
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 16 December 2016
Sec. Plant Metabolism and Chemodiversity
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.01898
Plastoquinone (PQ) and ubiquinone (UQ) are two important prenylquinones, functioning as electron transporters in the electron transport chain of oxygenic photosynthesis and the aerobic respiratory chain, respectively, and play indispensable roles in plant growth and development through participating in the biosynthesis and metabolism of important chemical compounds, acting as antioxidants, being involved in plant response to stress, and regulating gene expression and cell signal transduction. UQ, particularly UQ10, has also been widely used in people’s life. It is effective in treating cardiovascular diseases, chronic gingivitis and periodontitis, and shows favorable impact on cancer treatment and human reproductive health. PQ and UQ are made up of an active benzoquinone ring attached to a polyisoprenoid side chain. Biosynthesis of PQ and UQ is very complicated with more than thirty five enzymes involved. Their synthetic pathways can be generally divided into two stages. The first stage leads to the biosynthesis of precursors of benzene quinone ring and prenyl side chain. The benzene quinone ring for UQ is synthesized from tyrosine or phenylalanine, whereas the ring for PQ is derived from tyrosine. The prenyl side chains of PQ and UQ are derived from glyceraldehyde 3-phosphate and pyruvate through the 2-C-methyl-D-erythritol 4-phosphate pathway and/or acetyl-CoA and acetoacetyl-CoA through the mevalonate pathway. The second stage includes the condensation of ring and side chain and subsequent modification. Homogentisate solanesyltransferase, 4-hydroxybenzoate polyprenyl diphosphate transferase and a series of benzene quinone ring modification enzymes are involved in this stage. PQ exists in plants, while UQ widely presents in plants, animals and microbes. Many enzymes and their encoding genes involved in PQ and UQ biosynthesis have been intensively studied recently. Metabolic engineering of UQ10 in plants, such as rice and tobacco, has also been tested. In this review, we summarize and discuss recent research progresses in the biosynthetic pathways of PQ and UQ and enzymes and their encoding genes involved in side chain elongation and in the second stage of PQ and UQ biosynthesis. Physiological functions of PQ and UQ played in plants as well as the practical application and metabolic engineering of PQ and UQ are also included.
Plastoquinone (PQ) and ubiquinone (UQ) are two important prenylquinones functioning as electron transporters in plants. They are involved in photophosphorylation and oxidative phosphorylation located in chloroplast thylakoids and mitochondrial inner membrane, respectively (Swiezewska, 2004). PQ and UQ are both made up of an active benzoquinone ring attached to a polyisoprenoid side chain. The length of polyisoprenoid side chain determines the type of PQ and UQ. Difference between PQ and UQ in chemical structure mainly lies in diverse substituent groups of benzoquinone ring (Figure 1). Plant PQ and UQ usually include nine or ten units of isoprenoid side chain. For instance, the main PQ and UQ in Arabidopsis thaliana have nine such units, known as PQ9 and UQ9, respectively. PQ and UQ are localized in different organelles of plant cells. PQ is located on the thylakoids of chloroplasts, while UQ is located on the inner membrane of mitochondria. The locations of PQ and UQ are consistent with their roles in photophosphorylation and oxidative phosphorylation. Lifespan of PQ and UQ is very short in cells. The half-time of PQ and UQ in spinach cells is about 15 and 30 h, respectively (Wanke et al., 2000). Therefore, to maintain the concentration stable and dynamic balance for normal plant photosynthesis and respiration, PQ and UQ need to be continuously synthesized in cells. In addition to the significance of PQ and UQ in plants, UQ10 has also been used in people’s life. Significant progress has been made recently in PQ and UQ biosynthetic pathways and genes associated with PQ and UQ production. The biosynthesis and functions of UQ in Escherichia coli, Saccharomyces cerevisiae and animals (Clarke, 2000; Meganathan, 2001; Tran and Clarke, 2007; Bentinger et al., 2010; Aussel et al., 2014), UQ10 production in plants (Parmar et al., 2015) and metabolic engineering of UQ10 in microbes (de Dieu Ndikubwimana and Lee, 2014) have been reviewed. Here we mainly summarize and discuss recent advances in biosynthetic pathways, key enzymes and their encoding genes, physiological functions, and metabolic engineering of PQ and UQ in plants.
The biosynthetic pathways of PQ and UQ can be generally divided into two stages (Figures 2 and 3). The first stage leads to the biosynthesis of precursors of benzene quinone ring and prenyl side chain. The second stage includes the condensation of ring and side chain and subsequent modification. The benzene quinone ring precursor for UQ is 4-hydroxybenzoic acid (4HB) synthesized from tyrosine or phenylalanine under the catalysis of various enzymes known as phenylalanine ammonia-lyase (PAL), cinnamic acid 4-hydroxylase (C4H), 4-coumarate CoA ligase (4CL), and other unknown enzymes. The benzene quinone ring precursor for PQ is homogentisic acid (HGA). It is synthesized from tyrosine under the catalysis of tyrosine aminotransferase (TAT) and 4-hydroxyphenylpyruvate reductase (HPPR). The prenyl side chains of PQ and UQ are derived from glyceraldehyde 3-phosphate (G3P) and pyruvate through the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway and/or acetyl-CoA and acetoacetyl-CoA through the mevalonate (MVA) pathway. The universal isoprene precursor isopentenyl diphosphate (IPP, C5) and its isomer dimethylallyl diphosphate (DMAPP) synthesized through the MEP and MVA pathways are converted into intermediate diphosphate precursors, including geranyl diphosphate (GPP, C10), farnesyl diphosphate (FPP, C15), and geranylgeranyl diphosphate (GGPP, C20). Enzymes catalyzing this conversion are a group of polyprenyl diphosphate synthases (PPSs), including geranyl diphosphate synthase (GPPS), farnesyl diphosphate synthase (FPPS), and geranylgeranyl diphosphate synthase (GGPPS) (Ma et al., 2012; Zhang and Lu, 2016). Both PQ and UQ share the prenyl side chain precursors. Side chain may be elongated to solanesyl diphosphate (SPP, C45) and decaprenyl diphospate (DPS, C50) under the catalysis of solanesyl diphosphate synthase (SPS) and decaprenyl diphosphate synthase (DPS), respectively. In the second stage, SPP is attached to HGA by homogentisate solanesyltransferase (HST) to produce intermediate 2-dimethyl-plastoquinone, which is then catalyzed by a methytransferase to form the end-product PQ9 in plants. The condensation of PHB and the corresponding prenyl side chain is catalyzed by PHB polyprenyltransferase (PPT). It leads to the formation of 3-polyprenyl-4-hydroxybenzoate. After three methylations, three hydroxylations and one decarboxylation, ubiquinonol-n and ubiquinone-n are formed eventually. Enzymes catalyzing these modification steps are currently not well understood.
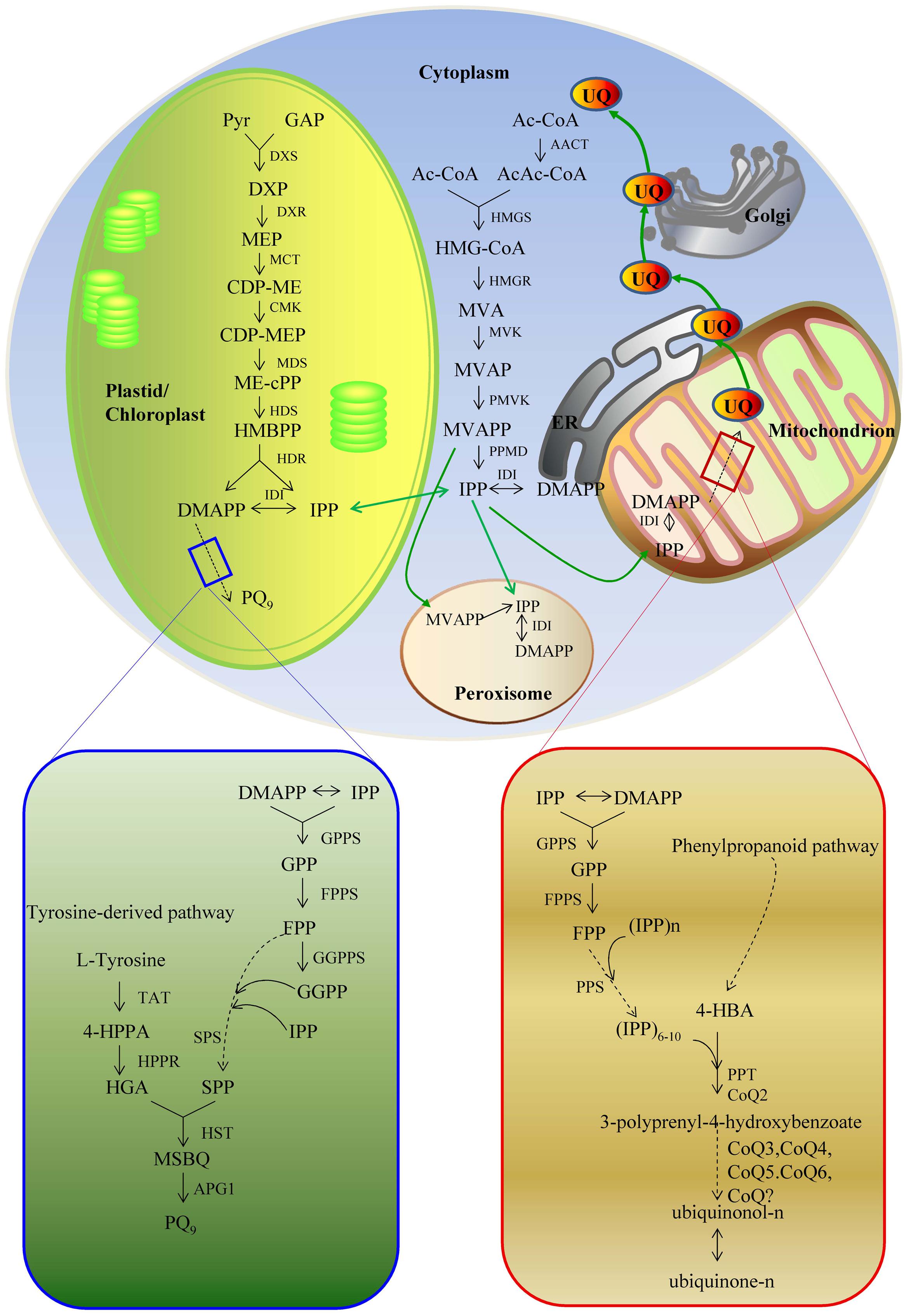
FIGURE 2. The biosynthetic pathways of PQ9 and UQ. 4-HBA, 4-hydroxybenzoate acid; 4-HPPA, 4-hydroxyphenylpyruvic acid; AACT, acetoacetyl-CoA thiolase; AcAc-CoA, acetyloacetyl-coenzyme A; Ac-CoA, acetyl-coenzyme A; APG1, MSBQ/MPBQ methyltransferase; CDP-ME, 4-diphosphocytidyl-2C-methyl-D-erythritol; CDP-MEP, 4-diphosphocytidyl-2C-methyl-D-erythritol 2-phosphate; CMK, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; Coq?, unknown enzymes; Coq3 and Coq5, O-methyltransferase and C-methyltransferase; Coq6, monooxygenase; DMAPP, dimethylallyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; DPS, decaprenyl diphosphate synthese; DXP, 1-deoxy-D-xylulose-5 phosphate; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; DXS, 1-deoxy-D-xylulose-5-phosphate; FPP, farnesyl diphosphate; FPPS, farnesyl diphosphate synthase; GAP, D-glyceraldehyde 3-phosphate; GGPP, geranylgeranyl diphosphate; GGPPS, geranylgeranyl diphosphate synthase; GPP, geranyl diphosphate; GPPS, geranyl diphosphate synthase; HDR, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase; HDS, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase; HGA, homogentisate acid; HMBPP, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate; HMG-CoA, 3-hydroxy-3-methylglutaryl-coenzyme A; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; HMGR, hydroxymethylglutaryl-CoA reductase; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; HMGS, hydroxymethylglutaryl-CoA synthetase; HPPR, 4-hydroxyphenylpyruvate reductase; HST, homogentisate solanesyltransferase; IDI, isopentenyl-diphosphate delta-isomerase; IPP, isopentenyl diphosphate isomerase; IPP, isopentenyl diphosphate; MCT, 2C-methyl-D-erythritol 4-phosphate cytidyl transferase; MDS, 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; ME-cPP, 2C-methyl-D-erythritol 2,4-cyclodiphosphate; MEP, 2C-methyl-D-erythritol-4-phosphate; MSBQ, methyl-solanesyl-benzoquinone; MVA, mevalonic acid; MVAP, mevalonate-5-phosphate; MVAPP, mevalonate-5-diphosphate; MVK, mevalonate kinase; PMVK, phosphomevalonate kinase; PPMD, diphosphomevalonate decarboxylase; PPS, diphosphate synthese; PPT/Coq2, 4-hydroxybenzoate polyprenyl transferase; Pyr, pyruvate; SPP, solanesyl diphosphate; SPS, solanesyl diphosphate synthase; TAT, tyrosine aminotransferase.
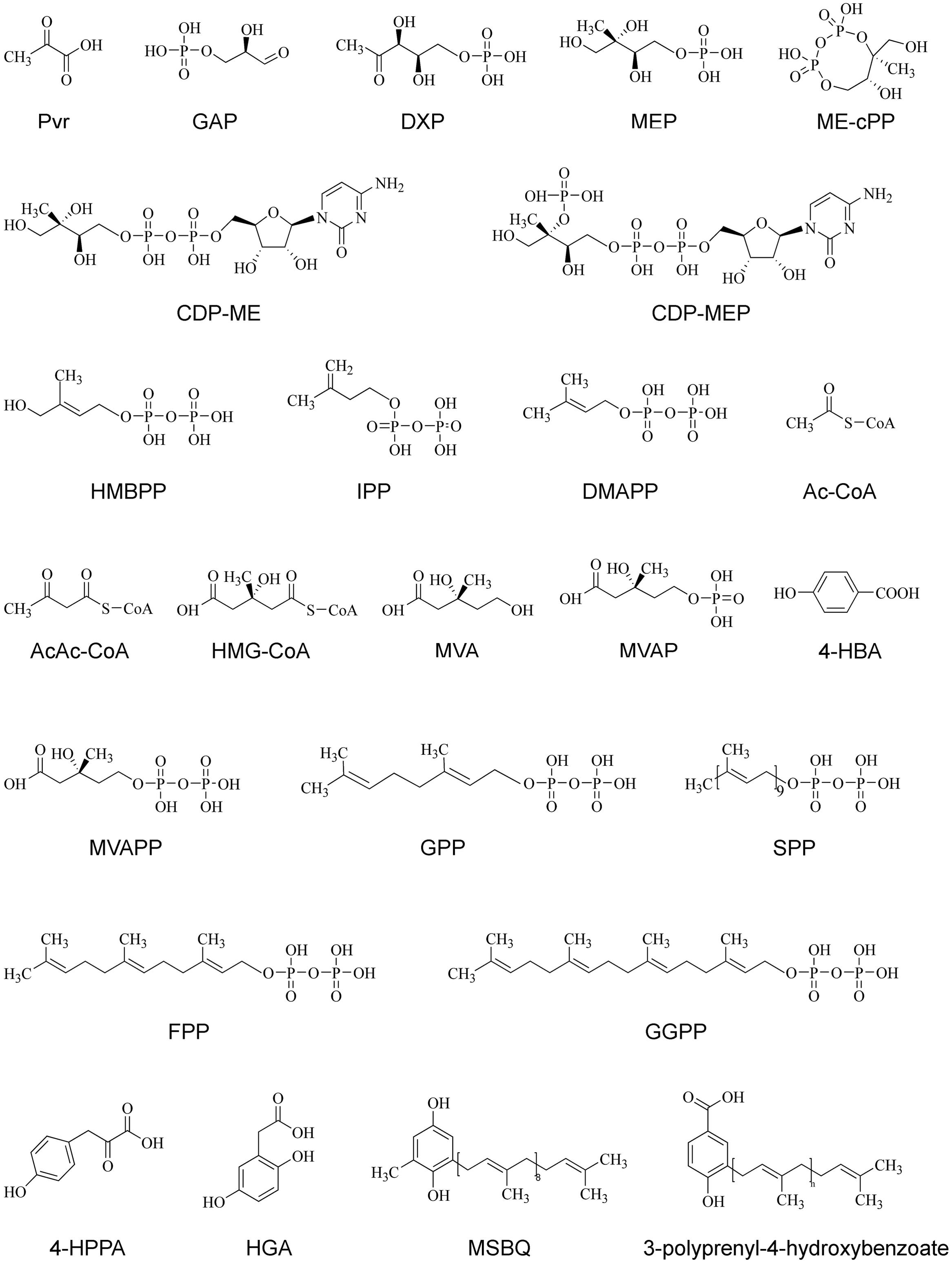
FIGURE 3. The chemical structures of various compounds in PQ and UQ biosynthetic pathways. 4-HBA, 4-hydroxybenzoate acid; 4-HPPA, 4-hydroxyphenylpyruvic acid; AcAc-CoA, acetyloacetyl-coenzyme A; Ac-CoA, acetyl-coenzyme A; CDP-ME, 4-diphosphocytidyl-2C-methyl-D-erythritol; CDP-MEP, 4-diphosphocytidyl-2C-methyl-D-erythritol 2-phosphate; DMAPP, dimethylallyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; DXP, 1-deoxy-D-xylulose-5 phosphate; DXS, 1-deoxy-D-xylulose-5-phosphate; FPP, farnesyl diphosphate; GAP, D-glyceraldehyde 3-phosphate; GGPP, geranylgeranyl diphosphate; GPP, geranyl diphosphate; HGA, homogentisate acid; HMBPP, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate; HMG-CoA, 3-hydroxy-3-methylglutaryl-coenzyme A; IPP, isopentenyl diphosphate; ME-cPP, 2C-methyl-D-erythritol 2,4-cyclodiphosphate; MEP, 2C-methyl-D-erythritol-4-phosphate; MSBQ, methyl-solanesyl-benzoquinone; MVA, mevalonic acid; MVAP, mevalonate-5-phosphate; MVAPP, mevalonate-5-diphosphate; Pyr, pyruvate; SPP, solanesyl diphosphate.
Recently, genes involved in the MEP and MVA pathways have been intensively studied. Chemical compounds synthesized through these pathways are precursors for many terpenoids and their derivatives. They are not specific to PQ and UQ production. Here, we are not going to summarize the advances in these genes. Instead, genes involved in side chain elongation and in the second stage of PQ and UQ biosynthesis are reviewed in detail (Table 1).
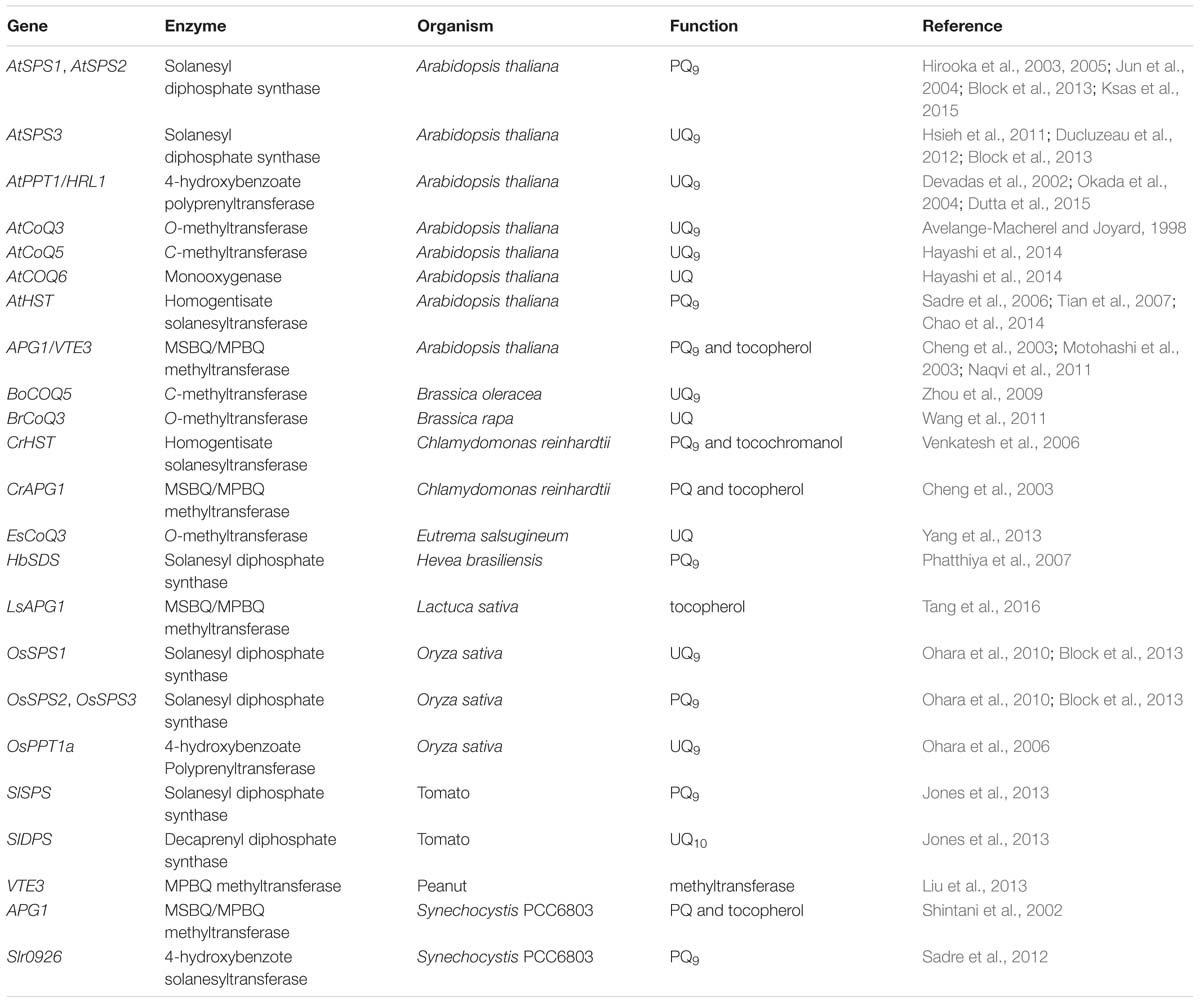
TABLE 1. Genes involved in side chain elongation and in the second stage of PQ and UQ biosynthesis in photosynthetic organisms.
Polyprenyl diphosphate synthases, also known as prenyltransferases (PTs) and isoprenyl diphosphate synthases (IDSs), are a group of enzymes widely distributed in organisms. They catalyze the formation of polyprenyl diphosphates with various chain lengths through consecutive condensation of IPP and are key enzymes involved in the biosynthesis of isoprenoid compounds, including monoterpenes, diterpenes, triterpenoids, carotenoids, natural rubber and many derivatives, such as PQ and UQ, vatamin E, prenylflavonoids, and shikonnin (Kellogg and Poulter, 1997).
Polyprenyl diphosphate synthases possess seven common conserved domains, I–VII, of which domain II is characterized with the first aspartate-rich motif (FARM), DDX2-4D, while domain VI is characterized with the second aspartate-rich motif (SARM), DDXXD (Wang and Ohnuma, 1999; Phatthiya et al., 2007). These aspartate-rich motifs are involved in substrate binding and catalysis via chelating Mg2+, a cofactor required for enzyme activity (Wang and Ohnuma, 1999). Based on the chain length of final products, PPSs can be divided into three subfamilies: short- (C15–C25), medium- (C30–C35) and long-chain (C40–C50) PTs (Hemmi et al., 2002). The most common PQ and UQ in plants have a C45 or C50 prenyl side chain moiety. For instance, Oryza sativa PQ9 and UQ9 contain a C45 prenyl side chain (Ohara et al., 2010), while cauliflower and pea UQ10 has a C50 prenyl side chain (Mattila and Kumpulainen, 2001). Thus, long chain PPSs, such as SPS catalyzing the formation of solanesyl diphosphate (SPP, C45) and decaprenyl diphosphate synthase (DPS) involved in decaprenyl diphosphate (DPP, C50) production, are significant to PQ and UQ biosynthesis in plants.
Database mining of fully sequenced genomes showed that Chlamydomnas reinhardtii, Cyanidioschyzon merolae, Cucumis sativus, Vitis vinifera and Hordeum vulgare contained two SPSs, Physcomitrella patens, Arabidopsis, Glycine max, Oryza sativa and Zea mays had three, and Brachypodium distachyon contained four (Block et al., 2013). Among the three Arabidopsis SPS genes, AtSPS1 (At1g78510) and AtSPS2 (At1g17050) are highly expressed in leaves with the level of AtSPS1 transcripts higher than AtSPS2 (Hirooka et al., 2003, 2005), whereas AtSPS3 (At2g34630) is ubiquitously expressed with peaks in seeds and shoot apical meristems (Ducluzeau et al., 2012). Although AtSPS1 had been shown previously to be localized in the ER (Hirooka et al., 2003, 2005; Jun et al., 2004), recent analysis have clearly demonstrated that both AtSPS1 and AtSPS2 is targeted exclusively to plastids and contribute to the biosynthesis of PQ9 (Block et al., 2013). Overexpression of AtSPS1 resulted in the accumulation of PQ9 and its derivative plastochromanol-8 (PC8) (Ksas et al., 2015). Moreover, the enzymatic activity of AtSPS1 and AtSPS2 is stimulated by a member of the lipid-associated fibrillin protein family, fibrillin 5 (FNB5-B), which physically binds to the hydrophobic solanesyl moiety and helps to release the moiety from the enzymes in Arabidopsis cells (Kim et al., 2015). AtSPS3 had been shown previously to be targeted to plastids and to contribute to gibberellin biosynthesis (Bouvier et al., 2000). However, recent results suggest that it is actually dual targeted to mitochondria and plastids and appears very likely bifunctional (Ducluzeau et al., 2012). AtSPS3 is able to complement a yeast coq1 knockout lacking mitochondrial hexaprenyl diphosphate synthase. Silence of AtSPS3 using RNAi technology led to 75–80% reduction of the UQ pool size. AtSPS3 overexpression resulted in a 40% increase in UQ content. No significant alternation of PQ levels was observed in AtSPS3 silenced or overexpressing lines. Therefore, AtSPS3 seems to be the main contributor to SPS activity required for UQ9 biosynthesis in Arabidopsis (Hsieh et al., 2011; Ducluzeau et al., 2012).
Similarly, three SPS genes exist in O. sativa (Ohara et al., 2010; Block et al., 2013). OsSPS1 is highly expressed in roots, whereas OsSPS2 is highly expressed in both leaves and roots. TargetP prediction and transient expression of GFP fusion protein showed the localization of OsSPS1 in mitochondria and OsSPS2 in plastids. Recombinant proteins of both OsSPS1 and OsSPS2 catalyzed the formation of solanesyl diphosphates. The enzyme activity of OsSPS1 was stronger than OsSPS2. OsSPS1 complemented the yeast coq1 disruptant and produced UQ9 in yeast cells, whereas OsSPS2 weakly complemented the growth defect of the coq1 mutant (Ohara et al., 2010). The results suggest that OsSPS1 and OsSPS2 are involved in the supply of solanesyl diphosphate for UQ9 production in mitochondria and PQ9 biosynthesis in chloroplasts, respectively. OsSPS3 is less studied compared with OsSPS1 and OsSPS2. Since it shows high homology with OsSPS2 (Block et al., 2013), OsSPS3 appears to be also involved in PQ9 formation.
Analysis of the fully sequenced tomato genomes showed that Solanum lycopersicum contained two long-chain PPSs genes (Block et al., 2013). Jones et al. (2013) cloned and termed them SlSPS and SlDPS, respectively. SlSPS is targeted to plastids, whereas the fluorescence signal of SlDPS-GFP may resemble the mitochondrial localization reported for rice OsSPS1 (Ohara et al., 2010; Jones et al., 2013). In E. coli, SlSPS and SlDPS extend the side chain of endogenous UQ to nine and ten isoprene units, respectively (Jones et al., 2013). Overexpression of SlSPS elevated the content of PQ in immature tobacco leaves. Silence of SlSPS resulted in photobleached phenotype and accumulated phytoene. SlSPS and SIDPS could not complement silencing of each other. SlDPS can use GPP, FPP or GGPP in SPP and DPP biosynthesis. Silence of SlDPS did not affect leaf appearance, but impacted on primary metabolism (Jones et al., 2013). The roles of SlSPS and SlDPS in PQ and UQ biosynthesis need to be further investigated.
Although long chain PPSs play significant roles in PQ and UQ biosynthesis, the corresponding genes have only been identified from various plants, such as Arabidopsis (Hirooka et al., 2003, 2005; Jun et al., 2004; Ducluzeau et al., 2012; Block et al., 2013), rice (Ohara et al., 2010), tomato (Jones et al., 2013), and Hevea brasiliensis (Phatthiya et al., 2007). Greater efforts are required for molecular cloning and functional characterization of the genes in other plant species and utilization of plant source long chain PPSs in PQ and UQ production through biotechnology.
HSTs catalyze the condensation of HGA and SPP to form 2-demethylplastoquinol-9 leading to PQ9 biosynthesis (Sadre et al., 2010). Although HSTs are members of the homogentisate PT family, they differ from other homogentisate PT members, such as the homogentisate phytyltransferases (HPTs) isolated from Synechocystis (slr1736) and Arabidopsis (VTE2), in enzyme properties (Collakova and DellaPenna, 2001; Savidge et al., 2002). HPTs showed the highest enzyme activities with phytyl diphosphate, whereas HSTs had the highest activities with solanesyl diphosphate and were hardly active with phytyl diphosphate (Sadre et al., 2006).
Compared with other genes involved in the upstream of PQ biosynthesis, HSTs are less studied, although they play an important role in the biosynthesis of PQ9, GAs and carotenoids, of which carotenoids are precursors of ABA and strigolactones. Genes encoding HSTs have only reported in C. reinhardtii and Arabidopsis (Sadre et al., 2006; Venkatesh et al., 2006; Tian et al., 2007; Sadre et al., 2010; Chao et al., 2014). The deduced C. reinhardtii CrHST and Arabidopsis AtHST proteins contain a chloroplast targeting sequence (Sadre et al., 2006; Tian et al., 2007). Confocal images of the AtHST-GFP fusion protein showed that AtHST protein was located in the chloroplast, most likely on the envelope membrane (Tian et al., 2007). Overexpression of AtHST caused modest elevation of PQ9 levels (Sadre et al., 2006). The pds2 mutant with an in-frame 6 bp deletion in AtHST showed an albino phenotype and the mutation can be functionally complemented by constitutive expression of AtHST (Norris et al., 1995; Tian et al., 2007). Similarly, T-DNA insertion mutant of HST gene displayed albino, dwarf and early flowering phenotypes with chloroplast development arrested, chlorophyll (Chl) absent and stomata closure defected (Chao et al., 2014). The GA and ABA levels were very low in the mutant. Exogenous GA could partially rescue the dwarf phenotype and the root development defects. Exogenous ABA could rescue the stomata closure defects (Chao et al., 2014). The expression levels of many genes involved in flowering time regulation and PQ, Chl, GA, ABA and carotenoid biosynthesis were changed in the mutant, suggesting the key roles of AtHST played in chloroplast development and plant hormone biosynthesis (Chao et al., 2014). HST genes in other plant species remain to be isolated and characterized.
PPT is a rate-limiting enzyme in UQ biosynthesis. It catalyzes the condensation of benzoquinone ring and polyisoprenoid side chain to form the intermediate, 3-polyprenyl-4-hydroxybenzoate. All of the reported eukaryotic PPTs contain a mitochondrial targeting signal at N-terminal sequence. Most PPTs have a broad specificity for prenyl diphosphates, accepting substrates of different chain lengths, but show a high specificity for 4HB (Forsgren et al., 2004; Okada et al., 2004; Ohara et al., 2006). PPT genes have been identified from various organisms, such as E. coli (Siebert et al., 1992), S. cerevisiae (Ashby et al., 1992), Schizosaccharomyces pombe (Uchida et al., 2000), human (Forsgren et al., 2004), and two plant species, including Arabidopsis (Okada et al., 2004) and rice (Ohara et al., 2006).
PPTs are members of the polyprenyl diphosphate transferase family. Although proteins in this family have sequence homology and relatively close phylogenetic relationship, only a small subset catalyzing the condensation of 4HB and prenyl chain are involved in UQ biosynthesis (Ohara et al., 2006). Other members may be involved in other metabolic pathways. For instance, Cyanobacteria hydroxybenzoate solanesyltransferase, Slr0926, which exhibits highly specific for 4-hybroxybenzoate and a broad specificity with regard to the prenyl donor substrate, including SPP and a number of shorter-chain prenyl diphosphates, is actually involved in PQ9 biosynthesis (Sadre et al., 2012). Lithospermum erythrorhizon 4HB geranyltransferases (LePGT1 and LePGT2) with a strict specificity for GPP as a prenyl substrate are involved in shikonin biosynthesis and not relevant to UQ formation (Heide and Berger, 1989; Yazaki et al., 2002; Ohara et al., 2006, 2009).
Arabidopsis AtPPT1 is the first PPT gene identified in plants (Okada et al., 2004). It is predominantly expressed in the flower cluster. The deduced AtPPT1 protein is localized in mitochondria and can complement the yeast coq2 disruptant. AtPPT1 has broad substrate specificity in terms of the prenyl donor. The T-DNA insertion mutant of AtPPT1 shows arrest of embryo development at an early stage of zygotic embryogenesis (Okada et al., 2004). The other mutant, known as hypersensitive response-like lesions 1 (hrl1), was identified in an ethyl methanesulfonate (EMS)-mutagenesis screen (Devadas et al., 2002). The hrl1 mutant spontaneously develops HR-like lesions and shows enhanced resistance against bacterial pathogens (Devadas et al., 2002). Positional cloning and subsequent DNA sequencing showed that the mutant had a single base change in an exon of AtPPT1. The mutation results in a leucine to phenylalanine substitution at position 228. Leucine 228 is not a part of the active site of the enzyme but is conserved across PPT sequences from various organisms (Dutta et al., 2015). Overexpression of HRL1 in A. thaliana leads to elevated UQ and decreased ubiquinol levels (Dutta et al., 2015).
The O. sativa genome contains three PPT genes, including OsPPT1a, OsPPT1b and OsPPT1c. However, only OsPPT1a was found to be expressed (Ohara et al., 2006). The deduced OsPPT1a proteins contain a putative mitochondrial sorting signal at the N-terminus. Consistently, GFP-PPT fusion proteins are mainly localized in mitochondria (Ohara et al., 2006). Same as AtPPT1 and other PPT proteins, OsPPT1a can complement the yeast coq2 mutant, accepts prenyl diphosphates of various chain lengths as prenyl donors, and shows strict substrate specificity for the aromatic substrate 4HB as a prenyl acceptor (Ohara et al., 2006).
2-demethylplastoquinol formed through the condensation of HGA and SPP by HSTs is a key intermediate in PQ synthesis. This intermediate can be converted to the final product PQ under the catalyzing of a modification enzyme, termed albino or pale green 1 (APG1) or methy-phytyl-benzoquinone (MPBQ)/methyl-solanesyl-benzoquinone (MSBQ) methyltransferase (Shintani et al., 2002; Motohashi et al., 2003; Cheng et al., 2003).
Arabidopsis APG1 was identified through characterization of the Ds-tagged albino or pale green mutant 1 (apg1) (Motohashi et al., 2003). This mutant lacks PQ, cannot survive beyond the seedling stage when germinated on soil, and contains decreased numbers of lamellae with reduced levels of chlorophyll (Motohashi et al., 2003). The insertion of Ds transposon disrupts a gene encoding a 37 kDa polypeptide precursor of the chloroplast inner envelope membrane. The 37 kDa protein had partial sequence similarity to the S-adenosylmethionine-dependent methyltransferase (Motohashi et al., 2003). Because of the lack of PQ in apg1 mutant and the putative methyltrasferase activity of the 37 kDa protein, APG1 appears to be involved in the methylation step of PQ biosynthesis (Motohashi et al., 2003). Almost at the same time, using a combined genomic, genetic and biochemical approach, Cheng et al. (2003) isolated and characterized the Arabidopsis VTE3 (vitamin E defective) locus. This locus is actually identical to APG1. In vitro enzyme assays showed that VTE3 was the plant functional equivalent of MPBQ/MSBQ methyltransferase from Synechocystis sp PCC6803 (Shintani et al., 2002).
In cyanobacterium, various genes involved in the PQ biosynthetic pathway and the vitamin E biosynthetic pathway are highly conserved. The biosynthesis of PQ and vitamin E share common precursor, HGA, which is prenylated with different substrates in the biosynthesis of different products. HGA is prenylated with phytyl diphosphate (PDP) or GGPP in tocochromanol formation, whereas it is prenylated with SPP in PQ9 biosynthesis. Thus, the MPBQ/MSBQ methyltransferase identified from Synechocystis sp PCC6803 has a dual function in the final methylation step of PQ and vitamin E biosynthesis (Shintani et al., 2002). Most genes involved in PQ and vitamin E synthesis are homologous, but MPBQ/MSBQ methyltransferase from Synechocystis sp PCC6803 and VTE3 from Arabidopsis are highly divergent in primary sequence (Cheng et al., 2003). The orthologs of Synechocystis MPBQ/MSBQ methyltransferase exist in C. reinhardtii and Thalassiosira pseudonana but absent from vascular and non-vascular plants. VTE3 orthologs exist in C. reinhardtii and vascular and non-vascular plants but absent from cyanobacteria (Cheng et al., 2003). It suggests that VTE3 is evolutionarily originated from archea rather than cyanobacteria. Interestingly, two mutants of VTE3, vte3-1 and vte3-2, which show partial or total disruption of MPBQ/MSBQ methyltransferase activity, respectively, have different effects on PQ and vitamin E biosynthesis. vte3-1 mainly impairs the methylation of tocopherol substrates. It has little effect on the methylation of MSBQ to PQ. On the contrary, vte3-2 completely disrupts MPBQ/MSBQ methyltransferase (Shintani et al., 2002). The underlying mechanism remains to be elucidated.
Although genes encoding MPBQ/MSBQ methyltransferase are largely unknown in plant species other than Arabidopsis, engineering of VTE3 has been tested (Van Eenennaam et al., 2003; Naqvi et al., 2011). Seed-specific expression of Arabidopsis VTE3 in transgenic soybean reduced seed delta-tocopherol from 20 to 2%. Coexpression of Arabidopsis VTE3 and VTE4 (gamma-tocopherol methyltransferase gene) resulted in a greater than eightfold increase of alpha-tocopherol and an up to fivefold increase in vitamin E activity in transgenic soybean seeds (Van Eenennaam et al., 2003). Simultaneous expression of Arabidopsis ρ-hydroxyphenylpyruvate dioxygenase and VTE3 in transgenic corn kernels triples the tocopherol content (Naqvi et al., 2011). It is currently unknown whether it is possible to increase the content of PQ in plants through VTE3 overexpression.
Compared with PQ benzene quinine ring modification, the process of UQ aromatic ring modification is complex. It includes three methylations (two O-methylations and one C-methylation), three hydroxylations, and one decarboxylation. UQ biosyntheses in prokaryotes and eukaryotes are similar. The difference lies in the reaction order of modifications. In eukaryotes, proposed modifications start with hydroxylation, followed by O-methylation, decarboxylation, two additional hydroxylations, C-methylation and one additional O-methylation (Tran and Clarke, 2007), whereas the reaction order of modifications in prokaryotes is decarboxylation, three hydroxylations, O-methylation, C-methylation and then an additional O-methylation (Meganathan, 2001).
The modification of UQ aromatic ring requires a total of three hydroxylations. The first hydroxylation of 3-polyprenyl-4-hydroxybenzoate in eukaryotes occurs before decarboxylation (Goewert et al., 1977). Two additional hydroxylations occur after decarboxylation in eukaryotes. Enzymes involved in hydroxylation are cytochrome P450 monooxygenases, known as COQ6 and COQ7 in yeast (Olson and Rudney, 1983). Genes encoding these enzymes have been identified from various organisms, such as S. cerevisiae (Marbois and Clarke, 1996; Kawamukai, 2000; Ozeir et al., 2011), rat (Jonassen et al., 1996), Caenorhabditis elegans (Ewbank et al., 1997), human (Vajo et al., 1999; Lu T.T. et al., 2013), and E. coli (Hajj Chehade et al., 2013). Homologs of the enzymes involved in UQ aromatic ring hydroxylations have also been found in various plants, such as Arabidopsis (Lange and Ghassemian, 2003) and alga (Blanc et al., 2010, 2012). Arabidopsis contains a COQ6 homolog, but lacks COQ7 (Hayashi et al., 2014). Although AtCOQ6 cannot complement the yeast coq6 disruptant, addition of a mitochondrial targeting signal to AtCOQ6 will enable the production of UQ10 (Hayashi et al., 2014). Except AtCOQ6, other plant COQ6 and COQ7 homologs were identified based on genome sequence search and computational annotation, their roles in UQ biosynthesis need to be further confirmed using experimental approaches.
The three methylations of UQ aromatic ring include two O-methylations and one C-methylation. The two O-methylation steps are catalyzed by COQ3, whereas the C-methylation step is catalyzed by COQ5 in yeast (Clarke et al., 1991; Barkovich et al., 1997; Poon et al., 1999; Hsu et al., 2000; Baba et al., 2004). COQ3 homologs have been identified in various organisms, such as E. coli (Hsu et al., 1996), yeast (Clarke et al., 1991; Poon et al., 1999; Hsu et al., 2000), rat (Marbois et al., 1994a,b) and human (Jonassen and Clarke, 2000). The gene encoding plant COQ3 was first identified in Arabidopsis (Avelange-Macherel and Joyard, 1998). AtCOQ3 gene product is localized within mitochondrial membranes and can complement Saccharomyces cerevisiae or Schizosaccharomyces pombe coq3 disruptant (Avelange-Macherel and Joyard, 1998; Hayashi et al., 2014). With the decoding of plant genomes, COQ3 has also been found in other plant species, such as Brassica rapa (Wang et al., 2011), Eutrema salsugineum (Yang et al., 2013). Similar to COQ3, COQ5 is associated with other COQ proteins on the inner mitochondrial membrane at the matrix side (Baba et al., 2004; Nguyen et al., 2014). COQ5 genes have been identified in yeast (Barkovich et al., 1997; Baba et al., 2004), human (Chen et al., 2013; Nguyen et al., 2014), broccoli (Zhou et al., 2009) and Arabidopsis (Hayashi et al., 2014). S. cerevisiae COQ5 displays a typical class I S-adenosyl methionine-dependent methyltransferase crystal structure (Dai et al., 2014). The broccoli BoCOQ5 can complement a yeast coq5 mutant and increases cellular UQ levels in bacteria (Zhou et al., 2009). Similarly, AtCOQ5 can complement the Schizosaccharomyces pombe coq5 disruptant (Hayashi et al., 2014).
The proposed modifications of UQ aromatic ring include a decarboxylation step. It occurs before the three hydroxylation steps in prokaryotes, whereas in eukaryotes, decarboxylation occurs after a hydroxylation step and an O-methylation step. It has been shown that ubiX and ubiD genes are involved in the decarboxylation of UQ aromatic ring in bacteria (Meganathan, 2001; Gulmezian et al., 2007; Aussel et al., 2014). ubiX encodes a flavin mononucleotide (FMN)-binding protein with no decarboxylase activity detected in vitro (Gulmezian et al., 2007; Aussel et al., 2014), and UbiX proteins are metal-independent and require dimethylallyl-monophosphate as substrate (White et al., 2015). During the biosynthesis of UQ, UbiX acts as a flavin PT, producing a flavin-derived cofactor required for the decarboxylase activity of UbiD (Payne et al., 2015; White et al., 2015). pad1 and fdc1 are two fungal genes related to bacterial ubiX and ubiD (Mukai et al., 2010; Lin et al., 2015; Payne et al., 2015). Although Pad1 and Fdc1 proteins are homologous with UbiX and UbiD, respectively, and UbiX can activate Fdc1, they are not essential for UQ synthesis in yeast (Mukai et al., 2010; Lin et al., 2015). Thus, the decarboxylation step is largely unknown in eukaryotes. Feeding the isolated mitochondria from potato tubers with 14C-labeled IPP, 4HB and S-adenosylmethionine shows the accumulation of methoxy-4-hydroxy-5-decaprenylbenzoate. It indicates the occurrence of decarboxylation after the first hydroxylation and subsequent O-methylation in plants; however, the immediate methoxy-4-hydroxy-5-decaprenylbenzoate cannot be detected in potato tubers (Lütke-Brinkhaus et al., 1984; Lütke-Brinkhaus and Kleinig, 1987), and no plant genes and enzymes involved in the decarboxylation of UQ aromatic ring have been identified.
In addition to the enzymes mentioned above, various other enzymes, such as yeast COQ4, COQ8, and COQ9, are potentially involved in UQ benzene quinone ring modification, although their actual functions are currently unknown (Tran and Clarke, 2007). Moreover, it seems that modification enzymes involved in modification of UQ benzene quinone ring form a multi-subunit complex. The interaction among subunits guarantees the normal function of enzymes.
Both PQ and UQ are functionally important electron transporters in plants. PQ is involved in the electron transport chain of oxygenic photosynthesis, whereas UQ works exclusively as an electron carrier in the aerobic respiratory chain (Cramer et al., 2011; de Dieu Ndikubwimana and Lee, 2014; Parmar et al., 2015). In addition to their basal functions in photophosphorylation and oxidative phosphorylation, PQ and UQ also play indispensable roles in plant growth and development through participating in the biosynthesis or metabolism of various important chemical compounds, acting as antioxidants, being involved in plant response to stress, and regulating gene expression and cell signal transduction.
It has been shown that PQ and UQ are involved in the biosynthesis or metabolism of various important chemical compounds in plants. For instance, PQ participates in the biosynthesis of carotenoids (Norris et al., 1995), abscisic acid (ABA) (Rock and Zeevaart, 1991) and gibberellin (GA) (Nievelstein et al., 1995), whereas UQ is involved in branch-chain amino acid metabolism (Ishizaki et al., 2006; Araújo et al., 2010).
Carotenoids are C40 tetraterpenoids functioned as accessory light-harvesting pigments in photosynthetic tissues. In non-photosynthetic tissues, such as fruits and flowers, high levels of carotenoids often bring intense orange, yellow and red colors (Pfander, 1992). During carotenoid biosynthesis, the phytoene desaturation reaction is a rate-limiting step. A certain quinone/hydroquinone balance is necessary for optimal phytoene desaturation (Mayer et al., 1990). In an anaerobic environment, the oxidized quinones rather than reduced quinones are involved in the desaturation of phytoene (Mayer et al., 1990). pds1 and pds2 are two Arabidopsis mutants showing albino phenotype (Norris et al., 1995). The mutations affect phytoene desaturation and cause accumulation of phytoene, but they are not occurred in the phytoene desaturation enzyme. Analysis of pds1 and pds2 shows that pds1 is 4-hydroxypheylpyruvate dioxygenase deficient (Norris et al., 1998), whereas pds2 is deficient in HST, a critical enzyme involved in PQ biosynthesis (Tian et al., 2007). Both of the mutations lead to plastoquionone/tocopherol absence from different aspects in Arabidopsis, providing conclusive evidence that PQ is an essential component in phytoene desaturation (Nievelstein et al., 1995; Norris et al., 1995).
Since the plant hormone ABA is synthesized by oxidative cleavage of epoxy-carotenoids (Rock and Zeevaart, 1991), it is reasonable that PQ is also important for ABA biosynthesis. In a T-DNA insertion mutant of HST gene (pds2-1), not only PQ but also carotenoids, ABA and GA3 levels are dramatically reduced (Chao et al., 2014). PQ works as the co-factor of phytoene desaturase and ζ-carotene desaturase and is the immediate electron acceptor in carotenoid and ABA biosynthesis (Chao et al., 2014). Disruption of HST gene results in PQ content decrease, which subsequently affects carotenoid and ABA biosynthesis. On the other hand, the biosynthesis of PQ, carotenoid, ABA and GA shares the common precursor, GGPP (Ma et al., 2012; Chao et al., 2014; Du et al., 2015; Zhang and Lu, 2016). In the pds2-1 mutant, expression of GA biosynthesis genes, such as GA1, GA2, and GA3, is significantly down-regulated (Chao et al., 2014). Consistently, in the Arabidopsis phytoene desaturase gene mutant (pds3), gibberellin biosynthesis is impaired (Qin et al., 2007). It indicates that PQ may affect the biosynthesis of other chemical compounds through negative feedback regulation or other indirect mechanisms.
The relationship between UQ and amino acids lies in two aspects: (1) The precursors of UQ biosynthesis are derived from amino acids, including phenylalanine and tyrosine; and (2) UQ is involved in catabolism of some branched-chain amino acids in mitochondria. It is well known that mitochondrion is an integration point of cellular metabolism and signaling. Amino acids are not only metabolized in peroxisomes but also broken down in mitochondria, which provide carbon skeletons for biosynthesis of many important compounds, such as vitamins, amino acids, and lipids (Sweetlove et al., 2007). In Arabidopsis, leucine is catabolized to form isovaleryl-CoA in mitochondrial matrix, and then the intermediate of the leucine catabolic pathway, isovaleryl-CoA, is dehydrogenated to 3-methylcrotonyl-CoA by isovaleryl-CoA dehydrogenase (IVD). This process occurs on the matrix face of the inner mitochondrial membrane and an electron is transferred through the electron-transfer flavoprotein/electron-transfer falvoprotein:ubiquinone oxidoreductase (ETF/ETFQO) system, first to flavoprotein and then to flavoprotein:ubiquione oxidoreductase (Ishizaki et al., 2006; Araújo et al., 2010). UQ is the final acceptor of electrons in the decomposition of leucine. Similarly, electrons produced during the catabolism of lysine can also be channeled to the mitochondrial electron transport chain (Araújo et al., 2010).
Plastoquinone and ubiquinone can scavenge free radicals to prevent lipid peroxidation, protein oxidation and DNA damage in plant response to biotic and abiotic stresses. They exert antioxidant activity in the reduced forms, plastoquinol and uibiquinol, located in chloroplast thylakoid membrane and mitochondrial membrane, respectively. Analysis of the antioxidant effect of reduced PQ in isolated spinach thylakoid membranes showed that the reduced PQ acted as a scavenger of toxic oxygen species generated in the thylakoid membranes under strong illumination stress (Hundal et al., 1995). Reduced PQ inhibits lipid peroxidation and pigment bleaching, whereas oxidized PQ plays an opposite role (Hundal et al., 1995). In PQ-depleted spinach PSII membranes, exogenously added plastoquinol serves as an efficient scavenger of singlet oxygen (Yadav et al., 2010). Similarly, in C. reinhardtii cells, the level of reduced PQ markedly increased under high-light stress. When pyrazolate, an inhibitor of PQ and tocopherol biosynthesis, was added, the content of reduced PQ quickly decreases (Kruk et al., 2005). Further analyzing the turnover of plastoquinol showed that, due to scavenging of singlet oxygen, the reduced PQ underwent high turnover rate under high-light conditions (Kruk and Trebst, 2008). Moreover, the redox state of PQ pool was found to be an upstream master switch associated with programmed cell death in Arabidopsis leaves in response to excess excitation energy and may be play a central role in the light acclimation of diatoms (Mühlenbock et al., 2008; Lepetit et al., 2013). PSII photoinhibition occurred as a consequence of more reduced PQ pool (Darwish et al., 2015).
In addition to abiotic stress, PQ is also involved in plant response to biotic stress. When Solanum nigrum was treated with the pathogen Phytophthora infestans-derived elicitor, reactive oxygen species (ROS) production, lipid peroxidation and lipoxygenase activity were elevated (Maciejewska et al., 2002). These events were accompanied by a significant increase in PQ level. The increase of PQ level was more significant in plants growing in darkness than under continuous light. It suggests that PQ may be involved in maintaining a tightly controlled balance between the accumulation of ROS and antioxidant activity (Maciejewska et al., 2002). Mesembryanthemum crystalinum performs C3 and CAM carbon metabolism. Analysis of M. crystalinum plants infected with pathogen Botrytis cinere showed that the redox state of PQ pool modifies plant response to biotic stress and hypersensitive-like response is accelerated when PQ pool is in the reduced state (Nosek et al., 2015).
Ubiquinone is an obligatory element of mitochondrial functions in both animals and plants. The antioxidant activity of UQ has been extensively characterized in animals (Littarru and Tiano, 2007, 2010). It prevents DNA damage and cell membrane lipid peroxidation through the elimination of ROS. Same as PQ, UQ also has two forms: the reduced type (ubiquinol) and the oxidized one (ubiquinone), of which ubiquinol is the form exerting antioxidant activity. Overexpression of yeast coq2 (p-hydroxybenzoate poliprenyltransferase) in tobacco resulted in the increase of UQ in transgenic lines (Ohara et al., 2004). Transgenics with the higher UQ level showed the greater tolerance to oxidative stresses caused by methyl viologen or high salinity (Ohara et al., 2004). Analysis of the suspension-cultured Chorispora bungeana cells showed that the redox transition of UQ played key roles in adaptation of cellular regulations under chilling stress (Chang et al., 2006). In addition, the redox state of UQ determines the levels of ROS and plays a key regulatory role in Arabidopsis basal resistance against bacterial pathogens and in response to high oxidative stress environments (Dutta et al., 2015).
Actually, the PQ and UQ pools play a dual role: (1) reducing O2 to superoxide by semiquinone; and (2) reducing superoxide to hydrogen peroxide by hydroquinone. In plant cells, the predominant ROS involved in plant defense includes superoxide and hydrogen peroxide, which are distributed in different pools. Moreover, ROS are generated in two ways. One is elicited by external stresses, such as environmental stresses and biotic stresses. The other way is produced through metabolic processes in the cells, such as the electron transport chains in mitochondria and chloroplasts (Mubarakshina and Ivanov, 2010; Tripathy and Oelmuller, 2012). An unknown interaction may be existed between different pools in modulation of ROS generation and plant response to stress.
Plastoquinone and ubiquinone may regulate cell signaling and gene expression indirectly through the generation of hydrogen peroxide, an important signaling molecule in plant resistance and cell metabolism. They can also directly regulate the expression of genes involved in cell metabolism. For instance, UQ10 influences the expression of hundreds of human genes involved in different cellular pathways. Among them, seventeen are functionally connected by signaling pathways of G-protein coupled receptor, JAK/STAT, integrin and β-arrestin (Schmelzer et al., 2007). These UQ10-inducible genes possess a common promoter framework with binding domains of transcription factor families EVII, HOXF, HOXC and CLOX (Schmelzer et al., 2007). Moreover, UQ10–mediated gene-gene network are involved in inflammation, cell differentiation, and peroxisome proliferator-activated receptor signaling (Schmelzer et al., 2011).
In plants, Chla/b-binding protein complex II (LHC II) and NADPH dehydrogenase complex are two important protein complexes in photosynthesis. The cytochrome b6f deficient mutant of lemna perpusilla maintains a low level of the light-harvesting chl a/b-binding protein complex II (LHC II) at low-light intensities (Yang et al., 2001). Inhibiting the reduction of PQ pool increases the level of LHC II in the mutant at both low- and high-light intensities, whereas the level of LHC II is increased in wild-type plants only under high-light conditions (Yang et al., 2001). It suggests that the redox state of PQ is an important signal-transducing component in plant photoacclimation process (Yang et al., 2001). Analysis of gene expression using DNA microarray technology showed that 663 genes were differentially expressed in A. thaliana under low, medium, high and excessive irradiances, of which 50 genes were reverted by 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), an inhibitor of the photosynthetic electron transport chain (Adamiec et al., 2008). It indicates that the expression of the 50 genes is regulated by the redox state of PQ pool. Hierarchical clustering and promoter motif analysis showed that the promoter regions of PQ-regulated genes contain conserved cis-acting elements involved in signal transduction from the redox state of the PQ pool (Adamiec et al., 2008). Active NADPH dehydrogenase complex is necessary for cyclic electron transport in photosystem I (PSI) and respiration. In glucose-treated cyanobacteria Synechocystis sp. strain PCC 6803 cells, NADPH dehydrogenase complex activity was inhibited and the cyclic PSI rate was decreased. In contrast, when the cells were treated with DCMU, the activity of NADPH dehydrogenase was significantly stimulated (Ma et al., 2008). Glucose treatment causes partial reduction of the PQ pool, whereas DCMU results in overoxidation. Differential responses of enzyme activity and cyclic PSI rate to glucose and DCMU treatments indicate that the redox state of PQ pool controls the NADPH dehydrogenase complex activity and further influences on cyclic PSI (Ma et al., 2008).
In addition to PQ, UQ is also involved in signal transduction in plants. Comparative analysis of hypersensitive tobacco Nicotiana tabacum L. variety Samsun NN treated with UQ10 and TMV and those treated with TMV only showed that UQ10 and TMV-treated tobbaco had less number of lesions and TMV and greater change of plant hormone levels, including the decrease of ABA and increase of IAA level (Rozhnova and Gerashchenkov, 2006). It indicates that UQ10 has a protective antiviral effect through controlling plant hormonal status (Rozhnova and Gerashchenkov, 2008). On the other hand, it has been reported that the ROS level generated by UQ redox state is a threshold for successful basal resistance response in plants (Dutta et al., 2015). In plant defenses, ROS acts as signaling molecules directly or mediates the generation of phytoalexins or serves as a source for activation of further defenses indirectly (Kovtun et al., 2000; Thoma et al., 2003; Mur et al., 2008). Plants employ both pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) in basal and R gene-mediated defense response. ROS is induced rapidly and transiently and then mediates signaling during PTI and ETI (Frederickson Matika and Loake, 2013). The induction of ROS is considered as a defining hallmark of identification and subsequent defense activation against pathogens (Torres et al., 2006; Dutta et al., 2015). Moreover, UQ is involved in the mitochondrial glycerol-3-phosphate shuttle for redox homeostasis in plants (Shen et al., 2007) and serves as mitochondrial permeability transition pore in cell metabolism (Amirsadeghi et al., 2007; Reape et al., 2007).
Plastoquinone is specific to plants. It has not been directly utilized for human. However, various synthesized PQ derivatives, such as SkQ1 (plastoquinonyl-decyl-triphenylphosphonium), SkQR1 (the rhodamine- containing analog of SkQ1) and SkQ3 (methylplastoquinonyl-decyl-triphenylphosphonium), were reported to show antioxidant and protonophore activity (Skulachev et al., 2011). They are able to penetrate cell membranes and potentially used in anti-aging treatment (Anisimov et al., 2008, 2011; Obukhova et al., 2009). SkQ1 is currently under clinical trials for glaucoma treatment in Russia (Iomdina et al., 2015). The phase 2 clinical trial indicates that SkQ1 is safe and efficacious in treating dry eye signs and symptoms (Petrov et al., 2016). In plants, SkQ1 and SkQ3 can retard the senescence of Arabidopsis rosette leaves and their death, increase the vegetative period, and improve crop structure of wheat (Dzyubinskaya et al., 2013). In addition, SkQ1 effectively suppresses the development of p50-induced PCD in tobacco plants through inhibiting ROS production (Solovieva et al., 2013; Samuilov and Kiselevsky, 2015). The role of SkQ1 and SkQ3 played in cells is mainly based on its antioxidant activity.
Compared with PQ, the practical application of UQ, particularly UQ10, has attracted more attention. UQ10 is effective in treating cardiovascular diseases, particularly in preventing and treating hypertension, hyperlipidemia, coronary artery disease and heart failure (Tran et al., 2001; Moludi et al., 2015). In the past decades, many studies have reported the remarkable clinical benefits of UQ10 and illuminated its antioxidant activity as the basis of pathology (Lee et al., 2012). Moreover, UQ10 controls energy metabolism and regulates cell death via redox signaling, indicating its potential in cancer treatment (Chai et al., 2010). Moderate UQ10 levels have favorable impact on breast cancer (Folkers et al., 1993; Lockwood et al., 1994; Premkumar et al., 2008). In addition, UQ10 can enhance viral immunity and affect the development of AIDS (Folkers et al., 1988, 1991). UQ10 is also closely related to human reproductive health. Moderate UQ10 supplement can effectively reduce the risk of spontaneous abortion and develop pre-eclampsia in pregnant women (Palan et al., 2004; Teran et al., 2009). Exogenous administration of UQ10 can increase sperm cell motility and the mean pregnancy rate. Its positive role in the treatment of male infertility also relies on the antioxidant properties and bioenergetics (Balercia et al., 2004; Mancini et al., 2005; Balercia et al., 2009; Mancini and Balercia, 2011; Safarinejad, 2012). As a good immunomdulator, UQ10 has been used to treat chronic gingivitis and periodontitis (Chatterjee et al., 2012; Hans et al., 2012). A series of toothpaste containing UQ10 are developed and sold in the market. Other UQ10-containing chemical products include cleanser, cosmetic products, and healthy foods. Similar to PQ derivatives, the role of UQ played in cells relied on the antioxidant activity. As an antioxidant, UQ effectively scavenge ROS and prevent ROS-induced damage to membrane lipids, DNA, and proteins.
Plastoquinone plays significant roles in plants; however, it is not directly used in human life. Moreover, PQ derivatives are mainly synthesized by chemical methods. Metabolic engineering of PQ and its derivatives is rarely reported. Differing from PQ, metabolic engineering of UQ, particularly UQ10, has been conducted in prokaryotes and eukaryotes, including bacterial, yeast and plants. Currently, the majority of commercially available UQ10 comes from yeast fermentation and chemical synthesis. Compared with microbial fermentation, chemical synthesis of UQ10 is more expensive and produces environmentally harmful waste products. Additionally, the scalability of both yeast fermentation and chemical synthesis is limited. Thus, plants are thought to be an attractive alternative source of UQ10.
The natural UQ producers, such as Agrobacterium tumefaciens, Paracoccus denitrificans, Rhodobacter sphaeroides, and their chemical mutants have been successfully used for commercial production of UQ; while, with the increase of knowledge about enzymes involved in UQ biosynthesis and regulatory mechanisms modulating UQ production, opportunities have arisen for UQ metabolic engineering in other organisms. For instance, overexpression of some key genes, such as ubiA encoding p-hydroxybenzoate-polyprenyl pyrophosphate transferase, ispB encoding polyprenyl pyrophosphate synthetase and ubiCA, in E. coli may achieve the level of UQ content 3-4 times to that of wild-type cells (Zhu et al., 1995; Jiang et al., 2006). Even though, UQ production using these methods does not meet industrial needs, which require a yield of higher than 500 mg/L (Cluis et al., 2007). Therefore, in addition to the highly efficient microbial system, growth condition optimization and alteration of cellular regulatory mechanisms are important for UQ production (Sakato et al., 1992; Zhang et al., 2007a,b). Recently, multiple strategies have been employed in improving UQ production. One hundred and eighty percent increase of UQ8 content is achieved in E. coli (ΔmenA) through a comprehensive approach, including blocking menaquinone pathway, coexpressing dxs-ubiA, and supplementing PYR and pHBA (Xu et al., 2014). The highest UQ10 titer and yield, 433 mg/L, is obtained in engineered E. coli through integrating dps into chromosome of E. coli ATCC8739, modulating dxs and idi genes of the MEP pathway and ubiCA genes, and recruiting the glucose facilitator protein of Zymomonas mobilis was to replace native phosphoenolpyruvate: carbohydrate phosphotransferase systems (PTS) (Dai et al., 2015).
Metabolic engineering of UQ in plants mainly concentrated on UQ10 production. Although UQ widely exists in plant cells, most cereal crops produce mainly UQ9. Tomato, Datura tatula and tobacco BY-2 cells can produce UQ10 naturally; however, its yield is very limited (Ikeda and Kagei, 1979; Ikeda et al., 1981; Matsumoto et al., 1981). It has been shown that overexpression of rate-limiting genes and increase of UQ precursors can improve UQ production in plants. Expression of ddsA from Gluconobacter suboxydans in rice leads to efficient production of UQ10 in rice seeds (Sakiko et al., 2006, 2010). Since PPT plays the catalyzing role in a rate-limiting step of the UQ biosynthesis pathway, it is a significant target for UQ metabolic engineering in plants (Ohara et al., 2004; Stiff, 2010; Parmar et al., 2015). For instance, expression of yeast coq2 gene resulted in a sixfold increase of UQ10 levels in transgenic tobacco plants (Ohara et al., 2004). Compared with wild type plants, coq2 transgenic tobacco with high UQ10 level are more resistant to oxidative stresses caused by methyl viologen or high salinity (Ohara et al., 2004). Similarly, overexpression of AtPPT1 in tobacco increases UQ10 content and enhances oxidative stress tolerance caused by high NaCl (Stiff, 2010). Increase of UQ precursors, such as 4HB and/or PPS, may potentially improve UQ production in plants (Sommer and Heide, 1998; Viitanen et al., 2004). However, due to the complex relationship among precursors, UQ production and many other intersecting metabolic pathways, the expected goal of improving UQ production is difficult to achieve. Further improvement of UQ content in plant cells may be expected using comprehensive approaches (Kumar et al., 2012), such as improving the amount of UQ precursors combined with overexpression of rate-limiting genes.
Plastoquinone and ubiquinone are two important compounds in plants. They function as electron transporters in the electron transport chain of oxygenic photosynthesis and the aerobic respiratory chain, respectively, and play indispensable roles in plant growth and development. UQ, particularly UQ10, has also been widely used in people’s life. Great efforts have been done to elucidate their biosynthetic pathways and genes associated with PQ and UQ production. As shown in Figure 2 and Table 1, significant achievements have been made. However, there are still several issues related to the biosynthetic pathways, regulatory mechanisms and metabolic engineering need to be addressed.
Although a great amount of studies have been done to the MEP and MVA pathways and various enzymes, such as PPT and HST, involved in the attachment of isoprenoid side chain to the benzoquinone ring, pathways and enzymes involved in isoprenoid side chain elongation, UQ benzoquinone ring biosynthesis, and PQ and UQ benoquinone ring modification are largely unknown. The key enzyme responsible for 4HB production has not been identified (Block et al., 2014). PPSs are a group of enzymes converting IPP and DMAPP to diphosphate precursors. Each PPS may play different role in isoprenoid side chain elongation, which needs to be clarified. Although various enzymes involved in benoqunone ring modifications have been identified in bacteria and yeast, few achievements have been made in plants. With more and more transcriptome and whole genome sequence available, gene network reconstruction becomes possible and can be used to address these problems.
Non-coding RNAs, including small RNAs and long non-coding RNAs, play significant regulatory roles in many aspects of plants (Lu et al., 2005, 2007, 2008; Lu S. et al., 2013; Li D. et al., 2015; Wang et al., 2015). Various microRNAs have been identified to be associated with secondary metabolism (Wu et al., 2012; Lu S. et al., 2013; Fan et al., 2015; Wei et al., 2015). However, the regulatory roles of non-coding RNAs in PQ and UQ biosynthesis have not been revealed. In addition to non-coding RNAs, transcription factors, such as MYB, WRKY and SPL, potentially play significant regulatory roles in PQ and UQ biosynthesis (Li and Lu, 2014a,b; Zhang et al., 2014; Li C. et al., 2015), which needs to be further demonstrated. Understanding the regulatory mechanisms of PQ and UQ are important for manipulating the content of PQ and UQ in plants.
Metabolic engineering of UQ10 has been successfully performed in bacteria and yeast. However, they were found to be low yield and high production cost. UQ10 metabolic engineering in plants has various advantages and great perspectives, whereas current efforts are limited to a few plant species (Parmar et al., 2015). Increasing UQ10 content in UQ10-producing plant species and engineering UQ10 in non-UQ10-producing plant species are two routes for UQ10 metabolic engineering in plants. With more and more genes involved in UQ10 biosynthesis and regulation to be identified, great achievements may be expected for UQ10 production in plants.
Both authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the National Key Research and Development Program of China (2016YFD0600104); the Natural Science Foundation of China (31370327, 31570667); the Beijing Natural Science Foundation (5152021); the CAMS Innovation Fund for Medical Sciences (CIFMS) (2016-I2M-3-016); and the Fund for Postgraduate Innovation in Peking Union Medical College (2015-1007-14).
Adamiec, M., Drath, M., and Jackowski, G. (2008). Redox state of plastoquionone pool regulates expression of Arabidopsis thaliana genes in response to elevated irradiance. Acta Biochim. Pol. 55, 161–173.
Amirsadeghi, S., Robson, C. A., and Vanlerberghe, G. C. (2007). The role of the mitochondrion in plant responses to biotic stress. Physiol. Plant. 129, 253–266. doi: 10.1111/j.1399-3054.2006.00775.x
Anisimov, V. N., Bakeeva, L. E., Egormin, P. A., Filenko, O. F., Isakova, E. F., Manskikh, V. N., et al. (2008). Mitochondria-targeted plastoquinone derivatives as tools to interrupt execution of the aging program. 5. SkQ1 prolongs lifespan and prevents development of traits of senescence. Biochemistry 73, 1329–1342. doi: 10.1134/S0006297908120055
Anisimov, V. N., Egorov, M. V., Krasilshchikova, M. S., Lyamzaev, K. G., Manskikh, V. N., Moshkin, M. P., et al. (2011). Effects of the mitochondria-targeted antioxidant SkQ1 on lifespan of rodents. Aging 3, 1110–1119. doi: 10.18632/aging.1000404
Araújo, W. L., Ishizaki, K., Nunes-Nesi, A., Larson, T. R., Tohge, T., Krahnert, I., et al. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22, 1549–1563. doi: 10.1105/tpc.110.075630
Ashby, M. N., Kutsunai, S. Y., Ackerman, S., Tzagoloff, A., and Edwards, P. A. (1992). COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate: polyprenyltransferase. J. Biol. Chem. 267, 4128–4136.
Aussel, L., Pierrel, F., Loiseau, L., Lombard, M., Fontecave, M., and Barras, F. (2014). Biosynthesis and physiology of coenzyme Q in bacteria. Biochim. Biophys. Acta 1837, 1004–1011. doi: 10.1016/j.bbabio.2014.01.015
Avelange-Macherel, M. H., and Joyard, J. (1998). Cloning and functional expression of AtCOQ3, the Arabidopsis homologue of the yeast COQ3 gene, encoding a methyltransferase from plant mitochondria involved in ubiquinone biosynthesis. Plant J. 14, 203–213. doi: 10.1046/j.1365-313X.1998.00109.x
Baba, S. W., Belogrudov, G. I., Lee, J. C., Lee, P. T., Strahan, J., Shepherd, J. N., et al. (2004). Yeast Coq5 C-methyltransferase is required for stability of other polypeptides involved in coenzyme Q biosynthesis. J. Biol. Chem. 279, 10052–10059. doi: 10.1074/jbc.M313712200
Balercia, G., Buldreghini, E., Vignini, A., Tiano, L., Paggi, F., Amoroso, S., et al. (2009). Coenzyme Q10 treatment in infertile men with idiopathic asthenozoospermia: a placebo-controlled, double-blind randomized trial. Fertil. Steril. 91, 1785–1792. doi: 10.1016/j.fertnstert.2008.02.119
Balercia, G., Mosca, F., Mantero, F., Boscaro, M., Mancini, A., Ricciardo-Lamonica, G., et al. (2004). Coenzyme Q(10) supplementation in infertile men with idiopathic asthenozoospermia: an open, uncontrolled pilot study. Fertil. Steril. 81, 93–98. doi: 10.1016/j.fertnstert.2003.05.009
Barkovich, R. J., Shtanko, A., Shepherd, J. A., Lee, P. T., Myles, D. C., Tzagoloff, A., et al. (1997). Characterization of the COQ5 gene from Saccharomyces cerevisiae. Evidence for a C-methyltransferase in ubiquinone biosynthesis. J. Biol. Chem. 272, 9182–9188. doi: 10.1074/jbc.272.14.9182
Bentinger, M., Tekle, M., and Dallner, G. (2010). Coenzyme Q–biosynthesis and functions. Biochem. Biophys. Res. Commun. 396, 74–79. doi: 10.1016/j.bbrc.2010.02.147
Blanc, G., Agarkova, I., Grimwood, J., Kuo, A., Brueggeman, A., Dunigan, D. D., et al. (2012). The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 13:R39. doi: 10.1186/gb-2012-13-5-r39
Blanc, G., Duncan, G., Agarakova, I., Borodovsky, M., Gumon, J., Kuo, A., et al. (2010). The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22, 2943–2955. doi: 10.1105/tpc.110.076406
Block, A., Fristedt, R., Rogers, S., Kumar, J., Barnes, B., Barnes, J., et al. (2013). Functional modeling identifies paralogous solanesyl-diphosphate synthases that assemble the side chain of plastoquinone-9 in plastids. J. Biol. Chem. 288, 27594–27606. doi: 10.1074/jbc.M113.492769
Block, A., Widhalm, J. R., Fatihi, A., Cahoon, R. E., Wamboldt, Y., Elowsky, C., et al. (2014). The origin and biosynthesis of the benzenoid moiety of ubiquinone (Coenzyme Q) in Arabidopsis. Plant Cell 26, 1938–1948. doi: 10.1105/tpc.114.125807
Bouvier, F., Suire, C., d’Harlingue, A., Backhaus, R. A., and Camara, B. (2000). Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. Plant J. 24, 241–252. doi: 10.1046/j.1365-313x.2000.00875.x
Chai, W., Cooney, R. V., Franke, A. A., Shvetsov, Y. B., Caberto, C. P., Wilkens, L. R., et al. (2010). Plasma coenzyme Q10 levels and postmenopausal breast cancer risk: the multiethnic cohort study. Cancer Epidemiol. Biomarkers Prev. 19, 2351–2356. doi: 10.1158/1055-9965.EPI-10-0396
Chang, J., Fu, X., An, L., and Chen, T. (2006). Properties of cellular ubiquinone and stress-resistance in suspension-cultured cells of Chorispora bungeana during early chilling. Environ. Exp. Bot. 57, 116–122. doi: 10.1016/j.envexpbot.2005.05.001
Chao, Y., Kang, J., Zhang, T., Yang, Q., Gruber, M. Y., and Sun, Y. (2014). Disruption of the homogentisate solanesyltransferase gene results in albino and dwarf phenotypes and root, trichome and stomata defects in Arabidopsis thaliana. PLoS ONE 9:e94031. doi: 10.1371/journal.pone.0094031
Chatterjee, A., Kandwal, A., Singh, N., and Singh, A. (2012). Evaluation of Co-Q10 anti-gingivitis effect on plaque induced gingivitis: a randomized controlled clinical trial. J. Indian Soc. Periodontol. 16, 539–542. doi: 10.4103/0972-124X.106902
Chen, S. W., Liu, C. C., and Yen, H. C. (2013). Detection of suppressed maturation of the human COQ5 protein in the mitochondria following mitochondrial uncoupling by an antibody recognizing both precursor and mature forms of COQ5. Mitochondrion 13, 143–152. doi: 10.1016/j.mito.2013.01.007
Cheng, Z., Sattler, S., Maeda, H., Sakuragi, Y., Bryant, D. A., and DellaPenna, D. (2003). Highly divergent methyltransferases catalyze a conserved reaction in tocopherol and plastoquinone synthesis in cyanobacteria and photosynthetic eukaryotes. Plant Cell 15, 2343–2356. doi: 10.1015/tpc.013656
Clarke, C. F. (2000). New advances in coenzyme Q biosynthesis. Protoplasma 213, 134–147. doi: 10.1007/BF01282151
Clarke, C. F., Williams, W., and Teruya, J. H. (1991). Ubiquinone biosynthesis in Saccharomyces cerevisiae. Isolation and sequence of COQ3, the 3,4-dihydroxy-5-hexaprenylbenzoate methyltransferase gene. J. Biol. Chem. 266, 16636–16644.
Cluis, C. P., Burja, A. M., and Martin, V. J. (2007). Current prospects for the production of coenzyme Q10 in microbes. Trends Biotechnol. 25, 514–521. doi: 10.1016/j.tibtech.2007.08.008
Collakova, E., and DellaPenna, D. (2001). Isolation and functional analysis of homogentisate phytyltransferase from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol. 127, 1113–1124. doi: 10.1104/pp.127.3.1113
Cramer, W. A., Hasan, S. S., and Yamashita, E. (2011). The Q cycle of cytochrome bc complexes: a structure perspective. Biochim. Biophys. Acta 1807, 788–802. doi: 10.1016/j.bbabio.2011.02.006
Dai, D., Miao, L., Sun, T., Li, Q., Xiao, D., and Zhang, X. (2015). Production of coenzyme Q(10) by metabolically engineered Escherichia coli. Chin. J. Biotechnol. 31, 206–219. doi: 10.13345/j.cjb.140211
Dai, Y. N., Zhou, K., Cao, D. D., Jiang, Y. L., Meng, F., Chi, C. B., et al. (2014). Crystal structures and catalytic mechanism of the C-methyltransferase Coq5 provide insights into a key step of the yeast coenzyme Q synthesis pathway. Acta Crystallogr. D Biol. Crystallogr. 70(Pt 8), 2085–2092. doi: 10.1107/S1399004714011559
Darwish, M., Vidal, V., Lopez-Lauri, F., Alnaser, O., Junglee, S., El Maataoui, M., et al. (2015). Tolerance to clomazone herbicide is linked to the state of LHC, PQ-pool and ROS detoxification in tobacco (Nicotiana tabacum L.). J. Plant Physiol. 175, 122–130. doi: 10.1016/j.jplph.2014.11.009
de Dieu Ndikubwimana, J., and Lee, B. H. (2014). Enhanced production techniques, properties and uses of coenzyme Q10. Biotechnol. Lett. 36, 1917–1926. doi: 10.1007/s10529-014-1587-1
Devadas, S. K., Enyedi, A., and Raina, R. (2002). The Arabidopsis hrl1 mutation reveals novel overlapping roles for salicylic acid, jasmonic acid and ethylene signalling in cell death and defence against pathogens. Plant J. 30, 467–480. doi: 10.1046/j.1365-313X.2002.01300.x
Du, Q., Li, C., Li, D., and Lu, S. (2015). Genome-wide analysis, molecular cloning and expression profiling reveal tissue-specifically expressed, feedback-regulated, stress-responsive and alternatively spliced novel genes involved in gibberellin metabolism in Salvia miltiorrhiza. BMC Genomics 16:1087. doi: 10.1186/s12864-015-2315-5
Ducluzeau, A. L., Wamboldt, Y., Elowsky, C. G., Mackenzie, S. A., Schuurink, R. C., and Basset, G. J. (2012). Gene network reconstruction identifies the authentic trans-prenyl diphosphate synthase that makes the solanesyl moiety of ubiquinone-9 in Arabidopsis. Plant J. 69, 366–375. doi: 10.1111/j.1365-313X.2011.04796.x
Dutta, A., Chan, S. H., Pauli, N. T., and Raina, R. (2015). HYPERSENSITIVE RESPONSE-LIKE LESIONS 1 codes for AtPPT1 and regulates accumulation of ROS and defense against bacterial pathogen Pseudomonas syringae in Arabidopsis thaliana. Antioxid. Redox Signal. 22, 785–796. doi: 10.1089/ars.2014.5963
Dzyubinskaya, E. V., Ionenko, I. F., Kiselevsky, D. B., Samuilov, V. D., and Samuilov, F. D. (2013). Mitochondria-addressed cations decelerate the leaf senescence and death in Arabidopsis thaliana and increase the vegetative period and improve crop structure of the wheat Triticum aestivum. Biochemistry 78, 68–74. doi: 10.1134/S0006297913010082
Ewbank, J. J., Barnes, T. M., Lakowski, B., Lussier, M., Bussey, H., and Hekimi, S. (1997). Structural and functional conservation of the Caenorhabditis elegans timing gene clk-1. Science 275, 980–983. doi: 10.1126/science.275.5302.980
Fan, R., Li, Y., Li, C., and Zhang, Y. (2015). Differential microRNA analysis of glandular trichomes and young leaves in Xanthium strumarium L. reveals their putative roles in regulating terpenoid biosynthesis. PLoS ONE 10:e0139002. doi: 10.1371/journal.pone.0139002
Folkers, K., Brown, R., Judy, W. V., and Morita, M. (1993). Survival of cancer patients on therapy with coenzyme Q10. Biochem. Biophys. Res. Commun. 192, 241–245. doi: 10.1006/bbrc.1993.1405
Folkers, K., Hanioka, T., Xia, L. J., McRee, J. T. Jr., and Langsjoen, P. (1991). Coenzyme Q10 increases T4/T8 ratios of lymphocytes in ordinary subjects and relevance to patients having the AIDS related complex. Biochem. Biophys. Res. Commun. 176, 786–791. doi: 10.1016/S0006-291X(05)80254-2
Folkers, K., Langsjoen, P., Nara, Y., Muratsu, K., Komorowski, J., Richardson, P. C., et al. (1988). Biochemical deficiencies of coenzyme Q10 in HIV-infection and exploratory treatment. Biochem. Biophys. Res. Commun. 153, 888–896. doi: 10.1016/S0006-291X(88)81179-3
Forsgren, M., Attersand, A., Lake, S., Grünler, J., Swiezewska, E., Dallner, G., et al. (2004). Isolation and functional expression of human COQ2, a gene encoding a polyprenyl transferase involved in the synthesis of CoQ. Biochem. J. 382, 519–526. doi: 10.1042/BJ20040261
Frederickson Matika, D. E., and Loake, G. (2013). Redox regulation in plant immune function. Antioxid. Redox Signal. 21, 1373–1388. doi: 10.1089/ars.2013.5679
Goewert, R. R., Sippel, C. J., and Olson, R. E. (1977). The isolation and identification of a novel intermediate in ubiquinone-6 biosynthesis by Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 77, 599–605. doi: 10.1021/bi00517a041
Gulmezian, M., Hyman, K. R., Marbois, B. N., Clarke, C. F., and Javor, G. T. (2007). The role of UbiX in Escherichia coli coenzyme Q biosynthesis. Arch. Biochem. Biophys. 467, 144–153. doi: 10.1016/j.abb.2007.08.009
Hajj Chehade, M., Loiseau, L., Lombard, M., Pecqueur, L., Ismail, A., Smadja, M., et al. (2013). ubiI, a new gene in Escherichia coli coenzyme Q biosynthesis, is involved in aerobic C5-hydroxylation. J. Biol. Chem. 288, 20085–20092. doi: 10.1074/jbc.M113.480368
Hans, M., Prakash, S., and Gupta, S. (2012). Clinical evaluation of topical application of perio-Q gel (Coenzyme Q10) in chronic periodontitis patients. J. Indian Soc. Periodontol. 16, 193–199. doi: 10.4103/0972-124X.99261
Hayashi, K., Ogiyama, Y., Yokomi, K., Nakagawa, T., Kaino, T., and Kawamukai, M. (2014). Functional conservation of coenzyme Q biosynthetic genes among yeasts, plants, and humans. PLoS ONE 9:e99038. doi: 10.1371/journal.pone.0099038
Heide, L., and Berger, U. (1989). Partial purification and properties of geranyl pyrophosphate synthase from Lithospermum erythrorhizon cell cultures. Arch. Biochem. Biophys. 273, 331–338. doi: 10.1016/0003-9861(89)90491-8
Hemmi, H., Ikejiri, S., Yamashita, S., and Nishino, T. (2002). Novel medium-chain prenyl diphosphate synthase from the thermoacidophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 184, 615–620. doi: 10.1128/JB.184.3.615-620.2002
Hirooka, K., Bamba, T., Fucusaki, E., and Kobayashi, A. (2003). Cloning and kinetic characterization of Arabidopsis thaliana solanesyl diphosphate synthase. Biochem. J. 370(Pt 2), 679–686. doi: 10.1042/BJ20021311
Hirooka, K., Izumi, Y., An, C. I., Nakazawa, Y., Fukusaki, E., and Kobayashi, A. (2005). Functional analysis of two solanesyl diphosphate synthases from Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 69, 592–601. doi: 10.1271/bbb.69.592
Hsieh, F. L., Chang, T. H., Ko, T. P., and Wang, A. H. J. (2011). Structure and mechanism of an Arabidopsis medium/long-chain-length prenyl pyrophosphate synthase. Plant Physiol. 155, 1079–1090. doi: 10.1104/pp.110.168799
Hsu, A. Y., Do, T. Q., Lee, P. T., and Clarke, C. F. (2000). Genetic evidence for a multi-subunit complex in the O-methyltransferase steps of coenzyme Q biosynthesis. Biochim. Biophys. Acta 1484, 287–297. doi: 10.1016/S1388-1981(00)00019-6
Hsu, A. Y., Poon, W. W., Shepherd, J. A., Myles, D. C., and Clarke, C. F. (1996). Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry 35, 9797–9806. doi: 10.1021/bi9602932
Hundal, T., Forsmark-Andrée, P., Ernster, L., and Andersson, B. (1995). Antioxidant activity of reduced plastoquinone in chloroplast thylakoid membranes. Arch. Biochem. Biophys. 324, 117–122. doi: 10.1016/j.bbabio.2007.10.008
Ikeda, M., and Kagei, K. (1979). Ubiquinone content of eight plant species in cell culture. Phytochemistry 18, 1577–1578. doi: 10.1016/S0031-9422(00)98506-6
Ikeda, T., Matsumoto, T., Obi, Y., Kisaki, T., and Noguchi, M. (1981). Characteristics of cultured tobacco cell strains producing high levels of ubiquinone-10 selected by a cell cloning technique. Agric. Biol. Chem. 45, 2259–2263. doi: 10.1080/00021369.1981.10864865
Iomdina, E. N., Khoroshilovamaslova, I. P., Robustova, O. V., Averina, O. A., Kovaleva, N. A., Aliev, G., et al. (2015). Mitochondria-targeted antioxidant SkQ1 reverses glaucomatous lesions in rabbits. Front. Biosci. (Landmark Ed.) 20, 892–901. doi: 10.2741/4343
Ishizaki, K., Schauer, N., Larson, T. R., Graham, I. A., Fernie, A. R., and Leaver, C. J. (2006). The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J. 47, 751–760. doi: 10.1111/j.1365-313X.2006.02826.x
Jiang, Y. E., Lin, M. A., Wu, H., Liu, X., and Zhang, H. (2006). Effect of overexpressing ubiCA genes responsible for ubiquinone biosynthesis on ubiquinone production in Escherichia coli. J. East China Univ. Sci. Technol. (Nat. Sci. Ed.) 2006, 269–273.
Jonassen, T., and Clarke, C. F. (2000). Isolation and functional expression of human COQ3, a gene encoding a methyltransferase required for ubiquinone biosynthesis. J. Biol. Chem. 275, 12381–12387. doi: 10.1074/jbc.275.17.12381
Jonassen, T., Marbois, B. N., Kim, L., Chin, A., Xia, Y. R., Lusis, A. J., et al. (1996). Isolation and sequencing of the rat Coq7 gene and the mapping of mouse Coq7 to chromosome 7. Arch. Biochem. Biophys. 330, 285–289. doi: 10.1006/abbi.1996.0255
Jones, M. O., Perez-Fons, L., Robertson, F. P., Bramley, P. M., and Fraser, P. D. (2013). Functional characterization of long-chain prenyl diphosphate synthases from tomato. Biochem. J. 449, 729–740. doi: 10.1042/BJ20120988
Jun, L., Saiki, R., Tatsumi, K., Nakagawa, T., and Kawamukai, M. (2004). Identification and subcellular localization of two solanesyl diphosphate synthases from Arabidopsis thaliana. Plant Cell Physiol. 45, 1882–1888. doi: 10.1093/pcp/pch211
Kawamukai, M. (2000). Biosynthesis, bioproduction and novel roles of ubiquinone. J. Biosci. Bioeng. 294, 511–517. doi: 10.1263/jbb.94.511
Kellogg, B. A., and Poulter, C. D. (1997). Chain elongation in the isoprenoid biosynthetic pathway. Curr. Opin. Chem. Biol. 1, 570–578. doi: 10.1016/S1367-5931(97)80054-3
Kim, E. H., Lee, Y., and Kim, H. U. (2015). Fibrillin 5 is essential for plastoquinone-9 biosynthesis by binding to solanesyl diphosphate synthases in Arabidopsis. Plant Cell 27, 2956–2971. doi: 10.1105/tpc.15.00707
Kovtun, Y., Chiu, W. L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activatedprotein kinase cascade in plants. Proc. Natl. Acad. Sci. U.S.A. 97, 2940–2945. doi: 10.1073/pnas.97.6.2940
Kruk, J., Holländer-Czytko, H., Oettmeier, W., and Trebst, A. (2005). Tocopherol as singlet oxygen scavenger in photosystem II. J. Plant Physiol. 162, 749–757. doi: 10.1016/j.jplph.2005.04.020
Kruk, J., and Trebst, A. (2008). Plastoquinol as a singlet oxygen scavenger in photosystem II. Biochim. Biophys. Acta 177, 154–162. doi: 10.1016/j.bbabio.2007.10.008
Ksas, B., Becuwe, N., Chevalier, A., and Havaux, M. (2015). Plant tolerance to excess light energy and photooxidative damage relies on plastoquinone biosynthesis. Sci. Rep. 5:10919. doi: 10.1038/srep10919
Kumar, S., Hahn, F. M., Baidoo, E., Kahlon, T. S., Wood, D. F., McMahan, C. M., et al. (2012). Remodeling the isoprenoid pathway in tobacco by expressing the cytoplasmic mevalonate pathway in chloroplasts. Metab. Eng. 14, 19–28. doi: 10.1016/j.ymben.2011.11.005
Lange, B. M., and Ghassemian, M. (2003). Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol. Biol. 51, 925–948. doi: 10.1023/A:1023005504702
Lee, B. J., Huang, Y. C., Chen, S. J., and Lin, P. T. (2012). Coenzyme Q10 supplementation reduces oxidative stress and increases antioxidant enzyme activity in patients with coronary artery disease. Nutrition 28, 250–255. doi: 10.1016/j.nut.2011.06.004
Lepetit, B., Sturm, S., Rogato, A., Gruber, A., Sachse, M., Falciatore, A., et al. (2013). High light acclimation in the secondary plastids containing diatom Phaeodactylum tricornutum is triggered by the redox state of the plastoquinone pool. Plant Physiol. 161, 853–865. doi: 10.1104/pp.112.207811
Li, C., Li, D., Shao, F., and Lu, S. (2015). Molecular cloning and expression analysis of WRKY transcription factor genes in Salvia miltiorrhiza. BMC Genomics 16:200. doi: 10.1186/s12864-015-1411-x
Li, C., and Lu, S. (2014a). Genome-wide characterization and comparative analysis of R2R3-MYB transcription factors shows the complexity of MYB-associated regulatory networks in Salvia miltiorrhiza. BMC Genomics 15:277. doi: 10.1186/1471-2164-15-277
Li, C., and Lu, S. (2014b). Molecular characterization of the SPL gene family in Populus trichocarpa. BMC Plant Biol. 14:131. doi: 10.1186/1471-2229-14-131
Li, D., Shao, F., and Lu, S. (2015). Identification and characterization of mRNA-like noncoding RNAs in Salvia miltiorrhiza. Planta 241, 1131–1143. doi: 10.1007/s00425-015-2246-z
Lin, F., Ferguson, K. L., Boyer, D. R., Lin, X. N., and Marsh, E. N. (2015). Isofunctional enzymes PAD1 and UbiX catalyze formation of a novel cofactor required by ferulic acid decarboxylase and 4-hydroxy-3-polyprenylbenzoic acid decarboxylase. ACS Chem. Biol. 10, 1137–1144. doi: 10.1021/cb5008103
Littarru, G. P., and Tiano, L. (2007). Bioenergetic and antioxidant properties of coenzyme Q10: recent developments. Mol. Biotechnol. 37, 31–37. doi: 10.1007/s12033-007-0052-y
Littarru, G. P., and Tiano, L. (2010). Clinical aspects of coenzyme Q10: an update. Nutrition 26, 250–254. doi: 10.1016/j.nut.2009.08.008
Liu, F. Z., Guo, A. Q., and Wan, Y. S. (2013). Cloning and polymorphism analysis of the 2-methyl-6-phytyl-1,4-benzoquinol methyltransferase gene (VTE3) in Arachis hypogaea, A. duranensis, and A. ipaënsis. Genet. Mol. Res. 12, 1859–1871. doi: 10.4238/2013.January.4.5
Lockwood, K., Moesgaard, S., and Folkers, K. (1994). Partial and complete regression of breast cancer in patients in relation to dosage of coenzyme Q10. Biochem. Biophys. Res. Commun. 199, 1504–1508. doi: 10.1006/bbrc.1994.1401
Lu, S., Li, Q., Wei, H., Chang, M. J., Tunlaya-Anukit, S., Kim, H., et al. (2013). Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl. Acad. Sci. U.S.A. 110, 10848–10853. doi: 10.1073/pnas.1308936110
Lu, S., Sun, Y. H., Amerson, H., and Chiang, V. L. (2007). MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J. 51, 1077–1098. doi: 10.1111/j.1365-313X.2007.03208.x
Lu, S., Sun, Y. H., and Chiang, V. L. (2008). Stress-responsive microRNAs in Populus. Plant J. 55, 131–151. doi: 10.1111/j.1365-313X.2008.03497.x
Lu, S., Sun, Y. H., Shi, R., Clark, C., Li, L., and Chiang, V. L. (2005). Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17, 2186–2203. doi: 10.1105/tpc.105.033456
Lu, T. T., Lee, S. J., Apfel, U. P., and Lippard, S. J. (2013). Aging-associated enzyme human clock-1: substrate-mediated reduction of the diiron center for 5-demethoxyubiquinone hydroxylation. Biochemistry 52, 2236–2244. doi: 10.1021/bi301674p
Lütke-Brinkhaus, F., and Kleinig, H. (1987). Ubiquinone biosynthesis in plant mitochondria. Methods Enzymol. 148, 486–490. doi: 10.1016/0076-6879(87)48047-6
Lütke-Brinkhaus, F., Liedvogel, B., and Kleinig, H. (1984). On the biosynthesis of ubiquinones in plant mitochondria. Eur. J. Biochem. 141, 537–541. doi: 10.1111/j.1432-1033.1984.tb08226.x
Ma, W., Deng, Y., and Mi, H. (2008). Redox of plastoquinone pool regulates the expression and activity of NADPH dehydrogenase supercomplex in Synechocystis sp. strain PCC 6803. Curr. Microbiol. 56, 189–193. doi: 10.1007/s00284-007-9056-x
Ma, Y., Yuan, L., Wu, B., Li, X., Chen, S., and Lu, S. (2012). Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J. Exp. Bot. 63, 2809–2823. doi: 10.1093/jxb/err466
Maciejewska, U., Polkowska-Kowalczyk, L., Swiezewska, E., and Szkopinska, A. (2002). Plastoquinone: possible involvement in plant disease resistance. Acta Biochim. Pol. 49, 775–780.
Mancini, A., and Balercia, G. (2011). Coenzyme Q(10) in male infertility: physiopathology and therapy. Biofactors 37, 374–380. doi: 10.1002/biof.164
Mancini, A., De Marinis, L., Littarru, G. P., and Balercia, G. (2005). An update of Coenzyme Q10 implications in male infertility: biochemical and therapeutic aspects. Biofactors 25, 165–174. doi: 10.1002/biof.5520250119
Marbois, B. N., and Clarke, C. F. (1996). The COQ7 gene encodes a protein in Saccharomyces cerevisiae necessary for ubiquinone biosynthesis. J. Biol. Chem. 271, 2995–3004. doi: 10.1074/jbc.271.6.2995
Marbois, B. N., Hsu, A., Pillai, R., Colicelli, J., and Clarke, C. F. (1994a). Cloning of a rat cDNA encoding dihydroxypolyprenylbenzoate methyltransferase by functional complementation of a Saccharomyces cerevisiae mutant deficient in ubiquinone biosynthesis. Gene 138, 213–217. doi: 10.1016/0378-1119(94)90810-9
Marbois, B. N., Xia, Y. R., Lusis, A. J., and Clarke, C. F. (1994b). Ubiquinone biosynthesis in eukaryotic cells: tissue distribution of mRNA encoding 3,4-dihydroxy-5-polyprenylbenzoate methyltransferase in the rat and mapping of the COQ3 gene to mouse chromosome 4. Arch. Biochem. Biophys. 313, 83–88. doi: 10.1006/abbi.1994.1362
Matsumoto, T., Kanno, N., Ikeda, I., Obi, Y., Kisaki, T., and Noguchi, M. (1981). Selection of cultured tobacco cell strains producing high levels of ubiquinone 10 by a cell cloning technique. Agric. Biol. Chem. 45, 1627–1633. doi: 10.1271/bbb1961.45.2259
Mattila, P., and Kumpulainen, J. (2001). Coenzymes Q9 and Q10: contents in foods and dietary intake. J. Food Comp. Anal. 14, 409–417. doi: 10.1006/jfca.2000.0983
Mayer, M. P., Beyer, P., and Kleinig, H. (1990). Quinone compounds are able to replace molecular oxygen as terminal electron acceptor in phytoene desaturation in chromoplasts of Narcissus pseudonarcissus L. Eur. J. Biochem. 191, 359–363. doi: 10.1111/j.1432-1033.1990.tb19130.x
Meganathan, R. (2001). Ubiquinone biosynthesis in microorganisms. FEMS Microbiol. Lett. 203, 131–139. doi: 10.1016/S0378-1097(01)00330-5
Moludi, J., Keshavara, S., Javad, H. M., Frooshani, A. R., Sadeghpour, A., Salakia, S., et al. (2015). Coenzyme Q10 effect in prevention of atrial fibrillation after coronnary artery bypass graft: double-blind randomized clinical trial. Tehran Univ. Med. J. 73, 79–85.
Motohashi, R., Ito, T., Kobayashi, M., Taji, T., Nagata, N., Asami, T., et al. (2003). Functional analysis of the 37 kDa inner envelope membrane polypeptide in chloroplast biogenesis using a Ds-tagged Arabidopsis pale-green mutant. Plant J. 34, 719–731. doi: 10.1046/j.1365-313X.2003.01763.x
Mubarakshina, M. M., and Ivanov, B. N. (2010). The production and scavenging of reactive oxygen speciesin the plastoquinone pool of chloroplast thylakoid membranes. Physiol. Plant. 140, 103–110. doi: 10.1111/j.1399-3054.2010.01391.x
Mühlenbock, P., Szechynska-Hebda, M., Plaszczyca, M., Baudo, M., Mateo, A., Mullineaux, P. M., et al. (2008). Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20, 2339–2356. doi: 10.1105/tpc.108.059618
Mukai, N., Masaki, K., Fujii, T., Kawamukai, M., and Iefuji, H. (2010). PAD1 and FDC1 are essential for the decarboxylation of phenylacrylic acids in Saccharomyces cerevisiae. J. Biosci. Bioeng. 109, 564–569. doi: 10.1016/j.jbiosc.2009.11.011
Mur, L. A., Kenton, P., Lloyd, A. J., Ougham, H., and Prats, E. (2008). The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59, 501–520. doi: 10.1093/jxb/erm239
Naqvi, S., Farré, G., Zhu, C., Sandmann, G., Capell, T., and Christou, P. (2011). Simultaneous expression of Arabidopsis ρ-hydroxyphenylpyruvate dioxygenase and MPBQ methyltransferase in transgenic corn kernels triples the tocopherol content. Transgenic Res. 20, 177–181. doi: 10.1007/s11248-010-9393-6
Nguyen, T. P., Casarin, A., Desbats, M. A., Doimo, M., Trevisson, E., Santos-Ocaña, C., et al. (2014). Molecular characterization of the human COQ5 C-methyltransferase in coenzyme Q10 biosynthesis. Biochim. Biophys. Acta 1841, 1628–1638. doi: 10.1016/j.bbalip.2014.08.007
Nievelstein, V., Vandekerckhove, J., Tadros, M. H., Lintig, J. V., Nitschke, W., and Beyer, P. (1995). Carotene desaturation is linked to a respiratory redox pathway in Narcissus pseudonarcissus chromoplast membranes. Eur. J. Biochem. 233, 864–872. doi: 10.1111/j.1432-1033.1995.864_3.x
Norris, S. R., Barrette, T. R., and DellaPenna, D. (1995). Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 7, 2139–2149. doi: 10.1105/tpc.7.12.2139
Norris, S. R., Shen, X., and DellaPenna, D. (1998). Complementation of the Arabidopsis pds1 mutation with the gene encoding p-hydroxyphenylpyruvate dioxygenase. Plant Physiol. 117, 1317–1323. doi: 10.1104/pp.117.4.1317
Nosek, M., Kornaš, A., Kuzniak, E., and Miszalski, Z. (2015). Plastiquinone redox state modified plant response to pathogen. Plant Physiol. Biochem. 96, 163–170. doi: 10.1016/j.plaphy.2015.07.028
Obukhova, L. A., Skulachev, V. P., and Kolosova, N. G. (2009). Mitochondria-targeted antioxidant SkQ1 inhibits age-dependent involution of the thymus in normal and senescence-prone rat. Aging 1, 389–401. doi: 10.18632/aging.100043
Ohara, K., Kokado, Y., Yamamoto, H., Sato, F., and Yazaki, K. (2004). Engineering of ubiquinone biosynthesis using the yeast coq2 gene confers oxidative stress tolerance in transgenic tobacco. Plant J. 40, 734–743. doi: 10.1111/j.1365-313X.2004.02246.x
Ohara, K., Muroya, A., Fukushima, N., and Yazaki, K. (2009). Functional characterization of LePGT1, a membrane-bound prenyltransferase involved in the geranylation of p-hydroxybenzoic acid. Biochem. J. 421, 231–241. doi: 10.1042/BJ20081968
Ohara, K., Sasaki, K., and Yazaki, K. (2010). Two solanesyl diphosphate synthases with different subcellular localizations and their respective physiological roles in Oryza sativa. J. Exp. Bot. 61, 2683–2692. doi: 10.1093/jxb/erq103
Ohara, K., Yamamoto, K., Hamamoto, M., Sasaki, K., and Yazaki, K. (2006). Functional characterization of OsPPT1, which encodes p-hydroxybenzoate polyprenyltransferase involved in ubiquinone biosynthesis in Oryza sativa. Plant Cell Physiol. 47, 581–590. doi: 10.1093/pcp/pcj025
Okada, K., Ohara, K., Yazaki, K., Nozaki, K., Uchida, N., Kawamukai, M., et al. (2004). The AtPPT1 gene encoding 4-hydroxybenzoate polyprenyl diphosphate transferase in ubiquinone biosynthesis is required for embryo development in Arabidopsis thaliana. Plant Mol. Biol. 57, 567–577. doi: 10.1007/s11103-004-1298-4
Olson, R. E., and Rudney, H. (1983). Biosynthesis of ubiquinone. Vitam. Horm. 40, 1–43. doi: 10.1016/S0083-6729(08)60431-8
Ozeir, M., Mühlenhoff, U., Webert, H., Lill, R., Fontecave, M., and Pierrel, F. (2011). Coenzyme Q biosynthesis: Coq6 is required for the C5-hydroxylation reaction and substrate analogs rescue Coq6 deficiency. Chem. Biol. 18, 1134–1142. doi: 10.1016/j.chembiol.2011.07.008
Palan, P. R., Shaban, D. W., Martino, T., and Mikhail, M. S. (2004). Lipid-soluble antioxidants and pregnancy: maternal serum levels of coenzyme Q10, α-tocopherol and γ-tocopherol in preeclampsia and normal pregnancy. Gynecol. Obstet. Invest. 58, 8–13. doi: 10.1159/000077011
Parmar, S. S., Jaiwal, A., Dhankher, O. P., and Jaiwal, P. K. (2015). Coenzyme Q10 production in plants: current status and future prospects. Crit. Rev. Biotechnol. 35, 152–164. doi: 10.3109/07388551.2013.823594
Payne, K. A., White, M. D., Fisher, K., Khara, B., Bailey, S. S., Parker, D., et al. (2015). New cofactor supports α,β-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition. Nature 522, 497–501. doi: 10.1038/nature14560
Petrov, A., Perekhvatova, N., Skulachev, M., Stein, L., and Ousler, G. (2016). SkQ1 Ophthalmic solution for dry eye treatment: results of a phase 2 safety and efficacy clinical study in the environment and during challenge in the controlled adverse environment model. Adv. Ther. 33, 96–115. doi: 10.1007/s12325-015-0274-5
Pfander, H. (1992). Carotenoids: an overview. Methods Enzymol. 213, 3–13. doi: 10.1016/0076-6879(92)13105-7
Phatthiya, A., Takahashi, S., Chareonthiphakorn, N., Koyama, T., Wititsuwannakul, D., Wititsuwannakul, R., et al. (2007). Cloning and expression of the gene encoding solanesyl diphosphate synthase from Hevea brasiliensis. Plant Sci. 172, 824–831. doi: 10.1016/j.plantsci.2006.12.015
Poon, W. W., Barkovich, R. J., Hsu, A. Y., Frankel, A., Lee, P. T., Shepherd, J. N., et al. (1999). Yeast and rat Coq3 and Escherichia coli UbiG polypeptides catalyze both O-methyltransferase steps in coenzyme Q biosynthesis. J. Biol. Chem. 274, 21665–21672. doi: 10.1074/jbc.274.31.21665
Premkumar, V. G., Yuvaraj, S., Sathish, S., Shanthi, P., and Sachdanandam, P. (2008). Anti-angiogenic potential of coenzyme Q10, riboflavin and niacin in breast cancer patients undergoing tamoxifen therapy. Vascul. Pharmacol. 48, 191–201. doi: 10.1016/j.vph.2008.02.003
Qin, G., Gu, H., Ma, L., Peng, Y., Deng, X. W., Chen, Z., et al. (2007). Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 17, 471–482. doi: 10.1038/cr.2007.40
Reape, T. J., Molony, E. M., and McCabe, P. F. (2007). Programmed cell death in plants: distinguishing between different modes. J. Exp. Bot. 59, 435–444. doi: 10.1093/jxb/erm258
Rock, C. D., and Zeevaart, J. (1991). The aba mutant of Arabidopsis thaliana is impaired in epoxy-carotenoid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 88, 7496–7499. doi: 10.1073/pnas.88.17.7496
Rozhnova, N. A., and Gerashchenkov, G. A. (2006). Hormonal status of tobacco variety Samsun NN exposed to synthetic coenzyme Q10 (ubiquinone 50) and TMV infection. Izv. Akad. Nauk Ser. Biol. 33, 581–590. doi: 10.1134/S1062359006050074
Rozhnova, N. A., and Gerashchenkov, G. A. (2008). Effect of ubiquinone 50 and viral infection on phytohemagglutinin activity in development of induced resistance in tobacco plants. Izv. Akad. Nauk Ser. Biol. 35, 442–447. doi: 10.1134/S1062359008040080
Sadre, R., Frentzen, M., Saeed, M., and Hawkes, T. (2010). Catalytic reactions of the homogentisate prenyl transferase involved in plastoquinone-9 biosynthesis. J. Biol. Chem. 285, 18191–18198. doi: 10.1074/jbc.M110.117929
Sadre, R., Gruber, J., and Frentzen, F. (2006). Characterization of homogentisate prenyltransferases involved in plastoquinone-9 and tocochromanol biosynthesis. FEBS Lett. 580, 5357–5362. doi: 10.1016/j.febslet.2006.09.002
Sadre, R., Pfaff, C., and Buchkremer, S. (2012). Plastoquinone-9 biosynthesis in cyanobacteria differs from that in plants and involves a novel 4-hydroxybenzoate solanesyltransferase. Biocheml J. 442, 621–629. doi: 10.1042/BJ20111796
Safarinejad, M. R. (2012). The effect of coenzyme Q10 supplementation on partner pregnancy rate in infertile men with idiopathic oligoasthenoteratozoospermia: an open-label prospective study. Int. Urol. Nephrol. 44, 689–700. doi: 10.1007/s11255-011-0081-0
Sakato, K., Tanaka, H., Shibata, S., and Kuratsu, Y. (1992). Agitation-aeration studies on coenzyme Q10 production using Rhodopseudomonas spheroides. Biotechnol. Appl. Biochem. 16, 19–22.
Sakiko, T., Ohtani, T., Saton, H., Nakamura, Y., Kawamukai, M., and Kadowaki, K. (2006). Metabolic engineering of Coenzyme Q by modification of isoprenoid side chain in plant. FEBS Lett. 580, 955–959. doi: 10.1016/j.febslet.1006.01.023
Sakiko, T., Ohtani, T., Saton, H., Nakamura, Y., Kawamukai, M., and Kadowaki, K. (2010). Development of coenzyme Q10-enriched rice using sugary and shrunken mutants. Biosci. Biotechnol. Biochem. 74, 182–184. doi: 10.1271/bbb.90562
Samuilov, V. D., and Kiselevsky, D. B. (2015). Effect of cationic plastoquinone SkQ1 on electron transfer reactions in chloroplasts and mitochondria from pea seedlings. Biochemistry 80, 417–423. doi: 10.1134/S0006297915040045
Savidge, B., Weiss, J. D., Wong, Y. H., Lassner, M. W., Mitsky, T. A., Shewmaker, C. K., et al. (2002). Isolation and characterization of tocopherol phytyltransferase genes from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol. 129, 321–332. doi: 10.1104/pp.010747
Schmelzer, C., Lindner, I., Vock, C., Fujii, K., and Döring, F. (2007). Functional connections and pathways of coenzyme Q10-inducible genes: an in-silico study. IUBMB Life 59, 628–633. doi: 10.1080/15216540701545991
Schmelzer, C., Niklowitz, P., Jürgen, G., Okun, J. G., Haas, D., Menke, T., et al. (2011). Ubiquinol-induced gene expression signatures are translated into altered parameters of erythropoiesis and reduced low density lipoprotein cholesterol levels in humans. IUBMB Life 63, 42–48. doi: 10.1002/iub.413
Shen, W., Wei, Y., Dauk, M., Tan, Y., Taylor, D. C., Selvaraj, G., et al. (2007). Involvement of a glycerol-3-phosphate dehydrogenase in modulating the NADH/NAD+ ratio provides evidence of a mitochondrial glycerol-3-phosphate shuttle in Arabidopsis. Plant Cell 18, 422–441. doi: 10.1105/tpc.105.039750
Shintani, D. K., Cheng, Z., and DellaPenna, D. (2002). The role of 2-methyl-6-phytylbenzoquinone methyltransferase in determining tocopherol composition in Synechocystis sp. FEBS Lett. 511, 1–5. doi: 10.1016/S0014-5793(01)0323-9
Siebert, M., Bechthold, A., Melzer, M., May, U., Berger, U., Schröder, G., et al. (1992). Ubiquinone biosynthesis: cloning of the genes coding for chorismate pyruvate-lyase and 4-hydroxybenzoate octaprenyl transferase from Escherichia coli. FEBS Lett. 307, 347–350. doi: 10.1016/0014-5793(92)80710-X
Skulachev, M. V., Antonenko, Y. N., Anisimov, V. N., Chernyak, B. V., Cherepanov, D. A., Chistyakov, V. A., et al. (2011). Mitochondrial-targeted plastoquinone derivatives. Effect on senescence and acute age-related pathologies. Curr. Drug Targets 12, 800–826. doi: 10.2174/138945011795528859
Solovieva, A. D., Frolova, O. Y., Solovyev, A. G., Morozov, S. Y., and Zamyatnin, A. A. (2013). Effect of mitochondria-targeted antioxidant SkQ1on programmed cell death induced by viral proteins in tobacco plants. Biochemistry 78, 1006–1012. doi: 10.1134/S000629791309006X
Sommer, S., and Heide, L. (1998). Expression of bacterial chorismate pyruvate-lyase in tobacco: evidence of the presence of chorismate in the plant cytosol. Plant Cell Physiol. 39, 1240–1244. doi: 10.1093/oxfordjournals.pcp.a029326
Stiff, M. R. (2010). Coenzyme Q10 Biosynthesis in Plants: Is the Polyprenyltransferase an Appropriate Gene Target for the Increased Production of CoQ?. Ph.D. thesis, North Carolina State University, Raleigh, NC.
Sweetlove, L. J., Fait, A., Nunes-Nesi, A., Williams, T., and Fernie, A. R. (2007). The mitochondrion: an integration point of cellular metabolism and signalling. Crit. Rev. Plant Sci. 26, 17–43. doi: 10.1080/07352680601147919
Swiezewska, E. (2004). Ubiquinone and plastoquinone metabolism in plants. Methods Enzymol. 378, 124–131. doi: 10.1016/S0076-6879(04)78007-6
Tang, Y., Fu, X., Shen, Q., and Tang, K. (2016). Roles of MPBQ-MT in promoting α/γ-tocopherol production and photosynthesis under high light in lettuce. PLoS ONE 11:e0148490. doi: 10.1371/journal.pone.0148490
Teran, E., Hernandez, I., Nieto, B., Tavara, R., Ocampo, J. E., and Calle, A. (2009). Coenzyme Q10 supplementation during pregnancy reduces the risk of pre-eclampsia. Int. J. Gynecol. Obstet. 105, 43–45. doi: 10.1016/j.ijgo.2008.11.033
Thoma, I., Loeffler, C., Sinha, A. K., Gupta, M., Krischke, M., Steffan, B., et al. (2003). Cyclopentenoneisoprostanes induced by reactive oxygen species triggerdefense gene activation and phytoalexin accumulation in plants. Plant J. 34, 363–375. doi: 10.1046/j.1365-313X.2003.01730.x
Tian, L., DellaPenna, D., and Dixon, R. A. (2007). The pds2 mutation is a lesion in the Arabidopsis homogentisate solanesyltransferase gene involved in plastoquinone biosynthesis. Planta 226, 1067–1073. doi: 10.1007/s00425-007-0564-5
Torres, M. A., Jones, J. D., and Dangl, J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. doi: 10.1104/pp.106.079467
Tran, M. T., Mitchell, T. M., Kennedy, D. T., and Giles, J. T. (2001). Role of coenzyme Q10 in chronic heart failure, angina, and hypertension. Pharmacotherapy 21, 797–806. doi: 10.1592/phco.21.9.797.34564
Tran, U. C., and Clarke, C. F. (2007). Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 7, S62–S71. doi: 10.1016/j.mito.2007.03.007
Tripathy, B. C., and Oelmuller, R. (2012). Reactive oxygen species generation and signaling in plants. Plant Signal. Behav. 7, 1621–1633. doi: 10.4161/psb.22455
Uchida, N., Suzuki, K., Saiki, R., Kainou, T., Tanaka, K., Matsuda, H., et al. (2000). Phenotypes of fission yeast defective in ubiquinone production due to disruption of the gene for p-hydroxybenzoate polyprenyl diphosphate transferase. J. Bacteriol. 182, 6933–6939. doi: 10.1128/JB.182.24.6933-6939.2000
Vajo, Z., King, L. M., Jonassen, T., Wilkin, D. J., Ho, N., Munnich, A., et al. (1999). Conservation of the Caenorhabditis elegans timing gene clk-1 from yeast to human: a gene required for ubiquinone biosynthesis with potential implications for aging. Mamm. Genome 10, 1000–1004. doi: 10.1007/s003359901147
Van Eenennaam, A. L., Lincoln, K., Durrett, T. P., Valentin, H. E., Shewmaker, C. K., Thorne, G. M., et al. (2003). Engineering vitamin E content: from Arabidopsis mutant to soy oil. Plant Cell 15, 3007–3019. doi: 10.1105/tpc.015875
Venkatesh, T. V., Karunanandaa, B., Free, D. L., Rottnek, J. M., Baszis, S. R., and Valentin, H. E. (2006). Identification and characterization of an Arabidopsis homogentisate phytyltransferase paralog. Planta 223, 1134–1144. doi: 10.1007/s00425-005-0180-1
Viitanen, P. V., Devine, A. L., Khan, M. S., Deuel, D. L., Dyk, D. E. V., and Daniell, H. (2004). Metabolic engineering of the chloroplast genome using the Escherichia coli ubiC gene reveals that chorismate is a readily abundant plant precursor for p-hydroxybenzoic acid biosynthesis. Plant Physiol. 136, 4048–4060. doi: 10.1104/pp.104.050054
Wang, K., and Ohnuma, S. (1999). Chain-length determination mechanism of isoprenyl diphosphate synthases and implications for molecular evolution. Trends Biochem. Sci. 24, 445–451. doi: 10.1016/S0968-0004(99)01464-4
Wang, M., Wu, B., Chen, C., and Lu, S. (2015). Identification of mRNA-like non-coding RNAs and validation of a mighty one named MAR in Panax ginseng. J. Integr. Plant Biol. 57, 256–270. doi: 10.1111/jipb.12239
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Wanke, M., Swiezewska, E., and Dallner, G. (2000). Half-life of ubiquinone and plastoquinone in spinach cells. Plant Sci. 154, 183–187. doi: 10.1016/S0168-9452(00)00200-4
Wei, R., Qiu, D., Wilson, I. W., Zhao, H., Lu, S., Miao, J., et al. (2015). Identification of novel and conserved microRNAs in Panax notoginseng roots by high-throughput sequencing. BMC Genomics 16:835. doi: 10.1186/s12864-015-2010-6
White, M. D., Payne, K. A., Fisher, K., Marshall, S. A., Parker, D., Rattray, N. J., et al. (2015). UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis. Nature 522, 502–506. doi: 10.1038/nature14559
Wu, B., Li, Y., Yan, H., Ma, Y., Luo, H., Yuan, L., et al. (2012). Comprehensive transcriptome analysis reveals novel genes involved in cardiac glycoside biosynthesis and mlncRNAs associated with secondary metabolism and stress response in Digitalis purpurea. BMC Genomics 13:15. doi: 10.1186/1471-2164-13-15
Xu, W., Yang, S., Zhao, J., Su, T., Zhao, L., and Liu, J. (2014). Improving coenzyme Q8 production in Escherichia coli employing multiple strategies. J. Ind. Microbiol. Biotechnol. 41, 1297–1303. doi: 10.1007/s10295-014-1458-8
Yadav, D. K., Kruk, J., Sinha, R. K., and Pospíšil, P. (2010). Singlet oxygen scavenging activity of plastoquinol in photosystem II of higher plants: electron paramagnetic resonance spin-trapping study. Biochim. Biophys. Acta 1797, 1807–1811. doi: 10.1016/j.bbabio.2010.07.003
Yang, D. H., Andersson, B., Aro, E. M., and Ohad, I. (2001). The redox state of the plastoquinone pool controls the level of the light-harvesting chlorophyll a/b binding protein complex II (LHC II) during photoacclimation. Photosynth. Res. 68, 163–174. doi: 10.1023/A:1011849919438
Yang, R., Jarvis, D. E., Chen, H., Beilstein, M. A., Grimwood, J., Jenkins, J., et al. (2013). The reference genome of halophytic plant Eutrema salsugineum. Front. Plant Sci. 4:46. doi: 10.3389/fpls.2013.00046
Yazaki, K., Kunihisa, M., Fujisaki, T., and Sato, F. (2002). Geranyl diphosphate:4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon. Cloning and characterization of a ket enzyme in shikonin biosynthesis. J. Biol. Chem. 277, 6240–6246. doi: 10.1016/S0014-5793(01)03223-9
Zhang, D., Li, Z., Wang, F., Shrestha, B., Tian, P., and Tan, T. (2007a). Expression of various genes to enhance ubiquinone metabolic pathway in Agrobacterium tumefaciens. Enzyme Microb. Technol. 41, 772–779. doi: 10.1016/j.enzmictec.2007.06.014
Zhang, D., Shrestha, B., Niu, W., Tian, P., and Tan, T. (2007b). Phenotypes and fed-batch fermentation of ubiquinone-overproducing fission yeast using ppt1 gene. J. Biotechnol. 128, 120–131. doi: 10.1016/j.jbiotec.2006.09.012
Zhang, L., and Lu, S. (2016). Overview of medicinally important diterpenoids derived from plastids. Mini Rev. Med. Chem. 16. doi: 10.2174/1389557516666160614005244 [Epub ahead of print].
Zhang, L., Wu, B., Zhao, D., Li, C., Shao, F., and Lu, S. (2014). Genome-wide analysis and molecular dissection of the SPL gene family in Salvia miltiorrhiza. J. Integr. Plant Biol. 56, 38–50. doi: 10.1111/jipb.12111
Zhou, X., Yuan, Y., Yang, Y., Rutzke, M., Thannhauser, T. W., Kochian, L. V., et al. (2009). Involvement of a broccoli COQ5 methyltransferase in the production of volatile selenium compounds. Plant Physiol. 151, 528–540. doi: 10.1104/pp.109.142521
Keywords: biosynthetic pathway, isoprenoid, metabolic engineering, plastoquinone, photosynthesis, respiration, secondary metabolism, ubiquinone
Citation: Liu M and Lu S (2016) Plastoquinone and Ubiquinone in Plants: Biosynthesis, Physiological Function and Metabolic Engineering. Front. Plant Sci. 7:1898. doi: 10.3389/fpls.2016.01898
Received: 13 July 2016; Accepted: 30 November 2016;
Published: 16 December 2016.
Edited by:
Basil J. Nikolau, Iowa State University, USACopyright © 2016 Liu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shanfa Lu, c2ZsdUBpbXBsYWQuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.