
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 09 February 2016
Sec. Plant Biotechnology
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.00067
This article is part of the Research Topic Mechanisms of abiotic stress responses and tolerance in plants: physiological, biochemical and molecular interventions View all 121 articles
Agricultural production and quality are adversely affected by various abiotic stresses worldwide and this will be exacerbated by the deterioration of global climate. To feed a growing world population, it is very urgent to breed stress-tolerant crops with higher yields and improved qualities against multiple environmental stresses. Since conventional breeding approaches had marginal success due to the complexity of stress tolerance traits, the transgenic approach is now being popularly used to breed stress-tolerant crops. So identifying and characterizing the critical genes involved in plant stress responses is an essential prerequisite for engineering stress-tolerant crops. Far beyond the manipulation of single functional gene, engineering certain regulatory genes has emerged as an effective strategy now for controlling the expression of many stress-responsive genes. Transcription factors (TFs) are good candidates for genetic engineering to breed stress-tolerant crop because of their role as master regulators of many stress-responsive genes. Many TFs belonging to families AP2/EREBP, MYB, WRKY, NAC, bZIP have been found to be involved in various abiotic stresses and some TF genes have also been engineered to improve stress tolerance in model and crop plants. In this review, we take five large families of TFs as examples and review the recent progress of TFs involved in plant abiotic stress responses and their potential utilization to improve multiple stress tolerance of crops in the field conditions.
Agricultural production and quality are adversely affected by a broad range of abiotic stresses including drought, salinity, heat, and cold. Especially when these stresses occur in combination, it can have devastating effects on plant growth and productivity. It is estimated that more than 50% of worldwide yield loss for major crop are caused by abiotic stresses (Shao et al., 2009; Ahuja et al., 2010; Lobell et al., 2011). According to the current climate prediction models, the deterioration of global climate will inevitably cause an increased frequency of drought, heat wave, and salinization (Easterling et al., 2000; Ipcc, 2007, 2008). This means that agricultural productivity will face a greater challenge in fighting against environmental stresses. Meanwhile, the growing world population will reach close to ten billion by the year 2050 and then almost two times of current agricultural productivity is needed to feed the large population (Bengtsson et al., 2006; United Nations, 2015). Moreover, such a tremendous increase of crop productivity must be achieved with no increase in arable land and in the face of multiple environmental stresses. Where is the way to solve this problem? Many scholars and experts worldwide have reached a consensus that breeding stress-tolerant crops with higher yields and improved qualities against multiple environmental stresses is an effective strategy, as well as one of the greatest challenges faced by modern agriculture (Takeda and Matsuoka, 2008; Newton et al., 2011; Liu J.-H. et al., 2014). In the past few decades, a great deal of efforts has been devoted to breeding crop cultivars with various stress-tolerant traits. Two main approaches have been employed to this process. One is traditional breeding methods such as wide-cross hybridization and mutation breeding, which often brings about unpredictable results. Another is modern transgenic technology by introducing novel exogenous genes or altering the expression levels of endogenous genes to improve stress tolerance. Since conventional breeding approaches have marginal success due to the complexity of stress tolerance traits, the transgenic approach is now being popularly used to develop transgenic crops tolerant to abiotic stresses (Yamaguchi and Blumwald, 2005). Therefore, deciphering the molecular mechanisms by which plants perceive and transduce stress signals to cellular machinery to initiate adaptive responses is an essential prerequisite for identification of the key genes and pathways to engineer stress-tolerant crop plants (Ray et al., 2009; Heidarvand and Amiri, 2010; Sanchez et al., 2011).
Substantial progress has been made to unravel the molecular mechanisms of abiotic stress responses in plants by means of high throughput sequencing and functional genomics tools. To date, a number of critical genes involved in abiotic stress tolerance have been identified and validated, which are generally classified into two types: functional genes and regulatory genes (Shinozaki et al., 2003). The former encodes important enzymes and metabolic proteins (functional proteins), such as detoxification enzyme, water channel, ion transporter, heat shock protein (HSP), and late embryogenesis abundant (LEA) protein, which directly function to protect cells from stresses. The latter encodes various regulatory proteins including TFs, protein kinases and protein phosphatases, which regulate signal transduction and gene expression in the stress responses. Although there have been numerous studies on functional genes, most of these studies pay more attention to single gene or several genes encoding enzymes and protective proteins by imposing a given stress. Due to the complexity of stress responses regulated by multi-genes, little success has been achieved by a single functional gene approach to significantly enhance plant stress tolerance (Mittler and Blumwald, 2010; Varshney et al., 2011). Given the complexity and variability of field conditions where crops are often simultaneously subjected to multiple abiotic stresses or some in combination (Ahuja et al., 2010), more and more studies have paid close attention to regulatory genes and found that some regulatory genes including TFs play essential roles in multiple abotic stress responses by regulating a large spectrum of downstream stress-responsive genes. Thus, genetically modifying the expression of certain regulatory genes can greatly influence plant stress tolerance because it mimics or enhances stress signals to regulate many downstream stress-responsive genes at a time (Century et al., 2008; Yang et al., 2011). Among the regulatory genes, stress-responsive TFs have attracted particular attention due to their important roles in plant stress responses (Chen and Zhu, 2004; Xu et al., 2008a). In this paper, we mainly review the recent progress of TFs involved in plant abiotic stress responses and their potential utilization to improve multiple stress tolerance of crops in the field conditions.
As sessile organisms, plants have evolved various defense mechanisms at multiple levels to respond to unfavorable environment including diverse abiotic stresses. As stated before, it is imperative to dissect regulatory mechanisms of stress response and identify the key regulators involved in this process to breed or genetically engineer stress-tolerant plants. With the availability of plant genomes and various omics tools including genomics, transcriptomics, and proteomics tools, major progress has been made in deciphering the stress signaling pathways and relevant components involved in plant abiotic stress response, but there is still much more to be determined (Liu J.-H. et al., 2014). According to our current knowledge about stress signaling pathways, the generic signaling pathway for any given abiotic stress can be divided into the following major steps: signal perception, signal transduction, stress responsive gene expression, in turn the activation of physiological, and metabolic responses (Chaves et al., 2003; Yamaguchi-Shinozaki and Shinozaki, 2006; Pérez-Clemente et al., 2013). In this process, plant cells first perceive stress stimulus through sensors or receptors located in the cell wall or membrane. Then the captured extracellular signals are converted into intracellular ones through second messengers including calcium ions, inositol phosphate, reactive oxygen species (ROS), cyclic nucleotides (cAMP and cGMP), sugars, and nitric oxide. Subsequently, these second messengers initiate the corresponding signaling pathways to transduce the signals (Chaves et al., 2009; Bhargava and Sawant, 2013). In many signal transduction pathways, the phosphorylation, and dephosphorylation of proteins mediated by protein kinase and phosphatases, respectively, is an important and effective mechanism for signal relay (Singh et al., 2003). For example, the mitogen activated protein kinases (MAPKs) pathway and calcium-dependent protein kinases (CDPKs) pathway are known to be involved in plant abiotic stress responses (Schaller et al., 2008; Huang G.T. et al., 2012). At the end of the phosphorylation cascade, TFs are activated or suppressed by protein kinases or phosphatases, and they further bind specifically to cis-elements in the promoters of stress-responsive genes and regulate their transcription (Danquah et al., 2014). Meanwhile, TFs themselves are regulated at the transcription level by other upstream components (Hirayama and Shinozaki, 2010) and also subjected to various tiers of modifications at the post-transcription level, such as ubiquitination and sumoylation, thus forming a complex regulatory network to modulate the expression of stress responsive genes, which in turn determine the activation of physiological and metabolic responses (Dong et al., 2006; Miura et al., 2007; Mizoi et al., 2013). All the components mentioned above, from the foremost receptors to the downstream functional genes, constitute the generic pathway for plant abiotic stress signal transduction (Figure 1). As one of the most important regulators, TFs function as terminal transducers and directly regulate the expression of an array of downstream genes by interacting with the specific cis-elements in their promoter region (Yamaguchi-Shinozaki and Shinozaki, 2006). In the last few decades, considerable research has been conducted to identify and characterize various TFs involved in plant abiotic stress responses either in abscisic acid (ABA)-dependent pathway or ABA-independent pathway, such as AP2/EREBP, MYB, WRKY, NAC, bZIP, and so on (Vinocur and Altman, 2005; Umezawa et al., 2006; Golldack et al., 2011). Numerous efforts have been also made to improve plant stress tolerance by engineering these TF genes, and some promising results have been reported in succession (Table 1). In the following sections, we mainly summarize current information on several major TF families including their features, roles, and biotechnological uses for improving the abiotic stress tolerance in plants.
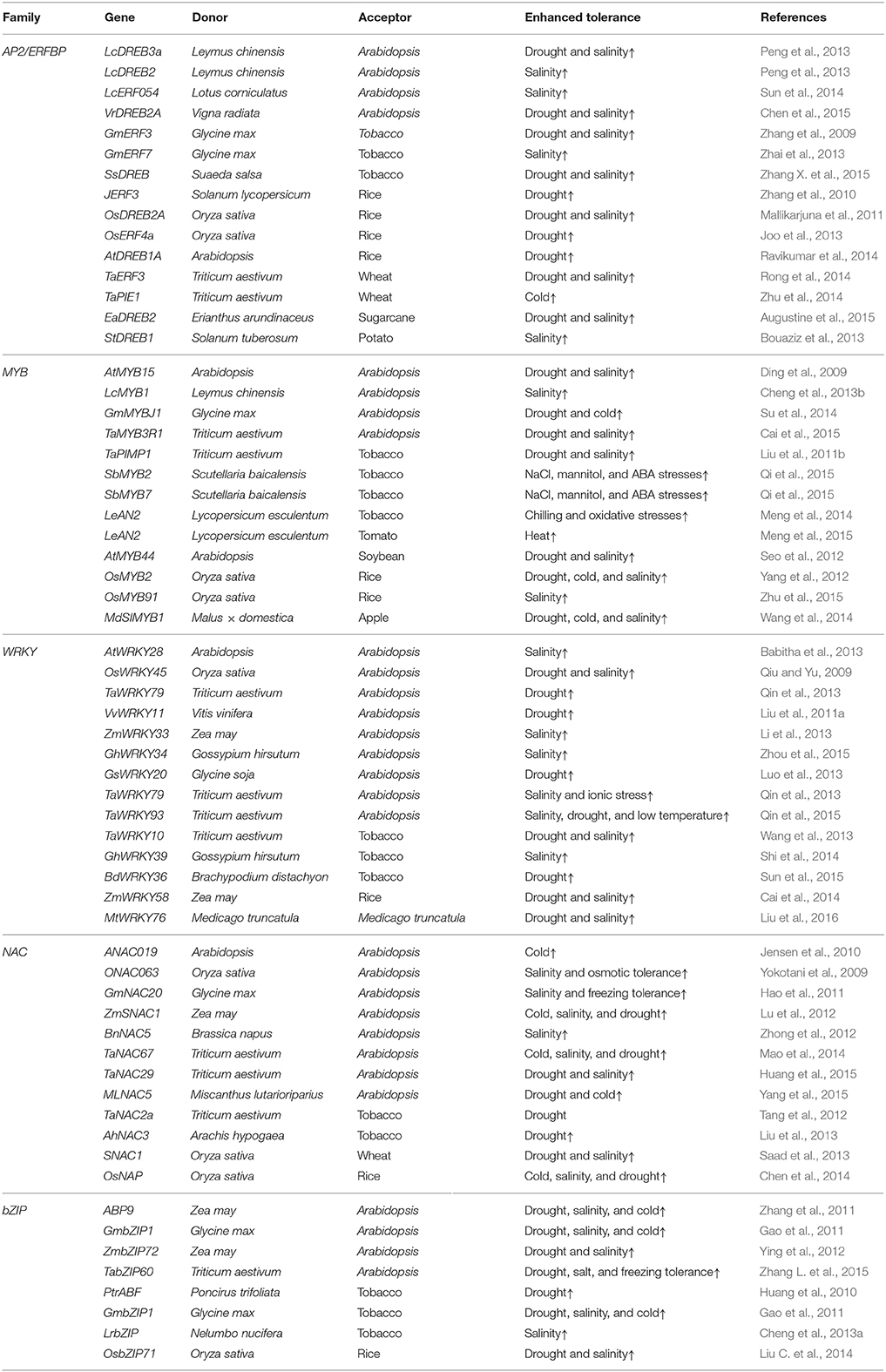
Table 1. Some examples of transgenic plants over-expressing transcription factor genes in recent years.
AP2/ERFBP family includes a large group of plant-specific TFs and is characterized by the presence of the highly conserved AP2/ethylene-responsive element-binding factor (ERF) DNA-binding domain that directly interact with GCC box and/or dehydration-responsive element (DRE)/C-repeat element (CRT) cis-acting elements at the promoter of downstream target genes (Riechmann and Meyerowitz, 1998). AP2/ERFBP TFs perform a variety of roles in plant developmental processes and stress responses, such as vegetative and reproductive development, cell proliferation, abiotic and biotic stress responses, and plant hormone responses (Nakano et al., 2006; Licausi et al., 2010; Sharoni et al., 2011). Presently, a multitude of AP2/ERFBP members have been identified in various species by means of genome-wide analysis, such as 145 in Arabidopsis (Riechmann and Meyerowitz, 1998), 163 in rice (Sharoni et al., 2011), 200 in poplar (Zhuang et al., 2008), 291 in Chinese cabbage (Song et al., 2013), 171 in foxtail millet (Lata et al., 2014), 116 in moso bamboo (Wu et al., 2015). Based on the number and similarity of AP2/ERF domains, these AP2/EREBP TFs are grouped into four major subfamilies: AP2 (Apetala 2), RAV (related to ABI3/VP1), DREB (dehydration-responsive element-binding protein), and ERF (Sakuma et al., 2002; Sharoni et al., 2011). Among these, both ERF and DREB subfamilies have been extensively studied due to their involvement in plant responses to biotic and abiotic stresses.
The DREB subfamily can regulate the expression of multiple dehydration/cold-regulated (RD/COR) genes by interacting with DRE/CRT cis-elements (A/GCCGAC) located in the promoters of RD/COR genes that are responsive to water deficit and low-temperature, such as COR15A, RD29A/COR78, and COR6.6 (Stockinger et al., 1997; Liu et al., 1998; Lucas et al., 2011). A lot of DREB-type TFs have been tested in many plants including Arabidopsis, wheat, tomato, soybean, rice, maize, and barley (Agarwal et al., 2006; Lata and Prasad, 2011; Mizoi et al., 2012). According to the variation in some conserved motifs and biological functions in divergent species, DREB TFs are further classified into two major subgroups: DREB1/C-repeat-binding factor (DREB1/CBF) and DREB2, and each of them is involved in separate signal transduction pathway under abiotic stresses (Dubouzet et al., 2003). Commonly, DREB1/CBF genes are involved in low temperature stress responses in Arabidopsis and rice, while DREB2 genes respond to dehydration, high salinity and heat shock (Liu et al., 1998; Sakuma et al., 2002; Lucas et al., 2011). For example, three major DREB1/CBF members in Arabidopsis, DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 are rapidly induced in response to cold stress (Stockinger et al., 1997; Gilmour et al., 1998; Liu et al., 1998; Shinwari et al., 1998). Over-expressing any one of these three DREB1s/CBFs displayed significantly improved tolerance to freezing, drought, and high salinity in transgenic Arabidopsis (Gilmour et al., 1998; Jaglo-Ottosen et al., 1998; Liu et al., 1998). Further, over-expressing Arabidopsis DREB1/CBF genes improved freezing tolerance in oilseed rape (Jaglo et al., 2001) and chilling tolerance in tomato, tobacco and rice (Tsai-Hung et al., 2002; Kasuga et al., 2004; Ito et al., 2006). Some DREB1/CBF homologous genes have also been isolated from many other plant species including tomato, oilseed rape, wheat, rye, rice, and maize, and some of them have been used to produce transgenic plants with improved tolerance to abiotic stress (Jaglo et al., 2001; Dubouzet et al., 2003; Qin et al., 2004). In contrast, DREB2 genes have been studied in a limited number of plant species, but the existing studies have shown that DREB2 genes are also involved in abiotic stress responses in plants. In Arabidopsis, DREB2A and DREB2B are major DREB2s induced by dehydration, high salinity, and heat, while DREB2C is induced by heat later than them (Liu et al., 1998; Nakashima et al., 2000; Sakuma et al., 2006b; Lim et al., 2007). Over-expression of the constitutively active form of AtDREB2A from Arabidopsis improved the tolerance to drought and osmotic stress in transgenic Arabidopsis plants (Sakuma et al., 2006a; Xu et al., 2008b). Over-expression of ZmDREB2A from maize enhanced drought tolerance in transgenic Arabidopsis plants (Qin et al., 2007). The transgenic plants harboring GmDREB2 from soybean also showed enhanced tolerance to drought and high salinity without any growth retardation (Chen et al., 2007), as was observed in transgenic rice by over-expressing OsDREB2A under control of stress-inducible RD29A promoter (Mallikarjuna et al., 2011).
The ERF subfamily is the largest group of the AP2/EREBP TF family (Dietz et al., 2010) and functions in plant stress tolerance by regulating the stress-responsive genes through interacting with the cis-element GCC boxes with core sequence of AGCCGCC (Ohme-Takagi and Shinshi, 1995; Hao et al., 1998). An array of ERF genes are induced by various abiotic stresses, such as drought, high salinity, osmotic stress, and cold (Xu et al., 2008a). Over-expression of these ERF genes resulted in improvement of abiotic stress tolerance in transgenic plants, as summarized in Table 1. It is worth mentioning that some ERFs function in both biotic and abiotic stress tolerance, and this is partly due to their involvement in various hormonal signaling pathways including ethylene, JA, or SA (Liang et al., 2008). For example, over-expressing TaPIE1 in wheat significantly enhanced resistance to both pathogen and freezing stress (Zhu et al., 2014). Over-expressing GmERF3 in tobacco not only enhanced resistance against infection by pathogen and tobacco mosaic virus (TMV) but also improved tolerance to high salinity and dehydration (Zhang et al., 2009). So far, functions of a limited number of ERFs have been well characterized, but most of ERF family members have yet to be identified.
The MYB TFs are widely distributed in plants and form a large family characterized by a highly conserved MYB domain for DNA-binding, which contains from 1 to 4 imperfect repeats (MYB repeat) at the N-terminus. In contrast, the activation domain is located at the C-terminus and varies significantly among MYBs, leading to versatile regulatory roles of MYB family. According to the number of MYB domain repeats, the MYB TFs are divided into four groups: 1R-MYB (MYB-related type), R2R3-MYB, R1R2R3-MYB, and 4R-MYB, containing one, two, three, and four MYB repeats, respectively. Among them, the R2R3-MYBs are more prevalent in plants (Dubos et al., 2010; Ambawat et al., 2013; Li et al., 2015). So far, large numbers of MYB members have been identified in different plant species, such as 198 in Arabidopsis (Yanhui et al., 2006), 183 in rice (Yanhui et al., 2006), 229 in apple (Cao et al., 2013), 177 in sweet orange (Hou et al., 2014), 209 in foxtail millet (Muthamilarasan et al., 2014).
Numerous MYB TFs have been found to function in many significant physiological and biochemical processes including cell development and cell cycle, primary and secondary metabolism, hormone synthesis and signal transduction, as well as in plant responses to various biotic and abiotic stresses (Allan et al., 2008; Dubos et al., 2010; Ambawat et al., 2013). Recently, some abiotic stress-responsive MYB TFs in Arabidopsis and other plants have been well summarized by Li (Li et al., 2015). For example, AtMYB15 improved freezing tolerance by regulating CBF expression (Agarwal et al., 2006); AtMYB44, AtMYB60, and AtMYB61 improved drought tolerance by regulating stomatal movement (Cominelli et al., 2005; Liang et al., 2005; Jung et al., 2008). Especially, AtMYB96 improved drought tolerance either by integrating ABA and auxin signals (Seo et al., 2009) or by activating cuticular wax biosynthesis (Seo et al., 2011), and also improved freezing and drought tolerance by regulating a lipid-transfer protein LTP3. This fact shows that a MYB factor can regulate diverse target genes involved in various physiological processes under abiotic stresses. In addition, OsMYB2 from rice was induced by salt, cold, and dehydration stress. The transgenic plants with over-expressing OsMYB2 exhibited enhanced tolerance to various stresses by the alteration of expression levels of numerous genes involving diverse functions in stress response (Yang et al., 2012). Salt and freezing tolerance in Arabidopsis was significantly enhanced by over-expressing either GmMYB76 or GmMYB177 from soybean (Liao et al., 2008).
The WRKY family is also extensively distributed in plants and contains many members. WRKY TFs are characterized by the presence of one or two highly conserved WRKY domains of about 60 amino acid residues, which contains a conserved WRKYGQK motif at the N-terminus and a C2H2 or C2HC zinc-finger motif at the C-terminus The WRKY domains can specifically bind to W-box cis-elements with a core sequence of TTGACC/T, located at the promoters of many target genes. According to the number of WRKY domains and the feature of the zinc finger motif, the WRKY TFs can be categorized into three groups. Group I members contain two WRKY domains and a C2H2 zinc-finger motif, whereas group II and III members contain one WRKY domain with a C2H2 and C2HC zinc-finger motif, respectively (Eulgem et al., 2000; Ulker and Somssich, 2004; Pandey and Somssich, 2009; Rushton et al., 2010). Since the cloning of the first cDNA encoding a WRKY protein (SPF1) from sweet potato (Ishiguro and Nakamura, 1994), an increasing number of WRKY TFs have been identified in various plants, such as 74 in Arabidopsis (Ulker and Somssich, 2004), 102 in rice (Wu et al., 2005), 104 in poplar (He et al., 2012), 86 in Brachypodium distachyon (Wen et al., 2014), 182 in soybean (Bencke-Malato et al., 2014), and 116 and 102 genes in two different species of cotton (Dou et al., 2014).
WRKY TFs have been shown to participate in various processes in plants, including plant growth, seed development, leaf senescence, and responses to biotic and abiotic stresses (Rushton et al., 2010). Accumulating evidence has demonstrated that WRKY TFs play key roles in plant responses to a variety of abiotic stresses such as drought, salt, heat, cold, and osmotic pressure, and these topics have been extensively reviewed recently (Chen et al., 2012; Rushton et al., 2012; Tripathi et al., 2014; Banerjee and Roychoudhury, 2015). Over-expression of some stress-responsive WRKY genes showed enhanced tolerance to abiotic stresses in transgenic plants. For example, transgenic rice plants harboring OsWRKY11 gene showed significant heat and drought tolerance (Wu et al., 2009). Transgenic Arabidopsis plants over-expressing GmWRKY21 gene exhibited improved tolerance to cold stress, while over-expressing GmWRKY54 gene improved tolerance to drought and salt stress (Zhou et al., 2008). Transgenic Arabidopsis plants over-expressing VvWRKY11 improved to tolerance mannitol-induced osmotic stress (Liu et al., 2011a). Although some WRKYs in several plants have been functionally characterized, the vast majority of WRKYs in many plants, especially in non-model plants, are far from being functionally elucidated.
Like the transcription factor families mentioned above, the NAC TFs also comprise a large plant-specific superfamily present in a wide range of plant species. The typical features of a NAC TF contain a highly conserved NAC domain in the N-terminal region and a variable transcriptional regulatory region in the C-terminal region. The NAC domain is associated with DNA binding, nucleus-oriented localization, and the formation of homodimers or heterodimers with other NAC proteins, while the transcriptional regulatory functions as a transcriptional activator or repressor (Olsen et al., 2005; Puranik et al., 2012). NAC TFs can regulate the transcription of downstream target genes by interacting with NAC recognition sequence (NACRS) with the CACG core-DNA binding motif in the promoter of these genes. NAC TFs have been found to participate in various processes including flower development, formation of secondary walls and cell division, shoot apical meristem formation, leaf senescence, as well as biotic and abiotic stress responses (Olsen et al., 2005; Tran et al., 2010; Nakashima et al., 2012; Nuruzzaman et al., 2013; Banerjee and Roychoudhury, 2015). To date, a lot of putative NAC TFs have been identified in many sequenced species at genome-wide scale, such as 117 in Arabidopsis and 151 in rice (Nuruzzaman et al., 2010), 152 in soybean (Le et al., 2011), 204 in Chinese cabbage (Liu T.K. et al., 2014), 152 in maize (Shiriga et al., 2014), tomato (Su et al., 2015). Moreover, quite a lot of them have been found to be involved in abiotic stress responses. For instance, 33 NAC genes changed significantly in Arabdopsis under salt stress (Jiang and Deyholos, 2006), 40 NAC genes changed under drought or salt stress in rice (Fang et al., 2008), 38 NAC genes changed in soybean under drought stress (Le et al., 2011), 32 NAC genes responded to at least two kinds of treatments in Chrysanthemum lavandulifolium (Huang H. et al., 2012). These stress-responsive NAC genes showed differential expression patterns such as tissue-specific, developmental stage- or stress-specific expression, indicating their involvement in the complex signaling networks during plant stress responses. Some of these stress-responsive NAC genes have been over-expressed in Arabidopsis, rice and other plants and displayed positive effects, summarized in Table 1.
The basic leucine zipper (bZIP) family contains a conserved bZIP domain which is composed of a highly basic region for nuclear localization and DNA binding at the N-terminus and a leucine-rich motif for dimerization at the C-terminus (Landschulz et al., 1988; Hurst, 1994). Like other TFs, the bZIP TFs not only play pivotal roles in developmental processes but also respond to various abiotic stresses such as drought, high salinity, and cold stresses (Jakoby et al., 2002). Now, many members of the bZIP TF family have been identified or predicted at genome-wide level in some species. For example, it has been reported 75 in Arabidopsis (Jakoby et al., 2002), 89 in rice (Nijhawan et al., 2008), 125 in maize (Wei et al., 2012), 89 in barley (Pourabed et al., 2015), 55 in grapevine (Liu J. et al., 2014), 96 in Brachypodium distachyon (Liu and Chu, 2015). However, only a small portion of bZIP TFs has been well studied and most studies on their involvement in stress responses have shown that bZIP TFs are induced by ABA and regulate the expression of stress-related genes in ABA-dependent manner through interaction with specific ABA-responsive cis-acting elements (ABRE) in their promoter region (Uno et al., 2000; Kim et al., 2006; Zou et al., 2008). A lot of efforts have been made to improve abiotic stress tolerance in transgenic plants by over-expressing some stress-responsive bZIP genes and some successful example have been achieved, as listed in Table 1.
Taking five large families of TFs as examples, this review emphasizes the promising roles of TFs as tools to improve plant responses to multiple abiotic stresses. In addition to the above-mentioned several TF families, there are still other TF families such as DNA binding with one finger (Dof) TFs, basic helix-loop-helix (bHLH) TFs, homeodomain-leucine zipper (HD-Zip) TFs, heat shock TFs (HSFs), etc. How to select the key TFs in such a huge gene families and fully display its potential is still an important issue before us. Although a great deal of information about TFs has been accumulated on their involvement in response to diverse abiotic stresses and a good number of promising candidate TF genes have been validated, but there are still some problems to be solved. First, functional redundancy between different TF members may hinder the dissection of the functions of an individual member. Second, most of transgenic studies based on TFs focused on plant growth and tolerance to a given stress at a given developmental stage rather than whole stage. Moreover, the evaluation of transgenic plants was conducted in controlled laboratory or greenhouse conditions rather than field conditions. Third, the constitutive over-expression of some TF genes may improve the stress tolerance, but occasionally lead to negative effects in transgenic plants such as dwarfing, late flowering, and lower yields. Finally, the complete regulation mechanism of individual transcription factor including its upstream and downstream co-regulators, as well as their interactions remains largely unknown.
Abiotic stress response in plants is an extremely complicated process because of the huge gene families and the complex interactions between TFs and cis-elements on the promoters of target genes. Moreover, one transcription factor may regulate a vast array of target genes with the corresponding cis-elements on the promoters, whereas one gene with several types of cis-elements may be regulated by different families of TFs. Thus, the stress-responsive TFs not only function independently but also cross-talk between each other in response to various abiotic stress responses, which indicates the complexity of signaling networks involved in plant stress responses. In the future research, we should first identify multiple stress-responsive TF genes by comparing their expression patterns and the identification of commonly regulated genes which have been proposed to be required for universal stress responses or represent points of cross-talk between signaling pathways (Prasch and Sonnewald, 2015). Genetic manipulation of these identified genes should be a more powerful approach for improving plant tolerance to multiple stresses than manipulation of individual functional genes. Then, the selected TF genes should be validated not only in model plant species but also in crop plants by use of stress-inducible promoter which can minimize the negative effects caused by over-expressing some TF genes. Furthermore, the critical field trials are required to evaluate the transgenetic plants, especially focusing on their growth and tolerance in the whole life period. which is a necessary step in many strategies to develop stress-tolerant crops. Taken together, we still need to struggle for a complete understanding the precise regulatory mechanisms involved in plant abiotic stress responses, which helps to obtain the promising candidate TF genes for breeding multiple abiotic stress-tolerant crops with better yields and qualities.
HYW, HLW, XT wrote the paper. HS provided the paper frame and revised the final paper. All authors reviewed the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work has been jointly supported by the National Natural Science Foundation of China (41171216), Jiangsu Autonomous Innovation of Agricultural Science and Technology [CX(15)1005], National Basic Research Program of China (2013CB430403; 2013CB430400), Open Foundation of Jiangsu Key Laboratory for Bioresources of Saline Soils (JKLBS2014005), Jiangsu Province Science and Technology Support Plan (BE2013429), and Yantai Double-hundred Talent Plan (XY-003-02), and Shuangchuang Talent Plan of Jiangsu Province.
Agarwal, M., Hao, Y., Kapoor, A., Dong, C. H., Fujii, H., Zheng, X., et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281, 37636–37645. doi: 10.1074/jbc.M605895200
Ahuja, I., de Vos, R. C., Bones, A. M., and Hall, R. D. (2010). Plant molecular stress responses face climate change. Trends Plant Sci. 15, 664–674. doi: 10.1016/j.tplants.2010.08.002
Allan, A. C., Hellens, R. P., and Laing, W. A. (2008). MYB transcription factors that colour our fruit. Trends Plant Sci. 13, 99–102. doi: 10.1016/j.tplants.2007.11.012
Ambawat, S., Sharma, P., Yadav, N. R., and Yadav, R. C. (2013). MYB transcription factor genes as regulators for plant responses: an overview. Physiol. Mol. Biol. Plants 19, 307–321. doi: 10.1007/s12298-013-0179-1
Augustine, S. M., Ashwin Narayan, J., Syamaladevi, D. P., Appunu, C., Chakravarthi, M., Ravichandran, V., et al. (2015). Overexpression of EaDREB2 and pyramiding of EaDREB2 with the pea DNA helicase gene (PDH45) enhance drought and salinity tolerance in sugarcane (Saccharum spp. hybrid). Plant Cell Rep. 34, 247–263. doi: 10.1007/s00299-014-1704-6
Babitha, K. C., Ramu, S. V., Pruthvi, V., Mahesh, P., Nataraja, K. N., and Udayakumar, M. (2013). Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 22, 327–341. doi: 10.1007/s11248-012-9645-8
Banerjee, A., and Roychoudhury, A. (2015). WRKY proteins: signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015:807560. doi: 10.1155/2015/807560
Bencke-Malato, M., Cabreira, C., Wiebke-Strohm, B., Bücker-Neto, L., Mancini, E., Osorio, M. B., et al. (2014). Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. BMC Plant Biol. 14:236. doi: 10.1186/s12870-014-0236-0
Bengtsson, M., Shen, Y., and Oki, T. (2006). A SRES-based gridded global population dataset for 1990–2100. Popul. Environ. 28, 113–131. doi: 10.1007/s11111-007-0035-8
Bhargava, S., and Sawant, K. (2013). Drought stress adaptation: metabolic adjustment and regulation of gene expression. Plant Breeding 132, 21–32. doi: 10.1111/pbr.12004
Bouaziz, D., Pirrello, J., Charfeddine, M., Hammami, A., Jbir, R., Dhieb, A., et al. (2013). Overexpression of StDREB1 transcription factor increases tolerance to salt in transgenic potato plants. Mol. Biotechnol. 54, 803–817. doi: 10.1007/s12033-012-9628-2
Cai, H., Tian, S., Dong, H., and Guo, C. (2015). Pleiotropic effects of TaMYB3R1 on plant development and response to osmotic stress in transgenic Arabidopsis. Gene 558, 227–234. doi: 10.1016/j.gene.2014.12.066
Cai, R., Zhao, Y., Wang, Y., Lin, Y., Peng, X., Li, Q., et al. (2014). Overexpression of a maize WRKY58 gene enhances drought and salt tolerance in transgenic rice. Plant Cell Tiss. Org. Cult. 119, 565–577. doi: 10.1007/s11240-014-0556-7
Cao, Z. H., Zhang, S. Z., Wang, R. K., Zhang, R. F., and Hao, Y. J. (2013). Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS ONE 8:e69955. doi: 10.1371/journal.pone.0069955
Century, K., Reuber, T. L., and Ratcliffe, O. J. (2008). Regulating the regulators: the future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol. 147, 20–29. doi: 10.1104/pp.108.117887
Chaves, M. M., Flexas, J., and Pinheiro, C. (2009). Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann. Bot. 103, 551–560. doi: 10.1093/aob/mcn125
Chaves, M. M., Maroco, J., and Joao, S. P. (2003). Understanding plant responses to drought - from genes to the whole plant. Funct. Plant Biol. 30, 239–264. doi: 10.1071/FP02076
Chen, H., Liu, L., Wang, L., Wang, S., and Cheng, X. (2015). VrDREB2A, a DREB-binding transcription factor from Vigna radiata, increased drought and high-salt tolerance in transgenic Arabidopsis thaliana. J. Plant Res. doi: 10.1007/s10265-015-0773-0. [Epub ahead of print].
Chen, L., Song, Y., Li, S., Zhang, L., Zou, C., and Yu, D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819, 120–128. doi: 10.1016/j.bbagrm.2011.09.002
Chen, M., Wang, Q. Y., Cheng, X. G., Xu, Z. S., Li, L. C., Ye, X. G., et al. (2007). GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 353, 299–305. doi: 10.1016/j.bbrc.2006.12.027
Chen, W. J., and Zhu, T. (2004). Networks of transcription factors with roles in environmental stress response. Trends Plant Sci. 9, 591–596. doi: 10.1016/j.tplants.2004.10.007
Chen, X., Wang, Y., Lv, B., Li, J., Luo, L., Lu, S., et al. (2014). The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 55, 604–619. doi: 10.1093/pcp/pct204
Cheng, L., Li, S., Hussain, J., Xu, X., Yin, J., Zhang, Y., et al. (2013a). Isolation and functional characterization of a salt responsive transcriptional factor, LrbZIP from lotus root (Nelumbo nucifera Gaertn). Mol. Biol. Rep. 40, 4033–4045. doi: 10.1007/s11033-012-2481-3
Cheng, L., Li, X., Huang, X., Ma, T., Liang, Y., Ma, X., et al. (2013b). Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 70, 252–260. doi: 10.1016/j.plaphy.2013.05.025
Cominelli, E., Galbiati, M., Vavasseur, A., Conti, L., Sala, T., Vuylsteke, M., et al. (2005). A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 15, 1196–1200. doi: 10.1016/j.cub.2005.05.048
Danquah, A., de Zelicourt, A., Colcombet, J., and Hirt, H. (2014). The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 32, 40–52. doi: 10.1016/j.biotechadv.2013.09.006
Dietz, K. J., Vogel, M. O., and Viehhauser, A. (2010). AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245, 3–14. doi: 10.1007/s00709-010-0142-8
Ding, Z., Li, S., An, X., Liu, X., Qin, H., and Wang, D. (2009). Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genomics 36, 17–29. doi: 10.1016/S1673-8527(09)60003-5
Dong, C. H., Agarwal, M., Zhang, Y., Xie, Q., and Zhu, J. K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. U.S.A. 103, 8281–8286. doi: 10.1073/pnas.0602874103
Dou, L., Zhang, X., Pang, C., Song, M., Wei, H., Fan, S., et al. (2014). Genome-wide analysis of the WRKY gene family in cotton. Mol. Genet. Genomics 289, 1103–1121. doi: 10.1007/s00438-014-0872-y
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., Miura, S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Easterling, D. R., Meehl, G. A., Parmesan, C., Changnon, S. A., Karl, T. R., and Mearns, L. O. (2000). Climate extremes: observations, modeling, and impacts. Science 289, 2068–2074. doi: 10.1126/science.289.5487.2068
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Fang, Y., You, J., Xie, K., Xie, W., and Xiong, L. (2008). Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics 280, 547–563. doi: 10.1007/s00438-008-0386-6
Gao, S. Q., Chen, M., Xu, Z. S., Zhao, C. P., Li, L., Xu, H. J., et al. (2011). The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol. Biol. 75, 537–553. doi: 10.1007/s11103-011-9738-4
Gilmour, S. J., Zarka, D. G., Stockinger, E. J., Salazar, M. P., Houghton, J. M., and Thomashow, M. F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442. doi: 10.1046/j.1365-313x.1998.00310.x
Golldack, D., Luking, I., and Yang, O. (2011). Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 30, 1383–1391. doi: 10.1007/s00299-011-1068-0
Hao, D., Ohme-Takagi, M., and Sarai, A. (1998). Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem. 273, 26857–26861. doi: 10.1074/jbc.273.41.26857
Hao, Y. J., Wei, W., Song, Q. X., Chen, H. W., Zhang, Y. Q., Wang, F., et al. (2011). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 68, 302–313. doi: 10.1111/j.1365-313X.2011.04687.x
He, H., Dong, Q., Shao, Y., Jiang, H., Zhu, S., Cheng, B., et al. (2012). Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 31, 1199–1217. doi: 10.1007/s00299-012-1241-0
Heidarvand, L., and Amiri, R. M. (2010). What happens in plant molecular responses to cold stress? Acta Physiol. Plant. 32, 419–431. doi: 10.1007/s11738-009-0451-8
Hirayama, T., and Shinozaki, K. (2010). Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 61, 1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x
Hou, X. J., Li, S. B., Liu, S. R., Hu, C. G., and Zhang, J. Z. (2014). Genome-wide classification and evolutionary and expression analyses of citrus MYB transcription factor families in sweet orange. PLoS ONE 9:e112375. doi: 10.1371/journal.pone.0112375
Huang, G. T., Ma, S. L., Bai, L. P., Zhang, L., Ma, H., Jia, P., et al. (2012). Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 39, 969–987. doi: 10.1007/s11033-011-0823-1
Huang, H., Wang, Y., Wang, S., Wu, X., Yang, K., Niu, Y., et al. (2012). Transcriptome-wide survey and expression analysis of stress-responsive NAC genes in Chrysanthemum lavandulifolium. Plant Sci. 193–194, 18–27. doi: 10.1016/j.plantsci.2012.05.004
Huang, Q., Wang, Y., Li, B., Chang, J., Chen, M., Li, K., et al. (2015). TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 15:268. doi: 10.1186/s12870-015-0644-9
Huang, X. S., Liu, J. H., and Chen, X. J. (2010). Overexpression of PtrABF gene, a bZIP transcription factor isolated from Poncirus trifoliata, enhances dehydration and drought tolerance in tobacco via scavenging ROS and modulating expression of stress-responsive genes. BMC Plant Biol. 10:230. doi: 10.1186/1471-2229-10-230
Ipcc (2007). “Climate change 2007: the physical science basis,” in Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change, eds S. Solomon, D. Qin, M. Manning, Z. Chen, M. Marquis, K. B. Averyt, M. Tignor, and H. L. Miller (Geneva: Ipcc Secretariat).
Ipcc (2008). “Climate change and water,” in Technical Paper of the Intergovernmental Panel on Climate Change, eds B. C. Bates, Z. W. Kundzewicz, J. Palutikof, and S. Wu (Geneva: Ipcc Secretariat).
Ishiguro, S., and Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5' upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol. Gen. Genet. 244, 563–571. doi: 10.1007/BF00282746
Ito, Y., Katsura, K., Maruyama, K., Taji, T., Kobayashi, M., Seki, M., et al. (2006). Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47, 141–153. doi: 10.1093/pcp/pci230
Jaglo, K. R., Kleff, S., Amundsen, K. L., Zhang, X., Haake, V., Zhang, J. Z., et al. (2001). Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 127, 910–917. doi: 10.1104/pp.010548
Jaglo-Ottosen, K. R., Gilmour, S. J., Zarka, D. G., Schabenberger, O., and Thomashow, M. F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. doi: 10.1126/science.280.5360.104
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Jensen, M. K., Kjaersgaard, T., Nielsen, M. M., Galberg, P., Petersen, K., O'shea, C., et al. (2010). The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426, 183–196. doi: 10.1042/BJ20091234
Jiang, Y., and Deyholos, M. K. (2006). Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 6:25. doi: 10.1186/1471-2229-6-25
Joo, J., Choi, H. J., Lee, Y. H., Kim, Y. K., and Song, S. I. (2013). A transcriptional repressor of the ERF family confers drought tolerance to rice and regulates genes preferentially located on chromosome 11. Planta 238, 155–170. doi: 10.1007/s00425-013-1880-6
Jung, C., Seo, J. S., Han, S. W., Koo, Y. J., Kim, C. H., Song, S. I., et al. (2008). Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 146, 623–635. doi: 10.1104/pp.107.110981
Kasuga, M., Miura, S., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 45, 346–350. doi: 10.1093/pcp/pch037
Kim, Y. S., Kim, S. G., Park, J. E., Park, H. Y., Lim, M. H., Chua, N. H., et al. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18, 3132–3144. doi: 10.1105/tpc.106.043018
Landschulz, W. H., Johnson, P. F., and McKnight, S. L. (1988). The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240, 1759–1764. doi: 10.1126/science.3289117
Lata, C., Mishra, A. K., Muthamilarasan, M., Bonthala, V. S., Khan, Y., and Prasad, M. (2014). Genome-wide investigation and expression profiling of AP2/ERF transcription factor superfamily in foxtail millet (Setaria italica L.). PLoS ONE 9:e113092. doi: 10.1371/journal.pone.0113092
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Le, D. T., Nishiyama, R., Watanabe, Y., Mochida, K., Yamaguchi-Shinozaki, K., Shinozaki, K., et al. (2011). Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 18, 263–276. doi: 10.1093/dnares/dsr015
Li, C., Ng, C. K. Y., and Fan, L.-M. (2015). MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 114, 80–91. doi: 10.1016/j.envexpbot.2014.06.014
Li, H., Gao, Y., Xu, H., Dai, Y., Deng, D., and Chen, J. (2013). ZmWRKY33, a WRKY maize transcription factor conferring enhanced salt stress tolerances in Arabidopsis. Plant Growth Regul. 70, 207–216. doi: 10.1007/s10725-013-9792-9
Liang, H., Lu, Y., Liu, H., Wang, F., Xin, Z., and Zhang, Z. (2008). A novel activator-type ERF of Thinopyrum intermedium, TiERF1, positively regulates defence responses. J. Exp. Bot. 59, 3111–3120. doi: 10.1093/jxb/ern165
Liang, Y. K., Dubos, C., Dodd, I. C., Holroyd, G. H., Hetherington, A. M., and Campbell, M. M. (2005). AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr. Biol. 15, 1201–1206. doi: 10.1016/j.cub.2005.06.041
Liao, Y., Zou, H. F., Wang, H. W., Zhang, W. K., Ma, B., Zhang, J. S., et al. (2008). Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 18, 1047–1060. doi: 10.1038/cr.2008.280
Licausi, F., Giorgi, F. M., Zenoni, S., Osti, F., Pezzotti, M., and Perata, P. (2010). Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genomics 11:719. doi: 10.1186/1471-2164-11-719
Lim, C. J., Hwang, J. E., Chen, H., Hong, J. K., Yang, K. A., Choi, M. S., et al. (2007). Over-expression of the Arabidopsis DRE/CRT-binding transcription factor DREB2C enhances thermotolerance. Biochem. Biophys. Res. Commun. 362, 431–436. doi: 10.1016/j.bbrc.2007.08.007
Liu, C., Mao, B., Ou, S., Wang, W., Liu, L., Wu, Y., et al. (2014). OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 84, 19–36. doi: 10.1007/s11103-013-0115-3
Liu, H., Yang, W., Liu, D., Han, Y., Zhang, A., and Li, S. (2011a). Ectopic expression of a grapevine transcription factor VvWRKY11 contributes to osmotic stress tolerance in Arabidopsis. Mol. Biol. Rep. 38, 417–427. doi: 10.1007/s11033-010-0124-0
Liu, H., Zhou, X., Dong, N., Liu, X., Zhang, H., and Zhang, Z. (2011b). Expression of a wheat MYB gene in transgenic tobacco enhances resistance to Ralstonia solanacearum, and to drought and salt stresses. Funct. Integr. Genomics 11, 431–443. doi: 10.1007/s10142-011-0228-1
Liu, J., Chen, N., Chen, F., Cai, B., Dal Santo, S., Tornielli, G. B., et al. (2014). Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genomics 15:281. doi: 10.1186/1471-2164-15-281
Liu, J.-H., Peng, T., and Dai, W. (2014). Critical cis-acting elements and interacting transcription factors: key players associated with abiotic stress responses in plants. Plant Mol. Biol. Report. 32, 303–317. doi: 10.1007/s11105-013-0667-z
Liu, L., Zhang, Z., Dong, J., and Wang, T. (2016). Overexpression of MtWRKY76 increases both salt and drought tolerance in Medicago truncatula. Environ. Exp. Bot. 123, 50–58. doi: 10.1016/j.envexpbot.2015.10.007
Liu, T. K., Song, X. M., Duan, W. K., Huang, Z. N., Liu, G. F., Li, Y., et al. (2014). Genome-wide analysis and expression patterns of NAC transcription factor family under different developmental stages and abiotic stresses in Chinese Cabbage. Plant Mol. Biol. Report. 32, 1041–1056. doi: 10.1007/s11105-014-0712-6
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Liu, X., and Chu, Z. (2015). Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genomics 16:227. doi: 10.1186/s12864-015-1457-9
Liu, X., Liu, S., Wu, J., Zhang, B., Li, X., Yan, Y., et al. (2013). Overexpression of Arachis hypogaea NAC3 in tobacco enhances dehydration and drought tolerance by increasing superoxide scavenging. Plant Physiol. Biochem. 70, 354–359. doi: 10.1016/j.plaphy.2013.05.018
Lobell, D. B., Schlenker, W., and Costa-Roberts, J. (2011). Climate trends and global crop production since 1980. Science 333, 616–620. doi: 10.1126/science.1204531
Lu, M., Ying, S., Zhang, D. F., Shi, Y. S., Song, Y. C., Wang, T. Y., et al. (2012). A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis. Plant Cell Rep. 31, 1701–1711. doi: 10.1007/s00299-012-1284-2
Lucas, S., Durmaz, E., Akpinar, B. A., and Budak, H. (2011). The drought response displayed by a DRE-binding protein from Triticum dicoccoides. Plant Physiol. Biochem. 49, 346–351. doi: 10.1016/j.plaphy.2011.01.016
Luo, X., Bai, X., Sun, X., Zhu, D., Liu, B., Ji, W., et al. (2013). Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 64, 2155–2169. doi: 10.1093/jxb/ert073
Mallikarjuna, G., Mallikarjuna, K., Reddy, M. K., and Kaul, T. (2011). Expression of OsDREB2A transcription factor confers enhanced dehydration and salt stress tolerance in rice (Oryza sativa L.). Biotechnol. Lett. 33, 1689–1697. doi: 10.1007/s10529-011-0620-x
Mao, X., Chen, S., Li, A., Zhai, C., and Jing, R. (2014). Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS ONE 9:e84359. doi: 10.1371/journal.pone.0084359
Meng, X., Wang, J. R., Wang, G. D., Liang, X. Q., Li, X. D., and Meng, Q. W. (2015). An R2R3-MYB gene, LeAN2, positively regulated the thermo-tolerance in transgenic tomato. J. Plant Physiol. 175, 1–8. doi: 10.1016/j.jplph.2014.09.018
Meng, X., Yin, B., Feng, H. L., Zhang, S., Liang, X. Q., and Meng, Q. W. (2014). Overexpression of R2R3-MYB gene leads to accumulation of anthocyanin and enhanced resistance to chilling and oxidative stress. Biol. Plant. 58, 121–130. doi: 10.1007/s10535-013-0376-3
Mittler, R., and Blumwald, E. (2010). Genetic engineering for modern agriculture: challenges and perspectives. Annu. Rev. Plant Biol. 61, 443–462. doi: 10.1146/annurev-arplant-042809-112116
Miura, K., Jin, J. B., Lee, J., Yoo, C. Y., Stirm, V., Miura, T., et al. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403–1414. doi: 10.1105/tpc.106.048397
Mizoi, J., Ohori, T., Moriwaki, T., Kidokoro, S., Todaka, D., Maruyama, K., et al. (2013). GmDREB2A;2, a canonical DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol. 161, 346–361. doi: 10.1104/pp.112.204875
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Muthamilarasan, M., Khandelwal, R., Yadav, C. B., Bonthala, V. S., Khan, Y., and Prasad, M. (2014). Identification and molecular characterization of MYB Transcription Factor Superfamily in C4 model plant foxtail millet (Setaria italica L.). PLoS ONE 9:e109920. doi: 10.1371/journal.pone.0109920
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Nakashima, K., Shinwari, Z. K., Sakuma, Y., Seki, M., Miura, S., Shinozaki, K., et al. (2000). Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol. Biol. 42, 657–665. doi: 10.1023/A:1006321900483
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Newton, A. C., Johnson, S. N., and Gregory, P. J. (2011). Implications of climate change for diseases, crop yields and food security. Euphytica 179, 3–18. doi: 10.1007/s10681-011-0359-4
Nijhawan, A., Jain, M., Tyagi, A. K., and Khurana, J. P. (2008). Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 146, 333–350. doi: 10.1104/pp.107.112821
Nuruzzaman, M., Manimekalai, R., Sharoni, A. M., Satoh, K., Kondoh, H., Ooka, H., et al. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene 465, 30–44. doi: 10.1016/j.gene.2010.06.008
Nuruzzaman, M., Sharoni, A. M., and Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4:248. doi: 10.3389/fmicb.2013.00248
Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. doi: 10.1105/tpc.7.2.173
Olsen, A. N., Ernst, H. A., Leggio, L. L., and Skriver, K. (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. doi: 10.1016/j.tplants.2004.12.010
Pandey, S. P., and Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Peng, X., Zhang, L., Zhang, L., Liu, Z., Cheng, L., Yang, Y., et al. (2013). The transcriptional factor LcDREB2 cooperates with LcSAMDC2 to contribute to salt tolerance in Leymus chinensis. Plant Cell Tissue Organ Cult. 113, 245–256. doi: 10.1007/s11240-012-0264-0
Pérez-Clemente, R. M., Vives, V., Zandalinas, S. I., López-Climent, M. F., Munoz, V., and Gomez-Cadenas, A. (2013). Biotechnological approaches to study plant responses to stress. Biomed Res. Int. 2013:654120. doi: 10.1155/2013/654120
Pourabed, E., Ghane Golmohamadi, F., Soleymani Monfared, P., Razavi, S. M., and Shobbar, Z. S. (2015). Basic leucine zipper family in barley: genome-wide characterization of members and expression analysis. Mol. Biotechnol. 57, 12–26. doi: 10.1007/s12033-014-9797-2
Prasch, C. M., and Sonnewald, U. (2015). Signaling events in plants: stress factors in combination change the picture. Environ. Exp. Bot. 114, 4–14. doi: 10.1016/j.envexpbot.2014.06.020
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Qi, L., Yang, J., Yuan, Y., Huang, L., and Chen, P. (2015). Overexpression of two R2R3-MYB genes from Scutellaria baicalensis induces phenylpropanoid accumulation and enhances oxidative stress resistance in transgenic tobacco. Plant Physiol. Biochem. 94, 235–243. doi: 10.1016/j.plaphy.2015.06.007
Qin, F., Kakimoto, M., Sakuma, Y., Maruyama, K., Osakabe, Y., Tran, L. S., et al. (2007). Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 50, 54–69. doi: 10.1111/j.1365-313X.2007.03034.x
Qin, F., Sakuma, Y., Li, J., Liu, Q., Li, Y. Q., Shinozaki, K., et al. (2004). Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol. 45, 1042–1052. doi: 10.1093/pcp/pch118
Qin, Y., Tian, Y., Han, L., and Yang, X. (2013). Constitutive expression of a salinity-induced wheat WRKY transcription factor enhances salinity and ionic stress tolerance in transgenic Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 441, 476–481. doi: 10.1016/j.bbrc.2013.10.088
Qin, Y., Tian, Y., and Liu, X. (2015). A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 464, 428–433. doi: 10.1016/j.bbrc.2015.06.128
Qiu, Y., and Yu, D. (2009). Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environmental and experimental botany 65. Environ. Exp. Bot. 65, 35–47. doi: 10.1016/j.envexpbot.2008.07.002
Ravikumar, G., Manimaran, P., Voleti, S. R., Subrahmanyam, D., Sundaram, R. M., Bansal, K. C., et al. (2014). Stress-inducible expression of AtDREB1A transcription factor greatly improves drought stress tolerance in transgenic indica rice. Transgenic Res. 23, 421–439. doi: 10.1007/s11248-013-9776-6
Ray, S., Dansana, P. K., Bhaskar, A., Giri, J., Kapoor, S., Khurana, J. P., et al. (2009). “Emerging trends in functional genomics for stress tolerance in crop plants,” in Plant Stress Biology, ed H. Hirt (Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA), 37–63.
Riechmann, J. L., and Meyerowitz, E. M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379, 633–646.
Rong, W., Qi, L., Wang, A., Ye, X., Du, L., Liang, H., et al. (2014). The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 12, 468–479. doi: 10.1111/pbi.12153
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Saad, A. S., Li, X., Li, H. P., Huang, T., Gao, C. S., Guo, M. W., et al. (2013). A rice stress-responsive NAC gene enhances tolerance of transgenic wheat to drought and salt stresses. Plant Sci. 203–204, 33–40. doi: 10.1016/j.plantsci.2012.12.016
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K., et al. (2006a). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18, 1292–1309. doi: 10.1105/tpc.105.035881
Sakuma, Y., Maruyama, K., Qin, F., Osakabe, Y., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006b). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. U.S.A. 103, 18822–18827. doi: 10.1073/pnas.0605639103
Sanchez, D. H., Pieckenstain, F. L., Szymanski, J., Erban, A., Bromke, M., Hannah, M. A., et al. (2011). Comparative functional genomics of salt stress in related model and cultivated plants identifies and overcomes limitations to translational genomics. PLoS ONE 6:e17094. doi: 10.1371/journal.pone.0017094
Schaller, G. E., Kieber, J. J., and Shiu, S. H. (2008). Two-component signaling elements and histidyl-aspartyl phosphorelays. Arabidopsis Book 6:e0112. doi: 10.1199/tab.0112
Seo, J., Sohn, H., Noh, K., Jung, C., An, J., Donovan, C., et al. (2012). Expression of the Arabidopsis AtMYB44 gene confers drought/salt-stress tolerance in transgenic soybean. Mol. Breed. 29, 601–608. doi: 10.1007/s11032-011-9576-8
Seo, P. J., Lee, S. B., Suh, M. C., Park, M. J., Go, Y. S., and Park, C. M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23, 1138–1152. doi: 10.1105/tpc.111.083485
Seo, P. J., Xiang, F., Qiao, M., Park, J. Y., Lee, Y. N., Kim, S. G., et al. (2009). The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151, 275–289. doi: 10.1104/pp.109.144220
Shao, H. B., Chu, L. Y., Jaleel, C. A., Manivannan, P., Panneerselvam, R., and Shao, M. A. (2009). Understanding water deficit stress-induced changes in the basic metabolism of higher plants - biotechnologically and sustainably improving agriculture and the ecoenvironment in arid regions of the globe. Crit. Rev. Biotechnol. 29, 131–151. doi: 10.1080/07388550902869792
Sharoni, A. M., Nuruzzaman, M., Satoh, K., Shimizu, T., Kondoh, H., Sasaya, T., et al. (2011). Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 52, 344–360. doi: 10.1093/pcp/pcq196
Shi, W., Liu, D., Hao, L., Wu, C. A., Guo, X., and Li, H. (2014). GhWRKY39, a member of the WRKY transcription factor family in cotton, has a positive role in disease resistance and salt stress tolerance. Plant Cell Tissue Organ Cult. 118, 17–32. doi: 10.1007/s11240-014-0458-8
Shinozaki, K., Yamaguchi-Shinozakiy, K., and Sekiz, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. doi: 10.1016/S1369-5266(03)00092-X
Shinwari, Z. K., Nakashima, K., Miura, S., Kasuga, M., Seki, M., Yamaguchi-Shinozaki, K., et al. (1998). An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem. Biophys. Res. Commun. 250, 161–170. doi: 10.1006/bbrc.1998.9267
Shiriga, K., Sharma, R., Kumar, K., Yadav, S. K., Hossain, F., and Thirunavukkarasu, N. (2014). Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta Gene 2, 407–417. doi: 10.1016/j.mgene.2014.05.001
Singh, R., Usha, Rizvi, S. M. H., and Sonia, Jaiwal, P. (2003). “Genetic engineering for enhancing abiotic stress tolerance,” in Improvement Strategies of Leguminosae Biotechnology, eds P. Jaiwal and R. Singh (Dordrecht: Springer), 223–243.
Song, X., Li, Y., and Hou, X. (2013). Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 14:573. doi: 10.1186/1471-2164-14-573
Stockinger, E. J., Gilmour, S. J., and Thomashow, M. F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U.S.A. 94, 1035–1040. doi: 10.1073/pnas.94.3.1035
Su, H. Y., Zhang, S. Z., Yin, Y. L., Zhu, D. Z., and Han, L. Y. (2015). Genome-wide analysis of NAM-ATAF1, 2-CUC2 transcription factor family in Solanum lycopersicum. J. Plant Biochem. Biotechnol. 24, 176–183. doi: 10.1007/s13562-014-0255-9
Su, L. T., Li, J. W., Liu, D. Q., Zhai, Y., Zhang, H. J., Li, X. W., et al. (2014). A novel MYB transcription factor, GmMYBJ1, from soybean confers drought and cold tolerance in Arabidopsis thaliana. Gene 538, 46–55. doi: 10.1016/j.gene.2014.01.024
Sun, J., Hu, W., Zhou, R., Wang, L., Wang, X., Wang, Q., et al. (2015). The Brachypodium distachyon BdWRKY36 gene confers tolerance to drought stress in transgenic tobacco plants. Plant Cell Rep. 34, 23–35. doi: 10.1007/s00299-014-1684-6
Sun, Z. M., Zhou, M. L., Xiao, X. G., Tang, Y. X., and Wu, Y. M. (2014). Genome-wide analysis of AP2/ERF family genes from Lotus corniculatus shows LcERF054 enhances salt tolerance. Funct. Integr. Genomics 14, 453–466. doi: 10.1007/s10142-014-0372-5
Takeda, S., and Matsuoka, M. (2008). Genetic approaches to crop improvement: responding to environmental and population changes. Nat. Rev. Genet. 9, 444–457. doi: 10.1038/nrg2342
Tang, Y., Liu, M., Gao, S., Zhang, Z., Zhao, X., Zhao, C., et al. (2012). Molecular characterization of novel TaNAC genes in wheat and overexpression of TaNAC2a confers drought tolerance in tobacco. Physiol. Plant. 144, 210–224. doi: 10.1111/j.1399-3054.2011.01539.x
Tran, L. S., Nishiyama, R., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2010). Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 1, 32–39. doi: 10.4161/gmcr.1.1.10569
Tripathi, P., Rabara, R. C., and Rushton, P. J. (2014). A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 239, 255–266. doi: 10.1007/s00425-013-1985-y
Tsai-Hung, H., Jent-Turn, L., Pei-Tzu, Y., Li-Hui, C., Yee-Yung, C., Yu-Chie, W., et al. (2002). Heterology expression of the ArabidopsisC-repeat/dehydration response element binding Factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 129, 1086–1094. doi: 10.1104/pp.003442
Ulker, B., and Somssich, I. E. (2004). WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7, 491–498. doi: 10.1016/j.pbi.2004.07.012
Umezawa, T., Fujita, M., Fujita, Y., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 17, 113–122. doi: 10.1016/j.copbio.2006.02.002
United Nations (2015). The World Population Prospects. New York, NY: United Nations Department of Economic and Social Affairs.
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U.S.A. 97, 11632–11637. doi: 10.1073/pnas.190309197
Varshney, R. K., Bansal, K. C., Aggarwal, P. K., Datta, S. K., and Craufurd, P. Q. (2011). Agricultural biotechnology for crop improvement in a variable climate: hope or hype? Trends Plant Sci. 16, 363–371. doi: 10.1016/j.tplants.2011.03.004
Vinocur, B., and Altman, A. (2005). Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr. Opin. Biotechnol. 16, 123–132. doi: 10.1016/j.copbio.2005.02.001
Wang, C., Deng, P., Chen, L., Wang, X., Ma, H., Hu, W., et al. (2013). A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS ONE 8:e65120. doi: 10.1371/journal.pone.0065120
Wang, R. K., Cao, Z. H., and Hao, Y. J. (2014). Overexpression of a R2R3 MYB gene MdSIMYB1 increases tolerance to multiple stresses in transgenic tobacco and apples. Physiol. Plant. 150, 76–87. doi: 10.1111/ppl.12069
Wei, K., Chen, J., Wang, Y., Chen, Y., Chen, S., Lin, Y., et al. (2012). Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 19, 463–476. doi: 10.1093/dnares/dss026
Wen, F., Zhu, H., Li, P., Jiang, M., Mao, W., Ong, C., et al. (2014). Genome-wide evolutionary characterization and expression analyses of WRKY family genes in Brachypodium distachyon. DNA Res. 21, 327–339. doi: 10.1093/dnares/dst060
Wu, H., Lv, H., Li, L., Liu, J., Mu, S., Li, X., et al. (2015). Genome-wide analysis of the AP2/ERF transcription factors family and the expression patterns of DREB genes in Moso bamboo (Phyllostachys edulis). PLoS ONE 10:e0126657. doi: 10.1371/journal.pone.0126657
Wu, K. L., Guo, Z. J., Wang, H. H., and Li, J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12, 9–26. doi: 10.1093/dnares/12.1.9
Wu, X., Shiroto, Y., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Xu, Z.-S., Chen, M., Li, L.-C., and Ma, Y.-Z. (2008a). Functions of the ERF transcription factor family in plants. Botany 86, 969–977. doi: 10.1139/B08-041
Xu, Z.-S., Ni, Z. Y., Liu, L., Nie, L. N., Li, L. C., Chen, M., et al. (2008b). Characterization of the TaAIDFa gene encoding a CRT/DRE-binding factor responsive to drought, high-salt, and cold stress in wheat. Mol. Genet. Genomics 280, 497–508. doi: 10.1007/s00438-008-0382-x
Yamaguchi, T., and Blumwald, E. (2005). Developing salt-tolerant crop plants: challenges and opportunities. Trends Plant Sci. 10, 615–620. doi: 10.1016/j.tplants.2005.10.002
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yang, A., Dai, X., and Zhang, W. H. (2012). A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 63, 2541–2556. doi: 10.1093/jxb/err431
Yang, W., Liu, X. D., Chi, X. J., Wu, C. A., Li, Y. Z., Song, L. L., et al. (2011). Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta 233, 219–229. doi: 10.1007/s00425-010-1279-6
Yang, X., Wang, X., Lu, J., Yi, Z., Fu, C., Ran, J., et al. (2015). Overexpression of a Miscanthus lutarioriparius NAC gene MlNAC5 confers enhanced drought and cold tolerance in Arabidopsis. Plant Cell Rep. 34, 943–958. doi: 10.1007/s00299-015-1756-2
Yanhui, C., Xiaoyuan, Y., Kun, H., Meihua, L., Jigang, L., Zhaofeng, G., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60, 107–124. doi: 10.1007/s11103-005-2910-y
Ying, S., Zhang, D. F., Fu, J., Shi, Y. S., Song, Y. C., Wang, T. Y., et al. (2012). Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 235, 253–266. doi: 10.1007/s00425-011-1496-7
Yokotani, N., Ichikawa, T., Kondou, Y., Matsui, M., Hirochika, H., Iwabuchi, M., et al. (2009). Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta 229, 1065–1075. doi: 10.1007/s00425-009-0895-5
Zhai, Y., Wang, Y., Li, Y., Lei, T., Yan, F., Su, L., et al. (2013). Isolation and molecular characterization of GmERF7, a soybean ethylene-response factor that increases salt stress tolerance in tobacco. Gene 513, 174–183. doi: 10.1016/j.gene.2012.10.018
Zhang, G., Chen, M., Li, L., Xu, Z., Chen, X., Guo, J., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60, 3781–3796. doi: 10.1093/jxb/erp214
Zhang, H., Liu, W., Wan, L., Li, F., Dai, L., Li, D., et al. (2010). Functional analyses of ethylene response factor JERF3 with the aim of improving tolerance to drought and osmotic stress in transgenic rice. Transgenic Res. 19, 809–818. doi: 10.1007/s11248-009-9357-x
Zhang, L., Zhang, L., Xia, C., Zhao, G., Liu, J., Jia, J., et al. (2015). A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiol. Plant. 153, 538–554. doi: 10.1111/ppl.12261
Zhang, X., Liu, X., Wu, L., Yu, G., Wang, X., and Ma, H. (2015). The SsDREB transcription factor from the succulent halophyte suaeda salsa enhances abiotic stress tolerance in transgenic tobacco. Int. J. Genomics 2015, 875497. doi: 10.1155/2015/875497
Zhang, X., Wang, L., Meng, H., Wen, H., Fan, Y., and Zhao, J. (2011). Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol. Biol. 75, 365–378. doi: 10.1007/s11103-011-9732-x
Zhong, H., Guo, Q. Q., Chen, L., Ren, F., Wang, Q. Q., Zheng, Y., et al. (2012). Two Brassica napus genes encoding NAC transcription factors are involved in response to high-salinity stress. Plant Cell Rep. 31, 1991–2003. doi: 10.1007/s00299-012-1311-3
Zhou, L., Wang, N. N., Gong, S. Y., Lu, R., Li, Y., and Li, X. B. (2015). Overexpression of a cotton (Gossypium hirsutum) WRKY gene, GhWRKY34, in Arabidopsis enhances salt-tolerance of the transgenic plants. Plant Physiol. Biochem. 96, 311–320. doi: 10.1016/j.plaphy.2015.08.016
Zhou, Q. Y., Tian, A. G., Zou, H. F., Xie, Z. M., Lei, G., Huang, J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. doi: 10.1111/j.1467-7652.2008.00336.x
Zhu, N., Cheng, S., Liu, X., Du, H., Dai, M., Zhou, D. X., et al. (2015). The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 236, 146–156. doi: 10.1016/j.plantsci.2015.03.023
Zhu, X., Qi, L., Liu, X., Cai, S., Xu, H., Huang, R., et al. (2014). The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol. 164, 1499–1514. doi: 10.1104/pp.113.229575
Zhuang, J., Cai, B., Peng, R. H., Zhu, B., Jin, X. F., Xue, Y., et al. (2008). Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 371, 468–474. doi: 10.1016/j.bbrc.2008.04.087
Keywords: abiotic stress, transcription factors, transgenic plant, stress-responsive, stress tolerance
Citation: Wang H, Wang H, Shao H and Tang X (2016) Recent Advances in Utilizing Transcription Factors to Improve Plant Abiotic Stress Tolerance by Transgenic Technology. Front. Plant Sci. 7:67. doi: 10.3389/fpls.2016.00067
Received: 20 November 2015; Accepted: 14 January 2016;
Published: 09 February 2016.
Edited by:
Shabir Hussain Wani, Sher-e-Kashmir University of Agricultural Sciences and Technology, IndiaReviewed by:
Hemant Ritturaj Kushwaha, International Centre for Genetic Engineering and Biotechnology, IndiaCopyright © 2016 Wang, Wang, Shao and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Honglei Wang, d2hsMTk3NzQ5QDE2My5jb20=;
Hongbo Shao, c2hhb2hvbmdib2NodUAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.