
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Plant Sci. , 25 February 2015
Sec. Plant Physiology
Volume 6 - 2015 | https://doi.org/10.3389/fpls.2015.00116
This article is part of the Research Topic ROS regulation during plant abiotic stress responses View all 27 articles
Peroxisomes are ubiquitous organelles with a notable oxidative metabolism. In plants, these subcellular compartments have been shown to be involved in the metabolism of reactive oxygen and nitrogen species (ROS and RNS), whose components, hydrogen peroxide and nitric oxide (NO), are important molecules involved in signaling processes. The presence of new elements in plant peroxisomes such as glutathione reductase (GR), sulfite oxidase (SO), glutathione (GSH), and S-nitrosoglutathione (GSNO) indicates the involvement of these organelles in the sulfur metabolism. This could suggest the participation of a new family of molecules designated as reactive sulfur species (RSS) which will possibly provide new functions for peroxisomes.
Peroxisomes are remarkable subcellular compartments given their simple morphology (granular/fibrillar matrix bounded by a single membrane) which does not reflect the complexity of their enzymatic composition (Hayashi et al., 2000; Baker and Graham, 2002; del Río et al., 2006). In plant cells, peroxisomes are involved in the photorespiration cycle, fatty acid β-oxidation, the glyoxylate cycle, and the metabolism of ureides (Corpas et al., 1997; Baker and Graham, 2002; del Río et al., 2006; Hu et al., 2012), thus indicating that these organelles play a role in key physiological processes such as seed germination, plant development, fruit ripening, and senescence. Plant peroxisomes have been shown to be a source of ROS including molecules such as superoxide radicals (O·−2), hydrogen peroxide (H2O2), and RNS (del Río et al., 2006; del Río, 2011; Corpas et al., 2013; Corpas and Barroso, 2014b). RNS include NO and related molecules such as peroxynitrite (ONOO−) and GSNO which are characterized by a broad spectrum of physiological/pathological activities. Both these molecular families (ROS and RNS) include radical molecules containing an unpaired electron as well as non-radical molecules and can also have dual effects depending on their cellular concentration. Thus, H2O2 and NO at low concentrations can function as signal molecules in the cell or may cause damage to cell components when overproduced as a consequence of adverse conditions (Valderrama et al., 2007; Chaki and Luque, 2011; Signorelli et al., 2013).
Sulfur (S) is an essential mineral for plant growth and development (Leustek and Saito, 1999; Rausch and Wachter, 2005; Hawkesford and De Kok, 2006). It is present in thiamin (B1) and pantothenic acid (B5) vitamins, amino acids (cysteine and methionine), biotin and molybdenum cofactors, and prosthetic groups (Leustek and Saito, 1999) and also in secondary sulfur compounds (polysulfides, glucosinolates, and phytochelatins). In addition, other organic sulfur compounds, such as thiols, GSH, GSNO, and sulfolipids, play an important role in physiological processes and plant stress conditions (Brychkova et al., 2007; Münchberg et al., 2007). In animal cells, the gas hydrogen sulfide (H2S) has been shown to be generated from L-cysteine by the pyridoxal-5′-phosphate-dependent enzyme. Thus, endogenous H2S can act as a neuromodulator in rat brain (Abe and Kimura, 1996). In higher plants, recent evidence indicates that H2S is actively involved in the regulation of ethylene-induced stomatal closure and also interacts with H2O2 to regulate the plasma membrane Na+/H+ antiporter system under salinity stress (Hou et al., 2013; Li et al., 2014). The term RSS has been proposed in order to designate a group of sulfur-related molecules that are formed in vivo under oxidative stress conditions in animal systems (Giles et al., 2001, 2002; Jacob et al., 2004). These molecules include thiyl radicals (RS·), disulfide-S-oxides [RS(O)2SR] and sulfenic acids (RSOH). Thus, high cellular GSH concentrations in an oxidative environment and the decomposition of S-nitrosothiols generate disulfide-S-oxides (Tao and English, 2004). These mechanisms can modulate the function of sulfur proteins throughout the redox status of biological thiols (Jacob and Anwar, 2008). Accordingly, disulfide formation is an important cysteine redox reaction in many proteins that affects its function, with thioredoxins and peroxiredoxins being good examples.
In plant peroxisomes, the presence of important sulfur compounds such as GSH (non-enzymatic antioxidants) (Jiménez et al., 1997; Müller et al., 2004) and GSNO (transport and storage vehicle for NO) has been demonstrated (Barroso et al., 2013). Furthermore, the presence of enzymes such as GR (Jiménez et al., 1997; Romero-Puertas et al., 2006), S-nitrosoglutathione reductase (GSNOR) (Reumann et al., 2007; Barroso et al., 2013) and SO (Eilers et al., 2001; Nakamura et al., 2002; Nowak et al., 2004; Hänsch and Mendel, 2005) involved in the sulfur metabolism has also been reported. These new insights lead us to suggest that peroxisomes may play a role in the RSS metabolism, as has been demonstrated for ROS and RNS. Figure 1 shows the potential interactions among the different ROS, RNS, and sulfur-containing compounds in peroxisomes. NO is generated by L-arginine-dependent nitric oxide synthase (NOS) activity (Corpas and Barroso, 2014a) which can react with superoxide radicals generated by xanthine oxidase to form peroxynitrite (ONOO−). This RNS is a highly oxidant compound capable of catalyzing the conversion of xanthine dehydrogenase to xanthine oxidase (Corpas et al., 2008) or inducing protein nitration (Radi, 2013). NO can also react with GSH to form GSNO which can be decomposed by GSNOR activity through the generation of GSSG (oxidized form) and NH3. GSSG is reduced by GR as a component of the ascorbate-glutathione cycle. H2O2, which is mainly generated by flavin-oxidases, is decomposed either by catalase or ascorbate peroxidase (APX). SO catalyzes the conversion of sulfite to sulfate with the concomitant generation of H2O2 (Hänsch et al., 2006). It has been reported that low concentrations of sulfite inhibit catalase activity (Veljović-Jovanović et al., 1998), which could therefore be a means of regulating both enzymes.
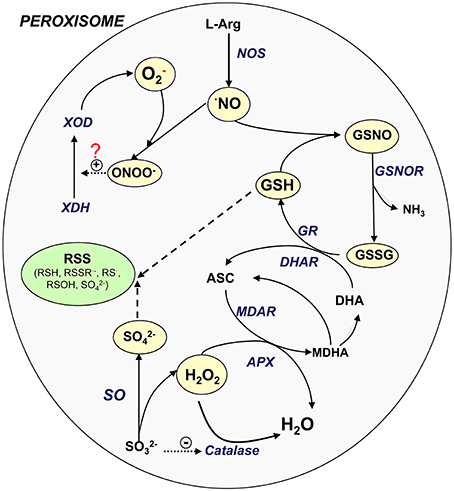
Figure 1. Signaling cross-talk between NO, ROS, and RSS. Model of the interaction amongst the different ROS, RNS, and sulfur metabolism into plant peroxisomes. ASC, ascorbate, reduced form; DHA, ascorbate, oxidized form (dehydroascorbate); GSH, glutathione, reduced form; GSNOR, nitrosoglutathione reductase; GSSG, glutathione, oxidized form; NO, nitric oxide; NOS, L-arginine-dependent nitric oxide synthase; MDAR, monodehydroascorbate reductase; ONOO−, peroxinitrite; SO, sulfite oxidase; XDH, xanthine dehydrogenase; XOD, xanthine oxidase; RSS, reactive sulfur species; RSH, thiol; RSSR·−, disulfide radical; RS·, thyl radical; ROSH, sulfenic acid; SO2−4, sulfate.
In this context, the interactions of ROS, RNS and possibly RSS components in plant peroxisomes open up new challenges and a new area of research to determine the biochemical interactions and potential functions of these reactive species of oxygen, nitrogen and sulfur in peroxisomes, some of which play a very important role as signaling molecules in physiological and phyto-pathological processes (Yamasaki, 2005).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Work in our laboratories is supported by an ERDF-cofinanced grant from the Ministry of Science and Innovation (BIO2012-33904 and RECUPERA2020) and Junta de Andalucía (Groups BIO192 and BIO286).
Abe, K., and Kimura, H. (1996). The possible role of hydrogen sulfide as an endogenous neuromodulator. J. Neurosci. 16, 1066–1071.
Baker, A., and Graham, I. A. (2002). “Plant Peroxisomes,” in Biochemistry, Cell Biology and Biotechnological Applications (Dordrecht: Kluwer Academic Publishers). doi: 10.1007/978-94-015-9858-3
Barroso, J. B., Valderrama, R., and Corpas, F. J. (2013). Immunolocalization of S-nitrosoglutathione, S-nitrosoglutathione reductase and tyrosine nitration in pea leaf organelles. Acta Physiol. Plant. 35, 2635–2640. doi: 10.1007/s11738-013-1291-0
Brychkova, G., Xia, Z., Yang, G., Yesbergenova, Z., Zhang, Z., Davydov, O., et al. (2007). Sulfite oxidase protects plants against sulfur dioxide toxicity. Plant J. 50, 696–709. doi: 10.1111/j.1365-313X.2007.03080.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chaki, M., Valderrama, R., Fernández-Ocaña, A. M., Carreras, A., Gómez-Rodríguez, M. V., Pedrajas, J. R., et al. (2011). Mechanical wounding induces a nitrosative stress by down-regulation of GSNO reductase and an increase in S-nitrosothiols in sunflower (Helianthus annuus) seedlings. J. Exp. Bot. 62, 1803–1813. doi: 10.1093/jxb/erq358
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Corpas, F. J., and Barroso, J. B. (2014a). Functional implications of peroxisomal nitric oxide (NO) in plants. Front Plant Sci. 5:97. doi: 10.3389/fpls.2014.00097
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Corpas, F. J., and Barroso, J. B. (2014b). Peroxynitrite (ONOO−) is endogenously produced in Arabidopsis peroxisomes and is overproduced under cadmium stress. Ann. Bot. 113, 87–96. doi: 10.1093/aob/mct260
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Corpas, F. J., Leterrier, M., Begara-Morales, J. C., Valderrama, R., Chaki, M., López-Jaramillo, J., et al. (2013). Inhibition of peroxisomal hydroxypyruvate reductase (HPR1) by tyrosine nitration. Biochim. Biophys. Acta 1830, 4981–4989. doi: 10.1016/j.bbagen.2013.07.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Corpas, F. J., de la Colina, C., Sánchez-Rasero, F., and del Río, L. A. (1997). A role of leaf peroxisomes in the catabolism of purines. J. Plant Physiol. 151, 246–250. doi: 10.1016/S0176-1617(97)80161-7
Corpas, F. J., Palma, J. M., Sandalio, L. M., Valderrama, R., Barroso, J. B., and del Río, L. A. (2008). Peroxisomal xanthine oxidoreductase: characterization of the enzyme from pea (Pisum sativum L.) leaves. J. Plant Physiol. 165, 1319–1330. doi: 10.1016/j.jplph.2008.04.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
del Río, L. A. (2011). Peroxisomes as a cellular source of reactive nitrogen species signal molecules. Arch. Biochem. Biophys. 506, 1–11. doi: 10.1016/j.abb.2010.10.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
del Río, L. A., Sandalio, L. M., Corpas, F. J., Palma, J. M., and Barroso, J. B. (2006). Reactive oxygen species and reactive nitrogen species in peroxisomes. Production, scavenging, and role in cell signaling. Plant Physiol. 141, 330–335. doi: 10.1104/pp.106.078204
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eilers, T., Schwarz, G., Brinkmann, H., Witt, C., Richter, T., Nieder, J., et al. (2001). Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase. A new player in plant sulfur metabolism. J. Biol. Chem. 276, 46989–46994. doi: 10.1074/jbc.M108078200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Giles, G. I., Tasker, K. M., and Jacob, C. (2001). Hypothesis: the role of reactive sulphur species in oxidative stress. Free Radic. Biol. Med. 31, 1279–1283. doi: 10.1016/S0891-5849(01)00710-9
Giles, G. I., Tasker, K. M., Collins, C., Giles, N. M., O'rourke, E., and Jacob, C. (2002). Reactive sulphur species: an in vitro investigation of the oxidation properties of disulphide S-oxides. Biochem. J. 364, 579–585. doi: 10.1042/BJ20011882
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jacob, C., and Anwar, A. (2008). The chemistry behind redox regulation with a focus on sulphur redox systems. Physiol. Plantarum 133, 469–480. doi: 10.1111/j.1399-3054.2008.01080.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jacob, C., Lancaster, J. R., and Giles, G. I. (2004). Reactive sulphur species in oxidative signal transduction. Biochem. Soc. Trans. 32(Pt 6), 1015–1017. doi: 10.1042/BST0321015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hänsch, R., Lang, C., Riebeseel, E., Lindigkeit, R., Gessler, A., Rennenberg, H., et al. (2006). Plant sulfite oxidase as novel producer of H2O2: combination of enzyme catalysis with a subsequent non-enzymatic reaction step. J. Biol. Chem. 281, 6884–6888. doi: 10.1074/jbc.M513054200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hänsch, R., and Mendel, R. R. (2005). Sulfite oxidation in plant peroxisomes. Photosynth. Res. 86, 337–343. doi: 10.1007/s11120-005-5221-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hawkesford, M. J., and De Kok, L. J. (2006). Managing sulfur metabolism in plants. Plant Cell Environ. 29, 382–395. doi: 10.1111/j.1365-3040.2005.01470.x
Hayashi, M., Toriyama, K., Kondo, M., Kato, A., Mano, S., De Bellis, L., et al. (2000). Functional transformation of plant peroxisomes. Cell Biochem. Biophys. 32, 295–304. doi: 10.1385/CBB:32:1-3:295
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hou, Z., Wang, L., Liu, J., Hou, L., and Liu, X. (2013). Hydrogen sulfide regulates ethylene-induced stomatal closure in Arabidopsis thaliana. J. Integr. Plant Biol. 55, 277–289. doi: 10.1111/jipb.12004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, J., Baker, A., Bartel, B., Linka, N., Mullen, R. T., Reumann, S., et al. (2012). Plant peroxisomes: biogenesis and function. Plant Cell 24, 2279–2303. doi: 10.1105/tpc.112.096586
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jiménez, A., Hernández, J. A., del Rio, L. A., and Sevilla, F. (1997). Evidence for the presence of the ascorbate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiol. 114, 275–284. doi: 10.1104/pp.114.1.275
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leustek, T., and Saito, K. (1999). Sulfate transport and assimilation in plants. Plant Physiol. 120, 637–644. doi: 10.1104/pp.120.3.637
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, J., Jia, H., Wang, J., Cao, Q., and Wen, Z. (2014). Hydrogen sulfide is involved in maintaining ion homeostasis via regulating plasma membrane Na+/H+ antiporter system in the hydrogen peroxide-dependent manner in salt-stress Arabidopsis thaliana root. Protoplasma 251, 899–912. doi: 10.1007/s00709-013-0592-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Müller, M., Zechmann, B., and Zellnig, G. (2004). Ultrastructural localization of glutathione in Cucurbita pepo plants. Protoplasma 223, 213–219. doi: 10.1007/s00709-003-0035-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Münchberg, U., Anwar, A., Mecklenburg, S., and Jacob, C. (2007). Polysulfides as biologically active ingredients of garlic. Org. Biomol. Chem. 5, 1505–1518. doi: 10.1039/b703832a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nakamura, T., Meyer, C., and Sano, H. (2002). Molecular cloning and characterization of plant genes encoding novel peroxisomal molybdoenzymes of the sulphite oxidase family. J. Exp. Bot. 53, 1833–1836. doi: 10.1093/jxb/erf042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nowak, K., Luniak, N., Witt, C., Wüstefeld, Y., Wachter, A., Mendel, R. R., et al. (2004). Peroxisomal localization of sulfite oxidase separates it from chloroplast-based sulfur assimilation. Plant Cell Physiol. 45, 1889–1894. doi: 10.1093/pcp/pch212
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Radi, R. (2013). Peroxynitrite, a stealthy biological oxidant. J. Biol. Chem. 288, 26464–26472. doi: 10.1074/jbc.R113.472936
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rausch, T., and Wachter, A. (2005). Sulfur metabolism: a versatile platform for launching defence operations. Trends Plant Sci. 10, 503–509. doi: 10.1016/j.tplants.2005.08.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reumann, S., Babujee, L., Ma, C., Wienkoop, S., Siemsen, T., Antonicelli, G. E., et al. (2007). Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19, 3170–3193. doi: 10.1105/tpc.107.050989
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Romero-Puertas, M. C., Corpas, F. J., Sandalio, L. M., Leterrier, M., Rodríguez-Serrano, M., del Río, L. A., et al. (2006). Glutathione reductase from pea leaves: response to abiotic stress and characterization of the peroxisomal isozyme. New Phytol. 170, 43–52. doi: 10.1111/j.1469-8137.2005.01643.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Signorelli, S., Corpas, F. J., Borsani, O., Barroso, J. B., and Monza, J. (2013). Water stress induces a differential and spatially distributed nitro-oxidative stress response in roots and leaves of Lotus japonicus. Plant Sci. 201–202, 137–146. doi: 10.1016/j.plantsci.2012.12.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tao, L., and English, A. M. (2004). Protein S-glutathiolation triggered by decomposed S-nitrosoglutathione. Biochemistry 43, 4028–4038. doi: 10.1021/bi035924o
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Valderrama, R., Corpas, F. J., Carreras, A., Fernández-Ocaña, A., Chaki, M., Luque, F., et al. (2007). Nitrosative stress in plants. FEBS Lett. 581, 453–461. doi: 10.1016/j.febslet.2007.01.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Veljović-Jovanović, S., Oniki, T., and Takahama, U. (1998). Detection of monodehydroascorbic acid radical in sulfite-treated leaves and mechanism of its formation. Plant Cell Physiol. 39, 1203–1208. doi: 10.1093/oxfordjournals.pcp.a029321
Keywords: nitric oxide, nitrosative stress, oxidative stress, peroxisomes, reactive nitrogen species
Citation: Corpas FJ and Barroso JB (2015) Reactive sulfur species (RSS): possible new players in the oxidative metabolism of plant peroxisomes. Front. Plant Sci. 6:116. doi: 10.3389/fpls.2015.00116
Received: 12 December 2014; Paper pending published: 27 January 2015;
Accepted: 11 February 2015; Published online: 25 February 2015.
Edited by:
Ken Yokawa, University of Bonn, GermanyReviewed by:
Vasileios Fotopoulos, Cyprus University of Technology, CyprusCopyright © 2015 Corpas and Barroso. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence:amF2aWVyLmNvcnBhc0BlZXouY3NpYy5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.