
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 23 January 2017
Sec. Plant Nutrition
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.00029
This article is part of the Research Topic Integrating plant genetics and genomics for delineating climate resilience and health benefitting characteristics from millets View all 23 articles
Nutritional insecurity is a major threat to the world’s population that is highly dependent on cereals-based diet, deficient in micronutrients. Next to cereals, millets are the primary sources of energy in the semi-arid tropics and drought-prone regions of Asia and Africa. Millets are nutritionally superior as their grains contain high amount of proteins, essential amino acids, minerals, and vitamins. Biofortification of staple crops is proved to be an economically feasible approach to combat micronutrient malnutrition. HarvestPlus group realized the importance of millet biofortification and released conventionally bred high iron pearl millet in India to tackle iron deficiency. Molecular basis of waxy starch has been identified in foxtail millet, proso millet, and barnyard millet to facilitate their use in infant foods. With close genetic-relatedness to cereals, comparative genomics has helped in deciphering quantitative trait loci and genes linked to protein quality in finger millet. Recently, transgenic expression of zinc transporters resulted in the development of high grain zinc while transcriptomics revealed various calcium sensor genes involved in uptake, translocation, and accumulation of calcium in finger millet. Biofortification in millets is still limited by the presence of antinutrients like phytic acid, polyphenols, and tannins. RNA interference and genome editing tools [zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR)] needs to be employed to reduce these antinutrients. In this review paper, we discuss the strategies to accelerate biofortification in millets by summarizing the opportunities and challenges to increase the bioavailability of macro and micronutrients.
Nutritional security is the key to improve the health status of the world’s population as mankind is primarily dependent on plant-based diets. Plants are the major source of nutrients essential for normal growth and development. However, half of the global population, especially people from Asia and Africa suffer from nutrition deficiency as they rely on cereal crops for food (White and Broadley, 2005; Hirschi, 2009; Zhao and McGrath, 2009). Biofortification is a food-based approach to overcome the nutrient starvation by delivering nutrient-dense crops at the door steps of poor populations (Bouis et al., 2011). Biofortification Challenge Program (BCP) under HarvestPlus-Consultative Group for International Agricultural Research (CGIAR) Micronutrients project has focused primarily on seven major staple crops (rice, beans, cassava, maize, sweet potato, pearl millet, and wheat) targeting three important micronutrients (Fe, Zn, and vitamin A) (Welch and Graham, 2004). In resource-poor countries of Asia and Africa, millets provide 75% of total calorie intake next to cereal grains with an average annual production of 14.2 and 12.4 million tons (Belton and Taylor, 2004; O’Kennedy et al., 2006). India is the leading producer of millets accounting for about 80% of the global millet production (Food and Agricultural Organization [FAO], 2015).
Millets are commonly referred as “small seeded grasses” which include pearl millet [Pennisetum glaucum (L.) R. Br.], finger millet [Eleusine coracana (L.) Gaertn], foxtail millet [Setaria italica (L.) Beauv], proso millet (Panicum miliaceum L.), barnyard millet (Echinochloa spp.), kodo millet (Paspalum scrobiculatum), and little millet (Panicum sumatrense). Among the millets, pearl millet occupies 95% of the production (Yadav et al., 2012; Yadav and Rai, 2013; Agricultural Statistics, Government of India, 2014; Nedumaran et al., 2014). Foxtail millet [S. italica (L.) P. Beauv] is the second largest crop among the millets, cultivated for food in semi-arid tropics of Asia and as forage in Europe, North America, Australia, and North Africa (Austin, 2006). Finger millet is the sixth largest crop under cultivation serving as the primary food for rural populations of East and Central Africa and southern India (Vijayakumari et al., 2003). Proso millet is a short-season crop cultivated in drier regions of Asia, Africa, Europe, Australia, and North America (Baltensperger, 2002; Kimata and Negishi, 2002). Barnyard millet is the fastest growing among the millets with a harvesting period of 6 weeks (Sood et al., 2015). It is predominantly cultivated in India, China, Japan, and Korea for food as well as fodder. Kodo millet is native to the tropical and sub-tropical regions of South America and domesticated in India 3,000 years ago (de Wet et al., 1983b). Little millet was domesticated in the Eastern Ghats of India occupying a major portion of diet amongst the tribal people and spread to Sri Lanka, Nepal, and Myanmar (de Wet et al., 1983a).
Millets are nutritionally superior to rice and wheat as they contain a high amount of proteins, dietary fibers, iron, zinc, calcium, phosphorus, potassium, vitamin B, and essential amino acids (Hegde et al., 2005; Saleh et al., 2013). But the presence of antinutrients like phytates, polyphenols, and tannins reduce the mineral bioavailability by chelating multivalent cations like Fe2+, Zn2+, Ca2+, Mg2+, and K+ (Oberleas, 1973; Gupta, 1980; Kumar and Chauhan, 1993; Abdalla et al., 1998; AbdelRahman et al., 2005). In addition, high amounts of protease and amylase inhibitors affect the digestibility of millet grains (Shivaraj and Pattabiraman, 1981; Pattabiraman, 1986; Joshi et al., 1999). The predominance of the antinutritional factors has thus rendered the orphan status to millets in terms of global economic importance.
Biofortified crops have been primarily developed through conventional breeding exploiting the natural genetic variation, with the exception of Golden rice (www.harvestplus.org). Millets exhibit vast genetic variability for key mineral elements like, iron, zinc, and calcium when compared to other cereal crops (Muthamilarasan and Prasad, 2015). Moreover, millets are drought tolerant crops (O’Kennedy et al., 2009), resistant to pests and diseases offering good insurance against crop failure in developing countries (Tsehaye et al., 2006; Reddy et al., 2011). In spite of the superior quality of millets, only pearl millet has been prioritized as crop of choice for iron biofortification in India. Therefore, vast potential exists to utilize the minor millets for biofortification. Biofortification in millets can be achieved through two strategies: (1) by enhancing the accumulation of nutrients in milled grains and (2) by reducing the antinutrients to increase the bioavailability of minerals. This review highlights the importance of germplasm characterization of millets to develop biofortified varieties and the use of omics approaches to enhance grain-nutrient density. Taking the leads from other cereal crops, we emphasize the application of genetic engineering and genome editing tools to facilitate nutrient accumulation in edible portions and to block the biosynthesis of antinutrients.
Millets are highly nutritious being rich source of proteins, vitamins, and minerals. About 80% of millet grains are used for food, while the rest is used as animal fodder and in brewing industry for alcoholic products (for detailed review, see Saleh et al., 2013; Shivran, 2016). The grains are ground into flour and consumed as cakes or porridges. In Asian countries, street food vendors serve less expensive, ready-to-eat millet-based foods for poor consumers. Millets are recommended for well-being of infants, lactating mothers, elderly, and convalescents. The grains release sugar slowly into the blood stream and thus considered “gluten-free” (Taylor and Emmambux, 2008). With high fiber and protein content, millets are preferred as dietary foods for people with diabetes and cardiovascular diseases (Muthamilarasan et al., 2016). In addition, they contain health promoting phenolic acids and flavonoids, that play a vital role in combating free-radical mediated oxidative stress and in lowering blood glucose levels (Hegde et al., 2005; Dykes and Rooney, 2006, 2007; Chandrasekara and Shahidi, 2010, 2011; Kim et al., 2011; Kunyanga et al., 2012). Pearl millet is rich in Fe, Zn, and lysine (17–65 mg/g of protein) compared to other millets (McDonough et al., 2000; Hadimani et al., 2001). Foxtail millet contains a high amount of protein (11%) and fat (4%). The protein fractions are represented by albumins and globulins (13%), prolamins (39.4%), and glutelins (9.9%). It is thus recommended as an ideal food for diabetics. It also contains significant amounts of potential antioxidants like phenols, phenolic acids, and carotenoids (Saleh et al., 2013; Zhang and Liu, 2015). Finger millet grains contain higher levels of minerals like Ca, Mg, and K (Saleh et al., 2013; Devi et al., 2014). Positive calcium content maintains healthy bones (Pettifor, 2004), while potassium prevents the onset of diabetes, renal and cardiovascular diseases (He and MacGregor, 2008). It also has high levels of amino acids like methionine, lysine and tryptophan (Bhatt et al., 2011), and polyphenols (Chandrasekara and Shahidi, 2011; Devi et al., 2014). Proso millet contains the highest amount of proteins (12.5%) while barnyard millet is the richest source of crude fiber (13.6%) and Fe (186 mg/kg dry matter) (Saleh et al., 2013). Barnyard millet grains possess other functional constituents’ viz. γ-amino butyric acid (GABA) and β-glucan, used as antioxidants and in reducing blood lipid levels (Kofuji et al., 2012; Sharma et al., 2016). With lowest carbohydrate content among the millets, barnyard millet is recommended as an ideal food for type II diabetics (Ugare et al., 2011). Kodo millet is bestowed with high magnesium content (1.1 g/kg dry matter). Millets are therefore consumed as multi-grains to reap the collective health benefits of nutrients.
Conservation of plant genetic resources (PGRs) provides a continuous supply of raw material for crop improvement. Success of biofortification program lies in the sustainable utilization of PGRs for nutritional enhancement (Muthamilarasan and Prasad, 2015). International Crop Research Institute for Semi-Arid Tropics (ICRISAT) contains the largest collection of millet germplasm representing 27.4% of total crop accessions in the genebank (Figure 1). Of this, pearl millet constitutes the vast majority of germplasm represented by 23,092 accessions including landraces, cultivars, genetic stocks, breeding lines, and wild relatives (Upadhyaya et al., 2016a). Finger millet germplasm consisting of 6,084 accessions is grouped under two subspecies, africana and coracana on the basis of morphology of inflorescence (Vetriventhan et al., 2016). Foxtail millet is a self-fertilizing species including 1,542 accessions from 23 different countries. Foxtail millet accessions are classified into three races, namely indica, maxima, and moharia and 10 subraces (Vetriventhan et al., 2016). Barnyard millet germplasm comprises of 749 accessions mainly from Japan and India (Upadhyaya et al., 2016c). The major collections of kodo millet from India and USA account for 665 accessions (Upadhyaya et al., 2016c). India is the prime contributor of little millet germplasm with 473 accessions (Upadhyaya et al., 2016c). Despite holding the largest millet germplasm, scientific community from India has made very few attempts to utilize the millet genomic resources for biofortification. This is mainly because of the scarcity of information on germplasm characterization for nutritional traits.
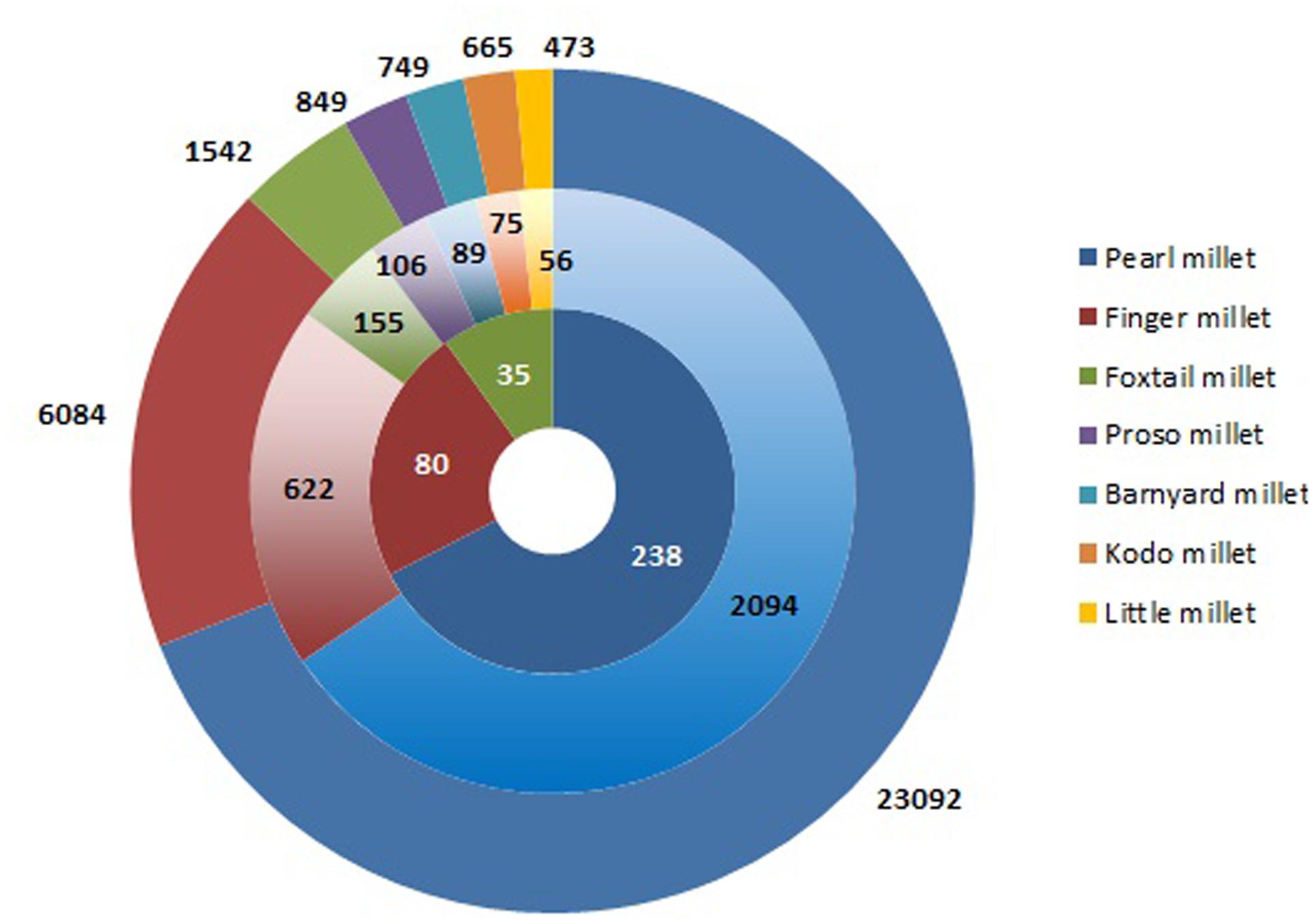
FIGURE 1. Germplasm collection of millet accessions in ICRISAT genebank. The outer concentric circle represents the entire collection of millets followed by reduced subsets of core and minicore collections in the inner circles.
Characterization of entire germplasm for economically important traits is a daunting task for breeders. For the past two decades, germplasm characterization at ICRISAT has led to the establishment of core and minicore collections for pearl millet (Bhattacharjee et al., 2007; Upadhyaya et al., 2009, 2011a), finger millet (Upadhyaya et al., 2006), and foxtail millet (Upadhyaya et al., 2008, 2011b) while only core collections was established for other small millets (Upadhyaya et al., 2014) (Figure 1).
Trait-specific germplasm characterization is a prerequisite to identify genotypes contrasting for desirable traits. All India Coordinated Small Millets Improvement Project implemented by 1986 has focused mainly on varietal development for high yield and disease resistance with due negligence on nutritional quality (Shivran, 2016). Only by the start of the 21st century, millet germplasm received scientific attention for nutrition traits. Screening of pearl millet, foxtail millet, and finger millet accessions for grain nutrients revealed sufficient genetic variability. Multi-location on farm trials identified nutritionally superior lines with farmer preferred traits such as earliness to flowering and grain yield, adapted to local environments (Upadhyaya et al., 2011c; Muthamilarasan and Prasad, 2015). This process accelerates the pace of breeding in millets by studying the inheritance pattern and genotype–environment interaction for grain nutrients. Hybridization between diverse genotypes will generate mapping populations for DNA marker-assisted tagging of genomic regions linked to grain nutrients. However, molecular characterization of other minor millets for nutritional traits is limited and needs special attention. A thorough evaluation of small millets core collections for nutritional traits is the need of the hour to bring genetically diverse parents into mainstream breeding for generating recombinant inbred lines suitable for biofortification. The following section details the utilization of millet germplasm in biofortification in millets under the major headings: macronutrients, micronutrients, and antinutrients.
Millets are the primary source of carbohydrates in tropics and semi-arid tropics of India and sub-Saharan Africa (Shivran, 2016). Grain starch typically comprises of two polymers, amylose (15–30%) and amylopectin (70–85%). Based on the amylose content, millet accessions are classified into two major phenotypes, waxy and non-waxy. Waxy grains containing 0% amylose and nearly 100% amylopectin are glutinous in nature, easily digestible and therefore recommended as food for infants under 6 years of age (Dreher et al., 1984; Englyst et al., 1992). Waxy mutants in staple crops have evolved during the domestication of landraces by human selection (Olsen et al., 2006; Fan et al., 2008). They have been identified in cereals and millets including rice (Oryza sativa; Isshiki et al., 1998), barley (Hordeum vulgare; Domon et al., 2002), sorghum (Sorghum bicolor; McIntyre et al., 2008), maize (Zea mays), foxtail millet (S. italica), proso millet (P. miliaceum), and barnyard millet (Echinochloa sp.) (Fukunaga et al., 2002a; Kawase et al., 2005; Kim et al., 2009). Amylose synthesis in millets is controlled by a single dominant waxy allele (Wx), while the recessive loss-of-function allele (wx) leads to the waxy phenotype with near 0% amylose content (Nakayama et al., 1998; Fukunaga et al., 2002a). In polyploid crops, mutations in different alleles of Wx loci produce low amylose, non-waxy and waxy phenotypes. Low amylose lines contain <3% amylose content due to the residual activity of non-mutant alleles (Fukunaga et al., 2002b; Graybosch and Baltensperger, 2009). Precise identification of mutations in low amylose and waxy mutants have led to the development of waxy starch foods.
The waxy gene product named as granule-bound starch synthase 1 (GBSS 1) is the key enzyme catalyzing the formation of amylose (Sano, 1984). Mutations in GBSS 1 result from insertions/deletions (InDels), transposable elements, and single base pair mutations (Isshiki et al., 1998; Domon et al., 2002; Van et al., 2008). In millets, Wx gene was found to contain 14 exons and 13 introns (Figure 2). Foxtail millet with waxy grains are cultivated in Japan, Taiwan, Philippines, and Indonesia (Takei and Sakamoto, 1989). Molecular analysis of foxtail millet waxy landraces by polymerase chain reaction (PCR) covering 4,006 bp of Wx gene using exon–intron primer sets (ex1/ex2 or ex2int1/ex4r) identified the insertion of transposable elements [transcriptionally silent information (TSI)-2 and TSI-7] in intron 1 or exon 3. All the waxy phenotypes harboring transposable elements in Wx loci differed in their geographical origin of domestication (Van et al., 2008). Next-generation sequencing technology has accelerated the re-sequencing of various crops to identify genome-wide nucleotide variations (Huang et al., 2009). High throughput analysis of genes and transcripts using genotyping by sequencing, transcriptome, and epigenetic analysis has been well documented as a powerful tool for identifying the genetic basis of nutritional traits in millets (Muthamilarasan and Prasad, 2015). The release of draft genome sequences of two foxtail millet cultivars, Yugu1 and Zhang gu has advanced the research for genetic improvement in foxtail millet (Bennetzen et al., 2012; Zhang et al., 2012). A foxtail millet waxy landrace, Shi-Li-Xiang (SLX) was re-sequenced using Solexa sequencing technology and Genome Analyzer II (GA II) to explore the nucleotide variations covering agronomic traits-related genes. Alignment with the reference genomes identified SNPs (single nucleotide polymorphisms), InDels, and SVs (structural variants). Novel markers obtained by re-sequencing helped in genome mapping of starch synthase encoding GBSS 1 peptide. Sequencing of GBSS 1 gene found the insertion of transposable elements confirming its waxy nature (Bai et al., 2013).
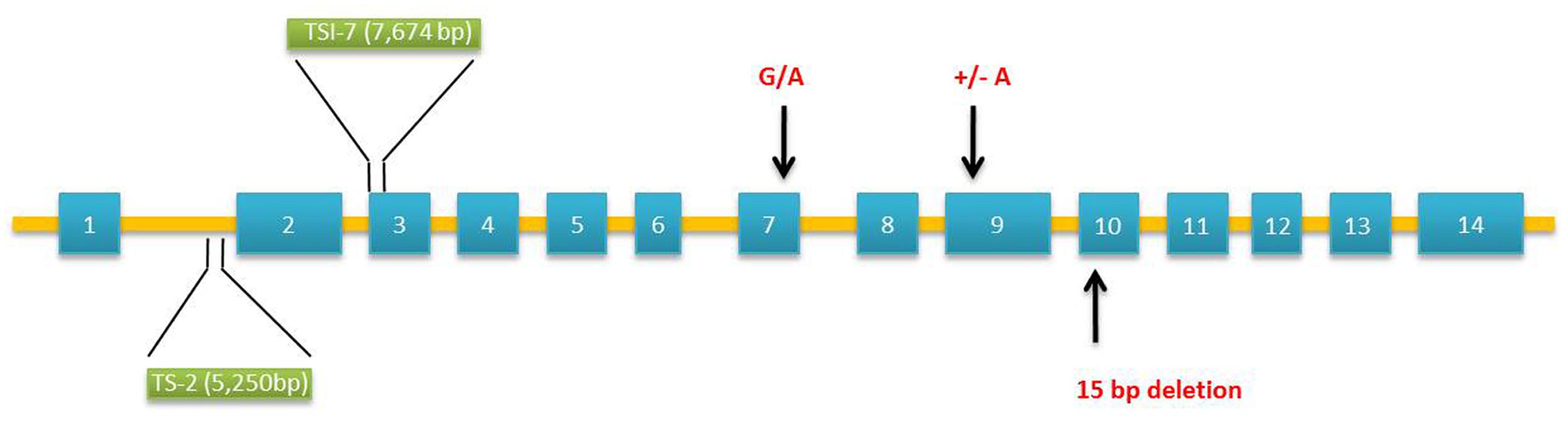
FIGURE 2. Molecular structure of granule-bound starch synthase 1 (GBSS 1) gene in Setaria italica and Panicum miliaceum associated with waxy phenotypes. Two polymorphisms occur in the wx allele of Setaria: insertion of transposable elements, TSI-2 (5,250 bp – Type IV) in intron 1 and TSI-7 (7,674 bp – Type V) in exon 3. Three SNPs occur in the exon sequence of waxy Panicum: 15-bp deletion [GCGCTGAACAAGGA GGCGCTG] in exon 10 (S type), insertion of an adenine residue in exon 9 leading to reading frameshift mutation (L type) and G/A substitution in exon 7 converting cysteine residue to tyrosine (L type). Exons and introns are denoted by boxes and bars, respectively.
Waxy proso millet varieties are in existence since 1885 (Hixon and Brimhall, 1968). Graybosch and Baltensperger (2009) identified six waxy accessions of proso millet from the germplasm collection of United States Department of Agriculture-Agricultural Research Service (USDA-ARS) North Central Regional Plant Introduction (PI) Station, Ames, IA, USA. Based on the crosses between two (PI 436625 and PI 436626) waxy accessions and several wild type lines, two recessive alleles designated as wx-1 and wx-2 were found to be responsible for waxy phenotype. Grain starch amylose concentration was found to be reduced by 7.2-folds in waxy lines. However, a low level of amylose (3.5%) was present in some waxy types. This is most likely due to the low level of GBSS 1 activity produced by one of the alleles. The molecular basis of waxy endosperm in proso millet was further investigated using 38 landraces. Of these, 14 waxy phenotypes from China and Korea had little or no GBSS 1 activity. Sequencing of GBSS 1 in waxy lines revealed two different forms designated as “L type” and “S type” (Figure 2). These two forms differ in their exon sequence coding for mature GBSS 1 peptides resulting from three polymorphisms. The “S type” contains 15-bp deletion leading to the loss of five amino acids from glucosyltransferase domain 1. The “L type” contains a frame shift mutation caused by the insertion of an adenine residue and by substitution of a cysteine to tyrosine amino acid (Hunt et al., 2010). Based on these mutations, Araki et al. (2012) developed a PCR-based marker system (length polymorphism and derived cleaved amplified polymorphic sequences (dCAPS)) to identify waxy landraces amongst 83 accessions from Japan and 15 accessions from other countries. Sequencing of wx gene confirmed the ability of dCAPS markers in locating the single-bp mutations in exon 7 and 9 of waxy landraces.
Japanese barnyard millet is an allohexaploid and mutations in three alleles are considered to confer waxy nature. The probability of spontaneous mutations in all three alleles to obtain waxy cultivars is difficult. Wheat, also an allohexaploid was found to carry spontaneous mutations at one or two waxy loci (Nakamura et al., 1993a,b). Mutagenesis by chemical treatment on a low amylose wheat cultivar with non-functional Wx-A1 and Wx-B1 genes produced a waxy mutant (Yasui et al., 1997). With wheat as the model system, Hoshino et al. (2010) attempted to mutagenize barnyard millet landrace (Noge-Hie) into waxy phenotypes by treatment with Co60 gamma ray irradiation. One of the mutant plants showed significant reduction in amylose content. PCR analysis of waxy genes from this low amylose line using primers designed from the consensus sequences of foxtail millet, pearl millet, sorghum, and wheat detected the absence of a gene sequence specific for waxy phenotypes. Contrary to waxy wheat with more than one non-functional Wx allele, loss-of-function mutation in one allele was found to produce waxy cultivars in barnyard millet (Hoshino et al., 2010).
Traditional breeding in millets for waxy trait is a labor intensive and time consuming process. It took nearly 15 years to transform waxy trait into non-waxy elite foxtail millet cultivar Yugu1 through cross breeding (Quan et al., 2010). Marker-assisted selection accelerates the process of conventional breeding. But the information on molecular markers linked to waxy traits in millets lags far behind cereals. Henceforth, genotyping of diverse millet germplasm by high throughput re-sequencing will facilitate the development of new molecular markers to map the waxy trait. Molecular analysis of waxy gene in foxtail millet, proso millet, and barnyard millet has identified mutations in one or more alleles. With recent advancements in genome editing, application of programmable site-specific nucleases is a straightforward approach to induce genetic mutations in non-waxy elite cultivars for transforming them into waxy phenotypes. Thus genomics approaches will speed up the genetic improvement in millets in a cost effective manner to produce biofortified varieties.
High quality proteins are essential for physical and mental well-being of humans, especially children (Heine et al., 1995; Tomé and Bos, 2007). Cereal proteins deficient in essential amino acids like methionine, lysine, and tryptophan lead to malnutrition in developing countries (Nirgude et al., 2014). Cereal proteins contain 1.5–2% lysine and 0.25–0.5% tryptophan while estimated average requirement is 5% and 1.1% for lysine and tryptophan (Young et al., 1998). Finger millet on the other hand is high in essential amino acids than cereals (Mbithi-Mwikya et al., 2000). It is therefore a suitable model system to elucidate the genetic control of protein quality. High lysine and tryptophan in finger millet is attributed to the transcriptional regulation of amino acid catabolism genes by Opaque2 (o2), a basic leucine zipper (bZIP) transcription factor. o2 modifiers (Opm) downregulate lysine ketoglutarate reductase dehydrogenase (Kemper et al., 1999) and upregulate aspartate kinase (Brennecke et al., 1996) resulting in free lysine and tryptophan in endosperm. Genetic differences in Opm alleles of finger millet germplasm remained largely uncharacterized until 2012. In the preliminary study, finger millet genotypes were evaluated for diversity in seed protein content using PCR-based markers. Random amplified polymorphic DNA (RAPD) and simple sequence repeat (SSR) profiles revealed few unique bands discriminating high and low protein genotypes. This study laid the foundation to select superior genotypes for use in traditional breeding to produce high quality proteins (Kumar et al., 2012).
Molecular characterization of Opm alleles using SSRs and SNPs can effectively identify quantitative trait loci (QTLs) influencing amino acid content (Goron and Raizada, 2015). Genic SSRs are powerful tools to link the genetic maps of related species and alleles of genic SSRs are associated with structural variations in the gene that affect transcription and translation of proteins (Kashi and Soller, 1999; Andersen and Lübberstedt, 2003). Nirgude et al. (2014) reported higher level of polymorphism in Opm genes using genic SSRs than RAPD, genomic SSRs and Cytochrome P450 markers as reported by Panwar et al. (2010b). Utilizing the functional potential of comparative genomics, high tryptophan finger millet genotypes were identified from global collection using genic SSRs for Opm genes derived from expressed sequence tag (EST) sequences of rice, maize, and sorghum. Association mapping of SSR loci found two QTLs for tryptophan and one QTL for protein content. Interestingly, a 220-bp allele of SSR locus OM5 marker designed from the 27-kDa γ-zein gene of Opm was present mostly in the high tryptophan-containing genotypes (Babu et al., 2014). This marker could be employed in marker-assisted breeding for introgression of Opm allele into high yielding cultivars. Fine mapping of Opm genes linked to QTLs could lead to genetic enhancement of seed protein quality in cereals and small millets. Recently, 16 prolamin encoding genes called setarins have been identified in foxtail millet using computational approaches (Muthamilarasan and Prasad, 2015). Sequence alignment of setarin genes with other millets and cereals revealed least homology indicating their uniqueness in improved protein quality. Quantitative RT-PCR analysis of setarin genes confirmed their role in seed protein accumulation by overexpression in developing spikes (Muthamilarasan and Prasad, 2015). Preliminary clues of such crop-specific genes for high protein could be useful in engineering other millets and cereals for protein enrichment.
Iron (Fe) deficiency in India report 79% of pre-school children and 56% of women to be anemic (Krishnaswamy, 2009). Fe supplementation program in India since 1970 failed to address the issue of iron deficiency (Anand et al., 2014). Recognizing biofortification as a feasible alternative for Fe delivery, HarvestPlus has developed high Fe pearl millet by conventional breeding (HarvestPlus, 2009). The first step in breeding crops for better nutrition is to evaluate the genetic diversity of available germplasm for target nutritional trait. ICRISAT, a member of HarvestPlus undertook the process of screening pearl millet germplasm for sources of high Fe density. Early reports revealed positive correlation of Fe and Zn grain content with no significant correlation on grain yield and seed size (Velu et al., 2007, 2011; Gupta et al., 2009; Govindaraj et al., 2013). This suggested increase in Zn grain content as an associated trait while breeding for high Fe pearl millet.
Prospects of breeding pearl millet for high Fe content began in the year 2004 with screening of germplasm accessions, seed parents, open pollinated varieties (OPV), improved populations, and population progenies. Progenies derived from AIMP 92901, a high-yielding OPV, exhibited large intra-population variability with highest levels of Fe (about 76 mg/kg) and Zn (about 65 mg/kg) than their parental control (Velu et al., 2007). All the improved progenies were derived from iniadi germplasm. Iniadi landraces from African subcontinent have farmer preferred agronomic traits like early maturing, large seed size, compact panicles, disease resistance, and drought tolerance (Andrews and Anand Kumar, 1996; Rai et al., 2008). Hence, iniadi germplasm was exploited for further hybridization experiments to breed for high Fe content. Velu et al. (2008) identified ICTP 8203, a commercial OPV (Rai et al., 1990) released in India by 1998 and still under cultivation to possess highest Fe and Zn content. By the year 2012, high Fe biofortified variety “ICTP 8203 Fe 10-2” developed by progeny testing was commercially released in India. ICTP 8203 Fe 10-2 had 9% more Fe (71 mg/kg) and 11% more grain yield than the parental control (Rai et al., 2013).
Taking advantage of cross-pollination in pearl millet, breeders are in continuous search for nutritionally elite varieties suited for local environments. Rai et al. (2014) screened seed parent progenies and restorer parent progenies for Fe and Zn variability using X-ray fluorescence spectroscopy. The mean Fe density of these progenies increased by 5–66% than the control cultivars. As Iniadi germplasm is a promising source for high grain Fe and Zn densities, Rai et al. (2015) and Upadhyaya et al. (2016b) evaluated germplasm accessions and landraces using the inductively coupled plasma atomic emission spectroscopy. Significant variability was observed for Fe (51–121 mg/kg) and Zn (46–87 mg/kg) with positive correlation. Novel sources for developing nutrient-rich pearl millet include IP 17521 of Gnali (106.9 mg/kg), IP 11523 of Idiyouwe (106.5 mg/kg) and IP 17518 of Gnali (79.1 mg/kg), IP 11535 of Iniadi (78.4 mg/kg) for iron (Fe) content and zinc (Zn) content, respectively. These germplasm accessions with greater nutrient density than the commercial cultivar ICTP 8203 are valuable sources for introgressing high Fe and Zn into elite breeding lines. Continuation of breeding program with planned crosses will help in identifying nutrient-dense parental lines suited for rapidly changing climatic conditions.
High Fe biofortified pearl millet provides twofolds higher iron than modern wheat varieties. This led to increase in iron absorption by 5–10% in around 35 million people consuming biofortified pearl millet (Cercamondi et al., 2013; Kodkany et al., 2013). Feeding trial by Haas et al. (2013) revealed that consumption of 232 g iron biofortified pearl millet flour/day resolved 65% more iron deficiency in Indian school children. Similarly, a randomized efficacy trial of iron-biofortified pearl millet was conducted for 6 months on secondary school children in Maharashtra to assess the improvement in Fe status. The study population included children with high risk of iron deficiency, low-iron diets, and regular consumption of pearl millet. Iron intake of children consuming biofortified pearl millet far exceeded the EAR within 4 months. There was a phenomenal increase in their serum ferritin and total body iron levels. The dose of iron delivered was comparable to that achieved by supplementation trials (Finklestein et al., 2015). Promising results from bioefficacy studies is the clear evidence for the use of biofortified millets as a sustainable intervention in high risk populations to overcome the iron deficiency.
Zinc (Zn) deficiency affects 50% of the world population resulting in diarrhea, impairment of physical growth, and suppressed immune function (Hotz and Brown, 2004; Gibson, 2006; Prasad, 2007; Qaim et al., 2007; Gibson et al., 2008). Breeding approach to improve the Zn grain content is discussed in the previous section. Genetic enhancement of grain Zn content is possible by modulating the metal transporters that facilitate their uptake, translocation, and storage. Members of Zn-regulated transporters and Iron (Fe) regulated transporter-like protein (ZIP) family contribute to Zn homeostasis by either uptake or remobilization in intracellular compartments (Guerinot, 2000; Mäser et al., 2001; Colangelo and Guerinot, 2006; Figure 3). ZIP transporters increase Zn uptake in several higher plants including Arabidopsis thaliana (Plaza et al., 2007), rice (O. sativa; Ishimaru et al., 2005; Bashir et al., 2012), barley (H. vulgare; Pedas et al., 2008), soybean (Glycine max; Moreau et al., 2002), and tomato (Solanum lycopersicum; Eckhardt et al., 2001). ZIP transporters are differentially regulated in various tissues under Zn deprivation and abundance in soils (Ramesh et al., 2003). Cereals and millets with high Zn seeds can be engineered by seed-specific expression of ZIP transporters. Initial success in transgenic development for seed Zn accumulation was recorded in rice (Ramesh et al., 2004). Recently, high zinc accumulating finger millet transgenic plants were produced by overexpression of OsZIP1 driven by constitutive (35S) and endosperm-specific promoters (Bx17). Seeds of T1 transgenic plants showed 10–15 mg/kg higher Zn than wild type and the difference further increased to 20 mg/kg in T2 generation. Interestingly, higher Mn (5–10 mg/kg higher than wild type) accumulation was also recorded in the seeds of transgenic plants (Ramegowda et al., 2013). The in planta evidence of Zn accumulation in seeds by upregulation of ZIP transporters is the key to develop high Zn biofortified millets. As millets exhibit high synteny with cereals, expression of heterologous Zn transporters need further investigation to enhance grain Zn content.
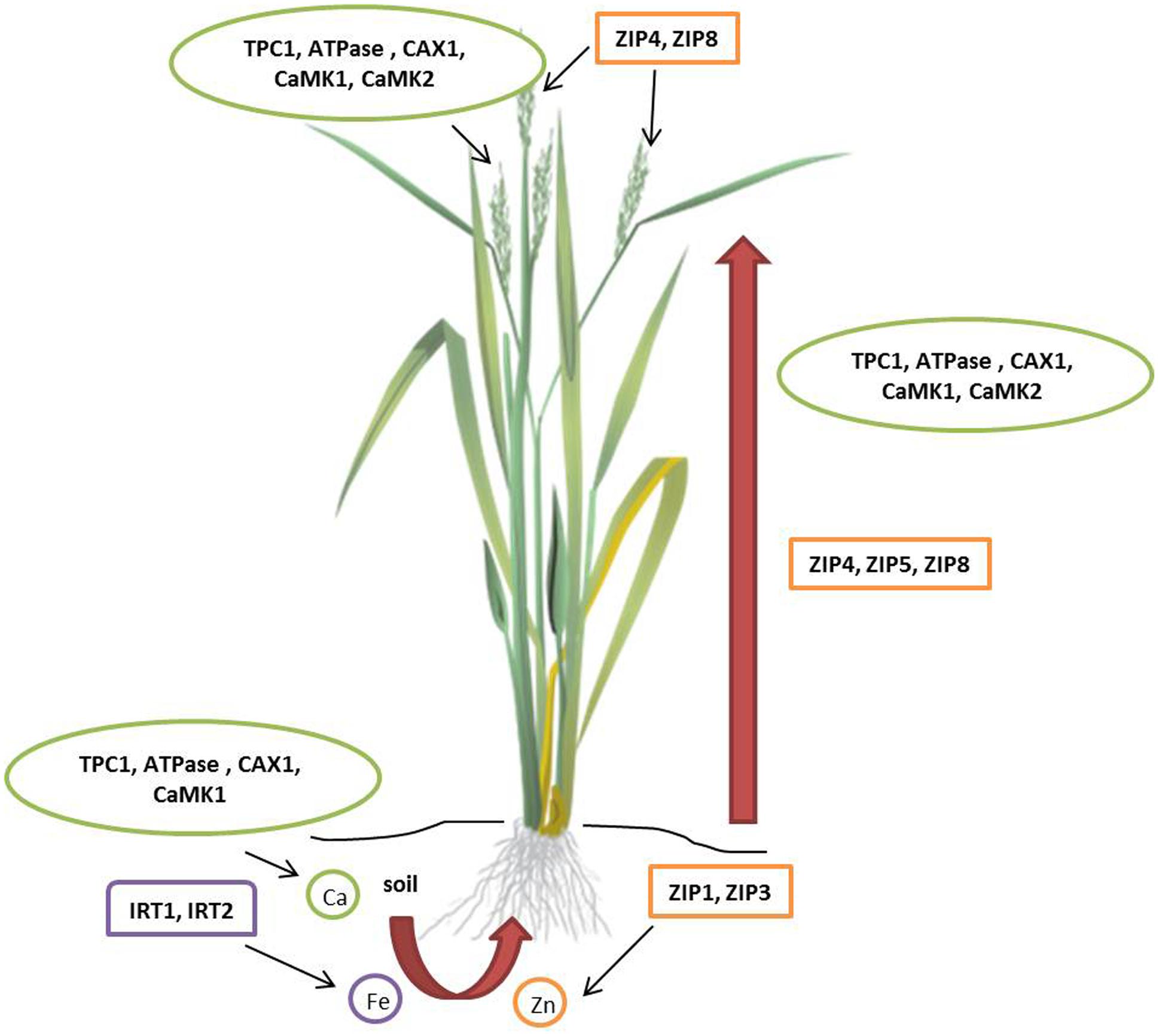
FIGURE 3. Molecular targets for micronutrient accumulation in millets. In rice (Oryza sativa), eight ZIPs facilitate iron and zinc uptake. OsZIP1 and OsZIP3 help in zinc uptake from soil; OsZIP4, OsZIP5, and OsZIP8 translocate zinc from roots to shoots; OsZIP4 and OsZIP8 are involved in grain filling. Two Fe2+ transporters, OsIRT1 and OsIRT2 help in iron uptake from the soil. In finger millet (Eleusine coracana), calcium uptake involves three Ca2+ transporters and two calmodulin-dependent protein kinases (CaMKs). TPC1, ATPase, CAX1, CaMK1, and CaMK2 play a vital role in uptake, translocation, and accumulation of calcium. ATPase, CaM stimulated type IIB Ca2+ pump; CAX1, Ca2+/H+ antiporter or exchanger; TPC1, two-pore channel; ZIP, Zn-regulated transporters and Iron (Fe) regulated transporter-like protein.
Calcium (Ca) deficiency resulting from high intake of cereals leads to osteoporosis in women (Ross et al., 2011). Plants absorb calcium ions from the soil solution and translocate to different organs via xylem transport predominantly controlled by transpiration pull (Grusak, 2002). Non-exposed structures like developing seeds exhibit negligible rate of transpiration and are low in calcium as they acquire minerals through phloem. In reproductive tissues, ion transporter proteins direct calcium transport (White and Broadley, 2003; White, 2005; Panwar et al., 2010a). Henceforth, elucidation of role of calcium transporters in plants favors the development of Ca biofortified cereals. Finger millet containing about 5–30 times higher Ca than wheat and rice serves as a model system to understand seed calcium accumulation. Singh et al. (2014) undertook transcriptomics approach to characterize calcium sensor gene family from the developing spikes of finger millet using Illumina paired-end sequencing methods. This study included characterization, identification, classification, phylogeny, and pathway analysis of calcium sensor genes of two genotypes, GP-1(low calcium) and GP-45 (high calcium). In total, 82 calcium sensor proteins identified in the transcriptome of finger millet spikes were grouped into 25-calmodulin (CaM) and calmodulin-like proteins (CaML), 9-CDPK-related protein kinases (CRK), 9-calcineurin B-like protein (CBL), 23-CBL interacting protein kinases (CIPK), and 14-Ca2+-dependent and CaM-independent protein kinases (CDPK) genes. Comparative phylogenetic analysis of calcium sensor gene family in finger millet identified 12 calcium sensor genes diverse from the rice orthologs. In addition, abundance of calcium sensor gene expression in GP-45 compared to GP-1 proved the polygenic nature of calcium accumulation. Quantitative real-time PCR analysis revealed higher expression of EcCIPK2, EcCIPK9, and EcCIPK11 genes in GP-45 in response to external calcium supplied to the rhizosphere resulting in increased seed calcium content. In a similar study, Mirza et al. (2014) identified higher expression of Ca2+/H+ antiporter (CAX1), two pore channel (TPC1), CaM-stimulated type IIB Ca2+ ATPase and two CaM-dependent protein kinase (CaMK1 and CaMK2) genes in GP-45 resulting in greater uptake, translocation, and accumulation of calcium (Figure 3). Higher expression of CAX1 genes and CaM isoform in developing spikes strongly influence positive seed calcium content. Multigene engineering for co-expression of calcium sensor genes in cereal crops and small millets will play a significant role in the production of transgenic biofortified varieties.
Vitamin A deficiency is a serious threat to well-being of children and pregnant women in developing countries (World Health Organization [WHO], 2011). Biofortification of staple crops such as sweet potato, cassava, and maize for high provitamin A by conventional breeding has achieved great success (Bouis et al., 2011). Millet collections on the other hand lack sufficient genetic variation for beta carotene content (Buerkert et al., 2001; Upadhyaya et al., 2011a). Hence traditional breeding for high provitamin A in millets is not feasible. In such cases, transgenic approach favors metabolic engineering of vitamin biosynthesis in plants. Golden rice is a notable example for genetic modification of provitamin A in grains (Paine et al., 2005). Regulatory issues concerning the genetically modified (GM) crops are being revisited with scientific evidences to facilitate their commercial production. Therefore, research on GM millets with enriched vitamins is gaining momentum.
Antinutrients like phytic acid, polyphenols, and tannins in millet grains greatly reduce the bioavailability of minerals. Phytic acid (myo-inositol-1,2,3,4,5,6-hexakisphosphate or IP6) the major form of phosphorus in seeds chelates the mineral cations in protein storage vacuoles (Raboy et al., 2000; Raboy, 2009). Bioefficacy studies on biofortified crops is a clear evidence for the antagonistic effect of IP6 on mineral absorption. Hama et al. (2012) carried out bioefficacy studies of biofortified and traditional pearl millet varieties of Africa subjected to abrasive decortication. Iron content of biofortified varieties (Tabi and GB8735) was 72 and 67 mg/kg dry matter, respectively. Though it corresponds to target biofortification levels, high phytate content reduced the iron bioavailability in these varieties. On the other hand, Zn content in biofortified varieties was 56 and 41 mg/kg dry matter, respectively. Low phytate to zinc ratio did not affect the zinc absorption to a greater level. Likewise, increased levels of polyphenols in biofortified iron pearl millet and black beans also reduced the Fe bioavailability (Tako et al., 2014, 2015). This study confirmed the negative impact of polyphenols on mineral absorption using in vivo chicken model and in vitro digestion/Caco2-cell model. Antinutrients are commonly removed by decortication, malting, fermentation, roasting, flaking, and grinding. However large-scale industrial methods for processing of millets to produce novel functional foods are not well developed as that for other cereal crops (Food and Agricultural Organization [FAO], 2012). Reduction in antinutrients during plant growth and development is therefore a promising strategy to improve the bioavailability of minerals from nutrient-rich millets.
Phytic acid is greatly reduced in low phytate (lpa) mutants of rice, wheat, and maize (Larson et al., 2000; Pilu et al., 2003; Guttieri et al., 2004; Kim et al., 2008). In most of the cases, lpa mutants had negative effects on crop yield and other agronomic traits. Henceforth, genetic engineering was considered as a safer alternative to generate lpa mutants (Feng and Yoshida, 2004). Genes controlling phytic acid biosynthetic pathway has been well characterized in major cereal crops (Raboy and Bowen, 2006). Three enzymes (MIPS, myo-inositol-3-phosphate synthase; MIK, myo-inositol-3-phosphate 5/6-kinase; IPK1, Inositol 1,3,4,5,6-pentakisphosphate 2-kinase) expressed in different levels of the biosynthetic pathway are the molecular targets for producing low phytate crops (Stevenson-Paulik et al., 2002; Shi et al., 2005; Sweetman et al., 2006; Kuwano et al., 2009). Successful silencing of IPK1 gene by RNAi technology produced low phytate rice with no significant effect on yield parameters (Ali et al., 2009). Recently, insertional mutagenesis of IPK1 gene by site-specific nucleases such as zinc finger nucleases (ZFNs) resulted in low phytate maize (Shukla et al., 2009). Rapidly emerging genome editing tools thus have enormous potential to develop biofortified millets by precise engineering of phytic acid biosynthetic pathway.
Millets are highly nutritious crops feeding poor populations in Asia and Africa. Scientific research to utilize the highly nutritious millet crops to combat micronutrient malnutrition is still meager. With good grain qualities and significant amounts of essential amino acids, minerals, and vitamins, bioavailability of nutrients need further improvement by reduction of antinutrients or by the use of novel promoters. Elucidation of role of various transporters in nutrient uptake, translocation, and storage could help in localizing the macro and micronutrients in edible parts of millets. Establishment of minicore collection of other millets will accelerate the molecular characterization of genetically diverse germplasm for new sources of variation in nutritional traits. Identification of molecular markers such as SNPs and InDels linked to nutritional traits will decipher the information on candidate genes controlling these traits. As millets exhibit cross-genera transferability, introgression of nutrient-linked genes into other cereals can become feasible by the use of molecular breeding or genetic engineering. Advent of next-generation sequencing platforms favors rapid sequencing of millet genome. Omics information on millets should advance more rapidly as cereal crops in order to enhance their utilization in the fight against micronutrient malnutrition. Thus integration of knowledge on genomics, transcriptomics, proteomics, and metabolomics could promote millets as model systems for advancements in biofortification of staple crops.
AV conceptualized and wrote the manuscript. RR edited the manuscript and critically revised the manuscript for publication.
This work is published under the major research project funded by Loyola College – Times of India, Chennai, India (Project approval code: 4LCTOI14PBB001) and UGC Research Award Scheme (F.30-1/2014/RA-2014-16-GE-TAM-5825 SA-II).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer [TB] and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
The authors thank the Loyola College management for providing the laboratory and infrastructure facilities to carry out the research work. We also extend our gratitude to Dr. G. Ganesan for his critical comments on the manuscript.
Abdalla, A. A., El Tinay, A. H., Mohamed, B. E., and Abdalla, A. H. (1998). Proximate composition, starch, phytate and mineral contents of 10 pearl millet genotypes. Food Chem. 63, 243–246. doi: 10.1016/S0308-8146(97)00228-8
AbdelRahman, S. M., Babiker, E. E., and El Tinay, A. H. (2005). Effect of fermentation on antinutritional factors and HCl extractability of minerals of pearl millet cultivars. J. Food Tech. 3, 516–522.
Agricultural Statistics, Government of India (2014). “Pearl Millet,” in Agricultural Statistics at a Glance–2014, Government of India (New Delhi: Oxford University Press), 85–86.
Ali, N., Paul, S., Gayen, D., Sarkar, S. N., Datta, K., and Datta, S. K. (2009). Development of low phytate rice by RNAi mediated seed-specific silencing of inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene (IPK1). PLoS ONE 8:e68161. doi: 10.1371/journal.pone.0068161
Anand, T., Rahi, M., Sharma, P., and Ingle, G. K. (2014). Issues in prevention of iron deficiency anemia in India. Nutrition 30, 764–770. doi: 10.1016/j.nut.2013.11.022
Andersen, J. R., and Lübberstedt, T. (2003). Functional markers in plants. Trends Plant Sci. 8, 554–560. doi: 10.1016/j.tplants.2003.09.010
Andrews, D. J., and Anand Kumar, K. (1996). Use of the West African pearl millet landrace iniadi in cultivar development. Plant Genet. Res. Newsl. 105, 15–22.
Araki, M., Numaoka, A., Kawase, M., and Fukunaga, K. (2012). Origin of waxy common millet, Panicum miliaceum L. in Japan. Genet. Res. Crop Evol. 59, 1303–1308. doi: 10.1007/s10722-011-9755-9
Austin, D. (2006). Fox-tail millets (Setaria: Poaceae): abandoned food in two hemispheres. Econ. Bot. 60, 143–158. doi: 10.1663/0013-0001200660[143:FMSPFI]2.0.CO;2
Babu, B. K., Agrawal, P. K., Pandey, D., and Kumar, A. (2014). Comparative genomics and association mapping approaches for opaque2 modifier genes in finger millet accessions using genic, genomic and candidate gene-based simple sequence repeat markers. Mol. Breed. 34, 1261–1279. doi: 10.1007/s11032-014-0115-2
Bai, H., Cao, Y., Quan, J., Dong, L., Li, Z., Zhu, Y., et al. (2013). Identifying the genome-wide sequence variations and developing new molecular markers for genetics research by re-sequencing a landrace cultivar of foxtail millet. PLoS ONE 8:e73514. doi: 10.1371/journal.pone.0073514
Baltensperger, D. D. (2002). “Progress with proso, pearl and other millets,” in Trends in New Crops and New Uses, eds J. Janick and A. Whipkey (Alexandria, VA: ASHS Press), 100–103.
Bashir, K., Ishimaru, Y., and Nishizawa, N. K. (2012). Molecular mechanisms of zinc uptake and translocation in rice. Plant Soil 361, 189–201. doi: 10.1007/s11104-012-1240-5
Belton, P. S., and Taylor, J. R. N. (2004). Sorghum and millets: protein sources for Africa. Trends Food Sci. Technol. 15, 94–98. doi: 10.1016/j.tifs.2003.09.002
Bennetzen, J. L., Schmutz, J., Wang, H., Percifield, R., Hawkins, J., Pontaroli, A. C., et al. (2012). Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30, 555–561. doi: 10.1038/nbt.2196
Bhatt, D., Negi, M., Sharma, P., Saxena, S. C., Dobriyal, A. K., and Arora, S. (2011). Responses to drought induced oxidative stress in five finger millet varieties differing in their geographical distribution. Physiol. Mol. Biol. Plants 17, 347–353. doi: 10.1007/s12298-011-0084-4
Bhattacharjee, R., Khairwal, I. S., Bramel, P. J., and Reddy, K. N. (2007). Establishment of a pearl millet [Pennisetum glaucum (L.) R. Br.] core collection based on geographical distribution and quantitative traits. Euphytica 155, 35–45. doi: 10.1007/s10681-006-9298-x
Bouis, H. E., Hotz, C., McClafferty, B., Meenakshi, J. V., and Pfeiffer, W. H. (2011). Biofortification: a new tool to reduce micronutrient malnutrition. Food Nutr. Bull. 32, S31–S40. doi: 10.1177/15648265110321S105
Brennecke, K., Souza Neto, A. J., Lugli, J., Lea, P. J., and Azevedo, R. A. (1996). Aspartate kinase in maize mutants ask1-lt19 and opaque-2. Phytochemistry 41, 707–712. doi: 10.1016/0031-9422(95)00634-6
Buerkert, A., Moser, M., Kumar, A. K., Furst, P., and Becker, K. (2001). Variation in grain quality of pearl millet from Sahelian West Africa. Field Crops Res. 69, 1–11. doi: 10.1016/S0378-4290(01)00166-6
Cercamondi, C. I., Egli, I. M., Mithchikpe, E., Tossou, F., Zeder, C., Hounhouigan, J. D., et al. (2013). Total iron absorption by young women from iron-biofortified pearl millet composite meals is double that from regular millet meals but less than that from post-harvest iron-fortified millet meals. J. Nutr. 143, 1376–1382. doi: 10.3945/jn.113.176826
Chandrasekara, A., and Shahidi, F. (2010). Content of insoluble bound phenolics in millets and their contribution to antioxidant capacity. J. Agric. Food Chem. 58, 6706–6714. doi: 10.1021/jf100868b
Chandrasekara, A., and Shahidi, F. (2011). Inhibitory activities of soluble and bound millet seed phenolics on free radicals and reactive oxygen species. J. Agric. Food Chem. 59, 428–436. doi: 10.1021/jf103896z
Colangelo, E. P., and Guerinot, M. L. (2006). Put the metal to the petal: metal uptake and transport throughout plants. Curr. Opin. Plant Biol. 9, 322–330. doi: 10.1016/j.pbi.2006.03.015
de Wet, J. M. J., Rao, K. E. P., and Brink, D. E. (1983a). Systematics and domestication of Panicum sumatrense (Gramineae). J. Agric. Trad. Bot. Appl. 30, 159–168. doi: 10.3406/jatba.1983.3898
de Wet, J. M. J., Rao, K. E. P., Mengesha, M. H., and Brink, D. E. (1983b). Diversity in kodo millet, Paspalum scrobiculatum. Econ. Bot. 37, 159–163. doi: 10.1007/BF02858779
Devi, P. B., Vijayabharathi, R., Sathyabama, S., Malleshi, N. G., and Priyadarisini, V. B. (2014). Health benefits of finger millet (Eleusine coracana L.) polyphenols and dietary fiber: a review. J. Food Sci. Technol. 51, 1021–1040. doi: 10.1007/s13197-011-0584-9
Domon, E., Fujita, M., and Ishikawa, N. (2002). The insertion/deletion polymorphisms in the waxy gene of barley genetic resources from East Asia. Theor. Appl. Genet. 104, 132–138. doi: 10.1007/s001220200016
Dreher, M. L., Dreher, C. J., and Berry, J. W. (1984). Starch digestibility of foods: a nutritional perspective. Crit. Rev. Food Sci. Nutr. 20, 47–71. doi: 10.1080/10408398409527383
Dykes, L., and Rooney, L. W. (2006). Review: sorghum and millet phenols and antioxidants. J. Cereal Sci. 44, 236–251. doi: 10.1016/j.jcs.2006.06.007
Dykes, L., and Rooney, L. W. (2007). Phenolic compounds in cereal grains and their health benefits. Cereal Food World 52, 105–111. doi: 10.1094/CFW-52-3-0105
Eckhardt, U., Mas Marques, A., and Buckhout, T. J. (2001). Two iron-regulated cation transporters from tomato complement metal uptake deficient yeast mutants. Plant Mol. Biol. 45, 437–448. doi: 10.1023/A:1010620012803
Englyst, H. N., Kingman, S. M., and Cummings, J. H. (1992). Classification and measurement of nutritionally important starch fractions. Eur. J. Clin. Nutr. 46, S33–S50.
Fan, L. J., Quan, L. Y., Leng, X. D., Guo, X. Y., Hu, W. M., Ruan, S. L., et al. (2008). Molecular evidence for post-domestication selection in the Waxy gene of Chinese waxy maize. Mol. Breed. 22, 329–338. doi: 10.1007/s11032-008-9178-2
Feng, X., and Yoshida, K. T. (2004). Molecular approaches for producing low-phytic–acid grains in rice. Plant Biotechnol. 21, 183–189. doi: 10.5511/plantbiotechnology.21.183
Finklestein, J. L., Mehta, S., Udipi, S. A., Ghugre, P., Luna, S. V., Wenger, M. J., et al. (2015). A randomized trial of iron-biofortified pearl millet in school children in India. J. Nutr. 145, 1576–1581. doi: 10.3945/jn.114.208009
Food and Agricultural Organization [FAO] (2012). Economic and Social Department: The Statistical Division. Statistics Division 2012. Available at: http://faostat.fao.org/site/567/DesktopDefault.aspx?PageID=567#ancor. [accessed December 03, 2016].
Food and Agricultural Organization [FAO] (2015). Available at: http://faostat3.fao.org/browse/Q/QC/E [accessed September 12, 2016].
Fukunaga, K., Kawase, M., and Kato, K. (2002a). Structural variation of waxy gene and differentiation in foxtail millet, Setaria italica (L.) P. Beauv. Implication for multiple origins of the waxy phenotype. Mol. Genet. Genomics 268, 214–222. doi: 10.1007/s00438-002-0728-8
Fukunaga, K., Wang, Z. M., Kato, K., and Kawase, M. (2002b). Geographical variation of nuclear genome RFLPs and genetic differentiation in foxtail millet, Setaria italica (L.) P. Beauv. Genet. Res. Crop Evol. 49, 95–101. doi: 10.1023/A:1013852007770
Gibson, R. S. (2006). Zinc: the missing link in combating micronutrient malnutrition in developing countries. Proc. Nutr. Soc. 65, 51–60. doi: 10.1079/PNS2005474
Gibson, R. S., Hess, S. Y., Hotz, C., and Brown, K. H. (2008). Indicators of zinc status at the population level: a review of the evidence. Br. J. Nutr. 99, S14–S23. doi: 10.1017/S0007114508006818
Goron, T. L., and Raizada, M. N. (2015). Genetic diversity and genomic resources available for the small millet crops to accelerate a New Green Revolution. Front. Plant. Sci. 6:157. doi: 10.3389/fpls.2015.00157
Govindaraj, M., Rai, K. N., Shanmugasundaram, P., Dwivedi, S. L., Sahrawat, K. L., Muthaiah, A. R., et al. (2013). Combining ability and heterosis for grain iron and zinc densities in pearl millet. Crop Sci. 53, 505–517. doi: 10.1186/2193-1801-3-763
Graybosch, R. A., and Baltensperger, D. D. (2009). Evaluation of the waxy endosperm trait in proso millet (Panicum miliaceum). Plant Breed. 128, 70–73. doi: 10.1111/j.1439-0523.2008.01511.x
Grusak, M. A. (2002). Enhancing mineral content in plant food products. J. Am. Coll. Nutr. 21, 178S–183S. doi: 10.1080/07315724.2002.10719263
Guerinot, M. L. (2000). The ZIP family of metal transporters. Biochim. Biophys. Acta 1465, 190–198. doi: 10.1016/S0005-2736(00)00138-3
Gupta, S. K., Velu, G., Rai, K. N., and Sumalini, K. (2009). Association of grain iron and zinc content with grain yield and other traits in pearl millet (Pennisetum glaucum (L.) R. Br.). Crop Improv. 36, 4–7. doi: 10.1186/2193-1801-3-763
Gupta, V. P. (1980). “Genetics of quality improvement,” in Trends in Genetical research on Pennisetums, eds V. P. Gupta and J. L. Minnocha (Ludhiana: Punjab Agricultural University), 291–294.
Guttieri, M., Bowen, D., Dorsch, J. A., Raboy, V., and Souza, E. (2004). Identification and characterization of a low phytic acid wheat. Crop Sci. 44, 418–424. doi: 10.2135/cropsci2004.4180
Haas, J. D., Finkelstein, J. L., Udipi, S. A., Ghugre, P., and Mehta, S. (2013). Iron biofortified pearl millet improves iron status in Indian school children: results of a feeding trial. FASEB J. 27, 355.2.
Hadimani, N. A., Muralikrishna, G., Tharanathan, R. N., and Malleshi, N. G. (2001). Nature of carbohydrates and proteins in three pearl millet varieties varying in processing characteristics and kernel texture. J. Cereal Sci. 33, 17–25. doi: 10.1006/jcrs.2000.0342
Hama, F., Vernière, C. I., Guyot, J. P., Rochette, I., Diawara, B., and Mouquet-Rivier, C. (2012). Potential of non-GMO biofortified pearl millet (Pennisetum glaucum) for increasing iron and zinc content and their estimated bioavailability during abrasive decortication. Int. J. Food Sci. Tech. 47, 1–9. doi: 10.1111/j.1365-2621.2012.03017.x
HarvestPlus (2009). Breeding Crops for Better Nutrition: General Brochure. Washington, DC: HarvestPlus.
He, F. J., and MacGregor, G. A. (2008). Beneficial effects of potassium on human health. Physiol. Plant. 133, 725–735. doi: 10.1111/j.1399-3054.2007.01033.x
Hegde, P. S., Rajasekaran, N. S., and Chandra, T. S. (2005). Effects of the antioxidant properties of millet species on oxidative stress and glycemic status in alloxan induced rats. Nutr. Res. 25, 1109–1120. doi: 10.1016/j.nutres.2005.09.020
Heine, W., Radke, M., and Wutzke, K. D. (1995). The significance of tryptophan in human nutrition. Amino Acids 9, 191–205. doi: 10.1007/BF00805951
Hirschi, K. D. (2009). Nutrient biofortification of food crops. Ann. Rev. Nutr. 29, 401–421. doi: 10.1146/annurev-nutr-080508-141143
Hixon, R. M., and Brimhall, R. (1968). “Waxy cereals and red iodine starches,” in Starch and its Derivatives, ed. J. A. Radley (London: Chapman and Hall), 247–281.
Hoshino, T., Nakamura, T., Seimiya, Y., Kamada, T., Ishikawa, G., Ogasawara, A., et al. (2010). Production of a fully waxy line and analysis of waxy genes in the allohexaploid crop, Japanese barnyard millet. Plant Breed. 129, 349–355.
Hotz, C., and Brown, K. H. (2004). Assessment of the risk of zinc deficiency in populations and options for its control. Food Nutr. Bull. 25, 94–204. doi: 10.1177/15648265040251S201
Huang, X., Feng, Q., Qian, Q., Zhao, Q., Wang, L., Wang, A., et al. (2009). High-throughput genotyping by whole-genome resequencing. Genome. Res 19, 1068–1076. doi: 10.1101/gr.089516.108
Hunt, H. V., Denyer, K., Packman, L. C., Jones, M. K., and Howe, C. J. (2010). Molecular basis of the waxy endosperm starch phenotype in broomcorn millet. Mol. Biol. Evol. 27, 1478–1494. doi: 10.1093/molbev/msq040
Ishimaru, Y., Suzuki, M., Kobayashi, T., Takahashi, M., Nakanishi, H., Mori, S., et al. (2005). OsZIP4, a novel zinc-regulated zinc transporter in rice. J. Exp. Bot. 56, 3207–3214. doi: 10.1093/jxb/eri317
Isshiki, K., Morino, K., Nakajima, M., Okagaki, R. J., Wessler, S. R., Izawa, T., et al. (1998). A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the intron. Plant J. 15, 133–138. doi: 10.1046/j.1365-313X.1998.00189.x
Joshi, B. N., Sainani, M. N., Bastawade, K. B., Deshpande, V. V., Gupta, V. S., and Ranjekar, P. K. (1999). Pearl millet cysteine protease inhibitor. Evidence for the presence of two distinct sites responsible for anti-fungal and anti-feedant activities. Eur. J. Biochem. 265, 556–563. doi: 10.1046/j.1432-1327.1999.00764.x
Kashi, Y., and Soller, M. (1999). “Functional roles of microsatellites and minisatellites,” in Microsatellites: Evolution and Applications, eds D. B. Goldstein and C. Schlötterer (New York, NY: Oxford University Press), 10–23.
Kawase, M., Fukunaga, K., and Kato, K. (2005). Diverse origins of waxy foxtail millet crops in East and Southeast Asia mediated by multiple transposable elements insertions. Mol. Genet. Genomics 274, 131–140. doi: 10.1007/s00438-005-0013-8
Kemper, E. L., Neto, G. C., Papes, F., Martinez Moraes, K. C., Leite, A., and Arrudaa, P. (1999). The role of Opaque2 in the control of lysine degrading activities in developing maize endosperm. Plant Cell 11, 1981–1993. doi: 10.1105/tpc.11.10.1981
Kim, J. S., Hyun, T. K., and Kim, M. J. (2011). The inhibitory effects of ethanol extracts from sorghum, foxtail millet and proso millet on α-glucosidase and α-amylase activities. Food Chem. 124, 1647–1651. doi: 10.1016/j.foodchem.2010.08.020
Kim, S. I., Andaya, C. B., Newman, J. W., Goyal, S. S., and Tai, T. H. (2008). Isolation and characterization of a low phytic acid rice mutant reveals a mutation in the rice orthologue of maize MIK. Theor. Appl. Genet. 117, 1291–1301. doi: 10.1007/s00122-008-0863-7
Kim, S. K., Sohn, E. Y., and Lee, I. J. (2009). Starch properties of native foxtail millet, Setaria italica Beauv. J. Crop Sci. Biotech. 12, 59–62. doi: 10.1007/s12892-009-0073-0
Kimata, M., and Negishi, M. (2002). Geographical distribution of proso millet, Panicum miliaceum L. on iodostarch and phenol reactions; with special references to a northern propagation route into Japanese Islands. Environ. Edu. Stud. Tokyo Gakugei University 12, 15–22.
Kodkany, B. S., Bellad, R. M., Mahantshetti, N. S., Westcott, J. E., Krebs, N. F., Kemp, J. F., et al. (2013). Biofortification of pearl millet with iron and zinc in a randomized controlled trial increases absorption of these minerals above physiologic requirements in young children. J. Nutr. 143, 1489–1493. doi: 10.3945/jn.113.176677
Kofuji, K., Aoki, A., Tsubaki, K., and Konishi, M. (2012). Antioxidant activity of β-glucan. ISRN Pharm. 2012, 125864. doi: 10.5402/2012/125864
Krishnaswamy, K. (2009). “The problem of double burden of disease and its consequences-a brief overview,” in Proceedings of the Programs and Abstracts, Symposium on Nutritional Security for India-Issues and way Forward (New Delhi: Indian National Science Academy), 5–6.
Kumar, A., and Chauhan, B. M. (1993). Effect of phytic acid on protein digestibility (in vitro) and HCl-extractability of minerals in pearl millet sprouts. Cereal Chem. 70, 504–506.
Kumar, A., Sharma, N., Panwar, P., and Gupta, A. K. (2012). Use of SSR, RAPD markers and protein profiles based analysis to differentiate Eleusine coracana genotypes differing in their protein content. Mol. Biol. Rep. 39, 4949–4960. doi: 10.1007/s11033-011-1291-3
Kunyanga, C. N., Imungi, J. K., Okoh, M. W., and Biesalski, H. K. (2012). Total phenolic content, antioxidant and antidiabetic properties of methanolic extract of raw and traditionally processed Kenyan indigenous food ingredients. LWT Food Sci. Technol. 45, 269–276. doi: 10.1016/j.lwt.2011.08.006
Kuwano, M., Mimura, T., Takaiwa, F., and Yoshida, K. T. (2009). Generation of stable ‘low phytic acid’ transgenic rice through antisense repression of the 1 D–myoinositol 3–phosphate synthase gene using the 18-kDa oleosin promoter. Plant Biotechnol. J. 7, 96–105. doi: 10.1111/j.1467-7652.2008.00375.x
Larson, S. R., Rutger, J. N., Young, K. A., and Raboy, V. (2000). Isolation and genetic mapping of a non-lethal rice (Oryza sativa L.) low phytic acid 1 mutation. Crop Sci. 40, 1397–1405. doi: 10.2135/cropsci2000.4051397x
Mäser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K., Sze, H., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126, 1646–1667. doi: 10.1104/pp.126.4.1646
Mbithi-Mwikya, S., Camp, J. V., Yiru, Y., and Huyghebaert, A. (2000). Nutrient and antinutrient changes in finger millet (Eleusine coracana) during sprouting. LWT Food Sci. Technol. 33, 9–14. doi: 10.1006/fstl.1999.0605
McDonough, C. M., Rooney, L. W., and Serna-Saldivar, S. O. (2000). “The millets,” in Handbook of Cereal Science and Technology (Food Science and Technology), eds K. Kulp and J. G. Ponte (New York, NY: Marcel Dekker Inc), 177–202.
McIntyre, C. L., Drenth, J., Gonzalez, N., Henzell, R. G., and Jordan, D. R. (2008). Molecular characterization of the waxy locus in sorghum. Genome 51, 524–533. doi: 10.1139/G08-035
Mirza, N., Taj, G., Arora, S., and Kumar, A. (2014). Transcriptional expression analysis of genes involved in regulation of calcium translocation and storage in finger millet (Eleusine coracana L. Gartn.). Gene 550, 171–179. doi: 10.1016/j.gene.2014.08.005
Moreau, S., Thomson, R. M., Kaiser, B. N., Trevaskis, B., Guerinot, M. L., Udvardi, M. K., et al. (2002). GmZIP1 encodes asymbiosis-specific zinc transporter in soybean. J. Biol. Chem. 15, 4738–4746. doi: 10.1074/jbc.M106754200
Muthamilarasan, M., Dhaka, A., Yadav, R., and Prasad, M. (2016). Exploration of millet models for developing nutrient rich graminaceous crops. Plant Sci. 242, 89–97. doi: 10.1016/j.plantsci.2015.08.023
Muthamilarasan, M., and Prasad, M. (2015). Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theor. Appl. Genet. 128, 1–14. doi: 10.1007/s00122-014-2399-3
Nakamura, T., Yamamori, M., Hirano, H., and Hidaka, S. (1993a). Identification of three Wx protein in wheat (Triticum aestivum L.). Biochem. Genet. 31, 75–86. doi: 10.1007/BF02399821
Nakamura, T., Yamamori, M., Hirano, H., and Hidaka, S. (1993b). Decrease of waxy (Wx) protein in two common wheat cultivars with low amylose content. Plant Breed. 111, 99–105. doi: 10.1111/j.1439-0523.1993.tb00614.x
Nakayama, H., Afzal, M., and Okuno, K. (1998). Intraspecific differentiation and geographical distribution of Wx alleles for low amylose content in endosperm of foxtail millet, Setaria italica (L.) Beauv. Euphytica 102, 289–293. doi: 10.1023/A:1018344819531
Nedumaran, S., Bantilan, M. C. S., Gupta, S. K., Irshad, A., and Davis, J. S. (2014). Potential Welfare Benefit of Millets Improvement Research at ICRISAT: Multi Country-Economic Surplus Model Approach, Socioeconomics Discussion Paper Series Number 15. Hyderabad: ICRISAT.
Nirgude, M., Babu, B. K., Shambhavi, Y., Singh, U. M., Upadhyaya, H. D., and Kumar, A. (2014). Development and molecular characterization of genic molecular markers for grain protein and calcium content in finger millet (Eleusine coracana (L.) Gaertn.). Mol. Biol. Rep. 41, 1189–1200. doi: 10.1007/s11033-013-2825-7
Oberleas, D. P. (1973). “Phytates,” in Toxicants Occurring Naturally in Foods, ed. F. M. Strong (Washington DC: National Academy of Sciences), 363–371.
O’Kennedy, M. M., Crampton, B. G., and Ozias-Akins, P. (2009). “Pearl millet,” in Compendium of Transgenic Crop Plants: Transgenic Cereals and Forage Grasses, Vol. 1, eds C. Kole and T. C. Hall (Chichester: John Wiley & Sons), 177–190.
O’Kennedy, M. M., Grootboom, A., and Shewry, P. R. (2006). Harnessing sorghum and millet biotechnology for food and health. J. Cereal Sci. 44, 224–235. doi: 10.1016/j.jcs.2006.08.001
Olsen, K. M., Caicedo, A. L., Polato, N., McClung, A., McCouch, S., and Purugganan, M. D. (2006). Selection under domestication: evidence for a sweep in the rice Waxy genomic region. Genetics 173, 975–983. doi: 10.1534/genetics.106.056473
Paine, J. A., Shipton, C. A., Chaggar, S., Howells, R. M., Kennedy, M. J., Vernon, G., et al. (2005). Improving the nutritional value of Golden rice through increased pro-vitamin A content. Nat. Biotechnol. 23, 482–487. doi: 10.1038/nbt1082
Panwar, P., Nath, M., Kumar, V., and Kumar, A. (2010a). Comparative evaluation of genetic diversity using RAPD, SSR and Cytochrome P450 gene based markers with respect to calcium content in finger millet (Eleusine coracana L. Gaertn.). J. Genet. 89, 121–133. doi: 10.1007/s12041-010-0052-8
Panwar, P., Saini, R. K., Sharma, N., Yadav, D., and Kumar, A. (2010b). Efficiency of RAPD, SSR and Cytochrome P450 gene based markers in accessing genetic variability amongst finger millet (Eleusine coracana) accessions. Mol. Biol. Rep. 37, 4075–4082. doi: 10.1007/s11033-010-0067-5
Pattabiraman, T. N. (1986). Trypsin/chymotrypsin inhibitors from millets. Adv. Exp. Med. Biol. 199, 439–448. doi: 10.1007/978-1-4757-0022-0_25
Pedas, P., Ytting, C. K., Fuglsang, A. T., Jahn, T. P., Schjoerring, J. K., and Husted, S. (2008). Manganese efficiency in barley: identification and characterization of the metal ion transporter HvIRT1. Plant Physiol. 148, 455–466. doi: 10.1104/pp.108.118851
Pettifor, J. M. (2004). Nutritional rickets: deficiency of vitamin D, calcium, or both? Am. J. Clin. Nutr. 80, 1725S–1729S. doi: 10.1007/978-1-59745-464-3_14
Pilu, R., Panzeri, D., Gavazzi, G., Rasmussen, S. K., Consonni, G., and Nielsen, E. (2003). Phenotypic, genetic and molecular characterization of a maize low phytic acid mutant (Lpa 241). Theor. Appl. Genet. 107, 980–987. doi: 10.1007/s00122-003-1316-y
Plaza, S., Tearall, K. L., Zhao, F. J., Buchner, P., McGrath, S. P., and Hawkesford, M. J. (2007). Expression and functional analysis of metal transporter genes in two contrasting ecotypes of the hyperaccumulator Thlaspi caerulescens. J. Exp. Bot. 58, 1717–1728. doi: 10.1093/jxb/erm025
Qaim, M., Stein, A. J., and Meenakshi, J. V. (2007). Economics of Biofortification. Washington, DC: HarvestPlus.
Quan, J., Dong, L., Ma, J., Li, Z., and Zheng, Z. (2010). Breeding of leaf rust resistance and waxy millet new variety Jichuang 1. J. Hebei. Agric. Sci. 14, 127–128.
Raboy, V. (2009). Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 177, 281–296. doi: 10.1016/j.plantsci.2009.06.012
Raboy, V., and Bowen, D. (2006). “Genetics of inositol polyphosphates,” in Biology of Inositols and Phosphoinositides, eds A. Lahiri Majumder and B. B. Biswas (Netherlands: U.S. Government), 71–102.
Raboy, V., Gerbasi, P. F., Young, K. A., Stoneberg, S. D., Pickett, S. G., Bauman, A. T., et al. (2000). Origin and seed phenotype of maize low phytic acid 1–1 and low phytic acid 2–1. Plant Physiol. 124, 355–368. doi: 10.1104/pp.124.1.355
Rai, K. N., Anand Kumar, K., Andrews, D. J., Rao, A. S., Raj, A. G. B., and Witcombe, J. R. (1990). Registration of ICTP 8203 pearl millet. Crop Sci. 30, 959. doi: 10.2135/cropsci1990.0011183X003000040048x
Rai, K. N., Hash, C. T., Singh, A. K., and Velu, G. (2008). Adaptation and quality traits of a germplasm-derived commercial seed parent of pearl millet. Plant Genet. Resour. Newsl. 154, 20–24.
Rai, K. N., Patil, H. T., Yadav, O. P., Govindaraj, M., Khairwal, I. S., Cherian, B., et al. (2014). Dhanashakti: a high-iron pearl millet variety. Ind. Farm. 64, 32–34.
Rai, K. N., Velu, G., Govindaraj, M., Upadhyaya, H. D., Rao, A. S., Shivade, H., et al. (2015). Iniadi pearl millet germplasm as a valuable genetic resource for high grain iron and zinc densities. Plant Genet. Resour. 13, 75–82. doi: 10.1017/S1479262114000665
Rai, K. N., Yadav, O. P., Rajpurohit, B. S., Patil, H. T., Govindaraj, M., Khairwal, I. S., et al. (2013). Breeding pearl millet cultivars for high iron density with zinc density as an associated trait. J. SAT Agric. Res. 11, 1–7.
Ramegowda, Y., Venkategowda, R., Jagadish, P., Govind, G., Hanumanthareddy, R. R., Makarla, U., et al. (2013). Expression of a rice Zn transporter, OsZIP1, increases Zn concentration in tobacco and finger millet transgenic plants. Plant Biotechnol. Rep. 7, 309–319. doi: 10.1007/s11816-012-0264-x
Ramesh, S. A., Choimes, S., and Schachtman, D. P. (2004). Over-expression of an Arabidopsis zinc transporter in Hordeum vulgare increases short-term zinc uptake after zinc deprivation and seed zinc content. Plant Mol. Biol. 54, 373–385. doi: 10.1023/B:PLAN.0000036370.70912.34
Ramesh, S. A., Shin, R., Eide, D. J., and Schachtman, D. P. (2003). Deferential metal selectivity and gene expression of two zinc transporters from rice. Plant Physiol. 133, 126–134. doi: 10.1104/pp.103.026815
Reddy, I. N. B. L., Reddy, D. S., Narasu, M. L., and Sivaramakrishnan, S. (2011). Characterization of disease resistance gene homologues isolated from finger millet (Eleusine coracana L. Gaertn). Mol. Breed. 27, 315–328. doi: 10.1007/s11032-010-9433-1
Ross, A. C., Manson, J. E., Abrams, S. A., Aloia, J. F., Brannon, P. M., Clinton, S. K., et al. (2011). The 2011 report on dietary reference intakes for calcium and vitamin D from the institute of medicine: what clinicians need to know. J. Clin. Endocrin. Metabol. 96, 53–58. doi: 10.1210/jc.2010-2704
Saleh, A. S. M., Zhang, Q., Chen, J., and Shen, Q. (2013). Millet grains: nutritional quality, processing, and potential health benefits. Compr. Rev. Food Sci. Food Saf. 12, 281–295. doi: 10.1111/1541-4337.12012
Sano, Y. (1984). Differential regulation of waxy gene expression in rice endosperm. Theor. Appl. Genet. 68, 467–473. doi: 10.1007/BF00254822
Sharma, S., Saxena, D. C., and Riar, C. S. (2016). Isolation of functional components β-glucan and γ-amino butyric acid from raw and germinated barnyard millet (Echinochloa frumentacea) and their characterization. Plant Foods Hum. Nutr. 71, 231–238. doi: 10.1007/s11130-016-0545-6
Shi, J., Wang, H., Hazebroek, J., Ertl, D. S., and Harp, T. (2005). The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J. 42, 408–419. doi: 10.1111/j.1365-313X.2005.02412.x
Shivaraj, B., and Pattabiraman, T. N. (1981). Natural plant enzyme inhibitors. Characterization of an unusual alpha-amylase/trypsin inhibitor from ragi (Eleusine coracana Geartn.). Biochem. J. 193, 29–36. doi: 10.1042/bj1930029
Shivran, A. C. (2016). “Biofortification for nutrient-rich millets,” in Biofortification of Food Crops, eds U. Singh, C. S. Praharaj, S. S. Singh, and N. P. Singh (New Delhi: Springer), 409–420. doi: 10.1007/978-81-322-2716-8_30
Shukla, V. K., Doyon, Y., Miller, J. C., DeKelver, R. C., Moehle, E. A., Worden, S. E., et al. (2009). Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nat. Lett. 459, 437–443. doi: 10.1038/nature07992
Singh, U. M., Chandra, M., Shankhdhar, S. C., and Kumar, A. (2014). Transcriptome wide identification and validation of calcium sensor gene family in the developing spikes of finger millet genotypes for elucidating its role in grain calcium accumulation. PLoS ONE 9:e103963. doi: 10.1371/journal.pone.0103963
Sood, S., Khulbe, R. K., Gupta, A. K., Agrawal, P. K., Upadhyaya, H. D., and Bhatt, J. C. (2015). Barnyard millet – a potential food and feed crop of future. Plant Breed. 134, 135–147. doi: 10.1111/pbr.12243
Stevenson-Paulik, J., Odom, A. R., and York, J. D. (2002). Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. J. Biol. Chem. 277, 42711–42718. doi: 10.1074/jbc.M209112200
Sweetman, D., Johnson, S., Caddick, S. E., Hanke, D. E., and Brearley, C. A. (2006). Characterization of an Arabidopsis inositol 1,3,4,5,6-pentakisphosphate 2-kinase (AtIPK1). Biochem. J. 394, 95–103. doi: 10.1042/BJ20051331
Takei, E., and Sakamoto, S. (1989). Further analysis of geographical variation of heading response to daylength in foxtail millet (Setaria italica P. Beauv.). Jpn. J. Breed. 39, 285–298. doi: 10.1270/jsbbs1951.39.285
Tako, E., Beebe, S. E., Reed, S., Hart, J. J., and Glahn, R. P. (2014). Polyphenolic compounds appear to limit the nutritional benefit of biofortified higher iron black bean (Phaseolus vulgaris L.). Nutr. J. 13, 28. doi: 10.1186/1475-2891-13-28
Tako, E., Beebe, S. E., Reed, S., Hart, J. J., and Glahn, R. P. (2015). Higher iron pearl millet (Pennisetum glaucum L.) provides more absorbable iron that is limited by increased polyphenolic content. Nutr. J. 14, 11. doi: 10.1186/1475-2891-14-11
Taylor, J. R. N., and Emmambux, M. N. (2008). “Gluten-free foods and beverages from millets,” in Gluten-Free Cereal Products and Beverages (Food Science and Technology), eds E. K. Arendt and F. Dal Bello (Cambridge, MA: Academic Press), 119–148.
Tomé, D., and Bos, C. (2007). Lysine requirement through the human life cycle. J. Nutr. 137, 1642S–1645S.
Tsehaye, Y., Berg, T., Tsegaye, B., and Tanto, T. (2006). Farmers’ management of finger millet (Eleusine coracana L.) diversity in Tigray, Ethiopia and implications for on-farm conservation. Biodivers. Conserv. 15, 4289–4308. doi: 10.1007/s10531-005-3581-3
Ugare, R., Chimmad, B., Naik, R., Bharathi, P., and Itagi, S. (2011). Glycemic index and significance of barnyard millet (Echinochloa frumentacae) in type II diabetics. J. Food Sci. Technol. 51, 392–395. doi: 10.1007/s13197-011-0516-8
Upadhyaya, H. D., Dwivedi, S. L., Singh, S. K., Singh, S., Vetriventhan, M., and Sharma, S. (2014). Forming core collections in barnyard, kodo, and little millets using morphoagronomic descriptors. Crop Sci. 54, 1–10. doi: 10.2135/cropsci2014.03.0221
Upadhyaya, H. D., Gowda, C. L. L., Pundir, R. P. S., Reddy, G. V., and Singh, S. (2006). Development of core subset of finger millet germplasm using geographical origin and data on 14 quantitative traits. Genet. Resour. Crop Evol. 53, 679–685. doi: 10.1007/s10722-004-3228-3
Upadhyaya, H. D., Gowda, C. L. L., Reddy, K. N., and Singh, S. (2009). Augmenting the pearl millet core collection for enhancing germplasm utilization in crop improvement. Crop Sci. 49, 573–580. doi: 10.2135/cropsci2008.06.0378
Upadhyaya, H. D., Pundir, R. P. S., Gowda, C. L. L., Reddy, G. V., and Singh, S. (2008). Establishing a core collection of foxtail millet to enhance the utilization of germplasm of an underutilized crop. Plant Genet. Resour. 7, 177–184. doi: 10.1017/S1479262108178042
Upadhyaya, H. D., Ramesh, S., Sharma, S., Singh, S. K., Varshney, S. K., Sarmad, N. D. R. K., et al. (2011a). Genetic diversity for grain nutrient contents in a core collection of finger millet (Eleusine coracana (L.) Gaertn.) germplasm. Field Crops Res. 121, 42–52. doi: 10.1016/j.fcr.2010.11.017
Upadhyaya, H. D., Ravishankar, C. R., Narasimhudu, Y., Sarma, N. D. R. K., Singh, S. K., Varshney, S. K., et al. (2011b). Identification of trait-specific germplasm and developing a minicore collection for efficient use of foxtail millet genetic resources in crop improvement. Field Crop. Res. 124, 459–467. doi: 10.1016/j.fcr.2011.08.004
Upadhyaya, H. D., Reddy, K. N., Ahmed, M. I., Kumar, V., Gumma, M. K., and Ramachandran, S. (2016a). Geographical distribution of traits and diversity in the world collection of pearl millet [Pennisetum glaucum (L.) R. Br., synonym: Cenchrus americanus (L.) Morrone] landraces conserved at the ICRISAT genebank. Genet. Resour. Crop Evol. 1–17. doi: 10.1007/s10722-016-0442-8
Upadhyaya, H. D., Reddy, K. N., Ahmed, M. I., Ramachandran, S., Kumar, V., and Singh, S. (2016b). Characterization and genetic potential of African pearl millet named landraces conserved at the ICRISAT genebank. Plant Genet. Resour. 1, 1–15. doi: 10.1017/S1479262116000113
Upadhyaya, H. D., Vetriventhan, M., Dwivedi, S. L., Pattanashetti, S. K., and Singh, S. K. (2016c). “Proso, barnyard, little, and kodo millets,” in Genetic and Genomic Resources for Grain Cereals Improvement, eds M. Singh and H. D. Upadhyaya (Cambridge, MA: Academic Press), 321–343.
Upadhyaya, H. D., Yadav, D., Reddy, K. N., Gowda, C. L. L., and Singh, S. (2011c). Development of pearl millet minicore collection for enhanced utilization of germplasm. Crop Sci. 51, 217–223. doi: 10.2135/cropsci2010.06.0336
Van, K., Onoda, S., Kim, M. Y., Kim, K. D., and Lee, S. H. (2008). Allelic variation of the Waxy gene in foxtail millet [Setaria italica (L.) P. Beauv.] by single nucleotide polymorphisms. Mol. Genet. Genomics 279, 255–266. doi: 10.1007/s00438-007-0310-5
Velu, G., Rai, K. N., Muralidharan, V., Kulkarni, V. N., Longvah, T., and Raveendran, T. S. (2007). Prospects of breeding biofortified pearl millet with high grain iron and zinc content. Plant Breed. 126, 182–185. doi: 10.1111/j.1439-0523.2007.01322.x
Velu, G., Rai, K. N., Muralidharan, V., Longvah, T., and Crossa, J. (2011). Gene effects and heterosis for grain iron and zinc density in pearl millet (Pennisetum glaucum (L.) R. Br). Euphytica 180, 251–259. doi: 10.1007/s10681-011-0387-0
Velu, G., Rai, K. N., and Sahrawat, K. L. (2008). Variability for grain iron and zinc content in a diverse range of pearl millet populations. Crop Improv. 35, 186–191.
Vetriventhan, M., Upadhyaya, H. D., Dwivedi, S. L., Pattanashetti, S. K., and Singh, S. K. (2016). “Finger and foxtail millets,” in Genetic and Genomic Resources for Grain Cereals Improvement, eds M. Singh and H. D. Upadhyaya (Cambridge, MA: Academic Press), 291–319.
Vijayakumari, J., Mushtari Begum, J., Begum, S., and Gokavi, S. (2003). “Sensory attributes of ethnic foods from finger millet (Eleusine coracana),” in Proceeding of the National Seminar on Processing and Utilization of Millet for Nutrition Security: Recent Trends in Millet Processing and Utilization (Hisar: CCSHAV), 7–12.
Welch, R. M., and Graham, R. D. (2004). Breeding for micronutrients in staple food crops from a human nutrition perspective. J. Exp. Bot. 55, 353–364. doi: 10.1093/jxb/erh064
White, P. J. (2005). “Calcium,” in Plant Nutritional Genomics, eds M. R. Broadley and P. J. White (Oxford: Blackwell), 66–86.
White, P. J., and Broadley, M. R. (2003). Calcium in plants. Ann. Bot. 92, 487–511. doi: 10.1093/aob/mcg164
White, P. J., and Broadley, M. R. (2005). Biofortifying crops with essential mineral elements. Trends Plant Sci. 10, 586–593. doi: 10.1016/j.tplants.2005.10.001
World Health Organization [WHO] (2011). Global Database on Vitamin A Deficiency. Available at: http://www.who.int/vmnis/database/vitamina/en/ [accessed December 03, 2016].
Yadav, O. P., and Rai, K. N. (2013). Genetic improvement of pearl millet in India. Agric. Res. 2, 275–292. doi: 10.1007/s40003-013-0089-z
Yadav, O. P., Rai, K. N., Bidinger, F. R., Gupta, S. K., Rajpurohit, B. S., and Bhatnagar, S. K. (2012). Pearl millet (Pennisetum glaucum) restorer lines for breeding dual-purpose hybrids adapted to arid environments. Ind. J. Agric. Sci. 82, 922–927.
Yasui, T., Sasaki, T., Matsuki, J., and Yamamori, M. (1997). Waxy endosperm mutants of bread wheat (Triticum aestivum L.) and their starch properties. Breed. Sci. 47, 161–163. doi: 10.1002/jsfa.5574
Young, V. R., Scrimshaw, N. S., and Pellet, P. (1998). “Significance of dietary protein source in human nutrition: animal and/or plant proteins?,” in Feeding a World Population of More than Eight Billion People, eds J. C. Waterlow, D. G. Armstrong, L. Fowden, and R. Riley (New York, NY: Oxford University Press).
Zhang, G., Liu, X., Quan, Z., Cheng, S., Xu, X., Pan, S., et al. (2012). Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 30, 549–554. doi: 10.1038/nbt.2195
Zhang, L. Z., and Liu, R. H. (2015). Phenolic and carotenoid profiles and antiproliferative activity of foxtail millet. Food Chem. 174, 495–501. doi: 10.1016/j.foodchem.2014.09.089
Keywords: millets, biofortification, macronutrients, micronutrients, nutritional security
Citation: Vinoth A and Ravindhran R (2017) Biofortification in Millets: A Sustainable Approach for Nutritional Security. Front. Plant Sci. 8:29. doi: 10.3389/fpls.2017.00029
Received: 22 September 2016; Accepted: 05 January 2017;
Published: 23 January 2017.
Edited by:
Manoj Prasad, National Institute of Plant Genome Research, IndiaReviewed by:
Salej Sood, Indian Council of Agricultural Research (ICAR), IndiaCopyright © 2017 Vinoth and Ravindhran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: R. Ravindhran, cmF2aWxveW9sYTE5OThAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.