- 1RIKEN Center for Sustainable Resource Science, Yokohama, Japan
- 2Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Tokyo, Japan
- 3Bio-Next Project, Okazaki Institute for Integrative Bioscience, National Institutes of Natural Sciences, Okazaki, Japan
A major source of diversity in flowering plant form is the extensive variability of leaf shape and size. Leaf formation is initiated by recruitment of a handful of cells flanking the shoot apical meristem (SAM) to develop into a complex three-dimensional structure. Leaf organogenesis depends on activities of several distinct meristems that are established and spatiotemporally differentiated after the initiation of leaf primordia. Here, we review recent findings in the gene regulatory networks that orchestrate leaf meristem activities in a model plant Arabidopsis thaliana. We then discuss recent key studies investigating the natural variation in leaf morphology to understand how the gene regulatory networks modulate leaf meristems to yield a substantial diversity of leaf forms during the course of evolution.
Introduction
A leaf is a flat lateral organ of the stem, and grows along three-dimensional axes: proximal-distal, medial-lateral, and adaxial-abaxial axes (Steeves and Sussex, 1989). In general, the leaf consists of a leaf blade and a leaf petiole, and the leaf is composed of different cell types including epidermal cells, palisade cells, spongy mesophyll cells, and xylem/phloem cells (Esau, 1977). Because cell proliferation and cell differentiation occur concurrently during leaf development (Donnelly et al., 1999), a single leaf maintains cells in different developmental stages such as mitotic cells, differentiating cells, and endoreduplicating cells. Therefore, an elaborated spatiotemporal regulation of organ and cellular morphologies should underpin the leaf formation.
Plants acquire the bulk of their energy from light capture by leaves, and for this reason the leaf is specialized for photosynthesis, respiration, and photoperception. Leaf shape has direct consequences on the efficiency of light capture, photosynthetic carbon fixation, and gas exchange (Nicotra et al., 2011; Chitwood et al., 2012). As a result, leaf morphology must be optimized in response to variations in environmental conditions. In addition, plant leaves are equipped with an array of structural, chemical and protein-based defenses against herbivores and pathogens, which often target leaves (Agrios, 2005). These multiple functions are accomplished by the heterologous organ and cellular morphologies in a single leaf.
Due to their sessile lifestyle, plants exhibit a variety of morphological and physiological leaf traits that have allowed adaptation to different natural habitats. Indeed, leaf structural traits such as shape, size, and venation pattern, and physiological traits such as photosynthetic mechanisms are diversified in angiosperms (Flood et al., 2011; Sack et al., 2012; Tsukaya, 2014a). There are pressing research questions regarding leaf formation and variation: How do cellular activities cause leaf formation? What are the gene regulatory networks controlling leaf development? How were the gene regulatory networks altered during evolution? In this review, we highlight recent findings on the activities of leaf meristem and their gene regulatory networks in the model plant Arabidopsis thaliana. Knowledge gained from studies of A. thaliana has facilitated evolutionary developmental studies of leaf morphology, and we discuss recent investigations of the natural variation in leaf morphology.
Leaf Meristems
What Are Leaf Meristems?
Plants have three major organs: leaves, stems, and roots. Stems and roots are directly derived from the shoot apical meristem (SAM) and the root apical meristem (RAM), respectively. The SAM and RAM maintain stem cells and exhibit indeterminate growth, which is an open-ended growth plan. On the other hand, leaves exhibit determinate growth, which is growth with a finite period of development. Meristems are defined, in a broad sense, as proliferating tissues regardless of presence of self-renewing stem cells, although the meristems have been controversial concept: Most molecular developmental biologists narrowly adopt a definition of meristems as proliferating tissues that maintain self-renewing stem cells, while meristematic tissues in leaves or stems are excluded by this definition at present because no stem cells have been observed in these tissues. (Esau, 1977; Tsukaya, 2014b). Notably WOX genes, which are key for sustaining stem cells both in SAM and RAM, are also important for the meristemactic activities in leaf primordia (Nardmann and Werr, 2013). The proliferative activity in leaf primordia is much stronger than that in the SAM, but cells that make a leaf come from restricted area of the primordium in angiosperms. Cell differentiation occurred subsequent to cell division makes the proliferative region in leaf separated spatially from SAM (Figure 1A). Therefore, leaf meristems producing leaf mesophyll cells as well as initial cells of stomata and veins are classified into intercalary meristems, that are meristematic tissues reside in a differentiating organ. Botanist Katherine Esau described in her textbook Plant Anatomy that a series of organogenesis steps in the leaf primordium depends on several distinct meristematic tissues including the plate meristem and the marginal meristem (Esau, 1977). The plate meristem consists of parallel layers of cells dividing anticlinally to play a major role in leaf growth. The marginal meristem, which is located at the edge of the leaf between the adaxial and abaxial surfaces, contributes to the establishment of tissue layers within the leaf.
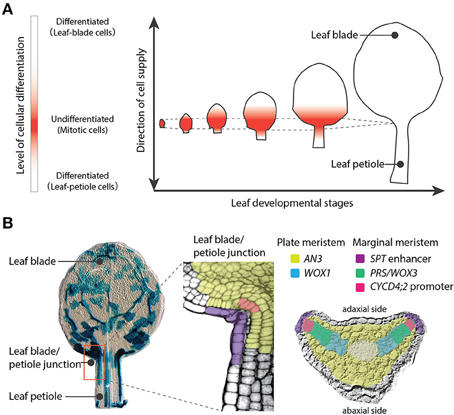
Figure 1. Leaf meristems of Arabidopsis thaliana. (A) Leaf developmental stages showing the proliferative region (red). This meristematic region localizes at the leaf blade/petiole junction and produces both leaf-blade and leaf-petiole cells in a bidirectional manner. The region maintains a constant size over a limited time period. (B) Left image: A leaf primordium at 7 days after sowing with cell lineages indicated by blue staining (sectors were induced at 4 days after sowing). The middle and right images indicate the spatial differentiation of leaf meristems. The plate meristem is marked by AN3 and WOX1 gene expression domains and the marginal meristem is marked by the SPT enhancer, PRS/WOX3, and the promoter of CYCD4;2.
These leaf meristems coordinate the orientation of the cell division plate, produce the main compartments of the leaf (the leaf blade and leaf petiole), and determine the proximal-distal gradient where the switch from cell proliferation to cell differentiation occurs in developing leaves (Donnelly et al., 1999; Ichihashi et al., 2011). This boundary is known as the cell cycle arrest front, and was assumed to progress with basipetal polarity, from the tip to the base of the leaf blade (Donnelly et al., 1999; Nath et al., 2003; White, 2006). However, recent detailed characterizations of the kinetics of cell division during leaf development have indicated that the cell-cycle arrest front does not progress gradually, but rather remains at an almost constant position, and is abolished abruptly (Kazama et al., 2010; Andriankaja et al., 2012; Figure 1A). Like the SAM or RAM, the leaf meristematic region maintains a constant size, but unlike the apical meristems, cell division ceases in the leaf after a certain time period. This leaf meristematic region is localized at the junction between the leaf blade and leaf petiole, and produces both the leaf-blade and leaf-petiole cells via cell divisions in a bidirectional manner (Ichihashi et al., 2011). In addition, leaf meristematic activity differs between tissue layers, and cell divisions directly related to the formation of veins and stomata occur throughout the period of leaf development (Donnelly et al., 1999; White, 2006; Ichihashi et al., 2011). Taken together, these studies reveal that leaves maintain their own meristems, and that the tightly controlled activity of these meristems directs the complex process of leaf tissue development.
Regulatory Mechanisms of Leaf Meristems
Molecular markers have been used to identify distinct regions within the leaf proliferative region of A. thaliana. (Figure 1B). The ANGUSTIFOLIA3 (AN3) gene promoter is active in mesophyll cells just above the leaf blade/petiole junction within the leaf proliferating region (Horiguchi et al., 2005; Ichihashi et al., 2011; Kawade et al., 2013). AN3 encodes a putative transcriptional coactivator homologous to human synovial sarcoma translocation protein, and is a positive regulator of cell proliferation in the leaf blade and leaf petiole (Kim and Kende, 2004; Horiguchi et al., 2005; Ichihashi et al., 2011). AN3 transcripts accumulate only in mesophyll cells, but the AN3 protein moves across different leaf layers to coordinate proliferation between clonally independent leaf cells (Kawade et al., 2013). Although the exact spatiotemporal distribution of AN3 protein has to be characterized, AN3 could mark the position of the plate meristem in leaf primordia. On the other hand, an enhancer trap line with T-DNA insertion in the 5′ region of SPATULA (SPT), along with other studies of the SPT promoter, show that SPT is expressed at the margin of the proliferative region in leaf primordia (Groszmann et al., 2010; Ichihashi et al., 2010). The promoter of a D-type cyclin gene, CYCD4;2, is active in a small number of cells directly adjacent to the marginal cells of the leaf primordia. Although the actual expression pattern of CYCD4;2 is not identical to that observed in pCYCD4;2 promoter studies (Kono et al., 2007), a specific cis-element seems to allow expression in this small cell population. SPT limits the size of the leaf proliferative region independently of AN3 activity (Ichihashi et al., 2010), and overexpression of CYCD4;2 promotes cell proliferation in leaves (Kono et al., 2007). Therefore, it appears that the promoter activities of SPT and CYCD4;2 mark the position of the marginal meristem in leaf primordia. In addition, two WUSCHEL-RELATED HOMEOBOX (WOX) genes, PRESSED FLOWER (PRS)/WOX3 and WOX1, which encode homeobox transcription factors, also redundantly promote leaf blade outgrowth (Nakata et al., 2012). PRS/WOX3 is expressed mainly near the leaf margin (Nardmann et al., 2004) and may play a role in marginal meristem activity. WOX1 is expressed within the two middle mesophyll layers located exactly between the adaxial and abaxial sides of the leaf blade (Nakata et al., 2012), and might be involved in plate meristem activity. Thus, leaf development depends on multiple leaf meristem activities with local controls of gene expression.
In addition to the local regulation factors, organ-level regulation of leaf meristem activity has also been identified in A. thaliana. KLUH (KLU) encodes the cytochrome P450 enzyme CYP78A5, which promotes organ growth, including growth of leaves, in a non-cell autonomous manner (Anastasiou et al., 2007; Adamski et al., 2009; Eriksson et al., 2010). This suggests that KLU is involved in generating a mobile growth factor. Computer simulation predicts that the KLU-dependent mobile growth factor might have less permeability or be regulated at the physical/biochemical level (Kazama et al., 2010). Computational modeling and time-lapse clonal analyses suggest that growth orientations are specified by a tissue polarity system that changes during leaf development, and that a basic pattern of growth rates across the leaf is established from an early developmental stage (Kuchen et al., 2012). Thus, organ-level regulation coordinates growth patterns at the cellular level to form leaf shape. Taken together, the studies discussed above indicate that leaf meristem activities are tightly controlled by both the local regulation systems in the plate and marginal meristem, and by mobile growth factor and tissue polarity information that functions at the organ level.
Gene Regulatory Networks of Leaf Meristems
Genes Functioning in Cell Proliferation
A number of genes responsible for cell proliferation in leaf primordia have been identified in studies of A. thaliana mutants (Gonzalez et al., 2012; Kalve et al., 2014; Figure 2A). As previously mentioned, AN3 functions at the plate meristem to produce cells of both the leaf blade and the leaf petiole (Kim and Kende, 2004; Horiguchi et al., 2005; Ichihashi et al., 2011; Kawade et al., 2013). AN3 shows protein-protein interaction with GROWTHREGULATING FACTOR5 (GRF5) to promote cell proliferation (Horiguchi et al., 2005). AN3 is also known as GRF-INTERACTING FACTOR1 (GIF1), and other members of the GIF family, GIF2 and GIF3, also promote cell proliferation in a redundant fashion (Lee et al., 2009). AN3 binds to the SWITCH/SUCROSE NONFERMENTING (SWI/SNF) chromatin remodeling complexes to regulate transcription during leaf development (Vercruyssen et al., 2014). AN3 is also involved in the establishment of leaf identity in cotyledons via the repression of root fate during embryogenesis (Kanei et al., 2012). On the other hands KLU is expressed in the basal region of leaf primordia and generates a mobile growth factor (Anastasiou et al., 2007). PRS/WOX3 and WOX1 are also classified as activators of cell proliferation (Nakata et al., 2012). The auxin inducible gene AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE (ARGOS) increases the expression level of the D-type cyclin CYCD3;1 gene through the regulation of the AINTEGUMENTA genes (Krizek, 1999; Mizukami and Fischer, 2000; Hu et al., 2003; Nole-Wilson et al., 2005). APC10 and CDC27a are subunits of the anaphase-promoting complex/cyclosome (APC/C), which functions at the G2 to M transition of the cell cycle and is also reported to regulate leaf cell proliferation (Rojas et al., 2009; Eloy et al., 2011). In addition, the C2H2 zinc finger protein JAGGED (JAG) and a subunit of the Mediator complex STRUWWELPETER (SWP) are also constituent factors that positively control cell proliferation in leaves (Autran et al., 2002; Ohno et al., 2004). All of these genes function mainly in the control of lateral organ growth and not in the SAM and RAM. Therefore, a specialized set of genes is utilized to maintain leaf meristem activities.
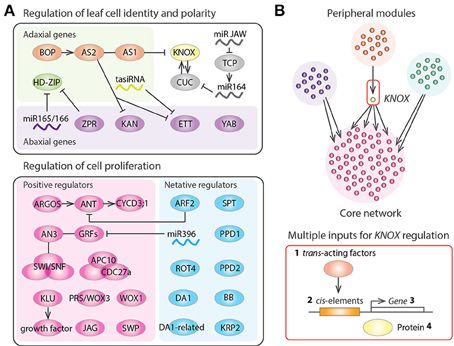
Figure 2. Gene regulatory networks of leaf development. (A) Regulators of leaf structural identification and leaf cell proliferation in Arabidopsis thaliana. Arrows, T bars, and lines indicate positive regulation, negative regulation, and protein-protein interactions, respectively. (B) Schematic diagram representing the gene regulatory networks controlling tomato leaf development, which consists of several peripheral gene network modules and a core network having highly interconnected genes. KNOX appears as a bottleneck in the network, suggesting that KNOX was an evolutionary hot spot that was repeatedly recruited for generating natural variation in leaf shape. KNOX regulation occurs at multiple levels including (1) modulation of trans-acting factors regulating KNOX (Ichihashi et al., 2014), (2) promoter changes at KNOX (Piazza et al., 2010), (3) changes in KNOX expression patterns (Bharathan et al., 2002), and (4) changes in effective KNOX protein concentration (Kimura et al., 2008).
Negative regulators of cell proliferation are important for conferring determinate growth in the leaves. As previously mentioned, SPT is expressed in the marginal meristem of leaf primordia to restrict the size of the leaf proliferative region (Ichihashi et al., 2010). Given that SPT may also help to restrict the size of the RAM (Makkena and Lamb, 2013), SPT might play a general role in the control of meristematic sizes in roots and leaves. MicroRNA396 represses cell proliferation through regulation of the GRF family (Jones-Rhoades and Bartel, 2004; Liu et al., 2009; Rodriguez et al., 2010). The AUXIN RESPONSE FACTOR2 (ARF2) is a repressor of auxin signaling that represses ANT gene expression to inhibit cell proliferation (Horiguchi et al., 2006; Schruff et al., 2006; Lim et al., 2010). The short polypeptide ROTUNDIFOLIA4 (ROT4) also functions to repress cell proliferation, especially along the proximal-distal axis (Narita et al., 2004; Ikeuchi et al., 2011; Guo et al., 2015). Therefore, a microRNA, auxin and a short peptide, all of which can potentially move across cell layers, underlie the delicate mechanisms needed to shut down leaf meristem activities. In addition, the RING-finger protein BIG BROTHER (BB); two putative ubiquitin receptors, DA and DA-RELATED (DAR); two TIFY-type transcription factors, PEAPOD1 and PEAPOD2; and the cyclin-dependent kinase inhibitor gene KIP-RELATED PROTEIN2 (KRP2) are also known to negatively regulate leaf cell proliferation (De Veylder et al., 2001; Disch et al., 2006; White, 2006; Ferjani et al., 2007; Li et al., 2008).
Genes Functioning in Leaf Cell Identity and Polarity
Several genes playing a role in the coordination of the above cell proliferation regulators have been identified in A. thaliana (Figure 2A). The BLADE-ON-PETIOLE (BOP) genes and their direct target ASYMMETRIC LEAVES2 (AS2), which interacts with AS1, are involved in the recruitment of the leaf founder cells from the SAM and in the establishment of all three-dimensional axes of the leaf (Semiarti et al., 2001; Iwakawa et al., 2002, 2007; Ha et al., 2003, 2004, 2007, 2010; Xu et al., 2003; Hepworth et al., 2005; Norberg et al., 2005; Zgurski et al., 2005; Fu et al., 2007; Ikezaki et al., 2010; Jun et al., 2010; Ichihashi et al., 2011; Kojima et al., 2011; Ishibashi et al., 2012; Chen et al., 2013). BOP and AS1/2 repress the expression of class I KNOTTED-like homeobox (KNOX) genes, which help maintain the indeterminate growth of the SAM (Semiarti et al., 2001; Byrne et al., 2002; Ha et al., 2003; Lin et al., 2003; Phelps-Durr et al., 2005; Guo et al., 2008). Interestingly, the chromatin-remodeling protein HIRA and the Polycomb-repressive complex2 interact with AS1/2 to bring about KNOX gene silencing (Phelps-Durr et al., 2005; Ueno et al., 2007; Guo et al., 2008; Lodha et al., 2013). In addition, the microRNA JAW regulates the class II TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) genes, which are heterochronic regulators of the leaf maturation schedule and determine the developmental window for organogenesis (Nath et al., 2003; Palatnik et al., 2003; Efroni et al., 2008). Like AN3, the TCPs also interact with SWI/SNF chromatin remodeling complex components (Efroni et al., 2013), suggesting that dynamic reorganization of chromatin architecture might play an important role throughout leaf development.
An array of genes responsible for the regulation of the adaxial-abaxial axis has been identified. These genes competitively regulate of adaxial and abaxial identity, required for flat outgrowth of the lamina. The regulators of adaxial identity are BOP, AS1/2, a family of class III HOMEODOMAIN LEUCINE ZIPPER (HD-ZIP) transcription factors, and a trans-acting small-interfering RNA (tasiRNA) (McConnell et al., 2001; Garcia et al., 2006). The regulators of abaxial identity are microRNA165/166 and LITTLE ZIPPER (ZPR), which both repress HD-ZIP (Mallory et al., 2004; Wenkel et al., 2007); KANADI (KAN), which is repressed by AS2 (Wu et al., 2008); ETTIN/ARF3, which is repressed by AS2 and tasiRNA (Garcia et al., 2006; Iwasaki et al., 2013; Takahashi et al., 2013). YABBY (YAB) genes also interact with the abaxial identity systems, but they are essential in switching from the SAM program to the leaf-specific program (Sawa et al., 1999; Siegfried et al., 1999; Sarojam et al., 2010). Moreover, various metabolites, ribosomal proteins, and plastid signals have been reported to affect the establishment of the leaf adaxial-abaxial axis (Pinon et al., 2008; Yao et al., 2008; Horiguchi et al., 2011; Toyokura et al., 2011, 2015; Tameshige et al., 2013). Thus, housekeeping genes working in basic metabolic and cellular functions might play specific roles in leaf development (Tsukaya et al., 2013).
In addition to three-dimensional axes of whole leaf, additional growth axes are formed to develop leaf serrations in case of Arabidopsis thaliana. Auxin maxima along leaf margins are required for the outgrowth of serrations, and automatically formed through the activity of auxin efflux carrier PIN-FORMED1 (Kawamura et al., 2010; Bilsborough et al., 2011). CUP-SHAPED COTYLEDON (CUC) genes, which are generally required for boundary formation between two organs, are also key players in the serration formation (Nikovics et al., 2006). CUC2 is essential for robustly positioning and indenting individual serrations (Bilsborough et al., 2011). TCP fine-tunes the expressional patterns of CUC through the regulation of microRNA164 to shape the serrations (Nikovics et al., 2006; Koyama et al., 2007, 2010; Kawamura et al., 2010).
Leaf Morphological Evolution
Despite of the diversity in leaf morphology, the molecular mechanisms that give rise to developmental variation are incompletely understood. Evolutionary developmental biology (evo-devo) studies of plants and animals have revealed the importance of gene regulation in determining developmental variation (Blein et al., 2008; Kimura et al., 2008; Rebeiz et al., 2009; Yamaguchi et al., 2010; Loehlin and Werren, 2012). This suggests that the rewiring of developmental gene regulatory networks is a crucial causal factor driving morphological evolution (Peter and Davidson, 2011). Current evo-devo studies of plant morphology suggest that KNOX expression was recruited repeatedly to generate natural variation in leaf shape in several plant lineages (Bharathan et al., 2002; Kimura et al., 2008; Hay and Tsiantis, 2010; Piazza et al., 2010; Nakayama et al., 2012, 2014). Changes in the activity of other homeobox genes REDUCED COMPLEXITY also cause the repeated evolutionary modification of leaf morphology in Brassicaceae (Sicard et al., 2014; Vlad et al., 2014), suggesting that the regulation of homeobox genes was recruited repeatedly to influence leaf diversity, similar to homeobox genes contributing to animal body plan evolution (Pick and Heffer, 2012).
As we have seen, a number of leaf development genes have been identified, but the exact interactions between these genes has not been fully elucidated. The development of new genomic tools has enabled the generation of large datasets, which can be used to determine exactly how developmental gene modules are organized into a network hierarchy (Fischer and Smith, 2012). Genome-wide gene expression analyses have been performed for several plant species, to reveal the dynamic changes in gene expression during leaf development (Beemster et al., 2005; Li et al., 2010; Andriankaja et al., 2012; Ichihashi et al., 2014; Palmer et al., 2015). Ichihashi et al. (2014) used cross-species, tissue-specific, and large-scale RNA-seq data to reveal the gene regulatory networks controlling leaf development in the domesticated tomato and its wild relatives. Comparisons of the gene networks among species showed that a module regulating KNOX at the protein level had significant differences across species in a manner correlating with leaf complexity (Kimura et al., 2008; Macalister et al., 2012; Ichihashi et al., 2014). Interestingly, KNOX serves as a bridge connecting a peripheral gene network module to the core network that includes leaf cell proliferation regulators (Ichihashi et al., 2014). Therefore, it appears that KNOX is repeatedly co-opted to generate plant morphological diversity by virtue of its bottleneck location in the gene regulatory network (Figure 2B).
Conclusion
The leaf is a complex three-dimensional photochemical reactor whose form and function are determined by gene regulatory networks. Recent technical advances are being used to unravel the mysteries of the molecular mechanisms behind leaf development and evolution. For example, gene expression studies using tissue-specific promoters will further reveal the detailed functions of leaf meristems. The key leaf development genes KNOX, TCP, and AN3 are involved in epigenetic regulation (Phelps-Durr et al., 2005; Ueno et al., 2007; Guo et al., 2008; Efroni et al., 2013; Lodha et al., 2013; Vercruyssen et al., 2014). Therefore, next-generation sequencing will be useful for characterizing the genome-wide changes in chromatin structure, DNA methylation, and histone modifications during leaf development and between plant species. Novel leaf structures such as the pitcher leaves of carnivorous plants and the unifacial leaves of monocots, are generated through tissue-specific changes in cell division (Yamaguchi et al., 2010; Fukushima et al., 2015). Future studies of leaf meristem activity in determining leaf shape will undoubtedly provide greater insights into the molecular mechanisms behind the substantial diversity of leaf forms in nature.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to colleagues whose work could not be highlighted because of space limitations. This work was partially supported by the RIKEN Special Postdoctoral Researchers Program; Grant-in-Aid for Young Scientists (B; grant no. 15K18589) from the Ministry of Education, Culture, Sports, Science and Technology, Japan to YI; and by the Japan Society for the Promotion of Science (Grants-in-Aid for Creative Scientific Research and Scientific Research A), the Ministry of Education, Culture, Sports, Science and Technology, Japan (Scientific Research on Priority Areas and Scientific Research on Innovative Areas) to HT.
References
Adamski, N. M., Anastasiou, E., Eriksson, S., O'Neill, C. M., and Lenhard, M. (2009). Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 20115–20120. doi: 10.1073/pnas.0907024106
Anastasiou, E., Kenz, S., Gerstung, M., MacLean, D., Timmer, J., Fleck, C., et al. (2007). Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 13, 843–856. doi: 10.1016/j.devcel.2007.10.001
Andriankaja, M., Dhondt, S., De Bodt, S., Vanhaeren, H., Coppens, F., De Milde, L., et al. (2012). Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev. Cell 22, 64–78. doi: 10.1016/j.devcel.2011.11.011
Autran, D., Jonak, C., Belcram, K., Beemster, G. T., Kronenberger, J., Grandjean, O., et al. (2002). Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J. 21, 6036–6049. doi: 10.1093/emboj/cdf614
Beemster, G. T., De Veylder, L., Vercruysse, S., West, G., Rombaut, D., Van Hummelen, P., et al. (2005). Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 138, 734–743. doi: 10.1104/pp.104.053884
Bharathan, G., Goliber, T. E., Moore, C., Kessler, S., Pham, T., and Sinha, N. R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296, 1858–1860. doi: 10.1126/science.1070343
Bilsborough, G. D., Runions, A., Barkoulas, M., Jenkins, H. W., Hasson, A., Galinha, C., et al. (2011). Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. U.S.A. 108, 3424–3429. doi: 10.1073/pnas.1015162108
Blein, T., Pulido, A., Vialette-Guiraud, A., Nikovics, K., Morin, H., Hay, A., et al. (2008). A conserved molecular framework for compound leaf development. Science 322, 1835–1839. doi: 10.1126/science.1166168
Byrne, M. E., Simorowski, J., and Martienssen, R. A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129, 1957–1965.
Chen, X., Wang, H., Li, J., Huang, H., and Xu, L. (2013). Quantitative control of ASYMMETRIC LEAVES2 expression is critical for leaf axial patterning in Arabidopsis. J. Exp. Bot. 64, 4895–4905. doi: 10.1093/jxb/ert278
Chitwood, D. H., Headland, L. R., Filiault, D. L., Kumar, R., Jiménez-Gómez, J. M., Schrager, A. V., et al. (2012). Native environment modulates leaf size and response to simulated foliar shade across wild tomato species. PLoS ONE 7:e29570. doi: 10.1371/journal.pone.0029570
De Veylder, L., Beeckman, T., Beemster, G. T., Krols, L., Terras, F., Landrieu, I., et al. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1668. doi: 10.1105/tpc.13.7.1653
Disch, S., Anastasiou, E., Sharma, V. K., Laux, T., Fletcher, J. C., and Lenhard, M. (2006). The E3 ubiquitin ligase BIG BROTHER controls arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16, 272–279. doi: 10.1016/j.cub.2005.12.026
Donnelly, P. M., Bonetta, D., Tsukaya, H., Dengler, R. E., and Dengler, N. G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215, 407–419. doi: 10.1006/dbio.1999.9443
Efroni, I., Blum, E., Goldshmidt, A., and Eshed, Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20, 2293–2306. doi: 10.1105/tpc.107.057521
Efroni, I., Han, S. K., Kim, H. J., Wu, M. F., Steiner, E., Birnbaum, K. D., et al. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24, 438–445. doi: 10.1016/j.devcel.2013.01.019
Eloy, N. B., de Freitas Lima, M., Van Damme, D., Vanhaeren, H., Gonzalez, N., De Milde, L., et al. (2011). The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. Plant J. 68, 351–363. doi: 10.1111/j.1365-313X.2011.04691.x
Eriksson, S., Stransfeld, L., Adamski, N. M., Breuninger, H., and Lenhard, M. (2010). KLUH/CYP78A5-dependent growth signaling coordinates floral organ growth in Arabidopsis. Curr. Biol. 20, 527–532. doi: 10.1016/j.cub.2010.01.039
Ferjani, A., Horiguchi, G., Yano, S., and Tsukaya, H. (2007). Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol. 144, 988–999. doi: 10.1104/pp.107.099325
Fischer, A. H., and Smith, J. (2012). Evo-devo in the era of gene regulatory networks. Integr. Comp. Biol. 52, 842–849. doi: 10.1093/icb/ics112
Flood, P. J., Harbinson, J., and Aarts, M. G. (2011). Natural genetic variation in plant photosynthesis. Trends Plant Sci. 16, 327–335. doi: 10.1016/j.tplants.2011.02.005
Fu, Y., Xu, L., Xu, B., Yang, L., Ling, Q., Wang, H., et al. (2007). Genetic interactions between leaf polarity-controlling genes and ASYMMETRIC LEAVES1 and 2 in Arabidopsis leaf patterning. Plant Cell Physiol. 48, 724–735. doi: 10.1093/pcp/pcm040
Fukushima, K., Fujita, H., Yamaguchi, T., Kawaguchi, M., Tsukaya, H., and Hasebe, M. (2015). Oriented cell division shapes carnivorous pitcher leaves of Sarracenia purpurea. Nat. Commun. 6, 6450. doi: 10.1038/ncomms7450
Garcia, D., Collier, S. A., Byrne, M. E., and Martienssen, R. A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16, 933–938. doi: 10.1016/j.cub.2006.03.064
Gonzalez, N., Vanhaeren, H., and Inzé, D. (2012). Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 17, 332–340. doi: 10.1016/j.tplants.2012.02.003
Groszmann, M., Bylstra, Y., Lampugnani, E. R., and Smyth, D. R. (2010). Regulation of tissue-specific expression of SPATULA, a bHLH gene involved in carpel development, seedling germination, and lateral organ growth in Arabidopsis. J. Exp. Bot. 61, 1495–1508. doi: 10.1093/jxb/erq015
Guo, M., Thomas, J., Collins, G., and Timmermans, M. C. (2008). Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20, 48–58. doi: 10.1105/tpc.107.056127
Guo, P., Yoshimura, A., Ishikawa, N., Yamaguchi, T., Guo, Y., and Tsukaya, H. (2015). Comparative analysis of the RTFL peptide family on the control of plant organogenesis. J. Plant Res. 128, 497–510. doi: 10.1007/s10265-015-0703-1
Ha, C. M., Jun, J. H., and Fletcher, J. C. (2010). Control of Arabidopsis leaf morphogenesis through regulation of the YABBY and KNOX families of transcription factors. Genetics 186, 197–206. doi: 10.1534/genetics.110.118703
Ha, C. M., Jun, J. H., Nam, H. G., and Fletcher, J. C. (2004). BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 45, 1361–1370. doi: 10.1093/pcp/pch201
Ha, C. M., Jun, J. H., Nam, H. G., and Fletcher, J. C. (2007). BLADE-ON-PETIOLE 1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell 19, 1809–1825. doi: 10.1105/tpc.107.051938
Ha, C. M., Kim, G. T., Kim, B. C., Jun, J. H., Soh, M. S., Ueno, Y., et al. (2003). The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 130, 161–172. doi: 10.1242/dev.00196
Hay, A., and Tsiantis, M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153–3165. doi: 10.1242/dev.030049
Hepworth, S. R., Zhang, Y., McKim, S., Li, X., and Haughn, G. W. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17, 1434–1448. doi: 10.1105/tpc.104.030536
Horiguchi, G., Fujikura, U., Ferjani, A., Ishikawa, N., and Tsukaya, H. (2006). Large-scale histological analysis of leaf mutants using two simple leaf observation methods: identification of novel genetic pathways governing the size and shape of leaves. Plant J. 48, 638–644. doi: 10.1111/j.1365-313X.2006.02896.x
Horiguchi, G., Kim, G. T., and Tsukaya, H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43, 68–78. doi: 10.1111/j.1365-313X.2005.02429.x
Horiguchi, G., Mollá-Morales, A., Pérez-Pérez, J. M., Kojima, K., Robles, P., Ponce, M. R., et al. (2011). Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 65, 724–736. doi: 10.1111/j.1365-313X.2010.04457.x
Hu, Y., Xie, Q., and Chua, N. H. (2003). The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15, 1951–1961. doi: 10.1105/tpc.013557
Ichihashi, Y., Aguilar-Martínez, J. A., Farhi, M., Chitwood, D. H., Kumar, R., Millon, L. V., et al. (2014). Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proc. Natl. Acad. Sci. U.S.A. 111, E2616–E2621. doi: 10.1073/pnas.1402835111
Ichihashi, Y., Horiguchi, G., Gleissberg, S., and Tsukaya, H. (2010). The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant Cell Physiol. 51, 252–261. doi: 10.1093/pcp/pcp184
Ichihashi, Y., Kawade, K., Usami, T., Horiguchi, G., Takahashi, T., and Tsukaya, H. (2011). Key proliferative activity in the junction between the leaf blade and leaf petiole of Arabidopsis. Plant Physiol. 157, 1151–1162. doi: 10.1104/pp.111.185066
Ikeuchi, M., Yamaguchi, T., Kazama, T., Ito, T., Horiguchi, G., and Tsukaya, H. (2011). ROTUNDIFOLIA4 regulates cell proliferation along the body axis in Arabidopsis shoot. Plant Cell Physiol. 52, 59–69. doi: 10.1093/pcp/pcq138
Ikezaki, M., Kojima, M., Sakakibara, H., Kojima, S., Ueno, Y., Machida, C., et al. (2010). Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J. 61, 70–82. doi: 10.1111/j.1365-313X.2009.04033.x
Ishibashi, N., Kanamaru, K., Ueno, Y., Kojima, S., Kobayashi, T., Machida, C., et al. (2012). ASYMMETRIC-LEAVES2 and an ortholog of eukaryotic NudC domain proteins repress expression of AUXIN-RESPONSE-FACTOR and class 1 KNOX homeobox genes for development of flat symmetric leaves in Arabidopsis. Biol. Open 1, 197–207. doi: 10.1242/bio.2012406
Iwakawa, H., Iwasaki, M., Kojima, S., Ueno, Y., Soma, T., Tanaka, H., et al. (2007). Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 51, 173–184. doi: 10.1111/j.1365-313X.2007.03132.x
Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., et al. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43, 467–478. doi: 10.1093/pcp/pcf077
Iwasaki, M., Takahashi, H., Iwakawa, H., Nakagawa, A., Ishikawa, T., Tanaka, H., et al. (2013). Dual regulation of ETTIN (ARF3) gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in Arabidopsis. Development 140, 1958–1969. doi: 10.1242/dev.085365
Jones-Rhoades, M. W., and Bartel, D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14, 787–799. doi: 10.1016/j.molcel.2004.05.027
Jun, J. H., Ha, C. M., and Fletcher, J. C. (2010). BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell 22, 62–76. doi: 10.1105/tpc.109.070763
Kalve, S., De Vos, D., and Beemster, G. T. (2014). Leaf development: a cellular perspective. Front. Plant Sci. 5:362. doi: 10.3389/fpls.2014.00362
Kanei, M., Horiguchi, G., and Tsukaya, H. (2012). Stable establishment of cotyledon identity during embryogenesis in Arabidopsis by ANGUSTIFOLIA3 and HANABA TARANU. Development 139, 2436–2446. doi: 10.1242/dev.081547
Kawade, K., Horiguchi, G., Usami, T., Hirai, M. Y., and Tsukaya, H. (2013). ANGUSTIFOLIA3 signaling coordinates proliferation between clonally distinct cells in leaves. Curr. Biol. 23, 788–792. doi: 10.1016/j.cub.2013.03.044
Kawamura, E., Horiguchi, G., and Tsukaya, H. (2010). Mechanisms of leaf tooth formation in Arabidopsis. Plant J. 62, 429–441. doi: 10.1111/j.1365-313X.2010.04156.x
Kazama, T., Ichihashi, Y., Murata, S., and Tsukaya, H. (2010). The mechanism of cell cycle arrest front progression explained by a KLUH/CYP78A5-dependent mobile growth factor in developing leaves of Arabidopsis thaliana. Plant Cell Physiol. 51, 1046–1054. doi: 10.1093/pcp/pcq051
Kim, J. H., and Kende, H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 13374–13379. doi: 10.1073/pnas.0405450101
Kimura, S., Koenig, D., Kang, J., Yoong, F. Y., and Sinha, N. (2008). Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 18, 672–677. doi: 10.1016/j.cub.2008.04.008
Kojima, S., Iwasaki, M., Takahashi, H., Imai, T., Matsumura, Y., Fleury, D., et al. (2011). Asymmetric leaves2 and Elongator, a histone acetyltransferase complex, mediate the establishment of polarity in leaves of Arabidopsis thaliana. Plant Cell Physiol. 52, 1259–1273. doi: 10.1093/pcp/pcr083
Kono, A., Umeda-Hara, C., Adachi, S., Nagata, N., Konomi, M., Nakagawa, T., et al. (2007). The Arabidopsis D-type cyclin CYCD4 controls cell division in the stomatal lineage of the hypocotyl epidermis. Plant Cell 19, 1265–1277. doi: 10.1105/tpc.106.046763
Koyama, T., Furutani, M., Tasaka, M., and Ohme-Takagi, M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19, 473–484. doi: 10.1105/tpc.106.044792
Koyama, T., Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22, 3574–3588. doi: 10.1105/tpc.110.075598
Krizek, B. A. (1999). Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev. Genet. 25, 224–236.
Kuchen, E. E., Fox, S., de Reuille, P. B., Kennaway, R., Bensmihen, S., Avondo, J., et al. (2012). Generation of leaf shape through early patterns of growth and tissue polarity. Science 335, 1092–1096. doi: 10.1126/science.1214678
Lee, B. H., Ko, J. H., Lee, S., Lee, Y., Pak, J. H., and Kim, J. H. (2009). The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151, 655–668. doi: 10.1104/pp.109.141838
Li, P., Ponnala, L., Gandotra, N., Wang, L., Si, Y., Tausta, S. L., et al. (2010). The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42, 1060–1067. doi: 10.1038/ng.703
Li, Y., Zheng, L., Corke, F., Smith, C., and Bevan, M. W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22, 1331–1336. doi: 10.1101/gad.463608
Lim, P. O., Lee, I. C., Kim, J., Kim, H. J., Ryu, J. S., Woo, H. R., et al. (2010). Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J. Exp. Bot. 61, 1419–1430. doi: 10.1093/jxb/erq010
Lin, W. C., Shuai, B., and Springer, P. S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15, 2241–2252. doi: 10.1105/tpc.014969
Liu, D., Song, Y., Chen, Z., and Yu, D. (2009). Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant. 136, 223–236. doi: 10.1111/j.1399-3054.2009.01229.x
Lodha, M., Marco, C. F., and Timmermans, M. C. (2013). The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes Dev. 27, 596–601. doi: 10.1101/gad.211425.112
Loehlin, D. W., and Werren, J. H. (2012). Evolution of shape by multiple regulatory changes to a growth gene. Science 335, 943–947. doi: 10.1126/science.1215193
MacAlister, C. A., Park, S. J., Jiang, K., Marcel, F., Bendahmane, A., Izkovich, Y., et al. (2012). Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Nat. Genet. 44, 1393–1398. doi: 10.1038/ng.2465
Makkena, S., and Lamb, R. S. (2013). The bHLH transcription factor SPATULA regulates root growth by controlling the size of the root meristem. BMC Plant Biol. 13:1. doi: 10.1186/1471-2229-13-1
Mallory, A. C., Reinhart, B. J., Jones-Rhoades, M. W., Tang, G., Zamore, P. D., Barton, M. K., et al. (2004). MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 23, 3356–3364. doi: 10.1038/sj.emboj.7600340
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. doi: 10.1038/35079635
Mizukami, Y., and Fischer, R. L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. U.S.A. 97, 942–947. doi: 10.1073/pnas.97.2.942
Nakata, M., Matsumoto, N., Tsugeki, R., Rikirsch, E., Laux, T., and Okada, K. (2012). Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 24, 519–535. doi: 10.1105/tpc.111.092858
Nakayama, H., Nakayama, N., Seiki, S., Kojima, M., Sakakibara, H., Sinha, N., et al. (2014). Regulation of the KNOX-GA gene module induces heterophyllic alteration in North American Lake Cress. Plant Cell. 26, 4733–4748. doi: 10.1105/tpc.114.130229
Nakayama, H., Yamaguchi, T., and Tsukaya, H. (2012). Acquisition and diversification of cladodes: leaf-like organs in the genus Asparagus. Plant Cell 24, 929–940. doi: 10.1105/tpc.111.092924
Nardmann, J., Ji, J., Werr, W., and Scanlon, M. J. (2004). The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 131, 2827–2839. doi: 10.1242/dev.01164
Nardmann, J., and Werr, W. (2013). Symplesiomorphies in the WUSCHEL clade suggest that the last common ancestor of seed plants contained at least four independent stem cell niches. New Phytol. 199, 1081–1092. doi: 10.1111/nph.12343
Narita, N. N., Moore, S., Horiguchi, G., Kubo, M., Demura, T., Fukuda, H., et al. (2004). Overexpression of a novel small peptide ROTUNDIFOLIA4 decreases cell proliferation and alters leaf shape in Arabidopsis thaliana. Plant J. 38, 699–713. doi: 10.1111/j.1365-313X.2004.02078.x
Nath, U., Crawford, B. C., Carpenter, R., and Coen, E. (2003). Genetic control of surface curvature. Science 299, 1404–1407. doi: 10.1126/science.1079354
Nicotra, A., Leigh, A., Boyce, K. C., Jones, C. S., Niklas, K. J., Royer, D. L., et al. (2011). The evolution and functional significance of leaf shape in the angiosperms. Funct. Plant Biol. 38, 535–552. doi: 10.1071/FP11057
Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H., Aida, M., et al. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18, 2929–2945. doi: 10.1105/tpc.106.045617
Nole-Wilson, S., Tranby, T. L., and Krizek, B. A. (2005). AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol. Biol. 57, 613–628. doi: 10.1007/s11103-005-0955-6
Norberg, M., Holmlund, M., and Nilsson, O. (2005). The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132, 2203–2213. doi: 10.1242/dev.01815
Ohno, C. K., Reddy, G. V., Heisler, M. G., and Meyerowitz, E. M. (2004). The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131, 1111–1122. doi: 10.1242/dev.00991
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Palmer, N. A., Donze-Reiner, T., Horvath, D., Heng-Moss, T., Waters, B., Tobias, C., et al. (2015). Switchgrass (Panicum virgatum L) flag leaf transcriptomes reveal molecular signatures of leaf development, senescence, and mineral dynamics. Funct. Integr. Genomics 15, 1–16. doi: 10.1007/s10142-014-0393-0
Peter, I. S., and Davidson, E. H. (2011). Evolution of gene regulatory networks controlling body plan development. Cell 144, 970–985. doi: 10.1016/j.cell.2011.02.017
Phelps-Durr, T. L., Thomas, J., Vahab, P., and Timmermans, M. C. (2005). Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17, 2886–2898. doi: 10.1105/tpc.105.035477
Piazza, P., Bailey, C. D., Cartolano, M., Krieger, J., Cao, J., Ossowski, S., et al. (2010). Arabidopsis thaliana leaf form evolved via loss of KNOX expression in leaves in association with a selective sweep. Curr. Biol. 20, 2223–2228. doi: 10.1016/j.cub.2010.11.037
Pick, L., and Heffer, A. (2012). Hox gene evolution: multiple mechanisms contributing to evolutionary novelties. Ann. N.Y. Acad. Sci. 1256, 15–32. doi: 10.1111/j.1749-6632.2011.06385.x
Pinon, V., Etchells, J. P., Rossignol, P., Collier, S. A., Arroyo, J. M., Martienssen, R. A., et al. (2008). Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135, 1315–1324. doi: 10.1242/dev.016469
Rebeiz, M., Pool, J. E., Kassner, V. A., Aquadro, C. F., and Carroll, S. B. (2009). Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326, 1663–1667. doi: 10.1126/science.1178357
Rodriguez, R. E., Mecchia, M. A., Debernardi, J. M., Schommer, C., Weigel, D., and Palatnik, J. F. (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112. doi: 10.1242/dev.043067
Rojas, C. A., Eloy, N. B., Lima Mde, F., Rodrigues, R. L., Franco, L. O., Himanen, K., et al. (2009). Overexpression of the Arabidopsis anaphase promoting complex subunit CDC27a increases growth rate and organ size. Plant Mol. Biol. 71, 307–318. doi: 10.1007/s11103-009-9525-7
Sack, L., Scoffoni, C., McKown, A. D., Frole, K., Rawls, M., Havran, J. C., et al. (2012). Developmentally based scaling of leaf venation architecture explains global ecological patterns. Nat. Commun. 3, 837. doi: 10.1038/ncomms1835
Sarojam, R., Sappl, P. G., Goldshmidt, A., Efroni, I., Floyd, S. K., Eshed, Y., et al. (2010). Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 22, 2113–2130. doi: 10.1105/tpc.110.075853
Sawa, S., Watanabe, K., Goto, K., Liu, Y. G., Shibata, D., Kanaya, E., et al. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13, 1079–1088. doi: 10.1101/gad.13.9.1079
Schruff, M. C., Spielman, M., Tiwari, S., Adams, S., Fenby, N., and Scott, R. J. (2006). The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133, 251–261. doi: 10.1242/dev.02194
Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128, 1771–1783.
Sicard, A., Thamm, A., Marona, C., Lee, Y. W., Wahl, V., Stinchcombe, J. R., et al. (2014). Repeated evolutionary changes of leaf morphology caused by mutations to a homeobox gene. Curr. Biol. 24, 1880–1886. doi: 10.1016/j.cub.2014.06.061
Siegfried, K. R., Eshed, Y., Baum, S. F., Otsuga, D., Drews, G. N., and Bowman, J. L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128.
Steeves, T. A., and Sussex, I. M. (1989). Patterns in Plant Development. Cambridge: Cambridge University Press.
Takahashi, H., Iwakawa, H., Ishibashi, N., Kojima, S., Matsumura, Y., Prananingrum, P., et al. (2013). Meta-analyses of microarrays of Arabidopsis asymmetric leaves1 (as1), as2 and their modifying mutants reveal a critical role for the ETT pathway in stabilization of adaxial-abaxial patterning and cell division during leaf development. Plant Cell Physiol. 54, 418–431. doi: 10.1093/pcp/pct027
Tameshige, T., Fujita, H., Watanabe, K., Toyokura, K., Kondo, M., Tatematsu, K., et al. (2013). Pattern dynamics in adaxial-abaxial specific gene expression are modulated by a plastid retrograde signal during Arabidopsis thaliana leaf development. PLoS Genet. 9:e1003655. doi: 10.1371/journal.pgen.1003655
Toyokura, K., Watanabe, K., Oiwaka, A., Kusano, M., Tameshige, T., Tatematsu, K., et al. (2011). Succinic semialdehyde dehydrogenase is involved in the robust patterning of Arabidopsis leaves along the adaxial-abaxial axis. Plant Cell Physiol. 52, 1340–1353. doi: 10.1093/pcp/pcr079
Toyokura, K., Yamaguchi, K., Shigenobu, S., Fukaki, H., Tatematsu, K., and Okada, K. (2015). Mutations in plastidial 5-aminolevulinic acid biosynthesis genes suppress a pleiotropic defect in shoot development of a mitochondrial gaba shunt mutant in Arabidopsis. Plant Cell Physiol. 56, 1229–1238. doi: 10.1093/pcp/pcv050
Tsukaya, H. (2014a). Comparative leaf development in angiosperms. Curr. Opin. Plant Biol. 17, 103–109. doi: 10.1016/j.pbi.2013.11.012
Tsukaya, H. (2014b). “Meristems,” in Atlas of Plant Cell Structure, eds T. Noguchi, S. Kawano, H. Tsukaya, S. Matsunaga, A. Sakai, I. Karahara, and Y. Hayashi (Tokyo: Springer), 187–202.
Tsukaya, H., Byrne, M. E., Horiguchi, G., Sugiyama, M., Van Lijsebettens, M., and Lenhard, M. (2013). How do ‘housekeeping’ genes control organogenesis?–Unexpected new findings on the role of housekeeping genes in cell and organ differentiation. J. Plant Res. 126, 3–15. doi: 10.1007/s10265-012-0518-2
Ueno, Y., Ishikawa, T., Watanabe, K., Terakura, S., Iwakawa, H., Okada, K., et al. (2007). Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19, 445–457. doi: 10.1105/tpc.106.042325
Vercruyssen, L., Verkest, A., Gonzalez, N., Heyndrickx, K. S., Eeckhout, D., Han, S. K., et al. (2014). ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26, 210–229. doi: 10.1105/tpc.113.115907
Vlad, D., Kierzkowski, D., Rast, M. I., Vuolo, F., Dello Ioio, R., Galinha, C., et al. (2014). Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343, 780–783. doi: 10.1126/science.1248384
Wenkel, S., Emery, J., Hou, B. H., Evans, M. M., and Barton, M. K. (2007). A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell 19, 3379–3390. doi: 10.1105/tpc.107.055772
White, D. W. (2006). PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 13238–13243. doi: 10.1073/pnas.0604349103
Wu, G., Lin, W. C., Huang, T., Poethig, R. S., Springer, P. S., and Kerstetter, R. A. (2008). KANADI1 regulates adaxial-abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proc. Natl. Acad. Sci. U.S.A. 105, 16392–16397. doi: 10.1073/pnas.0803997105
Xu, L., Xu, Y., Dong, A., Sun, Y., Pi, L., Xu, Y., et al. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130, 4097–4107. doi: 10.1242/dev.00622
Yamaguchi, T., Yano, S., and Tsukaya, H. (2010). Genetic framework for flattened leaf blade formation in unifacial leaves of Juncus prismatocarpus. Plant Cell 22, 2141–2155. doi: 10.1105/tpc.110.076927
Yao, Y., Ling, Q., Wang, H., and Huang, H. (2008). Ribosomal proteins promote leaf adaxial identity. Development 135, 1325–1334. doi: 10.1242/dev.017913
Keywords: Arabidopsis thaliana, gene regulatory network, leaf development, leaf meristems, natural variation
Citation: Ichihashi Y and Tsukaya H (2015) Behavior of Leaf Meristems and Their Modification. Front. Plant Sci. 6:1060. doi: 10.3389/fpls.2015.01060
Received: 21 May 2015; Accepted: 13 November 2015;
Published: 01 December 2015.
Edited by:
Simon Gilroy, University of Wisconsin - Madison, USAReviewed by:
Javier Palatnik, Instituto de Biologia Molecular y Celular de Rosario, ArgentinaMichael Lenhard, Universität Potsdam, Germany
Copyright © 2015 Ichihashi and Tsukaya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasunori Ichihashi, eWFzdW5vcmkuaWNoaWhhc2hpQHJpa2VuLmpw
 Yasunori Ichihashi
Yasunori Ichihashi Hirokazu Tsukaya
Hirokazu Tsukaya