
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 11 August 2014
Sec. Plant Pathogen Interactions
Volume 5 - 2014 | https://doi.org/10.3389/fpls.2014.00326
This article is part of the Research Topic Agrobacterium biology and its application to transgenic plant production View all 13 articles
 Tatiana V. Matveeva*
Tatiana V. Matveeva* Ludmila A. Lutova
Ludmila A. LutovaMost genetic engineering of plants uses Agrobacterium mediated transformation to introduce novel gene content. In nature, insertion of T-DNA in the plant genome and its subsequent transfer via sexual reproduction has been shown in several species in the genera Nicotiana and Linaria. In these natural examples of horizontal gene transfer from Agrobacterium to plants, the T-DNA donor is assumed to be a mikimopine strain of A. rhizogenes. A sequence homologous to the T-DNA of the Ri plasmid of Agrobacterium rhizogenes was found in the genome of untransformed Nicotiana glauca about 30 years ago, and was named “cellular T-DNA” (cT-DNA). It represents an imperfect inverted repeat and contains homologs of several T-DNA oncogenes (NgrolB, NgrolC, NgORF13, NgORF14) and an opine synthesis gene (Ngmis). A similar cT-DNA has also been found in other species of the genus Nicotiana. These presumably ancient homologs of T-DNA genes are still expressed, indicating that they may play a role in the evolution of these plants. Recently T-DNA has been detected and characterized in Linaria vulgaris and L. dalmatica. In Linaria vulgaris the cT-DNA is present in two copies and organized as a tandem imperfect direct repeat, containing LvORF2, LvORF3, LvORF8, LvrolA, LvrolB, LvrolC, LvORF13, LvORF14, and the Lvmis genes. All L. vulgaris and L. dalmatica plants screened contained the same T-DNA oncogenes and the mis gene. Evidence suggests that there were several independent T-DNA integration events into the genomes of these plant genera. We speculate that ancient plants transformed by A. rhizogenes might have acquired a selective advantage in competition with the parental species. Thus, the events of T-DNA insertion in the plant genome might have affected their evolution, resulting in the creation of new plant species. In this review we focus on the structure and functions of cT-DNA in Linaria and Nicotiana and discuss their possible evolutionary role.
Horizontal gene transfer (HGT) takes place widely in prokaryotes, where its ecological and evolutionary effects are well-studied (Koonin et al., 2001). Comparative and phylogenetic analyses of eukaryotic genomes show that considerable numbers of genes have been acquired by HGT. Gene acquisition by HGT is therefore a potential creative force in both eukaryotic and prokaryotic genome evolution. However, mechanisms of HGT are poorly understood in the Eukaryota in comparison to gene transfer among the Procaryotae. The persistence of horizontally transferred genes in some organisms may confer selective advantages (Koonin et al., 2001; Richardson and Palmer, 2007). Most examples of HGT in higher plants involve the transfer of chloroplast or mitochondrial DNA and have been the subject of numerous reviews (Dong et al., 1998; Richardson and Palmer, 2007). There are few descriptions of horizontal transfer of nuclear genes between species. One example is transfer of the gene that codes for the cytosolic enzyme phosphoglucose isomerase predicted to have occurred between Festuca ovina and some species from the genus Poa (Ghatnekar et al., 2006; Vallenback et al., 2008, 2010). Evidence of gene transfer from bacteria to the nuclei of multi-cellular eukaryotes is rare (Richards et al., 2006; Acuna et al., 2012). HGT from bacteria to plants has been restricted to Agrobacterium rhizogenes and representatives of genera Nicotiana and Linaria, and represents some of the most recent transfers in evolution (White et al., 1983; Intrieri and Buiatti, 2001; Matveeva et al., 2012; Pavlova et al., 2013).
A. rhizogenes, and the related bacterium A. tumefaciens, transform a wide variety of host plants by transferring a segment of the large tumor-inducing plasmid, called T-DNA, into host cells (White et al., 1982; Otten et al., 1992; Veena et al., 2003; Tzfira and Citovsky, 2006; Vain, 2007). The T-DNA is integrated through non-homologous recombination into the host cell genome where it is expressed. Expression of T-DNA genes results in the formation of hairy roots or crown galls, that are transgenic tissues, formed on a non-transgenic plant. This phenomenon is called “genetic colonization,” one of the examples of the host-parasite relationship (Tzfira and Citovsky, 2006). It is unclear whether or not colonized plants have received benefits from such colonization, however, we could expect that it is beneficial in some cases since there are footprints of HGT from Agrobacterium to plants in the genomes of several present day plant species.
In early investigations of Agrobacterium mediated transformation of plants, most researchers assumed that there was no significant homology to the T-DNA in untransformed plant genomes. White et al. (1982) attempted to detect pRiA4b T-DNA sequences in the genome of Nicotiana glauca, transformed in laboratory conditions by Agrobacterium rhizogenes strain A4. Southern analysis detected a fragment of pRiA4 in the transgenic tissue. Surprisingly, a hybridization signal was also detected in uninfected tissues of N. glauca. Further analysis confirmed the presence of DNA homologous to T-DNA in the N. glauca genome. This homologous DNA was referred to as “cellular T-DNA” (cT-DNA) (White et al., 1983).
Furner et al. (1986) investigated Nicotiana glauca plants, collected in geographically separated territories. Southern analyses showed the presence of cT-DNA in all studied varieties of N. glauca. Sequencing of the N. glauca cT-DNA demonstrated that it was organized as an imperfect inverted repeat. The left arm of cT-DNA, containing rolB and rolC homologs (NgrolB and NgrolCL) was more extended than the right arm, which contained only the rolC homolog (NgrolCR). The coding sequences of NgrolB and NgrolCR were found to contain early stop codons.
Subsequent analysis of the nucleotide sequence of this cT-DNA identified open reading frame 13 (ORF13) and ORF14 homologs in both the left and right arms, called NgORF13L, NgORF14L, NgORF13R, and NgORF14R, respectively (Aoki et al., 1994).
In 2001 Suzuki et al. characterized A. rhizogenes strain MAFF301724 and described a new opine synthase gene (mikimopine synthase gene mis). A part of the mis gene displayed strong homology to distal fragments of N. glauca cT-DNA, called NgmisL and NgmisR, respectively (Suzuki et al., 2002). Suzuki et al. (2002) suggested that the complete cT-DNA region of N.glauca is comprised of the 7968 bp left arm and 5778 bp right arm that were derived from the T-DNA of a mikimopine Ri plasmid similar to pRi1724. The level of nucleotide sequence similarity between the left and right arms is greater than 96% and the gene order is conserved suggesting a duplication event. The structure of the N.glauca cT-DNA is summarized in Figure 1. Since cT-DNA has been identified in all studied varieties of N. glauca (Furner et al., 1986), it is reasonable to suggest that the transformation event occurred before the formation of this species. This suggests that other related species may contain cT-DNA.
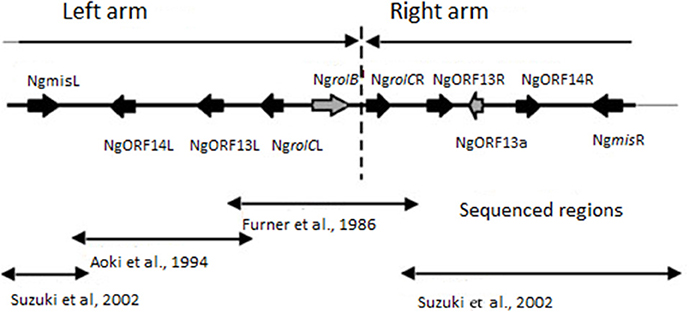
Figure 1. Structure of cT-DNA in the Nicotiana glauca genome (based on Suzuki et al., 2002). The cT-DNA and its flanking regions are indicated. Lines with a single arrowhead indicate the imperfect inverted repeat. Lines with arrowheads at both ends indicate regions sequenced by each of three groups.
The genus Nicotiana is one of the largest genera in the Solanaceae and contains 75 species that are characterized by a wide range of variations among their floral and vegetative morphology (Clarkson et al., 2004). The different Nicotiana species evidence interspecific crosses which complicates Nicotiana phylogeny. Goodspeed hypothesized that there are two distinct lineages in Nicotiana which arose from two ancestral pre-petunioid and pre-cestroid lineages. He supposed that the base chromosome number of the genus was 12 and stressed the role of doubling and hybridization in Nicotiana evolution. Goodspeed divided Nicotiana into three sub-genera Rustica, Tabacum, and Petunioides and 14 sections (Goodspeed, 1954). Since then, the number of subgenera of Nicotiana has remained constant, while the number and composition of the sections has been revised (Clarkson et al., 2004). 75% of tobacco species originate from the Americas and 25% of species are from Australia (Goodspeed, 1954; Clarkson et al., 2004).
Identification of T-DNA in N. glauca raises two questions: what other Nicotiana species contain cT-DNA, and what was the pattern of dissemination within the group?
To answer the first question Furner et al. (1986) examined the genomes of 17 species of the genus Nicotiana. Using Southern analyses he showed that only six species from the subgenera Rustica and Tabacum contained sequences homologous to the rol genes of Agrobacterium rhizogenes. These species are N. glauca, N. otophora, N. tomentosiformis, N. tomentosa, N. benavidesii, N. tabacum. Examination of T-DNA- like sequences in N. tabacum has shown that it contains a rolC homolog and two ORF13 homologs (trolC, tORF13-1 and tORF13-2, respectively) (Meyer et al., 1995; Frundt et al., 1998). Intrieri and Buiatti studied the distribution and evolution of Agrobacterium rhizogenes genes in the genus Nicotiana. Forty two species representing all Nicotiana sections were examined for the presence of rolB, rolC, ORF13, and ORF14 homologs in their genomes. T-DNA-like sequences detected were compared with each other and with contemporary sequences of Agrobacterium. The results demonstrated the presence of at least one T-DNA gene in each of 15 Nicotiana species representing all three subgenera. All currently available data on the distribution of cT-DNA among Nicotiana species are summarized in Table 1.
It is important to note, that there are some inconsistencies among the data by Furner et al. (1986) and Intrieri and Buiatti (2001). For example, Intrieri and Buiatti (2001) showed that T-DNA is present in N. debneyi and N. cordifolia. Furner et al. (1986) found no T-DNA in these species. This contradiction requires additional studies.
Thus, to date, T-DNA was found in every Nicotiana subgenus which include species, native to America and Australia (Goodspeed, 1954).
Phylogenetic analyses were performed by Intrieri and Buiatti (2001) to compare nucleotide sequences of cT-DNA in several Nicotiana species with the T-DNA of Agrobacterium. The following species were used in the analyses and represented all three subgenera: N. cordifolia (subgenus Rustica sec. Paniculatae); N. tomentosiformis and N. otophora (subgenus Tabacum), N. tomentosiformis (participated in N. tabacum speciation together with N. sylvestris); N. glauca used to be included in the subgenus Rustica sec. Paniculatae (Goodspeed, 1954), but later it was moved to the sec. Noctiflorae of the subgenus Petunioides (Knapp et al., 2004); and N. debneyi [Suaveolentes, an Australian section of Nicotiana, and a polyploid species of the subgenus Petunioides (Knapp et al., 2004)].
Analysis of nucleotide sequences revealed that N. cordifolia and N. glauca rolB, rolC, ORF13, and ORF14 genes show a high level of sequence similarity (93.5–98.5%). These data indicate that N. cordifolia and N. glauca are related species and are consistent with the proposal of Goodspeed (1954) that both species should be included in subgenus Rustica sec. Paniculatae. Similar clustering was found between the representatives of the subgenus Tabacum. Sequence similarities were lower between Rustica and Tabacum species, ranging from 66.3 to 68.6% for rolC and from 70.2 to 82.9% for ORF13 but was higher for ORF14 (94–97%). Surprisingly, the petunioid N. debneyi rolC gene demonstrates high sequence similarity (93.4%) with the N. glauca rolC gene, but lower similarity (around 67%) with those found in species belonging to the subgenus Tabacum. It was speculated that the polyploid species N. debneyi got cT-DNA from an ancestor of sec. Paniculatae. The homologies suggest that the Nicotiana rol genes parallel Nicotiana spp. evolution, being divided into two clusters, one that includes N. glauca, N. cordifolia, and N. debneyi, the second comprising species from the subgenus Tabacum (Intrieri and Buiatti, 2001).
Present day cT-DNA genes clustered with each other, making it difficult to predict which Ri-plasmid would be the source of the cT-DNA in each Nicotiana species. Since the pace of evolution differs between bacteria and plants, and since the pRi T-DNAs may have undergone rearrangements with each other (Moriguchi et al., 2001), it is difficult to define which ancient T-DNA was the origin of cT-DNA in different Nicotiana species using such phylogenetic analysis (Tanaka, 2008).
Another option for exploring the origin of cT-DNA is opine typing which was performed by Suzuki et al. (2002). They identified opine gene homologs in N. glauca (NgmisL and NgmisR, respectively) and screened 12 Nicotiana species for mis homologs using Southern blot hybridization. The analyses included five species from the subgenus Rustica (N. glauca, N. benavidesii, N. paniculata, N. knightiana, N. rustica), five species from the subgenus Tabacum (N. tomentosa, N. tomentosiformis, N. otophora, N. tabacum and N. glutinosa), and two species from the subgenus Petunioides (N. langsdorfii and N. sylvestris). Homologs of gene mis were detected in the genomes of N. glauca, N. tomentosa, N. tomentosiformis and N. tabacum, however, the size of the hybridized fragments was different between N. glauca and species in the subgenus Tabacum and the hybridization pattern in N. tomentosa was different from that of the two species in the subgenus Tabacum (N. tabacum and N. tomentosiformis). Since T-DNA fragments of N. tabacum were identical to those of N.tomentosiformis and were not detected in the genome of N. sylvestris, the mis gene of N. tabacum likely came from N. tomentosiformis.
Suzuki et al. (2002) sequenced DNA in N. glauca adjacent to the cT-DNA. To investigate regions adjacent to the cT-DNA in other species, Southern hybridization was carried out using either a DNA fragment outside the left or right arms of the cT-DNA of N. glauca as a probe. All examined Nicotiana genomes showed the presence of sequences homologous to both sides of the cT-DNA suggesting that these are original sequences existing in the genomes of Nicotiana plants. Similar size fragments were found in most species of the subgenus Rustica and Tabacum, which likely represent subgenus-specific restriction fragments. Interestingly, the signals using NgL and NgR as probes fell into the same fragment in the genomes of N. tomentosa, N. tomentosiformis, N. tabacum. Although the same DNA sequences bordering the cT-DNA in N. glauca were found in the genome of these three species, the sequences were not contiguous to the cT-DNA therefore the location of the cT-DNA in N. glauca is different from that in the species of the subgenus Tabacum.
Thus, the phylogenetic analysis undertaken by Intrieri and Buiatti (2001), and the study of opine genes and T-DNA integration sites performed Suzuki et al. (2002), suggest that there have been no less than two acts of Agrobacterium mediated transformation in the evolution of Nicotiana species (Figure 2). While the comparison of DNA sequences and the detection of mis homologs clearly demonstrates that the origin of the cT-DNA in N. glauca, N. tabacum, N. tomentosa, and N. tomentosiformis is derived from a mikimopine-type Ri plasmid similar to pRi1724, the origins of the cT-DNA in other species are still unknown.
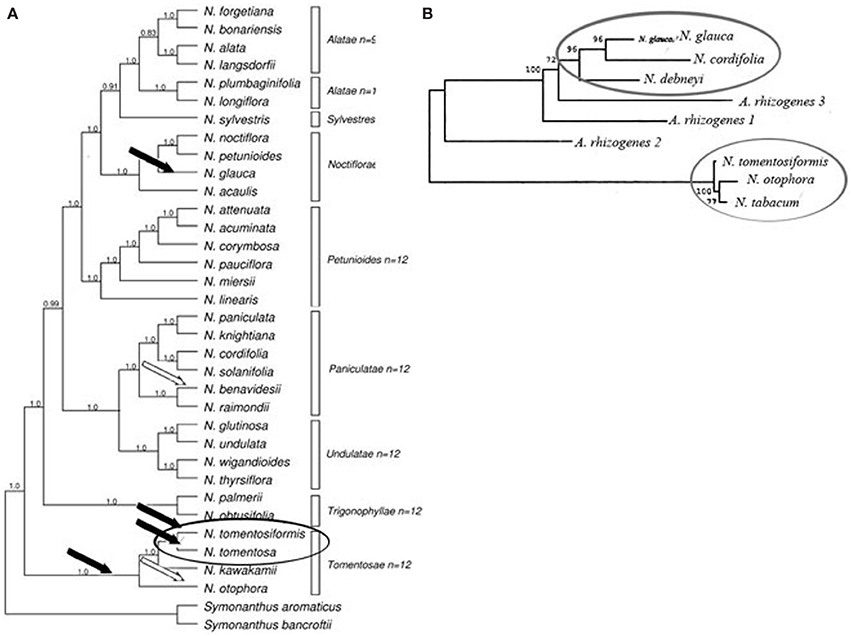
Figure 2. Phylogenetic analysis of Nicotiana (Clarkson et al., 2004), using ITS and chloroplast sequences together with T-DNA marker. (A) Bayesian analysis of diploids only combined dataset (plastid and ITS). Consensus of 40,001 trees with posterior probabilities shown above branches. Bars indicate Nicotiana sections according to Knapp et al. (2004), Black and white arrows indicate the deduced insertion events by mikimopine-type or unknown opine-type pRi T-DNAs correspondingly (Suzuki et al., 2002). (B) Phylogenetic analysis of rolC, gene (Intrieri and Buiatti, 2001) by neighbor-joining method. Ovals show results of possible independent transformation events.
pRi transgenic plants exhibit a specific phenotype (dwarfing, loss of apical dominance, increased root mass, and decreased rate of fertilization) (Tepfer, 1984). However, Nicotiana species that contain cT-DNA in their genomes show no such phenotype. Are these genes expressed and functional, or they are pseudogenes?
Early experiments by Taylor et al. (1985) did not detect transcripts of the cT-DNA genes in Nicotiana glauca. Other researchers were able to detect transcripts of NgrolB, NgrolC, NgORF13, NgORF14 in callus tissues of N. glauca (Aoki and Syono, 1999a) as well as in genetic tumors of F1 N. glauca × N. langsdorffii hybrids, but not in leaf tissues of the same hybrid (Ichikawa et al., 1990; Aoki et al., 1994). Northern analyses of N. tabacum oncogenes showed that a trolC transcript accumulated in shoot tips and young leaves. The expression pattern of tORF13 was similar to trolC, however, tORF13 expression was detected in flowers (Meyer et al., 1995; Frundt et al., 1998).
Intrieri and Buiatti studied transcription of the cT-DNA genes using RT-PCR in a number of species, using N. langsdorffii as a negative control. For their analyses authors used leaves from young in vitro-grown plantlets and hormone autotrophic (habituated) callus tissues grown on a hormone-free cultural medium as previously described by Bogani et al. (1985).
In this study rolB transcripts were found to be present in all cases in habituated callus tissues, but not in leaves; rolC was expressed in calli as well as in leaves of Rustica species (N. glauca and N. cordifolia), and only in calli in species representing the subgenus Petunioides; no expression was demonstrated to occur in species from the subgenus Tabacum. ORF13 and 14 mRNA was always detected in calli and ORF13 transcripts were found in leaf tissues of N. tabacum and N. tomentosiformis.
Aoki and Syono (1999b, 2000) analyzed the function of Ngrol genes by transforming leaf explants of N. tabacum and N. debneyi with A. tumefaciens that harbored either a rolB-rolC-ORF13-ORF14 fragment from pRi or cT-DNA of N. glauca. Nearly all of the leaf segments inoculated with pRi fragment developed hairy roots. No significant root growth, however, appeared on the explants treated with A. tumefaciens that harbored Ng cT-DNA.
A comparison of the nucleotide sequences of NgrolB and RirolB indicates that these oncogenes have different length coding regions. Each ORF starts at the same position, but NgrolB ends at early termination codon 633 bp from the initiation site. The authors suggested that N. glauca plants do not exhibit the hairy-root phenotype due to the truncation of the NgrolB reading frame.
A comparison of the DNA sequences of NgrolC and RirolC indicates that the reading frame of NgrolC begins and terminates at the same positions as RirolC. Nicotiana tabacum leaf disks were transformed with the P35S-NgrolC chimeric construction, yielded transformants that expressed a dramatically dwarfed phenotype, probably because of the reduced length of internodes. The leaves of these P35S-NgrolC transgenic plants were lanceolated and pale green, with floral organs that were thin and small. These characteristics were identical to the phenotype of the P35S-RirolC transgenic plants, described earlier (Schmülling et al., 1988). Transgene expression was detected only in transformants that demonstrated these characteristic morphological shifts, no transcripts were detected in leaf tissues from a comparable T0 plant demonstrating a normal phenotype (Aoki and Syono, 1999b).
To compare the expression patterns between the Ngrol genes of N. glauca and the Rirol genes of Agrobacterium rhizogenes, Nagata et al. (1996) carried out fluorometric and histochemical analyses of the tissues from transgenic genetic tumors, growing on the hybrid of Nicotiana glauca × N. langsdorffii (F1) that contained a beta-glucuronidase (GUS) reporter gene fused to the promoter of (NgrolB, NgrolC, RirolB, or RirolC. In all constructs they studied, significantly higher GUS activity was found in tumors than in the other organs (roots, stems, and leaves) of transgenic plants. The tendency toward higher GUS activities in tumors than in normal tissues seen with the RirolB and RirolC promoters was also seen with the NgrolB and NgrolC promoters. GUS activities from the rolB promoter expressed in normal F1 plants were, however, different from those seen from the rolC promoter. The expression of the RirolB and NgrolB promoters in stems, roots, and leaves were 10–100 fold lower than in genetic tumors. Almost no activity was detected in leaves. By contrast, expression from the RirolC and NgrolC promoters was only 1.5–10 fold lower than in genetic tumors and a significant activity was detected in leaves.
Histochemical analysis of transgenic normal Fl plant tissues showed that NgrolB-GUS and NgrolC-GUS, as well as RirolB-GUS and RirolC-GUS, had common tissue-specific expression patterns. NgrolB-GUS normal Fl transgenic plant tissues displayed high GUS activity in the meristematic zones of roots and in the apexes of shoots. A similar pattern of staining was ob-served in the RirolB-GUS transgenic plants. In the case of NgrolC-GUS and RirolC-GUS normal Fl transgenic plant tissues, GUS activity was observed primarily in the apices, vascular bundles of leaves, stems, and roots.
Expression of the mis gene homologs in N. glauca was detected by RT-PCR (Suzuki et al., 2002). It was shown that both homologs of the mis gene were amplified by RT-PCR using separate ortholog specific primers. These data support the hypothesis that the mis homologs are not pseudogenes. Transgenic plants transformed by the T-DNA of the wild type plasmid Ri1724 or by the mis gene alone, synthesize mikimopine in different organs (Suzuki et al., 2001), although no mikimopine accumulation was detected in wild-type N. glauca by paper electrophoresis. It is therefore likely that Ngmis homologs are transcribed at a very low level.
A full-length NgmisR homolog was isolated and integrated into an expression vector in Escherichia coli. The purified Mis protein was able to catalyze synthesis of mikimopine from L-histidine and α-ketoglutaric acid in a reaction buffer supplemented with NADH as a co-factor (Suzuki et al., 2002).
Thus, the oncogenes of Nicotiana cT-DNA are expressed in different tissues of present day tobacco plants at a low level and are therefore not pseudogenes.
Genetic tumors appear in certain genotypes spontaneously without being induced by any detectable environmental factor, and the tumor state is hereditary. Spontaneous genetic tumors in Nicotiana were first reported by Tanaka (2008). They have been detected throughout the plant and in whole progeny populations of certain crosses of Nicotiana species (Kehr and Smith, 1954). In some hybrids, genetic tumors have been reported to be formed irregularly in some of the offspring or limited to certain organs of the plants (Smith, 1958). It has been proposed that certain genes appropriately combined in a hybrid promote the development of these genetic tumors (Naf, 1958; Ahuja, 1968). Naf (1958) divided Nicotiana species into two groups, so called “plus” and “minus” groups. The “plus” group consists mainly of the species of the section Alatae whereas the minus group contains species from several sections. Crosses between the species within “plus” or “minus” groups do not produce tumorous progeny, while crosses between species from “plus” and “minus” groups do. Ahuja (1968) hypothesized that the species belonging to the “plus” group have a gene or a locus defined as initiator (I) and the species belonging to the “minus” group have a number of genes or loci (ee) for tumor enhancement and expression. For tumor formation both I and ee loci must be present.
Fujita (1994) expect that most species belonging to the minus group contain cT-DNA and that its genes could somehow be associated with the formation of genetic tumors on the Nicotiana hybrids. However, since there are no reports showing a connection between the ee genes and cT-DNA genes so far, this promising hypothesis has not yet been validated (Tanaka, 2008). As already mentioned, NgrolB, NgrolC, NgORF13 and NgORF14 genes are transcribed in genetic tumors on N. glauca × N. langsdorffii F1 hybrids (Ichikawa et al., 1990; Aoki et al., 1994). Some of these genes function in several organs of non-tumorous hybrid plants, like their counterparts in pRi T-DNA (Nagata et al., 1995, 1996; Udagawa et al., 2004). As soon as tumorigenesis is initiated by aging or stress, these genes are active in the developing outgrowth in a regulated manner. This means that a high level of expression of Ngrol genes is correlated with tumor formation on an F1 hybrid. However, it has not been determined if the formation of tumors is caused by the expression of Ngrol genes (Tanaka, 2008). Moreover, the stem and leaf tissues of Nicotiana species accumulate transcripts of the Ngrol genes (Meyer et al., 1995; Frundt et al., 1998). These observations suggest that the expression of these Ngrol genes might be unrelated to the induction of tumors. Overexpression of NgrolC, NgORF13, or tORF13 cause the proliferation of cells on carrot disks (Frundt et al., 1998), and morphological alterations of tobacco explants, similar to hairy root syndrome on transgenic plants (Aoki and Syono, 1999a,b). Therefore, cT-DNA oncogenes may be responsible for enhancing the development of genetic tumors (Tanaka, 2008).
It is widely discussed that phytohormones contribute to genetic tumor formation. The role of auxin and cytokinins in genetic tumor formation in Nicotiana, however, has been disputed. On the one hand, in the light-grown tissues of genetic tumors, indole acetic acid (IAA) was found to be the predominant auxin and its level increased during tumor initiation (Bayer, 1967; Ichikawa et al., 1989). On the other hand, in dark conditions endogeneous IAA remained at a constant, low level throughout the tumorigenetic process (Fujita et al., 1991). A higher cytokinin level was associated with tumorigenesis in tumor-prone hybrid tissues. While analyzing the role of cytokinins in Nicotiana genetic tumor formation, Feng et al. (1990) have shown that tumor formation of X-ray-induced non-tumorous mutants of N. glauca × N. langsdorffii was restored either by the insertion of the A. tumefaciens ipt gene, which encodes the key enzyme of cytokinin biosynthesis, or by the addition of cytokinin. Nandi et al. (1990) determined the profile of endogenous cytokinins in genetic tumors of N. glauca (Grah.) × N. langsdorffii (Weinm.) hybrids. They showed that while zeatin is predicted to be the predominant endogenous cytokinin in tissues of all ages, the genetic tumor tissue derived from this hybrid does not contain notably high endogenous cytokinin levels.
Since tumor growth may be caused not only by high concentrations of hormones, but also by enhanced sensitivity to them, this may explain the contradictory data on the content of hormones in tumors. Even if hormone levels are increased in tumors, it is not necessarily caused by the expression of the cT-DNA genes: rol genes can be regulated by hormones. For example, it was shown that expression of NgrolB was induced by auxin, as was RirolB, probably through the presence of the auxin-responsive cis-element ACTTTA found in the promoters of NgrolB and RirolB which is acted upon by the trans-factor NtBBF1 (Tanaka, 2008).
It is clear that further work will be needed to establish the relationship between cT-DNA oncogenes and genetic tumorigenesis in Nicotiana.
The Solanaceae is a large angiosperm family containing many economically important crops. A. rhizogenes is known to infect species belonging to different Solanaceae genera. Intrieri and Buiatti (2001) attempted to identify T-DNA-like sequences in species belonging to genera Cestrum, Petunia, and Solanum (C. parqui, C. foetidus, P. hybrida, S. tuberosa, S. melongena, C. annuum, and S. lycopersicon) using the same screening procedure, as they did for Nicotiana. None of the species screened showed amplification by PCR and no hybridization was obtained using A. rhizogenes and N. glauca probes. Kulaeva et al. (2013) extended the analysis to species from genus Solanum looking for T-DNA-like sequences. The authors used TaqMan real-time PCR with degenerate primers and probes for rolB, rolC, ORF13, ORF14 to analyze the following species: S. chmielewskii, S. esculentum var. cerasiforme, S. glabratum, S. habrochaites, S. peruvianum, S. pimpinellifolium, S. cheesmanii, S. parviflorum, S. chilense, S. acaule, S. ajanhuiri, S. albicans, S. andigenum, S. berthaultii, S. boyacense, S. boyacense, S. canarense, S. canarense, S. cardiophyllum, S. chacoense, S. chaucha, S. chocclo, S. curtilobum S. demissum, S. demissum, S. doddsii, S. dulcamara, S. fendleri, S. goniocalyx, S. hjertingii, S. hondelmanii, S. hougassi, S. jamesii, S. juzepczukii, S. kurtzianum, S. mamilliferum, S. phureja, S. pinnatisectum, S. pinnatisectum, S. polytrichon, S. riobambense, S. rybinii, S. sparsipilum, S. spegazzinii, S. stenotomum, S. stoloniferum, S. tarijense, S. tenuifilamentum, S. tuberosum, S. vernei, S. verrucosum, S. oplocense. They used N. tabacum DNA as positive control and N. langsdorffii as negative control. Amplification of specific sequences was not detected in any of the tested species.
This data shows that the presence of cT-DNA is not a feature of whole Solanaceae family, but has only been described to date for members of the genus Nicotiana.
Given the documented occurrence of cT-DNA, it is reasonable to hypothesize that other plant species, outside of the family Solanaceae, would have been transformed by Agrobacterium and contain at least remnants of cT-DNA.
The existence of cT-DNAs in species outside of the family Solanaceae has been reported by several groups. Using Southern analyses, sequences similar to pRi T-DNA were found in the genomic DNA of normal carrot (Daucus carota) (Spano et al., 1982), field bindweed (Convolvulus arvensis) (Tepfer, 1982); and carpet bugleweed (Ajuga reptans) (Tanaka, 2008). These studies did not involve DNA sequencing so they were not able to confirm whether there is T-DNA in the analyzed species.
Prior work in our group (Matveeva et al., 2012) attempted to clarify whether fixed T-DNA is present in other species outside the genus Nicotiana, and to evaluate the evolutionary relevance of natural T-DNA transfer. We sought to quickly screen a large number of plant genomes for the presence of T-DNA from both A. tumefaciens and A. rhizogenes using a modification of TaqMan-based real-time PCR (Livak et al., 1995) that combines the positive features of PCR and DNA blot hybridization in a single reaction (Matveeva et al., 2006). The search was limited to dicotyledonous plants native to temperate zones (mild winter, warm summer, and sufficient rainfall) due to the common occurrence of Agrobacterium in soils under these climate conditions.
This work analyzed 127 dicotyledonous plant species belonging to 38 different families. Species included carrot (Daucus carota) and field bindweed (Convolvulus arvensis), mentioned above. Each of the plants was screened for DNA sequences homologous to two different sets of T-DNA oncogenes: The first set included sequences homologous to A. rhizogenes oncogenes (rolB, rolC, ORF13, and ORF14), and the second contained sequences homologous to A. tumefaciens oncogenes (tms1 and tmr). Plant DNA samples from 126 species did not display detectable fluorescent signals for the T-DNA genes from either A. rhizogenes or A. tumefaciens. However, DNA samples isolated from several plants of L. vulgaris gave a positive result. In contrast, amplification was not observed using primers for the tms1 and tmr genes of A. tumefaciens (Matveeva et al., 2012).
The absence of T-DNA homologs in most of the plant species investigated leads us to the conclusion that HGT from Agrobacterium is a rare event in the plants. However, finding cT-DNA sequences in the genomes of species other plants than Nicotiana indicates that HGT from Agrobacterium to plants occurs outside of this genus.
It is interesting to note that the only examples of HGT demonstrated thus far occur in plants transformed by A. rhizogenes, but not A. tumefaciens. This may suggest that infection induced by A. rhizogenes is more efficient than that induced by A. tumefaciens (Tepfer, 1984).
Sequences homologous to T-DNA oncogenes in Linaria vulgaris, were identified by real time PCR and named LvrolB, LvrolC, LvORF13, LvORF14. BLAST analyses demonstrated the highest level of sequence identity (93%) between LvrolC and RirolC from the pRiA4 of A. rhizogenes. LvORF14 had the lowest similarity to the corresponding Agrobacterium oncogene (85%). A homolog of a gene for mikimopine synthase (mis) was also identified. To define the full extent of the cT-DNA integrated into Linaria vulgaris genome, a chromosome walking approach was performed to identify the upstream fragment of the Lv cT-DNA. This work indicated that the L. vulgaris genome contains two copies of cT-DNA which are organized as an imperfect direct repeat. Analysis of the cT-DNA copies demonstrated that both of them contain sequences similar to the following genes: ORF2, ORF3, ORF8, rolA, rolB, ORF 13, ORF1, and mis. The left side of the repeat contains additional sequence, homologous to part of the agrocinopine synthase (acs) gene. Analysis of the flanking regions of the Lv T-DNA was performed by real-time thermal asymmetrical interlaced (TAIL)-PCR with primers and probes to the Lvmis gene. The flanking plant DNA identified in this analysis was found to be similar to the Ty3/gypsy-like retrotransposon (Matveeva et al., 2012).
Samples of L. vulgaris were collected in the European part of Russia in Moscow, Voronezh and Krasnodar regions, and in the Asian part of Russia in the Novosibirsk, Tumen, and Chelyabinsk regions. The distance between the most western to the most eastern points was about 4000 km. The distance from the most northern to the most southern points was about 2000 km. Two to three plants were analyzed from each of these collection points. All of the samples contained T-DNA-like sequences, however, there was polymorphism among their nucleotide sequences (Matveeva et al., 2012).
Analysis of the sequences for both L. vulgaris homologs of rolC demonstrated that they contained intact open reading frames. The fragments corresponding to the coding regions of genes LvrolB, LvORF13, LvORF14, and Lvmis contain several stop codons or frameshifts that alter the ORFs. An analysis of the expression of these genes was carried out in tissues from the internodes, leaves, and roots of 1 month old, in vitro aseptically grown plants using RT real-time PCR. No mRNA corresponding to LvrolB, LvrolC, LvORF13, LvORF14 and Lvmis genes was amplifiable from these samples, therefore, the rol genes do not appear to be transcribed in L. vulgaris (Matveeva et al., 2012).
Toadflaxes (Linaria Mill.) form the largest genus within the tribe Antirrhineae. Linaria includes about 150 species that are widely spread in the Palearctic region, but the representatives of the genus are the most variable in the Mediterranean basin. The origin of the genus has been placed in the Miocene era (Fernandez-Mazuecos and Vargas, 2011) predating the Messinian Salinity Crisis (Hsu et al., 1977). The latest classification of the genus Linaria accepts seven sections (Linaria, Speciosae, Diffusae, Supinae, Pelisserianae, Versicolores, and Macrocentrum) (Sutton, 1988).
Studies indicate that cT-DNA exists in a number of Linaria species belonging to the sections Linaria and Speciosae (Matveeva and Kosachev, 2013; Pavlova et al., 2013). No cT-DNA was detected in Linaria species outside these sections. It appears that rolC is concerved among studied Linaria species based on the sequencing analysis of rolC homologs in L. genistifolia subsp. dalmatica (sec. Speciosae) and L. acutiloba (sec. Linaria) (Matveeva and Kosachev, 2013; Pavlova et al., 2013).
Thus, HGT of T-DNA from Agrobacterium to plants is not limited to Nicotiana spp, it has also occurred in the genus Linaria. The rolC homolog is the most conserved gene among the cT-DNA genes in Linaria and Nicotiana spp. In both genera plants were transformed by a mikimopine strain of A. rhizogenes.
The existence of several independent acts of Agrobacterium mediated transformation of plants and the maintenance of the cT-DNA in plant genomes during the process of evolution propose, that T-DNA-like sequences may give some selective advantages to the transformed plants (Ichikawa et al., 1990; Matveeva et al., 2012).
Suzuki et al. (2002) mentioned two possible functions of cT-DNA: increasing root mass leading to tolerance to drought, and changing the biological environment, particularly the soil microbiome represented by root-associated bacterial populations. Increasing root mass would seem beneficial for tolerance to dry conditions. Hence, ancient transformed plants with increased root mass might have demonstrated increased tolerance to dry environments surviving in arid conditions (Tanaka, 2008). However, no phenotype of the hairy root disease is observed in Nicotiana and L. vulgaris plants. In contrast, L. vulgaris explants show in vitro a shooty phenotype and in representatives of both genera rolB is mutated. Among the oncogenes of pRi T-DNA, rolB gene function seems to be the most important for hairy root induction because transformation of plants by the RirolB gene alone can induce hairy root formation. In contrast to the pRirolB, the NgrolB gene alone or in combination with other N. glauca homologs of A. rhizogenes oncogenes did not induce adventitious roots (Aoki and Syono, 1999a,b).
Aoki and Syono (1999b) performed base substitutions at two nucleotide positions, using site-directed mutagenesis, with the aim of producing a full-length form of NgrolB capable of stimulating adventitious root induction. Transgenic plants overexpressing this altered NgrolB demonstrated typical morphogenetic abnormalities. This experiment shows the possibility that a functional rolB gene may have operated during early steps of the evolution of transgenic Nicotiana.
Identification and sequencing of the mis homologs in Nicotiana and Linaria suggests that the origin of their cT-DNA is probably the mikimopine Ri plasmid. The presence of this gene may be related to plant–microbe interactions. Oger et al. (1997, 2000) reported that producing opines in genetically modified plants alters their ecological environment, in particular, changing the soil microbiome and root-associated microbe populations. If the synthesis of opines were beneficial for a plant species (even at a low level, in a specific tissue, or at a specific stage of oncogenesis), it may impact the appearance of advantageous plant–bacterium interactions. Plants maintaining cT-DNA in the genome could potentially maintain certain species of microorganisms in their rhizosphere via the secretion of opines in the root zone. Such potentially beneficial bacteria in the rhizosphere may in turn influence the root microbiome and convey nutritional and/or defensive features.
Early flowering or a shift from biennial to annual lifecycle without vernalization can take place on pRi transgenic Cichorium intybus and Daucus carota plants (Limami et al., 1998). These flowering features are beneficial when propagating such transgenic plants over the untransformed parentals. When considering the adaptational potential of natural transformation, the authors focused on the occurrence of flowering in the absence of a cold treatment. Given the mobility of seeds by wind, animals, and water, it is likely that biennial varieties or ecotypes may be transported to the southern latitudes where annualism would be beneficial. However, Nicotiana and Linaria species are not biennials (Goodspeed, 1947; Sutton, 1988; Blanco-Pastor et al., 2012). In addition, cT-DNA containing Linaria species from the sections Linaria and Speciosae are perennial, while other sections contain annual species. It is interesting to note that the cT-DNA containing sections are found worldwide while other sections are in the Mediterranean region and the Pyrenees (Table 2) (Sutton, 1988). It is unclear if this observation is due to the rarity by which plants acquire permanent cT-DNA, or if its foundation is related to some fitness benefit conferred by the cT-DNA. It can be speculated, however, that the ecological plasticity of species within the sections Linaria and Speciosae is somehow associated with the presence of cT-DNA in their genomes.
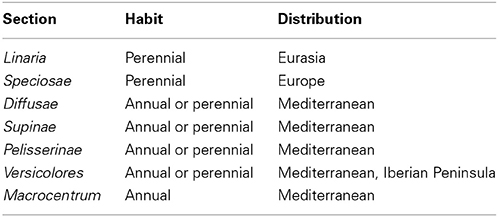
Table 2. Major features of infrageneric taxa of the genus Linaria (according to Sutton, 1988).
It would appear, therefore, that annualism is not related to natural transformation in Nicotiana and Linaria.
It is interesting to note that rolC is the most conserved gene among the cT-DNA oncogenes found in Nicotiana and Linaria (Intrieri and Buiatti, 2001; Mohajjel-Shoja et al., 2011). In some representatives of the Nicotiana, only rolC is able to encode a functional product (Mohajjel-Shoja et al., 2011). The same trend was observed for Linaria T-DNA-like sequences (Matveeva et al., 2012; Matveeva and Kosachev, 2013). The function of rolC, however, is poorly understood. It has been speculated that the product of rolC releases cytokinins from conjugates (Estruch et al., 1991). Other researchers demonstrated that the RolC protein participates in the processes of sucrose metabolism and/or transport (Nilsson and Olsson, 1997; Mohajjel-Shoja et al., 2011). RolC has also been proposed to promote somatic embryogenesis in plants (Gorpenchenko et al., 2006). Such data are consistent with a cytokinin function of the gene. Constitutive expression of rolC in cultured plant tissues activates secondary metabolism: the rolC gene alone increases production of tropane alkaloids, pyridine alkaloids, ginsenosides, and anthraquinones among others (Bulgakov et al., 1998, 2002; Palazon et al., 1998a; Bonhomme et al., 2000a,b; Bulgakov, 2008) and stimulates the expression of pathogenesis-related proteins (Kiselev et al., 2007). It is unclear how the rolC gene product mediates such pleiotropic effects, further biochemical characterization of RolC is required. This is complicated by the fact that rolC has no significant homology with any other genes (of prokaryotic or eukaryotic organisms) whose function is known (Bulgakov, 2008).
Activation of secondary metabolism in transformed cells may be due to the action of other rol genes (Chandra, 2012). Shkryl et al. (2008) studied the influence of rol genes products on secondary metabolism of Rubia cordifolia. They investigated rol genes individually and studied their combined action. They found that individual rolA, rolB, and rolC genes were able to stimulate biosynthesis of anthraquinones in transformed calli. The strongest anthraquinone—stimulating activity was detected for an R. cordifolia culture overproducing RolB where they saw a 15-fold increase of anthraquinone accumulation as compared to untransformed calli. The rolA- and rolC-expressing calli produced 2.8- and 4.3-fold higher amounts of anthraquinones, correspondingly. Palazon et al. (1998b) reported that the rolA gene stimulated production of nicotine.
Thus, increasing the amount of secondary metabolites is a characteristic of tissues where rol genes are expressed. This property can be useful for plants, because secondary metabolites may contribute to the resistance of plants to pests. It seems likely that a possible function cT-DNA is to mediate how plants interact with their environment by secreting opines and/or by changing the amounts of secondary metabolites. It will be essential to confirm such hypotheses through additional experimentation that might include silencing or excision experiments that are now possible using CRISPR technology (Qi et al., 2013).
The study of the long term impacts of HGT by Agrobacterium in plant lineages is in the early stages. However, we can note some trends:
– HGT of T-DNA from Agrobacterium to plants occurred in the evolution of several genera, at least Nicotiana (family Solanaceae) and Linaria (family Plantaginaceae);
– in both genera plants were transformed by a mikimopine strain of A. rhizogenes;
– a rolC homologe is the most conserved gene among the T-DNA genes in Linaria and Nicotiana spp;
– In Linaria vulgaris and Nicotiana glauca there are more than one copy of T-DNA per genome.
Continued studies of the genetic and biochemical effects of cT-DNA integration in naturally transgenic plants are important and will continue to provide insights into the impact of such rare acquisitions on plant evolution.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Derek Wood (Seattle Pacific University) for useful discussion and critical reading of the manuscript. This paper was prepared within the framework of the thematic plan of St. Petersburg State University ## 0.37.526.2013; 1.39.315.2014 and supported by a grant to Tatiana V. Matveeva from the Russian Foundation for Basic Research #14-04-01480.
Acuna, R., Padilla, B. E., Florez-Ramos, C. P., Rubio, J. D., Herrera, J. C., Benavides, P., et al. (2012). Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proc. Natl. Acad. Sci. U.S.A. 109, 4197–4202. doi: 10.1073/pnas.1121190109
Ahuja, M. A. (1968). An hypothesis and evidence concerning the genetic components controlling tumor formation in Nicotiana. Mol. Gen. Genet. 103, 176–184. doi: 10.1007/BF00427144
Aoki, S., Kawaoka, A., Sekine, M., Ichikawa, T., Fujita, T., Shinmyo, A., et al. (1994). Sequence of the cellular T-DNA in the untransformed genome of Nicotiana glauca that is homologous to ORFs 13 and 14 of the Ri plasmid and analysis of its expression in genetic tumors of N. glauca × N. langsdorffii. Mol. Gen. Genet. 243, 706–710.
Aoki, S., and Syono, K. (1999a). Function of Ngrol genes in the evolution of Nicotiana glauca: conservation of the function of NgORF13 and NgORF14 after ancient infection by an Agrobacterium rhizogenes-like ancestor. Plant Cell Physiol. 40, 222–230.
Aoki, S., and Syono, K. (1999b). Horizontal gene transfer and mutation: Ngrol genes in the genome of Nicotiana glauca. Proc. Natl. Acad. Sci. U.S.A. 96, 13229–13234.
Aoki, S., and Syono, K. (2000). The roles of Rirol and Ngrol genes in hairy root induction in Nicotiana debneyi. Plant Sci. 159, 183–189. doi: 10.1016/S0168-9452(00)00333-2
Bayer, M. H. (1967). Thin-layer chromatography of auxin and inhibitors in Nicotiana glauca, N. langsdorffii and three of their tumor-forming hybrids. Planta. 72, 329–337.
Blanco-Pastor, J. L., Vargas, P., and Pfeilm, B. E. (2012). Coalescent Simulations Reveal Hybridization and Incomplete Lineage Sorting in Mediterranean Linaria. PLoS ONE 7:e39089. doi: 10.1371/journal.pone.0039089
Bogani, P., Buiatti, M., Tegli, S., Pellegrini, M. G., Bettini, P., and Scala, A. (1985). Interspecific differences in differentiation and dedifferentiation patterns in the genus Nicotiana. Plant Syst. Evol. 151, 19–29. doi: 10.1007/BF02418016
Bonhomme, V., Laurain, M. D., and Fliniaux, M. A. (2000a). Effects of the rolC gene on hairy root: induction development and tropane alkaloid production by Atropa belladonna. J. Nat. Prod. 63, 1249–1252. doi: 10.1021/np990614l
Bonhomme, V., Laurain-Mattar, D., Lacoux, J., Fliniaux, M., and Jacquin-Dubreuil, A. (2000b). Tropane alkaloid production by hairy roots of Atropa belladonna obtained after transformation with Agrobacterium rhizogenes 15834 and Agrobacterium tumefaciens containing rol A, B, C genes only. J. Biotechnol. 81, 151–158. doi: 10.1016/S0168-1656(00)00287-X
Bulgakov, V. P. (2008). Functions of rol genes in plant secondary metabolism. Biotechnol. Adv. 26, 318–324. doi: 10.1016/j.biotechadv.2008.03.001
Bulgakov, V. P., Khodakovskaya, M. V., Labetskaya, N. V., Chernoded, G. K., and Zhuravlev, Y. N. (1998). The impact of plant rolC oncogene on ginsenoside production by ginseng hairy root cultures. Phytochemistry 49, 1929–1934. doi: 10.1016/S0031-9422(98)00351-3
Bulgakov, V. P., Tchernoded, G. K., Mischenko, N. P., Khodakovskay, M. V., Glazunov, V. P., Zvereva, E. V., et al. (2002). Effects of salicylic acid, methyl jasmonate, etephone and cantharidin on anthraquinone production by Rubia cordifolia callus cultures transformed with rolB and rolC genes. J. Biotechnol. 97, 213–221. doi: 10.1016/S0168-1656(02)00067-6
Chandra, S. (2012). Natural plant genetic engineer Agrobacterium rhizogenes: role of T-DNA in plant secondary metabolism. Biotechnol. Lett. 34, 407–415. doi: 10.1007/s10529-011-0785-3
Clarkson, J. J., Knapp, S., Garcia, V. F., Olmstead, R. G., Leitch, A. R., and Chase, M. W. (2004). Phylogenetic relationships in Nicotiana (Solanaceae) inferred from multiple plastid DNA regions. Mol. Phylogenet. Evol. 33, 75–90. doi: 10.1016/j.ympev.2004.05.002
Dong, F., Wilson, K. G., and Makaroff, C. A. (1998). Analysis of the four cox2 genes found in turnip (Brassica campestris, Brassicaceae) mitochondria. Am. J. Bot. 85, 153–161. doi: 10.2307/2446303
Estruch, J. J., Chriqui, D., Grossmann, K., Schell, J., and Spena, A. (1991). The plant oncogene rolC is responsible for the release of cytokinins from glucoside conjugates. EMBO J. 10, 2889–2895.
Feng, X. H., Dube, S. K., Bottino, P. J., and Kung, S. D. (1990). Restoration of shooty morphology of a nontumorous mutant of Nicotiana glauca × N. langsdorffii by cytokinin and the isopentenyltransferase gene. Plant Mol. Biol. 15, 407–420.
Fernandez-Mazuecos, M., and Vargas, P. (2011). Historical Isolation versus recent long-distance connections between Europe and Africa in Bifid Toadflaxes (Linaria sect. Versicolores). PLoS ONE 6:e22234 doi: 10.1371/journal.pone.0022234
Frundt, C., Meyer, A. D., Ichikawa, T., and Meins, F. Jr. (1998). A tobacco homologue of the Ri-plasmid orf13 gene causes cell proliferation in carrot root discs. Mol. Gen. Genet. 259, 559–568. doi: 10.1007/s004380050849
Fujita, T. (1994). Screening of genes related to tumor formation in tobacco genetic tumors. Plant Tiss. Cult. Lett. 11, 171–177. doi: 10.5511/plantbiotechnology1984.11.171
Fujita, T., Ichikawa, T., and Syono, K. (1991). Changes in morphology, levels of endogenous IAA and protein composition in relation to the development of tobacco genetic tumor induced in the dark. Plant Cell Physiol. 32, 169–177.
Furner, I. J., Huffman, G. A., Amasino, R. M., Garfinkel, D. J., Gordon, M. P., and Nester, E. W. (1986). An Agrobacterium transformation in the evolution of the genus Nicotiana. Nature 329, 422–427. doi: 10.1038/319422a0
Ghatnekar, L., Jaarola, M., and Bengtsson, B. O. (2006). The introgression of a functional nuclear gene from Poa to Festuca ovina. Proc. Biol. Sci. 273, 395–399. doi: 10.1098/rspb.2005.3355
Goodspeed, T. H. (1947). On the evolution of the genus Nicotiana. Proc. Natl. Acad. Sci. U.S.A. 33, 158–171. doi: 10.1073/pnas.33.6.158
Gorpenchenko, T. Y., Kiselev, K. V., Bulgakov, V. P., Tchernoded, G. K., Bragina, E. A., Khodakovskaya, M. V., et al. (2006). The Agrobacterium rhizogenes rolC-gene induced somatic embryogenesis and shoot organogenesis in Panax ginseng transformed calluses. Planta 223, 457–467. doi: 10.1007/s00425-005-0102-2
Hsu, K. J., Montadert, L., Bernoulli, D., Cita, M. B., and Erickson, A. (1977). History of the Mediterranean salinity crisis. Nature 267, 399–403. doi: 10.1038/267399a0
Ichikawa, T., Kobayashi, M., Nakagawa, S., Sakurai, A., and Syono, K. (1989). Morphological observations and qualitative and quantitative studies of auxins after induction of tobacco genetic tumor. Plant Cell. Physiol. 30, 57–63.
Ichikawa, T., Ozeki, Y., and Syono, K. (1990). Evidence for the expression of the rol genes of Nicotiana glauca in genetic tumors of N. glauca × N. langsdorffii. Mol. Gen. Genet. 220, 177–180.
Intrieri, M. C., and Buiatti, M. (2001). The horizontal transfer of Agrobacterium rhizogenes genes and evolution of the genus Nicotiana. Mol. Phylogenet. Evol. 20, 100–110. doi: 10.1006/mpev.2001.0927
Kehr, A. E., and Smith, H. H. (1954). Genetic tumors in Nicotiana hybrids. Brookhaven Symposia Biol. 6, 55–78.
Kiselev, K. V., Dubrovina, A. S., Veselova, M. V., Bulgakov, V. P., Fedoreyev, S. A., and Zhuravlev, Y. N. (2007). The rolB gene induced overproduction of resveratrol in Vitis amurensis transformed cells. J. Biotechnol. 128, 681–692. doi: 10.1016/j.jbiotec.2006.11.008
Knapp, S., Chase, M. W., and Clarkson, J. J. (2004). Nomenclatural changes and a new sectional classification in Nicotiana (Solanaceae). Taxon 53, 73–82. doi: 10.2307/4135490
Koonin, E. V., Makarova, K. S., and Aravind, L. (2001). Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55, 709–742. doi: 10.1146/annurev.micro.55.1.709
Kulaeva, O. A., Matveeva, T. V., Lutova, L. A., Kulaeva, O. A., Matveeva, T. V., and Lutova, L. A. (2013). Study of the possibility of horizontal gene transfer from Agrobacterium to some representatives of family Solanaceae. Ecol. Genet. 11, 3–9. Available online at: http://ecolgenet.ru/ru/node/1625
Limami, M. A., Sun, L. Y., Douatm, C., Helgeson, J., and Tepfer, D. (1998). Natural genetic transformation by Agrobacterium rhizogenes. Annual flowering in two biennials, belgian endive and carrot. Plant Physiol. 118, 543–550.
Livak, K. J., Flood, S. J., Marmaro, J., Giusti, W., and Deetz, K. (1995). Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4, 357–362. doi: 10.1101/gr.4.6.357
Matveeva, T. V., Bogomaz, D. I., Pavlova, O. A., Nester, E. W., and Lutova, L. A. (2012). Horizontal Gene Transfer from Genus Agrobacterium to the Plant Linaria in Nature. Mol. Plant Microbe Interact. 25, 1542–1551. doi: 10.1094/MPMI-07-12-0169-R
Matveeva, T. V., and Kosachev, P. A. (2013). “Sequences homologous to Agrobacterium rhizogenes rolC in the genome of Linaria acutiloba,” in International Conference on Frontiers of Environment, Energy and Bioscience (ICFEEB 2013). (Lancaster, PA: DEStech Publications, Inc.), 541–546.
Matveeva, T. V., Lutova, L. A., and Bogomaz, D. I. (2006). “Search for TDNA- like sequences in plant genomes, using real-time PCR with degenerate primers and probe,” in Biotechnology in the Agriculture and Food Industry, ed G. E. Zaikov (New York, NY: Nova Science Publishers), 101–104.
Meyer, A. D., Ichikawa, T., and Meins, F. (1995). Horizontal gene transfer: regulated expression of a tobacco homologue of the Agrobacterium rhizogenes rolC gene. Mol. Gen. Genet. 249, 265–273. doi: 10.1007/BF00290526
Mohajjel-Shoja, H., Clément, B., Perot, J., Alioua, M., and Otten, L. (2011). Biological activity of the Agrobacterium rhizogenes-derived trolC gene of Nicotiana tabacum and its functional relation to other plast genes. Mol. Plant Microbe Interact. 24, 44–53. doi: 10.1094/MPMI-06-10-0139
Moriguchi, K., Maeda, Y., Satou, M., Hardayani, N. S., Kataoka, M., Tanaka, N., et al. (2001). The complete nucleotide sequence of a plant root-inducing (Ri) plasmid indicates its chimeric structure and evolutionary relationship between tumor-inducing (Ti) and symbiotic (Sym) plasmids in Rhizobiaceae. J. Mol. Biol. 307, 771–784. doi: 10.1006/jmbi.2001.4488
Naf, U. (1958). Studies on tumor formation in Nicotiana hybrids. I. The classification of the parents into two etiologically significant groups. Growth 22, 167–180.
Nagata, N., Kosono, S., Sekine, M., Shinmyo, A., and Syono, K. (1995). The regulatory functions of the rolB and rolC genes of Agrobacterium rhizogenes are conserved in the homologous genes (Ng rol) of Nicotiana glauca in tobacco genetic tumors. Plant Cell. Physiol. 36, 1003–1012.
Nagata, N., Kosono, S., Sekine, M., Shinmyo, A., and Syono, K. (1996). Different expression patterns of the promoter of the NgrolB and NgrolC genes during the development of tobacco genetic tumors. Plant Cell. Physiol. 37, 489–493. doi: 10.1093/oxfordjournals.pcp.a028971
Nandi, S. K., Palni, L. S. M., and Parker, C. W. (1990). Dynamic of endogenous cytokinin during the growth cycle of a hormone-autotrophic genetic tumor line of tobacco. Plant Physiol. 94, 1084–1089. doi: 10.1104/pp.94.3.1084
Nilsson, O., and Olsson, O. (1997). Getting to the root: The role of the Agrobacterium rhizogenes rol genes in the formation of hairy roots. Physiol. Plant. 100, 463–473. doi: 10.1111/j.1399-3054.1997.tb03050.x
Oger, P., Mansouri, H., and Dessaux, Y. (2000). Effect of crop rotation and soil cover on alteration of the soil microflora generated by the culture of transgenic plants producing opines. Mol. Ecol. 9, 881–890. doi: 10.1046/j.1365-294x.2000.00940.x
Oger, P., Petit, A., and Dessaux, Y. (1997). Genetically engineered plants producing opines alter their biological environment. Nat. Biotechnol. 15, 369–372. doi: 10.1038/nbt0497-369
Otten, L., Canaday, J., Gerard, J. C., Fournier, P., Crouzet, P., and Paulus, F. (1992). Evolution of agrobacteria and their Ti plasmids. Mol. Plant Microbe Interact. 5, 279–287. doi: 10.1094/MPMI-5-279
Palazon, J., Cusido, R. M., Gonzalo, J., Bonfill, M., Morales, S., and Pinol, M. T. (1998a). Relation between the amount the rolC gene product and indole alkaloid accumulation in Catharanthus roseus transformed root cultures. J. Plant Physiol. 153, 712–718.
Palazon, J., Cusido, R. M., Roig, C., and Pino, M. T. (1998b). Expression of the rolC gene and nicotine production in transgenic roots and their regenerated plants. Plant Cell Rep. 17, 384–390.
Pavlova, O. A., Matveeva, T. V., and Lutova, L. A. (2013). Linaria dalmatica genome contains a homologue of rolC gene of Agrobacterium rhizogenes. Ecol. Genet. 11, 10–15. Available online at: http://ecolgenet.ru/ru/node/1629
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-Guided platform for sequence-specific control of gene expression. Cell 152, 1173 doi: 10.1016/j.cell.2013.02.022
Richards, T. A., Dacks, J. B., Campbell, S. A., Blanchard, J. L., Foster, P. G., McLeod, R., et al. (2006). Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot. Cell 5, 1517–1531. doi: 10.1128/EC.00106-06
Richardson, A. O., and Palmer, J. D. (2007). Horizontal gene transfer in plants. J. Exp. Bot. 58, 1–9. doi: 10.1093/jxb/erl148
Schmülling, T., Schell, J., and Spena, A. (1988). Single genes from Agrobacterium rhizogenes influence plant development. EMBO J. 7, 2621–2629.
Shkryl, Y. N., Veremeichik, G. N., and Bulgakov, V. P. (2008). Individual and combined effects of the rolA, B, and C genes on anthraquinone production in Rubia cordifolia transformed calli. Biotechnol. Bioeng. 100, 118–125. doi: 10.1002/bit.21727
Smith, H. H. (1958). Genetic plant tumors in Nicotiana. Ann. N.Y Acad. Sci. 71, 1163–1177. doi: 10.1111/j.1749-6632.1958.tb46832.x
Spano, L., Pomponi, M., Costantino, P., van Slogteren, G. M. S., and Tempé, J. (1982). Identification of T-DNA in the root-inducing plasmid of the agropine type Agrobacterium rhizogenes 1855. Plant. Mol. Biol. 1, 291–304. doi: 10.1007/BF00027560
Suzuki, K., Tanaka, N., Kamada, H., and Yamashita, I. (2001). Mikimopine synthase (mis) gene on pRi1724. Gene 263, 49–58. doi: 10.1016/S0378-1119(00)00578-3
Suzuki, K., Yamashita, I., and Tanaka, N. (2002). Tobacco plants were transformed by Agrobacterium rhizogenes infection during their evolution. Plant J. 32, 775–787. doi: 10.1046/j.1365-313X.2002.01468.x
Tanaka, N. (2008). Horizontal gene transfer in Agrobacterium: from Biology to Biotechnology, eds T. Tzfira and V. Citovsky (New York, NY: Springer), 623–647.
Taylor, B. H., White, F. F., Nester, E. W., and Gordon, M. P. (1985). Transcription of Agrobacterium rhizogenes A4 T-DNA. Mol. Gen. Genet. 201, 546–553. doi: 10.1007/BF00331354
Tepfer, D. (1982). “La transformation genetique de plantes superieures par Agrobacterium rhizogenes,” in 2e Colloque sur les Recherches Fruitieres (Bordeaux: Cent Tech Interprofessionnel des Fruits et Legumes), 47–59.
Tepfer, D. (1984). Transformation of several species of higher plants by Agrobacterium rhizogenes: sexual transmission of the transformed genotype and phenotype. Cell 37, 959–967. doi: 10.1016/0092-8674(84)90430-6
Tzfira, T., and Citovsky, V. (2006). Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr. Opin. Biotechnol. 17, 147–154. doi: 10.1016/j.copbio.2006.01.009
Udagawa, M., Aoki, S., and Syono, K. (2004). Expression analysis of the NgORF13 promoter during the development of tobacco genetic tumors. Plant Cell Physiol. 45, 1023–1031. doi: 10.1093/pcp/pch123
Vain, P. (2007). Thirty years of plant transformation technology development. Plant Biotechnol. J. 5, 221–229. doi: 10.1111/j.1467-7652.2006.00225.x
Vallenback, P., Ghatnekar, L., and Bengtsson, B. O. (2010). Structure of the natural transgene PgiC2 in the common grass Festuca ovina. PLoS ONE 5:e13529. doi: 10.1371/journal.pone.0013529
Vallenback, P., Jaarola, M., Ghatnekar, L., and Bengtsson, B. O. (2008). Origin and timing of the horizontal transfer of a PgiC gene from Poa to Festuca ovina. Mol. Phylogenet. Evol. 46, 890–896. doi: 10.1016/j.ympev.2007.11.031
Veena, H.-J., Doerge, R. W., and Gelvin, S. B. (2003). Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. Plant J. 5, 219–236. doi: 10.1046/j.1365-313X.2003.01796.x
White, F. F., Garfinkel, D. J., Huffman, G. A., Gordon, M. P., and Nester, E. W. (1983). Sequence homologous to Agrobacterium rhizogenes TDNA in the genomes of uninfected plants. Nature 301, 348–350. doi: 10.1038/301348a0
Keywords: Agrobacterium, T-DNA, horizontal gene transfer, Nicotiana, Linaria
Citation: Matveeva TV and Lutova LA (2014) Horizontal gene transfer from Agrobacterium to plants. Front. Plant Sci. 5:326. doi: 10.3389/fpls.2014.00326
Received: 31 January 2014; Accepted: 23 June 2014;
Published online: 11 August 2014.
Edited by:
Stanton B. Gelvin, Purdue University, USAReviewed by:
Leon Otten, Université de Strasbourg, FranceCopyright © 2014 Matveeva and Lutova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tatiana V. Matveeva, Department of Genetics and Biotechnology, St. Petersburg State University, University emb., 7/9, St. Petersburg 199034, Russia e-mail:cmFkaXNobGV0QGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.