- Key Laboratory of Insect Developmental and Evolutionary Biology, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
Specification of floral organ identity is critical for the establishment of floral morphology and inflorescence architecture. Although multiple genes are involved in the regulation of floral organogenesis, our understanding of the underlying regulating network is still fragmentary. MADs-box genes are principle members in the ABCDE model that characterized floral organs. OsMADS1 specifies the determinacy of spikelet meristem and lemma/palea identity in rice. However, the pathway through which OsMADS1 regulates floral organs remains elusive; here, we identified the microRNA172 (miR172) family as possible regulators downstream of OsMADS1. Genetic study revealed that overexpression of each miR172 gene resulted in elongated lemma/palea and indeterminacy of the floret, which resemble the phenotype of osmads1 mutant. On the contrary, overexpression of each target APETALA2 (AP2) genes resulted in shortened palea/lemma. Expression level and specificity of miR172 was greatly influenced by OsMADS1, as revealed by Northern blot analysis and In situ hybridization. Genetically, AP2-3 and AP2-2 over expression rescued the elongation and inconsistent development of the lemma/palea in OsMADS1RNAi transgenic plants. Our results suggested that in rice, OsMADS1 and miR172s/AP2s formed a regulatory network involved in floral organ development, particularly the elongation of the lemma and the palea.
Introduction
Specification of floral meristem fate and floral organ identity is pivotal for the reproductive development of plants, molecular, and genetic studies led to the establishment of the classic ABC and the modified ABCDE model to explain the development of floral organs (Coen and Meyerowitz, 1991; Angenent et al., 1995; Pelaz et al., 2000; Ditta et al., 2004). MADs-box genes are characterized by the presence of an approximately 60 amino acids DNA-binding MADS-box domain in the N-terminal (Schwarz-Sommer et al., 1990; Theissen et al., 2000; Arora et al., 2007). A considerable number of MADs-box genes exist in plants which are major players in the control of flower architecture, flower induction, and vegetative development, the phylogeny of MADs-box genes is tightly correlated with the evolution of plant reproductive structures (Theissen et al., 2000; Ferrario et al., 2004). Quite a lot of A/B/C/D/E class genes have been identified, and all of them except for AP2 genes are MADs-box family genes. Rice belongs to the grass family of monocots, the florets of which contain carpels and stamens, but lack petals and sepals, instead, lodicules surround the sex organs and the lemma/palea envelop the inner floral organs. Similarly, MADs-box genes, such as OsMADS1, OsMADS3 and OsMADS58 (Yamaguchi et al., 2006), OsMADS6 (Ohmori et al., 2009; Li et al., 2010), OsMADS7 and OsMADS8 (Cui et al., 2010), and OsMADS15 (Wang K. et al., 2010) characterize floral organ identities in rice.
Among them, the function of OsMADS1 is extensively studied due to identification of several mutants (Jeon et al., 2000; Agrawal et al., 2005; Chen et al., 2006; Wang K. et al., 2010). The naked seed rice (nsr) mutant displayed overdeveloped lemma and palea, the transformation of lodicules into palea/lemma-like organs, and decreased number of stamens (Chen et al., 2006). The leafy hull sterile1 (lsh1) mutant produces spikelets consisting of elongated leafy paleae and lemmas, two pairs of leafy palea-like and lemma-like lodicules, a decrease in stamen number and an increase in the number of carpels, some spikelets generate an additional floret from the same rachilla (Jeon et al., 2000). The afo mutant is an epigenetic mutation in OsMADS1 that showed pleiotropic defects in lemmas and the inner three whorls and the distinct “flower-in-flower” structure (Wang K. et al., 2010). Altogether, OsMADS1 mutations result in over developed lemma/palea, transition of the inner three whorls into lemma/palea structures and loss of flower determinacy, suggesting a role for OsMADS1 in specifying the determinacy of the flower meristem and influencing development of all floral organs. In accordance, sequence and function conservation allotted OsMADS1 gene to the E-function gene (Agrawal et al., 2005; Cui et al., 2010).
OsMADS34 and OsMADS55 are two downstream genes of OsMADS1; with OsMADS34 characterizing the spikelet meristem and OsMADS55 function in organ differentiation (Khanday et al., 2013). OsMGH3 might be an indirect downstream gene of OsMADS1 (Prasad et al., 2005). However, OsMADS1 is a regulator of genetic networks that orchestrate transcriptional and signaling pathways to promote rice floret specification and development, the molecular mechanism downstream of OsMADAS1 is still not so clear, and complex physical and genetic interaction might exist between OsMADS1 and other floral organ characterizing genes, most of which are also MADS-box genes (Hu et al., 2015; Khanday et al., 2016).
In recent years, microRNAs (miRNAs) have been shown to play pivotal regulative roles in many developmental and physiological processes in a wide variety of organisms (Chapman and Carrington, 2007; Molnar et al., 2007; Zhao T. et al., 2007; Xie and Qi, 2008; Zhou et al., 2013). In plants, miR172 is involved in the regulation of flowering time and floral organ identity through targeting AP2 genes (Chen, 2004; Mlotshwa et al., 2006; Zhao L. et al., 2007; Martin et al., 2009; Mathieu et al., 2009). AP2 genes encode plant specific transcriptional factors which are characterized by the AP2 DNA-binding domain (Yaish et al., 2010). AP2 genes form a big super-family in plants, and play various roles in plant development and physiology (Chen, 2004; Nakano et al., 2006), such as in floral organ development (Chen, 2004), in response to biotic and abiotic stress (Tang et al., 2005; Shukla et al., 2006), and in seed size control (Fu and Xue, 2010). In rice, one target of miR172, supernumerary bract (SNB), influences the floral organ identity and floral determinacy (Lee et al., 2007). Another target of miR172, Osindeterminate spikelet 1 (OsIDS1) functions in the establishment of the floral meristems and formation of the floral organs (Lee and An, 2012). miR172 expresses in the late vegetative stage and panicle, and over expression of miR172b could approximately pheno-copy the snb mutant (Zhu et al., 2009). Although miR172 is functionally conserved, and several studies strongly suggest the participation of miR172/AP2s in floral organ development in rice, a comprehensive understanding of their roles is still missing.
In the present study, the functions of all four miR172s and the five target AP2s in rice were analyzed. Over expression of the respective miR172s caused elongation of the lemma/palea and loss of floral determinacy, resembling the phenotype of the osmads1 mutant. Consistently, overexpression of each target AP2 gene resulted shortened lemma/palea to various degrees, suggesting that miR172/AP2s regulate the elongation of the lemma/palea. In OsMADS1 RNAi transgenic plants, the four miR172s were up-regulated, whereas in OsMADS1 overexpression transgenic plants, the four miR172s were down-regulated. Furthermore, the timing and specificity of miR172 expression were both influenced by OsMADS1. Genetically, overexpression of AP2-3 and AP2-2 partially rescued the phenotype of OsMADS1RNAi. These results strongly suggested that miR172/AP2s regulated palea/lemma development and floral determinacy in rice, and OsMADS1 was an upstream suppressor of miR172.
Materials and Methods
Plant Materials
Oryza sativa japonica variety Zhonghua No.11 (abbreviated as ZH11) was used as wild-type. All the plants used in this study were grown in the green house with an 8 h light and 16 h dark cycle; or in a paddy field under natural conditions in summer.
Construction of Transgenic Plasmids and Genetic Transformation
For overexpression of miR172s, genomic fragments containing the Osa_miR172s coding regions were, respectively, cloned into the pCAMBIA1301 under the 35S promoter and NOS terminator (p130135SNOS).
For overexpression of AP2s and OsMADS1, the full length cDNA of the respective genes were cloned into the p130135SNOS vector.
For overexpression of AP2-2 in the OsMADS1RNAi plants, the whole expression cassette of 35s-cDNA-nos was digested from the p130135SNOS-AP2-2 and cloned in the p230135SNOS vector, using HindIII and EcoRI.
For OsMADS1 RNAi construction, a gene-specific cDNA fragment was cloned into the p1301RNAi vector in the sense orientation using BamHI and KpnI and antisense orientation using SacI and SpeI.
Plasmid p2301-AP2-2 was transformed into OsMADS1RNAi plants with G418 selection, other plasmids were transformed into ZH11 using Agrobacterium-mediated genetic transformation with hygromycin selection, respectively (Hiei et al., 1994).
Reverse Transcription-Polymerase Chain Reaction (RT-PCR) and Real Time RT-PCR Analysis
Total RNAs were extracted from the leaves or young panicles of the plants using the TRIzol (Invitrogen), and then reverse transcribed using ReverAce (TOYOBO). cDNA was synthesized from 2 μg of total RNA treated with DNase I (TOYOBO) and used as template.
In situ Hybridization and miRNA In situ Hybridization
Young panicles were fixed in 4% paraformaldehyde PBS solution (0.1% Triton-X-100, 0.1% Tween-20, 4% formaldehyde, 25% glutaraldehyde) overnight at 4°C, dehydrated through a concentration grade of ethanol, cleared through a dimethylbenzene series, infiltrated through a series of paraffin (Sigma-Aldrich), and finally embedded in 100% paraffin melted at 60°C. The samples were sectioned longitudinally at 7 μm and then mounted on RNase-free glass slides (Sigma). A gene-specific region of OsMADS1 was cloned into the pBSK(-) vector, linearized, and used as template for amplifying digoxigenin-labeled sense and antisense RNA probes using a DIG RNA labeling kit (Promega). In situ hybridization was performed as previously described (Coen et al., 1990).
For miRNA in situ hybridization, materials were prepared as usual. miR172 was detected with Locked Nucleic Acid (LNA) probes which were Digoxin 5′-end labeled (Exiqon).
miRNA Northern Blot Hybridization
Approximately 30 μg of total RNA was separated on 15% polyacrylamide denaturing gels. RNAs were transferred to Amersham Hybond®-N+ membranes and cross-linked by UV irradiation; the membranes were hybridized with biotin-labeled DNA probes complementary to the miRNA sequences at 42°C overnight. The membranes were then washed and incubated with a stabilized streptavidin-horseradish peroxidase at 42°C. After washing with substrate equilibration buffer and adding stable peroxide solution and enhancer solution, the membranes were imaged using an FLA-5000 Phosphorimager. The DNA probes were synthesized and biotin-labeled using a 3′ end DNA labeling method.
Scanning Electron Microscope (SEM) Analysis
Shoot apical meristems and IMs and young florets were decorticated under light microscope and fixed quickly in 50% FAA at 4°C overnight after vacuuming, and dehydrated through a graded concentration of ethanol. For SEM analysis, the samples were then critical point dried using liquid carbon dioxide and mounted on SEM stubs, sputter coated with gold and palladium (4:1) and examined using a SEM (Hitachi S-2460, Japan). For paraffin analysis, samples were embedded in epoxide resin and cut into slices 2–3 μm slices; strips of these slices were spread at 42°C on a hot platform overnight, stained using 0.5% toluidine Blue O and sealed for observation under the microscope (Wang J.et al., 2010).
Yeast One-Hybrid Assay
The full length cDNA of OsMADS1 gene was cloned in frame into vector pPC86.
PCR fragments containing the binding motifs of OsMADS1 were, respectively, cloned into the p178 vector using the XhoI restriction site, which contains the CYC1 core promoter and the lacZ gene.
Yeast strain EGY48 (MATtrp1his3ura3leu2::6lexAop-LEU2) was used for transformation. The yeast assays were performed according to the manufacturer’s protocol with the substrate chlorophenol red-D-galactopyranoside (Matchmaker One-hybrid System; Clontech).
Yeast Two-Hybrid Assay
The open reading frame (ORF) of OsMADS1 was amplified and cloned into the prey vector pGAD-T7. The ORFs of AP2s were amplified and cloned into the bait vector pGBK-T7. The yeast two-hybrid assay was performed according to the manufacturer’s instructions (Clontech).
Sequence Information
Sequence data used in this study can be found in the rice genome annotation database1 and NCBI2 under the following accession numbers: LOC_Os03g11614 (OsMADS1), LOC_Os05g03040 (AP2-1), LOC_Os03g60430 (AP2-2), LOC_Os07g13170 (AP2-3), LOC_Os06g43220 (AP2-4), LOC_Os04g55560 (AP2-5), LM379345 (Osa-miR172a), LM379346 (Osa-miR172b); LM379347 (Osa-miR172c), LM383079 (Osa-miR172d), AC091532 (actin).
Primer sequences used in this study are listed in Supplementary Table S1.
Results
Overexpression of Each miR172 Gene Caused Elongated Lemma/Palea and Indeterminacy
Four miR172 genes (miR172a-d) have been identified in rice genome3. To study their function, we, respectively, over expressed them under the 35S promoter through genetic transformation. For each miR172 over expression, 30–50 independent transgenic plants were gotten and over 90% of them showed the phenotypes described below. The expression levels of respective miR172s in the transgenic lines were verified by Northern blotting (Supplementary Figure S1). According to the severity of the phenotype, the transgenic plants from different miR172s overexpression could be grouped into two classes: miR172aOE, miR712cOE, and miR172dOE were similar and showed severely abnormal floral organs (Figure 1A as compared with Figure 1B), they were tentatively represented as miR172aOEs unless specified; whereas phenotypic abnormality of miR172bOE was moderate (see the following).
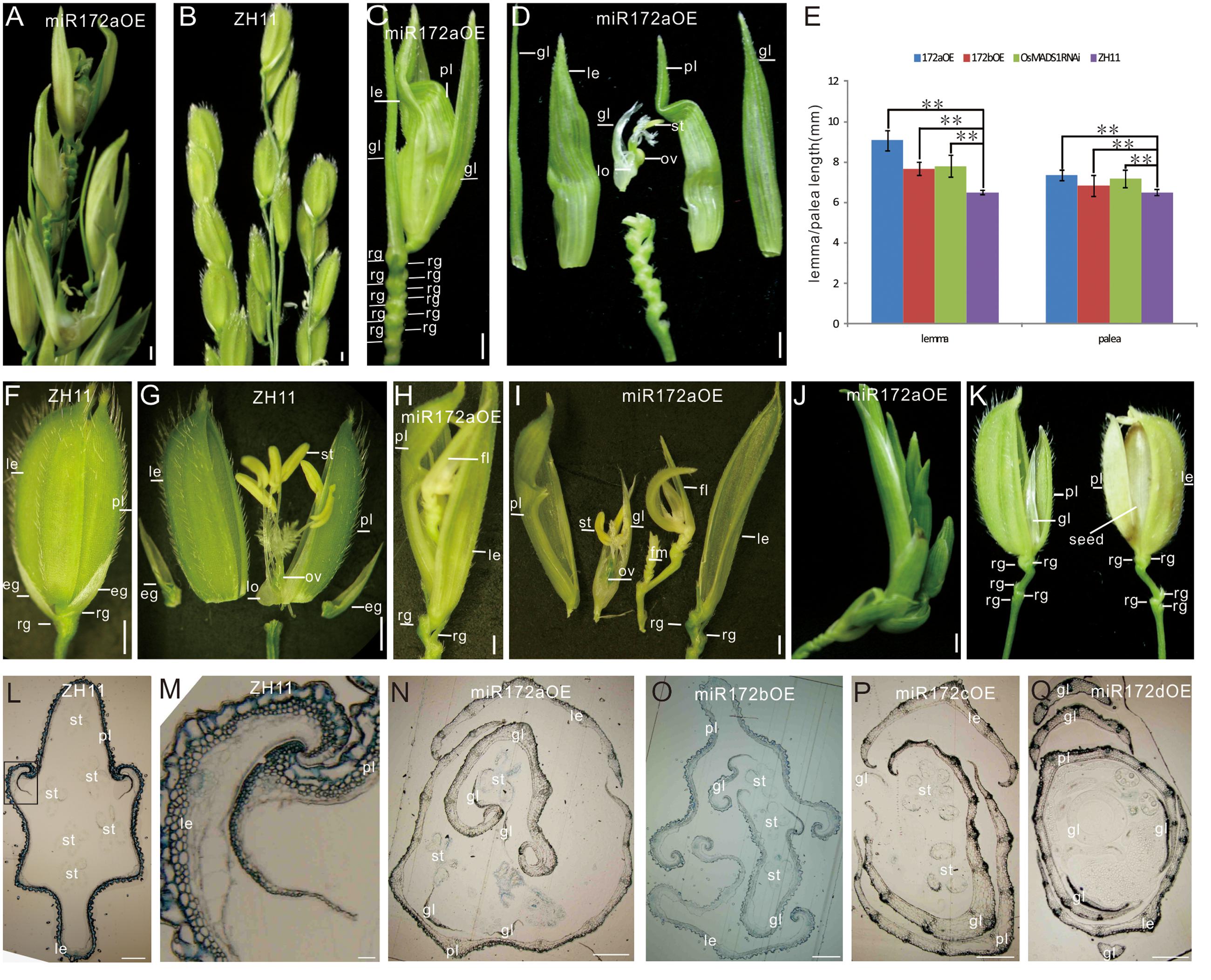
FIGURE 1. Phenotypes of miR172OEs plants. (A,B) Half panicles of the miR172aOE and ZH11 plants, respectively. (C,F) A spikelet of the miR172aOE plant and ZH11 plant, respectively. (D,G) Dissection of the spikelet in (C,F), respectively. (E) Statistical analysis of the length of the lemma and palea in the transgenic plants. Double asterisks represent significant difference determined by the Student’s t-test at ∗∗P < 0.01. (H,I) Full view and dissection of the spikelet of miR172aOE plants. (J) A plantlet-like spikelet of miR172aOE plants. (K) A spikelet (left) and a grain (right) of miR172bOE plants. (L) Transverse section of ZH11 floret. (M) Zoom in of the palea/lemma interlocking region in (L) (rectangle). (N–Q) Transverse section of the florets of miR172a, miR172b, miR172c, and miR172dOE plants. Bars in (A–D,F–J) were 1 mm, bars in (L–Q) were 0.5 cm. Abbreviations: le, lemma; gl, glume; pl, palea; rg, rudimentary glume; eg, empty glume; lo, lodicule; ov, ovule; st, stamen; sti, stigma; fl, floret; fm, floral meristem.
The miR172aOEs plants showed elongated lemma and palea, with the lemma longer than the palea (Figures 1C–E). Typically, a wild-type rice floret consists of a lemma and a palea in whorl1, and two lodicules at the lemma side in whorl2, six stamens in whorl3, and a carpel with a pair of feather-like stigmas and a green ovary in whorl4. A floret together with two pairs of sterile glumes (empty glumes and rudimentary glumes) constitutes a spikelet (Figure 1F). In the greenhouse, the lemma/palea of wild-type averaged 6.5 mm, whereas the lemma and palea of the miR172aOE was 9.07 and 7.34 mm, respectively, the lemma and palea of miR172bOE averaged 7.68 and 6.82 mm, respectively (Figure 1E).
miR172aOEs plants produced many rudimentary glumes (Figure 1C) instead of one pair in the wild-type (Figure 1F); and showed “flower-in-flower” structures (Figures 1H,I); the extreme phenotype was an overall plantlet-like structure instead of a flower (Figure 1J). All these characters indicated that miR172aOEs plants showed indeterminacy in flower development.
Phenotype of miR172bOE plants was more moderate than that of miR172aOEs. The lemma of miR172bOE was less elongated (Figures 1E,K); fewer ectopic glumes and rudimentary glumes were formed; and some flowers were fertile (Figure 1K).
Normally, the lemma and palea of wild-type plants were closed (Figure 1F) except for the short opening time during flowering. However, in the four miR172OE plants, the lemma and palea could not close due to severe distortion (Figures 1N–Q as compared with Figure 1L). As a result, the normal interlocking structure between the lemma and palea in the wild-type (Figure 1M) disappeared in the four miR172OE plants (Figures 1N–Q).
Furthermore, SEM analysis revealed that the transition from spikelet meristem to floral meristem was delayed in the miR172aOEs plants. In wild-type floret, after differentiation of a pair of rudimentary glumes and empty glumes, the floral meristem begins to development inner floral organs (Figures 2A–D). In the miR172aOEs plants, the meristem produced many rudimentary glumes before forming a floret (Figures 1C and 2E). In the process of floral meristem development, before the formation of the stamen, no visible phenotypic changes were observed (Figure 2F compared with Figure 2A, Figure 2G compared with Figure 2B), but after that, the palea/lemma elongated differently (Figure 2H); and ectopic glumes inside the palea/lemma (Figure 2H as compared with Figure 2C) and malformed stamens (Figure 2I) developed. In most of the florets, stamens were decreased and malformed (Figures 1D,I), miR172aOEs plants were infertile.
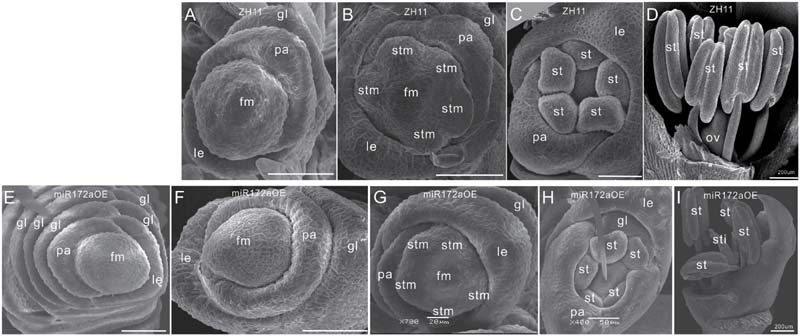
FIGURE 2. Floret development in miR172aOEs plants. (A–D) and (F–I) Spikelet meristem, stamen meristem, floret meristem, and floret of ZH11 and miR172aOE plants, respectively. (E) Disturbed determination of the miR172aOE spikelet meristem. Bars were 50 μm unless specified. Abbreviations: SAM, shoot apical meristem; P0, leaf meristem; gl, glume; pa, palea; le, lemma; fm, floral meristem; stm, stamen meristem; st, stamen; ov, ovule; sti, stigma; ca, carpel.
We further cloned AtmiR172a precursor from Arabidopsis and overexpressed it in rice. Mature AtmiR172a was the same as the OsmiR172a although their precursor differs (Supplementary Figure S2). The AtmiR172aOE lines showed elongated and distorted palea/lemma (indicated by stars) and indeterminacy of flowers, as indicated by increased number of rudimentary glumes (arrows), similar to the phenotype of OsmiR172aOEs (Supplementary Figure S3). This further illustrated the conservation of miR172.
Overexpression of Each Target AP2 Gene Resulted in Shortened Lemma/Palea
To study the function of miR172 in detail, we further analyzed the function of miR172 targets genetically. In rice, five AP2 genes are predicted as targets of miR1724 (Zhu et al., 2009), and they were tentatively designated as AP2-1, AP2-2, AP2-3, AP2-4, and AP2-5, respectively, in this study. All the five AP2 genes have two AP2 DNA-binding domains in a similar mode (Supplementary Figure S4). To mimic the knock-down function of miR172, these AP2 genes were, respectively, overexpressed. 30–50 individual transgenic plants were gotten for each gene, and over 90% of them showed the respective phenotype illustrated below, which proved the successful genetic transformation.
In contrast to miR172aOEs plants, all AP2OE plants showed shortened lemma and palea, and so that reduced grain size (Figure 3A), except that AP2-5OE plants were infertile (Figures 3C,D). As a result, the 1000-grain-weight of AP2-1OE, AP2-2OE, AP2-3OE, and AP2-4OE decreased (Supplementary Figure S5A), with the correspondingly reduced starch granules (Supplementary Figures S5B–F). AP2-2OE and AP2-5OE showed the most severe abnormality in floral organs. The lemma/palea was shortened appropriately one third in AP2-2OE and more in AP2-5OE (Figures 3B,C). Furthermore, in the AP2-2OE, the lemma and palea developed inconsistently, with the palea longer than the lemma (Figure 3C), contrasting to those in the miR172OEs (Figure 1A); in the AP2-5OE plants, the lemma/palea was malformed (Figure 3D). So that miR172/AP2 regulated floral organ identity and flower determinacy, especially elongation of the lemma/palea.
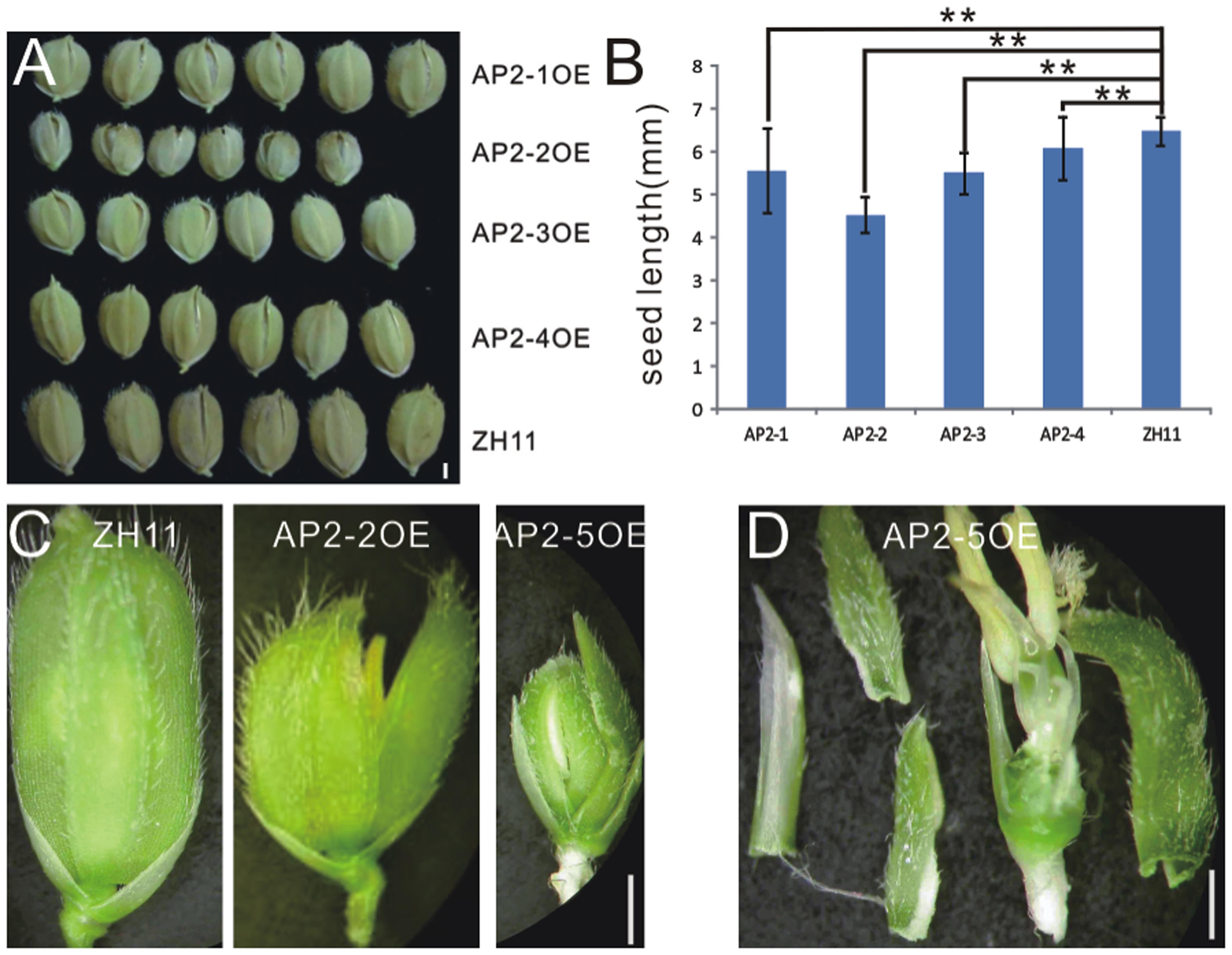
FIGURE 3. Phenotypes in the grain and spikelet of AP2s overexpressing plants. (A) Grains of the AP2OEs plants. Bar was 1 mm. (B) The grain length of the AP2OEs. Double asterisks represent significant difference determined by the Student’s t-test at ∗∗P < 0.01, respectively. (C) Spikelet of ZH11, AP2-2OE, and AP2-5OE plants. (D) Dissection of a spikelet of the AP2-5OE plant. Bars in (C,D) were 1 mm.
Furthermore, miR172s were down-regulated in the AP2-1OE, AP2-2OE, and AP2-5OE plants (Figure 4), indicating the possibility of negative feedback regulation between these AP2s and miR172.
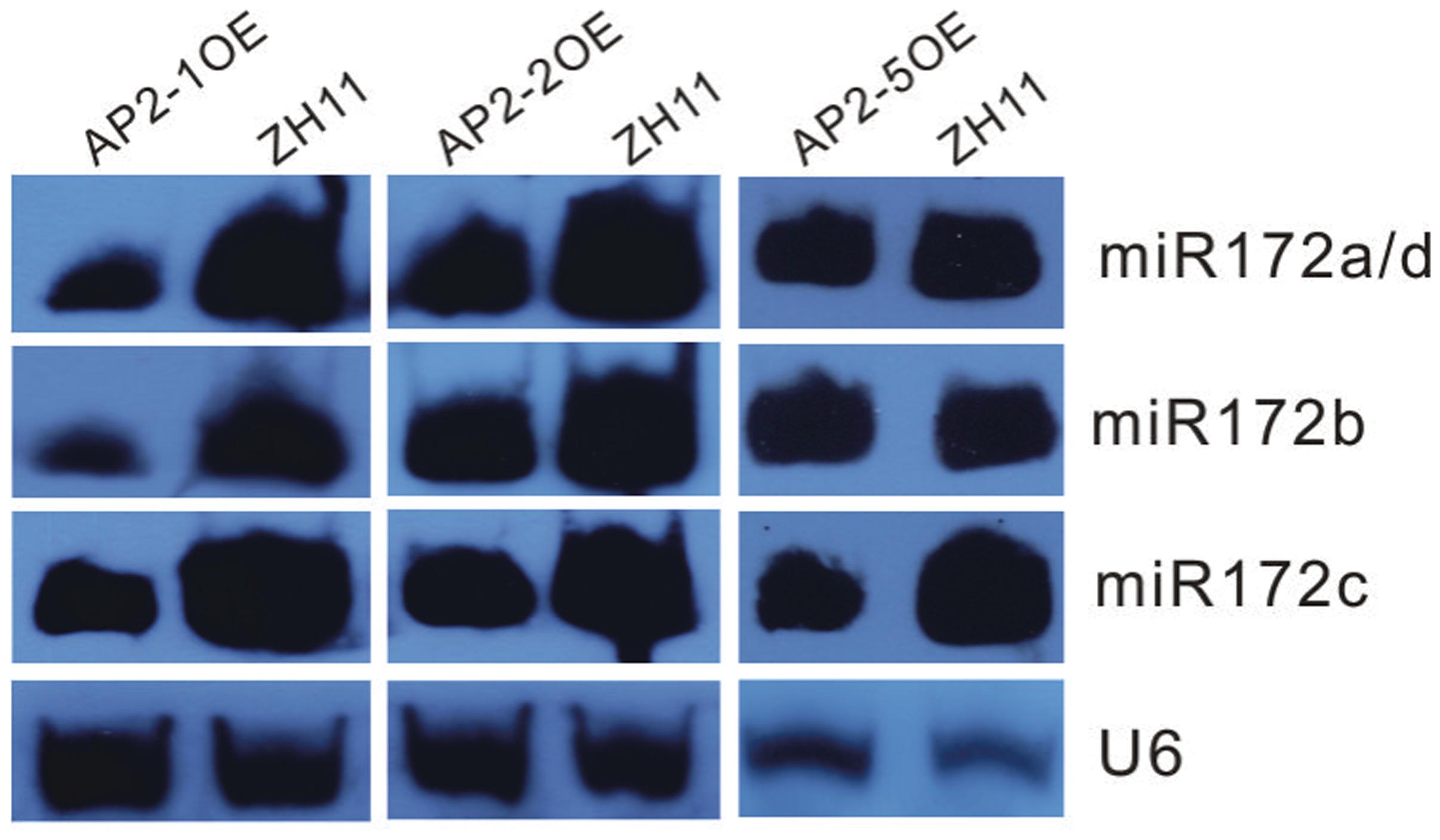
FIGURE 4. Northern blot analysis of miR172s in several AP2OE lines. Expression of respective miR172s was examined in the young panicles of the AP2-1OE, AP2-2OE, and AP2-5OE plants, respectively.
Expression of miR172s Was Inhibited by the OsMADS1 Gene
The OsMADS1 gene regulates floral organ identity, affecting lemma/palea development and spikelet determinacy (Jeon et al., 2000; Prasad et al., 2005; Wang K. et al., 2010). The phenotypic similarity of miR172OEs and the osmads1 mutant suggests a potential interaction between OsMADS1 and miR172.
To obviate the influence of the genetic background on the phenotype, we further constructed OsMADS1 over expression (OsMADS1OE) and RNAi transgenic plants (OsMADS1RNAi) in the ZH11 background; for each transgenic events, over 90% transformation efficiency were got. In the OsMADS1RNAi plants, the paleas and lemmas were slightly longer than those of the wild-type, and the lemma was longer than the palea (Figure 5A). Similarly, OsMADS1RNAi showed open hull and some ectopic glumes developed between whorl1 and whorl2 (Figure 5B, arrowhead). So that OsMADS1RNAi could mimic the osmads1 mutant, although to a milder degree.
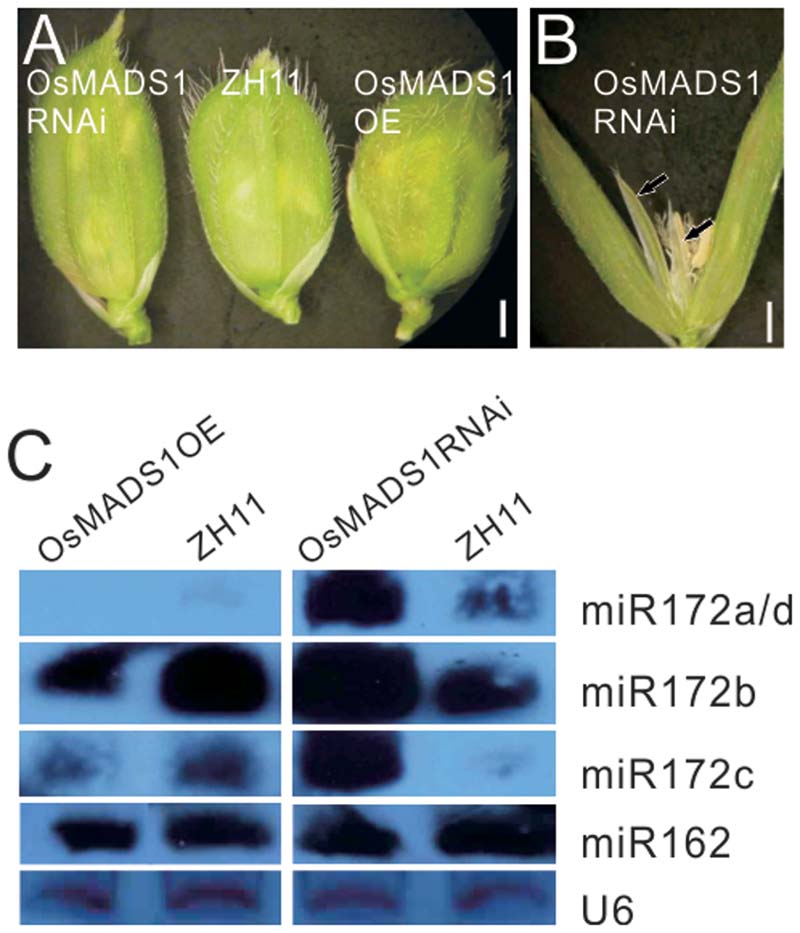
FIGURE 5. Analysis of OsMADS1OE and OsMADS1RNAi lines. (A) Phenotype of OsMADS1RNAi, ZH11, and Osmads1OE plants. (B) A floret of OsMADS1RNAi plants. (C) Expression of miR172s in the Osmads1OE and OsMADS1RNAi plants.
All miR172s were down-regulated in the Osmads1OE plants, but up-regulated in the OsMADS1RNAi plants (Figure 5C). Expression of miR162 in these plants was basically the same (Figure 5C), indicating that the expression of miR172s was specifically modulated by OsMADS1.
MADS-box genes regulate flower organ identity by binding to the cis-regulatory elements in the target genes termed “CArG-boxes” [consensus 5′CC(A/T)6GG3′] (Riechmann et al., 1996; Cui et al., 2010). In the 3 Kb promoter region of miR172a, miR172b, miR172c, and miR172d, there are 6, 4, 3, and 3 CArG-boxes, respectively (Supplementary Figure S6A). We carried out yeast one-hybrid and did not detect the direct binding of OsMADS1 protein to the motifs in the promoters of miR172 (Supplementary Figure S6B). However, OsMADS1 could interact with AP2-2, AP2-3, and AP2-5 in yeast two-hybrid systems (Supplementary Figure S7).
Expression Character of miR172 and OsMADS1 during Floral Organ Development.
To further examine the relationship between miR172a and OsMADS1, we performed in situ hybridization of them in flower development. Before differentiation of the floral organs, both OsMADS1 and miR172a expressed highly at the floret meristem (Figures 6A,F); thereafter, miR172a gradually accumulated on all the floral organs, with higher expression in the inner stamens and carpels, and lower expression in the outer palea and lemma (Figures 6B–E). However, OsMADS1 mRNA gradually concentrated on the palea and lemma (Figures 6G–I). So that the expression region of OsMADS1 and miR172 showed complementary character, OsMADS1 might repress excessive accumulation of miR172 in the lemma and the palea.
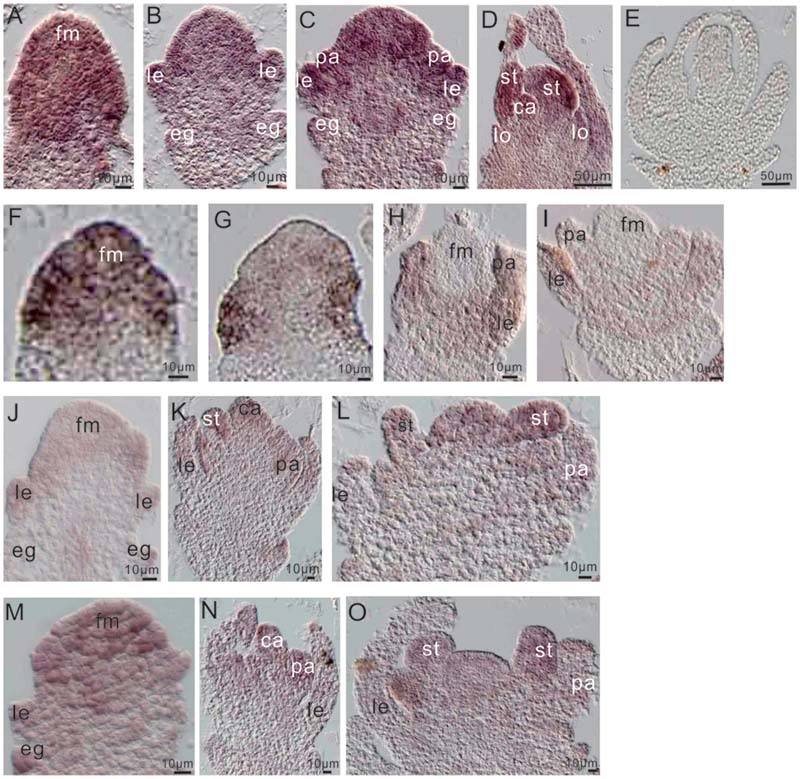
FIGURE 6. Expression of miR172 and OsMADS1 in the process of flower development. (A–D) and (F–I) Respective miR172a and OsMADS1 expression during floral organ development. (E) Sense probe. (J–L) and (M–O) miR172a expression in Osmads1OE and OsMADS1RNAi plants, respectively, during floral organ development. Abbreviations: fm, floret meristem; pa, palea; le, lemma; lo, lodicules; st, stamen; ca, carpel.
Assessment of the expression of miR172a in the Osmads1OE and OsMADS1RNAi lines showed that during development of the empty glume and palea/lemma, miR172a was down-regulated in the OsMADS1OE lines (Figure 6J as compared with Figure 6M); during the development of inner floral organs, miR172a expressed similarly in the OsMADS1OE and OsMADS1RNAi lines (Figure 6K as compared with Figure 6N, Figure 6L as compared with Figure 6O). Therefore, during the developmental process of palea/lemma, OsMADS1 exhibited inhibition to miR172, which may further explain the longer palea/lemma of miR172OEs plants.
Overexpression of AP2-2 and AP2-3 Rescued the Elongated Palea/Lemma in the OsMADS1RNAi Plants
Since the miR172aOE, miR172cOE, and miR172dOE were totally infertile, and miR172bOE barely produced any seeds, these lines were not amenable to genetic manipulation, we used AP2 genes to perform genetic complementation.
First, we made a cross between AP2-3OE and OsMADS1RNAi plants. Among the 11 hybrids of AP2-3OE/OsMADS1RNAi in the F1 generation, six individuals showed shortened and closed hull relatively to the OsMADS1RNAi plants (Figures 7A,B), the elongated and inconsistent palea/lemma in OsMADS1RNAi plants could be rescued by AP2-3 over expression (Figures 7A,B).
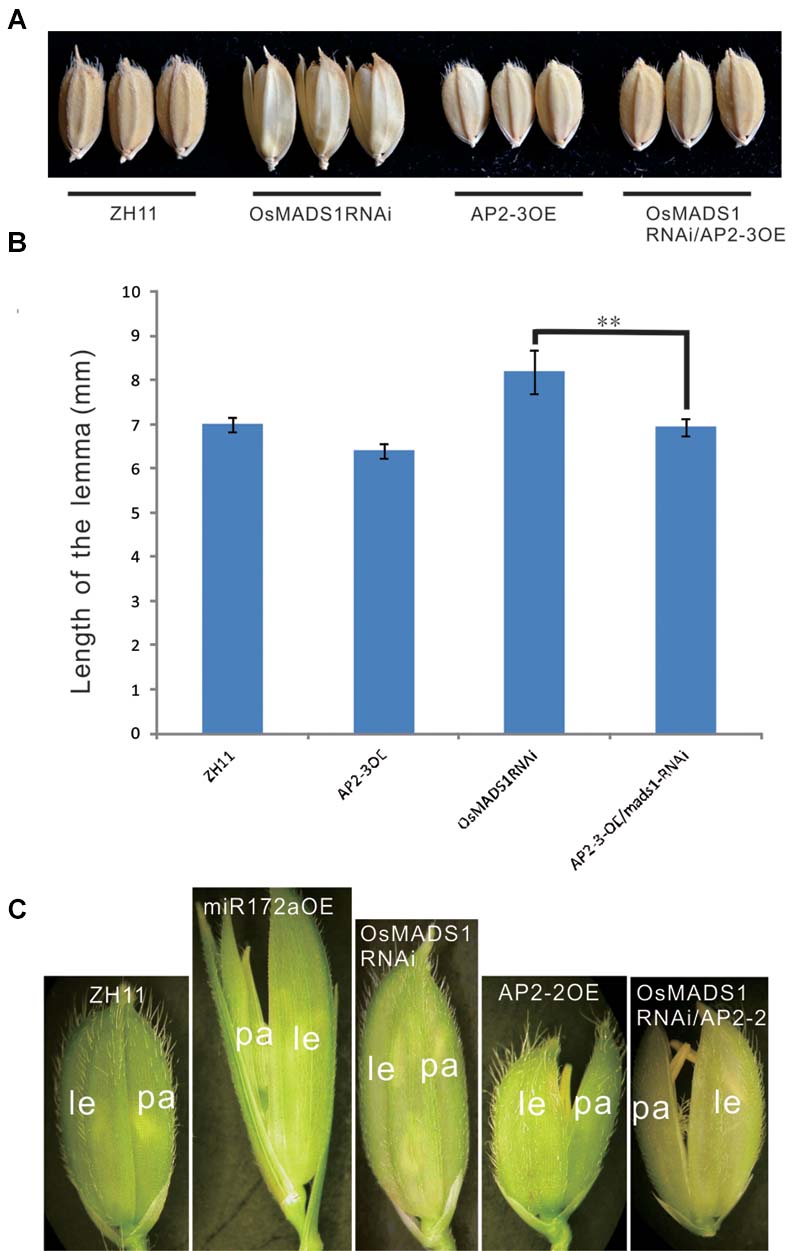
FIGURE 7. Phenotypic rescue of OsMADS1RNAi plants by AP2-3OE and AP2-2OE. (A) Phenotype of the grains of the AP2-3OE/OsMADS1RNAi hybrids. (B) The grain length of the OsMADS1RNAi, AP2-3OE/OsMADS1RNAi, and the wild-type. Double asterisks represent significant difference determined by the Student’s t-test at ∗∗P < 0.01, respectively. (C) Florets of AP2-2OE transformed into the OsMADS1RNAi plants. Pictures were arranged according to the relative size of the florets. Abbreviations: le, lemma; pa, palea.
Next, the AP2-2OE plasmid was genetically transformed into the OsMADS1RNAi plants. The lemma was longer than the palea in both the miR172aOE and OsMADS1RNAi plants, but shorter than the palea in the AP2-2OE plants. In the AP2-2/OsMADS1RNAi plants, the palea/lemma developed synchronously, and they were shorter than those of the OsMADS1RNAi plants but similar to those of the wild-type (Figure 7C). These results indicated the elongated and inconsistent palea/lemma in the OsMADS1RNAi plants could be rescued by AP2-2 over expression.
Discussion
miR172 was first reported to regulate floral organ development by negatively regulating the AP2 gene at the post-translation level in Arabidopsis (Chen, 2004), and miR172 could cooperate with miR156 to regulate flowering time sequentially (Wu et al., 2009). Various studies showed that miR172 is a pivotal regulator of reproductive development in plants. Here, we studied the function of miR172 in regulating rice floral organ development. In rice, miR172 expressed in each whorl of the floral organs (Figures 6B,C), suggesting its function in all these whorls, as manifested by the phenotype of miR172s over expression (Figure 1). In miR172aOEs plants, both the palea and the lemma were malformed and elongated, with the lemma longer than the palea, while in the AP2-2OE lines, the lemma was shorter than the lemma, overexpression of all target AP2 genes resulted in shortened lemma/palea, indicating miR172/AP2s module regulated the elongation and synchronous development of the palea and the lemma in rice. In some cases, miR172aOEs resulted in a “flower-in-flower” phenotype (Figure 1I) and a totally inverted process of flower development (Figure 1J), indicating an indeterminacy state in flower development. Correspondingly, mutation in the targets of miR172, such as the SNB gene and the OsIDS1 gene resulted in loss of flower determinacy (Lee et al., 2007; Lee and An, 2012).
In Arabidopsis, miR172 regulates the elongation of the valve under the modulation of the FUL gene (a MADS-box gene) and the ARF6/8 gene (José Ripoll et al., 2015). Here we showed that miR172 regulated floral organ development in rice, especially the elongation of the palea and lemma, under the modulation of OsMADS1. Therefore, the upstream regulatory pathway of miR172 in Arabidopsis and rice showed some similarity.
Previous studies revealed that plant miRNAs can act either upstream or downstream of transcription factors. For example, in Arabidopsis the transcription factors SQUAMOSA BINDING PROTEIN-like 9 (SPL9) and the MADS-box SHORT VEGETATIVE PHASE (SVP) act as a direct activator and a direct repressor, respectively, of miR172 (Wu et al., 2009; Cho et al., 2012). OsMADS1 expressed mainly in the palea/lemma and lodicules (Figure 6H), while miR172 expressed highly in the newly formed stamens and carpels (Figure 6D). Therefore, the expression domains of OsMADS1 and miR172 showed some degree of complementation, implying the inhibition of OsMADS1 to miR172. In OsMADS1RNAi plants, the lemma and palea showed inconsistent development, and some ectopic glumes formed between the palea/lemma and the lodicules (Figure 5B), which was similar to the pattern in miR172OEs lines. Furthermore, miR172OEs lines showed “flower-in-flower” structures similar to those in the osmads1 mutant (Wang K. et al., 2010). These phenotypic resemblances indicated the genetic connection between OsMADS1 and miR172, and our study indicated the inhibition of OsMADS1 to miR172. However, we did not find direct binding of OsMADS1 to any of the miR172 promoters by yeast one-hybrid system. OsMADS1 was reported to form homo-dimer and hetero-dimer with OsMADS7 and OsMADS8 (Cui et al., 2010). Different OsMADS proteins might form functional complex. Therefore, the binding of OsMADS1 to the promoter of miR172 might require the involvement of several other OsMADS proteins, making it more difficult to be detected. Another possibility is that, OsMADS1 might interact with AP2, and AP2 regulate the expression of miR172 at the transcription level, just as that in Arabidopsis, the miR172 repression orchestrated by LUG and SEU co-repressors is dependent on the miR172 target gene AP2 itself, by a positive-feedback loop allowing AP2 to maintain its own expression in the outer floral whorls (Grigorova et al., 2011), or by binding to the miR172b promoter (Yant et al., 2010).
Floral organ development is a fundamental event in plant development, especially for plant reproduction. In addition to OsMADS1, many genes are involved in this process. Among them, Extra Glume1 (EG1) showed a phenotype with high similarity to that of osmads1, and further analysis revealed that EG1 gene is required for the maintenance of OsMADS1 expression in the floral meristem and act as an upstream regulator of OsMADS1 (Li et al., 2009). Also, the polycomb group gene EMF2B is a direct repressor of OsMADS1 (Conrad et al., 2014). A LBD-like transcription factor, OsIG1, which mainly regulate female gametophyte development, might be another upstream regulator of OsMADS1 (Zhang et al., 2015). In the present study, we found that OsMADS1 functioned by modulating miR172s, indicating one primary pathway consisting of OsMADS1-miR172-AP2 involved in the regulation of floral organ development (Figure 8).
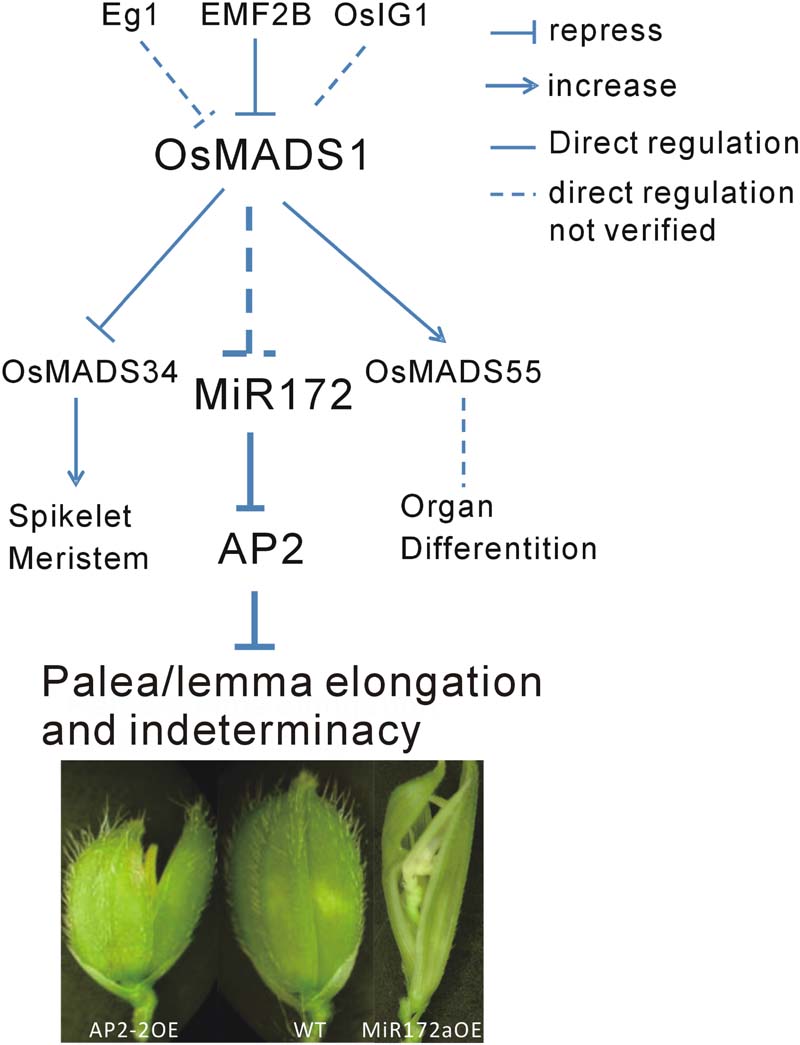
FIGURE 8. Diagram of the possible pathway of OsMADS1-miR172-AP2 regulation on palea/lemma. OsMADS1 might negatively regulated miR172s, when OsMADS1 was down-regulated or mutated, miR172s were up-regulated, and the flower showed elongated palea/lemma and indeterminacy. Meanwhile, miR172 negatively regulated its target AP2 genes, and up-regulation of AP2 showed shortened palea/lemma (exemplified by AP2-2OE).
Author Contributions
Experimental design: XM and ZS; Experiments: ZD, JW, and MZ; Data analysis: JW and ZS; Manuscript preparation: ZS and XM; Supervision, funding and reagents: ZS and XM.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFD0100600), Ministry of Science and Technology of China (2012AA10A302-2), the National Special Program on Research and Commercialization of Transgenic Plant (2014ZX08009-003-003), Scholarship Foundation from Shanghai Institutes for Biological Sciences (2007KIP206), and the SA-SIBS 2009 Young Faculty Award.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We would like to thank Professor Dabing Zhang from Shanghai Jiaotong University for helpful discussion, and Xiaoyan Gao from Shanghai Institutes for Biological Sciences for his help in SEM analysis and Anatomical analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01891/full#supplementary-material
Footnotes
- ^ http://rice.plantbiology.msu.edu/
- ^ https://www.ncbi.nlm.nih.gov/
- ^ http://sundarlab.ucdavis.edu/smrnas/
- ^ http://bioinformatics.cau.edu.cn/PMRD/
References
Agrawal, G. K., Abe, K., Yamazaki, M., Miyao, A., and Hirochika, H. (2005). Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol. Biol. 59, 125–135. doi: 10.1007/s11103-005-2161-y
Angenent, G. C., Franken, J., Busscher, M., Van Dijken, A., Van Went, J. L., Dons, H. J., et al. (1995). A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell 7, 1569–1582. doi: 10.1105/tpc.7.10.1569
Arora, R., Agarwal, P., Ray, S., Singh, A. K., Singh, V. P., Tyagi, A. K., et al. (2007). MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8:242. doi: 10.1186/1471-2164-8-242
Chapman, E. J., and Carrington, J. C. (2007). Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 8, 884–896. doi: 10.1038/nrg2179
Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. doi: 10.1126/science.1088060
Chen, Z. X., Wu, J. G., Ding, W. N., Chen, H. M., Wu, P., and Shi, C. H. (2006). Morphogenesis and molecular basis on naked seed rice, a novel homeotic mutation of OsMADS1 regulating transcript level of AP3 homologue in rice. Planta 223, 882–890. doi: 10.1007/s00425-005-0141-8
Cho, H. J., Kim, J. J., Lee, J. H., Kim, W., Jung, J. H., Park, C. M., et al. (2012). SHORT VEGETATIVE PHASE (SVP) protein negatively regulates miR172 transcription via direct binding to the pri-miR172a promoter in Arabidopsis. FEBS Lett. 586, 2332–2337. doi: 10.1016/j.febslet.2012.05.035
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Coen, E. S., Romero, J. M., Doyle, S., Elliott, R., Murphy, G., and Carpenter, R. (1990). floricaula: a homeotic gene required for flower development in antirrhinum majus. Cell 63, 1311–1322. doi: 10.1016/0092-8674(90)90426-F
Conrad, L. J., Khanday, I., Johnson, C., Guiderdoni, E., An, G., Vijayraghavan, U., et al. (2014). The polycomb group gene EMF2B is essential for maintenance of floral meristem determinacy in rice. Plant J. 80, 883–894. doi: 10.1111/tpj.12688
Cui, R., Han, J., Zhao, S., Su, K., Wu, F., Du, X., et al. (2010). Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 61, 767–781. doi: 10.1111/j.1365-313X.2009.04101.x
Ditta, G., Pinyopich, A., Robles, P., Pelaz, S., and Yanofsky, M. F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935–1940. doi: 10.1016/j.cub.2004.10.028
Ferrario, S., Immink, R. G., and Angenent, G. C. (2004). Conservation and diversity in flower land. Curr. Opin. Plant Biol. 7, 84–91. doi: 10.1016/j.pbi.2003.11.003
Fu, F. F., and Xue, H. W. (2010). Coexpression analysis identifies rice starch regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 154, 927–938. doi: 10.1104/pp.110.159517
Grigorova, B., Mara, C., Hollender, C., Sijacic, P., Chen, X., and Liu, Z. (2011). LEUNIG and SEUSS co-repressors regulate miR172 expression in Arabidopsis flowers. Development 138, 2451–2456. doi: 10.1242/dev.058362
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Hu, Y., Liang, W., Yin, C., Yang, X., Ping, B., Li, A., et al. (2015). Interactions of OsMADS1 with floral homeotic genes in rice flower development. Mol. Plant 8, 1366–1384. doi: 10.1016/j.molp.2015.04.009
Jeon, J. S., Jang, S., Lee, S., Nam, J., Kim, C., Lee, S. H., et al. (2000). leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12, 871–884. doi: 10.1105/tpc.12.6.871
José Ripoll, J., Bailey, L. J., Mai, Q.-A., Wu, S. L., Hon, C. T., Chapman, E. J., et al. (2015). microRNA regulation of fruit growth. Nat. Plants 1: 15036. doi: 10.1038/nplants.2015.36
Khanday, I., Das, S., Chongloi, G. L., Bansal, M., Grossniklaus, U., and Vijayraghavan, U. (2016). Genome-wide targets regulated by the OsMADS1 transcription factor reveals its DNA recognition properties. Plant Physiol. 172, 372–388. doi: 10.1104/pp.16.00789
Khanday, I., Yadav, S. R., and Vijayraghavan, U. (2013). Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways. Plant Physiol. 161, 1970–1983. doi: 10.1104/pp.112.212423
Lee, D. Y., and An, G. (2012). Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 69, 445–461. doi: 10.1111/j.1365-313X.2011.04804.x
Lee, D. Y., Lee, J., Moon, S., Park, S. Y., and An, G. (2007). The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J. 49, 64–78. doi: 10.1111/j.1365-313X.2006.02941.x
Li, H., Liang, W., Jia, R., Yin, C., Zong, J., Kong, H., et al. (2010). The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res. 20, 299–313. doi: 10.1038/cr.2009.143
Li, H., Xue, D., Gao, Z., Yan, M., Xu, W., Xing, Z., et al. (2009). A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice. Plant J. 57, 593–605. doi: 10.1111/j.1365-313X.2008.03710.x
Martin, A., Adam, H., Diaz-Mendoza, M., Zurczak, M., Gonzalez-Schain, N. D., and Suarez-Lopez, P. (2009). Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 136, 2873–2881. doi: 10.1242/dev.031658
Mathieu, J., Yant, L. J., Murdter, F., Kuttner, F., and Schmid, M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7:e1000148. doi: 10.1371/journal.pbio.1000148
Mlotshwa, S., Yang, Z., Kim, Y., and Chen, X. (2006). Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana. Plant Mol. Biol. 61, 781–793. doi: 10.1007/s11103-006-0049-0
Molnar, A., Schwach, F., Studholme, D. J., Thuenemann, E. C., and Baulcombe, D. C. (2007). miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature 447, 1126–1129. doi: 10.1038/nature05903
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Ohmori, S., Kimizu, M., Sugita, M., Miyao, A., Hirochika, H., Uchida, E., et al. (2009). MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21, 3008–3025. doi: 10.1105/tpc.109.068742
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Prasad, K., Parameswaran, S., and Vijayraghavan, U. (2005). OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 43, 915–928. doi: 10.1111/j.1365-313X.2005.02504.x
Riechmann, J. L., Krizek, B. A., and Meyerowitz, E. M. (1996). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. U.S.A. 93, 4793–4798. doi: 10.1073/pnas.93.10.4793
Schwarz-Sommer, Z., Huijser, P., Nacken, W., Saedler, H., and Sommer, H. (1990). Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250, 931–936. doi: 10.1126/science.250.4983.931
Shukla, R. K., Raha, S., Tripathi, V., and Chattopadhyay, D. (2006). Expression of CAP2, an APETALA2-family transcription factor from chickpea, enhances growth and tolerance to dehydration and salt stress in transgenic tobacco. Plant Physiol. 142, 113–123. doi: 10.1104/pp.106.081752
Tang, W., Charles, T. M., and Newton, R. J. (2005). Overexpression of the pepper transcription factor CaPF1 in transgenic Virginia pine (Pinus Virginiana Mill.) confers multiple stress tolerance and enhances organ growth. Plant Mol. Biol. 59, 603–617. doi: 10.1007/s11103-005-0451-z
Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J. T., Munster, T., et al. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42, 115–149. doi: 10.1023/A:1006332105728
Wang, J., Gao, X., Li, L., Shi, X., Zhang, J., and Shi, Z. (2010). Overexpression of Osta-siR2141 caused abnormal polarity establishment and retarded growth in rice. J. Exp. Bot. 61, 1885–1895. doi: 10.1093/jxb/erp378
Wang, K., Tang, D., Hong, L., Xu, W., Huang, J., Li, M., et al. (2010). DEP and AFO regulate reproductive habit in rice. PLoS Genet. 6:e1000818. doi: 10.1371/journal.pgen.1000818
Wu, G., Park, M. Y., Conway, S. R., Wang, J. W., Weigel, D., and Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. doi: 10.1016/j.cell.2009.06.031
Xie, Z., and Qi, X. (2008). Diverse small RNA-directed silencing pathways in plants. Biochim. Biophys. Acta 1779, 720–724. doi: 10.1016/j.bbagrm.2008.02.009
Yaish, M. W., El-Kereamy, A., Zhu, T., Beatty, P. H., Good, A. G., Bi, Y. M., et al. (2010). The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 6:e1001098. doi: 10.1371/journal.pgen.1001098
Yamaguchi, T., Lee, D. Y., Miyao, A., Hirochika, H., An, G., and Hirano, H. Y. (2006). Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18, 15–28. doi: 10.1105/tpc.105.037200
Yant, L., Mathieu, J., Dinh, T. T., Ott, F., Lanz, C., Wollmann, H., et al. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22, 2156–2170. doi: 10.1105/tpc.110.075606
Zhang, J., Tang, W., Huang, Y., Niu, X., Zhao, Y., Han, Y., et al. (2015). Down-regulation of a LBD-like gene, OsIG1, leads to occurrence of unusual double ovules and developmental abnormalities of various floral organs and megagametophyte in rice. J. Exp. Bot. 66, 99–112. doi: 10.1093/jxb/eru396
Zhao, L., Kim, Y., Dinh, T. T., and Chen, X. (2007). miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J. 51, 840–849. doi: 10.1111/j.1365-313X.2007.03181.x
Zhao, T., Li, G., Mi, S., Li, S., Hannon, G. J., Wang, X. J., et al. (2007). A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 21, 1190–1203. doi: 10.1101/gad.1543507
Zhou, M., Li, D., Li, Z., Hu, Q., Yang, C., Zhu, L., et al. (2013). Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 161, 1375–1391. doi: 10.1104/pp.112.208702
Keywords: OsMADS1, miR172, floral organs, lemma, palea, determinacy
Citation: Dai Z, Wang J, Zhu M, Miao X and Shi Z (2016) OsMADS1 Represses microRNA172 in Elongation of Palea/Lemma Development in Rice. Front. Plant Sci. 7:1891. doi: 10.3389/fpls.2016.01891
Received: 22 August 2016; Accepted: 30 November 2016;
Published: 20 December 2016.
Edited by:
Keqiang Wu, National Taiwan University, TaiwanReviewed by:
Mingyong Zhang, South China Botanical Garden, Chinese Academy of Sciences, ChinaDayong Li, Chinese Academy of Sciences, China
Copyright © 2016 Dai, Wang, Zhu, Miao and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenying Shi, enlzaGlAc2licy5hYy5jbg==
 Zhengyan Dai
Zhengyan Dai Zhenying Shi
Zhenying Shi