
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 31 July 2014
Sec. Plant Systems and Synthetic Biology
Volume 5 - 2014 | https://doi.org/10.3389/fpls.2014.00362
This article is part of the Research TopicLeaves: Regulatory networks underlying development and maintenance of the photosynthetic organView all 5 articles
Through its photosynthetic capacity the leaf provides the basis for growth of the whole plant. In order to improve crops for higher productivity and resistance for future climate scenarios, it is important to obtain a mechanistic understanding of leaf growth and development and the effect of genetic and environmental factors on the process. Cells are both the basic building blocks of the leaf and the regulatory units that integrate genetic and environmental information into the developmental program. Therefore, to fundamentally understand leaf development, one needs to be able to reconstruct the developmental pathway of individual cells (and their progeny) from the stem cell niche to their final position in the mature leaf. To build the basis for such understanding, we review current knowledge on the spatial and temporal regulation mechanisms operating on cells, contributing to the formation of a leaf. We focus on the molecular networks that control exit from stem cell fate, leaf initiation, polarity, cytoplasmic growth, cell division, endoreduplication, transition between division and expansion, expansion and differentiation and their regulation by intercellular signaling molecules, including plant hormones, sugars, peptides, proteins, and microRNAs. We discuss to what extent the knowledge available in the literature is suitable to be applied in systems biology approaches to model the process of leaf growth, in order to better understand and predict leaf growth starting with the model species Arabidopsis thaliana.
Understanding the regulation of plant growth and its constituent organs is an important objective in biology. It forms the basis for crop yield, turn-over in ecosystems and the means for the plant to adapt to environmental conditions and experimental treatments. The development of leaves in dicotyledonous plant species is an intriguing process, resulting from a complex interplay of a multitude of regulatory pathways. On the one hand it is so strictly regulated that the resultant leaf morphology is a reliable characteristic for taxonomic classification. On the other hand however, the process is so plastic that environmental factors can affect mature leaf size by an order of magnitude. Curiously, leaf shape is often largely conserved between related species with genetic variations in thousands of genes, while a single mutation can sometimes induce morphological differences similar to those that distinguish species and even families (e.g., Barkoulas et al., 2008). Due to these intriguing characteristics and the importance of leaves for plant performance and function, many aspects of leaf development have been extensively studied.
In recent decades, remarkable progress has been made in understanding the regulation of leaf development via molecular/genetic approaches. Moreover, increasing use of high-throughput technologies is constantly providing new biological information at various organizational levels. In this context, systems biology provides a means to integrate the accumulating knowledge into holistic mechanistic models to get a complete understanding of biological processes. These models are often implemented through computer simulations of normal and/or experimentally perturbed systems to test how well they resemble the real situation and increase our understanding of its mechanistic basis.
A mechanistic understanding of leaf development should encompass an integrated view on the regulatory networks that control developmental decisions and processes of cells as they migrate in space and time from the shoot apical meristem (SAM) to their final position in the leaf (Figure 1). Therefore, we review the subsequently acting developmental networks that guide individual cells on their way from the SAM to their differentiated state somewhere in a fully differentiated leaf. Based on this description we delineate to what extent we understand how variations in the regulation at the cell level affect the shape and size of the leaf as a whole, and what are the implications for implementing this knowledge into fully fledged simulation models.
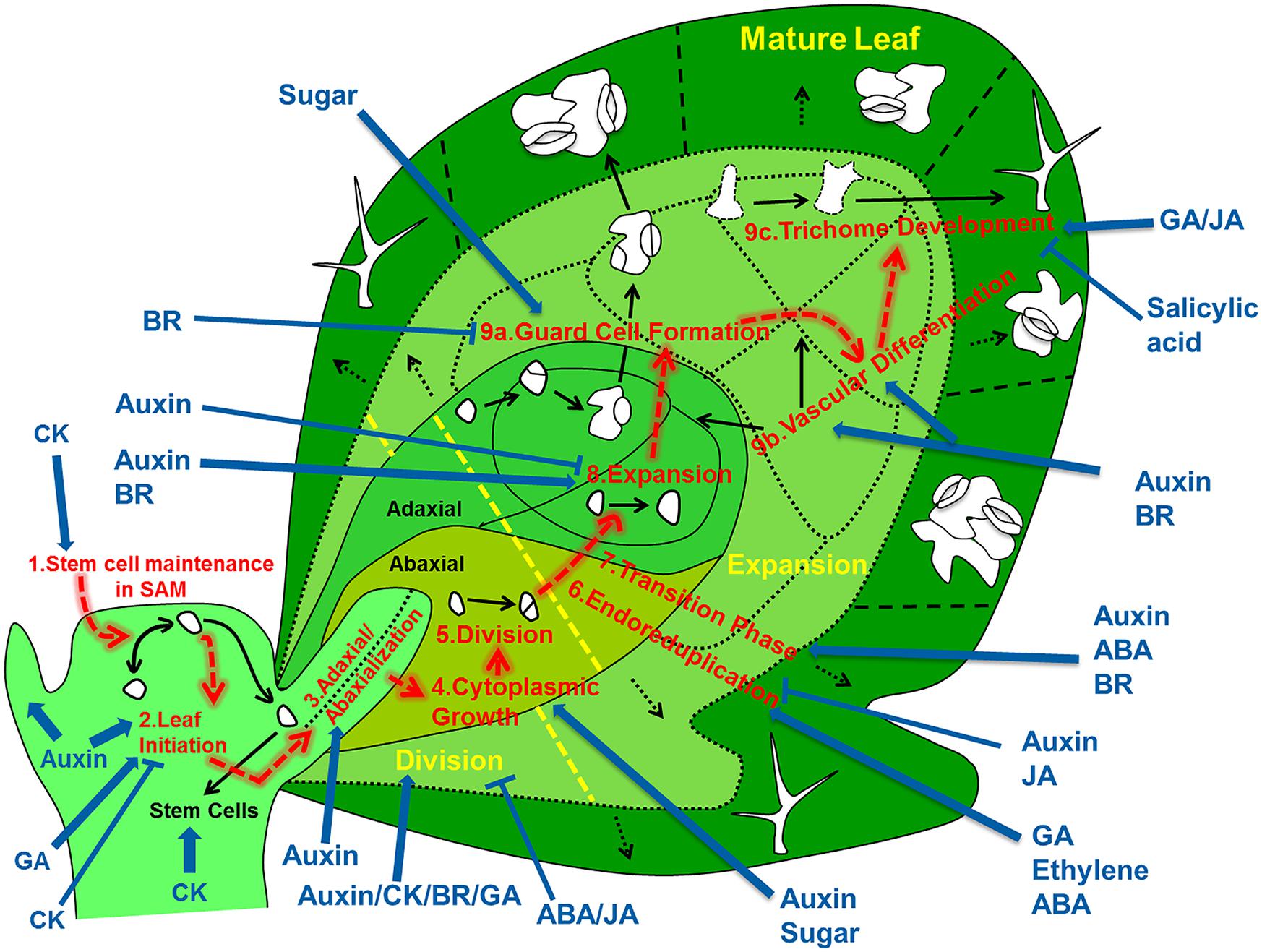
FIGURE 1. Overview of the regulatory processes that determine the development of a leaf. The cells that form the leaf originate from the stem cell niche at the shoot apical meristem. As a first step in their development, cells need to loose stem cell identity (1). A leaf primordium is initiated in groups of cells that migrate into the lateral regions of the SAM (2), which further acquires upper (adaxial) and lower (abaxial) sides through leaf-polarity control (3). Afterward, the transformation of the small leaf primordium to a mature leaf is controlled by at least six distinct processes: cytoplasmic growth (4), cell division (5), endoreduplication (6), transition between division and expansion (7), cell expansion (8) and cell differentiation (9) into stomata (9a), vascular tissue (9b), and trichomes (9c). Most of these processes are tightly controlled by different signaling molecules, including phytohormones. The developmental path of cells is indicated with red arrows, key regulatory processes are numbered and indicated and regulation of these processes by phytohormones/sugar is shown by blue arrows (pointed and T shaped arrows indicate positive and negative regulation, respectively).
The development of a leaf is a dynamic process where independent regulatory pathways instruct component cells at different stages of their development to make differentiation switches and to regulate the rate at which developmental processes are executed. Each of these regulatory control points is essential to steer the development of individual cells. When integrated over the entire cell population of a leaf, its growth and ultimately size and shape are emergent properties that can be compared to real leaves. Because developmental signals are perceived and executed at the level of individual cells, it is essential to understand how these signals are integrated in the leaf developmental process, which can be achieved by modeling the path of an individual cell (and its progeny) from SAM to the mature leaf. Although many of the pathways involved have been extensively reviewed, to our mind the perspective of the individual cells has not been explored systematically. Therefore the main aim of the present review is to provide this cellular perspective to leaf development.
The SAM is the source of all cells that ultimately form the shoot, including the subset that ends up building the leaves. Generally, cells in the central zone (CZ) of the SAM divide at a relatively low rate and remain in an undifferentiated state, whereas cells at the peripheral zone (PZ) divide faster and differentiate into organs such as leaves, axillary nodes, and floral parts (Veit, 2004; Braybrook and Kuhlemeier, 2010). In dicots, the SAM consists of three layers L1, L2, and L3; epidermal (L1) and subepidermal (L2) layers are known as tunica and the inner layer (L3) is called the corpus (Satina et al., 1940).
From the cellular perspective, on-going (slow) division in the stem cell niche will cause cells to become displaced away from the quiescent center, where at some well-defined place they lose their stem cell fate and acquire the actively dividing state. This transition is controlled by the interplay of a regulatory loop involving the homeodomain transcription factor WUSCHEL (WUS) in the rib zone (RZ) and CLAVATA gene products (CLV1, CLV2, and CLV3) expressed in the CZ of the SAM (Brand et al., 2000; Schoof et al., 2000; Carles and Fletcher, 2003; Yadav and Reddy, 2011). The WUS and CLV based pathway operates through two mobile signals: CLV3 and a hypothetical WUS mediated signal (Figure 2). CLV3 encodes a small secreted ligand that is produced specifically in L1 and L2 cells, and moves into the underlying L3 cells where it binds with receptor like proteins CLV1 (LRR receptor kinase) and/or CLV2 (receptor-like protein), which in turn inhibit WUS activity (Clark, 2001; Carles and Fletcher, 2003). WUS activity in the L3 cells induces the production of a non-cell-autonomous signal that moves to the stem cells and activates the expression of CLV3 there (Haecker and Laux, 2001; Braybrook and Kuhlemeier, 2010). It was proposed that the L1 produced miR394 signal is necessary for spatial organization of the SAM. This mobile microRNA regulates WUS mediated stem cell maintenance by inhibition of F box protein LEAF CURLING RESPONSIVENESS (LCR; Knauer et al., 2013).
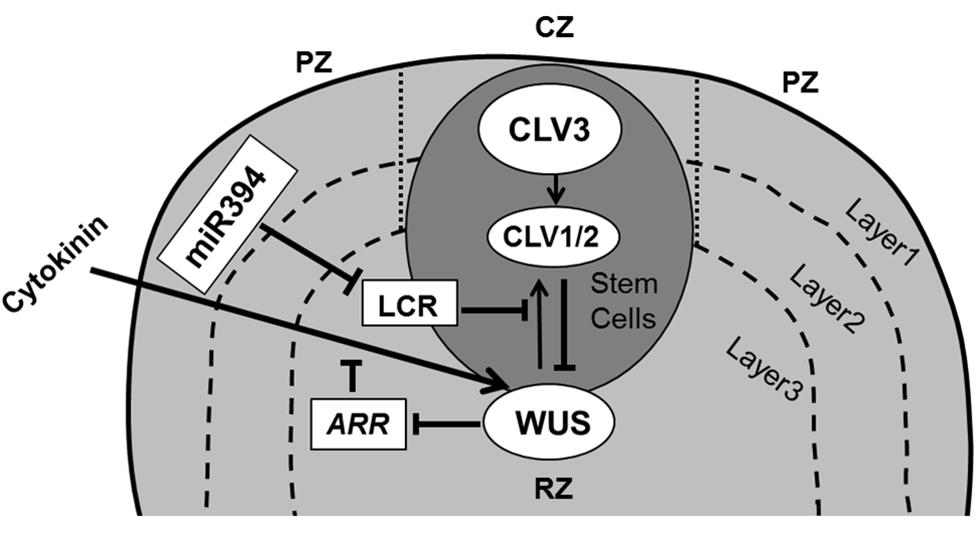
FIGURE 2. Maintenance of stem cells in shoot apical meristem. The SAM is organized in three functional zones [central zone (CZ), peripheral zone (PZ), and rib zone (RZ)] and three layers where the antagonistic relation between WUS and CLV is essential to preserve cells in the meristem. WUS activates CLV3, which further binds with CLV1/2 and in turn inhibits expression of WUS. Cytokinin positively controls WUS expression where ARRs are negative regulators of cytokinin and are inhibited by WUS. The L1 specific miR394 negatively affects the LCR protein, which interferes in WUS/CLV based stem cell maintenance (pointed and T shaped arrows indicate positive and negative regulation, respectively).
Upon mutation in WUS the stem cells precociously transit into the peripheral actively dividing zone, ultimately consuming the stem cell niche and thereby the meristem. Inversely, in clv1 and clv3 mutants WUS activity of SAM cells is maintained much longer, whereby the stem cell niche and consequently the SAM as a whole enlarges dramatically (Clark et al., 1993, 1995; Laux et al., 1996). Several mathematical models have focused on the WUS–CLV interaction, predicting to various degrees how their expression domains are modulated through mutation or misexpression (Jonsson et al., 2005; Nikolaev et al., 2007; Hohm et al., 2010). Recent experimental studies supported by mathematical modeling have shown that WUS movement is essential for direct transcriptional repression of the differentiation program (Yadav et al., 2013) as well as in restricting its own accumulation through activating its negative regulator CLV3 (Yadav et al., 2011).
It has been postulated that signaling by the plant hormone cytokinin (CK) regulates WUS expression via CLV-dependent and CLV-independent mechanisms (Gordon et al., 2009) to promote SAM growth and maintenance with WUS repressing the transcription of ARABIDOPSIS type-A RESPONSE REGULATORS (ARRs), which are the negative regulators of CK signaling (Leibfried et al., 2005; Sablowski, 2007; Figure 2). Indeed, mutations in CK receptors (Higuchi et al., 2004) and over-expression of the CK dehydrogenase gene family of Arabidopsis (AtCKX; Werner et al., 2003) reduce meristem size and leaf area, indicating a relation between the SAM and leaf size. It appears however that that the number of leaf founder cells is not an important determinant of the final leaf size. For instance, a meta-analysis across a wide range of cactus species indicates that the size of the SAM correlates closely to the number of leaves formed and has only minor implications for their ultimate size (Mauseth, 2004).
Once progenitor cells are outside the stem cell niche, they need to decide whether they will contribute to the main axis or will differentiate into lateral appendices such as leaf primordia. This decision is primarily governed by the accumulation of the plant hormone auxin its influx carrier [AUXIN RESISTANT (AUX1) and its PIN-FORMED1 (PIN1)] efflux transporter (Bayer et al., 2009; Guenot et al., 2012). The efflux carriers orient the transport of auxin toward neighboring cells with a higher auxin concentration, leading to the formation of accumulation patterns across the cell population. Several mathematical modeling studies (reviewed in De Vos et al., 2012) have simulated phyllotactic patterning based on feedback interactions between auxin and PIN distribution. Some models postulate that AUX1 creates auxin accumulation mainly in L1 layer cells, whereas PIN1 is initially localized in the protodermal (L1) layer cells and causes drainage of auxin toward the base of the shoot by inducing vascular strand differentiation in L2/3 layer cells of the SAM (Reinhardt et al., 2003; de Reuille et al., 2006; Figure 3).
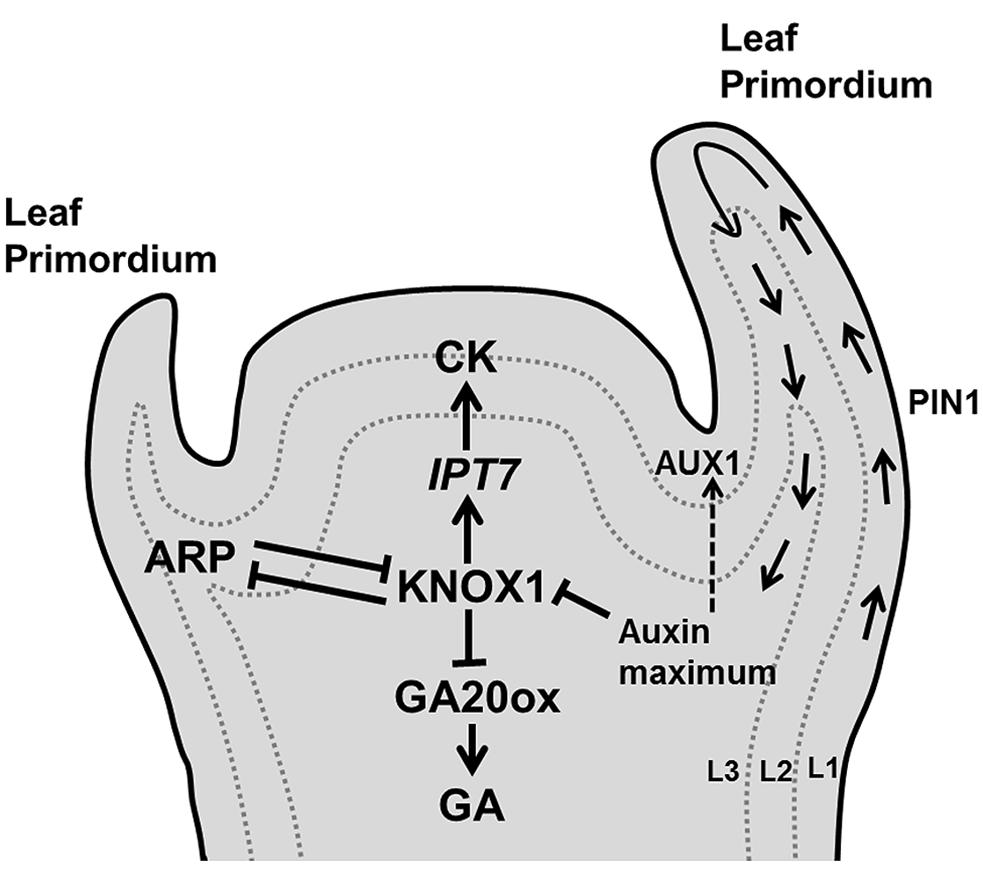
FIGURE 3. Decision of leaf initiation. Accumulation peaks of auxin at the flank of the SAM through PIN1/AUX1 mediate polar auxin transport, triggers development of a primordium where KNOX1 plays key role in stem cell maintenance. Additionally, KNOX1 positively regulates CK whereas it negatively affects GA signaling through IPT7 and GA20 oxidases, respectively. Opposite to it, ARP regulates the emergence of a young primordium (pointed and T shaped arrows indicate positive and negative regulation, respectively).
Arabidopsis leaf differentiation from the apical meristem is abolished in the auxin biosynthetic triple mutant yuc1 yuc4 pin1 (Cheng et al., 2007), whereas longer plastochron and irregular vegetative growth occurs in the pin1 mutant (Guenot et al., 2012). Cells in the SAM, which serve as a stem cell population, differ from the cells in the leaf primordium. This distinction is controlled by complex and still poorly characterized regulatory networks in which the antagonistic relation between two families of transcription factors, KNOTTED-like homeobox (KNOX1) and ASYMMETRIC LEAF1/ROUGH SHEATH2/PHANTASTICA (ARP) proteins (Byrne et al., 2002; Hay and Tsiantis, 2006, 2010) plays a crucial role (Figure 3). KNOX is expressed in all meristem cells except those at the site of the organ initiation (Jackson et al., 1994; Long et al., 1996), whereas the (ARP family) AS1 mRNA is expressed in the primordia forming cells, but not in the meristem (Byrne et al., 2000). KNOX1 is required to maintain undifferentiated cells in the SAM (Scofield and Murray, 2006) and increases CK biosynthesis (Yanai et al., 2005), whereas ARP initiates differentiation in the leaf primordium (Byrne et al., 2002). High levels of auxin restrain CK biosynthesis by the repression of KNOX1 activity (Su et al., 2011; Figure 3). High auxin and AS1 expression also suppress the expression of the KNOX gene BREVIPEDICELLUS (BP), which is required for leaf initiation (Hay et al., 2006). In Arabidopsis, the KNOX1 gene SHOOTMERISTEMLESS (STM) acts antagonistically with ARP gene products to control the induction of leaf primordia. The loss of function mutant stm fails to produce a SAM, also preventing leaf formation (Long et al., 1996), whereas the as1 and as2 mutants have small and round leaves (Byrne et al., 2002).
It has been demonstrated that the KNOX proteins trigger CK biosynthesis through the activation of IPT7 (encodes the CK biosynthetic enzyme isopentenyl transferase) and repress the transcription of gibberellin (GA) biosynthetic genes that encode GA20-oxidases (Sakamoto et al., 2001; Jasinski et al., 2005; Figure 3). Thus, high CK and low GA maintain stem cell identity in SAM cells by preventing cell differentiation (Gordon et al., 2009; Veit, 2009).
After acquiring “leaf” identity, the cells in the primordium have to develop a polarity gradient along the dorso-ventral axis. Once the position of the leaf primordium is established, a further increase in cell proliferation rates stimulates primordium outgrowth from the SAM. If this growing primordium is removed by tangential incision, another primordium arises which is cylindrical and abaxialized (lacking a flat leaf blade). This highlights the importance of signals originating in the SAM and received by cells in the primordium to determine polarity. This so called Sussex signal is yet to be identified (Sussex, 1951). Waites and Hudson (1995) proposed that the dorsal and ventral sides of the leaf are specified in the early development of the leaf primordium, when it is still located within the SAM. They showed that the PHAN gene (encoding a MYB type transcription factor) in Antirrhinum majus is involved in ab/ad-axial leaf polarity. Subsequently the phabulosa-1 (phb-1d) mutant was characterized in Arabidopsis whose leaves were unable to develop a blade and were radially symmetrical (McConnell and Barton, 1998).
Our knowledge of the regulation of antagonistic transcription factors specifying upper and lower sides has greatly increased, but the molecular signals exchanged between cells on both sides of the primordium to create this polarity are yet to be identified. Adaxial domain identity is determined by the expression of PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV) genes, which encode class III homeodomain-leucine zipper (HD-ZIPIII) proteins (McConnell et al., 2001). The identity of cells in the abaxial domain depends on the expression of KANADI [KAN; which encodes a Golden2/Arabidopsis response-regulator/P starvation/acclimatization response (Psr1; GARP) transcription factor; Eshed et al., 2001; Kerstetter et al., 2001] and the YABBY gene family (Siegfried et al., 1999; Eshed et al., 2004). These two classes of genes produce signals that suppress each other’s expression: the expression of PHB/PHV/REV genes in cells located at the abaxial side is inhibited de by KAN and inversely KAN expression in abaxially located cells in inhibited by the activity of PHB/PHV/REV genes, providing a feedback communication between the two sides (Tsukaya, 2013b; Figure 4).
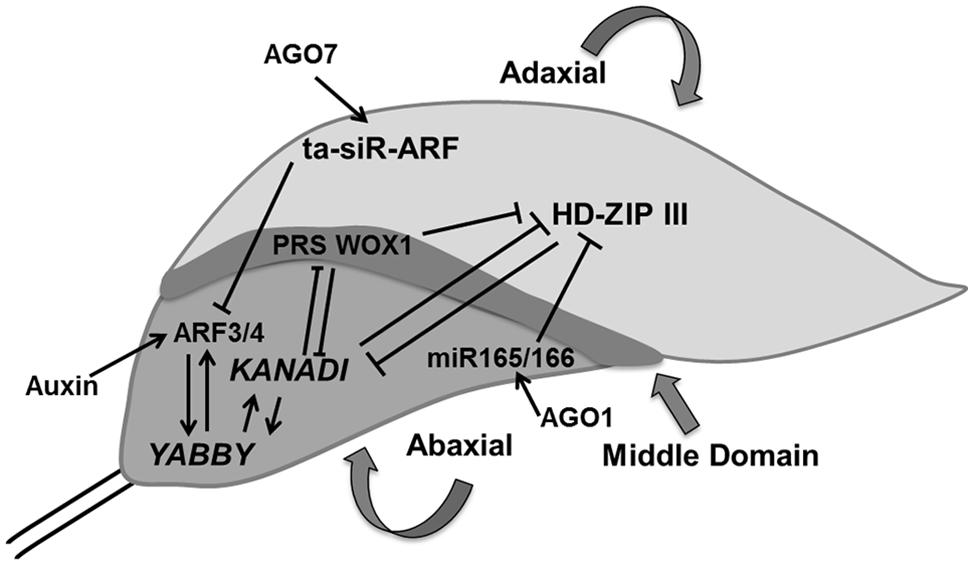
FIGURE 4. Polarity control. The young leaf primordium has three domains which are determined by domain specific transcription factors such as HD-ZIP III, KANADI, and PRS WOX1 for adaxial, abaxial, and middle regions, respectively. These transcription factors inhibit expression of each other and thereby control their expression in another domain. AGO1 regulates miR165/66 which inhibits HD-ZIP III whereas AGO7 stabilizes ta-siR-ARF which causes the degradation of ARF3/4, which itself is controlled by auxin. YABBY determines the abaxial side in cross talk with KANADI (pointed and T shaped arrows indicate positive and negative regulation, respectively).
Two small RNAs, the 21-nucleotide microRNA (miR165/166) and the 24-nucleotide transacting small interfering RNA (ta-siRNA), ta-siR-ARF, are also involved in determining leaf polarity (Chitwood et al., 2007, 2009; Nogueira et al., 2007). ARGONAUTE1 (AGO1) affects the regulation of miR165/166, which stimulates the cleavage of HD-ZIPIIIs transcripts in cells located on the adaxial side (Kidner and Martienssen, 2004) whereas, AGO7/ZIPPY (ZIP) stabilizes ta-siR-ARF, which further targets the degradation of auxin-related transcription factors, ETTIN (ETT)/ARF3 and ARF4 on the abaxial side (Adenot et al., 2006; Hunter et al., 2006). FILAMENTOUS FLOWER/YABBY3 (FIL/YAB3), a member of the YABBY family, up-regulates KAN1 and ARF4, which establishes a positive feedback loop (Bonaccorso et al., 2012; Figure 4).
Differences in cell growth rates along the principal developmental axes are crucial in determining final leaf shape. In addition to specification of adaxial and abaxial side of the leaf, margin specific cell fate is induced in cells residing at the boundary between these two surfaces (McHale, 1993). In contrast to regulation of leaf blade outgrowth, the influence of ad/abaxial specific genes on marginal cells is yet to be explored. Recently, middle leaf domain specific WUS-RELATED HOMEOBOX (WOX) genes were reported, which affect leaf blade outgrowth and margin specific development (Figure 4). These transcription factors (WOX1 and PRESSED FLOWER (PRS), i.e., WOX3) are repressed by KAN. The loss of function of WOX1 and PRS causes instable organization of ad-abaxial polarity (Nakata and Okada, 2012; Nakata et al., 2012).
A relatively simple computational model supported by time-lapse data and clonal analysis by Kuchen et al. (2012) accounts for local differences in cell growth rates and direction driving organ level shape changes during early Arabidopsis leaf development. A central model assumption is a yet uncharacterized early tissue polarity system that deforms during growth. CUP-SHAPED COTYLEDON (CUC) genes are emerging as prime candidate organizers of tissue polarity (Hasson et al., 2011). Correspondence of cell polarity and PIN1 auxin transporter patterns points to their involvement in such an organizer-based model (Scarpella et al., 2006). Moreover, it was reported that the outgrowth of lobes at the leaf margin is specified by a local auxin maximum as a result of the polar distribution of the PIN1 transporter (Hay et al., 2006). The transcription factor CUC2 which is expressed at the leaf sinuses and is negatively regulated by miR164 (Nikovics et al., 2006), promotes generation of these PIN1-dependent auxin maxima, which was supported by computer simulations (Bilsborough et al., 2011). Recently, a homeodomain protein REDUCED COMPEXITY (RCO) was reported to enhance serration by repressing growth at the flanks of initiating leaves (Vlad et al., 2014). Interestingly, CUC2, PIN1, and TEOSINTE BRANCHED/CYCLOIDEA/PCF (TCP), and KNOX have been implicated in compound leaf development illustrating that this regulatory system has the capacity to take more extreme forms than observed in Arabidopsis leaves (Barkoulas et al., 2008; Hay and Tsiantis, 2010; Koyama et al., 2010). A better understanding of the regulatory mechanisms operating at the cell level during the early phases of leaf outgrowth will likely provide invaluable insights into how diverse leaf morphologies are established.
In contrast to the morphology of the leaf primordium, the final size and shape of the leaf differ widely among species. Differences in leaf outgrowth are often interpreted as the result of cell division producing a certain number of cells and subsequent cell expansion determining their mature size. However, this is an overly simplistic view. Firstly, the relationship between cell division and expansion is complex and the two processes can mutually compensate each other (Tsukaya, 2002; Beemster et al., 2003). A theoretical framework to understand this phenomenon was provided by Green (1976), who proposed that cell growth and partitioning (division sensu-strictu) are two processes that co-occur in proliferating cells, whereas in expanding cells cell-growth continues in absence of partitioning. Clearly, this framework allows for continued growth irrespective of inhibited or stimulated cell division activity, at least until cells get too small or too large to function normally. However, the view that cell growth in proliferating cells is equivalent to that in expanding cells is overly simplistic. It is clear that whereas dividing cells grow by increasing cytoplasmic volume, expanding cells primarily increase their internal volume by expanding their vacuolar volume.
Cytoplasmic growth is mainly based on macromolecular synthesis and therefore consumes a lot of energy. A crucial role in ensuring a sufficient supply of elementary building blocks is played by the Target of Rapamycin (TOR) pathway. TOR, a Serine/Threonine kinase of the phosphatidylinositol-3-kinase-related kinase (PIKK) family is an essential controller of cytoplasmic growth and metabolism in plant cells. It controls a range of cellular responses such as ribosome biogenesis, translational initiation, cell proliferation, cell expansion and autophagy (Zhang et al., 2013; Sablowski and Dornelas, 2014; Figure 5).
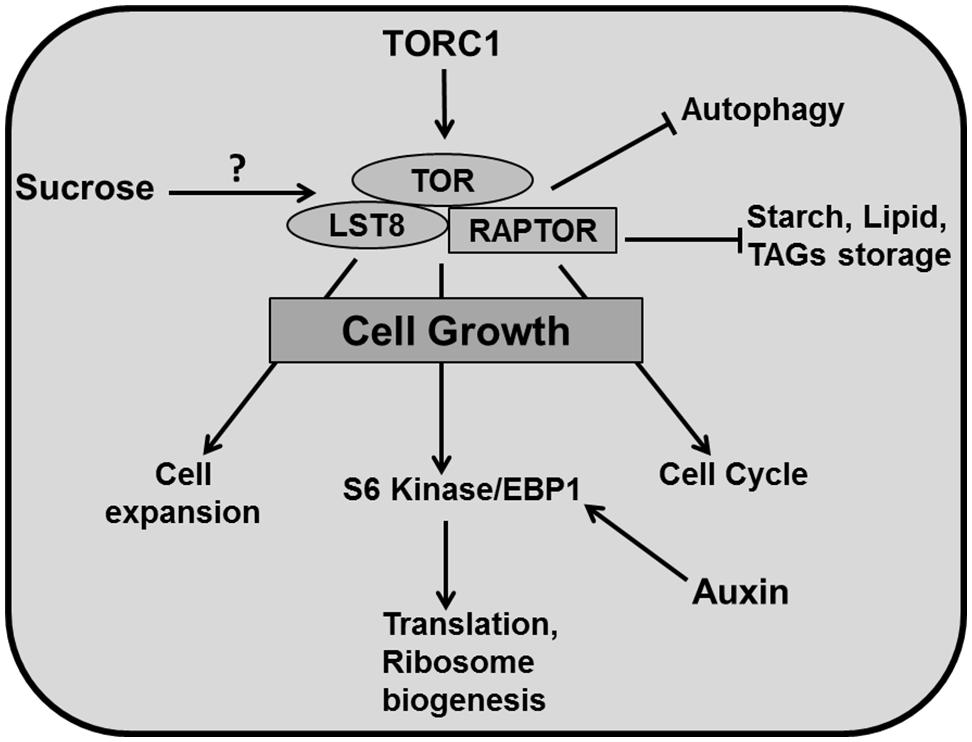
FIGURE 5. Cytoplasmic growth. TOR is the central regulator of diverse growth processes. TOR, RAPTOR, and LST8 are major components of TORC1 in plants. TOR has been reported to regulate different metabolic processes and positively controls cell expansion, cell cycle, translation, ribosome biogenesis (through phosphorylation of S6 kinase/EBP1). TOR activity inhibits autophagy and accumulation of carbon resources such as starch and lipids like triacylglycerides (TAGs). Auxin positively regulates EBP1 proteins. It has been reported that sucrose positively affects TOR activity (pointed and T shaped arrows indicate positive and negative regulation and question mark indicates an unknown mechanism, respectively).
In yeast and animals, there are two TOR complexes: TORC1 and TORC2 whereas in plants there is only evidence for TORC1. TOR, Regulatory associated protein of TOR (RAPTOR), and Lethal with Sec13 protein 8 (LST8) are three key components of TORC1 in Arabidopsis (Moreau et al., 2012). In contrast to other eukaryotes, much less is known about TOR signaling in plants. Arabidopsis thaliana is insensitive to the drug rapamycin, which is extensively used to study TOR function in yeast and animal systems, which has formed a major obstacle to study TOR in plants (Menand et al., 2002; Anderson et al., 2005; Deprost et al., 2007; Sormani et al., 2007). However, a recent study shows that plants do respond to rapamycin at the concentration of 10 μM, which is 100 times more than the concentration used for yeast and mammals (Xiong and Sheen, 2012). Mutation in TOR is embryo-lethal in plants (Menand et al., 2002; Ren et al., 2011) and therefore, an alternative approach of inducible knockout for conditional inhibition of TOR has to be used for functional studies (Deprost et al., 2007; Caldana et al., 2013).
An Arabidopsis T-DNA insertion line which overexpresses AtTOR produces bigger leaves containing larger cells (Deprost et al., 2007). Accordingly, down-regulation of TOR by inducible artificial microRNA produced smaller leaves with fewer cells along with up-regulation of metabolic pathways like the Krebs cycle. These lines also accumulated storage products such as starch and lipids like triacylglycerides (TAGs) displaying reduced growth (Caldana et al., 2013). Mutation of LST8, a member of the TOR complex (TORC1) decreased plant size by producing fewer and smaller leaves alongside a higher amount of starch and amino acids and reduced sucrose concentrations. Moreover, down-regulation of genes involved in cell wall formation like expansins (EXPs) and CELLULOSE SYNTHASE-LIKE G3 in this mutant demonstrates a role for TOR in cell expansion (Moreau et al., 2012). The TORC1 component RAPTOR accumulates in dividing and expanding cells whereas mutation in AtRaptor1B slows down the leaf initiation. A double mutant of AtRaptor1A and AtRaptor1B exhibited normal seedling growth, but was unable to maintain post embryonic development (Anderson et al., 2005).
TORC1 promotes the phosphorylation of S6 kinase (S6K) and eIF4E-binding proteins (E-BP1), which controls translation and ribosome biogenesis (Krizek, 2009). Again, auxin is involved through the regulation of EBP1 protein stability in Arabidopsis (Horvath et al., 2006; Figure 5). TORC1 also regulates autophagocytosis which ensures synthesis, degradation and recycling of cellular components (Sablowski and Dornelas, 2014). Recent studies have revealed that the TOR pathway is an essential controller for cellular development, which regulates cell expansion and cell cycle simultaneously. The TOR pathway is directly connected to cell cycle regulation by mediating E2Fa phosphorylation and activation of DNA synthesis genes (Xiong et al., 2013) and regulates cell wall modification and degradation processes like senescence and autophagy (Caldana et al., 2013). Importantly, there is a link between the TOR pathway and nutrient status. TOR plays a central role in connecting growth related genes to glucose signaling (Xiong et al., 2013).
Given this central role in connecting growth regulation to nutritional status, the TOR pathway is a key regulatory hub in organ development. Although molecular insight of its functioning in animal systems is rapidly increasing, currently we are not aware of mechanistic models that include this knowledge, a void that we expect to be filled in the coming years.
In addition to cell volume increase by cytoplasmic growth, cell proliferation exponentially increases the number of cells in the developing leaf. In general cell growth needs to be sufficiently balanced by cell division for stable tissue growth (Sablowski and Dornelas, 2014). The cell division cycle is a unidirectional process, tightly regulated by a molecular mechanism that is largely conserved between all eukaryotes (Inze et al., 1999; Dewitte and Murray, 2003; Inze and De Veylder, 2006; Figure 6).
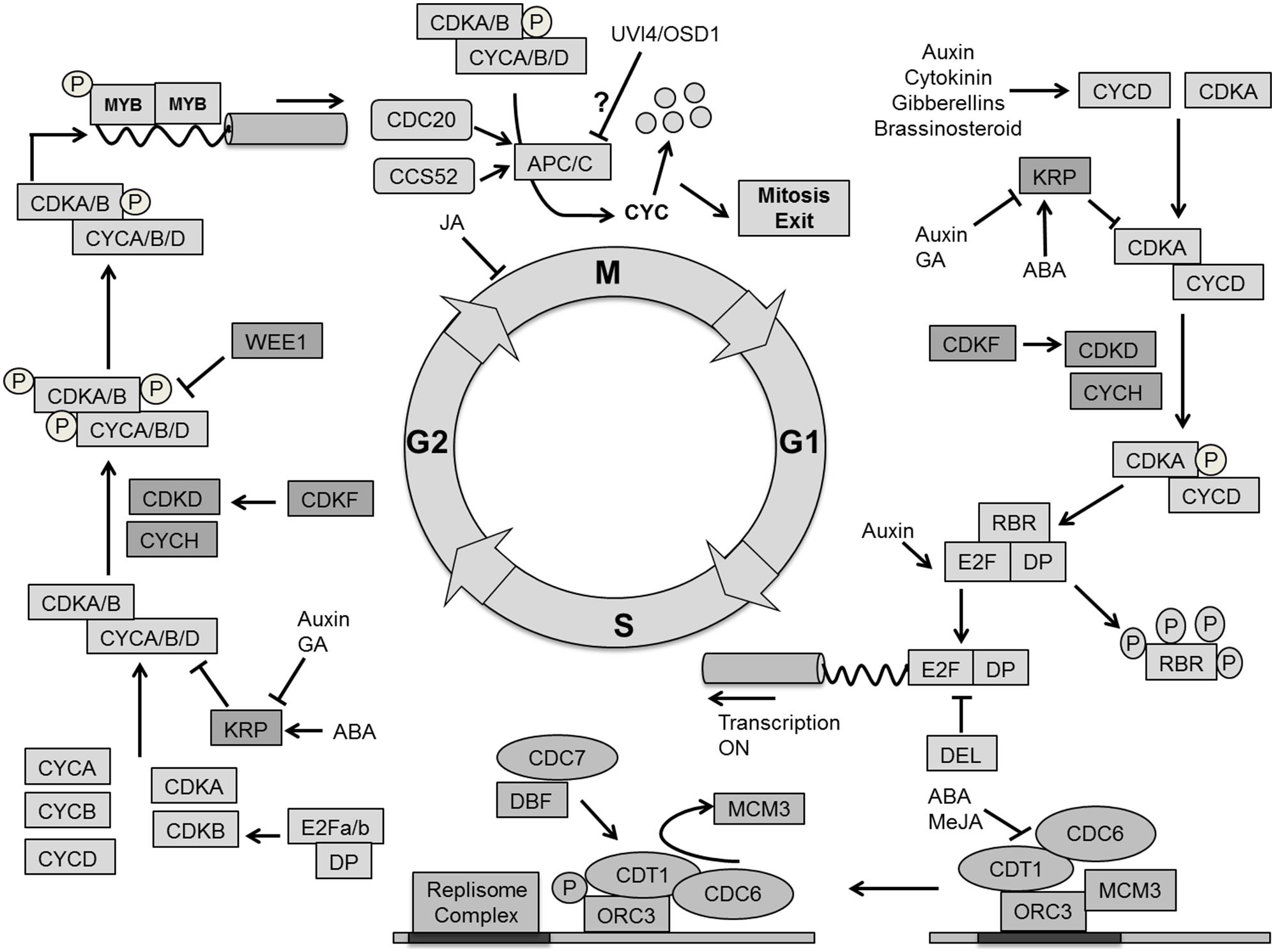
FIGURE 6. Molecular mechanism for cell cycle regulation. Four phases of cell cycle (G1, S, G2, and M) are operated by successive activation and deactivation of cyclin dependent kinases (CDKs). During the cell cycle these kinases bind with cyclins and get activated through phosphorylation by CDK activating kinases (CDKD and CDKF) whereas KRPs inhibit the complexes. G1 to S transition is controlled by CDKA–CYCD which phosphorylates the RBR proteins and releases the E2F transcription factor, which activates S phase related genes. The G2–M transition is dependent on CDKA/B and CYCA/B/D. The CDK complex is inactivated by phosphorylation through WEE1. The exit from mitosis requires proteolytic degradation of CYCs which as mediated by the Anaphase-Promoting Complex/Cyclosome (APC/C) bind with CCS52 and CDC20. Phytohormones like auxin, cytokinin, gibberellins (GA), brassinosteroids, abscisic acid (ABA) and methyl jasmonate (MeJA) impact cell cycle regulation at different points (pointed and T shaped arrows indicate positive and negative regulation and question mark indicates unknown regulation, respectively).
The plant cell cycle is controlled by the activity of complexes consisting of a cyclin-dependent kinase as the catalytic subunit and a cyclin as the regulatory subunit. A-type cyclin dependent kinase (CDKA) and D-type cyclin (CYCD) are central to the G1/S phase transition in which the cell activates DNA duplication. CDKA is a key protein to control cell division in A. thaliana, and is present at a constant level throughout the cell cycle (Porceddu et al., 2001; Joubes et al., 2004; Gaamouche et al., 2010). Overexpression of a dominant negative CDKA;1 of A. thaliana in tobacco plants inhibited cell division rate, resulting in the formation of fewer, but larger cells resulting in an overall reduction of leaf area (Hemerly et al., 1995). It has also been demonstrated that the CDKA;1 activity maintains the SAM cells in an undifferentiated state. Expressing a dominant negative allele of CDKA;1 in the STM domain of the shoot apex produces smaller leaves with a reduced number of epidermal cells (Gaamouche et al., 2010). Interestingly, irregularly shaped epidermal cells observed in a CDKA;1 dominant negative mutant expressed from the STM promoter point toward CDKA influencing cell wall and cytoskeleton properties (Borowska-Wykret et al., 2013). A triple cycd3;1–3 loss of function mutant in the Arabidopsis leaf shows a decreased cell number and reduced CK response (Dewitte et al., 2007). The plant hormones auxin, CK, brassinosteroid (BR), and GAs increase the level of CYCD, thereby activating CDKA (Riou-Khamlichi et al., 1999; Francis and Sorrell, 2001; Inze and De Veylder, 2006; Perrot-Rechenmann, 2010). The repression of ABP1 (AUXIN BINDING PROTEIN1) negatively influences transcript levels of CYCDs, which results in impaired cell division in the leaf (Braun et al., 2008), whereas BRs up-regulate the expression of CYCD3 and promote cell division through a mechanism that requires de novo protein synthesis (Hu et al., 2000). The BR biosynthesis mutant det2 (de-etiolated2 = cro1) and dwf1 (dwarf1 = cro2) produce fewer cells and a smaller leaf, which can be reversed by brassinolide application, indicating a dual role of BR in division and expansion (Nakaya et al., 2002).
The activity of CDKA/CYCD complexes is itself controlled by CDK activating kinases CDKD and CDKF coupled with CYCH, which activates the complex through a phosphorylation cascade. CDKF;1 was also found to activate CDKD;2 and CDKD;3 by T-loop phosphorylation (Umeda et al., 2005; Takatsuka et al., 2009). The active CDKA/CYCD complex triggers the dissociation of E2F/DP heterodimeric complex from RBR (retinoblastoma-related protein) through phosphorylation. Additionally, it initiates the destruction of E2Fc/DP/RBR transcriptional repressor complex by the Skp-Cullin1-F-Box (SCF) E3-ubiquitin protein ligase (Inze and De Veylder, 2006). The RBR protein regulates the activity of E2F transcription factors to control cell proliferation. It is an essential regulator of the cell cycle to coordinate between cell division, differentiation and homeostasis (Desvoyes et al., 2006; Borghi et al., 2010). Once the E2F/DP complex is separated from RBR, it initiates G1 to S transition by activating the transcription of genes required for DNA duplication. Furthermore, the E2F-like DEL transcription factors compete with E2F/DP for DNA binding sites (Figure 6).
Arabidopsis has three typical E2Fs (E2Fa, E2Fb, and E2Fc), two dimerization proteins (DPa and DPb) and three atypical E2Fs (E2F/DEL2, E2FE/DEL1, and E2Ff/DEL3; Mariconti et al., 2002). E2Fa and E2Fb stimulate entry into and progression of the S-phase and overexpression of these transcription factors leads to an enlarged phenotype due to enhanced cell proliferation (De Veylder et al., 2002; Sozzani et al., 2006). Auxin positively regulates E2Fb protein levels (Magyar et al., 2005). Additionally, the AXR1 transcript level was found to be high in an E2Fb overexpression line (Sozzani et al., 2006). On the other hand, E2Fc is a negative regulator of the S-phase where a decreased level leads to leaves and cotyledons with more but smaller cells (del Pozo et al., 2006). Auxin affects the E2Fc protein level in Arabidopsis. Mutation in the AXR1 gene leads to impaired modification of the CUL1 protein, a structural component of the E3–SCF complex, with the Ub-related protein RUB, and shows increased E2Fc protein levels (del Pozo et al., 2002). Atypical E2Fs/DELs are transcriptional regulators for endoploidization, which act independently of DPs and RBR. It is still unclear if they compete with typical E2Fs for binding sites or actively repress gene transcription (Vlieghe et al., 2005; Berckmans and De Veylder, 2009).
Activated E2Fs in Arabidopsis target the expression of genes involved in DNA repair and chromatin dynamics such as CDC6, CDT1, MCM3, ORC1 and ORC3, RNR, and PCNA. However, they also influence the expression of genes responsible for G2-M transition like CDKB1, MYB, and ANAPHASE-PROMOTING COMPLEX/CYCLOSOME (APC/C; de Jager et al., 2001; Ramirez-Parra et al., 2003; Boudolf et al., 2004b; Vandepoele et al., 2005; Lammens et al., 2008; Naouar et al., 2009). Replication origin sites are silenced in the G1 phase where CDC6 and CDT1 together with ORC allow the loading of MCM to the replication origins; hence promoting the complex for activation of the S phase. Later, the DBF–CDC7 complex phosphorylates ORC which then moves and exposes the replication initiation site for the replisome complex, allowing replication to start (Blow and Dutta, 2005; Francis, 2007). The plant hormone ABA negatively regulates the expression of the CDT1a gene (Castellano Mdel et al., 2004). Methyl jasmonate (MeJA) was also reported to affect initiation of DNA replication by inhibiting the pre-replication complex (Noir et al., 2013; Figure 6).
The Kip related proteins (KRPs) are direct inhibitors of CDK activity (ICKs). ICK1 inhibits the CDKA/CYCD complex in response to negative stimuli like abscisic acid (ABA; Wang et al., 1998; Van Leene et al., 2011). Kinematic analysis showed that the overexpression of KRP genes inhibits cell division rate, resulting in serrated leaves of reduced size with fewer but enlarged cells (De Veylder et al., 2001; Kang et al., 2007). Beemster et al. (2006) could simulate the effect on cell numbers using a computational model. Mutation of a single KRP gene does not have any effect on leaf growth, while down-regulation of multiple KRP genes increases final leaf area and cell proliferation (Cheng et al., 2013). The mechanism behind this enhanced leaf growth is yet to be explained. Auxin and CK activate CDKA and CYCD, while KRP4 transcription is down-regulated (Cho et al., 2010). It has been reported that auxin signaling is translated into modified KRP expression through PRZ1-mediated chromatin remodeling (Anzola et al., 2010). The mutual antagonistic effect of CDKA;1 and KRPs was highlighted in a model of the G1/S transition control in pollen (Zhao et al., 2012). The plant-specific F-box protein F-BOX LIKE 17 (FBL-17) is central in the proposed regulatory network, in particular by mediating the degradation of KRP1,3,4,6,7 (Kim et al., 2008). The plant-specific SIAMESE (SIM)/SIAMESE RELATED (SMR) family also inhibits CDK activity in a number of specific tissues, for instance the repression of the mitotic cycle in trichomes (Walker et al., 2000). GA is proposed to promote mitotic cycles by lowering expression of both KRP and SIM. CKIs act in a DELLA-dependent manner (Achard et al., 2009) and enhance expression of E2Fe/DEL1 (Claeys et al., 2012).
After DNA duplication, cells enter the G2 phase to prepare them for division through mitosis. CDKA and CDKB as well as CYCA, CYCB, and CYCD are involved in this process. The plant-specific B-type CDKs are subdivided into two groups: CDKB1 (with CDKB1;1 and CDKB1;2) and CDKB2 (with CDKB2;1 and CDKB2;2). CDKB1 accumulates from late S to M phase while CDKB2 is specifically expressed from G2 to M phase (Menges et al., 2005). Overexpression of a dominant negative CDKB1;1 causes early exit from the M phase, which increases the ploidy level in the leaf (Boudolf et al., 2004b). It has been reported that CDKB1;1 forms a functional complex with CYCA2;3 to trigger mitosis in Arabidopsis (Boudolf et al., 2009). Inhibition of CDKB2;1 and CDKB2;2 via the expression of an amiRNA leads to a dwarf phenotype with an abnormal SAM (Andersen et al., 2008). Jasmonates (JAs) cause G2 arrest in tobacco (Nicotiana tabacum) Bright Yellow-2 cell cultures by reducing CYCB1;1 and CDKB (Swiatek et al., 2004). Similarly, inhibition of mitotic phase genes by methyl JA causes G2 arrest in Arabidopsis cell cultures (Pauwels et al., 2008).
An additional level of regulation of CDKA/CYCD complex activity involves inactivation through phosphorylation by the WEE1 protein kinase. Overexpression of WEE1 inhibits plant growth by arresting cell division (De Schutter et al., 2007). However, in plants WEE1 is probably not a core cell cycle regulator, but rather a DNA damage checkpoint kinase (Dissmeyer et al., 2009).
Dissmeyer et al. (2009) have proposed and implemented alternative G2 phase modules in the form of mathematical models starting from an existing generic model. The primitive unicellular algae, Ostreococcus tauri, contain a bona fide CDC25, which antagonizes WEE1 phosphorylation (Inze and De Veylder, 2006). In Arabidopsis a small CDC25 like phosphatase can counteract the role of WEE1 kinase in vitro (Landrieu et al., 2004), but this CDC25-like protein has arsenate reductase activity and is most likely not involved in cell cycle regulation (Dissmeyer et al., 2010). The Arabidopsis genome therefore lacks a functional copy of the CDC25 gene, which means that generic cell cycle models (Novak and Tyson, 1993) need to be adapted to reflect the situation in plants.
Similar to the G1 phase, the CDK/CYC complex can be activated in G2 by a CDK-activating kinase pathway, involving CDKF and CDKD coupled with CYCH. The activated CDK/CYC complex promotes MYB repeat (MYB3R) transcription factors to bind with M phase Specific Activators (MAS) elements at the promoter region of the target genes. Afterward, MYB3R phosphorylation activates the expression of M phase specific genes such as KNOLLE, CDC20, CYCA, and CYCB and NACK1 (Berckmans and De Veylder, 2009; Figure 6).
There are two E3 ubiquitin ligase complexes involved in cell cycle control, SCF-PROTEIN and APC/C, which mark targets for degradation by the 26S proteasome. In analogy to the SCF complex playing an important role in G1 to S phase by degrading cell cycle inhibitors (KRPs; Fulop et al., 2005; Hershko, 2005; Ren et al., 2008) the APC/C is essential for the G2 to M transition. The exit from the M phase is regulated by the degradation of cyclins through ubiquitination by the APC/C in association with the activators CELL CYCLE SWITCH 52 (CCS52) and CDC20 (Fulop et al., 2005; Sullivan and Morgan, 2007; Marrocco et al., 2010).
Overexpression of APC10 promotes the cell division rate by degradation of CYCB1;1 which causes enlarged leaves (Eloy et al., 2011). Mutation of HOBBIT, a CDC27 subunit of the APC, in the SAM leads to accumulation of high levels of the auxin response inhibitor AXR3/IAA17, indicating that its activity would be involved in targeting AUX/IAA proteins for degradation (Blilou et al., 2002). CCS52 is an important regulator for controlling exit of mitosis. There are two classes of CCS52 in A. thaliana, CCS52A (CCS52A1 and CCS52A2) and CCS52B (Fulop et al., 2005). A-type CCS52 activators are typically expressed from late M to late S-G2 and regulate the onset of endoreduplication in leaves (Fulop et al., 2005; Lammens et al., 2008). CCS52B, like the APC/C activator CDC20, peaks from early G2 to M phase exit (Fulop et al., 2005; Kevei et al., 2011). Induced expression of CCS52B affects branching in trichomes where CCS52BOE line forms four to five branches, while the wild type has only three branches (Engler et al., 2012). However, more study is needed in particular to understand role of CCS52B in leaf development. It has been suggested that complementary phase-dependent expression of A and B-type CCS52 activators enable a fine-tuned APC/C regulation during the cell cycle (Tarayre et al., 2004). Expression of the negative regulator of CCS52A1 activity, ULTRAVIOLET-B-INSENSITIVE 4 (UVI4) peaks at the G1-to-S transition (Heyman et al., 2011) and determines cell number and size of leaves, likely through stabilization of CYCA2;3 required for mitotic cell divisions (Imai et al., 2006; Boudolf et al., 2009; Heyman et al., 2011). Its homolog UVI-Like/OMISSION OF SECOND DIVISION 1 (OSD1) influences meiosis and does not directly impact leaf size. However, it is expressed during the mitotic cell cycle peaking at the G2-to-M transition, possibly preventing endomitosis (=incomplete mitosis; Heyman and De Veylder, 2012). Besides the mechanism of the inhibitory action of the UVI4 and OSD1 regulators, not much is known about the specific targets of the APC/C (Heyman and De Veylder, 2012). Apart from a recent study pointing to CK up-regulating CCS52A1 in the Arabidopsis root (Takahashi et al., 2013), not much is known about the role of hormone signaling on APC/C regulation. However, in stress-conditions GA likely modulates APC/C activity through DELLA dependent down-regulation of the UVI4 and DEL1 negative regulators (Claeys et al., 2012).
Despite of the high level of conservation of the core cell cycle machinery (Harashima et al., 2013), many plant-specific features exist. Plants are characterized for instance by a remarkably broad cyclin family with many species-specific isoforms (at least 49 in Arabidopsis, cf. Sablowski and Dornelas, 2014). The involvement of different orthologous of many core cell cycle genes goes together with a functional diversity which manifests itself in gene expression differences between species, developmental and environmental conditions. Indeed, Beemster et al. (2005) showed by means of a microarray study that the expression profile of roughly half of all cell cycle genes differed between roots and leaf primordia. de Almeida Engler et al. (2009) found generally high expression of core cell cycle genes in leaf primordia, in the lamina of young leaves and in vascular tissue of expanding leaves. A number of cases of developmental stage-specific expression are described in the relevant sections throughout this review. However, for many others the functional significance has yet to be clarified.
Generally cells go through a regular cell cycle with the S phase (DNA duplication) followed by the M phase (mitosis). Endoreduplication, endoreplication, endoploidization or, in short, the endocycle is the process whereby DNA replicates repeatedly without alternating divisions through mitosis, causing a high ploidy level in the cell.
To establish endoreduplication, the CDK activity essentially has to be kept low enough and several ways have been proposed to achieve this (Berckmans and De Veylder, 2009; De Veylder et al., 2011). For instance, CYCD3;1 which is specifically expressed in proliferating tissues, reduces endoploidization (Dewitte et al., 2003). CDKB1 activity is also essential for the G2 to M transition (Beemster et al., 2005). Overexpression of a dominant negative CDKB1;1 interferes with cell cycle progression causing G2 arrest (Boudolf et al., 2004a). CDKB1;1 forms an active complex with CYCA2;3 to suppress endoreplication in the leaf (Boudolf et al., 2009). Loss of CYCA2;3 function increases ploidy in mature leaves (Imai et al., 2006). The INCREASE LEVELS OF PLOIDY (ILP1) gene, which encodes a protein homologous to the C terminal region of mammalian GC binding factor, is proposed to be involved in transcriptional repression of A2-type cyclins (Yoshizumi et al., 2006). Expression of B-type cyclins, on the other hand, was repressed by decreased phosphorylation of three-repeat MYB proteins (MYB3Rs; Ito et al., 2001; De Veylder et al., 2011). The E3 ubiquitin ligase complex, APC/C coupled with CCS52 influences endocycle onset by controlling proteolytic degradation of G2-M specific cyclins like CYCB1;1 and CYCB1;2 (Kasili et al., 2010) as well as CYCA2;3 (Boudolf et al., 2009; Figure 7). CCS52A1 and CCS52A2 knockout plants have reduced DNA ploidy levels in leaves (Lammens et al., 2008; Kasili et al., 2010). The previously mentioned plant-specific CCS52A1 inhibitor UVI4 is likely involved in securing the G2-to-M transition and therefore preventing endocycle onset (Heyman and De Veylder, 2012). Cells with increased ploidy levels in osd1 cotyledons and the developmentally severely compromised uvi4 osd1 suggest some functional redundancy between UVI4 and its homolog OSD1 (Iwata et al., 2011; Cromer et al., 2012). Mutation in SAMBA, a plant specific subunit of the APC complex which probably activates A2-type cyclin degradation, induces enhanced endoreplication in Arabidopsis leaves (Eloy et al., 2012). Other factors such as the DP/E2F like transcription factor E2Fe/DEL1 are involved in controlling APC/C activity. Down-regulation of DEL1 triggers the expression of the CCS52A2 gene, forcing cells to enter endoreduplication (Lammens et al., 2008).
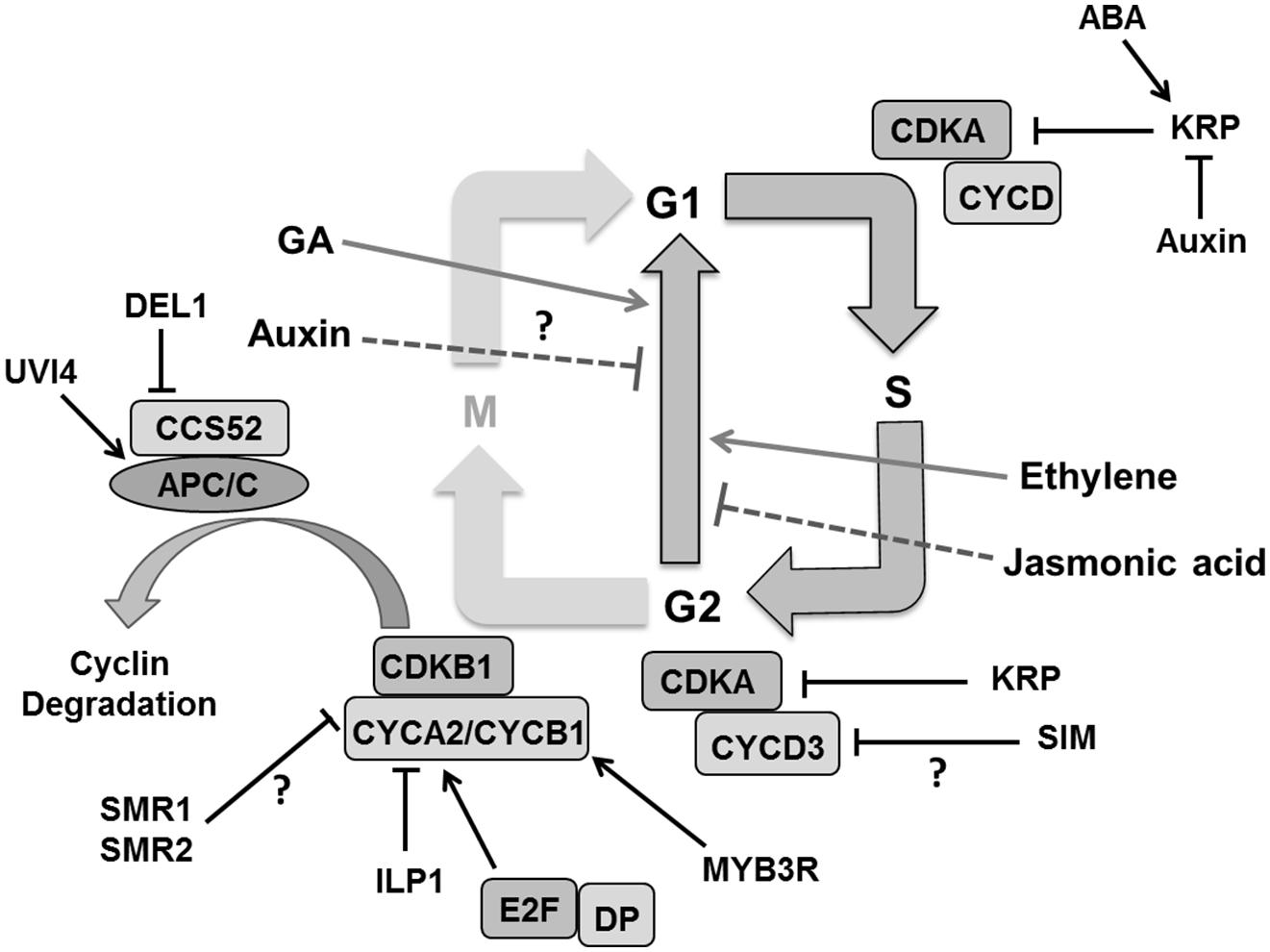
FIGURE 7. Regulation of endoreduplication. CDKA/B and CYC activity is inhibited by cell cycle inhibitors KRPs/SIM/SMRs that induce endoploidization. Cyclin A is inhibited by the ILP1 proteins whereas down-regulation of MYB3R causes decreased CYCB and ultimately induces endoploidization. Proteolytic degradation of G2–M cyclins by the APC/C complex causes endocycle onset. Other factors like, UVI4 and DEL1 suppress the endocycle by inhibiting the APC/C complex. Plant hormones like auxin and jasmonic acid suppress the endocycle whereas gibberellins (GA), abscisic acid (ABA), and ethylene stimulate it by regulating expression and activity of different components (pointed and T shaped arrows indicate positive and negative regulation and question mark shows the unknown regulation, respectively).
The plant specific cell cycle inhibitor SIM gene that encodes for a member of the SMR family also plays a role in endoreduplication (Walker et al., 2000; Churchman et al., 2006). A mutation in SIM causes repressed endoreplication leading instead to mitotic divisions in leaf trichomes. SIM interacts with CDKA;1 and D type CYCs (Churchman et al., 2006), and it was suggested that inhibition of CDKA:CYCD3 complexes might be the mechanism responsible for its role in endoreduplication onset (De Veylder et al., 2011). Indeed, CYCD3;1 overexpression inhibits endoreduplication in Arabidopsis leaves (Dewitte et al., 2003), whereas a cycd3 triple mutant displays premature onset of endoreduplication in young leaves (Dewitte et al., 2007). Other SIM family members, such as SMR1/LGO, might also promote polyploidization (Roeder et al., 2010). In fact, SMR1 and SMR2 interact with CDKB1;1 and its interactor CYCB2;4 associates with SMR11 (Van Leene et al., 2010).
Contrary to the emerging insights into endoreduplication onset, it is only poorly understood how the endocycle is sustained. It has been envisaged that the cell cycle inhibitor KRP controls CDKA activity by inhibiting the CDKA/CYCD complex to maintain the CDK oscillations needed for DNA replication in the endocycle. KRPs were reported to regulate mitosis and endoreplication in a dose dependent manner where low concentrations promote the endocycle while high levels cause cell cycle arrest (Verkest et al., 2005b). On the one hand, overexpression of KRP2/KRP5 in mitotically active cells inhibits cell division and enhances endoreplication (Verkest et al., 2005a; Jegu et al., 2013) while on the other hand its overexpression in postmitotic cells inhibits endocycle in Arabidopsis leaves (Schnittger et al., 2003). This implicates that KRPs are an important candidate for the regulation of rate and duration of endoreduplication in expanding leaf cells.
Arabidopsis leaves, cells enter into the endoreduplication process as a consequence of decreasing auxin concentrations. It has been observed that the mutants in auxin signaling, biosynthesis and transport show a rapid transition from mitotis to endocycle causing increased ploidy level in cotyledons (Ishida et al., 2010) but the detail of this mechanism is still not known. Ethylene and GAs are hypothesized to positively affect endoreduplication (Gendreau et al., 1999; Perazza et al., 1999; Swain et al., 2002). JAs were shown to inhibit cell proliferation as well as endoploidization in a COI1 (encoding an F-box protein which is a part of the SCF complex) dependent manner in Arabidopsis leaves (Noir et al., 2013). It also negatively regulates the expression of key determinants of DNA replication like CDC6A (Noir et al., 2013).
Despite of our increasing knowledge on the molecular mechanism of endoreduplication, its actual function remains ambiguous with proposed roles in promoting cell expansion, stress resistance or DNA damage protection (De Veylder et al., 2011). In any case, modulating CDK–CYC activity levels in various ways remains a central principle. This pertains to the ubiquitin dependent degradation of KRPs or the mechanism by which plant specific cell cycle inhibitors (SIM/SMR) as well as developmental and environmental signals influence the endocycle. Roodbarkelari et al. (2010) have modeled endocycle onset in Arabidopsis trichomes with KRP and the CULLIN4 ubiquitin ligase controlling G1/S and SIM and APC/C controlling G2/M transitions. Computational approaches to predict tissue distributions of cell ploidy combined with in vivo ploidy maps (Boudolf et al., 2004b) would provide powerful insights to better understand its regulation and relationship to cell expansion.
Leaf development involves two major phases. The first phase is dominated by proliferative activity and the second phase by cell expansion (Figure 8). There is a correlation between cell division activity and organ growth, so the timing of cell division has a large influence on the final leaf size (Korner et al., 1989; Meyerowitz, 1997; Gonzalez et al., 2012). As cell division ceases the cell continues expanding. This transition from division to expansion is manifested as a cell cycle arrest front which remains fixed at some position for a particular time period and then moves rapidly toward the base of the leaf blade (Andriankaja et al., 2012). Several regulators appear to control the transition from proliferation to expansion. Auxin plays an important role in the transition phase. It induces the expression of AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE (ARGOS) gene, encoding for an ER localized protein of unknown function (Hu et al., 2003). Overexpression and down-regulation of ARGOS increases and decreases leaf size, respectively. It regulates the action of a DNA-binding protein ANT (AINTEGUMENTA) and of CYCD3;1 (Hu et al., 2003; Figure 8). Loss of function of ANT blocks the increase in leaf growth in ARGOS overexpressing plants. The Arabidopsis ORGAN SIZE RELATED1 (ORS1) shares a conserved domain with ARGOS and ARGOS LIKE (ARL), and positively regulates cell division and expansion in the leaf (Hu et al., 2003; Feng et al., 2011). Like the ANT family proteins, the GROWTH REGULATING FACTOR (GRF) and TCP transcription factors are essential regulators of leaf growth (Figure 8). The Arabidopsis GRF family comprises nine members. Overexpression of AtGRF1 and AtGRF2 results in larger leaves whereas the grf1/2/3 triple mutant reduces leaf size, both as a result of alterations in cell size (Kim et al., 2003). Overexpression of GRF5 increased cell number with prolonged growth, whereas the grf5-1 mutant shows narrow leaves with reduced cell numbers. GIF1 (GRF-INTERACTING FACTOR1), also known as ANGUSTIFOLIA3 (AN3) interacts with GRF5 (Horiguchi et al., 2005). GIF1 overexpression increases leaf size with leaves having more cells, whereas its absence reduces cell proliferation (Lee et al., 2009).
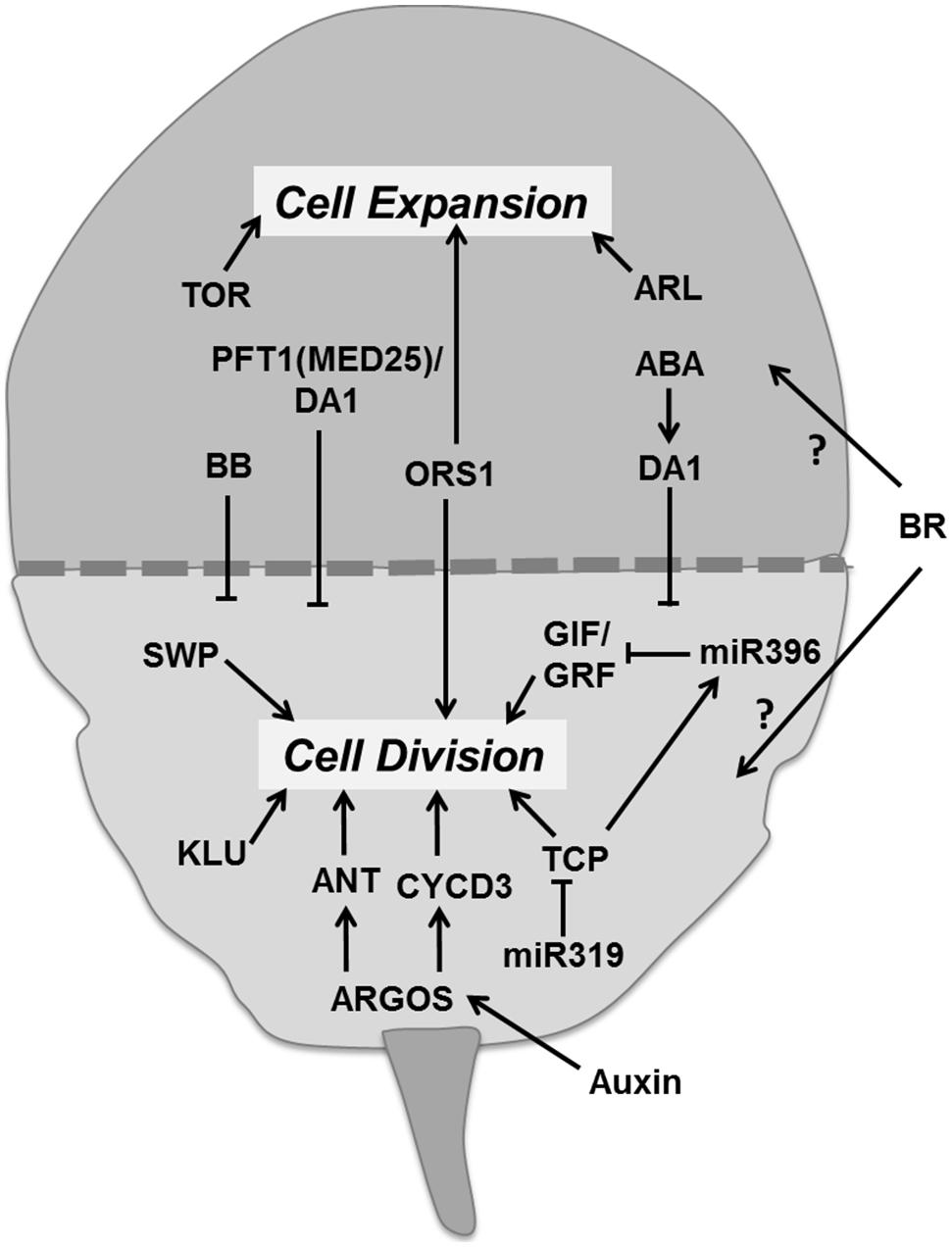
FIGURE 8. Regulation of the transition between proliferation and cell expansion. The transition between division and expansion is shown by a dashed line which separates these two growth processes according to their regulators. ARGOS promotes cell division via DNA binding protein ANT and CYCD3 which is regulated by auxin. TCP and GIF/GRF transcription factors promote division and are negatively regulated by miRNAs. Other factors like KLU and SWP also promote proliferation. Some factors like ORS1 have a positive influence on division as well as expansion. Cell expansion is directly controlled by the TOR pathway and ARL. Whereas, other regulators like BB, MED25, and DA1 control the timing of proliferation. Abscisic acid promotes transition at least in part by regulating DA1 whereas brassinosteroids with unknown molecular mechanism (pointed and T shaped arrows indicate positive and negative regulation of the particular process and question marks show unknown mechanisms, respectively).
miR396 negatively regulates six members of Arabidopsis GRF together with GIF1 (Liu et al., 2009; Figure 8). Interestingly, overexpression of miR396 in a mutant deficient for GRF1 reduces SAM size (Rodriguez et al., 2010). The miR396 targeted GRFs are also essential for leaf polarity (Wang et al., 2011). The TCP family of transcription factors regulates the expression of miR396 and miR319 (Palatnik et al., 2003; Rodriguez et al., 2010). A point mutation in the miR319 target site of TCP4 induces miR396 which in turn decreases GRF expression and results in smaller leaves (Figure 8). Similarly, overexpression of TPC4 decreases leaf size (Rodriguez et al., 2010). Transcription factors from the TCP family such as CINCINNATA (CIN) of Antirrhinum, LANCEOLATE (LA) of tomato and CIN-TCPs of A. thaliana control cell cycle arrest (Nath et al., 2003; Palatnik et al., 2003; Ori et al., 2007). In Arabidopsis, up-regulation of miR319 in the jaw-D mutant reduces the expression of TCP2, TCP3, TCP4, TCP10, and TCP24 producing large and wrinkled leaves (Palatnik et al., 2003). Down-regulation of single, double, and triple TPC genes resulted in proportional increase in leaf size and crinkliness (Schommer et al., 2008).
In addition to these transcription factors, other genes are also essential to promote the transition from division to expansion. The putative ubiquitin binding protein DA1 and the E3 ubiquitin ligase BIG BROTHER (BB) also known as ENHANCER OF DA1-1 (EOD1) controls organ size by restricting the duration of cell proliferation (Figure 8). In the da1-1 mutant, the production of a dominant negative protein negatively affects both DA1 and the DA1-related (DAR) protein and the overexpression of DA1 results in large leaves with increased cell numbers. ABA induces the expression of DA1, whereas the da1-1 mutant was less sensitive to ABA, implicating a role for ABA in determining final leaf size through control of mitotic exit (Li et al., 2008). Mediator complex subunit 25 (MED25 also known as PFT1), functions together with DA1 in controlling leaf growth by restricting cell proliferation (Figure 8). Overexpression of MED25 causes smaller leaves with reduced cell numbers and cell sizes, whereas a loss of function mutant enhances organ size with increased duration of cell proliferation and expansion (Xu and Li, 2011). Loss of function mutation of the RING-finger protein encoding BB leads to enlarged leaves and small changes in expression levels substantially alter organ size suggesting it controls cell division and leaf size in a dose dependent manner (Disch et al., 2006). The KLUH (KLU)/CYP78A5 gene, encoding for a cytochrome P450, required for generating a mobile growth signal distinct from the classical phytohormones, is also an essential regulator for leaf size control. Overexpression of KLU induces enlarged leaves having more cells whereas in the klu mutant premature arrest of cell proliferation causes smaller leaves (Anastasiou et al., 2007; Stransfeld et al., 2010). The SWP gene encodes a protein with similarities to subunits of the Mediator transcriptional regulatory complex of RNA polymerase II. It also plays a role in defining the period of cell proliferation. In the swp mutant leaf size was reduced due to less cells, which was partially compensated by an increase in final cell size (Autran et al., 2002; Figure 8).
Like auxin, sugar signaling controls leaf growth possibly via the ARGOS pathway (Hu et al., 2003; Wang and Ruan, 2013). BR also regulates leaf growth by controlling cell division and expansion. The BR deficient mutant constitutive photomorphogenesis and dwarfism (cpd) produces smaller leaves with fewer cells of reduced size (Zhiponova et al., 2013); however, the molecular mechanism controlling this process is yet to be clarified. The progressive general cell proliferation arrest front of epidermal and mesophyll cells is followed by a second cell cycle arrest front for dispersed meristematic cells (DMCs) that is controlled by the putative transcription factors PPD1 (PEAPOD1) and PPD2 (White, 2006).
The transition to the expansion phase is essentially dependent on the regulators of cell cycle arrest. Many factors have been implicated in the regulation of the cell division arrest front. An important question is how the spatiotemporal dynamics of the arrest front could be explained. Coordination through one or more gradients of (non-cell autonomous) growth regulators appears to be the most likely mechanism. However, Efroni et al. (2008) hypothesized a mechanism for organ differentiation through an internal self-advancing sequential maturation program, where rate and time of advancement is regulated by a cell-autonomous developmental clock (timed) program. CIN-TCPs would play a governing role in this mechanism for the leaf. Sufficient details for building simulation models of the transition phase still appear to be lacking in Arabidopsis. However, in the monocotyledonous maize leaf a clearer picture is arising, where a peak in the activity of GA is instrumental in regulating the spatial location of the transition (Nelissen et al., 2012). Possibly models that will be developed for this monocotyledonous system can be adapted to better understand the same process in the Arabidopsis leaf.
Cell expansion is an essential step in determining final leaf size that is governed by different mechanisms in each stage of cellular development. Expansion in meristematic cells is determined by both increases in cytoplasmic and nuclear volume whereas in differentiated tissues it is mainly determined by turgor-driven vacuolar enlargement that allows the accumulation of water and solutes (Wolf et al., 2012; Sablowski and Dornelas, 2014). Turgor driven cell expansion is the result of multiple steps like cell wall relaxation to accommodate water uptake, wall extension by turgor pressure, dehydration/cell wall stiffening and the accumulation of cell wall components (Cosgrove, 2005; Wolf et al., 2012; Figure 9).
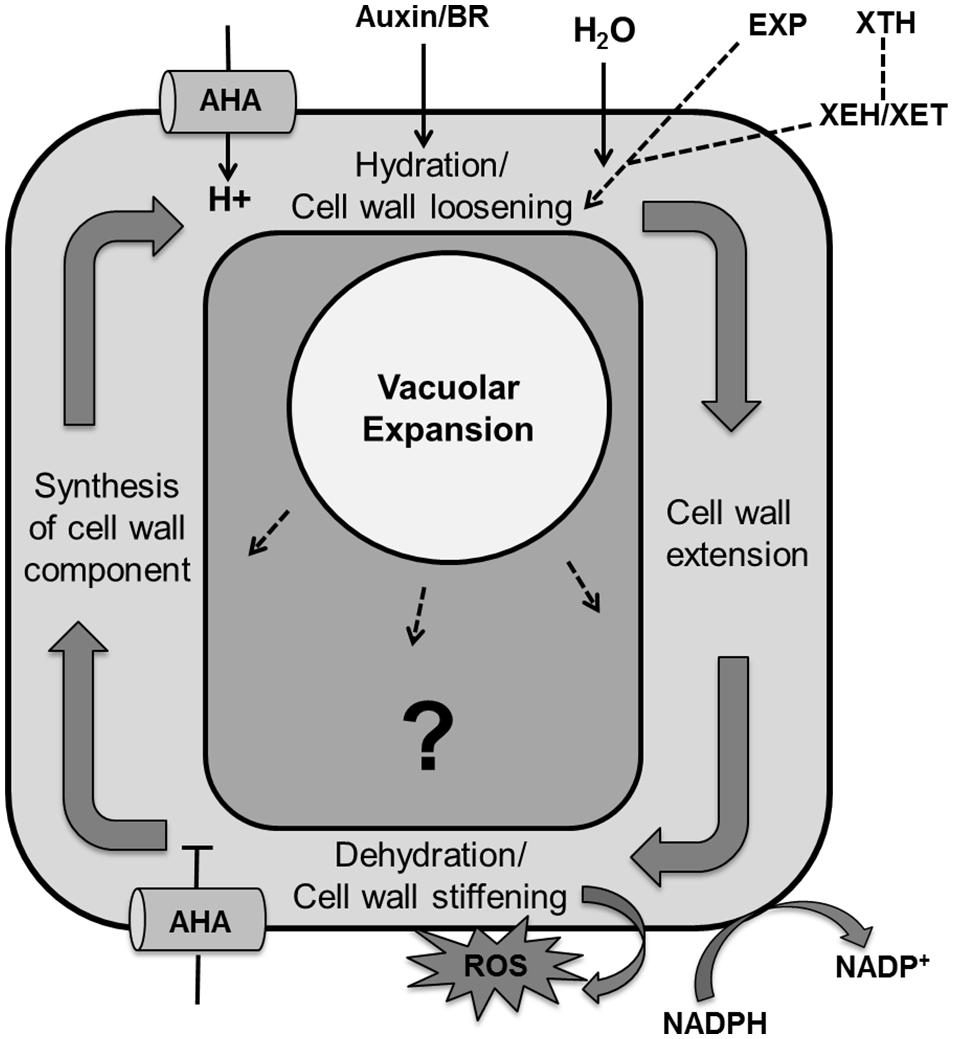
FIGURE 9. Regulation of the cell expansion process with unknown molecular mechanism. Cell expansion is the result of vacuolar enlargement as well turgor driven cell wall yielding. The vacuole expands while taking up water and solutes whereas turgor driven cell wall yielding is the result of multiple steps, including hydration and cell wall loosening, cell wall extension by turgor pressure, dehydration/cell wall stiffening by release of apoplastic reactive oxygen species, cross-linking and dehydration and lastly synthesis and accumulation of cell wall components. Cell wall loosening is controlled by expansin (EXP) proteins and the xyloglucan endohydrolase (XEH) and xyloglucan endotransglucosylase (XET) activities of xyloglucan endotransglucosylase/hydrolases (XTHs). Auxin and brassinosteroid (BR) enhance activity of P-type plasma membrane proton ATPase (AHA; pointed and T shaped arrows indicate positive and negative regulation and question mark shows the unknown regulation, respectively).
In the plant cell wall cellulose microfibrils are associated through hemicellulose tethers to form the cellulose–hemicellulose network, which is embedded in a pectin matrix. Primarily, auxin or brassinolide induce activity of P-type plasma membrane proton ATPase (AHA) that causes acidification of the apoplast and in turn activates hydration and cell wall loosening by EXP proteins and xyloglucanendotransglucosylase/hydrolases (XTHs), xyloglucan endohydrolase (XEH), and xyloglucan endotransglucosylase (XET; Yokoyama and Nishitani, 2001; Rose et al., 2002; Caesar et al., 2011; Wolf et al., 2012). Dyson et al. (2012) developed a model of hemicellulose dynamics in an expanding cell wall, showing how the action of XTH and EXP family enzymes determine yield and extensibility of the wall as encapsulated by the classical Lockhart equation (Lockhart, 1965). The mechanism controlling cell wall swelling/hydration is yet to be clarified. Antisense and sense constructs of the Arabidopsis EXP10 gene produce smaller leaves with altered morphology and larger leaves having bigger cells, respectively (Cho and Cosgrove, 2000). Wall hydration allows cell wall extension through structural alterations. Cell wall relaxation stretches the plasma membrane which promotes opening of Ca2+ channels. The resulting increase in cytoplasmic calcium affects growth by inhibiting P-ATPases that cause alkalization of the apoplast and inhibition of EXP activity. It also activates NADPH-oxidase which promotes secretion of superoxide into the cell wall, which is further converted into hydrogen peroxide. These reactive oxygen species promote cross linking of cell wall components, which causes cell wall dehydration and strengthening. At the end, wall thickness is reinstated by biosynthesis of membrane lipids, cell wall components and proteins, and appropriate channelization of these materials to their final cellular destination (Wolf et al., 2012).
Auxin does not always promote cell expansion as its concentration has also been observed to fall during leaf expansion (Braun et al., 2008), suggesting a more complicated dose–response relation. The yucca and sur mutant of Arabidopsis have elevated auxin levels and smaller leaves (Boerjan et al., 1995; Zhao et al., 2001). Mutation in EXIGUA (EXI) genes, which encode for different subunits of cellulose synthase complex required for secondary cell wall biosynthesis, produces small leaves having defects in cell expansion (Rubio-Díaz et al., 2012). Growth anisotropy, the existence of directions with distinct growth properties is determined by the orientation of the stiff cellulose microfibrils, which in turn is controlled by the orientation of cortical microtubule (CMT) arrays guiding cellulose synthase (Paredez et al., 2006). Uyttewaal et al. (2012) showed by experimental and modeling approaches that the microtubule severing protein katanin mediates the response of cells to mechanical stress in the Arabidopsis SAM. The alignment between PIN1 polarity and microtubule orientation in the SAM indicates a tight biophysical coupling between morphogenesis and auxin transport as further corroborated by mathematical modeling (Heisler et al., 2010).
Cell ploidy level is strongly correlated with mature cell size in many plant species (Sugimoto-Shirasu and Roberts, 2003). Alteration in genes specific for G2–M transition affects the onset of endocycle with earlier onset typically leading to enhanced ploidy. A notable exception is down-regulation of Arabidopsis REGULATORY PARTICLE AAA-ATPASE (RPT2a), which encodes a subunit of the 26S proteasome that causes enlarged plant organs having less but bigger cells. DNA content was higher in some organs but not in all which suggests that the increase in organ size was not the result of endoploidization (Kurepa et al., 2009). Another exception is KRP2 overexpression in Arabidopsis, which does not alter timing of cell cycle exit, but induces fewer and enlarged cells in combination with lower endoploidy levels (De Veylder et al., 2001). A recent study of different Arabidopsis mutant and transgenic lines with altered cell sizes showed strong differences in the effect of a same doubling of nuclear ploidy levels, by tetraploidization, on mature cell size (Tsukaya, 2013a). This indicates that genetic factors strongly affect and complicate the general relationship between endoploidy and size, by thus far unknown mechanisms.
In the process of leaf development, cells have the ability to differentiate into distinct cell types such as guard cells, vascular tissue cells, and trichomes, enabling them to perform diverse specialized functions. All these cell types develop from undifferentiated proliferating cells in the young primordium under the control of regulatory pathways that are increasingly being elucidated.
In Arabidopsis, guard cell development is initiated by an asymmetric cell division of a protodermal cell. The two daughter cells obtain different identities; the larger one maintains protodermal cell identity, whereas the smaller one becomes a meristemoid mother cell (MMC). The MMC divides asymmetrically to produce a larger stomatal lineage ground cell (SLGC) and smaller meristemoid. Subsequently these SLGCs give rise to new meristemoids by asymmetric division. The meristemoid can differentiate into a guard mother cell (GMC), which divides symmetrically to form a pair of guard cell precursors, which further differentiate into guard cells (Vaten and Bergmann, 2012). Two closely related two-MYB-repeat transcription factors, FOUR LIPS (FLP) and MYB88 restrict this final symmetric division to one (Lai et al., 2005). Interestingly, termination of the final division happens through transcriptional repression of the core cell cycle genes CYCA2;3 and CDKB1;1 (Xie et al., 2010; Vanneste et al., 2011). Transition of individual cell into the stomatal lineage is regulated by three helix–loop–helix (bHLH) transcription factors: SPEECHLESS (SPCH), MUTE, and FAMA (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007; Figure 10).
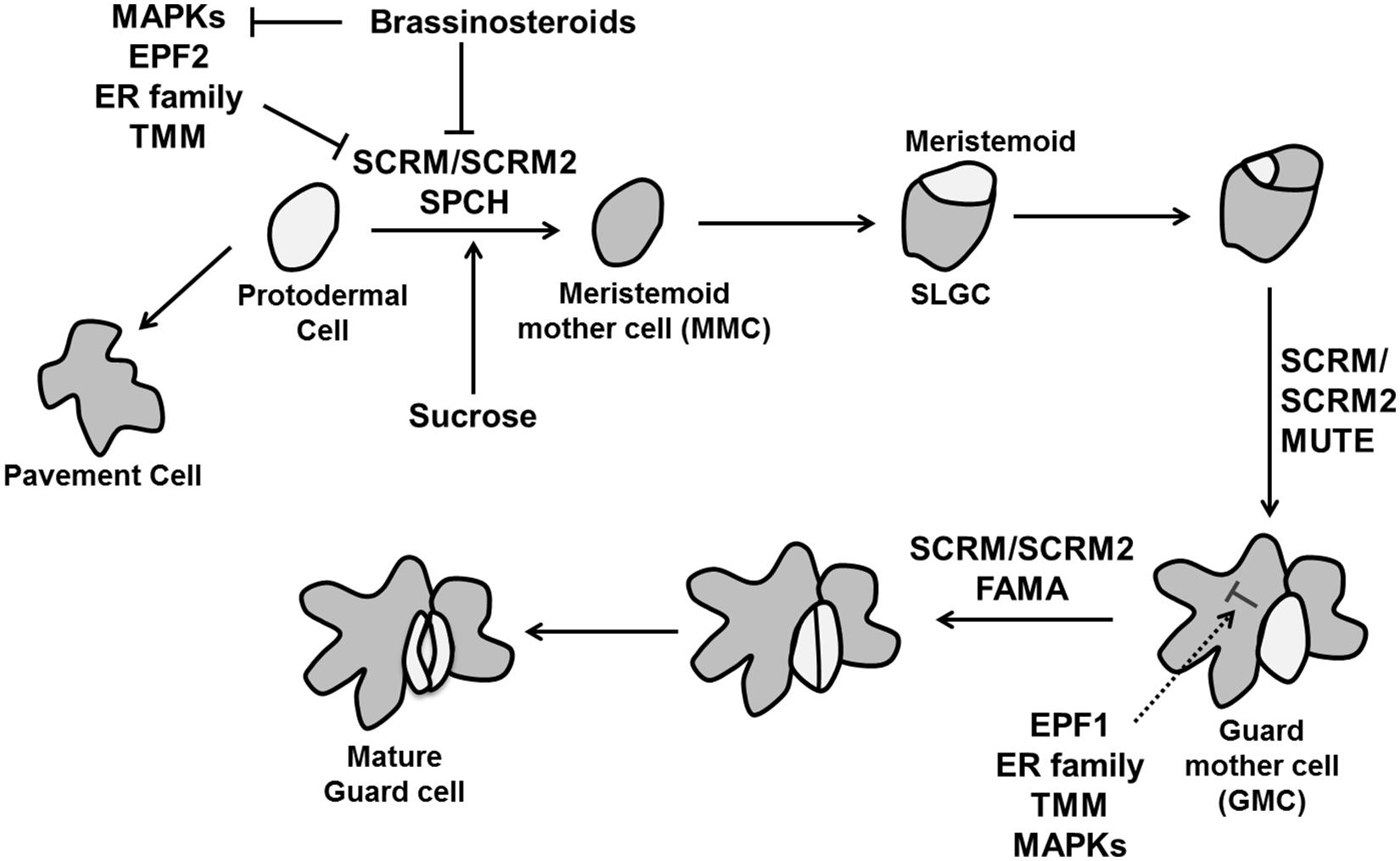
FIGURE 10. The control of stomatal development. Stomatal fate is determined by three transcription factors, SPEECHLESS (SPCH), MUTE, and FAMA. Specification of stomatal lineage where conversion of a protodermal cell into a meristemoid mother cell (MMC) is regulated by SPCH, MUTE controls the transition from meristemoid to guard mother cell (GMC) and FAMA is essential to make functional guard cells from GMC. The MAPK signaling cascade including the MAPK kinase YODA, MPKK4/5/7/9 and MAPKs (MPK3/6), EPIDERMAL PATTERNING FACTORs (EPF1 and EPF2) perceived by TMM and the ER family inhibit stomatal identity in non-stomatal cells. Brassinosteroids negatively regulate SPCH as well MAPKs simultaneously (pointed and T shaped arrows indicate positive and negative regulation, respectively).
The initial asymmetric division when protodermal cells enter the stomatal lineage is controlled by SPCH. Overexpression of SPCH initiates extra asymmetric cell divisions while no stomatal lineage was found in the spch mutant. MUTE is essential to transform a meristemoid into a GMC. Loss of function mutation of MUTE leads to the production of stomatal precursors but no stomata, whereas its overexpression converts the whole epidermis into stomata (MacAlister et al., 2007; Pillitteri et al., 2007). Lastly, FAMA is required for the conversion of GMCs into guard cells. The GMC divides rapidly in fama mutants, but the daughter cells do not differentiate, producing a row of parallel cells (Ohashi-Ito and Bergmann, 2006). A second group of bHLH proteins are INDUCER OF CBF EXPRESSION1/SCREAM (ICE1/SCRM) and SCRM2, which associate with SPCH, MUTE, and FAMA to activate sequential stomatal fate transition (Figure 10). Gain of function scrm-D causes conversion of epidermal into stomatal cell identity and loss of SCRM and SCRM2 resembles spch, mute, and fama mutant (Kanaoka et al., 2008).
Intracellular signaling pathway analysis revealed that stomatal patterning is regulated by interaction among three leucine-rich repeat receptor kinases (LRR-RLKs): ERECTA (ER), ERECTA-LIKE1 (ERL1), and ERL2 (Shpak et al., 2005), peptides of the EPIDERMAL PATTERNING FACOR-LIKE (EPFL) family (Hara et al., 2009) and the LRR-receptor-like protein, TOO MANY MOUTHS (TMM; Nadeau and Sack, 2002; Figure 10). EPF1 and EPF2 expressed in GMC and MMC, respectively (Peterson et al., 2010) control the number of guard and non-guard cells. Loss of function mutants of either EPF1 or EPF2 produces more stomata, whereas overexpression inhibits stomatal development (Hara et al., 2009). In contrast, another member of the EPF family EPFL9/STOMAGEN is a positive intercellular signaling factor involved in stomatal development (Sugano et al., 2010).
Members of the ER family also work as negative regulators with their down-regulation causing over-proliferation of stomata (Shpak et al., 2005). TMM affects stomatal spacing and density and its loss of function mutant tmm forms clusters of stomata in leaves (Nadeau and Sack, 2002). These intracellular signals in turn activate a mitogen-activated protein kinase (MAPK) signaling cascade including the MAPK kinase YODA, MPKK4/5/7/9, and MAPKs (MPK3/6) to inhibit stomatal development in neighboring cells (Bergmann et al., 2004; Lampard et al., 2009; Figure 10). MAPK mediated phosphorylation negatively regulates SPCH activity (Lampard et al., 2008), whereas the target in later stage of stomatal development is unknown. A recent study adds to the complexity of this network since the BR pathway phosphorylates YODA (Kim et al., 2012) and SPCH (Gudesblat et al., 2012; Figure 10). Thus, it is essential to understand the regulation of MAPK pathway in later stages of the stomatal development and shed light on the complex interaction between YODA and SPCH with BR. It is an interesting question how the stomatal lineage is established and which regulators cause the initiation of SPCH expression. In relation to that, a polarity-switching model for individual lineage behavior was able to predict the location of the polarity determinant BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL) over multiple divisions leading to stereotypical spatial patterns of stomata lineages (Robinson et al., 2011). Sugar signaling is also involved as an early signal, as sucrose, glucose and fructose all induce ectopic stomatal formation by inducing stomatal lineage markers in non-stomatal lineage cells (Akita et al., 2013).
At the time of leaf initiation, high local concentrations of auxin induce provascular identity leading to the differentiation of midvein and lateral veins preceded by enhanced expression of early markers for vascularization, e.g., ATHB8 (Arabidopsis homeobox transcription factor; Scarpella et al., 2004, 2006; Bayer et al., 2009). A Dual Polarization model proposed by Bayer et al. (2009) explains PIN1 protein localization at the time of leaf initiation and midvein formation. Generally, vasculature development begins with the formation of pre-procambium cells, which later differentiate into procambium cells under control of increased auxin flow (Kang and Soh, 2001; Scarpella et al., 2006). Xylem and phloem cells are produced by the vascular meristem with xylem produced on the dorsal (adaxial) side and phloem produced on the ventral (abaxial) side of the procambium. The radial patterning of the vascular bundle is the result of an antagonistic relation between Class III HD-ZIP (Class III Homeodomain Leucine Zipper) in the xylem domain and KAN transcription factors in phloem precursor cells (Jung and Park, 2007; Figure 11).
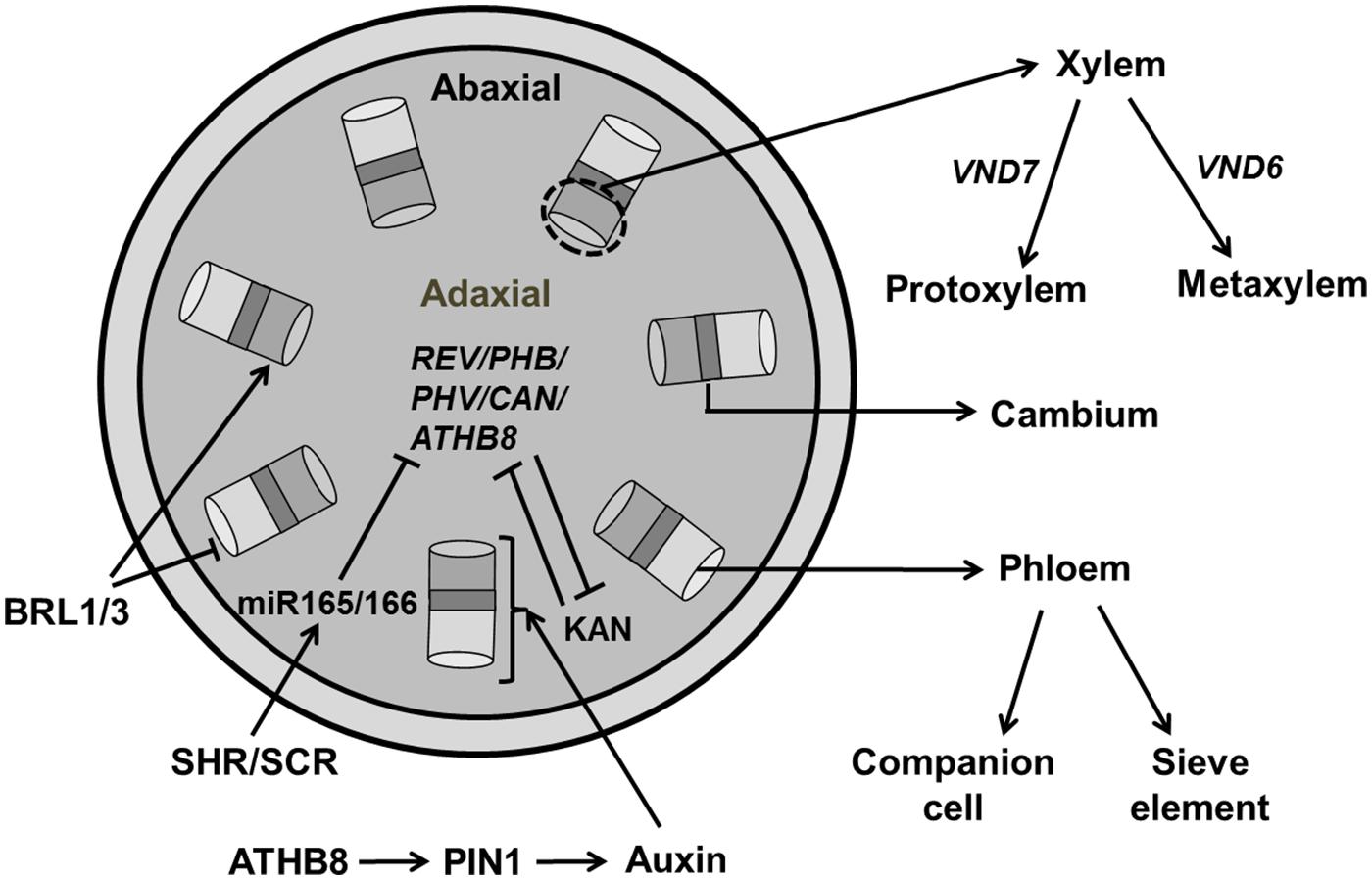
FIGURE 11. Regulation of vascular development. Central regulators for vascular development involves the REV/PHB/PHV/CAN/ATHB8 genes which are members of HD-ZIP III family and KAN (KANADI). These regulators act antagonistically to maintain xylem and phloem, respectively. Transcription factors, SHR (SHORT ROOT) and SCR (SCARECROW) activate miR165/166, which further inhibits HD-ZIP III. Auxin plays an essential role in regulating vascular formation through PIN1 transporter by early markers like ATHB8. BRASSINOSTEROID INSENSITIVE 1 (BRI1) family, BRI1-LIKE (BRLs) inhibit phloem formation while inducing xylem formation. Expression of VND6/7 affects the formation of proto/metaxylem (pointed and T shaped arrows indicate positive and negative regulation, respectively).
Of all five members of the HD-ZIP III family, PHV, PHB, and REV are expressed in vasculature, apical, and floral meristems, and the adaxial domain of lateral organs (McConnell et al., 2001; Emery et al., 2003) whereas ATHB8 and ATHB15 are exclusively expressed in vascular tissue (Baima et al., 2001; Ohashi-Ito and Fukuda, 2003) and these factors are negatively regulated by microRNA 165/166 (Emery et al., 2003). HD ZIP-III transcription factors are regulated by two members of GARS family of transcription factors, SHR (SHORT ROOT) and SCARECROW (SCR) which activate the genes encoding miR165/166 (Miyashima et al., 2013). Recently, it has been reported that the synchronous expression of SHR and ATHB8 is important for the transition to the pre-procambial cell state that precedes vein formation in leaf (Gardiner et al., 2011). The phb-6 phv-5 rev-9 loss of function mutant produces abaxialized radial cotyledons in which phloem surrounds xylem (Emery et al., 2003). The quintuple mutant rev-6 phb-13 phv-11 cna-2 athb8-11/athb8-12 has a severely compromised vascular phenotype similar to the phb phv rev triple mutant (Prigge et al., 2005). Loss of ATHB8 and ATHB15 has no evident phenotypic effects, though vascular development is slightly perturbed in athb15.
The KAN family that belongs to the GARP [Golden2, ARR, and Chlamydomonas regulatory protein of Psr1-type transcription factors], is also essential for vasculature development. The kan1 kan2 kan3 kan4 quadruple mutant makes abnormal vascular bundles where xylem is surrounded by phloem (Kerstetter et al., 2001; Emery et al., 2003). The transcription factor encoding genes ALTERED/PHLOEM DEVELOPMENT (APL; which encodes an MYB coiled-coil transcription factor), VASCULAR-RELATED NACDOMAIN6 (VND6), and VND7 have a direct effect on xylem identity (Bonke et al., 2003; Kubo et al., 2005). Next to the molecular mechanism regulating vascular development that has been extensively investigated (Scarpella et al., 2006), knowledge of the regulation of these processes by spatial signals such as growth hormones in order to explain the establishment of their spatial distribution in simulation models is also emerging. Various mathematical models were constructed to explore the role of auxin in vasculature development (Scarpella et al., 2006; De Vos et al., 2012). However, a model proposed by Cano-Delgado et al. (2004) highlights the role of BRs in vascular patterning in Arabidopsis. BRs is perceived by BRASSINOSTEROID INSENSITIVE 1 (BRI1), a membrane localized LRR-RL kinase which increase xylem and reduced phloem differentiation. The loss of function of members of BRI1 family, BRI1-LIKE1 (BRL1) and BRI1-LIKE3 (BRL3) produces a phenotype of reduced xylem and increased phloem (Cano-Delgado et al., 2004). A mathematical model by Ibanes et al. (2009) shows that BR interacts with auxin for spatial regulation of vascular bundles in shoot inflorescence.
During leaf development specific epidermal cells convert into leaf hairs or trichomes. Trichomes generally go through three stages for their developmental-cell fate determination, specification and morphogenesis (Hulskamp et al., 1994). Gene products related to trichome formation can be subdivided into positive and negative regulators. The R2R3 MYB transcription factor GLABRA1 (GL1), the bHLH factor GLABRA3 (GL3), and the WD40-repeat factor TRANSPARENT TESTA GLABRA1 (TTG1) are positive regulators for trichome formation (Figure 12). The Null mutant gl1-1 is not fully glabrous, a few trichomes develop at the edges of the late rosette leaf (Oppenheimer et al., 1991; Kirik et al., 2005). The gl3 mutant shows the same phenotype as gl1-1 whereas overexpression of GL3 overcomes the trichome defect of ttg1 (Zhang et al., 2003). GL1 and TTG1 bind with GL3, forming a MYB/bHLH/WD-repeat complex that activates the expression of its downstream activators GL2 and TTG2, causing trichome differentiation (Zhao et al., 2008; Grebe, 2012; Yang and Ye, 2013).
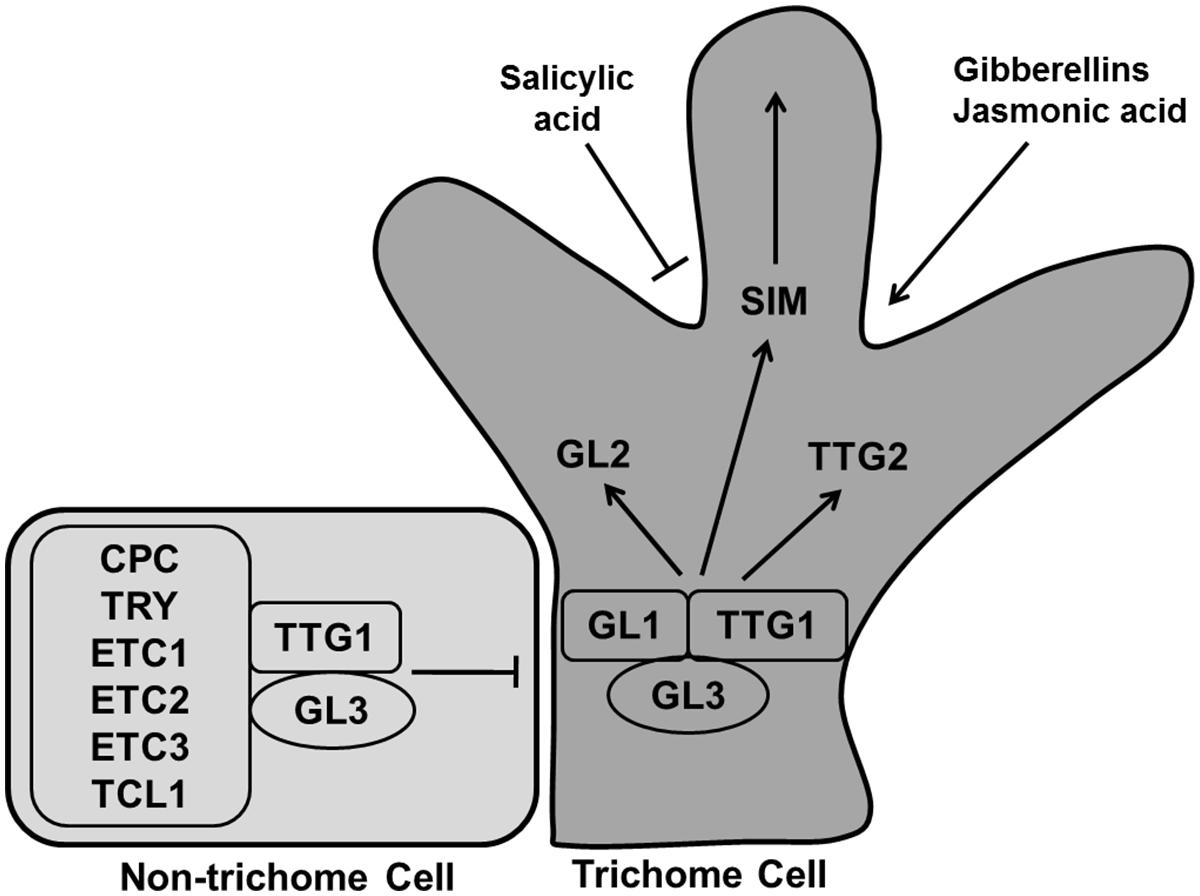
FIGURE 12. Regulation of trichome differentiation. Transcription factors GLABRA1 (GL1), GLABRA3 (GL3), and TRANSPARENT TESTA GLABRA1 (TTG1) forming the MYB/bHLH/WD-repeat complex activates trichome development whereas CAPRICE (CPC), TRIPTYCHON (TRY), ENHANCER OF TRY AND CPCs (ETC1, ETC2, and ETC3), and TRICHOMELESS1 (TCL1) inhibit the process. The MYB/bHLH/WD-repeat complex causes the activation of GL2, TTG2, and SIM to induce trichome differentiation. Trichome production is enhanced by gibberellins and jasmonic acid, while salicylic acid inhibit it (pointed and T shaped arrows indicate positive and negative regulation, respectively).
CAPRICE (CPC), TRIPTYCHON (TRY), ENHANCER OF TRY AND CPCs (ETC1, ETC2, and ETC3), and TRICHOMELESS1 (TCL1) are negative regulators, encoding for R3 MYB proteins (Tominaga et al., 2008; Figure 12). A loss of function mutant of tcl1-1 induces trichome formation and overexpression repressed trichome formation completely in Arabidopsis (Wang et al., 2007). The triple mutant etc2 try cpc produces trichome at the edges of the leaves. All these small MYB proteins replace GL1 in the MYB/bHLH/WD-repeat complex, rendering it inactive so that the cell remains in the undifferentiated state (Kirik et al., 2004).
Generally, trichome cells go through four endoreduplication cycles for their development, reaching an average DNA content of 32C, whereas other epidermal cells continue to divide (Schnittger and Hulskamp, 2002). It has been observed that the cell cycle related genes like SIM, TRY, SlCycB2, and genes involved in the endoreduplication process also regulate trichome formation. A sim mutant was found to have altered ploidy levels affecting trichome development (Schnittger et al., 1999; Walker et al., 2000; Pesch and Hulskamp, 2011). SIM is indeed directly targeted by the trichome initiation factors GL1 and GL3 (Morohashi and Grotewold, 2009). Plant hormones also regulate trichome formation with GA and jasmonic acid enhancing trichome number and density while salicylic acid reducing trichome number (Traw and Bergelson, 2003). Still, more study is needed to explore the role of phytohormone signaling pathways in trichome formation.
Two main theoretical models have been proposed to explain trichome patterning in Arabidopsis leaves: an activator–inhibitor model and an activator–depletion model. In the activator–inhibitor model, the activator (trimer complex of WD40, bHLH, and MYB factors) triggers its own inhibitor (R3MYB) which moves into the neighboring cell and impedes activation of the complex whereas, the activator–depletion model explains GL3 dependent depletion of TTG1 in non trichome cells (Pesch and Hulskamp, 2009). Computational modeling of the trichome pattern was used by Bouyer et al. (2008) to evaluate these conceptual models indicating that both models may act in concert.
In Systems Biology the aim is to acquire a mechanistic understanding of biological processes. In most cases the detailed knowledge is formulated in mathematical models that can simulate the behavior of the system and predict the effect of environmental and genetic perturbations. Such predictions can then be experimentally tested and the results used to improve the models further. This way models and experiments reinforce each other leading to increased understanding of the system (Kitano, 2002).
Here we adopted the view that cells are the units that direct development by integrating local signals into developmental decisions. Therefore, to build a mechanistic model for leaf growth we need to be able to model a single cell and its progeny as it progresses from the stem cell niche into the various positions in the mature leaf. The outcome of the integrated behavior of all cells ultimately forming the leaf is an organ of realistic size and morphology. This is currently still a very ambitious goal, but as a first step we addressed here the question what the current state of knowledge is with regards to regulatory networks that operate in cells as they progress in their individual developmental pathway.
During their developmental journey plant cells or rather their cell lineages are exposed to diverse biochemical and biophysical conditions, despite being tied into a symplastic mesh-work. Whereas, their final fate can be very different, ending up as a light harvesting mesophyll cell versus an epidermal hair cell for instance, many similarities exist in the events along their paths starting from the stem cell niche of the SAM. We have associated the different processes described above in separate sections with separate regulatory networks. However, by comparing the corresponding network diagrams, it readily becomes clear that many regulators and relations are shared; indicating that considering them as isolated systems is a radical assumption.
All presented networks are subject to intense investigation and some are far from the finished article, yet from a structural or topological perspective there are some recurring themes or motifs that emerge. A first case is the negative feedback loop which is a typical control structure that works like a thermostat: one factor stimulates a second factor which switches the first one off above a certain threshold. Not surprisingly this motif is active in the SAM where the WUS–CLV interaction ensures that sufficient stem cells are maintained for indeterminate growth, at the same time avoiding over-proliferation. A second case we have encountered is the CDK–APC/C interaction of the cell cycle. Here, the CDK–CYC activity required for cell proliferation eventually turns on the degradation machinery that inactivates CDK–CYC. Rather than providing spatial bounds the latter mechanism confers temporal control on cell proliferation. A somewhat related type of periodicity is a result of the auxin–PIN interaction crucial to phyllotactic patterning but likely also for determining leaf venation and serration (Bilsborough et al., 2011). By polarizing PINs toward auxin maxima, auxin levels are depleted in the surroundings. Another recurring motif is that of mutual negative feedback inhibition which can lead to switch like (bistable) behavior. An example is the proposed role of CDKA–KRP in the G1/S module of the cell cycle (Zhao et al., 2012). Such a motif can also provide a strong basis to support two spatially distinct and stable developmental domains. The antagonistic relation between ARP family and KNOX family transcription factors for instance appears to operate as a mechanism that enables primordium outgrowth while keeping the surrounding regions of the SAM undifferentiated. We have encountered other cases where such a duality appears to be crucial: determination of ab/adaxial leaf polarity on the one hand and vascular differentiation on the other hand are both governed by the antagonistic relation between HD-ZIPIII and KAN family transcription factors. Importantly, here to exert such spatial inhibitory effects additional mobile signals are in principle required, since the before mentioned TFs as far as we know are immobile. Various small RNAs are prime candidates for such a role (Braybrook and Kuhlemeier, 2010). In fact, many superimposed interactions with other factors are typically present to further increase robustness to deleterious perturbations or in contrast to increase the response to important developmental or environmental cues.
As we have seen, how well the described regulatory networks are understood varies considerably. Despite many plant-specific features and intricacies, the universal role and conserved character of the cell cycle has helped in uncovering its regulation to a considerable extent. Nevertheless, the precise functioning of KRPs and ubiquitin mediated degradation in cell cycle transitions remains to be elucidated. For the regulation of the transition between division and expansion for instance the coherency in the corresponding network diagram is weaker indicating that our knowledge is still more scattered and circumstantial. This lack of conceptual understanding is reflected in the absence of published computational models for this process and similarly for others. Our understanding of the regulation of cell growth is also relatively limited, in particular its relation to cell division (Sablowski and Dornelas, 2014). Whereas, the core machinery is relatively well understood, little is known about the way that primary growth determinants such as water or nutrient availability are translated into cell growth differences. The role of the TOR pathway in the regulation of growth and division, crucial in other eukaryotes, is only starting to emerge for plants (Xiong et al., 2013).
Because of the symplastic nature of plant tissue and the lack of a central nervous system, organ growth is more dependent on mobile growth signals that produce local gradients, such as phytohormones, mobile proteins and miRNAs. Since evolution tends to take a parsimonious approach it is not surprising that several growth signals are shared by different processes (and organisms as well). As illustrated above (see Figure 1), auxin has indeed been implicated in many developmental stages. If CKs are involved then they typically act antagonistically with auxin and GAs (cf. primordium initiation). BRs at one hand positively regulate many growth processes, while on the other hand they negatively regulate guard cell development. Some of the stress induced hormones such as ABA and ethylene modulate cellular processes for example division, endoreplication and the transition phase. Other growth hormones like JA and salicylic acid exert a negative control on the endocycle and on leaf hair development, respectively. These phytohormones have complex interactions where one affects other’s synthesis, transport, and signaling cascades.
Obviously a generalization of the role of specific hormones would imply over-simplification given the complexity of the regulatory interactions involved. Vernoux et al. (2011) demonstrated indeed that the control of gene expression by auxin not only depends on its distribution but also the expression patterns of the signaling network which consists of over 50 potentially interacting transcriptional activators and repressors. This study further highlights the importance of an integrative strategy which mathematical modeling supported by detailed expression maps, live imaging of biosensors, and high-throughput (interactome) data analysis. Given the crucial and complex role of non-cell-autonomous signals such as phytohormones the development of sensitive (fluorescent) biosensors to monitor their spatial and temporal distribution is an important trend (Brunoud et al., 2012; Shani et al., 2013; Wells et al., 2013).
Other experimental data becoming invaluable for developing improved mathematical models of leaf growth are quantitative growth data ranging from kinematic output (Nelissen et al., 2013) to cellular-resolution digital data extracted form confocal images (Kierzkowski et al., 2012). As repeatedly indicated above, multiple connections exist between the discussed developmental stages, suggesting that an important challenge will be to construct computational models that can reproduce these stages in a spontaneous way. This will likely require a more advanced geometrical representation than a flat plane or a simple sphere or cylinder. Developing coupled dynamical models of different tissues or organs interacting through an interface might provide a useful first step. However, eventually fully integrated three-dimensional models will be developed to grasp the complex cross-talk between various internal and external signals. Next to biological insight, increased computing power, for instance through improved parallelization algorithms, will likely become the limiting factor in that process. Ultimately, a mechanistic model for leaf development should integrate the regulatory networks that control developmental decisions and processes of cells as they migrate in space and time from the SAM to their final position in the leaf. Besides spatially and temporally highly resolved experimental techniques combined with advanced top down data-extraction techniques an important aspect will still remain to apply Ockham’s razor in a sensible way. Choosing a minimal set of variables to produce the desired behavior will present a challenge given the number of factors that are known to be involved or that are still to be discovered. As we have attempted to demonstrate, in a number of cases it is already clear which are the central regulators of the respective regulatory networks and some are indeed central to existing computational models. Furthermore, the non-cell autonomous signals and their gradients will inevitably be part of those future leaf developmental models and connect them to the rest of the plant and even the environment.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by a concerted research activity (GOA) research grant, “A Systems Biology Approach of Leaf Morphogenesis” from the research council of the University of Antwerp and an Interuniversity Attraction Poles (IUAP) from the Belgian Federal Science Policy Office (BELSPO). Dirk De Vos was funded by a return grant and grant IAP7/29 from BELSPO and Shweta Kalve by a training grant from the Department of Science and Education of the Flemish Government.
Achard, P., Gusti, A., Cheminant, S., Alioua, M., Dhondt, S., Coppens, F., et al. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19, 1188–1193. doi: 10.1016/j.cub.2009.05.059
Adenot, X., Elmayan, T., Lauressergues, D., Boutet, S., Bouche, N., Gasciolli, V., et al. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16, 927–932. doi: 10.1016/j.cub.2006.03.035
Akita, K., Hasezawa, S., and Higaki, T. (2013). Breaking of plant stomatal one-cell-spacing rule by sugar solution immersion. PLoS ONE 8:e72456. doi: 10.1371/journal.pone.0072456
Anastasiou, E., Kenz, S., Gerstung, M., Maclean, D., Timmer, J., Fleck, C., et al. (2007). Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 13, 843–856. doi: 10.1016/j.devcel.2007.10.001
Anderson, G. H., Veit, B., and Hanson, M. R. (2005). The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 3:12. doi: 10.1186/1741-7007-3-12
Andersen, S. U., Buechel, S., Zhao, Z., Ljung, K., Novak, O., Busch, W., et al. (2008). Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 20, 88–100. doi: 10.1105/tpc.107.054676
Andriankaja, M., Dhondt, S., De Bodt, S., Vanhaeren, H., Coppens, F., De Milde, L., et al. (2012). Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev. Cell 22, 64–78. doi: 10.1016/j.devcel.2011.11.011
Anzola, J. M., Sieberer, T., Ortbauer, M., Butt, H., Korbei, B., Weinhofer, I., et al. (2010). Putative Arabidopsis transcriptional adaptor protein (PROPORZ1) is required to modulate histone acetylation in response to auxin. Proc. Natl. Acad. Sci. U.S.A. 107, 10308–10313. doi: 10.1073/pnas.0913918107
Autran, D., Jonak, C., Belcram, K., Beemster, G. T., Kronenberger, J., Grandjean, O., et al. (2002). Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J. 21, 6036–6049. doi: 10.1093/emboj/cdf614
Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M. M., Ruberti, I., et al. (2001). The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126, 643–655. doi: 10.1104/pp.126.2.643
Barkoulas, M., Hay, A., Kougioumoutzi, E., and Tsiantis, M. (2008). A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 40, 1136–1141. doi: 10.1038/ng.189
Bayer, E. M., Smith, R. S., Mandel, T., Nakayama, N., Sauer, M., Prusinkiewicz, P., et al. (2009). Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev. 23, 373–384. doi: 10.1101/gad.497009
Beemster, G. T., De Veylder, L., Vercruysse, S., West, G., Rombaut, D., Van Hummelen, P., et al. (2005). Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 138, 734–743. doi: 10.1104/pp.104.053884
Beemster, G. T., Fiorani, F., and Inze, D. (2003). Cell cycle: the key to plant growth control? Trends Plant Sci. 8, 154–158. doi: 10.1016/S1360-1385(03)00046-3
Beemster, G. T. S., Vercruysse, S., De Veylder, L., Kuiper, M., and Inze, D. (2006). The Arabidopsis leaf as a model system for investigating the role of cell cycle regulation in organ growth. J. Plant Res. 119, 43–50. doi: 10.1007/s10265-005-0234-2
Berckmans, B., and De Veylder, L. (2009). Transcriptional control of the cell cycle. Curr. Opin. Plant Biol. 12, 599–605. doi: 10.1016/j.pbi.2009.07.005
Bergmann, D. C., Lukowitz, W., and Somerville, C. R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494–1497. doi: 10.1126/science.1096014
Bilsborough, G. D., Runions, A., Barkoulas, M., Jenkins, H. W., Hasson, A., Galinha, C., et al. (2011). Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. U.S.A. 108, 3424–3429. doi: 10.1073/pnas.1015162108
Blilou, I., Frugier, F., Folmer, S., Serralbo, O., Willemsen, V., Wolkenfelt, H., et al. (2002). The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes Dev. 16, 2566–2575. doi: 10.1101/gad.237302
Blow, J. J., and Dutta, A. (2005). Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 6, 476–486. doi: 10.1038/nrm1663
Boerjan, W., Cervera, M. T., Delarue, M., Beeckman, T., Dewitte, W., Bellini, C., et al. (1995). Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7, 1405–1419. doi: 10.1105/tpc.7.9.1405
Bonaccorso, O., Lee, J. E., Puah, L., Scutt, C. P., and Golz, J. F. (2012). FILAMENTOUS FLOWER controls lateral organ development by acting as both an activator and a repressor. BMC Plant Biol. 12:176. doi: 10.1186/1471-2229-12-176
Bonke, M., Thitamadee, S., Mahonen, A. P., Hauser, M. T., and Helariutta, Y. (2003). APL regulates vascular tissue identity in Arabidopsis. Nature 426, 181–186. doi: 10.1038/nature02100
Borghi, L., Gutzat, R., Futterer, J., Laizet, Y., Hennig, L., and Gruissem, W. (2010). Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22, 1792–1811. doi: 10.1105/tpc.110.074591
Borowska-Wykret, D., Elsner, J., De Veylder, L., and Kwiatkowska, D. (2013). Defects in leaf epidermis of Arabidopsis thaliana plants with CDKA;1 activity reduced in the shoot apical meristem. Protoplasma 250, 955–961. doi: 10.1007/s00709-012-0472-9
Boudolf, V., Barroco, R., Engler Jde, A., Verkest, A., Beeckman, T., Naudts, M., et al. (2004a). B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16, 945–955. doi: 10.1105/tpc.021774
Boudolf, V., Vlieghe, K., Beemster, G. T., Magyar, Z., Torres Acosta, J. A., Maes, S., et al. (2004b). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16, 2683–2692. doi: 10.1105/tpc.104.024398
Boudolf, V., Lammens, T., Boruc, J., Van Leene, J., Van Den Daele, H., Maes, S., et al. (2009). CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol. 150, 1482–1493. doi: 10.1104/pp.109.140269
Bouyer, D., Geier, F., Kragler, F., Schnittger, A., Pesch, M., Wester, K., et al. (2008). Two-dimensional patterning by a trapping/depletion mechanism: the role of TTG1 and GL3 in Arabidopsis trichome formation. PLoS Biol. 6:e141. doi: 10.1371/journal.pbio.0060141
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. doi: 10.1126/science.289.5479.617
Braun, N., Wyrzykowska, J., Muller, P., David, K., Couch, D., Rechenmann, C. P., et al. (2008). Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco. Plant Cell 20, 2746–2762. doi: 10.1105/tpc.108.059048
Braybrook, S. A., and Kuhlemeier, C. (2010). How a plant builds leaves. Plant Cell 22, 1006–1018. doi: 10.1105/tpc.110.073924
Brunoud, G., Wells, D. M., Oliva, M., Larrieu, A., Mirabet, V., Burrow, A. H., et al. (2012). A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103–106. doi: 10.1038/nature10791
Byrne, M. E., Barley, R., Curtis, M., Arroyo, J. M., Dunham, M., Hudson, A., et al. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. doi: 10.1038/35050091
Byrne, M. E., Simorowski, J., and Martienssen, R. A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129, 1957–1965.
Caesar, K., Elgass, K., Chen, Z., Huppenberger, P., Witthoft, J.,Schleifenbaum, F., et al. (2011). A fast brassinolide-regulated response pathway in the plasma membrane of Arabidopsis thaliana. Plant J. 66, 528–540. doi: 10.1111/j.1365-313X.2011.04510.x
Caldana, C., Li, Y., Leisse, A., Zhang, Y., Bartholomaeus, L., Fernie, A. R., et al. (2013). Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 73, 897–909. doi: 10.1111/tpj.12080
Cano-Delgado, A., Yin, Y. H., Yu, C., Vafeados, D., Mora-Garcia, S., Cheng, J. C., et al. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131, 5341–5351. doi: 10.1242/Dev.01403
Carles, C. C., and Fletcher, J. C. (2003). Shoot apical meristem maintenance: the art of a dynamic balance. Trends Plant Sci. 8, 394–401. doi: 10.1016/S1360-1385(03)00164-X
Castellano Mdel, M., Boniotti, M. B., Caro, E., Schnittger, A., and Gutierrez, C. (2004). DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. Plant Cell 16, 2380–2393. doi: 10.1105/tpc.104.022400
Cheng, Y., Cao, L., Wang, S., Li, Y., Shi, X., Liu, H., et al. (2013). Downregulation of multiple CDK inhibitor ICK/KRP genes upregulates the E2F pathway and increases cell proliferation, and organ and seed sizes in Arabidopsis. Plant J. 75, 642–655. doi: 10.1111/tpj.12228
Cheng, Y., Dai, X., and Zhao, Y. (2007). Auxin synthesized by the YUCCA Flavin Monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430–2439. doi: 10.1105/tpc.107.053009
Chitwood, D. H., Guo, M., Nogueira, F. T., and Timmermans, M. C. (2007). Establishing leaf polarity: the role of small RNAs and positional signals in the shoot apex. Development 134, 813–823. doi: 10.1242/dev.000497
Chitwood, D. H., Nogueira, F. T., Howell, M. D., Montgomery, T. A., Carrington, J. C., and Timmermans, M. C. (2009). Pattern formation via small RNA mobility. Genes Dev. 23, 549–554. doi: 10.1101/gad.1770009
Cho, H. J., Kwon, H. K., and Wang, M. H. (2010). Expression of Kip-related protein 4 gene (KRP4) in response to auxin and cytokinin during growth of Arabidopsis thaliana. BMB Rep. 43, 273–278. doi: 10.5483/BMBRep.2010.43.4.273
Cho, H. T., and Cosgrove, D. J. (2000). Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 97, 9783–9788. doi: 10.1073/pnas.160276997
Churchman, M. L., Brown, M. L., Kato, N., Kirik, V., Hulskamp, M., Inze, D., et al. (2006). SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 18, 3145–3157. doi: 10.1105/tpc.106.044834
Claeys, H., Skirycz, A., Maleux, K., and Inze, D. (2012). DELLA signaling mediates stress-induced cell differentiation in Arabidopsis leaves through modulation of anaphase-promoting complex/cyclosome activity. Plant Physiol. 159, 739–747. doi: 10.1104/pp.112.195032
Clark, E., Running, S. E., and Meyerowitz, M. P. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418.
Clark, E., Running, S. E., and Meyerowitz, M. P. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067.
Clark, S. E. (2001). Cell signalling at the shoot meristem. Nat. Rev. Mol. Cell Biol. 2, 276–284. doi: 10.1038/35067079
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861. doi: 10.1038/nrm1746
Cromer, L., Heyman, J., Touati, S., Harashima, H., Araou, E., Girard, C., et al. (2012). OSD1 promotes meiotic progression via APC/C inhibition and forms a regulatory network with TDM and CYCA1;2/TAM. PLoS Genet. 8:e1002865. doi: 10.1371/journal.pgen.1002865
de Almeida Engler, J., De Veylder, L., De Groodt, R., Rombauts, S., Boudolf, V., De Meyer, B., et al. (2009). Systematic analysis of cell-cycle gene expression during Arabidopsis development. Plant J. 59, 645–660. doi: 10.1111/j.1365-313X.2009.03893.x
de Jager, S. M., Menges, M., Bauer, U. M., and Murra, J. A. (2001). Arabidopsis E2F1 binds a sequence present in the promoter of S-phase-regulated gene AtCDC6 and is a member of a multigene family with differential activities. Plant Mol. Biol. 47, 555–568. doi: 10.1023/A:1011848528377
del Pozo, J. C., Boniotti, M. B., and Gutierrez, C. (2002). Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCFAtSKP2 pathway in response to light. Plant Cell 14, 3057–3071. doi: 10.1105/Tpc.006791
del Pozo, J. C., Diaz-Trivino, S., Cisneros, N., and Gutierrez, C. (2006). The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18, 2224–2235. doi: 10.1105/tpc.105.039651
Deprost, D., Yao, L., Sormani, R., Moreau, M., Leterreux, G., Nicolai, M., et al. (2007). The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 8, 864–870. doi: 10.1038/sj.embor.7401043
de Reuille, P. B., Bohn-Courseau, I., Ljung, K., Morin, H., Carraro, N., Godin, C., et al. (2006). Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 1627–1632. doi: 10.1073/pnas.0510130103
De Schutter, K., Joubes, J., Cools, T., Verkest, A., Corellou, F., Babiychuk, E., et al. (2007). Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19, 211–225. doi: 10.1105/tpc.106.045047
Desvoyes, B. N. D., Ramirez-Parra, E., Xie, Q., Chua, N.-H., and Gutierrez, C. (2006). Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 140, 67–80. doi: 10.1104/pp.105.071027
De Veylder, L., Beeckman, T., Beemster, G. T., De Almeida Engler, J., Ormenese, S., Maes, S., et al. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21, 1360–1368. doi: 10.1093/emboj/21.6.1360
De Veylder, L., Beeckman, T., Beemster, G. T. S., Krols, L., Terras, P., Landrieu, I., et al. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1667. doi: 10.1105/tpc.13.7.1653
De Veylder, L., Larkin, J. C., and Schnittger, A. (2011). Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 16, 624–634. doi: 10.1016/j.tplants.2011.07.001
De Vos, D., Dzhurakhalov, A., Draelants, D., Bogaerts, I., Kalve, S., Prinsen, E., et al. (2012). Towards mechanistic models of plant organ growth. J. Exp. Bot. 63, 3325–3337. doi: 10.1093/jxb/ers037
Dewitte, W., and Murray, J. A. (2003). The plant cell cycle. Annu. Rev. Plant Biol. 54, 235–264. doi: 10.1146/annurev.arplant.54.031902.134836
Dewitte, W., Riou-Khamlichi, C., Scofield, S., Healy, J. M. S., Jacqmard, A., Kilby, N. J., et al. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15, 79–92. doi: 10.1105/tpc.004838
Dewitte, W., Scofield, S., Alcasabas, A. A., Maughan, S. C., Menges, M., Braun, N., et al. (2007). Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. U.S.A. 104, 14537–14542. doi: 10.1073/pnas.0704166104
Disch, S., Anastasiou, E., Sharma, V. K., Laux, T., Fletcher, J. C., and Lenhard, M. (2006). The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16, 272–279. doi: 10.1016/j.cub.2005.12.026
Dissmeyer, N., Weimer, A. K., Pusch, S., De Schutter, K., Kamei, C. L. A., Nowack, M. K., et al. (2009). Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. Plant Cell 21, 3641–3654. doi: 10.1105/tpc.109.070417
Dissmeyer, N., Weimer, A. K., Veylder, L. D., Novak, B., and Schnittger, A. (2010). The regulatory network of cell cycle progression is fundamentally different in plants versus yeast or metazoans. Plant Signal. Behav. 5, 1613–1618. doi: 10.4161/psb.5.12.13969
Dyson, R. J., Band, L. R., and Jensen, O. E. (2012). A model of crosslink kinetics in the expanding plant cell wall: yield stress and enzyme action. J. Theor. Biol. 307, 125–136. doi: 10.1016/j.jtbi.2012.04.035
Efroni, I., Blum, E., Goldshmidt, A., and Eshed, Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20, 2293–2306. doi: 10.1105/tpc.107.057521
Eloy, N. B., De Freitas Lima, M., Van Damme, D., Vanhaeren, H., Gonzalez, N., De Milde, L., et al. (2011). The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. Plant J. 68, 351–363. doi: 10.1111/j.1365-313X.2011.04691.x
Eloy, N. B., Gonzalez, N., Van Leene, J., Maleux, K., Vanhaeren, H., De Milde, L., et al. (2012). SAMBA, a plant-specific anaphase-promoting complex/cyclosome regulator is involved in early development and A-type cyclin stabilization. Proc. Natl. Acad. Sci. U.S.A. 109, 13853–13858. doi: 10.1073/pnas.1211418109
Emery, J. F., Floyd, S. K., Alvarez, J., Eshed, Y., Hawker, N. P., Izhaki, A., et al. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. doi: 10.1016/j.cub.2003.09.035
Engler, J. D., Kyndt, T., Vieira, P., Van Cappelle, E., Boudolf, V., Sanchez, V., et al. (2012). CCS52 and DEL1 genes are key components of the endocycle in nematode-induced feeding sites. Plant J. 72, 185–198. doi: 10.1111/j.1365-313X.2012.05054.x
Eshed, Y., Baum, S. F., Perea, J. V., and Bowman, J. L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251–1260. doi: 10.1016/S0960-9822(01)00392-X
Eshed, Y., Izhaki, A., Baum, S. F., Floyd, S. K., and Bowman, J. L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131, 2997–3006. doi: 10.1242/dev.01186
Feng, G., Qin, Z., Yan, J., Zhang, X., and Hu, Y. (2011). Arabidopsis ORGAN SIZE RELATED1 regulates organ growth and final organ size in orchestration with ARGOS andARL. New Phytol. 191, 635–646. doi: 10.1111/j.1469-8137.2011.03710.x
Francis, D. (2007). The plant cell cycle – 15 years on. New Phytol. 174, 261–278. doi: 10.1111/j.1469-8137.2007.02038.x
Francis, D., and Sorrell, D. A. (2001). The interface between the cell cycle and plant growth regulators: a mini review. Plant Growth Regul. 33, 1–12. doi: 10.1023/A:1010762111585
Fulop, K., Tarayre, S., Kelemen, Z., Horvath, G., Kevei, Z., Nikovics, K., et al. (2005). Arabidopsis anaphase-promoting complexes: multiple activators and wide range of substrates might keep APC perpetually busy. Cell Cycle 4, 1084–1092. doi: 10.4161/cc.4.8.1856
Gaamouche, T., Manes, C. L., Kwiatkowska, D., Berckmans, B., Koumproglou, R., Maes, S., et al. (2010). Cyclin-dependent kinase activity maintains the shoot apical meristem cells in an undifferentiated state. Plant J. 64, 26–37. doi: 10.1111/j.1365-313X.2010.04317.x
Gardiner, J., Donner, T. J., and Scarpella, E. (2011). Simultaneous activation of SHR and ATHB8 expression defines switch to preprocambial cell state in Arabidopsis leaf development. Dev. Dyn. 240, 261–270. doi: 10.1002/dvdy.22516
Gendreau, E., Orbovic, V., Hofte, H., and Traas, J. (1999). Gibberellin and ethylene control endoreduplication levels in the Arabidopsis thaliana hypocotyl. Planta 209, 513–516.
Gonzalez, N., Vanhaeren, H., and Inze, D. (2012). Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 17, 332–340. doi: 10.1016/j.tplants.2012.02.003
Gordon, S. P., Chickarmane, V. S., Ohno, C., and Meyerowitz, E. M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U.S.A. 106, 16529–16534. doi: 10.1073/pnas.0908122106
Grebe, M. (2012). The patterning of epidermal hairs in Arabidopsis–updated. Curr. Opin. Plant Biol. 15, 31–37. doi: 10.1016/j.pbi.2011.10.010
Green, P. B. (1976). Growth and cell pattern formation on an axis: critique of concepts, terminology and modes of study. Bot. Gaz. 137, 187–202. doi: 10.1086/336858
Gudesblat, G. E., Schneider-Pizon, J., Betti, C., Mayerhofer, J., Vanhoutte, I., Van Dongen, W., et al. (2012). SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14, 548–554. doi: 10.1038/ncb2471
Guenot, B., Bayer, E., Kierzkowski, D., Smith, R. S., Mandel, T., Zadnikova, P., et al. (2012). Pin1-independent leaf initiation in Arabidopsis. Plant Physiol. 159, 1501–1510. doi: 10.1104/pp.112.200402
Haecker, A., and Laux, T. (2001). Cell–cell signaling in the shoot meristem. Curr. Opin. Plant Biol. 4, 441–446. doi: 10.1016/S1369-5266(00)00198-9
Hara, K., Yokoo, T., Kajita, R., Onishi, T., Yahata, S., Peterson, K. M., et al. (2009). Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50, 1019–1031. doi: 10.1093/Pcp/Pcp068
Harashima, H., Dissmeyer, N., and Schnittger, A. (2013). Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 23, 345–356. doi: 10.1016/j.tcb.2013.03.002
Hasson, A., Plessis, A., Blein, T., Adroher, B., Grigg, S., Tsiantis, M., et al. (2011). Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell 23, 54–68. doi: 10.1105/tpc.110.081448
Hay, A., Barkoulas, M., and Tsiantis, M. (2006). ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133, 3955–3961. doi: 10.1242/dev.02545
Hay, A., and Tsiantis, M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38, 942–947. doi: 10.1038/Ng1835
Hay, A., and Tsiantis, M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153–3165. doi: 10.1242/dev.030049
Heisler, M. G., Hamant, O., Krupinski, P., Uyttewaal, M., Ohno, C., Jonsson, H., et al. (2010). Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol. 8:e1000516. doi: 10.1371/journal.pbio.1000516
Hemerly, A., Engler Jde, A., Bergounioux, C., Van Montagu, M., Engler, G., Inze, D., et al. (1995). Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 14, 3925–3936.
Hershko, A. (2005). The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 12, 1191–1197. doi: 10.1038/sj.cdd.4401702
Heyman, J., and De Veylder, L. (2012). The anaphase-promoting complex/cyclosome in control of plant development. Mol. Plant 5, 1182–1194. doi: 10.1093/mp/sss094
Heyman, J., Van Den Daele, H., De Wit, K., Boudolf, V., Berckmans, B., Verkest, A., et al. (2011). Arabidopsis ULTRAVIOLET-B-INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase-promoting complex/cyclosome. Plant Cell 23, 4394–4410. doi: 10.1105/tpc.111.091793
Higuchi, M., Pischke, M. S., Mahonen, A. P., Miyawaki, K., Hashimoto, Y., Seki, M., et al. (2004). In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. U.S.A. 101, 8821–8826. doi: 10.1073/pnas.0402887101
Hohm, T., Zitzler, E., and Simon, R. (2010). A dynamic model for stem cell homeostasis and patterning in Arabidopsis meristems. PLoS ONE 5:e9189. doi: 10.1371/journal.pone.0009189
Horiguchi, G., Kim, G. T., and Tsukaya, H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43, 68–78. doi: 10.1111/j.1365–313X.2005.02429.x
Horvath, B. M., Magyar, Z., Zhang, Y., Hamburger, A. W., Bako, L., Visser, R. G., et al. (2006). EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J. 25, 4909–4920. doi: 10.1038/sj.emboj.7601362
Hu, Y., Bao, F., and Li, J. (2000). Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J. 24, 693–701. doi: 10.1046/j.1365-313x.2000.00915.x
Hu, Y., Xie, Q., and Chua, N. H. (2003). The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15, 1951–1961. doi: 10.1105/tpc.013557
Hulskamp, M., Misra, S., and Jurgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. doi: 10.1016/0092-8674(94)90118-X
Hunter, C., Willmann, M. R., Wu, G., Yoshikawa, M., De La Luz Gutierrez-Nava, M., and Poethig, S. R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133, 2973–2981. doi: 10.1242/dev.02491
Ibanes, M., Fabregas, N., Chory, J., and Cano-Delgado, A. I. (2009). Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc. Natl. Acad. Sci. U.S.A. 106, 13630–13635. doi: 10.1073/pnas.0906416106
Imai, K. K., Ohashi, Y., Tsuge, T., Yoshizumi, T., Matsui, M., Oka, A., et al. (2006). The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18, 382–396. doi: 10.1105/tpc.105.037309
Inze, D., and De Veylder, L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40, 77–105. doi: 10.1146/annurev.genet.40.110405.090431
Inze, D., Gutierrez, C., and Chua, N. H. (1999). Trends in plant cell cycle research. Plant Cell 11, 991–994. doi: 10.1105/tpc.11.6.991
Ishida, T., Adachi, S., Yoshimura, M., Shimizu, K., Umeda, M., and Sugimoto, K. (2010). Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 137, 63–71. doi: 10.1242/dev.035840
Ito, M., Araki, S., Matsunaga, S., Itoh, T., Nishihama, R., Machida, Y., et al. (2001). G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13, 1891–1905. doi: 10.1105/tpc.13.8.1891
Iwata, E., Ikeda, S., Matsunaga, S., Kurata, M., Yoshioka, Y., Criqui, M. C., et al. (2011). GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant Cell 23, 4382–4393. doi: 10.1105/tpc.111.092049
Jackson, D., Veit, B., and Hake, S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120, 405–413.
Jasinski, S., Piazza, P., Craft, J., Hay, A., Woolley, L., Rieu, I., et al. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15, 1560–1565. doi: 10.1016/j.cub.2005.07.023
Jegu, T., Latrasse, D., Delarue, M., Mazubert, C., Bourge, M., Hudik, E., et al. (2013). Multiple functions of Kip-related protein5 connect endoreduplication and cell elongation. Plant Physiol. 161, 1694–1705. doi: 10.1104/pp.112.212357
Jonsson, H., Heisler, M., Reddy, G. V., Agrawal, V., Gor, V., Shapiro, B. E., et al. (2005). Modeling the organization of the WUSCHEL expression domain in the shoot apical meristem. Bioinformatics 21(Suppl. 1), i232–i240. doi: 10.1093/bioinformatics/bti1036
Joubes, J., De Schutter, K., Verkest, A., Inze, D., and De Veylder, L. (2004). Conditional, recombinase-mediated expression of genes in plant cell cultures. Plant J. 37, 889–896. doi: 10.1111/j.1365-313X.2004.02004.x
Jung, J. H., and Park, C. M. (2007). Vascular development in plants: specification of xylem and phloem tissues. J. Plant Biol. 50, 301–305. doi: 10.1007/BF03030658
Kanaoka, M. M., Pillitteri, L. J., Fujii, H., Yoshida, Y., Bogenschutz, N. L., Takabayashi, J., et al. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20, 1775–1785. doi: 10.1105/tpc.108.060848
Kang, J., Mizukami, Y., Wang, H., Fowke, L., and Dengler, N. G. (2007). Modification of cell proliferation patterns alters leaf vein architecture in Arabidopsis thaliana. Planta 226, 1207–1218. doi: 10.1007/s00425-007-0567-2
Kang, J. S., and Soh, W. Y. (2001). The origin and development of vascular cambium in girdled stems of Eucommia ulmoides Oliv. J. Plant Biol. 44, 148–156. doi: 10.1007/BF03030232
Kasili, R., Walker, J. D., Simmons, L. A., Zhou, J., Veylder, L. D., and Larkin, J. C. (2010). SIAMESE cooperates with the CDH1-like protein CCS52A1 to establish endoreplication in Arabidopsis thaliana trichomes. Genetics 185, 257–268. doi: 10.1534/genetics.109.113274
Kerstetter, R. A., Bollman, K., Taylor, R. A., Bomblies, K., and Poethig, R. S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411, 706–709. doi: 10.1038/35079629
Kevei, Z., Baloban, M., Da Ines, O., Tiricz, H., Kroll, A., Regulski, K., et al. (2011). Conserved CDC20 cell cycle functions are carried out by two of the five isoforms in Arabidopsis thaliana. PLoS ONE 6:e20618. doi: 10.1371/journal.pone.0020618
Kidner, C. A., and Martienssen, R. A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428, 81–84. doi: 10.1038/Nature02366
Kierzkowski, D., Nakayama, N., Routier-Kierzkowska, A. L., Weber, A., Bayer, E., Schorderet, M., et al. (2012). Elastic domains regulate growth and organogenesis in the plant shoot apical meristem. Science 335, 1096–1099. doi: 10.1126/science.1213100
Kim, H. J., Oh, S. A., Brownfield, L., Hong, S. H., Ryu, H., Hwang, I., et al. (2008). Control of plant germline proliferation by SCF(FBL17) degradation of cell cycle inhibitors. Nature 455, 1134–1137. doi: 10.1038/nature07289
Kim, J. H., Choi, D., and Kende, H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104. doi: 10.1046/j.1365-313X.2003.01862.x
Kim, T. W., Michniewicz, M., Bergmann, D. C., and Wang, Z. Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419–422. doi: 10.1038/nature10794
Kirik, V., Lee, M. M., Wester, K., Herrmann, U., Zheng, Z., Oppenheimer, D., et al. (2005). Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132, 1477–1485. doi: 10.1242/dev.01708
Kirik, V., Simon, M., Wester, K., Schiefelbein, J., and Hulskamp, M. (2004). ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 55, 389–398. doi: 10.1007/s11103-004-0893-8
Knauer, S., Holt, A. L., Rubio-Somoza, I., Tucker, E. J., Hinze, A., Pisch, M., et al. (2013). A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 24, 125–132. doi: 10.1016/j.devcel.2012.12.009
Korner, C., Menendezriedl, S. P., and John, P. C. L. (1989). Why are bonsai plants small? A consideration of cell size. Aust. J. Plant Physiol. 16, 443–448. doi: 10.1071/PP9890443
Koyama, T., Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22, 3574–3588. doi: 10.1105/tpc.110.075598
Krizek, B. A. (2009). Making bigger plants: key regulators of final organ size. Curr. Opin. Plant Biol. 12, 17–22. doi: 10.1016/j.pbi.2008.09.006
Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., et al. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860. doi: 10.1101/gad.1331305
Kuchen, E. E., Fox, S., De Reuille, P. B., Kennaway, R., Bensmihen, S., Avondo, J., et al. (2012). Generation of leaf shape through early patterns of growth and tissue polarity. Science 335, 1092–1096. doi: 10.1126/science.1214678
Kurepa, J., Wang, S., Li, Y., Zaitlin, D., Pierce, A. J., and Smalle, J. A. (2009). Loss of 26S proteasome function leads to increased cell size and decreased cell number in Arabidopsis shoot organs. Plant Physiol. 150, 178–189. doi: 10.1104/pp.109.135970
Lai, L. B., Nadeau, J. A., Lucas, J., Lee, E.-K., Nakagawa, T., Zhao, L., et al. (2005). The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 17, 2754–2767. doi: 10.1105/tpc.105.034116
Lammens, T., Boudolf, V., Kheibarshekan, L., Zalmas, L. P., Gaamouche, T., Maes, S., et al. (2008). Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc. Natl. Acad. Sci. U.S.A. 105, 14721–14726. doi: 10.1073/pnas.0806510105
Lampard, G. R., Lukowitz, W., Ellis, B. E., and Bergmann, D. C. (2009). Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell 21, 3506–3517. doi: 10.1105/tpc.109.070110
Lampard, G. R., Macalister, C. A., and Bergmann, D. C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322, 1113–1116. doi: 10.1126/science.1162263
Landrieu, I., Da Costa, M., De Veylder, L., Dewitte, F., Vandepoele, K., Hassan, S., et al. (2004). A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 101, 13380–13385. doi: 10.1073/pnas.0405248101
Laux, T., Mayer, K. F., Berger, J., and Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96.
Lee, B. H., Ko, J.-H., Lee, S., Lee, Y., Pak, J.-H., and Kim, J. H. (2009). The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151, 655–668. doi: 10.1104/pp.109.141838
Leibfried, A., To, J. P., Busch, W., Stehling, S., Kehle, A., Demar, M., et al. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438, 1172–1175. doi: 10.1038/nature04270
Li, Y., Zheng, L., Corke, F., Smith, C., and Bevan, M. W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22, 1331–1336. doi: 10.1101/gad.463608
Liu, D., Song, Y., Chen, Z., and Yu, D. (2009). Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant. 136, 223–236. doi: 10.1111/j.1399-3054.2009.01229.x
Lockhart, J. A. (1965). An analysis of irreversible plant cell elongation. J. Theor. Biol. 8, 264–275. doi: 10.1016/0022-5193(65)90077-9
Long, J. A., Moan, E. I., Medford, J. I., and Barton, M. K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69. doi: 10.1038/379066a0
MacAlister, C. A., Ohashi-Ito, K., and Bergmann, D. C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537–540. doi: 10.1038/nature05491
Magyar, Z., De Veylder, L., Atanassova, A., Bako, L., Inze, D., and Bogre, L. (2005). The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17, 2527–2541. doi: 10.1105/tpc.105.033761
Mariconti, L., Pellegrini, B., Cantoni, R., Stevens, R., Bergounioux, C., Cella, R., et al. (2002). The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J. Biol. Chem. 277, 9911–9919. doi: 10.1074/jbc.M110616200
Marrocco, K., Bergdoll, M., Achard, P., Criqui, M. C., and Genschik, P. (2010). Selective proteolysis sets the tempo of the cell cycle. Curr. Opin. Plant Biol. 13, 631–639. doi: 10.1016/j.pbi.2010.07.004
Mauseth, J. D. (2004). Giant shoot apical meristems in cacti have ordinary leaf primordia but altered phyllotaxy and shoot diameter. Ann. Bot. 94, 145–153. doi: 10.1093/aob/mch121
McConnell, J. R., and Barton, M. K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942.
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. doi: 10.1038/35079635
McHale, N. A. (1993). LAM-l and FAT genes control development of the leaf blade in Nicotiana sylwestris. Plant Cell 5, 1029–1038. doi: 10.1105/tpc.5.9.1029
Menand, B., Desnos, T., Nussaume, L., Berger, F., Bouchez, D., Meyer, C., et al. (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. U.S.A. 99, 6422–6427. doi: 10.1073/pnas.092141899
Menges, M., De Jager, S. M., Gruissem, W., and Murray, J. A. H. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41, 546–566. doi: 10.1111/j.1365-313X.2004.02319.x
Meyerowitz, E. M. (1997). Control of cell division patterns in developing shoots and flowers of Arabidopsis thaliana. Cold Spring Harb. Symp. Quant. Biol. 62, 369–375. doi: 10.1101/SQB.1997.062.01.044
Miyashima, S., Sebastian, J., Lee, J. Y., and Helariutta, Y. (2013). Stem cell function during plant vascular development. EMBO J. 32, 178–193. doi: 10.1038/emboj.2012.301
Moreau, M., Azzopardi, M., Clement, G., Dobrenel, T., Marchive, C., Renne, C., et al. (2012). Mutations in the Arabidopsis homolog of LST8/GbetaL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24, 463–481. doi: 10.1105/tpc.111.091306
Morohashi, K., and Grotewold, E. (2009). A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 5:e1000396. doi: 10.1371/journal.pgen.1000396
Nadeau, J. A., and Sack, F. D. (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700. doi: 10.1126/science.1069596
Nakata, M., Matsumoto, N., Tsugeki, R., Rikirsch, E., Laux, T., and Okada, K. (2012). Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 24, 519–535. doi: 10.1105/tpc.111.092858
Nakata, M., and Okada, K. (2012). The three-domain model: a new model for the early development of leaves in Arabidopsis thaliana. Plant Signal. Behav. 7, 1423–1427. doi: 10.4161/psb.21959
Nakaya, M., Tsukaya, H., Murakami, N., and Kato, M. (2002). Brassinosteroids control the proliferation of leaf cells of Arabidopsis thaliana. Plant Cell Physiol. 43, 239–244. doi: 10.1093/pcp/pcf024
Naouar, N., Vandepoele, K., Lammens, T., Casneuf, T., Zeller, G., Van Hummelen, P., et al. (2009). Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J. 57, 184–194. doi: 10.1111/j.1365-313X.2008.03662.x
Nath, U., Crawford, B. C., Carpenter, R., and Coen, E. (2003). Genetic control of surface curvature. Science 299, 1404–1407. doi: 10.1126/science.1079354
Nelissen, H., Rymen, B., Coppens, F., Dhondt, S., Fiorani, F., and Beemster, G. T. (2013). Kinematic analysis of cell division in leaves of mono- and dicotyledonous species: a basis for understanding growth and developing refined molecular sampling strategies. Methods Mol. Biol. 959, 247–264. doi: 10.1007/978-1-62703-221-6_17
Nelissen, H., Rymen, B., Jikumaru, Y., Demuynck, K., Van Lijsebettens, M., Kamiya, Y., et al. (2012). A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Curr. Biol. 22, 1183–1187. doi: 10.1016/j.cub.2012.04.065
Nikolaev, S. V., Penenko, A. V., Lavrekha, V. V., Melsness, E. D., and Kolchanov, N. A. (2007). A model study of the role of proteins CLV1, CLV2, CLV3, and WUS in regulation of the structure of the shoot apical meristem. Ontogenez 38, 457–462.
Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H., Aida, M., et al. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18, 2929–2945. doi: 10.1105/tpc.106.045617
Nogueira, F. T., Madi, S., Chitwood, D. H., Juarez, M. T., and Timmermans, M. C. (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 21, 750–755. doi: 10.1101/gad.1528607
Noir, S., Bomer, M., Takahashi, N., Ishida, T., Tsui, T. L., Balbi, V., et al. (2013). Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol. 161, 1930–1951. doi: 10.1104/pp.113.214908
Novak, B., and Tyson, J. J. (1993). Numerical analysis of a comprehensive model of M-phase control in Xenopus oocyte extracts and intact embryos. J. Cell Sci. 106, 1153–1168.
Ohashi-Ito, K., and Bergmann, D. C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18, 2493–2505. doi: 10.1105/tpc.106.046136
Ohashi-Ito, K., and Fukuda, H. (2003). HD-Zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol. 44, 1350–1358. doi: 10.1093/pcp/pcg164
Oppenheimer, D. G., Herman, P. L., Sivakumaran, S., Esch, J., and Marks, M. D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67, 483–493. doi: 10.1016/0092-8674(91)90523-2
Ori, N., Cohen, A. R., Etzioni, A., Brand, A., Yanai, O., Shleizer, S., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39, 787–791. doi: 10.1038/Ng2036
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Paredez, A. R., Somerville, C. R., and Ehrhardt, D. W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312, 1491–1495. doi: 10.1126/science.1126551
Pauwels, L., Morreel, K., De Witte, E., Lammertyn, F., Van Montagu, M., Boerjan, W., et al. (2008). Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. U.S.A. 105, 1380–1385. doi: 10.1073/pnas.0711203105
Perazza, D., Herzog, M., Hulskamp, M., Brown, S., Dorne, A. M., and Bonneville, J. M. (1999). Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics 152, 461–476.
Perrot-Rechenmann, C. (2010). Cellular responses to auxin: division versus expansion. Cold Spring Harb. Perspect. Biol. 2:a001446. doi: 10.1101/cshperspect.a001446
Pesch, M., and Hulskamp, M. (2009). One, two, three...models for trichome patterning in Arabidopsis? Curr. Opin. Plant Biol. 12, 587–592. doi: 10.1016/j.pbi.2009.07.015
Pesch, M., and Hulskamp, M. (2011). Role of TRIPTYCHON in trichome patterning in Arabidopsis. BMC Plant Biol. 11:130. doi: 10.1186/1471-2229-11-130
Peterson, K. M., Rychel, A. L., and Torii, K. U. (2010). Out of the mouths of plants: the molecular basis of the evolution and diversity of stomatal development. Plant Cell 22, 296–306. doi: 10.1105/tpc.109.072777
Pillitteri, L. J., Sloan, D. B., Bogenschutz, N. L., and Torii, K. U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501–505. doi: 10.1038/nature05467
Porceddu, A., Stals, H., Reichheld, J., Segers, G., De Veylder, L., Barroco, R., et al. (2001). A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J. Biol. Chem. 276, 36354–36360. doi: 10.1074/jbc.M011060200
Prigge, M. J., Otsuga, D., Alonso, J. M., Ecker, J. R., Drews, G. N., and Clark, S. E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17, 61–76. doi: 10.1105/tpc.104.026161
Ramirez-Parra, E., Frundt, C., and Gutierrez, C. (2003). A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J. 33, 801–811. doi: 10.1046/J.1365-313x.2003.01662.X
Reinhardt, D., Pesce, E. R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., et al. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260. doi: 10.1038/nature02081
Ren, H., Santner, A., Del Pozo, J., Murray, J., and Estelle, M. (2008). Degradation of the cyclin-dependent kinase inhibitor KRP1 is regulated by two different ubiquitin E3 ligases. Plant J. 53, 705–716. doi: 10.1111/j.1365-313X.2007.03370.x
Ren, M., Qiu, S., Venglat, P., Xiang, D., Feng, L., Selvaraj, G., et al. (2011). Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol. 155, 1367–1382. doi: 10.1104/pp.110.169045
Riou-Khamlichi, C., Huntley, R., Jacqmard, A., and Murray, J. A. H. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283, 1541–1543. doi: 10.1126/science.283.5407.1541
Robinson, S., De Reuille, P. B., Chan, J., Bergmann, D., Prusinkiewicz, P., and Coen, E. (2011). Generation of spatial patterns through cell polarity switching. Science 333, 1436–1440. doi: 10.1126/science.1202185
Rodriguez, R. E., Mecchia, M. A., Debernardi, J. M., Schommer, C., Weigel, D., and Palatnik, J. F. (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112. doi: 10.1242/dev.043067
Roeder, A. H., Chickarmane, V., Cunha, A., Obara, B., Manjunath, B. S., and Meyerowitz, E. M. (2010). Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana. PLoS Biol. 8:e1000367. doi: 10.1371/journal.pbio.1000367
Roodbarkelari, F., Bramsiepe, J., Weinl, C., Marquardt, S., Novak, B., Jakoby, M. J., et al. (2010). Cullin 4-ring finger-ligase plays a key role in the control of endoreplication cycles in Arabidopsis trichomes. Proc. Natl. Acad. Sci. U.S.A. 107, 15275–15280. doi: 10.1073/pnas.1006941107
Rose, J. K., Braam, J., Fry, S. C., and Nishitani, K. (2002). The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol. 43, 1421–1435. doi: 10.1093/pcp/pcf171
Rubio-Díaz, S., Pérez-Pérez, J. M., González-Bayón, R., Muñoz-Viana, R., Borrega, N., Mouille, G., et al. (2012). Cell expansion-mediated organ growth is affected by mutations in three EXIGUA genes. PLoS ONE 7:e36500. doi: 10.1371/journal.pone.0036500
Sablowski, R. (2007). The dynamic plant stem cell niches. Curr. Opin. Plant Biol. 10, 639–644. doi: 10.1016/j.pbi.2007.07.001
Sablowski, R., and Dornelas, M. (2014). Interplay between cell growth and cell cycle in plants. J. Exp. Bot. 65, 2703–2714. doi: 10.1093/jxb/ert354
Sakamoto, T., Kamiya, N., Ueguchi-Tanaka, M., Iwahori, S., and Matsuoka, M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15, 581–590. doi: 10.1101/gad.867901
Satina, S., Blakeslee, A., and Avery, A. (1940). Demonstration of the three germ layers in the shoot apex of Datura by means of induced polyploidy in periclinal chimeras. Am. J. Bot. 27, 895–905. doi: 10.2307/2436558
Scarpella, E., Francis, P., and Berleth, T. (2004). Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development 131, 3445–3455. doi: 10.1242/dev.01182
Scarpella, E., Marcos, D., Friml, J., and Berleth, T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20, 1015–1027. doi: 10.1101/gad.1402406
Schnittger, A., Folkers, U., Schwab, B., Jurgens, G., and Hulskamp, M. (1999). Generation of a spacing pattern: the role of triptychon in trichome patterning in Arabidopsis. Plant Cell 11, 1105–1116. doi: 10.1105/tpc.11.6.1105
Schnittger, A., and Hulskamp, M. (2002). Trichome morphogenesis: a cell-cycle perspective. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 823–826. doi: 10.1098/rstb.2002.1087
Schnittger, A., Weinl, C., Bouyer, D., Schobinger, U., and Hulskamp, M. (2003). Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15, 303–315. doi: 10.1105/tpc.008342
Schommer, C., Palatnik, J. F., Aggarwal, P., Chetelat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. doi: 10.1371/journal.pbio.0060230
Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F. X., Jürgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. doi: 10.1016/S0092-8674(00)80700-X
Scofield, S., and Murray, J. A. (2006). KNOX gene function in plant stem cell niches. Plant Mol. Biol. 60, 929–946. doi: 10.1007/s11103-005-4478-y
Shani, E., Weinstain, R., Zhang, Y., Castillejo, C., Kaiserli, E., Chory, J., et al. (2013). Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A. 110, 4834–4839. doi: 10.1073/pnas.1300436110
Shpak, E. D., Mcabee, J. M., Pillitteri, L. J., and Torii, K. U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309, 290–293. doi: 10.1126/science.1109710
Siegfried, K. R., Eshed, Y., Baum, S. F., Otsuga, D., Drews, G. N., and Bowman, J. L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128.
Sormani, R., Yao, L., Menand, B., Ennar, N., Lecampion, C., Meyer, C., et al. (2007). Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol. 7:26. doi: 10.1186/1471-2229-7-26
Sozzani, R., Maggio, C., Varotto, S., Canova, S., Bergounioux, C., Albani, D., et al. (2006). Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol. 140, 1355–1366. doi: 10.1104/pp.106.077990
Stransfeld, L., Eriksson, S., Adamski, N. M., Breuninger, H., and Lenhard, M. (2010). KLUH/CYP78A5 promotes organ growth without affecting the size of the early primordium. Plant Signal. Behav. 5, 982–984. doi: 10.4161/psb.5.8.12221
Su, Y. H., Liu, Y. B., and Zhang, X. S. (2011). Auxin-cytokinin interaction regulates meristem development. Mol. Plant 4, 616–625. doi: 10.1093/mp/ssr007
Sugano, S. S., Shimada, T., Imai, Y., Okawa, K., Tamai, A., Mori, M., et al. (2010). Stomagen positively regulates stomatal density in Arabidopsis. Nature 463, 241–244. doi: 10.1038/nature08682
Sugimoto-Shirasu, K., and Roberts, K. (2003). “Big it up”: endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6, 544–553. doi: 10.1016/j.pbi.2003.09.009
Sullivan, M., and Morgan, D. O. (2007). Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 8, 894–903. doi: 10.1038/Nrm2276
Sussex, I. M. (1951). Experiments on the cause of dorsiventrality in leaves. Nature 167, 651–652. doi: 10.1038/167651a0
Swain, S. M., Tseng, T. S., Thornton, T. M., Gopalraj, M., and Olszewski, N. E. (2002). SPINDLY is a nuclear-localized repressor of gibberellin signal transduction expressed throughout the plant. Plant Physiol. 129, 605–615. doi: 10.1104/Pp.020002
Swiatek, A., Azmi, A., Stals, H., Inze, D., and Van Onckelen, H. (2004). Jasmonic acid prevents the accumulation of cyclin B1;1 and CDK-B in synchronized tobacco BY-2 cells. FEBS Lett. 572, 118–122. doi: 10.1016/j.febslet.2004.07.018
Takatsuka, H., Ohno, R., and Umeda, M. (2009). The Arabidopsis cyclin-dependent kinase-activating kinase CDKF;1 is a major regulator of cell proliferation and cell expansion but is dispensable for CDKA activation. Plant J. 59, 475–487. doi: 10.1111/j.1365-313X.2009.03884.x
Takahashi, N., Kajihara, T., Okamura, C., Kim, Y., Katagiri, Y., Okushima, Y., et al. (2013). Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Curr. Biol. 23, 1812–1817. doi: 10.1016/j.cub.2013.07.051
Tarayre, S., Vinardell, J. M., Cebolla, A., Kondorosi, A., and Kondorosi, E. (2004). Two classes of the Cdh1-type activators of the anaphase-promoting complex in plants: novel functional domains and distinct regulation. Plant Cell 16, 422–434. doi: 10.1105/Tpc.018952
Tominaga, R., Iwata, M., Sano, R., Inoue, K., Okada, K., and Wada, T. (2008). Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135, 1335–1345. doi: 10.1242/dev.017947
Traw, M. B., and Bergelson, J. (2003). Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol. 133, 1367–1375. doi: 10.1104/pp.103.027086
Tsukaya, H. (2002). Interpretation of mutants in leaf morphology: genetic evidence for a compensatory system in leaf morphogenesis that provides a new link between cell and organismal theories. Int. Rev. Cytol. 217, 1–39. doi: 10.1016/S0074-7696(02)17011-2
Tsukaya, H. (2013a). Does ploidy level directly control cell size? Counterevidence from Arabidopsis genetics. PLoS ONE 8:e83729. doi: 10.1371/journal.pone.0083729
Umeda, M., Shimotohno, A., and Yamaguchi, M. (2005). Control of cell division and transcription by cyclin-dependent kinase-activating kinases in plants. Plant Cell Physiol. 46, 1437–1442. doi: 10.1093/pcp/pci170
Uyttewaal, M., Burian, A., Alim, K., Landrein, B., Borowska-Wykret, D., Dedieu, A., et al. (2012). Mechanical stress acts via katanin to amplify differences in growth rate between adjacent cells in Arabidopsis. Cell 149, 439–451. doi: 10.1016/j.cell.2012.02.048
Vandepoele, K., Vlieghe, K., Florquin, K., Hennig, L., Beemster, G. T. S., Gruissem, W., et al. (2005). Genome-wide identification of potential plant E2F target genes. Plant Physiol. 139, 316–328. doi: 10.1104/Pp.105.066290
Van Leene, J., Boruc, J., De Jaeger, G., Russinova, E., and De Veylder, L. (2011). A kaleidoscopic view of the Arabidopsis core cell cycle interactome. Trends Plant Sci. 16, 141–150. doi: 10.1016/j.tplants.2010.12.004
Van Leene, J., Hollunder, J., Eeckhout, D., Persiau, G., Van De Slijke, E., Stals, H., et al. (2010). Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6:397. doi: 10.1038/msb.2010.53
Vanneste, S., Coppens, F., Lee, E., Donner, T. J., Xie, Z., Van Isterdael, G., et al. (2011). Developmental regulation of CYCA2s contributes to tissue-specific proliferation in Arabidopsis. EMBO J. 30, 3430–3441. doi: 10.1038/emboj.2011.240
Vaten, A., and Bergmann, D. C. (2012). Mechanisms of stomatal development: an evolutionary view. Evodevo 3:11. doi: 10.1186/2041-9139-3-11
Veit, B. (2004). Determination of cell fate in apical meristems. Curr. Opin. Plant Biol. 7, 57–64. doi: 10.1016/j.pbi.2003.11.009
Veit, B. (2009). Hormone mediated regulation of the shoot apical meristem. Plant Mol. Biol. 69, 397–408. doi: 10.1007/s11103-008-9396-3
Verkest, A., Manes, C. L. D., Vercruysse, S., Maes, S., Van Der Schueren, E., Beeckman, T., et al. (2005a). The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17, 1723–1736. doi: 10.1105/tpc.105.032383
Verkest, A., Weinl, C., Inze, D., De Veylder, L., and Schnittger, A. (2005b). Switching the cell cycle. Kip-related proteins in plant cell cycle control. Plant Physiol. 139, 1099–1106. doi: 10.1104/pp.105.069906
Vernoux, T., Brunoud, G., Farcot, E., Morin, V., Van Den Daele, H., Legrand, J., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7, 508. doi: 10.1038/msb.2011.39
Vlad, D., Kierzkowski, D., Rast, M. I., Vuolo, F., Ioio, R. D., Galinha, C., et al. (2014). Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343, 780–783. doi: 10.1126/science.1248384
Vlieghe, K., Boudolf, V., Beemster, G. T., Maes, S., Magyar, Z., Atanassova, A., et al. (2005). The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr. Biol. 15, 59–63. doi: 10.1016/j.cub.2004.12.038
Waites, R., and Hudson, A. (1995). Phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121, 2143–2154.
Walker, J. D., Oppenheimer, D. G., Concienne, J., and Larkin, J. C. (2000). SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development 127, 3931–3940.
Wang, H., Qi, Q., Schorr, P., Cutler, A., Crosby, W., and Fowke, L. (1998). ICK1, a cyclin dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15, 501–510. doi: 10.1046/j.1365-313X.1998.00231.x
Wang, L., Gu, X., Xu, D., Wang, W., Wang, H., Zeng, M., et al. (2011). miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J. Exp. Bot. 62, 761–773. doi: 10.1093/jxb/erq307
Wang, L., and Ruan, Y. L. (2013). Regulation of cell division and expansion by sugar and auxin signaling. Front. Plant Sci. 4:163. doi: 10.3389/fpls.2013.00163
Wang, S., Kwak, S. H., Zeng, Q., Ellis, B. E., Chen, X. Y., Schiefelbein, J., et al. (2007). TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134, 3873–3882. doi: 10.1242/dev.009597
Wells, D. M., Laplaze, L., Bennett, M. J., and Vernoux, T. (2013). Biosensors for phytohormone quantification: challenges, solutions, and opportunities. Trends Plant Sci. 18, 244–249. doi: 10.1016/j.tplants.2012.12.005
Werner, T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., and Schmulling, T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15, 2532–2550. doi: 10.1105/tpc.014928
White, D. W. (2006). PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 13238–13243. doi: 10.1073/pnas.0604349103
Wolf, S., Hematy, K., and Hofte, H. (2012). Growth control and cell wall signaling in plants. Annu. Rev. Plant Biol. 63, 381–407. doi: 10.1146/annurev-arplant-042811-105449
Xie, Z., Lee, E., Lucas, J. R., Morohashi, K., Li, D., Murray, J. A., et al. (2010). Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. Plant Cell 22, 2306–2321. doi: 10.1105/tpc.110.074609
Xiong, Y., Mccormack, M., Li, L., Hall, Q., Xiang, C., and Sheen, J. (2013). Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186. doi: 10.1038/nature12030
Xiong, Y., and Sheen, J. (2012). Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J. Biol. Chem. 287, 2836–2842. doi: 10.1074/jbc.M111.300749
Xu, R., and Li, Y. (2011). Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 138, 4545–4554. doi: 10.1242/dev.071423
Yadav, R. K., Perales, M., Gruel, J., Girke, T., Jonsson, H., and Reddy, G. V. (2011). WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 25, 2025–2030. doi: 10.1101/gad.17258511
Yadav, R. K., Perales, M., Gruel, J., Ohno, C., Heisler, M., Girke, T., et al. (2013). Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol. Syst. Biol. 9, 654. doi: 10.1038/msb.2013.8
Yadav, R. K., and Reddy, G. V. (2011). WUSCHEL-mediated cellular feedback network imparts robustness to stem cell homeostasis. Plant Signal. Behav. 6, 544–546. doi: 10.4161/psb.6.4.14838
Yanai, O., Shani, E., Dolezal, K., Tarkowski, P., Sablowski, R., Sandberg, G., et al. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15, 1566–1571. doi: 10.1016/j.cub.2005.07.060
Yang, C. X., and Ye, Z. B. (2013). Trichomes as models for studying plant cell differentiation. Cell Mol. Life Sci. 70, 1937–1948. doi: 10.1007/s00018-012-1147-6
Yokoyama, R., and Nishitani, K. (2001). A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 42, 1025–1033. doi: 10.1093/Pcp/Pce154
Yoshizumi, T., Tsumoto, Y., Takiguchi, T., Nagata, N., Yamamoto, Y. Y., Kawashima, M., et al. (2006). Increased level of polyploidy1, a conserved repressor of CYCLINA2 transcription, controls endoreduplication in Arabidopsis. Plant Cell 18, 2452–2468. doi: 10.1105/tpc.106.043869
Zhang, F., Gonzalez, A., Zhao, M., Payne, C. T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869. doi: 10.1242/dev.00681
Zhang, Y., Persson, S., and Giavalisco, P. (2013). Differential regulation of carbon partitioning by the central growth regulator target of rapamycin (TOR). Mol. Plant 6, 1731–1733. doi: 10.1093/mp/sst094
Zhao, M., Morohashi, K., Hatlestad, G., Grotewold, E., and Lloyd, A. (2008). The TTG1–bHLH–MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135, 1991–1999. doi: 10.1242/Dev.016873
Zhao, X., Harashima, H., Dissmeyer, N., Pusch, S., Weimer, A. K., Bramsiepe, J., et al. (2012). A general G1/S-phase cell-cycle control module in the flowering plant Arabidopsis thaliana. PLoS Genet. 8:e1002847. doi: 10.1371/journal.pgen.1002847
Zhao, Y., Christensen, S. K., Fankhauser, C., Cashman, J. R., Cohen, J. D., Weigel, D., et al. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309. doi: 10.1126/science.291.5502.306
Keywords: leaf growth, developmental pathway, plant hormones, stress, modeling
Citation: Kalve S, De Vos D and Beemster GTS (2014) Leaf development: a cellular perspective. Front. Plant Sci. 5:362. doi: 10.3389/fpls.2014.00362
Received: 15 February 2014; Paper pending published: 25 May 2014;
Accepted: 07 July 2014; Published online: 31 July 2014.
Edited by:
Bernd Mueller-Roeber, University of Potsdam, GermanyReviewed by:
Camila Caldana, Brazilian Bioethanol Science and Technology Laboratory – O Centro Nacional de Pesquisa em Energia e Materiais/da Associação Brasileira de Tecnologia de Luz Síncrotron, BrazilCopyright © 2014 Kalve, De Vos and Beemster. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerrit T. S. Beemster, Laboratory for Molecular Plant Physiology and Biotechnology, Department of Biology, University of Antwerp, 171 Groenenborgerlaan, Antwerp 2020, Belgium e-mail:Z2Vycml0LmJlZW1zdGVyQHVhbnR3ZXJwZW4uYmU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.