- 1Hainan Key Laboratory for Sustainable Utilization of Tropical Bioresources, College of Agriculture, Hainan University, Haikou, China
- 2Key Laboratory of Biology and Genetic Resources of Tropical Crops, Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, Haikou, China
- 3Institute of Apicultural Research, Chinese Academy of Agricultural Sciences, Beijing, China
As a well-known animal hormone, melatonin (N-acetyl-5-methoxytryptamine) is also involved in multiple plant biological processes, especially in various stress responses. Rice is one of the most important crops, and melatonin is taken in by many people everyday from rice. However, the transcriptional profiling of melatonin-related genes in rice is largely unknown. In this study, the expression patterns of 11 melatonin related genes in rice in different periods, tissues, in response to different treatments were synthetically analyzed using published microarray data. These results suggest that the melatonin-related genes may play important and dual roles in rice developmental stages. We highlight the commonly regulation of rice melatonin-related genes by abscisic acid (ABA), jasmonic acid (JA), various abiotic stresses and pathogen infection, indicating the possible role of these genes in multiple stress responses and underlying crosstalks of plant hormones, especially ABA and JA. Taken together, this study may provide insight into the association among melatonin biosynthesis and catabolic pathway, plant development and stress responses in rice. The profile analysis identified candidate genes for further functional characterization in circadian rhythm and specific stress responses.
Introduction
Melatonin (N-acetyl-5-methoxytryptamine) was first discovered in the cow’s pineal gland (Lerner et al., 1958). Dubbels et al. (1995) and Hattori et al. (1995), melatonin was identified in plants by two research groups. Until now, melatonin has been found in multiple plant species, including alfalfa, almond, anise, apples, Arabidopsis, banana, beetroot, bermudagrass, black mustard, cabbage, celery, cherry, coriander, cucumber, fennel, fenugreek, flax, green cardamom, milk thistle, oranges, poppy, potato, rice, sunflower, tobacco, tomato, white mustard, wolf berry, etc. (Manchester et al., 2000; Zhao et al., 2013; Shi and Chan, 2014). In addition, the endogenous melatonin concentration can also be modulated through genetic transformation in tomato and rice (Okazaki and Ezura, 2009; Okazaki et al., 2009, 2010; Byeon et al., 2012, 2013, 2014; Byeon and Back, 2014a,b).
To date, the biosynthesis and metabolic pathways of melatonin in plants have been established (Figure 1). Melatonin in plants can be synthesized by four sequential enzymes from tryptophan (Kang et al., 2011), including TDC, T5H, SNAT, and N-aceylserotonin O-methyltransferase (ASMT) (Arnao and Hernández-Ruiz, 2014, 2015; Zuo et al., 2014). Thereafter, melatonin is catabolized by M2H into 2-hydroxymelatonin (Byeon and Back, 2015). In rice, gene families of TDC, T5H, SNAT, and ASMT contain 3, 1, 1, and 3 known members, respectively (Kang et al., 2007; Fujiwara et al., 2010; Kang et al., 2013; Park et al., 2013a). However, OsASMT3 is barely detectable in any of the plant organs (Park et al., 2013b). OsM2H genes belong to 2-ODD family and at least 4 of 2-ODD genes show M2H activities in rice (Byeon and Back, 2015).
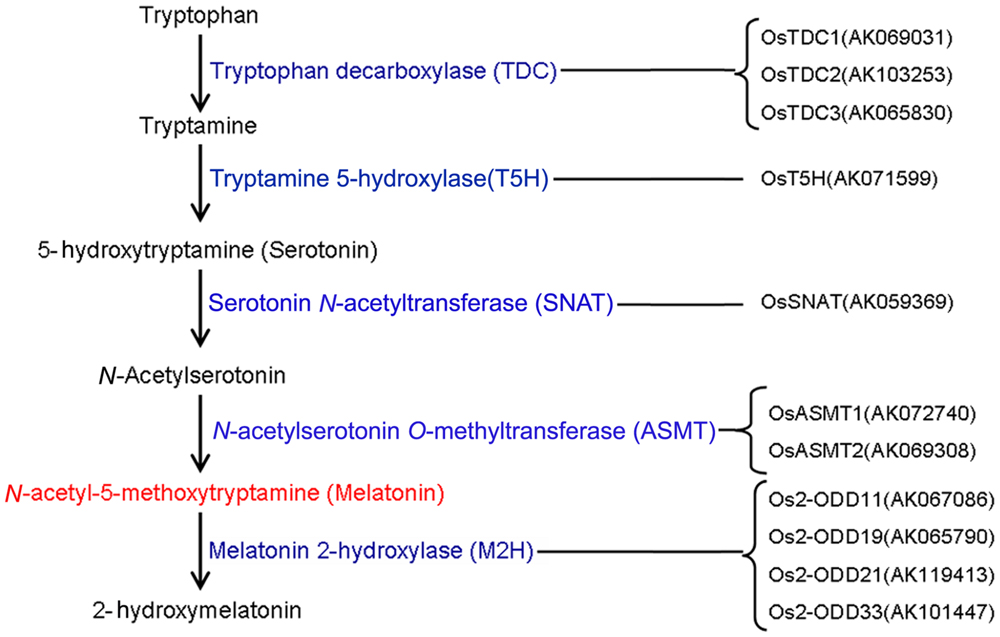
FIGURE 1. The genes responsible for melatonin synthesis from tryptophan and melatonin degradation in rice.
Solid evidence implicates that melatonin is involved in multiple plant biological processes and various stress responses (Hardeland, 2015; Zhan et al., 2015), including circadian rhythm (Kolár and Machácková, 2005; Arnao and Hernández-Ruiz, 2015), delayed senescence of leaves (Byeon et al., 2012; Wang et al., 2012, 2013a,b), leaf morphology (Okazaki et al., 2010), root development (Hernández-Ruiz et al., 2005; Pelagio-Flores et al., 2012; Zhang N. et al., 2014), coleoptile growth (Hernández-Ruiz et al., 2004, 2005), grain yield (Byeon and Back, 2014a), fruit ripening (Sun et al., 2015), drought stress (Wang et al., 2013a, 2014; Zhang et al., 2013; Meng et al., 2014; Zuo et al., 2014; Shi et al., 2015b), salt stress (Wei et al., 2014; Zhang H.J. et al., 2014; Liang et al., 2015; Shi et al., 2015b), cold stress (Posmyk et al., 2009a; Arnao and Hernández-Ruiz, 2014; Bajwa et al., 2014; Shi and Chan, 2014; Turk et al., 2014; Shi et al., 2015b), high temperature (Tiryaki and Keles, 2012), copper stress (Posmyk et al., 2008, 2009b), oxidative stress (Park et al., 2013b; Shi et al., 2015d), cadmium stress (Byeon et al., 2015) and pathogen infection (Yin et al., 2013; Lee et al., 2014, 2015; Reiter et al., 2015; Shi et al., 2015a; Zhao et al., 2015).
Melatonin plays protective roles in the regulation of plant tolerance to abiotic stress and biotic stress (Yin et al., 2013; Lee et al., 2014, 2015; Zhan et al., 2015). Overexpression of OsTDC increases endogenous melatonin level and delays leaf senescence in rice (Kang et al., 2007, 2009; Byeon et al., 2014). The transcript of OsT5H can be induced by Magnaporthe grisea infection (Fujiwara et al., 2010). Exogenous application of serotonin, the penultimate substrate for melatonin biosynthesis, induces defense gene expression and increases resistance to rice blast infection (Fujiwara et al., 2010). Transgenic rice plants ectopically expressing the AANAT regulates cold stress resistance (Kang et al., 2010), seminal root elongation (Park and Back, 2012), oxidative stress resistance (Park et al., 2013b), and seedling growth (Byeon and Back, 2014a). The transcript of OsASMT can be induced by ABA and methyl JA treatments, and OsASMT overexpressing plants result in higher level of melatonin (Park et al., 2013b). Exogenous application of melatonin improved apple resistance to Marssonina apple blotch (Diplocarpon mali) (Yin et al., 2013), enhanced disease defense against Pseudomonas syringae DC3000 in Arabidopsis and tobacco (Lee et al., 2014; Shi et al., 2015a).
Rice is one of the most important crops around the world, and melatonin is also taken in from rice by many people everyday. Thus, it is very useful and important to dissect the distribution and regulation of endogenous melatonin in rice. Melatonin is widely involved in plant development, multiple abiotic and biotic stress responses in Arabidopsis (Shi and Chan, 2014), and Bermudagrass (Shi et al., 2015b). However, transcriptional profiling of rice melatonin synthesis and catabolic genes has not been systematically carried out. In this study, we analyzed the expression profiling of 11 rice melatonin synthesis and catabolic genes in development, various tissues, and in response to hormone, pathogen infection, drought, salt, and cold stresses. These results may provide insight into the link among melatonin biosynthesis and catabolic pathway, plant development and stress responses in rice. Further functional characterization of identified candidate genes with potential involvement in circadian rhythm and stress responses through overexpressing, knocking down or knocking out will give more clues to melatonin-mediated signaling as well as underlying molecular mechanism.
Materials and Methods
Plant materials and Growth Conditions
Rice (Oryza sativa L. ssp. japonica cv. Nipponbare) seeds were sown in germinating boxes. At 30 days after germination, the seedlings were transplanted in a paddy field under normal conditions of the cultivation season. Thereafter, 56 DAT, 58 DAT, and 90 DAT were considered as the stage of panicle initiation, the early stage of panicle development indicating a complete reproductive transition, the stages of flowering and early stages of seed development corresponding to the ripening-stage transition, respectively.
For hormone treatments, rice seeds were germinated, and grown hydroponically in a growth chamber at 28°C under continuous light. Seven-day old seedlings were transferred in culture solution containing 50 μM ABA, or 10 μM GA, or 10 μM IAA, or 1 μM brassinolide (BL), or 1 μM tZ, or 100 μM JA, or in culture solution without hormone to serve as control (mock treatment). Samples were collected after 0, 0.25, 0.5, 1, 3, and 6 h incubation for root, and after 0, 1, 3, 6, and 12 h incubation for shoot.
Pathogen Infection
Rice (O. sativa cv Nipponbare) plants grown in the greenhouse for 42 days were inoculated with Xoo T7174R, a wild-type strain, and 74HrcV::Km, a T3S-defective mutant by the leaf-clipping method. Plants treated with water were used as control. Leaf sections (3–5 mm) that included the inoculation site were collected at 3, 6, and 12 hpi and 1, 2, 4, 6 dpi.
For the blast fungus infection, three lines of rice cultivar Nipponbare carrying the blast resistance genes (Pia, Pish) were inoculated with two strains of Magnaporthe oryzae harboring AVR-Pia and AVR-Pish. Rice seedlings at the 4-leaf stage were placed in moist chamber and sprayed with a conidial suspension of M. oryzae (1 × 106 conidia/ml). The seedlings were incubated in a moist chamber at 25°C for 24 h under dark condition, then grown in hydroponic culture under 14 h light (28°C) and 10 h dark (24°C). Leaf samples (4th leaf) from 3 individual experiments were harvested at 1, 2, 3, and 5 days post inoculation (dpi). Rice seedlings sprayed with water were used as control.
Development- and Pathogen Infection-Related Data Analysis
The data of spatio-temporal transcript levels in various tissues or organs (RXP_0001), leaf and root transcriptional profile in light (RXP_003 and RXP_007) and dark (RXP_004 and RXP_008) throughout entire growth in the field, diurnal, and circadian leaf (RXP_002) and root (RXP_009) transcriptional profile throughout entire growth, plant hormone profile (RXP_001 to RXP_012), Xoo-treated profile (RXP_3002), and M. oryzae-treated profile (RXP_3001) were downloaded from RiceXPro1 (Sato et al., 2011a,b, 2013). All samples were used for hybridization using the Agilent one-color (Cy3) microarray-based gene analysis system. As detailed described in Sato et al. (2013), all the above data were deposited in GEO through the following accession numbers: GSE21396, GSE21397, GSE36040, GSE36042, GSE36043, GSE36044, GSE39423, GSE39424, GSE39425, GSE39426, GSE39427, GSE39429, and GSE39432. All the raw data were downloaded and re-analyzed for cluster analysis of expression profile that shown as normalized data (log2).
Abiotic Stress-Related Data Analysis
As described in Jain et al. (2007), 7-day-old light-grown rice seedlings were transferred to control condition and 200 mM NaCl solution as salt stress for 3 h, were dried on tissue paper as dehydration stress for 3 h, and were kept at 4°C as cold stress for 3 h. Then the seedlings were sampled in triplicate. GEO series accession no. GPL2025 were used for microarray analysis as Jain et al. (2007) described. All the normalized data were obtained from Rice eFP Browser2 (Jain et al., 2007).
Cluster Analysis
The original data from RiceXPro and Rice eFP Browser were listed in Supplementary Table S1. Hierarchical cluster analysis of transcriptional profile was performed using CLUSTER program3 (Larkin et al., 2007), and the heatmap was obtained using Java Treeview4 (Saldanha, 2004) according to the instructions.
Results
The Spatio-Temporal Transcript Levels of Rice Melatonin Synthesis and Catabolic Genes in Various Tissues or Organs
To investigate the expression profiles of rice melatonin synthesis and catabolic genes in various tissues or organs, we analyzed the expression of these genes using published microarray data (Sato et al., 2013). Eleven of rice melatonin-related genes have the corresponding probe sets in the dataset (Figure 1). As shown in Figure 2, all genes showed different expression pattern in various tissues, indicating that these genes may play different roles in plant growth and development. Interestingly, the expression patterns of melatonin-related genes could be divided into two groups (Figure 2). One group contained six genes (OsT5H, Os2-ODD11, -19, -21,-33, and OsTDC1), and most of them showed high expression levels in endosperm tissues. The other group contained five genes (OsASMT1, -2, OsTDC2, -3, and OsSNAT), and most of them showed high expression levels in leaf blade and leaf sheath tissues. Moreover, Os2-ODD19 showed high expression level in four time points of anther, while other four genes (OsTDC1, -3, Os2-ODD11, and OsASMT1) exhibited lower expression level. Similarly, six genes (Os2-ODD11, -19, -21,-33, OsTDC1, and OsASMT1) showed high expression level in five time points of endosperm, while two genes (OsASMT2 and OsSNAT) with a relative low level of expression. These melatonin-related genes showed high expression levels in a special tissue indicated their possible roles of melatonin in special tissue.
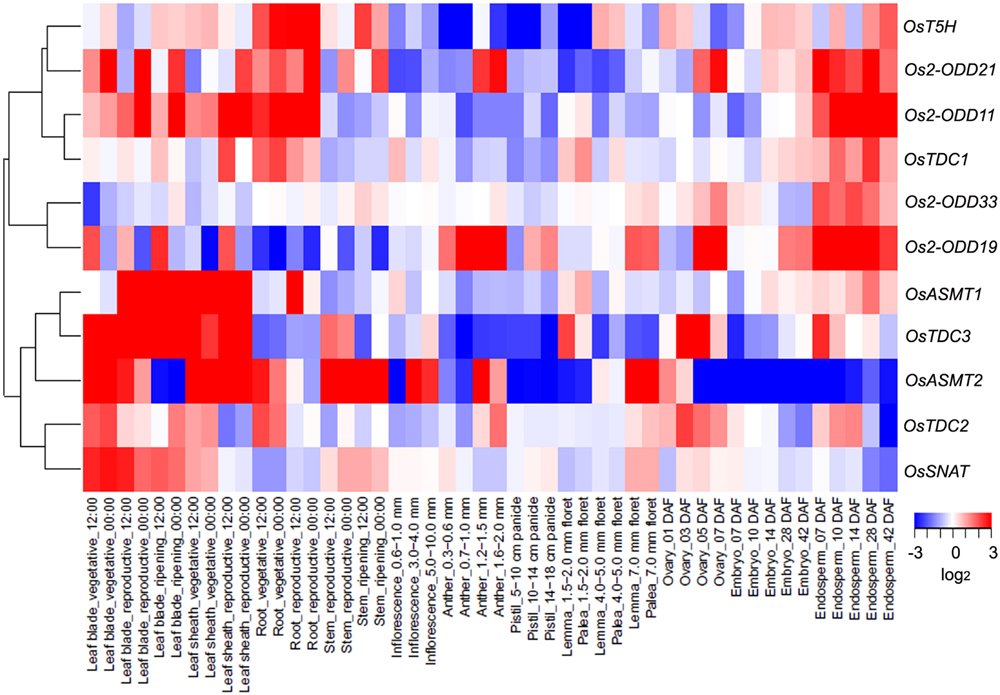
FIGURE 2. The spatio-temporal transcript levels of rice melatonin synthesis and catabolic genes in various tissues or organs. The transcriptional expression data were obtained from The Rice Expression Profile Database (RiceXPro) (http://ricexpro.dna.affrc.go.jp/). Rice (O. sativa L. ssp. japonica cv. Nipponbare) seeds were sown in germinating boxes. At 30 days after germination, the seedlings were transplanted in a paddy field under normal conditions during the cultivation season. Thereafter, 56 DAT, 58 DAT and 90 DAT were considered as the stage of panicle initiation, the early stage of panicle development indicating a complete reproductive transition, the stages of flowering and early stages of seed development corresponding to the ripening-stage transition, respectively. In various tissues or organs at different stages, a total of 48 samples in three replicates were used for hybridization using the Agilent one-color (Cy3) microarray-based gene analysis system. The cluster analysis of expression profile for each gene in various tissues is shown as normalized data (log2).
Moreover, three genes (Os2-ODD19, -21, and OsTDC2) showed different expression level in day and night at 9 tissues (leaf blade-vegetative, leaf blade-reproductive, leaf blade-ripening, leaf sheath-vegetative, leaf sheath-reproductive, root-vegetative, root-reproductive, stem-reproductive, and stem-ripening). The results indicate that Os2-ODD19, -21, and OsTDC2 may play some roles in circadian rhythm and may be used in further functional characterization.
Transcriptional Profile of Rice Melatonin Synthesis and Catabolic Genes throughout Entire Growth in the Field
In rice leaves at day, the transcript levels of OsTDC3 and OsASMT2 were increased at vegetative stages, while that of OsT5H was decreased (Figure 3A). At reproductive stages, the transcripts of seven genes (OsTDC1, -3, OsASMT1, -2, Os2-ODD11, -33, and OsSNAT) and two genes (OsTDC2 and Os2-ODD21) showed up-regulation and down-regulation, respectively (Figure 3A). At ripening stages, the transcripts of three genes (Os2-ODD11, -33, and OsASMT1) and two genes (OsTDC2 and Os2-ODD21) showed up-regulation and down-regulation, respectively (Figure 3A). In the leaves at night, the transcript of Os2-ODD11 showed up-regulation at all time points, while the transcripts of OsTDC2 and Os2-ODD19 were obviously down-regulated (Figure 3B). At vegetative stages, the transcripts of three genes (OsTDC3, OsT5H, and Os2-ODD11) and two genes (OsTDC2 and Os2-ODD19) showed up-regulation and down-regulation, respectively (Figure 3B). At reproductive stages, the transcripts of four genes (OsTDC3, OsT5H, Os2-ODD11,-33) and two genes (OsTDC2 and Os2-ODD19) showed up-regulation and down-regulation, respectively (Figure 3B). At ripening stages, the transcripts of two genes (Os2-ODD11, -33) and five genes (OsTDC1, -2, Os2-ODD19, OsSNAT, and OsASMT2) showed up-regulation and down-regulation, respectively (Figure 3B).
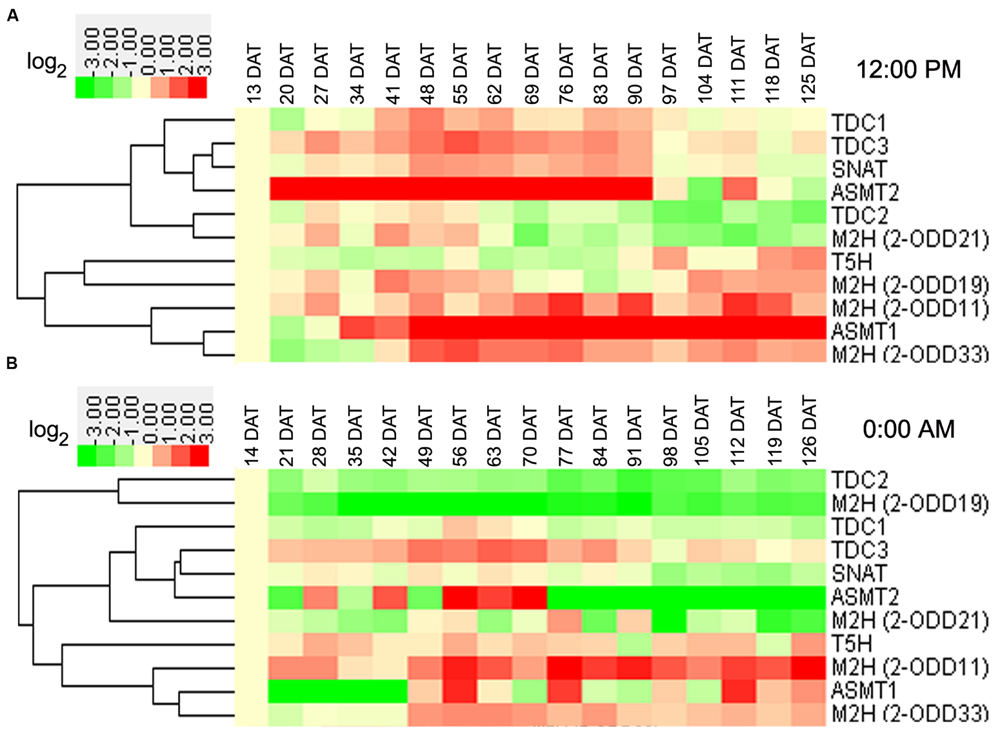
FIGURE 3. Leaf transcriptional profile of rice melatonin synthesis and catabolic genes in light (A) and dark (B) throughout entire growth in the field. The transcriptional expression data were obtained from RiceXPro (http://ricexpro.dna.affrc.go.jp/). Rice (Oryza sativa L. ssp. japonica cv. Nipponbare) seeds were sown in germinating boxes. At 30 days after germination, the seedlings were transplanted in a paddy field under normal conditions during the cultivation season. Samples corresponding to the uppermost fully expanded leaves were collected every 12:00 PM at weekly intervals from 13 to 125 DAT and every 0:00 AM at weekly intervals from 14 to 126 DAT. A total of 51 samples at 12:00 PM in three replicates and 34 samples at 0:00 AM in two replicates were used for hybridization using the Agilent one-color (Cy3) microarray-based gene analysis system. The cluster analysis of expression profile for each gene in various tissues is shown as normalized data (log2).
In rice roots at day, transcript of OsASMT2 increased in the entire growth stages in the field (Figure 4A). At vegetative stages, the transcripts of eight genes (Os2-ODD11, -19,-33, OsTDC1, -2, OsT5H, OsSNAT, and OsASMT2) showed up-regulation (Figure 4A). At reproductive stages, the transcripts of two genes (OsASMT1, -2) and four genes (OsTDC1, -3, Os2-ODD11,-21) showed up-regulation and down-regulation, respectively (Figure 4A). At ripening stages, the transcripts of two genes (OsASMT1 and -2) and eight genes (OsTDC1, -2,-3, Os2-ODD19, -21,-33, OsT5H and OsSNAT) showed up-regulation and down-regulation, respectively (Figure 4A). Interestingly, the transcripts of six genes (OsTDC1, -2, Os2-ODD19,-33, OsT5H, and OsSNAT) were obviously up-regulated at vegetative stages, but down-regulated at ripening stages (Figure 4A). In the roots at night, transcript of Os2-ODD21 decreased throughout entire growth stages in the field (Figure 4B). At vegetative stages, the transcripts of six genes (OsTDC1, -2, OsT5H, OsSNAT, Os2-ODD33, and OsASMT2) and two genes (Os2-ODD11, -21) showed up-regulation and down-regulation, respectively (Figure 4B). Additionally, the transcripts of two genes (OsTDC3 and Os2-ODD21) and five genes (Os2-ODD11, -21,-33, OsTDC2, and OsT5H) showed down-regulation at reproductive and ripening stages, respectively (Figure 4B). The transcripts of three genes (OsTDC2, OsT5H, and Os2-ODD33) showed up-regulation at vegetative stages, but down-regulation at ripening stages (Figure 4B).
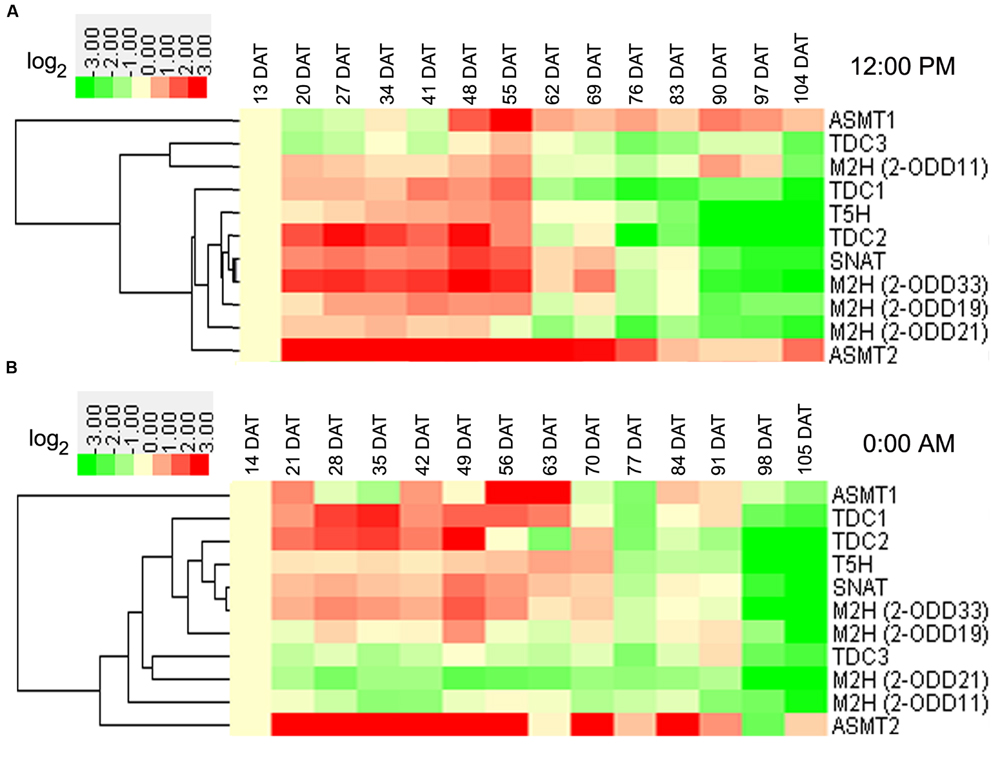
FIGURE 4. Root transcriptional profile of rice melatonin synthesis and catabolic genes in light (A) and dark (B) throughout entire growth in the field. The transcriptional expression data were obtained from RiceXPro (http://ricexpro.dna.affrc.go.jp/). Rice (O. sativa L. ssp. japonica cv. Nipponbare) seeds were sown in germinating boxes. At 30 days after germination, the seedlings were transplanted in a paddy field under normal conditions during the cultivation season. Root samples at various growth stages encompassing the vegetative, reproductive, and ripening stages were collected every 12:00 PM at weekly intervals from 13 to 104 DAT and every 0:00 AM at weekly intervals from 14 to 105 DAT. The cluster analysis of expression profile for each gene in various tissues is shown as normalized data (log2).
Some genes shared similar expression patterns at day and night at the same stage (Figures 3 and 4). The transcripts of Os2-ODD11 and -33 showed up-regulation at reproductive and ripening stages in leaves, the transcripts of six genes (OsTDC1, -2, OsT5H, OsSNAT, OsASMT2, and Os2-ODD33) showed up-regulation at vegetative stages in roots, the transcripts of OsTDC1 and Os2-ODD21 shared down-regulation at reproductive and ripening stages of roots. However, there were also some genes shared different expression patterns. For example, the transcript of OsT5H shared down-regulation in day at vegetative stages in leaves, but showed up-regulation in night. Moreover, some genes also shared similar expression patterns in different tissues at the same stage. At day, the transcripts of OsASMT2 and OsASMT1 showed up-regulation at vegetative and ripening stages in leaves and roots, respectively, while the transcript of Os2-ODD21 shared down-regulation at reproductive and ripening stages. On the contrary, some genes shared different expression patterns in different tissues at the same stage. The transcripts of OsT5H showed down-regulation in day at vegetative stages in leaves, but showed up-regulation in roots. The transcripts of Os2-ODD11 and Os2-ODD33 showed up-regulation at ripening stages of leaves, but showed down-regulation at roots. These results suggest that the melatonin-related genes as well as endogenous melatonin may play important and dual roles in rice developmental stages.
Diurnal and Circadian Transcriptional Profile of Rice Melatonin Synthesis and Catabolic Genes throughout Entire Growth
As shown in Figure 5A, Os2-ODD11 expression was induced throughout entire growth stages, and the transcripts of four genes (OsASMT1,-2 OsTDC1 and Os2-ODD33) were induced at most time points of growth stages, while those of Os2-ODD19 and Os2-ODD21 were intermittent. Before reproductive 1 stage, Os2-ODD21 showed significant induction at night. On the contrary, Os2-ODD19 was obviously down-regulated. This result suggested that Os2-ODD19 and Os2-ODD21 may play dual and important roles in the regulation of circadian rhythm. Moreover, the transcripts of four genes (Os2-ODD11, -33, OsASMT1, and -2) and two genes (OsASMT1 and Os2-ODD11) displayed significant up-regulation at four stages (vegetative 3, vegetative-reproductive, reproductive 1, and reproductive-ripening stages) and two stages (ripening 1 and 2 stages), respectively. However, OsTDC2, OsTDC3, and OsSNAT expressions were repressed during ripening 1 and 2 stages. Additionally, the transcripts of most genes were induced during reproductive 1 and reproductive-ripening stages. At the last two stages (ripening 1 and 2 stages), most of genes were obviously down-regulated. Interestingly, the transcripts of five genes (OsTDC2, -3, OsT5H, OsSNAT, and OsASMT2) were induced during reproductive 1 and reproductive-ripening stages, but exhibited down-regulation at ripening 1 and 2 stages. As shown in Figure 5B, the expression of Os2-ODD19 and Os2-ODD21 displayed obviously regular change throughout entire growth.
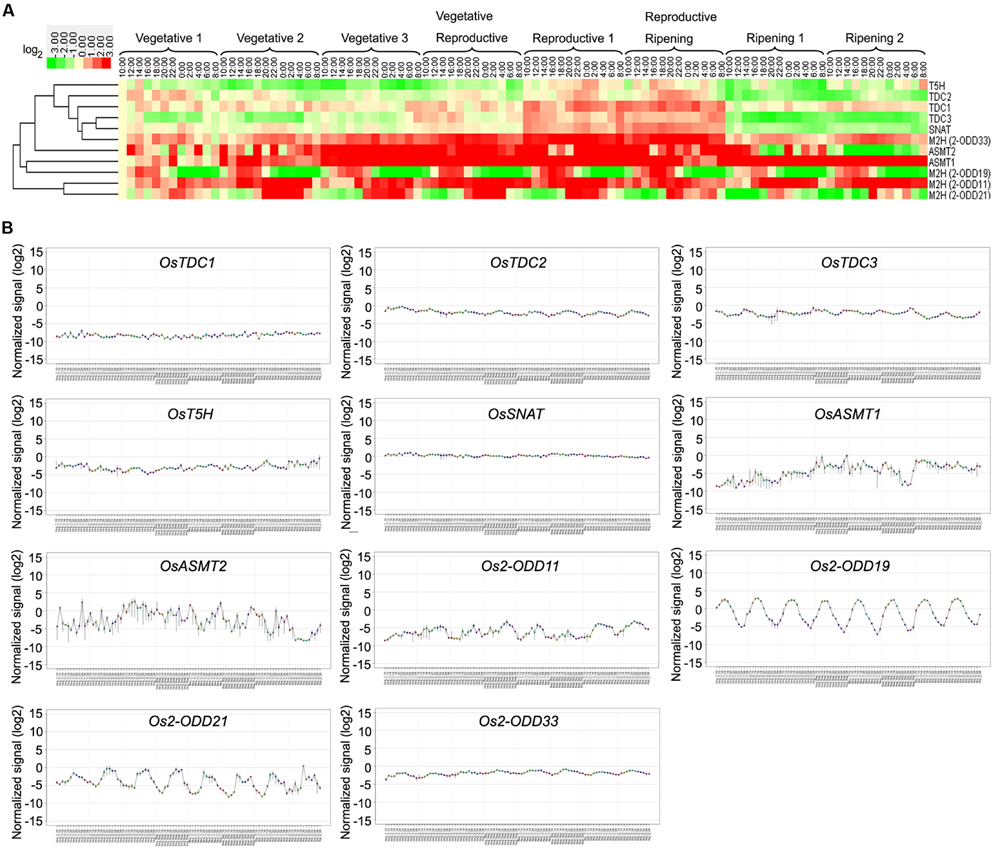
FIGURE 5. Diurnal and circadian leaf transcriptional profile of rice melatonin synthesis and catabolic genes throughout entire growth as shown by heatmap (A) and line chart (B). The transcriptional expression data were obtained from RiceXPro (http://ricexpro.dna.affrc.go.jp/). Rice (O. sativa L. ssp. japonica cv. Nipponbare) seeds were sown in germinating boxes. Gene expression profile of rice plants grown under natural field conditions based on microarray analysis of leaf samples at various growth stages encompassing the vegetative, reproductive and ripening stages. Samples corresponding to the uppermost fully expanded leaves were collected in a 48-h period at 2-h intervals at eight different growth stages. The cluster analysis and normalized signal of expression profile for each gene in various tissues is shown as normalized data (log2).
In roots, the transcript of Os2-ODD11 was up-regulated at all time-points, while those of OsTDC3, Os2-ODD21, and OsTDC2 were down-regulated (Figure 6). The transcriptional profile of Os2-ODD19 was intermittent, which was consistent with the result in leaves. During 15–17 DAT, the expressions of Os2-ODD21, OsTDC2, and OsASMT2 were repressed. The transcript of Os2-ODD11 was significantly induced during 43–45 DAT, whereas the transcripts of OsTDC1, -2, -3, OsT5H, and Os2-ODD21 were repressed.
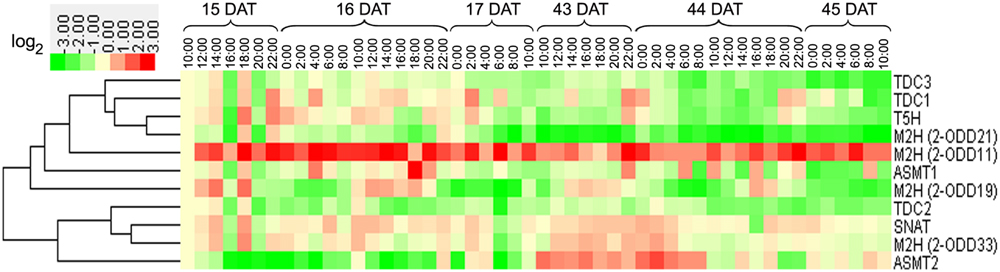
FIGURE 6. Diurnal and circadian root transcriptional profile of rice melatonin synthesis and catabolic genes throughout entire growth. The transcriptional expression data were obtained from RiceXPro (http://ricexpro.dna.affrc.go.jp/). Rice (O. sativa L. ssp. japonica cv. Nipponbare) seeds were sown in germinating boxes. Root samples corresponding were collected in a 48-h period at 2-h intervals at two different growth stages. The cluster analysis of expression profile for each gene in various tissues is shown as normalized data (log2).
The Transcriptional Profile of Rice Melatonin Synthesis and Catabolic Genes in Response to Plant Hormones
In response to ABA and JA treatments, the transcripts of OsT5H, OsTDC2, -3, and Os2-ODD19 displayed significantly up-regulation in root or shoot (Figure 7). The transcript of Os2-ODD11 was significant up-regulated after IAA, BL, and JA treatments in root, but was strongly down-regulated after IAA, BL, and tZ treatments in shoot. The transcripts of OsTDC1 and OsASMT1 showed up-regulation after ABA, GA3, IAA, BL, and tZ treatments in shoot, while OsTDC3 expression was induced after ABA, IAA, BL, tZ, and JA treatments in shoot (Figure 7).
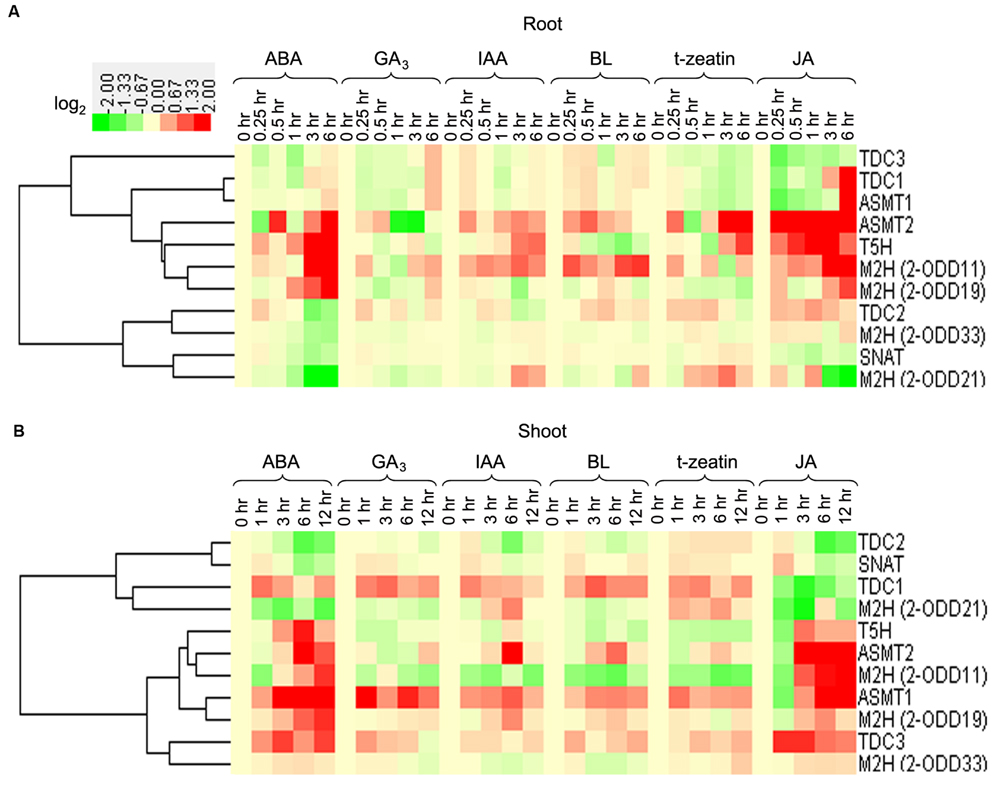
FIGURE 7. The transcriptional profile of rice melatonin synthesis and catabolic genes in root (A) and shoot (B) in response to plant hormones. The transcriptional expression data were obtained from RiceXPro (http://ricexpro.dna.affrc.go.jp/). The cluster analysis of expression profile for each gene in various tissues is shown as normalized data (log2) in relative to the 0 h of treatment which was set as 0.
Generally, melatonin-related genes showed different expression profiles in root or shoot tissues for the same treatment (Figure 7). The transcripts of OsTDC1, -3, Os2-ODD33, and OsASMT1 were up-regulated in the shoots, but were not significantly regulated or down-regulated in the roots after ABA and tZ treatments. The expression of Os2-ODD11 was increased in roots, but was decreased in shoots after ABA and BL treatments. Although some melatonin-related genes were from the same family, they exhibited different responses to plant hormones treatments, such as OsTDC2 and OsTDC3 in roots, Os2-ODD11 and Os2-ODD21 in roots, OsASMT1 and OsASMT2 in shoots. Thus, the transcriptional response of melatonin-related genes to plant hormones treatments in roots and shoots may provide new insight into crosstalk between melatonin and plant hormones, as well as mechanism underlying melatonin-mediated signaling in rice.
Gene Expression Profile in Whole Leaf of Rice Melatonin Synthesis and Catabolic Genes Inoculated with Pathogen Infection
Because melatonin plays important roles in response to pathogen infection (Yin et al., 2013; Lee et al., 2014, 2015; Reiter et al., 2015; Shi et al., 2015a; Zhao et al., 2015), so we analyzed the expression profile of rice melatonin synthesis and catabolic genes in response to pathogen inoculation to identify the candidate genes for further analysis.
After inoculation with Xoo, OsASMT2 expression was induced during almost all the time-points, while OsASMT1 transcript was decreased at these time points (Figure 8). The transcripts of OsT5H and Os2-ODD19 were induced during 1 to 6 dpi and 6 to 12 hpi, respectively (Figure 8). OsTDC1, -2, -3, Os2-ODD11, -21,-33, and OsSNAT, expressions were decreased during all the treated time points (Figure 8). Interestingly, the transcripts of OsASMT1 and OsASMT2 were increased after 6 dpi of wild-type strain T7114R, but were decreased after 6 dpi of ΔahrcV(III) strain. T3S is essential for XooT7174R conferred plant disease, and ΔahrcV(III) resulted in less plant disease in rice leaves (Sato et al., 2011b, 2013). Thus, the results indicate the possible role of OsASMT1 and OsASMT2 in immune response to Xoo.
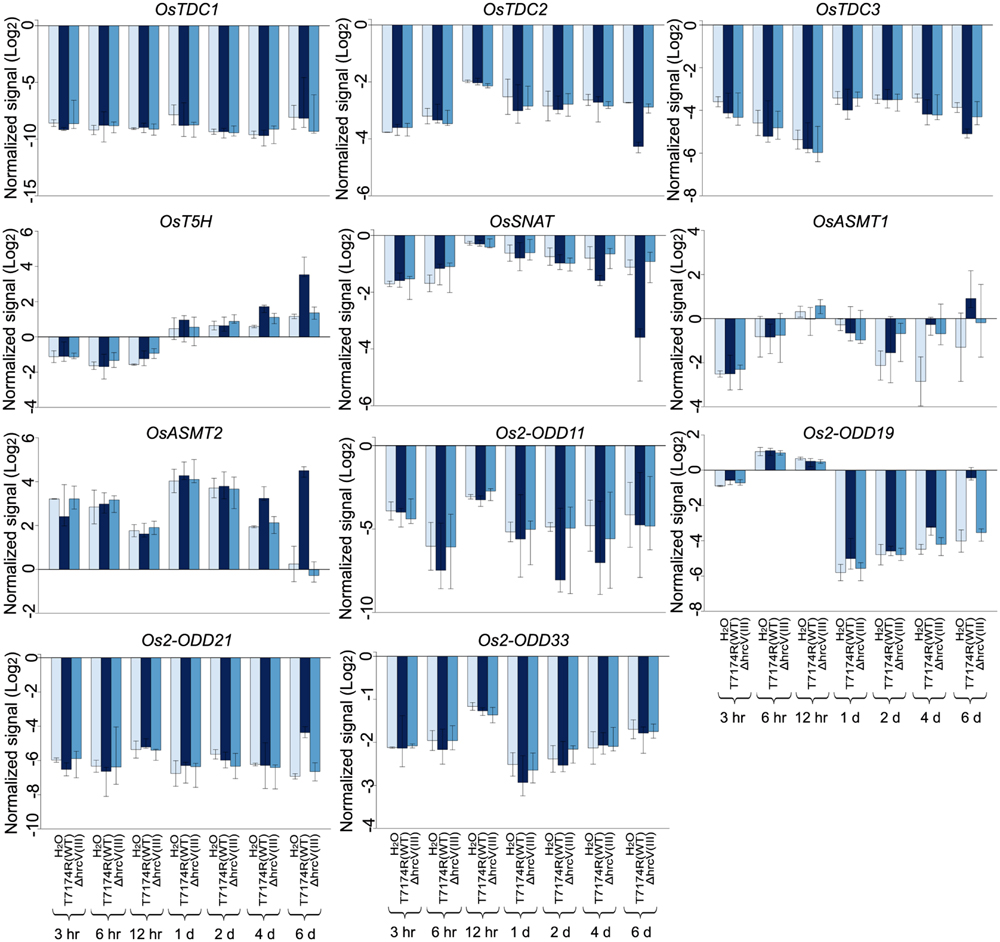
FIGURE 8. Gene expression profile in whole leaf of rice melatonin synthesis and catabolic genes inoculated with Xoo, the causal agent of bacterial blight disease. The transcriptional expression data were obtained from RiceXPro (http://ricexpro.dna.affrc.go.jp/). Rice leaves inoculated with T7114R, a wild-type strain, or 74HrcV::Km, ahrcV mutant deficient in type III secretion (T3S) system, were harvested at 3 hpi, 6 hpi, 12 hpi, 1 dpi, 2 dpi, 4 dpi, and 6 dpi. The expression profile for each gene in various tissues is shown as normalized data (log2).
After inoculation with the blast fungus (M. oryzae), the transcript levels of three genes (OsT5H, OsASMT1, and Os2-ODD11) showed up-regulation at most treated time points, while those of four genes (OsTDC1, -3, OsSNAT, and Os2-ODD21) showed down-regulation (Supplementary Figure S1). In response to inoculation with the three strains of M. oryzae harboring AVR-Pia and AVR-Pish (Pia/Pish × P91-15B, Pish × P91-15B, and Pish × Kyu77-07A), the transcript levels of OsASMT1, Os2-ODD11, -19, -33, and OsT5H showed significant up-regulation, while those of OsTDC3, OsSNAT, and Os2-ODD21 were seriously down-regulated at all time points (Supplementary Figure S1). OsASMT2 expression showed no obvious trends in response to inoculation with the two former, while was induced at all the treated time-points after inoculation with Pish × Kyu77-07A strain (Supplementary Figure S1). In response to inoculation with the pish mutant of M. oryzae (ΔPish × Kyu77-07A), the expressions of six genes (OsTDC1, OsT5H, OsASMT1, -2, Os2-ODD11, and 19) were significantly induced at all time points, while four genes (OsTDC2, -3, OsSNAT, and Os2-ODD33) were seriously down-regulated (Supplementary Figure S1). Os2-ODD21 expression was strongly repressed at 2 dpi. Notably, Os2-ODD33 expression was induced at all the treated time-points after inoculation with the three strains of M. oryzae harboring AVR-Pia and AVR-Pish, but was repressed in response to inoculation with ΔPish × Kyu77-07A strain (Supplementary Figure S1).
The Transcriptional Profile of Rice Melatonin Synthesis and Catabolic Genes in Response to Abiotic Stress Treatments
Melatonin is widely involved in plant stress responses (Shi et al., 2015b,d). Thus, investigation of the expression profiles of melatonin-related genes of rice in response to various abiotic stresses is needed. After drought treatment, the transcripts of OsTDC1, OsASMT1, and Os2-ODD19 were found to be up-regulated between 1.2 and 1.8-folds, whereas those of OsTDC2, -3, Os2-ODD11, -21, OsT5H, and OsSNAT were strongly repressed in comparison to the control (Figure 9). After salt treatment, the transcripts of OsTDC1, -3, OsASMT1, and Os2-ODD19 were increased between 1.3 and 4.5-fold, whereas NaCl strongly repressed OsTDC2, OsT5H, OsSNAT, and Os2-ODD11 expressions (Figure 9). After 4°C treatment, OsTDC1 and OsASMT1 transcript levels were slightly increased, whereas the expressions of six genes (Os2-ODD11, -19, -21, OsTDC3, OsT5H, and OsSNAT) were obviously down-regulated (Figure 9). Notably, the transcript of OsTDC3 was significantly increased after salt treatment, but was seriously decreased in response to drought and cold treatments (Figure 9). The result indicated that OsTDC1 and OsTDC3 may be involved in salt stress response.
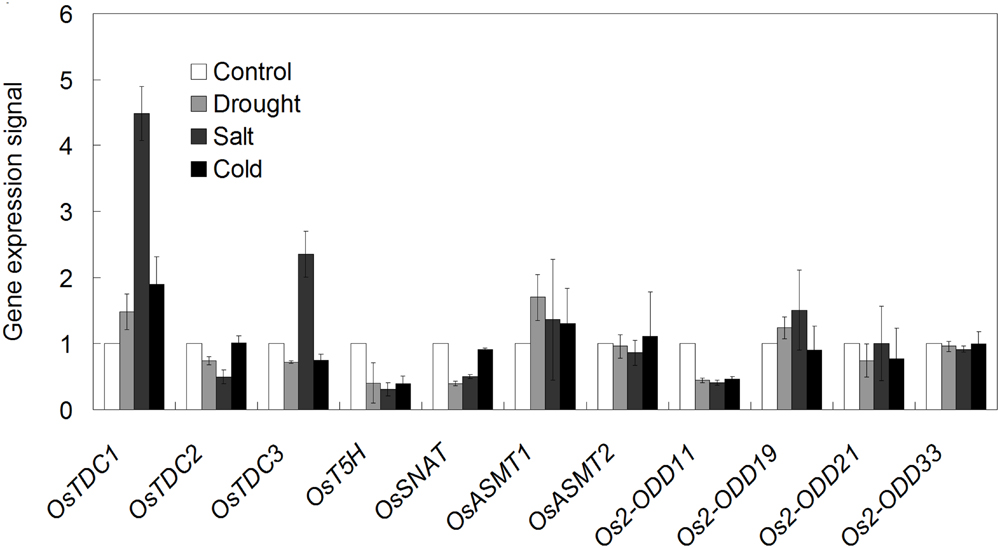
FIGURE 9. The transcriptional profile of rice melatonin synthesis and catabolic genes in response to abiotic stress treatments. The transcriptional expression data were obtained from Rice eFP Browser (http://bar.utoronto.ca/efprice/cgi-bin/efpWeb.cgi).
Discussion
To our knowledge, this is the first study systematically analyzed the diurnal and circadian transcriptional profile of melatonin synthesis and catabolic genes throughout the entire growth stages in rice. Our study confirmed that OsTDCs showed higher expression level at reproductive 1 and reproductive ripening stages then other stages (Figure 5). In cherry fruit (Prunus avium), the expression level of PaTDC was positively correlated to melatonin concentration throughout the entire period, and showed regularly circadian rhythm during a 24 h period with two peaks at 5:00 and 14:00 (Zhao et al., 2013), indicating that the melatonin concentration was higher in that two stages then others. The expression pattern of OsTDC3 also showed circadian rhythm on 16 DAT (Figure 6). Similarly, this expression pattern was also existed at Os2-ODD19 during the entire development (Figure 5). These results suggested that OsTDC3 and Os2-ODD19 may be involved in modulating endogenous during the entire development in rice.
Melatonin is widely involved in plant growth and development, as well as stress responses (Bajwa et al., 2014; Meng et al., 2014; Wang et al., 2014; Zuo et al., 2014; Liang et al., 2015; Shi et al., 2015b). In apple (Malus prunifolia), the transcripts of melatonin synthesis genes (MdTDC1, MdAANAT2, MdT5H4, and MdASMT1) was induced after drought treatment (Li et al., 2014). The concentration of melatonin was increased in barley roots and lupin after cold, drought and salt treatments (Arnao and Hernández-Ruiz, 2009, 2013). Consistently, OsTDC1 and OsASMT1 transcript levels were increased after drought, salt and cold treatments (Figure 9), indicating their possible involvement in abiotic stress response.
Recently, melatonin was shown to function as positive modulator against plant pathogen infection (Yin et al., 2013; Lee et al., 2014, 2015; Reiter et al., 2015; Shi et al., 2015a; Zhao et al., 2015). Exogenous application of melatonin improved apple resistance to D. mali, the pathogen of Marssonina apple blotch (Yin et al., 2013), enhanced Arabidopsis and tobacco defense against P. syringae pv. tomato DC3000 (Lee et al., 2014). Moreover, Arabidopsis snat knockout mutants exhibited increased susceptibility to the avirulent pathogen P. syringae pv. tomato DC3000 with decreased SA levels and reduced defense genes expression compared with wild-type (Lee et al., 2015). However, whether OsSNAT also involves in the regulation of plant immunity remains unknown. Treatments with melatonin significantly enhanced antioxidant protection in rice, suggesting that melatonin plays a major role in regulating pathogen infection (Liang et al., 2015). In response to bacterial pathogen infection, some genes showed similar expression profiles (Figure 8). The differential response of melatonin-related genes to pathogen infection in different kinds implied the dual mechanisms underlying melatonin-related genes mediated pathogens responses.
Previous studies have revealed that melatonin had significant effect in regulating ABA and GA4 in plant response to salinity and drought stress (Li et al., 2014; Zhang H.J. et al., 2014). Additionally, melatonin shared the common substrate (tryptophan) with IAA (Arnao and Hernández-Ruiz, 2014), and AXR3/IAA17 is involved in Arabidopsis melatonin signaling underlying senescence (Shi et al., 2015c). Thus, genome-wide expression analysis of melatonin-related genes in response to plant hormones may provide new insight into crosstalk between melatonin and plant hormones. Plant hormones, such as ABA, SA, and GA, related with most of the plant physiological responses, including water logging, drought and salt stress responses (Yang et al., 2004; Kim et al., 2011, 2014; Shimamura et al., 2014). Melatonin is also a regulator of ABA and GA4 in plant response to salinity and drought stress (Li et al., 2014; Zhang H.J. et al., 2014). Moreover, the expression of OsASMT2 and OsASMT3 were induced after ABA and JA treatments at 1-month-old detached rice leaves, while were down-regulated in response to ethephone, zeatin, and SA stress. OsASMT1 also showed up-regulation upon ABA and JA stress treatment, but did not display obvious trends during ethephone, zeatin, and SA treatments (Park et al., 2013a). In this study, the transcripts of four genes (Os2-ODD11,-19, OsASMT2, and OsT5H) and five genes (OsASMT1, -2, OsT5H, Os2-ODD19, and OsTDC3) were increased in response to ABA stress during 3 h to 6 h treatment in root and during 3 to 12 h in shoot, respectively. Under JA stress, the transcripts of three genes (OsASMT2, OsT5H, and Os2-ODD11) and three genes (OsASMT1, -2, and Os2-ODD11) were significantly increased during 0.25 to 6 h in root and 3 h to 12 h in shoot, respectively. Thus, different transcriptional responses of melatonin-related genes in hormone specific manners, suggested the dual role and crosstalk between melatonin and various hormones.
It is widely known that ABA is the most important regulator of abiotic stress (Kim et al., 2011, 2014; Shimamura et al., 2014), and JA serves as the major defense hormone that are associated with pathogen infection (Van der Ent et al., 2009; Ballaré, 2011; Xie et al., 2011; Yamada et al., 2012; Yang et al., 2013; Campos et al., 2014). More recently, the crucial role of ABA in virulence of rice blast fungus M. oryzae is confirmed (Spence et al., 2015), the involvement of JA in abiotic stress response is also largely confirmed (Riemann et al., 2015; Wasternack and Strnad, 2016). We highlight the commonly regulation of rice melatonin-related genes by ABA, JA, pathogen infection and various abiotic stresses (Figures 7–9), indicating the possible role of these genes in multiple stress responses and underlying crosstalks of plant hormones, especially ABA and JA. Weeda et al. (2014) identified 1308 differentially expressed genes (566 up-regulated genes and 742 down-regulated genes) exhibiting at least of twofold change by exogenous melatonin treatment in Arabidopsis, and many of them are enriched in plant hormone signaling. These differentially expressed genes include 52 genes in auxin signaling, 50 genes in ABA signaling, 67 genes in JA pathway, and 42 genes in ET pathway. Our studies together with the data of Weeda et al. (2014) further indicate the interaction among melatonin, ABA and JA pathways.
We have to pointed out the possible limitation of this study, since the different changes in expression levels of the various genes do not always explain in a simple way why melatonin concentrations increase or decrease under the different conditions. On one hand, there may be difference between expression level and enzyme activity, such as the posttranslational regulation of AANAT in primates via phosphorylation/dephosphorylation and association/dissociation of a 14-3-3 protein, which is decisive for the melatonin rhythm in those organisms (Ganguly et al., 2001, 2005). On the other hand, incomplete knowledge of rate-limiting enzymes or isoenzymes may also lead to the difference. Further studies by other methods may give more clues.
Taken together, the expression patterns of 11 melatonin related genes from rice were synthetically analyzed at different periods and after different treatments in this study. These information may provide abundant resources for functional characterization of melatonin related genes. The differential expression patterns of melatonin related genes in different tissues throughout entire growth stages and stress responses will be useful to investigate in vivo role of specific gene in rice development and circadian rhythm. Thus, this study will contribute to better understand the melatonin biosynthesis and catabolic pathway as well as their association with development and stress responses in rice. Further functional analysis of identified candidate genes with potential involvement in circadian rhythm and stress responses will give shed more lights in melatonin-mediated signaling as well as underlying molecular mechanism.
Author Contributions
HS conceived and directed this study, analyzed the data, wrote, and revised the manuscript; YW and HZ performed the experiments, analyzed the data, wrote, and revised the manuscript; WH and LC analyzed the data and revised the manuscript; CH provided suggestions and revised the manuscript. All authors approved the manuscript and the version to be published, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (No.31570249 and No.31260418), the Special Fund for Agro-scientific Research in the Public Interest of China (No.201403075), the Department of Science and Technology of Hainan Province (No.ZDZX2013023), the scientific research foundation of higher education in Hainan province (the education curriculum reform program, title: Research on the overall optimization of the curriculum system and teaching content of Hainan province excellent course-gene engineering, No.Hnjg2016-10), a central financial support to enhance the comprehensive strength of the central and western colleges and universities, the startup funding and the scientific research foundation of Hainan University (No.kyqd1531) and the education curriculum reform program of Hainan University (No.hdjy1601) to HS.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00676
TABLE S1 | The detailed data used for Figures 3–7 and Figure 9. The original data were obtained from RiceXPro (http://ricexpro.dna.affrc.go.jp/) and Rice eFP Browser (http://bar.utoronto.ca/efprice/cgi-bin/efpWeb.cgi).
FIGURE S1 | Gene expression profile in whole leaf of rice melatonin synthetic and degradated genes inoculated with the blast fungus, M. oryzae. The transcriptional expression data were obtained from RiceXPro (http://ricexpro.dna.affrc.go.jp/). Three lines of rice cultivar Nipponbare carrying the blast resistance genes (Pia, Pish) were inoculated with two strains of M. oryzae harboring AVR-Pia and AVR-Pish. Rice seedlings at the 4-leaf stage were inoculated with the conidial suspension of M. oryzae and the leaves were harvested at 1, 2, 3, and 5 dpi. The expression profile for each gene in various tissues is shown as normalized data (log2).
Abbreviations
2-ODD, 2-oxoglutarate-dependent dioxygenase; AANAT, arylalkylamine N-acetyltransferase; ABA, abscisic acid; ASMT, N-aceylserotonin methyltransferase; AXR3, Auxin Resistant 3; BL, indole-3-acetic acid; DAT, days after transplanting; GA, gibberellic acid; GEO, Gene Expression Omnibus; hpi, hour post inoculation; IAA, indole-3-acetic acid; JA, jasmonic acid; M2H, melatonin 2-hydroxylase; SNAT, serotonin N-acetyltransferase; T3S, type III secretion system; T5H, tryptamine 5-hydroxylase; TDC, tryptophan decarboxylase; tZ, trans-zeatin; Xoo, Xanthomonas oryzae pv. oryzae.
Footnotes
- ^http://ricexpro.dna.affrc.go.jp/
- ^http://bar.utoronto.ca/efprice/cgi-bin/efpWeb.cgi
- ^http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm
- ^http://jtreeview.sourceforge.net/
References
Arnao, M. B., and Hernández-Ruiz, J. (2009). Chemical stress by different agents affects the melatonin content of barley roots. J. Pineal Res. 46, 295–299. doi: 10.1111/j.1600-079X
Arnao, M. B., and Hernández-Ruiz, J. (2013). Growth conditions determine different melatonin levels in Lupinus albus L. J. Pineal Res. 55, 149–155. doi: 10.1111/jpi.12055
Arnao, M. B., and Hernández-Ruiz, J. (2014). Melatonin: plant growth regulator and/or biostimulator during stress? Trends Plant Sci. 19, 789–797. doi: 10.1016/j.tplants.2014.07.006
Arnao, M. B., and Hernández-Ruiz, J. (2015). Functions of melatonin in plants: a review. J. Pineal Res. 59, 133–150. doi: 10.1111/jpi.12253
Bajwa, V. S., Shukla, M. R., Sherif, S. M., Murch, S. J., and Saxena, P. K. (2014). Role of melatonin in alleviating cold stress in Arabidopsis thaliana. J. Pineal Res. 56, 238–245. doi: 10.1111/jpi.12115
Ballaré, C. L. (2011). Jasmonate-induced defenses: a tale of intelligence, collaborators and rascals. Trends Plant Sci. 16, 249–257. doi: 10.1016/j.tplants.2010.12.001
Byeon, Y., and Back, K. (2014a). An increase in melatonin in transgenic rice causes pleiotropic phenotypes, including enhanced seedling growth, delayed flowering, and low grain yield. J. Pineal Res. 56, 408–414. doi: 10.1111/jpi.12129
Byeon, Y., and Back, K. (2014b). Melatonin synthesis in rice seedlings in vivo is enhanced at high temperatures and under dark conditions due to increased serotonin N-acetyltransferase and N-acetylserotonin methyltransferase activities. J. Pineal Res. 56, 189–195. doi: 10.1111/jpi.12111
Byeon, Y., and Back, K. (2015). Molecular cloning of melatonin 2-hydroxylase responsible for 2-hydroxymelatonin production in rice (Oryza sativa). J. Pineal Res. 58, 343–351. doi: 10.1111/jpi.12220
Byeon, Y., Lee, H. Y., Hwang, O. J., Lee, H. J., Lee, K., and Back, K. (2015). Coordinated regulation of melatonin synthesis and degradation genes in rice leaves in response to cadmium treatment. J. Pineal Res. 58, 470–478. doi: 10.1111/jpi.12232
Byeon, Y., Park, S., Kim, Y. S., and Back, K. (2013). Microarray analysis of genes differentially expressed in melatonin-rich transgenic rice expressing a sheep serotonin N-acetyltransferase. J. Pineal Res. 55, 357–363. doi: 10.1111/jpi.12077
Byeon, Y., Park, S., Kim, Y. S., Park, D. H., Lee, S., and Back, K. (2012). Light-regulated melatonin biosynthesis in rice during the senescence process in detached leaves. J. Pineal Res. 53, 107–111. doi: 10.1111/j.1600-079X
Byeon, Y., Park, S., Lee, H. Y., Kim, Y. S., and Back, K. (2014). Elevated production of melatonin in transgenic rice seeds expressing rice tryptophan decarboxylase. J. Pineal Res. 56, 275–282. doi: 10.1111/jpi.12120
Campos, M. L., Kang, J. H., and Howe, G. A. (2014). Jasmonate-triggered plant immunity. J. Chem. Ecol. 40, 657–675. doi: 10.1007/s10886-014-0468-3
Dubbels, R., Reiter, R. J., Klenke, E., Goebel, A., Schnakenberg, E., Ehlers, C., et al. (1995). Melatonin in edible plants identified by radioimmunoassay and by high performance liquid chromatography-mass spectrometry. J. Pineal Res. 18, 28–31. doi: 10.1111/j.1600-079X.1995.tb00136.x
Fujiwara, T., Maisonneuve, S., Isshiki, M., Mizutani, M., Chen, L., Wong, H., et al. (2010). Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J. Biol. Chem. 285, 11308–11318. doi: 10.1074/jbc.M109.091371
Ganguly, S., Gastel, J. A., Weller, J. L., Schwartz, C., Jaffe, H., Namboodiri, M. A., et al. (2001). Role of a pineal cAMP-operated arylalkylamine N-acetyltransferase/14-3-3-binding switch in melatonin synthesis. Proc. Natl. Acad. Sci. U.S.A. 98, 8083–8088. doi: 10.1073/pnas.141118798
Ganguly, S., Weller, J. L., Ho, A., Chemineau, P., Malpaux, B., and Klein, D. C. (2005). Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc. Natl. Acad. Sci. U.S.A. 102, 1222–1227. doi: 10.1073/pnas.0406871102
Hardeland, R. (2015). Melatonin in plants and other phototrophs: advances and gaps concerning the diversity of functions. J. Exp. Bot. 66, 627–646. doi: 10.1093/jxb/eru386
Hattori, A., Migitaka, H., Iigo, M., Itoh, M., Yamamoto, K., Ohtani-Kaneko, R., et al. (1995). Identification of melatonin in plants and its effects on plasma melatonin levels and binding to melatonin receptors in vertebrates. Biochem. Mol. Biol. Inter. 35, 627–634.
Hernández-Ruiz, J., Cano, A., and Arnao, M. B. (2004). Melatonin: a growth-stimulating compound present in lupin tissues. Planta 220, 140–144. doi: 10.1007/s00425-004-1317-3
Hernández-Ruiz, J., Cano, A., and Arnao, M. B. (2005). Melatonin acts as a growth-stimulating compound in some monocot species. J. Pineal Res. 39, 137–142. doi: 10.1111/j.1600-079X.2005.00226.x
Jain, M., Nijhawan, A., Arora, R., Agarwal, P., Ray, S., Sharma, P., et al. (2007). F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143, 1467–1483. doi: 10.1104/pp.106.091900
Kang, K., Kim, Y. S., Park, S., and Back, K. W. (2009). Senescence-induced serotonin biosynthesis and its role in delaying senescence in rice leaves. Plant Physiol. 150, 1380–1393. doi: 10.1104/pp.109.138552
Kang, K., Kong, K., Park, S., Natsagdorj, U., Kim, Y., and Back, K. (2011). Molecular cloning of a plant N-acetylserotonin methyltransferase and its expression characteristics in rice. J. Pineal Res. 50, 304–309. doi: 10.1111/j.1600-079X.2010.00841.x
Kang, K., Lee, K., Park, S., Byeon, Y., and Back, K. (2013). Molecular cloning of rice serotonin N-acetyltransferase, the penultimate gene in plant melatonin biosynthesis. J. Pineal Res. 55, 7–13. doi: 10.1111/jpi.12011
Kang, K., Lee, K., Park, S., Kim, Y. S., and Back, K. W. (2010). Enhanced production of melatonin by ectopic overexpression of human serotonin N-acetyltransferase plays a role in cold resistance in transgenic rice seedlings. J. Pineal Res. 49, 176–182. doi: 10.1111/j.1600-079X.2010.00783.x
Kang, S., Kang, K., Lee, K., and Back, K. (2007). Characterization of rice tryptophan decarboxylases and their direct involvement in serotonin biosynthesis in transgenic rice. Planta 227, 263–272. doi: 10.1007/s00425-007-0614-z
Kim, Y. H., Khan, A. L., Hamayun, M., Kang, S. M., Beom, Y. J., and Lee, I. J. (2011). Influence of short-term silicon application on endogenous physiohormonal levels of Oryza sativa L. under wounding stress. Biol. Trace Elem. Res. 144, 1175–1185. doi: 10.1007/s12011-011-9047-4
Kim, Y. H., Khan, A. L., Kim, D. H., Lee, S. Y., Kim, K. M., Waqas, M., et al. (2014). Silicon mitigates heavy metal stress by regulating P-type heavy metal ATPases, Oryza sativa low silicon genes, and endogenous phytohormones. BMC Plant Biol. 14:13. doi: 10.1186/1471-2229-14-13
Kolár, J., and Machácková, I. (2005). Melatonin in higher plants: occurrence and possible functions. J. Pineal Res. 39, 333–341. doi: 10.1111/j.1600-079X.2005.00276.x
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, H. Y., Byeon, Y., and Back, K. (2014). Melatonin as a signal molecule triggering defense responses against pathogen attack in Arabidopsis and tobacco. J. Pineal Res. 57, 262–268. doi: 10.1111/jpi.12165
Lee, H. Y., Byeon, Y., Tan, D. X., Reiter, R. J., and Back, K. (2015). Arabidopsis serotonin N-acetyltransfase knockout mutant plants exhibit decreased melatonin and salicylic acid levels resulting in susceptibility to an avirulent pathogen. J. Pineal Res. 58, 291–299. doi: 10.1111/jpi.12214
Lerner, A. B., Case, J. D., Takahashi, Y., Lee, Y., and Mori, W. (1958). Isolation of melatonin, the pineal gland factor that lightening melanocytes. J. Am. Chem. Soc. 81:2587. doi: 10.1021/ja01543a060
Li, C., Tan, D. X., Liang, D., Chang, C., Jia, D., and Ma, F. (2014). Melatonin mediates the regulation of ABA metabolism, free-radical scavenging, and stomatal behaviour in two Malus species under drought stress. J. Exp. Bot. 66, 669–680. doi: 10.1093/jxb/eru476
Liang, C., Zheng, G., Li, W., Wang, Y., Hu, B., Wang, H., et al. (2015). Melatonin delays leaf senescence and enhances salt stress tolerance in rice. J. Pineal Res. 59, 91–101. doi: 10.1111/jpi.12243
Manchester, L. C., Tan, D. X., Reiter, R. J., Park, W., Monis, K., and Qi, W. (2000). High levels of melatonin in the seeds of edible plants: the seed of edible plants: possible function in germ tissue protection. Life Sci. 67, 3023–3029. doi: 10.1016/S0024-3205(00)00896-1
Meng, J. F., Xu, T. F., Wang, Z. Z., Fang, Y. L., Xi, Z. M., and Zhang, Z. W. (2014). The ameliorative effects of exogenous melatonin on grape cuttings under water-deficient stress: antioxidant metabolites, leaf anatomy, and chloroplast morphology. J. Pineal Res. 57, 200–212. doi: 10.1111/jpi.12159
Okazaki, M., and Ezura, H. (2009). Profiling of melatonin in model tomato (Solanum lycopersicum L.) cultivar micro-tom. J. Pineal Res. 43, 338–343. doi: 10.1111/j.1600-079X.2009.00668.x
Okazaki, M., Higuchi, K., Aouini, A., and Ezura, H. (2010). Lowering intercellular melatonin levels by transgenic analysis of indoleamine 2, 3-dioxygenase from rice in tomato plants. J. Pineal Res. 49, 239–247. doi: 10.1111/j.1600-079X.2010.00788.x
Okazaki, M., Higuchi, K., Hanawa, Y., Shiraiwa, Y., and Ezura, H. (2009). Cloning and characterization of a Chlamydomonas reinhardtii cDNA arylalkylamine N -acetyltransferase and its use in the genetic engineering of melatonin content in the Micro-Tom tomato. J. Pineal Res. 43, 373–382. doi: 10.1111/j.1600-079X.2009.00673.x
Park, S., and Back, K. W. (2012). Melatonin promotes seminal root elongation and root growth in transgenic rice after germination. J. Pineal Res. 53, 385–389. doi: 10.1111/j.1600-079X.2012.01008.x
Park, S., Byeon, Y., and Back, K. (2013a). Function analyses of three ASMT gene family members in rice plants. J. Pineal Res. 55, 409–415. doi: 10.1111/jpi.12088
Park, S., Lee, D. E., Jang, H., Byeon, Y., Kim, Y. S., and Back, K. (2013b). Melatonin-rich transgenic rice plants exhibit resistance to herbicide-induced oxidative stress. J. Pineal Res. 54, 258–263. doi: 10.1111/j.1600-079X.2012.01029.x
Pelagio-Flores, R., Muňoz-Parra, E., Ortiz-Castro, R., and López-Bucio, J. (2012). Melatonin regulates Arabidopsis root system architecture likely acting independently of auxin signaling. J. Pineal Res. 53, 279–288. doi: 10.1111/j.1600-079X.2012.00996.x
Posmyk, M. M., Bałabusta, M., Wieczorek, M., Sliwinska, E., and Janas, K. M. (2009a). Melatonin applied to cucumber (Cucumis sativus L.) seeds improves germination during chilling stress. J. Pineal Res. 46, 214–223. doi: 10.1111/j.1600-079X.2008.00652.x
Posmyk, M. M., Janas, K. M., and Kontek, R. (2009b). Red cabbage anthocyanin extract alleviates copper-induced cytological disturbances in plant meristematic tissue and human lymphocytes. Biometals 22, 479–490. doi: 10.1007/s10534-009-9205-8
Posmyk, M. M., Kuran, H., Marciniak, K., and Janas, K. M. (2008). Presowing seed treatment with melatonin protects red cabbage seedlings against toxic copper ion concentrations. J. Pineal Res. 45, 24–31. doi: 10.1111/j.1600-079X.2007.00552.x
Reiter, R. J., Tan, D. X., Zhou, Z., Cruz, M. H., Fuentes-Broto, L., and Galano, A. (2015). Phytomelatonin: assisting plants to survive and thrive. Molecules 20, 7396–7437. doi: 10.3390/molecules20047396
Riemann, M., Dhakarey, R., Hazman, M., Miro, B., Kohli, A., and Nick, P. (2015). Exploring jasmonates in the hormonal network of drought and salinity responses. Front. Plant Sci. 6:1077. doi: 10.3389/fpls.2015.01077
Saldanha, A. J. (2004). Java treeview–extensible visualization of microarray data. Bioinformatics 20, 3246–3248. doi: 10.1093/bioinformatics/bth349
Sato, Y., Antonio, B. A., Namiki, N., Motoyama, R., Sugimoto, K., Takehisa, H., et al. (2011a). Field transcriptome revealed critical developmental and physiological transitions involved in the expression of growth potential in japonica rice. BMC Plant Biol. 11:10. doi: 10.1186/1471-2229-11-10
Sato, Y., Antonio, B., Namiki, N., Takehisa, H., Minami, H., Kamatsuki, K., et al. (2011b). RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 39, D1141–D1148. doi: 10.1093/nar/gkq1085
Sato, Y., Namiki, N., Takehisa, H., Kamatsuki, K., Minami, H., Ikawa, H., et al. (2013). Rice FREND: a platform for retrieving coexpressed gene networks in rice. Nucleic Acids Res. 41, D1214–D1221. doi: 10.1093/nar/gks1122
Shi, H., and Chan, Z. (2014). The cysteine2/histidine2-type transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA 6-activated C-REPEAT-BINDING FACTOR pathway is essential for melatonin-mediated freezing stress resistance in Arabidopsis. J. Pineal Res. 57, 185–191. doi: 10.1111/jpi.12155
Shi, H., Chen, Y., Tan, D. X., Reiter, R. J., Chan, Z., and He, C. (2015a). Melatonin induces nitric oxide and the potential mechanisms relate to innate immunity against bacterial pathogen infection in Arabidopsis. J. Pineal Res. 59, 102–108. doi: 10.1111/jpi.12244
Shi, H., Jiang, C., Ye, T., Tan, D. X., Reiter, R. J., Zhang, H., et al. (2015b). Comparative physiological, metabolomic, and transcriptomic analyses reveal mechanisms of improved abiotic stress resistance in bermudagrass [Cynodon dactylon (L). Pers.] by exogenous melatonin. J. Exp. Bot. 66, 681–694. doi: 10.1093/jxb/eru373
Shi, H., Reiter, R. J., Tan, D. X., and Chan, Z. (2015c). INDOLE-3-ACETIC ACID INDUCIBLE 17 positively modulates natural leaf senescence through melatonin-mediated pathway in Arabidopsis. J. Pineal Res. 58, 26–33. doi: 10.1111/jpi.12188
Shi, H., Wang, X., Tan, D. X., Reiter, R. J., and Chan, Z. (2015d). Comparative physiological and proteomic analyses reveal the actions of melatonin in the reduction of oxidative stress in Bermuda grass (Cynodon dactylon (L). Pers.). J. Pineal Res. 59, 120–131. doi: 10.1111/jpi.12246
Shimamura, S., Yoshioka, T., Yamamoto, R., Hiraga, S., Nakamura, T., Shimada, S., et al. (2014). Role of abscisic acid in flood-induced secondary aerenchyma formation in soybean (Glycine max) hypocotyls. Plant Prod. Sci. 17, 131–137. doi: 10.1626/pps.17.131
Spence, C. A., Lakshmanan, V., Donofrio, N., and Bais, H. P. (2015). Crucial roles of abscisic acid biogenesis in virulence of rice blast fungus Magnaporthe oryzae. Front. Plant Sci. 6:1082. doi: 10.3389/fpls.2015.01082
Sun, Q., Zhang, N., Wang, J., Zhang, H., Li, D., Shi, J., et al. (2015). Melatonin promotes ripening and improves quality of tomato fruit during postharvest life. J. Exp. Bot. 66, 657–668. doi: 10.1093/jxb/eru332
Tiryaki, I., and Keles, H. (2012). Reversal of the inhibitory effect of light and high temperature on germination of Phacelia tanacetifolia seeds by melatonin. J. Pineal Res. 52, 332–339. doi: 10.1111/j.1600-079X.2011.00947.x
Turk, H., Erdal, S., Genisel, M., Atici, O., Demir, Y., and Yanmis, D. (2014). The regulatory effect of melatonin on physiological, biochemical and molecular parameters in cold-stressed wheat seedlings. Plant Growth Regul. 74, 139–152. doi: 10.1007/s10725-014-9905-0
Van der Ent, S., Van Wees, S. C., and Pieterse, C. M. (2009). Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 70, 1581–1588. doi: 10.1016/j.phytochem.2009.06.009
Wang, L., Zhao, Y., Reiter, R. J., He, C., Liu, G., Lei, Q., et al. (2014). Change in melatonin levels in transgenic ‘Micro-Tom’ tomato over-expressing ovine AANAT and ovine HIOMT genes. J. Pineal Res. 56, 126–133. doi: 10.1111/jpi.12105
Wang, P., Sun, X., Chang, C., Feng, F., Liang, D., Cheng, L., et al. (2013a). Delay in leaf senescence of Malus hupehensis by long-term melatonin application is associated with its regulation of metabolic status and protein degradation. J. Pineal Res. 55, 424–434. doi: 10.1111/jpi.12091
Wang, P., Sun, X., Li, C., Wei, Z., and Liang, D. (2013b). Long-term exogenous application of melatonin delays drought-induced leaf senescence in apple. J. Pineal Res. 54, 292–302. doi: 10.1111/jpi.12017
Wang, P., Yin, L., Liang, D., Li, C., Ma, F., and Yue, Z. (2012). Delayed senescence of apple leaves by exogenous melatonin treatment: toward regulating the ascorbate–glutathione cycle. J. Pineal Res. 53, 11–20. doi: 10.1111/j.1600-079X.2011.00966.x
Wasternack, C., and Strnad, M. (2016). Jasmonate signaling in plant stress responses and development-active and inactive compounds. N. Biotechnol. doi: 10.1016/j.nbt.2015.11.001 [Epub ahead of print].
Weeda, S., Zhang, N., Zhao, X., Ndip, G., Guo, Y., Buck, G. A., et al. (2014). Arabidopsis transcriptome analysis reveals key roles of melatonin in plant defense systems. PLoS ONE 9:e93462. doi: 10.1371/journal.pone.0093462
Wei, W., Li, Q. T., Chu, Y. N., Reiter, R. J., Yu, X. M., Zhu, D. H., et al. (2014). Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J. Exp. Bot. 66, 695–707. doi: 10.1093/jxb/eru392
Xie, X. Z., Xue, Y. J., Zhou, J. J., Zhang, B., Chang, H., and Takano, M. (2011). Phytochromes regulate SA and JA signaling pathways in rice and are required for developmentally controlled resistance to Magnaporthe grisea. Mol. Plant 4, 688–696. doi: 10.1093/mp/ssr005
Yamada, S., Kano, A., Tamaoki, D., Miyamoto, A., Shishido, H., Miyoshi, S., et al. (2012). Involvement of OsJAZ8 in jasmonate-induced resistance to bacterial blight in rice. Plant Cell Physiol. 53, 2060–2072. doi: 10.1093/pcp/pcs145
Yang, D. L., Yang, Y., and He, Z. (2013). Roles of plant hormones and their interplay in rice immunity. Mol. Plant 6, 675–685. doi: 10.1093/mp/sst056
Yang, Y., Qi, M., and Mei, C. (2004). Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 40, 909–919. doi: 10.1111/j.1365-313X.2004.02267
Yin, L., Wang, P., Li, M., Ke, X., Li, C., Liang, D., et al. (2013). Exogenous melatonin improves Malus resistance to Marssonina apple blotch. J. Pineal Res. 54, 426–434. doi: 10.1111/jpi.12038
Zhan, N., Sun, Q., Zhang, H., Cao, Y., Weeda, S., Ren, S., et al. (2015). Roles of melatonin in abiotic stress resistance in plants. J. Exp. Bot. 66, 647–656. doi: 10.1093/jxb/eru336
Zhang, H. J., Zhang, N., Yang, R. C., Wang, L., Sun, Q. Q., Li, D. B., et al. (2014). Melatonin promotes seed germination under high salinity by regulating antioxidant systems, ABA and GA interaction in cucumber (Cucumis sativus L.). J. Pineal Res. 57, 269–279. doi: 10.1111/jpi.12167
Zhang, N., Zhang, H. J., Zhao, B., Sun, Q. Q., Cao, Y. Y., Li, R., et al. (2014). The RNA-seq approach to discriminate gene expression profiles in response to melatonin on cucumber lateral root formation. J. Pineal Res. 56, 39–50. doi: 10.1111/jpi.12095
Zhang, N., Zhao, B., Zhang, H. J., Weeda, S., Yang, C., Yang, Z. C., et al. (2013). Melatonin promotes water-stress tolerance, lateral root formation, and seed germination in cucumber (Cucumis sativus L.). J. Pineal Res. 54, 15–23. doi: 10.1111/j.1600-079X
Zhao, H., Xu, L., Su, T., Jiang, Y., Hu, L., and Ma, F. (2015). Melatonin regulates carbohydrate metabolism and defenses against Pseudomonas syringae pv. tomato DC3000 infection in Arabidopsis thaliana. J. Pineal Res. 59, 109–119. doi: 10.1111/jpi.12245
Zhao, Y., Tan, D. X., Lei, Q., Chen, H., Wang, L., Li, T., et al. (2013). Melatonin and its potential biological functions in the fruits of sweet cherry. J. Pineal Res. 55, 79–88. doi: 10.1111/jpi.12044
Keywords: melatonin, rice, gene expression, circadian rhythm, development, immunity, stress response
Citation: Wei Y, Zeng H, Hu W, Chen L, He C and Shi H (2016) Comparative Transcriptional Profiling of Melatonin Synthesis and Catabolic Genes Indicates the Possible Role of Melatonin in Developmental and Stress Responses in Rice. Front. Plant Sci. 7:676. doi: 10.3389/fpls.2016.00676
Received: 23 February 2016; Accepted: 02 May 2016;
Published: 18 May 2016.
Edited by:
Jin-Gui Chen, Oak Ridge National Laboratory, USAReviewed by:
Ruediger Hardeland, University of Goettingen, GermanyKyoungwhan Back, Chonnam National University, South Korea
Copyright © 2016 Wei, Zeng, Hu, Chen, He and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haitao Shi, aGFpdGFvc2hpQGhhaW51LmVkdS5jbg==; Chaozu He, Y3poZUBoYWludS5lZHUuY24=
†These authors have contributed equally to this work.
 Yunxie Wei1†
Yunxie Wei1† Wei Hu
Wei Hu Lanzhen Chen
Lanzhen Chen Chaozu He
Chaozu He Haitao Shi
Haitao Shi