
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 21 June 2016
Sec. Plant Pathogen Interactions
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.00906
This article is part of the Research Topic Biotrophic plant-microbe interactions View all 34 articles
Extracellular recognition of pathogens by plants constitutes an important early detection system in plant immunity. Microbe-derived molecules, also named patterns, can be recognized by pattern recognition receptors (PRRs) on the host cell membrane that trigger plant immune responses. Most knowledge on extracellular pathogen detection by plants comes from research on bacterial and fungal pathogens. For oomycetes, that comprise some of the most destructive plant pathogens, mechanisms of extracellular pattern recognition have only emerged recently. These include newly recognized patterns, e.g., cellulose-binding elicitor lectin, necrosis and ethylene-inducing peptide 1-like proteins (NLPs), and glycoside hydrolase 12, as well as their receptors, e.g., the putative elicitin PRR elicitin response and the NLP PRR receptor-like protein 23. Immunity can also be triggered by the release of endogenous host-derived patterns, as a result of oomycete enzymes or damage. In this review we will describe the types of patterns, both pathogen-derived exogenous and plant-derived endogenous ones, and what is known about their extracellular detection during (hemi-)biotrophic oomycete infection of plants.
Most plant pathogens are able to penetrate host tissues but essentially grow in the plant apoplast or extracellular space. Even haustoria, feeding structures formed by many biotrophic fungi and oomycetes that invaginate host cells, remain separated from the plant cell cytoplasm by the plant-derived extrahaustorial membrane (Parniske, 2000). It, therefore, comes as no surprise that a first line of pathogen recognition is extracellular and mediated by membrane-bound receptors that detect microbe- or host damage-derived molecules or patterns. Over the last decades, many pattern-recognition receptors mediating immunity to patterns of bacteria and fungi have been reported. Well known examples include the Arabidopsis receptor-like kinase (RLK) FLAGELLIN-SENSITIVE 2 (FLS2) that mediates recognition of bacterial flagellin, and the RLK, CHITIN ELICITOR RECEPTOR KINASE 1 (CERK1) involved in detection of fungal chitin (Zipfel, 2014). Flagellin and chitin are considered microbe-associated molecular patterns (MAMPs), while their cognate receptors are termed pattern-recognition receptors (PRRs; Jones and Dangl, 2006; Hein et al., 2009; Dodds and Rathjen, 2010). MAMPs are generally considered conserved molecules that occur in all species of a given taxon. There are, however, many examples of patterns that are species-specific or that are less well conserved, e.g., apoplastic effectors that are recognized by cognate resistance gene-encoded membrane-bound receptors (Thomma et al., 2011). In this review we, therefore, refer to all extracellular molecules that trigger immunity as patterns (Cook et al., 2015). In older papers the term “elicitor” is most often used, but many of these can be regarded as patterns too (Boller and Felix, 2009; Cook et al., 2015). Although numerous oomycete patterns have been described, knowledge on the mechanism of their extracellular recognition has only emerged recently for some of them.
Oomycetes are filamentous organisms that belong to the Stramenopiles, a taxon that also encompasses the diatoms and brown algae. Many oomycetes are free-living saprobes in soils or aquatic environments. The best known oomycetes, or the most infamous ones, are species that are pathogenic on plants, e.g., the potato late blight pathogen Phytophthora infestans and the grape downy mildew Plasmopara viticola (Haas et al., 2009; Kamoun et al., 2015). Five main taxa of phytopathogenic oomycetes can be distinguished: (i) the genus Phytophthora, (ii) the downy mildews, (iii) the white blister rusts, (iv) the genus Pythium, and (v) the genus Aphanomyces (Thines and Kamoun, 2010).
In this review, we focus on the extracellular recognition of (hemi-)biotrophic oomycetes, on patterns that trigger immunity, and on mechanisms of pattern recognition. A broad range of molecules or patterns are released during oomycete infection of plants, either exogenous ones derived from the pathogen, or endogenous ones that are released from the plant host (Figure 1). The distinction between exogenous and endogenous signals can also be referred to as non-self and modified-self patterns (Schwessinger and Zipfel, 2008). Endogenous patterns, also known as damage-associated molecular patterns (DAMPs), either result from oomycete enzyme activities, or from lysis or disruption of host cells during the infection process. Oomycete patterns and other elicitors can be grouped based on their cellular origin (oomycete cell wall/membrane, or pathogen secreted). We will review the different patterns, their cellular origin, and what is known about the detection mechanisms that have evolved to recognize such patterns, and trigger the plant immune system.
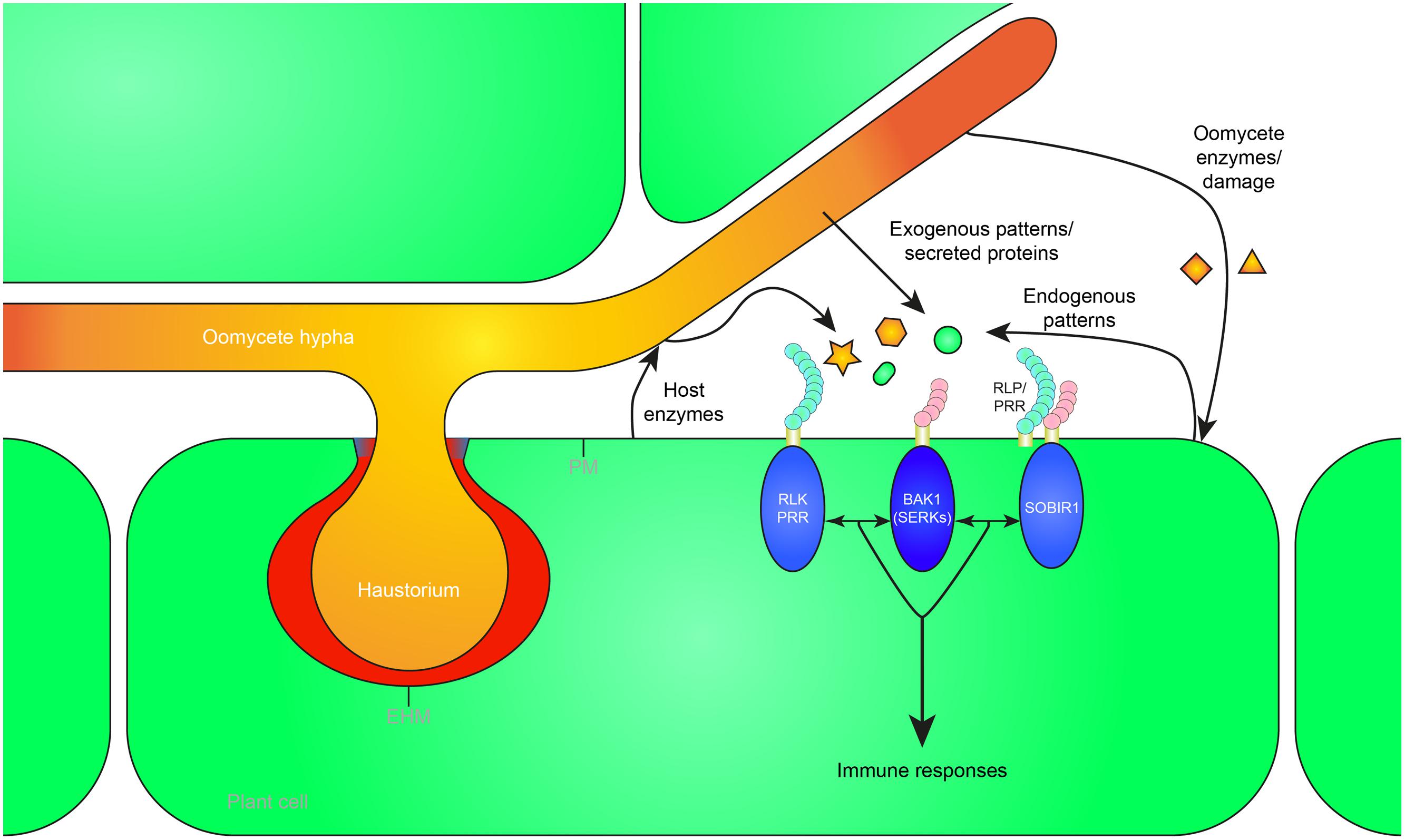
FIGURE 1. Recognition of exogenous and endogenous patterns during oomycete infection leads to the activation of plant immunity. Oomycete pathogens secrete proteins in the apoplast (white) and extrahaustorial matrix (red) which can be perceived as exogenous patterns by pattern recognition receptors (PRRs) in the plant plasma membrane (PM) or extrahaustorial membrane (EHM). Furthermore, pathogen-derived cell wall or membrane fragments are released during infection, possibly by host enzymes, and recognized as patterns by the host. Mechanical damage or damage caused by oomycete secreted enzymes can release endogenous patterns that trigger immunity. The receptor-like kinase (RLK) BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1), a member of the SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) family, functions as a central hub of RLK and receptor-like protein (RLP) triggered immunity. RLPs form a bimolecular receptor kinase with the RLK SUPPRESSOR OF BIR1 1 (SOBIR1). RLKs and RLPs bound to SOBIR1 associate with BAK1 to activate pattern-triggered immunity upon perception of exogenous or endogenous patterns. The haustorial callosic neckband that is sometimes formed in oomycete–plant interactions is depicted in blue. Oomycete-derived patterns and proteins are depicted in orange, plant-derived patterns in green.
Plants can sense a wide variety of extracellular oomycete-derived patterns. These molecules can be secreted by oomycetes during infection, or released from the invading pathogens by host-derived enzymes (Table 1). Several oomycete patterns are derived from the pathogen’s cell wall or membrane, whereas others are secreted to the extracellular environment before being detected by the plant immune system. Below we discuss the different extracellular patterns, where they derive from, and what is known about their function.
The most abundant constituents of oomycete cell walls are glucans, polysaccharides that consist of linked glucose units (Aronson et al., 1967; Sietsma et al., 1969). β-1,3 and β-1,6-glucan are the major components of oomycete cell walls, whereas cellulose, a β-1,4-glucan, forms a relatively small fraction (Aronson et al., 1967). β-1,6-Glucan is only found in oomycetes and fungi, whereas cellulose and β-1,3-glucan are present in plant cell walls too (Fesel and Zuccaro, 2015).
A β-glucan-triggered response, i.e., the accumulation of the phytoalexin glyceollin, was first observed when soybean (Glycine max) was treated with glucans isolated from cell walls of Phytophthora sojae (previously P. megasperma f. sp. glycinea and P. megasperma var. sojae) (Ayers et al., 1976). β-Glucans also trigger phytoalexin production in several other fabaceous species, and in potato (Solanum tuberosum), although this is a weaker response (Cline et al., 1978; Cosio et al., 1996). A purified β-1,3/1,6-glucan heptaglucoside was found to be one of the active molecules in eliciting production of phytoalexins in soybean (Sharp et al., 1984a,b). Laminarin, an oligomeric β-1,3-glucan with β-1,6-glucan branches isolated from the marine brown alga Laminaria digitata, is another pattern that can induce a plethora of defense-associated responses in Nicotiana tabacum, grapevine (Vitis vinifera), and the monocots rice (Oryza sativa) and wheat (Triticum aestivum; Inui et al., 1997; Klarzynski et al., 2000; Aziz et al., 2003). Furthermore, Arabidopsis thaliana is responsive to the β-glucan laminarin, although it does not respond to the oomycete-derived heptaglucoside elicitor. Arabidopsis responses to laminarin are mediated by the plant hormone ethylene, and do not seem to involve the well-known defense hormone salicylic acid (SA). In contrast, when Arabidopsis or tobacco plants are treated with a sulfated form of laminarin the expression of the SA-induced marker gene PR1 is induced (Ménard et al., 2004). Taken together, responses to β-glucans vary greatly depending on the specific β-glucan and plant species. Therefore, different plant species might have different receptors involved in the recognition of different β-glucan patterns.
Phytophthora-derived β-1,3-glucan was shown to bind soybean membranes (Yoshikawa et al., 1983). The glucan-binding protein (GBP) from soybean was identified and it was demonstrated that, when expressed in tobacco and Escherichia coli, GBP conferred β-glucan-binding activity. Furthermore, an antibody raised against GBP inhibited β-glucan-binding activity in soybean and reduced phytoalexin accumulation (Umemoto et al., 1997). Interestingly, GBP also shows β-glucanase activity and might release β-glucans from the pathogen’s cell wall (Fliegmann et al., 2004). After heterologous expression of soybean GBP in tomato, high affinity binding of the β-1,3/1,6-glucan heptaglucoside was observed. However, this did not result in activation of downstream defense responses in tomato (Mithöfer et al., 2000; Fliegmann et al., 2004). These data suggest that additional, probably membrane-bound, proteins are required to recognize the β-glucan patterns (Mithöfer et al., 2000).
Recently, glucan-chitosaccharides were isolated from the cell wall of the root oomycete Aphanomyces euteiches and were found as novel patterns that triggered calcium oscillations in the nucleus of root cells and induced defense genes in Medicago truncatula (Nars et al., 2013). How these molecules are perceived is not yet known, but there is a role for the nod factor perception (NFP) protein, a lysin motif (LysM)-RLK. NFP is involved in the recognition of microbial N-acetylglucosamine patterns and is required for nodule formation in interaction with Rhizobium bacteria. An nfp mutant was more susceptible to A. euteiches, whereas overexpression of NFP led to increased resistance, demonstrating its involvement in the perception of A. euteiches by M. truncatula (Rey et al., 2013). However, NFP was not required for the glucan-chitosaccharide-induced calcium oscillations, suggesting a regulatory function in defense for NFP rather than direct recognition (Nars et al., 2013).
Transglutaminases (TGases) are a widespread family of enzymes, found in prokaryotes and eukaryotes, that facilitate cross-linking between glutamine and lysine residues in proteins, thereby strengthening structures, e.g., cell walls (Lorand and Graham, 2003; Martins et al., 2014). The formation of a covalent bond between amino acid residues confers high resistance to proteolysis (Reiss et al., 2011). In oomycetes, TGases could protect cell walls from hydrolytic host enzymes. A 42-kDa TGase cell wall glycoprotein (GP42) of P. sojae functions as a potent elicitor of phytoalexin synthesis in the non-host parsley (Petroselinum crispum) (Parker et al., 1991). A 13-amino acid peptide fragment (Pep-13) derived from GP42 was found responsible for triggering immunity and was shown to bind to purified plasma membranes of parsley. Furthermore, Pep-13 elicits a multitude of defense responses, e.g., expression of defense-related genes and phytoalexin production (Nürnberger et al., 1994, 1995; Hahlbrock et al., 1995). Interestingly, Pep-13 treatment of potato resulted in a similar defense activation, with the distinct difference that it induced a hypersensitive response (HR; Halim et al., 2004).
GP42 homologs are only found in oomycetes and some marine bacteria belonging to the genus Vibrio that are pathogenic on fish and several marine invertebrates (Reiss et al., 2011). It is thought that an ancestral oomycete, from which species of Phytophthora, Pythium and downy mildews have evolved, acquired GP42 from Vibrio bacteria through horizontal gene transfer, giving an selective advantage over oomycetes that lack this TGase (Reiss et al., 2011). A 100 kDa monomeric plasma membrane protein from parsley was shown to bind to the Pep-13 ligand and thus may be part of the putative receptor complex (Nennstiel et al., 1998).
Application of mycelial extracts from P. infestans to potato tubers led to necrosis and accumulation of phytoalexins, predominantly rishitin, and lubimin. The molecules responsible for triggering this response were identified as the eicosapolyenoic acids (EPs), arachidonic acid (AA), and eicosapentaenoic acid (EPA; Bostock et al., 1981). Treating potato tuber slices with AA greatly reduced or even arrested growth of P. infestans (Bostock et al., 1982). EPs are components of Phytophthora cells and are seemingly not present in other microbial classes nor are they produced by higher plants (Robinson and Bostock, 2015). Interestingly, the downy mildew Hyaloperonospora arabidopsidis has lost the genes required for AA synthesis (Baxter et al., 2010). It is tempting to speculate that H. arabidopsidis has lost this ability through evolution as a way to avoid recognition.
Eicosapolyenoic acids induce the accumulation of antimicrobial compounds in many plant species, ranging from many solanaceous species, e.g., potato and tomato, to bean (Phaseolus vulgaris) and avocado (Persea americana) (Longland et al., 1987; Romero-Correa et al., 2014; Robinson and Bostock, 2015). Furthermore, in potato application of AA induced accumulation of reactive oxygen species (ROS), that could be involved in mediating synthesis of the phytoalexin rishitin from lubimin (Yoshioka et al., 2001).
EPs are able to trigger systemic acquired resistance in several plants species to different pathogens. The hormonal regulation of these responses seems to differ among plant species; in some the SA pathway is elicited, whereas in other species responses seem to rely on jasmonic acid (JA) or ethylene. It is postulated that this may be due to the concentration of EPs in the treatment (Robinson and Bostock, 2015). For example, Arabidopsis plants made to produce low levels of EP showed increased resistance to Botrytis cinerea, P. capsici and aphid feeding, but higher susceptibility to Pseudomonas syringae pv. tomato (DC3000). This was associated with higher levels of JA and enhanced expression of JA-related genes, but decreased SA levels and reduced expression of SA-related genes. Furthermore, low levels of AA administered to tomato leaves resulted in increased JA levels and decreased SA levels and higher resistance against Botrytis cinerea (Savchenko et al., 2010).
How, exactly, EPs are perceived remains to be resolved. EPs could be recognized directly by a membrane-bound receptor, leading to the activation of plant immunity. Another possibility is that plant membranes that readily incorporate AA (Ricker and Bostock, 1992), are perturbed leading to the release of endogenous patterns from the host cell cytoplasm. Or alternatively, AA can be used as a substrate for lipoxygenases, e.g., the potato LOX1, thereby producing oxylipin signals that trigger plant immunity. In the latter two scenarios recognition would be independent of plant PRRs (Robinson and Bostock, 2015).
Interestingly, treating potato with a combination of AA and β-1,3-glucans strongly increased the response to AA. β-glucans alone, however, did not trigger a response in potato (Preisig and Kuć, 1988).
Recently, the XEG1 (xyloglucanspecific endo-β-1,4-glucanase) protein was isolated from P. sojae culture filtrates (Ma et al., 2015). This secreted protein elicits cell death in N. benthamiana, N. tabacum, pepper (Capsicum annuum), tomato (S. lycopersicon) and soybean but not in maize (Zea mays) and cotton (Gossypium hirsutum). Analysis of the XEG1 protein sequence revealed that it belongs to the glycoside hydrolase GH12 family that is widespread amongst prokaryotic and eukaryotic microbes, especially in plant-associated microorganisms. Within the Phytophthora genus many GH12 proteins are found of which half trigger cell death in N. benthamiana. The downy mildew H. arabidopsidis also has three GH12 genes, however, none of them encode a protein that elicits cell death (Ma et al., 2015). Previously, it was demonstrated that fungal GH12 proteins are able to degrade β-glucan (Karlsson et al., 2002) and xyloglucan, a hemicellulose found in the plant cell wall (Master et al., 2008). Recombinant XEG1 protein partially released reducing sugars from both glucans, but was most active with a xyloglucan substrate. Mutations in the catalytic site of XEG1 strongly decreased xyloglucanase activity and abolished β-glucanase activity. In contrast, XEG1 enzyme activity was not required for the induction of cell death in N. benthamiana and soybean. Moreover, active and inactive recombinant XEG1 were able to induce resistance against P. sojae and Phytophthora parasitica var. nicotianae to a similar extent in soybean and N. benthamiana, respectively. Silencing as well as overexpression of XEG1 in P. sojae both led to reduced virulence on soybean through distinct mechanisms. Silenced P. sojae lines showed reduced virulence, but did not activate a stronger defense response in soybean, suggesting that XEG1 has a role in virulence, possibly through breakdown of cell wall components. XEG1 overexpression transformants induced more ROS accumulation and callose deposition compared to wildtype P. sojae, confirming the idea that XEG1 acts as a MAMP. A XEG1 PRR has not been identified but XEG1 requires the co-receptor SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 3/BRI1-ASSOCIATED RECEPTOR KINASE (SERK3/BAK1) for triggering cell death, suggesting that a SERK3/BAK1-associated RLK or RLP recognizes XEG1 (Ma et al., 2015).
Necrosis and ethylene-inducing peptide 1 (Nep1)-like proteins (NLPs) form a family of secreted proteins mainly found in plant-associated microorganisms, and cytotoxic members are well known to induce necrosis and ethylene production in dicot plants (Bailey, 1995; Oome and Van den Ackerveken, 2014). Three types of NLPs have been identified: type 1 NLPs are found in bacteria, oomycetes and fungi, type 2 NLPs are found in fungi and bacteria and the newly identified type 3 NLPs are only present in fungi (Oome and Van den Ackerveken, 2014). Although many members of the NLP family are cytotoxic to plants, in recent years many non-cytoxic NLPs have been identified in fungal and oomycete species with a (hemi-)biotrophic lifestyle (Cabral et al., 2012; Dong et al., 2012; Zhou et al., 2012). In search of the function of 10 non-cytotoxic NLPs of the obligate biotrophic downy mildew H. arabidopsidis (HaNLPs) it was found that NLPs activate plant immunity in Arabidopsis (Oome et al., 2014). Expression of HaNLPs in Arabidopsis led to a severe growth reduction and increased resistance to H. arabidopsidis for 7 out of 10 HaNLPs. Only a small fragment of the tested HaNLP3 protein was sufficient to activate plant defense responses and immunity to downy mildew. This 20–24 amino acid fragment (nlp20/nlp24) contains two conserved regions. The second region is the heptapeptide motif GHRHDWE which is highly conserved in all NLPs (Oome and Van den Ackerveken, 2014). The first motif that starts with the AIMY amino acid sequence is highly conserved in type 1 NLPs (Oome et al., 2014). Treatment of Arabidopsis plants with synthetic nlp24 peptides corresponding to an oomycete, fungal and bacterial type 1 NLP resulted in the increased production of the defense-related phytohormone ethylene and high resistance to downy mildew. Conversely, a synthetic peptide of a type 2 NLP from the bacterial pathogen Pectobacterium carotovorum that lacks the AIMY motif was unable to elicit a response in Arabidopsis. Taken together, this demonstrated that the first motif contains the immunogenic part of nlp24 (Oome et al., 2014). Furthermore, nlp20, a peptide based on PpNLP, a cytotoxic P. parasitica type 1 NLP, was sufficient for MAPK activation, production of ROS, and increased callose deposition in Arabidopsis, but did not have any cytotoxic effect (Böhm et al., 2014). Other plant species were tested for their ability to respond to nlp peptides, revealing that nlp-triggered ethylene production was observed in several closely related Brassicaceae species, and also in more distantly related lettuce plants (Lactuca sativa), but not in solanaceous species such as tomato, potato, and N. benthamiana (Böhm et al., 2014).
In a screen for nlp20 sensitivity, a collection of T-DNA insertion mutants corresponding to 29 RLKs and 44 RLPs were tested for loss of nlp20-induced ethylene production. Furthermore, 135 natural accessions of Arabidopsis were also tested for the loss of nlp20 sensitivity. Two T-DNA insertion alleles of RLP23, rlp23-1, and rlp23-2 that were unable to express the receptor-like protein as well as three Arabidopsis accessions that carried a frameshift mutation resulting in a premature stop codon in RLP23 coding sequence were insensitive to nlp20. It was shown that the RLP23 LRR domain physically interacts with nlp20 in vitro and in planta (Albert et al., 2015). RLP23 lacks a cytoplasmic signaling domain but was shown to require the RLK SUPPRESSOR OF BIR1 1 (SOBIR1) for signaling. RLP23 and SOBIR1 interact in the absence of nlp peptides (Bi et al., 2014; Albert et al., 2015), whereas a second RLK, BAK1, was recruited only in presence of the ligand (Albert et al., 2015). Arabidopsis sobir1 and bak1-5/bkk1 mutants lost nlp20-responsiveness, indicating that SOBIR1 and BAK1 are required for RLP23 to function. Moreover, it was demonstrated that RLP23 is required for nlp peptide-induced resistance. Unlike wildtype Arabidopsis, nlp24-treatment of rlp23 mutants did not result in an increased resistance to H. arabidopsidis (Albert et al., 2015).
Many oomycete pathogens secrete small 10 kDa proteins called elicitins. The first proteins from this family identified were cryptogein and capsicein from Phytophthora cryptogea and Phytophthora capsici, respectively. These proteins were found to elicit necrosis, induce resistance, and cause increased production of ethylene as well as the phytoalexin capsidiol in tobacco plants (Ricci et al., 1989; Milat et al., 1991). Elicitin responses were observed in all tested Nicotiana spp., but not in other solanaceous species, such as tomato and eggplant. Furthermore, some Brassicaceae species also respond to elicitin; most radish cultivars (Raphanus sativus) and one turnip cultivar (Brassica campestris), but not Arabidopsis, showed necrosis after elicitin treatment (Kamoun et al., 1993). The gene encoding for P. infestans elicitin INF1 was found to be downregulated during early infection of potato. However, in the necrotrophic phase of infection inf1 expression was upregulated (Kamoun et al., 1997). Interestingly, N. benthamiana, a nonhost of P. infestans, gained susceptibility after silencing of inf1, demonstrating that the recognition of INF1 contributes to resistance (Kamoun et al., 1998).
Members of the Peronosporales, e.g., Phytophthora spp. and downy mildews are unable to synthesize sterols and must, therefore, acquire them during pathogenesis. Elicitin and elicitin-like sequences are also found in downy mildew pathogens, but no functional analysis has been performed on these proteins (Baxter et al., 2010; Cabral et al., 2011; Stassen et al., 2012; Sharma et al., 2015). Dehydroergosterol binding activity was shown for several elicitins in vitro. Furthermore, elicitins are able to catalyze sterol transfer between liposomes (Mikes et al., 1998). However, in vivo sterol-binding activity of elicitins has not been demonstrated. Interestingly, the oomycete pathogen A. euteiches is able to synthesize sterols and seems to lack elicitin genes (Gaulin et al., 2008, 2010).
The putative elicitin receptor was recently cloned from a wild potato (Solanum) that responds to the P. infestans elicitin INF1. A S. microdontum ecotype showed a clear cell-death response when inf1 was transiently expressed. Crosses with an unresponsive S. microdontum subspecies and further screening and genetic mapping resulted in the identification of the RLP ELR (elicitin response). Stable expression of ELR in S. tuberosum cv. Désirée conferred the cell death response after expression of inf1. Furthermore, ELR mediated a broad-spectrum response to elicitins of oomycetes: most tested elicitins induced a cell-death response in transgenic ELR potato, even though there is often low sequence similarity between elicitins (Du et al., 2015). Recognition might therefore be based on structural similarity rather than a small conserved peptide. ELR was shown to bind to SERK3/BAK1, but binding of the putative receptor to the RLP adaptor protein SOBIR1 or the elicitin ligand was not tested (Du et al., 2015). Intracellular perception, however, cannot be ruled out as elicitins have, anecdotally, been reported to be detected inside plant cells, e.g., the immunocytochemical localization of the elicitin quercinin in oak (Quercus robur) root cells infected with P. quercina (Brummer et al., 2002). ELR is thought to mediate extracellular recognition of elicitins, but direct binding to confirm the receptor function of ELR still needs to be demonstrated (Du et al., 2015). Previously, studies in tobacco suggested that INF1 binds to the cytoplasmic domain of a lectin RLK from N. benthamiana, NbLRK1 (Kanzaki et al., 2008). Silencing of NbLRK1 resulted in reduced INF1 responsiveness suggesting the RLK contributes to defense signaling. Although no ELR has been identified in tobacco yet, SERK3/BAK1 and SOBIR1 were found to be required for elicitin-triggered cell death in N. benthamiana (Chaparro-Garcia et al., 2011; Peng et al., 2015). It is, therefore, likely that ELR acts similar to RLP23 (Albert et al., 2015) and tomato Cf-4 (Postma et al., 2016), in that it requires both a BAK1-like RLK and SOBIR1-like RLK for pattern-triggered immunity.
A 34 kDa glycoprotein was isolated from P. parasitica var. nicotianae mycelium that triggered enhanced lipoxygenase activity as well as accumulation the defense-related cell wall hydroxyproline-rich glycoproteins in tobacco. This protein was localized to the internal and external layers of the hyphal cell wall (Séjalon-Delmas et al., 1997). The protein sequence revealed two cellulose-binding domains belonging to the carbohydrate binding module 1 (CBM1) family similar to that of fungal glycanases (Mateos et al., 1997; Gaulin et al., 2006). This putative function was corroborated by demonstrating protein binding to fibrous cellulose and plant cell walls. Furthermore, the protein was shown to have lectin-like activities; human red blood cells were readily agglutinated by this protein. Therefore, it was designated cellulose-binding elicitor lectin (CBEL). Moreover, CBEL was able to elicit necrosis, activate defense gene expression, and trigger immunity to P. parasitica var. nicotianae. No enzymatic activities for CBEL were observed, suggesting it acts as a pattern (Mateos et al., 1997).
Silencing of CBEL resulted in a severe reduction of adhesive abilities of P. parasitica var. nicotianae to cellulosic surfaces, but did not affect pathogenicity. Interestingly, knockdown mutants showed dispersed abnormal cell wall thickenings, indicating that CBEL might be involved in cell wall deposition in the pathogen (Gaulin et al., 2002). CBEL activity as a pattern is not limited to tobacco, as infiltration of CBEL in Arabidopsis leaves resulted in defense responses differentially dependent on the phytohormones SA, JA, and ethylene (Khatib et al., 2004). CBEL-induced necrosis was lost in JA-insensitive coi1 and ethylene-insensitive ein2 mutant plants, whereas PR1 and WAK1 expression, accumulation hydroxyproline-rich glycoproteins, and peroxidase activity was greatly reduced or abolished in an Arabidopsis NahG mutant that metabolizes SA (Khatib et al., 2004). Transient expression of CBEL as well as infiltration of recombinant CBEL in tobacco leaves resulted in rapid development of necrotic lesions. Immunocytochemistry revealed that the delivered CBEL was bound to the plant cell wall. Substitution of aromatic residues in CBEL that are possibly involved in cellulose binding reduced the necrosis-inducing activity. Necrosis-induction in tobacco was lost for three recombinant CBEL proteins (Y52A, Y188A, and Y52A_Y188A), that were also unable to induce defense-related genes at similar concentrations as native CBEL. Recently, it was shown that CBM1-1 is the main determinant in the interaction with cellulose; a mutation in CBM1-2 (Y188A) only showed a slight decrease in cellulose binding compared to wild type CBEL, whereas a mutation in CBM1-1 (Y52A) strongly decreased the binding capacity of CBEL and the double mutant (Y52A_Y188A) entirely lost the ability to bind cellulose (Martinez et al., 2015). Taken together, these data show amino acids in the two CBM1s, that were predicted to be important for cellulose binding, are important for elicitor activity.
To define the minimum CBEL pattern that triggers immunity, synthetic peptides of CBM1-1 and CBM1-2 were generated. CBM1-1synt and CBM1-2synt were sufficient to activate plant defense in tobacco and Arabidopsis, respectively. Intriguingly, recombinant CBEL but not recombinant CBEL_Y52A_Y188A, induced calcium fluxes in tobacco cells but not in protoplasts. This demonstrates that the plant cell wall and unmodified CBM1s are important for CBEL perception (Gaulin et al., 2006).
CBM1s are probably not essential for pathogens with an obligate biotrophic lifestyle; only one was detected in the Albugo laibachii genome and no clear CBM1-encoding genes were found in H. arabidopsidis, whereas Pythium ultimum and Phytophthora spp. contain multiple CBM1-encoding genes (Larroque et al., 2012). It has been proposed that adhesion of CBEL or its CBM1s perturb the cellulose status, and the perception of this disturbance leads to defense activation, but this remains to be proven (Dumas et al., 2008). The fact that BAK1 and RESPIRATORY BURST OXIDASE HOMOLOGUE (RBOH) D and F proteins are required for some of the CBEL-induced defense responses suggests that a PRR might be involved (Larroque et al., 2013). The oxidative burst triggered by pattern recognition is mediated by the NADPH oxidases RBOH D and F (Suzuki et al., 2011). Necrosis-induction by CBEL in bak1-4 and the rbohD/F double mutant was similar to the Col-0 Arabidopsis wildtype. However, no ROS production was detected in bak1-4 and rbohD/F and activation of MAP kinases was reduced in bak1-4 and delayed in rbohD/F compared to Col-0. The expression of JA-responsive genes WRKY11 and PDF1.2, but not the expression of the SA-responsive gene PR1, was also reduced in these mutant lines (Larroque et al., 2013). The dependence of some CBEL-induced responses on BAK1 suggests a role for an RLK or RLP in the perception of CBEL. Three Arabidopsis accessions were found that are unresponsive to CBEL, and may therefore offer a way to decipher CBEL-triggered immunity (Larroque et al., 2013).
A secreted apoplastic protein from P. parasitica called OPEL was recently discovered to trigger a plant immune response (Chang et al., 2015). OPEL contains a thaumatin-like domain, a glycine-rich domain, and a glycosyl hydrolase (GH) domain that has a putative laminarinase active site. OPEL seems to be oomycete specific; homologues were only found in Phytophthora spp. and other oomycetes such as H. arabidopsidis, Py. ultimum and A. laibachii. OPEL is expressed during early infection stages of P. parasitica, rapidly increasing transcript levels within 12 hours after inoculation on N. benthamiana. Furthermore, infiltration of N. tabacum with recombinant OPEL protein resulted in cell death, increased callose deposition, ROS accumulation, induction of defense-related genes and systemic acquired resistance against several pathogens. Moreover, transient expression of OPEL in N. benthamiana enhanced resistance to P. parasitica. It was shown that the GH domain was essential for the increased callose deposition and increased accumulation of ROS in N. tabacum. Although the OPEL GH domain contains a laminarinase signature active site motif, no laminarin or β-1,3-glucan enzymatic activity was detected in OPEL recombinant protein. Mutation of the putative laminarinase active site motif in the predicted GH domain abolished elicitor activity of OPEL, which suggests enzymatic activity of OPEL is required for triggering the defense response (Chang et al., 2015). The OPEL substrate has not been identified but is likely a polysaccharide in the plant cell wall. OPEL-released degradation products might, therefore, be perceived by plants as DAMPs.
Next to exogenous patterns, host-derived molecules that are released upon pathogen infection can serve as danger signals (Table 2). Several endogenous patterns, also known as DAMPs, have been described that are plant cell wall derived or that are released from the host cytosol (Boller and Felix, 2009; Yamaguchi and Huffaker, 2011). The release of these patterns is promoted by a plethora of hydrolytic enzymes that are produced by pathogens (Baxter et al., 2010; Blackman et al., 2015). Interestingly, the downy mildew H. arabidopsidis has fewer hydrolases than the hemibiotrophic Phytophthora spp., probably as adaptation to its obligate biotrophic lifestyle (Baxter et al., 2010).
Oligogalacturonides (OGs) are released from the plant cell wall after mechanical damage or by pathogen-secreted hydrolytic enzymes through degradation of homogalacturonan (Ferrari et al., 2013). OGs bind to several members of the cell wall-associated kinase (WAK) family, which consequently leads to the activation of immunity (Brutus et al., 2010; Ferrari et al., 2013). Also cutin, the main constituent of the plant cuticle (Heredia, 2003), can be degraded to cutin monomers by pathogen released cutinases. Cutin monomers are potent elicitors of defense in several plant species (Schweizer et al., 1996; Fauth et al., 1998). However, it remains unknown how cutin monomers are recognized by plants.
Damage patterns could also be released from the plant cytosol during oomycete infection. These include members of the plant elicitor peptide (Pep) family. The cytosolic precursors of Peps, PROPEPS are released and cleaved when the plant cell is damaged, resulting in the production of endogenous patterns. The receptors for Peps have been identified, the RLKs PEP1 RECEPTOR 1 (PEPR1) and PEP1 RECEPTOR 2 (PEPR2) recognized Peps and contributed to immune responses against several pathogens (Yamaguchi et al., 2006, 2010; Krol et al., 2010; Yamaguchi and Huffaker, 2011; Albert, 2013; Bartels et al., 2013; Bartels and Boller, 2015).
Furthermore, extracellular adenosine triphosphate (eATP) could be perceived as a damage pattern. Treatment of Arabidopsis with ATP induced a similar set of genes as wounding did (Choi et al., 2014). In a screen for ATP-insensitivity, a dorn1 (Does Not Respond to Nucleotides 1) mutant was identified that is defective in the lectin receptor kinase LecRK-I.9. LecRK-I.9 binds to ATP with high affinity and is required for the activation of several ATP-induced responses, demonstrating it is an ATP receptor (Choi et al., 2014). Previously, lecrk-I.9 mutants were shown to be more susceptible to two Phytophthora species than wildtype Arabidopsis. Conversely, overexpression of LecRK-1.9 led to increased resistance to P. brassicae (Bouwmeester et al., 2011).
Finally, it has been proposed that recognition of the exogenous pattern β-1,3-glucan could have evolved as an endogenous danger signal; callose could be degraded by host or pathogen-derived β-1,3-glucanases, thereby eliciting a defense response (Klarzynski et al., 2000).
Plant genomes encode many RLKs and RLPs. The Arabidopsis genome, for example, encodes more than 600 RLKs and 57 RLPs (Shiu et al., 2004; Wang et al., 2008). For most of these proteins the function is unknown. We expect that several of these receptor proteins have a role in the perception of oomycete pathogens. Recently, it was shown that many RLP genes are upregulated after treatment with P. infestans and the P. infestans NLP NPP1, suggesting a role for these RLPs during oomycete infection (Wu et al., 2016). Several RLKs are also reported to affect the interaction with oomycete pathogens. For example, other LecRKs, next to the aforementioned LecRK-I.9 and NbLRK1, influence the defense response against Phytophthora in Arabidopsis, tomato and N. benthamiana (Wang et al., 2014, 2015b,a). Silencing of several LecRKs in tomato and N. benthamiana led to increased susceptibility to P. capsici and P. infestans, respectively (Wang et al., 2015b). Two Arabidopsis LecRKs from the same clade (IX) were shown to affect Phytophthora resistance in a similar way (Wang et al., 2015a). Finally, the Arabidopsis LecRK-VI.2A positively regulates the MAMP-triggered immunity response (Singh et al., 2012). Although, some RLKs and RLPs partly regulate the defense response against oomycetes, the patterns or molecules that are recognized by these proteins are still largely unknown.
Recent discoveries in extracellular recognition of oomycete patterns have provided new insight in how plants detect early infection of these (hemi-)biotrophic pathogens. Novel PRRs for elicitins and NLPs have been identified and mechanisms of how these exogenous patterns are perceived by plants have been elucidated. The scientific progress described in this review provides interesting leads for resistance breeding of crops. For example, transgenic expression of the PRRs ELR and RLP23 in cultivated potato resulted in increased resistance to the late blight pathogen P. infestans that is known to produce elicitins and NLPs (Haas et al., 2009; Albert et al., 2015; Du et al., 2015). Classical resistance breeding has mainly focused on the introgression of resistance genes encoding cytoplasmic NB-LRR receptors, which are rapidly broken by new emerging strains of the pathogen. The use of PRRs, many of which recognize conserved microbial patterns, for breeding a new generation of disease resistant crops could offer a more durable solution, especially if PRRs and resistance genes are stacked (Dangl et al., 2013; Schwessinger et al., 2015). A great example is the expression of the Arabidopsis PRR EFR in tomato that resulted in broad spectrum resistance to different bacterial pathogens that all produce the EF-Tu pattern that is recognized by EFR (Lacombe et al., 2010). As many of the described oomycete patterns are broadly distributed, expression of the cognate PRRs in crops could reduce plant disease and aid in securing our future food.
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors of this review are supported by the Less is More grant no. 847.13.006 of the Netherlands Organization for Scientific Research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Albert, I., Böhm, H., Albert, M., Feiler, C. E., Imkampe, J., Wallmeroth, N., et al. (2015). An RLP23–SOBIR1–BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1:15140. doi: 10.1038/nplants.2015.140
Albert, M. (2013). Peptides as triggers of plant defence. J. Exp. Bot. 64, 5269–5279. doi: 10.1093/jxb/ert275
Aronson, J. M., Cooper, B. A., and Fuller, M. S. (1967). Glucans of oomycete cell walls. Science 155, 332–335. doi: 10.1126/science.155.3760.332
Ayers, A. R., Ebel, J., Valent, B., and Albersheim, P. (1976). Host-pathogen interactions X. fractionation and biological activity of an elicitor isolated from the mycelial walls of Phytophthora megasperma var. sojae. Plant Physiol. 57, 760–765. doi: 10.1104/pp.57.5.760
Aziz, A., Poinssot, B., Daire, X., Adrian, M., Bézier, A., Lambert, B., et al. (2003). Laminarin elicits defense responses in grapevine and induces protection against Botrytis cinerea and Plasmopara viticola. Mol. Plant Microbe Interact. 16, 1118–1128. doi: 10.1094/MPMI.2003.16.12.1118
Bailey, B. A. (1995). Purification of a protein from culture filtrates of Fusarium oxysporum that induces ethylene and necrosis in leaves of Erythroxylum coca. Phytopathology 85:1250. doi: 10.1094/Phyto-85-1250
Bartels, S., and Boller, T. (2015). Quo vadis, pep? Plant elicitor peptides at the crossroads of immunity, stress, and development. J. Exp. Bot. 66, 5183–5193. doi: 10.1093/jxb/erv180
Bartels, S., Lori, M., Mbengue, M., van Verk, M., Klauser, D., Hander, T., et al. (2013). The family of Peps and their precursors in Arabidopsis: differential expression and localization but similar induction of pattern-triggered immune responses. J. Exp. Bot. 64, 5309–5321. doi: 10.1093/jxb/ert330
Baxter, L., Tripathy, S., Ishaque, N., Boot, N., Cabral, A., Kemen, E., et al. (2010). Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 330, 1549–1551. doi: 10.1126/science.1195203
Bi, G., Liebrand, T. W., Cordewener, J. H., America, A. H., Xu, X., and Joosten, M. H. (2014). Arabidopsis thaliana receptor-like protein AtRLP23 associates with the receptor-like kinase AtSOBIR1. Plant Signal. Behav. 9:e27937. doi: 10.4161/psb.27937
Blackman, L. M., Cullerne, D. P., Torreña, P., Taylor, J., and Hardham, A. R. (2015). RNA-seq analysis of the expression of genes encoding cell wall degrading enzymes during infection of lupin (Lupinus angustifolius) by Phytophthora parasitica. PLoS ONE 10:e0136899. doi: 10.1371/journal.pone.0136899
Böhm, H., Albert, I., Oome, S., Raaymakers, T. M., Van den Ackerveken, G., and Nürnberger, T. (2014). A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog. 10:e1004491. doi: 10.1371/journal.ppat.1004491
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Bostock, R. M., Kuc, J. A., and Laine, R. A. (1981). Eicosapentaenoic and arachidonic acids from Phytophthora infestans elicit fungitoxic sesquiterpenes in the potato. Science 212, 67–69. doi: 10.1126/science.212.4490.67
Bostock, R. M., Laine, R. A., and Kuć, J. A. (1982). Factors affecting the elicitation of sesquiterpenoid phytoalexin accumulation by eicosapentaenoic and arachidonic acids in potato. Plant Physiol. 70, 1417–1424. doi: 10.1104/pp.70.5.1417
Bouwmeester, K., de Sain, M., Weide, R., Gouget, A., Klamer, S., Canut, H., et al. (2011). The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 7:e1001327. doi: 10.1371/journal.ppat.1001327
Brummer, M., Arend, M., Fromm, J., Schlenzig, A., and Oßwald, W. (2002). Ultrastructural changes and immunocytochemical localization of the elicitin quercinin in Quercus robur L. roots infected with Phytophthora quercina. Physiol. Mol. Plant Pathol. 61, 109–120. doi: 10.1006/pmpp.2002.0419
Brutus, A., Sicilia, F., Macone, A., Cervone, F., and De Lorenzo, G. (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U.S.A. 107, 9452–9457. doi: 10.1073/pnas.1000675107
Cabral, A., Oome, S., Sander, N., Küfner, I., Nürnberger, T., and Van den Ackerveken, G. (2012). Nontoxic Nep1-like proteins of the downy mildew pathogen Hyaloperonospora arabidopsidis: repression of necrosis-inducing activity by a surface-exposed region. Mol. Plant Microbe Interact. 25, 697–708. doi: 10.1094/MPMI-10-11-0269
Cabral, A., Stassen, J. H. M., Seidl, M. F., Bautor, J., Parker, J. E., and Van den Ackerveken, G. (2011). Identification of Hyaloperonospora arabidopsidis transcript sequences expressed during infection reveals isolate-specific effectors. PLoS ONE 6:e19328. doi: 10.1371/journal.pone.0019328
Chang, Y.-H., Yan, H.-Z., and Liou, R.-F. (2015). A novel elicitor protein from Phytophthora parasitica induces plant basal immunity and systemic acquired resistance. Mol. Plant Pathol. 16, 123–136. doi: 10.1111/mpp.12166
Chaparro-Garcia, A., Wilkinson, R. C., Gimenez-Ibanez, S., Findlay, K., Coffey, M. D., Zipfel, C., et al. (2011). The receptor-like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen Phytophthora infestans in Nicotiana benthamiana. PLoS ONE 6:e16608. doi: 10.1371/journal.pone.0016608
Choi, J., Tanaka, K., Cao, Y., Qi, Y., Qiu, J., Liang, Y., et al. (2014). Identification of a plant receptor for extracellular ATP. Science 343, 290–294. doi: 10.1126/science.343.6168.290
Cline, K., Wade, M., and Albersheim, P. (1978). Host-pathogen interactions: XV. Fungal glucans which elicit phytoalexin accumulation in soybean also elicit the accumulation of phytoalexins in other plants. Plant Physiol. 62, 918–921. doi: 10.1104/pp.62.6.918
Cook, D. E., Mesarich, C. H., and Thomma, B. P. H. J. (2015). Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 53, 541–563. doi: 10.1146/annurev-phyto-080614-120114
Cosio, E. G., Feger, M., Miller, C. J., Antelo, L., and Ebel, J. (1996). High-affinity binding of fungal ß-glucan elicitors to cell membranes of species of the plant family Fabaceae. Planta 200, 92–99. doi: 10.1007/BF00196654
Dangl, J. L., Horvath, D. M., and Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. doi: 10.1126/science.1236011
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Dong, S., Kong, G., Qutob, D., Yu, X., Tang, J., Kang, J., et al. (2012). The NLP toxin family in Phytophthora sojae includes rapidly evolving groups that lack necrosis-inducing activity. Mol. Plant Microbe Interact. 25, 896–909. doi: 10.1094/MPMI-01-12-0023-R
Du, J., Verzaux, E., Chaparro-Garcia, A., Bijsterbosch, G., Keizer, L. C. P., Zhou, J., et al. (2015). Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat. Plants 1:15034. doi: 10.1038/nplants.2015.34
Dumas, B., Bottin, A., Gaulin, E., and Esquerré-Tugayé, M.-T. (2008). Cellulose-binding domains: cellulose associated-defensive sensing partners? Trends Plant Sci. 13, 160–164. doi: 10.1016/j.tplants.2008.02.004
Fauth, M., Schweizer, P., Buchala, A., Markstädter, C., Riederer, M., Kato, T., et al. (1998). Cutin monomers and surface wax constituents elicit H2O2 in conditioned cucumber hypocotyl segments and enhance the activity of other H2O2 elicitors. Plant Physiol. 117, 1373–1380. doi: 10.1104/pp.117.4.1373
Ferrari, S., Savatin, D. V., Sicilia, F., Gramegna, G., Cervone, F., and Lorenzo, G. D. (2013). Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 4:49. doi: 10.3389/fpls.2013.00049
Fesel, P. H., and Zuccaro, A. (2015). β-glucan: crucial component of the fungal cell wall and elusive MAMP in plants. Fungal Genet. Biol. 90, 53–60. doi: 10.1016/j.fgb.2015.12.004
Fliegmann, J., Mithöfer, A., Wanner, G., and Ebel, J. (2004). An ancient enzyme domain hidden in the putative ß-glucan elicitor receptor of soybean may play an active part in the perception of pathogen-associated molecular patterns during broad host resistance. J. Biol. Chem. 279, 1132–1140. doi: 10.1074/jbc.M308552200
Gaulin, E., Bottin, A., and Dumas, B. (2010). Sterol biosynthesis in oomycete pathogens. Plant Signal. Behav. 5, 258–260. doi: 10.4161/psb.5.3.10551
Gaulin, E., Dramé, N., Lafitte, C., Torto-Alalibo, T., Martinez, Y., Ameline-Torregrosa, C., et al. (2006). Cellulose binding domains of a Phytophthora cell wall protein are novel pathogen-associated molecular patterns. Plant Cell 18, 1766–1777. doi: 10.1105/tpc.105.038687
Gaulin, E., Jauneau, A., Villalba, F., Rickauer, M., Esquerré-Tugayé, M.-T., and Bottin, A. (2002). The CBEL glycoprotein of Phytophthora parasitica var. nicotianae is involved in cell wall deposition and adhesion to cellulosic substrates. J. Cell Sci. 115, 4565–4575. doi: 10.1242/jcs.00138
Gaulin, E., Madoui, M.-A., Bottin, A., Jacquet, C., Mathé, C., Couloux, A., et al. (2008). Transcriptome of Aphanomyces euteiches: new oomycete putative pathogenicity factors and metabolic pathways. PLoS ONE 3:e1723. doi: 10.1371/journal.pone.0001723
Haas, B. J., Kamoun, S., Zody, M. C., Jiang, R. H. Y., Handsaker, R. E., Cano, L. M., et al. (2009). Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461, 393–398. doi: 10.1038/nature08358
Hahlbrock, K., Scheel, D., Logemann, E., Nürnberger, T., Parniske, M., Reinold, S., et al. (1995). Oligopeptide elicitor-mediated defense gene activation in cultured parsley cells. Proc. Natl. Acad. Sci. U.S.A. 92, 4150–4157. doi: 10.1073/pnas.92.10.4150
Halim, V. A., Hunger, A., Macioszek, V., Landgraf, P., Nürnberger, T., Scheel, D., et al. (2004). The oligopeptide elicitor Pep-13 induces salicylic acid-dependent and -independent defense reactions in potato. Physiol. Mol. Plant Pathol. 64, 311–318. doi: 10.1016/j.pmpp.2004.10.003
Hein, I., Gilroy, E. M., Armstrong, M. R., and Birch, P. R. J. (2009). The zig-zag-zig in oomycete-plant interactions. Mol. Plant Pathol. 10, 547–562. doi: 10.1111/j.1364-3703.2009.00547.x
Heredia, A. (2003). Biophysical and biochemical characteristics of cutin, a plant barrier biopolymer. Biochim. Biophys. Acta 1620, 1–7. doi: 10.1016/S0304-4165(02)00510-X
Inui, H., Yamaguchi, Y., and Hirano, S. (1997). Elicitor actions of N-acetylchitooligosaccharides and laminarioligosaccharides for chitinase and L-phenylalanine ammonia-lyase induction in rice suspension culture. Biosci. Biotechnol. Biochem. 61, 975–978. doi: 10.1271/bbb.61.975
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kamoun, S., Furzer, O., Jones, J. D. G., Judelson, H. S., Ali, G. S., Dalio, R. J. D., et al. (2015). The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434. doi: 10.1111/mpp.12190
Kamoun, S., van West, P., de Jong, A. J., de Groot, K. E., Vleeshouwers, V. G. A. A., and Govers, F. (1997). A gene encoding a protein elicitor of Phytophthora infestans is down-regulated during infection of potato. Mol. Plant Microbe Interact. 10, 13–20. doi: 10.1094/MPMI.1997.10.1.13
Kamoun, S., van West, P., Vleeshouwers, V. G. A. A., de Groot, K. E., and Govers, F. (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10, 1413–1426. doi: 10.2307/3870607
Kamoun, S., Young, M., Glascock, C. B., and Tyler, B. M. (1993). Extracellular protein elicitors from Phytophthora: host-specificity and induction of resistance to bacterial and fungal phytopathogens. Mol. Plant Microbe Interact. 6, 15–25. doi: 10.1094/MPMI-6-015
Kanzaki, H., Saitoh, H., Takahashi, Y., Berberich, T., Ito, A., Kamoun, S., et al. (2008). NbLRK1, a lectin-like receptor kinase protein of Nicotiana benthamiana, interacts with Phytophthora infestans INF1 elicitin and mediates INF1-induced cell death. Planta 228, 977–987. doi: 10.1007/s00425-008-0797-y
Karlsson, J., Siika-aho, M., Tenkanen, M., and Tjerneld, F. (2002). Enzymatic properties of the low molecular mass endoglucanases Cel12A (EG III) and Cel45A (EG V) of Trichoderma reesei. J. Biotechnol. 99, 63–78. doi: 10.1016/S0168-1656(02)00156-6
Khatib, M., Lafitte, C., Esquerré-Tugayé, M.-T., Bottin, A., and Rickauer, M. (2004). The CBEL elicitor of Phytophthora parasitica var. nicotianae activates defence in Arabidopsis thaliana via three different signalling pathways. New Phytol. 162, 501–510. doi: 10.1111/j.1469-8137.2004.01043.x
Klarzynski, O., Plesse, B., Joubert, J. M., Yvin, J. C., Kopp, M., Kloareg, B., et al. (2000). Linear ß-1,3 glucans are elicitors of defense responses in tobacco. Plant Physiol. 124, 1027–1038. doi: 10.1104/pp.124.3.1027
Krol, E., Mentzel, T., Chinchilla, D., Boller, T., Felix, G., Kemmerling, B., et al. (2010). Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 285, 13471–13479. doi: 10.1074/jbc.M109.097394
Lacombe, S., Rougon-Cardoso, A., Sherwood, E., Peeters, N., Dahlbeck, D., van Esse, H. P., et al. (2010). Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 28, 365–369. doi: 10.1038/nbt.1613
Larroque, M., Barriot, R., Bottin, A., Barre, A., Rougé, P., Dumas, B., et al. (2012). The unique architecture and function of cellulose-interacting proteins in oomycetes revealed by genomic and structural analyses. BMC Genomics 13:605. doi: 10.1186/1471-2164-13-605
Larroque, M., Belmas, E., Martinez, T., Vergnes, S., Ladouce, N., Lafitte, C., et al. (2013). Pathogen-associated molecular pattern-triggered immunity and resistance to the root pathogen Phytophthora parasitica in Arabidopsis. J. Exp. Bot. 64, 3615–3625. doi: 10.1093/jxb/ert195
Longland, A. C., Slusarenko, A. J., and Friend, J. (1987). Arachidonic and Linoleic acids elicit isoflavonoid phytoalexin accumulation in Phaseolus vulgaris (French bean). J. Phytopathol. 120, 289–297. doi: 10.1111/j.1439-0434.1987.tb00492.x
Lorand, L., and Graham, R. M. (2003). Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 4, 140–156. doi: 10.1038/nrm1014
Ma, Z., Song, T., Zhu, L., Ye, W., Wang, Y., Shao, Y., et al. (2015). A Phytophthora sojae Glycoside Hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27, 2057–2072. doi: 10.1105/tpc.15.00390
Martinez, T., Texier, H., Nahoum, V., Lafitte, C., Cioci, G., Heux, L., et al. (2015). Probing the functions of carbohydrate binding modules in the CBEL protein from the Oomycete Phytophthora parasitica. PLoS ONE 10:e0137481. doi: 10.1371/journal.pone.0137481
Martins, I. M., Matos, M., Costa, R., Silva, F., Pascoal, A., Estevinho, L. M., et al. (2014). Transglutaminases: recent achievements and new sources. Appl. Microbiol. Biotechnol. 98, 6957–6964. doi: 10.1007/s00253-014-5894-1
Master, E. R., Zheng, Y., Storms, R., Tsang, A., and Powlowski, J. (2008). A xyloglucan-specific family 12 glycosyl hydrolase from Aspergillus niger: recombinant expression, purification and characterization. Biochem. J. 411, 161–170. doi: 10.1042/BJ20070819
Mateos, F. V., Rickauer, M., and Esquerré-Tugayé, M. T. (1997). Cloning and characterization of a cDNA encoding an elicitor of Phytophthora parasitica var. nicotianae that shows cellulose-binding and lectin-like activities. Mol. Plant Microbe Interact. 10, 1045–1053. doi: 10.1094/MPMI.1997.10.9.1045
Ménard, R., Alban, S., de Ruffray, P., Jamois, F., Franz, G., Fritig, B., et al. (2004). ß-1,3 glucan sulfate, but not ß-1,3 glucan, induces the salicylic acid signaling pathway in tobacco and Arabidopsis. Plant Cell 16, 3020–3032. doi: 10.1105/tpc.104.024968
Mikes, V., Milat, M. L., Ponchet, M., Panabières, F., Ricci, P., and Blein, J. P. (1998). Elicitins, proteinaceous elicitors of plant defense, are a new class of sterol carrier proteins. Biochem. Biophys. Res. Commun. 245, 133–139. doi: 10.1006/bbrc.1998.8341
Milat, M. L., Ricci, P., Bonnet, P., and Blein, J. P. (1991). Capsidiol and ethylene production by tobacco cells in response to cryptogein, an elicitor from Phytophthora cryptogea. Phytochemistry 30, 2171–2173. doi: 10.1016/0031-9422(91)83608-N
Mithöfer, A., Fliegmann, J., Neuhaus-Url, G., Schwarz, H., and Ebel, J. (2000). The hepta-ß-glucoside elicitor-binding proteins from legumes represent a putative receptor family. Biol. Chem. 381, 705–713. doi: 10.1515/BC.2000.091
Nars, A., Lafitte, C., Chabaud, M., Drouillard, S., Mélida, H., Danoun, S., et al. (2013). Aphanomyces euteiches cell wall fractions containing novel glucan-chitosaccharides induce defense genes and nuclear calcium oscillations in the plant host Medicago truncatula. PLoS ONE 8:e75039. doi: 10.1371/journal.pone.0075039
Nennstiel, D., Scheel, D., and Nürnberger, T. (1998). Characterization and partial purification of an oligopeptide elicitor receptor from parsley (Petroselinum crispum). FEBS Lett. 431, 405–410. doi: 10.1016/S0014-5793(98)00800-X
Nürnberger, T., Nennstiel, D., Hahlbrock, K., and Scheel, D. (1995). Covalent cross-linking of the Phytophthora megasperma oligopeptide elicitor to its receptor in parsley membranes. Proc. Natl. Acad. Sci. U.S.A. 92, 2338–2342. doi: 10.1073/pnas.92.6.2338
Nürnberger, T., Nennstiel, D., Jabs, T., Sacks, W. R., Hahlbrock, K., and Scheel, D. (1994). High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell 78, 449–460. doi: 10.1016/0092-8674(94)90423-5
Oome, S., Raaymakers, T. M., Cabral, A., Samwel, S., Böhm, H., Albert, I., et al. (2014). Nep1-like proteins from three kingdoms of life act as a microbe-associated molecular pattern in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 16955–16960. doi: 10.1073/pnas.1410031111
Oome, S., and Van den Ackerveken, G. (2014). Comparative and functional analysis of the widely occurring family of Nep1-like proteins. Mol. Plant Microbe Interact. 27, 1081–1094. doi: 10.1094/MPMI-04-14-0118-R
Parker, J. E., Schulte, W., Hahlbrock, K., and Scheel, D. (1991). An extracellular glycoprotein from Phytophthora megasperma f. sp. glycinea elicits phytoalexin synthesis in cultured parsley cells and protoplasts. Mol. Plant Microbe Interact. 4, 19–27. doi: 10.1094/MPMI-4-019
Parniske, M. (2000). Intracellular accommodation of microbes by plants: a common developmental program for symbiosis and disease? Curr. Opin. Plant Biol. 3, 320–328. doi: 10.1016/S1369-5266(00)00088-1
Peng, K.-C., Wang, C.-W., Wu, C.-H., Huang, C.-T., and Liou, R.-F. (2015). Tomato SOBIR1/EVR homologs are involved in elicitin perception and plant defense against the oomycete pathogen Phytophthora parasitica. Mol. Plant Microbe Interact. 28, 913–926. doi: 10.1094/MPMI-12-14-0405-R
Postma, J., Liebrand, T. W. H., Bi, G., Evrard, A., Bye, R. R., Mbengue, M., et al. (2016). Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytol. 210, 627–642. doi: 10.1111/nph.13802
Preisig, C. L., and Kuć, J. A. (1988). Metabolism by potato tuber of arachidonic acid, an elicitor of hypersensitive resistance. Physiol. Mol. Plant Pathol. 32, 77–88. doi: 10.1016/S0885-5765(88)80007-9
Reiss, K., Kirchner, E., Gijzen, M., Zocher, G., Löffelhardt, B., Nürnberger, T., et al. (2011). Structural and phylogenetic analyses of the GP42 transglutaminase from Phytophthora sojae reveal an evolutionary relationship between oomycetes and marine Vibrio bacteria. J. Biol. Chem. 286, 42585–42593. doi: 10.1074/jbc.M111.290544
Rey, T., Nars, A., Bonhomme, M., Bottin, A., Huguet, S., Balzergue, S., et al. (2013). NFP, a LysM protein controlling Nod factor perception, also intervenes in Medicago truncatula resistance to pathogens. New Phytol. 198, 875–886. doi: 10.1111/nph.12198
Ricci, P., Bonnet, P., Huet, J. C., Sallantin, M., Beauvais-Cante, F., Bruneteau, M., et al. (1989). Structure and activity of proteins from pathogenic fungi Phytophthora eliciting necrosis and acquired resistance in tobacco. Eur. J. Biochem. 183, 555–563. doi: 10.1111/j.1432-1033.1989.tb21084.x
Ricker, K. E., and Bostock, R. M. (1992). Evidence for release of the elicitor arachidonic acid and its metabolites from sporangia of Phytophthora infestans during infection of potato. Physiol. Mol. Plant Pathol. 41, 61–72. doi: 10.1016/0885-5765(92)90049-2
Robinson, S. M., and Bostock, R. M. (2015). β-glucans and eicosapolyenoic acids as MAMPs in plant-oomycete interactions: past and present. Front. Plant Sci. 5:797. doi: 10.3389/fpls.2014.00797
Romero-Correa, M. T., Villa-Gómez, R., Castro-Mercado, E., and García-Pineda, E. (2014). The avocado defense compound phenol-2,4-bis (1,1-dimethylethyl) is induced by arachidonic acid and acts via the inhibition of hydrogen peroxide production by pathogens. Physiol. Mol. Plant Pathol. 87, 32–41. doi: 10.1016/j.pmpp.2014.05.003
Savchenko, T., Walley, J. W., Chehab, E. W., Xiao, Y., Kaspi, R., Pye, M. F., et al. (2010). Arachidonic acid: an evolutionarily conserved signaling molecule modulates plant stress signaling networks. Plant Cell 22, 3193–3205. doi: 10.1105/tpc.110.073858
Schweizer, P., Felix, G., Buchala, A., Müller, C., and Métraux, J. P. (1996). Perception of free cutin monomers by plant cells. Plant J. 10, 331–341. doi: 10.1046/j.1365-313X.1996.10020331.x
Schwessinger, B., Bahar, O., Thomas, N., Holton, N., Nekrasov, V., Ruan, D., et al. (2015). Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to ligand-dependent activation of defense responses. PLoS Pathog. 11:e1004809. doi: 10.1371/journal.ppat.1004809
Schwessinger, B., and Zipfel, C. (2008). News from the frontline: recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol. 11, 389–395. doi: 10.1016/j.pbi.2008.06.001
Séjalon-Delmas, N., Mateos, F. V., Bottin, A., Rickauer, M., Dargent, R., and Esquerré-Tugayé, M. T. (1997). Purification, elicitor activity, and cell wall localization of a glycoprotein from Phytophthora parasitica var. nicotianae, a fungal pathogen of tobacco. Phytopathology 87, 899–909. doi: 10.1094/PHYTO.1997.87.9.899
Sharma, R., Xia, X., Cano, L. M., Evangelisti, E., Kemen, E., Judelson, H., et al. (2015). Genome analyses of the sunflower pathogen Plasmopara halstedii provide insights into effector evolution in downy mildews and Phytophthora. BMC Genomics 16:741. doi: 10.1186/s12864-015-1904-7
Sharp, J. K., Albersheim, P., Ossowski, P., Pilotti, A., Garegg, P., and Lindberg, B. (1984a). Comparison of the structures and elicitor activities of a synthetic and a mycelial-wall-derived hexa(ß-D-glucopyranosyl)-D-glucitol. J. Biol. Chem. 259, 11341–11345.
Sharp, J. K., McNeil, M., and Albersheim, P. (1984b). The primary structures of one elicitor-active and seven elicitor-inactive hexa(ß-D-glucopyranosyl)-D-glucitols isolated from the mycelial walls of Phytophthora megasperma f. sp. glycinea. J. Biol. Chem. 259, 11321–11336.
Shiu, S.-H., Karlowski, W. M., Pan, R., Tzeng, Y.-H., Mayer, K. F. X., and Li, W.-H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234. doi: 10.1105/tpc.020834
Sietsma, J. H., Eveleigh, D. E., and Haskins, R. H. (1969). Cell wall composition and protoplast formation of some oomycete species. Biochim. Biophys. Acta 184, 306–317. doi: 10.1016/0304-4165(69)90033-6
Singh, P., Kuo, Y.-C., Mishra, S., Tsai, C.-H., Chien, C.-C., Chen, C.-W., et al. (2012). The lectin receptor kinase-VI.2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. Plant Cell 24, 1256–1270. doi: 10.1105/tpc.112.095778
Stassen, J. H. M., Seidl, M. F., Vergeer, P. W. J., Nijman, I. J., Snel, B., Cuppen, E., et al. (2012). Effector identification in the lettuce downy mildew Bremia lactucae by massively parallel transcriptome sequencing. Mol. Plant Pathol. 13, 719–731. doi: 10.1111/j.1364-3703.2011.00780.x
Suzuki, N., Miller, G., Morales, J., Shulaev, V., Torres, M. A., and Mittler, R. (2011). Respiratory burst oxidases: the engines of ROS signaling. Curr. Opin. Plant Biol. 14, 691–699. doi: 10.1016/j.pbi.2011.07.014
Thines, M., and Kamoun, S. (2010). Oomycete-plant coevolution: recent advances and future prospects. Curr. Opin. Plant Biol. 13, 427–433. doi: 10.1016/j.pbi.2010.04.001
Thomma, B. P. H. J., Nürnberger, T., and Joosten, M. H. A. J. (2011). Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23, 4–15. doi: 10.1105/tpc.110.082602
Umemoto, N., Kakitani, M., Iwamatsu, A., Yoshikawa, M., Yamaoka, N., and Ishida, I. (1997). The structure and function of a soybean ß-glucan-elicitor-binding protein. Proc. Natl. Acad. Sci. U.S.A. 94, 1029–1034. doi: 10.1073/pnas.94.3.1029
Wang, G., Ellendorff, U., Kemp, B., Mansfield, J. W., Forsyth, A., Mitchell, K., et al. (2008). A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 147, 503–517. doi: 10.1104/pp.108.119487
Wang, Y., Bouwmeester, K., Beseh, P., Shan, W., and Govers, F. (2014). Phenotypic analyses of Arabidopsis T-DNA insertion lines and expression profiling reveal that multiple L-type lectin receptor kinases are involved in plant immunity. Mol. Plant Microbe Interact. 27, 1390–1402. doi: 10.1094/MPMI-06-14-0191-R
Wang, Y., Cordewener, J. H. G., America, A. H. P., Shan, W., Bouwmeester, K., and Govers, F. (2015a). Arabidopsis lectin receptor kinases LecRK-IX.1 and LecRK-IX.2 are functional analogs in regulating Phytophthora resistance and plant cell death. Mol. Plant Microbe Interact. 28, 1032–1048. doi: 10.1094/MPMI-02-15-0025-R
Wang, Y., Weide, R., Govers, F., and Bouwmeester, K. (2015b). L-type lectin receptor kinases in Nicotiana benthamiana and tomato and their role in Phytophthora resistance. J. Exp. Bot. 66, 6731–6743. doi: 10.1093/jxb/erv379
Wu, J., Liu, Z., Zhang, Z., Lv, Y., Yang, N., Zhang, G., et al. (2016). Transcriptional regulation of receptor-like protein genes by environmental stresses and hormones and their overexpression activities in Arabidopsis thaliana. J. Exp. Bot. 67, 3339–3351. doi: 10.1093/jxb/erw152
Yamaguchi, Y., and Huffaker, A. (2011). Endogenous peptide elicitors in higher plants. Curr. Opin. Plant Biol. 14, 351–357. doi: 10.1016/j.pbi.2011.05.001
Yamaguchi, Y., Huffaker, A., Bryan, A. C., Tax, F. E., and Ryan, C. A. (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22, 508–522. doi: 10.1105/tpc.109.068874
Yamaguchi, Y., Pearce, G., and Ryan, C. A. (2006). The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. U.S.A. 103, 10104–10109. doi: 10.1073/pnas.0603729103
Yoshikawa, M., Keen, N. T., and Wang, M. C. (1983). A receptor on soybean membranes for a fungal elicitor of phytoalexin accumulation. Plant Physiol. 73, 497–506. doi: 10.1104/pp.73.2.497
Yoshioka, H., Sugie, K., Park, H. J., Maeda, H., Tsuda, N., Kawakita, K., et al. (2001). Induction of plant gp91 phox homolog by fungal cell wall, arachidonic acid, and salicylic acid in potato. Mol. Plant Microbe Interact. 14, 725–736. doi: 10.1094/MPMI.2001.14.6.725
Zhou, B.-J., Jia, P.-S., Gao, F., and Guo, H.-S. (2012). Molecular characterization and functional analysis of a necrosis- and ethylene-inducing, protein-encoding gene family from Verticillium dahliae. Mol. Plant Microbe Interact. 25, 964–975. doi: 10.1094/MPMI-12-11-0319
Keywords: oomycete pathogens, pattern recognition, MAMP/DAMP, plant disease resistance, secreted proteins, extracellular recognition
Citation: Raaymakers TM and Van den Ackerveken G (2016) Extracellular Recognition of Oomycetes during Biotrophic Infection of Plants. Front. Plant Sci. 7:906. doi: 10.3389/fpls.2016.00906
Received: 03 May 2016; Accepted: 08 June 2016;
Published: 21 June 2016.
Edited by:
Pietro Daniele Spanu, Imperial College London, UKReviewed by:
Sebastian Schornack, University of Cambridge, UKCopyright © 2016 Raaymakers and Van den Ackerveken. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guido Van den Ackerveken, Zy52YW5kZW5hY2tlcnZla2VuQHV1Lm5s
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.