
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 07 January 2016
Sec. Plant Biotechnology
Volume 6 - 2015 | https://doi.org/10.3389/fpls.2015.01203
This article is part of the Research Topic Mechanisms of abiotic stress responses and tolerance in plants: physiological, biochemical and molecular interventions View all 121 articles
The gas molecule nitric oxide (NO) can cooperate with ethylene to tightly modulate plant growth and stress responses. One of the mechanism of their crosstalk is that NO is able to activate ethylene biosynthesis, possibly through post-translational modification of key enzymes such as ACC synthase and oxidase by S-nitrosylation. In this paper, we focus on the crosstalk of NO with ethylene signaling transduction transcription factor EIN3 (Ethylene Insensitive 3) and downstream gene expression in alleviating germination inhibition and growth damage induced by high salt. The Arabidopsis lines affected in ethylene signaling (ein3eil1) and NO biosynthesis (nia1nia2) were employed to compare with the wild-type Col-0 and overexpressing line EIN3ox. Firstly, the obviously inhibited germination, greater ratio of bleached leaves and enhanced electrolyte leakage were found in ein3eil1 and nia1nia2 lines than in Col-0 plants upon high salinity. However, the line EIN3ox obtained a notably elevated ability to germinate and improved seedling resistance. The experiment with SNP alone or plus high salt mostly enhanced the expression of EIN3 transcripts, compared with ACO4 and ACS2. The western blot and transcript analysis found that high-salt-induced EIN3 stabilization and EIN3 transcripts were largely attenuated in the NO biogenesis mutant nia1nia2 plants than in Col-0 ones. This observation was confirmed by simulation experiments with NO scavenger cPTIO to block NO emission. Taken together, our study provides insights that NO promotes seed germination and seedlings growth under salinity may depend on EIN3 protein.
Nitric oxide (NO) is a small ubiquitous molecule, which is not only involved in the promotion of seed germination and cell death, shaping of root architecture, senescence, flowering, but also involved in responses to abiotic and biotic stresses, such as drought, salt, heat stress, disease resistance and apoptosis (Zhao et al., 2004; Bethke et al., 2006, 2007; Zhang et al., 2006; Hebelstrup et al., 2012; Arc et al., 2013a,b; Sanz et al., 2015). Two main enzyme-based pathways have been proposed to be functional for NO biosynthesis in plants (Lozano-Juste and Leon, 2010). One is based on the activity of nitrate reductase, which primarily catalyses the reduction of nitrate to nitrite and is encoded by two genes in Arabidopsis, designated NIA1 and NIA2 (Meyer et al., 2005; Modolo et al., 2005). Another is based on AtNOS1, although AtNOS1 was originally identified as a NOS-like enzyme that produces NO from L-arginine (Guo and Crawford, 2005), further studies confirmed that AtNOS1 lacked its originally reported NOS activity (Zemojtel et al., 2006).
The gaseous phytohormone ethylene serves as a diffusible hormone in plants (Bleecker and Kende, 2000). Ethylene responses are mediated through a signal transduction cascade in Arabidopsis thaliana, including five receptors to the single negative regulator CTR1 and two downstream positive components, EIN2 and EIN3 (Ethylene Insensitive 3) (Chao et al., 1997; Johnson and Ecker, 1998; Alonso et al., 1999; Chang and Stadler, 2001; Guo and Ecker, 2003). EIN3 is a plant-specific nuclear transcription factor that initiates downstream transcriptional cascades for ethylene responses (Yoo et al., 2008). EIN3 was reported to be both necessary and sufficient for the activation of the ethylene pathway. ERF1 (Ethylene Response Factors) is the target of EIN3, which is sufficient and indispensable for ERF1 expression (Solano et al., 1998). Ethylene has long been regarded as a stress hormone. It not only regulates developmental processes such as inhibition of cell expansion, induction of fruit ripening, petal and leaf abscission, organ senescence, but also adapt to stress conditions including salt stress and pathogen responses (Bleecker and Kende, 2000; Chang and Stadler, 2001; Achard et al., 2006).
Salinity is one of the major abiotic stresses that adversely affect crop productivity and quality (Munns, 2002). It was reported that salt stress not only delayed the germination process, reduced the germination rate, but also inhibited the formation of plant seedling after germination (Kim and Park, 2008; Lee et al., 2010). Plants have a complex and sophisticated network to antagonize the seed germination inhibition caused by salt stress and many plant hormones and signaling molecules are involved in the process (Achard et al., 2006; Tanou et al., 2009; Mur et al., 2013). The previous researchers proved that the small signaling molecules ABA, GA, SA, NO, and ETH were all directly involved in the regulation of seed germination inhibition or promotion under salt stress (Gniazdowska et al., 2007; Kim and Park, 2008; Lee et al., 2010; Arc et al., 2013a,b; Jiang et al., 2013; Lin et al., 2013a). Ethylene was proposed to release the impaired seed germination and seedling growth through crosstalk with ABA, ROS, and NO during plant response to stress conditions (Linkies et al., 2009; Asensi-Fabado et al., 2012; Lin et al., 2012; Arc et al., 2013b). NO could decrease ABA and GA ratio in seeds necessary to release inhibition of seed germination induced by salinity or break the dormancy (Bethke et al., 2007). Recent report showed that NO regulates hydrogen peroxide homeostasis in plants through interplaying with ascorbate peroxidase activity during plant stress responses (Lindermayr and Durner, 2015; Yang et al., 2015).
The interaction between ethylene biogenesis and NO has been increasingly attractive (Lin et al., 2013a; Lockhart, 2013; Sanz et al., 2015; Tanou et al., 2015). It has been reported that NO signaling in seeds could principally rely on PTM of specific proteins such as ACS and ACO by S-nitrosylation (Hebelstrup et al., 2012). On the other hand, ethylene biosynthesis can be reversibly inhibited by NO through S-nitrosylation of methionine adenosyltransferase, leading to the reduction of the S-AdoMet pool (Lindermayr et al., 2006; Mur et al., 2013). These observations indicated that there was janus face of NO action on ethylene, inductive or suppresive (Mur et al., 2013). We previously reported that application of exogenous ACC (a precursor of ethylene biosynthesis) or sodium nitroprusside (SNP, an NO donor) largely overcame the inhibition of germination induced by salinity (Lin et al., 2013a). Exogenous nitric oxide was reported to improve seed germination in wheat against mitochondrial oxidative damage induced by high salinity (Zheng et al., 2009). Salt stress causes in-planta accumulation of reactive oxygen species (ROS), which can result in oxidative stress and cellular damage (Lockhart, 2013). These reports highlighted a common property that either NO or ethylene could release the inhibited germination induced by salt-produced ROS stress. In addition, EIN3 had been reported to be sufficient to enhance Arabidopsis salt tolerance by decreasing ROS accumulation (Peng et al., 2014). The enhanced oxidative stress in the ethylene-insensitive (ein3-1) mutant of Arabidopsis thaliana exposed to salt stress supported the predominant role of EIN3 (Asensi-Fabado et al., 2012) in the control of ethylene-mediated plant salt responses. In this study, we hypothesize and provide evidence from both genetic and molecular views that NO modulates seed germination and seedling growth under high salinity through regulating EIN3 protein as well as downstream gene responses.
Salinity delayed seed germination of all four lines (Col-0, nia1nia2, ein3eil1, and EIN3ox) 3 days after treatment. High salinity (150 and 200 mM NaCl) significantly inhibited seed germination rate of ein3eil1 and nia1nia2, however, EIN3ox seeds showed obviously higher germination rate after 3 days of salt stress. Under MS and mild salinity, all four lines normally germinated, as the concentration of salt increased, seed germination rate obviously declined. Compared with Col-0 and EIN3ox, nearly 35% of nia1nia2 and ein3eil1 seeds germinated on the medium containing 100 mM NaCl, suggesting a significant delay in germination for the nia1nia2 and ein3eil1 mutant.
When grown on MS and mild salinity (50 and 100 mM NaCl) 7 days after light, no differences in germination rate were observed in the wild-type, nia1nia2, ein3eil1, and EIN3ox. Under high salt stressed conditions (150 and 200 mM NaCl), large differences in germination rate occurred between Col-0 and mutants (ein3eil1 and nia1nia2). High salt stress significantly inhibited ein3eil1 and nia1nia2 seed germination, however, nearly all seeds of EIN3ox germinated on 200 mM NaCl after 7 days (Figures 1A,C).
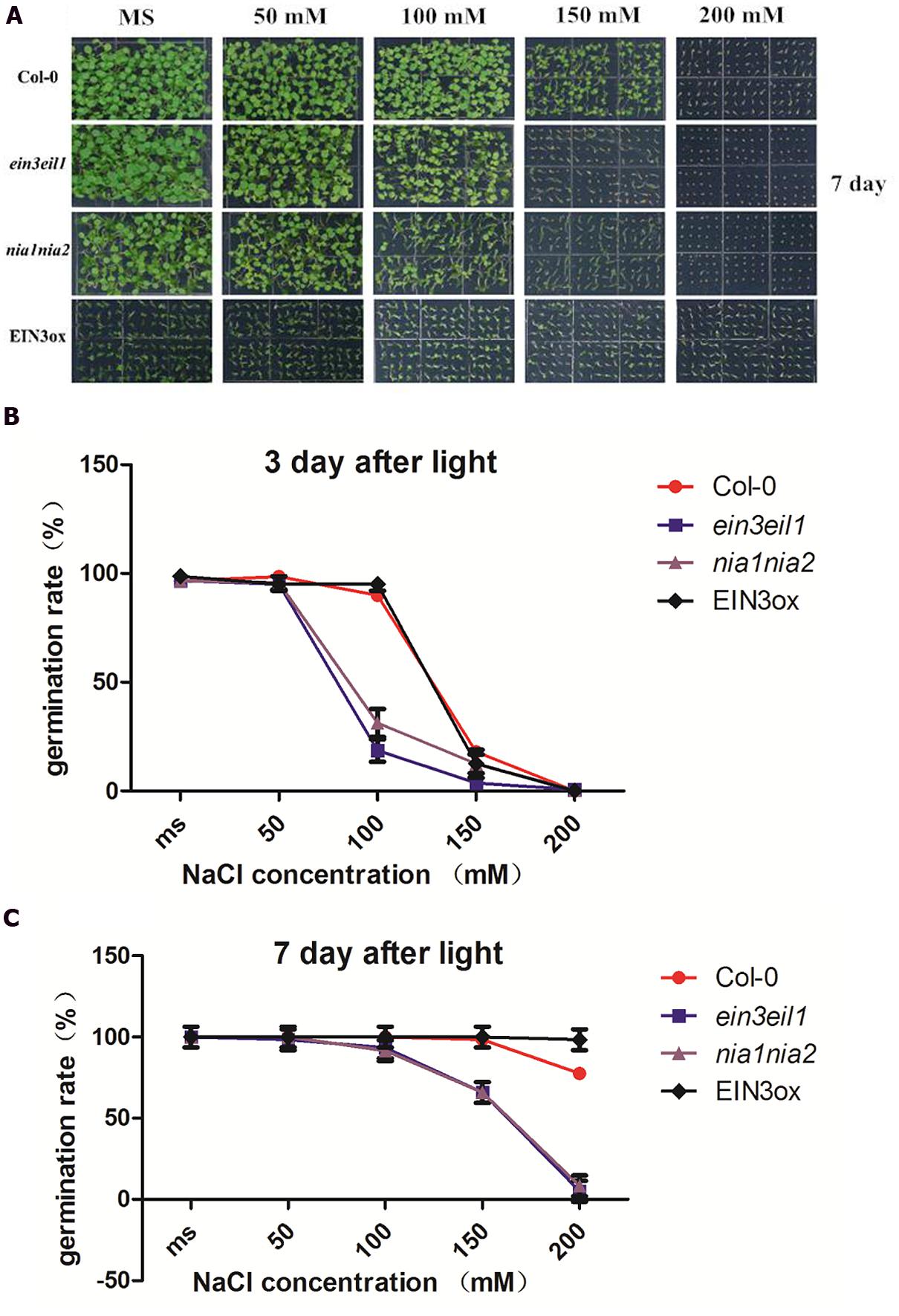
FIGURE 1. Comparative analysis of seed germination rate under salt stress among four lines of Arabidopsis. (A) Germinating status of Columbia (Col-0), ein3-1eil1-1, nia1-1nia2-5, and EIN3ox seeds from 0 to 200 mM NaCl concentration plate 7 days after transfer to light. (B) Germination rate of Col-0, ein3-1eil1-1, nia1-1nia2-5, and EIN3ox seeds on 0, 50, 100 150 200 mM NaCl conditions 3 days after transfer to light. (C) Germination rate of Col-0, ein3-1eil1-1, nia1-1nia2-5, and EIN3ox seeds on 0, 50, 100 150 200 mM NaCl conditions 7 days after transfer to light. Data are mean values (SE) of three independent experiments.
For phenotypic and physiological analysis under salt stress, the 5-day-old seedlings of Col-0, ein3-1eil1-1, nia1-1nia2-5, and EIN3ox were transferred onto MS agar plates containing 200 mM NaCl and their subsequent appearance was recorded photographically 3 days after transfer (Peng et al., 2014). The salt stress damages of seedlings were indicated by visibly bleached leaves and relative electrolyte leakage (REL). The phenotypic observation of Col-0 seedlings after transferred to salinity alone or plus NO scavenger cPTIO showed that the ratio of bleached seedlings obviously increased when cPTIO was supplemented (Figure 2A). We then compared the bleached response of four lines of seedlings 3 days after transferred and found that nia1nia2 and ein3eil1 mutant showed largely higher ratio of bleached leaves than Col-0 or EIN3ox seedlings (Figure 2B). The quantitative analysis according to the ratio of bleached to green leaves confirmed the above observations (Figure 2C). The REL of leaves under control or salt conditions was further measured to support phenotypic observation. The results showed that salt stress resulted in a higher enhancement of REL in the two mutant seedlings than in the wild-type and EIN3ox plants.
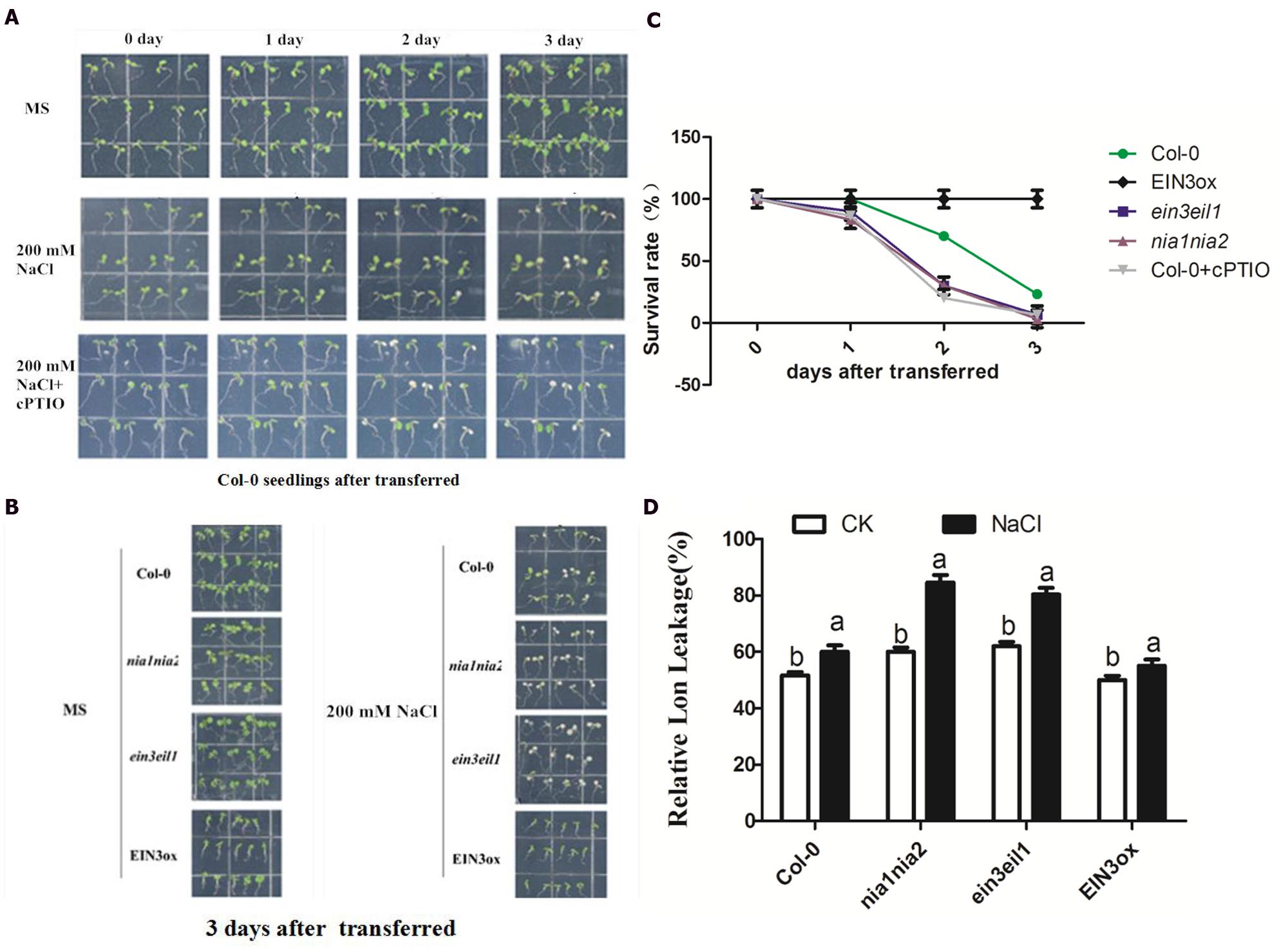
FIGURE 2. Phenotypic and physiological analysis of seedling tolerance upon high salt alone or plus NO scavenger cPTIO. (A) High salt generate visible bleached damage in wild type. 5-day-old Col-0 seedlings were continuously treated with 0 mM, 200 mM NaCl and 200 mM NaCl plus 200 μM cPTIO for 3 days. (B) High salt alone generates visible bleached damage in Col-0, ein3-1eil1-1, nia1-1nia2-5, and EIN3ox lines. 5-day-old seedlings were continuously treated with 0 mM NaCl or 200 mM NaCl 3 days. (C) The survival rate evaluated by visible bleaching rate. 5-day-old seedlings were treated with 200 mM NaCl for 3 days. (D) The comparison of relative electrolyte leakage (REL). 5-day-old seedlings were treated with 200 mM NaCl for 3 days. Data are mean values (SE) of three independent experiments. Within each set of experiments, bars with different letters were significantly different at the 0.05 level (Duncan’s test).
The ethylene signaling pathway transcription factor EIN3 is a critical regulator of plant salt responses. We found that the nia1nia2 and ein3eil1 seeds showed the similar germination rate and survival rate when transferred to MS medium supplemented with salinity (Figures 1 and 2). Consequently, EIN3 protein was detected in wild type and both nia1nia2 and ein3eil1 mutant. Western blotting results showed that EIN3 protein was dramatically increased after 3 h and 6 h of high salt treatment in wild-type Col-0, and EIN3 protein could not be detected in ein3eil1 mutant. At the same time, EIN3 protein level in nia1nia2 plants was slightly increased after 3 h of high salinity, but the abundance accumulated were still far less than Col-0 (Figure 3A). EIN3 mRNA level was also checked and qRT-PCR results showed that there was no obvious change after 3 h of salt treatment and only mild increase after 6 h of salt treatment in Col-0 (Figure 3B). The EIN3 mRNA in nia1nia2 even decreased after 3 and 6 h of salt treatment (Figure 3C).
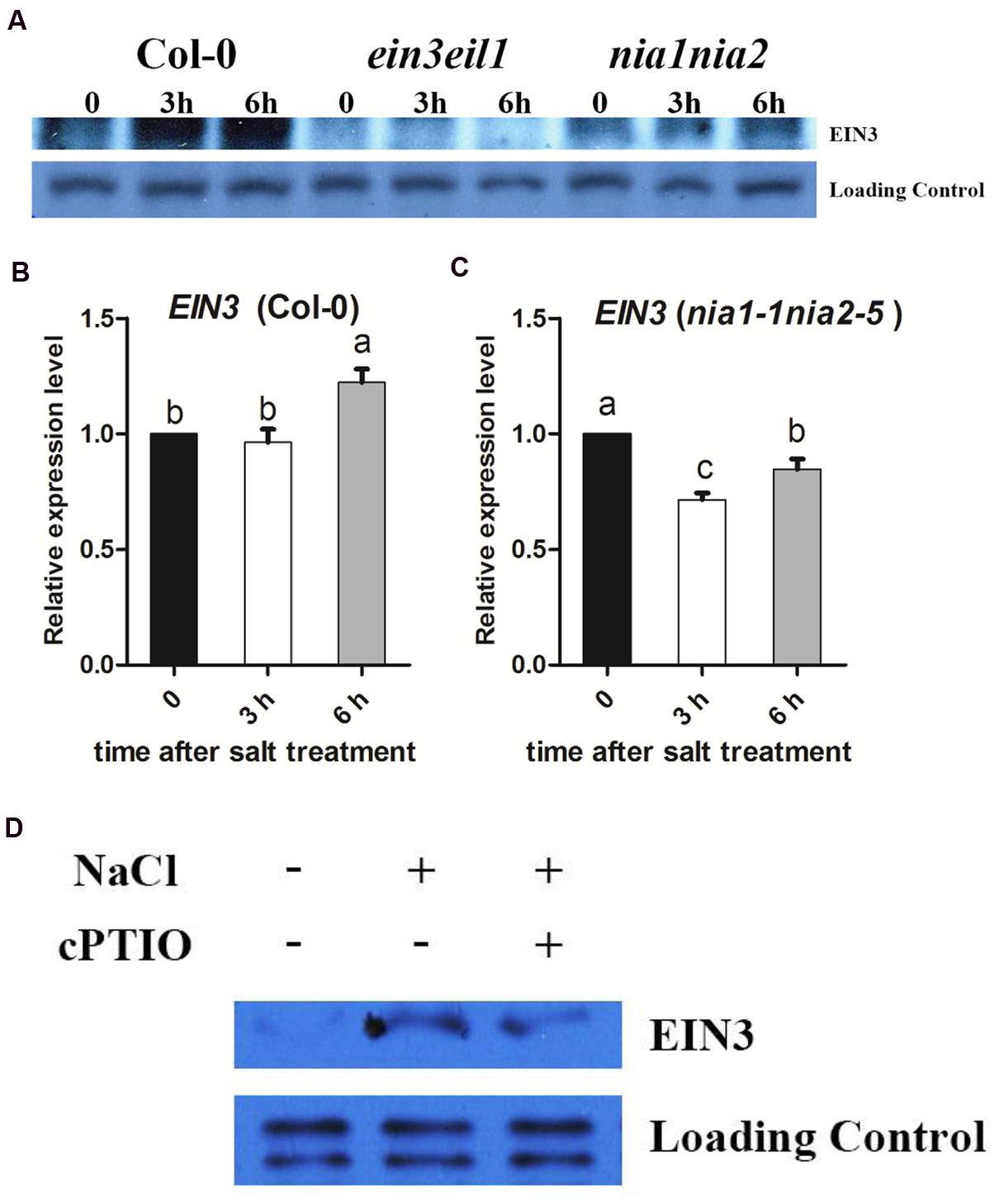
FIGURE 3. The salt induced EIN3 protein and transcript levels were modulated by NO release. (A) High salt promotes EIN3 protein accumulation. 5-day-old seedlings of Col-0, ein3-1eil1-1, and nia1-1nia2-5 were treated with 200 mM NaCl for 3 and 6 h. Protein was extracted and subjected to immunoblots using anti-EIN3 antibody. The non-specific band was used as a loading control. (B) EIN3 transcript analysis in wild type. 5-day-old seedlings were treated with 200 mM NaCl for 3 and 6 h. (C) EIN3 transcript analysis in nia1-1nia2-5. 5-day-old seedlings were treated with 200 mM NaCl for 3 and 6 h. (D) cPTIO impairs high salt induced EIN3 accumulation in wild type Col-0. 5-day-old seedlings were treated with 200 mM NaCl alone or plus 200 μM cPTIO for 6 h. The non-specific band was used as a loading control. Data are mean values (SE) of three independent experiments. Within each set of experiments, bars with different letters were significantly different at the 0.05 level (Duncan’s test).
On this basis, EIN3 protein was also detected to check whether NO scavenger cPTIO affected EIN3 protein accumulation or not under salt stress. Western blotting results showed that EIN3 protein in Col-0 dramatically increased after 6 h of high salt treatment, however, the EIN3 protein level was much lower when treated with high salt plus 200 μM cPTIO (Figure 3D).
To check whether the accumulation EIN3 protein is due to transcriptional function or not, the expression levels of the representative ethylene responsive genes ERF1 were measured. ERF1 transcript was up-regulated by high salt both in wild type Col-0 and nia1nia2 mutant, however, the increased amount of ERF1 expression in nia1nia2 is much lower than that in Col-0. The expression level of ERF1 increased eightfold after 6 h of high salt treatment, which coincided with the EIN3 protein level, however, in nia1nia2 mutants, 6 h high salt treatment only increased ERF1 expression about twofold (Figure 4A).
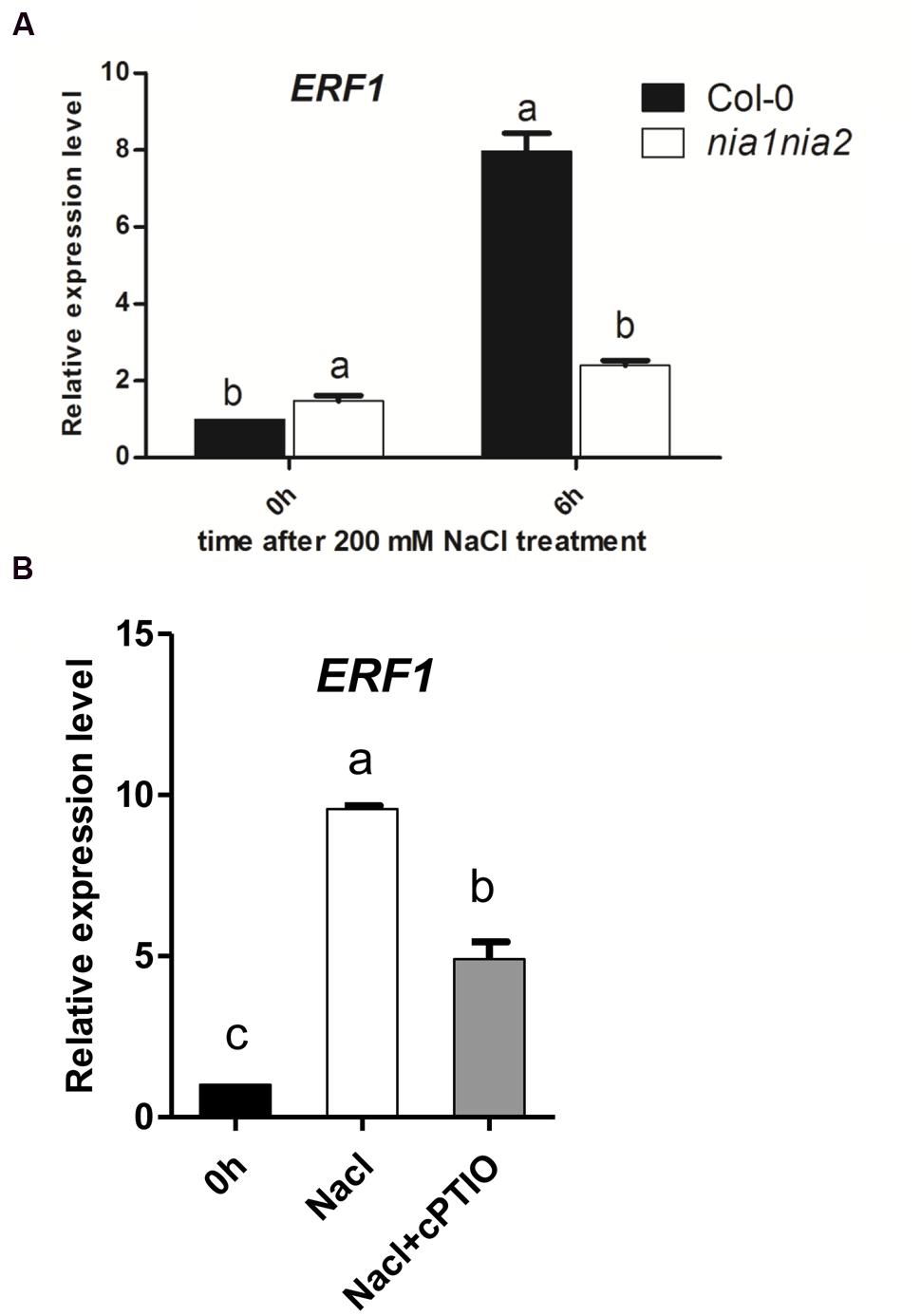
FIGURE 4. The salt induced ERF1 transcripts were modulated by NO release. (A) ERF1 transcript in Col-0 and nia1-1nia2-5. 5-day-old seedlings were treated with 200 mM NaCl for 6 h. (B) ERF1 transcript in Col-0 affected by salt and cPTIO. 5-day-old seedlings were treated with 200 mM NaCl alone or plus cPTIO for 6 h. Data are mean values (SE) of three independent experiments. Within each set of experiments, bars with different letters were significantly different at the 0.05 level (Duncan’s test).
Nitric oxide scavenger cPTIO were also used to treat wild type Col-0 and then check the ERF1 expression in seedlings affected by NO release under salt stress. The results showed that ERF1 gene expression was up-regulated by 200 mM NaCl treatment in wild type plants, which was consistent with the above-mentioned results (Figure 4A). The expression level of ERF1 increased approximately ninefold after 6 h of high salt treatment, however, the expression level of ERF1 decreased when seedlings were treated with high salt plus 200 μM cPTIO compared with high salt alone. In this case, the expression level of ERF1 only increased fourfold, which coincided with the EIN3 protein level observed (Figure 4B).
Compared to the control, no significant changes were observed in transcript levels of ethylene-synthesis related genes (ACO4 and ACS2) in Col-0 seedlings treated with SNP (50 μM) alone. However, the expression of ACO4 and ACS2 genes were increased markedly following salt stress. Furthermore, when SNP (50 μM) was applied to NaCl stressed seedlings at the same time, the expression of ACO4 and ACS2 genes were greatly induced compared to that under salinity alone (Figures 5A,B). On the contrary, treatment with SNP alone specifically enhanced the expression of EIN3 gene compared to ACO4 and ACS2. Treatment with salt and SNP together further increased EIN3 gene expression compared with the control (Figure 5C).
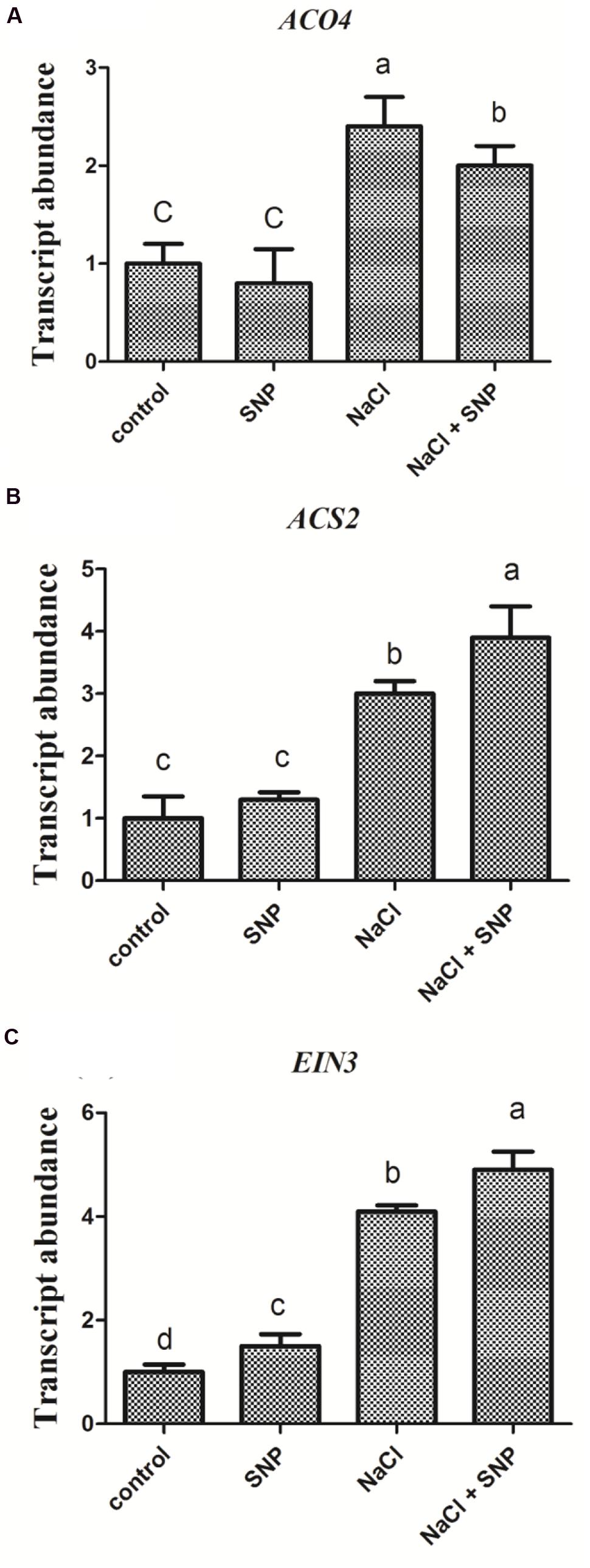
FIGURE 5. Transcriptional comparison of ACO4, ACS2, and EIN3 in Col-0 seedlings affected by high salt and NO donor. 5-day-old seedlings were exposed to four treatments 0 mM NaCl, 50 μM SNP, 200 mM NaCl, and 200 mM NaCl plus 50 μM SNP for 6 h. Then the seedlings were subjected to qRT-PCR analysis to compare the transcriptional abundance of ACO4 (A), ACS2 (B), and EIN3 (C). Data are mean values (SE) of three independent experiments. Within each set of experiments, bars with different letters were significantly different at the 0.05 level (Duncan’s test).
A critical role of NO in dormancy break or germination promotion has been demonstrated in many plants under optimal or stressed conditions (Bewley, 1997; Bethke et al., 2006, 2007; Libourel et al., 2006; Liu et al., 2009; Lin et al., 2013a; Lindermayr and Durner, 2015). NO is considered as a likely player of a signaling pathway that promotes loss of dormancy and has been suggested to behave as an endogenous regulator of this process (Arc et al., 2013a). The involved mechanisms had been widely investigated. A great deal of work proposed that NO-induced rapid decrease of abscisic acid concentration was required in breaking seed dormancy in Arabidopsis (Liu et al., 2009; Arc et al., 2013b; Wang et al., 2015). NO was demonstrated to act with ABA 8′-hydroxylase, resulting in ABA catabolism and breaking seed dormancy in Arabidopsis (Liu et al., 2009; Arc et al., 2013b). Increasing work indicated that NO could also interact with ethylene biosynthesis or signaling to break seed dormancy or improve salt tolerance. Dormancy removal in apple embryos by nitric oxide or cyanide involves modifications in ethylene biosynthetic pathway (Gniazdowska et al., 2007, 2010). In addition, the up-accumulation of NO under hypoxia can stimulate ethylene biosynthesis, possibly through PTM of key enzymes such as ACS and ACO by S-nitrosylation (Hebelstrup et al., 2012). Our previous study showed that NO and ethylene cooperated in enhancing seed germination of Arabidopsis under salinity (Lin et al., 2013a). All these evidence suggested a close crosstalk between NO and ethylene during plant growth and stress response. However, the directly functioning targets of NO in ethylene signaling pathway remain unclear.
Ethylene is a major phytohormone that regulates plant development in response to adverse environments (Wang et al., 2002; Guo and Ecker, 2003; Potuschak et al., 2003; Zhu et al., 2011). In ethylene signaling transduction pathway, EIN3 protein is the key transcription factor and EIN3/EIL1 (EIN3-LIKE 1) are both necessary and sufficient in the activation of transcription of ERF1 and other downstream genes (Guo and Ecker, 2003). The ein3-1eil1-1 double mutant exhibited remarkably reduced tolerance to high concentration of salt (Peng et al., 2014) and so this mutant was employed in this report. The experiment with EIN3-overexpressing (EIN3ox) line plants revealed that EIN3 was sufficient to enhance transcript levels of salt-related genes and salt tolerance (Zhang et al., 2011). Another important component in this pathway is EIN2 protein, which is in the upstream of EIN3 and positively regulates the functions of EIN3/EIL1. We previously reported that loss-of-function mutation of EIN2 protein in Arabidopsis exaggerated oxidative stress induced by salinity (Lin et al., 2013b). However, EIN2 was recently reported to be not necessary during high salinity, since that high salinity enhances EIN3 protein accumulation and transcriptional activity in both EIN2-dependent and EIN2-independent manners (Peng et al., 2014). NO was proposed to participate in diverse biological process through S-nitrosylation of nuclear proteins (Kovacs and Lindermayr, 2013; Mengel et al., 2013). It is coincident to find that EIN3/EIL1 are also stabilized and accumulated in the nucleus (Chao et al., 1997). These findings raise the assumption in this report that EIN3 is another NO targeted nuclear transcriptional factor by S-nitrosylation during their interplaying to release salt-induced seed germination inhibition.
Based on the above-mentioned four lines of Arabidopsis, we firstly compared the germination rate and seedling tolerance of ein3-1eil1-1, nia1-1nia2-5, and EIN3ox plants with the wild-type under salt conditions. Comparatively, the obviously decreased germination rate, greater ratio of bleached leaves and enhanced electrolyte leakage were found in ein3eil1 and nia1nia2 lines than in Col-0 plants upon high salinity. However, the line EIN3ox obtained a notably elevated ability to totally germinate and improved seedling resistance under high salt conditions. We then treated wild type Col-0 with NO scavenger cPTIO to mimic nia1nia2 mutant and the consistent results were achieved. It was reported that NO donor SNP greatly induced the expression of the ACS2 gene (Garcia et al., 2010; Lin et al., 2013a). Our experiment with SNP alone or plus high salt mostly enhanced the expression of EIN3 transcripts, compared with ACO4 and ACS2. These observations and reports then displayed a close crosstalk of NO with EIN3 protein to improve high salt resistance in Arabidopsis.
EIN3 protein level and downstream gene expression were then checked in the salt-stressed seedlings. We found that the high salt stabilized EIN3 protein accumulation and EIN3 transcripts were largely attenuated in the NO biogenesis mutant nia1nia2 plants than in Col-0 ones. For ein3eil1 line, EIN3 protein could not be detected under control or salt condition. Our data showed NO biogenesis mutant nia1nia2 impair EIN3 protein accumulation and transcript, especially induced by high salt. Our simulation experiments with NO scavenger cPTIO to block NO emission confirmed the above results with mutant plants. This evidence verifies that loss of NO will inhibit EIN3 level and gene transcripts under salt stress. The function of ethylene is finally accomplished by the signaling transduction especially the key transcription factor EIN3 protein level and downstream gene expression (Rieu et al., 2003; Asensi-Fabado et al., 2012). The classic EIN3 downstream ethylene responsive gene ERF1 transcript was simultaneously detected and the changes coincided with EIN3 protein and its transcript in salt-stressed Col-0 and nia1nia2 plants. All above data clearly indicates that NO modulates the ethylene signaling pathway transcription factor EIN3 accumulation to counteract salt stress.
Nitric oxide had been increasingly reported to modulate numerous cellular functions in plants through protein post-translational modifications (PTMs) of nuclear enzyme activities (Lindermayr et al., 2006; Tanou et al., 2009; Kovacs and Lindermayr, 2013; Mengel et al., 2013; Wang et al., 2015). The ethylene biosynthesis enzyme ACS (Lindermayr et al., 2006), ascorbate peroxidase (Yang et al., 2015) and OST1 (open stomata 1) (Wang et al., 2015) had all been successfully identified to be the S-nitrosylated substrates. In this way, NO establishes a comprehensive crosstalk network with other signaling molecules, such as ABA, JA, ethylene, ROS to regulate plant growth and defense (Abat and Deswal, 2009; Tanou et al., 2009; Manjunatha et al., 2012; Kovacs and Lindermayr, 2013; Wang et al., 2015). Here, we propose that NO promotes seed germination and seedling growth under high salt condition may depend on EIN3 protein accumulation and the downstream targeted responsive gene in Arabidopsis. Further study need to investigate whether NO also modify EIN3 activity through S-nitrosylation, which had been found in the way of NO acting on ACS, ACO activities in plants.
The wild type was Columbia ecotype (Col-0), an ein3 and eil1 double mutant (ein3-1eil1-1) (Alonso et al., 1999), and a transgenic line overexpressing EIN3 (EIN3ox) (Chao et al., 1997) and SALK line nia1nia2 (nia1-1nia2-5) were used in the study. Surface-sterilized seeds were sown on Murashige and Skoog (MS) medium (4.3 g/L MS salts, 10 g/L Suc, pH 5.7 to 5.8, and 8 g/L agar) imbibed at 4°C for 3 days in the dark and then germinated at 23°C under a 16/8 light/dark regime.
For germination assays, at least 120 seeds per plate from Col-0, ein3-1eil1-1, nia1nia2, and EIN3ox plants and three replicates of each were sowed onto MS medium supplemented with various concentrations of NaCl (50, 100, 150, and 200 mM). Germination was assessed from 3rd days to 7th days after transfer to light. A seed was considered as germinated when the radical protruded through the envelopes.
For bleaching analysis of salt-stressed leaves, 5 days seedlings were transferred onto MS agar plates containing 200 mM NaCl alone or plus 200 μM cPTIO and their subsequent appearance was recorded photographically 3 days after transferred. The seedling survival rate was simultaneously scored as bleaching ratio of leaves (Peng et al., 2014). Leaf REL were measured as described previously (Peever and Higgins, 1989).
Five days seedlings of Col-0, ein3-1eil1-1 and nia1-1nia2-5 were treated with 200 mM NaCl for 3 and 6 h. Protein was extracted and subjected to immunoblots using anti-EIN3 antibody. Plant samples were ground in liquid N2 and soluble protein extracts were made by homogenization in 50 mM Tris–HCl (pH 8.0), 10 mM NaCl, 0.1 M PMSF, and 0.1 M DTT, with subsequent centrifugation at 13000 g for 30 min at 4°C The protein in the supernatants was quantified by Bradford’s assay (Bradford, 1976). Western blot analysis was performed as described previously (Alonso et al., 1999) with anti-EIN3 antibodies (Guo and Ecker, 2003).
For expression profiling of ethylene-related genes in seedlings under salt stress in the presence of SNP or cPTIO was used as the NO donor and inhibitor. The seedlings were transferred onto MS agar plates containing 200 mM NaCl or 200 mM NaCl plus 50 μM SNP for 3–6 h. There were three replicates of each type of seed treatment plate. The qRT-PCR assay has been designed according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (Remans et al., 2014).
Total RNA was prepared using the TRIzol reagent (Invitrogen). Two micrograms of total RNA treated with DNase I (Promega) was added in a 20 μL reverse transcription reaction using the M-MLV reverse transcription system (Promega). Real-time PCR was performed using SYBR Green Mix (Takara) and the specific primers:
UBQ10-F: TCTCGTCTCTGTTATGCTTAAGAAG,
UBQ10-R: AGAAAGAAAGAGATAACAGGAACGG,
ERF1-F: ACCGCTCCGTGAAGTTAGATAATG,
ERF1-R: ATCCTAATCTTTCACCAAGTCCCAC,
EIN3-F: TGGAGAGACAAAATGCGGCT,
EIN3-R: ATAGCCGCAGGACCATTACG,
ACS2-F: AGGCAATTGCACATTTCATGG,
ACS2-R: CTGTCCGCCACCTCAAGTCT,
ACO4-F: CCGATGTCCCTGATCTCGAC,
ACO4- R: AGTCGGTCTTTTCGACCCGTA,
The expression level was normalized to that of the UBIQUITIN10 (UBQ10) control. The primer efficiency was calculated from qRT-PCR of the serial dilution of total cDNA, and the specificity of the primers was confirmed by the dissociation curve for each primer set in accordance with the MIQE guidelines.
Statistical treatment was carried out by analysis of variance (ANOVA) using SPSS-19 statistical software package. Results were represented as the means ± SE. Differences between treatments were separated by Duncan multiple range test at a 0.05 probability level.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Guo Hongwei (Peking University, China) for providing Arabidopsis seeds ein3eil1 and EIN3ox used in this report. This work was supported by National Natural Science Foundation of China (31370007).
Abat, J. K., and Deswal, R. (2009). Differential modulation of S-nitrosoproteome of Brassica juncea by low temperature: change in S-nitrosylation of Rubisco is responsible for the inactivation of its carboxylase activity. Proteomics 9, 4368–4380. doi: 10.1002/pmic.200800985
Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. doi: 10.1126/science.1118642
Alonso, J. M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J. R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152. doi: 10.1126/science.284.5423.2148
Arc, E., Galland, M., Godin, B., Cueff, G., and Rajjou, L. (2013a). Nitric oxide implication in the control of seed dormancy and germination. Front. Plant Sci. 4:346. doi: 10.3389/fpls.2013.00346
Arc, E., Sechet, J., Corbineau, F., Rajjou, L., and Marion-Poll, A. (2013b). ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 4:63. doi: 10.3389/fpls.2013.00063
Asensi-Fabado, M. A., Cela, J., Muller, M., Arrom, L., Chang, C., and Munne-Bosch, S. (2012). Enhanced oxidative stress in the ethylene-insensitive (ein3-1) mutant of Arabidopsis thaliana exposed to salt stress. J. Plant Physiol. 169, 360–368. doi: 10.1016/j.jplph.2011.11.007
Bethke, P. C., Libourel, I. G., Aoyama, N., Chung, Y. Y., Still, D. W., and Jones, R. L. (2007). The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 143, 1173–1188. doi: 10.1104/pp.106.093435
Bethke, P. C., Libourel, I. G., and Jones, R. L. (2006). Nitric oxide reduces seed dormancy in Arabidopsis. J. Exp. Bot. 57, 517–526. doi: 10.1093/jxb/erj060
Bewley, J. D. (1997). Seed germination and dormancy. Plant Cell 9, 1055–1066. doi: 10.1105/tpc.9.7.1055
Bleecker, A. B., and Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Chang, C., and Stadler, R. (2001). Ethylene hormone receptor action in Arabidopsis. Bioessays 23, 619–627. doi: 10.1002/bies.1087
Chao, Q., Rothenberg, M., Solano, R., Roman, G., Terzaghi, W., and Ecker, J. R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144. doi: 10.1016/S0092-8674(00)80300-1
Garcia, M. J., Lucena, C., Romera, F. J., Alcantara, E., and Perez-Vicente, R. (2010). Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. J. Exp. Bot. 61, 3885–3899. doi: 10.1093/jxb/erq203
Gniazdowska, A., Dobrzynska, U., Babanczyk, T., and Bogatek, R. (2007). Breaking the apple embryo dormancy by nitric oxide involves the stimulation of ethylene production. Planta 225, 1051–1057. doi: 10.1007/s00425-006-0384-z
Gniazdowska, A., Krasuska, U., and Bogatek, R. (2010). Dormancy removal in apple embryos by nitric oxide or cyanide involves modifications in ethylene biosynthetic pathway. Planta 232, 1397–1407. doi: 10.1007/s00425-010-1262-2
Guo, F. Q., and Crawford, N. M. (2005). Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. Plant Cell 17, 3436–3450. doi: 10.1105/tpc.105.037770
Guo, H., and Ecker, J. R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115, 667–677. doi: 10.1016/S0092-8674(03)00969-3
Hebelstrup, K. H., van Zanten, M., Mandon, J., Voesenek, L. A. C. J., Harren, F. J. M., Cristescu, S. M., et al. (2012). Haemoglobin modulates NO emission and hyponasty under hypoxia-related stress in Arabidopsis thaliana. J. Exp. Bot. 63, 5581–5591. doi: 10.1093/jxb/ers210
Jiang, C., Belfield, E. J., Cao, Y., Smith, J. A., and Harberd, N. P. (2013). An Arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell 25, 3535–3552. doi: 10.1105/tpc.113.115659
Johnson, P. R., and Ecker, J. R. (1998). The ethylene gas signal transduction pathway: a molecular perspective. Annu. Rev. Genet. 32, 227–254. doi: 10.1146/annurev.genet.32.1.227
Kim, S. G., and Park, C. M. (2008). Gibberellic acid-mediated salt signaling in seed germination. Plant Signal. Behav. 3, 877–879. doi: 10.4161/psb.3.10.6247
Kovacs, I., and Lindermayr, C. (2013). Nitric oxide-based protein modification: formation and site-specificity of protein S-nitrosylation. Front. Plant Sci. 4:137. doi: 10.3389/fpls.2013.00137
Lee, S., Kim, S. G., and Park, C. M. (2010). Salicylic acid promotes seed germination under high salinity by modulating antioxidant activity in Arabidopsis. New Phytol. 188, 626–637. doi: 10.1111/j.1469-8137.2010.03378.x
Libourel, I. G., Bethke, P. C., De Michele, R., and Jones, R. L. (2006). Nitric oxide gas stimulates germination of dormant Arabidopsis seeds: use of a flow-through apparatus for delivery of nitric oxide. Planta 223, 813–820. doi: 10.1007/s00425-005-0117-8
Lin, Y., Yang, L., Paul, M., Zu, Y., and Tang, Z. (2013a). Ethylene promotes germination of Arabidopsis seed under salinity by decreasing reactive oxygen species: evidence for the involvement of nitric oxide simulated by sodium nitroprusside. Plant Physiol. Biochem. 73, 211–218. doi: 10.1016/j.plaphy.2013.10.003
Lin, Y. C., Chen, D. D., Paul, M., Zu, Y. G., and Tang, Z. H. (2013b). Loss-of-function mutation of EIN2 in Arabidopsis exaggerates oxidative stress induced by salinity. Acta Physiol. Plant. 35, 1319–1328. doi: 10.1007/s11738-012-1172-y
Lin, Y. C., Wang, J. J., Zu, Y. G., and Tang, Z. H. (2012). Ethylene antagonizes the inhibition of germination in Arabidopsis induced by salinity by modulating the concentration of hydrogen peroxide. Acta Physiol. Plant. 34, 1895–1904. doi: 10.1007/s11738-012-0989-8
Lindermayr, C., and Durner, J. (2015). Interplay of reactive oxygen species and nitric oxide: nitric oxide coordinates reactive oxygen species homeostasis. Plant Physiol. 167, 1209–1210. doi: 10.1104/pp.15.00293
Lindermayr, C., Saalbach, G., Bahnweg, G., and Durner, J. (2006). Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-Nitrosylation. J. Biol. Chem. 281, 4285–4291. doi: 10.1074/jbc.M511635200
Linkies, A., Muller, K., Morris, K., Tureckova, V., Wenk, M., Cadman, C. S., et al. (2009). Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 21, 3803–3822. doi: 10.1105/tpc.109.070201
Liu, Y., Shi, L., Ye, N., Liu, R., Jia, W., and Zhang, J. (2009). Nitric oxide-induced rapid decrease of abscisic acid concentration is required in breaking seed dormancy in Arabidopsis. New Phytol. 183, 1030–1042. doi: 10.1111/j.1469-8137.2009.02899.x
Lockhart, J. (2013). Salt of the earth: ethylene promotes salt tolerance by enhancing Na/K homeostasis. Plant Cell 25:3150. doi: 10.1105/tpc.113.250911
Lozano-Juste, J., and Leon, J. (2010). Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR- and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiol. 152, 891–903. doi: 10.1104/pp.109.148023
Manjunatha, G., Gupta, K. J., Lokesh, V., Mur, L. A., and Neelwarne, B. (2012). Nitric oxide counters ethylene effects on ripening fruits. Plant Signal. Behav. 7, 476–483. doi: 10.4161/psb.19523
Mengel, A., Chaki, M., Shekariesfahlan, A., and Lindermayr, C. (2013). Effect of nitric oxide on gene transcription - S-Nitrosylation of nuclear proteins. Front. Plant Sci. 4:293. doi: 10.3389/fpls.2013.00293
Meyer, C., Lea, U. S., Provan, F., Kaiser, W. M., and Lillo, C. (2005). Is nitrate reductase a major player in the plant NO (nitric oxide) game? Photosynth. Res. 83, 181–189. doi: 10.1007/s11120-004-3548-3
Modolo, L. V., Augusto, O., Almeida, I. M., Magalhaes, J. R., and Salgado, I. (2005). Nitrite as the major source of nitric oxide production by Arabidopsis thaliana in response to Pseudomonas syringae. FEBS Lett. 579, 3814–3820. doi: 10.1016/j.febslet.2005.05.078
Munns, R. (2002). Comparative physiology of salt and water stress. Plant Cell Environ. 25, 239–250. doi: 10.1046/j.0016-8025.2001.00808.x
Mur, L. A., Prats, E., Pierre, S., Hall, M. A., and Hebelstrup, K. H. (2013). Integrating nitric oxide into salicylic acid and jasmonic acid/ ethylene plant defense pathways. Front. Plant Sci. 4:215. doi: 10.3389/fpls.2013.00215
Peever, T. L., and Higgins, V. J. (1989). Electrolyte leakage, lipoxygenase, and lipid peroxidation induced in tomato leaf tissue by specific and nonspecific elicitors from Cladosporium fulvum. Plant Physiol. 90, 867–875. doi: 10.1104/pp.90.3.867
Peng, J., Li, Z., Wen, X., Li, W., Shi, H., Yang, L., et al. (2014). Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 10:e1004664. doi: 10.1371/journal.pgen.1004664
Potuschak, T., Lechner, E., Parmentier, Y., Yanagisawa, S., Grava, S., Koncz, C., et al. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115, 679–689. doi: 10.1016/S0092-8674(03)00968-1
Remans, T., Keunen, E., Bex, G. J., Smeets, K., Vangronsveld, J., and Cuypers, A. (2014). Reliable gene expression analysis by reverse transcription-quantitative PCR: reporting and minimizing the uncertainty in data accuracy. Plant Cell 26, 3829–3837. doi: 10.1105/tpc.114.130641
Rieu, I., Mariani, C., and Weterings, K. (2003). Expression analysis of five tobacco EIN3 family members in relation to tissue-specific ethylene responses. J. Exp. Bot. 54, 2239–2244. doi: 10.1093/jxb/erg240
Sanz, L., Albertos, P., Mateos, I., Sánchez-Vicente, I., Lechón, T., Fernández-Marcos, M., et al. (2015). Nitric oxide (NO) and phytohormones crosstalk during early plant development. J. Exp. Bot. 66, 2857–2868. doi: 10.1093/jxb/erv213
Solano, R., Stepanova, A., Chao, Q., and Ecker, J. R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. doi: 10.1101/gad.12.23.3703
Tanou, G., Job, C., Rajjou, L., Arc, E., Belghazi, M., Diamantidis, G., et al. (2009). Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J. 60, 795–804. doi: 10.1111/j.1365-313X.2009.04000.x
Tanou, G., Minas, I. S., Karagiannis, E., Tsikou, D., Audebert, S., Papadopoulou, K. K., et al. (2015). The impact of sodium nitroprusside and ozone in kiwifruit ripening physiology: a combined gene and protein expression profiling approach. Ann. Bot. 116, 649–662. doi: 10.1093/aob/mcv107
Wang, K. L., Li, H., and Ecker, J. R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14(Suppl.), S131–S151.
Wang, P., Du, Y., Hou, Y. J., Zhao, Y., Hsu, C. C., Yuan, F., et al. (2015). Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. U.S.A. 112, 613–618. doi: 10.1073/pnas.1423481112
Yang, H., Mu, J., Chen, L., Feng, J., Hu, J., Li, L., et al. (2015). S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 167, 1604–1615. doi: 10.1104/pp.114.255216
Yoo, S. D., Cho, Y. H., Tena, G., Xiong, Y., and Sheen, J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451, 789–795. doi: 10.1038/nature06543
Zemojtel, T., Frohlich, A., Palmieri, M. C., Kolanczyk, M., Mikula, I., Wyrwicz, L. S., et al. (2006). Plant nitric oxide synthase: a never-ending story? Trends Plant Sci. 11, 524–525. doi: 10.1016/j.tplants.2006.09.008
Zhang, L., Li, Z., Quan, R., Li, G., Wang, R., and Huang, R. (2011). An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol. 157, 854–865. doi: 10.1104/pp.111.179028
Zhang, Y., Wang, L., Liu, Y., Zhang, Q., Wei, Q., and Zhang, W. (2006). Nitric oxide enhances salt tolerance in maize seedlings through increasing activities of proton-pump and Na+/H+ antiport in the tonoplast. Planta 224, 545–555. doi: 10.1007/s00425-006-0242-z
Zhao, L., Zhang, F., Guo, J., Yang, Y., Li, B., and Zhang, L. (2004). Nitric oxide functions as a signal in salt resistance in the calluses from two ecotypes of reed. Plant Physiol. 134, 849–857. doi: 10.1104/pp.103.030023
Zheng, C., Jiang, D., Liu, F., Dai, T., Liu, W., Jing, Q., et al. (2009). Exogenous nitric oxide improves seed germination in wheat against mitochondrial oxidative damage induced by high salinity. Environ. Exp. Bot. 67, 222–227. doi: 10.1016/j.envexpbot.2009.05.002
Keywords: Arabidopsis, seed germination, salt stress, nitric oxide, ethylene
Citation: Li X, Pan Y, Chang B, Wang Y and Tang Z (2016) NO Promotes Seed Germination and Seedling Growth Under High Salt May Depend on EIN3 Protein in Arabidopsis. Front. Plant Sci. 6:1203. doi: 10.3389/fpls.2015.01203
Received: 21 August 2015; Accepted: 14 December 2015;
Published: 07 January 2016.
Edited by:
Mohammad Anwar Hossain, Bangladesh Agricultural University, BangladeshReviewed by:
Françoise Corbineau, University Pierre et Marie Curie, FranceCopyright © 2016 Li, Pan, Chang, Wang and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhonghua Tang, dGFuZ3poQG5lZnUuZWR1LmNu; Yucheng Wang, d2FuZ3l1Y2hlbmcxMDI5QDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.