
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 01 October 2015
Sec. Plant Biotechnology
Volume 6 - 2015 | https://doi.org/10.3389/fpls.2015.00818
This article is part of the Research Topic Carbon allocation View all 5 articles
Terpenoid indole alkaloid (TIA) biosynthesis in Catharanthus roseus is a complex and highly regulated process. Understanding the biochemistry and regulation of the TIA pathway is of particular interest as it may allow the engineering of plants to accumulate higher levels of pharmaceutically important alkaloids. Toward this end, we generated a transgenic C. roseus hairy root line that overexpresses the CrBPF1 transcriptional activator under the control of a β-estradiol inducible promoter. CrBPF1 is a MYB-like protein that was previously postulated to help regulate the expression of the TIA biosynthetic gene STR. However, the role of CrBPF1 in regulation of the TIA and related pathways had not been previously characterized. In this study, transcriptional profiling revealed that overexpression of CrBPF1 results in increased transcript levels for genes from both the indole and terpenoid biosynthetic pathways that provide precursors for TIA biosynthesis, as well as for genes in the TIA biosynthetic pathway. In addition, overexpression of CrBPF1 causes increases in the transcript levels for 11 out of 13 genes postulated to act as transcriptional regulators of genes from the TIA and TIA feeder pathways. Interestingly, overexpression of CrBPF1 causes increased transcript levels for both TIA transcriptional activators and repressors. Despite the fact that CrBPF1 overexpression affects transcript levels of a large percentage of TIA biosynthetic and regulatory genes, CrBPF1 overexpression has only very modest effects on the levels of the TIA metabolites analyzed. This finding may be due, at least in part, to the up-regulation of both transcriptional activators and repressors in response to CrBPF1 overexpression, suggesting that CrBPF1 may serve as a “fine-tune” regulator for TIA biosynthesis, acting to help regulate the timing and amplitude of TIA gene expression.
Madagascar periwinkle Catharanthus roseus (L.) G. Don is of substantial pharmaceutical interest as it produces over 130 terpenoid indole alkaloids (TIAs). Several of these TIAs are used to treat different medical conditions. For example, vinblastine and vincristine are widely used as anticancer agents in the treatment of lymphoma and leukemia (Gidding et al., 1999) and ajmalicine and serpentine may be used to treat hypertension. Unfortunately these plant-derived pharmaceutical compounds are very expensive as periwinkle produces TIAs in very low amounts. In addition, due to their complex chemical structures, TIAs tend to be difficult, and therefore expensive, to synthesize in vitro. To lower the costs of producing TIAs for use as pharmaceuticals, many efforts have been made to increase TIA production using plant tissue and cell cultures or bacterial cultures. However, despite efforts since the early 1980s to develop cost-effective methods for large-scale production of TIAs in cultures or in vitro, progress has been limited (van der Heijden et al., 2004; Shanks, 2005).
A major reason why these efforts have not been more successful is that the TIA biosynthetic pathway and the regulation of the TIA pathway and TIA feeder pathways are not sufficiently well understood. Those deficiencies are beginning to be addressed, thanks to progress in identifying most of the genes encoding TIA biosynthetic enzymes and transcriptional regulators (Liu et al., 2007; Memelink and Gantet, 2007; Zhou et al., 2010a; Giddings et al., 2011; Asada et al., 2013; Besseau et al., 2013; Salim et al., 2013; Zhao et al., 2013; De Luca et al., 2014; Kellner et al., 2015; Qu et al., 2015; Van Moerkercke et al., 2015). TIA biosynthesis is a closely coordinated process involving many enzymatic steps that occur in several intra- and inter-cellular compartments (Burlat et al., 2004; van der Heijden et al., 2004; Murata et al., 2008; De Luca et al., 2014; Qu et al., 2015). TIA biosynthesis in C. roseus starts with the formation of strictosidine from tryptamine and secologanin, the two precursor molecules that are produced by the indole and the monoterpenoid pathways, respectively (Figure 1). The condensation process is catalyzed by strictosidine synthase (STR). Strictosidine is then deglucosylated by strictosidine β-D-glucosidase (SGD) to form strictosidine aglycone. Further enzymatic steps result in the formation of numerous TIAs via several specific branches. For example, one branch of the TIA pathway produces ajmalicine and serpentine, a second branch leads to production of lochnericine and hörhammericine, a third branch produces vindoline and a fourth branch produces catharanthine. The production of vindoline from tabersonine occurs via seven reactions. Genes encoding enzymes catalyzing all seven steps have now been identified (Vazquez-Flota et al., 1997; St-Pierre et al., 1998; Schröder et al., 1999; Levac et al., 2008; Liscombe et al., 2010; Qu et al., 2015). Vindoline and catharanthine are the substrates for a major class III peroxidase (PRX1), which catalyzes formation of α-3′,4′-anhydrovinblastine (Costa et al., 2008). Vinblastine and vincristine, two of the most pharmaceutically important TIAs, are formed through multiple steps from α-3′,4′-anhydrovinblastine.
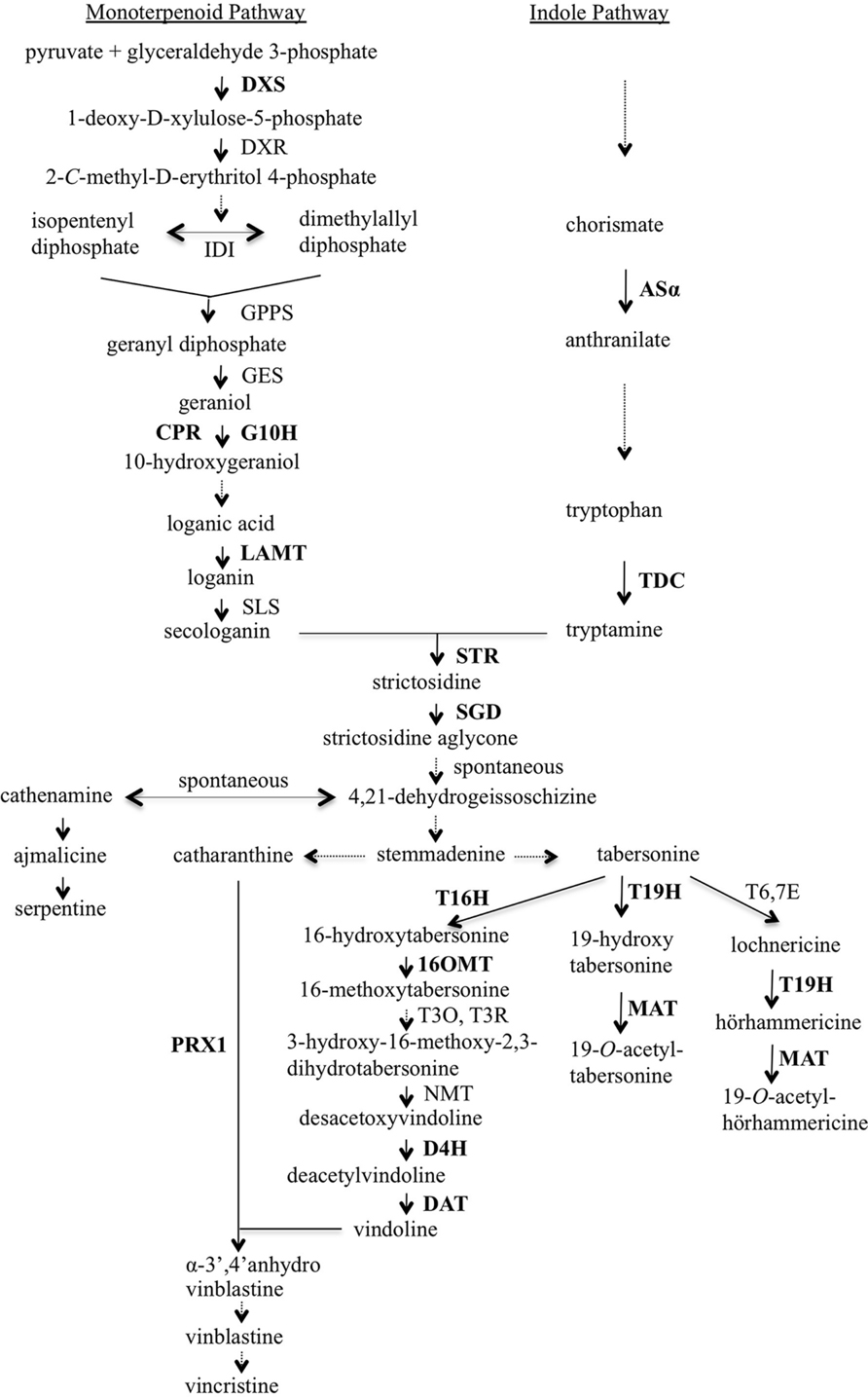
FIGURE 1. Overview of terpenoid indole alkaloid (TIA) biosynthesis in Catharanthus roseus. Enzymes are indicated using capital letters and metabolites using lowercase letters. Solid arrows indicate single enzymatic conversions; dashed arrows indicate multiple enzymatic reactions. The genes analyzed in this work are shown in bold type. 16OMT, 16-hydroxytabersonine-O-methyl-transferase; 19H, 19-hydroxylase; AS, anthranilate synthase; CPR, cytochrome P450 reductase; D4H, desacetoxyvindoline 4-hydroxylase; DAT, deacetylvindoline 4-O-acetyltransferase; DXR, 1-deoxy-D-xylulose-5-phosphate reductoisomerase; DXS, 1-deoxy-D-xylulose-5-phosphate synthase; G10H, geraniol 10-hydroxylase; GES, geraniol synthase; GPPS, geranyl diphosphate synthase; IDI, isopentenyl diphosphate isomerase; LAMT, loganic acid O-methyltransferase; MAT, minovincinine-19-O-acetyltransferase; NMT, N-methyltransferase; PRX1, vacuolar class III peroxidase; SGD, strictosidine glucosidase; SLS, secologanin synthase; STR, strictosidine synthase; T3O, tabersonine 3-oxygenase; T3R, tabersonine 3-reductase; T6,7E, tabersonine 6,7-epoxidase; T16H, tabersonine 16-hydroxylase; T19H, tabersonine/lochnericine19-hydroxylase; TDC, tryptophan decarboxylase.
Transcriptional activators and repressors play important roles in regulating TIA biosynthesis. Expression of both Octadecanoid-Responsive Catharanthus AP2-domain protein 2 (ORCA2) and ORCA3 increases rapidly upon fungal elicitation (Menke et al., 1999; van der Fits and Memelink, 2000). ORCA2 and ORCA3 are AP2-domain transcription factors that activate STR expression through a jasmonate signal transduction pathway by binding to the jasmonate and elicitor-responsive element (JERE) in the STR promoter (Menke et al., 1999; van der Fits and Memelink, 2001). ORCA2 (Li et al., 2013) and ORCA3 (van der Fits and Memelink, 2000; Peebles et al., 2009; Wang et al., 2010; Zhou et al., 2010b; Tang et al., 2011; Pan et al., 2012; Van Moerkercke et al., 2015) have also been shown to regulate many additional TIA biosynthetic and regulatory genes. The levels of some TIAs are also affected by overexpression of ORCA2 (Liu et al., 2011; Li et al., 2013) and/or ORCA3 (van der Fits and Memelink, 2000; Peebles et al., 2009; Wang et al., 2010; Zhou et al., 2010b; Tang et al., 2011; Pan et al., 2012). BIS1, a basic Helix-Loop-Helix transcription factor, regulates expression of the genes encoding all of the enzymes necessary for the conversion of geranyl diphosphate to loganic acid (Van Moerkercke et al., 2015). The CrMYC2 (basic Helix-Loop-Helix) transcription factor helps regulate TIA production by controlling the jasmonate-responsive expression of the ORCA genes (Zhang et al., 2011). Two AT-hook DNA binding proteins, 2D173 and 2D7, also help regulate ORCA3 expression (Vom Endt et al., 2007). The CrMYC1 transcription factor helps regulate STR expression and is responsive to both jasmonate and elicitor treatments (Chatel et al., 2003). Similarly, the CrWRKY1 and CrWRKY2 transcription factors exert positive regulatory effects on multiple TIA biosynthetic genes (Suttipanta, 2011; Suttipanta et al., 2011). However, overexpression of CrWRKY1 and CrWRKY2 has contrasting effects on expression of specific TIA regulatory genes. Overexpression of CrWRKY1 results in decreased transcript levels for the ORCA2, ORCA3, and CrMYC2 TIA transcriptional activators and increased transcript levels for the ZCT1, ZCT2, and ZCT3 TIA transcriptional repressors (Suttipanta, 2011), whereas overexpression of CrWRKY2 results in increased transcript levels for both the ORCA2, ORCA3, and CrWRKY1 TIA transcriptional activators and the ZCT1 and ZCT3 TIA transcriptional repressors (Suttipanta et al., 2011). In addition to transcriptional activators, three zinc finger proteins, ZCT1, ZCT2, and ZCT3 (Pauw et al., 2004; Chebbi et al., 2014) and two G-box-binding factors, GBF1 and GBF2 (Sibéril et al., 2001) act as transcriptional repressors of specific genes from the TIA or TIA feeder pathways.
CrBPF1 was identified as a protein that binds an element in the C. roseus STR promoter that is distinct from the element bound by the ORCAs (van der Fits et al., 2000). CrBPF1 contains a single MYB-like DNA binding domain near its C-terminus. The presence of a single MYB-like sequence places CrBPF1 in the R-MYB family of proteins (Feller et al., 2011). Sequence analysis showed that CrBPF1 has high homology to the parsley box P-binding factor-1 (BPF-1; da Costa e Silva et al., 1993), which accumulates rapidly in elicitor-treated parsley cells and around fungal infection sites on parsley leaves. A strong correlation between BPF-1 transcript levels and phenylalanine ammonia-lyase (PAL) expression in parsley cell cultures suggests that BPF-1 might play an important role in disease resistance by helping regulate expression of plant defense genes (da Costa e Silva et al., 1993). In C. roseus CrBPF1 was found to bind specifically to the BA element within the STR promoter. CrBPF1 promotes STR transcription through a signal transduction pathway that is responsive to elicitors but not jasmonate and acts downstream of protein phosphorylation and calcium influx (van der Fits et al., 2000). However, current evidence indicates that CrBFP1 activity is not sufficient for elicitor-induced STR expression (van der Fits et al., 2000). Deletion of the BA fragment did not eliminate the ability of the STR promoter to respond to elicitor or jasmonate, whereas alteration or deletion of the JERE fragment rendered the STR promoter unable to respond to either of these compounds (Menke et al., 1999).
Information regarding whether CrBPF1 plays a role in the regulation of TIA-related genes other than STR has not previously been reported, leaving the role of CrBPF1 in regulation of TIA metabolism unknown. To address this issue, a transgenic hairy root line of C. roseus that overexpresses CrBPF1 under the control of a β-estradiol inducible promoter was generated. The transcript levels of 31 TIA biosynthetic and regulatory genes were tracked over a 72-h period under β-estradiol-induced and un-induced condition. The levels of 14 metabolites from the TIA and TIA feeder pathways were also investigated over the same time course, with nine of those metabolites being present at detectable levels in the majority of the samples analyzed. The results of these transcriptional and metabolic profiling experiments have revealed the role of CrBPF1 in regulation of TIA metabolism.
Catharanthus roseus, Vinca Little Bright Eye1, was used for this work. Seeds were surface sterilized and then germinated on B5 medium (Sigma, St. Louis, MO, USA) supplemented with Gamborg’s vitamins (Sigma, St. Louis, MO, USA). Seeds were germinated in the dark at 26°C for 2 weeks. The seedlings were then transferred to a 16-h-light/8-h-dark cycle with a light intensity of approximately 44 μmol m-2 s-1 for 4 weeks before inoculation with Agrobacterium tumefaciens.
The pERKT vector (Tsuda et al., 2012) is a modified version of the pMDC32 Gateway vector (Curtis and Grossniklaus, 2003) that expresses the XVE chimeric transcriptional activator (Zuo et al., 2000) under the control of a strong constitutive promoter. XVE allows estradiol-inducible expression of genes cloned behind an appropriate promoter sequence (Zuo et al., 2000). For this work, the full-length CrBPF1 open reading frame was inserted into pERKT behind a promoter that allows estradiol-inducible expression by XVE. DNA containing the CrBPF1 open reading was obtained by PCR amplification of C. roseus cDNAs using KOD Hot Start DNA polymerase (Novagen, Madison, WI, USA) and the following primer pair: 5′ ATGGTGTTGAAGAGAAGGC 3′ and 5′ TTAATCCGCCTGAGCATCC 3′. The resulting PCR fragment was cloned into the pCR8/GW/TOPO entry vector (Invitrogen, Grand Island, NY, USA) and then transferred to pERKT by an LR reaction to form pERKT-CrBPF1 (Supplemental Figure S1).
Transformation of 6-weeks old C. roseus was carried out using an equal mixture of A. tumefaciens cultures transformed with pERKT-CrBPF1 or the pPZPROL plasmid that carries the rol ABC genes (Hong et al., 2006), as previously described (Hong et al., 2006). Hairy roots appeared on infection sites approximately 4 weeks after inoculation. These hairy roots were allowed to grow to a length of approximately 1 cm and then excised and transferred to solid medium supplemented with 30 g L-1 sucrose, 6 g L-1 agar, 250 mg L-1 cefotaxime, half-strength Gamborg’s B5 salts, and full-strength Gamborg’s vitamins (pH 5.8). One week after transfer to the above media, 30 mg L-1 hygromycin was used to select for roots carrying the pERKT-CrBPF1 construct. Hygromycin-resistant hairy roots were further screened by PCR using primers with sequences 5′ ATGATCACAAGCTGATCCCC 3′ and 5′ GTGCGTTCGGAAAAAGAATC 3′ to amplify a DNA sequence that spans CrBPF1 and adjacent vector sequences within pERKT-CrBPF1. Transgenic hairy root lines exhibiting hygromycin-resistance and a positive reaction in the PCR test were transferred to 50 mL of liquid media containing half-strength Gamborg’s B5 liquid solution supplemented with full-strength Gamborg’s vitamins and 30 g L-1 sucrose in a 150-mL flask. Hairy root cultures were incubated in the dark on a shaker at 225 rpm and sub-cultured every 5 weeks.
One transgenic hairy root line carrying the pERKT-CrBPF1 overexpression construct (CrBPF1-OE) and one control line carrying only the pPZPROL transformation construct (Li et al., 2013) were used for time course analyses. Each transgenic hairy root culture was started from five hairy roots that were 3–4 cm in length. These cultures were grown for 35 days and then transferred to fresh liquid media with 20-μM β-estradiol (induced cultures) or without β-estradiol (un-induced cultures) for 0, 6, 12, 24, 48, or 72 h before being harvested. Three separate cultures were harvested for each hairy root line, time point and treatment condition. Hairy roots were flash frozen immediately after harvest using liquid nitrogen and then stored at –80°C. Aliquots of the same tissue samples were used for analyses of both transcript and metabolite levels.
Total RNA was isolated using the Spectrum Total RNA Isolation Kit with on-column DNase 1 digestion (Sigma, St. Louis, MO, USA), as described (Huang et al., 2010). For quantification of transcripts produced by BIS1, the endogenous and trans CrBPF1 genes, CrMYC1, CrMYC2, CrWRKY1, CrWRKY2, DXS1, DXS2B, MAT, T16H2, and T19H the GoScript Reverse Transcription System (Promega, Madison, WI, USA) was used to produce cDNAs. These cDNAs were analyzed by qPCR using the LightCycler 480 SYBR Green I Master mix by Roche and a Roche LightCycler 480 II (Roche Diagnostics, Indianapolis, IN, USA). For quantification of transcripts produced by all other genes (including the total transcripts produced by the CrBPF1 endogenous and transgenes combined), SuperScript II reverse transcriptase (Invitrogen, Grand Island, NY, USA) was used for cDNA synthesis and qPCR was performed on an ABI 7900 HT (Applied Biosystems, Grand Island, NY, USA) with a 384-well ABI optical plate using Roche Universal Probes (Roche Applied Science, USA) and the Homebrew master mix (University of Minnesota Genomics Center, Minneapolis, MN, USA). PCR primers and Roche Universal Probe numbers are described in Supplemental Table S1. qPCR data were normalized using the geometric average (Vandesompele et al., 2002) of two control genes, EF1 and UBQ11, which exhibit particularly stable expression patterns in C. roseus (Wei, 2010). Differences in EF1 and UBQ11 ΔCT levels were typically minor (Supplemental Table S2). Relative mRNA levels for each gene were converted to ΔΔCt. ΔΔCt = ΔCtun-induced control line at 0 h - ΔCtother. ΔCtun-induced control line at 0 h = CTindicated gene in un-induced control line at 0 h - CTEF1/UBQ11in un-induced control line at 0 h. ΔCtother = CTindicated gene - CTEF1/UBQ11 for the indicated line, growth condition, and time point. The lone exception is that mRNA levels for the CrBPF1 transgene were normalized versus mRNA levels from the endogenous CrBPF1 gene in the un-induced control line at 0 h rather than against CrBPF1 transgene mRNA levels in the un-induced control line at 0 h because the control line lacks the CrBPF1 transgene. A positive ΔΔCt value indicates that mRNA levels for the indicated gene are higher in the indicated hairy root line grown for the indicated time under the indicated conditions than in the un-induced control line at 0 h. Negative ΔΔCt values indicate the reverse situation. As the amount of PCR product approximately doubles with each reaction cycle, a ΔΔCt of one corresponds to approximately a twofold difference in transcript levels. The genes analyzed in this study are: 16OMT [GenBank: EF444544], ASα [GenBank: AJ250008], BIS1 [GenBank: KM409646], CPR [GenBank: X69791], CrBPF1 [GenBank: AJ251686], CrMYC1 [GenBank: AF283506], CrMYC2 [GenBank: AF283507], CrWRKY1 [GenBank: HQ646368], CrWRKY2 [GenBank accession number not available], D4H [GenBank: U71605], DAT [GenBank: AF053307], DXS1 [GenBank: KC625536], DXS2A [GenBank: AJ011840], DXS2B [GenBank: DQ8486762], EF1 [GenBank: EU007436], G10H [GenBank: AJ251269], GBF1 [GenBank: AF084971], GBF2 [GenBank: AF084972], LAMT [GenBank: EU057974], MAT [GenBank: AF253415], ORCA2 [GenBank: AJ238740], ORCA3 [GenBank: EU072424], PRX1 [GenBank: AM236087], SGD [GenBank: EU072423], STR [GenBank: X53602], T16H1 [GenBank: FJ647194], T16H2 [GenBank: JF742645], T19H [GenBank: HQ901597], TDC [GenBank: X67662], UBQ11 [GenBank: EU007433], ZCT1 [GenBank: AJ632082], ZCT2 [GenBank: AJ632083], ZCT3 [GenBank: AJ632084].
Frozen hairy root tissue samples were lyophilized and ground to a fine powder. Fifty milligram aliquots of the ground tissue samples were extracted on ice for 10 min using 10 mL of methanol and a Model VC 130PB sonicating probe (Sonics & Materials, Inc., Newton, CT, USA), as previously described (Sander, 2009). The extracts were centrifuged for 12 min at 4000 rpm at 15°C. The biomass was re-extracted as above and the supernatants combined and passed through a 0.45 μm nylon filter (25 mm, PJ Cobert, St Louis, MO, USA). The supernatants were then dried using a nitrogen evaporator (Organomation Associates, Inc., Berlin, MA, USA). The residues were dissolved in 2 mL of methanol, filtered using a 0.22 μm nylon filter (13 mm, PJ Cobert) and then stored at –25°C.
Twenty-microliter aliquots of the extracted alkaloid concentrate were run on a Phenomenex Luna C18(2) column (250 mm × 4.6 mm) connected to a Waters high performance liquid chromatography system [1525 binary pump, 717plus Autosampler, 996 Photo Diode Array (PDA) detector] using three different solvent systems. TIAs were examined following a previously described method (Sander, 2009). PDA data extracted at 254 nm were compared to standards for the quantification of strictosidine (gift from Dr. O’Connor, John Innes Centre, UK), ajmalicine (Fluka/Sigma, St. Louis, MO, USA), serpentine (Sigma–Aldrich, St. Louis, MO, USA), vindoline (ChemPacific Corp., Baltimore, MD, USA), vincristine (Sigma, St. Louis, MO, USA), vinblastine (Sigma, St. Louis, MO, USA), and catharanthine (Qventas, Branford, CT, USA). PDA data extracted at 329 nm were compared to standards for the quantification of tabersonine, lochnericine, and hörhammericine (all in-house standards). Tryptophan (Sigma, St. Louis, MO, USA) and tryptamine (Sigma, St. Louis, MO, USA) were investigated using a previously described method and PDA data extracted at 218 nm (Peebles et al., 2005). Loganin (Fluka/Sigma, St. Louis, MO, USA) and secologanin (Fluka/Sigma, St. Louis, MO, USA) were measured using a previously describe method and PDA data extracted at 239 nm (Li et al., 2013).
Catharanthus roseus DNA sequences available in the Medicinal Plant Genomics Resource2 and NCBI3 databases were screened using BLASTN for sequences similar to the 16 (CAAAAGTATTATGATT) and 42 (CGCTATTTATCATATAATTATTTTACAATAATTAGTATTAGG) nucleotide CrBPF1 binding sites (van der Fits et al., 2000).
A two-tailed Student’s t-test was employed for statistical analyses. To identify statistically significant differences, results from β-estradiol induced hairy roots carrying the CrBPF1 overexpression construct were compared with results from un-induced hairy roots carrying the CrBPF1 overexpression construct. For qRT-PCR experiments, (∗) was used to represent p ≤ 0.05 and (∗∗) was used to represent p ≤ 0.01. For analyses of metabolite levels, (∗) was used to represent p ≤ 0.1 and (∗∗) was used to represent p ≤ 0.05.
CrBPF1 was identified by screening for proteins that bind the STR promoter. However, information regarding whether CrBPF1 plays a role in the regulation of other TIA or TIA-related genes is lacking, preventing determination of the role of CrBPF1 in regulation of these pathways. To address this deficiency, C. roseus transgenic hairy root lines that express CrBPF1 under the control of an estradiol-inducible promoter (Zuo et al., 2000) were generated. Use of an estradiol-inducible promoter provides several advantages. An inducible promoter allows the timing and level of transgene expression to be controlled. This ability allows transgenic cultures to be grown without transgene expression, avoiding the possible deleterious consequences of long-term transgene expression. An inducible system also allows studies on the transient effects of transgene expression.
To generate transgenic hairy roots, C. roseus seedlings were inoculated with a mixture of A. tumefaciens cells transformed with either pERKT-CrBPF1 (Supplemental Figure S1) or pPZPROL (Hong et al., 2006). A total of 62 hairy root lines were screened for hygromycin resistance and seven lines were found to be resistant. These seven hairy root lines were confirmed to carry the CrBPF1 transgene by using PCR to demonstrate the presence of a DNA fragment spanning part of the CrBPF1 gene and an adjacent sequence from the pERKT-CrBPF1 vector. These positive lines were transferred to liquid culture. Lines with good adaption to growth in liquid culture were screened for β-estradiol inducible expression of CrBPF1. As expression of the CrBPF1 transgene is particularly strongly induced by β-estradiol in the CrBPF1-OE line (Figure 2), and CrBPF1-OE also adapted well to growth in liquid media, further studies utilized this line.
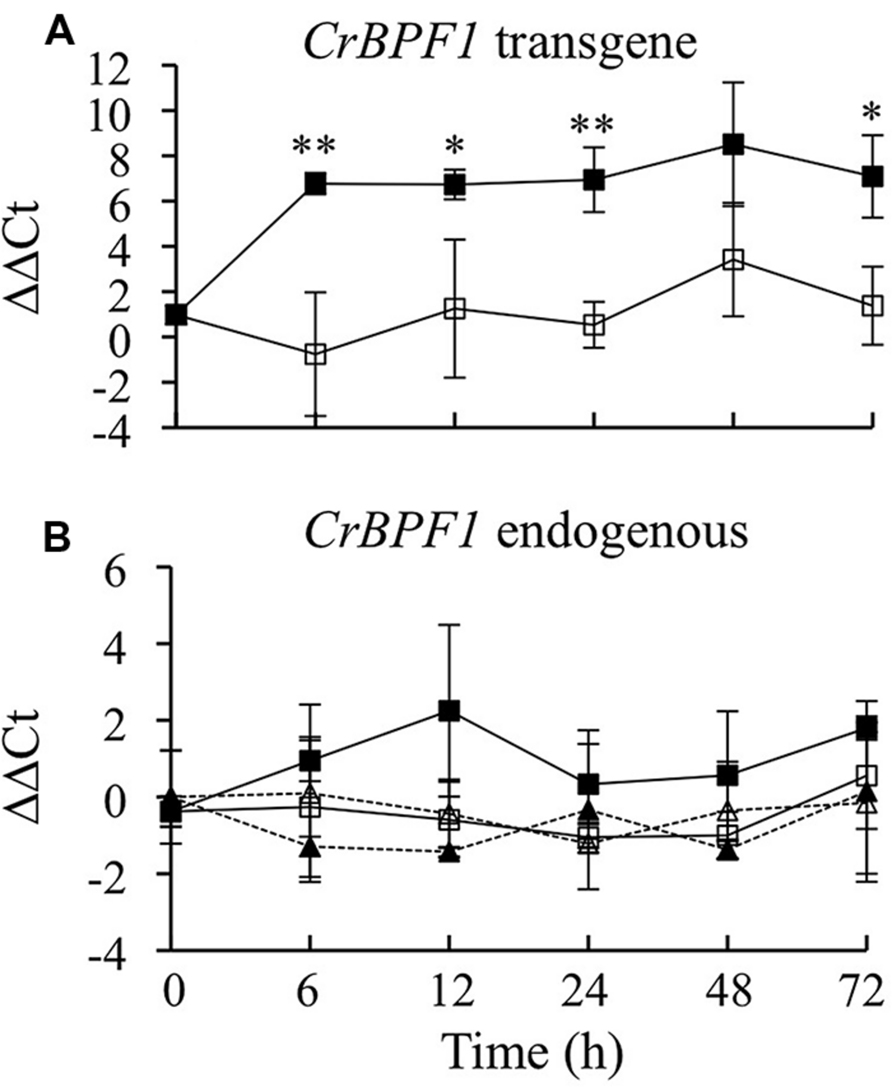
FIGURE 2. Time course analysis of CrBPF1 endogenous and transgene mRNA levels. CrBPF1 transcripts produced from the endogenous CrBPF1 gene and the CrBPF1 transgene were quantified independently by qRT-PCR using primers specific for each gene (Supplemental Table S1). As expected, the primer pair specific for transcripts produced by the CrBPF1 transgene yields only a very low background signal for the control line (data not shown), which lacks the CrBPF1 transgene. Results depicted are the following: un-induced CrBPF1-OE line ( , solid line), β-estradiol induced CrBPF1-OE line (
, solid line), β-estradiol induced CrBPF1-OE line ( , solid line), un-induced control line (Δ, dashed line) and β-estradiol induced control line (
, solid line), un-induced control line (Δ, dashed line) and β-estradiol induced control line ( , dashed line). (A) Relative CrBPF1 transgene mRNA levels are indicated as ΔΔCt. Note that CrBPF1 transgene mRNA levels were normalized versus CrBPF1 endogenous gene mRNA levels in the un-induced control line at 0 h rather than against CrBPF1 transgene mRNA levels in the un-induced control line at 0 h as the control line lacks the CrBPF1 transgene. (B) Relative CrBPF1 endogenous gene mRNA levels are indicated as ΔΔCt. A positive ΔΔCt value indicates that CrBPF1 endogenous gene mRNA levels are higher in the indicated hairy root line grown for the indicated time under the indicated conditions than in the un-induced control line at 0 h. Negative ΔΔCt values indicate the reverse situation. Results are the average ΔΔCt of three biological replicates, with two technical replicates per biological replicate. Error bars indicate SD. CrBPF1 mRNA levels in the un-induced versus induced CrBPF1-OE cultures differed at the same time point with: ∗p ≤ 0.05, ∗∗p ≤ 0.01 according to a Student’s t-test. The results of Student’s t-tests for the un-induced versus induced cultures of the control line are not depicted.
, dashed line). (A) Relative CrBPF1 transgene mRNA levels are indicated as ΔΔCt. Note that CrBPF1 transgene mRNA levels were normalized versus CrBPF1 endogenous gene mRNA levels in the un-induced control line at 0 h rather than against CrBPF1 transgene mRNA levels in the un-induced control line at 0 h as the control line lacks the CrBPF1 transgene. (B) Relative CrBPF1 endogenous gene mRNA levels are indicated as ΔΔCt. A positive ΔΔCt value indicates that CrBPF1 endogenous gene mRNA levels are higher in the indicated hairy root line grown for the indicated time under the indicated conditions than in the un-induced control line at 0 h. Negative ΔΔCt values indicate the reverse situation. Results are the average ΔΔCt of three biological replicates, with two technical replicates per biological replicate. Error bars indicate SD. CrBPF1 mRNA levels in the un-induced versus induced CrBPF1-OE cultures differed at the same time point with: ∗p ≤ 0.05, ∗∗p ≤ 0.01 according to a Student’s t-test. The results of Student’s t-tests for the un-induced versus induced cultures of the control line are not depicted.
To characterize β-estradiol inducible expression of CrBPF1, a time course experiment was performed using the CrBPF1-OE and control transgenic lines. The transgenic hairy root control line was generated previously (Li et al., 2013) by transforming C. roseus with pPZPROL alone, and thus lacks the β-estradiol-inducible CrBPF1 transgene. The CrBPF1-OE and control lines were grown for 35 days and then transferred to fresh media with 0 μM (un-induced) or 20 μM (induced) β-estradiol. Tissue samples were collected 0, 6, 12, 24, 48, and 72 h after addition of β-estradiol. CrBPF1 transcripts produced by the CrBPF1 endogenous gene and by the CrBPF1 transgene were quantified separately using qRT-PCR (Figure 2). CrBPF1 transgene mRNA levels increased rapidly in the CrBPF1-OE line after addition of 20 μM β-estradiol, rising approximately 50 fold within 6 h, and remained high for at least 72 h. Un-induced cultures of the CrBPF1-OE line exhibited much lower transcript levels for the CrBPF1 transgene than the induced cultures, indicating that β-estradiol is necessary for high-level expression of the CrBPF1 transgene (Figure 2A). As expected, qRT-PCR reactions using a primer pair specific for the CrBPF1 transgene produced only a very low signal from RNA isolated from the control line (data not shown), which lacks the CrBPF1 transgene. Transcript levels for the CrBPF1 endogenous gene were not significantly affected by treatment with β-estradiol and were similar in the CrBF1-OE and control lines (Figure 2B).
CrBPF1 is one of only a few putative activators of the TIA pathway identified to date. Understanding the role played by CrBPF1 in regulation of the TIA pathway is important for developing strategies for in vivo manipulation of TIA production. Toward this end, the effects of CrBPF1 overexpression on transcript levels of 31 genes were analyzed. The genes chosen for analysis include two genes from the indole pathway (ASα and TDC), six genes from the monoterpenoid pathway (DXS1, DXS2A, DXS2B, G10H, CPR, and LAMT) and ten genes from the TIA pathway (STR, SGD, T16H1, T16H2, 16OMT, D4H, DAT, PRX1, T19H, and MAT). In addition, transcript levels of all of the cloned genes currently postulated to play a role in regulation of the TIA pathway were characterized. These genes include eight TIA transcriptional activators (ORCA2, ORCA3, CrBPF1, CrMYC1, CrMYC2, CrWRKY1, CrWRKY2, and BIS1) and five TIA transcriptional repressors (ZCT1, ZCT2, ZCT3, GBF1, and GBF2). In addition, the concentrations of 14 TIA metabolites were investigated, with nine of those metabolites being present at detectable levels. Both transcript and metabolite levels were tracked over a 72-h period in the CrBPF1-OE and control lines grown in the absence or presence of β-estradiol.
The production of TIAs is dependent on the synthesis of tryptamine in the indole pathway, the synthesis of secologanin in the terpenoid pathway and their subsequent coupling to form strictosidine. Thus, it was of interest to determine whether overexpression of CrBPF1 affects expression of genes in the indole or terpenoid pathways. To characterize the effects of overexpressing CrBPF1 on the indole pathway, ASα and TDC transcripts were quantified. ASα encodes the alpha subunit of anthranilate synthase, which catalyzes the first committed step in tryptophan synthesis. Overexpression of CrBPF1 caused a significant increase in ASα transcript levels. ASα transcript levels were slightly higher in the induced versus un-induced CrBPF1-OE cultures 6 h after addition of β-estradiol, and reached a peak at the 12-h time point before beginning to decline. In contrast, addition of β-estradiol to the media had no significant effect on ASα transcript levels in the control line (Figure 3A). TDC encodes tryptophan decarboxylase, which catalyzes the formation of tryptamine from tryptophan. TDC transcript levels were higher in the induced versus un-induced CrBPF1-OE cultures 12 and 72 h after addition of β-estradiol. However, overexpression of CrBPF1 caused only slight differences in TDC transcript levels, with a maximum difference of 75% higher transcript levels in the induced versus un-induced CrBPF1-OE cultures at the 12-h time point. Addition of β-estradiol to the media had no significant effect on TDC transcript levels in the control line (Figure 3B). An attempt was also made to analyze tryptophan and tryptamine levels in aliquots of the same tissue samples used to analyze gene expression. However, tryptophan and tryptamine levels were below the detection threshold in many of the samples.
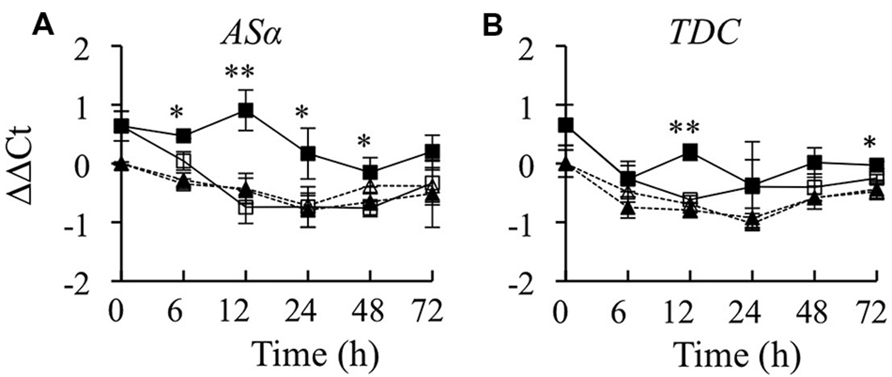
FIGURE 3. Time course analysis of ASα and TDC mRNA levels. Results depicted are the following: un-induced CrBPF1-OE line ( , solid line), β-estradiol induced CrBPF1-OE line (
, solid line), β-estradiol induced CrBPF1-OE line ( , solid line), un-induced control line (Δ, dashed line) and β-estradiol induced control line (
, solid line), un-induced control line (Δ, dashed line) and β-estradiol induced control line ( , dashed line). Relative transcript levels are presented as ΔΔCt. Results shown are the following: (A) ASα mRNA levels and (B) TDC mRNA levels. Results are the average ΔΔCt values of three biological replicates with two technical replicates per biological replicate. Error bars indicate SD. Transcript levels in the un-induced versus induced CrBPF1-OE cultures differed at the same time point with: ∗p ≤ 0.05, ∗∗p ≤ 0.01 according to a Student’s t-test. The results of Student’s t-tests for the un-induced versus induced cultures of the control line are not depicted.
, dashed line). Relative transcript levels are presented as ΔΔCt. Results shown are the following: (A) ASα mRNA levels and (B) TDC mRNA levels. Results are the average ΔΔCt values of three biological replicates with two technical replicates per biological replicate. Error bars indicate SD. Transcript levels in the un-induced versus induced CrBPF1-OE cultures differed at the same time point with: ∗p ≤ 0.05, ∗∗p ≤ 0.01 according to a Student’s t-test. The results of Student’s t-tests for the un-induced versus induced cultures of the control line are not depicted.
The expression levels of six genes in the terpenoid pathway were determined. Three of these genes (DXS1, DXS2A, and DXS2B) encode different isoforms of 1-deoxy-D-xylulose 5-phosphate synthase. DXS2A and DXS2B are induced by ORCA3 overexpression, whereas DXS1 is not regulated by ORCA3 (Han et al., 2013). Overexpression of CrBPF1 had only a very modest effect on expression of the DXS genes. DXS1 expression was 70% lower in the induced versus un-induced CrBPF1-OE cultures at the 72-h time point, but was not significantly altered at the other time points assayed. Addition of β-estradiol had a very slight effect on DXS1 expression in the control line, causing a 6 to 30% increase in DXS1 transcript levels at the 6, 12, and 72-h time points (Figure 4A). DXS2A expression was slightly induced by β-estradiol at the 12-h time point in the CrBPF1-OE line (Figure 4B). DXS2B transcript levels were increased 1.6 and 2.0 fold in the induced versus un-induced CrBPF1-OE cultures at the 6 and 12-h time points, respectively. However, DXS2B expression was also induced 1.5X in the induced versus un-induced control cultures at the 6-h time point, suggesting that application of β-estradiol, rather than increased CrBPF1 expression, might be responsible for the alterations in DXS2B transcript levels (Figure 4C). G10H transcript levels were significantly higher in the induced versus un-induced CrBPF1-OE cultures at the 6, 12, and 72-h time points, but the differences in transcript levels were slight, reaching a maximum difference of less than twofold at the 12-h time point (Figure 4D). CPR transcript levels were significantly higher in the induced versus un-induced CrBPF1-OE cultures at the 12, 48, and 72-h time points, but the differences in transcript levels were slight, with a maximum difference of approximately 1.5 fold at the 48-h time point (Figure 4E). LAMT transcript levels were approximately 2.5 fold higher in the induced versus un-induced CrBPF1-OE cultures at the 12-h time point, but were not significantly different at the other time points analyzed (Figure 4F). Addition of β-estradiol had no significant effects on transcript levels of G10H, CPR, or LAMT in the control line. The levels of loganin and secologanin were also determined in aliquots of the same tissue samples used for gene expression analyses. The addition of 20-μM β-estradiol to the media caused no substantial alterations in the levels of either of these metabolites over the time period analyzed (Figures 4G,H).
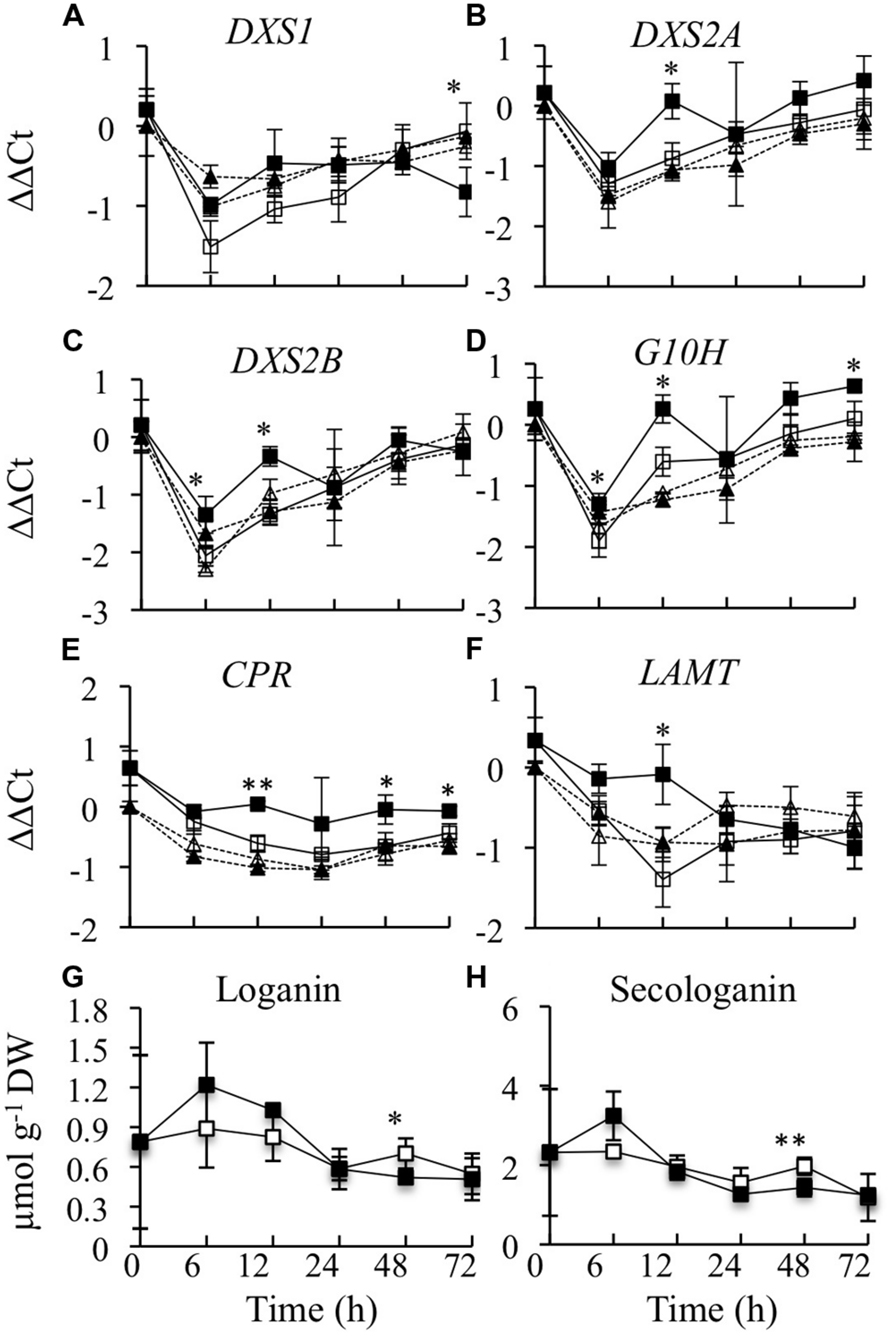
FIGURE 4. Time course analyses of transcript and metabolite levels from the terpenoid pathway. Results depicted are the following: un-induced CrBPF1-OE line ( , solid line), β-estradiol induced CrBPF1-OE line (
, solid line), β-estradiol induced CrBPF1-OE line ( , solid line), un-induced control line (
, solid line), un-induced control line ( , dashed line) and β-estradiol induced control line (
, dashed line) and β-estradiol induced control line ( , dashed line). Relative transcript levels are presented as ΔΔCt. Results shown are the following: (A) DXS1 mRNA levels, (B) DXS2A mRNA levels, (C) DXS2B mRNA levels, (D) G10H mRNA levels (E) CPR mRNA levels, (F) LAMT mRNA levels, (G) loganin levels, and (H) secologanin levels. Results for transcript levels are the average ΔΔCt values of three biological replicates with two technical replicates per biological replicate. Results for metabolite levels are the averages of three biological replicates. Metabolite levels for the control line are not depicted. Error bars indicate SD. Transcript levels in the un-induced versus induced CrBPF1-OE cultures differed at the same time point with: ∗p ≤ 0.05, ∗∗p ≤ 0.01 according to a Student’s t-test. The results of Student’s t-tests for the un-induced versus induced cultures of the control line are not depicted. Metabolite levels in the un-induced versus induced CrBPF1-OE cultures differed at the same time point with: ∗p ≤ 0.1, ∗∗p ≤ 0.05 according to a Student’s t-test.
, dashed line). Relative transcript levels are presented as ΔΔCt. Results shown are the following: (A) DXS1 mRNA levels, (B) DXS2A mRNA levels, (C) DXS2B mRNA levels, (D) G10H mRNA levels (E) CPR mRNA levels, (F) LAMT mRNA levels, (G) loganin levels, and (H) secologanin levels. Results for transcript levels are the average ΔΔCt values of three biological replicates with two technical replicates per biological replicate. Results for metabolite levels are the averages of three biological replicates. Metabolite levels for the control line are not depicted. Error bars indicate SD. Transcript levels in the un-induced versus induced CrBPF1-OE cultures differed at the same time point with: ∗p ≤ 0.05, ∗∗p ≤ 0.01 according to a Student’s t-test. The results of Student’s t-tests for the un-induced versus induced cultures of the control line are not depicted. Metabolite levels in the un-induced versus induced CrBPF1-OE cultures differed at the same time point with: ∗p ≤ 0.1, ∗∗p ≤ 0.05 according to a Student’s t-test.
To characterize the effects of overexpressing CrBPF1 on the TIA pathway, transcript levels of ten TIA biosynthetic genes were characterized. STR and SGD encode the enzymes that catalyze the first two steps in TIA biosynthesis. Overexpression of CrBPF1 had no significant effects on STR transcript levels (Figure 5A). SGD transcript levels were 75% higher in the induced versus un-induced CrBPF1-OE cultures at the 12-h time point, but were not significantly altered by CrBPF1 overexpression at the other time points assayed (Figure 5B). T16H, 16OMT, D4H, and DAT encode enzymes that catalyze different steps in the pathway leading from tabersonine to formation of vindoline. Overexpression of CrBPF1 had no significant effects on T16H1 (Figure 5C) or T16H2 (Figure 5D) transcript levels. 16OMT transcript levels were 2.6-fold higher in induced versus un-induced CrBPF1-OE cultures at the 12-h time point, but were not significantly altered by CrBPF1 overexpression at the other time points assayed (Figure 5E). D4H transcript levels were significantly higher in the induced versus un-induced CrBPF1-OE cultures at the 12 and 48-h time points, but not at the other time points analyzed (Figure 5F). The effects of CrBPF1 overexpression on DAT transcript levels were more complex. DAT transcript levels were approximately 3-fold higher in the induced versus un-induced CrBPF1-OE cultures at the 12-h time point, but were almost twofold lower in the induced versus un-induced CrBPF1-OE cultures at the 48-h time point. Interestingly, DAT transcript levels in the CrBPF1-OE line were consistently below those in the control line (Figure 5G). PRX1 encodes a vacuolar class III peroxidase that catalyzes the synthesis of α-3′, 4′-anhydrovinblastine from catharanthine and vindoline. Overexpression of CrBPF1 had no significant effects on PRX1 transcript levels (Figure 5H). T19H transcript levels were 2.3-fold higher in induced versus un-induced CrBPF1-OE cultures at the 48-h time point, but were not significantly altered by CrBPF1 overexpression at the other time points assayed (Figure 5I). Overexpression of CrBPF1 had no significant effects on MAT transcript levels (Figure 5J). Addition of β-estradiol to the media had little effect on TIA biosynthetic gene expression in the control line. Where TIA biosynthetic gene transcript levels did vary between control cultures grown in the presence of 0 versus 20 μM β-estradiol, transcript levels were typically higher in the cultures grown on 0 μM β-estradiol. For example, T16H1 transcript levels at the 48-h time point and DAT and PRX1 transcript levels at the 24-h time point were higher in control cultures grown on 0 μM β-estradiol than on 20 μM β-estradiol (Figure 5). These results are in contrast to the results observed for the CrBPF1-OE cultures, where addition of 20-μM β-estradiol to the media tended to cause increased transcript levels.
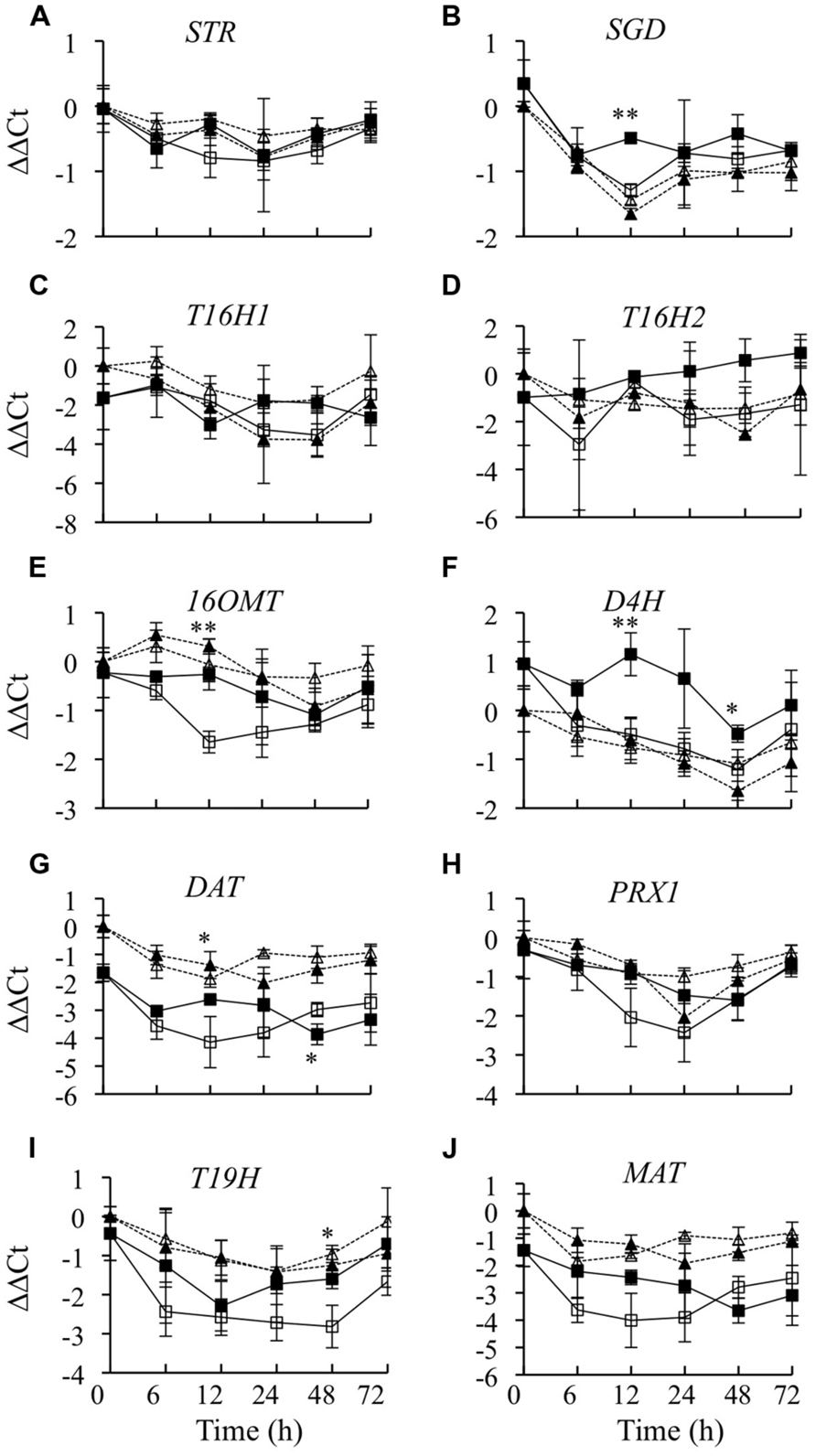
FIGURE 5. Time course analysis of TIA biosynthetic gene mRNA levels. Results depicted are the following: un-induced CrBPF1-OE line ( , solid line), β-estradiol induced CrBPF1-OE line (
, solid line), β-estradiol induced CrBPF1-OE line ( , solid line), un-induced control line (Δ, dashed line) and β-estradiol induced control line (
, solid line), un-induced control line (Δ, dashed line) and β-estradiol induced control line ( , dashed line). Relative transcript levels are presented as ??Ct. Results shown are the following: (A) STR mRNA levels, (B) SGD mRNA levels (C) T16H1 mRNA levels, (D) T16H2 mRNA levels, (E) 16OMT mRNA levels, (F) D4H mRNA levels, (G) DAT mRNA levels, (H) PRX1 mRNA levels, (I) T19H mRNA levels and (J) MAT mRNA levels. Results are the average ΔΔCt values of three biological replicates with two technical replicates per biological replicate. Error bars indicate SD. Transcript levels in the un-induced versus induced CrBPF1-OE cultures differed at the same time point with: ∗p ≤ 0.05, ∗∗p ≤ 0.01 according to a Student’s t-test. The results of Student’s t-tests for the un-induced versus induced cultures of the control line are not depicted.
, dashed line). Relative transcript levels are presented as ??Ct. Results shown are the following: (A) STR mRNA levels, (B) SGD mRNA levels (C) T16H1 mRNA levels, (D) T16H2 mRNA levels, (E) 16OMT mRNA levels, (F) D4H mRNA levels, (G) DAT mRNA levels, (H) PRX1 mRNA levels, (I) T19H mRNA levels and (J) MAT mRNA levels. Results are the average ΔΔCt values of three biological replicates with two technical replicates per biological replicate. Error bars indicate SD. Transcript levels in the un-induced versus induced CrBPF1-OE cultures differed at the same time point with: ∗p ≤ 0.05, ∗∗p ≤ 0.01 according to a Student’s t-test. The results of Student’s t-tests for the un-induced versus induced cultures of the control line are not depicted.
As CrBPF1 overexpression affects the transcript levels of many of the genes involved in synthesis of TIAs or TIA precursors, it was of interest to determine whether overexpression of CrBPF1 affects TIA metabolite levels. Toward that end, the levels of ten TIA metabolites were analyzed over a 72-h period in the CrBPF1-OE line grown in the presence or absence of 20-μM β-estradiol, with seven of those metabolites being present at detectable levels (Figure 6). The metabolites analyzed were tabersonine, lochnericine, hörhammericine, catharanthine, serpentine, ajmalicine, strictosidine, vindoline, vincristine, and vinblastine, with the levels of the last three being below the detection threshold. Overexpression of CrBPF1 had only modest effects on the levels of the other seven metabolites, with the largest statistically significant effect being ∼40% lower serpentine levels in the induced versus un-induced CrBPF1-OE cultures at the 12-h time point. The levels of the same metabolites were also analyzed in the control line, at 0 and 24 h after transfer to fresh media with 0 or 20 μM β-estradiol. Addition of 20-μM β-estradiol to the media had little effect on the levels of any of the TIA metabolites analyzed in the control line (data not shown).
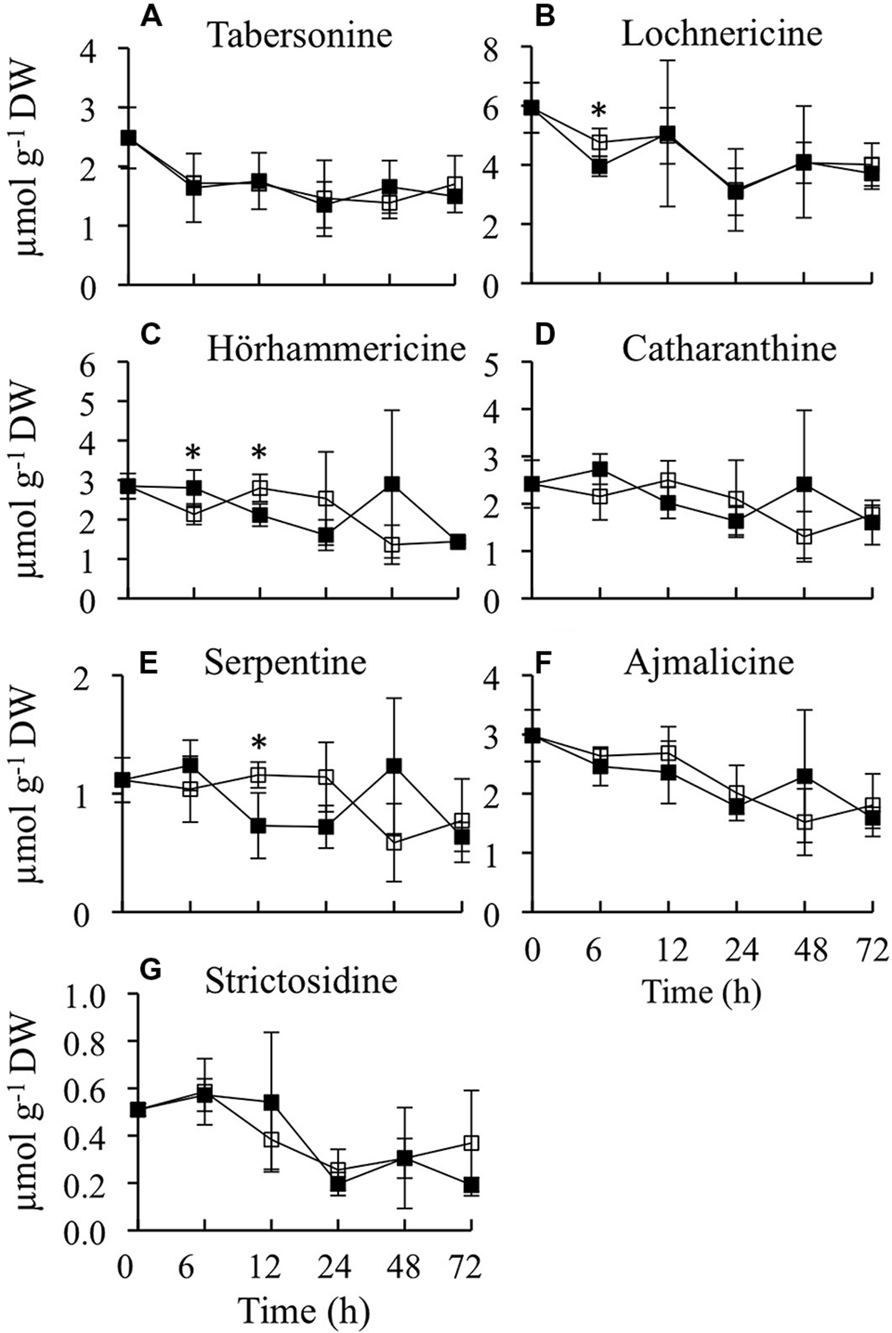
FIGURE 6. Time course analysis of TIA metabolite levels. Results depicted are the following: un-induced CrBPF1-OE line ( , solid line) and β-estradiol induced CrBPF1-OE line (
, solid line) and β-estradiol induced CrBPF1-OE line ( , solid line). Metabolite levels shown are the following: (A) tabersonine levels, (B) lochnericine levels (C) hörhammericine levels, (D) catharanthine levels, (E) serpentine levels, (F) ajmalicine levels and (G) strictosidine levels. Results are the average values of three biological replicates. Error bars indicate SD. Metabolite levels in the un-induced versus induced CrBPF1-OE cultures differed at the same time point with: ∗p ≤ 0.1 according to a Student’s t-test.
, solid line). Metabolite levels shown are the following: (A) tabersonine levels, (B) lochnericine levels (C) hörhammericine levels, (D) catharanthine levels, (E) serpentine levels, (F) ajmalicine levels and (G) strictosidine levels. Results are the average values of three biological replicates. Error bars indicate SD. Metabolite levels in the un-induced versus induced CrBPF1-OE cultures differed at the same time point with: ∗p ≤ 0.1 according to a Student’s t-test.
In addition to regulating expression of biosynthetic genes directly, a transcriptional regulator may affect expression of biosynthetic genes by altering the activities of other transcriptional regulators. To determine whether overexpression of CrBPF1 affects the activities of TIA transcriptional regulators, transcript levels for all the postulated TIA regulators for which genes have been cloned were analyzed in CrBPF1-OE and control cultures grown on 0 or 20 μM β-estradiol. Currently, eight transcriptional activators (ORCA2, ORCA3, CrBPF1, CrMYC1, CrMYC2, CrWRKY1, CrWRKY2, and BIS1) and five transcriptional repressors (ZCT1, ZCT2, ZCT3, GBF1, and GBF2) are postulated to function in regulation of the TIA pathway. Overexpression of CrBPF1 had little effect on ORCA2 transcript levels, with the only statistically significant effect being an approximately 60% increase in ORCA2 transcript levels in the induced versus un-induced CrBPF1-OE cultures at the 48-h time point (Figure 7A). Overexpression of CrBPF1 had a more consistent effect on ORCA3 expression, with ORCA3 transcript levels being significantly higher in the induced versus un-induced CrBPF1-OE cultures at the 6, 12, and 48-h time points, reaching a maximum difference of 2.4 fold at the 48-h time point (Figure 7B). To determine whether CrBPF1 affects its own expression, transcripts produced by the endogenous CrBPF1 gene were analyzed in the CrBPF1-OE and control lines grown on 0 or 20 μM β-estradiol. Increased expression of the CrBPF1 transgene had no significant effect on CrBPF1 endogenous gene transcript levels, indicating that CrBPF1 does not regulate its own expression at the steady-state transcriptional level (Figure 2B). CrMYC1 transcript levels were almost fourfold higher in induced versus un-induced CrBPF1-OE cultures at the 6 and 12-h time points, but were not significantly altered at later time points (Figure 7C). Overexpression of CrBPF1 had smaller, but more consistent effects on CrMYC2 transcript levels, with 30–60% higher CrMYC2 transcript levels in the induced versus un-induced CrBPF1-OE cultures at all time points assayed, except for the latest time point (Figure 7D). Overexpression of CrBPF1 also had a modest effect on CrWRKY1 transcript levels, which exhibited statistically significant 30% increases in the induced versus un-induced CrBPF1-OE cultures at the 12 and 48-h time points (Figure 7E). In contrast, overexpression of CrBPF1 had no statistically significant effects on CrWRKY2 transcript levels (Figure 7F). Overexpression of CrBPF1 caused a statistically significant 60% increase in BIS1 transcript levels at the 12-h time point and a 20% increase at the 72-h time point (Figure 7G).
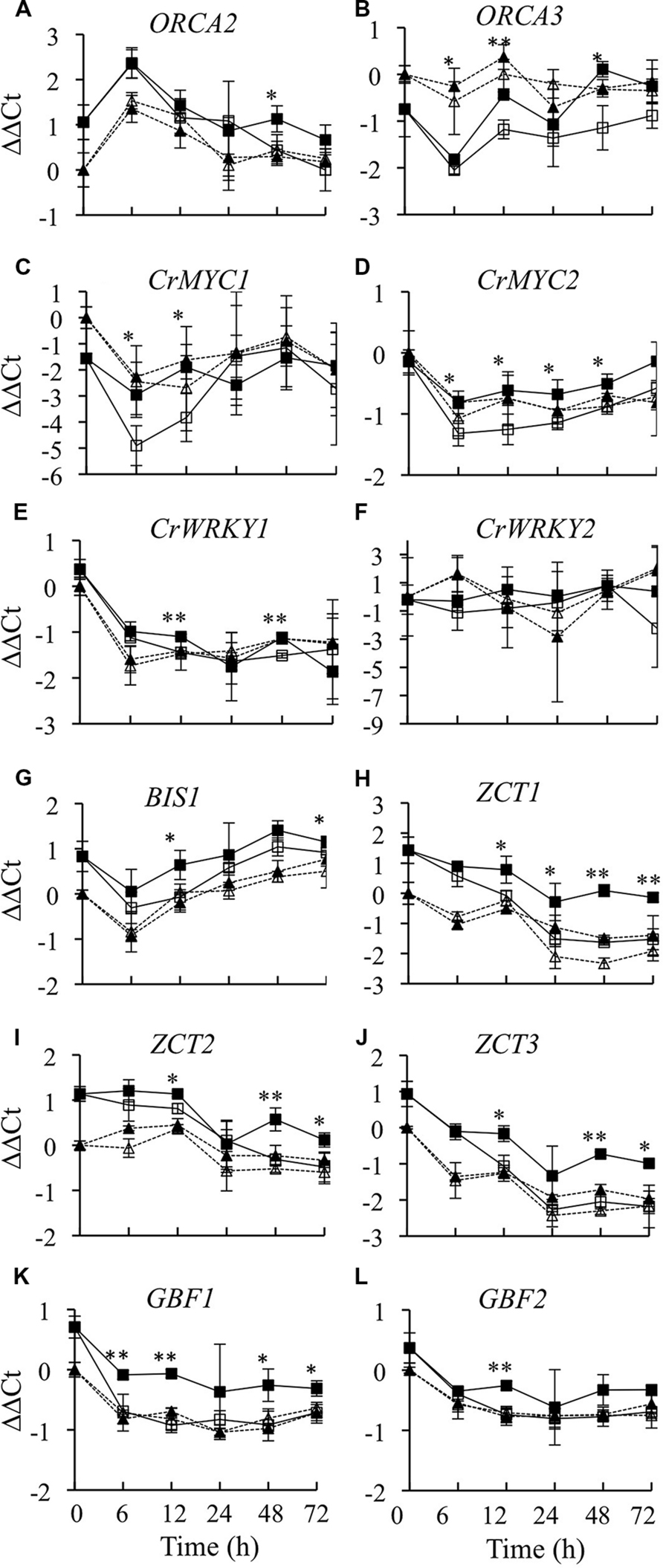
FIGURE 7. Time course analysis of TIA regulatory gene mRNA levels. Results depicted are the following: un-induced CrBPF1-OE line ( , solid line), β-estradiol induced CrBPF1-OE line (
, solid line), β-estradiol induced CrBPF1-OE line ( , solid line), un-induced control line (Δ, dashed line) and β-estradiol induced control line (
, solid line), un-induced control line (Δ, dashed line) and β-estradiol induced control line ( , dashed line). Relative transcript levels are presented as ΔΔCt. Results shown are the following: (A) ORCA2 mRNA levels, (B) ORCA3 mRNA levels (C) CRMYC1 mRNA levels, (D) CrMYC2 mRNA levels, (E) CrWRKY1 mRNA levels, (F) CrWRKY2 mRNA levels, (G) BIS1 mRNA levels, (H) ZCT1 mRNA levels, (I) ZCT2 mRNA levels, (J) ZCT3 mRNA levels, (K) GBF1 mRNA levels and (L) GBF2 mRNA levels. Results are the average ΔΔCt values of three biological replicates with two technical replicates per biological replicate. Error bars indicate SD. Transcript levels in the un-induced versus induced CrBPF1-OE cultures differed at the same time point with: ∗p ≤ 0.05, ∗∗p ≤ 0.01 according to a Student’s t-test. The results of Student’s t-tests for the un-induced versus induced cultures of the control line are not depicted.
, dashed line). Relative transcript levels are presented as ΔΔCt. Results shown are the following: (A) ORCA2 mRNA levels, (B) ORCA3 mRNA levels (C) CRMYC1 mRNA levels, (D) CrMYC2 mRNA levels, (E) CrWRKY1 mRNA levels, (F) CrWRKY2 mRNA levels, (G) BIS1 mRNA levels, (H) ZCT1 mRNA levels, (I) ZCT2 mRNA levels, (J) ZCT3 mRNA levels, (K) GBF1 mRNA levels and (L) GBF2 mRNA levels. Results are the average ΔΔCt values of three biological replicates with two technical replicates per biological replicate. Error bars indicate SD. Transcript levels in the un-induced versus induced CrBPF1-OE cultures differed at the same time point with: ∗p ≤ 0.05, ∗∗p ≤ 0.01 according to a Student’s t-test. The results of Student’s t-tests for the un-induced versus induced cultures of the control line are not depicted.
Overexpression of CrBPF1 had a comparatively large effect on expression of the ZCT1 transcriptional repressor. ZCT1 transcript levels were twofold to threefold higher in the induced versus un-induced CrBPF1-OE cultures at all time points assayed, except for the earliest, 6-h, time point (Figure 7H). Overexpression of CrBPF1 also caused significant increases in ZCT2 (Figure 7I) and ZCT3 (Figure 7J) expression levels at the 12, 48, and 72-h time points. Overexpression of CrBPF1 caused a larger increase in ZCT3 than in ZCT2 transcript levels, with ZCT3 transcript levels being approximately 2–2.5 fold higher in the induced versus un-induced CrBPF1-OE cultures, as opposed to approximately 1.25–2 fold increases in ZCT2 transcript levels. Overexpression of CrBPF1 had modest, but fairly consistent, effects on GBF1 expression. GBF1 transcript levels were significantly higher in the induced versus un-induced CrBPF1-OE cultures at all time points assayed, except for the 24-h time point, where the mean GBF1 transcript levels were higher in the induced than in the un-induced CrBPF1-OE cultures, but the difference was not statistically significant (Figure 7K). Overexpression of CrBPF1 had only a very slight effect on GBF2 transcript levels, with the only statistically significant difference being an approximately 40% increase in GBF2 transcript levels in the induced versus the un-induced CrBPF1-OE cultures at the 12-h time point (Figure 7L). Addition of 20-μM β-estradiol to the media had little effect on expression of TIA regulatory genes in the control cultures.
DNase1 fingerprinting resulted in the identification of 16 and 42 nt CrBPF1 binding sites within the BA fragment of the C. roseus STR promoter (van der Fits et al., 2000). C. roseus DNA sequences available in the Medicinal Plant Genomics Resource2 and NCBI3 databases were searched using BLASTN for sequences similar to these 16 and 42-nucleotide CrBPF1 binding sites. Examination of the top 50 matches for the 16 and 42-nt sequences from the Medicinal Plant Genomics Resource did not reveal any matches to promoter sequences from genes believed to be involved in TIA biosynthesis, with the exception of STR. In contrast, searches of the sequences available in GenBank revealed several partial matches to the 5′ regions of genes involved in TIA biosynthesis. The best matches to sequences lying within approximately 1,500 bp 5′ of a transcription start site are listed in Supplemental Table S3. In addition to STR, partial matches for both the 16 and 42-nt sequences were found for ORCA3 and TDC. However, the spacing between these partial matches was much larger for ORCA3 and TDC than for STR. The 5′ ends of the 16 and 42 nt sequences are 70 bp apart in the STR promoter, but are 365 and 552 bp apart in the ORCA3 and TDC promoters, respectively. Partial matches to the 16 nt sequence, but not to the 42 nt sequence, were found for CPR, PRX1, BIS1, and CrWRKY1. A partial match to the 42 nt sequence, but not to the 16 nt sequence, was found for DXS2B.
CrBPF1 was identified as a MYB-like protein that binds the BA region of the C. roseus STR promoter (van der Fits et al., 2000). Although thirteen transcriptional regulators have been postulated to act in regulation of the TIA pathway, the effects of most of these transcriptional regulators on expression of the majority of the known TIA biosynthetic and regulatory genes have not yet been determined. To address this deficiency, a C. roseus transgenic hairy root line that expresses CrBPF1 under the control of a β-estradiol inducible promoter was generated and characterized. Addition of β-estradiol to the medium causes a large and rapid induction of CrBPF1 transgene expression in the CrBPF1-OE line but does not affect CrBPF1 expression in a control line, indicating that the presence of the transgene is necessary for increased CrBPF1 transcript levels.
Terpenoid indole alkaloid production is dependent on the synthesis of precursors by both the indole and terpenoid pathways, the combination of these precursors by STR and subsequent reactions carried out by different branches of the TIA pathway. Overexpression of CrBPF1 caused increased expression of the majority of the genes analyzed from these pathways. However, these effects were typically transient and of limited magnitude. Interestingly, CrBPF1 overexpression may cause decreases in DAT expression. Although overexpression of CrBPF1 caused increased DAT transcript levels after 12 h, DAT transcript levels decreased after 48 h. In addition, DAT transcript levels were consistently lower in the CrBPF1-OE line than in the control line over the entire time course, suggesting that increased CrBPF1 expression tends to have a negative effect on DAT activity. Interestingly, DAT expression has been shown to be strongly decreased in response to overexpression of ORCA2 (Li et al., 2013). The finding that CrBPF1 overexpression does not have a significant effect on STR transcript levels might appear somewhat unexpected, given that CrBPF1 is known to bind the STR promoter (van der Fits et al., 2000). However, this finding is consistent with previous work suggesting that CrBPF1 might have only a limited effect on STR expression (van der Fits et al., 2000). Binding of CrBPF1 to the STR promoter is only part of the process promoting STR transcription, with binding of additional factors to other cis sequences playing an important role in regulation of STR activity (van der Fits et al., 2000; Chatel et al., 2003). Consequently, overexpression of CrBPF1 alone is insufficient to cause significant alterations in STR transcript levels.
To determine whether overexpression of CrBPF1 affects the expression of other regulators, transcript levels for all genes postulated to encode TIA regulators were analyzed. Overexpression of CrBPF1 caused increased expression of all of the other TIA transcriptional activators except CrWRKY2. In addition, expression of the CrBPF1 transgene did not affect expression of the CrBPF1 endogenous gene, indicating that CrBPF1 does not regulate its own expression. Interestingly, overexpression of CrBPF1 also caused increased transcript levels for all five of the genes postulated to encode TIA transcriptional repressors. These results suggest that CrBPF1 overexpression could be altering the transcript levels of some of the biosynthetic genes characterized indirectly, by altering the activities of other TIA regulatory genes. The results of a promoter analysis are consistent with this possibility. Partial matches for both the 16 and 42-nt CrBPF1 binding sites from the STR promoter were found in the promoters of only ORCA3 and TDC, among the genes analyzed as part of this study. However, the spacing between these partial matches was much greater for ORCA3 and TDC than for STR. Partial matches to the 16-nt CrBPF1 binding sequence were also found for the CPR, PRX1, BIS1, and CrWRKY1 promoters and to the 42-nt sequence for DXS2B.
As CrBPF1 overexpression affects the activities of several TIA biosynthetic genes and of most of the TIA regulatory genes analyzed, it was of interest to determine whether CrBPF1 overexpression also affects TIA metabolite levels. Toward that end the levels of 14 metabolites from different parts of the TIA and feeder pathways were analyzed, with the levels of nine of those metabolites being above the detection threshold. The results of these analyses indicate that CrBPF1 overexpression causes only slight, and transient, alterations in lochnericine, hörhammericine and serpentine levels, typically causing decreased levels of these metabolites. Lochnericine is the precursor for synthesis of hörhammericine, suggesting that flux to this branch of the TIA pathway may be altered by CrBPF1 overexpression. Serpentine is synthesized via a different branch of the TIA pathway. The relatively minor effects of CrBPF1 overexpression on TIA metabolite levels may be due to the fact that, although CrBPF1 overexpression increases the activities of many TIA and related genes, these increases in gene expression are typically of limited magnitude and duration.
The results of this work indicate that although CrBPF1 regulates a high percentage of the genes analyzed, the effects of CrBPF1 overexpression on gene activity levels tend to be of comparatively limited magnitude and are often transient. These modest alterations in TIA biosynthetic gene expression may help explain the limited effects of CrBPF1 overexpression on the levels of the TIA metabolites analyzed. The relatively minor increases in gene expression may be due to the fact that CrBPF1 overexpression causes increased expression of all five TIA transcriptional repressors, in addition to causing increased activity of most of the TIA transcriptional activators. In contrast, overexpression of ORCA2 (Li et al., 2013) and ORCA3 (Peebles et al., 2009) causes increased expression of the ZCT transcriptional repressors, but not of the GBF transcriptional repressors. As overexpression of CrBPF1 has comparatively large effects on expression of TIA transcriptional repressors, future characterization of these repressors is expected to yield further insight into the mechanism by which CrBPF1 helps regulate the TIA and related pathways. The simultaneous activation of transcriptional repressors and activators has been proposed to act as part of a “fine tune” mechanism for regulating TIA production (Memelink and Gantet, 2007). The findings reported here support this model and suggest that CrBPF1 plays a role in the “fine tune” regulation of TIA metabolism.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Prof. Christie A. M. Peebles at Colorado University for advice on induction of hairy root production, Dr. Sarah O’Connor of the John Innes Centre for the gift of the strictosidine standard, Dr. Kenneth Beckman and Trianna Full at the University of Minnesota Genomics Center for helping with qRT-PCR analysis and Dr. Kenichi Tsuda at the Max Planck Institute for Plant Breeding Research for supplying the pERKT vector. Financial support for conducting the research was provided by NSF CBET-0729753 (JVS) and NSF CBET-0729625 (SIG) and for preparing the manuscript by NSF CBET-1064903.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00818
Asada, K., Salim, V., Masada-Atsumi, S., Edmunds, E., Nagatoshi, M., Terasaka, K., et al. (2013). A 7-deoxyloganetic acid glucosyltransferase contributes a key step in secologanin biosynthesis in Madagascar periwinkle. Plant Cell 25, 4123–4134. doi: 10.1105/tpc.113.115154
Besseau, S., Kellner, F., Lanoue, A., Thamm, A. M. K., Salim, V., Schneider, B., et al. (2013). A pair of tabersonine 16-hydroxylases initiates the synthesis of vindoline in an organ-dependent manner in Catharanthus roseus. Plant Physiol. 163, 1792–1803. doi: 10.1104/pp.113.222828
Burlat, V., Oudin, A., Courtois, M., Rideau, M., and St-Pierre, B. (2004). Co-expression of three MEP pathway genes and geraniol 10-hydroxylase in internal phloem parenchyma of Catharanthus roseus implicates multicellular translocation of intermediates during the biosynthesis of monoterpene indole alkaloids and isoprenoid-derived primary metabolites. Plant J. 381, 131–141.
Chatel, G., Montiel, G., and Metal, P. (2003). CrMYC1, a Catharanthus roseus elicitor- and jasmonate- responsive bHLH transcription factor that binds the G-box element of the strictosidine synthase gene promoter. J. Exp. Bot. 54, 2587–2588. doi: 10.1093/jxb/erg275
Chebbi, M., Ginis, O., Courdavault, V., Glévarec, G., Lanoue, A., Clastre, M., et al. (2014). ZCT1 and ZCT2 transcription factors repress the activity of a gene promoter from the methyl erythritol phosphate pathway in Madagascar periwinkle cells. J. Plant Physiol. 171, 1510–1513. doi: 10.1016/j.jplph.2014.07.004
Costa, M. M. R., Hilliou, F., Duarte, P., Pereira, L. G., Almeida, I., Leech, M., et al. (2008). Molecular cloning and characterization of a vacuolar class III peroxidase involved in the metabolism of anticancer alkaloids in Catharanthus roseus. Plant Physiol. 146, 403–417. doi: 10.1104/pp.107.107060
Curtis, M. D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
da Costa e Silva, O., Klein, L., Schmelzer, E., Trezzini, G. F., and Hahlbrock, K. (1993). BPF-1, a pathogen-induced DNA-binding protein involved in the plant defense response. Plant J. 4, 125–135. doi: 10.1046/j.1365-313X.1993.04010125.x
De Luca, V., Salim, V., Thamm, A., Masada, S. A., and Yu, F. (2014). Making iridoids/secoiridoids and monoterpenoid indole alkaloids: progress on pathway elucidation. Curr. Opin. Plant Biol. 19, 35–42. doi: 10.1016/j.pbi.2014.03.006
Feller, A., Machemer, K., Braun, E. L., and Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Gidding, C. E. M., Kellie, S. J., Kamps, W. A., and de Graaf, S. S. N. (1999). Vincristine revisited. Crit. Rev. Oncol. Hematol. 29, 267–287. doi: 10.1016/S1040-8428(98)00023-7
Giddings, L.-A., Liscombe, D. K., Hamilton, J. P., Childs, K. L., DellaPenna, D., Buell, C. R., et al. (2011). A stereoselective hydroxylation step of alkaloid biosynthesis by a unique cytochrome P450 in Catharanthus roseus. J. Biol. Chem. 286, 16751–16757. doi: 10.1074/jbc.M111.225383
Han, M., Heppel, S. C., Su, T., Bogs, J., Zu, Y., An, Z., et al. (2013). Enzyme inhibitor studies reveal complex control of methy-D-erythritol 4-phosphate (MEP) pathway enzyme expression in Catharanthus roseus. PLoS ONE 8:e62467. doi: 10.1371/journal.pone.0062467
Hong, S. B., Peebles, C. A. M., Shanks, J. V., San, K. Y., and Gibson, S. I. (2006). Terpenoid indole alkaloid production by Catharanthus roseus hairy roots induced by Agrobacterium tumefaciens harboring rol ABC genes. Biotechnol. Bioeng. 93, 386–390. doi: 10.1002/bit.20699
Huang, Y. D., Li, C. Y., Pattison, D. L., Gray, W. M., Park, S., and Gibson, S. I. (2010). SUGAR-INSENSITIVE3, a RING E3 ligase, is a new player in plant sugar response. Plant Physiol. 152, 1889–1900. doi: 10.1104/pp.109.150573
Kellner, F., Geu-Flores, F., Sherden, N. H., Brown, S., Foureau, E., Courdavault, V., et al. (2015). Discovery of a P450-catalyzed step in vindoline biosynthesis: a link between aspidosperma and eburnamine alkaloids. Chem. Commun. 51, 7626–7628. doi: 10.1039/c5cc01309g
Levac, D., Murata, J., Kim, W. S., and De Luca, V. (2008). Application of carborundum abrasion for investigating the leaf epidermis: molecular cloning of Catharanthus roseus 16-hydroxytabersonine-16-O-methyltransferase. Plant J. 53, 225–236. doi: 10.1111/j.1365-313X.2007.03337.x
Li, C. Y., Leopold, A. L., Sander, G. W., Shanks, J. V., Zhao, L., and Gibson, S. I. (2013). The ORCA2 transcription factor plays a key role in regulation of the terpenoid indole alkaloid pathway. BMC Plant Biol. 13:155. doi: 10.1186/1471-2229-13-155
Liscombe, D. K., Usera, A. R., and O’Connor, S. E. (2010). Homolog of tocopherol C methyltransferases catalyzes N methylation in anticancer alkaloid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 18793–18798. doi: 10.1073/pnas.1009003107
Liu, D. H., Jin, H. B., Chen, Y. H., Cui, L. J., Ren, W. W., Gong, Y. F., et al. (2007). Terpenoid indole alkaloids biosynthesis and metabolic engineering in Catharanthus roseus. J. Int. Plant Biol. 49, 961–974. doi: 10.1111/j.1672-9072.2007.00457.x
Liu, D. H., Ren, W. W., Cui, L. J., Zhang, L. D., Sun, X. F., and Tang, K. X. (2011). Enhanced accumulation of catharanthine and vindoline in Catharanthus roseus hairy roots by overexpression of transcriptional activator ORCA2. Afr. J. Biotechnol. 10, 3260–3268.
Memelink, J., and Gantet, P. (2007). Transcription factors involved in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Phytochem. Rev. 6, 353–362. doi: 10.1007/s11101-006-9051-z
Menke, F. L. H., Champion, A., Kijne, J. W., and Memelink, J. (1999). A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor. ORCA2. EMBO J. 18, 4455–4463. doi: 10.1093/emboj/18.16.4455
Murata, J., Roepke, J., Gordon, H., and De Luca, V. (2008). The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell 20, 524–542. doi: 10.1105/tpc.107.056630
Pan, Q., Wang, Q., Yuan, F., Xing, S., Zhao, J., Choi, Y. H., et al. (2012). Overexpression of ORCA3 and G10H in Catharanthus roseus plants regulated alkaloid biosynthesis and metabolism revealed by NMR-metabolomics. PLoS ONE 7:e43038. doi: 10.1371/journal.pone.0043038
Pauw, B., Hilliou, F. A. O., Martin, V. S., Chatel, G., de Wolf, C. J. F., Champion, A., et al. (2004). Zinc finger proteins act as transcriptional repressors of alkaloid biosynthesis genes in Catharanthus roseus. J. Biol. Chem. 279, 52940–52948. doi: 10.1074/jbc.M404391200
Peebles, C. A. M., Hong, S.-B., Gibson, S. I., Shanks, J. V., and San, K.-Y. (2005). Transient effects of over-expressing anthranilate synthase α and ß subunits in Catharanthus roseus hairy roots. Biotechnol. Prog. 21, 1572–1576. doi: 10.1021/bp050210l
Peebles, C. A. M., Hughes, E. H., Shanks, J. V., and San, K. Y. (2009). Transcriptional response of the terpenoid indole alkaloid pathway to the overexpression of ORCA3 along with jasmonic acid elicitation of Catharanthus roseus hairy roots over time. Metabol. Eng. 11, 76–86. doi: 10.1016/j.ymben.2008.09.002
Qu, Y., Easson, M. L. A. E., Froese, J., Simionescu, R., Hudlicky, T., and De Luca, V. (2015). Completion of the seven-step pathway from tabersonine to the anticancer drug precursor vindoline and its assembly in yeast. Proc. Natl. Acad. Sci. U.S.A. 112, 6224–6229. doi: 10.1073/pnas.1501821112
Salim, V., Yu, F., Altarejos, J., and De Luca, V. (2013). Virus-induced gene silencing identifies Catharanthus roseus 7-deoxyloganic acid-7-hydroxylase, a step in iridoid and monoterpene indole alkaloid biosynthesis. Plant J. 76, 754–765. doi: 10.1111/tpj.12330
Sander, G. W. (2009). Quantitative Analysis of Metabolic Pathways in Catharanthus roseus Hairy Roots Metabolically Engineered for Terpenoid Indole Alkaloid Overproduction. Ph.D. thesis, Department of Chemical and Biological Engineering, Iowa State University, Ames, IA.
Schröder, G., Unterbusch, E., Kaltenbach, M., Schmidt, J., Strack, D., De Luca, V., et al. (1999). Light-induced cytochrome P450-dependent enzyme in indole alkaloid biosynthesis: tabersonine 16-hydroxylase. FEBS Lett. 458, 97–102. doi: 10.1016/S0014-5793(99)01138-2
Shanks, J. V. (2005). Phytochemical engineering: combining chemical reaction engineering with plant science. AIChE J. 51, 2–7. doi: 10.1002/aic.10418
Sibéril, Y., Benhamron, S., Memelink, J., Giglioli-Guivarc’h, N., Thiersault, M., Boisson, B., et al. (2001). Catharanthus roseus G-box binding factors 1 and 2 act as repressors of strictosidine synthase gene expression in cell cultures. Plant Mol. Biol. 45, 477–488. doi: 10.1023/A:1010650906695
St-Pierre, B., Lalfamme, P., Alarco, A. M., and De Luca, V. (1998). The terminal O-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible for coenzyme A-dependent acyl transfer. Plant J. 14, 703–713. doi: 10.1046/j.1365-313x.1998.00174.x
Suttipanta, N. (2011). Characterization of G10H Promoter and Isolation of WRKY Transcription Factors Involved in Catharanthus terpenoid Indole Alkaloid Biosynthesis Pathway. Ph.D. thesis, Plant Physiology Department, University of Kentucky, Kentucky.
Suttipanta, N., Pattanaik, S., Kulshrestha, M., Patra, B., Singh, S. K., and Yuan, L. (2011). The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 157, 2081–2093. doi: 10.1104/pp.111.181834
Tang, K. X., Liu, D. H., Wang, Y. L., Cui, L. J., Ren, W. W., and Sun, X. F. (2011). Overexpression of transcriptional factor ORCA3 increases the accumulation of catharanthine and vindoline in Catharanthus roseus hairy roots. Russ. J. Plant Physiol. 58, 415–422. doi: 10.1134/S1021443711030125
Tsuda, K., Qi, Y., Nguyen, L. V., Bethke, G., Tsuda, Y., Glazebrook, J., et al. (2012). An efficient agrobacterium-mediated transient transformation of Arabidopsis. Plant J. 69, 713–719. doi: 10.1111/j.1365-313X.2011.04819.x
van der Fits, L., and Memelink, J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289, 295–297. doi: 10.1126/science.289.5477.295
van der Fits, L., and Memelink, J. (2001). The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 25, 43–53. doi: 10.1111/j.1365-313X.2001.00932.x
van der Fits, L., Zhang, H., Menke, F. L. H., Deneka, M., and Memelink, J. (2000). A Catharanthus roseus BPF-1 homologue interacts with an elicitor-responsive region of the secondary metabolite biosynthetic gene Str and is induced by elicitor via a JA-independent signal transduction pathway. Plant Mol. Biol. 44, 675–685. doi: 10.1023/A:1026526522555
van der Heijden, R., Jacobs, D. I., Snoeijer, W., Hallared, D., and Verpoorte, R. (2004). The Catharanthus alkaloids: pharmacognosy and biotechnology. Curr. Med. Chem. 11, 607–628. doi: 10.2174/0929867043455846
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
Van Moerkercke, A., Steensma, P., Schweizer, F., Pollier, J., Gariboldi, I., Payne, R., et al. (2015). The bHLH transcription factor BIS1 control the iridoid branch of the monoterpenoid alkaloid pathway in Catharanthus roseus. Proc. Natl. Acad. Sci. U.S.A. 112, 8130–8135. doi: 10.1073/pnas.1504951112
Vazquez-Flota, F., De Carolis, E., Alarco, A. M., and De Luca, V. (1997). Molecular cloning and characterization of desacetoxyvindoline-4-hydroxylase, a 2-oxoglutarate dependent dioxygenase involved in the biosynthesis of vindoline in Catharanthus roseus (L) G. Don. Plant Mol. Biol. 34, 935–948. doi: 10.1023/A:1005894001516
Vom Endt, D., Soares e Silva, M., Kijne, J. W., Pasquali, G., and Memelink, J. (2007). Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-hook DNA-binding proteins. Plant Physiol. 144, 1680–1689. doi: 10.1104/pp.107.096115
Wang, C. T., Liu, H., Gao, X. S., and Zhang, H. X. (2010). Overexpression of G10H and ORCA3 in the hairy roots of Catharanthus roseus improves catharanthine production. Plant Cell Rep. 29, 887–894. doi: 10.1007/s00299-010-0874-0
Wei, S. (2010). Methyl jasmonic acid induced expression pattern of terpenoid indole alkaloid pathway genes in Catharanthus roseus seedlings. Plant Growth Regul. 61, 243–251. doi: 10.1007/s10725-010-9468-7
Zhang, H., Hedhili, S., Montiel, G., Zhang, Y., Chatel, G., Pré, M., et al. (2011). The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 67, 61–71. doi: 10.1111/j.1365-313X.2011.04575.x
Zhao, L., Sander, G. W., and Shanks, J. V. (2013). Perspectives of the metabolic engineering of terpenoid indole alkaloids in Catharanthus roseus hairy roots. Adv. Biochem. Eng. Biotechnol. 134, 23–54. doi: 10.1007/10_2013_182
Zhou, M. L., Hou, H. L., Zhu, X. M., Shao, J. R., Wu, Y. M., and Tang, Y. X. (2010a). Molecular regulation of terpenoid indole alkaloids pathway in the medicinal plant, Catharanthus roseus. J. Med. Plants Res. 4, 2760–2772.
Zhou, M. L., Zhu, X. M., Shao, J. R., Wu, Y. M., and Tang, Y. X. (2010b). Transcriptional response of the catharanthine biosynthesis pathway to methyl jasmonate/nitric oxide elicitation in Catharanthus roseus hairy root culture. Appl. Microbiol. Biotechnol. 88, 737–750. doi: 10.1007/s00253-010-2822-x
Keywords: Catharanthus roseus, CrBPF1, GBF, ORCA2, ORCA3, terpenoid indole alkaloid, transgenic hairy roots, ZCT
Citation: Li C, Leopold AL, Sander GW, Shanks JV, Zhao L and Gibson SI (2015) CrBPF1 overexpression alters transcript levels of terpenoid indole alkaloid biosynthetic and regulatory genes. Front. Plant Sci. 6:818. doi: 10.3389/fpls.2015.00818
Received: 12 June 2015; Accepted: 18 September 2015;
Published: 01 October 2015.
Edited by:
Henrik Toft Simonsen, University of Copenhagen, DenmarkReviewed by:
Kevin Davies, New Zealand Institute for Plant and Food Research, New ZealandCopyright © 2015 Li, Leopold, Sander, Shanks, Zhao and Gibson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susan I. Gibson, Department of Plant Biology, University of Minnesota Twin Cities, 110 Cargill Building, 1500 Gortner Avenue, Saint Paul, MN 55108, USA, gibso043@umn.edu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.