
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 19 December 2016
Sec. Plant Breeding
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.01906
This article is part of the Research TopicResponse of grass (Poaceae) species to abiotic stressView all 44 articles
Cereal crop species including bread wheat (Triticum aestivum L.), barley (Hordeum vulgare L.), rice (Oryza sativa L.), and maize (Zea mays L.) provide the bulk of human nutrition and agricultural products for industrial use. These four cereals are central to meet future demands of food supply for an increasing world population under a changing climate. A prerequisite for cereal crop production is the transition from vegetative to reproductive and grain-filling phases starting with flower initiation, a key developmental switch tightly regulated in all flowering plants. Although studies in the dicotyledonous model plant Arabidopsis thaliana build the foundations of our current understanding of plant phenology genes and regulation, the availability of genome assemblies with high-confidence sequences for rice, maize, and more recently bread wheat and barley, now allow the identification of phenology-associated gene orthologs in monocots. Together with recent advances in next-generation sequencing technologies, QTL analysis, mutagenesis, complementation analysis, and RNA interference, many phenology genes have been functionally characterized in cereal crops and conserved as well as functionally divergent genes involved in flowering were found. Epigenetic and other molecular regulatory mechanisms that respond to environmental and endogenous triggers create an enormous plasticity in flowering behavior among cereal crops to ensure flowering is only induced under optimal conditions. In this review, we provide a summary of recent discoveries of flowering time regulators with an emphasis on four cereal crop species (bread wheat, barley, rice, and maize), in particular, crop-specific regulatory mechanisms and genes. In addition, pleiotropic effects on agronomically important traits such as grain yield, impact on adaptation to new growing environments and conditions, genetic sequence-based selection and targeted manipulation of phenology genes, as well as crop growth simulation models for predictive crop breeding, are discussed.
An essential foundation of agriculture was the domestication of grasses from the Poaceae family about 15,000 years ago in the Fertile Crescent of the Eastern Mediterranean region (Gill et al., 2004). Since then, grasses have diversified across a range of ecological niches and are now cultivated in each of the different world climates. Barley (Hordeum vulgare L.) and rye (Secale cereale L.) are adapted to cooler temperate regions, wheat (Triticum aestivum L.) and oats (Avena sativa L.) to temperate regions, and rice (Oryza sativa L.) and maize (Zea mays L.) to tropical and warmer temperate zones. Wheat, rice, and maize provide about two-thirds of all energy in human diets, whereas barley grain contributes to the human diet indirectly as animal feed, and directly as a substrate for malting, brewing, and distilling industries (Cassman, 1999). These four cereal crops are central in current efforts to increase arable crop yield and other agronomic outputs in an attempt to secure future food supply security for an increasing world population under a changing climate (Rötter et al., 2015).
Flowering sets the switch from vegetative to reproductive development. The genetic regulation of flowering time is more sensitive to environmental cues than many other agriculturally relevant traits. A crucial phenological developmental step of crops is the floral initiation and timing of flowering, also known as heading or ear emergence date and defined as first anther bust on spikes in cereals. Heading date and photoperiod sensitivity are the fundamental traits that determine adaptation to geographic environments and different cropping systems, and are controlled by an endogenous genetic network as well as environmental cues including day length (photoperiod) and temperature (Andres and Coupland, 2012). The optimal timing of this transition directly affects grain yield as it needs to occur during specific seasons to avoid abiotic (such as cold, frost, heat, and drought) and biotic stresses (including fungi, bacteria, viruses, nematodes, or insects).
Considerable differences exist between cereals grown in temperate and tropical climates, as these climates have different seasons best suited for reproduction. In tropical regions, the majority of plants flower when the days become shorter during the cooler seasons of the year to avoid hot temperatures during summer and are described as short-day (SD) plants. By contrast, among many temperate species, flower development is controlled in response to changes in both photoperiod and prolonged exposure to cold temperature (vernalization). This ensures that autumn-sown crops flower during spring when growth conditions are more favorable than during winter (Greenup et al., 2009). Floral induction is delayed under short day conditions (8 h of light), and enhanced under long day conditions (16 h of light); therefore, many temperate species are described as long-day (LD) plants. Compared to autumn-sown winter cultivars, spring cultivars are sensitive to low temperatures, insensitive to photoperiod, and do not require vernalization (von Zitzewitz et al., 2005). In day-neutral plants, such as temperate maize (Zea mays ssp mays) cultivars, there is no specific photoperiod requirement as flowering is promoted almost exclusively through the coordinated action of autonomous regulatory pathway genes (Mascheretti et al., 2015).
The topic of this review is to summarize, compare and contrast the current understanding of flowering time regulation in the four major cereal crop species wheat, barley, rice, and maize. We will highlight how different flowering time regulatory mechanisms have evolved between SD, LD, and day-neutral plants and how evidence is accumulating that miRNA and epigenetic regulation play a major role in controlling phenology gene action in cereals. We will describe how genetic variation in flowering time genes paved the way for adaptation to new growing environments. A detailed understanding of flowering mechanisms is the cornerstone of future developments for genetic sequence-based selection and targeted manipulation of phenology genes. In the final part of this review, we will discuss how this knowledge can inform crop simulation models as well as gene-based models developed to predict phenology, the current status of development and use of these models, and how their findings can translate into an improved yield on the field.
Like the cereal crops wheat and barley, Arabidopsis is an LD plant sensitive to vernalization and photoperiod and adapted to temperate climates (Greenup et al., 2009). With some marked exceptions, genes and proteins with analogous function and similar molecular mechanisms found in Arabidopsis also control seasonal flowering responses in temperate grasses. These genes and proteins are part of complex signaling pathways that revolve around a set of main flowering time integrator genes that react to endogenous triggers including genes belonging to the circadian clock, autonomous, age, and gibberellin pathways, as well as environmental signals such as ambient temperature and photoperiod. Since recent reviews give comprehensive descriptions of flowering genes and mechanisms in Arabidopsis, the model plant for flowering time control (Song J. et al., 2013; Ó’Maoiléidigh et al., 2014; Johansson and Staiger, 2015; Song et al., 2015), only a brief overview of the main integrator genes is given in this section. This will provide a basis for a more detailed discussion of gene networks controlling cereal phenology in bread wheat, barley, rice, and maize, particularly for components absent in Arabidopsis.
For nearly 80 years, researchers were trying to characterize the elusive florigen, a hormone-like substance believed to be responsible for promotion or stimulation of flowering in plants. The term was first coined by Russian scientist Chailakhyan (1936) while experimenting on phototropism. Early grafting experiments demonstrated that this floral signal is produced in the leaves, and transferred to the shoot apical meristem (SAM) through the phloem (Zeevaart, 1976). However, after these initial successes, the exact identity of florigen remained unknown until the beginning of the 21st century when the first plant genome sequences were made publicly available and provided the genetic tools to investigate flowering time in more detail. In 2005, three independent groups from Germany (Wigge et al., 2005), Japan (Abe et al., 2005), and Sweden (Huang et al., 2005) reported having identified a gene that controls flowering time named FLOWERING LOCUS T (AtFT). Published in the journal Science, Huang et al. (2005) reported AtFT mRNA as the transmissible signal required for flowering. However, the journal article was retracted after 2 years (Bohlenius et al., 2007) as the results could neither be supported by other research groups (Lifschitz et al., 2006) nor could be replicated in their laboratory (Huang et al., 2005). Many more genetic components of flowering time were identified in the following years (Conti and Bradley, 2007; Sawa et al., 2007), but none provided evidence of how FT is transported from leaves to the shoot apex. In 2007, studies in Arabidopsis (Corbesier et al., 2007) and rice (Tamaki et al., 2007) independently reported that the FT protein itself is the mobile flowering signal, supporting the previous results found in tomato (Lifschitz et al., 2006).
According to the current status of research in Arabidopsis and rice, FT protein travels via the phloem to the SAM, where it binds to bZIP transcription factors FLOWERING LOCUS D (AtFD) and FD PARALOG (FDP) (Abe et al., 2005; Wigge et al., 2005) (Figure 1). This activates the expression of several floral meristem identity genes that induce the transition toward reproductive development. A central floral meristem identity gene is LEAFY (AtLFY), which is activated by transcription factors AGAMOUS-LIKE24 (AtAGL24), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (AtSOC1), and SHORT VEGETATIVE PHASE (AtSVP). AGL24 and SVP were shown to enhance expression of a MADS-box transcription factor encoding gene APETALA 1 (AtAP1), as well as two genes closely related to AP1, CAULIFLOWER (AtCAL) and FRUITFUL (AtFUL) (Ferrandiz et al., 2000; Wigge et al., 2005; Grandi et al., 2012; Jaeger et al., 2013). In addition to activating the expression of floral meristem identity genes, AtLFY and AtAP1 also repress negative regulators of AtFT, such as TERMINAL FLOWER1 (AtTFL1), TEMPRANILLO1 (AtTEM1) and AtTEM2 (Kaufmann et al., 2010).
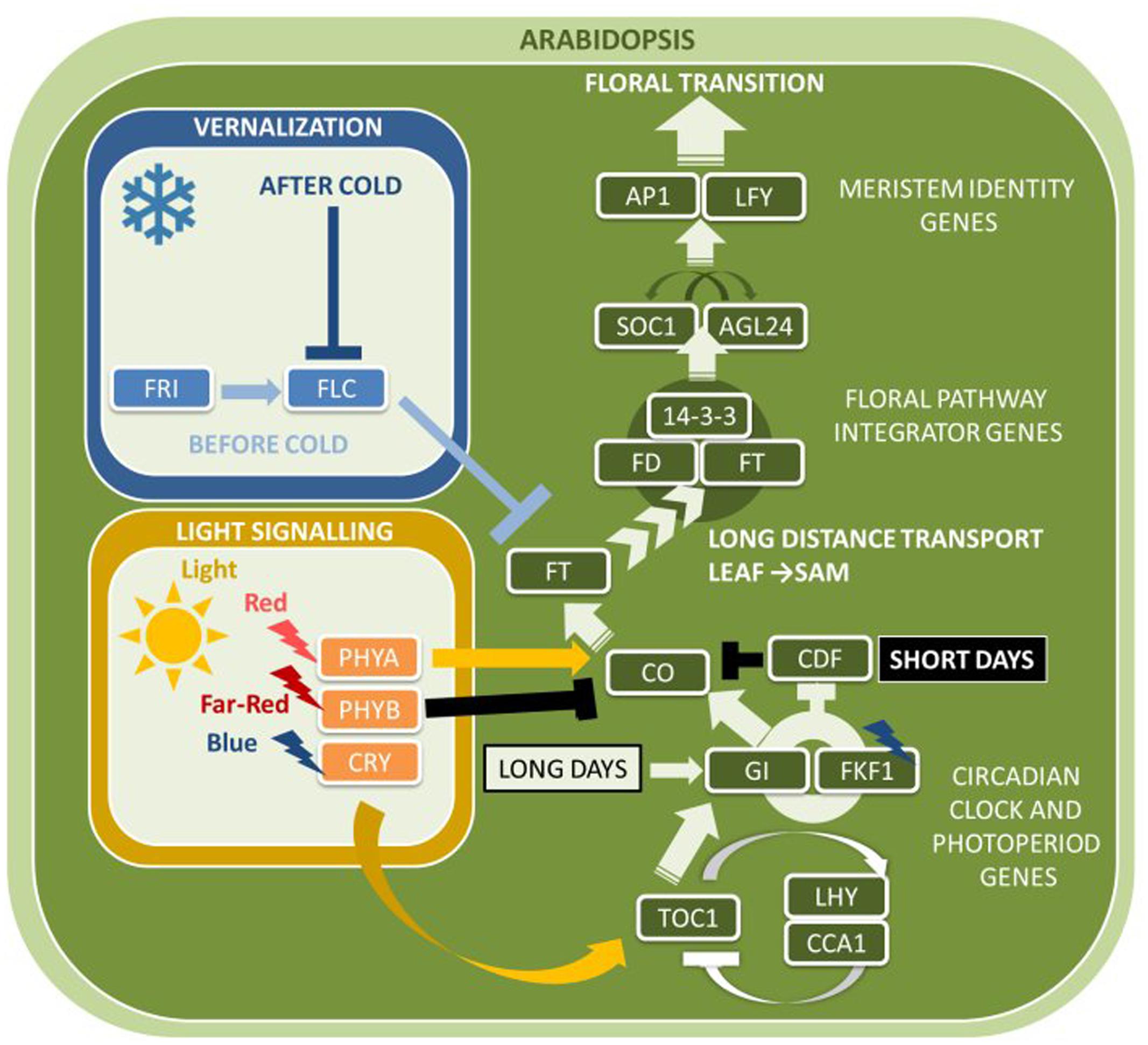
FIGURE 1. Major flowering pathway genes of Arabidopsis thaliana. Positive and negative regulatory connections are indicated by arrows and lines with T-ends, respectively. White and black arrows or T-ends indicate regulatory connections occurring primarily under long days and short days, respectively. Gene name abbreviations are explained in Supplementary Table 1.
Gibberellins (GAs) are diterpene phytohormones that regulate multiple aspects of plant growth, including flower and seed development (Yamaguchi, 2008). Many GA metabolism enzymes are multifunctional, accepting several substrates to produce a diverse set of GAs. Only a subset of all GAs has a bioactive function, including GA1, GA3, and GA4. Different forms of GA are converted via the action of a series of various enzymes, most notably encoded by AtGA20OX genes which catalyze the conversion of non-bioactive to bioactive forms of GA. In Arabidopsis, GA signaling is required at the apex during floral induction and involves binding of GA to the GIBBERELLIN INSENSITIVE DWARF1 (AtGID1) receptors to promote degradation of DELLA proteins via the ubiquitin proteasome pathway (Murase et al., 2008). GA further influences flowering time by regulating the expression of AtSOC1 and AtLFY (Moon et al., 2003). Very recently, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 15 (AtSPL) was demonstrated to be regulated by miR156 and to promote flowering under non-inductive conditions (Hyun et al., 2016).
Genetic regulatory mechanisms of flowering time consist of a series of feedback loops through which individual components of the circadian clock sequentially or reciprocally repress each other (Johansson and Staiger, 2015). Putterill et al. (1995) identified a late-flowering mutant under LD conditions and cloned the corresponding CONSTANS (AtCO) gene. The corresponding protein CO is a nuclear B-Box zinc-finger (BBX) protein with a C-terminal CCT (CO, CO-like, and TIMING OF CAB EXPRESSION 1, TOC1) DNA binding domain, located in the leaves, and responsible for regulating flowering in response to day length.
In addition to genetic regulation via feedback circuits, post-translational processes contribute to adjusting clock protein oscillations to LDs. Under SDs, CO expression is repressed in the morning by CYCLING DOF FACTOR (AtCDF) proteins that bind to its promoter (Fornara et al., 2009). During the day, AtCO mRNA levels increase with a peak before dusk but are ultimately degraded at night so that AtFT expression remains repressed. In SDs, GIGANTEA (AtGI) and FLAVIN BINDING, KELCH REPEAT, F-BOX1 (AtFKF1) clock proteins are asynchronously expressed and do not interact, but synchronize their expression under LDs (Sawa et al., 2007). AtFKF1 recognizes AtCDF proteins via its Kelch-repeat domain, whereas the LOV domain absorbs blue light, crucial for the interaction with AtGI. Only in response to LDs, AtCDF degrades via the ubiquitin-proteasome pathway through the action of the GI-FKF1 complex. As a result, AtCO mRNA can accumulate during the day to induce transcription of AtFT, and the AtFT protein is then transferred via the phloem to the SAM (Corbesier et al., 2007).
AtCO is regulated at the protein-level by phytochromes and cryptochromes that either stabilize or destabilize AtCO mRNA (Devlin, 2002). The blue-light-dependent interaction of AtFKF1with AtGI to antagonistically control CO transcript stability is one example of a plant molecular mechanism that adjusts the period of clock protein oscillations to a 24-h rhythm. Depending on the intensity, periodicity, and spectral quality (such as blue, red and far-red light) of incoming sunlight, photosensory pigments trigger transduction chains that alter gene activity of clock genes further downstream (Fankhauser and Staiger, 2002). In addition to AtFKFI, its homologs ZEITLUPE (AtZTL) and LOV KELCH PROTEIN 2 (AtLKP2), as well as the red/far-red-absorbing PHYTOCHROMES A–E (AtPHYA–PHYE) and the blue light-absorbing CRYPTOCHROMES (AtCRY1, AtCRY2) modulate gene activity to influence plant growth and development (Devlin, 2002).
Changes in light and temperature at dawn and dusk reset the circadian clock, thus allowing the plant to adjust physiologically and metabolically to changing day lengths during different seasons of the year, crucial for ensuring flowering occurs at the optimal time. Very recently, a molecular mechanism was proposed by which endogenous circadian clocks converge with light-signaling pathways through the interaction of AtTOC1 and PHYTOCHROME-INTERACTING FACTORS (AtPIFs), which belong to a subfamily of basic helix–loop–helix transcription factors (Soy et al., 2016). Arabidopsis phytochromes AtPHYA–PHYE regulate the PIF pathway and downstream targets of PIF by inducing degradation of AtPIF1, AtPIF3, AtPIF4, and AtPIF5 (Leivar and Monte, 2014). While the phytochromes remain photo-activated during the day, PIF levels remain low, but increase when the levels of photoactivated phytochromes decline particularly during long nights (Soy et al., 2012). AtPIF protein level oscillations facilitate timing of hypocotyl elongation growth rates to the optimum time just before dawn when AtPIF is most abundant in the circadian cycle. AtTOC1 directly represses the transcriptional activator activity of AtPIF3 protein in SDs after dusk by binding to the AtPIF3 promoter despite high AtPIF3 abundance (Soy et al., 2016). This mechanism regulates dawn-phased growth-related and hormone-associated genes in controlling early hypocotyl growth in diurnal SD conditions.
The major repressor of flowering is the MADS-box transcription factor gene FLOWERING LOCUS C (AtFLC) (Song Y.H. et al., 2013). AtFLC is expressed in mitotically active regions such as the SAM and the root apical meristem, the primary tissues of cold perception. When active alleles of the flowering time regulator gene FRIGIDA (AtFRI) are present, AtFLC expression is enhanced in a dose-dependent manner up to a level that prevents flowering (Michaels and Amasino, 1999). The promotion of flowering after exposure to prolonged cold temperatures, a process termed vernalization which occurs during winter or can be induced artificially, silences AtFLC and allows flowering in the following spring. Such silencing was shown to involve a Polycomb-based epigenetic memory system and requires polycomb repressive complex 2 (AtPRC2)-mediated methylation of H3K27 to form facultative heterochromatin, which allows for the maintained AtFLC repression even after warmer conditions have returned (Bastow et al., 2004; Wood et al., 2006). The extensive natural variation present for both the vernalization requirement and response to cold has played a key role in the adaptation of many plant species over a wide latitudinal range. For example, naturally occurring AtFLC, AtFRI, and AtPHYC polymorphisms result in varying flowering times and are involved in adaptations to different climates in Arabidopsis (Mendez-Vigo et al., 2011).
Although studies in the model dicot Arabidopsis were fundamental to deciphering flowering time mechanisms, recent research in rice has extended our knowledge of flowering processes to a model monocot species. However, considerable differences between cereals grown in temperate and tropical climates exist, thus compromising the relevance of rice as a model cereal for phenology. As an example, key regulatory genes for long day and short day photoperiodic control of flowering are conserved in the SD plant rice and LD plants wheat and barley, but the regulation of these genes is often reversed (Hayama et al., 2003).
The release of genome sequences for rice (Goff et al., 2002), maize (Schnable et al., 2009), barley (International Barley Genome Sequencing Consortium, 2012), and bread wheat (Mayer et al., 2014), coupled with advancements in molecular biology techniques [including DNA sequencing, RNA interference, and quantitative trait locus (QTL) analysis] as well as comparative genetics approaches, have enabled the isolation and functionally characterization of many phenology genes in cereal crops. Blast searches and phylogenetic analyses between Arabidopsis, rice, wheat, barley, and maize sequences identified many conserved as well as functionally divergent genes involved in flowering (Calixto et al., 2015). Several key pathways linked to specific environmental signals were uncovered and characterized, including temperature (thermoperiod and vernalization), day length (photoperiod), as well as autonomous and gibberellin-responsive pathways. More recently, miRNA-based regulation of gene expression at the post-transcriptional level has come into the focus of research efforts on flowering-related pathways (Teotia and Tang, 2015). In this section, we will summarize phenology genes and genetic regulation, and highlight how different flowering time regulatory mechanisms have evolved between SD, LD, and day-neutral plants.
Since originating from the Eastern Mediterranean region, the closely related Triticeae cereal crops bread wheat and barley have been adapted to a wide range of agricultural environments. Traditional breeding and phenotypic selection of natural genetic variants at flowering loci was used to optimize flowering time within a given production environment to achieve greater yields (Jung and Müller, 2009). This selection was also often associated with improved adaptations to abiotic and biotic stress factors.
Bread wheat and barley are facultative LD plants adapted to short growing seasons, with LDs promoting flowering in spring, while SDs delay reproductive development. Photoperiod and vernalization regulate the initiation of the reproductive phase and are the two main seasonal signals in temperate cereal crops. Many Pooid grasses, including wheat and barley, are vernalization responsive and photoperiod sensitive, which is believed to have evolved early during diversification as a crucial adaptation to allow for their transition into the temperate zone (Fjellheim et al., 2014). Temperate regions with long growing seasons and sufficient water supply allow cereal crops to flower late in the year and accumulate more biomass, whereas early flowering avoids abiotic and biotic stresses such as drought and pathogen attack late in the season. To date, only a relatively small number of major flowering time genes and pathways have been identified in wheat and barley (Figure 2). The following two sections on wheat (Flowering Time in Bread Wheat) and barley (Flowering Time in Barley) will give a detailed account of the current understanding of flowering time regulation in these two temperate cereal crops.
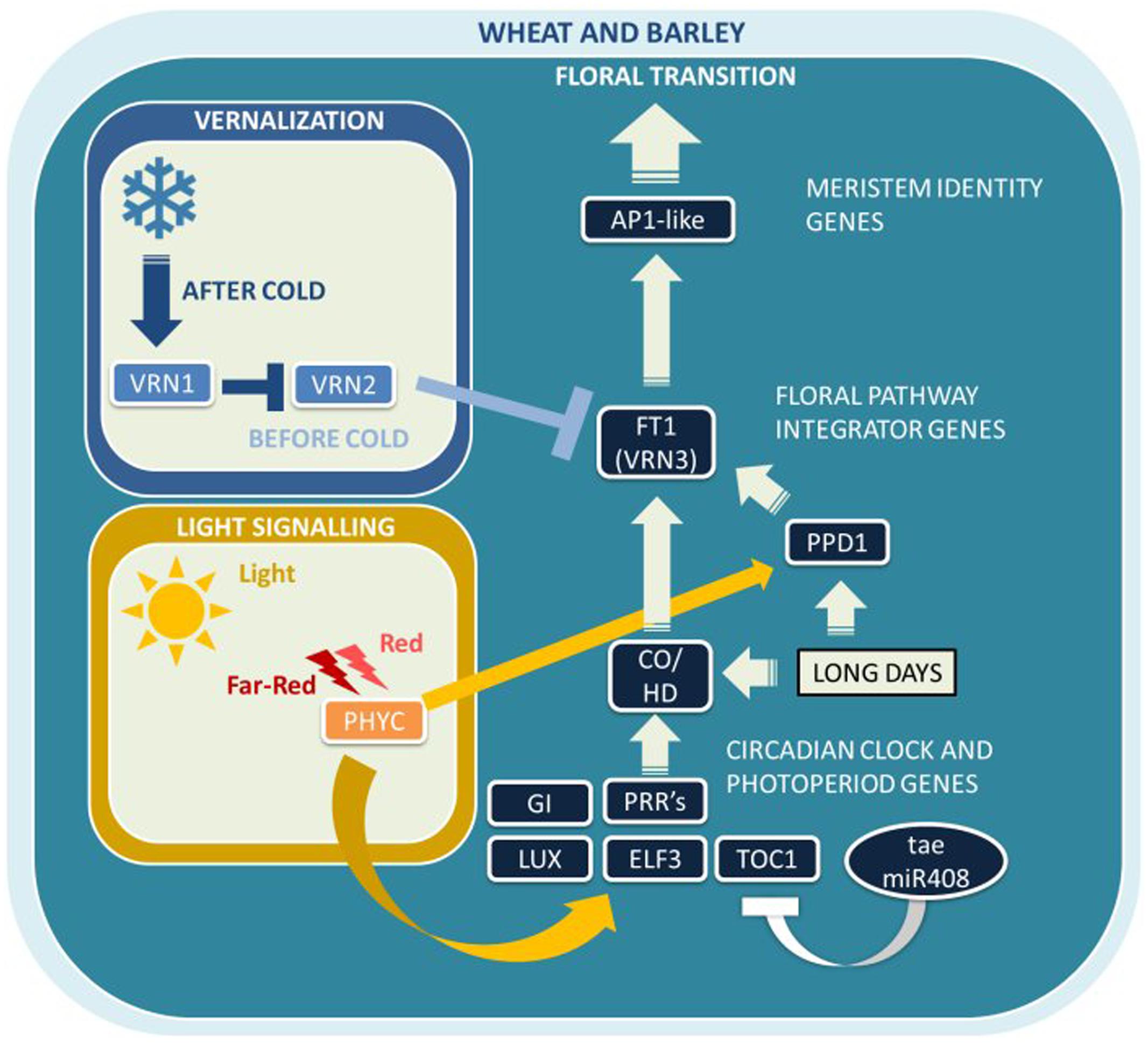
FIGURE 2. Major flowering pathway genes of bread wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.). Positive and negative regulatory connections are indicated by arrows and lines with T-ends, respectively. White arrows or T-ends indicate regulatory connections occurring primarily under long days. Gene name abbreviations are explained in Supplementary Table 1.
The process of wheat domestication involved several hybridization events between tetraploid progenitor species Triticum turgidum L. (AABB genomes) and diploid Aegilops tauschii (DD genome) that gave rise to hexaploid bread wheat (AABBDD genomes) about 8,000 years ago (Wang et al., 2013). As a hexaploid, bread wheat displays a high level of gene redundancy (one copy of the homoeologous gene in each genome), and therefore the precise function of many flowering-related genes and the current understanding of the complexity of the gene networks controlling flowering is still incomplete. Table 1 provides a summary of the current understanding of wheat phenology genes which are either functional orthologs or homologs of Arabidopsis thaliana genes.
Rapid genomic changes in polyploid wheat facilitated wheat adaptation to different growing environments. In particular, wheat crop expansion is associated with the exploitation of natural variation in the photoperiod (PPD), vernalization (VRN), and EARLINESS PER SE (EPS) genes. In bread wheat, PPD genes are relatively well understood and molecular markers have been developed to aid selection for breeders (Beales et al., 2007; Cane et al., 2013). Three copies of the gene (homeologs) that control photoperiod response in wheat, namely TaPPD-A1, TaPPD-B1 and TaPPD-D1, are located on the short arm of the homoeologous group 2 chromosomes. The region containing TaPPD-D1 in wheat is colinear with the barley photoperiod gene HvPPD-H1 on chromosome 2HS (Laurie, 1997). TaPPD-D1 encodes a pseudo-response regulator (PRR) family protein gene orthologous to the Arabidopsis PRR7 gene, and is more distantly related to AtCO and AtVRN2 (Yan et al., 2004; Turner et al., 2005).
The wild ancestors of wheat were photoperiod-sensitive, and photoperiod insensitivity in many cultivated wheat varieties is the result of mutations in PPD genes (Thomas and Vince-Prue, 1997). Dominant PPD alleles induce constitutive activation of the photoperiod pathway irrespective of day length and significantly reduce sensitivity to photoperiod. This leads to an early flowering phenotype in both SDs and LDs, which has been associated with pleiotropic effects in certain agricultural environments including in southern Europe (Worland, 1996) and Australia (Richards et al., 2014) resulting in increased grain yields. The potencies of the homoeologous group-2 PPD genes for insensitivity are ranked in the order TaPPD-D1 > TaPPD-B1 > TaPPD-A1 (Worland et al., 1998).
Pseudo-response regulator proteins are commonly found in plants and involved in circadian clock-associated pathways, including PRR1 (TOC1) (Mizuno and Nakamichi, 2005). The mRNA of TOC1 starts to accumulate at the beginning and reaching its maximum level at the end of the light period, where it directly represses the transcription of the myeloblastosis (MYB) transcription factors LATE ELONGATED HYPOCOTYL (LHY) and CCA1. In wheat, Zhao et al. (2016) recently demonstrated that the microRNA tae-miR408 targets and down-regulates expression levels of all three wheat paralogs of TaTOC1 (TaTOC-A1, TaTOC-B1, and TaTOC-D1) under both LDs and SDs, thereby regulating heading time and associated agronomic traits.
The extensive natural variation for both the requirement and responsiveness to cold temperature has played a major role in the adaptation of many species over a wide latitudinal range. Unlike the model monocot crop rice, wheat and barley flower in response to LDs. The photoperiod and vernalization pathways intersect at the regulation of TaVRN3, which is a gene encoding a phosphatidylethanolamine-binding protein (PEBP) homologous to the Arabidopsis flowering time gene AtFT1 (Yan et al., 2006). Similarly, VRN3 also encodes a mobile protein that is transported from the leaves to the SAM, where it becomes part of a flowering complex that induces TaVRN1 transcription by binding to its promoter. TaVRN1 is an AP1 clade MADS-box transcription factor, homologous to the Arabidopsis meristem identity gene AtAP1, and present on the long arms of chromosomes 5A, 5B, and 5D, respectively (TaVRN-A1, TaVRN-B1, and TaVRN-D1) (Yan et al., 2003). While vernalization in Arabidopsis results in silencing of the flowering repressor AtFLC, it induces transcription of the flowering activator TaVRN1 in wheat and other monocots. TaVRN1 is expressed in the leaves where it represses transcription of TaVRN2, which encodes a protein with a putative zinc finger and a CCT protein–protein interaction domain (Yan et al., 2004; Kippes et al., 2015). TaVRN2 represses TaVRN3 expression in LDs to prevent flowering during unfavorable conditions in autumn before vernalization. In the absence of TaVRN2, the increase in day length during spring results in the up-regulation of TaVRN3 expression, which leads to a further increase of TaVRN1 transcription trough a positive feedback loop, and ultimately to an irreversible acceleration of flowering.
A fourth vernalization locus, TaVRN-D4, was recently identified on the short arm of chromosome 5D (Kippes et al., 2015). It was shown to be paralogous to TaVRN-A1, as it originated from an insertion of a large segment on chromosome 5AL containing the TaVRN-A1 gene into chromosome 5DS. The first intron of TaVRN-A1 contains binding sites for the RNA-binding protein GLYCINE-RICH RNA-BINDING PROTEIN 2 (TaGRP2), which binds to TaVRN-A1 pre-mRNA and represses TaVRN-A1 expression (Xiao et al., 2014). The inserted copy of TaVRN-A1 carries mutations in its coding and regulatory regions including single nucleotide polymorphisms (SNPs) in the first intron that impedes binding of TaGRP2. Protein modification of TaGRP2 by O-linked β-N-acetyl glucosamine (O-GlcNAcylation) mediates interaction with VERNALIZATION-RELATED 2 (TaVER2), a carbohydrate-binding jacalin lectin, which relieves repression of TaVRN1 expression via TaGRP2 when vernalization occurs.
The genetic regulation of vernalization response in wheat (and other monocots) has some distinct differences from Arabidopsis. Comparative genomics studies detected Arabidopsis FLC-like genes in monocots, but they have yet to be shown to produce proteins of similar function in wheat or other temperate cereals (Ruelens et al., 2013). AtFLC is positively regulated by AtFRI, but to date also no functional AtFRI homologs were detected in wheat. The AtFT1 and AtFRI promoters and first introns were shown to contain cis-regulatory sites important for the transcriptional regulation of these genes in Arabidopsis (Tiwari et al., 2010; Sanchez-Bermejo and Balasubramanian, 2015).
Only comparatively little is currently known about genetic sequence variations and non-gene elements that regulate phenology gene expression in wheat. Autumn-sown winter cultivars that contain the ancestral VRN1 allele require vernalization during winter. By contrast, spring wheat varieties carrying mutations in regulatory regions including in the TaVRN1 promoter, first intron of TaVRN1, or in the TaVRN4 region (Fu et al., 2005; Kippes et al., 2015; Muterko et al., 2015), do not require vernalization and are sown in spring. Very recently, Kippes et al. (2016) developed a triple TaVRN2 mutant that contained three non-functional TaVRN2 alleles. This mutant flowered early, had a limited vernalization response, and spring growth habit. In a different study, an insertion in the promoter of TaVRN3 led to increased TaVRN3, bypassing the delay of flowering by TaVRN2 (Yan et al., 2006). Sequence variations in TaVRN3 genes were shown to cause early flowering in wheat (Ó’Maoiléidigh et al., 2014). As another recent example, copy number variation (CNV) was shown to play a major role in global wheat adaptation. Wheat lines with an increased copy number of TaVRN-A1 showed an increased requirement for vernalization, and lines with an increased copy number of TaPPD-B1 led to early flowering (Würschum et al., 2015).
The third class of genes responsible for fine-tuning of wheat flowering time are EPS genes, which can be defined as the least number of days to reproductive growth independent of vernalization and photoperiod. Also known as earliness in the narrow sense, intrinsic earliness, or basic development rate, EPS genes are hypothesized to function as a fine tune adjustment of flowering time (Zikhali and Griffiths, 2015). They are often considered as polygenic and of small genetic effect, and can be detected in both winter and spring wheats. Variation in EPS genes was shown to lead to minor or major differences in flowering time under field conditions, reported in the range of a few days up to a few weeks (Appendino and Slafer, 2003; Zikhali et al., 2014). To date, no EPS genes have been cloned, there is only a limited understanding of gene function, and furthermore, no known homologs in Arabidopsis exist that could help to fill the knowledge gaps. Instead, several studies used near isogenic lines (NILs) to narrow down the location of EPS loci on the wheat genome, and to identify several potential candidate genes (Zikhali et al., 2014, 2015). The genes MOLYBDENUM TRANSPORTER 1 (TaMOT1), FILAMENTATION TEMPERATURE SENSITIVE H (TaFtsH4) (Faricelli et al., 2010), and EARLY FLOWERING 3 (TaELF3) (Zikhali et al., 2014) were proposed as candidate genes underlying the EPS -Am1 locus. Gawroński et al. (2014) suggested a cereal ortholog of Arabidopsis circadian clock gene LUX ARRHYTHMO/PHYTOCLOCK 1 (AtLUX/PCL1) as the candidate gene underlying the EPS -3Am locus in einkorn wheat (Triticum monococcum L.). These recent findings question the previous assumption that EPS acts independently of the environment, but the exact nature of the environmental impact on EPS gene function remains unclear.
Previously, Dubcovsky et al. (2006) showed that an inductive LD photoperiod is needed to induce additional genes for normal spike development as the induction of TaVRN1 alone proved insufficient. Pearce et al. (2013) hypothesized that GAs may be involved in wheat spike development during LDs, which points to a functionally different role compared with Arabidopsis where GAs function in non-inductive SD conditions (Hyun et al., 2016). Pearce et al. (2013) applied exogenous GA to in wheat lines grown under non-inductive SDs, which resulted in an accelerated spike development only wheat lines expressing TaVRN1. Both GA and TaVRN1 are required for the up-regulation of the floral meristem identity genes TaSOC1 and TaLFY and the development of the wheat spike. Furthermore, GA biosynthetic gene expression was found to be elevated in the apices of plants transferred from SDs to LDs as well as in photoperiod-insensitive and transgenic wheat plants with increased TaFT transcription under SDs. Wheat genes for components of the GA biosynthetic pathway were identified based on homology with Arabidopsis, rice, and Brachypodium distachyon (Pearce et al., 2015), but due to the lack of a fully sequenced wheat genome, it is currently not possible to accurately determine the final number of genes in each family.
GA-sensitive and insensitive dwarfing genes have had an impact on all the main cereal crop species (Jia et al., 2009). Semi-dwarfing wheat varieties containing alleles of the REDUCED HEIGHT (RHT) DELLA genes were key factors to increasing yield during the “Green Revolution” (Flintham et al., 1997; Peng et al., 1999). Reduced crop height protected against lodging and improved harvest index, high spikelet fertility, and grain numbers per ear. RHT encodes a negative regulator of the GA signaling pathway (Pearce et al., 2013; Boden et al., 2014).
It is estimated that the common ancestor of barley and Arabidopsis shared about two-thirds of the circadian clock and clock-associated genes (Calixto et al., 2015) (Table 2). After separation into monocot and dicot classes, these genes further diversified due to extensive gene duplication and deletion events (Campoli et al., 2012a). Although some core clock genes were identified in barley based on map-based cloning (Yan et al., 2003), positional cloning (Yan et al., 2004), or homology with Arabidopsis (Yan et al., 2006), only a few are well characterized. Some of these genes were found to perform similar functions as their orthologs, whereas there is currently little evidence for other genes that would support their involvement in flowering.
The major genes controlling flowering time in barley in response to environmental cues are HvVRN-H1, HvVRN-H2, HvVRN-H3 (HvFT1), HvPPD-H1, and HvPPD-H2 (HvFT3) (Figure 2). The epistatic relationship of three genes involved in flowering regulation is well established: HvVRN-H1 and HvVRN-H2, with roles in vernalization, as well as the floral pathway integrator gene VRN-H3, which is synonymous to HvFT1 (Kikuchi et al., 2009).
Similarly as for Arabidopsis and wheat, FT-like genes in barley such as HvFT1 trigger flowering in response to LDs. HvFT1 plays a key role in promoting flowering and integrates the photoperiod and vernalization pathways: Under LDs, HvFT1 expression is low until induced by low temperatures in winter varieties that have a vernalization requirement (Yan et al., 2006). HvFT1 expression is primarily regulated by the major photoperiod response genes HvPPD-H1 and HvPPD -H2 (Laurie et al., 1994). Photoperiodic flowering under LDs is up-regulated by HvPPD-H1, which is a homolog of the Arabidopsis clock gene PRR7 (Turner et al., 2005). A mutation in the CCT domain of HvPPD-H1 is associated with lower transcript levels of HvFT1 and delayed flowering under LDs compared with the wild-type HvPPD-H1 allele, but it is not related to flowering variation under SDs (Turner et al., 2005; Hemming et al., 2008). HvPPD-H2 (with its proposed candidate gene HvFT3) enhances the expression of HvFT1 and promotes flowering under non-inductive SDs, whereas recessive mutant alleles confer delayed flowering under SDs, aiding the flowering repression over winter (Kikuchi et al., 2009).
In contrast to Arabidopsis, HvFT1 expression is induced via HvCO1 (the closest ortholog of Arabidopsis CO in barley) in both SDs and LDs (Campoli et al., 2012a). There is a high level of redundancy in both CO-like and FT-like genes, with currently nine and five members in barley, respectively (Faure et al., 2007). HvCO1 and HvCO2 are believed to be paralogs that exist due to a duplication event in temperate cereals. This functional diversification is important for the modulating of flowering responses and adaptation to different growing environments. Several studies demonstrated that functional polymorphisms in the HvFT1 gene alter its regulation, including mutations in the first intron that differentiate dominant from recessive alleles (Yan et al., 2006), and polymorphisms in the HvFT1 promoter region that lead to distinct phenotypic effects (Casas et al., 2011).
In barley, the VRN-H2 locus consists of three homologous CO-like genes, HvZCCT-Ha, HvZCCT-Hb, and HvZCCT-Hc (Dubcovsky et al., 2005; Karsai et al., 2005). VRN-H2 functions as a floral repressor of HvFT1 (VRN-H3) to delay flowering in plants that have not been vernalized (Trevaskis et al., 2006; Hemming et al., 2008). VRN-H2 is only expressed under LD conditions controlled by components of the circadian clock. Mutations in HvELF3 resulted in the expression of VRN-H2 under SD conditions due to an increased expression of PPD-H1 and, consequently, HvFT1 (Faure et al., 2012; Turner et al., 2013).
In winter barley cultivars, vernalization induces VRN-H1 to repress VRN-H2 which promotes the transition from vegetative to reproductive development (Trevaskis et al., 2006; Hemming et al., 2008). Sequence variations in the first intron of VRN-H1 alter the vernalization requirement for activation and, consequently, the repression of VRN-H2 and overall flowering behavior (Hemming et al., 2008, 2009). A recent survey showed that the down-regulation of VRN2 by cold is exclusively found in Pooid grasses including wheat and barley (Woods et al., 2016).
By contrast, spring barley cultivars are characterized by natural mutations at PPD-H1 and by deletions of the VRN-H2 locus and thus do not require vernalization (Dubcovsky et al., 2005). HvCO2 overexpression in spring barley was shown to induce flowering due to an increased expression of PPD-H1 and, as a direct result, HvFT1 (Mulki and von Korff, 2016). By contrast, overexpression of HvCO1/CO2 increased HvVRN-H2 expression in winter barley carrying the VRN-H2 locus, which led to lower HvFT1 levels and delayed flowering independently of photoperiod. Putative additional epistatic interactions of HvVRN-H2 with HvGI and HvCO1 point toward a role of HvVRN-H2 as an integrator of photoperiod and vernalization signals (Maurer et al., 2015).
In addition to the photoperiod and vernalization pathway genes, flowering time in barley is controlled by the EARLY MATURITY (EAM) loci referred to as EARLINESS PER SE (EPS) in wheat. The red/far-red light photoreceptor HvPHYC, an ortholog of Arabidopsis PHYC, was identified to underlie the EARLY MATURITY 5 (EAM5) QTL (Pankin et al., 2014). HvPHYC was found to promote light signal transmission to the circadian clock, modulating photoperiodic regulation of floral transition. It was also shown to interact with the PPD-H1 pathway and to increase HvFT1 expression not associated with the circadian clock (Nishida et al., 2013; Pankin et al., 2014). Similar mechanisms were discovered in wheat, where TaPHYC was found to activate TaPPD1 and TaVRN3 in inductive LDs (Chen et al., 2014).
The circadian clock gene HvLUX1, an ortholog of the Arabidopsis circadian gene LUX ARRHYTHMO, was detected as a candidate underlying the barley EAM10 locus (Campoli et al., 2013). As shown for barley plants carrying a mutation in EAM5 (HvPHYC), mutations in EAM10 (HvLUX1) altered the expression of HvPPD-H1 (as well as CCA1 in barley) and accelerated flowering under both LDs and SDs independently of the circadian clock. Furthermore, as shown for barley EAM8 mutants (Faure et al., 2012), early flowering of EAM10 mutants was linked with an up-regulation of HvFT1 transcription under SDs. In both cases this led to an induction of the LD photoperiod pathway under non-inductive (SD) conditions, suggesting that EAM10 acts as a repressor of HvPPD-H1.
In barley, HvCEN, a homolog of Antirrhinum majus CENTRORADIALIS, was identified at the EARLINESS PER SE 2 (EPS2) locus on chromosome 2H (Comadran et al., 2012). Antirrhinum cen mutants produce terminal flowers instead of indeterminate inflorescences common for wild-type plants without, affecting flowering time (Bradley et al., 1996). Analysis of the HvCEN alleles of a diverse set of spring and winter barley cultivars revealed that HvCEN facilitated the geographic range extension as well as the gradual separation between spring and winter cultivars (Comadran et al., 2012). A collection of flowering mutants was used to confirm sequence variations within HvCEN to be responsible for the observed variation in flowering time. While CEN homologs have been identified and characterized in rice (Nakagawa et al., 2002) and maize (Danilevskaya et al., 2008a), none have been identified to date in hexaploid wheat.
The circadian clock gene HvELF3, an ortholog of Arabidopsis ELF3, was identified to underlie the EARLY MATURITY 8 (EAM8) QTL (Faure et al., 2012; Zakhrabekova et al., 2012). Mutations in EAM8, also known as praematurum.a-8 (mat-a-8), were used since 1961 to facilitate short growing season adaptation and expansion of the geographic range in commercial barley varieties (Zakhrabekova et al., 2012). A loss-of-function mutation of HvELF3 led to an increased expression of HvFT1 compared with wild-type plants, causing a day-neutral early flowering phenotype (Faure et al., 2012). Boden et al. (2014) investigated barley plants carrying mutations in the HvELF3 gene and characterized them as early flowering under non-inductive SD photoperiods. The enhanced expression of HvGA20OX2 increased GA biosynthesis and expression of HvFT1. In spring barley, HvELF3 is necessary to maintain photoperiodic sensitivity through repression of HvFT1 and production of active GA via HvGA20OX2, a link not previously shown for AtELF3.
The phytohormone GA promotes flowering in barley and is essential for normal flowering of spring barley under inductive photoperiods (Boden et al., 2014). One of the gibberellin 20-oxidase genes, HvGA20OX2, was identified as the candidate gene underlying the allelic dwarfing gene sdw1/denso. sdw1/denso is an important locus selected for in barley breeding programs as the resulting semi-dwarf phenotype has reduced plant height and improved lodging resistance associated with higher yields and quality traits. The sdw1 mutant was distinguished from the denso mutants based on the more strongly reduced expression of HvGA20OX2 which was also linked to reduced plant height, enhanced grain yield, and lower grain quality (Jia et al., 2011).
Rice and maize are tropical SD plants and, unlike wheat and barley, do not require vernalization. However, a high level of conservation of photoperiod pathway genes with more distantly related plants like Arabidopsis and temperate cereals is found in both rice and maize. If similar genes controlling flowering are involved in plants that flower under LD photoperiods (such as temperate cereals and Arabidopsis), which regulatory mechanisms generate the reverse response to photoperiod in rice?
Two independent pathways act in response to different photoperiods to control flowering time in rice. Day-light periods of less than 13.5 h accelerate flowering of rice (Itoh et al., 2010). Under inductive SD conditions, a HEADING DATE 1 (HD1)-dependent pathway is activated in which the AtCO homolog HD1 accelerates flowering via activation of the AtFT ortholog HEADING DATE 3A (HD3A) (Kojima et al., 2002). This pathway is conserved between Arabidopsis and rice (Table 3). For heading initiation under non-inductive LD conditions, an HD1-independent pathway is activated (Yano et al., 2000). The activity of genes unique to rice play a major role in conferring the different seasonal flowering responses when compared to LD plants such as Arabidopsis and are highlighted in the following sections.
As rice does not require vernalization, the photoperiodic pathway is of great significance to control flowering, often referred to as heading date. Similarly as for Arabidopsis, the mobile flowering signal HD3A is expressed in leaves and the protein is translocated to the SAM where it accelerates floral development (Kojima et al., 2002). The key difference between the genetic regulation of Arabidopsis and rice flowering time is that unlike its ortholog AtFT, HD3A is only expressed under SD conditions (Figure 3).
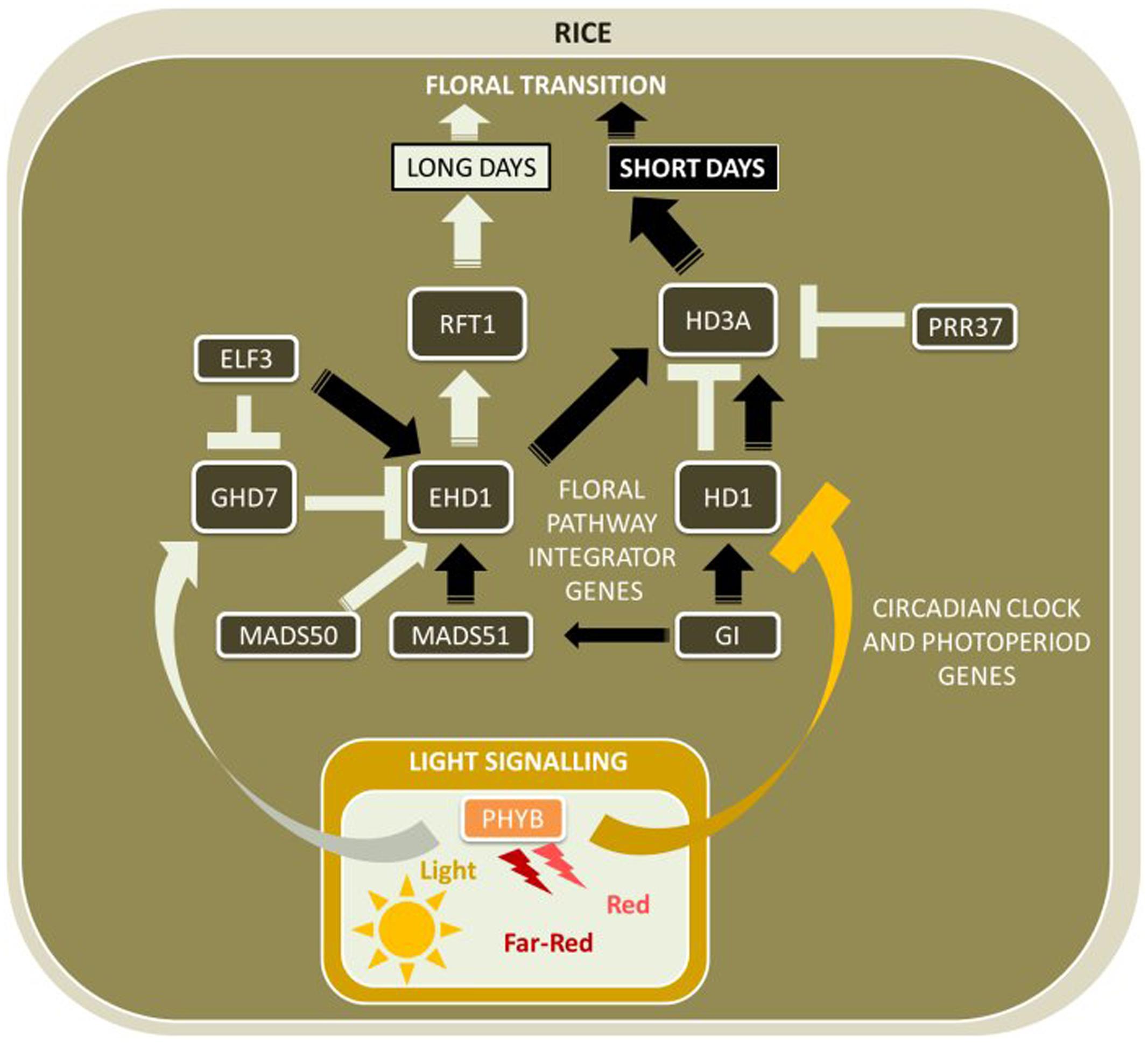
FIGURE 3. Major flowering pathway genes of rice (Oryza sativa). Positive and negative regulatory connections are indicated by arrows and lines with T-ends, respectively. White and black arrows or T-ends indicate regulatory connections occurring primarily under long days and short days, respectively. Gene name abbreviations are explained in Supplementary Table 1.
HD1 is expressed with a diurnal rhythm similar to AtCO, with a peak in expression in the afternoon in LD photoperiods (Yano et al., 2000). However, unlike AtCO, which activates AtFT in LDs, high HD1 activity in LDs is associated with low expression of HD3A (Hayama et al., 2003). OsGI is also expressed with a diurnal rhythm similar to its ortholog AtGI, and likely has a similar role in regulating HD1 expression (Hayama et al., 2002, 2003).
Other components of the circadian clock modify the activity of genes acting in the photoperiod pathway in rice. For example, the transcript of the VRN2-like CCT-motif gene GRAIN NUMBER, PLANT HEIGHT, AND HEADING DATE 7 (GHD7) is activated by red light signaling via phytochromes in the morning of LDs (Xue et al., 2008). GHD7 represses the expression peak of the B-type response regulator EARLY HEADING DATE 1 (EHD1), a gene without a known homolog in Arabidopsis, at the start of the light period to delay flowering under LD. EHD1 is known to confer promotion of flowering under SDs (the exact mechanism is currently unknown), and mutations in the DNA-binding domain or gene suppression via RNAi decreased HD3A expression under SD photoperiods (Doi et al., 2004; Kim et al., 2007). Another gene with a CCT motif, OsPRR37, is an ortholog to the circadian clock gene AtPRR3/7, and although expressed independently of photoperiod, is only functional to repress HD3A under LDs (Murakami et al., 2007). GHD7 was also found to suppress the expression of HD3A and to delay flowering in LDs but does not affect HD3A expression under SDs, indicating that GHD7 is further upstream of both HD3A and EHD1 in the gene network that controls rice flowering.
Cultivated rice is widely grown throughout Asia, including geographical areas in northern Asia with long light periods of up to 14.5 h per day during the growth period (Izawa, 2007). To avoid cold temperatures during grain filling common to these regions, photoperiod insensitive, early flowering and rice cultivars are required (Izawa, 2007). Although flowering in rice is normally restricted to SDs, rice cultivars were developed with the ability to flower under LD conditions. The pathway involved in floral activation under LD conditions involves OsMADS50, a homolog of AtSOC1, which activates EHD1 expression, which in turn activates expression of RICE FLOWERING LOCUS T 1 (OsRFT1), the closest homolog of HD3A (Komiya et al., 2009; Ryu et al., 2009). HD3A and OsRFT1 are both mobile flowering signals, located very closely on the same chromosome, but with inverse functions - HD3A is activated under LD, whereas OsRFT1 expression is activated under SD conditions. Also, natural variation of OsPRR37 (Koo et al., 2013) and major-effect HD-QTLs (HD1, HD2, HD4, and HD5) (Li X. et al., 2015), were found to modulate photoperiod sensitivity and to adapt rice varieties to cultivation at a wide range of latitudes. To date, no homologs in Arabidopsis or temperate cereals are known for any genes of this alternate pathway.
Inductive SD conditions activate a conserved pathway between rice and Arabidopsis, in which HD1 promotes flowering via activation of HD3A (Kojima et al., 2002). Therefore, HD1 has two opposing functions in the flowering network – to promote flowering under SD, but also to inhibit flowering under LD photoperiods via interaction with HD3A. Similarly, OsELF3, orthologous to AtELF3, has a dual role, as it promotes flowering in SDs via activation of EHD1, but also represses GHD7 to promote flowering in LDs (Zhao et al., 2012). Very recently, a novel flowering time pathway unique to monocot plants was discovered with HD1, GHD7, and EHD1 as its key genes: HD1 was shown to directly interact with GHD7 to form a complex that specifically bind to a cis-regulatory region in EHD1 resulting in repression of EHD1 gene expression only under non-inductive LD conditions (Nemoto et al., 2016).
As a gene unique to rice, EHD1 is a key enabler of flowering transition under SD conditions by enhancing expression of HD3A and OsRFT1 independently of HD1 (Doi et al., 2004). EHD1 expression is activated by blue light in the morning and controlled via OsGI.EHD1 was found to be up-regulated by the MADS-box gene OsMADS51 (Kim et al., 2007), which itself is also positively regulated by OsGI (Kim et al., 2007). The gene INDETERMINATE1 (OsID1) triggers the expression of HD1 and HD3A for flowering under SDs (Matsubara et al., 2008), and as for OsMADS51, no known homologs in Arabidopsis were detected.
The heterotrimeric heme activator protein (HAP) complex regulates flowering in Arabidopsis through binding to the CCAAT box, a cis-acting element (Ben-Naim et al., 2006). The gene GRAIN NUMBER, PLANT HEIGHT, AND HEADING DATE 8 (GHD8) encodes a CCAAT box-binding protein belonging to the HAP3 subfamily in rice, and was recently cloned (Yan et al., 2011). GHD8 is homologous to AtHAP3b and acts upstream of EHD1, HD3A, and OsRFT1 in the rice flowering pathway. Under SD conditions, GHD8 enhances, and under LD conditions inhibits gene expression of EHD1, HD3A, and OsRFT1 via a coordinated interaction with GHD7.1, a PSEUDO-RESPONSE REGULATOR (PRR) gene (Yan et al., 2013). GHD7, GHD8, and GHD7.1 have pleiotropic effects on heading date, grain yield and plant height, and together with HD1 are the key rice genes that defines grain yield and environmental adaptability to rice growing regions.
GA signaling affects floral organ development and pathways are largely conserved between rice and Arabidopsis. The casein kinase I (CKI) gene EARLY FLOWERING1 (OsEL1) is associated with the negative regulation of GA-responsive signaling via post-translational modification of SLENDER RICE 1 (OsSLR1) protein. Mutations in OsEL1 lead to an enhanced response to GA signaling, and as a consequence to early flowering, lower spikelet fertility and thus lower grain yield under non-inductive LDs. However, a few extremely early flowering rice cultivars contain the mutation in OsEL1 but can maintain normal GA signaling through a currently unknown mechanism.
The rice gene SEMI-DWARF1 (OsSD1) contributed to the major increases in rice yield during the “Green Revolution” and is involved in GA signaling and biosynthesis (Peng et al., 1999). A similar reduced-height phenotype is the result of a loss-of-function mutation in OsGA20OX2, orthologous to the GA biosynthetic gene HvGA20OX2 in barley (Spielmeyer et al., 2002). HvGA20OX2 was proposed as the candidate gene underlying sdw1/denso locus and confers a reduced plant height phenotype in barley (Jia et al., 2011).
Maize was domesticated about 9,000 years ago from the wild grass teosinte (Zea mays ssp. parviglumis), a native to tropical Central America (Matsuoka et al., 2002). Teosinte requires SD photoperiods to flower, and domesticated maize has been adapted since then to cooler temperate regions of North America and Europe (Chardon et al., 2004). Maize post-domestication breeding was particularly driven by selection for genes and loci to adapt flowering time to new growth environments (Mascheretti et al., 2015). Photoperiod insensitive (day-neutral) maize varieties are cultivated in temperate climates, whereas photoperiod sensitive maize varieties are grown in tropical regions.
Flowering time in maize can range from only 35 up to 120 days, highlighting the high level of genetic diversity in maize phenology genes (Colasanti and Muszynski, 2009). Only a few mutations in flowering time genes have been identified so far, which contributes to the lack of current understanding of genetic and regulatory factors that determine maize phenology. Our current knowledge about maize flowering time is mainly based on results from QTL meta-analysis, (transposon) mutagenesis, and comparative genomics studies with the Arabidopsis and rice genomes (Buckler et al., 2009; Wei et al., 2015). Key phenology genes are listed in Table 4.
Maize contains a large family of Zea CENTRORADIALIS (ZCN) genes and related to both AtFT and AtTFL (Danilevskaya et al., 2008a). The most likely candidate for the FT ortholog in maize is Zea CENTRORADIALIS8 (ZmZCN8), a gene encoding a PEBP with high sequence similarity to the Arabidopsis flowering time gene AtFT1. In addition, a second maize florigen, the ZmZCN8 paralog ZmZCN7, was proposed (Mascheretti et al., 2015). Displaying florigen-like characteristics, ZmZCN8 is expressed only in the leaf phloem of mature leaves and interacts with the FD-like bZIP protein DELAYED FLOWERING1 (ZmDLF1) in the SAM (Figure 4). ZmZCN8 expression is elevated following a diurnal cycle under SDs in photoperiod-sensitive tropical maize, whereas ZmZCN8 levels remained unchanged in day-neutral temperate maize (Muszynski et al., 2006; Meng et al., 2011). Ectopic expression of ZmDLF1 resulted in an early flowering phenotype whereas silencing led to a severe delay flowering, as found for its functional equivalent OsID1 in rice (Matsubara et al., 2008). A putative AtCO ortholog in maize, ZmCONZ1, was located at a chromosome region syntenic with the rice ortholog HD1 and exhibits a diurnal expression pattern dependent on photoperiod (Miller et al., 2008). However, and there is currently no evidence that ZmCONZ1 can directly activate ZmZCN8 expression in SDs.
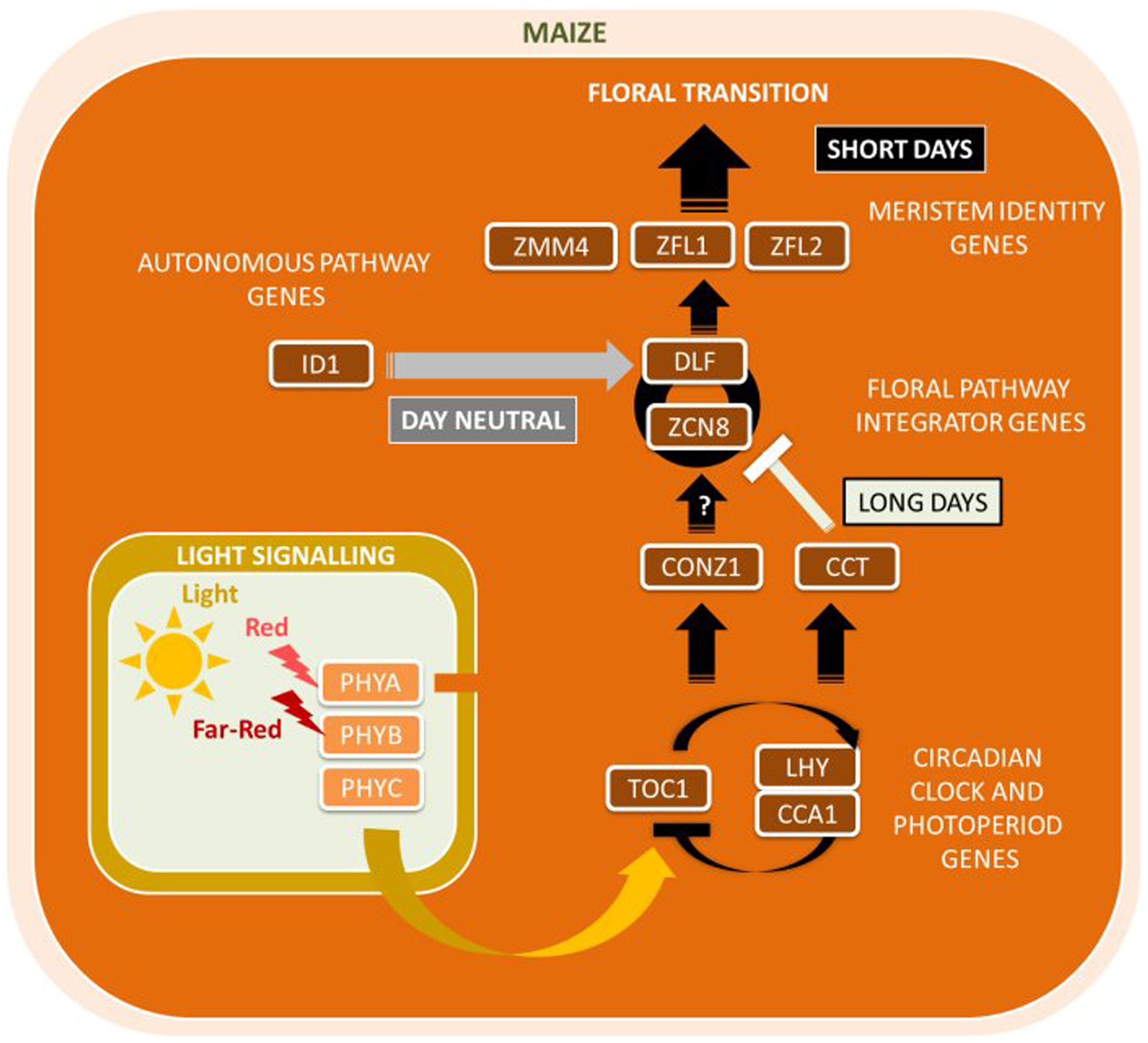
FIGURE 4. Major flowering pathway genes of maize (Zea mays). Positive and negative regulatory connections are indicated by arrows and lines with T-ends, respectively. White, black, and gray arrows or T-ends indicate regulatory connections occurring primarily under long days, short days, or independently of day length, respectively. Gene name abbreviations are explained in Supplementary Table 1.
Other genes in the maize flowering time network include Zea FLORICAULA/LEAFY genes (ZmZFL1 and ZmZFL2) (Bomblies et al., 2003). ZmZFL1 and ZmZFL2 are orthologous to the meristem identity genes Antirrhinum majus FLORICAULA (AmFLO) and AtLFY and share conserved roles in controlling inflorescence architecture and patterning (Bomblies et al., 2003). Among other genes with likely conserved functions between maize and Arabidopsis are phytochromes ZmPHYA, ZmPHYB, and ZmPHYC (Sheehan et al., 2004). Unique for maize are Miniature Inverted-Repeat Transposable Element (MITE) sequence insertions detected in ZmPHYB2 that showed that the absence of the MITE correlated with enhanced ZmPHYB2 mRNA expression compared to ZmPHYB2 with MITE (Zhao et al., 2014). This evidence points toward a role of MITEs to inhibit ZmPHYB2 mRNA expression which ultimately leads to delayed flowering. Similarly, a MITE insertion into the VEGETATIVE TO GENERATIVE TRANSITION (VGT1) locus was associated with early flowering (Castelletti et al., 2014). The CCT domain-containing protein ZmCCT was found to be associated with photoperiod sensitivity and to regulate maize flowering time under LD conditions (Yang et al., 2013). The insertion of a CACTA-like transposon into its promoter represses ZmCCT gene transcription and thus reduced maize photoperiod sensitivity under LDs. Yang et al. (2013) provided evidence that the CACTA-like transposon was originally inserted in a tropical maize plant sensitive to SDs and was then accumulated selectively post-domestication as maize adapted to a range of LD environments.
Compared to flowering sensitive to vernalization as found in the temperate grasses wheat and barley or photoperiod-induced pathways as found in tropical rice, endogenous cues are more crucial for flowering than environmental signals in day-neutral temperate maize. The floral promoter INDETERMINATE1 (ZmID1) is a transcriptional regulator of the maize autonomous flowering pathway that functions in developing leaves to promote flowering (Wong and Colasanti, 2007). ZmID1 encodes a monocot-specific C2H2-type zinc finger protein without any known Arabidopsis ortholog and is hypothesized to act independently of the photoperiod pathway (Colasanti et al., 2006). Recent reports show that ZmID1 expression levels are high in developing leaves, and transcript and protein levels remain stable throughout the light cycle (Wong and Colasanti, 2007; Coneva et al., 2007). Mutations in ZmID1 lead to an extended vegetative phase (Colasanti et al., 2006). Genetic and expression data has shown that ZmID1 activates DELAYED FLOWERING1 (ZmDLF1) in the SAM, most likely indirectly (Muszynski et al., 2006; Meng et al., 2011), which is necessary for flowering through its interaction with ZmZCN8. Subsequent expression of floral identity genes, such as the floral transition MADS box gene ZmZMM4, then initiate reproductive development (Danilevskaya et al., 2008b).
Very recently, transcription and chromatin modifications of ZmZCN8 and its paralog ZmZCN7, a potential second maize florigen, were analyzed during floral transition in day-neutral maize and tropical teosinte (Mascheretti et al., 2015). This study led to the proposal of an alternative epigenetic mechanism of ZmID1-mediated regulation of ZmZCN8 expression in which ZmID1 establishes chromatin modifications in developing leaves of day-neutral maize to enable the expression of florigen genes ZmZCN8 and ZmZCN7 in the mature leaf later in development (Mascheretti et al., 2015). By contrast, a different set of ZmZCN8 chromatin modification patterns were detected in teosinte in response to inductive SDs, which highlights both conserved and unique features of epigenetically controlled flowering time mechanisms between autonomous and photoperiod-dependent pathways in maize.
GA accumulation and signaling has a direct positive affect on flowering time in maize (Thornsberry et al., 2001). The maize gene DWARF8 (ZmD8) encodes a DELLA protein orthologous to both Arabidopsis GIBBERELLIN INSENSITIVE (AtGAI) and the wheat REDUCED HEIGHT mutations TaRHT-B1 and TaRHT-D1, which were used to develop the high-yielding semidwarf varieties of the “Green Revolution” (Peng et al., 1999). Polymorphisms in ZmD8 were associated with differences in flowering time (Thornsberry et al., 2001). Very recently, ZEA MAYS GA REGULATORY FACTOR (ZmGRF) was identified as a new member of the bZIP protein transcription factor family in maize (Xu et al., 2015). ZmGRF transgenic Arabidopsis plants had enhanced GA levels indicating that ZmGRF has a function in plant morphology and development, and Arabidopsis has a currently unknown ortholog of ZmGRF.
Global food production is not increasing fast enough to meet the needs of the rapidly growing human population, and at the same time, global warming as a result of climate change threatens the productivity of existing agricultural land (United Nations Food and Agriculture Organization, 2009). In cereal crops, the correct targeting of flowering time to a narrow seasonal window is directly linked with an increase in grain yield. Climate change and global warming strongly alter phenological cycles and decrease yields of cereals and other crops through rising temperatures and elevated atmospheric carbon dioxide (CO2) levels, and more frequent extreme weather events such as drought. This poses a significant challenge to crop growers as they have to adjust crop management practices including sowing dates to achieve flowering at the optimal time. Optimal timing of flowering has also implications for a variety of other traits such as plant vigor, water-use efficiency, and yield (Ni et al., 2009; Kenney et al., 2014). Also, susceptibility to abiotic and biotic stresses and fertilizer requirements vary during the course of plant development. A major challenge for crop breeding is how to plan and adapt breeding efforts to future climate change scenarios.
The Intergovernmental Panel on Climate Change Reports (IPCC) predicts that the current CO2 concentrations will more than double by 2050, and by 2100 more than triple (IPCC, 2013). Plants play a central role in mitigating the effects of rising atmospheric CO2 through photosynthesis which converts solar energy into energy stored in starch and other carbohydrates. Elevated CO2 was shown to increase photosynthesis and growth of many plant species (Long et al., 2004; Wang et al., 2015). Changes in phenology and plant size due to high CO2 is believed to be an indirect consequence of the effect of CO2 on higher photosynthetic rate resulting in enhanced plant growth. These changes can be highly variable depending on other environmental factors, plant species, and length and level of CO2 exposure, with some studies showing that elevated CO2 accelerates flowering particularly of crop species with a weaker or no effect on many wild plant species (Springer and Ward, 2007). An early study by Reekie et al. (1994) found that elevated CO2 enhanced flowering in LD plans, but delayed flowering in all SD plants grown under inductive photoperiods. By contrast, an experiment that exposed a species-rich temperate grassland to both pure-CO2 free-air CO2 enrichment (FACE) and infrared warming only detected an effect of higher temperature on phenology but no effect of elevated CO2 without experimental warming (Hovenden et al., 2008).
In a meta-analysis conducted by Taub et al. (2008) elevated CO2 was found to reduce grain protein concentration of wheat, barley, and rice of 10–15% compared to levels detected at ambient CO2 levels. A reduction in protein concentration was also detected for other crops including potato and soybean, but the magnitude of the effect varied was dependent on the experimental facilities and procedures. Such changes in grain nutritional composition have important implications for the malting industry for malting barley varieties, as well as the consumers of crop plant material.
Little research has been conducted so far trying to address the molecular pathways and mechanisms that drive high-CO2-mediated changes of flowering time. Elevated CO2 was shown to alter the accumulation of photosynthates including sugars and starch (Springer and Ward, 2007). Elevated CO2 was shown to interact with both sensing and subsequent transduction of light signals in the Arabidopsis photoperiod pathway, which varied with the direction and magnitude of photoperiod (Song et al., 2009). The impact of elevated CO2 on time of flowering was linked to enhanced plant growth and an increased number of leaves at flowering. Ward et al. (2012) identified a QTL that affects flowering time at elevated CO2 levels, also in Arabidopsis, with no connection to photoreceptors. Instead, the gene MOTHER OF FT AND TFL1 (AtMFT), a homolog of the flowering inducer AtFT and the inflorescence architecture gene AtTFL1, was identified as the underlying candidate gene.
Several reports on the impact of elevated CO2 on plant phenology often provide ambiguous results or are even contradicting - elevated CO2 leads to early flowering in some species, delays flowering in others, or shows no effect. Clearly, more work is needed to elucidate the mechanisms and causes of rising CO2 levels on flowering time and quality traits of plants including crops.
Temperature affects plant development rates in many plant species as well as vernalization in winter crops. Plants require a certain number of degrees accumulated per day above a defined temperature threshold called thermal time, also known as growing degree days (GDD) or units (GDU), to reach the next developmental stage (Bonhomme, 2000). Warmer temperatures often increase the rate of development (Craufurd and Wheeler, 2009). It is forecast that rising CO2 levels will also invariably drive temperatures higher, and their combined effects were shown to be more severely affecting flowering time than the individual effects (Hovenden et al., 2008; Johnston and Reekie, 2008). In wheat, elevated CO2 had a positive effect on root and shoot biomass, which was alleviated when plants were additionally exposed to high temperatures (Benlloch-Gonzalez et al., 2014). Field-based experiments using elevated CO2 combined with supplementary heating (temperature FACE, T-FACE) and different sowing date treatments exposed wheat plants to temperatures ranging from below 0°C to above 40°C (White et al., 2011). This resulted in significantly early heading date as a response to increasing temperature.
Current knowledge on molecular pathways and mechanisms impacted by temperature-mediated changes of flowering time is poor. Recent studies suggest a role of MADS-box genes in response to high temperatures in Arabidopsis: AtFLC, the best studied temperature-responsive MADS-box gene, was shown to prolong the circadian period at higher temperatures as a means to compensate the circadian clock (Edwards et al., 2006).
Flowering time genes and regulators often have pleiotropic effects on multiple agronomically important traits, including the number and size of seeds, spikelet fertility, growth vigor, and stress tolerance (Xue et al., 2008; Ni et al., 2009; Andres and Coupland, 2012; Kwon et al., 2015). A major driver of crop evolution and adaptation, flowering time genes are key selection factors for crop breeding.
The optimization of flowering time of cereal crops to target environments under different environmental conditions is key to adapting future crops to changing environments for increased crop productivity, and to meet the goal to increase world food supply by 70% by 2015 (Anonymous, 2009). When extreme weather events coincide with sensitive growth stages in crops, it can have devastating effects on plant development and yield. Crop models have been developed with the goal to simulate interactions between genotype × environment × cropping system to predict and optimize phenology, crop yields, and other agronomic parameters in response to climate variations and to support ideotype plant breeding (Rötter et al., 2015). Phenological stages of crop plant are simulated in response to photoperiod and temperature, which impacts the development rate and vernalization requirement, as well as their interaction (Keating et al., 2003).
Model-based predictions of these interactions can be used for
(i) ideotyping: identifying breeding targets that define crop growth and development in a given environment,
(ii) agronomic diagnosis: characterizing the growth environment, and
(iii) cultivar choice: facilitating cultivar selection for a given environment and cropping system (Jeuffroy et al., 2014).
The most frequently used genotypic characteristics in crop models relate to phenology traits. For example, in a study representing wheat growing regions in Europe with the aim to optimize wheat yields for future climate scenarios, Semenov et al. (2014) listed nine cultivar parameters, with three of them directly related to plant phenology (phyllochron, day length response, and duration of grain filling). Stresses at booting were also found to impact yield more severely than at anthesis.
In environments characterized by a scarcity of water during growth periods, crop phenology can be the main factor that limits yield potential and can explain much of the yield variability. For example, in Australia, which is characterized by warm winters, hot and dry spring-summers, low-fertility soils and highly variable rainfall, increasingly earlier maturity in the original wheat cultivars introduced from Europe have been a key adaptation to increase yields in these predominantly dry rainfed growing environments (Richards et al., 2014). This allowed crops to flower and fill grain sufficiently early to escape terminal drought stress and high temperatures that were favorable for the spread of diseases. By contrast, in temperate and more humid environments as found for example in Western Europe, biotic stresses including crop diseases are the main contributors of yield limitation and are rarely addressed by model simulations (Jeuffroy et al., 2014).
The model performance of individual models was shown to have limited capability to estimate yield across different experiments consistently (Li T. et al., 2015). To improve the quality of the modeling of climate change impacts on agriculture, long-term empirical data sets of high quality for model calibrations and testing need to be made available. The Agricultural Model Intercomparison and Improvement Project (AgMIP) is composed of international teams and linkages of climate, crop, and economic modeling research that work together to improve crop simulation models1. The Knowledge Hub FACCE MACSUR is based in Europe and brings together 18 participating European institutions to facilitate the modeling of climate change impacts on European agriculture2.
The potential for crop improvement using phenology-related traits has become a major research focus in recent years (Turner et al., 2005; Cockram et al., 2007; Greenup et al., 2009; Bendix et al., 2015; Müller et al., 2016). A clear understanding of the natural variation at the loci and underlying genes is a key prerequisite to enable the development of varieties adapted to future climates. Phenology gene and allelic information have demonstrated value for breeding improved cereal crops and were used to fine-tune adaptation to different geographic regions and climatic conditions (Porker et al., 2015). This has allowed growers and breeders in the past to produce elite varieties with optimal flowering time for various target environments that achieved high yields at least for current climates (Table 5). For example, polymorphisms in vernalization requirement (VRN) genes, photoperiod sensitivity (PPD) genes, EAM loci, as well as EPS loci were selected in wheat and barley which together account for much of the genetic variation in flowering time in these crops (Zakhrabekova et al., 2012).
Although considerable progress regarding model improvement has been made, difficulties remain that are often linked to high genotype × environment interactions. Gene-based models are a new concept as part of the future direction of crop modeling that utilizes a large amount of genetic data generated by molecular genetics techniques in the laboratory. They have the potential to allow in silico identification of the best allelic combination for a given set of environments. For example, Zheng et al. (2013) recently presented a gene-based model to predict wheat heading time across different environments along the Australian wheat belt. The effects of TaPPD-D1 genes were combined with three homoeologous TaVRN1 genes (TaVRN1-A1, TaVRN1-B1 and TaVRN1-D1) to explain variation for heading time. In this study, the winter allele of TaVRN-A1 had the strongest effect on delaying heading time compared with the effects of the winter alleles of TaVRN-B1 and TaVRN-D1. The gene-based model was built to predict wheat phenology based on the modeling framework Agricultural Production Systems Simulator (APSIM) using gene parameters for TaVRN1 and TaPPD-D1, showing an improved performance over existing gene- and QTL-based models.
Simulations of crop yield using their allelic values at QTL as an input in multi-environment crop models are promising to enable predictions for the beneficial or adverse effect of a given combination of alleles on plant performance and yield (Tardieu and Tuberosa, 2010). Bogard et al. (2014) proposed a QTL-based model to predict heading time in wheat, also using genotype vernalization requirements and photoperiod sensitivity as model parameters. The main difference to other gene-based approaches is that this model did not make a priori assumptions about which exact genes determine model parameters. Earlier examples of gene and QTL-based model prediction of flowering also exist for maize (Reymond et al., 2003), rice (Nakagawa et al., 2005), and barley (Yin et al., 2005).
The degree of conservation and functional diversification of phenology genes between Arabidopsis thaliana and cereal crops has been researched extensively since the release of cereal crops reference genomes. As a consequence, major discoveries have been made in recent years for wheat, barley, rice, and maize, but further developments are needed to translate these findings into improved yield in the field, particularly in the following areas:
(i) Identification of all genomic regions containing phenology and related genes for agronomically important cereal crops: The next phase of model development and validation is to incorporate genome-wide association mapping (GWAS) and genotyping-by-sequencing (GBS) technologies to maximize the rate of trait discovery and improve phenotypic prediction under diverse environments.
(ii) More extensive genetic characterization of cereal crop germplasms to determine allelic diversity of phenology and related genes and their effects on flowering time.
(iii) Development of diagnostic markers to capture the range of allelic variation for major phenology genes in cereal crops: The utilization of existing phenology gene alleles to combine traditional breeding techniques with modern biotechnology using marker-assisted selection will further increase the efficiency of introgression of favorable alleles into elite crop cultivars.
(iv) Improvements in accurate field phenotyping of phenology-related and agronomic traits across a range of latitudes: Crop responses to a combination of elevated CO2 and high temperature can vary depending on the experimental setup, and conclusions are often based on experiments conducted in controlled environment chambers. The development of field facilities to test several environmental factors on field crops remains a major challenge, and comparably little information is currently available on crops responses to elevated CO2 × high-temperature interactions under field conditions.
(v) Advancements in simulation methodologies to link gene sequence information and environmental parameters with performance in the field: Crop simulation models have been developed to provide new insights into how cereal crop varieties may respond under different climate change scenarios and associated stresses, and to design model-aided ideotypes tailored to specific cropping environments. A better understanding of the interactions between photoperiod and high temperature is required to predict responses to of future climates. Also, the significant impact of phenology-related genes on grain yield remains to be integrated into such models.
CH: reviewed the literature and wrote the paper; CL: developed the project idea and finalized the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer YX and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
This work was supported by funding from the Grains Research and Development Corporation (GRDC) of Australia (DAW00240), Department of Agriculture and Food Western Australia (DAFWA), and Western Australian State Agricultural Biotechnology Centre (SABC).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01906/full#supplementary-material
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Alter, P., Bircheneder, S., Zhou, L. Z., Schlüter, U., Gahrtz, M., Sonnewald, U., et al. (2016). Flowering time regulated genes in maize iinclude the transcription factor ZmMADS1. Plant Physiol. 172, 389–404. doi: 10.1104/pp.16.00285
Andres, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Anonymous (2009). How to Feed the World in 2050. High-level Experts Forum. Rome: Food and Agriculture Organization, 35.
Appendino, M. L., and Slafer, G. A. (2003). Earliness per se and its dependence upon temperature in diploid wheat lines differing in the major gene Eps – Am1 alleles. J. Agri. Sci. 141, 149–154. doi: 10.1017/S0021859603003472
Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. doi: 10.1038/nature02269
Beales, J., Turner, A., Griffiths, S., Snape, J. W., and Laurie, D. A. (2007). A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 115, 721–733. doi: 10.1007/s00122-007-0603-4
Bendix, C., Marshall, C. M., and Harmon, F. G. (2015). Circadian clock genes universally control key agricultural traits. Mol. Plant 8, 1135–1152. doi: 10.1016/j.molp.2015.03.003
Benlloch-Gonzalez, M., Bochicchio, R., Berger, J., Bramley, H., and Palta, J. A. (2014). High temperature reduces the positive effect of elevated CO2 on wheat root system growth. Field Crop Res. 165, 71–79. doi: 10.1016/j.fcr.2014.04.008
Ben-Naim, O., Eshed, R., Parnis, A., Teper-Bamnolker, P., Shalit, A., Coupland, G., et al. (2006). The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 46, 462–476. doi: 10.1111/j.1365-313X.2006.02706.x
Boden, S. A., Weiss, D., Ross, J. J., Davies, N. W., Trevaskis, B., Chandler, P. M., et al. (2014). EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell 26, 1557–1569. doi: 10.1105/tpc.114.123794
Bogard, M., Ravel, C., Paux, E., Bordes, J., Balfourier, F., Chapman, S., et al. (2014). Predictions of heading date in bread wheat (Triticum aestivum L.) using QTL-based parameters of an ecophysiological model. J. Exp. Bot. 65, 5849–5865. doi: 10.1093/jxb/eru328
Bohlenius, H., Eriksson, S., Parcy, F., and Nilsson, O. (2007). Retraction. Science 316:367. doi: 10.1126/science.316.5823.367b
Bomblies, K., Wang, R. L., Ambrose, B. A., Schmidt, R. J., Meeley, R. B., and Doebley, J. (2003). Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 130, 2385–2395. doi: 10.1242/dev.00457
Bonhomme, R. (2000). Bases and limits to using ‘degree day’ units. Eur. J. Agron. 13, 1–10. doi: 10.1016/S1161-0301(00)00058-7
Bradley, D., Carpenter, R., Copsey, L., Vincent, C., Rothstein, S., and Coen, E. (1996). Control of inflorescence architecture in Antirrhinum. Nature 379, 791–797. doi: 10.1038/379791a0
Brown, H. E., Jamieson, P. D., Brooking, I. R., Moot, D. J., and Huth, N. I. (2013). Integration of molecular and physiological models to explain time of anthesis in wheat. Ann. Bot. 112, 1683–1703. doi: 10.1093/aob/mct224
Buckler, E. S., Holland, J. B., Bradbury, P. J., Acharya, C. B., Brown, P. J., Browne, C., et al. (2009). The genetic architecture of maize flowering time. Science 325, 714–718. doi: 10.1126/science.1174276
Calixto, C. P., Waugh, R., and Brown, J. W. (2015). Evolutionary relationships among barley and Arabidopsis core circadian clock and clock-associated genes. J. Mol. Evol. 80, 108–119. doi: 10.1007/s00239-015-9665-0
Campoli, C., Drosse, B., Searle, I., Coupland, G., and von Korff, M. (2012a). Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. Plant J. 69, 868–880. doi: 10.1111/j.1365-313X.2011.04839.x
Campoli, C., Pankin, A., Drosse, B., Casao, C. M., Davis, S. J., and Korff, M. (2013). HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol. 199, 1045–1059. doi: 10.1111/nph.12346
Campoli, C., Shtaya, M., Davis, S. J., and von Korff, M. (2012b). Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol. 12:97. doi: 10.1186/1471-2229-12-97
Cane, K., Eagles, H. A., Laurie, D. A., Trevaskis, B., Vallance, N., Eastwood, R. F., et al. (2013). Ppd-B1and Ppd-D1 and their effects in southern Australian wheat. Crop Pasture Sci. 64, 100–114. doi: 10.1071/CP13086
Casas, A. M., Djemel, A., Yahiaoui, S., Ponce, L. J., Contreras-Moreira, B., Gracia, M. P., et al. (2011). HvFT1 (VrnH3) drives latitudinal adaptation in Spanish barleys. Theor. Appl. Genet. 122, 1293–1304. doi: 10.1007/s00122-011-1531-x
Cassman, K. G. (1999). Ecological intensification of cereal production systems: yield potential, soil quality, and precision agriculture. Proc. Natl. Acad. Sci. U.S.A. 96, 5952–5959. doi: 10.1073/pnas.96.11.5952
Castelletti, S., Tuberosa, R., Pindo, M., and Salvi, S. (2014). A MITE transposon insertion is associated with differential methylation at the maize flowering time QTL Vgt1. G3 4, 805–812. doi: 10.1534/g3.114.010686
Chailakhyan, M. Kh (1936). New facts in support of the hormonal theory of plant development. C. R. Acad. Sci. URSS 13, 79–83.
Chardon, F., Virlon, B., Moreau, L., Falque, M., Joets, J., Decousset, L., et al. (2004). Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome. Genetics 168, 2169–2185. doi: 10.1534/genetics.104.032375
Chen, A., Li, C., Hu, W., Lau, M. Y., Lin, H., Rockwell, N. C., et al. (2014). PHYTOCHROME C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proc. Natl. Acad. Sci. U.S.A. 111, 10037–10044. doi: 10.1073/pnas.1409795111
Cockram, J., Jones, H., Leigh, F. J., O’Sullivan, D., Powell, W., Laurie, D. A., et al. (2007). Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. J. Exp. Bot. 58, 1231–1244. doi: 10.1093/jxb/erm042
Colasanti, J., and Muszynski, M. (2009). “The maize floral transition,” in Handbook of Maize: Its Biology, eds J. Bennetzen and S. C. Hake (New York: Springer), 41–55.
Colasanti, J., Tremblay, R., Wong, A. Y., Coneva, V., Kozaki, A., and Mable, B. K. (2006). The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7:158. doi: 10.1186/1471-2164-7-158
Comadran, J., Kilian, B., Russell, J., Ramsay, L., Stein, N., Ganal, M., et al. (2012). Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 44, 1388–1392. doi: 10.1038/ng.2447
Coneva, V., Zhu, T., and Colasanti, J. (2007). Expression differences between normal and indeterminate1 maize suggest downstream targets of ID1, a floral transition regulator in maize. J. Exp. Bot. 58, 3679–3693. doi: 10.1093/jxb/erm217
Conti, L., and Bradley, D. (2007). TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell 19, 767–778. doi: 10.1105/tpc.106.049767
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. doi: 10.1126/science.114752
Craufurd, P. Q., and Wheeler, T. R. (2009). Climate change and the flowering time of annual crops. J. Exp. Bot. 60, 2529–2539. doi: 10.1093/jxb/erp196
Cuesta-Marcos, A., Casas, A. M., Hayes, P. M., Gracia, M. P., Lasa, J. M., Ciudad, F., et al. (2009). Yield QTL affected by heading date in Mediterranean grown barley. Plant Breed. 128, 46–53. doi: 10.1111/j.1439-0523.2008.01510.x
Danilevskaya, O. N., Meng, X., and Ananiev, E. V. (2011). Concerted modification of flowering time and inflorescence architecture by ectopic expression of TFL1-like genes in maize. Plant Physiol. 153, 238–251. doi: 10.1104/pp.110.154211
Danilevskaya, O. N., Meng, X., Hou, Z., Ananiev, E. V., and Simmons, C. R. (2008a). A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 146, 250–264. doi: 10.1104/pp.107.109538
Danilevskaya, O. N., Meng, X., Selinger, D., Deschamps, S., Hermon, P., Vansant, G., et al. (2008b). Involvement of the MADS-box gene ZMM4 in floral induction and inflorescence development in maize. Plant Physiol. 147, 2054–2069. doi: 10.1104/pp.107.115261
Devlin, P. F. (2002). Signs of the time: environmental input to the circadian clock. J. Exp. Bot. 53, 1535–1550. doi: 10.1093/jxb/erf024
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., et al. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18, 926–936. doi: 10.1101/gad.1189604
Dubcovsky, J., Chen, C., and Yan, L. (2005). Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Mol. Breed. 15, 395–407. doi: 10.1007/s11032-005-0084-6
Dubcovsky, J., Loukoianov, A., Fu, D. L., Valarik, M., Sanchez, A., and Yan, L. L. (2006). Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol. Biol. 60, 469–480. doi: 10.1007/s11103-005-4814-2
Edwards, K. D., Anderson, P. E., Hall, A., Salathia, N. S., Locke, J. C. W., Lynn, J. R., et al. (2006). FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18, 639–650. doi: 10.1105/tpc.105.038315
Fankhauser, C., and Staiger, D. (2002). Photoreceptors in Arabidopsis thaliana: light perception, signal transduction and entrainment of the endogenous clock. Planta 216, 1–16. doi: 10.1007/s00425-002-0831-4
Faricelli, M. E., Valárik, M., and Dubcovsky, J. (2010). Control of flowering time and spike development in cereals: the earliness per se Eps-1 region in wheat, rice, and Brachypodium. Funct. Integr. Genomic. 10, 293–306. doi: 10.1007/s10142-009-0146-7
Faure, S., Higgins, J., Turner, A., and Laurie, D. A. (2007). The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176, 599–609. doi: 10.1534/genetics.106.069500
Faure, S., Turner, A. S., Gruszka, D., Christodoulou, V., Davis, S. J., von Korff, M., et al. (2012). Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. U.S.A. 109, 8328–8333. doi: 10.1073/pnas.1120496109
Ferrandiz, C., Gu, Q., Martienssen, R., and Yanofsky, M. F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127, 725–734.
Fjellheim, S., Boden, S., and Trevaskis, B. (2014). The role of seasonal flowering responses in adaptation of grasses to temperate climates. Front. Plant Sci. 5:431. doi: 10.3389/fpls.2014.00431
Flintham, J. E., Börner, A., Worland, A. J., and Gale, M. D. (1997). Optimizing wheat grain yield: effects of Rht (gibberellin-insensitive) dwarfing genes. J. Agric. Sci. 128, 11–25. doi: 10.1017/S0021859696003942
Fornara, F., Panigrahi, K. C., Gissot, L., Sauerbrunn, N., Ruhl, M., Jarillo, J. A., et al. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86. doi: 10.1016/j.devcel.2009.06.015
Fu, D., Szûcs, P., Yan, L., Helguera, M., Skinner, J. S., Von Zitzewitz, J., et al. (2005). Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol. Genet. Genomic. 273, 54–65. doi: 10.1007/s00438-004-1095-4
Gawroński, P., Ariyadasa, R., Himmelbach, A., Poursarebani, N., Kilian, B., Stein, N., et al. (2014). A distorted circadian clock causes early flowering and temperature-dependent variation in spike development in the Eps-3Am mutant of einkorn wheat. Genetics 196, 1253–1261. doi: 10.1534/genetics.113.158444
Gill, B. S., Appels, R., Botha-Oberholster, A.-M., Buell, C. R., Bennetzen, J. L., Chalhoub, B., et al. (2004). Workshop report on wheat genome sequencing: international genome research on wheat consortium. Genetics 168, 1087–1096. doi: 10.1534/genetics.104.034769
Goff, S. A., Ricke, D., Lan, T. H., Presting, G., Wang, R., Dunn, M., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. doi: 10.1126/science.1068275
Grandi, V., Gregis, V., and Kater, M. M. (2012). Uncovering genetic and molecular interactions among floral meristem identity genes in Arabidopsis thaliana. Plant J. 69, 881–893. doi: 10.1111/j.1365-313X.2011.04840.x
Greenup, A., Peacock, W. J., Dennis, E. S., and Trevaskis, B. (2009). The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann. Bot. 103, 1165–1172. doi: 10.1093/aob/mcp063
Hayama, R., Izawa, T., and Shimamoto, K. (2002). Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol. 43, 494–504. doi: 10.1093/pcp/pcf059
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
Hemming, M. N., Fieg, S., Peacock, W. J., Dennis, E. S., and Trevaskis, B. (2009). Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol. Genet. Genomic. 282, 107–117. doi: 10.1007/s00438-009-0449-3
Hemming, M. N., Peacock, W. J., Dennis, E. S., and Trevaskis, B. (2008). Low temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol. 147, 355–366. doi: 10.1104/pp.108.116418
Hovenden, M. J., Wills, K. E., Vander Schoor, J. K., Williams, A. L., and Newton, P. C. (2008). Flowering phenology in a species-rich temperate grassland is sensitive to warming but not elevated CO2. New Phytol. 178, 815–822. doi: 10.1111/j.1469-8137.2008.02419.x
Huang, T., Böhlenius, H., Eriksson, S., Parcy, F., and Nilsson, O. (2005). The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science 309, 1694–1696. doi: 10.1126/science.1117768
Hyun, Y., Richter, R., Vincent, C., Martinez-Gallegos, R., Porri, A., and Coupland, G. (2016). Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev. Cell 37, 254–266. doi: 10.1016/j.devcel.2016.04.001
International Barley Genome Sequencing Consortium (2012). A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716. doi: 10.1038/nature11543
IPCC (2013). “Summary for policymakers,” in Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds T. F. Stocker, D. Qin, G.-K. Plattner, M. Tignor, S. K. Allen, J. Boschung, et al. (Cambridge, UK: Cambridge University Press).
Itoh, H., Nonoue, Y., Yano, M., and Izawa, T. (2010). A pair of floral regulators sets critical day length for expression in rice. Nat. Genet. 42, 635–638. doi: 10.1038/ng.606
Izawa, T. (2007). Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 58, 3091–3097. doi: 10.1093/jxb/erm159
Jaeger, K. E., Pullen, N., Lamzin, S., Morris, R. J., and Wigge, P. A. (2013). Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell 25, 820–833. doi: 10.1105/tpc.113.109355
Jeuffroy, M. H., Casadebaig, P., Debaeke, P., Loyce, C., and Meynard, J. M. (2014). Agronomic model uses to predict cultivar performance in various environments and cropping systems. A review. Agron. Sustain. Dev. 34, 121–137. doi: 10.1007/s13593-013-0170-9
Jia, Q., Zhang, J., Westcott, S., Zhang, X.-Q., Bellgard, M., Lance, R., et al. (2009). GA-20 oxidase as a candidate for the semidwarf gene sdw1/denso in barley. Funct. Integr. Genomics 9, 255–262. doi: 10.1007/s10142-009-0120-4
Jia, Q., Zhang, X. Q., Westcott, S., Broughton, S., Cakir, M., Yang, J., et al. (2011). Expression level of a gibberellin 20-oxidase gene is associated with multiple agronomic and quality traits in barley. Theor. Appl. Genet. 122, 1451–1460. doi: 10.1007/s00122-011-1544-5
Johansson, M., and Staiger, D. (2015). Time to flower: interplay between photoperiod and the circadian clock. J. Exp. Bot. 66, 719–730. doi: 10.1093/jxb/eru441
Johnston, A., and Reekie, E. (2008). Regardless of whether rising atmospheric carbon dioxide levels increase air temperature, flowering phenology will be affected. Int. J. Plant Sci. 169, 1210–1218. doi: 10.1086/591978
Jung, C., and Müller, A. E. (2009). Flowering time control and applications in plant breeding. Trends Plant Sci. 14, 563–573. doi: 10.1016/j.tplants.2009.07.005
Karsai, I., Szücs, P., Mészáros, K., Filichkina, T., Hayes, P. M., Skinner, J. S., et al. (2005). The Vrn-H2 locus is a major determinant of flowering time in a facultative × winter growth habit barley (Hordeum vulgare L.) mapping population. Theor. Appl. Genet. 110, 1458–1466. doi: 10.1007/s00122-005-1979-7
Kaufmann, K., Wellmer, F., Muino, J. M., Ferrier, T., Wuest, S. E., Kumar, V., et al. (2010). Orchestration of floral initiation by APETALA1. Science 328, 85–89. doi: 10.1126/science.1185244
Keating, B. A., Carberry, P. S., Hammer, G. L., Probert, M. E., Robertson, M. J., Holzworth, D., et al. (2003). An overview of APSIM, a model designed for farming systems simulation. Eur. J. Agron. 18, 267–288. doi: 10.1016/S1161-0301(02)00108-9
Kenney, A. M., McKay, J. K., Richards, J. H., and Juenger, T. E. (2014). Direct and indirect selection on flowering time, water-use efficiency (WUE, δ 13C), and WUE plasticity to drought in Arabidopsis thaliana. Ecol. Evol. 4, 4505–4521. doi: 10.1002/ece3.1270
Kikuchi, R., Kawahigashi, H., Ando, T., Tonooka, T., and Handa, H. (2009). Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 149, 1341–1353. doi: 10.1104/pp.108.132134
Kim, S. L., Lee, S., Kim, H. J., Nam, H. G., and An, G. (2007). OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 145, 1484–1494. doi: 10.1104/pp.107.103291
Kippes, N., Chen, A., Zhang, X., Lukaszewski, A. J., and Dubcovsky, J. (2016). Development and characterization of a spring hexaploid wheat line with no functional VRN2 genes. Theor. Appl. Genet. 129, 1417–1428. doi: 10.1007/s00122-016-2713-3
Kippes, N., Debernardi, J. M., Vasquez-Gross, H. A., Akpinar, B. A., Budak, H., Kato, K., et al. (2015). Identification of the VERNALIZATION 4 gene reveals the origin of spring growth habit in ancient wheats from South Asia. Proc. Natl. Acad. Sci. U.S.A. 112, E5401–E5410. doi: 10.1073/pnas.1514883112
Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., et al. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105. doi: 10.1093/pcp/pcf156
Komiya, R., and Shimamoto, K. (2008). Genetic and epigenetic regulation of flowering in rice. Plant Biotechnol. 25, 279–284. doi: 10.5511/plantbiotechnology.25.279
Komiya, R., Yokoi, S., and Shimamoto, K. (2009). A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136, 3443–3450. doi: 10.1242/dev.040170
Koo, B. H., Yoo, S. C., Park, J. W., Kwon, C. T., Lee, B. D., An, G., et al. (2013). Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 6, 1877–1888. doi: 10.1093/mp/sst088
Kwon, C. T., Kim, S. H., Kim, D., and Paek, N. C. (2015). The rice floral repressor Early flowering1 affects spikelet fertility by modulating gibberellin signaling. Rice 8:58. doi: 10.1186/s12284-015-0058-1
Laurie, D. A. (1997). Comparative genetics of flowering time. Plant Mol. Biol. 35, 167–177. doi: 10.1023/A:1005726329248
Laurie, D. A., Pratchett, N., Bezant, J. H., and Snape, J. W. (1994). Genetic analysis of a photoperiod response gene on the short arm of chromosome 2 (2H) of Hordeum vulgare. Heredity 72, 619–627. doi: 10.1038/hdy.1994.85
Lazakis, C. M., Coneva, V., and Colasanti, J. (2011). ZCN8 encodes a potential orthologue of Arabidopsis FT florigen that integrates both endogenous and photoperiod flowering signals in maize. J. Exp. Bot. 62, 4833–4842. doi: 10.1093/jxb/err129
Leivar, P., and Monte, E. (2014). PIFs: systems integrators in plant development. Plant Cell 26, 56–78. doi: 10.1105/tpc.113.120857
Li, T., Hasegawa, T., Yin, X., Zhu, Y., Boote, K., Adam, M., et al. (2015). Uncertainties in predicting rice yield by current crop models under a wide range of climatic conditions. Glob. Chang. Biol. 21, 1328–1341. doi: 10.1111/gcb.12758
Li, X., Liu, H., Wang, M., Liu, H., Tian, X., Zhou, W., et al. (2015). Combinations of Hd2 and Hd4 genes determine rice adaptability to Heilongjiang Province, northern limit of China. J. Integr. Plant Biol. 57, 698–707. doi: 10.1111/jipb.12326
Lifschitz, E., Eviatar, T., Rozman, A., Shalit, A., Goldshmidt, A., Amsellem, Z., et al. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. U.S.A. 103, 6398–6403. doi: 10.1073/pnas.0601620103
Long, S. P., Ainsworth, E. A., Rogers, A., and Ort, D. R. (2004). Rising atmospheric carbon dioxide: plants FACE the future. Annu. Rev. Plant Biol. 55, 591–628. doi: 10.1146/annurev.arplant.55.031903.141610
Mascheretti, I., Turner, K., Brivio, R. S., Hand, A., Colasanti, J., and Rossi, V. (2015). Focus on chromatin/epigenetics: florigen-encoding genes of day-neutral and photoperiod-sensitive maize are regulated by different chromatin modifications at the floral transition. Plant Physiol. 168:1351. doi: 10.1104/pp.15.00535
Matsubara, K., Yamanouchi, U., Wang, Z.-X., Minobe, Y., Izawa, T., and Yano, M. (2008). Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by upregulating Ehd1. Plant Physiol. 148, 1425–1435. doi: 10.1104/pp.108.125542
Matsuoka, Y., Vigouroux, Y., Goodman, M. M., Sanchez, J., Buckler, E., and Doebley, J. (2002). A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. U.S.A. 99, 6080–6084. doi: 10.1073/pnas.052125199
Maurer, A., Draba, V., Jiang, Y., Schnaithmann, F., Sharma, R., Schumann, E., et al. (2015). Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genomics 16:1. doi: 10.1186/s12864-015-1459-7
Mayer, K. F., Rogers, J., Doležel, J., Pozniak, C., Eversole, K., Feuillet, C., et al. (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788. doi: 10.1126/science.1251788
Mendez-Vigo, B., Pico, F. X., Ramiro, M., Martinez-Zapater, J. M., and Alonso-Blanco, C. (2011). Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiol. 157, 1942–1955. doi: 10.1104/pp.111.183426
Meng, X., Muszynski, M. G., and Danilevskaya, O. N. (2011). The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23, 942–960. doi: 10.1105/tpc.110.081406
Michaels, S. D., and Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. doi: 10.1105/tpc.11.5.949
Miller, T. A., Muslin, E. H., and Dorweiler, J. E. (2008). A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 227, 1377–1388. doi: 10.1007/s00425-008-0709-1
Mizuno, T., and Nakamichi, N. (2005). Pseudo-response regulators (PRRs) or true oscillator components (TOCs). Plant Cell Physiol. 46, 677–685. doi: 10.1093/pcp/pci087
Moon, J., Suh, S. S., Lee, H., Choi, K. R., Hong, C. B., Paek, N. C., et al. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35, 613–623. doi: 10.1046/j.1365-313X.2003.01833.x
Mulki, M. A., and von Korff, M. (2016). CONSTANS controls floral repression by up-regulating VERNALIZATION2 (VRN-H2) in Barley. Plant Physiol. 170, 325–337. doi: 10.1104/pp.15.01350
Müller, N. A., Wijnen, C. L., Srinivasan, A., Ryngajllo, M., Ofner, I., Lin, T., et al. (2016). Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat. Genet. 48, 89–93. doi: 10.1038/ng.3447
Murakami, M., Tago, Y., Yamashino, T., and Mizuno, T. (2007). Characterization of the rice circadian clock-associated pseudo-response regulators in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 71, 1107–1110. doi: 10.1271/bbb.70048
Murase, K., Hirano, Y., Sun, T. P., and Hakoshima, T. (2008). Gibberellin induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463. doi: 10.1038/nature07519
Muszynski, M. G., Dam, T., Li, B., Shirbroun, D. M., Hou, Z., Bruggemann, E., et al. (2006). Delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 142, 1523–1536. doi: 10.1104/pp.106.088815
Muterko, A., Balashova, I., Cockram, J., Kalendar, R., and Sivolap, Y. (2015). The new wheat vernalization response allele Vrn-D1 is caused by DNA transposon insertion in the first intron. Plant Mol. Biol. Rep. 33, 294–303. doi: 10.1007/s11105-014-0750-0
Nakagawa, H., Yamagishi, J., Miyamoto, N., Motoyama, M., Yano, M., and Nemoto, K. (2005). Flowering response of rice to photoperiod and temperature: a QTL analysis using a phenological model. Theor. Appl. Genet. 110, 778–786. doi: 10.1007/s00122-004-1905-4
Nakagawa, M., Shimamoto, K., and Kyozuka, J. (2002). Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 29, 743–750. doi: 10.1046/j.1365-313X.2002.01255.x
Nemoto, Y., Kisaka, M., Fuse, T., Yano, M., and Ogihara, Y. (2003). Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. Plant J. 36, 82–93. doi: 10.1046/j.1365-313X.2003.01859.x
Nemoto, Y., Nonoue, Y., Yano, M., and Izawa, T. (2016). Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 86, 221–233. doi: 10.1111/tpj.13168
Ni, Z., Kim, E.-D., Ha, M., Lackey, E., Liu, J., Zhang, Y., et al. (2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457, 327–331. doi: 10.1038/nature07523
Nishida, H., Ishihara, D., Ishii, M., Kaneko, T., Kawahigashi, H., Akashi, Y., et al. (2013). Phytochrome C is a key factor controlling long-day flowering in barley. Plant Physiol. 163, 804–814. doi: 10.1104/pp.113.222570
Nitcher, R., Pearce, S., Tranquilli, G., Zhang, X., and Dubcovsky, J. (2014). Effect of the hope FT-B1 allele on wheat heading time and yield components. J. Hered. 105, 666–675. doi: 10.1093/jhered/esu042
Ó’Maoiléidigh, D. S., Graciet, E., and Wellmer, F. (2014). Gene networks controlling Arabidopsis thaliana flower development. New Phytol. 201, 16–30. doi: 10.1111/nph.12444
Pankin, A., Campoli, C., Dong, X., Kilian, B., Sharma, R., Himmelbach, A., et al. (2014). Mapping-by-sequencing identifies HvPHYTOCHROME C as a candidate gene for the early maturity 5 locus modulating the circadian clock and photoperiodic flowering in barley. Genetics 198, 383–396. doi: 10.1534/genetics.114.165613
Pearce, S., Huttly, A. K., Prosser, I. M., Li, Y. D., Vaughan, S. P., Gallova, B., et al. (2015). Heterologous expression and transcript analysis of gibberellin biosynthetic genes of grasses reveals novel functionality in the GA3ox family. BMC Plant Biol. 15:1. doi: 10.1186/s12870-015-0520-7
Pearce, S., Vanzetti, L. S., and Dubcovsky, J. (2013). Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1. Plant Physiol. 163, 1433–1445. doi: 10.1104/pp.113.225854
Peng, J., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. doi: 10.1038/22307
Porker, K., Eglinton, J., Coventry, S., and Fettell, N. (2015). “Improvement of yield and adaptation by manipulating phenology genes,” in Exploration, Identification and Utilization of Barley Germplasm, eds G. Zhang and C. Li (Amsterdam: Elsevier), 241–264.
Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. doi: 10.1016/0092-8674(95)90288-0
Reekie, J. Y. C., Hicklenton, P. R., and Reekie, E. G. (1994). Effects of elevated CO2 on time of flowering in four short-day and four long-day species. Can. J. Bot. 72, 533–538. doi: 10.1139/b94-071
Reymond, M., Muller, B., Leonardi, A., Charcosset, A., and Tardieu, F. (2003). Combining quantitative trait loci analysis and an ecophysiological model to analyze the genetic variability of the responses of maize leaf growth to temperature and water deficit. Plant Physiol. 131, 664–675. doi: 10.1104/pp.013839
Richards, R. A., Hunt, J. R., Kirkegaard, J. A., and Passioura, J. B. (2014). Yield improvement and adaptation of wheat to water-limited environments in Australia—a case study. Crop Pasture Sci. 65, 676–689.
Rollins, J. A., Drosse, B., Mulki, M. A., Grando, S., Baum, M., Singh, M., et al. (2013). Variation at the vernalisation genes Vrn-H1 and Vrn-H2 determines growth and yield stability in barley (Hordeum vulgare) grown under dryland conditions in Syria. Theor. Appl. Genet. 126, 2803–2824. doi: 10.1007/s00122-013-2173-y
Rötter, R. P., Tao, F., Höhn, J. G., and Palosuo, T. (2015). Use of crop simulation modelling to aid ideotype design of future cereal cultivars. J. Exp. Bot. 66, 3463–3476. doi: 10.1093/jxb/erv098
Ruelens, P., de Maagd, R. A., Proost, S., Theissen, G., Geuten, K., and Kaufmann, K. (2013). FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat. Commun. 4:2280. doi: 10.1038/ncomms3280
Ryu, C. H., Lee, S., Cho, L. H., Kim, S. L., Lee, Y. S., Choi, S. C., et al. (2009). OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 32, 1412–1427. doi: 10.1111/j.1365-3040.2009.02008.x
Sanchez-Bermejo, E., and Balasubramanian, S. (2015). Natural variation involving deletion alleles of FRIGIDA modulate temperature-sensitive flowering responses in Arabidopsis thaliana. Plant Cell Environ 39, 1353–1365. doi: 10.1111/pce.12690
Sawa, M., Nusinow, D. A., Kay, S. A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. doi: 10.1126/science.1146994
Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F., Pasternak, S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. doi: 10.1126/science.1178534
Semenov, M. A., Stratonovitch, P., Alghabari, F., and Gooding, M. J. (2014). Adapting wheat in Europe for climate change. J. Cereal Sci. 59, 245–256. doi: 10.1016/j.jcs.2014.01.006
Sheehan, M. J., Farmer, P. R., and Brutnell, T. P. (2004). Structure and expression of maize phytochrome family homeologs. Genetics 167, 1395–1405. doi: 10.1534/genetics.103.026096
Shimada, S., Ogawa, T., Kitagawa, S., Suzuki, T., Ikari, C., Shitsukawa, N., et al. (2009). A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/ FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. Plant J. 58, 668–681. doi: 10.1111/j.1365-313X.2009.03806.x
Song, J., Irwin, J., and Dean, C. (2013). Remembering the prolonged cold of winter. Curr. Biol. 23, R807–R811. doi: 10.1016/j.cub.2013.07.027
Song, X., Kristie, D. N., and Reekie, E. G. (2009). Why does elevated CO2 affect time of flowering? An exploratory study using the photoperiodic flowering mutants of Arabidopsis thaliana. New Phytol. 181, 339–346. doi: 10.1111/j.1469-8137.2008.02669.x
Song, Y. H., Ito, S., and Imaizumi, T. (2013). Flowering time regulation: photoperiod-and temperature-sensing in leaves. Trends Plant Sci. 18, 575–583. doi: 10.1016/j.tplants.2013.05.003
Song, Y. H., Shim, J. S., Kinmonth-Schultz, H. A., and Imaizumi, T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Ann. Rev. Plant Biol. 66, 441–464. doi: 10.1146/annurev-arplant-043014-115555
Soy, J., Leivar, P., González-Schain, N., Martín, G., Diaz, C., Sentandreu, M., et al. (2016). Molecular convergence of clock and photosensory pathways through PIF3–TOC1 interaction and co-occupancy of target promoters. Proc. Natl. Acad. Sci. U.S.A. 113, 4870–4875. doi: 10.1073/pnas.1603745113
Soy, J., Leivar, P., González-Schain, N., Sentandreu, M., Prat, S., Quail, P. H., et al. (2012). Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J. 71, 390–401. doi: 10.1111/j.1365-313X.2012.04992.x
Spielmeyer, W., Ellis, M. H., and Chandler, P. M. (2002). Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. U.S.A. 99, 9043–9048. doi: 10.1073/pnas.132266399
Springer, C. J., and Ward, J. K. (2007). Flowering time and elevated atmospheric CO2. New Phytol. 176, 243–255. doi: 10.1111/j.1469-8137.2007.02196.x
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S., and Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. doi: 10.1126/science.1141753
Tardieu, F., and Tuberosa, R. (2010). Dissection and modelling of abiotic stress tolerance in plants. Curr. Opin. Plant Biol. 13, 206–212. doi: 10.1016/j.pbi.2009.12.012
Taub, D. R., Miller, B., and Allen, H. (2008). Effects of elevated CO2 on the protein concentration of food crops: a meta-analysis. Glob. Chang. Biol. 14, 565–575. doi: 10.1111/j.1365-2486.2007.01511.x
Teotia, S., and Tang, G. (2015). To bloom or not to bloom: role of microRNAs in plant flowering. Mol. Plant 8, 359–377. doi: 10.1016/j.molp.2014.12.018
Thomas, B., and Vince-Prue, D. (1997). “Photoperiodic control of flower initiation: some general principles,” in Photoperiodism in Plants, eds B. Thomas and D. Vince-Prue (San Diego, CA: Academic Press), 3–28.
Thornsberry, J. M., Goodman, M. M., Doebley, J., Kresovich, S., Nielsen, D., and Buckler, E. S. (2001). Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28, 286–289. doi: 10.1038/90135
Tiwari, S. B., Shen, Y., Chang, H. C., Hou, Y., Harris, A., Ma, S. F., et al. (2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187, 57–66. doi: 10.1111/j.1469-8137.2010.03251.x
Trevaskis, B., Hemming, M. N., Peacock, W. J., and Dennis, E. S. (2006). HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol. 140, 1397–1405. doi: 10.1104/pp.105.073486
Turner, A., Beales, J., Faure, S., Dunford, R. P., and Laurie, D. A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034. doi: 10.1126/science.1117619
Turner, A. S., Faure, S., Zhang, Y., and Laurie, D. A. (2013). The effect of day-neutral mutations in barley and wheat on the interaction between photoperiod and vernalization. Theor. Appl. Genet. 126, 2267–2277. doi: 10.1007/s00122-013-2133-6
United Nations Food and Agriculture Organization (2009). How to Feed the World in 2050. Rome: United Nations Food and Agriculture Organization.
von Zitzewitz, J., Szücs, P., Dubcovsky, J., Yan, L., Pecchioni, N., Francia, E., et al. (2005). Molecular and structural characterization of barley vernalization genes. Plant Mol. Biol. 59, 449–467. doi: 10.1007/s11103-005-0351-2
Wang, J., Luo, M. C., Chen, Z., You, F. M., Wei, Y., Zheng, Y., et al. (2013). Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 198, 925–937. doi: 10.1111/nph.12164
Wang, M., Xie, B., Fu, Y., Dong, C., Hui, L., Guanghui, L., et al. (2015). Effects of different elevated CO2 concentrations on chlorophyll contents, gas exchange, water use efficiency, and PSII activity on C3 and C4 cereal crops in a closed artificial ecosystem. Photosynth. Res. 126, 351–362. doi: 10.1007/s11120-015-0134-9
Ward, J. K., Roy, D. S., Chatterjee, I., Bone, C. R., Springer, C. J., and Kelly, J. K. (2012). Identification of a major QTL that alters flowering time at elevated [CO2] in Arabidopsis thaliana. PLoS ONE 7:e49028. doi: 10.1371/journal.pone.0049028
Wei, F. J., Tsai, Y. C., Wu, H. P., Huang, L. T., Chen, Y. C., Chen, Y. F., et al. (2016). Both Hd1 and Ehd1 are important for artificial selection of flowering time in cultivated rice. Plant Sci. 242, 187–194. doi: 10.1016/j.plantsci.2015.09.005
Wei, K., Pan, S., and Li, Y. (2015). Functional characterization of maize C2H2 zinc-finger gene family. Plant Mol. Biol. Rep. 34, 1–16.
White, J. W., Herndl, M., Hunt, L. A., Payne, T. S., and Hoogenboom, G. (2008). Simulation-based analysis of effects of and loci on flowering in wheat. Crop Sci. 48, 678–687. doi: 10.2135/cropsci2007.06.0318
White, J. W., Kimball, B. A., Wall, G. W., Ottman, M. J., and Hunt, L. A. (2011). Responses of time of anthesis and maturity to sowing dates and infrared warming in spring wheat. Field Crops Res. 124, 213–222. doi: 10.1016/j.fcr.2011.06.020
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. doi: 10.1126/science.1114358
Wong, A. Y., and Colasanti, J. (2007). Maize floral regulator protein INDETERMINATE1 is localized to developing leaves and is not altered by light or the sink/source transition. J. Exp. Bot. 58, 403–414. doi: 10.1093/jxb/erl206
Wood, C. C., Robertson, M., Tanner, G., Peacock, W. J., Dennis, E. S., and Helliwell, C. A. (2006). The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc. Natl. Acad. Sci. U.S.A. 103, 14631–14636. doi: 10.1073/pnas.0606385103
Woods, D., Mckeown, M., Dong, Y., Preston, J. C., and Amasino, R. M. (2016). Evolution of VRN2/GhD7-like genes in vernalization-mediated repression of grass flowering. Plant Physiol. 170, 2124–2135. doi: 10.1104/pp.15.01279
Worland, A. J. (1996). The influence of flowering time genes on environmental adaptability in European wheats. Euphytica 89, 49–57. doi: 10.1007/BF00015718
Worland, A. J., Korzun, V., Röder, M. S., Ganal, M. W., and Law, C. N. (1998). Genetic analysis of the dwarfing gene Rht8 in wheat. Part II. The distribution and adaptive significance of allelic variants at the Rht8 locus of wheat as revealed by microsatellite screening. Theor. Appl. Genet. 96, 1110–1120. doi: 10.1007/s001220050846
Würschum, T., Boeven, P. H., Langer, S. M., Longin, C. F., and Leiser, W. L. (2015). Multiply to conquer: copy number variations at Ppd-B1 and Vrn-A1 facilitate global adaptation in wheat. BMC Genet. 16:96. doi: 10.1186/s12863-015-0258-0
Xiao, J., Xu, S., Li, C., Xu, Y., Xing, L., Niu, Y., et al. (2014). O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat. Commun. 5: 4572. doi: 10.1038/ncomms5572
Xu, M., Lu, Y., Yang, H., He, J., Hu, Z., Hu, X., et al. (2015). ZmGRF, a GA regulatory factor from maize, promotes flowering and plant growth in Arabidopsis. Plant Mol. Biol. 87, 157–167. doi: 10.1007/s11103-014-0267-9
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Yan, L., Fu, D., Li, C., Blechl, A., Tranquilli, G., Bonafede, M., et al. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. U.S.A. 103, 19581–19586. doi: 10.1073/pnas.0607142103
Yan, L., Loukoianov, A., Blechl, A., Tranquilli, G., Ramakrishna, W., SanMiguel, P., et al. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303, 1640–1644. doi: 10.1126/science.1094305
Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T., and Dubcovsky, J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. U.S.A. 100, 6263–6268. doi: 10.1073/pnas.0937399100
Yan, W., Liu, H., Zhou, X., Li, Q., Zhang, J., Lu, L., et al. (2013). Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 23, 969–971. doi: 10.1038/cr.2013.43
Yan, W. H., Wang, P., Chen, H. X., Zhou, H. J., Li, Q. P., Wang, C. R., et al. (2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4, 319–330. doi: 10.1093/mp/ssq070
Yang, Q., Li, Z., Li, W., Ku, L., Wang, C., Ye, J., et al. (2013). CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl. Acad. Sci. U.S.A. 110, 16969–16974. doi: 10.1073/pnas.1310949110
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2483. doi: 10.2307/3871242
Yin, X., Struik, P. C., Van Eeuwijk, F. A., Stam, P., and Tang, J. (2005). QTL analysis and QTL-based prediction of flowering phenology in recombinant inbred lines of barley. J. Exp. Bot. 56, 967–976. doi: 10.1093/jxb/eri090
Zakhrabekova, S., Gough, S. P., Braumann, I., Müller, A. H., Lundqvist, J., Ahmann, K., et al. (2012). Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc. Natl. Acad. Sci. U.S.A. 109, 4326–4331. doi: 10.1073/pnas.1113009109
Zeevaart, J. A. D. (1976). Physiology of flower formation. Annu. Rev. Plant Physiol. 27, 321–348. doi: 10.1146/annurev.pp.27.060176.001541
Zhang, J., Zhou, X., Yan, W., Zhang, Z., Lu, L., Han, Z., et al. (2015). Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol. 208, 1056–1066. doi: 10.1111/nph.13538
Zhang, Z. H., Wang, K., Guo, L., Zhu, Y. J., Fan, Y. Y., Cheng, S. H., et al. (2012). Pleiotropism of the photoperiod-insensitive allele of Hd1 on heading date, plant height and yield traits in rice. PLoS ONE 7:e52538. doi: 10.1371/journal.pone.0052538
Zhao, J., Huang, X., Ouyang, X., Chen, W., Du, A., Zhu, L., et al. (2012). OsELF3-1, an ortholog of Arabidopsis early flowering 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS ONE 7:e43705. doi: 10.1371/journal.pone.0043705
Zhao, X., Liu, H., Wei, X., Wu, L., Cheng, F., Ku, L., et al. (2014). Promoter region characterization of ZmPhyB2 associated with the photoperiod-dependent floral transition in maize (Zea mays L.). Mol. Breed. 34, 1413–1422. doi: 10.1007/s11032-014-0125-0
Zhao, X. Y., Hong, P., Wu, J. Y., Chen, X. B., Ye, X. G., Pan, Y. Y., et al. (2016). The tae-mir408-mediated control of TaTOC1 genes transcription is required for the regulation of heading time in wheat. Plant Physiol. 170, 1578–1594. doi: 10.1104/pp.15.01216
Zhao, X. Y., Liu, M. S., Li, J. R., Guan, C. M., and Zhang, X. S. (2005). The wheat TaGI1, involved in photoperiodic flowering, encodes an Arabidopsis GI ortholog. Plant Mol. Biol. 58, 53–64. doi: 10.1007/s11103-005-4162-2
Zheng, B., Biddulph, B., Li, D., Kuchel, H., and Chapman, S. (2013). Quantification of the effects of VRN1 and Ppd-D1 to predict spring wheat (Triticum aestivum) heading time across diverse environments. J. Exp. Bot. 64, 3747–3761. doi: 10.1093/jxb/ert209
Zikhali, M., and Griffiths, S. (2015). “The effect of Earliness per se (Eps) genes on flowering time in bread wheat,” in Advances in Wheat Genetics: From Genome to Field, Proceedings of the 12th International Wheat Genetics Symposium, eds Y. Ogihara, S. Takumi, and H. Handa (Japan: Springer), 339–345.
Zikhali, M., Leverington-Waite, M., Fish, L., Simmonds, J., Orford, S., Wingen, L. U., et al. (2014). Validation of a 1DL earliness per se (eps) flowering QTL in bread wheat (Triticum aestivum). Mol. Breed. 34, 1023–1033. doi: 10.1007/s11032-014-0094-3
Keywords: flowering time, phenology, photoperiod, yield, barley, wheat, maize, rice
Citation: Hill CB and Li C (2016) Genetic Architecture of Flowering Phenology in Cereals and Opportunities for Crop Improvement. Front. Plant Sci. 7:1906. doi: 10.3389/fpls.2016.01906
Received: 06 September 2016; Accepted: 01 December 2016;
Published: 19 December 2016.
Edited by:
Zhulong Chan, Huazhong Agricultural University, ChinaReviewed by:
Meixue Zhou, University of Tasmania, AustraliaCopyright © 2016 Hill and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengdao Li, Yy5saUBtdXJkb2NoLmVkdS5hdQ== Camilla B. Hill, Y2FtaWxsYS5oaWxsQG11cmRvY2guZWR1LmF1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.