- School of Biological Sciences, The University of Sydney, Sydney, NSW, Australia
The ribosome in higher eukaryotes is a large macromolecular complex composed of four rRNAs and eighty different ribosomal proteins. In plants, each ribosomal protein is encoded by multiple genes. Duplicate genes within a family are often necessary to provide a threshold dose of a ribosomal protein but in some instances appear to have non-redundant functions. Here, we addressed whether divergent members of the RPL9 gene family are dosage sensitive or whether these genes have non-overlapping functions. The RPL9 family in Arabidopsis thaliana comprises two nearly identical members, RPL9B and RPL9C, and a more divergent member, RPL9D. Mutations in RPL9C and RPL9D genes lead to delayed growth early in development, and loss of both genes is embryo lethal, indicating that these are dosage-sensitive and redundant genes. Phylogenetic analysis of RPL9 as well as RPL4, RPL5, RPL27a, RPL36a, and RPS6 family genes in the Brassicaceae indicated that multicopy ribosomal protein genes have been largely retained following whole genome duplication. However, these gene families also show instances of tandem duplication, small scale deletion, and evidence of gene conversion. Furthermore, phylogenetic analysis of RPL9 genes in angiosperm species showed that genes within a species are more closely related to each other than to RPL9 genes in other species, suggesting ribosomal protein genes undergo convergent evolution. Our analysis indicates that ribosomal protein gene retention following whole genome duplication contributes to the number of genes in a family. However, small scale rearrangements influence copy number and likely drive concerted evolution of these dosage-sensitive genes.
Introduction
The 80S ribosome of higher eukaryotes is a macromolecular complex composed of two subunits, a large 60S subunit and a small 40S subunit. The 60S subunit comprises 28S, 5.8S, and 5S rRNA and 47 ribosomal proteins. The 40S subunit is composed of 18S rRNA and 33 proteins (Melnikov et al., 2012). Ribosomes are produced through a cascade of events involving coordinated processing of precursor rRNA, progressive association of individual ribosomal proteins with rRNA, export of pre-ribosome particles from the nucleolus to the cytoplasm, and assembly to form mature subunits. The two subunits join at translation initiation to form a ribosome, which carries out protein synthesis.
In Arabidopsis thaliana, each ribosomal protein is encoded by multiple genes (Barakat et al., 2001). Duplicate ribosomal protein genes may serve to provide a critical dose of a ribosomal protein or may provide distinct functions through differential expression or through diversification of protein function (Horiguchi et al., 2012; Xue and Barna, 2012). In Arabidopsis, mutations in members of a ribosomal protein family may have different phenotypic outcomes but more often show similar dose-dependent phenotypes (Degenhardt and Bonham-Smith, 2008; Yao et al., 2008; Fujikura et al., 2009; Creff et al., 2010; Rosado and Raikhel, 2010; Rosado et al., 2010; Horiguchi et al., 2011; Stirnberg et al., 2012; Casanova-Sáez et al., 2014; Zsögön et al., 2014). Mutations in A. thaliana ribosomal protein genes are generally recessive, and only two semi-dominant mutants have been described (Byrne, 2009; Horiguchi et al., 2012). Arabidopsis Minute-like1 (aml1) has a mutation in the RPS5B gene and homozygous mutants arrest during early stages of embryo development. Hemizygous plants are viable and have a range of phenotypes including reduced seedling size and altered organ vascular patterning (Weijers et al., 2001). rpl27ac-1d is a dominant-negative mutation in the RPL27aC gene and homozygous plants have abnormal development of embryos and pleiotropic defects in the plant shoot. Heterozygous plants are slow growing with distinct developmental phenotypes, including pointed and serrated leaves (Szakonyi and Byrne, 2011a,b). Increasing the ratio of rpl27ac-1d relative to wild type results in a progressive increase in the range and severity of phenotypes consistent with plant growth and development being sensitive to the dose of RPL27a (Zsögön et al., 2014).
RPL27a is encoded by two redundant genes. Loss-of-function mutations in RPL27aC and RPL27aB have mild and no leaf phenotype, respectively, whereas double heterozygote plants have a pointed and serrated leaf shape phenotype (Zsögön et al., 2014). Mutations in both RPL27aB and RPL27aC genes are not transmitted through gametes indicating dramatically reduced levels of RPL27a is haploid lethal (Zsögön et al., 2014). Likewise ribosomal proteins RPL4, RPL5, RPL36a, and RPS6, are each encoded by two functional genes. For each of these duplicate genes, single mutants are viable and plants display a pointed and serrated leaf phenotype that is characteristic of mutations in A. thaliana ribosomal protein genes. Double heterozygous mutants for both genes within a family also display these leaf phenotypes and mutant alleles in duplicate genes are not transmitted through gametes (Yao et al., 2008; Fujikura et al., 2009; Creff et al., 2010; Rosado et al., 2010; Casanova-Sáez et al., 2014). These phenotypes indicate that members of these ribosomal protein families are redundant and that the duplicate genes in a family are required for production of sufficient levels of a ribosomal protein for viability of haploid gametes and for plant growth.
Duplicate genes may arise through whole or partial genome duplication, or through tandem gene duplication. Many flowering plants are ancient polyploids and retain evidence of past genome duplications (Van de Peer et al., 2009). Duplicate genes created through genome duplication either diverge in function or one duplicate is lost from the genome. However, gene loss is biased and dosage sensitive genes appear to be preferentially retained following genome duplication. According to the gene balance hypothesis, following whole genome duplication, an unfavorable imbalance in the optimum ratio of proteins may arise from loss of genes that code for components of a protein complex or components in a molecular pathway. As such dosage-sensitive genes may be retained following whole genome duplication in order to maintain a balance in the concentration of proteins in complex or in a molecular pathway (Veitia, 2002; Papp et al., 2003; Birchler and Veitia, 2012). Consistent with the gene balance hypothesis, multiple plant species display evidence of over-retention of genes within the ontology category of “ribosome” following genome duplication (Blanc and Wolfe, 2004; Maere et al., 2005; Rizzon et al., 2006; Thomas et al., 2006; Wang et al., 2011; Jiang et al., 2013).
Although there is an overall trend toward retention of ribosomal protein genes post-genome duplication, there has been limited analysis of the evolution of specific cytoplasmic ribosomal protein gene families within plants. Here we demonstrate that two divergent members of the ribosomal protein family RPL9, RPL9C, and RPL9D, are dosage-sensitive and redundant, indicating that these duplicate ribosomal protein genes serve to maintain adequate levels of a ribosomal protein for sufficient ribosome production. Analysis of RPL9 family genes in Brassicaceae species, and more broadly within eudicots and monocots revealed limited RPL9 copy number variation between species. In the Brassicaceae, RPL9 copy number appears to be the outcome of multiple genome rearrangements including whole genome duplication, tandem duplication and gene loss. Furthermore nucleotide sequence variation between RPL9 genes within a species appears to be driven toward homogenization, likely through gene conversion. Analysis of RPL4, RPL5, RPL27a, RPL36a, and RPS6 genes in the Brassicaceae reveals dynamic evolution of ribosomal protein gene families.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis mutant rpl9c-1 (formerly published as piggyback2-1 (pgy2-1)) has been described previously (Pinon et al., 2008). rpl9d-1 (SALK_111804) was obtained from The European Arabidopsis Stock Center (Scholl et al., 2000) and was backcrossed five times to Landsberg erecta prior to genetic analysis. Plants were grown in soil at 22°C with a day length of 16 h. Growth measurement data from eight plants of each genotype were analyzed using SPSS Statistics for Macintosh, Version 22.0 (IBM Corporation). One-way or repeated measures analysis of variance (ANOVA) tests were performed, followed by Scheffe’s multiple comparison post hoc test and P < 0.05 were considered as significant.
Molecular Biology
The genotype of wild type and mutant rpl9c and rpl9d alleles was determined by PCR using gene specific primers. RPL9D:RPL9D was generated by PCR amplification of an 4.5 kb genomic region encompassing RPL9D and cloning into the binary vector pMDC123 (Curtis and Grossniklaus, 2003). The construct was transformed into rpl9c by floral dip (Clough and Bent, 1998).
Phylogenetic and Synteny Analysis
Gene sequences were obtained from Phytozome (Goodstein et al., 2012). Designated gene names used in phylogenetic analysis and corresponding genomic unique locus identifiers are listed in Supplementary Tables S1–S8. Brassicaceae species included A. thaliana, Arabidopsis lyrata, Capsella rubella, and Capsella grandiflora, within the Camelineae, and Eutrema salsugineum (formerly Thellungiella halophila) and B. rapa. For Brassicaceae species, genome sequence assembly into chromosomes was incomplete for several species. Therefore designated RPL9, RPL4, RPL5, RPL27a, RPL36a, and RPS6 gene names within a species were based on phylogenetic relationships. RPL9 gene names for other dicot species and for monocot species that had complete genome assemblies were assigned according to map location. This included the dicot species Gossypium raimondii, Medicago truncatula, Phaseolus vulgaris, Poplar trichocarpa, Solanum lycopersicum, S. tuberosum, and Vitis vinifera, and the monocot species Brachypodium distachyon, Oryza sativa, Sorghum bicolor, and Zea mays. RPL9 gene names for species where genome assembly was incomplete were arbitrarily assigned. This included the dicot species Aquilegia coerulea, Carica papaya, Citrus clementina, Citrus sinensis, Linum usitatissimum, and Mimulus guttatus, and the monocot species Panicum virgatum and Setaria italica. The CDS sequences were used to estimate phylogenetic relationships. Orthologous ribosomal proteins from Drosophila melanogaster were selected as the outgroup. Full-length sequences were aligned using ClustalW and phylogenetic relationships were inferred using MEGA6.06 (Tamura et al., 2013). Trees were constructed with the Maximum-Likelihood algorithm and default settings with 1000 bootstrap replications. Synteny analysis was carried out using CoGepedia (Lyons and Freeling, 2008; Lyons et al., 2008) with a sequence distance set to 100 kb.
Results
RPL9C and RPL9D have Redundant Functions in Plant Growth
Arabidopsis thaliana has three RPL9 genes, RPL9B, RPL9C, and RPL9D (Barakat et al., 2001). The proteins encoded by RPL9B and RPL9C share 100% amino acid identity whereas RPL9B/RPL9C and RPL9D encoded proteins are more divergent and share 89% amino acid identity. Although all three genes are ubiquitously expressed, transcript levels of RPL9C are approximately twofold higher than RPL9D and threefold higher than RPL9B (Laubinger et al., 2008; Pinon et al., 2008). To determine whether divergent members of the RPL9 gene family are redundant we compared phenotypes resulting from mutation in RPL9C and RPL9D. rpl9c (previously named pgy2) is a weak allele and has a splice-donor site point mutation that reduces the level of wild type transcript. rpl9c leaves are pointed and have more prominent marginal serrations compared to wild type (Pinon et al., 2008) (Figure 1). A T-DNA mutant rpl9d had an insertion in the first exon of RPL9D and was predicted to be a null allele. The leaf shape of rpl9d was not distinct from that of wild type (Figure 1). This indicated either RPL9D has no function in leaf development or the contribution of RPL9D to leaf development is not significant in the presence of functional RPL9B and RPL9C. To determine whether or not RPL9D contributes to leaf development, the effect of reduced levels of RPL9C in the rpl9d mutant was examined. The double homozygous mutant is embryo lethal (see below). Therefore rpl9c/+ rpl9d plants were examined. rpl9c/+ rpl9d plants had mildly serrated leaves compared with rpl9d single mutants indicating partial loss of RPL9C function slightly modifies leaf development in the rpl9d background (Figure 1). The effect of reduced RPL9D on the rpl9c mutant was also examined. The leaf phenotype of rpl9c rpl9d/+ plants was more severe and leaves were smaller and more pointed than rpl9c (Figure 1). This enhanced phenotype indicates RPL9D acts redundantly with RPL9C in leaf growth.
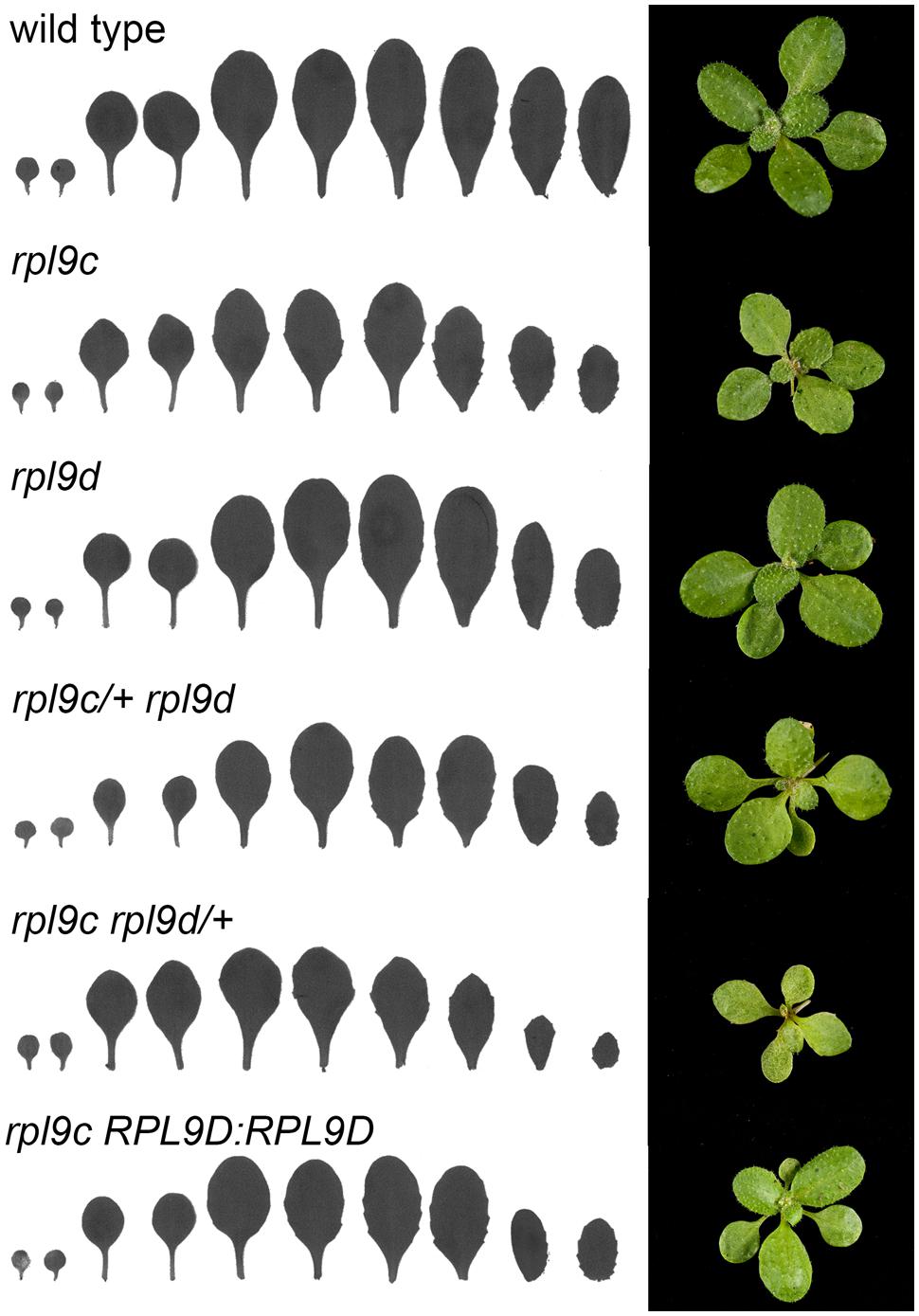
FIGURE 1. RPL9C and RPL9D have redundant functions in leaf development. Silhouettes (left) and rosettes (right) of wild type, rpl9c, rpl9d, rpl9c/+ rpl9d, rpl9c rpl9d/+, and rpl9c homozygous for the transgene RPL9D:RPL9D.
To confirm that RPL9D is redundant with RPL9C, a genomic clone RPL9D:RPL9D, encompassing the gene promoter and coding region, was transformed into the rpl9c mutant to test for complementation. The leaf shape of progeny from 10 independent transformants was examined. All ten lines segregated plants that had a wild-type phenotype. Progeny from phenotypically wild type plants in two lines were confirmed as homozygous for RPL9D:RPL9D (Figure 1). RPL9D:RPL9D is therefore able to replace the function of RPL9C.
In addition to leaf shape, mutations in ribosomal proteins result in slow growth, although there is limited information quantifying this growth defect. To compare the rate of growth of rpl9 mutants relative to wild type, we measured the rate of leaf production, the time to flower and the rate of inflorescence elongation for wild type, rpl9c, rpl9d, rpl9c/+ rpl9d, and rpl9c rpl9d/+ plants. Plants from two independent rpl9c RPL9D:RPL9D lines were also included in this analysis. All genotypes produced approximately 11 rosette and cauline leaves and transitioned to flowering on average 25.25–27.5 days after sowing, except for rpl9c/+ rpl9d and rpl9c rpl9d/+. Both of these genotypes produced more leaves (average 12.75 and 14.25 leaves, respectively, P < 0.05) and rpl9c rpl9d/+ flowered later (average 32.1 days after sowing, P < 0.05) than wild type (Figures 2A,B). Despite these differences, the rate of vegetative leaf initiation was similar for all genotypes (Figure 2C). During growth of the inflorescence rpl9d plants were not affected but rpl9c (P = 0.04), rpl9c/+ rpl9d (P < 0.005) and rpl9c rpl9d/+ (P < 0.005) plants appeared to be shorter than wild type at any given time point. The growth of the two independent rpl9c RPL9D::RPL9D plants were not significantly different from either wild type or rpl9c plants indicating that RPL9D can partially complement the rpl9c mutation (Figure 2D).
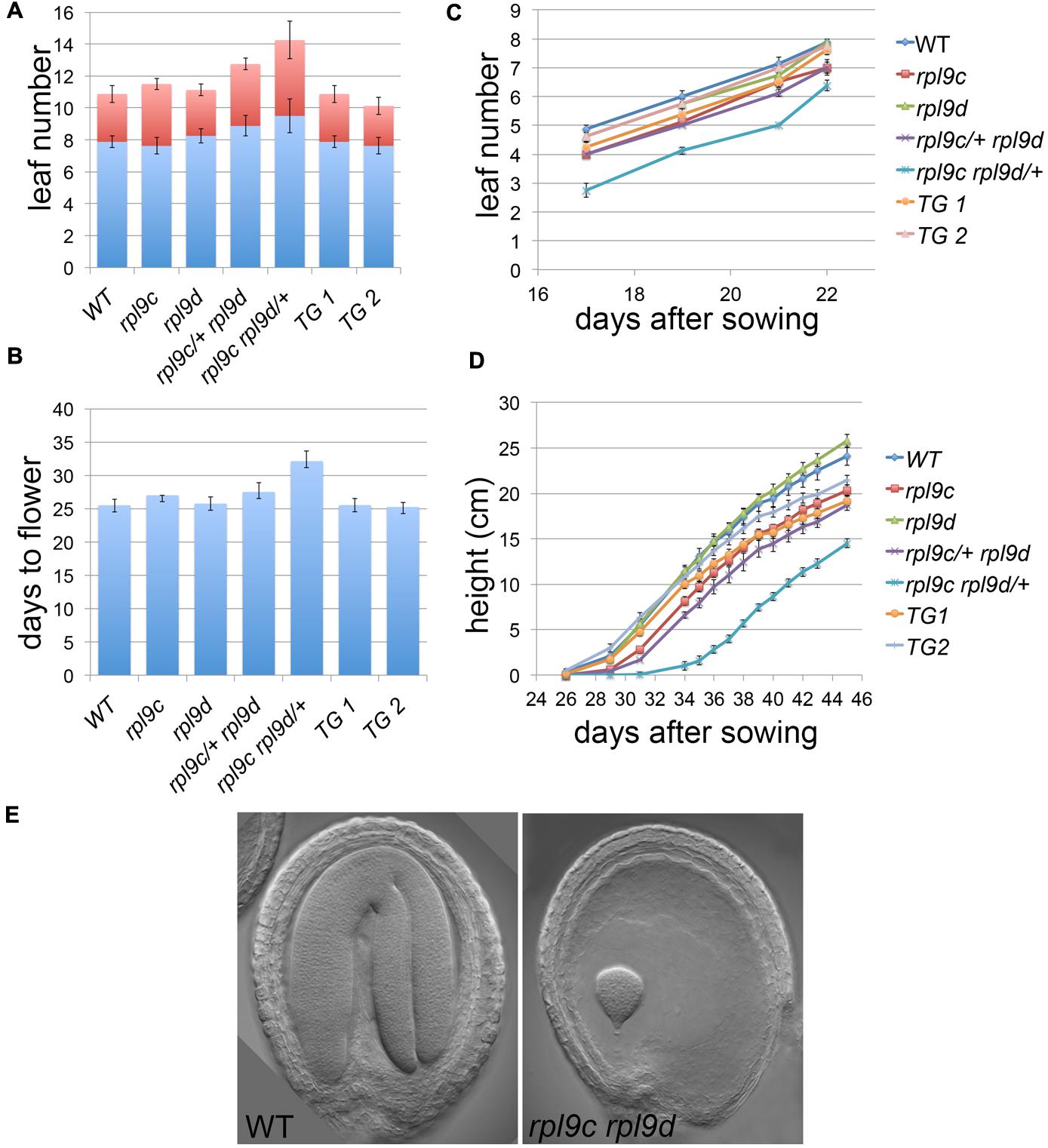
FIGURE 2. Mutations in RPL9 genes delay growth. The number of rosette (blue) and cauline (red) leaves (A), days to flower (B), rate of rosette leaf emergence (C), and rate of inflorescence growth (D) are shown for the genotypes wild type, rpl9c, rpl9d, rpl9c/+ rpl9d, rpl9c rpl9d/+, and for two independent lines rpl9c homozygous for the transgene RPL9D:RPL9D (TG1 and TG2). Data is the mean ± SE (n = 8). Embryos from a silique of an rpl9c/+ rpl9d plant (E). A mature wild type embryo (left) and a putative homozygous rpl9c rpl9d embryo arrested at a late globular stage of development (right).
The rpl9c rpl9d mutant was not identified in progeny of rpl9c rpl9d/+ or rpl9c/+ rpl9d. Plants of these two genotypes showed 24.3% (n = 236) and 22.6% (n = 260) white seed, respectively, consistent with the double homozygous mutant being embryo lethal. By comparison, siliques of the single rpl9c and rpl9d mutants had 0% (n = 205) and 0.6% (n = 309) white seed, respectively. Examination of siliques from rpl9c/+ rpl9d plants showed embryos that were arrested at a late globular stage of development (Figure 2E). Thus RPL9 levels in the rpl9c rpl9d double homozygous mutant are sufficient to support early stages of embryogenesis but are not sufficient to maintain growth throughout embryogenesis.
RPL9 Genes in the Brassicaceae
RPL9B and RPL9C are tandem genes, separated by 11,136 bp on Chromosome 1. Comparison of RPL9B and RPL9C nucleotide sequence revealed a region of identical sequence extending from -927 bp upstream to +623 bp downstream of the AUG initiation codon (Figure 3A). This sequence included the first exon, first intron and the 5′ half of the second exon. Nucleotide sequences of the 3′ half of the second exon and the third exon diverged and were 97% identical, whereas there was no significant sequence identity between the third intron of RPL9B and RPL9C. Comparison of these two genes with RPL9D, on Chromosome 4, showed that RPL9D is more divergent with the CDS sequence sharing 80% nucleotide sequence identity with RPL9B and RPL9C (Figure 3A). RPL9B/RPL9C and RPL9D are located in syntenic regions of A. thaliana Chromosomes 1 and 4 (Supplementary Figure S1A). These two regions are part of a recent whole genome duplication, the α duplication, which occurred 24–40 million years ago (Simillion et al., 2002; Blanc et al., 2003; Bowers et al., 2003).
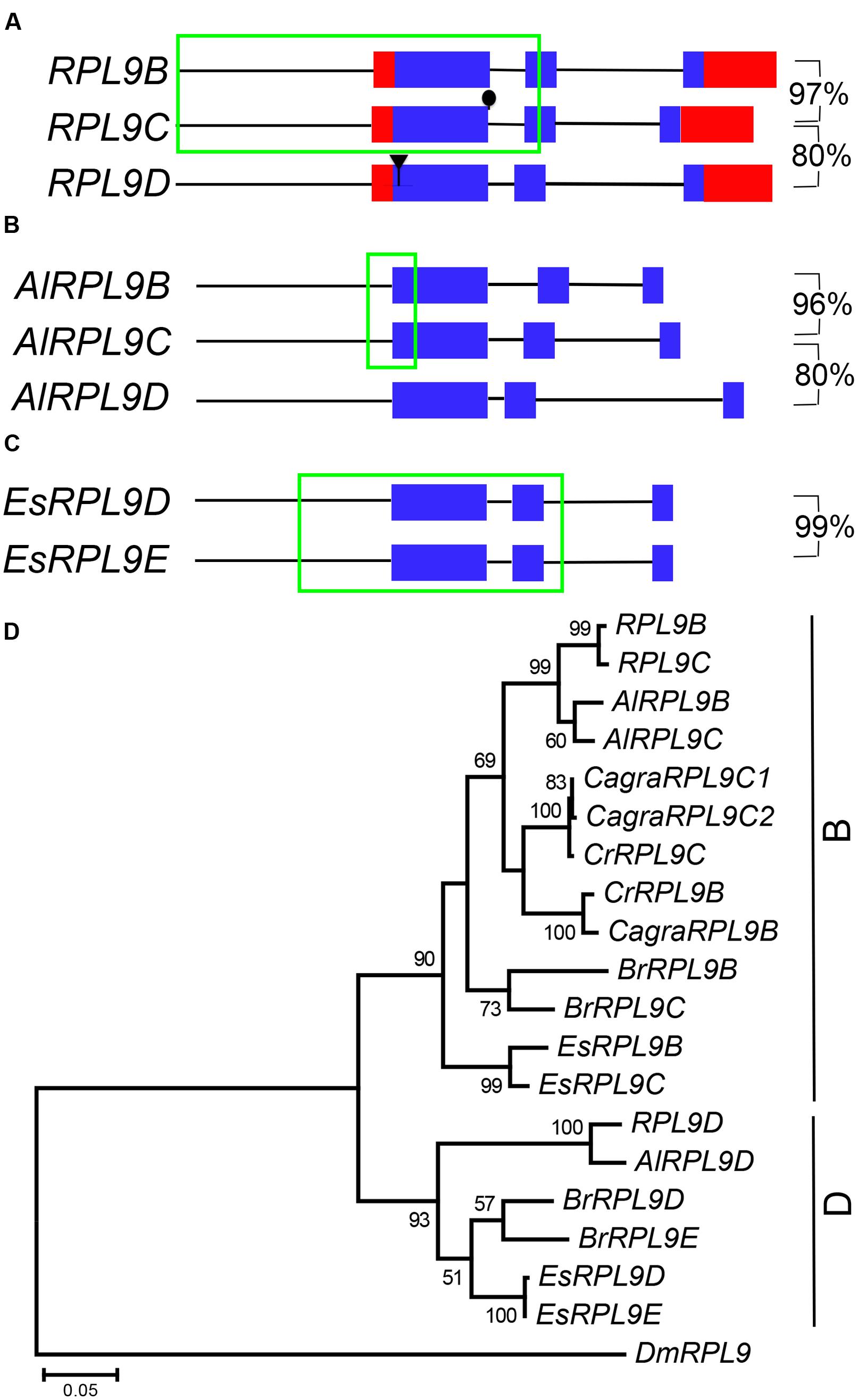
FIGURE 3. RPL9 genes in Brassicaceae. Diagrammatic representation of A. thaliana (A), Arabidopsis lyrata (B), and Eutrema salsugineum (C) genes showing UTR’s (red), exons (blue), non-coding upstream, and intron sequences (black line), and regions of 100% nucleotide sequence identity (green). Percentage nucleotide sequence identity of exons (excluding region in green box) is shown for gene pairs. Phylogenetic tree shown in (D) includes A. thaliana, A. lyrata (AlRPL9), Capsella rubella (CrRPL9), C. grandiflora (CagraRPL9), E. salsuginum (EsRPL9), B. rapa (BrRPL9), and Drosophila melanogaster (DmRPL9) genes.
To determine whether duplicate RPL9 genes in A. thaliana are conserved in closely related species we compared RPL9 family genes from A. lyrata, C. rubella, C. grandiflora, B. rapa, and E. salsugineum (Hu et al., 2011; Wang et al., 2011; Slotte et al., 2013; Yang et al., 2013). A. lyrata, which is most closely related to A. thaliana, has three RPL9 genes. We notionally designated these genes AlRPL9B, AlRPL9C, and AlRPL9D according to the most closely related A. thaliana gene (Supplementary Table S1). As in A. thaliana, AlRPL9B, and AlRPL9C are linked by 8,119 bp and share a region of identical nucleotide sequence, extending from –167 to +245 bp, which includes the 5′ region of the first exon (Figure 3B). The remaining coding regions of these two genes are 96% identical with no significant identity between the introns. AlRPL9D is more divergent and shares 80% sequence identity with AlRPL9B. Phylogenetic analysis showed the three genes in A. thaliana and A. lyrata fall into two distinct clades, which we named B and D group genes (Figure 3D).
Capsella rubella and C. grandiflora were found to have two and three RPL9 genes, respectively. In contrast to A. thaliana and A. lyrata, the two Capsella species had B group but not D group genes (Figure 3D). The additional gene in C. grandiflora appeared to be due to a recent gene duplication. CagraRPL9C1 and CagraRPL9C2 were adjacent direct repeats differing in a single nucleotide that altered the AUG initiation codon of CagraRPL9C2 to TTG (Supplementary Figure S2). This change to a non-canonical initiation codon suggests CagraRPL9C2 may not encode a functional protein (Tikole and Sankararamakrishnan, 2006). E. salsugineum and B. rapa each had two B and two D group RPL9 genes (Figure 3D). E. salsugineum genes occurred as pairs of tandem genes. The B group genes EsRPL9B and EsRPL9C were linked by 12,130 bp, and the D group genes EsRPL9D and EsRPL9E were linked by 9,155 bp (Supplementary Table S1). Unlike the A. thaliana and A. lyrata B group genes, EsRPL9B and EsRPL9C did not share an extended region of identical sequence. However, EsRPL9D and EsRPL9E shared identical sequence extending from -287 bp upstream to +713 bp downstream of the AUG initiation codon. The 3′ exon and intron sequences had only four base pair differences, and the third exon in these two genes shared 99% identity (Figure 3C).
The relationship between A. thaliana, A. lyrata, C. rubella, and E. salsugineum RPL9 family genes was further investigated by determining whether these genes map to syntenic regions between each genome. Consistent with the phylogeny, B group genes of A. lyrata, C. rubella and E. salsugineum were in regions of synteny with A. thaliana Chromosome 1 carrying RPL9B and RPL9C genes (Supplementary Figure S1A). Synteny was also identified between chromosomal regions carrying D group genes of A. thaliana, A. lyrata and E. salsugineum (Supplementary Figure S1B). Notably the tandem gene pair EsRPL9D/EsRPL9E in E. salsugineum was located in a region of synteny with the single gene RPL9D in A. thaliana (Supplementary Figure S1B). This suggests that there has been either a single gene loss or gain following the divergence of Arabidopsis and E. salsugineum. Potentially the pre-α duplication genome had two tandem RPL9 genes. Subsequent to the α duplication the Arabidopsis lineage has lost one RPL9 gene.
RPL9 Genes in Eudicots and Monocots
To further investigate the evolution of RPL9 families within plants we analyzed RPL9 genes from thirteen dicot and six monocot species. These represent a diverse range of plant taxa for which whole genome sequence was available. The number of RPL9 genes in the dicot and monocot species ranges from 2 to 4 copies (Figure 4). As in the Brassicaceae, tandem genes were found in the dicot species P. tichocarpa, P. vulgaris, and Carica papaya but were not found in the monocot species (Supplementary Table S1). Phylogenetic analysis showed that RPL9 genes within a species tended to be more closely related to each other than to orthologs in distantly related species. The exceptions were for closely related species. The two Citrus and two Solanum species had one gene in each of three clusters (Figure 4A). The closely related monocots Z. mays and S. bicolor, had RPL9 genes in two separate clusters (Figure 4B). The estimated gene trees suggest that RPL9 family members undergo homogenization over time, leading to limited variation between genes within a species and greater variation of gene families between species. This lack of concordance between species and gene trees is a hallmark of concerted evolution (Arguello and Connallon, 2011).
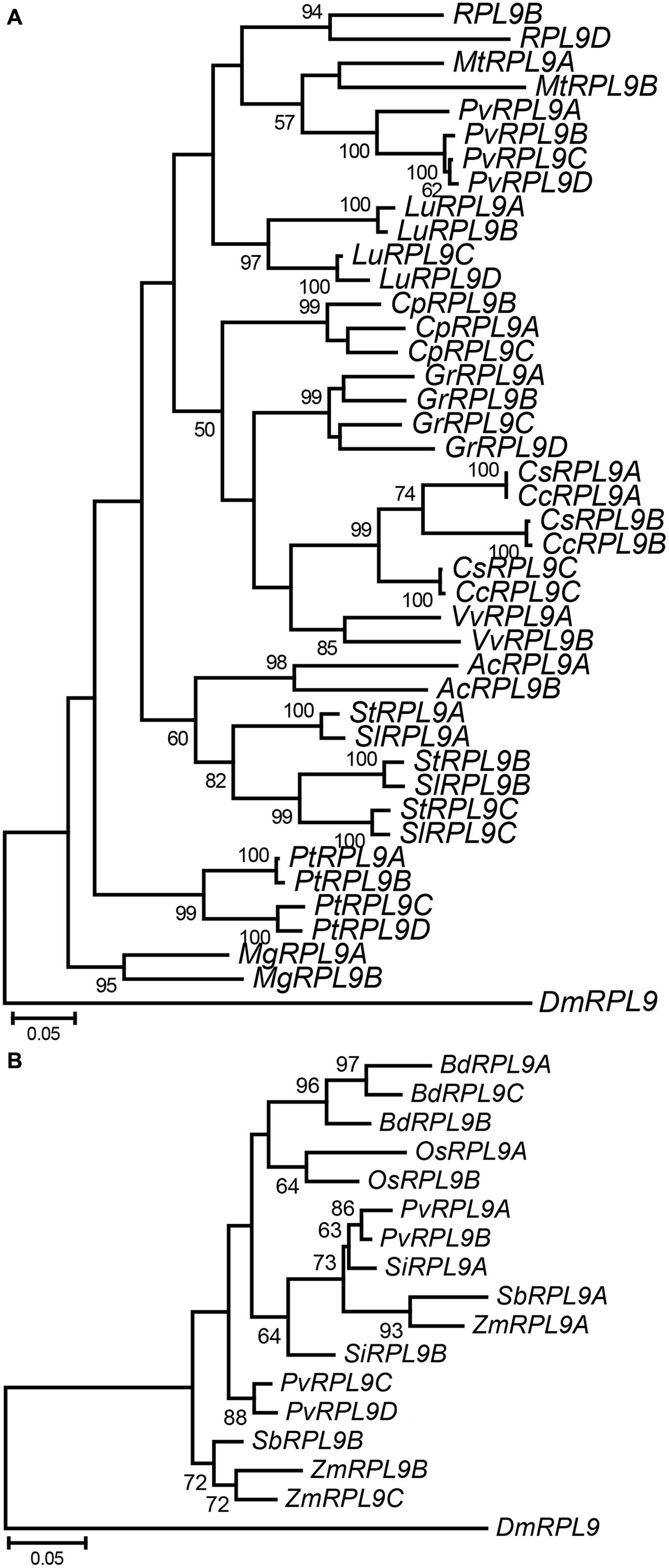
FIGURE 4. Phylogeny of RPL9 genes from angiosperm species. Tree for dicot species (A) Aquilegia coerulea (AcRPL9), Carica papaya (CpRPL9), Citrus clementina (CcRPL9), C. sinensis (CsRPL9), Gossypium raimondii (GrRPL9), Linum usitatissimum (LuRPL9), Medicago truncatula (MtRPL9), Mimulus guttatus (MgRPL9), P. vulgaris (PhvRPL9), Poplar trichocarpa (PtRPL9), Solanum lycopersicum (SlRPL9), S. tuberosum (StRPL9), and Vitis vinifera (VvRPL9). Tree for monocot species (B) Brachypodium distachyon (BdRPL9), Oryza sativa (OsRPL9), Panicum virgatum (PvRPL9), Setaria italica (SiRPL9), S. bicolor (SbRPL9), and Zea mays (ZmRPL9). D. melanogaster RPL9 gene (DmRPL9) is the outgroup.
RPL4, RPL5, RPL27a, RPL36a, and RPS6 Genes in the Brassicaceae
We have shown that RPL9 family genes have largely been retained during the divergence of the Brassicaceae but there is also evidence of rearrangements since divergence of different species. To investigate whether such rearrangements are common to other ribosomal protein gene families we examined phylogenetic relationships between members of five other ribosomal protein gene families in the Brassicaceae. The gene families selected for analysis included genes encoding RPL4, RPL5, RPL27a, RPL36a, and RPS6. In A. thaliana, each of these ribosomal proteins is encoded by two functional and redundant genes (Yao et al., 2008; Fujikura et al., 2009; Creff et al., 2010; Rosado et al., 2010; Casanova-Sáez et al., 2014; Zsögön et al., 2014).
The RPL4 family in A. thaliana comprises two functional genes, RPL4A and RPL4D, which are in syntenic regions retained from the α duplication (Bowers et al., 2003; Rosado et al., 2010) (Figure 5A, Supplementary Figure S3a). Phylogenetic analysis showed that A. lyrata, C. rubella, C. grandiflora, and E. salsuginum had one gene that clustered with RPL4A and another gene that clustered with RPL4D, forming A and D groups (Figure 5C). B. rapa had two genes in each of these groups. Analysis of A. thaliana, A. lyrata, and C. rubella chromosomal regions carrying A or D group genes showed these genes were in regions sharing synteny, indicating retention of A and D group genes in these species following the α duplication (Supplementary Figure S3a). The RPL4 family in A. thaliana also includes two pseudogenes, RPL4B and RPL4C, which comprise partial sequences (Barakat et al., 2001) (Figure 5A). RPL4 pseudogenes were also found in A. lyrata and C. rubella species (Figure 5B). The pseudogenes formed distinct clusters, which we designated P group genes (Figure 5C). In the P group, a single gene, EsRPL4C, was full length and was predicted to encode a functional protein (Supplementary Figure S4). Although some P group genes occurred in regions of synteny, RPL4B, RPL4C, AlRPL4B, and CrRPL4C retained different RPL4 sequences, and AlRPL4C had a base change that generated a premature stop codon (Supplementary Figures S3b and S4). This suggests the pseudogenes were derived from independent events.
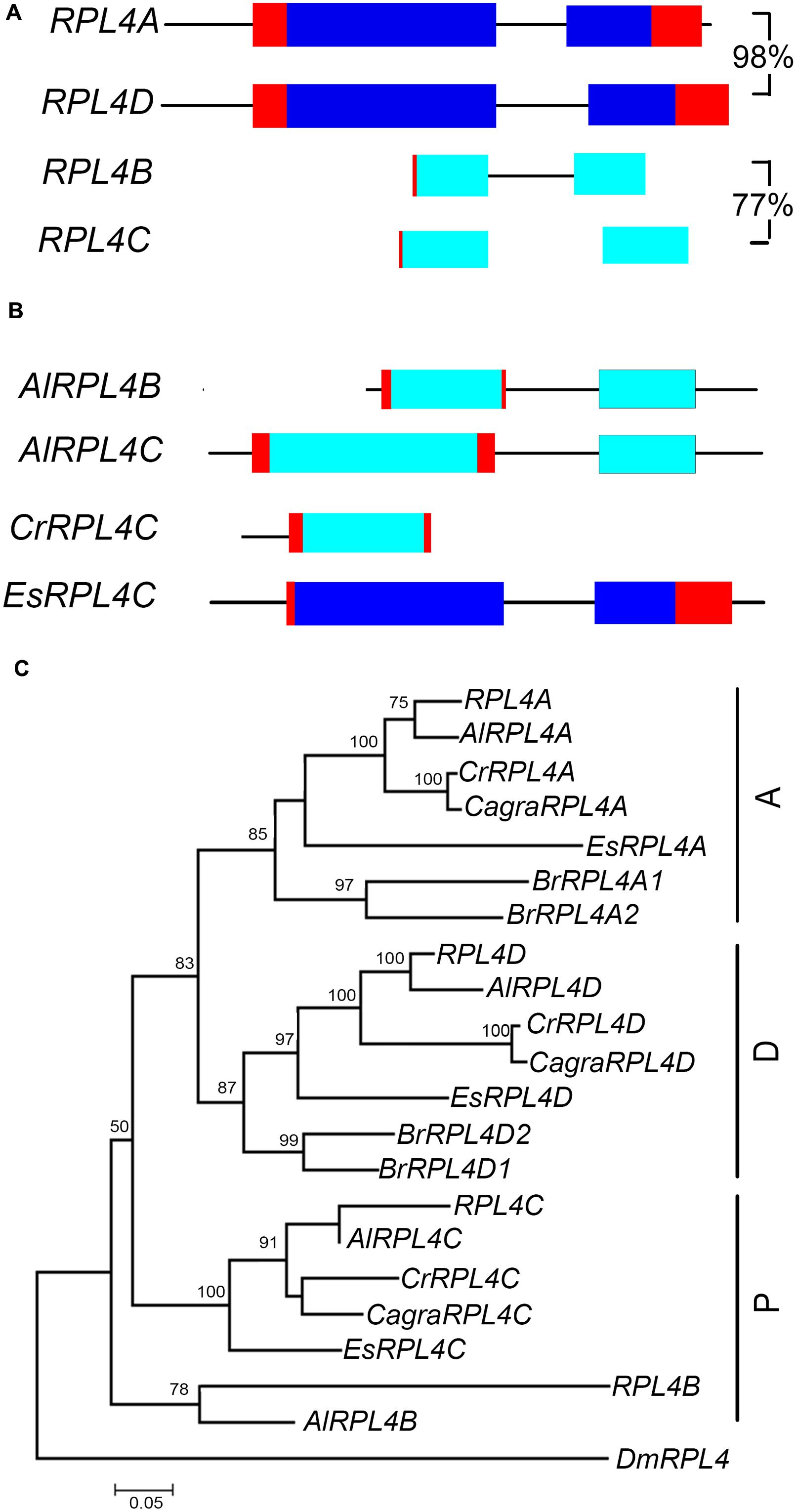
FIGURE 5. RPL4 genes in Brassicaceae. Diagrammatic representation of A. thaliana A group (A) and P group genes (B) showing UTR’s (red), exons (blue), non-coding upstream, and intron sequences (black line), and putative non-functional coding sequences (light blue). Percentage nucleotide sequence identity of exons is shown for A. thaliana genes and pseudogene pair. Phylogenetic tree of RPL4 genes in Brassicaceae (C) includes genes of species listed in Figure 3. The C. grandiflora RPL4C gene sequence used in phylogenetic analysis is likely incomplete.
A. thaliana has two functional RPL5 genes, RPL5A (also known as ATL5, PIGGYBACK3, ASYMMETRIC LEAVES1/2 ENHANCER6, OLIGOCELLULA5) and RPL5B (also known as OLIGOCELLULA7), as well as one pseudogene, RPL5C (Barakat et al., 2001; Pinon et al., 2008; Yao et al., 2008; Fujikura et al., 2009). Comparison of RPL5B and the pseudogene showed that they shared a 109 bp region of identical nucleotide sequence (Figure 6A). Unlike RPL9 and RPL4, the RPL5 genes were not in syntenic regions of the A. thaliana genome, indicating these genes have not been retained from a recent genome duplication or that synteny in these chromosomal regions has been lost since genome duplication. Phylogenetic analysis showed that A. lyrata, C. rubella, and C. grandiflora had one gene that clustered with RPL5A and one gene that clustered with RPL5B, forming A and B groups (Figure 6B). The A. thaliana, A. lyrata, and C. rubella genes in each group were in regions of synteny indicating a common origin (Supplementary Figures S5A,B). A third A. lyrata gene, AlRPL5E, two E. salsuginum, and three B. rapa genes clustered into a group that was distinct from genes in Arabidopsis and Capsella species (Figure 6B). AlRPL5E had no apparent ortholog in A. thaliana although AlRPL5E was located in a region of synteny with RPL5A (Supplementary Figure S5A).
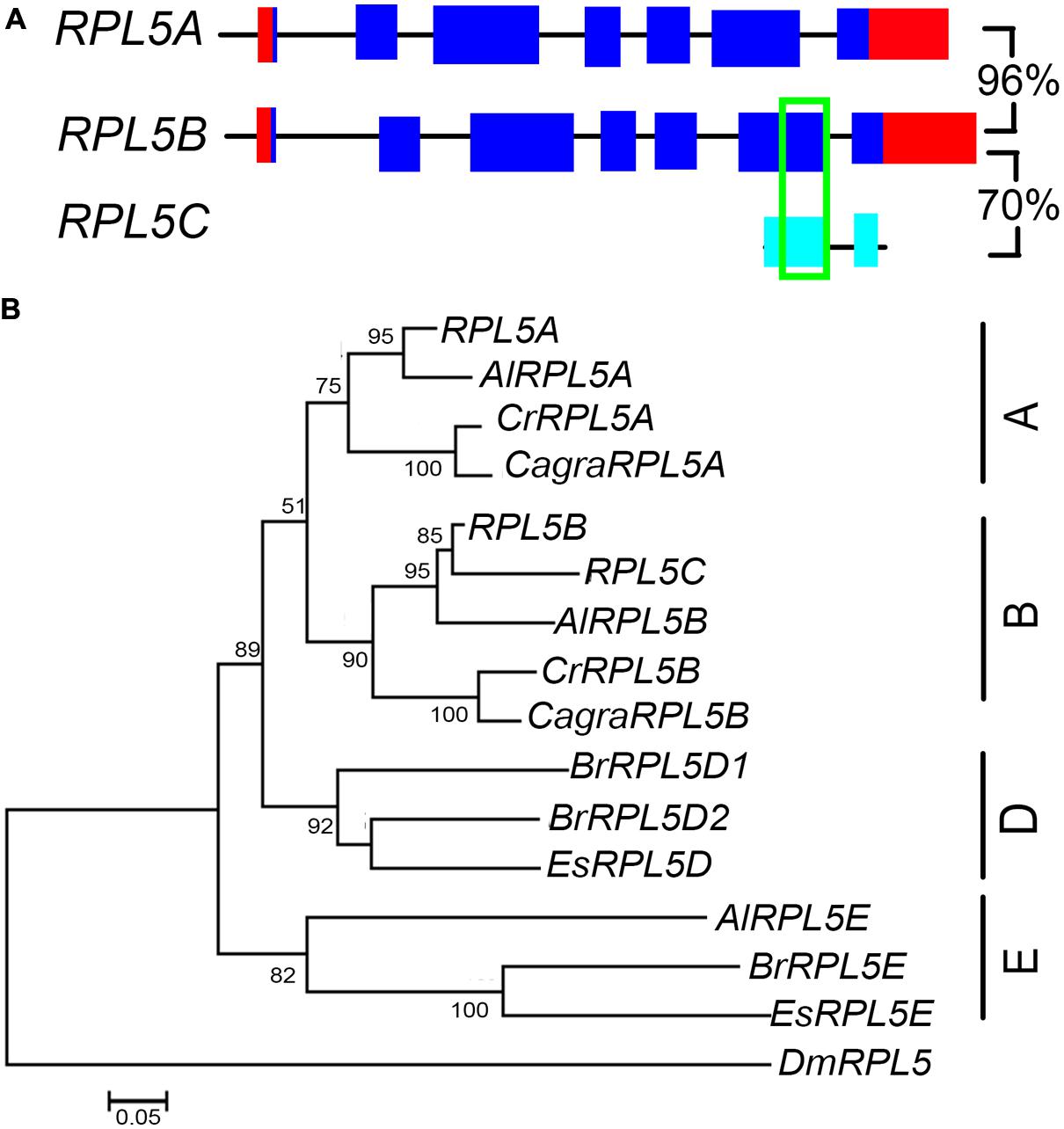
FIGURE 6. RPL5 genes in Brassicaceae. Diagrammatic representation of A. thaliana RPL5 genes (A) showing UTR’s (red), exons (blue), non-coding upstream, and intron sequences (black line), exons of pseudogenes (light blue) and regions of 100% nucleotide identity (green). Percentage nucleotide sequence identity of exons is shown for genes and pseudogene. The phylogenetic tree of RPL5 genes in Brassicaceae (B) includes genes of species as listed in Figure 3.
The phylogeny of RPL27a showed two gene clusters. All species had a single gene in each cluster, except B. rapa where RPL27a genes were more closely related to each other than to RPL27a genes in the other Brassicaceae species (Supplementary Figure S6A). In A. thaliana, the RPL36a family comprises RPL36aA and RPL36aB (also known as APICULATA2) (Barakat et al., 2001; Casanova-Sáez et al., 2014). Each Brassicaceae species had a RPL36a family member that clustered with RPL36aA and a member that clustered with RPL36aB, except for C. grandiflora, which only had one RPL36a gene, and B. rapa, which had several genes in each cluster (Supplementary Figure S6B). Likewise, RPS6 genes clustered into A and B groups and all species had a single gene in each group, with two exceptions. A. lyrata had no A group and two B group genes, and B. rapa had multiple genes in each group (Supplementary Figure S6C).
In summary, the phylogenies of RPL4, RPL5, RPL27a, RPL36a, and RPS6 ribosomal protein gene families show evidence of retention of genes following whole genome duplication. However, there is also evidence of gene gain and loss, partial gene loss, and some instances concerted evolution of genes within a species.
Discussion
All ribosomal proteins in A. thaliana are encoded by small gene families. Typically members of a family encode proteins sharing 95–100% identity, although there are several exceptions where family members encode proteins that show as little as 70% amino acid identity (Barakat et al., 2001). Family members may be required to maintain the dose of a ribosomal protein or each member may encode a variant of a ribosomal protein that contributes to production of functionally heterogeneous ribosome populations (Chang et al., 2005; Carroll et al., 2007; Byrne, 2009; Horiguchi et al., 2012; Xue and Barna, 2012). RPL9C encodes a protein that shares 89% amino acid identity to the RPL9D encoded protein. Despite this divergence, genetic analysis indicates RPL9C and RPL9D are redundant. Firstly, mutation in RPL9D enhanced rpl9c, and conversely, mutation in RPL9C enhanced the phenotype of rpl9d. Secondly, increasing expression of RPL9D, by an RPL9D:RPL9D transgene, repressed the rpl9c mutant. Thirdly, the rpl9c rpl9d double homozygous mutant arrested at the globular stage of development. Embryo arrest at the globular stage is similar to the phenotype of aml1, and to embryo-defective (emb) mutants emb2167 and emb2296, which correspond to mutations in cytoplasmic ribosomal protein genes RPL8A and RPL40B (Weijers et al., 2001; Tzafrir et al., 2004; Meinke, 2013). Deficiency of any one ribosomal protein impairs ribosome assembly or results in inefficient formation of translation-competent ribosomes (de la Cruz et al., 2015). As such it is predicted that inadequate levels of RPL9 lead to a reduction in ribosome production and impairment of translation.
Whole genome duplication is common in flowering plants and several duplications have occurred in dicot and monocot lineages (Bowers et al., 2003; Paterson et al., 2004). Genes encoding proteins that are dosage sensitive, such as transcription factors, signaling pathway, proteasome and ribosomal protein genes are preferentially retained following whole genome duplication (Freeling, 2009). The most recent genome duplication in the rosids, the α duplication, occurred prior to the Arabidopsis-Brassica split (Simillion et al., 2002; Blanc et al., 2003; Bowers et al., 2003). The phylogenetic trees of RPL9, RPL4, RPL5, RPL27a, RPL36a, and RPS6 families in the Brassicaceae showed that genes clustered into two main groups, consistent with retention of ribosomal protein genes following the α duplication. Furthermore, RPL9, RPL4, and RPL5 genes within a group map to regions of synteny indicating retention following genome duplication. However, there were exceptions in which closely related species varied in ribosomal protein gene copy number indicating recent gain or loss of family members. For example, an additional RPL9 gene in E. salsugineum suggested loss of one RPL9 gene after the divergence of the Camelineae species and E. salsugineum. Further RPL9 gene loss and gene duplication appears to have occurred in Capsella species. Compared with A. thaliana, A. lyrata had an additional RPL5 gene. B. rapa has undergone a recent whole genome triplication following divergence from A. thaliana (Wang et al., 2011). Retention of ribosomal protein genes following genome triplication would predict B. rapa to have six members of each ribosomal protein compared to two members in other Brassicaceae. All ribosomal protein families examined showed B. rapa genes occurred in higher copy number indicating that these genes have been retained following genome triplication. However, all families had fewer than six genes indicating a tendency toward loss of ribosomal protein genes. Pseudogenes resulting from partial gene deletion were present in several families and were most notable in the RPL4 family. Surprisingly RPL4 pseudogenes appeared to have been generated through independent deletion events. Potentially these genes are in chromosomal regions subject to frequent rearrangements.
Phylogenies of RPL9 genes in distantly related dicot and monocot species showed clustering of genes within a species rather than between species. A trend where genes within a species are closely related and cluster in a phylogenetic tree was also evident for some RPL5 and RPL27a genes in the Brassicaceae. Such gene relationships indicate ribosomal protein genes in plants undergo concerted evolution (Arguello and Connallon, 2011). Furthermore RPL9 genes in A. thaliana, A. lyrata, and E. salsuginum showed extended regions of identical nucleotide sequence characteristic of recent gene conversion events through homologous recombination between tandem copies of ribosomal protein genes (Chen et al., 2007). Concerted evolution of ribosomal protein genes is also observed in Saccharomyces cereviseae and closely related yeast species (Gao and Innan, 2004). Gene conversion could serve to maintain conservation of proteins that contribute to a complex macromolecule. In this case, amino acids that have low functional significance in a ribosomal protein would vary between between plant species. Interestingly, tandemly arrayed ribosomal RNA genes also undergo concerted evolution and maintain a high level of sequence homogeneity in eukaryotes (Brown et al., 1972; Copenhaver and Pikaard, 1996; Gonzalez and Sylvester, 2001; Stage and Eickbush, 2007). Potentially, ribosomal protein genes and rRNA co-evolve in order to maintain optimal RNA-protein interactions in the ribosome and limit synthesis of inefficient ribosomes (Roberts et al., 2008).
Ribosomal protein gene copy number in plants appears to be under constraint consistent with the gene balance hypothesis. Mechanisms maintaining gene copy number involves retention of paralogs following whole genome duplication. However, partial or whole gene deletion, tandem duplication and gene conversion are prominent features of ribosomal protein gene families across species, reflecting dynamic evolution of these genes.
Author Contributions
DD, SF, ZL, and MB carried out the experiments, prepared the figures, and reviewed the manuscript. MB wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Simon Ho for advice on phylogenetic analyses. This work was supported by the University of Sydney and Australian Research Council Discovery Project grant DP130101186.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01102
FIGURE S1 |RPL9 genes in Brassicaceae occur in regions of synteny. CoGe outputs showing chromosomal regions of synteny. Region of A. thaliana Chromosome 1, around RPL9B and RPL9C, aligned with the region of Chromosome 4 carrying RPL9D, A. lyrata region around AlRPL9B and AlRPL9C, and E. salsuginum region around EsRPL9B and EsRPL9C (A). Region of A. thaliana RPL9D aligned with A. lyrata region around AlRPL9D and E. salsuginum region around EsRPL9D and EsRPL9E (B). Chromosomes are indicated as dotted black lines with genes (green) indicated on both Watson and Crick strands. Unsequenced regions are marked (orange). Regions of identified sequence similarity (pink) are shown above the chromosomes. Interconnecting lines between chromosomes (pink) mark RPL9 genes.
FIGURE S2 | Tandem C. grandiflora RPL9 genes. Nucleotide sequence of CagraRPL9C1 (solid underline) and CagraRPL9C2 (dotted underline) genes. Exons (orange) and introns (black, lower case) are shown with initiation codons highlighted in green and stop codons highlighted in yellow.
FIGURE S3 |RPL4 genes in Brassicaceae occur in regions of synteny. CoGe outputs showing regions of synteny around A. thaliana RPL4D and RPL4A, A. thaliana RPL4D and A. lyrata AlRPL4D and A. thaliana RPL4D, and C. rubella CrRPL4D (A), and regions of synteny around A. thaliana RPL4C and A. lyrata AlRPL4C, and A. thaliana RPL4C and C. rubella CrRPL4C (B). Chromosomes are indicated as dotted black lines with genes (green) indicated on both Watson and Crick strands. Regions of identified sequence similarity (pink) are shown above the chromosomes. Interconnecting lines between chromosomes (pink) mark RPL4 genes.
FIGURE S4 | RPL4 pseudogenes in Brassicaceae. Alignment of nucleotide sequences of P group genes. Start and stop codons are highlighted in red. The C. grandiflora RPL4C gene sequence is likely incomplete.
FIGURE S5 |RPL5 genes in Brassicaceae occur in regions of synteny. CoGe outputs showing regions of synteny around A. thaliana RPL5A (A) and RPL5B (B) genes with RPL5 genes in A. lyrata and C. rubella. Chromosomes are indicated as dotted black lines with genes (green) indicated on both Watson and Crick strands. Regions of identified sequence similarity (pink) are shown above the chromosomes. Interconnecting lines between chromosomes (pink) mark RPL5 genes.
FIGURE S6 | Phylogenetic trees of RPL27a, RPL36a, and RPS6 genes in Brassicaceae. Phylogenetic trees of RPL27a (A), RPL36a (B), and RPS6 (C) includes A. thaliana, A. lyrata, C. rubella, C. grandiflora, E. salsuginum, B. rapa, and D. melanogaster genes.
References
Arguello, J. R., and Connallon, T. (2011). Gene duplication and ectopic gene conversion in Drosophila. Genes 2, 131–151. doi: 10.3390/genes2010131
Barakat, A., Szick-Miranda, K., Chang, I. F., Guyot, R., Blanc, G., Cooke, R., et al. (2001). The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 127, 398–415. doi: 10.1104/pp.010265
Birchler, J. A., and Veitia, R. A. (2012). Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc. Natl. Acad. Sci. U.S.A. 109, 14746–14753. doi: 10.1073/pnas.1207726109
Blanc, G., Hokamp, K., and Wolfe, K. H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13, 137–144. doi: 10.1101/gr.751803
Blanc, G., and Wolfe, K. H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16, 1679–1691. doi: 10.1105/tpc.021410
Bowers, J. E., Chapman, B. A., Rong, J., and Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. doi: 10.1038/nature01521
Brown, D. D., Wensink, P. C., and Jordan, E. (1972). A comparison of the ribosomal DNA’s of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. J. Mol. Biol. 63, 57–73. doi: 10.1016/0022-2836(72)90521-9
Byrne, M. E. (2009). A role for the ribosome in development. Trends Plant Sci. 14, 512–519. doi: 10.1016/j.tplants.2009.06.009
Carroll, A. J., Heazlewood, J. L., Ito, J., and Millar, A. H. (2007). Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol. Cell. Proteomics 7, 347–369. doi: 10.1074/mcp.M700052-MCP200
Casanova-Sáez, R., Candela, H., and Micol, J. L. (2014). Combined haploinsufficiency and purifying selection drive retention of RPL36a paralogs in Arabidopsis. Sci. Rep. 4:4122. doi: 10.1038/srep04122
Chang, I. F., Szick-Miranda, K., Pan, S., and Bailey-Serres, J. (2005). Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol. 137, 848–862. doi: 10.1104/pp.104.053637
Chen, J. M., Cooper, D. N., Chuzhanova, N., Férec, C., and Patrinos, G. P. (2007). Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 8, 762–775. doi: 10.1038/nrg2193
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Copenhaver, G. P., and Pikaard, C. S. (1996). Two-dimensional RFLP analyses reveal megabase-sized clusters of rRNA gene variants in Arabidopsis thaliana, suggesting local spreading of variants as the mode for gene homogenization during concerted evolution. Plant J. 9, 273–282. doi: 10.1046/j.1365-313X.1996.09020273.x
Creff, A., Sormani, R., and Desnos, T. (2010). The two Arabidopsis RPS6 genes, encoding for cytoplasmic ribosomal proteins S6, are functionally equivalent. Plant Mol. Biol. 73, 533–546. doi: 10.1007/s11103-010-9639-y
Curtis, M. D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
Degenhardt, R. F., and Bonham-Smith, P. C. (2008). Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are desperately required for normal development. Plant Physiol. 147, 128–142. doi: 10.1104/pp.107.111799
de la Cruz, J., Karbstein, K., and Woolford, J. L. (2015). Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 84, 93–129. doi: 10.1146/annurev-biochem-060614-033917
Freeling, M. (2009). Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60, 433–453. doi: 10.1146/annurev.arplant.043008.092122
Fujikura, U., Horiguchi, G., Ponce, M. R., Micol, J. L., and Tsukaya, H. (2009). Coordination of cell proliferation and cell expansion mediated by ribosome-related processes in the leaves of Arabidopsis thaliana. Plant J. 59, 499–508. doi: 10.1111/j.1365-313X.2009.03886.x
Gao, L. Z., and Innan, H. (2004). Very low gene duplication rate in the yeast genome. Science 306, 1367–1370. doi: 10.1126/science.1102033
Gonzalez, I. L., and Sylvester, J. E. (2001). Human rDNA: evolutionary patterns within the genes and tandem arrays derived from multiple chromosomes. Genomics 73, 255–263. doi: 10.1006/geno.2001.6540
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Horiguchi, G., Mollá-Morales, A., Pérez-Pérez, J. M., Kojima, K., Robles, P., Ponce, M. R., et al. (2011). Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 65, 724–736. doi: 10.1111/j.1365-313X.2010.04457.x
Horiguchi, G., Van Lijsebettens, M., Candela, H., Micol, J. L., and Tsukaya, H. (2012). Ribosomes and translation in plant developmental control. Plant Sci. 19, 24–34. doi: 10.1016/j.plantsci.2012.04.008
Hu, T. T., Pattyn, P., Bakker, E. G., Cao, J., Cheng, J. F., Clark, R. M., et al. (2011). The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43, 476–481. doi: 10.1038/ng.807
Jiang, W. K., Liu, Y. L., Xia, E. H., and Gao, L. Z. (2013). Prevalent role of gene features in determining evolutionary fates of whole-genome duplication duplicated genes in flowering plants. Plant Physiol. 161, 1844–1861. doi: 10.1104/pp.112.200147
Laubinger, S., Zeller, G., Henz, S. R., Sachsenberg, T., Widmer, C. K., Naouar, N., et al. (2008). At-TAX: a whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome Biol. 9:R112. doi: 10.1186/gb-2008-9-7-r112
Lyons, E., and Freeling, M. (2008). How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 53, 661–673. doi: 10.1111/j.1365-313X.2007.03326.x
Lyons, E., Pedersen, B., Kane, J., Alam, M., Ming, R., Tang, H., et al. (2008). Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 148, 1772–1781. doi: 10.1104/pp.108.124867
Maere, S., De Bodt, S., Raes, J., Casneuf, T., Van Montagu, M., Kuiper, M., et al. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 5454–5459. doi: 10.1073/pnas.0501102102
Meinke, D. W. (2013). A survey of dominant mutations in Arabidopsis thaliana. Trends Plant Sci. 18, 84–91. doi: 10.1016/j.tplants.2012.08.006
Melnikov, S., Ben-Shem, A., Garreau De Loubresse, N., Jenner, L., Yusupova, G., and Yusupov, M. (2012). One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 19, 560–567. doi: 10.1038/nsmb.2313
Papp, B., Pal, C., and Hurst, L. D. (2003). Dosage sensitivity and the evolution of gene families in yeast. Nature 424, 194–197. doi: 10.1038/nature01771
Paterson, A. H., Bowers, J. E., and Chapman, B. A. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. U.S.A. 101, 9903–9908. doi: 10.1073/pnas.0307901101
Pinon, V., Etchells, J. P., Rossignol, P., Collier, S. A., Arroyo, J. M., Martienssen, R. A., et al. (2008). Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135, 1315–1324. doi: 10.1242/dev.016469
Rizzon, C., Ponger, L., and Gaut, B. S. (2006). Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput. Biol. 2:e115. doi: 10.1371/journal.pcbi.0020115
Roberts, E., Sethi, A., Montoya, J., Woese, C. R., and Luthey-Schulten, Z. (2008). Molecular signatures of ribosomal evolution. Proc. Natl. Acad. Sci. U.S.A. 105, 13953–13958. doi: 10.1073/pnas.0804861105
Rosado, A., and Raikhel, N. V. (2010). Application of the gene dosage balance hypothesis to auxin-related ribosomal mutants in Arabidopsis. Plant Signal. Behav. 5, 450–452. doi: 10.4161/psb.5.4.11341
Rosado, A., Sohn, E. J., Drakakaki, G., Pan, S., Swidergal, A., Xiong, Y., et al. (2010). Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. Plant Cell 22, 143–158. doi: 10.1105/tpc.109.068320
Scholl, R. L., May, S. T., and Ware, D. H. (2000). Seed and molecular resources for Arabidopsis. Plant Physiol. 124, 1477–1480. doi: 10.1104/pp.124.4.1477
Simillion, C., Vandepoele, K., Van Montagu, M. C., Zabeau, M., and Van De Peer, Y. (2002). The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 99, 13627–13632. doi: 10.1073/pnas.212522399
Slotte, T., Hazzouri, K. M., Ågren, J. A., Koenig, D., Maumus, F., Guo, Y. L., et al. (2013). The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45, 831–835. doi: 10.1038/ng.2669
Stage, D. E., and Eickbush, T. H. (2007). Sequence variation within the rRNA gene loci of 12 Drosophila species. Genome Res. 17, 1888–1897. doi: 10.1101/gr.6376807
Stirnberg, P., Liu, J. P., Ward, S., Kendall, S. L., and Leyser, O. (2012). Mutation of the cytosolic ribosomal protein-encoding RPS10B gene affects shoot meristematic function in Arabidopsis. BMC Plant Biol. 12:160. doi: 10.1186/1471-2229-12-160
Szakonyi, D., and Byrne, M. E. (2011a). Involvement of ribosomal protein RPL27a in meristem activity and organ development. Plant Signal. Behav. 6, 712–714. doi: 10.4161/psb.6.5.15070
Szakonyi, D., and Byrne, M. E. (2011b). Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J. 65, 269–281. doi: 10.1111/j.1365-313X.2010.04422.x
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thomas, B. C., Pedersen, B., and Freeling, M. (2006). Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16, 934–946. doi: 10.1101/gr.4708406
Tikole, S., and Sankararamakrishnan, R. (2006). A survey of mRNA sequences with a non-AUG start codon in RefSeq database. J. Biomol. Struct. Dyn. 24, 33–42. doi: 10.1080/07391102.2006.10507096
Tzafrir, I., Pena-Muralla, R., Dickerman, A., Berg, M., Rogers, R., Hutchens, S., et al. (2004). Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135, 1206–1220. doi: 10.1104/pp.104.045179
Van de Peer, Y., Fawcett, J. A., Proost, S., Sterck, L., and Vandepoele, K. (2009). The flowering world: a tale of duplications. Trends Plant Sci. 14, 680–688. doi: 10.1016/j.tplants.2009.09.001
Veitia, R. A. (2002). Exploring the etiology of haploinsufficiency. Bioessays 24, 175–184. doi: 10.1002/bies.10023
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Weijers, D., Franke-Van Dijk, M., Vencken, R. J., Quint, A., Hooykaas, P., and Offringa, R. (2001). An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128, 4289–4299.
Xue, S., and Barna, M. (2012). Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13, 355–369. doi: 10.1038/nrm3359
Yang, R., Jarvis, D. E., Chen, H., Beilstein, M. A., Grimwood, J., Jenkins, J., et al. (2013). The reference genome of the halophytic plant Eutrema salsugineum. Front. Plant Sci. 4:46. doi: 10.3389/fpls.2013.00046
Yao, Y., Ling, Q., Wang, H., and Huang, H. (2008). Ribosomal proteins promote leaf adaxial identity. Development 135, 1325–1334. doi: 10.1242/dev.017913
Keywords: concerted evolution, dosage sensitive, gene redundancy, plant, ribosomal protein
Citation: Devis D, Firth SM, Liang Z and Byrne ME (2015) Dosage Sensitivity of RPL9 and Concerted Evolution of Ribosomal Protein Genes in Plants. Front. Plant Sci. 6:1102. doi: 10.3389/fpls.2015.01102
Received: 15 September 2015; Accepted: 22 November 2015;
Published: 16 December 2015.
Edited by:
Maria Eugenia Zanetti, Consejo Nacional de Investigaciones Científicas y Técnicas, and Universidad Nacional de LA Plata, ArgentinaReviewed by:
Paula Casati, Centro de Estudios Fotosinteticos–Consejo Nacional de Investigaciones Científicas y Técnicas, ArgentinaAdriana Garay, Universidad Nacional Autónoma de México, Mexico
Abel Rosado, The University of British Columbia, Canada
Copyright © 2015 Devis, Firth, Liang and Byrne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mary E. Byrne, bWFyeS5ieXJuZUBzeWRuZXkuZWR1LmF1
 Deborah Devis
Deborah Devis Sue M. Firth
Sue M. Firth Zhe Liang
Zhe Liang Mary E. Byrne
Mary E. Byrne