
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 01 November 2016
Sec. Plant Breeding
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.01584
This article is part of the Research TopicMechanisms of abiotic stress responses and tolerance in plants: physiological, biochemical and molecular interventionsView all 121 articles
The challenge to produce more food for a rising global population on diminishing agricultural land is complicated by the effects of climate change on agricultural productivity. Although great progress has been made in crop improvement, so far most efforts have targeted above-ground traits. Roots are essential for plant adaptation and productivity, but are less studied due to the difficulty of observing them during the plant life cycle. Root system architecture (RSA), made up of structural features like root length, spread, number, and length of lateral roots, among others, exhibits great plasticity in response to environmental changes, and could be critical to developing crops with more efficient roots. Much of the research on root traits has thus far focused on the most common cereal crops and model plants. As cereal yields have reached their yield potential in some regions, understanding their root system may help overcome these plateaus. However, root and tuber crops (RTCs) such as potato, sweetpotato, cassava, and yam may hold more potential for providing food security in the future, and knowledge of their root system additionally focuses directly on the edible portion. Root-trait modeling for multiple stress scenarios, together with high-throughput phenotyping and genotyping techniques, robust databases, and data analytical pipelines, may provide a valuable base for a truly inclusive ‘green revolution.’ In the current review, we discuss RSA with special reference to RTCs, and how knowledge on genetics of RSA can be manipulated to improve their tolerance to abiotic stresses.
Roots are essential for plant productivity and serve a variety of functions, such as water and nutrient uptake, forming symbioses with other microorganisms in the rhizosphere, anchoring the plant to the soil, and acting as storage organs. The different interactions of a root with its environment depend on its organization and structure, from the cellular to whole-plant level. The root contains a stele, comprised of the xylem, the phloem, and the pericycle (Smith and De Smet, 2012). The stele is encircled by concentric layers of epidermal, cortical, and endodermal tissues. The root apical meristem forms the basic stem cell pool from which other cell types develop. This root apical meristem also holds the quiescent center (QC), with rarely dividing cells that signals the surrounding cells to organize and maintain the initial stem cells (Dolan et al., 1993). There are generally two types of roots: (i) those that are formed in the embryo, such as the primary and seminal roots in maize (Hochholdinger, 2009), tap or primary root in common bean (Lynch and Brown, 2012); (ii) those formed post-embryonically from consecutive nodes on shoots, normally referred to as adventitious roots (ARs). These include basal roots in beans, nodal roots in maize, ARs of sweetpotato, potato, cassava, as well as yam, and lateral roots (LRs; Lynch and Brown, 2012). LRs are formed post-embryonically from the pericycle of all root classes through auxin-dependent cell cycle activation. This cell cycle forms the LR founder cells that undergo several rounds of cell division to initiate LRs (Overvoorde et al., 2010). The elongation, growth angles from the main axis, lateral branching and longevity of all root classes forms the root system which is determined by genetic, physiological, and environmental factors (Lynch and Brown, 2012).
Root system architecture (RSA) therefore refers to the spatial configuration of the root system or the explicit deployment of root axes (Lynch, 1995). Under poorly understood genetic control, RSA exhibits plasticity and responds to external environmental conditions such as soil moisture, nutrients, temperature, pH, and microbial communities (Bao et al., 2014). The study of RSA is important for agricultural productivity because most soils have uneven distribution of resources and/or localized depletions that make spatial distribution of the root system an important determinant of a plant’s ability to exploit these resources (Lynch, 1995). Progress in the study of RSA in agricultural crops has consequently been realized, especially for cereals, and evidence for the genetic control of RSA and its relationship to increased productivity under stress is currently well-documented. Despite these achievements, information on RSA in root and tuber crops (RTCs), which form the second largest group of crops for global food security after cereals, is still lacking. A recent review by Villordon et al. (2014b) on root architecture and RTC productivity clearly indicates this gap. In the current review, we discuss RSA with special reference to RTCs, the genetic control of RSA, the relationship between RSA and abiotic stresses, and how RSA can be manipulated to confer tolerance to abiotic stresses. We then draw conclusions on the way forward for RSA studies in RTCs.
Understanding RSA and the mechanisms of its development will allow manipulation and exploitation of different root traits to improve plants’ adaptation to changing climates and increase yields for the growing global human population (Smith and De Smet, 2012). Vegetatively propagated RTCs such as potato (Solanum tuberosum), sweetpotato (Ipomoea batatas), cassava (Manihot esculenta), and yam (Dioscorea spp.) provide food security for vast populations, especially in sub-Saharan Africa where many resource-poor small holder farmers provide the majority of food. Of the four major RTCs, cassava and sweetpotato are storage roots, while potato and yam are tubers. Little literature is specifically targeted to root growth and development in RTCs compared to cereals. The little literature available also mainly focuses on the growth and development of the storage root or tuber, primarily at harvest and post-harvest evaluations, rather than the entire root system. In this section we describe the root systems in the four major RTCs, based on available literature (Figure 1).
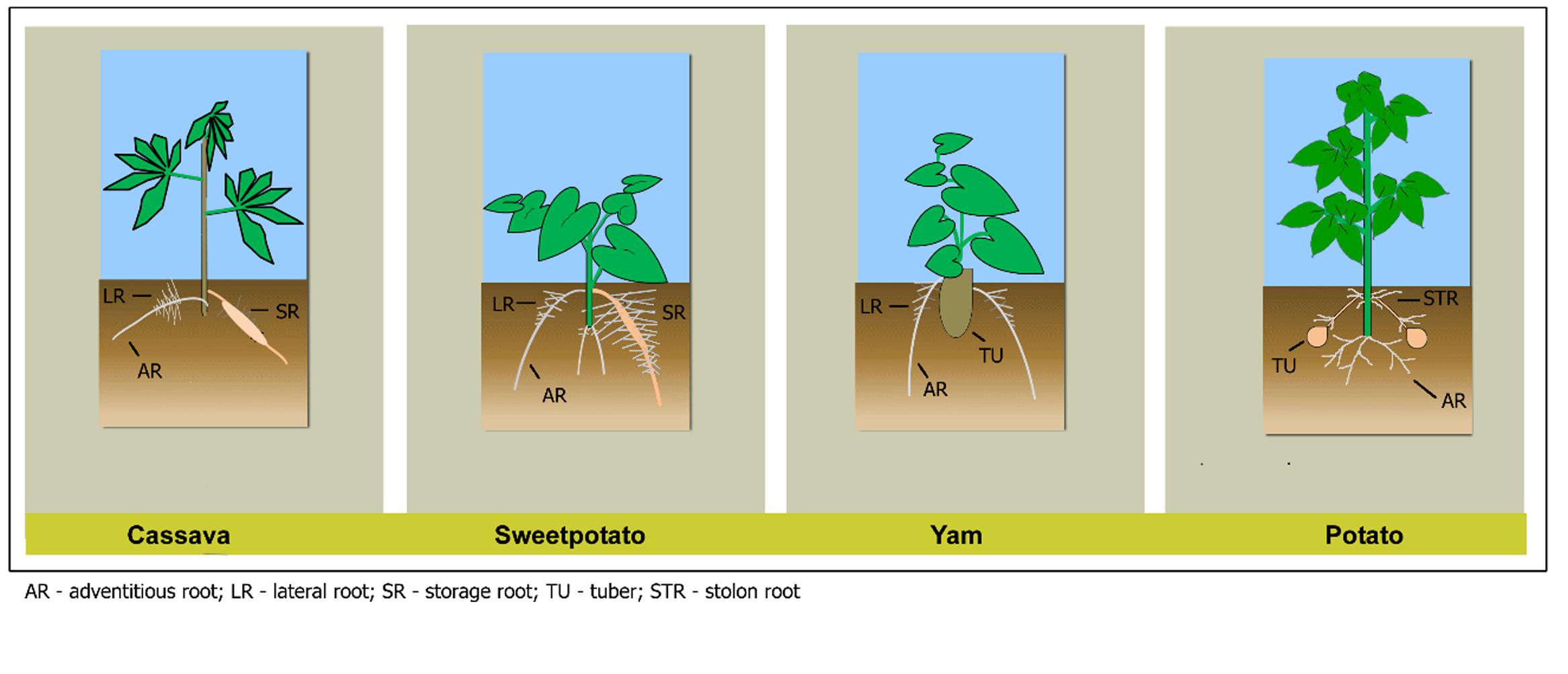
FIGURE 1. Root system architecture of cassava, sweetpotato, yam, and potato showing different root types (potato and sweetpotato figures adapted from Villordon et al., 2014b).
Cassava, potato, sweetpotato, and yam have ARs originating from the shoot or subterranean stem, in contrast with the primary root in seed-propagated crops which originates from the embryo. In sweetpotato and cassava, RSA is composed of ARs, LRs and storage roots (SRs), whereas in potato, the ARs can be divided into basal (ARs in Figure 1) and stolon roots (STR). In yam, the ARs root system is the most pronounced. The simple recognition of the main AR axis and its spatial and temporal relationship to LRs and their initiation in RTCs would enable systematic investigations to further understand the mechanisms that trigger LR emergence and morphogenesis.
A comparative survey of reports published in the last 25 years on the subject of root architecture among cassava, potato, sweetpotato, and yam is shown in Table 1. In general, current knowledge is at the level of classical morphology, with relatively little on the genetic, hormonal, and molecular control of root architecture development among RTCs. The first available documented attempt to specifically describe RSA development of several vegetable species, including sweetpotato, across different developmental stages was by Weaver and Bruner (1927). In sweetpotato, the pericyclic development of LRs and its connection to protoxylem poles, where the number of protoxylem poles correspond to the number of LRs on enlarged storage roots, was made in the early 1900s (Hayward, 1938). Yasui (1944) later reported that the protoxylem in ARs of sweetpotato was generally either pentarch or hexarch, and that adventitious buds arose from five or six longitudinal rows of LR “scars.” Relatively recent work documenting the pericyclic origin of cassava LRs noted that xylem poles ranged from four to eight and LRs developed from the pericycle opposite the xylem pole (Medina et al., 2007; Bonfim et al., 2011). Chaweewan and Taylor (2015) found that the roots developing from stem cut end of cassava (basal ARs) did not develop into storage roots. Such roots were also initiated from the cambium. However, roots developing from buried nodes (nodal ARs) at the boundary between the xylem and the stele had the capacity to develop into storage roots. Only one reference was found for yams, the second most important root crop in Sub-Saharan Africa1, which described two distinct well-organized root systems: the seminal root system and the adventitious, more definitive root system (Charles-Dominique et al., 2009). Other studies in yam only focused on root morphology at crop harvest stage in response to fertilization (Melteras et al., 2008; O’Sullivan, 2008; Hgaza et al., 2012). Iwama et al. (1977) specifically analyzed root systems and the relationship between root systems and tuber yield in potato (Iwama et al., 1981). The effect of environment on RSA has been examined from different angles, for example Asfary et al. (1983) measured average root length under different nitrogen (N) fertilization, Vos and Groenwold (1986) studied root growth of potato on a marine clay soil, while Parker et al. (1989) studied the properties of subsoil loosening and irrigation on soil physical characteristics, root distribution, and water uptake in potato. More recently, Iwama (2008) studied the physiology and morphology of potato roots, specifically root length distribution, and examined their relationship with tuber growth.
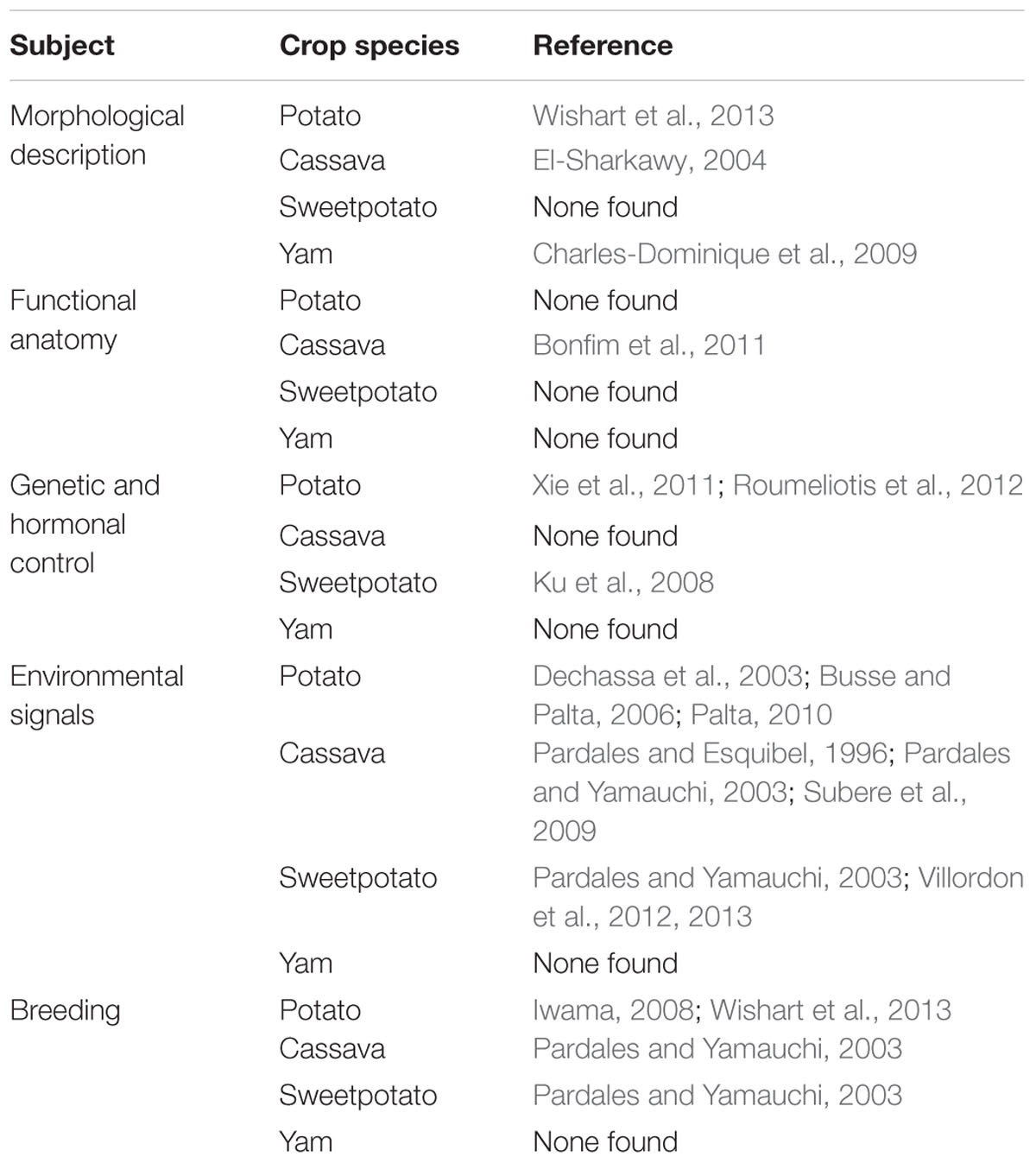
TABLE 1. Summary of articles published within the last 25 years that address root architecture development in cassava, sweetpotato, potato, and yams.
In sweetpotato, storage root formation is marked by the formation of cambia around the protoxylem and secondary xylem elements, but lignification in the stele region reduces storage root formation (Togari, 1950; Wilson and Lowe, 1973). The first evidence of a link between sweetpotato LR development and storage root yield suggested that LRs may be essential in supplying “internal growth elements” for storage root formation (Koshimizu and Nishida, 1949). Recent work demonstrated the link between LR development and lignification. In ARs with a prevalence of arrested or non-emerged LR primordia, the adjacent stelar tissue becomes lignified thus rendering it incapable of undergoing swelling due to the absence of vascular and anomalous cambia development (Villordon et al., 2012). The precise relationship between stele lignification and LR development is still not clear in sweetpotato. However, proteomics work with maize lrt1 (lateral rootless1) mutants showed the detection of proteins associated with lignin metabolism in the primary root, providing evidence that LRs influenced the proteome of the primary root (Hochholdinger et al., 2004). These findings suggest that intrinsic and external stimuli which promote LR development preclude stele lignification, rendering the juvenile AR competent for storage root formation. In cassava, LRs are responsible for root system plasticity during the critical storage root formation stage (Pardales and Yamauchi, 2003). There is currently a lack of evidence to suggest a relationship between LR development and the capacity of an AR to become a storage root. Early work describing anatomical changes associated with storage root formation in cassava did not mention LRs (Indira and Kurian, 1977). Related work examined the branching pattern of LRs and reported that LRs increased root surface area and compensated for the decrease in the main root length (Izumi et al., 1999). It was concluded that roots with a well-developed branching pattern likely absorbed water and essential nutrients for storage root growth better. In potato, root mass is positively correlated with shoot mass and tuber bulking, but negatively correlated with early tuber bulking. Final tuber yield is related to RSA component traits such as specific root length of basal roots and total root weight for various root classes of potato under field grown conditions. Basal roots are important for water uptake and anchorage, whereas stolon roots are connected with nutrient acquisition and tuber formation (Wishart et al., 2013). Despite these efforts, the link between storage root/tuber yield and the carbon partition to other root types as well as the regulatory networks involved in RTCs is yet to be established. Understanding the genetic, physiological and environmental factors influencing these components of RSA in RTCs is therefore critical in adapting genotypes to changing climates.
Most genetic studies on RSA have been carried out using ‘model’ plant Arabidopsis and a few ‘model’ cereals including maize and rice. In this section we review the hormonal/genetic control of RSA as reported in Arabidopsis and/or cereals followed by what is known in RTCs for storage root and tuber development. Formation of LRs as an important aspect of RSA which is a result of cell division of a specific subset of pericyle cells (Casimiro et al., 2001). The cell division process is under the control of both root- and shoot-derived auxin, where cellular levels of auxin contribute to the regulation of gene expression, which then impacts root branching (Overvoorde et al., 2010). Auxin activates Cyclin-dependent-kinases (CDKs) (Himanen et al., 2002) and D-type cyclin (CYCD) (Nieuwland et al., 2009). These two are cell-cycle genes involved in pericycle division during the LR initiation process and whose inhibition leads to a reduced number of LRs. Also related to the cell cycle initiation is the E2Fa/Dpa transcription factor, which promotes the G1-S transition by controlling the expression of genes required for DNA replication (Boudolf et al., 2004). However, initiation of the cell cycle alone is not enough to initiate LR formation but, as Vanneste et al. (2005) showed, LR initiation requires fine tuning by both negative and positive mechanisms regulating auxin homeostasis and signal transduction in the pericycle. These processes are under the control of auxin-responsive genes dependent on Auxin/indole-3-acetic acid-auxin response factors (AUX/IAA-ARFs) auxin signaling pathways. Genes containing auxin-responsive elements (AREs) in the promoter region are directly regulated by ARFs. In the absence of auxin, the ARFs combine with AUX/IAA proteins (AUX/IAA-ARFs) and are therefore not active. In the presence of auxin, however, the AUX/IAA proteins are degraded by auxin-receptor proteins TIR1 and AFBs through the SCF TIR1/AFBs complexes and 26S proteasomes (Goh et al., 2012). This degradation leaves the ARFs active to either positively or negatively regulate auxin responsive transcription. There are several of these AUX/IAA-ARF modules which are proposed to successively coordinate different developmental processes by regulating distinct targets (De Smet et al., 2010). The exact number of such modules involved in LR development is however still unknown. De Smet et al. (2010) showed a bimodal auxin response where they found that in addition to the Solitary root/indole-3-acetic acid14 – auxin response factors7 and 19 (SLR/IAA14-ARF7-ARF19), the Bodenlos/indole-3-acetic acid12/monopteros-auxin response factor5 (BDL/IAA12/MP-ARF5), acting downstream of SLR/IAA14, was required to guarantee organized LR patterning. Goh et al. (2012) listed several modules responsible for different stages of LR initiation, including the IAA28-ARFs module, which regulates the specification of LR founder cells; the SLR/IAA14-ARF7-ARF19, which regulates nuclear migration and asymmetric cell division of the LR founder cells for LR initiation and the BDL/IAA12/MP-ARF5, which regulates LR initiation and organogenesis; the Short hypocotyl2/IAA3-ARF (SHY2/IAA3-ARF), which regulates primordia development and emergence after SLR/IAA14-ARF dependent LR initiation, and which also inhibits LR initiation. Each of these modules have target genes. Okushima et al. (2007), for example, showed that the SLR/IAA14-ARF7-ARF19 module regulates LR formation by directly activating lateral organ boundaries domain/ asymmetric leaves2-like (LBD/ASL) genes. Many other hormones interact with the auxin signaling pathways during LR initiation Cytokinin (CK) and exogenous abscisic acid (ABA) negatively affect LR development whereas Brassinosteroid (BR) positively affects LR formation. The pericycle founder cell cycling is blocked in the G2 to M transition phase by CK thereby inhibiting LR formation. In the presence of exogenous ABA, emergence of LR primordia from the parent root is inhibited before the LR meristem is activated. Despite this negative regulation of LR development by exogenous ABA, ABA signaling also has cross talks with auxin action via the ABA insensitive 3 (ABI3) and the enhanced response to ABA 1 (ERA1) genes which enhance auxin-regulated LR formation. Cross-talk is also indicated between BR and auxin-dependent LR formation, where it is thought to promote acropetal auxin transport (reviewed by Fukaki and Tasaka, 2009). Although most of these studies were carried out in Arabidopsis, Orman-Ligeza et al. (2013) compared some of these molecular control pathways in cereals and Arabidopsis and found that the AUX/IAA-ARF and the LBD/ASL regulatory pathways were conserved. Several reviews on genetic and hormonal control of RSA are available. Smith and De Smet (2012) have reviewed the genetic control of root branching, giving insights from Arabidopsis and cereals. Jung and McCouch (2013) also provide a comprehensive review on the genetic and hormonal control of RSA.
In sweetpotato, the only study found specifically referring to genetic control of RSA is by Villordon et al. (2014a) who showed evidence that orthologs of genes associated with RSA in model crops were present in sweetpotato. They found increased expression of a putative nitrogen transporter and deceased expression of a high affinity nitrogen transporter as well as decreased expression of a MAD-box gene under low nitrogen (N) conditions. A substantial amount of information is however available for storage root formation which is part of RSA in sweetpotato and cassava. Cytokinin is important in regulating storage root development in sweetpotato (Hashizume et al., 1982). Zeatin Riboside (ZR), Trans-Zeatin Riboside (t-ZR) and 9-glucosyl-n-6-2-isopentenyl adenosine (i6Ado) are the major CK involved in developing and activating the primary cambium. Besides hormones, several genes are involved in storage root formation and development in sweetpotato. Tanaka et al. (2005) found SRF1 through SRF10 developmentally regulated genes to be involved in storage root formation. SRF1, SRF2, SRF3, SRF5, SRF6, SRF7, and SRF9 were upregulated while SRF4, SRF8, and SRF10 were downregulated during storage root formation. Tanaka et al. (2008) showed that knotted1-like homeobox (KNOX1) genes, Ibkn1, Ibkn2 and Ibkn3, are associated with storage root development in sweetpotato. Ibkn1 and Ibkn2 were upregulated in developing and mature storage roots relative to fibrous roots. Ibkn1 is homologous to shoot meristemless (STM) gene of Arabidopsis whose overexpression leads to higher CK levels, while Ibkn2 and Ibkn3 are homologous to Brevipedicellus gene of Arabidopsis which negatively regulates lignin biosynthesis. A group of MAD-box genes, IbMADS genes such as IbMADS3, IbMADS4, and IbMADS79 are also found in fibrous roots before thickening, mainly in the vascular cambium region where rapid cell division occurs during storage root thickening (Kim et al., 2002). Noh et al. (2010) found that a MADS-box protein copy DNA, SRD1 enhances the proliferation of the metaxylem and cambium cells during the auxin-dependent initial thickening and growth of storage roots. Storage root development in sweetpotato is enhanced when an expansin gene (IbEXP1) is down-regulated (Noh et al., 2012), but lignin biosynthesis is inhibited as starch biosynthesis is enhanced during early storage root formation (Firon et al., 2013). Details on the molecular regulation of storage root formation in sweetpotato have been reviewed by Ravi et al. (2009, 2014). No literature was available on the genetic control of ARs and LRs in cassava. However, de Souza et al. (2004) showed overexpression of the Mec1 gene which codes for a Pt2L4 glutamic acid-rich protein and a RING Zinc Finger and LEA protein genes in the secondary xylem tissue of storage roots relative to fibrous roots. Based on a correlation network, the relationship between KNOX1 genes, phytohormone biosynthesis and phytohormone-signaling genes was established, and it was hypothesized that phytohormones are involved in the initiation of storage root development in cassava (Sojikul et al., 2015). Both potato and cassava storage organs have been substantially studied, but not the genetic and hormonal control of RSA for either crop.
In addition to hormones, signaling components, and transcription factors, micro-interfering RNAs (miRNAs) and small-interfering RNAs (siRNAs) have been shown to affect RSA in plants, as reviewed by Meng et al. (2010) and Khan et al. (2011). The miRNAs and siRNAs are thought to be involved in auxin signaling, nutrition metabolism and stress response by mediating signal interactions. They have been identified in embryonic root development, radial patterning, formation of ARs and LRs. However, their role in RTCs has not yet been studied.
Root system architecture has a central role in crop plants’ response to abiotic stresses. Since roots grow underground, they are the first to sense abiotic stresses and adjust their genetic program for post-embryonic development to survive the stress (Lynch, 1995). Plant roots obtain water and nutrients from the soil, which is a complex system with intrinsic properties, abiotic and biotic interactions. Modulation of RSA is therefore affected when changes in the plant nutritional status and external nutrient supply over time are perceived and integrated into the intrinsic root development program. The degree of root plasticity is based on variations in the number, extension, placement, and growth direction of individual components of the root system (Giehl et al., 2014). These changes in RSA consequently affect the growth and development of above-ground biomass (Paez-Garcia et al., 2015) by altering carbon allocation to shoots and/or triggering signaling pathways involving hormones, proteins, RNAs, among others (DoVale and Fritsche-Neto, 2015). In this case therefore, roots indirectly regulate leaf stomatal conductance and affect leaf blade posture and photosynthetic rate when exposed to abiotic stress.
Different abiotic stresses affect RSA in varied ways. Table 2 summarizes the root traits necessary for adaptation to different abiotic stresses. Deeper roots are associated with increased acquisition of water and mobile nutrients like N that may leach to lower soil layers (Lynch and Wojciechowski, 2015). LRs, the main determinants of ultimate RSA, are influenced strongly by moisture and nutrient distribution in the soil (Postma et al., 2014). Deak and Malamy (2005) showed that LR formation from LR primordia in Arabidopsis is repressed under drought stress when ABA and lateral root development (LRD2) gene, interact with auxin. Since ABA, LRD2 and auxin are also involved in RSA even without drought stress, it appears that such genes like LRD2 regulate the formation of LRs through promotive and repressive hormone signaling pathways depending on the environmental conditions. Repression of LR development under abiotic stress is of particular importance in root crops. In sweetpotato for example, the final storage root yield depends on the capacity of a genotype to develop LRs on the main ARs. Those with arrested or non-emerged LRs develop lignified steles, which inhibit localized swelling into storage roots (Villordon et al., 2012). Other Important contributors to RSA include single-cell projections from root epidermal cells referred to as root hairs (Tanaka et al., 2014). A high density of both root hairs and LRs is associated with increased nutrient uptake, especially in the top soil (Postma et al., 2014) but increased metabolic costs is a trade-off here (Zhan et al., 2015). There are other trade-offs associated with crop adaptation to individual abiotic stresses. Primary root length is inhibited under low-soil phosphorus (P) conditions, while LR development is promoted thereby leading to a shallower root system. This has negative effects under drought stress where deeper roots are necessary in order to have better access to water (Wasson et al., 2012). Reduced frequency of LR branching improves N uptake where genotypes with fewer but longer LRs have greater axial root elongation, deeper roots and better N uptake than those with a higher number of LRs (Zhan and Lynch, 2015). On the other hand, a larger number of LRs is required under P-limited conditions for topsoil foraging (Lynch and Brown, 2001). Since abiotic stresses normally occur in combination under field conditions, it is therefore evident that there is ‘no size fits all’ if adaptation to abiotic stress conditions is done considering each stress individually.
In RTCs, it is known that the root system is made up of ARs and LRs which presumably are involved in water and nutrient uptake and hence respond to abiotic stress. However, some RTCs have complex RSA because the harvestable part is also underground with several root classes, e.g., in potato, which may have different functions with regard to adaptation to abiotic stress. The potato root system is known to be shallow, with poor ability to penetrate soils thereby being drought susceptible (Porter et al., 1999). Despite having a shallow root system, potato is still not efficient in P and N uptake because the larger root system has a respiration carbon cost (Balemi and Schenk, 2009). Furthermore, most findings studied the root system as a whole without identifying possible roles for different root classes. An attempt at this was done by Wishart et al. (2013) who reported genetic variation for potato root traits without any specific abiotic stress. They suggested that basal roots were responsible for water uptake and anchorage while stolon roots were responsible for nutrient uptake and tuberization. Cassava and sweetpotato have less root classes compared to potato because the harvestable part is also a root. Pardales et al. (1999) studied the effects of high root zone temperature on root systems of cassava and sweetpotato. They showed a reduction in the total length of ARs, number and total length of first order LRs, under high root zone temperature in both crops. Pardales and Yamauchi (2003) also showed a suppression of AR and LR formation and development under drought stress in both sweetpotato and cassava. Recent studies in sweetpotato indicate direct influence on RSA of spatial and temporal availability of water and N availability similar to model systems (Villordon et al., 2014a). In yam, Hgaza et al. (2012) found no fertilizer response of tuber yield but a positive correlation between thinner, longer roots with tuber yield.
The aim of carrying out RSA studies in crop plants is to understand areas of interest within the root system and incorporate this information in crop improvement programs. Several approaches may be applied in manipulating RSA in order to adapt crops to changing climates.
Most of the reported studies above were carried out for stress-specific responses. However, stresses always occur in combination, with different scenarios and complexity, to which responses are also varied. In this case therefore, combined stress scenarios may be considered in relation to RSA. The complexity of the root growing environment and the limitations associated with studying only one trait could be lessened by ideotype or trait-based breeding, originally proposed by Donald (1968). Ideotype breeding requires consideration of relationships between multiple traits in addition to each individual trait (Rasmusson, 1987). A potato root ‘ideotype’ as summarized by Wishart et al. (2013) for example, is hypothesized to be one which is architecturally plastic, long, deep, and thin, as well as being able to self-protect against parasites and pathogen. Such a system should ideally be responsive to water deficits, able to transport ABA to shoots, enable efficient stomatal closure, and have minimum carbon costs, either by producing aerenchyma or increasing specific root length, increasing root surface area, or desirable root exudates and symbioses in the rhizosphere. In sweetpotato, Villordon and Clark (2014) found an association among RSA, virus resistance and availability of N thereby indicating the necessity of a more systematic approach toward determining and managing yield constraints. They showed that AR and LR formation was decreased by about 51% under deficiency and virus infection. On the other hand, storage root formation is known to fail under excess moisture and high N as more biomass is partitioned to above-ground biomass (Villordon et al., 2012). An ideotype for sweetpotato therefore, in addition to being efficient in water and mineral uptake and utilization, and being pathogen resistance as indicated above for potato, should also be tolerant to excess moisture and have balanced biomass partitioning between storage root and above-ground biomass. Trait -based breeding does not always result in accumulation of additive effects, rather synergies among traits need to be considered. It is proposed to incorporate root phenes into breeding programs targeting RSA. A phene, which has variants referred to as phene states, is the basic functional unit of a phenotype, with a phenotype being defined as the particular combination of states of all phenes of an individual (York et al., 2013). With regard to RSA, Lynch and Brown (2012) propose ‘elementary’ and ‘unique’ root phenes which cannot be decomposed further and which are a result of only one set of genes and processes. Root growth angle may for example be considered one of the phenes of root depth as it is only one of the factors determining root depth, while root depth is referred to as a phene aggregate, being a result of several phenes. A group of interacting phenes that may be selected together are referred to as a phene module. York et al. (2013) proposed hypotheses for integrating root phenes in a breeding program. They considered synergies within a phene module which increase as the number of positively acting phene-state combination increases. Metabolic costs are to be expected with such synergies except in metabolically neutral, positively acting, phene-state combinations. The interactions between phenes within plants, between plants and with the environment are expected to result in genetic variation in RSA.
In breeding approaches such as ideotype or phene-integrated, structural-functional plant modeling and simulation may offer a robust way of understanding the complexity of the non-linear signals and transduction pathways involved in the roots’ responses to multiple abiotic stresses. This is expected to provide new mechanistic insights into the regulation of root growth and development (Chickarmane et al., 2010). In addition to advances in high-throughput phenotyping techniques that allow rapid evaluation of a large number of genotypes, model-assisted phenotyping enhances prediction of difficult traits such as those that vary with environmental conditions. It also allows precise prediction of genotype × environment × management interaction over a large number of environments thereby allowing the estimation of comparative advantage of a given phene state in different environments (Tardieu and Tuberosa, 2010). Multi-scale modeling which examines behavior at subcellular, cellular, tissue, organ, and whole organism states may allow the prediction of the effect of a given phene, phene state, phene module, or phenotype in a complex abiotic stress environment (Band et al., 2012). Leitner et al. (2014) showed that functional-structural root models were appropriate to better comprehend the role of roots in whole-plant adaptation to diverse drought scenarios, in addition to their contribution to distinct drought scenarios. Using a dynamic root architecture model and root xylem hydraulic properties model, they showed that plants which transpired more had root axes which matched the available water distribution. They also found that water saving genotypes had lower root conductance than the water spending genotypes. Despite these advantages, encouragement for the adoption of root models needs to be accompanied with realistic and more explicit plant regulatory networks, in addition to integration with phenomic databases (Dunbabin et al., 2013) in order to be more representative and applicable to actual field performance of genotypes. This approach has not been applied in RTCs yet.
Manipulating root traits has been carried out in several crops through the use of molecular markers. Several specific genes related with RSA were identified in crop plants, either from gene mutants with quantifiable characteristics, or from QTL analyses. The genetic control of LR formation as reviewed above was elucidated based on gene mutants with quantifiable characteristics. In rice, a gene controlling root angle, Deeper Rooting 1 (DRO1), was identified using QTL mapping and introgressed into an elite line through backcrossing, whereas Phosphorus Starvation 1 (PSTOL1), a pup-1-specific protein kinase gene confers early root growth for P-acquisition in rice (Gamuyao et al., 2012) and sorghum (Hufnagel et al., 2014). However, information on how these genes/QTL affect the phenotypes and/or their performance in different genetic backgrounds and/or different environments is still largely lacking. This is because RSA response to environmental conditions is normally quite different under field conditions, given the broader spectrum of stresses the roots find themselves in Rich and Watt (2013). Extrapolating results obtained from a response to a specific abiotic stress is not therefore adequate (Jung and McCouch, 2013). Most of these QTLs are small-effect QTLs, i.e., they are normally conditioned by a single gene and therefore not stable across environments. Kitomi et al. (2015) for example, established that genotypes having the same functional allele of DRO1 could have different rooting angles. Small-effect QTLs therefore are assumed to be part of a set of minor QTLs. In such cases, it is necessary to carry out comparative data analysis and integration across controlled environments and field studies to establish target variants for further investigation and introgression into genotypes of interest. Alternatively, identification of large-effect QTLs which are more stable across environments and genetic backgrounds seems to be the most promising way of ensuring impact from genomics-assisted breeding methods. Dixit et al. (2015) confirmed the multi-genic and multi-environment effectiveness of qDTY12.1, a large-effect QTL identified on chromosome 12 of the rice genome. They confirmed the central role of the no apical meristem (OsNAM12.1) transcription factor in the activity of qDTY12.1 together with promoters of six intra-QTL genes with NAM binding sites as well as three co-localized and/or partially co-expressed genes of OsNAM12.1. These findings suggested that identification and proper analysis of large-effect QTLs together with their component genes could lead to a more efficient breeding process for complex traits such as those involved in adaptation and abiotic stress tolerance.
These reports are mainly based on Arabidopsis and cereal crops with simple genetic make-ups. Most RTCs on the other hand are polyploid with very complex genetic backgrounds. Genetic analysis of these crops is complicated by multiple alleles and loci, mixed inheritance patterns, association between ploidy and variation in mating system, among others (Dufresne et al., 2014). Marker-based procedures developed in diploid species therefore present difficulty to apply in most polyploid RTCs and adoption of these approach is not therefore straight forward. Application of the most commonly used genotyping methods, including new generation sequencing techniques, in RTCs present problems in allele dosage determination, presence of null alleles, distinguishing orthologs from paralogs, and copy number variation (Dufresne et al., 2014). As a consequence, although new techniques such as genomic selection offer great potential in marker-based breeding, they are currently still difficult to adopt. Additive, dominant and epistatic genetic effects are all important in RTCs due to heterozygosity (Ceballos et al., 2015) while models developed for genomic selection in cereals mainly consider additive genetic effects. Marker-based approaches in this class of crops therefore require re-thinking on the methods and pipelines available so far.
In addition to identifying QTLs and genes of interest from a species, another approach for adapting roots to abiotic stress is through transgenic technology. For example, spermidine synthase genes (FSPD1) confer higher antioxidant enzyme activities to plants. Under abiotic stress, plants with higher antioxidant enzyme activities are generally more tolerant, as they are better able to remove by-product reactive oxygen species (ROS) that are harmful to the plant if allowed to accumulate. Using transgenic technology, sweetpotato transformation with spermidine synthase genes (FSPD1) from Cucurbita ficifolia increased their multiple stress tolerance, with a higher concentration of FSPD1 in leaves and storage roots (Kasukabe et al., 2006). Estrada-Melo et al. (2015) used a 9-cis-epoxycarotenoid dioxygenase gene from tomato (LeNCED1) overexpressed in petunia and confirmed that NCED increased drought resistance of the transgenic plants. A calcium-dependent protein kinase (OsCDPK7) conferred tolerance to cold and salt/drought in rice transgenics. Two distinct pathways for cold and salt/drought tolerance using a single CDPK were implied which showed that manipulation of CDPK has great potential to adaptation and abiotic stress tolerant crop improvement. ‘Gene stacking,’ a form of ideotype breeding, could be a good alternative to transgenic technology that relies on a single gene. However, this has only been successful in pest control engineering such as the Bt toxin resistance (York et al., 2013). Gene stacking for complex traits is therefore still a challenge due to trait interaction.
Genome editing, a new approach that involves targeted DNA sequence modification through creation of double-strand breaks using sequence specific nucleases, provides possibilities to change a protein’s amino acid sequence through specific nucleotide substitutions, delete genes or chromosome segments, and introduce foreign DNA at desired genomic regions (Voytas, 2013). Several nucleases are available for targeted genome engineering (reviewed by Esvelt and Wang, 2013), but the system receiving most attention recently is CRISPR/Cas9, which involves the use of a guided RNA to create targeted mutations in candidate genes of key pathways in order to identify their effects and create new variation within a relatively short time (Cong et al., 2013), among other potential uses. These methods are gaining application in crop plants including RTCs. Clasen et al. (2016) used a TALENs approach to improve cold storage and processing traits in potato. The vacuolar invertase (VINV)gene (VINV) encoding a protein that breaks down sucrose to glucose and fructose was silenced in order to minimize the accumulation of reducing sugars which turn into anti-nutrients upon processing.
No matter which manipulation method is followed, proper phenotypic evaluation prior to and after such manipulations is important in order to realize and quantify genetic gains from RSA manipulation in a breeding program.
Since roots grow below ground, studying the entire root system naturally requires digging it out, a complex process in itself, and it is difficult to extract the entire system without breaking off the finer parts. For this reason, studies have mainly dwelled on above-ground traits related to abiotic stress tolerance. However, given the pressures on crop productivity caused by global climate change, with the associated abiotic stresses, and the notion that food production needs to double in the next few years to accommodate the growing global population, root manipulation seems to hold the key toward sustainable food production. Villordon et al. (2014a) suggested that a paradigm shift toward RSA studies would enable a truly inclusive green revolution and allow food-insecure, resource-poor farmers who depend on RTCs in developing countries to also benefit. With this mindset, plant biologists, geneticists, and breeders have now shifted some focus toward studying of root traits. Due to the aforementioned complexity of studying roots under the soil, plant scientists are now set on finding minimally intrusive, non-destructive, whole-root system evaluating platforms. Hydroponics and gels are the most widely used systems to phenotype root systems (Jung and McCouch, 2013). Although, they offer a simple way to study different root traits and have given insight into root growth and development, both are controlled and do not represent actual field environments, and correlation of the findings from such experiments with actual performance of a plant in its natural environment are limited. To address this bottleneck, plant scientists are continually seeking to develop methods that will allow study of RSA in a more natural environment. Several methods have been proposed and applied in various studies including ‘shovelomics’ (Trachsel et al., 2011), soil coring (Wasson et al., 2014), rhizolysimeters (Eberbach et al., 2013) and minirhizotrons (Maeght et al., 2013), which are all soil-based. However, these methods are also low throughput, slow, and not amenable to large numbers of genotypes like those required for genome-wide association mapping studies. Image-based systems have also been developed and proposed to study roots in their natural environments, including X-ray computed tomography (Tracy et al., 2010) where x-rays are used to acquire 3-D cross-sectional images of the roots within the soil, Laser (Braga et al., 2009) which allows collection of bio-speckle patterns of gel-grown roots, nuclear magnetic resonance (NMR: Menzel et al., 2007), ground penetrating radar (GPR; Hirano et al., 2012), infra-red (IR) imaging (Dokken and Davis, 2007), and near-infra-red (NIR) imaging (Tirlapur and König, 1999), among others. However, application of some of these methods is still limited by the costs involved and to a few genotypes. Another bottleneck associated with imaging methods is image analysis. Several root image analysis platforms have been developed to address this limitation2. With these large numbers of imaging and image analysis platforms, the need for sharing and use of data requires establishment of trait ontology across them to allow development of root ideotypes for different environments. Efforts by Lobet et al. (2015) to develop a unified root architecture development language are therefore right on time. This, combined with scaling up of the image analysis methods mentioned above, will be able to provide further knowledge required to adapt crops to their highly variable environments.
The increasing global population requires increased food production on the same or even less agricultural land as used currently, if the effects of climate change render some of the available marginal lands unfit for agricultural production. Most of the present and past crop improvement efforts have focused on above-ground traits to adapt crop plants to different production constraints. Although great progress has been made, and food production significantly increased, by manipulating above-ground traits, an estimated 800 million people are still food insecure, whereas yields, especially in cereal, have reached their yield potential and are plateauing in certain regions of the globe. It is therefore time for crop scientists to tap into un-explored and less exploited diversity within RSA traits to ensure rapid genetic gains, and stable and enhanced productivity of agricultural systems for future environmental conditions and climate change scenarios. Due to the quantitative nature of RSA traits and complex interaction of several underlying pathways that control them, response of RSA to multiple individual stresses or combination of stresses is variable. Modeling of the responses of root traits to multiple stress scenarios in a combination of high-throughput root-trait phenotyping techniques, alongside a robust database and data analytical pipeline, could be a way to go. This proposed strategy is applicable to all crops, but is more urgent in RTCs, as the second largest source of food security after cereals, mainly growing in marginal areas where many cereals cannot survive. Also, for RTCs, the harvestable organs are part of the RSA. It is recommended to increase focus on RSA research by investing more resources. RTCs can learn from what has been found so far in cereals and adopt some of their methods, while developing high-throughput techniques to quantify RSA traits under optimal and stressful conditions.
Review was conceptualized and written by MK, DG, and AV.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Asfary, A. F., Wild, A., and Harris, P. M. (1983). Growth, mineral nutrition and water use by potato crops. J. Agric. Sci. 100, 87–101. doi: 10.1017/S0021859600032470
Balemi, T., and Schenk, M. K. (2009). Genotypic difference of potato in carbon budgeting as a mechanism of phosphorus utilization efficiency. Plant Soil 322, 91–99. doi: 10.1007/s11104-009-9897-0
Band, L. R., Fozard, J. A., Godin, C., Jensen, O. E., Pridmore, T., Bennett, M. J., et al. (2012). Multiscale systems analysis of root growth and development: modeling beyond the network and cellular scales. Plant Cell 24, 3892–3906. doi: 10.1105/tpc.112.101550
Bao, Y., Aggarwal, P., Robbins, N. E., Sturrock, C. J., Thompson, M. C., Tan, H. Q., et al. (2014). Plant roots use a patterning mechanism to position lateral root branches toward available water. Proc. Natl. Acad. Sci. U.S.A. 111, 9319–9324. doi: 10.1073/pnas.1400966111
Bonfim, N. N., Graciano-Ribeiro, D., and Nassar, N. M. A. (2011). Genetic diversity of root anatomy in wild and cultivated Manihot species. Genet. Mol. Res. 10, 544–551. doi: 10.4238/vol10-2gmr1093
Boudolf, V., Vlieghe, K., Beemster, G. T. S., Magyar, Z., Acosta, J. A. T., Maes, S., et al. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16, 2683–2692. doi: 10.1105/tpc.104.024398
Braga, R. A., Dupuy, L., Pasqual, M., and Cardoso, R. R. (2009). Live biospeckle laser imaging of root tissues. Eur. Biophys. J. 38, 679–686. doi: 10.1007/s00249-009-0426-0
Busse, J. S., and Palta, J. P. (2006). Investigating the in vivo calcium transport path to developing potato tuber using 45Ca: a new concept in potato tuber calcium nutrition. Physiol. Plant. 128, 313–323. doi: 10.1111/j.1399-3054.2006.00741.x
Casimiro, I., Marchant, A., Bhalerao, R. P., Beeckman, T., Dhooge, S., Swarup, R., et al. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13, 843–852. doi: 10.2307/3871344
Ceballos, H., Kawuki, R. S., Gracen, V. E., Yencho, G. C., and Hershey, C. H. (2015). Conventional breeding, marker-assisted selection, genomic selection and inbreeding in clonally propagated crops: a case study for cassava. Theor. Appl. Genet. 128, 1647–1667. doi: 10.1007/s00122-015-2555-4
Charles-Dominique, T., Mangenet, T., Rey, H., Jourdan, C., and Edelin, C. (2009). Architectural analysis of root system of sexually vs vegetated yam (Diocorea rotundata Poir.), a tuber monocot. Plant Soil 317, 61–77. doi: 10.1007/s11104-008-9788-9
Chaweewan, Y., and Taylor, N. (2015). Anatomical assessment of root formation and tuberization in cassava (Manihot esculenta Crantz). Trop. Plant Biol. 8, 1–8. doi: 10.1007/s12042-014-9145-5
Chickarmane, V., Roeder, A. H. K., Tarr, P. T., Cunha, A., Tobin, C., and Meyerowitz, E. M. (2010). Computational morphodynamics: a modeling framework to understand plant growth. Annu. Rev. Plant Biol. 61, 65–87. doi: 10.1146/annurev-arplant-042809-112213
Clasen, B. M., Stoddard, T. J., Luo, S., Demorest, Z. L., Li, J., Cedrone, F., et al. (2016). Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biol. J. 14, 169–176. doi: 10.1111/pbi.12370
Comas, L. H., Becker, S. R., Von Mark, V. C., Byrne, P. F., and Dierig, D. A. (2013). Root traits contributing to plant productivity under drought. Front. Plant Sci. 4:442. doi: 10.3389/fpls.2013.00442
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
De Smet, I., Lau, S., Voß, U., Vanneste, S., Benjamins, R., Rademacher, E. H., et al. (2010). Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 2705–2710. doi: 10.1073/pnas.0915001107
de Souza, C. R., Carvalho, L. J., and de Mattos Cascardo, J. C. (2004). Comparative gene expression study to identify genes possibly related to storage root formation in cassava. Protein Pept. Lett. 11, 577–582. doi: 10.2174/0929866043406319
Deak, K. I., and Malamy, J. (2005). Osmotic regulation of root system architecture. Plant J. 43, 17–28. doi: 10.1111/j.1365-313X.2005.02425.x
Dechassa, N., Schenk, M. K., Claassen, N., and Steingrobe, B. (2003). Phosphorus efficiency of cabbage (Brassica oleracea L. var. capitata), carrot (Daucus carota L.), and potato (Solanum tuberosum L.). Plant Soil 250, 215–224. doi: 10.1023/A:1022804112388
Dixit, S., Biswal, A. K., Min, A., Henry, A., Oane, R. H., Raorane, M. L., et al. (2015). Action of multiple intra-QTL genes concerted around a co-localised transcription factor underpins a large-effect QTL. Sci. Rep. 5:15183. doi: 10.1038/srep15183
Dokken, K. M., and Davis, L. C. (2007). Infrared imaging of sunflower and maize root anatomy.J. Agric. Food Chem. 55, 10517–10530. doi: 10.1021/jf072052e
Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., et al. (1993). Cellular organization of the Arabidopsis thaliana root. Development 119, 71–84.
Donald, C. M. (1968). The breeding of crop ideotypes. Euphytica 17, 385–403. doi: 10.1007/BF00056241
DoVale, J., and Fritsche-Neto, R. (2015). “Root phenomics,” in Phenomics, eds R. Fritsche-Neto and A. Borém (Cham: Springer International Publishing), 49–66. doi: 10.1007/978-3-319-13677-6_4
Dufresne, F., Stift, M., Vergilino, R., and Mable, B. K. (2014). Recent progress and challenges in population genetics of polyploid organisms: an overview of current state-of-the-art molecular and statistical tools. Mol. Ecol. 23, 40–69. doi: 10.1111/mec.12581
Dunbabin, V. M., Postma, J. A., Schnepf, A., Pagès, L., Javaux, M., Wu, L., et al. (2013). Modelling root–soil interactions using three-dimensional models of root growth, architecture and function. Plant Soil 372, 93–124. doi: 10.1007/s11104-013-1769-y
Eberbach, P. L., Hoffmann, J., Moroni, S. J., Wade, L. J., and Weston, L. A. (2013). Rhizo-lysimetry: facilities for the simultaneous study of root behavior and resource use by agricultural crop and pasture systems. Plant Methods 9:3. doi: 10.1186/1746-4811-9-3
El-Sharkawy, M. A. (2004). Cassava biology and physiology. Plant Mol. Biol. 56, 481–501. doi: 10.1007/s11103-005-2270-7
Estrada-Melo, A. C., Ma, C., Reid, M. S., and Jiang, C.-Z. (2015). Overexpression of an ABA biosynthesis gene using a stress inducible promoter enhances drought resistance in petunia. Hortc. Res. 2:15013. doi: 10.1038/hortres.2015.13
Esvelt, K. M., and Wang, H. H. (2013). Genome-scale engineering for systems and synthetic biology. Mol. Syst. Biol. 9:641. doi: 10.1038/msb.2012.66
Firon, N., LaBonte, D., Villordon, A., Kfir, Y., Solis, J., Lapis, E., et al. (2013). Transcriptional profiling of sweetpotato (Ipomoea batatas) roots indicates down-regulation of lignin biosynthesis and up-regulation of starch biosynthesis at an early stage of storage root formation. BMC Genomics 14:460. doi: 10.1186/1471-2164-14-460
Forde, B. G. (2014). Nitrogen signaling pathways shaping root system architecture: an update. Curr. Opin. Plant Biol. 21, 30–36. doi: 10.1016/j.pbi.2014.06.004
Fukaki, H., and Tasaka, M. (2009). Hormone interactions during lateral root formation. Plant Mol. Biol. 69, 437–449. doi: 10.1007/s11103-008-9417-2
Gamuyao, R., Chin, J. H., Pariasca-Tanaka, J., Pesaresi, P., Catausan, S., Dalid, C., et al. (2012). The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488, 535–539. doi: 10.1038/nature11346
Giehl, R. F., Gruber, B. D., and von Wirén, N. (2014). It’s time to make changes: modulation of root system architecture by nutrient signals. J. Exp. Bot. 65, 769–778. doi: 10.1093/jxb/ert421
Goh, T., Kasahara, H., Mimura, T., Kamiya, Y., and Fukaki, H. (2012). Multiple AUX/IAA–ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signaling. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1461–1468. doi: 10.1098/rstb.2011.0232
Gruber, B. D., Giehl, R. F., Friedel, S., and von Wirén, N. (2013). Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 163, 161–179. doi: 10.1104/pp.113.218453
Gupta, B., and Huang, B. (2014). Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int. J. Genomics 2014, 701596. doi: 10.1155/2014/701596
Hashizume, T., Suye, S. I., Soeda, T., and Sugiyama, T. (1982). Isolation and characterization of a new glucopyranosyl derivative of 6-(3-methyl-2-butenylamino) purine from sweet potato tubers. FEBS Lett. 144, 25–28. doi: 10.1016/0014-5793(82)80561-9
Hgaza, V. K., Diby, L. N., Herrera, J. M., Sangakkara, U. R., and Frossard, E. (2012). Root distribution patterns of white yam (Dioscorea rotundata Poir.): a field study. Acta Agric. Scand. B Soil Plant Sci. 62, 616–626. doi: 10.1080/09064710.2012.682734
Himanen, K., Boucheron, E., Vanneste, S., De Almeida Engler, J., Inze, D., and Beeckman, T. (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14, 2339–2351. doi: 10.1105/tpc.004960
Hirano, Y., Yamamoto, R., Dannoura, M., Aono, K., Igarashi, T., Ishii, M., et al. (2012). Detection frequency of Pinus thunbergii roots by ground-penetrating radar is related to root biomass. Plant Soil 360, 363–373. doi: 10.1007/s11104-012-1252-1
Hochholdinger, F. (2009). “The Maize Root System: morphology, Anatomy, and Genetics,” in Handbook of Maize: Its Biology, eds J. L. Bennetzen and S. C. Hake (New York, NY: Springer), 145–160.
Hochholdinger, F., Guo, L., and Schnable, P. S. (2004). Lateral roots affect the proteome of the primary root of maize (Zea mays L.). Plant Mol. Biol. 56, 397–412. doi: 10.1007/s11103-004-3476-9
Hufnagel, B., de Sousa, S. M., Assis, L., Guimaraes, C. T., Leiser, W., Azevedo, G. C., et al. (2014). Duplicate and conquer: multiple homologs of PHOSPHORUS-STARVATION TOLERANCE1 enhance phosphorus acquisition and sorghum performance on low-phosphorus soils. Plant Physiol. 166, 659–677. doi: 10.1104/pp.114.243949
Indira, P., and Kurian, T. (1977). A study on the comparative anatomical changes undergoing tuberization in the roots of cassava and sweet potato. J. Root Crops 3, 29–32.
Iwama, K. (2008). Physiology of the potato: new insights into root system and repercussions for crop management. Potato Res. 51, 333–353. doi: 10.1007/s11540-008-9120-3
Iwama, K., Nakaseko, K., and Gotoh, K. (1977). Analysis of the root system in potato (Solanum tuberosum L.). 1. Varietal differences in root dry weight. Crop Sci. Soc. 17, 28.
Iwama, K., Nakaseko, K., Isoda, A., Gotoh, K., and Nishibe, S. (1981). Relations between root system and tuber yield in the hybrid population of the potato plants. Jpn. J. Crop Sci. 50, 233–238. doi: 10.1626/jcs.50.233
Izumi, Y., Sunyoto, Y. E., and Iijima, M. (1999). Root system development including root branching in cuttings of cassava with reference to shoot growth and tuber bulking. Plant Prod. Sci. 2, 267–272. doi: 10.1626/pps.2.267
Julkowska, M. M., Hoefsloot, H. C. J., Mol, S., Feron, R., de Boer, G.-J., Haring, M. A., et al. (2014). Capturing Arabidopsis root architecture dynamics with ROOT-FIT reveals diversity in responses to salinity. Plant Physiol. 166, 1387–1402. doi: 10.1104/pp.114.248963
Jung, J. K. H., and McCouch, S. (2013). Getting to the roots of it: genetic and hormonal control of root architecture. Front. Plant Sci. 4:186. doi: 10.3389/fpls.2013.00186
Kasukabe, Y., He, L., Watakabe, Y., Otani, M., Shimada, T., and Tachibana, S. (2006). Improvement of environmental stress tolerance of sweet potatoby introduction of genes for spermidine synthase. Plant Biotechnol. 23, 75–83. doi: 10.5511/plantbiotechnology.23.75
Katori, T., Ikeda, A., Iuchi, S., Kobayashi, M., Shinozaki, K., Maehashi, K., et al. (2010). Dissecting the genetic control of natural variation in salt tolerance of Arabidopsis thaliana accessions. J. Exp. Bot. 61, 1125–1138. doi: 10.1093/jxb/erp376
Khan, G., Declerck, M., and Sorin, C. (2011). MicroRNAs as regulators of root development and architecture. Plant Mol. Biol. 77, 47–58. doi: 10.1007/s11103-011-9793-x
Kim, S. H., Mizuno, K., and Fujimura, T. (2002). Isolation of MADS-box genes from sweet potato (Ipomoea batatas L.) Lam.) expressed specifically in vegetative tissues. Plant Cell Physiol. 43, 314–322. doi: 10.1093/pcp/pcf043
Kitomi, Y., Kanno, N., Kawai, S., Mizubayashi, T., Fukuoka, S., and Uga, Y. (2015). QTLs underlying natural variation of root growth angle among rice cultivars with the same functional allele of DEEPER ROOTING 1. Rice (N. Y.) 8, 16. doi: 10.1186/s12284-015-0049-2
Koshimizu, T., and Nishida, M. (1949). On the relation between the distribution of free-auxin in the young sweet potato plant and its root tuber formation. Bot. Mag. Tokyo. 62, 735–736. doi: 10.15281/jplantres1887.62.146
Ku, A. T., Huang, Y.-S., Wang, Y.-S., Ma, D., and Yeh, K. W. (2008). IbMADS1 (Ipomoea batatas MADS-box 1 gene) is involved in tuberous root initiation in sweet potato (Ipomoea batatas). Ann. Bot. 102, 57–67. doi: 10.1093/aob/mcn067
Leitner, D., Felderer, B., Vontobel, P., and Schnepf, A. (2014). Recovering root system traits using image analysis exemplified by two-dimensional neutron radiography images of lupine. Plant Physiol. 164, 24–35. doi: 10.1104/pp.113.227892
Lobet, G., Pound, M. P., Diener, J., Pradal, C., Draye, X., Godin, C., et al. (2015). Root system markup language: toward a unified root architecture description language. Plant Physiol. 167, 617–627. doi: 10.1104/pp.114.253625
Lynch, J. (1995). Root architecture and plant productivity. Plant Physiol. 109, 7–13. doi: 10.1104/pp.109.1.7
Lynch, J. P. (2013). Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 112, 347–357. doi: 10.1093/aob/mcs293
Lynch, J. P. (2015). Root phenes that reduce the metabolic costs of soil exploration: opportunities for 21st century agriculture. Plant Cell Environ. 38, 1775–1784. doi: 10.1111/pce.12451
Lynch, J. P., and Brown, K. M. (2001). Topsoil foraging-An architectural adaptation of plants to low phosphorus availability. Plant Soil 237, 225–237. doi: 10.1023/A:1013324727040
Lynch, J. P., and Brown, K. M. (2012). New roots for agriculture: exploiting the root phenome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1598–1604. doi: 10.1098/rstb.2011.0243
Lynch, J. P., Chimungu, J. G., and Brown, K. M. (2014). Root anatomical phenes associated with water acquisition from drying soil: targets for crop improvement. J. Exp. Bot. 65, 6155–6166. doi: 10.1093/jxb/eru162
Lynch, J. P., and Wojciechowski, T. (2015). Opportunities and challenges in the subsoil: pathways to deeper rooted crops. J. Exp. Bot. 66, 2199–2210. doi: 10.1093/jxb/eru508
Maeght, J. L., Rewald, B., and Pierret, A. (2013). How to study deep roots-And why it matters. Front. Plant Sci. 4:299. doi: 10.3389/fpls.2013.00299
Medina, R. D., Faloci, M. M., Gonzalez, A. M., and Mroginski, L. A. (2007). In vitro cultured primary roots derived from stem segments of cassava (Manihot esculenta) can behave like storage organs. Ann. Bot. 99, 409–423. doi: 10.1093/aob/mcl272
Melteras, M., Lebot, V., Asher, C. J., and O’Sullivan, J. N. (2008). Crop development and root distribution in lesser yam (Dioscorea esculenta): implications for fertilization. Exp. Agric. 44, 209–221. doi: 10.1017/S0014479708006339
Meng, Y., Ma, X., Chen, D., Wu, P., and Chen, M. (2010). MicroRNA mediated signaling involved in plant root development. Biochem. Biophys. Res. Commun. 393, 345–349. doi: 10.1016/j.bbrc.2010.01.129
Menzel, M. I., Oros-Peusquens, A., Pohlmeier, A., Shah, N. J., Schurr, U., and Schneider, U. H. (2007). 1H-NMR imaging and relaxation mapping - a tool to distinguish the geographical origin of German white asparagus? J. Plant Nutr. Soil Sci. 170, 24–38. doi: 10.1002/jpln.200625114
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nielsen, K. L., Bouma, T. J., Lynch, J. P., and Eissenstat, D. M. (1998). Effects of phosphorus availability and vesicular arbuscular mycorrhizas on the carbon budget of common bean (Phaseolus vulgaris). New Phytol. 139, 647–656. doi: 10.1046/j.1469-8137.1998.00242.x
Nielsen, K. L., Eshel, A., and Lynch, J. P. (2001). The effect of phosphorus availability on the carbon economy of contrasting common bean (Phaseolus vulgaris L.) genotypes. J. Exp. Bot. 52, 329–339. doi: 10.1093/jexbot/52.355.329
Nieuwland, J., Maughan, S., Dewitte, W., Scofield, S., Sanz, L., and Murray, J. A. (2009). The D-type cyclin CYCD4;1 modulates lateral root density in Arabidopsis by affecting the basal meristem region. Proc. Natl. Acad. Sci. U.S.A. 106, 22528–22533. doi: 10.1073/pnas.0906354106
Noh, S. A., Lee, H.-S., Huh, E. J., Huh, G. H., Paek, K.-H., Shin, J. S., et al. (2010). SRD1 is involved in the auxin-mediated initial thickening growth of storage root by enhancing proliferation of metaxylem and cambium cells in sweetpotato (Ipomoea batatas). J. Exp. Bot. 61, 1337–1349. doi: 10.1093/jxb/erp399
Noh, S. A., Lee, H.-S., Kim, Y.-S., Paek, K.-H., Shin, J. S., and Bae, J. M. (2012). Down-regulation of the IbEXP1 gene enhanced storage rootdevelopment in sweetpotato. J. Exp. Bot. 63, 695–709. doi: 10.1093/jxb/ers236
Okushima, Y., Fukaki, H., Onoda, M., Theologis, A., and Tasaka, M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19, 118–130. doi: 10.1105/tpc.106.047761
Orman-Ligeza, B., Parizot, B., Gantet, P. P., Beeckman, T., Benett, M. J., and Draye, X. (2013). Post-embryonic root organogenesis in cereals: branching out from model plants. Trends Plant Sci. 18, 459–467. doi: 10.1016/j.tplants.2013.04.010
O’Sullivan, J. N. (2008). Root distribution of yam (Dioscorea alata) determined by strontium tracer. Exp. Agric. 44, 223–233. doi: 10.1017/S0014479708006169
Overvoorde, P., Fukaki, H., and Beeckman, T. (2010). Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2010:a001537. doi: 10.1101/cshperspect.a001537
Paez-Garcia, A., Motes, C. M., Scheible, W., Chen, R., Blancaflor, E. B., and Monteros, M. J. (2015). Root traits and phenotyping strategies for plant improvement. Plants 4, 334–355. doi: 10.3390/plants4020334
Palta, J. P. (2010). Improving potato tuber quality and production by targeted calcium nutrition: the discovery of tuber roots leading to a new concept in potato nutrition. Potato Res. 53, 267–275. doi: 10.1007/s11540-010-9163-0
Pardales, J. R., Baiioc, D. M., Yamauchi, A., Iijima, M., and Kono, Y. (1999). Root system development of cassava and sweetpotato during early growth stage as affected by high root zone temperature. Plant Prod. Sci. 2, 247–251. doi: 10.1626/pps.2.247
Pardales, J. R., and Esquibel, C. B. (1996). Effect of drought at different time during establishment period on the root system development of cassava. Jpn. J. Crop Sci. 65, 93–97. doi: 10.1626/jcs.65.93
Pardales, J. R., and Yamauchi, A. (2003). Regulation of root development in sweetpotato and cassava by soil moisture during their establishment period. Plant Soil 255, 201–208. doi: 10.1023/A:1026160309816
Parker, C. J., Carr, M. K. V., Jarvis, N. J., Evans, M. T. B., and Lee, V. H. (1989). Effects of subsoil loosening and irrigation on soil physical properties, root distribution and water uptake of potatoes (Solanum tuberosum). Soil Tillage Res. 13, 267–285. doi: 10.1016/0167-1987(89)90003-2
Porter, G. A., Opena, G. B., Bradbury, W. B., McBurnie, J. C., and Sisson, J. A. (1999). Soil management and supplemental irrigation effects on potato: I. Soil properties, tuber yield, and quality. Agron. J. 91, 416–425.
Postma, J. A., Dathe, A., and Lynch, J. P. (2014). The optimal lateral root branching density for maize depends on nitrogen and phosphorus availability. Plant Physiol. 166, 590–602. doi: 10.1104/pp.113.233916
Postma, J. A., and Lynch, J. P. (2011). Root cortical aerenchyma enhances the growth of maize on soils with suboptimal availability of nitrogen, phosphorus, and potassium. Plant Physiol. 156, 1190–1201. doi: 10.1104/pp.111.175489
Rasmusson, D. C. (1987). An evaluation of ideotype breeding. Crop Sci. 27, 1140–1146. doi: 10.2135/cropsci1987.0011183X002700060011x
Ravi, V., Chakrabarti, S. K., Makeshkumar, T., and Saravanan, R. (2014). Molecular regulation of storage root formation and development in sweet potato. Hortic. Rev. 42, 157–208. doi: 10.1002/9781118916827.ch03
Ravi, V., Naskar, S. K., Makeshkumar, T., Babu, B., and Prakash Krishnan, B. S. (2009). Molecular physiology of storage root formation and development in sweet potato (Ipomoea batatas (L.) Lam.). J. Root Crops 35, 1–27.
Rich, S. M., and Watt, M. (2013). Soil conditions and cereal root system architecture: review and considerations for linking Darwin and Weaver. J. Exp. Bot. 64, 1193–1208. doi: 10.1093/jxb/ert043
Richardson, A. E., Hocking, P. J., Simpson, R. J., and George, T. S. (2009). Plant mechanisms to optimize access to soil phosphorus. Crop Pasture Sci. 60, 124–143. doi: 10.1071/CP07125
Roumeliotis, E., Kloosterman, B., Oortwijn, M., Kohlen, W., Bouwmeester, H. J., Visser, R. G., et al. (2012). The effects of auxin and strigolactones on tuber initiation and stolon architecture in potato. J. Expt. Bot. 63, 4539–4547. doi: 10.1093/jxb/ers132
Roy, S. J., Huang, W., Wang, X. J., Evrard, A., Schmöckel, S. M., Zafar, Z. U., et al. (2013). A novel protein kinase involved in Na(+) exclusion revealed from positional cloning. Plant Cell Environ. 36, 553–568. doi: 10.1111/j.1365-3040.2012.02595.x
Rus, A., Baxter, I., Muthukumar, B., Gustin, J., Lahner, B., Yakubova, E., et al. (2006). Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2:e210. doi: 10.1371/journal.pgen.0020210
Smith, S., and De Smet, I. (2012). Root system architecture: insights from Arabidopsis and cereal crops. Philos. Trans. R. Soc. B. Biol. Sci. 367, 1441–1452. doi: 10.1098/rstb.2011.0234
Sojikul, P., Saithong, T., Kalapanulak, S., Pisuttinusart, N., Limsirichaikul, S., Tanaka, M., et al. (2015). Genome-wide analysis reveals phytohormone action during cassava storage root initiation. Plant Mol. Biol. 88, 531–543. doi: 10.1007/s11103-015-0340-z
Subere, J. O., Bolatete, D., Bergantin, R., Pardales, A., Belmonte, J. J., Mariscal, A., et al. (2009). Genotypic variation in responses of cassava (Manihot esculenta Crantz) to drought and rewatering: root system development. Plant Prod. Sci. 12, 462–474. doi: 10.1626/pps.12.462
Tanaka, M., Kato, N., Nakayama, H., Nakatani, M., and Takahata, Y. (2008). Expression of class I knotted1-like homeobox genes in the storage roots of sweet potato. J. Plant Physiol. 165, 1726–1735. doi: 10.1016/j.jplph.2007.11.009
Tanaka, M., Takahata, Y., and Nakatani, M. (2005). Analysis of genes developmentally regulated during storage root formation of sweetpotato. J. Plant Physiol. 162, 91–102. doi: 10.1016/j.jplph.2004.06.003
Tanaka, N., Kato, M., Tomioka, R., Kurata, R., Fukao, Y., Aoyama, T., et al. (2014). Characteristics of a root hair-less line of Arabidopsis thaliana under physiological stresses. J. Exp. Bot. 65, 1497–1512. doi: 10.1093/jxb/eru014
Tardieu, F., and Tuberosa, R. (2010). Dissection and modelling of abiotic stress tolerance in plants. Curr. Opin. Plant Biol. 13, 206–212. doi: 10.1016/j.pbi.2009.12.012
Tirlapur, U. K., and König, K. (1999). Near-infrared femtosecond laser pulses as a novel non-invasive means for dye-permeation and 3D imaging of localised dye-coupling in the Arabidopsis root meristem. Plant J. 20, 363–370. doi: 10.1046/j.1365-313X.1999.t01-1-00603.x
Togari, Y. (1950). A study of tuberous root formation in sweet potato. Bul. Nat. Agr. Expt. Sta. Tokyo 68, 1–96.
Trachsel, S., Kaeppler, S. M., Brown, K. M., and Lynch, J. P. (2011). Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 341, 75–87. doi: 10.1007/s11104-010-0623-8
Tracy, S. R., Roberts, J. A., Black, C. R., McNeill, A., Davidson, R., and Mooney, S. J. (2010). The X-factor: visualizing undisturbed root architecture in soils using X-ray computed tomography. J. Exp. Bot. 61, 311–313. doi: 10.1093/jxb/erp386
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102.
Vanneste, S., De Rybel, B., Beemster, G. T., Ljung, K., De Smet, I., Van Isterdael, G., et al. (2005). Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17, 3035–3050. doi: 10.1105/tpc.105.035493
Villordon, A., LaBonte, D., Firon, N., and Carey, E. (2013). Variation in nitrogen rate and local availability alter root architecture attributes at the onset of storage root initiation in ‘Beauregard’ sweetpotato. HortScience 48, 808–815. doi: 10.1371/journal.pone.0107384
Villordon, A., LaBonte, D., Solis, J., and Firon, N. (2012). Characterization of lateral root development at the onset of storage root initiation in ‘Beauregard’ sweetpotato adventitious roots. HortScience 47, 961–968.
Villordon, A. Q., and Clark, C. A. (2014). Variation in virus symptom development and root architecture attributes at the onset of storage root initiation in ‘Beauregard’ sweetpotato plants grown with or without nitrogen. PLoS ONE 9:e107384. doi: 10.1371/journal.pone.0107384
Villordon, A. Q., Firon, N., Clark, C. A., and Smith, A. (2014a). Manipulating Root System Architecture in Sweetpotato for Global Food Security: Progress Prospects And Applications. Paper Presented at the Society for Experimental Biology meeting, Roots for Global Food Security session, Manchester. doi: 10.6084/m9.figshare.1462655
Villordon, A. Q., Ginzberg, I., and Firon, N. (2014b). Root architecture and root and tuber crop productivity. Trend. Plant Sci. 19, 419–425. doi: 10.1016/j.tplants.2014.02.002
Vos, J., and Groenwold, J. (1986). Root growth of potato crops on a marine-clay soil. Plant Soil 94, 17–33. doi: 10.1007/BF02380587
Voytas, D. F. (2013). Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 64, 327–350. doi: 10.1146/annurev-arplant-042811-105552
Walch-Liu, P., Ivanov, I. I., Filleur, S., Gan, Y., Remans, T., and Forde, B. G. (2006). Nitrogen regulation of root branching. Ann. Bot. 97, 875–881. doi: 10.1093/aob/mcj601
Wasson, A., Rebetzke, G., Kirkegaard, J., Christopher, J., Richards, R., and Watt, M. (2014). Soil coring at multiple field environments can directly quantify variation in deep root traits to select wheat genotypes for breeding. J. Exp. Bot. 65, 6231–6249. doi: 10.1093/jxb/eru250
Wasson, A., Richards, R., Chatrath, R., Misra, S., Prasad, S. S., Rebetzke, G., et al. (2012). Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. J. Exp. Bot. 63, 3485–3498. doi: 10.1093/jxb/ers111
Weaver, J. E., and Bruner, W.E. (eds) (1927). Root Development of Vegetable Crops. New York, NY: McGraw-Hill Book Company.
Wilson, L. A., and Lowe, S. B. (1973). The anatomy of the root system in West Indian sweet potato [Ipomoea batatas (L.) Lam.] cultivars. Ann. Bot. 37, 633–643.
Wishart, J., George, T. S., Brown, L. K., Ramsay, G., Bradshaw, J. E., White, P. J., et al. (2013). Measuring variation in potato roots in both field and glasshouse: the search for useful yield predictors and a simple screen for root traits. Plant Soil 368, 231–249. doi: 10.1007/s11104-012-1483-1
Wishart, J., George, T. S., Brown, L. K., White, P. J., Ramsay, G., Jones, H., et al. (2014). Field phenotyping of potato to assess root and shoot characteristics associated with drought tolerance. Plant Soil 378, 351–363. doi: 10.1007/s11104-014-2029-5
Xie, F., Frazier, T. P., and Zhang, B. (2011). Identification, characterization and expression analysis of microRNAs and their targets in the potato (Solanum tuberosum). Gene 473, 8–22. doi: 10.1016/j.gene.2010.09.007
Yasui, K. (1944). Notes on the propagation of sweet potato, Ipomoea batatas Lam. I. The adventives bud formation in the root-tuber. Proc. Imperial Acad. 20, 41–44.
York, L. M., Nord, E. A., and Lynch, J. P. (2013). Integration of root phenes for soil resource acquisition. Front. Plant Sci. 4:355. doi: 10.3389/fpls.2013.00355
Zhan, A., and Lynch, J. P. (2015). Reduced frequency of lateral root branching improves N capture from low-N soils in maize. J. Exp. Bot. 66, 2055–2065. doi: 10.1093/jxb/erv007
Keywords: root system architecture (RSA), abiotic stress tolerance, root and tuber crops, drought tolerance, sweetpotato, potato, yam, cassava
Citation: Khan MA, Gemenet DC and Villordon A (2016) Root System Architecture and Abiotic Stress Tolerance: Current Knowledge in Root and Tuber Crops. Front. Plant Sci. 7:1584. doi: 10.3389/fpls.2016.01584
Received: 10 May 2016; Accepted: 07 October 2016;
Published: 01 November 2016.
Edited by:
Shabir Hussain Wani, SKUAST-Kashmir, IndiaReviewed by:
Anca Macovei, University of Pavia, ItalyCopyright © 2016 Khan, Gemenet and Villordon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. A. Khan, YXdhaXMua2hhbkBjb3JuZWxsLmVkdQ==
†Present address: M. A. Khan, Section of Plant Pathology and Plant-Microbe Biology, School of Integrative Plant Science, Cornell University, New York State Agricultural Experiment Station, Geneva, NY, USA
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.