
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 22 March 2016
Sec. Plant Pathogen Interactions
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.00328
This article is part of the Research Topic Advances in plant microbe interactions View all 30 articles
Plants have developed complex defense mechanisms to cope with microbial pathogens. Pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) are perceived by pattern recognition receptors (PRRs), leading to the activation of defense. While substantial progress has been made in understanding the activation of plant defense by PAMPs and DAMPs recognition in tracheophytes, far less information exists on related processes in early divergent plants like mosses. The aim of this study was to identify genes that were induced in P. patens in response to elicitors of Pectobacterium carotovorum subsp. carotovorum, using a cDNA suppression subtractive hybridization (SSH) method. A total of 239 unigenes were identified, including genes involved in defense responses related to the shikimate, phenylpropanoid, and oxylipin pathways. The expression levels of selected genes related to these pathways were analyzed using quantitative RT-PCR, confirming their rapid induction by P.c. carotovorum derived elicitors. In addition, P. patens induced cell wall reinforcement after elicitor treatment by incorporation of phenolic compounds, callose deposition, and elevated expression of Dirigent-like encoding genes. Small molecule defense markers and phytohormones such as cinnamic acid, 12-oxo-phytodienoic acid, and auxin levels all increased in elicitor-treated moss tissues. In contrast, salicylic acid levels decreased while abscisic acid levels remained unchanged. P. patens reporter lines harboring an auxin-inducible promoter fused to β-glucuronidase revealed GUS activity in protonemal and gametophores tissues treated with elicitors of P.c. carotovorum, consistent with a localized activation of auxin signaling. These results indicate that P. patens activates the shikimate, phenylpropanoid, oxylipins, and auxin pathways upon treatment with P.c. carotovorum derived elicitors.
Plants employ complex defense mechanisms against microbial infection that involves recognition of the invader, activation of signal transduction pathways, and the production of proteins and metabolites with different roles in defense. Plant cells perceive the presence of a pathogen by sensing pathogen-associated molecular patterns (PAMPs), such as fungal chitin or bacterial lipopolysaccharides and flagellin, by surface-localized pattern recognition receptors (PRRs; Bittel and Robatzek, 2007; Humphrey et al., 2007; Boller and Felix, 2009). Perception of PAMPs, also known as elicitors, by PRRs activates the first layer of inducible plant defenses, referred to as PAMP-triggered immunity (PTI), which provides protection against non-adapted pathogens (Jones and Dangl, 2006). In addition, some PRRs recognize host-derived “danger” signals, which are endogenous elicitors (damage-associated molecular patterns; DAMPs) such as plant cell wall and cutin fragments that are released by the enzymatic action of pathogens or wounding (Lotze et al., 2007; Boller and Felix, 2009). The ability of plant cells to monitor the presence of pathogens at the cell surface is essential for the activation of an effective defense response. Cellular responses of flowering plants to PAMPs and DAMPs include the accumulation of reactive oxygen species (ROS), the reinforcement of plant cell walls through deposition of callose and lignin, the synthesis of hormones, the expression of defense genes and the production of defensive proteins and metabolites (Hückelhoven, 2007).
While substantial progress has been made in understanding the activation of plant defense by PAMPs and DAMPs recognition in tracheophytes, little information exists on the recognition of pathogens and activation of defense responses in basal land plants like mosses. The moss Physcomitrella patens (P. patens) is a useful model system to analyze plant functions, including the activation of defense mechanisms after pathogen assault (Ponce de León, 2011; Ponce de León and Montesano, 2013). P. patens has several interesting features, including the fact that it can be easily cultivated in vitro, it has a relatively simple developmental pattern, and it responds to environmental stimuli and plant growth regulators similarly as other land plants (Cove et al., 1997, 2006; Schaefer and Zrÿd, 2001). In addition, P. patens can be transformed and targeted disruption of genes with possible roles in defense can be performed due to its high rate of homologous recombination, comparable to yeast cells (Schaefer, 2001). Detection of mutant phenotypes in primary transformants is facilitated by the presence of a dominant haploid gametophytic phase (Cove, 2005). P. patens genome (http://www.cosmoss.org/ and http://www.phytozome.net/; Rensing et al., 2008; Zimmer et al., 2013), ESTs and full-length cDNAs (http://moss.nibb.ac.jp/) are available, and P. patens microarray based expression data can be found at Genevestigator (Zimmermann et al., 2008; https://www.genevestigator.com). P. patens is infected by several pathogens that cause diseases in crop plants, including Botrytis cinerea (B. cinerea), Pythium irregulare, Pythium debaryanum, and Colletotrichum gloeosporioides (C. gloeosporioides) (Ponce de León et al., 2007, 2012; Oliver et al., 2009; Reboledo et al., 2015). In response to infection by these pathogens, P. patens activates defense responses that are conserved among plants, like the accumulation of ROS, the activation of an hypersensitive response (HR)-like response, the reinforcement of the cell wall, the accumulation of the defense hormone salicylic acid and the activation of defense genes (Ponce de León and Montesano, 2013). However, P. patens lacks key defense signals present in flowering plants, such as jasmonic acid, which is an important hormone involved in defense against necrotrophic pathogens (Ponce de León et al., 2012, 2015). Interestingly, while P. patens has a homolog of the fungal chitin receptor (CERK1), no homologs to the flagellin receptor FLS2, and the elongation factor Tu receptor EFR1 are present in its genome (Boller and Felix, 2009). As an evolutionary link between green algae and angiosperms (Lewis and McCourt, 2004), P. patens is an ideal non-vascular plant useful in the comparative analysis of different defense mechanisms associated with the evolution of plants.
The soft rot Pectobacterium carotovorum subsp. carotovorum (P.c. carotovorum; ex Erwinia carotovora subsp. carotovora) is a large-scale producer of plant cell wall degrading enzymes (PCWDEs), including cellulases, proteases, and pectinases (Pérombelon and Kelman, 1980; Davis et al., 1984; Palva et al., 1993; Toth and Birch, 2005). Treatments of flowering plants with cell-free culture filtrate (CF) containing P.c. carotovorum elicitors such as PCWDEs, mimic symptoms caused by pathogen infection, and release cell wall fragments, including oligogalacturonides, that act as endogenous elicitors activating a defense response evidenced by the accumulation of phytoalexin and activation of defense-related genes (Davis et al., 1984; Vidal et al., 1997; Norman-Setterblad et al., 2000; Montesano et al., 2001, 2005). We have previously shown that the strain SCC1 of P.c. carotovorum, harboring the harpin-encoding hrpN gene, which is an elicitor of the HR (Rantakari et al., 2001), infects and causes maceration of P. patens tissues. Similarly, treatments with CF from SCC1 also mimic symptoms development in P. patens (Ponce de León et al., 2007). P. patens activates defense-related gene expression that encode for lipoxygenase (LOX), phenylalanine ammonia-lyase (PAL), chalcone synthase (CHS), and pathogenesis-related-1 (PR-1) proteins (Ponce de León et al., 2007). In order to identify a broader array of genes involved in the defense responses of P. patens against P.c. carotovorum elicitor treatment, a suppression subtractive hybridization cDNA library (SSH) enriched in plant genes induced by P.c. carotovorum elicitors was generated. Here, we show that several genes involved in the shikimate, phenylpropanoid, and oxylipin pathways are induced, as well as genes encoding proteins related to cell wall reinforcement. In addition, auxin levels increased, and auxin signaling was activated in P. patens tissues treated with P.c. carotovorum elicitors.
Physcomitrella patens Gransden wild type isolate was grown axenically on cellophane overlaid BCDAT medium (1.6 g L−1 Hoagland's, 1 mM MgSO4, 1.8 mM KH2PO4 pH 6.5, 10 mM KNO3, 45 μM FeSO4, 1 mM CaCl2, 5 mM ammonium tartrate, and 10 g L−1 agar) as described by Ashton and Cove (1977). Moss colonies were generated and grown at 22°C under a photoperiod of 16 h light as described previously (Oliver et al., 2009). Pectobacterium carotovorum subsp. carotovorum strain SCC1 (Rantakari et al., 2001) was propagated on LB medium at 28°C and culture filtrates (CF) containing the elicitors were prepared according to Ponce de León et al. (2007). The CF was applied by spraying the moss colonies.
For cell death measurement, moss colonies were incubated for 30 min in 0.1% Evans blue and washed four times with water to remove unbound dye. Dye bound to dead cells was solubilized in 50% methanol with 1% SDS for 30 min at 60°C and the absorbance was measured at 600 nm (Levine et al., 1994). Each sample consisted of four colonies incubated in 6 ml of the mixture methanol/SDS. Six samples, corresponding to 24 colonies, were analyzed per experiment. Cell death measurement was repeated thrice and data expressed as OD/mg dry weight. Dry weight was measured after drying plant colonies on cellophane for 18 h at 65°C.
P. patens elicitor-induced cDNAs were generated by suppression subtractive hybridization (SSH; Diatchenko et al., 1996) using the PCR-select cDNA subtraction kit (BD Biosciences Clontech), according to the manufacturer's instructions. Briefly, 2 mg of total poly A+ RNA from 2, 4, 8, and 24 h water-treated moss colonies and 2, 4, 8, and 24 h P.c. carotovorum CF-treated moss colonies were used in equal quantities for synthesis of the driver and tester cDNA pools, respectively. Driver and tester cDNAs were digested with RsaI, phenol/chloroform extracted, ethanol precipitated and resuspended in water. Tester cDNA was then split into two pools, each of which was ligated to a different adapter (supplied in the cDNA subtraction kit). Subtracted cDNA fragments were cloned into the pGEM T-easy vector and transformed into competent Escherichia coli cells. Randomly selected clones were used for insert sequencing, and the cDNA sequences obtained were submitted to the National Center for Biotechnology Information (NCBI).
Total RNA was extracted from plant tissues treated with CF (2, 4, 8, and 24 h) or with water (mock; 2, 4, 8, and 24 h) as a control, using RNeasy Plant Mini Kits according to manufacturer's instructions (Qiagen, Germany). For cDNA synthesis, 4 μg of total RNA were treated with DNase I (Thermo Scientific) and cDNA was synthesized using RevertAid Reverse transcriptase (Thermo Scientific) and oligo (dT) according to the manufacturer's protocol. To validate the results of the SSH library, RT-qPCR amplification of the selected genes was performed using specific oligonucleotide Primers (Supplementary Table 2) designed by Primer3. qPCR was performed in a 96 well thermocycler (C1000 TouchTM Thermal Cycler, CFX96 TM Real-Time System, BioRad) using the Quantitect SYBR Green PCR Kit (Qiagen, Germany). Each 20 μL reactions contained 10 μL of SYBR Green PCR Master mix (2X), 0.5 μM primers mix and 1 μL of template cDNA. The thermocycler was programmed to run for 15 min at 95°C, followed by 40 cycles of 15 s at 94°C, 30 s at 60°C, and 30 s at 72°C. Transcript accumulation of each gene was normalized to the quantity of constitutively expressed EF1α (Le Bail et al., 2013). The amplification efficiencies of the different primer combinations were analyzed and all were >90%. Relative expression was determined using the 2−ΔΔCt method (Livak and Schmittgen, 2001), and the values of the CF-treated samples were expressed relative to the corresponding water-treated samples at the indicated time points. Each data point is the mean value of three biological replicates. Two technical replicates were used for each sample.
Three-week-old moss colonies were treated with P.c. carotovorum elicitors and tissues were collected at 2, 4, 8, and 24 h after treatment. Eight moss colonies were pooled and 200 mg of ground tissue were homogenized, derivatized, vapor phase extracted, and analyzed by gas chromatography/isobutene chemical ionization mass spectrometry (GC/CI-MS), as described previously (Schmelz et al., 2004). Control plants were sprayed with water.
To detect incorporation of phenolic compounds into cell walls, tissues were incubated with 0.01% safranin-O in 50% ethanol for 5 min and rinsed in water according to Oliver et al. (2009). For callose detection, tissues were fixed in ethanol, rinsed in water, and stained with 0.01% methyl blue in phosphate buffer pH 7.0 for 30 min and observed with epifluorescence. Bright field and fluorescence microscopy were performed with an Olympus 1X2-UCB inverted fluorescent microscope and images captured with Cell F software package (Olympus).
In situ localization of GUS activity was performed according to Peleman et al. (1989). Tissues were stained at 37°C for 24 h before destaining in an increasing serial dilution of ethanol, mounted in water, visualized in an Olympus BX61 microscope (Shinjuku-ku, Tokyo, Japan), and images were captured with the Cell F software package (Olympus).
Students t-test (RT-qPCR data), and one-way analysis of variance (ANOVA) followed by Dunnett's multiple comparison post-hoc test (hormone data), were used to evaluate the statistical significance of the difference in values using the GraphPad Prism Software (La Jolla, CA, USA).
CF of P.c. carotovorum contains harpin and PCWDEs that degrade plant tissues and cause cell death. In P. patens, moss tissues treated with CF of P.c. carotovorum develop disease symptoms after 24 h, evidenced by browning of cell walls in protonemal tissues and basal cauloid and rhizoids of gametophore tissues, which correlates with an increase of dead cells (Figure 1). No symptoms could be detected visually at 2, 4, or 8 h after CF treatment (data not shown). In order to identify more genes involved in the defense response of P. patens to P.c. carotovorum derived elicitors, we generated a suppression subtractive hybridization (SSH) cDNA library. In order to identify genes induced at early stages of tissue maceration, we decided to use an mRNA mixture with equal quantities of 2, 4, 8, and 24 h elicitor-treated samples. We only focused on genes up-regulated by P.c. carotovorum derived elicitors.
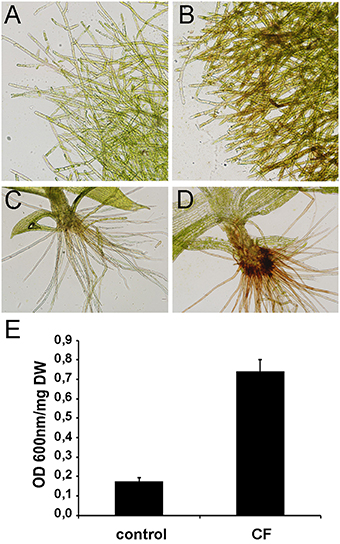
Figure 1. Symptom development in elicitor-treated plants. (A) Water-treated protonemal tissues, (B) elicitor-treated protonemal tissues, (C) water-treated gametophore, (D) elicitor-treated gametophore. (E) Measurement of cell death by Evans blue staining 24 h after treating moss colonies with water (control) or CF. Data were expressed as the optical density (OD) at 600 nm per milligram of dry weight (DW). Values are means with standard deviations of six independent replicate moss samples.
Subtracted cDNA fragments were cloned into a plasmid vector, randomly selected for sequencing (384 clones), and BLAST searched against public databases Phytozome v10.3 (http://www.phytozome.net). This resulted in the identification of expressed sequence tags (ESTs) corresponding to a total of 239 unigenes. A table containing all 239 genes and the predicted functions based on sequence similarities is given in Supplementary Table 1. Based on putative functions, the genes were classified into 11 groups: energy and metabolism (39), cell growth and maintenance (21), oxidation-reduction processes (14), signaling (5), transcription (15), phenylpropanoid (12), protein metabolism (38), defense related (11), others (31), unknown function (46), and no significant similarity (7), (Figure 2). The predicted functions of the different genes were determined manually, based upon their Gene Ontology terms, inspection of their UniProt record (Wu et al., 2006), and by searching published data. Among the genes related to defense responses, we identified genes encoding a putative chitinase, a pathogenesis-related-10 protein, several dirigent proteins, a betaine-aldehyde dehydrogenase, a lipoxygenase, an alpha-dioxygenase, several putative transcription factors with an APETALA2 (AP2) domain and a cyclic nucleotide gated channel protein.
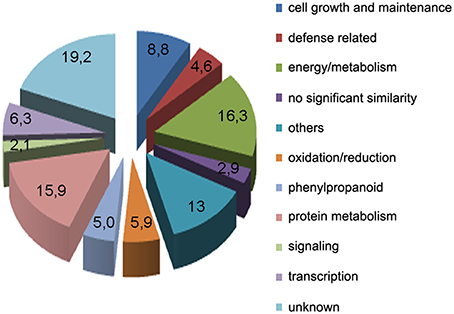
Figure 2. Functional classification of unigenes obtained from the SSH library within biological function categories.
In addition to the previously mentioned genes related to defense responses, several genes induced after elicitor treatment were associated with the shikimate and phenylpropanoid pathways, which are important defense pathways activated in plants under biotic stress. Chorismate is the final product of the shikimate pathway and two genes involved in chorismate synthesis (chorismate synthase; CS, transcript name; Phpat.001G145300.1), and chorismate metabolism (chorismate mutase; CM, Phpat.013G057800.1), were identified in the cDNA library. In addition, 11 differentially expressed unigenes related to the phenylpropanoid pathway were present in the SSH library, including genes encoding two phenylalanine ammonia-lyase (PAL; PAL.1; Phpat.001G070300.1, PAL.2; Phpat.021G025100.1), a cinnamic acid 4-hydroxylase (C4H; Phpat.004G084000.1), a 4-coumarate-CoA ligase (4CL; Phpat.022G056000.1), three chalcone synthases (CHS.1; Phpat.021G025500.1, CHS.2; Phpat.024G048200.1 and CHS.3; Phpat.002G126000.1), two chalcone isomerases (CHI.1; Phpat.026G016200.1 and CHI.2; Phpat.004G098400.1), a flavanone 3-hydroxylase (F3H; Phpat.020G078400.1), and a cinnamyl-alcohol dehydrogenase (CAD; Phpat.011G011800.1). The different genes involved in these pathways and identified in the present study are indicated with blue letters in Figure 3, which supports a large-scale coordinated regulation of phenylpropanoid biosynthesis following elicitation. To verify that the genes identified in the subtracted cDNA library were induced in elicitor-treated tissues, we performed quantitative real-time PCR to analyze the expression of 13 genes related to these defense pathways. Expression analysis was carried out at 2, 4, 8, and 24 h after elicitor and water (mock) treatments of different biological replicates. Levels of the accumulated transcripts were normalized against the expression of the elongation factor 1α (EF1α), which was shown earlier to be a good P. patens reference gene during different conditions, including hormonal treatments (Le Bail et al., 2013). Values of the CF-treated samples were expressed relative to the corresponding water-treated samples at the indicated time points (Figure 4). The obtained qPCR expression data of the selected genes are consistent with the results obtained in the SSH analysis. The results show that all genes were rapidly induced after P.c. carotovorum derived elicitor treatment. Transcripts were all upregulated at 2 and/or 4 h after elicitor treatment, corresponding to tissues harvested prior to visible disease symptoms (Figure 4). In addition, PAL.1, PAL.2, CHS.2, CHS.3, and CAD were also induced at 8 h after elicitor treatment, and C4H, 4CL, CHS.1, CHS.2, and CHS.3 transcript levels increased after 24 h of treatment. All genes showed a peak of expression at 2 or 4 h after elicitor treatment, except for C4H which showed its highest expression at 24 h after treatment (Figure 4). A strong increase in expression levels in tissues treated with elicitors compared to control tissues was observed for PAL.2, CHS.1, and CHS.2. Transcript levels encoding F3H also increased to very high levels (250 fold) in elicitor-treated tissues. Additionally, transcript including CS, CM, PAL.1, C4H, 4CL, CHS.3, CHI.1, CHI.2, and CAD increase approximately 8–70 fold in elicitor-treated tissues compared to control tissues. Since cinnamic acid (CA) is an important intermediate of the phenylpropanoid pathway, we measured CA levels in control and elicitor-treated moss tissues. The results show that cinnamic acid increases significantly after 4 and 8 h of elicitor treatment, reaching maximum levels at 4 h (Figure 4). Taken together, these results show that in response to P.c. carotovorum derived elicitors, P. patens activates very rapidly the expression of defense genes encoding enzymes involved in the shikimate and phenylpropanoid pathways, leading to the production of different metabolites with possible roles in defense, including cinnamic acid.
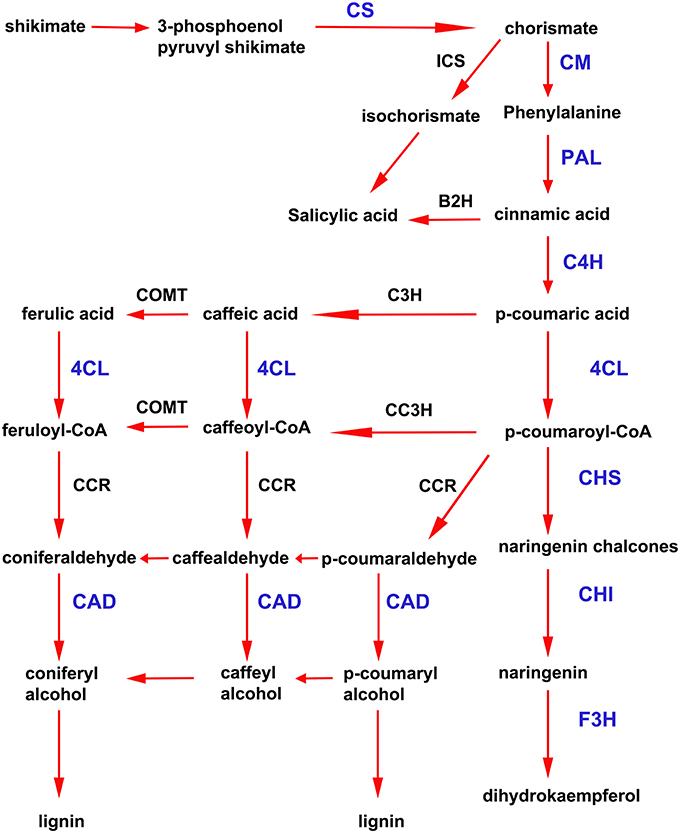
Figure 3. Activation of shikimate and phenylpropanoid pathways in elicitor-treated moss tissues. Simplified scheme of the shikimate and phenylpropanoid pathways. CS, chorismate synthase; CM, chorismate mutase; ICS, isochorismate synthase; B2H, benzoic acid-2-hydroxylase; PAL, phenylalanine ammonia lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4 coumarate CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; C3H, p-coumarate-3-hydroxylase; COMT, caffeate O-methyltransferase; CCR, cinnamoyl-CoA reductase; and CAD, cinnamyl alcohol dehydrogenase. Proteins encoded by the genes that were present in the cDNA library enriched in P.c. carotovorum elicitor inducible genes are indicated in blue letters. Adapted from Zhao et al. (2013) and Yeh et al. (2014).
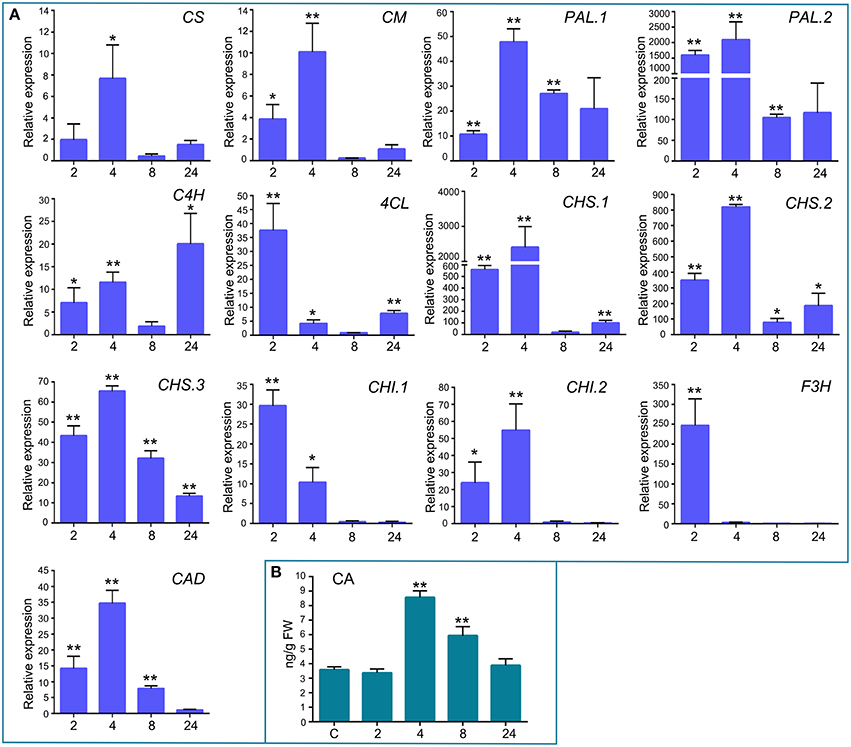
Figure 4. Validation of differentially expressed genes by RT-qPCR, and cinnamic acid accumulation in elicitor-treated tissues. (A) RT-qPCR analysis of selected genes at 2, 4, 8, and 24 h after elicitor-treatment. EF1α gene was used as the reference gene. The expression levels in CF-treated plants are relative to the corresponding level of expression in water-treated plants at the indicated time points. Results are reported as means ± standard deviation (SD) of three samples for each treatment. CS, chorismate synthase; CM, chorismate mutase; PAL, phenylalanine ammonia lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4 coumarate CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; and CAD, cinnamyl alcohol dehydrogenase. Asterisks indicate a statistically significant difference between the elicitor-treated and the water-treated plants (Students t-test, *P < 0.05; **P < 0.005). (B) Endogenous cinnamic acid levels [ng/g fresh weight (FW)] in water-treated and elicitor-treated tissues were analyzed at the indicated time points. Values are means and standard errors of three independent experiments. Statistical differences were evaluated according to one-way ANOVA followed by Dunnett's test relative to control condition, **P < 0.001.
The cell wall is the first barrier to invading pathogens, and plant cells perceive changes in cell wall integrity occurring due to damage caused by wounding and pathogen attack (Humphrey et al., 2007). Several defense genes related to cell wall reinforcement were identified in the cDNA library, including four genes encoding putative dirigent-like proteins, DIR.1 (Phpat.005G083400.1), DIR.2, (Phpat.006G024400.1), DIR.3 (Phpat.006G024500.1), and DIR.4 (Phpat.006G024300.1). DIR proteins are involved in plant defense against pathogens and they are proposed to mediate the free radical coupling of monolignol plant phenols to yield the cell wall polymers lignans and lignins (Davin and Lewis, 2000). Expression analysis confirmed that the DIR genes are induced by P.c. carotovorum derived elicitors (Figure 5A). DIR-1 is induced at 2 and 4 h, while DIR-2 and DIR-3 are induced at 2, 4, 8, and 24 h, and DIR-4 at 2, 4, and 24 h after elicitor treatment (Figure 5A). In addition, while DIR-1 and DIR-3 showed maximum expression levels at 4 h, DIR-2 expression peaked at 24 h and DIR-4 at 2 h after elicitor treatment. Since PCWDEs present in the CF of P.c. carotovorum cause cell wall damage, we evaluated cell wall modification with Safranin-O staining. Elicitor-treated protonemal tissues and leaves show the incorporation of phenolic compounds visualized as pink-red staining of cell walls (Figures 5C–E), while water-treated tissues were not stained (Figure 5B). The fortification mechanism after elicitor treatment was visible after 24 h, which correlates with visual tissue maceration. Callose is a β-1,3-glucan polymer synthesized between the cell wall and the plasma membrane, and deposition of callose makes the cell wall less vulnerable to pathogen infection and degradation by PCWDEs (Jacobs et al., 2003; Ton and Mauch-Mani, 2004). In order to analyze if P.c. carotovorum elicitor treatment induces the deposition of callose in P. patens tissues, protonemal tissue was stained with methyl blue 24 h after elicitor treatment. The results show that protonemal filaments treated with elicitors have callose depositions visualized as bright fluorescent spots, while water treated moss tissues did not (Figures 5F,G). Thus, P. patens induce cell wall reinforcement after P.c. carotovorum elicitor treatment.
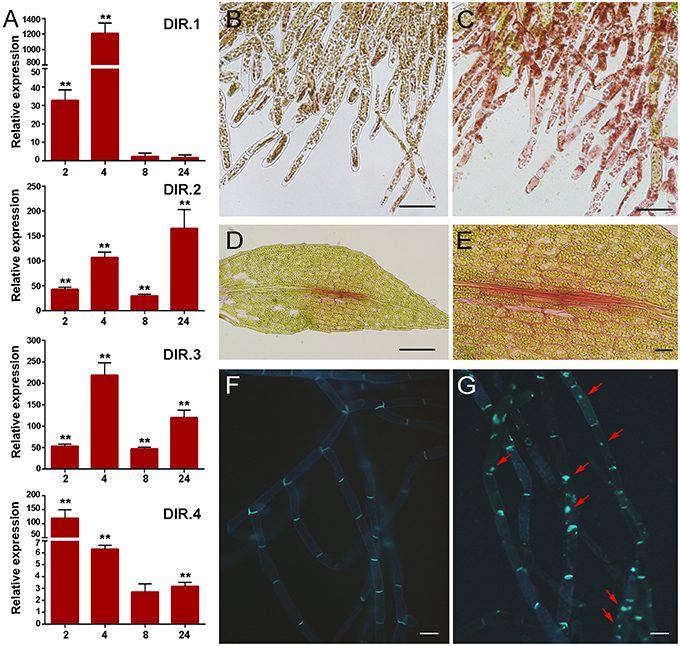
Figure 5. Cell wall fortification after elicitor treatment. (A) Expression levels of dirigent-encoding genes. Expression analyses by RT-qPCR were performed in samples collected at 2, 4, 8, and 24 h after elicitor-treatment. EF1α gene was used as the reference gene. The expression levels in CF-treated plants are relative to the corresponding level of expression in water-treated plants at the indicated time points. Results are reported as means ± standard deviation (SD) of three samples for each treatment. Asterisks indicate a statistically significant difference between the elicitor-treated and the water-treated plants (Student's t-test, **P < 0.005). Incorporation of phenolic compounds by safranin-O staining in water- (B) and CF-treated protonemal tissues (C), and in elicitor-treated leaves (D–E). Methyl blue staining for detection of callose depositions in water-treated (F) and elicitor-treated protonemal tissues (G). Callose deposits are indicated with red arrows. The scale bar represents 100 μm in (B–D) and (E–G) 20 μm.
Oxylipins are oxygenated fatty acids involved in defense responses against pathogens (Kachroo and Kachroo, 2009). Three genes encoding enzymes involved in oxylipin biosynthesis were identified in the cDNA library. One gene encodes an α-Dioxygenase (α-DOX), previously shown to be induced by P.c. carotovorum elicitors and with predicted involvement in protecting P. patens tissues against damage caused by the PCWDEs (Machado et al., 2015). The other genes encode a 13S-lipoxygenase (13-LOX; Phpat.015G051800.1) and a 12-oxo-phytodienoic acid reductase (OPR; Phpat.003G009100.1). In flowering plants, 13-LOX and OPR3 are involved in the synthesis of 12-oxo-phytodienoic acid (OPDA), the defense hormone jasmonic acid (JA), and other oxylipins with different roles in defense (Blée, 2002). The OPR encoding gene was induced after 4 h of elicitor treatment (Figure 6), while no significant increase in LOX transcript levels could be detected in elicitor-treated tissues compared to control tissues (data not shown). Since JA is not produced in P. patens (Ponce de León et al., 2012), we only measured OPDA content in control and elicitor-treated tissues. A small increase in OPDA content was detected in P. patens tissues treated with P.c. carotovorum-derived elicitors compared to control tissues (Figure 6).
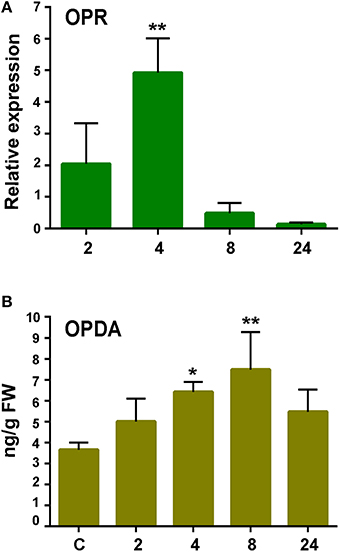
Figure 6. Expression analysis of 12-oxophytodienoic acid reductase (OPR) gene and OPDA accumulation in response to elicitor treatment. (A) RT-qPCR analysis of OPR at 2, 4, 8, and 24 h after elicitor-treatment. EF1α gene was used as the reference gene. The expression levels in CF-treated plants are relative to the corresponding level of expression in water-treated plants at the indicated time points. Results are reported as means ± standard deviation (SD) of three samples for each treatment. Asterisks indicate a statistically significant difference between the elicitor-treated and the control plants (Student's t-test; **P < 0.005). (B) Endogenous OPDA levels in elicitor treated tissues were analyzed at the indicated time points (h). Values are means and standard errors of three independent experiments. Statistical differences were evaluated according to one-way ANOVA followed by Dunnett's test relative to control condition, *P < 0.05, and **P < 0.01.
To further analyze moss defense responses induced after treatment with P.c. carotovorum derived elicitors, we measured defense hormones, including SA, abscisic acid (ABA), and indole-3-acetic acid (IAA). The results show that only auxin levels increase after elicitor treatment, while SA decreases at 2, 4, and 24 h of treatment, and no significant change in ABA levels were detected compared to control tissues (Figure 7). Auxin levels increase six-fold at 2 h after treatment, and four-, two-, and four-fold at 4, 8, and 24 h, respectively. We also analyzed a P. patens reporter line harboring an auxin inducible promoter from soybean (GmGH3) fused to β-glucuronidase (GUS; Bierfreund et al., 2003) in response to P.c. carotovorum derived elicitors. The results show that while in control tissues GUS staining was detected in the basal part of gametophores visualized as spots around the whole colony (Figure 8A), elicitor-treated colonies exhibited an overall GUS staining (Figures 8B,C). Protonemal tissue consists of chloronemal cells with perpendicular cross walls and a high density of chloroplasts and caulonemal cells with oblique cross walls and low density of chloroplasts (Cove et al., 2006). When tissues were analyzed in more detail, control protonemal filaments were not stained (Figure 8D), while elicitor-treated caulonemal filaments accumulated GUS and chloronemal cells were less stained (Figure 8F). In addition, young elicitor-treated gametophores expressed GUS in all the tissues, including rhizoids, cauloid, and leaves (Figure 8G), while control young gametophores accumulated GUS at the base of the cauloid (Figure 8E). GUS expression in leaves was detected in P.c. carotovorum elicitor-treated older gametophores and not in control gametophores (Figures 8H,I). Thus, P. patens activates auxin signaling after P.c. carotovorum elicitor treatment.
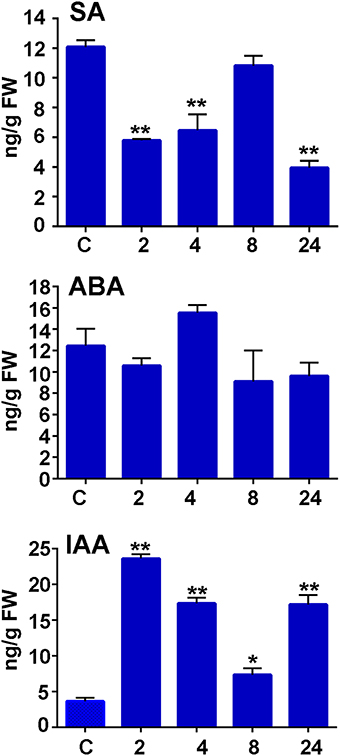
Figure 7. Salicylic acid, abscisic acid, and auxin accumulation in response to P.c. carotovorum elicitor treatment. Endogenous salicylic acid (SA), abscisic acid (ABA), and indole-3-acetic acid (IAA) levels [ng/g fresh weight (FW)] in control and elicitor-treated tissues were analyzed at the indicated time points (h). Values are means and standard errors of three independent experiments. Statistical differences were evaluated according to one-way ANOVA followed by Dunnett's test relative to control condition, *P < 0.005 and **P < 0.001.
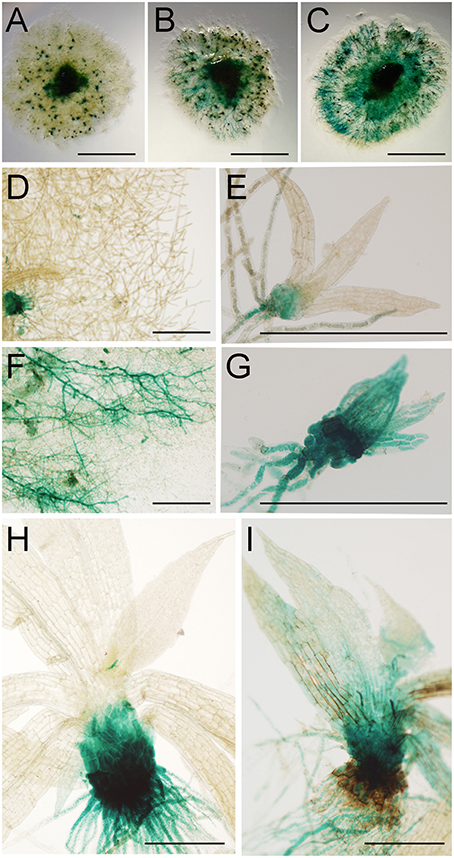
Figure 8. Activation of auxin signaling in elicitor-treated moss tissues. GUS staining of GH3::GUS reporter line in: (A) water-treated colony, (B,C) elicitor-treated colonies, (D) water-treated protonemal tissues, (E) water-treated young gametophore, (F) elicitor-treated protonemal tissues, (G) elicitor-treated young gametophore, (H) water-treated gametophore and (I) elicitor-treated gametophore. Panel (C) was taken at 2 days after treatment and (A,B,D–I) at 1 days after treatment. Scale bars represent 0.5 cm in (A–C) and 0.5 mm in (D–I).
P. patens is a basal land plant with interesting features to study plant-pathogen interactions and activation of defenses in response to elicitor treatment. However, only few studies related to defense against biotic stress in this moss has been performed. Further studies are needed to understand the resistance mechanisms developed by this primitive plant. The results presented here show that P. patens activates defense responses against elicitors of P.c. carotovorum, evidenced by a very rapid expression of defense related genes. Among the defense genes present in our cDNA library enriched in elicitor inducible genes, we identified genes encoding a putative chorismate synthase (CS) and a chorismate mutase (CM). Chorismate is the final product of the shikimate pathway, and it is formed by CS, which converts 5-enolpyruvylshikimate-3-phosphate (EPSP) into chorismate. CM catalyzes the first step of phenylalanine and tyrosine biosynthesis, and it converts chorismate to prephenate (Silber et al., 2008). P. patens has one and two putative gene encoding a CS and CM, respectively (Silber et al., 2008). Here, we show that two of these genes are induced upon elicitor treatment, suggesting that CS and CM could play a role in moss defenses against pathogens. Genes encoding CS and CM contribute to hyphae penetration resistance in barley during attack by powdery mildew (Hu et al., 2009). CS and CM overexpressing barley plants are more resistant, while silenced plants showed increased fungal penetration (Hu et al., 2009). We also show that several genes encoding enzymes involved in the phenylpropanoid pathway are induced by elicitors of P.c. carotovorum, including genes encoding two PALs (PAL.1 and PAL.2), a C4H, a 4CL, three CHS (CHS.1, CHS.2 and CHS.3), two CHI (CHI.1 and CHI.2), a F3H and a CAD. These enzymes are key components required to produce different classes of phenolic secondary metabolites with different roles in defense, including phenolic acids, flavonoids, isoflavonoids, coumarins, stilbenes, lignans, and lignins (Weisshaar and Jenkins, 1998; Vogt, 2010). PAL catalyzes the conversion of L-phenylalanine to trans-cinnamic acid. Previously, we have shown by Northern blot analysis, using the PAL.2 gene as a probe, that PAL was induced by P.c. carotovorum elicitor treatment (Ponce de León et al., 2007). However, due to the high sequence similarities between several PAL genes and the probe used, and the similar sizes of the transcripts, Northern blot results could represent the expression pattern of several of the PAL-encoding genes. Here, we show by qPCR analysis that PAL.2 expression is highly induced by P.c. carotovorum elicitor treatment. PAL.2 is also induced after B. cinerea, C. gloeosporoides and Pythium inoculations (Oliver et al., 2009; Ponce de León et al., 2012; Reboledo et al., 2015). In addition, we show that a second PAL gene, PAL.1, is also induced by P.c. carotovorum elicitors. P. patens has a PAL multigene family composed of 14 members and several of these genes are induced during abiotic UV stress (Wolf et al., 2010). Consistent with the increased expression of PAL.1 and PAL.2, the product derived from the enzymatic activity, cinnamic acid, increases in P. patens tissues in response to elicitors of P.c. carotovorum. Similarly, in potato plants treated with elicitors of Pectobacterium wasabiae (P. wasabiae), cinnamic acid also increased (Montesano et al., 2005). Cinnamic acid is converted in flowering plants by different enzymes to diverse defense compounds, and it can induce the expression of defense-related genes (Montesano et al., 2005). In plants, SA is synthesized from cinnamic acid via benzoic acid (Yalpani et al., 1993) or from isochorismate by the action of an isochorismate synthase (Wildermuth et al., 2001). SA is an important defense hormone against several pathogens in flowering plants (Vlot et al., 2009). However, in P. patens tissues treated with P.c. carotovorum elicitors (Figure 5), or with P. wasabiae elicitors (data not shown), SA levels decreased, suggesting that SA is not central in moss responses to these Pectobacterium strains. Interestingly, SA levels also decreased in P. wasabiae elicitor-treated potato plants (Montesano et al., 2005), while in Arabidopsis SA levels increased significantly after P.c. carotovorum elicitor treatment (strain SCC1; Li et al., 2004). Taken together, these results suggest differential regulation or roles for SA in different plants and/or important strains differences in Pectobacterium. Cinnamic acid is converted to coumaric acid by the action of C4H, which catalyzes aromatic ring-4 hydroxylation of cinnamic acid (Russell, 1971). 4CL catalyze the activation of 4-coumarate and a number of structurally related substrates to the corresponding CoA thioesters (Silber et al., 2008). Both C4H and 4CL moss genes are rapidly induced by elicitors of P.c. carotovorum. P. patens has four genes encoding 4CL and the encoded proteins of three of the genes can utilize 4-coumarate, caffeate, ferulate, and cinnamate as substrates (Silber et al., 2008). CHS condenses a phenylpropanoid CoA ester such as p-coumaroyl-CoA with three acetate units from malonyl-CoA molecules, and cyclizes the resulting intermediate to produce a chalcone like naringenin chalcone (Schröder, 1997). Similarly to the expression of PAL genes, we observed an increased expression of CHS by P.c. carotovorum elicitors by Northern blot analysis, using CHS.3 as a probe (Ponce de León et al., 2007), which could represent the expression of several CHSs. Here, we show that CHS.1, CHS.2, and CHS.3 are induced after P.c. carotovorum elicitor treatment. CHSs belong to a multigene family composed of 17 members (Koduri et al., 2010). Jiang et al. (2006) have demonstrated that CHS.2 is active with a substrate preference for p-Coumaroyl-CoA, suggesting that CHS.2 is a naringenin chalcone producing CHS. We also identified two putative CHI encoding genes and a F3H, which are induced by P.c. carotovorum elicitors. CHI converts chalcone to flavanone and F3H converts flavanone to dihydroflavonol in flowering plants (Morita et al., 2014). The P. patens genome contains two genes for CHI and five genes for F3H. However, more recently these CHI genes were classified as enhancer of flavonoid production (EFP) genes, which encode type IV CHI proteins with no CHI activity (Ngaki et al., 2012). The conversion of flavanone to flavone by flavone synthase seems to be absent in mosses (Koduri et al., 2010).
Lignins are complex aromatic polymers resulting from the oxidative polymerization of hydroxycinnamyl alcohols (p-coumaryl, coniferyl, and sinapyl alcohols). Increased lignification protect plant tissues against pathogen infection and damage (Moura et al., 2010). In addition, precursors of lignin, such as p-coumaric acid, p-coumaroyl-CoA, and p-coumaraldehyde, and its monomers like the monolignols p-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol have antimicrobial activities (Keen and Littlefield, 1979; Tronchet et al., 2010). p-Coumaroyl-CoA is transformed to H monolignol (p-coumaryl alcohol), through a series of reductions (Bonawitz and Chapple, 2010). Production of the more structurally complex G (coniferyl alcohol) and S (sinapyl alcohol) monolignols requires additional enzymes (Bonawitz and Chapple, 2010). 4CL, cinnamoyl-CoA reductase (CCR), and CAD catalyze consecutive steps in lignin biosynthesis, producing p-coumaryl alcohol from coumaric acid. CAD is involved in the reduction of cinnamaldehydes into cinnamyl alcohols, which is the last step of monolignol biosynthesis before oxidative polymerization in the cell wall (Tronchet et al., 2010). Here, we show that a putative CAD encoding gene is induced by elicitors of P.c. carotovorum. In Arabidopsis CADs are key player in the defense response against Pseudomonas syringae and plants with depleted levels of these enzymes are more susceptible to this pathogen (Tronchet et al., 2010). In addition, Montesano et al. (2003) have shown that a gene encoding a potato alcohol dehydrogenase, capable of using different aldehydes as substrates, including cinnamaldehyde, is induced by elicitors of P. wasabiae. The presence of lignin in mosses is still controversial (Xu et al., 2009). From an evolutionary point of view, mosses represent the first land plants with the complete lignin biosynthesis pathway, with the exception of ferulate 5-hydroxylase (F5H; Xu et al., 2009). In this study, we show that DIR.1, DIR.2, DIR.3, and DIR.4 genes are induced upon elicitor treatment, suggesting that monolignol coupling by these enzymes could lead to the production of lignans and/or lignin-like compounds. We have previously shown that DIR.4 is induced by fungal pathogens including B. cinerea and C. gloeosporioides (Ponce de León et al., 2012; Reboledo et al., 2015). The results presented here suggest that induced expression of DIR genes could lead to the modification of the cell wall and contribute to the reinforcement of P. patens cell walls after P.c. carotovorum elicitor treatment. DIR-like proteins could contribute to the formation of lignin-like polyphenols which has been detected in P. patens cells (Espiñeira et al., 2011). These polyphenolic compounds could play roles in defense mechanisms in mosses. Other types of phenolic compounds involved in cell wall strengthening could also be produced, given the safranin-O staining detection of phenolics incorporated into the cell wall of protonemal filaments and leaves treated with P.c. carotovorum elicitors. DIR encoding genes of Nicotiana benthamiana are induced after PAMPs treatment (Chakravarthy et al., 2010) and DIR-overexpressing transgenic plants exhibit enhanced resistance upon fungal assault (Wang and Fristensky, 2001). In addition, PAMPs such as flg22 or chitin induces production of lignin in flowering plants (Nicaise et al., 2009; Millet et al., 2010).
Until present, only few studies related to the phenylpropanoid pathway has been performed in mosses. The cinnamate conjugate, caffeoylquinic acid, has been found in P. patens tissues, as well as chlorogenic acid, which is an intermediate of lignin biosynthesis (Erxleben et al., 2012). Caffeic acid, which is a subunit in the synthesis of lignans has been detected in P. patens, liverworts, and hornworts (Mues, 2000; Scher et al., 2003; Erxleben et al., 2012). P. patens has several caffeic acid O-methyltransferases (Erxleben et al., 2012), suggesting the presence of lignans in this moss. Interesting, caffeoylquinic acid, and caffeic acid were found in gametophores, which are erect structures with leaves, cauloid, and rhizoids, and not in protonemal tissues (Erxleben et al., 2012). The fact that many of the genes encoding enzymes of the phenylpropanoid pathway are induced in response to elicitors of P.c. carotovorum suggests a physiological role of this pathway in P. patens defense responses. Several evidences indicate the importance of phenylpropanoids in flowering plants resistance to Pectobacterium. First, phenolic compounds accumulate after Pectobacterium atrosepticum inoculation, including chlorogenic acid, which inhibits bacterial growth (Kröner et al., 2011). Second, several phenolic compounds including cinnamic, coumaric, syringic, and salicylic acids directly affect virulence factors in a wide range of Pectobacterium strains, including P.c. carotovorum (Joshi et al., 2015). Third, exposure of the bacteria to these phenolic compounds affects motility, biofilm formation and extracellular enzyme activities reducing disease severity in vascular plants (Joshi et al., 2015). Finally, 4-methoxy-cinnamic acid and benzoic acid alter the type III secretion system in Erwinia amylovora, leading to a weakened HR response in tobacco plants (Khokhani et al., 2013). Further studies are needed to understand which phenolic compounds, in addition to cinnamic acid, are produced after P.c. carotovorum elicitor treatment in P. patens.
Oxylipins are involved in defense responses of flowering plants against pathogens, either by their antimicrobial activities, capacity to induce gene expression or to protect cells from cellular damage caused by pathogens (Blée, 2002; Ponce de León et al., 2002). P. patens has only one gene encoding an α-DOX (Ppα-DOX), which catalyzed the conversion of fatty acids into 2-hydroperoxy derivatives (Machado et al., 2015). Ppα-DOX expression increases after P.c. carotovorum elicitor treatment and spores of B. cinerea, and the oxylipins produced by these enzymes are involved in protecting moss tissues against cellular damage caused by pathogens (Machado et al., 2015). In addition, P. patens has eight LOX-encoding genes, some of which produce oxylipins that are not produced in flowering plants, like arachidonic acid-derived oxylipins (Anterola et al., 2009). Previously, we have shown that elicitors of P.c. carotovorum and B. cinerea induce the expression of a 12-LOX encoding gene (LOX1; Ponce de León et al., 2007). LOX1 uses preferentially arachidonic acid as a substrate (Anterola et al., 2009). In response to B. cinerea infection, P. patens also induces the expression of a linolenate 13-LOX-encoding gene (LOX6; Anterola et al., 2009) and an OPR encoding gene (Phpat.003G009100.1; Ponce de León et al., 2012). Here, we show that this OPR encoding gene is induced after P.c. carotovorum elicitor treatment and that OPDA levels increase. In flowering plants OPDA is active as a defense signal and it can activate defense gene expression (Taki et al., 2005; Browse, 2009). Consistently, P. patens induces the expression of PAL.2 in response to OPDA treatment (Oliver et al., 2009). Although, we could not detect an increase in transcript levels of the LOX gene (LOX2; Anterola et al., 2009) identified in the cDNA library after elicitor treatment, we have previously observed induced expression of LOX6 after elicitation with P.c. carotovorum CF (data not shown). Taken together, these results suggest that both the 13-LOX and the 12-LOX pathways are activated after P.c. carotovorum elicitor treatment, predictably leading to the production of oxylipins with different roles in defense responses.
When the accumulation of phytohormones was evaluated in P.c. carotovorum elicitor-treated P. patens plants, including SA, auxin, and ABA, only auxin levels increased. Interestingly, when SA decreased, auxin levels increased. Wang et al. (2007) have previously shown that SA causes global repression of auxin-related genes in flowering plant. Further studies are needed to examine whether or not SA may inhibit auxin biosynthesis in moss tissues. In addition, we show that auxin signaling increases in P.c. carotovorum treated moss tissues. In P. patens auxin signaling is induced in response to other pathogens, including Pythium irregulare, Pythium debaryanum, and C. gloeosporoides (Mittag et al., 2015; Reboledo et al., 2015). The involvement of auxin responses has been associated to flowering plant defense against P.c. carotovorum. Arabidopsis mutants in MAX2 (More Axillary Growth 2), which encodes a negative regulator of polar auxin transport are more susceptible to P.c. carotovorum (Piisilä et al., 2015). Similarly, gdsl lipase Arabidopsis mutants exhibited enhanced auxin responses and were more susceptible to P.c. carotovorum compared to wild type plants (Lee et al., 2009).
In this study, we show that genes encoding enzymes involved in important defense pathways such as the shikimate, phenylpropanoid, oxylipins, and auxin pathways are induced in P. patens in response to P.c. carotovorum elicitors. These defense genes are rapidly induced, suggesting that moss cells sense the presence of PAMPs and/or DAMPs generated by the action of elicitors such as PCWDE and harpin contained in the CF of P.c. carotovrum. Future studies are needed to broader our knowledge on the regulation of the shikimate and phenylpropanoid pathway, the secondary metabolites produced by the corresponding enzymes and their contribution to the adaptation of moss to biotic and abiotic stress. P.c. carotovorum derived elicitors activate auxin signaling in moss suggesting that this hormonal pathway has emerged early in evolution to mediate plant responses to pathogens. As an evolutionary link between green algae and flowering plants, P. patens is an ideal model plant for evolutionary studies on plant defense pathways, including hormonal signaling and activation of defense responses in land plants.
AA generated the construction of the SSH cDNA library and performed the expression analysis. ES performed the hormone and cinnamic acid analysis. MM participated in the discussions and drafting of this work. IP designed and supervised the study, performed the GUS accumulation studies and histological analysis, contributed to the analysis of the data, and wrote the manuscript. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Eva Decker for the GH3::GUS reporter line. This work was supported by Agencia Nacional de Investigación e Innovación (ANII), grant FCE2011_6095, and Programa de Desarrollo de las Ciencias Básicas (PEDECIBA) Uruguay.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00328
Supplementary Table 1. Classification of genes identified in the cDNA SSH library according to their predicted putative functions. Description of categories and selection criteria: energy and metabolism (photosynthesis and electron transport, carbohydrates, amino acids, lipid metabolism), cell growth and maintenance (cell wall, cell division, cytoskeleton, chaperones, membrane-related), oxidation-reduction processes (related to oxidative stress), signaling (signal transduction-related), transcription (transcription factors, DNA binding), phenylpropanoid, protein metabolism (ribosomal proteins, protein degradation), defense related (defense and stress-related), others (do not fit in the other categories; phosphatases, ion channels, transporters, etc), unknown function, and no significant similarity with other proteins.
Supplementary Table 2. Primer sequences for qPCR.
Anterola, A., Göbel, C., Hornung, E., Sellhorn, G., Feussner, I., and Grimes, H. (2009). Physcomitrella patens has lipoxygenases for both eicosanoid and octadecanoid pathways. Phytochemistry 70, 40–52. doi: 10.1016/j.phytochem.2008.11.012
Ashton, N. W., and Cove, D. J. (1977). The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss, Physcomitrella patens. Molec. Gen. Genet. 154, 87–95. doi: 10.1007/BF00265581
Bierfreund, N. M., Reski, R., and Decker, E. L. (2003). Use of an inducible reporter gene system for the analysis of auxin distribution in the moss Physcomitrella patens. Plant Cell Rep. 21, 1143–1152. doi: 10.1007/s00299-003-0646-1
Bittel, P., and Robatzek, S. (2007). Microbe-associated molecular patterns (MAMPs) probe plant immunity. Curr. Opin. Plant Biol. 10, 335–341. doi: 10.1016/j.pbi.2007.04.021
Blée, E. (2002). Impact of phytooxylipins in plant defense. Trends Plant Sci. 7, 315–322. doi: 10.1016/S1360-1385(02)02290-2
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Bonawitz, N. D., and Chapple, C. (2010). The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363. doi: 10.1146/annurev-genet-102209-163508
Browse, J. (2009). Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205. doi: 10.1146/annurev.arplant.043008.092007
Chakravarthy, S., Velásquez, A. C., Ekengren, S. K., Collmer, A., and Martin, G. B. (2010). Identification of Nicotiana benthamiana genes involved in pathogen associated molecular pattern-triggered immunity. Mol. Plant Microbe Interact. 23, 715–726. doi: 10.1094/MPMI-23-6-0715
Cove, D. (2005). The moss Physcomitrella patens. Annu. Rev. Genet. 39, 339–358. doi: 10.1146/annurev.genet.39.073003.110214
Cove, D., Bezanilla, M., Harries, P., and Quatrano, R. (2006). Mosses as model systems for the study of metabolism and development. Annu. Rev. Plant Biol. 57, 497–520. doi: 10.1146/annurev.arplant.57.032905.105338
Cove, D. J., Knight, C. D., and Lamparter, T. (1997). Mosses as model systems. Trends Plant Sci. 2, 99–105. doi: 10.1016/S1360-1385(96)10056-X
Davin, L. B., and Lewis, N. G. (2000). Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol. 123, 453–462. doi: 10.1104/pp.123.2.453
Davis, K. R., Lyon, G. D., Darvill, A. G., and Albersheim, P. (1984). Host pathogen interactions. XXV. Endopolygalacturonic acid lyase from Erwinia carotovora elicits phytoalexin accumulation by releasing plant cell wall fragments. Plant Physiol. 74, 52–60. doi: 10.1104/pp.74.1.52
Diatchenko, L., Lau, Y. F., Campbell, A. P., Chenchik, A., Moqadam, F., Huang, B., et al. (1996). Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. U.S.A. 93, 6025–6030. doi: 10.1073/pnas.93.12.6025
Erxleben, A., Gessler, A., Vervliet-Scheebaum, M., and Reski, R. (2012). Metabolite profiling of the moss Physcomitrella patens reveals evolutionary conservation of osmoprotective substances. Plant Cell Rep. 31, 427–436. doi: 10.1007/s00299-011-1177-9
Espiñeira, J. M., Novo, U. E., Gómez Ros, L. V., Carrión, J. S., Merino, F., Ros Barceló, A., et al. (2011). Distribution of lignin monomers and the evolution of lignification among lower plants. Plant Biol. 13, 59–68. doi: 10.1111/j.1438-8677.2010.00345.x
Hückelhoven, R. (2007). Cell wall-associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45, 101–127. doi: 10.1146/annurev.phyto.45.062806.094325
Hu, P., Meng, Y., and Wise, R. P. (2009). Functional contribution of chorismate synthase, anthranilate synthase, and chorismate mutase to penetration resistance in barley-powdery mildew interactions. Mol. Plant Microbe. Interact. 22, 311–320. doi: 10.1094/MPMI-22-3-0311
Humphrey, T. V., Bonetta, D. T., and Goring, D. R. (2007). Sentinels at the wall: cell wall receptors and sensors. New Phytol. 176, 7–21. doi: 10.1111/j.1469-8137.2007.02192.x
Jacobs, A. K., Lipka, V., Burton, R. A., Panstruga, R., Strizhov, N., Schulze-Lefert, P., et al. (2003). An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15, 2503–2513. doi: 10.1105/tpc.016097
Jiang, C., Schommer, C. K., Kim, S. Y., and Suh, D. Y. (2006). Cloning and characterization of chalcone synthase from the moss, Physcomitrella patens. Phytochemistry 67, 2531–2540. doi: 10.1016/j.phytochem.2006.09.030
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Joshi, J. R., Burdman, S., Lipsky, A., Yariv, S., and Yedidia, I. (2015). Plant phenolic acids affect the virulence of Pectobacterium aroidearum and P. carotovorum subsp brasiliense via quorum-sensing regulation. Mol. Plant Pathol. doi: 10.1111/mpp.12295.[Epub ahead of print].
Kachroo, A., and Kachroo, P. (2009). Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 47, 153–176. doi: 10.1146/annurev-phyto-080508-081820
Keen, N. T., and Littlefield, L. J. (1979). The possible association of phytoalexins with resistance gene expression in flax to Melampsora lini. Physiol. Plant Pathol. 14, 265–280. doi: 10.1016/0048-4059(79)90048-1
Khokhani, D., Zhang, C., Li, Y., Wang, Q., Zeng, Q., Yamazaki, A., et al. (2013). Discovery of plant phenolic compounds that act as type III secretion system inhibitors or inducers of the fire blight pathogen, Erwinia amylovora. Appl. Environ. Microbiol. 79, 5424–5436. doi: 10.1128/AEM.00845-13
Koduri, P. K., Gordon, G. S., Barker, E. I., Colpitts, C. C., Ashton, N. W., and Suh, D. Y. (2010). Genome-wide analysis of the chalcone synthase superfamily genes of Physcomitrella patens. Plant Mol. Biol. 272, 247–263. doi: 10.1007/s11103-009-9565-z
Kröner, A., Hamelin, G., Andrivon, D., and Val, F. (2011). Quantitative resistance of potato to Pectobacterium atrosepticum and Phytophthora infestans: integrating PAMP-triggered response and pathogen growth. PLoS ONE 6:e23331. doi: 10.1371/journal.pone.0023331
Le Bail, A., Scholz, S., and Kost, B. (2013). Evaluation of reference genes for RT qPCR analyses of structure-specific and hormone regulated gene expression in Physcomitrella patens gametophytes. PLoS ONE 8:e70998. doi: 10.1371/journal.pone.0070998
Lee, D. S., Kim, B. K., Kwon, S. J., Jin, H. C., and Park, O. K. (2009). Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling. Biochem. Biophys. Res. Commun. 379, 1038–1042. doi: 10.1016/j.bbrc.2009.01.006
Levine, A., Tenhaken, R., Dixon, R., and Lamb, C. (1994). H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79, 583–593. doi: 10.1016/0092-8674(94)90544-4
Lewis, L. A., and McCourt, R. M. (2004). Green algae and the origin of land plants. Am. J. Bot. 91, 1535–1556. doi: 10.3732/ajb.91.10.1535
Li, J., Brader, G., and Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lotze, M. T., Zeh, H. J., Rubartelli, A., Sparvero, L. J., Amoscato, A. A., Washburn, N. R., et al. (2007). The grateful dead: damage associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunol. Rev. 220, 60–81. doi: 10.1111/j.1600-065X.2007.00579.x
Machado, L., Castro, A., Hamberg, M., Bannenberg, G., Gaggero, C., Castresana, C., et al. (2015). The Physcomitrella patens unique alpha-dioxygenase participates in both developmental processes and defense responses. BMC Plant Biol. 15:45. doi: 10.1186/s12870-015-0439-z
Millet, Y. A., Danna, C. H., Clay, N. K., Songnuan, W., Simon, M. D., Werck-Reichhart, D., et al. (2010). Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22, 973–990. doi: 10.1105/tpc.109.069658
Mittag, J., Šola, I., Rusak, G., and Ludwig-Müller, J. (2015). Physcomitrella patens auxin conjugate synthetase (GH3) double knockout mutants are more resistant to Pythium infection than wild type. J. Plant Physiol. 183, 75–83. doi: 10.1016/j.jplph.2015.05.015
Montesano, M., Brader, G., Ponce de Leon, I., and Palva, E. T. (2005). Multiple defense signals induced by Erwinia carotovora ssp. carotovora in potato. Mol. Plant Pathol. 6, 541–549. doi: 10.1111/j.1364-3703.2005.00305.x
Montesano, M., Hyytiäinen, H., Wettstein, R., and Palva, E. T. (2003). A novel potato defence-related alcohol:NADP+ oxidoreductase induced in response to Erwinia carotovora. Plant Mol. Biol. 52, 177–189. doi: 10.1023/A:1023981631596
Montesano, M., Kõiv, V., Mäe, A., and Palva, E. T. (2001). Novel receptor-like protein kinases induced by Erwinia carotovora and short oligogalacturonides in potato. Mol. Plant Pathol. 2, 339–346. doi: 10.1046/j.1464-6722.2001.00083.x
Morita, Y., Takagi, K., Fukuchi-Mizutani, M., Ishiguro, K., Tanaka, Y., Nitasaka, E., et al. (2014). A chalcone isomerase-like protein enhances flavonoid production and flower pigmentation. Plant J. 78, 294–304. doi: 10.1111/tpj.12469
Moura, J. C. M. S., Bonine, C. A. V., de Oliveira Fernandes Viana, J., Dornelas, M. C., and Mazzafera, P. (2010). Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 52, 360–376. doi: 10.1111/j.1744-7909.2010.00892.x
Mues, R. (2000). “Chemical constituents and biochemistry,” in Bryophyte Biology, eds A. J. Shaw and B. Goffinet (Cambridge: Cambridge University Press), 150–181.
Ngaki, M. N., Louie, G. V., Philippe, R. N., Manning, G., Pojer, F., Bowman, M. E., et al. (2012). Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature 485, 530–533. doi: 10.1038/nature11009
Nicaise, V., Roux, M., and Zipfel, C. (2009). Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol. 150, 1638–1647. doi: 10.1104/pp.109.139709
Norman-Setterblad, C., Vidal, S., and Palva, E. T. (2000). Interacting signal pathways control defense gene expression in Arabidopsis in response to the plant pathogen Erwinia carotovora. Mol. Plant Microbe Interact. 13, 430–438. doi: 10.1094/MPMI.2000.13.4.430
Oliver, J. P., Castro, A., Gaggero, C., Cascón, T., Schmelz, E. A., Castresana, C., et al. (2009). Pythium infection activates conserved plant defense responses in mosses. Planta 230, 569–579. doi: 10.1007/s00425-009-0969-4
Palva, T. K., Holmström, K. O., Heino, P., and Palva, E. T. (1993). Induction of plant defense response by exoenzymes of Erwinia carotovora ssp. carotovora. Mol. Plant-Microbe Interact. 6, 190–196. doi: 10.1094/MPMI-6-190
Peleman, J., Boerjan, W., Engler, G., Seurinck, J., Botterman, J., Alliotte, T., et al. (1989). Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-adenosylmethionine synthetase. Plant Cell 1, 81–93. doi: 10.1105/tpc.1.1.81
Pérombelon, M. C. M., and Kelman, A. (1980). Ecology of the soft-rot Erwinia. Annu. Rev. Phytopathol. 12, 361–387. doi: 10.1146/annurev.py.18.090180.002045
Piisilä, M., Keceli, M. A., Brader, G., Jakobson, L., Jõesaar, I., Sipari, N., et al. (2015). The F-box protein MAX2 contributes to resistance to bacterial phytopathogens in Arabidopsis thaliana. BMC Plant Biol. 15:53. doi: 10.1186/s12870-015-0434-4
Ponce de León, I. (2011). The moss Physcomitrella patens as a model system to study interactions between plants and phytopathogenic fungi and oomycetes. J. Pathog. 2011:719873. doi: 10.4061/2011/719873
Ponce de León, I., Hamberg, M., and Castresana, C. (2015). Oxylipins in moss development and defense. Front Plant Sci. 6:483. doi: 10.3389/fpls.2015.00483
Ponce de León, I., and Montesano, M. (2013). Activation of defense mechanisms against pathogens in mosses and flowering plants. Int. J. Mol. Sci. 14, 3178–3200. doi: 10.3390/ijms14023178
Ponce de León, I., Oliver, J. P., Castro, A., Gaggero, C., Bentancor, M., and Vidal, S. (2007). Erwinia carotovora elicitors and Botrytis cinerea activate defense responses in Physcomitrella patens. BMC Plant Biol. 7:52. doi: 10.1186/1471-2229-7-52
Ponce de León, I., Sanz, A., Hamberg, M., and Castresana, C. (2002). Involvement of the Arabidopsis α-DOX1 fatty acid dioxygenase in protection against oxidative stress and cell death. Plant J. 29, 61–72. doi: 10.1046/j.1365-313x.2002.01195.x
Ponce de León, I., Schmelz, E. A., Gaggero, C., Castro, A., Álvarez, A., and Montesano, M. (2012). Physcomitrella patens activates reinforcement of the cell wall, programmed cell death and accumulation of evolutionary conserved defence signals, such as salicylic acid and 12-oxo-phytodienoic acid, but not jasmonic acid, upon Botrytis cinerea infection. Mol. Plant Pathol. 13, 960–974. doi: 10.1111/j.1364-3703.2012.00806.x
Rantakari, A., Virtaharju, O., Vähämiko, S., Taira, S., Palva, E. T., Saarilahti, H. T., et al. (2001). Type III secretion contributes to the pathogenesis of the soft-rot pathogen Erwinia carotovora partial characterization of the hrp gene cluster. Mol. Plant-Microbe Interact. 14, 962–968. doi: 10.1094/MPMI.2001.14.8.962
Reboledo, G., Del Campo, R., Alvarez, A., Montesano, M., Mara, H., and Ponce de León, I. (2015). Physcomitrella patens activates defense responses against the pathogen Colletotrichum gloeosporioides. Int. J. Mol. Sci. 16, 22280–22298. doi: 10.3390/ijms160922280
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. doi: 10.1126/science.1150646
Russell, D. W. (1971). The metabolism of aromatic compounds in higer plants. X. Properties of the cinnamic acid 4-hydroxylase of pea seedlings and some aspects of its metabolic and developmental control. J Biol Chem. 246, 3870–3878.
Schaefer, D. G. (2001). Gene targeting in Physcomitrella patens. Curr. Opin. Plant Biol. 4, 143–150. doi: 10.1016/S1369-5266(00)00150-3
Schaefer, D. G., and Zrÿd, J. P. (2001). The moss Physcomitrella patens, now and then. Plant Physiol. 27, 1430–1438. doi: 10.1104/pp.010786
Scher, J. M., Zapp, J., and Becker, H. (2003). Lignan derivatives from the liverwort Bazzania trilobata. Phytochemistry 62, 769–777. doi: 10.1016/S0031-9422(02)00626-X
Schmelz, E. A., Engelberth, J., Tumlinson, J. H., Block, A., and Alborn, H. T. (2004). The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J. 39, 790–808. doi: 10.1111/j.1365-313X.2004.02168.x
Schröder, J. (1997). A family of plant-specific polyketide synthases: facts and predictions. Trends Plant Sci. 2, 373–378. doi: 10.1016/S1360-1385(97)87121-X
Silber, M. V., Meimberg, H., and Ebel, J. (2008). Identification of a 4-coumarate:CoA ligase gene family in the moss, Physcomitrella patens. Phytochemistry 69, 2449–2456. doi: 10.1016/j.phytochem.2008.06.014
Taki, N., Sasaki-Sekimoto, Y., Obayashi, T., Kikuta, A., Kobayashi, K., Ainai, T., et al. (2005). 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 139, 1268–1283. doi: 10.1104/pp.105.067058
Ton, J., and Mauch-Mani, B. (2004). Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 38, 119–130. doi: 10.1111/j.1365-313X.2004.02028.x
Toth, I. K., and Birch, P. R. (2005). Rotting softly and stealthily. Curr. Opin. Plant Biol. 8, 424–429. doi: 10.1016/j.pbi.2005.04.001
Tronchet, M., Balagué, C., Kroj, T., Jouanin, L., and Roby, D. (2010). Cinnamyl alcohol dehydrogenases-C and D key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 11, 83–92. doi: 10.1111/j.1364-3703.2009.00578.x
Vidal, S., Ponce de León, I., Denecke, J., and Palva, E. T. (1997). Salicylic acid and the plant pathogen Erwinia carotovora induce defense genes via antagonistic pathways. Plant J. 11, 115–123. doi: 10.1046/j.1365-313X.1997.11010115.x
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wang, D., Pajerowska-Mukhtar, K., Culler, A. H., and Dong, X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17, 1784–1790. doi: 10.1016/j.cub.2007.09.025
Wang, Y. P., and Fristensky, B. (2001). Transgenic canola lines expressing pea defense gene DRR206 have resistance to aggressive blackleg isolates and to Rhizoctonia solani. Mol. Breed. 8, 263–271. doi: 10.1023/A:1013706400168
Weisshaar, B., and Jenkins, G. I. (1998). Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1, 251–257. doi: 10.1016/S1369-5266(98)80113-1
Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
Wolf, L., Rizzini, L., Stracke, R., Ulm, R., and Rensing, S. A. (2010). The molecular and physiological responses of Physcomitrella patens to ultraviolet-B radiation. Plant Physiol. 153, 1123–1134. doi: 10.1104/pp.110.154658
Wu, C. H., Apweiler, R., Bairoch, A., Natale, D. A., Barker, W. C., Boeckmann, B., et al. (2006). The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 34, D187–D191. doi: 10.1093/nar/gkj161
Xu, Z., Zhang, D., Hu, J., Zhou, X., Ye, X., Reichel, K. L., et al. (2009). Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinformatics 10:S3. doi: 10.1186/1471-2105-10-S11-S3
Yalpani, N., Leon, J., Lawton, M. A., and Raskin, I. (1993). Pathway of salicylic acid biosynthesis in healthy and virus-inoculated tobacco. Plant Physiol. 103, 315–321.
Yeh, S. Y., Huang, F. C., Hoffmann, T., Mayershofer, M., and Schwab, W. (2014). FaPOD27 functions in the metabolism of polyphenols in strawberry fruit (Fragaria sp.). Front Plant Sci. 5:518. doi: 10.3389/fpls.2014.00518
Zhao, S., Tuan, P. A., Li, X., Kim, Y. B., Kim, H., Park, C. G., et al. (2013). Identification of phenylpropanoid biosynthetic genes and phenylpropanoid accumulation by transcriptome analysis of Lycium chinense. BMC Genomics 14:802. doi: 10.1186/1471-2164-14-802
Zimmer, A. D., Lang, D., Buchta, K., Rombauts, S., Nishiyama, T., Hasebe, M., et al. (2013). Reannotation and extended community resources for the genome of the non-seed plant Physcomitrella patens provide insights into the evolution of plant gene structures and functions. BMC Genomics 14:498. doi: 10.1186/1471-2164-14-498
Keywords: Physcomitrella patens, defense, Pectobacterium carotovorum, phenylpropanoid, auxin, cell wall
Citation: Alvarez A, Montesano M, Schmelz E and Ponce de León I (2016) Activation of Shikimate, Phenylpropanoid, Oxylipins, and Auxin Pathways in Pectobacterium carotovorum Elicitors-Treated Moss. Front. Plant Sci. 7:328. doi: 10.3389/fpls.2016.00328
Received: 30 December 2015; Accepted: 03 March 2016;
Published: 22 March 2016.
Edited by:
Teresa Rebecca De Kievit, University of Manitoba, CanadaReviewed by:
Kemal Kazan, Commonwealth Scientific and Industrial Research Organization (CSIRO), AustraliaCopyright © 2016 Alvarez, Montesano, Schmelz and Ponce de León. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Inés Ponce de León, JiN4MDAwRUM7cG9uY2VAaWliY2UuZWR1LnV5
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.