
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 11 August 2015
Sec. Plant Physiology
Volume 6 - 2015 | https://doi.org/10.3389/fpls.2015.00600
This article is part of the Research TopicAbiotic Stress Signaling in Plants: Functional Genomic InterventionView all 50 articles
Transient changes in intracellular Ca2+ concentration have been well recognized to act as cell signals coupling various environmental stimuli to appropriate physiological responses with accuracy and specificity in plants. Calmodulin (CaM) and calmodulin-like proteins (CMLs) are major Ca2+ sensors, playing critical roles in interpreting encrypted Ca2+ signals. Ca2+-loaded CaM/CMLs interact and regulate a broad spectrum of target proteins such as channels/pumps/antiporters for various ions, transcription factors, protein kinases, protein phosphatases, metabolic enzymes, and proteins with unknown biochemical functions. Many of the target proteins of CaM/CMLs directly or indirectly regulate plant responses to environmental stresses. Basic information about stimulus-induced Ca2+ signal and overview of Ca2+ signal perception and transduction are briefly discussed in the beginning of this review. How CaM/CMLs are involved in regulating plant responses to abiotic stresses are emphasized in this review. Exciting progress has been made in the past several years, such as the elucidation of Ca2+/CaM-mediated regulation of AtSR1/CAMTA3 and plant responses to chilling and freezing stresses, Ca2+/CaM-mediated regulation of CAT3, MAPK8 and MKP1 in homeostasis control of reactive oxygen species signals, discovery of CaM7 as a DNA-binding transcription factor regulating plant response to light signals. However, many key questions in Ca2+/CaM-mediated signaling warrant further investigation. Ca2+/CaM-mediated regulation of most of the known target proteins is presumed based on their interaction. The downstream targets of CMLs are mostly unknown, and how specificity of Ca2+ signaling could be realized through the actions of CaM/CMLs and their target proteins is largely unknown. Future breakthroughs in Ca2+/CaM-mediated signaling will not only improve our understanding of how plants respond to environmental stresses, but also provide the knowledge base to improve stress-tolerance of crops.
As sessile organisms, plants encounter various types of environmental stresses, which are generally classified into biotic stresses such as insect and pathogen attacks, and abiotic stresses such as unfavorable temperature, lack of or excessive amounts of water, salinity, heavy metal toxicity, chemical toxicity, and nutrient deficiency. On the other hand, the process of industrialization inevitably brings many detrimental effects to the environment. Poorly controlled release of wastes from industrial processes and human life not only adds various toxic chemicals to our water and soil but also release harmful gasses into the atmosphere. Obviously, human activities are creating environmental challenges, making sustained crop production difficult. Classic agricultural technologies such as irrigation, applications of fertilizer, insecticides, fungicides, and chemical phytoprotectants have helped to improve crop yield, but the effects are limited, the costs are high and the impacts on the ecosystems and human health are undesirable and dangerous. Understanding how plants perceive and respond to various environmental stresses provides the necessary platform to create crop varieties which could fit better into the challenging environments, and has become one of the most important tasks for plant scientists around the world.
Calcium is one of the most abundant elements on earth. Ca2+ concentration outside the plasma membrane is usually at millimolar level. Since Ca2+ can form insoluble compounds with phosphate derivatives and complex with macromolecules, high levels of cytosolic Ca2+ are toxic to cells. Ca2+ concentration in the cytoplasm and nucleus is usually maintained at 50–100 nM under resting conditions (Reddy, 2001; Yang and Poovaiah, 2003). Ca2+ gradient across the plasma membrane as well as inner membrane system are involved in cell signaling process controlled by stimulus responsive Ca2+ permeable channels, Ca2+ pumps and Ca2+/H+ exchangers (Reddy, 2001; Kudla et al., 2010). Accumulating evidence reveals that various external stimuli such as gravity, light, cold, heat, drought, water-logging (hypoxia), salt, wind, touch, wounding, and pathogen attacks can quickly induce elevations in cytosolic Ca2+ concentration (Poovaiah and Reddy, 1993; Evans et al., 2001; Reddy, 2001; Snedden and Fromm, 2001; Zhu, 2001). Signal-induced nuclear Ca2+ changes have also been documented (Van Der Luit et al., 1999; Pauly et al., 2000), but they are not as well studied as cytosolic Ca2+ transients (Reddy et al., 2011). The excessive amount of Ca2+ in cytoplasm is quickly moved out of the cell or pumped back into the endogenous Ca2+ reservoirs such as vacuole and endoplasmic reticulum (ER) through the involvement of Ca2+ pumps and Ca2+/H+ exchangers distributed on the plasma membrane and inner membrane system (Reddy, 2001; Kudla et al., 2010). Interestingly, the transient changes of intracellular Ca2+ concentration triggered by various stimuli differ from each other in terms of amplitude, duration, frequency, and spatial distribution inside the cell; and these stimulus-specific Ca2+ transients are named calcium signatures by Webb et al. (1996). Stimulus-specific signals are decoded by downstream effector proteins to generate specific or overlapping responses (Poovaiah et al., 2013). These effectors include Ca2+ sensor proteins which are represented by three major types in plants, namely calmodulin (CaM)/CaM-like (CML) proteins, calcium-dependent protein kinases (CDPKs) and calcineurin B like (CBL) proteins (Luan, 2009). In this review, our primary focus will be limited to CaM/CMLs and their important roles in plant abiotic stress signaling and responses.
Calmodulin is a ubiquitous Ca2+-binding protein which exists in all eukaryotes (Snedden and Fromm, 1998; Yang and Poovaiah, 2003; McCormack et al., 2005; Kim et al., 2009; Du et al., 2011). It is a small acidic protein composed of two pairs of EF-hands located at both the N- and C-terminus. In Arabidopsis, seven genes encode four CaM isoforms (CaM1/4; CaM2/3/5; CaM6; CaM7), which differ only in one to five amino acid residues (McCormack and Braam, 2003; McCormack et al., 2005). It has been reported that different CaM isoforms differ in binding and regulating downstream effectors (Lee et al., 2000; Yoo et al., 2005). The slight differences in their structural features may have considerable impacts on their binding to targets (Yamniuk and Vogel, 2005).
In addition to canonical CaM, there are 50 genes coding for CaM-like proteins in the Arabidopsis genome which are made of varying number of EF hands and share at least 16% of overall sequence identity with canonical CaM (McCormack and Braam, 2003). Similarly, five CaM and 32 CML genes, respectively are reported in the rice genome (Boonburapong and Buaboocha, 2007). Despite having four EF hands, most CMLs show low (less than 50%) overall similarity to CaMs (McCormack and Braam, 2003; Boonburapong and Buaboocha, 2007; Perochon et al., 2011). Several Arabidopsis CMLs, including CML37, 38, 39, and 42 displayed an electrophoretic mobility shift in the presence of Ca2+, indicating that, like CaMs, CMLs also act as Ca2+ sensors (Vanderbeld and Snedden, 2007; Dobney et al., 2009). Besides EF-hands, CaMs and CMLs do not carry any known functional domain, and hence usually have no enzymatic or biochemical functions. So far the only exception is CaM7 from Arabidopsis which was reported to specifically bind Z-/G-box in a Ca2+-dependent manner and act as a transcription factor to regulate light-responsive gene expression and light morphogenesis (Kushwaha et al., 2008). Therefore, identifying CaM/CML targets and understanding the impacts of CaM/CML-binding on their functional behaviors are the major challenges in deciphering the functional significance of CaM/CMLs at molecular, biochemical, and physiological levels.
It is well-documented that Ca2+-binding-induced conforma tional changes in CaMs and CMLs usually increase their binding affinity to downstream targets through hydrophobic and electrostatic interactions (Snedden and Fromm, 1998; Hoeflich and Ikura, 2002). A stretch of 16–35 amino acids in the target proteins called CaM-binding domain (CaMBD) is usually necessary and sufficient for its interaction with CaM (Rhoads and Friedberg, 1997; Hoeflich and Ikura, 2002). In some cases, CaM interacts with its target proteins in a Ca2+-independent manner, and this kind of interaction requires that the target proteins carry an IQ motif, a stretch of amino acids fitting a conserved pattern of IQXXX(R/K)GXXXR where I could be replaced with “FLV” and “X” represents any amino acid residue (Hoeflich and Ikura, 2002; Yamniuk and Vogel, 2004). CMLs could follow similar models to interact with their targets; however, this assumption requires experimental verification. Usually, CaMBDs are not conserved in their primary structure, however, most of the Ca2+-dependent CaMBD peptides share a conserved secondary structure, a basic amphipathic helix with hydrophobic residues arranged on one side and positively charged residues arranged on the other side (Snedden and Fromm, 2001; Du and Poovaiah, 2004). Hence, most CaM and CML target proteins have to be identified empirically.
As mentioned above, the interactions between CaM/CMLs and target proteins are usually Ca2+-dependent; regular strategies used for detection of protein–protein interaction including yeast-two-hybrid and coimmunoprecipitation are not effective and fruitful in identifying CaM/CML-binding proteins. The majority of the CaM-binding proteins (CBPs) from plants were identified by screening cDNA expression libraries with labeled CaM as probes (usually 35S-labled; Fromm and Chua, 1992; Reddy et al., 1993; Yang and Poovaiah, 2000b). Another effective approach to identify CBPs is utilizing protein microarray (Popescu et al., 2007); however, false positive identification is still a major concern and making protein chips with adequate coverage is currently a challenge. Accumulated results indicated that CaM bind to a variety of CBPs in plants, which include kinases, phosphatases, transcription factors, receptors, metabolic enzymes, ion channels and pumps, and cytoskeletal proteins (Snedden and Fromm, 2001; Bouche et al., 2005; Kim et al., 2009; Du et al., 2011; Reddy et al., 2011). Hence, it is reasonable to conclude that, in most cases, CaMs and CMLs act as multifunctional regulatory proteins, and their functional significance is materialized through the actions of their downstream target proteins. CBPs with well-defined CaM-binding domain, CaM-binding property and involved in plant responses to abiotic stresses are listed in Table 1.
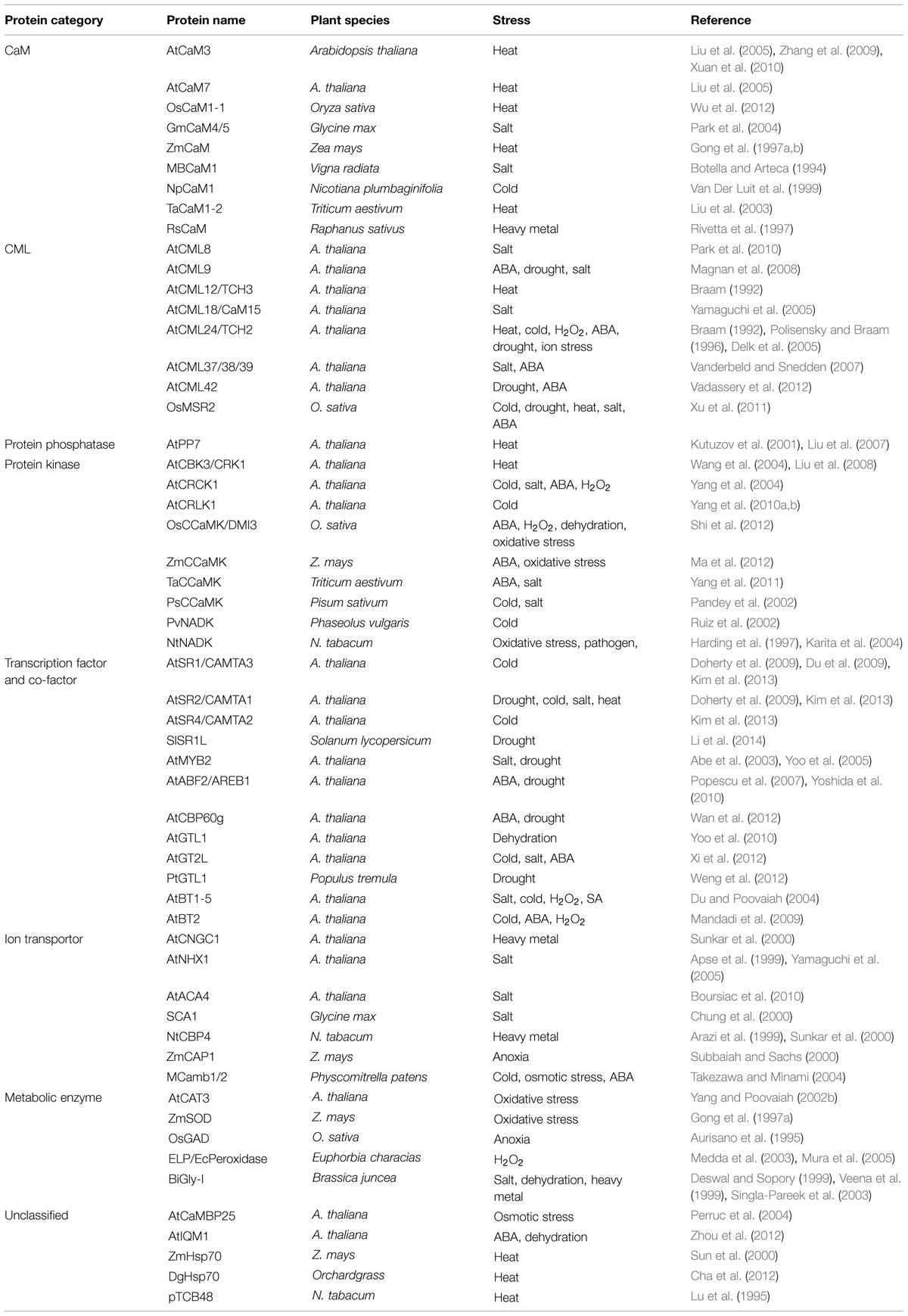
TABLE 1. Involvement of calmodulins (CaMs), CaM-like proteins (CMLs), and CaM-binding proteins (CBPs) in plant responses to diverse abiotic stresses.
Reactive oxygen species (ROS) such as hydrogen peroxide (H2O2), superoxide anion (O2-), and hydroxyl radical (⋅OH) are usually produced in various physiological processes and serve as a class of second messengers (Van Breusegem et al., 2001; Apel and Hirt, 2004). While controlled production of ROS is essential to signal appropriate actions to protect plants from various environmental stresses, excessive accumulation of ROS causes damages to plant cells. Oxidative stress is defined as disruption of the cellular redox balance, which could be triggered by a wide range of biotic and abiotic stimuli (Rentel and Knight, 2004). Because of its long half-life and excellent permeability, H2O2 is broadly accepted as the major form of ROS in plant cells. It is well known that H2O2 can trigger increases in cytosolic Ca2+ by activating the Ca2+-permeable channels (Price et al., 1994; Rentel and Knight, 2004). On the other hand, H2O2 production during oxidative burst is also dependent on continuous Ca2+ influx, which activates not only the NADPH oxidase, an EF-hand containing enzyme on the plasma membrane (Xing et al., 1997), but also the CaM-binding NAD kinase (NADK), which supplies NADP cofactor for ROS production through NADPH oxidase (Harding et al., 1997; Karita et al., 2004; Turner et al., 2004).
In addition, early studies from heat-stressed maize seedling suggested that ROS homeostasis and the entire antioxidant system including catalase, superoxide dismutase (SOD) and ascorbate peroxidase, could be regulated by Ca2+ influx and intracellular CaM (Gong et al., 1997a). Later plant catalases, a class of H2O2 scavenger enzymes catalyzing its degradation to water and oxygen was found to bind CaM in a Ca2+-dependent manner (Yang and Poovaiah, 2002b). The activity of the Arabidopsis CAT3 is stimulated by Ca2+/CaM rather than Ca2+ or CaM alone, but catalases from other organisms such as Aspergillus niger, human and bovine, do not interact with CaM (Yang and Poovaiah, 2002b). A peroxidase from Euphorbia latex, was also reported to be a CBP activated by Ca2+/CaM (Medda et al., 2003; Mura et al., 2005). Evidence also suggests that another class of ROS-scavenging enzyme SOD could be regulated by CaM in maize, although the specific SOD gene has not been cloned (Gong and Li, 1995). The critical role of Ca2+/CaM in balancing ROS actions was further supported by the observation that the oxidative damage caused by heat stress in Arabidopsis seedlings is exacerbated by pretreatment with CaM inhibitors (Larkindale and Knight, 2002).
In addition to these direct regulations on ROS homeostasis, Ca2+/CaM-mediated signaling is also well known to regulate ROS-related signal transduction at various stages. Maize CAP1 encoding a novel form of CaM-regulated Ca2+-ATPase was shown to be induced only during early anoxia, indicating its possible role in oxygen-deprived maize cells (Subbaiah and Sachs, 2000). CaM may also participate indirectly in regulating ROS content through the CaM-regulated -aminobutyrate (GABA) synthesis and the GABA shunt metabolic pathway (Bouche et al., 2003). Recently, it was demonstrated that ZmCCaMK and OsDMI3 (also called OsCCaMK) from maize (Zea mays) and rice (Oryza sativa), respectively, play a critical role in ABA-induced antioxidant actions (Ma et al., 2012; Shi et al., 2012), suggesting a role for CCaMK in plant oxidative stress response. Arabidopsis MPK8 was found to be activated by CaM and activated MPK8 suppresses wound-induced ROS accumulation via transcriptional control of RbohD expression, revealing a novel mechanism for CaM-mediated signaling to fine-tune ROS homeostasis under wounding stress (Takahashi et al., 2011). Interestingly, some genes encoding CaM-binding transcription factors (CAMTAs) and co-factor (AtBTs) are responsive to H2O2, suggesting that CaM-mediated signaling could directly regulate gene expression in plant responses to oxidative cues (Yang and Poovaiah, 2002a; Du and Poovaiah, 2004; Wang et al., 2015).
Prolonged high temperature is usually lethal to all organisms; fluctuations in temperature above optimal level, usually called heat shock (HS), impose major stress affecting plant growth and productivity. Almost all organisms including plants synthesize HS proteins (HSPs), a class of chaperons to assure normal function of various client proteins under adversely high temperature conditions. It was observed long ago that HS induced a quick and strong increase in cytosolic Ca2+ in tobacco (Gong et al., 1998). Expression of CaM in the maize coleoptiles was found to be remarkably induced during HS and was affected by Ca2+ level, suggesting that Ca2+ and CaM may be involved in the acquisition of HS-induced thermotolerance (Gong et al., 1997b). Liu et al. (2003) observed an increase in intracellular Ca2+ within one min after wheat was subjected to 37°C HS. Expression of CaM mRNA and protein was both induced by HS in the presence of Ca2+, and expression of HSP26 and HSP70 was stimulated by exogenous application of Ca2+. HS-induced expression of CaM was 10 min earlier than that of HSPs, and both were suspended by pharmacological reagents which interfere with Ca2+ signaling. These results indicate that Ca2+ and CaM are directly involved in HS signaling (Liu et al., 2003). The Ca2+/CaM signaling system was also proposed to be involved in the induction of HSP genes in Arabidopsis (Liu et al., 2005). Using molecular and genetic tools, Zhang et al. (2009) found that Arabidopsis AtCaM3 was involved in the Ca2+/CaM-mediated HS signal transduction pathway. atcam3 loss-of-function mutant showed a pronounced decrease in thermotolerance after 50 min of incubation at 45°C. The compromised thermotolerance of atcam3 mutant could be rescued by functional complementation with 35S promoter driven AtCaM3, and overexpression of AtCaM3 in wild-type (WT) background increased thermotolerance of the transgenic plants. Furthermore, the DNA-binding activity of HS transcription factors and the expression of tested HS genes at both mRNA and protein levels were shown to be down-regulated in atcam3 null mutant and up-regulated in its overexpressing lines upon HS treatment (Zhang et al., 2009). A role for CaM in HS signaling was also demonstrated in rice (Wu and Jinn, 2012; Wu et al., 2012). HS was reported to induce biphasic cytosolic Ca2+ transients, and this signature feature was found to be reflected in the HS-induced expression of OsCaM1-1. OsCaM1-1 was observed to localize to the nucleus and overexpression of OsCaM1-1 in Arabidopsis resulted in enhanced thermotolerance which coincided with elevated expression of HS-responsive AtCBK3, AtPP7, AtHSF, and AtHSP at a non-inducing temperature. Nitric oxide (NO) level in plants was found to be elevated by high temperatures (Gould et al., 2003), and exogenous application of NO donor provides effective protection to plants under heat stress (Uchida et al., 2002; Song et al., 2006). However, for a long time it was unknown how NO is involved in protecting plants from damage by HS. Recently, Arabidopsis CaM3 was reported to act as a downstream factor of NO in activation of HS transcription factors, accumulation of HSPs and establishment of thermotolerance (Xuan et al., 2010).
Calmodulin-binding proteins have also been shown to play a crucial role in mediating plant responses to heat stress. pTCB48 encoding a CBP was isolated by screening a cDNA expression library constructed from tobacco cell cultures subjected to HS, and its expression was strongly induced by HS treatment, suggesting a role in HS response (Lu et al., 1995). Maize cytosolic Hsp70 was identified to bind CaM in the presence of Ca2+ and could inhibit the activity of CaM-dependent NADK in a concentration-dependent manner, but its possible function in HS response has not been elucidated (Sun et al., 2000). Recently, DgHsp70, a homolog of cytosolic Hsp70 from orchardgrass (Dactylis glomerata) was also found to bind AtCaM2 in the presence of Ca2+, and the binding of Ca2+/CaM decreased the ATPase and foldase activities of this chaperon protein (Cha et al., 2012). PP7 is a ser/thr protein phosphatase which interacts with CaM in a Ca2+-dependent manner (Kutuzov et al., 2001). Arabidopsis AtPP7 was induced by HS and its knockout mutant is impaired in thermotolerance, while the overexpression of AtPP7 results in increased thermotolerance and increased expression of AtHSP70 and AtHSP101 following HS treatment (Liu et al., 2007). Interestingly, AtPP7 was also found to interact with HS transcription factor AtHSF1 implying that AtPP7 could also regulate the expression of HSP genes via AtHSF1 (Liu et al., 2007). However, the mechanistic detail by which AtPP7 dephosphorylates and regulates downstream substrates such as AtHSF1 is not clear. Arabidopsis AtCRK1 (CDPK-related protein kinase 1, also called AtCBK3) was identified as a Ca2+-dependent CBP (Wang et al., 2004). Liu et al. (2008) found that AtCBK3 activates HSFs which further regulate HS gene expression by binding to HS elements.
Ca2+ has been recognized as a vital second messenger coupling cold stress to specific plant responses (Knight et al., 1991; Dodd et al., 2010). Researchers found that cold shock and wind initiate Ca2+ transients in both cytosol and nucleus in transgenic tobacco (Nicotiana plumbaginifolia) seedlings expressing aequorin, and the expression of NpCaM-1 is induced by both cold shock and wind but mediated by distinct Ca2+ signaling pathways operating predominantly in the cytoplasm or in the nucleus (Van Der Luit et al., 1999). Transgenic Arabidopsis plants overexpressing CaM3 showed decreased levels of COR (cold regulated) transcripts suggesting CaM may function as a negative regulator of cold-induced gene expression (Townley and Knight, 2002). Genes encoding CMLs, such as AtCML24/TCH2 and OsMSR2 (O. sativa Multi-Stress-Responsive gene2, a novel CML gene), were also found to be induced by cold treatment and thus, likely participate in transducing cold-induced Ca2+ signals (Polisensky and Braam, 1996; Delk et al., 2005; Xu et al., 2011).
In addition, as downstream effectors of Ca2+/CaM-mediated signaling, CBPs are also known to be involved in plant responses to cold stress. Ca2+/CaM-regulated receptor-like kinase CRLK1, which is mainly localized in the plasma membrane, was found to be involved in cold tolerance (Yang et al., 2010b). CRLK1 carries two CaM-binding sites in both N- and C-termini with affinities for Ca2+/CaM of 25 and 160 nM, respectively (Yang et al., 2010b). crlk1 knockout mutant plants grow and behave like WT plants under regular conditions, but are more sensitive to chilling and freezing treatments than WT plants (Yang et al., 2010b). In addition, cold response genes such as CBF1, RD29A, COR15a, and KIN1 showed delayed responses to cold in crlk1, suggesting a positive role for CRLK1 in regulating cold tolerance. MEKK1, which is a member of the MAP kinase kinase kinase family, was shown to interact with CRLK1 both in vitro and in planta (Yang et al., 2010a). Knockout mutation of CRLK1 abolished the cold-triggered MAP kinase activities, and altered cold-induced expression of genes involved in MAP kinase signaling (Yang et al., 2010a). Therefore, Ca2+/CaM-regulated CRLK1 may modulate cold acclimation through MAP kinase cascade in plants. Other CaM-binding kinases are also suggested to be involved in cold acclimation. The expression of PsCCaMK in pea (Pisum sativa) roots was found to be up-regulated by low temperature or salinity stress (Pandey et al., 2002), and the activity of the Ca2+/CaM-dependent NADK was found to be increased by cold shock (Ruiz et al., 2002).
AtSRs/CAMTAs belong to one of the best characterized classes of CaM-binding transcription factors in plants and animals (Reddy et al., 2000; Yang and Poovaiah, 2000a, 2002a; Bouche et al., 2002; Choi et al., 2005; Du et al., 2011; Reddy et al., 2011). In an attempt to understand transcriptional control of CBF2 (a critical regulator of cold acclimation), Doherty et al. (2009) compared the promoter sequence of three CBFs and found seven conserved DNA motifs CM1 to 7 in their promoters. CM2 is a typical AtSR1/CAMTA3 recognition motif, and importantly, the expression of CBF2 was found to be positively regulated by AtSR1/CAMTA3. Although camta3 knockout mutant had no phenotypic change under cold stress, camta1/camta3 double mutant was found to have reduced freezing tolerance (Doherty et al., 2009). Recently, AtCAMTA1, AtCAMTA2, and AtCAMTA3 were shown to participate in cold tolerance by cooperatively inducing CBF genes and repressing SA biosynthesis (Kim et al., 2013). These results filled a long-standing knowledge gap between cold induced Ca2+ transients and cold-regulated gene expression.
Several Arabidopsis MADS box transcription factors were identified as putative CBPs by a high throughput proteomics approach (Popescu et al., 2007). Expression of some of these MADS box genes, including AGL3, AGL8, AGL15, and AGL32, was reported to be suppressed by cold stress (Hannah et al., 2005), implying a role for Ca2+ signal to regulate cold responses through the MADS proteins, however, whether and how Ca2+/CaM regulates MADS box transcription factors remain to be addressed. The expression of CBF2 is down-regulated in transgenic Arabidopsis plants constitutively expressing AGL15, in comparison to WT plants (Hill et al., 2008). GT factors are plant-specific transcription factors sharing a conserved trihelix DNA-binding domain that specifically interacts with GT cis-elements (Wang et al., 2014). Recently, AtGT2L, a classic member of the GT-2 subfamily, was identified to encode a Ca2+-dependent CBP and it is responsive to cold, salt and ABA treatments (Xi et al., 2012). Furthermore, overexpression of AtGT2L resulted in elevated expression levels of cold- and salt-specific marker genes RD29A and ERD10, both in basal and chilling- or salt-treated conditions. These results indicated that Ca2+/CaM-binding AtGT2L is involved in plant responses to cold and salt stresses (Xi et al., 2012).
High salinity and drought are the major environmental stresses frequently experienced by plants, and both impose osmotic stress on plant cells. Osmotic stress induces a series of responses at the molecular and cellular levels and a primary event is an increase in the cytosolic Ca2+ concentration and subsequent transduction of Ca2+ signals that promotes appropriate cellular responses in an effort to mitigate potential damages (Xiong and Zhu, 2002; Zhu, 2002). In addition to the well documented salt-overly-sensitive (SOS) pathway (Chinnusamy et al., 2004; Mahajan et al., 2008), CaM-mediated signaling is also actively involved in plant response to osmotic stress (Bouche et al., 2005). Overexpression of a salt-induced CaM gene from soybean, GmCaM4, in Arabidopsis confers salt stress tolerance through the up-regulation of DNA-binding activity of a MYB transcription factor MYB2. Interestingly, MYB2 was also reported to interact with CaM in a Ca2+-dependent manner and regulate salt and dehydration responsive genes (Abe et al., 2003; Yoo et al., 2005). AtCML8, an ortholog of GmCaM4, was also found to be induced by salt treatment (Park et al., 2010). Another similar CML protein AtCML9 was found to be involved in osmotic stress tolerance through ABA-mediated pathways (Magnan et al., 2008). AtCML9 was readily induced by abiotic stress and ABA; knock-out mutant atcml9 showed a hypersensitive response to ABA during seed germination and seedling growth stages, and exhibited enhanced tolerance to salt and dehydration stresses. Furthermore, expression of several stress and ABA-responsive genes including RAB18, RD29A, and RD20 was altered in atcml9. The rice CML gene OsMSR2 was also suggested to be involved in ABA-mediated salt and drought tolerance (Xu et al., 2011). As the most abundant vacuolar Na+-proton exchanger in Arabidopsis, Na+/H+ exchanger 1 (AtNHX1) regulates various cellular activities such as maintaining pH, ion homeostasis, and protein trafficking. Yamaguchi et al. (2005) found that AtCaM15 (also called AtCML18) is localized in the vacuolar lumen and interacts with the C-terminus of AtNHX1. The interaction between AtCaM15 and AtNHX1 is affected by both Ca2+ and pH, and the binding of AtCaM15 to AtNHX1 alters the Na+/K+ selectivity of the exchanger by decreasing its Na+/H+ exchange speed. The interaction between AtCaM15 and AtNHX1 suggests the presence of Ca2+-pH-dependent signaling components in the vacuole, which are involved in mediating plant responses to salt stress. In addition to the above mentioned CaM/CMLs, CML37, CML38, and CML39 are also responsive to various stimuli, including salt, drought, and ABA (Vanderbeld and Snedden, 2007), but whether they are also involved in osmotic stress tolerance remains to be identified.
A few CaMBPs are involved in the signaling pathways triggered by salt, drought or osmotic stresses. Wheat (Triticum aestivum) TaCCaMK was down-regulated by ABA, salt and PEG treatments, and overexpression of TaCCaMK reduces ABA sensitivity of Arabidopsis, indicating that TaCCaMK is a negative regulator of ABA-mediated signaling (Yang et al., 2011). Arabidopsis AtACA4 encoding a CaM-regulated Ca2+-ATPase was found to be localized to small vacuoles, which is similar to PMC1, the yeast vacuolar Ca2+-ATPase, and AtACA4 confers tolerance against osmotic stresses imposed by high NaCl, KCl, and mannitol, when expressed in the yeast K616 strain lacking Ca2+ transporter PMC1 (Geisler et al., 2000). A CaM-regulated Ca2+-ATPase gene from soybean, SCA1, was found to be induced by salt stress (Chung et al., 2000). Methylglyoxal (MG), a byproduct of carbohydrate and lipid metabolism and a potent mutagenic chemical known to arrest growth, reacts with DNA and protein and increases sister chromatid exchange; and glyoxalase enzymes, including glyoxalase I (gly-I) and glyoxalase II (gly-II), catalyze the detoxification of MG with the involvement of glutathione (GSH; Thornalley, 1990). Glyoxalase I from Brassica juncea (BjGly-I) was reported to be a Ca2+/CBP, and its enzymatic function is significantly stimulated by Ca2+/CaM binding (Deswal and Sopory, 1999). The expression of BjGly-I is induced by salt, dehydration and heavy metal stresses; ectopic expression of BjGly-I in tobacco conferred remarkable tolerance to exogenous MG and high salt stress (Veena et al., 1999). AtCaMBP25 was identified to be a CaM-binding nuclear protein and is induced by dehydration, low temperature or high salinity. Overexpression of AtCaMBP25 compromised the tolerance of transgenic plants to osmotic stress, and silencing AtCaMBP25 via antisense approach increased plant tolerance to osmotic stress. These results suggested that AtCaMBP25 functions as a negative regulator in plant tolerance to osmotic stress, revealing a connection coupling Ca2+ signals to plant responses to osmotic stresses (Perruc et al., 2004).
Ca2+/CaM-regulated transcription factors are also involved in plant response to salt and drought stresses. A few CAMTA genes from Arabidopsis and soybean are up-regulated by salt and dehydration treatments (Yang and Poovaiah, 2002a; Galon et al., 2010; Wang et al., 2015). Arabidopsis CAMTA1 is involved in drought stress response (Pandey et al., 2013). Knockout mutant camta1 was shown to be more sensitive to drought stress, and expression of many drought responsive genes was affected in this mutant. Similar to AtCAMTA1, tomato CAMTA homolog SlSR1L was also positively involved in drought stress tolerance (Li et al., 2014). In addition to regulating salicylic acid (SA)-induced defense response and systemic acquired resistance (Wang et al., 2009; Zhang et al., 2010), AtCBP60g, a CaM-binding transcription factor from Arabidopsis was found to positively regulate drought stress response (Wan et al., 2012). Transgenic plants overexpressing CBP60g displayed hypersensitivity to ABA and enhanced tolerance to drought stress. AtGTL1 (GT-2 LIKE1), a CaM-binding member of the GTL transcription factor family, was found to be a negative regulator of drought tolerance (Yoo et al., 2010). AtGTL1 expression was down-regulated by dehydration stress, and loss-of-function mutant gtl1 showed better survival under drought stress by reducing transpiration, due to lower stomata density on the abaxial surface and higher expression of SDD1, which is a negative regulator of stomatal development and is repressed by AtGTL1 (Yoo et al., 2010). Similarly, PtaGTL1 identified from Populus tremula × P. alba could bind to CaM and regulate water use efficiency and drought tolerance (Weng et al., 2012). Another transcription factor AtABF2/AREB1, which was identified as CBP through protein microarray analysis (Popescu et al., 2007), was also found to be up-regulated by ABA, dehydration, and salinity stresses (Yoshida et al., 2010). Single and multiple mutants of ABF2, 3, and 4 showed varying degrees of reduced survival rate under drought conditions, implying functional redundancy among these three ABFs and Ca2+/CaM could regulate drought tolerance through ABF2/AREB1 (Yoshida et al., 2010).
Elevated concentration of both essential (e.g., Cu and Zn) and non-essential (e.g., Cd, Hg, Pb, and Ni) heavy metals in the soil can cause toxicity and inhibit plant growth. It was reported that Ca2+/CaM is involved in radish (Raphanus sativus L.) responses to Cd2+ toxicity during the early phases of seed germination (Rivetta et al., 1997). Ca2+ added in the medium could partially reverse the Cd2+-induced growth inhibition of the germinating embryo, and this coincides with decreased Cd2+ uptake. An equilibrium dialysis study revealed that Cd2+ compete with Ca2+ for CaM-binding, hence Cd2+ could significantly reduce the binding of Ca2+/CaM to its target proteins. Apparently, supplementation of Ca2+ in the medium counteracts the toxicity of Cd2+ by restoring the Ca2+-dependent interaction between CaM and its targets during the radish seed germination. A tobacco (N. tabacum) cyclic nucleotide gated ion channel (CNGC) called NtCBP4 was identified to be a CBP through protein–protein interaction-based library screening, and shown to be localized to plasma membrane. Transgenic tobacco plants with elevated expression of NtCBP4 displayed tolerance to Ni2+ and hypersensitivity to Pb2+, and consistently showed decreased Ni2+ and increased Pb2+ accumulation, suggesting that NtCBP4 is involved in heavy metal uptake across the plant plasma membrane (Arazi et al., 1999). However, transgenic plants expressing a truncated version of NtCBP4 lacking the C-terminal stretch covering the CaMBD and part of the putative cyclic nucleotide-binding domain showed improved tolerance to Pb2+ and lower accumulation of Pb2+, and loss-of-function mutation of AtCNGC1, a homolog of NtCBP4 in Arabidopsis, also resulted in Pb2+ tolerance. These results suggested that CaM-binding is required for the normal function of both AtCNGC1 and NtCBP4 for the transport of heavy metals (Sunkar et al., 2000).
Ca2+ is a critical second messenger coupling diverse stimuli to various physiological responses in plants. CaM, as well as CML, is one of the most extensively studied Ca2+ sensors, which mediate interpretation of Ca2+ signals in all aspects of plant life, especially in responses to environmental stresses, through interaction with and regulation of various downstream target proteins. Figure 1 depicts an overview of generation and interpretation of Ca2+ signals which are regulated by CaM/CMLs during plant responses to abiotic stresses. One of the most actively regulated class of target proteins are calcium permeable channels, pumps, and antiporters which are actively involved in the generation of intracellular Ca2+ transients. This indicates that the preciseness and accuracy of Ca2+ signal itself is closely monitored by CaM-mediated regulation. Although more than 50 proteins from different plant species have been identified as CBPs with well-defined CaM-binding properties (Table 1), the CaM-mediated regulations of these target proteins are frequently presumptive including SAURs, PCBP, AtBTs, and WRKYIIds (Yang and Poovaiah, 2000a; Reddy et al., 2002; Du and Poovaiah, 2004; Park et al., 2005). Only a few cases of CaM-mediated regulation in planta are supported with empirical evidences, such as GAD, CCaMK, AtCAT3, MLO, DWF1, AtSRs/CaMTAs, CRLK1, and CBP60g (Snedden et al., 1996; Kim et al., 2002; Yang and Poovaiah, 2002b; Du and Poovaiah, 2005; Gleason et al., 2006; Tirichine et al., 2006; Du et al., 2009; Wang et al., 2009; Yang et al., 2010b). Hence, more emphasis should be placed on studying the CaM-mediated regulation of target proteins to further improve our understanding of CaM-mediated signaling. Currently, most of the CBPs are targets of canonic CaMs which count for only 10% of the CaM/CML family. The targets of most of the CMLs are not reported yet, let alone the CML-mediated regulation of downstream targets and associated signal transduction. Identification of novel target proteins of CaMs and CMLs especially those interact with CMLs deserve special attentions. Environmental cues are known to trigger stimulus specific Ca2+ transients. In an effort to explain how the simple Ca2+ ion could act as a messenger to couple various environmental stimuli to appropriate physiological responses with astonishing accuracy, Webb et al. (1996) proposed the theory of “Ca2+ signature” which hypothesized that stimulus-triggered increases in intracellular Ca2+ concentration vary in terms of duration, frequency, amplitude, and spatial distribution, and these carry specific information when they are interpreted into different physiological responses. An obvious support for this hypothesis is that the different Ca2+ spikes triggered by Nod factor from rhizobia and Myc factor from arbuscular mycorrhizal fungi could be interpreted through the action of the same Ca2+, Ca2+/CaM dependent protein kinase, CCaMK, into different physiological responses to support the establishment of root nodulation symbiosis or arbuscular mycorrhization (Kosuta et al., 2008). Although exciting progress on how Ca2+ signals are interpreted into various physiological responses has been made in the last decade, what we know so far may be very limited in scope when one considers the complicated Ca2+ signaling network. Many issues such as specificity, preference and flexibility of interaction between various CaM/CMLs and target proteins in planta are barely understood. The dynamics of Ca2+/CaM mediated regulation, the mechanistic details by which a particular effector detects a difference in Ca2+ signature and initiates distinct signaling pathways, are basically unknown. Progress in addressing these issues will help in understanding the most amazing properties, the versatility, efficiency and accuracy of Ca2+-mediated signaling in plant responses to environmental stresses.
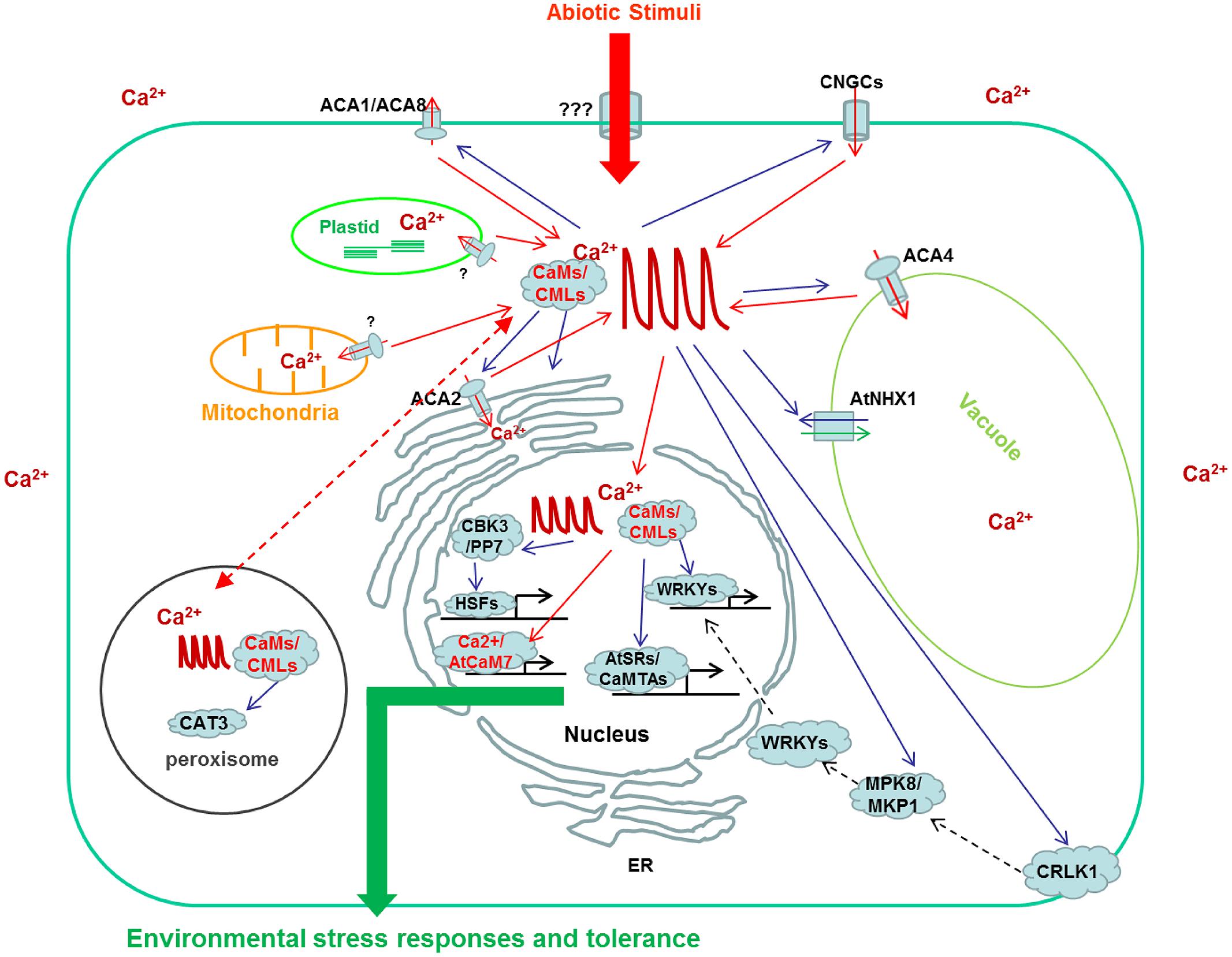
FIGURE 1. Schematic representation of Ca2+ transients and their modification and interpretation by CaM/CMLs as well as their target proteins in plant cells under abiotic stresses. This model is not exhaustive and only includes the actions of a limited number of CaM/CMLs and target proteins; CaMs/CMLs/CBPs involved in biotic stresses and Ca2+ signal interpretation by other sensors such as CBLs and CDPKs are not included. Actions modifing Ca2+ transients or CaM/CMLs are presented by red arrows and actions regulated by Ca2+/CaMs or Ca2+/CMLs are presented by blue arrows. The dashed arrows imply multiple regulations extended to nucleus.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Research of the authors is supported by the National Natural Science Foundation of China grants (U1130304 and 31201679), Zhejiang Provincial Natural Science Foundation of China grant (LY15C020006) and US National Science Foundation grant (1021344).
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Apse, M. P., Aharon, G. S., Snedden, W. A., and Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258. doi: 10.1126/science.285.5431.1256
Arazi, T., Sunkar, R., Kaplan, B., and Fromm, H. (1999). A tobacco plasma membrane calmodulin-binding transporter confers Ni2+ tolerance and Pb2+ hypersensitivity in transgenic plants. Plant J. 20, 171–182. doi: 10.1046/j.1365-313x.1999.00588.x
Aurisano, N., Bertani, A., and Reggiani, R. (1995). Involvement of calcium and calmodulin in protein and amino acid metabolism in rice roots under anoxia. Plant Cell Physiol. 36, 1525–1529. doi: 10.1080/11263509509440949
Boonburapong, B., and Buaboocha, T. (2007). Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biology 7:4. doi: 10.1186/1471-2229-7-4
Botella, J. R., and Arteca, R. N. (1994). Differential expression of two calmodulin genes in response to physical and chemical stimuli. Plant Mol. Biol. 24, 757–766. doi: 10.1007/BF00029857
Bouche, N., Fait, A., Bouchez, D., Moller, S. G., and Fromm, H. (2003). Mitochondrial succinic-semialdehyde dehydrogenase of the gamma-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proc. Natl. Acad. Sci. U.S.A. 100, 6843–6848. doi: 10.1073/pnas.1037532100
Bouche, N., Scharlat, A., Snedden, W., Bouchez, D., and Fromm, H. (2002). A novel family of calmodulin-binding transcription activators in multicellular organisms. J. Biol. Chem. 277, 21851–21861. doi: 10.1074/jbc.M200268200
Bouche, N., Yellin, A., Snedden, W. A., and Fromm, H. (2005). Plant-specific calmodulin-binding proteins. Annu. Rev. Plant Biol. 56, 435–466. doi: 10.1146/annurev.arplant.56.032604.144224
Boursiac, Y., Lee, S. M., Romanowsky, S., Blank, R., Sladek, C., Chung, W. S., et al. (2010). Disruption of the vacuolar calcium-ATPases in Arabidopsis results in the activation of a salicylic acid-dependent programmed cell death pathway. Plant Physiol. 154, 1158–1171. doi: 10.1104/pp.110.159038
Braam, J. (1992). Regulated expression of the calmodulin-related TCH genes in cultured Arabidopsis cells: induction by calcium and heat shock. Proc. Natl. Acad. Sci. U.S.A. 89, 3213–3216. doi: 10.1073/pnas.89.8.3213
Cha, J.-Y., Su’udi, M., Kim, W.-Y., Kim, D. R., Kwak, Y.-S., and Son, D. (2012). Functional characterization of orchardgrass cytosolic Hsp70 (DgHsp70) and the negative regulation by Ca2+/AtCaM2 binding. Plant Physiol. Biochem. 58, 29–36. doi: 10.1016/j.plaphy.2012.06.006
Chinnusamy, V., Schumaker, K., and Zhu, J.-K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 55, 225–236. doi: 10.1093/jxb/erh005
Choi, M. S., Kim, M. C., Yoo, J. H., Moon, B. C., Koo, S. C., Park, B. O., et al. (2005). Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.). J. Biol. Chem. 280, 40820–40831. doi: 10.1074/jbc.M504616200
Chung, W. S., Lee, S. H., Kim, J. C., Heo, W. D., Kim, M. C., Park, C. Y., et al. (2000). Identification of a calmodulin-regulated soybean Ca(2+)-ATPase (SCA1) that is located in the plasma membrane. Plant Cell 12, 1393–1407. doi: 10.2307/3871138
Delk, N. A., Johnson, K. A., Chowdhury, N. I., and Braam, J. (2005). CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol. 139, 240–253. doi: 10.1104/pp.105.062612
Deswal, R., and Sopory, S. K. (1999). Glyoxalase I from Brassica juncea is a calmodulin stimulated protein. Biochim. Biophys. Acta 1450, 460–467. doi: 10.1016/S0167-4889(99)00047-6
Dobney, S., Chiasson, D., Lam, P., Smith, S. P., and Snedden, W. A. (2009). The calmodulin-related calcium sensor CML42 plays a role in trichome branching. J. Biol. Chem. 284, 31647–31657. doi: 10.1074/jbc.M109.056770
Dodd, A. N., Kudla, J., and Sanders, D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620. doi: 10.1146/annurev-arplant-070109-104628
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., and Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. doi: 10.1105/tpc.108.063958
Du, L., Ali, G. S., Simons, K. A., Hou, J., Yang, T., Reddy, A. S. N., et al. (2009). Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158. doi: 10.1038/nature07612
Du, L., and Poovaiah, B. W. (2004). A novel family of Ca2+/calmodulin-binding proteins involved in transcriptional regulation: interaction with fsh/Ring3 class transcription activators. Plant Mol. Biol. 54, 549–569. doi: 10.1023/B:PLAN.0000038269.98972.bb
Du, L., and Poovaiah, B. W. (2005). Ca2+/calmodulin is critical for brassinosteroid biosynthesis and plant growth. Nature 437, 741–745. doi: 10.1038/nature03973
Du, L., Yang, T., Puthanveettil, S. V., Poovaiah, B. W., and Luan, S. (2011). “Decoding of calcium signal through calmodulin: calmodulin-binding proteins in plants coding and decoding of calcium signals in plants,” in Coding and Decoding of Calcium Signals in Plants, ed. S. Luan (Berlin: Springer), 177–233.
Evans, N. H., Mcainsh, M. R., and Hetherington, A. M. (2001). Calcium oscillations in higher plants. Curr. Opin. Plant Biol. 4, 415–420. doi: 10.1016/S1369-5266(00)00194-1
Fromm, H., and Chua, N.-H. (1992). Cloning of plant cDNAs encoding calmodulin-binding proteins using 35S-labeled recombinant calmodulin as a probe. Plant Mol. Biol. Rep. 10, 199–206. doi: 10.1007/BF02668347
Galon, Y., Aloni, R., Nachmias, D., Snir, O., Feldmesser, E., Scrase-Field, S., et al. (2010). Calmodulin-binding transcription activator 1 mediates auxin signaling and responds to stresses in Arabidopsis. Planta 232, 165–178. doi: 10.1007/s00425-010-1153-6
Geisler, M., Frangne, N., Gomes, E., Martinoia, E., and Palmgren, M. G. (2000). The ACA4 gene of Arabidopsis encodes a vacuolar membrane calcium pump that improves salt tolerance in yeast. Plant Physiol. 124, 1814–1827. doi: 10.1104/pp.124.4.1814
Gleason, C., Chaudhuri, S., Yang, T., Munoz, A., Poovaiah, B. W., and Oldroyd, G. E. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441, 1149–1152. doi: 10.1038/nature04812
Gong, M., Chen, S.-N., Song, Y.-Q., and Li, Z.-G. (1997a). Effect of calcium and calmodulin on intrinsic heat tolerance in relation to antioxidant systems in maize seedlings. Funct. Plant Biol. 24, 371–379. doi: 10.1071/pp96118
Gong, M., Li, Y.-J., Dai, X., Tian, M., and Li, Z.-G. (1997b). Involvement of calcium and calmodulin in the acquisition of heat-shock induced thermotolerance in maize seedlings. J. Plant Physiol. 150, 615–621. doi: 10.1016/S0176-1617(97)80328-8
Gong, M., and Li, Z.-G. (1995). Calmodulin-binding proteins from Zea mays germs. Phytochemistry 40, 1335–1339. doi: 10.1016/0031-9422(95)00381-G
Gong, M., Van Der Luit, A. H., Knight, M. R., and Trewavas, A. J. (1998). Heat-shock-induced changes in intracellular Ca2+ level in tobacco seedlings in relation to thermotolerance. Plant Physiol. 116, 429–437. doi: 10.1104/pp.116.1.429
Gould, K. S., Lamotte, O., Klinguer, A., Pugin, A., and Wendehenne, D. (2003). Nitric oxide production in tobacco leaf cells: a generalized stress response? Plant Cell Environ. 26, 1851–1862. doi: 10.1046/j.1365-3040.2003.01101.x
Hannah, M. A., Heyer, A. G., and Hincha, D. K. (2005). A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet 1:e26. doi: 10.1371/journal.pgen.0010026
Harding, S. A., Oh, S. H., and Roberts, D. M. (1997). Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J. 16, 1137–1144. doi: 10.1093/emboj/16.6.1137
Hill, K., Wang, H., and Perry, S. E. (2008). A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 53, 172–185. doi: 10.1111/j.1365-313X.2007.03336.x
Hoeflich, K. P., and Ikura, M. (2002). Calmodulin in action: diversity in target recognition and activation mechanisms. Cell 108, 739–742. doi: 10.1016/S0092-8674(02)00682-7
Karita, E., Yamakawa, H., Mitsuhara, I., Kuchitsu, K., and Ohashi, Y. (2004). Three types of tobacco calmodulins characteristically activate plant NAD kinase at different Ca2+ concentrations and pHs. Plant Cell Physiol. 45, 1371–1379. doi: 10.1093/pcp/pch158
Kim, M. C., Chung, W. S., Yun, D. J., and Cho, M. J. (2009). Calcium and calmodulin-mediated regulation of gene expression in plants. Mol. Plant 2, 13–21. doi: 10.1093/mp/ssn091
Kim, M. C., Panstruga, R., Elliott, C., Muller, J., Devoto, A., Yoon, H. W., et al. (2002). Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416, 447–451. doi: 10.1038/416447a
Kim, Y., Park, S., Gilmour, S. J., and Thomashow, M. F. (2013). Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 75, 364–376. doi: 10.1111/tpj.12205
Knight, M. R., Campbell, A. K., Smith, S. M., and Trewavas, A. J. (1991). Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352, 524–526. doi: 10.1038/352524a0
Kosuta, S., Hazledine, S., Sun, J., Miwa, H., Morris, R. J., Downie, J. A., et al. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. U.S.A. 105, 9823–9828. doi: 10.1073/pnas.0803499105
Kudla, J., Batistic, O., and Hashimoto, K. (2010). Calcium signals: the lead currency of plant information processing. Plant Cell 22, 541–563. doi: 10.1105/tpc.109.072686
Kushwaha, R., Singh, A., and Chattopadhyay, S. (2008). Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell 20, 1747–1759. doi: 10.1105/tpc.107.057612
Kutuzov, M. A., Bennett, N., and Andreeva, A. V. (2001). Interaction of plant protein Ser/Thr phosphatase PP7 with calmodulin. Biochem. Biophys. Res. Commun. 289, 634–640. doi: 10.1006/bbrc.2001.6020
Larkindale, J., and Knight, M. R. (2002). Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 128, 682–695. doi: 10.1104/pp.010320
Lee, S. H., Johnson, J. D., Walsh, M. P., Van Lierop, J. E., Sutherland, C., Xu, A. D., et al. (2000). Differential regulation of Ca2+/calmodulin-dependent enzymes by plant calmodulin isoforms and free Ca2+ concentration. Biochem. J. 350, 299–306. doi: 10.1042/0264-6021:3500299
Li, X., Huang, L., Zhang, Y., Ouyang, Z., Hong, Y., Zhang, H., et al. (2014). Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant Biol. 14:286. doi: 10.1186/s12870-014-0286-3
Liu, H. T., Gao, F., Li, G. L., Han, J. L., Liu, D. L., Sun, D. Y., et al. (2008). The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana. Plant J. 55, 760–773. doi: 10.1111/j.1365-313X.2008.03544.x
Liu, H. T., Li, B., Shang, Z. L., Li, X. Z., Mu, R. L., Sun, D. Y., et al. (2003). Calmodulin is involved in heat shock signal transduction in wheat. Plant Physiol. 132, 1186–1195. doi: 10.1104/pp.102.018564
Liu, H. T., Li, G. L., Chang, H., Sun, D. Y., Zhou, R. G., and Li, B. (2007). Calmodulin-binding protein phosphatase PP7 is involved in thermotolerance in Arabidopsis. Plant Cell Environ. 30, 156–164. doi: 10.1111/j.1365-3040.2006.01613.x
Liu, H.-T., Sun, D.-Y., and Zhou, R.-G. (2005). Ca2+ and AtCaM3 are involved in the expression of heat shock protein gene in Arabidopsis. Plant Cell Environ. 28, 1276–1284. doi: 10.1111/j.1365-3040.2005.01365.x
Lu, Y. T., Dharmasiri, M. A., and Harrington, H. M. (1995). Characterization of a cDNA encoding a novel heat-shock protein that binds to calmodulin. Plant Physiol. 108, 1197–1202. doi: 10.1104/pp.108.3.1197
Luan, S. (2009). The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 14, 37–42. doi: 10.1016/j.tplants.2008.10.005
Ma, F. F., Lu, R., Liu, H. Y., Shi, B., Zhang, J. H., Tan, M. P., et al. (2012). Nitric oxide-activated calcium/calmodulin-dependent protein kinase regulates the abscisic acid-induced antioxidant defence in maize. J. Exp. Bot. 63, 4835–4847. doi: 10.1093/jxb/ers161
Magnan, F., Ranty, B., Charpenteau, M., Sotta, B., Galaud, J. P., and Aldon, D. (2008). Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 56, 575–589. doi: 10.1111/j.1365-313X.2008.03622.x
Mahajan, S., Pandey, G. K., and Tuteja, N. (2008). Calcium- and salt-stress signaling in plants: shedding light on SOS pathway. Arch. Biochem. Biophys. 471, 146–158. doi: 10.1016/j.abb.2008.01.010
Mandadi, K. K., Misra, A., Ren, S., and Mcknight, T. D. (2009). BT2, a BTB protein, mediates multiple responses to nutrients, stresses, and hormones in Arabidopsis. Plant Physiol. 150, 1930–1939. doi: 10.1104/pp.109.139220
McCormack, E., and Braam, J. (2003). Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol. 159, 585–598. doi: 10.1046/j.1469-8137.2003.00845.x
McCormack, E., Tsai, Y.-C., and Braam, J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389. doi: 10.1016/j.tplants.2005.07.001
Medda, R., Padiglia, A., Longu, S., Bellelli, A., Arcovito, A., Cavallo, S., et al. (2003). Critical role of Ca2+ ions in the reaction mechanism of Euphorbia characias peroxidase. Biochemistry 42, 8909–8918. doi: 10.1021/bi034609z
Mura, A., Medda, R., Longu, S., Floris, G., Rinaldi, A. C., and Padiglia, A. (2005). A Ca2+/calmodulin-binding peroxidase from Euphorbia latex: novel aspects of calcium-hydrogen peroxide cross-talk in the regulation of plant defenses. Biochemistry 44, 14120–14130. doi: 10.1021/bi0513251
Pandey, N., Ranjan, A., Pant, P., Tripathi, R. K., Ateek, F., Pandey, H. P., et al. (2013). CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genomics 14:216. doi: 10.1186/1471-2164-14-216
Pandey, S., Tiwari, S. B., Tyagi, W., Reddy, M. K., Upadhyaya, K. C., and Sopory, S. K. (2002). A Ca2+/CaM-dependent kinase from pea is stress regulated and in vitro phosphorylates a protein that binds to AtCaM5 promoter. Eur. J. Biochem. 269, 3193–3204. doi: 10.1046/j.1432-1033.2002.02994.x
Park, C. Y., Lee, J. H., Yoo, J. H., Moon, B. C., Choi, M. S., Kang, Y. H., et al. (2005). WRKY group IId transcription factors interact with calmodulin. FEBS Lett. 579, 1545–1550. doi: 10.1016/j.febslet.2005.01.057
Park, H. C., Kim, M. L., Kang, Y. H., Jeon, J. M., Yoo, J. H., Kim, M. C., et al. (2004). Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 135, 2150–2161. doi: 10.1104/pp.104.041442
Park, H. C., Park, C. Y., Koo, S. C., Cheong, M. S., Kim, K. E., Kim, M. C., et al. (2010). AtCML8, a calmodulin-like protein, differentially activating CaM-dependent enzymes in Arabidopsis thaliana. Plant Cell Rep. 29, 1297–1304. doi: 10.1007/s00299-010-0916-7
Pauly, N., Knight, M. R., Thuleau, P., Van Der Luit, A. H., Moreau, M., Trewavas, A. J., et al. (2000). Control of free calcium in plant cell nuclei. Nature 405, 754–755. doi: 10.1038/35015671
Perochon, A., Aldon, D., Galaud, J.-P., and Ranty, B. (2011). Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie 93, 2048–2053. doi: 10.1016/j.biochi.2011.07.012
Perruc, E., Charpenteau, M., Ramirez, B. C., Jauneau, A., Galaud, J. P., Ranjeva, R., et al. (2004). A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant J. 38, 410–420. doi: 10.1111/j.1365-313X.2004.02062.x
Polisensky, D. H., and Braam, J. (1996). Cold-shock regulation of the Arabidopsis TCH genes and the effects of modulating intracellular calcium levels. Plant Physiol. 111, 1271–1279. doi: 10.1104/pp.111.4.1271
Poovaiah, B. W., Du, L., Wang, H., and Yang, T. (2013). Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant Physiol. 163, 531–542. doi: 10.1104/pp.113.220780
Poovaiah, B. W., and Reddy, A. S. (1993). Calcium and signal transduction in plants. CRC Crit. Rev. Plant Sci. 12, 185–211. doi: 10.1080/713608046
Popescu, S. C., Popescu, G. V., Bachan, S., Zhang, Z., Seay, M., Gerstein, M., et al. (2007). Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. U.S.A. 104, 4730–4735. doi: 10.1073/pnas.0611615104
Price, A. H., Taylor, A., Ripley, S. J., Griffiths, A., Trewavas, A. J., and Knight, M. R. (1994). Oxidative signals in tobacco increase cytosolic calcium. Plant Cell 6, 1301–1310. doi: 10.1105/tpc.6.9.1301
Reddy, A. S. N. (2001). Calcium: silver bullet in signaling. Plant Sci. 160, 381–404. doi: 10.1016/S0168-9452(00)00386-1
Reddy, A. S., Ali, G. S., Celesnik, H., and Day, I. S. (2011). Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23, 2010–2032. doi: 10.1105/tpc.111.084988
Reddy, A. S. N., Reddy, V. S., and Golovkin, M. (2000). A calmodulin binding protein from Arabidopsis is induced by ethylene and contains a DNA-binding motif. Biochem. Biophys. Res. Commun. 279, 762–769. doi: 10.1006/bbrc.2000.4032
Reddy, A. S. N., Takezawa, D., Fromm, H., and Poovaiah, B. W. (1993). Isolation and characterization of 2 cdnas that encode for calmodulin-binding proteins from corn root-tips. Plant Sci. 94, 109–117. doi: 10.1016/0168-9452(93)90012-O
Reddy, V. S., Ali, G. S., and Reddy, A. S. (2002). Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J. Biol. Chem. 277, 9840–9852. doi: 10.1074/jbc.M111626200
Rentel, M. C., and Knight, M. R. (2004). Oxidative stress-induced calcium signaling in Arabidopsis. Plant Physiol. 135, 1471–1479. doi: 10.1104/pp.104.042663
Rhoads, A. R., and Friedberg, F. (1997). Sequence motifs for calmodulin recognition. FASEB J. 11, 331–340.
Rivetta, A., Negrini, N., and Cocucci, M. (1997). Involvement of Ca2+-calmodulin in Cd2+ toxicity during the early phases of radish (Raphanus sativus L.) seed germination. Plant Cell Environ. 20, 600–608. doi: 10.1111/j.1365-3040.1997.00072.x
Ruiz, J. M., Sanchez, E., Garcia, P. C., Lopez-Lefebre, L. R., Rivero, R. M., and Romero, L. (2002). Proline metabolism and NAD kinase activity in greenbean plants subjected to cold-shock. Phytochemistry 59, 473–478. doi: 10.1016/S0031-9422(01)00481-2
Shi, B., Ni, L., Zhang, A., Cao, J., Zhang, H., Qin, T., et al. (2012). OsDMI3 is a novel component of abscisic acid signaling in the induction of antioxidant defense in leaves of rice. Mol. Plant 5, 1359–1374. doi: 10.1093/mp/sss068
Singla-Pareek, S. L., Reddy, M. K., and Sopory, S. K. (2003). Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc. Natl. Acad. Sci. U.S.A. 100, 14672–14677. doi: 10.1073/pnas.2034667100
Snedden, W. A., and Fromm, H. (1998). Calmodulin, calmodulin-related proteins and plant responses to the environment. Trends Plant Sci. 3, 299–304. doi: 10.1016/S1360-1385(98)01284-9
Snedden, W. A., and Fromm, H. (2001). Calmodulin as a versatile calcium signal transducer in plants. New Phytol. 151, 35–66. doi: 10.1046/j.1469-8137.2001.00154.x
Snedden, W. A., Koutsia, N., Baum, G., and Fromm, H. (1996). Activation of a recombinant Petunia glutamate decarboxylase by calcium/calmodulin or by a monoclonal antibody which recognizes the calmodulin binding domain. J. Biol. Chem. 271, 4148–4153. doi: 10.1074/jbc.271.8.4148
Song, L. L., Ding, W., Zhao, M. G., Sun, B. T., and Zhang, L. X. (2006). Nitric oxide protects against oxidative stress under heat stress in the calluses from two ecotypes of reed. Plant Sci. 171, 449–458. doi: 10.1016/j.plantsci.2006.05.002
Subbaiah, C. C., and Sachs, M. M. (2000). Maize cap1 encodes a novel SERCA-type calcium-ATPase with a calmodulin-binding domain. J. Biol. Chem. 275, 21678–21687. doi: 10.1074/jbc.M001484200
Sun, X. T., Li, B., Zhou, G. M., Tang, W. Q., Bai, J., Sun, D. Y., et al. (2000). Binding of the maize cytosolic Hsp70 to calmodulin, and identification of calmodulin-binding site in Hsp70. Plant Cell Physiol. 41, 804–810. doi: 10.1093/pcp/41.6.804
Sunkar, R., Kaplan, B., Bouche, N., Arazi, T., Dolev, D., Talke, I. N., et al. (2000). Expression of a truncated tobacco NtCBP4 channel in transgenic plants and disruption of the homologous Arabidopsis CNGC1 gene confer Pb2+ tolerance. Plant J. 24, 533–542. doi: 10.1046/j.1365-313x.2000.00901.x
Takahashi, F., Mizoguchi, T., Yoshida, R., Ichimura, K., and Shinozaki, K. (2011). Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis. Mol. Cell. 41, 649–660. doi: 10.1016/j.molcel.2011.02.029
Takezawa, D., and Minami, A. (2004). Calmodulin-binding proteins in bryophytes: identification of abscisic acid-, cold-, and osmotic stress-induced genes encoding novel membrane-bound transporter-like proteins. Biochem. Biophys. Res. Commun. 317, 428–436. doi: 10.1016/j.bbrc.2004.03.052
Thornalley, P. J. (1990). The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 269, 1–11.
Tirichine, L., Imaizumi-Anraku, H., Yoshida, S., Murakami, Y., Madsen, L. H., Miwa, H., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441, 1153–1156. doi: 10.1038/nature04862
Townley, H. E., and Knight, M. R. (2002). Calmodulin as a potential negative regulator of Arabidopsis COR gene expression. Plant Physiol. 128, 1169–1172. doi: 10.1104/pp.010814
Turner, W. L., Waller, J. C., Vanderbeld, B., and Snedden, W. A. (2004). Cloning and characterization of two NAD kinases from Arabidopsis. identification of a calmodulin binding isoform. Plant Physiol. 135, 1243–1255. doi: 10.1104/pp.104.040428
Uchida, A., Jagendorf, A. T., Hibino, T., Takabe, T., and Takabe, T. (2002). Effects of hydrogen peroxide and nitric oxide on both salt and heat stress tolerance in rice. Plant Sci. 163, 515–523. doi: 10.1016/S0168-9452(02)00159-0
Vadassery, J., Reichelt, M., Hause, B., Gershenzon, J., Boland, W., and Mithofer, A. (2012). CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 159, 1159–1175. doi: 10.1104/pp.112.198150
Van Breusegem, F., Vranov, E., Dat, J. F., and Inz, D. (2001). The role of active oxygen species in plant signal transduction. Plant Sci. 161, 405–414. doi: 10.1016/S0168-9452(01)00452-6
Vanderbeld, B., and Snedden, W. A. (2007). Developmental and stimulus-induced expression patterns of Arabidopsis calmodulin-like genes CML37, CML38 and CML39. Plant Mol. Biol. 64, 683–697. doi: 10.1007/s11103-007-9189-0
Van Der Luit, A. H., Olivari, C., Haley, A., Knight, M. R., and Trewavas, A. J. (1999). Distinct calcium signaling pathways regulate calmodulin gene expression in tobacco. Plant Physiol. 121, 705–714. doi: 10.1104/pp.121.3.705
Veena, A., Reddy, V. S., and Sopory, S. K. (1999). Glyoxalase I from Brassica juncea: molecular cloning, regulation and its over-expression confer tolerance in transgenic tobacco under stress. Plant J. 17, 385–395. doi: 10.1046/j.1365-313X.1999.00390.x
Wan, D. L., Li, R. L., Zou, B., Zhang, X., Cong, J. Y., Wang, R. G., et al. (2012). Calmodulin-binding protein CBP60g is a positive regulator of both disease resistance and drought tolerance in Arabidopsis. Plant Cell Rep. 31, 1269–1281. doi: 10.1007/s00299-012-1247-7
Wang, G., Zeng, H., Hu, X., Zhu, Y., Chen, Y., Shen, C., et al. (2015). Identification and expression analyses of calmodulin-binding transcription activator genes in soybean. Plant Soil 386, 205–221. doi: 10.1007/s11104-014-2267-6
Wang, L., Tsuda, K., Sato, M., Cohen, J. D., Katagiri, F., and Glazebrook, J. (2009). Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog 5:e1000301. doi: 10.1371/journal.ppat.1000301
Wang, X.-H., Li, Q.-T., Chen, H.-W., Zhang, W.-K., Ma, B., Chen, S.-Y., et al. (2014). Trihelix transcription factor GT-4 mediates salt tolerance via interaction with TEM2 in Arabidopsis. BMC Plant Biol. 14:339. doi: 10.1186/s12870-014-0339-7
Wang, Y., Liang, S., Xie, Q. G., and Lu, Y. T. (2004). Characterization of a calmodulin-regulated Ca2+-dependent-protein-kinase-related protein kinase, AtCRK1, from Arabidopsis. Biochem. J. 383, 73–81. doi: 10.1042/BJ20031907
Webb, A. A. R., Mcainsh, M. R., Taylor, J. E., and Hetherington, A. M. (1996). “Calcium ions as intracellular second messengers in higher plants,” in Advances in Botanical Research, ed. J. A. Callow (Waltham, MA: Academic Press), 45–96.
Weng, H., Yoo, C. Y., Gosney, M. J., Hasegawa, P. M., and Mickelbart, M. V. (2012). Poplar GTL1 is a Ca2+/calmodulin-binding transcription factor that functions in plant water use efficiency and drought tolerance. PLoS ONE 7:e32925. doi: 10.1371/journal.pone.0032925
Wu, H. C., and Jinn, T. L. (2012). Oscillation regulation of Ca (2+) /calmodulin and heat-stress related genes in response to heat stress in rice (Oryza sativa L.). Plant Signal Behav. 7, 1056–1057. doi: 10.4161/psb.21124
Wu, H. C., Luo, D. L., Vignols, F., and Jinn, T. L. (2012). Heat shock-induced biphasic Ca2+ signature and OsCaM1-1 nuclear localization mediate downstream signalling in acquisition of thermotolerance in rice (Oryza sativa L.). Plant Cell Environ. 35, 1543–1557. doi: 10.1111/j.1365-3040.2012.02508.x
Xi, J., Qiu, Y., Du, L., and Poovaiah, B. W. (2012). Plant-specific trihelix transcription factor AtGT2L interacts with calcium/calmodulin and responds to cold and salt stresses. Plant Sci. 185–186, 274–280. doi: 10.1016/j.plantsci.2011.11.013
Xing, T., Higgins, V. J., and Blumwald, E. (1997). Race-specific elicitors of Cladosporium fulvum promote translocation of cytosolic components of NADPH oxidase to the plasma membrane of tomato cells. Plant Cell 9, 249–259. doi: 10.1105/tpc.9.2.249
Xiong, L., and Zhu, J. K. (2002). Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ. 25, 131–139. doi: 10.1046/j.1365-3040.2002.00782.x
Xu, G. Y., Rocha, P. S., Wang, M. L., Xu, M. L., Cui, Y. C., Li, L. Y., et al. (2011). A novel rice calmodulin-like gene, OsMSR2, enhances drought and salt tolerance and increases ABA sensitivity in Arabidopsis. Planta 234, 47–59. doi: 10.1007/s00425-011-1386-z
Xuan, Y., Zhou, S., Wang, L., Cheng, Y. D., and Zhao, L. Q. (2010). Nitric oxide functions as a signal and acts upstream of AtCaM3 in thermotolerance in Arabidopsis seedlings. Plant Physiol. 153, 1895–1906. doi: 10.1104/pp.110.160424
Yamaguchi, T., Aharon, G. S., Sottosanto, J. B., and Blumwald, E. (2005). Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 102, 16107–16112. doi: 10.1073/pnas.0504437102
Yamniuk, A. P., and Vogel, H. J. (2004). Calmodulin’s flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 27, 33–57. doi: 10.1385/MB:27:1:33
Yamniuk, A. P., and Vogel, H. J. (2005). Structural investigation into the differential target enzyme regulation displayed by plant calmodulin isoforms. Biochemistry 44, 3101–3111. doi: 10.1021/bi047770y
Yang, C., Li, A. L., Zhao, Y. L., Zhang, Z. L., Zhu, Y. F., Tan, X. M., et al. (2011). Overexpression of a wheat CCaMK gene reduces ABA sensitivity of Arabidopsis thaliana during seed germination and seedling growth. Plant Mol. Biol. Rep. 29, 681–692. doi: 10.1007/s11105-010-0275-0
Yang, T., Ali, G. S., Yang, L., Du, L., Reddy, A. S., and Poovaiah, B. W. (2010a). Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal. Behav. 5, 991–994. doi: 10.4161/psb.5.8.12225
Yang, T., Chaudhuri, S., Yang, L., Du, L., and Poovaiah, B. W. (2010b). A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J. Biol. Chem. 285, 7119–7126. doi: 10.1074/jbc.M109.035659
Yang, T., Chaudhuri, S., Yang, L., Chen, Y., and Poovaiah, B. W. (2004). Calcium/Calmodulin Up-regulates a cytoplasmic receptor-like kinase in plants. J. Biol. Chem. 279, 42552–42559. doi: 10.1074/jbc.M402830200
Yang, T., and Poovaiah, B. W. (2000a). An early ethylene up-regulated gene encoding a calmodulin-binding protein involved in plant senescence and death. J. Biol. Chem. 275, 38467–38473. doi: 10.1074/jbc.M003566200
Yang, T. B., and Poovaiah, B. W. (2000b). Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J. Biol. Chem. 275, 3137–3143. doi: 10.1074/jbc.275.5.3137
Yang, T., and Poovaiah, B. W. (2002a). A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 277, 45049–45058. doi: 10.1074/jbc.M207941200
Yang, T., and Poovaiah, B. W. (2002b). Hydrogen peroxide homeostasis: activation of plant catalase by calcium/calmodulin. Proc. Natl. Acad. Sci. U.S.A. 99, 4097–4102. doi: 10.1073/pnas.052564899
Yang, T., and Poovaiah, B. W. (2003). Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci. 8, 505–512. doi: 10.1016/j.tplants.2003.09.004
Yoo, C. Y., Pence, H. E., Jin, J. B., Miura, K., Gosney, M. J., Hasegawa, P. M., et al. (2010). The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 22, 4128–4141. doi: 10.1105/tpc.110.078691
Yoo, J. H., Park, C. Y., Kim, J. C., Do Heo, W., Cheong, M. S., Park, H. C., et al. (2005). Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J. Biol. Chem. 280, 3697–3706. doi: 10.1074/jbc.M408237200
Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi: 10.1111/j.1365-313X.2009.04092.x
Zhang, W., Zhou, R.-G., Gao, Y.-J., Zheng, S.-Z., Xu, P., Zhang, S.-Q., et al. (2009). Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiol. 149, 1773–1784. doi: 10.1104/pp.108.133744
Zhang, Y., Xu, S., Ding, P., Wang, D., Cheng, Y. T., He, J., et al. (2010). Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 107, 18220–18225. doi: 10.1073/pnas.1005225107
Zhou, Y. P., Duan, J., Fujibe, T., Yamamoto, K. T., and Tian, C. E. (2012). AtIQM1, a novel calmodulin-binding protein, is involved in stomatal movement in Arabidopsis. Plant Mol. Biol. 79, 333–346. doi: 10.1007/s11103-012-9915-0
Zhu, J. K. (2001). Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 4, 401–406. doi: 10.1016/S1369-5266(00)00192-8
Keywords: calcium signal, calmodulin, calmodulin-like protein, calmodulin-binding protein, signal transduction, abiotic stress
Citation: Zeng H, Xu L, Singh A, Wang H, Du L and Poovaiah BW (2015) Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 6:600. doi: 10.3389/fpls.2015.00600
Received: 05 June 2015; Accepted: 20 July 2015;
Published: 11 August 2015.
Edited by:
Girdhar Kumar Pandey, University of Delhi South Campus, IndiaReviewed by:
Wayne Snedden, Queen’s University, CanadaCopyright © 2015 Zeng, Xu, Singh, Wang, Du and Poovaiah. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liqun Du, College of Life and Environmental Sciences, Hangzhou Normal University, Hangzhou 310036, China,bGlxdW5kdUBoem51LmVkdS5jbg==; B. W. Poovaiah, Laboratory of Molecular Plant Science, Department of Horticulture, Washington State University, Pullman, WA 99164-6414, USA,cG9vdmFpYWhAd3N1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.