
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 March 2016
Sec. Plant Breeding
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.00256
 Laura Toppino1
Laura Toppino1 Lorenzo Barchi2
Lorenzo Barchi2 Roberto Lo Scalzo3
Roberto Lo Scalzo3 Eristanna Palazzolo4
Eristanna Palazzolo4 Gianluca Francese5
Gianluca Francese5 Marta Fibiani3
Marta Fibiani3 Antonietta D'Alessandro5
Antonietta D'Alessandro5 Vincenza Papa3
Vincenza Papa3 Vito A. Laudicina4
Vito A. Laudicina4 Leo Sabatino4
Leo Sabatino4 Laura Pulcini6
Laura Pulcini6 Tea Sala1
Tea Sala1 Nazzareno Acciarri6
Nazzareno Acciarri6 Ezio Portis2
Ezio Portis2 Sergio Lanteri2
Sergio Lanteri2 Giuseppe Mennella5
Giuseppe Mennella5 Giuseppe L. Rotino1*
Giuseppe L. Rotino1*Eggplant berries are a source of health-promoting metabolites including antioxidant and nutraceutical compounds, mainly anthocyanins and chlorogenic acid; however, they also contain some anti-nutritional compounds such as steroidal glycoalkaloids (SGA) and saponins, which are responsible for the bitter taste of the flesh and with potential toxic effects on humans. Up to now, Quantitative Trait Loci (QTL) for the metabolic content are far from being characterized in eggplant, thus hampering the application of breeding programs aimed at improving its fruit quality. Here we report on the identification of some QTL for the fruit metabolic content in an F2 intraspecific mapping population of 156 individuals, obtained by crossing the eggplant breeding lines “305E40” × “67/3.” The same population was previously employed for the development of a RAD-tag based linkage map and the identification of QTL associated to morphological and physiological traits. The mapping population was biochemically characterized for both fruit basic qualitative data, like dry matter, °Brix, sugars, and organic acids, as well as for health-related compounds such chlorogenic acid, (the main flesh monomeric phenol), the two peel anthocyanins [i.e., delphinidin-3-rutinoside (D3R) and delphinidin-3-(p- coumaroylrutinoside)-5-glucoside (nasunin)] and the two main steroidal glycoalkaloids, solasonine, and solamargine. For most of the traits, one major QTL (PVE ≥10%) was spotted and putative orthologies with other Solanaceae crops are discussed. The present results supply valuable information to eggplant breeders on the inheritance of key fruit quality traits, thus providing potential tools to assist future breeding programs.
Eggplant (Solanum melongena L.) is a member of the Solanaceae, a large plant family comprising over 3000 species including important crops such as tomato (Solanum lycopersicum L.), potato (Solanum tuberosum L.), pepper (Capsicum annuum L.), and tobacco (Nicotiana tabacum L.). Eggplant is represented by three cultivated species: S. macrocarpon L. and S. aethiopicum L., which are indigenous to a vast area of Africa and are locally cultivated, and the worldwide-cultivated S. melongena L. Unlike most of the other Solanaceous crops, which are native of the New World (Fukuoka et al., 2010; Albert and Chang, 2014; Hirakawa et al., 2014), eggplant has a phylogenetic uniqueness, due to its exclusive Old World origin (Lester and Hasan, 1991) located in Asia as a result of two/three separate domestication events (Daunay and Hazra, 2012; Meyer et al., 2012; Cericola et al., 2013; Knapp et al., 2013). Eggplant was introduced to the Mediterranean Basin by Muslim invaders in the Seventh-to Eighth-century CE (Daunay, 2008) and, at present, it is one of the most consumed vegetables in the world. Its global production is estimated to be around 49.5 Mt and is mainly concentrated in Asia (93% of both the world production and harvested area), with China, India, Indonesia, Iran, representing the major producers. Egypt, Turkey, and Italy are the main producers of the Mediterranean countries (FAO, 2013; http://faostat3.fao.org/browse/Q/QC/E).
Despite the economic importance of S. melongena, genetic mapping studies on this species have been rather limited compared to other Solanaceae crop species. The first eggplant linkage map was constructed from an interspecific (eggplant × S. linnaeanum) F2 population by Doganlar et al. (2002a) and exploited for QTL analyses of several breeding traits (Doganlar et al., 2002b; Frary et al., 2003). The same map was later on improved with the addition 115 PCR-based tomato orthologous markers (Wu et al., 2009) and used for the identification of several QTL affecting morphological traits (Frary et al., 2014). A further inter-specific map (eggplant x S. incanum) was recently developed by Gramazio et al. (2014) and exploited for locating polyphenol oxidase genes as well as genes involved in the chlorogenic acid biosynthetic pathway.
The first intra-specific genetic map was published by Nunome et al. (2001), and was subsequently integrated with SSR markers (Nunome et al., 2003, 2009) as well as orthologous Solanum gene sets (Fukuoka et al., 2012). Miyatake et al. (2012) spotted QTL underpinning parthenocarpy in F2 populations obtained by crossing one parthenocarpic with two non-parthenocarpic breeding lines; the same populations were recently employed to explore QTL for the resistance trait to Fusarium oxysporum (Miyatake et al., 2015) taking advantage from sequence information retrieved from the first eggplant draft genome released (Hirakawa et al., 2014). Barchi et al. (2010) developed a further intra-specific linkage map from an F2 population, afterwards densely populated with RAD-tag derived marker and used to characterize the genetic basis of traits associated with anthocyanin content (Barchi et al., 2012). More recently, the same map was used for detecting QTL of key horticultural traits (Portis et al., 2014). Furthermore, through a GWAs approach, the previously identified loci were validated and a number of new marker/trait associations detected (Cericola et al., 2014; Portis et al., 2015).
In the last decades the eggplant breeding goals have been mainly focused on the improvement of agronomic traits, including fruit size, weight, color, and shape (Kashyap et al., 2003; Frary et al., 2006), prickliness, yield potential and adaptation to climatic conditions (Daunay et al., 2001). Resistance or tolerance to biotic stresses have been also central breeding objectives (Rotino et al., 2014) Only recently organoleptic and nutritional properties, bioactive and anti-nutritional compounds, post-harvest and processing-related traits of eggplant fruits have been studied in a genetic/genomic perspective (Prohens et al., 2007, 2013; Gajewski et al., 2009; Prohens et al., 2013; Plazas et al., 2013; Zhang et al., 2014; Docimo et al., 2016; Lo Scalzo et al., 2016).
Eggplant berries are a source of vitamins (Grubben, 1977) and other health-promoting metabolites including anthocyanins (delphinidin glycosides) and chlorogenic acid (Stommel and Whitaker, 2003; Mennella et al., 2010) with nutraceutical and anti-oxidant properties (Cao et al., 1996; Kwon et al., 2008; Akanitapichat et al., 2010). Otherwise, they also contain some anti-nutritional compounds, like saponins and steroidal glycoalkaloids (SGA), which are responsible of the bitter taste of the flesh (Aubert et al., 1989; Sánchez-Mata et al., 2010) and with a potential toxic effects on humans. The accumulation of these metabolites in the fruit and the total amount of all these metabolites appears developmentally regulated (Mennella et al., 2012) and, although strongly affected by the environment, a wide variation among different eggplant cultivars was highlighted (Plazas et al., 2013).
To date a very limited number of studies have been aimed at identifying QTL affecting the eggplant content in bioactive and antinutrional compounds as well as other fruit quality traits (Shetty et al., 2011; Gramazio et al., 2014). Here we report the first QTL associated to the eggplant fruit content in sugars, organic acids, dry matter and soluble solid, the steroidal glycoalkaloids solamargine, the two anthocyanic pigments delphinidin-3-rutinoside (D3R) and delphinidin-3-(p-coumaroylrutinoside)-5-glucoside (nasunin), chlorogenic acid, as well as fruit skin color.
A population of 156 F2 plants, previously obtained by crossing the two eggplant breeding lines “305E40” and “67/3,” contrasting for a wide number of key agronomic traits (Barchi et al., 2012; Portis et al., 2014) was employed (Figure 1). The double haploid female parent “305E40” was derived from an interspecific somatic hybrid Solanum aethiopicum gr. gilo(+)S. melongena cv. Dourga (Rizza et al., 2002), which was repeatedly backcrossed with the recurrent lines DR2 and Tal1/1, prior to selfing and anther culture. The female parent carries the resistance locus Rfo-sa1 conferring resistance to the soil-borne fungus F. oxysporum f. sp. melongenae (Toppino et al., 2008). “305E40” produces pink flowers and long, highly pigmented dark purple fruits characterized by the presence of the anthocyanin D3R as well as a higher glycoalkaloids and organic acid content than the line “67/3.” The latter is the result of an F8 selection from the intra-specific cross cv. “Purpura” × cv. “CIN2,” displays higher anthocyanic pigmentation than “305E40” in leaves and stems, and produces violet flowers and round, violet colored fruits with white peel color both under and next to calyx. The fruits are characterized by the presence of the anthocyanin nasunin in the peel, higher soluble solids, sugars, and chlorogenic acid content in the flesh with respect to “305E40”.

Figure 1. Phenotypical characterization of the parental lines and of the F1 progeny. (A) Features of the parental lines “305E40” and “67/3” and of the hybrid F1 at different ripening stages. For each line, from left to right are represented: stage A- (early immature, ~14 DAP), stage A (immature, ~21 DAP), stage B- (early commercial ripe, ~30 DAP), stage B (commercial ripe, ~38 DAP), stage C (physiological ripe, ~55 DAP scale bar represents 30 cm. Fruit features: (B1): Detail of fruits differing for peel color. (B2): Detail of fruits differing for color under and next to calyx.
The mapping population was field grown, along with both parents and the F1 hybrid, at two sites, namely ML (Montanaso Lombardo 45°20′N; 9°26′E) and MT (Monsampolo del Tronto 42°53′N; 13°47′E). Each F2 individual was replicated by establishing vegetative cuttings. To this end, cuttings were obtained both from apical and lateral shoots, by using a sharp scalpel, from greenhouse-grown plantlets at the 6–8 leaves stage. To stimulate rooting the cut surface of the cutting was immersed for 10 min in a solution containing 2 mg/l of IBA (indol 3 butyric acid) and 5 mg/l of NAA (α-Naphthaleneacetic acid) buffered to pH 6.0 using 500 mg/l of MES [2-(N-morpholino)ethanesulfonic acid] and then grown in small pot (9 × 9 cm) filled with peat and perlite (70:30 v/v). The pots were maintained in the shadow under high humidity to preserve leaf viability until rooting. This procedure was repeated three times to clone the entire F2 population. At both sites, the material was arranged as a set of two randomized complete blocks with two replicate plants per entry per block. The traits scored are detailed in Table 1.
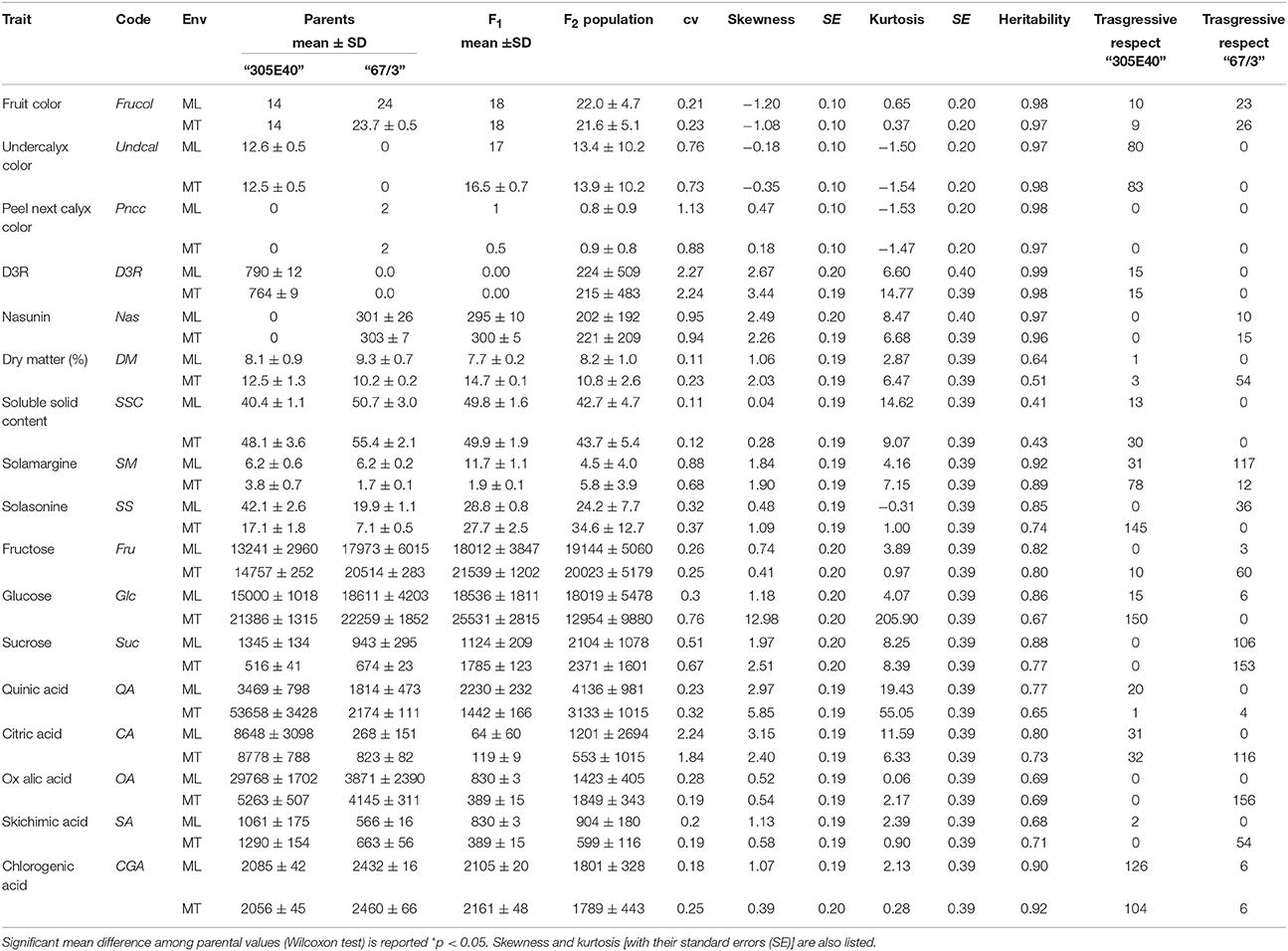
Table 1. List of the traits (unit of measurement for all the biochemical compounds is mg/100 dw) analyzed and their code, means, standard deviations (SD), coefficients of variation (cv), and broad sense heritability for the traits.
The fruit color was evaluated in two representative fruits per plant, collected at the commercial stage (stage B, as in Mennella et al., 2012; Figure 1) between the first and fourth harvest. A discrete phenotypic scale was used to characterize the skin color both as a whole and under the calyx, taking into consideration both the tonality (0 = white, 5 = reddish, 10 = purple, 15 = intermediate, 20 = violet, 25 = brilliant lavender) and the intensity (+0 = whitish, +1 = light +2 = normal, +3 = dark, +4 = blackish). For the presence of a lighter peel edge next to calyx, a scale from 0 to 2 (0 = absent, 1 = intermediate, 2 = present) was utilized.
Samplings for biochemical analyses were carried out at two fruit ripening stages, unripe [A, approximately 21 DAF (days after flowering), and commercial (B, approximately 38 DAF)]. According to Mennella et al. (2012), fruits at the stage A are still actively growing, close to half of the final size, with glossy skin color, calyx, and peduncle quite tender and flexible, flesh still soft and greenish or white and seeds having not reached final size and displaying a white tegument. Besides, fruits at the stage B have reached their commercial final size, with brighter or less brilliant skin (and eventually little dull in some lines); the flesh is less greenish and, in the 305E40 like-typology shows the characteristic green ring next to the skin (absent in “67/3”), seeds have almost reached their final size but are still immature.
The developing stages A was chosen as reference for the flesh biochemical characterization because, in a preliminary study, it was the stage in which the parental lines showed the highest significant differences with regard to the content of the majority of the compounds in study (Supplementary Table 1). Only for the D3R and nasunin content, the peel from fruits at developing stage B were analyzed.
The experimental sample was constituted by portions obtained from 5−8 fruits, sampled in duplicate. For fruits at stage A, fruit cubes were obtained within 2 h after harvest; while from fruits at stage B, peel slices of about 4 cm2 were collected from the same fruit analyzed for skin color. All the samples were immediately frozen in liquid N2 and lyophilized.
The fresh weight of all the samples and the correspondent weight at the end of freeze-drying process were detected for dry matter (DM) value calculation. The freeze-dried tissues were powdered and held at −80°C. All results were referred to as dry weight (dw).
Peel anthocyanins, glycoalkaloids (solamargine and solasonine) and phenolic acids of the fruit flesh were extracted and analyzed according to Mennella et al. (2012). The extraction and the analysis of anthocyanins was carried out on 200 mg of lyophilized and powdered peel, diluted in 10 mL of methanol containing 3% trifluoroacetic acid (TFA). RP-HPLC analysis was performed through a Waters E-Alliance HPLC system constituted by a 2695 separations module with quaternary pump, autosampler, and a 2996 photodiode array detector; data were acquired and analyzed with Waters Empower software on a PC. The chromatographic separations were performed at a flow rate of 0.8 mL/min and at 0.1 AUFS. Purified D3R (Polyphenols Laboratories AS, Sandnes, Norway) was used as external standard in RP-HPLC analyses. As for nasunin quantification, a partially purified standard was used according to Lo Scalzo et al. (2010). The results were expressed as mg/100 g of peel dw; the limit of detection was 1.3 mg/100 g of dw.
The extraction of chlorogenic acid (CGA) started from 200 mg of lyophilized flesh. The quantification carried out after a RP-HPLC separation, was based on absorbance at 325 nm relative to the sesamol internal standard and an external standard of authentic CGA (Sigma-Aldrich, St. Louis, MO). The results were expressed as mg/100 g of dw. Soluble solid content (SSC) was measured on the centrifuged extract of 300 mg of eggplant powder with 10 mL of 1 mM HCl (5 min at 25000 × g at room temperature), and it was expressed as percent substance on dw.
Glycoalkaloids, solamargine, and solasonine, were extracted from 0.5 g samples of lyophilized and powdered flesh tissue by 95% ethanol. The analyses were performed by means of RP-HPLC using partially purified solasonine and solamargine as the external standards. The data were expressed as mg/100 g dw; the limit of detection was 0.03 mg/100 g of dw.
Organic acids and simple sugars content of eggplant extracts were measured by HPLC, slightly modifying the methods proposed and validated by Caccamo et al. (1986) and by López-Tamames et al. (1996). The extracts were the same used for SRR determination (see above).
As for organic acids, the separation was carried out on an Inertsil ODS-3 column 5 μm of particle diameter and 0.46 × 25 cm dimension. The elution was carried out at 30°C with H3PO4 0.02 M in milli-Q water as mobile phase at 0.6 mL/min. The detection was spectrophotometrically made at 214 nm. The extracts from eggplant (3 mL) were filtered through 0.45 μm PTFE filters and 3-fold diluted with the mobile phase before injection (20 μL).
Oxalic, quinic, shikimic, and citric acid solutions were used as external standards (retention times 5.4, 6.7, 8.8, and 12.7 min, respectively), and the results were expressed as mg/100 g dw.
Sucrose, glucose and fructose were separated with a CarboSep Coregel 87C carbohydrate column with a 0.78 × 30 cm bed packed with a cation-exchange resin in the Ca2+ ionic form. The mobile phase was milli-Q water at 0.7 mL/min, the elution was performed at 85°C, and the signals were revealed by a refractive index detector. Samples were five-fold diluted with mobile phase before injection (20 μL). Sucrose, glucose and fructose solutions were used as external standards (retention times 9.0, 10.1, and 12.4 min, respectively) and the results were expressed as mg/100 g dw.
Statistical analyses were performed using R software (R Development Core Team, 2009). A conventional analysis of variance was applied to estimate genotype and environment effects based on the linear model Yij = μ + gi + bj + eij, where μ, g, b, and e represent, respectively, the overall mean, the genotypic effect, the block effect and the error. Broad-sense heritability values were given by /([ + ]/n), where represented the genetic variance, the residual variance and n the number of blocks. Correlations between traits were estimated using the Spearman coefficient, and normality, kurtosis and skewness were assessed with the Shapiro-Wilks test (α = 0.05). Segregation was considered as transgressive when at least one F2 individual recorded a trait value higher or lower by at least two standard deviations than the higher or lower scoring parental line. QTL detection was based on the Barchi et al. (2012) map, constituted of 415 markers (339 SNPs, 2 HRMs, 3 CAPSs, 11 RFLPs, 33 SSRs, and 27 COSII) and spanning 1390 cM. Putative QTL location was determined by both interval (Lander and Botstein, 1989) and MQM (Jansen, 1993; Jansen and Stam, 1994) mapping, as implemented in MapQTL v5 software (Van Ooijen, 2004). QTL were initially identified using interval mapping, after which one linked marker per putative QTL was treated as a co-factor in the approximate multiple QTL model. Co-factor selection and MQM analysis were repeated until no new QTL could be identified. LOD thresholds for declaring a QTL to be significant at the 5% genome-wide probability level were established empirically by applying 1000 permutations per trait (Churchill and Doerge, 1994). Additive and dominance genetic effects, as well as the percentage of the phenotypic variance explained (PVE) by each QTL were obtained from the final multiple QTL model. The program QTLNetwork 2.1 (Yang et al., 2007) was used to analyze each set of environment's data separately to identify epistasis, and was then extended across both environments to identify any QTL x environment interactions present. QTL effects were estimated on the basis of the Markov Chain Monte Carlo (MCMC) method. A type I error level of 0.05 was applied. The genome scan employed a 10 cM window and a 1 cM walk speed. Critical F values were obtained by 1000 permutations and a threshold of 0.05 was applied to assign significance to a QTL or to an epistatic effect. Individual QTL were prefixed by a trait abbreviation, followed by the relevant chromosome designation, and were suffixed as “a” or “b” where more than one QTL mapped to a single linkage group; ML or MT was added as a suffix where the QTL was expressed in a site-specific manner. Epistatic effects were indicated by adding “*” to the label of a major established QTL, while “ep” was added to a newly detected QTL. MapChart v2.1 software (Voorrips, 2002) was draw the resulting maps. Syntenic regions of the tomato genome (sequence build 2.50; http://solgenomics.net/organism/Solanum_lycopersicum/genome) were accessed to identify candidate genes co-localizing with the eggplant QTL. Initial searches were conducted using 20-kb sections and, for sections of interest, additional searches were performed using 10 kb sections. Putative tomato orthologs of the eggplant genes were identified by Blast search in the tomato gene indices at DFCI.
Trait codes, their values and broad sense heritability are shown in Table 1. The parental lines significantly differ each other for most of the traits in study at both sites (Table 1). Compared to “67/3” plants, “305E40” produces purple dark fruits reaching a score of 14 (10+4, see methods), with the same peel color under (Undcal) and next (Pncc) to the calyx, contains exclusively the D3R pigment and has a greater organic acids content than the line “67/3”. On the contrary, the latter is characterized by a dark/blackish violet color rated at 23–24 (20+3−4) given by the exclusive presence of the nasunin pigment, and absence of pigmentation under calyx, together with a higher amount of sugars and chlorogenic acid. At both sites, the F1 hybrid was intermediate for the fruit color (rating 18 given probably by the presence of both anthocyanin nasunin and green ring in the flesh next to the peel), the content of soluble solid, solasonine, and chlorogenic acid (Table 1). For several traits, F1 performance was significantly superior to the better performing parent, as for Undcal (both environments), DM (MT), SM (ML), and sugar contents; on the contrary, as an example, CA and QA (MT) and DM (ML) contents were lower to the worst performing parent. In the F2 progeny transgressive segregation (as calculated from the raw phenotypic data (Table 1) was observed for several traits. As an example, the traits Frucol (23 plants), SM (117 plants), Suc (106 plants) compared to “67/3” in ML and Undcal (80 plants), and CA (31 plants) with respect to “305E40.” In MT, transgressive phenotypes were found, among the others, for Undcal (83 plants) and SS (145 plants) compared to “305E40” and for DM (54 plants), Suc (153 plants), and CA (116 plants) toward “67/3” parent. The broad sense heritability values were generally uniform between ML and MT. The range was from 0.41 (SSC at ML) to 0.99 (D3R at ML) (Table 1). Significant inter-trait correlations (p < 0.05) were detected both within and across sites (Table 2). In both ML and MT, Frucol was positively correlated with Undcal and Nas and negatively correlated with D3R, Undcal was negatively correlated with Pncc, and Fru was positively correlated with Glc. Finally QA, SA, and OA were positively correlated between each other and negatively correlated with CA. The significant correlations across sites ranged from + 0.224 for SA to + 0.926 for Undcal.
In all, 29 QTL (of which 22 explained at least 10% of the PVE, hereafter referred to as “major” QTL) were identified and mapped onto 10 of the 12 eggplant chromosomes (Table 3, Figure 2), with the exclusion of E07 and E12. At ML and MT a set of 14 (10 major) and 15 (12 major) QTL were identified, respectively. Fourteen major QTL were uncovered at both sites, while three were exclusively detected in ML and four only in MT; finally, one was a major QTL in one of the sites and a minor one in the other. The genomic locations of QTL are shown in Figure 2. Clustering of the QTL was found in chromosomes E04, E05, and E10. The high inter-trait correlations (positive or negative) between the traits falling in QTL clusters on chromosome E05 and E10 (Table 2) suggest that they reflect either a set of closely linked loci or, more likely, a single pleiotropic locus. In the cluster on chromosome E05, the QTL were associated with fruit and under calyx color, as well as with D3R and Nas content. The other cluster which includes QTL for Undcal and Pncc was spotted on chromosome E10.
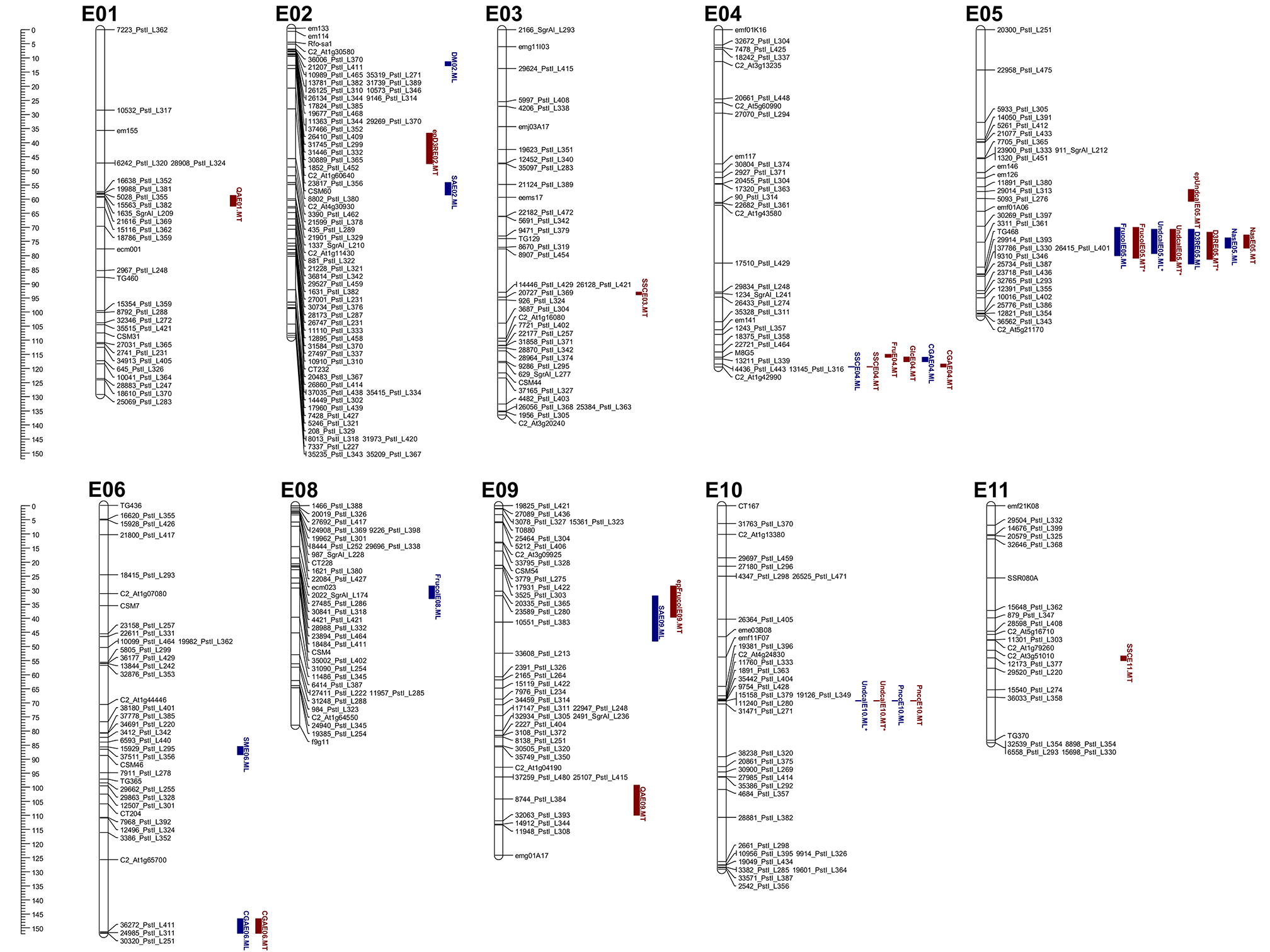
Figure 2. Eggplant linkage map with graphical representation of the QTL detected. Marker names are shown to the right of each chromosome, with map distances (in cM) shown on the left. Map positions of the QTL identified on each chromosome (or LG) are also given on the right. The length of the vertical bars represents the confidence interval of the QTL (LODmax−1 interval). QTL shown in blue were detected only at ML, and those in red only at MT. Epistatic interacting QTL are indicated by the prefix ^ where the QTL was recognized by MapQTL software.
Two Frucol QTL were mapped in ML and one in MT. A particularly large effect QTL (FrucolE05) explained 56.3% in ML and 69.9% in MT of the PVE and mapped to the same E5 region of QTL for Undcal, D3R, and Nas. A further minor QTL (FrucolE08) was detected only in ML and explained 5.5% of the PVE. The allele for the violet color (violet vs. purple) derived from “67/3.”
Two major QTL were detected in both environments and located in E05 and E10. The largest effect locus (UndcalE10) explained 77% of the PVE in ML and 82.5% in MT; the second major QTL (UndcalE05) explained 13.8% of the PVE at both the sites. For UndcalE05, the allele conferring the presence of under calyx pigmentation derives from “305E40” while absence of pigmentation comes from “67/3.”
One major QTL at locus PnccE10 was found in both the environments, with a PVE of 86.9% and 82.3% in ML and MT, respectively. The segment containing this E10 locus also influenced the major QTL for Undcal. The positive allele (presence of a lighter peel-next to calyx color vs. no different color) was contributed by “67/3.”
One QTL (D3RE05) was detected in both ML and MT, mapping to E05 and explaining 49.7% and 52% of the PVE in ML and MT respectively. The positive allele was inherited from “305E40.”
One QTL (NasE05) was detected in both ML and MT, mapping to E05 and explaining 28% and 28.4% of the PVE in ML and MT, respectively. The positive allele was inherited from “67/3.”
The major QTL DME02 was detected only in ML environment, explaining 11.9% of the PVE. The positive allele was transmitted from “305E40.”
One SSC QTL was mapped in ML and three in MT. The major QTL SSCE04 was detected in both environments and explained 10.7% of the PVE at ML and 10.1% at MT. The other two QTL, a major (SSCE03) and a minor one (SSCE11), were spotted only in MT, explaining the 14.75 and 7.8% of PVE. The “67/3” allele at SSCE04 was associated with an increment in soluble solid content, while “305E40” alleles contribute to increase soluble solid content at SSCE03 and SSCE11.
No QTL exceeding the minimum threshold was identified for this trait.
SME06 was the only QTL detected in ML, with 13.9% of PVE explained. The allele responsible for increased solamargine content derived from “67/3.”
A single major QTL (FruE04) was identified only in MT, explaining 10.7% of the PVE. The “67/3” allele was associated with an increase of fructose content.
A single major QTL (GlcE04) was identified only in MT, near FruE04, explaining 10.7% of the PVE. The “67/3” allele was associated with an increase of glucose content.
No QTL exceeding the minimum threshold was identified for this trait.
A major (QAE01) and a minor (QAE09) QTL were identified only in MT location, explaining 16.4 and 8.9% of the PVE, respectively. The “67/3” allele was associated with an increase in quinic acid.
No QTL exceeding the minimum threshold were identified for these traits.
A major (SAE02) and a minor (SAE09) QTL were identified only in ML, explaining 12.1% and 8.2% of the PVE, respectively. The positive alleles derived from “305E40.”
Two conserved QTL in both the environments were identified. The QTL CGAE06 was major in MT and minor in ML, explaining 15.4 and 7% of the PVE, respectively. QTL CGAE04 was minor in both environments and explained 7.3 and 9.5% of the PVE in ML and MT, respectively. All the positive alleles derived from “305E40.”
Epistatic interactions were evaluated by considering the two sites as independent replicates (Figure 2). In ML, epistatic interaction was observed for Undcal: a pair of previously detected QTL (UndcalE05.ML* and UndcallE10.ML*) displayed a significant level of additive x additive, additive x dominant and dominant x additive epistasis, with an individual variance of 0.7, 0.5, and 1.3%, respectively. In MT, epistatic interactions were observed for Frucol, Undcal, and D3R. For Frucol, an already identified QTL FrucolE05.MT* together with a de novo QTL (epFrucolE09.MT) displayed a significant degree of additive x additive and dominant x additive epistasis, explaining 3.4 and 0.4% of the PVE. The previously detected UndcalE05.MT* and UndcallE10.MT* showed additive × additive, additive × dominant, dominant × additive, and dominant × dominant epistasis, explaining 0.5, 0.7, 1.6, and 0.7% of the PVE. In addition, the previously identified UndcallE10.MT* showed additive × dominant and dominant × dominant epistatic interactions with a de novo QTL (epUndcalE05.MT), explaining 0.3 and 1% of PVE. Finally the brand new epD3RE02.MT QTL epistatically interact (additive × additive, additive × dominant, dominant × additive, and dominant × dominant) with the already identified D3RE05.MT region, explain 4.5, 2.4, 1.4, and 0.8% of the PVE. The combined site analysis showed that none of the additive effect × site or dominance effect × site interactions were statistically significant at p < 0.05.
The tomato QTL identified by Fulton et al. (2002) affecting the content in fructose (fru4.1), glucose (glu4.1), and °Brix (brx4.1), as well as the QTL fru-4I, glu-4I, and brx-4I reported by Causse et al. (2004) lie in a region which is syntenic to E04 region in which clustered the eggplant QTL controlling fructose, glucose and soluble solid content (FruE04.MT, SSCE04.ML, SSCE04.MT, GlcE04.MT).
The eggplant QAE01.MT and QAE09.MT regions are syntenic to the IL1-1 and IL9-3 reported to contain metabolic QTL for quinate. Similarly, the CGAE04 and CGAE06 QTL are syntenic to the IL4-1 and IL 6-3 containing QTL for the quinate (Schauer et al., 2006).
A blastX search of the NCBI non-redundant protein database carried out for the marker loci linked to the QTL, failed to highlight any known genes or transcription factors involved in anthocyanin, sugar, acid, and glycoalkaloid pathways.
To identify candidate genes underlying the eggplant QTL identified in the present study, we investigated (the tomato syntenic regions sequence (build 2.50; http://solgenomics.net/organism/Solanum_lycopersicum/genome) to search for genes and transcription factors putatively involved in the control of the traits in study. Among the tomato genes/transcription factors involved in anthocyanin biosynthesis, the gene encoding UDP glucose anthocyanidin 3-0 glucosyltansferase (3GT), an2 and ant1 are all located on chromosome T10, proximal to the E10 region containing 15158_PstI_L379 (linked to QTL for Undcal and Pncc). The anthocyanin synthesis-associated enzyme UDP glucose anthocyanidin 5–0 glucosyltransferase is located on T12, in a region distal to E05 region containing 3311_PstI_l361 (linked to QTL for Frucol, Undcol, D3R, and Nas).
Among the genes/transcription factors involved in the fructose/glucose pathways, we identified a UDP-glucosyltransferase (Solyc04g080010.2.1) and a Glycosyltransferase family 77 protein (Solyc04g080080) in the IL4-4 region (syntenic to FruE04.MT, SSCE04.ML, SSCE04.MT, GlcE04.MT), which was reported as containing QTL controlling the fructose and glucose content.
Nowadays, consumers are aware of the beneficial effect of consuming fruits and vegetables as many evidences now exist concerning their protective effect against a number of diseases. However, consumers do not choose their foods exclusively for the nutrients they provide but also on the basis of their taste, texture, and appearance (Causse et al., 2010). In recent years a wide number of studies have been focused in assessing the content in phytonutrients of vegetables (Daunay et al., 2001; Gupta and Prakash, 2014) but a limited number of studies have been carried out in identifying the genetic basis influencing their content and their interaction with the environment.
Within the Solanaceae family, efforts toward the characterization of QTL controlling the biochemical properties and nutritional value of the berries have been mainly carried out in tomato (Fulton et al., 2002; Causse et al., 2004; Sacco et al., 2013), while in potato the genetic basis of tuber quality and glycoalkaloids content (Bradshaw et al., 2008; D'hoop et al., 2008; Sørensen et al., 2008; Valcarcel et al., 2014) have been investigated. In eggplant, studies have been focused on polyphenols, anthocyanins, chlorogenic acid (Stommel and Whitaker, 2003; Mennella et al., 2010) as well as glycoalkaloids content (Aubert et al., 1989; Sánchez-Mata et al., 2010). However, QTL controlling basic qualitative traits, such as °Brix, simple sugars, organic acids, DM, and health-related metabolites have not been so far characterized, in spite of their key role in breeding programs for developing eggplants with improved content of phenolics, and other health-related promoting compounds.
Evidences that season, environment and genotype strongly affect eggplant fruit composition of carbohydrates and starch, vitamin C and phenolics has been recently ascertained (San José et al., 2014; Stommel et al., 2015). Similar results were also reported for dry matter, proteins, total phenolics, and mineral contents (Raigón et al., 2010) as well as solasonine, D3R and solamargine (Mennella et al., 2010). In the present work, the high value associated with the traits recorded along with the correlation between the two sites highlight that the environment played a moderate influence on the phenotypic outcome of the segregating F2 in study. The h values were higher than 60% for all the traits (Table 3) with the exception of SSC in both locations (0.41% in ML and 0.43% in MT) and DM in MT (0.51%). Collard et al. (2005) suggested that a QTL should only be classified as “major” if it can account for more than 10% of the PVE. A more stringent definition of “major” implies the conservation of a QTL in multiple seasons/locations (Li et al., 2001; Lindhout, 2002; Pilet-Nayel et al., 2002). Among the 14 QTL discovered in ML and the 15 in MT, at least one major QTL per each trait in study was identified. The less and the most convincing LOD scores associated with these QTL were 3.85 (SSCE04.ML and GlcE04.MT) and 72.35 (UndcalE10.MT) respectively. The explained PVE varied from ~10% (SSCE04.MT) up to ~86.9% (PnccE10.ML) and most of the identified QTL were stable across the two environments making them useful in the context of marker-assisted selection. On the other side the identification of QTL in just one environment (i.e., DME02.ML or FruE04.MT), supported the strong environment effect on some metabolic traits in eggplant, as described above.
Finally, some traits were QTL orphan: presumably, the genetic variance between the parents and/or the limited variation in the mapping population were not sufficient to dissect the genetic basis of these traits (Lander and Botstein, 1989).
The genetic basis of anthocyanin synthesis and accumulation has been widely studied in the Solanaceae (van Eck et al., 1993, 1994; Chaim et al., 2003; Borovsky et al., 2004; De Jong et al., 2004; Bovy et al., 2007; Gonzali et al., 2009). The genetic control of their accumulation and distribution in eggplant was for long time thought to be complex, involving at least three major and five minor loci with assumed epistatic interactions and/or pleiotropic effects (Tatebe, 1939; Tigchelaar et al., 1968). Recently, QTL-related studies using a family based and association mapping approach allowed to furtherly clarify the genetic basis of anthocyanin distribution in eggplant tissues and organs as well as to identify its syntenic relationships with tomato (Barchi et al., 2012; Ge et al., 2013; Cericola et al., 2014). The QTL cluster identified on chromosome E05, which controls Frucol, Undcal, the content of the anthocyanins D3R and Nas responsible of the peel color together with the detected inter-trait correlations, suggests the presence of a putative pleiotropic locus. Using the same mapping population, Barchi et al. (2012) identified QTL controlling several anthocyanin-related traits and the color of the corolla in the same chromosome region, while Ge et al. (2013) and Cericola et al. (2014) identified similar QTL in the distal portion of E05. Our data confirm that the bottom part of the E05 is clearly involved in anthocyanin production and distribution among the eggplant tissues and organ. A second QTL cluster for Pncc and Undcal was identified on E10, and is collinear with the one previously identified in several plant tissues by using the same F2 mapping population (Barchi et al., 2012) as well as with GWAS approach (Cericola et al., 2014). This chromosome region may be ascribable to the same QTL previously identified by Doganlar et al. (2002b) and involved in anthocyanin accumulation.
The QTL located on E5 and E10 are possibly involved in two different aspects of the anthocyanic determination and distribution in eggplant.The QTL on E05 is involved both in the alternative production of the nasunin or D3R and it is responsible of the fruit and corolla color. This hypothesis is supported by the following considerations: (i) the two QTL co-localize at the same molecular marker; (ii) nasunin completely replaced D3R in the parental line “67/3” and in the hybrid, which suggest that it originates from an enzymatic modification of D3R (Kroon et al., 1994; Fujiwara et al., 1997; Yonekura-Sakakibara et al., 2000; Ichiyanagi et al., 2005; Azuma et al., 2008). However, no genetic locus encoding the 5-5-O-glucosyltransferase gene or of a cumaryltransferase has been to date identified. Conversely, the QTL on chromosome E10 appears mainly involved in the color determination of the peel next to the calyx (Pncc) and in anthocyanin intensity in all the other tissues. The joint effect of both E05 and E10 QTL influence Undcal, alluding to an interaction between genes influencing both D3R/Nas and Pncc in controlling this trait.
The steroidal glycoalkaloids are distinctive Solanaceae secondary metabolites; they are warehoused in all tissues, including fruits (tomato and eggplant) and tubers (potato) (Friedman, 2002, 2006; Kozukue et al., 2008; Mennella et al., 2010; Sawai et al., 2014) and provide the plant with a chemical barrier against a broad range of pathogens (Chan and Tam, 1985; Hoagland, 2009). Their biosynthesis starts from cholesterol, which is converted firstly in steroidal alkaloids (SAs) and subsequently decorated with various sugar moieties to generate steroidal glycoalkaloids (SGAs, Itkin et al., 2013), and require genes encoding uridine 5′-diphosphate (UDP)–glycosyltransferases (UGTs), like GAME1 in tomato (Itkin et al., 2011) or SGT1 in potato (McCue et al., 2005). Recently, Sawai et al. (2014) identified St SSR2 as a key enzyme in the biosynthesis of toxic SGAs derived from cholesterol in potato. SGAs deeply influence the organoleptic properties of the flesh, by conferring a typical bitter taste (Aubert et al., 1989; Sánchez-Mata et al., 2010) and are potentially toxic to animals and humans, although some recent studies support also their anti-cancer and anti-microbial properties (Blankemeyer et al., 1997; Friedman et al., 2009; Milner et al., 2011). Reduction of the SGAs eggplant content represent a key breeding aspect for the crop improvement. The QTL analyses conducted over the two main eggplant SGAs, solasonine and solamargine, revealed the presence of one major QTL, SME06.ML only in ML. The blastX results did not identify any putative gene.
Sugars and acids, involved in soluble solid content and dry matter, are key quality components of the fruit quality and influence its nutritional values and organoleptic properties. QTL related for sugar and acids content were found in only one of the sites in study (ML for SA and MT for Fru, Glc and QA) as confirmed by their medium-low inter-site correlations. This finding suggests that environment influenced these traits although they had a high heritability, good inter-traits correlations (Fru with Glc and SA with QA) and a PV >10%; similar results were reported by San Josè et al. (2014). This phenomenon may also partially explain why QTL for sucrose, oxalic acid and citric acid content were not identified.
The chlorogenic acid in eggplant berries plays a well-documented anti-oxidant activity. The chlorogenic synthesis pathway in Solanaceae is quite well-known (Clé et al., 2008) and the sequences of the six genes codifying for the enzymes involved in this pathway have been studied. In a recent study, most of the genes involved in the polyphenols and chlorogenic acid biosynthetic pathway were positioned in an interspecific map by using a candidate gene approach (Gramazio et al., 2014). It has been demonstrated that the growing environment and its interaction with the genotype influences phenolic acid content; nonetheless, a reduced variability in phenolics may be obtained through selection for stability (Stommel et al., 2015). The two CGA QTL, identified in both locations, however did not contain any putative gene involved in the chlorogenic acid pathway. Thus, the genetic basis of the CGA related trait, as well as that of glycoalkaloids, deserve to be further investigated (e.g., by means of a candidate gene approach on the basis of the already identified genes in tomato and potato).
For some traits the parental origin of the QTL alleles reflected the performance of the parents; thus, for example, the positive alleles at D3R and SA were inherited from “305E40” while Frucol, Nas and Glc derived from “67/3.” On the contrary, Pncc and SSC were traits increased by positive alleles coming from both the parents. Finally, positive alleles for CGA and QA derived from the less performant parent.
Transgressive segregation, which derives from the combination of alleles from both parents having effect on the same direction (deVicente and Tanksley, 1993), was found for several traits, and in most cases displayed the same transgressive trend in both the environment (as Frucol, or Undcal). On the contrary, some traits (e.g., SS and OA), showed a different transgressive trend due to the different performances of the parents at the two sites. This phenomenon is common in QTL analysis and already observed in eggplant (Barchi et al., 2012; Portis et al., 2014).
In addition to a conserved epistatic interaction for Undcal between the two sites, a number of environmental-specific examples of epistasis were noticed, none of them explained a substantial proportion of the PV (0.3–4.5%). Such phenomenon previously detected in the same mapping population (Barchi et al., 2012; Portis et al., 2014) may be explained by invoking interference from other QTL in the background (Al-sane et al., 2011). Although the analysis carried out with QTLNetwork 2.1 (Yang et al., 2007) on the combined data set produced no significant QTL x Environment interactions, some QTL identified with the MQM were location-specific, for this reason we cannot rule out the presence of QTL x Environment interaction.
Tomato represents a model species in the Solanaceae family and many studies have been carried out to better understand the genetic basis underlying yield (Brix and SSC), fruit flavor, and taste (sensory quality, glycoalkaloids) by means of deep metabolic profiling and candidate gene survey (Fridman et al., 2000; Fulton et al., 2002; Causse et al., 2004; Schauer et al., 2006; Bermúdez et al., 2008; Itkin et al., 2011; Iijima et al., 2013).
The extensive synteny between the tomato and eggplant genomes enables genetic inferences to be made in eggplant exploiting the much greater knowledge of tomato genome (Wu et al., 2009; Barchi et al., 2012; Fukuoka et al., 2012; Tomato genome consortium, 2012). Furthermore, the availability of the eggplant genome (Hirakawa et al., 2014) confirmed the synteny level existing between eggplant and tomato.
Schauer et al. (2006) conducted a deep metabolic QTL analysis using the tomato ILs population developed by Liu and Zamir (1999). The IL4-4 was reported as containing QTL controlling the fructose and glucose content; in the corresponding eggplant genomic region we identified two QTL involved in the accumulation of fructose and glucose (FruE04.MT and GlcE04.MT). In this tomato region, a glycosyltransferase family GT8 protein, an UDP-glucosyltransferase, and a glycosyltransferase family 77 protein were identified, which might be involved in the sugar pathway. The eggplant QAE01.MT and QAE09.MT QTL are syntenic to ILs 1-1/1-1-3 and IL9.3 but no putative genes involved in the quinic acid pathway have been identified. Finally, the two QTL identified for CGA lie in tomato syntenic regions of the ILs 4-1 and 6-3, involved in quinic acid accumulation. The search for genes involved in chlorogenic pathway did not highlight any putative gene, although the CGAs pathway is well-known and eggplant and tomato genes have been identified in the work of Hirakawa et al. (2014).
In summary, the QTL here detected, provide important information on the genomic region associated to fruit biochemical and phenotypical properties in eggplant which may be usefully exploited in view of breeding programs aimed at releasing of new eggplant varieties with superior commercial, organoleptic and compositional characteristics.
GLR, SL, NA, RL, GM conceived and designed the research; LT, LB, RL, ErP, GF, MF, AD, VP, VL, LS, LP, TS, NA, GM performed fruit phenotypical and biochemical characterization; LT, LB and EzP analyzed the data; LT, LB, GLR wrote the manuscript. All authors read and approved the manuscript.
This research was partially supported by the Italian Ministry of Agriculture in the framework of the project “IT-IL-NUTRISOL ITalian-IsraeLi initiative for the NUTRItional improvement of SOLanaceus crops” (DM 27723/7303/11).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MP and Handling Editor declared their shared affiliation, and the Handling Editor states that the process nevertheless met the standards of a fair and objective review.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00256
Akanitapichat, P., Phraibung, K., Nuchklang, K., and Prompitakkul, S. (2010). Antioxidant and hepatoprotective activities of five eggplant varieties. Food Chem. Toxicol. 48, 3017–3021. doi: 10.1016/j.fct.2010.07.045
Albert, V. A., and Chang, T. H. (2014). Evolution of a Hot Genome. Proc. Natl. Acad. Sci. U.S.A. 111, 5069–5070. doi: 10.1073/pnas.1402378111
Al-sane, K. O., Povero, G., and Perata, P. (2011). Anthocyanin tomato mutants: overview and characterization of an anthocyanin-less somaclonal mutant. Plant Biosyst. 145, 436–444. doi: 10.1080/11263504.2010.548914
Aubert, S., Daunay, M. C., and Pochard, E. (1989). Saponosides stéroïdiques de l'aubergine (Solanum melongena L.) I. Intérêt alimentaire, méthodologie d'analyse, localisation dans le fruit. Agronomie 9, 641–651. doi: 10.1051/agro:19890701
Azuma, K., Ohyama, A., Ippoushi, K., Ichiyanagy, T., Takeuchi, A., Saito, T., et al. (2008). Structures and antioxidant activity of anthocyanins in many accessions of eggplant and its related species. J. Agric. Food Chem. 56, 10154−10159. doi: 10.1021/jf801322m
Barchi, L., Lanteri, S., Portis, E., Stagel, A., Valè, G., Toppino, L., et al. (2010). Segregation distortion and linkage analysis in eggplant (Solanum melongena L.). Genome 53, 805–815. doi: 10.1139/G10-073
Barchi, L., Lanteri, S., Portis, E., Valè, G., Volante, A., Pulcini, L., et al. (2012). A RAD Tag derived marker based eggplant linkage map and the location of QTLs determining anthocyanin pigmentation. PLoS ONE 7:e43740. doi: 10.1371/journal.pone.0043740
Bermúdez, L., Urias, U., Milstein, D., Kamenetzky, L., Asis, R., Fernie, A. R., et al. (2008). A candidate gene survey of quantitative trait loci affecting chemical composition in tomato fruit. J. Exp. Bot. 59, 2875–2890. doi: 10.1093/jxb/ern146
Blankemeyer, J. T., White, J. B., Stringer, B. K., and Friedman, M. (1997). Effect of α-tomatine and tomatidine on membrane potential of frog embryos and active transport of ions in frog skin. Food Chem. Toxicol. 35, 639–646.
Borovsky, Y., Oren-Shamir, M., Ovadia, R., Jong, W., and Paran, I. (2004). The A locus that controls anthocyanin accumulation in pepper encodes a MYB transcription factor homologous to Anthocyanin2 of Petunia. Theor. Appl. Genetics 109, 23–29. doi: 10.1007/s00122-004-1625-9
Bovy, A., Schijlen, E., and Hall, R. (2007). Metabolic engineering of flavonoids in tomato (Solanum lycopersicum): the potential for metabolomics. Metabolomics 3, 399–412. doi: 10.1007/s11306-007-0074-2
Bradshaw, J. E., Hackett, C. A., Pande, B., Waugh, R., and Bryan, G. J. (2008). QTL mapping of yield, agronomic and quality traits in tetraploid potato (Solanum tuberosum subsp. tuberosum). Theor. Appl. Genetics 116, 193–211. doi: 10.1007/s00122-007-0659-1
Caccamo, F., Carfagnini, G., Di Corcia, A., and Samperi, R. (1986). Improved high-performance liquid chromatographic assay for determining organic acids in wines. J. Chromatogr. A 362, 47–53 doi: 10.1016/S0021-9673(01)86949-1
Cao, G., Sofic, E., and Prior, R. L. (1996). Antioxidant capacity of tea and common vegetables. J. Agric. Food Chem. 44, 3426–3431. doi: 10.1021/jf9602535
Causse, M., Duffe, P., Gomez, M. C., Buret, M., Damidaux, R., Zamir, D., et al. (2004). A genetic map of candidate genes and QTLs involved in tomato fruit size and composition. J. Exp. Bot. 55, 1671–1685. doi: 10.1093/jxb/erh207
Causse, M., Friguet, C., Coiret, C., Lépicier, M., Navez, B., Lee, M., et al. (2010). Consumer preferences for fresh tomato at the European scale: a common segmentation on taste and firmness. J. Food Sci. 75, S531–S541. doi: 10.1111/j.1750-3841.2010.01841.x
Cericola, F., Portis, E., Toppino, L., Barchi, L., Acciarri, N., Ciriaci, T., et al. (2013). The population structure and diversity of eggplant from Asia and the Mediterranean Basin. PLoS ONE 8:e73702. doi: 10.1371/journal.pone.0073702
Cericola, F., Portis, E., Lanteri, S., Toppino, L., Barchi, L., Acciarri, N., et al. (2014). Linkage disequilibrium and genome-wide association analysis for anthocyanin pigmentation and fruit color in eggplant. BMC Genomics 15:896 doi: 10.1186/1471-2164-15-896
Chaim, A. C., Borovsky, Y. B., De Jong, W. D. J., and Paran, I. (2003). Linkage of the A locus for the presence of anthocyanin and fs10.1, a major fruit-shape QTL in pepper. Theor. Appl. Genetics 106, 889–894. doi: 10.1007/s00122-002-1132-9
Chan, H. T., and Tam, S. Y. T. (1985). Toxicity of tomatine to larvae of the mediterranean fruit fly (Diptera: Tephritidae). J. Econ. Entomol. 78, 305–307. doi: 10.1093/jee/78.2.305
Churchill, G. A., and Doerge, R. W. (1994). Empirical threshold values for quantitative trait mapping. Genetics 138, 963–971.
Clé, C., Hill, L. M., Niggeweg, R., Martin, C. R., Guisez, Y., Prinsen, E., et al. (2008). Modulation of chlorogenic acid biosynthesis in Solanum lycopersicum; consequences for phenolic accumulation and UV-tolerance. Phytochemistry 69, 2149–2156. doi: 10.1016/j.phytochem.2008.04.024
Collard, B. C. Y., Jahufer, M. Z. Z., Brouwer, J. B., and Pang, E. C. K. (2005). An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142, 169–196. doi: 10.1007/s10681-005-1681-5
D'hoop, B., Paulo, M., Mank, R., van Eck, H., and van Eeuwijk, F. (2008). Association mapping of quality traits in potato (Solanum tuberosum L.). Euphytica 161, 47–60. doi: 10.1007/s10681-007-9565-5
Daunay, M. C., and Hazra, P., (2012). “Eggplant,” in Handbook of Vegetables, eds K. V. Peter and P. Hazra (Houston, TX: Studium Press), 257–322.
Daunay, M. C., Lester, R., and Ano, G. (2001). “Cultivated eggplants,” in Tropical Plant Breeding, eds A. Charrier, M. Iacquot, S. Hamon and D. Nicolas (Enfield, NH: Science), 220–225.
Daunay, M. C. (2008). “Eggplant,”in Handbook of Plant Breeding - Vegetables II. Fabeceae, Liliaceae, Solanaceae and Umbrelliferae, eds J. Prohens and F. Nuez (New York, NY: Springer), 163–220.
De Jong, W. S., Eannetta, N. T., Jong, D. M., and Bodis, M. (2004). Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theor. Appl. Genetics 108, 423–432. doi: 10.1007/s00122-003-1455-1
deVicente, M. C., and Tanksley, S. D. (1993). QTL Analysis of transgressive segregation in an interspecific tomato cross. Genetics 134, 585–596.
Docimo, T., Francese, G., Ruggiero, A., Batelli, G., De Palma, M., Bassolino, L., et al. (2016). Phenylpropanoids accumulation in eggplant fruits: characterization of biosynthetic genes and regulation by a MYB transcription factor. Front. Plant Sci. 6:1233. doi: 10.3389/fpls.2015.01233
Doganlar, S., Frary, A., Daunay, M. C., Lester, R., and Tanksley, S. (2002a). A comparative genetic linkage map of eggplant (Solanum melongena) and its implications for genome evolution in the Solanaceae. Genetics 161, 1697–1711. doi: 10.1007/s00122-008-0950-9
Doganlar, S., Frary, A., Daunay, M. C., Lester, R., and Tanksley, S. (2002b). Conservation of gene function in the Solanaceae as revealed by comparative mapping of domestication traits in eggplant. Genetics 161, 1713–1726.
Frary, A., Doganlar, S., and Daunay, M. C. (2006). “Eggplant,” in Genome Mapping & Molecular Breeding in Plants, Vol. 5. Vegetables, ed C. Kole (Heidelberg: Springer Publishing), 287–313.
Frary, A., Doganlar, S., Daunay, M. C., and Tanksley, S. (2003). QTL analysis of morphological traits in eggplant and implications for conservation of gene function during evolution of solanaceous species. Theor. Appl. Genetics 107, 359–370. doi: 10.1007/s00122-003-1257-5
Frary, A., Frary, A., Daunay, M. C., Huvenaars, K., Mank, R., and Doğanlar, S. (2014). QTL hotspots in eggplant (Solanum melongena) detected with a high resolution map and CIM analysis. Euphytica 197, 211–228. doi: 10.1007/s10681-013-1060-6
Fridman, E., Pleban, T., and Zamir, D. (2000). A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc. Natl. Acad. Sci. U.S.A. 97, 4718–4723. doi: 10.1073/pnas.97.9.4718
Friedman, M., Levin, C. E., Lee, S. U., Kim, H. J., Lee, I. S., Byun, J. O., et al. (2009). Tomatine-containing green tomato extracts inhibit growth of human breast, colon, liver, and stomach cancer cells. J. Agric. Food Chem. 57, 5727–5733. doi: 10.1021/jf900364j
Friedman, M. (2002). Tomato glycoalkaloids: role in the plant and in the diet. J. Agric. Food Chem. 50, 5751–5780. doi: 10.1021/jf020560c
Friedman, M. (2006). Potato glycoalkaloids and metabolites: roles in the plant and in the diet. J. Agric. Food Chem. 54, 8655–8681. doi: 10.1021/jf061471t
Fujiwara, H., Tanaka, Y., Fukui, Y., Nakao, M., Ashikari, T., and Kusumi, T. (1997). Anthocyanin 5-aromatic acyltransferase from Gentiana triflora. Purification, characterization and its role in anthocyanin biosynthesis. Eur. J. Biochem. 249, 45–51. doi: 10.1111/j.1432-1033.1997.t01-1-00045.x
Fukuoka, H., Miyatake, K., Nunome, T., Negoro, S., Shirasawa, K., Isobe, S., et al. (2012). Development of gene-based markers and construction of an integrated linkage map in eggplant by using Solanum orthologous (SOL) gene sets. Theor. Appl. Genetics 125, 47–56. doi: 10.1007/s00122-012-1815-9
Fukuoka, H., Yamaguchi, H., Nunome, T., Negoro, S., Miyatake, K., and Ohyama, A. (2010). Accumulation, functional annotation, and comparative analysis of expressed sequence tags in eggplant (Solanum melongena L.), the third pole of the genus Solanum species after tomato and potato. Gene 450, 76–84. doi: 10.1016/j.gene.2009.10.006
Fulton, T. M., Bucheli, P., Voirol, E., López, J., Pétiard, V., and Tanskley, S. D. (2002). Quantitative trait loci (QTL) affecting sugars, organic acids and other biochemical properties possibly contributing to flavor, identified in four advanced backcross populations of tomato. Euphytica 127, 163–177. doi: 10.1023/A:1020209930031
Gajewski, M., Kowalczyk, K., Bajer, M., and Radzanowska, J. (2009). Quality of eggplant fruits in relation to growing medium used in greenhouse cultivation and to a cultivar. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 37, 229–234.
Ge, H. Y., Liu, Y., Zhang, J., Han, H. Q., Li, H. Z., Shao, W. T., et al. (2013). Simple sequence repeat-based association analysis of fruit traits in eggplant (Solanum melongena). Genetics and Molecular Research, 12, 5651–5663. doi: 10.4238/2013.November.18.14
Gonzali, S., Mazzucato, A., and Perata, P. (2009). Purple as a tomato: towards high anthocyanin tomatoes. Trends Plant Sci. 14, 237–241. doi: 10.1016/j.tplants.2009.02.001
Gramazio, P., Prohens, J., Plazas, M., Andújar, I., Herraiz, F. J., Castillo, E., et al. (2014). Location of chlorogenic acid biosynthesis pathway and polyphenol oxidase genes in a new interspecific anchored linkage map of eggplant. BMC Plant Biol. 14:350. doi: 10.1186/s12870-014-0350-z
Grubben, G. J. H. (1977). “Tropical vegetables and their genetic resources,” in International Board for Plant Genetic Resources, AGPE: IBPGR/77/23, eds H. D. Tindall and J. T. Williams (Rome: International Board for Plant Genetic Resources), 34–37.
Gupta, C., and Prakash, D. (2014). Phytonutrients as therapeutic agents. J. Complement. Integr. Med. 11, 151–169. doi: 10.1515/jcim-2013-0021
Hirakawa, H., Shirasawa, K., Miyatake, K., Nunome, T., Negoro, S., Ohyama, A., et al. (2014). Draft Genome Sequence of Eggplant (Solanum melongena L.): the representative Solanum species indigenous to the Old World. DNA Res. 21, 649–660. doi: 10.1093/dnares/dsu027
Hoagland, R. E. (2009). Toxicity of tomatine and tomatidine on weeds, crops and phytopathogens fungi. Allelopathy J. 23, 425–436.
Ichiyanagi, T., Kashiwada, Y., Shida, Y., Ikeshiro, Y., Kaneyuki, T., and Konishi, T. (2005). Nasunin from eggplant consists of cis-trans isomers of delphinidin 3-[4-(p-coumaroyl)-L-rhamnosyl (1 → 6) glucopyranoside]-5-glucopyranoside. J. Agric. Food Chem. 53, 9472–9477. doi: 10.1021/jf051841y
Iijima, Y., Watanabe, B., Sasaki, R., Takenaka, M., Ono, H., Sakurai, N., et al. (2013). Steroidal glycoalkaloid profiling and structures of glycoalkaloids in wild tomato fruit. Phytochemistry 95, 145–157. doi: 10.1016/j.phytochem.2013.07.016
Itkin, M., Heinig, U., Tzfadia, O., Bhide, A. J., Shinde, B., Cardenas, P. D., et al. (2013). Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341, 175–179. doi: 10.1126/science.1240230
Itkin, M., Rogachev, I., Alkan, N., Rosenberg, T., Malitsky, S., Masini, L., et al. (2011). GLYCOALKALOID METABOLISM1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell 23, 4507–4525. doi: 10.1105/tpc.111.088732
Jansen, R., and Stam, P. (1994). High-resolution of quantitative traits into multiple loci via interval mapping. Genetics 136, 1447–1455.
Kashyap, V., Kumar, S. V., Collonnier, C., Fusari, F., Haicour, R., Rotino, G. L., et al. (2003). Biotechnology of eggplant. Sci. Hortic. 97, 1–25. doi: 10.1016/S0304-4238(02)00140-1
Knapp, S., Vorontsova, M. S., and Prohens, J. (2013). Wild relatives of the eggplant (Solanum melongena L.: Solanaceae): new understanding of species names in a complex group. PLoS ONE 8:e57039. doi: 10.1371/journal.pone.0057039
Kozukue, N., Yoon, K. S., Byun, G. I., Misoo, S., Levin, C. E., and Friedman, M. (2008). Distribution of glycoalkaloids in potato tubers of 59 accessions of two wild and five cultivated Solanum species. J. Agric. Food Chem. 56, 11920–11928. doi: 10.1021/jf802631t
Kroon, J., Souer, E., de Graaff, A., Xue, Y., Mol, J., and Koes, R. (1994). Cloning and structural analysis of the anthocyanin pigmentation locus Rt of Petunia hybrida: characterization of insertion sequences in two mutant alleles. Plant J. 5, 69–80. doi: 10.1046/j.1365-313X.1994.5010069.x
Kwon, Y. I., Apostolidis, E., and Shetty, K. (2008). In vitro studies of eggplant (Solanum melongena) phenolics as inhibitors of key enzymes relevant for type 2 diabetes and hypertension. Bioresour. Technol. 99, 2981–2988. doi: 10.1016/j.biortech.2007.06.035
Lander, E., and Botstein, D. (1989). Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121, 185–199.
Lester, R. N. Hasan, S. M. Z. (1991). “Origin and domestication of the brinjal eggplant, Solanum melongena, from S. incanum, in Africa and Asia,” in Solanaceae III: Taxonomy, Chemistry, Evolution, eds J. G Hawkes, R. N. Lester, M. Nee and N. Estrada (London: The Linnean Society of London), 369–387.
Li, Z., Jakkula, L., Hussey, R. S., Tamulonis, J. P., and Boerma, H. R. (2001). SSR mapping and confirmation of the QTL from PI96354 conditioning soybean resistance to southern root-knot nematode. Theor. Appl. Genetics 103, 1167–1173. doi: 10.1007/s001220100672
Lindhout, P. (2002). The perspectives of polygenic resistance in breeding for durable disease resistance. Euphytica 124, 217–226. doi: 10.1023/A:1015686601404
Liu, Y. S., and Zamir, D. (1999). Second generation L. pennellii introgression lines and the concept of bin mapping. Rep. Tomato Genet. Coop. 49, 26–30.
Lo Scalzo, R., Fibiani, M., Francese, G., D'Alessandro, A., Rotino, G. L., Conte, P., et al. (2016). Cooking influence on physico-chemical fruit characteristics of eggplant (Solanum melongena L.). Food Chem. 194, 835–842. doi: 10.1016/j.foodchem.2015.08.063
Lo Scalzo, R., Fibiani, M., Mennella, G., Rotino, G. L., Dal Sasso, M., Culici, M., et al. (2010). Thermal treatment of eggplant (Solanum melongena L.) increases the antioxidant content and the inhibitory effect on human neutrophil burst. J. Agric. Food Chem. 58, 3371–3379. doi: 10.1021/jf903881s
López-Tamames, E., Buxaderas, S., Puig-Deu, M. A., and Teixeira, E. (1996). Organic Acids, Sugars, and Glycerol content in white winemaking products determined by HPLC: relationship to climate and varietal factors. Am. J. Enol. Viticolture 47, 193–198.
McCue, K. F., Shepherd, L. V. T., Allen, P. V., Maccree, M. M., Rockhold, D. R., Corsini, D. L., et al. (2005). Metabolic compensation of steroidal glycoalkaloid biosynthesis in transgenic potato tubers: using reverse genetics to confirm the in vivo enzyme function of a steroidal alkaloid galactosyltransferase. Plant Sci. 168, 267–273. doi: 10.1016/j.plantsci.2004.08.006
Mennella, G., Lo Scalzo, R., Fibiani, M., D'Alessandro, A., Francese, G., Toppino, L., et al. (2012). Chemical and bioactive quality traits during fruit ripening in eggplant (S. melongena L.) and allied species. J. Agric. Food Chem. 60, 11821–11831. doi: 10.1021/jf3037424
Mennella, G., Rotino, G. L., Fibiani, M., D'Alessandro, A., Francese, G., Lo Scalzo, R., et al. (2010). Characterization of health-related compounds in eggplant (Solanum melongena L.) lines derived from introgression of allied species. J. Agric. Food Chem. 58, 7597–7603. doi: 10.1021/jf101004z
Meyer, R. S., Karol, K. G., Little, D. P., Nee, M. H., and Litt, A. (2012). Phylogeographic relationships among Asian eggplants and new perspectives on eggplant domestication. Mol. Phylogenet. Evol. 63, 685–701. doi: 10.1016/j.ympev.2012.02.006
Milner, S. E., Brunton, N. P., Jones, P. W., O'Brien, N. M., Collins, S. G., and Maguire, A. R. (2011). Bioactivities of glycoalkaloids and their aglycones from Solanum species. J. Agric. Food Chem. 59, 3454–3484. doi: 10.1021/jf200439q
Miyatake, K., Saito, T., Negoro, S., Yamaguchi, H., Nunome, T., Ohyama, A., et al. (2012). Development of selective markers linked to a major QTL for parthenocarpy in eggplant (Solanum melongena L.). Theor. Appl. Genetics 124, 1403–1413. doi: 10.1007/s00122-012-1796-8
Miyatake, K., Saito, T., Negoro, S., Yamaguchi, H., Nunome, T., Ohyama, A., et al. (2015). Detailed mapping of a resistance locus against Fusarium wilt in cultivated eggplant (Solanum melongena) Theor. Appl. Genetics 129, 357–367. doi: 10.1007/s00122-015-2632-8
Nunome, T., Ishiguro, K., Yoshida, T., and Hirai, M. (2001). Mapping of fruit shape and color development traits in eggplant (Solanum melongena L.) based on RAPD and AFLP markers. Breed. Sci. 51, 19–26. doi: 10.1270/jsbbs.51.19
Nunome, T., Negoro, S., Kono, I., Kanamori, H., Miyatake, K., Yamaguchi, H., et al. (2009). Development of SSR markers derived from SSR-enriched genomic library of eggplant (Solanum melongena L.). Theor. Appl. Genetics 119, 1143–1153. doi: 10.1007/s00122-009-1116-0
Nunome, T., Suwabe, K., Iketani, H., and Hirai, M. (2003). Identification and characterization of microsatellites in eggplant. Plant Breed. 122, 256–262. doi: 10.1046/j.1439-0523.2003.00816.x
Pilet-Nayel, M., Muehlbauer, F., McGee, R., Kraft, J., Baranger, A., and Coyne, C. (2002). Quantitative trait loci for partial resistance to Aphanomyces root rot in pea. Theor. Appl. Gen. 106, 28–39. doi: 10.1007/s00122-002-0985-2
Plazas, M., Lopez-Gresa, M. P., Vilanova, S., Torres, C., Hurtado, M., Gramazio, P., et al. (2013). Diversity and relationships in Key traits for functional and apparent quality in a collection of eggplant: fruit phenolics content, antioxidant activity, polyphenol oxidase activity, and browning. J. Agric. Food Chem. 61, 8871–8879. doi: 10.1021/jf402429k
Portis, E., Barchi, L., Toppino, L., Lanteri, S., Acciarri, N., Felicioni, N., et al. (2014). QTL Mapping in eggplant reveals clusters of yield-related loci and orthology with the tomato genome. PLoS ONE 9:e89499. doi: 10.1371/journal.pone.0089499
Portis, E., Cericola, F., Barchi, L., Toppino, L., Acciarri, N., Pulcini, L., et al. (2015). Association Mapping for Fruit, plant and leaf morphology traits in eggplant PLoS ONE 10:E0135200. doi: 10.1371/journal.pone.0135200
Prohens, J., Rodríguez-Burruezo, A., Raigón, M. D., and Nuez, F. (2007). Total phenolic concentration and browning susceptibility in a collection of different varietal types and hybrids of eggplant: implications for breeding for higher nutritional quality and reduced browning. J. Am. Soc. Hortic. Sci. 132, 638–646.
Prohens, J., Whitaker, B. D., Plazas, M., Vilanova, S., Hurtado, M., Blasco, M., et al. (2013). Genetic diversity in morphological characters and phenolic acids content resulting from an interspecific cross between eggplant, Solanum melongena, and its wild ancestor (S. incanum). Ann. Appl. Biol. 162, 242–257. doi: 10.1111/aab.12017
Raigón, M. D., RodriÌĄguez-Burruezo, A., and Prohens, J. (2010). Effects of organic and conventional cultivation methods on composition of eggplant fruits. J. Agric. Food Chem. 58, 6833–6840. doi: 10.1021/jf904438n
Rizza, F., Mennella, G., Collonnier, C., Shiachakr, D., Kashyap, V., Rajam, M. V., et al. (2002). Androgenic dihaploids from somatic hybrids between Solanum melongena and S. aethiopicum group gilo as a source of resistance to Fusarium oxysporum f. sp melongenae. Plant Cell Rep. 20, 1022–1032. doi: 10.1007/s00299-001-0429-5
Rotino, G. L., Sala, T., and Toppino, L. (2014). “Eggplant,” in Alien Gene Transfer in Crop Plants, Vol. 2, eds A. Pratap and J. Kumar (NewYork, NY: Springer), 381–409.
Sacco, A., Di Matteo, A., Lombardi, N., Trotta, N., Punzo, B., Mari, A., et al. (2013). Quantitative trait loci pyramiding for fruit quality traits in tomato. Mol. Breed. 31, 217–222. doi: 10.1007/s11032-012-9763-2
San José, R., Sánchez-Mata, M. C., Cámara, M., and Prohens, J. (2014). Eggplant fruit composition as affected by the cultivation environment and genetic constitution. J. Sci. Food Agric. 94, 2774–2784. doi: 10.1002/jsfa.6623
Sánchez-Mata, M. C., Yokoyama, W. E., Hong, Y. J., and Prohens, J. (2010). α-Solasonine and α-solamargine contents of gboma (Solanum macrocarpon L.) and scarlet (Solanum aethiopicum L.) eggplants. J. Agric. Food Chem. 58, 5502–5508. doi: 10.1021/jf100709g
Sawai, S., Ohyama, K., Yasumoto, S., Seki, H., Sakuma, T., Yamamoto, T., et al. (2014). Sterol side chain reductase 2 is a key enzyme in the biosynthesis of cholesterol, the common precursor of Toxic Steroidal Glycoalkaloids in potato. Plant Cell 26, 3763–3774. doi: 10.1105/tpc.114.130096
Schauer, N., Semel, Y., Roessner, U., Gur, A., Balbo, I., Carrari, F., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24, 447–454. doi: 10.1038/nbt1192
Shetty, S. M., Chandrashekar, A., and Venkatesh, Y. P. (2011). Eggplant polyphenol oxidase multigene family: cloning, phylogeny, expression analyses and immunolocalization in response to wounding. Phytochemistry 72, 2275–2287. doi: 10.1016/j.phytochem.2011.08.028
Sørensen, K., Kirk, H., Olsson, K., Labouriau, R., and Christiansen, J. (2008). A major QTL and an SSR marker associated with glycoalkaloid content in potato tubers from Solanum tuberosum × S. sparsipilum located on chromosome I. Theor. Appl. Genetics 117, 1–9. doi: 10.1007/s00122-008-0745-z
Stommel, J. R., and Whitaker, B. D. (2003). Phenolic acid content and composition of eggplant fruit in a germplasm core subset. J. Am. Soc. Hortic. Sci. 128, 704–710. doi: 10.1007/s10681-015-1415-2
Stommel, J. R., Whitaker, B. D., Haynes, K. G., and Prohens, J. (2015). Genotype x environment interactions in eggplant for fruit phenolic acid content. Euphytica 205, 823–836 doi: 10.1007/s10681-015-1415-2
Tatebe, T. (1939). On inheritance of color in Solanum melongena L. Jpn. J. Genetics 15, 261–271. doi: 10.1266/jjg.15.261
Tigchelaar, E. C., Janick, J., and Erickson, H. T. (1968). The genetics of anthocyanin coloration in eggplant (Solanum melongena L.). Genetics 60, 475–491.
Tomato genome consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Toppino, L., Valè, G., and Rotino, G. L. (2008). Inheritance of Fusarium wilt resistance introgressed from Solanum aethiopicum Gilo and Aculeatum groups into cultivated eggplant (S. melongena) and development of associated PCR-based markers. Mol. Breed. 22, 237–250. doi: 10.1007/s11032-008-9170-x
Valcarcel, J., Reilly, K., Gaffney, M., and O'Brien, N. (2014). Effect of genotype and environment on the glycoalkaloid content of rare, heritage, and commercial potato varieties. J. Food Sci. 79, T1039–T1048. doi: 10.1111/1750-3841.12443
van Eck, H. J., Jacobs, J. M. E., Dijk, J., Stiekema, W. J., and Jacobsen, E. (1993). Identification and mapping of three flower colour loci of potato (S. tuberosum L.) by RFLP analysis. Theor. Appl. Genetics 86, 295–300. doi: 10.1007/BF00222091
van Eck, H. J., Jacobs, J. M. E., van den Berg, P. M. M. M., Stiekema, W. J., and Jacobsen, E. (1994). The inheritance of anthocyanin pigmentation in potato (Solanum tuberosum 1.) and mapping of tuber skin colour loci using RFLPs. Heredity 73, 410–421.
Van Ooijen, J. W. (2004). MapQTL 5, Software for the Mapping of Quantitative Trait Loci in Experimental Populations. Wageningen: Kyazma B V, 1–63.
Voorrips, R. (2002). Mapchart: software for the graphical presentation of linkage maps and QTLs. J. Heredity 93, 77–78. doi: 10.1093/jhered/93.1.77
Wu, F., Eannetta, N., Xu, Y., and Tanksley, S. (2009). A detailed synteny map of the eggplant genome based on conserved ortholog set II (COSII) markers. Theor. Appl. Genetics 118, 927–935. doi: 10.1007/s00122-008-950-9
Yang, J., Zhu, J., and Williams, R. W. (2007). Mapping the genetic architecture of complex traits in experimental populations. Bioinformatics 23, 1527–1536. doi: 10.1093/bioinformatics/btm143
Yonekura-Sakakibara, K., Tanaka, Y., Fukuchi-Mizutani, M., Fujiwara, H., Fukui, Y., Ashikari, T., et al. (2000). Molecular and biochemical characterization of a Novel Hydroxycinnamoyl-CoA: Anthocyanin 3-0-Glucoside-6”-0-Acyltransferase from Perilla frutescens. Plant Cell Physiol. 41, 495–502. doi: 10.1093/pcp/41.4.495
Keywords: QTL, Solanaceae, fruit quality, bioactive compounds, anthocyanins, chlorogenic acid, glycoalkaloid
Citation: Toppino L, Barchi L, Lo Scalzo R, Palazzolo E, Francese G, Fibiani M, D'Alessandro A, Papa V, Laudicina VA, Sabatino L, Pulcini L, Sala T, Acciarri N, Portis E, Lanteri S, Mennella G and Rotino GL (2016) Mapping Quantitative Trait Loci Affecting Biochemical and Morphological Fruit Properties in Eggplant (Solanum melongena L.). Front. Plant Sci. 7:256. doi: 10.3389/fpls.2016.00256
Received: 24 December 2015; Accepted: 15 February 2016;
Published: 04 March 2016.
Edited by:
Jaime Prohens, Universitat Politècnica de València, SpainReviewed by:
Emidio Albertini, University of Perugia, ItalyCopyright © 2016 Toppino, Barchi, Lo Scalzo, Palazzolo, Francese, Fibiani, D'Alessandro, Papa, Laudicina, Sabatino, Pulcini, Sala, Acciarri, Portis, Lanteri, Mennella and Rotino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giuseppe L. Rotino, Z2l1c2VwcGVsZW9uYXJkby5yb3Rpbm9AY3JlYS5nb3YuaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.