- 1Animal, Plant and Soil Sciences Department, AgriBio, AgriBiosciences Research Centre, La Trobe University, Melbourne, VIC, Australia
- 2School of BioSciences, University of Melbourne, Parkville, VIC, Australia
- 3Plant Sciences Division, Research School of Biology, The Australian National University, Canberra, ACT, Australia
- 4The New Zealand Institute for Plant and Food Research Limited, Auckland, New Zealand
- 5Life Sciences Computation Centre, Victorian Life Sciences Computation Initiative, Melbourne, VIC, Australia
Venturia inaequalis and V. pirina are Dothideomycete fungi that cause apple scab and pear scab disease, respectively. Whole genome sequencing of V. inaequalis and V. pirina isolates has revealed predicted proteins with sequence similarity to AvrLm6, a Leptosphaeria maculans effector that triggers a resistance response in Brassica napus and B. juncea carrying the resistance gene, Rlm6. AvrLm6-like genes are present as large families (>15 members) in all sequenced strains of V. inaequalis and V. pirina, while in L. maculans, only AvrLm6 and a single paralog have been identified. The Venturia AvrLm6-like genes are located in gene-poor regions of the genomes, and mostly in close proximity to transposable elements, which may explain the expansion of these gene families. An AvrLm6-like gene from V. inaequalis with the highest sequence identity to AvrLm6 was unable to trigger a resistance response in Rlm6-carrying B. juncea. RNA-seq and qRT-PCR gene expression analyses, of in planta- and in vitro-grown V. inaequalis, has revealed that many of the AvrLm6-like genes are expressed during infection. An AvrLm6 homolog from V. inaequalis that is up-regulated during infection was shown (using an eYFP-fusion protein construct) to be localized to the sub-cuticular stroma during biotrophic infection of apple hypocotyls.
Introduction
Effectors are generally small proteins secreted by plant pathogens that can interact with the host during infection. They may serve to facilitate infection, often by manipulating the host, or may be recognized by host receptor proteins, either directly or indirectly, leading to a resistance response (Chisholm et al., 2006; Jones and Dangl, 2006; Dodds and Rathjen, 2010). The outcome of the plant–pathogen interaction depends on the evolutionary context; the pathogen is under selection pressure to evade detection by the host, while the host must maintain the ability to detect the pathogen. These gene-for-gene relationships are at play in the host specificity of Venturia inaequalis races to different Malus cultivars, where so far 17 gene-for-gene relationships have been described (Bus et al., 2011). While two resistance (R) genes governing race-cultivar specificity have been identified in Malus (Belfanti et al., 2004; Schouten et al., 2013) none of the cognate fungal avirulence genes have been isolated. Genomic (BioProject ID PRJNA261633 for V. inaequalis and JEMP01000000 for the V. pirina genome) and transcriptomic (Thakur et al., 2013) sequences are now available for several isolates of V. inaequalis and a V. pirina isolate, providing a promising new avenue of bioinformatic discovery to identify effectors; however, identifying specific race-governing effectors of V. pirina and V. inaequalis among these large predicted secretomes remains a challenge. Most effectors identified to date, in other fungi, are small, secreted proteins (SSPs) with high cysteine content (Stergiopoulos and de Wit, 2009) and in silico prediction of effector candidates has been informed by these attributes (Saunders et al., 2012; Guyon et al., 2014; Sperschneider et al., 2015). Effectors are often race- or species-specific (Stergiopoulos and de Wit, 2009); however, it has been reported that some effectors can be more widely conserved, even across genera as shown with the Cladosporium fulvum effector Avr4 and its homolog in Mycosphaerella fijiensis, both of which induce R-gene (Cf-4)-mediated resistance responses on tomato (Stergiopoulos et al., 2010). More recently, an Avr4 homolog from Dothistroma septosporum was also shown to trigger a hypersensitive response (HR) when transiently expressed in Cf-4 transgenic Nicotiana tabacum (Mesarich et al., 2015).
AvrLm6 is a secreted, proteinaceous effector of Leptosphaeria maculans, the causal agent of canola black leg disease, that has been shown to trigger a resistance response on host plants carrying the Rlm6 resistance gene (Fudal et al., 2007). AvrLm6 was first identified genetically, with the phenotypic observation of segregation of differential resistance responses of Brassica napus and B. juncea cultivars to the progeny of B. juncea-virulent and -avirulent L. maculans isolates (Balesdent et al., 2002). Subsequently, the gene encoding AvrLm6 was identified using map based cloning. Southern blot analyses led to the conclusion that AvrLm6 homologs were restricted to L. maculans (Fudal et al., 2007). Since then, AvrLm6 homologs have been detected in the whole genome sequences of additional Leptosphaeria species. It is likely that these were not identified in the Southern blot analysis by Fudal et al. (2007) due to the low nucleotide sequence identity. Homologs are also found in more distantly related fungi, such as Colletotrichum and Fusarium species (Grandaubert et al., 2014), as well as in Venturia species (Bowen et al., 2011). Nothing is known about the function of AvrLm6 or orthologous proteins in any of these fungi. However, it has been demonstrated that two other L. maculans avirulence genes; AvrLm4-7 (Huang et al., 2006) and AvrLm1 (Huang et al., 2010) contribute to fungal fitness. Expression of Avrlm6 is highly up-regulated in L. maculans during primary infection of B. napus, compared with in vitro growth (Fudal et al., 2007; Van de Wouw et al., 2010). Expression levels were recorded to be highest at 7 days post inoculation on canola and then, after falling slightly, are maintained at high levels during stem necrosis. Similar expression patterns are seen in other L. maculans effectors AvrLm1, AvrLm4-7, and AvrLm11 (Gout et al., 2006; Parlange et al., 2009; Balesdent et al., 2013). Avrlm6 has been observed at high frequency among L. maculans populations in Europe, in the absence of Rlm6 resistance (Balesdent et al., 2006; Stachowiak et al., 2006), but when selection pressure from Rlm6 resistance has been applied in experimental trials, the resistance was overcome after just 3 years (Brun et al., 2000). AvrLm1 (Gout et al., 2007) and AvrLm4-7 (Daverdin et al., 2012) alleles have also been reported at high levels in the absence of selection pressure from the cognate resistance genes in the field, but, as seen with AvrLm6, virulent alleles increase rapidly once resistance genes are introduced, despite the fitness costs to the pathogens (Huang et al., 2006, 2010).
A diverse array of mechanisms have been shown to generate new virulent AvrLm6 alleles; including deletion, point mutation, and repeat induced point mutation (RIP; Fudal et al., 2009; Van de Wouw et al., 2010). RIP is a type of mutation, that only occurs in fungi, during sexual crossing and alters duplicated DNA sequences and produces CpA to TpA and TpG to TpA mutations (Cambareri et al., 1989). RIP can also impact sequences neighboring duplicated DNA sequences, such is the case with AvrLm6 in L. maculans where RIP has “leaked” from nearby repetitive DNA sequences into the Avrlm6 coding sequence (Van de Wouw et al., 2010; Rouxel et al., 2011). AvrLm6 inhabits a gene poor, AT-rich region of the L. maculans genome and is surrounded by large numbers of repetitive sequences comprising predominantly long terminal repeat (LTR) retrotransposons (Pholy, Olly, Polly, and Rolly), which appear to have been RIP-affected (Gout et al., 2006; Fudal et al., 2007).
Research into the AvrLm6-like genes from V. inaequalis (ALVis) and V. pirina (ALVps) was conducted to determine a possible role for these SSPs in scab disease development. The predicted secretomes and WGS of a single V. pirina and 4 V. inaequalis strains were mined for sequences with similarity to AvrLm6 and their surrounding genomic contexts were examined to further understand the evolution and nature of the gene family expansion. The large size of the gene families of AvrLm6-like genes and probable functional redundancy hindered the possibility of gene disruption or silencing to determine function. Timing and location of AvrLm6-like gene expression was investigated to further understand the role of these proteins in Venturia. An L. maculans isolate lacking the avirulent AvrLm6 allele was also transformed with a V. inaequalis ALVi gene to determine whether the phenotype of avirulence on the Rlm6-containing B. napus could be restored (i.e., by complementation of the heterologous gene) indicating conservation of function.
Materials And Methods
Venturia Isolates and Genomic Resources used in This Work
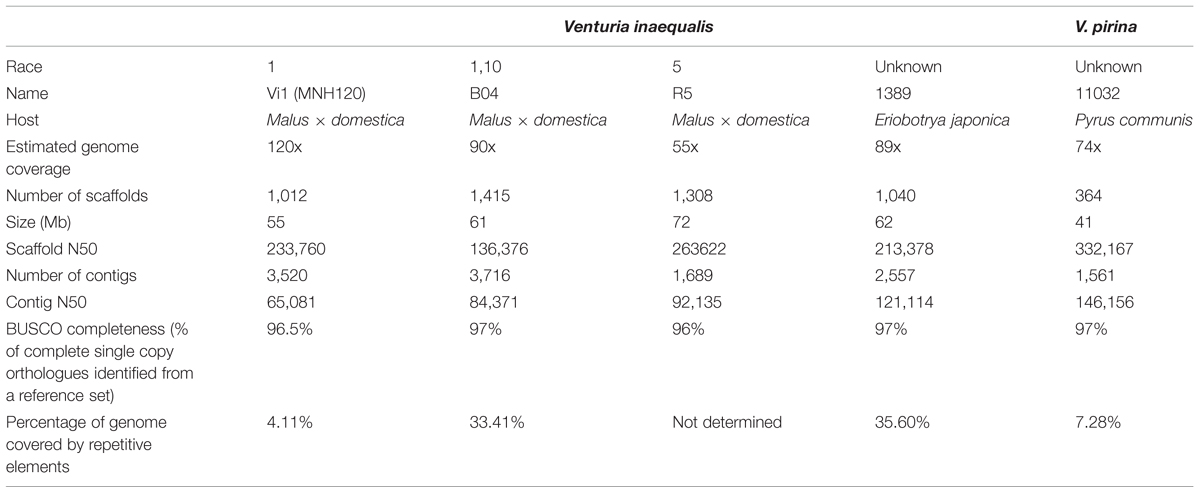
The genomic resources used in this study are detailed below in Table 1. The V. inaequalis Vi1 genome was used to investigate the genomic environment relating to the ALVi gene family as it is the genome that is annotated the most completely, and we have transcriptome data for various time points and growth conditions [BioProject (ID PRJNA261633): in vitro: SRR1586226, 2 dpi in planta: SRR1586224, 7 dpi in planta: SRR1586223J] which has been used to inform gene prediction.
The V. inaequalis Vi1 and the V. pirina genomes (Cooke et al., 2014) are publicly available via the MycoCosm genome portal at JGI1,2 . All V. inaequalis and V. pirina isolates used in this study have been reported previously (Stehmann et al., 2001; Le Cam et al., 2002; Win et al., 2003; Bowen et al., 2009; Broggini et al., 2011; Cooke et al., 2014; Caffier et al., 2015). The rest of the genomes were only used to extract the ALVi predicted protein sequences, these are included in the Supplementary Material.
Identification of AvrLm6 Homologs in Public Sequence Databases
The NCBI (National Centre for Biotechnology Information3) non-redundant (nr) public sequence database was queried with the AvrLm6 (GenBank: CAJ90695) predicted protein sequence using PSI-Blast with default settings and an e-value cut-off of 1E-5. Predicted proteins returned from the PSI-Blast search were collected and used in blastp analysis of protein databases at JGI (Joint Genome Institute4) and the Fungal Genomes proteins available at the Broad Institute5 also using 1E-5 e-value cut-off. In order to identify homologs which had not been predicted, tblastn searches were performed, with the same stringency as above.
Identifying ALVp and ALVi Gene Families
Two criteria were used to decide which genes to include in the ALVi and ALVp gene families. Firstly, standalone PSI-Blast (blast + 2.2.29) was used with AvrLm6 (GenBank: CAJ90695) protein sequence as the query with an e-value cut-off of 1E-5 against the protein catalogs of each Venturia isolate. The predicted proteins identified that met this first criterion were then used in back-blast searches to screen the NCBI (National Centre for Biotechnology Information6) nr public sequence database. If the e-value relating to the most similar protein was less than, or equal to 1E-2 to AvrLm6 or one of its homologs from another fungal species (defined above) the sequence was considered an ALVi or ALVp. All ALVp and ALVi amino acid sequences are included in the Supplementary Material (Data Sheet 1).
Phylogenetic Analysis
Predicted amino acid sequences of all ALVi and ALVp genes from V. inaequalis isolate Vi1 and V. pirina isolate 11032 were aligned using MUSCLE multiple sequence alignment implemented in the MEGA6 program (Tamura et al., 2013) and a maximum likelihood tree was derived from the alignment using the WAG substitution model tested with bootstrapping (500). Another tree was constructed in an identical manner with the inclusion of amino acid sequences from additional species including; AvrLm6 (GenBank: CAJ90695) and other homologs from; L. maculans (GenBank:XP_003843096.1), Colletotrichum gloeosporioides (GenBank:XP_007281214.1 and XP_007275717.1), C. higginsianum (GenBank:CCF39162.1), C. obliculare (GenBank:ENH83850.1 and ENH87246.1) and Fusarium oxysporum (GenBank:EXK76127.1) using a WAG substitution model and 1000 bootstrap tests. The trees were drawn with Figtree V1.4.2.
Quantification of Gene Expression of Selected ALVis in V. inaequalis Isolate Vi1
Expression analysis of eight ALVi genes from Vi1 (ALVi_Vi1_4, ALVi_Vi1_5, ALVi_Vi1_7, ALVi_Vi1_9, ALVi_Vi1_14, ALVi_Vi1_15, ALVi_Vi1_17 and ALVi_Vi1_22) was carried out by qRT-PCR out on a LightCycler® 480 instrument (Roche) using LightCycler® 480 SYBR Green I Master reagents (Roche). Each 10 μl reaction contained 5 μl of mastermix (2X) 0.5 μl of each primer (at 5 μM) and 4 μl of cDNA (see below). The PCR cycling conditions were as follows; Initial denaturation for 5 min 95°C, followed by 45 cycles of 95°C 10 s, 60°C 10 s, and 72°C 8 s. Primers used in this experiment are detailed in the Supplementary Table S2. β-tubulin and the 60 s ribosomal L12 gene were used as reference genes as previously described (Kucheryava et al., 2008). These ALVis were chosen for qRT-PCR analysis based on preliminary RNA-seq data which showed they were up-regulated during infection when compared with growth in vitro. Three biological replicates were included for each time point and each biological replicate was triplicated for technical replicates.
RNA used in qRT-PCR experiments was extracted from Vi1 cultures growing on cellophane amended potato dextrose agar (PDA) plates 10 days post inoculation to test in vitro gene expression and from detached leaf infections at 3, 7, and 14 days post inoculation (dpi). These infections were performed as previously described (Win et al., 2003). RNA was extracted from all samples following a published protocol (Chang et al., 1993).
To grow apple leaves for the detached leaf assays, apple seeds were sourced from open pollinated Malus × domestica ‘Royal Gala.’ Surface-sterilized seeds were stratified at 4°C in water-saturated vermiculite for 6–12 weeks to enable germination. Germinated seeds were planted in compost at 21°C with a 12 h light period/day under 4C W SHP-TS lights (Sylvania, Danvers, MA, USA) inoculations were performed between 4 and 6 weeks after planting in compost.
Localisation of an ALVi Protein during Infection
The ALVi_Vi1_5 gene from V. inaequalis Vi1 was chosen for localisation analysis using fluorescent protein (YFP) tagging. The enhanced yellow fluorescent protein (eYFP) gene had been adapted for V. inaequalis codon usage. The ALVi_Vi1_5 gene was chosen as preliminary RNA-seq data and qRT-PCR analysis showed that its expression was up-regulated during infection, compared with growth in vitro. The PJK4 plasmid containing a fusion of ALVi_Vi1_5:eYFP (PJK4: ALVi_Vi1_5:eYFP) was used as a binary vector for Agrobacterium-mediated transformation of V. inaequalis isolate Vi1 (Fitzgerald et al., 2003). The expression cassette was constructed with overlap extension PCR (Heckman and Pease, 2007), joining the ALVi_Vi1_5 predicted promoter (1003 bp upstream of start codon) and coding sequence in frame with the eYFP gene and the predicted ALVi_Vi1_5 terminator (1001 bp downstream of the stop codon), and cloned into PJK4 at the Spe1 and Bgl2 cloning sites. The predicted promoter and terminator contained no predicted open reading frames. Primers used in the construction are detailed in the SupplementaryTable S3. The sequence of the insert was validated by Sanger sequencing (Australian Genome Research Facility).
To observe localization of the fluorescent fusion protein in planta, apple hypocotyls were inoculated with 5 μl droplets of spore suspension (1 × 105 spores/ml) prepared from PJK4: ALVi_Vi1_5:eYFP transformants or the wild type, untransformed Vi1 isolate. Infected hypocotyls were incubated at 20°C in darkness for up to 14 days. PDA plates amended with cellophane membranes were also inoculated with the spore suspensions and stored under the same conditions. Infections were observed at 2, 7, and 14 dpi using a Leica TCS SP2 Confocal Microscope (Leica Wetzlar Germany) using the 100× magnification, oil immersion lens. Hypocotyls were grown from Malus × domestica ‘Royal Gala’ seed as described above for detached leaf assay.
Complementation Assays using ALVi-Vi1_8 Transformed into an L. maculans Virulent Isolate
A complementation assay was carried out to see if L. maculans expressing ALVi_Vi1_8 driven by the AvrLm6 promoter could confer avirulence toward the Rlm6-containing B. napus cultivar ‘Aurea’ which is resistant to L. maculans isolates expressing the AvrLm6 gene. ALVi_Vi1_8 was chosen because it had the most similar predicted sequence to AvrLm6 based on the blastp analysis (32% identity). The complementation cassette contained ALVi_Vi1_8 (477 bp) flanked by regions upstream (955 bp) and downstream (1255 bp) of AvrLm6 amplified from L. maculans isolate v23.1.3. This fragment was constructed using overlap extension PCR (Heckman and Pease, 2007) and cloned into the binary vector pZP-Nat containing the nourseothricin acetyltransferase gene (Elliott and Howlett, 2006). Agrobacterium-mediated transformation (Gardiner and Howlett, 2004) was used to transform L. maculans isolate M1, which lacks the AvrLm6 gene, with the construct. Pathogenicity testing were performed on cotyledons of B. napus ‘Westar’(lacking Rlm6) and B. juncea ‘Aurea’ (carrying Rlm6) as previously described (Van de Wouw et al., 2014). Expression of the transgene during infection was confirmed by RT-PCR. Primers used in vector construction and RT-PCR are listed in the Supplementary Table S4.
Genomic Context, RIP, and Repeat Regions
The prediction suite REPET 2.2 (Flutre et al., 2011) was used for the detection and annotation of transposable elements (TEs). RipCrawl6 was used to predict areas of the genome that had undergone RIP based on composite RIP indices (CRI) with the same restraints that have previously been reported (de Wit et al., 2012). Bedtools (Quinlan and Hall, 2010) was used to calculate distances from ALVi genes to features of interest. The above analysis was also done on a set of 439 core eukaryotic reference genes identified using the NCBI eukaryotic clusters of orthologous groups (KOGs). The KOG numbers and the corresponding gene names in V. inaequalis isolate Vi1 are listed in the Supplementary Table S5.
Results
AvrLm6 Homologs are Found Across Multiple Fungal Genera and Form Expanded Gene Families in V. pirina and V. inaequalis
In addition to the AvrLm6 homologs that were previously reported in the genomes of C. higginsianum, C. gloeosporioides, F. oxysporum, and L. biglobosa (Grandaubert et al., 2014), we report additional AvrLm6 homologs in C. orbiculare, C. fioriniae, and F. oxysporum f. sp. raphani (Figure 1). We observed differences in the copy number of homologs identified in each genome which varied, even within species (Figure 1). Venturia genomes had up to 30 copies, whereas no other genera had more than 3. In the case of F. oxysporum and Leptosphaeria species there are also isolates which appear to have no homologs. When all ALVi and ALVp predicted protein sequences were used as blast queries of the NCBI nr database, the levels of sequence identity to AvrLm6 homologs ranged from 24 to 41%. Only two sequences returned the L. maculans AvrLm6 protein as the most similar (ALVp_11032_13 from V. pirina isolate 11032 and ALVi_1389_1 from Vi1389). All other V. pirina and V. inaequalis homologs were more similar to AvrLm6 homologs from either Fusarium or Colletotrichum species than to AvrLm6, and none had similarity to any other L. maculans proteins other than to AvrLm6 or related homologs. The multiple sequence alignment revealed four cysteine residues that are conserved in all homologs (Figure 2), despite low sequence similarity overall. ALVis and ALVps also have different gene structure when compared to the homologs found in all the other species, which are generally well conserved with four exons in all homologs except EGU73747.1 from F. oxysporum Fo5176 and XP_003843096.1 from L. maculans which both have seven exons (Figure 1). The gene structures of ALVps and ALVis vary, in that they either have no introns, one intron in the five prime un-translated region (5′ UTR), or either one or two introns in the coding sequence of the gene. The most common structure found in the ALVis is a single intron in the 5′ UTR present in 19 of the 24 ALVis identified in Vi1. We were able to confirm these predicted gene structures in 11 of the 24 genes with reference to mapped RNA-seq reads. All of the AvrLm6 homologs identified in public databases, (with the exception of F. oxysporum EGU73747.1) and all of the ALVi and ALVp members we have described here, are predicted to contain a signal peptide sequence at their N terminus and so are expected to be secreted by the classical secretory pathway.
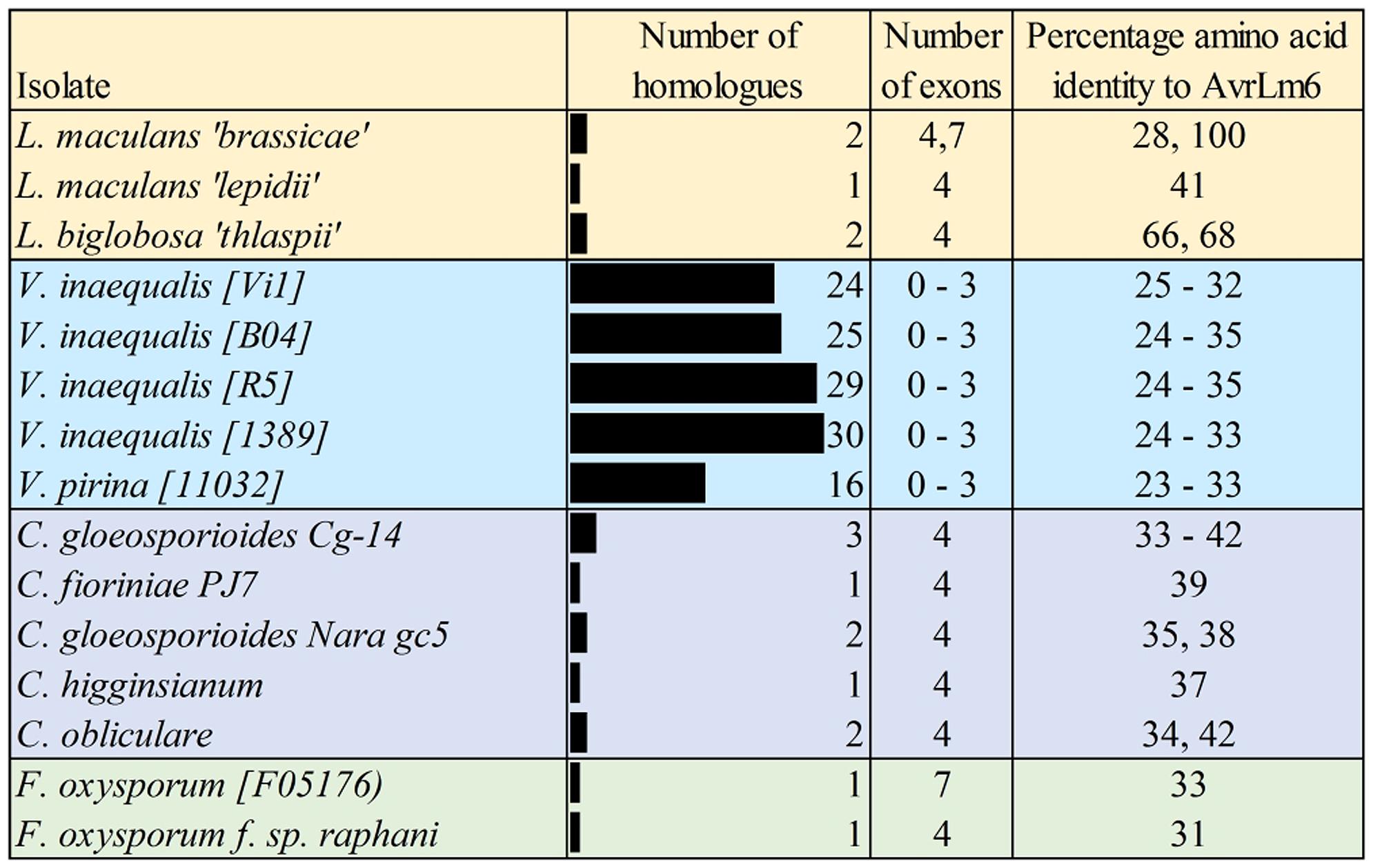
FIGURE 1. Summary of the AvrLm6 homologs identified in public and private sequence databases. The number of exons predicted for each gene are shown as well as the percentage amino acid identity to the AvrLm6 predicted protein (CAJ90695) from blastp searches. Accession numbers and further details of homologs are detailed in the Supplementary Table S1.
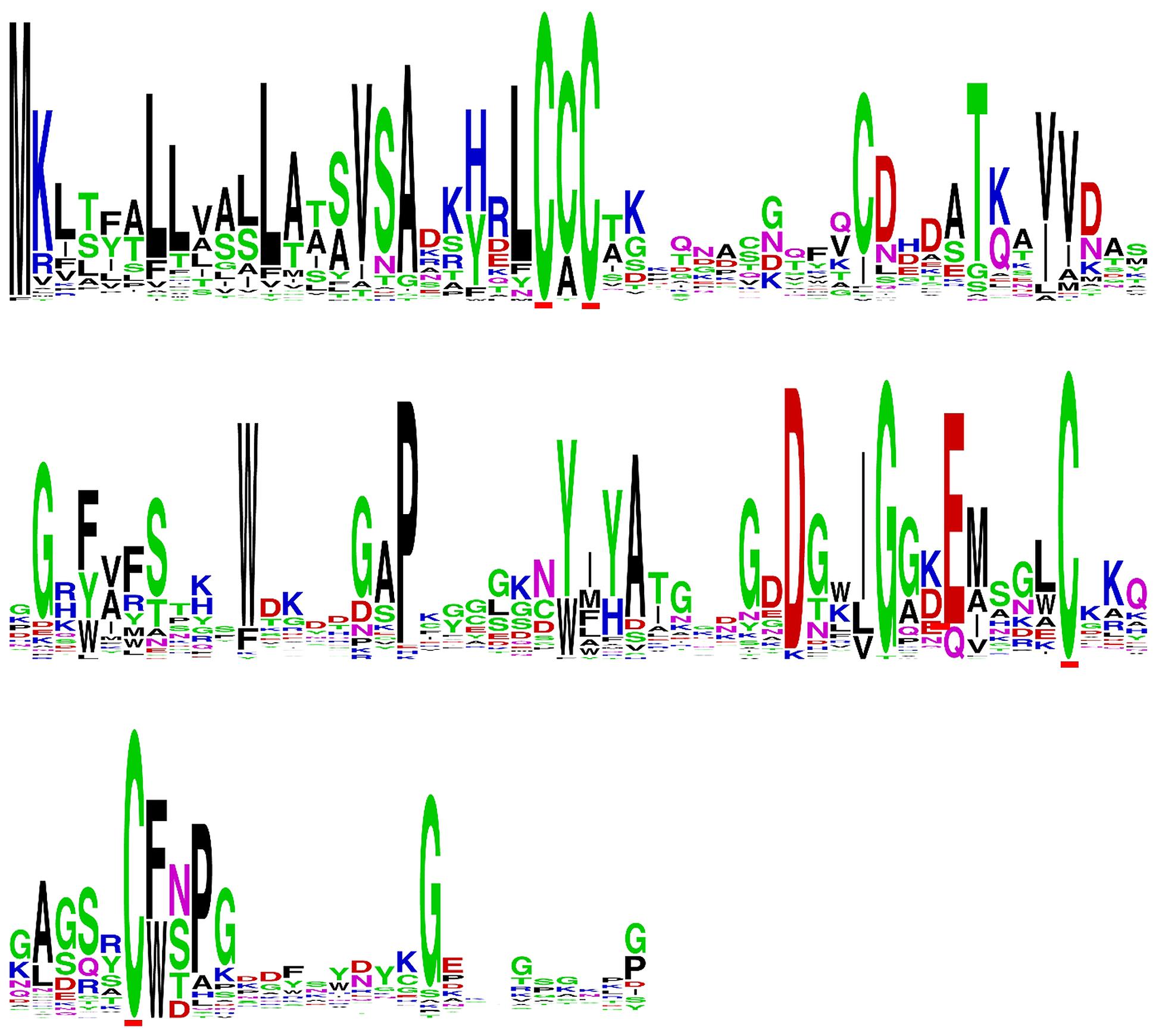
FIGURE 2. Sequence logo of a multiple sequence alignment of predicted protein sequences including AvrLm6 and a paralogue from Leptosphaeria maculans (GenBank:CAJ90695 and XP_003843096.1) homologs from Colletotrichum gloeosporioides (GenBank:XP_007281214 and XP_007275717.1), Fusarium oxysporum (GenBank:EXK76127.1), C. higginsianum (GenBank:CCF39162.1), C. fiorinae (GenBank:XP_007599415.1) as well as ALVI_1 to ALVi_24 from Venturia inaequalis Vi1 and ALVp_1 to ALVp_16 from V. pirina isolate 11032.
Expansion of ALVp and ALVi Gene Families
The maximum likelihood tree derived from the multiple sequence alignment of the ALVi and ALVp amino acid sequences (Figure 3) formed four strongly supported clades with 95, 93, 100, and 94% support that included only ALVp sequences, and one large clade, with 77% support, containing only ALVi sequences. Mixed clades containing sequences from both species did not have strong statistical support (Figure 3). When AvrLm6 from L. maculans and orthologues from Fusarium and Colletotrichum species are included in the tree, there were many clades with low support (Figure 4) making it difficult to resolve the evolutionary relationship between ALVi and ALVp families with orthologues in those species, however, a two well supported clades of ALVp and ALVi predicted proteins can be resolved supporting the recent expansion of the gene family in Venturia sp. AvrLm6 clustered in a strongly supported clade with the homologs from L. biglobosa, but there was not strong statistical support for clustering between these Leptosphaeria predicted proteins and those of any other species.
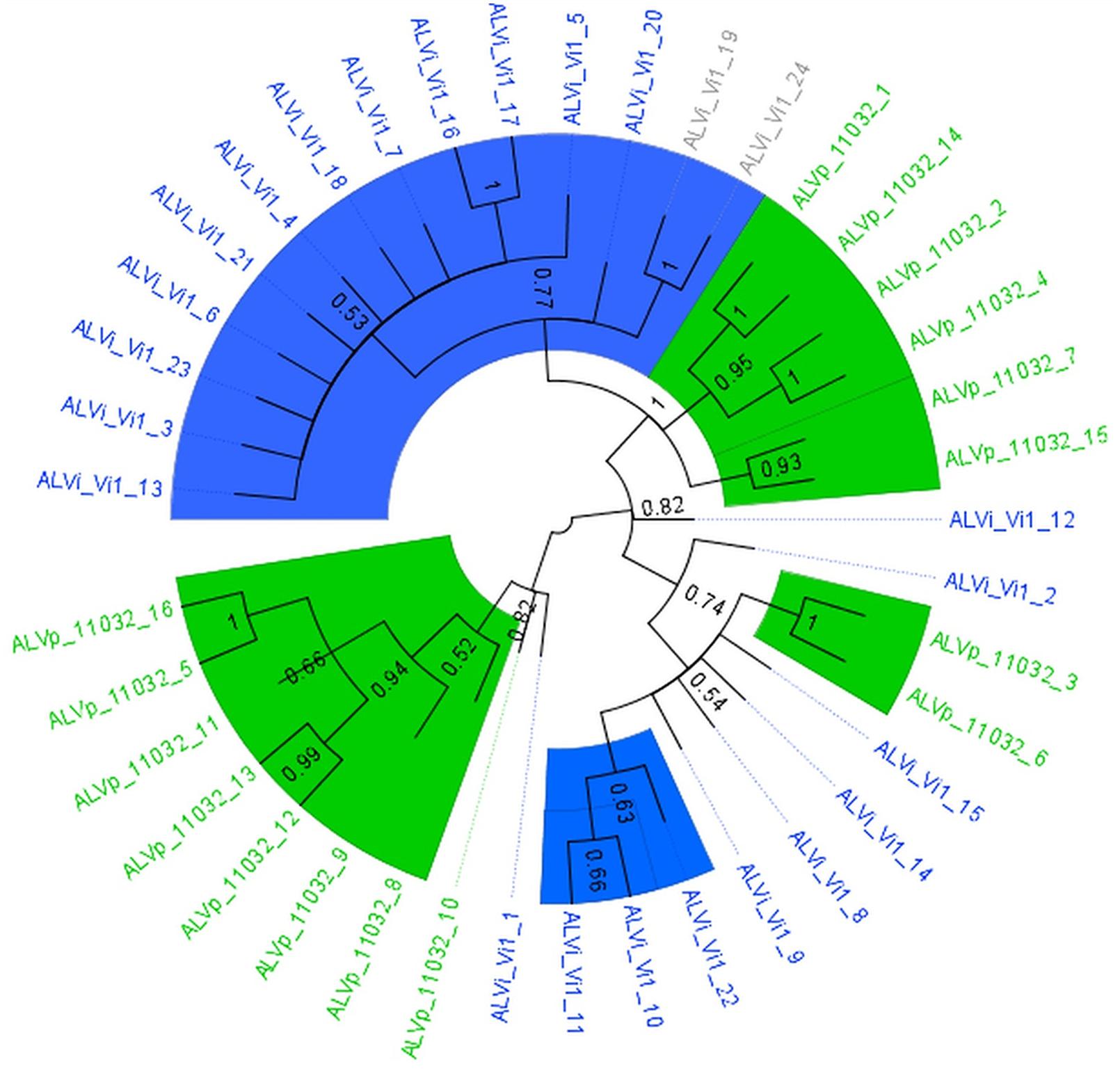
FIGURE 3. Maximum likelihood tree calculated on multiple sequence alignment of ALVi and ALVp predicted protein sequences from V. inaequalis isolate Vi1 and V. pirina isolate 11032 using a WAG substitution model and 500 bootstrap tests. Species-specific clades with over 75% bootstrap support are highlighted with colored wedges. Sequence names in green are from V. pirina isolate 11032 and blue from V. inaequalis Vi1. Gray sequence labels represent Vi1 genes for which we have no evidence for gene expression from in planta or in vitro growth.
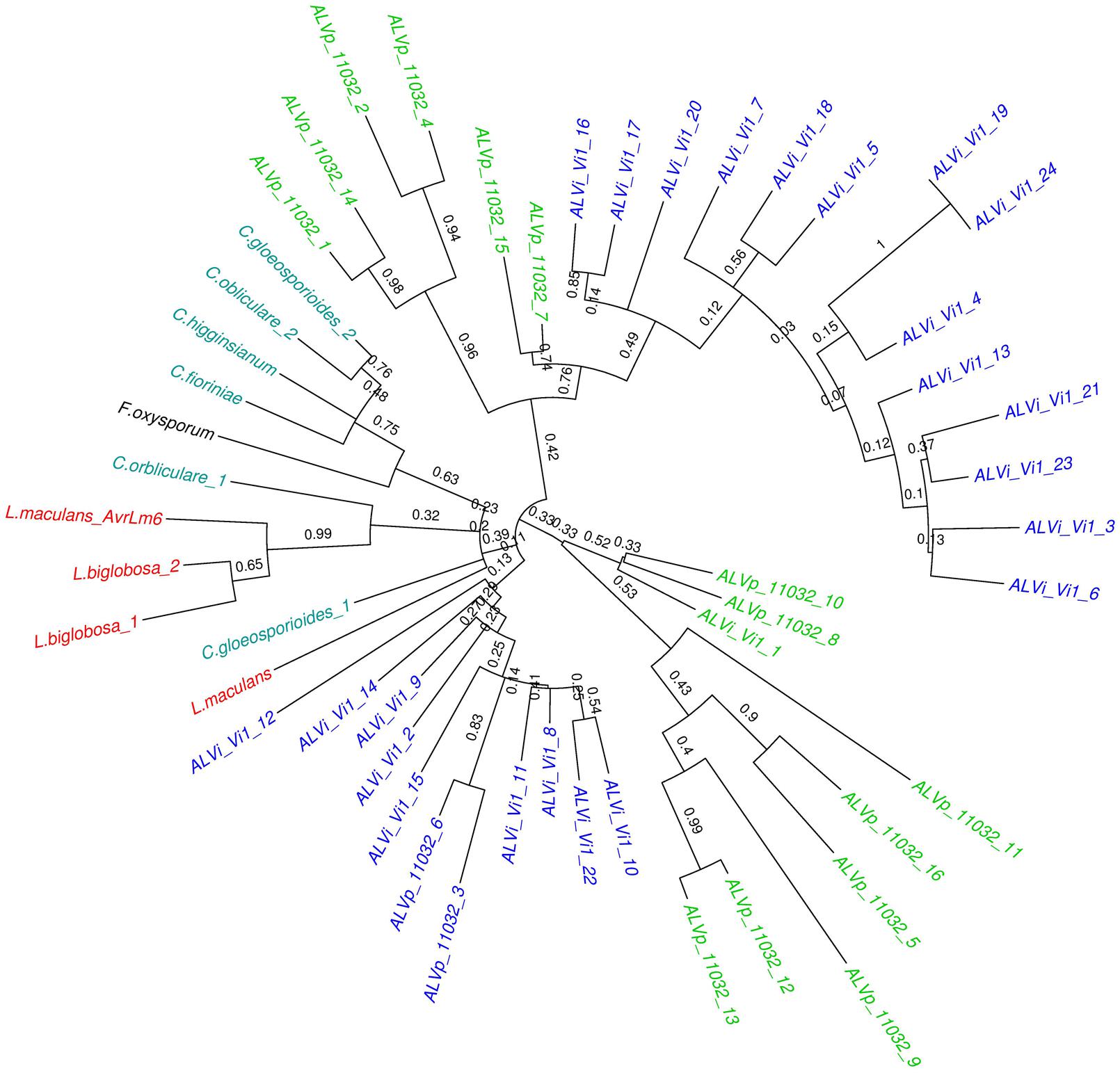
FIGURE 4. Maximum likelihood tree calculated on multiple sequence alignment of all ALVi and ALVp predicted protein sequences from V. inaequalis isolate Vi1 and V. pirina isolate 11032 as well as AvrLm6 (GenBank: CAJ90695) and a paralogue from L. maculans and homologs from C. gloeosporioides (GenBank:XP_007281214.1 and XP_007275717.1), C. higginsianum (CCF39162.1), C. orbliculare (ENH83850.1) and (ENH87246.1) and F. oxysporum (EXK76127.1) using a WAG substitution model and 1000 bootstrap tests. Sequence names in green are from V. pirina isolate 11032 and blue from V. inaequalis Vi1, red are from Leptosphaeria species, teal are Colletotrichum species, and black is Fusarium oxysporum.
ALVis are Found in Gene Poor Regions Associated with Transposable Elements
ALVi genes in the Vi1 genome are found in gene poor regions (Figure 5) when compared with a reference set of 439 core eukaryotic genes (Supplementary Table S5). The mean distance to the nearest gene was 1,370 bp for the ALVi-coding loci and 484 bp for the core eukaryotic genes, the difference between the means was found to be statistically significant by a Student’s t-test (p-value < 0.001). ALVi sequences are found in close proximity to TEs predicted with the REPET 2.2 pipeline. The mean distance to the nearest TE from an ALVi gene in isolate Vi1 (203 bp) was significantly less (Student’s t-test p-value < 0.00001), than that of the reference set of core genes (5,882 bp). The most common class of TE associated with the ALVi genes was the large retrotransposon derivative (LARD) class, 10 of these occurred within 260 bp of an ALVi gene and of those, six overlapped ALVi-coding loci. Terminal-repeat retrotransposons in miniature (TRIMs) were the second most common element identified at ALVi loci, overlapping with eight ALVi-coding sequences. Using the CRI index criteria that has been previously defined (de Wit et al., 2012) we could not find evidence of RIP in any of the ALVi coding sequences in isolate Vi1. However, evidence of RIP was found throughout the genome, with 72% of the TEs predicted to be RIP-affected. None of the TEs overlapping the ALVi sequences were shown to be RIP affected, but seven ALVis loci had evidence of RIP in the nearest neighboring repetitive elements. These elements included two predicted TRIMs, one LARD, as well as fragments of; long terminal repeats (x2) a terminal inverted repeat and a long interspersed nuclear element.
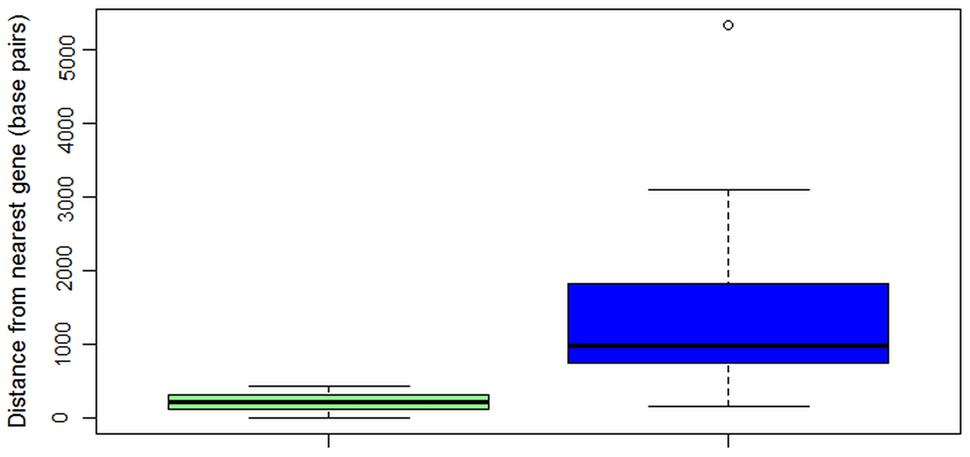
FIGURE 5. Box and whisker plot showing distances in base pairs from nearest gene to core eukaryotic genes (n = 439, green), or to ALVi genes (n = 24, blue) in V. inaequalis Vi1.
ALVi_Vi1_5 Localizes to the Stroma during Infection
Fluorescent signal from the ALVi_Vi1_5 fusion protein was visible at 7 and 14 dpi in infected apple hypocotyls (Figure 6) but not visible at the earlier infection time points or at any time-points in vitro. The signal appeared to localize to the periphery of the sub-cuticular stroma and was not visible in surface hyphae, conidiophores or spores. The similar levels of fluorescence at seven and 14 dpi reflected the qRT-PCR data (Supplementary Figure S2) which showed that expression was similar at these time points and higher than that observed at 3 dpi.
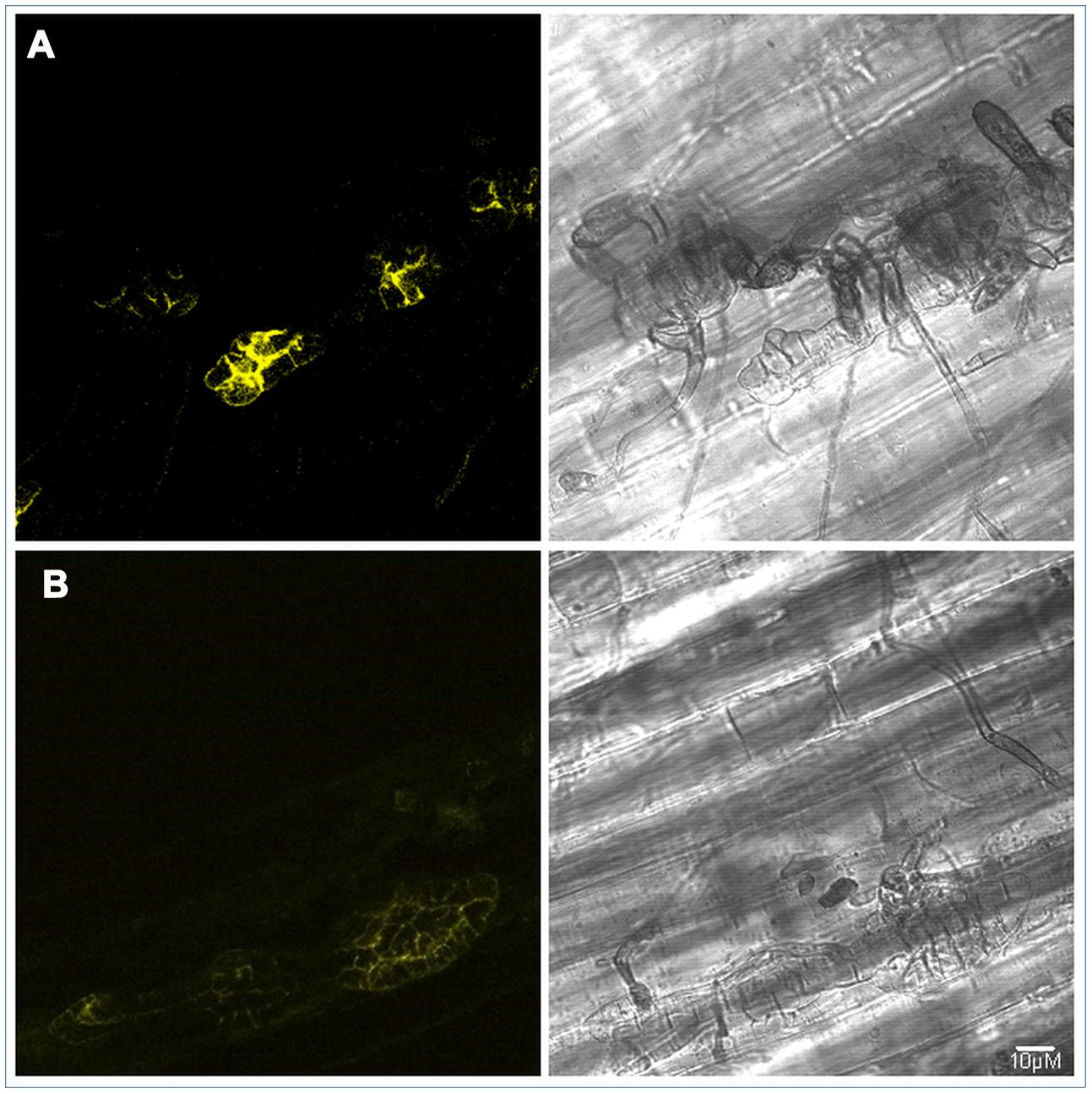
FIGURE 6. PJK4:ALVi_Vi1_5: eYFP transformed isolate V. inaequalis Vi1 growing on apple hypocotyl viewed with bright field microscopy (right panels) and confocal fluorescent (left panels) 7 days post inoculation (A) and 14 days post inoculation (B). YFP fluorescence observed in multicellular, sub-cuticular stroma only. Fluorescent images are z stacks.
ALVi_Vi_1_8 does not Trigger Rlm6-mediated Resistance in Canola
Expression of ALVi_Vi1_8 was confirmed by RT-PCR in the L. maculans (Isolate M1) transformants containing the complementation vector (Supplementary Figure S2), but none of these transformants triggered a resistance response in B. juncea ‘Aurea,’ expressing the Rlm6 resistance gene (Supplementary Figure S3). The expected resistance response was observed when B. juncea ‘Aurea’ was infected with L. maculans (Isolate M1) transformed with the AvrLm6 avirulence gene. The pathogenicity of the ALVi_Vi1_8 transformed isolates on the susceptible B. napus ‘Westar’ cultivar was indistinguishable from the wild type L. maculans expressing the native AvrLm6 gene and the wild type L. maculans M1 isolate.
Discussion
Fungal effector proteins have long been characterized by their lineage-specific distribution, and were generally thought to be conserved at the species or race level. A rapid rise in the availability of plant pathogen whole genome sequences, especially for those classified within the Dothideomycetes and the Sordariomycetes, has enabled this dogma to be tested by sequence similarity searches. In this way, new homologs of the L. maculans effector, AvrLm6, have been identified. The taxonomic distribution of AvrLm6 orthologues is interesting, because while they are found across two different classes of fungi, the Dothideomycetes and the Sordariomycetes, they have apparently only been conserved in a few species within these classes. Furthermore, AvrLm6 from L. maculans has higher sequence similarity to orthologues in the Sordariomycetes class than to orthologues from the more closely related Venturia species (Supplementary Figure S1). One possible explanation of the unusual taxonomic distribution of these genes could be horizontal gene transfer (HGT). There are now many examples of HGT in fungal and oomycete plant pathogens (Soanes and Richards, 2014). The most compelling evidence of HGT is ToxA, found in two wheat pathogens. The ToxA gene in Parastagonospora nodorum appears to have been horizontally transferred from Pyrenophora tritici-repentis. The ToxA genes in both species had very high nucleotide sequence identity (99.7%) which did not concur with the phylogenetic relationship inferred when comparing the ITS regions and glyceraldehyde-3-phosphate genes (83 and 80% respectively; Friesen et al., 2006). In the case of AvrLm6 though, the homologs have a high level of sequence divergence and while AvrLm6 did cluster with the Colletotrichum homologs in the phylogenetic tree, this relationship was not supported with bootstrapping. Even within the same species, the presence of these genes is not conserved, for example, we could only identify homologs in two isolates of F. oxysporum with blast searches, despite the 25 searchable genomes in the NCBI database. This finding is not simply due to poor gene models in the WGS of fungi as six frame similarity searches also did not reveal orthologues. The AvrLm6 allele is also highly polymorphic at the population level and can also be absent in some L. maculans isolates (Van de Wouw et al., 2010). The deletion of avirulence effector loci is one way that pathogens avoid recognition by resistant plants. The function of AvrLm6 is unknown, however, it is not essential for pathogenicity in L. maculans. This may be why it is so poorly conserved in the Dothideomycetes in general. The expansion of the AvrLm6-like genes in Venturia sp. is clearly against the trend in this group.
In V. inaequalis and V. pirina, copy number of AvrLm6-like genes varies between isolates and species. The ALVi and ALVp gene family expansions appear to have occurred independently after speciation. This can be seen on the gene tree (Figure 3), where strongly supported clades are composed of clusters of paralogues rather than orthologues. V. inaequalis and V. pirina species are restricted to and separated spatially on different host ranges, but otherwise have very similar lifestyles and biology, hence, differences in the evolution of these gene families may reflect adaptation to a new host.
The mechanism/s responsible for the expansion of the ALVp and ALVi gene families remains unclear. However, the association between ALVis and TEs, predominantly LARDs and TRIMs, suggests that the multiple gene duplications could have been mediated by these elements. Indeed the expansion of the AVRk1 effector gene family in Blumeria graminis has been hypothesized to be driven by the association with LINE1 retrotransposons (Sacristán et al., 2009).
Large retrotransposon derivatives are non-autonomous long terminal repeat (LTR) retrotransposons which have been described in Triticeae (Kalendar et al., 2004), rice (Nagaki et al., 2005; Vitte et al., 2007) and fungi (Labbé et al., 2012; Pereira et al., 2015); they consist of LTRs flanking a large non-coding internal domain, and lack the coding domains found in other LTR retrotransposons, which are responsible for self-replication (Kalendar et al., 2004). TRIMs, like LARDs, are non-autonomous retrotransposons, but differ structurally from LARDs, in that they have much shorter internal domains and terminal direct repeats (Witte et al., 2001). TRIMs have been observed within coding and non-coding regions of plant (Witte et al., 2001; Sampath and Yang, 2014) and animal genes (Zhou and Cahan, 2012). It has been observed that TRIMs can be involved in the transduction of host genes, a situation where a host gene or part of host gene becomes part of the replicating transposon, and can lead to gene duplication (Witte et al., 2001). A TRIM identified in Arabidopsis thaliana (Katydid-At1) was found to contain an open reading frame very similar to the host gene coding for the mRNA decay (NMD) trans-acting factor, as none of the intronic sequences were found in the transduced gene. It was hypothesized that this even occurred by the recombination of the host gene transcript and the TRIM (Witte et al., 2001). Such a method of gene family evolution could account for the variability in gene structure and expansion of the ALVps and ALVis in Venturia. Another explanation is that part of the TRIM may have been recruited by the ALVi genes and expansion occurred by another mechanism. The genomic context of the ALVis is similar to that of Avrlm6. AvrLm6 in the L. maculans genome (strain v23.1.3) is situated in a gene-poor and AT-rich isochore, adjacent to an abundance of repetitive elements and remnants of transposons (Fudal et al., 2007), however, TRIMs have not been reported in L. maculans. Gene sparse regions are a feature of some filamentous plant pathogens and can be niches for effector genes and their evolution (Raffaele and Kamoun, 2012). Despite sharing a similar genomic context to AvrLm6, RIP was not detected in any of the ALVi sequences in V. inaequalis. RIP was detected bioinformatically in other regions of the genome and the majority of TEs were predicted to be affected by RIP, so it appears that RIP machinery is or has been active in the V. inaequalis genome. It remains unclear how the ALVi gene family was able to expand without being affected by RIP. However, RIP has not been investigated experimentally in Venturia sp, so the constraints of the RIP machinery are unknown. It has been shown experimentally in Neurospora crassa that duplicate nucleotide sequences of less than 380 were not subject to RIP (Watters et al., 1999). While most ALVi sequences are longer than this, it is possible that the V. inaequalis RIP machinery has other constraints that preclude the ALVi genes from being targets for RIP.
Venturia species, upon infection, rapidly form stroma (multicellular, laterally dividing tissue) in the cuticle and subcuticular space (Bowen et al., 2011). It is thought that the stroma is the main feeding and conidia-producing structure in Venturia. The AvrLm6-like homolog, ALVi_VI1_5 was localized, as a fusion protein with eYFP, to the periphery of the subcuticular stroma cells during infection. This is consistent with the gene expression data with regards to the timing of appearance of stroma during infection. All of the ALVis and ALVps are predicted to be secreted by the classical secretory pathway, however, we still do not know the function of this protein or whether it is released into the apoplast to interface with the plant.
The ALVi_Vi1_8 protein failed to trigger a resistance response in canola giving no indication as to whether the protein behaves in a similar way to AvrLm6. R genes have been described previously which recognize interspecific effectors with relatively low sequence similarity; for example the Cf-4–mediated HR in tomato can be triggered by the Avr4 protein from C. fulvum as well as homologs from phylogenetically related species; the MfAvr4 protein from M. fijiensis which only share 42% amino acid identity (Stergiopoulos et al., 2010) and DsAvr4 from D. septosporum which shares 51.7% identity (de Wit et al., 2012). It has also been frequently demonstrated, however, that small variations in effector sequence can abolish avirulence functionality. Such is the case with the Cf-4 mediated resistance described above, which can be disrupted by exchanging a single conserved proline residue (Mesarich et al., 2015). An AvrLm6 allele with a single amino acid substitution has also been found in Rlm6-virulent isolates collected from the field (Van de Wouw et al., 2010).
The ALVp and ALVi predicted proteins have typical characteristics of effectors; i.e., they are all small, predicted to be secreted, cysteine rich, contain no known functional domains, and many are upregulated during infection compared to in vitro growth. Furthermore, these expanded protein families share sequence similarity to an avirulence effector, known to trigger a specific resistance response in canola to L. maculans. The variability in copy number and sequence divergence of ALVis and ALVps between different species and isolates of Venturia, also make these proteins strong candidates for effectors (possibly host specificity determinants). Elucidating the roles of the Venturia ALVp and ALVi proteins will be challenging, as there is likely to be a degree of functional redundancy among them. The availability of the whole genome and transcriptome sequences for these fungi will facilitate the determination of the genetic basis of physiological races, governing host and cultivar specificity in Venturia. This in turn will assist plant breeding efforts to select for more durable resistance against scab diseases.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
JS was supported by Scholarship from La Trobe University and a Victorian Life Sciences Computation Initiative PhD Top-up scholarship. AR is supported under the Victorian Life Sciences Computation Initiative’s (VLSCI) Life Sciences Computation Centre, a collaboration between LTU, Melbourne and Monash Universities (an initiative of the Victorian Government, Australia); We would also like to thank Dr Carl Mesarich for providing us with the eYFP gene optimized for Venturia inaequalis codon usage and Dr Rohan Lowe for the L. maculans lepidii sequence data. We would also like to thank Remmelt Groenwold (Wageningen UR, The Netherlands) and Bruno Le Cam (INRA, Angers, France) for the V. inaequalis isolates.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00980
Footnotes
- ^http://genome.jgi.doe.gov/Venin1/Venin1.home.html
- ^http://genome.jgi.doe.gov/Venpi1/Venpi1.home.html
- ^http://www.ncbi.nlm.nih.gov/
- ^http://www.jgi.doe.gov/
- ^http://www.broad.mit.edu/
- ^https://bitbucket.org/arobinson/sciencescripts
References
Balesdent, M. H., Attard, A., Kühn, M. L., and Rouxel, T. (2002). New avirulence genes in the phytopathogenic fungus Leptosphaeria maculans. Phytopathology 92, 1122–1133. doi: 10.1094/PHYTO.2002.92.10.1122
Balesdent, M.-H., Fudal, I., Ollivier, B., Bally, P., Grandaubert, J., Eber, F., et al. (2013). The dispensable chromosome of Leptosphaeria maculans shelters an effector gene conferring avirulence towards Brassica rapa. New Phytol. 198, 887–898. doi: 10.1111/nph.12178
Balesdent, M.-H., Louvard, K., Pinochet, X., and Rouxel, T. (2006). A large-scale survey of races of Leptosphaeria maculans occurring on oilseed rape in France. Eur. J. Plant Pathol. 114, 53–65. doi: 10.1007/s10658-005-2104-0
Belfanti, E., Silfverberg-Dilworth, E., Tartarini, S., Patocchi, A., Barbieri, M., Zhu, J., et al. (2004). The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety. Proc. Natl. Acad. Sci. U.S.A. 101, 886–890. doi: 10.1073/pnas.0304808101
Bowen, J. K., Mesarich, C. H., Bus, V. G. M., Beresford, R. M., Plummer, K. M., and Templeton, M. D. (2011). Venturia inaequalis: the causal agent of apple scab. Mol. Plant Pathol. 12, 105–122. doi: 10.1111/j.1364-3703.2010.00656.x
Bowen, J. K., Mesarich, C. H., Rees-George, J., Cui, W., Fitzgerald, A., Win, J., et al. (2009). Candidate effector gene identification in the ascomycete fungal phytopathogen Venturia inaequalis by expressed sequence tag analysis. Mol. Plant Pathol. 10, 431–448. doi: 10.1111/j.1364-3703.2009.00543.x
Broggini, G. A. L., Bus, V. G. M., Parravicini, G., Kumar, S., Groenwold, R., and Gessler, C. (2011). Genetic mapping of 14 avirulence genes in an EU-B04 × 1639 progeny of Venturia inaequalis. Fungal Genet. Biol. 48, 166–176. doi: 10.1016/j.fgb.2010.09.001
Brun, H., Levivier, S., Somda, I., Ruer, D., Renard, M., and Chèvre, A. M. (2000). A field method for evaluating the potential durability of new resistance sources: application to the Leptosphaeria maculans-Brassica napus pathosystem. Phytopathology 90, 961–966. doi: 10.1094/PHYTO.2000.90.9.961
Bus, V. G. M., Rikkerink, E. H. A., Caffier, V., Durel, C.-E., and Plummer, K. M. (2011). Revision of the nomenclature of the differential host-pathogen interactions of Venturia inaequalis and Malus. Annu. Rev. Phytopathol. 49, 391–413. doi: 10.1146/annurev-phyto-072910-095339
Caffier, V., Patocchi, A., Expert, P., Bellanger, M.-N., Durel, C.-E., Hilber-Bodmer, M., et al. (2015). Virulence characterization of Venturia inaequalis reference isolates on the differential set of Malus hosts. Plant Dis. 99, 370–375. doi: 10.1094/PDIS-07-14-0708-RE
Cambareri, E., Jensen, B., Schabtach, E., and Selker, E. (1989). Repeat-induced G-C to A-T mutations in Neurospora. Science 244, 1571–1575. doi: 10.1126/science.2544994
Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. doi: 10.1007/BF02670468
Chisholm, S. T., Coaker, G., Day, B., and Staskawicz, B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. doi: 10.1016/j.cell.2006.02.008
Cooke, I. R., Jones, D., Bowen, J. K., Deng, C., Faou, P., Hall, N. E., et al. (2014). Proteogenomic analysis of the Venturia pirina (Pear Scab Fungus) secretome reveals potential effectors. J. Proteome Res. 13, 363536–363544. doi: 10.1021/pr500176c
Daverdin, G., Rouxel, T., Gout, L., Aubertot, J.-N., Fudal, I., Meyer, M., et al. (2012). Genome structure and reproductive behaviour influence the evolutionary potential of a fungal phytopathogen. PLoS Pathog. 8:e1003020. doi: 10.1371/journal.ppat.1003020
de Wit, P. J. G. M., van der Burgt, A., Ökmen, B., Stergiopoulos, I., Abd-Elsalam, K. A., Aerts, A. L., et al. (2012). The genomes of the fungal plant pathogens Cladosporium fulvum and Dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. PLoS Genet. 8:e1003088. doi: 10.1371/journal.pgen.1003088
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Elliott, C. E., and Howlett, B. J. (2006). Overexpression of a 3-ketoacyl-CoA thiolase in Leptosphaeria maculans causes reduced pathogenicity on Brassica napus. Mol. Plant Microbe Interact. 19, 588–596. doi: 10.1094/MPMI-19-0588
Fitzgerald, A. M., Mudge, A. M., Gleave, A. P., and Plummer, K. M. (2003). Agrobacterium and PEG-mediated transformation of the phytopathogen Venturia inaequalis. Mycol. Res. 107, 803–810. doi: 10.1017/S0953756203008086
Flutre, T., Duprat, E., Feuillet, C., and Quesneville, H. (2011). Considering transposable element diversification in de novo annotation approaches. PLoS ONE 6:e16526. doi: 10.1371/journal.pone.0016526
Friesen, T. L., Stukenbrock, E. H., Liu, Z., Meinhardt, S., Ling, H., Faris, J. D., et al. (2006). Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956. doi: 10.1038/ng1839
Fudal, I., Ross, S., Brun, H., Besnard, A.-L., Ermel, M., Kuhn, M.-L., et al. (2009). Repeat-induced point mutation (RIP) as an alternative mechanism of evolution toward virulence in Leptosphaeria maculans. Mol. Plant Microbe Interact. 22, 932–941. doi: 10.1094/MPMI-22-8-0932
Fudal, I., Ross, S., Gout, L., Blaise, F., Kuhn, M. L., Eckert, M. R., et al. (2007). Heterochromatin-like regions as ecological niches for avirulence genes in the Leptosphaeria maculans genome: map-based cloning of AvrLm6. Mol. Plant Microbe Interact. 20, 459–470. doi: 10.1094/MPMI-20-4-0459
Gardiner, D. M., and Howlett, B. J. (2004). Negative selection using thymidine kinase increases the efficiency of recovery of transformants with targeted genes in the filamentous fungus Leptosphaeria maculans. Curr. Genet. 45, 249–255. doi: 10.1007/s00294-004-0488-6
Gout, L., Fudal, I., Kuhn, M.-L., Blaise, F., Eckert, M., Cattolico, L., et al. (2006). Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol. Microbiol. 60, 67–80. doi: 10.1111/j.1365-2958.2006.05076.x
Gout, L., Kuhn, M. L., Vincenot, L., Bernard-Samain, S., Cattolico, L., Barbetti, M., et al. (2007). Genome structure impacts molecular evolution at the AvrLm1 avirulence locus of the plant pathogen Leptosphaeria maculans. Environ. Microbiol. 9, 2978–2992. doi: 10.1111/j.1462-2920.2007.01408.x
Grandaubert, J., Lowe, R. G. T., Soyer, J. L., Schoch, C. L., Van de Wouw, A. P., Fudal, I., et al. (2014). Transposable element-assisted evolution and adaptation to host plant within the Leptosphaeria maculans-Leptosphaeria biglobosa species complex of fungal pathogens. BMC Genomics 15:891. doi: 10.1186/1471-2164-15-891
Guyon, K., Balagué, C., Roby, D., and Raffaele, S. (2014). Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics 15:336. doi: 10.1186/1471-2164-15-336
Heckman, K. L., and Pease, L. R. (2007). Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2, 924–932. doi: 10.1038/nprot.2007.132
Huang, Y. J., Balesdent, M. H., Li, Z. Q., Evans, N., Rouxel, T., and Fitt, B. D. L. (2010). Fitness cost of virulence differs between the AvrLm1 and AvrLm4 loci in Leptosphaeria maculans (phoma stem canker of oilseed rape). Eur. J. Plant Pathol. 126, 279–291. doi: 10.1007/s10658-009-9539-7
Huang, Y.-J., Li, Z.-Q., Evans, N., Rouxel, T., Fitt, B. D. L., and Balesdent, M.-H. (2006). Fitness cost associated with loss of the AvrLm4 Avirulence function in Leptosphaeria maculans (phoma stem canker of oilseed rape). Eur. J. Plant Pathol. 114, 77–89. doi: 10.1007/s10658-005-2643-4
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kalendar, R., Vicient, C. M., Peleg, O., Anamthawat-Jonsson, K., Bolshoy, A., and Schulman, A. H. (2004). Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics 166, 1437–1450. doi: 10.1534/genetics.166.3.1437
Kucheryava, N., Bowen, J. K., Sutherland, P. W., Conolly, J. J., Mesarich, C. H., Rikkerink, E. H. A., et al. (2008). Two novel Venturia inaequalis genes induced upon morphogenetic differentiation during infection and in vitro growth on cellophane. Fungal Genet. Biol. 45, 1329–1339. doi: 10.1016/j.fgb.2008.07.010
Labbé, J., Murat, C., Morin, E., Tuskan, G. A., Le Tacon, F., and Martin, F. (2012). Characterization of transposable elements in the ectomycorrhizal fungus Laccaria bicolor. PLoS ONE 7:e40197. doi: 10.1371/journal.pone.0040197
Le Cam, B., Parisi, L., and Arene, L. (2002). Evidence of two formae speciales in Venturia inaequalis. Responsible for Apple and Pyracantha Scab. Phytopathology 92, 314–320. doi: 10.1094/PHYTO.2002.92.3.314
Mesarich, C. H., Stergiopoulos, I., Beenen, H. G., Cordovez, V., Guo, Y., Karimi Jashni, M., et al. (2015). A conserved proline residue in Dothideomycete Avr4 effector proteins is required to trigger a Cf-4-dependent hypersensitive response. Mol. Plant Pathol. doi: 10.1111/mpp.12265 [Epub ahead of print].
Nagaki, K., Neumann, P., Zhang, D., Ouyang, S., Buell, C. R., Cheng, Z., et al. (2005). Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol. Biol. Evol. 22, 845–855. doi: 10.1093/molbev/msi069
Parlange, F., Daverdin, G., Fudal, I., Kuhn, M.-L., Balesdent, M.-H., Blaise, F., et al. (2009). Leptosphaeria maculans avirulence gene AvrLm4-7 confers a dual recognition specificity by the Rlm4 and Rlm7 resistance genes of oilseed rape, and circumvents Rlm4-mediated recognition through a single amino acid change. Mol. Microbiol. 71, 851–863. doi: 10.1111/j.1365-2958.2008.06547.x
Pereira, J. F., Araújo, E. F., Brommonschenkel, S. H., Queiroz, C. B., Costa, G. G. L., Carazzolle, M. F., et al. (2015). MpSaci is a widespread gypsy-Ty3 retrotransposon highly represented by non-autonomous copies in the Moniliophthora perniciosa genome. Curr. Genet. 61, 185–202. doi: 10.1007/s00294-014-0469-3
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
Raffaele, S., and Kamoun, S. (2012). Genome evolution in filamentous plant pathogens: why bigger can be better. Nat. Rev. Microbiol. 10, 417–430. doi: 10.1038/nrmicro2790
Rouxel, T., Grandaubert, J., Hane, J. K., Hoede, C., van de Wouw, A. P., Couloux, A., et al. (2011). Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nat. Commun. 2:202. doi: 10.1038/ncomms1189
Sacristán, S., Vigouroux, M., Pedersen, C., Skamnioti, P., Thordal-Christensen, H., Micali, C., et al. (2009). Coevolution between a family of parasite virulence effectors and a class of LINE-1 retrotransposons. PLoS ONE 4:e7463. doi: 10.1371/journal.pone.0007463
Sampath, P., and Yang, T.-J. (2014). Comparative analysis of Cassandra TRIMs in three Brassicaceae genomes. Plant Genet. Resour. 12, S146–S150. doi: 10.1017/S1479262114000446
Saunders, D. G. O., Win, J., Cano, L. M., Szabo, L. J., Kamoun, S., and Raffaele, S. (2012). Using hierarchical clustering of secreted protein families to classify and rank candidate effectors of rust fungi. PLoS ONE 7:e29847. doi: 10.1371/journal.pone.0029847
Schouten, H. J., Brinkhuis, J., Burgh, A., Schaart, J. G., Groenwold, R., Broggini, G. A. L., et al. (2013). Cloning and functional characterization of the Rvi15 (Vr2) gene for apple scab resistance. Tree Genet. Genomes 10, 251–260. doi: 10.1007/s11295-013-0678-9
Soanes, D., and Richards, T. A. (2014). Horizontal gene transfer in eukaryotic plant pathogens. Annu. Rev. Phytopathol. 52, 583–614. doi: 10.1146/annurev-phyto-102313-050127
Sperschneider, J., Dodds, P. N., Gardiner, D. M., Manners, J. M., Singh, K. B., and Taylor, J. M. (2015). Advances and challenges in computational prediction of effectors from plant pathogenic fungi. PLoS Pathog. 11:e1004806. doi: 10.1371/journal.ppat.1004806
Stachowiak, A., Olechnowicz, J., Jedryczka, M., Rouxel, T., Balesdent, M.-H., Happstadius, I., et al. (2006). Frequency of avirulence alleles in field populations of Leptosphaeria maculans in Europe. Eur. J. Plant Pathol. 114, 67–75. doi: 10.1007/s10658-005-2931-z
Stehmann, C., Pennycook, S., and Plummer, K. M. (2001). Molecular identification of a sexual interloper: the pear pathogen, Venturia pirina, has Sex on Apple. Phytopathology 91, 633–641. doi: 10.1094/PHYTO.2001.91.7.633
Stergiopoulos, I., and de Wit, P. J. G. M. (2009). Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. doi: 10.1146/annurev.phyto.112408.132637
Stergiopoulos, I., van den Burg, H. A., Okmen, B., Beenen, H. G., van Liere, S., Kema, G. H. J., et al. (2010). Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc. Natl. Acad. Sci. U.S.A. 107, 7610–7615. doi: 10.1073/pnas.1002910107
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thakur, K., Chawla, V., Bhatti, S., Swarnkar, M. K., Kaur, J., Shankar, R., et al. (2013). De novo transcriptome sequencing and analysis for Venturia inaequalis, the devastating apple scab pathogen. PLoS ONE 8:e53937. doi: 10.1371/journal.pone.0053937
Van de Wouw, A. P., Cozijnsen, A. J., Hane, J. K., Brunner, P. C., McDonald, B. A., Oliver, R. P., et al. (2010). Evolution of linked avirulence effectors in Leptosphaeria maculans is affected by genomic environment and exposure to resistance genes in host plants. PLoS Pathog. 6:e1001180. doi: 10.1371/journal.ppat.1001180
Van de Wouw, A. P., Lowe, R. G. T., Elliott, C. E., Dubois, D. J., and Howlett, B. J. (2014). An avirulence gene, AvrLmJ1, from the blackleg fungus, Leptosphaeria maculans, confers avirulence to Brassica juncea cultivars. Mol. Plant Pathol. 15, 523–530. doi: 10.1111/mpp.12105
Vitte, C., Chaparro, C., Quesneville, H., and Panaud, O. (2007). Spip and Squiq, two novel rice non-autonomous LTR retro-element families related to RIRE3 and RIRE8. Plant Sci. 172, 8–19. doi: 10.1016/j.plantsci.2006.07.008
Watters, M. K., Randall, T. A., Margolin, B. S., Selker, E. U., and Stadler, D. R. (1999). Action of repeat-induced point mutation on both strands of a duplex and on tandem duplications of various sizes in Neurospora. Genetics 153, 705–714.
Win, J., Greenwood, D. R., and Plummer, K. M. (2003). Characterisation of a protein from Venturia inaequalis that induces necrosis in Malus carrying the Vm resistance gene. Physiol. Mol. Plant Pathol. 62, 193–202. doi: 10.1016/S0885-5765(03)00061-4
Witte, C. P., Le, Q. H., Bureau, T., and Kumar, A. (2001). Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc. Natl. Acad. Sci. U.S.A. 98, 13778–13783. doi: 10.1073/pnas.241341898
Keywords: effector, Dothideomycete, gene families, RIP, WGS
Citation: Shiller J, Van de Wouw AP, Taranto AP, Bowen JK, Dubois D, Robinson A, Deng CH and Plummer KM (2015) A Large Family of AvrLm6-like Genes in the Apple and Pear Scab Pathogens, Venturia inaequalis and Venturia pirina. Front. Plant Sci. 6:980. doi: 10.3389/fpls.2015.00980
Received: 14 August 2015; Accepted: 26 October 2015;
Published: 17 November 2015.
Edited by:
Stéphane Hacquard, Max Planck Institute for Plant Breeding Research, GermanyReviewed by:
David L. Joly, Université de Moncton, CanadaIsabelle Fudal, Institut National de la Recherche Agronomique, France
Copyright © 2015 Shiller, Van de Wouw, Taranto, Bowen, Dubois, Robinson, Deng and Plummer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kim M. Plummer, ay5wbHVtbWVyQGxhdHJvYmUuZWR1LmF1
 Jason Shiller
Jason Shiller Angela P. Van de Wouw
Angela P. Van de Wouw Adam P. Taranto
Adam P. Taranto Joanna K. Bowen
Joanna K. Bowen David Dubois
David Dubois Andrew Robinson
Andrew Robinson Cecilia H. Deng4
Cecilia H. Deng4 Kim M. Plummer
Kim M. Plummer