- 1Faculty of Science, Hokkaido University, Sapporo, Japan
- 2Laboratory of Plant Molecular Physiology, Graduate School of Agricultural and Life Sciences, University of Tokyo, Tokyo, Japan
- 3Department of Advanced Bioscience, Faculty of Agriculture, Kinki University, Nara, Japan
- 4Institute of Plant Science and Resources, Okayama University, Kurashiki, Japan
- 5PRESTO, Japan Science and Technology Agency, Kawaguchi, Japan
Transposable elements (TEs) are key elements that facilitate genome evolution of the host organism. A number of studies have assessed the functions of TEs, which change gene expression in the host genome. Activation of TEs is controlled by epigenetic modifications such as DNA methylation and histone modifications. Several recent studies have reported that TEs can also be activated by biotic or abiotic stress in some plants. We focused on a Ty1/copia retrotransposon, ONSEN, that is activated by heat stress (HS) in Arabidopsis. We found that transcriptional activation of ONSEN was regulated by a small interfering RNA (siRNA)-related pathway, and the activation could also be induced by oxidative stress. Mutants deficient in siRNA biogenesis that were exposed to HS at the initial stages of vegetative growth showed transgenerational transposition. The transposition was also detected in the progeny, which originated from tissue that had differentiated after exposure to the HS. The results indicated that in some undifferentiated cells, transpositional activity could be maintained quite long after exposure to the HS.
Introduction
Plant genomes contain a large number of transposable elements (TEs) (Feschotte et al., 2002). In particular, retrotransposons that use RNA-mediated amplification constitute a large part of the plant genome (Kumar and Bennetzen, 1999). Retrotransposons are classified into two major subclasses on the basis of their sequence and structural similarity. The first class is long terminal repeat (LTR) retrotransposons, which contain LTRs on both sides. The second class is non-LTR retrotransposons, which include long interspersed nuclear elements and short interspersed nuclear elements (Schmidt, 1999). LTR retrotransposons are further divided into two families according to the order of the coding genes. Ty3/gypsy retrotransposons are enriched in intergenic regions that include centromeric heterochromatin in some plants (Jiang et al., 2003). The second family, Ty1/copia retrotransposons, are conserved in the euchromatic regions of some plant species (Kumar, 1996).
Although all plants contain various TEs, most of these are transcriptionally silenced through epigenetic regulation (Lippman et al., 2003). DNA methylation is one of the major regulators of transposon expression. TEs are activated in DNA hypomethylation mutants (Miura et al., 2001; Kato et al., 2003; Mirouze et al., 2009; Tsukahara et al., 2009), which represents a defense mechanism against ectopic transposon activation. One of the best-studied mechanisms for the regulation of TEs is RNA-directed DNA methylation (RdDM), which regulates TEs through small interfering RNA (siRNA)-mediated DNA methylation (Wierzbicki et al., 2008; Gao et al., 2010). Higher plants possess specific DNA-dependent RNA polymerases known as RNA polymerase IV (PolIV) and RNA polymerase V (PolV; Kanno et al., 2005; Onodera et al., 2005; Pontier et al., 2005). PolIV produces initial RNA transcripts for RNA silencing and PolV transcribes a messenger RNA for siRNA-induced DNA methylation on the target site.
In Arabidopsis, an ectopic transcript produced from TEs is transcribed to double-stranded RNA by RNA-dependent RNA polymerase 2 (RDR2) and subsequently processed in 24–26 nt siRNAs by DICER-LIKE 3 (DCL3; Zhang et al., 2007b; Mosher et al., 2008). The siRNAs bind to an RNA-induced silencing complex containing ARGONAUTE 4 (AGO4) that interacts with PolV to recruit the DNA methyltransferase DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2), leading to de novo DNA methylation of the target TEs (Cao and Jacobsen, 2002; Matzke and Birchler, 2005).
It remains unknown when the TEs are activated or what triggers TE activation in nature. The answer may lie in accumulated evidence that TEs are activated by environmental stress. Some TEs were activated by abiotic stress in combination with DNA demethylation. For example, in corn, cold stress induced hypomethylation and activated the ZmMI1 element that was similar at the LTR region of the putative retroelement (Steward et al., 2002).
In other cases, the activation of LTR retrotransposons is independent of DNA methylation and is under the control of cis-regulatory sequences in the LTRs. The best-characterized examples are the Tnt1 and Tto1 retrotransposons in tobacco. Tnt1 and Tto1 are activated by biotic stresses, such as tissue culturing, wounding, and pathogen infections, and by abiotic stresses, including salicylic acid and jasmonate (Peschke et al., 1987; Hirochika, 1993; Pouteau et al., 1994; Takeda et al., 1998). The stress-induced activation is controlled by cis-acting motifs in the U3 region of the LTR sequence that are similar to those of plant defense genes (Grandbastien et al., 1997; Mhiri et al., 1997; Vernhettes et al., 1997; Takeda et al., 1999).
Recently, a Ty1/copia retrotransposon named ONSEN was found to be activated by heat stress (HS) in Arabidopsis (Ito et al., 2011). The LTR of ONSEN contains a sequence that is recognized by the plant’s heat response transcription factor, HsfA2 (Cavrak et al., 2014). HsfA2 has a conserved N-terminal DNA-binding domain that binds a heat response element (HRE; Schramm et al., 2006). An electrophoretic mobility shift assay demonstrated that HsfA2 bound to an HRE in the ONSEN LTR (Cavrak et al., 2014), indicating that HsfA2 was directly involved in transcriptional activation. ONSEN is more strongly activated in a mutant that was deficient in a pathway for siRNA biogenesis than in the wild-type plant (WT), indicating that an siRNA-related pathway modulates transcriptional activation of ONSEN (Ito et al., 2011). Although heat-induced expression was enhanced by a mutant of DNA methyltransferase (Cavrak et al., 2014), ONSEN was not expressed in a hypomethylation mutant or a mutant lacking RdDM component without HS (Ito et al., 2011). This finding indicates that the initiation of the transcriptional activation is independent of DNA methylation.
In addition to transcriptional regulation, TEs may be controlled by transpositional processes in the host plant. A high frequency of new ONSEN insertions was observed in the progeny of stressed plants that were deficient in siRNA biogenesis (Ito et al., 2011). ONSEN was also expressed in the WT and in other epigenetic mutants subjected to HS; however, transgenerational transposition was not observed in the progeny (Ito et al., 2011). The results suggested that siRNA-mediated pathways regulated transgenerational transposition. Although the mechanism of ONSEN activation has been studied at the transcriptional level (Ito et al., 2011), the precise mechanism of transpositional regulation remains unknown. Here, we provide insight into siRNA-mediated regulation and the initiation of transgenerational transposition of ONSEN.
Materials and Methods
Plant Material and Growing Conditions
The Arabidopsis thaliana plants used in the experiments included WT plants, nrpd1 mutants (Herr et al., 2005), and transgenic plants that possessed a full-length LTR (genome position; 4212570–4213146) of ONSEN (AT5G13205) fused with a GFP gene. The plants were grown on Murashige and Skoog (MS) plates under continuous light at 21°C. For the analysis of high light and oxidative stress, WT, nrpd1, and hsfA2 T-DNA insertion mutant (Nishizawa et al., 2006) plants were grown on MS at 25°C under continuous light (irradiance 100 μmol m-2 s-1). In order to analyze their reactions to short days and long days, plants ware grown on MS in pots for 7 days with continuous light at 21°C and moved to a long-day chamber (16 h light and 8 h dark) or a short-day chamber (8 h light and 16 h dark). All WT and mutant plants were A. thaliana ecotype Columbia.
Stress Treatments
For HS treatment, 7-day-old seedlings were subjected to a temperature shift from 21°C to 37°C for 24 h except some experiments. After undergoing heat treatment, plants were transplanted to soil and allowed to grow at 21°C. For the high light and oxidative stress treatments, 7-day-old WT, nrpd1, and hsfa2 plants were exposed to high light at 800 μmol m-2 s-1 at 25°C or sprayed with 50 μM methylviologen.
Southern Blot Analysis
Arabidopsis genomic DNA was isolated using a Nucleon PhytoPure DNA extraction kit (GE Healthcare Life Science). Blotting of genomic DNA was performed as described (Miura et al., 2004). Hybridization signals were detected using a radio-labeled ONSEN-specific probe (Table 1) that was generated with the Megaprime DNA Labeling System (GE Healthcare Life Science) in a high-SDS hybridization buffer (Church and Gilbert, 1984).
Real-Time PCR
In order to analyze the TE expression resulting from heat treatment, total RNA was extracted from seedlings or leaves using TRI Reagent (Sigma T9424), according to the supplier’s recommendations. The five individual plants were pooled prior to RNA extraction. Around 3 to 5 μg of total RNA was treated with RQ1 RNase-free DNase (Promega) and reverse-transcribed using the ReverTraAce qPCR RT Kit (TOYOBO FSQ-101) with an oligo dT primer. In order to quantify the amount of ONSEN DNA, genomic DNA was extracted from seedlings or leaves using the DNeasy Plant Mini Kit (QIAGEN 69104), according to the supplier’s recommendations. Real-time PCR was performed using the Applied Biosystems 7300 Real Time PCR System with the THUNDERBIRD SYBR qPCR Mix (TOYOBO QPS-201). Three biological repetitions were performed and SD was determined (Figure 2). Similarly, three technical repetitions were performed and SD was determined (Figures 4 and 10). Following the high light and oxidative stress experiments, RNA was extracted using the Plant RNA Reagent (Life Technologies). A volume containing 2 μg of total RNA was subjected to RNase-free DNaseI (Thermo Scientific) and reverse-transcribed using the PrimeScript 1st strand cDNA Synthesis Kit (TaKaRa 6210A). Quantitative real-time PCR was performed with a LightCycler 96 System (Roche), using the FastStart Universal SYBR Green Master (ROX; Roche). Quantities of DNA were determined from a standard curve and were normalized to the amount of 18s rDNA. At least three biological repetitions were performed and SE were determined.
Laser Capture Microdissection and RNA Extraction
Arabidopsis seedlings were fixed in Farmer’s fixative (3:1 ethanol:acetic acid) overnight at 4°C. Subsequently, fixation, dehydration, and paraffin infiltration were performed using a microwave processor (H2850 EBS). Paraffin-embedded sections were cut to a thickness of 12 μm and mounted on a PEN membrane glass slide (Applied Biosystems LCM0522). To remove the paraffin, slides were immersed in Histo-Clear II (National Diagnostics HS-202; 2 × 5 min) and then air-dried at room temperature. Laser capture microdissection was performed using the ArcturusXT LCM system (Applied Biosystems). Selected areas were captured by an infrared laser onto Arcturus CapSure Macro LCM Cap (LCM0211 Applied Biosystems) and subsequently cut with a UV laser. Total RNA was extracted using the PicoPure RNA Isolation Kit (Applied Biosystems KIT0204) and quantified using the Agilent RNA 6000 Pico Kit (Agilent 5067-1513).
Results
Heat Activation of Transgene
The heat-activation of ONSEN is controlled by the promoter in the LTRs that involves a cis-regulatory sequence for HREs (Cavrak et al., 2014). HREs are bound by the heat shock transcription factor (HSF) that controls heat-induced genes (Nover et al., 2001). We produced a transgenic Arabidopsis that possessed an intact LTR of ONSEN (AT5G13205) fused with a gene for green fluorescent protein (GFP) (Figure 1A). A transgenic plant with a single-copy insert was analyzed for expression of the reporter gene. The insertion was mapped on an intergenic region between AT1G26850 and AT1G26860 on chromosome 1 (Figure 1B). The GFP signal was analyzed immediately after a group of 10 1-week-old seedlings was subjected to heat treatment and again 3 days after the treatment (Figure 2A). The GFP signals were detected throughout the plants that were subjected to the heat treatment, but gradually decayed and were below the detection limit at 3 days after heat treatment (Figure 2B).
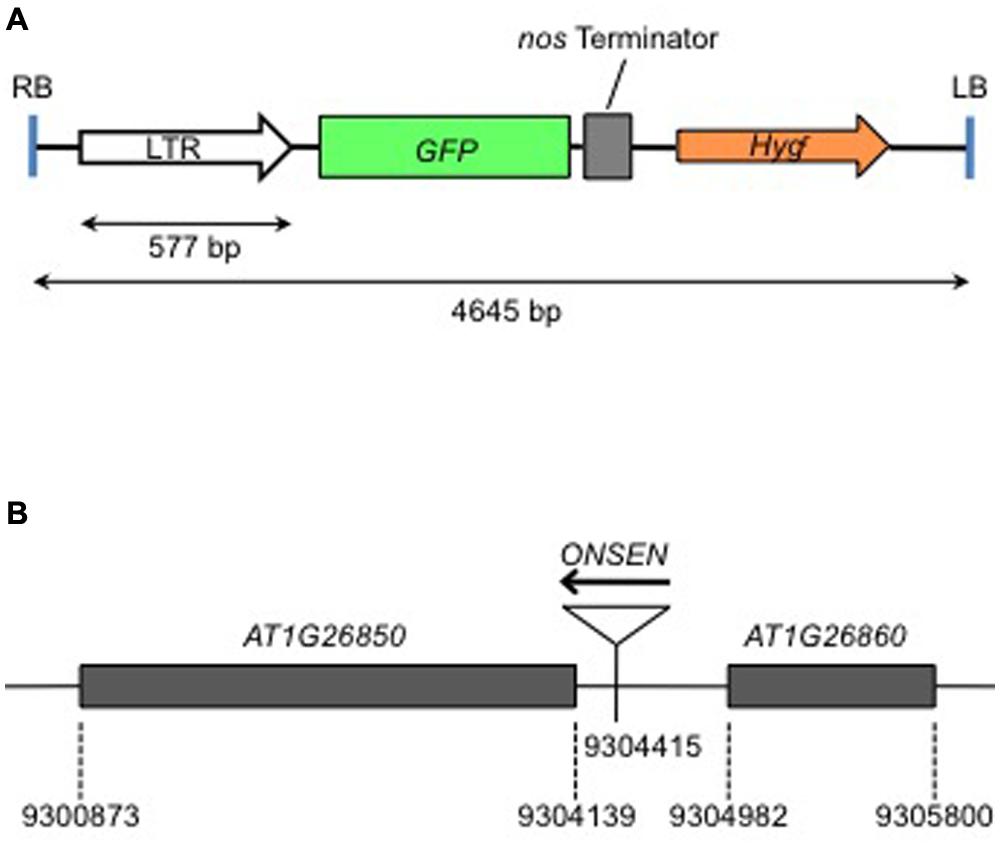
FIGURE 1. Structure of the gene construct used to produce transgenic plants, and the insertion position of the transgene in the genome. (A) The structure of the DNA sequence between the left border (LB) and the right border (RB) of the T-DNA. Intact long terminal repeat (LTR) of ONSEN (AT5G13205) was fused with the green fluorescent protein (GFP) gene. (B) The transgene was inserted in the intergenic region between At1g26850 and At1g26860, in the same direction. Numerals indicate the nucleotide number according to the AGI map.
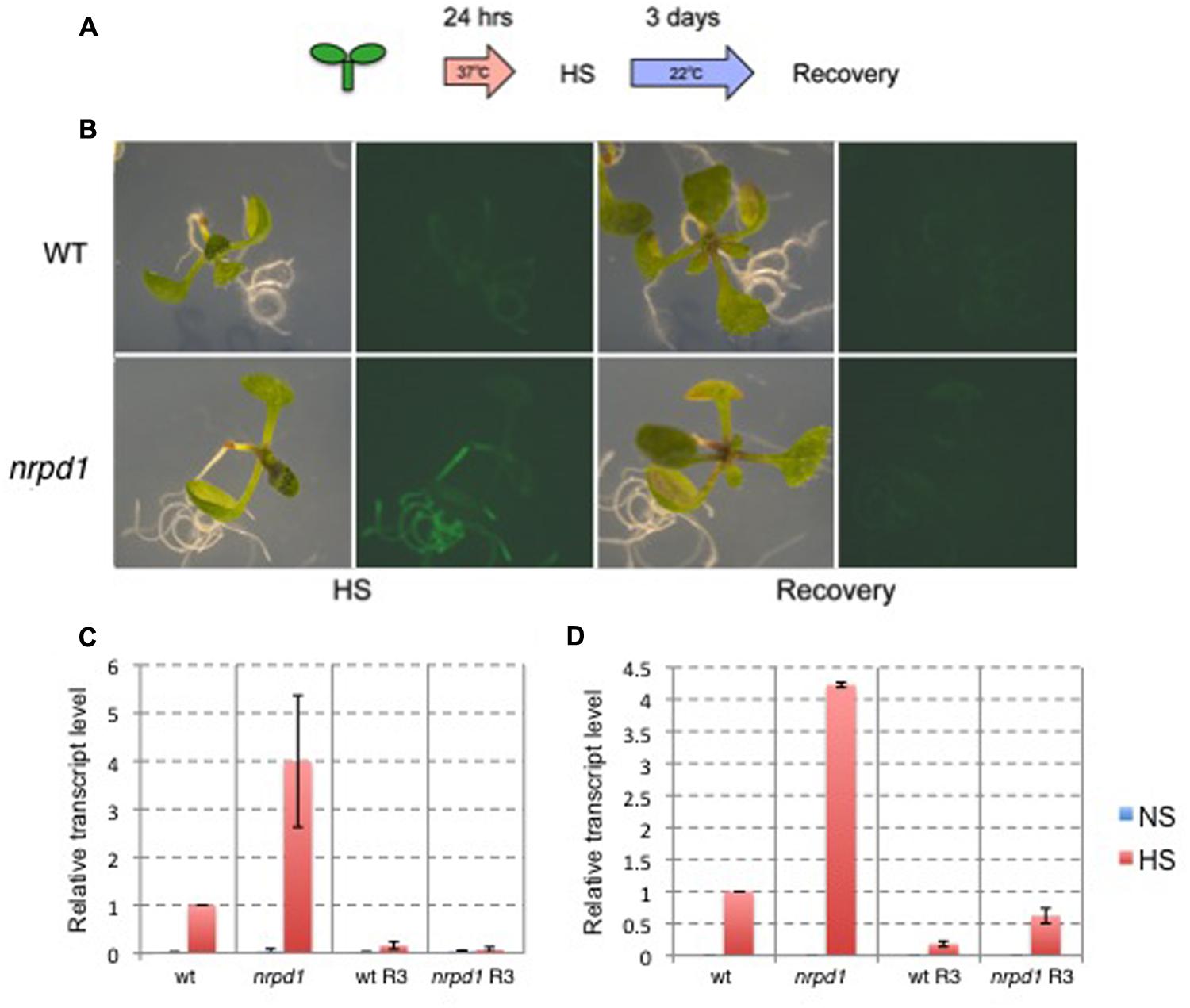
FIGURE 2. Expression analysis of GFP driven by the ONSEN promoter. (A) Scheme of the heat stress (HS) experiment. (B) Photographs of bright field and GFP fluorescence images. Wild-type plants (WT) and nrpd1 plants (nrpd1) were observed immediately after HS and after the recovery phase (Recovery). (C) Relative transcription level of GFP. WT and nrpd1 plants (nrpd1) were analyzed immediately after HS and after the recovery phase (R3). (D) Relative transcription level of endogenous ONSEN. The same samples as those used in c were used. NS, non-stress control samples. Error bars represent the mean ± SEM, n = 3; values are relative to heat-stressed wild-type (WT HS).
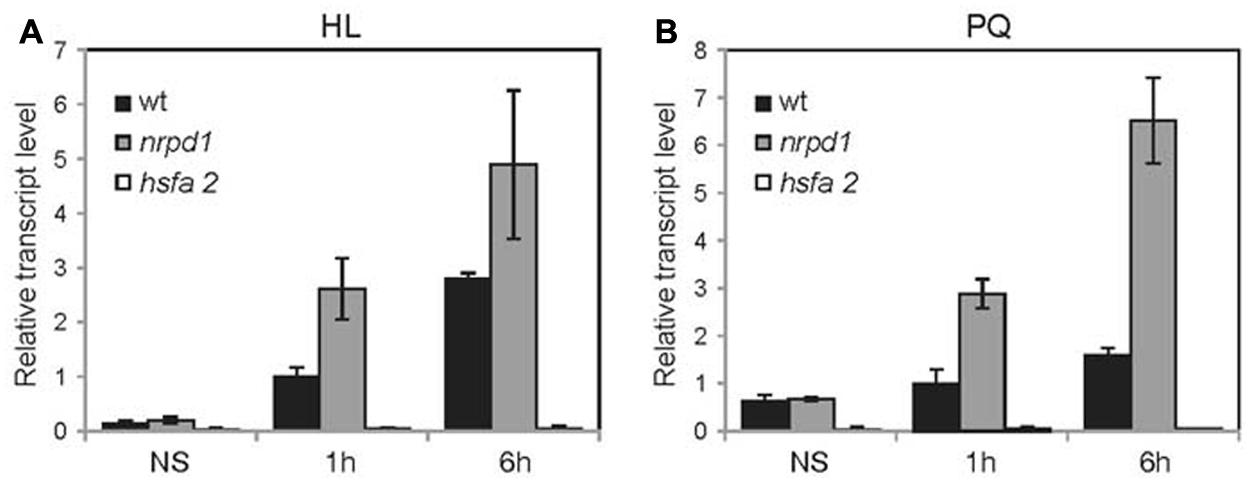
FIGURE 3. Activation of ONSEN by high light stress and oxidative stress. Plants before stress treatment, after continuous stress treatment for 1 h and after continuous stress treatment for 6 h were analyzed. (A) Relative transcription level of ONSEN immediately after high light stress (HL). (B) Relative transcription level of ONSEN immediately after Paraquat treatment (PQ). Error bars represent SDs (n = 3).
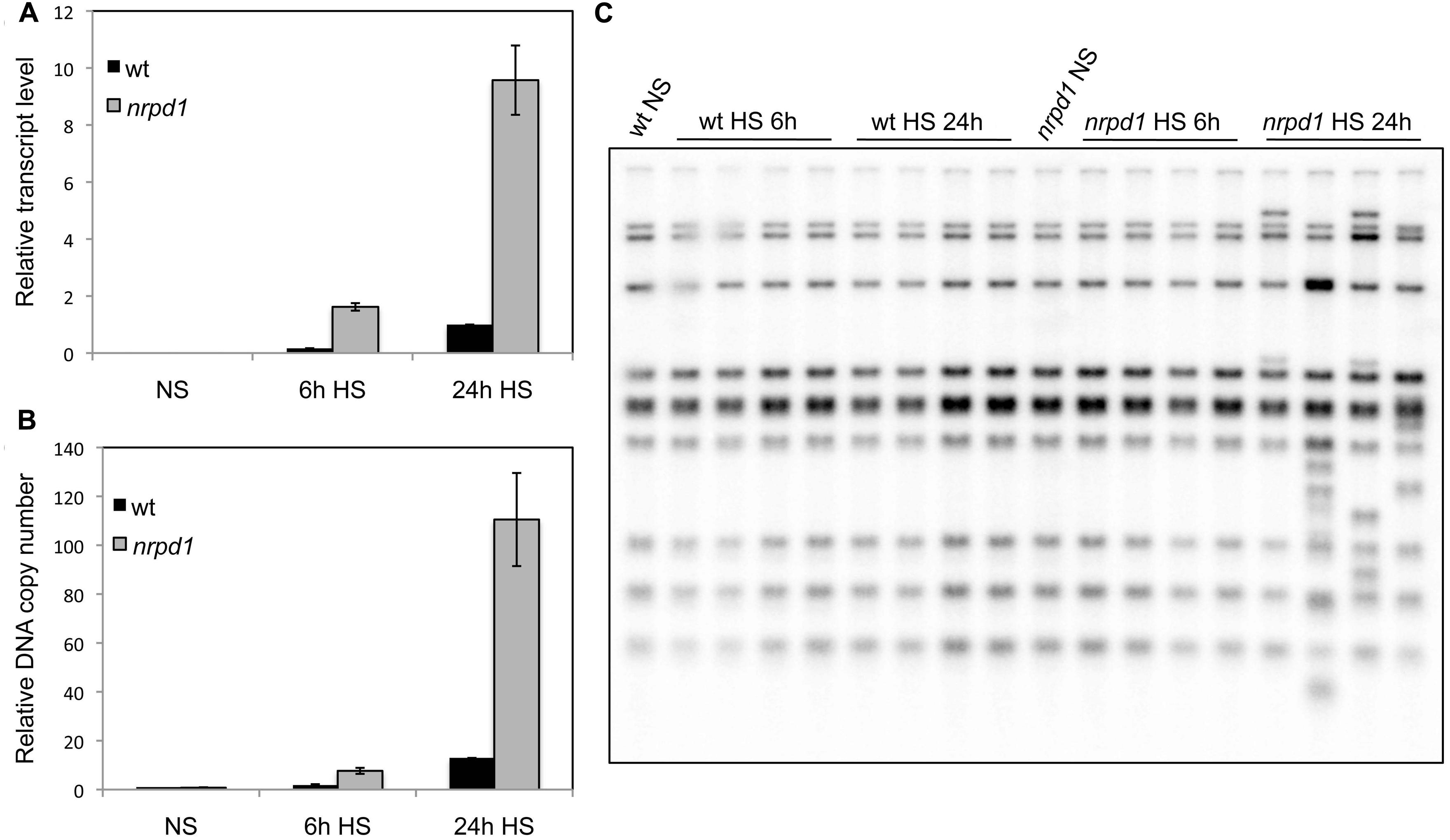
FIGURE 4. The quantitative analyses of transcripts, extrachromosomal DNAs, and newly transposed copies in progeny. WT and nrpd1 plants were exposed to HS for 6 h and for 24 h at 7 days after germination. NS, non-stressed samples. (A) Relative transcription level of ONSEN. Error bar represents the mean ± SEM, n = 3, values relative to 24 h heat-stressed WT. (B) Relative number of copies of extrachromosomal DNA of ONSEN. Error bar represents the mean ± SEM, n = 3; values are relative to the NS WT seedlings. (C) Southern blot analysis of ONSEN in progeny plants.
To understand the role of siRNA for transcriptional silencing of the transgene, the transgenic plant was crossed with nrpd1, a mutant that was deficient in siRNA biogenesis. The resulting transgenic line involved an LTR fused with GFP in an nrpd1 background. The intensity of the GFP signal was stronger in the nrpd1 transgenic line than in the WT line; however, the GFP signal fell below the detection level by 3 days after the heat treatment (Figure 2B). The intensity of the GFP signals was consistent with the transcription level of GFP (Figure 2C) and the endogenous ONSEN (Figure 2D). These results indicated that the transcriptional regulation of the transgene was controlled by an siRNA-related pathway that controls endogenous ONSEN copies.
ONSEN Activation by Stress
The LTR promoter of ONSEN contains a cis-regulatory sequence for HREs that binds to a heat-induced transcriptional factor, HsfA2 (Cavrak et al., 2014). HsfA2 was reported to play an important role in the response to environmental stresses, including high light stress (Nishizawa et al., 2006). To analyze the effect of high light stress on ONSEN, young seedlings were exposed to high light (800 μmol photons m-2s-1) for 6 h. The ONSEN mRNA level started to increase at 1 h and continued to gradually increase to the 6-h point, indicating that ONSEN was activated by high light stress (Figure 3A). The expression level of ONSEN was considerably higher in the nrpd1 mutant than in the WT (Figure 3A), suggesting that the transcriptional activation was controlled by siRNA-mediated mechanisms.
To analyze the response of high light stress on ONSEN more precisely, we applied N,N′-dimethyl-4,4′-bipyridinium dichloride (Paraquat) to young seedlings. Paraquat produces hydrogen peroxide, which causes oxidative stress to the plant, similar to that caused by high light stress. Approximately 50 7-day-old seedlings were exposed to 50 μM Paraquat for 6 h. ONSEN was activated and at 6 h after the treatment its mRNA levels were increased (Figure 3B). In the nrpd1 mutant, ONSEN was highly activated in response to oxidative stress (Figure 3B). Transcriptional activation was not detected in the hsfa2 mutant that was subjected to neither high light stress nor to oxidative stress (Figures 3A,B). The results indicated that ONSEN was activated by oxidative stress via the HsfA2 transcriptional factor and that the activation was regulated by an siRNA-mediated pathway.
The Quantity of Active ONSEN Affected Transgenerational Transposition
To verify the effect of transcriptional activity and transposition frequency in nrpd1, 1-week-old seedlings were exposed to 37°C for 6 h and 24 h, respectively. Quantitative analysis showed that after the 24-h HS, the transcription level had increased by approximately six times in both WT plants and nrpd1 (Figure 4A). In addition, the number of synthesized extrachromosomal DNA copies was six times as high in the WT and 14 times as high in nrpd1 plants (Figure 4B) as in these same seedling groups exposed to the 6-h treatment. New insertions of ONSEN were detected in the progeny of nrpd1 that were subjected to HS for 24 h; however, they were not detected in the progeny of nrpd1 that were subjected to HS for 6 h. A transposition was not detected in the progeny of WT plants subjected to HS (Figure 4C). These results suggested that a high amount of active ONSEN was important for transgenerational transposition to occur in the mutant that was impaired in the biogenesis of siRNAs.
Heat Stress-Induced Transgenerational Transposition in Seedlings
To investigate how the timing of the HS response in young seedlings affected transgenerational transposition, seedlings of the nrpd1 and WT plants, at ages ranging from 1 to 6 days after germination, were exposed to 37°C for 24 h (Figure 5A). Transgenerational transposition was analyzed in the progenies of heat-stressed plants. The new copies were not detected in the progeny of WT plants (Figure 5B), but they were detected in the progeny of stressed nrpd1 plants (Figure 5C). Interestingly, transgenerational transposition was observed in seedlings whose parents were stressed at a very early developmental stage, as young as 1 day old (Figure 5C).
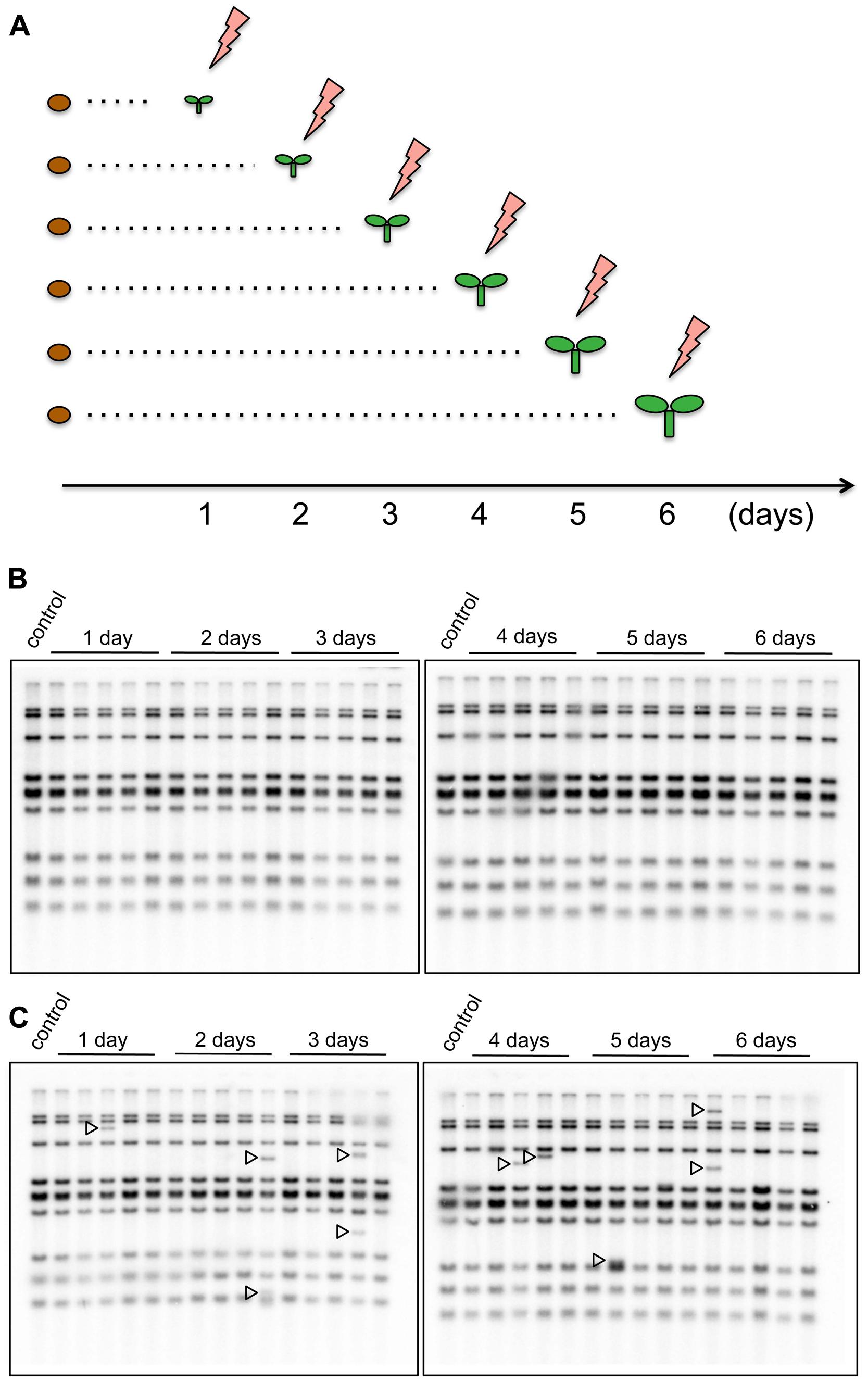
FIGURE 5. Transgenerational transposition of ONSEN in young seedling. (A) Scheme of the HS experiment. On each day after germination, seedlings were exposed to HS for 24 h. (B,C) Southern blot analysis of ONSEN using the progeny of WT (B) and nrpd1 (C) plants that were exposed to HS, as indicated in (A). Durations of HS are indicated over the lanes. NS plants were used as controls. Arrowheads indicate the transposed copies of ONSEN.
Transgenerational Transposition was Independent of Flowering Time
Next, we analyzed whether transposition frequency was affected by flowering time. In Arabidopsis, flowering is controlled by photoperiod and is promoted by long day lengths. Under long-day conditions, bolting was observed 30 days after germination in both WT and nrpd1 mutant plants. Under short-day conditions, bolting started approximately 50 days and 40 days after germination in WT and nrpd1 plant, respectively (Figure 6).
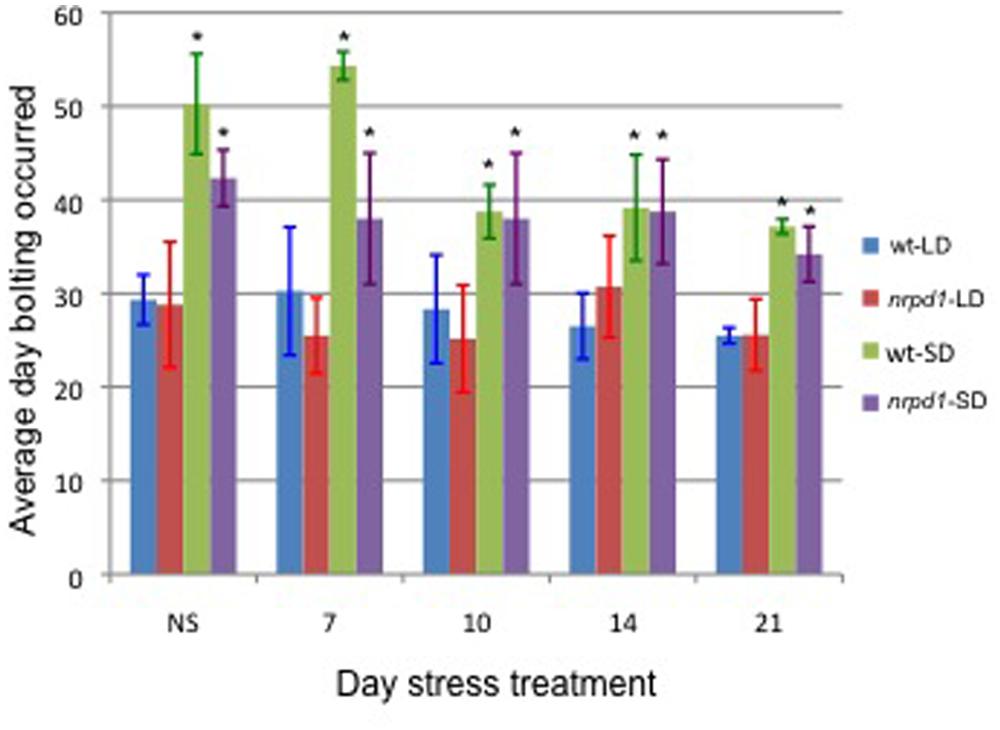
FIGURE 6. The effect of daylight conditions on flowering time. The average of the day on which bolting occurred was measured in 3 to 8 plants that grew under long-day (LD) or short-day (SD) conditions. Plants were exposed to HS for 24 h at 7, 10, 14, and 21 days after germination. The WT and nrpd1 plants grew under LD and SD were compared, respectively, using a t-test. An asterisk indicates significantly different, p < 0.05. NS control plants.
Seedlings that were growing under either long-day or short-day conditions were exposed to heat treatment 7, 10, 14, and 21 days after germination. The flowering time of the stressed nrpd1 was slightly affected by the treatment (Figure 6). No new copy of ONSEN was detected in the WT progeny, which were growing under conditions that were neither long day nor short day (Figure 7A). Transgenerational transposition was observed in the progeny of nrpd1 plants that were grown under either long-day or short-day conditions (Figure 7B). These results suggested that transposition frequency is not affected by flowering time and that the new insertions were transmitted into the reproductive tissue even when 3-week-old nrpd1 plants were exposed to HS.
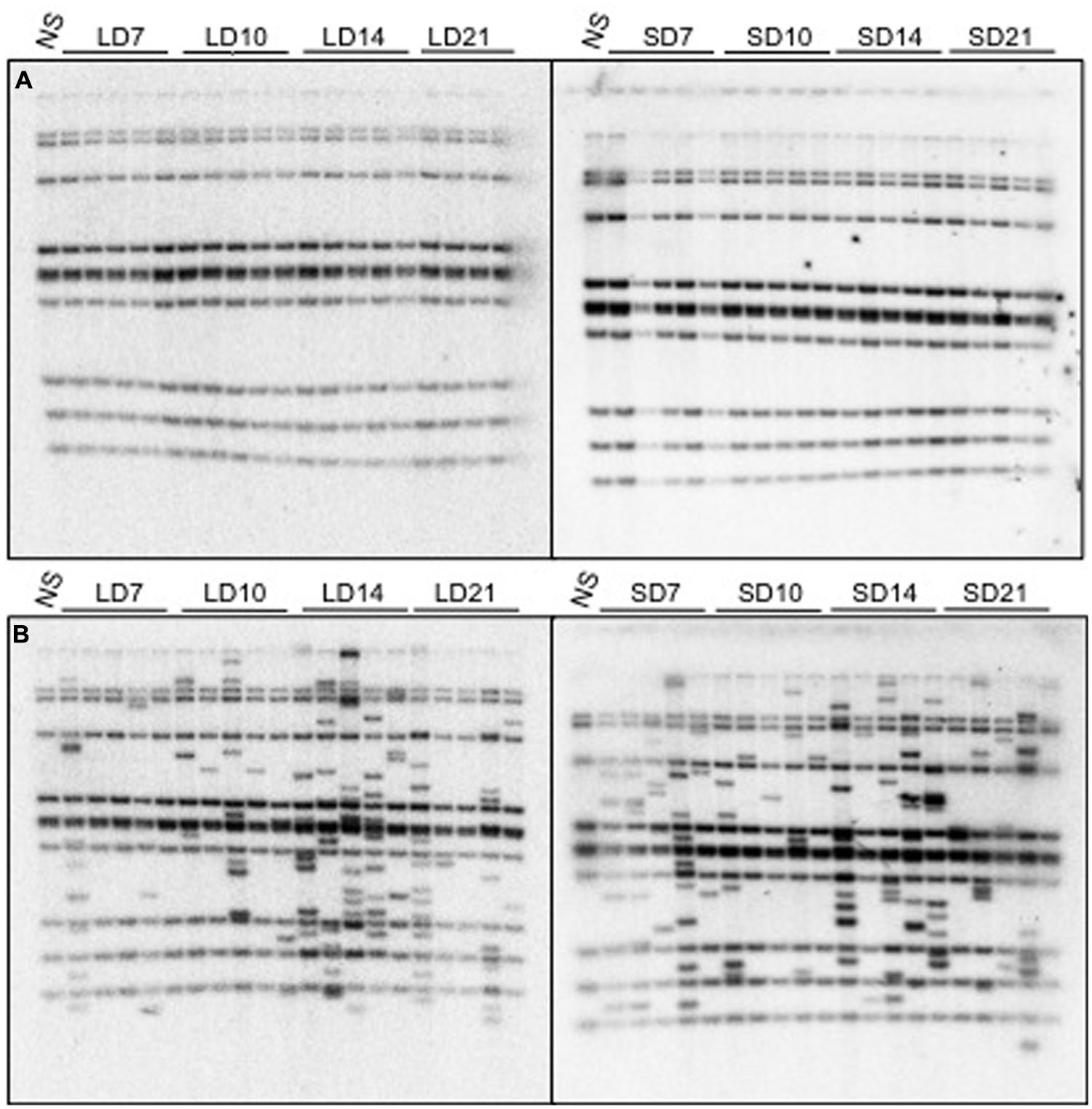
FIGURE 7. Transgenerational transpositions in the WT (A) and nrpd1 (B) plants. Data for plants grown under long-day (LD) and short-day (SD) conditions are shown in the left panels and right panels, respectively. 7, 10, 14, and 21 denote plants that were exposed to HS for 24 h at 7, 10, 14, and 21 days after germination, respectively. NS, non-stressed plants.
Stress Memory Could Be Regulated in Undifferentiated Tissues During Development
To better understand the regulation of ONSEN, we observed new insertions of ONSEN in the siblings of a single flower on a single branch of a heat-stressed nrpd1 plant. The pattern of new insertions was similar among the progeny within a single flower, although it differed among flowers on the same branch (Figures 8A,B). To understand the maintenance of heat activation of ONSEN during branch development, we analyzed transgenerational transposition in the secondarily produced branches. Surprisingly, new insertions were detected in an nrpd1 progeny originating from the side shoots that was produced after cutting the initial shoot that was subjected to HS (Figure 9). This result indicated that the transposition activity could be maintained for a long period of time after HS.
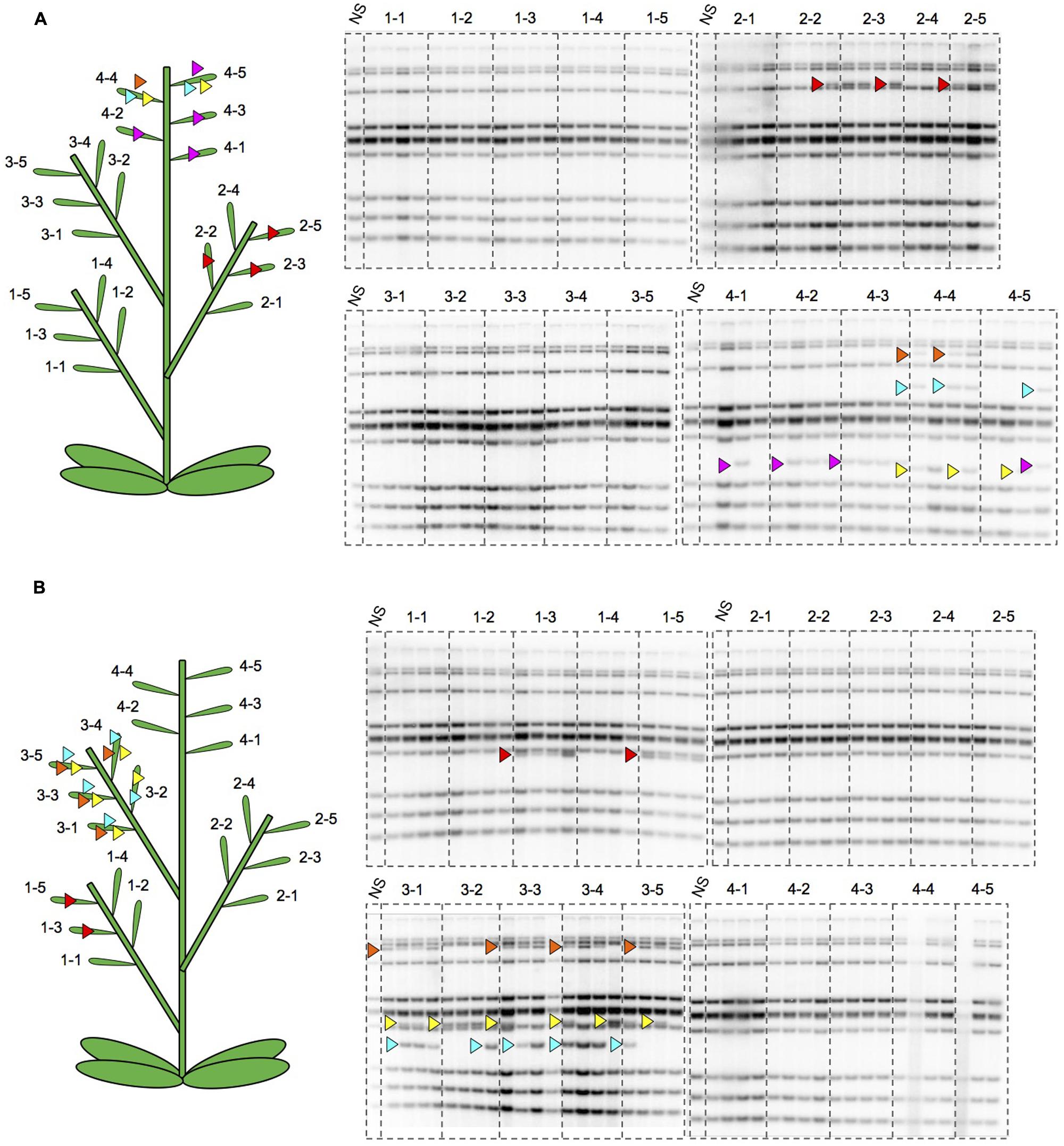
FIGURE 8. Southern hybridization of ONSEN using the progeny of heat-stressed plants. (A) DNAs were extracted from the progeny of one nrpd1 plant. Numbers over lanes indicate the seed pods having seeds to grow progeny. Four progeny from the same seed pod were analyzed. The location of each seed pod on the parent plant is shown on the illustrated plant. Arrowheads indicate the new copies of ONSEN and the same color of arrowhead denotes that they transposed to the same loci. (B) DNAs were extracted from the progeny of another nrpd1 plant. Symbols are the same as those used in (A).
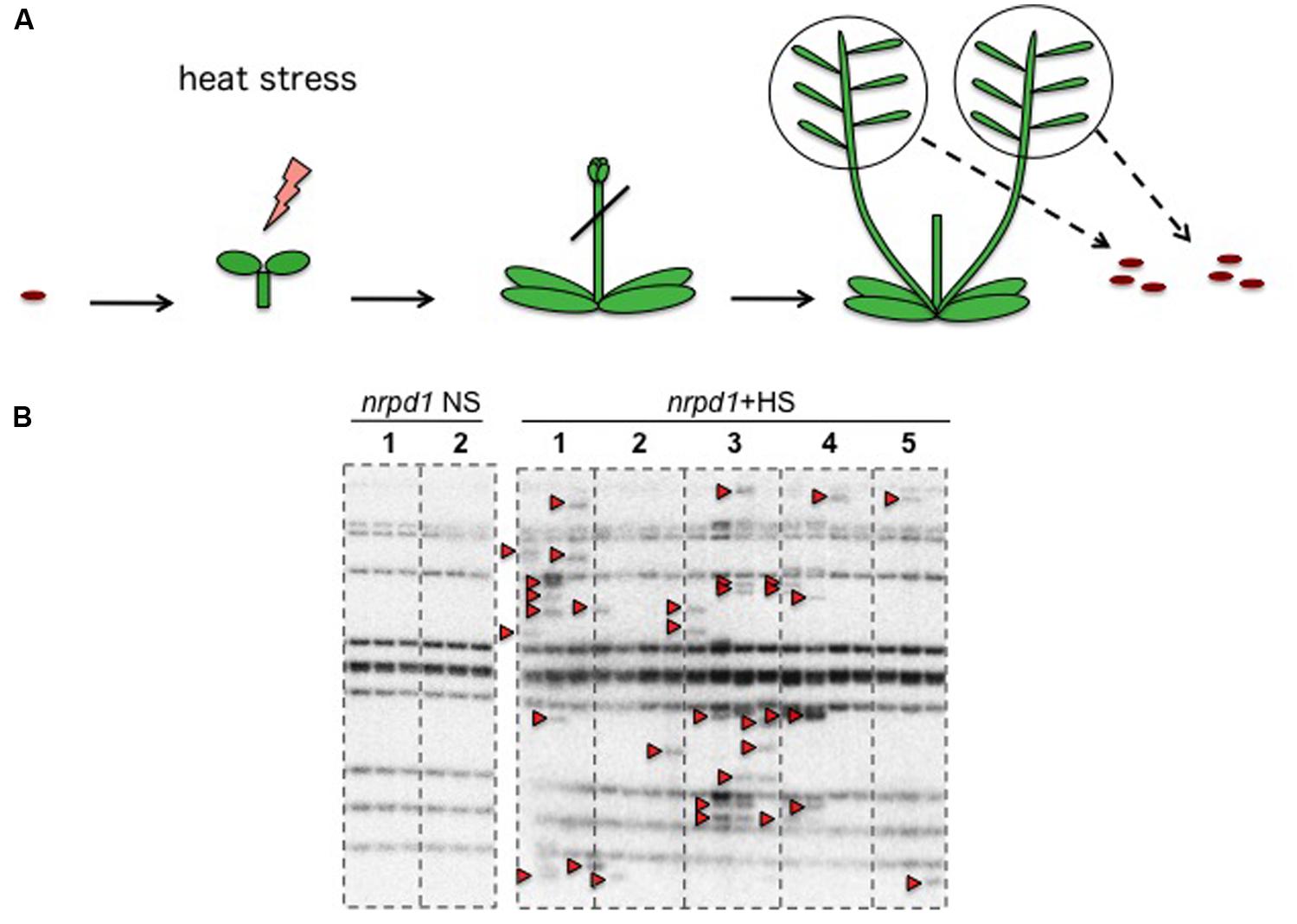
FIGURE 9. Transgenerational transposition in late-differentiated branches. (A) A scheme to obtain seeds for progeny. Parent nrpd1 plants were exposed to HS for 24 h at 7 days after germination, then the primary flower stalk was cut immediately after bolting, and seeds were harvested from secondary branches. (B) Southern hybridization of ONSEN. DNAs of progeny from two NS parents and five HS parents were analyzed. Numbers over the lanes indicate that DNAs were extracted from the progeny of different parents. Arrowheads indicate the new copies of ONSEN.
Regulation of ONSEN by an siRNA-Related Mechanism in Undifferentiated Tissues
To understand the mechanism of the transgenerational transposition of ONSEN, we analyzed the transcriptional activity of ONSEN in the shoot apex. The shoot apex, including the apical meristem, was fixed in paraffin (Figure 10A) and isolated by laser capture microdissection (Figure 10B). In nrpd1 plants, ONSEN was highly activated in the shoot apex after heat treatment; however, the activity level returned to the baseline value 5 days after HS. In the WT plant, ONSEN was expressed at relatively low levels even after heat treatment (Figure 10C). These results indicated that an siRNA-related pathway regulated transcriptional activation of ONSEN in the shoot apex.
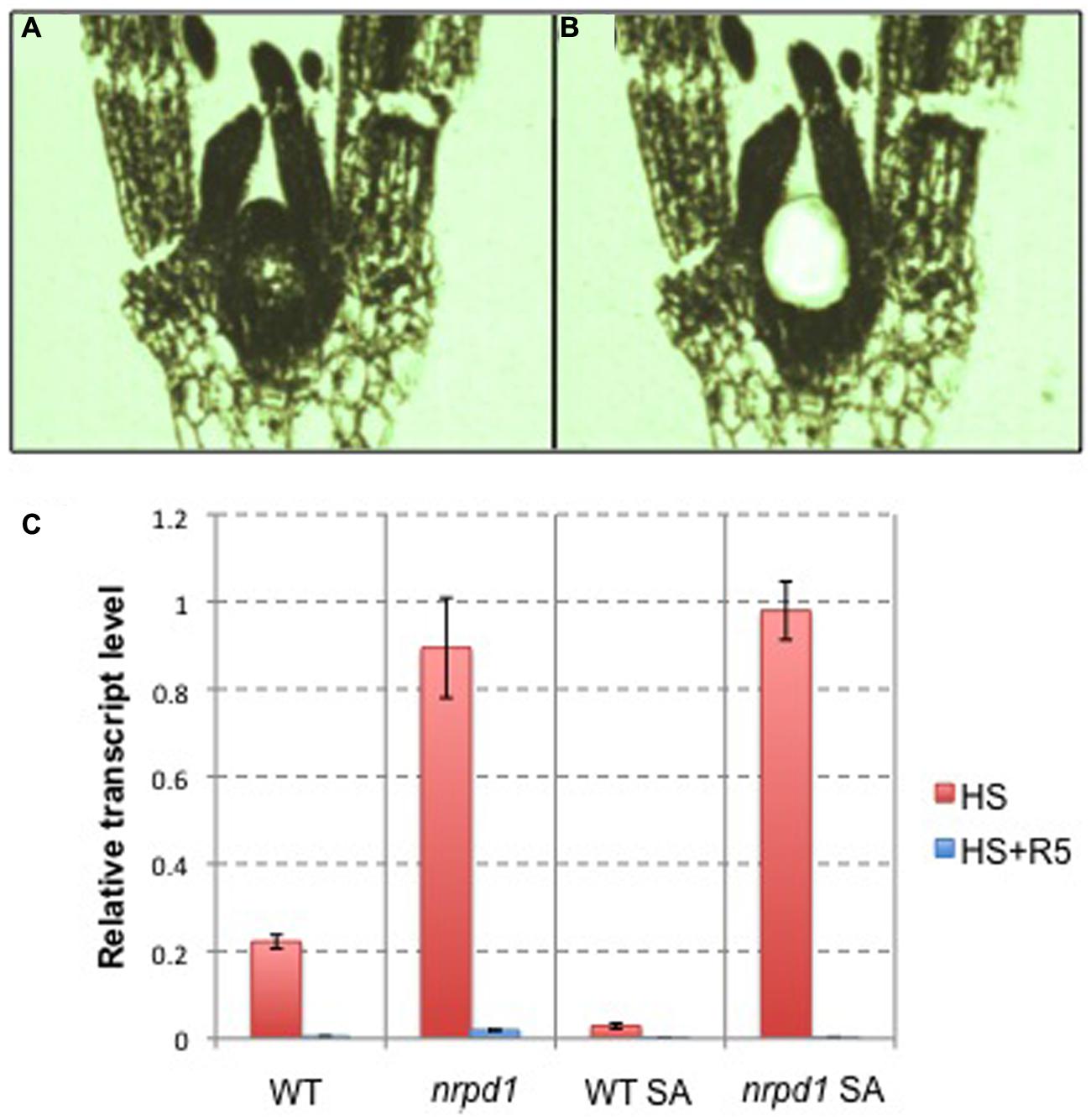
FIGURE 10. Tissue-specific expression of ONSEN. (A) A paraffin section of a young seedling. (B) A paraffin section of a young seedling after the shoot apex was isolated by laser capture microdissection. (C) Relative transcription level of ONSEN in young leaf tissue and the shoot apex immediately after being subjected to HS and 5 days after the heat-stressed (HS + R5). WT: hypocotyl tissue in WT plants. nrpd1: hypocotyl tissue in nrpd1 mutant plants. WT SA: the shoot apex in WT plants. nrpd1 SA: the shoot apex in nrpd1 mutant plants. Error bars represent the mean ± SEM, n = 3; values are relative to heat-stressed WT.
Discussion
Transposons are abundant in plant genomes and show great diversity among species. Large numbers of transposon copies may occur in a host genome as a result of activation of TEs by an external stimulus or environmental stress (Wessler, 1996; Grandbastien, 1998; Capy et al., 2000). We analyzed a Ty1/copia retrotransposon, ONSEN, as a model for stress-activated transposon. ONSEN is a relatively young element with conserved LTR sequences and functional coding genes in Arabidopsis (Ito et al., 2011). The LTR sequence contains conserved HREs that become functional stress-responding promoters of ONSEN (Cavrak et al., 2014). It is not known how ONSEN acquired the stress-responding promoter in the LTR; however, after a TE has gained a functional promoter that responds to environmental stress, the number of copies of activated TE may be amplified in the host genome when environmental changes occur in nature.
We analyzed the heat-induced promoter of ONSEN using a reporter gene assay. Gene expression was enhanced in a mutant that was deficient in siRNA biogenesis. Reduction of DNA methylation in the LTR has been shown to favor the HS response (Cavrak et al., 2014). However, we showed that the activity level of the transgene gradually declined and was no longer detected after 3 days, indicating that the activation of ONSEN was initially regulated by transcriptional factors and that the resilencing of ONSEN was controlled independently of RdDM. In Arabidopsis, heat-activation of several repetitive elements can occur without epigenetic changes such as DNA methylation and histone modifications but is accompanied by heterochromatin decondensation (Pecinka et al., 2010). The transcriptional silencing of the activated elements delayed in mutants of CAF-1 which loads nucleosomes onto replicated DNA suggested that CAF-1 was important for efficient restoration of silencing after HS (Pecinka et al., 2010). Recently, it is reported that a chromatin-remodeling factor, Decrease in DNA Methylation 1 (DDM1) and Morpheus Molecule 1 (MOM1) act redundantly to restore silencing after HS in Arabidopsis (Iwasaki and Paszkowski, 2014). The heat-induced transcriptional activation of some genes in ddm1mom1 double mutants persisted longer than in WT and was transgenerationally inherited. These findings indicated that heat-induced chromatin changes might play a role for the rapid resetting of ONSEN expression in nrpd1 with impaired siRNA repression.
Transposable elements could be activated in a harsh environment and could affect the host genome. For example, in Arabidopsis, activated Athila retrotransposons produce a small RNA that regulates a stress-response gene, OLIGOURIDYLATE BINDING PROTEIN 1 in trans (McCue et al., 2012). We determined that ONSEN was activated not only by HS but also by oxidative stress. Activity levels were enhanced in the mutant that was deficient in siRNA biogenesis, indicating that the activation was controlled by RdDM. The activation of ONSEN was not detected in the mutant lacking heat-inducible transcription factor, suggesting that activation of ONSEN induced by oxidative stress is regulated by the same pathway as heat-induced activation.
The mobility of ONSEN was regulated tightly by the siRNA-mediated pathway. In a mutant of siRNA biogenesis, a transgenerational transposition was observed, indicating that ONSEN transposition was transmitted to a reproductive cell. In this report, we analyzed whether transcriptional activation timing was an important factor for transposition frequency during plant development. Transposition occurred when the plant was heat-stressed during a period of the developmental phase. The same transposition pattern on the progeny within a single flower indicated that some transposition occurred prior to flower differentiation. Further, the same transposition pattern among the progenies from different branches indicated that some transposition occurred before branch development. Conversely, the different transposition pattern in the progeny of adjacent flowers indicated that transposition occurred just before flower bud initiation. The observation that transposition occurred in the progenies produced from flowers that were differentiated after HS indicated that transposition activation might be maintained over a long period of time, although the transcriptional activation was transient.
One question that remains to be resolved concerns the regulatory mechanism of the transposition that occurred during the period of flower differentiation. One possibility is that transposition requires HS during the period of inflorescence meristem formation. We found that transposition occurred when HS was applied to 1 day-old seedling in which the inflorescence meristem has not been formed, suggesting that the presence of inflorescence meristem cells is not necessary for HS-induced transposition. Another possibility is that the activity levels of ONSEN may be maintained in the shoot apical meristem and transposition may have occurred at the point when the cell’s fate changes during the reproductive growth of a stressed mutant. A further possibility is that ONSEN RNA was maintained in the shoot apical meristem during plant development. We found that the transcriptional activity levels of ONSEN returned to baseline levels within 5 days in the shoot apex of both WT and nrpd1 mutant plants. This result indicated that neither the activity level of ONSEN nor ONSEN RNA was maintained in the shoot apex during plant development. We could not exclude the possibility that the transcriptional activity of ONSEN was sustainable only in specific tissues, such as the shoot apical meristem of nrpd1 plants.
The transposition of ONSEN during flower differentiation might require an active mark in addition to the transient activation by HS. The transposition of ONSEN was independent of DNA methylation and ONSEN was not transposed in ddm1 hypomethylation mutation subjected to HS (Ito et al., 2011). The HS might induce active chromatin in ONSEN; however, subsequently, siRNA could induce ONSEN as a silenced chromatin. A transgenerational transposition of heat-activated ONSEN was not observed in a mutant deficient in histone H3K9 methyltransferases (SUVH4/KRYPTONITE) (Ito et al., 2011), indicating that the regulation of transposition was independent of histone H3K9 dimethylation. Histone H3K27 trimethylation (H3K27me3) is associated with gene repression and the target genes are tissue-specifically activated during differentiation processes or induced by abiotic or biotic stresses in Arabidopsis (Zhang et al., 2007a). H3K27me3 dynamically regulated the target gene expression during plant differentiation and the targets were enriched in TEs in meristem indicating that stem cells must be protected from TE activation to form germline (Lafos et al., 2011). ONSEN was a target of H3K27me3, however, the methylation level was not significantly different in meristem and leaves in WT (Lafos et al., 2011). It is interesting to know whether HS in nrpd1 mutant could affect the modification of H3K27me3 on ONSEN in a tissue-specific manner.
In nrpd1 mutant plants, HS could change chromatin structures, thereby releasing ONSEN transposition, and the active chromatin state could be maintained until the period of flower differentiation. A large-scale reorganization of chromatin was observed during floral transition in Arabidopsis (Tessadori et al., 2007). The pericentric heterochromatin reduced prior to bolting and recovered after elongation of the floral stem. Also decondensation of chromatin in gene-rich regions coincided with the floral transition. We still do not know the meristem-specific regulation of transposition during floral transition; however, transposon silencing in plant germ cells has been reported in Arabidopsis (Slotkin et al., 2009; Olmedo-Monfil et al., 2010; Ibarra et al., 2012). It is worth noting that chromatin decondensation was not observed in nuclei from meristematic tissue after HS (Pecinka et al., 2010). Recently, it has been reported that RdDM is function specific in the shoot apical meristem and reinforces the silencing of TEs during early vegetative growth (Baubec et al., 2014). These findings demonstrated the importance of TE regulation during vegetative growth prior to the formation of the next generation.
It is also worth noting that the progeny of heat-stressed Arabidopsis had fewer but large leaves and tended to bolt earlier to increased plant biomass (Migicovsky et al., 2014). In the stressed progeny, the expression of HsfA2 and ONSEN was elevated with decreasing of global DNA methylation. The transgenerational phenotypic and epigenetic changes were partially deficient in the Dicer-like mutant, however, ONSEN expression increased in the progeny of heat-stressed plants regardless of mutant type (Migicovsky et al., 2014). It will be worthwhile to investigate the mechanism by which the stress memory is maintained during plant development.
In this study, siRNA-mediated regulation functioned to control transposons that were ectopically activated by environmental stress. Plant has evolved the regulation mechanisms that were independent of suppression by constitutive heterochromatin. The regulation was important during the process of plant development and may have a function in undifferentiated cells.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Shigeru Shigeoka for helpful advice. This work was supported by a grant from the MEXT as part of Joint Research Program implemented at the Institute of Plant Science and Resources, Okayama University, the National Institute of Genetics Cooperative Research Program (2014-A), JST-PRESTO, Grants-in-Aid for JSPS Fellows (14J02452), and Grants-in-Aid for Scientific Research in Innovative Areas (2511970103).
References
Baubec, T., Finke, A., Mittelsten Scheid, O., and Pecinka, A. (2014). Meristem-specific expression of epigenetic regulators safeguards transposon silencing in Arabidopsis. EMBO Rep. 15, 446–452. doi: 10.1002/embr.201337915
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cao, X., and Jacobsen, S. E. (2002). Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12, 1138–1144. doi: 10.1016/S0960-9822(02)00925-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Capy, P., Gasperi, G., Biemont, C., and Bazin, C. (2000). Stress and transposable elements: co-evolution or useful parasites? Heredity 85, 101–106. doi: 10.1046/j.1365-2540.2000.00751.x
Cavrak, V. V., Lettner, N., Jamge, S., Kosarewicz, A., Bayer, L. M., and Scheid, O. M. (2014). How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet. 10:e1004115. doi: 10.1371/journal.pgen.1004115
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Church, G. M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. U.S.A. 81, 1991–1995. doi: 10.1073/pnas.81.7.1991
Feschotte, C., Jiang, N., and Wessler, S. R. (2002). Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3, 329–341. doi: 10.1038/nrg793
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gao, Z., Liu, H. L., Daxinger, L., Pontes, O., He, X., Qian, W.,et al. (2010). An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 465, 106–109. doi: 10.1038/nature09025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Grandbastien, M. A. (1998). Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 3, 181–187. doi: 10.1016/S1360-1385(98)01232-1
Grandbastien, M. A., Lucas, H., Morel, J. B., Mhiri, C., Vernhettes, S., and Casacuberta, J. M. (1997). The expression of the tobacco Tnt1 retrotransposon is linked to plant defense responses. Genetica 100, 241–252. doi: 10.1023/A:1018302216927
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Herr, A. J., Jensen, M. B., Dalmay, T., and Baulcombe, D. C. (2005). RNA polymerase IV directs silencing of endogenous DNA. Science 308, 118–120. doi: 10.1126/science.1106910
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hirochika, H. (1993). Activation of tobacco retrotransposons during tissue culture. EMBO J. 12, 2521–2528.
Ibarra, C. A., Feng, X., Schoft, V. K., Hsieh, T. F., Uzawa, R., Rodrigues, J. A.,et al. (2012). Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337, 1360–1364. doi: 10.1126/science.1224839
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ito, H., Gaubert, H., Bucher, E., Mirouze, M., Vaillant, I., and Paszkowski, J. (2011). An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119. doi: 10.1038/nature09861
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Iwasaki, M., and Paszkowski, J. (2014). Identification of genes preventing transgenerational transmission of stress-induced epigenetic states. Proc. Natl. Acad. Sci. U.S.A. 10, 8547–8552. doi: 10.1073/pnas.1402275111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jiang, J., Birchler, J. A., Parrott, W. A., and Dawe, R. K. (2003). A molecular view of plant centromeres. Trends Plant Sci. 8, 570–575. doi: 10.1016/j.tplants.2003.10.011
Kanno, T., Huettel, B., Mette, M. F., Aufsatz, W., Jaligot, E., Daxinger, L.,et al. (2005). Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 37, 761–765. doi: 10.1038/ng1580
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kato, M., Miura, A., Bender, J., Jacobsen, S. E., and Kakutani, T. (2003). Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr. Biol. 13, 421–426. doi: 10.1016/S0960-9822(03)00106-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kumar, A. (1996). The adventures of the Ty1-copia group of retrotransposons in plants. Trends Genet. 12, 41–43. doi: 10.1016/0168-9525(96)81393-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kumar, A., and Bennetzen, J. L. (1999). Plant retrotransposons. Annu. Rev. Genet. 33, 479–532. doi: 10.1146/annurev.genet.33.1.479
Lafos, M., Kroll, P., Hohenstatt, M. L., Thorpe, F. L., Clarenz, O., and Schubert, D. (2011). Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 7:e1002040. doi: 10.1371/journal.pgen.1002040
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lippman, Z., May, B., Yordan, C., Singer, T., and Martienssen, R. (2003). Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1:e67. doi: 10.1371/journal.pbio.0000067
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Matzke, M. A., and Birchler, J. A. (2005). RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6, 24–35. doi: 10.1038/nrg1500
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McCue, A. D., Nuthikattu, S., Reeder, S. H., and Slotkin, R. K. (2012). Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genet. 8:e1002474. doi: 10.1371/journal.pgen.1002474
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mhiri, C., Morel, J. B., Vernhettes, S., Casacuberta, J. M., Lucas, H., and Grandbastien, M. A. (1997). The promoter of the tobacco Tnt1 retrotransposon is induced by wounding and by abiotic stress. Plant Mol. Biol. 33, 257–266. doi: 10.1023/A:1005727132202
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Migicovsky, Z., Yao, Y., and Kovalchuk, I. (2014). Transgenerational phenotypic and epigenetic changes in response to heat stress in Arabidopsis thaliana. Plant Signal. Behav. 9:e27971. doi: 10.4161/psb.27971
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mirouze, M., Reinders, J., Bucher, E., Nishimura, T., Schneeberger, K., Ossowski, S.,et al. (2009). Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461, 427–430. doi: 10.1038/nature08328
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miura, A., Kato, M., Watanabe, K., Kawabe, A., Kotani, H., and Kakutani, T. (2004). Genomic localization of endogenous mobile CACTA family transposons in natural variants of Arabidopsis thaliana. Mol. Genet. Genomics 270, 524–532. doi: 10.1007/s00438-003-0943-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miura, A., Yonebayashi, S., Watanabe, K., Toyama, T., Shimada, H., and Kakutani, T. (2001). Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411, 212–214. doi: 10.1038/35075612
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mosher, R. A., Schwach, F., Studholme, D., and Baulcombe, D. C. (2008). PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 3145–3150. doi: 10.1073/pnas.0709632105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nishizawa, A., Yabuta, Y., Yoshida, E., Maruta, T., Yoshimura, K., and Shigeoka, S. (2006). Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48, 535–547. doi: 10.1111/j.1365-313X.2006.02889.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nover, L., Bharti, K., Doring, P., Mishra, S. K., Ganguli, A., and Scharf, K. D. (2001). Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6, 177–189. doi: 10.1379/1466-1268(2001)006<0177:AATHST>2.0.CO;2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Olmedo-Monfil, V., Duran-Figueroa, N., Arteaga-Vazquez, M., Demesa-Arevalo, E., Autran, D., Grimanelli, D.,et al. (2010). Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464, 628–632. doi: 10.1038/nature08828
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Onodera, Y., Haag, J. R., Ream, T., Costa Nunes, P., Pontes, O., and Pikaard, C. S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120, 613–622. doi: 10.1016/j.cell.2005.02.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pecinka, A., Dinh, H. Q., Baubec, T., Rosa, M., Lettner, N., and Mittelsten Scheid, O. (2010). Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22, 3118–3129. doi: 10.1105/tpc.110.078493
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Peschke, V. M., Phillips, R. L., and Gengenbach, B. G. (1987). Discovery of transposable element activity among progeny of tissue-culture derived maize plants. Science 238, 804–807. doi: 10.1126/science.238.4828.804
Pontier, D., Yahubyan, G., Vega, D., Bulski, A., Saez-Vasquez, J., Hakimi, M. A.,et al. (2005). Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 19, 2030–2040. doi: 10.1101/gad.348405
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pouteau, S., Grandbastien, M. A., and Boccara, M. (1994). Microbial elicitors of plant defense responses activate transcription of a retrotransposon. Plant J. 5, 535–542. doi: 10.1046/j.1365-313X.1994.5040535.x
Schmidt, T. (1999). LINEs, SINEs and repetitive DNA: non-LTR retrotransposons in plant genomes. Plant Mol. Biol. 40, 903–910. doi: 10.1023/A:1006212929794
Schramm, F., Ganguli, A., Kiehlmann, E., Englich, G., Walch, D., and von Koskull-Doring, P. (2006). The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 60, 759–772. doi: 10.1007/s11103-005-5750-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Slotkin, R. K., Vaughn, M., Borges, F., Tanurdzic, M., Becker, J. D., Feijo, J. A.,et al. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472. doi: 10.1016/j.cell.2008.12.038
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Steward, N., Ito, M., Yamaguchi, Y., Koizumi, N., and Sano, H. (2002). Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J. Biol. Chem. 277, 37741–37746. doi: 10.1074/jbc.M204050200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takeda, S., Sugimoto, K., Otsuki, H., and Hirochika, H. (1998). Transcriptional activation of the tobacco retrotransposon Tto1 by wounding and methyl jasmonate. Plant Mol. Biol. 36, 365–376. doi: 10.1023/A:1005911413528
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takeda, S., Sugimoto, K., Otsuki, H., and Hirochika, H. (1999). A 13-bp cis-regulatory element in the LTR promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant J. 18, 383–393. doi: 10.1046/j.1365-313X.1999.00460.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tessadori, F., Chupeau, M. C., Chupeau, Y., Knip, M., Germann, S., van Driel, R.,et al. (2007). Large-scale dissociation and sequential reassembly of pericentric heterochromatin in dedifferentiated Arabidopsis cells. J. Cell. Sci. 120, 1200–1208. doi: 10.1242/jcs.000026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsukahara, S., Kobayashi, A., Kawabe, A., Mathieu, O., Miura, A., and Kakutani, T. (2009). Bursts of retrotransposition reproduced in Arabidopsis. Nature 461, 423–426. doi: 10.1038/nature08351
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vernhettes, S., Grandbastien, M. A., and Casacuberta, J. M. (1997). In vivo characterization of transcriptional regulatory sequences involved in the defence-associated expression of the tobacco retrotransposon Tnt1. Plant Mol. Biol. 35, 673–679. doi: 10.1023/A:1005826605598
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wessler, S. R. (1996). Turned on by stress. Plant retrotransposons. Curr. Biol. 6, 959–961. doi: 10.1016/S0960-9822(02)00638-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wierzbicki, A. T., Haag, J. R., and Pikaard, C. S. (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635–648. doi: 10.1016/j.cell.2008.09.035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, X., Clarenz, O., Cokus, S., Bernatavichute, Y. V., Pellegrini, M., Goodrich, J.,et al. (2007a). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5:e129. doi: 10.1371/journal.pbio.0050129
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, X., Henderson, I. R., Lu, C., Green, P. J., and Jacobsen, S. E. (2007b). Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. U.S.A. 104, 4536–4541. doi: 10.1073/pnas.0611456104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: transposon, small RNA, environmental stress, Arabidopsis thaliana, ONSEN
Citation: Matsunaga W, Ohama N, Tanabe N, Masuta Y, Masuda S, Mitani N, Yamaguchi-Shinozaki K, Ma JF, Kato A and Ito H (2015) A small RNA mediated regulation of a stress-activated retrotransposon and the tissue specific transposition during the reproductive period in Arabidopsis. Front. Plant Sci. 6:48. doi: 10.3389/fpls.2015.00048
Received: 29 October 2014; Accepted: 17 January 2015;
Published online: 09 February 2015.
Edited by:
Mahmoud W. Yaish, Sultan Qaboos University, OmanReviewed by:
Dierk Wanke, Tuebingen University, GermanyPrakash P. Kumar, National University of Singapore, Singapore
Copyright © 2015 Matsunaga, Ohama, Tanabe, Masuta, Masuda, Mitani, Yamaguchi-Shinozaki, Ma, Kato and Ito. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hidetaka Ito, Faculty of Science, Hokkaido University, Kita10 Nishi 8, Kita-ku, Sapporo, Hokkaido 060-0810, Japan e-mail:aGl0b0BtYWlsLnNjaS5ob2t1ZGFpLmFjLmpw
 Wataru Matsunaga
Wataru Matsunaga Naohiko Ohama
Naohiko Ohama Noriaki Tanabe
Noriaki Tanabe Yukari Masuta1
Yukari Masuta1 Namiki Mitani
Namiki Mitani Kazuko Yamaguchi-Shinozaki
Kazuko Yamaguchi-Shinozaki Jian F. Ma
Jian F. Ma Atsushi Kato
Atsushi Kato Hidetaka Ito
Hidetaka Ito