
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 16 March 2016
Sec. Plant Genetics and Genomics
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.00307
 Xueyuan Lou1,2†
Xueyuan Lou1,2† Xin Li1†
Xin Li1† Aixia Li1,2
Aixia Li1,2 Mingyu Pu1,2
Mingyu Pu1,2 Muhammad Shoaib1
Muhammad Shoaib1 Dongcheng Liu1
Dongcheng Liu1 Jiazhu Sun1
Jiazhu Sun1 Aimin Zhang1,3*
Aimin Zhang1,3* Wenlong Yang1*
Wenlong Yang1*The extensive use of two alleles (Rht-B1b and Rht-D1b) at the Rht-1 locus in wheat allowed dramatic increases in yields, triggering the so-called “Green Revolution.” Here, we found that a new natural allelic variation (Rht-B1i) containing a single missense SNP (A614G) in the coding region significantly increased plant height against the genetic background of both Rht-D1a (11.68%) and Rht-D1b (7.89%). To elucidate the molecular mechanism of Rht-B1i, we investigated the promoter region. Sequence analysis showed that the Rht-B1i promoter could be divided into two classes depending on the presence or absence of a specific 160 bp insertion: Rht-B1i-1 (with the 160 bp insertion) and Rht-B1i-2 (without the 160 bp insertion). The promoter of Rht-B1i-1 contained 32 more possible cis-acting elements than Rht-B1a, including a unique auxin response element AUXREPSIAA4. Quantitative RT-PCR analysis indicated that the 160 bp insertion is likely to promote the transcription of the Rht-B1i-1 gene. The coleoptile lengths of wheat varieties treated with IAA, GA3, and IAA/GA3, combined with the histochemical staining of transgenic Arabidopsis containing the Rht-B1i-1 promoter, showed that the height-increasing effect of Rht-B1i-1 may be due to the synergistic action of IAA and GA3. These results augment our understanding of the regulatory mechanisms of Rht-1 in wheat and provide new genetic resources for wheat improvement.
The decrease in the stem stature of common wheat has resulted in improved lodging resistance and increased harvest index (Gale and Youssefian, 1985; Youssefian et al., 1992; Miralles and Slafer, 1995). As a result, plant height has made a significant contribution to high and stable yields in common wheat (Triticum aestivum L.).
Several wheat loci are or were once associated with decreased plant height and these are termed as Reduced height (Rht). These loci include the three homoeologs of Rht-1 (Rht-A1; Rht-B1; Rht-D1) and 19 other loci (Rht4–9; Rht11–23; McIntosh et al., 2013; Chen et al., 2015). These Rht genes are divided into two subgroups, GA-sensitive genes and GA-insensitive genes, depending on their response to exogenous gibberellic acid (GA; Börner et al., 1996; Pestsova et al., 2008; Chen et al., 2015).
Among GA-insensitive semi-dwarfing genes, two genes in Norin 10, originally named Rht1 and Rht2, were re-designated as Rht-B1b and Rht-D1b, respectively, as they were found to be mutant alleles of the Rht-1 loci on the short arms of chromosomes 4B and 4D (Gale et al., 1975; Gale and Marshall, 1976; McVittie et al., 1978; Börner et al., 1996). Under near-isogenic genetic backgrounds, Rht-B1b and Rht-D1b both have the ability to reduce plant height by 15–20% (Gale and Youssefian, 1985; Flintham et al., 1997; Miedaner and Voss, 2008; Blake et al., 2009). In combination, they can reduce plant height by 40% (Flintham et al., 1997). This effect is caused by a nucleotide substitution that creates a premature stop codon, resulting in truncated DELLA proteins in the region responsible for the GA response (Peng et al., 1999). In the 1960s, the introduction of the Rht-B1b and Rht-D1b alleles in wheat resulted in a great increase in yield during the ‘Green Revolution’ (Hedden, 2003), which abated a major worldwide food shortage.
At the Rht-B1 and Rht-D1 loci, five Rht-B1 alleles (Rht-B1a–e) and four Rht-D1 alleles (Rht-D1a–d) have been isolated and characterized in hexaploid wheat (Peng et al., 1999; Pearce et al., 2011; Wu et al., 2011; Li A.X. et al., 2012; Li Y.Y. et al., 2012; Li et al., 2013; Wen et al., 2013). Compared with the wild-type alleles Rht-B1a and Rht-D1a, the dwarfism conferred by Rht-B1b, Rht-B1d, Rht-B1e, Rht-D1b, and Rht-D1d is due to a single nucleotide change that introduces a premature stop codon in the DELLA region of encoded proteins (Peng et al., 1999; Pearce et al., 2011; Li A.X. et al., 2012; Li et al., 2013). Rht-B1c derived from “Tom Thumb” differs from Rht-B1a by one 2 kb Veju retrotransposon insertion, three SNPs in the coding region and one 197-bp insertion and four SNPs in the 1-kb upstream sequence (Wen et al., 2013). The intragenic insertion, which results in an in-frame 90-bp insertion in the transcript and a predicted 30-amino acid insertion within the DELLA domain, is primarily responsible for severe dwarfism (Pearce et al., 2011; Wu et al., 2011; Wen et al., 2013). In contrast, as the strongest height-reducing allele in wheat, Rht-D1c derived from “Aibian 1” generates a tandem segmental duplication (TSD) of a 1-Mb region, resulting in two copies of Rht-D1b (Pearce et al., 2011; Li Y.Y. et al., 2012).
To facilitate the understanding of the diversity of Rht-1 and the utilization of new resources in breeding programs, Li et al. (2013) previously identified six new Rht-A1 allelic variations (Rht-A1b–g), eight new Rht-B1 allelic variations (Rht-B1h–o), and six new Rht-D1 allelic variations (Rht-D1e–j) in Chinese wheat germplasm by a modified EcoTILLING method. Li et al. (2013) found that Rht-B1i (E205G) have frequencies of 4.2% in Chinese wheat MCC and 2.28% in 1,537 important Chinese wheat cultivars and germplasms, respectively. These frequencies are relatively high among the 10 Rht-B1 allelic variations (Rht-B1a, b, h–o). Recently, Wilhelm et al. (2013b) discovered new haplotypes of Rht-1 in western wheat cultivars and tetraploid and diploid wheat, and found that three haplotypes (Rht-B1a_2, 3, 4) also contained an E205G substitution in the poly S/T/V region. Moreover, there are five Rht-B1a_2, one Rht-B1a_3, and two Rht-B1a_4 in the 66 bread wheat accessions (nine accessions for bread wheat set 1, 12 accessions for a subset of BW1 accessions widely grown in the UK, and 45 accessions from the INRA worldwide bread wheat core collection of 372 accessions), the frequency of E205G substitution is 12.12%. Sequence alignment revealed that the open reading frame (ORF) sequences of the three haplotypes (Rht-B1a_2, 3, 4) in Wilhelm et al. (2013b) is identical to Rht-B1i in Li et al. (2013), indicating that Rht-B1i widely exist in the Chinese and western wheat cultivars.
Interestingly, the new natural allelic variation Rht-B1i, which contains a single missense SNP (A614G) in the coding region resulting in E205G, may increase plant height (Li et al., 2013). However, the underlying mechanism for this effect is unclear. Current studies mainly focus on alleles with dwarfing effects. Moderate dwarfing can increase grain yield in wheat breeding, but excessive dwarfing reduces production owing to decreased biomass. Reports of alleles with height-increasing effects are extremely rare, and elucidating this effect of Rht-B1i may enable the allele to contribute to ameliorating excessive dwarfism in wheat. In this study, we isolated and characterized the promoter of Rht-B1i. According to the presence or absence of 160 bp insertion, Rht-B1i could be divided into Rht-B1i-1 (with 160 bp insertion in promoter) and Rht-B1i-2 (without 160 bp insertion in promoter) using molecular markers and sequencing analysis. Quantitative RT-PCR analysis revealed that the 160 bp insertion may promote the transcription of the Rht-B1i-1 gene. The coleoptile lengths of wheat varieties treated with IAA, GA3, and IAA/GA3, combined with the histochemical staining of transgenic Arabidopsis containing the Rht-B1i-1 promoter, indicated that the height-increasing effect of Rht-B1i-1 may be due to the synergistic action of IAA and GA3.
The Chinese wheat micro-core collections (MCC) consist of Chinese Spring (CS), 155 landraces, 89 Chinese wheat-bred cultivars, and 17 introduced foreign accessions. The MCC is representative of 1% of the national wheat collection but over 70% of the genetic diversity (Wang et al., 2008; Li et al., 2013). We planted accessions containing five genotypes (Rht-B1a, b, h–j) against the genetic background of Rht-D1a and Rht-D1b in the field site of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences (Beijing, China) in 2012 and 2013. There are one accession ‘Chinese Spring’ of B1a/D1a, eleven accessions of B1b/D1a, eight accessions of B1h/D1a, 10 accessions of B1i/D1a (8 Rht-B1i-1 and 2 Rht-B1i-2), 11 accessions of B1j/D1a, five accessions of B1a/D1b, five accessions of B1b/D1b, three accessions of B1h/D1b, four accessions of B1i/D1b, and two accessions of B1j/D1b. Each accession was planted in 2-m two-row plot with 25 cm between rows and 40 seeds per row, and repeated for three times. Plants were managed using standard wheat production management. We measured plant height at 20 days after flowering using 10 randomly selected plants from each plot, and calculated the means and standard deviations of plant height using Microsoft Office Excel (2013). Differences among groups were analyzed with analysis of variance (ANOVA) procedure using SAS software (version 9.1).
We chose CS and four varieties containing Rht-B1i (three varieties of Rht-B1i-1 genotype H8, H83, and H215 containing a promoter with the 160 bp insertion as well as one variety of Rht-B1i-2 genotype H251 containing a promoter without the 160 bp insertion) to assess the effects of IAA and GA3 on coleoptile elongation. Seeds were treated with 1% (w/v) H2O2 in 4°C for 24 h and rinsed three times with sterile deionized water, then cultured individually with ddH2O (control), IAA (10 μM), GA3 (10 μM), or IAA/GA3 (1:1) in a greenhouse under controlled conditions (23°C, dark) for 7 days. We measured coleoptile length for each treatment. Each experiment used 20 plants, and all experiments were carried out in triplicate. We calculated the means and standard deviations of coleoptile length using Microsoft Office Excel (2013). Differences among groups were analyzed with analysis of variance (ANOVA) procedure using SAS software (version 9.1).
We grew 70 wheat lines containing different allelic variations at the Rht-1 locus (Supplementary Table S1), including CS, Mercia-carrying Rht-B1e, 68 Chinese wheat MCC varieties (including 23 Chinese wheat landraces, 37 Chinese wheat bread cultivars, and eight introduced foreign accessions), and 35 Chinese wheat-leading cultivars and important germplasm (Supplementary Table S2). All plants were grown in a greenhouse under controlled conditions (23°C, 16:8 photoperiod) for 2 weeks. We collected young leaves for genomic DNA extraction using the hexadecyl trimethyl ammonium bromide (CTAB) method (Aldrich, 1993).
We randomly selected 30 wheat lines from 46 Rht-B1i varieties (11 Chinese wheat MCC lines and 35 Chinese wheat-leading cultivars and important germplasm lines). All plants were grown in a greenhouse under controlled conditions (23°C, 16:8 photoperiod) for 2 weeks, and we collected samples of their young leaves to isolate total RNA using the RNeasy Plant Mini Kit (QIAGEN, Germany). Flag leaves and the fourth internodes from four varieties of Rht-B1i-1 genotype H8, H83, H213, H215, and one variety of Rht-B1i-2 genotype H251 at the heading stage were also sampled for RNA extraction. CS was used as control variety. The first strand of cDNA was synthesized using FastQuant RT Kit (with gDNase; TIANGEN, Beijing) according to the manufacturer’s instructions.
To clone the promoters of the Rht-1 allelic variations, we designed two primers, PB-CF and PB-CR (Supplementary Table S3), according to the sequence of Rht-B1a (FR719732, Triticum aestivum Rht-B1a gene for DELLA protein). We performed PCR in 20-μL reaction volumes containing 200 ng DNA template, 0.4 μM forward and reverse primers, 1 U TaKaRa LA Taq (Takara, Biotechnology Co., Ltd, Dalian, China), 1 × GC Buffer I, and 0.4 mM of each dNTP under the following conditions: 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 2 min; a final extension was performed at 72°C for 10 min. PCR products were cloned into pGEM-T easy vectors (Promega, Madison, WI, USA), introduced into Escherichia coli, and 10 positive independent clones were commercially sequenced. The promoter sequences and their GC characteristics were analyzed using DNAMAN software, and their cis-acting elements were predicted using PLACE and PlantCARE.
To facilitate the use of Rht-B1i in wheat-breeding programs, we developed two allele-specific PCR markers, B1i-MF1/MR1 and B1i-MF2/MR2 (Supplementary Table S3), based on the 160 bp insertion of the promoter. PCR was performed on an ABI 9700 thermal cycler (Applied Biosystems) as described above. The PCR products were separated on a 2% agarose gel and visualized using a UV spectrometer after ethidium bromide staining.
We carried out quantitative real-time PCR (qRT-PCR) to determine the transcript level of Rht-B1i using the LightCycler 480 system (Roche, Indianapolis, IN, USA) with SYBR Green I Master (Roche, Indianapolis, IN, USA). The gene-specific primers Rht-B1.EF and Rht-B1.ER (Supplementary Table S3) were used for gene expression analysis with the Ta4045 gene (Paolacci et al., 2009) as a reference. All qRT-PCR experiments were performed independently with three biological and three technical replicates, respectively. We calculated the relative expression of each gene according to the comparative CT method (ΔΔCT; Livak and Schmittgen, 2001) and calculated the means and standard deviations using Microsoft Office Excel (2013). Differences among groups were analyzed with analysis of variance (ANOVA) procedure using SAS software (version 9.1).
To construct the plasmid for promoter analysis, we cloned promoters of Rht-B1a and Rht-B1i-1 with the primers PB-PstI-F/PB-NcoI-R to introduce PstI and NcoI sites (Supplementary Table S3). The amplified fragment was inserted into the pGEM-T vector and recombined with pCAMBIA 3301 (35S::GUS) by replacing the 35S promoter before the GUS reporter gene to generate pB1a::GUS and pB1i-1::GUS. The plant binary vectors were introduced into Agrobacterium tumefaciens strain GV3101 using the freezing and thawing method and then transformed into Arabidopsis using the flower-dipping method (Clough and Bent, 1998, modified from Bechtold et al., 1993). Transgenic lines were selected on MS (Murashige and Skoog, 1962) plates with PPT (phosphinothricin, 5 mg/L), and more than 20 transgenic lines were obtained. Histochemical detection of GUS activity (Jefferson et al., 1987) was carried out on the T3 transgenic plants. T3 plants were grown in MS medium with PPT (5 mg/L) for 7 days, then moved to MS medium with GA3 (10 μM), IAA (10 μM), or IAA/GA3 (1:1) and grown for 5 days. Seedlings were fixed in 90% (v/v) acetone on ice for 10 min, then infiltrated in GUS staining solution [50 mM sodium phosphate buffer, pH 7.0, 10 mM EDTA, 0.01% Triton X-100, 1 mM potassium ferricyanide, 1 mM potassium ferrocyanide, 500 μg/mL 5-bromo-4-chloro-3-indoyl-β-D-glucuronic acid (X-gluc)], and incubated overnight at 37°C. Later, the GUS solution was replaced with 100% (v/v) ethanol at room temperature for 6 h and kept in 70% (v/v) ethanol at 4°C. The stained seedlings were observed and photographed using light microscopy (Olympus SZX16, Olympus Co., Japan).
We analyzed the effects of five genotypes (Rht-B1a, b, h–j) on wheat plant height against the backgrounds of Rht-D1a and Rht-D1b using 2 years (2012, 2013) of agronomic trait data (Figure 1). Against the background of Rht-D1a, Rht-B1b significantly reduced plant height by 24.27% as compared with Rht-B1a. The plant heights for Rht-B1h and Rht-B1j were roughly the same as those for Rht-B1a, whereas the height of Rht-B1i was significantly increased, by 11.68%, compared with that of Rht-B1a. The plant heights for all genotypes were smaller against the background of Rht-D1b than against the background of Rht-D1a. Rht-B1b reduced plant height by 10.91% compared with Rht-B1a, whereas Rht-B1i increased plant height by 7.89% compared with Rht-B1a. These results indicate that Rht-B1i significantly increased wheat plant height against both the Rht-D1a and Rht-D1b backgrounds.
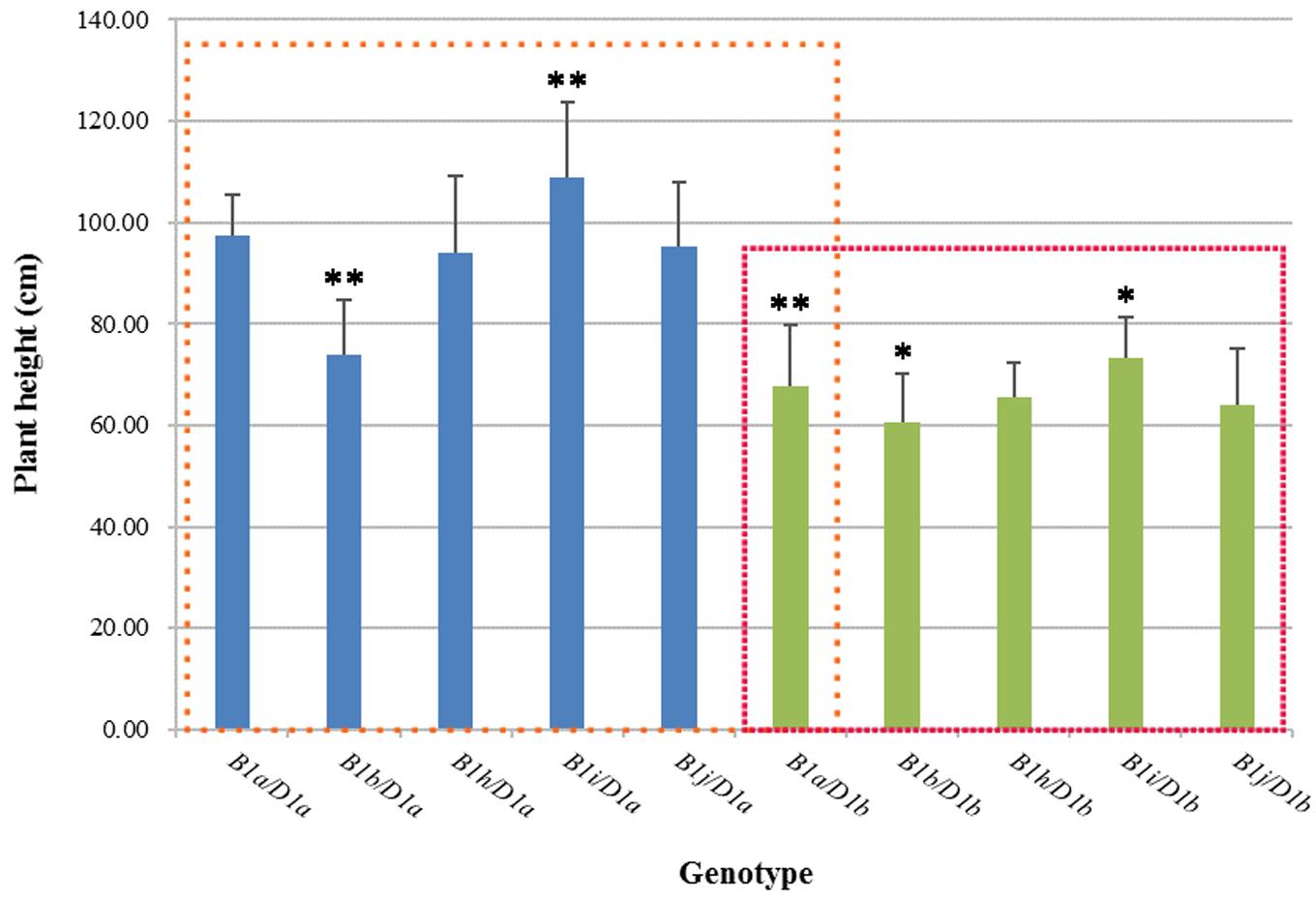
FIGURE 1. Plant height in different Rht-B1 genotypes of wheat. The blue column represents Rht-B1 allelic variations against the Rht-D1a background, and the green column represents Rht-B1 allelic variations against the Rht-D1b background; the left box compares plant height for different genotypes using B1a/D1a as a control, and the right box compares plant height for different genotypes using B1a/D1b as a control. ∗, ∗∗Denotes significant differences at 5 and 1% probability levels, respectively.
To further investigate the height-increasing effects of Rht-B1i, we analyzed the entire coding region of Rht-B1a and Rht-B1i. Sequence alignment showed that the missense SNP occurring in Rht-B1i was A614G, giving rise to an E205G amino acid change in the Poly S/T/V region of the encoded DELLA protein (Li et al., 2013). The SIFT (Sorting Intolerant from Tolerant, Sim et al., 2012) score of Rht-B1i was 0.41, indicating that the variation had no obvious effect on the protein: non-synonymous SNPs are predicted to be damaging to the encoded protein if the SIFT score is <0.05. Therefore, the height-increasing effect of Rht-B1i is unlikely to result directly from the missense mutation in the coding region. Next, we cloned the promoter of Rht-B1i using PB-CF/CR primers (Supplementary Table S3). Sequence alignment revealed a 160 bp insertion at 365 bp upstream of the start codon in the promoter of Rht-B1i, whereas this insertion was not found in the promoter of Rht-B1a.
To identify whether the 160 bp insertion was specific to Rht-B1i, we cloned the promoters of Rht-B1 allelic variations from Rht-B1e (Li A.X. et al., 2012) and 10 other Rht-B1 allelic variations (Rht-B1a, b, h–o; Li et al., 2013). The promoters of Rht-B1a (P0, 2040 bp) and seven other Rht-B1 allelic variations (Rht-B1b, e, j, k, m–o; P1, 2040 bp, GenBank accession no. LN907868) were identical; the promoter of Rht-B1h (P2, 2237 bp, the GenBank accession no. LN907869) contained seven SNPs and a 197 bp insertion at 596 bp upstream of the start codon; the promoter of Rht-B1i (P3, 2200 bp, GenBank accession no. LN907870) carried a 160 bp insertion at 365 bp upstream of the start codon; and the promoter of Rht-B1l (P4, 2020 bp, GenBank accession no. LN907871) had a 20-bp deletion at 3 bp upstream of the start codon (Figure 2, Supplementary Table S1). The 160 bp insertion can only be found in Rht-B1i, nevertheless, two of 11 MCC accessions containing Rht-B1i have no 160 bp insertion (Supplementary Table S1).

FIGURE 2. Sequence alignment for promoters of Rht-B1 allelic variations in wheat. Four promoters cloned from seventy wheat lines containing different allelic variations at the Rht-B1 loci. Compared with the promoter of Rht-B1a (P0), P1 was coincide exactly with it, P2 had a 197 bp insertion (sequences on blue lines), P3 had a 160 bp insertion (sequences on red lines), and P4 had a 20 bp deletion (sequences on green line).
To facilitate the use of Rht-B1i in wheat-breeding programs, and ascertain whether all the accessions containing Rht-B1i have 160 bp insertion in the promoter, we developed two allele-specific PCR markers (Supplementary Table S3), B1i-MF1/MR1 (dominant) and B1i-MF2/MR2 (codominant), based on the 160 bp insertion of Rht-B1i. The amplification products were 330 bp and 586 bp, respectively, in most Rht-B1i varieties (Figure 3). The PCR products of B1i-MF1/MR1 (Figure 3A) showed no amplification among the Rht-B1 allelic variations Rht-B1a, Rht-B1b, Rht-B1e, and Rht-B1h, whereas nine of the 12 varieties carrying Rht-B1i produced a positive 330-bp band. The PCR products of B1i-MF2/MR2 (Figure 3B) displayed 426-bp bands from Rht-B1a, Rht-B1b, Rht-B1e, and Rht-B1h, whereas 586 bp bands were observed in nine of 12 varieties containing Rht-B1i. Hence, the primers B1i-MF1/MR1 and B1i-MF2/MR2 are allele-specific for most Rht-B1i accessions. However, there were three accessions (Wumangchunmai, Kashi 1, and Jimai 37) that contained Rht-B1i but could not be identified by these two markers. We therefore amplified and sequenced their promoters and found that they lacked a 160 bp insertion. Using B1i-MF1/MR1 and B1i-MF2/MR2 to screen 262 Chinese wheat MCC and 1,537 Chinese wheat-leading cultivars and important germplasms, we found 3.4 and 2.02% frequencies of Rht-B1i, respectively. This is lower than the 4.2 and 2.28%, respectively, reported by Li et al. (2013), indicating that the Rht-B1i genotype might have other haplotypes.
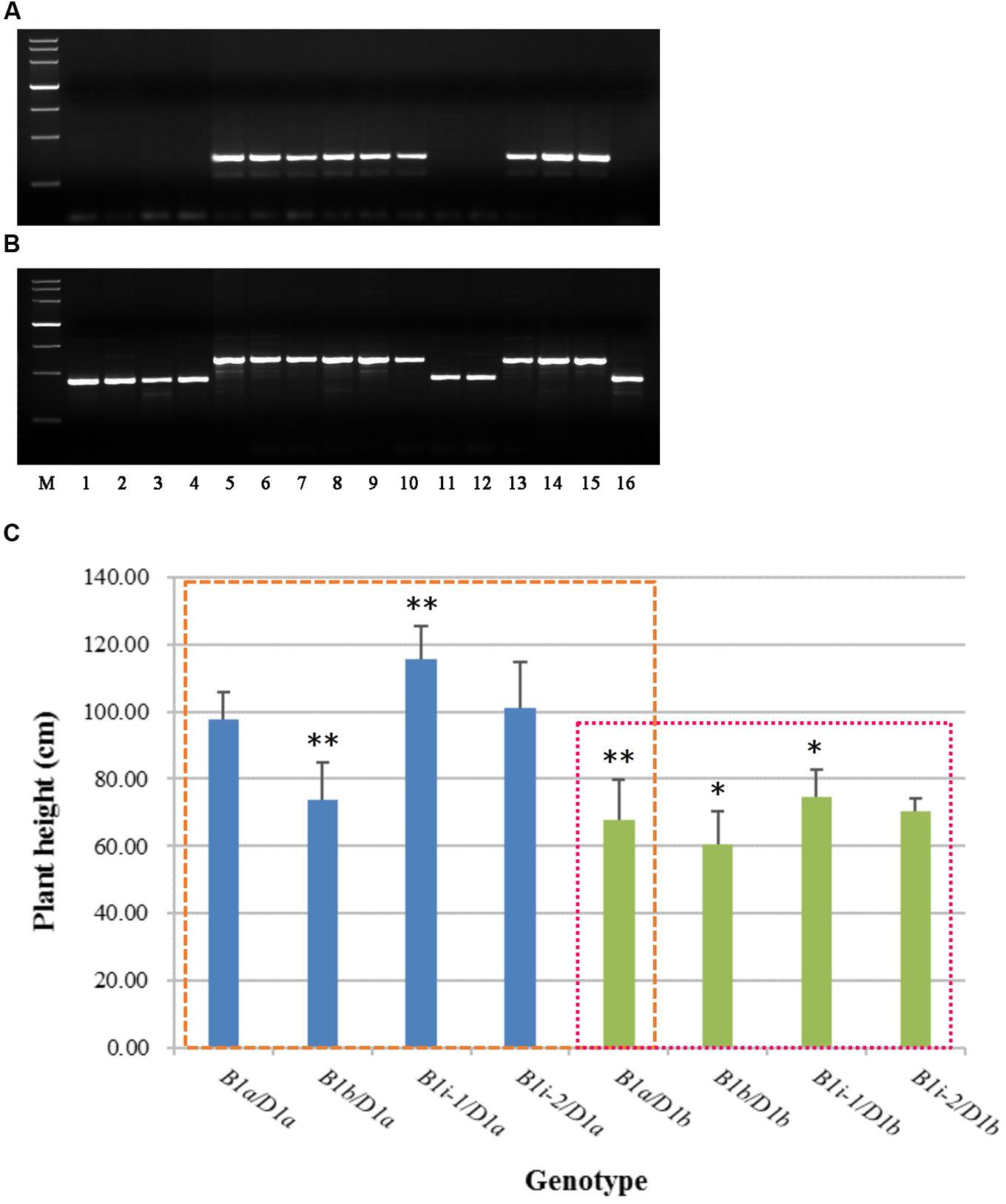
FIGURE 3. Rht-B1i-1 significantly increases the plant height of wheat. (A) PCR with B1i-MF1/MR1, (B) PCR with B1i-MF2/MR2, (C) Plant height in Rht-B1i-1 and Rht-B1i-2 genotypes of wheat. Lanes: M, molecular weight markers (DNA marker III, Tiangen Biotech Co. Ltd, China), the bands are 4,500, 3,000, 2,000, 1,200, 800, 500, and 200 bp, respectively; 1, CS (Rht-B1a), 2, Jinchun 3 (Rht-B1b), 3, Mercia (Rht-B1e), 4, Zhongyou 9507 (Rht-B1h), 5, Guangtou (Rht-B1i), 6, KaBka3 (Rht-B1i), 7, Triumph (Rht-B1i), 8, Dalibanmang (Rht-B1i), 9, Huzhuhong (Rht-B1i), 10, Dingxi 24 (Rht-B1i), 11, Wumangchunmai (Rht-B1i), 12, Kashi 1 (Rht-B1i), 13, Xinshuguang 6 (Rht-B1i), 14, Jinmai 56 (Rht-B1i), 15, Jinan24 (Rht-B1i), 16, Jimai 37 (Rht-B1i). The blue column represents Rht-B1 allelic variations against the Rht-D1a background, and the green column represents Rht-B1 allelic variations against the Rht-D1b background; the left box compares plant height for different genotypes using B1a/D1a as a control, and the right box compares plant height for different genotypes using B1a/D1b as a control. ∗, ∗∗Denotes significant differences at 5 and 1% probability levels, respectively.
Therefore, we cloned and sequenced the promoters from 46 lines, which were validated as carrying Rht-B1i from 262 Chinese wheat MCC (11 lines) and 1,537 Chinese wheat-leading cultivars and important germplasms (35 lines). Of 11 Chinese wheat MCC, nine accessions had the 160 bp insertion; among 35 Chinese wheat-leading cultivars and important germplasms, 25 lines had the 160 bp insertion (Supplementary Table S2). Consequently, we subclassified the Rht-B1i genotype into Rht-B1i-1 (75.8% had a 160 bp insertion in the promoter) and Rht-B1i-2 (24.2% were missing a 160 bp insertion in the promoter). To identify the effect of the 160 bp insertion, we analyzed the plant height of Rht-B1i-1 and Rht-B1i-2 (Figure 3C). Whether under the background of Rht-D1a or Rht-D1b, Rht-B1i-1 significantly increased plant height by 10.18 and 9.89% as compared with Rht-B1a, meanwhile, the height of Rht-B1i-2 was increased, by 3.64 and 3.54%, with no significance, indicating that Rht-B1i-1 significantly increased wheat plant height under both Rht-D1a and Rht-D1b backgrounds.
Structural analysis of promoters has wide implications in molecular biology, because it allows the prediction of gene expression profiles. We analyzed the GC content in the 1,500-bp section upstream from the translation initiation codon of the Rht-B1 allelic variations and found that the AT-content of the four Rht-B1 promoters (P1, 51.47%; P2, 52.53%; P3, 51.60%; P4, 51.80%) was greater than the corresponding GC content (48.53, 47.47, 48.40, and 48.20%, respectively), which is consistent with a role as an AT-rich plant gene-promoter element. To study the contribution of the 160 bp insertion to gene transcription, we performed quantitative real-time PCR (qRT-PCR) using the seedlings of CS and 30 other varieties containing Rht-B1i, flag leaves and the fourth internodes from four varieties of Rht-B1i-1 genotype H8, H83, H213, H215, and one variety of Rht-B1i-2 genotype H251 at the heading stage, and using Ta4045 as the reference gene. For seedlings (Figure 4A), all eight varieties containing Rht-B1i-2 (without the 160 bp insertion) expressed lower levels than wild-type Rht-B1a, but these differences were not statistically significant. Among the 22 varieties carrying Rht-B1i-1 (with the 160 bp insertion), 13 expressed higher levels than Rht-B1a (statistically significant in 2/13), whereas nine varieties expressed lower levels than Rht-B1a. This indicates that the 160 bp insertion can have differential effects on gene expression. For flag leaves (Figure 4B), the variety containing Rht-B1i-2 also expressed lower level than Rht-B1a, whereas three of the four varieties containing Rht-B1i-1 expressed significantly higher levels than Rht-B1a. For the fourth internodes (Figure 4B), the expression level of the variety containing Rht-B1i-2 was significantly lower than Rht-B1a, however, the expression levels of three varieties containing Rht-B1i-1 were significantly higher than Rht-B1a. Overall, the 160 bp insertion can affect the expression of Rht-B1i-1.
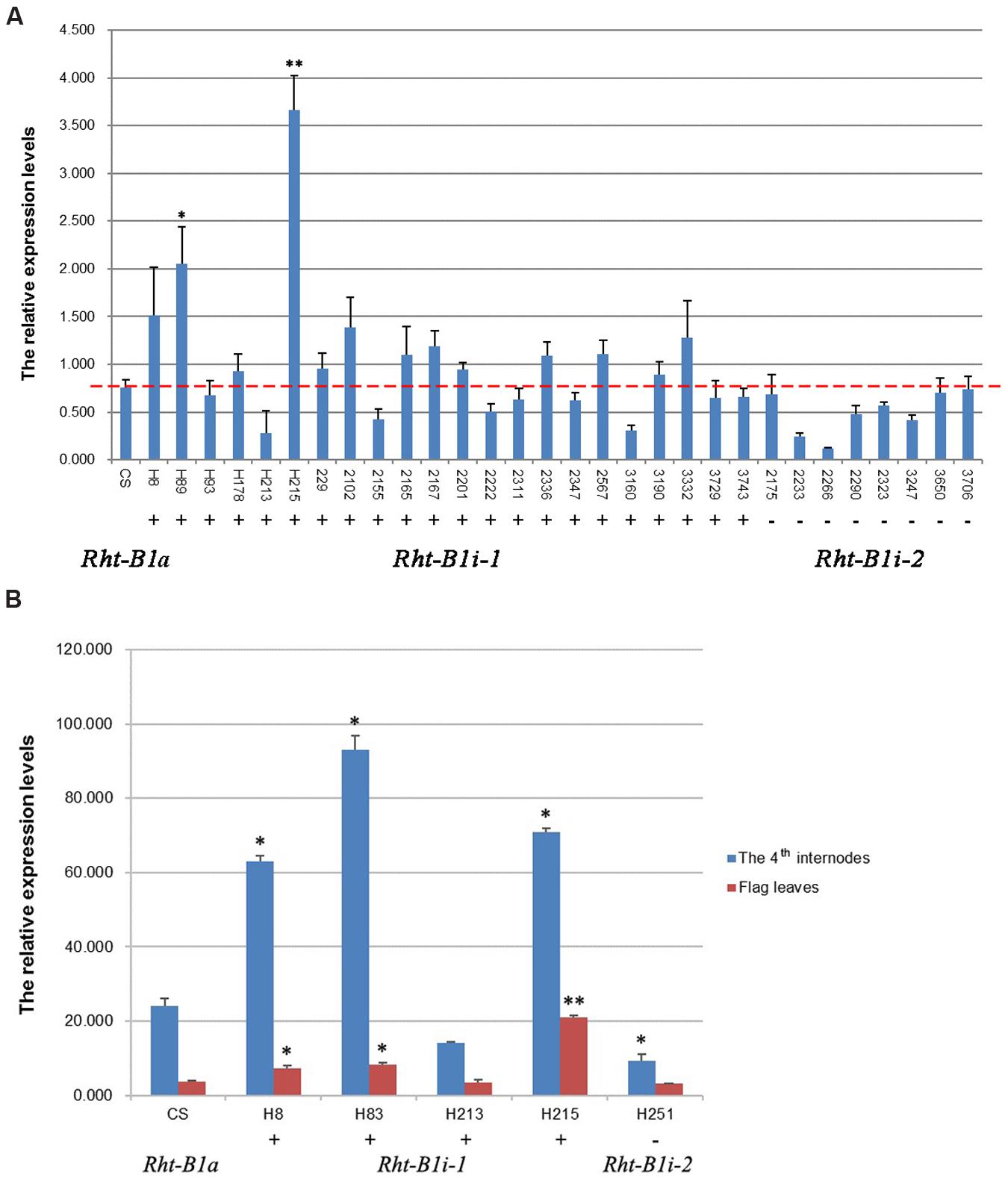
FIGURE 4. Expression analysis of Rht-B1i-1 and Rht-B1i-2. (A) Expression analysis in seedlings, (B) Expression analysis in flag leaves and the fourth internodes. The red line shown the relative expression levels of Rht-B1 in CS which containing Rht-B1a (control). ∗, ∗∗Denotes significant differences at 5 and 1% probability levels, respectively. +, – Denote Rht-B1i-1 containing the 160 bp insertion and Rht-B1i-2 missing the 160 bp insertion, respectively.
We carried out predictive analysis of the cis-acting elements in the promoters of Rht-B1a and Rht-B1i-1 using PLACE1 and PlantCARE2. The analysis revealed various possible cis-acting elements in the two promoters: in addition to basic cis-regulatory elements, such as TATA-box, CAAT-box, and so on, there were many possible cis-acting elements that were related to phyto-hormone response, tissue-specific expression, and stress induction. This suggests that Rht-B1 may participate in the regulation of multiple phyto-hormones and environmental signaling pathways. There were 453 and 485 possible cis-acting elements in the promoters of Rht-B1a and Rht-B1i-1, respectively, with the 32 additional possible cis-acting elements located in the 160 bp insertion in the promoter of Rht-B1i-1 (Table 1). Among these 32 additional elements, the promoter of Rht-B1i-1 contained four unique cis-acting elements, including an auxin response element (AUXREPSIAA4). This indicates that the regulatory network of Rht-B1i-1 is highly complex and that the height-increasing effect of Rht-B1i-1 may be related to auxin.
To study the effects of the 160 bp insertion on the function of Rht-B1i, we treated CS, three varieties containing Rht-B1i-1 (H8, H83, and H215 which had significantly higher expression levels), and one variety containing Rht-B1i-2 (H251) with IAA, GA3, and IAA/GA3 (Figure 5). The coleoptile length of CS carrying Rht-B1a was significantly reduced after IAA treatment, but it clearly increased after GA3 and IAA/GA3 treatment, indicating that Rht-B1a has a strong response to IAA, GA3, and IAA/GA3. However, we observed no significant difference in the coleoptile length of variety containing Rht-B1i-2 after IAA, GA3, or IAA/GA3 treatment, indicating that Rht-B1i-2 was insensitive to IAA, GA3, and IAA/GA3. The coleoptile length of varieties containing Rht-B1i-1 showed no significant difference after GA3 treatment, but it significantly increased after IAA and, particularly, IAA/GA3 treatment. This suggests that Rht-B1i-1 is also insensitive to GA3 and that the height-increasing effect of Rht-B1i-1 might be caused by the synergistic action of IAA and GA3.
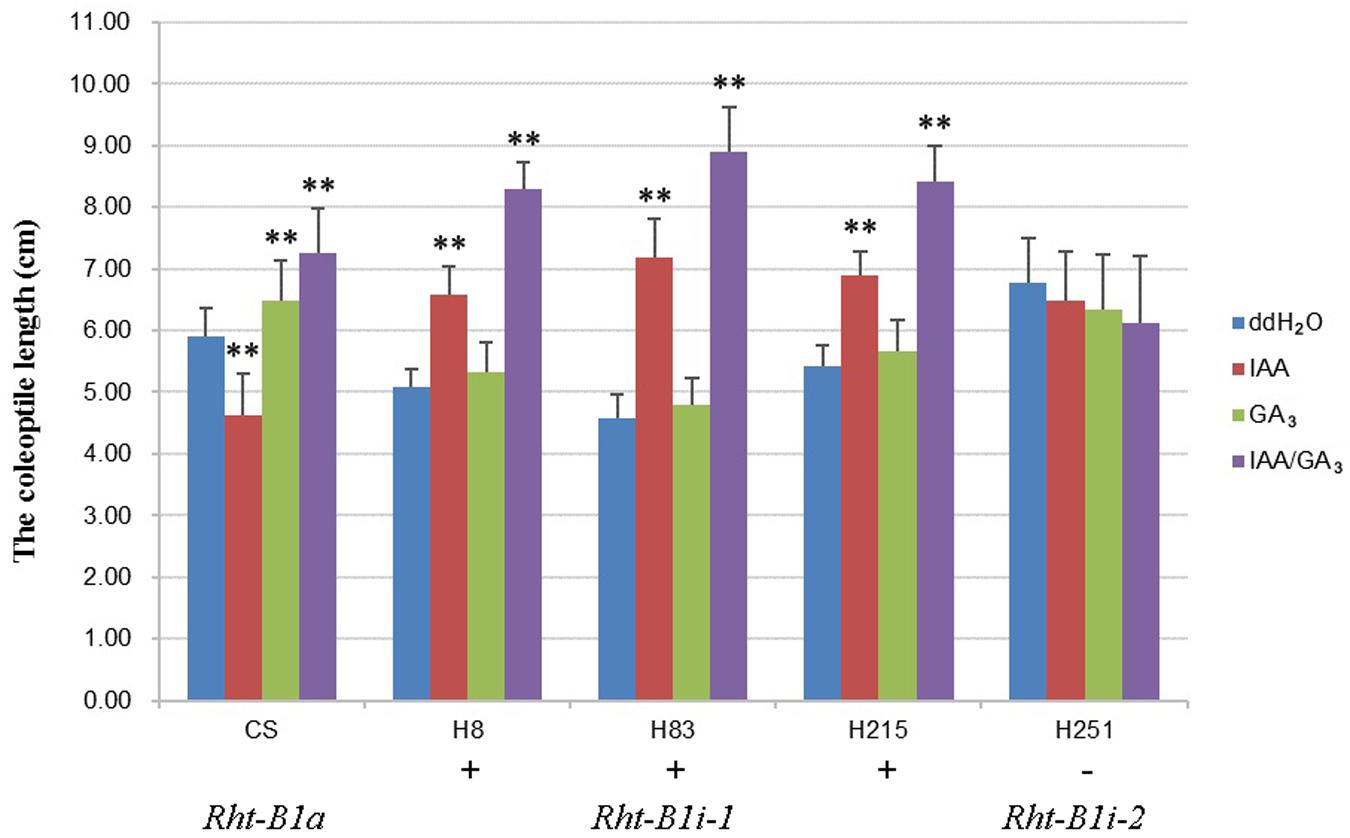
FIGURE 5. The coleoptile length of wheat varieties with Rht-B1i-1 and Rht-B1i-2. The phyto-hormone treatments of three genotypes, Rht-B1a, Rht-B1i-1, and Rht-B1i-2, with ddH2O as a control. ∗∗Denote significant differences at the 1% probability level. +, – Denote Rht-B1i-1 containing the 160 bp insertion and Rht-B1i-2 missing the 160 bp insertion, respectively.
To study the response of the promoters to exogenous IAA and GA3, we fused the promoters of Rht-B1a (pB1a, 2,040 bp) and Rht-B1i-1 (pB1i-1, 2,200 bp) with the GUS (β-glucuronidase) reporter gene using the binary vector pCAMBIA 3301-GUS. Vectors were then transformed into Arabidopsis ecotype Col-0. The transgenic Arabidopsis seedlings treated with GA3 had slender roots (Figures 6A–C), those treated with IAA had more lateral roots (Figures 6D–F), and those treated with IAA/GA3 had slender lateral roots (Figures 6G–I). The 35S::GUS transgenic Arabidopsis seedlings expressed strong GUS activity in the entire plant after GA3 (Figure 6A), IAA (Figure 6D), and IAA/GA3 (Figure 6G) treatment. After GA3 treatment, the pB1a::GUS (Figure 6B) and pB1i-1::GUS (Figure 6C) transgenic Arabidopsis seedlings both expressed strong GUS activity in leaves and stems and weak GUS activity in roots, although the difference was non-significant. After treated with IAA and IAA/GA3, the pB1a::GUS transgenic Arabidopsis seedlings (Figures 6E,H) primarily expressed GUS activity in leaves and stems. However, the pB1i-1::GUS transgenic Arabidopsis seedlings (Figures 6F,I) strongly expressed GUS activity not only in leaves and stems but also in roots, illustrating that the promoter of Rht-B1i-1 played a role in the response to IAA and IAA/GA3. The above results are in accordance with the predictive analysis of cis-acting elements in promoters and with the coleoptile lengths of wheat varieties treated with IAA, GA3, and IAA/GA3, demonstrating that the height-increasing effect of Rht-B1i-1 may be caused by the synergistic effects of IAA and GA3.
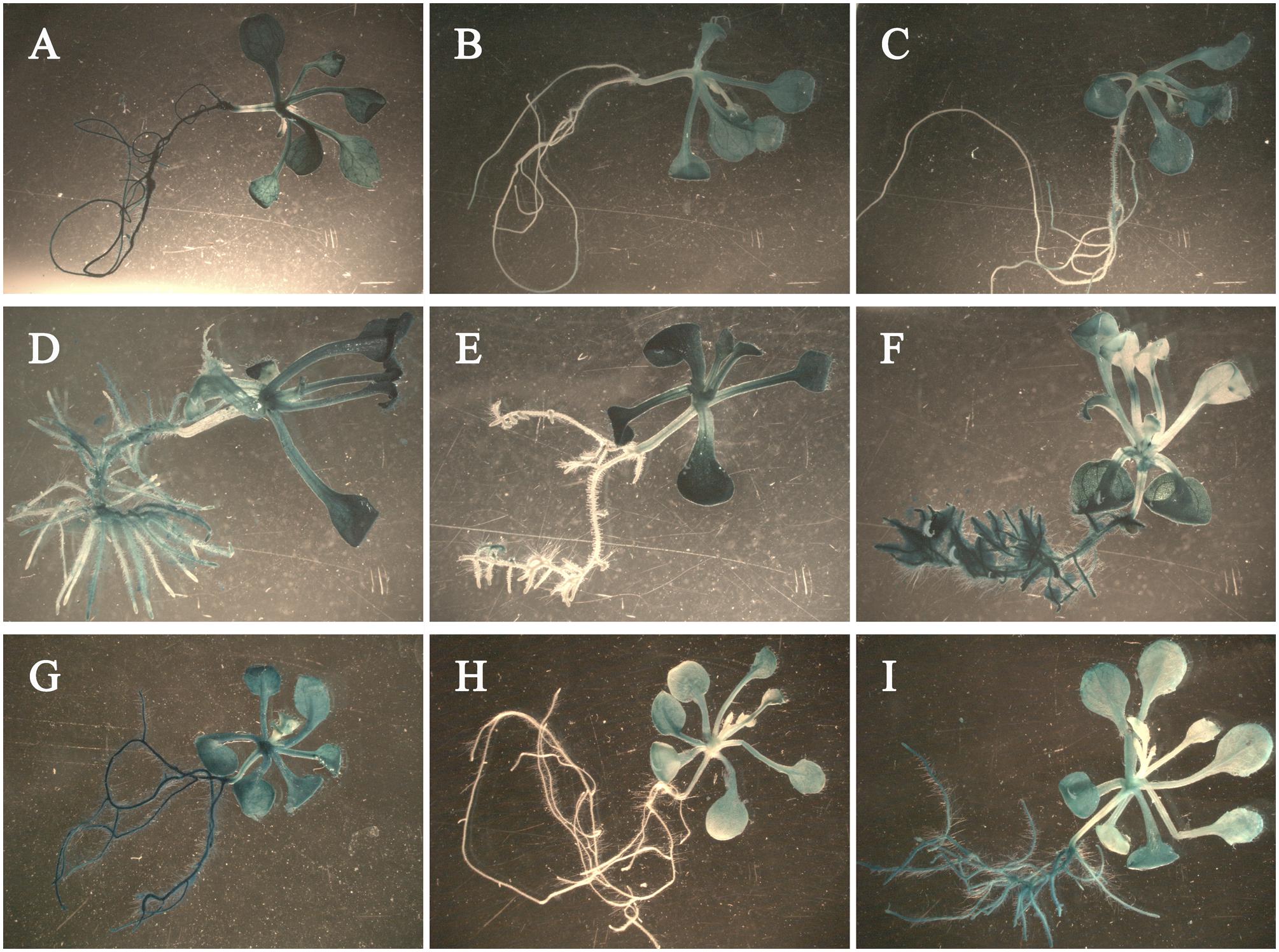
FIGURE 6. The histochemical staining of phyto-hormone-treated transgenic Arabidopsis seedlings. pB1a represents promoter of Rht-B1a, pB1i-1 represents promoter of Rht-B1i-1. (A–C) Arabidopsis seedlings treated with GA3: (A) 35S::GUS transgenic Arabidopsis seedling, (B) pB1a::GUS transgenic Arabidopsis seedling, (C) pB1i-1::GUS transgenic Arabidopsis seedling. (D–F) Arabidopsis seedlings treated with IAA: (D) 35S::GUS transgenic Arabidopsis seedling, (E) pB1a::GUS transgenic Arabidopsis seedling, (F) pB1i-1::GUS transgenic Arabidopsis seedling. (G–I) Arabidopsis seedlings treated with IAA/GA3: (G) 35S::GUS transgenic Arabidopsis seedling, (H) pB1a::GUS transgenic Arabidopsis seedling, (I) pB1i-1::GUS transgenic Arabidopsis seedling.
The primary function of the Rht-1 gene is to reduce the plant height of wheat. Börner et al. (1996) found that the relative strength of this effect among alleles at Rht-1 loci is Rht-B1a < Rht-B1d < Rht-B1b < Rht-B1e < Rht-B1c, and Rht-D1a < Rht-D1b < Rht-D1d < Rht-D1c. Among these Rht alleles, the two ‘green revolution’ genes, Rht-B1b and Rht-D1b, are the most economically important and, by far, the most commonly used. Therefore we analyzed the effects of Rht-B1b on the plant height of wheat against the genetic backgrounds of Rht-D1a and Rht-D1b using 2 years (2012, 2013) of agronomic traits data. We quantified the effect in terms of the percentage reduction in plant height after the introduction of the Rht-1 gene. Compared with the wild-type gene Rht-B1a, Rht-B1b significantly reduced plant height by 24.27 and 11.68% against the backgrounds of Rht-D1a and Rht-D1b, respectively. Rht-B1b+Rht-D1b reduced plant height by a statistically significant 38%. Flintham et al. (1997) also reported that Rht-B1b has the ability to reduce plant height by 14% but that Rht-B1b + Rht-D1b can reduce plant height by 42%, as can two copies of either Rht-B1b or Rht-D1b against near-isogenic backgrounds. Current studies are focusing on alleles with dwarfing effects; Rht-B1b and Rht-D1b are also associated with increased harvestable yield under favorable conditions (Gale and Youssefian, 1985; Flintham et al., 1997; Chapman et al., 2007). However, Rht-B1b and Rht-D1b also contribute to reduced coleoptile length and seedling vigor (Rebetzke et al., 2007; Addisu et al., 2009) and may reduce crop water-use efficiency in unfavorable environments (Butler et al., 2005; Chapman et al., 2007).
The discovery of new beneficial Rht-1 alleles is very important for wheat-breeding programs. Our previous study (Li et al., 2013) identified six new Rht-A1 allelic variations (Rht-A1b–g), eight new Rht-B1 allelic variations (Rht-B1h–o), and six new Rht-D1 allelic variations (Rht-D1e–j) by a modified EcoTILLING method. Among these, Rht-B1h, Rht-B1i, and Rht-B1j have relatively high frequencies in Chinese wheat MCC and Chinese wheat-leading cultivars and important germplasms. Interestingly, Rht-B1i containing a single missense SNP (A614G) in the coding region significantly increased wheat plant height against the background of both Rht-D1a (11.68%) and Rht-D1b (7.89%), suggesting that Rht-B1i could counteract the effects of Rht-D1b. Elucidating the height-increasing effect of Rht-B1i will contribute to our understanding of the regulatory mechanism of Rht-1 and aid in the use of Rht-B1i for ameliorating excessive dwarfism in wheat.
The missense SNP occurring in Rht-B1i results in an E205G amino acid change in the Poly S/T/V region of the encoded DELLA protein. However, the SIFT score (0.41) indicates that the mutation has no obvious effect on the protein (Li et al., 2013), suggesting that the height-increasing effect does not result directly from the missense mutation in the coding region. The expression of a protein depends on the transcription induction of the corresponding gene; therefore, the analysis of promoter regions is highly relevant. Li et al. (2011) found that base mutants in the pyrimidine box upstream of the BnGID1 sequence probably explain the low expression level of the GID1 protein. Similarly, Park et al. (2013) showed that the DELLA and BOI proteins inhibit GA responses by binding to the promoters of GA-responsive genes. Recognizing the importance of these regions, we isolated the promoters of Rht-B1 alleles, found a 160 bp insertion, and classified Rht-B1i into Rht-B1i-1 (with the 160 bp insertion in the promoter) and Rht-B1i-2 (without the 160 bp insertion in the promoter) genotypes. Wilhelm et al. (2013b) directly sequenced the ORF and 5′ and 3′ flanking regions in western bread cultivars and tetraploid and diploid wheat and also identified three haplotypes (Rht-B1a_2, 3, 4) that have a 160 bp insertion in the 5′ flanking regions. Sequence alignment revealed several variants of Rht-B1i: Rht-B1a_2, Rht-B1a_3, and Rht-B1a_4. Rht-B1a_2 is identical to Rht-B1i-1; Rht-B1a_3 has two SNPs in the 5′ flanking region; and Rht-B1a_4 has a CTA insertion in the 3′ flanking region. The Rht-B1 160 bp insertion is also present in a high frequency in western wheat accessions, and has a wide geographic distribution, suggesting that they are prevalent in modern bread wheat varieties. But the 160 insertion was not identified in any tetraploid and diploid wheat accessions, indicating that it is a recent event. In addition, the 160 bp insertion occurs in the middle of a highly conserved non-coding sequences among three wheat Rht-1 homoeologs and in Rht-1 Poaceae orthologs, could be important Rht-1 regulatory regions (Wilhelm et al., 2013a,b).
Rht-B1i-1 with the 160 bp insertion in the promoter significantly increases the plant height of wheat against both the Rht-D1a and Rht-D1b backgrounds. To study the contribution of the 160 bp insertion to gene transcription, we analyzed the expression profiles of Rht-B1i-1 and Rht-B1i-2 with Rht-B1a as a control. qRT-PCR analysis of CS and varieties containing Rht-B1i at the seedling and heading stage indicated that the 160 bp insertion might affect gene expression. At the seedling stage, nine of 22 varieties containing Rht-B1i-1 and all eight varieties containing Rht-B1i-2 expressed lower levels than wild-type Rht-B1a, whereas other 13 of 22 varieties carrying Rht-B1i-1 expressed higher levels than Rht-B1a (statistically significant in 2/13). Furthermore, at the heading stage, the variety H251 containing Rht-B1i-2 expressed significant lower level than Rht-B1a in the fourth internodes, whereas, the expression levels of three varieties H8, H83, and H215 containing Rht-B1i-1 were significantly higher than Rht-B1a in flag leaves and the fourth internodes. Although the Rht-B1i gene was expressed at different levels in different varieties at seedling and heading stage, the general trend is relatively consistent, indicating the 160 bp insertion is likely to promote the transcription of the Rht-B1i-1 gene. Compared with the three spring wheat varieties H8, H83, and H215, the expression levels of Rht-B1i-1 in H213 (which is a weak winter wheat variety from Qinghai province, China) were lower than that of CS at seedling and heading stage. Their difference in the expression levels of Rht-B1i-1 may be caused by the genetic backgrounds, this needs further investigation. Wilhelm et al. (2013b) reported that the insertions may have a minor effect on Rht-B1 transcript abundance in seedling tissue, although the normalized Rht-B1 transcript levels were slightly (non-significantly) reduced. The discrepancy among these varieties may be explained by their different genetic backgrounds, and Wilhelm et al. (2013b) used only two lines with the 160 bp insertion (‘Mercia’ and ‘Paragon’) for qRT-PCR. Moreover, the Rht-1 gene was expressed at different levels in different tissues and developmental stages (Pearce et al., 2011), our previous study showed that Rht-1 preferentially expressed in the fourth internodes than flag leaves at the heading stage (unpublished data). The effect of the 160 bp insertion on transcript needs more analysis in more tissues and developmental stages.
Wilhelm et al. (2013a) found that Rht-B1a-160 was associated with a small reduction in GA sensitivity relative to Rht-B1a-0 accessions, but the difference was not statistically significant. In the present study, the coleoptile lengths of varieties containing Rht-B1i-1 and Rht-B1i-2 showed no significant effect of GA3 treatment, indicating that Rht-B1i was insensitive to GA3. Rht-B1b and Rht-D1b contain a single nucleotide change introducing a premature stop codon in the DELLA domain of encoded proteins which conferred dwarfism and GA insensitivity, as well as Rht-B1d, Rht-B1e, and Rht-D1d (Peng et al., 1999; Hedden, 2003; Pearce et al., 2011; Li A.X. et al., 2012; Li et al., 2013). It is very surprise that Rht-B1i-1 who could increases the plant height of wheat was insensitive to GA3, the mechanism needs further investigation. Since the promoter of Rht-B1i-2 is the same as Rht-B1a, while their response to GA3 are different, suggesting the missense SNP occurring in Rht-B1i resulted in an E205G amino acid change in the Poly S/T/V region has an effect on the activity of Rht-B1i protein. Itoh et al. (2002) revealed that the polyS/T/V region had a strong suppression function for the activity of SLR protein. The function of this missense SNP in Rht-B1i needs further investigation.
Rht-B1 may participate in the regulation of multiple phyto-hormones and environmental signaling pathways because of the various cis-acting elements in its promoters. The 160 bp insertion in the promoter of Rht-B1i-1 contained 32 possible cis-acting elements not found in Rht-B1a. Among these 32 elements, four, including the auxin response element AUXREPSIAA4, are unique to Rht-B1i-1. This indicates that the height-increasing effect of Rht-B1i-1 may be related to auxin. GA can promote the transport of auxin, whereas auxin can promote the biosynthesis of GA (Frigerio et al., 2006). In root growth and differentiation, GA and DELLA proteins control the transport of auxin and coordinate cell division and differentiation (Moubayidin et al., 2010). In the present study, the coleoptile lengths of wheat varieties with the Rht-B1i-1 genotype responded to IAA, and IAA/GA3, while that of Rht-B1i-2 genotype did not respond to IAA, and IAA/GA3, and the transgenic Arabidopsis seedlings with promoter of Rht-B1i-1 also responded to IAA, and IAA/GA3, that of Rht-B1i-2 did not, illustrating the height-increasing effect of Rht-B1i-1 may be caused by the synergistic effects of IAA and GA3. Further comprehensive study should be launch to elucidate the mechanism of the height-increasing effect for Rht-B1i-1.
XYL, XL, AL, MP, and WY participated in experiments, drafting the manuscript and proposal writing. DL and JS participated in material preparation, AZ and WY participated in experimental design and corrected the manuscript. MS corrected the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported financially by the National Basic Research Program of China (2014CB138101) and CAS Strategic Priority Program (XDA08010104). We thank all the colleagues in our laboratory for constructive discussion and technical assistance.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00307
Addisu, M., Snape, J. W., Simmonds, J. R., and Gooding, M. J. (2009). Reduced height (Rht) and photoperiod insensitivity (Ppd) allele associations with establishment and early growth of wheat in contrasting production systems. Euphytica 166, 249–267. doi: 10.1007/s10681-008-9838-7
Aldrich, C. (1993). CTAB DNA extraction from plant tissues. Plant Mol. Biol. Rep. 11, 128–141. doi: 10.1007/BF02670471
Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1194–1199.
Blake, N. K., Lanning, S. P., Martin, J. M., Doyle, M., Sherman, J. D., Naruoka, Y., et al. (2009). Effect of variation for major growth habit genes on maturity and yield in five spring wheat populations. Crop Sci. 49, 1211–1220. doi: 10.2135/cropsci2008.08.0505
Börner, A., Plaschke, J., Korzun, V., and Worland, A. J. (1996). The relationship between the dwarfing genes of wheat and rye. Euphytica 89, 69–75. doi: 10.1111/plb.12237
Butler, J. D., Byrne, P. F., Mohammadi, V., Chapman, P. L., and Haley, S. D. (2005). Agronomic performance of Rht alleles in a spring wheat population across a range of moisture levels. Crop Sci. 45, 939–947. doi: 10.2135/cropsci2004.0323
Chapman, S. C., Mathews, K. L., Trethowan, R. M., and Singh, R. P. (2007). Relationsips between height and yield in near-isogenic spring wheats that contrast for major reduced height genes. Euphytica 157, 391–397. doi: 10.1007/s10681-006-9304-3
Chen, S., Gao, R., Wang, H., Wen, M., Xiao, J., Bian, N., et al. (2015). Characterization of a novel reduced height gene (Rht23) regulating panicle morphology and plant architecture in bread wheat. Euphytica 203, 583–594. doi: 10.1007/s10681-014-1275-1
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Flintham, J. E., Börner, A., Worland, A. J., and Gale, M. D. (1997). Optimizing wheat grain yield: effects of Rht (gibberellin-insensitive) dwarfing genes. J. Agric. Sci. 128, 11–25. doi: 10.1007/BF00223795
Frigerio, M., Alabadí, D., Pérez-Gómez, J., García-Cárcel, L., Phillips, A. L., Hedden, P., et al. (2006). Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 142, 553–563. doi: 10.1104/pp.106.084871
Gale, M. D., Law, C. N., and Worland, A. J. (1975). The chromosomal location of a major dwarfing gene from Norin 10 in new British semi-dwarf wheats. Heredity 35, 417–421. doi: 10.1038/hdy.1975.112
Gale, M. D., and Marshall, G. A. (1976). The chromosomal location of Gai1 and Rht1 genes for Gibberellin insensitivity and semi-dwarfism, in a derivative of Norin 10 wheat. Heredity 37, 283–289. doi: 10.1038/hdy.1976.88
Gale, M. D., and Youssefian, S. (1985). “Dwarfing genes in wheat,” in Progress in Plant Breeding, ed. G. E. Russell (London: Butterworths), 1–35.
Hedden, P. (2003). The genes of the green revolution. Trends Genet. 19, 5–9. doi: 10.1016/S0168-9525(02)00009-4
Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57–70. doi: 10.1105/tpc.010319
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Li, A. X., Yang, W. L., Guo, X. L., Liu, D. C., Sun, J. Z., and Zhang, A. M. (2012). Isolation of a gibberellin-insensitive dwarfing gene, Rht-B1e, and development of an allele-specific PCR marker. Mol. Breed. 30, 1443–1451. doi: 10.1007/s11032-012-9730-y
Li, A. X., Yang, W. L., Lou, X. Y., Liu, D. C., Sun, J. Z., Guo, X. L., et al. (2013). Novel natural allelic variations at the Rht-1 loci in wheat. J. Integr. Plant Biol. 55, 1026–1037. doi: 10.1111/jipb.12103
Li, H. P., Wang, Y., Li, X. C., Gao, Y., Wang, Z. J., Zhao, Y., et al. (2011). A GA-insensitive dwarf mutant of Brassica napus L. correlated with mutation in pyrimidine box in the promoter of GID1. Mol. Biol. Rep. 38, 191–197. doi: 10.1007/s11033-010-0094-2
Li, Y. Y., Xiao, J. H., Wu, J. J., Duan, J. L., Liu, Y., Ye, X. G., et al. (2012). A tandem segmental duplication (TSD) in green revolution gene Rht-D1b region underlies plant height variation. New Phytol. 196, 282–291. doi: 10.1111/j.1469-8137.2012.04243.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
McIntosh, R. A., Yamazaki, Y., Dubcovsky, J., Rogers, W. J., Morris, C., Appels, R., et al. (2013). “Catalogue of gene symbols for wheat,” in Proceeding of the 12th International wheat genetics Symposium, Yokohama.
McVittie, J. A., Gale, M. D., Marshall, G. A., and Westcott, B. (1978). The intrachromosomal mapping of the Norin 10 and Tom Thumb dwarfing genes. Heredity 40, 67–70. doi: 10.1038/hdy.1978.8
Miedaner, T., and Voss, H. H. (2008). Effect of dwarfing Rht genes on Fusarium head blight resistance in two sets of near-isogenic lines of wheat and check cultivars. Crop Sci. 48, 2115–2122. doi: 10.2135/cropsci2008.02.0107
Miralles, D. J., and Slafer, G. A. (1995). Yield, biomass and yield components in dwarf, semi-dwarf and tall isogenic lines of spring wheat under recommended and late sowing dates. Plant Breed. 114, 392–396. doi: 10.1111/j.1439-0523.1995.tb00818.x
Moubayidin, L., Perilli, S., Dello Ioio, R., Di Mambro, R., Costantino, P., and Sabatini, S. (2010). The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr. Biol. 20, 1138–1143. doi: 10.1016/j.cub.2010.05.035
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Paolacci, A. R., Tanzarella, O. A., Porceddu, E., and Ciaffi, M. (2009). Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 10:11. doi: 10.1186/1471-2199-10-11
Park, J., Nguyen, K. T., Park, E., Jeon, J.-S., and Choia, G. (2013). DELLA proteins and their interacting RING finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell 25, 927–943. doi: 10.1105/tpc.112.108951
Pearce, S., Saville, R., Vaughan, S. P., Chandler, P. M., Wilhelm, E. P., Spark, C. A., et al. (2011). Molecular characterization of Rht-1 dwarfing genes in hexaploid wheat. Plant Physiol. 157, 1820–1831. doi: 10.1104/pp.111.183657
Peng, J. R., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). “Green revolution” genes encode mutant gibberellin response modulators. Nature 400, 256–261. doi: 10.1038/22307
Pestsova, E. G., Korzun, V., and Börner, A. (2008). Validation and utilisation of Rht dwarfing gene specific markers. Cereal Res. Commun. 36, 235–246. doi: 10.1556/CRC.36.2008.2.4
Rebetzke, G. J., Ellis, M. H., Bonnett, D. G., and Richards, R. A. (2007). Molecular mapping of genes for coleoptile growth in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 114, 1173–1183. doi: 10.1007/s00122-007-0509-1
Sim, N. L., Kumar, P., Hu, J., Henikoff, S., Schneider, G., and Ng, P. C. (2012). SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40, W452–W457. doi: 10.1093/nar/gks539
Wang, J., Sun, J. Z., Liu, D. C., Yang, W. L., Wang, D. W., Tong, Y. P., et al. (2008). Analysis of Pina and Pinb alleles in the micro-core collections of Chinese wheat germplasm by Ecotilling and identification of a novel Pinb allele. J. Cereal Sci. 48, 836–842. doi: 10.1016/j.jcs.2008.06.005
Wen, W., Deng, Q. Y., Jia, H. Y., Wei, L. Z., Wei, J. B., Wan, H. S., et al. (2013). Sequence variations of the partially dominant DELLA gene Rht-B1c in wheat and their functional impacts. J. Exp. Bot. 64, 3299–3312. doi: 10.1093/jxb/ert183
Wilhelm, E. P., Boulton, M. I., Al-Kaff, N., Balfourier, F., Bordes, J., and Greenland, A. J. (2013a). Rht-1 and Ppd-D1 associations with height, GA sensitivity, and days to heading in a worldwide bread wheat collection. Theor. Appl. Genet. 126, 2233–2243. doi: 10.1007/s00122-013-2130-9
Wilhelm, E. P., Mackay, I. J., Saville, R. J., Korolev, A. V., Balfourier, F., Greenland, A. J., et al. (2013b). Haplotype dictionary for the Rht-1 loci in wheat. Theor. Appl. Genet. 126, 1733–1747. doi: 10.1007/s00122-013-2088-7
Wu, J., Kong, X. Y., Wan, J. M., Liu, X. Y., Zhang, X., Guo, X. P., et al. (2011). Dominant and pleiotropic effects of a GAI gene in wheat results from lack of interaction between DELLA and GID1. Plant Physiol. 157, 2120–2130. doi: 10.1104/pp.111.185272
Keywords: Triticum aestivum L., Rht-B1i, height-increasing effect, promoter, 160 bp insertion
Citation: Lou X, Li X, Li A, Pu M, Shoaib M, Liu D, Sun J, Zhang A and Yang W (2016) The 160 bp Insertion in the Promoter of Rht-B1i Plays a Vital Role in Increasing Wheat Height. Front. Plant Sci. 7:307. doi: 10.3389/fpls.2016.00307
Received: 07 September 2015; Accepted: 27 February 2016;
Published: 16 March 2016.
Edited by:
Keqiang Wu, National Taiwan University, TaiwanReviewed by:
Yuri Shavrukov, University of Adelaide, AustraliaCopyright © 2016 Lou, Li, Li, Pu, Shoaib, Liu, Sun, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aimin Zhang, YW16aGFuZ0BnZW5ldGljcy5hYy5jbg==; Wenlong Yang, d2x5YW5nQGdlbmV0aWNzLmFjLmNu
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.