) was investigated in OsBIHD1 transgenic plants using qRT-PCR. The results showed the transcript level of CYP734A4 was only slightly enhanced in both OsBIHD1-OX and osbihd1-ko lines and that of CYP734A6 was also not significantly altered (Figure 5A). However, CYP734A2 expression was significantly increased in OsBIHD1-OX plants, which might result in a decrease in bioactive BR levels (Figure 5A). As a consequence, the accumulation of CYP734A2 displayed dwarfing and abnormal leaf morphologies. To further determine whether OsBIHD1 activates CYP734A2 expression, a yeast one-hybrid assay and an EMSA were performed. The results showed OsBIHD1 directly bound to CYP734A2 promoter in vitro and in vitro (Figures 5B,C; Supplementary Figure S7). Our results revealed that OsBIHD1 promotes the expression of BR catabolic gene CYP734A2 through directly binding to CYP734A2’s promoter region.
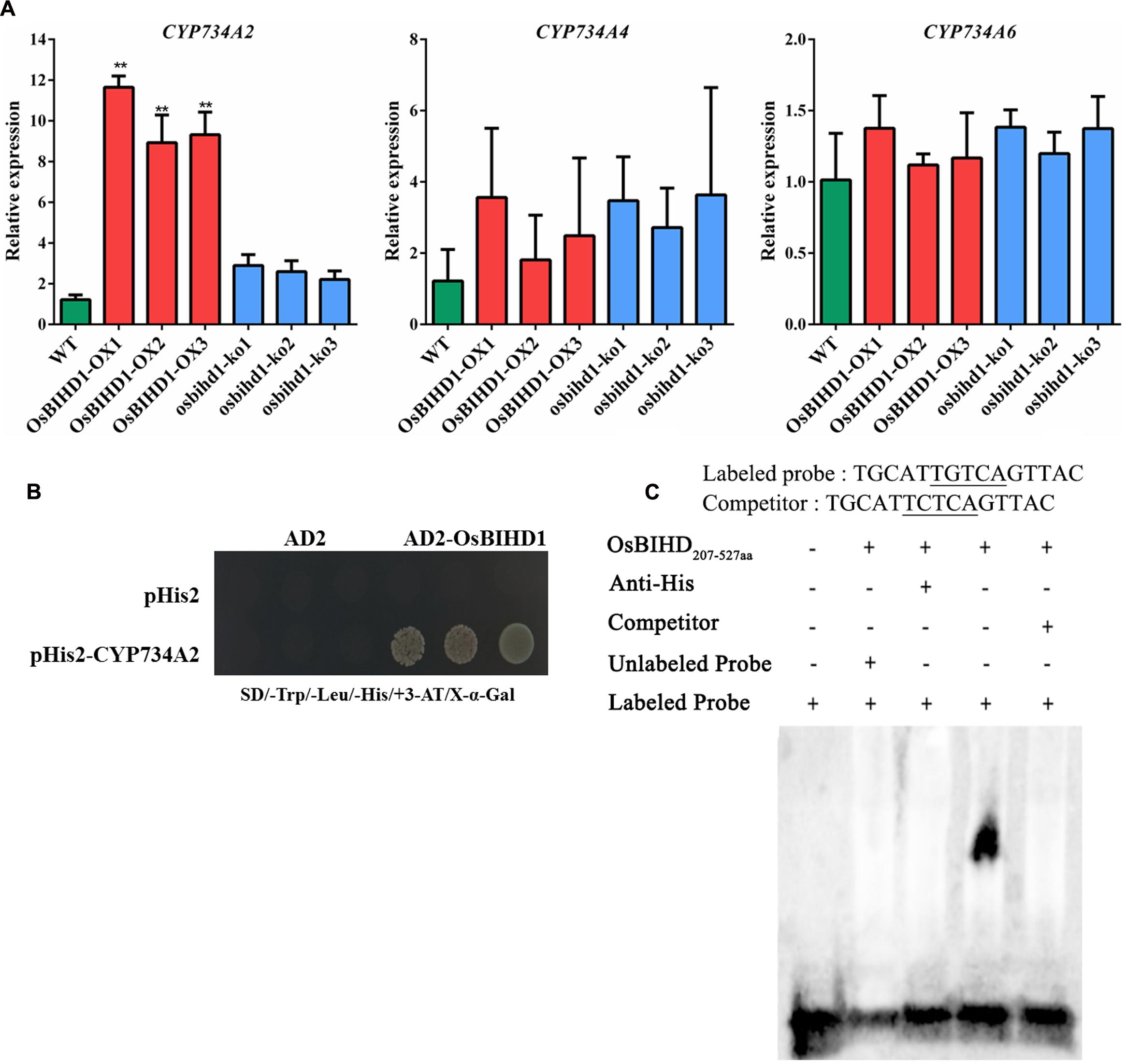
FIGURE 5. OsBIHD1 binds to the promoter of the BR catabolic gene CYP734A2. (A) Relative expression of BR catabolism genes in wild-type, OsBIHD1-OX, and osbihd1-ko plants. Values shown are means ± SD from three independent replicates, and asterisks indicate a significant difference according to the t-test (P < 0.05) compared with WT. (B) Yeast one-hybrid assay identifies OsBIHD1 binding the CYP734A2 promoter sequence in vivo. Reactions were score using an X-Gal assay. (C) EMSA analysis of recombinant OsBIHD1207-527 aa protein binding to the promoter cis-element TGTCA of CYP734A2 in vitro.
OsBIHD1 Activates the Hormone-Related Genes Expression
In spite of the directly bindings between the OsBIHD1 and hormone-related genes have been proved, but the concerned question is whether the OsBIHD1 activates the OsACO3 and CYP734A2 expression. To overcome this issue, we thought the transcriptional activity assay could make it convinced that OsBIHD1 activates the transcription of OsACO3 and CYP734A2. Therefore, the last constructs PromoterOsACO3:GUS and PromoterCY P734A2:GUS (contain the TGTCA cis-element) were co-transformed into tobacco leaves with 35S:OsBIHD1, respectively (Supplementary Figure S9). The results of GUS staining showed that OsBIHD1 was able to activate the GUS expression when co-transformed with the OsACO3 and CYP734A2 promoters region (Figure 6A). Moreover, the results obtained from ChIP-qPCR also proved the same conclusion in rice protoplast system (Figures 6B–E). Taken together, we concluded that the OsBIHD1 received the upstream signaling transduction from the Pik1-H4, and then directly binding the hormone-related genes promoter region and activating their expression.
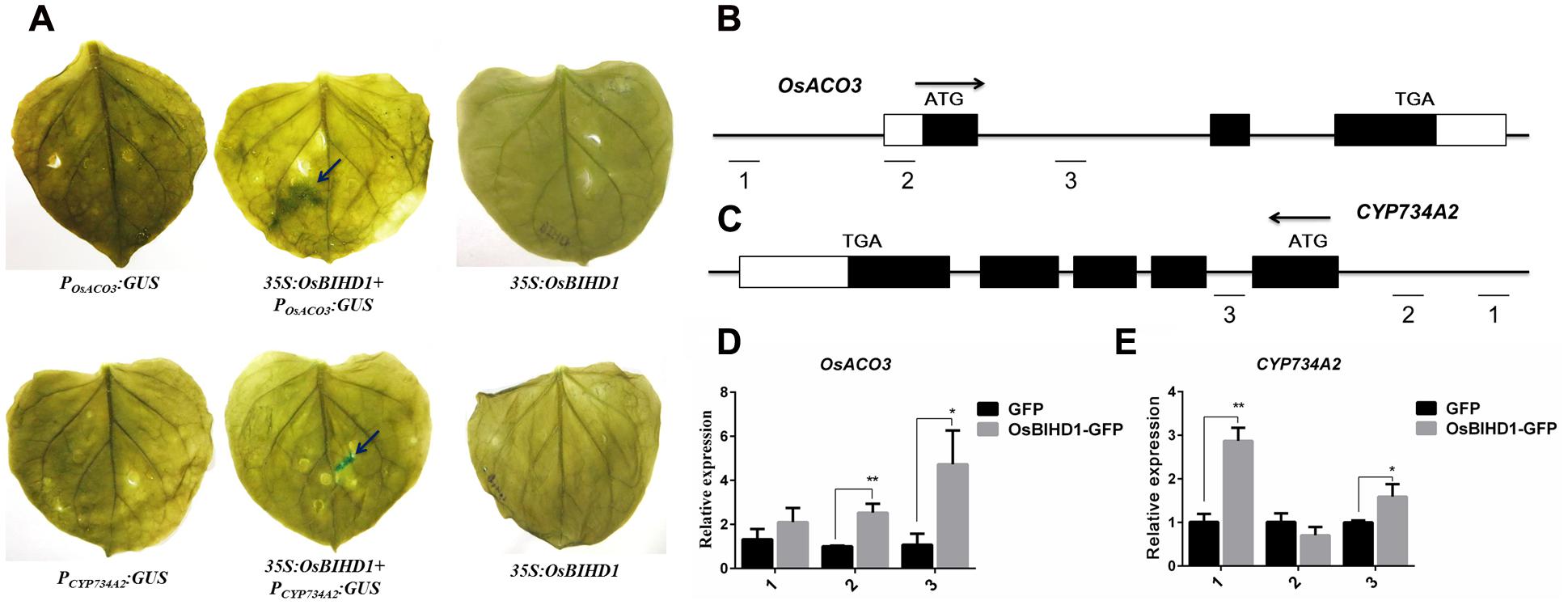
FIGURE 6. OsBIHD1 activates the hormone-related genes expression. (A) OsBIHD1 activated the GUS expression when co-transformed with OsACO3 and CYP734A2 promoter sequence into the tobacco leaves. (B–E) Results of chromatin immunoprecipitation (ChIP) assays at OsACO3 and CYP734A2 loci in rice protoplast system. ChIP analyses at OsACO3 and CYP734A2 chromatin regions were performed using antibodies against GFP in rice protoplast system. The reference gene Ubq13 was used as an internal standard for normalization. Values shown are means ± standard deviation from three parallel biological replicates, and asterisks indicate a significant difference according to the t-test (P < 0.05) compared with control group.
Pik-H4 Slightly Alters OsBIHD1 Expression
We showed that OsBIHD1 specifically interacted with Pik1-H4, and adjusted the ethylene and BR hormone pathway. But it needed to make it clear that whether the OsBIHD1-mediated transcriptional regulation is Pik-H4 activation dependent. So a Pik-H4 comprised M. oryzae race GDYJ8 (carrying the Avr-Pita) was used in this experiment. The results showed that the expression of Pik1-H4 and OsBIHD1 was decreased over 24 h after inoculated with GDYJ8 (Figure 7A), which was different from that inoculated with GDYJ7 (carrying the Avr-PikH4). In additional, we further detected the transcription levels of OsBIHD1 and hormone-related genes in Pik-H4 NILs and Pik-H4 null background rice (Figures 7B–E). Totally, these results indicated that Pik-H4 slightly up-regulates the expression of OsBIHD1, ET- and BR- related genes, but the values didn’t exhibit significantly difference between the Pik-H4 NILs and susceptible variety LTH.
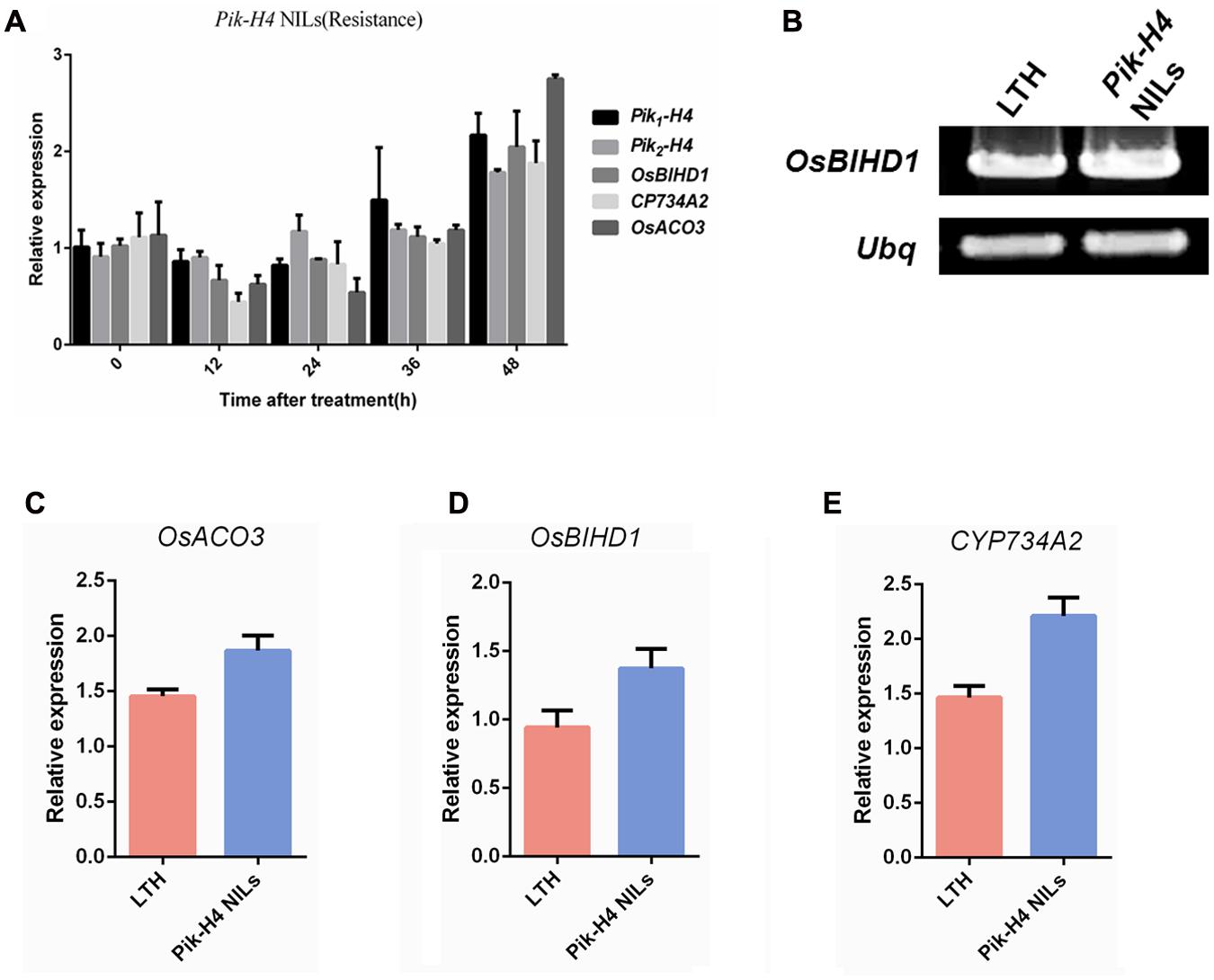
FIGURE 7. Pik-H4 positively regulates OsBIHD1 expression. (A) Relative expression of Pik-H4, OsBIHD1, CYP734A2, and OsACO3 over a time course of 48 h after inoculation with Moryzae oryzae GDYJ8 in Pik-H4 NILs. (B–E) Relative expression of OsACO3, OsBIHD1, and CYP734A2 in Pik-H4 NILs and LTH plants. Values shown are means ± standard deviation from three parallel biological replicates.
Since of the expression of OsBIHD1 could be up-regulated after inoculation with M. oryzae in BTH-treatment seedlings (Luo et al., 2005a), expression of BR catabolic genes should also be induced with challenge by M. oryzae. As we expected, the transcript levels of all BR catabolic genes were significantly up-regulated and peaked in wild type leaves at 36 h, and decreased in osbihd1-ko leaves after M. oryzae treatment (Supplementary Figure S10), indicating BR catabolic genes depend upon OsBIHD1 to activate their expression to suppress plant growth under pathogen invasion.
Discussion
In a previous yeast two-hybrid screening for Pik-H4 binding proteins, we identified a homeodomain (HD)-containing protein, OsBIHD1, which may be positively involved in activating expression of the defense-related genes in disease resistance responses (Luo et al., 2005b). We showed in this study that the NBS-LRR protein Pik-H4 interacts with OsBIHD1 and the blast resistance by Pik-H4 depends on OsBIHD1. Importantly, OsBIHD1 directly activates ET pathway and positively regulates the expression of BR catabolic genes to coordinate growth-resistance crosstalk. Therefore, OsBIHD1 is a key regulator for the crosstalk between growth and resistance (Figure 8).
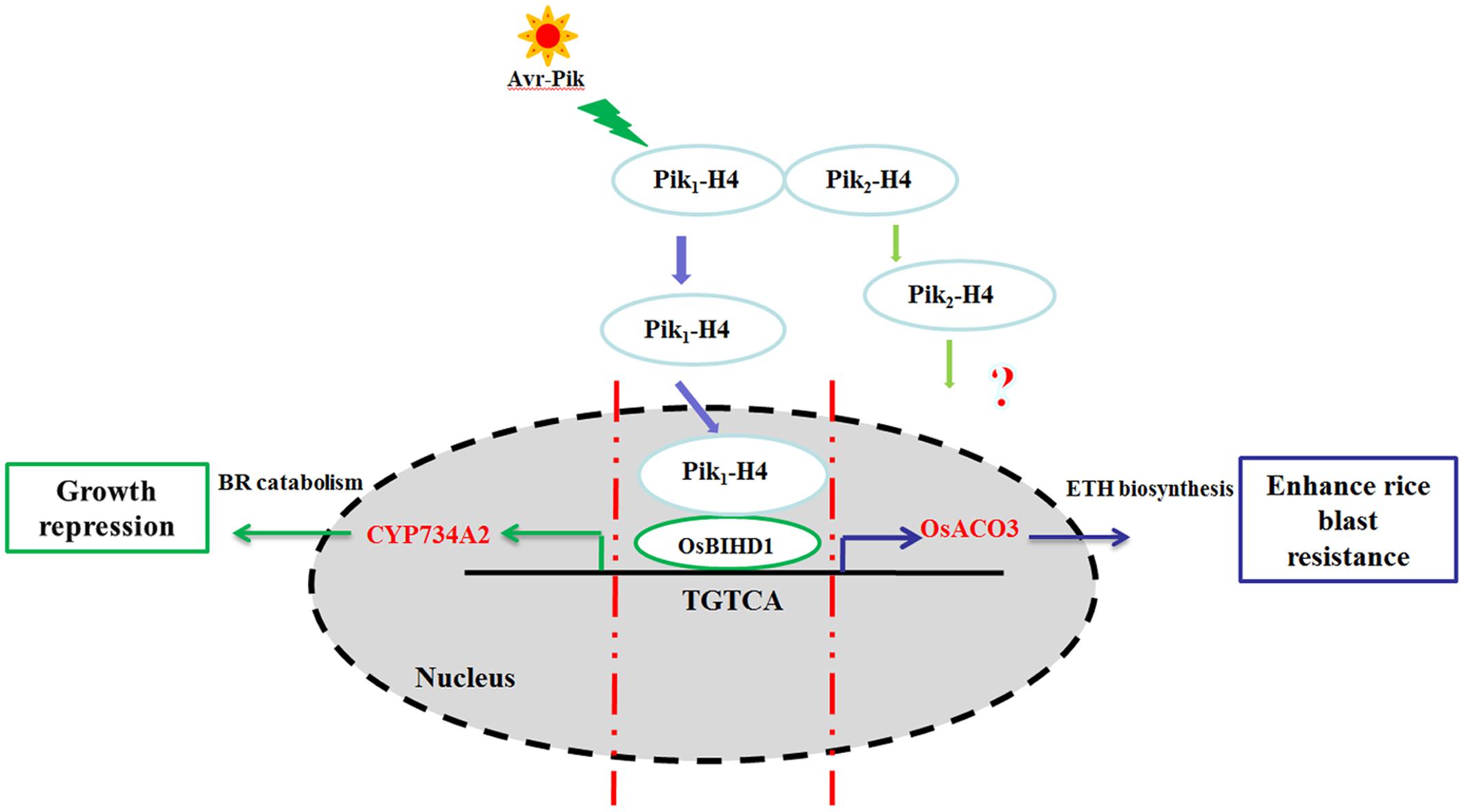
FIGURE 8. Model of the Pik-H4 interact OsBIHD1 to regulate rice blast and growth. The signaling initiating from the M. oryzae elicitor Avr-Pik is recognized by the Pik1-H4 CC domain and the nuclear Pik1-H4 interacts with the OsBIHD1 homeodomain. After signaling from Pik1-H4, OsBIHD1 binds the cis-element of the OsACO3 promoter and positively stimulates rice blast resistance via the ET defense pathway OsBIHD1 overexpression has an inhibitory effect on plant growth which promotes the expression of the BR catabolicgene CYP734A2. This results in a decrease in bioactive BR. As a consequence, OsBIHD1 mediates an important trade-off between rice blast resistance and growth.
Pik-H4 Interacts with OsBIHD1 to Regulate Blast Resistance
The majority of plant R genes encode nucleotide-binding leucine-rich repeat (NBS-LRR) proteins that mediate recognition of diverse effectors (Avirulence proteins, Avr). The NBS domain (also called the NB, NB-ARC and Nod domain) is involved in signaling by activation of a kinase or other proteins in the resistance response (DeYoung and Innes, 2006; Ting et al., 2008). LRR domain is thought to be the primary determinant of pathogen recognition specificity or downstream events (Belkhadir et al., 2004). In Plant NBS-LRR proteins, there is a toll/interleukin-1 receptor (TIR) domain or a coiled coil domain (CC), which influences the requirement for distinct downstream signaling components or direct recognition with pathogen effectors (Jia et al., 2000). Currently, more than 24 major R genes that confer resistance against M. oryzae in rice have been identified including Pi-ta (Bryan et al., 2000), Pi-k (Zhai et al., 2011), and Pb1 (Hayashi et al., 2010), the downstream signaling activated by the recognition of R-Avr remains little known. In rice, the important roles of the transcription factors in R gene-mediated disease resistance have been broadly reported, such as the blast resistance of Pb1 depends on its interaction with a transcription factor WRKY45 (Inoue et al., 2013), and the pattern recognition receptor Xa21 interacts with WRKY62 to regulate Xoo resistance (Peng et al., 2008; Park and Ronald, 2012). In this study we showed that Pik-H4 physically interacts with HD transcription factor OsBIHD1 through its CC domain in the nucleus. Knockout of OsBIHD1 gene down-regulates the expression levels of four PR genes, including PR1a, PR1b, PR10, and PAL, which are tightly correlated with the onset of defense responses against a variety of fungal, viral, and bacterial pathogens (Riviere et al., 2008; Zhang and Wang, 2013; Huang et al., 2016). Thus, the blast resistance of Pik-H4 was also reduced in osbihd1-ko mutants. These results indicated OsBIHD1 is required for Pik-H4-mediated blast resistance through protein–protein interaction and OsBIHD1 is a positive regulator of immunity.
OsBIHD1 Directly Activates the ET Pathway to Defend against M. oryzae
Effector-triggered immunity is usually accompanied by rapid ET production and a programmed cell death at the site of infection to prevent further infection by the invading pathogens (Yang C. et al., 2015; Zdarska et al., 2015). In Arabidopsis, avrRpt2 (effector)-triggered response caused higher ethylene production, which is dependent on RPS2, the R protein corresponding to avrRpt2 (Kunkel et al., 1993; Guan et al., 2015). Higher level of ET production indicates ET signaling pathway may be activated during plant-pathogen interaction through unknown ways. Besides ET, several other plant hormones such as JA, SA, and ABA are also involved in this defense. Blast resistance by Pb1 partially depends on the SA signaling pathway, which is mainly regulated bythe Pb1-interacting transcription factor, WRKY45 (Inoue et al., 2013). Overexpression of OsBIHD1 led to elevation of the expression of ET biosynthesis ACO family genes, suggesting that OsBIHD1 plays important roles in ET signaling pathway, which is consistent with the evidence that ET is accompanied by elevation of PR genes expression (Yang Y.X. et al., 2015). Yeast one-hybrid and EMSA confirmed that OsBIHD1 directly bounds to TGTCA motif in the cis-element sequence of OsACO3, indicating that resistance gene Pik-H4 may depend on OsBIHD1 to directly regulate the ET defense pathway.
OsBIHD1 Plays a Critical Role in the Crosstalk between Plant Growth and Immunity
Previous studies showed that rice KNOX-HD OSH1 proteins are involved in the determination of the state of the shoot apical meristem (SAM) (Tsuda et al., 2014). OSH1 represses the BR phytohormone pathway through activation of BR catabolism genes. These findings provided the evidence that HD containing protein, OsBIHD1, may participate in the regulation of BR phytohormone pathway. Our results demonstrated thatOsBIHD1 overexpression showed an increased lamina joint angle and erect leaves, and this phenotype is in accordance with the BR-deficient and -insensitive rice mutants (Sato et al., 1999).
We next test the expression of the BR catabolic genes CYP734A2, CYP734A4, and CYP734A6 in OsBIHD1 transgenic plants. CYP734A2 expression was significantly increased in OsBIHD1-OX plants and it also was induced by infection with M. oryzae, in consistent with OsBIHD1 expression in M. oryzae treatment seedlings (Sakamoto et al., 2011), suggesting the expression of CYP734A2 is regulated by OsBIHD1. Further yeast one-hybrid assay and EMSA showed OsBIHD1 directly bound to CYP734A2’s promoter region in vivo. These results suggest OsBIHD1 possibly plays a key role in the crosstalk between plant growth and immunity.
The crosstalk between the BR and PTI immunity clearly revealed that BR signaling transcription factor HBI1 represses the immunity response against Pseudomonas syringae in Arabidopsis (Fan et al., 2014). Our findings proved Pik-H4 interacts with OsBIHD1 to modulate BR catabolism and plant resistance so that the plants can focus its energy to fend off the pathogen invasion (Figure 8). Therefore, we conclude that OsBIHD1 is most likely HBI1 to coordinate growth-resistance crosstalk. Whether OsBIHD1 was involved in the PTI immunity response remains unknown. Though the BR-activated transcription factor BZR1 directly regulates many defense-related genes, BZR1 itself is not affected by PAMP signaling (Guo et al., 2013; Lozano-Duran et al., 2013). Further characterization of whether OsBIHD1 interacts with BZR1 and HBI1 and the OsBIHD1-interacting network are required to be demonstrated, which will be of great importance for understanding the trade-off between growth and immunity.
However, OsBIHD1 knock-out transgenic plants also showed dwarf and large lamina joint angle phenotype. We suspect that the putative roles of OsBIHD1 are similar to the dwarfism gene d6. Loss of function mutant of D6 in rice exhibited defects in internode elongation and repression in SAM formation (Sato et al., 1999; Nagasaki et al., 2001). In addition, our results suggested that the abnormal phenotype caused by the OsBIHD1 deficient in rice was attributed to the endogenous BR disorder. Whether the OsBIHD1 involves in other hormones pathway to regulate the plant growth, such as homeodomain containing proteins induce cytokinin (CK) biosynthesis and directly suppress gibberellin (GA) biosynthesis should be illustrated in future work (Jasinski et al., 2005; Yanai et al., 2005).
OsBIHD1 May Be Modified by Phosphorylation and Ubiquitination
Because OsBIHD1 overexpression caused abnormal plant phenotypes, plants require OsBIHD1 expression at a modest level to maintain growth under normal conditions. Once the pathogen has infected, the elicitor AvrPik was recognized by NLR protein Pik-H4 and the conserved CC domain of Pik-H4 maintained an interaction with OsBIHD1 in the nucleus. We do not know whether Pik1-H4 contains another domain (LRR domain) responsible for a direct phosphorylation of OsBIHD1 or whether this is dependent upon the CC domain. We suspect that phosphorylated OsBIHD1 further activates the expression of a battery of target genes, including the ET biosynthetic gene OsACO3 and the BR catabolic gene CYP734A2.
We wondered whether OsBIHD1 was involved in the PTI immunity response and induced crosstalk between immunity and plant growth pathways. If this was the case there would by other transcription factors that regulate OsBIHD1 expression after the PTI immunity activated by M. oryzae. At the conclusion of the immunity response, plants gradually recover from the conflict with M. oryzae. Thus, growth prevailed during the remainder of the reproductive stages and the plants took measures to eliminate the repression of OsBIHD1-induced BR catabolism. We speculate that the OsBIHD1-like protein WRKY45 relies on ubiquitination and proteasome protein degradation after fulfilling the task of immunity (Matsushita et al., 2013).
Future work will concentrate on the regulation of phosphorylation and ubiquitination of OsBIHD1 and to identify the transcriptional regulator of OsBIHD1. This study extends our knowledge concerning the mechanism whereby the R gene Pik-H4 associates with OsBIHD1 to balance growth and immunity involved in rice blast resistance.
Author Contributions
YL, TG, and MH conceived the original screening and research plans. HW, SD, and GY supervised the experiments. HL, FG, and WX performed most of the experiments. ZC provided technical assistance to HL, JW designed the experiments and analyzed the data. WL conceived the project and wrote the article with contributions of all the authors. HL supervised and complemented the writing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31401722) and the“863” High Technology Research and Development Program of China (863 Program) (2011AA10A101). The authors thank Prof. Yaoguang Liu (South China Agricultural University, Guangzhou 510642, China) for providing the plant binary vector pOX, pYLCRISPR/Cas9PUbi-Hi, BiFC vectors, and subcellular localization vector GFP.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00127/full#supplementary-material
References
Belkhadir, Y., Subramaniam, R., and Dangl, J. L. (2004). Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol. 7, 391–399. doi: 10.1016/j.pbi.2004.05.009
Bryan, G. T., Wu, K. S., Farrall, L., Jia, Y., Hershey, H. P., McAdams, S. A., et al. (2000). TA single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12, 2033–2046. doi: 10.1105/tpc.12.11.2033
Chae, H. S., Cho, Y. G., Park, M. Y., Lee, M. C., Eun, M. Y., Kang, B. G., et al. (2000). Hormonal cross-talk between auxin and ethylene differentially regulates the expression of two members of the 1-aminocyclopropane-1-carboxylate oxidase gene family in rice (Oryza sativa L.). Plant Cell Physiol. 41, 354–362. doi: 10.1093/pcp/41.3.354
Chandran, D., Rickert, J., Huang, Y., Steinwand, M. A., Marr, S. K., and Wildermuth, M. C. (2014). Atypical E2F transcriptional repressor DEL1 acts at the intersection of plant growth and immunity by controlling the hormone salicylic acid. Cell Host Microbe 15, 506–513. doi: 10.1016/j.chom.2014.03.007
Chen, Q., Xie, Q., Gao, J., Wang, W., Sun, B., Liu, B., et al. (2015). Characterization of rolled and erect leaf 1 in regulating leave morphology in rice. J. Exp. Bot. 66, 6047–6058. doi: 10.1093/jxb/erv319
DeYoung, B. J., and Innes, R. W. (2006). Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7, 1243–1249. doi: 10.1038/ni1410
Fan, M., Bai, M. Y., Kim, J. G., Wang, T., Oh, E., Chen, L., et al. (2014). The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 26, 828–841. doi: 10.1105/tpc.113.121111
Guan, R., Su, J., Meng, X., Li, S., Liu, Y., Xu, J., et al. (2015). Multilayered regulation of ethylene induction plays a positive role in Arabidopsis resistance against Pseudomonas syringae. Plant Physiol. 169, 299–312. doi: 10.1104/pp.15.00659
Guo, R., Qian, H., Shen, W., Liu, L., Zhang, M., Cai, C., et al. (2013). BZR1 and BES1 participate in regulation of glucosinolate biosynthesis by brassinosteroids in Arabidopsis. J. Exp. Bot. 64, 2401–2412. doi: 10.1093/jxb/ert094
Hayashi, N., Inoue, H., Kato, T., Funao, T., Shirota, M., Shimizu, T., et al. (2010). Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 64, 498–510. doi: 10.1111/j.1365-313X.2010.04348.x
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hong, Z., Ueguchi-Tanaka, M., Shimizu-Sato, S., Inukai, Y., Fujioka, S., Shimada, Y., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32, 495–508. doi: 10.1046/j.1365-313X.2002.01438.x
Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S., Takatsuto, S., et al. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15, 2900–2910. doi: 10.1105/tpc.014712
Huang, L. F., Lin, K. H., He, S. L., Chen, J. L., Jiang, J. Z., Chen, B. H., et al. (2016). Multiple patterns of regulation and overexpression of a ribonuclease-like pathogenesis-related protein gene, OsPR10a, conferring disease resistance in rice and Arabidopsis. PLoS ONE 11:e0156414. doi: 10.1371/journal.pone.0156414
Inoue, H., Hayashi, N., Matsushita, A., Xinqiong, L., Nakayama, A., Sugano, S., et al. (2013). Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc. Natl. Acad. Sci. U.S.A. 110, 9577–9582. doi: 10.1073/pnas.1222155110
Ito, M., Sato, Y., and Matsuoka, M. (2002). Involvement of homeobox genes in early body plan of monocot. Int. Rev. Cytol. 218, 1–35. doi: 10.1016/S0074-7696(02)18010-7
Iwai, T., Miyasaka, A., Seo, S., and Ohashi, Y. (2006). Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol. 142, 1202–1215. doi: 10.1104/pp.106.085258
Iwamoto, M., Baba-Kasai, A., Kiyota, S., Hara, N., and Takano, M. (2010). ACO1, a gene for aminocyclopropane-1-carboxylate oxidase: effects on internode elongation at the heading stage in rice. Plant Cell Environ. 33, 805–815. doi: 10.1111/j.1365-3040.2009.02106.x
Jasinski, S., Piazza, P., Craft, J., Hay, A., Woolley, L., Rieu, I., et al. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15, 1560–1565. doi: 10.1016/j.cub.2005.07.023
Jia, Y., McAdams, S. A., Bryan, G. T., Hershey, H. P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. doi: 10.1093/emboj/19.15.4004
Kunkel, B. N., Bent, A. F., Dahlbeck, D., Innes, R. W., and Staskawicz, B. J. (1993). RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5, 865–875. doi: 10.1105/tpc.5.8.865
Liu, W., Liu, J., Triplett, L., Leach, J. E., and Wang, G. L. (2014). Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 52, 213–241. doi: 10.1146/annurev-phyto-102313-045926
Lozano-Duran, R., Macho, A. P., Boutrot, F., Segonzac, C., Somssich, I. E., and Zipfel, C. (2013). The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. eLife 2:e00983. doi: 10.7554/eLife.00983
Lu, D., Wu, S., Gao, X., Zhang, Y., Shan, L., and He, P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 496–501. doi: 10.1073/pnas.0909705107
Luo, D., Xu, H., Liu, Z., Guo, J., Li, H., Chen, L., et al. (2013). A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat. Genet. 45, 573–577. doi: 10.1038/ng.2570
Luo, H., Song, F., Goodman, R. M., and Zheng, Z. (2005a). Up-regulation of OsBIHD1, a rice gene encoding BELL homeodomain transcriptional factor, in disease resistance responses. Plant Biol. (Stuttg) 7, 459–468. doi: 10.1055/s-2005-865851
Luo, H., Song, F., and Zheng, Z. (2005b). Overexpression in transgenic tobacco reveals different roles for the rice homeodomain gene OsBIHD1 in biotic and abiotic stress responses. J. Exp. Bot. 56, 2673–2682. doi: 10.1093/jxb/eri260
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Matsushita, A., Inoue, H., Goto, S., Nakayama, A., Sugano, S., Hayashi, N., et al. (2013). Nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. Plant J. 73, 302–313. doi: 10.1111/tpj.12035
Nagasaki, H., Sakamoto, T., Sato, Y., and Matsuoka, M. (2001). Functional analysis of the conserved domains of a rice KNOX homeodomain protein, OSH15. Plant Cell 13, 2085–2098. doi: 10.1105/tpc.13.9.2085
Park, C. J., and Ronald, P. C. (2012). Cleavage and nuclear localization of the rice XA21 immune receptor. Nat. Commun. 3, 920. doi: 10.1038/ncomms1932
Peng, Y., Bartley, L. E., Chen, X., Dardick, C., Chern, M., Ruan, R., et al. (2008). OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol. Plant 1, 446–458. doi: 10.1093/mp/ssn024
Riviere, M. P., Marais, A., Ponchet, M., Willats, W., and Galiana, E. (2008). Silencing of acidic pathogenesis-related PR-1 genes increases extracellular beta-(1- > 3)-glucanase activity at the onset of tobacco defence reactions. J. Exp. Bot. 59, 1225–1239. doi: 10.1093/jxb/ern044
Saini, S., Sharma, I., and Pati, P. K. (2015). Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks. Front. Plant Sci. 6:950. doi: 10.3389/fpls.2015.00950
Sakamoto, T., Kawabe, A., Tokida-Segawa, A., Shimizu, B., Takatsuto, S., Shimada, Y., et al. (2011). Rice CYP734As function as multisubstrate and multifunctional enzymes in brassinosteroid catabolism. Plant J. 67, 1–12. doi: 10.1111/j.1365-313X.2011.04567.x
Sakamoto, T., Morinaka, Y., Ohnishi, T., Sunohara, H., Fujioka, S., Ueguchi-Tanaka, M., et al. (2006). Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24,k105–109. doi: 10.1038/nbt1173
Sato, Y., Sentoku, N., Miura, Y., Hirochika, H., Kitano, H., and Matsuoka, M. (1999). Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J. 18, 992–1002. doi: 10.1093/emboj/18.4.992
Takeuchi, K., Gyohda, A., Tominaga, M., Kawakatsu, M., Hatakeyama, A., Ishii, N., et al. (2011). RSOsPR10 expression in response to environmental stresses is regulated antagonistically by jasmonate/ethylene and salicylic acid signaling pathways in rice roots. Plant Cell Physiol. 52, 1686–1696. doi: 10.1093/pcp/pcr105
Tanabe, S., Ashikari, M., Fujioka, S., Takatsuto, S., Yoshida, S., Yano, M., et al. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17, 776–790. doi: 10.1105/tpc.104.024950
Ting, J. P., Willingham, S. B., and Bergstralh, D. T. (2008). NLRs at the intersection of cell death and immunity. Nat. Rev. Immunol. 8, 372–379. doi: 10.1038/nri2296
Tsuda, K., and Hake, S. (2015). Diverse functions of KNOX transcription factors in the diploid body plan of plants. Curr. Opin. Plant Biol. 27, 91–96. doi: 10.1016/j.pbi.2015.06.015
Tsuda, K., Kurata, N., Ohyanagi, H., and Hake, S. (2014). Genome-wide study of KNOX regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell 26, 3488–3500. doi: 10.1105/tpc.114.129122
Wang, W., and Wang, Z. Y. (2014). At the intersection of plant growth and immunity. Cell Host Microbe 15, 400–402. doi: 10.1016/j.chom.2014.03.014
Xiao, W., Yang, Q., Wang, H., Guo, T., Liu, Y., Zhu, X., et al. (2011). Identification and fine mapping of a resistance gene to Magnaporthe oryzae in a space-induced rice mutant. Mol. Breed. 28, 303–312. doi: 10.1007/s11032-010-9481-6
Yanai, O., Shani, E., Dolezal, K., Tarkowski, P., Sablowski, R., Sandberg, G., et al. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15, 1566–1571. doi: 10.1016/j.cub.2005.07.060
Yang, C., Lu, X., Ma, B., Chen, S. Y., and Zhang, J. S. (2015). Ethylene signaling in rice and Arabidopsis: conserved and diverged aspects. Mol. Plant 8, 495–505. doi: 10.1016/j.molp.2015.01.003
Yang, J. W., Fu, J. X., Li, J., Cheng, X. L., Li, F., Dong, J. F., et al. (2014). A Novel Co-immunoprecipitation protocol based on protoplast transient gene expression for studying protein–protein interactions in rice. Plant Mol. Biol. Rep. 32, 153–161. doi: 10.1007/s11105-013-0633-9
Yang, Y. X., Ahammed, G. J., Wu, C., Fan, S. Y., and Zhou, Y. H. (2015). Crosstalk among jasmonate, salicylate and ethylene signaling pathways in plant disease and immune responses. Curr. Protein Pept. Sci. 16, 450–461. doi: 10.2174/1389203716666150330141638
Yun, J., Kim, Y. S., Jung, J. H., Seo, P. J., and Park, C. M. (2012). The AT-hook motif-containing protein AHL22 regulates flowering initiation by modifying FLOWERING LOCUS T chromatin in Arabidopsis. J. Biol. Chem. 287, 15307–15316. doi: 10.1074/jbc.M111.318477
Zdarska, M., Dobisova, T., Gelova, Z., Pernisova, M., Dabravolski, S., and Hejatko, J. (2015). Illuminating light, cytokinin, and ethylene signalling crosstalk in plant development. J. Exp. Bot. 66, 4913–4931. doi: 10.1093/jxb/erv261
Zhai, C., Lin, F., Dong, Z., He, X., Yuan, B., Zeng, X., et al. (2011). The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 189, 321–334. doi: 10.1111/j.1469-8137.2010.03462.x
Zhai, C., Zhang, Y., Yao, N., Lin, F., Liu, Z., Dong, Z., et al. (2014). Function and interaction of the coupled genes responsible for Pik-h encoded rice blast resistance. PLoS ONE 9:e98067. doi: 10.1371/journal.pone.0098067
Keywords: NBS-LRR resistance gene, homeodomain proteins, ethylenes, brassinosteroid signaling, fungal resistance.
Citation: Liu H, Dong S, Gu F, Liu W, Yang G, Huang M, Xiao W, Liu Y, Guo T, Wang H, Chen Z and Wang J (2017) NBS-LRR Protein Pik-H4 Interacts with OsBIHD1 to Balance Rice Blast Resistance and Growth by Coordinating Ethylene-Brassinosteroid Pathway. Front. Plant Sci. 8:127. doi: 10.3389/fpls.2017.00127
Received: 07 November 2016; Accepted: 20 January 2017;
Published: 06 February 2017.
Edited by:
Jens Staal, Ghent University, BelgiumReviewed by:
Nan Yao, Sun Yat-sen University, ChinaZonghua Wang, Fujian Agriculture and Forestry University, China
Copyright © 2017 Liu, Dong, Gu, Liu, Yang, Huang, Xiao, Liu, Guo, Wang, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiqiang Chen, chenlin@scau.edu.cn Jiafeng Wang, bcjfwang@gmail.com Hao Liu, liuhao2054@stu.scau.edu.cn
 Hao Liu
Hao Liu Shuangyu Dong
Shuangyu Dong