- 1Graduate School of Biological Sciences, Nara Institute of Science and Technology, Nara, Japan
- 2Japan Science and Technology, Core Research for Evolutional Science and Technology Agency, Ikoma, Japan
Epigenetics is defined as heritable changes in gene expression and genome integrity that are accompanied by no alteration in DNA sequence. Throughout plant life cycle, many processes, including genome imprinting, stress responses, and cellular differentiation, are known to be determined by epigenetic regulation. The root apex is also considered to be under the control of epigenetic regulation for optimal growth under variable environments. Recent reports reveal that epigenetic control is especially important in the stem cell niche and the meristematic zone where both cell production and cell specification occur. DNA methylation, histone methylation, and histone acetylation are well-known epigenetic modifications, and each epigenetic modification has distinct roles in roots. Here, we review the updated findings that demonstrate the significance of epigenetic regulation in root apex of Arabidopsis.
Epigenetic Modifications in Plants
The word “Epigenetics” was coined by C. H. Waddington in 1942 as a combination of the words “epigenesis” and “genetics”. In current parlance, epigenetics is defined as the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence (Riggs et al., 1996). In this paper, we highlight the key events that play a significant role in plant epigenetics, such as DNA methylation, histone modification, and histone chaperones.
DNA methylation is a process by which methyl groups are attached to DNA. It is involved in various biological processes in plants, such as transcriptional repression of transposable elements (TEs) and repetitive sequences, and genomic imprinting. DNA methylation occurs in both the promoter and the gene body. DNA methylation at the promoter and gene body has different effects on the gene expression. DNA methylation in promoter regions usually represses gene expression (Bell and Felsenfeld, 2000; Suzuki and Bird, 2008). In contrast, modest methylation in gene body can promote gene expression, and extremely low or high levels of DNA methylation lead to lower gene expression (Takuno and Gaut, 2012, 2013). In plants, cytosine in three sequence contexts, CG, CHG, and CHH (H = A, C, or T), can be methylated [for a review, see Henderson and Jacobsen (2007)]. DNA methylation is established de novo by DRM2, and maintained by DRM2, MET1, and CMT3, all of which encode DNA methyltransferases [for a review, see Matzke et al. (2007)]. Demethylation enzymes, such as DME and ROS1, actively remove the methyl group attached to DNA (Choi et al., 2002; Gong et al., 2002).
Histones are highly conserved proteins in eukaryotes that package DNA into structural units called nucleosomes, which provide sites for two copies each of histone H2A, H2B, H3, and H4 proteins. Histone tails are the sites for covalent modifications, such as acetylation, methylation, phosphorylation, ubiquitination, SUMOylation, ribosylation, and biotinylation (Berger, 2007). Amino acids on N-peptide tails of H3 and H4 protrude from nucleosomes and are easily modified. Among various kinds of histone modifications, acetylation, and methylation of H3 and H4 are the best characterized in plants. Gene expression is generally upregulated by acetylation, mono-, di-, or tri-methylation of histone H3 Lysine 4 (H3K4me1, H3K4me2, or H3K4me3), and di- or tri-methylation of histone H3 Lysine 36 (H3K36me2 or H3K36me3), while it is repressed by dimethylation of histone H3 Lysine 9 (H3K9me2) and trimethylation of histone H3 Lysine 27 (H3K27me3) [for a review, see Chen et al. (2010)]. Histone modifications have essential roles in plant development, such as seed development, vegetative growth, floral induction, and flower morphogenesis [for a review, see Wagner (2003)].
Chromatin structures are controlled not only at the level of histone modification, but also of histone dynamics, which are regulated by histone chaperones. Histone chaperones are highly conserved through evolution. The chaperone anti-silencing function 1 (ASF1) binds H3–H4 dimers (English et al., 2006; Natsume et al., 2007) in the cytoplasm, and is involved in histone import into the nucleus (Campos et al., 2010). ASF1 then transfers histones to chaperone complexes involved in nucleosome assembly. The Arabidopsis genome contains two genes encoding ASF1 orthologs, ASF1A and ASF1B (Zhu et al., 2011). In mammals, two distinct pathways control deposition of either the canonical histone H3.1 or the variant H3.3. Chromatin Assembly Factor 1 (CAF-1), consisting of three subunits p150, p60, and p48, promotes histone deposition in a DNA synthesis-dependent manner (Smith and Stillman, 1989; Gaillard et al., 1996). CAF-1 specifically deposits H3.1 interacting with ASF1 (Tyler et al., 2001; Tagami et al., 2004). In Arabidopsis, p150, p60, and p48 are referred to as FASCIATA1 (FAS1), FASCIATA2 (FAS2), and MULTICOPY SUPPRESSOR OF IRA1 (MSI1), respectively (Kaya et al., 2001). Plants with mutant copies of ASF1s, FAS1, and FAS2 show dramatic and pleiotropic abnormalities during their life cycle, demonstrating the essential roles of histone chaperones in plant development. On the other hand, deposition of histone H3.3 is promoted by Histone Regulator complex (the HIR complex), independent of DNA synthesis and throughout the entire cell cycle (Ray-Gallet et al., 2002; Tagami et al., 2004). HIR complex is constituted by HISTONE REGULATOR A (HIRA), UBINUCLEIN (UBN), and CAlCINEURIN BINDING PROTEIN 1 (CABIN1). In Arabidopsis, loss-of-function mutants of HIRA display reduced fertility (Duc et al., 2015). Although other types of histone chaperones have been identified in mammals [for a review, see Filipescu et al. (2014)], their orthologs have not been identified in plants thus far.
Four Distinct Zones in Roots
Roots of Arabidopsis are composed of four distinct zones; namely, the stem cell niche (SCN), the meristematic zone (MZ), the transition zone (TZ), and the elongation/differentiation zone (EDZ; Figure 1A) (Takatsuka and Umeda, 2014). The SCN is important to orchestrate the fine balance of stem cell maintenance and supply of differentiating daughter cells. The stem cells surrounding the approximately four quiescent center (QC) cells give rise to each cell lineage, such as columella, lateral root cap, epidermis, cortex, endodermis, and provascular tissues (Figure 1B). In the MZ, cells generated from the SCN continue to divide a few times, contributing to root growth through increasing cell number. After leaving the MZ, cells enter the TZ and initiate endoreplication. In the TZ, cells slowly elongate in the direction of both width and length. Although the significance of TZ is not well understood, recent reports shed light on endoreplication-mediated cell elongation in the TZ (Adachi et al., 2011; Takahashi et al., 2013; Takatsuka and Umeda, 2014). The EDZ mainly has two important roles; one is to elongate roots by increasing each cell’s volume, and the other one is to develop root hairs for effective uptake of water and mineral nutrients from the soil (Figures 1A,C). Below we summarize the role of epigenetic control in establishment and maintenance of each root zone.
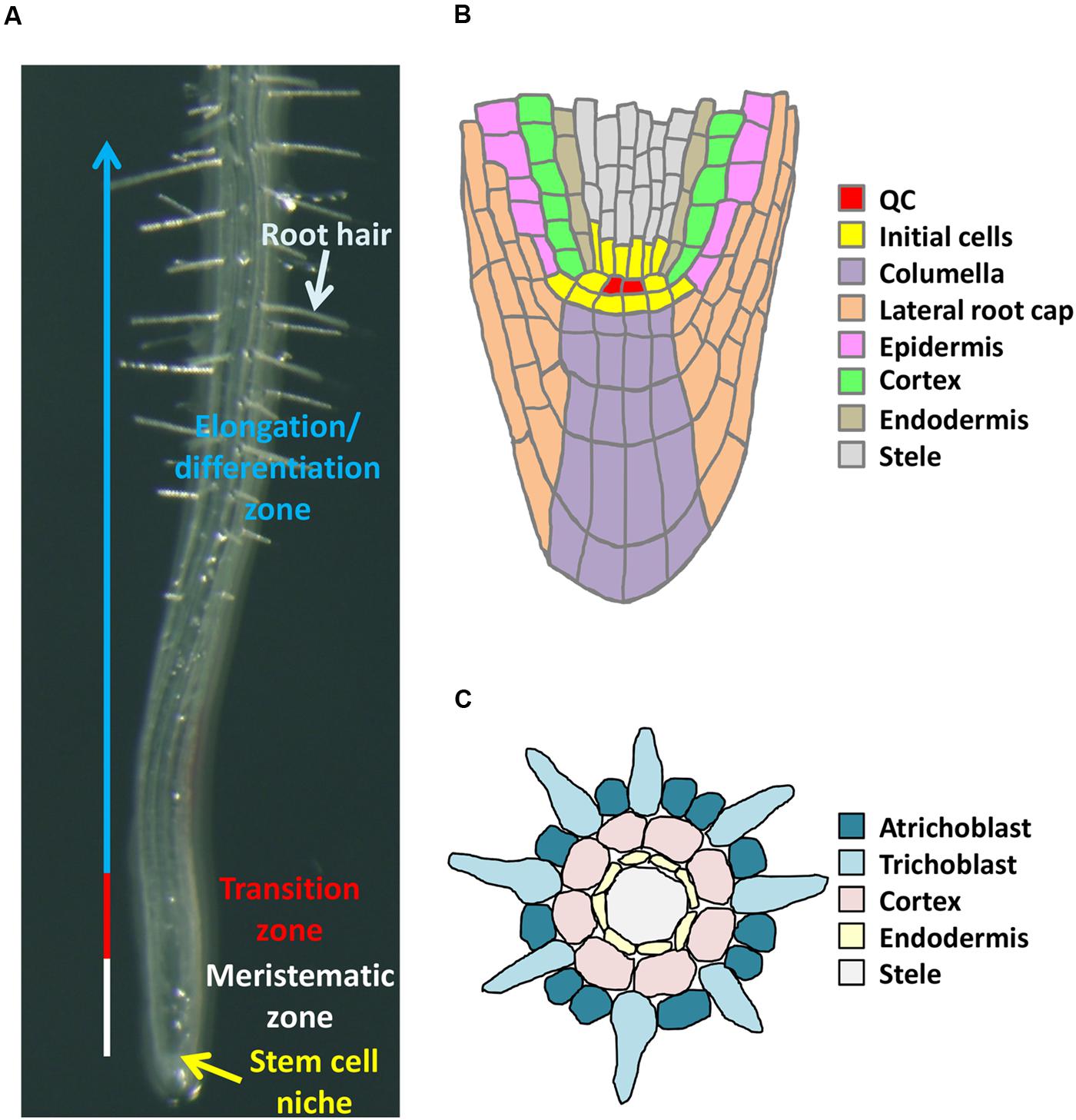
FIGURE 1. Arabidopsis root structure. (A) Four distinct zones. (B) Organization of cells in the root tip. (C) Transverse section of a root at an early stage of root hair formation.
Specification of the Stem Cell Niche and Control of Cell Division in the Meristematic Zone
The SCN consists of the QC and surrounding initial cells. For indeterminate growth of roots, it is indispensable to maintain QC cells intact (Figure 1A). PLETHORAs (PLTs) and WUSCHEL RELATED HOMEOBOX 5 (WOX5) play crucial roles in specifying QC cells in Arabidopsis (Aida et al., 2004; Sarkar et al., 2007). Both genes encode transcriptional factors; PLTs (PLT1 to PLT6) are AP2 domain-containing transcription factors, among which PLT1 and PLT2 play important roles in roots, and WOX5 is a WUSCHEL-related homeobox transcription factor. QC misspecification and resultant dysfunction of initial cells are observed in plt and wox5 mutants, indicating essential functions of these genes in SCN maintenance (Aida et al., 2004; Sarkar et al., 2007). WOX5 expression is confined to the QC (Sarkar et al., 2007) (Figure 2A), while PLTs are expressed over the MZ, forming a gradient with a maximum near the root tip (Figure 2A). This gradient has been reported to be prerequisite for the zonation of Arabidopsis roots (Galinha et al., 2007; Mähönen et al., 2014). Namely, high levels of PLT expression promote stem cell maintenance; intermediate levels of expression enhance mitotic activity in the MZ; PLT expression under the threshold level is required for cell differentiation and cell elongation in the EDZ (Galinha et al., 2007; Mähönen et al., 2014). Some reports demonstrate that expression of PLTs and WOX5 is under the control of epigenetic modifiers.
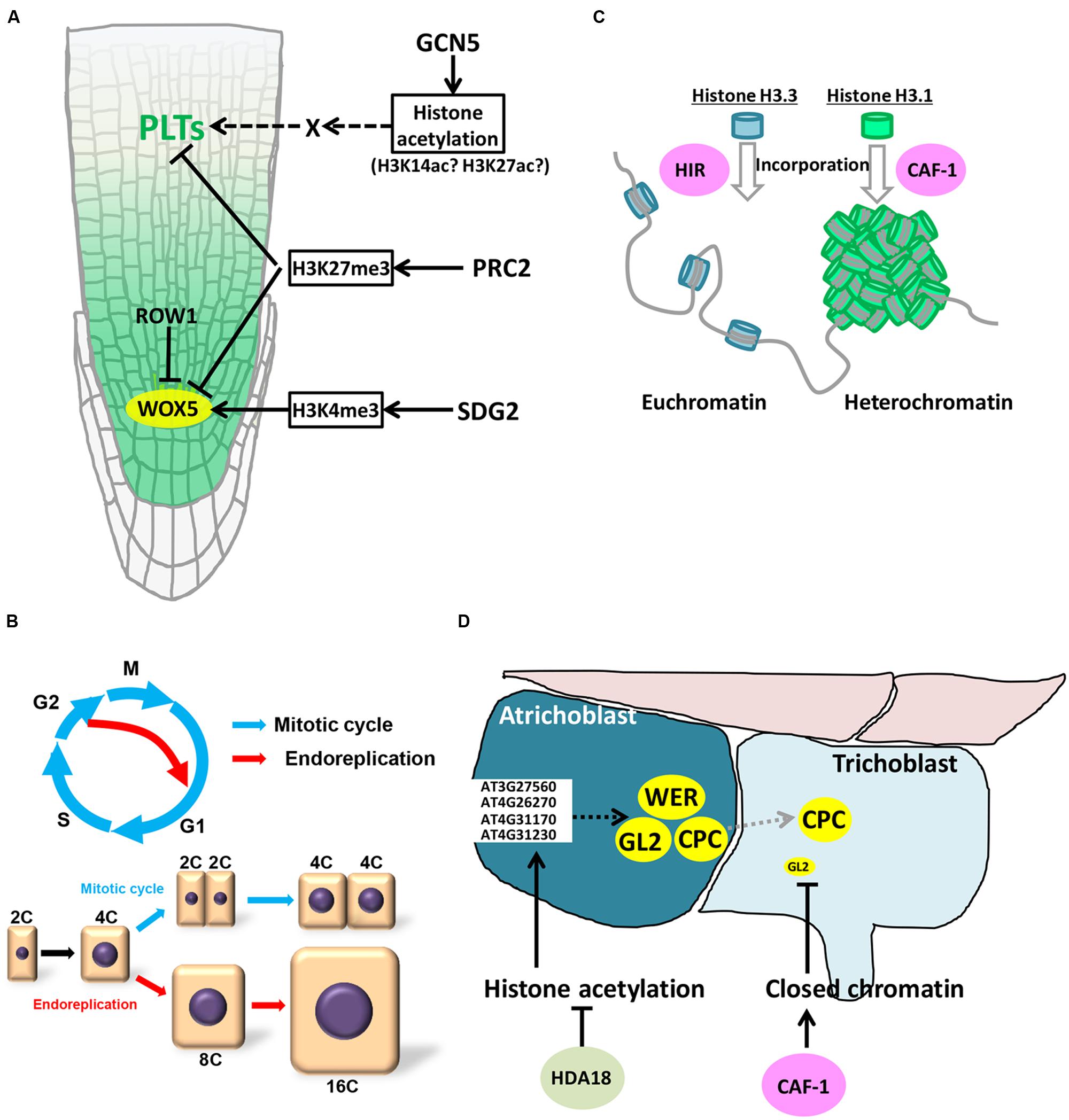
FIGURE 2. Epigenetic control in roots. (A) Epigenetic control of WOX5 and PLTs expression. (B) Endoreplication. (C) HIR- and CAF-1-mediated formation of euchromatin and heterochromatin. (D) Roles of CAF-1 and histone acetylation in root hair development.
Histone Acetylation
Histone acetylation catalyzed by histone acetyltranferases (HATs) is generally correlated with active transcription. Arabidopsis has four HAT families: GNAT (GCN5-related N-terminal acetyltransferases), MYST (MOZ, Ybt2, Sas2, Tip60-like), p300/CREB-binding protein (CBP), and TAFII250 families (Pandey et al., 2002). The GCN5 ortholog of the GNAT family is the best-characterized HAT in Arabidopsis (Pandey et al., 2002; Chen and Tian, 2007). Similar to yeast and mammals, Arabidopsis GCN5 interacts with ADA2a and ADA2b, and acetylates histone H3 (Stockinger et al., 2001; Mao et al., 2006; Earley et al., 2007). Histone H3 lysine 14 (H3K14) and H3K27 acetylation marks are reduced at defined loci in gcn5 mutants (Benhamed et al., 2006). Globally, the GCN5 complex regulates a subset of genes (∼5%) in the Arabidopsis genome (Vlachonasios et al., 2003; Benhamed et al., 2008). In the mutants of both GCN5 (also called HAG1) and ADA2b, expression of PLT1 and PLT2 was dramatically reduced, resulting in perturbed QC specification and a smaller meristem (Kornet and Scheres, 2009) (Figure 2A). Overexpression of PLT2 partially rescued the reduced meristem size of gcn5 mutants, suggesting that GCN5 acts in the PLT pathway (Kornet and Scheres, 2009) (Figure 2A). This indicates that the histone acetylation controlling PLT expression is essential for root zonation. However, it is still ambiguous whether the expression of PLT genes is directly regulated by the GCN5 complex. Anzola et al. (2010) showed that H3K9ac/K14ac was detected on the PLT2 promoter, although the acetylation level was sustained in ada2b mutants, suggesting a possibility of GCN5-independent histone acetylation. ChIP-chip analysis with anti-GCN5 antibodies also revealed that GCN5 did not associate with PLT genes (Benhamed et al., 2008) (Figure 2A). In support of this idea, both GCN5 and ADA2b are expressed ubiquitously in the MZ, whereas PLT expression is higher in the SCN and lower in the MZ, indicating that the gradient of PLT expression is regulated by factors downstream of GCN5. Since auxin is known to accumulate in the SCN and is involved in PLT expression, the GCN5 complex may control expression of some auxin-related factors, such as AUX/IAAs and ARFs, which then display the PLT gradient in the root tip. The recent report demonstrated that, under auxin-enriched conditions, bZIP11-related basic leucine zipper (bZIP) transcription factors bind the promoters of auxin-responsive genes and recruit histone acetylation machinery by interacting with ADA2b, thereby boosting the expression of auxin-responsive genes (Weiste and Dröge-Laser, 2014). Such a link between the GCN5 complex and auxin-related factors may explain consequent formation of the PLT gradient in the root tip.
Histone Methylation
The evolutionarily conserved Polycomb group (PcG) and trithorax group (trxG) proteins are central regulators of cell identity, maintaining the balance between cell proliferation and cell differentiation (Schuettengruber and Cavalli, 2009). PcG proteins form multimeric complexes, which repress transcription of target genes. In Arabidopsis, PcG proteins are incorporated into two complexes: Polycomb Repressive Complex1 (PRC1) and Polycomb Repressive Complex2 (PRC2). PRC2 is the best-studied PcG complex that catalyzes trimethylation of histone H3 lysine 27 (H3K27me3). The core component of PRC2 is composed of the SET-domain H3K27 methyltransferase, CURLY LEAF (CLF), and SWINGER (SWN), the latter of which has a partially overlapping function with CLF. In the Arabidopsis endosperm, MEDEA (MEA) functions as a H3K27 methyltransferase. Some proteins, such as EMBRYONIC FLOWER2 (EMF2), assist methyltransferases as a component of PRC2. It has been reported that, in Arabidopsis, around 4400 genes are modified by H3K27me3 (Zhang et al., 2007). Hence the loss of H3K27me3 in the mutant of PRC2 complex causes multiple developmental defects, such as in leaf and endosperm formation, in transition from embryo to seedlings, and in callus regeneration. Recently, Aichinger et al. (2011) reported the role of PRC2 in root development. Loss of CLF function increased meristematic activity, leading to larger MZ and longer roots. Moreover, additional undifferentiated cell layers were formed in columella initials in the clf mutant, demonstrating that the mitotic activity is elevated not only in the MZ but also in initial cells in the SCN. In agreement with this observation, WOX5 expression was upregulated in the mutant, and ChIP-PCR analysis revealed that WOX5 and other important regulators of the SCN and the MZ, such as PLTs and AGAMOUS-LIKEs(AGLs), are trimethylated at H3K27 (Aichinger et al., 2011) (Figure 2A). On the other hand, in the mutant of chromatin remodeling factor PICKLE (PKL), levels of H3K27me3 are substantially increased, suggesting an antagonistic interaction between PRC2 and PKL in determining the root zonation. However, PKL is ubiquitously expressed throughout the MZ and the SCN, and CLF is highly expressed in the MZ but poorly in the EDZ (Aichinger et al., 2011; Gu et al., 2014). These overlapping expression patterns indicate that root zonation is defined through reciprocal activities, rather than expression patterns, of PRC2 and PKL. Further analyses will reveal how their activities are controlled in distinct zones of roots.
Histone H3 lysine 4 di- and trimethylation variants (H3K4me2 and H3K4me3) are abundant in euchromatin and, in general, are associated with transcriptional activation in eukaryotes. Several Arabidopsis SET DOMAIN GROUP (SDG) proteins (ATX1, ATX2, ATXR7, and SDG2) have been shown to exhibit TrxG-like H3K4-methyltransferase activity. Root growth of atx1 and sdg2 mutants is severely impaired due to abnormal SCN organization and reduced mitotic activity in the MZ (Yao et al., 2013; Napsucialy-Mendivil et al., 2014). PLT1 expression is decreased in sdg2 mutants, suggesting a possibility that SDG2-mediated H3K4me2/3 controls root development via regulating PLT1 expression (Yao et al., 2013). Both ATX1 and SDG2 are strongly expressed in the MZ, and a gradient of expression, such as that of the PLTs, has not been reported so far. Therefore, ATX1 and SDG2, as well as GCN5 and ADA2b as mentioned above, likely contribute to the gradient formation of PLT expression in collaboration with other unidentified factors.
As for WOX5, it has been recently shown that H3K4me3 on the WOX5 promoter region has a crucial role in QC-specific expression of WOX5 (Zhang et al., 2015). ROW1 is a PHD-containing protein with two tandem BRCA1 C-terminal domains and a RING domain, and is expressed in the MZ, but not in the QC (Figure 2A). In the MZ, ROW1 binds to H3K4me3 on the WOX5 promoter to sequester the promoting function of H3K4me3 in transcription. However, in the QC, ROW1 expression is absent, thus H3K4me3 can induce WOX5 expression (Figure 2A). Consistent with these observations, the row1 mutation causes ectopic expression of WOX5 in the root tip, leading to a WOX5 overexpression-like phenotype, such as loss of starch granules in mature columella cells (Sarkar et al., 2007; Zhang et al., 2015).
CAF-1
In mutants of the CAF-1 components, FAS1 and FAS2, roots grow more slowly than the wild-type due to lower mitotic activity, and SCN organization is perturbed (Leyser and Furner, 1992; Kaya et al., 2001; Ramirez-Parra and Gutierrez, 2007). The expression pattern of SCARECROW (SCR), a key gene for SCN establishment and radial pattern formation, is not maintained in fas mutant roots. However, the relationship between CAF-1 and other SCN-related genes, such as WOX5 and PLTs, remains unknown.
DNA Methylation
All DNA methyltransferases, except for DRM1, are strongly expressed in the root tip of Arabidopsis (Jullien et al., 2012). However, to our knowledge, the effects of mutations of DNA methyltransferases on root phenotype have not been reported thus far. Loss of DNA methylation leads to movement of transposons, which frequently generates gene mutations and causes morphological abnormalities. Therefore, it is difficult to distinguish mutant phenotypes caused by global changes in DNA methylation from that of gene mutations induced by transposon insertion.
Cell Elongation and Differentiation in the Transition and Elongation/Differentiation Zones
After cells undergo a few rounds of mitosis in the MZ, they enter the TZ and start cell elongation (Figure 1A). This cell elongation is caused by endoreplication, a specialized mode of the cell cycle in which DNA replication is repeated without mitosis or cytokinesis. As a result, DNA ploidy increases in each elongating cell (Figure 2B). It is known that endoreplication is usually accompanied by cell enlargement and cell differentiation, while the underlying mechanisms remain unknown (Inzé and De Veylder, 2006; Kalve et al., 2014). The involvement of epigenetic regulation in endoreplication has been poorly understood, but here we discuss a possible link between epigenetic control and genome stability that is associated with the onset of endoreplication.
After leaving the TZ, cells enter the EDZ where rapid cell elongation and terminal differentiation occur (Figure 1A). Although cells produced in the MZ are no more than 10 μm in the longitudinal direction, they reach more than few hundred micrometers in the EDZ. Such a dramatic increase in cell volume, as well as cell division in the MZ, greatly contributes to root growth. Recently one report demonstrated the involvement of epigenetic control in cell elongation in the EDZ.
CAF-1
As in mammals, histone H3 of plants is classified into two types, H3.1 and H3.3. They differ from each other at positions 31, 41, 87, and 90 (T31Y41H87L90 for H3.3 and A31F41S87A90 for H3.1) (Ingouff and Berger, 2010). In spite of such a high similarity, they have distinct functions in controlling chromatin structure. H3.3 is incorporated into chromatin outside of the S phase, and its deposition is correlated with transcriptionally active genes, suggesting that H3.3 is involved in euchromatin formation (Mito et al., 2005). In contrast, H3.1 is incorporated into chromatin during DNA replication and is enriched in dimethylated H3 Lys9 (H3K9me2), which is associated with gene silencing and heterochromatin formation (Figure 2C) (Peters and Schübeler, 2005). As mentioned above, the CAF-1 complex plays an essential role in H3.1 incorporation in a DNA replication-dependent manner. Although plant CAF-1 subunits, FAS1 and FAS2, were originally identified from Arabidopsis mutants with abnormal meristem organization, endoreplication was later found to be abnormal in the mutants. Namely, the ploidy level of the fas1 and fas2 mutants was higher than that of the wild-type, leading to larger epidermal cells and increased trichome branching on leaves (Exner et al., 2006). In these mutants, expression levels of DNA double-strand break (DSB)-induced genes, such as DNA repair-related genes, were elevated (Schönrock et al., 2006; Ramirez-Parra and Gutierrez, 2007). Moreover, Endo et al. (2006) observed a small but significant increase in the extent of DSBs in the fas mutants as compared to the wild-type. Since it was reported that DSBs promote an early onset of endoreplication by inducing G2/M arrest (Adachi et al., 2011), lowered genome stability might result in enhanced endoreplication in the fas mutants. This suggests a possibility that CAF-1-mediated H3.1 incorporation is engaged in maintaining genome stability, thus protecting the genome from DNA damage.
The fas1 and fas2 mutants have a shorter root meristem (Kaya et al., 2001), indicating a possibility of an early transition from mitosis to endoreplication. If this is the case, CAF-1 may also be involved in the control of genome stability in roots. However, fas1 and fas2 mutants exhibit reduced mitotic activity (Ramirez-Parra and Gutierrez, 2007); thus it is also likely that the early transition to endoreplication is a consequence of lowered cyclin-dependent kinase activities, which inhibit the G2/M progression and promote the onset of endoreplication (De Veylder et al., 2011). On the other hand, Napsucialy-Mendivil et al. (2014) reported that in the atx1-1 mutant, the root meristem size is reduced, while the transition to cell elongation is suppressed. This suggests another possibility that fas1 and fas2 mutants have impaired cell division in the MZ, but no defect in endoreplication. Further studies will reveal how CAF-1 is associated with the control of the root meristem size, and whether CAF-1 is actively involved in the control of genome stability in the MZ, where endoreplication is suppressed.
Histone Acetylation
Rapid cell growth in the EDZ requires cell wall rearrangement, which is controlled by cell wall-related proteins like xyloglucan endotransglucosylase (XET), endo-1,4-β-D-endoglucanase (EGase), expansins (EXP), and the plasma membrane proton pump (PM-H+-ATPase, MHA) (Geilfus et al., 2011). In maize, high salinity stress makes roots swollen possibly due to up-regulation of cell wall-related genes (Li et al., 2014). Expression of ZmEXPB2 and ZmXET1 is regulated by H3K9 acetylation, and histone acetyltransferase (HAT) genes ZmHATB and ZmEXPB2 are highly expressed under high salinity conditions (Li et al., 2014). Therefore, it is likely that HAT-mediated acetylation of H3K9 is required for induction of ZmEXPB2 and ZmXET1 in the EDZ, and that salinity stress up-regulates the expression of ZmHATB and ZmGCN5, thus enhancing expression of ZmEXPB2 and ZmXET1 and leading to cell enlargement and resultant root swelling.
Root Hair Being Specified in the Meristematic Zone and Developed in the Elongation/Differentiation Zone
Recent reports showed that epigenetic regulation is required not only for determination of differentiation state along the apical-basal axis of roots but also for cell specification. Root epidermal cells consist of two cell types, trichoblast and atrichoblast, in Arabidopsis. The trichoblast lineage is in contact with two cortex cells and produces root hairs, whereas atrichoblast in contact with a single cortex cell never forms root hairs (Figure 1C). This indicates that positional cues are essential to trigger root hair development. Previous studies showed that position-dependent cellular pattern of epidermal cells is determined by interactions of six patterning genes; CAPRICE (CPC), ENHANCER OF TRY AND CPC 1 (ETC1), GLABRA 2 (GL2), GLABRA 3 (GL3), TRANSPARENT TESTA GLABRA 1 (TTG1), and WEREWOLF (WER) [for a review, see Schiefelbein et al. (2014)].
CAF-1
GL2 is expressed in atrichoblasts, but not in trichoblasts, and functions as a critical determinant specifying atrichoblast (Figure 2D). Indeed, ectopic root hairs are formed from the atrichoblast linage in the gl2 mutant due to misspecification of atrichoblasts. Costa and Shaw (2006) demonstrated that atrichoblast-specific expression of GL2 is controlled by CAF-1 complex (Figure 2D). The GL2 locus in the Arabidopsis genome is more condensed in trichoblasts as compared to atrichoblasts, resulting in higher accessibility of transcription machineries onto the promoter in atrichoblasts. In the fas2 mutant, GL2 is ubiquitously expressed in both trichoblasts and atrichoblasts, resulting in a reduced number of root hairs. This suggests that CAF-1-mediated control of chromatin structure is involved in determination of epidermal cell fate in roots (Figure 2D). However, it remains unknown how CAF-1 regulates the GL2 locus specifically in trichoblasts. Based on the previous report (Kaya et al., 2001) and the Arabidopsis expression database (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), it is unlikely that the FAS2 expression is specific to the trichoblast. Identifying mechanisms that control the CAF-1 activity in particular cell types will help discover a new regulatory system of cell differentiation during root development.
Histone Acetylation
Xu et al. (2005) found that treatment of Arabidopsis roots with trichostatin A (TSA), a specific inhibitor of histone deacetylase (HDAC), induces hair cell development ectopically in the atrichoblast lineage (Figure 2D). They showed that TSA treatment altered the expression of CPC, GL2, and WER in the atrichoblast, resulting in ectopic root hair development (Xu et al., 2005). Among the 18 HDAC genes of Arabidopsis (Hollender and Liu, 2008), HDA18 is specifically required for proper root hair development; in the hda18 mutant, the root hair was occasionally formed in the atrichoblast lineage, reminiscent of TSA-treated roots (Xu et al., 2005). Liu et al. (2013) reported that expression of some kinase genes (At3G27560, At4G26270, At4G31170, and At4G31230) is directly regulated by HDA18, and their mutants and overexpressors displayed root hair phenotypes. Therefore, it is probable that HDA18 controls the expression of CPC, GL2, and WER via these kinases in atrichoblast, suppressing root hair development in this cell lineage (Liu et al., 2013).
Summary and Future Perspectives
Many reports have demonstrated that epigenetic regulation plays an indispensable role in zonation of roots. However, as described in this article, epigenetic regulators are not expressed in a gradient or in a zone-specific manner. Rather, in many cases, they are expressed ubiquitously in roots, indicating that one epigenetic regulator cannot explain how to make the root zonation. One possibility to explain this issue is that interaction of multiple epigenetic regulators forms the gradient or zone-specific expression of key genes. For example, the PLT gradient may be formed as a result of combined action of the GCN5 complex, PRC2 and PKL as described above. In mouse embryonic stem cells, genes with H3K4me2 are generally highly expressed, whereas those with H3K27me3 are poorly expressed. In fact, many genes are bivalently marked with both H3K4me2 and H3K27me3, and their expression is lower as that of the genes with H3K27me3 marks only, demonstrating that the effect of H3K27me3 on gene expression is epistatic to that of H3K4me2 (Bernstein et al., 2006). There are many types of epigenetic modifications also in plants, thus it is probable that complex relationship among various epigenetic factors might be required to establish the root zonation. Another possibility is that unidentified factors regulating the activity of epigenetic regulators are expressed in a gradient or in a zone-specific manner. Although it is poorly understood how the activity of epigenetic regulators, such as histone methyltransferases and histone acetyltransferases, is regulated, ROW1 is a good example showing how epigenetic modifications are controlled by another factor. Further identification of such modifiers of epigenetic regulation will give a mechanistic insight into cell division, cell growth, and cell differentiation in each zone of roots.
Author Contributions
HT and MU wrote the paper cooperatively.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by MEXT KAKENHI Grant Numbers 26291061 and 26650099, and by JST, CREST.
References
Adachi, S., Minamisawa, K., Okushima, Y., Inagaki, S., Yoshiyama, K., Kondou, Y., et al. (2011). Programmed induction of endoreduplication by DNA double-strand breaks in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 10004–10009. doi: 10.1073/pnas.1103584108
Aichinger, E., Villar, C. B., Di Mambro, R., Sabatini, S., and Köhler, C. (2011). The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23, 1047–1060. doi: 10.1105/tpc.111.083352
Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., et al. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120. doi: 10.1016/j.cell.2004.09.018
Anzola, J. M., Sieberer, T., Ortbauer, M., Butt, H., Korbei, B., Weinhofer, I., et al. (2010). Putative Arabidopsis transcriptional adaptor protein (PROPORZ1) is required to modulate histone acetylation in response to auxin. Proc. Natl. Acad. Sci. U.S.A. 107, 10308–10313. doi: 10.1073/pnas.0913918107
Bell, A. C., and Felsenfeld, G. (2000). Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485. doi: 10.1038/35013100
Benhamed, M., Bertrand, C., Servet, C., and Zhou, D. X. (2006). Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18, 2893–2903. doi: 10.1105/tpc.106.043489
Benhamed, M., Martin-Magniette, M. L., Taconnat, L., Bitton, F., Servet, C., De Clercq, R., et al. (2008). Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J. 56, 493–504. doi: 10.1111/j.1365-313X.2008.03606.x
Berger, S. L. (2007). The complex language of chromatin regulation during transcription. Nature 447, 407–412. doi: 10.1038/nature05915
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. doi: 10.1016/j.cell.2006.02.041
Campos, E. I., Fillingham, J., Li, G., Zheng, H., Voigt, P., Kuo, W. H., et al. (2010). The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 17, 1343–1351. doi: 10.1038/nsmb.1911
Chen, Z., Wang, L., Wang, Q., and Li, W. (2010). Histone modifications and chromatin organization in prostate cancer. Epigenomics 2, 551–560. doi: 10.2217/epi.10.31
Chen, Z. J., and Tian, L. (2007). Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim. Biophys. Acta 1769, 295–307. doi: 10.1016/j.bbaexp.2007.04.007
Choi, Y., Gehring, M., Johnson, L., Hannon, M., Harada, J. J., Goldberg, R. B., et al. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110, 33–42. doi: 10.1016/S0092-8674(02)00807-3
Costa, S., and Shaw, P. (2006). Chromatin organization and cell fate switch respond to positional information in Arabidopsis. Nature 439, 493–496. doi: 10.1038/nature04269
De Veylder, L., Larkin, J. C., and Schnittger, A. (2011). Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 16, 624–634. doi: 10.1016/j.tplants.2011.07.001
Duc, C., Benoit, M., Le Goff, S., Simon, L., Poulet, A., Cotterell, S., et al. (2015). The histone chaperone complex HIR maintains nucleosome occupancy and counterbalances impaired histone deposition in CAF-1 complex mutants. Plant J. 81, 707–722. doi: 10.1111/tpj.12758
Earley, K. W., Shook, M. S., Brower-Toland, B., Hicks, L., and Pikaard, C. S. (2007). In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J. 52, 615–626. doi: 10.1111/j.1365-313X.2007.03264.x
Endo, M., Ishikawa, Y., Osakabe, K., Nakayama, S., Kaya, H., Araki, T., et al. (2006). Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 25, 5579–5590. doi: 10.1038/sj.emboj.7601434
English, C. M., Adkins, M. W., Carson, J. J., Churchill, M. E., and Tyler, J. K. (2006). Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508. doi: 10.1016/j.cell.2006.08.047
Exner, V., Taranto, P., Schönrock, N., Gruissem, W., and Hennig, L. (2006). Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development 133, 4163–4172. doi: 10.1242/dev.02599
Filipescu, D., Müller, S., and Almouzni, G. (2014). Histone H3 variants and their chaperones during development and disease: contributing to epigenetic control. Annu. Rev. Cell. Dev. Biol. 30, 615–646. doi: 10.1146/annurev-cellbio-100913-013311
Gaillard, P. H., Martini, E. M., Kaufman, P. D., Stillman, B., Moustacchi, E., and Almouzni, G. (1996). Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell 86, 887–896. doi: 10.1016/S0092-8674(00)80164-6
Galinha, C., Hofhuis, H., Luijten, M., Willemsen, V., Blilou, I., Heidstra, R., et al. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057. doi: 10.1038/nature06206
Geilfus, C. M., Zörb, C., Neuhaus, C., Hansen, T., Lüthen, H., and Mühling, K. H. (2011). Differential transcript expression of wall-loosening candidates in leaves of maize cultivars differing in salt resistance. J. Plant Growth Regul. 30, 387–395. doi: 10.1371/journal.pone.0118406
Gong, Z., Morales-Ruiz, T., Ariza, R. R., Roldán-Arjona, T., David, L., and Zhu, J. K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814. doi: 10.1016/S0092-8674(02)01133-9
Gu, X., Xu, T., and He, Y. (2014). A histone H3 lysine-27 methyltransferase complex represses lateral root formation in Arabidopsis thaliana. Mol. Plant. 7, 977–988. doi: 10.1093/mp/ssu035
Henderson, I. R., and Jacobsen, S. E. (2007). Epigenetic inheritance in plants. Nature 447, 418–424. doi: 10.1038/nature05917
Hollender, C., and Liu, Z. (2008). Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50, 875–885. doi: 10.1111/j.1744-7909.2008.00704.x
Ingouff, M., and Berger, F. (2010). Histone3 variants in plants. Chromosoma 119, 27–33. doi: 10.1007/s00412-009-0237-1
Inzé, D., and De Veylder, L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40, 77–105. doi: 10.1146/annurev.genet.40.110405.090431
Jullien, P. E., Susaki, D., Yelagandula, R., Higashiyama, T., and Berger, F. (2012). DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr. Biol. 22, 1825–1830. doi: 10.1016/j.cub.2012.07.061
Kalve, S., De Vos, D., and Beemster, G. T. (2014). Leaf development: a cellular perspective. Front. Plant Sci. 5:362. doi: 10.3389/fpls.2014.00362
Kaya, H., Shibahara, K. I., Taoka, K. I., Iwabuchi, M., Stillman, B., and Araki, T. (2001). FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104, 131–142. doi: 10.1016/S0092-8674(01)00197-0
Kornet, N., and Scheres, B. (2009). Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 21, 1070–1079. doi: 10.1105/tpc.108.065300
Leyser, H. M. O., and Furner, I. J. (1992). Characterization of three shoot apical meristem mutants of Arabidopsis thaliana. Development 116, 397–403.
Li, H., Yan, S., Zhao, L., Tan, J., Zhang, Q., Gao, F., et al. (2014). Histone acetylation associated up-regulation of the cell wall related genes is involved in salt stress induced maize root swelling. BMC Plant Biol. 14:105. doi: 10.1186/1471-2229-14-105
Liu, C., Li, L. C., Chen, W. Q., Chen, X., Xu, Z. H., and Bai, S. N. (2013). HDA18 affects cell fate in Arabidopsis root epidermis via histone acetylation at four kinase genes. Plant Cell 25, 257–269. doi: 10.1105/tpc.112.107045
Mähönen, A. P., ten Tusscher, K., Siligato, R., Smetana, O., Díaz-Triviño, S., Salojärvi, J., et al. (2014). PLETHORA gradient formation mechanism separates auxin responses. Nature 515, 125–129. doi: 10.1038/nature13663
Mao, Y., Pavangadkar, K. A., Thomashow, M. F., and Triezenberg, S. J. (2006). Physical and functional interactions of Arabidopsis ADA2 transcriptional coactivator proteins with the acetyltransferase GCN5 and with the cold-induced transcription factor CBF1. Biochim. Biophys. Acta 1759, 69–79. doi: 10.1016/j.bbaexp.2006.02.006
Matzke, M., Kanno, T., Huettel, B., Daxinger, L., and Matzke, A. J. (2007). Targets of RNA-directed DNA methylation. Curr. Opin. Plant. Biol. 10, 512–519. doi: 10.1016/j.pbi.2007.06.007
Mito, Y., Henikoff, J. G., and Henikoff, S. (2005). Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 37, 1090–1097. doi: 10.1038/ng1637
Napsucialy-Mendivil, S., Alvarez-Venegas, R., Shishkova, S., and Dubrovsky, J. G. (2014). Arabidopsis homolog of trithorax1 (ATX1) is required for cell production, patterning, and morphogenesis in root development. J. Exp. Bot. 65, 6373–6384. doi: 10.1093/jxb/eru355
Natsume, R., Eitoku, M., Akai, Y., Sano, N., Horikoshi, M., and Senda, T. (2007). Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 446, 338–341. doi: 10.1038/nature05613
Pandey, R., Müller, A., Napoli, C. A., Selinger, D. A., Pikaard, C. S., Richards, E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic. Acids. Res. 30, 5036–5055. doi: 10.1093/nar/gkf660
Peters, A. H., and Schübeler, D. (2005). Methylation of histones: playing memory with DNA. Curr. Opin. Cell Biol. 17, 230–238. doi: 10.1016/j.ceb.2005.02.006
Ramirez-Parra, E., and Gutierrez, C. (2007). E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 144, 105–120. doi: 10.1104/pp.106.094979
Ray-Gallet, D., Quivy, J. P., Scamps, C., Martini, E. M., Lipinski, M., and Almouzni, G. (2002). HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9, 1091–1100. doi: 10.1016/S1097-2765(02)00526-9
Riggs, A. D., Martienssen, R. A., and Russo, V. E. A. (1996). “Introduction,” in Epigenetic Mechanisms of Gene Regulation, eds V. E. A. Russo, R. A. Martienssen, and A. D. Riggs (New York, NY: Cold Spring Harbor Laboratory Press), 1–4.
Sarkar, A. K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K., et al. (2007). Conserved factors regulate signaling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. doi: 10.1038/nature05703
Schiefelbein, J., Huang, L., and Zheng, X. (2014). Regulation of epidermal cell fate in Arabidopsis roots: the importance of multiple feedback loops. Front. Plant Sci. 5:47. doi: 10.3389/fpls.2014.00047
Schönrock, N., Exner, V., Probst, A., Gruissem, W., and Hennig, L. (2006). Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. J. Biol. Chem. 281, 9560–9568. doi: 10.1074/jbc.M513426200
Schuettengruber, B., and Cavalli, G. (2009). Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136, 3531–3542. doi: 10.1242/dev.033902
Smith, S., and Stillman, B. (1989). Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58, 15–25. doi: 10.1016/0092-8674(89)90398-X
Stockinger, E. J., Mao, Y., Regier, M. K., Triezenberg, S. J., and Thomashow, M. F. (2001). Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic. Acids Res. 29, 1524–1533. doi: 10.1093/nar/29.7.1524
Suzuki, M. M., and Bird, A. (2008). DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476. doi: 10.1038/nrg2341
Tagami, H., Ray-Gallet, D., Almouzni, G., and Nakatani, Y. (2004). Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61. doi: 10.1016/S0092-8674(03)01064-X
Takahashi, N., Kajihara, T., Okamura, C., Kim, Y., Katagiri, Y., Okushima, Y., et al. (2013). Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Curr. Biol. 23, 1812–1817. doi: 10.1016/j.cub.2013.07.051
Takatsuka, H., and Umeda, M. (2014). Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 65, 2633–2643. doi: 10.1093/jxb/ert485
Takuno, S., and Gaut, B. S. (2012). Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol. Biol. Evol. 29, 219–227. doi: 10.1093/molbev/msr188
Takuno, S., and Gaut, B. S. (2013). Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc. Natl. Acad. Sci. U.S.A. 110, 1797–1802. doi: 10.1073/pnas.1215380110
Tyler, J. K., Collins, K. A., Prasad-Sinha, J., Amiott, E., Bulger, M., Harte, P. J., et al. (2001). Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Cell. Biol. 21, 6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001
Vlachonasios, K. E., Thomashow, M. F., and Triezenberg, S. J. (2003). Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15, 626–638. doi: 10.1105/tpc.007922
Wagner, D. (2003). Chromatin regulation of plant development. Curr. Opin. Plant. Biol. 6, 20–28. doi: 10.1016/S1369526602000079
Weiste, C., and Dröge-Laser, W. (2014). The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat. Commun. 5:3883. doi: 10.1038/ncomms4883
Xu, C. R., Liu, C., Wang, Y. L., Li, L. C., Chen, W. Q., Xu, Z. H., et al. (2005). Histone acetylation affects expression of cellular patterning genes in the Arabidopsis root epidermis. Proc. Natl. Acad. Sci. U.S.A. 102, 14469–14474. doi: 10.1073/pnas.0503143102
Yao, X., Feng, H., Yu, Y., Dong, A., and Shen, W. H. (2013). SDG2-mediated H3K4 methylation is required for proper Arabidopsis root growth and development. PLoS ONE 8:e56537. doi: 10.1371/journal.pone.0056537
Zhang, X., Germann, S., Blus, B. J., Khorasanizadeh, S., Gaudin, V., and Jacobsen, S. E. (2007). The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 14, 869–871. doi: 10.1038/nsmb1283
Zhang, Y., Jiao, Y., Liu, Z., and Zhu, Y. X. (2015). ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nat. Commun. 6:6003. doi: 10.1038/ncomms7003
Keywords: epigenetics, root, Arabidopsis, histone modification, DNA methylation, histone chaperone
Citation: Takatsuka H and Umeda M (2015) Epigenetic Control of Cell Division and Cell Differentiation in the Root Apex. Front. Plant Sci. 6:1178. doi: 10.3389/fpls.2015.01178
Received: 26 October 2015; Accepted: 09 December 2015;
Published: 24 December 2015.
Edited by:
Frantisek Baluska, University of Bonn, GermanyReviewed by:
Joseph Dubrovsky, Universidad Nacional Autónoma de Mexico, MexicoLewis Feldman, University of California, USA
Copyright © 2015 Takatsuka and Umeda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masaaki Umeda, bXVtZWRhQGJzLm5haXN0Lmpw
 Hirotomo Takatsuka
Hirotomo Takatsuka Masaaki Umeda
Masaaki Umeda