
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 22 June 2016
Sec. Plant Proteomics and Protein Structural Biology
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.00900
This article is part of the Research Topic Biology of plant fruit, seed and tuberous tissues: An important topic in food security View all 21 articles
Sugar beet is a species of the Chenopodiaceae family. It is an important sugar crop that supplies approximately 35% of the sugar in the world. Sugar beet M14 line is a unique germplasm that contains genetic materials from Beta vulgaris L. and Beta corolliflora Zoss. And exhibits tolerance to salt stress. In this review, we have summarized OMICS technologies and applications in sugar beet including M14 for identification of novel genes, proteins related to biotic and abiotic stresses, apomixes and metabolites related to energy and food. An OMICS overview for the discovery of novel genes, proteins and metabolites in sugar beet has helped us understand the complex mechanisms underlying many processes such as apomixes, tolerance to biotic and abiotic stresses. The knowledge gained is valuable for improving the tolerance of sugar beet and other crops to biotic and abiotic stresses as well as for enhancing the yield of sugar beet for energy and food production.
Sugar beet (Beta vulgaris. L), a species of Chenopodiaceae family, is an important sugar crop that supplies approximately 35% of the sugar in the world (Liu et al., 2008). In the United States, sugar beet has provided about 55 percent of the total sugar produced domestically since the mid-1990s (Benoit et al., 2015). Sugar beet was introduced to China from Arabia about 1500 years ago and it is a dicotyledonous plant with high economic value in many countries. Therefore, how to grow the crop efficiently has been a priority and extensively investigated (Draycott, 2006). Sugar beet is a biennial crop which grows a sugar-rich tap root in the first year (the vegetative stage) and a flowering seed stalk in the second year (the reproductive stage; Chen et al., 2016). The types of sugar beet can be distinguished according to various internal and external features, such as economic characters, trait diversity, and chromosome ploidy. Beta corolliflora Zoss. (2n = 36) is a wild species of the beet Corollinae section that has many characteristics including tolerance to drought, cold, salt and against disease. Sugar beets (Beta vulgaris) are classified as salt-tolerant crops (Dunajska-Ordak et al., 2014). Scientists have studied the interspecific crossing of cultivated sugar beet (Beta vulgaris L. 2n = 19) and B. corolliflora Zoss. for decades (Dalke et al., 1971; Filutowicz and Dalke, 1976). In our lab, Guo et al. obtained the sugar beet monosomic addition line M14 (Figure 1), which contains the Beta vulgaris L. genome with the addition of No. 9 chromosome of B. corolliflora Zoss (Guo et al., 1994). It has several interesting characteristics including apomixes and tolerance to drought, cold and salt stress (Guo et al., 1994). Apomixis is a mode of asexual reproduction characterized by the production of clonal seeds via the parthenogenesis development of an unreduced egg. The apomictic process bypasses meiosis and egg cell fertilization, producing offspring that are exact copies of the mother plant (Nogler, 1984; Ozias-Akins, 2006). Sugar beet M14 therefore can function as a unique germplasm for studying the characteristics of apomixes and tolerance to abiotic stresses.
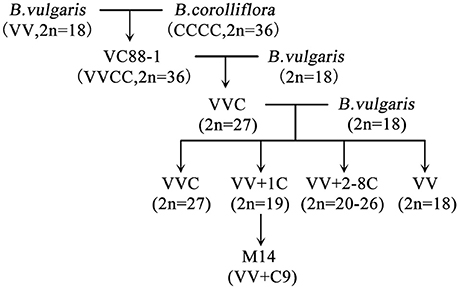
Figure 1. Generation of sugar beet M14 apomict (B. vulgaris genome plus chromosome No. 9 from B. corolliflora).
During evolution, plants have developed complex strategies that regulate biochemical and physiological acclimation in order to respond to biotic stress (viral, bacterial, fungal, and oomycete infections; Baum et al., 2007) and abiotic stress (salinity, drought, and low temperature; Barnabas et al., 2008; Yolcu et al., 2016). Biotic and abiotic stresses severely reduce agricultural productivity worldwide (Munns and Tester, 2008; Pinhero et al., 2011; Mishra et al., 2012). Therefore, understanding how plants respond and tolerate biotic and abiotic stresses is important for boosting plant (e.g., sugar beet) productivity under these challenging conditions. In order to minimize the negative impact of these stresses, studying how the sugar beet has evolved stress coping mechanisms will provide new insights and lead to novel strategies for improving the breeding of stress-resistant sugar beet and other crops.
In recent years, genomics knowledge based on Next Generation Sequencing (NGS), gene editing systems, gene silencing, and over-expression methods have provided a large amount of genetic information to help reveal the mechanisms of biotic and abiotic stress responses in plants (Saad et al., 2013; Shan et al., 2013; Yin et al., 2014; Luan et al., 2015). At the transcriptome level, technological innovations have made it possible to overview the changes that occur at the transcriptomic level under different environmental stress conditions. Microarrays and RNA sequencing techniques are employed to elucidate the differential expression of genes involved in biotic and abiotic stress responses in a variety of plant species (Kreps et al., 2002; Shinozaki and Yamaguchi, 2007; Ergen and Budak, 2009; Mitchell et al., 2014; Akpinar et al., 2015; Budak and Akpinar, 2015; Wang et al., 2016). Proteomics and metabolomics are two emerging “-omic” techniques in the post-genomic era (Fernandez-Garcia et al., 2011). Proteomics technologies allow the simultaneous identification and quantification of thousands of proteins that are an essential tool for understanding the biological systems and their regulations (Silva-Sanchez et al., 2015). Proteomics can be used to compare proteomes under varying stress conditions (Draycott, 2006; Liu et al., 2008; Benoit et al., 2015). Metabolomics focuses on the global profile of the low molecular weight (< 1000 Da) metabolites which are the end products of metabolisms in biofluids, tissues and even whole organism (Brosché et al., 2005). Metabolomics has recently been utilized in an increasing number of applications to investigate plant metabolite responses to abiotic stresses, particularly drought, flooding, salinity, and extreme temperatures (heat and cold; Jorge et al., 2015; Jia et al., 2016). Obviously, a combination of OMICS techniques including genomics, transcriptomics, proteomics and metabolomics could could serve to validate and complement one another in order to provide an efficient way capable of improving stress tolerance in plants.
Sugar beet is a good plant resource to explore and identify genes and proteins involved in stress resistance. Sugar beet is widely used in sugar industry (Liu et al., 2008). It is a source of the clean energy via hydrogen gas and bioethanol (Dhar et al., 2015). It contains abundant betaine and betalain metabolites. Betaine is used to improve the plant stress tolerance (Catusse et al., 2008). Betalains are natural pigments which have potential health benefits (anticarcinogenic and antioxidative) and have attracted both scientific and economic interest (Stintzing and Carle, 2007; Moreno et al., 2008). A rich and cheap source of betalains in red beet root (Beta vulgaris L.) is very attractive to the pharmaceutical and food industries (Wybraniec, 2005; Wybraniec et al., 2011, 2013). In this review, we have summarized OMICS applications and covered the recent discoveries in sugar beet research including the M14 for identification of novel genes and proteins related to biotic and abiotic stresses, apomixes, and metabolites related to energy and food production. The knowledge gained is valuable for improving sugar beet and other crops tolerance to biotic and abiotic stresses as well as for enhancing the yield of sugar beet for energy and food production.
In recent years, the use of OMICS tools has considerably increased for studying biotic and abiotic stresses in plants. The existing methods include genomics, transcriptomics, proteomics, metabolomics, and several others capable of discovering and characterizing the expression of genes or proteins during biotic and abiotic stresses with high efficiency shown in Figure 2. These highly sensitive tools can analyze plant tissues and help to improve our understanding of the tolerance mechanisms utilized by sugar beet.
The whole genome sequence of sugar beet has been reported by Dohm et al. (2014). A total of 27,421 protein-coding genes were predicted based on transcription data and annotated on the basis of sequence homology (Dohm et al., 2014). Compared to other flowering plants with the genome information, the sugar beet has a small number of genes encoding transcription factors. It has been suggested that the sugar beet may contain unknown genes associated with transcriptional control, and that the genetic interaction network of sugar beet may have evolved in unique ways compared with other species. Using the sugar beet genome sequence and related resources, we expected to find the molecular mechanisms underlying gene regulation and gene environment interaction. In addition, this information can help to develop crops with improved sugar and natural substance production and have an important role in future plant genomic research.
SMRT (Single Molecule Real-Time) is a third generation sequencing method, which offers much longer read length compared to NGS methods. It is well suited for de novo- or re-sequencing projects. It not only contains reads originating from the nuclear genome, but also lots of reads from the organelles of the target organism (Sanger et al., 1977; Liu et al., 2012). Stadermann et al. described a workflow for de novo assembly of the sugar beet chloroplast genome based on data originating from a SMRT sequencing dataset targeted on nuclear genome (Stadermann et al., 2015). They identified a total of 114 individual genes. Of these, 79 genes encode mRNA (i.e., proteins), 7 encode rRNA and 28 are tRNAs. Nine genes are located within the inverted repeat (IR) regions which encode 5 mRNAs, 1 rRNA, and 3 tRNAs. In comparison to the Illumina assembly, the annotation showed some differences due to changes in the underlying sequences.
miRNAs are small 19–23 nucleotides short non-coding RNAs, which play regulatory roles in many processes (Budak et al., 2015). miRNAs can act both at the transcriptional or post-transcriptional levels. miRNA mediated gene-silencing mechanism regulates the expression of transcription factors, phytohormones, and other developmental signaling pathways (Llave et al., 2002; Dalmay, 2006; Sunkar et al., 2007).
Earlier studies have shown that miRNAs mainly target transcription factors, but recent studies have revealed that miRNAs also target other development/stress signaling pathways, which are involved in various physiological processes, including root growth and development, response to stress, signal transduction, leaf morphogenesis, plant defenses, and biogenesis of sRNA (Curaba et al., 2014). Li J. L. et al. (2015) reported 13 mature miRNAs from 12 families using an in silico approach based on 29,857 expressed sequence tags and 279,223 genome survey sequences in B. vulgaris. The psRNA target server predicted 25 target genes for the 13 miRNAs. The target genes shown in Table 1 appeared to encode transcription factors or were involved in metabolism, signal transduction, stress response, growth, and development. However, there were no targets predicted from the current database of sugar beet for Bvu-miR4, Bvu-miR9, Bvu-miR10, Bvu-miR11, and Bvu-miR12.
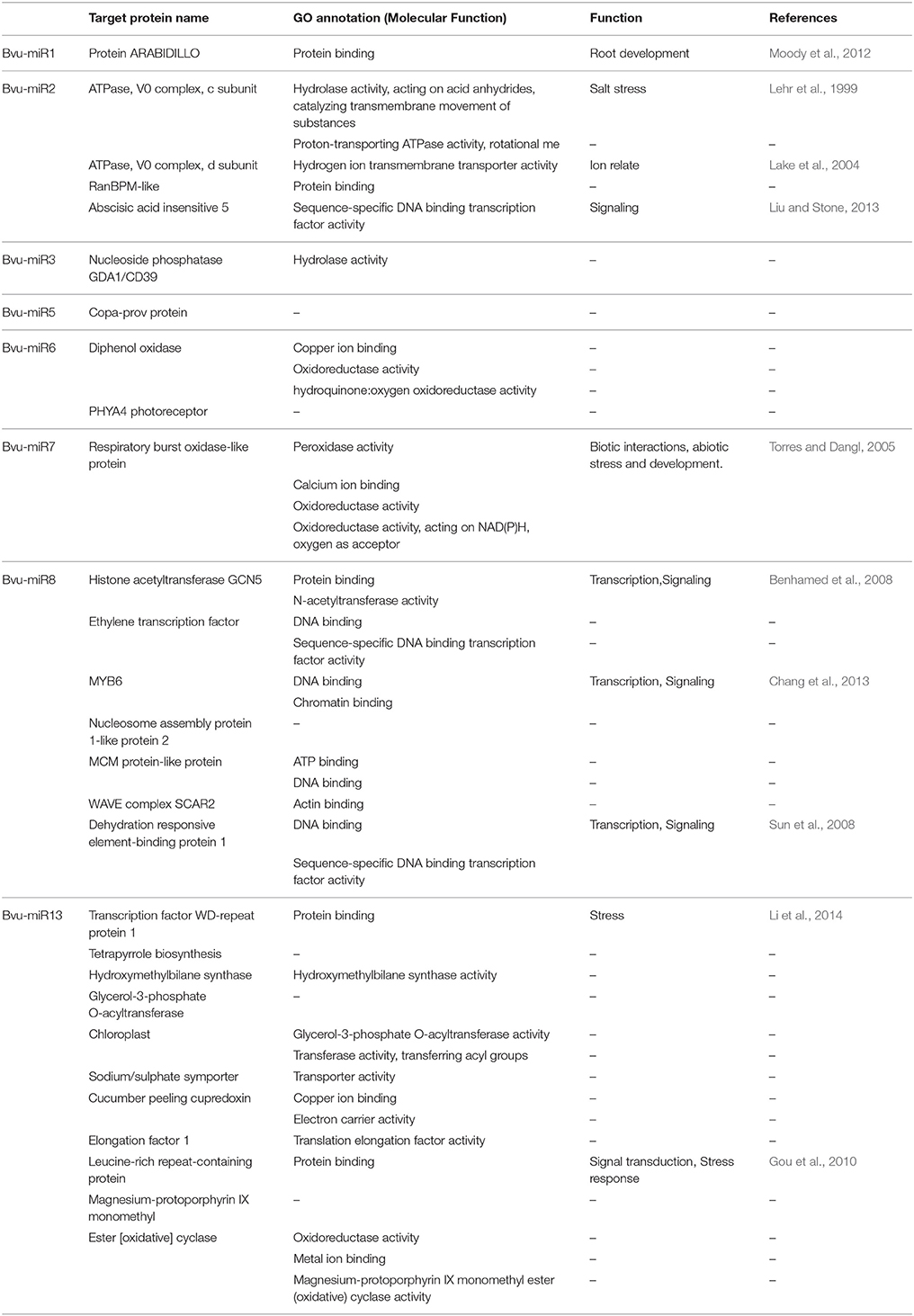
Table 1. Predicted functions of sugar beet miRNAs in plant development, signaling and stress responses.
Several miRNAs identified have been shown to have critical roles in plants. For example, the expression of Bvu-miR1 (Protein ARABIDILLO) in A. thaliana regulates multicellular root development (Moody et al., 2012). Bvu-miR2 regulates the expression of ATPase during plant development and coordinates its induction in response to high salinity (Lehr et al., 1999). Through transcriptional regulation, it also affects the ATPase activity of magnesium chelatase subunit I in Barley (Lake et al., 2004) and abscisic acid insensitive 5 required to delay growth of germinated seedlings under environmental stress (Liu and Stone, 2013). Bvu-miR7 targets the respiratory burst oxidase gene family, which encodes the key enzymatic subunit of the plant NADPH oxidase (Torres and Dangl, 2005). Bvu-miR8 activates the transcription of a histone acetyltransferase GCN5 in A. thalianaina (Benhamed et al., 2008). Another target protein MYB6 acts as an immediate and positive activation signaling component of the active state of MLA immune receptors during transcriptional reprogramming for defense responses (Chang et al., 2013). Dehydration-responsive element-binding proteins form a major AP2/ethylene- responsive element-binding protein family and play crucial roles in the regulation of abiotic stress responses (Sun et al., 2008). Bvu-miR13 targets WD-repeat proteins in this diverse family of regulatory proteins. To date, genome-wide characterization of this family has only been conducted in Arabidopsis and little is known about WD-repeat protein-coding genes in other species. Recently, it has become known that the WD-repeat protein plays an important role in cucumber stress resistance (Li et al., 2014). Other targets, e.g., leucine-rich repeat proteins, plays critical roles in both animal and plant signaling pathways regulating growth, development, differentiation, cell death, and pathogenic defense responses (Gou et al., 2010). These studies have provided insights into the molecular mechanisms of the miRNAs and may have great potential for sugar beet improvement. The functions of these interesting miRNAs in sugar beet need to be investigated in the future.
Leaf spot is one of the most serious and widespread foliar diseases of sugar beet. It causes necrotic lesions and progressive destruction of the plant's foliar structure and function (Holtschulte, 2000). The disease has greatly impacted on the yield and sugar contents of the crop. Doubled haploids (DHs), F2 populations of recombinant inbred lines (RILS), and near isogenic lines (NILS) are suitable populations for quantitative trait loci (QTL) mapping (Ibrahim et al., 2012). In order to deal with the complex inheritance of resistance to Cercospora leaf spot (CLS), Taguchi et al. (2011) used RILs of sugar beet, which were generated by a cross between a resistant line (“NK-310mm-O”) and a susceptible (“NK-184mm-O”) line. These RILs were then tested for their resistance to the CLS pathogen in the field (Taguchi et al., 2011). Composite interval mapping (CIM) showed four QTLs involved in CLS resistance that were consistently detected. There were two resistant QTLs (qcr1 on chromosome III, qcr4 on chromosome IX) that promoted resistance in the cross between lines (“NK-310mm-O”). There were two further QTLs (qcr2 on chromosome IV, qcr3 on chromosome VI) which promoted resistance in the susceptible line. In addition, a number of important resistance gene cluster have been mapped on the chromosome III in the sugar beet genome, for example: a CLS resistance QTL (Setiawan et al., 2000), the genes Rz1 toRz5 (Grimmer et al., 2007), gene Acr1 (Taguchi et al., 2010), gene RGAs (Lein et al., 2007), and gene X, a restorer of fertility for Owen CMS (Hagihara et al., 2005). These resistance-gene clusters on the chromosome III are mainly responsible for the disease resistance in sugar beet.
A plant-transformation-competent binary BAC library was constructed from the genomic DNA of the No. 9 chromosome in sugar beet M14 (Fang et al., 2004). A total of 2365 positive clones were obtained and arrayed into a sublibrary specific for the B. corolliflora chromosome 9 (designated bcBAC-IX). The bcBAC-IX sublibrary was further screened with a subtractive cDNA pool generated from the ovules of M14 and the floral buds of B. vulgaris by the suppression subtractive hybridization (SSH) method. One hundred and three positive binary BACs were obtained, which may potentially contain the genes of the alien No. 9 chromosome that is specifically expressed during the ovule and embryo development of M14, and which may be associated with apomictic reproduction. The binary BAC clones are useful for the identification of the genes responsible for apomixes by genetic transformation.
SSH is a technique used to identify differentially expressed genes in cells important for growth and differentiation (Lukyanov et al., 2007). This method has often been used to study molecular mechanisms of plants in biotic and abiotic stresses (Sahebi et al., 2015). The response to insect pests in the root of sugar beet is an interesting area of plant defense research.
Puthoff et al have identified more than 150 sugar beet root ESTs enriched for genes that respond to feeding by the sugar beet root maggot in both the moderately resistant genotype F1016 and the susceptible F1010 using SSH [49]. The differential expression of the root ESTs was confirmed via RT-PCR. The ESTs were further characterized using microarray-generated expression profiles from the F1016 sugar beet roots following mechanical wounding and treatment with the signaling molecules methyl jasmonate, salicylic acid and ethylene. Of the examined ESTs, 20% were regulated by methyl jasmonate, 17% by salicylic acid and 11% by ethylene, suggesting that these signaling pathways are involved in sugar beet root defense response. Identification of these sugar beet root ESTs provides knowledge concerning plant root defense and will likely lead to the development of novel strategies for the control of the sugar beet root maggot (Puthoff and Smigocki, 2007).
SSH was applied to isolating taproot expressed genes from sugar beet as well (Kloos et al., 2002). The taproot of sugar beet (Beta vulgaris L.) undergoes a specific developmental transition in order for it to function as a storage organ. SSH was utilized to isolate cDNA fragments of genes expressed in the taproot. Molecular analysis of six cDNAs that encoded complete gene products revealed that these genes comprise homologs of a drought-inducible linker histone, a major latex-like protein, a phosphoenolpyruvate carboxylase kinase, a putative vacuolar processing enzyme, a thaumatin-like protein and an alanine- and glutamic acid-rich protein. All of these genes are transcribed in taproots, while the expression in leaves is low or undetectable. SSH had also been used in the sugar beet M14 to identify differentially expressed genes. A subtractive cDNA library was prepared by SSH between the flower organ of M14 and that of B. vulgaris (Ma et al., 2011). A total of 190 unique sequences were identified in the library and their putative functions were analyzed using Gene Ontology (GO). All of the ESTs provide information about candidate genes useful for studying M14 reproductive development. One of the genes, designated as BvM14-MADS box, encodes a MADS box transcription factor. It was cloned from M14 and over-expressed in transgenic tobacco plants. Overexpression of BvM14-MADS box led to significant phenotypic changes in tobacco (Ma et al., 2011).
Li et al. (2009) reported a comparative proteomic and transcriptomic study of the sexual and apomictic processes in sugar beet. The cDNA libraries were constructed using SSH with the apomictic monosomic addition line M14 as the tester and B. vulgaris as the driver. Comparative analyses of proteomic data and transcriptomic data showed that eight proteins had significant agreement between protein and mRNA expression levels. Most of the matched proteins were associated with metabolism. Interestingly, two of the matched proteins, cystatin, and thioredoxin peroxidase, were found to be associated with disease and defense response, indicating that defense-related proteins may participate in the apomictic reproductive process. Yang et al. (2012) reported transcriptomic analysis of sugar beet M14 leaves and roots that were treated with 500 mM NaCl for 7 days. The SSH technology was used to produce a high quality subtractive cDNA library. A total of 600 positive clones were randomly selected and subjected to DNA sequencing, and 499 non-redundant ESTs were obtained. After assembly, 58 unigenes including 14 singletons and 44 contigs were obtained. Some salt-responsive genes were identified as important in metabolism (e.g., sadenosylmethionine synthase 2 (SAMS2) and nitrite reductase), photosynthesis (e.g., chloroplastic chlorophyll a–b binding protein 8), energy (e.g., phosphoglycerate kinase), protein synthesis (e.g., 60S ribosomal protein L19-3), and degradation (e.g., cysteine protease and carboxyl-terminal-processing protease), and stress and defense [e.g., glutathione S-transferase (GST)]. This study has revealed candidate genes for detailed functional characterization, and has set the stage for further investigation of salt tolerance mechanisms in sugar beet.
Comparative transcriptomics is used to identify differences in transcript abundance between different cultivars, organs, development stages and/or treatment conditions (Mardis, 2008; Schuster, 2008; Bräutigam and Gowik, 2010). Low-temperature stress is a significant factor effecting of crop quality and causing production losses in agriculture. The survival of young sugar beet seedlings and the subsequent sugar yield of mature plants are often seriously limited by low temperature, especially when the plants are exposed to freezing temperatures at early developmental stages. Moliterni et al. (2015) determined the transcriptomic changes using high-throughput sequencing of the leaves and root RNAs (RNA-Seq) from sugar beets which had been exposed to cold stress which mimicked the conditions of spring nights sometimes experienced by young seedlings (Moliterni et al., 2015). In the root tissue, CBF3 is up-regulated within a few minutes of cold stress. The authors suggested that CBF3 transcription in the stressed plants is either maintained for a longer period or begins earlier in roots compared to leaves. The AP2/ERF family genes were also found to be either activated or up-regulated in all the organs by cold stress. This is an expected result, as it is known that these TFs are rapidly induced upon exposure to low temperature in Arabidopsis (Lee et al., 2005). AP2/ERF TFs are involved in the regulation of primary and specialized metabolism and in a number of JA responses (Licausi et al., 2013). It has been reported that the lack of ADA2b TFs leads to an increase of freezing tolerance by affecting nucleosome occupancy in Arabidopsis (Vlachonasios et al., 2003). In addition, a putative histone acetylase and a lysine-specific demethylase are strongly up-regulated in the leaves under cold stress, implicating chromatin remodeling, and modification in the response.
These studies suggested that the metabolic pathway most affected by low temperature was carbohydrate metabolism. In addition, the authors found 13 differentially expressed sequences related to phospholipid secondary metabolism, none of which were common to leaves and roots, implicating this pathways as another important component in early cold signaling in sugar beet roots. The high degree of organ specificity is probably due to the repertoire of compounds synthesized by the two organs upon stress. This data has illuminated the transcriptome of young sugar beet during cold stress at night, and has detailed both organ-specificity and shared pathways in the physiological response to low temperatures. These RNA-Seq based transcriptomics techniques are an effective and powerful tool, with the analyses identifying novel genes for future studies.
Proteomic analysis has been carried out to address several important questions in many processes, such as: signaling, regulatory processes, and transport in plants (Zhang et al., 2008). Important knowledge of the proteomic response to stress has been mainly derived from studies of the model plants A. thaliana and rice (Janmohammadi et al., 2015; Liu et al., 2015; Xu et al., 2015). Proteomic analysis provides an important way to test the changes in protein levels to help identify novel proteins.
Zhu et al. (2009) compared the proteomes of the monosomic addition line M14 and B. vulgaris using 2-DE (two-dimensional gel electrophoresis). They have identified 27 protein spots using MALDI-TOF MS. Among them, only two protein spots were found in B. vulgaris and five protein spots were unique to M14. These proteins were involved in many biological pathways. The results may be useful for us to better understand how genotype differences relate to proteome and phenotype differences.
Li et al. (2009) reported a comparative proteomic and transcriptomic analysis of the sexual and apomictic processes in sugar beet. A total of 71 differentially expressed protein spots from the floral organs of the M14 were identified in the course of apomictic reproductive development using 2-DE and MS analysis. The differentially expressed proteins were involved in several processes which may work cooperatively to promote apomictic reproduction, generating new potential protein markers important for apomictic development.
To date, some proteomic studies concerning the response of sugar beet to salt stress have been reported. Wakeel et al. identified six proteins from sugar beet shoots and three proteins from roots that significantly changed under 125 mM salt treatment (Wakeel et al., 2011). Our group has performed proteomic analysis of the monosomic addition line M14 under 500 mM salt stress for 7 days. A total of 71 differentially expressed protein 2D spots were identified using LC-MS/MS. The largest functional group is represented by metabolism (28%), followed by energy (21%), protein synthesis (10%), stress and defense (10%), destination proteins (8%), unknown proteins (8%), secondary metabolism (5%), signal transduction (4%), transporters (1%), and cell division (1%). Of the identified proteins, only eight had corresponding transcriptomic data. This highlights the importance of expression profiling at the protein level (Li et al., 2009). On this basis, we focused on the functions of cystatin (Wang et al., 2012), glyoxalase I (Wu et al., 2013), CCoAOMT, and thioredoxin peroxidase. All of these proteins showed increased protein levels under salt stress. Transgenic plants exhibited enhanced tolerance to salt stress. This research has directly improved our understanding of mechanisms underlying the M14's high salt tolerance.
Another proteomics study aims to identify salt-responsive proteins in the M14 plants under 0, 200, and 400 mM NaCl mild salt stress conditions using 2D-DIGE to separate the proteins from control and salt-treated M14 leaves and roots (Yang et al., 2013). The differentially expressed proteins were identified using nanoflow liquid chromatography (LC)−MS/MS and Mascot database searching. As a complementary approach, iTRAQ LC−MS/MS was employed to identify and quantify differentially expressed proteins during salinity response in M14. We have identified 86 protein spots representing 67 unique proteins in leaves, and 22 protein spots representing 22 unique proteins in roots. In addition, 75 differentially expressed proteins were identified in leaves and 43 differentially expressed proteins were identified in roots, respectively. The proteins were mainly involved in photosynthesis, energy, metabolism, protein folding and degradation, and stress and defense. Compared to the transcriptomic data, 13 proteins in leaves and 12 proteins in roots showed significant correlation in gene expression and protein levels. These results suggest that there are several processes underlying the M14 tolerance to salt stress.
Our group also reported the changes in membrane proteome of the M14 plants in response to salt stress (0, 200, 400 mM NaCl; Li H. et al., 2015). We have used an iTRAQ two-dimensional LC–MS/MS technology for quantitative proteomic analysis. In total, 274 proteins were identified and mostly of them were membrane proteins. A total of 50 differential proteins were identified, with 40 proteins showing increased expression and 10 with decreased expression. The proteins were mainly involved in transport (17%), metabolism (16%), protein synthesis (15%), photosynthesis (13%), protein folding and degradation (9%), signal transduction (6%), stress and defense (6%), energy (6%), and cell structure (2%). These results have revealed that membrane proteins contribute to the salt stress tolerance observed in M14.
Hajheidari et al. (2005) studied the proteome changes of sugar beet in response to drought stress. Leaves from well-watered and drought treated plants at 157 days after sowing were collected. The changes of proteins were analyzed using 2D-DIGE followed by image analysis. More than 500 protein spots were detected, and 79 spots had significant changes under drought stress. Twenty protein spots were digested and subjected to LC-MS/MS, and 11 proteins involved in oxidative stress, signal transduction and redox regulation were identified. These proteins may be important targets for improving plant abiotic stress tolerance via breeding.
Metabolomics is an exciting technology, which was used to identify secondary metabolites important for physiological processes and different stress responses (Capuano et al., 2013). During plant development and interaction with the environment, the dynamic metabolome reflects the plant's physiological and biochemical processes, and can determines the phenotypes and traits (Fernie et al., 2004; Oksman-Caldentay and Saito, 2005). Now there are gas chromatography (GC)–MS, LC–MS, capillary electrophoresis (CE)–MS, and nuclear magnetic resonance (NMR) as the major analytical tools in metabolomics. Kazimierczak et al. (2014) determined the levels of metabolites in both raw beet root and naturally fermented beet root juices from organic (ORG) vs. conventional (CONV) products. The aim of the paper was to find out the value of the fermented beetroot juices in terms of anticancer properties. The results showed that ORG fresh beetroots contained more useful compounds than CONV beetroots, such as dry matter and vitamin C, more than CONV beetroots. Compared to the CONV juice, it was found that the ORG fermented juices have stronger anticancer activity. Metabolomics is still in its infancy with these analyses of sugar beet being rare, but future research can be expected to implement this powerful technology.
Many plants accumulate glycine betaine (betaine) to regulate biochemical and physiological processes under abiotic stresses (Takabe et al., 2006). For example, glycine betaine serves as a methyl donor in several biochemical pathways (Pummer et al., 2000). Sugar beet is a betaine-accumulating dicotyledonous plant with high economic value (Catusse et al., 2008). It has been reported that betaine is synthesized by the two-step oxidation of choline in which choline monooxygenase (CMO) catalyzes the first step, and betaine aldehyde dehydrogenase (BADH) performs the second step (Yamada et al., 2009). CMO is therefore a key enzyme to protect plants against abiotic stresses. It has been found in Chenopodiaceae and Amaranthaceae, but not in some betaine-accumulating plants such as mangrove (Bhuiyan et al., 2007). Unlike sugar beet, many plants do not have the betaine biosysthesis pathway. Therefore, genetic engineering of the betaine biosynthesis pathways represents a potential way to improve the plant stress tolerance (Hibino et al., 2001; Fitzgerald et al., 2009).
In addition to betaine, betalains are rich in red beets and exist only in 10 families of plants of the Caryophyllale. Red beetroot (B. vulgaris) is widely used as a food ingredient because of its beetroot red color. Therefore, most studies on red beetroot constituents have focused on the betalains. Currently, the yield of betalains extracted and purified from the beetroot red is only about 10%. Phenolics (such as betalains) have been shown to have nutritional value and there has been an increasing interest utilizing these plant constituents to improve food ingredients and as antioxidants. Betalains are important plant phenolics with many attractive properties, such as: stability, antioxidant activity, antitumor properties, and reduction of blood lipid and sugar levels. In addition, betalains are effective free-radical scavengers, which help to maintain health and protect from diseases such as cancer and coronary heart disease (Kujala et al., 2000; Han et al., 2015; Mikołajczyk-Bator et al., 2016).
Additonally, sugar beet provides approximately 30% of the world's annual sugar production and is a source of both bioethanol and animal feed. Dhar et al. (2015) have developed two highly efficient methods to produce hydrogen gas from sugar beet juice as a clean energy source (Dhar et al., 2015). Sugar beet byproducts (SBB) generated during industrial sugar extraction are mainly composed of pulp and molasses and the use of SBB as a renewable energy resource could add additional economic and environmental benefits (Aboudi et al., 2015).
OMICS research can greatly enhance potential applications of sugar beet in at least three ways. One is to improve our knowledge of molecular networks involving key metabolite synthesis, e.g., glyceine betaine and betalains. The knowledge will enable modeling and rationale engineering of the important metabolites. Additionally, we can utilize OMICS to investigate the global molecular changes that occur in response to stress and the tolerance of sugar beet to stress conditions. This information can help to improve stress tolerance and thereby yields of sugar beet, even under non-ideal conditions. Finally, research on unique sugar beet germplasms (e.g., M14 under salt stress) may be useful for enhancing yield, and food and bioenergy production in other crops.
In this review, we have summarized OMICS technologies and applications in sugar beet including: M14 for identification of novel genes, proteins related to biotic and abiotic stresses, apomixes, and metabolites related to energy, food and human health. Genomics is a powerful technology to provide the whole genome blueprint of sugar beet. Mechanisms underlying apomixes and stress tolerance have mainly been studied using transcriptomics and proteomics technologies, while metabolomics studies in sugar beet are still rare. To date, a lot of genes and proteins related to apomixes and salt stress were identified to reveal apomixes and salt tolerance mechanisms in a special germplasm sugar beet M14. The results have enhanced our understanding of the molecular mechanisms of sugar beet in response to tolerance to biotic and abiotic stresses and apomixes, which may be applied to improving stress tolerance of sugar beet and other crops to improve food production, energy output (e.g., hydrogen gas and bioethanol), and accumulation of health promoting chemicals (such as betalains). Despite the use of sugar beet to produce the clean energy of hydrogen gas and bioethanol and to isolate betalains used for natural food colorants, dietary supplements and medicines were not widely applied in market, sugar beet as a high economic value crop will have a prosperous perspective of application in the food, bioenergy and pharmacy industries.
YZ collected and analyzed references for this paper, and wrote the first draft, JN drew the figure and assisted in the reference organization. BY played a supervision role, and led the writing and organization. All three authors have edited the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was supported by the National Science Foundation of China (Project 31471552: The response of antioxidant enzymes to salt stress in sugar beet M14, and Project 31501359: Study on phosphorylation sites of a Ser/Thr protein kinase from sugar beet M14 line in response to salt stress) and 100 Academic Backbone Support Program for Young Teachers of Heilongjiang University in 2016 and the Common College Science and Technology Innovation Team of Heilongjiang Province (2014TD004). The paper represents serial 016 from our innovation team at the Heilongjiang University (Hdtd2010-05). We thank Joe Collins from Plant Molecular and Cellular Biology, University of Florida for critical reading of the manuscript.
Aboudi, K., Álvarez-Gallego, C. J., and Romero-García, L. I. (2015). Semi-continuous anaerobic co-digestion of sugar beet byproduct and pig manure: effect of the organic loading rate (OLR) on process performance. Bioresour. Technol. 194, 283–290. doi: 10.1016/j.biortech.2015.07.031
Akpinar, B. A., Kantar, M., and Budak, H. (2015). Root precursors of microRNAs in wild emmer and modern wheats show major differences in response to drought stress. Funct. Integr. Genomics 15, 587–598. doi: 10.1007/s10142-015-0453-0
Barnabás, B., Jäger, K., and Fehár, A. (2008). The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 31, 11–38. doi: 10.1111/j.1365-3040.2007.01727.x
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteraninsect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. doi: 10.1038/nbt1359
Benhamed, M., Martin-Magniette, M. L., Taconnat, L., Bitton, F., Servet, C., De Clercq, R., et al. (2008). Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J. 56, 493–504. doi: 10.1111/j.1365-313X.2008.03606.x
Benoit, I., Zhou, M., Vivas Duarte, A., Downes, D. J., Todd, R. B., Kloezen, W., et al. (2015). Spatial differentiation of gene expression in Aspergillus niger colony grown for sugar beet pulputilization. Sci Rep. 5:13592. doi: 10.1038/srep13592
Bhuiyan, N. H., Hamada, A., Yamada, N., Rai, V., Hibino, T., and Takabe, T. (2007). Regulation of betaine synthesis by precursor supply and choline monooxygenase expression in Amaranthus tricolor. J. Exp. Bot. 58, 4203–4212. doi: 10.1093/jxb/erm278
Bräutigam, A., and Gowik, U. (2010). What can next generation sequencing do for you? Next generation sequencing as a valuable tool in plant research. Plant Biol. 12, 831–841. doi: 10.1111/j.1438-8677.2010.00373.x
Brosché, M., Vinocur, B., Alatalo, E. R., Lamminmäki, A., Teichmann, T., Ottow, E. A., et al. (2005). Gene expression and metabolite profiling of Populus euphratica growing in the Negev desert. Genome Biol. 6:R101. doi: 10.1186/gb-2005-6-12-r101
Budak, H., and Akpinar, B. A. (2015). Plant miRNAs: biogenesis, organization and origins. Funct. Integr. Genomics 15, 523–531. doi: 10.1007/s10142-015-0451-2
Budak, H., Kantar, M., Bulut, R., and Akpinar, B. A. (2015). Stress responsive miRNAs and isomiRs in cereals. Plant Sci. 235, 1–13. doi: 10.1016/j.plantsci.2015.02.008
Capuano, E., Boerrigter-Eenling, R., van der Veer, G., and van Ruth, S. M. (2013). Analytical authentication of organic products: an overview of markers. J. Sci. Food Agric. 93, 12–28. doi: 10.1002/jsfa.5914
Catusse, J., Strub, J. M., Job, C., Dorsselaer, A., and Job, D. (2008). Proteome-wide characterization of sugarbeet seed vigor and its tissue specific expression. Proc. Natl. Acad. Sci. U.S.A. 105, 10262–10267. doi: 10.1073/pnas.0800585105
Chang, C., Yu, D., Jiao, J., Jing, S., Schulze-Lefert, P., and Shen, Q. H. (2013). Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell 25, 1158–1173. doi: 10.1105/tpc.113.109942
Chen, T., Li, Z., Yin, X., Hu, F., and Hu, C. (2016). Discrimination of genetically modified sugar beets based on terahertz spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 153, 586–590. doi: 10.1016/j.saa.2015.09.028
Curaba, J., Singh, M. B., and Bhalla, P. L. (2014). miRNAs in the crosstalk between phytohormone signalling pathways. J. Exp. Bot. 65, 1425–1438. doi: 10.1093/jxb/eru002
Dalke, L., Filutowicz, A., and Pawelska-Koziuska, K. (1971). Results of investigations on hybrids between tetraploid monogerm sugar beet and Beta corolliftora 2n-36. Biul. Inst. Hod. Aklim. Rośl. 6, 3–7.
Dalmay, T. (2006). Short RNAs in environmental adaptation. Proc. Biol. Sci. 273, 1579–1585. doi: 10.1098/rspb.2006.3516
Dhar, B. R., Elbeshbishy, E., Hafez, H., and Lee, H. S. (2015). Hydrogen production from sugar beet juice using an integrated biohydrogen process of dark fermentation and microbial electrolysis cell. Bioresour Technol. 198, 223–230. doi: 10.1016/j.biortech.2015.08.048
Dohm, J. C., Minoche, A. E., Holtgräwe, D., Capella-Gutiérrez, S., Zakrzewski, F., Tafer, H., et al. (2014). The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505, 546–549. doi: 10.1038/nature12817
Dunajska-Ordak, K., Skorupa-Kłaput, M., Kurnik, K., Tretyn, A., and Tyburski, J. (2014). Cloning and expression analysis of a gene encoding for ascorbate peroxidase and responsive to salt stress in Beet (Beta vulgaris). Plant Mol. Biol. Rep. 32, 162–175. doi: 10.1007/s11105-013-0636-6
Ergen, N. Z., and Budak, H. (2009). Sequencing over 13 000 expressed sequence tags from six subtractive cDNA libraries of wild and modern wheats following slow drought stress. Plant Cell Environ. 32, 220–236. doi: 10.1111/j.1365-3040.2008.01915.x
Fang, X., Gu, S., Xu, Z., Chen, F., Guo, D., Zhang, H. B., et al. (2004). Construction of a binary BAC library for an apomictic monosomic addition line of Beta corolliflora in sugar beet and identification of the clones derived from the alien chromosome. Theor. Appl. Genet. 108, 1420–1425. doi: 10.1007/s00122-003-1566-8
Fernandez-Garcia, N., Hernandez, M., Casado-vela, J., Bru, R., Elortza, F., Hedden, P., et al. (2011). Changes to the proteome and targeted metabolites of xylemsapin Brassica oleracea in response to salt stress. Plant Cell Environ. 34, 821–836. doi: 10.1111/j.1365-3040.2011.02285.x
Fernie, A. R., Trethewey, R. N., Krotzky, A. J., and Willmitzer, L. (2004). Metabolite profiling: from diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5, 763–769. doi: 10.1038/nrm1451
Filutowicz, A., and Dalke, L. (1976). Interspecific hybrids in the Corollinae section Genus Beta. Hod. Rosl. Aklim. Nas. 20, 1–17.
Fitzgerald, T. L., Waters, D. L. E., and Henry, R. J. (2009). Betaine aldehyde dehydrogenase in plants. Plant Biol. 11, 119–130. doi: 10.1111/j.1438-8677.2008.00161.x
Gou, X., He, K., Yang, H., Yuan, T., Lin, H., Clouse, S. D., et al. (2010). Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genomics 11:19. doi: 10.1186/1471-2164-11-19
Grimmer, M. K., Trybush, S., Hanley, S., Francis, S. A., Karp, A. Asher, M. J. C., et al. (2007). An anchored linkage map for sugar beet based on AFLP, SNP and RAPD markers and QTL mapping of a new source of resistance to Beet necrotic yellow vein virus. Theor. Appl. Genet. 114, 1151–1160. doi: 10.1007/s00122-007-0507-3
Guo, D., Wang, G., Kang, C., Jia, S., and Liu, L. (1994). Studies on the interspecific crossing of cultivated sugar beet and Beta corolliflora Zoss. China Sugarbeet 3, 2–7.
Hagihara, E., Itchoda, N., Habu, Y., Iida, S., Mikami, T., and Kubo, T. (2005). Molecular mapping of a fertility restorer gene for Owen cytoplasmic male sterility in sugar beet. Theor. Appl. Genet. 111, 250–255. doi: 10.1007/s00122-005-2010-z
Hajheidari, M., Abdollahian-Noghabi, M., Askari, H., Heidari, M., Sadeghian, S. Y., Ober, E. S., et al. (2005). Proteome analysis of sugar beet leaves under drought stress. Proteomics 5, 950–960. doi: 10.1002/pmic.200401101
Han, J., Tan, C., Wang, Y., Yang, S., and Tan, D. (2015). Betanin reduces the accumulation and cross-links of collagen in high-fructose-fed rat heart through inhibiting non-enzymatic glycation. Chem. Biol. Interact. 5, 37–44. doi: 10.1016/j.cbi.2014.12.032
Hibino, T., Meng, Y. L., Kawamitsu, Y., Uehara, N., Matsuda, N., Tanaka, Y., et al. (2001). Molecular cloning and functional characterization of two kinds of betaine-aldehyde dehydrogenase in betaine-accumulating mangrove Avicennia marina (Forsk). Plant Mol. Biol. 45, 353–363. doi: 10.1023/A:1006497113323
Holtschulte, B. (2000). “Cercospora beticola-worldwide distribution and incidence,” in Cercospora Beticola Sacc. Biology, Agronomic Influence and Control Measures in Sugar Beet, eds M. J. C. Asher, B. Holtschulte M. M. Richard, F. Rosso, G. Steinrücken, and R. Beckers (Belgium: International Institute for Beet Research), 5–16.
Ibrahim, S. E., Schubert, A., Pillen, K., and Léon, J. (2012). Comparison of QTLs for drought tolerance traits between two advanced backcross populations of spring wheat. Int. Acad. J. 2, 216–227.
Janmohammadi, M., Zolla, L., and Rinalducci, S. (2015). Low temperature tolerance in plants: changes at the protein level. Phytochemistry 117, 76–89. doi: 10.1016/j.phytochem.2015.06.003
Jia, X., Sun, C., Zuo, Y., Li, G., Li, G., Ren, L., et al. (2016). Integrating transcriptomics and metabolomics to characterise the response of Astragalus membranaceus Bge. var. mongolicus (Bge) to progressive drought stress. BMC Genomics 17:188. doi: 10.1186/s12864-016-2554-0
Jorge, T. F., Rodrigues, J. A., Caldana, C., Schmidt, R., van Dongen, J. T., Thomas-Oates, J., et al. (2015). Mass spectrometry-based plant metabolomics: metabolite responses to abiotic stress. Mass Spectrom. Rev. doi: 10.1002/mas.21449. [Epub ahead of print].
Kazimierczak, R., Hallmann, E., Lipowski, J., Drela, N., Kowalik, A., Püssa, T., et al. (2014). Beetroot (Beta vulgaris L.) and naturally fermented beetroot juices from organic and conventional production: metabolomics, antioxidant levels and anticancer activity. J. Sci. Food Agric. 94, 2618–2629. doi: 10.1002/jsfa.6722
Kloos, D. U., Oltmanns, H., Dock, C., Stahl, D., and Hehl, R. (2002). Isolation and molecular analysis of six taproot expressed genes from sugar beet. J. Exp. Bot. 53, 1533–1534. doi: 10.1093/jexbot/53.373.1533
Kreps, J. A., Wu, Y. J., Chang, H. S., Zhu, T., Wang, X., and Harper, J. F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130, 2129–2141. doi: 10.1104/pp.008532
Kujala, T. S., Loponen, J. M., Klika, K. D., and Pihlaja, K. (2000). Phenolics and betacyanins in red beetroot (Beta vulgaris) root: distribution and effect of cold storage on the content of total phenolics and three individual compounds. J. Agric. Food Chem. 48, 5338–5342. doi: 10.1021/jf000523q
Lake, V., Olsson, U., Willows, R. D., and Hansson, M. (2004). ATPase activity of magnesium chelatase subunit I is required to maintain subunit D in vivo. Eur. J. Biochem. 271, 2182–2188. doi: 10.1111/j.1432-1033.2004.04143.x
Lee, B., Henderson, D. A., and Zhu, J. (2005). The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17, 3155–3175. doi: 10.1105/tpc.105.035568
Lehr, A., Kirsch, M., Viereck, R., Schiemann, J., and Rausch, T. (1999). cDNA and genomic cloning of sugar beet V-type H+-ATPase subunit A and c isoforms: evidence for coordinate expression during plant development and coordinate induction in response to high salinity. Plant Mol. Biol. 39, 463–475. doi: 10.1023/A:1006158310891
Lein, J. C., Asbach, K., Tian, Y., Schulte, D., Li, C., Koch, G., et al. (2007). Resistance gene analogues are clustered on chromosome 3 of sugar beet and cosegregate with QTL for Rhizomania resistance. Genome 50, 61–71. doi: 10.1139/G06-131
Li, H., Cao, H., Wang, Y., Pang, Q., Ma, C., and Chen, S. (2009). Proteomic analysis of sugar beet apomictic monosomic addition line M14. J. Proteomics 73, 297–308. doi: 10.1016/j.jprot.2009.09.012
Li, H., Pan, Y., Zhang, Y., Wu, C., Ma, C., Yu, B., et al. (2015). Salt stress response of membrane proteome of sugar beet monosomic addition line M14. J. Proteomics 127(Pt A), 18–33. doi: 10.1016/j.jprot.2015.03.025
Li, J. L., Cui, J., and Cheng, D. Y. (2015). Computational identification and characterization of conserved miRNAs and their target genes in beet (Beta vulgaris). Genet. Mol. Res. 14, 9103–9108. doi: 10.4238/2015
Li, Q., Zhao, P., Li, J., Zhang, C., Wang, L., and Ren, Z. (2014). Genome-wide analysis of the WD-repeat protein family in cucumber and Arabidopsis. Mol. Genet. Genomics 289, 103–124. doi: 10.1007/s00438-013-0789-x
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Liu, H., and Stone, S. L. (2013). Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J. Biol. Chem. 288, 20267–20279. doi: 10.1074/jbc.M113.465369
Liu, H., Wang, Q., Yu, M., Zhang, Y., Wu, Y., and Zhang, H. (2008). Transgenic salt-tolerant sugar beet (Beta vulgaris L) constitutively expressing an Arabidopsis thaliana vacuolar Na/H antiporter gene, AtNHX3, accumulates more soluble sugar but less salt in storage roots. Plant Cell Environ. 31, 1325–1334. doi: 10.1111/j.1365-3040.2008.01838.x.
Liu, L., Li, Y., Li, S., Hu, N., He, Y., Pong, R., et al. (2012). Comparison of next-generation sequencing systems. J. Biomed. Biotechnol. 2012:251364. doi: 10.1155/2012/251364
Liu, Z., Li, Y., Cao, H., and Ren, D. (2015). Comparative phospho-proteomics analysis of salt-responsive phosphoproteins regulated by the MKK9-MPK6 cascade in Arabidopsis. Plant Sci. 241, 138–150. doi: 10.1016/j.plantsci.2015.10.005
Llave, C., Xie, Z., Kasschau, K. D., and Carrington, J. C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. doi: 10.1126/science.1076311
Luan, Y., Cui, J., Zhai, J., Li, J., Han, L., and Meng, J. (2015). High-throughput sequencing reveals differential expression of miRNAs in tomato inoculated with Phytophthora infestans. Planta 241, 1405–1416. doi: 10.1007/s00425-015-2267-7
Lukyanov, S. A., Rebrikov, D., and Buzdin, A. A. (2007). “Suppression subtractive hybridization,” in Nucleic Acids Hybridization Modern Applications eds A. A. Buzdin and S. A. Lukyanov (Berlin: Springer), 53–84.
Ma, C., Wang, Y., Wang, Y., Wang, L., Chen, S., and Li, H. (2011). Identification of a sugar beet BvM14-MADS box gene through differential gene expression analysis of monosomic addition line M14. J. Plant Physiol. 168, 1980–1986. doi: 10.1016/j.jplph.2011.05.027
Mardis, E. R. (2008). The impact of next-generation sequencing technology on genetics. Trends Genet. 24, 133–141. doi: 10.1016/j.tig.2007.12.007
Mikołajczyk-Bator, K., Błaszczyk, A., Czyzniejewski, M., and Kachlicki, P. (2016). Characterisation and identification of triterpene saponins in the rootsof red beets (Beta vulgaris L) using two HPLC–MS systems. Food Chem. 192, 979–990. doi: 10.1016/j.foodchem.2015.07.111
Mishra, K. B., Iannacone, R., Petrozza, A., Mishra, A., Armentano, N., La Vecchia, G., et al. (2012). Engineered drought tolerance in tomato plants is reflected in chlorophyll fluorescence emission. Plant Sci. 182, 79–86. doi: 10.1016/j.plantsci.2011.03.022
Mitchell, D. S., Laramy, S. E., Teresa, J. D. R., Frederick, P. B., Blair, D. S., and Tiffany, M. H. M. (2014). Transcriptional response of soybean to thiamethoxam seed treatment in the presence and absence of drought stress. BMC Genomics 15:1055. doi: 10.1186/1471-2164-15-1055
Moliterni, V. M., Paris, R., Onofri, C., Orrù, L., Cattivelli, L., Pacifico, D., et al. (2015). Early transcriptional changes in Beta vulgaris in response to low temperature. Planta 242, 187–201. doi: 10.1007/s00425-015-2299-z.
Moody, L. A., Saidi, Y., Smiles, E. J., Bradshaw, S. J., Meddings, M., Winn, P. J., et al. (2012). ARABIDILLO gene homologues in basal land plants: species-specific gene duplication and likely functional redundancy. Planta 236, 1927–1941. doi: 10.1007/s00425-012-1742-7
Moreno, D. A., Garcia-Viguera, C., Gil, J., and Gil-Izquierdo, A. (2008). Betalains in the era of global agri-food science, technology and nutritional health. Phytochem. Rev. 7, 261–280. doi: 10.1007/s11101-007-9084-y
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nogler, G. A. (1984). “Gametophytic apomixis,” in Embryology of Angiosperms, ed B. M. Johri (Berlin: Springer), 475–518.
Oksman-Caldentay, K. M., and Saito, K. (2005). Integrating genomics and metabolomics for engineering plant metabolic pathways. Curr. Opin. Biotechnol. 16, 174–179. doi: 10.1016/j.copbio.2005.02.007
Ozias-Akins, P. (2006). Apomixis: developmental characteristicsand genetics. Crit. Rev. Plant Sci. 25, 199–214. doi: 10.1080/07352680600563926
Pinhero, R., Pazhekattu, R., Marangoni, A. G., Liu, Q., and Yada, R. Y. (2011). Alleviation of low temperature sweetening in potato by expressing Arabidopsis pyruvate decarboxylase gene and stress-inducible rd29A: a preliminary study. Physiol. Mol. Biol. Plants 17, 105–114. doi: 10.1007/s12298-011-0056-8
Pummer, S., Dantzler, W. H., Lien, Y. H., Moeckel, G. W., Völker, K., and Silbernagl, S. (2000). Reabsorption of betaine in Henle's loops of rat kidney in vivo. Am. J. Physiol. Renal. Physiol. 278, 434–439.
Puthoff, D. P., and Smigocki, A. C. (2007). Insect feeding-induced differential expression of Beta vulgaris root genes and their regulation by defense-associated signals. Plant Cell Rep. 26, 71–84. doi: 10.1007/s00299-006-0201-yAN
Saad, A. S. I., Li, X., Li, H. P., Huang, T., Gao, C. S., Guo, M. W., et al. (2013). A rice stress-responsive NAC gene enhances tolerance of transgenic wheat to drought and salt stresses. Plant Sci. 203–204, 33–40. doi: 10.1016/j.plantsci.2012.12.016
Sahebi, M., Hanafi, M. M., Azizi, P., Hakim, A., Ashkani, S., and Abiri, R. (2015). Suppression subtractive hybridization versus next-generation sequencing in plant genetic engineering: challenges and perspectives. Mol. Biotechnol. 57, 880–903. doi: 10.1007/s12033-015-9884-z
Sanger, F., Nicklen, S., and Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467. doi: 10.1073/pnas.74.12.5463
Schuster, S. C. (2008). Next-generation sequencing transforms today's biology. Nat. Methods 5, 16–18. doi: 10.1038/nmeth1156
Setiawan, A., Koch, G., Barnes, S. R., and Jung, C. (2000). Mapping quantitative trait loci (QTLs) for resistance to Cercospora leaf spot disease (Cercospora beticola Sacc) in sugar beet (Beta vulgaris L.). Theor. Appl. Genet. 100, 1176–1182. doi: 10.1007/s001220051421
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Shinozaki, K., and Yamaguchi, S. K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Silva-Sanchez, C., Li, H., and Chen, S. (2015). Recent advances and challenges in plant phosphoproteomics. Proteomics 15, 1127–1141. doi: 10.1002/pmic.201400410
Stadermann, K. B., Weisshaar, B., and Holtgräwe, D. (2015). SMRT sequencing only de novo assembly of the sugar beet (Beta vulgaris) chloroplast genome. BMC Bioinformatics 16:295. doi: 10.1186/s12859-015-0726-6
Stintzing, F. C., and Carle, R. (2007). Betalains-emerging prospects for food scientists. Trends Food Sci. Technol. 18, 514–525. doi: 10.1016/j.tifs.2007.04.012
Sun, S., Yu, J. P., Chen, F., Zhao, T. J., Fang, X. H., Li, Y. Q., et al. (2008). TINY, a dehydration-responsive element (DRE)-binding protein-like transcription factor connecting the DRE- and ethylene-responsive element-mediated signaling pathways in Arabidopsis. J. Biol. Chem. 283, 6261–6271. doi: 10.1074/jbc.M706800200
Sunkar, R., Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12, 301–309. doi: 10.1016/j.tplants.2007.05.001
Taguchi, K., Kubo, T., Takahashi, H., and Abe, H. (2011). Identification and precise mapping of resistant QTLs of Cercospora leaf spot resistance in sugar beet (Beta vulgaris L). G3 (Bethesda) 1, 283–291. doi: 10.1534/g3.111.000513
Taguchi, K., Okazaki, K., Takahashi, H., Kubo, T., and Mikami, T. (2010). Molecular mapping of a gene conferring resistance to Aphanomyces root rot (black root) in sugar beet (Beta vulgaris L). Euphytica 173, 409–418. doi: 10.1007/s10681-010-0153-8
Takabe, T., Rai, V., and Hibino, T. (2006). “Metabolic engineering of glycinebetaine,” in Abiotic Stresss Tolerance in Plants, eds A. K. Rai and T. Takabe (Berlin: Springer), 137–151.
Torres, M. A., and Dangl, J. L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8, 397–403. doi: 10.1016/j.pbi.2005.05.014
Vlachonasios, K. E., Thomashow, M. F., and Triezenberg, S. J. (2003). Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development and gene expression. Plant Cell 15, 626–638. doi: 10.1105/tpc.007922
Wakeel, A., Asif, A. R., Pitann, B., and Schubert, S. (2011). Proteome analysis of sugar beet (Beta vulgaris L) elucidatas constitutive adaptation during the first phase of salt stress. J. Plant Physiol. 168, 519–526. doi: 10.1016/j.jplph.2010.08.016
Wang, Y. N., Tang, L., Hou, Y., Wang, P., Yang, H., and Wei, C. L. (2016). Differential transcriptome analysis of leaves of tea plant (Camellia sinensis) provides comprehensive insights into the defense responses to Ectropis oblique attack using RNA-Seq. Funct. Integr. Genomics. doi: 10.1007/s10142-016-0491-2. [Epub ahead of print].
Wang, Y., Zhan, Y., Wu, C., Gong, S., Zhu, N., Chen, S., et al. (2012). Cloning of a cystatin gene from sugar beet M14 that can enhance plant salt tolerance. Plant Sci. 191–192, 93–99. doi: 10.1016/j.plantsci.2012.05.001
Wu, C., Ma, C., Pan, Y., Gong, S., Zhao, C., Chen, S., et al. (2013). Sugar beet M14 glyoxalase I gene can enhance plant tolerance to abiotic stresses. J. Plant Res. 26, 415–425. doi: 10.1007/s10265-012-0532-4
Wybraniec, S. (2005). Formation of decarboxylated betacyanins in heated red beet (Betavulgaris L) root juice analysed by LC–MS/MS. J. Agric. Food Chem. 53, 3483–3487. doi: 10.1021/jf048088d
Wybraniec, S., Stalica, P., Spórna, A., Nemzer, B., Pietrzkowski, Z., and Michałowski, T. (2011). Antioxidant activity of betanidin: electrochemical study in aqueous media. J. Agric. Food Chem. 59, 12163–12170. doi: 10.1021/jf2024769
Wybraniec, S., Starzak, K., Skopi'nska, A., Nemzer, B., Pietrzkowski, Z., and Michałowski, T. (2013). Studies on nonenzymatic oxidation mechanisms in neobe-tanin, betanin, and decarboxylated betanins. J. Agric. Food Chem. 61, 6465–6476. doi: 10.1021/jf400818s
Xu, J., Lan, H., Fang, H., Huang, X., Zhang, H., and Huang, J. (2015). Quantitative proteomic analysis of the rice (Oryza sativa L) salt response. PLoS ONE 10:e0120978. doi: 10.1371/journal.pone.0120978
Yamada, N., Promden, W., Yamane, K., Tamagake, H., Hibino, T., Tanaka, Y., et al. (2009). Preferential accumulation of betaine uncoupled to choline monooxygenase in young leaves of sugar beet–importance of long-distance translocation of betaine under normal and salt-stressed conditions. J. Plant Physiol. 166, 2058–2070. doi: 10.1016/j.jplph.2009.06.016
Yang, L., Ma, C., Wang, L., Chen, S., and Li, H. (2012). Salt stress induced proteome and transcriptome changes in sugar beet monosomic addition line M14. J. Plant Physiol. 169, 839–850. doi: 10.1016/j.jplph.2012.01.023
Yang, L., Zhang, Y., Zhu, N., Koh, J., Ma, C., Pan, Y., et al. (2013). Proteomic analysis of salt tolerance in sugar beet monosomic addition line M14. J. Proteome Res. 12, 4931–4950. doi: 10.1021/pr400177m
Yin, F., Gao, J., Liu, M., Qin, C., Zhang, W., Yang, A., et al. (2014). Genome-wide analysis of water-stress-responsive microRNA expression profile in tobacco roots. Funct. Integr. Genomics 14, 319–332. doi: 10.1007/s10142-014-0365-4
Yolcu, S., Ozdemir, F., Güler, A., and Bor, M. (2016). Histone acetylation influences the transcriptional activation of POX in Beta vulgaris L. and Beta maritima L. under salt stress. Plant Physiol. Biochem. 100, 37–46. doi: 10.1016/j.plaphy.2015.12.019
Zhang, Y., Lai, J., Sun, S., Li, Y., Liu, Y., Liang, L., et al. (2008). Comparison analysis of transcripts from the halophyte Thellungiella halophila. J. Integr. Plant Biol. 50, 1327–1335. doi: 10.1111/j.1744-7909.2008.00740.x
Keywords: sugar beet, genomics, transcriptomics, proteomics, metabolomics
Citation: Zhang Y, Nan J and Yu B (2016) OMICS Technologies and Applications in Sugar Beet. Front. Plant Sci. 7:900. doi: 10.3389/fpls.2016.00900
Received: 31 March 2016; Accepted: 07 June 2016;
Published: 22 June 2016.
Edited by:
Pingfang Yang, Chinese Academy of Sciences, ChinaCopyright © 2016 Zhang, Nan and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Yu, eWJnaXJsMTIzNEBzaW5hLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.