
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 21 September 2016
Sec. Plant Biotechnology
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.01366
This article is part of the Research Topic Plant cell walls as extra cellular matrices (ECMs) and biosensors View all 20 articles
 Mingliang Zhang1,2,3
Mingliang Zhang1,2,3 Feng Wei1,2,3
Feng Wei1,2,3 Kai Guo1,2,4
Kai Guo1,2,4 Zhen Hu1,2,3
Zhen Hu1,2,3 Yuyang Li1,2,3
Yuyang Li1,2,3 Guosheng Xie1,2,3
Guosheng Xie1,2,3 Yanting Wang1,2,3
Yanting Wang1,2,3 Xiwen Cai5
Xiwen Cai5 Liangcai Peng1,2,3
Liangcai Peng1,2,3 Lingqiang Wang1,2,3*
Lingqiang Wang1,2,3*We report isolation and characterization of a fragile culm mutant fc116 that displays reduced mechanical strength caused by decreased cellulose content and altered cell wall structure in rice. Map-based cloning revealed that fc116 was a base substitution mutant (G to A) in a putative beta-1,6-N-acetylglucosaminyltransferase (C2GnT) gene (LOC_Os05g07790, allelic to BC10). This mutation resulted in one amino acid missing within a newly-identified protein motif “R, RXG, RA.” The FC116/BC10 gene was lowly but ubiquitously expressed in the all tissues examined across the whole life cycle of rice, and slightly down-regulated during secondary growth. This mutant also exhibited a significant increase in the content of hemicelluloses and lignins, as well as the content of pentoses (xylose and arabinose). But the content of hexoses (glucose, mannose, and galactose) was decreased in both cellulosic and non-cellulosic (pectins and hemicelluloses) fractions of the mutant. Transcriptomic analysis indicated that the typical genes in the fc116 mutant were up-regulated corresponding to xylan biosynthesis, as well as lignin biosynthesis including p-hydroxyphenyl (H), syringyl (S), and guaiacyl (G). Our results indicate that FC116 has universal function in regulation of the cell wall polymers in rice.
Plant cell walls are complex, dynamic cellular structures essential for plant growth, development, physiology, and adaptation. Cell walls can be classed as primary or secondary, depending upon whether they are extendable or non-extendable during organ growth (Lee et al., 2011). Primary cell wall is a flexible matrix that mainly composed of cellulose, hemicelluloses, pectins, and proteins, allowing directed cell growth. Specialized cell types differentiate by depositing a secondary cell wall, providing mechanical support for water and nutrient transport as well as the barrier against invading pathogens (Cantu et al., 2008; Wang and Dixon, 2012). In general, cellulose makes up almost 25–30% of dry matter in grasses (Zhang et al., 2013) and 40–45% in woody plants (Smook, 1992). Hemicelluloses include polysaccharides such as xyloglucans, xylans, mannans, and glucomannans, and β-(1,3-1,4)-glucans (Scheller and Ulvskov, 2010). Lignin is an amorphous polymer of phenylpropane units with three major monomers: p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S), laid down during secondary cell wall formation (Zhao and Dixon, 2011; Sun et al., 2012).
The polymers contained within the cell wall are an important renewable resource for humans as dietary fiber, as raw material for pulp manufacturing, and for biofuel production (Taylor-Teeples et al., 2015). Elucidation of the regulatory mechanisms of cell wall synthesis will facilitate the engineering of plant feedstocks suitable for biofuel production (Wang and Dixon, 2012). Although significant progress has been made in understanding the regulation of secondary cell wall formation in Arabidopsis thaliana (Zhong and Ye, 2007; Demura and Ye, 2010; Wang and Dixon, 2012; Taylor-Teeples et al., 2015), our knowledge of the precise regulatory mechanisms of cell wall biosynthesis is still limited especially in monocot plants. Thousands of genes have been reported to be responsible for plant cell wall biogenesis and modifications (Carpita et al., 2001; Torney et al., 2007; Xie and Peng, 2011). Among the products of the genes, glycosyltransferases (GTs) are one of the largest enzyme groups required for the synthesis of complex cell wall polysaccharides and glycoproteins in plants (http://www.cazy.org; Rosén et al., 2004; Scheible and Pauly, 2004). Further, the rapid increase in the availability of genomic resources and improvement in molecular techniques have led to additional GTs being identified through bioinformatic analysis. However, only a small portion of the genes involving in cell wall formation have been functionally characterized primarily in model organisms A. thaliana and rice (Somerville et al., 2004; Zhang and Zhou, 2011).
A powerful approach to probe the functions of individual components of cell walls is through the identification and characterization of relevant mutants (Reiter et al., 1993). Over the last two decades, dozens of genes have been identified to be involved in cell wall synthesis and modifications by characterizing cell wall mutants in Arabidopsis and rice (Turner and Somerville, 1997; Sun et al., 2012). Studies on the brittle culm (bc) mutants of rice have identified several genes responsible for secondary cell wall biosynthesis. These studies have revealed some mechanistic aspects of cell wall biosynthesis and identified associated genes in rice (Zhang and Zhou, 2011). BC7(t) and BC11 encoding cellulose synthases are assembled into the cellulose synthase complex (CSC) in Golgi and then translocated to the plasma membrane (Yan et al., 2007). BC1 encodes a COBRA-like protein that is released to the extracellular matrix and is essential for cellulose assembly (Li et al., 2003; Sato et al., 2010). BC3 encoding dynamin-related proteins (DRPs) functions in membrane vesiculation and trafficking (Hirano et al., 2010; Xiong et al., 2010). BC12/GDD1 encodes a kinesin-like protein secreted to the cytoplasm and functions in the cortical microtubule-dependent cellulose deposition and in regulation of GA biosynthesis (Zhang et al., 2010; Li et al., 2011). BC15 encodes a membrane-associated chitinase-like protein, a class of glycosyl hydrolases that are required for cellulose biosynthesis (Wu et al., 2012). More recently, OsMYB103L, an R2R3-MYB transcription factor, was found involved in GA-mediated regulation of secondary cell wall biosynthesis. It influences leaf rolling and mechanical strength in rice (Yang et al., 2014; Ye et al., 2015).
Among the brittle culm genes, BC10 is proposed to encode a protein that belongs to a newly-identified glycosyltransferase family in plants. It is located in Golgi or part of the structural and is regulatory protein for cell wall remodeling/formation (Zhou et al., 2009). Mutation in the “domain of unknown function 266” (DUF266) of BC10 resulted in a truncated protein that modifies the levels of cellulose and arabinogalactan proteins (AGPs) in cell walls and leads to brittleness of the plant parts and retarded growth. Since DUF266 is the only domain predicted using Pfam and is plant-specific in this protein family, it may be critical for the function of BC10 (Finn et al., 2006). It was predicted that the mutation in bc10 probably leads to perturbation of the biosynthesis of glycoprotein and/or non-cellulosic polysaccharides (Zhou et al., 2009). However, the precise functions of BC10 proteins and their roles in cellulose biosynthesis and secondary cell wall formation have not been fully elucidated. How the bc10 mutation influences biosynthesis of cellulose and AGP as well as other cell wall polymers remains elusive. Here, we reported the characterization and map-based gene cloning of a novel rice fragile culm mutant designated fc116 that exhibits abnormal sclerenchyma secondary cell walls in the internodes.
The fragile culm mutant fc116 was isolated from the EMS-induced mutant pool of the japonica rice cultivar “Zhonghua 11” (ZH11) in year 2010, and was crossed with two different rice cultivars “Minghui 63” (MH63) and “9311,” respectively. Two F2 mapping populations were generated and planted in the field in year 2013 and map-based cloning was conducted to identify the mutant allele responsible for the fc116 phenotype. The field experiments were conducted in the rice-growing seasons on the experimental farm of Huazhong Agricultural University, Wuhan, China. The sowing dates were May 25. Ten plants per line were transplanted in a single row with 16.5 cm between plants and 26.4 cm between rows. Field management essentially followed normal agricultural practice.
The second internodes of stem and flag leaves of wild-type and fc116 plants were tested for mechanical properties at the milk maturity stage. The stems and leaves were cut into segments of 5 cm in length. The stretching force of the samples before being broken was measured with a universal force/length testing device (model RH-K300, Guangzhou, China). The newton is used as the unit of extension force (EF). The stem tissues at 30 days after flowering were used to detect the plant lodging index. The breaking resistance of the third internode was detected using a Prostrate Tester (DIK 7401, Japan), with the distance between fulcra of the tester at 5 cm. The newton is used as the unit of breaking force (BF). Fresh weight (W) of the upper portion of the plant was measured including panicle and the three internodes, leaf, and leaf sheath. Bending moment (BM) and lodging index (LI) were calculated using the following formula: BM = Length from the third internode to the top of panicle × W, LI = BM/BF
The initial mapping of fc116 was performed using 289 SSR markers distributed throughout the rice genome in bulked pools and parents. The genomic region harboring the fc116 locus was saturated with newly developed molecular markers. Using 1917 F2 plants derived from the cross of the mutant fc116 with MH63, the mutated locus was finally narrowed down to a 202-kb region on chromosome 5. The corresponding DNA and cDNA fragments were amplified from fc116 and its wild type “Zhonghua 11” (ZH11) plants using KOD plus (Toyobo, www.toyobo-global.com) and sequenced with a 3730 sequencer (ABI).
Prediction of FC116 protein domain was performed by the SMART (http://smart.embl-heidelberg.de/). The protein transmembrane helices were predicted by TMHMM2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/). Three dimensional structure of FC116 was performed using SWISS-MODEL online prediction software (http://swissmodel.expasy.org/). The unrooted phylogenetic trees were constructed with the MEGA6 program and the neighbor joining method with 1000 bootstrap replicates. We performed exon-intron structure analysis using GSDS (http://gsds.cbi. pku.edu.cn/). FC116 protein sequences alignment was performed by ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi).
The expression profile data of OsFC116 in 33 tissue samples (Table S2) of Zhenshan 97 (ZS97) and Minghui 63 (MH63) were obtained from the CREP database (http://crep.ncpgr.cn). The expression values were log-transformed, and cluster analyses were performed using a software cluster with Euclidean distances and the hierarchical cluster method of “complete linkage clustering.”
For histochemical localization of lignin, a Wiesner reaction was performed according to Strivastava (1966) with minor modifications. Fresh hand-cut sections from the second culms were incubated for 1 min in phloroglucin solution [5% in ethanol: water (95:5, v/v)], mounted on slides with 37% HCl, and photographed using a microscope (Olympus BX61, Japan). For cellulose staining, fresh hand-cut sections were stained with 0.1% calcofluor (Fluka) for 2 min. Excess calcofluor was washed away with tap water and slides were visualized with a fluorescent microscope (Olympus BX61, Japan).
The sample preparation and immunohistochemical localization of xylan were performed as previously described by Cao (Cao et al., 2014). The main vein of leaves at the trefoil stage were cut into 0.2–1.5 cm pieces and subsequently fixed with glutaraldehyde. The above pieces were embedded by paraffin and sectioned into slices with 8.0 μm in thickness. Monoclonal antibody CCRC-M148 was used to detect xylan. The sections were incubated in PBS (phosphate buffer saline) contained 3% SMP (3% skim milk powder, w/v) containing 10 μg/ml monoclonal antibody CCRC-M148 for 2 h. The immunolabeled samples were washed three times (5 min each) with PBS and incubated with a 100-fold dilution of anti-mouse-IgG for the CCRC-M148 antibody in dark for 1 h. The anti-mouse-IgG antibody was labeled by fluorescein-isothiocyanate (FITC). Immunofluorescence sections were observed under an Olympus BX-61 microscope.
The method of plant cell wall fractionation was used to extract cellulose and hemicellulose, as described by Peng et al. (2000) and Li et al. (2015). We used potassium phosphate buffer (pH 7.0), chloroform-methanol (1: 1, v/v), and DMSO-water (9: 1, v/v) to remove soluble sugar, lipids, and starch from the samples. The remaining pellets as total crude cell wells were suspended in 0.5% (w/v) ammonium oxalate and heated for 1 h in a boiling water bath, and the supernatants were total pectins. The remaining pellets were suspended in 4 M KOH containing 1.0 mg/mL sodium borohydride for 1 h at 25°C. The mixtures were then centrifuged and the supernatants were collected and regarded as KOH- extractable hemicelluloses. The remaining pellets were reacted with 2 M trifluoroacetic acid (TFA) and the supernatants were collected and regarded as non-KOH- extractable hemicelluloses. The pellets were further extracted with acetic-nitric acids-water (8: 1: 2) for 1 h at 100°C, and the remaining materials were regarded as crystalline cellulose. The supernatants extracted from 4 M KOH treatment and from TFA reaction were separately transfered into 5 mL screw-cap test tubes and were neutralized, dialysed, and lyophilized according to the method described by Xu et al. (2012), then subjected to GC-MS measurements or stored −20°C until GC-MS measurements. During the treatments, the screw-cap test tubes were sealed with adhesive tape to avoid possible sample lost.
The hexose and pentose assays were performed using an UV-vis spectrometer according to Li et al. (2013) (V-1100D, Shanghai MAPADA Instruments Co., Ltd. Shanghai, China). Hexoses were measured by the anthrone/H2SO4 method and pentoses by orcinol/HCl (Dische, 1962). As the high pentose level affects the absorbance reading at 620 nm for the hexose assay by the anthrone/H2SO4 method, the deduction from pentoses was carried out for a final hexose calculation. A series of xylose concentrations were analyzed for plotting the standard curve referred for the deduction, which was verified by GC-MS analysis.
Total lignin content was measured by the two-step acid hydrolysis method according to the Laboratory Analytical Procedure of the National Renewable Energy Laboratory with minor modifications as described by Wu et al. (2013) and Li et al. (2015). The acid-insoluble lignin (AIL) was calculated gravimetrically as acid-insoluble residues after subtraction for ash, and the acid-soluble lignin (ASL) was detected by UV spectroscopy. All experiments were carried out in technological triplicate.
GC/MS analysis was conducted with SHIMADZU GCMSQP2010 Plus according to Xu et al. (2012). This analysis used a Restek Rxi-5 ms, 30 m × 0.25 mm ID × 0.25 um df column. Monosaccharide standards including L-rhamnose, L-arabinose, L-fucose, D-xylose, D-galactose, D-glucose, and D-mannose, were obtained from Sinopham Chemical Reagent Co., Ltd. The GC-MS Analytical Conditions are listed as the follows: Carrier gas: He. Injection Method: Split. Injection port: 250°C, Interface: 250°C. Injection Volume: 1.0 μL. The temperature program: from 170°C (held for12 min) to 220°C (held for 8 min) at 3°C/min. Ion source temperature: 200°C, ACQ Mode: SIM. The mass spectrometer was operated in the EI mode with ionization energy of 70 ev. Mass spectra were acquired with full scans based on the temperature program from 50 to 500 m/z in 0.45 s. Calibration curves of all analytes routinely yielded correlation coefficients 0.999 or better. Peaks were identified by mass profiles and/or retention times of standards. Monosaccharides were quantified based on standard curves.
Lignin monomers were determined by HPLC as previously described by Xu et al. (2012). Standard chemicals, including p-Hydroxybenzaldehyde (H), guaiacy (G), and syringaldehyde (S), were purchased from Sinopharm Chemical Reagent Co., Ltd. The samples after pretreatment were used to detect lignin monomers by HPLC. The solution was filtered with membrane filter (0.22 μm). The filtered solution was injected into HPLC with 20 μL (Waters 1525 HPLC) column Kromat Universil C18 (4.6 mm × 250 mm, 5 μm) operating at 28°C with CH3OH: H2O: HAc (25:74:1, v/v/v) carrier liquid (flow rate: 1.1 mL/min).
The sample preparation and cellulose crystallinity index (CrI) determination were conducted as previously described by Zhang et al. (2013). X-ray diffraction (XRD) method was used to determine cellulose crystallinity index (CrI) with Rigaku-D/MAX instrument (Uitima III, Japan). The raw biomass powder was laid on the glass sample holder (35 × 50 × 5 mm) and detected under plateau conditions. Ni-filtered Cu Kα radiation (λ = 0.154056 nm) generated at voltage of 40 kV and current of 18 mA, and scanned at speed of 0.0197°/s from 10 to 45°. The crystallinity index (CrI) was estimated using the intensity of the 200 peak (I200, θ = 22.5°) and the intensity at the minimum between the 200 and 110 peaks (Iam, θ = 18.5°) as the follow: CrI = 100 (I200-Iam)/I200 (Segal et al., 1959). I200 represents both crystalline and amorphous materials while Iam represents amorphous material. Standard error of the CrI method was detected at ±0.05–0.15 using five representative samples in triplicate.
RNA extraction was performed as previously described by Liu et al. (2016). The raw data were cleaned by removing reads containing the poly-N or adapter and low-quality reads, which obtained from Illumina sequencing. The quality of the raw reads was assessed using FastQC (version 0.10.1) (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). The high quality data was used in all downstream analyses. After trimming low-quality bases (Q < 20) from the 5′ and 3′ ends of the remaining reads, the resulting high-quality reads were mapped onto the Nipponbare reference genome collected from Ensemble v17 using tophat (Sigurgeirsson et al., 2014). HTseq version 0.5.3p9 was used to convert aligned reads into counts per gene using the Ensembl v17 annotation of the rice genome (Fatima et al., 2014). The above analysis used the number of reads mapping over the entire gene length to identify differentially expressed genes. The expression level for each transcript was calculated as RPKM (Reads Per Kilobase exon Model per Million mapped reads)-derived read counts based on the number of uniquely mapped reads that overlapped with exonic regions. We used the DESeq R package (1.12.0) to identify the differentially expressed genes (DEGs) between mutant and wide-type (Burden et al., 2014). Then P-values were adjusted using the Benjamini and Hochberg method. A corrected P-value of 0.01 and log2 (fold change) of one were set as the thresholds for significant differential expression.
One thousand-grain weight was converted by calculating on the weight of 200 grains. Head rice yield was calculated as the percentage of intact grains to the total number of grains after milling. The measurement of grain chalkiness rate was the percentage of chalky grains in the total number of dehulled grains. The area of chalkiness in the grain was used as the measurement of chalkiness degree (Li et al., 2014). Grain was ground to flour, which was used for measuring amylose content and gel consistency, the two parameters for eating and cooking qualities, following the methods of Wang et al. (2007).
The prominent phenotype of the fc116 plants is the brittle internodes, sheaths, and leaves after booting stage (Figures 1A–D). We quantitatively measured the extension force (EF), a criterion indicating the elasticity of the plant organs, in the wild and mutant types of the rice accession. The organ with lower EF is more brittle than those with higher EF. We found that EF of the fc116 mutant in culms was reduced by 67% comparing to its wild type at the heading stage (Figure 1F). However, the breaking force (BF), a parameter for lodging resistance, of the mutant was 36% higher than the wild type (Figure 1G). At the seedling stage, the mutant plants were not brittle, which was different from other brittle culm mutants that show brittle culms at seedling stage. The fc116 mutant plants had slightly shorter stature/roots and fewer tillers than wild-type plants (Figures 1A,B). The mutant showed decreased plant height and tillers at the maturity stage although the growth period of the mutant was similar to the wild type (Figures 1B,E). Therefore, the mutation caused not only reduction in mechanical strength of plant parts, but also abnormalities in the growth and development. However, fc116 showed no changes in seed weight, spikelet size, grain qualities, and lodging resistance (Table S1, Figure S1). We noticed that the mutant plants of BC14//OsNST1 encoding a UDP-glucose transporter showed additional morphological abnormalities including shortened stature and reduced seed size and fertility.
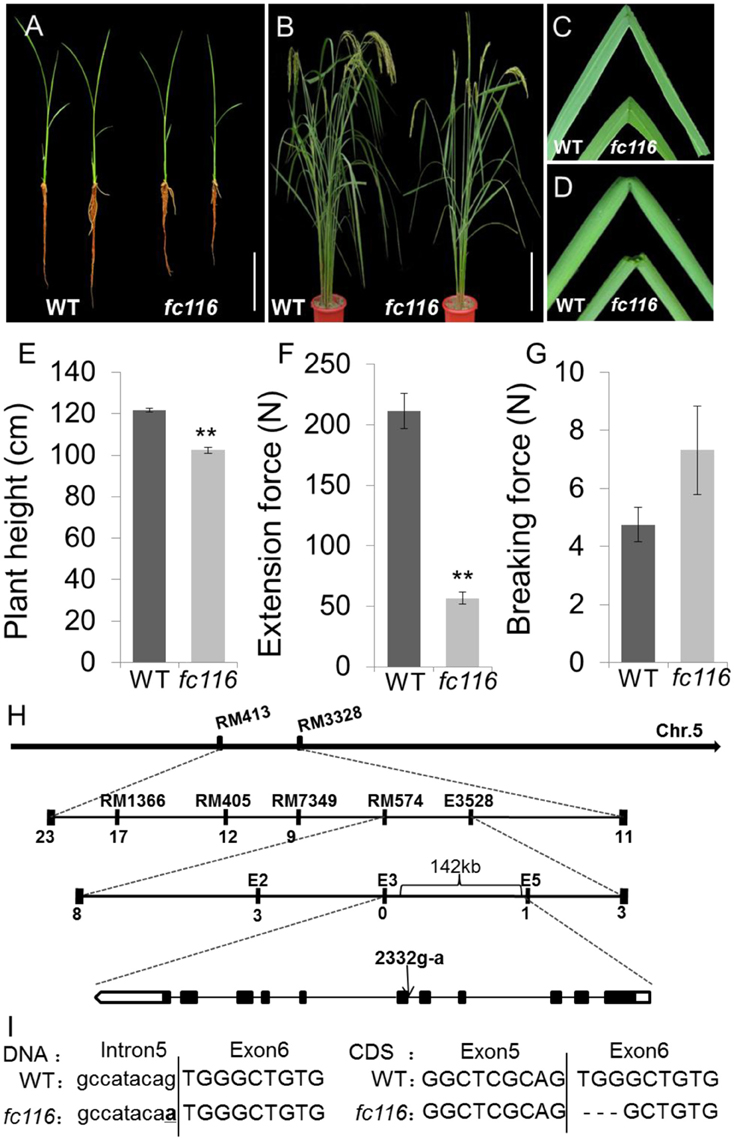
Figure 1. Phenotypes and map-based cloning of the gene for fc116. (A) Three-week-old wild-type and fc116 plants. (B) Three-month-old wild-type and fc116 plant. (C,D) The mutant leaves and internodes are easily broken. (E) Plant height of wild-type and fc116. (F,G) Measurement of the force required for pulling and breaking wild-type and fcll6 mutant internodes. (H) The fc116 locus was mapped to a 142 kb region on chromosome 5. Black boxes represent exons and black lines represent introns. (I) Sequencing analysis revealed a point mutation in genomic DNA that results 3 bp deletion in fc116 CDS. Bar = 5 cm (A,B). **Significantly different (t = test at P < 0.01) compared with wild-type (E,F).
All F1 progeny derived from the both crosses fc116/MH63 and fc116/9311 showed a normal phenotype similar to that of the wild-type. Tests of heterozygotes with F2 progeny yielded a segregation of 6012 normal and 1917 brittle plants in fc116/MH63, and 7939 normal and 1842 brittle plant in fc116/9311 [X2 (3:1) = 0.37 and 0.31, < p(0.05) = 4.16 and 3.98], indicating that the brittle phenotype of the fc116 mutant was due to a recessive mutation at a single nuclear gene locus. After screening with 289 simple sequence repeat (SSR) markers that distribute evenly across the 12 rice chromosomes, the mutant locus was mapped between markers RM413 and RM3328 on chromosome 5 (Figure 1H). Fine-mapping further placed this locus in the 132-kb genomic region between markers E3 and E5, which includes 9 predicted ORFs (Figure 1H).
After sequencing all of the candidate genes, a point mutation was found in the junction point of the 5th intron and 6th exon of an ORF (LOC_Os05g07790), which was annotated as DNA-binding protein by the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). The point mutation resulted in a 3 bp deletion at the beginning of the 6th exon of the coding sequence (CDS) of the mRNA of LOC_Os05g07790. That may be caused by a change in splicing pattern of the mRNA because the substitution mutation was in the recognition site for mRNA splicing (Figure 1I). This gene is allelic to bc10, that was reported had a mutation within the domain of a plant-specific domain with unknown function domain 266 (DUF266) located in a Core-2/I-Branching enzyme functional motif (PF02485) (Zhou et al., 2009; Figure 2A). The fc116 mutation occurs not in DUF266 domain (Sonnhammer et al., 1988; Krogh et al., 2001) (Figures 2B,C). The predicted three dimension structure of the protein was not changed (Figure 2D).
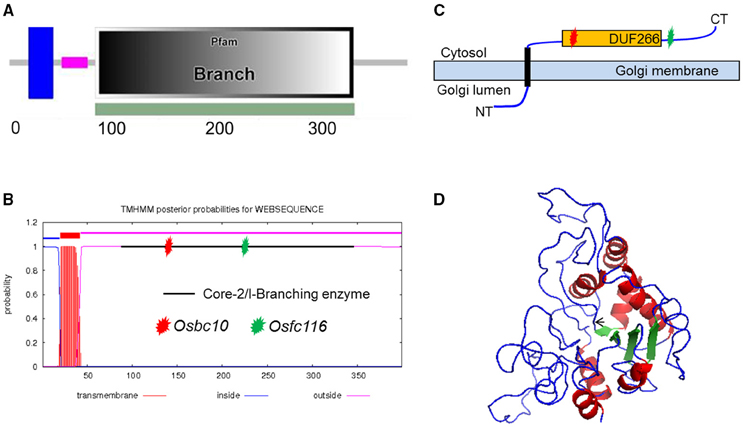
Figure 2. Analysis of FC116 protein. (A,B) Prediction of FC116 protein domain by the SMART and TMHMM2.0 program, showing a transmembrane helix domain between amino acid residues 20 and 42, a core-2/I-Branching enzyme between amino acid residues 86 and 345. Red star represents the mutation site of bc10; green star represents the mutation site of fc116. (C) Schematic diagram of the FC116 protein location in the Golgi membrane. (D) Three dimensional structure of FC116 protein. The black arrow was the deletion.
A BLASTX search for FC116 homologs identified 19 DUF266-containing proteins in the Oryza sativa (japonica) genome (Figure 3A). This is a family of two different beta-1, 6-N-acetylglucosaminyltransferase enzymes, I-branching enzyme and core-2 branching enzyme. I-branching enzyme is responsible for the production of the blood group I-antigen during embryonic development. Core-2 branching enzyme forms crucial side-chain branches in O-glycans. An unrooted phylogenetic tree was generated from the alignment of 19 DUF266-containing protein sequences into two distinct clusters (Figure 3A). Intron-exon pattern also revealed that these homologs are clustered into two major clades, implying that they may be related but have distinct functions (Figure 3A). FC116 is in Cluster I that have more introns than Cluster II and the closest homolog is LOC_Os01g50040. We found that the mutation occurred within a new highly conserved protein motif “R, RXG, RA” in this family (Figure 3B), The motif “R, RXG, RA” is also located in PF02485, but outside the “domain with unknown function 266” (DUF266) which has been proposed to be crucial for the protein function in a previous study (Zhou et al., 2009).
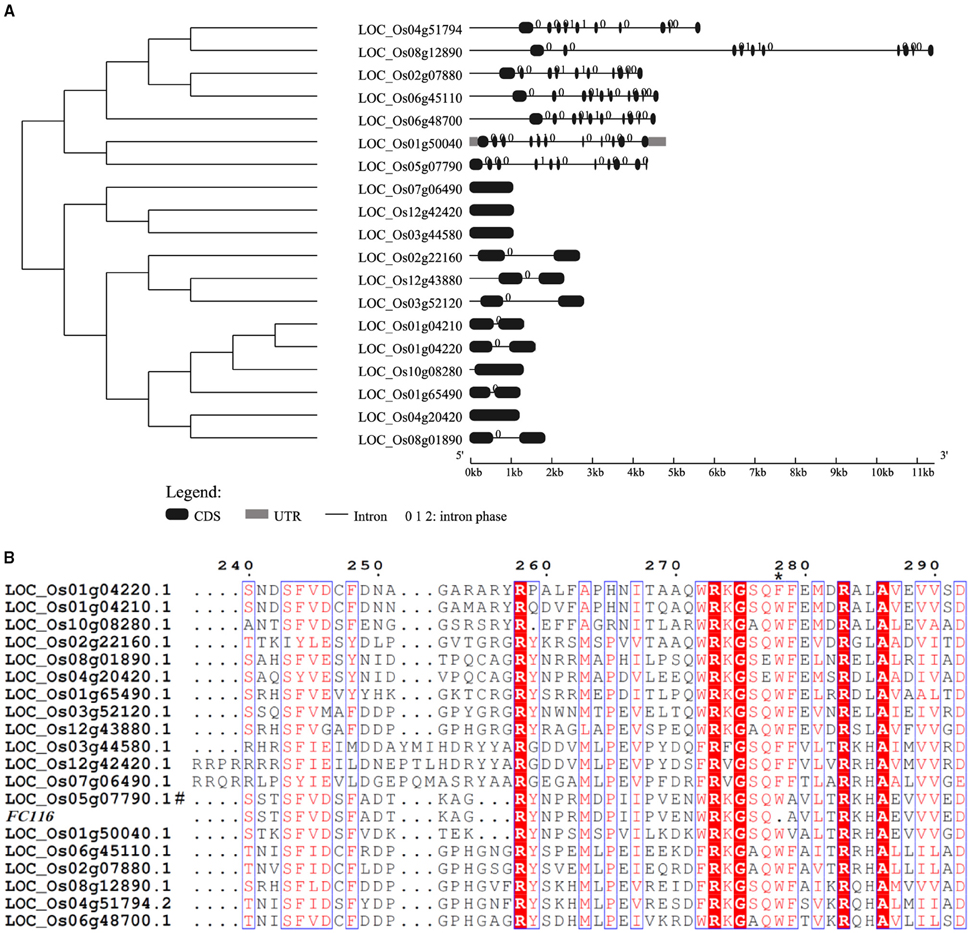
Figure 3. Phylogenetic tree and protein sequences alignment of FC116 protein family. (A) Unrooted tree of FC116 protein family and organization of exons and introns of the corresponding genes. (B) Black asterisk represents the deletion amino acid.
The data for expression pattern analysis of the FC116 gene downloaded from the CREP database (http://crep.ncpgr.cn) which was created during a rice transcriptome project using the Affymetrix Rice GeneChip microarray by rice group in our university (Wang et al., 2010a). FC116 exhibited a ubiquitous expression in most of the 33 tissues sampled from the entire life cycle of the rice plants (Figure 4A, Figure S2). It was moderately but ubiquitously expressed in almost all tissues with relatively high levels in the endosperm, plumule, and radicles, leaf, and root at three leaf stages, stem at heading stage (Figure 4A, Figure S2). These tissues were distinct in cell wall constitutions with the variation in contents of cellulose (2.3–26.6%), hemicelluloses (2.3–32.5%), and pectins (0.9–3.3%) as we measured in previous studies (Wang et al., 2010b; Guo et al., 2014).
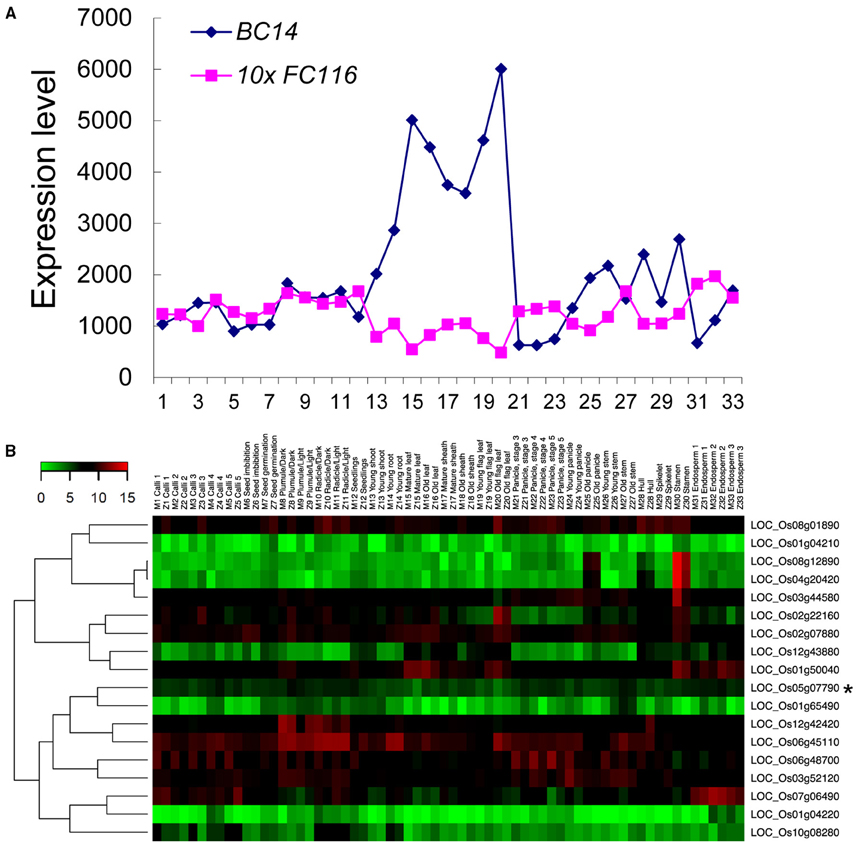
Figure 4. Co-expression profiling of the FC116 family in rice. (A) Expression patterns of the bc14 and fc116 in rice. The X-axis indicates the tissues at the developmental stages with 1, Calli (15 days after subculture); 2, Calli (5 days after regeneration); 3, Calli (Screening stage); 4, Calli (15 days after induction T2); 5, Calli (15 days after induction T3); 6, Seed imbibition; 7, Seed germination; 8, Plumule (48 h after emergence, Dark); 9, Plumule (48 h after emergence, Light); 10, Radicle (48 h after emergence, Dark); 11, Radicle (48 h after emergence, Light); 12, Seedling; 13, Young shoot; 14, Young root; 15, Mature leaf; 16, Old leaf; 17, Mature sheath; 18, Old sheath; 19, Young flag leaf; 20, Old flag leaf; 21, Young panicle stages 3 (secondary branch primordium differentiation stage); 22, Young panicle stages 4 (pistil/stamen primordium differentiation stage); 23, Young panicle stages 5 (pollen-mother cell formation stage); 24, Young panicle; 25, Old panicle; 26, Young stem; 27, Old stem; 28, Hull; 29, Spikelet; 30, Stamen; 31, Endosperm (7 days after pollination); 32, Endosperm (14 days after pollination); 33, Endosperm (21 days after pollination). The Y-axis represents the expression values obtained from the microarray analysis. (B) fc116 co-expression patterns in two rice varieties during the life cycles. The color scale representing the relative signal values is shown (green refers to low expression; black refers to medium expression and red refers to high expression). Black asterisk is the fc116.
We further examined the expression pattern of the members in the FC116 gene family. It was found that the absolute expression levels of the many other members in this family were much higher than FC116, even in stem (Table S3). Based on the hierarchical cluster analysis, this family can be classified into two major groups with ten distinct sub-groups that exhibited a complementary expression pattern in 33 tissues from the entire life cycle of two rice varieties (Figure 4B). The closest co-expressed gene was LOC_Os01g65490 that was classified into a different cluster with FC116 according to their protein sequence similarity, while the closest homolog LOC_Os01g50040 was quite different in expression. This may indicate that the function of FC116 is unique in this family. Thus, the loss of function in fc116 cannot be completely compensated by other members in the family.
Gene co-expression analyses can reveal functional relationships between gene products. Investigating any possible genes outside the family that are co-expressed with FC116 may provide evidence for the functional role of FC116. We identified the top 100 positively co-expressed genes and top 100 negatively co-expressed genes with FC116 (Table S4). We could not find highly co-expressed genes genome-wide with FC116 as we used the Pearson's correlation coefficient (PCC) to quantify the association between two genes in the genome (rmax = 0.53). Go analysis indicated that some of these genes may be involved in cell wall biosynthesis such as GPI-anchored protein, fasciclin-like arabinogalactan, and wall-associated receptor kinase (Table S4). Since the gene co-expression Modules were established for rice cell wall in a previous study (Guo et al., 2007), we investigated the Modules these co-expressed gene belong to. Interestingly, we found that most negatively co-expressed genes of FC116 belonged to Modules in close vicinity of modules 44 that typically were for secondary cell wall (SCW), whereas the positively co-expressed genes distributed to the modules including the modules 24 that typically were for primary cell wall. We also found that the expression of FC116 was negatively correlated with the contents of cellulose and xylose (r = −0.4 and −0.4). These results indicate that FC116 is not the gene that is typically for SCW although its mutation causes impaired cell wall thickening.
We also marked the CESA family and the brittle culm genes (bcs) to the modules and found that the CESA were distributed to the Modules 2, 10, 24, and 44, while the BC genes were distributed to the Modules 5, 10, 24, 27, 28, and 44 (Figure 5; Table S5). Strikingly, the genes contained in Module 44 included those for cellulose synthases CESA4, CESA7, CESA9, BC1 (OsCOBRA), BC6 (CESA9) (Kotake et al., 2011), BC7(t) (CEAS4), BC11 (CESA4), BC13 (CESA9) (Song et al., 2013), FC9 (OsMYB103), and FC16 (CESA9), while the Module 24 included the CESA1, CESA3, CESA6, CESA8, and BC15/OsCTL1 (chitinase-like). FC116/BC10 belongs to Modules 5 that in close vicinity of Modules 10 contain the CESA5 and BC14 (Figure 5). Interestingly, we found significant negative co-expression of FC116/BC10 with BC14 (r = −0.41). We noticed that FC116/BC10 and BC14 showed overlapped expression at the seedling stage; however, they showed divergence in expression tendency in many tissues that underwent secondary growth from the booting stage (Figure 4A). It is indicate that the causing of the brittle/fragile culms can be complicated and the overlapping or the close vicinity of these Modules imply their relationship that worth further investigation.
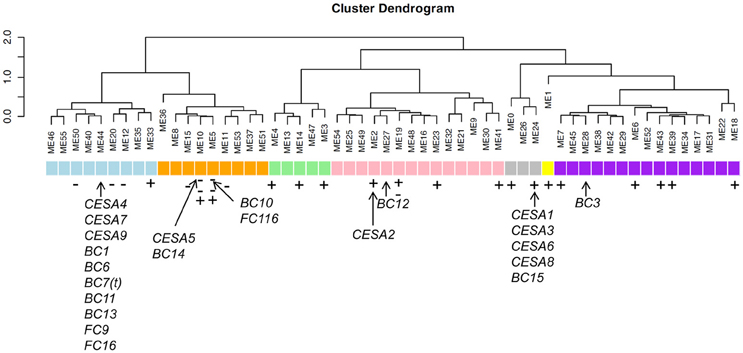
Figure 5. Certain modules correlate with specific cell wall content in rice modified from Guo et al. (2014). Arrows represent the corresponding modules of the brittle culm (BCs), fragile clum (FCs) and CESA genes. Minus and plus represent the down and up regulation genes in fc116, respectively. Two minuses represent more down regulation genes in the module. Blue represent the modules secondary cell wall biosynthesis. Pink represent the modules of cellulose, xylose, arabinose, and guaiacyl lignin biosynthesis. Gray represents the modules of primary cell wall biosynthesis. Purple represent the modules of arabinose and galactose biosynthesis.
In rice internodes, peripheral vascular tissue cells and sclerenchyma cells under the epidermal layer develop thickened cell walls, which are presumed to provide mechanical strength to the plants. To determine the alterations in cell wall structure and thickness that are responsible for the brittle phenotype, we compared the cell wall morphology in the internodes between fc116 and its wild type by microscopy. Cellulose and other cell wall components showed fluorescence staining with calcofluor, while phloroglucinol reacted with coniferaldehyde groups in lignin and the color intensity approximately reflects the total lignin content. In transection, the peripheral sclerenchyma in wild-type internodes showed substantial staining with calcofluor white, while fc116 internodes were slightly stained, and have an apparent defect in cell wall thickening (Figures 6A,B). The phloroglucinol staining of the transection indicated increased lignin content in the peripheral sclerenchyma and vascular bundles in fc116 and again decreased cell wall thickening compared with the wide type (Figures 6C,D). We further observed that fc116 generated stronger fluorescent signals in the vascular bundle cells than in wild-type plants using CCRC-M148 antibody recognizing xylan (Figures 6E,F). These results suggest that the fc116 mutation affects the formation of the secondary cell walls, particularly in the sclerenchyma tissues.
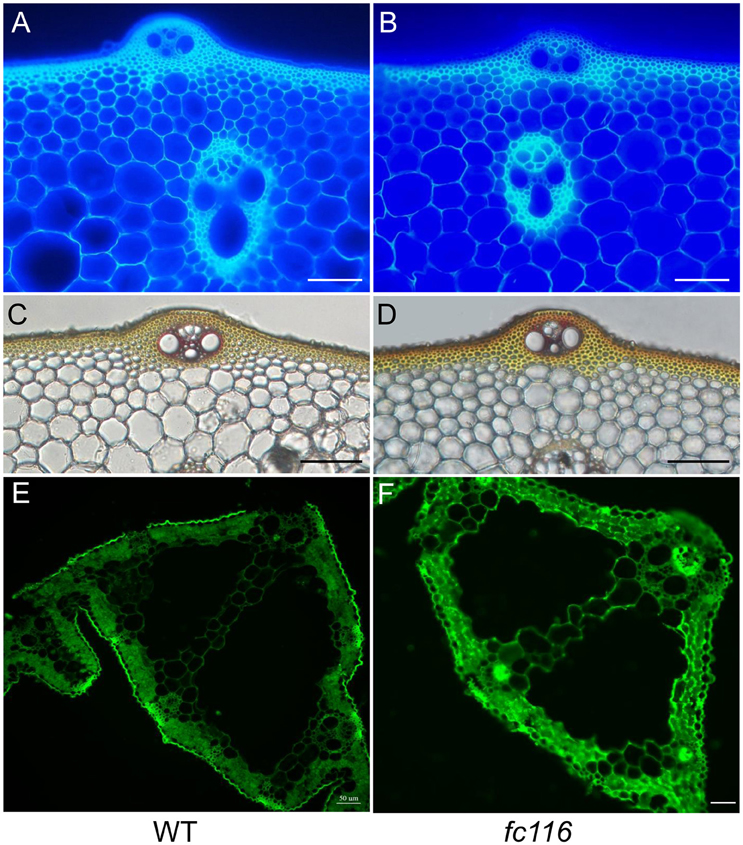
Figure 6. Staining of cellulose, lignin, and hemicellulose in wild-type and fc116 plants. (A) and (B) Calcofluor staining of the transverse culm sections of wild-type (A) and fc116 (B) plants, showing the decreased level of cellulose in the cell walls in the mutant culm. (C,D) Wiesner's staining of the transverse culm sections of wild-type (C) and fc116 (D), showing the increased level of lignin in the walls of sclerenchyma cells. (E,F) Immunohistochemical localization of xylan in leaf sections of wild-type and fc116 plants, showing the increased fluorescent signals in all cells visualized with xylan antibody CCRC-M148. Bar = 50 μm.
The culms of fc116 mutant plants showed 45% reduction in cellulose content and 34% increase in hemicellulose and 18% in lignin (Table 1). In addition, fc116 exhibited lower crystallinity index (CrI) than the wild type in both crude cell wall fraction and crude cellulose. However, the CrI changes in crude cellulose were not as significant as in the crude cell wall fraction. This was different from the mutant fc16/OsCESA9 (another mutant maintained in our lab) that showed significant decrease in CrI in crude cellulose but slightly decrease in CrI of the crude cell wall fraction (data not shown). The monosaccharide composition analysis showed that the amount of pentoses (xylose and arabinose) in the hemicelluloses was significantly increased in the mutant, while the all hexose (Glucose, Mannose, Galactose) were decreased. Compared with the wild type, fc116 had 10 and 14% increase of Ara and Xyl, respectively (Table 2). We also found the ratio of Ara/Xyl significantly decreased despite of increasing absolute value of Ara. Similar trend in the change of polysaccharides was also found with the mutant in the pectic fraction (Table 1). The changes in the cell wall content were consistent with the staining analysis. Our results were basically consistent with the previous study that reported significant decrease in cellulose and increase in lignin in bc10 (Zhou et al., 2009). Our results imply that both cellulosic and non-cellulosic polysaccharide synthesis is widely impaired in fc116. We noticed that the OsCESA4 mutant of Nipponbare, displayed 60% reduction in cellulose content and a twofold compensatory increase in arabinoxylan (Zhang et al., 2009). BC15 (chitinase-like) which was predicated to have binding sites to N-acetylglucosamine also showed that Ara and Xyl contents were increased by 58 and 77%, respectively by mutation despite the mutants did not show any changes in lignin content (Wu et al., 2012). The polyphenolic molecule lignin is an amorphous polymer of phenylpropane units with three monomers: p-hydroxyphenyl (H), syringyl (S), and guaiacyl (G). It was found that the contents of the all three monomers H, S and G in the mutant plants were significantly increased compared to the wider type. However, the relative amount of H monomer (% of the total monomers) slightly decreased, whereas the S and G% slightly increased (Table 3). These results suggest that fc116 mutation might cause equal increase of the every individual steps, but under such circumstances, the efficient of the branch leading to three different major monomers can be slightly different as they share and thus compete for limited precursors.
Since the dramatical and universal changes in cell wall composition were obseved, in fc116 plants, we detected the transcription alterations of the major genes associated with wall polymers using RNA-seq which reliably quantifies the expression levels of individual genes in multigene families. Relative to the wild type, the mutants exhibit pathway-wide up-regulation of lignin biosynthesis genes, including the 4-coumarate-CoA ligase (4CL), cinnamyl alcohol dehydrogenase (CAD), ferulate 5-hydroxylase (F5H), and cinnamoyl-CoA reductase (CCR), that catalyze specific branches of lignin biosynthesis leading to different lignin monomers, suggested possible alterations in lignin composition along with overall increase in response to FC116 mutation (Figure 7). Of the total 27 examined lignin units biosynthesis genes that showed high transcription levels in the culm at heading stage based on microarray data, 20 were identified with the expression levels beyond 200 in this RNA-seq and 18 with much higher expression levels in fc116 than in wild-type. The similar up-regulation tendency was found in typical SCW-related Modules 40 (19 up and 5 down) and 44 (58 up and 4 down), the modules for cellulose, xylan, H, S, and G. With regard to the increased hemicellulose, Xyl and Ara levels, the mutants were detected with higher transcription levels in all three glycosyltransferase genes LOC_Os03g08600 (GT8), LOC_Os02g22380 (GT61), LOC_Os06g47340 (GT43), which proposed to be participated in hemicellulose synthesis. LOC_Os06g47340 (OsIRX14), orthologs to the putative β-1,4-xylan backbone elongating Arabidopsis IRX14 gene, was demonstrated as one of three important genes involved in backbone chain synthesis of hemicelluloses (Scheller and Ulvskov, 2010; Chiniquy et al., 2013). The only enzymes characterized so far were belong to GT8 and GT61 that decorate the xylan backbone, as the members of the GT8 family (GUX1 and GUX2) are required for glucuronosyl substitution in Arabidopsis stem (Mortimer et al., 2010), while the members from GT61 family (xylan arabinosyltransferases, XATs) responsible for α-(1,3)-arabinosyl substitution of xylan (Anders et al., 2012). The data in expression of these genes were consistent with the extensive increase in the levels of hemicelluloses and lignins. Interestingly, that only CESA 4, 7, 9 that especially during SCW formation expression were slightly increased while the other CESAs remained unchanged or slightly decreased. It appears to differ from the fact that the CESA genes were all co-upregulated during culm development in the normal condition and even in the bc15 despite the dramatic decrease of the cellulose content in this mutant (Wang et al., 2010b; Wu et al., 2012). So, we conceive that in fc116 the pathway relative typical for SCW might be activated through an unknown mechanism. It was observed that the BC12/GDD1 (kinesin-like protein) can act as a transcriptional activator that might regulate KO2 gene expression in GA biosynthesis pathway, besides binding to MTs in an ATP-dependent manner (Li et al., 2011). fc116 differ from other rice brittle culm mutants such as bc1 and bc15 that show no obviously compensatory changes in lignin content and the expression profile of the relevant genes (Li et al., 2003; Wu et al., 2012). It is possible that there might be multiple knobs and switches that can be tuned to execute specific regulation of different cell wall pathways in order to optimize secondary cell wall composition.
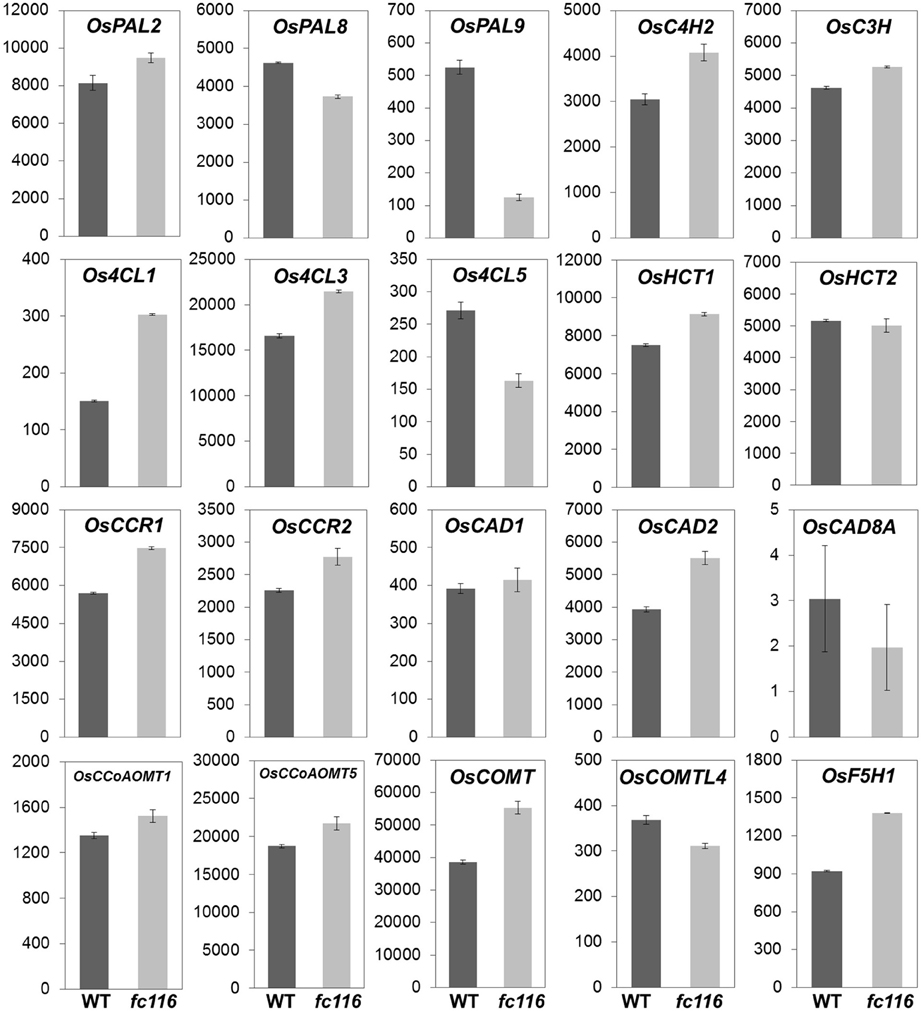
Figure 7. Expression of lignin monomers biosynthesis genes between wild-type and fc116. The y-axis represents the raw expression values obtained from the RNA-seq analysis. C3H, p-coumarate 3-hydroxylase; C4H, trans-cinnamate 4-hydroxylase; 4CL, 4-Coumarate-CoA ligase; CAD, Cinnamyl alcohol dehydrogenase; CCoAOMT, caffeoyl-CoA 3-O-methyltransferase; CCR, cinnamoyl-CoA reductase; COMT, caffeic acid O-methyltransferase; COMTL, caffeic acid O-methyltransferase like; F5H, ferulate 5-hydroxylase; HCT, hydroxycinnamoyltransferase; PAL, phe ammonialyase.
Plant cell walls are complex and dynamic structures composed mostly polysaccharides, highly glycosylated proteins, and lignin. In recent years, studies on the brittle culm mutants have identified several genes responsible for secondary cell wall formation, revealing many processes of monocot cell wall biosynthesis. Here, we report the isolation of a fragile culm gene fc116, allelic to bc10, which has sequence similarity with the domain to a core 2 beta-1,6-N-acetylglucosaminyltransferase (C2GnT). We investigated the changes in cell wall features, monosaccharide composition, phenolic monomers constitution, and the transcriptomic of the fc116. Our results are basically consistent with the previous study by Zhou et al. (2009), but have some interesting findings including: 1) fc116 mutation occurred within a newly identified protein motif “R, RXG, RA” located outside the “domain of unknown function 266” (DUF266) which was proposed to be crucial for the function of protein BC10 (Zhou et al., 2009); 2) The three major cell wall polymers of the mutant were all significantly changed in their content, constitution and features. Also, we observed contrary changes in pentoses (Xyl and Ara) and hexoses (Glc, Man, and Gal) in hemicellulosic polysaccharides. All these findings are of interesting for further biochemical and functional characterization of protein FC116; 3) The transcriptomic profiling of fc116 revealed the genetic regulation of the content and monomers of hemicelluloses and lignins.
The FC116/BC10 gene was lowly but ubiquitously expressed in many tissues and organs that showed distinct cell wall constitutions. It was expressed in the all tissues examined across the whole life cycle of rice, and slightly down-regulated during secondary growth. Co-expression analysis further indicated it belongs to the co-expression Modules 5 that in close vicinity of the Modules 10 containing the CESA5 and BC14, without obvious inclination to gene modules for typical PCW and SCW. In addition, the expression of gene FC116 was not regulated by OsMYB103, the transcription factor that typically regulates secondary cell wall biosynthesis in rice (Figure S3). We noticed that, other than FC116, many BC genes (BC3, BC12, BC14) also could distribute to the Modules that not typically involved in the biosynthesis of neither SCW nor PCW in spite of their prominent effects on the secondary cell wall (Figure 5). We noticed that BC3, BC12, and BC14 function in the membrane vesiculation, cortical microtubule-dependent cellulose deposition and nucleotide sugar transportation, respectively (Xiong et al., 2010; Zhang et al., 2010, 2011; Li et al., 2011). These biological processes are fundamental and may be common required for both PCW and SCW formation. Although the brittle culm mutants identified in rice have commonly reduced mechanical strength caused by decreased cellulose content and altered cell wall structure, the mechanism underlying that trait could be quite complex.
Our results revealed that fc116 caused a universal effect on the hexoses in the cellulosic and non-cellulosic (pectins and hemicelluloses) fractions of cell wall residues. That the dramatic decrease in crystallinity of raw material other than the crude cellulose also indicated the changes of non-cellulosic fraction. Compared to its allelic mutant bc10, fc116 exhibited significant increase in the hemicellulose content and alteration in its monosaccharide components (increased in Xyl and Ara, but decreased in Glc, Man and Gal) with up-regulated expression of genes for hemicelluloses and lignin. However, the soluble hexoses derived from matrix polysaccharides were much higher in fc116 plants compared to wild-type, indicating the source of the sugar for cell wall biosynthesis in this fragile culm mutant was not affected (Figure S4). Our RNA-seq data further indicated that the expression levels of various genes directly involved in cellulose biosynthesis (such as CESA- and BC-encoding genes) are not impaired in fc116 (Table S6). Thus, the alteration of the hexose polysaccharides might lie in the availability, transporting or/and delivery of the substrate for the cell wall polysaccharides biosynthesis as many studies have revealed the importance of substrate availability in polysaccharide biosynthesis (Zablackis et al., 1996; Bonin et al., 1997; Usadel et al., 2004; Diet et al., 2006; Oka et al., 2007). We noticed that the mutation of BC15/OsCTL1, a class II chitinase that may use GlcNAc-containing glycoproteins or chitooligosaccharides as the substrates, also causes universal effects on the hexoses in cellulosic and non-cellulosic polysaccharides (De Jong et al., 1993; Domon et al., 2000; Passarinho et al., 2001; Zhong et al., 2002; Hermans et al., 2010). BC15 belongs to Module 24 typically for PCW without an obvious effect on lignin biosynthesis, but has an overlap expression pattern with the FC116. The mutation in BC14/OsNST1, a Golgi-localized nucleotide sugar transporter generally work together with themselves or with glycosyltransferases (GTs), also causes a decrease in the glucose content not only in cellulose but also other glucoconjugated polymers. The mechanism for universal effects of FC116 on several kinds of hexoses in both cellulosic and non-cellulosic fractions remains to be answered. Some GTs are reported displaying not strict regiospecificities and substrate specificity (Thibodeaux et al., 2008; Gantt et al., 2011; Wang et al., 2012), as well as AtUTr1 (UDP-galactose transporter) and BC14/OsNST1 (UDP-glucose transporter) have been identified to have both UDP-galactose and UDP-glucose transport activity (Norambuena et al., 2002; Zhang et al., 2011). More studies are needed to further characterize the possible molecular mechanisms of FC116 protein function in plant cell wall formation process and regulation. First, the fc116 mutants could be characterized using more advanced tools such as immunological approaches like Glycome Profiling or ComPP (Sørensen and Willats, 2011; Pattathil et al., 2012), which would give more insights in to the glycome modification resulting from fc116 mutation. Secondary, the FC116 could be expressed and purified to validate the actual biochemical function of this protein. Overall, our results reported here can give a hint for the further characterization of biochemical function of the FC116 protein and its crucial roles in cell-wall formation.
MZ completed major experiments. FW and ZH screened the mutants and participated the genetic mapping of the FC116. MZ and KG analyzed the data. YL completed the immunofluorescence detection. GX, XC, and YW contributed in revision of the manuscript. LP and LW designed project, supervised experiments and finalized manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported in part by grants from the 111 Project (B08032), the projects (30900890, 31171524, & 31100140) of National Natural Science foundation of China, the China Postdoctoral Science Foundation (20090460964), and Fundamental Research Funds for the Central Universities (2011PY047, 2662015PY207, and 2662015PY009).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01366
Anders, N., Wilkinson, M. D., Lovegrove, A., Freeman, J., Tryfona, T., Pellny, T. K., et al. (2012). Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc. Natl. Acad. Sci. U.S.A. 109, 989–993. doi: 10.1073/pnas.1115858109
Bonin, C. P., Potter, I., Vanzin, G. F., and Reiter, W. D. (1997). The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4, 6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proc. Natl. Acad. Sci. U.S.A. 94, 2085–2090.
Burden, C. J., Qureshi, S. E., and Wilson, S. R. (2014). Error estimates for the analysis of differential expression from RNA-seq count data. PeerJ. 2:e576. doi: 10.7717/peerj.576
Cantu, D., Vicente, A. R., Labavitch, J. M., Bennett, A. B., and Powell, A. L. T. (2008). Strangers in the matrix: plant cell walls and pathogen susceptibility. Trends Plant Sci. 13, 610–617. doi: 10.1016/j.tplants.2008.09.002
Cao, Y. P., Li, J. L., Yu, L., Chai, G. H., He, G., Hu, R. B., et al. (2014). Cell wall polysaccharide distribution in Miscanthus lutarioriparius stem using immuno-detection. Plant Cell Rep. 33, 643–653. doi: 10.1007/s00299-014-1574-y
Carpita, N., Tierney, M., and Campbell, M. (2001). Molecular biology of the plant cell wall: searching for the genes that define structure architecture and dynamics. Plant Mol. Biol. 47, 1–5. doi: 10.1007/978-94-010-0668-2_1
Chiniquy, D., Varanasi, P., Oh, T., Harholt, J., Katnelson, J., Singh, S., et al. (2013). Three novel rice genes closely related to the Arabidopsis IRX9, IRX9L, and IRX14 genes and their roles in xylan biosynthesis. Front. Plant Sci. 4:83. doi: 10.3389/fpls.2013.00083
De Jong, A. J., Heidstra, R., Spaink, H. P., Hartog, M. V., Meijer, E. A., Hendriks, T., et al. (1993). Rhizobium lipooligosaccharides rescue a carrot somatic embryo mutant. Plant Cell. 5, 615–620. doi: 10.1105/tpc.5.6.615
Demura, T., and Ye, Z. H. (2010). Regulation of plant biomass production. Curr. Opin. Plant Biol. 13, 299–304. doi: 10.1016/j.pbi.2010.03.002
Diet, A., Link, B., Seifert, G. J., Schellenberg, B., Wagner, U., Pauly, M., et al. (2006). The Arabidopsis root hair cell wall formation mutant Irx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell. 18, 1630–1641. doi: 10.1105/tpc.105.038653
Dische, Z. (1962). “Color reactions of carbohydrates,” in Methods in Carbohydrate Chemistry, Vol. 1, eds R. L. Whistler and M. L. Wolfrom (New York, NY: Academic Press), 477–512.
Domon, J.-M., Neutelings, G., Roger, D., David, A., and David, H. (2000). A basic chitinase-like protein secreted by embryogenic tissues of Pinus carbaea acts on arabinogalactan proteins extracted from the same cell lines. J. Plant Physiol. 156, 33–39. doi: 10.1016/S0176-1617(00)80269-2
Fatima, A., Lynn, D. J., O'Boyle, P., Seoighe, C., and Morris, D. (2014). The miRNAome of the postpartum dairy cow liver in negative energy balance. BMC Genomics 15:279. doi: 10.1186/1471-2164-15-279
Finn, R. D., Mistry, J., Schuster-Böckler, B., Griffiths-Jones, S., Hollich, V., Lassmann, T., et al. (2006). Pfam: clans, web tools and services. Nucleic Acids Res. 34, D247–D251. doi: 10.1093/nar/gkj149
Gantt, R. W., Peltier-Pain, P., and Thorson, J. S. (2011). Enzymatic methods for glyco(diversification/randomization) of drugs and small molecules. Nat. Prod. Rep. 8, 1811–1853. doi: 10.1039/c1np00045d
Guo, A. Y., Zhu, Q. H., Chen, X., and Luo, J. C. (2007). GSDS: a gene structure display server. Yi Chuan. 29, 023–1026. doi: 10.1360/yc-007-1023
Guo, K., Zou, W. H., Feng, Y. Q., Zhang, M. L., Zhang, J., Tu, F., et al. (2014). An integrated genomic and metabolomic framework for cell wall biology in rice. BMC Genomics. 15:596. doi: 10.1186/1471-2164-15-596
Hermans, C., Porco, S., Verbruggen, N., and Bush, D. R. (2010). Chitinase-like protein CTL1 plays a role in altering root system architecture in response to multiple environmental conditions. Plant Physiol. 152, 904–917. doi: 10.1104/pp.109.149849
Hirano, K., Kotake, T., Kamihara, K., Tsuna, K., Aohara, T., Kaneko, Y., et al. (2010). Rice BRITTLE CULM 3 (BC3) encodes a classical dynamin OsDRP2B essential for proper secondary cell wall synthesis. Planta 232, 95–108. doi: 10.1007/s00425-010-1145-6
Kotake, T., Aohara, T., Hirano, K., Sato, A., Kaneko, Y., Tsumuraya, Y., et al. (2011). Rice Brittle culm 6 encodes a dominant-negative form of CesA protein that perturbs cellulose synthesis in secondary cell walls. J. Exp. Bot. 62, 2053–2062. doi: 10.1093/jxb/erq395
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/j.2000.4315
Lee, K. J., Marcus, S. E., and Knox, J. P. (2011). Cell wall biology: perspectives from cell wall imaging. Mol. Plant 4, 212–219. doi: 10.1093/mp/ssq075
Li, F. C., Ren, S. F., Zhang, W., Xu, Z. D., Xie, G. S., Chen, Y., et al. (2013). Arabinose substitution degree in xylan positively affects lignocellulose enzymatic digestibility after various NaOH/H2SO4pretreatments in Miscanthus. Bioresour. Technol. 130, 629–637. doi: 10.1016/j.biortech.2012.12.107
Li, F. C., Zhang, M. L., Guo, K., Hu, Z., Zhang, R., Feng, Y. Q., et al. (2015). High-level hemicellulosic arabinose predominately affects lignocellulose crystallinity for genetically enhancing both plant lodging resistance and biomass enzymatic digestibility in rice mutants. Plant Biotechnol. J. 13, 514–525. doi: 10.1111/pbi.12276
Li, J., Jiang, J. F., Qian, Q., Xu, Y. Y., Zhang, C., Xiao, J., et al. (2011). Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell 23, 628–640. doi: 10.1105/tpc.110.081901
Li, Y. H., Qian, Q., Zhou, Y. H., Yan, M. X., Sun, L., Zhang, M., et al. (2003). BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15, 2020–2031. doi: 10.1105/tpc.011775
Li, Y. B., Fan, C. C., Xing, Y. Z., Yun, P., Luo, L. J., Yan, B., et al. (2014). Chalk5 encodes a vacuolar H+-translocating pyrophosphatase influencing grain chalkiness in rice. Nat. Genet. 46, 398–404. doi: 10.1038/ng.2923
Liu, J. T., Zhou, Y. L., Luo, C. X., Xiang, Y., and An, L. (2016). De novo transcriptome sequencing of desert herbaceous Achnatherum splendens (Achnatherum) seedlings and identification of salt tolerance genes. Genes 7, 12. doi: 10.3390/genes7040012
Mortimer, J. C., Miles, G. P., Brown, D. M., Zhang, Z., Segura, M. P., Weimar, T., et al. (2010). Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc. Natl. Acad. Sci. U.S.A. 107, 17409–17414. doi: 10.1073/pnas.1005456107
Norambuena, L., Marchant, L., Berninsone, P., Hirschberg, C. B., Silva, H., and Orellana, A. (2002). Transport of UDP-galactose in plants identification and functional characterization of AtUTr1, an Arabidopsis thaliana UDP-galactos/UDP-glucose transporter. J. Biol. Chem. 277, 32923–32929. doi: 10.1074/jbc.M204081200
Oka, T., Nemoto, T., and Jigami, Y. (2007). Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-D-glucose to UDP-L-rhamnose conversion. J. Biol. Chem. 282, 5389–5403. doi: 10.1074/jbc.M610196200
Passarinho, P. A., van Hengel, A. J., Fransz, P. F., and De Vries, S. C. (2001). Expression pattern of the Arabidopsis thaliana AtEP3/AtchilV endochitinase gene. Planta. 210, 543–550. doi: 10.1007/s004250000464
Pattathil, S., Avci, U., Miller, J. S., and Hahn, M. G. (2012). “Biomass conversion: methods and protocols methods,” in Molecular Biology, Vol. 908, ed M. E. Himmel (Clifton, NJ: Humana Press), 61–72.
Peng, L. C., Hocart, C. H., Redmond, J. W., and Williamson, R. E. (2000). Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta 211, 406–414. doi: 10.1007/s004250000301
Reiter, W. D., Chapple, C. C. S., and Somerville, C. R. (1993). Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science 261, 1032–1035. doi: 10.1126/science.261.5124.1032
Rosén, M. L., Edman, M., Sjöström, M., and Wieslander, A. (2004). Recognition of fold and sugar linkage for glycosyltransferases by multivariate sequence analysis. J. Biol. Chem. 279, 38683–38692. doi: 10.1074/jbc.M402925200
Sato, K., Ito, S., Fujii, T., Suzuki, R., Takenouchi, S., Nakaba, S., et al. (2010). The carbohydrate-binding module (CBM)-like sequence is crucial for rice CWA1/BC1 function in proper assembly of secondary cell wall materials. Plant Signal Behav. 5, 1433–1436. doi: 10.1007/s00425-010-1171-4
Scheible, W. R., and Pauly, M. (2004). Glycosyltransferases and cell wall biosynthesis: novel players and insights. Curr. Opin. Plant Biol. 7, 285–295. doi: 10.1016/j.pbi.2004.03.006
Scheller, H. V., and Ulvskov, P. (2010). Hemicelluloses. Annu. Rev. Plant Biol. 61, 263–289. doi: 10.1146/annurev-arplant-042809-112315
Segal, L., Creely, J. J., Martin, A. E., and Conrad, C. M. (1959). An empirical method for estimating the degree of crystallinity of native cellulose using the X-Ray diffractometer. Text. Res. J. 29, 786–794.
Sigurgeirsson, B., Emanuelsson, O., and Lundeberg, J. (2014). Analysis of stranded information using an automated procedure for strand specific RNA sequencing. BMC Genomics 15:631. doi: 10.1186/1471-2164-15-631
Smook, G. A. (1992). Handbook for Pulp and Paper Technologists. Vancouver, BC: Angus Wilde Publications. 163–183.
Somerville, C. R., Bauer, S., Brininstool, G., Facette, M., Hamann, T., Milne, J., et al. (2004). Toward a systems approach to understanding plant cell walls. Science 306, 2206–2211. doi: 10.1126/science.1102765
Song, X. Q., Liu, L. F., Jiang, Y. J., Zhang, B. C., Gao, Y. P., Liu, X. L., et al. (2013). Disruption of secondary wall cellulose biosynthesis alters cadmium translocation and tolerance in rice plants. Mol. Plant 6, 768–780. doi: 10.1093/mp/sst025
Sonnhammer, E. L., von Heijne, G., and Krogh, A. (1988). A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182.
Sørensen, I., and Willats, W. G. T. (2011). “The plant cell wall: methods and protocols methods,” in Molecular Biology, Vol. 715, ed Z. A. Popper (Clifton, NJ: Humana Press), 115–121.
Sun, H. Y., Li, Y., Feng, S. Q., Zou, W. H., Guo, K., Fan, C. F., et al. (2012). Analysis of five rice 4-coumarate: coenzyme A ligase enzyme activity and stress response for potential roles in lignin and flavonoid biosynthesis in rice. Biochem. Biophys. Res. Commun. 430, 1151–1156. doi: 10.1016/j.bbrc.2012.12.019
Taylor-Teeples, M., Lin, L., Lucas, M. D., Turco, G., Toal, T. W., Gaudinier, A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. doi: 10.1038/nature14099
Thibodeaux, C. J., Melançon, C. E., and Liu, H. W. (2008). Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew. Chem. Int. Ed Engl. 47, 9814–9859. doi: 10.1002/anie
Torney, F., Moeller, L., Scarpa, A., and Wang, K. (2007). Genetic engineering approaches to improve bioethanol production from maize. Curr. Opin. Biotechnol. 18, 193–199. doi: 10.1016/j.copbio.2007.03.006
Turner, S. R., and Somerville, C. R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701. doi: 10.2307/3870425
Usadel, B., Kuschinsky, A. M., Rosso, M. G., Eckermann, N., and Pauly, M. (2004). RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiol. 134, 286–295. doi: 10.1104/pp.103.034314
Wang, G. J., Pahari, P., Kharel, M. K., Chen, J., Zhu, H. N., Van Lanen, S. G., et al. (2012). Cooperation of two bifunctional enzymes in the biosynthesis and attachment of deoxysugars of the antitumor antibiotic mithramycin. Angew. Chem. Int. Ed Engl. 51, 10638–10642. doi: 10.1002/anie.20120541
Wang, H. Z., and Dixon, R. A. (2012). On-off switches for secondary cell wall biosynthesis. Mol. Plant 5, 297–303. doi: 10.1093/mp/ssr098
Wang, L. Q., Guo, K., Li, Y., Tu, Y. Y., Hu, H. Z., Wang, B. R., et al. (2010b). Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 10:282. doi: 10.1186/1471-2229-10-282
Wang, L. Q., Liu, W. J., Xu, Y. B., He, Y. Q., Luo, L. J., and Zhag, Q. F. (2007). Genetic basis of 17 traits and viscosity parameters characterizing the eating and cooking quality of the rice grain. Theor. Appl. Genet. 115, 463–476. doi: 10.1007/s00122-007-0580-7
Wang, L., Xie, W. B., Chen, Y., Tang, W. J., Yang, J. Y., Ye, R. J., et al. (2010a). A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 61, 752–766. doi: 10.1111/j.1365-313X.2009.04100.x
Wu, B., Zhang, B. C., Dai, Y., Zhang, L., Shang-Guan, K., Peng, Y. G., et al. (2012). Brittle culm15 encodes a membrane-associated chitinase-like protein required for cellulose biosynthesis in rice. Plant Physiol. 159, 1440–1452. doi: 10.1104/pp.112.195529
Wu, Z. L., Zhang, M. L., Wang, L. Q., Tu, Y. Y., Zhang, J., Xie, G. S., et al. (2013). Biomass digestibility is predominantly affected by three factors of wall polymer features distinctive in wheat accessions and rice mutants. Biotechnol. Biofuels 6:183. doi: 10.1186/1754-6834-6-183
Xie, G. S., and Peng, L. C. (2011). Genetic engineering of energy crops: a strategy for biofuel production in China. J. Integr. Plant Biol. 53, 143–150. doi: 10.1111/j.1744-7909.2010.01022.x
Xiong, G. Y., Li, R., Qian, Q., Song, X. Q., Liu, X. L., Yu, Y. C., et al. (2010). The rice dynamin-related protein DRP2B mediates membrane trafficking, and thereby plays a critical role in secondary cell wall cellulose biosynthesis. Plant J. 64, 56–70. doi: 10.1111/j.1365-313X.2010.04308.x
Xu, N., Zhang, W., Ren, S. F., Liu, F., Zhao, C. Q., Liao, H. F., et al. (2012). Hemicelluloses negatively affect lignocellulose crystallinity for high biomass digestibility under NaOH and H2SO4 pretreatments in Miscanthus. Biotechnol. Biofuels 5:58. doi: 10.1186/1754-6834-5-58
Yan, C. J., Yan, S., Zeng, X. H., Zhang, Z. Q., and Gu, M. H. (2007). Fine mapping and isolation of Bc7(t), allelic to OsCesA4. J. Genet. Genomics 34, 1019–1027. doi: 10.1016/S1673-8527(07)60115-5
Yang, C. H., Li, D. Y., Liu, X., Ji, C. J., Hao, L. L., Zhao, X. F., et al. (2014). OsMYB103L, an R2R3-MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa L.). BMC Plant Biol. 14:158. doi: 10.1186/1471-2229-14-158
Ye, Y. F., Liu, B. M., Zhao, M., Wu, K., Cheng, W. M., Chen, X. B., et al. (2015). CEF1/OsMYB103L is involved in GA-mediated regulation of secondary wall biosynthesis in rice. Plant Mol. Biol. 89, 385–401. doi: 10.1007/s11103-015-0376-0
Zablackis, E., York, W. S., Pauly, M., Hantus, S., Reiter, W. D., Chapple, C. C., et al. (1996). Substitution of L-fucose by L-galactose in cell walls of Arabidopsis mur1. Science 272, 1808–1810. doi: 10.1126/science.272.5269.1808
Zhang, B. C., Deng, L. W., Qian, Q., Xiong, G. Y., Zeng, D. L., Li, R., et al. (2009). A missense mutation in the transmembrane domain of CESA4 affects protein abundance in the plasma membrane and results in abnormal cell wall biosynthesis in rice. Plant Mol. Biol. 71, 509–524. doi: 10.1007/s11103-009-9536-4
Zhang, B. C., Liu, X. L., Qian, Q., Liu, L. F., Dong, G. J., Xiong, G. Y., et al. (2011). Golgi nucleotide sugar transporter modulates cell wall biosynthesis and plant growth in rice. Proc. Natl. Acad. Sci. U.S.A. 108, 5110–5115. doi: 10.1073/pnas.1016144108
Zhang, B. C., and Zhou, Y. H. (2011). Rice brittleness mutants: a way to open the ‘black box’ of monocot cell wall biosynthesis. J. Integr. Plant Biol. 53, 136–142. doi: 10.1111/j.1744-7909.2010.01011.x
Zhang, M., Zhang, B. C., Qian, Q., Yu, Y. C., Li, R., Zhang, J. W., et al. (2010). Brittle Culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. Plant J. 63, 312–328. doi: 10.1111/j.1365-313X.2010.04238.x
Zhang, W., Yi, Z. L., Huang, J. F., Li, F. C., Hao, B., Li, M., et al. (2013). Three lignocellulose features that distinctively affect biomass enzymatic digestibility under NaOH and H2SO4pretreatments in Miscanthus. Bioresour. Technol. 130, 30–37. doi: 10.1016/j.biortech.2012.12.029
Zhao, Q., and Dixon, R. A. (2011). Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci. 16, 227–233. doi: 10.1016/j.tplants.2010.12.005
Zhong, R., Kays, S. J., Schroeder, B. P., and Ye, Z. H. (2002). Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14, 165–179. doi: 10.1105/tpc.010278
Zhong, R., and Ye, Z. H. (2007). Regulation of cell wall biosynthesis. Curr. Opin. Plant Biol. 10, 564–572. doi: 10.1016/j.pbi.2007.09.001
Keywords: rice, cell wall, cellulose, fragile culm, mechanical strength
Citation: Zhang M, Wei F, Guo K, Hu Z, Li Y, Xie G, Wang Y, Cai X, Peng L and Wang L (2016) A Novel FC116/BC10 Mutation Distinctively Causes Alteration in the Expression of the Genes for Cell Wall Polymer Synthesis in Rice. Front. Plant Sci. 7:1366. doi: 10.3389/fpls.2016.01366
Received: 13 April 2016; Accepted: 29 August 2016;
Published: 21 September 2016.
Edited by:
Huanzhong Wang, University of Connecticut, USAReviewed by:
Taras P. Pasternak, Institute of Biology II, GermanyCopyright © 2016 Zhang, Wei, Guo, Hu, Li, Xie, Wang, Cai, Peng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingqiang Wang, bHF3YW5nQG1haWwuaHphdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.