- 1School of Computational and Integrative Sciences, Jawaharlal Nehru University, New Delhi, India
- 2Synthetic Biology and Biofuels Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, India
- 3Stress Physiology and Molecular Biology Laboratory, School of Life Sciences, Jawaharlal Nehru University, New Delhi, India
- 4Plant Molecular Biology Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, India
Two-component system (TCS) is one of the key signal sensing machinery which enables species to sense environmental stimuli. It essentially comprises of three major components, sensory histidine kinase proteins (HKs), histidine phosphotransfer proteins (Hpts), and response regulator proteins (RRs). The members of the TCS family have already been identified in Arabidopsis and rice but the knowledge about their functional indulgence during various abiotic stress conditions remains meager. Current study is an attempt to carry out comprehensive analysis of the expression of TCS members in response to various abiotic stress conditions and in various plant tissues in Arabidopsis and rice using MPSS and publicly available microarray data. The analysis suggests that despite having almost similar number of genes, rice expresses higher number of TCS members during various abiotic stress conditions than Arabidopsis. We found that the TCS machinery is regulated by not only various abiotic stresses, but also by the tissue specificity. Analysis of expression of some representative members of TCS gene family showed their regulation by the diurnal cycle in rice seedlings, thus bringing-in another level of their transcriptional control. Thus, we report a highly complex and tight regulatory network of TCS members, as influenced by the tissue, abiotic stress signal, and diurnal rhythm. The insights on the comparative expression analysis presented in this study may provide crucial leads toward dissection of diverse role(s) of the various TCS family members in Arabidopsis and rice.
Introduction
Growth potential of the plants are severely affected under various abiotic stress conditions especially salinity and drought. Since plants are rooted to a place, they have to make adjustments in their genetic and metabolic machinery in order to survive under abiotic stress conditions. Under stress conditions, plants use specific signaling machineries to relay the stress signals in order to “switch on” the adaptive responses which assist plants in developing tolerance toward abiotic stress. Some of the signaling machineries are conserved across various genera. One such signaling machinery is the two-component system (TCS) or His-to-Asp phosphorelay which is well-known and conserved machinery for signal transduction in the cells (Mochida et al., 2010; Nongpiur et al., 2012). Apart from stress signaling, TCS has been one of the key regulators for many biological processes such as cell division, cell growth and proliferation, and responses to growth regulators in both prokaryotic and eukaryotic cells (Stock et al., 2000; Hwang et al., 2002; Mizuno, 2005; Pareek et al., 2006; Schaller et al., 2008; Pils and Heyl, 2009).
The TCS signaling system essentially comprise of sensory histidine kinases (HKs) and their cognate response regulators (RRs) substrates, which have been reported in almost all the sequenced bacterial genomes except for mycoplasma (Mascher et al., 2006; Laub and Goulian, 2007). In a simple prototypical TCS regulatory system, HK protein senses the environmental signals, autophosphorylates a histidine residue (H), and signals to its corresponding cytosolic RR protein by transferring the phosphate to an aspartate residue (D) (Figure 1). Phosphorylated RR further mediates the downstream signaling (Urao et al., 2000). Some of the bacteria, yeast, slime molds and plants possess a more complex form of TCS or His-to-Asp phosphorelay system. This is due to the presence of “hybrid” type of kinases which possess both His-kinase (HK) domain and a receiver domain (RD) in one protein. Another protein namely, His-containing phosphotransfer (Hpt) protein is involved which acts as a signaling module connecting to the final RRs (Schaller et al., 2008). Hpts allows species to have multistep phosphorelays which has a major advantage of having multiple regulatory checkpoints for signal crosstalk or negative regulation by specific phosphatases (Urao et al., 2000).
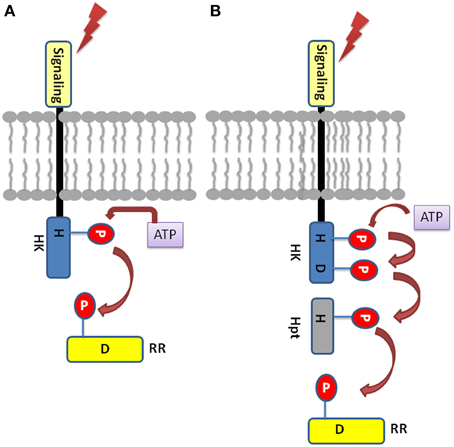
Figure 1. Basic Two-component system architecture (A) A simple TCS member: The sensory domain of histidine kinase sense the extracellular signal which leads to the phosphorylation of the conserved His in its transmitter domain. Further, the conserved Asp in the receiver domain of the RR is autophosphorylated resulting in signal output. (B) Hybrid-type TCS member: The conserved His and Asp are present in the same protein which is membrane bound. The Hpt protein acts as mediator for the transfer of the phosphoryl group between the HK and the RR.
Structurally, Histidine kinase (HK) protein is a dimeric protein and is regulated by receptor-ligand interactions (Grebe and Stock, 1999; Koretke et al., 2000). The HK proteins consist of highly conserved domains, the dimerization and histidine phosphotransfer domain (DHp), which contains the conserved histidine, and the catalytic and ATP binding (CA) domain (Cheung and Hendrickson, 2010). Apart from other functional domains, the HK protein has a sensory domains like HAMP (Histidine Kinases, adenyl cyclases, methyl accepting proteins, and phosphatases), GAF (cGMP-specific phosphodiesterases, adenyl cyclases and FhlA), PAS (Per Arnt Sim), and phytochrome domains for sensing wide range of environmental cues. Response regulators share a common, well conserved receiver domain RD that catalyzes phosphotransfer from its cognate HK (Capra and Laub, 2012). The differential gene expression is the consequence of protein-protein interaction or protein-DNA interaction which is mediated by the C-terminal effector (or output) domain of the RR thus giving rise to the appropriate cellular response (Mascher et al., 2006). In hybrid type kinases, phosphate is first transferred from the histidine residue in the transmitter to the aspartate residue of the attached RD, then to a histidine residue on a histidine phosphotransfer domain (Figure 1). Finally, the phosphate is relayed from the Hpt domain to the RD of a down-stream response regulator protein (RRs), which results in the output response. Based on highly conserved residues which HK proteins possess conserved sequence fingerprints, namely H, N, D, F, and G-boxes can be identified. The H-box bears the histidine that get phosphorylated while the N, D, F, and G-boxes are located at the ATP binding site (Kofoid and Parkinson, 1988; Stock et al., 1988, 1995).
Several plant species, including model plant Arabidopsis, are known to possess TCS signaling machinery (Hwang and Sheen, 2001; Grefen and Harter, 2004). Crucial processes such as cytokinin signaling, ethylene signaling, and light perception involves members of the TCS (Hwang et al., 2002). The presence of TCS system in eukaryotes was anticipated in Arabidopsis with the characterization of ethylene receptor ETR1 (Chang et al., 1993), photoreceptors (Schneider-Poetsch, 1992; Li et al., 2011) and yeast osmosensor SLN1 (Ota and Varshavsky, 1993) which was earlier considered to be restricted only to prokaryotes. The characterization of multi-step TCS machinery in Arabidopsis as the key element of plant cytokinin signaling revealed TCS machinery in plants (To and Kieber, 2008). In Arabidopsis, AtHK1 of the TCS family is indicated to be involved in the osmosensing mechanism (Urao et al., 1999). Earlier, we have performed whole genome analysis of the TCS members in rice in comparison to Arabidopsis (Pareek et al., 2006). Further, current advances have shown role of TCS machinery in various environmental stresses such as drought, cold, osmotic stress and abscisic acid (ABA) (Tran et al., 2010; Ha et al., 2011; Hwang et al., 2012).
The current investigation presents the comprehensive expression analysis of various members of TCS in Arabidopsis thaliana and Oryza sativa using massively parallel signature sequencing (MPSS) and publicly available microarray data under various abiotic stress conditions. The analysis would be able to enhance our understanding about the role of TCS members in the two plant species.
Materials and Methods
Search of TCS Members in Oryza sativa and Arabidopsis
Earlier, all the members of TCS signaling machinery were identified and characterized in rice (TIGR rice database version 4.0) and were compared to the TCS members present in Arabidopsis (Hwang and Sheen, 2001; Grefen and Harter, 2004). The TCS signaling members were retrieved for Arabidopsis and rice as done earlier (Pareek et al., 2006) using TIGR rice database version 7.0 and TAIR version 10 for Arabidopsis, in order to rule out any new member or deleted member protein from the updated genome database versions.
Analysis of MPSS Database for Expression Profiles
With the representation of more number of signature libraries in the MPSS database (Brenner et al., 2000), we have extracted expression evidence from the most recent MPSS tags for both Arabidopsis and Oryza gene models (Database release 2008). With high specificity, the signature sequence uniquely represents a gene and shows perfect match (100% identity over 100% length of the tag). The expression of the gene is estimated by the normalized abundance (tags per million, tpm) of specific signatures in a given library. Class 1, 2, 5, and 7 were used for sense coding sets while Class 3 and 6 were used for antisense coding sets. For both the genomes, 20-nt tags were used for determining the tissue specific expression of TCS members.
In Arabidopsis, the tissue specific signature libraries considered for analysis are as follows: for Callus—CAF, CAS; for inflorescence—INF, AP1, AP3, AGM, INS, SAP; for leaves—LEF, LES, S04, S52; for roots—ROF, ROS; for Silique—SIF, SIS; for seeds—GSE. These libraries were earlier considered for analysis of CBS family protein in Arabidopsis (Kushwaha et al., 2009).
In rice, the tissue specific signature libraries considered for analysis are as follows: for leaves—I9LA, I9LB, I9LC, I9LD, FLA, FLB, FLC, FLD, NDL, NCL, NLA, NLB, NLC, NLD, NYL, NSL, PLA, PLW, PLC; for meristem—NME, I9ME, FME; for roots—I9RO, I9RR, FRO, FRR, NYR, NRA, NRB, NDR, NCR, NSR; for callus—NCA; for panicle—NIP; for stigma—NOS; for pollen—NPO; for stem—NST; for seeds—NGD, NGS, PSC, PSI, PSL, PSN, PSY. These libraries were earlier considered for analysis of CBS family protein in rice (Kushwaha et al., 2009). The quantitative values obtained for respective TCS genes were used for making the heatmap using open source R software.
Expression Analysis using Microarrays
In order to analyze the gene expression for various abiotic stress conditions the latest microarray data for cold, UV, wound, heat, genotoxic, drought, osmotic, salt, and oxidative stress were retrieved from the Arabidopsis Information Resource (Lamesch et al., 2012). The tissue (root and shoot) specific datasets were obtained for different time sets namely 30 min, 1 h, 3 h, 6 h, 12 h and 24 h of various abiotic stresses and analyzed, as performed earlier (Kushwaha et al., 2009). The pre-normalized data thus obtained, was used for analysis of fold change expression in Arabidopsis. The expression datasets for the rice were obtained from NCBI-GEO database (Supplementary Table 1). The microarray expression data was downloaded using Bioconductor package. The array quality of the experiment was assessed by MA and RNA degradation plot for individual arrays. The individual GEO raw data sets were normalized using RMA method. The normalized datasets were integrated and differentially expressed genes were identified using RankProd package in Bioconductor (Hong et al., 2006). The expression matrix thus obtained, was used to extract expression values for TCS members. Fold increase in transcript abundance under stress conditions were calculated with respect to their respective controls. The expression with respect to the control was calculated using in-house PERL programs. The hierarchical clustering analysis and the heatmaps were made using R software.
Plant Material and Growth Conditions
Seeds of Oryza sativa L. cv “IR-64” were washed with deionized water and allowed to germinate in half Yoshida medium (Yoshida et al., 1972) under hydroponic system with continuous air bubbling for 48 h in dark and then transferred to light for further growth for 14 days under control conditions (28 ± 2°C, 12 h light and 12 h dark cycle) in plant growth chamber, having 70% relative humidity. To find out the rhythmic expression of TCS genes in rice, shoot samples were harvested for 2 days at an interval of 3 h starting from the dawn of 15th day from rice seedlings grown under 12 h light/12 h dark cycle. After harvesting, each sample was immediately frozen in liquid nitrogen and stored at −80°C till further use.
Isolation of Total RNA and cDNA Synthesis
Total RNA was isolated from the harvested plant samples using RaFlex™ solution I and solution II (GeNei, India) as per the manufacturer's protocol. Two hundred milligram of each sample was used for RNA extraction. The quantity and quality of RNA was estimated by determining absorbance at a wavelength of 260 nm. Concentration of RNA was calculated using OD260 nm formula (OD260 = 1, corresponds to 40 μg/ml of RNA). Quality of RNA was checked by A260/A280 ratio. Five microgram of total RNA of each sample was checked by electrophoresis. EtBr stained formaldehyde agarose gel showed the presence of two distinct bands of 28S rRNA and 18S rRNA in each sample of total RNA. It confirmed the integrity of RNA of each sample which was then used for subsequent cDNA synthesis. First strand cDNA was synthesized using first strand cDNA synthesis kit (Fermentas). Total RNA was treated with DNase to remove genomic DNA contamination. For DNase treatment, 5 μg RNA was incubated with 1 unit of DNase in 1X buffer for 30 min at 37°C. The DNase was then denatured by heating at 75°C for 5 min. Before heating 1 μl of 25 mM EDTA, which works as a chelating agent, was added to the reaction mixture to prevent RNA break down. After this treatment, RNA samples were used for first strand cDNA synthesis. The primers of TCS members were designed using Primer 3 express (Applied biosystem, USA) and NCBI primer BLAST software. The sequences for these primers are listed in Supplementary Table 2. All Primers were specific to the unique regions in the 3′-UTR of their respective genes. These primers were rechecked for their uniqueness via primer BLAST at NCBI database. For primer designing the transcript nucleotide sequences were downloaded from TIGR rice database. Designed primers were ordered to Sigma-Aldrich, India for synthesis.
Quantitative RT-PCR
The rice translation elongation factor 1α (eEF-1α) gene was taken as the reference gene for the analysis. The quantitative RT-PCR reaction mixture contained 5 μl of 10 fold diluted cDNA, 10 μl of 2X SYBR Green PCR Master Mix (Applied Biosystems, USA), and 100 nM of each gene-specific primers in a final volume of 20 μl. No template controls (NTCs) were also taken for each primer pair. The real-time PCR reactions were performed employing ABI Prism 7500 Sequence Detection System and software (PE Applied Biosystems, USA). All the reactions of quantitative real-time PCR were performed under following conditions: 10 min at 95°C, and 40 cycles of denaturation at 95°C for 25 s, annealing and extension at 59°C for 1 min in 48-well optical reaction plates (Applied Biosystems, USA). The specificity of amplification was tested by dissociation curve analysis. The experiment was repeated with two biological replicates, each of them having three technical replicates. Data analysis was performed using ddCT method (Livak and Schmittgen, 2001).
Results
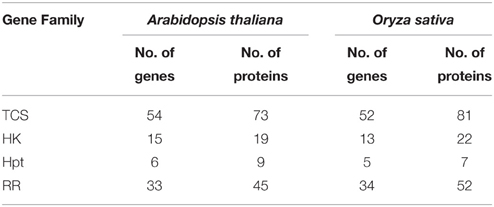
The analysis of TCS members has been carried out using latest version of genomes of Arabidopsis thaliana (TAIR ver. 10) and Oryza sativa (TIGR ver. 7) using pfam profiles (Pareek et al., 2006). In comparison to the earlier report (Pareek et al., 2006), some new members have been found in both Arabidopsis and rice, in the current analysis (Supplementary Tables 3A–F). Earlier analysis suggested 54 genes coding for 63 proteins in Arabidopsis while current analysis found 54 genes coding for 73 proteins. Similarly in O. sativa, 51 genes encoding 73 putative proteins was reported earlier but the current analysis showed 52 genes coding for 81 proteins (Table 1). The new members added to the list of histidine kinase have been named as histidine kinase like (OsHKL1) gene because of the presence of only histidine kinase domain in the protein sequence. The increase in the number of protein products has been attributed to the presence of more alternative spliced products. In order to avoid any ambiguity in nomenclature, the previous nomenclature has been retained as reported in Pareek et al. (2006).
Analysis of Expression Profiles for TCS Members in Arabidopsis and Rice using MPSS Database
Sensitive measures of expression of all genes in the genome can be assessed using MPSS (Brenner et al., 2000). MPSS has been used for the analysis of genome-level expression analysis in various plant systems including rice, Arabidopsis and grapes (Meyers et al., 2004). To extract information about the relative abundance of transcripts of TCS members in various tissues/organs of Arabidopsis and rice, analysis was carried out using MPSS database (http://mpss.udel.edu). Analysis of TCS-specific mRNA tags (measured as transcript per million; TPM) in various libraries showed considerable variability in their abundance in various tissues.
Among the HKs, ERS1 showed considerably large accumulation of transcripts in untreated 21-days roots while it showed moderate rise in transcripts in 28–48 h post fertilized silique, untreated 21-day leaf and actively growing callus in Arabidopsis (Figures 2A,B). Another HK, CRE1 showed moderate accumulation of transcripts in untreated 21-days roots. Low transcripts accumulation was observed in AHK2, AHK3, ETR1, PHYA, and PHYB in Arabidopsis. On the other hand, OsHK5 showed considerably high accumulation of transcripts in leaves. The transcripts accumulation was also observed in leaves, 60 days mature leaf for OsPHYc in rice. The accumulation of transcripts was also observed in 60 days mature roots for OsPHYA gene. Among the Hpts in Arabidopsis, AHP2 showed accumulation of transcripts in 24–48 h post fertilized silique while it shows moderate accumulation in callus. On the contrary, OsHpt3 in rice showed high accumulation in developing seeds (Figures 2C,D). For the RRs, in Arabidopsis, APRR1 showed high accumulation of transcripts in agamous inflorescence and leaves while ARR4 showed transcript abundance in callus. In rice, OsPRR3 and OsPRR1 showed accumulation of transcripts in beet armyworm damaged; water weevil damaged and mechanically damaged leaves. Another RR, OsRRA2 and OsRRA3 showed transcripts accumulation in 60 days mature leaves (Figures 2E,F). These observations suggest that the TCS-related transcriptome of Arabidopsis and rice is complex, showing a tissue-specific differential regulation.
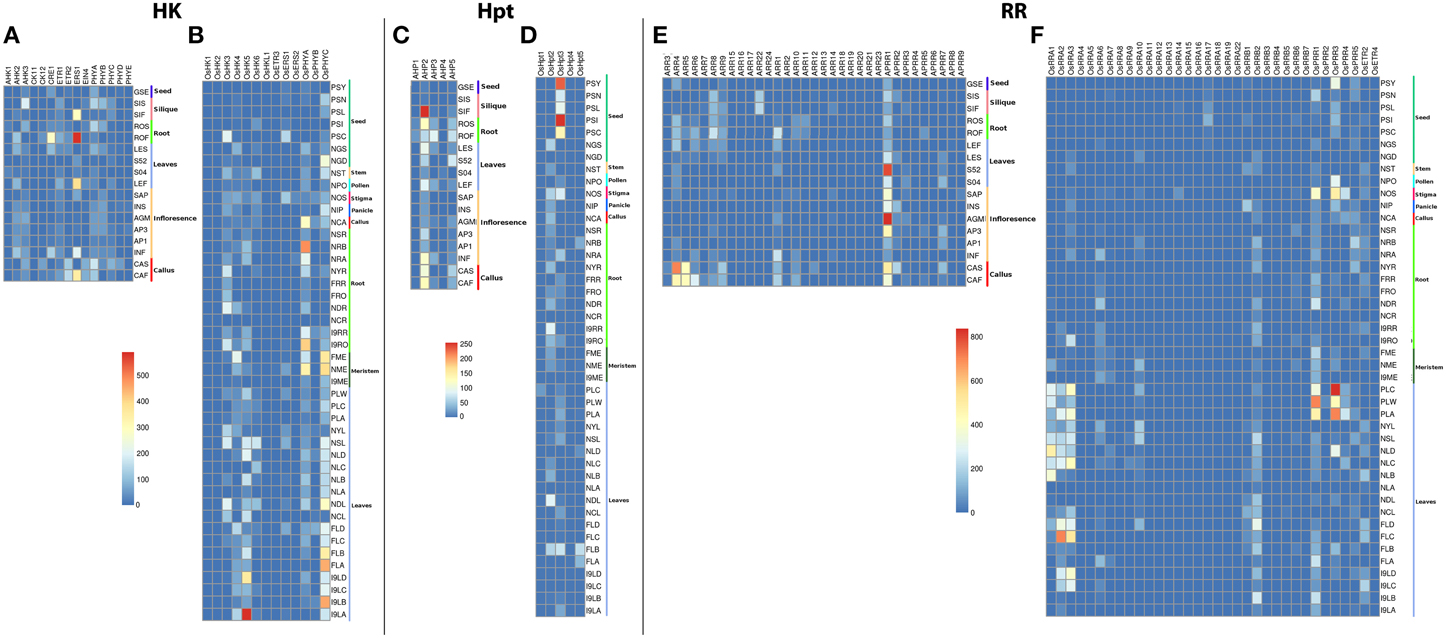
Figure 2. Heatmap depicting the tissue-specific transcript accumulation for Histidine Kinases (HKs) for (A) Arabidopsis and (B) rice, Histidine phosphotransfer proteins (Hpts) for (C) Arabidopsis and (D) rice, Response regulators (RRs) for (E) Arabidopsis and (F) rice. These heatmaps are based on the MPSS database. The heatmap was made using open source R software.
Analysis of Expression Profiles for TCS Members in Arabidopsis and Rice using Microarray
For analysis of expression of the TCS members in Arabidopsis, we used data available on the TAIR database consisting time series analysis performed in root and shoot tissues under various abiotic stress conditions while for rice, the expression was analyzed using various abiotic stress experiment data available at NCBI-GEO database.
Histidine Kinase Proteins (HKs)
The expression profile of the histidine kinase genes in root tissues of Arabidopsis under cold conditions showed downregulation of AHK1 and AHK2 genes at 24 h of stress, while at the other time spans, its expression remained unchanged. On the other hand, in shoots, AHK1 maintained a basal level of expression in all the cold stress time points but the transcripts for AHK2 showed two fold downregulation at 12 and 24 h of cold stress conditions (Figure 3A). Another cytokinin signaling gene, CK11 showed changing expression during all time-series. The gene was observed to be downregulated at 30 min of cold stress and two fold upregulated at 1 h of stress. The expression of this gene gets normalized only to get further upregulated by over two folds at 24 h of cold stress. Further, in shoots, CK11 gene showed similar behavior where the basal level of expression is maintained at 30 min of cold stress, which gets upregulated upto two folds in 1 h of stress. The expression further goes down two folds at 3 h of stress and again gets two fold upregulated at 6 h and 12 h of stress. Finally, it again shows downregulation at 24 h of cold stress (Figure 3A). CK11 and AHK1 have been found to play major role in the cytokinin signaling and osmosensing process respectively in Arabidopsis (Urao et al., 2000). In roots, ethylene receptor, ETR2 showed downregulated response as the time span of the cold stress increases from 30 min to 24 h while, in shoots, it shows an upregulated response till 12 h of cold stress and finally maintains a basal expression at 24 h of cold stress. In shoots, another member of TCS, namely AHK3, showed downregulated response at 24 h of cold stress and CK12 showed upregulation in 30 min, 1 h and 6 h of cold stress while it gets downregulated in 3 h of cold stress. ETR2 also showed upregulation in response to 12 h of cold stress. Among the photoreceptors, PHYC showed downregulation in 12 and 24 h of cold stress in both root and shoot tissues. In roots, photoreceptor PHYE showed upregulation in 6 h of cold stress, while it maintained basal level expression in the shoot tissues.
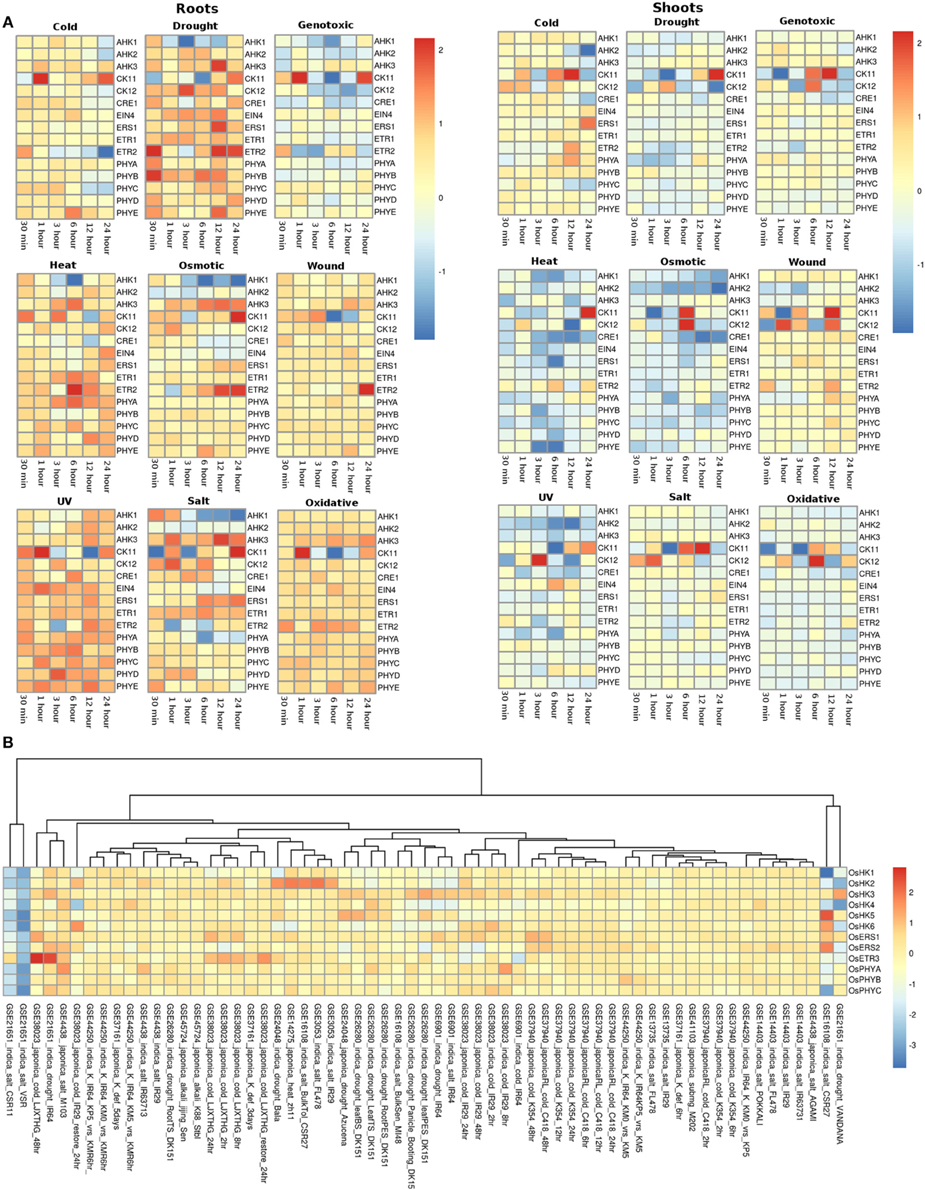
Figure 3. Heatmap depicting the stress-induced expression of Histidine Kinase (HKs) genes from root and shoot tissues obtained using microarray data for Arabidopsis (A) and O. sativa (B). The heatmap was made using open source R software.
Under drought conditions, in the root tissues, all the histidine kinase genes in Arabidopsis were found to be upregulated. AHK1 was found to be upregulated in 30 min of drought stress followed by its downregulation in 1, 3, 6, and 12 h of drought stress, after which it again got upregulated by two folds in 24 h. Ethylene receptor ETR2, showed two fold upregulation at 30 min, 12 h and 24 h of drought stress, photoreceptor PHYB showed upregulated response at 30 min of drought stress. On the other hand, in shoots of Arabidopsis, most of histidine kinase genes showed downregulation in response to drought stress. Only CK11 showed three fold upregulation in 12 h of drought stress and then got downregulated in continuation of drought stress for 24 h.
Under the genotoxic stress conditions, in root tissue, genes namely, AHK1, AHK2, CK12, CRE1, and ETR2 showed downregulated response. Rest all the members of the HK family showed minimal level of expression at all the time points in both root and shoot tissues. Only CK11 showed three fold upregulation in 1 h of genotoxic stress and got downregulated only to get upregulated again at 24 h of stress. In shoot tissues, CK11 gene showed upregulation at 6 and 12 h of genotoxic stress. Another gene, CK12 showed 1.5-fold upregulation at 6 h of stress.
Similar response of the members of the HK family was found in response to heat stress. In root tissues, all genes of HK family maintained minimal to high expression at all time points of the stress. Specifically, gene ETR2 showed three fold upregulation at 6 h of stress while AHK1 showed downregulation at 3 h and 6 h of heat stress condition. On the other hand, in shoot tissues, the entire HK family members showed exactly the reverse of the expression as it showed in root tissues, that is, mainly down expression for most of the members. Only, CK11 showed two fold upregulated expression at 24 h of heat stress.
Under osmotic stress, in root tissues, CK11 and ETR2 showed three fold upregulation during 24 h of osmotic stress while AHK1 showed downregulated expression during the 3, 6, 12, and 24 h of osmotic stress. CK11 and CK12 showed upregulation during 6 h of osmotic stress in the shoot tissues. All the other members showed similar level of expression as in the heat stress, that is, downregulation for most of the members. Under the wounding stress, CK11 and CK12 showed upregulated response in 1 h and 12 h of stress in the shoot tissues while in root tissues, only ETR2 showed two fold upregulation in the expression. Rest of the members of the HK family maintained unchanged expression levels at all time points in both root and shoot tissues.
Similar to heat and osmotic stress conditions, HK members showed minimal to high expression under UV stress in the root tissues while in shoot tissues, CK12 showed three fold upregulation in 3 h of stress. In roots, AHK1 showed downregulation after 6 h of UV stress condition. Further, under salt stress conditions, all the HK members were found to be upregulated except AHK1 and photoreceptor, PHYA in the root tissue while in shoot tissues, the expression of HK members remained unchanged with respect to the control conditions. CK11 and CK12 showed high expression in the salt stress condition in shoot tissues. Similar expression profile was observed in oxidative stress conditions in both root and shoot tissues. Overall, the HK family members in Arabidopsis appear to be more active in the root tissues than in the shoot tissues in all the stress conditions.
In rice (in all the experiments and genotypes of rice) histidine kinases (HKs) showed similar behavior of basal expression under cold conditions. Only in GEO dataset, GSE38023, ethylene receptors OsERS1, OsERS2, and OsETR3 showed one to two fold upregulation at various time points (Figure 3B). Under drought conditions, in indica genotype of rice Vandana (GSE21651), OsHK3 showed 1.5-fold upregulation while OsHK2 and OsHK3 showed similar fold downregulation in the expression. In IR64 genotype (GSE21651), OsETR3 showed two fold upregulation under drought stress. All the other members of histidine kinase gene family showed basal expression levels in all other experiments. In salt stress conditions, all the members of the HK gene family maintained basal expression in all the experimental conditions. In two of the rice genotypes, VSR and CSR11 all the HK members were found to be downregulated under salt stress conditions.
Histidine Phosphotransfer Proteins (Hpts)
The expression profiles of the Histidine phosphotransfer (Hpt) genes in Arabidopsis in root and shoot tissues appear to show reverse behavior as compared to the HK family members under all the stress conditions. Overall, Hpt family members appear to show higher expression in shoots than in roots under all the considered abiotic stress conditions (Figure 4A). Under osmotic stress conditions, in the root tissues, all the Hpt members appear to be downregulated except for AHP4 which showed three fold upregulation. Under osmotic stress, Hpt members namely AHP1, AHP2, and AHP3 showed downregulation at 30 min and 1 h of stress but they maintained a basic level of expression at other time points in the root tissues. While in shoot tissues, all the genes were observed to be expressed at all the time points especially, AHP4 and AHP6 showed high (two or three fold) upregulation under osmotic stress. Under drought stress conditions, all the Hpt members showed similar expression in root as well as shoot tissues. Again, AHP4 showed high upregulation at 1, 6, and 12 h of drought stress in roots and at 3 h in shoots. Under genotoxic conditions, all the members, with an exception of AHP4 showed downregulation in the root tissues. In shoot tissues, members of the Hpt family showed unchanged minimal expression with respect to the control conditions. Similar to cold stress, all the members showed downregulation all time points except for AHP4 which showed three fold upregulation during 1 h of stress in the root tissues. However, in the shoot tissues, all the Hpt members were observed to be upregulated except for AHP6 which showed two fold downregulation in 12 h cold stress (Figure 4A). On the other hand, all the members of Hpt family maintained minimal expression except for AHP4 which showed three fold upregulation at 30 min of heat stress. Under wounding stress, in roots, all the members were downregulated while in shoot tissues all the Hpt genes were upregulated. AHP4 was found to be downregulated under the wounding stress at all the time points in the shoot tissues. AHP1 showed higher expression under the wounding stress in the shoot tissues. Under the UV stress, Hpt members showed exactly reverse expression behavior in the root and shoot tissues with the expression of AHP4 showing periodic pattern in the expression. Under salt and oxidative stress conditions, the expression levels remained same in both root and shoot tissues showing that the salt stress nearly leads to the oxidative stress as well. Overall, it appears that the AHP4 gene of the Hpt family is most active members among all, showing expression in all the considered abiotic stress conditions.
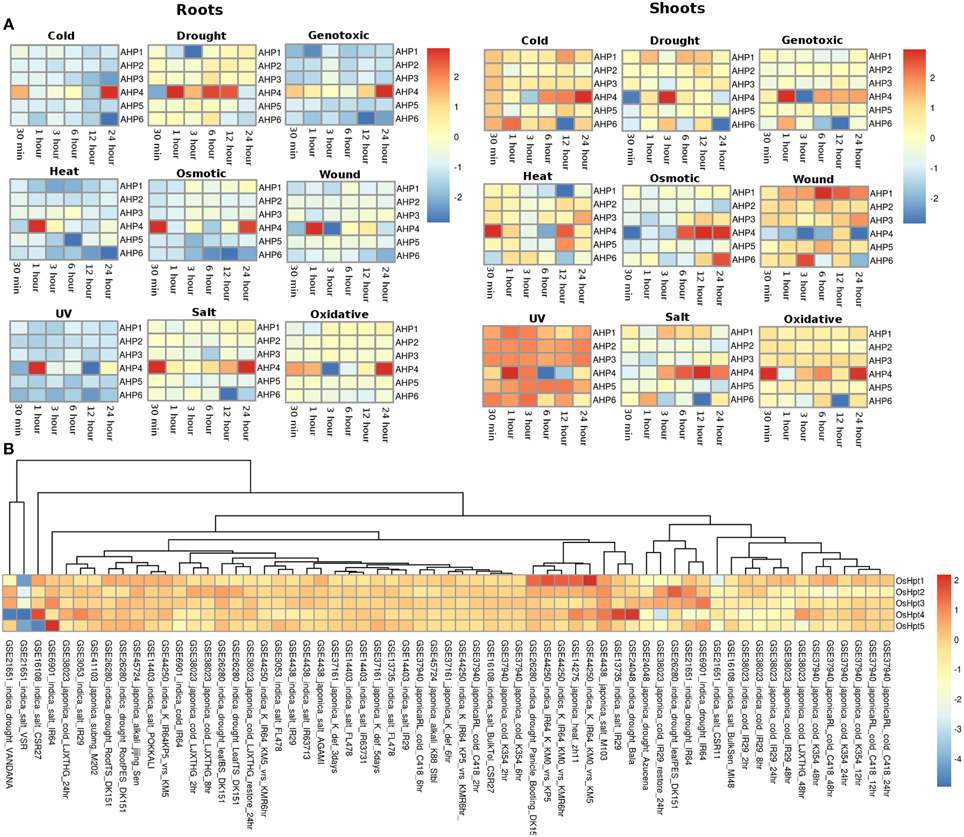
Figure 4. Heatmap depicting the stress-induced expression of Histidine phosphotransfer protein (Hpts) genes from root and shoot obtained using microarray data for Arabidopsis (A) and O. sativa (B). The heatmap was made using open source R software.
In rice, the members of the Hpt family showed expression in all the considered experimental conditions in all the considered genotype suggesting their potential importance in the rice plant (Figure 4B). OsHpt4 showed three fold expression in drought and salt stress conditions in Bala (GSE24048) and IR29 (GSE13735) genotypes of rice respectively. OsHpt4 showed three folds expression under salt stress in the CSR27 genotype of rice (GSE16108) while OsHpt5 showed three folds expression under similar conditions in IR64 genotype of rice (GSE6901). In other genotypes namely, VSR and Vandana, OsHpt4 showed two fold downregulation under salt and drought stress (GSE21651).
Response Regulator Proteins (RRs)
The RR gene family members in root and shoot tissues of Arabidopsis showed similar pattern of expression under various abiotic stresses considered in this investigation. Under the cold stress, APRR9 showed three fold upregulation in 12 h of stress in both root and shoot tissues. Most of the members of the RR family were found to be downregulated under cold stress conditions in both root and shoot tissues (Figure 5A). Under drought conditions, most genes of the RR family maintained minimal expression in both root and shoot conditions. Genes namely APRR4 (Arabidopsis pseudo response regulator), APRR9 and ARR15 were upregulated in the root tissues while APRR4, APRR9, ARR11, and ARR21 were upregulated in the shoot tissues under cold stress conditions at various time points. In genotoxic stress conditions, most gene members of the family were downregulated in the root tissues, while in shoots, most of the genes maintained unaltered expression at various time points. Similar expression pattern was observed in response to heat and osmotic stress. Only ARR23 in the root tissues was found to be upregulated while in shoots, it maintained unchanged expression with respect to control conditions. Under osmotic stress, APRR9 in the shoot tissues got upregulated in 12 h of stress. Under wounding stress, most of the members maintained unaltered expression at various time points in the root tissues while in shoot tissues, RR gene family members showed mixed response ranging from downregulation to unaltered expression. In response to UV stress, RR family members maintained unchanged expression with respect to the control conditions with only APRR4, APRR9, and ARR3 showing upregulated expression in the root tissues while in shoot, only APRR9 showed three fold upregulation. In salt stress conditions, RR gene family members in shoot tissues were found to have marginally higher expression levels than in the root tissues. Pseudo response regulator, APRR9 showed two fold and three fold high expression in the shoot and root tissues respectively. Similar pattern of expression was found in the oxidative stress of both root and shoot tissues of Arabidopsis.
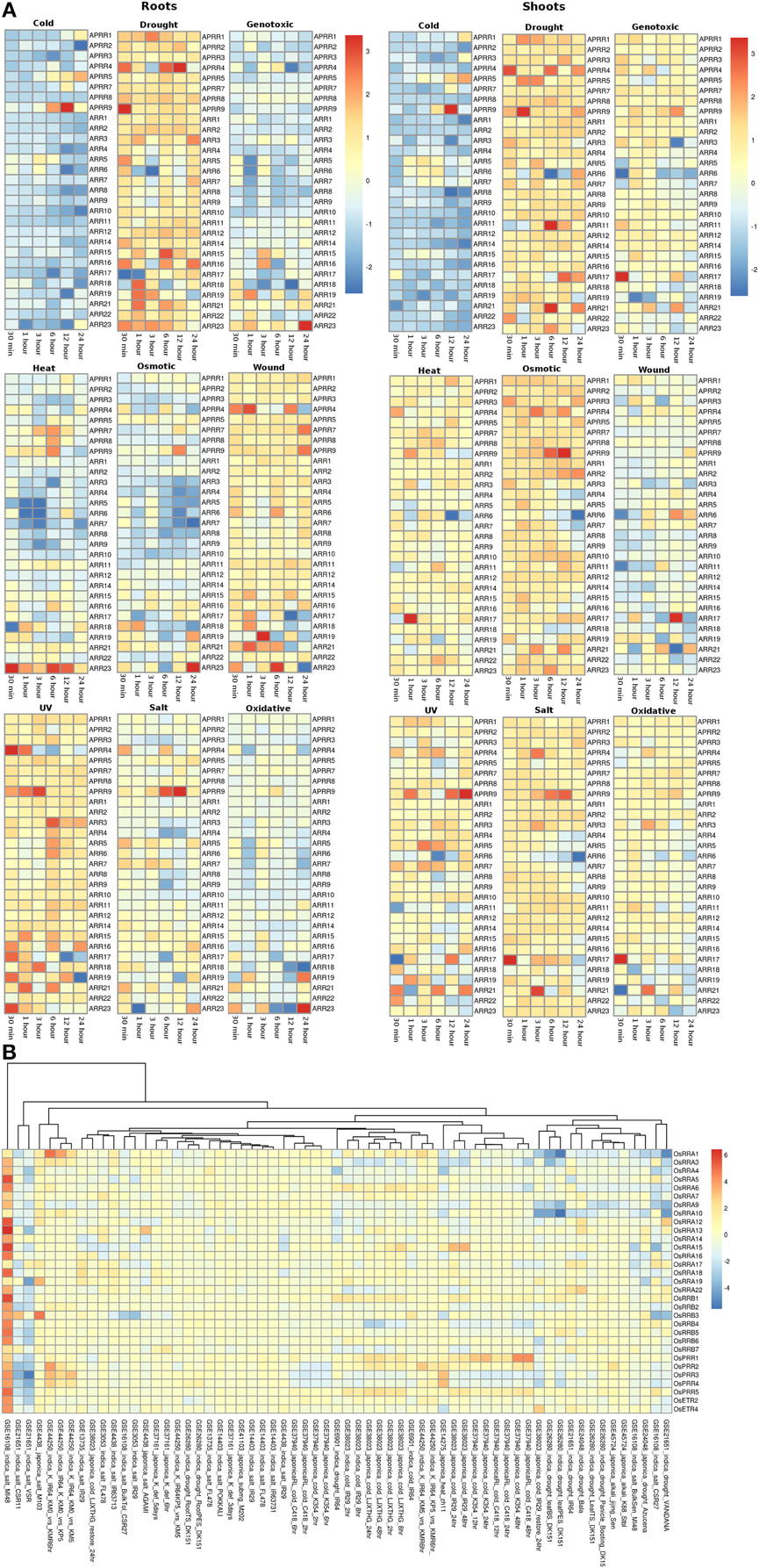
Figure 5. Heatmap depicting the stress-induced expression of Response regulator (RRs) genes from root and shoot obtained using microarray data for Arabidopsis (A) and O. sativa (B). The heatmap was made using open source R software.
In rice, similar to Arabidopsis, all the members of RR gene family showed unaltered expression under all the abiotic stress conditions in all the genotypes. OsRRA1 showed downregulation in the indica genotype under salt and drought conditions (Figure 5B). Some genes of the RR gene family showed downregulation in the indica genotype in VSR and CSR11 (GSE21651) variety of rice. Another variety of the indica genotype, MI48 showed two to three fold upregulation of various gene members of the RR gene family (GSE16108).
qRT-PCR-based Analysis of TCS Members Toward Diurnal Rhythm in Rice
The circadian clock controls many aspects of plant physiology such as flowering, photosynthesis and growth. The diurnal expression profiles of representative genes of TCS family were analyzed under conditions of 12 h light (L)/12 h dark (D) with a constant temperature 28 ± 2°C using IR64 genotype of rice at the seedling stage. Histidine kinases showed rhythmic expression in diurnal manner in response to light and dark cycle (Figure 6A). OsHK1, OsHK2, and OsHK5 genes showed comparatively low expression while OsHK4 and OsHK6 showed comparatively high expression. OsHK4 and OsHK6 showed same pattern of expression, with a phase of 24 h and peak at transition period of night to light during morning. It was evident from the expression analysis that the HKs followed diurnal cycle. Similar to the rhythmic cycle of HKs, Hpts, and RRs also showed expression in diurnal cycle (Figures 6B,C). Among the Hpts, OsHpt5 showed comparatively low expression than other members of the family. Changes in expression levels of Hpts also coincide with dark to light transition in diurnally regulated manner. Level of mRNAs of all OsHpts genes except OsHpt4 oscillated during the 24 h cycle of light-dark, peaking in the morning. OsHpt3 expression oscillated with higher amplitude in comparison to that of other Hpts.
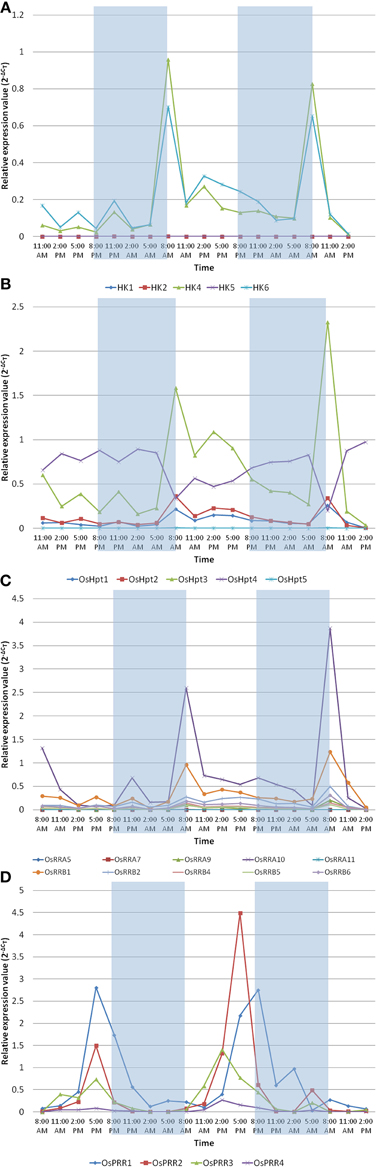
Figure 6. Expression analysis of the representative family members of two-component systems, (A) OsHKs, (B) OsHpts, (C) OsRRAs and OsRRBs, (D) OsPRRs in seedlings of Oryza sativa L. (cv IR-64) during day and night cycles. The rice seedlings were subjected to 12 h of dark followed by the 12 h of light period. The shaded area shows dark and non shaded area shows the light period.
Among the RRs, A type RR- OsRRA10 and B type RR- OsRRB1 showed comparatively high expression (Figure 6C). OsPRR1, OsPRR2, OsPRR3 and OsPRR4 belong to pseudo-response regulator family. Expression of each member of this family rhythmically oscillated in the given 24 h period (Figure 6D). Interestingly, the level of each mRNA reached its maximum at a distinctive time. OsPRR1 showed evening specific peak. mRNA of members of PRR family started accumulating after dawn sequentially at 3 h intervals in the order of OsPRR3→OsPRR4→OsPRR2→OsPRR1. OsPRR2>OsPRR1>OsPRR3>OsPRR4 is the order of their expression amplitude.
In summary, most of the considered TCS members except pseudo-response regulators, showed similar pattern of expression, with a phase of 24 h and peak at transition period of night to light during morning.
Discussion
TCS is considered as one of the most crucial signal transduction system in plants. Evidence suggest that TCS pathways are involved in sensing the environmental stimuli, ethylene signaling, light perception, circadian rhythm and cytokinin-dependent processes which include shoot and root development, vascular differentiation and leaf senescence (Hwang et al., 2002; Kakimoto, 2003; Tran et al., 2010). Cytokinin signaling has been associated with the variety of stress response (Hare et al., 1997). Histidine kinase of the TCS is known to function as an oxidative stress sensor (Singh, 2000). ERS1 gene provides ethylene sensitivity to the plants. Analysis has shown the accumulation of ERS1 in the leaves of Nicotiana tabacum L. on exogenous ethylene treatment, while transcripts were observed in root, shoot, and leaf of the plant (Terajima et al., 2001). MPSS analysis also showed the accumulation of transcripts in roots and leaves in Arabidopsis. Cytokinin receptor, CRE1 transcripts were observed to accumulate in root tissues. Recent analysis has shown that CRE1 cytokinin pathway is differentially recruited depending on the root environmental conditions in Medicago truncatula (Laffont et al., 2015). Further, the expression of AHK2, AHK3, and AHK4 was observed in several organs of the plant species (Ueguchi et al., 2001). In Arabidopsis, various histidine kinases namely AHK2, AHK3, and CRE1 (cytokinin response1/AHK4) are considered as principle cytokinin receptors. Mutant analysis of these cytokinin receptors confirmed their role in response toward low water potential and salt stress (Kumar and Verslues, 2015). Expression analysis showed little or no change in the expression of AHK2 and CRE1 genes in various abiotic stresses in both root and shoot tissues. In the present analysis, AHK3 showed two fold expression at 12 h of salt stress. AHK2 and AHK3 were found to be negatively controlling osmotic stress responses in Arabidopsis. CRE1 also negatively regulates osmotic stress in the presence of cytokinin (Tran et al., 2007). It was found that cold stress did not significantly induce AHK2 and AHK3 expression which indicates that these proteins may mediate cold temperatures for A-type RR expression (Jeon et al., 2010). Our expression analysis of the AHK2 and AHK3 genes in cold stress in both root and shoot tissues corroborated with the earlier result. Previously, AHP1 was shown to be expressed in roots; AHP2 and AHP3 were found to express more in roots, stems, leaves, flowers, and siliques (Suzuki et al., 1998; Hradilova and Brzobohaty, 2007). Our MPSS data analysis showed the accumulation of AHP2 and AHP3 transcripts in the root, silique and inflorescent tissues. Analysis shows that Hpt proteins in Arabidopsis namely AHP2, AHP3, and AHP5 control the response toward drought stress in negative and redundant manner. Also, the downregulated expression of these genes was observed under dehydrating conditions which is assumed to be due to the stress induced reduction of the endogenous cytokinin levels (Nishiyama et al., 2013). Analysis using microarray also showed downregulated expression of these genes under various abiotic stress pertaining to dehydrating conditions such as osmotic and salt stress. Recently, a knock-down analysis of two histidine phosphotransfer (OsHpt2 and OsHpt3) via RNA interference (RNAi) showed that OsHpts function as positive regulators of the cytokinin signaling pathway and play different roles in salt and drought tolerance in rice (Sun et al., 2014). A 1.5-fold expression of these genes in the various rice genotypes in microarray, as reported in the present analysis, also supports the earlier results. Earlier, type-A response regulator genes in rice were shown to have an overlapping/differential expression patterns in various organs and in response to light (Jain et al., 2006). Previously, under short day conditions, B-type RR, Ehd1 (Early heading date 1) from rice has been shown to be a floral inducer (Doi et al., 2004). In Arabidopsis, B-type RRs are involved in cytokinin and ethylene signaling (Hwang et al., 2002) while in rice they are involved in developmental and environmental signals mediated by light, cytokinin, and ethylene (Doi et al., 2004). Multiple A type ARRs were found to be upregulated by cold stress (Argueso et al., 2009). The upregulation of A type RR was also observed in the expression analysis in both Arabidopsis and rice. In Arabidopsis, the expression of ARR4 and ARR5 is found to be induced by the low temperature, dehydration and high salinity (Urao et al., 1998). Triple mutant analysis among the pseudo-RRs (APRRs) showed APRR5, APRR7 and APRR9 as the negative regulators in the abiotic stress conditions (Nakamichi et al., 2009).
All the organisms have a natural time keeping mechanism popularly known as circadian clocks that is used for the coordination of the physiology of organism with its surrounding environment. In plants, circadian clocks have been shown to play a major role in regulating numerous stress and growth response mechanism (Dodd et al., 2005). Regulation of signaling of phytohormones like auxin and ABA by circadian clock has been reported (Covington and Harmer, 2007; Seung et al., 2012). Earlier, in Arabidopsis, two Myb-related transcription factors, circadian clock associated (CCA1) and late elongated hypocotyls (LHY) have been shown to induce the expression of PRR7 and PPR9 in circadian rhythm (in morning cycles) and PRR1 (in evening cycles) which also, in turn bind and repress the expression of the formers (Alabadí et al., 2001; Nakamichi et al., 2010). Earlier, prr9/prr7/prr5 triple mutant analysis revealed the molecular link between metabolism and the circadian clock (Fukushima et al., 2009). In Arabidopsis, pseudoresponse regulators have been shown to be involved in circadian rhythms, control of flowering time and also photo-sensory signal transduction (Devlin and Kay, 2001; Mouradov et al., 2002). Our data show that the expression of OsPrr genes is under diurnal control in indica rice IR64. Murakami et al. (2003) did similar analysis in japonica O. sativa (var Nipponbare) and observed similar results. It again indicates both the dicotyledonous (e.g., Arabidopsis thaliana) and monocotyledonous (e.g., Oryza sativa) plants might share the evolutionarily conserved molecular mechanism underlying the circadian rhythm. Our expression analysis of representative members of the TCS family showed that not only PRRs, but also other members like HKs, Hpts and RRs are also regulated by the diurnal clock. The TCS members have been shown to play a major role in the abiotic stress response mechanism. The result provides a crucial input related to molecular link between abiotic stress response and diurnal clock.
Conclusions
The progress made over a decade has enhanced our understanding about the two-component signaling system and the crucial role played by its members in perceiving environmental stimuli. Even though the members of the TCS system have been characterized in many plant species but their functional involvement in various environmental stress conditions is still a conundrum. The current analysis has assembled all the expression data for all the TCS members, in order to understand their functional complexity. Further, MPSS data analysis presented an overview of the transcript abundance of the TCS members in various plant tissues under various stress conditions. Expression analysis suggest that rice involves more number of TCS members (HKs, Hpts, and RRs) in these responses, despite having comparable number of genes with respect to Arabidopsis (Table 2). Also, the diurnal and rhythmic expression of the TCS gene family members in response to the day and night cycle provides a crucial information about the complexities of the process that are regulated by various TCS members in response to various abiotic stress conditions. The analysis presented in this study provides interesting insights about the functional involvement of the TCS members in growth and stress response in plants.
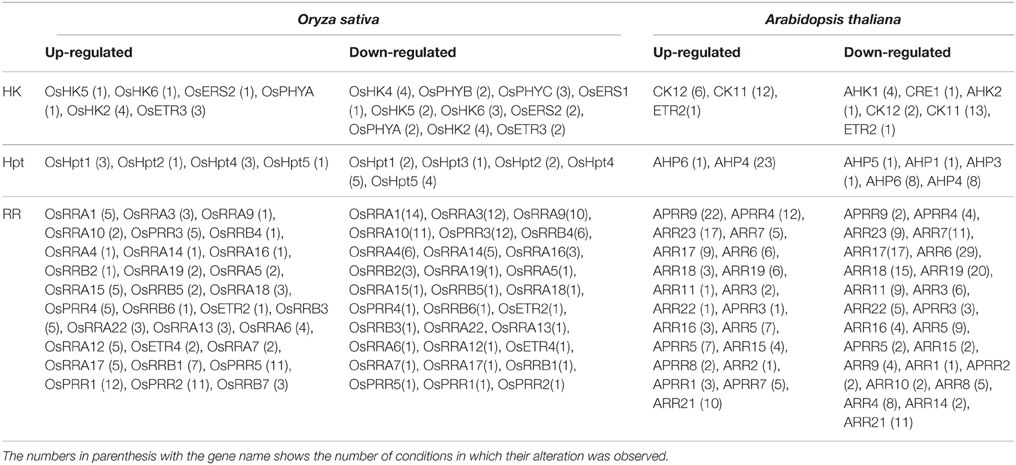
Table 2. Table showing genes of the TCS family which were found to be altered significantly (≥1.5 fold; upregulation/downregulation) under various abiotic stress conditions.
Author Contributions
AP and SLS-P conceived the idea and designed the experiments. PS and HG did the real time PCR work and its analysis. AS and HK performed the MPSS and microarray database analysis and wrote the manuscript. AP and SLS-P edited the manuscript. All the authors approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors would like to thank Jawaharlal Nehru University (JNU), International Centre for Genetic Engineering and Biotechnology (ICGEB) and Department of Biotechnology (DBT), Government of India for financial support. HK acknowledges DST for the grants received as DST-INSPIRE award.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00711
References
Alabadí, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Más, P., and Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. doi: 10.1126/science.1061320
Argueso, C. T., Fernando, J. F., and Joseph, J. K. (2009). Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ. 32, 1147–1160. doi: 10.1111/j.1365-3040.2009.01940.x
Brenner, S., Johnson, M., Bridgham, J., Golda, G., Lloyd, D. H., Johnson, D., et al. (2000). Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 18, 630–634. doi: 10.1038/76469
Capra, E. J., and Laub, M. T. (2012). Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66, 325–347. doi: 10.1146/annurev-micro-092611-150039
Chang, C., Kwok, S. F., Bleecker, A. B., and Meyerowitz, E. M. (1993). Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262, 539–544. doi: 10.1126/science.8211181
Cheung, J., and Hendrickson, W. A. (2010). Sensor domains of two-component regulatory systems. Curr. Opin. Microbiol. 13, 116–123. doi: 10.1016/j.mib.2010.01.016
Covington, M. F., and Harmer, S. L. (2007). The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 5:e222. doi: 10.1371/journal.pbio.0050222
Devlin, P. F., and Kay, S. A. (2001). Circadian photoperception. Annu. Rev. Physiol. 63, 677–694. doi: 10.1146/annurev.physiol.63.1.677
Dodd, A. N., Salathia, N., Hall, A., Kévei, E., Tóth, R., Nagy, F., et al. (2005). Plant circadian clocks increase photosynthesis, growth, survival and competitive advantage. Science 309, 630–633. doi: 10.1126/science.1115581
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., et al. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18, 926–936. doi: 10.1101/gad.1189604
Fukushima, A., Kusano, M., Nakamichi, N., Kobayashi, M., Hayashi, N., Sakakibara, H., et al. (2009). Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc. Natl. Acad. Sci. U.S.A. 106, 7251–7256. doi: 10.1073/pnas.0900952106
Grebe, T. W., and Stock, J. B. (1999). The histidine protein kinase superfamily. Adv. Microb. Physiol. 41, 139–227. doi: 10.1016/S0065-2911(08)60167-8
Grefen, C., and Harter, K. (2004). Plant two-component systems: principles, functions, complexity and cross talk. Planta 219, 733–742. doi: 10.1007/s00425-004-1316-4
Ha, S., Vankova, R., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. S. (2011). Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 17, 172–179. doi: 10.1016/j.tplants.2011.12.005
Hare, P. D., Cress, W. A., and van Staden, J. (1997). The involvement of cytokinins in plant responses to environmental stress. Plant Growth Regul. 23, 79–103. doi: 10.1023/A:1005954525087
Hong, F., Breitling, R., McEntee, C. W., Wittner, B. S., Nemhauser, J. L., and Chory, J. (2006). RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22, 2825–2827. doi: 10.1093/bioinformatics/btl476
Hradilova, J., and Brzobohaty, B. (2007). Expression pattern of the AHP gene family from Arabidopsis thaliana and organ specific alternative splicing in the AHP5 gene. Biol. Plant. 51, 257–267. doi: 10.1007/s10535-007-0051-7
Hwang, I., Chen, H. C., and Sheen, J. (2002). Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 129, 500–515. doi: 10.1104/pp.005504
Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389. doi: 10.1038/35096500
Hwang, I., Sheen, J., and Müller, B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63, 353–380. doi: 10.1146/annurev-arplant-042811-105503
Jain, M., Tyagi, A. K., and Khurana, J. P. (2006). Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol. 6:1. doi: 10.1186/1471-2229-6-1
Jeon, J., Kim, N. Y., Kim, S., Kang, N. Y., Novák, O., Ku, S. J., et al. (2010). A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 285, 23371–23386. doi: 10.1074/jbc.M109.096644
Kakimoto, T. (2003). Perception and signal transduction of cytokinins. Annu. Rev. Plant Biol. 54, 605–627. doi: 10.1146/annurev.arplant.54.031902.134802
Kofoid, E. C., and Parkinson, J. S. (1988). Transmitter and receiver modules in bacterial signaling proteins. Proc. Natl. Acad. Sci. U.S.A. 85, 4981–4985. doi: 10.1073/pnas.85.14.4981
Koretke, K. K., Lupas, A. N., Warren, P. V., Rosenberg, M., and Brown, J. R. (2000). Evolution of two-component signal transduction. Mol. Biol. Evol. 17, 1956–1970. doi: 10.1093/oxfordjournals.molbev.a026297
Kumar, M. N., and Verslues, P. E. (2015). Stress physiology functions of the Arabidopsis histidine kinase cytokinin receptors. Physiol. Plant 154, 369–380. doi: 10.1111/ppl.12290
Kushwaha, H. R., Singh, A. K., Sopory, S. K., Singla-Pareek, S. L., and Pareek, A. (2009). Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genomics 10:200. doi: 10.1186/1471-2164-10-200
Laffont, C., Rey, T., André, O., Novero, M., Kazmierczak, T., Debellé, F., et al. (2015). The CRE1 cytokinin pathway is differentially recruited depending on Medicago truncatula root environments and negatively regulates resistance to a pathogen. PLoS ONE 10:e0116819. doi: 10.1371/journal.pone.0116819
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, 1202–1210. doi: 10.1093/nar/gkr1090
Laub, M. T., and Goulian, M. (2007). Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41, 121–145. doi: 10.1146/annurev.genet.41.042007.170548
Li, J., Li, G., Wang, H., and Wang Deng, X. (2011). Phytochrome signaling mechanisms. Arabidopsis Book 9:e0148. doi: 10.1199/tab.0148
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mascher, T., Helmann, J. D., and Unden, G. (2006). Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70, 910–938. doi: 10.1128/MMBR.00020-06
Meyers, B. C., Vu, T. H., Tej, S. S., Ghazal, H., Matvienko, M., Agrawal, V., et al. (2004). Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat. Biotechnol. 22, 1006–1011. doi: 10.1038/nbt992
Mizuno, T. (2005). Two-component phosphorelay signal transduction systems in plants: from hormone responses to circadian rhythms. Biosci. Biotechnol. Biochem. 69, 2263–2276. doi: 10.1271/bbb.69.2263
Mochida, K., Yoshida, T., Sakurai, T., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. S. (2010). Genome-wide analysis of two-component systems and prediction of stress-responsive two-component system members in soybean. DNA Res. 17, 303–324. doi: 10.1093/dnares/dsq021
Mouradov, A., Cremer, F., and Coupland, G. (2002). Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 14, S111–S130.
Murakami, M., Ashikari, M., Miura, K., Yamashino, T., and Mizuno, T. (2003). The evolutionarily conserved OsPRR quintet: rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 44, 1229–1236. doi: 10.1093/pcp/pcg135
Nakamichi, N., Kiba, T., Henriques, R., Mizuno, T., Chua, N. H., and Sakakibara, H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7 and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22, 594–605. doi: 10.1105/tpc.109.072892
Nakamichi, N., Kusano, M., Fukushima, A., Kita, M., Ito, S., Yamashino, T., et al. (2009). Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50, 447–462. doi: 10.1093/pcp/pcp004
Nishiyama, R., Watanabe, Y., Leyva-Gonzalez, M. A., Ha, C. V., Fujita, Y., Tanaka, M., et al. (2013). Arabidopsis AHP2, AHP3, and AHP5 histidinephosphotransfer proteins function as redundant negative regulators of drought stress response. Proc. Natl. Acad. Sci. U.S.A. 110, 4840–4845. doi: 10.1073/pnas.1302265110
Nongpiur, R., Soni, P., Karan, R., Singla-Pareek, S. L., and Pareek, A. (2012). Histidine kinases in plants: cross talk between hormone and stress responses. Plant Signal. Behav. 7, 1230–1237. doi: 10.4161/psb.21516
Ota, I. M., and Varshavsky, A. (1993). A yeast protein similar to bacterial two-component regulators. Science 262, 566–569. doi: 10.1126/science.8211183
Pareek, A., Singh, A., Kumar, M., Kushwaha, H. R., Lynn, A. M., and Singla-Pareek, S. L. (2006). Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 142, 380–397. doi: 10.1104/pp.106.086371
Pils, B., and Heyl, A. (2009). Unraveling the evolution of cytokinin signaling. Plant Physiol. 151, 782–791. doi: 10.1104/pp.109.139188
Schaller, G. E., Kieber, J. J., and Shiu, H. (2008). Two-component signaling elements and histidyl-aspartyl phosphorelays. Arabidopsis Book 6:e0112. doi: 10.1199/tab.0112
Schneider-Poetsch, H. A. (1992). Signal transduction by phytochrome: phytochromes have a module related to the transmitter modules of bacterial sensor proteins. Photochem. Photobiol. 56, 839–846. doi: 10.1111/j.1751-1097.1992.tb02241.x
Seung, D., Risopatron, J. P. M., Jones, B. J., and Marc, J. (2012). Circadian clock-dependent gating in ABA signalling networks. Protoplasma 249, 445–457. doi: 10.1007/s00709-011-0304-3
Singh, K. K. (2000). The Saccharomyces cerevisiae SLN1P-SSK1P two component system mediates response to oxidative stress and in an oxidant-specific fashion. Free Rad. Biol. Med. 29, 1043–1050. doi: 10.1016/S0891-5849(00)00432-9
Stock, A., Chen, T., Welsh, D., and Stock, J. (1988). CheA protein, a central regulator of bacterial chemotaxis, belongs to a family of proteins that control gene expression in response to changing environmental conditions. Proc. Natl. Acad. Sci. U.S.A. 85, 1403–1407. doi: 10.1073/pnas.85.5.1403
Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215. doi: 10.1146/annurev.biochem.69.1.183
Stock, J. B., Surette, M. G., Levit, M., and Park, P. (1995). Two-component Signal Transduction Systems: Structure-function Relationships and Mechanisms of Catalysis. Washington, DC: ASM Press.
Sun, L., Zhang, Q., Wu, J., Zhang, L., Jiao, X., Zhang, S., et al. (2014). Two rice authentic histidinephosphotransfer proteins, OsAHP1 and OsAHP2, mediate cytokinin signaling and stress responses in rice. Plant Physiol. 165, 335–345. doi: 10.1104/pp.113.232629
Suzuki, T., Imamura, A., Ueguchi, C., and Mizuno, T. (1998). Histidine containing phosphotransfer (Hpt) signal transducers implicated in His-to-Asp phosphorelay in Arabidopsis. Plant Cell Physiol. 39, 1258–1268.
Terajima, Y., Nukui, H., Kobayashi, A., Fujimoto, S., Hase, S., Yoshioka, T., et al. (2001). Molecular cloning and characterization of a cDNA for a novel ethylene receptor, NT-ERS1, of tobacco (Nicotiana tabacum L.). Plant Cell Physiol. 42, 308–313. doi: 10.1093/pcp/pce038
To, J. P., and Kieber, J. J. (2008). Cytokinin signaling: two-components and more. Trends Plant Sci. 13, 85–92. doi: 10.1016/j.tplants.2007.11.005
Tran, L. S. P., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2010). Role of cytokinin responsive two-component system in ABA and osmotic stress signaling. Plant Signal. Behav. 5, 148–150. doi: 10.4161/psb.5.2.10411
Tran, L. S., Urao, T., Qin, F., Maruyama, K., Kakimoto, T., Shinozaki, K., et al. (2007). Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 20623–20628. doi: 10.1073/pnas.0706547105
Ueguchi, C., Koizumi, H., Suzuki, T., and Mizuno, T. (2001). Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 42, 231–235. doi: 10.1093/pcp/pce015
Urao, T., Yakubov, B., Satoh, R., Yamaguchi-Shinozaki, K., Seki, M., Hirayama, T., et al. (1999). A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11, 1743–1754. doi: 10.1105/tpc.11.9.1743
Urao, T., Yakubov, B., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Stress-responsive expression of genes for two-component response regulator-like proteins in Arabidopsis thaliana. FEBS Lett. 427, 175–178. doi: 10.1016/S0014-5793(98)00418-9
Urao, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2000). Two-component systems in plant signal transduction. Trends Plant Sci. 5, 67–74. doi: 10.1016/S1360-1385(99)01542-3
Keywords: abiotic stress, Arabidopsis, histidine kinase, histidine phosphotransfer protein, response regulator, rice, two-component system
Citation: Singh A, Kushwaha HR, Soni P, Gupta H, Singla-Pareek SL and Pareek A (2015) Tissue specific and abiotic stress regulated transcription of histidine kinases in plants is also influenced by diurnal rhythm. Front. Plant Sci. 6:711. doi: 10.3389/fpls.2015.00711
Received: 05 June 2015; Accepted: 25 August 2015;
Published: 11 September 2015.
Edited by:
Girdhar Kumar Pandey, Delhi University, IndiaReviewed by:
Mukesh Jain, National Institute of Plant Genome Research, IndiaOm Parkash Dhankher, University of Massachusetts Amherst, USA
Copyright © 2015 Singh, Kushwaha, Soni, Gupta, Singla-Pareek and Pareek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashwani Pareek, Stress Physiology and Molecular Biology Laboratory, School of Life Sciences, Jawaharlal Nehru University, New Delhi 110067, India, ashwanip@mail.jnu.ac.in
 Anupama Singh1
Anupama Singh1 Hemant R. Kushwaha
Hemant R. Kushwaha Praveen Soni
Praveen Soni Sneh L. Singla-Pareek
Sneh L. Singla-Pareek Ashwani Pareek
Ashwani Pareek