- 1State Key Laboratory of Crop Biology, Shandong Agricultural University, Taian, China
- 2College of Horticulture Science and Engineering, Shandong Agricultural University, Taian, China
- 3Shandong Collaborative Innovation Center for Fruit and Vegetable Production with High Quality and Efficiency, Taian, China
Dormancy inhibits seed and bud growth of perennial plants until the environmental conditions are optimal for survival. Previous studies indicated that certain co-regulation pathways exist in seed and bud dormancy. In our study, we found that seed and bud dormancy are similar to some extent but show different reactions to chemical treatments that induce breaking of dormancy. Whether the abscisic acid (ABA) regulatory networks are similar in dormant peach seeds and buds is not well known; however, ABA is generally believed to play a critical role in seed and bud dormancy. In peach, some genes putatively involved in ABA synthesis and catabolism were identified and their expression patterns were studied to learn more about ABA homeostasis and the possible crosstalk between bud dormancy and seed dormancy mechanisms. The analysis demonstrated that two 9-cis-epoxycarotenoid dioxygenase-encoding genes seem to be key in regulating ABA biosynthesis to induce seed and bud dormancy. Three CYP707As play an overlapping role in controlling ABA inactivation, resulting in dormancy-release. In addition, Transcript analysis of ABA metabolism-related genes was much similar demonstrated that ABA pathways was similar in the regulation of vegetative and flower bud dormancy, whereas, expression patterns of ABA metabolism-related genes were different in seed dormancy showed that ABA pathway maybe different in regulating seed dormancy in peach.
Introduction
Compared with traditional cultivation, protected cultivation of fruit trees is more economical and efficient, and it is an important way to promote farmers' income and rural economic development. Thus, protected cultivation of fruit trees has increased in China in recent years. The peach (Prunus persica) originated in China and has been cultivated for about 3000 years (Zheng et al., 2014). Peaches are important fruit trees in China, being cultivated in about 20,000 hectares. Generally, peach seedlings and fruit can be produced under protected cultivation all year. The production of peach fruit is influenced by blooming time; only after dormancy is broken, peach trees can proceed with the normal life cycle. Therefore, determining the mechanism of dormancy and using this knowledge to develop practical measures to break the seed and bud dormancy would be useful in the protected cultivation of peach trees.
Dormancy is defined as the inability to initiate growth from meristems under favorable conditions (Rohde and Bhalerao, 2007). Lang et al. (1987) distinguished bud dormancy into three types: paradormancy, inhibited by distal organs; endodormancy, inhibited by the dormant structure itself; and ecodormancy, inhibited by an unfavorable environment. Both seed and bud dormancy have been studied, and both types of dormant organs must undergo induction of dormancy, deep dormancy and dormancy breaking. Only after completing the whole process of dormancy can perennial plants proceed with the normal growth and development, otherwise, abnormal physiological phenomena might occur, such as dwarf seedlings, inconsistency of seed and bud germination and decreased fruit quality. Fu et al. (2014) proposed that there are certain similar molecular mechanisms between seed and bud dormancy; indeed, the hormonal regulations of bud and seed dormancy are similar (Cooke et al., 2012). Multiple physiological and transcriptomic studies have revealed the involvement of ABA in bud dormancy (Rohde et al., 2007; Shalom et al., 2014; Zhu et al., 2015); however, few genetic studies support this view. By contrast, the involvement of ABA in seed dormancy has been confirmed using Arabidopsis mutants and other plants mutants (Finkelstein, 2013). In general, ABA plays an important role in biotic stress and abiotic stress responses, controlling a wide range of essential physiological processes. Furthermore, bud and seed dormancy are regulated by ABA in perennial trees to cope with the more extreme environment conditions encountered during autumn and winter. The level of ABA is regulated by its biosynthesis and catabolism. Previous studies have revealed the ABA regulatory network in plants.
ABA Synthesis and Catabolism
ABA is synthesized de novo from a C40 carotenoid (Figure 1). The first step of ABA biosynthesis is epoxidation of zeaxanthin to all-trans-xanthophylls zeaxanthin and violaxanthin, catalyzed by zeaxanthin epoxidase (ZEP) in plastids (Koornneef et al., 1982; Marin et al., 1996). Violaxanthin is then converted to 9-cis-violaxanthin and 9-cis-neoxanthin, which are cleaved by 9-cis-epoxycarotenoid dioxygenase (NCED) to yield xanthoxin, the first C15 intermediate (Schwartz et al., 2003; Leng et al., 2014). Overexpression experiments in transgenic plants demonstrated that NCED is the key enzyme in limiting the rate of ABA synthesis (Thompson et al., 2000; Tung et al., 2008). Xanthoxin is transported to the cytosol, where it is converted to abscisic aldehyde by a short-chain alcohol dehydrogenase, which is encoded by SDR1 (Cheng et al., 2002; González-Guzmán et al., 2002). Finally, abscisic aldehyde is oxidized to ABA by aldehyde oxidase 3 (encoded by AAO3) in the presence of a molybdenum cofactor (MOCO) (Seo et al., 2000; Bittner et al., 2001). In addition, ABA inactivation is the crucial mechanism that controls ABA level though oxidation or conjugation. ABA can be hydroxylated at the C-7′, C-8′ and C-9′ positions, and C-8′ is the major position for the hydroxylation reaction catalyzed by ABA 8′-hydroxylase (encoded by CYP707A) to yield 8′-hydroxy-ABA (8′-OH-ABA), which is unstable and enzymatically isomerizes to form phaseic acid (PA) (Kushiro et al., 2004; Saito et al., 2004).
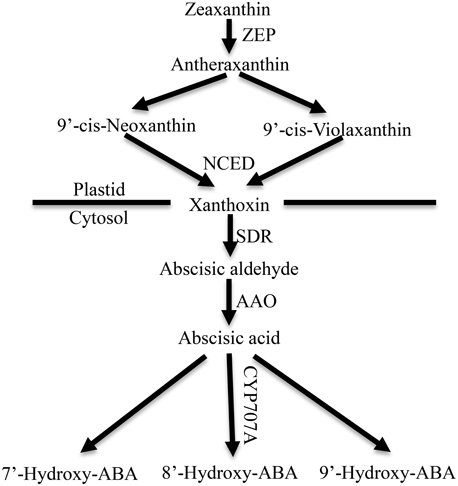
Figure 1. ABA metabolic pathway in higher plants. ZEP, zeaxanthin epoxidase; NCED, 9-cisepoxycarotenoid dioxygenase; SDR, short-chain dehydrogenase/reductase; AAO, aldehyde oxidase. CYP707A, abscisic acid-8′-hydroxylase (Leng et al., 2014).
ABA and Seed Dormancy
In Arabidopsis, the regulation network controlling seed maturation seems to control the induction of seed dormancy (Rikiishi and Maekawa, 2014). ABA produced in the mature seeds regulates the induction of seed dormancy, and changes in the expressions of genes related to ABA synthesis and catabolism have been verified in Arabidopsis, bean, barley, and wheat (Chono et al., 2006; Millar et al., 2006; Yang and Zeevaart, 2006; Shu et al., 2013).
In higher plants, overexpression of certain genes related to ABA synthesis resulted in an increase of dry seed ABA content and delayed germination (Frey et al., 1999; Thompson et al., 2000; Qin and Zeevaart, 2002; Holdsworth et al., 2008). By contrast, their corresponding mutants showed ABA deficiency during various stages of plant and seed development, and exhibited greatly reduced seed dormancy: fresh seeds germinated at high frequency (Nambara et al., 1998; Lefebvre et al., 2006). Carotenoid cleavage by NCED plays an important role in ABA biosynthesis, and affects seed dormancy and germination. In Arabidopsis, nine NCED-related sequences have been identified, but only five gene members (AtNCED2, AtNCED3, AtNCED5, AtNCED6, AtNCED9) encode xanthoxin-producing enzymes, and AtNCED5, AtNCED6, and AtNCED9 were reported to influence the development and dormancy of seeds (Tan et al., 2003; Lefebvre et al., 2006; Frey et al., 2012). By reporter gene analysis and in situ hybridization, AtNCED6 was observed to be expressed in the endosperm, whereas, AtNCED9 was expressed in both the endosperm and the embryo. ABA levels in nced6 and nced9 seeds were lower than in the wild-type, and nced6nced9 double mutants showed reduced seed dormancy (Lefebvre et al., 2006). Microarray data showed AtNCED5 expression increased at late seed maturation stages, which was also confirmed by analysis of developing seeds of pNCED5:GUS transgenic plants (Frey et al., 2012). Overexpression of NCED in tobacco resulted in an increase in seed ABA content, and delayed the germination of imbibed seeds (Qin and Zeevaart, 2002).
ABA 8′-hydroxylase (encoded by CYP707A) is considered the key catabolic enzyme in ABA inactivation for controlling ABA level. In Arabidopsis, expression analysis found that AtCYP707A1 was expressed in seeds at the mid-maturation stage and AtCYP707A2 was expressed at late-maturation stage. In addition, ABA levels in dry seeds of cyp707a1, cyp707a2, and cyp707a2 cyp707a3 double mutants were higher than in the wild-type, and the freshly harvest seeds of the cyp707a mutant showed a lower germination rate compared with the wild-type (Okamoto et al., 2006). In wheat, TM1833 (a double mutant of in TaABA8′OH1-A and TaABA8′OH1-D) showed lower TaABA8′OH1 expression, higher ABA content during seed development and lower germination than “Tamaizumi” (a single mutant in TaABA8′OH1-D) (Chono et al., 2013). In barley, lower expression in transgenic ABA8′OH RNAi barley grains resulted in a higher ABA content and increased dormancy compared with non-transgenic barley grains (Gubler et al., 2008).
ABA and Bud Dormancy
The role of ABA in controlling seed dormancy has been well established; however, its role in regulating bud dormancy is not well known. Until now, the progress of research on bud dormancy has been relatively slow because of its complex biological mechanisms.
The role of ABA in the regulation of bud endodormancy has been discussed in previous studies, and it is generally believed to be a key hormone in regulating bud dormancy (Cooke et al., 2012). In the autumn, the ABA content increases, resulting in cessation of shoot growth, promotion of apical buds set and induction of bud dormancy. In poplar, ABA levels reached their maximum after growth cessation in apical buds, accompanied by induction of genes related to ABA biosynthesis (ABA1, NCED3, and ABA2) (Arora et al., 2003; Ruttink et al., 2007). In the winter, buds remain dormant to inhibit bud growth to cope with the cold environment. During dormancy induction and maintenance of grape bud, ABA levels increased accompanied by increased expression of VvNCED1; however, the endogenous ABA levels decreased and ABA catabolites increased after buds achieved enough chilling accumulation, and VvNCED1 was down-regulated and VvCYP707A4 was up-regulated (Zheng et al., 2014). During peach bud dormancy release, ppa005059m and ppa005020m (highly similar to ABA 8′-hydroxylase genes CYP707A2 from Arabidopsis) were identified by suppression subtractive hybridization and microarray hybridization (Leida et al., 2010).
Using bud-breaking chemicals to study the mechanism of endogenous dormancy is an efficient way to demonstrate the relationship between ABA and bud dormancy. Although ABA is generally considered an important hormone for maintaining and releasing bud dormancy, there are conflicting results in some studies. In apple (Dutcher and Powell, 1972), and birch (Rinne et al., 1994), spraying ABA on buds resulted in delayed bud breaking; however, spraying ABA in grape buds had little effect on bud breaking (Hellman et al., 2006). Hydrogen cyanamide (HC) is a chemical that induces respiratory stress, leading to effective dormancy release. HC application reduced VvNCED1 transcript levels and increased levels of VvABA8′OH homologs in grape, which was accompanied by reduced ABA content that resulted in a higher percentage of bud breaking than that in the control (Zheng et al., 2015).
Gibberellins (GA) and ABA
In general, Gibberellin and ABA play an antagonistic role in regulating plant growth; Gibberellin is considered to promote plant growth, and ABA is considered to inhibit plant growth. In barely grains, addition of GA could promote germination during malting, and this was ascribed to increase of the enzymes relating to synthesis and secretion resulting in seed germination (Huang et al., 2016). In Arabidopsis, Cold stratification could increase GA level resulting in the release seed dormancy, and GA3ox1, a gene regulating GA biosynthesis, was induced by cold stratification (Nonogaki, 2014). Furthermore, previous studies had showed that the ratio of ABA and GA was relevant with seed dormancy and seed germination. In sweet cherry flower bud, ABA/GA increased with the process of endodormancy, reached maximum in the stage of deep dormancy, after that, it decreased with the broken of bud dormancy release (Duan et al., 2004).
Although ABA is confirmed to influence bud and seed dormancy, and the ABA pathway is quite clear, there have been few systematic studies of ABA metabolism in whole process of bud and seed dormancy, including their similarities and differences. In the current study, we monitored the expressions of genes related to ABA metabolism during the process of bud and seed dormancy in peach and demonstrated the relationship between seed dormancy and bud dormancy.
Materials and Methods
Plant Material
Peach trees (Prunus persica L. cv Zhong You Tao 4) were grown in an orchard located at the Shandong Institute of pomology in Taian, China under standard agricultural practices. As embryos appeared, 50 fruit were randomly collected from 30 distinct trees every 5 days and immediately dissected to remove the endocarp using special scissors. The seeds, seed coats and embryos were collected separately. Seed material was weighed and then frozen in liquid N2, and stored at −80°C until RNA extraction and quantitative real-time PCR (qRT-PCR) analysis.
Peach flower and vegetative buds (0.5 g) were sampled in late autumn (September) to mid-Spring (February) approximately every 15 days, from 9 a.m. to 11 a.m. The samples were immediately frozen in liquid N2 and stored at −80°C until RNA extraction and qRT-PCR analysis. Furthermore, at every time point, we randomly collected 30 1-year-old shoots and cultivated them in a vase containing 5% sucrose solution under a daily cycle of 25°C with artificial fluorescent light (200 μmol·m−2·s−1) for 16 h and 18°C in darkness for 8 h with constant 70% relative humidity to monitor the bud burst rate.
Seed Stratification at 4°C
Mature fruit (300) were collected and seeds with their endocarp were stratified at 4°C in wet gauze for 10 weeks to break dormancy. Stratified seeds (50 at each time point) were randomly brought out at a specified time interval and broken with a hammer to collect seeds, seed coats, and embryos. During the course of stratification, the seed germination rate was regularly examined at the optimum water supply and temperature (22°C) in the dark.
Gibberellic Acid (GA3) Treatments
Mature seeds were soaked in 1 mM GA3 (Solarbio, Beijing, China) solution for 4 days. During this time, seeds were collected separately, washed three times with ultrapure water, weighed, frozen in liquid N2, and stored at −80°C. In addition, the control was soaked in ultrapure water. After treatment, the germination rate was tested under optimum water supply and temperature (22°C) in the dark.
Analysis of the Effect of Hydrogen Cyanamide (HC), High Temperature (HT), Gibberellic Acid (GA3), and Thidiazuron (TDZ) on Bud Break
On 22 October 2014, long 1-year-old branches were cut from Zhong You Tao 4 trees. Their basal parts were placed in 5% sucrose solution and incubated in a illumination incubator for a natural day length and under controlled conditions (conditions were the same as above). Subsequently, these branches were separated with five groups for treatments and every group contained 50 cuttings. 100 mL of 1 mM GA3(Solarbio, Beijing, China), 0.2 m MTDZ (KAYON, Shanghai, China) and 1% (v/v) HC (a solution containing 50% (w/v) HC, Sigma-Aldrich, St Louis, USA) with addition of 0.02% (v/v) Triton X-100 (Solarbio, Beijing, China) added as a surfactant were sprayed onto the branches, and then these treatments were transferred to the illumination incubator for an additional 30 d for bud-break monitoring. For HT treatment, cuttings were immersed in 50°C water for 1 h. In the control treatment, 0.02% (v/v) Triton X-100 was used as a spray.
Identifying the Genes Related to ABA Metabolism
Peach genomic sequencing was accomplished in 2013 (Verde et al., 2013); however, the functions of most of the genes are not clear. Few genes relating to ABA synthesis and catabolism have been identified. We used Endo's method (2014) to deduce the NCEDs, CYP707As, AAOs, ZEP, SDR1 genes in the peach genome sequence (Endo et al., 2014). Firstly, a tblastp search was performed with one known amino acid sequence of each gene family from Arabidopsis in the peach genome at the Phytozome database (http://phytozome.jgi.doe.gov/pz/portal.html). This identified several candidate genes (e < 1e-10) of each family and their amino acid sequences were recorded. Secondly, a phylogenetic tree was constructed by Mega software version 6.0, using the neighbor-joining method and the protein sequences of candidate genes of each gene family. We then analyzed the phylogenetic tree and deleted those candidate genes whose genetic relationship was relatively distant. Thirdly, online SMART software at EMBL (http://smart.embl-heidelberg.de/) was used to determine whether each candidate gene's protein sequence had the typical conserved domains belonging to this family to further validate that these candidate genes belong to this family. Finally, the candidate gene protein sequences' similarities between Arabidopsis and peach were analyzed using DNAMAN software.
After these processes, PpCYP707A1 (ppa005059m), PpCYP707A2 (ppa005020m), PpCYP707A3 (ppa005226m), and PpCYP707A4 (ppa005234m) were chosen as candidate genes of the CYP707A gene family. PpNCED1 (ppa002804m), PpNCED2 (ppa002314m), PpNCED3 (ppa014647m), and PpNCED4 (ppa006109m) belonged to NCED gene family. PpAAO1 (ppa000263m) was the candidate gene of the AAO gene family. ZEP and SDR1 are single genes belonging to the LOS gene family and SDR gene family, respectively. After analysis of these two gene families, the genetic relationship of ppa002248m and ppa009814m was quite close to ZEP and ABA2; therefore ppa002248m and ppa009814m were chosen as PpZEP and PpSDR1 candidate peach genes.
RNA Extraction and qRT-PCR Analyses
Total RNA was extracted from 0.2 g seeds, seed coats, embryos, and buds using a rapid plant RNA extraction kit (Aidlab, Beijing, China). The single-stranded cDNAs were synthesized from 1 μg of RNA using a cDNA synthesis kit according to the manufacturer's instructions (Takara Biotechnology, Dalian, China). qRT-PCR was performed with a gene specific primer pair and a β-actin primer pair as an internal control (Table 1). Reactions were performed on a CFX96 real-time PCR detection system with SYBR Premix Ex Taq (Takara). The thermo cycling parameters were as follows: 10 min at 95°C, followed by 40 cycles of 15 s at 95°C for denaturation and 1 min at 60°C for annealing and extension. The specificity of the PCR was assessed by the presence of a single peak in the dissociation curve after the amplification and by size estimation of the amplified product. The comparative cycle threshold (CT) method (2-ΔΔCT) method was used to quantify cDNAs with amplification efficiencies equivalent to that of the reference actin gene. Each experiment was repeated at least three times using the same cDNA source. For every reaction, the mean and SE values of relative transcript abundance were calculated. All the primers were designed by Beacon Designer 7 software. Results presented are the average of three independent biological replicates repeated three times.

Table 1. Primers used to quantify the expression of the ABA metabolic genes and the reference genes.
Determination of Endogenous (±) ABA
Two hundred milligrams of plant material was extracted in 1.8 ml cold phosphate-buffered saline (0.01 M, pH 9.2) (1:9, m/v), adjusted to pH 8.5. After centrifugation at 10,000 g (4°C, 10 min), the supernatant was filtered through 0.22 μm filter membrane. The efflux was collected for (±) ABA qualification, and ABA levels were quantified by Plant hormone abscisic acid (ABA) ELISA Kit (Bangyi Biotechnology, Shanghai, China), following the manufacturer's protocol. Each treatment consisted of three biological replications and each sample was measured with two technical replicates, and data from a typical experiment were presented.
Statistical Analysis
Statistical analysis was performed using SPSS for Windows version 19 (SPSS, Chicago, IL, USA). Categorical variables were expressed as frequencies, and percentages and continuous variables as the mean ± standard errors, as appropriate. The data were analyzed by ANOVA and, when appropriate, Duncan's test was used. A significance level of p < 0.05 was applied.
Results
Identification of Peach ABA Metabolism Genes
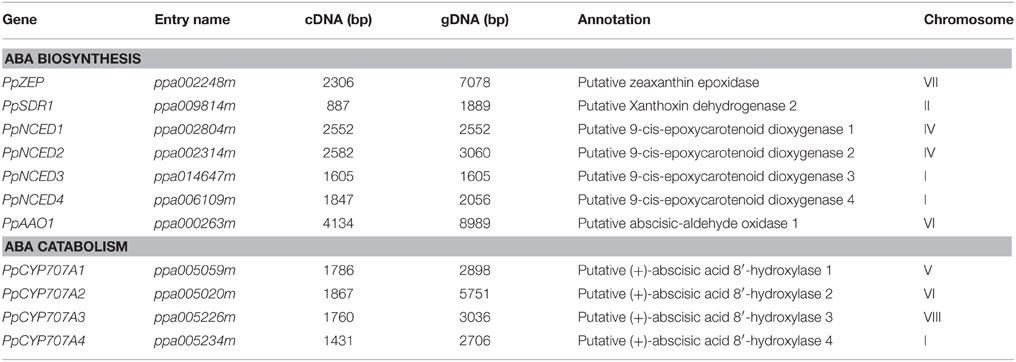
ABA synthesis and catabolism genes had not been well annotated in the peach genome; therefore, we extracted annotated Arabidopsis genes from public database and searched for orthologs and paralogs from the peach database. Phylogenetic and protein domain analyses identified four NCED, four CYP707A, and one AAO gene family members. Table 2 lists the sizes of the full-length cDNAs and gDNAs, as well as the chromosomal locations of the candidate genes. Analysis of the genomic structure of the candidate genes related to ABA metabolism showed that the number of exons in the ABA-related genes in the available plant genomes is remarkably well conserved (Table S1). The degree of identity between the predicted amino acid sequences of each ABA-related catabolism gene and its respective orthologs in Arabidopsis thaliana, Populus trichocarpa, and Oryza sativa was calculated (Table S2). The amino acid sequences of the peach genes are very similar to members of the eudicots (e.g., Arabidopsis and Populus) and less similar to the monocot species (Oryza) (Table S2).
Phylogenic Analysis of ABA Metabolism Genes
In Arabidopsis, NCED belongs to CCD gene family and there are nine AtCCDs (including five NCEDs). We identified four NCED genes in peach. PpNCED1 and PpNCED2 were located on chromosome 4 and PpNCED3 and PpNCED4 were located on chromosome 1 (Table 2). The amino acid sequence of Arabidopsis AtNCED3 showed high similarity to PpNCED1 and PpNCED2 (61–67%), but relatively lower identities with PpNCED3 and PpNCED4 (27 and 32%). Phylogenetic analysis revealed that PpNCED3 was quite similar to AtNCED6, and PpNCED4 was similar to AtNCED4 (Figure S1).
The CYP707A gene family encodes ABA 8′- hydroxylases that control ABA catabolism, including four CYP707A genes in Arabidopsis (Saito et al., 2004), three in rice (Saika et al., 2007), two in peanut (Liu et al., 2014), three in bean (Yang and Zeevaart, 2006), three in potato (Suttle et al., 2012), five in maize (Vallabhaneni and Wurtzel, 2010), and 10 in soybean (Zheng et al., 2012). In our study, four genes were identified as candidate CYP707A genes in peach, among which PpCYP707A1 and PpCYP707A2 had been identified in a previous study (Leida et al., 2012). Multiple sequence alignments of the deduced amino acids of PpCYP707A1 showed that it was quite similar to AtCYP707A1 and AtCYP707A2 (77 and 74% similarity, respectively) (Table S2 and Figure S1), and PpCYP707A2 was similar to AtCYP707A2 (66% similarity). PpCYP707A3 and PpCYP707A4 were similar to AtCYP707A4 (70 and 67% similarity, respectively). Multiple sequence alignments of the deduced amino acids of the CYP707A gene family in peach showed 68% similarity. All of PpCYP707A gene family proteins contain the highly conserved cysteine residue (PFGNGTHSCPG), which is the putative heme iron ligand, and it is essential for catalysis.
Four AAO genes are present in Arabidopsis; however, we could only identify one ortholog in peach (Figure S1). Multiple sequence alignments of the deduced amino acid sequence of PpAAO1 showed 60, 61, 62, and 61% sequence identity with Arabidopsis AtAAO1, AtAAO2, AtAAO3, and AtAAO4, respectively.
ABA2/SDR1 is a member of the SDR gene family in Arabidopsis; however, SDR1 is thought to encode a unique protein that catalyzes the conversion of Xan (Xanthoxin) to ABAld (abscisic aldehyde) in Arabidopsis. We identified 10 SDR-like gene family members in peach, and constructed a phylogenetic tree using 20 amino acid sequences of the SDR family from peach and Arabidopsis. Phylogenetic analysis showed that ppa009814m was the most likely SDR1-like gene in peach (Figure S1). The deduced protein showed 71% sequence identity with Arabidopsis AtABA2/AtSDR1, and contained the N-terminal motif (Gly-X-X-X-Gly-X-Gly) and the Tyr-X-X-X-Lys motif that are characteristic of SDRs.
Seasonal Dormancy Status of Vegetative and Flower Buds of Peach Trees
To study how ABA-related genes changed during the whole process of dormancy of buds, the stages of bud dormancy should be precisely identified. In this study, 30 6-year-old peach trees in an orchard located at Shandong Institute of pomology were selected to study bud development. We used two methods to identify flower bud dormancy stage: measuring the increase in bud weight, including the bud water content, caused by initiation of meristem growth; the other was testing the percentage of bud breaking every 15 days from September to February in the next year caused by removal of environmental effects. We only use the latter method to identify vegetative bud development because peach vegetative buds are quite small.
Figure 2 shows seasonal percentage of bud breaking in the 25-day after incubated in 5% sucrose solution. At the same time, the weather conditions and chilling accumulation (number of hours below 7.2°C) were recorded to understand the environmental effect on bud development (Figure 2B). From 27 September to 5 December, no flower buds or vegetative buds broke from 1-year-old shoots (accumulated chilling hours, 482 h), which we termed endodormancy, caused by inhibition by the dormant buds itself. After 22 November, the dormancy of the flower and vegetative buds started to be released. From 23 November to 15 January in the next year, the dormancy of flower buds and vegetative buds was fully released, with 80 and 63% of buds breaking, respectively. We termed this stage as the dormancy release period (transition stage), in which the weight of the flower bud increased (Figure 2C). After 15 January, the buds could burst, in theory; however, they did not burst because of the cold environment; this is known as ecodormancy, where bursting is inhibited by the disadvantageous environment. Figure 2A shows the buds sampled on 5 December that broke first; we considered this the key time point of bud dormancy release. In addition, flower bud dormancy release was earlier than vegetative bud dormancy release in peach trees, which could explain why peach blossoms emerge earlier than leaves under field conditions.
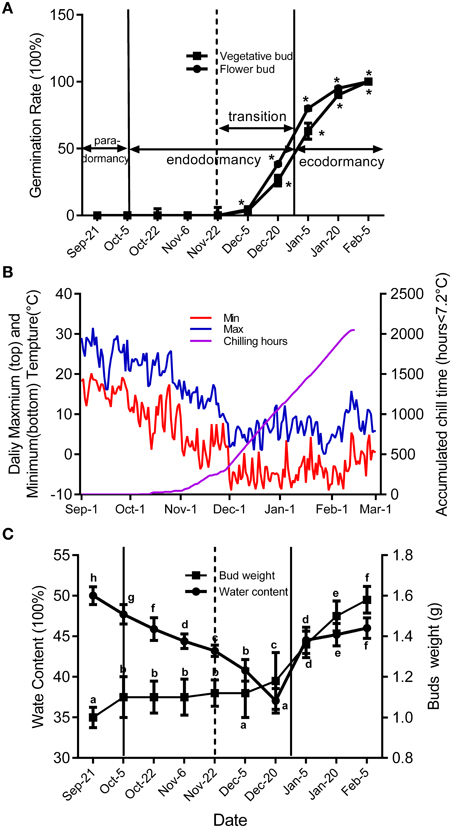
Figure 2. Characterization of vegetative and flower bud condition and seasonal changes of weather conditions around orchard located at Shandong Institute of pomology. Measurements are averages of three biological replicates with error bars representing the SEM. (A) Changes in dormancy status of the bud population throughout the dormancy cycle. Bud-break percentages at 25 d were presented to describe the seasonal changes in dormancy status of the bud population (*P < 0.05). (B) Daily maximum and minimum temperatures and cumulative chilling hours from September to the following February in 2014–2015. (C) Changes of average flower bud weight and water content from 21 September to 5 February. Different letters above bars indicate a significant difference among chilling periods according to ANOVA and Duncan's test (P < 0.05).
Seasonal Expression Changes of ABA Synthesis and Catabolism Genes in Vegetative and Flower Bud Dormancy
Seasonal expression analysis of ABA synthesis and catabolism genes demonstrated that these genes were expressed and showed similar change trends between vegetative and lateral flower bud dormancy (Figure 3). Meanwhile ABA concentration in vegetative and lateral flower buds showed the similar trends, and both were high during the early part of the rest period and then gradually declined (transition stage), reaching the minimum concentration in the period of ecodormancy. PpNCED1 and PpCYP707A1 transcript levels showed the same change trend: both steadily decreased from September to February. PpNCED2 was expressed at lower level on 21 September, and then increased dramatically to reach a peak on 4 October, after which it decreased gradually until February. The PpSDR1 transcript level was relatively higher initially, but decreased on 22 October and remained at a relatively low expression level until 22 November, after which it increased to a high expression level. Both PpZEP and PpCYP707A2 were expressed at low levels from 21 September to 22 October, and then increased suddenly on 5 December (the key time point of bud dormancy release), and then after 20 December, the expressions of PpZEP and PpCYP707A2 returned to relatively lower expression levels. The transcript level of PpAAO1 was always maintained at a high level during the period of vegetative and bud dormancy, and its expression showed no obvious change. The expression of PpCYP707A3 increased gradually from September to February in the next year, with a dramatic peak on 5 December. In addition, we found that all of the genes were expressed at their lowest levels on 22 November [the first phase before the key phase of bud release (5 December)]. This experiment was performed 2 years and showed the same expression trends (data not shown).
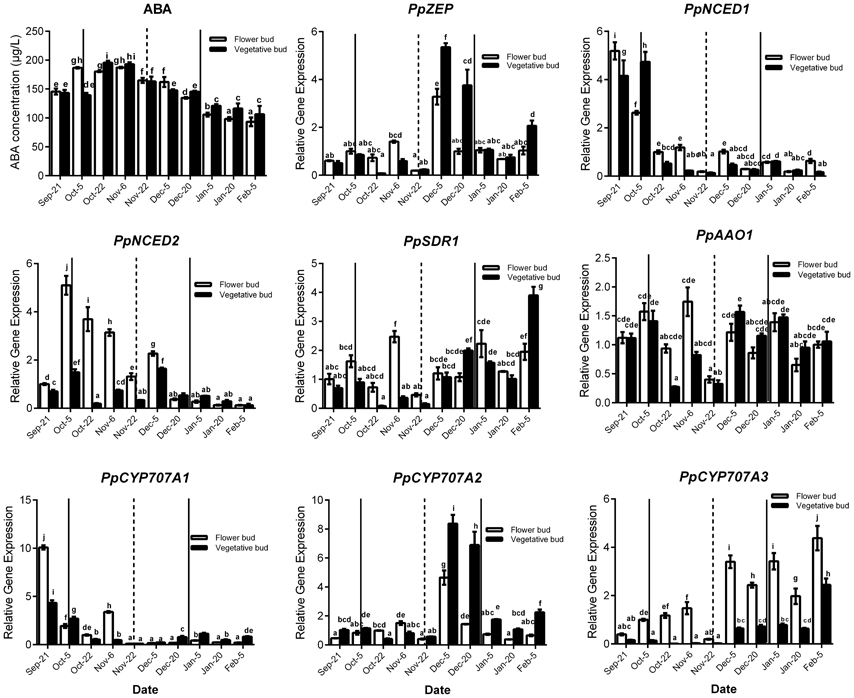
Figure 3. Relative gene expression of eight ABA metabolic genes during the period of vegetative and flower bud development stage. Each experiment was performed with three biological replicates with error bars representing the SEM. Different letters above bars indicate a significant difference among chilling periods according to ANOVA and Duncan's test (P < 0.05). The lines show buds development stage which has the same meaning with the lines in Figure 2.
Expression Patterns of ABA Synthesis and Catabolism Genes during Seed Development
Seeds were sampled once embryos appeared (7 June), and seeds were separated into seed coat and embryo for analysis. For ABA qualification, data showed that ABA was gradually increased in seed coat, seed and embryo during seeds development, and ABA concentration in seed coat was higher than embryo (Figure 4). In the embryo, the expressions of PpZEP, PpSDR1, PpAAO1, PpCYP707A1, PpCYP707A2, and PpCYP707A3 increased as the seeds developed (Figure 4). In seed coat, all these gene expression was higher than in seed and embryo. PpNCED1, PpNCED2, PpCYP707A1, and PpCYP707A2 was expressed at a low level at the beginning, and then increased to a high transcript level at six-fold, 20-fold, 11-fold, 20-fold higher than the baseline level in 3 July, respectively. The transcript levels of PpZEP, PpSDR1, and PpAAO1 gradually increased to a peak on 17 June, after which they decreased slightly. The expression of PpCYP707A3 was high in the beginning, and then decreased until 12 June, after which it increased as the seeds developed (Figure 4). In addition, we observed that endosperm disappeared by the 12 June. Figure 4 demonstrates that all the genes were expressed at a relatively low level in early seed development, and then gradually increased to high expression levels, except PpCYP707A2. PpCYP707A2 was expressed at a high level in the beginning, dramatically decreased to a low level, and then gradually increased to a peak at the end of seed development. In 3 June and 8 June, the endosperm was still present within seeds except embryo and seed coat; therefore, we measured these gene expressions during the process of endosperm abortive (data not shown). We found that PpCYP707A1, PpCYP707A3, PpNCED1, and PpNCED2 were not expressed in late endosperm development in peach. PpZEP, PpSDR1, and PpAAO1 expression increased from 3 to 8 June; however, PpCYP707A2 showed the reverse trend, which was why PpCYP707A2 transcript being at a higher level compared with the other genes in seeds from 3 to 8 June.
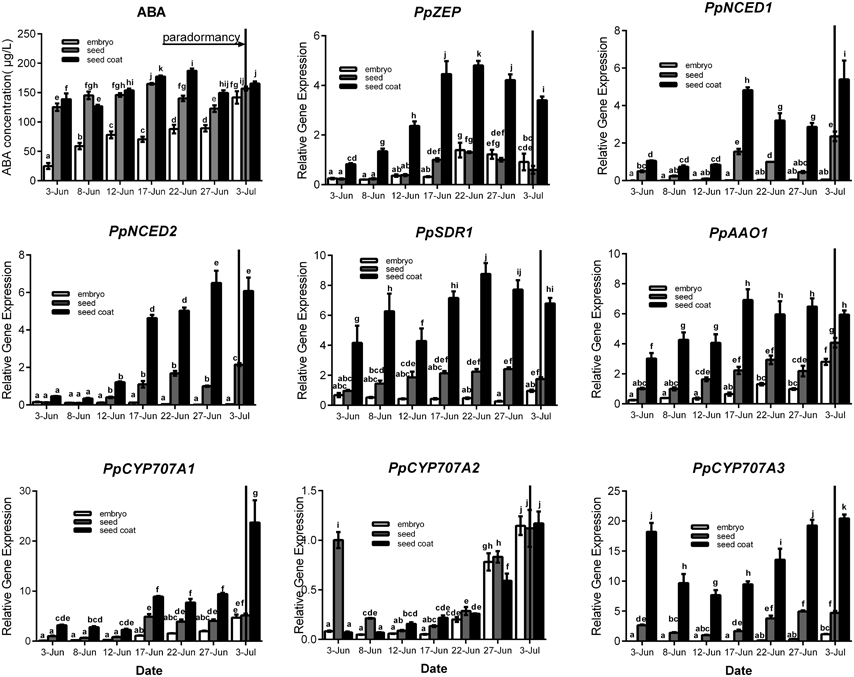
Figure 4. Relative expression levels of eight ABA metabolic genes in seeds, seeds coat, and embryos during the period of seed development from 3 June to 3 July in 2015. Seeds were sampled every 5 days. Expression levels were calculated relative to a peach β-actin as Figure 3, and data are presented as the means ± standard errors. Each experiment was performed three times. Different letters above bars indicate a significant difference among chilling periods according to ANOVA and Duncan's test (P < 0.05). In addition, the period of seeds development [paradormancy (induction of seed dormancy)] corresponds to buds paradormancy (induction of bud dormancy). The lines have the same meaning.
Effect of Stratification on Seed Germination and Expression Patterns of ABA Metabolic Genes during Seed Stratification
In production, insufficient chilling accumulation may be result in lower germination rate or disturbed emergence. In this study, a germination experiment was performed to characterize the response of seeds to different cold stratification periods. Seeds sampled from different stratification periods were placed in petri dish filled with wet gauze and cultured in the dark at 22°C. Figure 5B shows that the germination rate was low after 0, 1, and 3 weeks of stratification with 12, 16, and 50%, respectively. After 5 and 7 weeks of stratification, seed germination rate could reach 100%; however, the root length and germination time were not consistent (Figure 5A). After 10 weeks of stratification, seeds consistently germinated with 100% germination rate and root length was much the same on the 5th day (Figure 5A). In addition, we measured the difference between stratified seeds and non-stratified seeds through an in vitro culture experiment. After 10 weeks' stratification, the seedling formation rate reached 100%, whereas that of non-stratified seeds was much lower at 20% (Figure 5C). Stratification improved the seedling formation rate, shoot growth, the consistency of germination, and root length.
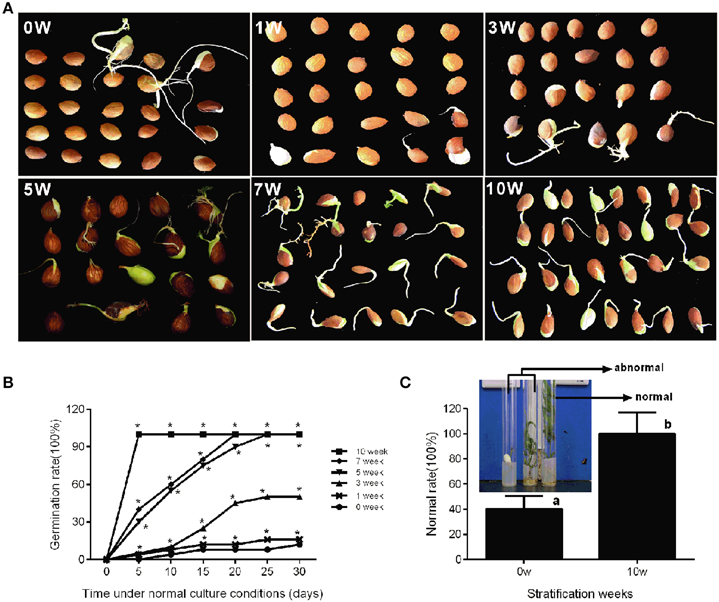
Figure 5. Characterization of seed conditioning and germination after discrete periods of stratification. The phenotype (A) and germination percentage of seed germination (B) were measured after discrete periods of stratification. (C) The normal rate of seedling after in vitro culture experiment of stratified embryo (10w) and non-stratified embryo (0w). The picture showed the normal seedlings and abnormal seedlings. Letters and asterisk above bars indicate a significant difference among chilling periods according to ANOVA and Duncan's test (*P < 0.05).
Seed stratification resembles the bud dormancy release process, and also requires sufficient chilling accumulation, otherwise, some physiological abnormalities could occur during cultivation. In the period of seeds stratification, ABA concentration was gradually decreased in seed, embryo, and seed coat (Figure 6) which were the same with ABA changes in vegetative and lateral flower bud in the period of dormancy release (Figure 3). In addition, we also analyzed expression during seed stratification to assess whether ABA synthesis and catabolism genes performed similarly in stratification compared with bud dormancy. The transcript levels of PpZEP, PpNCED1, and PpNCED2 were high at the start, and then decreased in seeds and seed coats during stratification (Figure 6). In seeds and embryos, PpSDR1 and PpAAO1 expression gradually decreased, but in seed coats their expression levels were relatively stable from 0 to 3 weeks, and then increased to a peak at 7 weeks (Figure 6). PpCYP707A gene members showed different expression patterns during seed stratification (Figure 6). The expression level of PpCYP707A1 showed the same trends during seed stratification in seeds and embryos. Initially, it decreased at 1 week of stratification, and then increased to a peak at 5 weeks (a key point in seed dormancy release), but in seed coat, the expression level of was steadily increased with seed stratification. The PpCYP707A2 transcript level declined with seed stratification in seed coats; however, in embryos, its expression level gradually increased to a peak at 3 week. In seeds, the expression of PpCYP707A2 was initially high, decreased in the first 3 weeks, and then dramatically increased to a peak at week 5. The expression of PpCYP707A3 showed the same trends in seeds and embryos during seed stratification: the transcript was relatively stable in the first 3 weeks, and then dramatically increased to a peak at 5 weeks, with 55- and 90-fold higher expression compared with the baseline; however, in seed coats, the expression level was relatively stable.
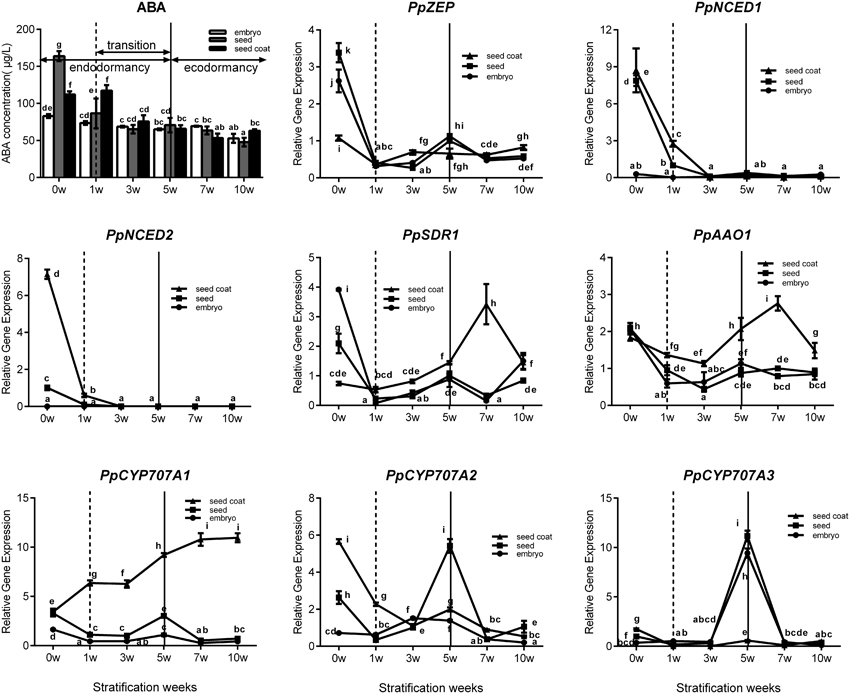
Figure 6. Expression analysis of eight ABA metabolic genes in seeds, seeds coat, and embryos during the period of seed stratification. Each experiment was performed with three biological replicates with error bars representing the SEM. Different letters above bars indicate a significant difference among chilling periods according to ANOVA and Duncan's test (P < 0.05). The lines have the same meaning.
Effect of Gibberellic Acid on Expression of Central Components of ABA Synthesis and Catabolism Genes in Seeds
In general, Gibberellin (GA3) and ABA play an antagonistic role in regulating seed dormancy and germination. In our study, mature peach seeds were soaked in 1 mM GA3 solution to study how Gibberellic acid affected the ABA metabolic network in regulating seed dormancy. We found that GA3 treatment resulted in seed dormancy release compared with the control (Figure 7A). Seeds treated with GA3 for 96 h showed 50% higher seed dormancy release compared with the control after 30 days of cultivation after treatment (Figure 7A). Even though GA3 treatment had an obvious role in breaking seed dormancy, it did not fully break seed dormancy, in contrast with 10 weeks' seed stratification (Figures 5A, 7A). Most seeds treated with GA3 showed the same phenotype as seeds with insufficient chilling accumulation. This illustrated that gibberellin could not fully break peach seed dormancy. In fruit production, a combination of Gibberellic acid treatment and stratification is usually used to shorten the peach seed dormancy period and fully break seed dormancy.
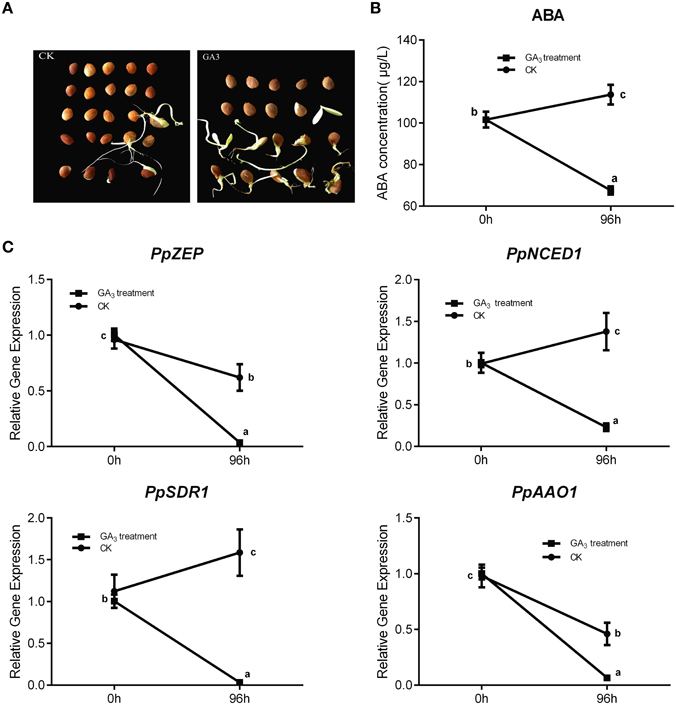
Figure 7. The effect of GA3 on seed dormancy release and its role in relating ABA metabolism. (A) Germination test was test after mature seeds treated with 1 mM GA3. (B) ABA changes after GA3 treatment in seed. (C) The expression analysis of ABA metabolic genes responding to GA3 treatment. Each experiment was performed with three biological replicates with error bars representing the SEM. Different letters above bars indicate a significant difference among chilling periods according to ANOVA and Duncan's test (P < 0.05).
After GA3 treatment, data showed that ABA level was decreased comparing with control (Figure 7B), and qRT-PCR analysis showed that the expressions of PpNCED1, PpZEP, PpSDR1, and PpAAO1 decreased compared with the control at 96 h (Figure 7C), whereas the transcript levels of all PpCYP707As showed no significant changes (data not shown).
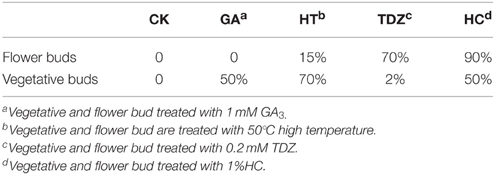
Effect of Different Measures on Dormancy Breaking of Peach Buds in the Period of Endodormancy
To test the effect of different dormancy breaking measures on peach bud dormancy, vegetative buds, and flower buds were treated with 1 m MGA3, high temperature (50°C), 1%HC, and 0.2 m MTDZ (thidiazuron). The data showed that different dormancy-breaking measures had different effects on flower bud and vegetative bud dormancy release (Table 3). For flower buds, HC treatment resulted in the earliest dormancy break and the highest bud burst rate at 25 days after treatment (90%), making it an effective way to break flower bud dormancy. 0.2 MTDZ treatment was less efficient (70% bud burst rate), and 50°C (15%) or 1 mM GA3 treatment (0%) were the least effective. For vegetative buds, 50°C treatment was the most effective way to break bud dormancy, with 70% of buds burst, and 1 mM GA3 and HC less effective (both 50% buds burst). 0.2 MTDZ had the lowest efficiency, with 2% buds burst at 25 days after treatment. In addition, 1 mM GA3and 1%HC also showed toxic effects on flower buds, which appeared as bark color changes, shoot tip drying and bud necrosis.
Discussion
Relationship Between Vegetative Bud and Flower Bud Dormancy with ABA Metabolic Crosstalk
Dormancy defends woody plants against cold winters, protecting young meristems from freeze injury, and is indispensable in the vital process of buds life activity under natural conditions. In addition, buds in different phases, from dormancy to reactivation of growth, have different responses to environmental factors to adapting to local climatic conditions. In previous studies, plant hormones were confirmed to play key roles in regulating the bud dormancy cycle in woody plants (Rohde and Bhalerao, 2007; Cooke et al., 2012). ABA is the main hormone that responds to various environmental stresses, which affects the circadian clock, which in turn regulates plants sensitivity to ABA (Robertson et al., 2009; Cooke et al., 2012). Zheng et al. (2015) supported the assumption that the effect of ABA was dependent on the developmental stage of the buds. This would explained by the following three scenarios, or a combination of them: (i) the total ABA level increases because of the increase in endogenous ABA levels; (ii) the removal of the added exogenous ABA is caused by an increase of ABA degradation capacity; and (iii) ABA no longer controls primordial growth activity after developmental phase transition.
Peach bud dormancy is induced by the photoperiod, and the change in ABA level seems to be related to seasonal changes. In peach, continuous growth proceeded at 23/17°C day/night temperature in a 16-h photoperiod, but growth ceased and entry into dormancy began with a photoperiod of ≤ 13 h at the same temperature (Heide, 2008; Barros et al., 2012). In our previous studies, we considered that lateral buds went into dormancy induction stage in mid-September in Taian, China. At the same time, the day length was ~12.5 h, with average temperatures ranging from 28°C (maximum) to 15°C (minimum) and the transcript levels of PpNCED1 and PpNCED2 were high in vegetative and flower bud dormancy induction periods, resulting in increased ABA synthesis (Figures 2B, 3). In peach, ABA concentration increased with the development of bud dormancy and it gradually decreased with dormancy release, and reached the minimum concentration in ecodormancy (Figure 3). This result was the same as poplar dormancy, in which SD induced growth cessation and ABA levels increased dramatically after 3–4 weeks of short days in apical buds, in parallel with up-regulation of NCEDs and genes encoding enzymes catalyzing ABA biosynthesis (Rohde et al., 2002). We observed that PpCYP707A1 was expressed at its highest level in the peach bud dormancy induction period (Figure 3); this might be relevant to water loss in the buds because CYP707A is involved in dehydration in many plants (Kushiro et al., 2004; Umezawa et al., 2006; Yang and Zeevaart, 2006). As the season progressed, the environmental temperature decreased and water loss continued in buds, resulting in deep dormancy of vegetative and flower buds in November; the increase in ABA level seemed to respond to the cold stress and waster loss. In deeply dormant potato tubers, the ABA content reached its highest level, which was also observed in grape and sweet cherry (Duan et al., 2004; Destefano-Beltrán et al., 2006; Zheng et al., 2015). When chilling accumulation reached about 300 h chilling requirements (CR) around 22 November), vegetative and flower buds burst under normal cultural environment in illumination incubator (Figure 2A). Thus, peach bud dormancy began to be released, and we considered that the period of bud dormancy release was from 22 November to 20 December, according to the bud germination test (Figure 2A). In this stage, the environmental temperature was lowest and living environment was poorest for buds, although they showed a potential to burst. In peach and grape, the ABA concentration declined during this period, which demonstrated that ABA inhibits bud burst (Ramina et al., 1994; Zheng et al., 2015). Our experimental data showed that the expressions of PpCYP707A2 and PpCYP707A3 were up-regulated during this stage to inactivate ABA, whereas PpNCED2 was down-regulated, which would inhibit ABA synthesis. This was similar to grape dormancy release: treatment with HC resulted in bud dormancy release accompanied by a decrease in ABA levels in grape buds that paralleled the induction of VvCYP707A4 expression and reduced VvNCED1 expression (Zheng et al., 2015). Besides, PpZEP expression also dramatically increased in vegetative and flower bud in this period, seemingly in to response to cold weather: overexpression of the ZEP gene enhanced the ability to resist chilling stress in tomatoes (Wang et al., 2008; Figure 3). After dormancy release, peach vegetative and flower buds entered the stage of ecodormancy after 20 December. During this period, the buds swelled and showed increased metabolic activity and water content, and ABA was no longer an inhibitor of bud burst: the ABA concentration gradually decreased after bud dormancy release, possibly because PpCYP707A3 was highly expressed to control the ABA content. At this stage, spraying ABA on peach vegetative and flower buds had no effect compared with the control, which were the same as the results with grape buds (data not shown).
In grape, ABA regulates bud dormancy and dormancy release by modifying ABA metabolism (Zheng et al., 2015). In our study, we found that NCED and CYP707A gene families are the key genes controlling ABA synthesis and catabolism. All the genes in our study showed similar expression patterns, which suggested that ABA crosstalk was comparable in vegetative and flower bud dormancy. The experiment was conducted in two different years (2013–2014 and 2014–2015) and all the genes related to ABA synthesis and catabolism were dramatically down-regulated on 22 November, before the buds entered the period of transition from endodormancy to ecodormancy (Figure 3); however, we cannot explain why this change occurred.
High Expression of ABA-Related Genes during Seed Development Might Induce Seed Dormancy
The central role of ABA in seed development and seed dormancy is well established and the key genes related to ABA synthesis and catabolism have been identified by studying plant mutants (Tan et al., 1997; Galpaz et al., 2008; Finkelstein, 2013). For most plants, ABA accumulates during seed development and declines during seed dormancy release (Kermode, 2005). To determine if the expressions of genes related to ABA metabolism correlated with seed ABA and dormancy levels, the expression patterns of certain ABA metabolic genes used to study bud dormancy were measured using qRT-PCR, and ABA concentration was measured by ELISA.
In Arabidopsis, the regulation network of the seed development program appears to control the induction of seed dormancy (Rikiishi and Maekawa, 2014). Generally, seed dormancy is classed into two major types: embryo dormancy and coat enhanced dormancy (Kermode, 2005). In peach seeds, the ABA concentration increased during the last developmental stage, and ABA accumulated in seed coats was higher compared with embryo (Figure 4). Our study found that all genes associated with ABA synthesis also increased during seed development, with the highest expression in the seed coat (Figure 4). Perhaps that was because a great quantity of ABA was synthesized in the seed coat to induce seed dormancy and prevent seed germination in late development. In Arabidopsis, AtNCED6 and AtNCED9 are required for ABA synthesis during seed development and show distinctive expression patterns. AtNCED6 is expressed exclusively in the endosperm, whereas AtNCED9 is expressed in testa and embryos (Lefebvre et al., 2006). In our study, PpNCED1 and PpNCED2 were found to be quite similar to AtNCED9 through similarity analysis (Figure 2) and both of them were highly expressed in seed coats, but they were barely expressed in the embryo, endosperm, and embryo, which was the same as AtNCED9. In barley, the expression of ABA2/SDR1, a member of the SDR gene family, indicated its broader role in ABA synthesis. HvSDR1, HvSDR2, and HvSDR4 were detected with high expression in embryos at various stages of seed maturation (Seiler et al., 2011); however, SDR1 was not detected in the Medicago truncatula seed coat (Verdier et al., 2013), whereas we found that PpSDR1 was expressed at low levels in the embryo and was highly expressed in the peach seed coat, which demonstrated that SDR1 shows species specific expression patterns. In Arabidopsis, AtAAO3 is the key gene catalyzing the final step of ABA biosynthesis in seed development, and ABA levels were low in the aao3 mutant, which showed reduced dormancy, compared with the wild-type (Seo et al., 2004). In barley, five putative AAO gene family members were detected that participate in seed development, indicating their potential importance in seed ABA synthesis. Each of them plays different roles in different seed tissues (Seiler et al., 2011). Interestingly, we identified only one AAO gene in our study. Our data showed that PpAAO1 expression increased in embryos, seeds and seed coats during late seed development (Figure 4) and increased during the period when the endosperm disappeared (data not shown). In addition to the genes related to ABA synthesis, ABA catabolic genes (CYP707A) were also induced during seed development to promote ABA inactivation (Figure 4). In Arabidopsis, in situ hybridization showed that AtCYP707A1 mRNA could be detected in the vascular tissue in the embryo, whereas AtCYP707A2 was found in the endosperm and vascular tissue in embryos during seed maturation (Okamoto et al., 2006). Similar to the PvCYP707A gene family in bean (Yang and Zeevaart, 2006), our data indicated that PpCYP707As also plays an overlapping role in peach seed development, Comparing the expression patterns of CYP707As in different seed tissues showed that PpCYP707A2 is the main gene in the CYP707A gene family that participates in embryo development.
In late seed development, the genes relating to ABA synthesis are activated resulting in high ABA synthesis (Figure 4). Their high expression in the seed coat suggested that high levels of ABA in the seed coat induce embryo dormancy during late seed maturation. Thus, peach seed dormancy seems to be seed coat enhanced dormancy. Generally, some CYP707A gene family members showed increased expression that mirrored the ABA increase during tissue development (Chono et al., 2006; Millar et al., 2006; Chono et al., 2013). These observations agreed with our study, and we believe that the highly expression of CYP707A family members was to control of ABA content at normal level functions via inducing ABA inactivation and antagonizing high ABA synthesis.
Peach Seed Dormancy Termination is Accompanied by Expression Changes in Genes Relating to ABA Biosynthesis and Catabolism
Stratification is an effective way to break seed dormancy in fruit trees. In a previous study, an in vitro culture experiment of peach seeds showed that seedlings would dwarf if chilling accumulation was insufficient (Leida et al., 2012). In general, seed dormancy release is accompanied by an obvious decrease in ABA content. The decline in ABA corresponded well with increasing germination capacity (Kermode, 2005). For example, the ABA content decreased in the embryo and megagametophyte during moist-chilling of Douglas fir seeds (Corbineau et al., 2002). In the dormancy termination of western white pine seeds, ABA and metabolite contents declined suddenly to relatively low levels in the seed coat after the pre-moist-chilling water soak, and in the embryo, the ABA content declined as ABA catabolism increased (Feurtado et al., 2004).
ABA catabolism is associated with dormancy release in some higher plants (Corbineau et al., 2002; Jacobsen et al., 2002; Feurtado et al., 2004); however, there are few reports on the molecular mechanism that regulates ABA metabolism to affect seed dormancy during moist-chilling. Previous studies on peach embryo dormancy showed that the ABA content dramatically declined after 1 week of stratification; however, the occurrence of seedling dwarfing did not improve even though ABA decreased to a quite a low level (Leida et al., 2012). In our study, we also found that ABA was decreased after at 1 week stratification, and then it gradually decreased and reach the minimum concentration after dormancy termination (Figure 6). All genes related to ABA synthesis were dramatically decreased after 1 week of stratification, and these changes corresponded to changes in ABA contents, especially for PpNCED1 and PpNCED2, whose expression declined in seeds and seed coats during stratification (Figure 6). However, the expression pattern of other genes relating to ABA synthesis was quite different in seed coats, but they declined along with stratification as a whole in seeds and embryos (Figure 6). In Arabidopsis, the expression of AtCYP707A regulated seed ABA levels and dormancy depth, as deduced from analysis of the cyp707a mutant (Okamoto et al., 2006). Furthermore, seed dormancy increase under cold-maturation is related to AtCYP707A2 down-regulation (Kendall et al., 2011). In our study, PpCYP707As showed different expression patterns in dormancy-release of peach seeds, which suggested that they have overlapping roles in ABA catabolism during bud dormancy release. PpCYP707A1 was highly expressed in the seed coat, whereas PpCYP707A2 and PpCYP707A3 were mainly expressed in the embryo, suggested that PpCYP707A1 and PpCYP707A3 contribute to the inactivation of ABA in the seed coat and embryo, respectively. In addition, their expressions were dramatically increased at 5 weeks of stratification (a key time point in seed dormancy release), and the burst rate of seeds reached 100% at the same time, which suggests that they play a critical role in seed germination. Gibberellin and ABA were generally believed to have antagonistic effects on the regulation of dormancy breakage and germination. However, there was no report on the molecular mechanism of ABA catabolism in seeds treated with GA3. We found that only the genes relating to ABA synthesis were down-regulated in seeds treated with GA3 for 96 h accompanied with ABA decrease (Figure 7); thus GA3 might regulate seed dormancy by regulating ABA synthesis.
Bud and Seed Dormancy are Similar but Different
Bud and seed dormancy are two different forms of dormancy that prevent pre-germination until environmental conditions become suitable for germination. Many studies have demonstrated similarities between seed dormancy and bud dormancy. For instance, in peach, expression analysis of genes identified by suppression subtractive hybridization during bud dormancy release showed similar expression patterns during embryo stratification (Leida et al., 2012). This demonstrated that there was a common regulation network in seed and bud dormancy. Moreover, expression of endoplasmic reticulum stress- and unfolded protein response-associated genes in bud endodormancy and seed stratification were similar in peach (Fu et al., 2014). ABA has been confirmed to control bud and seed dormancy as a stress hormone, and ABA accumulates during late seed development to induce seed dormancy in parallel with ABA accumulated in late autumn to induce bud dormancy. Furthermore, seed dormancy termination and bud dormancy release are associated with changes in ABA metabolism (Figures 3, 4). In the western white pine, dormancy termination of seeds is associated with changes in ABA metabolism (Feurtado et al., 2004), and grape bud dormancy is regulated by ABA through modification of ABA metabolism (Zheng et al., 2015). These observations supported the results of our study: peach seed and bud dormancy may be induced by two key genes contributing to ABA synthesis (PpNCED1 and PpNCED2) and terminated by up-regulation of CYP707A gene family, resulting in ABA inactivation.
In our study, we found that vegetative buds and flower buds showed different bud burst rates after different chemical treatments, which suggested different regulation pathways between them (Table 3); however, ABA pathway crosstalk was suggested by the similar expression patterns in the two. In addition, HC could break the dormancy of both flower and vegetative buds, and GA3 could break vegetative and seed dormancy, which demonstrated there must be crossover point between these three different types of dormancy.
Conclusions
Previous studies have demonstrated that ABA biosynthetic and catabolic genes play an important role in dormancy onset and release; however, there has been no systematic study on bud and seed dormancy that compared their differences. In our study, we found that PpCYP707As and PpNCEDs were the key genes in regulating peach seed and bud dormancy, which agreed with previous studies. The expression patterns of other genes indicated that their roles in regulating seed and bud dormancy are quite complex. During the induction of seed and bud dormancy, high expression of PpNCED1 and PpNCED2 might be associated with significant ABA synthesis, and the expressions of PpCYP707A2 and PpCYP707A3 suddenly increased during the period of dormancy-release maybe result in inactivation of ABA, which promotes seed germination and bud burst. Moreover, ABA crosstalk is similar between vegetative and flower bud dormancy, but different in seed dormancy, meanwhile the change trend of ABA concentration was much the same between seed dormancy and bud dormancy; this suggested that there might be another molecular mechanism contributing to ABA synthesis during bud dormancy. Even though we observed similarities and differences among the dormancy regulations of vegetative buds, flower buds, and seeds through studying ABA-related gene expressions, uncovering the integrated molecular network still requires further research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author Contributions
DW is the main author performed this work, and ZG contributed equally to this work. The others provided technical support and theoretical support to this work. LL and DG supervised the project.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31372050) and the Modern Agricultural Industry Technology System of Fruit Industry in Shandong province-Equipment of Cultivation and Facilities (SDAIT-03-022-05).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01248
References
Arora, R., Rowland, L. J., and Tanino, K. (2003). Induction and release of bud dormancy in woody perennials: a science comes of age. HortScience 38, 911–921.
Barros, P. M., Gonçalves, N., Saibo, N. J., and Oliveira, M. M. (2012). Cold acclimation and floral development in almond bud break: insights into the regulatory pathways. J. Exp. Bot. 63, 4585–4596. doi: 10.1093/jxb/ers144
Bittner, F., Oreb, M., and Mendel, R. R. (2001). ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J. Biol. Chem. 276, 40381–40384. doi: 10.1074/jbc.C100472200
Cheng, W. H., Endo, A., Zhou, L., Penney, J., Chen, H. C., Arroyo, A., et al. (2002). A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14, 2723–2743. doi: 10.1105/tpc.006494
Chono, M., Honda, I., Shinoda, S., Kushiro, T., Kamiya, Y., Nambara, E., et al. (2006). Field studies on the regulation of abscisic acid content and germinability during grain development of barley: molecular and chemical analysis of pre-harvest sprouting. J. Exp. Bot. 57, 2421–2434. doi: 10.1093/jxb/erj215
Chono, M., Matsunaka, H., Seki, M., Fujita, M., Kiribuchi-Otobe, C., Oda, S., et al. (2013). Isolation of a wheat (Triticum aestivum L.). mutant in ABA 8′-hydroxylase gene: effect of reduced ABA catabolism on germination inhibition under field condition. Breed. Sci. 63, 104–115. doi: 10.1270/jsbbs.63.104
Cooke, J. E. K., Eriksson, M. E., and Junttila, O. (2012). The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant Cell Environ. 35, 1707–1728. doi: 10.1111/j.1365-3040.2012.02552.x
Corbineau, F., Bianco, J., Garello, G., and Côme, D. (2002). Breakage of Pseudotsuga menziesii seed dormancy by cold treatment as related to changes in seed ABA sensitivity and ABA levels. Physiol. Plant. 114, 313–319. doi: 10.1034/j.1399-3054.2002.1140218.x
Destefano-Beltrán, L., Knauber, D., Huckle, L., and Suttle, J. (2006). Chemically forced dormancy termination mimics natural dormancy progression in potato tuber meristems by reducing ABA content and modifying expression of genes involved in regulating ABA synthesis and metabolism. J. Exp. Bot. 57, 2879–2886. doi: 10.1093/jxb/erl050
Duan, C., Li, X., Gao, D., Liu, H., and Li, M. (2004). Studies on regulations of endogenous ABA and GA3 in sweet cherry flower buds on dormancy. Acta Hortic. Sinica 31, 149–154. doi: 10.3321/j.issn:0513-353X.2004.02.002
Dutcher, R. D., and Powell, L. E. (1972). Culture of apple shoots from buds in vitro. J. Am. Soc. Hortic. Sci. 97, 511–514.
Endo, A., Nelson, K. M., Thoms, K., Abrams, S. R., Nambara, E., and Sato, Y. (2014). Functional characterization of xanthoxin dehydrogenase in rice. J. Plant Physiol. 171, 1231–1240. doi: 10.1016/j.jplph.2014.05.003
Feurtado, J. A., Ambrose, S. J., Cutler, A. J., Ross, A. R., Abrams, S. R., and Kermode, A. R. (2004). Dormancy termination of western white pine (Pinus monticola Dougl. Ex D Don) seeds is associated with changes in abscisic acid metabolism. Planta 218, 630–639. doi: 10.1007/s00425-003-1139-8
Finkelstein, R. (2013). Abscisic acid synthesis and response. Arabidopsis Book 11:e0166. doi: 10.1199/tab.0166
Frey, A., Audran, C., Marin, E., Sotta, B., and Marion-Poll, A. (1999). Engineering seed dormancy by the modification of zeaxanthin epoxidase gene expression. Plant Mol. Biol. 39, 1267–1274. doi: 10.1023/A:1006145025631
Frey, A., Effroy, D., Lefebvre, V., Seo, M., Perreau, F., Berger, A., et al. (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 70, 501–512. doi: 10.1111/j.1365-313X.2011.04887.x
Fu, X. L., Xiao, W., Wang, D. L., Chen, M., Tan, Q. P., Li, L., et al. (2014). Roles of endoplasmic reticulum stress and unfolded protein response associated genes in seed stratification and bud endodormancy during chilling accumulation in Prunus persica. PLoS ONE 9:e101808. doi: 10.1371/journal.pone.0101808
Galpaz, N., Wang, Q., Menda, N., Zamir, D., and Hirschberg, J. (2008). Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J. 53, 717–730. doi: 10.1111/j.1365-313X.2007.03362.x
González-Guzmán, M., Apostolova, N., Bellés, J. M., Barrero, J. M., Piqueras, P., Ponce, M. R., et al. (2002). The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14, 1833–1846. doi: 10.1105/tpc.002477
Gubler, F., Hughes, T., Waterhouse, P., and Jacobsen, J. (2008). Regulation of dormancy in barley by blue light and after-ripening: effects on abscisic acid and gibberellin metabolism. Plant Physiol. 147, 886–896. doi: 10.1104/pp.107.115469
Heide, O. M. (2008). Interaction of photoperiod and temperature in the control of growth and dormancy of prunus species. Sci. Hortic. 115, 309–314. doi: 10.1016/j.scienta.2007.10.005
Hellman, E., Shelby, S., and Lowery, C. (2006). Exogenously applied abscisic acid did not consistently delay budburst of deacclimating grapevines. J. Am. Pomol. Soc. 60, 178–186.
Holdsworth, M. J., Bentsink, L., and Soppe, W. J. J. (2008). Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179, 33–54. doi: 10.1111/j.1469-8137.2008.02437.x
Huang, Y., Cai, S., Ye, L., Hu, H., Li, C., and Zhang, G. (2016). The effects of GA and ABA treatments on metabolite profile of germinating barley. Food Chem. 192, 928–933. doi: 10.1016/j.foodchem.2015.07.090
Jacobsen, J. V., Pearce, D. W., Poole, A. T., Pharis, R. P., and Mander, L. N. (2002). Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol. Plant. 115, 428–441. doi: 10.1034/j.1399-3054.2002.1150313.x
Kendall, S. L., Hellwege, A., Marriot, P., Whalley, C., Graham, I. A., and Penfield, S. (2011). Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 23, 2568–2580. doi: 10.1105/tpc.111.087643
Kermode, A. R. (2005). Role of abscisic acid in seed dormancy. J. Plant Growth Regul. 24, 319–344. doi: 10.1007/s00344-005-0110-2
Koornneef, M., Jorna, M. L., Swan, B. V. D., and Karssen, C. M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 61, 385–393.
Kushiro, T., Okamoto, M., Nakabayashi, K., Yamagishi, K., Kitamura, S., Asami, T., et al. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J. 23, 1647–1656. doi: 10.1038/sj.emboj.7600121
Lang, G. A., Early, J. D., Martin, G. C., and Darnell, R. L. (1987). Endo-, para-, and ecodormancy: physiological terminology and classification for dormancy research. Hortic. Sci. 22, 371–377.
Lefebvre, V., North, H., Frey, A., Sotta, B., Seo, M., Okamoto, M., et al. (2006). Functional analysis of Arabidopsis nced6 and nced9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45, 309–319. doi: 10.1111/j.1365-313X.2005.02622.x
Leida, C., Conejero, A., Arbona, V., Gómez-Cadenas, A., Llácer, G., Badenes, M. L., et al. (2012). Chilling-dependent release of seed and bud dormancy in peach associates to common changes in gene expression. PLoS ONE 7:e35777. doi: 10.1371/journal.pone.0035777
Leida, C., Terol, J., Martí, G., Agustí, M., Llácer, G., Badenes, M. L., et al. (2010). Identification of genes associated with bud dormancy release in Prunus persica by suppression subtractive hybridization. Tree Physiol. 30, 655–666. doi: 10.1093/treephys/tpq008
Leng, P., Yuan, B., and Guo, Y. (2014). The role of abscisic acid in fruit ripening and responses to abiotic stress. J. Exp. Bot. 65, 4577–4588. doi: 10.1093/jxb/eru204
Liu, S., Lv, Y., Wan, X. R., Li, L. M., Hu, B., and Li, L. (2014). Cloning and expression analysis of cdnas encoding ABA 8′-hydroxylase in peanut plants in response to osmotic stress. PLoS ONE 9:e97025. doi: 10.1371/journal.pone.0097025
Marin, E., Nussaume, L., Quesada, A. M., Sotta, B., Hugueney, P., Frey, A., et al. (1996). Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 15, 2331–2342.
Millar, A. A., Jacobsen, J. V., Ross, J. J., Helliwell, C. A., Poole, A. T., Scofield, G., et al. (2006). Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. Plant J. 45, 942–954. doi: 10.1111/j.1365-313X.2006.02659.x
Nambara, E., Kawaide, H., Kamiya, Y., and Naito, S. (1998). Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol. 39, 853–858.
Nonogaki, H. (2014). Seed dormancy and germination—emerging mechanisms and new hypotheses. Front. Plant Sci. 5:233. doi: 10.3389/fpls.2014.00233
Okamoto, M., Kuwahara, A., Seo, M., Kushiro, T., Asami, T., Hirai, N., et al. (2006). CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 141, 97–107. doi: 10.1104/pp.106.079475
Qin, X., and Zeevaart, J. A. D. (2002). Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 128, 544–551. doi: 10.1104/pp.010663
Ramina, A., Colauzzi, M., Masia, A., Pitacco, A., Caruso, T., Messina, R., et al. (1994). Hormonal and climatological aspects of dormancy in peach buds. Acta Hortic. 395, 35–46.
Rikiishi, K., and Maekawa, M. (2014). Seed maturation regulators are related to the control of seed dormancy in wheat (Triticum aestivum L.). PLoS ONE 9:e107618. doi: 10.1371/journal.pone.0107618
Rinne, P., Saarelainen, A., and Junttila, O. (1994). Growth cessation and bud dormancy in relation to ABA level in seedlings and coppice shoots of Betula pubescens as affected by a short photoperiod, water stress and chilling. Physiol. Plant. 90, 451–458. doi: 10.1111/j.1399-3054.1994.tb08801.x
Robertson, F. C., Skeffington, A. W., Gardner, M. J., and Webb, A. A. (2009). Interactions between circadian and hormonal signalling in plants. Plant Mol. Biol. 69, 419–427. doi: 10.1007/s11103-008-9407-4
Rohde, A., and Bhalerao, R. P. (2007). Plant dormancy in the perennial context. Trends Plant Sci. 12, 217–223. doi: 10.1016/j.tplants.2007.03.012
Rohde, A., Prinsen, E., Rycke, R. D., Engler, G., Van Montagu, M., and Boerjan, W. (2002). PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. Plant Cell 14, 1885–1901. doi: 10.1105/tpc.003186
Rohde, A., Ruttink, T., Hostyn, V., Sterck, L., Vanessche, K., and Boerjan, W. (2007). Gene expression during the induction, maintenance, and release of dormancy in apical buds of poplar. J. Exp. Bot. 58, 4047–4060. doi: 10.1093/jxb/erm261
Ruttink, T., Arend, M., Morreel, K., Storme, V., Rombauts, S., Fromm, J. P., et al. (2007). A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 8, 2370–2390. doi: 10.1105/tpc.107.052811
Saika, H., Okamoto, M., Miyoshi, K., Kushiro, T., Shinoda, S., Jikumaru, Y., et al. (2007). Ethylene promotes submergence-induced expression of osABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol. 48, 287–298. doi: 10.1093/pcp/pcm003
Saito, S., Hirai, N., Matsumoto, C., Ohigashi, H., Ohta, D., Sakata, K., et al. (2004). Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134, 1439–1449. doi: 10.1104/pp.103.037614
Schwartz, S. H., Qin, X., and Zeevaart, J. A. (2003). Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 131, 1591–1601. doi: 10.1104/pp.102.017921
Seiler, C., Harshavardhan, V. T., Rajesh, K., Reddy, P. S., Strickert, M., Rolletschek, H., et al. (2011). ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J. Exp. Bot. 62, 2615–2632. doi: 10.1093/jxb/erq446
Seo, M., Aoki, H., Koiwai, H., Kamiya, Y., Nambara, E., and Koshiba, T. (2004). Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol. 45, 1694–1703. doi: 10.1093/pcp/pch198
Seo, M., Peeters, A. J., Koiwai, H., Oritani, T., Marion-Poll, A., Zeevaart, J. A., et al. (2000). The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. U.S.A. 97, 12908–12913. doi: 10.1073/pnas.220426197
Shalom, L., Samuels, S., Zur, N., Shlizerman, L., Doron-Faigenboim, A., Blumwald, E., et al. (2014). Fruit load induces changes in global gene expression and in abscisic acid (ABA) and indole acetic acid (IAA) homeostasis in citrus buds. J. Exp. Bot. 65, 3029–3044. doi: 10.1093/jxb/eru148
Shu, K., Zhang, H., Wang, S., Chen, M., Wu, Y., Tang, S., et al. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 9:e1003577. doi: 10.1371/journal.pgen.1003577
Suttle, J. C., Abrams, S. R., Stefano-Beltrán, L. D., and Huckle, L. L. (2012). Chemical inhibition of potato ABA-8′-hydroxylase activity alters in vitro and in vivo ABA metabolism and endogenous ABA levels but does not affect potato microtuber dormancy duration. J. Exp. Bot. 63, 5717–5725. doi: 10.1093/jxb/ers146
Tan, B. C., Joseph, L. M., Deng, W. T., Liu, L., Li, Q. B., Cline, K., et al. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35, 44–56. doi: 10.1046/j.1365-313X.2003.01786.x
Tan, B. C., Schwartz, S. H., Zeevaart, J. A., and McCarty, D. R. (1997). Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. U.S.A. 94, 12235–12240.
Thompson, A. J., Jackson, A. C., Symonds, R. C., Mulholland, B. J., Dadswell, A. R., Blake, P. S., et al. (2000). Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J. 23, 363–374. doi: 10.1046/j.1365-313x.2000.00789.x
Tung, S. A., Smeeton, R., White, C. A., Black, C. R., Taylor, I. B., Hilton, H. W., et al. (2008). Over-expression of LeNCED1 in tomato (Solanum lycopersicum L.) with the rbcS3C promoter allows recovery of lines that accumulate very high levels of abscisic acid and exhibit severe phenotypes. Plant Cell Environ. 31, 968–981. doi: 10.1111/j.1365-3040.2008.01812.x
Umezawa, T., Okamoto, M., Kushiro, T., Nambara, E., Oono, Y., Seki, M., et al. (2006). CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. 46, 171–182. doi: 10.1111/j.1365-313X.2006.02683.x
Vallabhaneni, R., and Wurtzel, E. T. (2010). From epoxycarotenoids to ABA: the role of ABA 8′-hydroxylases in drought-stressed maize roots. Arch. Biochem. Biophys. 504, 112–117. doi: 10.1016/j.abb.2010.07.005
Verde, I., Abbott, A. G., Scalabrin, S., Jung, S., Shu, S., Marroni, F., et al. (2013). The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet 45, 487–494. doi: 10.1038/ng.2586
Verdier, J., Dessaint, F., Schneider, C., and Abirached-Darmency, M. (2013). A combined histology and transcriptome analysis unravels novel questions on Medicago truncatula seed coat. J. Exp. Bot. 64, 459–470. doi: 10.1093/jxb/ers304
Wang, N., Fang, W., Han, H., Sui, N., Li, B., and Meng, Q. W. (2008). Overexpression of zeaxanthin epoxidase gene enhances the sensitivity of tomato PSII photoinhibition to high light and chilling stress. Physiol. Plant. 132, 384–396. doi: 10.1111/j.1399-3054.2007.01016.x
Yang, S. H., and Zeevaart, J. A. (2006). Expression of ABA 8′-hydroxylases in relation to leaf water relations and seed development in bean. Plant J. 47, 675–686. doi: 10.1111/j.1365-313X.2006.02815.x
Zheng, C., Halaly, T., Acheampong, A. K., Takebayashi, Y., Jikumaru, Y., Kamiya, Y., et al. (2015). Abscisic acid (ABA) regulates grape bud dormancy, and dormancy release stimuli may act through modification of ABA metabolism. J. Exp. Bot. 66, 1527–1542. doi: 10.1093/jxb/eru519
Zheng, Y., Crawford, G. W., and Chen, X. (2014). Archaeological evidence for peach (Prunus persica) cultivation and domestication in China. PLoS ONE 9:e106595. doi: 10.1371/journal.pone.0106595
Zheng, Y., Huang, Y., Xian, W., Wang, J., and Liao, H. (2012). Identification and expression analysis of the glycine max cyp707a gene family in response to drought and salt stresses. Ann. Bot. 110, 743–756. doi: 10.1093/aob/mcs133
Keywords: seed dormancy, bud dormancy, abscisic acid, ABA metabolic genes, peach
Citation: Wang D, Gao Z, Du P, Xiao W, Tan Q, Chen X, Li L and Gao D (2016) Expression of ABA Metabolism-Related Genes Suggests Similarities and Differences Between Seed Dormancy and Bud Dormancy of Peach (Prunus persica). Front. Plant Sci. 6:1248. doi: 10.3389/fpls.2015.01248
Received: 30 September 2015; Accepted: 21 December 2015;
Published: 11 January 2016.
Edited by:
Jaime Prohens, Universitat Politècnica de València, SpainReviewed by:
Hanaé Roman, Université d'Angers, FranceTakaya Moriguchi, NARO Institute of Fruit Tree Science, Japan
Copyright © 2016 Wang, Gao, Du, Xiao, Tan, Chen, Li and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Li, bGlsaW5nc2RhdUAxNjMuY29t;
Dongsheng Gao, ZHNnYW9Ac2RhdS5lZHUuY24=
†These authors have contributed equally to this work as co-first author.
 Dongling Wang
Dongling Wang Zhenzhen Gao1,2,3†
Zhenzhen Gao1,2,3†