
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Plant Sci. , 16 February 2016
Sec. Plant Pathogen Interactions
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.00096
This article is part of the Research Topic Biotrophic plant-microbe interactions View all 34 articles
Recent years have witnessed major advances in our knowledge of plant mutualistic symbioses such as the rhizobium-legume symbiosis (RLS) and arbuscular mycorrhizas (AM). Some of these findings caused the revision of longstanding hypotheses, but one of the most solid theories is that a conserved set of plant proteins rules the transduction of symbiotic signals from beneficial glomeromycetes and rhizobia in a so-called common symbiotic pathway (CSP). Nevertheless, the picture still misses several elements, and a few crucial points remain unclear. How does one common pathway discriminate between – at least – two symbionts? Can we exclude that microbes other than AM fungi and rhizobia also use this pathway to communicate with their host plants? We here discuss the possibility that our current view is biased by a long-lasting focus on legumes, whose ability to develop both AM and RLS is an exception among plants and a recent innovation in their evolution; investigations in non-legumes are starting to place legume symbiotic signaling in a broader perspective. Furthermore, recent studies suggest that CSP proteins act in a wider scenario of symbiotic and non-symbiotic signaling. Overall, evidence is accumulating in favor of distinct activities for CSP proteins in AM and RLS, depending on the molecular and cellular context where they act.
Our understanding of the major beneficial plant–microbe interactions – the rhizobium-legume symbiosis (RLS) and arbuscular mycorrhizas (AM) – has changed over the last decade in the light of breakthrough discoveries on the role of hormones, the exchange of symbiotic signals, or the lifetime of intraradical structures (Gutjahr and Parniske, 2013; Oldroyd, 2013; Schmitz and Harrison, 2014). AM fungi were once believed to open their way across the root apoplast thanks to cell wall degrading enzymes: genomic sequencing (Tisserant et al., 2013; Lin et al., 2014) suggests this is not the case and cellular evidence (Genre et al., 2005, 2008; Rich et al., 2014) has shown that host cell responses are critical for fungal colonization. Similarly, rhizobium entry in root hairs has been ascribed to the action of bacterial enzymes (Gage, 2004; Robledo et al., 2008); nevertheless, evidence is accumulating in favor of a plant-driven meltdown of the wall surrounding the ‘infection chamber,’ which then expands into the growing infection thread as one semi-solid compartment, where bacteria proliferate and slide (Fournier et al., 2008, 2015).
The demonstration that host plants have major control over such interactions has supported the results of genetic studies, where single plant gene mutations were shown to block both bacterial and fungal penetration of the root (Kistner et al., 2005). Such studies on legume mutants gave rise to the hypothesis that AM and RLS share one signal transduction pathway (Oldroyd, 2013). This common symbiotic pathway, or CSP, is proposed to act downstream of both fungal and rhizobial signal perception and upstream of the activation of the appropriate response to either symbiont (Figure 1).
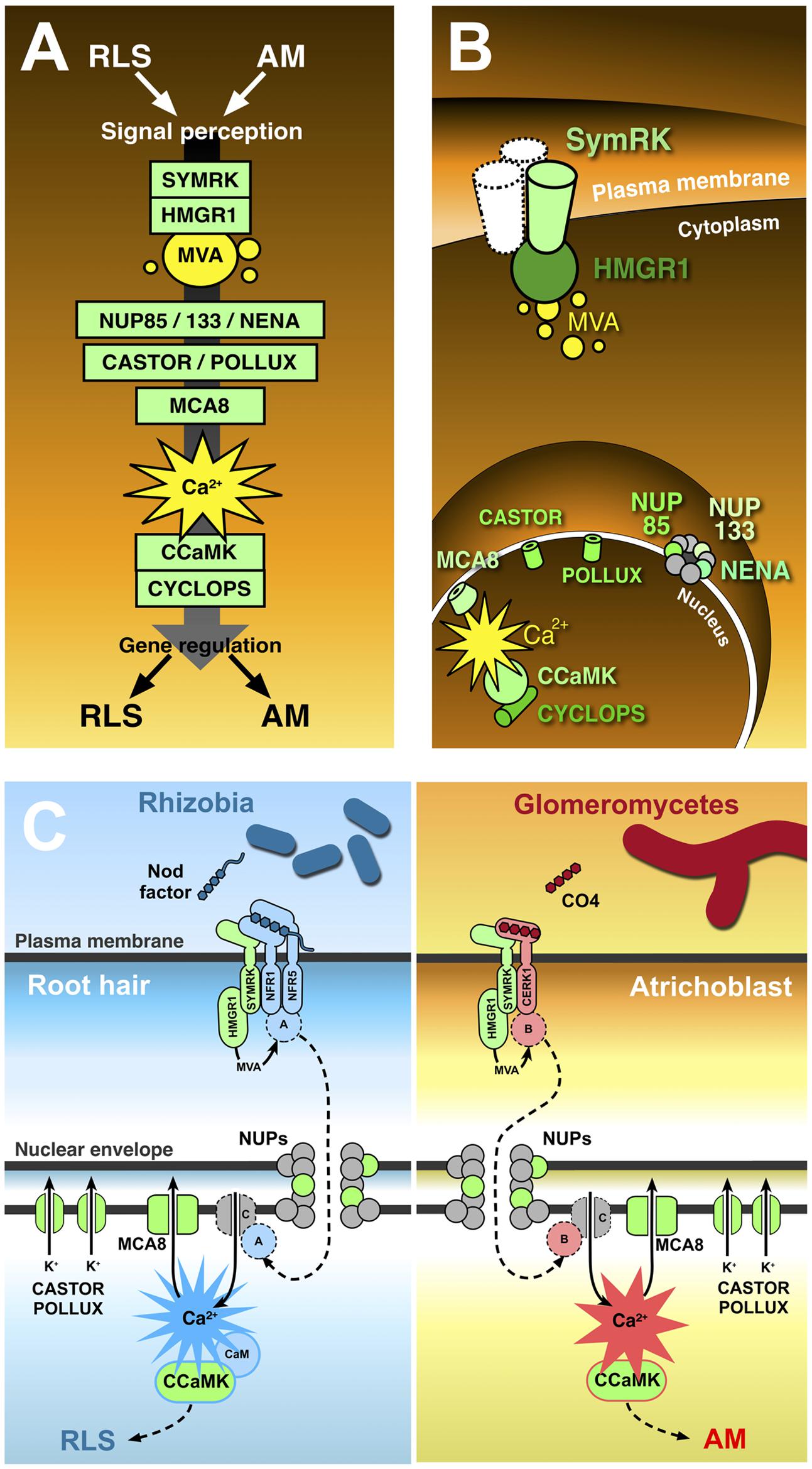
FIGURE 1. The common symbiotic pathway. Ten proteins (in green) have been identified as essential for both RLS and AM. In contrast with the conventional representation of signal transduction along the common symbiotic pathway (A), CSP protein location in different plant cell districts (B) highlights the existence of significant discontinuities between three subsets of CSP members. The receptor-like kinase SYMRK is believed to interact with other co-receptors (such as NFR1 and NFR5) at the cell membrane; its cytoplasmic domain binds the mevalonate (MVA) biosynthetic enzyme HMGR1(3-Hydroxy-3-Methylglutaryl CoA Reductase 1), linking SYMRK activation to MVA synthesis in the vicinity of the plasmalemma. Three nucleoporins (NUP85, NUP133, and NENA) localize at the nuclear pore complex, but their role in signal transduction as well as their functional connection with the previous set of CSP proteins remains unclear. The final stretch of the CSP localizes to the nucleoplasm: here, repeated oscillations in Ca2+ concentration (spiking) are generated with the contribution of MCA8 (a nuclear envelope-bound ATP-powered Ca2+ pump) and the potassium channels encoded by CASTOR and POLLUX genes. Such Ca2+-mediated signals are believed to activate the nuclear kinase CCaMK. In turn, CYCLOPS phosphorylation by CCaMK preludes to the regulation of gene expression. (C) Presents a proposal model of receptor complex assortment and CSP protein role in two separate pathways for the transduction of Nod factors or chito-tetraose in legume root hairs and atrichoblasts, respectively. Localized on the plasma membrane of root hairs (left), the extracellular LysM domain of NFR1 and NFR5 directly bind Nod factors (Broghammer et al., 2012). NFR1 and NFR5 form a complex with SYMRK (Ried et al., 2014) and HMGR1 (Kevei et al., 2007). MVA, produced upon HMGR1 activation (Venkateshwaran et al., 2015) is small enough to diffuse through the nuclear pore complex without the involvement of nucleoporins (NUPs); nevertheless, the inclusion of three NUPs in the CSP opens the possibility that at least one additional unknown protein (A) is involved, which could be activated as a consequence of MVA production and translocated to the nucleus through the action of NUPs. Ca2+ spiking activation in the nucleoplasm is generated by the recursive release of Ca2+ through unidentified channel proteins (C) in the nuclear envelope, combined with the continuous action of ATP-powered Ca2+ pumps like MCA8 (Engstrom et al., 2002; Capoen et al., 2011). To consider a minimal number of unidentified proteins, we can assume Ca2+ channels are directly activated by nuclear-imported A. CASTOR/POLLUX has been proposed to act in concert with Ca2+ channels (Oldroyd, 2013). The resulting Ca2+ spiking activates CCaMK through a RLS-specific conformational change requiring calmodulin (Shimoda et al., 2012; Poovaiah et al., 2013), which then modulates the activity of gene expression regulators, allowing the establishment of RLS (Oldroyd, 2013). A parallel pathway acts in atrichoblasts (right), where chito-tetraose (CO4) released by glomeromycetes is recognised by a complex possibly including CERK1 (Miyata et al., 2014), SYMRK, and HMGR1. Also in this case an additional protein (B) is proposed to be activated by MVA. B is then translocated to the nucleoplasm, where it activates Ca2+ spiking signals with a distinct, AM-specific signature (Kosuta et al., 2008; Russo et al., 2013). Consequently, CCaMK is activated in an AM-specific mode (Shimoda et al., 2012; Poovaiah et al., 2013), and its activity regulates AM-specific gene expression.
Research on plant symbioses has largely been focussed on legumes. The culturability of rhizobia and their amenable genetics – compared to far less manageable glomeromycetes – is probably the main reason why research has progressed more rapidly in the field of RLS than AM. When our knowledge of RLS was later applied to AM, legumes were the obvious biological system for such studies.
A number of legume mutants had been selected for their RLS-defective phenotype (Catoira et al., 2000; Sandal et al., 2006); some of them were later found to display a mycorrhizal phenotype too, either blocking fungal entry at the epidermis surface or altering fungal development inside the root tissues (Kistner et al., 2005; Parniske, 2008; Oldroyd, 2013). The corresponding genes have been characterized and, based on their functions, positioned along a signal transduction pathway, the CSP, transducing glomeromycotan or rhizobial signal perception from the plasma membrane into the nucleus (Figure 1).
In Lotus japonicus, CSP gene products include the receptor-like kinase SYMRK; three nucleoporins, NUP85, NUP133, and NENA; CASTOR and POLLUX, cationic channels located on the nuclear envelope; a nuclear calcium- and calmodulin-dependent kinase CCaMK; and a CCaMK substrate, CYCLOPS (Oldroyd, 2013 and references therein). Furthermore, HMGR1, a key enzyme in the mevalonate biosynthetic pathway, and MCA8, a SERCA-type Ca2+-ATPase localized on the nuclear envelope, have been characterized in Medicago truncatula as additional members of the CSP (Kevei et al., 2007; Capoen et al., 2011). Secondary messengers such as mevalonate and Ca2+ have also been demonstrated to act within the CSP, either as a product of HMGR1 or an activator of CCaMK, respectively (Levy et al., 2004; Venkateshwaran et al., 2015).
In spite of its reassuring name, though, not all evidence confirms that CSP genes actually encode a signal transduction pathway that is common and restricted to RLS and AM.
While a solid link between signal perception and gene expression is a frequent feature in CSP representations (Figure 1A), evidence only supports a connection between sub-sets of the CSP members (Figure 1B).
A first set of CSP proteins includes the membrane-bound receptor-like kinase SYMRK (interacting with other proteins, like the Nod factor receptors NFR1 and NFR5) and the enzyme HMGR1 (Madsen et al., 2003; Kevei et al., 2007; Lefebvre et al., 2010). As a consequence of HMGR1 activation, mevalonate production can also be localized in the vicinity of the cytoplasmic face of the plasma membrane (Venkateshwaran et al., 2015).
A second cluster of CSP proteins is located in the nuclear pore complex: it is composed of three nucleoporins (NUP133, NUP85, and NENA), each of which is responsible for a strong symbiotic phenotype (Kanamori et al., 2006; Saito et al., 2007; Groth et al., 2010). This suggests that the nuclear pore must be controlling the import of an unknown key CSP component. This is not likely to be mevalonate, whose small size should allow nucleoporin-independent diffusion across the nuclear pore (Evans et al., 2004).
Also bound to the nuclear envelope are the ATP-powered Ca2+ pump MCA8 (Capoen et al., 2011) and the cationic channel encoded by CASTOR and POLLUX genes in L. japonicus (Ané et al., 2004; Charpentier et al., 2008). Both proteins contribute to the intense oscillations in nuclear Ca2+ concentration (spiking) that are observed during both AM and RLS establishment (Ehrhardt et al., 1996; Kosuta et al., 2008; Chabaud et al., 2011; Sieberer et al., 2012). In detail, so far unidentified channels are hypothesized to release Ca2+ from the nuclear envelope lumen. This release is sustained by the opposite flow of potassium ions (K+) through CASTOR/POLLUX, in a charge compensation mechanism (Parniske, 2008; Venkateshwaran et al., 2012); concomitant MCA8 activity contributes to the re-establishment of basic nuclear Ca2+ concentration at the end of each peak. Intriguingly, the nuclear pore has been proposed to play a role in flipping membrane-bound proteins from the outer to the inner nuclear membrane (Capoen et al., 2011), shedding light on the possible function of CSP nucleoporins in the targeting of CASTOR, POLLUX, MCA8, and Ca2+ channels.
The last group of CSP proteins whose direct interaction has been demonstrated resides in the nucleoplasm: Ca2+ spiking is supposed to activate CCaMK with the help of calmodulin, through a complex conformational change (Shimoda et al., 2012; Miller et al., 2013; Poovaiah et al., 2013); the enzyme can thus phosphorylate CYCLOPS, a CCaMK-interacting protein (Yano et al., 2008). Phosphorylated CYCLOPS regulates gene expression either directly, as in the case of the NIN promoter (Singh et al., 2014) or through the action of other transcription factors like NSP1, NSP2, and RAM1 (Oldroyd, 2013).
This topological review of the CSP highlights the gaps that uncouple each set of proteins from the next one: with so much missing information, depicting signal transduction along the CSP requires some extrapolation.
On the front of secondary messengers, reactive species of oxygen (Salzer et al., 1999; Pauly et al., 2006) and nitrogen (Meilhoc et al., 2010; Calcagno et al., 2012; Zhang et al., 2013) have also been associated with RLS and AM signaling, although their role in relation to the CSP remains unclear. Furthermore, growing evidence hints at the existence of symbiotic signal transduction pathways that bypass or run parallel to the CSP (Gutjahr et al., 2008, 2009; Bonfante and Requena, 2011), indicating that the plant’s perception of rhizobia and glomeromycetes could rely on multiple signaling routes.
In this context, the information we are still missing will critically challenge the CSP hypothesis: new data will either demonstrate that the CSP is indeed a pathway, or show that the remaining elements differ for each interaction, and what we had imagined as a straight line is rather a core of conserved, yet disconnected, protein functions.
Even if the concept of symbiosis can be extended to any interaction between organisms that live together (De Bary, 1879), the CSP concept is mostly restricted to RLS and AM, where the requirement for CSP genes was initially described (Stougaard, 2001; Kistner et al., 2005). Nevertheless, a third symbiosis also requires SYMRK (Gherbi et al., 2008), nuclear Ca2+ signals (Granqvist et al., 2015; Chabaud et al., 2016) and CCaMK (Svistoonoff et al., 2013): nitrogen-fixing actinorrhizas (Supplementary Figure S1).
In addition, non-symbiotic interactions have been shown to depend on CSP members. Parasitic interactions with root-knot nematodes involve NFR1, NFR5, and SYMRK (Weerasinghe et al., 2005). An intriguing role for SYMRK and CCaMK was also described during the colonization of Pisum sativum by the parasitic plant Orobanche crenata (Fernández-Aparicio et al., 2009): in this case symrk and ccamk mutants were more severely infected, indicating an unprecedented role for these CSP genes in inhibiting (rather than allowing) root colonization.
Furthermore, a key role in the regulation of gene expression has been demonstrated for M. truncatula CCaMK during interaction with the rhizobacterium Pseudomonas fluorescens (Sanchez et al., 2005).
The infection of M. truncatula by the pathogens Phytophthora palmivora (Huisman et al., 2015; Rey et al., 2015), Aphanomyces euteiches, and Colletotrichum trifolii (Rey et al., 2013) has been proposed to partially depend on Nod factor receptors, albeit the involvement of the CSP core was not highlighted. Nevertheless, M. truncatula mutants in CCaMK ortholog DMI3 did not develop any cytoplasmic aggregation – a common defense response – upon Phoma medicaginis or C. trifolii attack (Genre et al., 2009), and displayed an anticipation of necrotrophic fungal growth.
Lastly, CSP homologs are found in mosses, Charophytes and Chlorophytes clades (Wang et al., 2010; Delaux et al., 2013b, 2015): the lack of known naturally occurring symbiotic interactions in such organisms (Field et al., 2015), and the tight phylogenetic relationship between Charales and land plants, might suggest the existence of conserved non-symbiotic functions for CSP proteins throughout the plant clade.
Overall, as research explores additional aspects of plant interactions, the functions of CSP genes appear to be growing in diversity, and extending well beyond the range of symbioses (Supplementary Figure S1).
There is no question CSP genes are essential for both legume endosymbiosis, but can we conclude that the signal transduction process involving CSP proteins is shared? Over the years, such a hypothesis has raised several questions (Bonfante and Requena, 2011), the most striking being: how can one pathway discern two signals, and activate distinct sets of downstream responses?
The foundations of the CSP hypothesis are built on the categorical results of forward genetic approaches: mutant phenotyping has shown that each CSP protein is indispensable for both RLS and AM establishment (Kistner et al., 2005). Nevertheless, signal transduction is not just a matter of protein presence/absence; finer aspects, such as the intensity of enzyme activation in response to each symbiont, could not be revealed by genetic investigations. In fact, recent biochemical analyses indicate that calmodulin binding is dispensable for CCaMK activation during mycorrhization, but essential for nodulation, suggesting that CCaMK is activated in two distinct modes during the perception of rhizobial versus glomeromycotan signals (Shimoda et al., 2012; Poovaiah et al., 2013). Along the same lines, evidence is accumulating in favor of different ‘signatures’ in the Ca2+-mediated signals, which can be responsible for such differential activation of CCaMK at the core of the CSP (Kosuta et al., 2008; Russo et al., 2013).
These studies strongly suggest that the same molecular actors can be playing different biochemical scripts in each interaction. The case of the GRAS-type transcription factor NSP2 (Kaló et al., 2005) is enlightening: NSP2 has been proposed to form a transcription regulator complex with other RLS- or AM-specific factors (NSP1 and RAM1, respectively); it has therefore been described as a CSP protein (Oldroyd, 2013). Nevertheless, in vitro interaction experiments support a model where NSP2 performs distinct symbiotic functions, depending on its interactors (Maillet et al., 2011; Gobbato et al., 2012, 2013; Lauressergues et al., 2012).
Importantly, cellular investigations on model legumes have demonstrated that rhizobia preferentially attach to and penetrate through root hair cells (Gage, 2004), whereas AM fungi contact and enter non-hair cells (atrichoblasts; Genre et al., 2005). Consequently, the study of early plant responses to rhizobial and glomeromycotan signaling only has a biological meaning when this specialization in root epidermal cell types is taken into account: while one pathway including all the necessary proteins could theoretically be designed based on legume genomes, it is likely that each cell type complements the expression of CSP genes with a set AM- or RLS-specific proteins, assembling two spatially and functionally distinct pathways (Figure 1C).
Even if legumes remain the most important model plants for the study of symbiotic interactions, a growing number of publications is providing significant advancements on AM signal perception in non-legumes (Gutjahr et al., 2009; Miyata et al., 2014; Sun et al., 2015; Zhang et al., 2015).
A particular interest has recently been raised by rice mutants in CERK1 (Miya et al., 2007; Sánchez-Vallet et al., 2015), a well-characterized LysM-type chitin receptor involved in defense responses: cerk1 mutants display a strong mycorrhizal phenotype, blocking AM hyphae at the surface of the root epidermis (Miyata et al., 2014). RNAi-based knock-down of CERK1 also induced a significant reduction in AM colonization (Zhang et al., 2015). Involving chitin receptors in symbiotic signaling is very intriguing: chitin-based molecules secreted by AM fungi activate the CSP and downstream responses (Maillet et al., 2011; Czaja et al., 2012; Genre et al., 2013; Giovannetti et al., 2015; Sun et al., 2015). Such Myc factors include lipo-chito-oligosaccharides (LCOs), structurally similar to Nod factors (Maillet et al., 2011), and short undecorated chito-oligosaccharides, or COs (Genre et al., 2013). LCOs appear to be particularly active in legumes, where they trigger a range of responses that are generally common to Nod Factor perception, such as lateral root formation or gene regulation (Maillet et al., 2011; Czaja et al., 2012; Sun et al., 2015), suggesting that some degree of overlap exists between legume perception of Nod and Myc factors. By contrast COs trigger CSP-dependent Ca2+ spiking in both legumes and non-legumes at concentrations as low as 10-8 M (Genre et al., 2013; Sun et al., 2015) and can be considered universal pre-symbiotic AM signals (Sun et al., 2015; Zhang et al., 2015).
Overall, a model is emerging where the assembly of different membrane-residing receptor complexes (Oldroyd, 2013; Liang et al., 2014; Gobbato, 2015; Limpens et al., 2015; Shinya et al., 2015) depends on which receptors are expressed by each cell type and possibly which signaling molecule is present. In the case of defense responses to pathogenic fungi, CERK1/CERK1 (as in Arabidopsis) or CERK1/CEBIP receptor dimers (as in rice) are hypothesized to bind long oligomers such as chito-octaose (CO8), half of the CO8 molecule fitting in to each receptor’s LysM domain (Wan et al., 2008; Shimizu et al., 2010; Liu et al., 2012; Shinya et al., 2015). The perception of shorter COs such as chito-tetraose could rather rely on monomeric receptors (Miyata et al., 2014; Shinya et al., 2015). In legumes, a further level of specificity results in their ability to respond to COs as well as LCOs (such as Nod factors), which fits with the proliferation of legume LysM and LysM-related receptor families through gene duplication events (Zhang et al., 2007).
It is reasonable to speculate that each receptor complex interacts with a corresponding set of cytoplasmic proteins (Asai et al., 2002). At present, information is very limited, but in analogy to SYMRK interaction with HMGR1, we can hypothesize that other proteins associate with each receptor, generating a signal-specific composition in the cytoplasmic moiety of the signaling complex. MAP kinases (Chen et al., 2012) may be playing a role; sensitivity to mevalonate appears as a stringent requisite for the selection of other candidates.
In such a scenario, CSP proteins would represent a conserved backbone in distinct AM and RLS signaling pathways that legumes localize in different cell types: atrichoblasts or root hairs, respectively. Symbiont-specific signal transduction and downstream responses would rather depend on the specific subsets of proteins that act in association with CSP members in each cell, as hypothesized in the schemes of Figure 1C.
The history of life is rich in examples of so-called evolutionary tinkering: processes in evolution where new functions are obtained through small modifications of a pre-existing biological mechanism. Evolutionary tinkering consists of two opposite processes: on the one hand, gene duplication and neo-functionalization produce new proteins for the novel functions; on the other hand, all those genes that play the same function in both conditions are conserved. The hypothesis that RLS has evolved by redirecting AM responses toward bacterial accommodation is widely accepted and explains the numerous similarities that exist between these two interactions (Bonfante and Genre, 2008; Parniske, 2008). In particular, several features of plant cell restructuring (e.g., symbiotic interface biogenesis) are strikingly similar (Parniske, 2008). In this context, conserved genes should be much more numerous than just the few currently listed in the CSP; not surprisingly, common symbiotic genes have already been identified which do not fit into the pathway. To mention just a few examples, VAPYRIN (Pumplin et al., 2010) is a partially characterized protein featuring a Major Sperm Protein domain and several ankyrin repeats, likely involved in membrane dynamics; CERBERUS (Yano et al., 2009) is an E3 ubiquitin ligase. Both are required for symbiont accommodation, but more likely in cellular remodeling and interface development than in signaling. On the same line, a group of SNARE proteins belonging to the VAMP72 family has been involved in symbiotic interface assembly for both interactions (Ivanov et al., 2012). It is reasonable to conclude that CSP proteins belong to this array of conserved genes, and act in a complex mix of common and interaction-specific processes, required for the establishment of each symbiosis.
Further indications may come from detailed analyses of AM phenotypes in available mutants. The legume transcription factor NSP1 was originally described as indispensable for rhizobial, but not fungal, colonization (Catoira et al., 2000; Smit et al., 2005). Nevertheless, a recent study showed that nsp1 mutation significantly slows down AM infection (Delaux et al., 2013a). Similarly, a partial involvement of Nod factor receptors NFR1 and NFR5 has been described in the induction of common symbiotic responses such as root branching and gene regulation (Maillet et al., 2011; Czaja et al., 2012; Zhang et al., 2015). Such studies suggest that fine phenotypic analyses, reaching cellular and molecular detail, can be crucial for completing the picture.
In conclusion, the CSP lives on as a precious genetic reference in our simplistic models of plant-microbe signaling. Nevertheless, we have sufficient clues to suspect the existence of a more complex scenario of CSP protein localization and activity. Studying symbiotic signaling in non-legumes appears today as a very promising approach to address such questions (Watts-Williams and Cavagnaro, 2015): working on a biological system that intrinsically excludes the cross-talk of two evolutionarily related interactions such as RLS and AM will deliver crucial knowledge that can then be applied to decipher the multiple symbiotic system of legumes.
AG conceived the general layout of the manuscript and was primarily involved in text writing and figure preparation. GR contributed to text writing, and gave a major contribution to literature search and figure elaboration. Both AG and GR contributed to critical literature reviewing and model elaboration.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are very grateful to David Barker and Mireille Chabaud for fruitful discussion; to Paola Bonfante for her constructive criticism of the manuscript; and to Dan Chamberlain for his revision of the English language.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00096
FIGURE S1 | The role of CSP proteins in different plant interactions. CSP and CSP-related proteins have been shown to play several roles beyond RLS and AM. SYMRK is required for the establishment of actinorhizal symbiosis and the parasitic interaction with root knot nematodes; furthermore it has been implicated in host defense against root colonization by parasitic plants. CCaMK is also necessary for actinorhizal symbiosis and defense responses to parasitic plants; in addition, it has a role in responses to pathogenic fungi. Beside CSP proteins, a few receptors that act upstream of the CSP have also been assigned with multiple roles. Nod factor receptors NFR1 and NFR5 are required for the establishment of actinorhizas and root knot nematode parasitism, and both have also been assigned a limited role in AM signaling. Finally, the chitin receptor CERK1 is required for both defense responses to pathogenic fungi and the accommodation of AM fungi. Overall, a survey of the literature data suggests that CSP and CSP-related genes have a diverse spectrum of functions in several plant interactions.
Ané, J. M., Kiss, G. B., Riely, B. K., Penmetsa, R. V., Oldroyd, G. E. D., Ayax, C., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303, 1364–1367. doi: 10.1126/science.1092986
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Bonfante, P., and Genre, A. (2008). Plants and arbuscular mycorrhizal fungi: an evolutionary-developmental perspective. Trends Plant Sci. 13, 492–498. doi: 10.1016/j.tplants.2008.07.001
Bonfante, P., and Requena, N. (2011). Dating in the dark: how roots respond to fungal signals to establish arbuscular mycorrhizal symbiosis. Curr. Opin. Plant Biol. 14, 451–457. doi: 10.1016/j.pbi.2011.03.014
Broghammer, A., Krusell, L., Blaise, M., Sauer, J., Sullivan, J. T., Maolanon, N., et al. (2012). Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. U.S.A. 109, 13859–13864. doi: 10.1073/pnas.1205171109
Calcagno, C., Novero, M., Genre, A., Bonfante, P., and Lanfranco, L. (2012). The exudate from an arbuscular mycorrhizal fungus induces nitric oxide accumulation in Medicago truncatula roots. Mycorrhiza 22, 259–269. doi: 10.1007/s00572-011-0400-4
Capoen, W., Sun, J., Wysham, D., Otegui, M. S., Venkateshwaran, M., Hirsch, S., et al. (2011). Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. U.S.A. 108, 14348–14353. doi: 10.1073/pnas.1107912108
Catoira, R., Galera, C., de Billy, F., Penmetsa, R. V., Journet, E. P., Maillet, F., et al. (2000). Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12, 1647–1666. doi: 10.1105/tpc.12.9.1647
Chabaud, M., Genre, A., Sieberer, B. J., Faccio, A., Fournier, J., Novero, M., et al. (2011). Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytol. 189, 347–355. doi: 10.1111/j.1469-8137.2010.03464.x
Chabaud, M., Gherbi, H., Pirolles, E., Vaissayre, V., Fournier, J., Moukouanga, D., et al. (2016). Chitinase-resistant hydrophilic symbiotic factors secreted by Frankia activate both Ca2+ spiking and NIN gene expression in the actinorhizal plant Casuarina glauca. New Phytol. 209, 86–93. doi: 10.1111/nph.13732
Charpentier, M., Bredemeier, R., Wanner, G., Takeda, N., Schleiff, E., and Parniske, M. (2008). Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20, 3467–3479. doi: 10.1105/tpc.108.063255
Chen, T., Zhu, H., Ke, D., Cai, K., Wang, C., Gou, H., et al. (2012). A MAP kinase kinase interacts with SymRK and regulates nodule organogenesis in Lotus japonicus. Plant Cell 24, 823–838. doi: 10.1105/tpc.112.095984
Czaja, L. F., Hogekamp, C., Lamm, P., Maillet, F., Martinez, E. A., Samain, E., et al. (2012). Transcriptional responses toward diffusible signals from symbiotic microbes reveal MtNFP- and MtDMI3-dependent reprogramming of host gene expression by arbuscular mycorrhizal fungal lipo-chitooligosaccharides. Plant Physiol. 159, 1671–1685. doi: 10.1104/pp.112.195990
Delaux, P. M., Bécard, G., and Combier, J. P. (2013a). NSP1 is a component of the Myc signaling pathway. New Phytol. 199, 59–65. doi: 10.1111/nph.12340
Delaux, P. M., Séjalon-Delmas, N., Bécard, G., and Ané, J. M. (2013b). Evolution of the plant-microbe symbiotic “toolkit.” Trends Plant Sci. 18, 298–304. doi: 10.1016/j.tplants.2013.01.008
Delaux, P. M., Radhakrishnan, G. V., Jayaraman, D., Cheema, J., Malbreil, M., Volkening, J. D., et al. (2015). Algal ancestor of land plants was preadapted for symbiosis. Proc. Natl. Acad. Sci. U.S.A 112, 13390–13395. doi: 10.1073/pnas.1515426112
Ehrhardt, D. W., Wais, R., and Long, S. R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85, 673–681. doi: 10.1016/S0092-8674(00)81234-9
Engstrom, E. M., Ehrhardt, D. W., Mitra, R. M., and Long, S. R. (2002). Pharmacological analysis of nod factor-induced calcium spiking in Medicago truncatula. Evidence for the requirement of type IIA calcium pumps and phosphoinositide signaling. Plant Physiol. 128, 1390–1401. doi: 10.1104/pp.010691
Evans, D. E., Hutchison, C. J., and Bryant, J. A. (2004). The Nuclear Envelope. Oxford: Taylor and Francis Bios.
Fernández-Aparicio, M., Rispail, N., Prats, E., Morandi, D., García-Garrido, J. M., Dumas-Gaudot, E., et al. (2009). Parasitic plant infection is partially controlled through the symbiotic pathways. Weed Res. 50, 76–82. doi: 10.1111/j.1365-3180.2009.00749.x
Field, K. J., Pressel, S., Duckett, J. G., Rimington, W. R., and Bidartondo, M. I. (2015). Symbiotic options for the conquest of land. Trends Ecol. Evol. 30, 477–486. doi: 10.1016/j.tree.2015.05.007
Fournier, J., Teillet, A., Chabaud, M., Ivanov, S., Genre, A., Limpens, E., et al. (2015). Remodeling of the infection chamber prior to infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol. 167, 1233–1242. doi: 10.1104/pp.114.253302
Fournier, J., Timmers, A. C. J., Sieberer, B. J., Jauneau, A., Chabaud, M., and Barker, D. G. (2008). Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol. 148, 1985–1995. doi: 10.1104/pp.108.125674
Gage, D. J. (2004). Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68, 280–300. doi: 10.1128/MMBR.68.2.280-300.2004
Genre, A., Chabaud, M., Balzergue, C., Puech-Pagès, V., Novero, M., Rey, T., et al. (2013). Short-chain chitin oligomers from arbus- cular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 198, 190–202. doi: 10.1111/nph.12146
Genre, A., Chabaud, M., Faccio, A., Barker, D. G., and Bonfante, P. (2008). Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungus within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20, 1407–1420. doi: 10.1105/tpc.108.059014
Genre, A., Chabaud, M., Timmers, T., Bonfante, P., and Barker, D. G. (2005). Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17, 3489–3499. doi: 10.1105/tpc.105.035410
Genre, A., Ortu, G., Bertoldo, C., Martino, E., and Bonfante, P. (2009). Biotic and abiotic stimulation of root epidermal cells reveals common and specific responses to arbuscular mycorrhizal fungi. Plant Physiol. 149, 1424–1434. doi: 10.1104/pp.108.132225
Gherbi, H., Markmann, K., Svistoonoff, S., Estevan, J., Autran, D., Giczey, G., et al. (2008). SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankia bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 4928–4932. doi: 10.1073/pnas.0710618105
Giovannetti, M., Mari, A., Novero, M., and Bonfante, P. (2015). Early Lotus japonicus root transcriptomic responses to symbiotic and pathogenic fungal exudates. Front. Plant Sci. 6:480. doi: 10.3389/fpls.2015.00480
Gobbato, E. (2015). Recent developments in arbuscular mycorrhizal signaling. Curr. Opin. Plant Biol. 26, 1–7. doi: 10.1016/j.pbi.2015.05.006
Gobbato, E., Marsh, J. F., Vernié, T., Wang, E., Maillet, F., Kim, J., et al. (2012). A GRAS-type transcription factor with a specific function in mycorrhizal signalling. Curr. Biol. 22, 2236–2241. doi: 10.1016/j.cub.2012.09.044
Gobbato, E., Wang, E., Higgins, G., Bano, S. A., Henry, C., Schultze, M., et al. (2013). RAM1 and RAM2 function and expression during arbuscular mycorrhizal symbiosis and Aphanomyces euteiches colonization. Plant Signal. Behav. 8:e26049. doi: 10.4161/psb.26049
Granqvist, E., Sun, J., den Camp, R., Pujic, P., Hill, L., Normand, P., et al. (2015). Bacterial-induced calcium oscillations are common to nitrogen-fixing associations of nodulating legumes and nonlegumes. New Phytol. 207, 551–558. doi: 10.1111/nph.13464
Groth, M., Takeda, N., Perry, J., Uchida, H., Dräxl, S., Brachmann, A., et al. (2010). NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22, 2509–2526. doi: 10.1105/tpc.109.069807
Gutjahr, C., Banba, M., Croset, V., An, K., Miyao, A., An, G., et al. (2008). Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20, 2989–3005. doi: 10.1105/tpc.108.062414
Gutjahr, C., Casieri, L., and Paszkowski, U. (2009). Glomus intraradices induces changes in root system architecture of rice independently of common symbiosis signaling. New Phytol. 182, 829–837. doi: 10.1111/j.1469-8137.2009.02839.x
Gutjahr, C., and Parniske, M. (2013). Cell and developmental biology of arbuscular mycorrhiza symbiosis. Ann. Rev. Cell Dev. Biol. 29, 593–617. doi: 10.1146/annurev-cellbio-101512-122413
Huisman, R., Bouwmeester, K., Brattinga, M., Govers, F., Bisseling, T., and Limpens, E. (2015). Haustorium formation in Medicago truncatula roots infected by Phytophthora palmivora does not involve the common endosymbiotic program shared by AM fungi and rhizobia. Mol. Plant Microbe Interact. 28, 1271–1280. doi: 10.1094/MPMI-06-15-0130-R
Ivanov, S., Fedorova, E. E., Limpens, E., De Mita, S., Genre, A., Bonfante, P., et al. (2012). Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc. Natl. Acad. Sci. U.S.A. 109, 8316–8321. doi: 10.1073/pnas.1200407109
Kaló, P., Gleason, C., Edwards, A., Marsh, J., Mitra, R. M., Hirsch, S., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308, 1786–1789. doi: 10.1126/science.1110951
Kanamori, N., Madsen, L. H., Radutoiu, S., Frantescu, M., Quistgaard, E. M., Miwa, H., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 103, 359–364. doi: 10.1073/pnas.0508883103
Kevei, Z., Lougnon, G., Mergaert, P., Horvath, G. V., Kereszt, A., Jayaraman, D., et al. (2007). 3-hydroxy-3-methylglutaryl coenzyme A reductase1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell 19, 3974–3989. doi: 10.1105/tpc.107.053975
Kistner, C., Winzer, T., Pitzschke, A., Mulder, L., Sato, S., Kaneko, T., et al. (2005). Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17, 2217–2229. doi: 10.1105/tpc.105.032714
Kosuta, S., Hazledine, S., Sun, J., Miwa, H., Morris, R. J., Downie, J. A., et al. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. U.S.A. 105, 9823–9828. doi: 10.1073/pnas.0803499105
Lauressergues, D., Delaux, P.-M., Formey, D., Lelandais-Brière, C., Fort, S., Cottaz, S., et al. (2012). The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting SP2. Plant J. 72, 512–522. doi: 10.1111/j.1365-313X.2012.05099.x
Lefebvre, B., Timmers, T., Mbengue, M., Moreau, S., Hervé, C., Tóth, K., et al. (2010). A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 107, 2343–2348. doi: 10.1073/pnas.0913320107
Levy, J., Bres, C., Geurts, R., Chalhoub, B., Kulikova, O., Duc, G., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303, 1361–1364. doi: 10.1126/science.1093038
Liang, Y., Tóth, K., Cao, Y., Tanaka, K., Espinoza, C., and Stacey, G. (2014). Lipochitooligosaccharide recognition: an ancient story. New Phytol. 204, 289–296. doi: 10.1111/nph.12898
Limpens, E., van Zeijl, A., and Geurts, R. (2015). Lipochitooligosaccharides modulate plant host immunity to enable endosymbioses. Annu. Rev. Phytopathol. 53, 311–334. doi: 10.1146/annurev-phyto-080614-120149
Lin, K., Limpens, E., Zhang, Z., Ivanov, S., Saunders, D. G. O., Desheng, M., et al. (2014). Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet. 10:e1004078. doi: 10.1371/journal.pgen.1004078
Liu, T., Liu, Z., Song, C., Hu, Y., Han, Z., She, J., et al. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164. doi: 10.1126/science.1218867
Madsen, E. B., Madsen, L. H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., et al. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640. doi: 10.1038/nature02045
Maillet, F., Poinsot, V., Andre, O., Puech-Pages, V., Haouy, A., Gueunier, M., et al. (2011). Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469, 58–63. doi: 10.1038/nature09622
Meilhoc, E., Cam, Y., Skapski, A., and Bruand, C. (2010). The response to nitric oxide of the nitrogen-fixing symbiont Sinorhizobium meliloti. Mol. Plant Microbe Interact. 23, 748–759. doi: 10.1094/MPMI-23-6-0748
Miller, J. B., Pratap, A., Miyahara, A., Zhou, L., Bornemann, S., Morris, R. J., et al. (2013). Calcium/Calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell 25, 5053–5066. doi: 10.1105/tpc.113.116921
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Miyata, K., Kozaki, T., Kouzai, Y., Ozawa, K., Ishii, K., Asamizu, E., et al. (2014). The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol. 55, 1864–1872. doi: 10.1093/pcp/pcu129
Oldroyd, G. E. D. (2013). Speak, friend, and enter: signalling systems that promote bene?cial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Parniske, M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6, 763–775. doi: 10.1038/nrmicro1987
Pauly, N., Pucciariello, C., Mandon, K., Innocenti, G., Jamet, A., Baudouin, E., et al. (2006). Reactive oxygen and nitrogen species and glutathione: key players in the legume–Rhizobium symbiosis. J. Exp. Bot. 57, 1769–1776. doi: 10.1093/jxb/erj184
Poovaiah, B. W., Du, L., Wang, H., and Yang, T. (2013). Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant: microbe interactions. Plant Physiol. 163, 531–542. doi: 10.1104/pp.113.220780
Pumplin, N., Mondo, S. J., Topp, S., Starker, C. G., Gantt, J. S., and Harrison, M. J. (2010). Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 61, 482–494. doi: 10.1111/j.1365-313X.2009.04072.x
Rey, T., Chatterjee, A., Buttay, M., Toulotte, J., and Schornack, S. (2015). Medicago truncatula symbiosis mutants affected in the interaction with a biotrophic root pathogen. New Phytol. 206, 497–500. doi: 10.1111/nph.13233
Rey, T., Nars, A., Bonhomme, M., Bottin, A., Huguet, S., Balzergue, S., et al. (2013). NFP, a LysM protein controlling Nod factor perception, also intervenes in Medicago truncatula resistance to pathogens. New Phytol. 198, 875–886. doi: 10.1111/nph.12198
Rich, M. K., Schorderet, M., and Reinhardt, D. (2014). The role of the cell wall compartment in mutualistic symbioses of plants. Front. Plant. Sci. 5:238. doi: 10.3389/fpls.2014.00238
Ried, M. K., Antolín-Llovera, M., and Parniske, M. (2014). Spontaneous symbiotic reprogramming of plant roots triggered by receptor-like kinases. eLife 3:e03891. doi: 10.7554/eLife.03891
Robledo, M., Jiménez-Zurdo, J., Velázquez, E., Trujillo, M. E., Zurdo-Piñeiro, J. L., Ramírez-Bahena, M. H., et al. (2008). Rhizobium cellulase CelC2 is essential for primary symbiotic infection of legume host roots. Proc. Natl. Acad. Sci. U.S.A. 105, 7064–7069. doi: 10.1073/pnas.0802547105
Russo, G., Spinella, S., Sciacca, E., Bonfante, P., and Genre, A. (2013). Automated analysis of calcium spiking profiles with CaSA software: two case studies from root-microbe symbioses. BMC Plant Biol. 13:224. doi: 10.1186/1471-2229-13-224
Saito, K., Yoshikawa, M., Yano, K., Miwa, H., Uchida, H., Asamizu, E., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19, 610–624. doi: 10.1105/tpc.106.046938
Salzer, P., Corbiere, H., and Boller, T. (1999). Hydrogen peroxide accumulation in Medicago truncatula roots colonized by the arbuscular mycorrhiza-forming fungus Glomus intraradices. Planta 208, 319–325. doi: 10.1007/s004250050565
Sanchez, L., Weidmann, S., Arnould, C., Bernard, A. R., Gianinazzi, S., and Gianinazzi-Pearson, V. (2005). Pseudomonas fluorescens and Glomus mosseae trigger DMI3-dependent activation of genes related to a signal transduction pathway in roots of Medicago truncatula. Plant Physiol. 139, 1065–1077. doi: 10.1104/pp.105.067603
Sánchez-Vallet, A., Mesters, J. R., and Thomma, B. P. H. J. (2015). The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol. Rev. 39, 171–183. doi: 10.1093/femsre/fuu003
Sandal, N., Petersen, T. R., Murray, J., Umehara, Y., Karas, B., Yano, K., et al. (2006). Genetics of symbiosis in Lotus japonicus: recombinant inbred lines, comparative genetic maps, and map position of 35 symbiotic loci. Mol. Plant Microbe Interact. 19, 80–91. doi: 10.1094/MPMI-19-0080
Schmitz, A. M., and Harrison, M. J. (2014). Signalling events during initiation of arbuscular mycorrhizal symbiosis. J. Integr. Plant Biol. 56, 250–261. doi: 10.1111/jipb.12155
Shimizu, T., Nakano, T., Takamizawa, D., Desaki, Y., Ishii-Minami, N., Nishizawa, Y., et al. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. doi: 10.1111/j.1365-313X.2010.04324.x
Shimoda, Y., Han, L., Yamazaki, T., Suzuki, R., Hayashi, M., and Imaizumi-Anraku, H. (2012). Rhizobial and fungal symbioses show different requirements for calmodulin binding to calcium calmodulin-dependent protein kinase in Lotus japonicus. Plant Cell 24, 304–321. doi: 10.1105/tpc.111.092197
Shinya, T., Nakagawa, T., Kaku, H., and Shibuya, N. (2015). Chitin-mediated plant–fungal hiding and handshaking. Curr. Opin. Plant Biol. 26, 64–71. doi: 10.1016/j.pbi.2015.05.032
Sieberer, B. J., Chabaud, M., Fournier, J., Timmers, A. C., and Barker, D. G. (2012). A switch in Ca2+ spiking signature is concomitant with endosymbiotic microbe entry into cortical root cells of Medicago truncatula. Plant J. 69, 822–830. doi: 10.1111/j.1365-313X.2011.04834.x
Singh, S., Katzer, K., Lambert, J., Cerri, M., and Parniske, M. (2014). CYCLOPS, A DNA-Binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15, 139–152. doi: 10.1016/j.chom.2014.01.011
Smit, P., Raedts, J., Portyanko, V., Debellé, F., Gough, C., Bisseling, T., et al. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308, 1789–1791. doi: 10.1126/science.1111025
Stougaard, J. (2001). Genetics and genomics of root symbiosis. Curr. Opin. Plant Biol. 4, 328–335. doi: 10.1016/S1369-5266(00)00181-3
Sun, J., Miller, J. B., Granqvist, E., Wiley-Kalil, A., Gobbato, E., Maillet, F., et al. (2015). Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell 27, 823–838. doi: 10.1105/tpc.114.131326
Svistoonoff, S., Benabdoun, F. M., Nambiar-Veetil, M., Imanishi, L., Vaissayre, V., Cesari, S., et al. (2013). The independent acquisition of plant root nitrogen-fixing symbiosis in Fabids recruited the same genetic pathway for nodule organogenesis. PLoS ONE 8:e64515. doi: 10.1371/journal.pone.0064515
Tisserant, E., Malbreil, M., Kuo, A., Kohlera, A., Symeonidid, A., Balestrini, R., et al. (2013). Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Acad. Sci. U.S.A. 110, 20117–20122. doi: 10.1073/pnas.1313452110
Venkateshwaran, M., Cosme, A., Han, L., Banba, M., Satyshur, K. A., Schleiff, E., et al. (2012). The recent evolution of a symbiotic ion channel in the legume family altered ion conductance and improved functionality in calcium signaling. Plant Cell 24, 2528–2545. doi: 10.1105/tpc.112.098475
Venkateshwaran, M., Jayaraman, D., Chabaud, M., Genre, A., Balloon, A. J., Maeda, J., et al. (2015). A role for the mevalonate pathway in early plant symbiotic signaling. Proc. Natl. Acad. Sci. U.S.A. 112, 9781–9786. doi: 10.1073/pnas.1413762112
Wan, J., Zhang, X.-C., and Stacey, G. (2008). Chitin signaling and plant disease resistance. Plant Signal. Behav. 3, 831–833. doi: 10.4161/psb.3.10.5916
Wang, B., Yeun, L. H., Xue, J. Y., Liu, Y., Ané, J.-M., and Qiu, Y. L. (2010). Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants. New Phytol. 186, 514–525. doi: 10.1111/j.1469-8137.2009.03137.x
Watts-Williams, S. J., and Cavagnaro, T. R. (2015). Using mycorrhiza-defective mutant genotypes of non-legume plant species to study the formation and functioning of arbuscular mycorrhiza: a review. Mycorrhiza 25, 587–597. doi: 10.1007/s00572-015-0639-2
Weerasinghe, R. R., Bird, D., and Allen, N. S. (2005). Root-knot nematodes and bacterial Nod factors elicit common signal transduction events in Lotus japonicus. Proc. Natl Acad. Sci. U.S.A. 102, 3147–3152. doi: 10.1073/pnas.0407926102
Yano, K., Shibata, S., Chen, W. L., Sato, S., Kaneko, T., Jurkiewicz, A., et al. (2009). CERBERUS, a novel U-box protein containing WD-40 repeats, is required for formation of the infection thread and nodule development in the legume-Rhizobium symbiosis. Plant J. 60, 168–180. doi: 10.1111/j.1365-313X.2009.03943.x
Yano, K., Yoshida, S., Muller, J., Singh, S., Banba, M., Vickers, K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. U.S.A. 105, 20540–20545. doi: 10.1073/pnas.0806858105
Zhang, R.-Q., Zhu, H.-H., Zhao, H.-Q., and Yao, Q. (2013). Arbuscular mycorrhizal fungal inoculation increases phenolic synthesis in clover roots via hydrogen peroxide, salicylic acid and nitric oxide signaling pathways. J. Plant Physiol. 170, 74–79. doi: 10.1016/j.jplph.2012.08.022
Zhang, X., Dong, W., Sun, J., Feng, F., Deng, Y., He, Z., et al. (2015). The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 81, 258–267. doi: 10.1111/tpj.12723
Keywords: plant–microbe interactions, symbiosis, arbuscular mycorrhiza, legume nodulation, signaling pathways
Citation: Genre A and Russo G (2016) Does a Common Pathway Transduce Symbiotic Signals in Plant–Microbe Interactions? Front. Plant Sci. 7:96. doi: 10.3389/fpls.2016.00096
Received: 02 November 2015; Accepted: 18 January 2016;
Published: 16 February 2016.
Edited by:
Pietro Daniele Spanu, Imperial College London, UKReviewed by:
Christian Staehelin, Sun Yat-sen University, ChinaCopyright © 2016 Genre and Russo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Genre, YW5kcmVhLmdlbnJlQHVuaXRvLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.