- Rice Research Institute, Fujian Academy of Agricultural Sciences, Fujian High Quality Rice Research and Development Center, Fuzhou, China
Common wild rice (Oryza rufipogon Griff.) represents an important resource for rice improvement. Genetic populations provide the basis for a wide range of genetic and genomic studies. In particular, chromosome segment substitution lines (CSSLs) are most powerful tools for the detection and precise mapping of quantitative trait loci (QTLs). In this study, 146 CSSLs were produced; they were derived from the crossing and back-crossing of two rice cultivars: Dongnanihui 810 (Oryza sativa L.), an indica rice cultivar as the recipient, and ZhangPu wild rice, a wild rice cultivar as the donor. First, a physical map of the 146 CSSLs was constructed using 149 molecular markers. Based on this map, the total size of the 147 substituted segments in the population was 1145.65 Mb, or 3.04 times that of the rice genome. To further facilitate gene mapping, heterozygous chromosome segment substitution lines (HCSSLs) were also produced, which were heterozygous in the target regions. Second, a physical map of the 244 HCSSLs was produced using 149 molecular markers. Based on this map, the total length of substituted segments in the HCSSLs was 1683.75 Mb, or 4.47 times the total length of the rice genome. Third, using the 146 CSSLs, two QTLs for plant height, and one major QTL for apiculus coloration were identified. Using the two populations of HCSSLs, the qPa-6-2 gene was precisely mapped to an 88 kb region. These CSSLs and HCSSLs may, therefore, provide powerful tools for future whole genome large-scale gene discovery in wild rice, providing a foundation enabling the development of new rice varieties. This research will also facilitate fine mapping and cloning of quantitative trait genes, providing for the development of superior rice varieties.
Introduction
Given the rapid increase in world population, the next century may witness serious global food shortage problems. Consequently, the need to increase grain yield is critical. Meanwhile, rice is one of the most important crops in the world, and in order to meet the growing demand for food driven by population growth and economic development, global rice production must double by 2050 (Arbelaez et al., 2015). This signals the importance of identifying, transferring, and utilizing beneficial allele genes from wild and cultivated rice. Over the past few decades, many different population types have been used to map QTLs, but some factors have impeded the fine mapping or cloning of more QTLs. Researchers have attributed this to several factors, including insufficient population size, unstable statistical thresholds for detecting putative loci, minimal number of molecular markers for analyses, and low heritability of target traits (Yano and Sasaki, 1997). For the most part, however, limited populations prevent fine mapping or cloning.
Early temporary primary mapping populations, such as F2:3 and BC1 families, have been used for genetic analysis and mapping of target QTLs (Li et al., 1995; Ray et al., 1996; Redona and Mackill, 1996). However, these populations are difficult to maintain, and trials cannot be repeated. Therefore, in order to confirm mapping results, doubled haploid (DH) and recombinant inbred lines (RILs) have been developed. However, neither method is suitable for further analysis, such as fine mapping and cloning of target QTLs (Yano, 2001). For example, separation distortion of RIL populations can occur in groups, actual building construction groups do not necessarily represent theory.
Advanced backcross populations, and NILs have also been developed and used. For example, genetic background noise can be eliminated, and a QTL can be visualized as a single Mendelian factor. Each NIL carries either one or more donor segments in the near-isogenic background of the recurrent parent, which has distinct advantages for QTL identification. Several QTLs have been fine-mapped or cloned on the basis of NILs (Ashikari et al., 2005; Fan et al., 2006; Song et al., 2007; Shomura et al., 2008; Xue et al., 2008; Huang et al., 2009; Fujita et al., 2010; Zhou et al., 2012; Henry et al., 2015). However, since development is trying and time-consuming, many researchers do not perform map-based cloning of QTLs (Xu et al., 2010).
The development of CSSLs, as suggested by Doi et al. (1997) and Kubo et al. (2002), allows QTL detection of complex agronomical traits in plants and may well resolve the issues of precise mapping of QTLs (Li et al., 2015). Specifically, CSSLs can be used for detecting and fine mapping of QTLs as a single Mendelian factor by blocking background genetic noise. So far, several CSSLs in rice have been developed and many QTLs for traits of biological and economic interest have been detected (Kubo et al., 2002; Ebitani et al., 2005; Mei et al., 2006; Takai et al., 2007; Zhu et al., 2009; Xu et al., 2010; Chen et al., 2014; Nagata et al., 2015; Subudhi et al., 2015). These achievements have undoubtedly enhanced the understanding of complex traits and promoted plant genomic studies.
However, rice breeding still faces the problem of yield plateaus and weak resistance, caused by the narrow genetic basis of parental materials (Tanksley and McCouch, 1997; Tian et al., 2006). Sun et al. (2002) compared the genetic diversity between common wild rice and cultivated rice, and the results showed that the number of alleles of cultivated rice was only 60% that of wild rice, indicating that many alleles were lost during the course of domestication from wild to cultivated rice. Therefore, exploitation and utilization of favorable alleles of wild rice previously lost in cultivated rice might overcome the yield plateaus.
The ZhangPu wild rice was found in Zhangpu County in 1982, and it was the only wild rice in Fujian province. However, since it is the easternmost distribution of wild rice in China, it may contain many elite genes, including those for disease resistance, high-yield and resistance to stresses (Li, 2010). This wild rice may contain many genes of use to breeders, such as novel resistance genes to biotic or abiotic stresses, because it was not exposed to selective pressure for all of these traits in its native environment (Furuta et al., 2014). Therefore, systematic and further research on ZhangPu wild rice is needed in order to discover favorable gene resources and enhance our understanding of the molecular basis. Dongnanihui 810 (Oryza sativa L.), an indica rice cultivar, is an excellent restorer. Ganyou 810, derived from the CMS line Ganxiang A and a restorer line Dongnanihui 810, was certified and released for commercial production in 2014 in Fujian province1.
In this study, we described the development of a novel population of CSSLs in rice. The population was derived from the crossing and back-crossing of two rice cultivars: Dongnanihui 810, an indica cultivar as the recipient and ZhangPu wild rice, as the donor. Meanwhile a physical map of 146 CSSLs and the 244 HCSSLs was produced on the basis of estimates of lengths and locations using 149 molecular markers. Using the bin map converted from the ultrahigh-quality physical map associated with the purple apiculus of the 146 CSSLs, the qPa-6-2 gene was precisely mapped to an 88 kb region that contained the OsC1 gene. This research will facilitate fine mapping and cloning of quantitative trait genes, leading to the development of superior rice varieties. Furthermore, it will be possible to illustrate the genetic mechanisms of complex traits in plant functional genomics.
Materials and Methods
Plant Materials
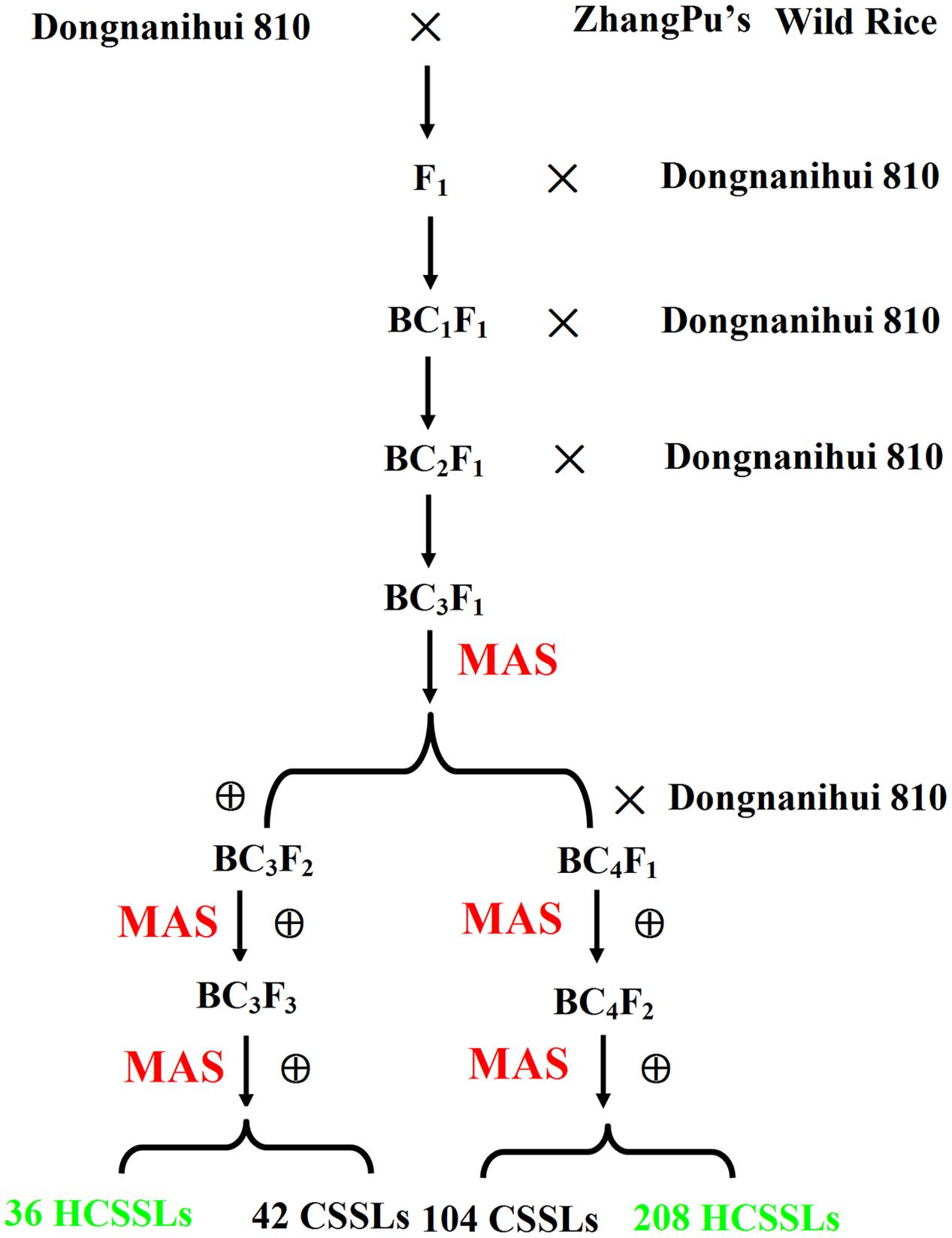
To develop CSSLs and HCSSLs, Dongnanihui 810, a restorer indica cultivar, was used as the recipient, and ZhangPu wild rice was used as the donor. The F1 plants were generated from Dongnanihui 810 as female and ZhangPu wild rice as male. The F1 plants were back-crossed with Dongnanihui 810 to produce the BC1F1 generation. These BC1F1 plants were backcrossed to Dongnanihui 810 to produce BC2F1. In the same way, 156 BC3F1 individuals were obtained. MAS with a whole-genome survey of 156 BC3F1 lines, which were selected at random by taking three from each line, identified 213 plants in which most genomic regions were homozygous for Dongnanihui 810 alleles. However, more than four heterozygous substituted segments from ZhangPu wild rice were excluded.
The Choice of Polymorphism Markers
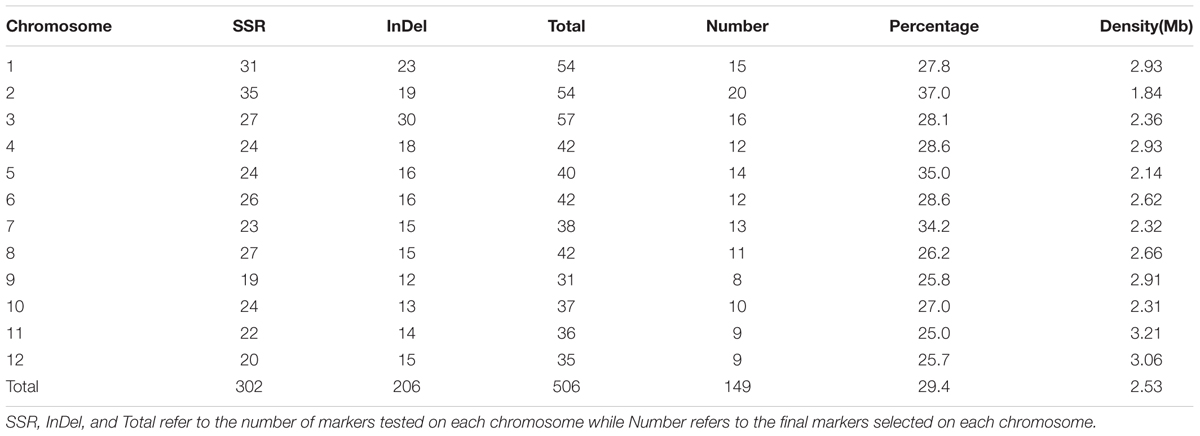
A lot of 302 SSR markers were selected from dense rice microsatellite maps (McCouch et al., 2002), and 206 InDel markers were developed using Primer Premier 5.0 software according to the publicly available rice genome sequence comparisons between Nipponbare and 93112. Of the 506 markers, 149 (29.4%) displayed better polymorphisms between the two parents.
PCR Amplification and Marker Detection
Plant DNA was extracted from the frozen leaves of rice plants using the CTAB method (Murray and Thompson, 1980) with minor modifications. The extracted DNA was dissolved in ddH2O. DNA amplification was performed by PCR with the following parameters: 5 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 60°C (for Indel) or 55°C (for SSR), and 50s at 72°C, with a final extension of 10 min at 72°C. For PCR amplification of markers, each 20 μL reaction mixture contained 50 ng DNA, 5 μmol of each primer, 10× PCR buffer [100 mM Tris (pH 8.3), 500 mM KCl, 15 mM MgCl2, 2 μg gelatin], 250 μM of each dNTP and 0.5 U of Taq polymerase. Amplified PCR products were separated through electrophoresis on a 6% non-denaturing polyacrylamide gel, and bands were revealed using a silver staining procedure.
Determination of the Length of Substituted Segment in CSSLs
The substituted segment was determined based on its location on the rice microsatellite map (McCouch et al., 2002). The length of substituted segment in CSSLs was estimated based on graphical genotypes (Young and Tanksley, 1989; Zhu et al., 2009). A chromosome segment flanked by two markers of donor genotype (DD) was considered to have a 100% donor genotype, while a chromosome segment flanked by two markers of recipient genotype (RR) was considered to be 0% donor genotype. A chromosome segment flanked by one marker of donor type and one marker of recipient type (DR) was considered as 50% donor genotype. Therefore, the length of DD plus the length of two half DR was considered to be the estimated length of a substituted chromosome segment.
Identification and Substitution Mapping of QTLs for Purple Apiculus and Plant Height
Dongnanihui 810, ZhangPu wild rice, and 146 CSSLs were grown in a paddy field under natural conditions at the experimental farm of Fujian Academy of Agricultural Sciences (Fuzhou, China), in early of 2015. The field experiment was designed in randomized plots with one plot per genotype. For parents and each CSSL, 60 plants were planted in six rows, and 12 plants in the center of each plot were selected to investigate the characters of purple apiculus. Plant height was the mean value of 6 plants from the middle section of each plot, and QTLs were identified on the basis of significant differences between parents and each CSSL, as determined by t-test. All plants were grown according to standard commercial practices, with spacing of 13.3 cm between plants within each row and 26.4 cm between rows. Field management essentially followed normal agricultural practices, and the amounts of N, P2O5 and K2O applied were 127.5 kg/hm2, 45.0 kg/hm2, and 30.0 kg/hm2, respectively.
Observed Agronomic Characteristics of 146 CSSLs
According to standard commercial practices, 146 CSSLs were grown in a paddy field under natural conditions at the experimental farm of Fujian Academy of Agricultural Sciences (Fuzhou, China). All materials were planted in the same field, the level height of field was consistent, N, P2O5 and K2O were applied in the following amounts: 127.5 kg/hm2, 45.0 kg/hm2, and 30.0 kg/hm2, respectively, and the amount and time of fertilization was uniform. Plant height, panicle length, number of effective panicles, spikelet number per panicle, seed setting rate and 1000-grain weight were all measured at maturity in 2015.
Physical Map Construction of Target Gene
The physical map of the qPa-6-2 gene was constructed by bioinformatics analysis using the published sequences of BAC and P1-derived artificial chromosome (PAC) clones of cv. Nipponbare released by the International Rice Genome Sequencing project (IRGSP3).
Results
Polymorphisms Detected between the Two Parents
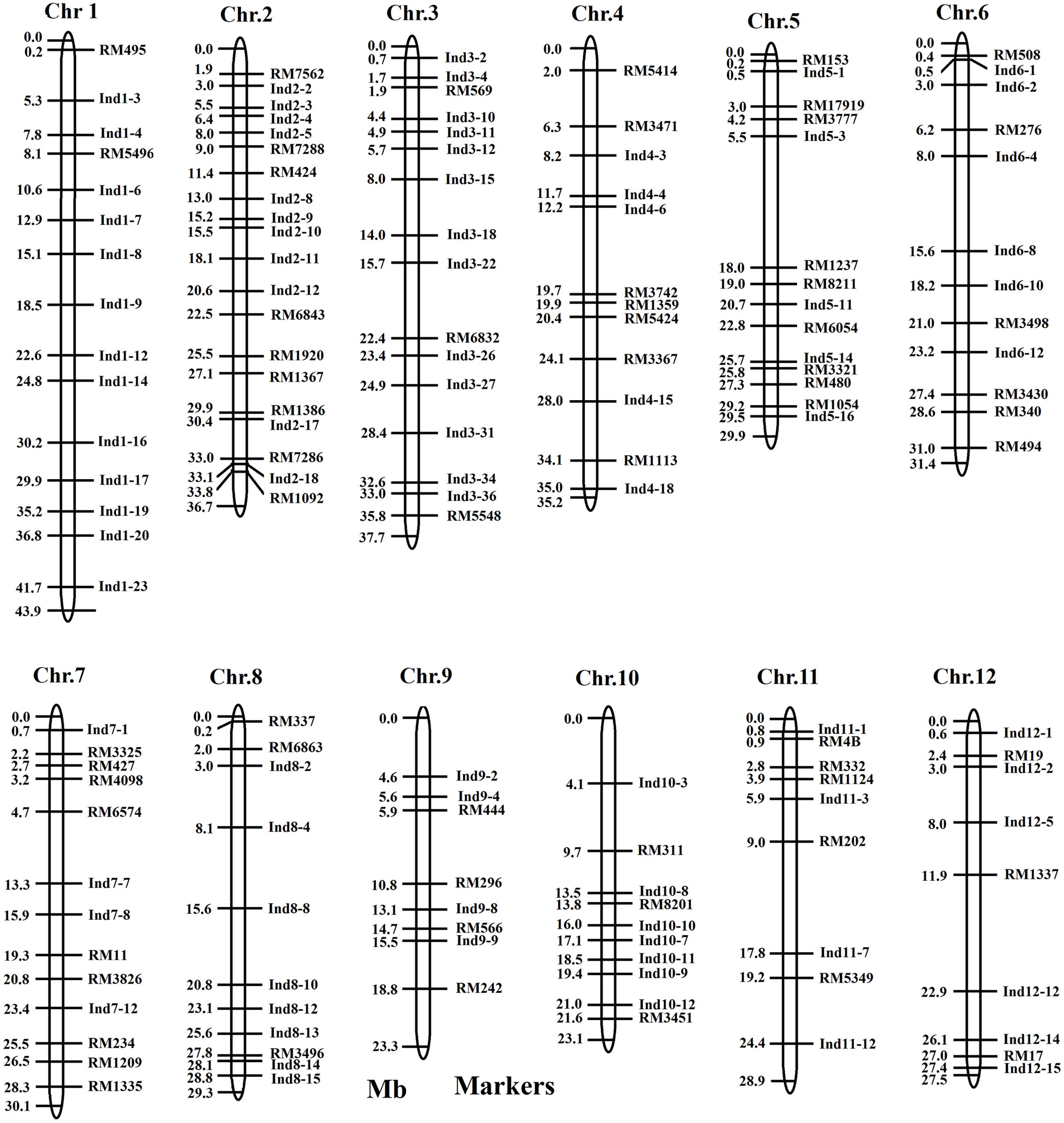
Polymorphisms were detected by SSR and InDel markers between the two parents, and were used in this study to survey the polymorphisms between the two parents (Table 1). 302 SSR markers were selected from dense rice microsatellite maps (McCouch et al., 2002). A total of 206 InDel markers were developed using Primer Premier 5.0 software according to the publicly available rice genome sequence comparisons between Nipponbare and 93114. Of the 506 markers, 149 (29.4%) displayed better polymorphisms between the two parents (Supplement Table S1). The length of the interval between two polymorphic markers ranged from 0.1Mb to 12.5 Mb, with an average of 2.53 Mb on the rice physical map (Figure 1; Table 1). The polymorphic markers were utilized further for the further development of CSSLs and HCSSLs.
CSSL and HCSSL Development
The CSSL and HCSSL development procedure was summarized in Figure 2. In total, we identified 213 BC3F1 plants in which the majority of genomic regions were homozygous for Dongnanihui 810 alleles, and 31 BC3F1 plants that had one substituted segment were self-pollinated to produce thirty-one BC3F2 lines. Then, 21 plants from each BC3F2 line were genotyped with the polymorphic markers on the target-substituted segments to select plants that had one homozygous substituted segment from ZhangPu wild rice. Heterozygous substituted segments from ZhangPu wild rice were also selected. As a result, 42 CSSLs were obtained, and 36 HCSSLs were obtained. The remaining 182 BC3F1 plants, which had two to four substituted segments, were back-crossed with Dongnanihui 810 to produce the BC4F1 lines, and these plants were self-pollinated to produce BC4F2 lines. Twenty-one plants from each line were genotyped on the target region to select plants that had one homozygous substituted segment. Forty-five plants from each line were genotyped on the target region to select plants that had two homozygous substituted segments. Ninety-three plants from each line were genotyped on the target region to select plants that had three homozygous substituted segments. At the same time, heterozygous substituted segments from ZhangPu wild rice were selected. As a result, 104 CSSLs were obtained, and 208 HCSSLs were selected. Using these techniques, a total of 146 CSSLs and 244 HCSSLs were selected. Of these CSSLs, eight carried two substituted segments and one carried three substituted segments. Nine HCSSLs, which contained the target region, were planted. Then nine CSSLs were obtained, but only one carried two substituted segments.
Physical Map of CSSLs and HCSSLs
According to the physical locations and genotypes of the 149 molecular markers in the 146 CSSLs, the lengths and locations of the substituted chromosome segments were estimated. Then, using the results of the estimation, a physical map of the 146 CSSLs was constructed (Figure 3). In addition, based on 149 molecular markers in the 244 HCSSLs, a physical map, which contained the 244 HCSSLs, was also constructed (Figure 4).
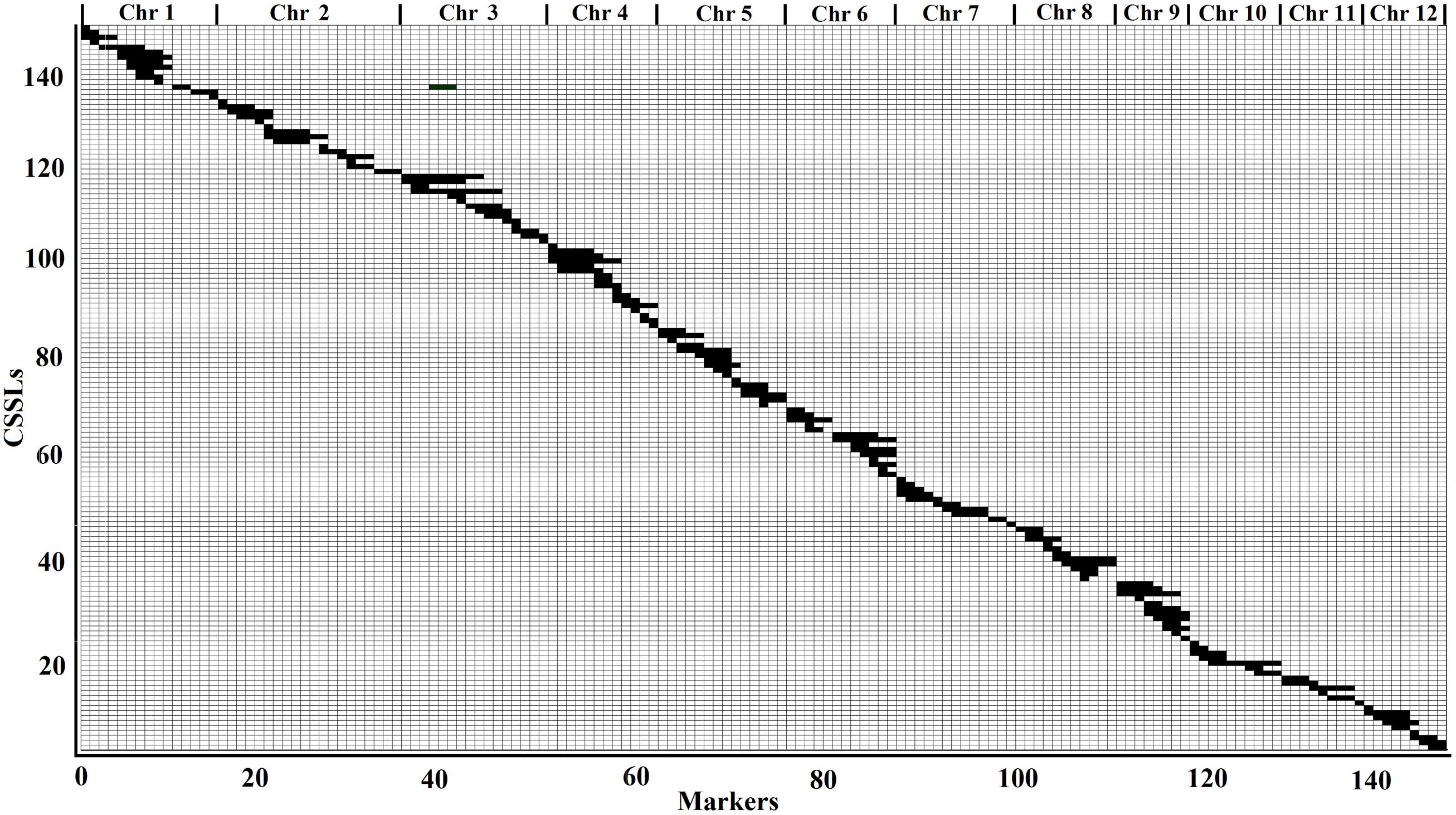
FIGURE 3. Physical map of the 146 CSSLs. Each row represented a CSSL and each column represented a molecular marker locus. The black areas indicate regions that were homozygous genotype for ZhangPu Wild Rice alleles: the white areas indicate regions homozygous for Dongnanihui 810 alleles.
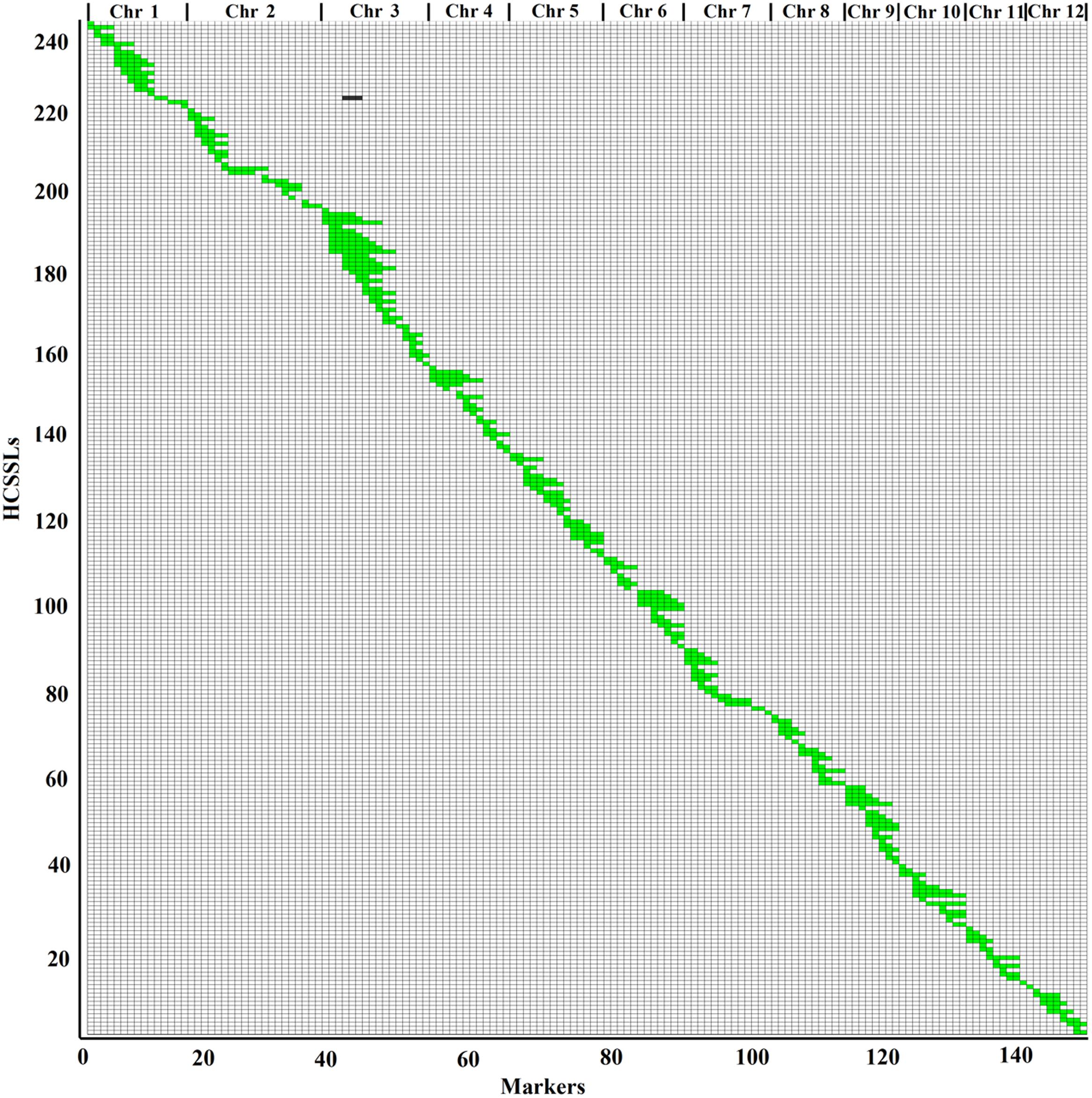
FIGURE 4. Physical map of the 244 HCSSLs. Each row represented a HCSSL and each column represented a molecular marker locus. The black areas indicate regions that were homozygous genotypes for ZhangPu Wild Rice alleles; the white areas indicate regions homozygous for Dongnanihui 810 alleles; the green areas indicate regions that were heterozygous genotypes for Dongnanihui 810 and ZhangPu Wild Rice.
Number, Length and Distribution of Chromosome Substituted Segments in the CSSLs and HCSSLs
From the physical map constructed with molecular markers, the 146 CSSLs carried 147 homozygous substituted segments. Only one CSSL on chromosome 1 carried two substituted segments and the remaining 145 carried only one substituted segment (Table 2). In the 146 CSSLs, the length of substituted segments ranged from 550 kb to 24.75 Mb, but averaging 7.79 Mb. The length of 81 substituted segments was shorter than 7.0 Mb and 15 were longer than 16.0 Mb (Figure 5). The average number of substitution segments per chromosome was 12.2, but the distribution of the segments was not random among the 12 chromosomes. Different substituted frequencies existed in different chromosomes. For example, 17 existed on chromosome 4, 14 existed on chromosome 1, 4, and 6, while only six existed on chromosome 11 (Table 2).
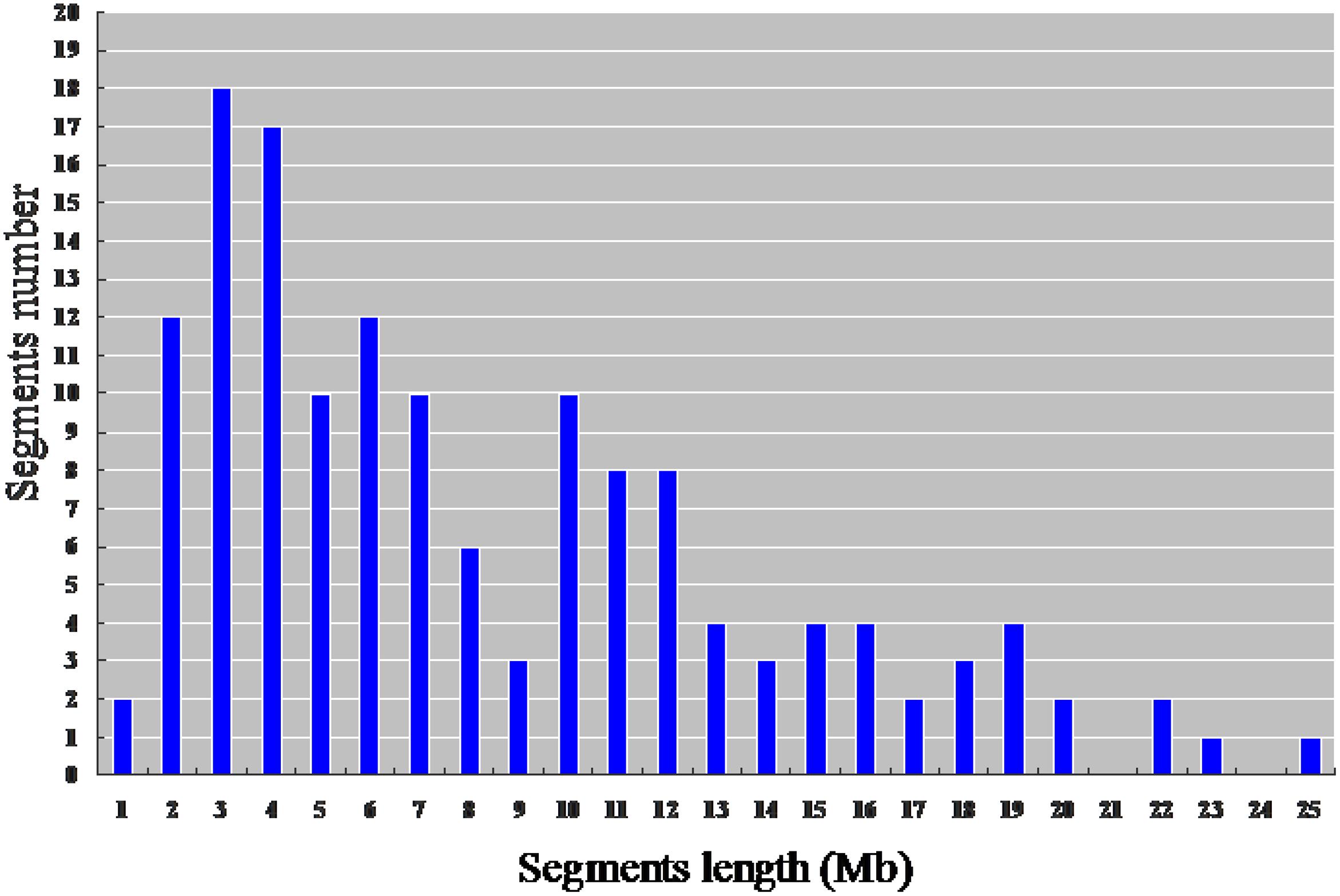
FIGURE 5. Distribution of the length of the substituted chromosome segments in the 146 CSSLs based on the physical map constructed with molecular markers.
The physical map constructed with molecular markers indicated that the 244 HCSSLs carried 244 heterozygous substituted segments and one homozygous substituted segment. Of these, only one HCSSL carried two substituted segments, which contained one heterozygous and one homozygous segment, while the remaining 243 HCSSLs carried only one heterozygous substituted segment (Table 2). The length of heterozygous substituted chromosome segments in the 244 HCSSLs ranged from 300 kb to 23.45 Mb, but averaging 6.90 Mb. Overall, 152 heterozygous segments were shorter than 7.0 Mb, and 21 heterozygous segments were longer than 16.0 Mb (Figure 6). The average number of substitution segments per chromosome was 20.3 in the 244 HCSSLs. However, different introgressed frequencies were identified among the 12 chromosomes in that 38 heterozygous segments existed on chromosome 3, while 12 heterozygous segments existed on chromosome 12 (Table 2).
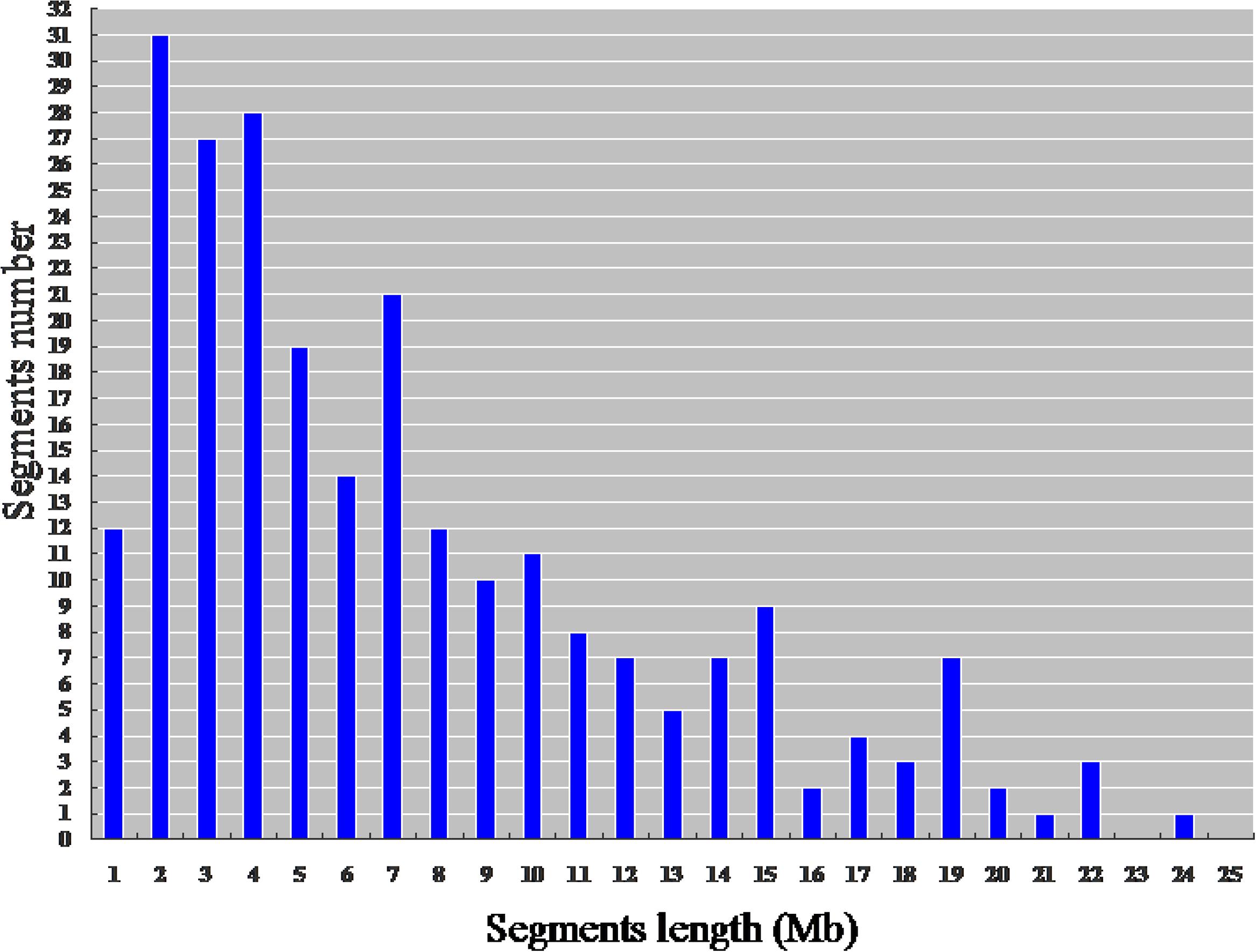
FIGURE 6. Distribution of the length of the substituted chromosome segments in the 244 HCSSLs based on the physical map constructed with molecular markers.
Genome Coverage of Substituted Segments in the CSSLs and HCSSLs
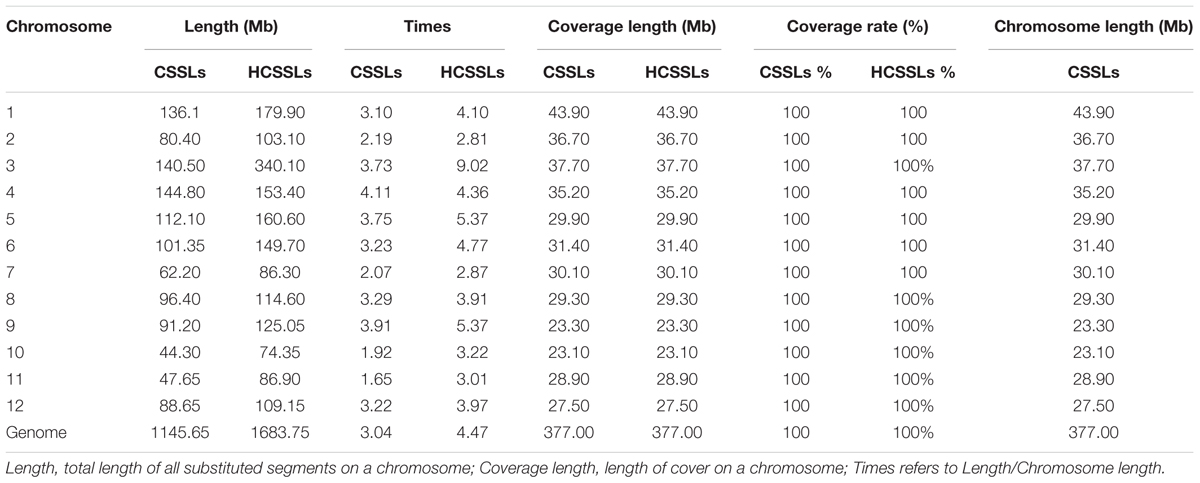
The physical map indicated that the average length of substituted segments per chromosome was 95.47 Mb in the CSSLs, ranging from 44.30 Mb on chromosome 10–144.80 Mb on chromosome 4. The total length of substituted segments in the CSSLs was 1145.65 Mb, which was 3.04 times the total length of the rice genome, and there were different in 12 chromosomes, ranging from 1.65 times on chromosome 11–4.11 times on chromosome 4. All of the chromosomes had 100% coverage in both cases (Table 3).
The physical map showed that the average length of substituted segments per chromosome was 140.31 Mb in the HCSSLs, ranging from 74.35 Mb on chromosome 10–340.10 Mb on chromosome 3. The total length of substituted segments in the HCSSLs was 1683.75 Mb, which was 4.47 times the total length of the rice genome, and different frequencies in rice chromosome, ranging from 2.81 times on chromosome 2 to 9.02 times on chromosome 3. The average coverage of substituted segments per chromosome was also 100% (Table 3).
Substitution Mapping of QTLs for Plant Height in the CSSLs
In order to evaluate the potential advantages of the CSSLs for QTL detection, phenotypic variations of plant height were observed in 146 CSSLs.
The two parents showed highly significant differences in plant height. Using six CSSLs, two QTLs were identified, in which qPH-1-1 was mapped in the marker intervals between Ind 1–20 and Ind 1–23 (7.1 Mb) on rice chromosome 1 (Figure 7A), while qPH-7-1 was mapped between Ind7-1 and RM427, which spanned 2.0 Mb in genetic distance on rice chromosome 7 (Figure 7B).
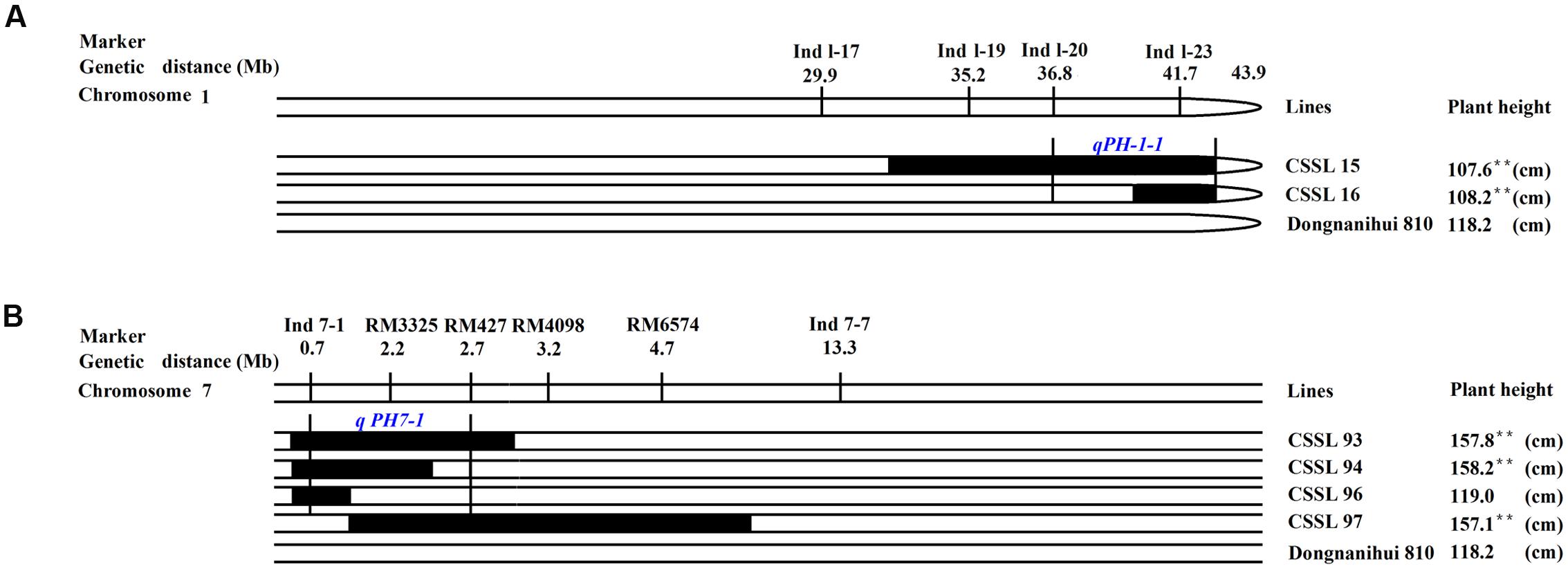
FIGURE 7. Substitution mapping of plant height. (A) Substitution mapping of the qPH-1-1 gene on rice chromosome 1; (B) Substitution mapping of the qPH-7-1 gene on rice chromosome 7. The substituted segments from ZhangPu wild rice were denoted by dark bars. The substituted segments from ZhangPu wild rice were denoted by black bars with the assumption that a segment flanked by one marker of donor type and one marker of recipient type was 50% donor genotype. Note: ∗∗Shows significant at 0.01 level. For the purposes of mapping, however, the full region between one marker of donor type and one marker of recipient type was used as the boundary on each end. The vertical bars through the CSSLs designate the region to which the gene was mapped.
Substitution Mapping of QTLs for Purple Apiculus in the CSSLs
For purple apiculus, ZhangPu wild rice displayed purple apiculus, while the indica variety, Dongnanihui 810, displayed green apiculus. Using four CSSLs, one major QTL for purple apiculus was identified, in which qPa-6-2 was located between Ind6-1 and RM276, which comprised 5.7 Mb in physical distance on rice chromosome 6 (Figure 8).
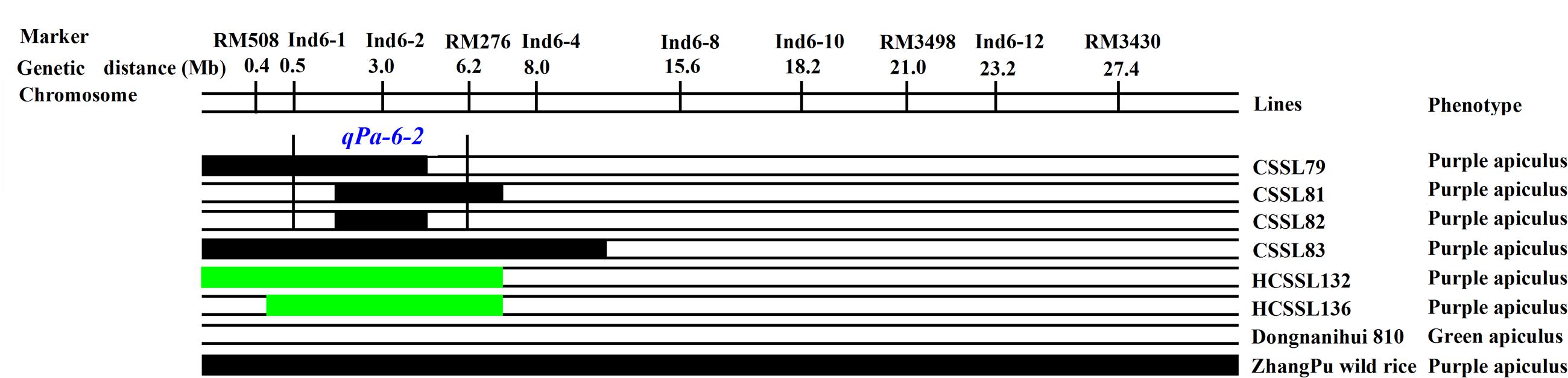
FIGURE 8. Substitution mapping of Pa-6-2 for purple apiculus on rice chromosome 6. The substituted segments from ZhangPu wild rice were denoted by dark bars; the green areas indicate regions that were heterozygous genotypes for Dongnanihui 810 and ZhangPu Wild Rice; the substituted segments from ZhangPu wild rice were denoted by black bars with the assumption that a segment flanked by one marker of donor type and one marker of recipient type was 50% donor genotype. For the purposes of mapping, however, the full region between one marker of donor type and one marker of recipient type was used as the boundary on each end. The vertical bars through the CSSLs designate the region to which the gene was mapped.
Genetic Analysis of the qPa-6-2 Gene for Purple Apiculus
The two segregating populations, HCSSL132 and HCSSL136, which were heterozygous in the target region and showed purple apiculus (Table 4), were grown in a paddy field under natural conditions at the experimental farm of Fujian Academy of Agricultural Sciences (Fuzhou, China), in late 2015. Segregating plants were therefore recorded as either purple or green apiculus. The segregation of purple to green apiculus based on 4932 F2 plants fitted a ratio of 3:1(χ2 = 0.330 ∼ 0.688, P > 0.05) (Table 4), indicating that the gene for purple was dominant over green color and was controlled by a major gene.
Fine Mapping of the qPa-6-2 Gene
To map the gene to a smaller region, 1241 recessive individuals were identified from the two HCSSL populations (Table 4). A higher precision map was constructed using published markers5 in the region between Ind6-1 and RM276 (Figure 9A). All recombinants were genotyped using seven polymorphic markers. The results showed that the qPa-6-2 gene was mapped between molecular markers RM19551 and RM19590 on chromosome 6 and that the physical distance between the two markers was 539 kb with a physical distance (Figure 9B; Table 5).
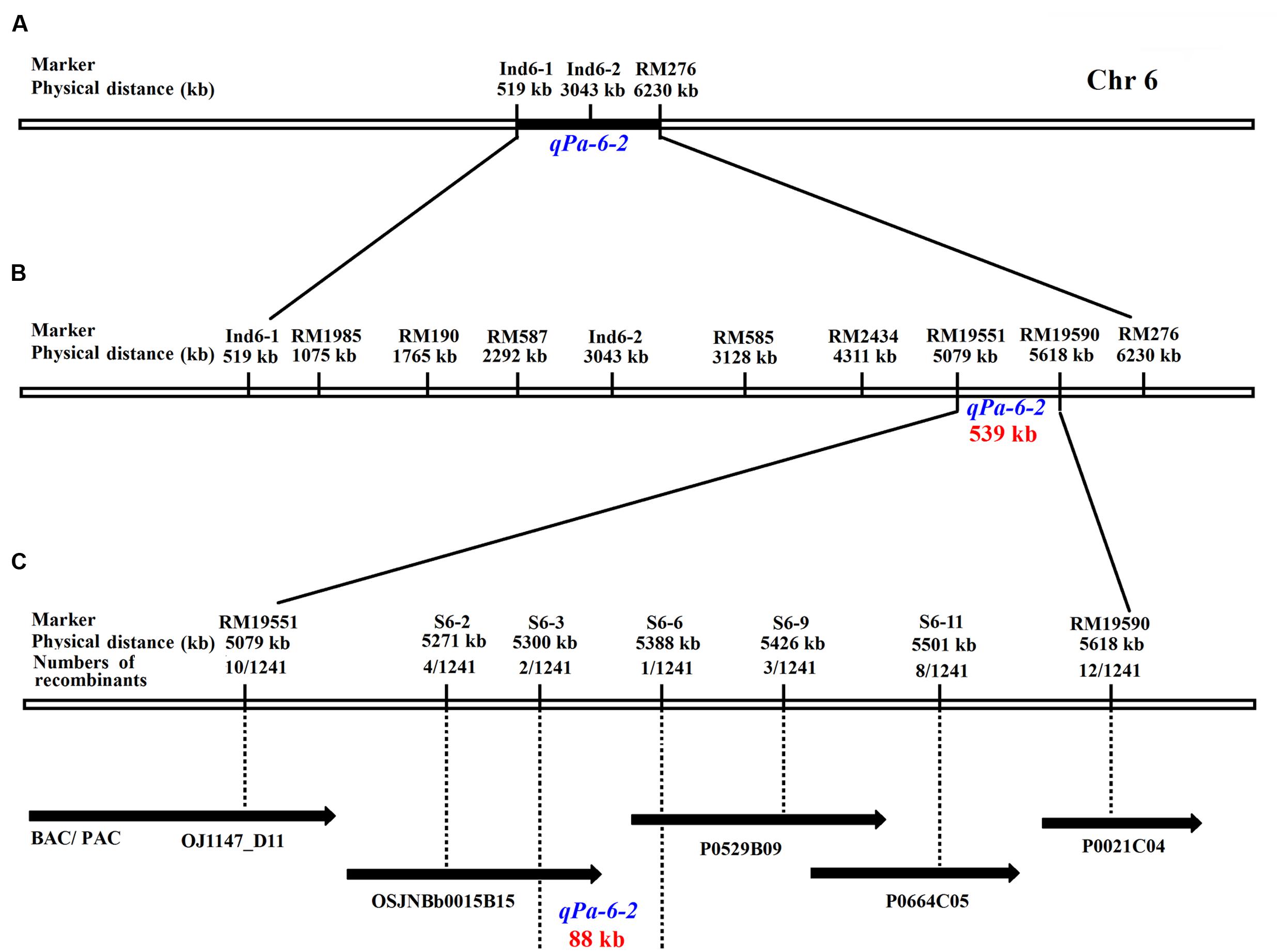
FIGURE 9. Genetic and physical maps of the qPa-6-2 gene. (A) Primary mapping of the qPa-6-2 gene. The gene was mapped to the region between markers Ind6-1 and RM276; (B) Further mapping of the qPa-6-2 gene. The gene was mapped to the region between markers RM19551 and RM19590; (C) Fine mapping of the qPa-6-2 gene. The qPa-6-2 gene was finally defined in an 88 kb region, and the recombinants number between markers and target gene was indicated under the physical distance.
To fine map the qPa-6-2 gene, five polymorphic InDels were selected from 12 new InDels (Table 5). The InDel markers were designed from the publicly available rice genome sequences, and the likelihood of detecting polymorphism between ZhangPu wild rice and Dongnanihui 810 was predicted by comparing sequences from Nipponbare6 and Indica cultivar 93-117. Firstly, the BAC clone’s sequences of japonica and indica were aligned, then primers were designed using Primer premier 5.0 based on the polymorphism region between the two rice subspecies and the polymorphic markers were used for gene mapping. Recombinant screening with five markers (S6-2, S6-3, S6-6, S6-9 and S6-11), which were more internal to the qPa-6-2 locus, detected four, two, one, three and eight recombinants, respectively. Thus, the qPa-6-2 gene was precisely mapping in an 88 kb region by S6-3 and S6-6 (Figure 9C).
Discussion
Excellent Resources in Wild Rice
Oryza Rufipogon Griff., a wild rice, is the progenitor of the cultivated rice O. sativa L. As such, it is well recognized as a natural gene bank that conserves many specific genes. During the course of domestication from common wild rice to cultivated rice, profound changes in morphology and molecular genetic profile occurred via natural and artificial selection. Over the years, polymorphisms have been identified between wild and cultivated rice, including some that affect yield, disease resistance, or insect resistance (Tian et al., 2006; Jena, 2014).
Major resistance genes have been transferred to cultivated rice from O. nivara and O. longistaminata (Khush, 1977; Khush et al., 1990). A new gene, Xa-23, which showed resistance to bacterial blight was discovered and cloned from common wild rice of Guangxi (Wang et al., 2014, 2015). The new resistance gene for Magnaporthe oryzae, Pi9, was identified from wild rice (Zhuang et al., 1998; Jiang et al., 2012). The major gene for Magnaporthe oryzae, Pi40, was discovered from the wild rice O. australiensis (Jeung et al., 2007). The new genes, Bph14 and Bph15, which showed resistance to the brown planthopper, were identified from the wild rice (Oryza officinalis Wall) (Yang et al., 2004; Du et al., 2009). Meanwhile, Li et al. (2002) identified two high-yielding QTLs from Dongxiang common wild rice of China. Tan et al. (2008) cloned the PROG1 gene from common wild rice, and prog1 variants identified in O. sativa disrupt prog1 function and inactivate prog1 expression, leading to erect growth, greater grain number and higher grain yield in cultivated rice. At the same time, the flowering time QTL dth3, was detected from wild rice species (Bian et al., 2011), and the seed dormancy QTL was also isolated in the genetic populations derived from wild rice species (Gu et al., 2004).
CSSLs as a Platform for QTL Mapping and Cloning
Chromosome segment substitution lines were a series of NILs in which the substituted segments of containing the entire information of the donor, and each CSSL carried one or more donor chromosome segments. Since the main characteristic of CSSLs is that the substituted segment of each CSSL was homozygous and stable, they were useful for genetic studies and fine mapping of QTLs for genome-wide target traits. In the present study, we developed 146 CSSLs in the genetic background of rice restorer Dongnanihui 810. The 146 CSSLs had different agronomic traits, including, for example, long grain, plant height, panicle length, effective panicle, and long flag leaf. Using these lines, we mapped two QTLs for plant height. The qPH-1-1 mapped in this study was probably an allele of sd1 (Sasaki et al., 2002; Ye et al., 2015), while qPH-7-1 might be a novel QTL. The qPH-1-1 mapped between the markers Ind1-20 and Ind1-23 in this study, which contained the sdl locus, a gene known to play a significant role in height in rice. These results demonstrated that the uniformity of the genetic background of the lines facilitated the identification of a number of QTLs by direct comparison between each line and the recipient parent. For further fine mapping and positional cloning of interesting QTLs, secondary F2 populations could be derived from a further back-cross between the target CSSL and the recurrent parent (Frary et al., 2000; Yano et al., 2000). Therefore, CSSLs simplified the process of data analysis and increased the accuracy of the results. In this study, using the 146 CSSLs, one major QTL for purple apiculus was identified. Here, the qPa-6-2 was located between two markers Ind6-1 and RM276 on rice chromosome 6. More importantly, the qPa-6-2 gene was precisely mapped to an 88 kb region, which contained the rice OsC1 gene (Saitoh et al., 2004). The occurrence of varying degrees of apiculus coloration (also shows purple apiculus) due to anthocyanin pigmentation, which was reported to be caused by a series of alleles at the C locus, and the C gene appears to be the rice homolog (OsC1) (Saitoh et al., 2004). In our study, we identified the qPa-6-2 for purple apiculus, which was also located in the same region. Meanwhile, the Pa-6 gene was also mapped to the same region with a genetic distance of 41.7 kb (Liu et al., 2012). It is well known that male sterile lines play very important in hybrid rice. Meanwhile, purple apiculus is helpful to increase outcrossing rate of male sterile lines in rice. Therefore, fine mapping and positional cloning of purple apiculus are of great significance in hybrid rice breeding.
The CSSLs as a Platform for Rice Molecular Breeding
Plant breeding, which aimed to improve the genetic basis of new varieties of crops with increased productivity and quality, combines art with science (Xi et al., 2006). In a general way, traditional breeding was predominantly based on phenotypic assays. Facilitated by the recent developments in genome sequencing, molecular markers and bioinformatics, plant breeding systems entered an era of molecular breeding, shortening the breeding period, improving efficiency, and overcoming the traditional shortcoming of low accuracy with broad applications. Thus, it became the standard in plant breeding programs to combine phenotype and genotype (Xu et al., 2010).
However, this approach only targeted the QTLs with very few traits for genetic improvement. On the other hand, CSSLs, which selected at the level of the whole genome with multi-trait breeding objectives, have expanded targets and thus become important in improving the properties of plants (Xu et al., 2010). Therefore, CSSLs for rice breeding were considered as a key tool in the ongoing technological innovation of plant breeding (Xu et al., 2010).
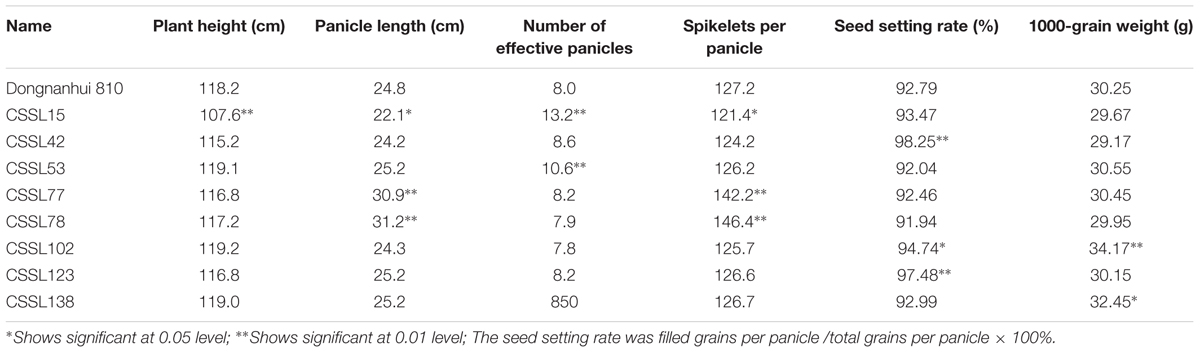
The indica cultivar Dongnanihui 810, used as the recipient in this study, has been planted on a large scale as an excellent restorer. Ganyou 810, derived from the CMS line Ganxiang A and a restorer line Dongnanihui 810, was certified and released for commercial production in 2014 in Fujian province. The ZhangPu wild rice, used as the donor in the present study, was the only wild rice in Fujian province. Interestingly, eight elite CSSLs in this wide population had a genetic background similar to that of Dongnanihui 810, but their agronomic traits were better than those of Dongnanihui 810 (Table 6). The effective panicle, each panicle contains at least five seeds (including 5) in per plant, was one of the important indicators of yield traits. For example, CSSL15 had lower plant height and more effective panicle than Dongnanihui 810. CSSL42, CSSL102 and CSSL123 had higher seed setting rate, and CSSL53 had more effective panicle. CSSL77 and CSSL78 had longer panicle and more spikelets, while CSSL102 and CSSL138 both had higher 1000-grain weight. None of these eight CSSLs had poor agronomic traits. Therefore, they could be used to create new varieties with direct marketing applications and, as a parent, create new hybrids. Furthermore, favorable alleles could be combined through MAS to improve the production of superior rice varieties.
Why Should We Develop the HCSSLs in This Paper?
HCSSLs showed technological innovation in rice genetic research. Up to now, after developing CSSLs in rice, many HCSSLs, which carried one or more donor heterozygous chromosome segments, were discarded. However, for further fine mapping and positional cloning of interesting QTLs, secondary F2 populations were reconstructed, which should be derived from a further back-cross between the target CSSL and the recurrent parent. Obviously, this not only increased experimental cost, but also reduced efficiency. In the present study, 244 HCSSLs were produced on the basis of estimates of lengths and locations, and the total length of substituted segments among HCSSLs was 1683.75 Mb, or 4.47 times the total length of the rice genome.
It was convenient to perform gene mapping and analysis of genetic characteristics for agronomic traits using the HCSSLs. In this paper, using the 146 CSSLs, the major qPa-6-2 for purple apiculus was identified. To confirm this, further analysis showed that both HCSSL132 and HCSSL136 were heterozygous in the target region and showed purple apiculus. Meanwhile, HCSSL132 and HCSSL136 were planted. Further observation of these two populations showed that the gene for purple apiculus was dominant over green and was controlled by a major gene. Importantly, without reconstructing secondary F2 populations, the qPa-6-2 was precisely mapping in an 88 kb region using the two HCSSLs populations.
It was easy to improve the development of CSSLs carrying overlapping chromosome segments of the whole rice genome using HCSSLs. From a genetics perspective, the number of heterozygous plants, as a separate population, was nearly twice that of homozygous plants. Regrettably, only homozygous substituted plants were selected, while many heterozygous substituted plants were discarded. As a result, CSSLs could barely cover the whole genome of donor parents. However, incomplete coverage of a donor genome in the CSSL population might, in turn miss some QTLs responsible for useful traits (Subudhi et al., 2015). In this paper, with the help of HCSSLs, CSSLs carrying one substituted segment could be obtained. At the early stage of constructing CSSLs, although 146 CSSLs were obtained in this paper, eight of these CSSLs carried two substituted segments and one carried three substituted segments. Using nine HCSSLs containing the target region, nine CSSLs were obtained, which only one CSSL carried two substituted segments, the rest of the eight carried one substituted segment.
Author Contribution
DY drafted the manuscript. DY and FH participated in the development of chromosome segment substitution lines. DY, XZ, CC, and NY contributed to data analysis. DY and XY participated in the design of the study and the interpretation of the results and wrote and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Special Fund for Agro-scientific Research in the Public Interest of Fujian Province (2017R1101022-5), Fujian Provincial Natural Science Foundation of China (No. 2015J01108), Technology Great Item of Fujian Province (2015NZ002-3-1), Special Fund for Agro-scientific Research in the Public Interest of Fujian Province (No. 2015R1021-10), and David Martin for language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01737/full#supplementary-material
Abbreviations
Ls, chromosome segment substitution lines; HCSSLs, heterozygous chromosome segment substitution lines; INDEL, insertion/deletion; MAB, marker-assisted breeding; MAS, marker-assisted selection; NILs, near-isogenic lines; QTLs, quantitative trait loci; SSR, simple sequence repeats.
Footnotes
- ^ http://www.ricedata.cn/variety/varis/614685.htm
- ^ http://rgp.dna.affrc.go.jp
- ^ http://rgp.dna.affrc.go.jp/IRGSP/index.html
- ^ http://rgp.dna.affrc.go.jp
- ^ http://archive.gramene.org/markers/
- ^ http://rgp.dna.affrc.go.jp/
- ^ http://rice.genomics.org.cn/
References
Arbelaez, J. D., Moreno, L. T., Singh, N., Tung, C. W., Maron, L. G., Ospina, Y., et al. (2015). Development and GBS-genotyping of introgression lines (ILs) using two wild species of rice, O. meridionalis and O. rufipogon, in a common recurrent parent, O. sativa cv. Curinga. Mol. Breed. 35:81. doi: 10.1007/s11032-015-0276-7
Ashikari, M., Sakakibara, H., Lin, S. Y., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. doi: 10.1126/science.1113373
Bian, X. F., Liu, X., Zhao, Z. G., Jiang, L., Gao, H., Zhang, Y. H., et al. (2011). Heading date gene, dth3 controlled late flowering in O. Glaberrima Steud. by down-regulating Ehd1. Plant Cell Rep. 30, 2243–2254. doi: 10.1007/s00299-011-1129-4
Chen, J. B., Li, X. Y., Cheng, C., Wang, Y. H., Qin, M., Zhu, H. T., et al. (2014). Characterization of epistatic interaction of QTLs LH8 and EH3 controlling heading date in rice. Sci. Rep. 4:4263. doi: 10.1038/srep04263
Doi, K., Iwata, N., and Yoshimura, A. (1997). The construction of chromosome substitution lines of African rice (Oryza glaberrima Steud.) in the background of Japonica rice (O. sativa L.). Rice Genet. News 14, 39–41.
Du, B., Zhang, W. L., Liu, B. F., Hu, J., Wei, Z., Shi, Z. Y., et al. (2009). Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl. Acad. Sci. U.S.A. 106, 22163–22168. doi: 10.1073/pnas.0912139106
Ebitani, T., Takeuchi, Y., Nonoue, Y., Yamamoto, T., Takeuchi, K., and Yano, M. (2005). Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar ‘Kasalath’ in a genetic background of japonica elite cultivar ‘Koshihikari’. Breed. Sci. 55, 65–73. doi: 10.1270/jsbbs.55.65
Fan, C. C., Xing, Y. Z., Mao, H. L., Lu, T. T., Han, B., Xu, C. G., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. doi: 10.1007/s00122-006-0218-1
Frary, A., Nesbitt, T. C., Grandillo, S., Knaap, E. V. D., Cong, B., Liu, J. P., et al. (2000). fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88. doi: 10.1126/science.289.5476.85
Fujita, D., Ebron, L. A., Araki, E., Kato, H., Khush, G. S., Sheehy, J. E., et al. (2010). Fine mapping of a gene for low-tiller number, Ltn, in japonica rice (Oryza sativa L.) variety Aikawa 1. Theor. Appl. Genet. 120, 1233–1240.
Furuta, T., Uehara, K., Rosalyn, B., Shim, A., Shim, J., Ashikari, A., et al. (2014). Development and evaluation of chromosome segment substitution lines (CSSLs) carrying chromosome segments derived from Oryza rufipogon in the genetic background of Oryza sativa L. Breed. Sci. 63, 468–475. doi: 10.1270/jsbbs.63.468
Gu, X. Y., Kianian, S. F., and Foley, M. E. (2004). Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa). Genetics 166, 1503–1516. doi: 10.1534/genetics.166.3.1503
Henry, A., Swamy, B. P. M., Dixit, S., Torres, R. D., Batoto, T. C., Manalili, M., et al. (2015). Physiological mechanisms contributing to the QTL-combination effects on improved performance of IR64 rice NILs under drought. J. Exp. Bot. 66, 1787–1799. doi: 10.1093/jxb/eru506
Huang, X. Z., Qian, Q., Liu, Z. B., Sun, H. Y., He, S. Y., Luo, D., et al. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497. doi: 10.1038/ng.352
Jena, K. K. (2014). The species of the genus Oryza and transfer of useful genes from wild species into cultivated rice. O. sativa. Breed. Sci. 60, 518–552. doi: 10.1270/jsbbs.60.518
Jeung, J. U., Kim, B. R., Cho, Y. C., Han, S. S., Moon, H. P., Lee, Y. T., et al. (2007). A novel gene, Pi40(t), linked to the DNA markers derived from NBS-LRR motifs confers broad spectrum of blast resistance in rice. Theor. Appl. Genet. 115, 1163–1177. doi: 10.1007/s00122-007-0642-x
Jiang, N., Li, Z. Q., Wu, J., Wang, Y., Wu, L. J., Wang, S. H., et al. (2012). Molecular mapping of the Pi2/9 allelic gene Pi2-2 conferring broad-spectrum resistance to Magnaporthe oryzae in the rice cultivar Jefferson. Rice 5:29. doi: 10.1186/1939-8433-5-29
Khush, G. S. (1977). Disease and insect resistance in rice. Adv. Agron. 29, 265–341. doi: 10.1016/S0065-2113(08)60221-7
Khush, G. S., Bacalangco, E., and Ogawa, T. (1990). A new gene for resistance to bacterial blight from O. longistaminata. Rice Genet. Newslett. 7, 121–122.
Kubo, T., Aida, Y., Nakamura, K., Tsunematsu, H., Doi, K., and Yoshimura, A. (2002). Reciprocal chromosome segment substitution series derived from japonica and indica cross of rice (Oryza sativa L.). Breed. Sci. 52, 319–325. doi: 10.1270/jsbbs.52.319
Li, D. J., Sun, C. Q., Fu, Y. C., Li, C., Zhu, Z. F., Chen, L., et al. (2002). Identification and mapping of genes for improving yield from Chinese common wild rice (O. rufipogon Griff.) using advanced backcross QTL analysis. Chin. Sci. Bul. 47, 1533–1537. doi: 10.1360/02tb9337
Li, S. K. (2010). Identification and Evaluation on Genetic Diversity of Common Wild Rice in Zhangpu County Fujian Province. Master Thesis, Fujian Agriculture and Forestry University, Fuzhou.
Li, X. N., Wang, W. K., Wang, Z., Li, K. N., Lim, Y. P., and Piao, Z. Y. (2015). Construction of chromosome segment substitution lines enables QTL mapping for flowering and morphological traits in Brassica rapa. Front. Plant Sci. 6:432. doi: 10.3389/fpls.2015.00432
Li, Z. K., Pinson, S. R. M., Stansel, J. W., and Park, W. D. (1995). Identification of quantitative trait loci (QTLs) for heading date and plant height in cultivated rice (Oryza sativa L.). Theor. Appl. Genet. 91, 374–381. doi: 10.1007/BF00220902
Liu, X., Sun, X., Wang, W. Y., Ding, H. F., Liu, W., Li, G. X., et al. (2012). Fine Mapping of Pa-6 gene for purple apiculus in rice. J. Plant Biol. 55, 218–225. doi: 10.1007/s12374-011-0276-z
McCouch, S. R., Teytelman, L., Xu, Y. B., Lobos, K. B., Clare, K., Walton, M., et al. (2002). Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9, 199–207. doi: 10.1093/dnares/9.6.199
Mei, H. W., Xu, J. L., Li, Z. K., Yu, Q., Guo, L. B., Wang, Y. P., et al. (2006). QTLs influencing panicle size detected in two reciprocal introgressive line (IL) populations in rice (Oryza sativa L.). Theor. Appl. Genet. 112, 648–656. doi: 10.1007/s00122-005-0167-0
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
Nagata, K., Ando, T., Nonoue, Y., Mizubayashi, T., Kitazawa, N., Shomura, A., et al. (2015). Advanced backcross QTL analysis reveals complicated genetic control of rice grain shape in a japonica × indica cross9. Breed. Sci. 65, 308–318. doi: 10.1270/jsbbs.65.308
Ray, J. D., Yu, L., McCouch, S. R., Champoux, M. C., Wang, G., and Nguyen, H. T. (1996). Mapping quantitative trait loci associated with root penetration ability in rice (Oryza sativa L.). Theor. Appl. Genet. 92, 627–636. doi: 10.1007/BF00226082
Redona, E. D., and Mackill, D. J. (1996). Mapping quantitative trait loci for seedling vigor in rice using RFLPs. Theor. Appl. Genet. 92, 395–402. doi: 10.1007/BF00223685
Saitoh, K., Onishi, K., Mikami, I., Thidar, K., and Sano, Y. (2004). Allelic diversification at the C (OsC1) locus of wild and cultivated rice: nucleotide changes associated with phenotypes. Genetics 168, 997–1007. doi: 10.1534/genetics.103.018390
Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., et al. (2002). A mutant gibberellin-synthesis gene in rice. Nature 416, 701–702. doi: 10.1038/416701a
Shomura, A., Ebana, K., Ebitani, T., Kanegae, H., Konishi, S., and Yano, M. (2008). Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40, 1023–1028. doi: 10.1038/ng.169
Song, X. J., Huang, W., Shi, M., Zhu, M. Z., and Lin, H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. doi: 10.1038/ng2014
Subudhi, P. K., De Leon, T., Singh, P. K., Parco, A., Cohn, M. A., and Sasaki, T. (2015). A chromosome segment substitution library of weedy rice for genetic dissection of complex agronomic and domestication traits. PLoS ONE 10:e0130650. doi: 10.1371/journal.pone.0130650
Sun, J., Tan, B. K., Huang, S. H., Whiteman, M., and Zhu, Y. Z. (2002). Effects of natural products on ischemic heart diseases and cardiovascular system. Acta Pharmacol. Sin. 23, 1142–1151.
Takai, T., Nonoue, Y., Yamamoto, S., Yamanouchi, U., Matsubara, K., Liang, Z. W., et al. (2007). Development of chromosome segment substitution lines derived from backcross between indica donor cultivar ‘Nona bokra’ and japonica recipient cultivar ‘Koshihikari’. Breed. Sci. 57, 257–261. doi: 10.1270/jsbbs.57.257
Tan, L. B., Li, X. R., Liu, F. X., Sun, X. Y., Li, C. G., Zhu, Z. F., et al. (2008). Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40, 1360–1364. doi: 10.1038/ng.197
Tanksley, S. D., and McCouch, S. R. (1997). Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277, 1063–1066. doi: 10.1126/science.277.5329.1063
Tian, F., Li, D. J., Fu, Q., Zhu, Z. F., Fu, Y. C., Wang, X. K., et al. (2006). Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor. Appl. Genet. 112, 570–580. doi: 10.1007/s00122-005-0165-2
Wang, C. L., Fan, Y. L., Zheng, C. K., Qin, T. F., Zhang, X. P., and Zhao, K. J. (2014). High-resolution genetic mapping of rice bacterial blight resistance gene Xa23. Mol. Genet. Genom. 289, 745–753. doi: 10.1007/s00438-014-0848-y
Wang, C. L., Zhang, X. P., Fan, Y. L., Gao, Y., Zhu, Q. L., Zheng, C. K., et al. (2015). Xa23 is an executor R protein and confers broad-spectrum disease resistance in Rice. Mol Plant 8, 290–302. doi: 10.1016/j.molp.2014.10.010
Xi, Z. Y., He, F. H., Zeng, R. Z., Zhang, Z. M., Ding, X. H., Li, W. T., et al. (2006). Development of a wide population of chromosome single segment substitution lines in the genetic background of an elite cultivar of rice (Oryza sativa L.). Genome 49, 476–484. doi: 10.1139/G06-005
Xu, J. J., Zhao, Q., Du, P. N., Xu, C. W., Wang, B. H., Feng, Q., et al. (2010). Developing high throughput genotyped chromosome segment substitution lines based on population whole-genome re-sequencing in rice (Oryza sativa L.). BMC Genomics 24:656. doi: 10.1186/1471-2164-11-656
Xue, W. Y., Xing, Y. Z., Weng, X. Y., Zhao, Y., Tang, W. J., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yang, H. Y., You, A. Q., Yang, Z. F., Zhang, F. T., He, R. F., Zhu, L. L., et al. (2004). High-resolution genetic mapping at the Bph15 locus for brown planthopper resistance in rice (Oryza sativa L.). Theor. Appl. Genet. 110, 182–191. doi: 10.1007/s00122-004-1844-0
Yano, M. (2001). Genetic and molecular dissection of naturally occurring variations. Curr. Opin. Plant Biol. 4, 130–135. doi: 10.1016/S1369-5266(00)00148-5
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in Rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2483. doi: 10.2307/3871242
Yano, M., and Sasaki, T. (1997). Genetic and molecular dissection of quantitative traits in rice. Plant Mol. Biol. 35, 145–153. doi: 10.1023/A:1005764209331
Ye, H., Feng, J. H., Zhang, L. H., Zhan, J. F., Mispan, M. S., Cao, Z. Q., et al. (2015). Map-based cloning of seed dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol. 169, 2152–2165.
Young, N. D., and Tanksley, S. D. (1989). Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theor. Appl. Genet. 77, 95–101. doi: 10.1007/BF00292322
Zhou, L., Zeng, Y. W., Hu, G. L., Pan, Y. H., Yang, S. M., You, A. Q., et al. (2012). Characterization and identification of cold tolerant near-isogenic lines in rice. Breed. Sci. 62, 196–201. doi: 10.1270/jsbbs.62.196
Zhu, W. Y., Lin, J., Yang, D. W., Zhao, L., Zhang, Y. D., Zhu, Z., et al. (2009). Development of chromosome segment substitution lines derived from backcross between two sequenced rice cultivars, indica recipient 93-11 and japonica donar Nipponbare. Plant Mol. Biol. Rep. 27, 126–131. doi: 10.1007/s11105-008-0054-3
Keywords: rice (Oryza sativa L. subsp. indica), the Zhangpu wild rice (Oryza rufipogon Griff.), chromosome segment substitution lines, quantitative trait loci, gene mapping
Citation: Yang D, Ye X, Zheng X, Cheng C, Ye N and Huang F (2016) Development and Evaluation of Chromosome Segment Substitution Lines Carrying Overlapping Chromosome Segments of the Whole Wild Rice Genome. Front. Plant Sci. 7:1737. doi: 10.3389/fpls.2016.01737
Received: 01 June 2016; Accepted: 04 November 2016;
Published: 24 November 2016.
Edited by:
Henry T. Nguyen, University of Missouri, USAReviewed by:
Matthew R. Willmann, Cornell University, USAMingyong Zhang, Chinese Academy of Sciences, China
Copyright © 2016 Yang, Ye, Zheng, Cheng, Ye and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinfu Ye, eWV4aW5mdUAxMjYuY29t
 Dewei Yang
Dewei Yang Xinfu Ye*
Xinfu Ye*