- 1Seaweed Biology and Cultivation Group, Division of Marine Biotechnology and Ecology, CSIR-Central Salt and Marine Chemicals Research Institute, Bhavnagar, India
- 2Academy of Scientific and Innovative Research (AcSIR), New Delhi, India
Epiphytic and endophytic bacteria associated with green macroalgae Ulva (U. fasciata and U. lactuca) and red macroalgae Gracilaria (G. corticata and G. dura) have been identified from three different seasons to evaluate the effect of quorum sensing (QS) molecules on carpospores liberation from Gracilaria dura. The bacterial isolates belonging to the orders Bacillales, Pseudomonadales, Alteromonadales, and Vibrionales were present in all seasons, whereas Actinomycetales and Enterobacteriales were confined to pre-monsoon and post-monsoon seasons, respectively. Among all the Gram-negative bacteria, seven isolates were found to produce different types of N-acyl homoserine lactones (AHLs). Interestingly, Shewanella algae produced five types of AHL: C4-HSL, HC4-HSL, C6-HSL, 3-oxo-C6-HSL, and 3-oxo-C12-HSL. Subsequently, the AHLs producing bacterial isolates were screened for carpospore liberation from G. dura and these isolates were found to positively induce carpospore liberation over the control. Also, observed that carpospore liberation increased significantly in C4- and C6-HSL treated cystocarps. Sodium dodecyl sulfate and native polyacrylamide gel electrophoresis of the total protein of the C4- and C6-HSL treated cystocarps showed two specific peptide bands of different molecular weights (50 kDa and 60 kDa) as compared to the control, confirming their indirect effect on carpospore liberation.
Introduction
Extracellular substances released from macroalgal communities serve as feed for diverse microorganisms in coastal ecosystems (Armstrong et al., 2001; Lane and Kubanek, 2008). Microbial communities living on macroalgal surfaces are highly diverse, complex and dynamic and they consist of a consortium of microorganisms (Holmström et al., 2002). However, bacteria are the most ubiquitous, occurring on the external surfaces and in the internal tissues of the algae (Hollants et al., 2011). Macroalgal bacterial communities have been found to play an important role in the growth, development, morphogenesis, and reproduction of the green macroalga Ulva (Patel et al., 2003; Matsuo et al., 2005; Tait et al., 2005; Joint et al., 2007; Singh and Reddy, 2014). The green macroalga Ulva forms an aberrant morphology instead of the typical foliose thallus morphology when cultured axenically (Provasoli and Pintner, 1980). This aberrant morphology is successfully reversed to the foliose thallus morphology following the inoculation of appropriate morphogenesis-inducing bacteria to the culture medium (Nakanishi et al., 1996; Singh et al., 2011a). Additionally, macroalgae-associated bacterial isolates of epi- and endophytic origin have been reported to produce indole-3-acetic acid (IAA) that regulates morphogenesis pattern and growth in Ulva spp. (Maruyama et al., 1988) and Gracilaria dura (Singh et al., 2011b). Several studies have revealed that bacterial groups belonging to Proteobacteria, Firmicutes, and Actinobacteria are commonly associated with the Ulva and Gracilaria species (Patel et al., 2003; Tait et al., 2005; Burke et al., 2011; Lachnit et al., 2011). Furthermore, it has been found that consistent detection of these bacterial communities may have a more important functional role in the life processes of the Ulva and Gracilaria species. Therefore, the characterization of epi- and endophytic bacterial communities and further evaluation of the effect, they have on their hosts is of paramount importance in the ecophysiology of macroalgae.
It has also been established that macroalgae-associated bacterial isolates produce quorum sensing (QS) signal molecules, such as N-acyl homoserine lactone (AHLs), thereby facilitating the settlement of zoospores in Ulva spp. (Joint et al., 2002, 2007; Williams, 2007). Joint et al. (2002) established that AHLs producing a Vibrio anguillarum biofilm positively enhanced the settlement of zoospores of the Enteromorpha species. Tait et al. (2005) studied the stability and diffusion rate of AHLs produced from V. anguillarum biofilm and found that AHLs with longer N-acyl side-chains tended to result in increased zoospore settlement of Ulva. Further investigation of zoospore settlement revealed that the orientation of zoospore does not change during settlement (Wheeler et al., 2006). The mechanism underlining this phenomenon has not yet been reported; however, it has been assumed that AHLs influence Ca2+ influx in zoospore which preferentially induces the settlement through chemokinesis (Wheeler et al., 2006). Interestingly, the effect of AHLs was also observed in the red alga Acrochaetium sp. (Weinberger et al., 2007). That study found that C4-HSL has the ability to induce the carpospores' liberation from Acrochaetium sp. (Weinberger et al., 2007). However, the study did not identify AHLs producing host-associated bacteria. Thus, there is limited knowledge about the significant role of cross-kingdom QS signaling between associated bacterial communities and carpospore liberation from red macroalgae.
Cross-kingdom QS signaling between plant roots and their rhizospheric bacteria has also been demonstrated (Hartmann et al., 2014). For example, AHLs produced from symbiotic bacteria elicited developmental changes in the root system (Ortíz-Castro et al., 2008) and root stimulatory effect in Arabidopsis (Jin et al., 2012; Liu et al., 2012). Götz et al. (2007) has found that C6-, C8- and C10-HSL altered root and shoot growth in Hordeum vulgare. Recently, Veliz-Vallejos et al. (2014) demonstrated that 3-oxo-C14-HSL from Sinorhizobium meliloti increased nodule numbers in Medicago truncatula. Some studies have also been carried out to understand the role of AHLs in plant defense (Hartmann et al., 2004; Schuhegger et al., 2006). Serratia liquefaciens MG1 produces C4- and C6-HSL and is found to induce specific systemic resistance proteins after the roots were inoculated with the bacterium (Hartmann et al., 2004). S. meliloti specifically enhances the resistance of A. thaliana toward the pathogens Pseudomonas syringae and Golovinomyces orontii and the resistance of H. vulgare and Blumera graminis (Schikora et al., 2011; Schenk et al., 2012; Zarkani et al., 2013).
Ulva and Gracilaria are the most common types of macroalgae and they grow abundantly in intertidal regions of coastal habitats worldwide. The present study has investigated the epi- and endophytic bacteria associated with the Ulva and Gracilaria species from two different locations and three different seasons in order to identify the bacterial isolates that play a significant role in carpospore liberation. Subsequently, all the isolated bacteria were preliminary screened for their ability to produce AHLs using ESI-MS and the positive isolates were further analyzed using LC-ESI-MS/MS-collision-induced dissociation (CID) to qualitatively analyse the type of AHL. The AHLs producing bacteria were then screened for their potential to liberate carpospores from the red macroalga G. dura. All the bacterial isolates obtained in this study were identified by 16S rRNA gene sequencing.
Materials and Methods
Chemicals
QS signaling molecules, such as N-acyl-homoserine-lactone, N-butanoyl- (C4-HSL), N-3-hydroxybutanoyl- (HC4-HSL), N-hexanoyl- (C6-HSL), N-heptanoyl- (C7-HSL), N-octanoyl- (C8-HSL), N-decanoyl- (C10-HSL), N-dodecanoyl- (C12-HSL), N-3-oxo-hexanoyl- (3-oxo-C6-HSL), N-3-oxo-octanoyl-(3-oxo-C8-HSL), and N-3-oxo-dodecanoyl-(3-oxo-C12-HSL) homoserine lactone, were procured from Sigma Aldrich (Buchs, Switzerland). Analytical grade acetonitrile and formaldehyde were purchased from Sisco Research Pvt. Lit. (India). Working concentrations of the AHLs were prepared by dissolving them in acetonitrile (CH3CN) at a concentration of 1 mg/ml and then storing them at −20°C.
Collection of Samples and Isolation of Epiphytic and Endophytic Bacterial Isolates
Ulva fasciata, U. lactuca, Gracilaria dura and G. corticata were collected from the Veraval coast of India (N 20° 54.87′, E 70° 20.83′). Two samples, U. fasciata and G. dura, were also collected from Okha Port sites in India (22° 28′ 22″ N and 69° 05′ 03″ E). Neither U. lactuca nor G. corticata were found at the Okha Port locations. Samples were collected during the low tide periods in three different seasons in 2011. Both sites are located 250 km from each other (Figure 1). The pH, temperature and salinity of the seawater were measured during each collection time (Supplementary Table 1). Three individual plantlets of each species were collected from different three intertidal tide pools spread at least <25 m away from each other. The collection of the macroalgal samples and the isolation of the associated bacteria were carried out using the same procedure as previously described by Singh et al. (2011a,b). In brief, the macroalgal fronds were gently cleaned in autoclaved seawater (ASW) and then a small portion of the frond was placed into different bacterial media [Zobell marine (ZM) agar 2216, Simmons citrate (SC), thiosulfate citrate bile salts sucrose (TCBS), xylose, lysine, deoxycholate (XLD) agar and pseudomonas agar] and incubated at 25 ± 1°C for 2–15 days to isolate the epiphytic bacteria. To isolate the endophytic bacteria, the fronds of Ulva and Gracilaria spp. were surface-sterilized with different concentrations of surfactant (liquid detergent, 1 and 2% in seawater for 10 min for Ulva and Gracilaria respectively), oxidizing agents (betadine, 1 and 2% in seawater for 2 min for Ulva and Gracilaria respectively) and an antibiotic mixture (penicillin-G- 1 g, gentamycin- 1 g, streptomycin sulfate- 2 g, kanamycin- 1 g, neomycin- 200 mg, nystatin- 50 mg) of 1% in seawater for 24 h for Ulva and Gracilaria, and then incubated at 25 ± 1°C (Singh, 2013). To test the efficacy of the treatment's ability to obtain the surface-sterilized material, the surface-sterilized macroalgal plantlets (four replicates for each sample) were individually placed on different bacterial media, as mentioned above. The surface-sterilized macroalgal plantlets were crushed to fine tissues using a mortar and pestle. Thereafter, up to 10 ml of fine slurry was made using ASW and 100 μl aliquots of it were spread onto the different bacterial media as mentioned above. Different colonies were picked off and re-streaked on the respective media in order to obtain a pure colony. The pure bacterial colonies were maintained at 4 ± 1°C in slants as stock for further experimentation.
Figure 1. Map of Gujarat showing macroalgal sampling locations. The land mass showing collection spots is flanked by Gulf of Kutch on northern part (Okha) and Gulf of Khambhat (Veraval) on southern part of Gujarat (Northern west coast), India.
16S rRNA Gene Amplification and Sequencing
The genomic DNA of different bacteria was extracted using the cetyltrimethylammonium bromide buffer [CTAB 2%, NaCl 1.4 mM, EDTA 50 mM, Tris 100 mM, PVP 20%] method (Chen and Kuo, 1993). Purification of genomic DNA was confirmed with 0.8% agarose gel electrophoresis. The universal 16S rRNA primers 27F and 1492R were used for PCR amplification and sequencing (Lane, 1991). The reaction mixture and PCR conditions were the same as previously described (Singh et al., 2011a). In brief, the PCR reaction mixture contained 2.5 μl 10 × PCR buffer with MgCl2, 25 mM of each deoxynucleoside triphosphate (dATP, dCTP, dGTP, dTTP), 100 ng of each of the forward and reverse primers, 1 unit of Taq DNA polymerase and 10 ng of template DNA. The PCR protocol included a 5-min initial denaturation at 95°C, followed by 30 cycles at 94°C for 40 s, 55°C for 40 s, and 72°C for 2 min, with a final cycle of 10 min at 72°C. The amplified products were analyzed on 1.2% (w/v) agarose gels stained with ethidium bromide and the bands were visualized under UV light. The PCR products were purified using a QIAquick PCR purification kit (QIAGEN, no. 28104). The sequences were manually trimmed and their sequence homology was checked against other sequences available at the NCBI GenBank. The sequence alignment of 16S rRNA was carried out by ClustalW2 software (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and the aligned sequences were clustered into operational taxonomic units (OTUs) at 0.03 cut off values using sequence homology. Finally, the aligned 16S rRNA bacterial sequences were used to construct the phylogenetic trees with the neighbor joining method using the MEGA-5 software (Tamura et al., 2011). The bootstrap test was performed with 1000 replicates in the phylogenetic trees. The sequences were taxonomically classified using the Ribosome Database Project (RDP) using Naive Bayesian rRNA classifier version 2.4 with an 80% confidence threshold (Wang et al., 2007).
AHL Production, Separation and Identification
For the AHL detection, a pure single colony of each Gram-negative bacteria was separately inoculated in a conical flask containing 150 ml Zobell Marine Broth and incubated at 25 ± 1°C overnight on an orbital shaker at 150 rpm. On the following day, an aliquot of 50 ml culture medium was centrifuged at 4000 rpm for 15 min, then the supernatant was collected and the pH was adjusted to 2.5 using 1 N HCl to prevent hydrolysis of the AHLs. The supernatants were mixed with an equal volume of ethyl acetate to extract the AHLs. This step was repeated again to recover the AHLs from the supernatant. The upper organic layer was separated and washed with an equal volume of Milli-Q water. Thereafter, the upper organic layer was again collected and concentrated under nitrogen gas (Shaw et al., 1997). The residues were finally dissolved in 1 ml of 25% methanol containing 0.1% acetic acid and used for analysing the samples with liquid chromatography electrospray ionization mass/mass spectrometry (LC-ESI-MS/MS) and ESI-MS.
The preliminary screening of the samples was first accomplished with ESI-MS, which was then followed by LC-ESI-MS/MS-CID. The characteristics of the ion products were proposed on the basis of low-resolution MS/MS spectra (Morin et al., 2003). The spectra of LC-ESI-MS/MS were recorded from 0 m/z to 300 m/z to obtain definite identification of these ion products for their accurate mass values. The theoretical masses of the most likely AHLs in the protonated form were calculated and compared with standards. ESI-MS and LC-ESI-MS/MS-CID were performed using a Waters® Micromass® Q-Tof micro™ mass spectrometer connected with a Waters alliance HPLC and equipped with an electrospray ionization source. For ESI-MS, the samples were directly injected into the mass spectroscopy and the flow rate was 20 μl/min. Throughout the analysis, the capillary voltage, sample cone and extraction cone were maintained at 2.5 KV, 25 V, and 1.5 V, respectively. For LC-ESI-MS/MS, 20 μl sample residues were injected onto a reverse phase C18 column (Phenomenex, 150 mm × 4.6 mm) and run with a different solvent gradient (Supplementary Table 2). Argon gas was used as the collision source.
Effect of the Bacterial Supernatant and the AHL Standard on Carpospore Liberation from G. dura
The healthy and mature cystocarpic thalli of G. dura were collected from the intertidal region of the Veraval coast on the western side of India and brought to the laboratory in cold seawater (Figure 1). The thallus-bearing cystocarpic structure was cleaned and surface-sterilized following the protocol aforementioned. Thereafter, the surface-sterilized thalli were maintained in conical flasks with sterilized MP 1 medium at 25 ± 1°C under daylight white fluorescent lamps at 15 μ mol photon m−2 s−1 irradiance with 12:12 h light: dark photoperiod. The plantlets bearing five cystocarps were placed into Petri dishes containing 15 ml of 30% ASW and they were allowed to liberate the carpospores naturally for 7 days. After the carpospores were naturally liberated, the cystocarp-bearing plantlets were treated with different standards of AHLs (C4-, C6-, C8-, C10- and 3-oxo-C12-HSL) at a concentration of 10 μM each. The different concentrations (2, 4, 6, 8, and 10 μM) of the effective C4- and C6-HSLstandards were also used to determine the dose dependency of the AHLs for carpospore liberation.
A culture filtrate of different AHLs producing Gram-negative bacteria and Bacillus flexus were also used to examine the effect on carpospore liberation. Culture supernatant was collected from an overnight cell culture (Zobell Marine Broth) after centrifugation at 10000 rpm for 2 min. Subsequently, the supernatant was filter sterilized (syringe filters, 0.22 μm, Millipore) and used for the experiments. The experimental set up and the culture condition were maintained in the same way as mentioned in above paragraph, but sterilized culture filtrates were added instead of standard AHLs. Petri dishes containing fronds but no supplementation of AHLs and without added bacterial culture filtrates were treated as the control. We used also used acetonitrile as negative control. All the experiments were carried out in triplicate. The plantlets were transferred to new Petri dishes every 24 h and the liberated carpospores were counted manually using an inverted microscope. The data were represented in average release per mm2. One Way ANOVA and Dunnett's post-hoc analysis were used to analyse the effect of bacterial culture filtrates and AHLs on carpospore liberation; significant differences were determined at p > 0.01. Bonferroni correction was also applied at p < 0.001 and p > 0.05. Letter designation format was carried out with Tukey's HSD (honestly significant difference) using JMP software, which means sharing the same letters were not different at p < 0.05.
Electrophoresis of Protein Profile of the AHL-Treated Cystocarps and the Cystocarp-Bearing Plantlets
To evaluate the effect of the C4-, C6-, C8-, C10-, and 3-oxo-C12-HSL on the protein profile of the surface-sterilized cystocarps and the cystocarpic plantlets of G. dura, the surface-sterilized cystocarps and cystocarpic plantlets were treated with different concentrations of AHLs in conical flasks and kept at 25 ± 1°C for 48 h. Thereafter, the total protein of the control and the different AHL-treated cystocarps and cystocarpic plantlets were extracted by homogenizing 0.2 g fresh weight in 1 ml of the extraction buffer containing 0.5 M Tris–HCl (pH 8.0), 0.7 M sucrose, 50 mM ethylenediaminetetraacetic acid (EDTA), 0.1 M KCl, 2% (v/v) β-mercaptoethanol, and 2 mM phenylmethylsulfonyl fluoride under cool conditions. The homogenates were centrifuged at 12,000 rpm for 20 min at 4°C. The total proteins extracted from the different sources were stored at −20°C for use in further experiments. The protein concentration was determined by Folin's phenol method (Lowry et al., 1951).
The extracted proteins were analyzed with 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) according to Laemmli (1970). The 20 μg of the total protein extracted from the different AHL-treated cystocarps and cystocarpic plantlets were loaded into gels along with the control. Next, 10% Native-PAGE was used to confirm the results of SDS- PAGE. The protein bands were developed by the silver staining method.
Accession Numbers
The bacterial sequences reported in the present study were submitted to GenBank with the following accession numbers: JQ665283-JQ665389, JN996469, JQ408391, JQ408396, JQ613503- JQ613504, and JQ613506, for the 16S rRNA gene sequences.
Results
Taxonomic Classification and Phylogenetic Analysis of the Bacteria
The present study did not include any short, chimeric or repeated nucleotide sequences. Thus, all the bacterial nucleotide sequences were used to construct the phylogenetic trees. A greater proportion of sequences belonged to the Gammaproteobacteria, particularly Vibrionales, followed by Bacillales, during the pre-monsoon and monsoon seasons. The 87.87% proportion of bacteria collected during the post-monsoon season only belonged to the Vibrionaceae family (Figures 2, 3). The phylogenetic trees of the 16S rRNA sequences revealed the proper affiliation of the bacteria that were not properly assigned by the RDP analysis (Figure 2A).
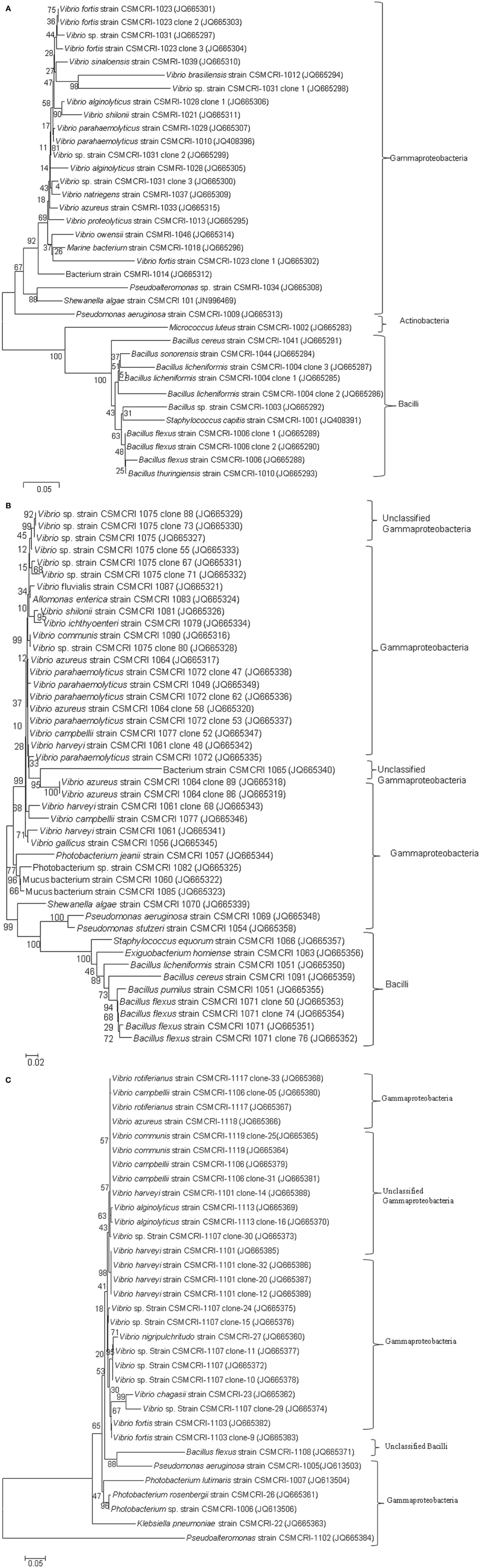
Figure 2. Phylogenetic relationships of bacterial communities isolated from Ulva and Gracilaria species during pre-monsoon (2A), monsoon (2B), and post-monsoon (2C) seasons in 2011. Neighbor-Joining method used for 16S rRNA analysis (Saitou and Nei, 1987). Bootstrap test was performed with 1000 replicates in the phylogenetic trees (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer phylogenetic trees. The evolutionary distances were computed using the Kimura 2-parameter method (Kimura, 1980) and are in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 5 for pre-monsoon post-monsoon and 2 for monsoon season). Phylogenetic analyses were conducted in MEGA5 (Tamura et al., 2011).
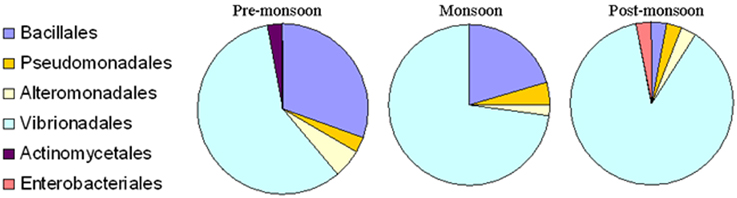
Figure 3. Percentage composition of different bacterial communities which were isolated from Ulva fasciata, U. lactuca, Gracilaria dura, and G. corticata. Samples were collected during low tide periods in three different seasons in 2011.
A total of 77 OTUs (≥97% sequence identity) were obtained from all the bacterial nucleotide sequences. The OTUs for the pre-monsoon, monsoon and post-monsoon seasons were 20, 32, and 27, respectively. All of the OTUs represent six orders from three bacterial phyla: Bacillales, Pseudomonadales, Alteromonadales, Actinomycetales, Enterobacteriales, and Vibrionales. Among these, the bacterial species belonging to Actinomycetales (Micrococcus luteus) and Enterobacteriales (Klebsiella pneumoniae) were only found during the pre-monsoon and post-monsoon seasons, respectively (Figure 3).
Epiphytic and Endophytic Bacterial Isolation
A number of epiphytic bacteria were isolated from seaweeds collected from different locations and during different seasons (Figures 4A,B, Supplementary Table 3). A total of 102 and 11 bacterial isolates were obtained as epiphytic and endophytic bacteria, respectively, based on their distinct morphological characteristics. Subsequently, the epiphytic and endophytic bacteria were phylogenetically identified. The epiphytic bacteria belonged to six orders: Actinomycetales, Alteromonadales, Bacillales, Enterobacteriales, Pseudomonadales, andVibrionales. Interestingly, the epiphytic bacteria that belonged to Vibrionales were commonly isolated from all of the macroalgal samples irrespective of the location and the season in which they were collected. Bacteria belonging to Bacillales were present only in the macroalgal samples that were collected during the pre-monsoon and monsoon seasons. Bacterial isolates belonging to Pseudomonadales and Alteromonadales were only isolated from G.dura collected from the Veraval coast while Actinomycetales and Enterobacteriales were only collected from G. corticata that was obtained from the Okha coast.
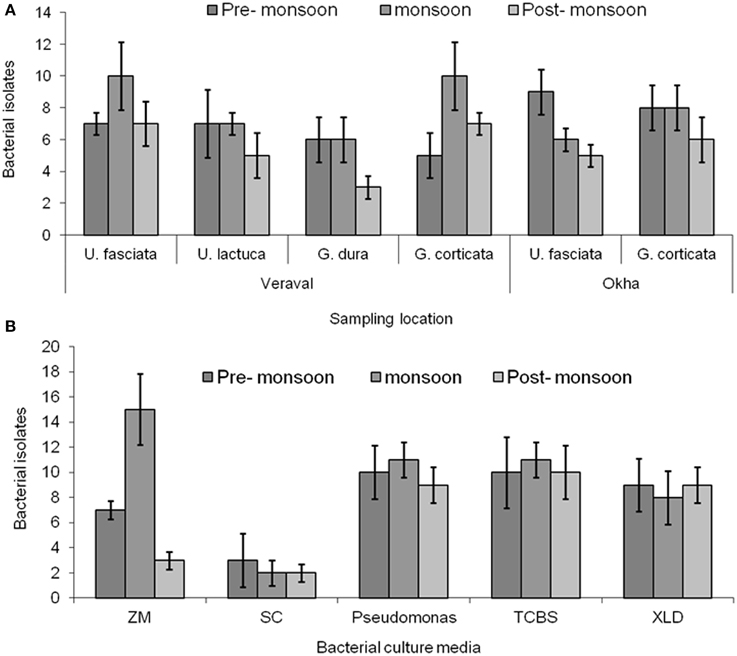
Figure 4. Bacterial isolation. (A) Enumeration of bacteria from different macroalgal samples such as Ulva fasciata, U. lactuca, Gracilaria dura and G. corticata. (B) Small plantlets of macroalgae were placed on the different culture media for isolating bacteria from them. Bars indicate, deviation of three independent replicates.
The endophytic bacteria are: Allomonas enterica (JQ665324), Vibrio parahaemolyticus (JQ665335), Shewanella algae (JN996469), Pseudomonas aeruginosa (JQ665348), P. stutzeri (JQ665358), Micrococcus luteus (JQ665283), Bacillus cereus (JQ665291), B. licheniformis (JQ665350), V. sinaloensis (JQ665310), V. nigripulchritudo (JQ665360), and V. rotiferianus (JQ665367). Among all of the endophytic bacteria, 10 bacterial isolates were isolated from the genus Gracilaria while B. cereus (JQ665291) was obtained from U. fasciata. V. parahaemolyticus was always found to be associated with G. corticata, whereas S. algae and P. aeruginosa were associated with G. dura, thereby showing evidence of algal host specificity.
Identification of the AHL Signals
In the MS/MS analysis, the activated natural compound [M + H]+ ion derived from the AHLs decomposed into specific ion products, including the [M + H- C4H7NO2 or M + H -101]+ that resulted from the neutral loss of homoserine lactone and an ion at m/z 102 corresponding to the protonated lactone (Decho et al., 2009). In the present study, seven different Gram-negative bacteria were found to produce different types of AHLs. The S. algae (JN996469) was found to produce several types of AHLs (C4-HSL, HC4-HSL, C6-HSL, 3-oxo-C6-HSL, and 3-oxo-C12-HSL), as shown in the Supplementary Datasheet, Figures S1A-D,H, (Table 1). Photobacterium lutimaris (JQ613504) was found to produce three types of AHLs (C4-HSL, HC4-HSL, C6-HSL) and each of the remaining bacterial isolates produced two types of AHLs, as shown in Table 1 and the Supplementary Datasheet 1, Figure 1. This experiment was repeated three times and the data were found to be reproducible.
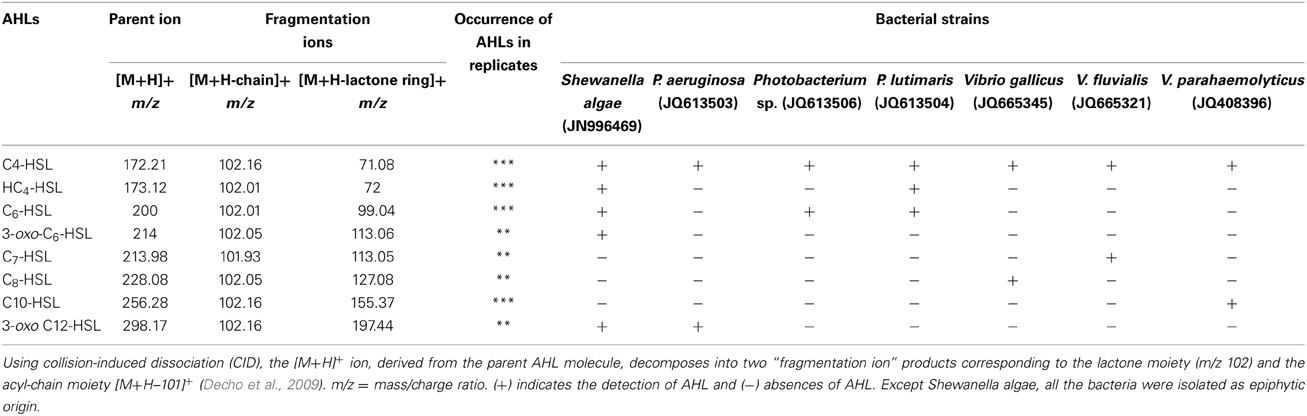
Table 1. Liquid chromatography–mass spectrometry/MS analysis of Gram-negative bacterial extracts for detecting N-acyl-homoserine lactone (AHL).
Effect of Different AHLs on Carpospore Liberation from G. dura
AHL containing culture filtrates of seven Gram-negative bacteria and the AHL standards of C4- and C6-HSL were found to induce the liberation of carpospores in G. dura as compared to the control and the C10-, 3-oxo-C12-HSL, and culture filtrates of B. flexus. There was a positive correlation between different concentrations (2, 4, 6, 8, and 10 μM) of the C4- and C6-HSL and carpospore liberation from the cystocarps (Figure 5A). The culture filtrates of S. algae showed the ability to enhance carpospore liberation up to 179.625 ± 3.6 mm2 carpospores as compared to P. aeruginosa, which produced 108.375 ± 21.62 mm2 carpospores. The carpospores that were liberated with culture filtrates of Photobacterium sp., P. lutimaris, V. gallicus, V. fluvialis, and V. parahaemolyticus were 76.66 ± 5.07 mm2, 66.87 ± 28.97 mm2, 44.26 ± 6.06 mm2, 50.58 ± 3.74 mm2, and 62.83 ± 6.34 mm2, respectively. On the other hand, the standard C4- and C6- HSL yielded 93.333 ± 15.33 mm2 and 99.448 ± 30.94 mm2 carpospores, respectively (Figure 5B). One Way ANOVA and Dunnett's post-hoc analysis showed significant differences at p > 0.01 for the AHL standards and the bacterial culture filtrates. Additionally, Bonferroni correction was used to determine effect of AHLs and bacterial culture filtrates on carpospores liberation. Effect of C4-HSL, C6-HSL and culture filtrates of AHLs producing bacterial isolates (except V. gallicus) were significant at p < 0.001 whereas others had no effect (P > 0.05) in Bonferroni correction.
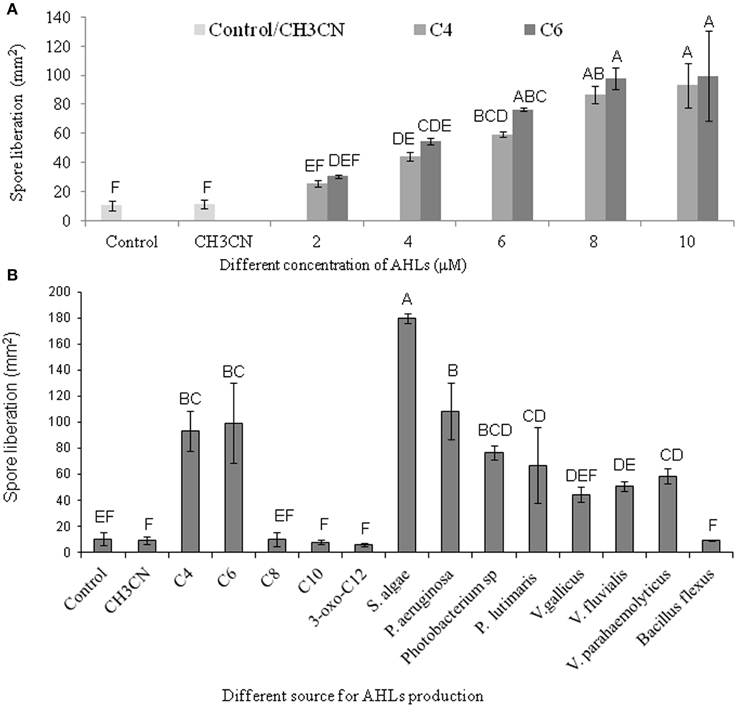
Figure 5. Effect of different standard AHLs and Gram-negative bacterial isolates on carpospores liberation from Gracilaria dura. (A) Effect of different concentrations (2, 4, 6, 8, and 10 μM) of C4- and C6-HSL on carpospores liberation. (B) Effect of different AHLs standard at 10 μM, culture filtrates of Gram-negative bacterial isolates and Bacillus flexus on carpospores liberation. Bars indicate minima and maxima of three replicates. One Way ANOVA and Dunnett's post-hoc analysis showed significant differences at p > 0.01 for the C4-HSL, the C6-HSL, and culture filtrates of AHLs producing bacterial isolates. Effect of C4-HSL, C6-HSL, and culture filtrates of AHLs producing bacterial isolates (except V. gallicus) were significant at p < 0.001 whereas others had no effect (P > 0.05) in Bonferroni correction. Letter designation format was carried out with Tukey's HSD using JMP software, which means sharing the same letters were not different at p < 0.05. AHLs were dissolved in 100% CH3CN and working concentration of control was fixed at 0.04%.
Electrophoresis of Protein Profile of the AHL-Treated Cystocarps and the Cystocarp-Bearing Plantlets
To understand the effect of different AHLs on carpospore liberation from the cystocarps of G. dura, the total protein profile of the AHL-treated cystocarps and the cystocarp-bearing plantlets were analyzed with polyacrylamide gel electrophoresis. Among all of the AHL-treated cystocarpic plantlets, those treated with C4- and C6-HSL showed three specific peptide bands with an approximate molecular weight of 45, 50, and 60 kDa, respectively (Figure 6A). In another experiment, the C4- and C6-HSL-treated cystocarps showed two specific peptide bands having an approximately molecular weight of 50 kDa and 60 kDa, respectively (Figure 6B). The C8-, C10-, and 3-oxo-C12-HSL-treated cystocarpic plantlets and the cystocarps and the control did not induce these specific protein bands. The specificity of the peptide bands was determined using Native-PAGE and it was found that these peptide bands represented three different proteins.
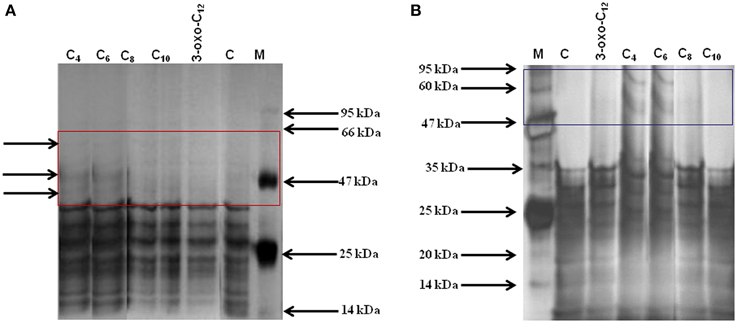
Figure 6. Total protein profiling of AHLs treated cystocarps and cystocarp bearing plantlets of G. dura with polyacrylamide gel electrophoresis. (A) SDS-PAGE analysis of AHLs treated cystocarpic plantlets. (B) SDS-PAGE analysis of AHLs treated cystocarps. Different types of AHLs (C4, C6,C8, C10, and 3-oxo-C12-HSL) were used for experiments at concentration of 10 μM.
Discussion
To obtain insight about the important role that seaweed-associated bacteria play in the host's life cycle, several types of epiphytic and endophytic bacteria were isolated from the Ulva and Gracilaria species. Subsequently, the isolated bacteria were screened for AHL production and their ability to liberate carpospores from the cyctocarp of G. dura was evaluated. The bacterial communities identified in this study were more or less similar to the bacterial communities identified from different seaweeds (Burke et al., 2011; Lachnit et al., 2011). Dominant bacterial members of Gammaproteobacteria were consistently encountered in all of the samples, seasons and locations thereby indicating their abundance in the marine environment. Similarly, Patel et al. (2003) and Tait et al. (2005) also reported Gammaproteobacteria as the dominant epiphytic bacteria associated with green macroalgae Enteromorpha and Ulva in samples taken from Wembury Beach, Devon, UK. The red macroalga Amphiroa anceps was also found to be a habitat for Gammaproteobacteria while Bacteroidetes and Gammaproteobacteria were found to be associated with another red alga Corallina officinalis (Huggett et al., 2006). The high abundance of Gammaproteobacteria on the surface of the seaweeds could be attributed to its tendency to form biofilms (Tait et al., 2009). Venter et al. (2004) and Giovannoni and Stingl (2005) analyzed planktonic communities found in seawater and they observed that Gammaproteobacteria, Actinobacteria, Planctomycetes, and Bacillales are commonplace in oceanic waters. Thus, phylogenetic studies of these epiphytic bacteria reveal that the recruitment of different bacterial communities that coexist with different seaweeds is of oceanic origin. A few previous reports have dealt with endophytic bacteria isolated from different macroalgae. In earlier studies, endophytic bacteria were isolated mainly for the chemical interactions from Caulerpa, Codium, Bryopsis, and Penicillus and those studies did not characterize their phylogenetic relevance (Please see the review of Goecke et al., 2010). Recently, Hollants et al. (2011) isolated endophytic bacteria belonging to Flavobacteriaceae, Bacteroidetes, and Phyllobacteriaceae from the siphonous green alga Bryopsis hypnoides, as well as, Xanthomonadaceae, Gammaproteobacteria, Epsilonproteobacteria and a new Arcobacter species isolated from B. pennata. Thus, limited information is available about the endophytic communities of seaweeds.
The age of the plantlets is also considered to be a significant inherent source of variation in seaweed-associated bacterial communities at spatial and temporal scales (Staufenberger et al., 2008; Goecke et al., 2010). It has been demonstrated that bacterial communities of young meristem and cauloid sections of different plantlets of the brown alga Laminaria saccharina were more similar to each other than the aging phyloid section of the same plantlets (Staufenberger et al., 2008). The present study has also confirmed the temporal variations of bacterial communities associated with macroalgal samples across seasons. We observed less seaweed-associated bacterial communities during the post-monsoon season as compared to the pre-monsoon and monsoon seasons (Figures 2, 3). During the pre-monsoon and monsoon seasons, the seaweed surfaces were also occupied by bacterial species of Firmicutes. Considering this fact, the present findings revealed that the bacterial species belonging to Firmicutes are highly variable while the bacterial species belonging to Gammaproteobacteria were found to be seaweed-philic but temporally variable. Despite these levels of variability, the epi- and endophytic communities are included in a sub-population of bacteria that were consistently associated with the Ulva and Gracilaria species. Such an observation provided evidence of core bacterial communities that have an important function in host macroalgae and will enhance our understanding of bacterial-host interactions in plant science.
In this study, S. algae was found to produce several types of AHLs (C4-, HC4-, C6-, 3-oxo-C6- and 3-oxo-C12-HSL); thus, its culture filtrates promoted carpospore liberation from G. dura as compared to the culture filtrates of P. aeruginosa, Vibrio and Photobacteria and the control. Weinberger et al. (2007) reported that C4-HSL potentially influenced the carpospore liberation capacity in Acrochaetium sp. While, the present study found that both C4- and C6-HSL equally contributed to carpospore liberation from G. dura. The positive correlation between different concentrations of C4- and C6-HSL and carpospore liberation from G. dura revealed that relative increases in the concentration of C4- and C6-HSL up to 10 μM also enhances carpospore liberation. The C8-, C10-, 3-oxo-C12-HSL and culture filtrates of Gram-positive bacterium B. flexus did not influence carpospore liberation thereby indicating selective regulation by C4- and C6-HSL (Figure 5). SDS-PAGE analysis of the total protein profile of the cystocarps of G. dura treated with C4- and C6-HSL revealed induction of specific peptide bands with an approximate molecular weight of 50 kDa and 60 kDa, whereas the cystocarpic plantlets treated with C4- and C6-HSL revealed three specific peptide bands with the approximate molecular weight of 45, 50, and 60 kDa as compared to the C8-, C10- and 3-oxo-C12-HSL-treated samples. Although, the AHLs identified from seven different bacteria in the present study are not quantified, a recent study reported 0.1–30 μM of AHLs are produced by Gram-negative biofilm-forming bacteria (Ahlgren et al., 2011). The effect of bacterial culture filtrates on the liberation of carpospores could not be limited to a particular species or even a specific genus level as different bacterial isolates of different orders showed dissimilar effects (Figure 5). The findings of the present study also suggested that the diffusion ability, stability and availability of AHLs around the cystocarpic plantlets are important factors for carpospore liberation in a natural environment and cross-talk between the seaweed-bacteria association. Tait et al. (2005) reported that short acyl chain molecules (C6-HSL and 3-hydroxy-C6-HSL) were diffused more quickly from agarose gel than longer acyl chain molecules, such as 3-oxo-C10-HSL. A similar finding was also observed in the higher plants. It has been reported that short side-chain AHL compounds are easily soluble in water and are actively taken up into plant roots as well as transported through shoots, as compared to long acyl side chain AHLs in barley and A. thaliana (Götz et al., 2007; von Rad et al., 2008; Hartmann et al., 2014; Sieper et al., 2014).
Macroalgal surfaces are living hosts and they perform an essential role in coastal ecosystems (Burke et al., 2011). Firmicutes have been found to be the second most abundant bacteria on these algal surfaces and they contribute to approximately 15–30% of dimethylsulfoniopropionate assimilation (Malmstrom et al., 2004). The high variability of bacterial communities associated with different species of Ulva and Gracilaria, or even among the same species, suggests that functional redundancy exists within these communities. This conclusion follows the redundancy hypothesis, which presumes that more than one species is capable of performing a specific role within an ecosystem (Naeem, 1998; Burke et al., 2011).
In conclusion, this study identified and characterized several epi- and endophytic bacterial communities associated with different taxa of Ulva and Gracilaria. It also demonstrated that some Gram-negative epi- and endophytic seaweed-associated bacteria produce different types of AHLs. The C4- and C6-HSL as well as the culture filtrate of seven AHL- producing Gram-negative bacteria were found to enhance carpospore liberation from the cyctocarps of G. dura. Thus, these bacterial isolates can effectively be used for mass carpospore liberation, even though the underpinning molecular mechanisms of this phenomenon are not well-understood yet. Additional biochemical and molecular studies are required to characterize their signaling mechanisms and those studies will serve to illuminate new avenues for further optimization of this phenomenon. Therefore, the evaluation of this molecule signaling cascade is our long-term goal and will be reflected in future publications.
Author Contributions
RPS, CRKR and BJ conceived and designed the work. RPS and RSB collected the samples and performed the experiments. CRKR and BJ analyzed the QS data. RPS and RSB conducted electrophoresis analysis and identified the bacteria associated with the seaweeds. RPS, CRKR and BJ wrote the manuscript. All of the authors contributed to the discussion and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank S. Thiruppathi, Technical Assistant for assisting in the carpospores liberation aspect of this study. The authors are also grateful to Arun K. Das, CSMCRI for helping with LC-MS analysis. The first author (RPS) gratefully acknowledges the CSIR, New Delhi (India) for awarding the Senior Research Fellowship. The financial support received from CSIR Network projects BioPros (BSC 0106) and BioEn (CSC 0116) is gratefully acknowledged. We also thank Prof. Bryan Bailey and Dr. Andrea Campisano for their critical comments which have improved the presentation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2015.00117/abstract
References
Ahlgren, N. A., Harwood, C. S., Schaefer, A. L., Giraud, E., and Greenberg, E. P. (2011). Aryl- homoserine lactone quorum sensing in stem-nodulating photosynthetic Bradyrhizobia. Proc. Nati. Acad. Sci. U.S.A. 108, 7183–7188. doi: 10.1073/pnas.1103821108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Armstrong, E., Yan, L., Boyd, K. G., Wright, P. C., and Burgess, J. G. (2001). The symbiotic role of marine microbes on living surfaces. Hydrobiologia 461, 37–40. doi: 10.1023/A:1012756913566
Burke, C., Thomas, T., Lewis, M., Steinberg, P., and Kjelleberg, S. (2011). Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 5, 590–600. doi: 10.1038/ismej.2010.164
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, W., and Kuo, T. (1993). A simple and rapid method for the preparation of gram negative bacterial genomic DNA. Nucleic Acids Res. 21:2260. doi: 10.1093/nar/21.9.2260
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Decho, A. W., Visscher, P. T., Ferry, J., Kawaguchi, T., He, L., Przekop, K. M., et al. (2009). Autoinducers extracted from microbial mats reveal a surprising diversity of N-acylhomoserine lactones (AHLs) and abundance changes that may relate to diel pH. Environm. Microbiol. 11, 409–420. doi: 10.1111/j.1462-2920.2008.01780.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Giovannoni, S. J., and Stingl, U. (2005). Molecular diversity and ecology of microbial plankton. Nature 437, 343–348. doi: 10.1038/nature04158
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Goecke, F., Labes, A., Wiese, J., and Imhoff, J. F. (2010). Chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Ser. 409, 267–300. doi: 10.3354/meps08607
Götz, C., Fekete, A., Gebefügi, I., Forczek, S., Fuksová, K., Li, X., et al. (2007). Uptake, degradation and chiral discrimination of N-acyl-D/L-homoserine lactones by barley (Hordeum vulgare) and yam bean (Pachyrhizus erosus) plants. Anal. Bioanal. Chem. 389, 1447–1457. doi: 10.1007/s00216-007-1579-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hartmann, A., Gantner, S., Schuhegger, R., Steidle, A., Dürr, C., Schmid, M., et al. (2004). “N-acyl-homoserine lactones of rhizosphere bacteria trigger systemic resistance in tomato plants,” in Biology of Molecular Plant-Microbe Interactions, Vol. 4, eds B. Lugtenberg, I. Tikhonovich, and N. Provorov (St. Paul, MN: MPMI-Press), 554–556
Hartmann, A., Rothballer, M., Hense, B. A., and Schröder, P. (2014). Bacterial quorum sensing compounds are important modulators of microbe-plant interactions. Front. Plant Sci. 5:131. doi: 10.3389/fpls.2014.00131
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hollants, J., Leroux, O., Leliaert, F., Decleyre, H., De-Clerck, O., and Willems, A. (2011). Who is in there? exploration of endophytic bacteria within the siphonous green seaweed Bryopsis (Bryopsidales, Chlorophyta). PLoS ONE 6:e26458. doi: 10.1371/journal.pone.0026458
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Holmström, C., Egan, S., Franks, A., McCloy, S., and Kjelleberg, S. (2002). Antifouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microbiol. Ecol. 41, 47–58. doi: 10.1016/S0168-6496(02)00239-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huggett, M. J., Williamson, J. E., de Nys, R., Kjelleberg, S., and Steinberg, P. D. (2006). Larval settlement of the common Australian sea urchin Heliocidaris erythrogramma in response to bacteria from the surface of coralline algae. Oecologia 149, 604–619. doi: 10.1007/s00442-006-0470-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jin, G., Liu, F., Ma, H., Hao, S., Zhao, Q., Bian, Z., et al. (2012). Two G-protein-coupled-receptor candidates, Cand2 and Cand7, are involved in Arabidopsis root growth mediated by the bacterial quorum-sensing signals N-acyl- homoserine lactones. Biochem. Biophys. Res. Commun. 417, 991–995. doi: 10.1016/j.bbrc.2011.12.066
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Joint, I., Tait, K., Callow, M. E., Callow, J. E., Milton, D., Williams, P., et al. (2002). Cell-to-cell communication across the procaryote – eucaryote boundary. Science 298, 1207. doi: 10.1126/science.1077075
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Joint, I., Tait, K., and Wheeler, G. (2007). Cross-kingdom signalling: exploitation of bacterial quorum sensing molecules by the green seaweed Ulva. Phil. Trans. R. Soc. B. 362, 1223–1233. doi: 10.1098/rstb.2007.2047
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kimura, M. (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lachnit, T., Meske, D., Wahl, M., Harder, T., and Schmitz, R. (2011). Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 13, 655–665. doi: 10.1111/j.1462-2920.2010.02371.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lane, A. L., and Kubanek, J. (2008). “Secondary metabolite defenses against pathogens and biofoulers,” in Algal Chemical Ecology, ed C. H. Amsler (Berlin: Springer), 229–243.
Liu, F., Bian, Z., Jia, Z., Zhao, Q., and Song, S. (2012). The GCR1 and GPA1 participate in promotion of Arabidopsis primary root elongation induced by N-acyl-homoserine lactones, the bacterial quorum-sensing system. Mol. Plant Microbe Interact. 25, 677–683. doi: 10.1094/MPMI-10-11-0274
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951). Protein measurement with folin phenol reagent. J. Biol. Chem. 193, 265–275.
Malmstrom, R. R., Kiene, R. P., and Kirchman, D. L. (2004). Identification and enumeration of bacteria assimilating dimethylsulfoniopropionate (DMSP) in the North Atlantic and Gulf of Mexico. Limnol. Oceanogr. 49, 597–606. doi: 10.4319/lo.2004.49.2.0597
Maruyama, A., Yamaguchi, I., Maeda, M., and Shimizu, U. (1988). Evidence of cytokinin production by a marine bacterium and its taxonomic characteristics. Can. J. Microbiol. 34, 829–833. doi: 10.1139/m88-142
Matsuo, Y., Imagawa, H., Nishizawa, M., and Shizuri, Y. (2005). Isolation of an algal morphogenesis inducer from a marine bacterium. Science 307, 1598. doi: 10.1126/science.1105486
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morin, D., Grasland, B., Vallée-Réhel, K., Dufau, C., and Haras, D. (2003). On-line high performance liquid chromatography-mass spectrometric detection and quantification of N-acylhomoserine lactones, quorum sensing signal molecules, in the presence of biological matrices. J. Chromatogr. A 1002, 79–92. doi: 10.1016/S0021-9673(03)00730-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Naeem, S. (1998). Species redundancy and ecosystem reliability. Conv. Biol. 12, 39–45. doi: 10.1046/j.1523-1739.1998.96379.x
Nakanishi, K., Nishijima, M., Nishimura, M., Kuwano, K., and Saga, N. (1996). Bacteria that induce morphogenesis in Ulva pertusa (Chlorophyceae) grown under axenic conditions. J. Phycol. 32, 479–482. doi: 10.1111/j.0022-3646.1996.00479.x
Ortíz-Castro, R., Martinez-Trujillo, M., and López-Bucio, J. (2008). N-acyl-homoserine lactones: a class of bacterial quorum-sensing signals alter post-embryonic root development in Arabidopsis thaliana. Plant Cell Environ. 31, 1497–1509. doi: 10.1111/j.1365-3040.2008.01863.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Patel, P., Callow, M. E., Joint, I., and Callow, J. A. (2003). Specificity in the settlement – modifying response of bacterial biofilms towards zoospores of the marine alga Enteromorpha. Environ. Microbiol. 5, 338–349. doi: 10.1046/j.1462-2920.2003.00407.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Provasoli, L., and Pintner, I. J. (1980). Bacteria induced polymorphism in an axenic laboratory strain of Ulva lactuca (Chlorophyceae). J. Phycol. 32, 479–482.
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Schenk, S. T., Stein, E., Kogel, K. H., and Schikora, A. (2012). Arabidopsis growth and defense are modulated by bacterial quorum sensing molecules. Plant Signal. Behav. 7, 178–181. doi: 10.4161/psb.18789
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schikora, A., Schenk, S. T., Stein, E., Molitor, A., Zuccaro, A., and Kogel, K. H. (2011). N-acyl-homoserine lactone confers resistance toward biotrophic and hemibiotrophic pathogens via altered activation of AtMPK6. Plant Physiol. 157, 1407–1418. doi: 10.1104/pp.111.180604
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schuhegger, R., Ihring, A., Gantner, S., Bahnweg, G., Knappe, C., Vogg, G., et al. (2006). Induction of systemic resistance in tomato plants by N-acyl-homoserine lactone–producing rhizosphere bacteria. Plant Cell Environ. 29, 909–918. doi: 10.1111/j.1365-3040.2005.01471.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shaw, P. D., Ping, G., Daly, S. L., Cha, C., Cronan, J. E., Rinehart, K. L., et al. (1997). Detecting and characterizing N-acyl homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. U.S.A. 94, 6036–6041. doi: 10.1073/pnas.94.12.6036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sieper, T., Forczek, S., Matucha, M., Krämer, P., Hartmann, A., and Schröder, P. (2014). N-acyl-homoserine lactone uptake and systemic transport in barley rest upon active parts of the plant. New Phytol. 201, 545–555. doi: 10.1111/nph.12519
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Singh, R. P. (2013). Studies on Certain Seaweed–Bacterial Interaction from Saurashtra Coast, Chapter 2. Ph.D. thesis. Available online at: http://ir.inflibnet.ac.in:8080/jspui/handle/10603/9191
Singh, R. P., Bijo, A. J., Baghel, R. S., Reddy, C. R. K., and Jha, B. (2011b). Role of bacterial isolates in enhancing the bud induction in the industrially important Red alga Gracilaria dura. FEMS Microbiol. Ecol. 76, 381–392. doi: 10.1111/j.1574-6941.2011.01057.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Singh, R. P., Mantri, V. A., Reddy, C. R. K., and Jha, B. (2011a). Isolation of seaweed associated bacteria and their morphogenesis inducing capability in axenic cultures of the green alga Ulva fasciata. Aquat. Biol. 12, 13–21. doi: 10.3354/ab00312
Singh, R. P., and Reddy, C. R. K. (2014). Seaweed–microbial interactions: key functions of seaweed-associated bacteria. FEMS Microbiol. Ecol. 88, 213–230. doi: 10.1111/1574-6941.12297
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Staufenberger, T., Thiel, V., Wiese, J., and Imhoff, J. F. (2008). Phylogenetic analysis of bacteria associated with Laminaria saccharina. FEMS Microbiol. Ecol. 64, 65–77. doi: 10.1111/j.1574-6941.2008.00445.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tait, K., Joint, I., Daykin, M., Milton, D., Williams, P., and Cámara, M. (2005). Disruption of quorum sensing in seawater abolished attraction of zoospores of the green alga Ulva to bacterial biofilms. Environ. Microbiol. 7, 229–240. doi: 10.1111/j.1462-2920.2004.00706.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tait, K., Williamson, H., Atkinson, S., Williams, P., Camara, M., and Joint, I. (2009). Turnover of quorum sensing signal molecules modulate cross-kingdom signalling. Environ. Microbiol. 11, 1792–1802. doi: 10.1111/j.1462-2920.2009.01904.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Veliz-Vallejos, D. F., van Noorden, G. E., Yuan, M., and Mathesius, U. (2014). A Sinorhizobium meliloti-specific N-acyl homoserine lactone quorum-sensing signal increases nodule numbers in Medicago truncatula independent of autoregulation. Front. Plant Sci. 5:551. doi: 10.3389/fpls.2014.00551
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Venter, J. C., Remington, K., Heidelberg, J. F., Halpern, A. L., Rusch, D., Eisen, J. A., et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74. doi: 10.1126/science.1093857
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
von Rad, U., Klein, I., Dobrev, P. I., Kottova, J., Zazimalova, E., Fekete, A., et al. (2008). The response of Arabidopsis thaliana to N-hexanoyl- DL-homoserine-lactone, a bacterial quorum sensing molecule produced in the rhizosphere. Planta 229, 73–85. doi: 10.1007/s00425-008-0811-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weinberger, F., Beltran, J., Correa, J. A., Lion, U., Pohnert, G., Kumar, N., et al. (2007). Spore release in Acrochaetium sp. (Rhodophyta) is bacterially controlled. J. Phycol. 43, 235–241. doi: 10.1111/j.1529-8817.2007.00329.x
Wheeler, G. L., Tait, K., Taylor, A., Brownlee, C., and Joint, I. (2006). Acyl-homoserine lactones modulate the settlement rate of zoospores of the marine alga Ulva intestinalis via a novel chemokinetic mechanism. Plant Cell Environ. 29, 608–618. doi: 10.1111/j.1365-3040.2005.01440.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Williams, P. (2007). Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153, 3923–3938. doi: 10.1099/mic.0.2007/012856-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zarkani, A. A., Stein, E., Röhrich, C. R., Schikora, M., Evguenieva-Hackenberg, E., Degekolb, T., et al. (2013). Homoserine lactones influence the reaction of plants to rhizobia. Int. J. Mol. Sci. 14, 17122–17146. doi: 10.3390/ijms140817122
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: quorum sensing, carpospores liberation, Gracilaria dura, Vibrio, Ulva spp.
Citation: Singh RP, Baghel RS, Reddy CRK and Jha B (2015) Effect of quorum sensing signals produced by seaweed-associated bacteria on carpospore liberation from Gracilaria dura. Front. Plant Sci. 6:117. doi: 10.3389/fpls.2015.00117
Received: 12 November 2014; Accepted: 12 February 2015;
Published online: 04 March 2015.
Edited by:
Anton Hartmann, Helmholtz Zentrum München - German Research Center for Environmental Health, GermanyReviewed by:
Bryan Bailey, United States Department of Agriculture, USAAndrea Campisano, Fondazione Edmund Mach, Italy
Copyright © 2015 Singh, Baghel, Reddy and Jha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: C. R. K. Reddy, Discipline of Marine Biotechnology and Ecology, CSIR-Central Salt and Marine Chemicals Research Institute, Bhavnagar 364002, India e-mail:Y3JrQGNzbWNyaS5vcmc=
†Present address: Ravindra Pal Singh, Laboratory of Microbial Technology, Department of Bioscience and Biotechnology, Faculty of Agriculture, Kyushu University, Kyushu, Japan
 Ravindra Pal Singh
Ravindra Pal Singh Ravi S. Baghel
Ravi S. Baghel C. R. K. Reddy
C. R. K. Reddy Bhavanath Jha
Bhavanath Jha