- 1Centro de Citricultura y Producción Vegetal, Instituto Valenciano de Investigaciones Agrarias, Valencia, Spain
- 2CIRAD, UMR AGAP, Petit-Bourg, France
Alternaria brown spot (ABS) is a serious disease affecting susceptible citrus genotypes, which is a strong concern regarding citrus breeding programs. Resistance is conferred by a recessive locus (ABSr) previously located by our group within a 3.3 Mb genome region near the centromere in chromosome III. This work addresses fine-linkage mapping of this region for identifying candidate resistance genes and develops new molecular markers for ABS-resistance effective marker-assisted selection (MAS). Markers closely linked to ABSr locus were used for fine mapping using a 268-segregating diploid progeny derived from a heterozygous susceptible × resistant cross. Fine mapping limited the genomic region containing the ABSr resistance gene to 366 kb, flanked by markers at 0.4 and 0.7 cM. This region contains nine genes related to pathogen resistance. Among them, eight are resistance (R) gene homologs, with two of them harboring a serine/threonine protein kinase domain. These two genes along with a gene encoding a S-adenosyl-L-methionine-dependent-methyltransferase protein, should be considered as strong candidates for ABS-resistance. Moreover, the closest SNP was genotyped in 40 citrus varieties, revealing very high association with the resistant/susceptible phenotype. This new marker is currently used in our citrus breeding program for ABS-resistant parent and cultivar selection, at diploid, triploid and tetraploid level.
Introduction
Alternaria brown spot (ABS) is a fungal disease caused by the tangerine pathotype of Alternaria alternata (Fr.) Keissl., that induces necrotic lesions on fruits and young leaves, defoliation and fruit drop in susceptible citrus genotypes (Akimitsu et al., 2003). The disease was first observed in Australia in 1903 on “Emperor” mandarin (Pegg, 1966) and was subsequently detected in citrus-growing regions all over the world (Vicent et al., 2000; Timmer et al., 2003; Golmohammadi et al., 2006; Wang et al., 2010). Currently, ABS control is primarily based on fungicide application. Depending on the climate of the region and the susceptibility of the cultivar, between four and ten fungicide sprays per year are needed to produce quality fruit for the fresh market. Even with this large number of sprays, damage reduction is not always satisfactory. These constraints force growers to remove susceptible cultivars such as “Fortune” or “Nova” mandarins (Cuenca et al., 2013). Thus, genetic resistance remains as the best option for disease control (Bhatia et al., 2003; Peres and Timmer, 2006; Vicent et al., 2007).
The tangerine pathotype of A. alternata is a necrotrophic pathogen, and carries a gene cluster (ACTT) located in a small chromosome responsible for ACT-toxin biosynthesis (Ajiro et al., 2010). This host-specific toxin is released during the germination of conidia, rapidly affecting the plasma membrane integrity of susceptible host cells (Kohmoto et al., 1993). There is also indirect evidence suggesting the presence of toxin receptors in susceptible citrus genotypes (Tsuge et al., 2012).
Due to constraints of the citrus reproductive system, such as polyembryony in most mandarins, specific characteristics or capacity to produce triploid hybrids (Aleza et al., 2010b), several ABS-susceptible cultivars are being used as parents in many mandarin breeding programs, both at diploid and triploid level (McCollum, 2008; Aleza et al., 2010a,b; Cuenca et al., 2010; Grosser et al., 2010; Aleza et al., 2012). This is the case of the monoembryonic cultivar “Fortune,” widely used as female parent in the triploid breeding programs in Spain (Navarro et al., 2012), France and Italy (Recupero et al., 2005; Froelicher et al., 2012); the susceptible cultivars “Murcott” and “Ponkan” are widely used as male parents in many diploid breeding programs carried out in Japan and Brazil (JinPing et al., 2009; Schinor et al., 2012). Also, this is the case for “Dancy,” “Minneola,” “Nova,” “Fairchild,” “Fremont,” “Page,” “Orlando,” “Pixie,” and “Daisy,” which are also used as male parents or for induced mutations in diploid and triploid breeding programs in Spain (Navarro et al., 2012) and USA (Williams, 2012). On the other hand, previous studies stated that the inheritance of ABS resistance in citrus is controlled by a single recessive allele (Dalkilic et al., 2005; Gulsen et al., 2010; Cuenca et al., 2013). Therefore, segregation is expected in progeny arising from crosses between resistant and heterozygous ABS-susceptible parents or even between two heterozygous ABS-susceptible ones.
Genetic mapping of agronomical traits is of principal importance in breeding programs, and its usefulness has been widely demonstrated for marker-assisted selection (MAS) and for candidate gene identification and cloning (Dirlewanger et al., 2004; MacKay and Powell, 2007; Sorkheh et al., 2008). In addition, defining the exact chromosomal position for genes of interest, i.e., fine-mapping, is critical for improving the effectiveness of MAS, since the smaller the distance from the gene controlling the trait, the more accurate will be the selection (Lörz and Wenzel, 2008).
Genetic mapping can be achieved mainly by linkage mapping and/or association mapping. Linkage mapping, in which a progeny segregating for the trait of interest is analyzed, has been a useful approach to map many agronomical traits in several crops (Würschum, 2012), including the mapping of resistance genes to fungal pathogens. In contrast, association mapping studies a collection of genotypes with a broader genetic variation in a more representative genetic background than in linkage-mapping (Ingvarsson and Street, 2011; Khan and Korban, 2012).
In a recent study (Cuenca et al., 2013), we located a region containing the so-called ABSr locus, near the centromere on chromosome III using bulked-segregant and half-tetrad analyses from triploid populations. The identified region was flanked by a Simple Sequence Repeat (SSR) marker (TTC8) and a Single Nucleotide Polymorphism (SNP) marker (CiC3248-06), found at 3.77 and 1.71 cM from the ABSr locus, respectively, delimiting a 3.3 Mb genome region. Moreover, no recombination was observed between another SSR marker (AT21) and the ABSr locus. This locus appears to be included in a genomic region very rich in disease resistance homologous genes.
In the present study, we have developed new SNP markers to perform fine-linkage mapping of the previously located region. The ABSr locus has now been restricted to 366 kb, containing several candidate genes that may be involved in ABS-resistance. Moreover, a new SNP marker with very little recombination frequency with the phenotype has been tested for a large set of genotypes used as breeding mandarin parents, demonstrating its usefulness in MAS for a wide range of breeding populations.
Materials and Methods
Linkage Mapping Population
A 268-diploid mapping population obtained from a cross between “Fortune” mandarin (C. clementina × C. tangerina; ABS heterozygous susceptible—“Aa”) and a hybrid between clementine and sweet orange –“C × SO-1” (C. clementina × C. sinensis; ABS resistant—“aa”) was used to perform linkage mapping. The cross was made in spring 2011 and seedlings were maintained in the greenhouse during the experiments.
Phenotyping Method for Resistance to Alternaria alternata
The evaluation of ABS response was assessed by in vitro inoculation of young detached leaves from the mapping population, their parental genotypes and controls, following the procedure described by Vicent et al. (2004).
Inoculum Production
A virulent single-spore isolate of A. alternata (IVIA-A005) isolated from an infected “Fortune” mandarin fruit was used for inoculations. Abundant conidia were obtained by a method adapted from Everts and Lacy (1996). The isolate was grown on potato dextrose agar (PDA) plates at 25°C in darkness for 8–10 days and then, illuminated with fluorescent lamps (Philips TLD 18 W/33) at 25°C for 8 h to initiate conidiophore formation, and placed in the dark at 18°C for 12 h. Conidial suspensions were prepared by pouring sterile water over the colonies and gently rubbing the surface with a sterile glass rod. The suspension was filtered through two layers of cheesecloth, and the spore concentration was adjusted to 105 conidia·ml−1 with a haemocytometer. Suspensions with conidial germination lower than 90% were discarded.
Leaf Inoculations
Bioassays were performed immediately after leaf harvest. Young leaves (about 50% developed) were inoculated with 105 conidia·ml−1. This suspension was sprayed over both upper and lower surfaces of each leaflet, using five leaves per genotype. Controls were inoculated by spraying sterile distilled water. Leaves were incubated in a moist chamber in the dark at 27°C. In susceptible genotypes, leaf symptoms appear during the second day after inoculations and very clear necrosis induced by the ACT-toxin can be observed after 48–72 h, when the results were evaluated (Figure 1). A genotype was considered resistant when no symptoms of ABS were observed in any leaf, whereas presence of infection was recorded when a clear symptom of ABS was observed in at least one leaf. The inoculations were repeated when there were doubts regarding interpretation. Experiments inoculating the whole population were carried out twice each spring in 2013, 2014, and 2015.
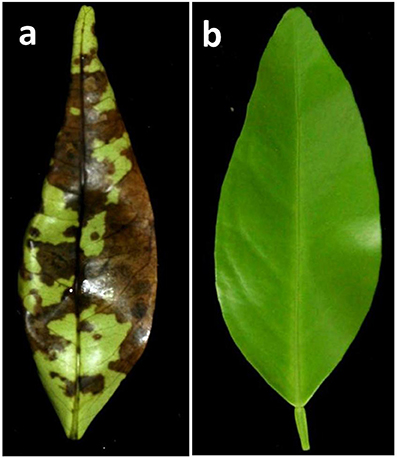
Figure 1. Symptoms observed 48 h after in vitro inoculation of young detached leaves from control genotypes. (A) Susceptible cultivar “Fortune.” (B) Resistant cultivar “CXS0-1.”
DNA Isolation
Genomic DNA of the mapping population and germplasm accessions was isolated using the Plant DNeasy kit from Qiagen, Inc. (Valencia, CA, USA), following the manufacturer's protocol. DNA concentrations were estimated with PicoGreen® and adjusted to 30 ng/μl for further analyses.
Simple Sequence Repeat (SSR) Genotyping
Polymerase chain reactions (PCRs) were performed with wellRED oligonucleotides (Sigma-Aldrich®, St Louis, MO, USA) using the following protocol: Mastercycler ep Gradient S (Eppendorf Scientific Inc., Westbury, NY, USA); reaction volume, 15 μl; 0.8 U Taq polymerase (Fermentas®, Burlington, VT, USA); reaction buffer [750 mMTris-HCl (pH 9), 50 mMKCl, 200 mM (NH4)2SO4, 0.001% bovine serum albumin], 0.1 mM of each dNTP, 5 mM MgCl2, 3 mM of each primer and 30 ng of total DNA. The PCR program was as follows: 94°C for 5 min; 40 cycles of 30 s at 94°C, 1 min at 55°C, and 30 s at 72°C; final elongation 10 min at 72°C. Separation was carried out by capillary gel electrophoresis (CEQ 8000 Genetic Analysis System; Beckman Coulter Inc., Fullerton, CA, USA). Data collection and analysis were carried out using the GenomeLabGeXP10.0 software (Beckman Coulter Inc.). The molecular markers used for SSR genotyping were AT21 and TTC8 (Cuenca et al., 2013).
Fragment Sequencing and SNP Mining
Fragment Sequencing
Twenty DNA 600 bp-fragments spanning the ABSr region were sequenced in “Fortune” and “C × SO-1” genotypes to find polymorphisms that could be heterozygous (Aa) in “Fortune” and homozygous (aa) in “C × SO-1.” Information on location of the corresponding sequences on the haploid clementine reference genome (www.phytozome.org/clementine) and annotation is given in Table 1. In order to obtain more information correlated with the expected polymorphisms in relation with the response to the disease, “Clemenules” clementine (C. clementina) and “Minneola” tangelo (C. paradisi × C. tangerina), which are homozygous resistant and homozygous susceptible, respectively (from available segregation data) were also included for fragment sequencing. Twenty-two diallelic SNPs were found useful (heterozygous in “Fortune,” homozygous in “C × SO-1” and “Clemenules” and the alternative homozygous type in “Minneola”). Nine SNPs, included in 9 sequenced fragments, were selected for subsequent analysis.
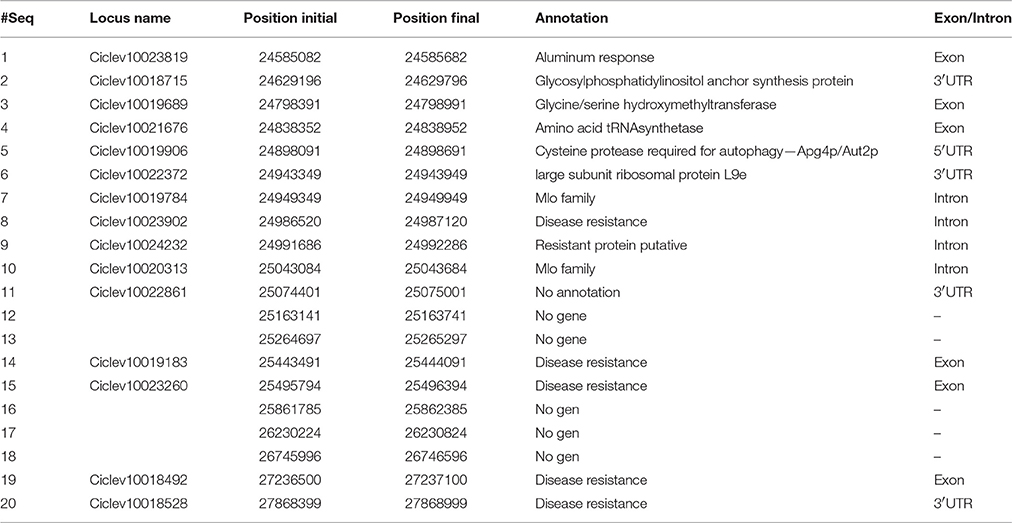
Table 1. Annotations on the haploid clementine's reference genome (http://www.phytozome.net/clementine) corresponding to the sequenced fragments.
SNP Analyses
Polymorphisms found as related to response to the disease were used to develop SNP markers. SNP genotyping was performed by Kbioscience® services, using the KASPar technique. Detailed explanation of specific conditions and reagents can be found in Cuppen (2007).
Linkage Mapping
The mapping population was genotyped with two previously developed SSRs flanking the ABSr locus (Cuenca et al., 2013) and nine new SNP markers developed from the sequences shown in Table 1. Besides, it was phenotyped as described above.
The fine linkage map was constructed using the JoinMap 4.1 software package (Stam, 1993), where markers were coded as lm × ll (i.e., pseudo-test cross), using Kosambi's mapping function. The recombination threshold was of 0.35, and default values were set for all other parameters. Markers were subjected to segregation distortion analysis.
Set of Parental Genotypes to Test the New SNP Marker Selected for MAS
Forty citrus genotypes from the IVIA citrus germplasm bank, either already or candidate to be used as parents in mandarin breeding programs were selected to test the association between the phenotype (resistance to ABS) and the genotype for the closest SNP to the ABS resistance gene (Supplementary Material—Table S1). The set includes 7 citrus species, according to Tanaka's classification (Tanaka, 1977) represented by 12 genotypes and 28 recent hybrids. The selection was done with the aim of spanning the majority of the diversity found within mandarins (Garcia-Lor et al., 2015).
Results
SNP Mining from Sequencing
With the objective to perform fine mapping, useful SNPs were searched in the previously defined 3.3 Mb region spanning the ABSr locus. In a first step, 20 DNA fragments within the ABSr region were Sanger-sequenced in the parents of the analyzed population and two additional genotypes (one resistant and one homozygous susceptible) to find informative polymorphisms potentially correlated with the response to the disease. A total of 9.7 kb corresponding to these sequences were finally obtained. Consensus sequences have been submitted to Genbank (accession numbers from KX368965 to KX368984). Twenty-two SNP were found to have useful polymorphisms (data not shown). An example for SNP08 development from sequencing (fragment #16) is shown in Figure 2. At this position, “Fortune” (heterozygous susceptible parent; Figure 2A) is heterozygous G/T and “C × SO-1” (resistant parent; Figure 2B) is homozygous T/T; additional genotypes are “Clemenules” (resistant, T/T; Figure 2C) and “Minneola” (homozygous susceptible G/G; Figure 2D). Finally, nine SNPs included in nine sequenced fragments spanning the entire region were selected to develop KASPar markers for subsequent analysis (Table 2). The absence of additional polymorphisms close to the selected SNP was a major criteria for this second step of selection.
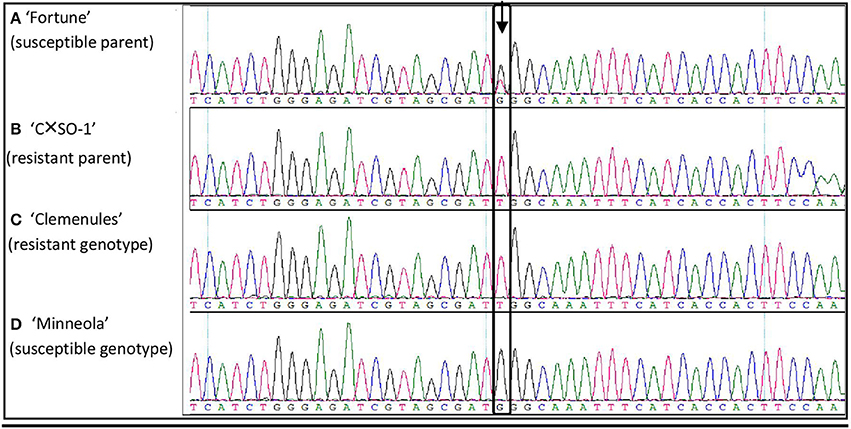
Figure 2. Sequence alignment of four genotypes revealing the SNP08. (A) “Fortune”; (B) “CxS0-1”; (C) “Clemenules”; (D) “Minneola.”
Fine Mapping of the ABSr Locus
The new map was constructed using two existing SSR markers and the nine newly developed SNPs markers. Missing data average was 2.0%, ranging from zero for SNP01, SNP02, SNP04, SNP05, SNP06, SNP07, and AT21 markers; 0.37% for SNP08; 0.75% for AAT9, and SNP03 markers, to 20.15% of missing data for the SNP09, (54 individuals). Resistant/susceptible phenotype segregation was 118 (45%)/150 (55%), showing a slightly distorted segregation toward susceptible individuals (χ2 = 3.821; p = 0.051; n.s.) according to the 1:1 hypothesis. Most of the analyzed markers resulted in similar non-significant bias toward the susceptible genotype. This is a logical observation because all these markers are closely linked. Only the AAT9 (χ2 = 5.429; p = 0.0198) and the SNP01 (χ2 = 4.313; p = 0.0378) showed significant distortion at a probability of 95%. Genotyping results for SNP05, SNP06, SNP07, and AT21 markers are identical for all analyzed individuals, so they were localized at the same position in the map. All markers maintained expected mapping order based on their physical location (www.phytozome.org/clementine; Wu et al., 2014). Figure 3 shows the region where the ABSr locus is located in the new map compared to the data from Cuenca et al. (2013).
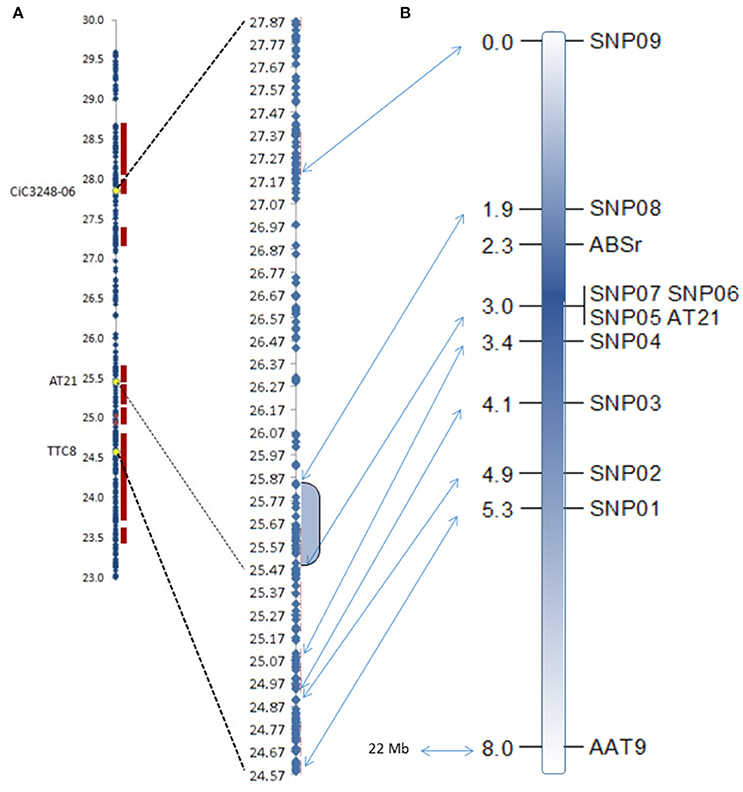
Figure 3. Physical maps of the ABSr locus from Cuenca et al. (2013) (A) compared with the fine genetic mapping of the region from the present study (B). Shadowed area in the physical map corresponds to the identified region flanked by the closest markers. Arrows indicate the physical position of each marker used in the fine mapping. Units are Mbases and centiMorgan in the physical and the genetic maps, respectively.
Fine mapping showed that the ABSr locus is located in a region of 1.1 cM between the markers SNP05/SNP06/SNP07/AT21 (at 0.7 cM) and SNP08 (at 0.4 cM), limiting the chromosome region to 365.991 bp between the positions 25.496.094 and 25.862.085 within the chromosome 3 in the clementine reference genome sequence. Therefore, this analysis allowed us to narrow the ABSr locus to a region containing a much more limited number of candidate genes (Table 3).
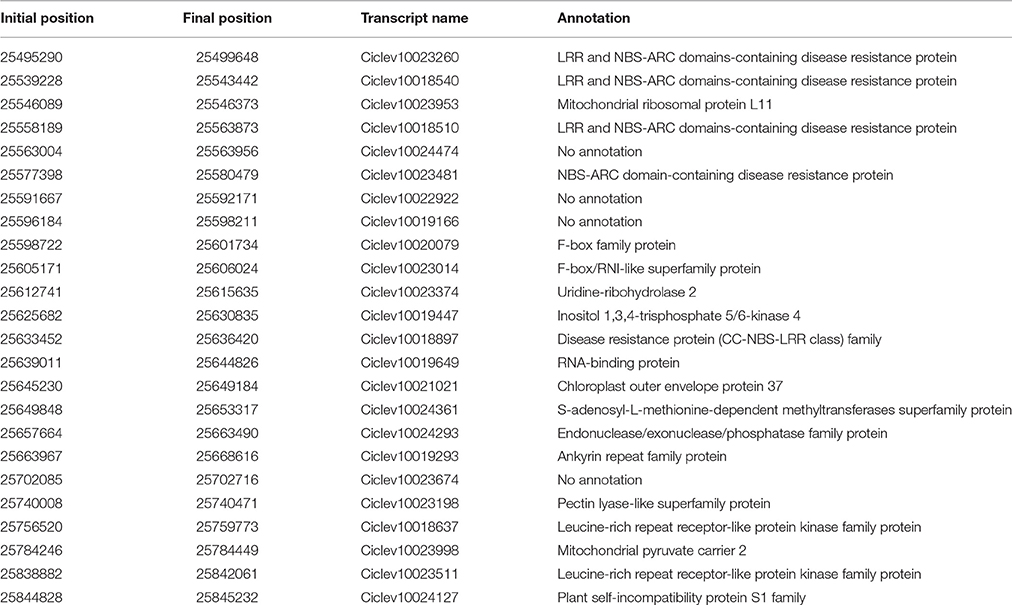
Table 3. Annotated genes in the region of interest, indicating their physical position and annotation.
Identification of Candidate Genes for ABS Resistance
Twenty-four genes are predicted in the clementine genome (www.phytozome.org/clementine) within the region delimited by fine-mapping (Table 3; Figure 4). Twenty of these genes have a functional annotation, and eight of them may be related to pathogen resistance. They include seven resistance gene homologs, harboring Leucine-Rich-Repeat (LRR) and Nucleotide-Binding-Site (NBS) domains (Ciclev10023260, Ciclev10018540, Ciclev10018510, Ciclev10023481, Ciclev10018897, Ciclev10018637, and Ciclev10023511). Among these resistance genes, Ciclev10018637 and Ciclev10023511 present LRR receptor-like and serine/threonine protein kinase domains. Another gen (Ciclev10024361) related to pathogen resistance encodes a S-adenosyl-L-methionine-dependent methyltransferases superfamily protein.
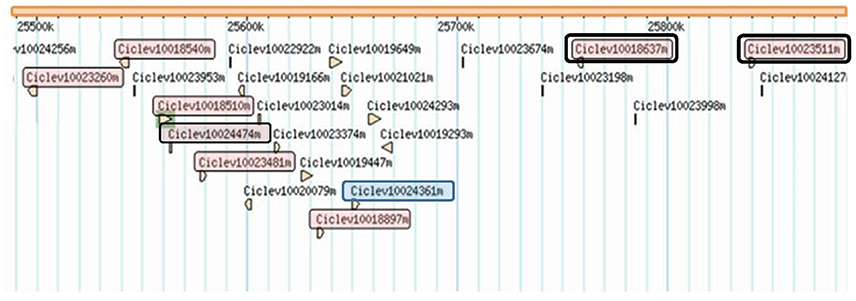
Figure 4. Physical position of candidate genes for ABS resistance found in the identified mapped within the chromosome 3, between 25.49 and 25.86 Kb. Genes in red box are NBS-LRR genes; Genes remarked in double box present LRR receptor-like and serine/threonine protein kinase domains; Gene in blue box belongs to the S-adenosyl-L-methionine-dependent methyltransferase family.
In addition to these genes, there are other four genes with no functional annotation in www.phytozome.org website. These genes were then blasted against the NCBI database (https://blast.ncbi.nlm.nih.gov), where no similarity with any previously characterized protein was found for Ciclev10019166, Ciclev10022922, and Ciclev10023674 genes. Blast results for the Ciclev10024474 gene showed high similarity with a predicted putative RPP13-like protein annotated in Citrus sinensis. Therefore, this gene was also considered as annotated for disease resistance.
Response to ABS Inoculations in Mandarin Breeding Parents
Detached leaves from 40 genotypes used or selected to be used as parents in mandarin breeding programs were inoculated with A. alternata spores as explained in Materials and Methods Section. Results indicate that 20 of them showed symptoms after inoculations, scored as susceptible varieties; 20 varieties did not show any symptom in any replicate, and then were scored as resistant genotypes. Results of ABS response are in agreement with most of the previous studies for all the genotypes for which information is available (Table 4).
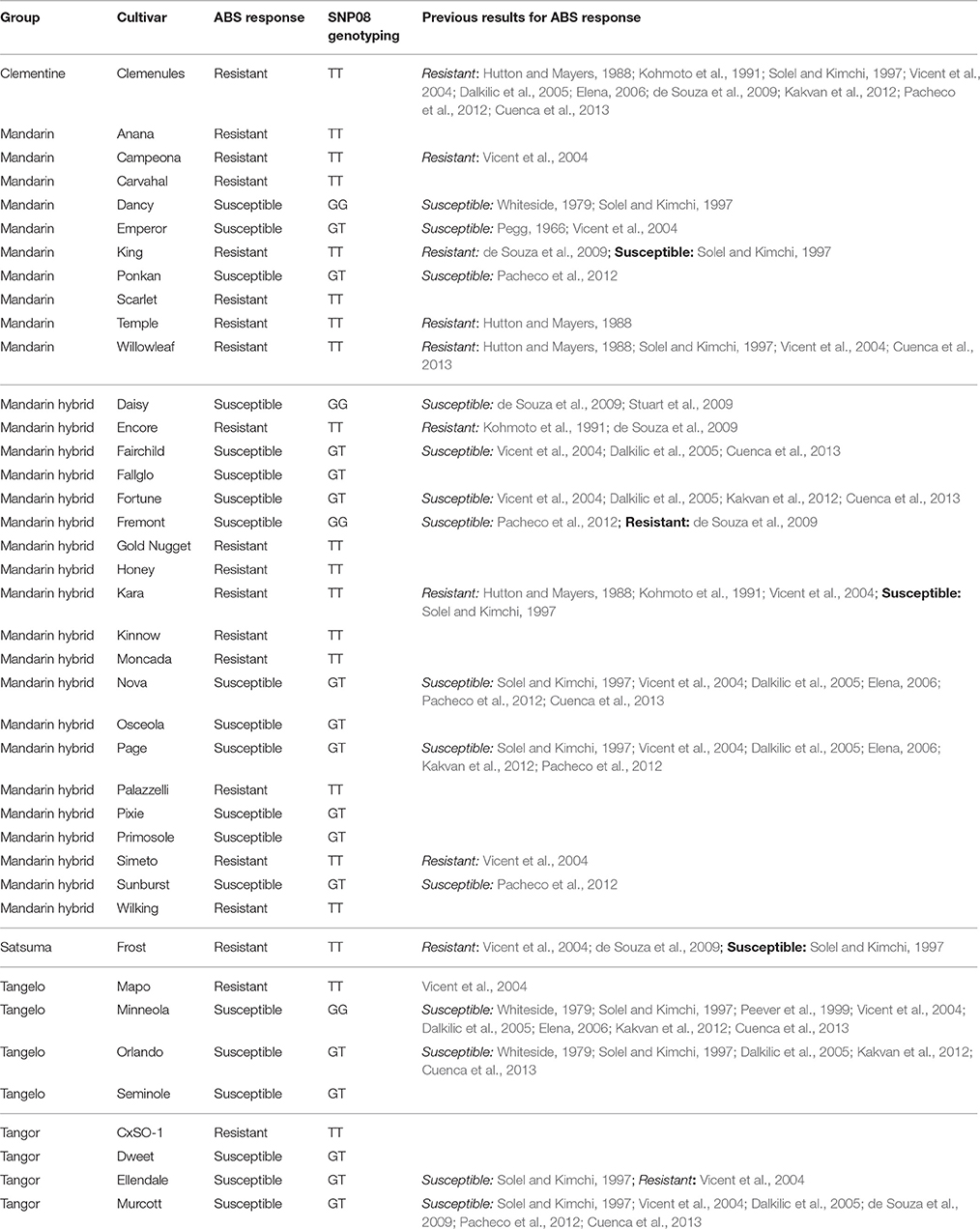
Table 4. ABS response, SNP08 genotyping results, and previous data for ABS response for the 40 cultivars analyzed in the present study.
New Reliable SNP Marker for MAS in Mandarin Breeding Programs
The closet SNP marker flanking the ABSr locus was SNP08, mapped at 0.4 cM from the responsible gene for ABS resistance. This marker was analyzed using the KASPar technique in 40 citrus genotypes, including 20 resistant, and 20 susceptible cultivars. Results from phenotyping and SNP08 genotyping for these 40 accessions are indicated in Table 4. As previously identified from the segregating populations, the allele of this SNP marker linked with the dominant susceptible allele of the ABSr locus is G. Therefore, considering identity by descent in the analyzed germplasm, without recombination between the marker and the ABSr locus, resistant genotypes are expected to be TT, whereas susceptible genotypes are expected to be GG or GT.
All genotypes included in the study were correctly classified as resistant or susceptible using this single SNP marker, highlighting its discriminating power between the two groups (Figure 5).
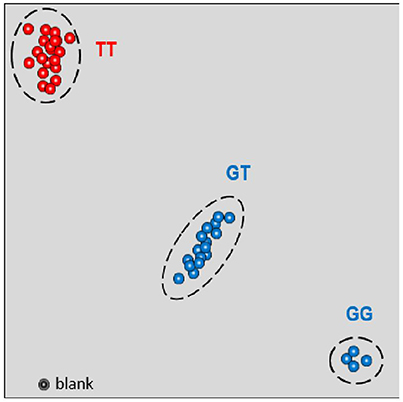
Figure 5. KASPar genotyping results for the SNP08 for the set of genotypes selected for this study. Resistant cultivars are represented by red points (TT); susceptible cultivars are represented by blue points (GT and GG).
Discussion
ABS is an important fungal disease affecting susceptible citrus genotypes. Currently, ABS control is primarily based on fungicide application, but despite a large number of sprays, disease control is not always satisfactory and cannot be considered as a sustainable crop management practice. As a consequence, cultivation of susceptible cultivars has declined significantly, as is the case of “Fortune” mandarin in Spain during recent years. Thus, genetic resistance remains as the best option for disease control [1]. The objective of this study was (i) to finely map the ABSr locus identified by Cuenca et al. (2013) on LG III of the clementine's genetic map (Ollitrault et al., 2012), (ii) to identify candidate genes for resistance, and (iii) to develop SNP molecular markers for efficient MAS in citrus breeding programs.
A 366 Kb Genomic Region Has Been Targeted for Alternaria Brown Spot Resistance in Citrus
In our previous work (Cuenca et al., 2013), the ABSr locus was located by using a bulked segregant analysis coupled with genome scan and targeted genetic mapping by half tetrad analysis in a triploid progeny. The region containing the ABSr locus was flanked by markers TTC8 and CiC3248-06 defining a 5.48 cM interval, corresponding to 3.3 Mb in the clementine reference genome. In the present work, the fine-mapping was carried out using a 268-diploid progeny arising from a heterozygous susceptible × resistant hybridization. As a result, we have limited the candidate region containing the ABSr locus to 1.5 cM flanked by two SNP markers at 1.1 and 0.4 cM, corresponding to 366 kb in the clementine reference genome (between positions 25.496.094 and 25.862.085 in the chromosome III).
Candidate Genes for ABS Resistance
Interactions between pathogens and their hosts are complex and dynamic. At the early stages of necrotrophic infections, host cell death is associated with the production of various secondary metabolites, antimicrobial peptides, hormones and also with the accumulation of reactive oxygen species, callose, and some other cell wall modifications (Wen, 2012). Cell death is a common phenomenon in both resistant and susceptible responses of plant–pathogen interactions to confine pathogens by abolishing nutrient supply. However, cell death generally enhances colonization by necrotrophic pathogens. Effectors produced by the fungus can be recognized by immune receptors in plants, generally belonging to the nucleotide–binding site–leucine–rich repeat (NBS–LRR) protein family. The identified region contains eight genes harboring NBS-LRR repeats, which should be considered as candidate genes for ABS resistance. Nevertheless, the RLM3, a Toll/interleukin 1 receptor domain R–protein in Arabidopsis is the only NBS-LRR gene found to be responsible for resistance to necrotrophs (Mengiste, 2012). A BLAST with this protein results in 200 hits in the citrus genome. Surprisingly, 170 of them are located within chromosome 3, showing the highest score (E = 8.1e-58), and 37% of them are physically mapped around the region of interest (i.e., from 21 to 28 Mb in chromosome 3); however, none of them is within the region identified by fine mapping.
Among the identified resistance genes, Ciclev10018637 and Ciclev10023511 encode for a Leucine-Rich Repeat (LRR) receptor-like protein with serine/threonine kinase domain. LRR receptor-like kinases (LRR-RLK) appear to play a central role in signaling during pathogen recognition, the subsequent activation of plant defense mechanisms, and developmental control (Afzal et al., 2008). Indeed, most of the RLK identified as being involved in plant defense are of the LRR-RLK class including the rice Xa21 protein and the Arabidopsis FLS2 and EFR receptors (Goff and Ramonell, 2007). Both genes were mapped very close to the most significant SNP related to ABS resistance (SNP08). Thus, they should be considered as strong candidates for resistance.
Another strong candidate for ABS resistance found within the region of interest and close to the SNP08 is the gene Ciclev10024361, encoding for an S-adenosyl-L-methionine-dependent methyltransferases superfamily protein, with thiopurine S-methyltransferase superfamily protein. The ACTTS3 gene from A. alternata, involved in the production of the ACT-toxin, encodes a polyketide synthase with putative β-ketoacyl synthase, acyltransferase, methyltransferase, β-ketoacyl reductase, and phosphopantetheine attachment site domains (Miyamoto et al., 2010). During infections of Brassica juncea with Alternaria brassicola, the transcriptional activation of jasmonic acid carboxyl methyltransferase was observed after 2 days post infection (Meur et al., 2015). This gene could be a good target for achieving resistance against necrotrophic pathogens, and therefore, it should also be considered as a strong candidate for resistance to ABS.
There is little information in other plant species about genes involved in resistance to necrotrophic pathogens in general, and to A. alternata in particular. In Solanum lycopersicum, the Asc gene mediates resistance to AAL toxin produced by A. alternata lycopersici (Spassieva et al., 2002). A BLAST for this gene in citrus indicates that there are orthologous genes in two regions of chromosome 3, but none of them within the region of interest; in addition, many other regions in other chromosomes contain Asc orthologous genes. In barley, mlo mutants are resistant to penetration by Blumeria graminis, overproducing p-coumaroyl-hydroxyagmatine; this compound has antifungal activity and inhibits haustorium formation in vivo (Büschges et al., 1997; von Röpenack et al., 1998). BLAST for mlo family genes in citrus indicates that seven of the nine citrus chromosomes contain orthologous to this family, including three genes found to be very close (but outside) to the region of interest in chromosome 3 (Ciclev10019784, Ciclev10023336, Ciclev10020313). In Arabidopsis, Coego et al. (2005) showed that the homeodomain transcription factor OCP3, mediates resistance to infection by necrotrophic pathogens. BLAST results for this gene showed that the only orthologous gene in the citrus genome (Ciclev10010768) is in the chromosome 1. Arabidopsis RFO1 gene, conferring quantitative resistance against Fusarium oxysporum (Diener and Ausubel, 2005), had significant BLAST in five regions of chromosome 3, but none of them within the region of interest; in addition, many other regions in other chromosomes contain RFO1 orthologous genes. Other genes and transcriptions factors have been found to be implicated in the resistance to necrotrophs in Arabidopsis, like the BOS1 and BOI, which appear to restrict necrosis triggered by diverse pathogens and stress factors (Luo et al., 2010). BLAST for these genes in citrus reveals orthologous genes in chromosomes 2 and 3, but outside the region of interest. The WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens in Arabidopsis (Zheng et al., 2006) and BLAST results in citrus indicate that there are orthologous genes in chromosome 3, but outside the region of interest. Also, the ATG18a gene, which interacts with WRKY33, has no orthologous within the region of interest.
The information generated from sequence comparison and annotations allows to discard 15 out of the 24 genes found within the region delimited by fine-mapping. Among the nine genes that may be related to pathogen resistance, the genes Ciclev10018637, Ciclev10023511, and Ciclev10024361 are the strongest candidates for ABS resistance in citrus. This limited number will now allow to make affordable although time-consuming approaches to determine whether these genes are really involved in the Alternaria-citrus interaction. Further experiments, including differential expression performed during fungal infection and functional approaches as genetic transformation or reverse genetics should be performed to accomplish this goal.
A New SNP Marker for Efficient MAS Has Been Developed
In fruit crops, several molecular markers have been found to be linked to agronomic traits of interest (van Nocker and Gardiner, 2014). Examples of successful early selection of interesting genotypes within breeding programs are found in grapes (www.vitisgen.org), Prunus and other Rosaceae species (Dirlewanger et al., 2004).
In citrus, a few characters of agronomic interest have been linked to molecular markers, such as RAPD markers linked to dwarfing (Cheng and Roose, 1995) or fruit acidity (Fang et al., 1997), SSR markers linked to CTV resistance from Poncirus trifoliata (Gmitter et al., 1996; Bernet et al., 2004), AFLP markers linked to nucellar embryony (García et al., 1999; Kepiro and Roose, 2010), and the dominant PCR assay for the anthocyan content of pulp from blood orange due to a transposable element in the 5′ extremity of the Ruby gene (Butelli et al., 2012). Other characters of interest have been tagged to QTLs, such as salinity tolerance (Ben-Hayyim and Moore, 2000) and nematode resistance (Ling et al., 2000). However, for citrus scion breeding programs, only the Alternaria resistance presented in this paper, along with the anthocyanin content, are currently subjected to MAS.
ABS resistance has also been previously tagged with molecular markers. Dalkilic et al. (2005) reported two RAPD markers with loose linkage with the locus (15.3 and 36.7 cM far from ABS resistance locus in the same side). More recently, Gulsen et al. (2010) identified two flanking SRAP markers at 3 and 13 cM. However, due to the distance between markers and the resistance gene as well as the dominant characters of RAPDs and SRAP, these studies did not resulted in concrete MAS application.
In the present work, we have developed a SNP marker (SNP08) mapped at 0.4 cM from the ABS resistance gene, which greatly improves the selection of resistant genotypes in early development stages and avoid growing and evaluating susceptible genotypes. The SNP08 marker is diallelic (G/T), in which the G base is phased with ABS dominant susceptibility. It means that when the G allele is present, i.e., GT and GG genotypes, the cultivar is expected to be susceptible and therefore, only TT genotypes are expected to be resistant. With 0.4 cM distance, in a progeny between a heterozygous susceptible parent and a resistant one, only 0.4% of GT hybrids are expected to be resistant and the same proportion is expected for TT hybrids to be susceptible. Flanking markers at 0.7 cM from the ABSr locus have also been identified on the other side of the gene. By coupling the SNP08 and one of these markers, the error rate would fall to 0.0028% (<3 false resistant hybrids per 100.000).
This SNP08 marker has been tested for association in 40 mandarin genotypes used as breeding parents and covering a large range of the mandarin diversity (Garcia-Lor et al., 2015). ABS inoculations revealed 20 resistant and 20 susceptible genotypes. Citrus clementina, C. nobilis, C. temple, C. deliciosa, and C. unshiu species resulted resistant, whereas the analyzed genotype from C. tangerina is susceptible. Citrus reticulata and the group of hybrid mandarins include both resistant and susceptible genotypes. In the latter group, hybrids between resistant genotypes are resistant, in agreement with the recessive inheritance of the ABS resistance.
Since SNP08 is very close to the ABS resistance gene, it is expected to obtain a very close correlation between the observed SNP08 genotyping and the actual allele configuration of the resistance gene in the germplasm. However, the development of this new marker is based on the segregation of only one progeny, and, a priori, the transferability between different citrus species or interspecific hybrids may not be efficient. In this work, we confirmed this transferability by comparing the results observed for some varieties with segregation progenies obtained from previous works. GG genotypes for the SNP08 are susceptible presumably homozygous for the gene, and no segregation is expected in their progeny. This has been demonstrated for “Minneola” by Cuenca et al. (2013), from which no resistant genotypes were obtained in a progeny of 127 hybrids. Moreover, segregations in the response to ABS were observed by the same authors in progenies arising from “Fortune,” “Nova,” “Orlando,” “Fairchild,” and “Murcott,” confirming that all these cultivars (genotyped as “GT” for the SNP08) are heterozygous for the ABSr locus. Therefore, this marker appears to be phased with the ABS resistance gene in the analyzed germplasm and it could be very useful not only for discriminating between resistant and susceptible cultivars, but also for inferring the allele configuration of the ABS resistance gene. It is therefore a valuable tool for selecting susceptible heterozygous genotypes that can be used as parents in breeding programs and conversely to discard homozygous susceptible varieties. The fact that susceptible yet heterozygous genotypes can be used as parents allows the exploitation of more genetic diversity in citrus breeding programs.
The SNP08 is currently used for the assisted selection of ABS-resistant genotypes in the citrus breeding programs carried out at IVIA and CIRAD. Since its development, it has been successfully applied to select 2187 resistant hybrids from 4517 total hybrids arising from 10 different parental combinations. This analysis avoided to keep growing for a long time more than 2000 susceptible genotypes, which were removed from the breeding program at a very early stage of development and, therefore, saving a considerable amount of time, personnel and money resources.
Author Contributions
JC performed the experiments, analyzed the data, and wrote the manuscript. AG performed molecular experiments. PA obtained and provided the plant material. PO and LN conceived and supervised the experiments. All authors revised and approved the manuscript.
Funding
This work was supported by a grant [AGL2011-26490] from the Ministry of “Economía y Competividad”—Fondo Europeo de Desarrollo Regional (FEDER) and a grant [Prometeo 2008/121] from the Generalitat Valenciana, Spain.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors thank the collaboration of Dr. Antonio Vicent in the inoculum production and leaf inoculations. Also, we thank Ana M. Moreno, Maria Hernandez, Frederique Ollitrault, and Juan Romero their laboratory work, and Jose A. Pina, Rafael Montalt, and Diego Conchilla their greenhouse work.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01948/full#supplementary-material
References
Afzal, A. J., Wood, A. J., and Lightfoot, D. A. (2008). Plant receptor-like serine threonine kinases: roles in signaling and plant defense. Mol. Plant Microbe Interact. 21, 507–517. doi: 10.1094/MPMI-21-5-0507
Ajiro, N., Miyamoto, Y., Masunaka, A., Tsuge, T., Yamamoto, M., Ohtani, K., et al. (2010). Role of the host-selective ACT-toxin synthesis gene ACTTS2 encoding an enoyl-reductase in pathogenicity of the tangerine pathotype of Alternaria alternata. Phytopathology 100, 120–126. doi: 10.1094/PHYTO-100-2-0120
Akimitsu, K., Peever, T. L., and Timmer, L. W. (2003). Molecular, ecological and evolutionary approaches to understanding Alternaria diseases of citrus. Mol. Plant Pathol. 4, 435–446. doi: 10.1046/j.1364-3703.2003.00189.x
Aleza, P., Cuenca, J., Juárez, J., Pina, J. A., and Navarro, L. (2010a). ‘Garbi’ mandarin: a new late-maturing triploid hybrid. HortScience 45, 139–141.
Aleza, P., Juárez, J., Cuenca, J., Ollitrault, P., and Navarro, L. (2010b). Recovery of citrus triploid hybrids by embryo rescue and flow cytometry from 2x × 2x sexual hybridisation and its application to extensive breeding programs. Plant Cell Rep. 29, 1023–1034. doi: 10.1007/s00299-010-0888-7
Aleza, P., Juárez, J., Cuenca, J., Ollitrault, P., and Navarro, L. (2012). Extensive citrus triploid hybrid production by 2x × 4x sexual hybridizations and parent-effect on the length of the juvenile phase. Plant Cell Rep. 31, 1723–1735. doi: 10.1007/s00299-012-1286-0
Ben-Hayyim, G., and Moore, G. A. (2000). “Recent advances in breeding citrus for drought and saline stress tolerance,” in Advances in Molecular Breeding Toward Drought and Salt Tolerant Crops, eds M. A. Jenks, P. M. Hasegawa, S. M. Jain (Dordrecht: Springer), 627–642.
Bernet, G. P., Bretó, M. P., and Asins, M. J. (2004). Expressed sequence enrichment for candidate gene analysis of citrus tristeza virus resistance. Theor. Appl. Genet. 108, 592–602. doi: 10.1007/s00122-003-1479-6
Bhatia, A., Roberts, P. D., and Timmer, L. W. (2003). Evaluation of the alter-rater model for timing of fungicide applications for control of Alternaria brown spot of citrus. Plant Dis. 87, 1089–1093. doi: 10.1094/PDIS.2003.87.9.1089
Büschges, R., Hollricher, K., Panstruga, R., Simons, G., Wolter, M., Frijters, A., et al. (1997). The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88, 695–705. doi: 10.1016/S0092-8674(00)81912-1
Butelli, E., Licciardello, C., Zhang, Y., Liu, J., MacKay, S., Bailey, P., et al. (2012). Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24, 1242–1255. doi: 10.1105/tpc.111.095232
Cheng, F. S., and Roose, M. L. (1995). Origin and inheritance of dwarfing by the citrus rootstock Poncirus trifoliata ‘Flying Dragon’. J. Am. Soc. Hortic. Sci. 120, 286–291.
Coego, A., Ramírez, V., Gil, M. J., Flors, V., Mauch-Mani, B., and Vera, P. (2005). An Arabidopsis homeodomain transcription factor, OVEREXPRESSOR OF CATIONIC PEROXIDASE 3, mediates resistance to infection by necrotrophic pathogens. Plant Cell 17, 2123–2137. doi: 10.1105/tpc.105.032375
Cuenca, J., Aleza, P., Juárez, J., Pina, J. A., and Navarro, L. (2010). ‘Safor’ mandarin: a new citrus mid-late triploid hybrid. HortScience 45, 977–980.
Cuenca, J., Aleza, P., Vicent, A., Brunel, D., Ollitrault, P., and Navarro, L. (2013). Genetically based location from triploid populations and gene ontology of a 3.3-Mb genome region linked to Alternaria brown spot resistance in citrus reveal clusters of resistance genes. PLoS ONE 8:e76755. doi: 10.1371/journal.pone.0076755
Cuppen, E. (2007). Genotyping by allele-specific amplification (KASPar). Cold Spring Harb. Protoc. 2007:pdb.prot4841. doi: 10.1101/pdb.prot4841
Dalkilic, Z., Timmer, L. W., and Gmitter, F. (2005). Linkage of an Alternaria disease resistance gene in mandarin hybrids with RAPD fragments. J. Am. Soc. Hortic. Sci. 130, 191–195.
de Souza, M. C., Sanches, E., and de Goes, A. (2009). Evaluation of tangerine hybrid resistance to Alternaria alternata. Sci. Hort. 123, 1–4. doi: 10.1016/j.scienta.2009.07.005
Diener, A. C., and Ausubel, F. M. (2005). RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171, 305–321. doi: 10.1534/genetics.105.042218
Dirlewanger, E., Graziano, E., Joobeur, T., Garriga-Calderé, F., Cosson, P., Howad, W., et al. (2004). Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc. Natl. Acad. Sci. U.S.A. 101, 9891–9896. doi: 10.1073/pnas.0307937101
Elena, K. (2006). Alternaria Brown Spot of Minneola in Greece; evaluation of Citrus species susceptibility. Eur. J. Plant Pathol. 115, 259–262. doi: 10.1007/s10658-006-9005-8
Everts, K. L., and Lacy, M. L. (1996). Factors influencing infection of onion leaves by Alternaria porri and subsequent lesion expansion. Plant Dis. 80, 276–280. doi: 10.1094/PD-80-0276
Fang, D. Q., Federici, C. T., and Roose, M. L. (1997). Development of molecular markers linked to a gene controlling fruit acidity in citrus. Genome 40, 841–849. doi: 10.1139/g97-809
Froelicher, Y., Bouffin, J., Dambier, D., and Ollitrault, P. (2012). “Triploid seedless mandarin breeding in France,” in XII International Citrus Congress, December, 2012. (Valence), 19–23.
García, R., Asíns, M. J., Forner, J., and Carbonell, E. A. (1999). Genetic analysis of apomixis in Citrus and Poncirus by molecular markers. Theor. Appl. Genet. 99, 511–518. doi: 10.1007/s001220051264
Garcia-Lor, A., Luro, F., Ollitrault, P., and Navarro, L. (2015). Genetic diversity and population-structure analysis of mandarin germplasm by nuclear, chloroplastic and mitochondrial markers. Tree Genet. Genomes 11:123. doi: 10.1007/s11295-015-0951-1
Gmitter, F. G. Jr., Xiao, S. Y., Huang, S. L., Hu, X. L., Garnsey, S. M., and Deng, Z. (1996). A localized linkage map of the citrus tristeza virus resistance gene region. Theor. Appl. Genet. 92, 688–695. doi: 10.1007/BF00226090
Goff, K. E., and Ramonell, K. M. (2007). The role and regulation of Receptor-Like Kinases in plant defense. Gene Regul. Syst. Biol. 1, 167–175.
Golmohammadi, M., Andrew, M., Peever, T. L., Peres, N. A., and Timmer, L. W. (2006). Brown spot of tangerine hybrid cultivars Minneola, Page and Fortune caused by Alternaria alternate in Iran. Plant Pathol. 55, 578. doi: 10.1111/j.1365-3059.2006.01427.x
Grosser, J., An, H., Calovi, M., Lee, D., Chen, C., Vasconcellos, M., et al. (2010). Production of new allotetraploid and autotetraploid citrus breeding parents: focus on zipperskin mandarins. Hortscience 45, 1160–1163.
Gulsen, O., Uzun, A., Canan, I., Seday, U., and Canihos, E. (2010). A new citrus linkage map based on SRAP, SSR, ISSR, POGP, RGA and RAPD markers. Euphytica 173, 265–277. doi: 10.1007/s10681-010-0146-7
Hutton, D. G., and Mayers, P. E. (1988). Brown spot of Murcott tangor caused by Alternaria alternata in Queensland. Aust. Plant Pathol. 17, 69–73. doi: 10.1071/APP9880069
Ingvarsson, P. K., and Street, N. R. (2011). Association genetics of complex traits in plants. New Phytol. 189, 909–922. doi: 10.1111/j.1469-8137.2010.03593.x
JinPing, X., LiGeng, C., Ming, X., HaiLin, L., and WeiQi, Y. (2009). Identification of AFLP fragments linked to seedlessness in Ponkan mandarin (Citrus reticulata Blanco) and conversion to SCAR markers. Sci. Hort. 121, 505–510. doi: 10.1016/j.scienta.2009.03.006
Kakvan, N., Zamanizadeh, H., Morid, B., Hajmansor, S., and Taeri, H. (2012). Evaluation of citrus cultivars resistance to Alternaria alternata, the causal agent of brown spot disease, using RAPD-PCR. J. Res. Agric. Sci. 8, 69–76.
Kepiro, J. L., and Roose, M. L. (2010). AFLP markers closely linked to a major gene essential for nucellar embryony (apomixis) in Citrus maxima × Poncirus trifoliata. Tree Genet. Genomes 6, 1–11. doi: 10.1007/s11295-009-0223-z
Khan, M. A., and Korban, S. S. (2012). Association mapping in forest trees and fruit crops. J. Exp. Bot. 63, 4045–4060. doi: 10.1093/jxb/ers105
Kohmoto, K., Akimitsu, K., and Otani, H. (1991). Correlation of resistance and susceptibility of citrus to Alternaria alternata with sensitivity to host-specific toxins. Phytopathology 81, 719–722. doi: 10.1094/Phyto-81-719
Kohmoto, K., Itoh, Y., Shimomura, N., Kondoh, Y., Otani, H., Kodama, M., et al. (1993). Isolation and biological-activities of 2 host-specific toxins from the tangerine pathotype of Alternaria alternata. Phytopathology 83, 495–502. doi: 10.1094/Phyto-83-495
Ling, P., Duncan, L. W., Deng, Z., Dunn, D., Hu, X., Huang, S., et al. (2000). Inheritance of citrus nematode resistance and its linkage with molecular markers. Theor. Appl. Genet. 100, 1010–1017. doi: 10.1007/s001220051382
Lörz, H., and Wenzel, G. (2008). “Molecular marker systems in plant breeding and crop improvement,” in Biotechnology in Agriculture and Forestry, eds T. Nagata, H. Lörz, and J. M. Widholm (Berlin: Springer).
Luo, H., Laluk, K., Lai, Z., Veronese, P., Song, F., and Mengiste, T. (2010). The Arabidopsis Botrytis Susceptible1 Interactor defines a subclass of RING E3 ligases that regulate pathogen and stress responses. Plant Physiol. 154, 1766–1782. doi: 10.1104/pp.110.163915
MacKay, I., and Powell, W. (2007). Methods for linkage disequilibrium mapping in crops. Trends Plant Sci. 12, 57–63. doi: 10.1016/j.tplants.2006.12.001
McCollum, T. G. (2008). Update of the USDA, ARS citrus scion improvement project. Proc. Fla. State Hort. Soc. 120, 285–287.
Mengiste, T. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50, 267–294. doi: 10.1146/annurev-phyto-081211-172955
Meur, G., Shukla, P., Dutta-Gupta, A., and Kirti, P. B. (2015). Characterization of Brassica juncea-Alternaria brassicicola interaction and jasmonic acid carboxyl methyl transferase expression. Plant Gene 3, 1–10. doi: 10.1016/j.plgene.2015.06.001
Miyamoto, Y., Masunaka, A., Tsuge, T., Yamamoto, M., Ohtani, K., Fukumoto, T., et al. (2010). ACTTS3 encoding a polyketide synthase is essential for the biosynthesis of act-toxin and pathogenicity in the tangerine pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 23, 406–414. doi: 10.1094/MPMI-23-4-0406
Navarro, L., Aleza, P., Cuenca, J., Juárez, J., Pina, J. A., Ortega, C., et al. (2012). “The triploid mandarin breeding program in Spain,” in XII International Citrus Congress, December, 2012 (Valence), 19–23.
Ollitrault, P., Terol, J., Chen, C., Federici, C. T., Lotfy, S., Hippolyte, I., et al. (2012). A reference genetic map of C. clementina hort. ex Tan.; citrus evolution inferences from comparative mapping. BMC Genomics 13:593. doi: 10.1186/1471-2164-13-593
Pacheco, C. A., Martelli, I. B., Polydoro, D. A., Schinor, E. H., Pio, R. M., Kupper, K. C., et al. (2012). Resistance and susceptibility of mandarins and their hybrids to Alternaria alternata. Sci. Agric. 69, 386–392. doi: 10.1590/S0103-90162012000600007
Pegg, K. G. (1966). Studies of a strain of Alternaria citri Pierce, the causal organism of brown spot of Emperor mandarin. Queensl. J. Agric. Anim. Sci. 23, 15–28.
Peres, N. A., and Timmer, L. W. (2006). Alternaria brown spot of citrus in Brazil: evaluation of the Alter-Rater for spray timing and effects on yield of Murcott tangor. Crop Prot. 25, 454–460. doi: 10.1016/j.cropro.2005.07.010
Peever, T. L., Canihos, L. Y., Olsen, L., Ibanez, A., Liu, Y. C., and Timmer, L. W. (1999). Population genetic structure and host specificity of Alternaria spp. causing brown spot of Minneola tangelo and rough lemon in Florida. Phytopathology 89, 851–860. doi: 10.1094/PHYTO.1999.89.10.851
Recupero, G. R., Russo, G., and Recupero, S. (2005). New promising citrus triploid hybrids selected from crosses between monoembryonic diploid female ant tetraploid male parents. HortScience 40, 516–520.
Schinor, E., Pacheco, C., Bastianel, M., Azevedo, F., and Cristofani-Yali, M. (2012). “New tangors for Brazilian citriculture,” in XII International Citrus Congress December, 2012 (Valence), 19–23.
Solel, Z., and Kimchi, M. (1997). Susceptibility and resistance of citrus genotypes to Alternaria alternata pv. citri. J. Phytopathol. 145, 389–391. doi: 10.1111/j.1439-0434.1997.tb00420.x
Sorkheh, K., Malysheva-Otto, L. V., Wirthensohn, M. G., Tarkesh-Esfahani, S., and Martínez-Gómez, P. (2008). Linkage disequilibrium, genetic association mapping and gene localization in crop plants. Genet Mol. Biol. 31, 805–814. doi: 10.1590/S1415-47572008000500001
Spassieva, S. D., Markham, J. E., and Hille, J. (2002). The plant disease resistance gene Asc-1 prevents disruption of sphingolipid metabolism during AAL-toxin-induced programmed cell death. Plant J. 32, 561–572. doi: 10.1046/j.1365-313X.2002.01444.x
Stam, P. (1993). Construction of integrated genetic linkage maps by means of a new computer package: joinmap. Plant J. 3, 739–744. doi: 10.1111/j.1365-313X.1993.00739.x
Stuart, R. M., Bastianel, M., Azevedo, F. A., and Machado, M. A. (2009). Alternaria brown spot. Laranja 30, 29–44.
Timmer, L. W., Peever, T. L., Solel, Z., and Akimitsu, K. (2003). Alternaria diseases of citrus-novel Pathosystems. Phytopathol. Mediterr. 42, 99–112.
Tsuge, T., Harimoto, Y., Akimitsu, K., Ohtani, K., Kodama, M., Akagi, Y., et al. (2012). Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol. Rev. 37, 44–66. doi: 10.1111/j.1574-6976.2012.00350.x
van Nocker, S., and Gardiner, S. E. (2014). Breeding better cultivars, faster: applications of new technologies for the rapid deployment of superior horticultural tree crops. Hort. Res. 1:14022. doi: 10.1038/hortres.2014.22
Vicent, A., Armengol, J., and García-Jimenez, J. (2007). Rain fastness and persistence of fungicides for control of Alternaria Brown Spot of citrus. Plant Dis. 91, 393–399. doi: 10.1094/PDIS-91-4-0393
Vicent, A., Armengol, J., Sales, R., García-Jiménez, J., and Alfaro-Lassala, F. (2000). First report of Alternaria brown spot of citrus in Spain. Plant Dis. 84:1044. doi: 10.1094/PDIS.2000.84.9.1044B
Vicent, A., Badal, J., Asensi, M. J., Sanz, N., Armengol, J., and Garcia-Jimenez, J. (2004). Laboratory evaluation of citrus cultivars susceptibility and influence of fruit size on Fortune mandarin to infection by Alternaria alternata pv. citri. Eur. J. Plant. Pathol. 110, 245–251. doi: 10.1023/B:EJPP.0000019794.00000.02
von Röpenack, E., Parr, A., and Schulze-Lefert, P. (1998). Structural analyses and dynamics of soluble and cell wall-bound phenolics in a broad spectrum resistance to the powdery mildew fungus in barley. J. Biol. Chem. 273, 9013–9022. doi: 10.1074/jbc.273.15.9013
Wang, X. F., Li, Z. A., Tang, K. Z., Zhou, C. Y., and Yi, L. (2010). First report of alternaria brown spot of citrus caused by Alternaria alternata in Yunnan Province, China. Plant Dis. 94, 375. doi: 10.1094/PDIS-94-3-0375C
Wen, L. (2012). Cell death in plant immune response to necrotrophs. J. Plant Biochem. Physiol. 1:e103. doi: 10.4172/2329-9029.1000e103
Whiteside, J. O. (1979). Alternaria brown spot of Dancy tangerines and its control. Proc. Fla. State Hort. Soc. 92, 34–37.
Williams, T. E. (2012). “Experiences in the development, release and commercialization of new irradiated citrus varieties from the citrus breeding program at the University of California, Riverside,” in XII International Citrus Congress, December, 2012 (Valence), 19–23.
Wu, G. A., Prochnik, S., Jenkins, J., Salse, J., Hellsten, U., Murat, F., et al. (2014). Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 32, 656–662. doi: 10.1038/nbt.2906
Würschum, T. (2012). Mapping QTL for agronomic traits in breeding populations. Theor. Appl. Genet. 125, 201–210. doi: 10.1007/s00122-012-1887-6
Keywords: mandarin, fungal disease resistance, gene mapping, molecular markers, Alternaria alternata
Citation: Cuenca J, Aleza P, Garcia-Lor A, Ollitrault P and Navarro L (2016) Fine Mapping for Identification of Citrus Alternaria Brown Spot Candidate Resistance Genes and Development of New SNP Markers for Marker-Assisted Selection. Front. Plant Sci. 7:1948. doi: 10.3389/fpls.2016.01948
Received: 04 November 2016; Accepted: 07 December 2016;
Published: 23 December 2016.
Edited by:
Manuel Acosta, University of Murcia, SpainReviewed by:
Miguel A. Aranda, Spanish National Research Council, SpainAbhishek Bohra, Indian Institute of Pulses Research, India
Copyright © 2016 Cuenca, Aleza, Garcia-Lor, Ollitrault and Navarro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick Ollitrault, ollitrault@cirad.fr
Luis Navarro, lnavarro@ivia.es
 Jose Cuenca
Jose Cuenca Pablo Aleza
Pablo Aleza Andres Garcia-Lor1
Andres Garcia-Lor1 Patrick Ollitrault
Patrick Ollitrault Luis Navarro
Luis Navarro