
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 03 November 2015
Sec. Plant Physiology
Volume 6 - 2015 | https://doi.org/10.3389/fpls.2015.00960
This article is part of the Research Topic Winter Survival in the Common Garden - Molecular Aspects of Frost Tolerance in Perennials View all 7 articles
 Jian Wu1†
Jian Wu1† Shanshan Seng1†
Shanshan Seng1† Juanjuan Sui1
Juanjuan Sui1 Eliana Vonapartis2,3
Eliana Vonapartis2,3 Xian Luo4
Xian Luo4 Benhe Gong1
Benhe Gong1 Chen Liu1
Chen Liu1 Chenyu Wu1
Chenyu Wu1 Chao Liu1
Chao Liu1 Fengqin Zhang1
Fengqin Zhang1 Junna He1*
Junna He1* Mingfang Yi1*
Mingfang Yi1*The phytohormone abscisic acid (ABA) regulates plant development and is crucial for abiotic stress response. In this study, cold storage contributes to reducing endogenous ABA content, resulting in dormancy breaking of Gladiolus. The ABA inhibitor fluridone also promotes germination, suggesting that ABA is an important hormone that regulates corm dormancy. Here, we report the identification and functional characterization of the Gladiolus ABI5 homolog (GhABI5), which is a basic leucine zipper motif transcriptional factor (TF). GhABI5 is expressed in dormant vegetative organs (corm, cormel, and stolon) as well as in reproductive organs (stamen), and it is up-regulated by ABA or drought. Complementation analysis reveals that GhABI5 rescues the ABA insensitivity of abi5-3 during seed germination and induces the expression of downstream ABA response genes in Arabidopsis thaliana (EM1, EM6, and RD29B). Down-regulation of GhABI5 in dormant cormels via virus induced gene silence promotes sprouting and reduces the expression of downstream genes (GhLEA and GhRD29B). The results of this study reveal that GhABI5 regulates bud dormancy (vegetative organ) in Gladiolus in addition to its well-studied function in Arabidopsis seeds (reproductive organ).
Dormancy has been described as one of the least understood phenomena in plant biology and remains confusing in spite of much recent progress (Finkelstein et al., 2008). This reflects the fact that dormancy is a complex process with many contributing factors. Since plants often face harsh environmental conditions, they have evolved a number of traits that promote survival. For example, many plants undergo a period of temporary growth cessation known as dormancy (Finkelstein et al., 2008). It can occur in both reproductive organs (seed) and vegetative organs (bud or tuber), and is also crucial for organ development and architecture (Horvath et al., 2003;Graeber et al., 2012). In cereal crops, dormancy is an essential trait because it prevents pre-harvest sprouting. The lack of harvest dormancy (vivipary) in cultivars of maize or wheat has resulted in large economic losses (Suzuki et al., 2001;Yang et al., 2007). However, deep dormancy can also be problematic: in barley, it prevents grains from germinating in a rapid and uniform manner that is required in the process of malting (Millar et al., 2006). In the horticultural industry, deep dormancy is an issue, and some treatments are necessary to promote germination and prevent extra storage costs (Gubler et al., 2005).
In seeds, various hormones regulate dormancy and germination. Specifically, while gibberellic acid (GA) and brassinosteroids are reported to promote dormancy breaking, ABA is the primary positive regulator of seed dormancy (Finch-Savage and Leubner-Metzger, 2006; Rentzsch et al., 2012). Compared with seed dormancy, little is known about bud dormancy in perennial plants (Horvath et al., 2004;Cooke et al., 2012). For perennials, bud dormancy is commonly divided into three consecutive stages: para-dormancy (inhibition regulated by the apical bud), endo-dormancy (inhibition regulated by internal signals), and eco-dormancy (inhibition regulated by environmental factors) (Horvath et al., 2003; Anderson et al., 2010). However, Gladiolus corm dormancy differs slightly from common bud dormancy in perennials: after flowering in the summer, a new corm emerges and expands on the top of mother corm. In late autumn, the new corm and its cormels are in a state of endo-dormancy. Following 3–4 months of cold storage, the endo-dormant corm or cormel, transitions into the eco-dormant stage. During the latter stage, germination is affected by environmental factors such as light, temperature and water. In order to reduce the costs associated with cold storage, weaker dormancy is required for cultivators in the flower industry.
Although the molecular mechanism underlying the regulation of bud dormancy is not well-understood, advances that shed light on this physiological process have been made. A gene involved in the regulation of flowering, FT (FLOWERING LOCUS T), is reported to be involved in establishing dormant buds and in dormancy state transitions in poplar. Knocking down the expression of FT results in growth cessation and bud set (Bohlenius et al., 2006; Horvath, 2009). Moreover, transcriptome analysis of bud dormancy in leafy spurge, peach and pear led to the proposition that DORMANCY ASSOCIATED MADS-BOX (DAM) genes regulate bud dormancy-related processes (Horvath et al., 2008; Leida et al., 2012; Liu et al., 2012). Ectopic expression of DAM in Arabidopsis down-regulated FT expression (Horvath et al., 2010). In woody plants, CBF (C-REPEAT/DRE BINDING FACTOR), DAM, RGL (RGA-LIKE), and EBB (EARLY BUD-BREAK) genes have been shown to be associated with freezing tolerance, dormancy, and bud breaking (Wisniewski et al., 2015).
Abscisic acid is also a key factor that regulates dormancy. Short day length impacts the induction and maintenance of endo-dormancy by triggering ABA accumulation (Heide, 1986; Li et al., 2005). In autumn, short-term cold treatment also inhibits the growth of perennials by inducing endo-dormancy through the action of ABA and GA (Horvath et al., 2003). However, reducing ABA content in potato tubers results in endo-dormancy breaking, which may be due to the altered expression of NCEDs (9-cis-epoxycarotenoid dioxygenases) and CYP707As (Cytochrome P450, Family 707, Subfamily A) (Destefano-Beltran et al., 2006). Cold temperature has been reported to reduce endogenous ABA content, and contributes to dormancy breaking in lily, grape and pear (Xu et al., 2006; Liu et al., 2012; Zheng et al., 2015). ABA is commonly regarded as a stress hormone since it is involved in the response to drought and osmotic stress, abscission and the induction of dormancy (Finkelstein, 2013). The signal transduction pathway of ABA in the plant is clear: in the absence of ABA, PP2Cs (Type 2C Protein Phosphatases) are active and inhibit the activity of SnRK2 (SNF1-Related Protein Kinase 2), rendering it unable to regulate downstream factors, blocking ABA responses. In the presence of ABA, ABA receptors interact with PP2Cs and repress their phosphatase activity, thereby activating SnRK2. SnRK2 then phosphorylates bZIP TFs (e.g., ABI5) which activate the expression of ABA-response genes (Hubbard et al., 2010; Melcher et al., 2010).
ABI5 belongs to the bZIP superfamily group A owing to its structural features including conserved regions C1–C2, basic regions, and leucine zippers (Finkelstein and Lynch, 2000). AtABI5 plays a central role in regulating ABA-responsive genes in seeds by binding to ABA-responsive-elements (ABREs, ACGTGG/TC) in the promoter region of genes such as AtEM6 (EARLY METHIONINE-LABELED 6), AtRD29A (RESPONSIVE TO DESICCATION 29A), AtRD29B and AtRAB18 (RAB GTPASE HOMOLOG B18) (Carles et al., 2002; Miura et al., 2009). Phosphorylation by SnRK2 affects AtABI5 activation and stability, whereas ABI5 instability is promoted by SER/THR PROTEIN PHOSPHATASE 6 (Dai et al., 2013; Feng et al., 2014). Furthermore, AtABI5 is degraded by KEEP ON GOING (KEG; Stone et al., 2006). The transcriptional activation and protein stability of AtABI5 are also positively regulated by ABA (Lopez-Molina et al., 2001). It has been previously reported that rice OsABI5-Like1 contributes to drought, salinity and osmotic stress responses mediated by ABA (Yang et al., 2011). However, research concerning the effects of ABI5 on bud dormancy is limited.
Through comparison of transcriptome data from dormant seed and bud in peach, some common features have been identified, including ABA response-related genes such as ABI5-like, ABI3-like, ABI5 binding protein (AFP)-like, and LATE EMBRYOGENESIS ABUNDANT (LEA)-like (Leida et al., 2012). AFP is believed to promote the degradation of AtABI5, and AtABI3 acts directly upstream of AtABI5 to positively regulate AtABI5 expression (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2002, 2003; Dai et al., 2013). Although the effects of ABI5 on seed dormancy have been well-characterized, no direct genetic evidence yet exists to support the idea that ABI5 also functions in bud dormancy (Piskurewicz et al., 2008; Rodriguez et al., 2009; Nonogaki, 2014).
The purpose of the present study was to characterize the expression of GhABI5 in vegetative dormant organs and to elucidate how it regulates corm dormancy.
Gladiolus hybridus (‘Rose supreme’) was planted in the Science Research Garden at China Agricultural University on April 30th, 2012 and corms were harvested on October 30th. The corms were washed and dried at 25°C for 6 weeks, after which the cormels were separated from the corms. The cormels were randomly divided into two batches: one was stored in nylon mesh bags at 22 ± 1°C and the other was stored at low temperature (4 ± 1°C) in the refrigeration house; relative humidity was kept at 60–70%. Cormels for qRT-PCR and phytohormone quantification were sampled at 14-days intervals (0–12 weeks) during drought (0–6 weeks) and storage (6–12 weeks). GA and ABA content were measured using the indirect enzyme linking immunosorbent assay (ELISA) method (Yang et al., 2001), following the manufacturer’s instructions (Chemical Control Laboratory of Agriculture and Biotechnology College, China Agricultural University). Cormel tissue (0.2 g fresh weight) was immediately frozen in liquid nitrogen and stored at -80°C prior to hormone extraction. The tissue was homogenized in extraction solution (80% methanol (v/v) including 1 mM butylated hydroxytoluene as an antioxidant) and centrifuged at 10,000 g for 20 min at 4°C. The supernatant was then extracted at 4°C for 8 h.
Dormant cormels used for hormone treatments measured 0.8–1.0 cm in diameter and had been stored at 4°C for 2 weeks. These cormels were treated with ABA (25 μM), fluridone (25 μM) or distilled water before being placed in a light homoeothermic incubator at 25°C with 16/8 h light/dark. The tunic was removed from cormels used for drought treatment, which were left to break dormancy at room temperature for 1, 3, 6 days. The experiment were repeated with three biological replicates (n = 48).
Dormant cormels stored at 4 or 22°C for different lengths of time were sterilized and plated on 0.6% agar. They were then placed in a light incubator with 16/8 h light/dark for sprouting. The sprouting percentage was counted on the 21st day after planting. Fifty cormels per sample were used for each sprouting test. Error bars represent the SE of three biological repeats.
Arabidopsis seeds were sterilized with 1% (v/v) NaClO for 30 min, washed five times with sterile distilled water, and plated on MS medium. Following stratification for 3 days at 4°C in the dark, the seeds were transferred into a light homoeothermic incubator (22°C) with 16 h/8 h light/dark. Seven day-old seedlings were transferred to forest soil and vermiculite (1:1, v/v) in a greenhouse at 22°C with 16 h light/8 h dark under 50 μmol m-2 s-1 light. Nicotiana benthamiana seeds were planted in sterilized forest soil and vermiculite (1:1, v/v) and cultured under the same conditions as the Arabidopsis seedlings.
Total RNA from dormant Gladiolus cormels was extracted using the Tiangen RNA extraction reagent kit (Tiangen, China). Subsequently, cDNA was synthesized by M-MLV reverse transcriptase (TaKaRa, Japan) with a 3′ adaptor (5′-CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGC(T)17-3′).
Degenerate primers, GhABI5-DP-for (5′-GGBTCCATGAACATGGACGAG-3′) and GhABI5-DP-rev (5′-KCCAVACCTCVTCVACVGTCTT-3′), were designed for GhABI5 cloning based on conserved regions of homologous ABI5 genes from Arabidopsis thaliana, Triticum aestivum, Hordeum vulgare, and Zea mays (Supplementary Table S2). The amplified product was used to clone the full-length GhABI5 sequences by the Rapid Amplification of cDNA Ends (RACE) method and nested PCR according to the manufacturer’s protocol (Clontech, Mountain View, CA, USA). All PCR products were subcloned into the pMD18-T vector (TaKaRa, Japan) and transformed into E. coli DH5α cells (Trans, China) and sequenced. Primer sequences are listed in Supplementary Table S1.
Protein characteristics were analyzed with the ExPASy Proteomics Tools.1 ClustalX1.8 and BioEdit7.0 were used to perform multiple amino acid alignments, and phylogenetic trees were constructed by the neighbor-joining method using the MEGA5.0 software (Tamura et al., 2011). Accession numbers for the public sequences used in the phylogenetic analysis are listed in Supplementary Table S2.
The upstream regulatory sequence of GhABI5 was isolated using high-efficiency thermal asymmetric interlaced PCR (hiTAIL-PCR; Liu and Chen, 2007). The position of the translation start site was designated as “+1”. The cis-regulatory elements were analyzed and annotated using the software programs from the Plant Cis-acting Regulatory DNA Elements database2 (PLACE). The 1688 bp upstream regulatory sequence of GhABI5 was inserted into the pCAMBIA1391 binary vector with HindIII and BamHI restriction sites. This GhABI5:GUS plasmid was then introduced into Agrobacterium tumefaciens strain GV3101 and injected into G. hybridus petals (Batoko et al., 2000). The petals were cultured with water or 50 μM ABA, and covered with preservative film. Petal disks were GUS-stained after 2 days, following a previously established protocol for GUS histochemical staining (Jefferson, 1987).
About 400 ng cDNA was used as the template for qRT-PCR using the Applied Biosystems StepOnePlusTM real-time PCR system with the KAPATM SYBR® FAST qPCR kit (Kapa Biosystems, Woburn, MA, USA). The Gladiolus actin gene (GenBank accession no. JF831193) and Arabidopsis Actin2 (GenBank accession no. NM-112764) were used as controls in Gladiolus and Arabidopsis, respectively. The PCR procedure was performed according to the manufacturer’s instructions. Each qRT-PCR assay was run in three biological replicates. Primer sequences are listed in Supplementary Table S1.
The open reading frame (ORF) of GhABI5 was cloned into pCAMBIA1300-GFP with XbaI and KpnI restriction sites. Both the fusion construct (GhABI5-GFP) and the control (GFP) were transformed into onion epidermal cells by particle bombardment. After incubation for 20 h at 25°C in the dark, the cells were observed under bright field and fluorescence using confocal microscopy (Nikon Inc., Melville, NY, USA). The vectors were also transformed into Agrobacterium tumefaciens GV3101 and used to agroinfiltrate N. benthamiana leaves for transient expression, as described previously (Batoko et al., 2000). After 3 days, the leaves were observed in the same manner as the onion cells.
Arabidopsis plants (abi5-3 and Col-0) were transformed using the floral dip method (Clough and Bent, 1998) with Agrobacterium tumefaciens GV3101 harboring GhABI5 fused with GFP at its N-terminus (GFP-GhABI5/pCAMBIA1300).
Infected seeds were collected and sterilized with 1% (v/v) NaClO for 30 min, washed with distilled water and plated on MS medium containing 50 μg l-1 hygromycin B (Roche, Germany) before stratification for 3 days at 4°C. The plates were then transferred into the growth chamber to select transformed seedlings. T3-generation homozygous seeds were used for all experiments.
To ensure all freshly harvested seeds were of similar maturity, we carefully selected early siliques of the same yellow color. Seeds were sown directly on water-saturated filter paper, with or without ABA, to allow for germination under 16/8 h light/dark conditions at 22°C without stratification. Seeds were considered germinated once green cotyledons emerged, and germinated seeds were scored at the indicated times. Statistical analysis was performed with three biological replicates.
Silencing of GhABI5 by virus-induced gene silencing (VIGS) was performed as described previously (Zhong et al., 2014), with some modifications. The pTRV1, pTRV2, and pTRV2-GhABI5 (specific fragment of about 344 bp) vectors were transformed individually into Agrobacterium tumefaciens GV3101. The transformed Agrobacterium lines were cultured overnight in Luria-Bertani (LB) medium supplemented with 50 μg ml-1 kanamycin and 100 μg ml-1 rifampicin. The next day, the cells were collected and re-suspended in infiltration buffer (10 mM MgCl2, 200 mM acetosyringone, 10 mM MES, pH 5.6) to a final OD600 of about 2.0. Equal volumes of TRV1 and TRV2 (used as the control), as well as TRV1 and TRV2-GhABI5 were mixed together and placed at room temperature for 3 h in the dark prior to vacuum infiltration.
Dormant corms (0.8–1.0 cm in diameter, stored for 2 months at 4°C) were first poked with pins on their top and bottom. Subsequently, they were subjected to vacuum infiltration under 0.7 MPa for 30 min. The cormels were then planted in pots and kept at 20°C for 7 days in darkness before cultivation in a growth chamber at 20°C with 16/8 h light/dark. The plants were measured with a vernier caliper after 10 days, and images were taken with a digital camera (Canon D500). The experiments were repeated three times using at least 96 cormels in each repetition. Duncan’s multiple range test (∗P < 0.05, ∗∗P < 0.01) was used for statistical analysis.
Abscisic acid is believed to be the main phytohormone that controls seed dormancy. Therefore, we wished to track fluctuations in ABA levels during Gladiolus dormancy breaking. We quantified ABA and GA levels during different time points throughout the desiccation and storage periods of freshly harvested corms. For the first 6 weeks, the freshly harvested cormels were dried in a well-ventilated and dry environment; during this period, endogenous ABA dramatically increased and reached its peak, while GA content somewhat decreased (Figures 1A,B) and the ratio of ABA to GA sustainably increased (Figure 1C). Meanwhile, we observed that after 6 weeks, the cormels were covered by a tunic that prevents further water loss and may protect internal organs.
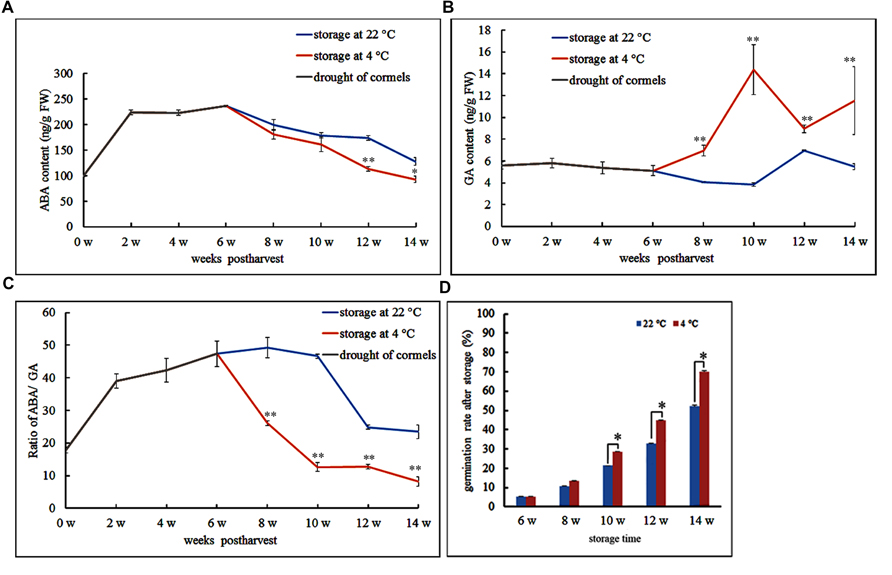
FIGURE 1. Endogenous abscisic acid (ABA) and gibberellic acid (GA) levels in Gladiolus cormels under different temperature storage. Postharvest storage is divided into two stages: drought of cormels (6 weeks) and then storage at 4°C or 22°C. (A) ABA changes in drought of cormels (0–6 w) and different storage temperature (6–14 w). (B) GA changes in drought of cormels (0–6 w) and different storage temperature (6–14 w). (C) Ratio of ABA/GA changes in drought of cormels (0–6 w) and different storage temperature (6–14 w). (D) The effect of temperature and duration on the sprouting of cormels. Error bars represent the SE of three replicates. (∗P < 0.05; ∗∗P < 0.01).
To compare the effects of different storage temperatures on dormancy breaking, we measured ABA and GA content under storage at 4 and 22°C. Following the first 6 weeks, one batch of cormels was stored at 22°C. During this storage stage, ABA content showed a steady decrease, whereas GA levels were lowest at 10 weeks and peaked at 12 weeks post-harvest (Figures 1A,B). The ABA/GA ratio was highest at 10 weeks and then decreased dramatically (Figure 1C). However, low temperature helped reduce ABA content and increased GA content in the cormels stored at 4°C (Figures 1A,B). Thus, the ratio of ABA to GA is higher at 22°C than at 4°C (Figure 1C). We also tracked cormel sprouting under different storage temperatures. Our data shows that the cormels sprouted with greater frequency when stored for a longer period of time. In this case, storage at 4°C can accelerate germination compared with 22°C (Figure 1D).
To further examine the role of ABA in corm dormancy (vegetative organ), corms were treated with exogenous ABA and fluridone. The results show that 62.5% of cormels treated with fluridone for 9 days germinated, compared with 18.75% of the control cormels (Figures 2A,D). Moreover, none of the ABA-treated cormels germinated. After 11 days, about 75% of fluridone-treated cormels germinated, while only 25% of the control and none of the cormels treated with ABA germinated (Figures 2B,D). After 13 days of fluridone treatment, almost all cormels had germinated. However, the germination rate for the control and ABA-treated cormels was roughly 62.5 and 15%, respectively (Figures 2C,D). These results show that ABA significantly inhibits the emergence and elongation of Gladiolus buds and roots (Figure 2E), thereby forcing Gladiolus to maintain dormancy. Fluridone effectively promotes sprouting but has an inhibitory effect on root elongation (Figure 2E). Taken together, the results illustrate that exogenous ABA is sufficient to repress germination and maintain Gladiolus corm dormancy.
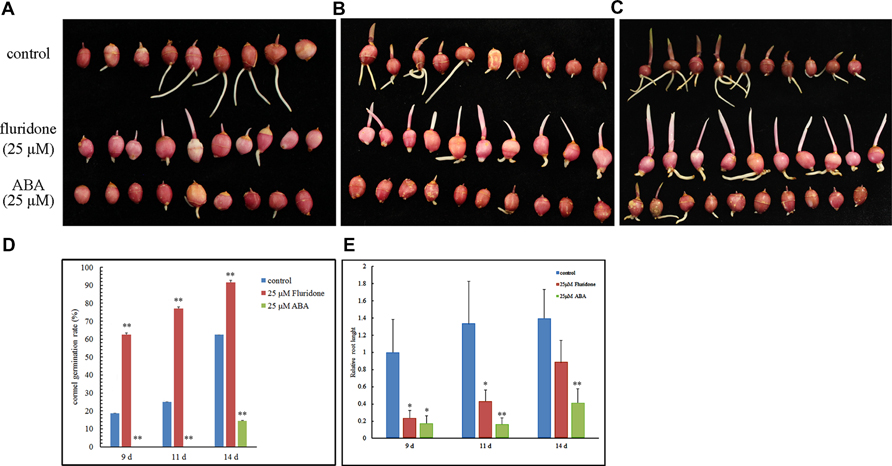
FIGURE 2. Effects of exogenous ABA and fluridone on releasing Gladiolus corm dormancy. (A–C) Cormels were submerged in water (control treatment), 25 μM fluridone or 25 μM ABA. Photos were taken at 9, 11, and 14 days post-treatment, respectively. (D) Germination (emergence of 1 cm buds) of cormels placed in water, 25 μM fluridone or 25 μM ABA. (E) The relative root length of cormels. Forty-eight cormels were used in each experiment. Error bars represent the SE of three biological replicates. (∗P < 0.05; ∗∗P < 0.01).
In order to better understand the role of ABA signaling in G. hybridus corm dormancy, we cloned a transcription factor belonging to the bZIP family. A conserved cDNA fragment was amplified using degenerate primers, and the full-length sequence was then isolated by the rapid amplification of cDNA ends (RACEs) method. The ORF of this bZIP transcription factor is 1221 bp in length, encoding a protein of 406 amino acid residues (AARs), with a calculated molecular mass of 44.53 KDa and an isoelectric point (pI) of 5.65.
To investigate the characteristics of this gene, we performed a phylogenetic analysis using MEGA 5. The results (Figure 3A) suggest that our gene clusters closely with AtABI5. In addition, it shares more similarity with AtABI5 than with the other 72 members of the Arabidopsis bZIP family3; therefore, we named the gene GhABI5 (GenBank accession: KC344270). Furthermore, GhABI5 forms its own clade and clusters more closely with the dicot genes than the monocot genes. (Figure 3B). GhABI5 is similar to GmABI5 from Glycine max (45% identity), HvABI5 from Hordeum vulgare (42% identity), WABI5 from T. aestivum (42% identity), FvABI5 from Fragaria vesca (41% identity), and AtABI5 from Arabidopsis thaliana (40% identity). Sequence alignment analysis (Figure 3C) shows that GhABI5 contains all distinguishing features of ABI5 proteins, including the conserved domains C1 (AARs 38 to 83), C2 (AARs 111 to 138), C3 (AARs 157 to 178), and basic domain (AARs 324 to 344) which is necessary for DNA binding (Finkelstein and Lynch, 2000; Zhou et al., 2013).
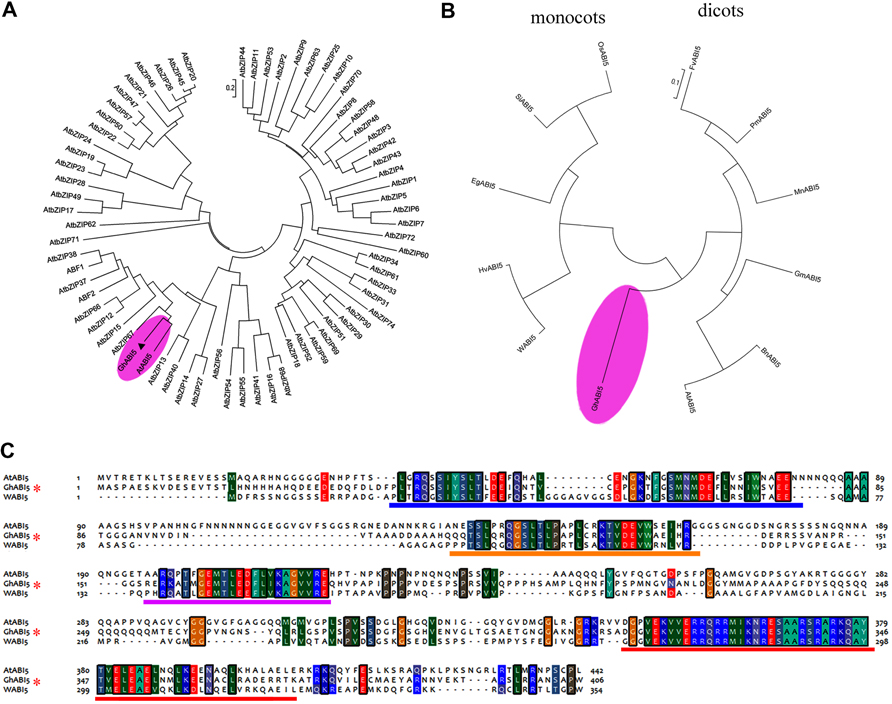
FIGURE 3. Phylogenetic analysis of GhABI5. (A) Phylogenetic relationships among GhABI5 and Arabidopsis bZIP proteins. (B) Phylogenetic analysis of GhABI5 and ABI5 homologous genes in other species. (C) Sequence alignment of GhABI5 with its homologs from Arabidopsis thaliana (AtABI5) and Triticum aestivum (WABI5). The conserved C1, C2,C3, and bZIP domains are indicated by a blue, orange, purple, and red line, respectively. Information on the homologous ABI5 proteins is listed in Supplementary Table S2.
Initially, AtABI5 was thought to be expressed specifically in the seed; however, recent research has shown that OsABI5 is also expressed in flowers (Zou et al., 2008) and in response to stresses (Lopez-Molina et al., 2003). Here, we analyzed the tissue-specific expression of GhABI5 by real-time PCR. We found high expression levels in the corm, suggesting that GhABI5 may play an important role in this vegetative organ (Figure 4A). Then, we tracked GhABI5 expression during the course of desiccation (0 to 6 weeks) and storage (6 to 14 weeks). The data (Figure 4B) showed increasing GhABI5 expression during desiccation, reaching a peak at the end of this water loss period. GhABI5 expression then gradually decreased during storage (Figure 4C). These results were consistent with the measured levels of ABA, suggesting that GhABI5 expression is likely regulated by endogenous ABA.
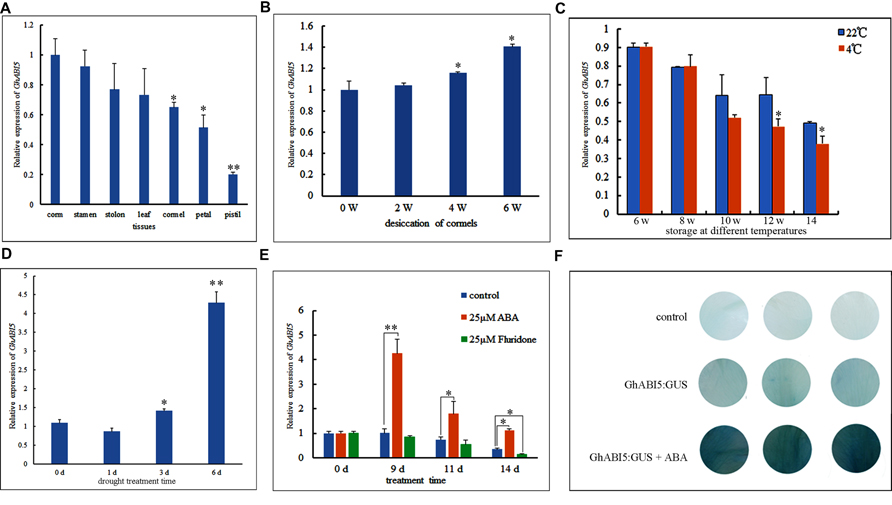
FIGURE 4. Quantitative real-time PCR analysis of GhABI5 expression levels and GhABI5 promoter analysis. (A) GhABI5 expression levels in various organs. (B) GhABI5 expression during storage at 22°C. (C) The effect of temperature and duration on the expression of GhABI5. (D) The expression of GhABI5 under drought treatment (0, 1, 3, 6-days); (E) The expression of GhABI5 in cormels treated with ABA and fluridone (0, 9, 11, 14-days). (F) Transient expression of GhABI5:GUS in Gladiolus petal disks with 50 μM ABA. Error bars represent the SE of three biological replicates. (∗P < 0.05; ∗∗P < 0.01).
In our study, we have shown that cold storage (4°C) helps Gladiolus break dormancy (Figure 1D). Therefore, we investigated the effect of temperature on GhABI5 expression. Cold storage significantly repressed GhABI5 expression at the two latest time points, compared with a higher temperature (22°C) (Figure 4C). This finding suggests that cold treatment can regulate GhABI5 expression for a relatively short period of time. Furthermore, we found that GhABI5 was highly responsive to drought, and its expression levels greatly increased 6 days following treatment (Figure 4D). Next, we analyzed the effect of ABA and the ABA inhibitor fluridone on the expression of GhABI5. Our data (Figure 4E) revealed that GhABI5 was sharply up-regulated by 25 μM ABA treatment after 9 days, but 25 μM fluridone down-regulated GhABI5 at 14 days. Fluridone may affect the metabolism of other hormones and indirectly influence GhABI5 expression. Alternatively, there may be feedback regulation between GhABI5 and ABA metabolism.
Around 1.7 kb of the 5′ regulatory region upstream of the GhABI5 transcription start site was isolated by hiTAIL-PCR. The acquired sequence was then scanned using the Plant cis-acting regulatory DNA elements (PLACE) database to obtain putative cis-regulatory elements potentially involved in the regulation of GhABI5 expression. The data revealed a large number of elements that are putative target motifs for transcription factors reported to respond to hormones and hormone-related stresses (Supplementary Table S3 and Figure S1). Among these elements, ABA-related motifs are the most abundant: E-box/ABRE, CTR/DRE, low-temperature-responsive elements (LTREs) and RY/G box (an ABI3-binding motif). Moreover, GA and auxin response elements were also present in the promoter region (Supplementary Table S3). To test whether the GhABI5 promoter is responsive to ABA, a transient expression in Gladiolus petal disks was performed through agroinfiltration (Batoko et al., 2000). The results show that ABA induces GhABI5:GUS expression (Figure 4F), confirming the result that GhABI5 is up-regulated by ABA (Figure 4E).
To learn more about GhABI5, we investigated its subcellular localization using both onion and N. benthamiana epidermal cells. For this purpose, we fused the ORF of GhABI5 to GFP. The recombined plasmid was transformed into onion epidermal cells by particle bombardment. In the epidermal cells expressing GFP-GhABI5, the GFP signal was present in the nucleus, while the 35S:GFP signal could be seen throughout the cells (Figure 5A). The same results were obtained in the N. benthamiana transient expression system (Figure 5B), which had been transformed using Agrobacterium. These results confirm that GhABI5 is a nuclear protein.
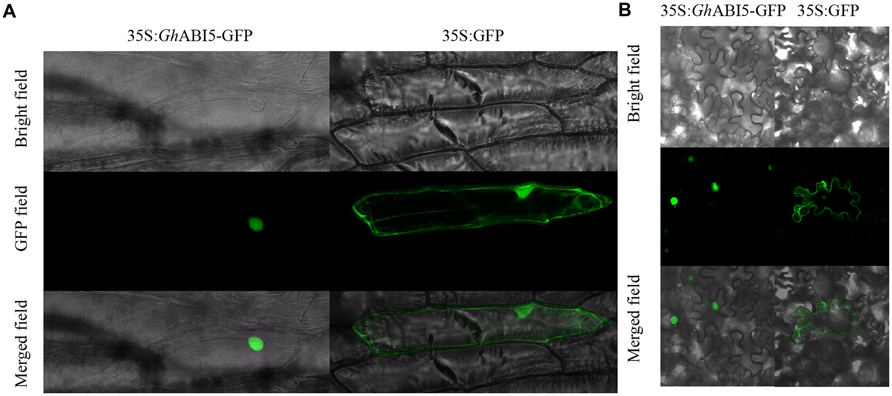
FIGURE 5. The nuclear localization of GhABI5. (A) The subcellular localization of GhABI5 in onion epidermal cells. (B) The subcellular localization of GhABI5 in Nicotiana benthamiana epidermal cells. 35S:GFP was used as the control.
The role of GhABI5 in dormancy was also examined by the ectopic expression of 35S:GhABI5 in abi5-3 and wild type (Col) (Supplementary Figure S2). Without ABA treatment, the transgenic seeds exhibited a somewhat lower germination rate compared with Col (Figures 6A,B). Furthermore, when grown on media containing 0.5 μM ABA for 6 days, 57% of GhABI5 complemented abi5-3 seeds were sensitive to ABA while abi5-3 was still insensitive to ABA (Figure 6B). In addition, on 1 μM ABA, seeds of 35S:GhABI5/Col and two complemented lines did not germinate, whereas seeds of the weakly complemented line showed similar ABA sensitivity to that of Col. Comparable results were observed with the 2 μM ABA treatment (Figure 6A). These results indicate that with exogenous ABA treatment, GhABI5 contributes to seed dormancy, thereby enhancing the dormancy of Arabidopsis seeds. GhABI5 also rescues the dormancy in complemented lines. Seeds overexpressing GhABI5 in the Col-0 background exhibit deeper dormancy than the abi5-3 lines complemented with GhABI5.
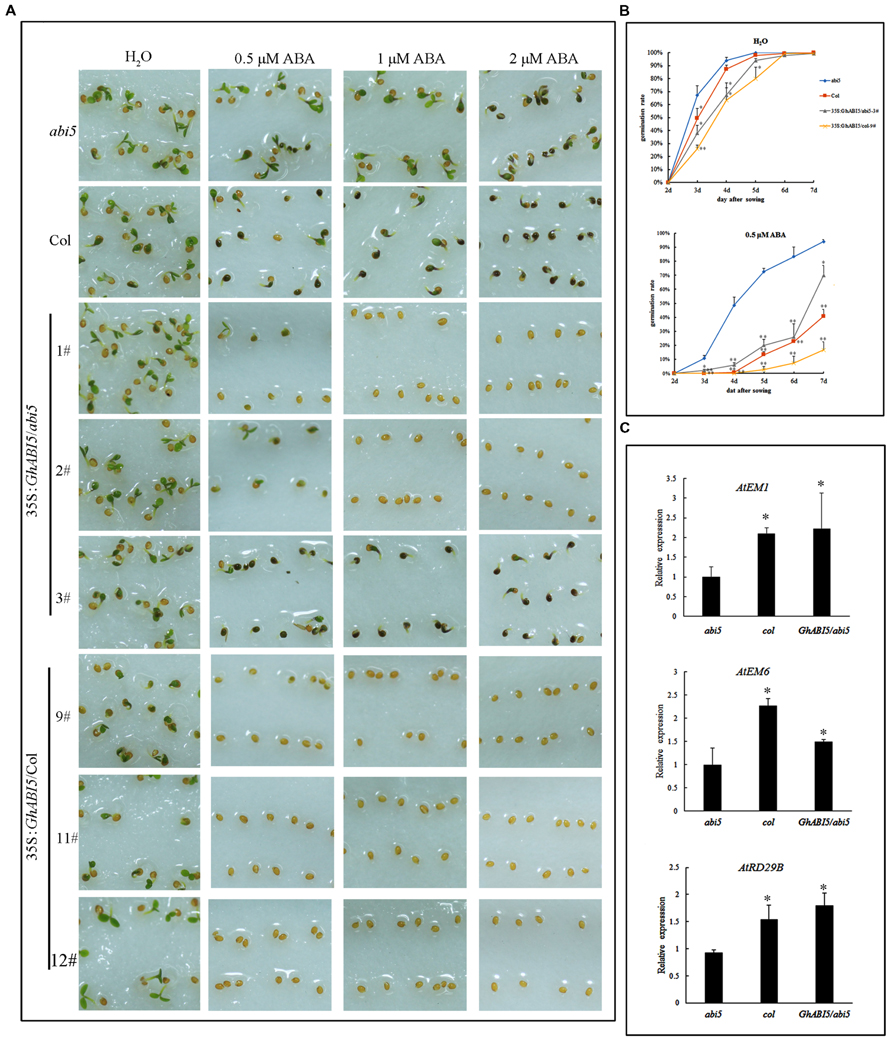
FIGURE 6. GhABI5 responds to ABA, inhibiting seed germination and inducing downstream gene expression. (A) Transgenic plants overexpressing GhABI5 are hypersensitive to ABA. Seeds of abi5-3(abi5), Col, GhABI5 complemented lines (1, 2, 3#) and overexpression lines (9, 11, 12#) were sown on filter paper with different concentrations of ABA without stratification, and photographed after 2 days. (B) Germination rate of transgenic seeds (35S:GhABI5/abi5-1# and 35S:GhABI5/Col-9#), abi5 and Col. Without ABA, the germination rate is similar among the lines, compared with the different rates resulting from 0.5 μM ABA treatment. A seed with open green cotyledons is considered germinated. (C) GhABI5 positively regulates the expression levels of ABA-responsive genes. The freshly harvested seeds, including complemented transgenic seeds (1, 2, 3#), abi5 and Col, were used for qRT- PCR. Bars represent the SE of three biological replicates. (∗P < 0.05; ∗∗P < 0.01).
To further confirm whether GhABI5 functions as an ABA response transcription factor and has conserved functions similar to AtABI5, we analyzed the genes downstream of AtABI5 in freshly harvested, complemented mature dormant seeds. The results of the qRT-PCR analysis show that transcript levels of EM1, EM6, and RD29B were up-regulated to various degrees in GhABI5 complemented lines (Figure 6C). EM1 is an ABA-inducible protein that accumulates during seed maturation; in the complemented lines, its expression recovered to a level similar to Col, and was 2.3-fold greater than in the abi5-3 mutant. The transcript level of EM6 increased 1.5-fold but could not recover to the wild type level. The expression of RD29B in complemented lines increased 1.9-fold compared with abi5-3. Overall, the transcript levels of genes downstream of AtABI5 were up-regulated in the abi5-3 lines complemented with GhABI5. These results are consistent with the notion that ABI5 positively regulates ABA response genes, and, like AtABI5, GhABI5 has conserved functions in seed dormancy.
To further investigate the role of GhABI5 in corm dormancy, GhABI5 silencing was carried out through VIGS. In order to specifically silence GhABI5, we chose specific 5′ fragments (344 bp) to construct a tobacco rattle virus (TRV)-GhABI5 vector (Figure 7E). Dormant cormels were infiltrated with Agrobacterium harboring TRV-GhABI5. After 10 days of cultivation, TRV1 and TRV2 were detected in the infiltrated cormels by RT-PCR (Figure 7A). We successfully detected the expression of TRV1 and TRV2 in plantlets. Then, we analyzed GhABI5 gene expression in infected plantlets. Compared with the control, silenced plantlets showed low GhABI5 expression, which was approximately 41% (silencing-1), 0.03% (silencing-2), 0.2% (silencing-3), and 22% (silencing-4) of the control’s expression (Figure 7B). Bud length was measured in the control and GhABI5-silenced plantlets. Silenced cormels had longer buds (23.62 ± 4.63 mm), approximately 1.34-fold longer than the buds of control cormels (17.61 ± 1.78 mm) (Figures 7C,D). The results indicate that GhABI5 plays an important role in corm dormancy and inhibits cormel germination.
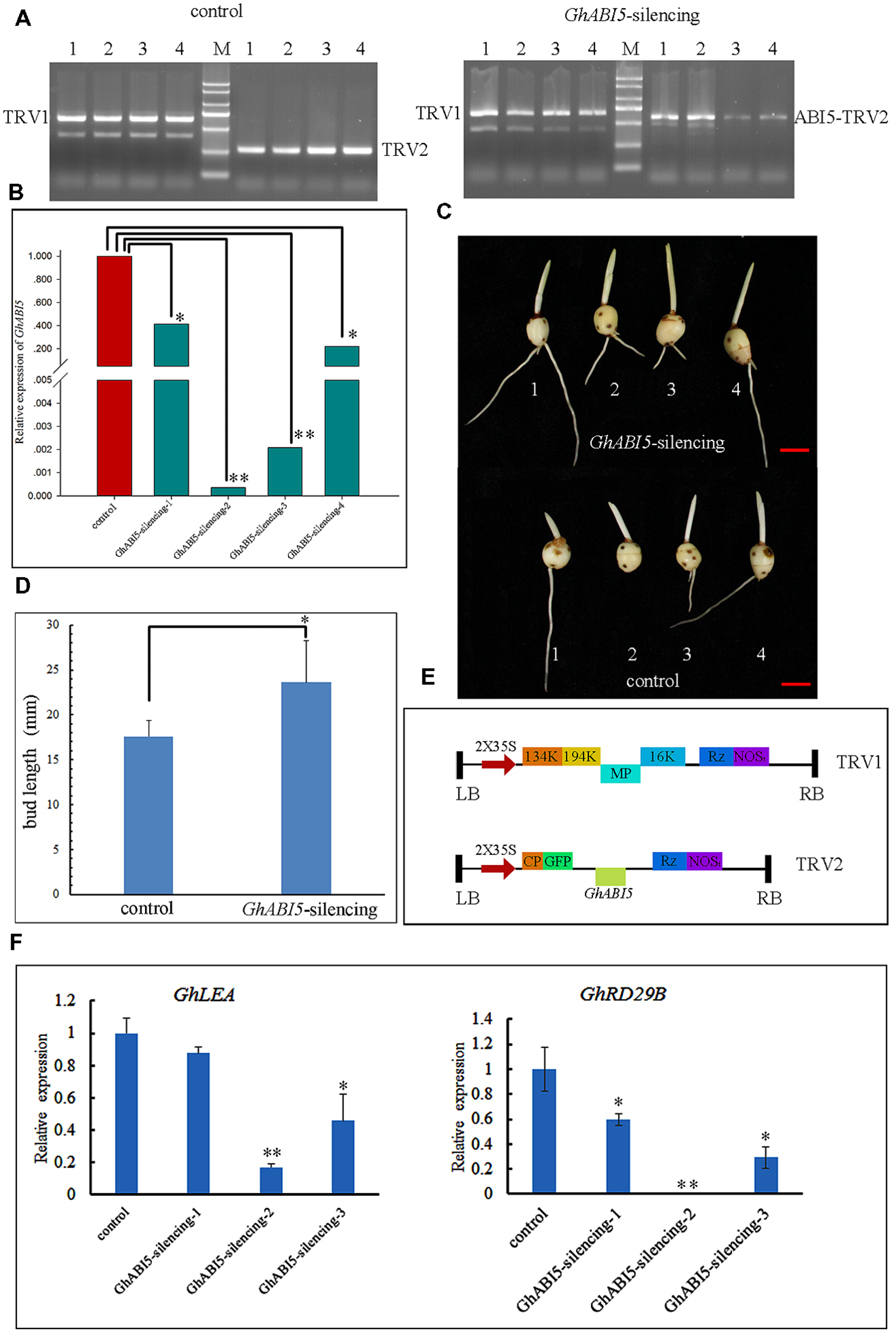
FIGURE 7. Silencing of GhABI5 promotes the germination of dormant cormels. (A) Detection of TRV1 (628 bp), TRV2 (263 bp) and ABI5-TRV2 (607 bp) by RT-PCR in silenced cormels. Left (the control), dormant cormels inoculated with Agrobacterium (TRV1 and TRV2); Right, dormant cormels inoculated with Agrobacterium (TRV1 and ABI5-TRV2). (B) Quantitative real-time PCR analysis of the expression level of GhABI5 in the control and silenced lines. (C) Phenotype of silenced lines and controls. (D) Bud length of the control and silenced cormels (n = 8). (E) Schematic diagram of the TRV1 and TRV2–GFP vectors. (F) The expression of GhLEA and GhRD29B in silencing cormels by Quantitative real-time PCR. (∗P < 0.05; ∗∗P < 0.01).
Gladiolus is one of the most commonly sold cut flowers around the world as well as in China, where Gladiolus sales rank second among bulb flowers (Wu et al., 2014). Gladiolus corms must go through a long period of growth cessation known as endo-dormancy, which is one of the main factors limiting its production value. In addition, endo-dormant corms require refrigeration storage, leading to high costs associated with electricity and labor. Therefore, elucidating the mechanism of corm dormancy will provide a theoretical basis for the production of weakly dormant Gladiolus cultivars through molecular breeding.
Endogenous hormones and other factors are responsible for inhibiting germination of the Gladiolus bud. However, this limitation can be overcome by cold storage, which reduces endogenous ABA content (Figure 1A) and results in earlier germination (Figure 1D). Exogenous ABA treatment maintains corm dormancy and inhibits bud germination, while fluridone promotes sprouting (Figure 2). Fluridone is a carotenoid biosynthesis inhibitor known to prevent ABA biosynthesis. However, it is not an ABA-specific inhibitor; our results show that fluridone inhibits root elongation, which has also been demonstrated in previous research (Hooker and Thorpe, 1998; Spollen et al., 2000). Although GA has proven to be important in seed dormancy, it may play a limited role in corm dormancy (Ram et al., 2002; Finkelstein et al., 2008). GA content increases only in the late dormancy-releasing period and fluctuates irregularly during dormancy breaking (Figure 1B). Previous studies have shown that GA has a limited effect on Gladiolus corm dormancy (Ram et al., 2002). In addition, corm dormancy and seed dormancy share a common physiological characteristic: they are both heavily regulated by ABA (Ahrazem et al., 2012; Finkelstein, 2013). This finding is also consistent with the transcriptome data of bud dormancy in perennials such as peach, leafy spurge and oak; in these plants, bud dormancy is also ABA-regulated (Horvath et al., 2008; Leida et al., 2012; Ueno et al., 2013). In the current study, VIGS was employed to knock down GhABI5 expression. Although a target-specific fragment was chosen for siRNA-mediated GhABI5 knockdown, it is also possible that other bZIP family members in the cormel were indirectly targeted. Therefore, the early sprouting phenotype and down-regulation of downstream gene expression (GhLEA and GhRD29B) in the silenced dormant cormels may be caused by down-regulation of multiple genes, including GhABI5 (Figure 7F). These results suggest that ABA signaling is also involved in the regulation of corm dormancy (a vegetative organ). -= In Arabidopsis, ABI5 is exclusively expressed in the seed (a reproductive organ). More specifically, it functions as an enhancer of seed dormancy as well as a repressor of seedling germination (Lopez-Molina et al., 2001; Garcia et al., 2008). Here, we have found that the expression of its homologous gene in Gladiolus (GhABI5) extends into dormant vegetative organs (dormant cormels, corms. and stolons), where it functions to enhance organ dormancy. Furthermore, GhABI5 is able to complement the ABA insensitivity of abi5-3 in Arabidopsis, can extend dormancy when overexpressed in transgenic lines (Figure 6) and induces the expression of downstream ABA response genes (EM1, EM6, and RD29B) (Figure 6C). It is clear that GhABI5 has characteristics in common with AtABI5, including the induction of an ABA response and dormancy maintenance.
GhABI5 belongs to the bZIP superfamily of transcription factors, since it possesses a typical basic leucine zipper that binds DNA. Sequence analysis revealed that, among all members of the bZIP superfamily, GhABI5 shows the highest sequence similarity to AtABI5 (Figure 3A). Similar to the ABI5s in other species, GhABI5 also has the conserved C1, C2, and C3 domains (Zou et al., 2008; Zhou et al., 2013). The GhABI5 protein is localized in the nucleus and functions as a transcription factor, which is consistent with other homologous ABI5 proteins (Lopez-Molina et al., 2003; Zou et al., 2008; Zhou et al., 2013).
Abscisic acid is able to induce the expression of GhABI5, suggesting that this phytohormone may have an important physiological function in corm development. In addition, both drought treatment (Figure 4D) and corm desiccation (ABA increases during this period, Figure 1A) could increase the expression of GhABI5. Exogenous ABA also significantly enhances the transcription of GhABI5. Previous research has shown that ABA not only increases the expression level of AtABI5 in Arabidopsis seeds, but also stabilizes the AtABI5 protein by relieving SnRK2 repression, which then phosphorylates AtABI5 (Brocard et al., 2002; Feng et al., 2014) and inhibits the E3 ligase KEG (Stone et al., 2006). In contrast, the ABA inhibitor fluridone could reduce the endogenous ABA content in dormant cormels, leading to down-regulation of ABI5 expression (Figure 4E) and earlier germination (Figure 2). A similar situation was apparent following cold treatment (Figures 1A and 4C). Cold storage has proven to be effective in breaking Gladiolus corm dormancy (Ginzburg, 1973). We hypothesize that this might occur through the modulation of ABA levels and ABA-associated signaling pathways within the plant. Long-term cold treatment diminishes endogenous ABA content, resulting in lower expression of GhABI5 as well as a weakened ABA response. However, we noticed that the fluridone-treated cormels had shorter roots compared with the control. This finding is similar to that obtained from Z. mays L. seedlings, in which endogenous ABA levels correlated positively with increased rates of root elongation in most cultivars whose root tips contain a certain amount of ABA (Rivier and Pilet, 1981; Ng and Moore, 1985). We also found that GhABI5 transcript levels are highly abundant in the stamen (reproductive organ). In rice, OsABI5 is expressed in mature pollen and contributes to plant fertility (Zou et al., 2008). In Brassica oleracea, BoABI5 is specifically expressed in the flower (Zhou et al., 2013).
The GhABI5 promoter harbors numerous cis-acting regulatory elements, which mediate plant responses to developmental cues and to stress-related hormones. ABA up-regulates GhABI5 expression, as evidenced by the transient expression of GhABI5:GUS in Gladiolus petals. Several conserved Ebox/ABRE (CANNTG) and DPBF (ACACNNG) motifs are likely responsible for the up-regulation of GhABI5 expression upon ABA treatment (Stalberg et al., 1996; Kim et al., 1997). It is reported that MYB and MYC transcription factors contribute to ABA-related responses such as drought and water stress (Abe et al., 1997, 2003; Cominelli et al., 2005). Therefore, MYBs and MYCs may regulate GhABI5 expression in an ABA-dependent manner during corm dormancy and drought stress. Of particular note, the RY/G box (CATGCA) motif occurs in the GhABI5 promoter sequence (Ezcurra et al., 2000). Previous research has shown that ABI3 specifically binds to the RY/G box motif and up-regulates AtABI5 expression, suggesting that other transcription factors involved in ABA signaling may regulate corm dormancy (Lopez-Molina et al., 2002). Moreover, our study suggests that cold treatment can reduce the expression of GhABI5. The LTRE motif, which is found in the promoter of cold-inducible genes, is present in the promoter of GhABI5; this suggests potential crosstalk between cold and ABA, mediated by GhABI5 (Baker et al., 1994). Future work will focus on the identification of hormone-regulated transcription factors that modulate GhABI5 expression and affect corm dormancy.
Taken together, our results provide novel insight into the molecular mechanisms underlying corm dormancy. This physiological process is regulated through ABA signal transduction, in which GhABI5 has a positive response to ABA and enhances corm dormancy. We suspect that similar cellular and molecular mechanisms exist among various types of dormant organs (seed, modified stem and modified root).
JW and MY conceived and designed the research. MY and JH conducted experiments. JW and SS performed the assays. JS, XL, and BG helped take the photographs. Chen L and CW analyzed data. CL and FZ helped take care of plants. JW, MY, JH, and EV wrote the manuscript. All authors read and approved the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Junping Gao (China Agricultural University) for kindly providing the vectors for the VIGS assay, Ruth R. Finkelstein (University of California–Santa Barbara) for providing abi5-3 mutant seeds, and Dr. Sonia Gazzarrini and Dr. Sheryl Stevenson (University of Toronto Scarborough) for revision. This work is supported by grants from Doctoral Fund of Ministry of Education of China (20120008110006), National Natural Science Foundation projects (31171991), Special Fund for Agro-scientific Research in the Public Interest (200903020), and the Foundation of China Scholarship Council (CSC).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00960
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/Tpc.006130
Abe, H., Yamaguchishinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought– and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. doi: 10.1105/tpc.9.10.1859
Ahrazem, O., Rubio-Moraga, A., Trapero, A., and Gomez-Gomez, L. (2012). Developmental and stress regulation of gene expression for a 9-cis-epoxycarotenoid dioxygenase, CstNCED, isolated from Crocus sativus stigmas. J. Exp. Bot. 63, 681–694. doi: 10.1093/jxb/err293
Anderson, J., Horvath, D., Chao, W., and Foley, M. (2010). “Bud dormancy in perennial plants: a mechanism for survival,” in Dormancy and Resistance in Harsh Environments, eds E. Lubzens, J. Cerda, and M. Clark (Berlin: Springer), 69–90.
Baker, S. S., Wilhelm, K. S., and Thomashow, M. F. (1994). The 5′-region of Arabidopsis thaliana Cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. doi: 10.1007/Bf00029852
Batoko, H., Zheng, H. Q., Hawes, C., and Moore, I. (2000). A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12, 2201–2217. doi: 10.1105/tpc.12.11.2201
Bohlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A. M., Jansson, S., Strauss, S. H., et al. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043. doi: 10.1126/science.1126038
Brocard, I. M., Lynch, T. J., and Finkelstein, R. R. (2002). Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 129, 1533–1543. doi: 10.1104/Pp.005793
Carles, C., Bies-Etheve, N., Aspart, L., Leon-Kloosterziel, K. M., Koornneef, M., Echeverria, M., et al. (2002). Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J. 30, 373–383. doi: 10.1046/j.1365-313X.2002.01295.x
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cominelli, E., Galbiati, M., Vavasseur, A., Conti, L., Sala, T., Vuylsteke, M., et al. (2005). A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 15, 1196–1200. doi: 10.1016/j.cub.2005.05.048
Cooke, J. E., Eriksson, M. E., and Junttila, O. (2012). The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant Cell Environ. 35, 1707–1728. doi: 10.1111/j.1365-3040.2012.02552.x
Dai, M. Q., Xue, Q., Mccray, T., Margavage, K., Chen, F., Lee, J. H., et al. (2013). The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 25, 517–534. doi: 10.1105/tpc.112.105767
Destefano-Beltran, L., Knauber, D., Huckle, L., and Suttle, J. C. (2006). Effects of postharvest storage and dormancy status on ABA content, metabolism, and expression of genes involved in ABA biosynthesis and metabolism in potato tuber tissues. Plant Mol. Biol. 61, 687–697. doi: 10.1007/s11103-006-0042-7
Ezcurra, I., Wycliffe, P., Nehlin, L., Ellerstrom, M., and Rask, L. (2000). Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J. 24, 57–66. doi: 10.1046/j.1365-313x.2000.00857.x
Feng, C. Z., Chen, Y., Wang, C., Kong, Y. H., Wu, W. H., and Chen, Y. F. (2014). Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 80, 654–668. doi: 10.1111/Tpj.12670
Finch-Savage, W. E., and Leubner-Metzger, G. (2006). Seed dormancy and the control of germination. New Phytol. 171, 501–523. doi: 10.1111/j.1469-8137.2006.01787.x
Finkelstein, R. (2013). Abscisic Acid synthesis and response. Arabidopsis Book 11, e0166. doi: 10.1199/tab.0166
Finkelstein, R. R., and Lynch, T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. doi: 10.1105/tpc.12.4.599
Finkelstein, R., Reeves, W., Ariizumi, T., and Steber, C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415. doi: 10.1146/annurev.arplant.59.032607.092740
Garcia, M. E., Lynch, T., Peeters, J., Snowden, C., and Finkelstein, R. (2008). A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol. 67, 643–658. doi: 10.1007/s11103-008-9344-2
Ginzburg, C. (1973). Hormonal regulation of cormel dormancy in Gladiolus-grandiflorus. J. Exp. Bot. 24, 558–566. doi: 10.1093/Jxb/24.3.558
Graeber, K., Nakabayashi, K., Miatton, E., Leubner-Metzger, G., and Soppe, W. J. (2012). Molecular mechanisms of seed dormancy. Plant Cell Environ. 35, 1769–1786. doi: 10.1111/j.1365-3040.2012.02542.x
Gubler, F., Millar, A. A., and Jacobsen, J. V. (2005). Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 8, 183–187. doi: 10.1016/j.pbi.2005.01.011
Heide, O. M. (1986). Effects of ABA application on cessation or shoot elongation in long-day grown Norway spruce seedlings. Tree Physiol. 1, 79–83. doi: 10.1093/treephys/1.1.79
Hooker, T., and Thorpe, T. (1998). Effects of fluridone and abscisic acid on lateral root initiation and root elongation of excised tomato roots cultured in vitro. Plant Cell Tissue Organ Cult. 52, 199–203. doi: 10.1023/A:1006033430898
Horvath, D. (2009). Common mechanisms regulate flowering and dormancy. Plant Sci. 177, 523–531. doi: 10.1016/j.plantsci.2009.09.002
Horvath, D. P., Anderson, J. V., Chao, W. S., and Foley, M. E. (2003). Knowing when to grow: signals regulating bud dormancy. Trends Plant Sci. 8, 534–540. doi: 10.1016/j.tplants.2003.09.013
Horvath, D. P., Chao, W. S., Suttle, J. C., Thimmapuram, J., and Anderson, J. V. (2008). Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genomics 9:536. doi: 10.1186/1471-2164-9-536
Horvath, D. P., Sung, S., Kim, D., Chao, W., and Anderson, J. (2010). Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol. Biol. 73, 169–179. doi: 10.1007/s11103-009-9596-5
Horvath, D. R., Anderson, J. V., Chao, W. S., and Foley, M. E. (2004). Knowing when to grow: signals regulating bud dormancy (vol 8, pg 534, 2003). Trends Plant Sci. 9, 12. doi: 10.1016/j.tplants.2003.11.005
Hubbard, K. E., Nishimura, N., Hitomi, K., Getzoff, E. D., and Schroeder, J. I. (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 24, 1695–1708. doi: 10.1101/Gad.1953910
Jefferson, R. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. doi: 10.1007/BF02667740
Kim, S. Y., Chung, H. J., and Thomas, T. L. (1997). Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J. 11, 1237–1251. doi: 10.1046/j.1365-313X.1997.11061237.x
Leida, C., Conejero, A., Arbona, V., Gomez-Cadenas, A., Llacer, G., Badenes, M. L., et al. (2012). Chilling-dependent release of seed and bud dormancy in peach associates to common changes in gene expression. PLoS ONE 7:35777. doi: 10.1371/journal.pone.0035777
Li, C. Y., Welling, A., Puhakainen, T., Vihera-Aarnio, A., Ernstsen, A., Junttila, O., et al. (2005). Differential responses of silver birch (Betula pendula) ecotypes to short-day photoperiod and low temperature. Tree Physiol. 25, 1563–1569. doi: 10.1093/treephys/25.12.1563
Liu, G., Li, W., Zheng, P., Xu, T., Chen, L., Liu, D., et al. (2012). Transcriptomic analysis of ‘Suli’ pear (Pyrus pyrifolia white pear group) buds during the dormancy by RNA-Seq. BMC Genomics 13:700. doi: 10.1186/1471-2164-13-700
Liu, Y. G., and Chen, Y. (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43, 649–650. doi: 10.2144/000112601
Lopez-Molina, L., Mongrand, B., Mclachlin, D. T., Chait, B. T., and Chua, N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317–328. doi: 10.1046/j.1365-313X.2002.01430.x
Lopez-Molina, L., Mongrand, S., and Chua, N. H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the AB15 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 98, 4782–4787. doi: 10.1073/pnas.081594298
Lopez-Molina, L., Mongrand, S., Kinoshita, N., and Chua, N. H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17, 410–418. doi: 10.1101/Gad.1055803
Melcher, K., Xu, Y., Ng, L. M., Zhou, X. E., Soon, F. F., Chinnusamy, V., et al. (2010). Identification and mechanism of ABA receptor antagonism. Nat. Struct. Mol. Biol. 17, 1102–1108. doi: 10.1038/Nsmb.1887
Millar, A. A., Jacobsen, J. V., Ross, J. J., Helliwell, C. A., Poole, A. T., Scofield, G., et al. (2006). Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8/prime-hydroxylase. Plant J. 45, 942–954. doi: 10.1111/j.1365-313X.2006.02659.x
Miura, K., Lee, J., Jin, J. B., Yoo, C. Y., Miura, T., and Hasegawa, P. M. (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 5418–5423. doi: 10.1073/pnas.0811088106
Ng, Y. K., and Moore, R. (1985). Root growth, secondary root formation and root gravitropism in carotenoid-deficient seedlings of Zea mays L. Ann. Bot. 55, 387–394.
Nonogaki, H. (2014). Seed dormancy and germination-emerging mechanisms and new hypotheses. Front. Plant Sci. 5:233. doi: 10.3389/fpls.2014.00233
Piskurewicz, U., Jikumaru, Y., Kinoshita, N., Nambara, E., Kamiya, Y., and Lopez-Molina, L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745. doi: 10.1105/tpc.108.061515
Ram, R., Mukherjee, D., and Manuja, S. (2002). Plant growth regulators affect the development of both corms and cormels in Gladiolus. HortScience 37, 343–344.
Rentzsch, S., Podzimska, D., Voegele, A., Imbeck, M., Muller, K., Linkies, A., et al. (2012). Dose– and tissue-specific interaction of monoterpenes with the gibberellin-mediated release of potato tuber bud dormancy, sprout growth and induction of alpha-amylases and beta-amylases. Planta 235, 137–151. doi: 10.1007/s00425-011-1501-1
Rivier, L., and Pilet, P. E. (1981). Abscisic acid levels in the root tips of seven Zea mays varieties. Phytochemistry 20, 17–19. doi: 10.1016/0031-9422(81)85209-0
Rodriguez, M. V., Mendiondo, G. M., Maskin, L., Gudesblat, G. E., Iusem, N. D., and Benech-Arnold, R. L. (2009). Expression of ABA signalling genes and ABI5 protein levels in imbibed Sorghum bicolor caryopses with contrasting dormancy and at different developmental stages. Ann. Bot. 104, 975–985. doi: 10.1093/aob/mcp184
Spollen, W. G., Lenoble, M. E., Samuels, T. D., Bernstein, N., and Sharp, R. E. (2000). Abscisic acid accumulation maintains maize primary root elongation at low water potentials by restricting ethylene production. Plant Physiol. 122, 967–976. doi: 10.1104/pp.122.3.967
Stalberg, K., Ellerstom, M., Ezcurra, I., Ablov, S., and Rask, L. (1996). Disruption of an overlapping E-box/ABRE motif abolished high transcription of the napA storage-protein promoter in transgenic Brassica napus seeds. Planta 199, 515–519. doi: 10.1007/BF00195181
Stone, S. L., Williams, L. A., Farmer, L. M., Vierstra, R. D., and Callis, J. (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415–3428. doi: 10.1105/tpc.106.046532
Suzuki, M., Kao, C. Y., Cocciolone, S., and Mccarty, D. R. (2001). Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J. 28, 409–418. doi: 10.1046/j.1365-313X.2001.01165.x
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Ueno, S., Klopp, C., Leple, J. C., Derory, J., Noirot, C., Leger, V., et al. (2013). Transcriptional profiling of bud dormancy induction and release in oak by next-generation sequencing. BMC Genomics 14:236. doi: 10.1186/1471-2164-14-236
Wisniewski, M., Norelli, J., and Artlip, T. (2015). Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant Sci. 6:85. doi: 10.3389/fpls.2015.00085
Wu, J., Liu, C., Seng, S., Khan, M., Sui, J., Gong, B., et al. (2014). Somatic embryogenesis and Agrobacterium-mediated transformation of Gladiolus hybridus cv. ‘Advance Red’. Plant Cell Tissue Organ Cult. 120, 717–728. doi: 10.1007/s11240-014-0639-5
Xu, R.-Y., Niimi, Y., and Han, D.-S. (2006). Changes in endogenous abscisic acid and soluble sugars levels during dormancy-release in bulbs of Lilium rubellum. Sci. Hortic. (Amsterdam) 111, 68–72. doi: 10.1016/j.scienta.2006.08.004
Yang, J. C., Zhang, J. H., Wang, Z. Q., Zhu, Q. S., and Wang, W. (2001). Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol. 127, 315–323. doi: 10.1104/Pp.127.1.315
Yang, X., Yang, Y. N., Xue, L. J., Zou, M. J., Liu, J. Y., Chen, F., et al. (2011). Rice ABI5-Like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes. Plant Physiol. 156, 1397–1409. doi: 10.1104/pp.111.173427
Yang, Y., Ma, Y. Z., Xu, Z. S., Chen, X. M., He, Z. H., Yu, Z., et al. (2007). Isolation and characterization of Viviparous-1 genes in wheat cultivars with distinct ABA sensitivity and pre-harvest sprouting tolerance. J. Exp. Bot. 58, 2863–2871. doi: 10.1093/Jxb/Erm073
Zheng, C., Halaly, T., Acheampong, A. K., Takebayashi, Y., Jikumaru, Y., Kamiya, Y., et al. (2015). Abscisic acid (ABA) regulates grape bud dormancy, and dormancy release stimuli may act through modification of ABA metabolism. J. Exp. Bot. 66, 1527–1542. doi: 10.1093/jxb/eru519
Zhong, X. H., Yuan, X., Wu, Z., Khan, M. A., Chen, J., Li, X. X., et al. (2014). Virus-induced gene silencing for comparative functional studies in Gladiolus hybridus. Plant Cell Rep. 33, 301–312. doi: 10.1007/s00299-013-1530-2
Zhou, X. N., Yuan, F. F., Wang, M. Y., Guo, A. G., Zhang, Y. F., and Xie, C. G. (2013). Molecular characterization of an ABA insensitive 5 orthologue in Brassica oleracea. Biochem. Biophys. Res. Commun. 430, 1140–1146. doi: 10.1016/j.bbrc.2012.12.023
Keywords: Gladiolus hybridus, corm, dormancy, GhABI5, ABA
Citation: Wu J, Seng S, Sui J, Vonapartis E, Luo X, Gong B, Liu C, Wu C, Liu C, Zhang F, He J and Yi M (2015) Gladiolus hybridus ABSCISIC ACID INSENSITIVE 5 (GhABI5) is an important transcription factor in ABA signaling that can enhance Gladiolus corm dormancy and Arabidopsis seed dormancy. Front. Plant Sci. 6:960. doi: 10.3389/fpls.2015.00960
Received: 28 July 2015; Accepted: 21 October 2015;
Published: 03 November 2015.
Edited by:
Jens Rohloff, Norwegian University of Science and Technology, NorwayReviewed by:
Richard G. H. Immink, Plant Research International, NetherlandsCopyright © 2015 Wu, Seng, Sui, Vonapartis, Luo, Gong, Liu, Wu, Liu, Zhang, He and Yi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junna He, aGVqdW5uYUBjYXUuZWR1LmNu; Mingfang Yi, eW1mYW5nQGNhdS5lZHUuY24=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.