
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 17 September 2015
Sec. Plant Physiology
Volume 6 - 2015 | https://doi.org/10.3389/fpls.2015.00757
This article is part of the Research Topic Abiotic Stress Signaling in Plants: Functional Genomic Intervention View all 50 articles
Basic transcriptional machinery in eukaryotes is assisted by a number of cofactors, which either increase or decrease the rate of transcription. Mediator complex is one such cofactor, and recently has drawn a lot of interest because of its integrative power to converge different signaling pathways before channeling the transcription instructions to the RNA polymerase II machinery. Like yeast and metazoans, plants do possess the Mediator complex across the kingdom, and its isolation and subunit analyses have been reported from the model plant, Arabidopsis. Genetic, and molecular analyses have unraveled important regulatory roles of Mediator subunits at every stage of plant life cycle starting from flowering to embryo and organ development, to even size determination. It also contributes immensely to the survival of plants against different environmental vagaries by the timely activation of its resistance mechanisms. Here, we have provided an overview of plant Mediator complex starting from its discovery to regulation of stoichiometry of its subunits. We have also reviewed involvement of different Mediator subunits in different processes and pathways including defense response pathways evoked by diverse biotic cues. Wherever possible, attempts have been made to provide mechanistic insight of Mediator's involvement in these processes.
The process of transcription in eukaryotic organism is an immensely complex and highly orchestrated phenomenon, and is mediated by a plethora of proteins wherein primary role is played by RNA polymerase II (RNAP II) (Lee and Young, 2000). The process is regulated both at the transcription initiation and elongation stages by a seemingly endless collections of regulatory proteins involved in different mechanisms (Woychik and Hampsey, 2002). Over the past 30 years, elegant biochemical, genetic, and structural biology works have established a core set of six general transcription factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) along with RNAP II as the core elements, which are obligatory to initiate and sustain any successful gene transcription event. On the other hand, among the numerous co-activators characterized till date to facilitate the initial recruitment of RNAP II to the core promoter and the subsequent transcript elongation, the Mediator complex has emerged as potentially the most crucial by virtue of its essentiality in RNAP II-mediated transcription (Myers and Kornberg, 2000; Conaway et al., 2005; Kornberg, 2005; Malik and Roeder, 2010). The Mediator complex is a highly conserved and integral part of RNAP II-mediated transcriptional machinery of the eukaryotes. In the past, the composition of the Mediator complex and the functions of different Mediator subunits have been reviewed several times focusing on yeasts and metazoans. In plant biology, the central role of Mediator complex in RNAP II-mediated transcriptional event has already been recognized by its discovery in Arabidopsis and other crop plants. The recent time has experienced a flush of interesting reports on the plant Mediator subunits detailing its quintessential role not only in growth and developmental processes, but also in biotic and abiotic stress responses (Figure 1). Realizing the need for an updated and critical analysis of roles of Mediator subunits in plants' life, this review summarizes the functions of Mediator subunits and provides insight about the depth and complexity of involvement of Mediator complex in transcriptional regulations of plant genes.
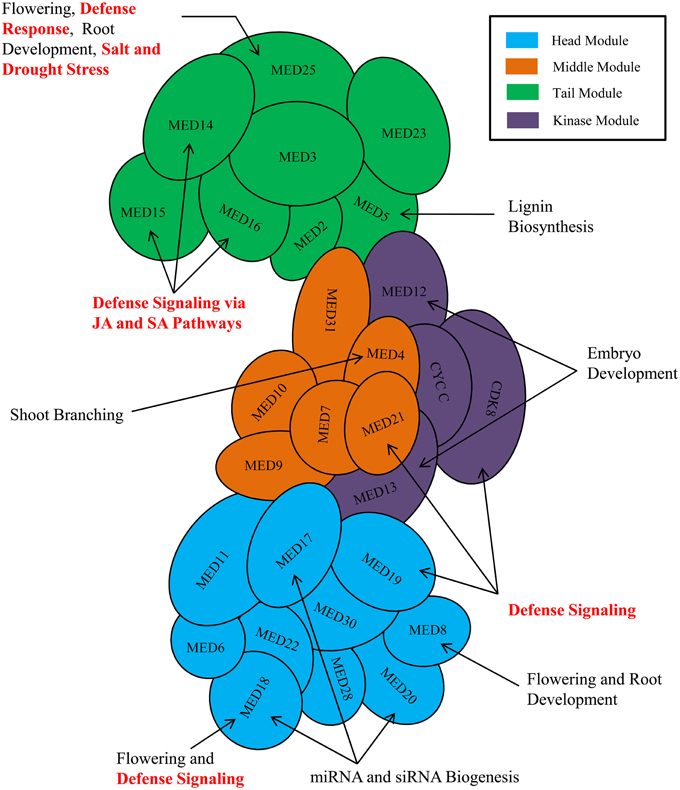
Figure 1. Involvement of plant Mediator subunits in development, and various biotic and abiotic stress responses. Arrangement of Mediator subunits is modular in nature. Mediator complex subunits are arranged into four modules-Head module (Cyan), Middle module (Orange), Tail module (Green), and a separable Kinase module (Purple). Only the known and important functions of plant Mediator complex are shown in the figure. MED26 and plant-specific Mediator subunits (MED34, MED35, MED36, and MED37) are not shown in the figure because of insufficient information about their module positions.
Mediator complex was first discovered in yeast in 1990, and within few years also reported in human (Kelleher et al., 1990; Flanagan et al., 1991; Thompson et al., 1993; Kim et al., 1994; Fondell et al., 1996; Ito et al., 1999). It took more than a decade to purify and characterize first Mediator complex from Arabidopsis cell suspension culture (Bäckström et al., 2007). Bioinformatics predictions including 16 plant species representing the entire plant kingdom ascertained its existence in major crop species including rice (Mathur et al., 2011). Despite low sequence similarity among the orthologs of the Mediator subunits in different organisms because of its rapid evolution, orthologs of all the yeast Mediator subunits reported to be present in plants too (Levine and Tjian, 2003; Bourbon, 2008; Mathur et al., 2011). MED1 is not found in higher plants, but is encoded by the genome of red algae. Also, many of the subunits which were earlier reported to be plant-specific, are actually present in organisms of other kingdom (Bäckström et al., 2007; Bourbon, 2008; Mathur et al., 2011). Thus, most of the Mediator subunits are conserved across the eukaryotic organisms. Structure of Mediator complexes from different organisms have been analyzed with the help of electron microscopy and seemed to be astonishingly similar (Cai et al., 2009; Tsai et al., 2014; Wang et al., 2014). Alignment of secondary structures of the individual plant Mediator subunits with orthologs in other organisms also suggests quite high structural resemblance (Mathur et al., 2011). However, as in the case of many other proteins, plant genomes code for more number of paralogs of several Mediator subunits. Four paralogs of MED15 are encoded by the genes present in MED15 cluster on chromosome 1 of Arabidopsis (Pasrija and Thakur, 2012). Though functional significance of the presence of multiple paralogs of a particular Mediator subunit is not demonstrated yet, they might help in broadening the regulatory capability of the complex. The spatio-temporal regulation of the expression level of different paralogs of a particular Mediator subunit can make the Mediator structure more dynamic depending upon the external milieu and the growth and developmental phase of the plant.
Mediator is a multi-protein conglomerate, which is enormous in size and complex in composition. The individual protein identity is termed as MED subunit, and the numbers can vary from 25 to 30 depending upon the species. Another salient feature of the Mediator complex is its modular structure. The entire array of subunits of the Mediator complex is arranged into three modular structures; head module, middle module, and tail module (Asturias et al., 1999; Dotson et al., 2000; Chadick and Asturias, 2005; Bourbon, 2008). These three modules together form the Mediator core. The RNAP II-bound Mediator complex is called “Holoenzyme.” In addition, there is also a separable kinase or CDK8 module in the Mediator complex, which consists of CDK8, cyclin C, MED12, and MED13 (Wang et al., 2001; Spahr et al., 2003; Elmlund et al., 2006). The Mediator core associates with RNAP II favoring transcription whereas the kinase module-bound Mediator complex dissociates from RNAP II to repress transcription. The Mediator can shuttle between these two different forms (Mediator core and Mediator core-kinase complex) depending upon the cellular contexts. It is worth mentioning here that the kinase or the CDK module subunits were characteristically absent from the first-ever Mediator complex purified from Arabidopsis (Bäckström et al., 2007). The mechanism of Mediator functioning is manifested in different ways. Mediator acts as a bridge between the cis-element bound transcription factors and the promoter bound RNAP II, hence recruits the RNAP II machinery to the promoter of the transcriptionally active genes. However, recent progress suggests that Mediator is not just an adaptor molecule between the transcription factor and the basic transcriptional machinery, but provides a platform for recruitment of other cofactors, GTFs, and TFs for the formation of Pre-Initiation Complex (PIC). These interactions can bring changes in the structure of the resultant complex which may affect transcription. Thus, Mediator acts as a docking site for many other transcriptional regulators and plays critical role in relaying regulatory signals from them to RNAP II machinery (Takahashi et al., 2011, 2015).
In the beginning, Mediator complex was thought to be involved only in the initiation step of transcription as evident by its interaction with the components of the transcription initiation complex (Mittler et al., 2001; Baek et al., 2002; Cantin et al., 2003; Johnson and Carey, 2003; Wang et al., 2005). However, in last few years, Mediator has been reported to be involved in the regulation of many other steps of transcription like promoter escape (Malik et al., 2007; Cheng et al., 2012; Jishage et al., 2012), elongation (Takahashi et al., 2011; Conaway and Conaway, 2013; Galbraith et al., 2013), termination (Mukundan and Ansari, 2011, 2013), as well as in other co-transcriptional RNA processing events (Kim et al., 2011; Huang et al., 2012; Oya et al., 2013). Mediator has also been implicated in epigenetic and architectural modification of chromatin leading to changes in gene expression (Kagey et al., 2010; Zhu et al., 2011; Fukasawa et al., 2012; Liu and Myers, 2012; Lai et al., 2013; Tsutsui et al., 2013; Zhang et al., 2013a). Thus, it seems that Mediator is critical for almost every aspect of transcription of eukaryotic genes.
Mediator complex was first discovered as an entity required for enhanced transcription of an in vitro transcription system which contained RNAP II and essential general transcription factors (Flanagan et al., 1991; Kim et al., 1994). Now, it is well-established that a number of transcription factors need Mediator to enhance the process of transcription. At this point of time, it is not clear if this positive effect in the activation of transcription is direct or indirect. However, the role of Mediator complex in the repression of gene expression has also been reported in many cases. The repressor activity of Mediator is primarily attributed to the kinase module. Association of this module with the core Mediator complex occludes the RNAP II from the PIC exerting a repressive role in the RNAP II-mediated transcriptional events (Holstege et al., 1998; Samuelsen et al., 2003; Elmlund et al., 2006; Knuesel et al., 2009). However, contrary to this, researchers from different laboratories have also reported the positive effect of kinase module on gene expression (Donner et al., 2007, 2010; Belakavadi and Fondell, 2010). Mechanistically, the activation property of CDK8 module may partly be attributed to its ability to recruit transcription elongation factor, P-TEFb, and to release the promoter-proximally paused RNAP II into productive elongation phase (Takahashi et al., 2011, 2015). The added complexity of the Mediator functions is brought about by the presence of multiple isoforms of the kinase module, which might help the Mediator complex to fine-tune the gene expression in a tissue, cell, or even pathway specific manner (Sato et al., 2004; Bourbon, 2008; Conaway and Conaway, 2011; Mathur et al., 2011).
Even after two decades of its discovery, it is still debatable whether Mediator complex is a general transcription factor or just a cofactor of gene expression. Although, initially identified as an entity that supports activator-dependent transcription, now, according to several evidences, Mediator complex can also be categorized as general transcription factor. The human Mediator complex can support basal level transcription of many genes by playing important roles in assembly of PIC and transcription initiation (Mittler et al., 2001; Baek et al., 2002). Mediator complex enhances the RNAP II recruitment to the protein coding genes and provides stability to the transcription machinery assembled at the promoter region (Cantin et al., 2003; Baek et al., 2006). In yeast, deletion of MED17 makes Mediator structurally unstable, and in the conditional yeast med17 knockout expression of protein-coding genes is severely compromised on a genome-wide scale (Thompson and Young, 1995; Ansari et al., 2009). In plants, comparison of transcriptomes between nrpb2-3 (mutant in second largest subunit of RNAP II) and med20a plants revealed 84% overlap in the down-regulated genes, implying that Mediator complex is as important as the RNAP II for gene expression (Kim et al., 2011). Thus, the Mediator complex should be regarded as an integral component of the basal transcriptional machinery in the eukaryotes, and the roles are manifested in the forms of RNAP II recruitment and activation, co-ordination of PIC assembly, control of TFIIH-dependent RNAP II CTD phosphorylation within the PIC, and sustained or transient repression of transcription initiation via Mediator-CDK8 module interactions (Taatjes, 2010). Nevertheless, increasing number of reports of deletions of certain Mediator subunits affecting particular phenotype suggest that several individual Mediator subunits possess specific functions as well (Tables 1–3). This dilemma could be explained by taking into account the modular nature of Mediator complex and by assigning the “division of labor” principles to each module. The head module subunits might be involved in the more basic functions of the Mediator complex, whereas the tail module subunits residing on the periphery might be controlling gene-specific functions by contacting the specific transcription factors.
Tight transcriptional regulation of gene expression is very important for proper growth and development of plant and its protection from adverse environmental conditions. After the basic transcriptional machinery, Mediator complex probably can be considered as the second most important regulatory hub for different signaling networks in response to different developmental as well as environmental changes both in animals and plants as suggested by the work of many laboratories including ours. Before we describe the functions of individual Mediator subunit genes, the following is an account of changes in the transcript level of Mediator subunit genes in different tissues, and also how they are affected by different stages of growth and development. We have also discussed the changes in the transcript level of Mediator subunit genes in response to different hormones and abiotic stress treatments.
In an attempt to answer the question of what affect the levels of expression of individual Mediator subunits, analyses of differential expression of MED genes in different tissues and during different stages of plant development were performed. Several MED genes were significantly regulated during panicle and seed development stages as compared to root and leaf of the rice plants (Mathur et al., 2011). The enrichment of seed storage-specific promoter elements in certain MED genes raises possibilities of important function of MED subunits during embryo development and seed maturation. The increased abundance of OsMED8 and OsMED11_1 at early panicle and seed stages implicates their probable roles in reproductive development of rice. Middle module subunit, AtMED21_1 showed approximately two-fold upsurge in the advanced stages of seed development which supports the reported role of AtMED21 in embryo development and cotyledon expansion. OsMED21 might be involved prominently in the early stages of panicle development (Dhawan et al., 2009; Mathur et al., 2011). OsMED31_1 is expressed more in leaf as compared to root. The tail module subunit, AtMED14 is significantly expressed in leaf as compared to other parts of the plants, and has been implicated in the control of cell cycle duration and root elongation (Autran et al., 2002; Krichevsky et al., 2009). In rice, OsMED26 is expressed more in root as compared to leaves. Significant up-regulation of OsMED15_1 during different stages of seed development in different rice cultivars supports its probable role in seed development (Thakur et al., 2013). In this process, interaction of OsMED15_1 with seed-specific transcription factors could be predicted. SNP analysis of this gene sequence among several rice cultivars significantly segregated long and short grain varieties. Thus, an important gene regulatory role of this Mediator subunit in seed development and size determination is highly anticipated (Thakur et al., 2013). Several plant-specific Mediator subunits like MED34, MED35, MED36, and MED37, which have not been assigned to any module yet, are expressed more in reproductive stages as compared to vegetative parts implying its tissue-specific functions. The Mediator subunit, MED36 is expressed more in the root of Arabidopsis and is anticipated to be involved in root-specific gene regulatory functions (Pasrija and Thakur, 2013). Thus, Mediator complex as a whole is a dynamic entity, and its composition may fluctuate in different tissues at different stages of growth and development.
In the process of delineating how hormones affect the expression of different MED subunits in Arabidopsis, it was found that brassinosteroid (BR) and abscissic acid (ABA) have more significant impact on the transcription of MED genes as compared to other hormones including auxin and jasmonic acid (JA) (Pasrija and Thakur, 2012). AtMED37, which was discovered as a plant-specific Mediator subunit, is the most highly up-regulated MED in response to BR treatment. The reported 2.5-fold increase of AtMED12 in response to BR treatment might also shed some light on its role in embryo development (Gillmor et al., 2010; Pasrija and Thakur, 2012). Among the other significant expression changes of Mediator subunits in response to phytohormone treatments, more than two-fold build-up in the transcript level of AtMED18 in response to JA deserves special mention. AtMED18 has been reported to be involved not only in flower development but also in disease signaling (Zheng et al., 2013; Lai et al., 2014). On the other hand, we noted significant down-regulation (>40%) of tail module subunit genes, like AtMED15, AtMED14, and AtMED5 in response to auxin treatment (Pasrija and Thakur, 2012). Although auxin and BR are known for their synergistic effects on plant growth and development, transcription of a set of Mediator genes was different in response to these hormones. For instance, AtMED15 was up-regulated by BR but severely down-regulated by auxin. It seems that these two hormones show their transcriptional effects by a combination of different set of Mediator subunits (Pasrija and Thakur, 2012). In rice, there was not much effect on the transcript abundance of MED genes in response to different stresses like drought, salt, and salinity, but one, OsMed37_6, exhibits around two-fold change in response to different stresses (Mathur et al., 2011). However, in Arabidopsis, significant transcriptomic reprogramming of the Mediator subunit genes in response to high light, dark, and high salinity conditions was documented (Pasrija and Thakur, 2012). Interestingly, MED16 has been reported to be involved in cold signaling pathways, but the expression level of both AtMED16 and OsMED16 remains unchanged in response to cold treatment (Warren et al., 1996; Knight et al., 1999; Mathur et al., 2011). Like its role in cold signaling, more than two-fold increase of MED16 transcript in response to salinity stress may imply its role as a converging point of both salt and cold signaling pathways. The important functions of AtMED12 in light and salt signaling pathways can not be ruled out because of its two-fold up-regulation in response to high light and salt conditions. Induction of AtMED37 in response to BR and low light suggests a probable link between shade and BR signaling, and the process may be mediated by Endoplasmic Reticulum-Associated Degradation (ERAD) (Hong et al., 2008; Pasrija and Thakur, 2012). The up-regulation of AtMED37 in response to cold and salinity stresses provokes an intriguing hypothesis that AtMED37 may act as an integrative hub of many different signaling pathways, which is supported by the near ubiquitous, high expression level of AtMED37 in all the tissues tested so far (Pasrija and Thakur, 2012, 2013).
Accumulating evidences suggest that Mediator complex is a dynamic and highly flexible entity, and its structural composition alters depending upon the context. Based on the spatio-temporal regulations of transcription of individual Mediator subunit genes in response to different stimuli, we predicted enrichment of specific structural arrangement composed of specific Mediator subunits during certain developmental stages (Pasrija and Thakur, 2013). However, as the Mediator stimulates basal transcription by participating in the recruitment of RNAP II at specific sites all over the genome, a basic, core structure should always be maintained irrespective of tissue, cell, development stage, or any environmental condition. That is the reason that transcription of a set of MED genes is not affected by hormones, stresses, or developmental cues. In animal cells, this has been well-illustrated by the presence of a simpler Mediator complex made of just 6–8 members in differentiated cells as compared to a 26-member Mediator complex in the cancerous and stem cells (Deato et al., 2008).
A large portion of total protein coding genes in eukaryotes requires the presence of Mediator complex even to sustain basal level of transcription. This proves unequivocally that Mediator constitutes important part of the basal transcriptional machinery. However, drastic morphological changes in mutants of individual Mediator subunits suggest that Mediator could also act as selective gene regulator both in metazoans and plants (Malik and Roeder, 2010; Taatjes, 2010; Kidd et al., 2011; Mathur et al., 2011; An and Mou, 2013; Poss et al., 2013; Allen and Taatjes, 2015). As the present review is plant specific, the following is an account and critical analyses of important functions of Mediator subunits reported from different plant species through mutational and genome-wide transcriptom analyses (Tables 1–3).
In Arabidopsis and other plants, different phases of embryo development and maturation are marked by specific patterns and shapes. The Mediator subunits, MED12 and MED13, also known as GRAND CENTRAL (GCT) and CENTER CITY (CCT), respectively, mediate the embryo pattern formation, albeit in a transient manner (Gillmor et al., 2010). Mutations in these two genes disrupt the central and peripheral identity of the embryo along with the inhibition of globular to heart transition (Gillmor et al., 2010). Further investigations led to the prediction that the aberrant pattern during early embryo development might be due to a transient transcriptional repression of important genes like those encoding KANADI 1 and KANADI 2 transcription factors. AtMED13, also known as Macchi Bou2 (MAB2), has also been reported to be involved in embryo patterning and cotyledon development (Ito et al., 2011). In this case, the mutant shows aberrations in auxin response. The inability of Atmed13 embryo to perceive and respond to auxin signals might account for its defective cotyledon formation. Recently, these two kinase module subunits have been shown to be involved in three more developmental transitions, i.e., germination, vegetative phase change, and flowering (Gillmor et al., 2014). Interestingly, the delay in vegetative phase change occurs largely due to over-expression of miR156 and the delay in flowering is caused by the increased production of FLC. On the whole, AtMED12 and 13 act as a global regulator of temporal genes making the developmental transitions a tightly controlled phenomenon.
One of the most well-characterized Mediator subunits in plants is AtMED25, which has been described earlier as PFT1. MED25/PFT1 was discovered as a positive regulator of shade avoidance in Arabidopsis (Cerdán and Chory, 2003). It was postulated to be involved in control of flowering by phytochrome B pathway, which is dependent on light quality. The Atmed25 plants flower late as compared to the wild type (Kidd et al., 2009). It has been demonstrated that AtMED25 positively regulates CONSTANT (CO) and FLOWERING LOCUS T (FT), two important flowering regulators in Arabidopsis (Inigo et al., 2012a). AtMED25 seems to be subjected to the phenomena of “activation by destruction.” AtMED25 is an unstable protein that is targeted by two RING H2 proteins, MBR1, and MBR2 for degradation by proteosomal pathway (Inigo et al., 2012b). The high turnover of AtMED25 is required for the activation of FT which promotes flowering. The phenomena elegantly demonstrate how a Mediator subunit follows “activation by destruction” principle to control a plant-specific event, and also adds a new dimension to Mediator function.
Few other Mediator subunits have also been implicated to be involved in flowering process (Table 1). Along with AtMED25, the delayed flowering phenotype was also observed in Atmed8 mutants both under short and long day conditions (Kidd et al., 2009). In Atmed8 mutants, the level of FT, a positive regulator of flowering, is low whereas FLC, a negative regulator of flowering, is expressed more. Enhanced phenotype in the double mutant of Atmed25/Atmed8 suggests that AtMED25 and AtMED8 work independently and they might be controlling flowering process by responding to two different signaling pathways in synergy.
Mediator subunit MED12 (also known as CRYPTIC PRECOCIOUS, CRP) is a positive regulator of flowering and affects multiple genes working upstream and downstream of FT (Imura et al., 2012). As AtMED12 is a part of the kinase module and could interact with histone H3K9 methyltransferase, there is a possibility that it is involved in the epigenetic regulation of FLC and FT genes (Ding et al., 2008).
Very recently, another Mediator subunit, AtMED18, has been reported to be involved in flowering (Zheng et al., 2013; Lai et al., 2014). The loss-of-function mutant showed delayed flowering, and has altered level of FLC and FT. AtMED18 has been reported to interact with SUPPRESSOR OF FRIGIDA 4 (SUF4), and together binds to the promoter of FLC gene (Lai et al., 2014). Normally, AtSUF4 is a positive regulator of FLC gene. AtMED18 probably acts as suppressor of AtSUF4 activity.
The process of flowering requires the transition of vegetative primordia to reproductive primordia, and the region is marked with constant cell division. As the Mediator complex is often connected with dynamic cellular activities, it is quite obvious that Mediator plays significant role in the process of flowering and that is why several subunits affect this process (Table 1). Mostly, the loss-of-function mutations of Mediator subunits led to late and abnormal floral development, which is attributed to the perturbation in the transcript level of important flowering regulators like FLC, FT, and floral identity regulators like AG. But the missing link is how Mediator subunits control the expression of these genes. The non-coding RNAs play important role in epigenetic regulation of FLC gene (Crevillén and Dean, 2011; De Lucia and Dean, 2011). Given the reported association of non-coding RNA with the Mediator complex, a similar kind of mechanism in flowering time control could be envisaged (Lai et al., 2013).
A search for the role of Mediator subunits in root morphogenesis revealed the pivotal role of MED25 and MED8 in the production of root hairs in Arabidopsis (Sundaravelpandian et al., 2013; Raya-González et al., 2014). The absence of root hairs in Atmed25 and Atmed8 is due the inappropriate distribution of hydrogen peroxides (H2O2) and superoxides () over the surface of tap root system. In fact, the comparison of the transcriptome of wild type and the Atmed25 plants revealed that class III peroxidases are the worst affected ones in the mutant, perturbing the ROS homeostasis across the root length. The more severe phenotype of med25/med8 double mutant eliminates the possibility of these two genes interacting in the same pathway. It will be interesting to find out if other MED subunits assist MED25 and MED8 in root hair development. Also, knowledge of transcription factors targeting these subunits will be helpful in understanding the mechanisms of transcriptional regulation of this process.
Mediator subunit CDK8 (or HEN3) of the kinase module plays important role in specification of stamen and carpel in Arabidopsis (Wang and Chen, 2004). Mechanistically, like in yeast and mammals, AtCDK8 phosphorylates the CTD domain of largest subunit of RNAP II and represses transcription. This leads to an enhanced expression of AG, AP1, and AP2 in cdk8 mutant. CDK8 is abundantly expressed in the proliferating tissues suggesting its involvement in mediating cell division and cell fate specification. Alternatively, as the RNA transcription and RNA processing are coupled and CDK8 interacts with CTD domain of RNAP II, the perturbed alternative transcript of AG1 in the mutant plant indicates its probable role in alternative splicing. What it warrants at this moment is to identify the transcription factors that interact with these Mediator subunits and the immediate target genes for a better understanding of the regulatory circuitry that controls cell number and size.
Another Mediator subunit AtMED18 contributes to the organ identity and number. Other than being short in stature and late flowering, Atmed18 plants have altered number of floral parts. In mutant plants, sepals and petals are more and anthers are less. There are two carpels, and the pollen maturation is delayed (Kim et al., 2011; Zheng et al., 2013). The down-regulation of floral homeotic genes like AP1, PI, and AG in Atmed18 mutant plants indicates crucial regulatory role of AtMED18 in homeotic gene expression (Zheng et al., 2013). Additionally, AtMED18 may control the organ identity genes through its association with HEN3/CDK8, which also controls organ identity and shows similar loss-of-function phenotypes (Wang and Chen, 2004).
Cell number over the entire arial parts of Arabidopsis is decreased if there is a mutation in another Mediator subunit MED14, more popularly known as STRUWWELPETER (SWP) (Autran et al., 2002). Both the leaf number and size in the heterozygous mutant lines are reduced whereas homozygous mutant lines are sterile. The importance of AtMED14 in leaf development is also evident by its strong expression in leaves. The mutant plant also carries a disorganized Shoot Apical Meristem (SAM). The arrest of cell division in Atmed14 plants may result from the endoreduplication of the chromosomal DNA. Mechanistically, AtMED14 may interact with SMP1, SMP2 which encode step II splicing factors as both the mutants show similar phenotypes (Clay and Nelson, 2005). LEUNIG, a GroTLE transcription corepressor, has been reported to interact with AtMED14 and controls multiple physiological processes (Gonzalez et al., 2007).
MED4 is a subunit in the middle module, and has recently been speculated to be involved in growth of the tillers in rice (Li et al., 2014). Its homozygous mutants are embryonic lethal. Surprisingly enough, it interacts with SAD1, an ortholog of RNA polymerase I subunit RPA 34.5 in rice, and is involved in rRNA biosynthesis. It is worth mentioning here that SAD1 was isolated as a component of Mediator complex during the complex purification study in Arabidopsis (Bäckström et al., 2007). It also interacts with the counter parts of the other RNA polymerases like pol II and pol III. Thus, this is the first example which shows the interaction between the Mediator complex and the RNA pol I and III, and thus extends the function of Mediator beyond RNAP II-mediated transcription.
Cell proliferation and cell expansion are two important basic processes in any organism, which ultimately determine the organ size, hence the entire body size. DA1 is an ubiquitin receptor and restricts cell proliferation to control final size of organs in Arabidopsis (Li et al., 2008). In a genetic screen to find the enhancer of DA1 mutation, AtMed25 mutant was characterized (Xu and Li, 2011). AtMED25 too negatively controls the cell proliferation and cell enlargement. Loss-of-function mutant of MED25 has large organs, with larger and slightly increased numbers of cells as a result of an increased period of cell proliferation and cell expansion. The observed phenotype in Atmed25 mutant plants may be partly because of the up-regulation of expansin genes like AtEXP1, AtEXP3, AtEXP5, AtEXP9, AtEXP11, and AtEXPB3. Consistent to this, plants over-expressing MED25 have small organs owing to decrease in both cell number and size. Further analysis eliminated the possibility of higher ploidy level in the mutant plants as the cause of larger organ size. The genetic and physiological data suggest that MED25 acts to limit cell and organ growth independently of its involvement in phytochrome and JA signaling pathways (Cerdán and Chory, 2003; Kidd et al., 2009; Xu and Li, 2011; Chen et al., 2012; Inigo et al., 2012a,b). Rather, MED25 functions synergistically with DA1 to control organ growth by restricting cell proliferation. In contrast to MED25, MED8 positively controls the organ size (Xu and Li, 2012). The mutant Atmed8 plants have shorter flowers because of reduced cell expansion. Analysis of med25med8 double mutants revealed the antagonistic behavior of MED25 and MED8, at least in the case of cell expansion and cell proliferation, hence in organ size determination.
Getting rid of lignins from the crops for its usage as forage, pulp, and paper production poses a significant challenge because most of the lignin related mutants are stunted and growth defective. Two such mutants, ref8-1 and ref8-2, which are deficient in lignin content, are short and display little vegetative growth. Two Mediator subunits, AtMED5a (REF4), and AtMED5b (RFR1), have been shown to negatively regulate plant height and lignin content (Bonawitz et al., 2012). Interestingly, the mutants of either of these subunits rescue the phenotype of ref8-1 or ref8-2 without any yield penalty on biomass production (Bonawitz et al., 2014). Importantly, the mutants are free from biomass recalcitrance. Thus, the domain of Mediator function also encompasses the regulation of cell wall biosynthesis, which is of great practical value.
Iron is one of the essential elements in plants, and its uptake and assimilation are tightly controlled. Two Mediator subunits, AtMED16 (YID), and AtMED25, have been reported to control iron homeostasis is plants (Yang et al., 2014; Zhang et al., 2014). The mutants of these Mediator subunits display hypersensitivity toward iron deficiency resulting in leaf chlorosis. AtMED16 directly interacts with FIT, the master regulator of iron homeostasis in plants. In chromatin immunoprecipitation analysis, AtMED16 was found to be present on the promoter of the iron acquisitions genes like FRO2 and IRT1, probably by interacting with FIT (Zhang et al., 2014). FIT also interacts with other bHLH proteins forming heterodimers and these heterodimers bind to FRO2 and IRT1 promoters. The binding of AtMED16 probably confers stability to the FIT/bHLH complex (Zhang et al., 2014). On the other hand, MED25 interacts with two transcription factors, EIN3 and EIL1, which are involved in ethylene signaling. EIN3 and EIL1 directly interact with FIT. FIT is a highly unstable protein and the interaction of MED25 with EIN3 and EIL1 provides stability to FIT enabling it to regulate downstream iron regulatory genes like FRO2 and IRT1 (Yang et al., 2014). Interaction between AtMED16 and AtMED25 has also been reported (Zhang et al., 2014). However, the effects of double mutations of these two genes are yet to be investigated. Probably, AtMED16, AtMED25, EIN3, EIL1, FIT, and other bHLH proteins form a stable activator complex on the promoter of FRO2 and IRT1 leading to their activation during iron deficient conditions.
Plants in its natural environments are being constantly challenged by myriad of insect pests and pathogens, which together constitute the biotic stresses. A survivor plant activates its defense arsenal quickly and efficiently in order to counter the invading and inflicting biotic agents. Such an orchestrated and rapid response is only achievable by the timely activation of key defense genes. Emerging reports have established Mediator complex as an essential component for regulation of genes involved in defense pathways (An and Mou, 2013). In comparison to other pathways, higher number of Mediator subunits has been shown to be involved in defense signaling (Table 2).
The first Mediator subunit reported to be involved in defense response was AtMED25 (Kidd et al., 2009). AtMED25 bears similarity with the mammalian MED25, which also plays important role in defense response (Leal et al., 2009). In Arabidopsis, MED25 directly affects JA-dependent gene expression (PDF1.2, HEL, CHIB, and ESP), and provides resistance against the leaf-infecting necrotrophic fungi, Alternaria brassicicola, and Botrytis cinerea (Kidd et al., 2009). The complementation of Atmed25 by its homologs from wheat strengthened the view that functions of some of the Mediator subunits may be conserved in higher plants (Kidd et al., 2009). A group of 12 transcription factors (TFs) have been shown to interact with AtMED25, which includes AP2/ERF, bHLH, MYB, WRKY, and bZIP. Among these transcription factors, many have previously been demonstrated to be involved in JA signaling pathway (Çevik et al., 2012). Furthermore, AtMED25 takes part in ERF1- and ORA59-dependent activation of PDF1.2 gene as well as MYC2-dependent activation of VSP1 gene, which are some important genes in the JA signaling pathway (Çevik et al., 2012). In fact, MED25 physically associates with the bHLH transcription factor, MYC2 in promoter regions of its target genes to elicit a positive effect on their transcription (Chen et al., 2012). The head module subunit mutant, Atmed8, behaves like Atmed25 but shows pronounced susceptibility toward A. brassicicola (Kidd et al., 2009). These two mutants, however, do not interact genetically, suggesting that AtMED25 and AtMED8 might be acting in two independent pathways controlling the same response and phenotype (Kidd et al., 2009).
The middle module subunit MED21 is an essential requirement for survival of Arabidopsis plants as its T-DNA insertional homozygous lines are embryonic lethal (Dhawan et al., 2009). The RNAi lines of MED21 are highly susceptible to A. brassicicola and B. cinerea. The detailed study revealed that MED21 interacts with RING E3 ligase, Histone Monoubiquitination1 (HUB1), which mediates the H2B ubiquitination, thus establishing a link between Mediator and the chromatin remodeling. The induced expression of both MED21 and HUB1 in response to chitin treatment, an important constituent of fungal cell wall, suggests their probable role in defense signaling (Dhawan et al., 2009).
The head module subunit, AtMed19a interacts with nuclear localized fungal effector (HaRxL44) of powdery mildew pathogen, Hyaloperonospora arabidopsidis (Hpa). This leads to proteasome-dependent degradation of AtMed19a and shift the balance from SA-mediated disease resistance to ET/JA-mediated transcriptomic changes making the plants more vulnerable to bitrophs (Caillaud et al., 2013). This highlights how pathogens can break plant immune barrier by hijacking the important resistance mechanisms offered by Mediator complex. Another head module subunit, AtMED18, plays a positive regulatory role toward necrotropic fungal infection by interacting with YYI keeping the expression of glutaredoxin and thioredoxin genes suppressed (Lai et al., 2014).
Three tail module subunits, AtMED14, AtMED15, and AtMED16 have been reported to be involved in defense signaling as well (Canet et al., 2012; Wathugala et al., 2012; Zhang et al., 2012, 2013b) (Table 2). The Arabidopsis plants carrying mutation in MED16 are compromised for SA- and JA-dependent defense responses (Wathugala et al., 2012). The Atmed16 mutant plants are more susceptible to Pseudomomas syringae attack, and exhibit lower expression of defense-related genes like those coding for PR (Pathogenesis Related) proteins and defensins. Moreover, the expression levels of the important SAR (systemic acquired resistance) markers like PR1, PR2, PR5, GST11, EDR11, SAG21 are severely reduced in Atmed16 mutant (Zhang et al., 2012). Hence, MED16 acts as a positive regulator of SA-induced gene expression. Similarly, the Atmed16 mutation also blocks the induction of the JA/ET-dependent gene expression making the plants vulnerable to necrotrophic fungi like A. brassicicola and B. cinerea (Zhang et al., 2012). Thus, MED16 seems to function as an integrative hub for both SA and JA signaling pathways. The tail module subunit, AtMED15, also dubbed as NRB4 (Non-recognition of BTH4, a salicylic acid analog), has recently been shown to be involved in defense signaling via its involvement in SA pathway (Canet et al., 2012). The mutant plants with defective MED15 do not show any noticeable phenotypic change except its attenuated response to SA, reminiscent of the effects of npr1 mutation in plants' defense signaling. NPR1 (non-expresser of PR genes) plays a pivotal role and takes the center stage in the SA-mediated defense pathways (Dong, 2004). However, neither a genetic nor a biochemical interaction has been reported between MED15 and NPR1. The additive phenotypes of Atmed15/npr1-70 plants indicate that they might work at different point of SA signaling pathway. Moreover, Atmed15 affects neither the localization of NPR1 nor its stability. Thus, mechanistically, MED15/NRB4 might be functioning downstream of NPR1 in the regulation of SA response pathway. The exact position of MED15 in SA signaling pathway is not known, and it warrants detailed molecular and genetic investigations. A mutation in AtMED14 subunit gene suppresses the SA-dependent expression of defense genes (Zhang et al., 2013b). AtMED14 prevents PR1 expression without interfering the binding of NPR1, the master regulator of defense gene expression, to its promoter. This leads to the speculation that AtMED14 might be responsible for the recruitment of RNAP II to the promoter of PR1 gene. Further investigation is needed to delineate the exact mechanism involved in the process. Thus, it seems that most of the subunits in the tail module play significant role in the regulation of defense gene expression during pathogen attack. However, the mechanisms employed by the three different Mediator subunits (MED14, MED15, and MED16) differ considerably toward controlling the expression of defense genes. The Atmed16 mutation differentially affects the expression of different positive and negative regulators of SAR, whereas Atmed14 mutation inhibits expression of similar genes. Moreover, defense-related transcriptomic change in the case of Atmed14 is much smaller as compared to that in the case of Atmed16.
The kinase module component, AtCDK8, has recently been reported to be a positive regulator of disease response (Zhu et al., 2014). The mutant plants are highly susceptible to A. brassicicola. Mechanistically, it interacts with another Mediator subunit, AtMED25, and regulates JA-mediated gene expression during pathogen signaling. Additionally, it binds with the promoter of AGMATINE COUMAROYLTRANSFERASE (AACT1) gene whose products are involved in the biosynthesis of defense active bio-compounds like hydroxycinnamic acid amides in plants.
Plants are sessile organisms. They cannot run away to safer places during inclement weather. On the other hand, growth and development of the plant is profoundly influenced by the environment. A robust, surviving plant must translate the vagaries of the surrounding environments into proper signals relaying them to the transcriptional machinery ensuring the adaptability of the plants to the changed milieu. Of late, Mediator has emerged as an integrative hub for the different signaling pathways leading to the transcription regulation by RNAP II. So it is highly anticipated that the Mediator will also play a crucial role in the integration of signals originated in response to stresses like drought, cold, salinity etc. So far two Mediator subunits (Table 2), which also play important roles in biotic stresses, have been reported to be involved in abiotic stress signaling. The Atmed25 mutant seeds display increased sensitivity toward salt stress during germination. The importance of MED25 in high salinity is conserved across the plant species (Elfving et al., 2011). In a yeast two hybrid screen, three stress-specific transcription factors, DREB2A, ZFHD1, and MYB like proteins were found to be interacting with the ACID (Activator Interacting Domain) domain of AtMED25. The plants carrying mutations in any of these genes also display severe salt sensitivity. Mechanistically, ACID domain of MED25 might be targeted by these transcription factors for communication with the RNAP II transcriptional machinery for effective salt-responsive transcriptomic changes in plants. Surprisingly, MED25 negatively regulates drought tolerance in plants (Elfving et al., 2011). The mutant plants display huge increase in the expression level of drought responsive marker genes like RD29A, RD29B, and DREB2A. AtMED25 has been projected as a co-repressor interacting with the repressor domain of DREB2A making the plants vulnerable to drought stress (Elfving et al., 2011). Thus, it is one of those examples, where the same Mediator subunit, AtMED25, controls salt and dehydration stresses in an antagonistic manner.
MED16, originally discovered as SFR6 in Arabidopsis before being identified as a part of Mediator complex, has been reported as an important component involved in acclimation to cold (Knight et al., 1999, 2008; Wathugala et al., 2011). The mutant plants fail to embrace freezing temperature following its exposure to subzero temperature. At the molecular level, the plants are incapable of switching on the COR (cold on regulation) regulon including the expression of LTI78, COR15A, and KIN1/2. Microarray analysis revealed that a subset of cold-responsive genes bearing CRT/DRE motifs in their promoter regions gets miss-regulated in Atmed16 mutant plants (Knight et al., 1999). These genes are involved in freezing tolerance and controlled by CBF transcription factors (Boyce et al., 2003). However, neither the expression of CBF nor its localization is affected in Atmed16 mutant plants (Knight et al., 2009). Thus, it provokes the intriguing speculation that MED16 might modulate the activity of CBFs through post-transcriptional modulation.
One of the most significant discoveries of Mediator function in plants is related to miRNA and siRNA biogenesis (Kim et al., 2011). The loss-of-function mutants of three Mediator subunits, Atmed17, Atmed18, and Atmed20a, are short in stature, late flowering, and bear small fruits as compared to the wild types. The in-depth, detailed analyses revealed that these mutants are defective in the regulation of miRNA and siRNA at the transcriptional level. The occupancy of RNAP II at the promoters of miRNA and siRNA genes was also highly reduced in these mutants. The role of these Mediator subunits has also been implicated in the silencing of transposons and repeat sequences. These elements normally undergo siRNA-mediated transcriptional gene silencing, and were de-repressed in med17, med18, and med20a. On the other hand, co-purification of MED36 with the largest subunit of RNA pol V led to the intriguing hypothesis that Mediator complex may act in cooperation with other RNA polymerases in the production of non-coding RNA (Huang et al., 2009). Although it is a matter of debate, the same study also advocated the role of the Mediator complex as a general transcription factor. The discovery brought a paradigm shift in the understanding of Mediator functions beyond the regulation of subunit specific functions (Table 3).
The newest entrants into the expanding list of plant Mediator subunits are MED34 to MED37 (Bäckström et al., 2007). The phenomenon that provokes curiosity is that a DNA helicase, AtRecQ2, which takes part in replication related phenomena like genome stability, D-loop and Holliday structure disruption, turned out to be MED34 (Kobbe et al., 2008). The Arabidopsis Mediator subunit, MED36/FIB2 has been shown to encode a Fibrillarin (FIB2), which is involved in rRNA processing (Barneche et al., 2000). It interacts with and is methylated by histone methyltransferases, AtPRMT1a and AtPRMT1b, and co-purified with RNA pol V (Yan et al., 2007; Huang et al., 2009). MED37a (also known as BiP) was first characterized as one of the HSP70 family members, and is homologous to yeast Ig-binding protein (Rose et al., 1989). It is involved in polar nuclei fusion during female gametophyte development, and is essential for the regulation of endosperm nuclei proliferation (Maruyama et al., 2010). In Arabidopsis, it also interacts with BR hormone receptor, BRI1, facilitating its proteasome-independent endoplasmic reticulum–associated degradation (ERAD) (Hong et al., 2008). Among the three AtPRP40s (Arabidopsis thaliana pre-mRNA processing protein 40), AtPRP40a has been recently named as AtMED35 of the Mediator complex. It interacts both with the phosphorylated and the unphosphorylated forms of the largest subunit of RNAP II. In Arabidopsis, it has its characteristic high expression level in roots and cauline leaves as compared to the other parts. The mutant does not show any phenotype, probably because of its redundancy with AtPRP40b and AtPRP40c (Kang et al., 2009).
The key importance of the Mediator complex lies in its ability to act as an adaptor molecule between transcription factors and the RNAP II, and hence the on-going research has so far been directed toward its role in the initial processes of transcription. The recent findings regarding its probable role in elongation and termination have not only expanded its arena of functionality, but have given fresh impetus toward the possibility of involvement of Mediator complex in other co-transcriptional processes like RNA processing (splicing, capping, polyadenylation), alternative splicing and epigenetic regulation (Table 3). Hence, the functional association of some Mediator subunits in these processes seems quite natural, and these issues need to be addressed more critically in future. As expected Mediator complex has critical control over miRNA and siRNA biogenesis as these are also transcribed by RNAP II. However, the association of other RNA polymerases with the Mediator complex, and its role in other RNA polymerase-mediated transcriptional events need to be examined further. Currently, we lack explanations for the Mediator subunits, which take part in phenomena like replication, protein degradation etc.
Mediator acts as an intermediary between the cis-element bound transcription factor and the RNAP II-mediated transcriptional machinery relaying the information from the transcription factor to the transcription apparatus. Recently, a couple of reports in plants have made the picture more complicated as the interaction between the transcription factor and the Mediator complex is not a simple binary one-one interaction. The Arabidopsis Mediator subunit, MED25 can interact with several transcription factors (DREB2A, ZFHD1, and MYB like proteins), that function in the same pathway. The mutants of all these three genes show increased sensitivity to salinity stress (Elfving et al., 2011). On the other hand, it has been also shown that AtMED25 can differentially control two seemingly different pathways, JA and ABA signaling, by interacting with two different transcription factors like MYC2 and ABI5, respectively (Chen et al., 2012). Similarly, AtMED18 has been shown controlling multiple plant responses by interacting with different transcription factors (Lai et al., 2014). An analogous situation also happens in yeast where different nuclear receptor-like transcription factors like Oaf1, Pdr1, and Pdr3 target the same Mediator subunit, MED15, to control different processes like fatty acid metabolism and multi-drug resistance (Thakur et al., 2008, 2009). On the contrary, a single characteristic/phenotype in plants can also be controlled by the concerted actions of more than one Mediator subunits (Tables 1, 2). Detailed investigation is needed to figure out whether these Mediator subunits do take part in the same developmental pathway while controlling a specific character or they control different developmental programs converging to a single phenotype. There are copious examples in animals where distinct Mediator subunits can control specific developmental and signaling pathways (Ito et al., 2000; Stevens et al., 2002; Ge et al., 2008). We suggest that the permutations and combinations of transcription factors with the Mediator subunits probably generate a Mediator code which dictates the downstream gene expression phenomena in co-ordination to the developmental stage and the prevailing environmental conditions. It might also involve Mediator complex undergoing a great deal of structural adjustment and alignment after binding with the transcription factors, which need to be studied in detail in the future.
The universality of the Mediator complex in the transcription of protein coding genes has ushered a new era in the understanding of transcriptional regulations in yeast and human. The plant science community is not lagging far behind in Mediator research. The achievement includes not only the first Mediator complex isolation from Arabidopsis but also the discovery of ubiquitous presence of Mediator complex in almost all the phyla of plant kingdom.
A general revelation from different studies is that the repertoire of Mediator subunits has been expanded in plant species to cope up with the increased number of plant transcription factors. This provides better resilience power to the sessile plants against the vagaries of the biotic and abiotic stresses. However, a note of caution should be shown regarding the discovery of new Mediator subunits. Until now there are no defined parameters to designate a protein as Mediator subunit. The Mediator acts as a scaffold for the interaction of a number of transcriptional regulatory proteins. Does mere co-purification with the Mediator complex qualify a protein to be regarded as Mediator subunit? Recently, six new plant-specific Mediator subunits (AtMED32–AtMED37) were discovered in Arabidopsis, but later AtMED32 and AtMED33 were found to be AtMED2 and AtMED5, respectively. As some of their functions are not directly related to Mediator functions or transcription (as for example, AtMED34 or AtRecQ2), concern has been expressed regarding how truly these proteins represent Mediator subunits.
Many of the Arabidopsis Mediator subunits were characterized earlier, but not in consideration of its Mediator membership. Over the time, several Mediator subunits have been characterized in Arabidopsis and many more may follow. In most of the cases, phenotypes of a particular Mediator subunit mutant has been described, but its association with transcription factors and the set of genes under its control are yet to be discovered in majority of the cases. What is lacking more is the understanding of how the Mediator subunits interact with components of the basic transcriptional machinery resulting in the regulated transcription.
Recently, many of the hitherto unknown but interesting functional aspects of Mediator has been unveiled in other organisms further broadening the horizon of its roles. Mediator not only takes part in the recruitment of RNAP II on the promoters of the active genes but also in transcription elongation and termination, chromatin remodeling, alternative splicing, small, and long non-coding RNA biogenesis, heterochromatin formation. All these developments are taking place in the arena of yeast and metazoan biology. Except characterization of few Mediator subunits, studies involving the Mediator complex as a whole or the mechanistic dexterity of Mediator complex in general or gene-specific regulation has not been addressed with proper emphasis and interest in plants. So, besides characterization of the every Mediator subunits in model species, attention should also be focused to address how the Mediator controls different steps of transcription in terms of mechanical intricacies.
Presence of more than one paralog has been reported for some Mediator subunits. Another level of complicacy may arise regarding which paralog remains with the complex, which most probably is controlled in a temporal and spatial manner. The presence of more than one paralog at a time in the Mediator complex has not been reported by any group. The more interesting question which has just been started to be answered is how stable is the Mediator structure in terms of its subunit composition. We postulate that the structure of Mediator complex changes depending on the composition of Mediator subunits, which again is controlled by different biotic and abiotic stimuli. Mediator complex isolation and its structural comparison from different stages of growth and development hold the key to the questions of how the structural shifts due to changes in Mediator composition are translated into transcriptomic changes of a species in response to intrinsic and extrinsic factors. Armed with the tools of modern molecular biology like TAP, MudPIT, LC-MS/MS, and HT-ChIP; the aforementioned questions are anticipated to be answered at an accelerated speed in near future.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Research in our lab is funded by NIPGR core grant and grants (IYBA grant BT/BI/12/045/2008 and BT/PR14519/BRB/10/869/2010) from Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India. SS acknowledges Research Associate Fellowship from DBT and Short-Term Research Fellowship from NIPGR. We would like to thank Dr. Pradipto Mukhopadhyay for his comments and suggestions which helped to improve the manuscript.
MED, Mediator; RNAP II, RNA Polymerase II; BR, Brassinosteroid; SA, Salicylic acid; JA, Jasmonic acid; ET, Ethylene; MudPIT, Multi-dimensional Protein Identification Technology; TAP, Tandem Affinity Purification; LC-MS/MS, Liquid Chromatography-Mass Spectrometry; HT-ChIP, High-Throughput Chromatin Immunoprecipitation.
Allen, B. L., and Taatjes, D. J. (2015). The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166. doi: 10.1038/nrm3951
An, C., and Mou, Z. (2013). The function of the Mediator complex in plant immunity. Plant Signal. Behav. 8:e23182. doi: 10.4161/psb.23182
Ansari, S. A., He, Q., and Morse, R. H. (2009). Mediator complex association with constitutively transcribed genes in yeast. Proc. Natl. Acad. Sci. U.S.A. 106, 16734–16739. doi: 10.1073/pnas.0905103106
Asturias, F. J., Jiang, Y. W., Myers, L. C., Gustafsson, C. M., and Kornberg, R. D. (1999). Conserved structures of Mediator and RNA polymerase II holoenzyme. Science 283, 985–987. doi: 10.1126/science.283.5404.985
Autran, D., Jonak, C., Belcram, K., Beemster, G. T. S., Kronenberger, J., Grandjean, O., et al. (2002). Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J. 21, 6036–6049. doi: 10.1093/emboj/cdf614
Backstrom, S., Elfving, N., Nilsson, R., Wingsle, G., and Bjorklund, S. (2007). Purification of a plant Mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26, 717–729. doi: 10.1016/j.molcel.2007.05.007
Baek, H. J., Kang, Y. K., and Roeder, R. G. (2006). Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J. Biol. Chem. 281, 15172–15181. doi: 10.1074/jbc.M601983200
Baek, H. J., Malik, S., Qin, J., and Roeder, R. G. (2002). Requirement of TRAP/Mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol. Cell. Biol. 22, 2842–2852. doi: 10.1128/MCB.22.8.2842-2852.2002
Barneche, F., Steinmetz, F., and Echeverría, M. (2000). Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 275, 27212–27220. doi: 10.1074/jbc.M002996200
Belakavadi, M., and Fondell, J. D. (2010). Cyclin-dependent kinase 8 positively cooperates with Mediator to promote thyroid hormone receptor-dependent transcriptional activation. Mol. Cell. Biol. 30, 2437–2448. doi: 10.1128/MCB.01541-09
Bonawitz, N. D., Kim, J. I., Tobimatsu, Y., Ciesielski, P. N., Anderson, N. A., Ximenes, E., et al. (2014). Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509, 376–380. doi: 10.1038/nature13084
Bonawitz, N. D., Soltau, W. L., Blatchley, M. R., Powers, B. L., Hurlock, A. K., Seals, L. A., et al. (2012). REF4 and RFR1, subunits of the transcriptional coregulatory complex Mediator, are required for phenylpropanoid homeostasis in Arabidopsis. J. Biol. Chem. 287, 5434–5445. doi: 10.1074/jbc.M111.312298
Bourbon, H.-M. (2008). Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional Mediator complex. Nucleic Acids Res. 36, 3993–4008. doi: 10.1093/nar/gkn349
Boyce, J. M., Knight, H., Deyholos, M., Openshaw, M. R., Galbraith, D. W., Warren, G., et al. (2003). The sfr6 mutant of Arabidopsis is defective in transcriptional activation via CBF/DREB1 and DREB2 and shows sensitivity to osmotic stress. Plant J. 34, 395–406. doi: 10.1046/j.1365-313X.2003.01734.x
Cai, G., Imasaki, T., Takagi, Y., and Asturias, F. J. (2009). Mediator structural conservation and implications for the regulation mechanism. Structure 17, 559–567. doi: 10.1016/j.str.2009.01.016
Caillaud, M. C., Asai, S., Rallapalli, G., Piquerez, S., Fabro, G., and Jones, J. D. (2013). A downy mildew effector attenuates salicylic Acid-triggered immunity in Arabidopsis by interacting with the host Mediator complex. PLoS Biol. 11:e1001732. doi: 10.1371/journal.pbio
Canet, J. V., Dobón, A., and Tornero, P. (2012). Non-recognition-of-BTH4, an Arabidopsis Mediator subunit homolog, is necessary for development and response to salicylic acid. Plant Cell 24, 4220–4235. doi: 10.1105/tpc.112.103028
Cantin, G. T., Stevens, J. L., and Berk, A. J. (2003). Activation domain-Mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc. Natl. Acad. Sci. U.S.A. 100, 12003–12008. doi: 10.1073/pnas.2035253100
Cerdán, P. D., and Chory, J. (2003). Regulation of flowering time by light quality. Nature 423, 881–885. doi: 10.1038/nature01636
Çevik, V., Kidd, B. N., Zhang, P., Hill, C., Kiddle, S., and Denby, K. J. (2012). MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 160, 541–555. doi: 10.1104/pp.112.202697
Chadick, J. Z., and Asturias, F. J. (2005). Structure of eukaryotic Mediator complexes. Trends Biochem. Sci. 30, 264–271. doi: 10.1016/j.tibs.2005.03.001
Chen, R., Jiang, H., Li, L., Zhai, Q., Qi, L., Zhou, W., et al. (2012). The Arabidopsis Mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24, 2898–2916. doi: 10.1105/tpc.112.098277
Cheng, B., Li, T., Rahl, P. B., Adamson, T. E., Loudas, N. B., Guo, J., et al. (2012). Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol. Cell 45, 38–50. doi: 10.1016/j.molcel.2011.10.022
Clay, N. K., and Nelson, T. (2005). The recessive epigenetic swellmap mutation affects the expression of two step II splicing factors required for the transcription of the cell proliferation gene STRUWWELPETER and for the timing of cell cycle arrest in the Arabidopsis leaf. Plant Cell 17, 1994–2008. doi: 10.1105/tpc.105.032771
Conaway, R. C., and Conaway, J. W. (2011). Function and regulation of the Mediator complex. Curr. Opin. Genet. Dev. 21, 225–230. doi: 10.1016/j.gde.2011.01.013
Conaway, R. C., and Conaway, J. W. (2013). The Mediator complex and transcription elongation. Biochim. Biophys. Acta 1829, 69–75. doi: 10.1016/j.bbagrm.2012.08.017
Conaway, R. C., Sato, S., Tomomori-Sato, C., Yao, T., and Conaway, J. W. (2005). The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30, 250–255. doi: 10.1016/j.tibs.2005.03.002
Crevillén, P., and Dean, C. (2011). Regulation of the floral repressor gene FLC: the complexity of transcription in a chromatin context. Curr. Opin. Plant Biol. 14, 38–44. doi: 10.1016/j.pbi.2010.08.015
Deato, M. D. E., Marr, M. T., Sottero, T., Inouye, C., Hu, P., and Tjian, R. (2008). MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol. Cell 32, 96–9105. doi: 10.1016/j.molcel.2008.09.009
De Lucia, F., and Dean, C. (2011). Long non-coding RNAs and chromatin regulation. Curr. Opin. Plant Biol. 14, 168–173. doi: 10.1016/j.pbi.2010.11.006
Dhawan, R., Luo, H., Foerster, A. M., Abuqamar, S., Du, H.-N., Briggs, S. D., et al. (2009). HISTONE MONOUBIQUITINATION1 Interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21, 1000–1019. doi: 10.1105/tpc.108.062364
Ding, N., Zhou, H., Esteve, P.-O., Chin, H. G., Kim, S., Xu, X., et al. (2008). Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol. Cell 31, 347–359. doi: 10.1016/j.molcel.2008.05.023
Dong, X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–552. doi: 10.1016/j.pbi.2004.07.005
Donner, A. J., Ebmeier, C. C., Taatjes, D. J., and Espinosa, J. M. (2010). CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 17, 194–201. doi: 10.1038/nsmb.1752
Donner, A. J., Szostek, S., Hoover, J. M., and Espinosa, J. M. (2007). CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol. Cell 27, 121–133. doi: 10.1016/j.molcel.2007.05.026
Dotson, M. R., Yuan, C. X., Roeder, R. G., Myers, L. C., Gustafsson, C. M., Jiang, Y. W., et al. (2000). Structural organization of yeast and mammalian Mediator complexes. Proc. Natl. Acad. Sci. U.S.A. 97, 14307–14310. doi: 10.1073/pnas.260489497
Elfving, N., Davoine, C., Benlloch, R., Blomberg, J., Brännström, K., Muller, D., et al. (2011). The Arabidopsis thaliana Med25 Mediator subunit integrates environmental cues to control plant development. Proc. Natl. Acad. Sci. U.S.A. 108, 8245–8250. doi: 10.1073/pnas.1002981108
Elmlund, H., Baraznenok, V., Lindahl, M., Samuelsen, C. O., Koeck, P. J. B., Holmberg, S., et al. (2006). The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 103, 15788–15793. doi: 10.1073/pnas.0607483103
Flanagan, P. M., Kelleher, R. J., Sayre, M. H., Tschochner, H., and Kornberg, R. D. (1991). A Mediator required for activation of RNA polymerase II transcription in vitro. Nature 350, 436–438. doi: 10.1038/350436a0
Fondell, J. D., Ge, H., and Roeder, R. G. (1996). Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. U.S.A. 93, 8329–8333.
Fukasawa, R., Tsutsui, T., Hirose, Y., Tanaka, A., and Ohkuma, Y. (2012). Mediator CDK subunits are platforms for interactions with various chromatin regulatory complexes. J. Biochem. 152, 241–249. doi: 10.1093/jb/mvs065
Galbraith, M. D., Allen, M. A., Bensard, C. L., Wang, X., Schwinn, M. K., Qin, B., et al. (2013). HIF1A employs CDK8-Mediator to stimulate RNAPII elongation in response to hypoxia. Cell 153, 1327–1339. doi: 10.1016/j.cell.2013.04.048
Ge, K., Cho, Y.-W., Guo, H., Hong, T. B., Guermah, M., Ito, M., et al. (2008). Alternative mechanisms by which Mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor gamma-stimulated adipogenesis and target gene expression. Mol. Cell. Biol. 28, 1081–1091. doi: 10.1128/MCB.00967-07
Gillmor, C. S., Park, M. Y., Smith, M. R., Pepitone, R., Kerstetter, R. A., and Poethig, R. S. (2010). The MED12-MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development 137, 113–122. doi: 10.1242/dev.043174
Gillmor, C. S., Silva-Ortega, C. O., Willmann, M. R., Buendía-Monreal, M., and Poethig, R. S. (2014). The Arabidopsis Mediator CDK8 module genes CCT (MED12) and GCT (MED13) are global regulators of developmental phase transitions. Development 141, 4580–4589. doi: 10.1242/dev.111229
Gonzalez, D., Bowen, A. J., Carroll, T. S., and Conlan, R. S. (2007). The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and Mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol. Cell. Biol. 27, 5306–5315. doi: 10.1128/MCB.01912-06
Hemsley, P. A., Hurst, C. H., Kaliyadasa, E., Lamb, R., Knight, M. R., De Cothi, E. A., et al. (2014). The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell 26, 465–484. doi: 10.1105/tpc.113.117796
Holstege, F. C., Jennings, E. G., Wyrick, J. J., Lee, T. I., Hengartner, C. J., Green, M. R., et al. (1998). Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728. doi: 10.1016/S0092-8674(00)81641-4
Hong, Z., Jin, H., Tzfira, T., and Li, J. (2008). Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20, 3418–3429. doi: 10.1105/tpc.108.061879
Huang, L., Jones, A. M. E., Searle, I., Patel, K., Vogler, H., Hubner, N. C., et al. (2009). An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat. Struct. Mol. Biol. 16, 91–93. doi: 10.1038/nsmb
Huang, Y., Li, W., Yao, X., Lin, Q.-J., Yin, J.-W., Liang, Y., et al. (2012). Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol. Cell 45:459–469. doi: 10.1016/j.molcel.2011.12.022
Imura, Y., Kobayashi, Y., Yamamoto, S., Furutani, M., Tasaka, M., Abe, M., et al. (2012). CRYPTIC PRECOCIOUS/MED12 is a novel flowering regulator with multiple target steps in Arabidopsis. Plant Cell Physiol. 53, 287–303. doi: 10.1093/pcp/pcs002
Iñigo, S., Alvarez, M. J., Strasser, B., Califano, A., and Cerdán, P. D. (2012a). PFT1, the MED25 subunit of the plant Mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J. 69, 601–612. doi: 10.1111/j.1365-313X.2011.04815.x
Iñigo, S., Giraldez, A. N., Chory, J., and Cerdán, P. D. (2012b). Proteasome-mediated turnover of Arabidopsis MED25 is coupled to the activation of FLOWERING LOCUS T transcription. Plant Physiol. 160, 1662–1673. doi: 10.1104/pp.112.205500
Ito, J., Sono, T., Tasaka, M., and Furutani, M. (2011). MACCHI-BOU 2 is required for early embryo patterning and cotyledon organogenesis in Arabidopsis. Plant Cell Physiol. 52, 539–552. doi: 10.1093/pcp/pcr013
Ito, M., Yuan, C. X., Malik, S., Gu, W., Fondell, J. D., Yamamura, S., et al. (1999). Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell 3, 361–370. doi: 10.1016/S1097-2765(00)80463-3
Ito, M., Yuan, C. X., Okano, H. J., Darnell, R. B., and Roeder, R. G. (2000). Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5, 683–693. doi: 10.1016/S1097-2765(00)80247-6
Jishage, M., Malik, S., Wagner, U., Uberheide, B., Ishihama, Y., Hu, X., et al. (2012). Transcriptional regulation by Pol II(G) involving Mediator and competitive interactions of Gdown1 and TFIIF with Pol II. Mol. Cell 45, 51–63. doi: 10.1016/j.molcel.2011.12.014
Johnson, K. M., and Carey, M. (2003). Assembly of a Mediator/TFIID/TFIIA complex bypasses the need for an activator. Curr. Biol. 13, 772–777. doi: 10.1016/S0960-9822(03)00283-5
Kagey, M. H., Newman, J. J., Bilodeau, S., Zhan, Y., Orlando, D. A., van Berkum, N. L., et al. (2010). Mediator and cohesin connect gene expression and chromatin architecture. Nature 467:430–435. doi: 10.1038/nature09380
Kang, C. H., Feng, Y., Vikram, M., Jeong, I. S., Lee, J. R., Bahk, J. D., et al. (2009). Arabidopsis thaliana PRP40s are RNA polymerase II C-terminal domain-associating proteins. Arch. Biochem. Biophys. 484, 30–38. doi: 10.1016/j.abb.2009.01.004
Kelleher, R. J., Flanagan, P. M., and Kornberg, R. D. (1990). A novel Mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61, 1209–1215. doi: 10.1016/0092-8674(90)90685-8
Kidd, B. N., Cahill, D. M., Manners, J. M., Schenk, P. M., and Kazan, K. (2011). Diverse roles of the Mediator complex in plants. Semin. Cell Dev. Biol. 22, 741–748. doi: 10.1016/j.semcdb.2011.07.012
Kidd, B. N., Edgar, C. I., Kumar, K. K., Aitken, E. A., Schenk, P. M., Manners, J. M., et al. (2009). The Mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21, 2237–2252. doi: 10.1105/tpc.109.066910
Kim, Y. J., Björklund, S., Li, Y., Sayre, M. H., and Kornberg, R. D. (1994). A multiprotein Mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599–608. doi: 10.1016/0092-8674(94)90221-6
Kim, Y. J., Zheng, B., Yu, Y., Won, S. Y., Mo, B., and Chen, X. (2011). The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 30, 814–822. doi: 10.1038/emboj.2011.3
Knight, H., Mugford, S. G., Ulker, B., Gao, D., Thorlby, G., and Knight, M. R. (2009). Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J. 58, 97–108. doi: 10.1111/j.1365-313X.2008.03763.x
Knight, H., Thomson, A. J. W., and McWatters, H. G. (2008). Sensitive to freezing6 integrates cellular and environmental inputs to the plant circadian clock. Plant Physiol. 148, 293–303. doi: 10.1104/pp.108.123901
Knight, H., Veale, E. L., Warren, G. J., and Knight, M. R. (1999). The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell 11, 875–886. doi: 10.1105/tpc.11.5.875
Knuesel, M. T., Meyer, K. D., Bernecky, C., and Taatjes, D. J. (2009). The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 23, 439–451. doi: 10.1101/gad.1767009
Kobbe, D., Blanck, S., Demand, K., Focke, M., and Puchta, H. (2008). AtRECQ2, a RecQ helicase homologue from Arabidopsis thaliana, is able to disrupt various recombinogenic DNA structures in vitro. Plant J. 55, 397–405. doi: 10.1111/j.0960-7412.2008.03511.x
Kornberg, R. D. (2005). Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30, 235–239. doi: 10.1016/j.tibs.2005.03.011
Krichevsky, A., Zaltsman, A., Kozlovsky, S. V., Tian, G.-W., and Citovsky, V. (2009). Regulation of root elongation by histone acetylation in Arabidopsis. J. Mol. Biol. 385:45–50. doi: 10.1016/j.jmb.2008.09.040
Lai, F., Orom, U. A., Cesaroni, M., Beringer, M., Taatjes, D. J., Blobel, G. A., et al. (2013). Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494, 497–501. doi: 10.1038/nature11884
Lai, Z., Schluttenhofer, C. M., Bhide, K., Shreve, J., Thimmapuram, J., Lee, S. Y., et al. (2014). MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat. Commun. 5, 3064. doi: 10.1038/ncomms4064
Leal, A., Huehne, K., Bauer, F., Sticht, H., Berger, P., Suter, U., et al. (2009). Identification of the variant Ala335Val of MED25 as responsible for CMT2B2: molecular data, functional studies of the SH3 recognition motif and correlation between wild-type MED25 and PMP22 RNA levels in CMT1A animal models. Neurogenetics 10, 275–287. doi: 10.1007/s10048-009-0183-3
Lee, T. I., and Young, R. A. (2000). Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34, 77–7137. doi: 10.1146/annurev.genet.34.1.77
Levine, M., and Tjian, R. (2003). Transcription regulation and animal diversity. Nature 424, 147–151. doi: 10.1038/nature01763
Li, W., Yoshida, A., Takahashi, M., Maekawa, M., Kojima, M., Sakakibara, H., et al. (2014). SAD1, an RNA polymerase I subunit A34.5 of rice, interacts with Mediator and controls various aspects of plant development. Plant J. 81, 282–291. doi: 10.1111/tpj.12725
Li, Y., Zheng, L., Corke, F., Smith, C., and Bevan, M. W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22, 1331–1336. doi: 10.1101/gad.463608
Liu, Z., and Myers, L. C. (2012). Med5(Nut1) and Med17(Srb4) are direct targets of Mediator histone H4 tail interactions. PLoS ONE 7:e38416. doi: 10.1371/journal.pone.0038416
Malik, S., Barrero, M. J., and Jones, T. (2007). Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Proc. Natl. Acad. Sci. U.S.A. 104, 6182–6187. doi: 10.1073/pnas.0608717104
Malik, S., and Roeder, R. G. (2010). The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772. doi: 10.1038/nrg2901
Maruyama, D., Endo, T., and Nishikawa, S. (2010). BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 1684–1689. doi: 10.1073/pnas.0905795107
Mathur, S., Vyas, S., Kapoor, S., and Tyagi, A. K. (2011). The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol. 157, 1609–1627. doi: 10.1104/pp.111.188300
Mittler, G., Kremmer, E., Timmers, H. T., and Meisterernst, M. (2001). Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2, 808–813. doi: 10.1093/embo-reports/kve186
Mukundan, B., and Ansari, A. (2011). Novel role for Mediator complex subunit Srb5/Med18 in termination of transcription. J. Biol. Chem. 286, 37053–37057. doi: 10.1074/jbc.C111.295915
Mukundan, B., and Ansari, A. (2013). Srb5/Med18-mediated termination of transcription is dependent on gene looping. J. Biol. Chem. 288, 11384–11394. doi: 10.1074/jbc.M112.446773
Myers, L. C., and Kornberg, R. D. (2000). Mediator of transcriptional regulation. Annu. Rev. Biochem. 69, 729–749. doi: 10.1146/annurev.biochem.69.1.729
Oya, E., Kato, H., Chikashige, Y., Tsutsumi, C., Hiraoka, Y., and Murakami, Y. (2013). Mediator directs co-transcriptional heterochromatin assembly by RNA interference-dependent and -independent pathways. PLoS Genet. 9:e1003677. doi: 10.1371/journal.pgen.1003677
Pasrija, R., and Thakur, J. K. (2012). Analysis of differential expression of Mediator subunit genes in Arabidopsis. Plant Signal. Behav. 7, 1676–1686. doi: 10.4161/psb.22438
Pasrija, R., and Thakur, J. K. (2013). Tissue specific expression profile of Mediator genes in Arabidopsis. Plant Signal. Behav. 8:e23983. doi: 10.4161/psb.23983
Poss, Z. C., Ebmeier, C. C., and Taatjes, D. J. (2013). The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 48, 575–608. doi: 10.3109/10409238.2013.840259
Raya-González, J., Ortiz-Castro, R., Ruíz-Herrera, L. F., Kazan, K., and López-Bucio, J. (2014). Phytochrome and flowering time1/Mediator25 regulates lateral root formation via auxin signaling in arabidopsis. Plant Physiol. 165, 880–894. doi: 10.1104/pp.114.239806
Rose, M. D., Misra, L. M., and Vogel, J. P. (1989). KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57, 1211–1221. doi: 10.1016/0092-8674(89)90058-5
Samuelsen, C. O., Baraznenok, V., Khorosjutina, O., Spahr, H., Kieselbach, T., Holmberg, S., et al. (2003). TRAP230/ARC240 and TRAP240/ARC250 Mediator subunits are functionally conserved through evolution. Proc. Natl. Acad. Sci. U.S.A. 100, 6422–6427. doi: 10.1073/pnas.1030497100
Sato, S., Tomomori-Sato, C., Parmely, T. J., Florens, L., Zybailov, B., Swanson, S. K., et al. (2004). A set of consensus mammalian Mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14, 685–691. doi: 10.1016/j.molcel.2004.05.006
Spahr, H., Khorosjutina, O., Baraznenok, V., Linder, T., Samuelsen, C. O., Hermand, D., et al. (2003). Mediator influences Schizosaccharomyces pombe RNA polymerase II-dependent transcription in vitro. J. Biol. Chem. 278, 51301–51306. doi: 10.1074/jbc.M306750200
Stevens, J. L., Cantin, G. T., Wang, G., Shevchenko, A., Shevchenko, A., and Berk, A. J. (2002). Transcription control by E1A and MAP kinase pathway via Sur2 Mediator subunit. Science 296, 755–758. doi: 10.1126/science.1068943
Sundaravelpandian, K., Chandrika, N. N. P., and Schmidt, W. (2013). PFT1, a transcriptional Mediator complex subunit, controls root hair differentiation through reactive oxygen species (ROS) distribution in Arabidopsis. New Phytol. 197, 151–161. doi: 10.1111/nph.12000
Taatjes, D. J. (2010). The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem. Sci. 35, 315–322. doi: 10.1016/j.tibs.2010.02.004
Takahashi, H., Parmely, T. J., Sato, S., Tomomori-Sato, C., Banks, C. A. S., Kong, S. E., et al. (2011). Human Mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–9104. doi: 10.1016/j.cell.2011.06.005
Takahashi, H., Takigawa, I., Watanabe, M., Anwar, D., Shibata, M., Tomomori-Sato, C., et al. (2015). MED26 regulates the transcription of snRNA genes through the recruitment of little elongation complex. Nat. Commun. 6, 5941. doi: 10.1038/ncomms6941
Thakur, J. K., Agarwal, P., Parida, S., Bajaj, D., and Pasrija, R. (2013). Sequence and expression analyses of KIX domain proteins suggest their importance in seed development and determination of seed size in rice, and genome stability in Arabidopsis. Mol. Genet. Genomics 288, 329–346. doi: 10.1007/s00438-013-0753-9
Thakur, J. K., Arthanari, H., Yang, F., Chau, K. H., Wagner, G., and Näär, A. M. (2009). Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J. Biol. Chem. 284, 4422–4428. doi: 10.1074/jbc.M808263200
Thakur, J. K., Arthanari, H., Yang, F., Pan, S.-J., Fan, X., Breger, J., et al. (2008). A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452, 604–609. doi: 10.1038/nature06836
Thompson, C. M., Koleske, A. J., Chao, D. M., and Young, R. A. (1993). A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73, 1361–1375. doi: 10.1016/0092-8674(93)90362-T
Thompson, C. M., and Young, R. A. (1995). General requirement for RNA polymerase II holoenzymes in vivo. Proc. Natl. Acad. Sci. U.S.A. 92, 4587–4590.
Tsai, K.-L., Tomomori-Sato, C., Sato, S., Conaway, R. C., Conaway, J. W., and Asturias, F. J. (2014). Subunit architecture and functional modular rearrangements of the transcriptional Mediator complex. Cell 157, 1430–1444. doi: 10.1016/j.cell.2014.05.015
Tsutsui, T., Fukasawa, R., Shinmyouzu, K., Nakagawa, R., Tobe, K., Tanaka, A., et al. (2013). Mediator complex recruits epigenetic regulators via its two cyclin-dependent kinase subunits to repress transcription of immune response genes. J. Biol. Chem. 288, 20955–20965. doi: 10.1074/jbc.M113.486746
Wang, G., Balamotis, M. A., Stevens, J. L., Yamaguchi, Y., Handa, H., and Berk, A. J. (2005). Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell 17, 683–694. doi: 10.1016/j.molcel.2005.02.010
Wang, G., Cantin, G. T., Stevens, J. L., and Berk, A. J. (2001). Characterization of Mediator complexes from HeLa cell nuclear extract. Mol. Cell. Biol. 21, 4604–4613. doi: 10.1128/MCB.21.14.4604-4613.2001
Wang, W., and Chen, X. (2004). HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131, 3147–3156. doi: 10.1242/dev.01187
Wang, X., Sun, Q., Ding, Z., Ji, J., Wang, J., Kong, X., et al. (2014). Redefining the modular organization of the core Mediator complex. Cell Res. 24, 796–808. doi: 10.1038/cr.2014.64
Warren, G., McKown, R., Marin, A. L., and Teutonico, R. (1996). Isolation of mutations affecting the development of freezing tolerance in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 111, 1011–1019. doi: 10.1104/pp.111.4.1011
Wathugala, D. L., Hemsley, P. A., Moffat, C. S., Cremelie, P., Knight, M. R., and Knight, H. (2012). The Mediator subunit SFR6/MED16 controls defense gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol. 195, 217–230. doi: 10.1111/j.1469-8137.2012.04138.x
Wathugala, D. L., Richards, S. A., Knight, H., and Knight, M. R. (2011). OsSFR6 is a functional rice orthologue of SENSITIVE TO FREEZING-6 and can act as a regulator of COR gene expression, osmotic stress and freezing tolerance in Arabidopsis. New Phytol. 191, 984–995. doi: 10.1111/j.1469-8137.2011.03759.x
Woychik, N. A., and Hampsey, M. (2002). The RNA polymerase II machinery: structure illuminates function. Cell 108, 453–463. doi: 10.1016/S0092-8674(02)00646-3
Xu, R., and Li, Y. (2011). Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 138, 4545–4554. doi: 10.1242/dev.071423
Xu, R., and Li, Y. (2012). The Mediator complex subunit 8 regulates organ size in Arabidopsis thaliana. Plant Signal. Behav. 7, 182–183. doi: 10.4161/psb.18803
Yan, D., Zhang, Y., Niu, L., Yuan, Y., and Cao, X. (2007). Identification and characterization of two closely related histone H4 arginine 3 methyltransferases in Arabidopsis thaliana. Biochem. J. 408, 113–121. doi: 10.1042/BJ20070786
Yang, Y., Ou, B., Zhang, J., Si, W., Gu, H., Qin, G., et al. (2014). The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant J. 77, 838–851. doi: 10.1111/tpj.12440
Zhang, A., Liu, Z., and Myers, L. C. (2013a). Differential regulation of white-opaque switching by individual subunits of Candida albicans mediator. Eukaryot. Cell 12, 1293–1304. doi: 10.1128/EC.00137-13
Zhang, X., Wang, C., Zhang, Y., Sun, Y., and Mou, Z. (2012). The Arabidopsis Mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24, 4294–4309. doi: 10.1105/tpc.112.103317
Zhang, X., Yao, J., Zhang, Y., Sun, Y., and Mou, Z. (2013b). The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant J. 75, 484–497. doi: 10.1111/tpj.12216
Zhang, Y., Wu, H., Wang, N., Fan, H., Chen, C., Cui, Y., et al. (2014). Mediator subunit 16 functions in the regulation of iron uptake gene expression in Arabidopsis. New Phytol. 203, 770–783. doi: 10.1111/nph.12860
Zheng, Z., Guan, H., Leal, F., Grey, P. H., and Oppenheimer, D. G. (2013). Mediator subunit18 controls flowering time and floral organ identity in Arabidopsis. PLoS ONE 8:e53924. doi: 10.1371/journal.pone.0053924
Zhu, X., Zhang, Y., Bjornsdottir, G., Liu, Z., Quan, A., Costanzo, M., et al. (2011). Histone modifications influence Mediator interactions with chromatin. Nucleic Acids Res. 39, 8342–8354. doi: 10.1093/nar/gkr551
Zhu, Y., Schluttenhoffer, C. M., Wang, P., Fu, F., Thimmapuram, J., Zhu, J. K., et al. (2014). Cyclin-dependent kinase8 differentially regulates plant immunity to fungal pathogens through kinase-dependent and -independent functions in Arabidopsis. Plant Cell 26, 4149–4170. doi: 10.1105/tpc.114.128611
Keywords: transcription, RNA polymerase II, mediator complex, development, defense signaling, abiotic stress, Arabidopsis, rice
Citation: Samanta S and Thakur JK (2015) Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Front. Plant Sci. 6:757. doi: 10.3389/fpls.2015.00757
Received: 26 June 2015; Accepted: 04 September 2015;
Published: 17 September 2015.
Edited by:
Amita Pandey, University of Delhi South Campus, IndiaReviewed by:
Hong-Qing Ling, Institute of Genetics and Developmental Biology, ChinaCopyright © 2015 Samanta and Thakur. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jitendra K. Thakur, Plant Mediator Lab, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi-110067, India,anRoYWt1ckBuaXBnci5hYy5pbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.