
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 24 August 2016
Sec. Plant Biotechnology
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.01284
This article is part of the Research TopicMechanisms of abiotic stress responses and tolerance in plants: physiological, biochemical and molecular interventionsView all 121 articles
Ribosomal proteins (RPs) are well-known for their role in mediating protein synthesis and maintaining the stability of the ribosomal complex, which includes small and large subunits. In the present investigation, in a genome-wide survey, we predicted that the large subunit of rice ribosomes is encoded by at least 123 genes including individual gene copies, distributed throughout the 12 chromosomes. We selected 34 candidate genes, each having 2–3 identical copies, for a detailed characterization of their gene structures, protein properties, cis-regulatory elements and comprehensive expression analysis. RPL proteins appear to be involved in interactions with other RP and non-RP proteins and their encoded RNAs have a higher content of alpha-helices in their predicted secondary structures. The majority of RPs have binding sites for metal and non-metal ligands. Native expression profiling of 34 ribosomal protein large (RPL) subunit genes in tissues covering the major stages of rice growth shows that they are predominantly expressed in vegetative tissues and seedlings followed by meiotically active tissues like flowers. The putative promoter regions of these genes also carry cis-elements that respond specifically to stress and signaling molecules. All the 34 genes responded differentially to the abiotic stress treatments. Phytohormone and cold treatments induced significant up-regulation of several RPL genes, while heat and H2O2 treatments down-regulated a majority of them. Furthermore, infection with a bacterial pathogen, Xanthomonas oryzae, which causes leaf blight also induced the expression of 80% of the RPL genes in leaves. Although the expression of RPL genes was detected in all the tissues studied, they are highly responsive to stress and signaling molecules indicating that their encoded proteins appear to have roles in stress amelioration besides house-keeping. This shows that the RPL gene family is a valuable resource for manipulation of stress tolerance in rice and other crops, which may be achieved by overexpressing and raising independent transgenic plants carrying the genes that became up-regulated significantly and instantaneously.
Ribosomes are tiny (200–300Å) ribonucleoprotein complexes typically existing as two unequal sized subunits in all organisms and constituting 25–30% of total cell mass (Alberts et al., 2002). The ribosome complex, as a whole, performs mRNA-directed protein synthesis. Specific interaction of RPs and rRNA with mRNA, tRNA, and other non-ribosomal protein cofactors ensure that the process of initiation of protein synthesis, amino acid assembly and termination occurs appropriately in the cells (Maguire and Zimmermann, 2001). Eukaryotic ribosomes have a sedimentation coefficient of 80S with the large 60S subunit having 25S, 5.8S, 5S rRNA, and the small 40S subunit consisting of 18S rRNA (Ben-Shem et al., 2010). The number of RPs in ribosomes varies between organisms, with eukaryotes having up to 80 RPs and prokaryotes possess a total of only 54 RPs in both the subunits (Doudna and Rath, 2002).
The ribosomal gene family has more than 200 genes, but less than 100 corresponding RPs are incorporated into the ribosomes in all organisms including yeast, animals, and plants (Ban et al., 2000; Barakat et al., 2001; Hanson et al., 2004). This supports the fact that each RP-gene exists as 2–5 identical members with 95–100% nucleotide and predicted protein similarity. An RP synthesized from only one gene copy of a group incorporates into a ribosome under a given condition/tissue (Guarinos et al., 2003; Schuwirth et al., 2005). For example, the Arabidopsis genome has 249 genes for 80 RPs (48-large subunit proteins, 32-small subunit proteins) with each gene having 3–4 expressed copies and none exists as a single gene copy (Wool et al., 1996). RPs, in addition to their universal roles of stabilizing the ribosomal complex and mediating polypeptide synthesis also have extra-ribosomal functions such as their involvement in response to the environmental stresses (Warner and McIntosh, 2009; Sormani et al., 2011).
Mutations in plant RP genes have been implicated in perturbed phenotypes as has been seen in animal systems including humans. Earlier studies with Arabidopsis showed that mutations in many RP genes (RPS18A, RPL24B, RPS5B, RPS13B, and RPL27A) resulted in a ‘pointed first leaf’ phenotype characterized by reduced cell division and growth, and genotoxic sensitive plants (Lijsebettens et al., 1994; Revenkova et al., 1999; Ito et al., 2000; Szakonyi and Byrne, 2011). A T-DNA insertion mutation in the Arabidopsis AtRPL10 gene caused lethal female gametophytes, while overexpression complemented the same with the recovery of the severe dwarf phenotype that resulted from the disruption of the ACL5 gene (Imai et al., 2008). A transposon insertion mutation in one of the three copies of the AtRPS13A gene resulted in reduced cell division, late flowering, retarded root and leaf growth (Ito et al., 2000). Similar effects of plant growth retardation and reduced fertility were observed after knockdown of AtRPL23aA resulting in reduced synthesis of the RPL23aA protein, while knockout of its paralog, RPL23aB, had no effect on growth (Degenhardt and Bonham-Smith, 2008a). RPL23aB is the only RP paralog that did not produce any visible phenotypic defects upon knockout (Degenhardt and Bonham-Smith, 2008b).
These RP-gene knockout studies clearly show that although RP genes exist as multiple gene copies, the maximum possible expression of all the gene copies is required for them to be incorporated into the ribosomes during specific stages of growth and development and under certain stress conditions (Schmid et al., 2005; Byrne, 2009). The variation in the composition of ribosomes by the incorporation of RPs derived from identical members could be a major factor in the translational regulation of transcripts in different cell types and under various specific conditions (Giavalisco et al., 2005; Carroll et al., 2008). The change in the composition of RPs upon feeding of Arabidopsis leaves with sucrose further supports the heterogeneity of ribosomes in response to external stimuli (Hummel et al., 2012).
The expression of RP genes has also been shown to be differentially regulated by signaling molecules and environmental stresses. The transcript levels of Arabidopsis RPS15a (RPS15aA, C, D and F) were up-regulated in response to phytohormone and heat treatments (Hulm et al., 2005). Similar transcript abundance under BAP treatment was detected for Arabidopsis RPS14, RPL13, and RPL30 genes (Cherepneva et al., 2003). Low temperature induced the expression of three RP genes; RPS6, RPS13, and RPL37 in soybean (Kim et al., 2004) and a homolog of RPL13, BnC24 in Brassica and E. coli (Sáez-Vásquez et al., 2000; Tanaka et al., 2001). The overexpression of RPL13 also resulted in tolerance against a fungal pathogen, Verticillium dahliae in transgenic potato with coordinated up-regulation of genes coding for defense and antioxidant enzymes (Yang et al., 2013) implying that RPs function in stress-response/tolerance through a network of multiple stress-related genes. In maize and Arabidopsis, RPL10A and RPL10C were shown to be significantly up-regulated under UV-B stress (Casati and Virginia, 2003; Ferreyra et al., 2010, 2013). RPL44 was found to be up-regulated under osmotic stresses, and the overexpression of Aspergillus glaucus RPL44 in yeast and tobacco ensured increased tolerance to salt and drought stresses (Liu et al., 2014). The majority of studies on RPs were undertaken in Arabidopsis largely because of the availability of insertion mutant lines and smaller genome size.
Until now, not much emphasis has been placed on the differential expression patterns of RP genes of rice in response to external stimuli. We had generated a large-scale enhancer based activation-tagged gain-of-function mutant population in indica rice, which was screened for water-use efficiency. Among the potential mutants with sustained productivity under prolonged water-limiting conditions, two of them were found to have enhanced expression of large subunit ribosomal genes because of their being tagged by the enhancers (Moin et al., 2016). This has prompted us to investigate the other rice RPL genes in the context of stress-responsiveness.
In the present study, we describe the genome-wide organization of predicted 123 RPL genes in rice including the individual gene copies. We investigated their overall expression pattern in selected tissues covering the major growth stages of rice. Also, we have provided an overview of their differential expression pattern under biotic and abiotic stress conditions that limit rice productivity. We identified specific RP genes, whose expression is unique or overlapping under native and treated conditions. In summary, the information presented in this study provides a resource for subsequent exploitation of RPL genes to ameliorate abiotic and biotic stress conditions in rice and also other crop plants in future.
To identify the total members of the large subunit ribosomal gene family, a keyword search using “ribosomal” was performed under the putative function search of Rice Genome Annotation Project Data Base (RGAP-DB v7)1 and Phytozome v112. The large subunit members were shortlisted by selecting the genes starting with prefix ‘L,’ for large subunit as opposed to ‘S’ that specifies small subunit genes. A total of 123 RPL genes were identified, and since the number of RPL genes in both the databases was same, the gene sequences were downloaded from RGAP-DB. When further looked for the presence of identical members or copies of each gene in RGAP-DB, we observed that each RPL gene has an average of 2–3 gene copies in the genome. From these 123 genes, we selected 34 candidate genes each representing one orthologous group excluding the identical copies for expression studies. All the identified 123 sequences were also confirmed through nucleotide and protein BLAST search in the NCBI3 and Hidden Markov Model (HMM) of Pfam4 databases, respectively. The predicted protein sequences of all the 123 RPL genes were also verified in NCBI conserved domain database5. To minimize the missing of potential RPL genes and to ensure that all the identified sequences belong to the ribosomal large subunit gene family, multiple databases were employed.
To determine the chromosomal distribution, the locus number of each of the 123 RPL genes obtained from RGAP-DB was submitted to the OryGenesDB6. Based on the output generated in OryGenesDB, the position of each gene at its corresponding locus on the chromosome was located manually.
The structure of each of the 34 RPL genes was determined to study the number and position of introns and exons, GC-content, gene orientation in the genome and alternative splice forms. The full-length sequences of each gene and cDNA were submitted to the Gene Structure Display Server (GSDSv2)7 to predict the structure.
The predicted sequences of 34 RPL proteins were obtained from the RGAP-DB and analyzed using an online tool, PSORT8 to predict the protein properties such as size, molecular weight and isoelectric point (pI). The amino acid sequences of these proteins were aligned in ClustalW9 and submitted to the Molecular Evolutionary Genetic Analysis (MEGAv6)10 program for constructing an unrooted phylogenetic tree to identify the protein similarities in the RPL family in rice. The domains and motifs in proteins were identified using SMART11 (Simple Modular Architecture Research Tool). The GRAVY (Grand average of hydropathicity) indices of RPL proteins, which are the determinants of the hydrophobicity of whole protein was calculated using ExPASy ProtParam12. The GRAVY values of most of the proteins are usually in the range of +2 to -2, and values in negative range or less than zero indicate that the proteins are hydrophilic in nature (Song et al., 2015).
Although the detailed crystal structure of ribosomal complex has been well-characterized (Ben-Shem et al., 2010), we tried to study the properties of individual RPs. To gain an insight into the secondary structure of RPL proteins and to characterize the presence of metal–ligand/protein/RNA interacting sites, the three-dimensional secondary structures of 34 RPL proteins were predicted using Phyre213 program (Protein Homology/AnalogY Recognition Engine v2; Kelley et al., 2015). Individual protein sequences were submitted in Phyre2 in FASTA format and after studying the properties such as α-helices and β-strands, they were directed to 3DLigandSite14 (Wass et al., 2010) to predict the metal/non-metal ligands and their binding sites in each protein.
To determine the presence of stress-responsive cis-regulatory elements, the nucleotide sequence ≤1 kb upstream of each RPL gene was retrieved from RGAP-DB and submitted to the Plant Cis-Acting Regulatory Elements15 database. The location and number of repeats of each cis-regulatory sequence in the putative promoter regions of each RPL gene were identified.
The seeds of Oryza sativa L. sp. indica var. Samba Mahsuri (BPT-5204) maintained in greenhouse conditions were surface sterilized with 70% ethanol for 50–60 s followed by 4% sodium hypochlorite for 20 min. Seeds were then washed thrice with sterile double-distilled water, blot dried and cultured on solid MS medium at 28 ± 2°C and 16 h light/8 h dark photoperiods.
To analyze the native tissue-specific expression pattern of 34 RPL genes at different stages of rice development, samples were collected from 13 different tissues covering major stages of rice growth. After sterilization, seeds were soaked in water in a rotary shaker and after 16 h of incubation, the embryonic portion was manually cut under a stereo-microscope to collect the embryos and endosperm. Some of these seeds were allowed to continue to germinate on MS medium. After 3 and 6 days of germination, the plumules, radicles, shoot, and leaf tissues were collected separately. After 2 weeks of growth on MS medium, some of the seedlings were transferred to pots containing alluvial soil and grown under greenhouse conditions (30 ± 2°C, 16 h light/8 h dark photoperiods). Plants were amply watered with RO (Reverse Osmosis) purified water up to 3 cm overlay in the pots as required for normal growth of rice. About 45 days after transfer (DAT) to the greenhouse, rice plants were uprooted to collect shoot, root, flag leaf, and root–shoot transition tissues. After 60 DAT, flowers, partially filled grains and spikes were collected.
To analyze the differential expression pattern of 34 RPL genes and to distinguish RPL genes that are up/down-regulated under abiotic conditions, 7-day-old seedlings were exposed to five different abiotic treatments such as MeJa, SA, cold stress (4°C), heat stress (42°C), and oxidative stress (H2O2). The 7-d-old seedlings were dipped in the solutions of 100 μM MeJa (Wang et al., 2007), 3 mM SA (Mitsuhara et al., 2008), and 10 μM H2O2 (Fuller, 2007). The root and shoot tissues were collected separately at 5 min, 3 h, 6 h, 24 h, and 60 h after treatments. For cold and heat induced stresses, seedlings in water were exposed to 4°C and 42°C (Jami et al., 2012), respectively, and root and shoot samples were collected at time intervals as described. Since WT rice seedlings started to wilt after 24 h of exposure to 42°C, heat stress samples were collected up to 24 h only. Seedlings in water at corresponding time intervals served as controls to normalize the expression patterns. Tissue samples were collected as three biological replicates after each time interval.
To check the expression pattern of rice ribosomal genes in response to biotic stress, we used the bacterial pathogen Xanthomonas oryzae pv. oryzae that causes Bacterial Leaf Blight (BLB) of rice, which is one of the most severe yield constraints of rice worldwide (Sundaram et al., 2014). At the seedling stage, the infected leaves start to roll-up, and as the disease progresses, the leaves turn yellow and wilt, leading to drying up and death. This drastically reduces the total seed yield of the plant. The yield loss may be as high as 70% when plants are grown in conditions favorable to the disease (Ryan et al., 2011). The bacterial suspension of Xanthomonas oryzae pv. oryzae was applied on the leaves of 2-month-old plants grown in greenhouse conditions, and leaf samples were collected after 11 days of infection. Leaf samples of untreated plants grown under similar conditions were used as a control to normalize the expression.
Because the transcript level of RPL10 was significantly up-regulated 11 days after treatment, we selected this gene in particular to analyze its expression at progressive time-points such as 3 h, 6 h, 1 day, 2 days, 3 days, up to 11 days post-infection of rice leaves with Xanthomonas oryzae pv. oryzae pathogen. The qRT-PCR was performed with Xanthomonas oryzae pv. oryzae treated and untreated samples collected as three biological and three technical replicates. Rice specific act1 and β-tub genes were used as controls for normalization and the mean of the fold change was represented as bar diagrams constructed using SigmaPlot v11.
Total RNA was isolated from stress-treated and untreated tissues using TriReagent (Takara Bio, UK) following the manufacturer’s protocol. The quality of extracted RNA was checked on 1.2% agarose gel prepared in TBE (Tris-borate-EDTA) buffer and quantified using Nanodrop. Total RNA (2 μg) was used to synthesize the first strand cDNA using reverse transcriptase (Takara Bio, UK). The cDNA was diluted in 1:7 proportions and 2 μl of it was used in qRT-PCR. Primers specific for each RPL gene sequence retrieved from RGAP-DB was designed using the primer-316 online tool and 10 μM of each was used per reaction. The reaction conditions for qRT-PCR included an initial denaturation at 94°C for 2 min followed by 40 cycles of 94°C for 30 s, an appropriate annealing temperature for 25 s and an extension of 72°C for 30 s. At the end of the reaction, a melting curve step was inserted to analyze the specificity of amplification of each gene. Rice specific actin (act1) and tubulin (β-tub) were used as internal reference genes to normalize the expression patterns. The mean values of relative fold change, which was calculated as per ΔΔCT method (Livak and Schmittgen, 2001) obtained from each reference gene was considered as the final fold change in the transcript levels. Each qRT-PCR reaction was performed as three biological and three technical replicates.
The relative fold change of the 34 genes in 13 tissues and under five abiotic treatments was illustrated in the form of heat maps. A dendrogram was constructed to represent the Hierarchical clustering of relative fold change of 34 genes under each treatment using the GENE-E17 program.
To explore the cytoplasmic large subunit (60S) ribosomal gene family members in rice, we used a keyword search “ribosomal” in the putative function search of RGAP-DB and Phytozome databases, which resulted in the identification of 428 and 754 genes, respectively, and these included genes belonging to cytoplasmic 60S and 40S subunits and 50S and 30S subunits of chloroplast and mitochondrial ribosomes. Keyword search and homology-based identification through HMM are widely used practices in identifying genome-wide copies of the annotated genes (Kapoor et al., 2008; Liang et al., 2016). We then searched for genes starting with the prefix ‘L’ to select large subunit genes. This process excluded small subunit genes and identified 215 genes that included large subunit members of cytoplasmic (60S) and chloroplast and mitochondrial (50S) ribosomal subunits. We then shortlisted the cytoplasmic 60S subunit genes by their putative cellular localization using the information available in RGAP-DB. A similar process was applied in shortlisting the 60S subunit genes from Phytozome. Both these approaches identified 123 genes belonging to the cytoplasmic 60S subunit. Each of these genes was then confirmed by a BLAST search of their nucleotide and predicted amino acid sequences in other rice databases like RAP-DB18 and OryGenesDB. BLASTn and BLASTp results in NCBI and HMM of Pfam and NCBI conserved domain databases, respectively, further confirmed that these genes belong to the 60S ribosomal family by the presence of ribosomal domains.
The locus numbers of 123 genes were submitted in OryGenesDB and, based on the output generated, the location of each gene on the corresponding chromosome was mapped manually using OryGenesDB. The location of these 123 genes was found on all chromosomes, indicating their wide distribution throughout the rice genome. Chromosome-7 has 19; chromosome-1, being the largest of rice chromosomes has 18; chromosome-2 has 16; chromosome-5 has 14; and chromosome-3 showed 13 genes. Chromosomes-9, 10, and 11 exhibited four genes each, while chromosomes-4, 8, and 9 evidenced 5, 10, and 9 RPL genes, respectively (Figure 1). The nucleotide sequence alignment of genes within an orthologous group exhibited 100% similarity, but their chromosomal locations are different. We selected the 34 candidate genes, each representing one orthologous group for a detailed characterization to understand their gene and protein structures, and comprehensive expression analysis in response to a wide range of stress treatments.
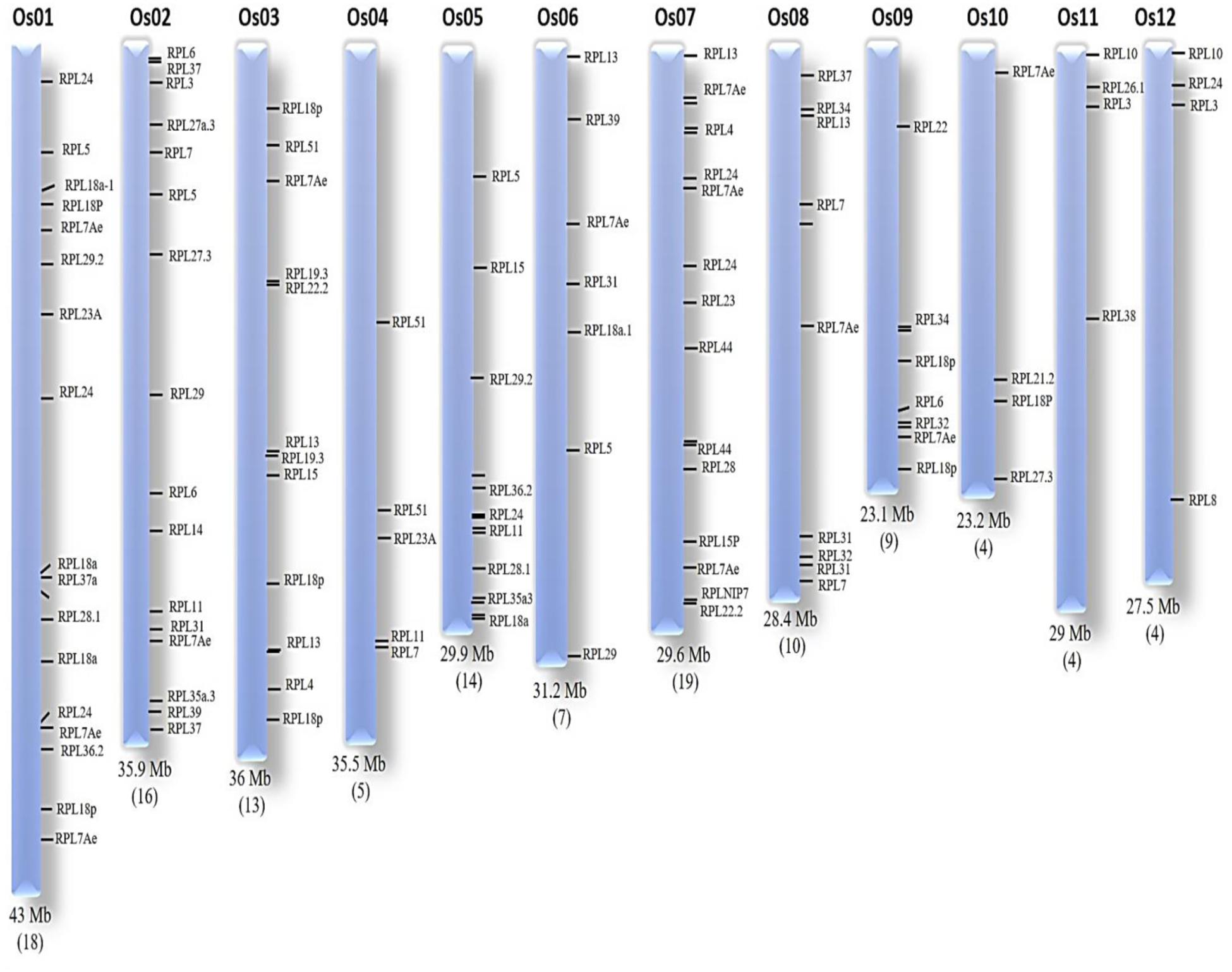
FIGURE 1. Chromosomal organization of RPL genes. The chromosomal number and size is represented at the top and bottom of each chromosome, respectively. The number of RPL genes is given within brackets at the bottom of each corresponding chromosome.
A comparative study between 34 RPL genes was performed to determine the number and position of introns and exons, GC-content and 5′ and 3′ untranslated regions. The number of introns varied from none to seven. The RPL10 and RPL18p have the highest number of introns (7), whereas RPL4, 8, 21.2, 23A, 26.1, 28, and 31 have only one intron. RPL7, 11, 24b, 27.3, and 38.2 do not contain any introns in their coding regions; these genes also have high GC-content. For example, RPL24b, 27.3 and 38 have 54, 60, and 51% GC-content, respectively. Genes with introns are reported to be crucial for gene expression, and a high number of introns is linked with increased expression of a gene (Callis et al., 1987; Karve et al., 2011). We observed that RPL18p and RPL10 with seven introns exhibited high expression in various tissues studied compared with other RPL members. Introns when particularly present at the start site or 5′ end of a gene have an enhanced ability for expression (Callis et al., 1987; Donath et al., 1995). RPL35a.3, 51, 32, 30e, 22, 19.3, 18a, 10, and 5 have their first introns within 500 bp regions from the transcription start site, which might be the reason for their constitutive expression in almost all the tissues studied. Similarly, the number of exons also varied among 34 RPL genes. Genes like RPL10 and RPL18p have the highest number of exons (8), whereas RPL7, 11, 24b, 27.3, and 38.2 have only one exon. Some of these RPL genes (RPL4, 13a, 14, 15, 23A, 28, 30e, 32, 35a, and 51) undergo alternative splicing to produce 2–5 splice-variants (Table 1) with similar nucleotide sequence. For expression analysis, we used the sequence of only one variant of a gene as it is difficult to design the primers for alternative splice forms exhibiting high nucleotide similarity. In addition, the 5′ and 3′ UTRs of these genes varied in size and positions. RPL28 has an unusually long UTR of 2.5 kb at the 3′ end (Supplementary Figure S1). Table 1 provides detailed information about their gene structure, site of location in the genome and copy number of individual genes.
Among the 34 proteins, RPL4 is the largest with a predicted molecular mass of 44.5 kDa. All the RPL proteins have similar isoelectric points ranging from 9.5 to 12. The maintenance of similar pI values in RPL proteins might be to reduce coulombic repulsions as these proteins are interactive in nature. They have a varied percentage of α, β, and distorted regions with RPL proteins interacting with other r– and non-r proteins exhibiting high content of α-helices. For example, RPL7, which interacts with elongation factors-Tu and -G, has 55% α-helical structure. RPL13a, existing at the interface of RPL3 has 57% of α-helix content, whereas RPL29 that interacts with RPL23A and initiation factors has 48% α-helix content. Furthermore, all the RPL proteins have a GRAVY value < 0, which indicate their high hydrophilicity. The proteins with high hydrophilic nature tend to undergo conformational changes and form flexible structures with other molecules and also contribute toward inducing tolerance during stress conditions (Fuxreiter et al., 2004; Liang et al., 2016).
All the RPs are characterized by the presence of ribosomal domain(s). They also have several other domains that participate in interaction with other proteins. RPL 14, 19.3, 21.2, 24b, 26, and 27 have KOW-SH3 motifs at their N-terminal regions, which are involved in protein–protein interactions. KOW-motifs link RPs with transcription factors (Kyrpides et al., 1996). RPL18p (135–226 amino acids) has an FCD domain (FadR C-terminal Domain) that is involved in the regulation of transcription of genes. RPL18p also has a XPGN domain (208–281 amino acids) that is associated with cancer and Xeroderma pigmentosum in humans. RPL21.2 has cheY motif at the C-terminus activated by phosphorylation through histidine kinases. RPL22 has a DUF1087 domain (amino acids 1–67) that is involved in chromatin remodeling and a WWE domain that is associated with poly-ADP-ribosylation and ubiquitin-mediated proteolysis. RPL29 has a carboxyl-terminal domain (CTD) and proteins with such domains are related to pre-mRNA processing by binding with mRNA capping enzymes (Schwer et al., 2009). It also has RQC, a DNA binding domain found in RecQ helicases.
Ligand-mediated signal transmission is essential for proper functioning of a majority of proteins including certain RPs. However, recognition of the core structures or amino acids involved in protein–ligand interactions is of paramount importance for understanding the dynamic and kinetic properties of the proteins. Analysis of RPL proteins for the presence of sites for ligand binding reveals that out of 34, 20 RPL proteins have sites for binding with ligands (that include metals ions and cofactors) whereas no such ligand binding sites were observed in 14 proteins (RPL3, 4, 6, 10, 11, 13b, 14, 15, 18a, 24a, 29, 34, 36, and 37). RPL8 (Lys198), 13a (Pro114, 115), 19.3 (Glu179, Arg180), 21.2 (Arg70), 22 (Ile52), 24b (Leu118, Lys121, Ala122), 26 (Leu105), 28 (Tyr49), 38 (Lys35), and 44 (Gly51) have Mg+2 ion binding sites. RPL7, 31, and 51 bind with Cu+2 and RPL5 (Asp30, Thr33) and 23A (Lys104) have sites for binding with Ca2+ ions. RPL19, 35, 38, and 51 also have Zn2+ binding sites. RPL29 present at the interface of RPL23A binds with the cofactor FAD. The RPL35 binds with cofactors FUC and FAD. Metal ions in RPs serve important biological functions by interacting with nucleic acids, particularly RNA. Non-metal ligands are cofactors involved in catalytic activities. Cofactors in RPs ensure that the processes of protein initiation, amino acid assembly and termination are correctly undertaken (Maguire and Zimmermann, 2001). Because of this, RPs might have binding sites for both metal and non-metal ligands. The details of the protein properties such as size, pI, the percentage of α-helices and β-sheets, GRAVY indices, the presence of metal/non-metal ligands and their binding sites are detailed in Supplementary Table S1. The secondary structures of selected RPL proteins with ligand binding properties are represented in Supplementary Figure S2.
To evaluate the evolutionary relationships within the RPL protein family of rice, three phylogenetic trees were constructed using full-length amino acid sequences and amino acid sequences derived from ribosomal domains and Low Complexity Regions (LCR; Supplementary Figure S3). The unrooted phylogenetic tree was constructed by using a ‘neighbor joining algorithm’ with a bootstrap value of 1000. The homologous proteins having significant bootstrap value (>95%) were considered as having the highest similarity with respect to others.
The phylogenetic tree of full-length RPL proteins was divided into four clades or groups (A, B, C, and D). The RPL proteins, RPL24b and 26.1 have the highest similarity indicating that these two genes might have become duplicated recently in the rice genome. The phylogenetic relationship has also been used for gene function identification and the proteins with the highest similarity perhaps exhibit similar functions and expression patterns (Lijavetzky et al., 2013). The expression of RPL24b and RPL26.1 was similar in shoots indicating their similar roles in shoot growth and development; their expression was also similar in oxidative stress indicating functional similarity.
The phylogenetic trees of two separate domains, the ribosomal domain and LCR were also divided into groups to check domain-wise similarity. The ribosomal-domain analysis showed maximum sequence similarity between RPL24b and 26, which exhibited similarity with RPL18a and 19.3 proteins. RPL18a and 19.3 showed similar expression patterns in different tissues like spikes, endosperm, plumules, radicles, 45 days shoot and root tissues and root–shoot transition indicating their functional similarity during growth and development. All the RPL proteins except RPL10, 11, 14, 21.2, 35, 36, 37, 38, and 51 have predicted LCR. The phylogenetic analysis of LCR showed that RPL6 and RPL26 exhibited the maximum sequence similarity followed by RPL5 and 31 and RPL27 and RPL8, whereas RPL3 formed a separate clade, which showed its possible divergence from the other RPL proteins.
The expression studies showed that many RPL genes are differentially regulated in various tissues and under various abiotic treatments. To assess whether this differential regulation is due to the presence of stress or signal-responsive elements in their regulatory regions, nucleotide sequences ≤1 kb upstream to each of the 34 genes were retrieved and searched using the PlantCARE database. This analysis resulted in the identification of multiple stress-responsive elements in the putative promoter regions of all the genes. Abiotic stress-responsive elements that are associated with heat and cold temperatures such as HSE (Heat Stress Elements) and LTR (Low-Temperature Response) and dehydration stress such as MBS (Myb Binding Site) are widely distributed within the putative promoter regions of RPL genes. MBS is a binding site for MYB-related transcription factors that are involved in the regulation of genes responsive to water-deficit conditions (Urao et al., 1993). The presence of these elements in the promoter regions suggests that the corresponding genes become activated under water stress or drought conditions. In addition to abiotic stresses, elements that respond to phytohormones such as ABA (ABRE-Abscisic acid responsive element and Motif IIb), MeJa (TGACG-motif and CGTCA-motif), SA (TCA-motif), Gibberellic acid (GARE-Gibberellic acid responsive element), and Auxin (TGA-motif and AuxR-Auxin responsiveness) are also present in multiple copies.
Except RPL37, which did not exhibit any abiotic-responsive element in its upstream region, the putative promoters of all other genes had one or the other stress-responsive elements. MBS, ABRE, TGACG, and CGTCA motifs are commonly found in multiple copies. RPL8 has five repeats of ABRE and two repeats of each MBS, TGACG, CGTCA and TGA elements. RPL10 exhibited five repeats of each TGACG and CGTCA motifs that respond to MeJa treatment and three copies of MBS elements. RPL14 has four repeats of dehydration responsive elements. RPL18a showed four and three copies of Motif IIb and ABRE, respectively, that respond to ABA and two copies of TGACG and CGTCA motifs. RPL28 showed five repeats of ABRE and four repeats of MeJa responsive elements. RPL29 has six copies of ABRE and three copies of TGA element. RPL31 showed three copies of TCA element and two repeats of TGACG and CGTCA motifs. RPL35 has four copies of ABRE and two copies of TCA element. RPL36 and 38 had three copies of ABRE and MBS elements, respectively. RPL44 has two copies of ABRE, CGTCA and TGACG motifs (Figure 2). In addition, TC-rich repeats that are involved in defense and stress-responsiveness (Diaz-De-Leon et al., 1993), W-box motifs which are the binding sites for stress-responsive WRKY transcription factors (Eulgem and Somssich, 2007), a WUN-motif, a wound-responsive element that is associated with biotic stress (Jiang et al., 2014), a Box-W1 motif, a fungal elicitor element that binds with WRKY33 transcription factor in response to phytopathogens (Rushton et al., 1996; Lippok et al., 2007) are present in single copies in the putative cis-elements. Supplementary Table S2 presents a detailed analysis of both abiotic and biotic responsive elements and their repeats in the putative promoter regions.
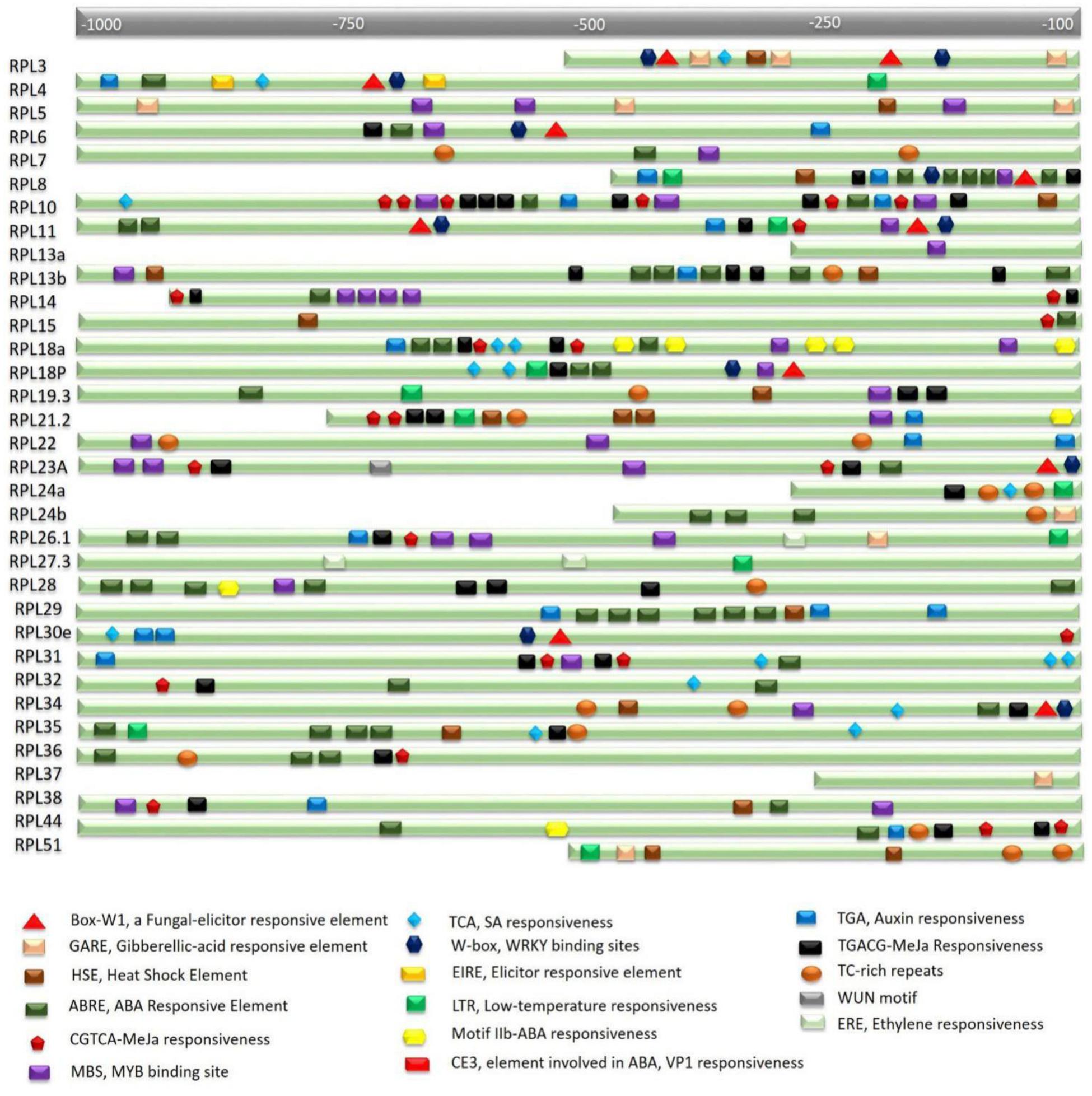
FIGURE 2. In silico analysis of rice RPL promoters for the identification of cis-regulatory elements. Nucleotide sequence ≤1 kb upstream of the transcription start site of each gene carries multiple stress and signal-responsive elements. Each element is represented with a different shape and color which is described at the bottom of the figure. A scale at the top indicates the putative localization of corresponding elements.
To obtain insights into the tissue-specific and native expression patterns of RPL genes, we studied the expression of 34 RPL genes in 13 different tissues including 16 h embryo and endosperm, plumule and radicle of 3-day-old seedlings, root and shoot tissues of 7-day-old seedlings, 2-month-old tissues of mature flag leaf, shoot, root, root–shoot transition, partially filled grains, flowers and spikes. The primer details of 34 RPL genes used in the expression analysis were provided in Supplementary Table S3.
Two-month-old shoot and root tissues induced an up-regulation of 30 and 22 RPL genes, respectively, which is larger than any other tissue. Out of 34, root–shoot transition and grains induced the expression of 17 RPL genes. Embryo, 6 days root and floral organs induced the expression of 12 RPL genes, whereas endosperm, plumule, radicle, 6 days shoot, flag leaf, and spikes induced the expression of a total of 16, 13, 14, 10, 21, and 19 RPL genes, respectively. RPL5 and RPL24a were highly up-regulated in all the tissues. The expression of RPL27 and RPL37 was detected only in three tissues; endosperm, 45 days shoot, flowers and 6 days shoot, 45 days root and flowers, respectively. RPL8 and RPL15 were expressive only in endosperm and 6 days roots, respectively, but non-expressive in the remaining tissues studied. Ten RPL genes viz., RPL5, 7, 8, 18P, 19.3, 21.2, 22, 24a, 31, and 34 were commonly up-regulated in embryo and endosperm indicating that they can be implicated in early embryonic development. RPL4-6, 13a, 19.3, 23A, and 24a were up-regulated in plumules and radicle suggesting their role in root and shoot initiation. RPL5, 6, 24a, 31, 34, and 51 were highly expressive in shoot and root tissues of 7-day-old seedlings. RPL4-6, 11, 13a, 14, 18, 19, 21.2, 22, 23A, 24a, 31, 34, 35, 38, and 44 were commonly up-regulated in root, shoot and flag leaf indicating that these genes are involved in vegetative growth and plant maturity. RPL13a, 14, 19.3, 22, 24a, 26, and 34 were highly expressive in spikes, flowers and partially filled grains indicating that these are likely associated with the development of reproductive organs and grain filling. RPL13a, 14 and 24a were expressive from the 6-day-old seedling stage to the grain filling stage in all the tissues studied indicating that they play a major role in the growth and development of both vegetative and reproductive organs such as root, shoot, flowers and grains. RPL5, 19.3, 23A, and 24a were expressive in mitotically active tissues like embryo, endosperm, plumule, and radicles suggesting that these genes are involved in the early maturity and emergence of shoot and root. RPL10 and RPL29 were specifically expressed only in endosperm and flowers, respectively (Supplementary Table S4). The spatial expression of 34 RPL genes in 13 different tissues has been represented in the form of heat maps generated by incorporating the qRT-PCR data obtained from tissue samples collected as three biological replicates (Figure 3).
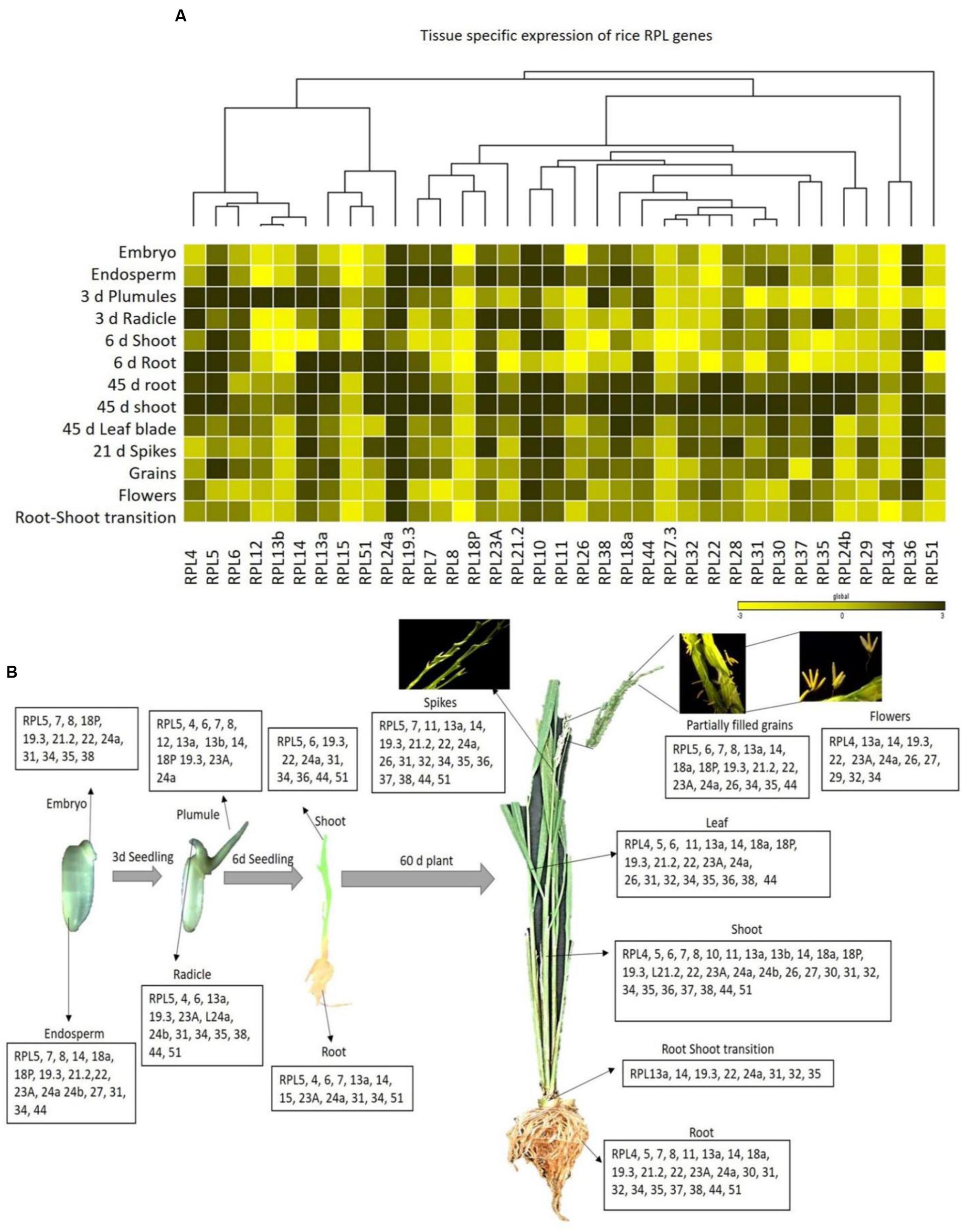
FIGURE 3. Tissue-specific expression of rice RPL genes. (A) The qRT-PCR of 34 RPL genes was performed in 13 different tissues and the level of expression was normalized with rice actin. The mean values of fold change of biological and technical triplicates were represented in the form of heat maps. A dendrogram was constructed to represent the Hierarchical clustering. (B) The RPL transcripts that are significantly up-regulated in each tissue are represented pictorially at the bottom.
The motivation for studying rice large subunit ribosomal genes in stress-response stems from our results on activation-tagged mutant population generated for an important agronomical trait called water-use efficiency (Moin et al., 2016). The mutants with sustained/improved seed productivity under the conditions of limited water availability were selected for flanking sequence analysis and subsequently for studying the expression pattern of the enhancer tagged genes. The seven short-listed mutant plants that appeared to have high productivity were then characterized with physiological parameters related to WUE such as measuring their photosynthetic efficiency and carbon isotope analysis. Among these, two mutants were found to have ribosomal large subunit genes, RPL6 and RPL23A activated by the integrated enhancers. The presence of multiple stress-responsive elements in their putative promoter regions and their significant up-regulation in response to ABA, NaCl, and dehydration stresses further corroborated our findings (Moin et al., 2016). This study not only suggested that RPL6 and RPL23A are potential candidates for abiotic-stress amelioration, but more importantly it provides a basis for the exploration of other members of large subunit ribosomal genes for stress-responsiveness.
Taking a cue from these observations, we assessed the abiotic stress responsive roles of other rice 60S ribosomal genes. For this, we selected 34 genes, one from each orthologous group as described earlier and comprehensively studied the differential transcriptional regulation of 34 genes under phytohormone (MeJa and SA), temperature (heat; 42°C and cold; 4°C) and oxidative stress treatments in shoot and root tissues at six different time intervals (5 min, 3 h, 6 h, 12 h, 24 h, and 60 h). After applying the abiotic treatments, tissue samples were collected as early as 5 min to check the immediate responsiveness of the RPL genes and continued up to 60 h. All the RPL genes responded to the treatments in the form of either up or down-regulation.
The genes that exhibited ≥3-fold transcript level on the log2 scale were considered as significantly up-regulated. MeJa, SA, and cold treatments induced the up-regulation of more genes (>60%) than heat and H2O2 treatments, which caused the down-regulation of 75% of the genes. In shoots, MeJa and SA-induced the up-regulation of 27 (79%) RPL genes each, cold treatment up-regulated 19 genes (55%), while heat and H2O2 treatments up-regulated 6 (17%) RPL genes each. In roots, MeJa, SA, cold, heat, and H2O2 treatments up-regulated 19 (55%), 22 (64%), 16 (47%), 6 (17%), and 14 (41%) RPL genes, respectively. Genes that were up-regulated in both the shoot and root tissues include; RPL7, 8, 12, 13b, 18P, 19.3, 24a, 32, 35, and 51 under MeJa treatment, whereas SA-induced the expression of RPL7, 8, 12, 13b, 19.3, 24a, 26, 32, 35, and 51. Genes such as RPL6, 7, 23A, 28, 32, 35, and 37 were up-regulated in cold treatment, while RPL6, 12, 23A, and RPL18a and 13a were up-regulated under heat and H2O2 treatments, respectively.
To study the detailed regulation of RPL genes at various time points, we categorized the genes that responded within 5 min to 3 h after treatment as immediate-early (IE), those that responded between 3 h to 12 h as early (E) and those that were regulated after 12 h of treatment up to 60 h were considered as late (L) responding genes. The majority of the genes that were up-regulated had responded immediately within 5 min to 3 h after the onset of the stress. In shoots, among the genes that were expressive, a total of 17, 21, 19, 6, and 1 genes belonged to the IE-responsive class with instantaneous up-regulation under MeJa, SA, cold, heat, and H2O2 treatments, respectively. Among this IE-responsive class of genes, some continued to maintain a high level of expression at all the time points observed, while others exhibited a split in the expression followed by again an increase in the level of their expression. These probably form an important set of genes that respond to environmental stresses and might function as an immediate defense after the onset of the stress (Kawasaki et al., 2001).
The other class of IE genes was down-regulated after an IE response. Under MeJa and SA treatments, RPL7, 8, 12, 13b, 19.3, 24a, 28, and 35 maintained a high level of expression throughout the duration of stress in both shoot and root tissues. However, the level of transcriptional up-regulation varied with some exhibiting a very high level of up-regulation up to 100-fold (RPL8, 12, 19.3, 24a, and 35), while some had moderate expression up to 30-fold (RPL7) and others showed low transcript levels with <10-fold (RPL28). RPL6, 7, 12, and 24a exhibited a consistent up-regulation under cold and heat treatments, of which RPL6 and 12 became up-regulated more than 50-fold. In H2O2 treatment, RPL18a was up-regulated in both roots and shoots up to 65-fold whereas 24a, 24b, 30, and 34 showed significant up-regulation in roots. Since stress signals are transmitted through the roots to other parts of the plant body, genes that were significantly up-regulated particularly in roots might have an important role in combating the stress and providing early defense.
The differential expression patterns in response to MeJa and SA (Figure 4) and cold, heat and oxidative treatments (Figure 5) have been represented in the form of heat maps. These were generated by incorporating the mean values of fold change normalized using ΔΔCT method obtained from three biological and three technical replicates. The overlap in the up-regulation (Figure 6) and down-regulation (Figure 7) of 34 RPL genes in both shoot and root tissues were represented as Venn diagrams. Supplementary Tables S5 and S6 provide a detailed list of genes that exhibited overlap in the up and down-regulation, respectively, in shoot and root tissues at each time point.
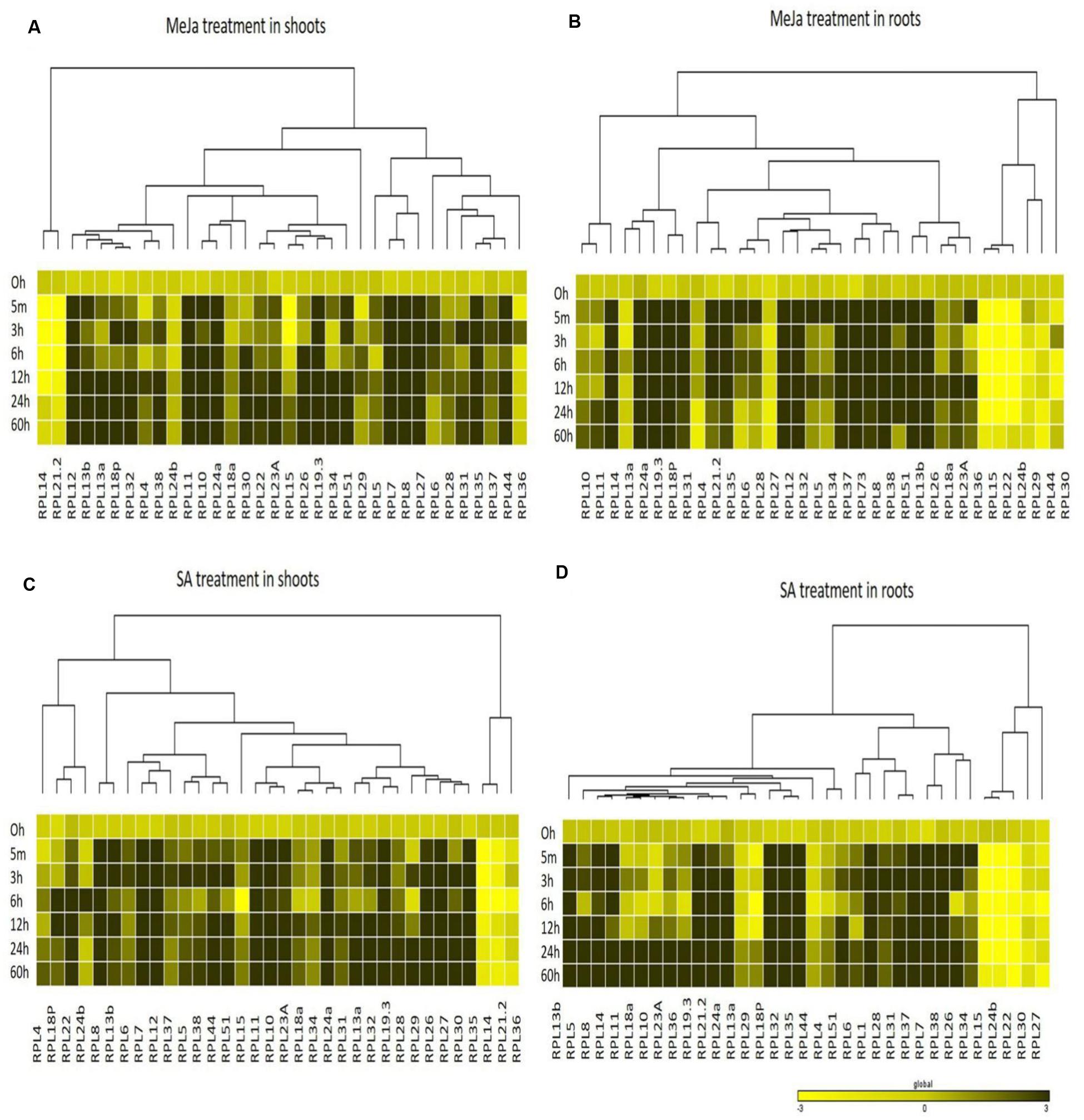
FIGURE 4. Heat map representation of RPL genes in rice treated with MeJa and SA. Seven-day-old rice seedlings were exposed to different abiotic stresses such as MeJa; 100 μM (A,B) and SA; 3 mM (C,D) at six different time points as indicated on the left. The qRT-PCR is used to determine the expression levels of RPL genes and the fold change was normalized using ΔΔCT method relative to that in unstressed seedlings dipped in water at corresponding time points. Rice actin (act1) and β-tub genes were used as internal controls. Three biological replicates and two technical replicates were included in the study. A dendrogram was constructed to represent the Hierarchical clustering of genes.
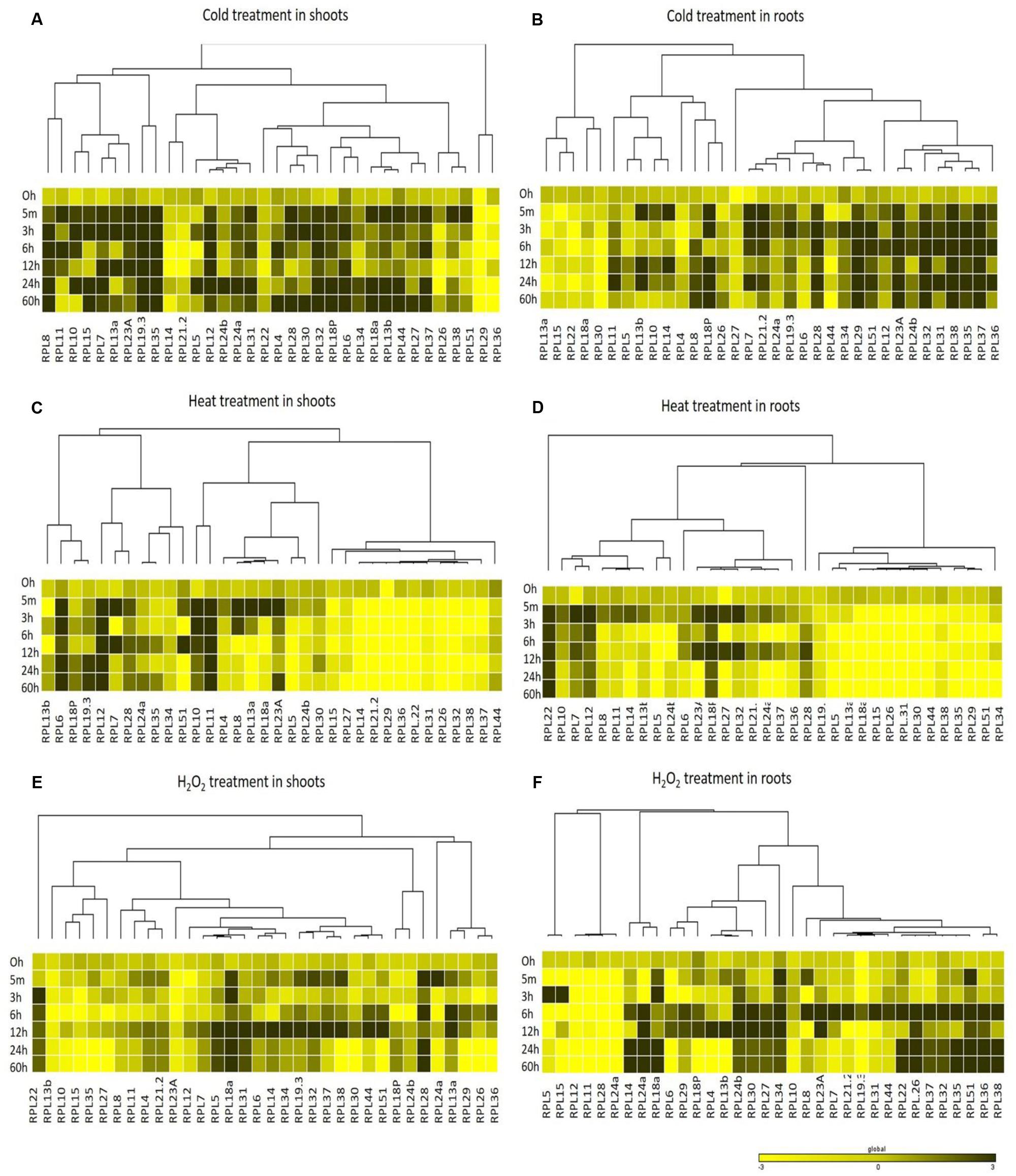
FIGURE 5. Heat map representation of RPL genes in rice in response to cold, heat and H2O2 treatments. Seven-day-old rice seedlings were exposed to different abiotic stresses such as cold stress at 4°C (A,B), heat stress at 42°C (C,D) and oxidative stress with H2O2; 10 μM (E,F) at six different time points as indicated on the left. The qRT-PCR is used to determine the expression levels of RPL genes and the fold change was normalized relative to that in unstressed seedlings dipped in water at corresponding time points. Rice actin (act1) and β-tubulin were used as internal reference genes. Three biological replicates and two technical replicates were included in the study. A dendrogram was constructed to represent the Hierarchical clustering of genes.
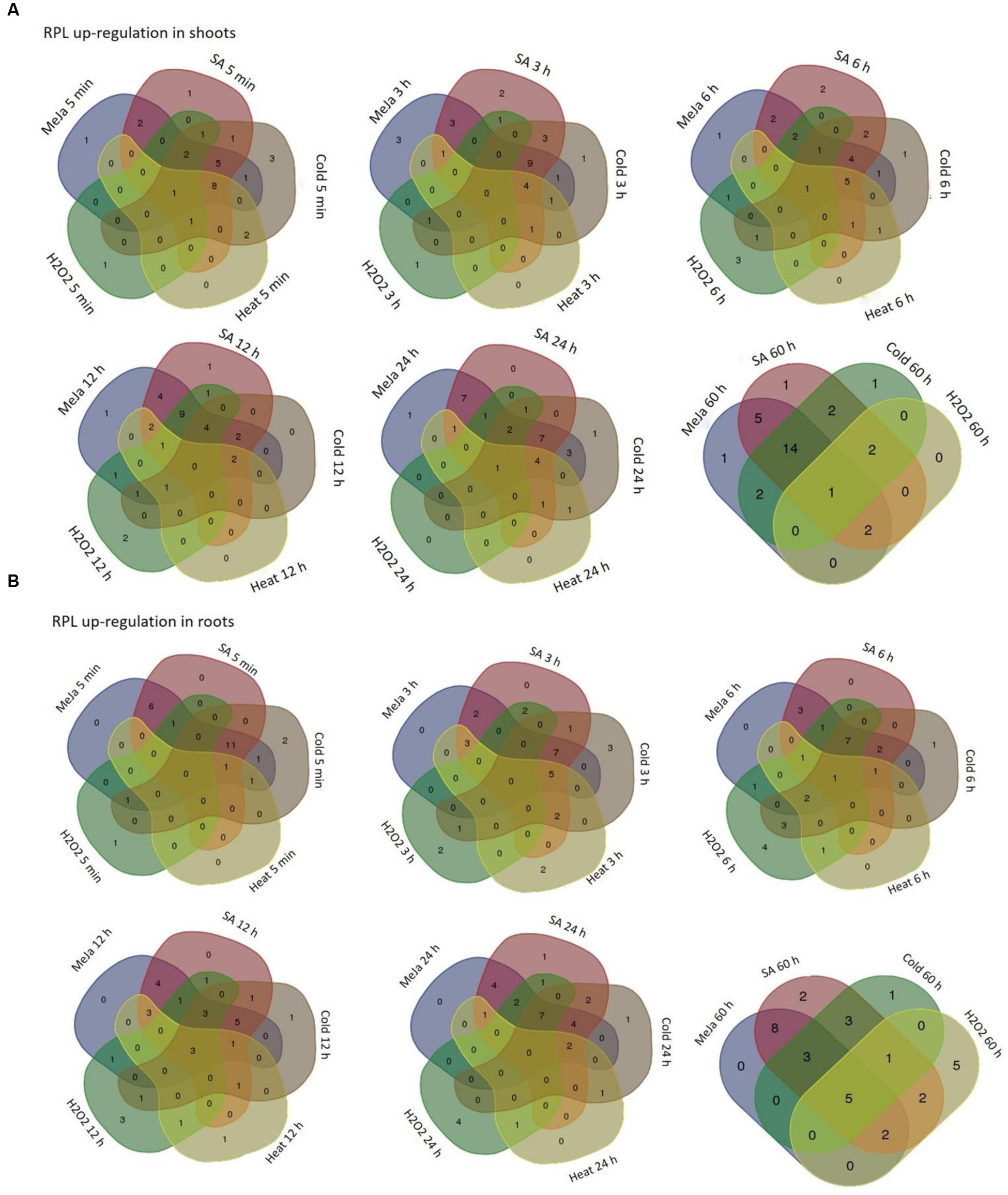
FIGURE 6. Overlap in the up-regulation of rice RPL genes under five abiotic conditions. The RPL genes that exhibited ≥3-fold transcript level on the log2 scale were considered as significantly up-regulated while others were considered as down-regulated or without any change in expression. Venn diagrams are used to show the overlap in the up-regulation in shoot (A) and root (B) tissues.
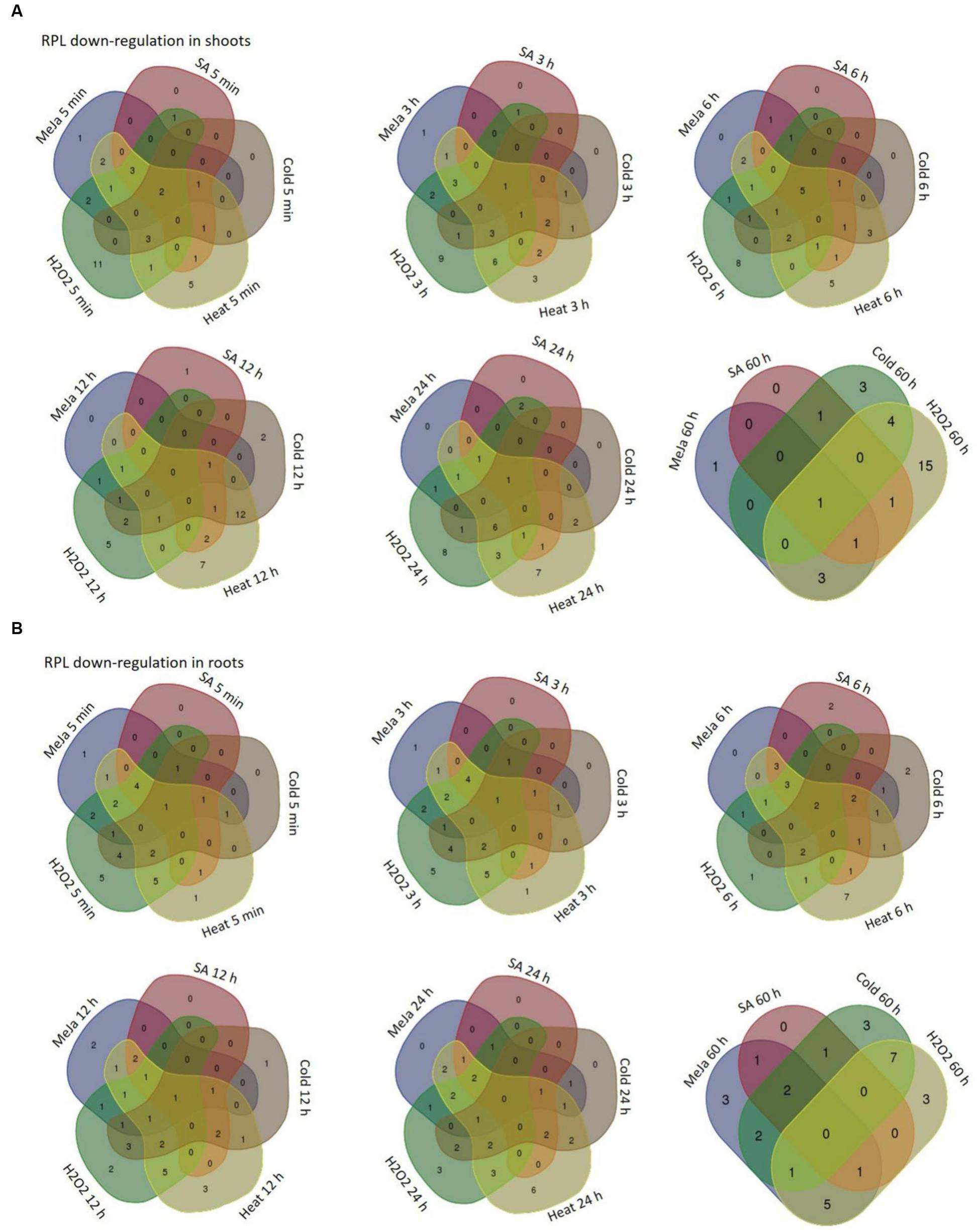
FIGURE 7. Overlap in the down-regulation of rice RPL genes under five abiotic conditions. The RPL genes that exhibited <3-fold transcript level on the log2 scale were considered as down-regulated with respect to others. Venn diagrams are used to show the overlap in the down-regulation in shoot (A) and root (B) tissues separately.
The qRT-PCR analysis of 34 genes showed that they are differentially regulated under abiotic treatments, with many of them becoming significantly and immediately up-regulated. Hence, we simultaneously examined the expression levels of 34 RPL genes in response to the Xanthomonas oryzae pv. oryzae pathogen that causes BLB of rice. Out of 34 genes, 6 (17%) were down-regulated, RPL38 was non-responsive, and the remaining genes became activated (80%). RPL12, 28, 30, 36, 44, and 51 were among those that were down-regulated, and the transcript level of RPL38 did not change significantly, while all other genes studied were up-regulated. Among those that were expressive, RPL10, 11, 15, 24a, 26, 27, and 37 up-regulated more than 10-fold. The transcript level of RPL10 was the highest with more than 75-fold up-regulation (Figure 8A). In addition to significant expression at 11 days, the transcript level of RPL10 also exhibited a gradual increase at 3 h, 6 h, 1 day, and 2 days up to 7 days post-infection with the Xanthomonas oryzae pv. oryzae pathogen (Figure 8B).
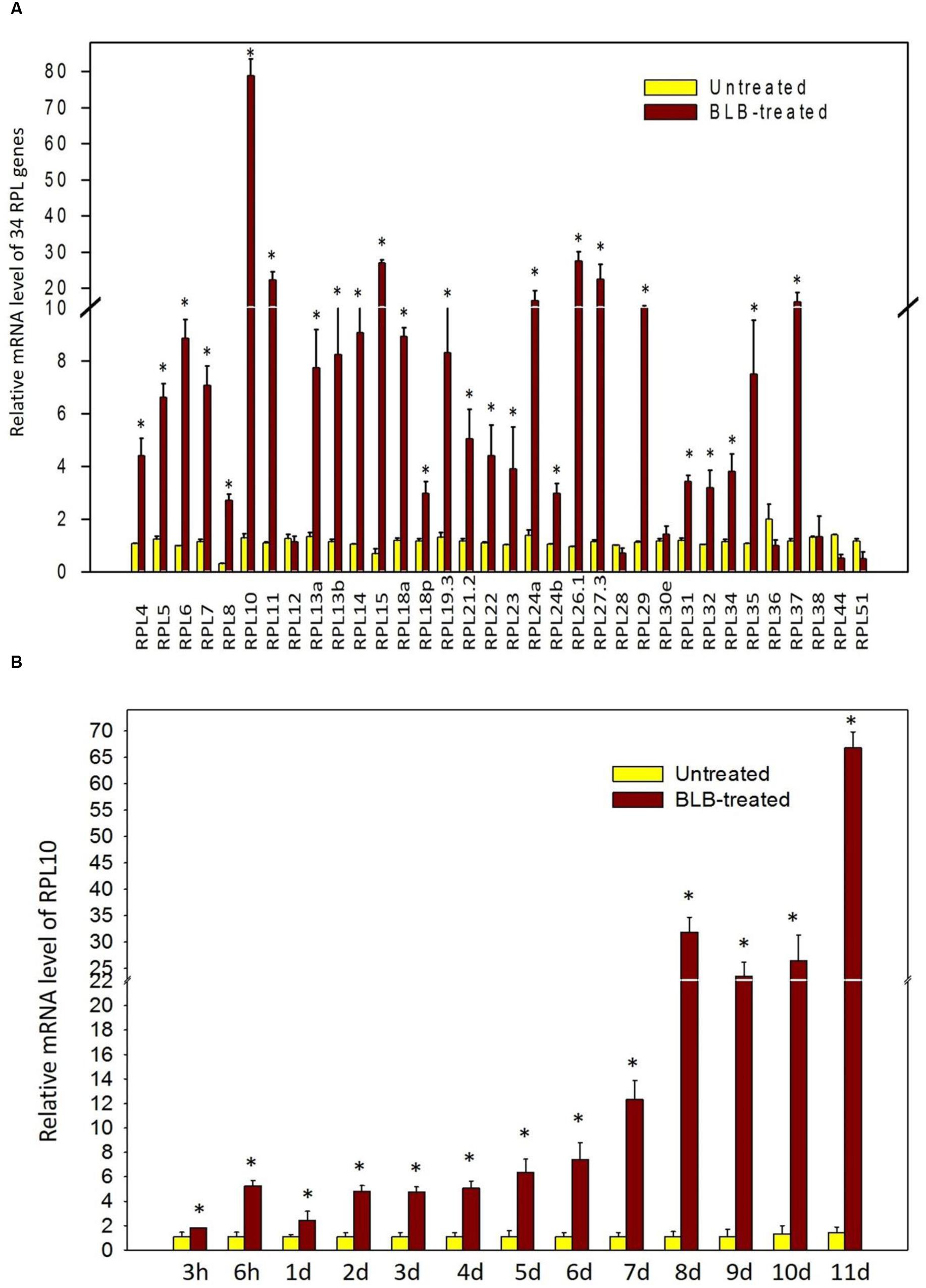
FIGURE 8. Expression of RPL genes upon infection of rice with the Xanthomonas oryzae pv. oryzae pathogen. The expression of RPL genes was determined in response to the bacterium, Xanthomonas oryzae, which causes leaf blight. (A) The bacterial suspension was applied on 60-day-old rice plants, and after 11 days of treatment, leaf samples were analyzed for differential transcript levels of 34 RPL genes. (B) Since the up-regulation of RPL10 was significant, we analyzed its expression at progressive time courses such as 3 h, 6 h, 1 day, 2 days, 3 days, 4 days, 5 days, 6 days, 7 days, 8 days, 9 days, 10 days, and 11 days post-infection with Xanthomonas oryzae pv. oryzae pathogen. The expression was normalized with untreated samples of same age grown under identical conditions. The statistical significance was calculated using one-way ANOVA at P < 0.05 and represented with asterisks.
Stress factors such as heat, cold, dehydration, and pathogen attack can exacerbate the global agriculture system with an estimated >50% crop yield loss per annum (Wang et al., 2003). Each of these stresses elicits a cascade of signaling pathways to ensure plant survival. It is important to identify the genes that contribute to sustainable plant productivity under conditions of stress to further improve their productivity potential. Although more than 500 genes have been overexpressed and characterized in rice for stress-tolerance, many remain to be identified and examined.
In the present work, we report on the comprehensive expression profiling of rice ribosomal large subunit genes under multiple abiotic and biotic stress treatments at progressive time points and also identified their putative promoter sequences. The information provided here can be exploited further in the functional characterization of these stress-responsive genes, which might help in augmenting rice yields by generating independent transgenic plants. We also identified the genes that exhibited an overlap in the expression patterns in response to two or more stresses. We propose that such genes are particularly promising in bringing about the tolerance to multiple stresses as the presence of a second stress factor can enhance the detrimental effects of the first one (Atkinson and Urwin, 2012).
Ribosomal genes encode proteins that are the components of the two-subunit ribosomal complex, which together with the members of the same group and other proteins participate in protein synthesis. There have been limited reports on the role of ribosomal genes in the stress-responses. We had generated an activation-tagged transgenic rice plant population carrying CaMV35S tetrameric enhancers. These rice mutants were screened for water-use efficiency by growing them under the provision of limited water supply compared to the level that required for normal growth of rice. Flanking sequence analysis of selected mutants having sustained growth and productivity under the condition of limited water availability revealed the activation of the two ribosomal genes (RPL6 and RPL23A) by the integrated enhancers (Moin et al., 2016). This has persuaded us further to analyze the importance of several of RPL genes in stress-responses. We therefore, performed a comprehensive native tissue-specific and differential expression of 34 RPL genes under various abiotic and biotic stress conditions at different time intervals.
The availability of full-length and high-quality rice genome sequence and databases, further helped us to exploit the information on these genes. Based on the information available in rice databases, we identified 123 genes that are the components of rice ribosomal large subunit, of which 2–3 genes exist as identical gene copies in the genome. These genes are distributed throughout the 12 chromosomes of rice genome with chromosome 7 and 1 having the highest number of genes. The present investigation that reports on the analysis of native and differential expression of the RPL gene family also corroborated the earlier reports that RPL genes are regulated spatio-temporally (Sormani et al., 2011; Carroll, 2013; Zheng et al., 2016). In rice, RPL genes appear to be developmentally regulated as they are widely expressed in all the 13 tissues studied starting from as early as embryonic initiation to plant maturity. Among the 19 RPL genes that were expressive in grain filling stage, the expression of RPL5, 7, 13a, 14, 19.3, 22, 24a, 34, and 35 were conspicuously detected. Further characterization of these genes would throw useful insights into their role in grain production, which is a significant yield-related trait in rice.
Although many of the RPL genes were expressive in all the tissues, they cannot be considered as house-keeping as their level of expression changed in response to environmental signals. Similar expression profiling of small and large subunit genes was reported in response to macro-elements deficiency in Arabidopsis in which about 244 among 249 RP genes became up-regulated (Wang et al., 2013). The up-regulation of the RPL genes is likely to maintain or improve protein synthesis and hence, proper functioning of ribosomes, the basic cellular moieties under the conditions of stress (Kim et al., 2004). Plants being sessile acclimate to environmental cues by undergoing many metabolic changes, one of them being increased protein turnover that includes both protein biosynthesis and ubiquitination (Kosová et al., 2014). Proteomic studies revealed variations in the levels of translation-related proteins such as initiation factors, elongation factors, and proteins of both small and large subunits during the process of acclimation particularly, to dehydration, salt and temperature stresses in cereals (Fatehi et al., 2012; Budak et al., 2013; Ghabooli et al., 2013; Gharechahi et al., 2014).
In addition to their significant up-regulation, the presence of multiple cis-regulatory elements in the putative promoter regions of RPL genes further corroborates our findings that these genes might also play a role in alleviating plant biotic and abiotic stress. RPL6 and RPL23A, in addition to their role in WUE, became up-regulated in almost all the stresses studied illustrating their possible involvement in inducing tolerance to abiotic stresses (Moin et al., 2016). Cold, MeJa, and SA treatments induced the up-regulation of a majority of RPL genes, while H2O2 and heat treatments down-regulated 75% of the genes.
The up-regulation of RPL genes by cold treatment is to enhance the process of polypeptide synthesis at low temperatures (Kim et al., 2004). RPL7, 8, 12, 13b, 19.3, 24a, 28, and 35 were up-regulated and constantly maintained a high level of expression throughout the duration of stress in response to SA and MeJa, the two phytohormones involved in plant defense against pathogen attack. These genes also contain TC-rich repeats, which are known for their involvement in plant defense and stress response (Diaz-De-Leon et al., 1993). RPL18a, 24a, 24b, 30, and 34 were expressed at higher levels when exposed to H2O2 treatment. This may reflect that these genes might have potential in combating oxidative stress. High temperature appeared to cause detrimental effects on the expression of RPL genes as heat stress had down-regulated >75% of the genes. RPL6, 12, and 23A were among those whose expression was detected. Down-regulation of RPL genes under high temperature might be because of decreased stability of RNA molecules with increasing temperatures.
Infection with Xanthomonas oryzae pv. oryzae, which causes BLB also up-regulated a large number of RPL genes. Among those that were expressive (RPL10, 11, 15, 24a, 26, 27, and 37), the expression of RPL10 was more evident as its transcript levels gradually increased from 3 h post-infection and reached a peak at 11 days after treatment. Also, RPL10 was activated in shoots under MeJa and SA up to 60 h after treatment, further suggesting its involvement in biotic stress response.
Our exploration on the detailed expression analysis underpins that these genes regulate tissue-specific development and respond rapidly to the environmental cues and might function as facilitators of immediate defense against stresses. The coordinated transcriptional up-regulation of translation-related genes is a necessity for the cells to maintain the crucial functions of ribosomes under the conditions of stress. The increase in the expression of RPL genes under a wide range of stress-treatments including both biotic and abiotic conditions demonstrate that these are potential targets for the manipulation of stress-tolerance in rice and other related cereal crops as well. However, the level of tolerance induced by each of these genes needs to be analyzed by their independent overexpression in the transgenic rice plants.
PK and MM designed the experiments. MM performed all the experiments. AB, MD, and AS helped in the analysis of qRT-PCR data. SM performed the Xanthomonas oryzae pv. oryzae infection on rice leaves. MM and PK prepared the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The funding for the current work has been obtained through a grant sponsored by DBT, GOI to PK with grant number BT/PR13105/AGR/02/684/2009. MM is grateful to DBT for proving a Research Fellowship. AB is also thankful to DBT for Project Fellowships.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01284
H2O2, hydrogen peroxide; MeJa, methyl jasmonate; RP, ribosomal protein; RPL, ribosomal protein large subunit; SA, salicylic acid.
Alberts, B., Jhonson, A., Lewis, J., Raff, M., Roberts, K., and Walter, P. (2002). Molecular Biology of the Cell. 4th ed. New York, NY: Garland Science.
Atkinson, N. J., and Urwin, P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3543. doi: 10.1093/jxb/ers100
Ban, N., Nissen, P., Hansen, J., Moore, P. B., and Steitz, T. A. (2000). The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 289, 905–920. doi: 10.1126/science.289.5481.905
Barakat, A., Szick-Miranda, K., Chang, I. F., Guyot, R., Blanc, G., Cooke, R., et al. (2001). The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 127, 398–415. doi: 10.1104/pp.010265
Ben-Shem, A., Jenner, L., Yusupova, G., and Yusupov, M. (2010). Crystal structure of the eukaryotic ribosome. Science 330, 1203–1209. doi: 10.1126/science.1194294
Budak, H., Akpinar, B. A., Unver, T., and Turktas, M. (2013). Proteome changes in wild and modern wheat leaves upon drought stress by two-dimensional electrophoresis and nanoLC-ESI–MS/MS. Plant Mol. Biol. 83, 89–103. doi: 10.1007/s11103-013-0024-5
Byrne, M. E. (2009). A role for the ribosome in development. Trends Plant Sci. 14, 512–519. doi: 10.1016/j.tplants.2009.06.009
Callis, J., Fromm, M., and Walbot, V. (1987). Introns increase gene expression in cultured maize cells. Genes. Dev. 1, 1183–1200. doi: 10.1101/gad.1.10.1183
Carroll, A. J. (2013). The Arabidopsis cytosolic ribosomal proteome: from form to function. Front. Plant Sci. 4:32. doi: 10.3389/fpls.2013.00032
Carroll, A. J., Heazlewood, J. L., Ito, J., and Millar, A. H. (2008). Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol. Cell. Proteomics. 7, 347–369. doi: 10.1074/mcp.M700052-MCP200
Casati, P., and Virginia, W. (2003). Gene expression profiling in response to ultraviolet radiation in maize genotypes with varying flavonoid content. Plant Physiol. 132, 1739–1754. doi: 10.1104/pp.103.022871
Cherepneva, G. N., Schmidt, K. H., Kulaeva, O. N., Oelmüller, R., and Kusnetsov, V. (2003). Expression of the ribosomal proteins S14, S16, L13a and L30 is regulated by cytokinin and abscisic acid: implication of the involvement of phytohormones in translational processes. Plant Sci. 165, 925–932. doi: 10.1016/S0168-9452(03)00204-8
Degenhardt, R. F., and Bonham-Smith, P. C. (2008a). Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol. 147, 128–142. doi: 10.1104/pp.107.111799
Degenhardt, R. F., and Bonham-Smith, P. C. (2008b). Evolutionary divergence of ribosomal protein paralogs in Arabidopsis. Plant Signal. Behav. 3, 493–495. doi: 10.4161/psb.3.7.5991
Diaz-De-Leon, F., Klotz, K. L., and Lagrimini, M. (1993). Nucleotide sequence of the tobacco (Nicotiana tabacum) anionic peroxidase gene. Plant Physiol. 101:1117. doi: 10.1104/pp.101.3.1117
Donath, M., Mendel, R., Cerff, R., and Martin, W. (1995). Intron-dependent transient expression of the maize GapA1 gene. Plant Mol. Biol. 28, 667–676. doi: 10.1007/BF00021192
Doudna, J. A., and Rath, V. L. (2002). Structure and function of the eukaryotic ribosome: the next frontier. Cell 109, 153–156. doi: 10.1016/S0092-8674(02)00725-0
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Fatehi, F., Hosseinzadeh, A., Alizadeh, H., Brimavandi, T., and Struik, P. C. (2012). The proteome response of salt-resistant and salt-sensitive barley genotypes to long-term salinity stress. Mol. Biol. Rep. 39, 6387–6397. doi: 10.1007/s11033-012-1460-z
Ferreyra, M. L. F., Casadevall, R., Luciani, M. D., Pezza, A., and Casati, P. (2013). New evidence for differential roles of l10 ribosomal proteins from Arabidopsis. Plant Physiol. 163, 378–391. doi: 10.1104/pp.113.223222
Ferreyra, M. L. F., Pezza, A., Biarc, J., Burlingame, A. L., and Casati, P. (2010). Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet-B stress. Plant Physiol. 153, 1878–1894. doi: 10.1104/pp.110.157057
Fuller, M. (2007). Abiotic Stress Tolerance in Plants: Towards the Improvement of Global Environment and food (2006). Berlin: Springer.
Fuxreiter, M., Simon, I., Friedrich, P., and Tompa, P. (2004). Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J. Mol. Biol. 338, 1015–1026. doi: 10.1016/j.jmb.2004.03.017
Ghabooli, M., Khatabi, B., Ahmadi, F. S., Sepehri, M., Mirzaei, M., Amirkhani, A., et al. (2013). Proteomics study reveals the molecular mechanisms underlying water stress tolerance induced by Piriformospora indica in barley. J. Proteomics. 94, 289–301. doi: 10.1016/j.jprot.2013.09.017
Gharechahi, J., Alizadeh, H., Naghavi, M., and Sharifi, G. (2014). A proteomic analysis to identify cold acclimation associated proteins in wild wheat (Triticum urartu L.). Mol. Biol. Rep. 41, 3897–3905. doi: 10.1007/s11033-014-3257-8
Giavalisco, P., Wilson, D., Kreitler, T., Lehrach, H., Klose, J., Gobom, J., et al. (2005). High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol. Biol. 57, 577–591. doi: 10.1007/s11103-005-0699-3
Guarinos, E., Santos, C., Sánchez, A., Qiu, D. Y., Remacha, M., and Ballesta, J. P. (2003). Tag-mediated fractionation of yeast ribosome populations p1roves the monomeric organization of the eukaryotic ribosomal stalk structure. Mol. Microbiol. 50, 703–712. doi: 10.1046/j.1365-2958.2003.03733.x
Hanson, C. L., Videler, H., Santos, C., Ballesta, J. P., and Robinson, C. V. (2004). Mass spectrometry of ribosomes from Saccharomyces cerevisiae: implications for assembly of the stalk complex. J. Biol. Chem. 279, 42750–42757. doi: 10.1074/jbc.M405718200
Hulm, J. L., McIntosh, K. B., and Bonham-Smith, P. C. (2005). Variation in transcript abundance among the four members of the Arabidopsis thaliana RIBOSOMAL PROTEIN S15a gene family. Plant Sci. 169, 267–278. doi: 10.1016/j.plantsci.2005.04.001
Hummel, M., Cordewener, J. H., De Groot, J., Smeekens, S., America, A. H., and Hanson, J. (2012). Dynamic protein composition of Arabidopsis thaliana cytosolic ribosomes in response to sucrose feeding as revealed by label free MSE proteomics. Proteomics 12, 1024–1038. doi: 10.1002/pmic.201100413
Imai, A., Komura, M., Kawano, E., Kuwashiro, Y., and Takahashi, T. (2008). A semi-dominant mutation in the ribosomal protein L10 gene suppresses the dwarf phenotype of the acl5 mutant in Arabidopsis thaliana. Plant J. 56, 881–890. doi: 10.1111/j.1365-313X.2008.03647.x
Ito, T., Gyung-Tae, K., and Kazuo, S. (2000). Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 22, 257–264. doi: 10.1046/j.1365-313x.2000.00728.x
Jami, S. K., Clark, G. B., Ayele, B. T., Roux, S. J., and Kirti, P. B. (2012). Identification and characterization of annexin gene family in rice. Plant Cell Rep. 31, 813–825. doi: 10.1007/s00299-011-1201-0
Jiang, Y., Duan, Y., Yin, J., Ye, S., Zhu, J., Zhang, F., et al. (2014). Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. Eru. 65, 6629–6644. doi: 10.1093/jxb/eru381
Kapoor, M., Arora, R., Lama, T., Nijhawan, A., Khurana, J. P., Tyagi, A. K., et al. (2008). Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics 9:451. doi: 10.1186/1471-2164-9-451
Karve, R., Liu, W., Willet, S. G., Torii, K. U., and Shpak, E. D. (2011). The presence of multiple introns is essential for ERECTA expression in Arabidopsis. RNA 17, 1907–1921. doi: 10.1261/rna.2825811
Kawasaki, S., Borchert, C., Deyholos, M., Wang, H., Brazille, S., Kawai K., et al. (2001). Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13, 889–905. doi: 10.1105/tpc.13.4.889
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kim, K. Y., Park, S. W., Chung, Y. S., Chung, C. H., Kim, J. I., and Lee, J. H. (2004). Molecular cloning of low-temperature-inducible ribosomal proteins from soybean. J. Exp. Bot. 55, 1153–1155. doi: 10.1093/jxb/erh125
Kosová, K., Vítámvás, P., and Prášil, I. T. (2014). Proteomics of stress responses in wheat and barley-search for potential protein markers of stress tolerance. Front. Plant Sci 5:711. doi: 10.3389/fpls.2014.00711
Kyrpides, N. C., Woese, C. R., and Ouzounis, C. A. (1996). KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci. 21, 425–426. doi: 10.1016/S0968-0004(96)30036-4
Liang, Y., Xiong, Z., Zheng, J., Xu, D., Zhu, Z., Xiang, J., et al. (2016). Genome-wide identification, structural analysis and new insights into late embryogenesis abundant (LEA) gene family formation pattern in Brassica napus. Sci. Rep. 6, 24265. doi: 10.1038/srep24265
Lijavetzky, D., Carbonero, P., and Vicente-Carbajosa, J. (2013). Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 3:1. doi: 10.1186/1471-2148-3-17
Lijsebettens, V. M., Vanderhaeghen, R., De Block, M., Bauw, G., Villarroel, R., and Van Montagu, M. (1994). An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J. 13, 3378.
Lippok, B., Birkenbihl, R. P., Rivory, G., Brümmer, J., Schmelzer, E., Logemann, E., et al. (2007). Expression of AtWRKY33 encoding a pathogen-or PAMP-responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol. Plant Microbe Interact. 20, 420–429. doi: 10.1094/MPMI-20-4-0420
Liu, X. D., Xie, L., Wei, Y., Zhou, X., Jia, B., Liu, J., et al. (2014). Abiotic stress resistance, a novel moonlighting function of ribosomal protein RPL44 in the halophilic fungus Aspergillus glaucus. Appl. Environ. Microbiol. 80, 4294–4300. doi: 10.1128/AEM.00292-14
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Maguire, B. A., and Zimmermann, R. A. (2001). The ribosome in focus. Cell 104, 813–816. doi: 10.1016/S0092-8674(01)00278-1
Mitsuhara, I., Iwai, T., Seo, S., Yanagawa, Y., Kawahigasi, H., Hirose, S., et al. (2008). Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol. Genet. Genomics. 279, 415–427. doi: 10.1007/s00438-008-0322-9
Moin, M., Bakshi, A., Saha, A., Kumar, M. U., Reddy, A. R., Rao, K. V., et al. (2016). Activation tagging in indica rice identifies ribosomal proteins as potential targets for manipulation of water-use efficiency and abiotic stress tolerance in plants. Plant Cell Environ. doi: 10.1111/pce.12796 [Epub ahead of print].
Revenkova, E., Masson, J., Koncz, C., Afsar, K., Jakovleva, L., and Paszkowski, J. (1999). Involvement of Arabidopsis thaliana ribosomal protein S27 in mRNA degradation triggered by genotoxic stress. EMBO J. 18, 490–499. doi: 10.1093/emboj/18.2.490
Rushton, P. J., Torres, J. T., Parniske, M., Wernert, P., Hahlbrock, K., and Somssich, I. E. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15:5690.
Ryan, R. P., Vorhölter, F. J., Potnis, N., Jones, J. B., Van Sluys, M. A., Bogdanove, A. J., et al. (2011). Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nat. Rev. Microbiol. 9, 344–355. doi: 10.1038/nrmicro2558
Sáez-Vásquez, J., Gallois, P., and Delseny, M. (2000). Accumulation and nuclear targeting of BnC24, a Brassica napus ribosomal protein corresponding to a mRNA accumulating in response to cold treatment. Plant Sci. 156, 35–46. doi: 10.1016/S0168-9452(00)00229-6
Schmid, M., Davison, T. S., Henz, S. R., Pape, U. J., Demar, M., Vingron, M., et al. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506. doi: 10.1038/ng1543
Schuwirth, B. S., Borovinskaya, M. A., Hau, C. W., Zhang, W., Vila-Sanjurjo, A., Holton, J. M., et al. (2005). Structures of the bacterial ribosome at 3.5 Å resolution. Science 310, 827–834. doi: 10.1126/science.1117230
Schwer, B., Schneider, S., Aronova, A., and Shuman, S. (2009). Characterization of the schizosaccharomyces pombe Spt5-Spt4 complex. RNA 15, 1241–1250. doi: 10.1261/rna.1572709
Song, Q., Wang, S., Zhang, G., Li, Y., Li, Z., Guo, J., et al. (2015). Comparative proteomic analysis of a membrane-enriched fraction from flag leaves reveals responses to chemical hybridization agent SQ-1 in wheat. Front. Plant Sci. 6:669. doi: 10.3389/fpls.2015.00669
Sormani, R., Masclaux-Daubresse, C., Daniele-Vedele, F., and Chardon, F. (2011). Transcriptional regulation of ribosome components are determined by stress according to cellular compartments in Arabidopsis thaliana. PLoS ONE 6:e28070. doi: 10.1371/journal.pone.0028070
Sundaram, R. M., Chatterjee, S., Oliva, R. L., Laha, G. S., Cruz, C. V., Leach, J. E., et al. (2014). Update on bacterial blight of rice: fourth. Rice 7:12. doi: 10.1186/s12284-014-0012-7
Szakonyi, D., and Byrne, M. E. (2011). Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J. 65, 269–281. doi: 10.1111/j.1365-313X.2010.04422.x
Tanaka, S., Ikeda, K., and Miyasaka, H. (2001). Enhanced tolerance against salt-stress and freezing-stress of Escherichia coli cells expressing algal bbc1 gene. Curr. Microbiol. 42, 173–177. doi: 10.1007/s002840010199
Urao, T., Yamaguchi-Shinozaki, K., Urao, S., and Shinozaki, K. (1993). An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5, 1529–1539. doi: 10.1105/tpc.5.11.1529
Wang, H., Hao, J., Chen, X., Hao, Z., Wang, X., Lou, Y., et al. (2007). Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol. 65, 799–815. doi: 10.1007/s11103-007-9244-x
Wang, J., Lan, P., Gao, H., Zheng, L., Li, W., and Schmidt, W. (2013). Expression changes of ribosomal proteins in phosphate-and iron-deficient Arabidopsis roots predict stress-specific alterations in ribosome composition. BMC Genom. 14:1. doi: 10.1186/1471-2164-14-783
Wang, W. X., Vinocur, B., and Altman, A. (2003). Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14. doi: 10.1007/s00425-003-1105-5
Warner, J. R., and McIntosh, K. B. (2009). How common are extraribosomal functions of ribosomal proteins? Mol. Cell 34, 3–11. doi: 10.1016/j.molcel.2009.03.006
Wass, M. N., Kelley, L. A., and Sternberg, M. J. (2010). 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. W469–W473. doi: 10.1093/nar/gkq406
Wool, I. G., Chan, Y. L., and Glück, A. (1996). Mammalian ribosomes: the structure and the evolution of the proteins. Cold Spring Harbor Monogr. Arch. 30, 685–732. doi: 10.1101/087969458.30.685
Yang, L., Xie, C., Li, W., Ruijie, Z., Dengwei, J., and Qing, Y. (2013). Expression of a wild eggplant ribosomal protein L13a in potato enhances resistance to Verticillium dahliae. Plant Cell Tissue Organ Cult. 115, 329–340. doi: 10.1007/s11240-013-0365-4
Keywords: ribosomal proteins, abiotic stress, biotic stress, gene expression, rice
Citation: Moin M, Bakshi A, Saha A, Dutta M, Madhav SM and Kirti PB (2016) Rice Ribosomal Protein Large Subunit Genes and Their Spatio-temporal and Stress Regulation. Front. Plant Sci. 7:1284. doi: 10.3389/fpls.2016.01284
Received: 09 June 2016; Accepted: 11 August 2016;
Published: 24 August 2016.
Edited by:
Shabir Hussain Wani, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, IndiaReviewed by:
Biswapriya Biswavas Misra, University of Florida, USACopyright © 2016 Moin, Bakshi, Saha, Dutta, Madhav and Kirti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: P. B. Kirti, cGJraXJ0aUB1b2h5ZC5hYy5pbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.