
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 10 February 2016
Sec. Technical Advances in Plant Science
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.00060
 Yonghui Dong1,2
Yonghui Dong1,2 Bin Li3
Bin Li3 Sergey Malitsky2
Sergey Malitsky2 Ilana Rogachev2
Ilana Rogachev2 Asaph Aharoni2
Asaph Aharoni2 Filip Kaftan4
Filip Kaftan4 Aleš Svatoš4
Aleš Svatoš4 Pietro Franceschi1*
Pietro Franceschi1*Mass spectrometry imaging (MSI) is a mass spectrometry based molecular ion imaging technique. It provides the means for ascertaining the spatial distribution of a large variety of analytes directly on tissue sample surfaces without any labeling or staining agents. These advantages make it an attractive molecular histology tool in medical, pharmaceutical, and biological research. Likewise, MSI has started gaining popularity in plant sciences; yet, information regarding sample preparation methods for plant tissues is still limited. Sample preparation is a crucial step that is directly associated with the quality and authenticity of the imaging results, it therefore demands in-depth studies based on the characteristics of plant samples. In this review, a sample preparation pipeline is discussed in detail and illustrated through selected practical examples. In particular, special concerns regarding sample preparation for plant imaging are critically evaluated. Finally, the applications of MSI techniques in plants are reviewed according to different classes of plant metabolites.
Mass spectrometric imaging (MSI) enables label-free detection and mapping of a wide range of molecules at complex surfaces, and it has become an attractive molecular histology tool in pharmaceutical and medical research. Since 2005 MSI has been gradually applied in plant research (Imai et al., 2005; Mullen et al., 2005). The information on the spatial organization of proteins and metabolites will greatly improve our understanding of plant metabolism and the biochemical functions of specific plant tissues (Lee et al., 2012; Matros and Mock, 2013).
The core of MSI experiment is the mass spectrometer, which consists of three major parts: ion source, mass analyzer and detector. In the ion source, analytes are desorbed and ionized; in the analyzer, they are separated on the basis of their mass to charge ratios (m/z); and, finally, the separated ions are detected in the detector. As a final output a mass spectrum is generated by displaying the intensity of the detected ions over a full m/z scale. The basic principle of microprobe MSI is straightforward: the instrument collects a series of mass spectra by rastering a certain area of a tissue sample. The distribution maps of the analytes over the sample surface are subsequently generated by plotting the intensity of their individual m/z peak in the mass spectra against the x-y coordinate (Goodwin et al., 2008; Svatos, 2010). The typical workflow of a MALDI imaging experiment is shown in Figure 1.
Several ion sources are available for MSI, among which secondary ion mass spectrometry (SIMS), matrix-assisted laser desorption ionization (MALDI) and desorption electrospray ionization (DESI) are the most popular (Amstalden van Hove et al., 2010; Svatos, 2010). Many other MSI sources are still emerging, and they are usually derivatives or modifications of the above mentioned approaches. Among them it is worth mentioning laser ablation electrospray ionization (LAESI), laser ablation inductively coupled plasma (LA-ICP), liquid extraction surface analysis (LESA), direct analysis in real time (DART), and nano-desorption electrospray ionization (Nano-DESI). The different ion sources differ greatly in the method of ion generation (and hence on the nature of the produced ions), pressure regime, spatial resolution, probing depth, and speed. Each of them has its own advantages and disadvantages; for a comprehensive overview on this topic the reader is kindly referred to the following papers (Bhardwaj and Hanley, 2014; Boughton et al., 2015; Li et al., 2015; Sumner et al., 2015).
Sample handling is the most crucial step in MSI, as only an appropriate sample preparation can maintain the origin, distribution and abundance of the molecules, ensuring high-quality signals and sufficient spatial resolution (Grassl et al., 2011; Kaspar et al., 2011; Peukert et al., 2012; Spengler, 2014). Although Sample preparation for proteins and peptides has been somewhat standardized (Goodwin et al., 2008), there are very few examples of protein or peptide imaging in plant-based research (see Section Proteins and Peptides). Conversely, a large percentage of publications have dealt with plant secondary metabolites. The challenges are not only relevant to secondary, but also primary metabolites which can be oxidized, can diffuse through tissues during preparation, or be degraded through enzymatic processes or through exposure to light, heat or atmosphere (Lee et al., 2012).
MSI sample preparation methods for plant tissues are more challenging than those for mammalian tissues (Boughton et al., 2015; Heyman and Dubery, 2015). Cuticles in higher plant bodies represent the first barrier for direct MS imaging of internal metabolites, since it is difficult for most soft ionization techniques such as MALDI and DESI to penetrate through them (Thunig et al., 2011). Plant cell walls form the second barrier for MS imaging of molecules within the cell. For instance, when a matrix solution is sprayed onto the surface of a plant body, the cell wall prevents the solution diffusing across the cell wall, leading to the reduced “analyte extraction” efficiency in MALDI imaging (Takahashi et al., 2015). The high water content in plant tissues poses another challenge during cryosectioning. Plant tissues are more fragile upon freezing, and additionally, it is more difficult to get thin and intact sections for water-rich plant samples. Sample shrinkage or partial flaking is often observed upon dehydration, and this phenomenon usually results in an uneven surface which further affects MSI analysis. It should be noted that the sample shrinkage could also lead to mismatch between the MS image and the optical image (Cha et al., 2008), making biological interpretation difficult. This review therefore discusses the sample preparation strategies for plant tissues, aiming at providing guidelines for researchers who are planning to image different plant tissues and metabolites. In the first part, all the essential steps of sample preparation as well as specific considerations for plant tissues are discussed in detail. In the second part, the application of MSI to the analysis of different classes of metabolites in plant samples is summarized and discussed referring to a selected list of publications.
In this section we discuss each sample preparation step, including tissue storage (2.1), sectioning (2.2), mounting (2.3) and ionization supporting treatments (2.4) (Pol et al., 2010; Goodwin, 2012). In addition, other considerations for plant sample handling (2.5) such as the use of fresh sample or dry sample (2.5.1), removal of plant cuticle (2.5.2), matrix effects (2.5.3), and morphological effect (2.5.4) are also covered here. It is worth noting that sample preparation steps vary depending on the MSI instrument, on the nature of the tissues and on the analytes to be imaged.
Samples are typically flash frozen in liquid nitrogen to prevent enzymatic degradation or analyte migration, and then stored at −80°C for up to 1 year with no reported significant degradation (Schwartz et al., 2003). However, water-rich plant samples may shrink upon long-term storage due to the gradual sublimation of water. When samples are too large and the storage space is limited, plant samples can be stored as section slides (Dill et al., 2011). For example, in our laboratory, several apple sections are mounted onto the glass slide, vacuum dried (~50 Torr, 4 h), vertically placed in a back-to-back manner into a 50 ml falcon/centrifuge tube with several small holes (~2 mm) drilled on its cap (i.e., corning® 50 mL PP centrifuge tubes, Sigma Aldrich). The tubes are subsequently vacuum sealed in a vacuum bag, and stored at −80°C. Vacuum sealing prevents the sample from contacting with air and water, and placing the section slide into the tube prevents the damaging of the section as it avoids direct contact with the bag during storage. When ready for use, sections can be recovered for 2 h under vacuum (~50 Torr). Our recent MALDI imaging study on the distribution of flavonoids in apple suggests that there are no significant quantitative differences in detection between long-term-stored (9 months) and fresh-prepared apple sections (Franceschi and Wehrens, 2014). A possible alternative is to preserve the tissue section as imprints on flat surfaces like PTFE sheet (Cabral et al., 2013) (details about imprinting are discussed in Section Removal of the Plant Cuticle). Imprinting is a method which can be useful for samples that could readily wither and fade even when stored under low temperature, as, for example, leaves and flowers, but the effect of storage on the quality of the imprints has to be still evaluated.
Plant cells have rigid cell walls, large intercellular spaces and they are often rich in water, thereby embedding materials are often used in conventional plant histology practice to maintain the tissue morphology and ensure precise sample sectioning. Unfortunately, many of the commonly used embedding media are incompatible with MSI. For example, optimum temperature cutting (OCT) compound, which is a mixture of polyethylene glycols, is strongly discouraged since its use has been associated with diffusion into tissue and smearing across the surface during sectioning (Zaima et al., 2010b). Carboxymethyl cellulose (CMC) (Goto-Inoue et al., 2012), gelatin (Chen et al., 2009; Gemperline et al., 2015), ice (Khatib-Shahidi et al., 2006; Gorzolka et al., 2014) or their combinations (Nelson et al., 2013) have been successfully employed as MSI-compatible embedding mediums. Zaima and coworkers found that with the assistance of adhesive film, CMC embedding offers good sectioning performance (Zaima et al., 2010a; Yoshimura et al., 2012b). The use of adhesive film reduced the distortion of the sections and the dislocation of the analytes, additionally facilitating the transfer and attachment of sections to the slides. This method is described in Figure 2 (Kawamoto, 2003). It is important to mention that this method is initially proposed for histology and tissue staining so steps such as fixation, washing and staining should be avoided in MSI.
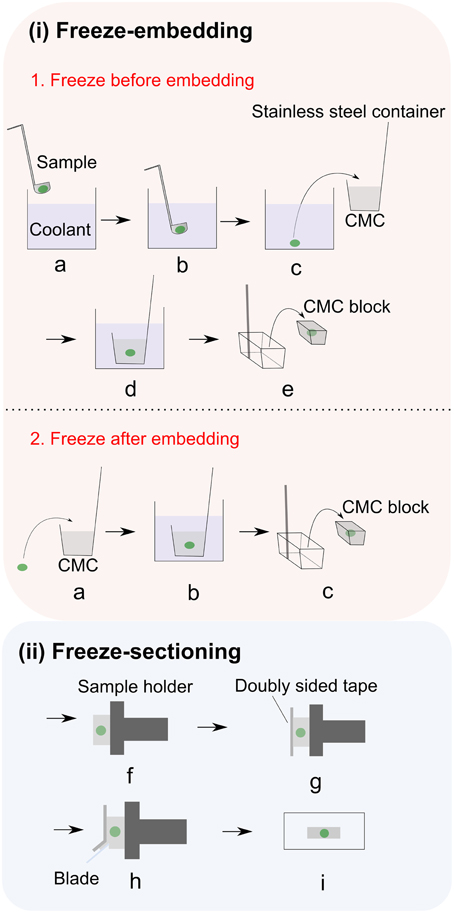
Figure 2. Adhesive film assisted CMC embedding method. (i) Freeze embedding can be achieved by either (1) freezing before embedding or (2) freezing after embedding. In freezing before embedding method: (a,b) Freezing of the sample in the coolant; (c) freeze-embedding of the frozen sample with 4–5% CMC gel; (d) completely freezing the CMC gel in coolant (i.e., hexane); (e) removing the CMC block from container;. In freezing after embedding method: (a) placing the sample into 4–5% CMC gel; (b) completely freezing the CMC gel in coolant (i.e., hexane); (c) removing the CMC block from container; (ii) After freeze embedding, the CMC block is subjected to freeze sectioning: (f) fixing the embedded sample to the sample holder with CMC gel and then attaching it to the cryomicrotome; (g) adhering the film to the exposed cutting surface; (h) cutting the sample; (i) placing sample section on the glass slide. Figure is adapted with permission from Kawamoto (2003).
Cryosectioning is the most commonly used method to prepare plant tissue slices as freezing well quenches metabolic processes. Before sectioning, samples are generally snap frozen in various low temperature conditions such as inside an ultra-low temperature freezer, or in powdered dry ice, liquid nitrogen or liquid nitrogen-chilled isopentane (Zaima et al., 2010b). Liquid nitrogen freezing usually makes plant sections brittle, and it results in ice crystal formation, thus rapid plunging of the tissue into liquid nitrogen is not recommended. Floating tissues in aluminum foil in liquid nitrogen (Schwartz et al., 2003; Nimesh et al., 2013) or freezing tissues on dry ice-chilled steel plate is more favored. Dry tissues like plant stems, can be also sectioned at room temperature (RT) using a microtome (Imai et al., 2005; Saito et al., 2008, 2012; Lunsford et al., 2011; Zhou et al., 2011) or vibratome (Lunsford et al., 2011). Sectioning at RT is only limited to some dry/dead plant tissues such as wood with dense and relatively hard structure. This is not suitable for most fresh tissues due to the dispersing of plant juice. Additionally, the enzyme-substrate reaction may still be active when tissues are sectioned at RT.
Section thickness is another important parameter to take into account because it affects the number and the intensity of the ion signals in MSI instruments. For example, poor ion intensity due to inefficient ionization can be expected in MALDI instruments where the “z” direction of the sample holder is not easy to adjust or the laser cannot be focused to the top of the thick tissue section. In addition, electrical non-conductivity (i.e., in non-orthogonal TOF-MS), high impurities, matrix absorption (in porous tissues) and tissue distortion (i.e., in linear/reflectron MALDI-TOF) in thick sections could also lead to the reduced signal intensity (Sugiura et al., 2006).
For mammalian tissues, section thickness between 5 and 20 μm is recommended for analysis of low molecular weight molecules, and < 5 μm thickness for high molecular weight proteins (m/z>9000) (Sugiura et al., 2006). In contrast, the relationship between tissue thickness and the quality of MS spectrum has been seldom studied in the MSI of plant sections. Generally it is difficult to cut thin slices of water rich plant tissues due to crumbling and fracturing of the tissues. The thickness of most plant sections in current MSI studies is about 50 μm, providing a good compromise between optimum MSI performance and practicality, especially when a large number of samples have to be prepared (Peukert et al., 2012).
Sample tissues can be either mounted onto a glass slide or on a MS-compatible plate (Schwartz et al., 2003). Different mounting surfaces are required depending on the ionization technique. For non-orthogonal MALDI TOF, conductive surfaces such as steel and metal-coated glass slides are required, while, for orthogonal MALDI TOF normal glass slides are sufficient. In contrast, in ambient ionization techniques like DESI, non-conductive surface is used in order to avoid the neutralization of charged spray solvent (Takats et al., 2005; Costa and Cooks, 2007, 2008).
Three mounting approaches are commonly used to attach plant sections onto the glass slide: the sample can be either secured by using double sided tape/epoxy glue or it can be thaw-mounted. The use of double sided tape is fast and easy, but care should be taken not to contaminate the sample. Epoxy glue is suitable for delicate samples and it does not produce extra mass signals (Kaftan et al., 2014). Thaw-mounting is usually used to attach tissue sections acquired by cryosectioning. This approach minimizes the risk of sample contamination, but relocation of water soluble analytes resulting from water condensation during thaw mounting is a major concern (Cha et al., 2008). In the case of water rich plant tissues, it is necessary to consider not only the condensation of atmospheric water, but also water originating from the sample itself. With this specific method, downstream sample processing steps should be minimized to avoid washing sample off the glass slide by any vigorous solution-based treatments (i.e., the washing steps in MSI of proteins for the purpose of removing, e.g., salts and lipids) (Goodwin et al., 2008). To alleviate this problem, thaw mounted samples are usually dried within a desiccator at a reduced pressure (Lee et al., 2012; Boughton et al., 2015).
As already discussed, molecules have to be ionized before MSI analysis, and in some case the ionization efficiency and its selectivity can be increased by using specific ionization aiding treatments. In SIMS, the ionization is direct: a focused high energy primary ion beam (e.g., Ar+, Ga+, In+) is used to strike the sample surface. The analyte molecules are then released from the surface and ionized upon collision with the primary ions (Amstalden van Hove et al., 2010). The high energy used in SIMS usually causes extensive secondary ion fragmentation, limiting its practical mass range to ~ m/z 1000 (Heeren et al., 2006). Recently, several strategies aiming at extending the potential of SIMS and at increasing the ionization efficiency of large intact biomolecule have been proposed, one of them is to coat the sample surface with common MALDI matrices, possible alternatives are metallization of samples with silver and gold (Delcorte et al., 2003; Altelaar et al., 2006).
In MALDI, the deposition of a matrix over the sample surface serves several functions, in particular: (i) extraction of analytes from the sample surface, (ii) co-crystallization of analytes and matrix, and (iii) absorption of the laser energy aiding desorption of the molecules from the surface into the gas phase, where ionization eventually occurs (Lewis et al., 2006). Earlier on, MALDI imaging was almost exclusively performed using conventional matrices such as α-cyano-4-hydroxycinnamic acid (CHCA) and 2,5-dihydroxybenzoic acid (DHB). However, the imaging of small molecules, especially those with molecular weights similar to the one of the matrix, is problematic due to the high degree of ion suppression caused by the presence of the dominant matrix ion signals. A variety of alternative matrices have been proposed to obviate this problem. The first possibility is to use ionless matrices which do not produce matrix-related interfering ions, i.e., 1,8-bis(dimethylamino) naphthalene (DMAN) (Shroff et al., 2009). However, it is important to point out that DMAN is instable in the high-vacuum MALDI (Thomas et al., 2012), which limits its application only in atmospheric pressure (AP)-MALDI. Alternatively, it is possible to choose high-molecule-weight matrices which do not generate ions in the low mass region [i.e., porphyrins (MW: 974.57) (van Kampen et al., 2009)]. Another possibility is to use inorganic matrices which show relatively clean background. This can be deposited over the sample surface (i.e., colloidal graphite, Zhang et al., 2007; Cha et al., 2008) or they can constitute the target plate, like in the case of electrochemically etched porous silicon (desorption/ionization on silicon MS, DIOS-MS) (Wei et al., 1999; Ronci et al., 2012). At the same time, it is also possible to use nanostructures coated with liquid-phase perfluorinated initiator molecules. This type of solution has been already used in LDI MS and NIMS (nanostructure initiator MS, NIMS) (Yanes et al., 2009).
MALDI matrix can be deposited on tissue sections in several ways. The most common methods include spraying, spotting, sublimation and dry coating. Spraying can be performed manually, i.e., with an airbrush, or automatically, using commercial devices like ImagePrep (Bruker) (Figure 3i) or TM-Sprayer™ (HTXImaging). Manual spraying requires skill because inhomogeneous matrix application and analyte delocalization can easily occur, while automatic solutions are more reproducible. In spotting, the matrix can also be delivered by micro-spotting to a specific sample surface location with robots like the CHIP-1000 chemical printer (Shimadzu). This method allows accurate deposition of matrix onto a tissue section, and this is good to attain a good quantitative MS signal generation in MSI (Vegvari et al., 2010). Sublimation allows fast and uniform matrix deposition (Figure 3ii). During sublimation, the matrix is placed inside a sublimation chamber, and the sample plate with the tissue is placed inverted over the top of the matrix and attached to a cold-finger (Norris and Caprioli, 2013). The matrix is heated at elevated temperature and under reduced pressure, while the sample itself remains cooled as heat could degrade the analytes. Sublimation is solvent free, therefore diffusion of most analyte molecules during matrix application is almost eliminated, even though it has been reported that lipids from very fatty tissues can to some degree diffuse by capillarity through dry matrix during storage (Berry et al., 2011). It is important to point out that, owing to the absence of solvent, the extraction efficiency can be poor for some compounds like peptides and proteins. Additional re-hydration has been demonstrated to improve the detection sensitivity (Bouschen et al., 2010; Yang and Caprioli, 2011). Other advantages of sublimation include high reproducibility, increased matrix purity, formation of fine matrix crystals and relatively low cost (Hankin et al., 2007). Finely ground matrices and nanomaterials can be also spread over the sample surface by using a fine mesh sieve (i.e., 20–50 μm) (Puolitaival et al., 2008; Chaurand et al., 2011). This method is simple, fast and meanwhile it avoids analyte delocalization, but also in this case the analyte extraction efficiency is reduced.
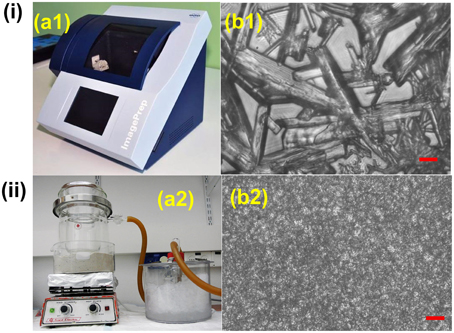
Figure 3. Comparison between (i) spray based (Bruker ImagePrep system) and (ii) vacuum sublimated DHB matrix in MALDI imaging. (a1) 30 gL−1 DHB (in 70% ACN solution) was used. Typical instrumental parameters include: 0.8 s spray, 10 s incubation, 70 s drying, and 60 spray cycles. (a2) 300 mg DHB was used. Major parameters include: ~120°C heating temperature, 0.05 Torr vacuum pressure, 10–11 min sublimation, matrix density 0.05 mgcm−2. (b1,b2): DHB crystal sizes resulting from Bruker ImagePrep (b1) and sublimation (b2), scale bar: 10 μm. Figures were adapted with the permission from Ref. (Hankin et al., 2007; Franceschi et al., 2012).
As far as DESI is concerned, the first element determining the ionization efficiency is the composition of the spray solvent, which should be optimized taking into account the metabolites under investigation and the specific characteristics of the samples (Badu-Tawiah et al., 2010; Green et al., 2010). In general, a higher fraction of water is used to have long lasting signals, while a higher proportion of methanol is used when higher spatial resolution is required (Manicke et al., 2008). When necessary, a reactive reagent can also be added to the spray solvent to selectively improve the ionization efficiency of analytes which are difficult to ionize under normal DESI conditions (Zhang and Chen, 2010; Muller et al., 2011; Lostun et al., 2015).
Fresh plant tissues would be ideal for MSI studies since they are chemically unmodified and treatment-free. In these conditions, the spatial arrangement of the molecules is preserved and the risk of chemical contamination during sample handling is minimized. Fresh tissues, however, can be directly analyzed only by using ambient ionization techniques, while they are likely to shrink during MSI analyses performed under vacuum (e.g., MALDI or SIMS). Sample shrinkage could result in mismatch between the MS image and the optical image (Cha et al., 2008), making biological interpretation difficult. Moreover, shrinkage during MSI analysis may cause unwanted mass shift during MSI analysis in non-orthogonal TOF-MS analyzer (as the flying time can be different for the same analyte located on an uneven surface), hampering molecule identification and reproducibility (Kulkarni et al., 2015). Another concern is that inside fresh samples the biological processes are still active and they may cause degradation and/or chemical modification during the analysis (Cha et al., 2009). In the case of infrared (IR)-MALDI and LAESI, since native water from the sample is employed as a matrix, the sample tissues have to be fresh or at least not totally dried out (Li et al., 2007a,b; Shrestha et al., 2011).
Most plant samples are vacuum- or freeze- dried prior to MSI. Vacuum desiccation can be applied for thin plant organs, such as leaves and flowers (Cha et al., 2008, 2009; Li et al., 2011; Korte et al., 2012), or for microtome or razor blade-sectioned samples, such as apple section (Franceschi et al., 2012). Vacuum pressure and drying time should be carefully optimized according to the nature of the sample and analytes of interest. It is likely, for example, that some compound (i.e., volatile essential oils, terpenes, alcohols and other small molecules) will be lost during vacuum desiccation, but the majority of molecules are expected to be unaffected due to their low vapor pressure (Franceschi et al., 2012). This has been confirmed by comparing MS profiles of target metabolites between fresh and dried Arabidopsis samples (Cha et al., 2008).
Land plant organs, such as leaves and flowers are covered with a lyophilic cuticular layer (0.1–10 μm in thickness) (Riederer and Schreiber, 2001). The cuticle serves for a variety of important biological functions, and MS imaging of plant cuticles is specifically discussed in Section Lipids and Fatty Acids. On the other hand, the cuticle poses a barrier for the MS imaging of the internal metabolites since soft ionization techniques such as MALDI and DESI cannot easily penetrate through it. LAESI-MSI is capable of depth profiling and it can be applied for imaging the internal metabolites, but its spatial resolution is still limited (typically 300 μm) (Nemes et al., 2009). Sample cuticle can be either physically removed or chemically washed off. For example, kaempferol and kaempferol rhamnoside were mostly detected in Arabidopsis leaf areas pre-treated with chloroform for 60 s (Figure 4ia,d,e) (Cha et al., 2008). However, those “aggressive methods” may delocalize and/or wash away the target compounds, as shown in the cases for C26 and C30 fatty acids (Figure 4ic,f). In addition, not all plant epidermis can be physically removed with ease as the in the case of barley leaf (Figure 4iia; Li et al., 2011), and scratching or grasping of the leaf cuticle will deter either the detection (Figure 4ie) or spatial resolution (Figure 4ia,c,d,f) of the analytes (Cha et al., 2008).
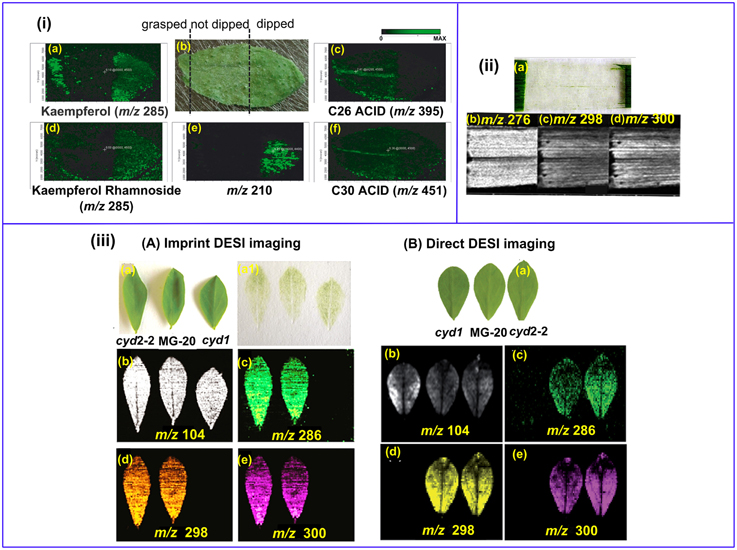
Figure 4. MS imaging of metabolites below the cuticle. (i) MALDI imaging of Arabidopsis leaf. The left area of the leaf was grasped using forceps, the middle part remained un-treated and the right area was chloroform-dipped (b); Kaempferol (a) and Kaempferol Rhamnoside (d) were readily detected in the forceps- grasped and chloroform-dipped areas, while C26 (c) and C30 (f) fatty acids were washed off in the chloroform-dipped area. An un-identified analyte m/z = 210 (d) showed high abundance in the chloroform-dipped area. (ii) DESI images of the hydroxynitrile glucosides of m∕z = 276 (b), 298 (c) and 300 (d) from barley leaf epidermis. The leaf abaxial epidermis strip was physically peeled off (a). (iii) Comparison of imprinting and direct DESI imaging of L. japonicus leaves. (A) Indirect DESI imaging of the leaf imprint on Teflon. (B) Direct DESI imaging of the leaf, with chloroform, methanol and water (1:1:0.4, v/v/v) as spray solvent. (a) Optical image of L. japonicus leaves. (a1) Optical image of the leaf imprints on Teflon. (b–e) DESI images of m/z 104, 286, 298, and 300, respectively. Figures were adapted with the permission from Cha et al. (2008), Li et al. (2011, 2013), and Bjarnholt et al. (2014), respectively.
Imprinting is a promising way to circumvent this problem. With this approach, plant tissues are pressed onto porous Teflon (Li et al., 2011, 2013; Thunig et al., 2011), porous polytetrafluoroethylene (PTFE) (Muller et al., 2011), TLC plate (Cabral et al., 2013; Hemalatha and Pradeep, 2013), print paper (Ifa et al., 2011), or tape (Tata et al., 2014) by applying a moderate pressure over the plant tissues, thereby transferring the plant metabolites onto a flat hard surface while keeping their spatial distribution. Li et al. successfully imaged the distribution of hydroxynitrile glucosides in leaves of L. japonicas with both imprinting (Li et al., 2013) and direct DESI imaging (Bjarnholt et al., 2014). The abundance of hydroxynitrile glucosides were rather even in imprinting DESI imaging (Figure 4iiiAb–e), while direct DESI imaging showed a decreased abundance of hydroxynitrile glucosides in the leaf midvein, which was possibly due to the reduced accessibility to the solvent spray in direct DESI imaging (Figure 4iiiBb–e). A recent publication demonstrated that the transfer efficiency could be improved with the assistance of solvent extraction and/or heating during TLC-imprinting (Cabral et al., 2013). However, imprinting is only efficient for relatively “fleshy” plant tissues and the spatial resolution is limited since analytes can be smeared during imprinting (Lee et al., 2012).
The chemical composition of the sample matrix can affect, in most cases negatively, the ionization efficiencies of the analytes of interest in MS analysis (this phenomenon is widely known as “matrix effect”). Although the limited sample pretreatment and the absence of separation are among the major advantages of MSI, they also make MS imaging prone to matrix effects, which alter the observed molecular distribution (Lanekoff et al., 2014). Matrix effects have been reported in SIMS (Jones et al., 2007), MALDI (Hankin et al., 2011; Janfelt et al., 2012; Wang et al., 2012), and nano-DESI imaging (Lanekoff et al., 2014). Since matrix effect is intrinsically associated with MS, it is also a major challenge in MSI of plant tissues even though it has not yet been well studied. Several strategies have been used to compensate for matrix effects in MSI, such as desalting the tissue sections prior to MSI analysis (Wang et al., 2012), and normalizing ion signals to the signals of their corresponding internal standards (Lanekoff et al., 2014).
Similarly to what happens in the case of the matrix effects, the physical properties of the sample can also influence the analyte ionization process. As a consequence, variations in the physical properties of a heterogeneous tissue surface can affect the ion yield, resulting in MS images misrepresenting the real distribution of the metabolites. This phenomenon has been reported in a MALDI imaging study of tobacco root, where a notable loss of ion signals was observed in the central root region due to the reason that the MALDI matrix was mostly absorbed in that region (Peukert et al., 2012). It has been proposed that spraying the sample surface with large amounts of matrix could reduce the surface effects at least in MALDI (Lee et al., 2012). In DESI, the sample surface effect can be more obvious. It has been shown that both the chemical (i.e., bond strength and polarity) (Takats et al., 2005; Ifa et al., 2008; Manicke et al., 2008; Volny et al., 2008; Benassi et al., 2009; Douglass et al., 2012) and the physical properties (i.e., conductivity and roughness) (Takats et al., 2005; Manicke et al., 2008) of the surface strongly affect the DESI results. In particular, they impact on the lower limits of detection/quantification (LOD/LOQ) of the analytes, on the signal stability, on the degree of carryover, but also on the reproducibility and on the linear dynamic range. Additionally, it has been shown that the DESI source acts as a direct current capacitor and the surface properties play an important role in the charge transfer (redox) process (Volny et al., 2008; Benassi et al., 2009; Dong et al., 2015). Imprinting can be a promising way to minimize sample surface effect in DESI imaging, since the analytes are transferred to a homogeneous surface.
Arguably, plant based MSI has historically been focused on methodological aspects (Matros and Mock, 2013), but now the technique has matured to a point where it has also been used to address biologically important questions (Lee et al., 2012) in fields like plant-environment interactions (Shroff et al., 2008, 2015; Klein et al., 2015; Ryffel et al., 2015; Soares et al., 2015; Tata et al., 2015), new compound identification (Jaeger et al., 2013; Debois et al., 2014), and functional genomics (Korte et al., 2012; Li et al., 2013). In the following section, we summarize the recent studies of MSI in plants organizing them on the bases of the class of molecules which were the subject of each specific study. The key characteristics of some selected applications are summarized in Table 1. Among the different aspects, we will focus on the choice of matrix for MALDI and of spray solvents for DESI since these elements are crucial in determining the quality of MSI analyses.
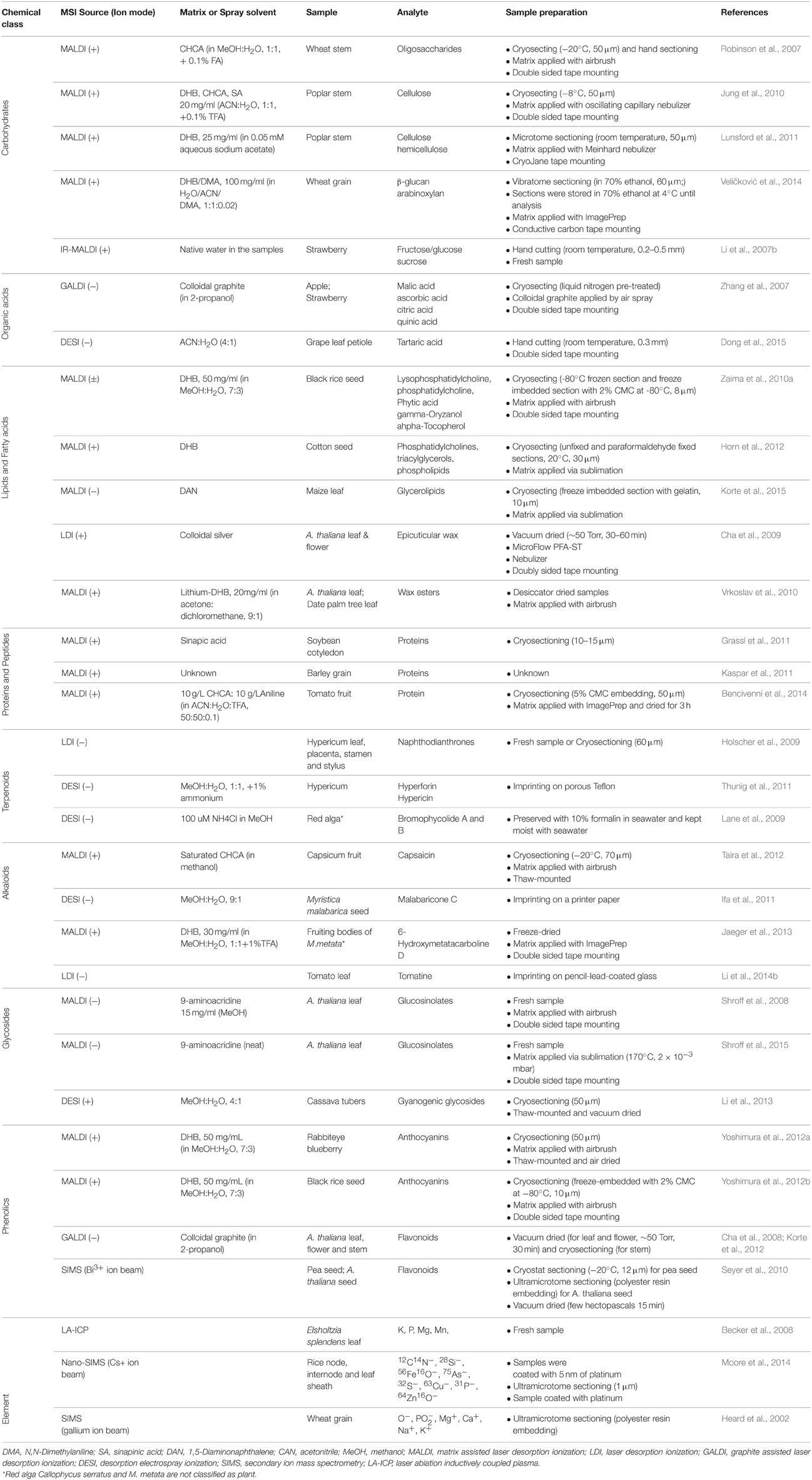
Table 1. Selected examples of studies in which MSI was used to detect different classes of plant metabolites.
Carbohydrates are initially synthesized through photosynthesis in plants, and they are well known for their essential roles. Additionally, they could also function as signaling molecules, in a way similar to hormones (Trouvelot et al., 2014).
Distribution of carbohydrates has been mapped by MALDI in several plant samples (Robinson et al., 2007; Jung et al., 2010; Yoshimura et al., 2012a; Veličković et al., 2014) and in these applications DHB and CHCA are the most common matrices. DHB has proved to be slightly better than CHCA in detecting small oligosaccharides such as glucose and sucrose (Zhang et al., 2007). Colloidal graphite (Graphite assisted laser desorption ionization, GALDI) has been proposed as an alternative matrix for imaging small oligosaccharides since it largely reduces the matrix interference in the small mass region (m∕z < 500) (Zhang et al., 2007). IR-MALDI has also been used to image carbohydrates distribution in different plants, such as strawberry (Li et al., 2007b) and lily flower (Li et al., 2008). In MALDI imaging, carbohydrates are mostly detected in positive ion mode. Application of DESI imaging in localization of carbohydrates in plants is rarely reported (Thunig et al., 2011), partially due to the low selectivity and sensitivity of this technique toward this class of molecules. Yet 3-nitrophenylboronic acid and N-methyl-4-pyridineboronic have been suggested as effective reagents added to the DESI spray solvent for in situ derivatization of sugars (reactive-DESI) to improve the ionization efficiency of intact sugars in complicated biological matrices (Zhang and Chen, 2010).
Lipids are a major component of plant tissues. They are present in all cells as constituents of the various cellular membranes. Other lipids such as waxes, fulfill important protective functions both in plant leaves and fruits. Pigmented lipids are involved in light harvesting and energy transduction, while others are concerned in electron transport processes. Certain plant lipids also represent energy stores that can be mobilized and consumed for growth and development of plants (Harwood, 1996).
The localization of various unsaturated lipids has been mapped in rice (Zaima et al., 2010a) and cotton seeds (Horn et al., 2012), Camelina sativa seed and avocado fruit (Horn et al., 2013) using MALDI imaging. In these tissues, lipids are readily detected as multiple adducted ions (primarily H+, Na+, and K+) in positive ion mode by using DHB as a matrix. MALDI imaging of saturated hydrocarbons (HCs) however, is more challenging because these species do not contain any polar groups neither susceptible to protonation nor to which cations or anions can be easily attached (Cvacka and Svatos, 2003). When monovalent cations of transition metals (e.g., Fe, Mn, Cu) are co-deposited on MALDI target with HCs, they give cationized species which can be detected in a mass spectrometer. However, due to the high reactivity of transition metals, HCs are easily fragmented during analysis, hampering molecular identification (Cvacka and Svatos, 2003). The reactivity of silver with HCs is lower than that of any other transition metals so it can be used to generate intact silver adduct ions. MALDI imaging of epicuticular wax in Arabidopsis has been successfully reported by using silver colloid as matrix. In this study, 14 cuticular wax compounds were identified in Arabidopsis wild-type (Ler) and genetic stock CS8 (which carries carried the mutant alleles cer2-2, ap2-1, and bp1) leaves (Cha et al., 2009). The pitfall of silver matrix is that silver is present with similar abundance for its two stable isotopes. Each molecule, then, produces a group of silver adduct ion peaks with two major ions [monoisotopic mass of the metabolite + 107Ag or 109Ag]+, making compound identification and quantification difficult (Cvacka and Svatos, 2003; Cha et al., 2009). Reports on MALDI imaging of saturated wax esters in Arabidopsis and date palm leaves suggested that the lithium salt of DHB (LiDHB) is the most versatile matrix for detection of a majority of neutral lipids and it can potentially replace the currently used silver salts (Vrkoslav et al., 2010).
Localization of lipids is the most frequent application of DESI imaging in mammalian tissues, while DESI imaging of lipids in plants has not yet been reported. Since lipids are more readily ionized by DESI, DESI would be an ideal complementary tool to MALDI for mapping lipids in plants where high spatial resolution is not required. Mixtures of water-methanol or water-acetonitrile, with or without an acidic modifier are the most commonly used spray solvents for DESI imaging of lipids (Eberlin et al., 2011).
The fundamental component of a protein is the polypeptide chain composed of amino acid residues. Proteins are highly ubiquitous either as plant storage proteins (e.g., legumin, vicilin, convicilin, albumin, and gliadin) or as functional proteins such as enzymes, membrane components or hormones (Aluko, 2015).
MSI studies of proteins and peptides are in general particularly challenging, and, up to now, they are well described in mammalian tissues. Sample preparation is the first critical point for this type of studies and it is more challenging than for other molecules. The different protocols include several additional washing steps to remove endogenous molecular species, such as salts and sugars, which may interfere with protein desorption/ionization efficiency, to ensure tissue dehydration and fixation, and to prevent proteolysis (Schwartz et al., 2003). The wash procedure varies in solvent composition, temperature and duration depending on the tissue, and it needs to be optimized accordingly. Even after careful optimization, proteins larger than 25kDa are not routinely detectable by MALDI MSI, as they are not efficiently stabilized in the matrix solution and are not extractable from the tissue (Franck et al., 2010). On-tissue digestion of large proteins can be used to detect and identify larger proteins, but the treatment with proteolytic enzymes enhances analyte diffusion thus reducing the spatial resolution in MSI studies (Kaspar et al., 2011). In the specific case of plants, protein identification is also challenging, due to the lack of extensive and reliable databases (Kaspar et al., 2011).
The application of MSI of proteins in plants has, however, been illustrated in few cases, as demonstrated by the MALDI imaging analysis of proteins in developing barley grains (Kaspar et al., 2011) and in soybean cotyledons (Grassl et al., 2011), where tissue-specific protein expression patterns have been revealed. A detailed analytical protocol is presented and discussed in the latter example, there sinapinic acid (SA) is suggested as matrix for MALDI imaging of proteins (>3000 Da), and CHCA and DHB for peptides (< 3000 Da).
Terpenoids are a large and diverse class of metabolites which are built up from isoprene. Plants employ terpenoids for a variety of functions in growth and development but use the majority of them for more specialized chemical interactions and protection in the abiotic and biotic environment (Tholl, 2015).
Distribution of terpenoids in Hypericum perforatum have been studied by LDI imaging on fresh tissues (Holscher et al., 2009) and by DESI imaging on leaves and their imprints (Thunig et al., 2011) respectively. The results of the two studies are in complete agreement showing that hyperforin and adhyperforin are found in translucent glands, and hypericin, pseudohypericin, protopseudohypericin, and protohypericin are exclusively located in dark glands in leaves. Since they are highly UV absorbing compounds, application of a matrix is not necessary and thereby LDI allows 10 μm spatial resolution with a 10 × 10 μm laser focus setting, removing the spatial limitations associated with the use of a matrix. It is important to point out that the understanding of the biological function of natural products requires direct fine-scale evaluation in the tissue of the producing organism (Esquenazi et al., 2009). One example is the DESI imaging of a tropical red alga tissue surface where bromophycolide A and B are found exclusively distributed in association with distinct surface patches at concentrations sufficient to inhibit the detrimental Lindra thalassiae fungus (Lane et al., 2009).
Alkaloids contain secondary, tertiary, or quaternary nitrogen atoms. In plants, they act as defense compounds against pathogenic organisms and herbivores, but they can also be used as protoxins by insects which further modify them before incorporating them into their own defense system or secretions (Ghosh, 2000). Alkaloids are typically found in particular medicinal plants organs, often defined as a “medicinal part” (Shitan and Yazaki, 2007). Knowing their localization is valuable not only to understand their metabolic origins, but also to optimize their isolation process from the compound containing parts.
Rapid profiling of alkaloids has been performed in several plant species by DESI and MALDI. Relevant examples include DESI profiling of alkaloids in Conium maculatum, Datura stramonium, and Atropa belladonna (Talaty et al., 2005), and MALDI profiling of alkaloids in Rhizoma Coptidis and Strychnos nux-vomica L. (Lu et al., 2010). This type of studies provided the basic protocols for MS imaging of alkaloids in plants. Distribution of alkaloids has been mapped for several purposes: for example MALDI imaging of capsaicin in capsicum fruits (Taira et al., 2012) and DESI imaging of malabaricone C. in Myristica malabarica seed (Ifa et al., 2011) to study their metabolic origin, and MALDI imaging of fruiting bodies of a mushroom to screen for new compounds (Jaeger et al., 2013). All the above examples were performed in positive ion mode, and alkaloids are mostly detected as [M+H]+, and in some cases as salt adducts, such as [M+K]+.
Phenolics are compounds possessing one or more aromatic rings with one or more hydroxyl groups. Plant phenolics include phenolics acids, flavonoids, tannins and the less common stilbenes and lignans. They are generally involved in defense against ultraviolet radiation or aggression by pathogens, parasites and predators, as well as contributing to plants' colors (Dai and Mumper, 2010).
Distribution of phenolics has been mapped in several plants including strawberry (Zhang et al., 2007), grapevine leaf (Becker et al., 2014), apple (Zhang et al., 2007; Franceschi et al., 2012), Arabidopsis (Cha et al., 2008; Holscher et al., 2009), rice (Yoshimura et al., 2012b), licorice (Li et al., 2014a) and blueberry (Yoshimura et al., 2012a) by MALDI, GALDI, and LDI imaging. Flavonols are mainly detected as [M-H]− in negative ion mode, while anthocyanins are mostly detected in positive mode as [M]+. Both CHCA and DHB are common matrices used for the analysis of phenolics in MALDI imaging. DESI imaging was recently applied to localize flavonols in ginkgo leaves in negative ionization mode, and anthocyanins in strawberry in positive mode (Cabral et al., 2013). Information on their distribution has been used to explain gene expression patterns in the wild-type and mutant (transparent testa; tt7) Arabidopsis flowers. In particular, the Arabidopsis tt7 mutant effectively blocks the production of quercetin, isorhamnetin and their glycoside derivatives, but leads to the accumulation of kaempferol and its glycoside glycosides. By imaging the distribution of several flavonoids in wild-type and the tt7 mutant flowers, the MSI results could explain the expression of the TT7 gene localization to the proximal part of the petal and the expression of the other genes of the upstream pathway that are evenly expressed throughout the petal (Korte et al., 2012).
The mapping of the biologically essential trace metals (i.e., Cu, Zn, Fe), metalloids (i.e., Se), or non-metals (i.e., S, P, I) is of increasing interest in modern bio-analytics (Becker et al., 2010). LA-ICP-MS and SIMS are two major MSI ion sources of choice for trace elements localization (Heard et al., 2002; Zhu et al., 2012; Choi et al., 2014; Hanć et al., 2014). In particular, the absence of charging-up effects combined with fewer matrix effects makes LA-ICP-MS a simple quantification tool in which is possible to apply certified standard reference materials or matrix-matched laboratory standards (Becker et al., 2010). For example, with the apple leaf SRM NIST 1515 as a certified standard reference material, distribution of K, P, Mg, and Mn in Elsholtzia splendens leaves has been quantitatively determined employing LA-ICP-MS imaging. The results showed that the four elements were predominantly located in the leaf veins, which highlighted the importance of the vein in transporting macro-essential elements (Becker et al., 2008). Other attractive feature of SIMS and LA-ICP-MS is that they require little or no sample preparation.
The goal and philosophy of sample handling in MSI is to preserve the integrity of the tissue samples, keeping the original localization of the analytes within the tissues, and increasing the amount of ions desorbed from the sample surface. The importance of sample preparation for plants has been highlighted in several recent excellent reviews (Fujimura and Miura, 2014; Horn and Chapman, 2014; Spengler, 2014; Boughton et al., 2015; Sturtevant et al., 2016). A number of factors are reported to be critical for successful sample preparation in MSI, and they range from tissue storage, sectioning and mounting to the selection of the optimal ionization aiding treatment. It is worth noting that each factor should be carefully optimized depending on the characteristics of the MSI instrument, the nature of the sample and the analytes of interest.
Among the different aspects, it is important to remark that further improvement in reproducibility in sample preparation is highly needed in order to allow reliable inter-sample comparison, and to improve serial-section based 3D MSI. In this regard, the setting-up of simplified and automatic sample preparation pipelines is required.
In addition, we believe that further development of sample preparation strategies able to account for tissue-specific ion suppression (i.e., local chemical and morphological suppression) is needed in order to increase molecular coverage and improve the quantitative potential of MSI.
It is anticipated that these advances in sample preparation will largely expand the potential of MSI in plant sciences.
Conceptualization, DY and FP; Methodology, DY, FP, and AA; Investigation, DY, MS, and RI; Writing—Original Draft, DY; Writing-Review and Editing, FP, LB, AA, MS, RI, KF, and SA; Visualization, DY and LB; Funding Acquisition, FP and AA; Supervision, FP and AA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Tom and Sondra Rykoff Family Foundation Research and the Israeli Centers of Research Excellence (i-CORE) program on Plant Adaptation to Changing Environment for supporting the AA lab activity. AA is the incumbent of the Peter J. Cohn Professorial Chair.
Altelaar, A. M., Klinkert, I., Jalink, K., de Lange, R. P., Adan, R. A., Heeren, R. M., et al. (2006). Gold-enhanced biomolecular surface imaging of cells and tissue by SIMS and MALDI mass spectrometry. Anal. Chem. 78, 734–742. doi: 10.1021/ac0513111
Aluko, R. E. (2015). Structure and function of plant protein-derived antihypertensive peptides. Curr. Opin. Food Sci. 4, 44–50. doi: 10.1016/j.cofs.2015.05.002
Amstalden van Hove, E. R., Smith, D. F., and Heeren, R. M. (2010). A concise review of mass spectrometry imaging. J. Chromatogr. A 1217, 3946–3954. doi: 10.1016/j.chroma.2010.01.033
Badu-Tawiah, A., Bland, C., Campbell, D. I., and Cooks, R. G. (2010). Non-aqueous spray solvents and solubility effects in desorption electrospray ionization. J. Am. Soc. Mass Spectrom. 21, 572–579. doi: 10.1016/j.jasms.2009.12.012
Becker, J. S., Zoriy, M., Matusch, A., Wu, B., Salber, D., Palm, C., et al. (2010). Bioimaging of metals by laser ablation inductively coupled plasma mass spectrometry (LA−ICP−MS). Mass Spectrom. Rev. 29, 156–175. doi: 10.1002/mas.20239
Becker, J. S., Zoriy, M., Wu, B., Matusch, A., and Becker, J. S. (2008). Imaging of essential and toxic elements in biological tissues by LA-ICP-MS. J. Anal. At. Spectrom. 23, 1275–1280. doi: 10.1039/b805228j
Becker, L., Carré, V., Poutaraud, A., Merdinoglu, D., and Chaimbault, P. (2014). MALDI mass spectrometry imaging for the simultaneous location of resveratrol, pterostilbene and viniferins on grapevine leaves. Molecules 19, 10587–10600. doi: 10.3390/molecules190710587
Benassi, M., Wu, C. P., Nefliu, M., Ifa, D. R., Volny, M., and Cooks, R. G. (2009). Redox transformations in desorption electrospray ionization. Int. J. Mass Spectrom. 280, 235–240. doi: 10.1016/j.ijms.2008.10.012
Bencivenni, M., Faccini, A., Zecchi, R., Boscaro, F., Moneti, G., Dossena, A., et al. (2014). Electrospray MS and MALDI imaging show that non-specific lipid-transfer proteins (LTPs) in tomato are present as several isoforms and are concentrated in seeds. J. Mass Spectrom. 49, 1264–1271. doi: 10.1002/jms.3454
Berry, K. A. Z., Hankin, J. A., Barkley, R. M., Spraggins, J. M., Caprioli, R. M., and Murphy, R. C. (2011). MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem. Rev. 11, 6491–6512. doi: 10.1021/cr200280p
Bhardwaj, C., and Hanley, L. (2014). Ion sources for mass spectrometric identification and imaging of molecular species. Nat. Prod. Rep. 31, 756–767. doi: 10.1039/c3np70094a
Bjarnholt, N., Li, B., D'Alvise, J., and Janfelt, C. (2014). Mass spectrometry imaging of plant metabolites-principles and possibilities. Nat. Prod. Rep. 31, 818–837. doi: 10.1039/C3NP70100J
Boughton, B. A., Thinagaran, D., Sarabia, D., Bacic, A., and Roessner, U. (2015). Mass spectrometry imaging for plant biology: a review. Phytochem. Rev. 14, 1–44. doi: 10.1007/s11101-015-9440-2
Bouschen, W., Schulz, O., Eikel, D., and Spengler, B. (2010). Matrix vapor deposition/recrystallization and dedicated spray preparation for high-resolution scanning microprobe matrix-assisted laser desorption/ionization imaging mass spectrometry (SMALDI-MS) of tissue and single cells. Rapid Commun. Mass Spectrom. 24, 355–364. doi: 10.1002/rcm.4401
Cabral, E. C., Mirabelli, M. F., Perez, C. J., and Ifa, D. R. (2013). Blotting assisted by heating and solvent extraction for DESI-MS imaging. J. Am. Soc. Mass Spectrom. 24, 956–965. doi: 10.1007/s13361-013-0616-y
Cha, S., Song, Z., Nikolau, B. J., and Yeung, E. S. (2009). Direct profiling and imaging of epicuticular waxes on Arabidopsis thaliana by laser desorption/ionization mass spectrometry using silver colloid as a matrix. Anal. Chem. 81, 2991–3000. doi: 10.1021/ac802615r
Cha, S., Zhang, H., Ilarslan, H. I., Wurtele, E. S., Brachova, L., Nikolau, B. J., et al. (2008). Direct profiling and imaging of plant metabolites in intact tissues by using colloidal graphite-assisted laser desorption ionization mass spectrometry. Plant J. 55, 348–360. doi: 10.1111/j.1365-313X.2008.03507.x
Chaurand, P., Cornett, D. S., Angel, P. M., and Caprioli, R. M. (2011). From whole-body sections down to cellular level, multiscale imaging of phospholipids by MALDI mass spectrometry. Mol. Cell. Proteomics 10:O110.004259. doi: 10.1074/mcp.o110.004259
Chen, R. B., Hui, L. M., Sturm, R. M., and Li, L. J. (2009). Three dimensional mapping of neuropeptides and lipids in crustacean brain by mass spectral imaging. J. Am. Soc. Mass Spectrom. 20, 1068–1077. doi: 10.1016/j.jasms.2009.01.017
Choi, S. H., Kim, J. S., Lee, J. Y., Jeon, J. S., Kim, J. W., Russo, R. E., et al. (2014). Analysis of arsenic in rice grains using ICP-MS and fs LA-ICP-MS. J. Anal. At. Spectrom. 29, 1233–1237. doi: 10.1039/c4ja00069b
Costa, A. B., and Cooks, R. G. (2007). Simulation of atmospheric transport and droplet-thin film collisions in desorption electrospray ionization. Chem. Commun. 38, 3915–3917. doi: 10.1039/b710511h
Costa, A. B., and Cooks, R. G. (2008). Simulated splashes: elucidating the mechanism of desorption electrospray ionization mass spectrometry. Chem. Phys. Lett. 464, 1–8. doi: 10.1016/j.cplett.2008.08.020
Cvacka, J., and Svatos, A. (2003). Matrix-assisted laser desorption/ionization analysis of lipids and high molecular weight hydrocarbons with lithium 2,5-dihydroxybenzoate matrix. Rapid. Commun. Mass Spectrom. 17, 2203–2207. doi: 10.1002/rcm.1178
Dai, J., and Mumper, R. J. (2010). Plant phenolics: extraction, analysis and their antioxidant and anticancer properties. Molecules 15, 7313–7352. doi: 10.3390/molecules15107313
Debois, D., Jourdan, E., Smargiasso, N., Philippe, T., De Pauw, E., and Ongena, M. (2014). Spatiotemporal monitoring of the antibiome secreted by Bacillus biofilms on plant roots using MALDI Mass Spectrometry Imaging. Anal. Chem. 86, 4431–4438. doi: 10.1021/ac500290s
Delcorte, A., Bour, J., Aubriet, F., Muller, J.-F., and Bertrand, P. (2003). Sample metallization for performance improvement in desorption/ionization of kilodalton molecules: quantitative evaluation, imaging secondary ion MS, and laser ablation. Anal. Chem. 75, 6875–6885. doi: 10.1021/ac0302105
Dill, A. L., Eberlin, L. S., Costa, A. B., Ifa, D. R., and Cooks, R. G. (2011). Data quality in tissue analysis using desorption electrospray ionization. Anal. Bioanal. Chem. 401, 1949–1961. doi: 10.1007/s00216-011-5249-z
Dong, Y., Guella, G., Mattivi, F., and Franceschi, P. (2015). High production of small organic dicarboxylate dianions by DESI and ESI. J. Am. Soc. Mass Spectrom. 26, 386–389. doi: 10.1007/s13361-014-1065-y
Douglass, K. A., Jain, S., Brandt, W. R., and Venter, A. R. (2012). Deconstructing desorption electrospray ionization: independent optimization of desorption and ionization by spray desorption collection. J. Am. Soc. Mass Spectrom. 23, 1896–1902. doi: 10.1007/s13361-012-0468-x
Eberlin, L. S., Ferreira, C. R., Dill, A. L., Ifa, D. R., and Cooks, R. G. (2011). Desorption electrospray ionization mass spectrometry for lipid characterization and biological tissue imaging. Biochim. Biophys. Acta 1811, 946–960. doi: 10.1016/j.bbalip.2011.05.006
Esquenazi, E., Dorrestein, P. C., and Gerwick, W. H. (2009). Probing marine natural product defenses with DESI-imaging mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 106, 7269–7270. doi: 10.1073/pnas.0902840106
Franceschi, P., Dong, Y., Strupat, K., Vrhovsek, U., and Mattivi, F. (2012). Combining intensity correlation analysis and MALDI imaging to study the distribution of flavonols and dihydrochalcones in Golden Delicious apples. J. Exp. Bot. 63, 1123–1133. doi: 10.1093/jxb/err327
Franceschi, P., and Wehrens, R. (2014). Self-organizing maps: a versatile tool for the automatic analysis of untargeted imaging datasets. Proteomics 14, 853–861. doi: 10.1002/pmic.201300308
Franck, J., Longuespee, R., Wisztorski, M., Van Remoortere, A., Van Zeijl, R., Deelder, A., et al. (2010). MALDI mass spectrometry imaging of proteins exceeding 30,000 daltons. Med. Sci. Monit. 16, BR293–BR299.
Fujimura, Y., and Miura, D. (2014). MALDI mass spectrometry imaging for visualizing in situ metabolism of endogenous metabolites and dietary phytochem 10.3390/metabo4020319 icals. Metabolites 4, 319–346. doi: 10.3390/metabo4020319
Gemperline, E., Jayaraman, D., Maeda, J., Ané, J.-M., and Li, L. (2015). Multifaceted investigation of metabolites during nitrogen fixation in medicago via high resolution MALDI-MS imaging and ESI-MS. J. Am. Soc. Mass Spectrom. 26, 149–158. doi: 10.1007/s13361-014-1010-0
Goodwin, R. J. (2012). Sample preparation for mass spectrometry imaging: small mistakes can lead to big consequences. J. Proteomics 75, 4893–4911. doi: 10.1016/j.jprot.2012.04.012
Goodwin, R. J., Pennington, S. R., and Pitt, A. R. (2008). Protein and peptides in pictures: imaging with MALDI mass spectrometry. Proteomics 8, 3785–3800. doi: 10.1002/pmic.200800320
Gorzolka, K., Bednarz, H., and Niehaus, K. (2014). Detection and localization of novel hordatine-like compounds and glycosylated derivates of hordatines by imaging mass spectrometry of barley seeds. Planta 239, 1321–1335. doi: 10.1007/s00425-014-2061-y
Goto-Inoue, N., Hayasaka, T., Zaima, N., Nakajima, K., Holleran, W. M., Sano, S., et al. (2012). Imaging mass spectrometry visualizes ceramides and the pathogenesis of dorfman-chanarin syndrome due to ceramide metabolic abnormality in the skin. PLoS ONE 7:e49519. doi: 10.1371/journal.pone.0049519
Grassl, J., Taylor, N. L., and Millar, A. H. (2011). Matrix-assisted laser desorption/ionisation mass spectrometry imaging and its development for plant protein imaging. Plant Methods 7:21. doi: 10.1186/1746-4811-7-21
Green, F. M., Salter, T. L., Gilmore, I. S., Stokes, P., and O'Connor, G. (2010). The effect of electrospray solvent composition on desorption electrospray ionisation (DESI) efficiency and spatial resolution. Analyst 135, 731–737. doi: 10.1039/b924208b
Hanć, A., Piechalak, A., Tomaszewska, B., and Barałkiewicz, D. (2014). Laser ablation inductively coupled plasma mass spectrometry in quantitative analysis and imaging of plant's thin sections. Int. J. Mass Spectrom. 363, 16–22. doi: 10.1016/j.ijms.2014.01.020
Hankin, J. A., Barkley, R. M., and Murphy, R. C. (2007). Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 18, 1646–1652. doi: 10.1016/j.jasms.2007.06.010
Hankin, J. A., Farias, S. E., Barkley, R. M., Heidenreich, K., Frey, L. C., Hamazaki, K., et al. (2011). MALDI mass spectrometric imaging of lipids in rat brain injury models. J. Am. Soc. Mass Spectrom. 22, 1014–1021. doi: 10.1007/s13361-011-0122-z
Harwood, J. L. (1996). Recent advances in the biosynthesis of plant fatty acids. Biochim. Biophys. Acta. 1301, 7–56. doi: 10.1016/0005-2760(95)00242-1
Heard, P. J., Feeney, K. A., Allen, G. C., and Shewry, P. R. (2002). Determination of the elemental composition of mature wheat grain using a modified secondary ion mass spectrometer (SIMS). Plant J. 30, 237–245. doi: 10.1046/j.1365-313X.2001.01276.x
Heeren, R. M. A., McDonnell, L. A., Amstalden, E., Luxembourg, S. L., Altelaar, A. F. M., and Piersma, S. R. (2006). Why don't biologists use SIMS? A critical evaluation of imaging MS. Appl. Surf. Sci. 252, 6827–6835. doi: 10.1016/j.apsusc.2006.02.134
Hemalatha, R. G., and Pradeep, T. (2013). Understanding the molecular signatures in leaves and flowers by desorption electrospray ionization mass spectrometry (DESI MS) imaging. J. Agric. Food Chem. 61, 7477–7487. doi: 10.1021/jf4011998
Heyman, H. M., and Dubery, I. A. (2015). The potential of mass spectrometry imaging in plant metabolomics: a review. Phytochem. Rev. 14, 1–20. doi: 10.1007/s11101-015-9416-2
Holscher, D., Shroff, R., Knop, K., Gottschaldt, M., Crecelius, A., Schneider, B., et al. (2009). Matrix-free UV-laser desorption/ionization (LDI) mass spectrometric imaging at the single-cell level: distribution of secondary metabolites of Arabidopsis thaliana and Hypericum species. Plant J. 60, 907–918. doi: 10.1111/j.1365-313X.2009.04012.x
Horn, P. J., and Chapman, K. D. (2014). Lipidomics in situ: insights into plant lipid metabolism from high resolution spatial maps of metabolites. Prog. Lipid Res. 54, 32–52. doi: 10.1016/j.plipres.2014.01.003
Horn, P. J., James, C. N., Gidda, S. K., Kilaru, A., Dyer, J. M., Mullen, R. T., et al. (2013). Identification of a new class of lipid droplet-associated proteins in plants. Plant Physiol. 162, 1926–1936. doi: 10.1104/pp.113.222455
Horn, P. J., Korte, A. R., Neogi, P. B., Love, E., Fuchs, J., Strupat, K., et al. (2012). Spatial mapping of lipids at cellular resolution in embryos of cotton. Plant Cell 24, 622–636. doi: 10.1105/tpc.111.094581
Ifa, D. R., Manicke, N. E., Rusine, A. L., and Cooks, R. G. (2008). Quantitative analysis of small molecules by desorption electrospray ionization mass spectrometry from polytetrafluoroethylene surfaces. Rapid Commun. Mass Spectrom. 22, 503–510. doi: 10.1002/rcm.3377
Ifa, D. R., Srimany, A., Eberlin, L. S., Naik, H. R., Bhat, V., Cooks, R. G., et al. (2011). Tissue imprint imaging by desorption electrospray ionization mass spectrometry. Anal. Methods 3, 1910–1912. doi: 10.1039/c1ay05295k
Imai, T., Tanabe, K., Kato, T., and Fukushima, K. (2005). Localization of ferruginol, a diterpene phenol, in Cryptomeria japonica heartwood by time-of-flight secondary ion mass spectrometry. Planta 221, 549–556. doi: 10.1007/s00425-004-1476-2
Jaeger, R. J., Lamshoft, M., Gottfried, S., Spiteller, M., and Spiteller, P. (2013). HR-MALDI-MS imaging assisted screening of beta-carboline alkaloids discovered from Mycena metata. J. Nat. Prod. 76, 127–134. doi: 10.1021/np300455a
Janfelt, C., Wellner, N., Leger, P.-L., Kokesch-Himmelreich, J., Hansen, S. H., Charriaut-Marlangue, C., et al. (2012). Visualization by mass spectrometry of 2-dimensional changes in rat brain lipids, including N-acylphosphatidylethanolamines, during neonatal brain ischemia. FASEB J. 26, 2667–2673. doi: 10.1096/fj.11-201152
Jones, E. A., Lockyer, N. P., and Vickerman, J. C. (2007). Mass spectral analysis and imaging of tissue by ToF-SIMS-The role of buckminsterfullerene, , primary ions. Int. J. Mass Spectrom. 260, 146–157. doi: 10.1016/j.ijms.2006.09.015
Jung, S., Chen, Y., Sullards, M. C., and Ragauskas, A. J. (2010). Direct analysis of cellulose in poplar stem by matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun. Mass Spectrom. 24, 3230–3236. doi: 10.1002/rcm.4757
Kaftan, F., Vrkoslav, V., Kynast, P., Kulkarni, P., Böcker, S., Cvačka, J., et al. (2014). Mass spectrometry imaging of surface lipids on intact Drosophila melanogaster flies. J. Mass Spectrom. 49, 223–232. doi: 10.1002/jms.3331
Kaspar, S., Peukert, M., Svatos, A., Matros, A., and Mock, H. P. (2011). MALDI-imaging mass spectrometry - an emerging technique in plant biology. Proteomics 11, 1840–1850. doi: 10.1002/pmic.201000756
Kawamoto, T. (2003). Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole-animals, insects and plants. Arch. Histol. Cytol. 66, 123–143. doi: 10.1679/aohc.66.123
Khatib-Shahidi, S., Andersson, M., Herman, J. L., Gillespie, T. A., and Caprioli, R. M. (2006). Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal. Chem. 78, 6448–6456. doi: 10.1021/ac060788p
Klein, A. T., Yagnik, G. B., Hohenstein, J. D., Ji, Z., Zi, J., Reichert, M. D., et al. (2015). Investigation of the chemical interface in the soybean-aphid and rice-bacteria interactions using MALDI-mass spectrometry imaging. Anal. Chem. 87, 5294–5301. doi: 10.1021/acs.analchem.5b00459
Korte, A. R., Song, Z., Nikolau, B. J., and Lee, Y. J. (2012). Mass spectrometric imaging as a high-spatial resolution tool for functional genomics: tissue-specific gene expression of TT7 inferred from heterogeneous distribution of metabolites in Arabidopsis flowers. Anal. Methods 4, 474–481. doi: 10.1039/C2AY05618F
Korte, A. R., Yandeau-Nelson, M. D., Nikolau, and, B. J., and Lee, Y. J. (2015). Subcellular-level resolution MALDI-MS imaging of maize leaf metabolites by MALDI-linear ion trap-Orbitrap mass spectrometer. Anal. Bioanal. Chem. 407, 2301–2309. doi: 10.1007/s00216-015-8460-5
Kulkarni, P., Kaftan, F., Kynast, P., Svatoš, A., and Böcker, S. (2015). Correcting mass shifts: a lock mass-free recalibration procedure for mass spectrometry imaging data. Anal. Bioanal. Chem. 407, 7603–7613. doi: 10.1007/s00216-015-8935-4
Lane, A. L., Nyadong, L., Galhena, A. S., Shearer, T. L., Stout, E. P., Parry, R. M., et al. (2009). Desorption electrospray ionization mass spectrometry reveals surface-mediated antifungal chemical defense of a tropical seaweed. Proc. Natl. Acad. Sci. U. S. A. 106, 7314–7319. doi: 10.1073/pnas.0812020106
Lanekoff, I., Stevens, S. L., Stenzel-Poore, M. P., and Laskin, J. (2014). Matrix effects in biological mass spectrometry imaging: identification and compensation. Analyst 139, 3528–3532. doi: 10.1039/c4an00504j
Lee, Y. J., Perdian, D. C., Song, Z., Yeung, E. S., and Nikolau, B. J. (2012). Use of mass spectrometry for imaging metabolites in plants. Plant J. 70, 81–95. doi: 10.1111/j.1365-313X.2012.04899.x
Lewis, J. K., and Wei, J., Siuzdak, G. (2006). “Matrix-assisted laser desorption/ionization mass spectrometry in peptide and protein analysis,” in Encyclopedia of Analytical Chemistry, ed R. A. Meyers (Chichester: John Wiley & Sons Ltd), 5880–5894.
Li, B., Bhandari, D. R., Janfelt, C., Römpp, A., and Spengler, B. (2014a). Natural products in licorice (Glycyrrhiza glabra) rhizome imaged at the cellular level by atmospheric pressure Matrix−Assisted Laser Desorption/Ionization Tandem Mass Spectrometry Imaging. Plant J. 80, 161–171. doi: 10.1111/tpj.12608
Li, B., Bjarnholt, N., Hansen, S. H., and Janfelt, C. (2011). Characterization of barley leaf tissue using direct and indirect desorption electrospray ionization imaging mass spectrometry. J. Mass Spectrom. 46, 1241–1246. doi: 10.1002/jms.2010
Li, B., Dunham, S. J., Dong, Y., Yoon, S., Zeng, M., and Sweedler, J. V. (2015). Analytical capabilities of mass spectrometry imaging and its potential applications in food science. Trends Food Sci. Tech. 47, 50–63. doi: 10.1016/j.tifs.2015.10.018
Li, B., Knudsen, C., Hansen, N. K., Jorgensen, K., Kannangara, R., Bak, S., et al. (2013). Visualizing metabolite distribution and enzymatic conversion in plant tissues by desorption electrospray ionization mass spectrometry imaging. Plant J. 74, 1059–1071. doi: 10.1111/tpj.12183
Li, C., Wang, Z., and Jones, A. D. (2014b). Chemical imaging of trichome specialized metabolites using contact printing and laser desorption/ionization mass spectrometry. Anal. Bioanal. Chem. 406, 171–182. doi: 10.1007/s00216-013-7444-6
Li, Y., Shrestha, B., and Vertes, A. (2007a). Atmospheric pressure infrared MALDI imaging mass spectrometry for plant metabolomics. Anal. Chem. 80, 407–420. doi: 10.1021/ac701703f
Li, Y., Shrestha, B., and Vertes, A. (2007b). Atmospheric pressure molecular imaging by infrared MALDI mass spectrometry. Anal. Chem. 79, 523–532. doi: 10.1021/ac061577n
Li, Y., Shrestha, B., and Vertes, A. (2008). Atmospheric pressure infrared MALDI imaging mass spectrometry for plant metabolomics. Anal. Chem. 80, 407–420. doi: 10.1021/ac701703f
Lostun, D., Perez, C. J., Licence, P., Barrett, D. A., and Ifa, D. R. (2015). Reactive DESI-MS imaging of biological tissues with dicationic ion-pairing compounds. Anal. Chem. 87, 3286–3293. doi: 10.1021/ac5042445
Lu, L., Yue, H., Song, F. R., Tsao, R., Liu, Z. Q., and Liu, S. Y. (2010). Rapid profiling of alkaloids in several medicinal herbs by matrix-assisted laser desorption/ionization mass spectrometry. Chem. Res. Chin. Univ. 26, 11–16.
Lunsford, K. A., Peter, G. F., and Yost, R. A. (2011). Direct matrix-assisted laser desorption/ionization mass spectrometric imaging of cellulose and hemicellulose in Populus tissue. Anal. Chem. 83, 6722–6730. doi: 10.1021/ac2013527
Manicke, N. E., Wiseman, J. M., Ifa, D. R., and Cooks, R. G. (2008). Desorption electrospray ionization (DESI) mass spectrometry and tandem mass spectrometry (MS/MS) of phospholipids and sphingolipids: ionization, adduct formation, and fragmentation. J. Am. Soc. Mass Spectrom. 19, 531–543. doi: 10.1016/j.jasms.2007.12.003
Matros, A., and Mock, H. P. (2013). Mass spectrometry based imaging techniques for spatially resolved analysis of molecules. Front. Plant Sci. 4:89. doi: 10.3389/fpls.2013.00089
Moore, K. L., Chen, Y., Meene, A. M., Hughes, L., Liu, W., Geraki, T., et al. (2014). Combined NanoSIMS and synchrotron X-ray fluorescence reveal distinct cellular and subcellular distribution patterns of trace elements in rice tissues. New Phytol. 201, 104–115. doi: 10.1111/nph.12497
Mullen, A. K., Clench, M. R., Crosland, S., and Sharples, K. R. (2005). Determination of agrochemical compounds in soya plants by imaging matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Commun. Mass Spectrom. 19, 2507–2516. doi: 10.1002/rcm.2078
Muller, T., Oradu, S., Ifa, D. R., Cooks, R. G., and Krautler, B. (2011). Direct plant tissue analysis and imprint imaging by desorption electrospray ionization mass spectrometry. Anal. Chem. 83, 5754–5761. doi: 10.1021/ac201123t
Nelson, K. A., Daniels, G. J., Fournie, J. W., and Hemmer, M. J. (2013). Optimization of whole-body zebrafish sectioning methods for mass spectrometry imaging. J. Biomol. Tech. 24, 119–127. doi: 10.7171/jbt.13-2403-002
Nemes, P., Barton, A. A., and Vertes, A. (2009). Three-dimensional imaging of metabolites in tissues under ambient conditions by laser ablation electrospray ionization mass spectrometry. Anal. Chem. 81, 6668–6675. doi: 10.1021/ac900745e
Nimesh, S., Mohottalage, S., Vincent, R., and Kumarathasan, P. (2013). Current status and future perspectives of mass spectrometry imaging. Int. J. Mol. Sci. 14, 11277–11301. doi: 10.3390/ijms140611277
Norris, J. L., and Caprioli, R. M. (2013). Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem. Rev. 113, 2309–2342. doi: 10.1021/cr3004295
Peukert, M., Matros, A., Lattanzio, G., Kaspar, S., Abadia, J., and Mock, H. P. (2012). Spatially resolved analysis of small molecules by matrix-assisted laser desorption/ionization mass spectrometric imaging (MALDI-MSI). New Phytol. 193, 806–815. doi: 10.1111/j.1469-8137.2011.03970.x
Pol, J., Strohalm, M., Havlicek, V., and Volny, M. (2010). Molecular mass spectrometry imaging in biomedical and life science research. Histochem. Cell Biol. 134, 423–443. doi: 10.1007/s00418-010-0753-3
Puolitaival, S. M., Burnum, K. E., Cornett, D. S., and Caprioli, R. M. (2008). Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J. Am. Soc. Mass Spectrom. 19, 882–886. doi: 10.1016/j.jasms.2008.02.013
Riederer, M., and Schreiber, L. (2001). Protecting against water loss: analysis of the barrier properties of plant cuticles. J. Exp. Bot 52, 2023–2032. doi: 10.1093/jexbot/52.363.2023
Robinson, S., Warburton, K., Seymour, M., Clench, M., and Thomas-Oates, J. (2007). Localization of water-soluble carbohydrates in wheat stems using imaging matrix-assisted laser desorption ionization mass spectrometry. New Phytol. 173, 438–444. doi: 10.1111/j.1469-8137.2006.01934.x
Ronci, M., Rudd, D., Guinan, T., Benkendorff, K., and Voelcker, N. H. (2012). Mass spectrometry imaging on porous silicon: investigating the distribution of bioactives in marine mollusc tissues. Anal. Chem. 84, 8996–9001. doi: 10.1021/ac3027433
Ryffel, F., Helfrich, E. J., Kiefer, P., Peyriga, L., Portais, J.-C., Piel, J., et al. (2015). Metabolic footprint of epiphytic bacteria on Arabidopsis thaliana leaves. ISME J. doi: 10.1038/ismej.2015.141. [Epub ahead of print].
Saito, K., Mitsutani, T., Imai, T., Matsushita, Y., and Fukushima, K. (2008). Discriminating the indistinguishable sapwood from heartwood in discolored ancient wood by direct molecular mapping of specific extractives using time-of-flight secondary ion mass spectrometry. Anal. Chem. 80, 1552–1557. doi: 10.1021/ac7021162
Saito, K., Watanabe, Y., Shirakawa, M., Matsushita, Y., Imai, T., Koike, T., et al. (2012). Direct mapping of morphological distribution of syringyl and guaiacyl lignin in the xylem of maple by time-of-flight secondary ion mass spectrometry. Plant J. 69, 542–552. doi: 10.1111/j.1365-313X.2011.04811.x
Schwartz, S. A., Reyzer, M. L., and Caprioli, R. M. (2003). Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass Spectrom. 38, 699–708. doi: 10.1002/jms.505
Seyer, A., Einhorn, J., Brunelle, A., and Laprévote, O. (2010). Localization of flavonoids in seeds by cluster time-of-flight secondary ion mass spectrometry imaging. Anal. Chem. 82, 2326–2333. doi: 10.1021/ac902528t
Shitan, N., and Yazaki, K. (2007). Accumulation and membrane transport of plant alkaloids. Curr. Pharm. Biotechnol. 8, 244–252. doi: 10.2174/138920107781387429
Shrestha, B., Patt, J. M., and Vertes, A. (2011). In situ cell-by-cell imaging and analysis of small cell populations by mass spectrometry. Anal. Chem. 83, 2947–2955. doi: 10.1021/ac102958x
Shroff, R., Rulisek, L., Doubsky, J., and Svatos, A. (2009). Acid-base-driven matrix-assisted mass spectrometry for targeted metabolomics. Proc. Natl. Acad. Sci. U.S.A. 106, 10092–10096. doi: 10.1073/pnas.0900914106
Shroff, R., Schramm, K., Jeschke, V., Nemes, P., Vertes, A., Gershenzon, J., et al. (2015). Quantification of plant surface metabolites by MALDI mass spectrometry imaging: glucosinolates on Arabidopsis thaliana leaves. Plant J. 81, 961–972. doi: 10.1111/tpj.12760
Shroff, R., Vergara, F., Muck, A., Svatos, A., and Gershenzon, J. (2008). Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc. Natl. Acad. Sci. U.S.A. 105, 6196–6201. doi: 10.1073/pnas.0711730105
Soares, M. S., da Silva, D. F., Forim, M. R., Fernandes, J. B., Vieira, P. C., Silva, D. B., et al. (2015). Quantification and localization of hesperidin and rutin in Citrus sinensis grafted on C. limonia after Xylella fastidiosa infection by HPLC-UV and MALDI imaging mass spectrometry. Phytochemistry 15, 161–170. doi: 10.1016/j.phytochem.2015.02.011
Spengler, B. (2014). Mass spectrometry imaging of biomolecular information. Anal. Chem. 87, 64–82. doi: 10.1021/ac504543v
Sturtevant, D., Lee, Y.-J., and Chapman, K. D. (2016). Matrix assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) for direct visualization of plant metabolites in situ. Curr. Opin. Biotechnol. 37, 53–60. doi: 10.1016/j.copbio.2015.10.004
Sugiura, Y., Shimma, S., and Setou, M. (2006). Thin sectioning improves the peak intensity and signal-to-noise ratio in direct tissue mass spectrometry. J. Mass Spectrom. Soc. Jpn. 54, 45–48. doi: 10.5702/massspec.54.45
Sumner, L. W., Lei, Z., Nikolau, B. J., and Saito, K. (2015). Modern plant metabolomics: advancednatural product gene discoveries, improved technologies, and future prospects. Nat. Prod. Rep. 32, 212–229. doi: 10.1039/C4NP00072B
Svatos, A. (2010). Mass spectrometric imaging of small molecules. Trends Biotechnol. 28, 425–434. doi: 10.1016/j.tibtech.2010.05.005
Taira, S., Shimma, S., Osaka, I., Kaneko, D., Ichiyanagi, Y., Ikeda, R., et al. (2012). Mass spectrometry imaging of the capsaicin localization in the capsicum fruits. Int. J. Biotech. Well. Indus. 1, 61–65. doi: 10.6000/1927-3037.2012.01.01.04
Takahashi, K., Kozuka, T., Anegawa, A., Nagatani, A., and Mimura, T. (2015). Development and application of a high-resolution imaging mass spectrometer for the study of plant tissues. Plant Cell Physiol. 56, 1329–1338. doi: 10.1093/pcp/pcv083
Takats, Z., Wiseman, J. M., and Cooks, R. G. (2005). Ambient mass spectrometry using desorption electrospray ionization (DESI): instrumentation, mechanisms and applications in forensics, chemistry, and biology. J. Mass Spectrom. 40, 1261–1275. doi: 10.1002/jms.922
Talaty, N., Takats, Z., and Cooks, R. G. (2005). Rapid in situ detection of alkaloids in plant tissue under ambient conditions using desorption electrospray ionization. Analyst 130, 1624–1633. doi: 10.1039/b511161g
Tata, A., Perez, C. J., Hamid, T. S., Bayfield, M. A., and Ifa, D. R. (2014). Analysis of metabolic changes in plant pathosystems by imprint imaging DESI-MS. J. Am. Soc. Mass Spectrom. 26, 641–648. doi: 10.1007/s13361-014-1039-0
Tata, A., Perez, C. J., Ore, M. O., Lostun, D., Passas, A., Morin, S., et al. (2015). Evaluation of imprint DESI-MS substrates for the analysis of fungal metabolites. RSC Adv. 5, 75458–75464. doi: 10.1039/C5RA12805F
Tholl, D. (2015). Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng. Biotechnol. 148, 63–106. doi: 10.1007/10_2014_295
Thomas, A. L., Charbonneau, J. L., Fournaise, E., and Chaurand, P. (2012). Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1, 5-diaminonapthalene deposition. Anal. Chem. 84, 2048–2054. doi: 10.1021/ac2033547
Thunig, J., Hansen, S. H., and Janfelt, C. (2011). Analysis of secondary plant metabolites by indirect desorption electrospray ionization imaging mass spectrometry. Anal. Chem. 83, 3256–3259. doi: 10.1021/ac2004967
Trouvelot, S., Héloir, M.-C., Poinssot, B., Gauthier, A., Paris, F., Guillier, C., et al. (2014). Carbohydrates in plant immunity and plant protection: roles and potential application as foliar sprays. Front Plant Sci. 5:592. doi: 10.3389/fpls.2014.00592
van Kampen, J. J., Luider, T. M., Ruttink, P. J., and Burgers, P. C. (2009). Metal ion attachment to the matrix meso-tetrakis(pentafluorophenyl)porphyrin, related matrices and analytes: an experimental and theoretical study. J. Mass Spectrom. 44, 1556–1564. doi: 10.1002/jms.1601
Vegvari, A., Fehniger, T. E., Gustavsson, L., Nilsson, A., Andren, P. E., Kenne, K., et al. (2010). Essential tactics of tissue preparation and matrix nano-spotting for successful compound imaging mass spectrometry. J. Proteomics 73, 1270–1278. doi: 10.1016/j.jprot.2010.02.021
Veličković, D., Ropartz, D., Guillon, F., Saulnier, L., and Rogniaux, H. (2014). New insights into the structural and spatial variability of cell-wall polysaccharides during wheat grain development, as revealed through MALDI mass spectrometry imaging. J. Exp. Bot. 65, 2079–2091. doi: 10.1093/jxb/eru065
Volny, M., Venter, A., Smith, S. A., Pazzi, M., and Cooks, R. G. (2008). Surface effects and electrochemical cell capacitance in desorption electrospray ionization. Analyst 133, 525–531. doi: 10.1039/b717693g
Vrkoslav, V., Muck, A., Cvacka, J., and Svatos, A. (2010). MALDI imaging of neutral cuticular lipids in insects and plants. J. Am. Soc. Mass Spectrom. 21, 220–231. doi: 10.1016/j.jasms.2009.10.003
Wang, H.-Y. J., Wu, H.-W., Tsai, P.-J., and Liu, C. B. (2012). MALDI-mass spectrometry imaging of desalted rat brain sections reveals ischemia-mediated changes of lipids. Anal. Bioanal. Chem. 404, 113–124. doi: 10.1007/s00216-012-6077-5
Wei, J., Buriak, J. M., and Siuzdak, G. (1999). Desorption-ionization mass spectrometry on porous silicon. Nature 399, 243–246. doi: 10.1038/20400
Yanes, O., Woo, H. K., Northen, T. R., Oppenheimer, S. R., Shriver, L., Apon, J., et al. (2009). Nanostructure initiator mass spectrometry: tissue imaging and direct biofluid analysis. Anal. Chem. 81, 2969–2975. doi: 10.1021/ac802576q
Yang, J., and Caprioli, R. M. (2011). Matrix sublimation/recrystallization for imaging proteins by mass spectrometry at high spatial resolution. Anal. Chem. 83, 5728–5734. doi: 10.1021/ac200998a
Yoshimura, Y., Enomoto, H., Moriyama, T., Kawamura, Y., Setou, M., and Zaima, N. (2012a). Visualization of anthocyanin species in rabbiteye blueberry Vaccinium ashei by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal. Bioanal. Chem. 403, 1885–1895. doi: 10.1007/s00216-012-5876-z
Yoshimura, Y., Zaima, N., Moriyama, T., and Kawamura, Y. (2012b). Different localization patterns of anthocyanin species in the pericarp of black rice revealed by imaging mass spectrometry. PLoS ONE 7:e31285. doi: 10.1371/journal.pone.0031285
Zaima, N., Goto-Inoue, N., Hayasaka, T., and Setou, M. (2010a). Application of imaging mass spectrometry for the analysis of Oryza sativa rice. Rapid Commun. Mass Spectrom. 24, 2723–2729. doi: 10.1002/rcm.4693
Zaima, N., Hayasaka, T., Goto-Inoue, N., and Setou, M. (2010b). Matrix-assisted laser desorption/ionization imaging mass spectrometry. Int. J. Mol. Sci. 11, 5040–5055. doi: 10.3390/ijms11125040
Zhang, H., Cha, S., and Yeung, E. S. (2007). Colloidal graphite-assisted laser desorption/ionization MS and MS(n) of small molecules. 2. Direct profiling and MS imaging of small metabolites from fruits. Anal. Chem. 79, 6575–6584. doi: 10.1021/ac0706170
Zhang, Y., and Chen, H. (2010). Detection of saccharides by reactive desorption electrospray ionization (DESI) using modified phenylboronic acids. Int. J. Mass Spectrom. 289, 98–107. doi: 10.1016/j.ijms.2009.09.015
Zhou, C., Li, Q., Chiang, V. L., Lucia, L. A., and Griffis, D. P. (2011). Chemical and spatial differentiation of syringyl and guaiacyl lignins in poplar wood via time-of-flight secondary ion mass spectrometry. Anal. Chem. 83, 7020–7026. doi: 10.1021/ac200903y
Keywords: mass spectrometry imaging, molecular histology, MALDI, DESI, SIMS, plant metabolites
Citation: Dong Y, Li B, Malitsky S, Rogachev I, Aharoni A, Kaftan F, Svatoš A and Franceschi P (2016) Sample Preparation for Mass Spectrometry Imaging of Plant Tissues: A Review. Front. Plant Sci. 7:60. doi: 10.3389/fpls.2016.00060
Received: 28 September 2015; Accepted: 14 January 2016;
Published: 10 February 2016.
Edited by:
Basil J. Nikolau, Iowa State University, USAReviewed by:
Sen Subramanian, South Dakota State University, USACopyright © 2016 Dong, Li, Malitsky, Rogachev, Aharoni, Kaftan, Svatoš and Franceschi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pietro Franceschi, cGlldHJvLmZyYW5jZXNjaGlAZm1hY2guaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.