
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 03 November 2015
Sec. Plant Physiology
Volume 6 - 2015 | https://doi.org/10.3389/fpls.2015.00944
This article is part of the Research Topic Fructans and RFOs in plants View all 22 articles
Global meat and milk production depends to a large extent on grazed pastures, with Lolium perenne being the major forage grass in temperate regions. Defoliation and subsequent regrowth of leaf blades is a major and essential event with respect to L. perenne growth and productivity. Following defoliation, carbohydrates (mainly fructans and sucrose) have to be mobilized from heterotrophic tissues to provide energy and carbon for regrowth of photosynthetic tissues. This mobilization of reserve carbohydrates requires a substantial change in the expression of genes coding for enzymes involved in carbohydrate metabolism. Here we tested the hypothesis that gibberellins (GA) are at the core of the processes regulating the expression of these genes. Thus, we examined the transcript profiles of genes involved in carbohydrate and GA metabolic pathways across a time course regrowth experiment. Our results show that following defoliation, the immediate reduction of carbohydrate concentrations in growing tissues is associated with a concomitant increase in the expression of genes encoding carbohydrate mobilizing invertases, and was also associated with a strong decrease in the expression of fructan synthesizing fructosyltransferase genes. We also show that the decrease in fructan levels is preceded by increased expression of the GA activating gene GA3-oxidase and decreased expression of the GA inactivating gene GA2-oxidase in sheaths. GA3-oxidase expression was negatively, while GA2-oxidase positively linked to sucrose concentrations. This study provides indicative evidence that gibberellins might play a role in L. perenne regrowth following defoliation and we hypothesize that there is a link between gibberellin regulation and sugar metabolism in L. perenne.
The global consumption of animal derived food (meat and milk) is projected to increase considerably in the coming decades, driven mainly by population growth, economic growth and urbanization in China and Southeast Asia (FAO, 2011). Currently, more than 60% of the global meat production and 90% of milk production is produced either exclusively on grazed pastures, or in mixed systems with a large proportion of grazed and conserved (silage) pasture. This high demand for pasture dry matter production is met by increased intensification with a concomitant increase in, mainly nitrogen (N), inputs. These additional inputs come at a high cost to the environment due to high levels of N losses either leached as nitrate and polluting waterways, or emitted from soils as nitrous oxide, a greenhouse gas almost 300 times more potent than carbon dioxide (Kebreab et al., 2001; Tas, 2007; Ellis et al., 2011; van Groenigen et al., 2011). The root cause of these high N losses from grazed pastures is the discrepancy between the relatively low ruminants' dietary N requirements for optimal performance (around 2.5% N) and the plants' requirements for very high N concentrations in photosynthetic tissues (4.5–5% N) for maximal photosynthetic capacity (Van Soest, 1982; Woledge and Pearse, 1985). Overcoming this dilemma would require the development of plants with an improved ability to grow at low N supply and a higher C/N ratio in photosynthetic active tissues, and we proposed previously that a prerequisite for this might be to change a plant's growth “strategy” (Parsons et al., 2011, 2013a,b).
Perennial ryegrass (Lolium perenne L.) is a cool-season grass and the main pasture plant in temperate zones such as Northern Europe and New Zealand (Parsons and Chapman, 2000). So far, breeding has made little progress in improving ryegrass yield at low N inputs (Parsons et al., 2011). Heterotrophic tissues in the meristematic cell division zone and the enclosed elongation zone of immature blades of this grass species generally depend on the supply of newly fixed carbon from autotrophic mature blades to support growth (Morvan-Bertrand et al., 1999; Parsons et al., 2013a). Regular defoliation of ryegrass by grazing animals or mowing removes most of the photosynthetic tissues capable of fixing CO2 and deprives the growing tissues of sugars. This depletion of sugars induces a shift from carbon storage [mainly high molecular weight (HMW) fructans in ryegrass] to low molecular weight (LMW) sugars to support rapid elongation of enclosed immature blades to form new photosynthetic tissue, a prerequisite for continued plant growth (Schnyder et al., 1990; Morvan-Bertrand et al., 1999, 2001; Schnyder and de Visser, 1999). These processes require a substantial change in the expression of genes encoding enzymes involved in carbohydrate mobilization and accumulation (Morvan-Bertrand et al., 1999, 2001; Lee et al., 2011). Sucrose mobilization is catalyzed by the action of invertases, which are enzymes that cleave sucrose into glucose and fructose, and different forms of invertases, including cell wall (CWInv), cytoplasmic (CytInv), and vacuolar invertase (VacInv), are distributed between cellular compartments (Kingston-Smith et al., 1999; Cairns and Gallagher, 2004). Fructan mobilization is catalyzed by fructan exohydrolases (FEHs), in L. perenne two FEHs have been characterized so far, 1-FEH (Lothier et al., 2007) and 6-FEH (Lothier et al., 2014).
Plant hormones have been shown to regulate changes from the accumulation to mobilization of carbohydrates, and vice versa (Morvan et al., 1997; Perata et al., 1997; Morvan-Bertrand et al., 2001; Gibson, 2004; Ranwala and Miller, 2008; Eveland and Jackson, 2012; Matsoukas, 2014). The important role of one of these classes of hormones, the gibberellins (GAs), for vegetative growth in cereals is obvious by the success of the “green revolution” which produced wheat, rice and other cereal cultivars with stunted growth beneficial for grain production by selecting mutants in the GA biosynthetic pathway or impaired in GA reception (Peng et al., 1999). GAs are plant hormones that promote stem and leaf growth and also activate dormant enzyme systems (see reviews by Yamaguchi, 2008; Colebrook et al., 2014). GA3-oxidase and GA20-oxidase are both genes encoding enzymes which activate gibberellin precursors in the GA biosynthetic pathway, while GA2-oxidase codes for a GA-inactivating enzyme (Yamaguchi, 2008), while DELLA is a protein that negatively regulates plant growth (Achard and Genschik, 2009). The role of GAs as mediators of environmental stimuli is well established (Gocal et al., 1999; MacMillan et al., 2005; Yamaguchi, 2008) and a number of reports have indicated that the GA-mediated elongation of shoots in various plants occurs as a result of an increase in cell division (Sauter et al., 1995) and cell elongation (Smith et al., 1996; Matsukura et al., 1998). Gibberellins are regulated by nutrient levels, particularly low soluble sugar levels appear to activate and/or induce GAs; these activated GAs subsequently induce the expression of a range of hydrolases involved in the enzymatic degradation and mobilization of storage macromolecules, for example starch and proteins to provide substrates for growth (Bewley and Black, 1985; Gibson, 2004; Hong et al., 2012; Paparelli et al., 2013).
Active GA3 has been demonstrated to be present in wheat (Triticum aestivum) and other cereals, but it is absent from the vegetative tissues of the dicots Arabidopsis thaliana and pea (Pisum sativum; Appleford et al., 2006), indicating that the types of active GAs vary in plant species and may respond to altering environmental stimuli (e.g., defoliation) differentially. Morvan et al. (1997) showed that defoliation induces a strong and rapid increase in fructan exohydrolase (FEH, EC 3.2.1.80) activity and a reduction of fructans in the immature elongating leaf bases of L. perenne. The increased activity of FEH was strongly inhibited by an inhibitor of GA biosynthesis, uniconazole, and this effect was reversed by treatment with active GA3, suggesting that the increased FEH activity following defoliation was mediated by the biosynthesis of GA3. However, neither in a subsequent study by the same researchers (Morvan-Bertrand et al., 2001), nor, to our knowledge, in any other studies has GA3 ever been reported to be present in L. perenne, even though it has been shown to induce stem and leaf elongation when exogenously applied (MacMillan et al., 2005; Parsons et al., 2013a). The above evidence indicates that GAs may be core to regulating vegetative growth of L. perenne in a tight interplay with carbohydrate metabolism, but little other information is available about the situation in forage grasses. Since GA biosynthesis is regulated by GA-related genes (see reviews by Yamaguchi, 2008; Colebrook et al., 2014), it is worth pursuing further evidence to test if GA and carbohydrate associated genes are differentially regulated during regrowth following defoliation in L. perenne. In an earlier experiment using exogenous gibberellin (GA3) we demonstrated that it is possible to increase the growth of L. perenne without increasing N supply and we suggested that GA3 might be at the core of limiting ryegrass growth at a given N availability (Parsons et al., 2013a). Here, we undertook to investigate a possible role of GA biosynthesis during regrowth of perennial ryegrass, and report on a time-course experiment in which carbohydrate concentrations in plant tissues and transcript profiles of sugar- and GA-related genes were analyzed during plant regrowth following defoliation.
Seeds of the L. perenne cultivar “Expo” (PGG Wrightson Seeds Ltd., NZ) were germinated and grown for 2 months in 9 × 9 × 18 (d × w × h) cm pots (one seedling per pot) containing commercial potting mixture (Midland Horticulture, Palmerston North NZ) before the study commenced. Plants were randomly distributed and grown in a glasshouse with supplementary lighting and heating (~17–22°C at day; 9–13°C at night) to ensure good plant growth. During this pre-study period, plants were cut back every 4 weeks to 6 cm above soil level and watered every second day to “pot field capacity.” No additional nutrients were applied during the period of the experiment.
The critical experiment commenced in July 2010. Two-month old plants (40 plants in total) were cut back to 6 cm above soil level, which removed nearly all emerged leaf material, see Figure 1. At this first time point, T0, and on seven subsequent occasions after 2, 4, 24, 48, 72, 168, and 336 h (h) regrowth, five individual plants (as experimental replicates) were harvested at the soil level; dead tissues removed; and plant shoots dissected into three categories of tissues (Figure 1): sheath tissues (S), elongating enclosed leaf tissues (EE), and emerged leaf tissue (EM). The latter included all tissue that had emerged beyond the (initially) cutting level, or (later) all tissue that had emerged beyond the ligule of the youngest ligulated leaf, if one was present. Plants harvested at 0 h represent “before defoliation controls.” The harvested tissues were immediately flash-frozen and ground in a pestle and mortar under liquid nitrogen. A portion of the powdered samples was freeze-dried for carbohydrate analysis, and the rest stored at −80°C for subsequent RNA isolation.
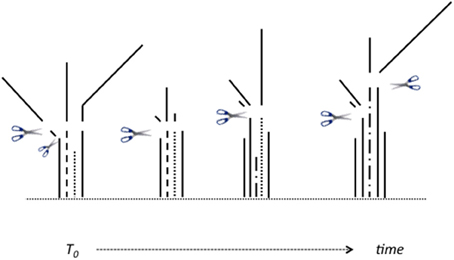
Figure 1. Schematic of how harvested plant parts were allocated to three categories of tissues, as plants regrew following defoliation. Tissues were divided between: emerged leaf tissues (“EM”); enclosed and elongating leaf tissues (“EE”), and sheaths (“S”) in text and graphs. Emerged tissue “EM” refers to all tissues removed above the initial 6 cm cutting height (e.g., at T0), and subsequently all emerged tissues either above that same height, or above/beyond the ligule of any ligulated leaves, as these became present (see solid lines above/beyond scissor icons). The fully enclosed bases of unligulated, and still elongating, leaves (“EE,” see broken lines in Figure) were dissected out from the enclosing older sheaths. The remaining tissue is sheath, “S,” being the non-elongating bases of older ligulated leaves (see solid lines below scissor icons). See Materials and Methods for further explanation/basis for this approach.
Water soluble carbohydrates (WSCs) were extracted and quantified as described previously (Hunt et al., 2005). Powdered, freeze-dried plant material (25 mg) was extracted with 2 ml 80% ethanol for the LMW WSC fraction and subsequently with 2 ml water for the HMW WSC fraction. The ethanol extracted LMW WSC fraction contains a mixture of glucose, fructose, sucrose and low DP (degree of polymerization) fructans and the water extracted HMW WSC fraction contains mainly high DP fructans (Rasmussen et al., 2008). Extracts were briefly centrifuged and WSCs determined in the supernatants using anthrone as a colorimetric reagent as described previously (Jermyn, 1956). Glucose, fructose, and sucrose concentrations were determined enzymatically in the LMW WSC extracts as described (Rasmussen et al., 2007).
During this study, we isolated two full length L. perenne mRNA sequences encoding candidates for a GA3-oxidase (LpGA3ox) and a key growth regulator, DELLA (LpDELLA). The corresponding gene sequences have been deposited in NCBI Genebank as accession numbers KP954695 (LpGA3ox) and KP954694 (LpDELLA) respectively.
To isolate the above GA related genes, we constructed L. perenne RACE cDNA libraries using plant shoot tissues (sheaths plus blades), following the manufacturer's recommendations (SMART™ RACE cDNA Amplification Kit, Clontech, Norrie Biotech, Auckland, NZ). Specific primers (Supplementary Table 1) for the amplification of 3′- and 5′-fragments were designed from the conserved regions of gene sequences from wheat, barley and rice (QD118250, AB189152, and AB054083 for LpGA3ox; AJ242531, AF460219, and AB262980 for LpDELLA). The 5′- and 3′- fragments of candidate genes were amplified using the Advantage 2 PCR Kit (Clontech, Norrie Biotech, Auckland, NZ). PCR products were cloned (TOPO TA Cloning kit, Invitrogen NZ Ltd, Auckland, NZ) and sequenced at the Allan Wilson DNA Centre, Massey University, Palmerston North, NZ. Full length cDNA sequences were amplified using primers designed from the termini of the target cDNA sequences and cloned and sequenced as described above.
Bioinformatic analyses of the sequences were performed with the software programmes BlastP (http://www.ncbi.nlm.nih.gov/) and ClustalO (http://www.ebi.ac.uk/). The secondary protein structure was determined using a PROSITE programme of Expasy (http://www.expasy.ch/prosite).
Total RNA was isolated from plant material using TRIzol Reagent (Invitrogen NZ Ltd., Auckland, NZ) and treated with RNase-free DNase I (Roche NZ Ltd., Auckland, NZ) to remove residual genomic DNA. DNase treated RNA was subsequently purified using the RNeasy Plant Mini Spin Kit (Qiagen, Biostrategy Ltd., Auckland, NZ) to remove enzymes, salts and degraded DNA fragments. RNA quality and integrity was checked by 1% agarose gel electrophoresis, and the absence of genomic DNA confirmed by PCR prior to reverse transcription. RNA was reverse transcribed and converted into cDNA using the SuperScript®VILO™ cDNA Synthesis Kit (Invitrogen NZ Ltd., Auckland, NZ) following the manufacturer's instructions. Synthesized cDNAs were then diluted 50-fold; and 5 μl diluted cDNA was used for subsequent qPCR analysis in a total PCR reaction volume of 20 μl.
Transcript profiling of L. perenne genes encoding the fructosyltransferases (Lp1-SST (sucrose: sucrose 1-fructosyltransferase, Acc#AY245431; Chalmers et al., 2003) and Lp6-SFT (sucrose: fructan 6-fructosyltransferase, Acc#AB186920; Lasseur et al., 2011), a fructan exohydrolase (Lp1-FEH, Acc#AY693396), invertases including a vacuolar invertase (LpVacInv, Acc#AY082350), cell wall invertase (LpCWInv, Acc#DQ073969) and cytosolic invertase (LpCytInv, Acc#AM489692), gibberellin synthesizing genes [LpGA3ox (LpGA3-oxidase, Acc# KP954695) and LpGA20ox (LpGA20-oxidase, Acc#DQ071620)], and gibberellin inactivating genes [LpGA2ox (LpGA2-oxidase, Acc#EF687858) and LpDELLA, Acc# KP954694] were quantified by qPCR using a 96-well LightCycler® 480 II system (Roche Diagnostics NZ Ltd., Auckland, NZ). qPCRs were assayed using LightCycler® 480 SYBR Green I Master mix following the manufacturers' protocols (Roche Diagnostics NZ Ltd., Auckland, NZ). For PCR amplification, after an initial 5 min at 95°C, a total of 45 cycles of 10 s at 95°C; 10 s at 60°C; and 10 s at 72°C were performed. The primers for qPCR were designed based either on sequences in the NCBI Genbank (http://www.ncbi.nlm.nih.gov/) or on the isolated genes from our L. perenne RACE cDNA libraries (Supplementary Table 1). Three technical replicates were analyzed for each time point and tissue.
Attempts to normalize the transcript data against the means of two L. perenne housekeeping genes [elongation factor (LpEF1α) and ubiquitin (LpUBQ)] previously reported to be stably expressed (Martin et al., 2008; Lee et al., 2010; Huang et al., 2014) were unsuccessful as neither of the reference genes were found to be stable in our experimental conditions (see Results). Consequently, an mRNA fragment encoding enhanced Green Fluorescent Protein (eGFP) was artificially synthesized and used as an external reference gene as described previously (Liu and Slininger, 2007; Ellefsen et al., 2008; Rehrig et al., 2011). In the present study, 2 pg eGFP RNA were spiked into 1 μg of sample RNA prior to cDNA synthesis. The gene transcript profiles were normalized against eGFP transcripts, and the results are presented as the ratio of target gene copies to eGFP copies. Copy numbers were calibrated against the corresponding standard plasmid DNA.
The experiment comprised five biological replicates and eight time points (one pre-defoliation and seven post-defoliation), i.e., a total of 40 plants. Three tissues were analyzed at each time point to give a total of 120 samples. All analyses were conducted by One-way ANOVA to exploit significant differences in individual tissues between the regrowth time points, using Minitab statistical software version 16. We used Box-Cox transformation to homogenize the error variances and, where appropriate, a Tukey's honestly significant different (HSD) mean-separation test was used to help interpret significant effects. We report here untransformed means as a measure of data dispersion in response to treatment effects with the regrowth times and also calculated standard errors (SE) using Minitab (v.16.22), which are shown as bars in Figures 2–5. The results of the Tukey's HSD tests are presented in Table 1.
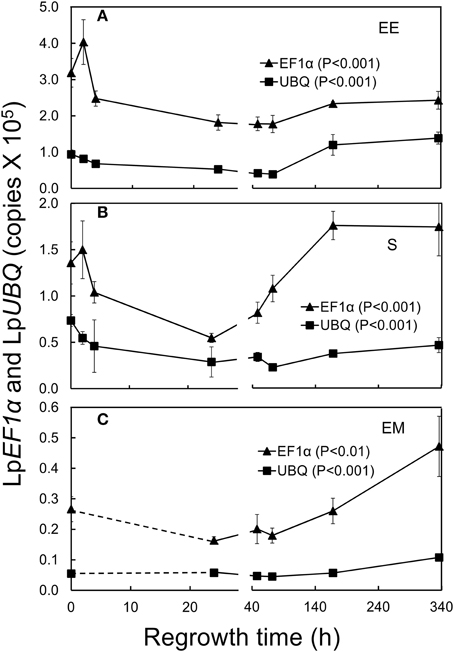
Figure 2. Changes in elongation factor (LpEF1α; triangles) and ubiquitin (LpUBQ; squares) gene expression in L. perenne enclosed elongating leaf tissues (EE; A), sheaths (S; B), and emerged leaf tissues (EM; C) after cutting and at subsequent regrowth intervals. Bars indicate standard errors. For details of Tukey HSD tests see Table 1.
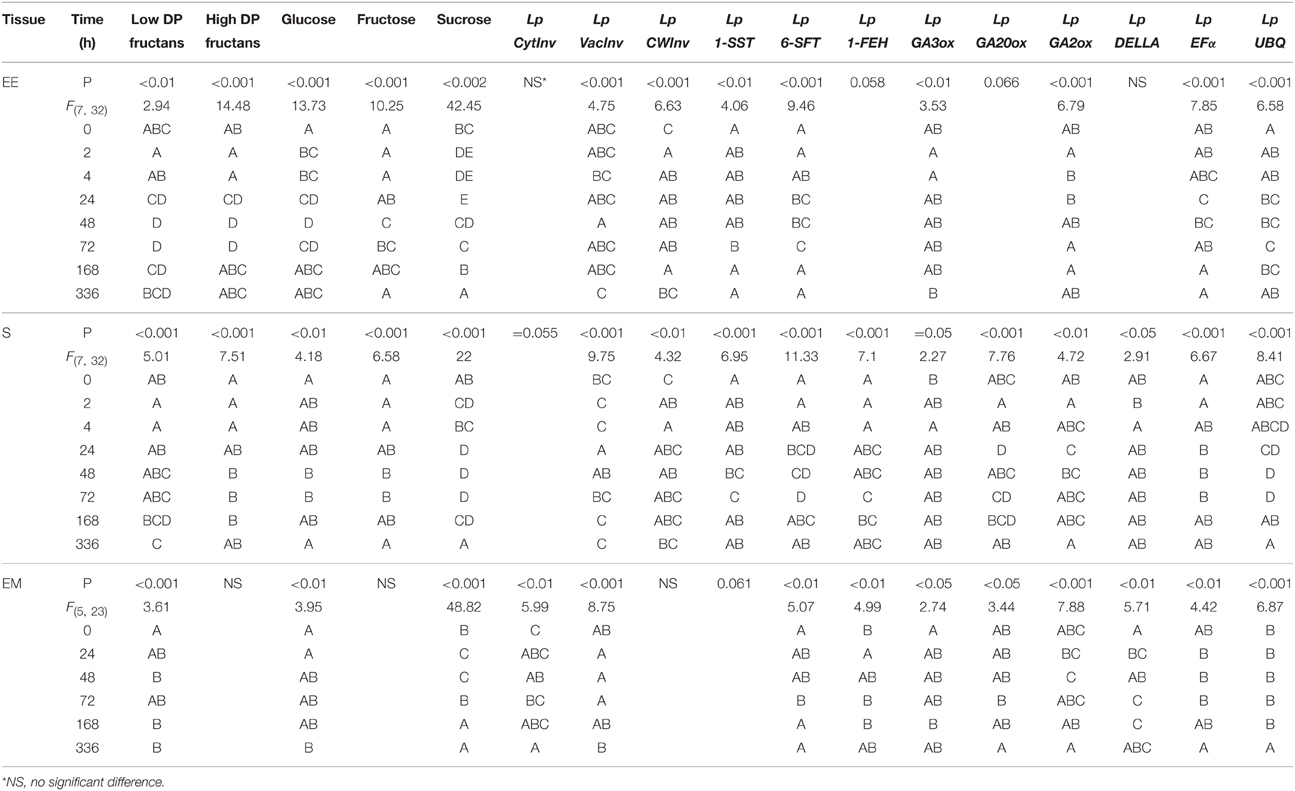
Table 1. Summary of ANOVA results and Tukey honestly significant different tests (different letters represent significant differences).
The regression analysis was conducted using Minitab (v.16.22) Fitted-Line-Plot.
The putative LpGA3-oxidase (LpGA3ox) mRNA contains an open reading frame (ORF) of 359 amino acids (Supplementary Figure 1), and the deduced amino acid sequence suggests that it belongs to a family of 2-oxoglutarate-dependent dioxygenases which contain conserved sequences, including two histidine residues and an aspartic acid at the cofactor binding sites. The L. perenne amino acid sequence shows greatest similarity to GA3-oxidases from barley, Hordeum vulgare (83%; Spielmeyer et al., 2004), wheat, T. aestivum (85%; Appleford et al., 2006), and rice, Oryza sativa (77%; Itoh et al., 2001).
LpDELLA encodes a putative GRAS-type transcriptional regulator. The sequence contains an ORF of 612 amino acids which includes the DELLA and TVHYNP motifs at the N-terminal end and a SAW motif at the C-terminal end (Supplementary Figure 2). Alignment of the LpDELLA amino acid sequence with DELLAs from rice (80%; Itoh et al., 2002), wheat (85%; Peng et al., 1999), and barley (84%; Chandler et al., 2002) shows sequence similarity is highly conserved at the C terminal end, while considerable sequence variation occurs at the N-terminus.
The expression of LpEF1α and LpUBQ has been shown previously to be relatively stable across a variety of experimental conditions in L perenne (Martin et al., 2008; Lee et al., 2010; Huang et al., 2014). Consequently, these genes are often used to “normalize” the expression of target genes in L. perenne, i.e., to correct mainly for inefficiencies in cDNA synthesis caused by reverse transcriptase inhibitors possibly present in the sample matrix. Our analysis of the transcript levels of these two genes clearly shows that the expression of LpEF1α and, to a lesser extent, LpUBQ was significantly affected by defoliation resulting in a strong decline in the expression of both of these genes at the early stages of regrowth (Figure 2; Table 1). We therefore normalized the transcript levels of our target genes below against an external control mRNA (eGFP) which was spiked into our quantified RNA samples essentially following the rationale discussed by Rehrig et al. (2011).
Please note that, to make immediate changes (0–24 h post-defoliation) in transcript levels and carbohydrate concentrations better visible, we have split the x-axes in Figures 2–5 into two parts representing 0–24 h and 48–336 h post-defoliation, respectively. Please also note here and in the following that the first two time points (2 and 4 h) after defoliation could not be analyzed in emerged leaf tissues due to a lack of material.
Transcript levels of LpGA3ox increased immediately after defoliation in sheaths (Figure 3B; Table 1). In emerged leaf tissues LpGA3ox was not significantly different compared to pre-defoliation levels except 1 week (168 h) after defoliation (Figure 3C; Table 1).
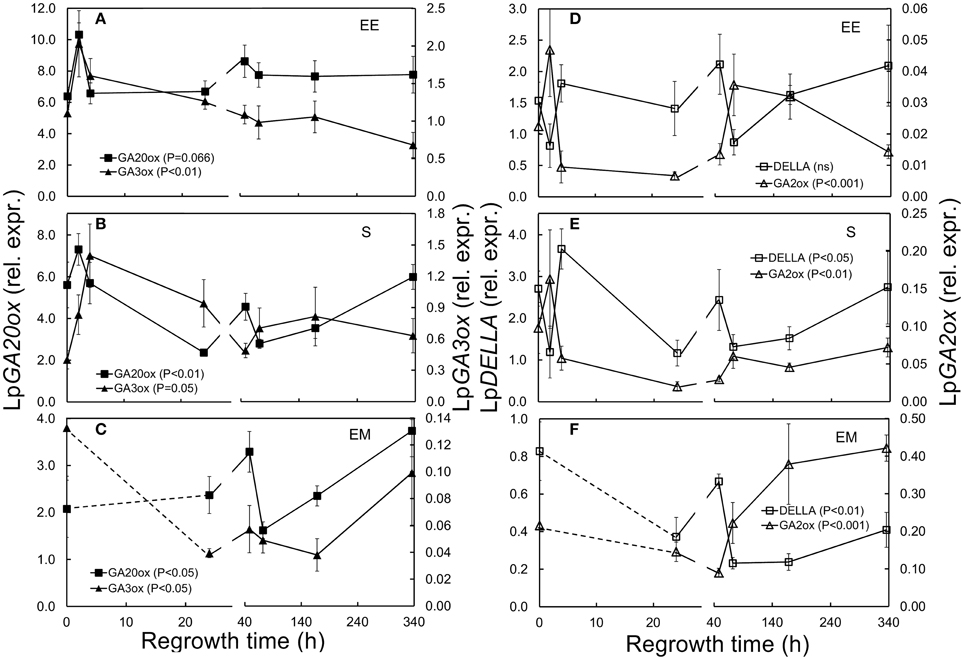
Figure 3. Expression of GA related genes in L. perenne enclosed elongating leaf tissues (EE; A,D), sheaths (S; B,E), and emerged leaf tissues (EM; C,F) after cutting and at subsequent regrowth intervals. Panels on the left show the relative expression of the GA synthesizing LpGA3ox (filled triangles) and LpGA20ox (filled squares). Panels on the right show the relative expression of the GA inactivating LpGA2ox (open triangles) and the GA regulator LpDELLA (open squares). Gene transcript levels were normalized relative to a spiked external reference gene eGFP and values presented here have been enlarged 100-fold.
Transcript levels of both LpGA20ox and LpGA2ox were significantly lower in sheaths 24 h after defoliation (0.4-fold LpGA20ox and 0.2-fold LpGA2ox, Table 1). Neither LpGA20ox nor LpGA2ox expression was significantly affected in enclosed elongating and emerged leaf tissues by defoliation (Figures 3C,F).
LpDELLA transcript levels were not significantly affected in enclosed elongating leaf tissues, while in sheaths LpDELLA levels were significantly higher 4 h compared to 2 h after defoliation (Figure 3E; Table 1). In emerged leaf tissues, LpDELLA expression was significantly lower at 24, 72, and 168 h after defoliation (Figure 3F; Table 1).
Overall, sucrose concentrations were lower compared to glucose and fructose in enclosed elongating leaf tissues, while they were higher compared to these monomeric sugars in both mature sheaths and emerged leaf tissues, with very high levels in the latter tissue (Figures 4A–C). Compared to sugar concentrations prior to defoliation, sucrose concentrations dropped sharply in the first 2 h after defoliation (P < 0.01) in enclosed elongating leaf tissues and sheaths (Figures 4A,B; Table 1) reaching lowest levels after 24 h. These low sucrose concentrations were also seen in emerged leaf tissues 24 and 48 h after defoliation (Figure 4C). Glucose concentrations were also significantly lower already 2 h after defoliation, but only in enclosed elongating leaf tissues (Figure 4A), while they dropped only slightly in sheaths (Figure 4B). Lowest glucose levels were observed in both these tissues 48 h after defoliation. The concentration of fructose in enclosed elongating leaf tissues and sheaths remained at pre-defoliation levels in the first 24 h and dropped to their lowest levels 48 h after defoliation (Figures 4A,B), while they were not significantly affected in emerged leaf tissues (Figure 4C).
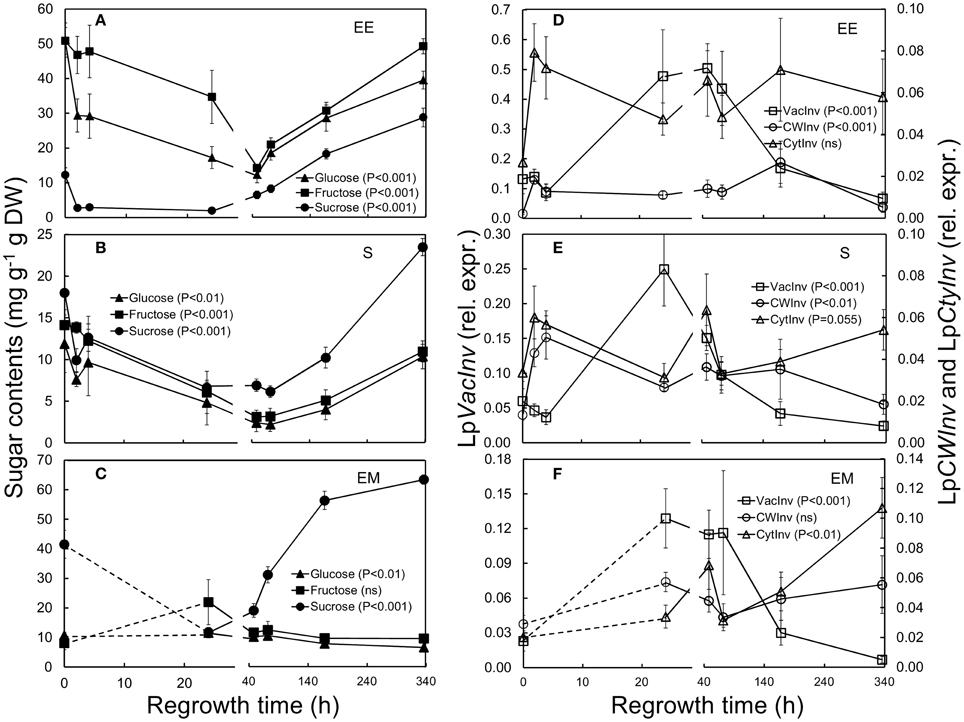
Figure 4. Sugar concentrations (left panels) and relative expression of invertases (right panels) in L. perenne enclosed elongating leaf tissues (EE; A,D), sheaths (S; B,E) and emerged leaf tissues (EM; C,F) after cutting and at subsequent regrowth intervals. Panels on the left show the concentrations of glucose (filled triangles), fructose (filled squares) and sucrose (filled cycles). Panels on the right show the relative expression of LpVacInv (open squares), LpCWInv (open circles), and LpCytInv (open triangles). Gene transcript levels were normalized relative to a spiked external reference gene eGFP.
The expression of the vacuolar invertase gene (LpVacInv) increased significantly in sheaths 24 h after defoliation (Figures 4D–F; Table 1). The expression of LpVacInv was logarithmically and negatively linked to sucrose concentrations for all tissues tested across the regrowth period [sucrose = −6.53 log(LpVacInv) + 0.3921, R2 = 0.316, P < 0.001; Supplementary Figure 3A].
An increase in LpCWInv expression was also observed and this occurred already 2 h after cutting in enclosed elongating leaf tissues and sheaths (P < 0.01, Figures 4D,E; Table 1), while no difference in emerged leaf tissues was detected. In contrast, increased expression of the cytosolic invertase (LpCytInv) was also detected, but only in emerged leaf tissues 48 h following defoliation (P < 0.01, Figure 4F; Table 1).
Interestingly, in this study a significant negative association was seen between sucrose concentrations and the expression of the GA-activating LpGA3ox [sucrose = −7.806 log(LpGA3ox) −24.931, R2 = 0.408, P < 0.001; Supplementary Figure 3B]; and a positive association between sucrose and the GA-inactivating LpGA2ox [sucrose = −983,886(LpGA2ox)2 + 14,371(LpGA2ox) + 6.5402, R2 = 0.574, P < 0.001] was observed for all tested tissues (Supplementary Figure 3C).
WSC concentrations were investigated by determining LMW [mixture of low degree of polymerisation (DP) fructans, sucrose, glucose, and fructose] and HMW WSC (high DP fructans) fractions using anthrone as the detecting reagent. To deduce the concentration of low DP fructans, the sum of glucose, fructose, and sucrose (quantified by enzymatic analysis, Rasmussen et al., 2008) was subtracted from the total LMW WSC concentration. Overall low DP fructans were lower compared to high DP fructans in all tissues analyzed (Figures 5A–C).
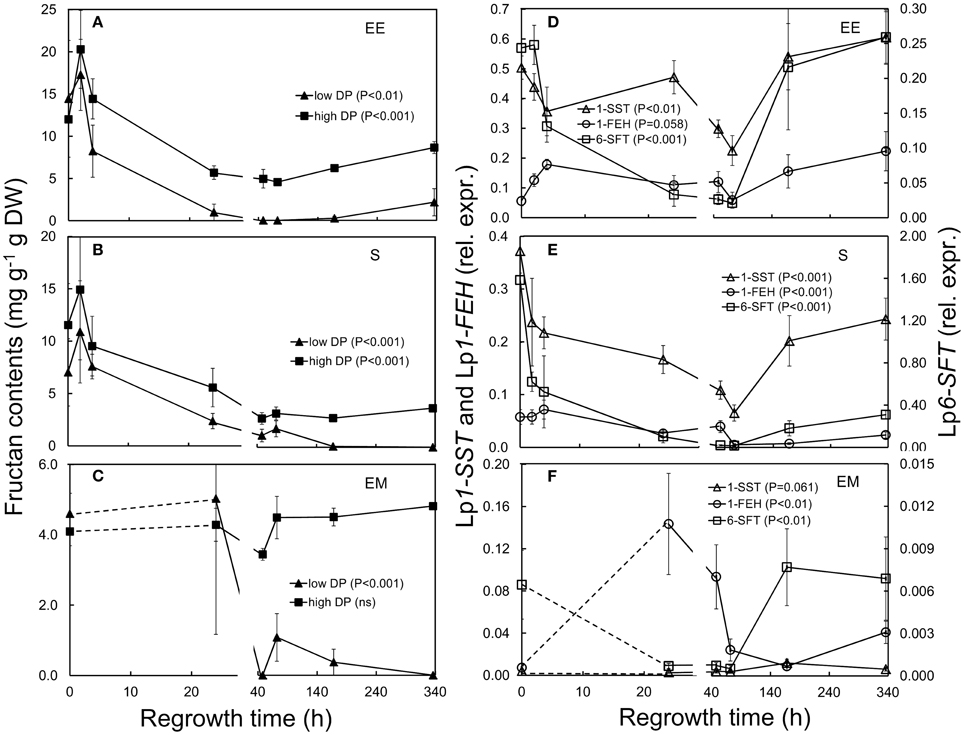
Figure 5. Fructan concentrations (left panels) and relative expression of fructan-regulating genes (right panels) in L. perenne enclosed elongating leaf tissues (EE; A,D), sheaths (S; B,E), and emerged leaf tissues (EM; C,F) after defoliation and at subsequent regrowth intervals. Panels on the left show the concentration of low DP fructans (filled triangles) and high DP fructans (filled squares). Panels on the right show the relative expression of Lp1-SST (open triangles), Lp6-SFT (open squares) and Lp1-FEH (open circles). Gene transcript levels were normalized relative to a spiked external reference gene eGFP.
In enclosed elongating leaf tissues concentrations of low and high DP fructans decreased significantly (low DP fructans 48–72 h; high DP fructans 24–72 h) after defoliation, and reached pre-defoliation levels after a week (168 h, Figure 5A; Table 1). In sheaths, high DP fructans were significantly lower 48–168 h post-defoliation (Figure 5B; Table 1). In emerged leaf tissues concentrations of low DP fructans also decreased significantly 48 h after defoliation (Figure 5C; Table 1) and remained low during the course of the experiment. Concentrations of high DP fructans were not significantly affected by defoliation in this tissue.
In association with the changes in fructans following defoliation, transcript levels of fructan synthesizing Lp1-SST and Lp6-SFT, were significantly reduced in enclosed elongating leaf tissues (P < 0.01, Figure 5D; Table 1) and sheaths (P < 0.001, Figure 5E; Table 1). In both tissues expression of Lp1-SST and Lp6-SFT was lowest 72 h after cutting, then generally increased to the levels seen prior to defoliation after 1 week. Significantly decreased Lp6-SFT transcript levels also occurred in emerged leaf tissues during the first 72 h after cutting (P < 0.01), but there was no significant change observed for Lp1-SST in this tissue. As expected, a significant positive association between high DP fructan concentrations and Lp6-SFT gene expression was observed in enclosed elongating leaf tissues and sheaths [in enclosed elongating leaf tissues: High DP fructans = 15.663(Lp6-SFT)0.2544, R2 = 0.4537, P < 0.001, Supplementary Figure 3D; in sheaths: High DP fructans = −3.2407(Lp6-SFT)2 + 14.596(Lp6-SFT) + 2.7459, R2 = 0.3939, P < 0.001, Supplementary Figure 3E].
In this study, the expression of Lp1-FEH was significantly higher in emerged leaf tissues 24 h following defoliation (Figure 5F). However, compared to emerged leaf tissues at T0, which consist mainly of mature blade tissue, the majority of this leaf category at 24 h post-defoliation had only recently emerged, i.e., represents very young tissues and might not be directly comparable to T0 of the same category. The increase in Lp1-FEH gene expression in enclosed elongating leaf tissues was marginally significant (P = 0.058; Figure 5D; Table 1). There was no change detected in sheaths in the earlier stages, but expression of Lp1-FEH was significantly reduced 72–168 h post-defoliation (Figure 5E).
In a recent review (Parsons et al., 2011) we highlighted that although there is an urgent need to increase dry matter production of forage grasses, while at the same time limiting adverse environmental impacts caused by excessive use of N fertilization, there are only a few examples of forage breeding strategies targeted at the very processes (photosynthesis, respiration, N uptake and utilization) acting at the core of plant growth. In a previous study (Parsons et al., 2013a) we used exogenous application of GA3 to successfully stimulate L. perenne growth without the need for additional resources (N), although this was dependant on seasonal development. We postulated that GA3 might act at the center of vegetative growth regulation in this important forage grass particularly during regrowth after defoliation. To test this hypothesis further, we analyzed in the present study the expression of GA metabolic pathway and regulatory genes, and their possible relationship to carbohydrate, especially fructan metabolism.
The plant hormone gibberellin (GA) acts as a key mediator between environmental cues and plant morphology in a variety of developmental processes, including stem and root elongation, seed germination, and floral development (see reviews Yamaguchi, 2008; Colebrook et al., 2014). Numerous studies, particularly in Arabidopsis, rice, barley, and wheat demonstrated that the biologically active GA1, GA3 and GA4 are synthesized by the 2-oxoglutarate-dependent dioxygenases GA20-oxidase and GA3-oxidase, but inactivated by the degrading enzyme GA2-oxidase (see reviews Yamaguchi, 2008; Harberd et al., 2009; Hedden and Thomas, 2012). GA activity is also regulated by transduction regulators of growth, the repressing DELLA protein (Peng et al., 1999) and GA receptors such as GID1 (gibberellin insensitive dwarf; Ueguchi-Tanaka et al., 2005; Chandler et al., 2008). To date, however, only an LpGA2- oxidase (EF687858; MacMillan et al., 2005) and an LpGA20- oxidase (DQ071620, King et al., 2008) have been isolated from L. perenne.
In the present study, we isolated genes encoding a putative LpGA3-oxidase, which catalyzes the latter steps of GA activation, and a putative LpDELLA which is a growth repressing regulator of the GA signaling pathway. Our study showed that there was a significant up-regulation of LpGA3ox in sheaths following defoliation. Expression of both LpGA20ox and LpGA2ox was significantly down-regulated in sheaths 24 h post-defoliation. Reduced levels of LpGA2ox were also observed in enclosed elongating blades, but the reduction was only significant comparing the 4 and 24 h time points to the 2 h post-defoliation time point. The expression of DELLA, a repressor of GA responses, was initially reduced followed by an up-regulation, but this was statistically significant in mature sheaths (when comparing the 4 h with the 2 h time point) and in emerged leaf tissues only. Taken together our results indicate that defoliation of ryegrass does result in a differential expression of GA activating and inactivating enzymes, but due to high between replicate variability of the expression of the tested genes, and therefore lack of statistical significance, our study provides only indicative evidence of defoliation effects on GA metabolism. The expression of GA-related genes in response to environmental triggers has been shown previously to be very rapid and transient (Yamaguchi et al., 1998), and the high variability seen in our study might have been a result of the relative long time (15–20 min per plant) it takes to separate enclosed elongating leaf tissues from surrounding sheaths.
It needs to be noted here that GA3 has never been demonstrated to be present in L. perenne, while GA1, another active gibberellin, has been shown to be decreased after defoliation (Morvan-Bertrand et al., 2001). On the other hand, exogenous application of GA3 has been shown to induce stem elongation (MacMillan et al., 2005) and to increase dry matter production (Parsons et al., 2013a), and it is possible that levels of GA3 are too low to be detected by common analytical methods or GA3 might be located in very small tissue complexes, such as the basal leaf meristems, only. Another possibility might be that exogenously applied GA3 is subsequently converted in planta to other growth-stimulating gibberellins, but clearly more research is needed.
Defoliation is a major event in terms of L. perenne growth in an agricultural context (Morvan et al., 1997; Pilon-Smits et al., 1999; Wei and Chatterton, 2001; Lee et al., 2011), because the removed photosynthetic tissue needs to be compensated for as soon as possible in order to restore plant maintenance and sugar storage to pre-defoliation levels. This can only be achieved by elongation and eventual emergence of the enclosed immature blades, which requires the supply of carbohydrates to this heterotrophic tissue. Our results demonstrate that glucose and sucrose are rapidly depleted in immature blades, already 2 h after defoliation sucrose levels decreased to almost zero in this tissue. Insufficient new photosynthetically active tissue has emerged at this time and, in the absence of de novo synthesized sugars, carbohydrates have to be mobilized from their storage forms in the remaining tissues.
In L. perenne the two major storage carbohydrates are sucrose and fructans (Pollock and Cairns, 1991; Pavis et al., 2001a,b). To provide glucose, sucrose needs to be cleaved by invertases into glucose and fructose and we show here that the expression of all three forms of invertases, i.e., vacuolar, cytosolic and cell wall invertases are up-regulated after defoliation. The first invertase gene to be significantly up-regulated after 2 h was LpCWInv in both enclosed immature blades and mature sheaths, and we suggest that the immediate drop in sucrose in these two tissues is caused by an increased activity of invertases located in the cell walls (Sherson et al., 2003; Lammens et al., 2008; Proels and Hückelhoven, 2014). A second invertase gene, i.e., LpVacInv, was up-regulated much later (24 h post-defoliation) with expression particularly high in newly emerged blades, and we hypothesize that this type of invertase serves to hydrolyse de novo synthesized sucrose. Both, the hydrolysis of sucrose and of fructans also releases fructose, and concentrations of this monomeric sugar remained relatively constant in the first 24 h after defoliation and only decreased at later time points possibly indicating that fructose cannot be used directly for growth and that isomerases transforming it into glucose are relatively slowly up-regulated.
The second major storage carbohydrate in L. perenne are fructans (Pollock and Cairns, 1991; Smouter and Simpson, 1991; Morvan-Bertrand et al., 2001) and we found high levels of fructans in enclosed elongating blades and in mature sheaths prior to defoliation, indicating that these tissues act as strong sinks for photosynthates (Schnyder et al., 1988; Spollen and Nelson, 1988; Allard and Nelson, 1991). Following defoliation we saw a reduction in both low and high DP fructans, but this took place much later than the reduction in sucrose, between 24 (enclosed blades) and 48 h (mature sheaths) post-defoliation.
Degradation of fructans after defoliation in both enclosed and emerged blades of L. perenne has been reported to be associated with an increased enzymatic activity of fructan exohydrolase (FEH) which catalyzes fructan breakdown (Morvan et al., 1997; Chalmers et al., 2005; Lothier et al., 2007, 2014). Lp1-FEH gene expression was also reported to be negatively correlated with fructan accumulation and the expression of fructosyltransferases in both mature blades and sheaths (Morvan et al., 1997; Rasmussen et al., 2014). In the present study, we observed 24 h post-defoliation a higher Lp1-FEH gene expression compared to T0 in the emerged leaf tissue category. However, this tissue consists mainly of very young tissue which had at the earlier time points been enclosed elongating leaf tissue and comparing the levels of 1-FEH transcripts with those found in enclosed emerging tissue 2 and 4 h post-defoliation, levels of 1-FEH were actually lower.
Strongly declined transcript levels of genes encoding enzymes responsible for synthesizing reserve carbohydrates (Lp1-SST and Lp6-SFT) were observed following defoliation, which is consistent with previous observations on fructan accumulation in perennial ryegrass (Morvan et al., 1997; Lidgett et al., 2002; Hisano et al., 2008). As anticipated, we found that the relative transcript levels of fructan biosynthesizing genes, particularly Lp6-SFT, correlated positively with the accumulation of high DP fructans in both enclosed elongating blades and mature sheaths.
Taken together, our results suggest that after defoliation in mature sheaths and enclosed blades the immediate glucose demand of elongating blades is met by hydrolysis of sucrose first through cell wall and subsequently vacuolar invertases. Degradation of any pre-formed fructans and inhibition of de novo fructan synthesis are more long term processes occurring during 24–72 h post-defoliation. A full recovery of sugar and sugar metabolism related gene transcript levels to pre-defoliation levels is achieved between 1 and 2 weeks post-defoliation.
Low nutrient levels including low sucrose and glucose concentrations have been shown to activate GA biosynthesis and repress its degradation in several plant systems including cereal seed germination (Bewley and Black, 1985; Gibson, 2004; Hong et al., 2012; Colebrook et al., 2014). Increased levels of active GAs in turn induce a plethora of macromolecule degrading enzymes such as proteases, lipases and α-amylases (in starch accumulating plants/tissues). In our study, defoliation of L. perenne also resulted in a dramatic and very rapid depletion of glucose and sucrose pools. This depletion might also have caused an increase in genes coding for GA-activating enzymes, but because this increase was statistically significant only for LpGA3ox in mature sheaths, our results are not conclusive. However, a strong negative association between sucrose and transcript levels of LpGA3ox as well as a strong positive association between sucrose and levels of LpGA2ox might be seen as additional evidence for signaling between low sucrose levels and GA-activation.
Induction of GA biosynthesis as well as exogenous supply of GA3 to barley and rice seeds were shown to induce the expression of more than 1000 genes encoding e.g., hydrolases, proteases, and lipases (Chen and An, 2006; Tsuji et al., 2006; Hong et al., 2012) and also sucrolytic activity (see review Roitsch and González, 2004). In our study we saw a strong and very rapid increase in LpCWInv (2 h post-defoliation) in both enclosed elongating blades and mature sheaths, from our data it is unlikely that this increase was induced by active GA as induction of LpGA3ox and LpGA20ox was seen at the same or later time points. Although it is possible that, at least for LpGA3ox, we might have missed the time of highest expression, which might have been earlier than 2 h post-defoliation, our results do not allow a clear statement regarding these events. However, we did see a strong increase in LpVacInv expression 24 h post-defoliation which might have prevented sucrose build-up in sheaths and emerged mature blades at the early stages of re-growth (after formation of new, photosynthetically active tissue). We saw a strong down-regulation of fructan biosynthesizing genes following defoliation, and it would be interesting to test if the expression of these genes is repressed by GAs. Future studies should therefore aim to isolate promoter regions of carbohydrate related genes from L. perenne and perform promoter binding studies (see Hong et al., 2012).
Summarizing, we propose that our study indicates that sugar depletion following defoliation might affect GA-biosynthesis, but that our results are not always conclusive and additional experiments are needed to unequivocally show a link between sugars and GA in perennial ryegrass during re-growth after defoliation.
Our results here and those of a previous study (Parsons et al., 2013a) provide indicative evidence that gibberellins might play a role in L. perenne re-growth after defoliation, but that further studies are needed to unambiguously demonstrate a link between defoliation, sugar depletion and GA activation. Increasing ryegrass vegetative (dry matter) production is at the center of efforts to increase pasture based animal production. Because a strong link between vegetative production and gibberellins has been shown in cereals, we propose that gibberellin signaling in perennial ryegrass should be studied in more detail to possibly improve our chances of achieving this goal.
All authors contributed to the research and manuscript equally; and read and approved the final version of the manuscript. All authors agree to be accountable for all aspects of the work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the New Zealand Agricultural Greenhouse Gas Research Centre (NZAGRC) and the Ministry of Science and Innovation, New Zealand (contract # C10X0903); and conducted at AgResearch Grasslands, Palmerston North, New Zealand.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00944
GA, gibberellin; WSC, water soluble carbohydrate; DP, degree of polymerisation; FEH, fructan exohydrolase; SST, sucrose: sucrose fructosyltransferase; SFT, sucrose: fructan fructosyltransferase.
Achard, P., and Genschik, P. (2009). Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J. Exp. Bot. 60, 1085–1092. doi: 10.1093/jxb/ern301
Allard, G., and Nelson, C. J. (1991). Photosynthate partitioning in basal zones of tall fescue leaf blades. Plant Physiol. 95, 663–668. doi: 10.1104/pp.95.3.663
Appleford, N. E. J., Evans, D. J., Lenton, J. R., Gaskin, P., Croker, S. J., Devos, K. M., et al. (2006). Function and transcript analysis of gibberellin-biosynthetic enzymes in wheat. Planta 223, 568–582. doi: 10.1007/s00425-005-0104-0
Bewley, J. D., and Black, M. (1985). Seeds: Physiology of Development and Germination. New York, NY: Plenum Press. doi: 10.1007/978-1-4615-1747-4_8
Cairns, A. L., and Gallagher, J. A. (2004). Absence of turnover and futile cycling of sucrose in leaves of Lolium temulentum L: implications for metabolic compartmentation. Planta 219, 836–846. doi: 10.1007/s00425-004-1281-y
Chalmers, J., Johnson, X., Lidgett, A., and Spangenberg, G. (2003). Isolation and characterization of a sucrose: sucrose 1-fructosyltransferase gene from perennial ryegrass (Lolium perenne). J. Plant Physiol. 160, 1385–1391. doi: 10.1078/0176-1617-01107
Chalmers, J., Lidgett, A., Cummings, N., Cao, Y., Forster, J., and Spangenberg, G. (2005). Molecular genetics of fructan metabolism in perennial ryegrass. Plant Biotechnol. J. 3, 459–474. doi: 10.1111/j.1467-7652.2005.00148.x
Chandler, P. M., Harding, C. A., Ashton, A. R., Mulcair, M. D., Dixon, N. E., and Mander, L. N. (2008). Characterization of gibberellin receptor mutants of barley (Hordeum vulgare L.). Mol. Plant 1, 285–294. doi: 10.1093/mp/ssn002
Chandler, P. M., Marion-Poll, A., Ellis, M., and Gubler, F. (2002). Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterisation. Plant Physiol. 129, 181–190. doi: 10.1104/pp.010917
Chen, K., and An, Y. Q. C. (2006). Transcriptional responses to gibberellin and abscisic acid in barley aleurone. J. Integr. Plant Biol. 48, 591-612. doi: 10.1111/j.1744-7909.2006.00270.x
Colebrook, E. H., Thomas, S. G., Phillips, A. L., and Hedden, P. (2014). The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217, 67–75. doi: 10.1242/jeb.089938
Ellefsen, S., Stensløkken, K. O., Sandvik, G. K., Kristensen, T. A., and Nilsson, G. E. (2008). Improved normalization of real-time reverse transcriptase polymerase chain reaction data using an external RNA control. Anal. Biochem. 376, 83–93. doi: 10.1016/j.ab.2008.01.028
Ellis, J. L., Dijkstra, J., Bannink, A., Parsons, A. J., Rasmussen, S., Edwards, G. R., et al. (2011). The effect of high-sugar grass on predicted nitrogen excretion and milk yield simulated using a dynamic model. J. Dairy Sci. 94, 3105–3118. doi: 10.3168/jds.2010-4059
Eveland, A. L., and Jackson, D. P. (2012). Sugars, signalling, and plant development. J. Exp. Bot. 63, 3367–3377. doi: 10.1093/jxb/err379
FAO. (2011). “Mapping supply and demand for animal-source foods to 2030,” in Animal Production and Health Working Paper. No. 2, eds T. P. Robinson and F. Pozzi (Rome).
Gibson, S. (2004). Sugar and phytohormone response pathways: navigating a signalling network. J. Exp. Bot. 55, 253–264. doi: 10.1093/jxb/erh048
Gocal, G. F., Poole, A. T., Gubler, F., Watts, R. J., Blundell, C., and King, R. W. (1999). Long-day up-regulation of a GAMYB gene during Lolium temulentum inflorescence formation. Plant Physiol. 119, 1271–1278. doi: 10.1104/pp.119.4.1271
Harberd, N. P., Belfield, E., and Yasumura, Y. (2009). The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of inhibitor” enables flexible response to fluctuating environments. Plant Cell 21, 1328–1339. doi: 10.1105/tpc.109.066969
Hedden, P., and Thomas, S. G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444, 11–25. doi: 10.1042/BJ20120245
Hisano, H., Kanazawa, A., Yoshida, M., Humphreys, M. O., Iizuka, M., Kitamura, K., et al. (2008). Coordinated expression of functionally diverse fructosyltransferase genes is associated with fructan accumulation in response to low temperature in perennial ryegrass. New Phytol. 178, 766–780. doi: 10.1111/j.1469-8137.2008.02409.x
Hong, Y. F., Ho, T. H., Wu, C. F., Ho, S. L., Yeh, R. H., Lu, C. A., et al. (2012). Convergent starvation signals and hormone crosstalk in regulating nutrient mobilization upon germination in cereals. Plant Cell 24, 2857–2873. doi: 10.1105/tpc.112.097741
Huang, L., Yan, H., Jiang, X., Yin, G., Zhang, X., Qi, X., et al. (2014). Identification of candidate reference genes in perennial ryegrass for quantitative RT-PCR under various abiotic stress conditions. PLoS ONE 9:e93724. doi: 10.1371/journal.pone.0093724
Hunt, M. G., Rasmussen, S., Newton, P. C. D., Parsons, A. J., and Newman, J. A. (2005). Near-term impacts of elevated CO2, nitrogen and fungal endophyte-infection on Lolium perenne L.: growth, chemical composition and alkaloid production. Plant Cell Environ. 28, 1345–1354. doi: 10.1111/j.1365-3040.2005.01367.x
Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signalling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57–70. doi: 10.1105/tpc.010319
Itoh, H., Ueguchi-Tanaka, M., Sentoku, N., Kitano, H., Matsuoka, M., and Kobayashi, M. (2001). Cloning and functional analysis of two gibberellin 3ß-hydroxylase genes that are differently expressed during growth of rice. Proc. Natl. Acad. Sci. U.S.A. 98, 8909–8914. doi: 10.1073/pnas.141239398
Jermyn, M. A. (1956). A new method for determining ketohexoses in the presence of aldohexoses. Nature 177, 38–39. doi: 10.1038/177038a0
Kebreab, E., France, J., Beever, D. E., and Castillo, A. R. (2001). Nitrogen pollution by dairy cows and its mitigation by dietary manipulation. Nutr. Cycl. Agroecosyst. 60, 275–285. doi: 10.1023/A:1012668109662
King, R. W., Mander, L. N., Asp, T., MacMillan, C. P., Blundell, C. A., and Evans, L. T. (2008). Selective deactivation of gibberellins below the shoot apex is critical to flowering but not to stem elongation of Lolium. Mol. Plant 1, 295–307. doi: 10.1093/mp/ssm030
Kingston-Smith, A. H., Walker, R. P., and Pollock, C. J. (1999). Invertase in leaves: conundrum or control point? J. Exp. Bot. 50, 735–743. doi: 10.1093/jxb/50.335.735
Lammens, W., Le Roy, K., Van Laere, A., Rabijns, A., and Van den Ende, W. (2008). Crystal structures of Arabidopsis thaliana cell-wall invertase mutants in complex with sucrose. J. Mol. Biol. 377, 378–385. doi: 10.1016/j.jmb.2007.12.074
Lasseur, B., Lothier, J., Wiemken, A., Van Laere, A., Morvan-Bertrand, A., Van den Ende, W., et al. (2011). Towards a better understanding of the generation of fructan diversity diversity in plants: molecular and functional characterization of a sucrose: fructan 6-fructosyltransferase (6-SFT) cDNA from perennial ryegrass (Lolium perenne). J. Exp. Bot. 62, 1871–1885. doi: 10.1093/jxb/erq388
Lee, J. M., Roche, J. R., Donaghy, D. J., Thrush, A., and Sathish, P. (2010). Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass (Lolium perenne L.). BMC Mol. Biol. 11:8. doi: 10.1186/1471-2199-11-8
Lee, J. M., Sathish, P., Donaghy, D. J., and Roche, J. R. (2011). Impact of defoliation severity on photosynthesis, carbon metabolism and transport gene expression in perennial ryegrass. Funct. Plant Biol. 38, 808–817. doi: 10.1071/FP11048
Lidgett, A., Jennings, K., Johnson, X., Guthridge, K., Jones, E., and Spangenberg, G. (2002). Isolation and characterisation of a fructosyltransferase gene from perennial ryegrass (Lolium perenne). J. Plant Physiol. 159, 1037–1043. doi: 10.1078/0176-1617-00808
Liu, Z. L., and Slininger, P. J. (2007). Universal external RNA controls for microbial gene expression analysis using microarray and qRT-PCR. J. Microbiol. Methods 68, 486–496. doi: 10.1016/j.mimet.2006.10.014
Lothier, J., Lasseur, B., Le Roy, K., Van Laere, A., Prud'homme, M. P., Barre, P., et al. (2007). Cloning, gene mapping, and functional analysis of a fructan 1-exohydrolase (1-FEH) from Lolium perenne implicated in fructan synthesis rather than in fructan mobilization. J. Exp. Bot. 58, 1969–1983. doi: 10.1093/jxb/erm053
Lothier, J., Van Laere, A., Prud'homme, M.-P., Van den Ende, W., and Morvan-Bertrand, A. (2014). Cloning and characterization of a novel fructans 6-exohydrolase strongly inhibited by sucrose in Lolium perenne. Planta 240, 629–643. doi: 10.1007/s00425-014-2110-6
MacMillan, C. P., Blundell, C. A., and King, R. W. (2005). Flowering of the grass Lolium perenne: effects of vernalization and long days on gibberellin biosynthesis and signalling. Plant Physiol. 138, 1794–1806. doi: 10.1104/pp.105.062190
Martin, R. C., Hollenbeck, V. G., and Dombrowski, J. E. (2008). Evaluation of reference genes for quantitative RT-PCR in Lolium perenne. Crop Sci. 48, 1881–1887. doi: 10.2135/cropsci2007.10.0597
Matsoukas, I. G. (2014). Interplay between sugar and hormone signaling pathways modulate floral signal transduction. Front. Genet. 5:218. doi: 10.3389/fgene.2014.00218
Matsukura, C., Itoh, S. I., Netomo, K., Tanimoto, E., and Yamaguchi, J. (1998). Promotion of leaf sheath growth by gibberellic acid in a dwarf mutant of rice. Planta 205, 145–152. doi: 10.1007/s004250050306
Morvan, A., Challe, G., Prud'homme, M. P., Le Saos, J., and Boucaud, J. (1997). Rise of fructan exohydrolase activity in stubble of Lolium perenne after defoliation is decreased by uniconazole, an inhibitor of the biosynthesis of gibberellins. New Phytol. 136, 81–88.
Morvan-Bertrand, A., Ernstsen, A., Lindgård, B., Koshioka, M., Le Saos, J., Boucaud, J., et al. (2001). Endogenous gibberellins in Lolium perenne and influence of defoliation on their contents in elongating leaf bases and in leaf sheaths. Physiol. Plant. 111, 225–231. doi: 10.1034/j.1399-3054.2001.1110214.x
Morvan-Bertrand, A., Pavis, N., Boucaud, J., and Prud'homme, M. P. (1999). Partitioning of reserve and newly assimilated carbon in roots and leaf tissues of Lolium perenne during regrowth after defoliation: assessment by 13C steady-state labelling and carbohydrate analysis. Plant Cell Environ. 22, 1097–1108.
Paparelli, E., Parlanti, S., Gonzali, S., Novi, G., Mariotti, L., Ceccarelli, N., et al. (2013). Night time sugar starvation orchestrates gibberellin biosynthesis and plant growth in Arabidopsis. Plant Cell 25, 3760–3769. doi: 10.1105/tpc.113.115519
Parsons, A. J., and Chapman, D. F. (2000). “The principles of pasture growth and utilisation,” in Grass: Its production and Utilisation, ed A. Hopkins (London: Blackwell Science), 31–89.
Parsons, A. J., Edwards, G. R., Newton, P. C. D., Chapman, D. F., Caradus, J. R., Rasmussen, S., et al. (2011). Past lessons and future prospects: plant breeding for yield and persistence in cool temperate pastures. Grass Forage Sci. 66, 153–172. doi: 10.1111/j.1365-2494.2011.00785.x
Parsons, A. J., Rasmussen, S., Liu, Q., Xue, H., Ball, C., and Shaw, C. (2013a). Plant growth – resource or strategy limited: insights from responses to gibberellin. Grass Forage Sci. 68, 577–588. doi: 10.1111/gfs.12035
Parsons, A. J., Thornley, J. H. M., Newton, P. C. D., Rasmussen, S., and Rowarth, J. S. (2013b). Soil carbon dynamics: the effects of nitrogen input, intake demand and off-take by animals. Sci. Total Environ. 465, 205–215. doi: 10.1016/j.scitotenv.2013.02.019
Pavis, N., Boucaud, J., and Prud'homme, M. P. (2001a). Fructans and fructan-metabolising enzymes in leaves of Lolium perenne. New Phytol. 150, 97–109. doi: 10.1046/j.1469-8137.2001.00068.x
Pavis, N., Chatterton, N. J., Harrison, P. A., Baumgartner, S., Praznik, W., Boucaud, J., et al. (2001b). Structure of fructans in roots and leaf tissues of Lolium perenne. New Phytol. 150, 83–95. doi: 10.1046/j.1469-8137.2001.00069.x
Peng, J., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. doi: 10.1038/22307
Perata, P., Matsukura, C., Vernieri, P., and Yamaguchi, J. (1997). Sugar repression of a gibberellin-dependent signalling pathway in barley embryos. Plant Cell 9, 2197–2208. doi: 10.1105/tpc.9.12.2197
Pilon-Smits, E. A. H., Ebskamp, M. J. M., van Dun, K., and Terry, N. (1999). Enhanced drought resistance in fructan-producing sugar beet. Plant Physiol. Biochem. 37, 313–317. doi: 10.1016/S0981-9428(99)80030-8
Pollock, C. J., and Cairns, A. J. (1991). Fructan metabolism in grasses and cereals. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 77–101. doi: 10.1146/annurev.pp.42.060191.000453
Proels, R. K., and Hückelhoven, R. (2014). Cell-wall invertases, key enzymes in the modulation of plant metabolism during defence responses. Mol. Plant Pathol. 15, 858–864. doi: 10.1111/mpp.12139
Ranwala, A. P., and Miller, W. B. (2008). Gibberellin-mediated changes in carbohydrate metabolism during flower stalk elongation in tulips. Plant Growth Regul. 55, 241–248. doi: 10.1007/s10725-008-9280-9
Rasmussen, S., Parsons, A. J., Bassett, S., Christensen, M. J., Hume, D. E., Johnson, L. J., et al. (2007). High nitrogen supply and carbohydrate content reduce fungal endophyte and alkaloid concentration in Lolium perenne. New Phytol. 173, 787–797. doi: 10.1111/j.1469-8137.2006.01960.x
Rasmussen, S., Parsons, A. J., Fraser, K., Xue, H., and Newman, J. A. (2008). Metabolic profiles of Lolium perenne are differentially affected by nitrogen supply, carbohydrate content, and fungal endophyte infection. Plant Physiol. 146, 1440–1453. doi: 10.1104/pp.107.111898
Rasmussen, S., Parsons, A. J., Xue, H., Liu, Q., Jones, C. S., Ryan, G. D., et al. (2014). Transcript profiling of fructan biosynthetic pathway genes reveals association of a specific fructosyltransferase isoform with the high sugar trait in Lolium perenne. J. Plant Physiol. 171, 475–485. doi: 10.1016/j.jplph.2013.12.008
Rehrig, E. M., Appel, H. M., and Schultz, J. C. (2011). Measuring ‘normalcy’ in plant gene expression after herbivore attack. Mol. Ecol. Resour. 11, 294–304. doi: 10.1111/j.1755-0998.2010.02929.x
Roitsch, T., and González, M. C. G. (2004). Function and regulation of plant invertases: sweet sensations. Trends Plant Sci. 9, 606–613. doi: 10.1016/j.tplants.2004.10.009
Sauter, M., Mekhedov, S. L., and Kende, H. (1995). Gibberellin promotes histone H1 kinase activity and expression of cdc2 and cyclin genes during the induction of rapid growth in deep water rice. Plant J. 7, 623–632. doi: 10.1046/j.1365-313X.1995.7040623.x
Schnyder, H., and de Visser, R. (1999). Fluxes of reserve-derived and currently assimilated carbon and nitrogen in perennial ryegrass recovering from defoliation. The regrowing tiller and its component functionally distinct zones. Plant Physiol. 119, 1423–1435. doi: 10.1104/pp.119.4.1423
Schnyder, H., Nelson, C. J., and Spollen, W. G. (1988). Diurnal growth of tall fescue leaf blades. II. Dry matter partitioning and carbohydrate metabolism in the elongation zone and adjacent expanded tissue. Plant Physiol. 86, 1077–1083. doi: 10.1104/pp.86.4.1077
Schnyder, H., Seo, S., Rademacher, I. F., and Kühlbauch, W. (1990). Spatial distribution of growth rates and of epidermal cell lengths in the elongation zone during leaf development in Lolium perenne L. Planta 181, 423–431. doi: 10.1007/BF00195897
Sherson, S. M., Alford, H. L., Forbes, S. M., Wallace, G., and Smith, S. M. (2003). Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. J. Exp. Bot. 54, 525–531. doi: 10.1093/jxb/erg055
Smith, R. C., Matthews, P. R., Schünmann, P. H. D., and Chandler, P. M. (1996). The regulation of leaf elongation and xyloglucan endotransglycosylase by gibberellin in ‘Himalaya’ barley (Hordeum vulgare L.). J. Exp. Bot. 47, 1395–1404. doi: 10.1093/jxb/47.9.1395
Smouter, H., and Simpson, R. J. (1991). Fructan metabolism in leaves of Lolium rigidum Gaudin. I. Synthesis of fructan. New Phytol. 119, 509–516. doi: 10.1111/j.1469-8137.1991.tb01042.x
Spielmeyer, W., Ellis, M., Robertson, M., Ali, S., Lenton, J. R., and Chandler, P. M. (2004). Isolation of gibberellin metabolic pathway genes from barley and comparative mapping in barley, wheat and rice. Theor. Appl. Genet. 109, 847–855. doi: 10.1007/s00122-004-1689-6
Spollen, W. G., and Nelson, C. J. (1988). Characterization of fructan from mature leaf blades and elongation zones of developing leaf blades of wheat, tall fescue and timothy. Plant Physiol. 88, 1349–1353. doi: 10.1104/pp.88.4.1349
Tas, B. M. (2007). “Nitrogen utilisation of perennial ryegrass in dairy cows,” in Fresh Herbage for Dairy Cattle, eds A. Elgersma, J. Dijkstra, and S. Tamminga (Wageningen: Springer Inc.), 125–140.
Tsuji, H., Aya, K., Ueguchi-Tanaka, M., Shimada, Y., Nakazono, M., Watanabe, R., et al. (2006). GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J. 47, 427–444. doi: 10.1111/j.1365-313X.2006.02795.x
Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., et al. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698. doi: 10.1038/nature04028
van Groenigen, K. J., Osenberg, C. W., and Hungate, B. A. (2011). Increased soil emissions of potent greenhouse gases under increased atmospheric CO2. Nature 475, 214–216. doi: 10.1038/nature10176
Van Soest, P. J. (1982). Nutritional Ecology of the Ruminant, 2nd Edn. Ithaca, NY: Cornell University Press.
Wei, J. Z., and Chatterton, N. J. (2001). Fructan biosynthesis and fructosyltransferase evolution: the expression of the 6-SFT (sucrose: fructan 6-fructosyltransferase) gene in crested wheatgrass (Agropyron cristatum). J. Plant Physiol. 158, 1203–1213. doi: 10.1078/0176-1617-00241
Woledge, J., and Pearse, J. P. (1985). The effect of nitrogen fertiliser on the photosynthesis of leaves of a ryegrass sward. Grass Forage Sci. 40, 305–309. doi: 10.1111/j.1365-2494.1985.tb01756.x
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Keywords: perennial ryegrass, defoliation, gibberellins, carbohydrates, gene expression, fructans
Citation: Liu Q, Jones CS, Parsons AJ, Xue H and Rasmussen S (2015) Does gibberellin biosynthesis play a critical role in the growth of Lolium perenne? Evidence from a transcriptional analysis of gibberellin and carbohydrate metabolic genes after defoliation. Front. Plant Sci. 6:944. doi: 10.3389/fpls.2015.00944
Received: 11 March 2015; Accepted: 17 October 2015;
Published: 03 November 2015.
Edited by:
Marie-Pascale Prud'Homme, University of Caen Normandy, FranceReviewed by:
Annette Morvan-Bertrand, University of Caen Normandy, FranceCopyright © 2015 Liu, Jones, Parsons, Xue and Rasmussen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susanne Rasmussen, cy5yYXNtdXNzZW5AbWFzc2V5LmFjLm56
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.