
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Plant Sci., 11 January 2017
Sec. Plant Cell Biology
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.02030
This article is part of the Research TopicProtein quality controlling systems in plant responses to environmental stressesView all 17 articles
Plants are sessile organisms that cannot escape from stressful environments, such as drought, high salinity, high temperature, and shortage of essential minerals in the soil. Hence, plants have evolved processes that protect them from these harmful conditions. One of these major processes is autophagy (which means, “self-eating”), a mechanism that destroys specific compounds that participate in efficient growth and requires extensive energy input and on the other hand stimulates biological processes that protects from the stress. Autophagy can be either a bulk process, turning over bulk amounts of various components in response to major stresses, such as serious accumulation of damaging compounds in the soil, or a selective process turning over specific components in response to specific and/or relatively minor environmental cues, such as minor shortage of rain and/or non-significant shortage of minerals in the soil (Han et al., 2011; Avin-Wittenberg et al., 2012; Liu and Bassham, 2012; Michaeli et al., 2016).
The identification of autophagy-related genes (ATGs) was an important milestone in the understanding of the mechanism of autophagy. Thus far, over 30 ATGs have been identified in Arabidopsis, rice, tobacco, and pepper based on comprehensive, genome-wide analysis (Xia et al., 2011; Zhou et al., 2015; Zhai et al., 2016). ATGs can be divided into three categories with respect to their function: (i) ATG1-ATG13 comprising the kinase complex, an upstream regulator that initiates autophagosome formation (Suttangkakul et al., 2011); (ii) The ATG9 and ATG6/vps30 complexes involved in vacuolar protein sorting in which ATG9 interacts with ATG2 and ATG18, boosting phagophore expansion. This operates via diverse shuttling of endomembranes, such as those of the endoplasmic reticulum (ER), Golgi, and mitochondria (Tooze and Yoshimori, 2010; Yang and Klionsky, 2010; Kang et al., 2011). The ATG6/vps30 complex recruited by ATG14, which localizes to the pre-autophagosomal structure (PAS) as well as the vacuolar membrane, to generate the autophgosomes (Tooze and Yoshimori, 2010); (iii) ubiquitin like conjugation systems (ATG5-ATG12 complex and ATG8-PE complex), which is essential for autophagosome formation. The ATG8–PE complex has been proven to recruit the cytoplasmic cargo to ensure autophagosome maturation and closure, and it is subsequently transported to the vacuole for degradation (Bassham, 2007; Michaeli et al., 2016). Most of complexes participating in each selective autophagy process have been identified in yeast and animal systems. However, it is still in infancy in plants (Michaeli et al., 2016).
Among the ATG genes, ATG8 is the central protein involved in autophagy and also a marker for the autophgosome (Shpilka et al., 2011). It has been shown to participate in various processes, such as diverse intracellular trafficking, post-mitotic Golgi reassembly, cargo receptor recognition, conjugation to phosphatidylethanolamine, etc. (Kwon and Park, 2008; Tooze and Yoshimori, 2010; Yang and Klionsky, 2010). Moreover, ATG8 plays an important role in the sensitivity of plants to abiotic stresses. There are nine isoforms of ATG8 in Arabidopsis (ATG8a to ATG8i). The over-expression of AtATG8f led transgenic Arabidopsis plants to be more sensitive to mild salt and/or osmotic stress. This kind of sensitivity was accompanied by the modification of root architecture (Sláviková et al., 2005). Furthermore, the overexpression of SiATG8a improved the tolerance to nitrogen starvation and drought stress in transgenic Arabidopsis plants (Li et al., 2015).
The other ATGs also play critical roles in the stress response, especially the response to carbon and nitrogen starvation (Han et al., 2011). Two classical autophagy-related mutants, atg5 and atg7, showed hypersensitivity to carbon and nitrogen starvation (Phillips et al., 2008; Yoshimoto et al., 2009). Autophagy-defective RNAi-AtATG18a plants displayed enhanced sensitive to salt, drought and methy viologen treatments compared with wild-type plants (Xiong et al., 2007; Liu et al., 2009). Moreover, atg13 double-knockout (atg13a atg13b) and atg11 knockout plants also showed a classical atg mutant phenotype, which exhibited increased sensitivity to carbon and nitrogen starvation (Suttangkakul et al., 2011). Rice Osatg10b mutants were sensitive to treatments with high salt (250mM) and methyl viologen (MV) (Shin et al., 2009). Additionally, a series of autophagy-deficient mutants, such as atg2-1, atg5-1, atg7-3, atg10-1, were hypersensitive to submergence stress (Chen et al., 2015).
Out of a number of autophagy cargo receptors, the cargo receptor NBR1 (NEIGHBOR OF BRCA1 GENE 1) is one of the critical components of the autophagy process. NBR1 was identified in yeast, mammals and plants (Johansen and Lamark, 2011). NBR1 was the first selective cargo receptor, which was found to be responsible for sequestration of ubiquitinated proteins to the vacuole for their degradation inside this organelle. However, the involvement of the cargo receptors in autophagy is poorly understood. Recently, functional homologs of NBR1 proteins were identified in Arabidopsis (AtNBR1) and tobacco (JOKA2) plants (Svenning et al., 2011; Zientara-Rytter et al., 2011).
The Arabidopsis NBR1 is a homolog of the mammalian autophagic adaptor NBR1, with an ubiquitin-association domain binds selectively to six of the nine Arabidopsis ATG8 protein isoforms. The nbr1 mutant exhibits sensitivity to a spectrum of abiotic stresses, similar to the autophagy-deficient atg5 and atg7 mutants, but had no obvious effect onthe response to carbon starvation and resistance to a necrotrophic fungal pathogen. This indicates that AtNBR1 participates in the response to abiotic stress (Zhou et al., 2013). Under high heat conditions, an insoluble highly ubiquitinated detergent-resistant protein was shown to be prone to aggregation, thereby enabling recognition by NBR1 and its subsequent transport to the vacuole for degradation. Moreover, Rubisco activase and a number of catalases are linked to the response of plants to a wide variety of abiotic stresses and these enzymes accumulated in the nbr1 mutant (Zhou et al., 2014). These findings suggest that NBR1-mediated selective autophagy pathway plays a critical role during abiotic stresses (Figure 1).
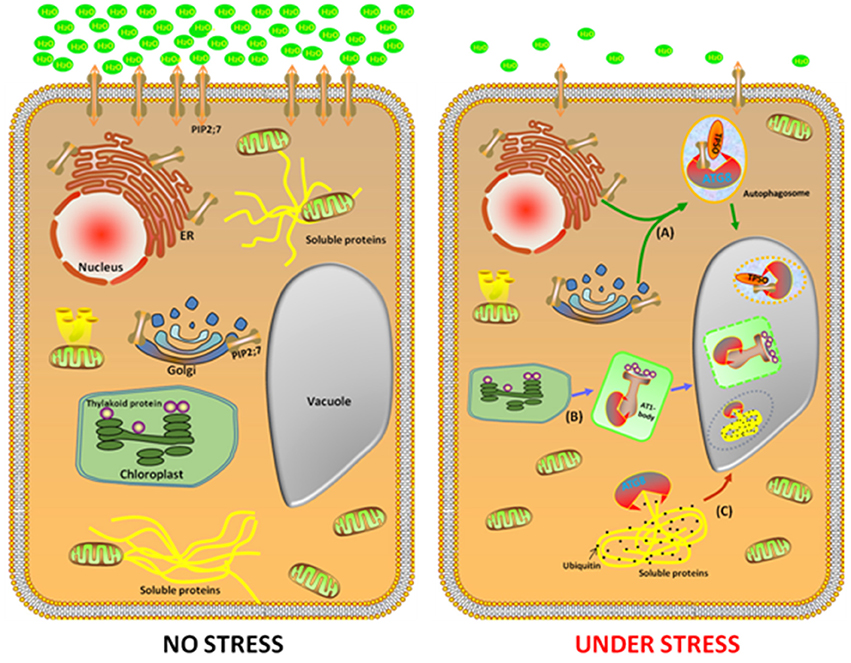
Figure 1. Identification of three potential cargo receptors in plants. In the favorable growth conditions (left cell), a large number of genes, such as NBR1, TPSO, and ATI1 were expressed with a low level to maintain homeostasis. Under abiotic stress conditions (right cell), NBR1, TPSO, and ATI1 were induced. In the process of TSPO-mediated selective autophagy pathway, TSPO is transcriptionally upregulated during heat and drought stress, then interacts with PIP2;7 and ATG8 to enable the degradation of porphyrins via an autophagy-dependent degradation mechanism (A). The second pathway is ATI1 interact with chloroplast protein in the ATI1-body, then this compound transport their cargo proteins to autophagosomes, which eventually transport to the vacuole for degradation (B). In the NBR1-mediated selective autophagy pathway, an insoluble highly ubiquitinated detergent-resistant protein and ATG8 were shown to be prone to aggregation to form autophgosome, thereby enabling recognition by NBR1 and its subsequent transport to the vacuole for degradation (C).
To decipher the involvement of autophagy in abiotic stress in plants, the identification of the cargo receptor in the process of autophagy is crucial. This is because of the following reasons (i) under normal conditions, autophagy maintains a basal level for homeostasis; (ii) under abiotic stress, cargo receptors are active to remove the damaged or unwanted materials or to recycle the materials for providing anabolic substrates and metabolites to the cells. In plants, one of the critical processes of autophagy is the transport of the unnecessary components to the vacuole for degradation, which is a selective process requiring the cargos.
To minimize the effects of abiotic stresses, plants have developed a sophisticated protein quality control system that maintains the protein homeostasis. When subjected to abiotic stresses, such as heat and drought, the earliest response is inhibition of protein synthesis and increase in protein folding and processing. Recently a new Arabidopsis cargo receptor was identified and was named TRYPTOPHAN-RICH SENSORY PROTEIN (TSPO). It is a multi-stress regulator that is transiently induced by abiotic stress and is originally described as a heme-binding protein, which interacts with ATG8 to enable the degradation of porphyrins via an autophagy-dependent degradation mechanism (Vanhee et al., 2011). Moreover, TSPO interacts intracellularly with the plasma membrane aquaporin PIP2;7 (PLASMA MEMBRANE INTRINSIC PROTEIN 2;7) and downregulates it in the cell. The coexpression of TSPO and PIP2;7 led to decreased levels of PIP2;7 in the plasma membrane and abolished the membrane water permeability mediated by the overexpression of PIP2;7 in transgenic seedlings. Furthermore, ABA treatment activates TSPO and triggers the degradation of PIP2;7 through the autophagic pathway. These findings suggest that TSPO acts as a selective plant-specific autophagy cargo receptor during abiotic stress (Hachez et al., 2014). Remarkably, the autophagy-mediated reduction in the quantity of PIP2;7 modulates the osmotic water permeability of membranes, which is important during heat and drought stress (Figure 1).
With the aim of elucidating the biological processes in plants that are associated with selective autophagy in plants, our lab identified a number of plant-specific ATG8f binding proteins based on a yeast two-hybrid analysis. One of these proteins, named “Autophagy Interacting Protein 1” (ATI1), was further subjected to detailed studies. When grown under regular, non-stress conditions, ATI1 was partially associated with the endoplasmic reticulum(ER) membrane. Furthermore, upon exposure of the plants to carbon or nitrogen starvation, ATI1 was assembled into two different types of novel bodies that were associated with either the ER or the plastids (Honig et al., 2012).
When the plants were exposed to carbon or nitrogen starvation, ATI1 was incorporated into novel bodies that were either moved along the ER network, or localized inside the plastids. These novel bodies were then transported into the central vacuole in which their contents were apparently being turned over inside the plastids (Figure 1). Interestingly, the seedlings of the over-expressing ATI1 plants germinated faster and showed increased tolerance to carbon starvation and salt stress, whereas the plants with suppressed expression of ATI1 showed reduced tolerance to carbon starvation and salt stress, indicating that the biological processes using ATI1 confer faster growth and increased stress tolerance to the germinating seedlings (Michaeli et al., 2014).
The above results imply the ATI1 is a multifunctional protein, which is associated with ER-to-vacuole and plastid-to-vacuole trafficking by ATG8-mediated selective autophagy. Moreover, ATI1 is also involved in an autophagic system that promotes the seedling organization under abiotic stress conditions.
Despite the identification of three potential cargo receptors in plants, a detailed understanding of these cargos is imperative. The questions that need to be addressed are whether there are links between the different cargo-mediated autophagy pathways, the mechanisms for the recognition and delivery of the misfolded and damaged proteins and organelles, and how the cargos are trafficked. We believe that with an in-depth research on the involvement of autophagy in abiotic stress, particularly in crop plants, it is possible to open new avenues for the enhancement of stress tolerance by using genetic and/or genetic engineering approaches, ultimately leading to enhanced production.
WW and GG conceived and designed the project, and wrote the manuscript. MX and GW helped with figure revision. The manuscript was approved by all other authors.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Avin-Wittenberg, T., Honig, A., and Galili, G. (2012). Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma 249, 285–299. doi: 10.1007/s00709-011-0296-z
Bassham, D. C. (2007). Plant autophagy—more than a starvation response. Curr. Opin. Plant Biol. 10, 587–593. doi: 10.1016/j.pbi.2007.06.006
Chen, L., Liao, B., Qi, H., Xie, L. J., Huang, L., Tan, W. J., et al. (2015). Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 11, 2233–2246. doi: 10.1080/15548627.2015.1112483
Hachez, C., Veljanovski, V., Reinhardt, H., Guillaumot, D., Vanhee, C., Chaumont, F., et al. (2014). The Arabidopsis abiotic stress-induced TSPO-related protein reduces cell-surface expression of the aquaporin PIP2; 7 through protein-protein interactions and autophagic degradation. Plant Cell 26, 4974–4990. doi: 10.1105/tpc.114.134080
Han, S., Yu, B., Wang, Y., and Liu, Y. (2011). Role of plant autophagy in stress response. Protein Cell 2, 784–791. doi: 10.1007/s13238-011-1104-4
Honig, A., Avin-Wittenberg, T., Ufaz, S., and Galili, G. (2012). A new type of compartment, defined by plant-specific ATG8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell 24, 288–303. doi: 10.1105/tpc.111.093112
Johansen, T., and Lamark, T. (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296. doi: 10.4161/auto.7.3.14487
Kang, R., Zeh, H. J., Lotze, M. T., and Tang, D. (2011). The Beclin 1 network regulates autophagy and apoptosis. Cell Death Diff. 18, 571–580. doi: 10.1038/cdd.2010.191
Kwon, S. I., and Park, O. K. (2008). Autophagy in plants. J. Plant Biol. 51, 313–320. doi: 10.1007/BF03036132
Li, W. W., Chen, M., Zhong, L., Liu, J. M., Xu, Z. S., Li, L. C., et al. (2015). Overexpression of the autophagy-related gene SiATG8a from foxtail millet (Setariaitalica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochem. Biophys. Res. Commun. 468, 800–806. doi: 10.1016/j.bbrc.2015.11.035
Liu, Y., and Bassham, D. C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63, 215–237. doi: 10.1146/annurev-arplant-042811-105441
Liu, Y., Xiong, Y., and Bassham, D. C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5, 954–963. doi: 10.4161/auto.5.7.9290
Michaeli, S., Galili, G., Genschik, P., Fernie, A. R., and Avin-Wittenberg, T. (2016). Autophagy in plants–what's new on the menu? Trends Plant Sci. 21, 134–144. doi: 10.1016/j.tplants.2015.10.008
Michaeli, S., Honig, A., Levanony, H., Peled-Zehavi, H., and Galili, G. (2014). Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 26, 4084–4101. doi: 10.1105/tpc.114.129999
Phillips, A. R., Suttangkakul, A., and Vierstra, R. D. (2008). The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178, 1339–1353. doi: 10.1534/genetics.107.086199
Shin, J. H., Yoshimoto, K., Ohsumi, Y., Jeon, J. S., and An, G. (2009). OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol. Cells 27, 67–74. doi: 10.1007/s10059-009-0006-2
Shpilka, T., Weidberg, H., Pietrokovski, S., and Elazar, Z. (2011). Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 12:1. doi: 10.1186/gb-2011-12-7-226
Sláviková, S., Shy, G., Yao, Y., Glozman, R., Levanony, H., Pietrokovski, S., et al. (2005). The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 56, 2839–2849. doi: 10.1093/jxb/eri276
Suttangkakul, A., Li, F., Chung, T., and Vierstra, R. D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23, 3761–3779. doi: 10.1105/tpc.111.090993
Svenning, S., Lamark, T., Krause, K., and Johansen, T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7, 993–1010. doi: 10.4161/auto.7.9.16389
Tooze, S. A., and Yoshimori, T. (2010). The origin of the autophagosomal membrane. Nat. Cell Biol. 12, 831–835. doi: 10.1038/ncb0910-831
Vanhee, C., Zapotoczny, G., Masquelier, D., Ghislain, M., and Batoko, H. (2011). The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 23, 785–805. doi: 10.1105/tpc.110.081570
Xia, K., Liu, T., Ouyang, J., Wang, R., Fan, T., and Zhang, M. (2011). Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 18, 363–377. doi: 10.1093/dnares/dsr024
Xiong, Y., Contento, A. L., Nguyen, P. Q., and Bassham, D. C. (2007). Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143, 291–299. doi: 10.1104/pp.106.092106
Yang, Z., and Klionsky, D. J. (2010). Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12, 814–822. doi: 10.1038/ncb0910-814
Yoshimoto, K., Jikumaru, Y., Kamiya, Y., Kusano, M., Consonni, C., Panstruga, R., et al. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914–2927. doi: 10.1105/tpc.109.068635
Zhai, Y., Guo, M., Wang, H., Lu, J., Liu, J., Zhang, C., et al. (2016). Autophagy, a conserved mechanism for protein degradation, responds to heat, and other abiotic stresses in Capsicum annuum L. Front. Plant Sci. 7:131. doi: 10.3389/fpls.2016.00131
Zhou, J., Wang, J., Cheng, Y., Chi, Y. J., Fan, B., Yu, J. Q., et al. (2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9:e1003196. doi: 10.1371/journal.pgen.1003196
Zhou, J., Zhang, Y., Qi, J., Chi, Y., Fan, B., Yu, J. Q., et al. (2014). E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 10:e1004116. doi: 10.1371/journal.pgen.1004116
Zhou, X. M., Zhao, P., Wang, W., Zou, J., Cheng, T. H., Peng, X. B., et al. (2015). A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res. 22, 245–257. doi: 10.1093/dnares/dsv012
Keywords: autophagy, protein degradation, abiotic stress, nbr1, TPSO, ATI1
Citation: Wang W, Xu M, Wang G and Galili G (2017) Autophagy: An Important Biological Process That Protects Plants from Stressful Environments. Front. Plant Sci. 7:2030. doi: 10.3389/fpls.2016.02030
Received: 01 November 2016; Accepted: 19 December 2016;
Published: 11 January 2017.
Edited by:
Minghui Lu, Northwest A&F University, ChinaReviewed by:
Shi Xiao, Sun Yat-sen University, ChinaCopyright © 2017 Wang, Xu, Wang and Galili. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoping Wang, Z3B3YW5nQHNjYXUuZWR1LmNu
Gad Galili, Z2FkLmdhbGlsaUB3ZWl6bWFubi5hYy5pbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.