- Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture, Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences, Wuhan, China
Here, we report the identification of the Brassica-specific gene MS5d, which is responsible for male sterility in Brassica napus. The MS5d gene is highly expressed in the microsporocyte and encodes a protein that localizes to the nucleus. Light microscopy analyses have demonstrated that the MS5d gene affects microsporocyte meiosis in the thermosensitive genic male sterility line TE5A. Sequence comparisons and genetic complementation revealed a C-to-T transition in MS5d, encoding a Leu-to-Phe (L281F) substitution and causing abnormal male meiosis in TE5A. These findings suggest arrested meiotic chromosome dynamics at pachytene. Furthermore, immunofluorescence analyses showed that double-strand break (DSB) formation and axial elements were normal but that DSB repair and spindle behavior were aberrant in TE5A meiocytes. Collectively, our results indicate that MS5d likely encodes a protein required for chromosomal DSB repair at early stages of meiosis in B. napus.
Introduction
Meiosis is a highly conserved developmental process that is essential for eukaryotic sexual reproduction and produces haploid cells for progeny formation. In flowering plants, microsporocytes undergo meiosis to produce microspores in the anther, and megasporocytes undergo meiosis to produce megaspores in the ovary. During anther development, microsporocytes initiate meiosis as they develop from primary sporogenous cells. During meiosis, two successive rounds of chromosomal segregation (meiosis I and meiosis II) occur after a single round of DNA replication, without an intervening S-phase. Meiosis I is a reductional division in which homologous chromosomes are separated from each other. Meiosis II is an equational division that results in segregation of sister chromatids (Ma, 2005). Both meiosis I and meiosis II are divided into prophase, metaphase, anaphase, and telophase.
Meiotic prophase I is a unique and highly organized process that includes sister chromatid cohesion (SCC), homologous chromosomal alignment, pairing, synapsis, and recombination (Hamant et al., 2006). Highly organized chromosomes and correct chromosomal architecture are critical to meiosis and sexual reproduction (Kleckner, 1996; Zickler and Kleckner, 1999). Previous studies in many organisms have suggested that the successful completion of homologous chromosome pairing is dependent upon the progression of meiotic recombination, which is universally initiated by the generation of double-strand breaks (DSBs; Franklin et al., 1999; Li et al., 2004; Pawlowski et al., 2004; Ronceret et al., 2009). In addition, strong and close connections between homologous chromosomal pairs is stabilized by a network of longitudinal and transverse protein fibers, which are termed the synaptonemal complex (SC; Moens, 1969; Westergaard and Wettstein, 1972; Gillies, 1975). The search for sequence homology prior to SC formation has been proposed to be facilitated by the presynaptic alignment of homologous chromosomes in many organisms (Zickler and Kleckner, 1999). A number of spontaneous or induced synapsis mutants have been reported in rice (Kitada and Omura, 1983; Kitada et al., 1983; Nonomura et al., 2004a,b; Yuan et al., 2009) and Arabidopsis (Caryl et al., 2000; Armstrong et al., 2002). Mutations in SC genes typically cause defects in the homologous pairing, synapsis, and univalent formation processes. Mutations in genes that are involved in meiotic chromosome function often show defects in SCC and chromosome pairing (Lam et al., 2005; Hamant et al., 2006), DSB formation and/or repair (Lohmiller et al., 2008), and chromosome condensation, finally resulting in abnormal homologous chromosome segregation (Cai et al., 2003; Houben et al., 2005; Andreuzza et al., 2015). For example, Arabidopsis plants transformed with an RNAi construct targeting AtZYP1 exhibit delayed meiosis and an absence of pairing and synapsis in most meiocytes (Higgins et al., 2005). In Arabidopsis, loss of SPO11-1 and/or SPO11-2 (conserved type-II topoisomerase-like enzyme SPO11) caused defective meiotic DSBs, homologous pairing and synapsis (Grelon et al., 2001).
Rapeseed, a crop belonging to the Brassica genus that is cultivated worldwide, is an important vegetable oil source for human consumption. Male sterility is the most effective and economical pollination control system for utilization of heterosis in rapeseed breeding. Previous studies have described the genetic male gene MS5, which has three different alleles (MS5a, MS5b, and MS5c) that lead to observed differences in fertility in Brassica napus (Lu et al., 2013; Xin et al., 2016). Plants carrying the BnMs5bBnMs5b or BnMs5bBnMs5c genotypes were sterile, whereas plants homozygous for the BnMs5c or BnMs5a allele were fertile (Lu et al., 2013). Relative to the sequence of MS5a, an 8,115-bp Mutator-like transposable element (MULE) insertion was detected in MS5b. Loss of MS5a function had no effect on normal meiotic chromosome configuration at leptotene, but it resulted in abnormal chromosome morphology and cytokinesis during the following stages in MS5bMS5b microspore mother cells (MMCs; Xin et al., 2016). Functional analyses demonstrated that MS5 was essential for homologous pairing in meiosis but not for the initiation of DNA DSBs. Further analysis revealed that different expression levels of MS5 and its allelic variants caused differences in fertility (Xin et al., 2016).
The thermosensitive genic male sterility line TE5A is a spontaneous mutant from the B. napus inbred line TE5 (Zeng et al., 2014). The fertility of TE5A is normal at low temperature, but it shows complete male sterility and partial female sterility at temperatures above 20°C. Previous genetic analysis revealed that the male sterility of TE5A is controlled by a single dominant gene, BntsMs (equivalent to MS5d; Zeng et al., 2014). Using a map-based cloning approach, the MS5d gene was successfully localized to a region containing 24 annotated genes (Zeng et al., 2014). In the current study, MS5d was successfully identified based on the microsynteny between Brassica and Arabidopsis and was confirmed by Agrobacterium tumefaciens-mediated genetic transformation assays. Further analyses demonstrated that the MS5d gene in the TE5A mutant caused defects in DSB repair. Thus, the identification of MS5d will contribute significantly to understanding the molecular machinery of plant meiosis and hence to crop breeding.
Materials and Methods
Plant Materials and Growth Conditions
The B. napus lines TE5A and TE5 were obtained from the Oil Crop Research Institute of the Chinese Academy of Agricultural Sciences; we described TE5A in a previous study (Zeng et al., 2014). Mutant lines were planted in a greenhouse under 25°C day/15°C night temperature conditions. All transgenic plants were grown under similar growth conditions.
Functional Verification of Candidate Genes
To verify the functions of candidate genes, a 3.9-kb candidate genomic fragment containing the entire MS5d coding region along with its 1.2-kb upstream and 0.5-kb downstream regions was amplified using the primer pair ZT2-1-F/R (Supplementary Table S1) with Pfu DNA polymerase (Fermentas) and cloned into the pCAMBIA2300 binary vector (Hajdukiewicz et al., 1994). The construct was confirmed by complete sequencing, introduced into Agrobacterium tumefaciens GV3101 host cells and transformed into calli with the homozygous MS5d genotype, as previously described (Dun et al., 2011). Transformed plants with roots were subsequently transplanted into experimental plots. Mature transgenic plants were assessed for male fertility/sterility. DNA from the transgenic plants was analyzed by PCR using primers targeting the coding sequences of 35S and NPTII present in the vector (primers NPT-F/R and 35S-F/R; Supplementary Table S1). Pollen grains from the anthers of transgenic plants collected before anthesis were stained with 1% iodine/potassium iodide solution (KI/I2). The stained pollen was observed under a microscope.
Histological Analysis
Paraffin and plastic sections were prepared to analyze flower development. Buds from various flower developmental stages (1.0–3.0 mm) were fixed in a solution containing 50% ethanol, 5% glacial acetic acid, and 3.7% formaldehyde for 24 h at room temperature; the tissues were then washed twice in 70% ethanol and stored in 70% ethanol at 4°C. For TEM analysis, fresh anthers from wild-type and MS5d mutant plants at various developmental stages were fixed in 2.5% (w/v) glutaraldehyde in 0.1 M phosphate buffer (pH 7.4). The analytical procedures were performed as previously described by Zhu et al. (2010) and Yi et al. (2010).
RT-PCR and qRT-PCR Analysis of Gene Expression
Total RNA was isolated from various tissues and buds of TE5A and homozygous TE5 plant lines using a plant RNA extraction kit (TIANGEN1) as recommended by the manufacturer. RNA (5 μg) was synthesized to first-strand cDNA using the ReverTra Ace-a-First-Strand cDNA Synthesis Kit (TIANGEN1). The reverse-transcription products from various tissues were then used as templates for RT-PCR and qRT-PCR. qRT-PCR was performed in 96-well optical plates containing SYBR Premix (TIANGEN) on an OPTICON 2 PCR instrument (MJ Research). Actin was used as a control for normalization. The primers RT1-F/R were used for the detection of MS5d transcripts (Supplementary Table S1). All reactions were performed in three independent experiments. To determine the full-length transcript of MS5d, 5’-RACE and 3’-RACE were performed using total RNA from young buds with the SMART RACE cDNA amplification kit (Takara; Supplementary Table S1).
Preparation of Meiotic Chromosome Spreads
Chromosome spreads were prepared from inflorescences that had been fixed in Carnoy’s solution (ethanol:glacial acetic acid, 3:1, v/v). Anthers containing microsporocytes undergoing meiosis were incubated with 3% cytohelicase, 3% pectolyase, and 3% cellulase in citric acid buffer for 90 min at 37°C and were then washed three times in PBS. Squashes were made in 45% acetic acid. The microscope slides were frozen in liquid nitrogen, and the coverslips were then removed. The chromosome spreads on air-dried slides were stained with 4′,6-diamidino-2-phenylindole (DAPI) in an antifade solution (Vector), and images were captured using a DM2500 microscope equipped with a DFC420C digital camera system (Leica).
Protein Localization
The full-length cDNA of MS5d without its termination codon (TGA) was PCR-amplified using the PM-F and PM-R primers (Supplementary Table S1) and inserted into the pM999GFP vector obtained from Dr. Jian Xu (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan, China). A cyan fluorescent protein (CFP) fusion with the chaperone binding protein GHD7 was employed as a nuclear marker protein; this fusion protein was obtained from Qifa Zhang (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University). The fusion constructs were introduced into Arabidopsis protoplasts prepared from whole seedlings via PEG/calcium-mediated transformation (Yi et al., 2010). Fluorescence microscopy was performed using a confocal laser microscope.
In situ Hybridization
Inflorescences undergoing meiosis were fixed in 50% FAA solution (50% ethanol, 5% glacial acetic acid, and 3.7% formaldehyde) for 16 h at 4°C before being washed twice with 70% ethanol. The tissue was dehydrated through a graded ethanol series, transferred into xylene, embedded in paraffin wax and sectioned at a thickness of 8 μm. The MS5d cDNA fragment was amplified with the primers in situ-F/R (Supplementary Table S1) and was then ligated into the linearized TA cloning vector pEASY-T3 (TransGen Biotech). The antisense and sense probes were subsequently transcribed in vitro from the T7 and SP6 promoters using polymerase with digoxigenin RNA labeling reagents (Roche). RNA hybridization and immunological detection of the hybridized probes were performed according to the protocol of Deblock and Debrouwer (1993). Images were captured with a DM2500 microscope using a DFC420C digital camera system (Leica).
Immunofluorescence
For immunofluorescence, B. napus anthers were harvested and fixed in 4% (w/v) paraformaldehyde for 30 min at room temperature. Anthers with microsporocytes at the appropriate meiotic stages were flattened onto poly-L-lysine coated slides. After cellular debris was removed, the slides were blocked in 1 × PBS containing 1% BSA for 60 min and then incubated overnight at 4°C in a moist chamber with different antibodies (monoclonal anti-α-tubulin antibody: Sigma MFCD00145891; anti-γH2AX polyclonal antibody: Trevigen 4418-APC-100; anti-BnASY: self-prepared antibody) diluted 1:200 in 1 × PBS containing 1% BSA. After three washes in 1 × PBS, the slides were incubated with DyLight 488-labeled goat anti-rabbit secondary antibody or DyLight 594-labeled goat anti-rabbit secondary antibody (1:1000 dilution) for 1 h. The chromosome spreads were then counterstained with DAPI in an antifade solution (Vector). For immunofluorescence analyses, primary images were captured using a confocal laser microscope.
Results
Mapping and Isolation of the MS5d Gene
The MS5d gene had been previously localized to a 103-kb region that is highly syntenic to the B. rapa linkage group A8, and 24 putative genes (from Bra018439 to Bra018465) were identified in the target region (Figure 1a) (Zeng et al., 2014). Based on homolog analyses in Arabidopsis, functional annotation of the B. rapa transcripts suggested that six of the 24 B. rapa genes (Bra018457, Bra018451, Bra018449, Bra018446, Bra018443, and Bra018441) were involved in anther development2. In addition, sequence comparisons revealed that three other B. rapa genes (Bra018452, Bra018455, and Bra018456) lacked homologs in the Arabidopsis syntenic region (Figure 1a). Based on the microsynteny of homologs in the B. napus linkage group N8 target region, these nine B. rapa genes were selected as candidate genes for MS5d. Next, we performed comparative sequence analysis of the nine candidate genes in the normal fertile line TE5 and the male sterile line TE5A. Interestingly, BnaA08g25920D (homologous to Bra018456) was the only gene in which a single-nucleotide polymorphism (SNP) was detected between TE5 and TE5A; no sequence differences were found in the other candidate genes (Figure 1a). Thus, we selected BnaA08g25920D as the target candidate gene.
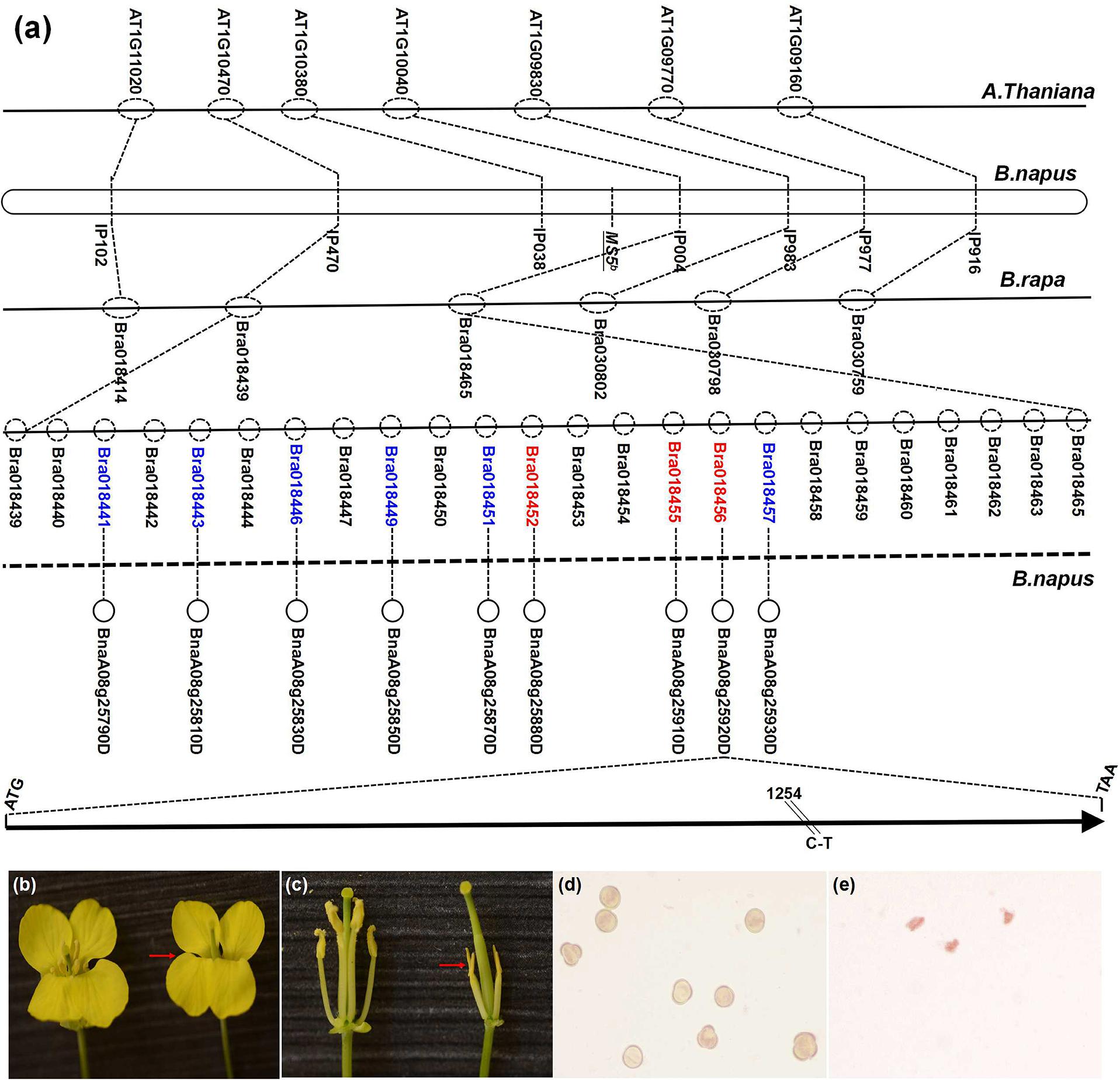
FIGURE 1. Mapping and positional cloning of the MS5d gene and phenotypic complementation testing with the MS5d gene. (a) The dotted circles indicate predicted genes of Brassica rapa; the red type indicates candidate genes that are not present in the corresponding Arabidopsis homologous region. (b,c) Flowers of a fertile non-transgenic TE5 plant (left) and of a sterile T0 transgenic plant in the TE5 background (right, red arrow). (d) Pollen grains of the non-transgenic TE5 plant stained with KI/I2 solution. (e) Pollen grains of the transgenic plant stained with KI/I2 solution.
To validate the candidate gene, functional verification experiments were performed by transforming wild-type B. napus lines with the full genomic fragment containing BnaA08g25920D from TE5A; the fragment contained a 1.2-kb native promoter region, the-1.39 kb MS5d coding region, and a 1.41-kb downstream region. The amplified sequence was cloned into the pCAMBIA2300 binary vector, and the complementation construct was introduced into TE5 and Zhongshuang11 (a de novo-sequenced rapeseed cultivar in China) via Agrobacterium tumefaciens-mediated transformation. Of 34 kanamycin-resistant T0 transgenic TE5 plants, 23 lines exhibited the wild-type fertile flower phenotype, and 11 lines showed a male sterile phenotype at temperatures above 20° (Figures 1b–e). In addition, in the Zhongshuang11 line, three out of nine kanamycin-resistant T0 transgenic plants showed a male sterile phenotype. The recapitulated male sterility of the transgenic TE5 and Zhongshuang11 T1 progeny plants was co-transmitted stably, and it co-segregated with the introduced DNA. These experiments indicated that the male sterile phenotype of TE5A was caused by the SNP in BnaA08g25920D, which is equivalent to MS5d.
MS5d Is Highly Expressed in Microsporocytes and the Tapetum during Anther Development
Various organs of TE5A plants were examined using real-time PCR to reveal the expression profile of the MS5d gene. Figure 2a shows that MS5d is expressed in various organs, including the roots, stems, leaf, pods, and young anthers. Expression of MS5d was detected in the anthers of TE5 and TE5A plants at both the permissive and restrictive temperatures. To further investigate the spatial and temporal patterns of MS5d expression during anther development, RNA in situ hybridization was performed using an MS5d-derived probe with sections of wild-type flower buds at different stages. In stages 5–12, MS5d RNA was clearly detectable in the tapetum, microsporocytes, tetrads, and microspores. Maximal expression was observed in the tapetum and microspores at stage 8 (Figures 2b–e). A sense probe was used as the negative control (Figure 2f). These results suggest that MS5d is highly expressed in microsporocytes during meiosis in the stamen of B. napus.
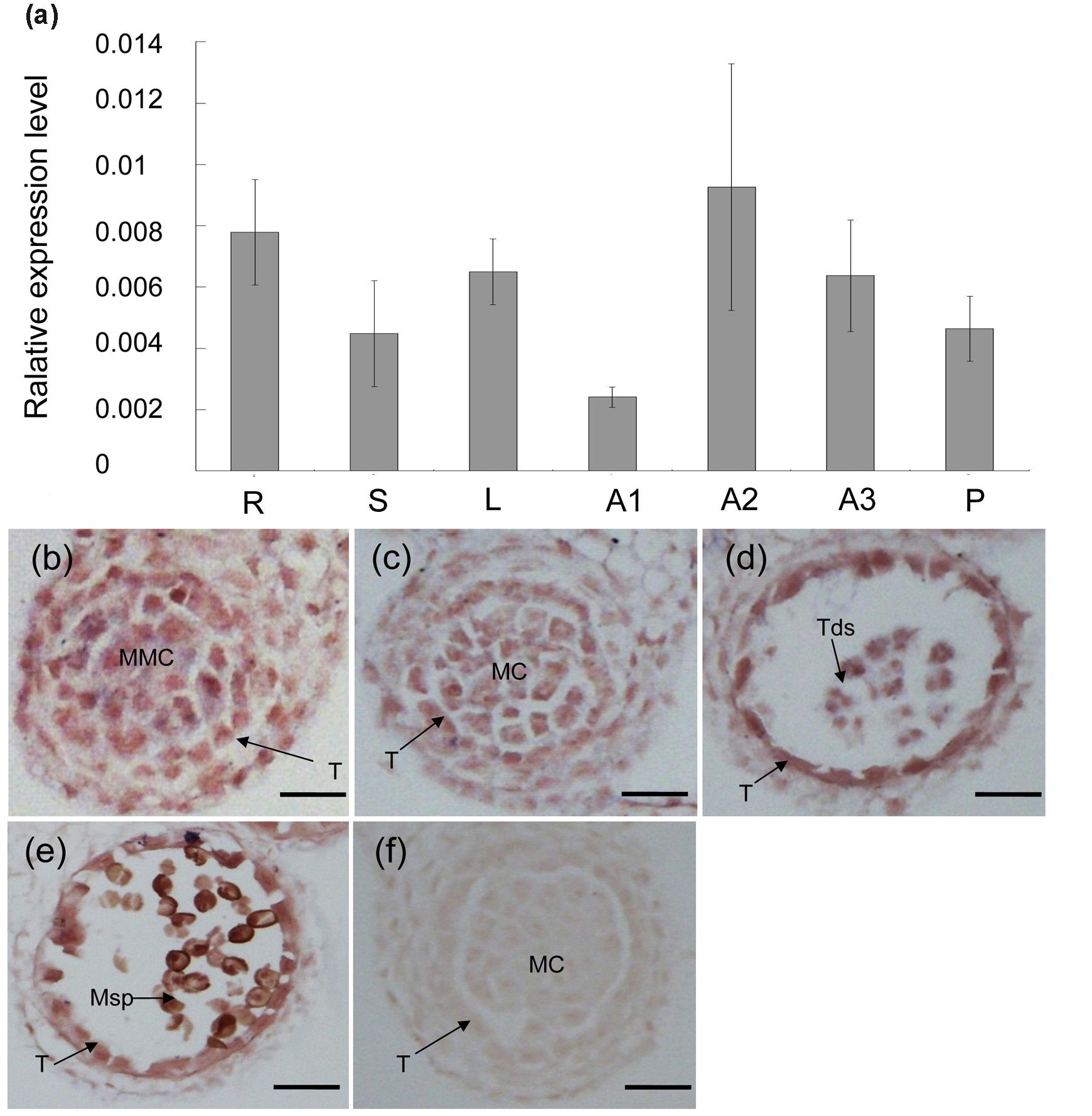
FIGURE 2. Expression analysis of the MS5d gene. (a) Real-time PCR analysis of MS5d in roots (R), leaves (L), stems (S), and young anthers (A) of TE5A. Young anthers of TE5A showed complete male sterility when grown at high temperatures (>20°C) (A1) and complete male fertility when grown at low temperatures (<20°C) (A2). Young anthers of the wild-type (A3) and pods (P) were also analyzed. Actin was used as a control for normalization. The data shown represent the mean ± SE of three independent experiments. (b–f) In situ hybridization assays of MS5d in a longitudinal section at the young anther stage and in transverse sections at the early meiosis stage (b), the meiosis stage (c), the tetrad stage (d), and the pollen mitosis stage (e). (f) Negative control with sense probe in anthers at the meiosis stage. T, tapetum; MMC, microspore mother cell; MC, meiotic cell; Tds, tetrads; Msp, microspore; bars = 25 μm.
MS5d Encodes a Novel Protein Located in the Nucleus
By performing alignments to the B. napus genome database (Chalhoub et al., 20143) and the Brassica database (BRAD4), the full-length coding sequence of MS5d was predicted to comprise 981 bp and to consist of six exons (Figure 3a). This predicted exon-intron structure was experimentally supported by 5’-RACE and 3’-RACE analyses and RT-PCR product sequencing. MS5d encodes a protein containing 326 amino acids with an estimated size of 37.8 kDa. The structure of the MS5d protein was predicted using network protein sequence analysis5, which indicated that it contains six helices, 19 coils, and 13 strands (Figure 3b). The SNP (C-to-T) in the MS5d gene occurred in the sixth exon and caused a substitution of Leu with Phe at position 281 (hereafter referred to as L281F) of the protein (Figure 3b). The substitution was located near the C-terminus of MS5d, and we speculated that the L281F mutation might modify the function of MS5d.
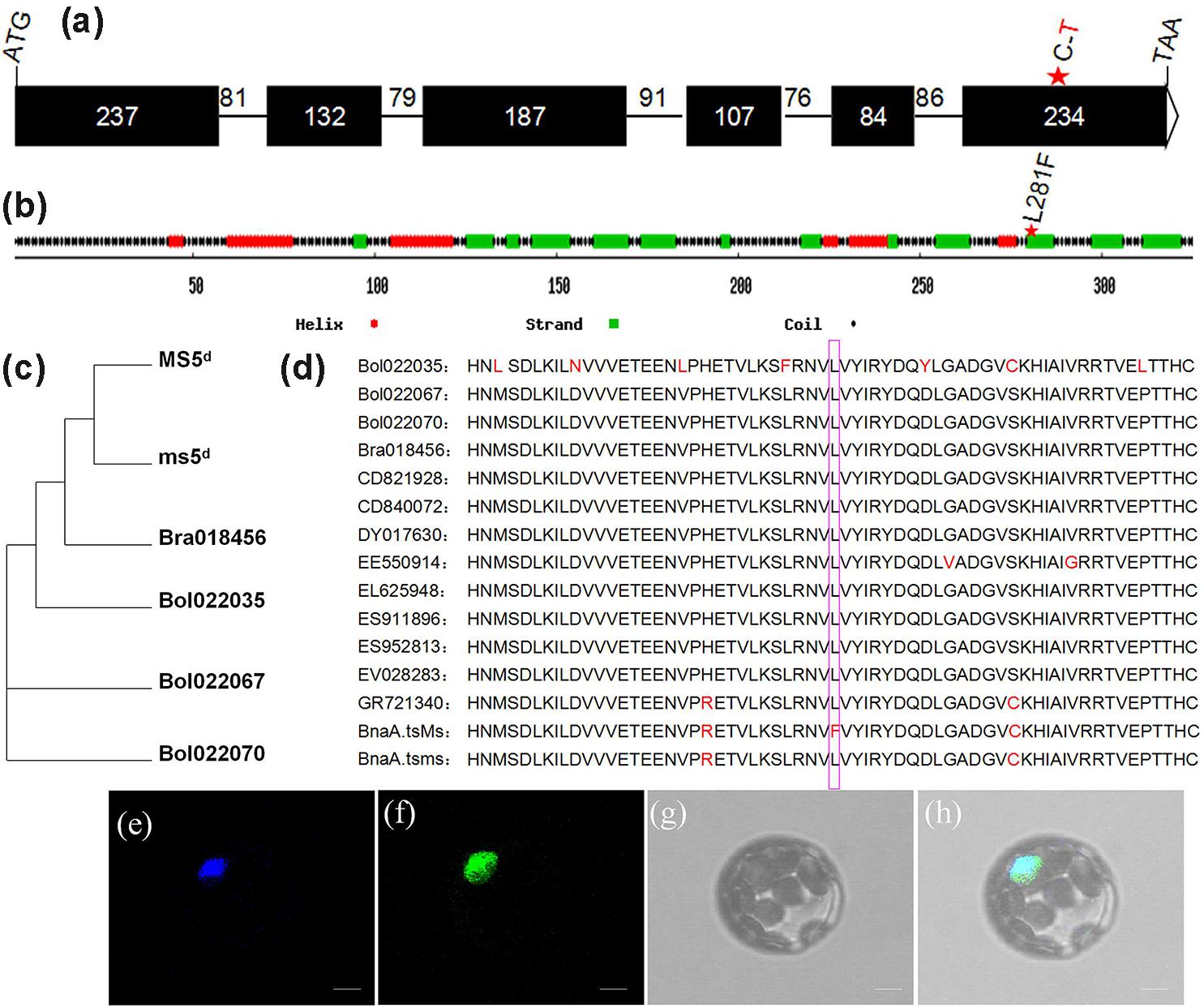
FIGURE 3. Sequence analysis and subcellular localization of the MS5d protein. (a) Exon/intron structure of MS5d, with six exons (black boxes) and five introns; their corresponding sizes are shown. (b) Deduced MS5d protein structure. (c) Phylogenetic analysis between MS5d homologs identified in Brassica oleracea and B. rapa. (d) Deduced amino acid sequences of MS5d homologs from different Brassica species were aligned with MEGA 4.0. The pink box indicates the L281F mutation. (e) The cyan fluorescent protein (CFP) signal was distributed throughout the protoplast cell after transformation with the control CFP construct. (f) The protoplast showed a green fluorescent signal after transformation with the MS5d-GFP fusion construct. (g) The same protoplast as (e) under bright field. (h) The images in (e–g) were merged. Bars = 10 μm in (e–h).
To investigate the subcellular localization of MS5d, we constructed a fusion protein of MS5d with green fluorescent protein (GFP) driven by the 35S promoter. As a positive control, the previously characterized protein GHD7, which is located in the nucleus, was fused to CFP (Xue et al., 2008). The GHD7-CFP and MS5d-GFP fusion constructs were simultaneously introduced into Arabidopsis protoplasts via PEG/calcium-mediated transformation. The GFP signal colocalized with the CFP signal, indicating that MS5d is a nuclear protein (Figures 3e–h).
To understand the origin of MS5d, BLAST was used with the full-length cDNA sequence of ms5d and public databases, including the B. napus genome database (Chalhoub et al., 2014), the Brassica database (BRAD4), the NCBI database6, and the Phytozome database7. The results showed that three copies of the gene are present in B. napus (BnaA08g25920D, BnaC08g14090D, and BnaC08g14440D). In comparison, one copy was found in B. rapa (Bra018456), and two copies were found in B. oleracea (Bol022067 and Bol022070). Interestingly, with the exception of homologs from B. rapa, B. oleracea, and B. napus, ms5d yielded no homologs in other Brassicaceae species. Moreover, ms5d displayed higher sequence identity with Bra018456 (96.5%) than with the other homologs (Figure 3c). Despite the observed sequence divergence, the MS5d L281F substitution was highly conserved in all of the analyzed Brassicaceae species (Figure 3d). Furthermore, a BLASTp search6 of the MS5d protein sequence against public databases also uncovered no proteins with functional similarity in other organisms, except in the Brassica genus. Thus, we propose that the ms5d gene, encoding a novel nucleoprotein, originated from B. rapa after its divergence from Arabidopsis and the Brassica species.
Defective Meiosis in TE5A Microsporocytes
To investigate the anther development defects in the TE5A mutant when grown at high temperatures (>20°C), semi-thin sections of anthers were used to compare the differences between TE5A and wild-type TE5. No obvious differences were detected between the mutant and the wild-type during early pre-meiosis (stages 1–5). The anther primordia of both TE5A and TE5 differentiated to form the characteristic anther structure, with microsporocytes, locules, walls, connective tissues, and vascular regions characteristic of mature anthers (Figures 4a,d). At anther developmental stage 6, both the mutant and wild-type microsporocytes entered normally into meiosis (Figures 4b,e). At stage 7, the tapetal cell cytoplasm in TE5 became condensed, indicating that the tapetal cells were developing into secretory cells. The microsporocytes eventually completed meiosis to form tetrads that had characteristic callose walls (Figure 4c). In the TE5A anther, similarly to wild-type, the tapetal cells transformed normally into secretory cells at stage 7. However, the microsporocytes stalled in meiosis (Figure 4f). From stages 8 to 12 in the wild-type, after the callose wall was dissolved, the microspores were released into the locule. Subsequently, microspores underwent two rounds of mitosis and developed into viable pollen grains (Figures 4g–i). However, in TE5A, the microsporocytes did not undergo mitosis and then disintegrated concomitantly with the tapetum (Figures 4j–l).
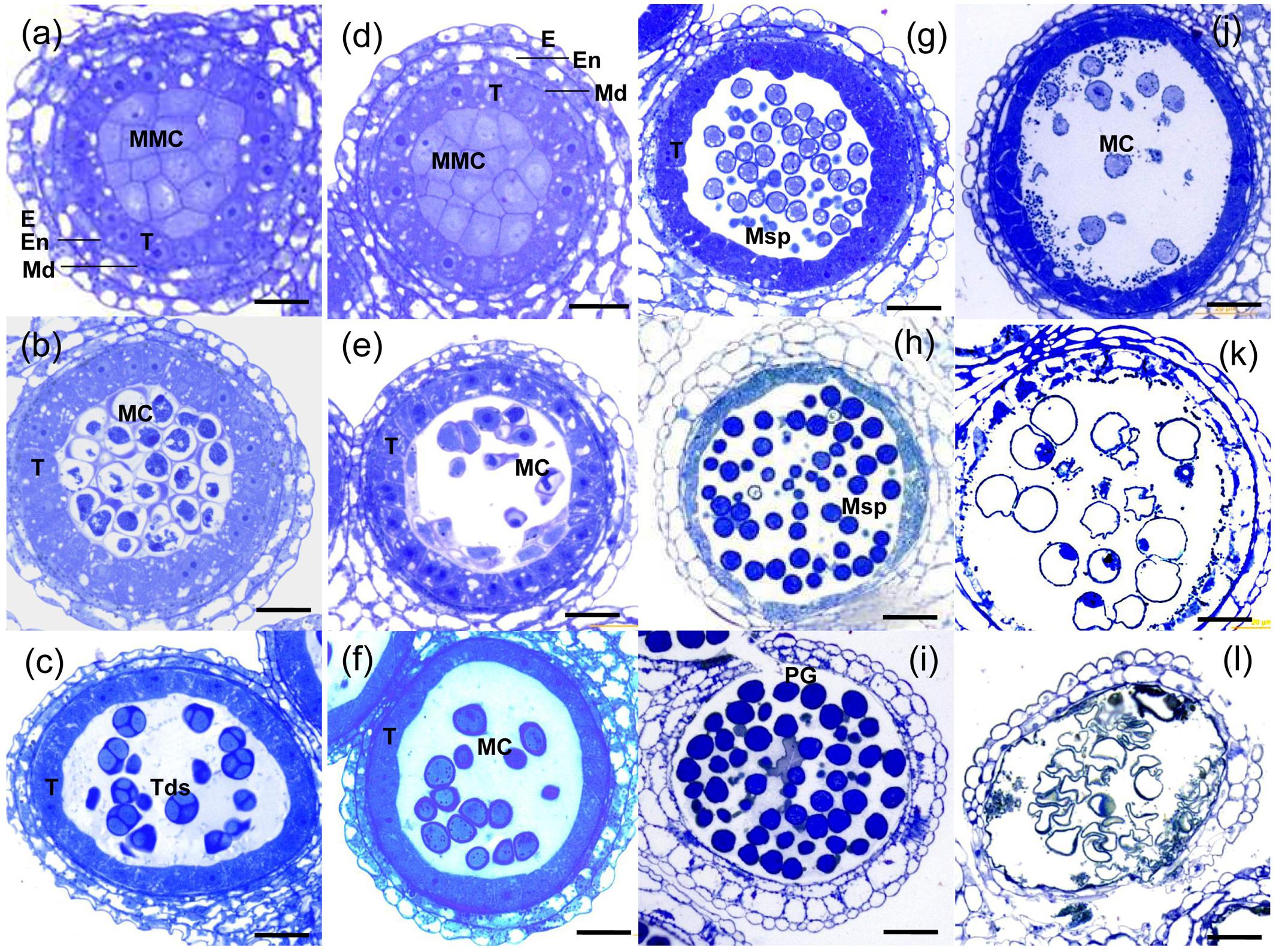
FIGURE 4. Histological analysis of anthers from wild-type and TE5A plants. (a) MMC stage of wild-type; (d) MMC stage of TE5A. (b) Meiosis of wild-type; (e) meiosis of TE5A. (c) Tetrad stage of wild-type; (f) tetrad stage of TE5A. (g) Vacuolated pollen stage of wild-type; (j) vacuolated pollen stage of TE5A. (h) Pollen mitosis stage of wild-type; (k) pollen mitosis stage of TE5A. (i) Mature pollen stage of wild-type; (l) mature pollen stage of TE5A. Arrows indicate the lipid bodies. E, epidermis; En, endothecium; ML, middle layer; T, tapetum; MMC, microspore mother cell; MC, meiotic cell; Tds, tetrads; Msp, microspore; PG pollen grain; Bars = 25 μm.
Next, we investigated the detailed chromosome behavioral defects of TE5A microsporocytes at different meiotic stages using DAPI staining analysis. From leptotene to pachytene, no obvious differences were observed between chromosome spread preparations in microsporocytes from TE5A mutant and wild-type TE5 plants (n = 26; Figures 5a,b,e,f,o,r). At diplotene, the homologous chromosomes of TE5 microsporocytes remained associated through chiasmata following breakdown of synapsis (n = 12; Figure 5c). However, in TE5A, unassociated chromosomes were present, and no condensed chromosomes or chiasmata were observed (n = 18; Figure 5g). At diakinesis, the chromosomes in TE5 microsporocytes condensed to produce very short pairs, and 19 bivalents could be identified (n = 14; Figure 5d). Subsequently, the bivalents aligned on the equatorial plate at metaphase I (n = 21; Figure 5i). Next, the chiasmata were released, and the homologous chromosomes segregated at anaphase I (n = 17; Figures 5j,p). Tetrads were finally produced after completion of the second meiotic division (n = 12; Figures 5k,q). In contrast to the wild-type, these chromosome behaviors were not observed in TE5A. Instead, unassociated and incompact chromosomes were present at diplotene (n = 20; Figure 5g). Crescent-shaped chromatin was present at diakinesis (n = 30; Figure 5h), and the characteristic metaphase I stage was missing. Furthermore, unusual condensed chromatin was observed (n = 25; Figure 5l), and it subsequently became highly condensed to form chromatin bodies at the dyad (Figures 5m,s) and tetrad stages (n = 32; Figures 5n,t) before disintegrating at the microspore stage (n = 16; Figure 4k). Collectively, these results suggest that the defective meiosis in the TE5A mutant is due to obviously abnormal chromosome dynamics that affect subsequent homologous chromosome segregation.
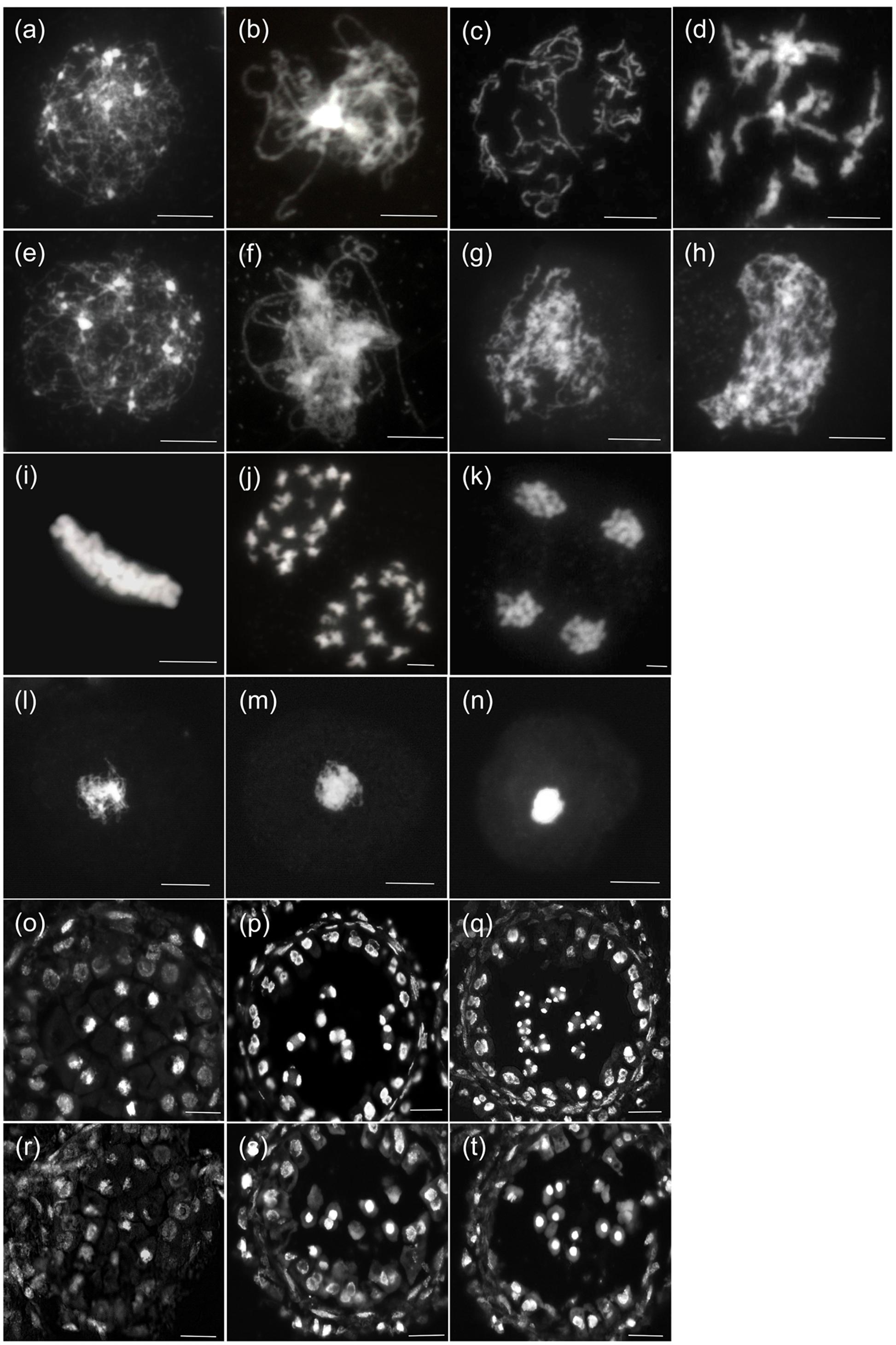
FIGURE 5. Meiosis of wild-type and TE5A microsporocytes. A male meiotic chromosome spread was stained with DAPI for the wild-type (a–d),(i–k),(o–q) and TE5A (e–h),(l–n),(r–t). (a,e) Show leptotene; (b,f,o,r) show pachytene; (c,g) show diplotene; (d,h) show diakinesis; (i,l) show metaphase I; (j,m,p,s) show telophase I; (k,n,q,t) show the tetrad stage. (a–n) Bars = 5 μm. (o–t) Bars = 20 μm.
Meiotic Chromosomal Axial Elements (AEs) Are Not Affected in TE5A
The plant protein ASY1 is localized to the nucleus and associates with chromosomal axes to function as a good marker of early prophase I of meiosis (Caryl et al., 2000; Armstrong et al., 2002; Mikhailova et al., 2006). To investigate whether MS5d affected AEs in TE5A, we performed immunolocalization analysis using a rabbit polyclonal antibody against the BnASY protein. The anti-BnASY antiserum clearly recognized both the recombinant protein and endogenous proteins extracted from the anthers of B. napus at the predicted 66-kDa size of BnASY (Supplementary Figure S1). Chromosomal axes in TE5 microsporocytes were first evident at leptotene (n = 25; Figure 6A), and short stretches of BnASY signals were present at early pachytene (n = 19; Figure 6B). Linear BnASY signals persisted until pachytene before disappearing as the homologs desynapsed (n = 20; Figure 6C). In TE5A, the immunolocalization signals appeared identical to those from leptotene to pachytene in TE5 microsporocytes (n > 15; Figures 6D–E), and the linear BnASY signals persisted until pachytene, when the chromatin started to disintegrate (n = 16; Figure 6F). The consistent distribution of BnASY1 along the chromosomal axes in both TE5A and TE5 microsporocytes indicates that meiotic AEs are not affected by MS5d in microsporocytes during early meiosis.
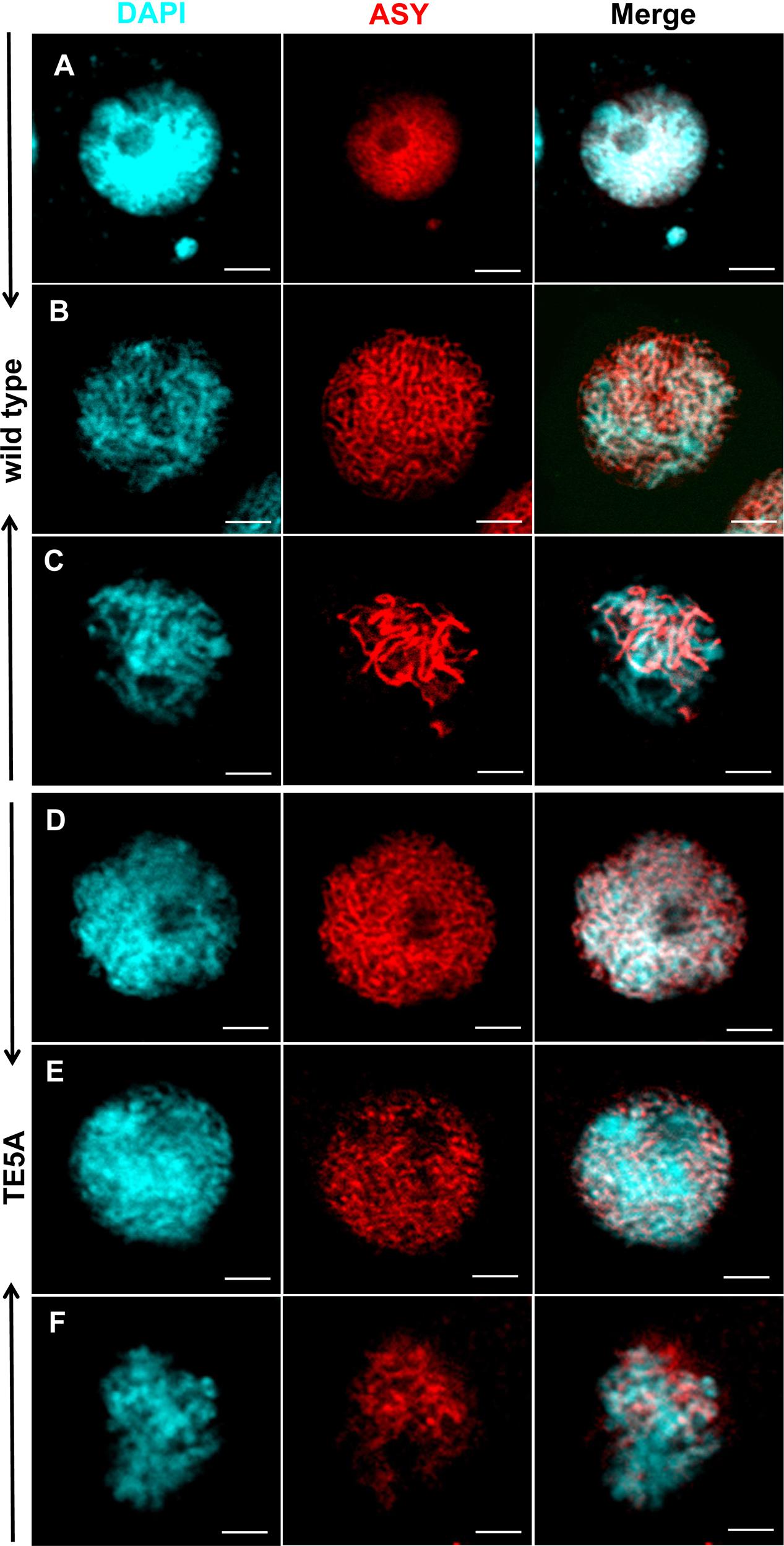
FIGURE 6. Immunolocalization of ASY in wild-type and TE5A cells. Immunolocalization using rabbit polyclonal antibody against ASY (red); chromosomal DNA was counterstained with DAPI (blue). Merged images show the overlap of blue and red fluorescence. (A,D) Show leptotene; (B,E) show early pachytene; (C,F) show pachytene. Bars = 5 μm.
Meiotic DSBs and Microtubule Behavior in TE5A Microsporocytes during Meiosis I
4′,6-diamidino-2-phenylindole staining analysis suggested that the presence of the MS5d allele resulted in the disintegration of chromatin after pachytene in TE5A microsporocytes. We thus sought to determine whether MS5d affects microtubule dynamics and the process of meiotic DSB formation and repair. Previous studies have reported that deficiency in DSB formation is most easily detected by cytological detection of γH2AX rather than by the direct monitoring of DSB in DNA molecules (Sanchez-Moran et al., 2007). Thus, spindle morphogenesis and homologous DSBs were examined in meiosis I using dual immunostaining with anti-α-tubulin and anti-γH2AX antibodies (Figure 7). In TE5 microsporocytes, numerous diffuse γH2AX signals were detected throughout the chromatin at early pachytene (n = 13; Figure 7B), indicating DSB formation. In TE5A microsporocytes, γH2AX signals showed the same chronology and distribution at early pachytene as in TE5 (n = 18; Figure 7G). Thus, DSB formation appeared normal in TE5A microsporocytes. Following prophase I progression, the γH2AX foci disappeared at late pachytene in TE5 microsporocytes, suggesting that the DSBs were repaired normally (n = 17; Figure 7C). However, strong γH2AX signals were observed until the chromatin disintegrated in TE5A cells (n = 19; Figures 7H–J). Our observations indicate that DSB formation occurred normally in TE5A microsporocytes but that DSB repair progression failed at prophase I.
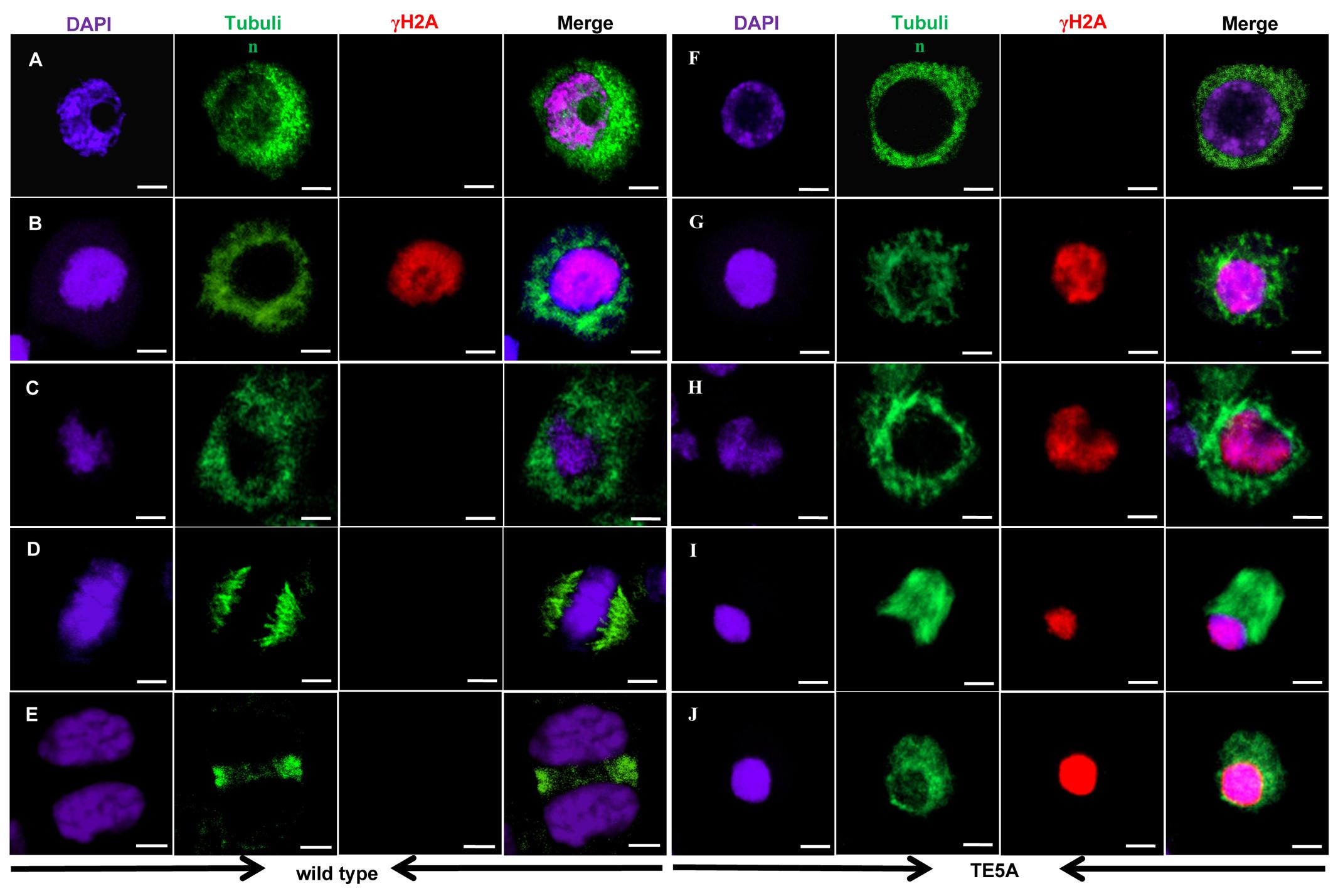
FIGURE 7. Immunolocalization of tubulin and γ-H2AX in wild-type and TE5A cells. Dual immunolocalization using a polyclonal antibody against tubulin (green) and rabbit anti-γ-H2AX antibody (Bethyl Laboratories) (red); chromosomal DNA was counterstained with DAPI (purple). Merged images show the overlap of green, purple, and red fluorescence. (A,F) Show leptotene; (B,G) show early pachytene; (C,H) show pachytene; (D,I) show metaphase I; (E,J) show telophase I. Bars = 10 μm.
Furthermore, immunostaining analyses suggested that, in TE5 microsporocytes, a network of cytoplasmic microtubules was present around the nucleus at prophase I (Figures 7A–C). At metaphase I, the microtubules displayed an obvious bipolar and highly fusiform configuration, with the chromosomes located at the equator (n = 12; Figure 7D). At telophase I, the homologs of each bivalent segregated toward opposite poles, and the interzonal microtubules showed a tight configuration between the recently separated homologous chromosomes (n = 16; Figure 7E). In TE5A, the prophase network of cytoplasmic microtubules in microsporocytes was similar to that in TE5 microsporocytes from leptotene to pachytene (n = 20; Figures 7F–H). However, at metaphase I, the microtubules in TE5A microsporocytes showed a less clear-cut bipolar configuration compared to TE5, and they displayed a diffuse fusiform configuration (n = 29; Figure 7I). In addition, abnormal attachment between the chromosomes and microtubules was observed in TE5A microsporocytes (n = 20; Figure 7I). The microtubules then depolymerized and scattered throughout the nucleus (n = 30; Figure 7J). Collectively, these observations indicate that the attachment between the chromosomes and meiotic microtubules and the movement of microtubules are specifically defective in TE5A microsporocytes.
Discussion
The MS5d Gene Functions in Gametophyte Development
This study identified a Brassica-specific gene, MS5d, and showed that it plays an important role in anther development. Our morphological and histological studies analyzing gametophyte development suggest that the MS5d mutant shows complete male sterility owing to a failure in gametophyte development. Combined with evidence that the MS5d gene is expressed in the anther during male meiosis (Figure 2), we conclude that MS5d functions in male meiosis. Previous studies have reported that male sterility is often associated with defects in tapetum development (Jung et al., 2005; Li et al., 2006). Notably, the PAIR3 gene, which is required for homologous chromosome pairing and synapsis in rice, is highly expressed in the degenerating tapetum. However, there was no obvious difference in the tapetum of the pair3 mutants, indicating that PAIR3 may not function in tapetal degeneration (Yuan et al., 2009). We observed a similar phenomenon in TE5A plants. Although the MS5d gene was highly expressed in the tapetum, there was no obvious difference in tapetum development between TE5A cells (Figure 4) and the wild-type, indicating that MS5d may not function in tapetal development. These results suggest that MS5d has no function in parietal layer cell development.
MS5d Affects Chromosomal DSB Repair in B. napus Microsporocytes
The accurate segregation of homologous chromosomes at the first meiotic division depends on correct chromosomal behaviors, including SCC, homologous chromosomal alignment, pairing, synapsis, and recombination during prophase I of meiosis (Higgins et al., 2008). The formation and repair of homologous DSBs plays important roles both in maintaining genome stability and in generating genetic variability during the process of homologous recombination. The reciprocal exchange of DNA segments occurs between homologs after the repair of deliberately induced DSBs at prophase I and from random homolog segregation at anaphase I. In the present study, compared to the wild-type, TE5A meiotic chromosomes showed a normal configuration at leptotene and pachytene but displayed distinct chromosome morphology during the subsequent stages. Analyses of γH2AX signals suggested that meiosis-specific DSB formation was normal, whereas DSB repair was disrupted in TE5A microsporocytes, thereby causing the abortive segregation of homologous chromosomes. In Arabidopsis, several genes involved in the repair of AtSPO11-induced DSBs have been identified (Ma, 2006; Mercier and Grelon, 2008; Chang et al., 2011). Gene mutations, including atrad50, atmre11, atrad51, atrad51c, and atxrcc3, exhibited similar meiotic chromosome behaviors, such as asynaptic homologs and/or chromosome fragmentation (Bleuyard and White, 2004; Bleuyard et al., 2004; Li et al., 2004, 2005; Puizina et al., 2004). Unlike the defects observed in these mutants, the chromosomes in TE5A microsporocytes exhibited normal chromosomal AEs, as suggested by immunostaining for BnASY. Moreover, the γH2AX signals were relatively strong after late pachytene in TE5A, indicating that DNA damage may be more severe during the process of meiosis in TE5A. Taken together, these data raise the possibility that MS5d controls DSB repair rather than DSB formation in B. napus.
MS5d Affects the Normal Attachment of Microtubules and Chromosomes in Male Meiosis I
Previous studies have shown that microtubules play a key role in telomere-mediated meiotic chromosome dynamics in a wide variety of organisms, including Schizosaccharomyces pombe, budding yeast (Saccharomyces cerevisiae), Caenorhabditis elegans, mice, rye, and wheat (Tepperberg et al., 1997; Ding et al., 1998; Yamamoto et al., 2001; Cowan and Cande, 2002; Cowan et al., 2002; Corredor and Naranjo, 2007; Sato et al., 2009). The separation of homologous chromosomes during anaphase I of meiosis is dependent on their correct association with the spindle. In the present study, microtubules showed abnormal movement and defective chromosomal attachment in TE5A microsporocytes. These results suggest that the MS5d gene most likely affects the organization and movement of microtubules in B. napus. In Arabidopsis, the mps1 atk1 mutant and ATK1atk1/ATK5atk5 double heterozygote have been reported to be defective in chromosome segregation and spindle formation during meiosis (Chen et al., 2002; Quan et al., 2008; Jiang et al., 2009). In these mutants, homologous chromosomes are condensed and associate with the spindle normally but then separate asynchronously at the transition from metaphase to anaphase. However, in TE5A, the remarkable defects in chromosome dynamics included the lack of a characteristic diplotene, as suggested by the lack of chromosome condensation, a diffuse fusiform configuration of microtubules and disrupted attachment between chromosomes and microtubules. These findings indicate that the abortive homologous chromosome segregation in TE5A is significantly different from that in the mps1 atk1 mutant or in the ATK1atk1/ATK5atk5 double heterozygote. On the basis of these observations, we propose that mutation of ms5d affects normal chromosome configuration, resulting in defective attachment between microtubules and chromosomes during B. napus meiosis.
MS5d and B. napus MS5b Display Divergent Mechanisms Despite Sharing the Same Ancestor
Xin et al. (2016) demonstrated that the loss of the MS5a gene resulted in abortive meiosis in the B. napus line Rs1046A; the phenotypic defects in microsporocytes resembled those of TE5A. We found that ms5d and MS5a, both of which originated from Bra018456, exhibit the same sequences. Interestingly, an 8,115-bp MULE insertion into the MS5a gene causes a recessive loss-of-function mutation known as MS5b, whereas an amino acid substitution in the C-terminus of ms5d resulted in a gain of function in the male sterility gene MS5d. These findings suggest that MS5d and MS5b display divergent mechanisms but share a common ancestor. Functional divergence of genes from common ancestors can result in three distinct evolutionary fates: gene silencing with negative mutations, or pseudogenization; partition of ancestral function, or subfunctionalization; and acquisition of new function, or neofunctionalization (Force et al., 1999; Conant and Wolfe, 2008; Liu and Adams, 2010). Thus, we propose that MS5d was derived from the neofunctionalization of its ancestor, whereas MS5b originated from gene silencing with negative mutations of the same ancestor of MS5d (Xin et al., 2016). These findings provide an interesting system in which to study the divergence of functional genes in paleoploid Brassica species.
A loss of function of the MS5a gene caused by an 8,115-bp MULE insertion resulted in abnormal chromosome morphology and cytokinesis after the leptotene stage in MS5bMS5b MMCs (Xin et al., 2016). MS5d, with an amino acid substitution in the C-terminus of ms5d, resulted in aberrant meiotic chromosome dynamics and pachytene arrest. We speculated that these differences might be caused by variations between MS5b and MS5d. The loss of MS5a gene function has been suggested to affect synapsis and homologous pairing in meiosis, but it has no detectable effect on ASY1 loading (Xin et al., 2016). In our study, a similar phenotype was observed by FISH (Yan et al., 2016) and anti-BnASY in TE5A. These results indicate that different mutations of MS5b and MS5d have no effect on chromosomal axis formation in meiosis. Further experiments are required to reveal the effect of MS5d on synapsis of homologs in meiosis. Previous results suggested that MS5 was essential for DSB repair, although it had little effect on DSB formation. In the present study, immunofluorescence experiments showed that MS5d affected DSB repair but was dispensable for DSB formation. These results indicate that different mutations of MS5b and MS5d might have a similar effect on homologous recombination. Collectively, our results provide valuable information for understanding the molecular mechanism of ms5d and the molecular machinery of plant meiosis.
Author Contributions
XZ, XY, RY, and GW designed and supervised the study; YW, FL, and JLuo participated in its design. RY participated in the mapping and transgenetic experiments. XY, KL, and JLi participated in the cytology and histology analysis. XZ and GW wrote the manuscript. All the authors discussed the results and contributed to the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the Natural Science Foundation of Hubei Province (2013CFB423; 2014CFB320), and National Natural Science Foundation of China (31671733).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Professor Qifa Zhang at Huazhong Agricultural University for providing the GHD7 vector, and to Professor Jian Xu at Huazhong Agricultural University for providing the pM999GFP vector.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01966/full#supplementary-material
FIGURE S1 | Western blot with BnaASY polyclonal antibodies.
Footnotes
- ^ http://www.tiangen.com/
- ^ http://www.genoscope.cns.fr/brassicanapus/data/
- ^ http://www.genoscope.cns.fr/brassicanapus/data/
- ^ http://brassicadb.org/brad/
- ^ http://zhanglab.ccmb.med.umich.edu/
- ^ http://www.ncbi.nlm.nih.gov/
- ^ http://www.phytozome.net/
References
Andreuzza, S., Nishal, B., Singh, A., and Siddiqi, I. (2015). The chromatin protein DUET MMD1 controls expression of the meiotic gene TDM1 during male meiosis in Arabidopsis. PLoS Genet. 11:e1005396. doi: 10.1371/journal.pgen.1005396
Armstrong, S. J., Caryl, A. P., Jones, G. H., and Franklin, F. C. (2002). Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115, 3645–3655. doi: 10.1242/jcs.00048
Bleuyard, J. Y., Gallego, M. E., and White, C. I. (2004). Meiotic defects in the Arabidopsis rad50 mutant point to conservation of the MRX complex function in early stages of meiotic recombination. Chromosoma 113, 197–203. doi: 10.1007/s00412-004-0309-1
Bleuyard, J. Y., and White, C. I. (2004). The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J. 23, 439–449. doi: 10.1038/sj.emboj.7600055
Cai, X., Dong, F. G., Edelmann, R. E., and Makaroff, C. A. (2003). The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J. Cell Sci. 116, 2999–3007. doi: 10.1242/jcs.00601
Caryl, A. P., Armstrong, S. J., Jones, G. H., and Franklin, F. C. (2000). A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109, 62–71. doi: 10.1007/s004120050413
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chang, F., Wang, Y., Wang, S., and Ma, H. (2011). Molecular control of microsporogenesis in Arabidopsis. Curr. Opin. Plant Biol. 2011, 66–73. doi: 10.1016/j.pbi.2010.11.001
Chen, C. B., Adam, M., Li, W. X., Hu, Y., and Vielle-Calzada, J. P. (2002). The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development 129, 2401–2409.
Conant, G. C., and Wolfe, K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi: 10.1038/nrg2482
Corredor, E., and Naranjo, T. (2007). Effect of colchicine and telocentric chromosome conformation on centromere and telomere dynamics at meiotic prophase I in wheat-rye additions. Chromosome Res. 15, 231–245. doi: 10.1007/s10577-006-1117-7
Cowan, C. R., and Cande, W. Z. (2002). Meiotic telomere clustering is inhibited by colchicine but does not require cytoplasmic microtubules. J. Cell Sci. 115, 3747–3756. doi: 10.1242/jcs.00055
Cowan, C. R., Carlton, P. M., and Cande, W. Z. (2002). Reorganization and polarization of the meiotic bouquetstage cell can be uncoupled from telomere clustering. J. Cell Sci. 115, 3757–3766. doi: 10.1242/jcs.00054
Deblock, M., and Debrouwer, D. (1993). RNA-RNA in situ hybridization using digoxigenin-labeled probes: the use of high-molecular-weight polyvinyl alcohol in the alkaline phosphatase indoxyl-nitroblue tetrazolium reaction. Anal. Biochem. 215, 86–89. doi: 10.1006/abio.1993.1558
Ding, D. Q., Chikashige, Y., Haraguchi, T., and Hiraoka, Y. (1998). Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci. 111, 701–712.
Dun, X. L., Zhou, Z. F., Xia, S. Q., Wen, J., Yi, B., Shen, J. X., et al. (2011). BnaC.Tic40, a plastid inner membrane translocon originating from Brassica oleracea, is essential for tapetal function and microspore development in Brassica napus. Plant J. 68, 532–545. doi: 10.1111/j.1365-313X.2011.04708.x
Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545.
Franklin, A. E., McElver, J., Sunjevaric, I., Rothstein, R., Bowen, B., and Cande, W. Z. (1999). Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell 11, 809–824. doi: 10.1105/tpc.11.5.809
Gillies, C. B. (1975). Synaptonemal complex and chromosome structure. Annu. Rev. Genet. 9, 91–109. doi: 10.1146/annurev.ge.09.120175.000515
Grelon, M., Vezon, D., Gendrot, G., and Pelletier, G. (2001). AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20, 589–600. doi: 10.1093/emboj/20.3.589
Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. doi: 10.1007/BF00014672
Hamant, O., Ma, H., and Cande, W. Z. (2006). Genetics of meiosis prophase I in plants. Annu. Rev. Plant Biol. 57, 267–302. doi: 10.1146/annurev.arplant.57.032905.105255
Higgins, J. D., Sanchez-Moran, E., Armstrong, S. J., Jones, G. H., and Frankin, F. C. (2005). The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19, 2488–2500. doi: 10.1101/gad.354705
Higgins, J. D., Vignard, J., Mercier, R., Pugh, A. G., Franklin, F. C. H., and Jones, G. H. (2008). AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 55, 28–39. doi: 10.1111/j.1365-313X.2008.03470.x
Houben, A., Demidov, D., Rutten, T., and Scheidtmann, K. H. (2005). Novel phosphorylation of histone H3 at threonine 11 that temporally correlates with condensation of mitotic and meiotic chromosomes in plant cells. Cytogenet. Genome Res 109, 148–155. doi: 10.1159/000082394
Jiang, H., Wang, F., Wu, Y., Zhou, X., Huang, X., Zhu, J., et al. (2009). MULTIPOLAR SPINDLE 1 (MPS1), a novel coiled-coil protein of Arabidopsis thaliana, is required for meiotic spindle organization. Plant J. 59, 1001–1010. doi: 10.1111/j.1365-313X.2009.03929.x
Jung, K. H., Han, M. J., Lee, Y. S., Kim, Y. W., Hwang, I., Kim, M. J., et al. (2005). Rice undeveloped tapetum1 is a major regulator of early tapetum development. Plant Cell 17, 2705–2722. doi: 10.1105/tpc.105.034090
Kitada, K., Kurata, N., Satoh, H., and Omura, T. (1983). Genetic control of meiosis in rice, Oryza sativa L. I. Classification of meiotic mutants induced by MNU and their cytogenetical characteristics. Jpn. J. Genet. 58, 231–240. doi: 10.1266/jjg.58.231
Kitada, K., and Omura, T. (1983). Genetic control of meiosis in rice Oryza sativa L. II. Cytogenetical analyses of desynaptic mutants. Jpn. J. Genet. 58, 567–577. doi: 10.1266/jjg.58.567
Kleckner, N. (1996). Meiosis: how could it work? Proc. Natl. Acad. Sci. U.S.A. 93, 8167–8174. doi: 10.1073/pnas.93.16.8167
Lam, W. S., Yang, X., and Makaroff, C. A. (2005). Characterization of Arabidopsis thaliana SMC1 and SMC3: evidence that AtSMC3 may function beyond chromosome cohesion. J. Cell Sci. 118, 3037–3048. doi: 10.1242/jcs.02443
Li, N., Zhang, D. S., Liu, H. S., Yin, C. S., Li, X. X., Liang, W. Q., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18, 2999–3014. doi: 10.1105/tpc.106.044107
Li, W., Chen, C., Markmann-Mulisch, U., Timofejeva, L., Schmelzer, E., Ma, H., et al. (2004). The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. U.S.A. 101, 10596–10601. doi: 10.1073/pnas.0404110101
Li, W., Yang, X., Lin, Z., Timofejeva, L., Xiao, R., Makaroff, C. A., et al. (2005). The AtRAD51C gene is required for normal meiotic chromosome synapsis and double-stranded break repair in Arabidopsis. Plant Physiol. 138, 965–976. doi: 10.1104/pp.104.058347
Liu, S. L., and Adams, K. L. (2010). Dramatic change in function and expression pattern of a gene duplicated by polyploidy created a paternal effect gene in the Brassicaceae. Mol. Biol. Evol. 27, 2817–2828. doi: 10.1093/molbev/msq169
Lohmiller, L. D., De Muyt, A., Howard, B., Offenberg, H. H., Heyting, C., Grelon, M., et al. (2008). Cytological analysis of MRE11 protein during early meiotic prophase I in Arabidopsis and tomato. Chromosoma 117, 277–288. doi: 10.1007/s00412-007-0147-z
Lu, W., Liu, J., Xin, Q., Wan, L., Hong, D., and Yang, G. (2013). A triallelic genetic male sterility locus in Brassica napus: an integrative strategy for its physical mapping and possible local chromosome evolution around it. Ann. Bot. 111, 305–315. doi: 10.1093/aob/mcs260
Ma, H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56, 393–434. doi: 10.1146/annurev.arplant.55.031903.141717
Ma, H. (2006). “A molecular portrait of Arabidopsis meiosis,” in The Arabidopsis Book, eds C. Somerville, E. Meyerowitz, J. Dangl, and M. Stitt (Rockville, MD: American Society of Plant Biologists), 1–39.
Mercier, R., and Grelon, M. (2008). Meiosis in plants: ten years of gene discovery. Cytogenet. Genome Res. 120, 281–290. doi: 10.1159/000121077
Mikhailova, E. I., Phillips, D., Sosnikhina, S. P., Lovtsyus, A. V., Jones, R. N., and Jenkins, G. (2006). Molecular assembly of meiotic proteins Asy1 and Zyp1 and pairing promiscuity in rye (Secale cereale L.) and its synaptic mutant sy10. Genetics 174, 1247–1258. doi: 10.1534/genetics.106.064105
Moens, P. B. (1969). The fine structure of meiotic chromosome polarization and pairing in Locusta migratoria spermatocytes. Chromosoma 28, 1–25. doi: 10.1007/BF00325986
Nonomura, K. I., Nakano, M., Fukuda, T., Eiguchi, M., Miyao, A., Hirochika, H., et al. (2004a). The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16, 1008–1020. doi: 10.1105/tpc.020701
Nonomura, K. I., Nakano, M., Murata, K., Miyoshi, K., Eiguchi, M., Miyao, A., et al. (2004b). An insertional mutation of rice PAIR2 gene, the ortholog of Arabidopsis ASY1, resulits in a defect in homologous chromosome pairing during meiosis. Mol. Genet. Genomics 271, 121–129. doi: 10.1007/s00438-003-0934-z
Pawlowski, W. P., Golubovskaya, I. N., Timofejeva, L., Meeley, R. B., Sheridan, W. F., and Cande, W. Z. (2004). Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science 303, 89–92. doi: 10.1126/science.1091110
Puizina, J., Siroky, J., Mokros, P., Schweizer, D., and Riha, K. (2004). Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16, 1968–1978. doi: 10.1105/tpc.104.022749
Quan, L., Xiao, R., Li, W., Oh, S. A., Kong, H., Ambrose, J. C., et al. (2008). Functional divergence of the duplicated AtKIN14a andAtKIN14b genes: critical roles in Arabidopsis meiosis and gametophyte development. Plant J. 53, 1013–1026. doi: 10.1111/j.1365-313X.2007.03391.x
Ronceret, A., Doutriaux, M. P., Golubovskaya, I. N., and Pawlowski, W. P. (2009). PHS1 regulates meiotic recombination and homologous chromosome pairing by controlling the transport of RAD50 to the nucleus. Proc. Natl. Acad. Sci. U.S.A. 106, 20121–20126. doi: 10.1073/pnas.0906273106
Sanchez-Moran, E., Santos, J. L., Jones, G. H., and Franklin, F. C. (2007). ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 21, 2220–2233. doi: 10.1101/gad.439007
Sato, A., Isaac, B., Phillips, C. M., Rillo, R., Carlton, P. M., Wynne, D. J., et al. (2009). Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139, 907–919. doi: 10.1016/j.cell.2009.10.039
Tepperberg, J. H., Moses, M. J., and Nath, J. (1997). Colchicine effects on meiosis in the male mouse. I. Meiotic prophase: synaptic arrest, univalents, loss of damaged spermatocytes and a possible checkpoint at pachytene. Chromosoma 106, 183–192. doi: 10.1007/s004120050238
Westergaard, M., and Wettstein, D. V. (1972). The synaptonemal complex. Annu. Rev. Genet. 6, 71–110. doi: 10.1146/annurev.ge.06.120172.000443
Xin, Q., Shen, Y., Li, X., Lu, W., Wang, X., Han, X., et al. (2016). MS5 mediates early meiotic progression and its natural variants may have applications for hybrid production in Brassica napus. Plant Cell 28, 1263–1278. doi: 10.1105/tpc.15.01018
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yamamoto, A., Tsutsumi, C., Kojima, H., Oiwa, K., and Hiraoka, Y. (2001). Dynamic behavior of microtubules during dynein-dependent nuclear migrations of meiotic prophase in fission yeast. Mol. Biol. Cell 12, 3933–3946. doi: 10.1091/mbc.12.12.3933
Yan, X., Zeng, X., Wang, S., Li, K., Yuan, R., Gao, H., et al. (2016). Aberrant meiotic prophase I leads to genic male sterility in the novel TE5A mutant of Brassica napus. Sci. Rep. 6:33955. doi: 10.1038/srep33955
Yi, B., Zheng, F. Q., Lei, S. L., Chen, Y. N., Yao, X. Q., Wen, J., et al. (2010). Two duplicate CYP704B1 homologous genes BnMs1 and BnMs2 are required for pollen exine formation and tapetal development in Brassica napus. Plant J. 63, 925–938. doi: 10.1111/j.1365-313X.2010.04289.x
Yuan, W., Li, X., Chang, Y., Wen, R., Chen, G., Zhang, Q., et al. (2009). Mutation of the rice gene PAIR3 results in lack of bivalent formation in meiosis. Plant J. 59, 303–315. doi: 10.1111/j.1365-313X.2009.03870.x
Zeng, X. H., Li, W. P., Wu, Y. H., Liu, F., Luo, J. L., Cao, Y. L., et al. (2014). Fine mapping of a dominant thermo-sensitive genic male sterility gene (BntsMs) in rapeseed (Brassica napus) with AFLP-and Brassica rapa-derived PCR markers. Theor. Appl. Genet. 127, 1733–1740. doi: 10.1007/s00122-014-2335-6
Zhu, Y., Dun, X. L., Zhou, Z. F., Xia, S. Q., Yi, B., Wen, J., et al. (2010). A separation defect of tapetum cells and microspore mother cells results in male sterility in Brassica napus: the role of abscisic acid in early anther development. Plant Mol. Biol. 72, 111–123. doi: 10.1007/s11103-009-9556-0
Keywords: Brassica napus, male sterility, meiosis, microtubule, double-strand breaks
Citation: Zeng X, Yan X, Yuan R, Li K, Wu Y, Liu F, Luo J, Li J and Wu G (2017) Identification and Analysis of MS5d: A Gene That Affects Double-Strand Break (DSB) Repair during Meiosis I in Brassica napus Microsporocytes. Front. Plant Sci. 7:1966. doi: 10.3389/fpls.2016.01966
Received: 07 September 2016; Accepted: 12 December 2016;
Published: 04 January 2017.
Edited by:
Changbin Chen, University of Minnesota, USACopyright © 2017 Zeng, Yan, Yuan, Li, Wu, Liu, Luo, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Wu, d3VnYW5nQGNhYXMuY24=
 Xinhua Zeng
Xinhua Zeng Gang Wu
Gang Wu