- The National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, China
Sharp eyespot, caused mainly by the necrotrophic fungus Rhizoctonia cerealis, is a destructive disease in hexaploid wheat (Triticum aestivum L.). In Arabidopsis, certain cinnamyl alcohol dehydrogenases (CADs) have been implicated in monolignol biosynthesis and in defense response to bacterial pathogen infection. However, little is known about CADs in wheat defense responses to necrotrophic or soil-borne pathogens. In this study, we isolate a wheat CAD gene TaCAD12 in response to R. cerealis infection through microarray-based comparative transcriptomics, and study the enzyme activity and defense role of TaCAD12 in wheat. The transcriptional levels of TaCAD12 in sharp eyespot-resistant wheat lines were significantly higher compared with those in susceptible wheat lines. The sequence and phylogenetic analyses revealed that TaCAD12 belongs to IV group in CAD family. The biochemical assay proved that TaCAD12 protein is an authentic CAD enzyme and possesses catalytic efficiencies toward both coniferyl aldehyde and sinapyl aldehyde. Knock-down of TaCAD12 transcript significantly repressed resistance of the gene-silenced wheat plants to sharp eyespot caused by R. cerealis, whereas TaCAD12 overexpression markedly enhanced resistance of the transgenic wheat lines to sharp eyespot. Furthermore, certain defense genes (Defensin, PR10, PR17c, and Chitinase1) and monolignol biosynthesis-related genes (TaCAD1, TaCCR, and TaCOMT1) were up-regulated in the TaCAD12-overexpressing wheat plants but down-regulated in TaCAD12-silencing plants. These results suggest that TaCAD12 positively contributes to resistance against sharp eyespot through regulation of the expression of certain defense genes and monolignol biosynthesis-related genes in wheat.
Introduction
Hexaploid wheat (Triticum aestivum L., AABBDD, common wheat) is one of the most widely cultivated and consumed food crops. The global demand for wheat and other foods will increase along with the constantly increasing world population. Environmental stresses and diseases often negatively affect wheat production. For example, sharp eyespot is a devastating soil-borne disease impacting wheat production globally (Chen et al., 2008, 2013; Hamada et al., 2011; Lemańczyk and Kwaśna, 2013). China is the largest epidemic region in the world, as exemplified by more than 8.1 million hectares of wheat affected by sharp eyespot since 2005 (Chen et al., 2013; Zhu et al., 2015). The necrotrophic fungus Rhizoctonia cerealis van der Hoeven is a major causal pathogen of sharp eyespot (Chen et al., 2013). Sharp eyespot manifests as “eye”-shaped lesions on basal stems and basal sheaths of infected wheat plants. The disease can destroy the transport tissues in stems of plants and block transportation of substances required for nutrition, leading to yield losses of ∼10–40%. Breeding resistant wheat varieties is an effective and environmentally safe approach to control diseases. However, the sharp eyespot resistance in wheat accessions is partial and quantitative (Cai et al., 2006; Chen et al., 2013). To improve wheat resistance to sharp eyespot, it is vital to identify genes that play important roles in the defense response and unravel their underlying functional mechanisms. However, the complex and huge genome as well as transformation difficulty of common wheat make genetic and functional analyses extremely challenging.
To combat against invading microbial pathogens, plants have evolved a multi-layered immunity system. After plant recognition events, an array of defense mechanisms are activated, which include the generation of a complex signaling network, synthesis of antimicrobial compounds, lignification of cell walls, and expression of pathogenesis-related (PR) proteins or defense genes (Glazebrook, 2005). Frequently, lignins are frequently major structural components of secondary cell walls in vascular plants. They are not only associated with plant growth and development (Rinaldi et al., 2007; Thévenin et al., 2011; Anderson et al., 2015), but also with defense responses to environmental and biotic stresses (Nicholson and Hammerschmidt, 1992; Lange et al., 1995; Schenk et al., 2000; Cheong et al., 2002; Tronchet et al., 2010). Lignification has the potential to act in several ways in plant defense against pathogen infection. It can establish mechanical barriers to pathogen invasion, chemically modify cell walls to be more resistant to cell wall-degrading enzymes, increase the resistance of walls to the diffusion of toxins from the pathogens to the hosts and of nutrients from the hosts to the pathogens, produce toxic precursors and free radicals, and lignify and entrap the pathogens (Nicholson and Hammerschmidt, 1992; Bhuiyan et al., 2009). Unpolymerized monolignols may also have antimicrobial activities (Keen and Littlefield, 1979). However, genetic evidence of CAD function in plant disease resistance is very limited (Tronchet et al., 2010).
Angiosperm lignins are composed of three main subunits (referred to as monolignols) named the hydroxyphenyl (H), guaiacyl (G), and syringyl (S) monolignols. These monolignols are produced with three main branches and 11 enzymes, such as cinnamyl alcohol dehydrogenase (CAD), cinnamoyl CoA reductase (CCR), caffeic acid O-methyltransferase (COMT), and caffeoyl CoA 3-O-methyltransferase (CCOMT) (Anderson et al., 2015). CAD is a key enzyme in monolignol biosynthesis before oxidative polymerization in the cell wall (Baucher et al., 1996; Kim et al., 2004; Bhuiyan et al., 2009). In angiosperm plant species, CAD proteins have significant affinities for coniferyl aldehyde and sinapyl aldehyde (Brill et al., 1999). Numbers of CAD genes in the family have been provided by the complete genome sequencing and annotation of Arabidopsis thaliana, rice (Oryza sativa), and sorghum (Sorghum bicolor) (Sasaki and Sederoff, 2003; Tobias and Chow, 2005; Saballos et al., 2009). In Arabidopsis, nine CAD homologs have identified from the genome sequence database (Sibout et al., 2003), but only three (AtCAD5/CAD-D, AtCAD4/CAD-C, and AtCAD1) have been confirmed to be the important enzymes involved in monolignol biosynthesis (Kim et al., 2004; Sibout et al., 2005; Eudes et al., 2006). In Arabidopsis, through assessment on mutants (cad-C cad-D), the genetic and functional analyses indicate that the CAD-C (At3g19450) and CAD-D (At4g34230) are the primary genes being involved in lignin biosynthesis (Sibout et al., 2005), and act as essential components of defense response against virulent and avirulent strains of the bacterial pathogen Pseudomonas syringae pv. tomato (Tronchet et al., 2010). Recently, 12 and 14 genes belonging to CAD family have been identified in genomes of rice (Tobias and Chow, 2005) and sorghum (Saballos et al., 2009), respectively. The rice OsCAD2 and sorghum SbCAD2 have been shown to be responsible for lignin biosynthesis in rice (Zhang and Cheng, 2006) and sorghum (Saballos et al., 2009; Sattler et al., 2009), respectively. The rice OsCAD7 mutant named flexible culm 1 displays a dramatic reduction in culm mechanical strength (Li et al., 2009). To date, through a genome-wide data mining in wheat EST database, 11 wheat CAD genes, namely TaCAD1 to TaCAD11, have been identified (Ma, 2010). However, only one wheat CAD named TaCAD1 was functionally analyzed through RNA blot and biochemical assay. TaCAD1 has been shown to be a CAD enzyme in wheat stem, and might participate in lodging resistance (Ma, 2010). However, in common wheat, no genetic evidence for defense roles of CAD genes has yet been reported.
In this study, based on comparative transcriptomics between the sharp eyespot-resistant wheat line CI12633 and the susceptible wheat line Wenmai 6 following inoculation with R. cerealis R0301, the probe A_99_P016444, being homologous to certain plant CADs, was identified. Subsequently, this gene, named as TaCAD12, was cloned, and its defense role and the mechanism underlying the function were characterized through TaCAD12-silencing and overexpressing wheat plants. The results of the CAD enzyme kinetic assay revealed that TaCAD12 protein possessed high catalytic efficiencies toward coniferyl aldehyde and sinapyl aldehyde. The functional dissection and preliminary mechanism analyses revealed that TaCAD12 positively contributes to wheat resistance response against the sharp eyespot disease caused by R. cerealis through regulation of the expression of certain defense genes (Defensin, PR10, PR17c, and Chitinase1) and monolignol biosynthesis-related genes (TaCAD1, TaCCR, and TaCOMT1).
Materials and Methods
Plant and Fungal Materials
Six wheat (T. aestivum) lines/cultivars (cv.), including sharp eyespot-resistant lines CI12633 and Shanhongmai, moderately resistant line Niavt14, moderately susceptible lines Yangmai 158 and Yangmai 6, and susceptible line Wenmai 6, were used in this research.
A major Jiangsu virulent strain of the pathogen R. cerealis isolate R0301, was kindly provided by Profs. Huaigu Chen and Shibin Cai (Jiangsu Academy of Agricultural Sciences, China). A North China high-virulence strain of the pathogen R. cerealis isolate WK207 was provided by Prof. Jinfen Yu (Shandong Agricultural University, China).
DNA or RNA Extraction and cDNA Synthesis
Genomic DNA for each sample was isolated from the wheat leaves using the CTAB method (Saghai-Maroof et al., 1984).
Total RNA was extracted using TRIzol (Invitrogen), and then subjected to Rnase-free Dnase I (Promega) digestion and purification.
The purified RNA sample (2 μg) was reverse-transcribed to cDNA using the FastQuant RT Kit with gDNase (Beijing Transgen Biotech, China).
Isolation and Characterization of the TaCAD12 Sequence
The microarray analysis using the Agilent wheat microarray indicated that a probe A_99_P016444, corresponding to 3′-terminal sequence of a wheat cDNA sequence with accession number BT008979, was expressed at a significantly higher level in the sharp eyespot-resistant wheat CI12633 than in the susceptible wheat Wenmai 6. This gene cloned from stems of the resistant wheat CI12633, based on BT008979 sequence, was designated as TaCAD12. The 1393-bp full-length cDNA sequence of TaCAD12 was amplified in 2 rounds of 3′-RACE from cDNA of CI12633 stems inoculated with R. cerealis R0301 for 4 d. The primers for the first round PCR were TaCAD12-U1 and AUAP; these for the second round PCR were TaCAD12-U2 and AUAP.
The deduced protein sequence was analyzed by Pfam database1 and smart software2 to predict conserved motifs. A phylogenetic tree was constructed using a neighbor-joining method implemented with MEGA 5.0 software.
CAD Enzyme Kinetic Assay of TaCAD12 In Vitro
The ORF (open reading frame) sequence of TaCAD12 was sub-cloned in frame to 3′-terminus of a GST (glutathione S-transferase) gene in the pGEX-4T-1 vector (GE Amersham), resulting in GST-TaCAD12 expression vector pGST-TaCAD12. The pGST-TaCAD12 DNA was transformed into competent cells of Escherichia coli (E. coli) strain BL21. The GST-TaCAD12 overexpression in the E. coli cells and the recombinant protein purification were conducted according to Dong et al. (2010).
The CAD enzyme activity of TaCAD12 was measured at OD340 according to the method of Goffner et al. (1992). Each reaction was monitored ten times with 1 min intervals. Following Ma (2010) and Saathoff et al. (2011), both Vmax and Km values were determined by extrapolation from the Lineweaver–Burk plots. The reactions were started by enzyme addition and terminated by holding at 85°C for 10 min. The final reaction mixture during the analysis of the reaction mechanism catalyzed by TaCAD12 contained 50 mM NADPH, 20 mM coniferyl aldehyde or sinapyl aldehyde, and 0.5 μg purified GST-TaCAD12 recombinant protein. Each reaction was prepared with two-component mixture and the third component were added later. Assay without NADPH was used as control.
Virus-Induced Gene Silencing (VIGS) Assay for TaCAD12 Function
In barley and wheat, the barley stripe mosaic virus (BSMV)-based VIGS (virus-induced gene silencing) assay has been shown to be an effective reverse genetic tool for rapidly investigating the functions of interest genes (Holzberg et al., 2002; Scofield et al., 2005; Zhou et al., 2007; Zhu et al., 2014). To generate the BSMV:TaCAD12 recombinant construct, a 190-bp sequence of TaCAD12 (from 1011 to 1200 nucleotides in TaCAD12 cDNA sequence) was sub-cloned in an antisense orientation into the Nhe I restriction site of the RNAγ of BSMV (Supplementary Figure S2). Following the protocols described by Holzberg et al. (2002) and Zhou et al. (2007), the tripartite cDNA chains of BSMV:TaCAD12 or the control BSMV:GFP virus genome were separately transcribed into RNAs, mixed, and used to inoculate the leaves of wheat CI12633 plants at the two-leaf stage. Then, the plants were grown in a 14 h light (25°C)/10 h dark (17°C) regime. To investigate if BSMV successfully infected CI12633 plants, and to test if TaCAD12 transcript was down-regulated, at 8 days after the virus inoculation, the fourth leaves of the inoculated seedlings were collected and subjected to quantitative real-time PCR (Q-RT-PCR) to analyze the transcription of the BSMV coat protein (CP) gene and the transcription of TaCAD12. At 25 days after BSMV infection, the BSMV-infected CI12633 plants were further inoculated with R. cerealis isolate WK207 mycelia following Chen et al. (2008). They were scored at 10 and 40 dpi with R. cerealis WK207, respectively.
TaCAD12-Overexpressing Construct and Transformation into Wheat
The TaCAD12 ORF sequence with the Spe I and Sac I restriction sites was amplified with the primers TaCAD12-SPEI-U and TaCAD12-SACI-L and then sub-cloned into the Spe I and Sac I sites of a modified monocot transformation vector pAHC20-RSS1P (Christensen and Quail, 1996; Li et al., 2011) with a c-myc epitope tag. In the resulting overexpression transformation vector pRSS1P:myc-TaCAD12 (Figure 5A), the transcript of the c-myc-TaCAD12 fusion gene is driven by a rice sucrose synthase-1 (RSS1) promoter (GenBank accession no. X64770.1) that was reported to be specifically expressing in the phloem tissue (Shi et al., 1994), and terminated by 3′-non-transcribed region of Agrobacterium tumefaciens nopaline synthase gene (Tnos). A total of 2,000 immature embryos of the wheat cultivar Yangmai 16 were transformed by biolistic bombardment using pRSS1P:myc-TaCAD12 plasmid DNA-contained golden powder following the protocol described by Chen et al. (2008).
PCR and Western Blotting Analyses on TaCAD12-Overexpressing Wheat
The presence of the TaCAD12-overexpressing transgene in the transformed wheat plants was monitored by PCR using the specific primers, TaCAD12-4166-ZJF (located in TaCAD12 coding sequence) and Tnos-3214L (located in Tnos of the transformation vector). PCR was performed in a 25 μl volume containing ∼200 ng genomic DNA, 12.5 μl 2x Taq MasterMix (Beijing ComWin Biotech Co. Ltd, China), 1 μl each primer (10 mM). The amplified product (277-bp size) specific to the introduced TaCAD12-Tnos chimera was resolved on a 1.2% agarose gel and visualized by ethidium bromide staining.
The c-myc-TaCAD12 fusion protein in the overexpressing wheat lines was visualized by Western blotting analysis. Total proteins were extracted from ∼0.5 g stems inoculated with R. cerealis R0301 for 40 days by using the tissue protein extraction kit (Beijing ComWin Biotech Co. Ltd, China). Total soluble proteins (∼10 μg) for each line were separated on 12% SDS-PAGE and transferred to polyvinyl difluoride membranes (Amersham). The blotting membranes were incubated with 2500-fold diluted Anti-c-myc Mouse Monoclonal Antibody (Beijing Transgen Biotech, China) at 4°C overnight, then incubated with 1500-fold diluted Goat Anti-Mouse IgG (H+L), HPR conjugated secondary antibody (Beijing Transgen Biotech, China) at 22-23°C for 1 h. The c-myc-TaCAD12 protein was visualized using the Pro-light HRP Chemiluminescent Kit (Beijing Transgen Biotech, China).
CAD Kinetic Activity and Western Blot Analyses of TaCAD12 in Wheat Stems
Total proteins in TaCAD12-overexpressing and wild-type (WT, non-transgenic) wheat Yangmai 16 (recipient) lines were extracted from ∼0.5 g stems inoculated with R. cerealis R0301 for 40 days as mentioned above. The CAD enzyme kinetic activities in both TaCAD12-overexpressing and WT Yangmai 16 wheat lines were examined in the soluble protein fraction from their stem tissues according to the methods described by Goffner et al. (1992) and Ma (2010).
The Western blot analysis was performed to examine TaCAD12 protein in wheat stems with GST-TaCAD12 antibody. Briefly, the total proteins were extracted from stems of TaCAD12-overexpressing or WT Yangmai 16 wheat lines inoculated with R. cerealis R0301 for 40 days, then separated on 12% SDS-PAGE, and transferred to polyvinyl difluoride membranes (Amersham). The GST-TaCAD12 polyclonal antibody was raised in mouse injected by purified GST-TaCAD12 protein. The immunoblots were developed with the polyclonal GST-TaCAD12 antibody (1:1500), then 1500-fold diluted Goat Anti-Mouse IgG (H+L), HPR conjugated secondary antibody, and visualized using the Pro-light HRP Chemiluminescent Kit (Beijing Transgen Biotech, China).
Assessment on Responses of Transgenic Wheat Plants to R. cerealis
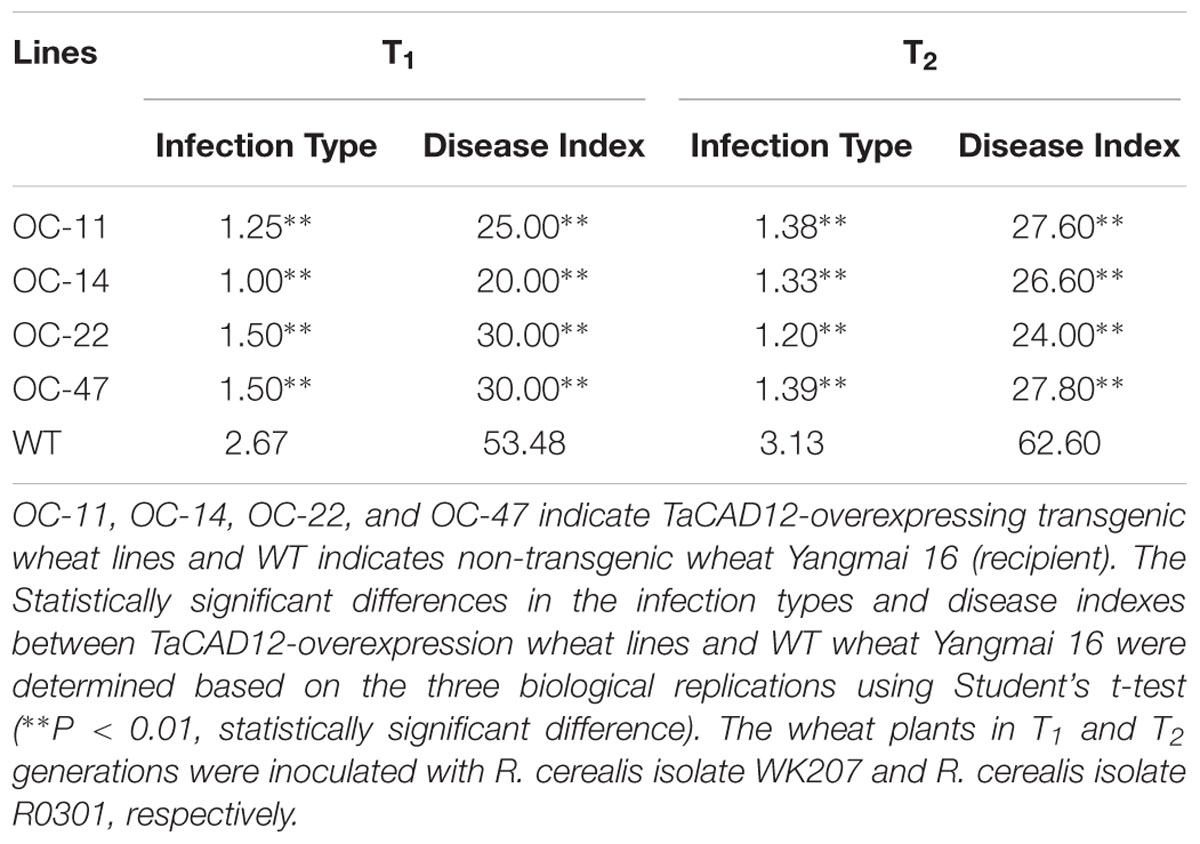
At least 10 plants for each line of the TaCAD12-overexpressing wheat lines in T1–T2 generations and WT wheat Yangmai 16 were inoculated with sterilized grains harboring the well-developed mycelia of R. cerealis isolates WK207 or R0301 following the protocol of Wei et al. (2015). Based on the disease lesion squares, the infection type (IT) of each plant and disease index (DI) for each wheat line were categorized at harvest according to Cai et al. (2006).
Analysis on Transcriptional Levels of Target Genes by Q-RT-PCR
Q-RT-PCR analysis with TaCAD12 specific primers TaCAD12-Q-265F265F and TaCAD12-Q-557R was used to investigate the relative transcriptional levels of TaCAD12 in various wheat plants. The tested wheat defense-marker genes include Defensin (NCBI accession no. CA630387), PR10 (NCBI accession no. CA613496), PR17c (NCBI accession no. TA65181), and chitinase1 (Chit1, NCBI accession no. CA665185). The tested wheat monolignol biosynthesis-related genes include TaCAD1 (NCBI accession no. GU563724), TaCCR (NCBI accession no. DQ449508), and TaCOMT1 (NCBI accession no. AY226581). Q-RT-PCR was performed using SYBR Green I Master Mix (TaKaRa) in a volume of 25 μl on an ABI 7300 RT-PCR system (Applied Biosystems). Reactions were set up using the following thermal cycling profile: 95°C for 15 min, followed by 41 cycles of 95°C for 10 s, 55°C for 20 s, and 72°C for 32 s. The relative transcriptional levels of the target genes was calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001), where the wheat Actin gene TaActin was used as the internal reference. The relative transcriptional levels of the tested genes in the TaCAD12-overexpressing wheat lines or in BSMV:TaCAD12-infected wheat plants were relative to those in WT recipient or in BSMV:GFP-infected wheat plants.
Primers
All primers used in this study are listed in Supplementary Table S1.
Results
TaCAD12 Transcriptional Level Is Higher and More Enhanced by R. cerealis in the Resistant Wheat Lines
Microarray-based comparative transcriptomic assay was used to identify defense-related genes of wheat in response to R. cerealis infection. Among the differentially expressed probes between sharp eyespot-resistant wheat line CI12633 and the susceptible wheat line Wenmai 6 at 4 and 21 day post inoculation (dpi) with R. cerealis R0301 (microarray raw data, GEO accession number GSE69245), the probe A_99_P016444, corresponding to 3′-terminal sequence of the wheat full insert mRNA sequence BT008979, showed 3.90-fold and 21.56-fold transcriptional increase in resistant wheat line CI12633 than in susceptible wheat line Wenmai 6 at 4 and 21 dpi with R. cerealis R0301, respectively. Blast search against the nucleotide acid sequence database in GenBank indicated that the sequence of BT008979 hit to Lolium perenne CAD2 mRNA (AF472592.1, sharing 89% identity), and Brachypodium distachyon putative CAD5 mRNA (XM_003573500.2, 88% identity). Further sequence alignment showed that the sequence of BT008979 is homologous to the previously reported wheat TaCAD3 gene (TC143265, 89.11% identity), but less than 49.11% identity with TaCAD1-TaCAD2 and TaCAD4-TaCAD11 (Ma, 2010). Thus, this gene, which was cloned from the resistant wheat line CI12633 based on BT008979, was designated as TaCAD12. Q-RT-PCR analysis results showed that transcriptional level of TaCAD12 was higher in the resistant wheat line CI12633 stems than in susceptible wheat line Wenmai 6 stems with R. cerealis R0301 inoculation or mock-inoculation without R. cerealis (Figure 1A). The Q-RT-PCR results were in agreement with the microarray analysis trend. The transcriptional level of TaCAD12 was markedly increased in the resistant wheat line CI12633 after R. cerealis R0301 inoculation for 4 and 21 days, whereas the transcriptional induction in susceptible wheat line Wenmai 6 was relatively weaker (Figure 1A). Moreover, following R. cerealis R0301 inoculation, TaCAD12 transcriptional levels were significantly higher in three sharp eyespot-resistant wheat lines (CI12633, Shanghongmai, and Niavt14) than in two susceptible lines (Wenmai 6 and Yangmai 158) (Figure 1B). Additionally, the tissue expression analysis showed that the transcriptional level of TaCAD12 was the highest in roots, intermediate in stems, and the lowest in the leaves of the wheat line Yangmai 16 at both 4 and 10 dpi with R. cerealis R0301 (Figure 1C). These results suggested that TaCAD12 might participate in wheat defense response to R. cerealis infection.
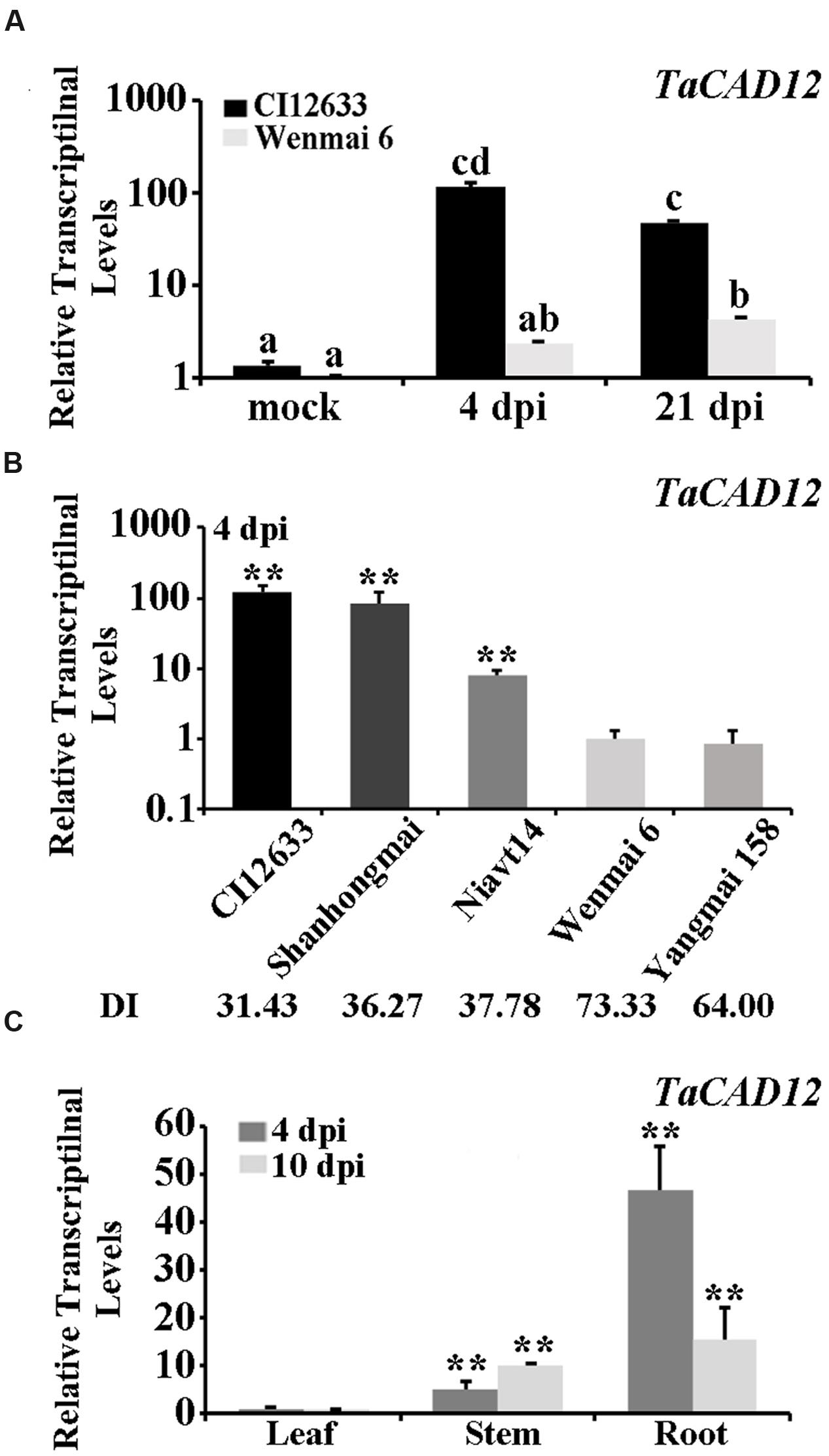
FIGURE 1. Transcriptional patterns of TaCAD12 in wheat stems after R. cerealis inoculation analyzed by Q-RT-PCR. (A) The relative transcriptional levels of TaCAD12 in the sharp eyespot-resistant wheat line CI12633 and the susceptible wheat line Wenmai 6 after inoculation with R. cerealis R0301 for 4 and 21 days or mock-treatment. The mock indicates an inoculation without R. cerealis. The transcriptional levels with different letters are significantly different from each other based on statistical comparisons (Student’s t-test, ∗∗P < 0.01). (B) The relative transcriptional levels of TaCAD12 in sharp eyespot-resistant, moderately resistant, susceptible, and moderately susceptible wheat lines were measured after inoculation with R. cerealis R0301 for 4 days. The relative transcriptional levels of TaCAD12 in different wheat lines were calculated relative to that of Wenmai 6. DI indicates the sharp eyespot disease index in each wheat line. (C) Transcription of TaCAD12 in roots, stems and leaves of the wild-type (WT) Yangmai 16 at 4 and 10 dpi with R. cerealis R0301. Three technical replicates were averaged and statistically analyzed using Student’s t-test (∗∗P < 0.01). Bars indicate standard error of the mean (SEM).
TaCAD12 Encodes a Cinnamyl Alcohol Dehydrogenase
The full-length cDNA sequence of TaCAD12 was cloned from cDNA of CI12633 stems inoculated with R. cerealis R0301, and was deposited in GenBank with the accession no. KX585233. The TaCAD12 cDNA sequence with 1393-bp contains an ORF with 1116-bp (from 79 to 1194 nt), the 5′-untranslated region (UTR) with 78-bp, and 3′-UTR with 200-bp (Figure 2). The TaCAD12 cDNA sequence shares an 87.53% identity with the previously reported wheat TaCAD3 gene (TC143265). The predicted protein of TaCAD12 is consisted of 371 amino acids with a molecular weight of 39.43 kDa and a theoretical iso-electric point of 7.39. As shown in Figure 2, the TaCAD12 protein contains one alcohol dehydrogenase domain (42–157 aa) with two Zn binding sites (41–63 and 76–90 aa) and one Zinc-binding dehydrogenase domain (199–323 aa) with one highly conserved motif (196GLGGLG201) in CAD. The motif GLGGLG has been shown to participate in binding the pyrophosphate group of NADP+ (Youn et al., 2006).
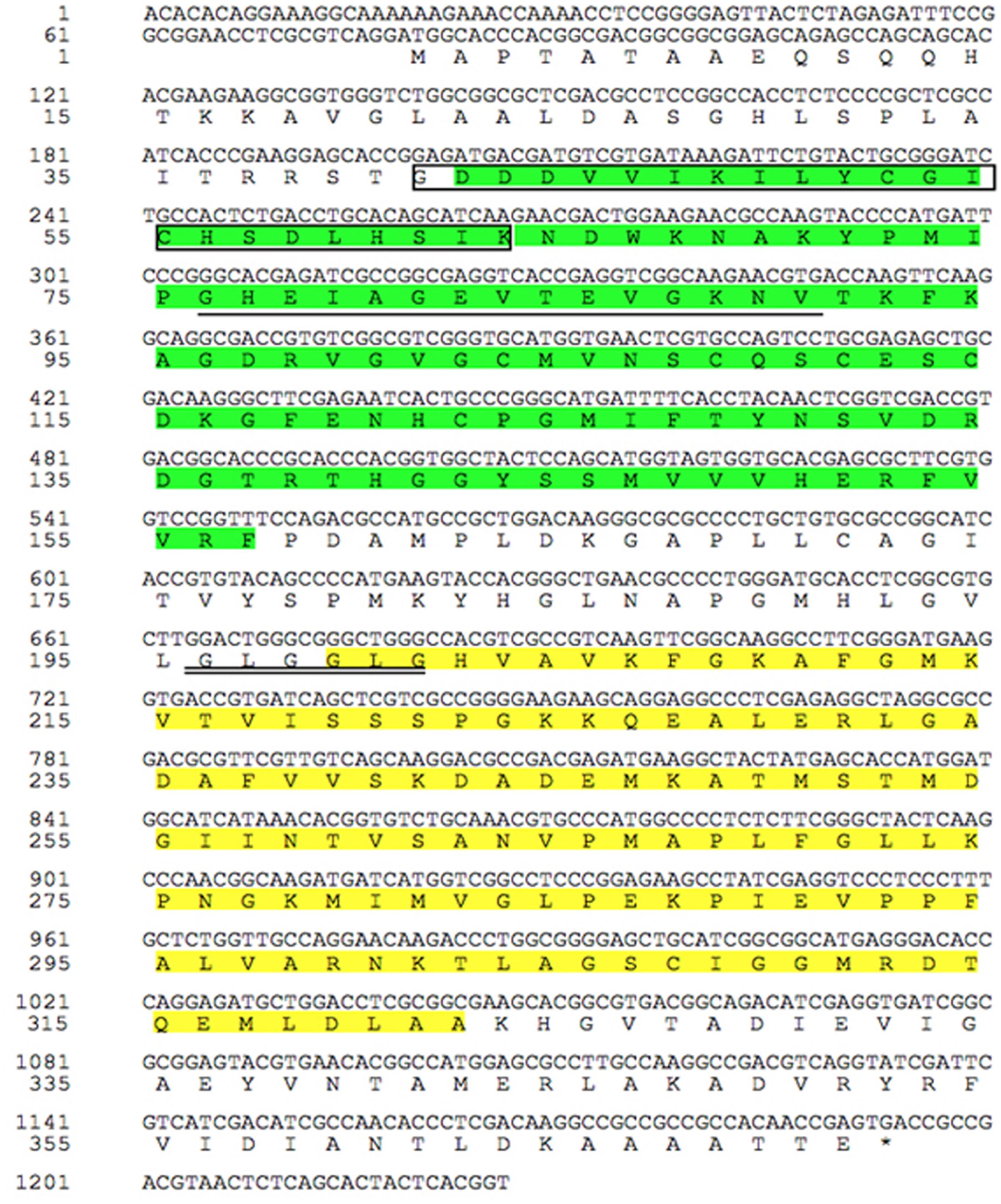
FIGURE 2. The analyses of nucleotide sequence and deduced amino acid sequence of TaCAD12. The green part represents alcohol dehydrogenase domain (42–157 aa), in which the amino acid sequence in box represents the first Zn binding site in TaCAD12, and the amino acid sequence with single underline is the second Zn binding site. The Zinc-binding dehydrogenase domain (199–323 aa) is marked by yellow spaces, in which the highly conserved motif in CAD is marked with double underlines (196–201 aa).
Following the classification method announced by Saballos et al. (2009), a phylogenetic tree, including 12 wheat CAD proteins and 35 CADs from other plant species, was constructed and divided into six groups (Figure 3). The proteins TaCAD12, TaCAD3, BdCAD5, LpCAD2, and OsCAD7 belonged to IV Group of CAD family (Figure 3). The protein sequence alignment indicated that the protein sequence of TaCAD12 shares 83.79, 88.44, 76.16, and 67.62% identities with TaCAD3, BdCAD5, LpCAD2, and OsCAD7, respectively. The functions of TaCAD3, BdCAD5, and LpCAD2 have not been studied yet. The OsCAD7 is confirmed to have strong activity toward coniferyl aldehyde and weak activity toward sinapyl aldehyde, and controls culm mechanical strength in rice (Li et al., 2009). Additionally, the protein sequence of TaCAD12 shares 47.81 and 47.39% identities with PviCAD1 and PviCAD2, two reported CADs in switchgrass (Panicum Virgatum L.), respectively (Saathoff et al., 2011, 2012). These results suggested that TaCAD12 encoded a CAD.
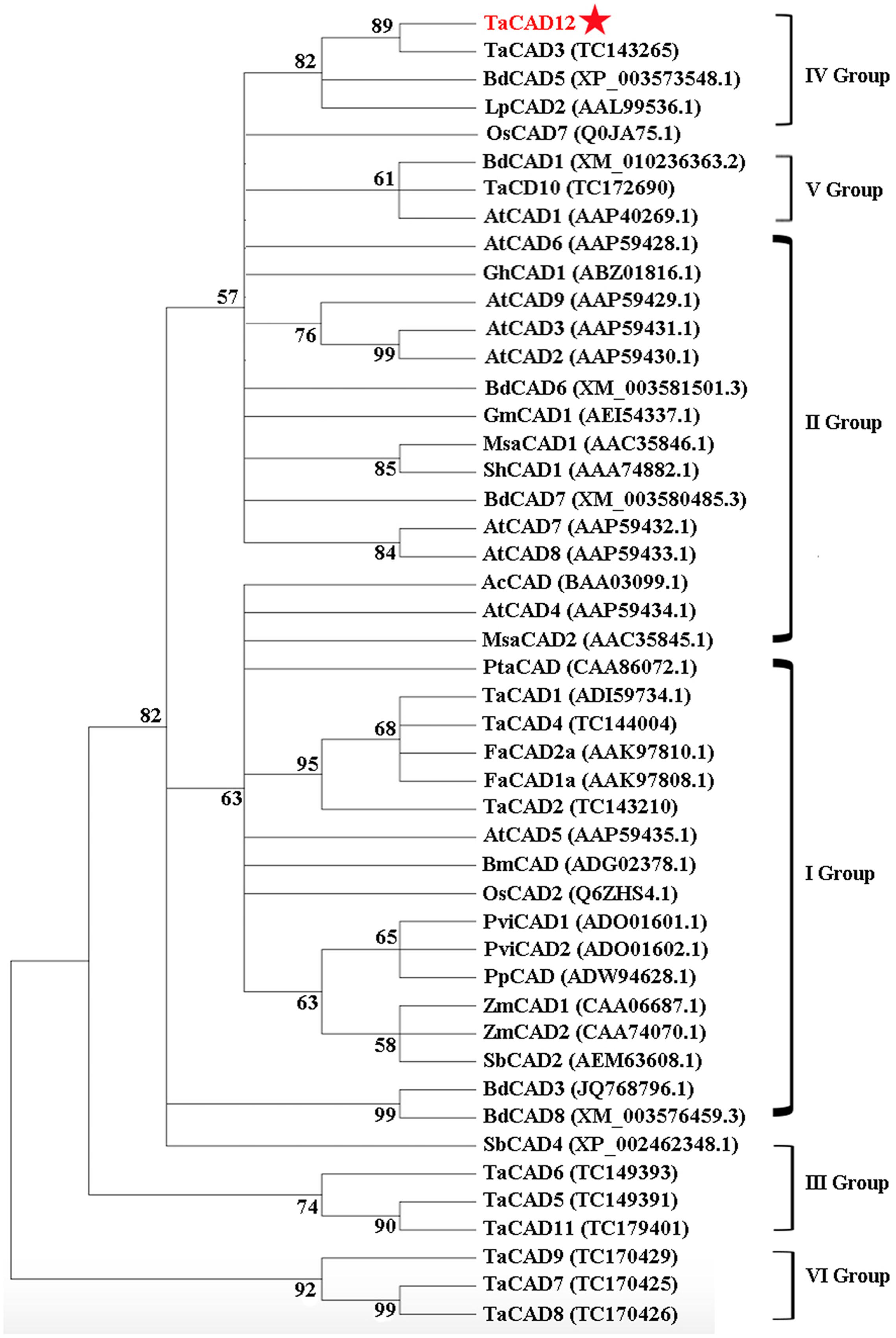
FIGURE 3. Phylogenetic relationships of TaCAD12 and 46 other CAD proteins in plants. The phylogenetic tree was constructed by using neighbor-joining phylogeny of MEGA 5.0 following ClustalW method, and is showed in bootstrapped manner. The GenBank accession number or TIGR number of each protein was marked in the bracket behind the corresponding proteins.
TaCAD12 Possesses Catalytic Activities toward Coniferyl Aldehyde and Sinapyl Aldehyde
To investigate if TaCAD12 has activities toward coniferyl aldehyde and sinapyl aldehyde, we analyzed the kinetics of recombinant GST-TaCAD12 protein toward two putative substrates (coniferyl aldehyde and sinapyl aldehyde). The catalytic properties of TaCAD12 were labeled by Vmax and Km values. The Vmax values of the GST-TaCAD12 protein toward coniferyl aldehyde and sinapyl aldehyde are 347.58 ± 17.38 and 315.01 ± 18.58 nkat mg-1 protein, respectively. These results indicated that TaCAD12 protein showed high catalytic efficiencies toward both coniferyl aldehyde and sinaphy aldehyde, even though a slightly higher efficiency was displayed for catalyzing coniferyl aldehyde. Meanwhile, the Km values of the GST-TaCAD12 protein toward coniferyl aldehyde and sinapyl aldehyde were 28.91 ± 3.67 and 34.18 ± 5.56 μM, respectively. Additionally, as shown in Supplementary Figure S1, the catalytic reactions of GST-TaCAD12 protein toward both coniferyl aldehyde and sinapyl aldehyde happened immediately, regardless of the component adding order. These results prove that TaCAD12 is an authentic CAD enzyme, and possesses catalytic activities toward both coniferyl aldehyde and sinapyl aldehyde.
Down-Regulation of TaCAD12 Represses Wheat Resistance to Sharp Eyespot
When the sharp eyespot-resistant wheat CI12633 plants had been infected with BSMV for 8 days, BSMV-infected symptom was present in both BSMV:GFP- and BSMV:TaCAD12-inoculated CI12633 plants (Figure 4A), and the expression of BSMV CP gene could be detected from stems of these plants (Figure 4B). The results indicated that these BSMV:GFP and BSMV:TaCAD12 viruses successfully infected these viruses-inoculated wheat plants. The TaCAD12 transcriptional level was substantially reduced in BSMV:TaCAD12-infected CI12633 plants, indicating that TaCAD12 was successfully knock-downed in BSMV:TaCAD12-infected CI12633 plants (Figure 4C). Then, these plants were further inoculated with R. cerealis isolate WK207 to evaluate the defense role of TaCAD12. At 10 dpi with R. cerealis WK207, the sharp eyespot symptom (brown lesion) was present on the fungal inoculated sheaths and stems of BSMV:TaCAD12-infected CI12633 plants, but absent on the fungal inoculated stems of BSMV:GFP-treated CI12633 plants (Figure 4D). At 40 dpi with R. cerealis WK207, more serious symptoms of sharp eyespot were present on the stems of BSMV:TaCAD12-infected CI12633 plants (average IT: 3.33), compared with those on the stems of BSMV:GFP-treated CI12633 plants (average IT: 1.46) (Figure 4D). These results indicated that the down-regulation of TaCAD12 transcript in CI12633 impaired host resistance to sharp eyespot caused by R. cerealis, and that TaCAD12 seems to be essential to the wheat resistance against R. cerealis infection.
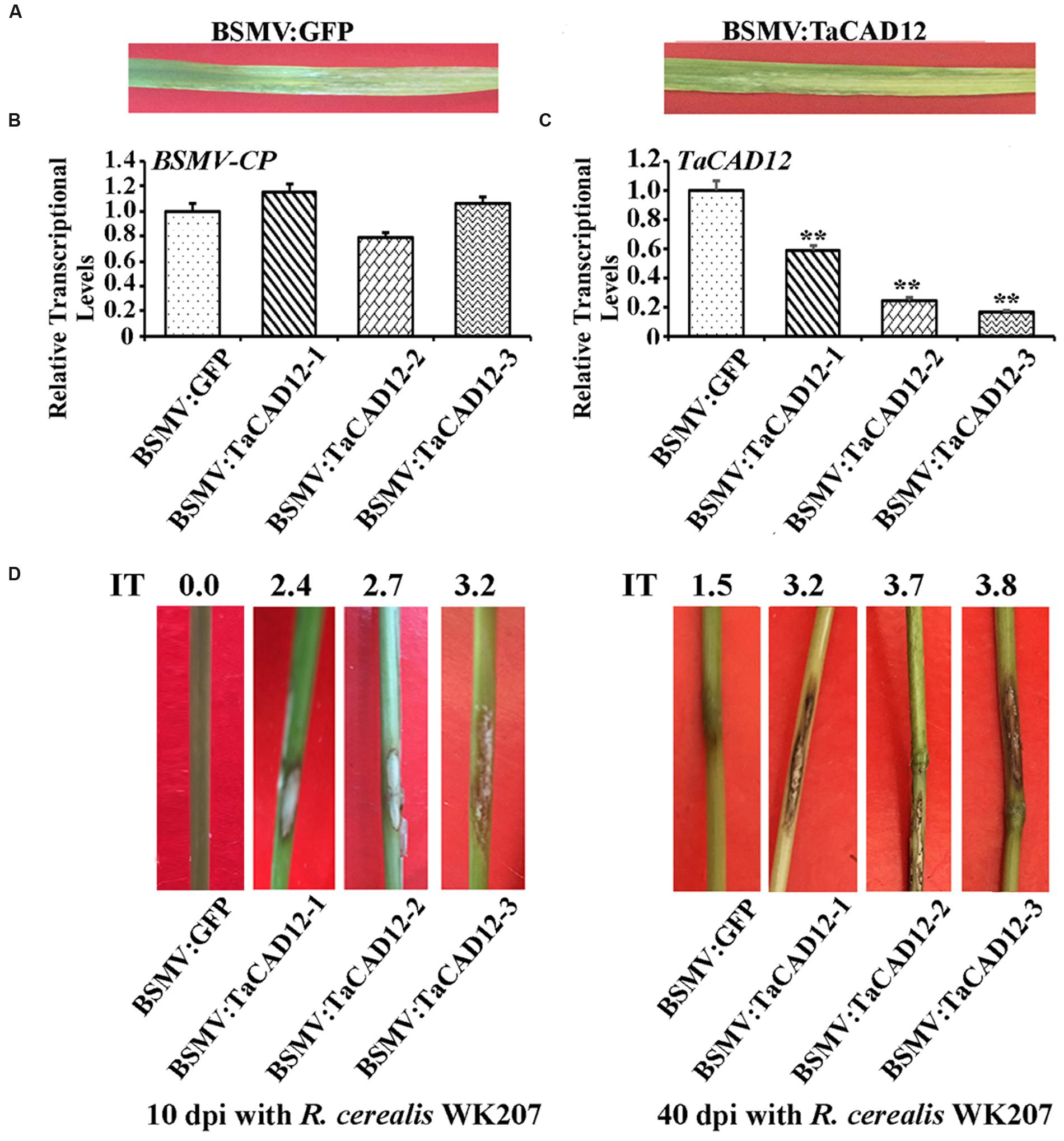
FIGURE 4. Molecular characterizations and R. cerealis responses of TaCAD12-silencing CI12633 plants. (A) BSMV symptoms on the leaves of BSMV:GFP- and BSMV:TaCAD12-infected wheat CI12633 plants. (B) Transcriptional levels of BSMV coat protein (CP) gene in the leaves of the BSMV:GFP- and BSMV:TaCAD12-infected wheat CI12633 plants at 8 days post virus inoculation. (C) Transcriptional levels of TaCAD12 in the leaves of the BSMV:GFP- and BSMV:TaCAD12-infected wheat CI12633 plants at 8 days post virus inoculation. Statistically significant differences between BSMV:TaCAD12-inoculated plants and BSMV:GFP-inoculated plants at the same time point based on three replications using Student’s t-test (∗∗P < 0.01). (D) Sharp eyespot symptoms of BSMV:GFP- and BSMV:TaCAD12-infected wheat CI12633 plants following R. cerealis WK207 inoculation for 10 and 40 days. IT indicates sharp eyespot infection type in each wheat line.
Overexpression of TaCAD12 Increases Wheat Resistance to Sharp Eyespot
The role of TaCAD12 in resistance response to R. cerealis was further studied by developing and evaluating TaCAD12-overexpressing wheat lines. By using the primers specific to the overexpression transformation vector pRSS1P:myc-TaCAD12 (Figure 5A), PCR analysis results showed that the introduced TaCAD12 transgene was present in four wheat lines (OC-11, OC-14, OC-22, and OC-47) in T0–T2 generations (Figure 5B), suggesting that the transgene could be inheritable in these four lines. Q-RT-PCR analyses indicated that the transcriptional levels of TaCAD12 in these four transgenic lines (OC-11, OC-14, OC-22, and OC-47) were markedly elevated than in WT Yangmai 16 (recipient) (Figure 5C), and the introduced TaCAD12 was overexpressed in these four lines. The Western blotting results proved that the introduced myc-TaCAD12 gene was translated into the fusion protein in these four overexpressing lines, but not in WT Yangmai 16 (Figure 5D). The sharp eyespot severity assessments in two successive (T1–T2) generations showed that compared with WT Yangmai 16, all these four TaCAD12-overexpressing wheat lines (OC-11, OC-14, OC-22, and OC-47) displayed significantly enhanced resistance to sharp eyespot caused by both R. cerealis R0301 and R. cerealis WK207 (Table 1; Figure 5E). For example, the ITs and DIs of these four TaCAD12-overexpressing lines in T2 generation were 1.20–1.39 and 24.00–27.80, respectively, whereas the average IT and DI of WT Yangmai 16 were 3.13 and 62.60, respectively (Table 1). These results revealed that TaCAD12 positively contributed to wheat resistance against sharp eyespot caused by R. cerealis infection.
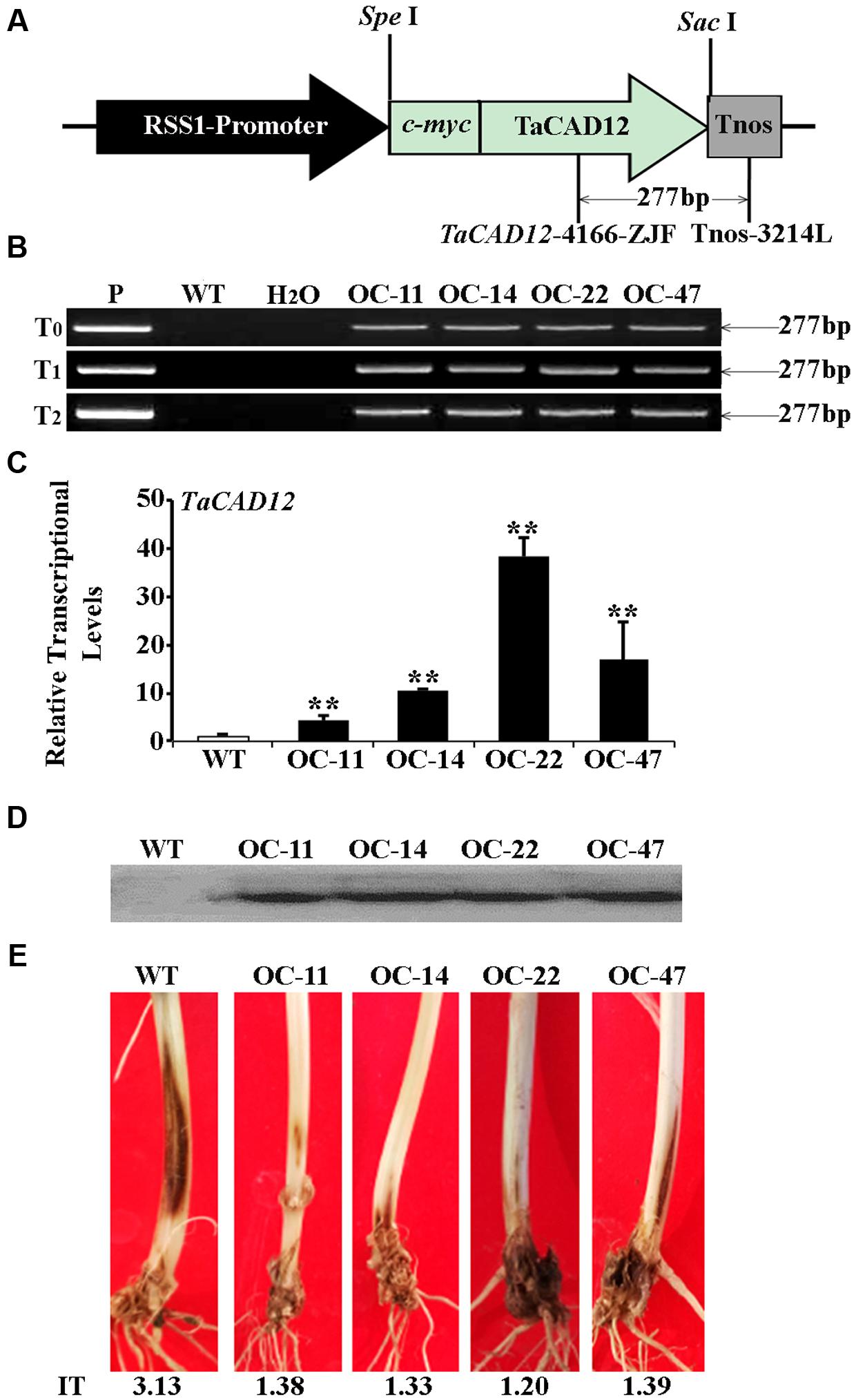
FIGURE 5. Molecular characterizations and R. cerealis responses of TaCAD12-overexpressing wheat lines. (A) Schema of the transformation vector pRSS1P:myc-TaCAD12. TaCAD12 in the transformation vector was driven by the rice sucrose synthase-1 promoter (RSS1P) and terminated by Tnos. The arrows region indicates the 277-bp fragment amplified in PCR assays using transgene-specific primers (TaCAD12-4166-ZJF/Tnos-3214L). (B) PCR patterns of TaCAD12-overexpressing transgenic lines in T0–T2 generations and WT (non-transgenic) Yangmai 16 using transgene-specific primers. P: transformed vector pRSS1P:myc-TaCAD12, WT: WT Yangmai 16, M: 100-bp DNA ladder. (C) The transcriptional levels of TaCAD12 in the stems of T2 TaCAD12-overexpressing lines and WT Yangmai 16 through Q-RT-PCR analysis. The transcriptional levels in the transgenic lines (OC-11, OC-14, OC-22, and OC-47) were quantified relative to that in WT Yangmai 16. (D) Western blot pattern in stems of these four TaCAD12-overexpressing lines and WT Yangmai 16 was probed with an anti-c-myc antibody. (E) Sharp eyespot symptoms of the TaCAD12-overexpressing lines and WT Yangmai 16 in16 in T2 generation after the 40 dpi with R. cerealis R0301. IT indicates sharp eyespot infection type in each wheat line.
Furthermore, we examined the enzyme catalytic activities toward coniferyl aldehyde and sinapyl aldehyde (Figure 6A) and the TaCAD12 protein levels (Figure 6B) in the stem tissues of both TaCAD12-overexpressing and WT Yangmai 16 wheat lines. The enzyme catalytic activity toward coniferyl aldehyde in these four TaCAD12-overexpressing wheat lines (OC-11, OC-14, OC-22, and OC-47) was significantly higher than in WT Yangmai 16 (Figure 6A). The catalytic activity toward sinapyl aldehyde was slightly higher in these four TaCAD12-overexpressing wheat lines (OC-11, OC-14, OC-22, and OC-47) than in WT wheat Yangmai 16 (Figure 6A), although the difference was not significant. Western blotting analysis showed that with the polyclonal GST-TACAD12 antibody, a single band was detected only in the stem tissues of these four TaCAD12-overexpressing wheat lines (OC-11, OC-14, OC-22, and OC-47), but not in WT Yangmai 16 (Figure 6B). These results indicated that compared with WT Yangmai 16, these four TaCAD12-overexpressing wheat lines (OC-11, OC-14, OC-22, and OC-47) possessed both higher TaCAD12 protein levels and higher catalytic efficiency of CAD.
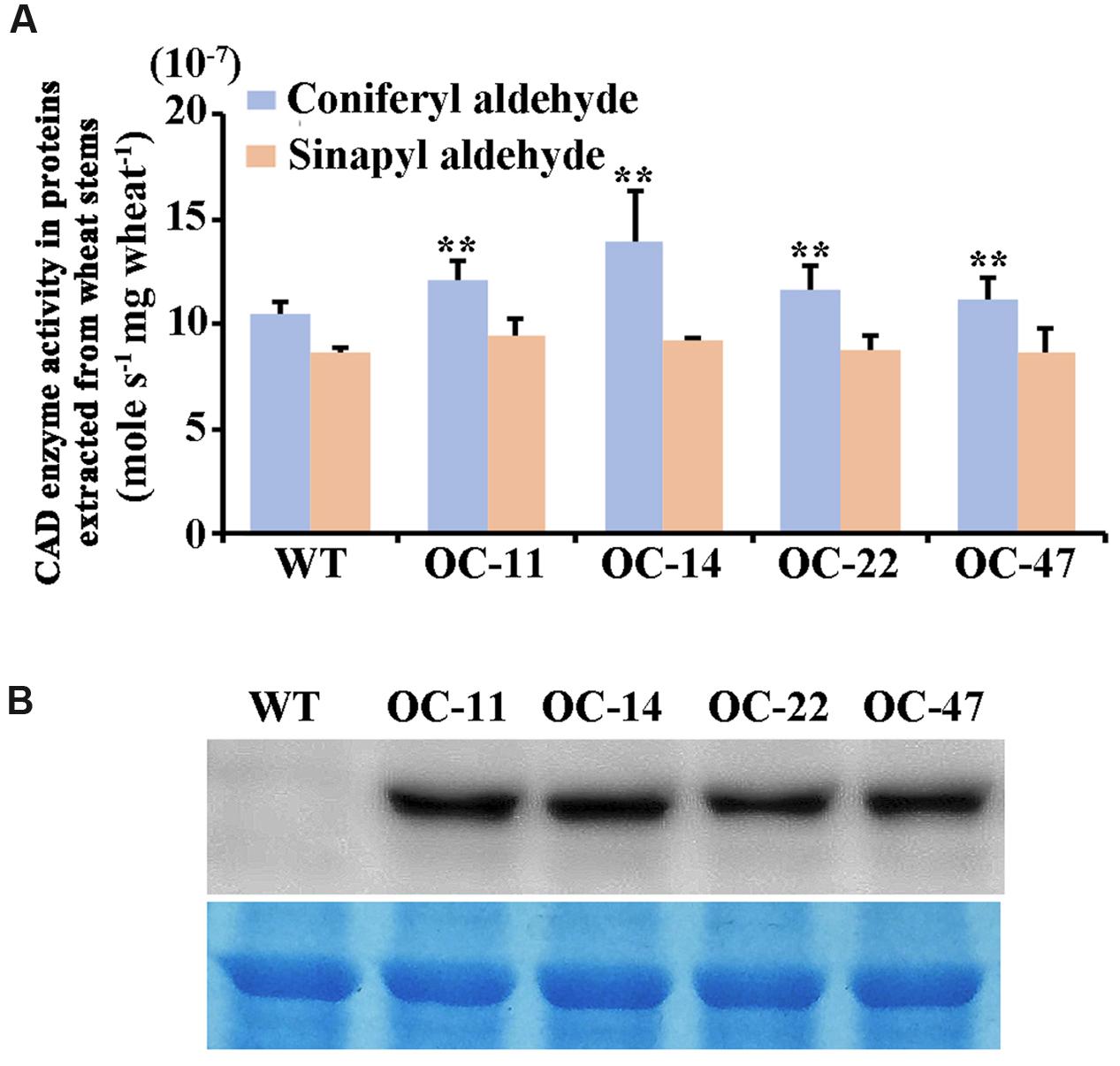
FIGURE 6. CAD kinetic activity and Western blot analyses of TaCAD12 in wheat stems. (A) Comparison of catalytic activities (mole s-1 mg wheat-1) toward both coniferyl aldehyde and sinapyl aldehyde in TaCAD12-overexpressing and non-transgenic Yangmai 16 wheat lines. Proteins were extracted from the stem tissues of TaCAD12-overexpressing and WT Yangmai 16 wheat lines after inoculated with R. cerealis R0301 for 40 d. Statistically significant differences between TaCAD12-overexpressing lines and WT Yangmai 16 were determined based on three replications using Student’s t-test (∗∗P < 0.01). (B) Western blot analysis of TaCAD12 protein from stem tissues of TaCAD12-overexpressing and WT Yangmai 16 wheat lines after 40 dpi with R. cerealis R0301. The blot was probed with the polyclonal GST-TACAD12 antibody (upper panel), and equal loading of protein samples was shown by coomassie brilliant blue staining (lower panel).
TaCAD12 Positively Regulates the Expression of Certain Defense- and Monolignol Biosynthesis-Related Genes
In plants, defense genes play important roles in the resistance to pathogens. The monolignol biosynthesis-related genes (CAD, CCR, and COMT) may also participate in pathogen-resistance (Bhuiyan et al., 2009; Tronchet et al., 2010). To explore if TaCAD12 regulates defense and monolignol biosynthesis-related genes in the resistance response to R. cerealis infection, we investigated the transcription changes of certain defense and monolignol biosynthesis-related genes (Defensin, PR10, PR17c, chitinase1, TaCAD1, TaCCR, and TaCOMT1) in TaCAD12-overexpressing wheat plants and TaCAD12-silencing wheat plants as well as their control plants. After 40 dpi with R. cerealis R0301, the transcriptional levels of these tested genes were significantly increased in enhanced-resistant TaCAD12-overexpressing wheat lines than in the susceptible WT Yangmai 16 plants (Figure 7), suggesting that the overexpression of TaCAD12 up-regulated the transcriptional levels of these defense and monolignol biosynthesis-related genes. On the contrary, the transcriptional levels of these tested genes were significantly decreased in reduced-resistant TaCAD12-silencing wheat plants than in the BSMV:GFP-infected CI12633 plants after inoculation with R. cerealis WK207 for 40 days (Figure 8). These results indicated that TaCAD12 positively regulated the expression of these defense and monolignol biosynthesis-related genes in wheat.
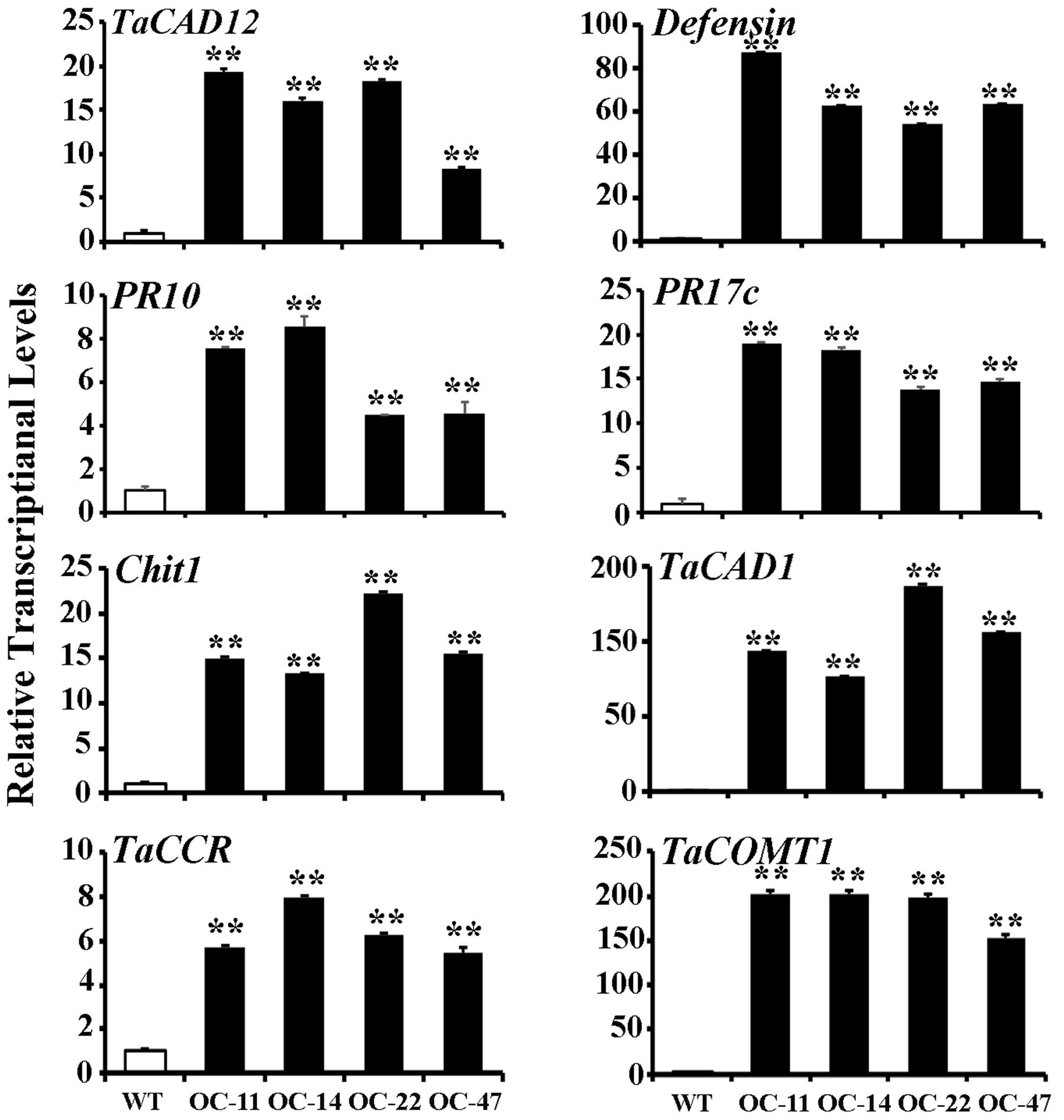
FIGURE 7. Transcriptional levels of TaCAD12, defense, and monolignol biosynthesis-related genes in TaCAD12-overexpressing and WT Yangmai 16 wheat lines after R. cerealis inoculation. Relative transcriptional abundances of the tested genes (TaCAD12, Defensin, PR10, PR17c, Chit1, TaCAD1, TaCCR, and TaCOMT1) in TaCAD12-overexpressing wheat lines (OC-11, OC-14, OC-22, and OC-47) were quantified relative to that in WT Yangmai 16 at 40 dpi with R. cerealis R0301. Statistically significant differences between TaCAD12-overexpressing lines and WT Yangmai 16 were determined based on three replications using Student’s t-test (∗∗P < 0.01).
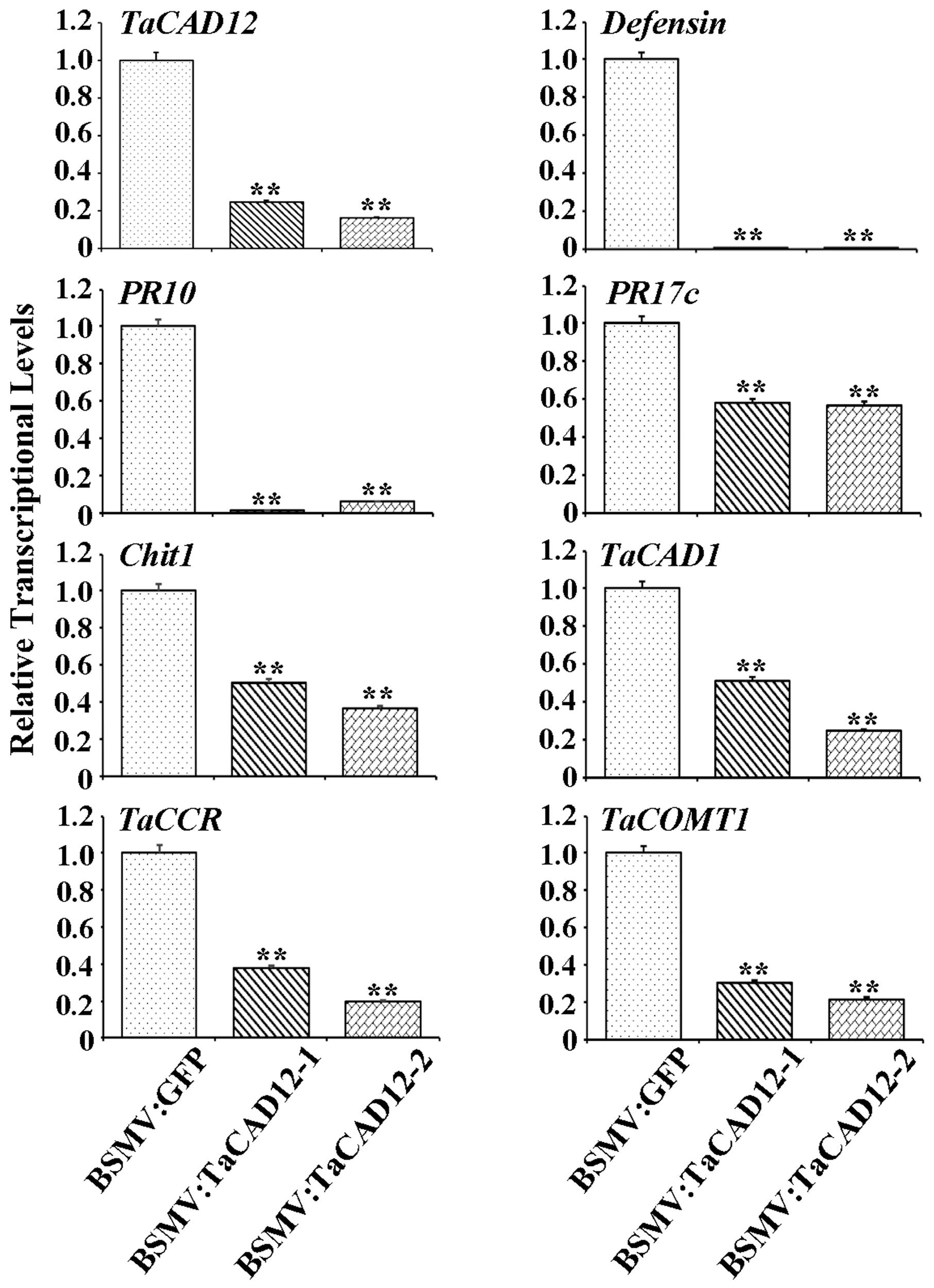
FIGURE 8. The transcriptional levels of TaCAD12, defense, and monolignol biosynthesis-related genes in BSMV:TaCAD12- and BSMV:GFP-infected CI12633 plants after R. cerealis inoculation. Relative transcriptional abundances of the tested genes (TaCAD12, Defensin, PR10, PR17c, Chit1, TaCAD1, TaCCR, and TaCOMT1) in BSMV:TaCAD12-infected CI12633 plants were quantified relative to that in BSMV:GFP-infected CI12633 plants after R. cerealis WK207 inoculation for 40 days. Statistically significant differences between BSMV:TaCAD12- and BSMV:GFP- inoculated CI12633 plants were determined based on three replications using Student’s t-test (∗∗P < 0.01).
Discussion
In this study, the wheat CAD gene TaCAD12 was identified based on comparative transcriptomics between sharp eyespot-resistant wheat line CI12633 and susceptible wheat line Wenmai 6. Comparing with susceptible wheat lines, the transcriptional level of TaCAD12 gene was higher and could be significantly enhanced in the sharp eyespot-resistant wheat lines (CI12633 and Shanhongmai) after R. cerealis infection. In Arabidopsis, the expression of CAD-D was obviously induced after inoculation with Pseudomonas syringae pv. tomato; two CAD proteins CAD-C and CAD-D not only act as key enzymes in lignin biosynthesis (Sibout et al., 2005), but also play essential roles in plant defense against infection of the bacterial (Pseudomonas syringae pv. tomato) pathogen (biotrophic pathogen, Tronchet et al., 2010). Additionally, the transcriptional level of TaCAD12 was the highest in roots that are the original infecting site of R. cerealis, and intermediate in stems that are the main occurring site of sharp eyespot symptom. These results imply that TaCAD12 may participate in defense response of wheat to R. cerealis infection. Importantly, silencing of TaCAD12 in the resistant wheat line CI12633 significantly impairs host resistance to R. cerealis, TaCAD12-overexpressing wheat lines displayed significantly increased resistance during whole growth stages. These results clearly reveal that TaCAD12 positively contributes to resistance against R. cerealis infection in wheat. To our knowledge, this is the first report about a CAD member positively participating in plant resistance responses to necrotrophic fungal pathogens through both partial loss-of-function (gene-silencing) analysis and partial gain-of-function (overexpressing transgene) analysis. To date, the mechanisms of plant responses to necrotrophic pathogens have been limited. Our results extend the current knowledge of plant defenses against pathogens.
The protein sequence and phylogenetic tree analyses suggest that TaCAD12 is closer to TaCAD3, than to BdCAD5, LpCAD2, and OsCAD7 in the same Group (IV), but far from the wheat TaCAD1 in I Group of CAD family. In IV Group, only OsCAD7 has been reported to have strong activity toward coniferyl aldehyde and weak activity toward sinapyl aldehyde, which influences lignin contents and mechanical strength of rice culm (Li et al., 2009). The results of CAD activity analysis in vitro prove that TaCAD12 protein is an authentic CAD enzyme, similar to the reported TaCAD1. TaCAD1 may be involved in monolignol biosynthesis and lodging resistance in wheat (Ma, 2010). TaCAD12 possesses catalytic activities toward both coniferyl aldehyde and sinapyl aldehyde. Moreover, our analysis results of CAD catalytic activity and TaCAD12 protein levels in wheat stems suggest that higher TaCAD12 expression levels could lead to relatively stronger CAD kinetic activities in TaCAD12-overexpressing wheat lines (OC-11, OC-14, OC-22, and OC-47) compared with WT Yangmai 16. The CAD catalytic activities play a key role in biosynthesis of monolignols and lignin abundance, and the mechanical properties of plants (Mellerowice et al., 2001; Sibout et al., 2003, 2005; Bhuiyan et al., 2009; Sattler et al., 2009; Li et al., 2009; Anderson et al., 2015).
The elevated expression levels of certain monolignol biosynthesis-related enzymes have been documented to occur during plant-microbe interaction (Nicholson and Hammerschmidt, 1992; Bhuiyan et al., 2009; Tronchet et al., 2010). Mutations or down-regulation of monolignol biosynthesis-related genes lead to reduction in lignin biosynthesis/concentration (Piquemal et al., 1998; Sibout et al., 2005; Do et al., 2007; Sattler et al., 2009; Tamasloukht et al., 2011; Thévenin et al., 2011; Scully et al., 2016). For example, in sorghum, brown midrib 6 mutants (mutations in the evolutionary conserved amino acids of SbCAD2) have been shown to have limited CAD activity and to reduce the abundance of lignin (Sattler et al., 2009; Scully et al., 2016). Importantly, the interruption of monolignol biosynthesis through CAD and COMT inhibitors or the gene-silencing of monolignol biosynthesis-related enzymes could increase the susceptibility of barley and diploid wheat to the pathogen Blumeria graminis (a biotrophic fungal pathogen), indicating that monolignol biosynthesis is critically important for host defense against biotrophic pathogen invasion (Kruger et al., 2002; Bhuiyan et al., 2009). In our study, following R. cerealis infection, comparing with their control wheat plants, the transcriptional levels of monolignol biosynthesis-related genes (TaCAD12, TaCAD1, TaCCR, and TaCOMT1) were reduced in more susceptible TaCAD12-silenced wheat plants, but were increased in TaCAD12-overexpressing wheat lines with enhanced-resistance to sharp eyespot. The results indicated that TaCAD12 positively regulated the expression of TaCAD1, TaCCR, and TaCOMT1, and might enhance monolignol biosynthesis, resulting in increased-resistance to sharp eyespot in wheat. These data suggest that TaCAD12 positively regulates host resistance to sharp eyespot possibly through elevating monolignol biosynthesis.
Defense genes played a vital role in plant resistance against pathogen infections. In Arabidopsis, cad-C/cad-D mutations negatively affected the expression of PR1 and PR5 (defense) genes after inoculation with virulent strain of P. syringae pv. tomato (Tronchet et al., 2010). In order to explore if TaCAD12 regulates defense genes in wheat resistance response to R. cerealis, we investigated the transcriptional levels of four wheat defense marker genes, including Defensin, PR10, PR17c, and Chit1, in TaCAD12-overexpressing and silencing wheat plants as well as their control plants. The results showed that transcriptional levels of Defensin, PR10, PR17c, and Chit1 in stems of TaCAD12-overexpression wheat plants were elevated than in WT wheat plants, whereas these genes exhibited transcriptional reduction in TaCAD12-silencing wheat plants compared with the control plants. These data suggested that TaCAD12 could up-regulate the transcription of these four defense genes tested, consequently leading to the increased resistance against R. cerealis infection.
Conclusion
The wheat CAD gene TaCAD12 was identified via comparative transcriptomics. The transcriptional levels of TaCAD12 are higher and markedly elevated after the infection of R. cerealis in resistant wheat lines. The TaCAD12 protein is an active CAD enzyme with catalytic activities toward both coniferyl aldehyde and sinapyl aldehyde. TaCAD12, acting as a positive contributor, appears to be essential to resistance response to R. cerealis infection through regulating the expression of certain defense genes and monolignol biosynthesis-related genes in wheat. This study provides insights into the roles of CAD members in plant defense responses. TaCAD12 is a candidate gene to improve wheat resistance to sharp eyespot.
Author Contributions
ZZ designed the research and wrote the paper. WR, ML, and TS performed the cloning, sequencing, enzyme kinetic and transcriptional analyses, and functional assays. XW analyzed the Q-RT-PCR data. HX and LD conducted wheat transformation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This study was supported by the programs of the Chinese Key Research & Development Plan Project (2016YFD0101004) and a National “Key Sci-Tech” Project in China (2016ZX08002001-4).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01723/full#supplementary-material
Figure S1 | The reaction patterns catalyzed by the GST-TaCAD12 recombinant protein. The reaction patterns catalyzed by TaCAD12 with (A) coniferyl aldehyde and (B) sinapyl aldehyde were measured under different mixture component adding orders. The third component added was (i) purified GST-TaCAD12 recombinant protein, (ii) NADPH, and (iii) substrates (coniferyl aldehyde and sinapyl aldehyde), respectively.
Figure S2 | Schematic representation of the barley strip mosaic virus (BSMV) genome organization and BSMV recombinant constructs engineered to express GFP and TaCAD12 fragments as inverted repeats. RNAs α, β, and γ were as described (Holzberg et al., 2002). The 190-bp TaCAD12 fragment (from 1011 to 1200 nt in TaCAD12 cDNA sequence) was sub-cloned in an antisense orientation into the Nhe I restriction site of the RNA γ of BSMV.
Table S1 | Sequences of primers used in this study.
Footnotes
References
Anderson, N. A., Tobimatsu, Y., Ciesielski, P. N., Ximenes, E., Ralph, J., Donohoe, B. S., et al. (2015). Manipulation of guaiacyl and syringyl monomer biosynthesis in an Arabidopsis cinnamyl alcohol dehydrogenase mutant results in atypical lignin biosynthesis and modified cell wall structure. Plant Cell 27, 2195–2209. doi: 10.1105/tpc.15.00373
Baucher, M., Chabbert, B., Pilate, G., Van Doorsselaere, J., Tollier, M. T., Petit-Conil, M., et al. (1996). Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in Poplar. Plant Physiol. 112, 1479–1490. doi: 10.1104/pp.112.4.1479
Bhuiyan, N. H., Selvaraj, G., Wei, Y. D., and King, J. (2009). Gene expression profiling and scilencing reveal that monolignol biosynthesis plays a critical role in pernetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 60, 509–521. doi: 10.1093/jxb/ern290
Brill, E. M., Abrahams, S., Hayes, C. M., Jenkins, C. L., and Watson, J. M. (1999). Molecular characterization and expression of a wound-inducible cDNA encoding a novel cinnamyl-alcohol dehydrogenase enzyme in Lucerne (Medicago sativa L.). Plant Mol. Biol. 41, 279–291. doi: 10.1023/A:1006381630494
Cai, S. B., Ren, L. J., Yan, W., Wu, J. Z., Chen, H. G., Wu, X. Y., et al. (2006). Germplasm development and mapping of resistance to sharp eyespot (Rhizoctonia cerealis) in wheat. Sci. Agric. Sin. 39, 928–934.
Chen, J., Li, G. H., Du, Z. Y., Quan, W., Zhang, H. Y., Che, M. Z., et al. (2013). Mapping of QTL conferring resistance to sharp eyespot (Rhizoctonia cerealis) in bread wheat at the adult plant growth stage. Theor. Appl. Genet. 126, 2865–2878. doi: 10.1007/s00122-013-2178-6
Chen, L., Zhang, Z. Y., Liang, H. X., Liu, H., Du, L. P., Xu, H. J., et al. (2008). Overexpression of TiERF1 enhances resistance to sharp eyespot in wheat. J. Exp. Bot. 59, 4195–4204. doi: 10.1093/jxb/ern259
Cheong, Y. H., Chang, H. S., Gupta, R., Wang, X., Zhu, T., and Luan, S. (2002). Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129, 661–677. doi: 10.1104/pp.002857
Christensen, A. H., and Quail, P. H. (1996). Ubiquitin promoter-based vectors for high levels of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5, 213–218.
Do, C. T., Pollet, B., Thévenin, J., Sibout, R., Denoue, D., Barrière, Y., et al. (2007). Both caffeoyl coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226, 1117–1129. doi: 10.1007/s00425-007-0558-3
Dong, N., Liu, X., Liu, Y., Du, L. P., Xu, H. J., Xin, Z. Y., et al. (2010). Overexpression of TaPIEP1, a pathogen-induced ERF gene of wheat, confers host-enhanced resistance to fungal pathogen Bipolaris sorokiniana. Funct. Integr. Genomics 10, 215–226. doi: 10.1007/s10142-009-0157-4
Eudes, A., Pollet, B., Sibout, R., Do, C. T., Séguin, A., Lapierre, C., et al. (2006). Evidence for a role of AtCAD1 in lignification of elongating stems of Arabidopsis thaliana. Planta 225, 23–39. doi: 10.1007/s00425-006-0326-9
Glazebrook, J. (2005). Constrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Goffner, D., Joffroy, I., Grima-Pettenati, J., Halpin, C., Knight, M. E., Schuch, W., et al. (1992). Purification and characterization of isoforms of cinnamyl alcohol dehydrogenase (CAD) from eucalyptus xylem. Planta 188, 48–53. doi: 10.1007/BF00198938
Hamada, M. S., Yin, Y., Chen, H., and Ma, Z. (2011). The escalating threat of Rhizoctonia cerealis, the causal agent of sharp eyespot in wheat. Pest Manag. Sci. 67, 1411–1419. doi: 10.1002/ps.2236
Holzberg, S., Brosio, P., Gross, C., and Pogue, G. P. (2002). Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 30, 315–327. doi: 10.1046/j.1365-313X.2002.01291.x
Keen, N. T., and Littlefield, L. J. (1979). The possible association of phytoalexins with resistance gene expression in flax to Melampsora lini. Physiol. Plant Pathol. 14, 265–280. doi: 10.1016/0048-4059(79)90048-1
Kim, S. J., Kim, M. R., Bedgar, D. L., Moinuddin, S. G., Cardenas, C. L., Davin, L. B., et al. (2004). Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 1455–1460. doi: 10.1073/pnas.0307987100
Kruger, W. M., Carver, T. L. W., and Zeyen, R. J. (2002). Effects of inhibiting phenolic biosynthesis on penetration resistance of barley isolines containing seven powdery mildew resistance genes for alleles. Physiol. Mol. Plant Pathol. 61, 41–51. doi: 10.1006/pmpp.2002.0415
Lange, B. M., Lapierre, C., and Sandermann, H. Jr. (1995). Elicitor-induced spruce stress lignin (structural similarity to early developmental lignins). Plant Physiol. 108, 1277–1287. doi: 10.1104/pp.108.3.1277
Lemańczyk, G., and Kwaśna, H. (2013). Effects of sharp eyespot (Rhizoctonia cerealis) on yield and grain quality of winter wheat. Eur. J. Plant Pathol. 135, 187–200. doi: 10.1007/s10658-012-0077-3
Li, X. J., Yang, Y., Yao, J. L., Chen, G. X., Li, X. H., Zhang, Q. F., et al. (2009). FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol. Biol. 69, 685–697. doi: 10.1007/s11103-008-9448-8
Li, Z., Zhuang, H. T., Du, L. P., Zhou, M. P., Cai, S. B., Jun, X. H., et al. (2011). Utilization of tissue specific expressing promoter RSS1P in TiERF1 wheat. Acta Agron. Sin. 37, 1897–1903. doi: 10.3724/SP.J.1006.2011.01897
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔC(T) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Q. H. (2010). Functional analysis of a cinnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat. J. Exp. Bot. 61, 2735–2744. doi: 10.1093/jxb/erq107
Mellerowice, E. J., Baucher, M., Sundberg, B., and Boerjan, W. (2001). Unravelling cell wall formation in the woody dicot stem. Plant Mol. Biol. 47, 239–274. doi: 10.1023/A:1010699919325
Nicholson, R. L., and Hammerschmidt, R. (1992). Phenolic-compounds and their role in disease resistance. Annu. Rev. Phytopathol. 30, 369–389. doi: 10.1146/annurev.py.30.090192.002101
Piquemal, J., Lapierre, C., Myton, K., O’Connell, A., Schuch, W., Grima-Pettenati, J., et al. (1998). Down-regulation of cinnamoyl-CoA reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J. 13, 71–83. doi: 10.1046/j.1365-313X.1998.00014.x
Rinaldi, T., Kulangara, K., Antoniello, K., and Markram, H. (2007). Elevated NMDA receptor levels and enhanced postsynaptic long-term potentiation induced by prenatal exposure to valproic acid. Proc. Natl. Acad. Sci. U.S.A. 104, 13501–13506. doi: 10.1073/pnas.0704391104
Saathoff, A. J., Hargrove, M. S., Haas, E. J., Tobias, C. M., Twigg, P., Sattler, S., et al. (2012). Swithcgrass PviCAD1: understanding residues important for substrate preferences and acitivity. Appl. Biochem. Biotechnol. 168, 1086–1100. doi: 10.1007/s12010-012-9843-0
Saathoff, A. J., Sarath, G., Chow, E. K., Dien, B. S., and Tobias, C. M. (2011). Downregulation of Cinnamyl-alcohol dehydrogenase in swichgrass by RNA silencing results in enhanced glucose release after cellulase treatment. PLoS ONE 6:e16416. doi: 10.1371/journal.pone.0016416
Saballos, A., Ejeta, G., Sanchez, E., Kang, C. H., and Vermerris, W. (2009). A genomewide analysis of the cinnamyl alcohol dehydrogenase family in Sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics 181, 783–795. doi: 10.1534/genetics.108.098996
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A., and Allard, R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. U.S.A. 81, 8014–8018. doi: 10.1073/pnas.81.24.8014
Sasaki, T., and Sederoff, R. R. (2003). Genome studies and molecular genetics: the rice genome and comparative genomics of higher plants. Curr. Opin. Plant Biol. 6, 97–100. doi: 10.1016/S1369-5266(03)00018-9
Sattler, S. E., Saathoff, A. J., Haas, E. J., Palmer, N. A., Funnell-Harris, D. L., Sarath, G., et al. (2009). A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the Sorghum brown midrib6 phenotype. Plant Physiol. 150, 584–595. doi: 10.1104/pp.109.136408
Schenk, P. M., Kanzan, K., Wilson, I., Anderson, J. P., Richmond, T., Somerville, S. C., et al. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. U.S.A. 97, 11655–11660. doi: 10.1073/pnas.97.21.11655
Scofield, S. R., Li, H., Brandt, A. S., and Gill, B. S. (2005). Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 138, 2165–2173. doi: 10.1104/pp.105.061861
Scully, E. D., Gries, T., Funnell-Harris, D. L., Xin, Z. G., Kovacs, F. A., Vermerris, W., et al. (2016). Characterization of novel Brown midrib 6 mutations affecting lignin biosynthesis in Sorghum. J. Integr. Plant Biol. 58, 136–149. doi: 10.1111/jipb.12375
Shi, Y., Wang, M. B., Powell, K. S., Van Damme, E., Hilder, V. A., Gatehouse, A. M. R., et al. (1994). Use of the rice sucrose synthase-1 promoter to direct phloem-specific expression of β-glucuronidase and snowdrop lectin genes in tobacco plants. J. Exp. Bot. 45, 623–631. doi: 10.1093/jxb/45.5.623
Sibout, R., Eudes, A., Mouille, G., Pollet, B., Lapierre, C., Jouanin, L., et al. (2005). Cinnamyl alcohol dehydrogenase-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17, 2059–2076. doi: 10.1105/tpc.105.030767
Sibout, R., Eudes, A., Pollet, B., Goujon, T., Mila, I., Granier, F., et al. (2003). Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiol. 132, 848–860.
Tamasloukht, B., Lam, M. S., Martinez, Y., Tozo, K., Barbier, O., Jourda, C., et al. (2011). Characterization of a cinnamoyl-CoA reductase 1 (CCR1) mutant in maize: effects on lignification, fibre development, and global gene expression. J. Exp. Bot. 62, 3837–3848. doi: 10.1093/jxb/err077
Thévenin, J., Pollet, B., Letarnec, B., Saulnier, L., Gissot, L., Maia-Grondard, A., et al. (2011). The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthesis pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol. Plant 4, 70–82. doi: 10.1093/mp/ssq045
Tobias, C. M., and Chow, E. K. (2005). Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification. Planta 220, 678–688. doi: 10.1007/s00425-004-1385-4
Tronchet, M., Balagué, C., Thomas, K. R. O. J., Jouanin, L., and Roby, D. (2010). Chinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 11, 83–92. doi: 10.1111/j.1364-3703.2009.00578.x
Wei, X. N., Shen, F. D., Hong, Y. T., Rong, W., Du, L. P., Liu, X., et al. (2015). The wheat calcium-dependent protein kinase TaCPK7-D positively regulates host resistance to sharp eyespot disease. Mol. Plant Pathol. 17, 1252–1264. doi: 10.1111/mpp.12360
Youn, B., Camacho, R., Moinuddin, S. G. A., Lee, C., Davin, L. B., Lewis, N. G., et al. (2006). Crystal structures and catalytic mechanism of the Arabidopsis cinnamyl alcohol dehydrogenases AtCAD5 and AtCAD4. Org. Biomol. Chem. 4, 1687–1697. doi: 10.1039/b601672c
Zhang, K., and Cheng, Z. (2006). GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol. 140, 972–983. doi: 10.1104/pp.105.073007
Zhou, H. B., Li, S. F., Deng, Z. Y., Wang, X. P., Chen, T., Zhang, J. S., et al. (2007). Molecular analysis of three new receptor-like kinase genes from hexaploid wheat and evidence for their participation in the wheat hypersensitive response to stripe rust fungus infection. Plant J. 52, 420–434. doi: 10.1111/j.1365-313X.2007.03246.x
Zhu, X. L., Qi, L., Liu, X., Cai, S. B., Xu, H. J., Huang, R. F., et al. (2014). The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediated host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol. 164, 1499–1514. doi: 10.1104/pp.113.229575
Keywords: hexaploid wheat, cinnamyl alcohol dehydrogenase (CAD), Rhizoctonia cerealis, sharp eyespot-resistance, defense genes, monolignol biosynthesis-related genes
Citation: Rong W, Luo M, Shan T, Wei X, Du L, Xu H and Zhang Z (2016) A Wheat Cinnamyl Alcohol Dehydrogenase TaCAD12 Contributes to Host Resistance to the Sharp Eyespot Disease. Front. Plant Sci. 7:1723. doi: 10.3389/fpls.2016.01723
Received: 27 July 2016; Accepted: 02 November 2016;
Published: 16 November 2016.
Edited by:
Soren K. Rasmussen, University of Copenhagen, DenmarkReviewed by:
Kanwarpal Singh Dhugga, Consultative Group on International Agricultural Research, USAQing-Hu Ma, Institute of Botany (CAS), China
Copyright © 2016 Rong, Luo, Shan, Wei, Du, Xu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zengyan Zhang, emhhbmd6ZW5neWFuQGNhYXMuY24=
†These authors have contributed equally to this work.
 Wei Rong†
Wei Rong† Zengyan Zhang
Zengyan Zhang