- 1College of Life Sciences, Xinyang Normal University, Xinyang, China
- 2Institute for Conservation and Utilization of Agro-bioresources in Dabie Mountains, Xinyang Normal University, Xinyang, China
- 3Center for Genomics and Biotechnology, Haixia Institute of Science and Technology, Fujian Agriculture and Forestry University, Fuzhou, China
- 4Key Laboratory of Genetics, Breeding and Multiple Utilization of Corps (Fujian Agriculture and Forestry University), Ministry of Education, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, Fujian Agriculture and Forestry University, Fuzhou, China
Histone modifications, such as methylation and demethylation, play an important role in regulating chromatin structure and gene expression. The JmjC domain-containing proteins, an important family of histone lysine demethylases (KDMs), play a key role in maintaining homeostasis of histone methylation in vivo. In this study, we performed a comprehensive analysis of the jumonji C (JmjC) gene family in the soybean genome and identified 48 JmjC genes (GmJMJs) distributed unevenly across 18 chromosomes. Phylogenetic analysis showed that these JmjC domain-containing genes can be divided into eight groups. GmJMJs within the same phylogenetic group share similar exon/intron organization and domain composition. In addition, 16 duplicated gene pairs were formed by a Glycine-specific whole-genome duplication (WGD) event approximately 13 million years ago (Mya). By investigating the expression profiles of these gene pairs in various tissues, we showed that the expression pattern is conserved in the polyploidy-derived JmjC duplicates, demonstrating that the majority of GmJMJs were preferentially retained after the most recent WGD event and suggesting important roles for demethylase duplications in soybean evolution. These results shed light on the evolutionary history of this family in soybean and provide insights into the JmjCs which will be helpful to reveal their functions in controlling soybean development.
Introduction
Histone methylation and demethylation have important roles in regulating transcription, genome integrity, and epigenetic inheritance (Klose et al., 2006; Klose and Zhang, 2007; Liu et al., 2010). Histone methylation can occur at various lysine and arginine residues, including K4, K9, K27, K36, and K79 in histone H3 and K20 in histone H4 (Allis et al., 2007). Histone methylation, which is mainly catalyzed by protein families that contain PRMT and SET domains, can have both activating and repressive effects on chromatin function (Ahmad and Cao, 2012; Zhang and Ma, 2012). Two kinds of demethylase are involved in the homeostasis of methylation in organisms. Lysine Specific Demethylase 1 (LSD1) was the first histone demethylase identified and is a member of the flavin-dependent amine oxidase family (Lee et al., 2005; Metzger et al., 2005; Chen et al., 2011). Genes in the second class of histone demethylases have a JmjC domain with which they catalyze histone lysine demethylation through oxidative reactions dependent on ferrous ion (Fe(II)) and α-ketoglutarate (α-KG) (Elkins et al., 2003; Trewick et al., 2005).
Plant JmjC proteins are known to play important roles in regulating epigenetic processes and in growth and development (Klose et al., 2006; Kouzarides, 2007). Many members of the JmjC gene family from different plant species have been characterized. In Arabidopsis, AtJMJ11/ELF6 (EARLY FLOWERING 6) is a repressor in the photoperiodic flowering pathway, and its loss-of-function mutation causes early flowering (Noh et al., 2004; Yu et al., 2008). Its relative, AtJMJ12/REF6 (RELATIVE OF EARLY FLOWERING 6) has an opposite effect in the regulation of flowering time (Noh et al., 2004; Yu et al., 2008; Lu et al., 2011a). Loss-of-function mutation of REF6 leads to increased expression of the flowering repressor FLC (FLOWERING LOCUS C) and hence late flowering (Lu et al., 2011a). In addition, AtJMJ14, an active histone H3K4 demethylase (Lu et al., 2010; Yang et al., 2010), was also implicated in preventing early flowering by repressing the expression of FLOWERING LOCUS T (FT) and its homologs. Recently, Ning reported that AtJMJ14 associates with two NAC transcription factors, NAC050 and NAC052, and co-occupies hundreds of common target genes, resulting in H3K4 demethylation and transcriptional repression (Ning et al., 2015). Apart from controlling flowering time, there is also evidence that AtJMJ14 functions in RNA silencing and cell-to-cell movement of an RNA silencing signal (Lu et al., 2010). The histone H3K9 demethylase AtJMJ25/IBM1 (INCREASE IN BONSAI METHYLATION 1) (Wang et al., 2013; Shen et al., 2014a) protects genes from CHG (H represents A, T, or G) hypermethylation by CMT3 (CHROMOMETHYLASE 3). Gain-of-function mutants of AtJMJ15 showed enhanced salt tolerance, in contrast with increased salt sensitivity in the loss-of-function mutant (Shen et al., 2014a). In addition, AtJMJ14 and AtJMJ15 have also been shown to be involved in the control of flowering time (Yang et al., 2012). AtJMJ30/JMJD5, an evening-expressed gene, is the sole AtJMJ protein to show a robust circadian rhythm of expression (Mockler et al., 2007; Michael et al., 2008; Jones and Harmer, 2011; Lu et al., 2011b). The role of AtJMJ30 as a genetic regulator of period length in the Arabidopsis circadian clock was confirmed by analysis of loss- and gain-of-function mutants (Lu et al., 2011b). In tomato, a similar role is played by JMJ524, which standardizes the circadian clock and also alters GA response to regulate stem elongation (Li et al., 2015). In Medicago truncatula, MtJMJC5 (Medtr4g066020), an ortholog of AtJMJ30/JMJD5, may play a role in epigenetic regulation of the link between the circadian clock and cold signaling (Shen et al., 2016). In rice, OsJMJ705 is a biotic stress-responsive H3K27me2/3 demethylase that may remove H3K27me3 from marked defense-related genes and increase their basal and induced expression during pathogen infection (Li et al., 2013). OsJMJ706, encodes a heterochromatin-associated H3K9 demethylase, is reported to involve in the regulation of flower development in rice (Sun and Zhou, 2008).
Soybean [Glycine max (L.) Merr.] is one of the most economically important crop species in the world. Its genome has undergone two rounds of whole-genome duplication (WGD; Schlueter et al., 2004; Schmutz et al., 2010; Vanneste et al., 2014; Liu et al., 2015); one occurred approximately 59 Mya and was shared by other legumes such as Medicago and Lotus, while the other was specific to Glycine and occurred around 13 Mya. Thus, about 75% of the genes in the soybean genome have multiple paralogs (Schmutz et al., 2010; Severin et al., 2011; Singh and Jain, 2015), making it an excellent model for studying the evolution of duplicate genes following polyploidy. Here, we systemically identify the JmjC gene family members in soybean (G. max), Medicago (M. truncatula), and Lotus (Lotus japonicus) and subsequently analyze the evolutionary relationships between these genes among the three legumes, Oryza sativa, and Arabidopsis. In addition, we study the GmJMJs in further detail, including subfamily classification, gene structures, chromosomal distribution, duplication patterns, conserved residues, and expression profiling. We propose that demethylases exhibit conservative functions through duplication events. Our data will facilitate future studies to elucidate the exact biological functions of the GmJMJs.
Materials and Methods
Identification of JmjC Domain-Containing Proteins in Soybean and Other Legumes
The G. max 2.0 genome database (https://phytozome.jgi.doe.gov/pz/#) was searched to identify JmjC domain-containing proteins using Basic Local Alignment Search Tool algorithms (BLASTP) with a threshold of e-value < 1e-10, using the published Arabidopsis (21) and O. sativa (20) JmjC domain-containing protein sequences (Table S1) as queries (Huang et al., 2016). All obtained protein sequences were examined for the presence of the JmjC (PF02373, SM00558) domain using the Hidden Markov Model (HMM) of Pfam (Finn et al., 2016) (http://pfam.sanger.ac.uk/search), and SMART (Letunic et al., 2015) (http://smart.embl-heidelberg.de/). Sequences with obvious errors and/or JmjC domain length of <90 amino acids were removed manually. Following the same approach, putative M. truncatula and L. japonicus JmjC domain-containing proteins were identified from Phytozome v10 (https://phytozome.jgi.doe.gov/pz/portal.html) and the L. japonicus genome assembly build 3.0 (http://www.kazusa.or.jp/lotus/), respectively.
Phylogenetic Analysis
Multi-species phylogenetic tree was constructed using MEGA 6.0 (Tamura et al., 2013) with the Neighbor-Joining (NJ) method, and bootstrap analysis was conducted using 1000 replicates with the p-distance model. The JmjC domain alone was used to set up the phylogenetic tree and define the groups (Figure S1). And then with the aim to obtain a better phylogeny within each group, we added additional conserved domains to JmjC domain (Table S2) to construct the phylogenetic tree (Figures 1A, 4A). Multiple sequences alignments were performed using ClustalW with default parameters in MEGA 6.0.
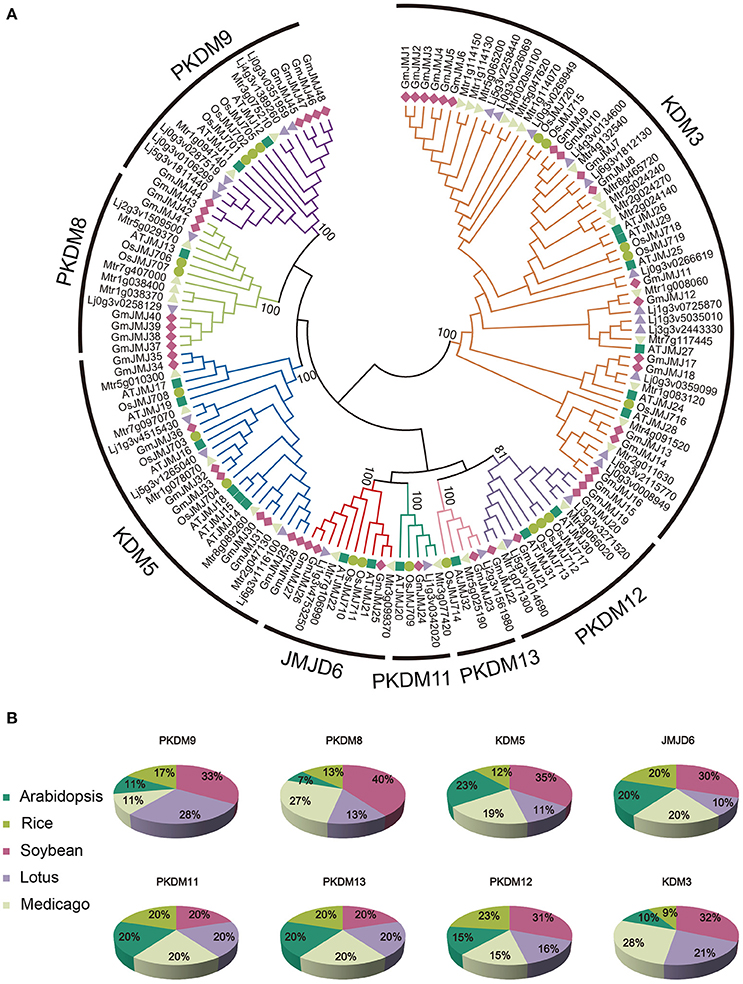
Figure 1. Phylogenetic relationship and distribution of JmjC-domain containing proteins from five plant species. (A) Phylogenetic relationship of JmjC-domain containing proteins among soybean, Medicago, Lotus, rice, and Arabidopsis. Both multiple sequences alignment and phylogenetic tree were performed by MEGA6.0. The value at the nodes represents bootstrap values from 1000 replicates. Different groups were shown by different color. (B) Percentage representation of JmjC-domain containing proteins across the five plant species within each group. Colors correspond to the plant taxa as listed in the left.
Chromosomal Locations and Gene Structure of JmjC Genes
The locations of the JmjC domain-containing genes on the soybean chromosomes were plotted using the MapChart software. The location information of each JmjC domain-containing gene on each chromosome was determined from the soybean genome annotation file (Gmax_275_Wm82.a2.v1.gene.gff3). The blocks regarded as recent duplications were obtained from SoyBase (Grant et al., 2010) (http://www.soybase.org/). The presence of introns and exons was also annotated according to the soybean genome annotation file. Schematic diagrams were pictured by using GSDS2.0 (Gene Structure Display Server http://gsds.cbi.pku.edu.cn/).
Conserved Domains and Conserved Residues in the JmjC Domain-Containing Proteins
To explore the full-length sequences of JmjC domain-containing proteins, NCBI CDD (Marchler-Bauer et al., 2015) (http://www.ncbi.nlm.nih.gov/cdd/), SMART (http://smart.embl-heidelberg.de/), and Pfam (http://pfam.xfam.org/) were performed with default parameters to search for conserved domains. To identify conserved amino acid residues for interaction with co-factors, the sequences of JmjC domain were aligned using the DNAMAN software.
Expression Analysis of Soybean JmjC Genes
To determine the expression patterns of the JmjC genes in soybean tissues, transcriptome data was downloaded from the NCBI Short Read Archive database under the following accession numbers: SRX474427, SRX474441, SRX474445, SRX474430, SRX474431, SRX474433, SRX474432, SRX474439, SRX474442, SRX474419, SRX474428, SRX474440, SRX474443, SRX474424, SRX474423, SRX474422, SRX474434, SRX474436, SRX474437, SRX474416, SRX474435, SRX474438, SRX474421, SRX474420, SRX474446, SRX474444, SRX474426, and SRX474429 (Shen et al., 2014b). Transcriptome analysis was performed to identify expression patterns in representative tissues, including roots, cotyledons, stems, shoot meristems, leaf buds, leaves, flowers, pods, pod and seeds, and seeds (Table S3). Finally, heatmaps of GmJMJ expression were produced using the pheatmap packages in R.
Calculation of Ka/Ks-Values and Evaluation Divergence Time
To investigate whether positive Darwinian selection was involved in GmJMJ divergence following duplication and to estimate the date of the duplication pairs, the non-synonymous (Ka) and synonymous substitution (Ks) rate ratios of the paralog pairs were calculated using the YN00 method of the PAML program (Yang, 2007). Based on a rate of 6.1 × 10−9 substitutions per site per year, we calculated the divergence time (T) as T = Ks/(2 × 6.1 × 10−9) × 10−6 Mya (Lynch and Conery, 2000).
Results
Identification of JmjC Gene Family in Soybean
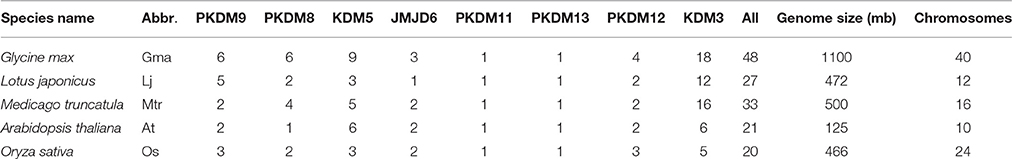
Using the combined methods, we identified a total of 48 GmJMJs, which is more than twice the number found in Arabidopsis (21) or rice (20) (Lu et al., 2008). To better understand the expansion and evolutionary history of GmJMJs, the same methods were used to search for JmjC genes in two other legumes, Medicago and Lotus. We identified 33 Medicago and 27 Lotus JmjC genes, which is still less than the number found in soybean.
A variety of information about GmJMJs, such as different version of gene codes, gene length, isoelectric point (pI), and molecular weight (Mw) and so on, were listed in Table S4. For example, the identified GmJMJs encode proteins ranging from 284 (GmJMJ2) to 1831 (GmJMJ35) amino acids, with the isoelectric point (pI) varying from 4.91 (GmJMJ23) to 9.25 (GmJMJ37) and the molecular weight (Mw) varying from 32.2 kD (GmJMJ2) to 209.2 kD (GmJMJ35). GmJMJ1 was excluded from further analyses, as there is no annotation data available for it.
Phylogenetic Analysis of JmjC Genes in Soybean
According to the phylogenetic analysis (Figure 1A), the JmjC genes can be divided into eight groups: PKDM9, PKDM8, KDM5, JMJD6, PKDM11, PKDM13, PKDM12, and KDM3. KDM3 has the most members, with 57 homologous JmjC genes, and KDM5 is the second largest group, containing 26 JmjC genes. The smallest clades are PKDM11 and PKDM13, which both consist of only five JmjC genes, one from each species. In general, most of the clades include genes from all five species, although the clades are also enriched in particular species. For example, doubled GmJMJ pairs are sister genes to AtJMJ28 in a clade which also includes doubled genes from the other legumes, forming a cluster of several legume JMJ genes with a single AtJMJ gene. Likewise, there is a larger of percentage of soybean (40%) than Arabidopsis (7%) genes in PKDM8 (Figure 1B). These findings indicate that different levels of gene duplication or lose may have been occurred among the five species after the divergence of eudicot and monocot.
The phylogenetic relationships of AtJMJ30, OsJMJ717, and MtJMJC5 (Mtr4g066020) in PKDM12 are consistent with a recent report (Shen et al., 2016). Shen et al. showed that MtJMJC5 is involved in regulating circadian rhythm (Shen et al., 2016). And recently, Li et al also reported that JMJ524, consistent with its counterparts AtJMJ30, is also involved in a circadian clock response in tomato (Li et al., 2015). Based on the phylogenetic analysis we hypothesis that soybean orthologs of MtJMJC5 and JMJ524 (GmJMJ19 and GmJMJ20) may also play similar roles in rhythm regulation.
Chromosome Location and Duplication of GmJmJ Genes
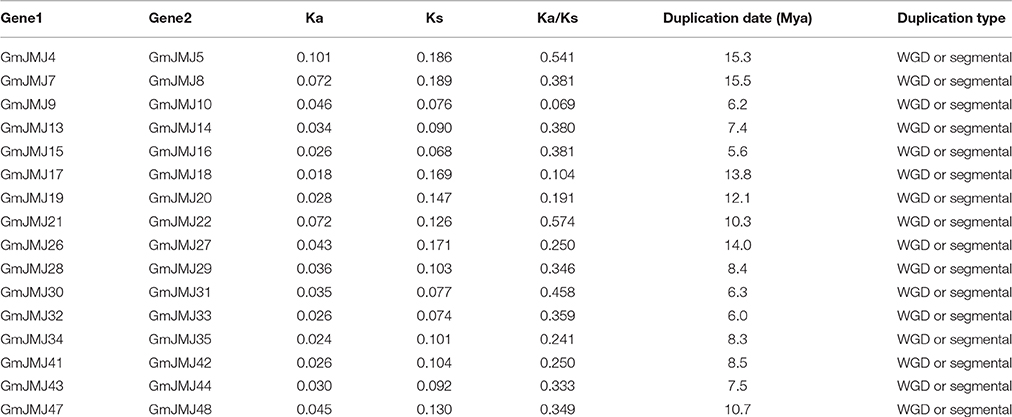
Compared to other species, soybean has an extensively expanded GmJMJ family with more than twice as many JmjCs as rice and Arabidopsis (Table 1). We carried out a comprehensive analysis of the GmJMJs with the aim of understanding their duplication status and identifying duplicated gene pairs. First, GmJMJ pairs located in a pair of paralogous blocks formed by Glycine-specific WGD were considered as candidate duplicate gene pairs. As shown in Figure 2, all 47 GmJMJs (except GmJMJ1) were randomly located on 18 of the 20 soybean chromosomes. For example, chromosome 10 possesses six GmJMJs, chromosomes 4, 6, 7, 8, and 15 each contain three GmJMJs, and chromosome 2, 3, 5, 12, 13, 14, and 17 each have only one GmJMJ. In total, we found that a large proportion (41 of 47) of the GmJMJs (linked by purple lines in Figure 2) was distributed preferentially in duplicated blocks. These 41 genes be considered as candidates of the most recent Glycine-specific WGD and used in the next analysis. Second, close phylogenetic relationships have been shown among all candidate GmJMJs (Figure S2). Third, duplication types of all GmJMJs have been obtained by MCscanX programs (Wang et al., 2012) (Table S5). Fourth, collinearity analysis (Figure 3) was carried out among each candidate duplicated gene pairs. The candidate pairs were considered as created by the recent Glycine-specific WGD duplications with at least three paralogous gene pairs along the flanking regions. In conclusion, we identified that 16 GmJMJ pairs were formed by the most recent Glycine-specific WGD (Table 2).
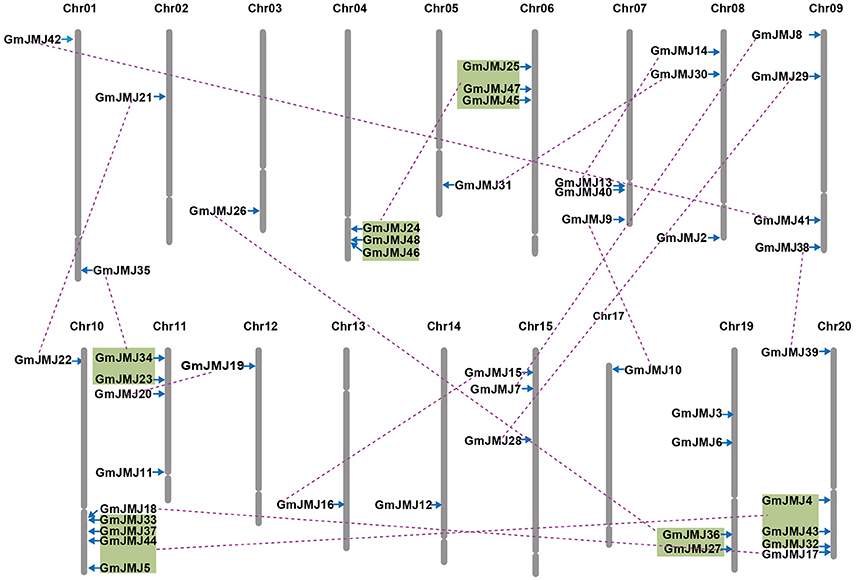
Figure 2. Chromosomal locations, region duplication, and predicted cluster for soybean JmjC genes. The chromosome number is indicated above each chromosome. Throat on the chromosomes indicate the position of centromere. Only those chromosomes bearing JmjC genes (18) were represented. Segmental duplicated genes are indicated by purple lines. Colored boxes indicate the gene blocks generated by the most recently WGD event. The figure was produced and modified using the MapChart and Adobe Illustrator, respectively.
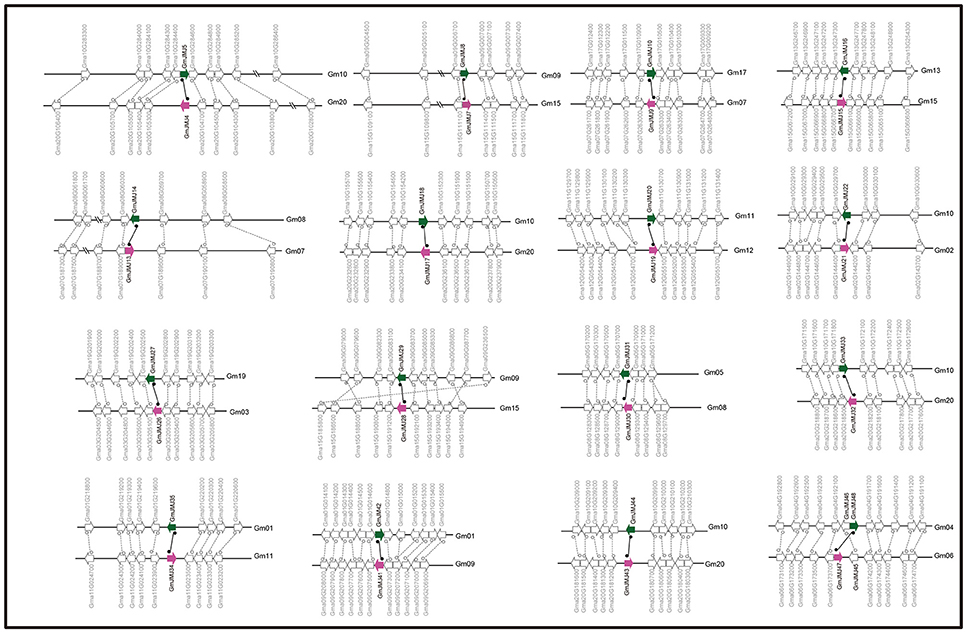
Figure 3. Collinearity analysis of duplicated gene pairs formed by Glycine-specific WGD event. Green and purple arrows represented duplicated JmjC gene pairs. The direction of the arrow indicates the position of the gene in the positive (to left) and negative (to right) chain of DNA.
Based on the divergence rate of 6.1 × 10−9 synonymous mutations per synonymous site per year which has been proposed for soybean (Lynch and Conery, 2000), among the 48 JmjCs in soybean, 73% (35 of 48) represented WGD/segmental duplication genes. Ks-value was calculated for estimating the separation time of each paralogous gene pair. All Ks-values ranged from 0.068 to 0.18, which was consisted with whole genome duplication events at round 13 Mya. In addition, our divergence time analyses showed that duplications among 16 paralogous pairs occurred between 5.6 and 15.5 Mya, with an average of 9.7 Mya (Table 2).
The history of selection acting on coding sequences can also be measured based on the ratio of non-synonymous to synonymous substitutions (Ka/Ks) (Li et al., 1981). Ka and Ks can be estimated using a number of substitution models and methods, and the estimates are sensitive to these choices and other complications such as the GC content of the sequences and their genomic context (Bustamante et al., 2002). Ka is usually much smaller than Ks, so a pair of sequences will have Ka/Ks << 1 if both sequences have been under purifying selection, Ka/Ks < 1 if one sequence has been under purifying selection but the other drifting naturally, and in rare, Ka/Ks > 1 when both sites are under positive seletion (Juretic et al., 2005). As shown in Table 2, the average Ka/Ks-values of the GmJMJ gene pairs were 0.36. Five paralog pairs have small Ka/Ks ratios (< 0.3), most Ka/Ks ratios in the range from 0.3 to 0.7, and none of them > 1.
Exon–Intron Structure and Domain Architecture of GmJmJ Genes
Structural divergence has been very prevalent in duplicate genes and, in many cases, has led to the generation of functionally distinct paralogs (Lynch and Conery, 2000). To better understand the structural diversity of the GmJMJs following duplication events, the exon/intron structures (Figure 4B) were compared using Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn/). Our analysis clearly revealed that most of the paralogs share a similar gene structure. For example, 12 gene pairs (GmJMJ9/-10, GmJMJ13/-14, GmJMJ15/-16, GmJMJ19/-20, GmJMJ21/-22, GmJMJ28/-29, GmJMJ30/-31, GmJMJ32/-33, GmJMJ34/-35, GmJMJ41/-42, GmJMJ43/-44, and GmJMJ47/-48) were found to have highly consistent gene structures, including the numbers of exons/introns and the length of exons. However, there were some differences in intron lengths and in the 5′ UTR region, which is related to the regulation of expression. For example, GmJMJ9/-10 and GmJMJ30/-31 both present a large divergence in the length of their 5′ UTR, implying that a subtle distinction in function in the development and growth of soybean may have appeared between the two paralogs. In addition, four gene pairs (GmJMJ4/-5, GmJMJ7/-8, GmJMJ17/-18, and GmJMJ26/-27) had greater changes in their structural organization, especially in the numbers of exons.
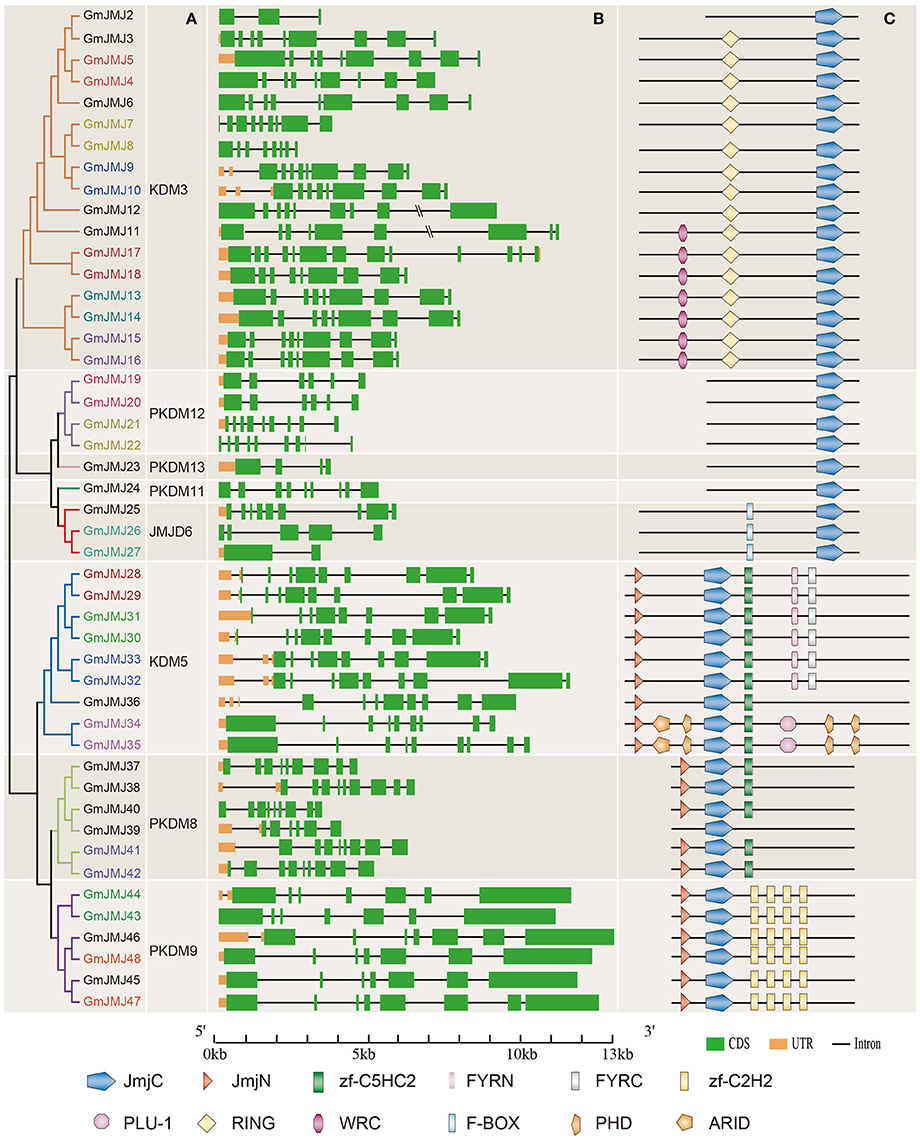
Figure 4. Phylogenetic analysis, gene structure, and domain architecture of GmJMJs. (A) Phylogenetic tree construction of GmJMJs based on the JmjC domain amino acid sequences. Name of genes marked in same color are a pair of paralogs. (B) Exon/intron structures of GmJMJs genes. The black line refers introns, the green box represents exons, and the orange box refers UTR. Over-longed introns were represented with slash–slash. The sizes of exons and introns can be estimated using the scale at the bottom. (C) The domain architecture of the full-length JmjC-domain containing proteins. JmjC, Jumonji C domain; JmjN, Jumonji N domain; PHD, plant homeobox domain; ARID, AT-rich interaction domain; zf-C2H2, Zinc finger of C2H2-type; zf-C5HC2, Zinc finger of C5HC2-type; FYRN, “FY-rich” domain N-terminal; FYRC, “FY-rich” domain C-terminal; WRC, Trp, Arg, and Cys domain; RING, (Really interesting new gene) finger domain.
We also studied the proteins encoded by the GmJMJs, using the full-length protein sequences of JmjCs as queries in CDD (Marchler-Bauer et al., 2015), SMART (Letunic et al., 2015), and Pfam (Finn et al., 2016) with default parameters in order to gain more insights into the diversity of the domain architecture, as shown in Figure 4C. These proteins all share a JmjC domain. The JmjN domain was the second most widespread domain, appearing in the majority of members of three groups, KDM5, PKDM8, and PKDM9. This domain, which is not adjacent with JmjC, was identified in the jumonji family (Balciunas and Ronne, 2000), and its interaction with the JmjC catalytic domain was found to be important for Jhd27 (also known as KDM5), a H3K4-specific demethylase in budding yeast (Huang et al., 2010; Quan et al., 2011). In PKDM9, the zf-C2H2 domain, which contains two cysteines and two histidines that coordinate a zinc atom to create a compact nucleic acid-binding domain (Chrispeels et al., 2000), was found in four tandem repeats. Furthermore, another zinc-finger domain, zf-C5HC2, was identified in PKDM8 and KDM5. Three groups, PKDM11, PKDM12 and PKDM13, all have only one domain (JmjC) in their full-length sequence, and can be grouped together as “JmjC domain-only proteins,” in keeping with previous studies (Klose et al., 2006; Lu et al., 2008; Huang et al., 2016). Two-thirds of the members of KDM5 have FYRN and FYRC domains, which may harbor chromatin-binding activity (Lu et al., 2008) or contribute to JmjC function by interacting with other proteins. For example, it has been reported that the functional specificity of AtJMJ14 in flowering time control is based on the specificity of its interaction with transcription factors through the FYRC domain (Ning et al., 2015). Two GmJMJ proteins, GmJMJ34 and GmJMJ35, have the ARID domain (AT-rich interaction domain), which has been implicated in sequence-specific DNA binding (Gregory et al., 1996).
Strangely, we found that not only the gene pairs which share a similar gene structure but also the four pairs which had greater differences in gene structural organization all had consistent domain architectures. For example, GmJMJ26/-27 share a relatively consistent functional domain in the full-length protein sequences. This implies that although the gene structure of JmjC family may change through evolution, their protein structures and functions were conserved.
Conserved Amino Acid Residues in Active Sites of GmJmJ
Fe(II) iron and α-KG are needed as cofactors by JmjC-domain proteins to carry out their demethylase activity (Chen et al., 2006; Huang et al., 2016). A total of five amino acid residues are needed to bind these cofactors; three residues (His188, Glu/Asp190, and His276) bind to the Fe (II) cofactor and two other residues (Thr/Phe185 and Lys206) bind to α-KG. With the aim of clarifying whether the conserved residues interacting with cofactors had diverged among GmJMJs, we aligned the domain sequences of JmjC proteins from soybean and Arabidopsis.
Based on the alignments, we grouped these proteins into two groups according to amino acids at the conserved sites. The first group, which includes PKDM8, PKDM9, and KDM5, has the conserved amino acids His (H), Glu (E), and His (H) for Fe(II) binding, and Phe (F) and Lys (K) for α-KG binding (Figure 5A), while the second group, which includes JMJD6, PKDM13, PKDM11, PKDM12, and KDM3, has conserved the residues His (H), Asp (D), and His (H) for Fe(II) and Thr (T) and Lys (K) for α-KG (Figure 5B). Both forms are compatible with histone demethylation activity (Lu et al., 2008). In general, most members have conserved residues for interacting with cofactors, although there are some exceptions. For example, a substitution can be seen in the first sites in PKDM12, where Thr (T) was changed into Ser (S) in GmJMJ21/22 and AtJMJ31. However, Thr and Ser have similar physical and chemical properties, so the ability to bind to cofactors may not have changed despite the presence of a different amino acid. Furthermore, the first site to interact with α-KG is absent in JMJD6, which is consistent with findings in rice (Lu et al., 2008). The detection of this change in all three plants, soybean, rice, and Arabidopsis, suggests that it may have occurred in the ancestor of these plants and is necessary for their common function. Overall, the high conservation in the interaction sites implies a significant role for these sites in the demethylase activity of the JmjC gene family.

Figure 5. Two alignment groups (A,B) of JmjC domain sequences. (Only the representative sequences were shown, other sequences can be seen in Figure S3). The conserved residues compatible with the demethylation activity within the Fe(II) binding site are highlighted in red and those in the αKG binding site are indicated in yellow. The sequences with black, gray, light gray background indicated identical 100%, conservative (75–99%), and block (50–74%) similarity of amino acid residues, respectively.
Expression Profiles of JmjC Genes in Soybean
To investigate the tissue-specific expression profiles of GmJMJs, transcriptome data (Shen et al., 2014b) were studied in 10 tissues at different developmental stages including roots, cotyledons, stems, shoot meristems, leaf buds, leaves, flowers, pods, pod and seeds, and seeds. As indicated in Figure 6, the expression patterns of the GmJMJs can be divided into three clusters, C1–C3. C1 genes show hardly any expression in almost all of the tissues except certain expression in flower, implying that these genes may possess certain specific function in flower after WGD events. C2 can be divided into two subgroups, C2-Sub1 and C2-Sub2, according to their expression levels, with lower expression levels in C2-Sub1 than C2-Sub2. Genes in C3 show a low expression level in all tissues investigated. We found that genes, which belong to the same clade in the phylogenetic tree (Figure 1A), were sometimes dispersed in different clusters based on expression (Figure 6). For example, the four genes in C1 (GmJMJ37, GmJMJ38, GmJMJ39, and GmJMJ40) grouped together with GmJMJ41 and GmJMJ42 in a clade, indicating that they may have been produced through evolution after WGD events, but clustered into two clusters and show different expression patterns, implying they have acquired different functions after the duplication event.
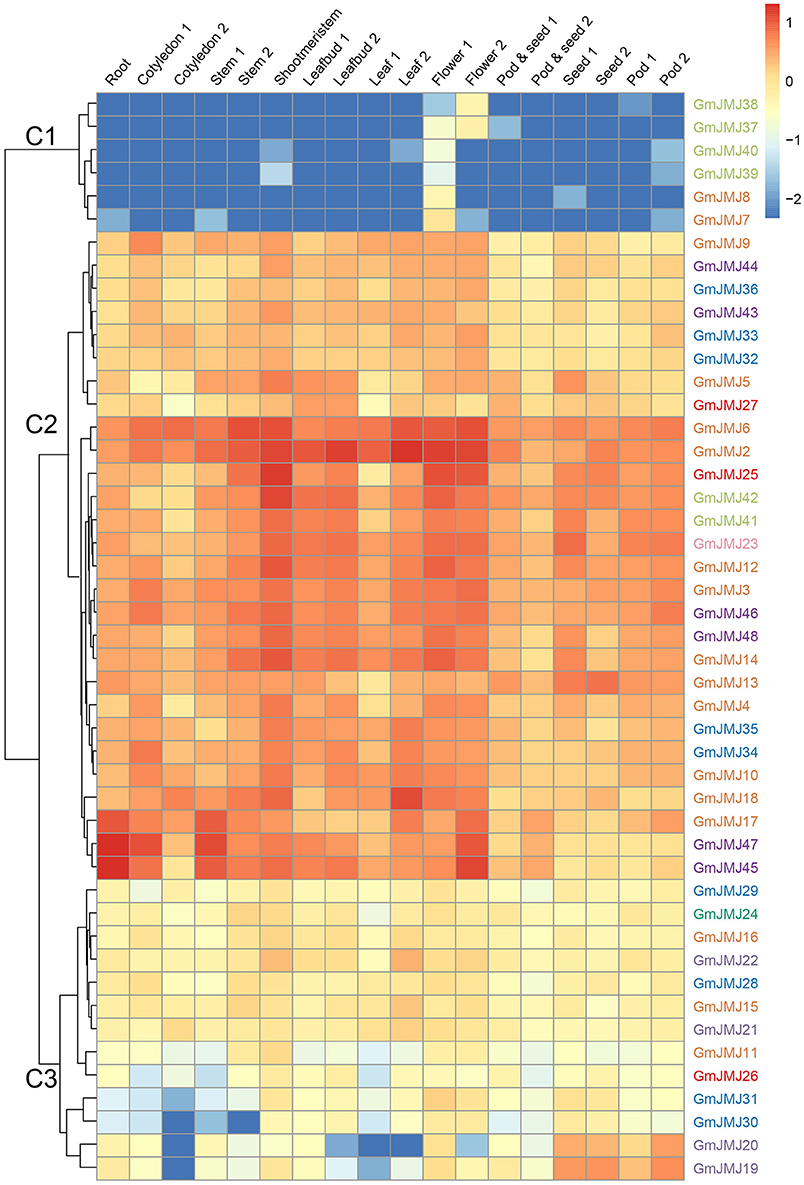
Figure 6. Heatmaps representing the expression profiles of GmJMJs in several tissues. The numbers following the tissues indicated different developmental stages. The color scale on the right indicates expression values, blue indicating low transcript abundance, and red indicating high levels of transcript abundance.
In addition, we determined the expression profiles of the recently duplicated JmjC gene pairs in 10 tissues. Most of the paralogs generally have the same expression pattern. For further analysis, we divided the duplicated genes into three types based on their detailed expression patterns, shown with a blue, green, and red box in Figure 7. The 12 paralogs in the blue box all have a relatively low expression level in four tissues (pods, podseeds, roots, and seeds) compared with other tissues. These paralogs also show a complex expression pattern in another six tissues examined, but almost all have high expression in the flower, leaf, and shoot meristem. For example, the two copies GmJMJ4 and GmJMJ5 both have high expression in flowers, leaf buds and shoot meristems. The expression pattern of these genes can therefore be pictured as similar to the shape of the letter “M.” The three paralogs in the green box only show high expression in one organ, such as the flower, seed, and root. For instance, GmJMJ7/-8 and GmJMJ41-42 both have a high expression level in flower and roots, respectively. The third box contains only one paralog pair (GmJMJ26 and GmJMJ27), and the expression level of one copy (GmJMJ27) is higher than the other copy (GmJMJ26) in all tissues. Overall, the paralogs show conserved expression profiles, demonstrating that the JmjC gene family has conserved its functions through duplication events.
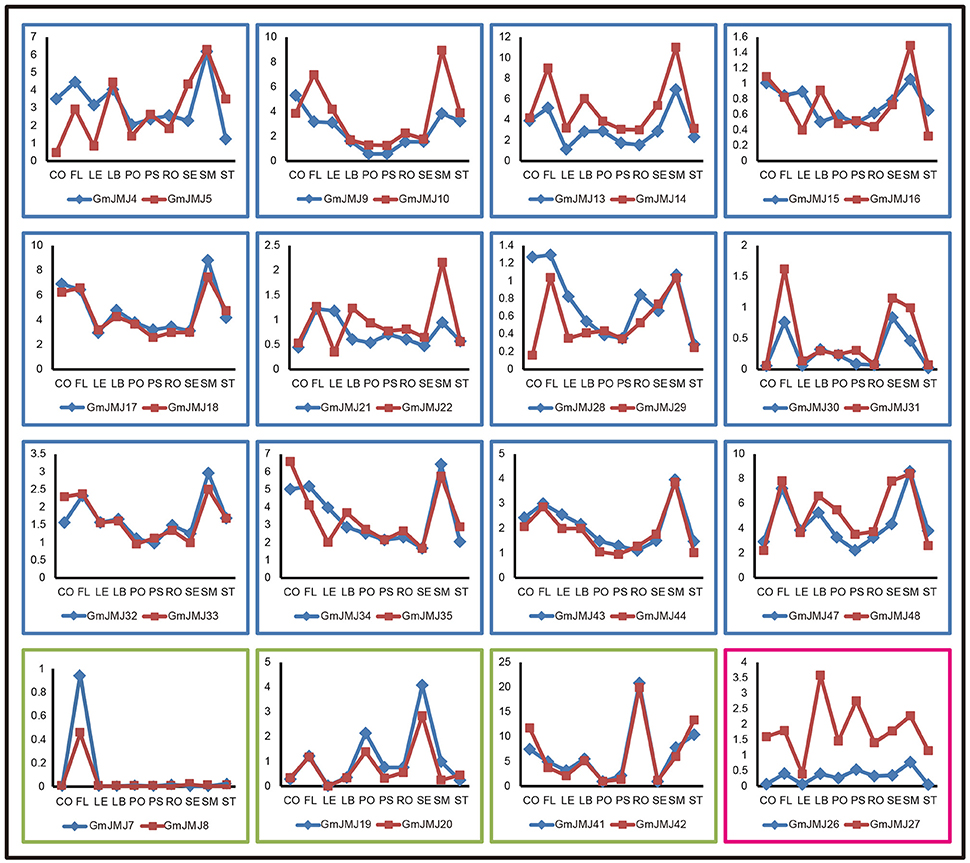
Figure 7. Three trends of expression patterns of duplicated GmJMJ gene pairs. X-axis indicates representative tissues and Y-axis represents scale. The sample used are Cotyledon1(CO), Flower1 (FL), Leaf1(LE), LeafBud1(LB), Pod1(PO), Pod and Seed1(PS), Root1(RO), Seed1(SE), ShootMeristem(SM), Stem1(ST).
Discussion
As histone demethylases, JmjC domain-containing proteins play essential roles in histone modification, which is a significant part of epigenetics (Klose et al., 2006; Chen et al., 2011). To date, many efforts on the JmjC gene family have been undertaken to elucidate their evolutionary history in a wide variety of plant species, such as Arabidopsis (Lu et al., 2008; Zhao et al., 2015), rice (Lu et al., 2008; Zong et al., 2013), and Fragaria vesca (Gu et al., 2016). However, little is known about the JmjC gene family in soybean. In this study, we performed a comprehensive analysis of GmJMJs, including their phylogenetic relationships, gene structure, domain architecture, chromosome location, duplication patterns, and expression profiles.
Phylogeny and Domain Architectures of JmjCs in Soybean
In total, 48 JmjC genes were identified in the soybean genome, which is larger than other model plants or the other two legumes examined. The number of JmjCs from each species in each group is summarized in Table 1. In most groups, there is still a larger number of JmjCs from soybean than any other species, indicating that these groups may have different evolutionary history among the five species. PKDM11 and PKDM13, two exception groups, both of which including a single gene from each species, may have no duplication or loss after divergence from Arabidopsis. The phylogenetic analysis of JmjC genes among five plant species showed that each group contains JmjCs from all species investigated, four eudicots and one monocot, revealing that the JmjC family may have already existed before the divergence of these two lineages. And combining the phylogeny with the time estimation, 16 GmJMJs pairs, formed by the most recent WGD of soybean, were identified.
Although many GmJMJ genes were produced by WGD events, there has been little differentiation in their gene structure, domain architecture and the conserved residues acting with cofactors. Another interesting phenomenon is that even outside the conserved coactive-sites we found GmJMJs to have higher amino acid similarity with AtJMJs from the same sub-cluster rather than with GmJMJs from other sub-clusters even though these genes were all grouped in one clade. For example, in PKDM8, GmJMJ41 and GmJMJ42 have the same residues as AtJMJ13 around the Lys (K) site, a Cys (C) and Ser (S), whereas GmJMJ37/-38/-39/-40 have different residues. The unconserved sites around the coactive-sites indicate that the JmjCs may use different ways to bind to the co-factors. However, further evidence is needed to demonstrate and understand this mechanism.
WGDs Contributed to JmjC Gene Expansion in Glycine Max
WGD/segmental duplication and tandem duplication might lead to duplicated gene pairs on the DNA level. Previous studies have shown that the soybean genome has undergone two rounds of WGD (Schmutz et al., 2010). In this study, we have demonstrated that 16 soybean paralogous pairs derived from the second WGD, which suggests that the WGD duplication might be the main mechanism of JmjC gene family expansion and functional diversity during the evolution of soybean. This result is consistent with some other gene families not only AAT gene family (Cheng et al., 2016), GST supergene family (Liu et al., 2015), and receptor-like kinase genes (Zhou et al., 2016) in soybean, but also SET domain family in Populus trichocar (Lei et al., 2012; Zhang and Ma, 2012), and 14-3-3 family genes and AP2/ERF superfamily in M. truncatula (Qin et al., 2016; Shu et al., 2016). And in Arabidopsis, previous studies have proposed that more than 90% regulatory genes increased due to WGD (Maere et al., 2005). However, the dispersed duplications and retro-transpositions played the most important role in the evolution of JmjC genes in F. vesca (Gu et al., 2016). Furthermore, KDM3 group is preferentially expanded in the soybean genome compared to other groups, consistent with F. vesca (Gu et al., 2016), indicating that KDM3 group genes may have evolved to meet some unique regulatory needs.
Expression Profiles of JmjC Genes and Functional Diversity of Duplicated Pairs in Soybean
In angiosperms and vertebrates, both of the JmjC and SET genes, maintaining homeostasis of the histones methylation, are the key regulators of chromatin structure, suggesting that the epigenetic modulation playing an important role in regulation of gene expression in developmental stages and responses to abiotic stresses (Lei et al., 2012; Zhang and Ma, 2012; Qian et al., 2015). We investigated the expression profiles of JmjC genes using public expression data and found that most JmjC genes are widely expressed (Figure 6), indicating that these genes, remained after WGD/segmental events, are likely functional. To further elucidate whether functional differentiation has occurred after the WGD event, we analyzed the expression patterns between the duplicated pairs (Figure 7). All the expression patterns of 16 duplicated gene pairs can be classed into three types according to their tendency in each tissues detected. The first type is that two copies have the complicated expression patterns in different tissues. We could hypothesize possible functions of GmJMJs by coupled their expression patterns with the functions of their Arabidopsis orthologs. For example, AtJMJ24, the ortholog of GmJMJ17 and GmJMJ18 in Arabidopsis, is a histone H3K9 demethylase (Lu et al., 2008). AtJMJ24 has been proved to promote basal level transcription of endogenous silenced loci by counteracting H3K9me (Deng et al., 2015). Therefore, GmJMJ17 and GmJMJ18 might have the functions similar to AtJMJ24 in reinforcing the silence. The second type showed that both copies have high expression in one organ, such as the flower, seed, and root. The third type is that one duplicate was expressed at higher levels than the other one nearly in all tissues, implying that the former one has stronger function than the latter, and implying that it may play important roles in regulating broad developmental or reproductive stages. Above all, the expression patterns among the duplicated pairs are relatively conserved, suggesting little functional differentiation has occurred following the WGD event. However, some JmjC genes were specific to soybean, for example, GmJMJ4 and GmJMJ5. All of these genes have abundant transcripts in soybean and are expressed at different levels in different tissues. These results indicate that in Arabidopsis their counterparts may be lost and the functions might have been performed by other genes.
Conclusions
Here, we performed comprehensive and evolutionary analyses of JmjC gene family in soybean, and provided detailed information on its members. A total of 48 putative JmjC genes were identified in the soybean genome, which represented non-random across all soybean genome chromosomes and majority of them expanded from WGD/segmental duplication rather than the dispersed duplications. The exon/intron compositions and domain arrangements were considerably conserved among members in the same groups or subgroups. Many duplicated genes present similar expression patterns in soybean tissues detected implying functional conservation. The close phylogenetic relationship between GmJMJs and AtJMJs in the same subgroup provided insights into their putative functions. Taken together, all of these results provided valuable clues in future efforts to identify specific gene functions of this gene family and gene diversity among different genotype of soybean and other plants in Leguminosae.
Author Contributions
LW and LZ designed the research; YH, XL, and LW performed phylogenetic analysis and wrote the manuscript; LC, YL, and HW annotated the JmjC genes on chromosomes and calculated the duplication date; LC, DK, and HY analyzed the expression data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the foundation and frontier technology research of Henan Province (No. 162300410257), the industry-university-research cooperation of Henan Province (No. 162107000032), Funding scheme for young core teachers of Xinyang Normal University (2015), Nanhu Scholars Program for Young Scholars of XYNU, and Funding scheme for young backbone teachers of Xinyang Normal University (2016GGJS-13). This work was also supported by the National Natural Science Funds of China (No. 31400213). Fujian-Taiwan Joint Innovative Center for Germplasm Resources and Cultivation of Crop [Fujian 2011 Program, (2015)75] to LZ.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01800/full#supplementary-material
Figure S1. Phylogenetic relationship of JmjC-domain containing proteins from five plant species by using the JmjC domain alone.
Figure S2. Phylogenetic relationships of GmJMJs.
Figure S3. Alignment of JmjC domain sequences. The conserved residues compatible with the demethylation activity within the Fe(II) binding site are highlighted in red and those in the αKG binding site are indicated in yellow. The sequences with black, gray, light gray background indicated identical 100%, conservative (75–99%), and block (50–74%) similarity of amino acid residues, respectively.
Table S1. Protein sequences of Arabidopsis, Oryza sativa JmjC domain-containing protein used as blast queries.
Table S2. The domain sequences used in the phylogenetic trees.
Table S3. The expression data of GmJmjC genes in various organs.
Table S4. Detailed information of soybean JmjC family genes.
Table S5. The duplication types of GmJmjC genes calculating by MCScanX program.
References
Ahmad, A., and Cao, X. (2012). Plant PRMTs broaden the scope of arginine methylation. J. Genet. Genomics 39, 195–208. doi: 10.1016/j.jgg.2012.04.001
Allis, C. D., Berger, S. L., Cote, J., Dent, S., Jenuwien, T., Kouzarides, T., et al. (2007). New nomenclature for chromatin-modifying enzymes. Cell 131, 633–636. doi: 10.1016/j.cell.2007.10.039
Balciunas, D., and Ronne, H. (2000). Evidence of domain swapping within the jumonji family of transcription factors. Trends Biochem. Sci. 25, 274–276. doi: 10.1016/S0968-0004(00)01593-0
Bustamante, C. D., Nielsen, R., and Hartl, D. L. (2002). A maximum likelihood method for analyzing pseudogene evolution: Implications for silent site evolution in humans and rodents. Mol. Biol. Evol. 19, 110–117.
Chen, X., Hu, Y., and Zhou, D. X. (2011). Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim. Biophys. Acta 1809, 421–426. doi: 10.1016/j.bbagrm.2011.03.004
Chen, Z., Zang, J., Whetstine, J., Hong, X., Davrazou, F., Kutateladze, T. G., et al. (2006). Structural insights into histone demethylation by JMJD2 family members. Cell 125, 691–702. doi: 10.1016/j.cell.2006.04.024
Cheng, L., Yuan, H. Y., Ren, R., Zhao, S. Q., Han, Y. P., Zhou, Q. Y., et al. (2016). Genome-wide identification, classification, and expression analysis of amino acid transporter gene family in Glycine max. Front. Plant Sci. 7:515. doi: 10.3389/fpls.2016.00515
Chrispeels, H. E., Oettinger, H., Janvier, N., and Tague, B. W. (2000). AtZFP1, encoding Arabidopsis thaliana C2H2 zinc-finger protein 1, is expressed downstream of photomorphogenic activation. Plant Mol. Biol. 42, 279–290. doi: 10.1023/A:1006352809700
Deng, S., Xu, J., Liu, J., Kim, S. H., Shi, S., and Chua, N. H. (2015). JMJ24 binds to RDR2 and is required for the basal level transcription of silenced loci in Arabidopsis. Plant J. 83, 770–782. doi: 10.1111/tpj.12924
Elkins, J. M., Hewitson, K. S., McNeill, L. A., Seibel, J. F., Schlemminger, I., Pugh, C. W., et al. (2003). Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1 alpha. J. Biol. Chem. 278, 1802–1806. doi: 10.1074/jbc.C200644200
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Grant, D., Nelson, R. T., Cannon, S. B., and Shoemaker, R. C. (2010). SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 38, D843–D846. doi: 10.1093/nar/gkp798
Gregory, S. L., Kortschak, R. D., Kalionis, B., and Saint, R. (1996). Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol. Cell. Biol. 16, 792–799. doi: 10.1128/MCB.16.3.792
Gu, T., Han, Y., Huang, R., McAvoy, R. J., and Li, Y. (2016). Identification and characterization of histone lysine methylation modifiers in Fragaria vesca. Sci. Rep. 6:23581. doi: 10.1038/srep23581
Huang, F., Chandrasekharan, M. B., Chen, Y.-C., Bhaskara, S., Hiebert, S. W., and Sun, Z.-W. (2010). The JmjN domain of Jhd2 is important for its protein stability, and the plant homeodomain (PHD) finger mediates its chromatin association independent of H3K4 methylation. J. Biol. Chem. 285, 24548–24561. doi: 10.1074/jbc.M110.117333
Huang, Y., Chen, D., Liu, C., Shen, W., and Ruan, Y. (2016). Evolution and conservation of JmjC domain proteins in the green lineage. Mol. Genet. Genomics 291, 33–49. doi: 10.1007/s00438-015-1089-4
Jones, M. A., and Harmer, S. (2011). JMJD5 functions in concert with TOC1 in the arabidopsis circadian system. Plant Signal. Behav. 6, 445–448. doi: 10.4161/psb.6.3.14654
Juretic, N., Hoen, D. R., Huynh, M. L., Harrison, P. M., and Bureau, T. E. (2005). The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 15, 1292–1297. doi: 10.1101/gr.4064205
Klose, R. J., Kallin, E. M., and Zhang, Y. (2006). JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727. doi: 10.1038/nrg1945
Klose, R. J., and Zhang, Y. (2007). Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 8, 307–318. doi: 10.1038/nrm2143
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi: 10.1016/j.cell.2007.02.005
Lee, M. G., Wynder, C., Cooch, N., and Shiekhattar, R. (2005). An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437, 432–435. doi: 10.1038/nature04021
Lei, L., Zhou, S. L., Ma, H., and Zhang, L. S. (2012). Expansion and diversification of the SET domain gene family following whole-genome duplications in Populus trichocarpa. BMC Evol. Biol. 12:51. doi: 10.1186/1471-2148-12-51
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Li, J., Yu, C., Wu, H., Luo, Z., Ouyang, B., Cui, L., et al. (2015). Knockdown of a JmjC domain-containing gene JMJ524 confers altered gibberellin responses by transcriptional regulation of GRAS protein lacking the DELLA domain genes in tomato. J. Exp. Bot. 66, 1413–1426. doi: 10.1093/jxb/eru493
Li, T., Chen, X., Zhong, X., Zhao, Y., Liu, X., Zhou, S., et al. (2013). Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 25, 4725–4736. doi: 10.1105/tpc.113.118802
Li, W.-H., Gojobori, T., and Nei, M. (1981). Pseudogenes as a paradigm of neutral evolution. Nature 292, 237–239. doi: 10.1038/292237a0
Liu, C., Lu, F., Cui, X., and Cao, X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61, 395–420. doi: 10.1146/annurev.arplant.043008.091939
Liu, H.-J., Tang, Z.-X., Han, X.-M., Yang, Z.-L., Zhang, F.-M., Yang, H.-L., et al. (2015). Divergence in enzymatic activities in the soybean gst supergene family provides new insight into the evolutionary dynamics of whole-genome duplicates. Mol. Biol. Evol. 32, 2844–2859. doi: 10.1093/molbev/msv156
Lu, F., Cui, X., Zhang, S., Jenuwein, T., and Cao, X. (2011a). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43, 715–719. doi: 10.1038/ng.854
Lu, F., Cui, X., Zhang, S., Liu, C., and Cao, X. (2010). JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res. 20, 387–390. doi: 10.1038/cr.2010.27
Lu, F., Li, G., Cui, X., Liu, C., Wang, X. J., and Cao, X. (2008). Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in arabidopsis and rice. J. Integr. Plant Biol. 50, 886–896. doi: 10.1111/j.1744-7909.2008.00692.x
Lu, S. X., Knowles, S. M., Webb, C. J., Celaya, R. B., Cha, C., Siu, J. P., et al. (2011b). The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 155, 906–915. doi: 10.1104/pp.110.167015
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Maere, S., De, Bodt, S., Raes, J., Casneuf, T., Van, Montagu,. M., Kuiper, M., et al. (2005). Modeling gene and genome duplications in Eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 5454–5459. doi: 10.1073/pnas.0501102102
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Metzger, E., Wissmann, M., Yin, N., Müller, J. M., Schneider, R., Peters, A. H., et al. (2005). LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439. doi: 10.1038/nature04020
Michael, T. P., Mockler, T. C., Breton, G., McEntee, C., Byer, A., Trout, J. D., et al. (2008). Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 4:e14. doi: 10.1371/journal.pgen.0040014
Mockler, T. C., Michael, T. P., Priest, H. D., Shen, R., Sullivan, C. M., Givan, S. A., et al. (2007). The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 72, 353–363. doi: 10.1101/sqb.2007.72.006
Ning, Y.-Q., Ma, Z.-Y., Huang, H.-W., Mo, H., Zhao, T.-T., Li, L., et al. (2015). Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids Res. 43, 1469–1484. doi: 10.1093/nar/gku1382
Noh, B., Lee, S.-H., Kim, H.-J., Yi, G., Shin, E.-A., Lee, M., et al. (2004). Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16, 2601–2613. doi: 10.1105/tpc.104.025353
Qian, S., Wang, Y., Ma, H., and Zhang, L. (2015). Expansion and functional divergence of Jumonji C-containing histone demethylases: significance of duplications in ancestral angiosperms and vertebrates. Plant Physiol. 168, 1321–1337. doi: 10.1104/pp.15.00520
Qin, C., Cheng, L. M., Shen, J., Zhang, Y., Cao, H., Lu, D., et al. (2016). Genome-wide identification and expression analysis of the 14-3-3 family genes in Medicago truncatula. Front. Plant Sci. 7:320. doi: 10.3389/fpls.2016.00320
Quan, Z., Oliver, S. G., and Zhang, N. (2011). JmjN interacts with JmjC to ensure selective proteolysis of Gis1 by the proteasome. Microbiology 157, 2694–2701. doi: 10.1099/mic.0.048199-0
Schlueter, J. A., Dixon, P., Granger, C., Grant, D., Clark, L., Doyle, J. J., et al. (2004). Mining EST databases to resolve evolutionary events in major crop species. Genome 47, 868–876. doi: 10.1139/g04-047
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Severin, A. J., Cannon, S. B., Graham, M. M., Grant, D., and Shoemaker, R. C. (2011). Changes in twelve homoeologous genomic regions in soybean following three rounds of polyploidy. Plant Cell 23, 3129–3136. doi: 10.1105/tpc.111.089573
Shen, Y., Silva, N., Audonnet, L., Servet, C., Wei, W., and Zhou, D.-X. (2014a). Over-expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in Arabidopsis. Front. Plant Sci. 5:290. doi: 10.3389/fpls.2014.00290
Shen, Y., Wu, X., Liu, D., Song, S., Liu, D., and Wang, H. (2016). Cold-dependent alternative splicing of a Jumonji C domain-containing gene MtJMJC5 in Medicago truncatula. Biochem. Biophys. Res. Commun. 474, 271–276. doi: 10.1016/j.bbrc.2016.04.062
Shen, Y., Zhou, Z., Wang, Z., Li, W., Fang, C., Wu, M., et al. (2014b). Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26, 996–1008. doi: 10.1105/tpc.114.122739
Shu, Y., Liu, Y., Zhang, J., Song, L., and Guo, C. (2016). Genome-wide analysis of the AP2/ERF superfamily genes and their responses to abiotic stress in Medicago truncatula. Front. Plant Sci. 6:1247. doi: 10.3389/fpls.2015.01247
Singh, V. K., and Jain, M. (2015). Genome-wide survey and comprehensive expression profiling of Aux/IAA gene family in chickpea and soybean. Front. Plant Sci. 6:918. doi: 10.3389/fpls.2015.00918
Sun, Q., and Zhou, D.-X. (2008). Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc. Natl. Acad. Sci. U.S.A. 105, 13679–13684. doi: 10.1073/pnas.0805901105
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Trewick, S. C., McLaughlin, P. J., and Allshire, R. C. (2005). Methylation: lost in hydroxylation? EMBO Rep. 6, 315–320. doi: 10.1038/sj.embor.7400379
Vanneste, K., Baele, G., Maere, S., and Van de Peer, Y. (2014). Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res. 24, 1334–1347. doi: 10.1101/gr.168997.113
Wang, X., Duan, C.-G., Tang, K., Wang, B., Zhang, H., Lei, M., et al. (2013). RNA-binding protein regulates plant DNA methylation by controlling mRNA processing at the intronic heterochromatin-containing gene IBM1. Proc. Natl. Acad. Sci. U.S.A. 110, 15467–15472. doi: 10.1073/pnas.1315399110
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Yang, H., Mo, H., Fan, D., Cao, Y., Cui, S., and Ma, L. (2012). Overexpression of a histone H3K4 demethylase, JMJ15, accelerates flowering time in Arabidopsis. Plant Cell Rep. 31, 1297–1308. doi: 10.1007/s00299-012-1249-5
Yang, W., Jiang, D., Jiang, J., and He, Y. (2010). A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J. 62, 663–673. doi: 10.1111/j.1365-313X.2010.04182.x
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yu, X., Li, L., Li, L., Guo, M., Chory, J., and Yin, Y. (2008). Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105, 7618–7623. doi: 10.1073/pnas.0802254105
Zhang, L., and Ma, H. (2012). Complex evolutionary history and diverse domain organization of SET proteins suggest divergent regulatory interactions. New Phytol. 195, 248–263. doi: 10.1111/j.1469-8137.2012.04143.x
Zhao, W., Shafiq, S., Berr, A., and Shen, W. H. (2015). Genome-wide gene expression profiling to investigate molecular phenotypes of Arabidopsis mutants deprived in distinct histone methyltransferases and demethylases. Genom. Data 4, 143–145. doi: 10.1016/j.gdata.2015.04.006
Zhou, F., Guo, Y., and Qiu, L. J. (2016). Genome-wide identification and evolutionary analysis of leucine-rich repeat receptor-like protein kinase genes in soybean. BMC Plant Biol. 16:58. doi: 10.1186/s12870-016-0744-1
Keywords: soybean (Glycine max L.), JmjC gene family, genome-wide analysis, phylogeny, gene structure, expression pattern
Citation: Han Y, Li X, Cheng L, Liu Y, Wang H, Ke D, Yuan H, Zhang L and Wang L (2016) Genome-Wide Analysis of Soybean JmjC Domain-Containing Proteins Suggests Evolutionary Conservation Following Whole-Genome Duplication. Front. Plant Sci. 7:1800. doi: 10.3389/fpls.2016.01800
Received: 11 August 2016; Accepted: 15 November 2016;
Published: 05 December 2016.
Edited by:
José M. Romero, University of Seville, SpainReviewed by:
David Smyth, Monash University, AustraliaNing Zhang, Food and Drug Administration, USA
Copyright © 2016 Han, Li, Cheng, Liu, Wang, Ke, Yuan, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Wang, d2FuZ2xlaWJpb0B4eW51LmVkdS5jbg==; d2FuZ2xlaWJpb0AxMjYuY29t
Liangsheng Zhang, emxzQHRvbmdqaS5lZHUuY24=
†These authors have contributed equally to this work.
 Yapeng Han
Yapeng Han Xiangyong Li1,2†
Xiangyong Li1,2† Danxia Ke
Danxia Ke Liangsheng Zhang
Liangsheng Zhang Lei Wang
Lei Wang