- 1Laboratorio de Bioingeniería, Facultad de Ingeniería y Ciencias, Universidad Adolfo Ibáñez, Santiago, Chile
- 2Center for Applied Ecology and Sustainability, Santiago, Chile
Protein complexes involved in epigenetic regulation of transcription have evolved as molecular strategies to face environmental stress in plants. SAGA (Spt–Ada–Gcn5 Acetyltransferase) is a transcriptional co-activator complex that regulates numerous cellular processes through the coordination of multiple post-translational histone modifications, including acetylation, deubiquitination, and chromatin recognition. The diverse functions of the SAGA complex involve distinct modules that are highly conserved between yeast, flies, and mammals. In this review, the composition of the SAGA complex in plants is described and its role in gene expression regulation under stress conditions summarized. Some of these proteins are likely involved in the regulation of the inducible expression of genes under light, cold, drought, salt, and iron stress, although the functions of several of its components remain unknown.
Introduction
Transcriptional coactivators are multi-protein complexes that can recognize histone markers, modify chromatin, and recruit the transcriptional machinery to control gene expression (Näär et al., 2001). In general, these complexes regulate eukaryotic gene expression by interacting with transcription factors and/or other regulatory components of the basal transcription machinery. SAGA (Spt–Ada–Gcn5–Acetyl transferase) is a transcriptional coactivator complex involved in the regulation of numerous cellular processes through the coordination of the post-translational modification of various histones. The yeast SAGA complex is thought to control transcription of approximately 10% of genes, particularly stress-related genes (Lee et al., 2000; Huisinga and Pugh, 2004). This complex is generally regarded as a coactivator complex (Kuo et al., 1998), but also has a negative role in gene expression (Belotserkovskaya et al., 2000; Ricci et al., 2002). The SAGA complex is involved in histone acetylation (HAT) (Grant et al., 1997), histone deubiquitination (Daniel et al., 2004), mRNA export (Rodríguez-Navarro et al., 2004), transcription elongation (Govind et al., 2007), chromatin recognition (Pray-Grant et al., 2005), and regulation of the basal transcription machinery (Sterner et al., 1999). Unraveling the modular composition of the SAGA complex has enabled interpretation of its multifunctional role (Wu et al., 2004), principally in regulating the transcription of many stress-inducible (Huisinga and Pugh, 2004) and developmentally regulated genes (reviewed in Wang and Dent, 2014). The diverse functions of SAGA involve the participation of modules that are highly conserved between yeast, flies, and mammals. The SAGA complex is composed of more than 20 polypeptide subunits, grouped in four modules: the deubiquitinating module, the histone acetyltransferase module, and the SPT and TAF modules, which are implicated in the recruitment and SAGA architecture, respectively (Reviewed in Daniel and Grant, 2007 and Koutelou et al., 2010). Despite the abundance of genetic information available for plants, little is known about the presence and role of SAGA in photosynthetic organisms. Recently, a study determined the genes encoding subunits of the SAGA complex across a number of plants species (Srivastava et al., 2015), suggesting conservation of the SAGA complex throughout evolution. The yeast SAGA is particularly important for stress-induced transcription, and this function seems to be conserved during evolution (Spedale et al., 2012). In this review, the composition and our current knowledge of the role of the SAGA complex in the control of gene expression under stress conditions in plants is summarized.
Histone Acetylation Module
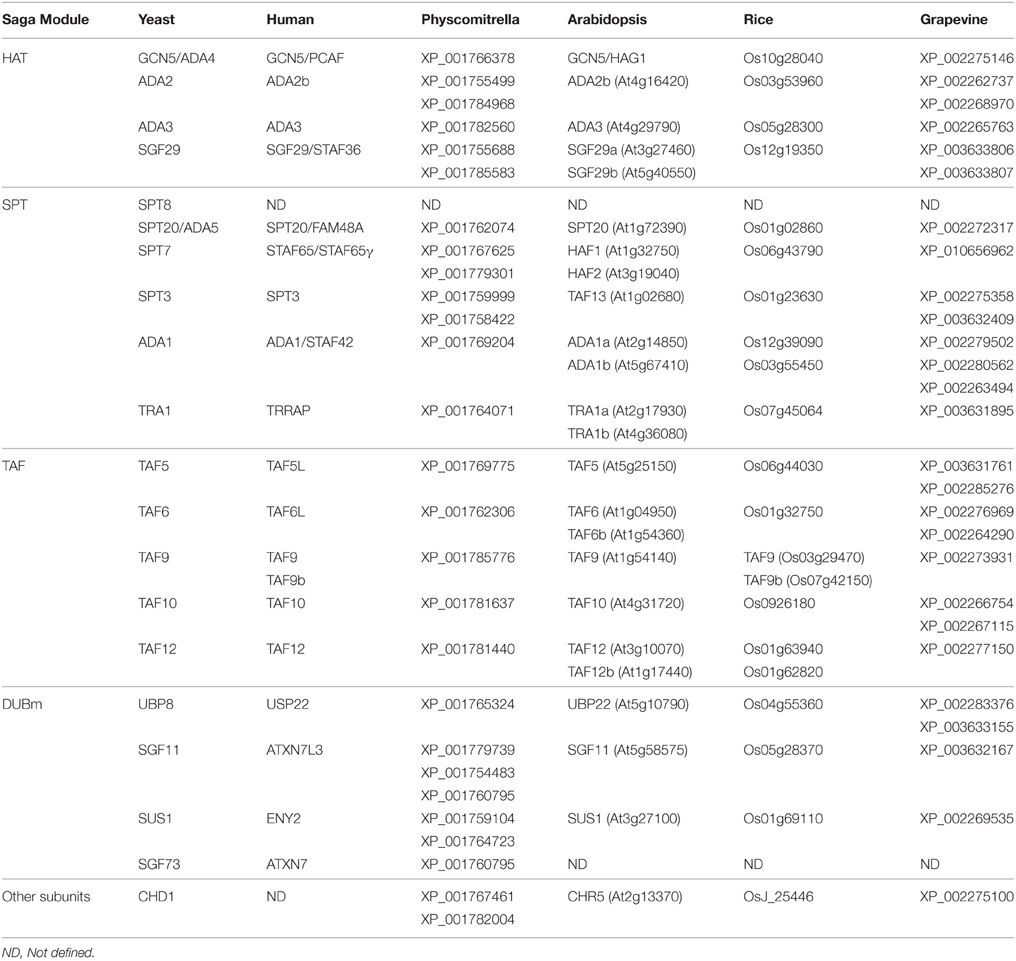
The histone acetylation (HAT) module contains the General Control Non-depressible 5 (GCN5) acetyltransferase in complex with ADA2, ADA3, and SGF29. This module is completely conserved in several photosynthetic organisms (Table 1). The GCN5 protein, which harbors a HAT domain has been identified in Arabidopsis thaliana (Pandey et al., 2002), Vitis vinifera (Aquea et al., 2010), and rice (Liu et al., 2012). The GCN5 protein mainly modifies Lys residue 14 in histone H3 in yeast (Kuo et al., 1996; Grant et al., 1999) and Arabidopsis (Benhamed et al., 2006; Earley et al., 2007). HAT by GCN5 has been shown to displace promoter nucleosomes (Barbaric et al., 2001), recruit RNA Polymerase II and coactivators to yeast promoter regions (Qiu et al., 2004; Govind et al., 2005), and increase the efficiency of trimethylation of H3-Lysine 4 in transcribed coding sequences (Govind et al., 2007). In Arabidopsis, GCN5 acetylates not only histones but also other proteins, as ADA2 (Mao et al., 2006), and appears to be a phosphorylated given that a phosphatase physically interacts and dephosphorylates GCN5 in vitro (Servet et al., 2008). In addition, GCN5 has a BROMO domain that recognizes acetylated lysine residues and increases the retention of the SAGA complex, promoting its HAT, and other functions (Mujtaba et al., 2007). The presence of the HAT and BROMO domain makes GCN5 a “reader” and “writer” of epigenetic marks. ADA2 (alteration/deficiency in activation 2) is an adaptor protein that physically associates with GCN5 (Grant et al., 1997). In Arabidopsis, two related ADA2 factors (ADA2a and ADA2b) have been identified (Stockinger et al., 2001), but only ADA2b is considered a member of the SAGA complex (Srivastava et al., 2015) Both proteins can bind directly to GCN5 through their N-terminal regions (Mao et al., 2006). This interaction enhances the ability of GCN5 to acetylate histones in vitro and enables GCN5 to acetylate nucleosomal histones (Mao et al., 2006). Maize homologs of GCN5 and ADA2 also interact with each other in vitro and in vivo (Bhat et al., 2003, 2004). SGF29 (SaGa associated Factor 29) is another component of the HAT module. In Arabidopsis, two homologous proteins of yeast SGF29 have been identified (Kaldis et al., 2011). In humans, SGF29 interacts with GCN5 but not with ADA2 (Nguyen-Huynh et al., 2015). Deletion of yeast SGF29 does not affect SAGA integrity or composition of the HAT module, indicating that SGF29 is a peripheral subunit in this complex (Shukla et al., 2012). In addition, SGF29 binds H3K4me2/3 via its double TUDOR domain (Bian et al., 2011), suggesting a critical role in mediating transcriptional regulation through subsequent chromatin modifications. In Humans, ADA3 is associated with GCN5 and ADA2 to form the catalytic module of the SAGA complex and cooperates to stimulate GCN5-mediated HAT of nucleosomal templates (Gamper et al., 2009). There is no evidence of a role for ADA3 in plants.
Recruiting Module
This module contains the proteins SPT8, SPT20, SPT7, SPT3, ADA1, and TRA1 and is conserved in several photosynthetic organisms with the exception of SPT8 (Srivastava et al., 2015, Table 1). Notably, orthologs of the SPT8 gene are also absent in the genomes of metazoans (Spedale et al., 2012). The SPT3 subunit recruits the TATA Binding-Protein (TBP) and contributes to the formation of the preinitiation transcription complex (Dudley et al., 1999). In plants, a homologous protein of SPT3/TAF13 has been described in Arabidopsis (Lago et al., 2004) and pepper (Wen et al., 2013). In Arabidopsis, TAF13 interacts physically with other TAFs (TBP-associated factor) proteins (Lawit et al., 2007) and with MEDEA and SWINGER, both members of a plant variant of Polycomb Repressive Complex 2 (PRC2; Lindner et al., 2013). PRC2 is involved in transcriptional repression through tri-methylation of lys27 of histone H3, suggesting a possible link between SAGA and other complexes involved in chromatin remodeling. SPT20 has a primordial function in the assembly of the SAGA complex, as no intact SAGA could be purified in spt20 yeast mutant strains (Sterner et al., 1999). In plants, an SPT20 domain containing protein has been reported by Endo et al. (2013) and is an interactor that bridges PHYTCHROME B (phyB) and CONSTANS (CO) proteins involved in the photoperiodic regulation of flowering (Endo et al., 2013). There is no evidence of such a molecular mechanism of SPT20 in plants.
On the other hand, the SPT7 protein works as a scaffold element that maintains and stabilizes the SAGA complex also (Wu et al., 2004). In Arabidopsis, their homologous proteins are HAF1 and HAF2, putative proteins that harbor a histone acetyltransferase, and BROMO domain that can interact with acetylated lysine (Jacobson et al., 2000). The SPT7 BROMO domain interacts weakly with individually acetylated lysine residues (Hassan et al., 2007), suggesting that the BROMO domain within GCN5 is perhaps more important for recognition and binding to acetylated lysine residues in the histone tails, whereas the SPT7 BROMO domain may have another function such as recognition of acetylated transcription factors or multiple lysine residues (Hassan et al., 2007). In Arabidopsis, genetic analysis has shown that HAF2 interacts with GCN5 to integrate light signals, regulating gene expression and growth (Bertrand et al., 2005; Benhamed et al., 2006). Both genes are required for H3K9, H3K27, and H4K12 acetylation on the target promoters (Benhamed et al., 2006).
The SAGA complex is recruited to gene loci by the interaction of yeast TRA1 protein or its mammalian ortholog TRRAP (Transformation/Transcription domain-Associated Protein) with specific transcriptional activators (Brown et al., 2001). These proteins are large and represent almost one quarter of the mass of the entire SAGA complex, suggesting that TRA1 may serve as a scaffold for complex assembly or for recruitment to chromatin in SAGA (Grant et al., 1998; Murr et al., 2007) or other complex (Allard et al., 1999; Knutson and Hahn, 2011). TRA1 and TRRAP proteins show a striking sequence similarity to the family of phosphatidylinositol-3-kinase. There are two genes homologous to TRA1 in Arabidopsis. The genes At2g17930 and At4g36080 encode for a 3858 and 3834 amino acid protein, respectively, with a FAT domain and predicted phosphatidylinositol 3-kinase activity. The recruitment of TRRAP precedes that of GCN5, suggesting that TRA1 and TRRAP function in targeting co-activator complexes to specific promoters during transcription (Memedula and Belmont, 2003). The function of At2g17930, At4g36080 or its homologous proteins in other species of plants has not yet been described.
Coactivator Architecture Module
The coactivator architecture of the TAF module contains the TBP-associated factors TAF5, TAF6, TAF9, TAF10, and TAF12. This module is completely conserved in plants (Table 1), and these proteins are shared with the general transcription factor TFIID (Lee et al., 2011). The amino acid sequences of TAFs are conserved from yeast to humans (Struhl and Moqtaderi, 1998; Albright and Tjian, 2000). Initial studies on in vitro transcription suggested that TAFs might act as general co-activators that mediate the transcriptional activation of different activators (Goodrich et al., 1996). However, several TAFs have shown tissue- and/or developmental stage-specific expression and are required for the expression of only a subset of genes (Hiller et al., 2001). The endogenous expression of TAF10 was monitored in transgenic Arabidopsis plants (pTAF10:GUS), yielding mostly vascular tissue preferential expression (Tamada et al., 2007). This expression pattern is closely similar to a TAF10 homologous gene in Flaveria trinervia, as been observed by in situ hybridization (Furumoto et al., 2005). The A. thaliana TAF6 gene is expressed in different tissues (Lago et al., 2005). A morphological analysis showed that T-DNA insertion in TAF6 specifically affects pollen tube growth, indicating that this TAF protein regulates the transcription of only a specific subset of genes in plants (Lago et al., 2005). In addition, TAF12 is required for proper hormone response, by negatively regulating cytokinin sensitivity (Kubo et al., 2011) and ethylene response in Arabidopsis (Robles et al., 2007) and TAF5 is an essential gene, required for male gametogenesis, pollen tube growth, and required in transcriptional mechanisms involved in the maintenance of indeterminate inflorescence (Mougiou et al., 2012).
Deubiquitination Module
The Deubiquitination (DUB) module comprises four proteins: UBP8, SGF11, SUS1, and SGF73. This module is conserved in plants with the exception of SGF73 (Table 1). In yeast, the central domain of SGF73 tethers the DUB module to the rest of the SAGA complex while the N-terminal domain forms an integral part of the DUB module (Lee et al., 2009). A homologous protein of SGF73 has only been identified in physcomitrella (Table 1), suggesting that other protein(s) could be involved in this function in higher plants. The homologous protein of UBP8 in Arabidopsis is UBP22 (At5g10790), a member of a family of Ubiquitin-specific proteases highly conserved in eukaryotes. The function of UBP22 has not been described in plants. UBP8 has been described as an ubiquitin protease that specifically removes monoubiquitin from lysine 123 of the H2B C-terminal tail (Henry et al., 2003; Daniel et al., 2004). In humans, biochemical analysis of the substrate specificity of USP22 reveals that it deubiquitylates histone H2A in addition to H2B (Zhang et al., 2008). Although UBP8 contains an ubiquitin-specific hydrolase domain, the protein is inactive unless in complex with the other three DUB module proteins (Weake et al., 2008; Lee et al., 2009). The loci At3g27100 and At5g58575 are the homologous genes of SUS1 and SGF11, respectively. It has been demonstrated that the interaction of SUS1 with the SAGA complex requires UBP88 and SGF11, suggesting that SGF11 could be the direct binding partner of SUS1 (Köhler et al., 2006). Interestingly, although there is no evidence for the role of both proteins in Arabidopsis, the physical interaction between At3g27100 and At5g58575 has been reported (Arabidopsis Interactome Mapping, 2011), suggesting a conserved role inside the SAGA complex in plants.
Other Subunits
The protein CHD1 has been identified as a component of the SAGA complex in yeast (Pray-Grant et al., 2005). This protein is involved in ATP-dependent chromatin remodeling and contains a CHROMO domain that binds methylated H3K4. In Arabidopsis, CHR5 is the homologous protein of CHD1. This gene is expressed during embryo development and seed maturation and is directly involved in the activation of ABI3 and FUS3 expression, key transcriptional regulators of zygotic embryo development (Shen et al., 2015). This protein might recruit SAGA to chromatin and coordinates different complexes implicated in chromatin remodeling, like the COMPASS complex involved in tri-methyl marks on histone 3 lysine4.
Role of the SAGA Complex in Control of Gene Expression Under Abiotic Stress
Plant growth is significantly affected by environmental stresses such as cold, salinity, drought, light quality, temperature, and excess or deficiency of nutrients (reviewed in Mahajan and Tuteja, 2005 and Hänsch and Mendel, 2009). Therefore, plants have developed diverse strategies to adapt their growth in response to environmental changes and ensure reproductive success (Franklin et al., 2005; Bäurle and Dean, 2006). Epigenetic mechanisms have been implicated in regulating the expression of stress related genes (Chinnusamy and Zhu, 2009). Dynamic and reversible HAT under abiotic stress enables the switch between permissive and repressive chromatin that regulates transcription. Different members of the SAGA complex play pivotal roles in the environmental stress response and in many developmental transitions in plants (Figure 1; Chen and Tian, 2007; Vlachonasios et al., 2011; Kim et al., 2015, reviewed in Kim et al., 2010 and Servet et al., 2010). Additionally, the gene expression of some components of the SAGA complex is induced under elevated salt concentration and high temperature, add weight to a potentially significant role of SAGA components gene expression in plants during abiotic stresses (Srivastava et al., 2015).
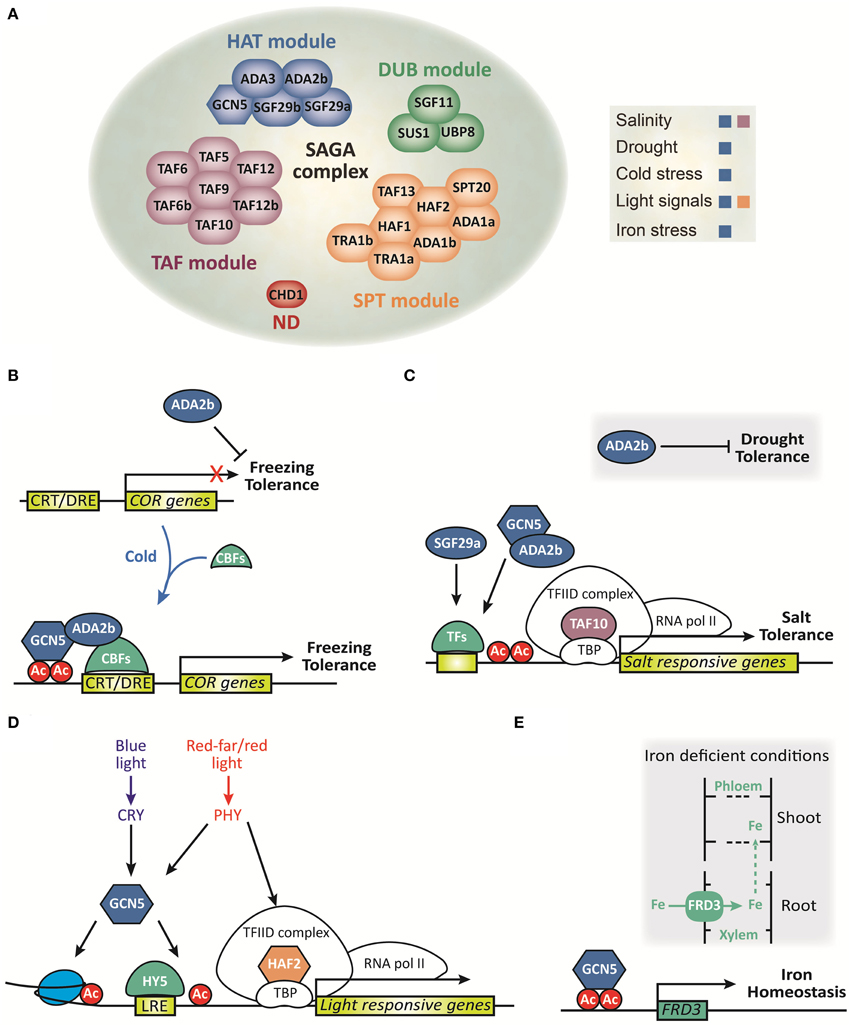
Figure 1. Composition and function of the SAGA complex in plants. (A) Schematic representation of each module that integrates into the SAGA complex and its role in abiotic stress response. ND, not defined. (B–E) Schematic representation of molecular functions of the SAGA complex under abiotic stress. (B) ADA2b represses freezing tolerance before cold exposure. During cold exposure, CBFs are induced and together with ADA2b and GCN5 promotes COR genes induction and consequently freezing tolerance. (C) ADA2b represses drought tolerance whereas it promotes histone acetylation of salt stress responsive genes and confers salt tolerance. TAF10 promotes salt tolerance during seed germination, while SGF29a plays a modest role in the expression of salt stress responsive genes in arabidopsis. TFs, transcription factors. (D) GCN5 integrates both blue and red/far-red light signals to induce histone acetylation and HY5-dependent gene activation of light responsive genes. TAF1 integrates red/far-red light signals to induce histone acetylation and gene activation of light-responsive genes (adapted from Servet et al., 2010). (E) Under deficient iron conditions GCN5 promotes histone acetylation of FRD3, which is involved in the transport of Fe into the xylem, to regulate iron homeostasis.
Salinity and Drought Stress
Plants have developed sophisticated signaling pathways that act in concert to counteract salinity and drought stress conditions through the action of transcription factors and histone modifications, thereby promoting the induction of many stress responsive genes and ultimately increasing stress tolerance (Reviewed in Chinnusamy and Zhu, 2009; Huang et al., 2012; Yuan et al., 2013; Golldack et al., 2014). The finding that Arabidopsis have homologs of both GCN5 and ADA2 genes (Stockinger et al., 2001) warrant additional study of how HAT-containing complexes related to SAGA complexes activate gene expression under abiotic stress conditions in plants. In Arabidopsis, ada2b and gcn5 mutants, but not ada2a mutants, demonstrate pleiotropic effects on plant growth, and development (Vlachonasios et al., 2003). Moreover, both mutants exhibit an altered response to low temperatures and hypersensitivity to salt and abscisic acid (Vlachonasios et al., 2003; Hark et al., 2009). In addition, the whole plant transpiration rate in ada2b mutants is lower in comparison to wild-type plants after water starvation, suggesting that drought tolerance arises from a reduction in transpiration water loss that probably occurs through stomata closure (Vlachonasios et al., 2011). Recently SGF29a has been identified as another component of the SAGA complex that is involved in stress response (Kaldis et al., 2011). While in root growth and seed germination assays the sgf29a-1 mutant plants are more resistant to salt stress, the reduction in transcript levels of salt stress responsive genes compared to wild-type plants suggests that SGF29a plays a modest role in the expression of salt-inducible genes (Kaldis et al., 2011). In contrast, the levels of salt stress responsive genes are dramatically reduced under salinity conditions in ada2b mutants. Interestingly, the reduction in transcript levels and the pattern of locus-specific acetylation of histones H3 and H4 of salt stress responsive genes in the ada2b mutant plants support the hypothesis that some transcription factors involved in salt stress response are capable of recruiting the SAGA complex to their target promoters (Kaldis et al., 2011).
On the other hand, a mutant screen from a chemical-inducible activation tagging allowed the identification of one mutant, designated stg1 (salt tolerance during germination1), which demonstrates an increased tolerance to salt and osmotic stress in comparison to wild-type plants during seed germination (Gao et al., 2006). STG1 encodes a putative Arabidopsis TBP-associated factor 10 (TAF10), which constitutes the TFIID complex involved in PIC assembly. The constitutive expression of TAF10 enhances seed tolerance to salt stress during germination, and the knocked-down mutant is more sensitive to salt stress (Gao et al., 2006). Together, this evidence suggests that TAF10 plays a role in mediating an adaptive response under adverse environmental conditions, but its direct interaction with SAGA complex has not yet been determined.
Cold Stress
The plant adaptive response to cold temperatures involves extensive physiological and biochemical changes such as stabilization of the integrity of cellular membranes and gene expression of inducible cold regulated (COR) genes (Reviewed in Thomashow, 1999; Lissarre et al., 2010). The inducible expression of COR genes is mediated mainly by a family of transcriptional activator proteins known as CBF/DREB1 which recognize the DNA regulatory element CRT/DRE present in the promoters of many COR and dehydration inducible genes (Yamaguchi-Shinozaki and Shinozaki, 1994; Park et al., 2015). The CBF transcription factors alter the expression of more than 100 genes that contribute to enhanced freezing tolerance (Fowler and Thomashow, 2002; Vogel et al., 2005). In Arabidopsis, protein interaction assays revealed that the DNA-binding domain of CBF1 binds directly to ADA2b-containing SAGA complexes (Mao et al., 2006). Additionally, the evidence that the transcriptional activity of Arabidopsis CBF1 in yeasts requires ADA2, ADA3, and GCN5 to activate the transcription of reporter genes carrying the CRT/DRE regulatory element (Stockinger et al., 2001), paired with the observation that the expression of CBFs are induced and COR genes are reduced in gcn5 and ada2b cold-acclimated mutant Arabidopsis plants, supports the notion that CBFs stimulate transcription through recruitment of SAGA transcriptional adaptor complexes to the promoters of COR genes (Vlachonasios et al., 2003). Remarkably, non-acclimated ada2b mutant plants are more tolerant to freezing temperatures than wild-type plants, indicating that freezing tolerance in non-acclimated ada2b mutant is achieved by a novel, undefined pathway that does not require the expression of CBF or COR genes (Vlachonasios et al., 2003). Thus, ADA2b and GCN5 proteins have similar yet distinct functions in gene expression and may be also components of separate co-activator complexes with different biological activities.
Light Signals
Plants perceive light by a set of wavelength-specific photoreceptors such as phytochromes (PHY) and cryptochromes (CRY) that direct adaptive changes in gene expression in response to environmental signals. Ultimately, these light signals are integrated by downstream DNA-binding transcription factors, which bind to several light responsive elements (LRE) present in the promoters of light-inducible genes (Reviewed in Casal and Yanovsky, 2005; Franklin et al., 2005; Jiao et al., 2007). It has been determined that HAF2 functions in concert with GCN5 to integrate light signals and acetylate the core promoter regions of light-inducible genes (Bertrand et al., 2005; Benhamed et al., 2006). Indeed, double mutations of HAF2 and HY5, a bZIP transcription factor that promotes the expression of light-inducible genes, have a synergic effect on hypocotyl length (a photomorphogenesis trait) and light-activated gene expression under different light wavelengths (Bertrand et al., 2005; Benhamed et al., 2006). This suggests that HAF2 is involved in the signaling pathways of both red/far-red and blue signals, and interacts with HY5 to rapidly activate the expression of light-responsive genes. Moreover, gcn5/taf1 double mutations result in a further loss of light-responsive genes and exert a cumulative effect on both plant growth and H3K9 acetylation (Benhamed et al., 2006). This evidence, together with the observation that GCN5 and HY5 share many genomic targets (Benhamed et al., 2008), indicates that GCN5 and HY5 might act cooperatively to activate the expression of light-inducible genes. Thus, HAF2 is presumably recruited to its target promoters by interacting with the TBPs, while GCN5 may be recruited to the target promoters by interacting either with specific DNA-binding transcription factors such as HY5 and/or with acetylated histone lysine residues of nearby nucleosomes. Recently, it has been reported that the expression of light-activated genes is considerably reduced in six SAGA subunits in Arabidopsis mutants (Srivastava et al., 2015), indicating that other components of SAGA are involved in the expression of light-inducible genes as well.
Nutritional Stress
Recently a report demonstrated that a mutation in GCN5 resulted in accumulation of manganese, zinc, and iron in the roots (Xing et al., 2015). Specifically, this mutant exhibited impaired iron translocation from the root to the shoot, and this retention was rescued by TSA treatment, a chemical inhibitor of histone deacetylase (Xing et al., 2015). These results suggest that HAT via GCN5 is an important mechanism for iron distribution in Arabidopsis. In addition, GCN5 is necessary for the expression of hundreds of genes involved in iron homeostasis (Xing et al., 2015). These observations, together with the fact that GCN5 directly binds to the promoters of FRD3, a key gene in iron homeostasis, and modulates the H3K9/14 global acetylation levels under iron deficient conditions, suggest that GCN5 plays a critical role in iron homeostasis through the regulation of target genes (Xing et al., 2015). There is no evidence for the role of other members of the SAGA complex in the regulation of nutrients homeostasis.
Concluding Remarks
The protein complexes involved in chromatin remodeling and epigenetic modifications are highly conserved in eukaryotes. The SAGA complex is no exception, and although highly conserved in plants, the physical and functional relationships between its different modules remain to be elucidated. Additional study is needed to identify the target genes of the SAGA complex in different environmental conditions and developmental stages, as well as which transcription factors interact with these complexes. Further characterization of the SAGA complex presents the opportunity to identify new actors that participate in the control of gene expression in plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant of CONICYT-Chile (FONDECYT N° 11130567) awarded to FA, the Center for Applied Ecology and Sustainability (CAPES FB-002-2014), and the Millennium Nucleus Center for Plant Systems and Synthetic Biology (NC130030). We thank to Angela Court for the support in this manuscript and Alyssa Grube for assistance in language support.
References
Albright, S. R., and Tjian, R. (2000). TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242, 1–13. doi: 10.1016/S0378-1119(99)00495-3
Allard, S., Utley, R. T., Savard, J., Clarke, A., Grant, P., Brandl, C. J., et al. (1999). NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM−related cofactor Tra1p. EMBO J. 18, 5108–5119. doi: 10.1093/emboj/18.18.5108
Aquea, F., Timmermann, T., and Arce-Johnson, P. (2010). Analysis of histone acetyltransferase and deacetylase families of Vitis vinifera. Plant Physiol. Biochem. 48, 194–199. doi: 10.1016/j.plaphy.2009.12.009
Arabidopsis Interactome Mapping, C. (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607. doi: 10.1126/science.1203877
Barbaric, S., Walker, J., Schmid, A., Svejstrup, J. Q., and Horz, W. (2001). Increasing the rate of chromatin remodeling and gene activation–a novel role for the histone acetyltransferase Gcn5. EMBO J. 20, 4944–4951. doi: 10.1093/emboj/20.17.4944
Bäurle, I., and Dean, C. (2006). The timing of developmental transitions in plants. Cell 125, 655–664. doi: 10.1016/j.cell.2006.05.005
Belotserkovskaya, R., Sterner, D. E., Deng, M., Sayre, M. H., Lieberman, P. M., and Berger, S. L. (2000). Inhibition of TATA binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 20, 634–647. doi: 10.1128/MCB.20.2.634-647.2000
Benhamed, M., Bertrand, C., Servet, C., and Zhou, D. X. (2006). Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18, 2893–2903. doi: 10.1105/tpc.106.043489
Benhamed, M., Martin-Magniette, M. L., Taconnat, L., Bitton, F., Servet, C., De Clercq, R., et al. (2008). Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J. 56, 493–504. doi: 10.1111/j.1365-313X.2008.03606.x
Bertrand, C., Benhamed, M., Li, Y. F., Ayadi, M., Lemonnier, G., Renou, J. P., et al. (2005). Arabidopsis HAF2 gene encoding TATA-binding protein (TBP)-associated factor TAF1, is required to integrate light signals to regulate gene expression and growth. J. Biol. Chem. 280, 1465–1473. doi: 10.1074/jbc.M409000200
Bhat, R., Borst, J., Riehl, M., and Thompson, R. (2004). Interaction of maize Opaque-2 and the transcriptional co-activators GCN5 and ADA2, in the modulation of transcriptional activity. Plant Mol. Biol. 55, 239–252. doi: 10.1007/s11103-004-0553-z
Bhat, R., Riehl, M., Santandrea, G., Velasco, R., Slocombe, S., Donn, G., et al. (2003). Alteration of GCN5 levels in maize reveals dynamic responses to manipulating histone acetylation. Plant J. 33, 455–469. doi: 10.1046/j.1365-313X.2003.01642.x
Bian, C., Xu, C., Ruan, J., Lee, K. K., Burke, T. L., Tempel, W., et al. (2011). Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 30, 2829–2842. doi: 10.1038/emboj.2011.193
Brown, C., Howe, L., Sousa, K., Alley, S., Carrozza, M., Tan, S., et al. (2001). Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292, 2333–2337. doi: 10.1126/science.1060214
Casal, J., and Yanovsky, M. (2005). Regulation of gene expression by light. Int. J. Dev. Biol. 49, 501–511. doi: 10.1387/ijdb.051973jc
Chen, Z., and Tian, L. (2007). Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim. Biophys. Acta 1769, 295–307. doi: 10.1016/j.bbaexp.2007.04.007
Chinnusamy, V., and Zhu, J. (2009). Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12, 133–139. doi: 10.1016/j.pbi.2008.12.006
Daniel, J. A., and Grant, P. A. (2007). Multi-tasking on chromatin with the SAGA coactivator complexes. Mutat. Res. 618, 135–148. doi: 10.1016/j.mrfmmm.2006.09.008
Daniel, J., Torok, M., Sun, Z., Schieltz, D., Allis, C., Yates, J. III, et al. (2004). Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279, 1867–1871. doi: 10.1074/jbc.C300494200
Dudley, A., Rougeulle, C., and Winston, F. (1999). The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13, 2940–2945. doi: 10.1101/gad.13.22.2940
Earley, K., Shook, M., Brower-Toland, B., Hicks, L., and Pikaard, C. (2007). In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J. 52, 615–626. doi: 10.1111/j.1365-313X.2007.03264.x
Endo, M., Tanigawa, Y., Murakami, T., Araki, T., and Nagatani, A. (2013). Phytochrome-dependent late-flowering accelerates flowering through physical interactions with phytochrome B and constans. Proc. Natl. Acad. Sci. U.S.A. 110, 18017–18022. doi: 10.1073/pnas.1310631110
Fowler, S., and Thomashow, M. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. doi: 10.1105/tpc.003483
Franklin, K., Larner, V., and Whitelam, G. (2005). The signal transducing photoreceptors of plants. Int. J. Dev. Biol. 49, 653–664. doi: 10.1387/ijdb.051989kf
Furumoto, T., Tamada, Y., Izumida, A., Nakatani, H., Hata, S., and Izui, K. (2005). Abundant expression in vascular tissue of plant TAF10, an orthologous gene for TATA box-binding protein-associated factor 10, in Flaveria trinervia and abnormal morphology of Arabidopsis thaliana transformants on its overexpression. Plant Cell Physiol. 46, 108–117. doi: 10.1093/pcp/pci006
Gamper, A. M., Kim, J., and Roeder, R. G. (2009). The STAGA subunit ADA2b is an important regulator of human GCN5 catalysis. Mol. Cell. Biol. 29, 266–280. doi: 10.1128/MCB.00315-08
Gao, X., Ren, F., and Lu, Y. T. (2006). The Arabidopsis mutant stg1 identifies a function for TBP-associated factor 10 in plant osmotic stress adaptation. Plant Cell Physiol. 47, 1285–1294. doi: 10.1093/pcp/pcj099
Golldack, D., Li, C., Mohan, H., and Probst, N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5:151. doi: 10.3389/fpls.2014.00151
Goodrich, J. A., Cutler, G., and Tjian, R. (1996). Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell 84, 825–830. doi: 10.1016/S0092-8674(00)81061-2
Govind, C. K., Yoon, S., Qiu, H., Govind, S., and Hinnebusch, A. G. (2005). Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol. Cell. Biol. 25, 5626–5638. doi: 10.1128/MCB.25.13.5626-5638.2005
Govind, C. K., Zhang, F., Qiu, H., Hofmeyer, K., and Hinnebusch, A. G. (2007). Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25, 31–42. doi: 10.1016/j.molcel.2006.11.020
Grant, P., Duggan, L., Cote, J., Roberts, S. M., Brownell, J. E., Candau, R., et al. (1997). Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11, 1640–1650. doi: 10.1101/gad.11.13.1640
Grant, P., Eberharter, A., John, S., Cook, R. G., Turner, B. M., and Workman, J. L. (1999). Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274, 5895–5900. doi: 10.1074/jbc.274.9.5895
Grant, P., Schieltz, D., Pray−Grant, M. G., Yates, J. R. III, and Workman, J. L. (1998). The ATM−related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell 2, 863–867. doi: 10.1016/S1097-2765(00)80300-7
Hänsch, R., and Mendel, R. (2009). Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 12, 259–266. doi: 10.1016/j.pbi.2009.05.006
Hark, A. T., Vlachonasios, K. E., Pavangadkar, K. A., Rao, S., Gordon, H., Adamakis, I. D., et al. (2009). Two Arabidopsis orthologs of the transcriptional coactivator ADA2 have distinct biological functions. Biochim. Biophys. Acta 1789, 117–124. doi: 10.1016/j.bbagrm.2008.09.003
Hassan, A. H., Awad, S., Al-Natour, Z., Othman, S., Mustafa, F., and Rizvi, T. A. (2007). Selective recognition of acetylated histones by bromodomains in transcriptional co-activators. Biochem. J. 402, 125–133. doi: 10.1042/BJ20060907
Henry, K. W., Wyce, A., Lo, W. S., Duggan, L. J., Emre, N. C., Kao, C. F., et al. (2003). Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17, 2648–2663. doi: 10.1101/gad.1144003
Hiller, M. A., Lin, T. Y., Wood, C., and Fuller, M. T. (2001). Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 15, 1021–1030. doi: 10.1101/gad.869101
Huang, G. T., Ma, S. L., Bai, L. P., Zhang, L., Ma, H., Jia, P., et al. (2012). Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 39, 969–987. doi: 10.1007/s11033-011-0823-1
Huisinga, K. L., and Pugh, B. F. (2004). A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13, 573–585. doi: 10.1016/S1097-2765(04)00087-5
Jacobson, R. H., Ladurner, A. G., King, D. S., and Tjian, R. (2000). Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425. doi: 10.1126/science.288.5470.1422
Jiao, Y., Lau, O. S., and Deng, X. W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8, 217–230. doi: 10.1038/nrg2049
Kaldis, A., Tsementzi, D., Tanriverdi, O., and Vlachonasios, K. E. (2011). Arabidopsis thaliana transcriptional co-activators ADA2b and SGF29a are implicated in salt stress responses. Planta 233, 749–762. doi: 10.1007/s00425-010-1337-0
Kim, J. M., To, T. K., Nishioka, T., and Seki, M. (2010). Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 33, 604–611. doi: 10.1111/j.1365-3040.2009.02076.x
Kim, J. Y., Oh, J. E., Noh, Y. S., and Noh, B. (2015). Epigenetic control of juvenile-to-adult phase transition by the Arabidopsis SAGA-like complex. Plant J. 83, 537–545. doi: 10.1111/tpj.12908
Knutson, B. A., and Hahn, S. (2011). Domains of Tra1 important for activator recruitment and transcription coactivator functions of SAGA and NuA4 complexes. Mol. Cell. Biol. 31, 818–831. doi: 10.1128/MCB.00687-10
Köhler, A., Pascual-Garcia, P., Llopis, A., Zapater, M., Posas, F., Hurt, E., et al. (2006). The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol. Biol. Cell 17, 4228–4236. doi: 10.1091/mbc.E06-02-0098
Koutelou, E., Hirsch, C. L., and Dent, S. Y. (2010). Multiple faces of the SAGA complex. Curr. Opin. Cell Biol. 22, 374–382. doi: 10.1016/j.ceb.2010.03.005
Kubo, M., Furuta, K., Demura, T., Fukuda, H., Liu, Y. G., Shibata, D., et al. (2011). The CKH1/EER4 gene encoding a TAF12-like protein negatively regulates cytokinin sensitivity in Arabidopsis thaliana. Plant Cell Physiol. 52, 629–637. doi: 10.1093/pcp/pcr021
Kuo, M. H., Brownell, J. E., Sobel, R. E., Ranalli, T. A., Cook, R. G., Edmondson, D. G., et al. (1996). Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383, 269–272. doi: 10.1038/383269a0
Kuo, M. H., Zhou, J., Jambeck, P., Churchill, M. E., and Allis, C. D. (1998). Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12, 627–639. doi: 10.1101/gad.12.5.627
Lago, C., Clerici, E., Dreni, L., Horlow, C., Caporali, E., Colombo, L., et al. (2005). The Arabidopsis TFIID factor AtTAF6 controls pollen tube growth. Dev. Biol. 285, 91–100. doi: 10.1016/j.ydbio.2005.06.006
Lago, C., Clerici, E., Mizzi, L., Colombo, L., and Kater, M. M. (2004). TBP-associated factors in Arabidopsis. Gene 342, 231–241. doi: 10.1016/j.gene.2004.08.023
Lawit, S. J., O'grady, K., Gurley, W. B., and Czarnecka-Verner, E. (2007). Yeast two-hybrid map of Arabidopsis TFIID. Plant Mol. Biol. 64, 73–87. doi: 10.1007/s11103-007-9135-1
Lee, K. K., Sardiu, M. E., Swanson, S. K., Gilmore, J. M., Torok, M., Grant, P. A., et al. (2011). Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol. Syst. Biol. 7, 503. doi: 10.1038/msb.2011.40
Lee, K., Swanson, S., Florens, L., Washburn, M., and Workman, J. (2009). Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik (SALSA) HAT complexes. Epigenetics Chromatin 2:2. doi: 10.1186/1756-8935-2-2
Lee, T. I., Causton, H. C., Holstege, F. C., Shen, W. C., Hannett, N., Jennings, E. G., et al. (2000). Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405, 701–704. doi: 10.1038/35015104
Lindner, M., Simonini, S., Kooiker, M., Gagliardini, V., Somssich, M., Hohenstatt, M., et al. (2013). TAF13 interacts with PRC2 members and is essential for Arabidopsis seed development. Dev. Biol. 379, 28–37. doi: 10.1016/j.ydbio.2013.03.005
Lissarre, M., Ohta, M., Sato, A., and Miura, K. (2010). Cold-responsive gene regulation during cold acclimation in plants. Plant Signal. Behav. 5, 948–952. doi: 10.4161/psb.5.8.12135
Liu, X., Luo, M., Zhang, W., Zhao, J., Zhang, J., Wu, K., et al. (2012). Histone acetyltransferases in rice (Oryza sativa L.): phylogenetic analysis, subcellular localization and expression. BMC Plant Biol. 12:145. doi: 10.1186/1471-2229-12-145
Mahajan, S., and Tuteja, N. (2005). Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 444, 139–158. doi: 10.1016/j.abb.2005.10.018
Mao, Y., Pavangadkar, K. A., Thomashow, M. F., and Triezenberg, S. J. (2006). Physical and functional interactions of Arabidopsis ADA2 transcriptional coactivator proteins with the acetyltransferase GCN5 and with the cold-induced transcription factor CBF1. Biochim. Biophys. Acta 1759, 69–79. doi: 10.1016/j.bbaexp.2006.02.006
Memedula, S., and Belmont, A. S. (2003). Sequential recruitment of HAT and SWI/SNF components to condensed chromatin by VP16. Curr. Biol. 13, 241–246. doi: 10.1016/S0960-9822(03)00048-4
Mougiou, N., Poulios, S., Kaldis, A., and Vlachonasios, K. (2012). Arabidopsis thaliana TBP-associated factor 5 is essential for plant growth and development. Mol Breeding 30, 355–366. doi: 10.1007/s11032-011-9626-2
Mujtaba, S., Zeng, L., and Zhou, M. M. (2007). Structure and acetyl-lysine recognition of the bromodomain. Oncogene 26, 5521–5527. doi: 10.1038/sj.onc.1210618
Murr, R., Vaissiere, T., Sawan, C., Shukla, V., and Herceg, Z. (2007). Orchestration of chromatin−based processes: mind the TRRAP. Oncogene 26, 5358–5372. doi: 10.1038/sj.onc.1210605
Näär, A. M., Lemon, B. D., and Tjian, R. (2001). Transcriptional coactivator complexes. Annu. Rev. Biochem. 70, 475–501. doi: 10.1146/annurev.biochem.70.1.475
Nguyen-Huynh, N. T., Sharov, G., Potel, C., Fichter, P., Trowitzsch, S., Berger, I., et al. (2015). Chemical cross-linking and mass spectrometry to determine the subunit interaction network in a recombinant human SAGA HAT subcomplex. Protein Sci. 24, 1232–1246. doi: 10.1002/pro.2676
Pandey, R., Muller, A., Napoli, C. A., Selinger, D. A., Pikaard, C. S., Richards, E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055. doi: 10.1093/nar/gkf660
Park, S., Lee, C. M., Doherty, C. J., Gilmour, S. J., Kim, Y., and Thomashow, M. F. (2015). Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 82, 193–207. doi: 10.1111/tpj.12796
Pray-Grant, M. G., Daniel, J. A., Schieltz, D., Yates, J. R. III, and Grant, P. A. (2005). Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433, 434–438. doi: 10.1038/nature03242
Qiu, H., Hu, C., Yoon, S., Natarajan, K., Swanson, M. J., and Hinnebusch, A. G. (2004). An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol. Cell. Biol. 24, 4104–4117. doi: 10.1128/MCB.24.10.4104-4117.2004
Ricci, A. R., Genereaux, J., and Brandl, C. J. (2002). Components of the SAGA histone acetyltransferase complex are required for repressed transcription of ARG1 in rich medium. Mol. Cell. Biol. 22, 4033–4042. doi: 10.1128/MCB.22.12.4033-4042.2002
Robles, L. M., Wampole, J. S., Christians, M. J., and Larsen, P. B. (2007). Arabidopsis enhanced ethylene response 4 encodes an EIN3-interacting TFIID transcription factor required for proper ethylene response, including ERF1 induction. J. Exp. Bot. 58, 2627–2639. doi: 10.1093/jxb/erm080
Rodríguez-Navarro, S., Fischer, T., Luo, M. J., Antunez, O., Brettschneider, S., Lechner, J., et al. (2004). Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116, 75–86. doi: 10.1016/S0092-8674(03)01025-0
Servet, C., Benhamed, M., Latrasse, D., Kim, W., Delarue, M., and Zhou, D. X. (2008). Characterization of a phosphatase 2C protein as an interacting partner of the histone acetyltransferase GCN5 in Arabidopsis. Biochim. Biophys. Acta 1779, 376–382. doi: 10.1016/j.bbagrm.2008.04.007
Servet, C., Conde E Silva, N., and Zhou, D. X. (2010). Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol. Plant 3, 670–677. doi: 10.1093/mp/ssq018
Shen, Y., Devic, M., Lepiniec, L., and Zhou, D. X. (2015). Chromodomain, Helicase and DNA-binding CHD1 protein, CHR5, are involved in establishing active chromatin state of seed maturation genes. Plant Biotechnol. J. 13, 811–820. doi: 10.1111/pbi.12315
Shukla, A., Lahudkar, S., Durairaj, G., and Bhaumik, S. R. (2012). Sgf29p facilitates the recruitment of TATA box binding protein but does not alter SAGA's global structural integrity in vivo. Biochemistry 51, 706–714. doi: 10.1021/bi201708z
Spedale, G., Timmers, H. T., and Pijnappel, W. W. (2012). ATAC-king the complexity of SAGA during evolution. Genes Dev. 26, 527–541. doi: 10.1101/gad.184705.111
Srivastava, R., Rai, K. M., Pandey, B., Singh, S. P., and Sawant, S. V. (2015). Spt-Ada-Gcn5-Acetyltransferase (SAGA) complex in plants: genome wide identification, evolutionary conservation and functional determination. PLoS ONE 10:e0134709. doi: 10.1371/journal.pone.0134709
Sterner, D. E., Grant, P. A., Roberts, S. M., Duggan, L. J., Belotserkovskaya, R., Pacella, L. A., et al. (1999). Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19, 86–98.
Stockinger, E. J., Mao, Y., Regier, M. K., Triezenberg, S. J., and Thomashow, M. F. (2001). Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29, 1524–1533. doi: 10.1093/nar/29.7.1524
Struhl, K., and Moqtaderi, Z. (1998). The TAFs in the HAT. Cell 94, 1–4. doi: 10.1016/S0092-8674(00)81213-1
Tamada, Y., Nakamori, K., Nakatani, H., Matsuda, K., Hata, S., Furumoto, T., et al. (2007). Temporary expression of the TAF10 gene and its requirement for normal development of Arabidopsis thaliana. Plant Cell Physiol. 48, 134–146. doi: 10.1093/pcp/pcl048
Thomashow, M. F. (1999). Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Vlachonasios, K. E., Kaldis, A., Nikoloudi, A., and Tsementzi, D. (2011). The role of transcriptional coactivator ADA2b in Arabidopsis abiotic stress responses. Plant Signal. Behav. 6, 1475–1478. doi: 10.4161/psb.6.10.17695
Vlachonasios, K. E., Thomashow, M. F., and Triezenberg, S. J. (2003). Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15, 626–638. doi: 10.1105/tpc.007922
Vogel, J. T., Zarka, D. G., Van Buskirk, H. A., Fowler, S. G., and Thomashow, M. F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41, 195–211. doi: 10.1111/j.1365-313X.2004.02288.x
Wang, L., and Dent, S. Y. (2014). Functions of SAGA in development and disease. Epigenomics 6, 329–339. doi: 10.2217/epi.14.22
Weake, V. M., Lee, K. K., Guelman, S., Lin, C. H., Seidel, C., Abmayr, S. M., et al. (2008). SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 27, 394–405. doi: 10.1038/sj.emboj.7601966
Wen, J. F., Huo, J. L., Chen, H. X., Ma, C. H., Jiang, H., Zhu, H. S., et al. (2013). Cloning and bioinformatic analysis of full-length novel pepper (Capsicum annuum) genes TAF10 and TAF13. Genet. Mol. Res. 12, 6947–6956. doi: 10.4238/2013.December.19.14
Wu, P. Y., Ruhlmann, C., Winston, F., and Schultz, P. (2004). Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15, 199–208. doi: 10.1016/j.molcel.2004.06.005
Xing, J., Wang, T., Liu, Z., Xu, J., Yao, Y., Hu, Z., et al. (2015). GCN5-mediated Histone Acetylation of FRD3 Contributes to Iron Homeostasis in Arabidopsis thaliana. Plant Physiol. 168, 1309–1320. doi: 10.1104/pp.15.00397
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. doi: 10.1105/tpc.6.2.251
Yuan, L., Liu, X., Luo, M., Yang, S., and Wu, K. (2013). Involvement of histone modifications in plant abiotic stress responses. J. Integr. Plant Biol. 55, 892–901. doi: 10.1111/jipb.12060
Keywords: SAGA complex, chromatin remodeling, transcriptional coactivator, abiotic stress, protein complex, histone acetyltransferase
Citation: Moraga F and Aquea F (2015) Composition of the SAGA complex in plants and its role in controlling gene expression in response to abiotic stresses. Front. Plant Sci. 6:865. doi: 10.3389/fpls.2015.00865
Received: 30 July 2015; Accepted: 30 September 2015;
Published: 14 October 2015.
Edited by:
Mahmoud W. Yaish, Sultan Qaboos University, OmanReviewed by:
Jorge E. Mayer, fAB Consult (freelance AgBiotech), AustraliaSamir Sawant, Council of Scientific and Industrial Research - National Botanical Research Institute, India
Copyright © 2015 Moraga and Aquea. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Felipe Aquea, ZmVsaXBlLmFxdWVhQHVhaS5jbA==
 Felipe Moraga
Felipe Moraga Felipe Aquea
Felipe Aquea