- Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China
Broccoli (Brassica oleracea var. italica) is an important commercial vegetable crop. As part of an efficient pollination system, cytoplasmic male sterility (CMS) has been widely used for broccoli hybrid production. Identifying the original sources of CMS in broccoli accessions has become an important part of broccoli breeding. In this study, the diversity of the CMS sources of 39 broccoli accessions, including 19 CMS lines and 20 hybrids, were analyzed using mitochondrial markers. All CMS accessions contained the ogu orf138-related DNA fragment and the key genes of nap CMS, pol CMS, and tour CMS were not detected. The 39 CMS accessions were divided into five groups using six orf138-related and two simple sequence repeat markers. We observed that ogu CMS R3 constituted 79.49% of the CMS sources. CMS6 and CMS26 were differentiated from the other accessions using a specific primer. CMS32 was distinguished from the other accessions based on a 78-nucleotide deletion at the same locus as the orf138-related sequence. When the coefficient was about 0.90, five CMS accessions (13CMS6, 13CMS23, 13CMS24, 13CMS37, and 13CMS39) exhibiting abnormal floral organs with poor seed setting were grouped together. The polymerase chain reaction amplification profiles for these five accessions differed from those of the other accessions. We identified eight useful molecular markers that can be used to detect CMS types during broccoli breeding. Our data also provide important information relevant to future studies on the possible origins and molecular mechanisms of CMS in broccoli.
Introduction
Broccoli (Brassica oleracea var. italica) is an economically important vegetable crop that displays heterosis (Mao et al., 2012; Walley et al., 2012). The use of cytoplasmic male sterility (CMS) to produce hybrid seed can prevent self-pollination of female parents and lead to hybridization rates of up to 100%. The application of CMS provides an alternative to using self-incompatible lines during crop breeding (Wang et al., 2012). Currently, CMS is widely used for hybrid breeding and seed production of important vegetable crops, including onion (Havey, 2000), cabbage (Fang et al., 2001, 2004), radish (Lee et al., 2008), and cauliflower (Dey et al., 2011; Kaminski et al., 2012; Bhatia et al., 2015). Similarly, CMS lines play a crucial role in broccoli breeding and hybrid production (Mao et al., 2012; Shu et al., 2014; Jing et al., 2015). However, the morphologies of CMS floral organs usually exhibit complex variations according to genetic backgrounds and CMS types during the transfer process (Zhu et al., 1998), and the variations could considerably affect the efficiency of pollination and seed production (Shu J. S. et al., 2015). Brassica species are a rich source of different types of CMS, including ogu CMS (Ogura, 1968), pol CMS (Fu et al., 1995), nap CMS (Thompson, 1972), nig CMS (Pearson, 1981), and hau CMS (Wan et al., 2008), which are all widely used. Domestic and international breeding organizations currently cross CMS materials with each other to transfer CMS to broccoli. However, a lack of information regarding the types of CMS has made the development of broccoli lines with ideal CMS relatively difficult. The rapid identification of CMS sources and types would provide valuable information relevant to the introduction of CMS into broccoli and genetic improvement of this important crucifer.
Expression of specific mitochondrial genes is highly correlated with CMS, exactly which mitochondrial genes are important depends on species in which CMS source originated (Schnable and Wise, 1998; Hanson and Bentolila, 2004). However, most of the genes exhibit a maternal inheritance pattern in Brassica crops (Reboud and Zeyl, 1994). orf138, orf224-atp6, orf263, orf222, and orf288 are key genes of ogu (Bonhomme et al., 1992), pol (Singh and Brown, 1993), tour (Landgren et al., 1996), nap (L'Homme et al., 1997), and hau (Heng et al., 2014) CMS, respectively. Therefore, specific primers designed based on these mitochondrial genomic regions can be used to distinguish CMS types. Complete mitochondrial genomes of Arabidopsis thaliana, B. napus (nap and pol), B. rapa (cam), B. oleracea, B. juncea, and B. carinata have been determined (Unseld et al., 1997; Handa, 2003; Chang et al., 2011; Chen et al., 2011; Heng et al., 2014), with the increasing number of mitochondrial nucleotide sequences, many mitochondrial markers have been developed to identify CMS types in B. juncea (Ashutosh et al., 2005; Wan et al., 2008; Yu et al., 2014), B. napus (Wei et al., 2005; Wan et al., 2008), B. oleracea (Li et al., 2009; Zhang et al., 2010, 2011; Wang et al., 2012), B. oleracea var. italica (Jing et al., 2015), and B. rapa (Zhang et al., 2007; Shi et al., 2012; Heng et al., 2015). The markers have also been used to analyze genetic diversity in varieties of B. napus (Handa, 2007; Zhao et al., 2010) and B. rapa (Zhang et al., 2013), and to differentiate between somatic hybrids of B. juncea and B. oleracea (Lian et al., 2011). Thus, far, mitochondrial markers have not been successfully used to analyze the different types of CMS and genetic diversity of original sources of CMS in broccoli.
We completed this study to address the issues involved in selecting suitable CMS sources for broccoli breeding. Our objectives were as follows: (1) distinguish the cytoplasm types of 19 CMS lines and 20 hybrids using mitochondrial molecular markers, (2) reveal the diversity in the cytoplasm types of CMS in broccoli, and (3) confirm the polymorphic loci by cloning and sequencing.
Materials and Methods
Plant Materials and DNA Extraction
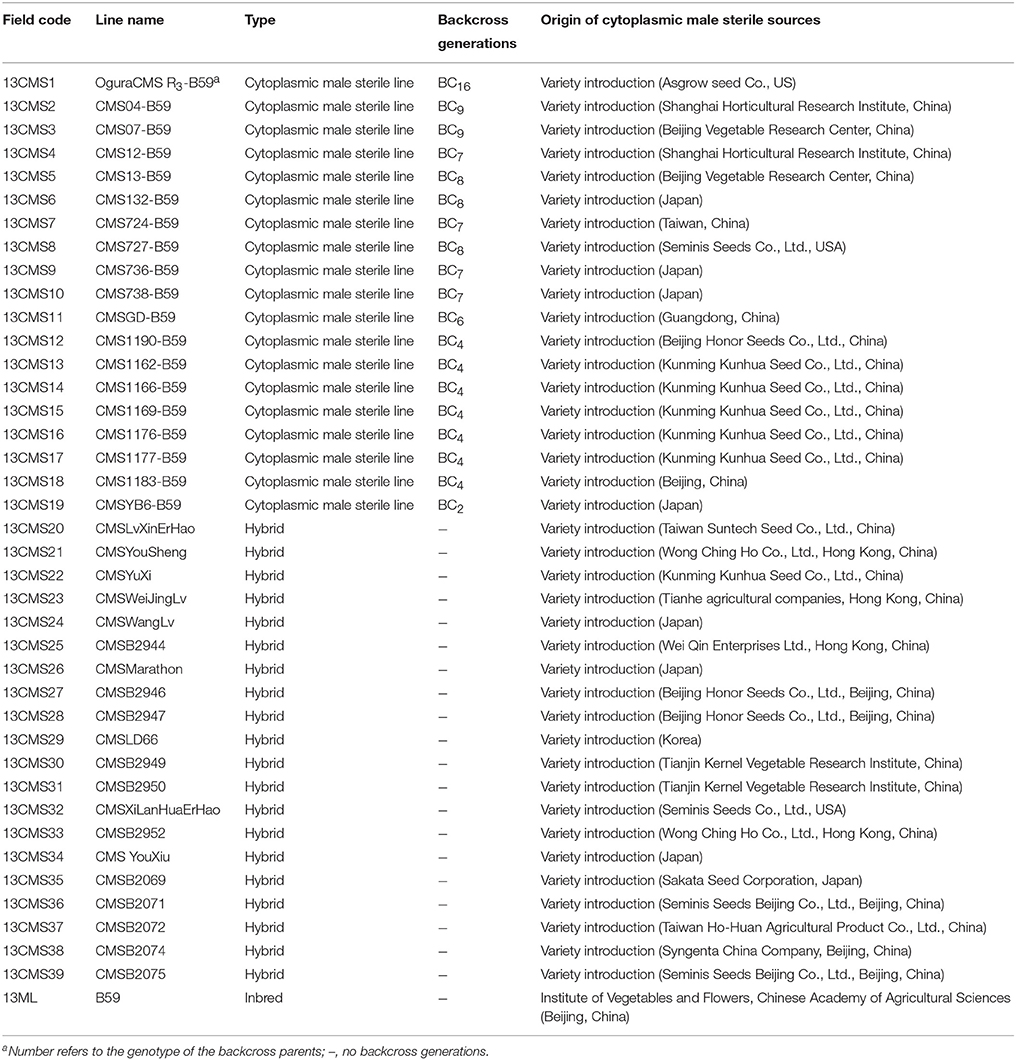
Details of the plant materials used in this study are provided in Table 1. We analyzed 40 broccoli CMS accessions (13CMS1–13CMS39, 13ML), including 19 CMS lines (13CMS1–13CMS19) and 20 hybrids (13CMS20–13CMS39) with different origins, and one inbred line (13ML) as a reference. The original CMS sources and hybrids were obtained from research institutes and seed companies from all over the world. 14CMS1-39 and 15CMS1-39 were obtained from 13CMS1-39 and 14CMS1-39 through backcrossing, respectively. The recurrent male parent was broccoli inbred line B59 (Supplementary Table 1). All accessions were grown in experimental greenhouses of the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China in 2013–2015. For each accession, total genomic DNA was isolated from eight to ten true leaves using a modified hexadecyltrimethylammonium bromide method (Murray and Thompson, 1980) and stored at −30°C.
Polymerase Chain Reaction and Sequence Analysis
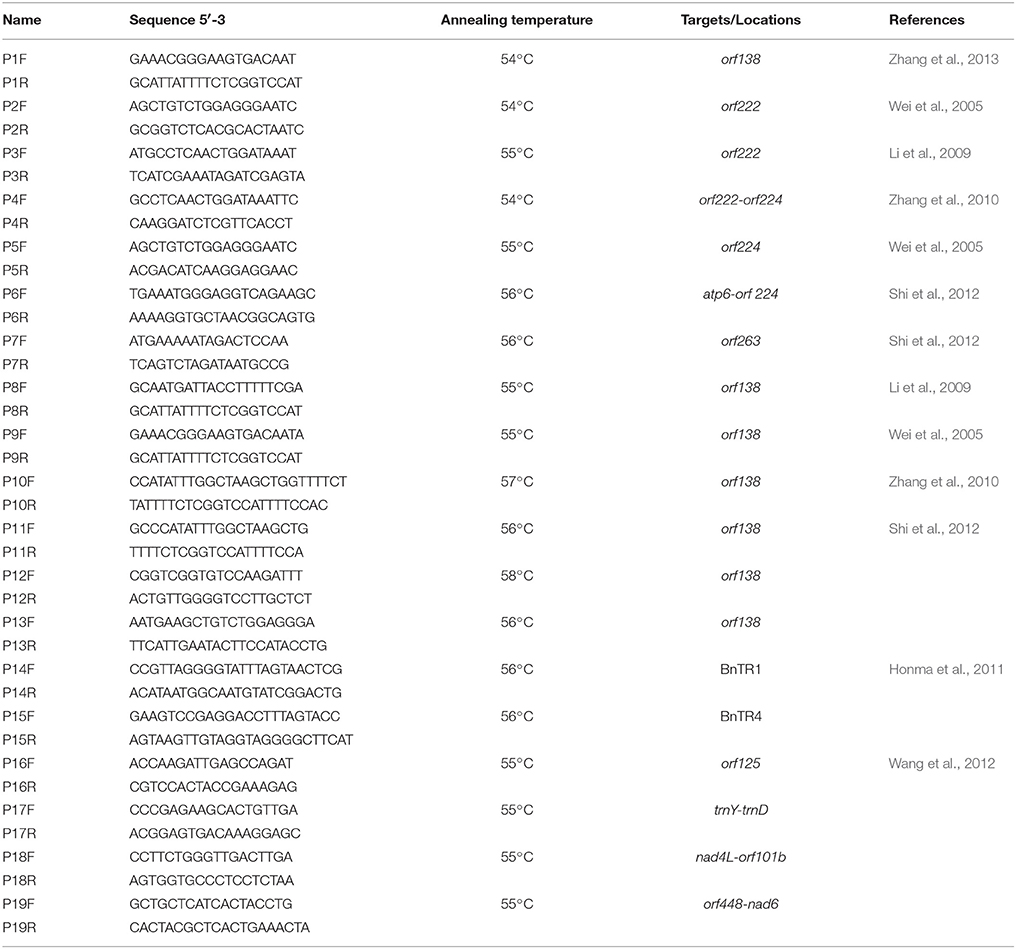
We used the following primers (Table 2): one pair of primers specific for orf138 (Zhang et al., 2013) and six pairs of primers designed based on orf 138-related genomic sequences (GenBank GI: 288879, Raphanus sativus mitochondrial orf138, orfB, and trnfM; P8–P13); one pair of primers specific for orf222–orf224 (Zhang et al., 2010), orf224 (Wei et al., 2005), atp6-orf 224 (Shi et al., 2012), or orf263 (Shi et al., 2012); two pairs of primers specific for orf222 (Wei et al., 2005; Li et al., 2009); and six pairs of mitochondrial simple sequence repeat (SSR) primers (Honma et al., 2011; Wang et al., 2012). All primers were synthesized by Bo Maide Biotech Co., Ltd. (Beijing, China).
Polymerase chain reaction (PCR) amplifications were performed in a final reaction volume of 20 μl, which consisted of 10 μl Dream Taq™ Green PCR Master Mix (Thermo Fisher Scientific Inc., Waltham, MA, USA), 1 μl primer (10 pmol/μl), and 2 μl (50 ng/μl) genomic DNA. The PCR program was as follows: 95°C for 5 min; 35 cycles of 95°C for 30 s, a suitable annealing temperature for 30 s, 72°C for 30–90 s; and a final 72°C for 10 min. The PCR was completed using an ABI Veriti 96-well PCR thermal cycler (Applied Biosystems, Foster City, CA, USA). The amplification products were analyzed on 3.0% (w/v) agarose gels in 0.5 × TBE buffer and visualized using the GoldView staining solution (Solarbio Technology Co., Ltd., Beijing, China). Stained gels were viewed and photographed under ultraviolet light using the Universal Hood II (Bio-Rad, Hercules, CA, USA).
Each polymorphic marker was tried to repeat amplification for three times, and for each polymorphic locus, the cloned products were separated and sequenced by Bo Maide Biotech Co., Ltd. Sequence alignment analyses were completed using the alignment tools of the Universal Protein Resource (http://www.uniprot.org/align/).
Cluster Analyses
We constructed a binary matrix by converting the polymorphic loci data into “1” or “0,” each DNA band generated was visually scored as an independent locus (1 for presence and 0 for absence). Qualitative differences in band intensities were not considered. Then completed the Unweighted Pair Group Method with Arithmetic Mean cluster analyses using NTSYSpc, version 2.11 software (Applied Biostatistics Inc., Exeter Software, Setauket, New York, USA). The genetic distances were calculated with the SM coefficient.
Results
Origins of CMS in Broccoli Accessions Determined Using Mitochondrial Molecular Markers
We screened 19 CMS lines (13CMS1–13CMS19; Table 1) and 20 CMS hybrids (13CMS20–13CMS39; Table 1) with thirteen pairs of mitochondrial DNA-specific primers (P1–P13; Table 2). Primer P1 exhibited polymorphism between CMS accessions and the inbred line (Figure 1). The fragment amplified by primer P1 was about 390 bp in 13CMS32 and 460 bp in 13CMS1–13CMS31 and 13CMS33–13CMS39. The results for primers P8–P11 were similar to those of primer P1. Primers P8–P11 did not generate an amplification product for 13ML, but did for the 39 CMS accessions. Additionally, the amplified product for 13CMS32 differed from those of the other 38 CMS accessions. Primer P12 amplified a 1200 bp product for 13CMS1–39, but did not amplify anything for 13ML (Figure 1). Primer P13 did not amplify any gene fragment for 13CMS6 and 13CMS26. However, an ~1000 bp fragment was amplified for the other accessions (Figure 1). Taken together, these results indicated the CMS of the 39 accessions was derived from ogu (Ogura) CMS, but there were differences in the nucleotide sequences among the accessions.

Figure 1. PCR amplification profiles for 40 broccoli accessions using primer P1, P12, P13, P15, P16. M, marker; 1–39, 13CMS1–13CMS39; 40, 13ML.
Origins of CMS in Broccoli Accessions Determined Using Mitochondrial SSR Molecular Markers
We used six pairs of mitochondrial SSR primers (P14–P19, Table 2) to amplify genomic DNA from the 40 broccoli accessions (13CMS1–13CMS39, 13ML; Table 1). The primers were developed based on the B. napus mitochondrial genome and effectively amplified Brassica fragments (Honma et al., 2011; Wang et al., 2012). All primers successfully amplified a gene fragment. However, only primers P15 and P16 exhibited clear polymorphisms (Figure 1). The amplicons produced by P15 for 13CMS6, 13CMS23, 13CMS24, 13CMS37, and 13CMS39 were about 220 bp, while those of the other broccoli accessions and 13ML were about 250 bp (Figure 1). The amplicons generated by P16 for 13CMS6, 13CMS23, 13CMS24, 13CMS37, and 13CMS39 were about 420 bp, while those of the other broccoli accessions and 13ML were ~470 bp (Figure 1). Our results indicated that the original CMS source for 13CMS6, 13CMS23, 13CMS24, 13CMS37, and 13CMS39 was the same, but differed from that of the other 34 CMS accessions.
Validation of the Polymorphic Markers
In 2014 and 2015, we used nine polymorphic primers (P1, P8–P13, P15, and P16; Table 2) to screen another 80 broccoli accessions (14CMS1–14CMS39, 14ML, 15CMS1–15CMS39, 15ML; Supplementary Table 1) to confirm the accuracy of the polymorphic markers. The results we obtained were the same as those from 2013, which demonstrated the stability of the molecular markers developed in this study.
Diversity of CMS origins in Broccoli Accessions
The 39 broccoli CMS accessions could be categorized into separate groups using ogu CMS mitochondrial DNA-specific markers or B. napus mitochondrial SSR markers. Combining the two types of analytical methods may be necessary to evaluate the diversity of CMS origins in broccoli accessions. A cluster analysis using all of the ogu CMS mitochondrial DNA-specific and B. napus mitochondrial SSR polymorphic markers revealed that the 39 CMS broccoli accessions could be divided into five groups (coefficient > 0.89) based on the origins of their CMS. We determined that CMS6, CMS26, and CMS32 each formed its own group, CMS23, CMS24, CMS37, and CMS39 belonged to one group, and the other 32 CMS accessions formed another group (coefficient > 0.89). When the coefficient was larger than 0.745, the 39 CMS accessions could be divided into three groups, CMS32 formed its own group, CMS6, CMS23, CMS24, CMS37, and CMS39 belonged to one group, and the other 33 CMS accessions formed another group (Figure 2).
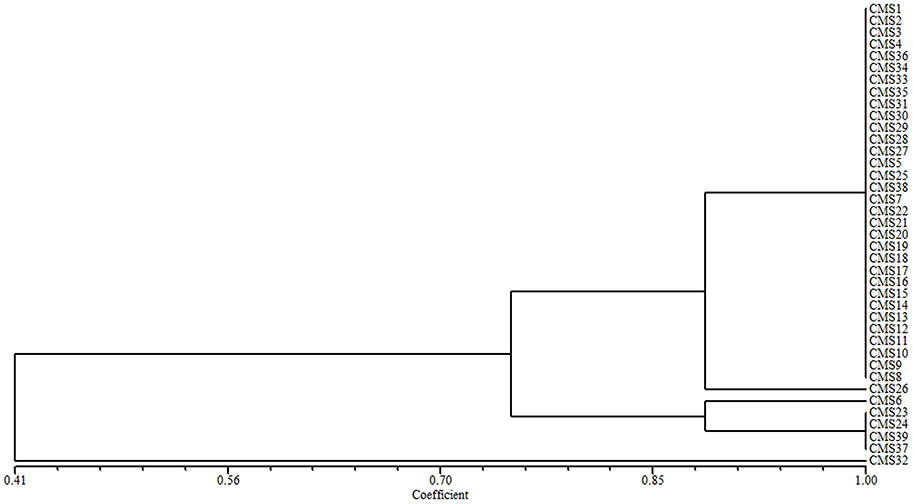
Figure 2. Cluster analysis of the CMS origins for 39 broccoli accessions based on ogu CMS mitochondrial DNA-specific markers and B. napus mitochondrial SSR markers.
Sequence Features of the Polymorphic Amplicons
Polymorphic amplicons obtained using seven polymorphic markers (P1, P8–P11, P15, and P16; Table 2) were sequenced and aligned. The amplicons generated by primer P1 for CMS32 were 387 bp, whereas those of the other CMS accessions were 465 bp. There was an 83.01% sequence similarity, with a single nucleotide polymorphism at position 28 (C/T). We also observed a 78-nucleotide deletion between positions 361 and 438 of 13CMS32 (Figure 3). The amplicons produced by primers P8–P11 also had a 78-nucleotide deletion at the same position as the orf138-related sequence in 13CMS32.
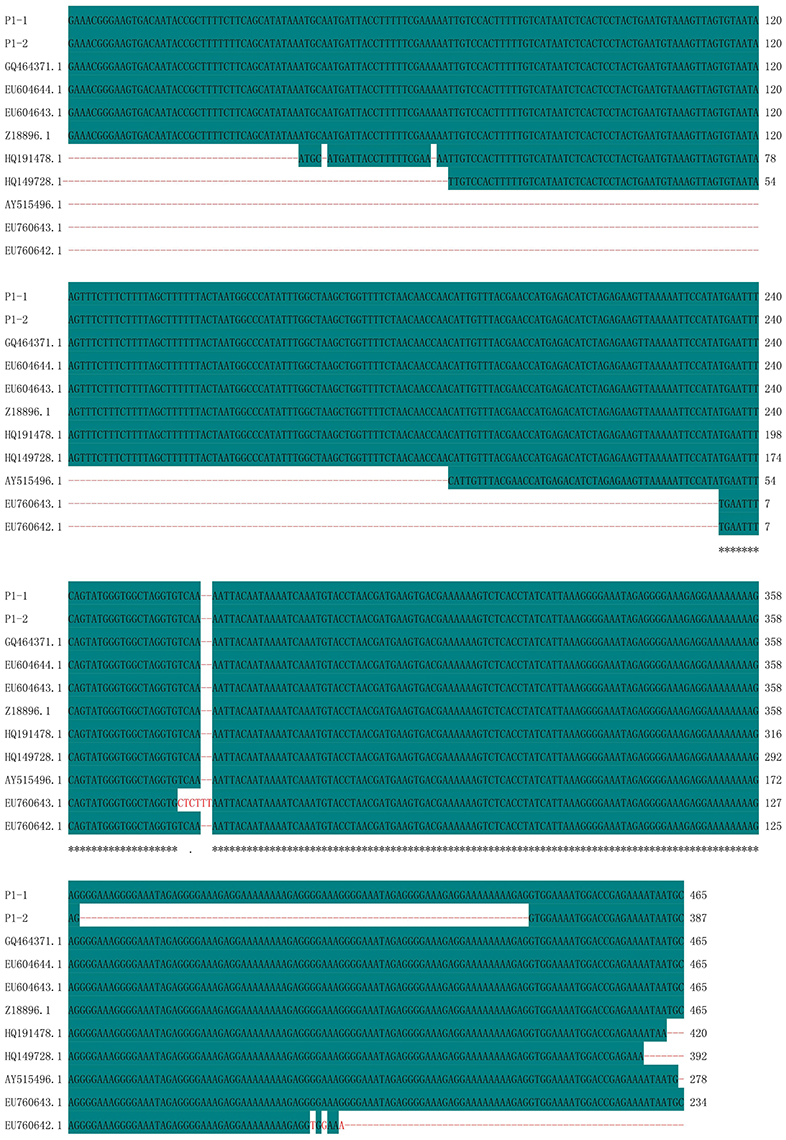
Figure 3. Sequence alignment of amplicons produced by primer P1. P1-1, 13CMS1–13CMS31 and 13CMS33–13CMS39; P1-2, 13CMS32. GQ464371.1 (B. oleracea), EU604644.1 (B. oleracea var. italica), EU604643.1 (B. oleracea var. italica), Z18896.1 (R. sativus), HQ191478.1 (B. oleracea var. acephala), HQ149728.1 (B. oleracea var. italica), AY515496.1 (B. oleracea var. botrytis), EU760643.1 (B. oleracea var. capitata), and EU760642.1 (B. oleracea var. capitata) were the related sequences in the GenBank. Asterisks means identical base.
The amplicons generated by primer P15 were 220 bp and identical for 13CMS6, 13CMS23, 13CMS24, 13CMS37, and 13CMS39. However, they were 250 bp for the other CMS accessions. We observed an 86.85% sequence similarity, with one nucleotide deleted at position 32 and a single nucleotide polymorphism at position 73 (T/C). Additionally, there was a 31-nucleotide deletion between positions 96 and 126 for 13CMS6, 13CMS23, 13CMS24, 13CMS37, and 13CMS39 (Figure 4).

Figure 4. Sequence alignment of amplicons produced by primer P15. P15-1, 13CMS1–13CMS5, 13CMS7–13CMS22, 13CMS25–13CMS36, and 13CMS38; P15-2, 13CMS6, 13CMS23, 13CMS24, 13CMS37, and 13CMS39. AP012988.1 (B. oleracea), KJ820683.1 (B. oleracea var. botrytis), JF920286.1 (B. oleracea), and AB627043.1 (B. oleracea) were the related sequences in the GenBank. Asterisks means identical base.
The amplicons produced by primer P16 were 420 bp and identical in 13CMS6, 13CMS23, 13CMS24, 13CMS37, and 13CMS39. They were 471 bp and identical in all other CMS accessions. The sequence similarity was 88.96%, with a single nucleotide polymorphism at position 100 (C/T). Additionally, there was a 51-nucleotide deletion between positions 309 and 359 for 13CMS6, 13CMS23, 13CMS24, 13CMS37, and 13CMS39 (Figure 5).

Figure 5. Sequence alignment of amplicons produced by primer P16. P16-1, 13CMS1–13CMS5, 13CMS7–13CMS22, 13CMS25–13CMS36, and 13CMS38; P16-2, 13CMS6, 13CMS23, 13CMS24, 13CMS37, and 13CMS39. AP012988.1 (B. oleracea), KJ820683.1 (B. oleracea var. botrytis) and JF920286.1 (B. oleracea) were the related sequences in the GenBank. Asterisks means identical base.
Similarity analysis of the sequences revealed that the amplicons of primer P1 have the highest similarity with ogura cytoplasmic male sterility-related protein gene in B. oleracea, B. oleracea var. italic, and R. sativus (Figure 3, Supplementary Table 2); the amplicons of primer P15 and P16 have high similarity with mitochondrial DNA in B. oleracea and B. oleracea var. botrytis (Figures 4, 5, Supplementary Tables 3, 4).
Discussion
CMS, which is usually caused by mutations in the mitochondrial genome (Jing et al., 2015), is prevalent in higher plants and can be used to increase heterosis and improve genetic resources. Mitochondrial markers can be used to quickly and accurately differentiate between the various types of CMS sources (Zhao et al., 2010; Wang et al., 2012; Zhang et al., 2013). Protoplast fusion (Yarrow et al., 1990) and distant hybridization (Jing et al., 2015) have been the main approaches used to obtain CMS materials in broccoli. However, to the best of our knowledge, there are few reports describing the application of mitochondrial markers to identify the types of CMS resources. Furthermore, the genetic diversity of the sources of CMS in broccoli has not been determined. Previous reports were all based on the development of markers to differentiate between ogu CMS materials and fertile broccoli accessions (Zhu et al., 2006; Yao et al., 2009; Jing et al., 2015).
In this study, we used primers specific for orf138, orf222, orf222-orf224, orf224, orf224-atp6, orf263, and mitochondrial SSR markers to systematically determine the CMS origins of 39 broccoli accessions, which were obtained from various countries and regions. All 39 CMS accessions contained the CMS-related orf138 fragment, and orf222, orf222-orf224, orf224, orf224-atp6, and orf263 fragments could not be detected, suggesting these accessions expressed ogu CMS. This is consistent with previous reports that ogu CMS occurs in Brassica and Raphanus crops (Makaroff et al., 1989; Zhang et al., 2006; Chen et al., 2009). Our results are also consistent with those of previous studies on CMS sources in broccoli (Yao et al., 2009; Jing et al., 2015) and indicate that the CMS origin is single in broccoli and more types of CMS from related species should be introduced into broccoli in future breeding programs.
CMS1 consisted of ogu CMS R3, which originated from a somatic hybridization and was transferred from a cabbage CMS line. The type of CMS in CMS2–CMS39 could not be confirmed. We developed eight polymorphic cytoplasmic markers (P1, P8–P11, P13, P15, and P16; Table 2) in 39 broccoli CMS accessions (CMS1–CMS39) which could distinguish their cytoplasm types. Using the eight cytoplasmic markers, we divided these accessions into five groups (coefficient > 0.89) or three groups (coefficient > 0.745) according to cluster analysis data. When the coefficient was larger than 0.745, CMS32 formed its own group, CMS6, CMS23, CMS24, CMS37, and CMS39 belonged to one group, and the other 33 CMS accessions formed another group. The results were difference when the coefficient was larger than 0.89, CMS6, CMS26, and CMS32 each formed its own group, CMS23, CMS24, CMS37, and CMS39 belonged to one group, and the other 32 CMS accessions formed another group. These results suggesting lines CMS2–CMS5, CMS7–CMS22, CMS25, CMS27–CMS31, CMS33–CMS36, and CMS38 were included in the same group as CMS1 with high coefficient, and ogu CMS R3was likely the original CMS. These lines constituted 79.49% of the total CMS resources used in our study, demonstrating that ogu CMS R3 is prevalent in broccoli. It is likely that the original CMS source developed from a different protoplast fusion event and the resulting CMS was subsequently transferred to broccoli. When the coefficient is larger than 0.89, CMS6, CMS23, CMS24, CMS26, CMS37, CMS32, and CMS39 didn't belong to the same group with CMS1, implying they weren't ogu CMS R3type.
In this study, the amplicons of primer P16 were 420 bp and identical in lines CMS6, CMS23, CMS24, CMS37, and CMS39, they were 471 bp and identical in other 34 CMS accessions, the length of the polymorphic products were same as reported in cabbage, which can distinguish ogu CMS HY and ogu CMS R1−2 (420 bp) from pol CMS and ogu CMS R3 (471 bp; Wang et al., 2012). So our results suggest that the original of CMS6, CMS23, CMS24, CMS37, and CMS39 could come from ogu CMS HY or ogu CMS R1−2. Sequences analysis showed that the deleted sequences of these broccoli CMS accessions were not at the same location as in cabbage. Unfortunately, as far as we know, the genetic diversity of original sources of CMS in broccoli has not been documented and we have no references. In our study, the polymorphic cytoplasmic markers were less and maybe some accessions could not be distinguished, further experiments with more polymorphic markers and extensive research materials will deepen our understanding of the diversity in the cytoplasm types of CMS in broccoli.
When the coefficient (Figure 2) was about 0.90, CMS6, CMS23, CMS24, CMS37, and CMS39 formed one group. The flowers of the original broccoli lines of CMS6, CMS37, and CMS39 had carpelloid stamens and exhibited poor seed setting (Shu J. et al., 2015), which suggests that CMS23 and CMS24 also contain carpelloid stamens. Therefore, the CMS source for CMS6, CMS23, CMS24, CMS37, and CMS39 should not be transferred into broccoli lines. Lines CMS26 and CMS32 each formed its own group, indicating their ogu CMS was different from those of the other 37 CMS accessions. Our findings suggest that some of the original sources of ogu CMS were better than others. Alternatively, existing fragments were lost during CMS transfer, resulting in differences among the observed ogu CMS.
In conclusion, we have identified eight useful mitochondrial molecular markers to detect ogu CMS in different broccoli CMS accessions. The 39 CMS accessions could be divided into five groups (coefficient > 0.89) or three groups (coefficient > 0.745), ogu CMS R3 is prevalent in broccoli, ogu CMS HY or ogu CMS R1−2 is likely exist in broccoli. Our findings may help to improve the efficiency of CMS selection during broccoli breeding and also form a solid basis for further studies into the molecular mechanisms of ogu CMS in broccoli.
Author Contributions
YL and JS conceived and designed the study. JS and LZ performed the experiments. JS, ZL, LZ, and HL analyzed the data. JS prepared the manuscript, YL improved the manuscript. YL, ZF, LY, MZ, and YZ contributed reagents and materials. YL provided guidance on the whole study. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by China Agriculture Research System (Grant no. CARS-25-A), National Natural Science Foundation of China (Grant No. 31372067), the Key Projects in the National Science and Technology Pillar Program of China (Grant No. 2013BAD01B04), the National High Technology Research and Development Program (863 Program) of China (Grant No. 2012AA100105), the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P. R. China, and the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (Grant No. CAAS-ASTIP-IVFCAAS). The work was done in the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P. R. China.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00927
References
Ashutosh, D. K. K., Kumar, V. D., Rakash, S., and Bhat, S. R. (2005). rep-PCR helps to distinguish different alloplasmic cytoplasmic male sterile lines of Brassica juncea. Plant Sci. 168, 1083–1087. doi: 10.1016/j.plantsci.2004.12.005
Bhatia, R., Dey, S. S., Sharma, K., Parkash, C., and Kumar, R. (2015). In vitro maintenance of CMS lines of Indian cauliflower: an alternative for conventional CMS-based hybrid seed production. J. Hortic. Sci. Biotech. 90, 171–179. doi: 10.1080/14620316.2015.11513169
Bonhomme, S., Burdar, F., Lancelin, D., Samll, I., Defrance, M. C., and Pelletier, G. (1992). Sequence and analysis of Nco 2.5 Ogura-specific fragment correlated with male sterility in Brassica cybrids. Mol. Gen. Genet. 235, 240–248. doi: 10.1007/BF00279379
Chang, S., Yang, T., Du, T., Huang, Y., Chen, J., Yan, J., et al. (2011). Mitochondrial genome sequencing helps show the evolutionary mechanism of mitochondrial genome formation in Brassica. BMC Genomics 12:497. doi: 10.1186/1471-2164-12-497
Chen, J., Guan, R., Chang, S., Du, T., and Zhang, H. (2011). Substoichiometrically different mitotypes coexist in mitochondrial genomes of Brassica napus L. PLoS ONE 6:e17662. doi: 10.1371/journal.pone.0017662
Chen, L., Liu, L., Jin, P., Gong, Y., Sun, X., and Ma, E. (2009). Cytological and molecular identification of cytoplasm in two male sterile lines in radish. Mol. Plant Breed. 7, 757–762. doi: 10.3969/mpb.007.000757
Dey, S. S., Sharma, S. R., Bhatia, R., Kumar, P. R., and Parkash, C. (2011). Development and characterization of “Ogura” based improved CMS lines of cauliflower (Brassica oleracea var. botrytis L). Indian J. Genet. Plant Breed. 71, 37–42.
Fang, Z. Y., Liu, Y. M., Yang, L. M., Wang, X. W., Zhuang, M., Zhang, Y. Y., et al. (2004). Breeding and seed production technique of dominant genic male sterile line and cytoplasmic male sterile line in cabbage. Sci. Agric. Sin. 37, 717–723.
Fang, Z. Y., Sun, P. T., Liu, Y. M., Yang, L. M., Wang, X. W., and Zhuang, M. (2001). Investigation of different types of male sterility and application of dominant male sterility in cabbage. China Veg. 1, 6–10.
Fu, T. D., Yang, G. S., Yang, X. N., and Ma, C. Z. (1995). The discovery, research and utilization of pol cytoplasmic male sterile in Brassica napus. Prog. Nat. Sci. Commun. State Key Lab. 5, 287–293.
Handa, H. (2003). The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic. Acids Res. 20, 5907–5916. doi: 10.1093/nar/gkg795
Handa, H. (2007). Investigation of the origin and transmission of linear mitochondrial plasmid based on phylogenetic analysis in Japanese rapeseed varieties. Genome 50, 234–240. doi: 10.1139/G06-150
Hanson, M. R., and Bentolila, S. (2004). Interactions of mitoehondrial and nuclear genes that affect male gametophyte development. Plant Cell 16, 154–169. doi: 10.1105/tpc.015966
Havey, M. J. (2000). Diversity among male-sterility-inducing and male-fertile cytoplasms of onion. Theor. Appl. Genet. 101, 778–782. doi: 10.1007/s001220051543
Heng, S. P., Shi, D. Y., Hu, Z. H., Huang, T., Li, J. P., Liu, L. Y., et al. (2015). Characterization and classification of one new cytoplasmic male sterility (CMS) line based on morphological, cytological and molecular markers in non-heading Chinese cabbage (Brassica rapa L). Plant Cell Rep. 34, 1529–1537. doi: 10.1007/s00299-015-1804-y
Heng, S. P., Wei, C., Jing, B., Wan, Z. J., Wen, J., Yi, B., et al. (2014). Comparative analysis of mitochondrial genomes between the hau cytoplasmic male sterility (CMS) line and its iso-nuclear maintainer line in Brassica juncea to reveal the origin of the CMS-associated gene orf288. BMC Genomics 15:322. doi: 10.1186/1471-2164-15-322
Honma, Y., Yoshida, Y., Terachi, T., Toriyama, K., Mikami, T., and Kubo, T. (2011). Polymorphic minisatellites in the mitochondrial DNAs of Oryza and Brassica. Curr. Genet. 57, 261–270. doi: 10.1007/s00294-011-0345-3
Jing, Z. G., Pei, X. L., Tang, Z., Liu, Q., Zhang, X. L., Luo, T. K., et al. (2015). Molecular identification of Ogu cytoplasmic male sterile and sequence analysis in broccoli (Brassica oleracea var. italica). Guihaia 35, 239–243. doi: 10.11931/guihaia.gxzw201309005
Kaminski, P., Dyki, B., and Stepowska, A. A. (2012). Improvement of cauliflower male sterile lines with Brassica nigra cytoplasm, phenotypic expression and possibility of practical application. J. Agric. Sci. 4, 190–200. doi: 10.5539/jas.v4n4p190
L'Homme, Y., Stahl, R. J., Li, X. Q., Hameed, A., and Brown, G. G. (1997). Brassica nap cytoplasmic male sterility is associated with expression of a mtDNA region containing a chimeric gene similar to the pol CMS-associated orf224 gene. Curr. Genet. 31, 325–335.
Landgren, M., Zetterstrand, M., Sundberg, E., and Glimelius, K. (1996). Alloplasmic male-sterile Brassica lines containing B. tournefortii mitochondria express an ORF 3′ of the atp6 gene and a 32 kDa protein. Plant Mol. Biol. 32, 879–890. doi: 10.1007/BF00020485
Lee, Y. P., Park, S., Lim, C., Kim, H., Lim, H., Ahn, Y. S., et al. (2008). Discovery of a novel cytoplasmic male-sterility and its restorer lines in radish (Raphanus sativus L). Theor. Appl. Genet. 117, 905–913. doi: 10.1007/s00122-008-0830-3
Li, J. B., Zhang, H. J., Yu, X. L., Wang, S. Y., and Cao, J. S. (2009). Molecular identification of the Cytoplasmic Male Sterile (CMS) type in common head cabbage. Mol. Plant Breed. 7, 1149–1153. doi: 10.3969/mpb.007.001149
Lian, Y. J., Lin, G. Z., Zhao, X. M., and Lim, H. T. (2011). Production and genetic characterization of somatic hybrids between leaf mustard (Brassica juncea) and broccoli (Brassica oleracea). In Vitro Cell. Dev. Biol. Plant 47, 289–296. doi: 10.1007/s11627-011-9355-6
Makaroff, C. A., Aepl, I. J., and Palmer, J. D. (1989). The atp6 coding region has been disrupted and a novel reading frame generated in the mitochondria genome of cytoplasm male-sterile radish. J. Biol. Chem. 264, 11706–11713.
Mao, Z. L., Pan, Y. F., Jin, Y. Q., Dai, Z. L., Wang, J. H., Zhang, Z. C., et al. (2012). Selection and utilization of cytoplasmic male sterile line of broccoli. Jiangsu J. Agric. Sci. 28, 1511–1512.
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic. Acid Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
Ogura, H. (1968). Studies on the new male sterility in Japanese radish with special reference to the Utilization of this sterility towards the practical raising of hybrid seeds. Mem. Fac. Agric. Kagoshima Univ. 6, 39–78.
Pearson, O. H. (1981). Nature and mechanisms of cytoplasmic male sterility in plants: a review. Hortscience 16, 482–487.
Reboud, X., and Zeyl, C. (1994). Organelle inheritance in plants. Heredity 72, 132–140. doi: 10.1038/hdy.1994.19
Schnable, P. S., and Wise, R. P. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3, 175–180. doi: 10.1016/S1360-1385(98)01235-7
Shi, Z., Wan, Z. J., Xu, Y. J., Li, X. H., and Zou, R. C. (2012). Characterization of the Chinese cabbage hau cytoplasmic male-sterile line and sequence analysis of the fertility-related gene. Acta Hortic. Sin. 39, 469–476.
Shu, J., Liu, Y., Fang, Z., Yang, L., Zhang, L., Zhuang, M., et al. (2014). Study on the floral characteristics and structure in two types of male sterile lines of broccoli (Brassica oleracea var. italica). J. Plant Genet. Resour. 15, 113–119. doi: 10.13430/j.cnki.jpgr.2014.01.015
Shu, J., Liu, Y., Li, Z., Zhang, L., Fang, Z., Yang, L., et al. (2015). Organelle simple sequence repeat markers help to distinguish carpelloid stamen and normal cytoplasmic male sterile sources in broccoli. PLoS ONE 10:e0138750. doi: 10.1371/journal.pone.0138750
Shu, J. S., Liu, Y. M., Li, Z. S., Zhang, L. L., Fang, Z. Y., Yang, L. M., et al. (2015). Effect of different pruning methods on flowering and fruiting characteristics between different types of male sterile lines in broccoli seed plants. Acta Hortic. Sin. 42, 689–696. doi: 10.16420/j.issn.0513-353x.2014-1107
Singh, M., and Brown, G. G. (1993). Characterization of expression of a mitochondrial gene region associated with the Brassica “Polima” CMS: developmental influences. Curr. Genet. 24, 316–322. doi: 10.1007/BF00336783
Thompson, K. F. (1972). Cytoplasmic male sterility in oil-seed rape. Heredity 29, 253–257. doi: 10.1038/hdy.1972.89
Unseld, M., Marienfeld, J. R., Brandt, P., and Brennicke, A. (1997). The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366, 924 nucleotides. Nat. Genet. 15, 57–61. doi: 10.1038/ng0197-57
Walley, P. G., Carder, J., Skipper, E., Mathas, E., Lynn, J., Pink, D., et al. (2012). A new broccoli × broccoli immortal mapping population and framework genetic map: tools for breeders and complex trait analysis. Theor. Appl. Genet. 124, 467–484. doi: 10.1007/s00122-011-1721-6
Wan, Z. J., Jing, B., Tu, J. X., Ma, C. Z., Shen, J. X., Yi, B., et al. (2008). Genetic characterization of a new cytoplasmic male sterility system (hau) in Brassica juncea and its transfer to B. napu. Theor. Appl. Genet. 116, 355–362. doi: 10.1007/s00122-007-0673-3
Wang, Q. B., Zhang, Y. Y., Fang, Z. Y., Liu, Y. M., Yang, L. M., and Zhuang, M. (2012). Chloroplast and mitochondrial SSR help to distinguish allo-cytoplasmic male sterile types in cabbage (Brassica oleracea L. var. capitata). Mol. Breed. 30, 709–716. doi: 10.1007/s11032-011-9656-9
Wei, W. L., Wang, H. Z., and Liu, G. H. (2005). Molecular Identification of the sterile cytoplasm of NCa of a cytoplasmic male sterile line in rapeseed (Brassica napus L). Sci. Agric. Sin. 38, 1965–1972.
Yao, X. Q., Li, Y., Xie, Z. J., and Liu, L. W. (2009). Identification of specific SRAP marker associated with cytoplasmic male sterility gene of broccoli. Mol. Plant Breed. 7, 941–947. doi: 10.3969/mpb.007.000941
Yarrow, S. A., Burnett, L. A., Wildeman, R. P., and Kemble, R. J. (1990). The transfer of ‘Polima’ cytoplasmic male sterility from oilseed rape (Brassica napus) to broccoli (B. oleracea) by protoplast fusion. Plant Cell Rep. 9, 185–188.
Yu, X. L., Liu, Y. P., Lv, Y. X., Liu, Z. N., Chen, Z. J., Lu, G., et al. (2014). Development of molecular markers specific to petaloid-type cytoplasmic male sterility in tuber mustard (Brassica juncea var. tumida Tsen et Lee). Mol. Biol. Rep. 41, 769–778. doi: 10.1007/s11033-013-2916-5
Zhang, D. S., Zhang, F. L., Wang, Y. J., Fang, Z. Y., Yu, Y. J., Zhao, X. Y., et al. (2006). Molecular ientification of ctoplasmic male sterile line CMS96 in Chinese cabbage. Mol. Plant Breed. 4, 545–552.
Zhang, D. S., Zhang, F. L., Wang, Y. J., Yu, Y. J., Zhao, X. Y., Xu, J. B., et al. (2007). Molecular identification and sequence analysis of Cytoplasmic MaleSterile types in Chinese cabbage. Acta Agric. Boreali-Sin. 22, 53–59.
Zhang, R. J., Hu, S. W., Yan, J. Q., and Sun, G. L. (2013). Cytoplasmic diversity in Brassica rapa L. investigated by mitochondrial markers. Genet. Resour. Crop Evol. 60, 967–974. doi: 10.1007/s10722-012-9892-9
Zhang, Y., Wang, X. J., Li, C. Q., Song, H. Y., Ren, X. S., and Si, J. (2010). Molecular identification of Brassica oleracea CMS and the morphology response of flower to nuclear background. Acta Hortic. Sin. 37, 915–922.
Zhang, Y. Y., Fang, Z. Y., Wang, Q. B., Liu, Y. M., Yang, L. M., Zhuang, M., et al. (2011). Molecular distinction of two Ogura CMS sources in Brassica oleracea var. capitata L. Sci. Agric. Sin. 44, 2959–2965. doi: 10.3864/j.issn.0578-1752.2011.14.013
Zhao, H. X., Li, Z. J., Hu, S. W., Sun, G. L., Chang, J. J., and Zhang, Z. H. (2010). Identification of cytoplasm types in rapeseed (Brassica napus L) accessions by a multiplex PCR assay. Theor. Appl. Genet. 121, 643–650. doi: 10.1007/s00122-010-1336-3
Zhu, Y. Y., Yang, X. F., Hou, R. X., Li, S. J., and Wu, Q. S. (2006). Extraction and RAPD analysis of mitochondrial DNA of cytoplasmic male sterile (CMS) line in broccoli. J. Wuhan Bot. Res. 24, 505–508.
Keywords: broccoli, cytoplasmic male sterility, origin, mitochondrial markers, diversity
Citation: Shu J, Liu Y, Li Z, Zhang L, Fang Z, Yang L, Zhuang M, Zhang Y and Lv H (2016) Detection of the Diversity of Cytoplasmic Male Sterility Sources in Broccoli (Brassica Oleracea var. Italica) Using Mitochondrial Markers. Front. Plant Sci. 7:927. doi: 10.3389/fpls.2016.00927
Received: 12 November 2015; Accepted: 10 June 2016;
Published: 24 June 2016.
Edited by:
Diego Rubiales, Consejo Superior de Investigaciones Cientificas, SpainReviewed by:
Anil Khar, Indian Agricultural Research Institute, IndiaAndrea Coppi, University of Florence, Italy
Copyright © 2016 Shu, Liu, Li, Zhang, Fang, Yang, Zhuang, Zhang and Lv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yumei Liu, bGl1eXVtZWlAY2Fhcy5jbg==
 Jinshuai Shu
Jinshuai Shu Yumei Liu
Yumei Liu Zhansheng Li
Zhansheng Li Lili Zhang
Lili Zhang Honghao Lv
Honghao Lv