
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 29 September 2015
Sec. Plant Nutrition
Volume 6 - 2015 | https://doi.org/10.3389/fpls.2015.00796
This article is part of the Research Topic Ethylene’s role in plant mineral nutrition View all 13 articles
Phosphorus is an essential macronutrient for plant growth and development. Phosphate (Pi), the major form of phosphorus that plants take up through roots, however, is limited in most soils. To cope with Pi deficiency, plants activate an array of adaptive responses to reprioritize internal Pi use and enhance external Pi acquisition. These responses are modulated by sophisticated regulatory networks through both local and systemic signaling, but the signaling mechanisms are poorly understood. Early studies suggested that the phytohormone ethylene plays a key role in Pi deficiency-induced remodeling of root system architecture. Recently, ethylene was also shown to be involved in the regulation of other signature responses of plants to Pi deficiency. In this article, we review how researchers have used pharmacological and genetic approaches to dissect the roles of ethylene in regulating Pi deficiency-induced developmental and physiological changes. The interactions between ethylene and other signaling molecules, such as sucrose, auxin, and microRNA399, in the control of plant Pi responses are also examined. Finally, we provide a perspective for the future research in this field.
Plants are sessile organisms that acquire essential mineral nutrients from soils through their roots. Plants often encounter nutrient deficiency in natural ecosystems and in agricultural lands. P (Phosphorus) is an essential macronutrient for plant growth, development, and metabolism. Inorganic phosphate, the major form of P that plants assimilate, however, is highly immobile in most soils because it is converted to organophosphates by microorganisms or is fixed with metals (Bieleski, 1973). Although P is abundant in most soils, the availability of Pi for plant uptake is quite low (Raghothama, 2000). Pi deficiency has become one of the most important constraints on agricultural productivity.
To cope with Pi deficiency, plants have evolved elaborate strategies to enhance acquisition and utilization of Pi from the environment and to conserve and reprioritize the internal use of Pi through recycling and redistribution processes. The major plant responses to Pi deficiency include: the active remodeling of RSA; the reduction in photosynthesis; the enhancement of high-affinity Pi transporter activities; the induction and secretion of APases, ribonucleases, and organic acids; the replacement of phospholipids in membranes with glycolipids and sulfolipids; and the accumulation of anthocyanin and starch (Vance et al., 2003; Yuan and Liu, 2008; Figure 1). These responses are modulated by sophisticated regulatory networks through both local and systemic signaling in which phytohormones play important roles (Chiou and Lin, 2011).
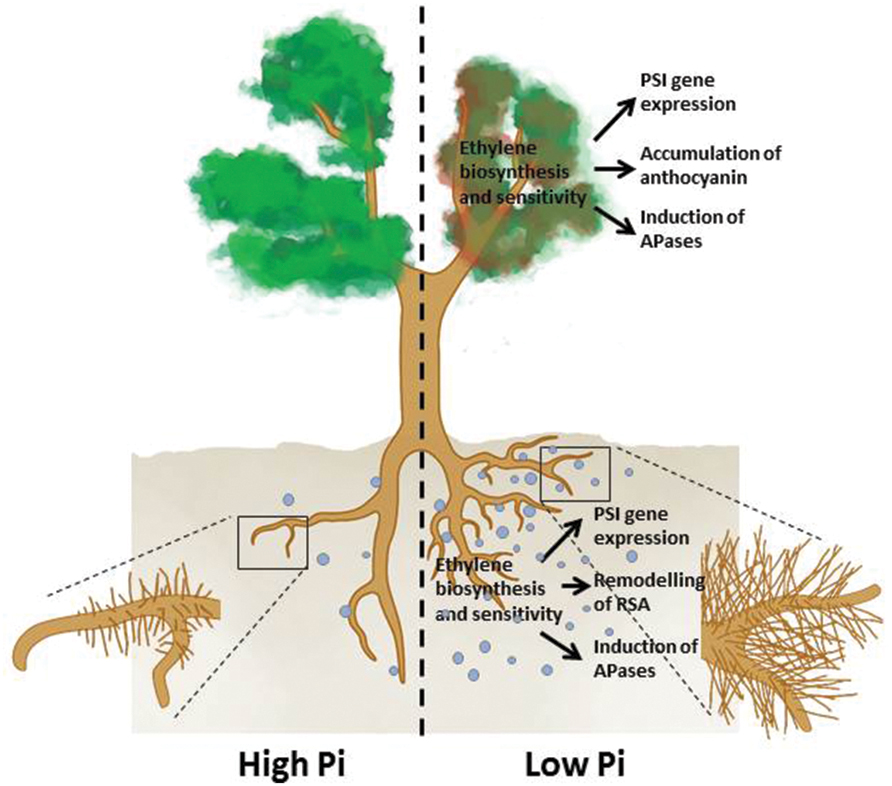
FIGURE 1. An overview of the plant responses to Pi deficiency that involve ethylene. Under Pi deficiency, primary root growth is inhibited, production of lateral roots and root hairs is enhanced (remodeling of RSA) APases are induced, expression of PSI genes is increased in both roots and shoots, and shoots accumulate more anthocyanins. The light blue dots denote root exudates, including APases, RNases, organic acids, and protons.
Ethylene, which is one of five classic phytohormones, regulates multiple aspects of plant development, such as seed germination, root growth, leaf abscission and senescence, and fruit ripening, as well as plant responses to biotic and abiotic stresses (Abeles et al., 1992). Ethylene was previously shown to be involved in Pi deficiency-induced remodeling of RSA. Recent research indicates that in addition to being a regulator of root growth, ethylene also participates in other plant responses to Pi deficiency (Nagarajan and Smith, 2012; Roldan et al., 2013). In this article, we will review the current knowledge about the roles of ethylene in plant responses to Pi deficiency. We will also provide a perspective about how research might increase our understanding of the molecular mechanisms by which ethylene regulates plant Pi responses.
The ethylene biosynthetic pathway has been well described in higher plants (Kende, 1993; Xu and Zhang, 2015; Figure 2A). Ethylene is produced from methionine, which is first converted to AdoMet by AdoMet synthetase. The next two steps are the conversion of AdoMet to ACC and the oxidative cleavage of ACC to form ethylene. The enzymes that catalyze these two reactions are ACS and ACO. Once ACC is formed in plant cells, it is automatically converted to ethylene by ACO in the presence of oxygen. The regulation of ethylene biosynthesis can be achieved by altering gene expression, protein stability, and enzymatic activity of ACS and ACO (McKeon et al., 1995; Bleecker and Kende, 2000).
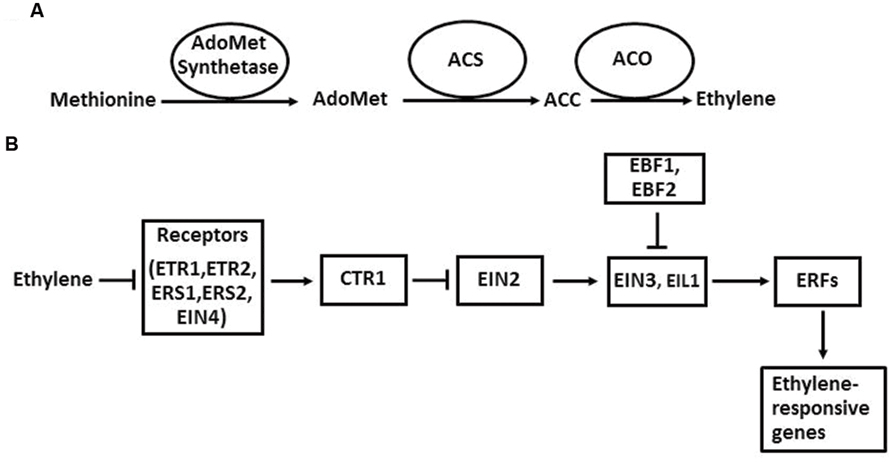
FIGURE 2. The ethylene biosynthesis pathway in plants in general (A) and the ethylene signaling pathway in Arabidopsis (B). Arrows indicate promotion, and perpendicular lines indicate inhibition.
The ethylene signaling pathway in Arabidopsis has provided a framework for the action mechanism of this hormone in higher plants (Zhao and Guo, 2011; Wen et al., 2015). In Arabidopsis, ethylene is perceived by its receptors, which are ETR1, ETR2, ERS1, ERS2, and EIN4 located in ER membrane. When ethylene levels are low, these five receptors activate the downstream component CTR1 through direct physical interaction. CTR1 belongs to the Raf-1 family of Ser/Thr protein kinases. The activated CTR1 suppresses the function of its target EIN2, an ER membrane-localized protein. When ethylene binds to the receptors, it disrupts the interaction between the receptors and CTR1, which inactivates CTR1. Therefore, the suppression of EIN2 by CTR1 is released. Next, a EIN2-C′ (C-terminal fragment of EIN2) is translocated to the nucleus. In the nucleus, EIN2-C′ enhances the levels of EIN3 and EIL1, two key transcription factors of the ethylene signaling pathway. EIN3 and EIL1 further activate the transcription of downstream target genes, such as ERFs, CHIB, PORA, and FLS2. Finally, the activation of these genes initiates a diverse array of plant responses to internal and external signals. In the absence of ethylene, EIN3 and EIL1 are degraded by the F-box proteins EBF1 (EIN3 BINDING F-BOX PROTEIN 1) and EBF2 through a 26S proteasome-mediated degradation pathway.
It has long been observed that Pi deficiency alters ethylene biosynthesis in plants. Some reports indicated that ethylene production was decreased in Pi-starved maize and tomato (Drew et al., 1989; Kim et al., 2008). In contrast, the increase of ethylene production was found in the roots of common bean under Pi deficiency (Phaseolus vulgaris), white lupin (Lupinus albus), and Medicago falcata (Borch et al., 1999; Gilbert et al., 2000; Li et al., 2009). Using quantitative reverse-transcription PCR (RT-qPCR), Lei et al. (2011b) showed that the expression of three members of the ACS gene family, ACS2, ACS4, and ACS6, was enhanced in Arabidopsis seedlings grown on a Pi-deficient medium. The enhanced expression of ethylene biosynthetic genes, mainly ACS and ACO, in Pi-starved Arabidopsis plants has also been demonstrated in several microarray and RNA-seq analyses (Misson et al., 2005; Morcuende et al., 2007; Calderon-Vazquez et al., 2008; Thibaud et al., 2010; Chacón-López et al., 2011; O’Rourke et al., 2013; Kang et al., 2014; Wang et al., 2014b). Morcuende et al. (2007) further found that the enhanced expression of ACS2 and ACS6 genes was reversed when Pi-deficient plants were resupplied with an adequate amount of Pi, indicating a causal relationship between the expression of these genes and the levels of Pi in the environment. The up-regulation of ACO genes by Pi deficiency was also detected in a variety of plant species, including common bean (Graham et al., 2006; Hernández et al., 2007), white lupin (Uhde-Stone et al., 2003; Wang et al., 2014b), and white clover (Trifolium repens; Roldan et al., 2013). In the plants examined, not all members of ACS and ACO families respond to Pi deficiency in the same manner. Thus, it is important to know how the transcription of different members of ACS and ACO gene families is affected by Pi deficiency in specific tissues and at specific developmental stages. Such information will increase our understanding of how ethylene biosynthesis is regulated in a spatiotemporal manner by Pi deficiency.
In Arabidopsis, PHR1 is a MYB-type transcription factor that binds to a cis-element with the imperfect palindromic sequence GNATATNC, a sequence that is prevalent in the promoters of PSI (Pi starvation-induced) genes (Rubio et al., 2001). PHR1 is regarded as a central regulator for transcriptional responses of plants to Pi starvation (Bustos et al., 2010). In Arabidopsis, PHL1 is a close relative of PHR1. In the phr1phl1 double mutant, the induction of most PSI genes, including ACS6 and ACS7, is impaired to different extents (Bustos et al., 2010). These results indicate that the increased expression of these two ACS genes is modulated by the central regulatory pathway of Pi responses. Interestingly, the expression of another two PSI ACS genes, ACS2 and ACS4, is independent of the PHR1 pathway (Bustos et al., 2010), suggesting that the regulation of different members of the ACS family is mediated by different signaling pathways.
In addition to altering ethylene biosynthesis, Pi deficiency also alters plant sensitivity to ethylene. Pi deficiency-enhanced ethylene sensitivity has been reported for adventitious roots of maize seedlings (He et al., 1992), basal roots of common bean (Basu et al., 2007), and lateral roots of white clover (Dinh et al., 2012). The enhanced ethylene sensitivity is also reflected in the induction of gene expression by Pi deficiency (Lei et al., 2011b) as discussed in more detail later in this article. The enhanced ethylene sensitivity in Pi-starved plants is probably achieved through the alteration of the expression of the genes that encode molecular components involved in the ethylene signaling pathway. ERFs are a group of AP2 (APETALA2) domain-containing transcription factors that bind to ethylene-responsive elements present in the promoters of many ethylene-responsive genes. These transcription factors serve either as activators or repressors of ethylene-mediated transcription. Some transcriptomic analyses have shown that the expression of several ERF genes, including ERF1, ERF2, and ERF5, is altered in Pi-starved Arabidopsis roots (Wang et al., 2002; Misson et al., 2005; Thibaud et al., 2010; Chacón-López et al., 2011; Kang et al., 2014). In the phr1phl1 double mutant, the expression of at least eight AP2/ERF genes is attenuated (Bustos et al., 2010). Another mechanism for enhancing ethylene sensitivity may involve a change of protein abundance of some key components in the ethylene signaling pathway. When plants were grown under salinity conditions, the accumulation of EIN3 protein was increased while that of EBFs, which mediate EIN3 protein degradation, was decreased (Peng et al., 2014). Whether this is also the case for Pi-deficient plants requires investigation.
When plants are grown under Pi deficiency, their RSA undergoes a dramatic change, i.e., a remodeling. The Pi deficiency-induced remodeling of RSA includes a cessation of primary root growth and an enhanced production of root hairs and lateral roots (López-Bucio et al., 2003). For maize and some species in the Proteaceae and Casuarinaceae families, the remodeling of RSA involves a production of adventitious roots and cluster-roots (CRs; He et al., 1992; Wang et al., 2015). Such remodeling of RSA results in an increase in root surface area for Pi absorption. The remodeling starts with a reduction of cell elongation followed by the progressive loss of meristematic cells, i.e., “meristem exhaustion” (Sánchez-Calderón et al., 2005). At later stages, cell proliferation is arrested, and cell differentiation takes place at the former meristematic and elongation regions of the primary root. In some plant species, the angle between the basal root and primary root is increased, which enhances the capacity of roots to forage Pi in the top soil (Basu et al., 2007). The remodeling of RSA triggered by Pi deficiency is thought to be an active developmental response controlled by internal genetic programs because Arabidopsis mutants lpi and lpr1 show normal growth of primary roots with an unexhausted meristem under Pi deficiency (Sánchez-Calderón et al., 2006; Svistoonoff et al., 2007).
Under normal growth conditions, application of ethylene (or ACC) to Arabidopsis plants inhibits primary root growth and enhances root hair production. This treatment results in meristem exhaustion of the primary root. Arabidopsis mutant that overproduces ethylene (eto1; Wang et al., 2004) or that has constitutive ctr1 (Kieber et al., 1993) also exhibit reduced primary root growth and increased production of root hairs. These ethylene-induced root growth phenotypes mimic the plant root responses triggered by low Pi, suggesting that ethylene biosynthesis and signaling are involved in the Pi deficiency-triggered remodeling of RSA.
The roles of ethylene in Pi deficiency-induced remodeling of RSA have been investigated using both pharmacological and genetic approaches. Regarding the effect of ethylene on primary root growth under Pi deficiency, Chacón-López et al. (2011) found that inhibiting ethylene biosynthesis with AVG or ethylene perception with Ag+ restricted the low Pi-induced meristem exhaustion of the primary root. This suggested that ethylene is involved in the Pi deficiency-induced inhibition of primary root growth. Our laboratory also found that application of Ag+ reduces the inhibition of primary root growth triggered by Pi deficiency (Yu et al., 2012). The Arabidopsis mutant hps4 (hypersensitive to Pi starvation 4) shows enhanced sensitivity to Pi deficiency in terms of Pi deficiency-induced inhibition of primary root growth and induction of APase activity. Under Pi sufficiency, the primary root of hps4 was about 80% as long as WT (wild type) plants. Under Pi deficiency, the primary root growth was reduced for both the WT and hps4; however, this reduction was much greater for hps4 than for the WT. HPS4 encodes the SABRE protein. Although the precise biochemical function of SABRE is unknown, SABRE has been shown to antagonistically interact with ethylene to regulate root cell expansion (Aeschbacher et al., 1995). The hypersensitivity to Pi deficiency-induced inhibition of primary root growth is diminished when the hps4 mutant is treated with Ag+ but not with AVG. Nagarajan et al. (2011) showed that overexpression of an Arabidopsis high-affinity Pi transporter, Pht1:5, reduced primary root growth and increased root hair production under both Pi sufficiency and deficiency. These phenotypes could be reversed by application of AVG or Ag+, indicating that ethylene biosynthesis had been altered in the Pht1:5-overexpressing lines. The Pht1:5-overexpressing lines also showed a disruption of the shoot to root Pi ratio, suggesting that an altered Pi homeostasis might enhance ethylene biosynthesis and/or signaling, which in turn, might modulate primary root growth. More experimental evidence is needed, however, to support this proposed link between altered Pi homeostasis and enhanced ethylene biosynthesis and/or signaling. Interestingly, when Ma et al. (2003) investigated the role of ethylene in Pi starvation-induced inhibition of primary root growth, they found that inhibition of ethylene production by AVG or inhibition of ethylene action by MCP increased primary root growth under high Pi but decreased primary root growth under low Pi. This result differs from the observations of López-Bucio et al. (2002) and Yu et al. (2012). The discrepancy could be due to the differences in experimental conditions used by the research groups. For example, the degree of low Pi stress in the different media differed, and whether the effect of ethylene on primary root growth is stimulatory or inhibitory could depend on a subtle difference in Pi concentrations, even though all concentrations were low. In fact, ethylene can be as both a promoter or an inhibitor for root growth depending on its concentration (Pierik et al., 2006). Thus, after inhibitors were applied in the experiments of Ma et al. (2003), the concentration of ethylene in Pi-deficient roots may have been far below the optimal level for sustained primary root growth under Pi deficiency.
In young maize seedlings, Pi deficiency induces the formation of aerenchyma (tissue with large cortical gas spaces) in their adventitious roots (He et al., 1992). When Ag+ or AVG was added to the nutrient solution, the formation of aerenchyma was blocked. Furthermore, when ethylene was added to the air of the growth chamber at a concentration as low as 1.0 μL/L, the aerenchyma formation was strongly promoted in Pi-starved roots relative to Pi-sufficient roots. Because the production of ethylene was decreased in Pi-starved maize seedlings in these experiments, it seemed that ethylene perception or sensitivity rather than ethylene production was involved in the formation of aerenchyma triggered by Pi deficiency. A similar case was found for tomato plants. Low Pi induced the formation of adventitious root in WT tomato plants but not in the ethylene-insensitive cultivar “Never-ripe” (Kim et al., 2008). Pi deficiency, however, reduced ethylene production in both tomato genotypes. This again indicated that it is ethylene perception rather than ethylene production that is involved in the response of roots to Pi availability.
The effects of ethylene on lateral root formation in Pi starved-plants have also been investigated. Pi deficiency stimulates the formation of lateral roots of white clover (Dinh et al., 2012). A low concentration of ACC had little effect on the development of lateral roots under Pi sufficiency but caused a super-stimulation of lateral roots under Pi deficiency. Unlike in white clover, Pi deficiency in common bean reduced lateral root number and did not inhibit primary root growth (Borch et al., 1999). This resulted in a reduction of lateral root density. AVG treatment increased lateral root density in Pi-deficient plants but reduced lateral root density in Pi-sufficient plants. These responses could be reversed by exogenous ethylene, suggesting an involvement of ethylene in modulating lateral root formation in common bean under Pi deficiency.
When grown under Pi deprivation, most members of the Proteaceae and Casuarinaceae form CR. The formation of CR densely covered with long root hairs dramatically increases the surface area for secretion of root exudates, including organic acids, protons, and APases. This root response helps plants mobilize Pi from organophosphates or Pi fixed with metals. Application of ethylene did not induce CR formation under Pi sufficiency; however, in Pi-deficient plants, the inhibition of ethylene production by Co2+ completely suppressed CR formation (Wang et al., 2015). Associated with this, the authors found a moderate increase in the expression of an ACO gene in the pre-emergent CR but a dramatic increase in ACO expression in the maturing CR.
In many soils, Pi availability is greatest in the upper layers and decreases with depth. Analyses of different genotypes of bean indicated that the degree of growth angle of basal roots was closely correlated with the extent of Pi acquisition of plants under low Pi availability (Basu et al., 2007). Ethylene sensitivity was higher for plants grown under Pi deficiency than under Pi sufficiency, and basal roots produced from the uppermost whorl were more sensitive to ethylene than those from the lower-most whorl. Thus, the growth angle of basal roots was strongly correlated with ethylene sensitivity but not with ethylene production.
The earliest visible change in the morphology of roots responding to Pi deficiency is the enhanced production of root hairs, including an increase in both root hair density and root hair length (Bates and Lynch, 1996). The increase in root hair length is due to an increase in both growth rate and growth duration. In Arabidopsis, root hairs are produced from H cells that are located over the intercellular space between two underlying adjacent cortical cells; however, not all H cells form root hairs. Under normal growth conditions, treatment with ACC or a mutation in the CTR1 and ETO1 genes causes a dramatic increase in root hair production that mimics the effect of Pi deficiency. Schmidt and Schikora (2001) found that low Pi could not fully restore the root hair density of ein2 and etr1 mutants to that of the WT. And, for the three ethylene biosynthesis inhibitors that they used (AVG, Co2+, and aminooxyacetic acid), only Co2+ significantly blocked the Pi deficiency-induced increase in the root hair density of the WT plants. Thus, the authors concluded that the canonical ethylene signaling pathway was not involved in the development of extra root hairs in response to Pi deficiency. However, the research of Zhang et al. (2003) strongly suggested that ethylene is involved in the Pi deficiency-enhanced production of root hairs. These authors found that under Pi deficiency, treatment with inhibitors of either ethylene biosynthesis or ethylene signaling significantly reduced root hair density and root hair length. Although all ethylene-insensitive mutants still responded to Pi deficiency with increased root hair density and length, the extent of the increase was much lower than that of the WT. These results indicated that ethylene is indeed involved in the enhanced production of root hairs induced by Pi deficiency.
Anatomic examination, however, revealed some similarities and differences in the effects of ethylene and Pi deficiency on root hair formation (Zhang et al., 2003). The similarity is that both low Pi and ethylene shortened the length of trichoblast cells and that AVG added to Pi-deficient plants increased the length of trichoblast cells. The differences include: (1) Low Pi increases the number of cortical cells, but ethylene does not; (2) Ethylene increase the percentage of H cells that form root hairs, but low Pi does not. These differences suggested that low Pi and ethylene may use the different gene activation mechanisms to regulate root hair formation. It will be interest to compare the transcriptomic changes in Pi-starved and ethylene-treated roots to identify the common and distinct targets of low Pi and ethylene. These information will help us further understand the molecular mechanisms of how ethylene mediates the root hair growth under Pi deficiency.
Further analysis demonstrated that there is an interaction between low Pi and ethylene in regulating both root hair length and root hair density (Zhang et al., 2003). The reduction in trichoblast length in ein2 and ein4 was stronger with low Pi than with high Pi, indicating that the degree to which ethylene affects extra root hair production depends on Pi availability. And, ein2 and ein4 mutants or the WT treated with AVG had greater reduction of H cells forming hairs under low Pi than under high Pi. In addition, under Pi deficiency, all ethylene-insensitive mutants clearly showed a reduction in root hair length, but the reduction varied for different mutants under high Pi. Cho and Cosgrove (2002) found that root hair elongation for ethylene-insensitive mutants or for plants treated with AVG was relatively normal under high Pi but was reduced under low Pi, providing additional evidence of an interaction between ethylene and Pi availability.
In searching for molecular components involved in transcriptional responses of plants to Pi starvation, Lei et al. (2011b) performed a screen for Arabidopsis mutants with altered transcriptional response. They used a transgenic line that carries a LUC gene fused to the promoter of the high-affinity Pi transporter AtPT2. The transcription of AtPT2 is induced by Pi starvation. Using this marker line, the authors identified the Arabidopsis mutant hps2 (hypersensitive to Pi starvation2), which showed hyper-induction of the AtPT2::LUC gene by Pi deficiency. hps2 is a new allele of the CTR1 gene (Figure 2B). Furthermore, under Pi deficiency, treatment of Arabidopsis plants with Ag+ suppressed the induction of AtPT2 whereas the addition of ACC dramatically enhanced its expression. Accordingly, the expression of AtPT2 was partially blocked in ein2 but was enhanced in eto1. A similar expression pattern was observed for several other PSI genes in the hps2 and ein2 mutants. These PSI genes included another high-affinity phosphate transporter, AtPT1 (Pht1; 1; Muchhal et al., 1996); a non-coding transcript, At4 (Burleigh and Harrison, 1999); an APase, ACP5 (del Pozo et al., 1999); a ribonuclease, RNS1 (Bariola et al., 1994; and miR399d, Fujii et al., 2005). The enhanced transcription of these PSI genes was also observed in the mutant hps3, which is another allele of ETO1 (Wang et al., 2012), and in the mutant hps4, which has enhanced ethylene signaling (Yu et al., 2012). ETO1 protein is a member of the broad complex/tramtrack/bric-a-brac (BTB) protein superfamily that participates in substrate recognition during ubiquitin-mediated protein degradation (Christians et al., 2009). It directly binds to the C-terminal of ACS5 and mediates its degradation. When ETO1 is mutated, it causes an overproduction of ethylene in young seedlings (Wang et al., 2004). These results provided the first genetic evidence that ethylene signaling is involved in the transcriptional responses of plants to Pi deficiency. In another study using M. falcata, ACC induced the expression of the Pi transporter genes MfPT1 and MfPT5 under Pi-sufficient conditions, whereas both AVG and Co2+ blocked the low Pi-induced expression of these genes (Li et al., 2011). Taken together, these results indicate that ethylene positively regulates transcription of a subset of PSI genes. The incomplete blockage of PSI gene expression in the ein2 mutant also indicates that ethylene is not the only mediator for the expression of these genes.
Interestingly, application of 25 μM ACC to young Arabidopsis seedlings under high Pi conditions barely induced the expression of AtPT2; under Pi deficiency, however, 0.5 μM ACC dramatically increased AtPT2 expression (Lei et al., 2011b). These results suggested a synergistic interaction between low Pi and ethylene in mediating transcriptional Pi responses. This also provided another example of plant cells being sensitized to ethylene by low Pi.
Induction and secretion of APases is a universal response of plants to Pi deficiency (Tran et al., 2010). The intracellular APases are believed to be involved in the remobilization of Pi from senescing tissues to young growing tissues whereas secreted APases are thought to be important for releasing Pi from organophosphates in the rhizosphere and thus increasing Pi availability for root uptake. The secreted APases are further classified into two groups: one group is released into the environment, and the other is tightly associated with the root surface after secretion. Members of the second group are called root-associated APases. Lei et al. (2011b) found that hps2 also had enhanced APase activity on the root surface. In contrast, ein2 exhibited reduced root-associated APase activity under Pi deficiency, indicating that ethylene is a positive regulator of PSI APase activity. The role of ethylene in regulating APase activity was further confirmed by the analyses of mutants hps3 and hps4 (Wang et al., 2012; Yu et al., 2012). Treatment with Ag+ suppressed the enhanced APase activity in hps3 and hps4. Similarly, Li et al. (2011) showed that the induction of root APase activity in M. falcata was stimulated by ACC under Pi sufficiency but was blocked by AVG under Pi deficiency.
In Arabidopsis, AtPAP10 is a major PSI APase that is predominantly associated with the root surface after secretion (Wang et al., 2011, 2014a). Zhang et al. (2014) investigated how ethylene affects root-associated AtPAP10 activity at different regulatory steps. The transcription of AtPAP10 was previously found to be increased by Pi starvation in the whole seedlings of hps3 and hps4 (Wang et al., 2012; Yu et al., 2012). The total root intracellular APase activity in hps3 and hps4, however, did not significantly differ from that of the WT. When the transcription of AtPAP10 was further analyzed using separated root and shoot tissues, Zhang et al. (2014) found that the transcription of AtPAP10 did not significantly increase in ACC-treated seedlings or the ctr1 mutant under Pi deficiency, nor did the accumulation of AtPAP10 proteins. Taken together, these results indicated that, in roots, ethylene mainly modulated the secretion of AtPAP10 protein or its enzymatic activity on the root surface. Some reports have shown that ethylene can increase H+-ATPase activity by up-regulating the expression of H+-ATPase genes (Waters et al., 2007; Wang et al., 2009; Staal et al., 2011), which may decrease the cytosolic pH or the pH on the root surface. In contrast, application of MCP blocked the change of cytosolic pH (Sundaresan et al., 2015). In humans and animals, cytosolic pH levels affect protein secretion (Paroutis et al., 2004). In addition, APase activity is sensitive to the change of pH. Thus, it is possible that ethylene may increase secretion of AtPAP10 proteins and stabilize AtPAP10 enzymatic activity on the root surface by modulating the cytosolic and root surface pH. More experimental evidence, however, is required to support this hypothesis.
Accumulation of anthocyanin is another hallmark response of plants to Pi starvation. The accumulation of anthocyanin is lower in hps2, hps3, and hps4 mutants than in the WT under low Pi conditions (Lei et al., 2011b; Wang et al., 2012; Yu et al., 2012). In contrast, the Pi-starved ein2 mutant shows increased anthocyanin content. Ag+-treated Arabidopsis seedlings also displayed increased accumulation of anthocyanin under Pi deficiency. Furthermore, Lei et al. (2011b) showed that the expression of four genes that encode three enzymes involved in anthocyanin biosynthetic pathway and one transcription factor regulating anthocyanin biosynthesis was increased in ein2 but reduced in ctr1. These results demonstrate that ethylene is a negative regulator of Pi starvation-induced anthocyanin accumulation and that this regulation is achieved at least partly through the regulation of the transcription of the genes involved in the biosynthesis of anthocyanin.
Plant responses to Pi deficiency are controlled by a complex regulatory network involving both local and systemic signaling. Whether a Pi response is controlled by local or systemic signaling can be determined by examining whether the degree of the response depends on the local Pi level or the Pi status of the whole plant. Using this approach, Ticconi et al. (2004) found that the remodeling of RSA triggered by Pi deficiency was regulated by the local, external Pi level. They first germinated Arabidopsis seeds on a Pi-sufficient medium for 5 days and then transferred the seedlings to an agar plate that contained a Pi-sufficient medium in the upper half and a Pi-deficient medium in the lower half, or vice versa. After the roots had grown for another 6 days, lateral root production had increased in the parts of the root that contacted the Pi-deficient medium but decreased in the parts that contacted the Pi-sufficient medium. The Arabidopsis phf1 mutant has defects in the ER exit of high-affinity Pi transporters, which greatly impairs Pi uptake by roots (González et al., 2005). When these plants were grown on a Pi-sufficient medium, their internal Pi level was only 20% of the WT, but no remodeling of RSA was observed (Thibaud et al., 2010). Furthermore, Thibaud et al. (2010) found that injection of a high concentration of Pi into the shoots of Arabidopsis plants grown on a Pi-deficient medium could not suppress the Pi deficiency-induced remodeling of RSA. Together, these results demonstrated that it is the local, external Pi level rather than the internal Pi status of the whole plant that regulates the remodeling of RSA. The requirement for ethylene in the Pi deficiency-induced remodeling of RSA indicates that ethylene is involved in local Pi signaling.
The Arabidopsis mutant defective in LPR1 and its close paralog LPR2 are insensitive to the Pi deficiency-induced inhibition of the primary root growth (Svistoonoff et al., 2007). In contrast, the Arabidopsis mutant with functional disruption of PDR2, which encodes a P5-type ATPase, exhibits an exaggerated short-root phenotype under Pi deficiency owing to meristem exhaustion (Ticconi et al., 2004, 2009). SCR and SHR are two key regulators of root patterning. PDR2 is required for maintaining the levels of SCR protein and SHR trafficking from stele into endodermis. Based on genetic analysis, PDR2 was proposed to act upstream of LPR1/LPR2 to adjust meristem activity in an ER-resident pathway. The roots of WT plants accumulated more Fe under Pi deficiency than under Pi sufficiency (Svistoonoff et al., 2007; Ward et al., 2008; Zheng et al., 2009; Lei et al., 2011a). On a Fe-free medium, the inhibition of primary root growth was abolished. Thus, Ward et al. (2008) hypothesized that Pi deficiency triggers the inhibition of primary root growth by enhancing the accumulation of Fe in the root meristem, which results in severe damage to root cells. In a recent study, Müller et al. (2015) demonstrated that LPR1 is a ferroxidase. The root meristem of lpr1 contains reduced levels of Fe3+ under Pi deficiency, which makes the root tip growth insensitive to inhibition caused by Pi deficiency. In contrast, pdr2 accumulates increased levels of Fe3+, which generates high levels of reactive oxygen species (ROS). The high level of ROS, in turn, causes the increased deposition of callose which impairs the trafficking of SHR, thus restricting root tip growth. Ethylene positively regulates Fe homeostasis in plants by up-regulating the expression of the genes involved in Fe acquisition (Waters et al., 2007; García et al., 2011). Thus, it is also possible that ethylene mediates Pi deficiency-induced inhibition of primary root growth by enhancing Fe accumulation in root tips.
As previously mentioned, AtPAP10 is a PSI APase that is predominantly associated with the root surface after secretion. Using split-root experiments, Zhang et al. (2014) demonstrated that although the transcription of AtPAP10 is systemically controlled (i.e., affected by the Pi status of whole plant), AtPAP10 protein accumulation and enzymatic activity on the root surface are regulated by local Pi levels. Once the mRNA of AtPAP10 is produced, the subsequent accumulation and secretion of AtPAP10 protein and perhaps also the protein’s enzymatic activity on the root surface are controlled only by local signaling. Because ethylene mainly participates in the secretion of AtPAP10 proteins or stabilization of AtPAP10 enzymatic activity on the root surface during the induction of root-associated AtPAP10 activity, ethylene could be regarded as a local signal in regulating the induction of AtPAP10 activity. This represents another example that ethylene functions in local signaling besides being involved in the control of remodeling of RSA.
Although it is now evident that ethylene is involved in local Pi sensing and signaling, the mechanism by which ethylene regulates local signaling is largely unknown. Thibaud et al. (2010) dissected the transcriptional responses controlled by local and systemic signaling. They found that the transcription of the genes involved in development, stress responses, and hormonal signaling is controlled by local signaling. The locally regulated genes include ERF1 and ERF2. These results provide further support for the role of ethylene in local Pi signaling. The next important task will be to identify the direct downstream genes that are targets of ethylene and that participate in local Pi signaling.
Researchers have hypothesized that when the external Pi level drops, the root tissues sense the change in Pi availability and send warning signals through the xylem to the shoots (Chiou and Lin, 2011; Figure 3). Once these signals arrive in shoots, they trigger Pi-deficiency responses in the shoots including enhanced PSI gene expression, reduced photosynthetic activity, inhibition of shoot growth, increased anthocyanin accumulation, and induction of intracellular APase activity. At the same time, the shoots send signals to the roots through the phloem to regulate Pi responses in roots including enhanced high-affinity Pi transporter activity and induction of APase activity. Cytokinins, strigolactones, and Pi itself have been proposed to be root-to-shoot signals, whereas sucrose and miRNA399 are believed to be the shoot-to-root signals (Lin et al., 2014). Enhanced ethylene biosynthesis and expression of ethylene biosynthetic genes in shoots have been observed under Pi deficiency (Misson et al., 2005; Kim et al., 2008), suggesting that ethylene is involved in the Pi responses in shoots. Whether the increase of ethylene biosynthesis in shoots is triggered by local sensing due to the drop of Pi levels in the shoots or by the signals from the roots, however, is not known.
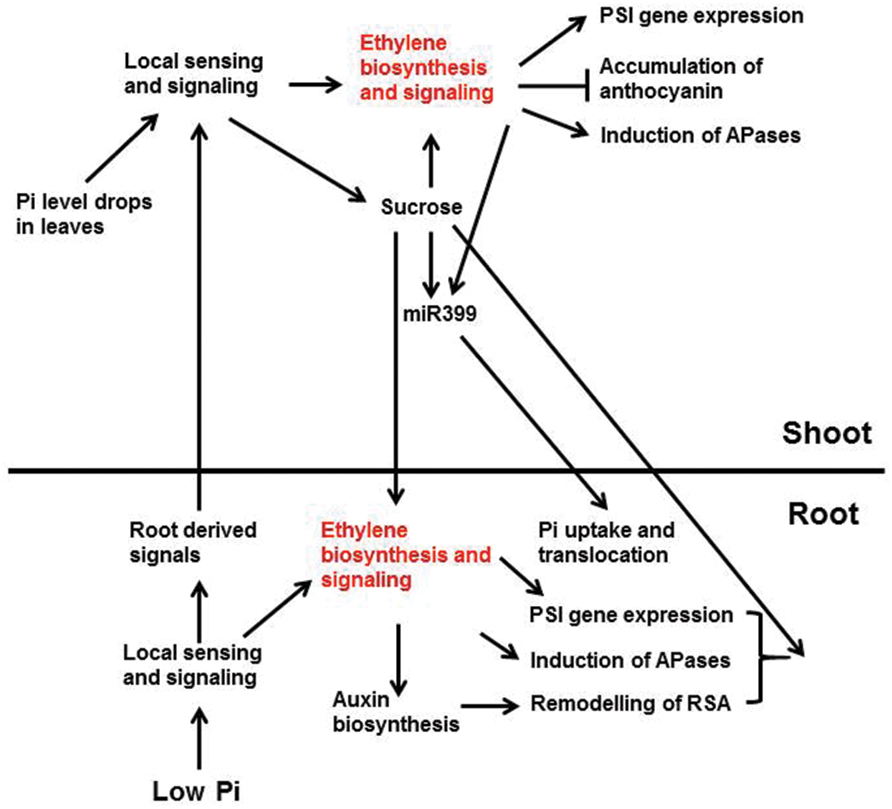
FIGURE 3. A schematic model showing the interactions between ethylene and other Pi signaling components and Pi deficiency responses that these interactions are involved. Low Pi is perceived by unidentified intracellular and/or extracellular sensors in root, which activates local signaling pathways and the synthesis of systemic signals. The activated local signaling pathways then trigger the expression of PSI genes, the induction of APases, and the remodeling of RSA. These response are egulated partly through ethylene biosynthesis and signaling. The ethylene-mediated remodeling of RSA may also be achieved by enhancing auxin biosynthesis and signaling. The root-derived systemic signals are translocated to shoot via xylem. These signals, together with the drop of Pi level in shoots, elicit the local sensing and signaling there. The local signaling in shoots increases ethylene biosynthesis and signaling, as well as the synthesis of systemic signals, such as sucrose. The enhanced ethylene biosynthesis and signaling induce the expression of PSI genes and APase activity, but suppress the accumulation of anthocyanin. The increased sucrose level, in turn, can reinforce the ethylene biosynthesis and signaling and induces the production of miR399. The induction of miR399 is also modulated by ethylene and transcription factor PHR1 (not show in the figure). The shoot-derived sucrose and miR399 then move down to roots through phloem transport. In roots, sucrose is critical for the expression of some PSI genes and the induction of APase activity, perhaps also the remodeling of RSA (not discussed in this article), and the accumulation of miR399 enhances Pi uptake and translocation to shoots. Arrows indicate promotion, and the perpendicular line indicate inhibition (Lei et al., 2011b).
Thibaud et al. (2010) showed that the transcription of PSI genes that participate in Pi transport, signaling, and recycling is regulated by systemic signaling. The PSI genes whose expression is regulated by ethylene also belong to these functional categories (Lei et al., 2011b; Wang et al., 2012; Yu et al., 2012). The transcription of the Pi transporters Pht1;1 and Pht1;4, the APase ACP5, the RNase RNS1, and the Pi signaling molecules miRNA399, At4, and IPS1 is enhanced in hps2, hps3, and hps4 but reduced in ein2 or in the plants treated with Ag+. Thibaud et al. (2010) further demonstrated that the promoters of most systemically regulated PSI genes contain the P1BS element, which is the binding site of the PHR1 transcription factor, indicating that PHR1 is an important component of the systemic signaling pathway. The reduced expression of one ACS gene and at least eight AP2/ERF genes in the phr1phl1 mutant suggests that ethylene biosynthesis and signaling themselves are also controlled by systemic signaling. Thus, ethylene might be part of a regulatory loop involved in the systemic control of PSI gene expression. Once activated, however, how ethylene signaling functions in systemic control of PSI gene expression is unknown.
In the phr1 mutant, anthocyanin accumulation is greatly attenuated, indicating that this response to low Pi is systemically controlled (Rubio et al., 2001). As discussed before, the accumulation of anthocyanin is enhanced in ctr1 and eto1 but reduced in ein2, indicating that ethylene is a negative regulator of this systemic response (Lei et al., 2011b; Wang et al., 2012). These results reveal another function of ethylene in the systemic control of plant responses to Pi deprivation.
The past studies have indicated that the regulation of plant responses to Pi deficiency is complex and involves crosstalk among different signaling pathways. The precise control and coordination of these multi-faceted responses undoubtedly depends on an efficient interaction among the different signals. Here, we discuss how ethylene interacts with sucrose, auxin, and miRNA399 to regulate Pi-deficiency responses.
Growing evidence indicates that sucrose is a key systemic signal that globally regulates Pi-starvation responses. In several plant species, sucrose biosynthesis increases under low Pi availability (Hammond and White, 2008). Also, the expression of the genes involved in the synthesis, translocation, and degradation of sucrose is altered when plants are grown under Pi deficiency (Hammond et al., 2003; Wu et al., 2003; Misson et al., 2005; Muller et al., 2007). In addition to functioning as a carbon source, the sucrose delivered to the roots from leaves acts as a signal to initiate changes in gene expression, metabolism, and development in roots (Hammond and White, 2011). Karthikeyan et al. (2007) found that the level of PSI gene expression was positively correlated with the concentrations of sucrose in the growth medium under Pi deficiency. When plants were grown in the dark, the expression of PSI genes was greatly reduced; this reduction in PSI gene expression, however, was prevented by adding sucrose to the growth medium. Similarly, Liu et al. (2005, 2010) observed that application of sucrose stimulated accumulation of LaPT1 (a Pi transporter), LaSAP1 (an APase) and miR399 transcripts in dark-grown white lupin and common bean under Pi sufficiency. Furthermore, the use of stem-girdling to disrupt the phloem transport of photosynthates to P-deficient roots resulted in the suppression of the expression of these genes. Jain et al. (2007) reported that sucrose is required for Pi deficiency-induced lateral root proliferation and root hair formation. In addition, exogenous application of sucrose, like Pi deficiency, induced CR formation and PSI gene expression in Pi-sufficient white lupin (Zhou et al., 2008). Definitive evidence for the role of sucrose in plant responses to Pi starvation came from the study of the Arabidopsis mutant hps1 (Lei et al., 2011a). hps1 overexpresses the SUC2 gene due to a T-DNA insertion in the promoter of the SUC2 gene. SUC2 is the only transporter involved in the phloem loading of sucrose in mesophyll cells. hps1 accumulates a high level of sucrose in both roots and shoots and is hypersensitive in almost all aspects of plant response to Pi starvation. Lei et al. (2011a) further showed that the suc2-5 mutant had enhanced expression of the Pht1:4 Pi transporter in shoots but reduced expression in roots, as well as reduced root-associated APase activity. These results were consistent with the high accumulation of sucrose in shoots and low level of sucrose in roots in this mutant. The reduced root-associated APase activity was also observed in another mutant allele of SUC2, pho3 (Zakhleniuk et al., 2001; Lloyd and Zakhleniuk, 2004).
In several plant species, sucrose increases ethylene production in a sucrose concentration-dependent manner (Philosoph-Hadas et al., 1985; Kobayashi and Saka, 2000; Jeong et al., 2010; Figure 3). The sucrose-induced formation of CR in white lupin was completely suppressed by application of Co2+ (Wang et al., 2015). In addition, Co2+ also suppressed Pi deficient-induced CR formation. Together, these results suggest that sucrose induces CR formation via the induction of ethylene biosynthesis (Figure 3).
Both sucrose and ethylene are positive regulators for the induction of AtPAP10 APase activity on the root surface (Lei et al., 2011a,b; Wang et al., 2012; Yu et al., 2012). Zhang et al. (2014) further investigated the relationship between sucrose and ethylene in the regulation of AtPAP10 activity. Under Pi deficiency, hps1 had enhanced APase activity while ein2 had reduced APase activity on the root surface. The double mutant hps1ein2 displayed root-associated AtPAP10 activity that was intermediate between that of hps1 and ein2. Also, when treated with Ag+, the AtPAP10 activity in WT plants was partially reduced. When ctr1 was grown in the dark on a sucrose-free Pi-deficient medium, however, the induction of the AtPAP10 activity on the root surface was completely blocked, although ethylene signaling was constitutively activated in the mutant. Thus, ethylene’s induction of AtPAP10 activity depends on sucrose, but sucrose’s function does not depend on ethylene. Further study indicated that sucrose was largely required for the induction of AtPAP10 transcription while ethylene only modulated the secretion of AtPAP10 protein or AtPAP10 enzymatic activity on the root surface (Zhang et al., 2014). This is understandable because if AtPAP10 mRNA is not transcribed due to the absence of sucrose, ethylene signaling, even if is constitutively activated, will not increase the AtPAP10 activity on the root surface. In addition, the above results also indicate that ethylene is not the only component that controls the induction of APase activity; even if the ethylene pathway is completely blocked by the ein2 mutation or by treatment with Ag+, the induction of APase is only partially abolished.
Auxin also plays an important role in controlling Pi deficiency-induced remodeling of RSA (López-Bucio et al., 2002; Nacry et al., 2005; Jain et al., 2007; Pérez-Torres et al., 2008). López-Bucio et al. (2002) showed that Pi-deprived plants were more sensitive to exogenously applied auxin than Pi-replete plants with respect to the arrest of primary root growth and enhanced formation of lateral roots. This enhanced auxin sensitivity was found to result from the enhanced expression of the auxin receptor TIR1 (Pérez-Torres et al., 2008). The enhanced TIR expression accelerates the degradation of AUX/IAA (AUXIN/INDOLE-3-ACETIC ACID) auxin response repressors, thus releasing repression of transcription factor ARF19 that is involved in the formation of lateral roots. Nacry et al. (2005) proposed that Pi deficiency causes: (1) an over-accumulation of auxin in the root apex of primary root and young lateral roots; (2) an over-accumulation of auxin or an increased auxin sensitivity in the lateral primordia; (3) a decrease in auxin concentration in the lateral primordia initiation zone of the primary roots and in old laterals. Using auxin transport inhibitor or the mutants with defects in auxin transport, the authors also showed that the changes in local auxin concentrations was achieved through the changes in auxin transport rather than auxin biosynthesis. Given that both ethylene and auxin are involved in the Pi deficiency-induced remodeling of RSA, ethylene may cooperate with auxin to regulate root growth. In fact, several lines of evidence have indicated that ethylene promotes auxin biosynthesis and transport to modulate root development (Osmont et al., 2007; Růzicka et al., 2007; Swarup et al., 2007; Stepanova and Alonso, 2009).
The Arabidopsis mutant hps4 is hypersensitive to the Pi deficiency-induced inhibition of primary root growth (Yu et al., 2012). In this mutant, the expression of several genes related to auxin biosynthesis is increased. In addition, the hps4 root tip produced twice as much auxin as that of the WT under Pi deficiency. The hypersensitivity of hps4 to Pi deficiency was suppressed when the plants were treated with Ag+ but not with AVG. These results suggested that the enhanced ethylene signaling in hps4 might increase auxin biosynthesis in the root tip, thus enhancing the inhibition of the primary root growth (Figure 3). In white clover, Pi deficiency increases the ethylene sensitivity in roots (Dinh et al., 2012). ACC treatment induced a high expression of the auxin-responsive DR5::GUS marker gene in the root apex. To separate the effect of ACC on auxin biosynthesis from auxin transport, the authors applied auxin transport inhibitor to Pi deficient-roots. The results showed that the ACC-enhanced DR5::GUS expression could not be suppressed by the auxin transport inhibitor, suggesting that the effect of ACC is on auxin biosynthesis, although the role of ACC on auxin transport cannot be strictly excluded. This result is also consistent with what observed by Yu et al. (2012).
The first and also the best characterized miRNA involved in Pi-deficiency response is miR399. The expression of miR399 is highly induced in both shoots and roots by a decrease in external Pi or copper levels, but is reduced by iron deficiency (Fujii et al., 2005; Chiou et al., 2006; Buhtz et al., 2010). A detailed time course study indicated that the increase of miR399 in the shoot occurs prior to that in the root (Lin et al., 2008). Reciprocal grafting experiments further showed that miR399 could move from shoot to root (Lin et al., 2008; Pant et al., 2008). Observation of the increased accumulation of miR399 in the phloem sap in Pi starved-Arabidopsis and Brassica plants (Buhtz et al., 2008; Pant et al., 2008) also supported that miR399 is synthesized in shoots and is translocated to roots in order to act as a systemic signal that regulates Pi uptake. miR399 enhances Pi acquisition by direct cleavage of the mRNA that encodes the ubiquitin E2 conjugase PHO2, which is involved in the ubiquitin-mediated protein degradation pathway (Aung et al., 2006; Bari et al., 2006; Chiou et al., 2006). Overexpression of miRNA399 or functional disruption of PHO2 leads to the over-accumulation of Pi in shoots. By screening for pho2 suppressors and using quantitative membrane proteomics, Liu et al. (2012) and Huang et al. (2013) identified PHO1 and a group of high-affinity Pi transporters (PHT1) as the substrates of PHO2. PHO1 and PHT1 transporters are responsible for the translocation of Pi from roots to shoots and for the uptake of Pi from the external environment. When plants are exposed to Pi deficiency, miR399 is rapidly induced and degrades the mRNA of PHO2. The down-regulation of PHO2 expression increases the stability of PHO1 and PHT1 proteins, thus enhancing Pi uptake in roots and Pi translocation from roots to shoots.
Transcription of the precursor of miR399d is enhanced in the Arabidopsis mutants hps2, hps3, and hps4, but the expression of miR399d is reduced in ein2 (Lei et al., 2011b; Wang et al., 2012; Yu et al., 2012). These results demonstrated that ethylene positively regulates the expression of miR399d. Interestingly, Liu et al. (2010) found that the induction of miR399 was completely blocked in dark-grown or stem-girdled white lupin, suggesting that sucrose is required for miR399 expression (Figure 3). Given that an increase in sucrose level can induce ethylene biosynthesis, ethylene might act downstream of sucrose to affect the expression of miR399. Also, because the expression of miR399 is completely abolished in the absence of sucrose but is only partially blocked in the ein2 mutant, it seems that ethylene functions in a branch of the pathway downstream of sucrose to modulate the expression of miR399 and thus to fine-tune Pi uptake by roots.
Based on genetic, pharmacological, biochemical, and physiological data, it is now evident that ethylene plays an important role in mediating plant responses to Pi deficiency. Pi deficiency increases ethylene biosynthesis and signaling in both roots and leaves. Early studies showed that ethylene is involved in Pi deficiency-induced inhibition of primary root growth and enhanced production of root hairs. Recent research indicates that ethylene also regulates expression of PSI genes, induction of APases, and accumulation of anthocyanin under Pi deficiency. Ethylene participates in both transcriptional and post-transcriptional regulation of plant Pi responses. Moreover, ethylene interacts with other signals, such as sucrose, auxin, and miRNA399, to regulate both local and systemic signaling. Also, it should be noted that although ethylene clearly plays an important role in mediating multiple plant responses to Pi deficiency, there is no single Pi response that is completely under the control of ethylene.
Although the role of ethylene in mediating plant responses to Pi deficiency is well established, the following important questions remain regarding the underlying mechanisms: (1) How do plants sense the change in Pi availability to elicit the production of ethylene or to enhance ethylene sensitivity? (2) Which specific cells are the targets for ethylene action? (3) Through which downstream components does ethylene mediate plant responses to Pi starvation? To answer these questions, we need to identify the transcription factors that bind to the promoters of ethylene biosynthetic genes, such as ACS and ACO, and to understand how the expression of these transcription factors is regulated by Pi deficiency. The expression of ACS and ACO genes and the stability of these proteins are also regulated by CDPK and MAPK signaling pathways (Kim et al., 2003; Hernández Sebastià et al., 2004; Liu and Zhang, 2004). A recent report indicated that Arabidopsis MKK9-MPK3/MPK6 pathway is involved in the maintenance of Pi homeostasis by regulating transcription of Pi acquisition-related genes (Lei et al., 2014). It is not known, however, whether this pathway regulates Pi homeostasis by affecting the expression or activity of ACS and ACO. Thus, it may also worth to investigate the molecular link between this MAPK pathway and ethylene biosynthesis/signaling under Pi deficiency.
To identify the cells that are the targets for ethylene effects, researchers might use a cell type-specific promoter to drive a mutated ethylene receptor gene etr1 to specifically block the action of ethylene in those cells; the change in Pi response would then indicate whether those cells are involved. This approach has been successfully used to dissect the role of ABA signaling in mediating the response of root growth to drought stress (Duan et al., 2013). To identify the downstream targets of ethylene signaling that are directly involved in plant Pi responses, researchers can combine genomic and genetic approaches. For example, EIN3 and EIL1 are two key transcription factors that regulate a suite of downstream ethylene-responsive genes. A transcriptomic analysis would indicate those genes whose expression is blocked in the Pi deficient-ein3eil1 mutant. Using such an approach, researchers could identify a battery of genes that directly participate in plant responses to Pi deficiency, and these might include, for example, the genes of cell wall proteins that are directly involved in the elongation of root hairs. This approach could also reveal the gene regulatory network that ethylene uses to control specific Pi responses. In addition, by using a Pi starvation-responsive marker line (such as the plant carrying the AtPT2::LUC marker gene), researchers could screen for a mutant that uncouples the interaction between ethylene and Pi; thus identifying the molecular components involved in the regulation of ethylene sensitivity under Pi deficiency. With increased knowledge in these areas, we will better understand how ethylene functions in plant responses to Pi deficiency and how overall plant responses to Pi deficiency are regulated at the molecular level.
LS drafted the manuscript. LS and DL revised the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Research in this laboratory was supported by funds from Natural Science Foundation of China (grant no. 31370290) and the Ministry of Agriculture of China (grant no. 2014ZX0800932B).
ACC, 1-aminocyclopropane-1-carboxylate; ACO, ACC OXIDASE; ACP5, ACID PHOSPHATASE TYPE 5; ACS, ACC SYNTHASE; AdoMet, S-adenosyl methionine; APase, acid phosphatase; AP2/ERF, APETALA2/ETHYLENE RESPONSE FACTOR; At4, Arabidopsis thaliana cDNA 4; ARF19, AUXIN RESPONSE FACTOR 19; AVG, aminoethoxyvinyl glycine; CDPK, CALCIUM-DEPENDENT PROTEIN KINASE; CHIB, CHITINASE B; CR, cluster root; CTR1, CONSTITUTIVE TRIPLE RESPONSE 1; EBF, EIN3-BINDING F-BOX; EIL1, EIN3-LIKE 1; EIN2, ETHYLENE INSENSITIVE 2; EIN3, ETHYLENE INSENSITIVE 3; EIN4, ETHYLENE INSENSITIVE 4; FLS2, FLAGELLIN SENSITIVE 2; ER, endoplasmic reticulum; ERS1, ETHYLENE RESPONSE SENSOR 1; ETR1, ETHYLENE RESPONSE 1; ETR1, ETHYLENE RESPONSE 1; ETO1, ETHYLENE OVERPRODUCTION 1; hps, hypersensitive to phosphate starvation; IPS1, INDUCED BY PHOSPHATE STARVATION 1; LPR1, LOW PHOSPHATE ROOT 1; LUC, luciferase; MAPK, MITOGEN-ACTIVATED PROTEIN KINASE; MCP, 1-methylcyclopropene; PAP10, PURPLE ACID PHOSPHATASE 10; PDR2, PHOSPHATE DEFICIENCY RESPONSE 2; PHL1, PHR1-LIKE 1; PHO2, PHOSPHATE 2; PHR1, PHOSPHATE STARVATION RESPONSE 1; Pht1(AtPT1), PHOSPHATE TRANSPORTER 1; Pi, phosphate; PORA, PROTOCHLOROPHYLLIDE OXIDOREDUCTASE A; PSI, phosphate starvation induced; RNS1, RIBONUCLEASE 1; RSA, root system architecture; SCR, SCARECROW; SHR, SHORT-ROOT; SUC2, SUCROSE TRANSPORTER 2; TIR1, TRANSPORT INHIBITOR RESPONSE 1.
Abeles, F. B., Morgan, P. W., and Saltveit, J. M. E. (1992). Ethylene in Plant Biology. San Diego: Academic Press.
Aeschbacher, R. A., Hauser, M. T., Feldmann, K. A., and Benfey, P. N. (1995). The SABRE gene is required for normal cell expansion in Arabidopsis. Genes Dev. 9, 330–340. doi: 10.1101/gad.9.3.330
Aung, K., Lin, S. I., Wu, C. C., Huang, Y. T., Su, C. L., and Chiou, T. J. (2006). pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141, 1000–1011. doi: 10.1104/pp.106.078063
Bari, R., Pant, B. D., Stitt, M., and Scheible, W. R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141, 988–999. doi: 10.1104/pp.106.079707
Bariola, P. A., Howard, C. J., Taylor, C. B., Verburg, M. T., Jaglan, V. D., and Green, P. J. (1994). The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 6, 673–685. doi: 10.1046/j.1365-313X.1994.6050673.x
Basu, P., Zhang, Y. J., Lynch, J. P., and Brown, K. M. (2007). Ethylene modulates genetic, positional, and nutritional regulation of root plagiogravitropism. Funct. Plant Biol. 34, 41–51. doi: 10.1071/FP06209
Bates, T., and Lynch, J. (1996). Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 19, 529–538. doi: 10.1111/j.1365-3040.1996.tb00386.x
Bieleski, R. L. (1973). Phosphate pools, phosphate transport, and phosphate availability. Annu. Rev. Plant Physiol. 24, 225–252. doi: 10.1146/annurev.pp.24.060173.001301
Bleecker, A. B., and Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell. Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Borch, K., Bouma, T., Lynch, J., and Brown, K. (1999). Ethylene: a regulator of root architectural responses to soil phosphorus availability. Plant Cell Environ. 22, 425–431. doi: 10.1046/j.1365-3040.1999.00405.x
Buhtz, A., Pieritz, J., Springer, F., and Kehr, J. (2010). Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 10:64. doi: 10.1186/1471-2229-10-64
Buhtz, A., Springer, F., Chappell, L., Baulcombe, D. C., and Kehr, J. (2008). Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 53, 739–749. doi: 10.1111/j.1365-313X.2007.03368.x
Burleigh, S. H., and Harrison, M. J. (1999). The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol. 119, 241–248. doi: 10.1104/pp.119.1.241
Bustos, R., Castrillo, G., Linhares, F., Puga, M. I., Rubio, V., Perez-Perez, J., et al. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 6:e1001102. doi: 10.1371/journal.pgen.1001102
Calderon-Vazquez, C., Ibarra-Laclette, E., Caballero-Perez, J., and Herrera-Estrella, L. (2008). Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant- and species-specific levels. J. Exp. Bot. 59, 2479–2497. doi: 10.1093/jxb/ern115
Chacón-López, A., Ibarra-Laclette, E., Sánchez-Calderón, L., Gutiérrez-Alanís, D., and Herrera-Estrella, L. (2011). Global expression pattern comparison between low phosphorus insensitive 4 and WT Arabidopsis reveals an important role of reactive oxygen species and jasmonic acid in the root tip response to phosphate starvation. Plant Signal. Behav. 6, 382–392. doi: 10.4161/psb.6.3.14160
Chiou, T. J., Aung, K., Lin, S. I., Wu, C. C., Chiang, S. F., and Su, C. L. (2006). Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 18, 412–421. doi: 10.1105/tpc.105.038943
Chiou, T. J., and Lin, S. I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206. doi: 10.1146/annurev-arplant-042110-103849
Cho, H. T., and Cosgrove, D. J. (2002). Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14, 3237–3253. doi: 10.1105/tpc.006437
Christians, M. J., Gingerich, D. J., Hansen, M., Binder, B. M., Kieber, J. J., and Vierstra, R. D. (2009). The BTB ubiquitin ligases ETO1. EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J. 57, 332–345. doi: 10.1111/j.1365-313X.2008.03693.x
del Pozo, J. C., Allona, I., Rubio, V., Leyva, A., de la Pena, A., Aragoncillo, C., et al. (1999). A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J. 19, 579–589. doi: 10.1046/j.1365-313X.1999.00562.x
Dinh, P. T., Roldan, M., Leung, S., and McManus, M. T. (2012). Regulation of root growth by auxin and ethylene is influenced by phosphate supply in white clover (Trifolium repens L.). Plant Growth Regul. 66, 179–190. doi: 10.1007/s10725-011-9642-6
Drew, M. C., He, C. J., and Morgan, P. W. (1989). Decreased ethylene biosynthesis, and induction of aerenchyma, by nitrogen- or phosphate-starvation in adventitious roots of Zea mays L. Plant Physiol. 91, 266–271. doi: 10.1104/pp.91.1.266
Duan, L., Dietrich, D., Ng, C. H., Chan, P. M., Bhalerao, R., Bennett, M. J., et al. (2013). Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25, 324–341. doi: 10.1105/tpc.112.107227
Fujii, H., Chiou, T. J., Lin, S. I., Aung, K., and Zhu, J. K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15, 2038–2043. doi: 10.1016/j.cub.2005.10.016
García, M. J., Suárez, V., Romera, F. J., Alcántara, E., and Pérez-Vicente, R. (2011). A new model involving ethylene, nitric oxide and Fe to explain the regulation of Fe-acquisition genes in Strategy I plants. Plant Physiol. Biochem. 49, 537–544. doi: 10.1016/j.plaphy.2011.01.019
Gilbert, G. A., Knight, J. D., Vance, C. P., and Allan, D. L. (2000). Proteoid root development of phosphorus deficient Lupin is mimicked by auxin and phosphonate. Ann. Bot. 85, 921–928. doi: 10.1006/anbo.2000.1133
González, E., Solano, R., Rubio, V., Leyva, A., and Paz-Ares, J. (2005). PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17, 3500–3512. doi: 10.1105/tpc.105.036640
Graham, M. A., Ramirez, M., Valdes-Lopez, O., Lara, M., Tesfaye, M., Vance, C., et al. (2006). Identification of candidate phosphorus stress induced genes in Phaseolus vulgaris through clustering analysis across several plant species. Funct. Plant Biol. 33, 789–797. doi: 10.1071/FP06101
Hammond, J. P., Bennett, M. J., Bowen, H. C., Broadley, M. R., Eastwood, D. C., May, S. T., et al. (2003). Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 132, 578–596. doi: 10.1104/pp.103.020941
Hammond, J. P., and White, P. J. (2008). Sucrose transport in the phloem: integrating root responses to phosphorus starvation. J. Exp. Bot. 59, 93–109. doi: 10.1093/jxb/erm221
Hammond, J. P., and White, P. J. (2011). Sugar signaling in root responses to low phosphorus availability. Plant Physiol. 156, 1033–1040. doi: 10.1104/pp.111.175380
He, C. J., Morgan, P. W., and Drew, M. C. (1992). Enhanced sensitivity to ethylene in nitrogen-or phosphate-starved roots of Zea mays L. during aerenchyma formation. Plant Physiol. 98, 137–142. doi: 10.1104/pp.98.1.137
Hernández, G., Ramirez, M., Valdes-Lopez, O., Tesfaye, M., Graham, M. A., Czechowski, T., et al. (2007). Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol. 144, 752–767. doi: 10.1104/pp.107.096958
Hernández Sebastià, C., Hardin, S. C., Clouse, S. D., Kieber, J. J., and Huber, S. C. (2004). Identification of a new motif for CDPK phosphorylation in vitro that suggests ACC synthase may be a CDPK substrate. Arch. Biochem. Biophys. 428, 81–91. doi: 10.1016/j.abb.2004.04.025
Huang, T. K., Han, C. L., Lin, S. I., Chen, Y. J., Tsai, Y. C., Chen, Y. R., et al. (2013). Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25, 4044–4060. doi: 10.1105/tpc.113.115998
Jain, A., Poling, M. D., Karthikeyan, A. S., Blakeslee, J. J., Peer, W. A., Titapiwatanakun, B., et al. (2007). Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol. 144, 232–247. doi: 10.1104/pp.106.092130
Jeong, S. W., Das, P. K., Jeoung, S. C., Song, J. Y., Lee, H. K., Kim, Y. K., et al. (2010). Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis. Plant Physiol. 154, 1514–1531. doi: 10.1104/pp.110.161869
Kang, J., Yu, H., Tian, C., Zhou, W., Li, C., Jiao, Y., et al. (2014). Suppression of photosynthetic gene expression in roots is required for sustained root growth under phosphate deficiency. Plant Physiol. 165, 1156–1170. doi: 10.1104/pp.114.238725
Karthikeyan, A. S., Varadarajan, D. K., Jain, A., Held, M. A., Carpita, N. C., and Raghothama, K. G. (2007). Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta 225, 907–918. doi: 10.1007/s00425-006-0408-8
Kende, H. (1993). Ethylene biosynthesis. Annu. Rev. Plant Biol. 44, 283–307. doi: 10.1146/annurev.arplant.44.1.283
Kieber, J. J., Rothenberg, M., Roman, G., Feldmann, K. A., and Ecker, J. R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72, 427–441. doi: 10.1016/0092-8674(93)90119-B
Kim, C. Y., Liu, Y., Thorne, E. T., Yang, H., Fukushige, H., Gassmann, W., et al. (2003). Activation of a stress-responsive mitogen-activated protein kinase cascade induces the biosynthesis of ethylene in plants. Plant Cell 15, 2707–2718. doi: 10.1105/tpc.011411
Kim, H. J., Lynch, J. P., and Brown, K. M. (2008). Ethylene insensitivity impedes a subset of responses to phosphorus deficiency in tomato and petunia. Plant Cell Environ. 31, 1744–1755. doi: 10.1111/j.1365-3040.2008.01886.x
Kobayashi, H., and Saka, H. (2000). Relationship between ethylene evolution and sucrose content in excised leaf blades of rice. Plant Prod. Sci. 3, 398–403. doi: 10.1626/pps.3.398
Lei, L., Li, Y., Wang, Q., Xu, J., Chen, Y., Yang, H., et al. (2014). Activation of MKK9-MPK3/MPK6 enhances phosphate acquisition in Arabidopsis thaliana. New. Phytol. 203, 1146–1160. doi: 10.1111/nph.12872
Lei, M., Liu, Y., Zhang, B., Zhao, Y., Wang, X., Zhou, Y., et al. (2011a). Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol. 156, 1116–1130. doi: 10.1104/pp.110.171736
Lei, M., Zhu, C., Liu, Y., Karthikeyan, A. S., Bressan, R. A., Raghothama, K. G., et al. (2011b). Ethylene signaling is involved in regulation of phosphate starvation-induced gene expression and production of acid phosphatases and anthocyanin in Arabidopsis. New Phytol. 189, 1084–1095. doi: 10.1111/j.1469-8137.2010.03555.x
Li, Y. S., Gao, Y., Tian, Q. Y., Shi, F. L., Li, L. H., and Zhang, W. H. (2011). Stimulation of root acid phosphatase by phosphorus deficiency is regulated by ethylene in Medicago falcata. Environ. Exp. Bot. 71, 114–120. doi: 10.1016/j.envexpbot.2010.11.007
Li, Y. S., Mao, X. T., Tian, Q. Y., Li, L. H., and Zhang, W. H. (2009). Phosphorus deficiency-induced reduction in root hydraulic conductivity in Medicago falcata is associated with ethylene production. Environ. Exp. Bot. 67, 172–177. doi: 10.1016/j.envexpbot.2009.05.013
Lin, S. I., Chiang, S. F., Lin, W. Y., Chen, J. W., Tseng, C. Y., Wu, P. C., et al. (2008). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 147, 732–746. doi: 10.1104/pp.108.116269
Lin, W. Y., Huang, T. K., Leong, S. J., and Chiou, T. J. (2014). Long-distance call from phosphate: systemic regulation of phosphate starvation responses. J. Exp. Bot. 65, 1817–1827. doi: 10.1093/jxb/ert431
Liu, J. Q., Allan, D. L., and Vance, C. P. (2010). Systemic signaling and local sensing of phosphate in common bean: cross-talk between photosynthate and microRNA399. Mol. Plant 3, 428–437. doi: 10.1093/mp/ssq008
Liu, J., Samac, D. A., Bucciarelli, B., Allan, D. L., and Vance, C. P. (2005). Signaling of phosphorus deficiency-induced gene expression in white lupin requires sugar and phloem transport. Plant J. 41, 257–268. doi: 10.1111/j.1365-313X.2004.02289.x
Liu, T. Y., Huang, T. K., Tseng, C. Y., Lai, Y. S., Lin, S. I., Lin, W. Y., et al. (2012). PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24, 2168–2183. doi: 10.1105/tpc.112.096636
Liu, Y., and Zhang, S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16, 3386–3399. doi: 10.1105/tpc.104.026609
Lloyd, J. C., and Zakhleniuk, O. V. (2004). Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J. Exp. Bot. 55, 1221–1230. doi: 10.1093/jxb/erh143
López-Bucio, J., Cruz-Ramírez, A., and Herrera-Estrella, L. (2003). The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6, 280–287. doi: 10.1016/S1369-5266(03)00035-9
López-Bucio, J., Hernandez-Abreu, E., Sanchez-Calderon, L., Nieto-Jacobo, M. F., Simpson, J., and Herrera-Estrella, L. (2002). Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 129, 244–256. doi: 10.1104/pp.010934
Ma, Z., Baskin, T. I., Brown, K. M., and Lynch, J. P. (2003). Regulation of root elongation under phosphorus stress involves changes in ethylene responsiveness. Plant Physiol. 131, 1381–1390. doi: 10.1104/pp.012161
McKeon, T. A., Fernández-Maculet, J. C., and Yang, S. F. (1995). “Biosynthesis and metabolism of ethylene,” in Plant Hormones, ed. P. J. Davies (Amsterdam: Springer), 118–139.
Misson, J., Raghothama, K. G., Jain, A., Jouhet, J., Block, M. A., Bligny, R., et al. (2005). A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. U.S.A. 102, 11934–11939. doi: 10.1073/pnas.0505266102
Morcuende, R., Bari, R., Gibon, Y., Zheng, W., Pant, B. D., Blasing, O., et al. (2007). Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 30, 85–112. doi: 10.1111/j.1365-3040.2006.01608.x
Muchhal, U. S., Pardo, J. M., and Raghothama, K. G. (1996). Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 93, 10519–10523. doi: 10.1073/pnas.93.19.10519
Müller, J., Toev, T., Heisters, M., Teller, J., Moore, K. L., Hause, G., et al. (2015). Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev. Cell 33, 216–230. doi: 10.1016/j.devcel.2015.02.007
Muller, R., Morant, M., Jarmer, H., Nilsson, L., and Nielsen, T. H. (2007). Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 143, 156–171. doi: 10.1104/pp.106.090167
Nacry, P., Canivenc, G., Muller, B., Azmi, A., Van Onckelen, H., Rossignol, M., et al. (2005). A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol. 138, 2061–2074. doi: 10.1104/pp.105.060061
Nagarajan, V. K., Jain, A., Poling, M. D., Lewis, A. J., Raghothama, K. G., and Smith, A. P. (2011). Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 156, 1149–1163. doi: 10.1104/pp.111.174805
Nagarajan, V. K., and Smith, A. P. (2012). Ethylene’s role in phosphate starvation signaling: more than just a root growth regulator. Plant Cell Physiol. 53, 277–286. doi: 10.1093/pcp/pcr186
O’Rourke, J. A., Yang, S. S., Miller, S. S., Bucciarelli, B., Liu, J., Rydeen, A., et al. (2013). An RNA-Seq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol. 161, 705–724. doi: 10.1104/pp.112.209254
Osmont, K. S., Sibout, R., and Hardtke, C. S. (2007). Hidden branches: developments in root system architecture. Annu. Rev. Plant Biol. 58, 93–113. doi: 10.1146/annurev.arplant.58.032806.104006
Pant, B. D., Buhtz, A., Kehr, J., and Scheible, W. R. (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53, 731–738. doi: 10.1111/j.1365-313X.2007.03363.x
Paroutis, P., Touret, N., and Grinstein, S. (2004). The pH of the secretory pathway: measurement, determinants, and regulation. Physiology 19, 207–215. doi: 10.1152/physiol.00005.2004
Peng, J., Li, Z., Wen, X., Li, W., Shi, H., Yang, L., et al. (2014). Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 10:e1004664. doi: 10.1371/journal.pgen.1004664
Pérez-Torres, C. A., López-Bucio, J., Cruz-Ramirez, A., Ibarra-Laclette, E., Dharmasiri, S., Estelle, M., et al. (2008). Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20, 3258–3272. doi: 10.1105/tpc.108.058719
Philosoph-Hadas, S., Meir, S., and Aharoni, N. (1985). Carbohydrates stimulate ethylene production in tobacco leaf discs: ii. Sites of stimulation in the ethylene biosynthesis pathway. Plant Physiol. 78, 139–143. doi: 10.1104/pp.78.1.139
Pierik, R., Tholen, D., Poorter, H., Visser, E. J., and Voesenek, L. A. (2006). The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci. 11, 176–183. doi: 10.1016/j.tplants.2006.02.006
Raghothama, K. G. (2000). Phosphate transport and signaling. Curr. Opin. Plant Biol. 3, 182–187. doi: 10.1016/S1369-5266(00)00062-5
Roldan, M., Dinh, P., Leung, S., and McManus, M. T. (2013). Ethylene and the responses of plants to phosphate deficiency. AoB Plants 5:lt013. doi: 10.1093/aobpla/plt013
Rubio, V., Linhares, F., Solano, R., Martin, A. C., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. doi: 10.1101/gad.204401
Růzicka, K., Ljung, K., Vanneste, S., Podhorska, R., Beeckman, T., Friml, J., et al. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19, 2197–2212. doi: 10.1105/tpc.107.052126
Sánchez-Calderón, L., López-Bucio, J., Chacón-López, A., Cruz-Ramirez, A., Nieto-Jacobo, F., Dubrovsky, J. G., et al. (2005). Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol. 46, 174–184. doi: 10.1093/pcp/pci011
Sánchez-Calderón, L., López-Bucio, J., Chacón-López, A., Gutierrez-Ortega, A., Hernandez-Abreu, E., and Herrera-Estrella, L. (2006). Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiol. 140, 879–889. doi: 10.1104/pp.105.073825
Schmidt, W., and Schikora, A. (2001). Different pathways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiol. 125, 2078–2084. doi: 10.1104/pp.125.4.2078
Staal, M., De Cnodder, T., Simon, D., Vandenbussche, F., Van der Straeten, D., Verbelen, J. P., et al. (2011). Apoplastic alkalinization is instrumental for the inhibition of cell elongation in the Arabidopsis root by the ethylene precursor 1-aminocyclopropane-1-carboxylic acid. Plant Physiol. 155, 2049–2055. doi: 10.1104/pp.110.168476
Stepanova, A. N., and Alonso, J. M. (2009). Ethylene signaling and response: where different regulatory modules meet. Curr. Opin. Plant Biol. 12, 548–555. doi: 10.1016/j.pbi.2009.07.009
Sundaresan, S., Philosoph-Hadas, S., Riov, J., Belausov, E., Kochanek, B., Tucker, M. L., et al. (2015). Abscission of flowers and floral organs is closely associated with alkalization of the cytosol in abscission zone cells. J. Exp. Bot. 66, 1355–1368. doi: 10.1093/jxb/eru483
Svistoonoff, S., Creff, A., Reymond, M., Sigoillot-Claude, C., Ricaud, L., Blanchet, A., et al. (2007). Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 39, 792–796. doi: 10.1038/ng2041
Swarup, R., Perry, P., Hagenbeek, D., Van Der Straeten, D., Beemster, G. T., Sandberg, G., et al. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19, 2186–2196. doi: 10.1105/tpc.107.052100
Thibaud, M. C., Arrighi, J. F., Bayle, V., Chiarenza, S., Creff, A., Bustos, R., et al. (2010). Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J. 64, 775–789. doi: 10.1111/j.1365-313X.2010.04375.x
Ticconi, C. A., Delatorre, C. A., Lahner, B., Salt, D. E., and Abel, S. (2004). Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J. 37, 801–814. doi: 10.1111/j.1365-313X.2003.02005.x
Ticconi, C. A., Lucero, R. D., Sakhonwasee, S., Adamson, A. W., Creff, A., Nussaume, L., et al. (2009). ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc. Natl. Acad. Sci. U.S.A. 106, 14174–14179. doi: 10.1073/pnas.0901778106
Tran, H. T., Hurley, B. A., and Plaxton, W. C. (2010). Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Sci. 179, 14–27. doi: 10.1016/j.plantsci.2010.04.005
Uhde-Stone, C., Zinn, K. E., Ramirez-Yáñez, M.,Li, A., Vance, C. P., and Allan, D. L. (2003). Nylon filter arrays reveal differential gene expression in proteoid roots of white lupin in response to phosphorous deficiency. Plant Physiol. 131, 1064–1079. doi: 10.1104/pp.102.016881
Vance, C. P., Uhde-Stone, C., and Allan, D. L. (2003). Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157, 423–447. doi: 10.1046/j.1469-8137.2003.00695.x
Wang, H., Liang, X., Wan, Q., Wang, X., and Bi, Y. (2009). Ethylene and nitric oxide are involved in maintaining ion homeostasis in Arabidopsis callus under salt stress. Planta 230, 293–307. doi: 10.1007/s00425-009-0946-y
Wang, K. L., Yoshida, H., Lurin, C., and Ecker, J. R. (2004). Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428, 945–950. doi: 10.1038/nature02516
Wang, L., Dong, J., Gao, Z., and Liu, D. (2012). The Arabidopsis gene HYPERSENSITIVE TO PHOSPHATE STARVATION3 encodes ETHYLENE OVERPRODUCTION1. Plant Cell Physiol. 53, 1093–1105. doi: 10.1093/pcp/pcs072
Wang, L., Li, Z., Qian, W., Guo, W., Gao, X., Huang, L., et al. (2011). The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol. 157, 1283–1299. doi: 10.1104/pp.111.183723
Wang, L., Lu, S., Zhang, Y., Li, Z., Du, X., and Liu, D. (2014a). Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10. AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J. Integr. Plant Biol. 56, 299–314. doi: 10.1111/jipb.12184
Wang, Z., Straub, D., Yang, H., Kania, A., Shen, J., Ludewig, U., et al. (2014b). The regulatory network of cluster-root function and development in phosphate-deficient white lupin (Lupinus albus) identified by transcriptome sequencing. Physiol. Plant. 151, 323–338. doi: 10.1111/ppl.12187
Wang, Y. H., Garvin, D. F., and Kochian, L. V. (2002). Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol. 130, 1361–1370. doi: 10.1104/pp.008854
Wang, Z., Rahman, A. B., Wang, G., Ludewig, U., Shen, J., and Neumann, G. (2015). Hormonal interactions during cluster-root development in phosphate-deficient white lupin (Lupinus albus L.). J. Plant Physiol. 177, 74–82. doi: 10.1016/j.jplph.2014.10.022
Ward, J. T., Lahner, B., Yakubova, E., Salt, D. E., and Raghothama, K. G. (2008). The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol. 147, 1181–1191. doi: 10.1104/pp.108.118562
Waters, B. M., Lucena, C., Romera, F. J., Jester, G. G., Wynn, A. N., Rojas, C. L., et al. (2007). Ethylene involvement in the regulation of the H(+)-ATPase CsHA1 gene and of the new isolated ferric reductase CsFRO1 and iron transporter CsIRT1 genes in cucumber plants. Plant Physiol. Biochem. 45, 293–301. doi: 10.1016/j.plaphy.2007.03.011
Wen, C.-K., Li, W. Y., and Guo, H. W. (2015). “Regulatory components of ethylene signal transduction,” in Ethylene in Plants, ed. C.-K. Wen (Dordrecht: Springer), 73–92. doi: 10.1007/978-94-017-9484-8
Wu, P., Ma, L., Hou, X., Wang, M., Wu, Y., Liu, F., et al. (2003). Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 132, 1260–1271. doi: 10.1104/pp.103.021022
Xu, J., and Zhang, S. (2015). “Ethylene biosynthesis and regulation in plants,” in Ethylene in Plants, ed. C.-K. Wen (Dordrecht: Springer), 1–25. doi: 10.1007/978-94-017-9484-8
Yu, H., Luo, N., Sun, L., and Liu, D. (2012). HPS4/SABRE regulates plant responses to phosphate starvation through antagonistic interaction with ethylene signaling. J. Exp. Bot. 63, 4527–4538. doi: 10.1093/jxb/ers131
Yuan, H., and Liu, D. (2008). Signaling components involved in plant responses to phosphate starvation. J. Integr. Plant Biol. 50, 849–859. doi: 10.1111/j.1744-7909.2008.00709.x
Zakhleniuk, O. V., Raines, C. A., and Lloyd, J. C. (2001). pho3: a phosphorus-deficient mutant of Arabidopsis thaliana (L.) Heynh. Planta 212, 529–534. doi: 10.1007/s004250000450
Zhang, Y. J., Lynch, J. P., and Brown, K. M. (2003). Ethylene and phosphorus availability have interacting yet distinct effects on root hair development. J. Exp. Bot. 54, 2351–2361. doi: 10.1093/jxb/erg250
Zhang, Y., Wang, X., Lu, S., and Liu, D. (2014). A major root-associated acid phosphatase in Arabidopsis. AtPAP10, is regulated by both local and systemic signals under phosphate starvation. J. Exp. Bot. 65, 6577–6588. doi: 10.1093/jxb/eru377
Zhao, Q., and Guo, H. W. (2011). Paradigms and paradox in the ethylene signaling pathway and interaction network. Mol. Plant 4, 626–634. doi: 10.1093/mp/ssr042
Zheng, L., Huang, F., Narsai, R., Wu, J., Giraud, E., He, F., et al. (2009). Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol. 151, 262–274. doi: 10.1104/pp.109.141051
Keywords: ethylene, phosphate responses, root architecture, transcriptional regulation, signaling, crosstalk
Citation: Song L and Liu D (2015) Ethylene and plant responses to phosphate deficiency. Front. Plant Sci. 6:796. doi: 10.3389/fpls.2015.00796
Received: 26 July 2015; Accepted: 13 September 2015;
Published: 29 September 2015.
Edited by:
Aaron Smith, Louisiana State University, USAReviewed by:
Soumitra Paul, Krishnagar Government College, IndiaCopyright © 2015 Song and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong Liu, Ministry of Education Key Laboratory of Bioinformatics, Center for Plant Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China,bGl1LWRAbWFpbC50c2luZ2h1YS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.