
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 12 June 2015
Sec. Plant Breeding
Volume 6 - 2015 | https://doi.org/10.3389/fpls.2015.00444
This article is part of the Research Topic Advances in genetic engineering strategies for fruit crop breeding View all 4 articles
Beside the well known nutritional and health benefits, strawberry (Fragaria X ananassa) crop draws increasing attention as plant model system for the Rosaceae family, due to the short generation time, the rapid in vitro regeneration, and to the availability of the genome sequence of F. X ananassa and F. vesca species. In the last years, the use of high-throughput sequence technologies provided large amounts of molecular information on the genes possibly related to several biological processes of this crop. Nevertheless, the function of most genes or gene products is still poorly understood and needs investigation. Transient transformation technology provides a powerful tool to study gene function in vivo, avoiding difficult drawbacks that typically affect the stable transformation protocols, such as transformation efficiency, transformants selection, and regeneration. In this review we provide an overview of the use of transient expression in the investigation of the function of genes important for strawberry fruit development, defense and nutritional properties. The technical aspects related to an efficient use of this technique are described, and the possible impact and application in strawberry crop improvement are discussed.
Strawberry is an important fruit crop worldwide, both for the impact on the economy of several countries and for its nutritional properties highly beneficial to human health. Strawberry is considered a plant model system for Rosaceae family due to its small genome size, the short generation time for a perennial species (Folta and Davis, 2006), the easy in vitro regeneration and transformation system (Barceló et al., 1998; Folta et al., 2006; Debnath and Teixeira da Silva, 2007; Qin et al., 2008) and the current availability of different genome sequences of Fragaria species. These include the diploid F. vesca genome (Shulaev et al., 2011) and a virtual strawberry reference genome, named ‘FANhybrid_r1.2,’ obtained by integrating the four homoeologous subgenomes sequences of the octoploid F. X ananassa (Hirakawa et al., 2014). In the past, different transcriptomic approaches have been used to highlight genes regulated during fruit development or upon pathogen interaction. These included microarray experiments, cDNA libraries sequencing, and suppressive subtractive hybridization (SSH) technique. These provided a large set of expressed sequence tags (ESTs) from F. vesca and F. X ananassa related to candidate genes involved in important biological processes for strawberry (Medina-Escobar et al., 1997; Aharoni and O’Connell, 2002; Aharoni et al., 2002; Salentijn et al., 2003; Casado-Dìaz et al., 2006; Bombarely et al., 2010; Folta et al., 2010; Guidarelli et al., 2011). More recently, the new RNA sequencing technologies offered the opportunity to uncover whole transcriptome changes underneath complex biological processes as, for example, the strawberry flower and fruit development (Kang et al., 2013; Hollender et al., 2014). The large amount of gene expression data derived by these techniques is generally thoroughly processed with the available bioinformatics tools that allow visualizing all the transcriptome networks controlling a specific function in comprehensive pictures. These are mostly based on the predictive gene functional classification derived by gene homology scores, and only for a little percentage on the experimental evidence of a specific gene function. Indeed, to fully understand the role of each gene involved in a biological process an in vivo genetic analysis should be performed. Before the advent of the whole genome sequencing, gene function analysis was carried out using ‘forward’ genetics, where the genetic basis of a phenotype was identified by associating the phenotypic effects of natural or induced mutations to a specific gene sequence. At present, thanks to the new sequencing technologies and the consequent massive availability of gene sequences, it is possible to explore gene function in the opposite direction, namely from gene to phenotype, through the so-called ‘reverse’ genetics. Here, a specific gene with a known sequence is disrupted or modified and the phenotype is analyzed to determine the corresponding gene function (Tierney and Lamour, 2005). For plants several approaches have been developed for this purpose, including overexpression, gene silencing or mutagenesis (Gilchrist and Haughn, 2010).
In particular for strawberry, reverse genetics has been successfully used to characterize gene function mainly through gene down-regulation, via post-transcriptional gene silencing (PTGS) RNA interference (RNAi). This can be achieved through the introduction of a double-stranded RNA (dsRNA) homologous in sequence to the gene of interest, resulting in the degradation of the target gene transcripts and leading to a knockout or a knockdown phenotype, depending on the specific silencing efficiency (Filipowicz et al., 2005). The silence-inducing dsRNA can be delivered in host plant cells either by the introduction of plasmid constructs encoding self-complementary ‘hairpin’ RNAs, or by Virus-induced gene silencing (VIGS). The latter is obtained by cloning a cDNA fragment of the gene of interest into a DNA copy of a RNA-virus genome, resulting in the production of an autonomous replicating virus, which, during its replication in plant, forms a dsRNA (Lu et al., 2003). However, most viruses used for VIGS have a limited number of hosts and the efficiency of silencing is strongly dependent on the virus-host affinity. Furthermore, the phenotypes introduced with this technique are not heritable and cannot be used for obtaining stable genetic lines (Gilchrist and Haughn, 2010). On the contrary, the phenotype introduced by hairpin RNAi-based silencing can be heritable, moreover the transcripts of multigene families can be silenced by a single construct (Alvarez et al., 2006), making this technique a powerful tool to study the loss of function phenotype of a target gene in plants with high levels of polyploidy such as strawberry.
Similarly, gain of function phenotypes, which are achievable simply by inducing the expression of a specific gene under the control of a strong promoter, have also been used to efficiently study gene function in strawberry (Agius et al., 2005; Hoffmann et al., 2011; Cumplido-Laso et al., 2012). On the other hand, the utility of the overexpression system is limited because the ectopic protein expression ceases after few days due to PTGS. In Nicotiana benthamiana, the viral protein P19 was shown to prevent the onset of PTGS allowing high level of transient expression of genes (Voinnet et al., 2003). Aragüez et al. (2013) used P19 to coexpress FaEGS2, a gene involved in the formation of eugenol and isoeugenol, and to evaluate its effect in the aroma of strawberry. In strawberry, the use of this factor could be very useful to reduce PTGS, however, is still very limited; more studies are needed to deeply evaluate the co-expression of P19 together with target gene over-expression, thus confirming its role as gene expression enhancer, and ultimately allowing overcoming the problem of the PTGS in this type of experiments.
Nevertheless, RNAi and overexpression systems, represent the most powerful tools to investigate gene function in plants and consequently to improve beneficial traits such as novel disease resistances, quality and nutritional improvements, and changes in metabolism which will increase crop productivity.
Reverse genetics can be accomplished through stable transformation or transient expression of a target gene. However, generation of stably transformed plants is difficult and labor-intensive, and represents a relatively low-throughput process lasting several months, depending on the plant species used. In comparison with stable transformation, the transient transformation has several advantages: (i) it does not interfere with the stability of the host genome (Lu et al., 2013), (ii) it does not require regeneration of a transformed cell in order to analyze the transformation, (iii) the expression of target genes can be analyzed shortly after DNA delivery, and (iv) is not influenced by positional effects. Moreover, transient transformation systems can greatly accelerate research timing, since many constructs can be analyzed in parallel within a short time frame (Li et al., 2009). These advantages make this system a powerful method especially in plants that are recalcitrant to regeneration (Kapila et al., 1997) and can be widely exploited for studying gene functions through physiological characterization of the phenotype and cellular localization of gene product (Koroleva et al., 2005; Kirienko et al., 2012). Routine transient assays include biolistic bombardment (Christou, 1995), protoplast tranfection (Sheen, 2001), and Agrobacterium-mediated transient assays (Kapila et al., 1997; Yang et al., 2000) each with advantages and disadvantages depending on the research goals: the biolistic approach can be used on various plant species but it requires an expensive particle bombardment equipment and a procedure relatively complex, in particular for the case of ripe fleshy fruits, anatomically not so suitable to bombardment. Similarly, protoplast transfection works well for several plant species and could be useful to investigate cell-autonomous regulatory processes and responses in a quantitative and high-throughput way (Yoo et al., 2007). However, protoplast assay has limited applicability for those cases where the cell wall or a tissue context is required (Li et al., 2009). On the contrary, Agrobacterium-mediated transformation represents one of the most facile and effective methods to transfer a gene in plant cells and to analyze its function (Gelvin, 2003). Here, the common gall-inducing bacterium Agrobacterium tumefaciens is used to transfer a DNA segment (T-DNA) from the bacterial tumour-inducing (Ti) plasmid to plant cell. The target gene (either for overexpression or for RNAi silencing), inserted into the T-DNA depleted of tumor-inducing genes, can be efficiently expressed under the control of a eukaryotic promoter. Once moved into the plant cell, the T-DNA migrates to the nucleus and only a tiny part is integrated into the host chromosomes; these not-integrated T-DNA copies persist in the nuclei of transfected cells remaining transcriptionally competent for several days and leading to non-stable, transient transcripition (Hellens et al., 2005). This technique, commonly known as agroinfiltration or agroinjection, provides fast and efficient ways to transiently express or silence a desired gene whose function must be investigated ‘in planta.’
In the past, functional genomics in strawberry was mainly confined to gene expression analysis and in silico prediction. Only recently, reverse genetics approaches were used to assign gene functions (Schwab et al., 2011). A number of genetically transformed lines of Fragaria sp. were obtained throughout genetic engineering to study gene functions or to improve traits such as resistance, ripening, and fruit production (Haymes and Davis, 1998; Mezzetti et al., 2004; Schaart et al., 2004, 2013; Oosumi et al., 2006; Jiwan et al., 2013; Pantazis et al., 2013; see also reviews: Folta and Davis, 2006; Aldwinckle and Malnoy, 2009). Indeed, in strawberry the efficiency of transformation and the regeneration capacity is highly dependent on the genetic background (Landi and Mezzetti, 2006) and several critical factors such as hormones concentrations, incubation conditions, explant source, antibiotic, and mode of transfections can affect the frequency of regeneration (Folta and Dhingra, 2006). In addition, these traditional ways of studying gene functions in strawberry are very time-consuming, especially for ripening-related genes, considering that, from the transformation experiment until the first ripe fruits become available for analyses, at least 15 months are needed (Hoffmann et al., 2006). For these reasons, more recently, transient transformation systems are emerging as most powerful tool to investigate the function of genes involved in important physiological processes, such as ripening related genes, resistance genes, and strawberry promoters (Table 1).
The first report of transient transformation in strawberry fruits goes back to 2001 when Spolaore et al. (2001), showed that strawberry fruit is suitable to transient gene expression mediated by Agrobacterium using the β-glucuronidase (GUS) reporter gene interrupted by an intron. Consistently, this system has been firstly used for the functional analysis of two strawberry β-1-4 glucanases promoters or, combined with a biolistic transformation protocol, to functionally characterize other homologous and heterologous promoters (Spolaore et al., 2003; Agius et al., 2005).
A number of transient transformation studies on strawberry fruits were subsequently performed, the majority of which were finalized to understand the role of genes involved in ripening processes (Table 1). Strawberry fruit ripening is accompanied by an increase of flavonoid biosynthesis resulting in anthocyanin accumulation at ripe red stage (Almeida et al., 2007). Chalcone synthase (CHS), the key gene in the flavonoid pathway, was chosen as a reporter gene to set up an efficient protocol for RNAi-based transient gene silencing in strawberry fruit: by agroinjecting receptacles with an inoculum of Agrobacterium carrying an intron-containing self-complementary hairpin construct, a drastic reduction in mRNA levels and enzymatic activity of CHS gene was obtained, leading to the loss of pigmentation in ripe strawberry fruits (Hoffmann et al., 2006). This transient RNAi method was successfully employed afterward for the silencing of other flavonoids related genes and found to be applicable also to strawberry fruits at postharvest stage (Schwab et al., 2011; Miyawaki et al., 2012).
With the aim to elucidate in planta function of Anthocyanidin 3-0-glycosyltransferase (FaGT1), a gene encoding for the enzyme involved in the anthocyanin biosynthesis, an Agrobacterium carrying a T-DNA expressing an RNAi-FaGT1 construct was injected into midsized ripening strawberry fruits. This resulted in reduced concentrations of anthocyanin pigments and in a significant increase in epiafzelechin, the compound sinthesized by the anthocyanidin reductase (ANR) enzyme, indicating competition between FaGT1 and FaANR for the common athocyanidin substrate and showing that FaGT1 is a key gene in the anthocyanin pathway (Griesser et al., 2008). More recently others RNAi studies were performed to clarify the role of other important genes involved in the strawberry fruit pigmentation and anthocyanin biosynthesis (Jiang et al., 2013; Lin et al., 2013; Kadomura-Ishikawa et al., 2014; Medina-Puche et al., 2014). These works greatly helped to clarify the flavonoid pathway and to highlight the contribution of each gene to the synthesis of key compounds, important not only for the agronomic but also for the medical and nutritional interest of this crop (Figure 1).
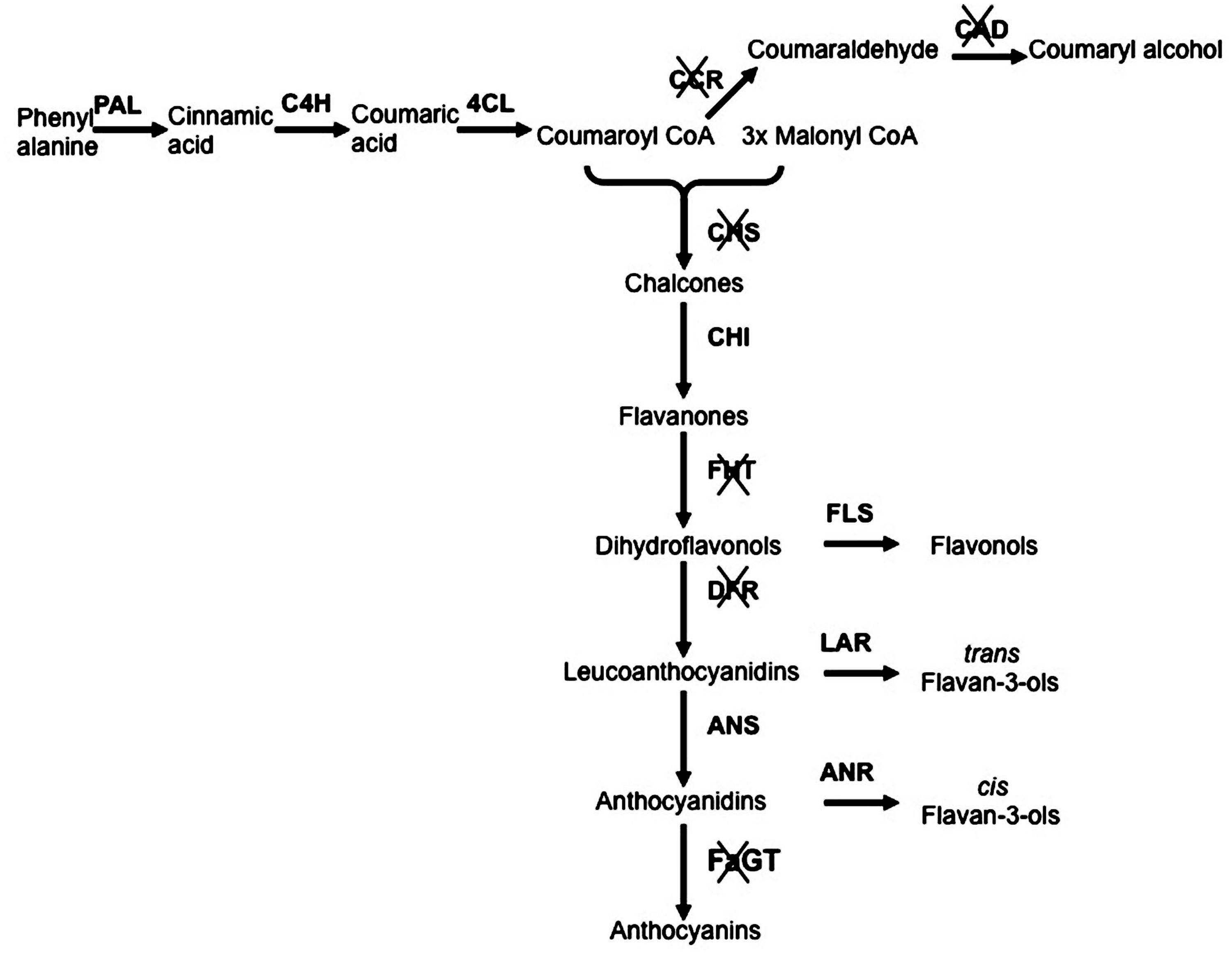
FIGURE 1. Schematic representation of phenylpropanoid/flavonoid pathway with silenced genes. The genes functionally investigated throughout RNA interference (RNAi)-transient transformation are crossed. Genes abbreviations: PAL, phenylalanine ammonia lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, p-coumarate:CoA ligase; CCR, Cinnamoyl-CoA reductase; CAD, Cinnamyl alcohol dehydrogenase; CHS, Calchone synthase; CHI, chalcone isomerase; FHT, Flavanone 3-Hydroxylase; FLS, flavonol synthase; DFR, Dihydroflavonol 4-reductase; LAR, leucoanthocyanidin reductase; ANS, anthocyanidin synthase; ANR, anthocyanidin reductase; FaGT, flavonoid glycosyltransferases.
Furthermore, Muñoz et al. (2010) found a correlation between a strawberry pathogenesis-related 10 (PR-10) gene and anthocyanin biosynthesis. In particular, a Fra a1 gene, was RNAi-silenced, leading to a decreased endogenous concentration of the main flavonoids responsible for the red color of fruits and to a decrease of the expression levels of genes encoding for phenylalanine ammonia lyase (PAL) and CHS. These results demonstrated that Fra a1 is directly linked to flavonoid biosynthesis and that its function may be regulatory (Muñoz et al., 2010). Consistent to this, very recently, Casañal et al. (2013) demonstrated that strawberry Fra proteins bind flavonoids, providing an important contribution to the function of these PR proteins in the control of flavonoid metabolism.
Gene function in strawberry has been investigated with transient transformation not only through gene silencing but also through gene overexpression of target genes or by combining, alternatively or simultaneously, overexpression with silencing and by observing the consequent phenotype (Hoffmann et al., 2011; Cumplido-Laso et al., 2012; Aragüez et al., 2013; Daminato et al., 2013; Jia et al., 2013; Kadomura-Ishikawa et al., 2013, 2014; Guidarelli et al., 2014; Yeh et al., 2014). For instance, the role of Eugenol synthase (EGS) genes in contributing to volatile compounds production responsible for strawberry flavor was studied by modifying the anthocyanin biosynthesis pathway in strawberry fruit using a simultaneous downregulation and ovexpression of CHS and EGS genes, respectively. This led to the deviation of the flavonoid pathway to the synthesis of phenylpropene, such as eugenol, confirming the role of EGS enzyme in the production of strawberry aroma (Hoffmann et al., 2011; Aragüez et al., 2013). Furthermore, in a recent study, Medina-Puche et al. (2015) used FaEOBII-ripe silenced strawberry fruits to prove that eugenol production is regulated by the R2R3-MYB transcription factor FaEOBII.
Similarly, by using independent transient overexpression/silencing methods or by combining them, the role of a FaPOD27 peroxidase gene was found to contribute to lignin biosynthesis and strawberry fruit firmness (Ring et al., 2013; Yeh et al., 2014).
If a large number of experiments were aimed at elucidating the role of genes involved in strawberry ripening, very little with transient transformation has been done for understanding the role of genes involved in strawberry resistance to stress.
By using a VIGS RNAi approach, a β-glucosidase gene (FaBG3) was found to be involved in the resistance of strawberry to Botrytis cinerea (Li et al., 2013); silencing of this gene led to a decrease of endogenous abscisic acid (ABA) and consequent fruit ripening inhibition, but also concomitantly to resistance to the B. cinerea fungus and increase in PAL activity and content of phenolic compounds. These results indicated that this gene plays roles not only in fruit growth regulation but also in the fruit response to pathogens. More recently, Agrobacterium transient transformation was successfully applied to study the role of the gene FaMBL1, encoding for a Mannose Binding Lectin. Expression of this gene was originally found upregulated exclusively in unripe strawberries upon infection with the pathogen Colletotrichum acutatum, but not in infected ripe ones. Since, contrary to red ripe fruits, white unripe fruits are reluctant to C. acutatum infection, a role for this lectin gene in the different susceptibility of strawberries during ripening was hypothesized. To test this hypothesis, Agrobaterium-mediated transient transformation was performed to silence and overexpress FaMBL1 gene in unripe and ripe strawberries, respectively, before C. acutatum infection. FaMBL1-silenced unripe fruits showed anthracnose symptoms, whereas FaMBL1-overexpressing ripe strawberries presented a lower susceptibility to C. acutatum. These experiments allowed concluding that this gene plays a crucial role in the resistance of unripe strawberry fruits to C. acutatum (Guidarelli et al., 2014).
Particular attention musts be paid while using Agrobacterium-mediated transient transformation for studying plant pathogen interactions: Agrobacterium itself is a plant pathogen and can interfere with plant defense response to stress, resulting in misleading interpretation of data. However, by using the appropriate controls, this technique can be a powerful tool for discovering function of genes involved in strawberry–pathogen interaction, and to unravel the complex network of defense signaling pathways in this important crop.
Despite the large amount of molecular information originated from the use of new high-throughput technologies, providing useful data to improve strawberry crop, the knowledge of the role of most genes involved in important physiological processes is still scarce.
The advances in the last two decades of high efficiency stable and transient transformation protocols for strawberry, open new frontiers to understand the role of genes regulating important biological processes and potentially exploitable for strawberry improvement through breeding or genetic engineering.
However, the high cost, also in terms of time, of developing transgenic over-expressing or silenced lines for determining gene function, limits their usefulness to the analysis of few candidate genes. The transient transformation approach represents a rapid alternative and a highly efficient method for gene function discovery, opening new scenarios at the service of genetic engineering for strawberry crop improvement. Future research focused on the development of valuable molecular tools capable to extent the effect of transient expression, and to overcome post-transcriptional gene silencing, such as, for example, the above mentioned P19 viral protein, would be very important to allow technical advances and promote wider applications of this investigation method.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Agius, F., Amaya, I., Botella, M. A., and Valpuesta, V. (2005). Functional analysis of homologous and heterologous promoters in strawberry fruits using transient expression. J. Exp. Bot. 56, 37–46. doi: 10.1093/jxb/eri004
Aharoni, A., Keizer, L. C., Van Den Broeck, H. C., Blanco-Portales, R., Muñoz-Blanco, J., Bois, G., et al. (2002). Novel insight into vascular, stress, and auxin-dependent and -independent gene expression programs in strawberry, a non-climacteric fruit. Plant Physiol. 129, 1019–1031. doi: 10.1104/pp.003558
Aharoni, A., and O’Connell, A. P. (2002). Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J. Exp. Bot. 53, 2073–2087. doi: 10.1093/jxb/erf026
Aldwinckle, H., and Malnoy, M. (2009). Plant regeneration and transformation in the Rosaceae. Transgeninc Plant J. 3, 1–39.
Almeida, J. R. M., D’Amico, E., Preuss, A., Carbone, F., de Vos, C. H. R., Deiml, B., et al. (2007). Characterization of major enzymes and genes involved in flavonoid and proanthocyanidin biosynthesis during fruit development in strawberry (Fragaria x ananassa). Arch. Biochem. Biophys. 465, 61–71. doi: 10.1016/j.abb.2007.04.040
Alvarez, J. P., Pekker, I., Goldshmidt, A., Blum, E., Amsellem, Z., and Eshed, Y. (2006). Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18, 1134–51. doi: 10.1105/tpc.105
Aragüez, I., Osorio, S., Hoffmann, T., Rambla, J. L., Medina-Escobar, N., Granell, A., et al. (2013). Eugenol production in achenes and receptacles of strawberry fruits is catalyzed by synthases exhibiting distinct kinetics. Plant Physiol. 163, 946–958. doi: 10.1104/pp.113.224352
Barceló, M., El-Mansouri, I., Mercado, J. A., Quesada, M., and Alfaro, F. P. (1998). Regeneration and transformation via Agrobacterium tumefaciens of the strawberry cultivar Chandler. Plant Cell Tissue Organ Cult. 54, 29–36. doi: 10.1023/A:1006031527413
Bombarely, A., Merchante, C., Csukasi, F., Cruz-Rus, E., Caballero, J., Medina-Escobar, N., et al. (2010). Generation and analysis of ESTs from strawberry (Fragaria x ananassa) fruits and evaluation of their utility in genetic and molecular studies. BMC Genomics 11:503. doi: 10.1186/1471-2164-11-503
Casado-Díaz, A., Encinas-Villarejo, S., de los Santos, B., Schilirò, E., Yubero- Serrano, E.-M., Amil-Ruiz, F., et al. (2006). Analysis of strawberry genes differentially expressed in response to Colletotrichum infection. Physiol. Plant. 128, 633–650. doi: 10.1093/jxb/erp152
Casañal, A., Zander, U., Muñoz, C., Dupeux, F., Luque, I., Botella, M. A., et al. (2013). The strawberry Pathogenesis-Related 10 (PR-10) Fra a proteins control flavonoid biosynthesis by binding to metabolic intermediates. J. Biol. Chem. 288, 35322–35332. doi: 10.1074/jbc.M113.501528
Chai, Y. M., Jia, H. F., Li, C. L., Dong, Q. H., and Shen, Y. Y. (2011). FaPYR1 is involved in strawberry fruit ripening. J. Exp. Bot. 62, 5079–5089. doi: 10.1093/jxb/err207
Christou, P. (1995). Strategies for variety-independent genetic transformation of important cereals, legumes and woody species utilizing particle bombardment. Euphytica 85, 13–27. doi: 10.1007/BF00023926
Cumplido-Laso, G., Medina-Puche, L., Moyano, E., Hoffmann, T., Sinz, Q., Ring, L., et al. (2012). The fruit ripening-related gene FaAAT2 encodes an acyl transferase involved in strawberry aroma biogenesis. J. Exp. Bot. 63, 4275–4290. doi: 10.1093/jxb/ers120
Daminato, M., Guzzo, F., and Casadoro, G. (2013). A SHATTERPROOF-like gene controls ripening in non-climacteric strawberries, and auxin and abscisic acid antagonistically affect its expression. J. Exp. Bot. 64, 3775–3786. doi: 10.1093/jxb/ert214
Debnath, S. C., and Teixeira da Silva, J. A. (2007). Strawberry culture in vitro: applications in genetic transformation and biotechnology. Fruit Veg. Cereal Sci. Biotechnol. 1, 1–2.
Filipowicz, W., Jaskiewicz, L., Kolb, F. A., and Pillai, R. S. (2005). Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 15, 331–341. doi: 10.1016/j.sbi.2005.05.006
Folta, K. M., Clancy, M. A., Chamala, S., Brunings, A. M., Dhingra, A., Gomide, L., et al. (2010). A transcript accounting from diverse tissues of a cultivated strawberry. Plant Genome 3, 90–105. doi: 10.3835/plantgenome2010.02.0003
Folta, K. M., and Davis, T. M. (2006). Strawberry genes and genomics. Crit. Rev. Plant Sci. 25, 399–415. doi: 10.1080/07352680600824831
Folta, K. M., and Dhingra, A. (2006). Transformation of strawberry: the basis for translational genomics in Rosaceae. In Vitro Cell. Dev. Biol. Plant 42, 482–490. doi: 10.1079/IVP2006807
Folta, K. M., Dhingra, A., Howard, L., Stewart, P. J., and Chadler, C. K. (2006). Characterization of LF9, an octoploid strawberry genotype selected for rapid regeneration and transformation. Planta 224, 1058–1067. doi: 10.1007/s00425-006-0278-0
Gelvin, S. B. (2003). Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67, 16–37. doi: 10.1128/MMBR.67.1.16-37.2003
Gilchrist, E., and Haughn, G. (2010). Reverse genetics techniques: engineering loss and gain of gene function in plants. Brief. Funct. Genomics 9, 103–110. doi: 10.1093/bfgp/elp059
Griesser, M., Hoffmann, T., Bellido, M. L., Rosati, C., Fink, B., Kurtzer, R., et al. (2008). Redirection of flavonoid biosynthesis through the down-regulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit. Plant Physiol. 146, 1528–1539. doi: 10.1104/pp.107.114280
Guidarelli, M., Carbone, F., Mourgues, F., Perrotta, G., Rosati, C., Bertolini, P., et al. (2011). Colletotrichum acutatum interactions with unripe and ripe strawberry fruits and differential responses at histological and transcriptional levels. Plant Pathol. 60, 685–697. doi: 10.1111/j.1365-3059.2010.02423.x
Guidarelli, M., Zoli, L., Orlandini, A., Bertolini, P., and Baraldi, E. (2014). The Mannose Binding Lectin gene FaMBL1 is involved in the resistance of unripe strawberry fruits to Colletotrichum acutatum. Mol. Plant Pathol. 15, 832–840. doi: 10.1111/mpp.12143
Haymes, K. M., and Davis, T. M. (1998). Agrobacterium mediated transformation of ‘Alpine’ Fragaria vesca, and transmission of transgenes to R1 progeny. Plant Cell Rep. 17, 279–283. doi: 10.1007/s002990050392
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 1–14. doi: 10.1186/1746-4811-1-13
Hirakawa, H., Shirasawa, K., Kosugi, S., Tashiro, K., Nakayama, S., Yamada, M., et al. (2014). Dissection of the octoploid strawberry genome by deep sequencing of the genomes of Fragaria species. DNA Res. 21, 169–181. doi: 10.1093/dnares/dst049
Hoffmann, T., Kalinowski, G., and Schwab, W. (2006). RNAi-induced silencing of gene expression in strawberry fruit (Fragaria x ananassa) by agroinfiltration: a rapid assay for gene function analysis. Plant J. 48, 818–826. doi: 10.1111/j.1365-313X.2006.02913.x
Hoffmann, T., Kurtzer, R., Skowranek, K., Kiessling, P., Fridman, E., Pichersky, E., et al. (2011). Metabolic engineering in strawberry fruit uncovers a dormant biosynthetic pathway. Metab. Eng. 13, 527–531. doi: 10.1016/j.ymben.2011.06.002
Hollender, C. A., Kang, C., Darwish, O., Geretz, A., Matthews, B. F., Slovin, J., et al. (2014). Floral transcriptomes in woodland strawberry uncover developing receptacle and anther gene networks. Plant Physiol. 165, 1062–1075. doi: 10.1104/pp.114
Jia, H. F., Chai, Y. M., Li, C. L., Lu, D., Luo, J. J., Qin, L., et al. (2011). Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 157, 188–199. doi: 10.1104/pp.111.177311
Jia, H., Wang, Y., Sun, M., Li, B., Han, Y., Zhao, Y., et al. (2013). Sucrose functions as a signal involved in the regulation of strawberry fruit development and ripening. New Phytol. 198, 453–465. doi: 10.1111/nph.12176
Jiang, F., Wang, J. Y., Jia, H. F., Jia, W. S., Wang, H. Q., and Xiao, M. (2013). RNAi-mediated silencing of the flavanone 3-hydroxylase gene and its effect on flavonoid biosynthesis in strawberry fruit. J. Plant Growth Regul. 32, 182–190. doi: 10.1007/s00344-012-9289-1
Jiwan, D., Roalson, E. H., Main, D., and Dhingra, A. (2013). Antisense expression of peach mildew resistance locus O(PpMlo1) gene confers cross-species resistance to powdery mildew in Fragaria x ananassa. Transgenic Res. 22, 1119–1131. doi: 10.1007/s11248-013-9715-6
Kadomura-Ishikawa, Y., Miyawaki, K., Noji, S., and Takahashi, A. (2013). Phototropin 2 is involved in blue light-induced anthocyanin accumulation in Fragaria x ananassa fruits. J. Plant Res. 126, 847–857. doi: 10.1007/s10265-013-0582-2
Kadomura-Ishikawa, Y., Miyawaki, K., Takahashi, A., Masuda, T., and Noji, S. (2014). Light and abscisic acid independently regulated FaMYB10 in Fragaria x ananassa fruit. Planta 241, 953–965. doi: 10.1007/s00425-014-2228-6
Kang, C., Darwish, O., Geretz, A., Shahan, R., Alkharouf, N., and Liu, Z. (2013). Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25, 1960–1978. doi: 10.1105/tpc.113.111732
Kapila, J., De Rycke, R., Van Montagu, M., and Angenon, G. (1997). An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 122, 101–108. doi: 10.1016/S0168-9452(96)04541-4
Kirienko, D. R., Luo, A., and Sylvester, A. W. (2012). Reliable transient transformation of intact maize leaf cells for functional genomics and experimental study. Plant Physiol. 159, 1309–1318. doi: 10.1104/pp.112.199737
Koroleva, O. A., Tomlinson, M. L., Leader, D., Shaw, P., and Doonan, J. H. (2005). High throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J. 41, 162–174. doi: 10.1111/j.1365-313X.2004.02281.x
Landi, L., and Mezzetti, B. (2006). TDZ, auxin and genotype effects on leaf organogenesis in Fragaria. Plant Cell Rep. 25, 281–288. doi: 10.1007/s00299-005-0066-5
Li, J., Park, E., von Arnim, A. G., and Nebenfu_hr, A. (2009). The FAST technique: a simplified Agrobacterium based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5, 6. doi: 10.1186/1746-4811-5-6
Li, Q., Ji, K., Sun, Y. F., Luo, H., Wang, H. Q., and Leng, P. (2013). The role of FaBG3 in fruit ripening and B. cinerea fungal infection of strawberry. Plant J. 76, 24–35. doi: 10.1111/tpj.12272
Lin, X., Xiao, M., Luo, Y., Wang, J., and Wang, H., (2013). The effect of RNAi-induced silencing of FaDFR on anthocyanin metabolism in strawberry (Fragaria x ananassa) fruit. Sci. Hortic. 160, 123–128. doi: 10.1016/j.scienta.2013.05.024
Lu, R., Martin-Hernandez, A. M., Peart, J. R., Malcuit, I., and Baulcombe, D. C. (2003). Virus-induced gene silencing in plants. Methods 30, 296–303. doi: 10.1016/S1046-2023(03)00037-9
Lu, Y., Chen, X., Wu, Y., Wang, Y., He, Y., and Wu, Y. (2013). Directly transforming PCR-amplified DNA fragments into plant cells is a versatile system that facilitates the transient expression assay. PLoS ONE 8:e57171. doi: 10.1371/journal.pone.0057171
Medina-Escobar, N., Cárdenas, J., Valpuesta, V., Muñoz-Blanco, J., and Caballero, J. L. (1997). Cloning and characterization of cDNAs from genes differentially expressed during the strawberry fruit ripening process by a MAST-PCR-SBDS method. Anal. Biochem. 248, 288–296. doi: 10.1006/abio.1997.2110
Medina-Puche, L., Cumplido-Laso, G., Amil-Ruiz, F., Hoffmann, T., Ring, L., Rodriguez-Franco, A., et al. (2014). MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria x ananassa fruits. J. Exp. Bot. 65, 401–417. doi: 10.1093/jxb/ert377
Medina-Puche, L., Molina-Hidalgo, F. J., Boersma, M. R., Schuurink, R. C., López-Vidriero, I., Solano, R., et al. (2015). A R2R3-MYB transcription factor (FaEOBII) regulates eugenol production in ripe strawberry (Fragaria x ananassa) fruit receptacles. Plant Physiol. doi: 10.1104/pp.114.252908 [Epub ahead of print].
Mezzetti, B., Landi, L., Pandolfini, T., and Spena, A. (2004). The defH9-iaaM auxin-synthesizing gene increases plant fecundity and fruit production in strawberry and raspberry. BMC Biotechnol. 4:4. doi: 10.1186/1472-6750-4-4
Miyawaki, K., Fukuoka, S., Kadomura, Y., Hamaoka, H., Mito, T., Ohuchi, H., et al. (2012). Establishment of a novel system to elucidate the mechanisms underlying light-induced ripening of strawberry fruit with an Agrobacterium-mediated RNAi technique. Plant Biotechnol. 29, 271–277. doi: 10.5511/plantbiotechnology.12.0406a
Molina-Hidalgo, F. J., Franco, A. R., Villatoro, C., Medina-Puche, L., Mercado, J. A., Hidalgo, M. A., et al. (2013). The strawberry (Fragaria x ananassa) fruit-specific rhamnogalacturonate lyase 1 (FaRGLyase1) gene encodes an enzyme involved in the degradation of cell walls middle lamellae. J. Exp. Bot. 64, 1471–1483. doi: 10.1093/jxb/ers386
Moyano-Cañete, E., Bellido, M. L., García-Caparrós, N., Medina-Puche, L., Amil-Ruiz, F., González-Reyes, J. A., et al. (2013). FaGAST2, a strawberry ripening-related gene, acts together with FaGAST1 to determine cell size of the fruit receptacle. Plant Cell Physiol. 54, 218–236. doi: 10.1093/pcp/pcs167
Muñoz, C., Hoffmann, T., Escobar, N. M., Ludemann, F., Botella, M. A., Valpuesta, V., et al. (2010). The strawberry fruit Fra a allergen functions in flavonoid biosynthesis. Mol. Plant 3, 113–124. doi: 10.1093/mp/ssp087
Oosumi, T., Gruszewski, H. A., Blischak, L. A., Baxter, A. J., Wadl, P. A., Shuman, J. L., et al. (2006). High-efficiency transformation of the diploid strawberry (Fragaria vesca) for functional genomics. Planta 223, 1219–1230. doi: 10.1007/s00425-005-0170-3
Pantazis, C. J., Fisk, S., Mills, K., Flinn, B. S., Shulaev, V., Veilleux, R. E., et al. (2013). Development of an efficient transformation method by Agrobacterium tumefaciens and high throughput spray assay to identify transgenic plants for woodland strawberry (Fragaria vesca) using NPTII selection. Plant Cell Rep. 32, 329–337. doi: 10.1007/s00299-012-1366-1
Qin, Y., Silva, J. T. D., Zhang, L., and Zhang, S. (2008). Transgenic strawberry: state of the art for improved traits. Biotechnol. Adv. 26, 219–232. doi: 10.1016/j.biotechadv.2007.12.004
Ring, L., Yeh, S. Y., Hu_cherig, S., Hoffmann, T., Blanco-Portales, R., Fouche, M., et al. (2013). Metabolic interaction between anthocyanin and lignin biosynthesis is associated with peroxidase FaPRX27 in strawberry fruit. Plant Physiol. 163, 43–60. doi: 10.1104/pp.113.222778
Salentijn, E. M. J., Aharoni, A., Schaart, J. G., Boone, M. J., and Krens, F. A. (2003). Differential gene expression analysis of strawberry cultivars that differ in fruit-firmness. Physiol. Plant. 118, 571–578. doi: 10.1034/j.1399-3054.2003.00138.x
Schaart, J. G., Dubos, C., De La Fuente, I. R., van Houwelingen, A. M. M. L., de Vos, R. C. H., Jonker, H. H., et al. (2013). Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria x ananassa) fruits. New Phytol. 197, 454–467. doi: 10.1111/nph.12017
Schaart, J. G., Krens, F. A., Pelgrom, K. T. B., Mendes, O., and Rouwendal, G. J. A. (2004). Effective production of marker-free transgenic strawberry plants using inducible site-specific recombination and a bifunctional selectable marker gene. Plant Biotechnol. J. 2, 233–240. doi: 10.1111/j.1467-7652.2004.00067.x
Schwab, W., Hoffmann, T., Kalinowski, G., and Preuβ, A. (2011). Functional genomics in strawberry fruit through RNAi-mediated silencing. Genes Genomes Genomics 5, 91–101.
Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127, 1466–1475. doi: 10.1104/pp.010820
Shulaev, V., Sargent, D. J., Crowhurst, R. N., Mockler, T. C., Folkerts, O., Delcher, A. L., et al. (2011). The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43, 109–116. doi: 10.1038/ng.740
Spolaore, S., Trainotti, L., and Casadoro, G. (2001). A simple protocol for transient gene expression in ripe fleshy fruit mediated by Agrobacterium. J. Exp. Bot. 52, 845–850. doi: 10.1093/jexbot/52.357.845
Spolaore, S., Trainotti, L., Pavanello, A., and Casadoro, G. (2003). Isolation and promoter analysis of two genes encoding different endo-β-1,4-glucanases in the non-climacteric strawberry. J. Exp. Bot. 54, 271–277. doi: 10.1093/jxb/erg025
Tierney, M. B., and Lamour, K. H. (2005). An introduction to reverse genetic tools for investigating gene function. Plant Health Instructor doi: 10.1094/PHI-A-2005-1025-1. Available at: http://www.apsnet.org/edcenter/advanced/topics/Pages/ReverseGeneticTools.aspx
Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on sup-pression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956. doi: 10.1046/j.1365-313X.2003.01676.x
Yang, Y., Li, R., and Qi, M. (2000). In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22, 543–551. doi: 10.1046/j.1365-313x.2000.00760.x
Yeh, S. Y., Huang, F. C., Hoffmann, T., Mayershofer, M., and Schwab, W. (2014). FaPOD27 functions in the metabolism of polyphenols in strawberry fruit (Fragaria sp.). Front. Plant Sci. 5:518. doi: 10.3389/fpls.2014.00518
Keywords: strawberry, Agrobacterium-mediated transient transformation, gene function discovery, RNA-interference, overexpression
Citation: Guidarelli M and Baraldi E (2015) Transient transformation meets gene function discovery: the strawberry fruit case. Front. Plant Sci. 6:444. doi: 10.3389/fpls.2015.00444
Received: 25 February 2015; Accepted: 29 May 2015;
Published online: 12 June 2015
Edited by:
Bruno Mezzetti, Università Politecnica delle Marche, ItalyReviewed by:
Shogo Matsumoto, Nagoya University, JapanCopyright © 2015 Guidarelli and Baraldi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elena Baraldi, Laboratory of Plant Pathology and Biotechnology, Department of Agricultural Sciences, University of Bologna, Viale Fanin 46, 40127 Bologna, Italy,ZWxlbmEuYmFyYWxkaUB1bmliby5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.