- 1Department of Biological Production Science, Graduate School of Agriculture, Tokyo University of Agriculture and Technology, Fuchu, Japan
- 2Institute of Global Innovation Research, Tokyo University of Agriculture and Technology, Fuchu, Japan
- 3Precursory Research for Embryonic Science and Technology, Japan Science and Technology Agency, Kawaguchi, Japan
- 4Institute of Crop Science, National Agriculture and Food Research Organization, Tsukuba, Japan
- 5Department of Frontier Research, Kazusa DNA Research Institute, Kisarazu, Japan
- 6Rice Research Institute, Shenyang Agricultural University, Shenyang, China
- 7Department of Ecology and Evolutionary Biology, University of Toronto, Toronto, ON, Canada
Increasing the rate of leaf photosynthesis is one important approach for increasing grain yield in rice (Oryza sativa). Exploiting the natural variation in CO2 assimilation rate (A) between rice cultivars using quantitative genetics is one promising means to identify genes contributing to higher photosynthesis. In this study, we determined precise location of Carbon Assimilation Rate 8 (CAR8) by crossing a high-yielding indica cultivar with a Japanese commercial cultivar. Fine mapping suggested that CAR8 encodes a putative Heme Activator Protein 3 (OsHAP3) subunit of a CCAAT-box-binding transcription factor called OsHAP3H. Sequencing analysis revealed that the indica allele of CAR8 has a 1-bp deletion at 322 bp from the start codon, resulting in a truncated protein of 125 amino acids. In addition, CAR8 is identical to DTH8/Ghd8/LHD1, which was reported to control rice flowering date. The increase of A is largely due to an increase of RuBP regeneration rate via increased leaf nitrogen content, and partially explained by reduced stomatal limitation via increased stomatal conductance relative to A. This allele also increases hydraulic conductivity, which would promote higher stomatal conductance. This indicates that CAR8 affects multiple physiological aspects relating to photosynthesis. The detailed analysis of molecular functions of CAR8 would help to understand the association between photosynthesis and flowering and demonstrate specific genetic mechanisms that can be exploited to improve photosynthesis in rice and potentially other crops.
Introduction
Rice (Oryza sativa) is one of the most valuable crops in the world, both in terms dollar value and contribution to the human food supply (FAO, 2015). Increasing its yield is a major challenge for improving global food security (Khush, 2013) and could be achieved by increasing the rate of net CO2 assimilation rate in individual leaves (A) (Long et al., 2006; Murchie et al., 2009). While photosynthetic improvement often emphasizes improving specific known traits within the photosynthetic apparatus (Suzuki et al., 2007; Takahara et al., 2010; von Caemmerer and Evans, 2010), or through introducing novel photosynthetic pathways such as the C4 pathway (Kajala et al., 2011; http://C4rice.irri.org), analysis of quantitative trait locus (QTL) through crossing experiments provide the opportunity to identify novel genetic elements that control photosynthetic performance in existing rice cultivars (Flood et al., 2011).
Most agronomic traits including A are controlled by multiple genetic factors, such traits are known as quantitative traits. QTL analyses can provide associations between quantitative traits and molecular markers (Tanksley, 1993). To conduct a QTL analysis, phenotypic values of interest are quantified in a segregating population whose genotypes have been determined by DNA markers. In rice, the complete genome sequence is available and many DNA markers have been identified (International Rice Genome Sequencing Project, 2005). Several advanced populations, including backcrossed inbred lines and chromosome segment substitution lines, have been developed to facilitate the QTL investigations in rice (Yamamoto et al., 2009). As a result, many genes associating with important agronomic traits have been identified using QTL methods (Yamamoto et al., 2014).
Wide variations in A among rice cultivars have been described (Takano and Tsunoda, 1971; Cook and Evans, 1983; Yeo et al., 1994; Kanemura et al., 2007; Jahn et al., 2011), and several QTL underlying this variation have been identified in populations derived from crosses between japonica and indica cultivars (Teng et al., 2004; Hu et al., 2009; Takai et al., 2010) and between japonica and indica/japonica cultivars (Gu et al., 2012). However, there is only one report that identified a causal gene controlling photosynthetic variation among rice cultivars (Takai et al., 2013). To understand the whole picture of the genetic control of A and to apply it in breeding aimed at increasing rice grain yield, it is necessary to identify the causal genes and understand their physiological aspects.
The CO2 assimilation rate in C3 species is considered to be limited by ribulose 1,5-bisphosphate (RuBP) carboxylation capacity of Rubisco or the RuBP regeneration capacity (Farquhar et al., 1980). Under low CO2 concentration and light-saturated conditions, A is commonly limited by the RuBP carboxylation capacity, while it is limited by RuBP regeneration capacity under elevated CO2 concentration and light-saturated conditions. The RuBP regeneration capacity reflects the capacity of electron transport, the Calvin cycle, and under high CO2 concentration, the ability of starch and sucrose synthesis to release inorganic phosphate (Sharkey, 1985). The CO2 diffusion from air into leaves is also important determinant of A (Farquhar and Sharkey, 1982). In healthy leaves, stomatal conductance (gs) is regulated to track the value of A such that the intercellular CO2 concentration (Ci) and the ratio of intercellular to ambient CO2 (Ci/Ca) vary little as A increases (Farquhar and Sharkey, 1982). In contrast, Kusumi et al. (2012) shows that the increase in gs relative to A can enhance A and Ci/Ca in a rice mutant with a defective anion channel in the guard cells. This suggested that we should consider both the stomatal control and the enzymatic control of the photosynthetic apparatus to know the physiological reasons relating to the difference in A.
During grain filling, the flag leaf is the most important leaf in the rice canopy because its position at the top of the canopy ensures maximum light availability and it has greater photosynthetic capacity than leaves lower in the canopy. In our previous research, we used chromosome segment substitution lines derived from “Habataki,” a high-yielding indica cultivar with high A, and the japonica variety “Koshihikari,” the most popular cultivar in Japan with lower A, to identify four QTLs affecting A in flag leaves (Adachi et al., 2011, 2014). One of the four QTLs was identified at ~1.2 Mb region on the short arm of chromosome 8 (Adachi et al., 2011). According to the rice annotation database, 124 genes are predicted in this region (Sakai et al., 2013, http://rapdb.dna.affrc.go.jp). To determine gene responsible for the increase in A, fine-scale mapping is required. In this study, we examined a region in the QTL that correlates with the increase in A, which we term Carbon Assimilation Rate 8 (CAR8). Our objective is to identify the gene underlying CAR8 via fine mapping and to evaluate the physiological mechanism by which it increases A.
Materials and Methods
Growth Conditions
We grew rice plants in three conditions—paddy fields, outdoors in pots, and in a controlled-environment cabinet in pots. We used the plants grown in paddy fields for QTL mapping, the plants grown outdoors in pots for evaluating the physiological effect of CAR8 on A, and the plants grown in a controlled environment cabinet in pots for evaluating the hydraulic conductance and root surface area. Plants in a paddy field were grown at the National Institute of Agrobiological Sciences in Tsukuba, Japan (36°03′N, 140°11′E). Seedlings at the fifth-leaf stage were transplanted (one plant per hill) into the field (alluvial clay loam). Each line was planted in a single row of 12 hills (18 cm between hills and 30 cm between rows) and fertilized with 56kg N, 176kg P2O5, and 56 kg K2O ha−1 with no top dressing was applied. Plants in pots were grown outdoors in 12-L pots filled with a 1:1 (v/v) mixture of paddy soil (alluvial clay loam) and upland soil (diluvial volcanic ash) at a density of three hills per pot (three plants per hill). Fertilizer (1.0 g each N, P2O5, and K2O per pot) was applied at planting, and additional fertilizer (0.3 g N per pot) was applied at 69 and 85 days after sowing (DAS). Plants grown in a controlled-environment cabinet (14.5 h light/9.5 h dark; 28°C for 12 h and 24°C for 12 h) were in 3-L pots filled with a flooded, granular culture soil. The relative humidity was 60%; the photosynthetic photon flux density (PPFD) at the top of the canopy was 500 μmol photons m−2 s−1. The soil contained 1.2 g N, 3.2 g P2O5, and 1.8 g K2O per pot.
Plant Materials for QTL Mapping
CAR8 mapping was carried out using self-pollinated progenies derived from a BC5F4 population (912 plants) of a “Koshihikari” × “Habataki” cross with “Koshihikari” as the recurrent parent. They have a single heterozygous region in chromosome 8 and most other regions were homozygous for “Koshihikari” alleles. We selected 23 plants from the BC5F4 population and used homozygous BC5F6 generation for phenotyping (Figure 1). The near isogenic line NIL(CAR8) was also selected from the BC5F6 generation. Subsequently, fine mapping was carried out using self-pollinated progenies derived from a BC5F5 population (144 plants) of the same “Koshihikari” × “Habataki” cross. We selected 6 plants from the BC5F5 generation and used homozygous BC5F7 generation for the phenotyping (Figure 2). Molecular markers used for mapping are listed in Table S1. These plants were grown in the paddy field. For both experiments, a randomized block design (three replicates) was used and 4~6 plants were evaluated in each replicate.
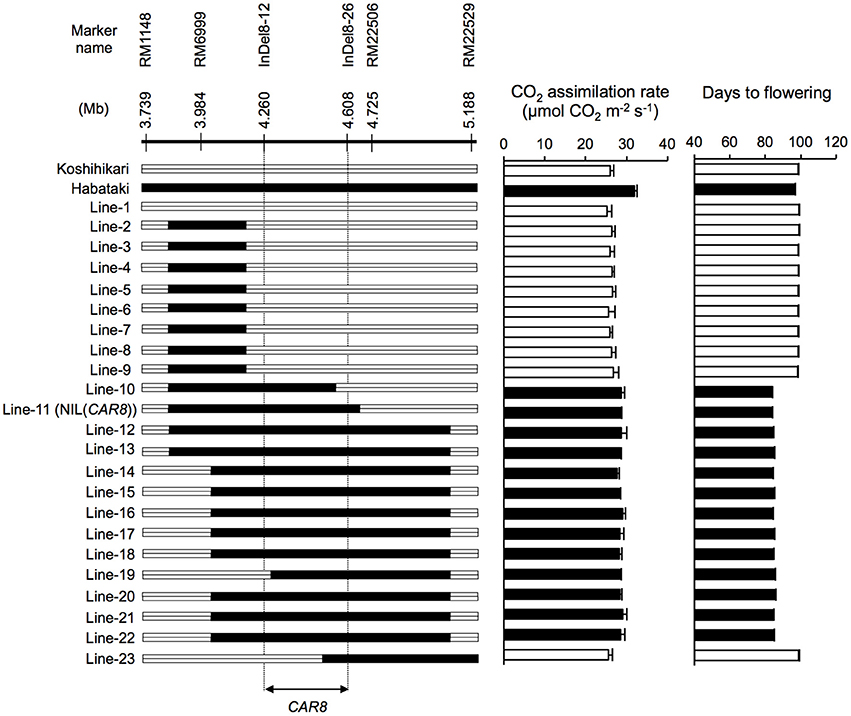
Figure 1. Substitution mapping of CAR8 using homozygous recombinant lines (BC5F6). Molecular markers are shown from the short arm (left) to the long arm (right) of chromosome 8. White segments, homozygous for “Koshihikari” alleles; black segments, homozygous for “Habataki” alleles. Field-grown plants were used. CO2 assimilation rate of flag leaves was measured at an ambient CO2 concentration of 370 μmol mol−1, a PPFD of 2000 μmol photons m−2 s−1, a leaf-to-air vapor pressure difference of 1.3–1.6 kPa, and an air temperature of 30°C. Black bars in graphs indicate significant difference from “Koshihikari” at the 5% level by Dunnett's test. Error bars indicate SD (n = 3).
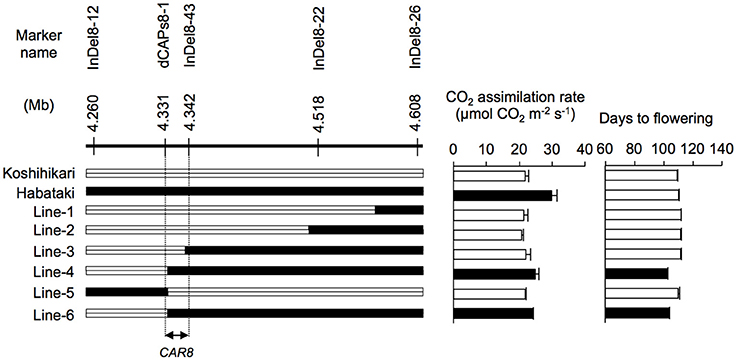
Figure 2. Fine mapping of CAR8 using homozygous recombinant lines (BC5F7). Molecular markers are shown from the short arm (left) to the long arm (right) of chromosome 8. White segments, homozygous for “Koshihikari” alleles; black segments, homozygous for “Habataki” alleles. Field-grown plants were used. CO2 assimilation rate of flag leaves was measured at an ambient CO2 concentration of 370 μmol mol−1, a PPFD of 2000 μmol photons m−2 s−1, a leaf-to-air vapor pressure difference of 1.3–1.6 kPa, and an air temperature of 30°C. Black bars in graphs indicate significant difference from “Koshihikari” at the 5% level by Dunnett's test. Error bars indicate SD (n = 3).
Gas Exchange and Nitrogen Measurements
Leaf gas exchange was measured with a portable gas-exchange system (LI-6400; LI-COR, Lincoln, NE, USA) and 2 × 3 cm cuvette with an LED irradiation source (LI-6400- 02B; LI-COR). The uppermost fully expanded leaves were used for the measurements before heading, and flag leaves after heading. A and gs were measured at an ambient CO2 concentration of 370 μmol mol−1, PPFD of 2000 μmol photons m−2 s−1, a leaf-to-air vapor pressure difference of 1.3–1.6 kPa, and a leaf temperature of 30°C. Plants were examined from 08:30 to 11:30, when the photosynthetic rate was close to the daily maximum (Hirasawa and Ishihara, 1992). The CO2 assimilation rate vs. Ci was examined at a light intensity of 2000 μmol photons m−2 s−1 and a leaf temperature of 30°C at full heading stage by changing the ambient CO2 concentration. To prevent potential leaks, we sealed the gaskets with vacuum grease. Rubisco-limited photosynthesis (Ac) was calculated from Farquhar et al. (1980) as:
where Γ* (μmol mol−1) is the CO2 compensation point in the absence of day respiration, Kc (μmol mol−1) and Ko (mmol mol−1) are the Michaelis constants for CO2 and O2 respectively, and Rd (μmol mol−1) is the day respiration rate. Photosynthetic rate limited by RuBP regeneration capacity (Ar) is calculated as;
The Kc, Ko, and Γ* at 30°C were calculated from the data of Makino et al. (1988) using the Arrhenius function described by von Caemmerer (2000). To convert the Kc and Ko from concentrations to partial pressures, solubilities of 0.0334 mol L−1 bar−1 for CO2 and 0.00126 mol L−1 bar−1 for O2 were used (von Caemmerer, 2000). A/Ci response curves were analyzed using the mathematical model developed by Sharkey et al. (2007) and the data were automatically fitted with the model fitting utility based on a Microsoft Excel program (http://www.blackwellpublishing.com/plantsci/pcecalculation/).
Immediately after the measurements of photosynthesis, 30-mm-long segment was cut from the center of the leaf of measured plants and stored at −80°C. The leaves were then dried at 80°C for 24 h and the nitrogen content was assayed using with a CN analyzer (MT700 Mark II, Yanako, Kyoto, Japan).
Determination of Stomatal Density and Pore Length
The middle part of flag leaves was fixed in solution containing (v/v) 5% formalin, 5% acetic acid, and 45% ethyl alcohol in distilled water. Abaxial and adaxial surfaces of the fixed leaves were photographed under a scanning electron microscope (TM3030; Hitachi, Tokyo, Japan). Stomatal number was counted using a touch screen (Flexscan T2351W; Eizo, Ishikawa, Japan) connected to a computer that was installed with original computer software that senses the number of contacts. Length of stomatal pores was analyzed with ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Determination of Hydraulic Conductance and Hydraulic Conductivity of Plants
The hydraulic conductance from roots to leaves (Cp, 10−8 m3 s−1 MPa−1) was calculated as Uw/(Ψs − Ψl; Hirasawa and Ishihara, 1991), where Uw (10−8 m3 s−1) is the water uptake rate of the whole plant, Ψs (MPa) is the water potential of the soil immediately outside the root, and Ψl (MPa) is the average water potential of the uppermost three leaves. Since plants were submerged the water potential of the soil solution, Ψs was regarded as 0. Plants grown in 3-L pots were used. Measurements were made in a controlled-environment cabinet [air temperature, 28°C; air vapor pressure deficit (VPD), 1.5 kPa; PPFD at the top leaves, 900 μmol m−2 s−1]. Uw was determined from the rate of weight loss of the pot over 20 min after a steady state had been reached. To prevent evaporation from the surface of the pot, the top was covered with polystyrene foam and the gap between the foam and the stem was sealed with oil clay. After measurement of Uw, Ψl was measured in a pressure chamber (model 3005; Soil Moisture Equipment, Santa Barbara, CA, USA). The transpiration rate and gs do not influence Cp when the transpiration rate is high (Fiscus, 1975; Hirasawa and Ishihara, 1991; Stiller et al., 2003). The Uw per leaf area was sufficiently high (>2.0 mmol m−2 s−1) to eliminate the effect of the difference in water uptake rate on Cp. After roots had been washed gently in water, root surface area (Sr) was measured with an image analyzer (WinRHIZO REG V 2004b; Regent Instruments, Quebec, Canada). The hydraulic conductivity (Lp,10−8 m s−1 MPa−1) was expressed as Cp per Sr (Steudle and Peterson, 1998).
Response to the Change of Vapor Pressure Deficit
Plants grown outdoors in 12-L pots until full heading stage were moved to a controlled-environment cabinet (KG-50HLA; Koito Manufacturing Co. Ltd, Tokyo, Japan) at a PPFD and temperature at the flag leaf surface of 900 μmol photons m−2 s−1 and 30°C. Air humidity was modified in steps to generate a range of VPD values inside the cabinet. The temperature and humidity near the flag leaf were monitored with a thermo-hygro sensor (Climomaster model 6531; Kanomax, Osaka, Japan). gs of the flag leaf was measured with the LI-6400 portable gas-exchange system after a steady state had been reached; the leaf chamber conditions were similar to those in the cabinet. After gas exchange measurements, water potential of each leaf was determined with the pressure chamber.
Statistical Analysis
For the fine mapping, Dunnett's test was applied in the mapping population. For comparisons of physiological traits, we analyzed ANOVA and least significant difference (LSD) test. All analyses were tested with JMP v.12 software (SAS Institute, Cary, NC, USA).
Results
Fine Mapping of CAR8
Using homozygous recombinant lines derived from a cross between “Koshihikari” and “Habataki,” we conducted fine mapping of CAR8 (Figures 1–3). These plants were grown in the paddy field. The A of the flag leaves was measured at full heading stage, which was 3–7 days after flowering, under light-saturated conditions and ambient CO2 concentration. Using lines of BC5F6 generation, we narrowed down the CAR8 region to 348.3 kb between insertion-deletion (InDel) marker InDel8-12 and InDel8-26 on the short arm of chromosome 8 (Figure 1). Among lines of BC5F7 generation, two of the six lines showed higher A-values than “Koshihikari” (Figure 2). This enabled us to delimit the CAR8 region to 11.0 kb between the derived cleaved amplified polymorphic sequence (dCAPS) marker dCAPS8-1 and InDel8-43 (Figure 3A). A single gene, Os08g0174500, was predicted in this region using the RAP-DB. Os08g0174500 encodes a Heme Activator Protein 3 (OsHAP3) subunit of CCAAT-box-binding transcription factor called OsHAP3H. This gene was same gene to DTH8, Ghd8, and LHD1 (days to heading 8, grain number, plant height and heading date 8, and Late Heading Date 1), which have been reported to regulate heading date (Wei et al., 2010; Yan et al., 2011; Dai et al., 2012). The time to heading was 7–10 days shorter in two homozygous recombinant lines with higher A than in “Koshihikari” (Figures 1, 2). Sequence analysis of Os08g0174500 revealed that “Koshihikari” had a reading frame totaling 894 bp that encodes a protein of 297 amino acids. In “Habataki,” a 1-bp deletion at 322 bp from the initiation codon caused a frameshift and premature termination of translation, resulting in a truncated protein of 125 amino acids (Figures 3B,C).
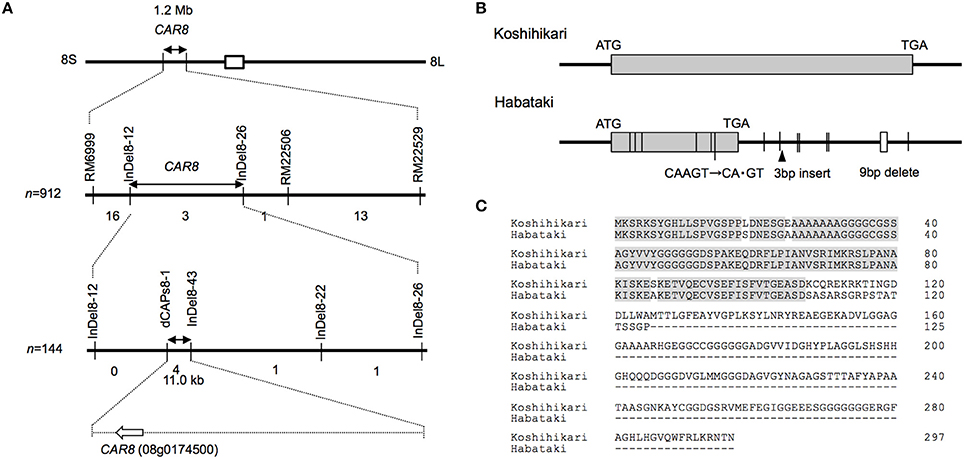
Figure 3. Map-based cloning of CAR8. (A) Fine mapping of CAR8. The number of recombinants between molecular markers is indicated below the each line. (B) Structure of CAR8. The exon is shown as a gray box. Vertical lines without labels represent single-base substitutions between “Koshihikari” and “Habataki.” Small open boxes represent deletions. (C) Alignment of CAR8 amino acid sequences.
Photosynthesis Response
The near isogenic line NIL(CAR8) was selected from the BC5F6 generation derived from a cross between “Koshihikari” and “Habataki” with DNA marker assisted selection (Figure 1). NIL(CAR8) carries a single chromosome segment of “Habataki,” which includes the CAR8 region, in the genetic background of “Koshihikari” (Figure S1). The length of the substituted region in NIL(CAR8) was approximately 1.0 Mb.
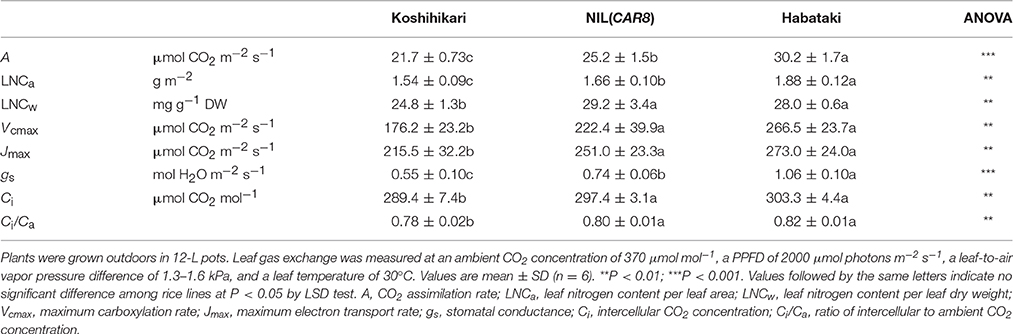
Using plants grown outdoors in 12-L pots, we evaluated several traits that affect A (Table 1). At full heading stage, which was 2–4 days after flowering, A of the flag leaves in NIL(CAR8) at CO2 concentration of 370 μmol mol−1 was 16% higher than that of the flag leaves in “Koshihikari.” Leaf nitrogen content per leaf area (LNCa) and leaf nitrogen content per leaf dry weight (LNCw) in NIL(CAR8) were also higher than in “Koshihikari.” Vcmax and Jmax estimated from A–Ci responses (Sharkey et al., 2007) were higher in NIL(CAR8) than in “Koshihikari.” gs was higher in NIL(CAR8) than in “Koshihikari,” such that Ci and Ci/Ca in NIL(CAR8) were also higher than those of “Koshihikari.” These values in “Habataki” were generally higher than those in NIL(CAR8), although the statistically significant differences were found in only A, LNCa and gs. We also found that NIL(CAR8) had slightly higher A regardless of Ci values than “Koshihikari” in the A-Ci curve (Figure 4).
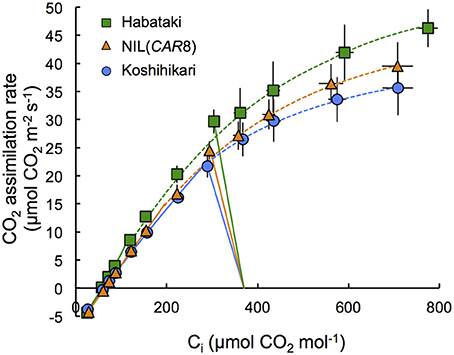
Figure 4. Response of CO2 assimilation rate of flag leaves at full heading to intercellular CO2 concentration. Plants of “Koshihikari” (circles), NIL(CAR8) (triangles), and “Habataki” (squares) were grown outdoors in 12-L pots. Leaf gas exchange was measured at a PPFD of 2000 μmol photons m−2 s−1 and an air temperature of 30°C. CO2 assimilation rate limited by RuBP carboxylation (solid line) and CO2 assimilation rate limited by RuBP regeneration (dotted line) were shown. Curve fitting was described in the Materials and Methods Section. The straight lines represent the measurement at ambient CO2 concentration of 370 μmol mol−1. Error bars indicate SD (n = 6).
Values of gs are affected by stomatal density, pore length, and aperture (Maruyama and Tajima, 1990; Ohsumi et al., 2007). Stomatal densities in the adaxial and abaxial epidermis were similar between NIL(CAR8) and “Koshihikari” (Figure 5A). There was no significant difference in the pore length between NIL(CAR8) and “Koshihikari” (Figure 5B). These values in “Habataki” were significantly higher than those in NIL(CAR8) and “Koshihikari.”
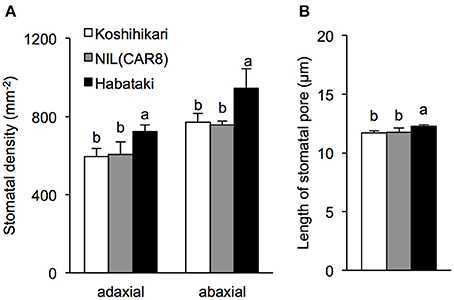
Figure 5. Stomatal density (A) and stomatal pore length (B) in flag leaves of field-grown plants at full heading. Error bars indicate SD (n = 3). Values followed by the same letters indicate no significant difference among rice lines at P < 0.05 by LSD test.
We monitored leaf gas exchange and LNC throughout the growth period (Table 2). The number of days from sowing to flowering was 100 in “Koshihikari,” 91 in NIL(CAR8), and 103 in “Habataki.” There was no difference in A, gs, LNCa, and LNCw between “Koshihikari” and NIL(CAR8) at 47 and 67 days after sowing (DAS). At 95 DAS, A, gs, LNCa, and LNCw in NIL(CAR8) were higher than those of “Koshihikari,” while only gs was higher in NIL(CAR8) at 105 DAS. “Habataki” showed higher A than NIL(CAR8) from 67 to 105 DAS, which was accompanied by the higher gs and in some cases higher LNCa and LNCw.
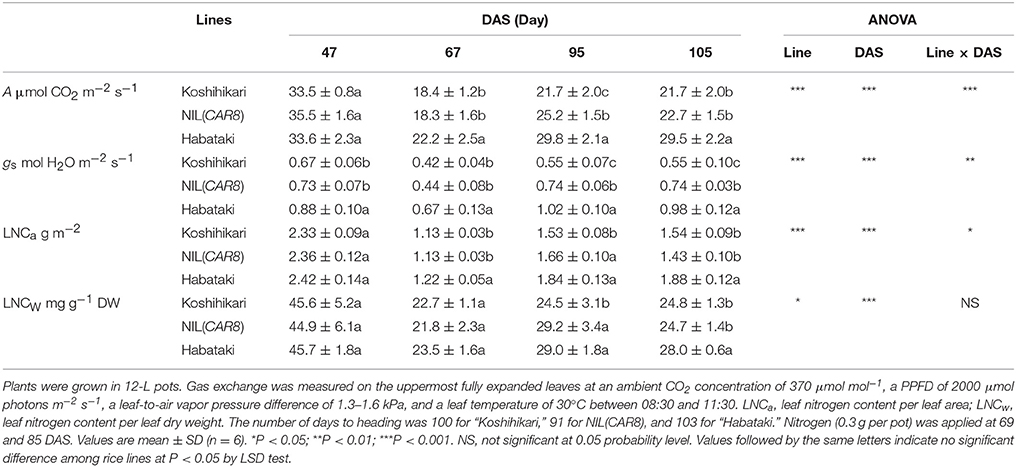
Table 2. CO2 assimilation rate (A), stomatal conductance (gs), and leaf nitrogen content (LNC) between 47 and 105 days after sowing (DAS).
Hydraulic Conductance
It is suggested that gs is influenced by the hydraulic conductance of a plant (Brodribb and Holbrook, 2003). When we compared the plants grown in 3-L pots in a controlled-environment cabinet at the full heading stage, Cp in NIL(CAR8) was significantly higher than in “Koshihikari” (Table 3). The Cp in “Habataki” was much higher than that in NIL(CAR8). Cp can be divided in root surface area (Sr) and hydraulic conductance per Sr, i.e., hydraulic conductivity (Lp) (Steudle and Peterson, 1998). NIL(CAR8) showed similar Sr but higher Lp in comparison with “Koshihikari,” while “Habataki” showed higher Sr but similar Lp in comparison with “Koshihikari.” We also determined that A and gs in NIL(CAR8) were higher than those of “Koshihikari” (data not shown).
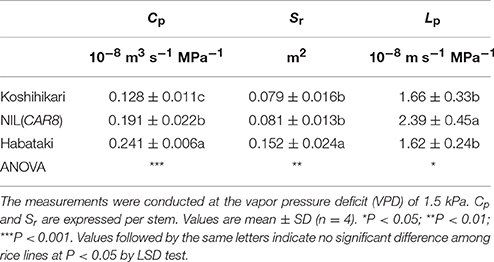
Table 3. Hydraulic conductance from roots to leaves (Cp), root surface area (Sr), and hydraulic conductivity (Lp) of plants grown in a controlled-environment cabinet.
To assess relationship between leaf water status and gs, we compared the responses of transpiration rate (T), gs, and leaf water potential (Ψl) to vapor pressure deficit (VPD) at the full heading stage with the plants grown in 12-L pots (Figure S2). In all genotypes, T increased and gs and Ψl declined with increasing VPD. In NIL(CAR8) and “Habataki,” T and gs were always higher than in “Koshihikari,” whereas Ψl was similar in all three genotypes irrespective to VPD conditions. These results indicate that NIL(CAR8) keeps Ψl at a certain level even though their T are significantly higher than “Koshihikari.”
Grain Yield
We examined the final grain yield of the plant grown in the paddy field (Figure S3). The brown rice yield in NIL(CAR8) was lower than that of “Koshihikari”, while the yield of “Habataki” was significantly higher than the others.
Discussion
The understanding of genetic factors and their physiological aspects that control the natural variation of rice photosynthesis are important for future rice breeding aimed at increasing grain yield. In this study, we narrowed down the genetic region of CAR8 located in the short arm of chromosome 8 and evaluated the physiological aspects of CAR8.
The result of the fine mapping suggests that the protein encoded by CAR8 is a putative OsHAP3 subunit of the HAP complex, OsHAP3H. HAP complex binds to CCAAT box and act either as a transcription activator or as a repressor (Laloum et al., 2013). The HAP complex consists of three subunits: HAP2, HAP3, and HAP5 (Mantovani, 1999). Each of the HAP subunits is encoded by a single gene in yeast (Saccharomyces cerevisiae) and mammals (Mantovani, 1999), while in rice, the genome encodes 10 OsHAP2, 11 OsHAP3, and seven OsHAP5 subunits (Thirumurugan et al., 2008). CAR8 might be identical to DTH8/Ghd8/LHD1, which was reported to control rice flowering date (Wei et al., 2010; Yan et al., 2011; Dai et al., 2012). According to the classification of Wei et al. (2010), the “Koshihikari” allele corresponds to type 1 and the “Habataki” allele to type 8. Under long-day conditions, the type 1 allele of DTH8 negatively influenced the expression of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a), resulting in repression of flowering (Wei et al., 2010). Recently, it is revealed that DTH8 binds to Heading date 1 (Hd1), which represses the expression of Ehd1 and control the heading date (Chen et al., 2014). The “Koshihikari” allele might suppress the expression of these genes and delay the heading date, while the allele of “Habataki” might not. Although it is well known that OsHAP3H regulates rice flowering, the association of this gene with photosynthesis has been little noticed. The detailed analysis of molecular mechanisms including complementation tests would contribute to understand how CAR8 controls both photosynthesis and flowering.
Two hypotheses can generally explain an increase in A in C3 plants: (1) increase in the biochemical activity of the leaf photosynthetic machinery and (2) enhancement of CO2 diffusion from air into leaves (Farquhar and Sharkey, 1982). While we didn't find any difference in A during vegetative stage (i.e., at 47 and 67 DAS in Table 2), we found the higher A by 16% in NIL(CAR8) than that in “Koshihikari” at full heading stage. A higher abundance of photosynthetic proteins is indicated by a corresponding increase in LNC (Makino et al., 1984). We found that NIL(CAR8) had higher LNCa and LNCw than “Koshihikari” at the full heading stage (Table 1). Biochemically, the rate of photosynthesis is generally limited by either RuBP carboxylation capacity, or RuBP regeneration capacity in the broad sense (which would include Calvin cycle capacity and Pi regeneration in addition to electron transport rate; Farquhar et al., 1980; Sharkey, 1985). Yamori et al. (2011a) reported that the value of A in rice (cv. Notohikari) at an ambient CO2 concentration of 380 μmol mol−1 was limited by RuBP regeneration rate. We applied the theoretical analysis of our results using the Farquhar and von Caemmerer model (as modified by Sharkey et al., 2007) and found the A of NIL(CAR8) and “Kohishikari” at 370 μmol CO2 mol−1 tended to be limited by RuBP regeneration rate, although it is close to the limiting region of Vcmax (Figure 4). The increase of RuBP regeneration rate corresponds to increase of ~2.0 μmol m−2 s−1 of A at the ambient CO2 concentration when we calculated from the A-Ci curve. Hence, we conclude that the higher A in NIL(CAR8) than “Koshihikari” is mainly due to an enhanced Jmax (Table 1). “Habataki” had even higher Jmax and A, indicating “Habataki” includes additional QTL for enhanced Jmax.
A number of possibilities could explain how Jmax is enhanced in NIL(CAR8) and “Habataki.” The simplest is that higher LNCa and LNCw in these lines provides more photosynthetic protein in both leaf area and leaf weight basis. This is probably the best explanation since both the Vcmax and Jmax were increased, indicating an across the board enhancement of photosynthetic protein. With respect to Jmax, it has been suggested that the electron flow through the Cytochrome b6/f complex is a rate-limiting step for RuBP regeneration (Yamori et al., 2011b). Therefore, the increased LNC in NIL(CAR8) likely increases the cytochrome b6/f, but could also the photosystems, quinones, and plastocyanin components of whole chain-electron transport. Enhanced Calvin cycle protein may also contribute to higher A should it share in the limitation of Jmax (Raines, 2011). Enzymes of starch and sucrose synthesis probably do not, as the CO2 responsiveness apparent in the A/Ci curve at 370 μmol mol−1 is much greater than would be expected under a Pi regeneration limitation (Sage, 1990). The higher LNC might be explained by the higher net accumulation of aboveground nitrogen and/or the higher rate of distribution of nitrogen to leaves (Mae and Ohira, 1981). This should be elucidated in future study. It is known that HAP3 genes are associated with chloroplast biosynthesis and photosynthesis. In rice, an RNA interference construct silencing OsHAP3A, OsHAP3B, and OsHAP3C resulted in reduced expression of nuclear-encoded photosynthesis genes and degenerated chloroplast (Miyoshi et al., 2003). Recently, Alam et al. (2015) showed the overexpression of OsHAP2E increased the leaf chlorophyll content and A in rice. These suggest that HAP members redundantly affect the leaf photosynthesis in rice. It is also reported that the overexpression of TaNF-YB3, a member of HAP3, led to increases in the leaf chlorophyll content and photosynthesis in wheat (Triticum aestivum, Stephenson et al., 2011). These reports imply the association to the increased Jmax in this study.
We then considered the second hypothesis that higher A results from enhancement of CO2 diffusion from air into leaves. NIL(CAR8) showed higher gs than “Koshihikari” at full heading stage (Table 1). While much of the gs response could reflect the regulation of gs to track A (Wong et al., 1979), there was a slight increases in Ci and Ci/Ca ratio in NIL(CAR8) relative to “Koshihikari” (Table 1). The higher Ci/Ca in NIL(CAR8) demonstrate a greater proportional increase in gs than A, such that the stomatal control over A has been relaxed. The 3% higher Ci in NIL(CAR8) than “Koshihikari” at the ambient CO2 concentration corresponds to increase of 1.5 μmol m−2 s−1 of A calculated from the A/Ci curve. These results indicate that CAR8 enhances gs independently of A.
In rice, gs is determined by stomatal density, pore length, and aperture (Maruyama and Tajima, 1990; Ohsumi et al., 2007). Our results show that CAR8 increases gs by increasing stomatal aperture rather than stomatal density (Figure 5). The increase in stomatal density may also increase gs in rice because stomatal density is higher in “Habataki” than in “Koshihikari” (Figure 5). This indicates that “Habataki” has alleles that enhance stomatal density, and a combination of these alleles and CAR8 may further enhance gs.
Stomatal conductance responds to changes in plant water status (Schulze and Hall, 1982), and several studies have shown that it is closely related to Cp (Meinzer and Grantz, 1990; Hirasawa and Ishihara, 1992; Hubbard et al., 2001; Cochard et al., 2002; Brodribb et al., 2007). NIL(CAR8) had higher Cp than “Koshihikari” due to higher Lp (Table 3). We also found that Ψl in NIL(CAR8) was similar to that of “Koshihikari” while gs in NIL(CAR8) remained high regardless of VPD conditions (Figure S2). This suggests that the higher water uptake of the root in NIL(CAR8) keeps Ψl high and decreases the risk of water stress even though T in NIL(CAR8) is significantly higher than “Koshihikari.” Therefore, the higher gs in NIL(CAR8) would be partially explained by the higher Lp. In contrast, Sakurai-Ishikawa et al. (2011) suggested the increase of water demand of shoots enhances root hydraulic conductivity via increase in gene expression of several aquaporins in the plasma membrane intrinsic protein family. This might explain the concomitant increases of gs and Lp in NIL(CAR8). To our knowledge, there has been no report that shows association between HAPs genes and stomatal conductance. The identification of molecular network of CAR8 would help to understand the regulations of stomatal conductance in rice. Kanemura et al. (2007) reported a weak negative relationship between gs of flag leaves and days to heading using the rice diversity research set of germplasm. This suggests that flowering time affects photosynthesis of flag leaves and the allelic variation of CAR8 would explain in part the natural variation of photosynthesis. This also implies the necessity to determine the association between flowering genes and photosynthesis, comprehensively.
The final grain yield in NIL(CAR8) was inferior to that in “Koshihikari” (Figure S3). This might be resulted from the short growth duration due to the “Habataki” allele of CAR8 gene. To enhance the grain yield in rice, we should extend the growth duration of NIL(CAR8) by adding genes which delay heading date or modifying the growth conditions such as planting time.
In conclusion, the “Habataki” allele of CAR8 associates to LNC under the 12-L pot condition. The higher LNC in NIL(CAR8), which relates to the higher RuBP regeneration rate, would mainly explain the enhanced A of the flag leaves. The “Habataki” allele of CAR8 also associates hydraulic conductivity and hydraulic conductance at full heading stage under the 3-L pot condition. This could allow for a higher gs in NIL(CAR8), which would partially explain the enhanced A. The fine mapping suggested that CAR8 encodes a putative OsHAP3 subunit of a CCAAT-box-binding transcription factor and is identical to DTH8/Ghd8/LHD1, which has been reported to regulate flowering date. Identification of its molecular function would help understanding the association between photosynthesis and flowering and demonstrate specific genetic mechanisms that can be exploited to improve photosynthesis in rice and potentially other crops.
Author Contributions
SA, TY, RS, and JY designed the experiments. SA, KY, UY, and JS performed the experiments. TT built the stomata counting system. SA, TO, RS, TH, and JY wrote the manuscript.
Funding
This work was supported in part by Grants-in-Aid from the Japan Society for the Promotion of Science (Postdoctoral Fellowship to SA), Japan Science and Technology Agency, Precursory Research for Embryonic Science and Technology to SA, the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics-based for Agricultural Innovation, RBS-2006 to TH), and the Institute of Global Innovation Research in TUAT to SA and TH.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AS and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
We are grateful to Dr. J. Wu and Dr. Y. Katayose for help in genome sequence analysis, Ms. H.J. Zhu, Ms. N. Iioka, Ms. M. Takahashi, Ms. M. Iizumi, Ms. Y. Shimazu, and Ms. E. Abe for their excellent technical support, and Dr. K. Hori, E. Yamamoto, and H. Omori for helpful advice on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00060/full#supplementary-material
Abbreviations
A, CO2 assimilation rate; Ca, ambient CO2 concentration; CAR8, carbon assimilation rate 8; Ci, intercellular CO2 concentration; Cp, hydraulic conductance from roots to leaves; dCAPS, derived cleaved amplified polymorphic sequence; DAS, days after sowing; DTH8, days to heading 8; Ehd1, Early heading date1; gs, stomatal conductance; Γ*, CO2 compensation point in the absence of day respiration; Ghd8, grain number, plant height and heading date 8; GPS, Green for Photosynthesis; HAP, heme activator protein; Hd1, Heading date 1; Hd3a, Heading date 3a; InDel, insertion-deletion; Jmax, maximum rate of electron transport; Kc, Michaelis constants for CO2; Ko,Michaelis constants for O2; LHD1, Late Heading Date 1; Lp, hydraulic conductivity; LNCa, leaf nitrogen content per leaf area; LNCw, leaf nitrogen content per leaf dry weight; NIL, near-isogenic line; O, intercellular O2 concentration; PPFD, Photosynthetic photon flux density; Ψl, leaf water potential; QTL, quantitative trait locus; Rd, day respiration rate; Rubisco, ribulose-1,5 bisphosphate carboxylase/oxygenase; RuBP, ribulose 1,5-bisphosphate; Sr, root surface area; T, transpiration rate; Vcmax, maximum rate of RuBP carboxylation; VPD, vapor pressure deficit.
References
Adachi, S., Baptista, L. Z., Sueyoshi, T., Murata, K., Yamamoto, T., Ebitani, T., et al. (2014). Introgression of two chromosome regions for leaf photosynthesis from an indica rice into the genetic background of a japonica rice. J. Exp. Bot. 65, 2049–2056. doi: 10.1093/jxb/eru047
Adachi, S., Nito, N., Kondo, M., Yamamoto, T., Arai-Sanoh, Y., Ando, T., et al. (2011). Identification of chromosomal regions controlling the leaf photosynthetic rate in rice by using a progeny from japonica and high-yielding indica varieties. Plant Prod. Sci. 14, 118–127. doi: 10.1626/pps.14.118
Alam, M. M., Tanaka, T., Nakamura, H., Ichikawa, H., Kobayashi, K., Yaeno, T., et al. (2015). Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant Biotech. J. 13, 85–96. doi: 10.1111/pbi.12239
Brodribb, T. J., Feild, T. S., and Jordan, G. J. (2007). Leaf maximum photosynthetic rate and venation are linked by hydraulics. Plant Physiol. 144, 1890–1898. doi: 10.1104/pp.107.101352
Brodribb, T. J., and Holbrook, N. M. (2003). Stomatal closure during leaf dehydration, correlation with other leaf physiological traits. Plant Physiol. 132, 2166–2173. doi: 10.1104/pp.103.023879
Chen, J., Li, X., Cheng, C., Wang, Y., Qin, M., Zhu, H., et al. (2014). Characterization of epistatic interaction of QTLs LH8 and EH3 controlling heading date in rice. Sci. Rep. 4:4263. doi: 10.1038/srep04263
Cochard, H., Coll, L., Le Roux, X., and Améglio, T. (2002). Unraveling the effects of plant hydraulics on stomatal closure during water stress in walnut. Plant Physiol. 128, 282–290. doi: 10.1104/pp.010400
Cook, M., and Evans, L. (1983). Some physiological aspects of the domestication and improvement of rice (Oryza spp.). Field Crops Res. 6, 219–238. doi: 10.1016/0378-4290(83)90062-X
Dai, X., Ding, Y., Tan, L., Fu, Y., Liu, F., Zhu, Z., et al. (2012). LHD1, an allele of DTH8/Ghd8, controls late heading date in common wild rice (Oryza rufipogon). J. Int. Plant Biol. 54, 790–799. doi: 10.1111/j.1744-7909.2012.01166.x
FAO (2015). FAOSTAT Agriculture Data. Available online at: http://faostat.fao.org/site/339/default.aspx (Accessed 1 October, 15).
Farquhar, G. D., von Caemmerer, S., and Berry, J. A. (1980). A biochemical-model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 149, 78–90. doi: 10.1007/BF00386231
Farquhar, G., and Sharkey, T. (1982). Stomatal conductance and photosynthesis. Ann. Rev. Plant Physiol. 33, 317–345. doi: 10.1146/annurev.pp.33.060182.001533
Fiscus, E. L. (1975). The interaction between osmotic-and pressure-induced water flow in plant roots. Plant Physiol. 55, 917–922. doi: 10.1104/pp.55.5.917
Flood, P. J., Harbinson, J., and Aarts, M. G. (2011). Natural genetic variation in plant photosynthesis. Trends Plant Sci. 16, 327–335. doi: 10.1016/j.tplants.2011.02.005
Gu, J., Yin, X., Struik, P. C., Stomph, T. J., and Wang, H. (2012). Using chromosome introgression lines to map quantitative trait loci for photosynthesis parameters in rice (Oryza sativa L.) leaves under drought and well-watered field conditions. J. Exp. Bot. 63, 455–469. doi: 10.1093/jxb/err292
Hirasawa, T., and Ishihara, K. (1991). On resistance to water transport in crop plants for estimating water uptake ability under intense transpiration. Jpn. J. Crop. Sci. 60, 174–183. doi: 10.1626/jcs.60.174
Hirasawa, T., and Ishihara, K. (1992). “Relationship between resistance to water transport and midday stomatal aperture,” in Research in Photosynthesis, Vol. IV, ed N. Murata (Dordrecht: Kluwer Academic Publishers), 283–286.
Hu, S. P., Zhou, Y., Zhang, L., Zhu, X. D., Li, L., Luo, L. J., et al. (2009). Correlation and quantitative trait loci analyses of total chlorophyll content and photosynthetic rate of rice (Oryza sativa) under water stress and well-watered conditions. J. Int. Plant Biol. 51, 879–888. doi: 10.1111/j.1744-7909.2009.00846.x
Hubbard, R., Ryan, M., Stiller, V., and Sperry, J. (2001). Stomatal conductance and photosynthesis vary linearly with plant hydraulic conductance in Ponderosa pine. Plant Cell Environ. 24, 113–121. doi: 10.1046/j.1365-3040.2001.00660.x
International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436, 793–800. doi: 10.1038/nature03895
Jahn, C. E., McKay, J. K., Mauleon, R., Stephens, J., McNally, K. L., Bush, D. R., et al. (2011). Genetic variation in biomass traits among 20 diverse rice varieties. Plant Physiol. 155, 157–168. doi: 10.1104/pp.110.165654
Kajala, K., Covshoff, S., Karki, S., Woodfield, H., Tolley, B. J., Dionora, M. J. A., et al. (2011). Strategies for engineering a two-celled C4 photosynthetic pathway into rice. J. Exp. Bot. 62, 3001–3010. doi: 10.1093/jxb/err022
Kanemura, T., Homma, K., Ohsumi, A., Shiraiwa, T., and Horie, T. (2007). Evaluation of genotypic variation in leaf photosynthetic rate and its associated factors by using rice diversity research set of germplasm. Photosynth. Res. 94, 23–30. doi: 10.1007/s11120-007-9208-7
Khush, G. S. (2013). Strategies for increasing the yield potential of cereals: case of rice as an example. Plant Breed. 132, 433–436. doi: 10.1111/pbr.1991
Kusumi, K., Hirotsuka, S., Kumamaru, T., and Iba, K. (2012). Increased leaf photosynthesis caused by elevated stomatal conductance in a rice mutant deficient in SLAC1, a guard cell anion channel protein. J. Exp. Bot. 63, 5635–5644. doi: 10.1093/jxb/ers216
Laloum, T., De Mita, S., Gamas, P., Baudin, M., and Niebel, A. (2013). CCAAT-box binding transcription factors in plants: y so many? Trends Plant Sci. 18, 157–166. doi: 10.1016/j.tplants.2012.07.004
Long, S. P., Zhu, X. G., Naidu, S. L., and Ort, D. R. (2006). Can improvement in photosynthesis increase crop yields? Plant Cell Environ. 29, 315–330. doi: 10.1111/j.1365-3040.2005.01493.x
Mae, T., and Ohira, K. (1981). The remobilization of nitrogen related to leaf growth and senescence in rice plants (Oryza sativa L.). Plant Cell Physiol. 22, 1067–1074.
Makino, A., Mae, T., and Ohira, K. (1984). Relation between nitrogen and ribulose-1, 5-bisphosphate carboxylase in rice leaves from emergence through senescence. Plant Cell Physiol. 25, 429–437.
Makino, A., Mae, T., and Ohira, K. (1988). Differences between wheat and rice in the enzymic properties of ribulose-1,5-bisphosphate carboxylase/oxygenase and the relationship to photosynthetic gas exchange. Planta 174, 30–38. doi: 10.1007/BF00394870
Mantovani, R. (1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239, 15–27. doi: 10.1016/S0378-1119(99)00368-6
Maruyama, S., and Tajima, K. (1990). Leaf conductance in japonica and indica rice varieties, 1: size, frequency, and aperture of stomata. Jpn. J. Crop. Sci. 59, 801–808. doi: 10.1626/jcs.59.801
Meinzer, F., and Grantz, D. (1990). Stomatal and hydraulic conductance in growing sugarcane: stomatal adjustment to water transport capacity. Plant Cell Environ. 13, 383–388. doi: 10.1111/j.1365-3040.1990.tb02142.x
Miyoshi, K., Ito, Y., Serizawa, A., and Kurata, N. (2003). OsHAP3 genes regulate chloroplast biogenesis in rice. Plant J. 36, 532–540. doi: 10.1046/j.1365-313X.2003.01897.x
Murchie, E. H., Pinto, M., and Horton, P. (2009). Agriculture and the new challenges for photosynthesis research. New Phytol. 181, 532–552. doi: 10.1111/j.1469-8137.2008.02705.x
Ohsumi, A., Kanemura, T., Homma, K., Horie, T., and Shiraiwa, T. (2007). Genotypic variation of stomatal conductance in relation to stomatal density and length in rice (Oryza sativa L.). Plant Prod. Sci. 10, 322–328. doi: 10.1626/pps.10.322
Raines, C. A. (2011). Increasing photosynthetic carbon assimilation in C3 plants to improve crop yield: current and future strategies. Plant Physiol. 155, 36–42. doi: 10.1104/pp.110.168559
Sage, R. F. (1990). A model describing the regulation of ribulose-1,5-bisphosphate carboxylase, electron transport, and triose phosphate use in response to light intensity and CO2 in C3 Plants. Plant Physiol. 94, 1728–1734. doi: 10.1104/pp.94.4.1728
Sakai, H., Lee, S. S., Tanaka, T., Numa, H., Kim, J., Kawahara, Y., et al. (2013). Rice annotation project database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 54, e61–e611. doi: 10.1093/pcp/pcs183
Sakurai-Ishikawa, J., Murai-Hatano, M., Hayashi, H., Ahamed, A., Fukushi, K., Matsumoto, T., et al. (2011). Transpiration from shoots triggers diurnal changes in root aquaporin expression. Plant Cell Environ. 34, 1150–1163. doi: 10.1111/j.1365-3040.2011.02313.x
Schulze, E. D., and Hall, A. (1982). “Stomatal responses, water loss and CO2 assimilation rates of plants in contrasting environments,” in Physiological Plant Ecology II, eds O. L. Lange, P. S. Nobel, C. B. Osmond, and H. Ziegler (Berlin: Springer), 181–230.
Sharkey, T. D. (1985). Photosynthesis in intact leaves of C3 plants: physics, physiology and rate limitations. Bot. Rev. 51, 53–105. doi: 10.1007/BF02861058
Sharkey, T. D., Bernacchi, C. J., Farquhar, G. D., and Singsaas, E. L. (2007). Fitting photosynthetic carbon dioxide response curves for C3 leaves. Plant Cell Environ. 30, 1035–1040. doi: 10.1111/j.1365-3040.2007.01710.x
Stephenson, T. J., McIntyre, C. L., Collet, C., and Xue, G. P. (2011). TaNF-YB3 is involved in the regulation of photosynthesis genes in Triticum aestivum. Funct. Integr. Genomics 11, 327–340. doi: 10.1007/s10142-011-0212-9
Steudle, E., and Peterson, C. A. (1998). How does water get through roots? J. Exp. Bot. 49, 775–788. doi: 10.1093/jxb/49.322.775
Stiller, V., Lafitte, H. R., and Sperry, J. S. (2003). Hydraulic properties of rice and the response of gas exchange to water stress. Plant Physiol. 132, 1698–1706. doi: 10.1104/pp.102.019851
Suzuki, Y., Ohkubo, M., Hatakeyama, H., Ohashi, K., Yoshizawa, R., Kojima, S., et al. (2007). Increased Rubisco content in transgenic rice transformed with the ‘sense’ rbcS gene. Plant Cell Physiol. 48, 626–637. doi: 10.1093/pcp/pcm035
Takahara, K., Kasajima, I., Takahashi, H., Hashida, S. N., Itami, T., Onodera, H., et al. (2010). Metabolome and photochemical analysis of rice plants overexpressing Arabidopsis NAD kinase gene. Plant Physiol. 152, 1863–1873. doi: 10.1104/pp.110.153098
Takai, T., Adachi, S., Taguchi-Shiobara, F., Sanoh-Arai, Y., Iwasawa, N., Yoshinaga, S., et al. (2013). A natural variant of NAL1, selected in high-yield rice breeding programs, pleiotropically increases photosynthesis rate. Sci. Rep. 3:2149. doi: 10.1038/srep02149
Takai, T., Kondo, M., Yano, M., and Yamamoto, T. (2010). A quantitative trait locus for chlorophyll content and its association with leaf photosynthesis in rice. Rice 3, 172–180. doi: 10.1007/s12284-010-9047-6
Takano, Y., and Tsunoda, S. (1971). Curvilinear regression of the leaf photosynthetic rate on leaf nitrogen content among strains of Oryza species. Jpn. J. Breed. 21, 69–76. doi: 10.1270/jsbbs1951.21.69
Tanksley, S. D. (1993). Mapping polygenes. Ann. Rev. Genet. 27, 205–233. doi: 10.1146/annurev.ge.27.120193.001225
Teng, S., Qian, Q., Zeng, D., Kunihiro, Y., Fujimoto, K., Huang, D., et al. (2004). QTL analysis of leaf photosynthetic rate and related physiological traits in rice (Oryza sativa L.). Euphytica 135, 1–7. doi: 10.1023/B:EUPH.0000009487.89270.e9
Thirumurugan, T., Ito, Y., Kubo, T., Serizawa, A., and Kurata, N. (2008). Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. Genomics 279, 279–289. doi: 10.1007/s00438-007-0312-3
von Caemmerer, S., and Evans, J. R. (2010). Enhancing C3 photosynthesis. Plant Physiol. 154, 589–592. doi: 10.1104/pp.110.160952
Wei, X., Xu, J., Guo, H., Jiang, L., Chen, S., Yu, C., et al. (2010). DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153, 1747–1758. doi: 10.1104/pp.110.156943
Wong, S. C., Cowan, I. R., and Farquhar, G. D. (1979). Stomatal conductance correlates with photosynthetic capacity. Nature 282, 424–426. doi: 10.1038/282424a0
Yamamoto, T., Uga, Y., and Yano, M. (2014). “Genomics-assisted allele mining and its integration into rice breeding,” in Genomics of Plant Genetic Resources, eds E. Tuberosa, A. Graner, and E. Frison (Berlin: Springer), 251–265.
Yamamoto, T., Yonemaru, J., and Yano, M. (2009). Towards the understanding of complex traits in rice: substantially or superficially? DNA Res. 16, 141–154. doi: 10.1093/dnares/dsp006
Yamori, W., Nagai, T., and Makino, A. (2011a). The rate-limiting step for CO2 assimilation at different temperatures is influenced by the leaf nitrogen content in several C3 crop species. Plant Cell Environ. 34, 764–777. doi: 10.1111/j.1365-3040.2011.02280.x
Yamori, W., Takahashi, S., Makino, A., Price, G. D., Badger, M. R., and von Caemmerer, S. (2011b). The roles of ATP synthase and the cytochrome b6/f complexes in limiting chloroplast electron transport and determining photosynthetic capacity. Plant Physiol. 155, 956–962. doi: 10.1104/pp.110.168435
Yan, W. H., Wang, P., Chen, H. X., Zhou, H. J., Li, Q. P., Wang, C. R., et al. (2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4, 319–330. doi: 10.1093/mp/ssq070
Keywords: leaf nitrogen content, Oryza sativa, photosynthesis, quantitative trait locus, RuBP regeneration, stomatal conductance
Citation: Adachi S, Yoshikawa K, Yamanouchi U, Tanabata T, Sun J, Ookawa T, Yamamoto T, Sage RF, Hirasawa T and Yonemaru J (2017) Fine Mapping of Carbon Assimilation Rate 8, a Quantitative Trait Locus for Flag Leaf Nitrogen Content, Stomatal Conductance and Photosynthesis in Rice. Front. Plant Sci. 8:60. doi: 10.3389/fpls.2017.00060
Received: 06 August 2016; Accepted: 11 January 2017;
Published: 31 January 2017.
Edited by:
Lisa Ainsworth, Agricultural Research Service (USDA), USAReviewed by:
Johannes Kromdijk, University of Illinois at Urbana–Champaign, USAAnthony J. Studer, University of Illinois Urbana-Champaign, USA
Copyright © 2017 Adachi, Yoshikawa, Yamanouchi, Tanabata, Sun, Ookawa, Yamamoto, Sage, Hirasawa and Yonemaru. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junichi Yonemaru, eW9uZW1hcnVAYWZmcmMuZ28uanA=
 Shunsuke Adachi
Shunsuke Adachi Kazuaki Yoshikawa1
Kazuaki Yoshikawa1 Takanari Tanabata
Takanari Tanabata Taiichiro Ookawa
Taiichiro Ookawa Toshio Yamamoto
Toshio Yamamoto Rowan F. Sage
Rowan F. Sage Junichi Yonemaru
Junichi Yonemaru